Note: Descriptions are shown in the official language in which they were submitted.
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
1
BRANCHED RECEPTOR BINDING MULTI-SUBUNIT PROTEIN
COMPLEXES FOR USE IN BACTERIAL DELIVERY VEHICLES
TECHNICAL FIELD
[I] The present disclosure relates generally to bacterial delivery vehicles
for use in
efficient transfer of a desired payload into a target bacterial cell.
BACKGROUND
[2] Bacteriophages are parasites that infect and multiply in bacteria. In
general, the
infection process can be divided in several stages: (i) adsorption
corresponding to
recognition and binding to the bacterial cell; (ii) injection of the DNA
genome into the
bacterial cell cytoplasm; (iii) production of a set of viral proteins that can
lead to
insertion in the host target genome (lysogenic phages) or to the production of
infective
particles (lytic phages) and (iv) release of mature virions from the infected
cell, usually
by controlled lysis [1].
[3] Being the first step necessary for a successful infection, recognition
and binding
to the target cell is an essential process in the bacteriophage life cycle.
Bacteriophages
can in some cases recognize several strains of the same species, having a
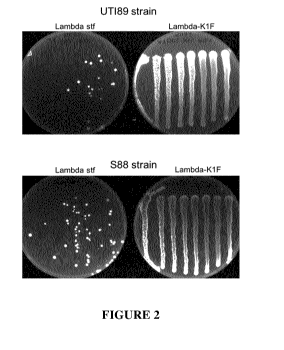
"broad host
range", but very commonly are able to recognize a specific antigen present
only on some
strains of the same species [2]. It is thus not surprising that this step of
the infection
process is central in the competition between bacteriophage and bacteria for
successful
infection.
[4] As a general mechanism, a bacteriophage encodes two main sets of
proteins that
are involved in the recognition process. The first set is able to attach to
the
bacteriophage's primary receptor on the cell surface, an event that triggers
DNA ejection
into the cytoplasm and is usually viewed as an "irreversible" binding process
[3].
Different bacteriophage genera differ in the organization of this set of
proteins, and
hence the naming can be different. In some Siphovirus, for example, they are
called the
"central tail fiber" or "tail tip", which binds irreversibly to the LamB
receptor in
Escherichia coli. In the siphoviridae lambda, the "central tail fiber" or
"tail tip" is
composed of the protein gpJ [4]. In some other Siphovirus, like T5, a protein
located at
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
2
the very tip of the tail mediates this process. In the case of T5, a protein
called pb5
recognizes the FhuA receptor [5]. This type of protein can be found in many
other
bacteriophages. In Myoviruses, like T4, the irreversible binding to the
primary receptor
or to the cell surface in general is mediated by the "short tail fibers",
which are also
located at the end of the tail tube [5].
[5] The second set of proteins in the bacteriophage (herein referred to as
"receptor
binding proteins") encodes recognition and binding activities to the so-called
"secondary
receptor" on the bacterium. This secondary receptor allows for transient
binding of the
phage particle on the cell surface in order to scan the surface and position
the first set of
proteins in contact with the primary receptor. Since this binding is
reversible, it allows
the phage to "walk" on the cell surface until a primary receptor is found and
the
infection process starts. These protein complexes are sometimes referred to as
"L-shape
fibers", such as in T5, "side tail fibers" such as in lambda, "long tail
fibers" as in T4, or
tailspikes such as in phage P22 [5]¨[8]. For some phages, the presence of this
second set
of proteins is necessary for the infection process to occur, such as T4 [5].
In some other
phages, like lambda, this second set of proteins is not strictly necessary for
the infection
process to happen, but it may allow for a more efficient binding to the target
cell [7].
[6] Since the adsorption process is strictly necessary for a successful
infection to
happen, bacteria can develop multiple ways to avoid being recognized by a
bacteriophage. For example, they can mutate the primary or secondary receptor
to which
the bacteriophage binds; they can mask this receptor by attaching proteins to
it (receptor
masking); or they can grow physical barriers around them in the form of
bacterial
capsules, thus blocking any access to the cell surface [9]. Bacteria can
produce many
different types of extracellular polymeric capsules [10]. In turn,
bacteriophages have
evolved different strategies to bypass these defense mechanisms. For instance,
mutating
the tail tip proteins allows them to use a different receptor [11]. However,
the presence
of a polymeric capsule around the bacterium requires a different approach, as
it blocks
all contact to any receptors on the cell surface. In these cases,
bacteriophages have
evolved specific proteins that can enzymatically degrade this capsule to gain
access to
the cells. These depolymerase activities are encoded in protein complexes that
are
CA 03120160 2021-05-17
WO
2020/128108 PCT/EP2019/086990
3
distinct to the primary receptor recognition machinery, in the form of side
tail fibers,
long tail fibers or tailspikes [12], [13], [14].
[7] The concept of a bacteriophage's host range needs to be redefined when
only the
adsorption and injection processes are taken into account. Since all
incompatibilities or
defense mechanisms related to the phage replication cycle are left out of the
picture, the
"adsorption host range" of a given phage is usually larger than the "classical
host range"
in which the infectious cycle leads to newly produced mature virions. The
concept of
host range becomes even more different to the classical definition when
packaged
phagemids based on a given bacteriophage capsid is used. Packaged phagemids do
not
contain the information necessary to replicate the viral particles, because
they do not
package their cognate viral genome. Thus, the host range of a packaged
phagemid tends
to be larger than that of the parental bacteriophage it derives
from.Therefore, for
development of novel bacterial delivery vehicles, designed for the efficient
delivery of
exogenous DNA payload into target strains, it is of utmost importance to be
able to
engineer delivery vehicles with desired host ranges as well as the ability to
bypass
bacterial mechanisms that can lead to unsuccessful binding of the packaged
phagemid to
the bacterial cell surface.
SUMMARY
[8] As a general mechanism, a bacteriophage encodes sets of proteins that
are
involved in the bacterial cell recognition process. Described herein are novel
approaches
to engineering synthetic bacterial delivery vehicles with desired target host
ranges.
[9] In some aspects, synthetic bacterial delivery vehicles are provided
that are
characterized by a chimeric receptor binding protein (RBP), wherein the
chimeric RBP
comprises a fusion between an N-terminal domain of a RBP from a lambda-like
bacteriophage, or lambda bacteriophage, and a C-terminal domain of a different
bacteriophage RBP. Such bacteriophage RBPs, from which the chimeric RBP are
derived, include, for example, and depending on phages families, "L-shape
fibers", "side
tail fibers (stfs)", "long tail fibers" or "tailspikes." As disclosed herein,
it has been
demonstrated that a significant portion of a lambda-like bacteriophage
receptor binding
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
4
protein (RBP), such as a stf protein, can be exchanged with a portion of a
different RBP.
Moreover, specific fusion positions in the RBPs have been identified which
allow one to
obtain functional chimeric RBPs.
[10] In additional aspects, the disclosure relates to bacterial delivery
vehicles with
desired host ranges based on the presence of an engineered branched receptor
binding
multi-subunit protein complex ("branched-RBP"). The branched-RBP comprises two
or
more associated receptor binding proteins derived from bacteriophages, wherein
said
RBPs contain "interaction domains" (IDs) that mediate association of the
different
subunits. The association of one subunit to another can be non-covalent or
covalent. The
two or more associated RBPs include, but are not limited to, the chimeric
receptor
binding proteins (RBPs) described herein that comprise a fusion between the N-
terminal
domain of a RBP derived from a lambda-like, or lambda bacteriophage and the C-
terminal domain of a different RBP.
[11] The chimeric receptor binding protein (RBP) is one wherein the chimeric
RBP
comprises a fusion between an N-terminal domain of a RBP derived from a lambda-
like
bacteriophage, or lambda bacteriophage, and a C-terminal domain of a different
RBP
wherein said N-terminal domain of the RBP is fused to said C-terminal domain
of a
different RBP within one of the amino acids regions selected from positions 1-
150, 320-
460, or 495-560 of the N-terminal RBP with reference to the lambda stf
sequence (SEQ
ID NO: 1) or a similar region of a RBP having homology with one or more of
three
amino acid regions ranging from positions 1-150, 320-460, and 495-560 of the
RBP with
reference to the lambda stf sequence. In one specific aspect of the invention,
the
different RBP domain of the chimeric receptor binding protein (RBP) is derived
from
any bacteriophage or from any bacteriocin. In one specific aspect, the RBP
from the
lambda-like bacteriophage, or the lambda bacteriophage, or the different RBP
contains
homology in one or more of three amino acid regions ranging from positions 1-
150, 320-
460, or 495-560 of the N-terminal RBP with reference to the lambda
bacteriophage stf
sequence (SEQ ID NO: 1). In certain aspects, the homology between the lambda-
like
bacteriophage, the lambda bacteriophage, or the different RBP and the one or
more of
three amino acids regions is around 35% identity for 45 amino acids or more,
around
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
50% identify for 30 amino acids or more, and around 90% identity for 18 amino
acids or
more with reference to the lambda bacteriophage stf sequence (SEQ ID NO:1).
Determination of homology can be performed using alignment tools such as the
Smith-
Waterman algorithm (Smith et al., 1981, J. Mol. Biol 147:195-197) or EMBOSS
5 Matcher (Rice, Longden, Bleasby 2000 EMBOSS Trends in Genetics 16: 276-277).
[12] In one aspect of the invention, the chimeric RBP comprises the N-terminal
domain of a RBP fused to the C-terminal domain of a different RBP within one
of the
amino acid regions selected from positions 80-150, 320-460, or 495-560 of the
N-
terminal RBP with reference to the lambda bacteriophage stf sequence (SEQ ID
NO:1).
In another embodiment of the invention, the chimeric RBP comprises an N-
terminal
domain and a C-terminal domain fused within one of the amino acids regions
selected
from positions 1-150, 320-460 or 495-560 at an insertion site having at least
80%,
preferably at least 85%, at least 90%, at least 95%, at least 98% or at least
99%, identity
with an insertion site selected from the group consisting of amino acids
SAGDAS (SEQ
ID NO:190), ADAKKS (SEQ ID NO:191), MDETNR (SEQ ID NO:192), SASAAA
(SEQ ID NO:193), GAGENS (SEQ ID NO:194), ATLKQI (SEQ ID NO:195), IIQLED
(SEQ ID NO:196), GNIIDL (SEQ ID NO:197), IATRV (SEQ ID NO:198), TPGEL
(SEQ ID NO:199), GAIIN (SEQ ID NO:200), NQIID (SEQ ID NO:201), GQIVN (SEQ
ID NO:202), and VDRAV (SEQ ID NO:203). In a more specific embodiment of the
invention, the chimeric RBP comprises an N-terminal domain and a C-terminal
domain
fused within one of the amino acids regions selected from positions 1-150, 320-
460 or
495-560 at an insertion site having at least 80%, preferably at least 85%, at
least 90%, at
least 95%, at least 98% or at least 99%, identity with an insertion site
selected from the
group consisting of amino acids SAGDAS (SEQ ID NO:190), ADAKKS (SEQ ID
NO:191), MDETNR (SEQ ID NO:192), SASAAA (SEQ ID NO:193) and GAGENS
(SEQ ID NO:194)
[13] In another aspect, the chimeric RBP comprises the N-terminal domain of a
RBP
fused to the C-terminal domain of different RBP wherein the different RBP is a
protein
or group a different proteins that confers an altered host range. In one
embodiment, the
different RBP is a T4-like or T4 long tail fiber composed of a proximal tail
fiber and a
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
6
distal tail fiber (DTF), and the C-terminal domain of a T4-like or T4 RBP is
the distal
tail fiber (DTF). In another embodiment, the N-terminal domain of a RBP is
fused to the
T4-like or T4 distal tail fiber at an insertion site within the T4-like or T4
DTF having at
least 80%, preferably at least 85%, at least 90%, at least 95%, at least 98%
or at least
99%, identity with an insertion site selected from the group consisting of
amino acids
ATLKQI (SEQ ID NO:195), IIQLED (SEQ ID NO:196), GNIIDL (SEQ ID NO:197),
IATRV (SEQ ID NO:198), TPGEL (SEQ ID NO:199), GAIIN (SEQ ID NO:200),
NQIID (SEQ ID NO:201), GQIVN (SEQ ID NO:202), and VDRAV (SEQ ID NO:203).
In a specific embodiment, the N-terminal domain of a RBP is fused to the T4-
like or T4
distal tail fiber within a region from amino acid 1 to 90, with a preferred
region from
amino acid 40 to 50 of the DTF.
[14] In specific embodiments, the disclosure provides specific chimeric RBPs.
SEQ
ID NOS 2-61, 135-165, 215-242, 271, 273, 282 and 283 disclose the amino acid
sequences of such chimeric RBPs as well as, in some instances, their
corresponding
natural chaperone proteins (designated "AP"). Such AP proteins assist in the
folding of
the chimeric RBPs. In a specific embodiment, the RBP comprises the amino acid
sequence of SEQ ID NO: 2, 4, 7, 9, 12, 15, 17, 20, 23, 24, 25, 27, 29, 31, 33,
35, 37, 39,
41, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53, 56, 59, 135 to 144, 147, 150, 151,
154, 157,
160, 163, 215, 216, 219, 221, 223, 225, 227, 229, 232, 325, 237, 239, 241, 282
or 283.
In a more specific embodiment, the RBP comprises the amino acid sequence of
SEQ ID
NO: 2, 4, 7, 9, 12, 15, 17, 20, 23, 24, 25, 27, 29, 31, 33, 35, 37, 39, 41,
42, 44, 46, 47,
48, 49, 50, 51, 52, 53, 56 or 59.
[15] In another aspect, the present disclosure provides nucleotide sequences
encoding
for the chimeric RBPs disclosed herein. In a specific embodiment, nucleic
acids
encoding such chimeric RBPs, as well as their corresponding AP proteins, are
depicted
in SEQ ID NOS 62-120, 166-189, 206-212, 243-270, 272, 274 and 284. In a
specific
embodiment, the nucleic acids encoding such chimeric RBPs comprise the
nucleotide
sequence of SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91,
93, 95,
97, 99, 101, 102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116, 119, 166,
167, 168,
171, 174, 175, 178, 181, 184, 187, 206, 207, 208, 209, 210, 211, 212, 243,
244, 247,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
7
249, 251, 253, 255, 257, 260, 263, 265, 267, 269 or 284. In a more specific
embodiment,
the nucleic acids encoding such chimeric RBPs comprise the nucleotide sequence
of
SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91, 93, 95, 97,
99, 101,
102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116 or 119.
[16] In one specific non-limiting aspect of the invention, it has been
demonstrated that
engineering the chimeric RBP to encode enzymatic activity such as depolymerase
activity can dramatically increase the delivery efficiency of the provided
bacterial
delivery vehicles comprising the chimeric RBP disclosed herein. In an
embodiment of
the invention, the different RBP domain of the chimeric RBP comprises
enzymatic
activity such as depolymerase activity against an encapsulated bacterial
strain. In a
specific embodiment, the depolymerase is an endosialidase such as, for
example, a K 1F
or K5 endosialidase.
[17] In another aspect, the present disclosure provides for engineered
branched-RBPs,
as well as bacterial delivery vehicles, with desired host ranges and/or
specific biological
functions, based on the presence of an engineered branched receptor binding
multi-
subunit protein complex ("branched-RBP"). The engineered branched-RBP
comprises
two or more associated receptor binding proteins, derived from bacteriophages
that
associated with one another based on the presence of interaction domains
(IDs). Each of
the protein complex subunits contain IDs that function as "anchors" for
association of
one subunit RBP with another. The association of one subunit with another can
be non-
covalent or covalent. The engineered branched RBP may comprise non-covalent
association of the different subunits; in some instance, the engineered
branched RBP
may comprise covalent association of the different subunits; and in further
instances, the
engineered branched RBP may comprise both covalent and non-covalent
associations of
the different subunits. In instances where the association is non-covalent,
the protein
subunits are assembled into the engineered branched-RBP as separate protein
subunits
each having their own ID. In a non-limiting example, where the interaction is
covalent,
the engineered branched-RBP may exist as a single fusion protein comprising
different
protein domains of interest fused to two or more ID domains. In specific
embodiments,
the branched-RBP may comprise multiple RBP subunits, including, for example,
two,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
8
three, four, etc. subunits. Each of the RBP subunits may bring different
biological
functions to the overall branched-RBP. Such functions include, but are not
limited to,
host recognition and enzymatic activity. Such enzymatic activity includes
depolymerase
activity.
[18] Disclosed herein are amino acid sequences that are able to function as
interaction
domains (IDs). Such IDs, for purposes of the present invention, are those
amino acid
sequences that provide for association of one subunit to another thereby
providing for
assembly of the engineered branched-RBPs. The IDs may be naturally occurring
bacteriophage IDs, IDs derived from non-bacteriophage polypeptides, or
recombinantly
derived IDs. The two or more of the associated receptor binding proteins of
the
engineered branched-RBP may be any bacteriophage RBP, or a functional domain
of a
bacteriophage RBP, e.g. a domain that provides desired host range or
biological
acitivity, wherein said RBP, or the domain of an RBP, are fused to an ID. The
associated receptor binding proteins may include, but are not limited to,
chimeric
receptor binding proteins (RBPs) described herein that comprise of a fusion
between the
N-terminal domain of a RBP derived from a lambda-like, or lambda bacteriophage
and
the C-terminal domain of a different RBP wherein said chimeric RBP also
comprises an
ID.
[19] In an embodiment of the invention, nucleic acid molecules encoding the
chimeric
RBPs disclosed herein, as well as the two or more subunit RBPs of the
engineered
branched-RBP, are provided. Such nucleic acids may be included in vectors such
as
bacteriophages, plasmids, phagemids, viruses, and other vehicles which enable
transfer
and expression of the chimeric RBP encoding nucleic acids. In instances where
the
subunits of a branched-RBP are to be expressed, it may be advantageous to
express the
subunits from a polycistronic expression unit containing multiple ribosomal
binding
sites (RBSs). The use of such an expression unit can be used to regulate the
expression
of each of the RBP subunits so that equal quantities of expression of each
subunit are
achieved.
[20] Bacterial delivery vehicles are provided which enable transfer of a
nucleic acid
payload, encoding a protein or nucleic acid of interest, into a desired target
bacterial host
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
9
cell. Such bacterial delivery vehicles are characterized by having a chimeric
RBP
comprising a fusion between the N-terminal domain of a RBP from a lambda-like
bacteriophage, or lambda bacteriophage, and the C-terminal domain of a
different RBP.
In an embodiment of the invention, the bacterial delivery vehicles contain a
chimeric
RBP comprising a fusion between an N-terminal domain of a RBP derived from a
lambda-like bacteriophage, or lambda bacteriophage, and a C-terminal domain of
a
different RBP wherein said N-terminal domain of the chimeric RBP is fused to
said C-
terminal domain of a different RBP within one of the amino acids regions
selected from
positions 1-150, 320-460, or 495-560 of the N-terminal domain with reference
to the
lambda stf sequence (SEQ ID NO: 1). In one aspect, the RBP from the lambda-
like
bacteriophage, the lambda bacteriophage, and the different RBP contain
homology in
one or more of three amino acids regions ranging from positions 1-150, 320-
460, and
495-560 of the RBP with reference to the lambda bacteriophage stf sequence
(SEQ ID
NO: 1). In certain aspects, the homology is around 35% identity for 45 amino
acids or
more, around 50% identify for 30 amino acids or more, or around 90% identity
for 18
amino acids or more within the one or more of three amino acids regions
ranging from
positions 1-150, 320-460, and 495-560 of the RBP with reference to the lambda
bacteriophage stf sequence (SEQ ID NO:1). In one specific aspect of the
invention, the
different RBP domain of the chimeric receptor binding protein (RBP) is derived
from a
bacteriophage or a bacteriocin. In one aspect of the invention, the chimeric
RBP
comprises an N-terminal domain of a RBP fused to a C-terminal domain of a RBP
within one of the amino acids regions selected from positions 80-150, 320-460,
or 495-
560 of the N-terminal RBP domain with reference to the lambda stf sequence
(SEQ ID
NO:1). In another embodiment of the invention, the chimeric RBP comprises an N-
terminal domain of a RBP and a C-terminal domain of a RBP fused within a site
of the
N-terminal RBP domain having at least 80%, preferably at least 85%, at least
90%, at
least 95%, at least 98% or at least 99%, identity with a site selected from
the group
consisting of amino acids SAGDAS (SEQ ID NO:190), ADAKKS (SEQ ID NO:191),
MDETNR (SEQ ID NO:192), SASAAA (SEQ ID NO:193), GAGENS (SEQ ID
NO:194), ATLKQI (SEQ ID NO:195), IIQLED (SEQ ID NO:196), GNIIDL (SEQ ID
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
NO:197), IATRV (SEQ ID NO:198), TPGEL (SEQ ID NO:199), GAIIN (SEQ ID
NO:200), NQIID (SEQ ID NO:201), GQIVN (SEQ ID NO:202), and VDRAV (SEQ ID
NO:203), preferably with a site selected from the group consisting of amino
acids
SAGDAS (SEQ ID NO:190), ADAKKS (SEQ ID NO:191), MDETNR (SEQ ID
5 NO:192), SASAAA (SEQ ID NO:193) and GAGENS (SEQ ID NO:194).
[21] In specific embodiments, the disclosure provides a bacterial delivery
vehicle
comprising a chimeric RBP. SEQ ID NOS 2-61, 135-165, 215-242, 271, 273, 282
and
283 disclose the amino acid sequences of such chimeric RBPs and in addition,
in some
instances, their corresponding natural chaperone proteins (designated "AP").
Such AP
10 proteins assist in the folding of the chimeric PBPs. In a specific
embodiment, the RBP
comprises the amino acid sequence of SEQ ID NO: 2, 4, 7, 9, 12, 15, 17, 20,
23, 24, 25,
27, 29, 31, 33, 35, 37, 39, 41, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53, 56,
59, 135 to 144,
147, 150, 151, 154, 157, 160, 163, 215, 216, 219, 221, 223, 225, 227, 229,
232, 325,
237, 239, 241, 282 or 283. In a more specific embodiment, the RBP comprises
the
amino acid sequence of SEQ ID NO: 2, 4, 7, 9, 12, 15, 17, 20, 23, 24, 25, 27,
29, 31, 33,
35, 37, 39, 41, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53, 56 or 59.
[22] In one aspect, the present disclosure also provides nucleotide sequences
encoding
for the chimeric RBPs disclosed herein. In a specific embodiment, nucleic
acids
encoding such chimeric RBPs, as well as corresponding AP proteins, are
depicted in
SEQ ID NOS 62-120, 166-189, 206-212, 243-270, 272, 274 and 284. In a specific
embodiment, the nucleic acids encoding such chimeric RBPs comprise the
nucleotide
sequence of SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91,
93, 95,
97, 99, 101, 102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116, 119, 166,
167, 168,
171, 174, 175, 178, 181, 184, 187, 206, 207, 208, 209, 210, 211, 212, 243,
244, 247,
249, 251, 253, 255, 257, 260, 263, 265, 267, 269 or 284. In a more specific
embodiment,
the nucleic acids encoding such chimeric RBPs comprise the nucleotide sequence
of
SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91, 93, 95, 97,
99, 101,
102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116, or 119.
[23] In other specific embodiments and to increase the delivery efficiency of
the
bacterial delivery vehicles disclosed herein the different RBP domain of the
chimeric
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
11
RBP comprises a domain having depolymerase activity against an encapsulated
bacterial
strain. In a specific embodiment, the depolymerase is an endosialidase, such
as for
example, a KlF or K5 endosialidase.
[24] In another aspect of the invention, bacterial delivery vehicles are
provided which
enable transfer of a nucleic acid payload, encoding a protein or nucleic acid
of interest,
into a desired target bacterial host cell wherein said bacterial delivery
vehicles are
characterized by having a branched-RBP as disclosed herein.
[25] The bacterial delivery vehicles provided herein enable transfer of a
nucleic acid
payload, encoding a protein or nucleic acid of interest, into a desired target
bacterial host
cell. In certain embodiments of the invention, the nucleic acid of interest is
selected
from the group consisting of a Cas nuclease gene, a Cas9 nuclease gene, a
guide RNA, a
CRISPR locus, a toxin gene, a gene expressing an enzyme such as a nuclease or
a
kinase, a TALEN, a ZFN, a meganuclease, a recombinase, a bacterial receptor, a
membrane protein, a structural protein, a secreted protein, a gene expressing
resistance
to an antibiotic or to a drug in general, a gene expressing a toxic protein or
a toxic factor,
and a gene expressing a virulence protein or a virulence factor, or any of
their
combination. In an embodiment of the invention, the nucleic acid payload
encodes a
therapeutic protein. In another embodiment, the nucleic acid payload encodes
an anti-
sense nucleic acid molecule.
[26] In one aspect, the bacterial delivery vehicle enables the transfer of a
nucleic acid
payload that encodes a nuclease that targets cleavage of a host bacterial cell
genome or a
host bacterial cell plasmid. In some aspects, the cleavage occurs in an
antibiotic resistant
gene. In another embodiment of the invention, the nuclease mediated cleavage
of the
host bacterial cell genome is designed to stimulate a homologous recombination
event
for insertion of a nucleic acid of interest into the genome of the bacterial
cell.
[27] The present invention also provides pharmaceutical or veterinary
compositions
comprising one or more of the bacterial delivery vehicles disclosed herein and
a
pharmaceutically-acceptable carrier. Also provided is a method for treating a
disease or
disorder caused by bacteria, preferably a bacterial infection, comprising
administering to
a subject having a disease or disorder caused by bacteria, preferably a
bacterial infection,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
12
in need of treatment the provided pharmaceutical or veterinary composition.
The present
invention also relates to a pharmaceutical or veterinary composition as
disclosed herein
for use in the treatment of a disease or disorder caused by bacteria,
preferably a bacterial
infection. It further relates to the use of a pharmaceutical or veterinary
composition as
disclosed herein for the manufacture of a medicament for treating a disease or
disorder
caused by bacteria, preferably a bacterial infection. A method for reducing
the amount of
virulent and/or antibiotic resistant bacteria in a bacterial population is
provided
comprising contacting the bacterial population with the bacterial delivery
vehicles
disclosed herein. The method may be an in vivo or in vitro method. The present
invention also relates to a pharmaceutical or veterinary composition as
disclosed herein
for use in reducing the amount of virulent and/or antibiotic resistant
bacteria in a
bacterial population, in particular in a subject having a bacterial infection.
It further
relates to the use of a pharmaceutical or veterinary composition as disclosed
herein for
the manufacture of a medicament for reducing the amount of virulent and/or
antibiotic
resistant bacteria in a bacterial population, in particular in a subject
having a bacterial
infection.
BRIEF DESCRIPTION OF FIGURES
[28] In order to better understand the subject matter that is disclosed herein
and to
exemplify how it may be carried out in practice, embodiments will now be
described, by
way of non-limiting example, with reference to the accompanying drawings. With
specific reference to the drawings, it is stressed that the particulars shown
are by way of
example and for purposes of illustrative discussion of embodiments of the
invention.
[29] FIG. 1 demonstrates delivery in wild-type E. coli strains with lambda and
OMPF-lambda packaged phagemids. Lambda packaged phagemids were diluted 1:5 in
LB plus 5mM CaCl2 and 10 lit added in each well. 90 lit of cells grown to an
0D600 of
around 0.5 were then added to each phagemid-containing well, incubated for 30
min at
37 C and 10 lit spotted on LB-agar supplemented with chloramphenicol. Left
panel,
wild type lambda packaged phagemids; right panel, OMPF-lambda variant. Circles
show
strains with modified delivery as compared to lambda wild-type.
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
13
[30] FIG. 2 depicts wild-type lambda and lambda-stf-K1F chimeric delivery
vehicles
on Kl+ strains. Lambda packaged phagemids were sequentially diluted 10X in LB
plus
5mM CaCl2 and 10 lit added in each well. Cells grown to an 0D600 of around 0.5
were
then added to each phagemid dilution, incubated for 30 min at 37 C and 10 lit
plated on
LB supplemented with chloramphenicol. Top panel, strain UTI89; bottom panel,
strain
S88. Left plates, wild type lambda packaged phagemids; right plates, stf-K1F
lambda
packaged phagemids.
[31] FIG. 3 depicts wild-type lambda and lambda-stf-K5 chimeric delivery
vehicles
on a K5+ strain. Lambda packaged phagemids were sequentially diluted 10X in LB
plus
5mM CaCl2 and 10 lit added in each well. ECOR55 grown to an 0D600 of around
0.5
were then added to each phagemid dilution, incubated for 30 min at 37 C and 10
lit
plated on LB supplemented with chloramphenicol. Left panel, wild type lambda
packaged phagemids; right panel, stf-K15 lambda packaged phagemids.
[32] FIG. 4 depicts wild-type lambda, lambda-stf-AG22 and lambda-stf-SIEA1 1
chimeric delivery vehicles on a variety of encapsulated strains (0 and K
capsules).
Lambda phagemids were diluted 1:5 in LB plus 5mM CaCl2 and 10 lit added in
each
well. 90 lit of cells grown to an 0D600 of around 0.5 were then added to each
phagemid-
containing well, incubated for 30 min at 37 C and 10 lit spotted on LB-agar
supplemented with chloramphenicol. Left panel, wild type lambda phagemids;
middle
panel, lambda stf-SIEAll variant; right panel, lambda-stf-AG22 variant.
Circles show
strains with modified delivery as compared to lambda wild-type.
[33] FIG. 5A-C demonstrates delivery of wild-type lambda and stf chimeras with
different insertion sites on a variety of encapsulated strains (0 and K
capsules). Lambda
packaged phagemids were diluted 1:5 in LB plus 5mM CaCl2 and 10 lit added in
each
well. 90 lit of cells grown to an 0D600 of around 0.5 were then added to each
phagemid-
containing well, incubated for 30 min at 37 C and 10 lit spotted on LB-agar
supplemented with chloramphenicol. FIG. 5A. Left panel, wild type lambda
packaged
phagemids; rest of panels, three different ADAKKS (SEQ ID NO:191)-stf
variants. FIG.
5B Left panel, wild type lambda packaged phagemids; rest of panels, three
different
SASAAA (SEQ ID NO: 193)-stf variants. FIG. 5C Left panel, wild type lambda
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
14
packaged phagemids; rest of panels, three different MDETNR (SEQ ID NO:192)-stf
variants. For all panels, red circles show strains with improved delivery
efficiency as
compared to lambda wild-type.
[34] FIG. 6 depicts a phmmer search that was performed with a 50aa sliding
window
(step 10) on the representative proteome database (rp75). The number of
significant hits
(E-value <0.01) is reported.
[35] FIG. 7A-B depicts branched stf architectures with 2 subunits. FIG. 7A is
a
schematic view of a delivery vehicle with a 2 subunits branched stf
architecture. ID:
"Interaction Domain". FIG. 7B is a schematic view of the genetic architecture
of an
engineered lambda stf construct.
[36] FIG. 8 demonstrates delivery of branched lambda stf packaged phagemids.
Lambda packaged lambda-stf-WW11.1 stf, lambda-stf-K1F or the branched
construct
shown in FIG. 7 (WW11.1-K1F) were produced and titrated against 057 and K1
strains.
[37] FIG. 9A-B depicts branched stf architectures with 4 subunits. FIG. 9A is
a
schematic view of a delivery vehicle with a 4 subunits branched stf
architecture. Actual
interactions among different ID may be different in the biological assembly
from the
graph depicted here. FIG. 9B depicts a genetic circuit encoding the 4 subunits
branched
stf under the control of an inducible promoter.
[38] FIG 10. depicts architecture of the engineered lambda stf-T4-like DTF
chimera.
The semicircles denote RBS sites; the T sign, a transcriptional terminator;
the arrow, a
promoter.
[39] FIG. 11. shows screening of phagemid particles with chimeric lambda stf-
T4-
like DTFs. A collection of 96 different wild type E. coli strains,
encompassing different
serotypes, was transduced with lambda-based phagemids and plated on Cm LB
agar.
Left panel, wild-type lambda stf; middle panel, chimeric lambda-stf-WW13;
right panel,
chimeric lambda- stf-PP-1.
[40] FIG 12. demonstrates screening of phagemid particles with chimeric lambda
stf-
T4-like DTFs. A collection of 96 different wild type E. coli strains,
encompassing
different serotypes, was transduced with lambda-based phagemids and plated on
Cm LB
agar. Left panel, wild-type lambda stf; middle panel, chimeric lambda-stf-
WW55; right
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
panel, chimeric lambda-stf-WW34.
[41] FIG. 13. depicts screening of phagemid particles with chimeric lambda stf-
T4-
like DTFs. All points shown refer to the universal insertion site of the DTF,
located
within amino acid range from position 1 to 90 with reference to WW13 amino
acid
5 sequence. A collection of 96 different wild type E. coli strains,
encompassing different
serotypes, was transduced with lambda-based phagemids and plated on Cm LB agar
(names on top).
[42] FIG. 14. depicts dot scoring system to quantify delivery efficiency.
Density 0, 5
or fewer colonies; density 1, more than 5 colonies but not enough to define a
clear
10 circular drop; density 2, several colonies, but the background is clearly
visible and some
colonies are still separated; density 3, many colonies, the background is
still visible but
the colonies are hardly discernible as separate; density 4, spot almost
completely dense,
the background can only be seen faintly in some parts of the drop; density 5,
spot looks
completely dense, background cannot be seen.
15 [43] FIG. 15 depicts raw dot titrations of delivery particles with chimeric
stf in 40
human strains of the ECOR collection. Below each panel, the name of the
chimeric stf.
Above each dot, the 1-2 letter code used to identify strains.
[44] FIG. 16 represents bar-formatted delivery data of Figure 15. From 0 (no
entry,
grey background) to 5 (maximum delivery). The bar length is proportional to
the entry
score from 1 (smallest bars) to 5 (longest bars).
DETAILED DESCRIPTION
[45] Disclosed herein are novel approaches to engineering synthetic bacterial
delivery
vehicles with desired target host ranges. The synthetic bacterial delivery
vehicles are
characterized by a chimeric receptor binding protein (RBP), wherein the
chimeric RBP
comprises a fusion between the N-terminal domain of a RBP from a lambda-like
bacteriophage, or lambda bacteriophage, and the C-terminal domain of a
different RBP.
It has been demonstrated herein that a significant portion of a lambda-like
RBP, such as
a stf protein, can be exchanged with a portion of a different RBP. Moreover,
specific
fusion positions of the receptor binding protein have been identified which
allow one to
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
16
obtain a functional chimeric RBP.
[46] Additionally, disclosed herein are synthetic bacterial delivery vehicles
that are
characterized by the presence of an engineered branched receptor binding multi-
subunit
protein complex ("branched-RBP"). The engineered branched-RBP comprises two or
more associated receptor binding proteins, derived from bacteriophages, which
associate
with one another based on the presence of interaction domains (IDs). Each of
the
polypeptide subunits are engineered to contain IDs that function as "anchors"
for
association of one subunit RBP with another. The association of one subunit
with
another can be non-covalent or covalent. In specific embodiments the branched-
RBP
may comprise multiple RBP subunits, including, for example, two, three, four,
etc.
subunits.
[47] As used herein, a receptor binding protein or RBP is a polypeptide that
recognizes, and optionally binds and/or modifies or degrades a substrate
located on the
bacterial outer envelope, such as, without limitation, bacterial outer
membrane, LPS,
capsule, protein receptor, channel, structure such as the flagellum, pili,
secretion system.
The substrate can be, without limitation, any carbohydrate or modified
carbohydrate,
any lipid or modified lipid, any protein or modified protein, any amino acid
sequence,
and any combination thereof. As used herein, a lambda-like bacteriophage
refers to any
bacteriophage encoding a RBP having amino acids sequence homology of around
35%
identity for 45 amino acids or more, around 50% identify for 30 amino acids or
more, or
around 90% identity for 18 amino acids or more in one or more of three amino
acids
regions ranging from positions 1-150, 320-460, and 495-560 with reference to
the
lambda bacteriophage stf sequence of SEQ ID NO: 1, independently of other
amino
acids sequences encoded by said bacteriophage.
[48] The present disclosure provides a chimeric receptor binding protein
(RBP),
wherein the chimeric RBP comprises a fusion between an N-terminal domain of a
RBP
from a lambda-like bacteriophage, or lambda bacteriophage, and a C-terminal
domain of
a different bacteriophage RBP. Such bacteriophage RBPs, from which the
chimeric RBP
are derived, include, for example, "L-shape fibers", "side tail fibers
(stfs)", "long tail
fibers" or "tailspikes." As disclosed herein, it has been demonstrated that a
significant
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
17
portion of a lambda-like bacteriophage receptor binding protein (RBP), such as
a stf
protein, can be exchanged with a portion of a different RBP. Moreover,
specific fusion
positions in the RBPs have been identified which allow one to obtain a
functional
chimeric RBP. Such chimeric RBPs include those having an altered host range
and/or
biological activity such as, for example, depolymerase activity.
[49] The chimeric receptor binding protein (RBP) is one wherein the chimeric
RBP
comprises a fusion between an N-terminal domain of a RBP derived from a lambda-
like
bacteriophage, or lambda bacteriophage, and a C-terminal domain of a different
RBP
wherein said N-terminal domain of the RBP is fused to said C-terminal domain
of a
different RBP within one of the amino acids regions selected from positions 1-
150, 320-
460, or 495-560 of the N-terminal RBP with reference to the lambda stf
sequence (SEQ
ID NO: 1) or a similar region of a RBP having homology with one or more of
three
amino acids regions ranging from positions 1-150, 320-460, and 495-560 of the
RBP
with reference to the lambda stf sequence (SEQ ID NO:1). In one specific
aspect of the
invention, the different RBP of the chimeric receptor binding protein (RBP) is
derived
from any bacteriophage or from any bacteriocin.
[50] In one specific aspect, the RBP from the lambda-like bacteriophage, the
lambda
bacteriophage, or the different RBP contain homology with one or more of three
amino
acids regions ranging from positions 1-150, 320-460, and 495-560 of the RBP
with
reference to the lambda bacteriophage stf sequence (SEQ ID NO:1). In certain
aspects,
the homology between the lambda-like bacteriophage, the lambda bacteriophage,
or the
different RBP and the one or more amino acids regions is around 35% identity
for 45
amino acids or more, around 50% identify for 30 amino acids or more, and
around 90%
identity for 18 amino acids or more. Determination of homology can be
performed
using alignment tools such as the Smith-Waterman algorithm (Smith et al.,
1981, J. Mol.
Biol 147:195-197) or EMBOSS Matcher (Rice, Longden, Bleasby 2000 EMBOSS
Trends in Genetics 16: 276-277). In one aspect of the invention, the chimeric
RBP
comprises the N-terminal domain of the chimeric RBP fused to the C-terminal
domain
of the chimeric RBP within one of the amino acids regions selected from
positions 80-
150, 320-460, or 495-560 with reference to the lambda bacteriophage stf
sequence (SEQ
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
18
ID NO: 1). In another embodiment of the invention, the chimeric RBP comprises
an N-
terminal domain and a C-terminal domain fused within one of the three amino
acids
regions at an insertion site having at least 80%, preferably at least 85%, at
least 90%, at
least 95%, at least 98% or at least 99%, identity with an insertion site
selected from the
group consisting of amino acids SAGDAS (SEQ ID NO:190), ADAKKS (SEQ ID
NO:191), MDETNR (SEQ ID NO:192), SASAAA (SEQ ID NO:193), GAGENS (SEQ
ID NO:194), ATLKQI (SEQ ID NO:195), IIQLED (SEQ ID NO:196), GNIIDL (SEQ
ID NO:197), IATRV (SEQ ID NO:198), TPGEL (SEQ ID NO:199), GAIIN (SEQ ID
NO:200), NQIID (SEQ ID NO:201), GQIVN (SEQ ID NO:202), and VDRAV (SEQ ID
NO:203), preferably from the group consisting of amino acids SAGDAS (SEQ ID
NO:190), ADAKKS (SEQ ID NO:191), MDETNR (SEQ ID NO:192), SASAAA (SEQ
ID NO:193) and GAGENS (SEQ ID NO:194). In a specific embodiment, where
branched-RBPs comprise such chimeric RBPs, IDs may be inserted at such
insertion
sites thereby acting to fuse the N-terminal domain to the C-terminal domain.
[51] In some instances, an ID domain may be fused to either an N-terminal
domain,
or C-terminal domain, of a bacteriophage RBP, to provide a non-chimeric
protein
subunit of an engineered branched RBP. The N-terminal domain, or C-terminal
domain,
may be chosen depending on the desired function of the domain, e.g. host range
or
biological function. Where such non-chimeric protein subunits are utilized for
production of an engineered branched-RBP, the ID domain may be fused at the
preferred
insertion sites disclosed herein, or alternatively, at insertion sites that
permit
maintainance of the function of the chosen domain.
[52] In specific embodiments, the disclosure provides chimeric RBPs. Such
chimeric
RBPs may function as protein subunits of an engineered branched-RBP protein
complex. SEQ ID NOS 2-61, 135-165, 215-242, 271, 273, 282 and 283 disclose the
amino acid sequences of such chimeric RBPs and in addition, in some instances,
their
corresponding natural chaperone proteins (designated "AP"). Such AP proteins
assist in
the folding of the chimeric RBPs. In a specific embodiment, the RBP comprises
the
amino acid sequence of SEQ ID NO: 2, 4, 7, 9, 12, 15, 17, 20, 23, 24, 25, 27,
29, 31, 33,
35, 37, 39, 41, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53, 56, 59,135 to 144,
147, 150, 151,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
19
154, 157, 160, 163, 215, 216, 219, 221, 223, 225, 227, 229, 232, 325, 237,
239, 241, 282
or 283. In a more specific embodiment, the RBP comprises the amino acid
sequence of
SEQ ID NO: 2, 4, 7, 9, 12, 15, 17, 20, 23, 24, 25, 27, 29, 31, 33, 35, 37, 39,
41, 42, 44,
46, 47, 48, 49, 50, 51, 52, 53, 56 or 59.
[53] In one aspect, the present disclosure also provides nucleotide sequences
encoding
for the chimeric RBPs disclosed herein. In a specific embodiment, nucleic
acids
encoding such chimeric RBPs, as well as corresponding AP proteins, are
depicted in
SEQ ID NOS 62-120, 166-189, 206-212, 243-270, 272, 274 and 284. In a specific
embodiment, the nucleic acids encoding the chimeric RBP comprise the
nucleotide
sequence of SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91,
93, 95,
97, 99, 101, 102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116, 119, 166,
167, 168,
171, 174, 175, 178, 181, 184, 187, 206, 207, 208, 209, 210, 211, 212, 243,
244, 247,
249, 251, 253, 255, 257, 260, 263, 265, 267, 269 or 284. In a more specific
embodiment,
the nucleic acids encoding such chimeric RBPs comprise the nucleotide sequence
of
SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91, 93, 95, 97,
99, 101,
102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116, or 119.
[54] In aspects where the above described chimeric RBPs are utilized as
subunits for
production of branched RBP protein complexes, said chimeric RBPs may be
further
engineered to contain ID domains that act to mediate the association of the
various
engineered branched-RBP protein subunits with one another.
[55] In one specific non-limiting aspect of the disclosure, it has been
demonstrated
that engineering the chimeric RBP to encode depolymerase activity can
dramatically
increase the delivery efficiency of the provided bacterial delivery vehicles
comprising
the chimeric RBP disclosed herein. In an embodiment of the disclosure, the
different
RBP domain of the chimeric RBP comprises depolymerase activity against an
encapsulated bacterial strain. In a specific embodiment, the depolymerase is
an
endosialidase such as, for example, a KlF or K5 endosialidase
[56] With regard to the engineered branched-RBPs disclosed herein, any of the
chimeric RBPs disclosed herein may be used as RBP subunits, wherein said RBPs
may
be further engineered to contain IDs. As disclosed in the Examples section, it
has been
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
demonstrated that engineering branched-RBPs can alter the host range of the
resulting
delivery particle.
[57] Nucleic acid molecules encoding the chimeric RBPs and branched-RBPs,
disclosed herein are provided. Such nucleic acids may be included in vectors
such as
5 bacteriophages, plasmids, phagemids, viruses, and other vehicles which
enable transfer
and expression of the chimeric RBP encoding nucleic acids.
[58] Bacterial delivery vehicles are provided which enable transfer of a
nucleic acid
payload, encoding a protein or nucleic acid of interest, into a desired target
bacterial host
cell. Such bacterial delivery vehicles are characterized by having a chimeric
RBP
10 comprising a fusion between the N-terminal domain of a RBP from a lambda-
like
bacteriophage, or lambda bacteriophage, and the C-terminal domain of a
different RBP.
In an embodiment of the invention, the bacterial delivery vehicles contain a
chimeric
RBP comprising a fusion between an N-terminal domain of a RBP derived from a
lambda-like bacteriophage, or lambda bacteriophage, and a C-terminal domain of
a
15 different RBP wherein said N-terminal domain of the chimeric RBP is fused
to said C-
terminal domain of a different RBP within one of the amino acids regions
selected from
positions 1-150, 320-460, or 495-560 of the N-terminal domain RBP with
reference to
the lambda stf sequence (SEQ ID NO: 1). In one aspect, the RBP from the lambda-
like
bacteriophage, the lambda bacteriophage, and the different RBP contain
homology in
20 one or more of three amino acids regions ranging from positions 1-150, 320-
460, and
495-560 of the N-terminal RBP with reference to the lambda bacteriophage stf
sequence.
In certain aspects, the homology is around 35% identity for 45 amino acids or
more,
around 50% identify for 30 amino acids or more, or around 90% identity for 18
amino
acids or more within the one or more of three amino acids regions ranging from
positions 1-150, 320-460, and 495-560 of the N-terminal RBP with reference to
the
lambda bacteriophage stf sequence (SEQ ID NO: 1). In one specific aspect of
the
invention, the different RBP domain of the chimeric receptor binding protein
(RBP) is
derived from a bacteriophage or a bacteriocin. In one aspect of the invention,
the
chimeric RBP comprises an N-terminal domain of a RBP fused to a C-terminal
domain
of a RBP within one of the amino acids regions selected from 80-150, 320-460,
or 495-
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
21
560 of the RBPs with reference to the lambda stf sequence (SEQ ID NO: 1). In
another
embodiment of the invention, the chimeric RBP comprises an N-terminal domain
of a
RBP and a C-terminal domain of a RBP fused within a site of the N-terminal
RBPs
having at least 80%, preferably at least 85%, at least 90%, at least 95%, at
least 98% or
at least 99%, identity with a site selected from the group consisting of amino
acids
SAGDAS (SEQ ID NO:190), ADAKKS (SEQ ID NO:191), MDETNR (SEQ ID
NO:192), SASAAA (SEQ ID NO:193), GAGENS (SEQ ID NO:194), ATLKQI (SEQ
ID NO:195), IIQLED (SEQ ID NO:196), GNIIDL (SEQ ID NO:197), IATRV (SEQ ID
NO:198), TPGEL (SEQ ID NO:199), GAIIN (SEQ ID NO:200), NQIID (SEQ ID
NO:201), GQIVN (SEQ ID NO:202), and VDRAV (SEQ ID NO:203), preferably
selected from the group consisting of amino acids SAGDAS (SEQ ID NO:190),
ADAKKS (SEQ ID NO:191), MDETNR (SEQ ID NO:192), SASAAA (SEQ ID
NO:193), and GAGENS (SEQ ID NO:194).
[59] In specific embodiments, the disclosure provides a bacterial delivery
vehicle
comprising a chimeric RBP. SEQ ID NOS 2-61, 135-165, 215-242, 271, 273, 282
and
283 disclose the amino acid sequences of such chimeric RBPs and in addition,
in some
instances, their corresponding natural chaperone proteins (designated "AP").
Such AP
proteins assist in the folding of the chimeric RBPs. In a specific embodiment,
the RBP
comprises the amino acid sequence of SEQ ID NO: 2, 4, 7, 9, 12, 15, 17, 20,
23, 24, 25,
27, 29, 31, 33, 35, 37, 39, 41, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53, 56,
59, 135 to 144,
147, 150, 151, 154, 157, 160, 163, 215, 216, 219, 221, 223, 225, 227, 229,
232, 325,
237, 239, 241, 282 or 283. In a more specific embodiment, the RBP comprises
the
amino acid sequence of SEQ ID NO: 2, 4, 7, 9, 12, 15, 17, 20, 23, 24, 25, 27,
29, 31, 33,
35, 37, 39, 41, 42, 44, 46, 47, 48, 49, 50, 51, 52, 53, 56 or 59.
[60] In one aspect, the present disclosure also provides nucleotide sequences
encoding
for the chimeric RBPs disclosed herein. In a specific embodiment, nucleic
acids
encoding such chimeric RBPs, as well as corresponding AP proteins, are
depicted in
SEQ ID NOS 62-120, 166-189, 206-212, 243-270, 272, 274 and 284. In a specific
embodiment, the nucleic acids encoding the chimeric RBPs comprise the
nucleotide
sequence of SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91,
93, 95,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
22
97, 99, 101, 102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116, 119, 166,
167, 168,
171, 174, 175, 178, 181, 184, 187, 206, 207, 208, 209, 210, 211, 212, 243,
244, 247,
249, 251, 253, 255, 257, 260, 263, 265, 267, 269 or 284. In a more specific
embodiment,
the nucleic acids encoding such chimeric RBPs comprise the nucleotide sequence
of
SEQ ID NO: 62, 64, 67, 69, 72, 75, 77, 80, 83, 84, 85, 87, 89, 91, 93, 95, 97,
99, 101,
102, 104, 106, 107, 108, 109, 110, 111, 112, 113, 116, or 119.
[61] In other specific embodiments and to increase the delivery efficiency of
the
bacterial delivery vehicles disclosed herein the different RBP domain of the
chimeric
RBP comprises a domain having depolymerase activity against an encapsulated
bacterial
strain. In a specific embodiment, the depolymerase is an endosialidase, such
as for
example, a KlF or K5 endosialidase.
[62] The present disclosure provides synthetic bacterial delivery vehicles
that are
characterized by the presence of an engineered branched receptor binding multi-
subunit
protein complex ("branched-RBP"). The engineered branched-RBP comprises two or
more associated receptor binding proteins, derived from bacteriophages, which
associate
with one another based on the presence of interaction domains (IDs). The
association of
one subunit with another can be non-covalent or covalent. Each of the
polypeptide
subunits contain IDs that function as "anchors" for association of one subunit
RBP with
another. In specific embodiments the branched-RBP may comprise multiple RBP
subunits, including, for example, two, three, four, etc. subunits.
[63] The individual RBP subunit may bring different biological functions to
the
overall engineered branched-RBP. Such functions include but are not limited to
host
recognition and enzymatic activity. Such enzymatic activity includes
depolymerase
activity.
[64] Disclosed herein are amino acid sequences that are able to function as ID
polypeptides. Such IDs, for purposes of the present invention, are those amino
acid
sequences that provide for non-covalent or covalent association of one
receptor binding
protein to another. An interaction domain is a polypeptide whose function
mediates the
association of one biological molecule, e.g., a protein, to another biological
molecule.
As a non limitating example, the biological molecule can be a protein, a part
of a
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
23
protein, a carbohydrate, a lipid and a nucleic acid.
[65] The IDs may be naturally occurring bacteriophage IDs, IDs derived from
non-
bacteriophage polypeptides that naturally associate with one another, or
recombinantly
derived IDs that function to mediate non-covalent or covalent association of
two
proteins or polypeptide domains.
[66] The two or more associated receptor binding proteins of the branched-RBP
include, but are not limited to, chimeric receptor binding proteins (RBPs)
described
herein that comprise a fusion between the N-terminal domain of a RBP derived
from a
lambda-like, or lambda bacteriophage and the C-terminal domain of a different
RBP
wherein said chimeric RBP further comprises an ID domain..
[67] With regard to IDs, such sequences are linked to receptor binding
proteins
(RBPs), e.g. can be fusion, can be coiled coil, can be a non-covalent
interaction or can
be natural sequence of the RBP. An RBP subunit of the branched-RBP may be a
polypeptide that recognizes, and optionally binds and/or modifies or degrades
a substrate
located on the bacterial outer envelope, such as, without limitation,
bacterial outer
membrane, LPS, capsule, protein receptor, channel, structure such as the
flagellum, pili,
secretion system. The substrate can be, without limitation, any carbohydrate
or modified
carbohydrate, any lipid or modified lipid.
[68] The bacterial delivery vehicles provided herein enable transfer of a
nucleic acid
payload, encoding a protein or nucleic acid of interest, into a desired target
bacterial host
cell. As used herein, the term "delivery vehicle" refers to any means that
allows the
transfer of a payload into a bacterium. There are several types of delivery
vehicles
encompassed by the present invention including, without limitation,
bacteriophage
scaffold, virus scaffold, chemical based delivery vehicle (e.g., cyclodextrin,
calcium
phosphate, cationic polymers, cationic liposomes), protein-based or peptide-
based
delivery vehicle, lipid-based delivery vehicle, nanoparticle-based delivery
vehicles, non-
chemical-based delivery vehicles (e.g., transformation, electroporation,
sonoporation,
optical transfection), particle-based delivery vehicles (e.g., gene gun,
magnetofection,
impalefection, particle bombardment, cell-penetrating peptides) or donor
bacteria
(conjugation).Any combination of delivery vehicles is also encompassed by the
present
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
24
invention. The delivery vehicle can refer to a bacteriophage derived scaffold
and can be
obtained from a natural, evolved or engineered capsid. In some embodiments,
the
delivery vehicle is the payload as bacteria are naturally competent to take up
a payload
from the environment on their own.
[69] Delivery vehicles as disclosed herein include packaged phagemids, as well
as
bacteriophage, comprising the chimeric and/or branched-RBPs disclosed herein.
The
engineering of such delivery vehicles are well known to those skilled in the
art. Such
engineering techniques may employ production cell lines engineered to express
the
chimeric RBPs or branched-RBP disclosed herein. Generation of packaged
phagemids
and bacteriophage particles are routine techniques well-known to one skilled
in the art.
A satellite phage and/or helper phage may be used to promote the packaging of
the
payload in delivery vehicles of the present invention. Helper phages provide
functions in
trans and are well known to the man skilled in the art. The helper phage
comprises all
the genes coding for the structural and functional proteins that are
indispensable for the
payload to be packaged, according to the invention (i.e. the helper phage
provides all the
necessary gene products for the assembly of the delivery vehicle). The helper
phage may
contain a defective origin of replication or packaging signal, or completely
lack the
latter, and hence it is uncapable of self-packaging, thus only bacterial
delivery particles
carrying the payload or plasmid will be produced. Helper phages may be chosen
so that
they cannot induce lysis of the host used for the delivery particle
production. One skilled
in the art would understand that some bacteriophages are defective and need a
helper
phage for payload packaging. Thus, depending on the bacteriophage chosen in
connection with the present invention to prepare the bacterial delivery
particles, the
person skilled in the art would know if a helper phage is required. Sequences
coding for
one or more proteins or regulatory processes necessary for the assembly or
production of
packaged payloads may be supplied in trans. For example, the RBPs of the
present
dislcosure may be provided in a plasmid under the control of an inducible
promoter or
expressed constitutively. In this case, the phage wild-type sequence may or
not contain a
deletion of the gene or sequence supplied in trans. Additionally, chimeric or
modified
phage sequences encoding a new function, like a RBP, may be directly inserted
into the
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
desired position in the genome of the helper phage, hence bypassing the
necessity of
providing the modified sequence in trans. Methods for both supplying a
sequence or
protein in trans in the form of a plasmid, as well as methods to generate
direct genomic
insertions, modifications and mutations are well known to those skilled in the
art.
5 [70] As used herein, the term "payload" refers to any nucleic acid sequence
or amino
acid sequence, or a combination of both (such as, without limitation, peptide
nucleic
acid or peptide-oligonucleotide conjugate) transferred into a bacterium with a
delivery
vehicle. The term "payload" may also refer to a plasmid, a vector or a cargo.
The
payload can be a phagemid or phasmid obtained from natural, evolved or
engineered
10 bacteriophage genome. The payload can also be composed only in part of
phagemid or
phasmid obtained from natural, evolved or engineered bacteriophage genome.
[71] As used herein, the term "nucleic acid" refers to a sequence of at least
two
nucleotides covalently linked together which can be single-stranded or double-
stranded
or contains portion of both single-stranded and double-stranded sequence.
Nucleic acids
15 of the present invention can be naturally occurring, recombinant or
synthetic. The
nucleic acid can be in the form of a circular sequence or a linear sequence or
a
combination of both forms. The nucleic acid can be DNA, both genomic or cDNA,
or
RNA or a combination of both. The nucleic acid may contain any combination of
deoxyribonucleotides and ribonucleotides, and any combination of bases,
including
20 uracil, adenine, thymine, cytosine, guanine, inosine, xathanine,
hypoxathanine,
isocytosine, 5-hydroxymethylcytosine and isoguanine. Other examples of
modified
bases that can be used in the present invention are detailed in Chemical
Reviews 2016,
116 (20) 12655-12687. The term "nucleic acid" also encompasses any nucleic
acid
analogs which may contain other backbones comprising, without limitation,
25 phosphoramide, phosphorothioate, phosphorodithioate, 0-
methylphophoroamidite
linkage and/or deoxyribonucleotides and ribonucleotides nucleic acids. Any
combination
of the above features of a nucleic acid is also encompassed by the present
invention.
[72] Origins of replication known in the art have been identified from species-
specific
plasmid DNAs (e.g. CoIE1, R1, pT181, pSC101, pMB1, R6K, RK2, p 15a and the
like),
from bacterial virus (e.g. yX174, M13, Fl and P4) and from bacterial
chromosomal
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
26
origins of replication (e.g. oriC). In one embodiment, the phagemid according
to the
disclosure comprises a bacterial origin of replication that is functional in
the targeted
bacteria.
[73] Alternatively, the plasmid according to the disclosure does not comprise
any
functional bacterial origin of replication or contain an origin of replication
that is
inactive in the targeted bacteria. Thus, the plasmid of the disclosure cannot
replicate by
itself once it has been introduced into a bacterium by the bacterial virus
particle.
[74] In one embodiment, the origin of replication on the plasmid to be
packaged is
inactive in the targeted bacteria, meaning that this origin of replication is
not functional
in the bacteria targeted by the bacterial virus particles, thus preventing
unwanted
plasmid replication.
[75] In one embodiment, the plasmid comprises a bacterial origin of
replication that is
functional in the bacteria used for the production of the bacterial virus
particles.
[76] Plasmid replication depends on host enzymes and on plasmid-controlled cis
and
trans determinants. For example, some plasmids have determinants that are
recognized
in almost all gram-negative bacteria and act correctly in each host during
replication
initiation and regulation. Other plasmids possess this ability only in some
bacteria
(Kues, U and Stahl, U 1989 Microbiol Rev 53:491-516).
[77] Plasmids are replicated by three general mechanisms, namely theta type,
strand
displacement, and rolling circle (reviewed by Del Solar et al. 1998 Microhio
and Molec
Biol. Rev 62:434-464) that start at the origin of replication. These
replication origins
contain sites that are required for interactions of plasmid and/or host
encoded proteins.
[78] Origins of replication used on the plasmid of the disclosure may be of
moderate
copy number, such as colE1 ori from pBR322 (15-20 copies per cell) or the R6K
plasmid
(15-20 copies per cell) or may be high copy number, e.g. pUC oris (500-700
copies per
cell), pGEM oris (300-400 copies per cell), pTZ oris (>1000 copies per cell)
or
pBluescript oris (300-500 copies per cell).
[79] In one embodiment, the bacterial origin of replication is selected in the
group
consisting of ColE1, pMB1 and variants (pBR322, pET, pUC, etc), pl5a, ColA,
ColE2,
pOSAK, pSC101, R6K, IncW (pSa etc), IncFII, pT181, P 1 , F IncP, IncC, IncJ,
IncN,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
27
IncP1, IncP4, IncQ, IncH11, RSF1010, CloDF13, NTP16, R1, f5, pPS10, pC194,
pE194, BBR1, pBC1, pEP2, pWV01, pLF1311, pAP1, pWKS 1, pLS 1, pLS 11,
pUB6060, pJD4, pll101, pSN22, pAMbetal, pIP501, pIP407, ZM6100(Sa), pCUl,
RA3, pM0L98, RK2/RP4/RP1/R68, pB 10, R300B, pR01614, pR01600, pECB 2,
pCM1, pFA3, RepFIA, RepFIB, RepFIC, pYVE439-80, R387, phasyl, RA1, TF-FC2,
pMV158 and pUB113.
[80] More preferably, the bacterial origin of replication is a E.coli origin
of
replication selected in the group consisting of ColE1, pMB1 and variants
(pBR322, pET,
pUC, etc), pl5a, ColA, ColE2, pOSAK, pSC101, R6K, IncW (pSa etc), IncFII,
pT181,
Pl, F IncP, IncC, IncJ, IncN, IncP1, IncP4, IncQ, IncH11, RSF1010, CloDF13,
NTP16,
R1, f5 and pPS10.
[81] More preferably, the bacterial origin of replication is selected in the
group
consisting of pC194, pE194, BBR1, pBC1, pEP2, pWV01, pLF1311, pAP1, pWKS1,
pLS1, pLS11, pUB6060, pJD4, pll101, pSN22, pAMbetal, pIP501, pIP407,
ZM6100(S a), pCUl, RA3, pM0L98, RK2/RP4/RP1/R68, pB 10, R300B, pR01614,
pR01600, pECB2, pCM1, pFA3, RepFIA, RepFIB, RepFIC, pYVE439-80, R387,
phasyl, RA1, TF-FC2, pMV158 and pUB113.
[82] Even more preferably, the bacterial origin of replication is ColEl.
[83] The delivered nucleic acid sequence according to the disclosure may
comprise a
phage replication origin which can initiate, with complementation of a
complete phage
genome, the replication of the delivered nucleic acid sequence for later
encapsulation
into the different capsids.
[84] A phage origin of replication comprised in the delivered nucleic acid
sequence of
the disclosure can be any origin of replication found in a phage.
[85] Preferably, the phage origin of replication can be the wild-type or non-
wildtype
sequence of the M13, fl, yX174, P4, lambda, P2, lambda-like, HK022, mEP237,
HK97,
HK629, HK630, mEP043, mEP213, mEP234, mEP390, mEP460, mEPxl, mEPx2,
phi80, mEP234, T2, T4, T5, T7, RB49, phiX174, R17, PRD1 P1-like, P2-like, P22,
P22-
like, N15 and N15-like bacteriophages.
[86] More preferably, the phage origin of replication is selected in the group
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
28
consisting of phage origins of replication of M13, fl, yX174, P4, and lambda.
[87] In a particular embodiment, the phage origin of replication is the lambda
or P4
origin of replication.
[88] The delivered nucleic acid of interest comprises a nucleic acid sequence
under
the control of a promoter. In certain embodiments of the invention, the
nucleic acid of
interest is selected from the group consisting of a Cas nuclease gene, a Cas9
nuclease
gene, a guide RNA, a CRISPR locus, a toxin gene, a gene expressing an enzyme
such as
a nuclease or a kinase, a TALEN, a ZFN, a meganuclease, a recombinase, a
bacterial
receptor, a membrane protein, a structural protein, a secreted protein, a gene
expressing
resistance to an antibiotic or to a drug in general, a gene expressing a toxic
protein or a
toxic factor, and a gene expressing a virulence protein or a virulence factor,
or any of
their combination. In an embodiment of the invention, the nucleic acid payload
encodes
a therapeutic protein. In another embodiment, the nucleic acid payload encodes
an anti-
sense nucleic acid molecule.
[89] In one embodiment, the sequence of interest is a programmable nuclease
circuit
to be delivered to the targeted bacteria. This programmable nuclease circuit
is able to
mediate in vivo sequence-specific elimination of bacteria that contain a
target gene of
interest (e.g. a gene that is harmful to humans). Some embodiments of the
present
disclosure relate to engineered variants of the Type II CRISPR-Cas (Clustered
Regularly
Interspaced Short Palindromic Repeats-CRISPR-associated) system of
Streptococcus
pyogenes. Other programmable nucleases that can be used include other CRISPR-
Cas
systems, engineered TALEN (Transcription Activator-Like Effector Nuclease)
variants,
engineered zinc finger nuclease (ZFN) variants, natural, evolved or engineered
meganuclease or recombinase variants, and any combination or hybrids of
programmable nucleases. Thus, the engineered autonomously distributed nuclease
circuits provided herein may be used to selectively cleave DNA encoding a gene
of
interest such as, for example, a toxin gene, a virulence factor gene, an
antibiotic
resistance gene, a remodeling gene or a modulatory gene (cf. W02014124226).
[90] Other sequences of interest, preferably programmable, can be added to the
delivered nucleic acid sequence so as to be delivered to targeted bacteria.
Preferably, the
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
29
sequence of interest added to the delivered nucleic acid sequence leads to
cell death of
the targeted bacteria. For example, the nucleic acid sequence of interest
added to the
plasmid may encode holins or toxins.
[91] Alternatively, the sequence of interest circuit added to the delivered
nucleic acid
sequence does not lead to bacteria death. For example, the sequence of
interest may
encode reporter genes leading to a luminescence or fluorescence signal.
Alternatively,
the sequence of interest may comprise proteins and enzymes achieving a useful
function
such as modifying the metabolism of the bacteria or the composition of its
environment.
[92] In a particular embodiment, the nucleic sequence of interest is selected
in the
group consisting of Cas9, a single guide RNA (sgRNA), a CRISPR locus, a gene
expressing an enzyme such as a nuclease or a kinase, a TALEN, a ZFN, a
meganuclease,
a recombinase, a bacterial receptor, a membrane protein, a structural protein,
a secreted
protein, resistance to an antibiotic or to a drug in general, a gene
expressing a toxic
protein or a toxic factor and a gene expressing a virulence protein or a
virulence factor.
[93] In a particular embodiment, the delivered nucleic acid sequence according
to the
disclosure comprises a nucleic acid sequence of interest that encodes a
bacteriocin,
which can be a proteinaceous toxin produced by bacteria to kill or inhibit
growth of
other bacteria. Bacteriocins are categorized in several ways, including
producing strain,
common resistance mechanisms, and mechanism of killing. Such bacteriocin had
been
described from gram negative bacteria (e.g. microcins, colicin-like
bacteriocins and
tailocins) and from gram positive bacteria (e.g. Class I, Class II, Class III
or Class IV
bacteriocins).
[94] In one embodiment, the delivered nucleic acid sequence according to the
disclosure further comprises a sequence of interest encoding a toxin selected
in the
group consisting of microcins, colicin-like bacteriocins, tailocins, Class I,
Class II, Class
III and Class IV bacteriocins.
[95] In a particular embodiment, the corresponding immunity polypeptide (i.e.
anti-
toxin) may be used to protect bacterial cells (Cotter et al., Nature Reviews
Microbiology
11: 95, 2013) for delivered nucleic acid sequence production and encapsidation
purpose
but is absent in the pharmaceutical composition and in the targeted bacteria
in which the
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
delivered nucleic acid sequence of the disclosure is delivered.
[96] In one aspect of the disclosure, the CRISPR system is included in the
delivered
nucleic acid sequence. The CRISPR system contains two distinct elements, i.e.
i) an
endonuclease, in this case the CRISPR associated nuclease (Cas or "CRISPR
associated
5 protein") and ii) a guide RNA. The guide RNA is in the form of a chimeric
RNA which
consists of the combination of a CRISPR (RNAcr) bacterial RNA and a RNAtracr
(trans-activating RNA CRISPR) (Jinek et al., 2012, Science 337: 816-821). The
guide
RNA combines the targeting specificity of the RNAcr corresponding to the
"spacing
sequences" that serve as guides to the Cas proteins, and the conformational
properties of
10 the RNAtracr in a single transcript. When the guide RNA and the Cas protein
are
expressed simultaneously in the cell, the target genomic sequence can be
permanently
modified or interrupted. The modification is advantageously guided by a repair
matrix.
In general, the CRISPR system includes two main classes depending on the
nuclease
mechanism of action. Class 1 is made of multi-subunit effector complexes and
includes
15 type I, III and IV. Class 2 is made of single-unit effector modules, like
Cas9 nuclease,
and includes type II (II-A,II-B,II-C,II-C variant), V (V-A,V-B,V-C,V-D,V-E,V-
U1,V-
U2,V-U3,V-U4,V-U5) and VI (VI-A,VI-B1,VI-B2,VI-C,VI-D)
[97] The sequence of interest according to the present disclosure comprises a
nucleic
acid sequence encoding Cas protein. A variety of CRISPR enzymes are available
for use
20 as a sequence of interest on the plasmid. In some embodiments, the CRISPR
enzyme is a
Type II CRISPR enzyme. In some embodiments, the CRISPR enzyme catalyzes DNA
cleavage. In some other embodiments, the CRISPR enzyme catalyzes RNA cleavage.
In
one embodiment, the CRISPR enzymes may be coupled to a sgRNA. In certain
embodiments, the sgRNA targets a gene selected in the group consisting of an
antibiotic
25 resistance gene, virulence protein or factor gene, toxin protein or factor
gene, a bacterial
receptor gene, a membrane protein gene, a structural protein gene, a secreted
protein
gene and a gene expressing resistance to a drug in general.
[98] Non-limiting examples of Cas proteins as part of a multi-subunit effector
or as a
single-unit effector include Cas 1, Cas1B, Cas2, Cas3, Cas4, Cas5, Cas6, Cas7,
Cas8,
30 Cas9 (also known as Csnl and Csx12), Cas10, Cash 1 (SS), Cas12a (Cpfl),
Cas12b
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
31
(C2c1), Cas12c (C2c3), Cas12d (CasY), Cas12e (CasX), C2c4, C2c8, C2c5, C2c10,
C2c9, Cas13a (C2c2), Cas13b (C2c6), Cas13c (C2c7), Cas13d, Csa5, Cscl, Csc2,
Csel,
Cse2, Csy 1, Csy2, Csy3, Csfl, Csf2, Csf3, Csf4, Csm2, Csm3, Csm4, Csm5, Csm6,
Cmrl, Cmr3, Cmr4, Cmr5, Cmr6, Csn2, Csbl, Csb2, Csb3, Csx17, Csx14, Csx10,
Csx16, CsaX, Csx13, Csxl, Csx15, SdCpfl, CmtCpfl, TsCpfl, CmaCpfl, PcCpfl,
ErCpfl, FbCpfl, UbcCpfl, AsCpfl, LbCpfl, homologues thereof, orthologues
thereof,
variants thereof, or modified versions thereof. In some embodiments, the
CRISPR
enzyme cleaves both strands of the target nucleic acid at the Protospacer
Adjacent Motif
(PAM) site.
[99] In a particular embodiment, the CRISPR enzyme is any Cas9 protein, for
instance any naturally-occurring bacterial Cas9 as well as any variants,
homologs or
orthologs thereof.
[100] By "Cas9" is meant a protein Cas9 (also called Csnl or Csx12) or a
functional
protein, peptide or polypeptide fragment thereof, i.e. capable of interacting
with the
guide RNA(s) and of exerting the enzymatic activity (nuclease) which allows it
to
perform the double-strand cleavage of the DNA of the target genome. "Cas9" can
thus
denote a modified protein, for example truncated to remove domains of the
protein that
are not essential for the predefined functions of the protein, in particular
the domains
that are not necessary for interaction with the gRNA(s).
[101] The sequence encoding Cas9 (the entire protein or a fragment thereof) as
used in
the context of the disclosure can be obtained from any known Cas9 protein
(Fonfara et
al., Nucleic Acids Res 42 (4), 2014; Koonin et al., Nat Rev Microbiol 15(3),
2017).
Examples of Cas9 proteins useful in the present disclosure include, but are
not limited
to, Cas9 proteins of Streptococcus pyo genes (SpCas9), Streptococcus
thermophilus
(St1Cas9, St3Cas9), Streptococcus mutans, Staphylococcus aureus (SaCas9),
Campylobacter jejuni (CjCas9), Francisella novicida (FnCas9) and Neisseria
meningitides (NmCas9).
[102] The sequence encoding Cpfl (Cas12a) (the entire protein or a fragment
thereof)
as used in the context of the disclosure can be obtained from any known Cpfl
(Cas12a)
protein (Koonin et al., Nat Rev Microbiol 15(3), 2017). Examples of
Cpfl(Cas12a)
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
32
proteins useful in the present disclosure include, but are not limited to,
Cpfl(Cas12a)
proteins of Acidaminococcus sp, Lachnospiraceae bacteriu and Francisella
novicida.
[103] The sequence encoding Cas13a (the entire protein or a fragment thereof)
can be
obtained from any known Cas13a (C2c2) protein (Abudayyeh et al., 2017, Nature
550:
280-284) . Examples of Cas13a (C2c2) proteins useful in the present disclosure
include,
but are not limited to, Cas13a (C2c2) proteins of Leptotrichia wadei
(LwaCas13a).
[104] The sequence encoding Cas13d (the entire protein or a fragment thereof)
can be
obtained from any known Cas13d protein (Yan et al., 2018, Mol Cell 70: 327-
339).
Examples of Cas13d proteins useful in the present disclosure include, but are
not limited
to, Cas13d proteins of Eubacterium siraeum and Ruminococcus sp.
[105] In a particular embodiment, the nucleic sequence of interest is a
CRISPR/Cas9
system for the reduction of gene expression or inactivation of a gene selected
in the
group consisting of an antibiotic resistance gene, virulence factor or protein
gene, toxin
factor or protein gene, a gene expressing a bacterial receptor, a membrane
protein, a
structural protein, a secreted protein, and a gene expressing resistance to a
drug in
general.
[106] In one embodiment, the CRISPR system is used to target and inactivate a
virulence factor. A virulence factor can be any substance produced by a
pathogen that
alter host-pathogen interaction by increasing the degree of damage done to the
host.
Virulence factors are used by pathogens in many ways, including, for example,
in cell
adhesion or colonization of a niche in the host, to evade the host's immune
response, to
facilitate entry to and egress from host cells, to obtain nutrition from the
host, or to
inhibit other physiological processes in the host. Virulence factors can
include enzymes,
endotoxins, adhesion factors, motility factors, factors involved in complement
evasion,
and factors that promote biofilm formation. For example, such targeted
virulence factor
gene can be E. coli virulence factor gene such as, without limitation, EHEC-
HlyA, Stx 1
(VT1), Stx2 (VT2), Stx2a (VT2a), Stx2b (VT2b), Stx2c (VT2c), Stx2d (VT2d),
Stx2e
(VT2e) and Stx2f (VT2f), Stx2h (VT2h), fimA, fimF, fimH, neuC, kpsE, sfa, foc,
iroN,
aer, iha, papC, papGI, papGII, papGIII, hlyC, cnfl, hra, sat, ireA, usp ompT,
ibeA,
malX, fyuA, irp2, traT, afaD, ipaH, eltB, estA, bfpA, eaeA, espA, aaiC, aatA,
TEM,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
33
CTX, SHY, csgA, csgB, csgC, csgD, csgE, csgF, csgG, csgH, T1SS, T2SS, T3SS,
T4SS, T5SS, T6SS (secretion systems). For example, such targeted virulence
factor gene
can be Shigella dysenteriae virulence factor gene such as, without limitation,
stx 1 and
stx2. For example, such targeted virulence factor gene can be Yersinia pestis
virulence
factor gene such as, without limitation, yscF (plasmid-borne (pCD1) T3SS
external
needle subunit). For example, such targeted virulence factor gene can be
Francisella
tularensis virulence factor gene such as, without limitation, fs1A. For
example, such
targeted virulence factor gene can be Bacillus anthracis virulence factor gene
such as,
without limitation, pag (Anthrax toxin, cell-binding protective antigen). For
example,
such targeted virulence factor gene can be Vibrio cholera virulence factor
gene such as,
without limitation, ctxA and ctxB (cholera toxin), tcpA (toxin co-regulated
pilus), and
toxT (master virulence regulator). For example, such targeted virulence factor
gene can
be Pseudomonas aeruginosa virulence factor genes such as, without limitation,
pyoverdine (e.g., sigma factor pvdS, biosynthetic genes pvdL, pvdl, pvdJ,
pvdH, pvdA,
pvdF, pvdQ, pvdN, pvdM, pvd0, pvdP, transporter genes pvdE, pvdR, pvdT, opmQ),
siderophore pyochelin (e.g., pchD, pchC, pchB, pchA, pchE, pchF and pchG, and
toxins
(e.g., exoU, exoS and exoT). For example, such targeted virulence factor gene
can be
Klebsiella pneumoniae virulence factor genes such as, without limitation, fimA
(adherence, type I fimbriae major subunit), and cps (capsular polysaccharide).
For
example, such targeted virulence factor gene can be Acinetobacter baumannii
virulence
factor genes such as, without limitation, ptk (capsule polymerization) and
epsA
(assembly). For example, such targeted virulence factor gene can be Salmonella
enterica
Typhi virulence factor genes such as, without limitation, MIA (invasion, SPI-1
regulator), ssrB (SPI-2 regulator), and those associated with bile tolerance,
including
efflux pump genes acrA, acrB and to1C. For example, such targeted virulence
factor
gene can be Fusobacterium nucleatum virulence factor genes such as, without
limitation,
FadA and TIGIT. For example, such targeted virulence factor gene can be
Bacteroides
fragilis virulence factor genes such as, without limitation, bft.
[107] In another embodiment, the CRISPR/Cas9 system is used to target and
inactivate
an antibiotic resistance gene such as, without limitation, GyrB, ParE, ParY,
AAC(1),
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
34
AAC(2'), AAC(3), AAC(6'), ANT(2"), ANT(3"), ANT(4'), ANT(6), ANT(9), APH(2"),
APH(3"), APH(3'), APH(4), APH(6), APH(7"), APH(9), ArmA, RmtA, RmtB, RmtC,
Sgm, AER, BLA1, CTX-M, KPC, SHY, TEM, BlaB, CcrA, IMP, NDM, VIM, ACT,
AmpC, CMY, LAT, PDC, OXA 13-lactamase, mecA, 0mp36, OmpF, PIB, bla (blaI,
blaR1) and mec (mecI, mecR1) operons, Chloramphenicol acetyltransferase (CAT),
Chloramphenicol phosphotransferase, Ethambutol-resistant arabinosyltransferase
(EmbB), MupA, MupB, Integral membrane protein MprF, Cfr 23S rRNA
methyltransferase, Rifampin ADP-ribosyltransferase (Arr),
Rifampin
glycosyltransferase, Rifampin monooxygenase, Rifampin phosphotransferase,
DnaA,
RbpA, Rifampin-resistant beta-subunit of RNA polymerase (RpoB), Erm 23S rRNA
methyltransferases, Lsa, MsrA, Vga, VgaB, Streptogramin Vgb lyase, Vat
acetyltransferase, Fluoroquinolone acetyltransferase, Fluoroquinolone-
resistant DNA
topoisomerases, Fluoroquinolone-resistant GyrA, GyrB, ParC, Quinolone
resistance
protein (Qnr), FomA, FomB, FosC, FosA, FosB, FosX, VanA, VanB, VanD, VanR,
VanS, Lincosamide nucleotidyltransferase (Lin), EreA, EreB, GimA, Mgt, Ole,
Macrolide phosphotransferases (MPH), MefA, MefE, Mel, Streptothricin
acetyltransferase (sat), Su11, Sul2, Sul3, sulfonamide-resistant FolP,
Tetracycline
inactivation enzyme TetX, TetA, TetB, TetC, Tet30, Tet31, TetM, Tet0, TetQ,
Tet32,
Tet36, MacAB-To1C, MsbA, MsrA,VgaB, EmrD, EmrAB-To1C, NorB, GepA, MepA,
AdeABC, AcrD, MexAB-OprM, mtrCDE, EmrE, adeR, acrR, baeSR, mexR, phoPQ,
mtrR, or any antibiotic resistance gene described in the Comprehensive
Antibiotic
Resistance Database (CARD https://card.mcmaster.ca/).
[108] In another embodiment, the CRISPR/Cas9 system is used to target and
inactivate
a bacterial toxin gene. Bacterial toxin can be classified as either exotoxins
or endotoxins.
Exotoxins are generated and actively secreted; endotoxins remain part of the
bacteria.
The response to a bacterial toxin can involve severe inflammation and can lead
to sepsis.
Such toxin can be for example Botulinum neurotoxin, Tetanus toxin,
Staphylococus
toxins, Diphteria toxin, Anthrax toxin, Alpha toxin, Pertussis toxin, Shiga
toxin, Heat-
stable enterotoxin (E. coli ST), colibactin, BFT (B. fragilis toxin) or any
toxin described
in Henkel et al., (Toxins from Bacteria in EXS. 2010; 100: 1-29).
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
[109] The bacteria targeted by bacterial delivery vehicles disclosed herein
can be any
bacteria present in a mammal organism. In a certain aspect, the bacteria are
targeted
through interaction of the chimeric RBPs and/or the branched-RBPs expressed by
the
delivery vehicles with the bacterial cell. It can be any commensal, symbiotic
or
5 pathogenic bacteria of the microbiota or microbiome.
[110] A microbiome may comprise of a variety of endogenous bacterial species,
any of
which may be targeted in accordance with the present disclosure. In some
embodiments,
the genus and/or species of targeted endogenous bacterial cells may depend on
the type
of bacteriophages being used for preparing the bacterial delivery vehicles.
For example,
10 some bacteriophages exhibit tropism for, or preferentially target, specific
host species of
bacteria. Other bacteriophages do not exhibit such tropism and may be used to
target a
number of different genus and/or species of endogenous bacterial cells.
[111] Examples of bacterial cells include, without limitation, cells from
bacteria of the
genus Yersinia spp., Escherichia spp., Klebsiella spp., Acinetobacter spp.,
Bordetella
15 spp., Neisseria spp., Aeromonas spp., Franciesella spp., Corynebacterium
spp.,
Citrobacter spp., Chlamydia spp., Hemophilus spp., Brucella spp.,
Mycobacterium spp.,
Legionella spp., Rhodococcus spp., Pseudomonas spp., Helicobacter spp., Vibrio
spp.,
Bacillus spp., Erysipelothrix spp., Salmonella spp., Streptomyces spp.,
Streptococcus
spp., Staphylococcus spp., Bacteroides spp., Prevotella spp., Clostridium
spp.,
20 Bifidobacterium spp., Clostridium spp., Brevibacterium spp., Lactococcus
spp.,
Leuconostoc spp., Actinobacillus spp., Selnomonas spp., Shigella spp., Zymonas
spp.,
Mycoplasma spp., Treponema spp., Leuconostoc spp., Corynebacterium spp.,
Enterococcus spp., Enterobacter spp., Pyrococcus spp., Serratia spp.,
Morganella spp.,
Parvimonas spp., Fusobacterium spp., Actinomyces spp., Porphyromonas spp.,
25 Micrococcus spp., Bartonella spp., Borrelia spp., Brucelia spp.,
Campylobacter spp.,
Chlamydophilia spp., Cutibacterium (formerly Propionibacterium) spp.,
Ehrlichia spp.,
Haemophilus spp., Leptospira spp., Listeria spp., Mycoplasma spp., Nocardia
spp.,
Rickettsia spp., Ureaplasma spp., and Lactobacillus spp, and a mixture
thereof.
[112] Thus, bacterial delivery vehicles may target (e.g., specifically target)
a bacterial
30 cell from any one or more of the foregoing genus of bacteria to
specifically deliver the
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
36
payload of interest according to the disclosure.
[113] Preferably, the targeted bacteria can be selected from the group
consisting of
Yersinia spp., Escherichia spp., Klebsiella spp., Acinetobacter spp.,
Pseudomonas spp.,
Helicobacter spp., Vibrio spp, Salmonella spp., Streptococcus spp.,
Staphylococcus spp.,
Bacteroides spp., Clostridium spp., Shigella spp., Enterococcus spp.,
Enterobacter spp.,
and Listeria spp.
[114] In some embodiments, bacterial cells of the present disclosure are
anaerobic
bacterial cells (e.g., cells that do not require oxygen for growth). Anaerobic
bacterial
cells include facultative anaerobic cells such as but not limited to
Escherichia coli,
Shewanella oneidensis and Listeria. Anaerobic bacterial cells also include
obligate
anaerobic cells such as, for example, Bacteroides and Clostridium species. In
humans,
anaerobic bacteria are most commonly found in the gastrointestinal tract. In
some
particular embodiment, the targeted bacteria are thus bacteria most commonly
found in
the gastrointestinal tract. Bacteriophages used for preparing the bacterial
virus particles,
and then the bacterial virus particles, may target (e.g., to specifically
target) anaerobic
bacterial cells according to their specific spectra known by the person
skilled in the art to
specifically deliver the plasmid.
[115] In some embodiments, the targeted bacterial cells are, without
limitation,
Bacteroides thetaiotaomicron, Bacteroides fragilis, Bacteroides distasonis,
Bacteroides
vulgatus, Clostridium leptum, Clostridium coccoides, Staphylococcus aureus,
Bacillus
subtilis, Clostridium butyricum, Brevibacterium lactofermentum, Streptococcus
agalactiae, Lactococcus lactis, Leuconostoc lactis,
Actinobacillus
actinobycetemcomitans, cyanobacteria, Escherichia coli, Helicobacter pylori,
Selnomonas ruminatium, Shigella sonnei, Zymomonas mobilis, Mycoplasma
mycoides,
Treponema denticola, Bacillus thuringiensis, Staphilococcus lugdunensis,
Leuconostoc
oenos, Corynebacterium xerosis, Lactobacillus plantarum, Lactobacillus
rhamnosus,
Lactobacillus casei, Lactobacillus acidophilus, Enterococcus faecalis,
Bacillus
coagulans, Bacillus cereus, Bacillus popillae, Synechocystis strain PCC6803,
Bacillus
liquefaciens, Pyrococcus abyssi, Selenomonas nominantium, Lactobacillus
hilgardii,
Streptococcus ferus, Lactobacillus pentosus, Bacteroides fragilis,
Staphylococcus
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
37
epidermidis, Streptomyces phaechromo genes, Streptomyces ghanaenis, Klebsiella
pneumoniae, Enterobacter cloacae, Enterobacter aero genes, Serratia
marcescens,
Morganella morganii, Citrobacter freundii, Pseudomonas aerigunosa, Parvimonas
micra, Prevotella intermedia, Fusobacterium nucleatum, Prevotella nigrescens,
Actinomyces israelii, Porphyromonas endodontalis, Porphyromonas gingivalis
Micrococcus luteus, Bacillus megaterium, Aeromonas hydrophila, Aeromonas
caviae,
Bacillus anthracis, Bartonella henselae, Bartonella Quintana, Bordetella
pertussis,
Borrelia burgdorferi, Borrelia garinii, Borrelia afzelii, Borrelia
recurrentis, Brucella
abortus, Brucella canis, Brucella melitensis, Brucella suis, Campylobacter
jejuni,
Campylobacter coli, Campylobacter fetus, Chlamydia pneumoniae, Chlamydia
trachomatis, Chlamydophila psittaci, Clostridium botulinum, Clostridium
difficile,
Clostridium perfringens, Clostridium tetani, Corynebacterium diphtheria,
Cutibacterium
acnes (formerly Propionibacterium acnes), Ehrlichia canis, Ehrlichia
chaffeensis,
Enterococcus faecium, Francisella tularensis, Haemophilus influenza,
Legionella
pneumophila, Leptospira interrogans, Leptospira santarosai, Leptospira weilii,
Leptospira noguchii, Listeria monocyto genes, Mycobacterium leprae,
Mycobacterium
tuberculosis, Mycobacterium ulcerans, Mycoplasma pneumonia, Neisseria
gonorrhoeae,
Neisseria meningitides, Nocardia asteroids, Rickettsia rickettsia, Salmonella
enteritidis,
Salmonella typhi, Salmonella paratyphi, Salmonella typhimurium, Shigella
flexnerii,
Shigella dysenteriae, Staphylococcus saprophyticus, Streptococcus pneumoniae,
Streptococcus pyo genes, Streptococcus viridans, Treponema pallidum,
Ureaplasma
urealyticum, Vibrio cholera, Vibrio parahaemolyticus, Yersinia pestis,
Yersinia
enterocolitica, Yersinia pseudotuberculosis, Actinobacter baumanii,
Pseudomonas
aerigunosa, and a mixture thereof, preferably the bacteria of interest are
selected from
the group consisting of Escherichia coli, Enterococcus faecium, Staphylococcus
aureus,
Klebsiella pneumoniae, Acinetobacter baumanii, Pseudomonas aeruginosa,
Enterobacter cloacae, and Enterobacter aero genes, and a mixture thereof.
[116] In one embodiment, the targeted bacteria are Escherichia co/i.
[117] Thus, bacteriophages used for preparing the bacterial delivery vehicles,
and then
the bacterial delivery vehicles, may target (e.g., specifically target) a
bacterial cell from
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
38
any one or more of the foregoing genus and/or species of bacteria to
specifically deliver
the payload of interest.
[118] In one embodiment, the targeted bacteria are pathogenic bacteria. The
targeted
bacteria can be virulent bacteria.
[119] The targeted bacteria can be antibacterial resistance bacteria,
preferably selected
from the group consisting of extended-spectrum beta-lactamase-producing (ESBL)
Escherichia coli, ESBL Klebsiella pneumoniae, vancomycin-resistant
Enterococcus
(VRE), methicillin-resistant Staphylococcus aureus (MRSA), multidrug-resistant
(MDR) Acinetobacter baumannii, MDR Enterobacter spp., and a combination
thereof.
Preferably, the targeted bacteria can be selected from the group consisting of
extended-
spectrum beta-lactamase-producing (ESBL) Escherichia coli strains.
[120] Alternatively, the targeted bacterium can be a bacterium of the
microbiome of a
given species, preferably a bacterium of the human microbiota.
[121] The present disclosure is directed to bacterial delivery vehicle
containing the
payload as described herein. The bacterial delivery vehicles are prepared from
bacterial
virus. The bacterial delivery vehicles are chosen in order to be able to
introduce the
payload into the targeted bacteria.
[122] Bacterial viruses, from which the bacterial delivery vehicles having
chimeric
receptor binding proteins and/or branched-RBPs may be derived, are preferably
bacteriophages. Optionally, the bacteriophage is selected from the Order
Caudovirales
consisting of, based on the taxonomy of Krupovic et al, 2015, Arch Virol,
161(1): 233-
247:
[123] Bacteriophages may be selected from the family Myoviridae (such as,
without
limitation, genus Cp220virus, Cp8virus, Ea214virus, Felixolvirus, Mooglevirus,
Suspvirus , Hplvirus, P2virus, Kayvirus , PlOOvirus, Silviavirus , Spolvirus,
Tsarbombavirus, Twortvirus, Cc31virus, Jd18virus, Js98virus, Kp15virus,
Moonvirus,
Rb49virus, Rb69virus, S16virus, Schizot4virus, Sp18virus, T4virus, Cr3virus,
Selvirus,
V5virus, Abouovirus, Agatevirus, Agrican357virus, Ap22virus, Arvlvirus,
B4virus,
B astillevirus, Bc431virus, Bcep78virus, Bcepmuvirus, Biquartavirus,
Bxzlvirus,
Cd119virus, Cp51virus, CvmlOvirus, Eah2virus, Elvirus, Hapunavirus,
Jimmervirus,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
39
KpplOvirus, M12virus, Machinavirus, Marthavirus, Ms w3virus , Muvirus,
Myohalovirus, Nitivirus, P1 virus, Pakpunavirus, Pbunavirus, Phikzvirus,
Rheph4virus,
Rs12virus, Rslunavirus, Secunda5virus, Seplvirus, 5pn3virus, Svunavirus,
Tglvirus,
Vhmlvirus and Wphvirus)
[124] Bacteriophages may be selected from the family Podoviridae (such as,
without
limitation, genus Frilvirus, Kp32virus, Kp34virus, Phikmvvirus, Pradovirus,
5p6virus,
T7virus, Cplvirus, P68virus, Phi29virus, Nona33virus, Pocj virus ,
T12011virus,
Bcep22virus, Bpplvirus, Cba41virus, Dfll2virus, Ea92virus, Epsilon15virus,
F116virus,
G7cvirus, Jwalphavirus, Kfl virus, Kpp25virus, Litl virus, Luz24virus,
Luz7virus,
N4virus, Nonanavirus, P22virus, Pagevirus, Phieco32virus, Prtbvirus,
5p58virus,
Una961virus and Vp5virus)
[125] - Bacteriophages may be selected from the family Siphoviridae (such as,
without
limitation, genus Camvirus, Likavirus, R4virus, Acadianvirus, Coopervirus,
Pglvirus,
Pipefishvirus, Rosebushvirus, Brujitavirus, Che9cvirus, Hawkeyevirus,
Plotvirus,
Jerseyvirus, Klgvirus, 5p31 virus, Lmdlvirus, Una4virus, Bongovirus, Reyvirus,
Buttersvirus, Charlievirus, Redivirus, Baxtervirus, Nymphadoravirus,
Bignuzvirus,
Fishburnevirus, Phayoncevirus, Kp36virus, Roguelvirus, Rtpvirus, Ti virus, Tls
virus ,
Abl8virus, Amigovirus, Anatolevirus, Andromedavirus, Attis virus , B
arnyardvirus,
B ernal 1 3virus, Biseptimavirus, Bronvirus, C2virus, C5virus, Cbal8lvirus,
Cbastvirus,
Cecivirus, Che8virus, Chivirus, Cjwlvirus, Corndogvirus, Cronusvirus,
D3112virus,
D3virus, Decurrovirus, Demosthenesvirus, Doucettevirus, E125virus, Eiauvirus,
Ff47virus, Gaiavirus, Gilesvirus, Gordonvirus, Gordtnkvirus, Harrisonvirus,
Hk578virus, Hk97virus, Jenstvirus, Jwxvirus, Kelleziovirus, Korravirus,
L5virus,
lambdavirus, Laroyevirus, Liefievirus, Marvinvirus, Mudcatvirus, N15virus,
Nonagvirus, Nplvirus, Omegavirus, P12002virus, P12024virus, P23virus,
P70virus,
Pa6virus, Pamx74virus, Patiencevirus, Phil virus, Pepy6virus, Pfrl virus,
Phic31virus,
Phicbkvirus, Phietavirus, Phifelvirus, Phijll virus, Pis4avirus, Psavirus,
Psimunavirus,
Rdjlvirus, Rer2virus, 5ap6virus, Send513virus, 5eptima3virus, Seuratvirus,
Sextaecvirus, Sfillvirus, 5fi21dt1virus, Sitaravirus, Sklvirus, Slashvirus,
Smoothievirus, Soupsvirus, Spbetavirus, Ssp2virus, T5virus, Tankvirus,
Tin2virus,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
Titanvirus, Tm4virus, Tp21virus, Tp84virus, Triavirus, Trigintaduovirus,
Vegasvirus,
Vendettavirus, Wbetavirus, Wildcatvirus, Wizardvirus, Woesvirus, XplOvirus,
Ydn12virus and Yuavirus)
[126] Bacteriophages may be selected from the family Ackermannviridae (such
as,
5 without limitation, genus Ag3virus, Limestonevirus, Cba120virus and
Vilvirus)
[127] Optionally, the bacteriophage is not part of the order Caudovirales but
from
families with unassigned order such as, without limitation, family
Tectiviridae (such as
genus Alphatectivirus, Betatectivirus), family Corticoviridae (such as genus
Corticovirus), family Inoviridae (such as genus Fibrovirus, Habenivirus,
Inovirus,
10 Lineavirus, Plectrovirus, Saetivirus, Vespertiliovirus), family Cy
stoviridae(such as
genus Cystovirus), family Leviviridae(such as genus Allolevivirus, Levivirus),
family
Microviridae (such as genus Alpha3microvirus, G4microvirus, Phix174microvirus,
Bdellomicrovirus, Chlamydiamicrovirus, Spiromicrovirus) and family
Plasmaviridae
(such as genus Plasmavirus).
15 [128] Optionally, the bacteriophage is targeting Archea not part of the
Order
Caudovirales but from families with Unassigned order such as, without
limitation,
Ampullaviridae, FuselloViridae, Globuloviridae, Guttaviridae,
Lipothrixviridae,
Pleolipoviridae, Rudiviridae, Salterprovirus and Bicaudaviridae.
[129] A non-exhaustive listing of bacterial genera and their known host-
specific
20 bacteria viruses is presented in the following paragraphs. The chimeric
RBPs and/or the
branched RBPs and the bacterial delivery vehicles disclosed herein may be
engineered,
as non-limiting examples, from the following phages. Synonyms and spelling
variants
are indicated in parentheses. Homonyms are repeated as often as they occur
(e.g., D, D,
d). Unnamed phages are indicated by "NN" beside their genus and their numbers
are
25 given in parentheses.
[130] Bacteria of the genus Actinomyces can be infected by the following
phages: Av-I,
Av-2, Av-3, BF307, CT1, CT2, CT3, CT4, CT6, CT7, CT8 and 1281.
[131] Bacteria of the genus Aeromonas can be infected by the following phages:
AA-I,
Aeh2, N, PM1, TP446, 3, 4, 11, 13, 29, 31, 32, 37, 43, 43-10T, 51, 54, 55R.1,
56,
30 56RR2, 57, 58, 59.1, 60, 63, Aehl, F, PM2, 1, 25, 31, 40RR2.8t, (syn= 44R),
(syn=
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
41
44RR2.8t), 65, PM3, PM4, PM5 and PM6.
[132] Bacteria of the genus Bacillus can be infected by the following phages:
A, aizl,
Al-K-I, B, BCJA1, BC1, BC2, BLL1, BL1, BP142, BSL1, BSL2, BS1, BS3, BS8, BS15,
BS18, BS22, BS26, BS28, BS31, BS104, BS105, BS106, BTB, B1715V1, C, CK-I,
Coll, Corl, CP-53, CS-I, CSi, D, D, D, D5, entl, FP8, FP9, FSi, F52, F53, F55,
F58,
F59, G, GH8, GT8, GV-I, GV-2, GT-4, g3, g12, g13, g14, g16, g17, g21, g23,
g24, g29,
H2, kenl, KK-88, Kuml, Kyul, J7W-1, LP52, (syn= LP-52), L7, Mexl, MJ-I, mor2,
MP-
7, MP10, MP12, MP14, MP15, Neol, N 2, N5, N6P, PBC1, PBLA, PBP1, P2, S-a, SF2,
SF6, Shal, Sill, 5P02, (syn= (I)5PP1), 5P13, STI, STi, SU-Il, t, TbI, Tb2,
Tb5, TbIO,
Tb26, Tb51, Tb53, Tb55, Tb77, Tb97, Tb99, Tb560, Tb595, Td8, Td6, Td15, TgI,
Tg4,
Tg6, Tg7, Tg9, TgIO, TgIl, Tg13, Tg15, Tg21, Tinl, Tin7, Tin8, Tin13, Tm3,
Tocl, Togl,
toll, TP-I, TP-10vir, TP-15c, TP-16c, TP-17c, TP-19, TP35, TP51, TP-84, Tt4,
Tt6, type
A, type B, type C, type D, type E, TO, VA-9, W, wx23, wx26, Yunl, a, y, pllõ
ymed-2,
(pT, (p11-4, (PT, (p75, (p105, (syn= y105), IA, IB, 1-97A, 1-97B, 2, 2, 3, 3,
3, 5, 12, 14,
20, 30, 35, 36, 37, 38, 41C, 51, 63, 64, 138D, I, II, IV, NN-Bacillus (13),
alel, AR1,
AR2, AR3, AR7, AR9, Bace-11, (syn= 11), Bastille, BL1, BL2, BL3, BL4, BLS,
BL6,
BL8, BL9, BP124, B528, B580, Ch, CP-51, CP-54, D-5, darl, denl, DP-7, entl,
FoSi,
FoS2, F54, F56, F57, G, gall, gamma, GE1, GF-2, GSi, GT-I, GT-2, GT-3, GT-4,
GT-5,
GT-6, GT-7, GV-6, g15, 19, 110, ISi, K, MP9, MP13, MP21, MP23, MP24, MP28,
MP29, MP30, MP32, MP34, MP36, MP37, MP39, MP40, MP41, MP43, MP44, MP45,
MP47, MP50, NLP-I, No.1, N17, N19, PBS1, PK1, PMB1, PMB12, PMJ1, S, SP01, 5P3,
5P5, 5P6, 5P7, 5P8, 5P9, SP10, SP-15, 5P50, (syn= SP-50), 5P82, SST, subl, SW,
Tg8,
Tg12, Tg13, Tg14, thul, thuA, thuS, Tin4, Tin23, TP-13, TP33, TP50, TSP-I,
type V, type
VI, V, Vx, 022, ye, (pNR2, (p25, (p63, 1, 1, 2, 2C, 3NT, 4, 5, 6, 7, 8, 9, 10,
12, 12, 17, 18,
19, 21, 138, III, 4 (B. megateriwn), 4 (B. sphaericus), AR13, BPP-I0, B532,
B5107, Bl,
B2, GA-I, GP-I0, GV-3, GV-5, g8, MP20, MP27, MP49, Nf, PPS, PP6, SF5, Tg18, TP-
I, Versailles, y15, (p29, 1-97, 837/IV, mi-Bacillus (1), Bat10, BSL10, BSLI 1,
B56, BSI 1,
B516, B523, BS101, B5102, g18, morl, PBL1, 5N45, thu2, thu3, TmI, Tm2, TP-20,
TP21, TP52, type F, type G, type IV, HN-BacMus (3), BLE, (syn= 0c), B52, B54,
B55,
B57, B10, B12, B520, B521, F, MJ-4, PBA12, AP50, AP50-04, AP50-11, AP50-23,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
42
AP50-26, AP50-27 and Bam35. The following Bacillus-specific phages are
defective:
DLP10716, DLP-11946, DPB5, DPB12, DPB21, DPB22, DPB23, GA-2, M, No. IM,
PBLB, PBSH, PBSV, PBSW, PBSX, PBSY, PBSZ, phi, SPa, type 1 and 1.t.
[133] Bacteria of the genus Bacteriodes can be infected by the following
phages: ad 12,
Baf-44, Baf-48B, Baf-64, Bf-I, Bf-52, B40-8, Fl, f31, yAl, yBrOl, yBr02, 11,
67.1, 67.3,
68.1, mt-Bacteroides (3), Bf42, Bf71, HN-Bdellovibrio (1) and BF-41.
[134] Bacteria of the genus Bordetella can be infected by the following
phages: 134
and NN-Bordetella (3).
[135] Bacteria of the genus Borrellia can be infected by the following phages:
NN-
Borrelia (1) and NN-Borrelia (2).
[136] Bacteria of the genus Brucella can be infected by the following phages:
A422,
Bk, (syn= Berkeley), BM29, F0i, (syn= F01), (syn= FQ1), D, FP2, (syn= FP2),
(syn=
FD2), Fz, (syn= Fz75/13), (syn= Firenze 75/13), (syn= Fi), Fi, (syn= F1), Fim,
(syn=
FIm), (syn= Fim), FiU, (syn= Flu), (syn= FiU), F2, (syn= F2), F3, (syn= F3),
F4, (syn=
F4), F5, (syn= F5), F6, F7, (syn= F7), F25, (syn= F25), (syn= 25), F25U,
(syn= F25u),
(syn= F25U), (syn= F25V), F44, (syn- F44), F45, (syn= F45), F48, (syn= F48),
I, Im, M,
MC/75, M51, (syn= M85), P, (syn= D), S708, R, Tb, (syn= TB), (syn= Tbilisi),
W,
(syn= Wb), (syn= Weybridge), X, 3, 6, 7, 10/1, (syn= 10), (syn= F8), (syn=
F8), 12m,
24/11, (syn= 24), (syn= F9), (syn= F9), 45/111, (syn= 45), 75, 84, 212/XV,
(syn= 212),
(syn= Fi0), (syn= F10), 371/XXIX, (syn= 371), (syn= Fn), (syn= F11) and 513.
[137] Bacteria of the genus Burkholderia can be infected by the following
phages:
CP75, NN-Burkholderia (1) and 42.
[138] Bacteria of the genus Campylobacter can be infected by the following
phages: C
type, NTCC12669, NTCC12670, NTCC12671, NTCC12672, NTCC12673,
NTCC12674, NTCC12675, NTCC12676, NTCC12677, NTCC12678, NTCC12679,
NTCC12680, NTCC12681, NTCC12682, NTCC12683, NTCC12684, 32f, 111c, 191,
NN-Campylobacter (2), Vfi-6, (syn= V19), VfV-3, V2, V3, V8, V16, (syn= Vfi-1),
V19,
V20(V45), V45, (syn= V-45) and NN-Campylobacter (1).
[139] Bacteria of the genus Chlamydia can be infected by the following phage:
Chpl.
[140] Bacteria of the genus Clostridium can be infected by the following
phages:
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
43
CAK1, CA5, Ca7, CEP, (syn= 1C), CEy, Cldl, c-n71, c-203 Tox-, DEP, (syn= ID),
(syn=
1Dt0X+), HM3, KM1, KT, Ms, NA1, (syn= Naltox+), PA1350e, Pf6, PL73, PL78,
PL81,
Pl, P50, P5771, P19402, 1Ct0X+, 2Ct0X\ 2D3 (syn= 2Dt0X+), 3C, (syn= 3Ctox+),
4C,
(syn= 4Ct0X+), 56, III-1, NN-Clostridium (61), NB1t0X+, al, CA1, HMT, HM2,
PF15 P-
23, P-46, Q-05, Q-oe, Q-16, Q-21, Q-26, Q-40, Q-46, S111, SA02, WA01, WA03,
Wm,
W523, 80, C, CA2, CA3, CPT1, CPT4, cl, c4, c5, HM7, H11/A1, H18/Ax, FWS23,
Hi58ZA1, K2ZA1, K21ZS23, ML, NA2t0X; Pf2, Pf3, Pf4, S9ZS3, S41ZA1, S44ZS23,
a2, 41, 112ZS23, 214/S23, 233/Ai, 234/S23, 235/S23, II-1, 11-2, 11-3, NN-
Clostridium
(12), CA1, Fl, K, S2, 1, 5 and NN-Clostridium (8).
[141] Bacteria of the genus Corynebacterium can be infected by the following
phages:
CGK1 (defective), A, A2, A3, A101, A128, A133, A137, A139, A155, A182, B, BF,
B17, B18, B51, B271, B275, B276, B277, B279, B282, C, capi, CC1, CG1, CG2,
CG33,
CL31, Cog, (syn= CGS), D, E, F, H, H-I, hqi, hq2, 11ZH33, Ii/31, J, K, K,
(syn= Ktox"),
L, L, (syn= Ltox+), M, MC-I, MC-2, MC-3, MC-4, MLMa, N, 0, ovi, ov2, ov3, P,
P, R,
RP6, RS29, S, T, U, UB1, ub2, UH1, UH3, uh3, uh5, uh6, f3, (syn= f3tox+),
f3hv64, Pvir,
y, (syn= ytox-), y19, 6, (syn= 6'ox+), p, (syn= ptox-),(1)9, (p984, w, IA,
1/1180, 2, 2/1180,
5/1180, 5ad/9717, 7/4465, 8/4465, 8ad/10269, 10/9253, 13Z9253, 15/3148,
21/9253, 28,
29, 55, 2747, 2893, 4498 and 5848.
[142] Bacteria of the genus Enterococcus can be infected by the following
phages:
DF78, Fl, F2, 1, 2, 4, 14, 41, 867, D1, SB24, 2BV, 182, 225, C2, C2F, E3, E62,
DS96,
H24, M35, P3, P9, SB101, S2, 2BII, 5, 182a, 705, 873, 881, 940, 1051, 1057,
21096C,
NN-Enterococcus (1), PE1, Fl, F3, F4, VD13, 1, 200, 235 and 341.
[143] Bacteria of the genus Erysipelothrix can be infected by the following
phage: NN-
Eiysipelothrix (1).
[144] Bacteria of the genus Escherichia can be infected by the following
phages:
BW73, B278, D6, D108, E, El, E24, E41, FI-2, FI-4, FI-5, HI8A, Ffl8B, i, MM,
Mu,
(syn= mu), (syn= MuI), (syn= Mu-I), (syn= MU-I), (syn= MuI), (syn= [t), 025,
PhI-5,
Pk, PSP3, Pl, P1D, P2, P4 (defective), Si, Wy, (K13, yR73 (defective), yl,
(p2, (p7, (p92,
w (defective), 7 A, 8y, 9y, 15 (defective), 18, 28-1, 186, 299, HH-Escherichia
(2),
AB48, CM, C4, C16, DD-VI, (syn= Dd-Vi), (syn= DDVI), (syn= DDVi), E4, E7, E28,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
44
FIl, FI3, H, H1, H3, H8, K3, M, N, ND-2, ND-3, ND4, ND-5, ND6, ND-7, Ox-I
(syn=
OX1), (syn= HF), Ox-2 (syn= 0x2), (syn= 0X2), Ox-3, Ox-4, Ox-5, (syn= 0X5), Ox-
6,
(syn= 66F), (syn= y66t), (syn= (66t-)5 0111, PhI-I, RB42, RB43, RB49, RB69, S,
Sai-
1, Sal-2, Sal-3, Sal-4, Sal-5, Sal-6, TC23, TC45, TuII*-6, (syn= TuII*), TuIP-
24,
TuII*46, TuIP-60, T2, (syn= ganuTia), (syn= y), (syn= PC), (syn= P.C.), (syn=
T-2),
(syn= T2), (syn= P4), T4, (syn= T-4), (syn= T4), T6, T35, al, 1, IA, 3, (syn=
Ac3), 3A,
3T+, (syn= 3), (syn= M1), 5y, (syn= (p5), 9266Q, CF0103, HK620, J, K, K1F,
m59, no.
A, no. E, no. 3, no. 9, N4, sd, (syn= Sd), (syn= SD), (syn= Sa)3 (syn= sd),
(syn= SD),
(syn= CD), T3, (syn= T-3), (syn= T3), T7, (syn= T-7), (syn= T7), WPK, W31, AH,
(pC3888, (pK3, (pK7, (pK12, yV-1, (I)04-CF, (I)05, (I)06, (I)07, yl, y1.2,
(p20, (p95, (p263,
y1092, yl, yll, (syn=cpW), S28, 1, 3, 7, 8, 26, 27, 28-2, 29, 30, 31, 32, 38,
39, 42, 933W,
NN-Escherichia (1), Esc-7-11, AC30, CVX-5, Cl, DDUP, EC1, EC2, E21, E29, Fl,
F265, F275, Hi, HK022, HK97, (syn= 4141K97), HK139, HK253, HK256, K7, ND-I,
no.D, PA-2, q, S2, Ti, (syn= a), (syn= P28), (syn= T-I), (syn= Tx), T3C, T5,
(syn= T-5),
(syn= T5), UC-I, w, (34, y2, 2\., (syn= lambda), (syn= (R), (I)D326, yy,
(I)06, (I)7, (MO,
y80, x, (syn= xi), (syn= yx), (syn= yxi), 2, 4, 4A, 6, 8A, 102, 150, 168, 174,
3000, AC6,
AC7, AC28, AC43, AC50, AC57, AC81, AC95, HK243, K10, ZG/3A, 5, 5A, 21EL,
H19-J and 933H.
[145] Bacteria of the genus Fusobacterium can be infected by the following
phages:
NN-Fusobacterium (2), fv83-554/3, fv88-531/2, 227, fv2377, fv2527 and fv8501.
[146] Bacteria of the genus Haemophilus can be infected by the following
phages: HP1,
52 and N3.
[147] Bacteria of the genus Helicobacter can be infected by the following
phages: HP1
and AA-Helicobacter (1).
[148] Bacteria of the genus Klebsiella can be infected by the following
phages: AI0-2,
KI4B, Kl6B, K19, (syn= K19), K114, K115, K121, K128, K129, KI32, K133, K135,
K1106B, K1171B, K1181B, K1832B, AIO-I, AO-I, A0-2, A0-3, FC3-10, K, K11, (syn=
KI1), K12, (syn= K12), K13, (syn= K13), (syn= K1 70/11), K14, (syn= K14), K15,
(syn=
K15), K16, (syn= K16), K17, (syn= K17), K18, (syn= K18), K119, (syn= K19),
K127,
(syn= K127), K131, (syn= K131), K135, K1171B, II, VI, IX, CI-I, Kl4B, K18,
K111,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
K112, K113, K116, K117, K118, K120, K122, K123, K124, K126, K130, K134,
K1106B,
KIi65B, K1328B, KLXI, K328, P5046, 11, 380, III, IV, VII, VIII, FC3-11, K12B,
(syn=
K12B), K125, (syn= K125), K142B, (syn= K142), (syn= K142B), K1181B, (syn= Kul
81), (syn= K1181B), K1765/!, (syn= K1765/1), K1842B, (syn= K1832B), K1937B,
(syn=
5 K1937B), Li, (p28, 7, 231, 483, 490, 632 and 864/100.
[149] Bacteria of the genus Lepitospira can be infected by the following
phages: LE1,
LE3, LE4 and -NN-Leptospira (1).
[150] Bacteria of the genus Listeria can be infected by the following phages:
A511,
01761, 4211, 4286, (syn= B054), A005, A006, A020, A500, A502, A511, Al 18,
A620,
10 A640, B012, B021, B024, B025, B035, B051, B053, B054, B055, B056, B101, BI
10,
B545, B604, B653, C707, D441, HS047, HlOG, H8/73, H19, H21, H43, H46, H107,
H108, HI 10, H163/84, H312, H340, H387, H391/73, H684/74, H924A, PSA, U153,
yMLUP5, (syn= P35), 00241, 00611, 02971A, 02971C, 5/476, 5/911, 5/939,
5/11302,
5/11605, 5/11704, 184, 575, 633, 699/694, 744, 900, 1090, 1317, 1444, 1652,
1806,
15 1807, 1921/959, 1921/11367, 1921/11500, 1921/11566, 1921/12460, 1921/12582,
1967,
2389, 2425, 2671, 2685, 3274, 3550, 3551, 3552, 4276, 4277, 4292, 4477, 5337,
5348/11363, 5348/11646, 5348/12430, 5348/12434, 10072, 11355C, 11711A, 12029,
12981, 13441, 90666, 90816, 93253, 907515, 910716 and NN-Lisferia (15).
[151] Bacteria of the genus Morganella can be infected by the following phage:
47.
20 [152] Bacteria of the genus Mycobacterium can be infected by the following
phages:
13, AG1, ALi, ATCC 11759, A2, B.C3, BG2, BK1, BK5, butyricum, B-I, B5, B7,
B30,
B35, Clark, Cl, C2, DNAIII, DSP1, D4, D29, GS4E, (syn= GS4E), GS7, (syn= GS-
7),
(syn= GS7), IPa, lacticola, Legendre, Leo, L5, (syn= (1L-5), MC-I, MC-3, MC-4,
minetti, MTPHI 1, Mx4, MyF3P/59a, phlei, (syn= phlei 1), phlei 4, Polonus II,
25 rabinovitschi, smegmatis, TM4, TM9, TM10, TM20, Y7, Y10, (p630, IB, IF, IH,
1/1, 67,
106, 1430, Bl, (syn= Bol), B24, D, D29, F-K, F-S, HP, Polonus I, Roy, R1,
(syn= R1-
Myb), (syn= Ri), 11, 31, 40, 50, 103a, 103b, 128, 3111-D, 3215-D and NN-
Mycobacterium (1).
[153] Bacteria of the genus Neisseria can be infected by the following phages:
Group I,
30 group II and NP1.
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
46
[154] Bacteria of the genus Nocardia can be infected by the following phages:
MNP8,
NJ-L, NS-8, N5 and TtiN-Nocardia.
[155] Bacteria of the genus Proteus can be infected by the following phages:
Pm5,
13vir, 2/44, 4/545, 6/1004, 13/807, 20/826, 57, 67b, 78, 107/69, 121, 9/0,
22/608,
30/680, PmI, Pm3, Pm4, Pm6, Pm7, Pm9, PmIO, PmI 1, Pv2, al, (pm, 7/549, 9B/2,
10A/31, 12/55, 14, 15, 16/789, 17/971, 19A/653, 23/532, 25/909, 26/219,
27/953,
32A/909, 33/971, 34/13, 65, 5006M, 7480b, VI, 13/3a, Clichy 12, n2600, (px7,
1/1004,
5/742, 9, 12, 14, 22, 24/860, 2600/D52, Pm8 and 24/2514.
[156] Bacteria of the genus Providencia can be infected by the following
phages:
PL25, PL26, PL37, 9211/9295, 9213/921 lb, 9248, 7/R49, 7476/322, 7478/325,
7479,
7480, 9000/9402 and 9213/921 Ia.
[157] Bacteria of the genus Pseudomonas can be infected by the following
phages: PH,
(syn= Pf-I), Pf2, Pf3, PP7, PRR1, 7s, im-Pseudomonas (1), AI-I, AI-2, B 17,
B89, CB3,
Col 2, Col 11, Col 18, Col 21, C154, C163, C167, C2121, E79, F8, ga, gb, H22,
Kl, M4,
N2, Nu, PB-I, (syn= PB1), pf16, PMN17, PP1, PP8, Psal, PsPl, PsP2, PsP3, PsP4,
PsP5,
PS3, PS17, PTB80, PX4, PX7, PY01, PY02, PY05, PY06, PY09, PY010, PY013,
PY014, PY016, PY018, PY019, PY020, PY029, PY032, PY033, PY035, PY036,
PY037, PY038, PY039, PY041, PY042, PY045, PY047, PY048, PY064, PY069,
PY0103, P1K, SLP1, SL2, S2, UNL-I, wy, Yai, Ya4, Yan, (BE, (pCTX, (pC17, (pKZ,
(syn=41)KZ), (p-LT, 410mu78, (pNZ, (pPLS-1, (ST-1, (2W-14, (p-2, 1/72, 2/79,
3, 3/DO,
4/237, 5/406, 6C, 6/6660, 7, 7v, 7/184, 8/280, 9/95, 10/502, 11/DE, 12/100,
12S, 16, 21,
24, 25F, 27, 31, 44, 68, 71, 95, 109, 188, 337, 352, 1214, HN-Pseudomonas
(23), A856,
B26, CI-I, CI-2, C5, D, gh-1, Fl 16, HF, H90, K5, K6, K1 04, K109, K166, K267,
N4,
N5, 06N-25P, PE69, Pf, PPN25, PPN35, PPN89, PPN91, PP2, PP3, PP4, PP6, PP7,
PP8, PP56, PP87, PP1 14, PP206, PP207, PP306, PP651, Psp231a, Pssy401,
Pssy9220,
psi, PTB2, PTB20, PTB42, PX1, PX3, PX10, PX12, PX14, PY070, PY071, R, SH6,
SH133, tf, Ya5, Ya7, (BS, 41)1(f77, (p-MC, 41)mnF82, (pPLS27, (pPLS743, (S-1,
1, 2, 2,
3, 4, 5, 6, 7, 7, 8, 9, 10, 11, 12, 12B, 13, 14, 15, 14, 15, 16, 17, 18, 19,
20, 20, 21, 21, 22,
23, 23, 24, 25, 31, 53, 73, 119x, 145, 147, 170, 267, 284, 308, 525, NN-
Pseudomonas
(5), af, A7, B3, B33, B39, BI-I, C22, D3, D37, D40, D62, D3112, F7, F10, g,
gd, ge, g
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
47
Hw12, Jb 19, KF1, L , OXN-32P, 06N-52P, PCH-I, PC13-1, PC35-1, PH2, PH51,
PH93,
PH132, PMW, PM13, PM57, PM61, PM62, PM63, PM69, PM105, PM1 13, PM681,
PM682, PO4, PP1, PP4, PP5, PP64, PP65, PP66, PP71, PP86, PP88, PP92, PP401,
PP711, PP891, Pssy41, Pssy42, Pssy403, Pssy404, Pssy420, Pssy923, PS4, PS40,
Pz,
SD1, SL1, SL3, SL5, SM, (pC5, (pC11, (pC11-1, (pC13, (pC15, yMO, yX, y04, yl
1, y240,
2, 2F, 5, 7m, 11, 13, 13/441, 14, 20, 24, 40, 45, 49, 61, 73, 148, 160, 198,
218, 222, 236,
242, 246, 249, 258, 269, 295, 297, 309, 318, 342, 350, 351, 357-1, 400-1, HN-
Pseudomonas (6), G101, M6, M6a, Li, PB2, Pssy15, Pssy4210, Pssy4220, PY012,
PY034, PY049, PY050, PY051, PY052, PY053, PY057, PY059, PY0200, PX2,
PX5, 5L4, y03, y06 and 1214.
[158] Bacteria of the genus Rickettsia can be infected by the following phage:
NN-
Rickettsia.
[159] Bacteria of the genus Salmonella can be infected by the following
phages: b,
Beccles, CT, d, Dundee, f, Fels 2, GI, GUI, GVI, GVIII, k, K, i, j, L, 01,
(syn= 0-1),
(syn= 01), (syn= 04), (syn= 7), 02, 03, P3, P9a, P10, 5ab3, 5ab5, San1S,
5an17, SI,
Taunton, Vii, (syn= Vii), 9, imSalmonella (1), N-I, N-5, N40, N-17, N-22, 11,
12, 16-
19, 20.2, 36, 449C/C178, 966A/C259, a, B.A.O.R., e, G4, GUI, L, LP7, M, MG40,
N-
18, P5A68, P4, P9c, P22, (syn= P22), (syn= PLT22), (syn= PLT22), P22al, P22-4,
P22-
7, P22-11, SNT-I, SNT-2, 5P6, Villi, ViIV, ViV, ViVI, ViVII, Worksop, 5j5,
634, 1,37,
1(40), (syn= yl[40]), 1,422, 2, 2.5, 3b, 4, 5, 6,14(18), 8, 14(6,7), 10, 27,
28B, 30, 31, 32,
33, 34, 36, 37, 39, 1412, SNT-3, 7-11, 40.3, c, C236, C557, C625, C966N, g,
GV, G5,
G1 73, h, IRA, Jersey, MB78, P22-1, P22-3, P22-12, Sabi, 5ab2, 5ab2, 5ab4,
Sanl, 5an2,
5an3, 5an4, 5an6, 5an7, 5an8, 5an9, 5an13, 5an14, 5an16, 5an18, 5an19, 5an20,
5an21,
5an22, 5an23, 5an24, 5an25, 5an26, SasL1, SasL2, SasL3, SasL4, SasL5, S1BL,
SIT,
Viii, yl, 1, 2, 3a, 3al, 1010, Ym-Salmonella (1), N-4, SasL6 and 27.
[160] Bacteria of the genus Serratia can be infected by the following phages:
A2P,
PS20, SMB3, SMP, SMP5, 5M2, V40, V56, ic, (I)CP-3, (I)CP-6, 3M, 10/1a, 20A,
34CC,
34H, 38T, 345G, 345P, 501B, SMB2, SMP2, BC, BT, CW2, CW3, CW4, CW5, Lt232,
L2232, L34, L.228, SLP, SMPA, V.43, a, (pCW1, (I)CP6-1, (I)CP6-2, (I)CP6-5,
3T, 5, 8,
9F, 10/1, 20E, 32/6, 34B, 34CT, 34P, 37, 41, 56, 56D, 56P, 60P, 61/6, 74/6,
76/4,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
48
101/8900, 226, 227, 228, 229F, 286, 289, 290F, 512, 764a, 2847/10, 2847/10a,
L.359
and SMB1.
[161] Bacteria of the genus Shigella can be infected by the following phages:
Fsa,
(syn=a), FSD2d, (syn= D2d), (syn= W2d), FSD2E, (syn= W2e), fv, F6, P.8, H-Sh,
PE5,
P90, SflI, Sh, SHm, SHrv, (syn= HIV), SHvi, (syn= HVI), SHVvm, (syn= HVIII),
SK766, (syn= gamma 66), (syn= yf3f3), (syn= y66b), SKm, (syn= SIIIb)5 (syn=
UI),
SKw, (syn= Siva), (syn= IV), SIC ' , (syn= SIVA.), (syn= IVA), SKvi, (syn=
KVI),
(syn= Svi), (syn= VI), SKvm, (syn= Svm), (syn= VIII), SKVHIA, (syn= SvmA),
(syn=
VIIIA), STvi, STK, STxl, STxn, S66, W2, (syn= D2c), (syn= D20), yl, (pIVb 3-SO-
R,
8368-SO-R, F7, (syn= F57), (syn= K29), F10, (syn= FS10), (syn= K31), IL (syn=
alfa),
(syn= FSa), (syn= K1 8), (syn= a), 12, (syn= a), (syn= K19), 5G33, (syn= G35),
(syn=
SO-35/G), 5G35, (syn= SO-55/G), 5G3201, (syn= SO-3201/G), SHn, (syn= HIT),
SHv,
(syn= SHV), SHx, SHX, SKn, (syn= K2), (syn= KII), (syn= Sn), (syn= SsII),
(syn= II),
SKrv, (syn= Sm), (syn= SsIV), (syn= IV), SK1Va, (syn= Swab), (syn= SsIVa),
(syn=
IVa), SKV, (syn= K4), (syn= KV), (syn= SV), (syn= SsV), (syn= V), SKx, (syn=
K9),
(syn= KX), (syn= SX), (syn= SsX), (syn= X), STV, (syn= T35), (syn= 35-50-R),
STvm,
(syn= T8345), (syn= 8345-SO-S-R), Wl, (syn= D8), (syn= FSD8), W2a, (syn= D2A),
(syn= FS2a), DD-2, Sf6, FSi, (syn= F1), SF6, (syn= F6), 5G42, (syn= SO-42/G),
5G3203, (syn= SO-3203/G), SKF12, (syn= SsF12), (syn= F12), (syn= F12), STn,
(syn=
1881-SO-R), y66, (syn= gamma 66a), (syn= Ssy66), (p2, BIl, DDVII, (syn= DD7),
FSD2b, (syn= W2B), F52, (syn= F2), (syn= F2), F54, (syn= F4), (syn= F4), F55,
(syn=
F5), (syn= F5), F59, (syn= F9), (syn= F9), Fl 1, P2-SO-S, 5G36, (syn= SO-
36/G), (syn=
G36), 5G3204, (syn= SO-3204/G), 5G3244, (syn= SO-3244/G), SHi, (syn= HI),
SHva,
(syn= HVII), SHK, (syn= HIX), SHxl, SHx7c, (syn= HXn), SKI, KI, (syn= Si),
(syn=
SsI), SKVII, (syn= KVII), (syn= Sva), (syn= SsVII), SKIX, (syn= KIX), (syn=
Six),
(syn= SsIX), SKXII, (syn= KXII), (syn= Sxn), (syn= SsXII), STi, STffl, STrv,
STVi,
STva, S70, S206, U2-S0-S, 3210-SO-S, 3859-SO-S, 4020-SO-S, (p3, (p5, (p7, (p8,
(p9,
y10, yl 1, y13, y14, y18, SHm, (syn= Hai), SHxi, (syn= HXt) and SKxI, (syn=
KXI),
(syn= Sri), (syn= SsXI), (syn= XI).
[162] Bacteria of the genus Staphylococcus can be infected by the following
phages: A,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
49
EW, K, Ph5, Ph9, PhIO, Ph13, Pl, P2, P3, P4, P8, P9, P10, RG, SB-i, (syn= Sb-
I), S3K,
Twort, (1)51(311, (p812, 06, 40, 58, 119, 130, 131, 200, 1623, STC1,
(syn=stc1), STC2,
(syn=stc2), 44AHJD, 68, Ad, AC2, A6"C", A9"C", b581, CA-I, CA-2, CA-3, CA-4,
CA-5, DI 1, L39x35, L54a, M42, Ni, N2, N3, N4, N5, N7, N8, N10, Ni 1, N12,
N13,
N14, N16, Ph6, Ph12, Ph14, UC-18, U4, U15, Si, S2, S3, S4, S5, X2, Z1, (B5-2,
(pD, 0),
11, (syn= yl 1), (syn= P11-M15), 15, 28, 28A, 29, 31, 31B, 37, 42D, (syn=
P42D), 44A,
48, 51, 52, 52A, (syn= P52A), 52B, 53, 55, 69, 71, (syn= P71), 71A, 72, 75,
76, 77, 79,
80, 80a, 82, 82A, 83 A, 84, 85, 86, 88, 88A, 89, 90, 92, 95, 96, 102, 107,
108, 111, 129-
26, 130, 130A, 155, 157, 157A, 165, 187, 275, 275A, 275B, 356, 456, 459, 471,
471A,
489, 581, 676, 898, 1139, 1154A, 1259, 1314, 1380, 1405, 1563, 2148, 2638A,
2638B,
2638C, 2731, 2792A, 2792B, 2818, 2835, 2848A, 3619, 5841, 12100, AC3, A8, A10,
A13, b594n, D, HK2, N9, N15, P52, P87, Si, S6, Z4, (RE, 3A, 3B, 3C, 6, 7, 16,
21,
42B, 42C, 42E, 44, 47, 47A5 47C, 51, 54, 54x1, 70, 73, 75, 78, 81, 82, 88, 93,
94, 101,
105, 110, 115, 129/16, 174, 594n, 1363/14, 2460 and mS-Staphylococcus (1).
[163] Bacteria of the genus Streptococcus can be infected by the following
phages: EJ-
I, NN-Streptococais (1), a, Cl, FLOThs, H39, Cp-I, Cp-5, Cp-7, Cp-9, Cp-I0,
AT298,
AS, a10/J1, a10/J2, a10/J5, a10/J9, A25, BTI 1, b6, CA1, c20-1, c20-2, DP-I,
Dp-4, DT1,
ET42, e10, FA101, FEThs, Fic, FKKIOI, FKLIO, FKP74, FKH, FLOThs, FyI01, fl,
F10,
F20140/76, g, GT-234, HB3, (syn= HB-3), HB-623, HB-746, M102, 01205, y01205,
PST, PO, Pl, P2, P3, P5, P6, P8, P9, P9, P12, P13, P14, P49, P50, P51, P52,
P53, P54,
P55, P56, P57, P58, P59, P64, P67, P69, P71, P73, P75, P76, P77, P82, P83,
P88, sc,
sch, sf, SfIl 1, (syn= SFiI 1), (syn= (pSFill), (syn= (I)Sfil 1), (syn= (pSfil
1), sfil9, (syn=
SFi19), (syn= (pSFi19), (syn= (p5fi19), 5fi21, (syn= SFi21), (syn= (pSFi21),
(syn= (p5fi21),
STO, STX, st2, 5T2, 5T4, S3, (syn= 03), s265, (1)17, (p42, (1)57, y80, (p81,
(p82, (p83,
(p84, (p85, (p86, (p87, (p88, (p89, y90, (p91, (p92, (p93, (p94, (p95, (p96,
(p97, (p98, (p99,
(p100, (p101, (p102, (p227, (1)7201, wl, w2, w3, w4, w5, w6, w8, w10, 1, 6, 9,
10F, 12/12,
14, 175R, 19S, 24, 50/33, 50/34, 55/14, 55/15, 70/35, 70/36, 71/ST15, 71/45,
71/46,
74F, 79/37, 79/38, 80/J4, 80/J9, 80/5T16, 80/15, 80/47, 80/48, 101, 103/39,
103/40,
121/41, 121/42, 123/43, 123/44, 124/44, 337/5T17 and mStreptococcus (34).
[164] Bacteria of the genus Treponema can be infected by the following phage:
NN-
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
Treponema (1).
[165] Bacteria of the genus Vibrio can be infected by the following phages:
CTX41), fs,
(syn= si), fs2, Ivpf5, Vfl2, Vf33, VPI41), VSK, v6, 493, CP-T1, ET25, kappa,
K139,
Labol, )XN-69P, OXN-86, 06N-21P, PB-I, P147, rp-1, SE3, VA-I, (syn= VcA-I),
VcA-
5 2, VP1, VP2, VP4, VP7, VP8, VP9, VP10, VP17, VP18, VP19, X29, (syn= 29
d'Herelle), t, (DHAWI-1, (DHAWI-2, (DHAWI-3, (DHAWI-4, (DHAWI-5, (DHAWI-6,
(DHAWI-7, XHAWI-8, (DHAWI-9, (DHAWI-10, (DHCl-1, HC1-2, HC1-3, HC1-4,
(DHC2-1, >HC2-2, HC2-3, HC2-4, HC3-1, HC3-2, HC3-3, (DHD1S-1, 41)HD1S-
2, (DHD2S-1, (DHD2S-2, (DHD2S-3, (DHD2S-4, (DHD2S-5, HDO-1, HDO-2, 41)HDO-
10 3, HDO-4, HDO-5, HDO-6, 41)KL-33, 41)KL-34, 41)KL-35, 41)KL-36, 41)KWH-2,
41)KWH-3, 41)KWH-4, (I)MARQ-1, (I)MARQ-2, (I)MARQ-3, (I)MOAT-1, 4100139,
(toPEL1A-1, (toPEL1A-2, (toPEL8A-1, (toPEL8A-2, (toPEL8A-3, (toPEL8C-1,
(toPEL8C-2,
(toPEL13A-1, (toPEL13B-1, (toPEL13B-2, (toPEL13B-3, (toPEL13B-4, (toPEL13B-5,
(toPEL13B-6, (toPEL13B-7, (toPEL13B-8, (toPEL13B-9, (toPEL13B-10, yVP143,
yVP253,
15 (1)16, (p138, 1- II, 5, 13, 14, 16, 24, 32, 493, 6214, 7050, 7227, II,
(syn= group II), (syn==
(p2), V, VIII, -m-Vibrio (13), KVP20, KVP40, nt-1, 06N-22P, P68, el, e2, e3,
e4, e5,
FK, G, I, K, nt-6, N1, N2, N3, N4, N5, 06N-34P, OXN-72P, OXN-85P, OXN-100P, P,
Ph-I, PL163/10, Q, S, T, (p92, 1-9, 37, 51, 57, 70A-8, 72A-4, 72A-10, 110A-4,
333,
4996, I (syn= group I), III (syn= group III), VI, (syn= A-Saratov), VII, IX,
X, HN-
20 Vibrio (6), pAl, 7, 7-8, 70A-2, 71A-6, 72A-5, 72A-8, 108A-10, 109A-6, 109A-
8, 110A-
1, 110A-5, 110A-7, hv-1, OXN-52P, P13, P38, P53, P65, P108, Pill, TP13 VP3,
VP6,
VP12, VP13, 70A-3, 70A-4, 70A-10, 72A-1, 108A-3, 109-B1, 110A-2, 149, (syn=
y149), IV, (syn= group IV), NN-Vibrio (22), VP5, VPI1, VP15, VP16, al, a2,
a3a, a3b,
353B and HN-Vibrio (7).
25 [166] Bacteria of the genus Yersinia can be infected by the following
phages: H, H-I,
H-2, H-3, H-4, Lucas 110, Lucas 303, Lucas 404, YerA3, YerA7, YerA20, YerA41,
3/M64-76, 5/G394-76, 6/C753-76, 8/C239-76, 9/F18167, 1701, 1710, PST, 1/F2852-
76,
D'Herelle, EV, H, Kotljarova, PTB, R, Y, YerA41, yYer03-12, 3, 4/C1324-76,
7/F783-
76, 903, 1/M6176 and Yer2AT.
30 [167] More preferably, the bacteriophage is selected in the group
consisting of
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
51
Salmonella virus SKML39, Shigella virus AG3, Dickeya virus Limestone, Dickeya
virus
RC2014, Escherichia virus CBA120, Escherichia virus PhaxI, Salmonella virus
38,
Salmonella virus Det7, Salmonella virus GG32, Salmonella virus PM10,
Salmonella
virus SFP10, Salmonella virus 5H19, Salmonella virus 5J3, Escherichia virus
ECML4,
Salmonella virus Marshall, Salmonella virus Maynard, Salmonella virus 5J2,
Salmonella
virus STML131, Salmonella virus Vii, Erwinia virus Ea2809, Klebsiella virus
0507KN21, Serratia virus IME250, Serratia virus MAM1, Campylobacter virus
CP21,
Campylobacter virus CP220, Campylobacter virus CPt10, Campylobacter virus
IBB35,
Campylobacter virus CP81, Campylobacter virus CP30A, Campylobacter virus CPX,
Campylobacter virus NCTC12673, Erwinia virus Ea214, Erwinia virus M7,
Escherichia
virus AY0145A, Escherichia virus EC6, Escherichia virus HY02, Escherichia
virus
JH2, Escherichia virus TP1, Escherichia virus VpaEl, Escherichia virus wV8,
Salmonella virus Felix01, Salmonella virus HB2014, Salmonella virus Mushroom,
Salmonella virus UAB87, Citrobacter virus Moogle, Citrobacter virus Mordin,
Escherichia virus SUSP1, Escherichia virus SUSP2, Aeromonas virus phi018P,
Haemophilus virus HP1, Haemophilus virus HP2, Pasteurella virus F108, Vibrio
virus
K139, Vibrio virus Kappa, Burkholderia virus phi52237, Burkholderia virus
phiE122,
Burkholderia virus phiE202, Escherichia virus 186, Escherichia virus P4,
Escherichia
virus P2, Escherichia virus Wphi, Mannheimia virus PHL101, Pseudomonas virus
phiCTX, Ralstonia virus RSA1, Salmonella virus Fels2, Salmonella virus PsP3,
Salmonella virus SopEphi, Yersinia virus L413C, Staphylococcus virus Gl,
Staphylococcus virus G15, Staphylococcus virus JD7, Staphylococcus virus K,
Staphylococcus virus MCE2014, Staphylococcus virus P108, Staphylococcus virus
Rodi, Staphylococcus virus S253, Staphylococcus virus S25-4, Staphylococcus
virus
5Al2, Listeria virus A511, Listeria virus P100, Staphylococcus virus Remus,
Staphylococcus virus SA1 1, Staphylococcus virus 5tau2, Bacillus virus
Camphawk,
Bacillus virus SP01, Bacillus virus BCP78, Bacillus virus TsarBomba,
Staphylococcus
virus Twort, Enterococcus virus phiEC24C, Lactobacillus virus Lb338-1,
Lactobacillus
virus LP65, Enterobacter virus PG7, Escherichia virus CC31, Klebsiella virus
JD18,
Klebsiella virus PK0111, Escherichia virus Bp7, Escherichia virus IME08,
Escherichia
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
52
virus JS10, Escherichia virus JS98, Escherichia virus QL01, Escherichia virus
VR5,
Enterobacter virus Eap3, Klebsiella virus KP15, Klebsiella virus KP27,
Klebsiella virus
Matisse, Klebsiella virus Miro, Citrobacter virus Merlin, Citrobacter virus
Moon,
Escherichia virus JSE, Escherichia virus phil, Escherichia virus RB49,
Escherichia virus
HX01, Escherichia virus JS09, Escherichia virus RB69, Shigella virus UTAM,
Salmonella virus S16, Salmonella virus STML198, Vibrio virus KVP40, Vibrio
virus
ntl, Vibrio virus ValKK3, Escherichia virus VR7, Escherichia virus VR20,
Escherichia
virus VR25, Escherichia virus VR26, Shigella virus 5P18, Escherichia virus
AR1,
Escherichia virus C40, Escherichia virus E112, Escherichia virus ECML134,
Escherichia virus HY01, Escherichia virus Ime09, Escherichia virus RB3,
Escherichia
virus RB14, Escherichia virus T4, Shigella virus Pss 1, Shigella virus 5hfl2,
Yersinia
virus D1, Yersinia virus PST, Acinetobacter virus 133, Aeromonas virus 65,
Aeromonas
virus Aehl, Escherichia virus RB16, Escherichia virus RB32, Escherichia virus
RB43,
Pseudomonas virus 42, Cronobacter virus CR3, Cronobacter virus CR8,
Cronobacter
virus CR9, Cronobacter virus PBES02, Pectobacterium virus phiTE, Cronobacter
virus
GAP31, Escherichia virus 4MG, Salmonella virus SE1, Salmonella virus 55E121,
Escherichia virus FFH2, Escherichia virus FV3, Escherichia virus JE52013,
Escherichia
virus V5, Brevibacillus virus Abouo, Brevibacillus virus Davies, Bacillus
virus Agate,
Bacillus virus Bobb, Bacillus virus Bp8pC, Erwinia virus Deimos, Erwinia virus
Ea35-
70, Erwinia virus RAY, Erwinia virus 5immy50, Erwinia virus SpecialG,
Acinetobacter
virus AB1, Acinetobacter virus AB2, Acinetobacter virus AbC62, Acinetobacter
virus
AP22, Arthrobacter virus ArV1, Arthrobacter virus Trina, Bacillus virus
AvesoBmore,
Bacillus virus B4, Bacillus virus Bigbertha, Bacillus virus Riley, Bacillus
virus Spock,
Bacillus virus Troll, Bacillus virus Bastille, Bacillus virus CAM003, Bacillus
virus
Bc431, Bacillus virus Bcpl, Bacillus virus BCP82, Bacillus virus BM15,
Bacillus virus
Deepblue, Bacillus virus JBP901, Burkholderia virus Bcepl, Burkholderia virus
Bcep43,
Burkholderia virus Bcep781, Burkholderia virus BcepNY3, Xanthomonas virus 0P2,
Burkholderia virus BcepMu, Burkholderia virus phiE255, Aeromonas virus 44RR2,
Mycobacterium virus Alice, Mycobacterium virus Bxzl, Mycobacterium virus
Dandelion, Mycobacterium virus HyRo, Mycobacterium virus 13, Mycobacterium
virus
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
53
Nappy, Mycobacterium virus Sebata, Clostridium virus phiC2, Clostridium virus
phiCD27, Clostridium virus phiCD119, Bacillus virus CP51, Bacillus virus JL,
Bacillus
virus Shanette, Escherichia virus CVM10, Escherichia virus ep3, Erwinia virus
Asesino,
Erwinia virus EaH2, Pseudomonas virus EL, Halomonas virus HAP1, Vibrio virus
VP882, Brevibacillus virus Jimmer, Brevibacillus virus Osiris, Pseudomonas
virus
Ab03, Pseudomonas virus KPP10, Pseudomonas virus PAKP3, Sinorhizobium virus
M7, Sinorhizobium virus M12, Sinorhizobium virus N3, Erwinia virus Machina,
Arthrobacter virus Brent, Arthrobacter virus Jawnski, Arthrobacter virus
Martha,
Arthrobacter virus Sonny, Edwardsiella virus MSW3, Edwardsiella virus PEi21,
Escherichia virus Mu, Shigella virus SfMu, Halobacterium virus phiH, Bacillus
virus
Grass, Bacillus virus NIT1, Bacillus virus SPG24, Aeromonas virus 43,
Escherichia
virus Pl, Pseudomonas virus CAbl, Pseudomonas virus CAb02, Pseudomonas virus
JG004, Pseudomonas virus PAKP1, Pseudomonas virus PAKP4, Pseudomonas virus
PaP1, Burkholderia virus BcepFl, Pseudomonas virus 141, Pseudomonas virus
Ab28,
Pseudomonas virus DL60, Pseudomonas virus DL68, Pseudomonas virus F8,
Pseudomonas virus JG024, Pseudomonas virus KPP12, Pseudomonas virus LBL3,
Pseudomonas virus LMA2, Pseudomonas virus PB1, Pseudomonas virus SN,
Pseudomonas virus PA7, Pseudomonas virus phiKZ, Rhizobium virus RHEph4,
Ralstonia virus RSF1, Ralstonia virus RSL2, Ralstonia virus RSL1, Aeromonas
virus 25,
Aeromonas virus 31, Aeromonas virus Aes12, Aeromonas virus Aes508, Aeromonas
virus A54, Stenotrophomonas virus IME13, Staphylococcus virus IPLAC1C,
Staphylococcus virus SEP1, Salmonella virus SPN3US, Bacillus virus 1,
Geobacillus
virus GBSV1, Yersinia virus R1RT, Yersinia virus TG1, Bacillus virus G,
Bacillus virus
PBS1, Microcystis virus Ma-LMM01, Vibrio virus MAR, Vibrio virus VHML, Vibrio
virus VP585, Bacillus virus BPS13, Bacillus virus Hakuna, Bacillus virus
Megatron,
Bacillus virus WPh, Acinetobacter virus AB3, Acinetobacter virus Abpl,
Acinetobacter
virus Fri 1, Acinetobacter virus IME200, Acinetobacter virus PD6A3,
Acinetobacter
virus PDAB9, Acinetobacter virus phiAB1, Escherichia virus K30, Klebsiella
virus K5,
Klebsiella virus K11, Klebsiella virus Kpl, Klebsiella virus KP32, Klebsiella
virus
KpV289, Klebsiella virus F19, Klebsiella virus K244, Klebsiella virus Kp2,
Klebsiella
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
54
virus KP34, Klebsiella virus KpV41, Klebsiella virus KpV71, Klebsiella virus
KpV475,
Klebsiella virus SU503, Klebsiella virus SU552A, Pantoea virus Limelight,
Pantoea
virus Limezero, Pseudomonas virus LKA1, Pseudomonas virus phiKMV, Xanthomonas
virus f20, Xanthomonas virus f30, Xylella virus Prado, Erwinia virus Era103,
Escherichia virus K5, Escherichia virus K1-5, Escherichia virus KlE,
Salmonella virus
5P6, Escherichia virus T7, Kluyvera virus Kvpl, Pseudomonas virus ghl,
Prochlorococcus virus PSSP7, Synechococcus virus P60, Synechococcus virus
5yn5,
Streptococcus virus Cpl, Streptococcus virus Cp7, Staphylococcus virus 44AHJD,
Streptococcus virus Cl, Bacillus virus B103, Bacillus virus GA1, Bacillus
virus phi29,
Kurthia virus 6, Actinomyces virus Avl, Mycoplasma virus Pl, Escherichia virus
24B,
Escherichia virus 933W, Escherichia virus Min27, Escherichia virus PA28,
Escherichia
virus 5tx2 II, Shigella virus 75025tx, Shigella virus POCJ13, Escherichia
virus 191,
Escherichia virus PA2, Escherichia virus TL2011, Shigella virus VASD,
Burkholderia
virus Bcep22, Burkholderia virus Bcepi102, Burkholderia virus Bcepmigl,
Burkholderia
virus DC1, Bordetella virus BPP1, Burkholderia virus BcepC6B, Cellulophaga
virus
Cba41, Cellulophaga virus Cba172, Dinoroseobacter virus DFL12, Erwinia virus
Ea9-2,
Erwinia virus Frozen, Escherichia virus phiV10, Salmonella virus Epsilon15,
Salmonella virus SPN1S, Pseudomonas virus F116, Pseudomonas virus H66,
Escherichia virus APEC5, Escherichia virus APEC7, Escherichia virus Bp4,
Escherichia
virus EC1UPM, Escherichia virus ECBP1, Escherichia virus G7C, Escherichia
virus
IME11, Shigella virus Sbl, Achromobacter virus Axp3, Achromobacter virus
JWAlpha,
Edwardsiella virus KF1, Pseudomonas virus KPP25, Pseudomonas virus R18,
Pseudomonas virus Ab09, Pseudomonas virus LIT1, Pseudomonas virus PA26,
Pseudomonas virus Ab22, Pseudomonas virus CHU, Pseudomonas virus LUZ24,
Pseudomonas virus PAA2, Pseudomonas virus PaP3, Pseudomonas virus PaP4,
Pseudomonas virus TL, Pseudomonas virus KPP21, Pseudomonas virus LUZ7,
Escherichia virus N4, Salmonella virus 9NA, Salmonella virus 5P069, Salmonella
virus
BTP1, Salmonella virus HK620, Salmonella virus P22, Salmonella virus 5T64T,
Shigella virus Sf6, Bacillus virus Page, Bacillus virus Palmer, Bacillus virus
Pascal,
Bacillus virus Pony, Bacillus virus Pookie, Escherichia virus 172-1,
Escherichia virus
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
ECB2, Escherichia virus NJ01, Escherichia virus phiEco32, Escherichia virus
Septimall, Escherichia virus SU10, Brucella virus Pr, Brucella virus Tb,
Escherichia
virus Pollock, Salmonella virus FSL SP-058, Salmonella virus FSL SP-076,
Helicobacter virus 1961P, Helicobacter virus KHP30, Helicobacter virus KHP40,
5 Hamiltonella virus APSE1, Lactococcus virus KSY1, Phormidium virus WMP3,
Phormidium virus WMP4, Pseudomonas virus 119X, Roseobacter virus SI01, Vibrio
virus VpV262, Vibrio virus VC8, Vibrio virus VP2, Vibrio virus VP5,
Streptomyces
virus Amela, Streptomyces virus phiCAM, Streptomyces virus Aaronocolus,
Streptomyces virus Caliburn, Streptomyces virus Danzina, Streptomyces virus
Hydra,
10 Streptomyces virus Izzy, Streptomyces virus Lannister, Streptomyces virus
Lika,
Streptomyces virus Sujidade, Streptomyces virus Zemlya, Streptomyces virus
ELB20,
Streptomyces virus R4, Streptomyces virus phiHau3, Mycobacterium virus
Acadian,
Mycobacterium virus Baee, Mycobacterium virus Reprobate, Mycobacterium virus
Adawi, Mycobacterium virus Banel, Mycobacterium virus BrownCNA, Mycobacterium
15 virus Chrisnmich, Mycobacterium virus Cooper, Mycobacterium virus JAMaL,
Mycobacterium virus Nigel, Mycobacterium virus Stinger, Mycobacterium virus
Vincenzo, Mycobacterium virus Zemanar, Mycobacterium virus Apizium,
Mycobacterium virus Manad, Mycobacterium virus Oline, Mycobacterium virus
Osmaximus, Mycobacterium virus Pgl, Mycobacterium virus Soto, Mycobacterium
20 virus Suffolk, Mycobacterium virus Athena, Mycobacterium virus Bernardo,
Mycobacterium virus Gadjet, Mycobacterium virus Pipefish, Mycobacterium virus
Godines, Mycobacterium virus Rosebush, Mycobacterium virus Babsiella,
Mycobacterium virus Brujita, Mycobacterium virus Che9c, Mycobacterium virus
Sbash,
Mycobacterium virus Hawkeye, Mycobacterium virus Plot, Salmonella virus AG11,
25 Salmonella virus Entl, Salmonella virus f18SE, Salmonella virus Jersey,
Salmonella
virus L13, Salmonella virus LSPA1, Salmonella virus 5E2, Salmonella virus
SETP3,
Salmonella virus SETP7, Salmonella virus SETP13, Salmonella virus SP101,
Salmonella virus 553e, Salmonella virus wks13, Escherichia virus K1G,
Escherichia
virus K1H, Escherichia virus Klindl, Escherichia virus Klind2, Salmonella
virus 5P31,
30 Leuconostoc virus Lmdl, Leuconostoc virus LN03, Leuconostoc virus LN04,
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
56
Leuconostoc virus LN12, Leuconostoc virus LN6B, Leuconostoc virus P793,
Leuconostoc virus 1A4, Leuconostoc virus Ln8, Leuconostoc virus Ln9,
Leuconostoc
virus LN25, Leuconostoc virus LN34, Leuconostoc virus LNTR3, Mycobacterium
virus
Bongo, Mycobacterium virus Rey, Mycobacterium virus Butters, Mycobacterium
virus
Michelle, Mycobacterium virus Charlie, Mycobacterium virus Pipsqueaks,
Mycobacterium virus Xeno, Mycobacterium virus Panchino, Mycobacterium virus
Phrann, Mycobacterium virus Redi, Mycobacterium virus Skinnyp, Gordonia virus
BaxterFox, Gordonia virus Yeezy, Gordonia virus Kita, Gordonia virus Zirinka,
Gorrdonia virus Nymphadora, Mycobacterium virus Bignuz, Mycobacterium virus
Brusacoram, Mycobacterium virus Donovan, Mycobacterium virus Fishburne,
Mycobacterium virus Jebeks, Mycobacterium virus Malithi, Mycobacterium virus
Phayonce, Enterobacter virus F20, Klebsiella virus 1513, Klebsiella virus
KLPN1,
Klebsiella virus KP36, Klebsiella virus PKP126, Klebsiella virus Sushi,
Escherichia
virus AHP42, Escherichia virus AH524, Escherichia virus AK596, Escherichia
virus
C119, Escherichia virus E41c, Escherichia virus Eb49, Escherichia virus Jk06,
Escherichia virus KP26, Escherichia virus Roguel, Escherichia virus ACGM12,
Escherichia virus Rtp, Escherichia virus ADB2, Escherichia virus JMPW1,
Escherichia
virus JMPW2, Escherichia virus Ti, Shigella virus PSf2, Shigella virus Shfll,
Citrobacter virus Stevie, Escherichia virus TLS, Salmonella virus 5P126,
Cronobacter
virus Esp2949-1, Pseudomonas virus Ab18, Pseudomonas virus Ab19, Pseudomonas
virus PaMx11, Arthrobacter virus Amigo, Propionibacterium virus Anatole,
Propionibacterium virus B3, Bacillus virus Andromeda, Bacillus virus Blastoid,
Bacillus
virus Curly, Bacillus virus Eoghan, Bacillus virus Finn, Bacillus virus
Glittering,
Bacillus virus Riggi, Bacillus virus Taylor, Gordonia virus Attis,
Mycobacterium virus
Barnyard, Mycobacterium virus Konstantine, Mycobacterium virus Predator,
Mycobacterium virus Bernal13, Staphylococcus virus 13, Staphylococcus virus
77,
Staphylococcus virus 108PVL, Mycobacterium virus Bron, Mycobacterium virus
Faithl,
Mycobacterium virus Joedirt, Mycobacterium virus Rumpelstiltskin, Lactococcus
virus
bIL67, Lactococcus virus c2, Lactobacillus virus c5, Lactobacillus virus Ld3,
Lactobacillus virus Ld17, Lactobacillus virus Ld25A, Lactobacillus virus LLKu,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
57
Lactobacillus virus phiLdb, Cellulophaga virus Cba121, Cellulophaga virus
Cba171,
Cellulophaga virus Cba181, Cellulophaga virus ST, Bacillus virus 250, Bacillus
virus
IEBH, Mycobacterium virus Ardmore, Mycobacterium virus Avani, Mycobacterium
virus Boomer, Mycobacterium virus Che8, Mycobacterium virus Che9d,
Mycobacterium virus Deadp, Mycobacterium virus Dlane, Mycobacterium virus
Dorothy, Mycobacterium virus Dotproduct, Mycobacterium virus Drago,
Mycobacterium virus Fruitloop, Mycobacterium virus Gumbie, Mycobacterium virus
Ibhubesi, Mycobacterium virus Llij, Mycobacterium virus Mozy, Mycobacterium
virus
Mutaforma13, Mycobacterium virus Pacc40, Mycobacterium virus PMC,
Mycobacterium virus Ramsey, Mycobacterium virus Rockyhorror, Mycobacterium
virus
SG4, Mycobacterium virus Shaunal, Mycobacterium virus Shilan, Mycobacterium
virus
Spartacus, Mycobacterium virus Taj, Mycobacterium virus Tweety, Mycobacterium
virus Wee, Mycobacterium virus Yoshi, Salmonella virus Chi, Salmonella virus
FSLSP030, Salmonella virus FSLSP088, Salmonella virus iEPS5, Salmonella virus
SPN19, Mycobacterium virus 244, Mycobacterium virus Bask21, Mycobacterium
virus
CJW1, Mycobacterium virus Eureka, Mycobacterium virus Kostya, Mycobacterium
virus Porky, Mycobacterium virus Pumpkin, Mycobacterium virus Sirduracell,
Mycobacterium virus Toto, Mycobacterium virus Corndog, Mycobacterium virus
Firecracker, Rhodobacter virus RcCronus, Pseudomonas virus D3112, Pseudomonas
virus DMS3, Pseudomonas virus FHA0480, Pseudomonas virus LPB1, Pseudomonas
virus MP22, Pseudomonas virus MP29, Pseudomonas virus MP38, Pseudomonas virus
PA1KOR, Pseudomonas virus D3, Pseudomonas virus PMG1, Arthrobacter virus
Decurro, Gordonia virus Demosthenes, Gordonia virus Katyusha, Gordonia virus
Kvothe, Propionibacterium virus B22, Propionibacterium virus Doucette,
Propionibacterium virus E6, Propionibacterium virus G4, Burkholderia virus
phi6442,
Burkholderia virus phi1026b, Burkholderia virus phiE125, Edwardsiella virus
eiAU,
Mycobacterium virus Ff47, Mycobacterium virus Muddy, Mycobacterium virus Gaia,
Mycobacterium virus Giles, Arthrobacter virus Captnmurica, Arthrobacter virus
Gordon,
Gordonia virus GordTnk2, Paenibacillus virus Harrison, Escherichia virus
EK99P1,
Escherichia virus HK578, Escherichia virus JL1, Escherichia virus SSL2009a,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
58
Escherichia virus YD2008s, Shigella virus EP23, Sodalis virus S01, Escherichia
virus
HK022, Escherichia virus HK75, Escherichia virus HK97, Escherichia virus
HK106,
Escherichia virus HK446, Escherichia virus HK542, Escherichia virus HK544,
Escherichia virus HK633, Escherichia virus mEp234, Escherichia virus mEp235,
Escherichia virus mEpX1, Escherichia virus mEpX2, Escherichia virus mEp043,
Escherichia virus mEp213, Escherichia virus mEp237, Escherichia virus mEp390,
Escherichia virus mEp460, Escherichia virus mEp505, Escherichia virus mEp506,
Brevibacillus virus Jenst, Achromobacter virus 83-24, Achromobacter virus JWX,
Arthrobacter virus Kellezzio, Arthrobacter virus Kitkat, Arthrobacter virus
Bennie,
Arthrobacter virus DrRobert, Arthrobacter virus Glenn, Arthrobacter virus
HunterDalle,
Arthrobacter virus Joann, Arthrobacter virus Korra, Arthrobacter virus
Preamble,
Arthrobacter virus Pumancara, Arthrobacter virus Wayne, Mycobacterium virus
Alma,
Mycobacterium virus Arturo, Mycobacterium virus Astro, Mycobacterium virus
Backyardigan, Mycobacterium virus BBPiebs31, Mycobacterium virus Benedict,
Mycobacterium virus Bethlehem, Mycobacterium virus Billknuckles, Mycobacterium
virus Bruns, Mycobacterium virus Bxbl, Mycobacterium virus Bxz2, Mycobacterium
virus Che12, Mycobacterium virus Cuco, Mycobacterium virus D29, Mycobacterium
virus Doom, Mycobacterium virus Ericb, Mycobacterium virus Euphoria,
Mycobacterium virus George, Mycobacterium virus Gladiator, Mycobacterium virus
Goose, Mycobacterium virus Hammer, Mycobacterium virus Heldan, Mycobacterium
virus Jasper, Mycobacterium virus JC27, Mycobacterium virus Jeffabunny,
Mycobacterium virus JHC117, Mycobacterium virus KBG, Mycobacterium virus
Kssjeb, Mycobacterium virus Kugel, Mycobacterium virus L5, Mycobacterium virus
Lesedi, Mycobacterium virus LHTSCC, Mycobacterium virus lockley, Mycobacterium
virus Marcell, Mycobacterium virus Microwolf, Mycobacterium virus Mrgordo,
Mycobacterium virus Museum, Mycobacterium virus Nepal, Mycobacterium virus
Packman, Mycobacterium virus Peaches, Mycobacterium virus Perseus,
Mycobacterium
virus Pukovnik, Mycobacterium virus Rebeuca, Mycobacterium virus Redrock,
Mycobacterium virus Ridgecb, Mycobacterium virus Rockstar, Mycobacterium virus
Saintus, Mycobacterium virus Skipole, Mycobacterium virus Solon, Mycobacterium
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
59
virus Switzer, Mycobacterium virus SWU1, Mycobacterium virus Ta17 a,
Mycobacterium virus Tiger, Mycobacterium virus Timshel, Mycobacterium virus
Trixie,
Mycobacterium virus Turbido, Mycobacterium virus Twister, Mycobacterium virus
U2,
Mycobacterium virus Violet, Mycobacterium virus Wonder, Escherichia virus DE3,
Escherichia virus HK629, Escherichia virus HK630, Escherichia virus lambda,
Arthrobacter virus Laroye, Mycobacterium virus Halo, Mycobacterium virus
Liefie,
Mycobacterium virus Marvin, Mycobacterium virus Mosmoris, Arthrobacter virus
Circum, Arthrobacter virus Mudcat, Escherichia virus N15, Escherichia virus
9g,
Escherichia virus JenKl, Escherichia virus JenPl, Escherichia virus JenP2,
Pseudomonas virus NP1, Pseudomonas virus PaMx25, Mycobacterium virus Baka,
Mycobacterium virus Courthouse, Mycobacterium virus Littlee, Mycobacterium
virus
Omega, Mycobacterium virus Optimus, Mycobacterium virus Thibault, Polaribacter
virus P12002L, Polaribacter virus P12002S, Nonlabens virus P12024L, Nonlabens
virus
P12024S, Thermus virus P23-45, Thermus virus P74-26, Listeria virus LP26,
Listeria
virus LP37, Listeria virus LP110, Listeria virus LP114, Listeria virus P70,
Propionibacterium virus ATCC29399BC, Propionibacterium virus ATCC29399BT,
Propionibacterium virus Attacne, Propionibacterium virus Keiki,
Propionibacterium
virus Kubed, Propionibacterium virus Lauchelly, Propionibacterium virus MrAK,
Propionibacterium virus Ouroboros, Propionibacterium virus P91,
Propionibacterium
virus P105, Propionibacterium virus P144, Propionibacterium virus P1001,
Propionibacterium virus P1.1, Propionibacterium virus P100A, Propionibacterium
virus
PlOOD, Propionibacterium virus P101A, Propionibacterium virus P104A,
Propionibacterium virus PA6, Propionibacterium virus
Pacnes 201215,
Propionibacterium virus PAD20, Propionibacterium virus PAS50,
Propionibacterium
virus PHLOO9M11, Propionibacterium virus PHL025M00, Propionibacterium virus
PHL037M02, Propionibacterium virus PHL041M10, Propionibacterium virus
PHL060L00, Propionibacterium virus PHL067M01, Propionibacterium virus
PHL070N00, Propionibacterium virus PHL071N05, Propionibacterium virus
PHL082M03, Propionibacterium virus PHL092M00, Propionibacterium virus
PHL095N00, Propionibacterium virus PHL111M01, Propionibacterium virus
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
PHL112N00, Propionibacterium virus PHL113M01, Propionibacterium virus
PHL114L00, Propionibacterium virus PHL116M00, Propionibacterium virus
PHL117M00, Propionibacterium virus PHL117M01, Propionibacterium virus
PHL132N00, Propionibacterium virus PHL141N00, Propionibacterium virus
5 PHL151M00, Propionibacterium virus PHL151N00, Propionibacterium virus
PHL152M00, Propionibacterium virus PHL163M00, Propionibacterium virus
PHL171M01, Propionibacterium virus PHL179M00, Propionibacterium virus
PHL194M00, Propionibacterium virus PHL199M00, Propionibacterium virus
PHL301M00, Propionibacterium virus PHL308M00, Propionibacterium virus Pirate,
10 Propionibacterium virus Procrass 1, Propionibacterium virus SKKY,
Propionibacterium
virus Solid, Propionibacterium virus Stormborn, Propionibacterium virus Wizzo,
Pseudomonas virus PaMx28, Pseudomonas virus PaMx74, Mycobacterium virus
Patience, Mycobacterium virus PBIl, Rhodococcus virus Pepy6, Rhodococcus virus
Poco6, Propionibacterium virus PFR1, Streptomyces virus phiBT1, Streptomyces
virus
15 phiC31, Streptomyces virus TG1, Caulobacter virus Karma, Caulobacter virus
Magneto,
Caulobacter virus phiCbK, Caulobacter virus Rogue, Caulobacter virus Swift,
Staphylococcus virus 11, Staphylococcus virus 29, Staphylococcus virus 37,
Staphylococcus virus 53, Staphylococcus virus 55, Staphylococcus virus 69,
Staphylococcus virus 71, Staphylococcus virus 80, Staphylococcus virus 85,
20 Staphylococcus virus 88, Staphylococcus virus 92, Staphylococcus virus 96,
Staphylococcus virus 187, Staphylococcus virus 52a, Staphylococcus virus
80a1pha,
Staphylococcus virus CNPH82, Staphylococcus virus EW, Staphylococcus virus
IPLA5,
Staphylococcus virus IPLA7, Staphylococcus virus IPLA88, Staphylococcus virus
PH15, Staphylococcus virus phiETA, Staphylococcus virus phiETA2,
Staphylococcus
25 virus phiETA3, Staphylococcus virus phiMR11, Staphylococcus virus phiMR25,
Staphylococcus virus phiNM1, Staphylococcus virus phiNM2, Staphylococcus virus
phiNM4, Staphylococcus virus 5AP26, Staphylococcus virus X2, Enterococcus
virus
FL1, Enterococcus virus FL2, Enterococcus virus FL3, Lactobacillus virus
ATCC8014,
Lactobacillus virus phiJL1, Pediococcus virus cIP1, Aeromonas virus pIS4A,
Listeria
30 virus LP302, Listeria virus PSA, Methanobacterium virus psiM1, Roseobacter
virus
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
61
RDJL1, Roseobacter virus RDJL2, Rhodococcus virus RER2, Enterococcus virus
BC611, Enterococcus virus IMEEF1, Enterococcus virus SAP6, Enterococcus virus
VD13, Streptococcus virus SPQS1, Mycobacterium virus Papyrus, Mycobacterium
virus
5end513, Burkholderia virus KL1, Pseudomonas virus 73, Pseudomonas virus Ab26,
Pseudomonas virus Kakheti25, Escherichia virus Cajan, Escherichia virus
Seurat,
Staphylococcus virus SEP9, Staphylococcus virus Sextaec, Streptococcus virus
858,
Streptococcus virus 2972, Streptococcus virus ALQ132, Streptococcus virus
01205,
Streptococcus virus Sfil 1, Streptococcus virus 7201, Streptococcus virus DT1,
Streptococcus virus phiAbc2, Streptococcus virus Sfi19, Streptococcus virus
Sfi21,
Paenibacillus virus Diva, Paenibacillus virus Hb10c2, Paenibacillus virus
Rani,
Paenibacillus virus Shelly, Paenibacillus virus Sitara, Paenibacillus virus
Willow,
Lactococcus virus 712, Lactococcus virus ASCC191, Lactococcus virus A5CC273,
Lactococcus virus ASCC281, Lactococcus virus A5CC465, Lactococcus virus
A5CC532, Lactococcus virus Bibb29, Lactococcus virus bIL170, Lactococcus virus
CB13, Lactococcus virus CB14, Lactococcus virus CB19, Lactococcus virus CB20,
Lactococcus virus jj50, Lactococcus virus P2, Lactococcus virus P008,
Lactococcus
virus ski, Lactococcus virus S14, Bacillus virus Slash, Bacillus virus Stahl,
Bacillus
virus Staley, Bacillus virus Stills, Gordonia virus Bachita, Gordonia virus
ClubL,
Gordonia virus OneUp, Gordonia virus Smoothie, Gordonia virus Soups, Bacillus
virus
SPbeta, Vibrio virus MARIO, Vibrio virus 55P002, Escherichia virus AKFV33,
Escherichia virus BF23, Escherichia virus DT57C, Escherichia virus EPS7,
Escherichia
virus FFH1, Escherichia virus H8, Escherichia virus s1ur09, Escherichia virus
T5,
Salmonella virus 118970sa12, Salmonella virus Shivani, Salmonella virus 5PC35,
Salmonella virus Stitch, Arthrobacter virus Tank, Tsukamurella virus TIN2,
Tsukamurella virus TIN3, Tsukamurella virus TIN4, Rhodobacter virus RcSpartan,
Rhodobacter virus RcTitan, Mycobacterium virus Anaya, Mycobacterium virus
Angelica, Mycobacterium virus Crimd, Mycobacterium virus Fionnbarth,
Mycobacterium virus Jaws, Mycobacterium virus Larva, Mycobacterium virus
Macncheese, Mycobacterium virus Pixie, Mycobacterium virus TM4, Bacillus virus
BMBtp2, Bacillus virus TP21, Geobacillus virus Tp84, Staphylococcus virus 47,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
62
Staphylococcus virus 3a, Staphylococcus virus 42e, Staphylococcus virus
IPLA35,
Staphylococcus virus phil2, Staphylococcus virus phiSLT, Mycobacterium virus
32HC,
Rhodococcus virus RGL3, Paenibacillus virus Vegas, Gordonia virus Vendetta,
Bacillus
virus Wbeta, Mycobacterium virus Wildcat, Gordonia virus Twister6, Gordonia
virus
Wizard, Gordonia virus Hotorobo, Gordonia virus Monty, Gordonia virus Woes,
Xanthomonas virus CP1, Xanthomonas virus OP1, Xanthomonas virus phi17,
Xanthomonas virus Xop411, Xanthomonas virus XplO, Streptomyces virus TP1604,
Streptomyces virus YDN12, Alphaproteobacteria virus phiJ1001, Pseudomonas
virus
LK04, Pseudomonas virus M6, Pseudomonas virus MP1412, Pseudomonas virus PAE1,
Pseudomonas virus Yua, Pseudoalteromonas virus PM2, Pseudomonas virus phi6,
Pseudomonas virus phi8, Pseudomonas virus phil2, Pseudomonas virus phil3,
Pseudomonas virus phi2954, Pseudomonas virus phiNN, Pseudomonas virus phiYY,
Vibrio virus fs 1, Vibrio virus VGJ, Ralstonia virus R5603, Ralstonia virus
RSM1,
Ralstonia virus RSM3, Escherichia virus M13, Escherichia virus 122, Salmonella
virus
IKe, Acholeplasma virus L51, Vibrio virus fs2, Vibrio virus VFJ, Escherichia
virus Ifl,
Propionibacterium virus B5, Pseudomonas virus Pfl, Pseudomonas virus Pf3,
Ralstonia
virus PE226, Ralstonia virus RSS1, Spiroplasma virus SVTS2, Stenotrophomonas
virus
PSH1, Stenotrophomonas virus SMA6, Stenotrophomonas virus SMA7,
Stenotrophomonas virus SMA9, Vibrio virus CTXphi, Vibrio virus KSF1, Vibrio
virus
VCY, Vibrio virus Vf33, Vibrio virus Vf03K6, Xanthomonas virus Cflc,
Spiroplasma
virus C74, Spiroplasma virus R8A2B, Spiroplasma virus SkV1CR23x, Escherichia
virus
Fl, Escherichia virus Qbeta, Escherichia virus BZ13, Escherichia virus M52,
Escherichia virus a1pha3, Escherichia virus ID21, Escherichia virus ID32,
Escherichia
virus ID62, Escherichia virus NC28, Escherichia virus NC29, Escherichia virus
NC35,
Escherichia virus phiK, Escherichia virus Stl, Escherichia virus WA45,
Escherichia
virus G4, Escherichia virus ID52, Escherichia virus Talmos, Escherichia virus
phiX174,
Bdellovibrio virus MAC1, Bdellovibrio virus MH2K, Chlamydia virus Chpl,
Chlamydia virus Chp2, Chlamydia virus CPAR39, Chlamydia virus CPG1,
Spiroplasma
virus SpV4, Acholeplasma virus L2, Pseudomonas virus PR4, Pseudomonas virus
PRD1, Bacillus virus AP50, Bacillus virus Bam35, Bacillus virus GIL16,
Bacillus virus
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
63
Wipl, Escherichia virus phi80, Escherichia virus RB42, Escherichia virus T2,
Escherichia virus T3, Escherichia virus T6, Escherichia virus VT2-Sa,
Escherichia virus
VT1-Sakai, Escherichia virus VT2-Sakai, Escherichia virus CP-933V, Escherichia
virus
P27, Escherichia virus Stx2phi-I, Escherichia virus Stxlphi, Escherichia virus
Stx2phi-
II, Escherichia virus CP-1639õ based on the Escherichia virus BP-4795,
Escherichia
virus 86, Escherichia virus Min27, Escherichia virus 2851, Escherichia virus
1717,
Escherichia virus YYZ-2008, Escherichia virus ECO26 PO6, Escherichia virus
EC0103 P15, Escherichia virus EC0103 P12, Escherichia virus EC0111 P16,
Escherichia virus EC0111 P11, Escherichia virus VT2phi 272, Escherichia virus
TL-
2011c, Escherichia virus P13374, Escherichia virus Sp5.
[168] In one embodiment, the bacterial virus particles target E. coli and
includes the
capsid of a bacteriophage selected in the group consisting of BW73, B278, D6,
D108, E,
El, E24, E41, FI-2, FI-4, FI-5, HI8A, Ffl8B, i, MM, Mu, 025, PhI-5, Pk, PSP3,
Pl, P1D,
P2, P4, Sl, Wy, yK13, yl, y2, y7, y92, 7 A, 8y, 9y, 18, 28-1, 186, 299, HH-
Escherichia
(2), AB48, CM, C4, C16, DD-VI, E4, E7, E28, FIl, FI3, H, H1, H3, H8, K3, M, N,
ND-
2, ND-3, ND4, ND-5, ND6, ND-7, Ox-I, Ox-2, Ox-3, Ox-4, Ox-5, Ox-6, PhI-I,
RB42,
RB43, RB49, RB69, S, Sal-I, Sal-2, Sal-3, Sal-4, Sal-5, Sal-6, TC23, TC45,
TuII*-6,
TuIP-24, TuII*46, TuIP-60, T2, T4, T6, T35, al, 1, IA, 3, 3A, 3T+, 5y, 9266Q,
CF0103,
HK620, J, K, K1F, m59, no. A, no. E, no. 3, no. 9, N4, sd, T3, T7, WPK, W31,
AH,
yC3888, 21(3, 21(7, 2K12, yV-1, 41)04-CF, 41005, 41006, 41007, yl, y1.2, y20,
y95, y263,
y1092, yl, yll, S28, 1, 3, 7, 8, 26, 27, 28-2, 29, 30, 31, 32, 38, 39, 42,
933W, NN-
Escherichia (1), Esc-7-11, AC30, CVX-5, Cl, DDUP, EC1, EC2, E21, E29, Fl,
F26S,
F27S, Hi, HK022, HK97, HK139, HK253, HK256, K7, ND-I, PA-2, q, S2, Tl, ), T3C,
T5, UC-I, w, (34, y2, 2\., , (I)D326, yy, 41006, 4107, (MO, y80, x, 2, 4, 4A,
6, 8A, 102, 150,
168, 174, 3000, AC6, AC7, AC28, AC43, AC50, AC57, AC81, AC95, HK243, K10,
ZG/3A, 5, 5A, 21EL, H19-J and 933H.
[169] As used herein, a "prebiotic" refers to an ingredient that allows
specific changes,
both in the composition and/or activity in the gastrointestinal microbiota
that may confer
benefits upon the host. A prebiotic can be a comestible food or beverage or
ingredient
thereof. A prebiotic may be a selectively fermented ingredient. Prebiotics may
include
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
64
complex carbohydrates, amino acids, peptides, minerals, or other essential
nutritional
components for the survival of the bacterial composition. Prebiotics include,
but are not
limited to, amino acids, biotin, fructo-oligosaccharide, galacto-
oligosaccharides,
hemicelluloses (e.g., arabinoxylan, xylan, xyloglucan, and glucomannan),
inulin, chitin,
lactulose, mannan oligosaccharides, oligofructose-enriched inulin, gums (e.g.,
guar gum,
gum arabic and carregenaan), oligofructose, oligodextrose, tagatose, resistant
maltodextrins (e.g., resistant starch), trans- galactooligosaccharide, pectins
(e.g.,
xylogalactouronan, citrus pectin, apple pectin, and rhamnogalacturonan-I),
dietary fibers
(e.g., soy fiber, sugarbeet fiber, pea fiber, corn bran, and oat fiber) and
xylooligosaccharides .
[170] As use herein, a "probiotic" refers to a dietary supplement based on
living
microbes which, when taken in adequate quantities, has a beneficial effect on
the host
organism by strengthening the intestinal ecosystem. Probiotic can comprise a
non-
pathogenic bacterial or fungal population, e.g., an immunomodulatory bacterial
population, such as an anti-inflammatory bacterial population, with or without
one or
more prebiotics. They contain a sufficiently high number of living and active
probiotic
microorganisms that can exert a balancing action on gut flora by direct
colonisation. It
must be noted that, for the purposes of the present description, the term
"probiotic" is
taken to mean any biologically active form of probiotic, preferably but not
limited to
lactobacilli, bifidobacteria, streptococci,
enterococci, propionibacteria or
saccharomycetes but even other microorganisms making up the normal gut flora,
or also
fragments of the bacterial wall or of the DNA of these microorganisms. These
compositions are advantageous in being suitable for safe administration to
humans and
other mammalian subjects and are efficacious for the treatment, prevention, of
a
bacterial infection. Probiotics include, but are not limited to lactobacilli,
bifidobacteria,
streptococci, enterococci, propionibacteria, saccharomycetes, lactobacilli,
bifidobacteria,
or proteobacteria.
[171] The antibiotic can be selected from the group consisting in penicillins
such as
penicillin G, penicillin K, penicillin N, penicillin 0, penicillin V,
methicillin,
benzylpenicillin, nafcillin, oxacillin, cloxacillin, dicloxacillin,
ampicillin, amoxicillin,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
pivampicillin, hetacillin, bacampicillin, metampicillin, talampicillin,
epicillin,
carbenicillin, ticarcillin, temocillin, mezlocillin, and piperacillin;
cephalosporins such as
cefacetrile, cefadroxil, cephalexin, cefaloglycin, cefalonium, cefaloridine,
cefalotin,
cefapirin, cefatrizine, cefazaflur, cefazedone, cefazolin, cefradine,
cefroxadine,
5 ceftezole, cefaclor, cefonicid, cefprozil, cefuroxime, cefuzonam,
cefmetazole, cefotetan,
cefoxitin, loracarbef, cefbuperazone, cefminox, cefotetan, cefoxitin,
cefotiam,
cefcapene, cefdaloxime, cefdinir, cefditoren, cefetamet, cefixime,
cefmenoxime,
cefodizime, cefotaxime, cefovecin, cefpimizole, cefpodoxime, cefteram,
ceftamere,
ceftibuten, ceftiofur, ceftiolene, ceftizoxime, ceftriaxone, cefoperazone,
ceftazidime,
10 latamoxef, cefclidine, cefepime, cefluprenam, cefoselis, cefozopran,
cefpirome,
cefquinome, flomoxef, ceftobiprole, ceftaroline, ceftolozane, cefaloram,
cefaparole,
cefcanel, cefedrolor, cefempidone, cefetrizole, cefivitril, cefmatilen,
cefmepidium,
cefoxazole, cefrotil, cefsumide, ceftioxide, cefuracetime, and nitrocefin;
polymyxins
such as polysporin, neosporin, polymyxin B, and polymyxin E, rifampicins such
as
15 rifampicin, rifapentine, and rifaximin; Fidaxomicin; quinolones such as
cinoxacin,
nalidixic acid, oxolinic acid, piromidic acid, pipemidic acid, rosoxacin,
ciprofloxacin,
enoxacin, fleroxacin, lomefloxacin, nadifloxacin, norfloxacin, ofloxacin,
pefloxacin,
rufloxacin, balofloxacin, grepafloxacin, levofloxacin, pazufloxacin,
temafloxacin,
tosufloxacin, clinafloxacin, gatifloxacin, gemifloxacin, moxifloxacin,
sitafloxacin,
20 trovafloxacin, prulifloxacin, delafloxacin, nemonoxacin, and zabofloxacin;
sulfonamides
such as sulfafurazole, sulfacetamide, sulfadiazine, sulfadimidine,
sulfafurazole,
sulfisomidine, sulfadoxine, sulfamethoxazole, sulfamoxole,
sulfanitran,
sulfadimethoxine, sulfametho-xypyridazine, sulfametoxydiazine, sulfadoxine,
sulfametopyrazine, and terephtyl; macrolides such as azithromycin,
clarithromycin,
25 erythromycin, fidaxomicin, telithromycin, carbomycin A, josamycin,
kitasamycin,
midecamycin, oleandomycin, solithromycin, spiramycin, troleandomycin, tylosin,
and
roxithromycin; ketolides such as telithromycin, and cethromycin;
lluoroketolides such as
solithromycin; lincosamides such as lincomycin, clindamycin, and pirlimycin;
tetracyclines such as demeclocycline, doxycycline, minocycline,
oxytetracycline, and
30 tetracycline; aminoglycosides such as amikacin, dibekacin, gentamicin,
kanamycin,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
66
neomycin, netilmicin, sisomicin, tobramycin, paromomycin, and streptomycin;
ansamycins such as geldanamycin, herbimycin, and rifaximin; carbacephems such
as
loracarbef; carbapenems such as ertapenem, doripenem, imipenem (or
cilastatin), and
meropenem; glycopeptides such as teicoplanin, vancomycin, telavancin,
dalbavancin,
and oritavancin; lincosamides such as clindamycin and lincomycin; lipopeptides
such as
daptomycin; monobactams such as aztreonam; nitrofurans such as furazolidone,
and
nitrofurantoin; oxazolidinones such as linezolid, posizolid, radezolid, and
torezolid;
teixobactin, clofazimine, dapsone, capreomycin, cycloserine, ethambutol,
ethionamide,
isoniazid, pyrazinamide, rifabutin, arsphenamine,chloramphenicol, fosfomycin,
fusidic
acid, metronidazole, mupirocin, platensimycin, quinupristin (or dalfopristin),
thiamphenicol, tigecycline, tinidazole, trimethoprim, alatrofloxacin,
fidaxomycin,
nalidixice acide, rifampin, derivatives and combination thereof.
[172] The present invention provides pharmaceutical or veterinary compositions
comprising one or more of the bacterial delivery vehicles disclosed herein and
a
pharmaceutically-acceptable carrier. Generally, for pharmaceutical use, the
bacterial
delivery vehicles may be formulated as a pharmaceutical preparation or
compositions
comprising at least one bacterial delivery vehicles and at least one
pharmaceutically
acceptable carrier, diluent or excipient, and optionally one or more further
pharmaceutically active compounds. Such a formulation may be in a form
suitable for
oral administration, for parenteral administration (such as by intravenous,
intramuscular
or subcutaneous injection or intravenous infusion), for topical
administration, for
administration by inhalation, by a skin patch, by an implant, by a
suppository, etc. Such
administration forms may be solid, semi-solid or liquid, depending on the
manner and
route of administration. For example, formulations for oral administration may
be
provided with an enteric coating that will allow the synthetic bacterial
delivery vehicles
in the formulation to resist the gastric environment and pass into the
intestines. More
generally, synthetic bacterial delivery vehicle formulations for oral
administration may
be suitably formulated for delivery into any desired part of the
gastrointestinal tract. In
addition, suitable suppositories may be used for delivery into the
gastrointestinal tract.
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
67
Various pharmaceutically acceptable carriers, diluents and excipients useful
in bacterial
delivery vehicle compositions are known to the skilled person.
[173] Also provided are methods for treating a bacterial infection using the
synthetic
bacterial delivery vehicles disclosed herein. The methods include
administering the
synthetic bacterial delivery vehicles or compositions disclosed herein to a
subject having
a bacterial infection in need of treatment. In some embodiments, the subject
is a
mammal. In some embodiments, the subject is a human.
[174] The pharmaceutical or veterinary composition according to the disclosure
may
further comprise a pharmaceutically acceptable vehicle. A solid
pharmaceutically
acceptable vehicle may include one or more substances which may also act as
flavouring
agents, lubricants, solubilisers, suspending agents, dyes, fillers, glidants,
compression
aids, inert binders, sweeteners, preservatives, dyes, coatings, or tablet-
disintegrating
agents. Suitable solid vehicles include, for example calcium phosphate,
magnesium
stearate, talc, sugars, lactose, dextrin, starch, gelatin, cellulose,
polyvinylpyrrolidine, low
melting waxes and ion exchange resins.
[175] The pharmaceutical or veterinary composition may be prepared as a
sterile solid
composition that may be suspended at the time of administration using sterile
water,
saline, or other appropriate sterile injectable medium. The pharmaceutical or
veterinary
compositions of the disclosure may be administered orally in the form of a
sterile
solution or suspension containing other solutes or suspending agents (for
example,
enough saline or glucose to make the solution isotonic), bile salts, acacia,
gelatin,
sorbitan monoleate, polysorbate 8o (oleate esters of sorbitol and its
anhydrides
copolymerized with ethylene oxide) and the like. The particles according to
the
disclosure can also be administered orally either in liquid or solid
composition form.
Compositions suitable for oral administration include solid forms, such as
pills,
capsules, granules, tablets, and powders, and liquid forms, such as solutions,
syrups,
elixirs, and suspensions. Forms useful for enteral administration include
sterile solutions,
emulsions, and suspensions.
[176] The bacterial delivery vehicles according to the disclosure may be
dissolved or
suspended in a pharmaceutically acceptable liquid vehicle such as water, an
organic
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
68
solvent, a mixture of both or pharmaceutically acceptable oils or fats. The
liquid vehicle
can contain other suitable pharmaceutical additives such as solubilisers,
emulsifiers,
buffers, preservatives, sweeteners, flavouring agents, suspending agents,
thickening
agents, colours, viscosity regulators, stabilizers or osmo-regulators.
Suitable examples of
liquid vehicles for oral and enteral administration include water (partially
containing
additives as above, e.g. cellulose derivatives, preferably sodium
carboxymethyl cellulose
solution), alcohols (including monohydric alcohols and polyhydric alcohols,
e.g.
glycols) and their derivatives, and oils (e.g. fractionated coconut oil and
arachis oil). For
parenteral administration, the vehicle can also be an oily ester such as ethyl
oleate and
isopropyl myristate. Sterile liquid vehicles are useful in sterile liquid form
compositions
for enteral administration. The liquid vehicle for pressurized compositions
can be a
halogenated hydrocarbon or other pharmaceutically acceptable propellant.
[177] For transdermal administration, the pharmaceutical or veterinary
composition
can be formulated into ointment, cream or gel form and appropriate penetrants
or
detergents could be used to facilitate permeation, such as dimethyl sulfoxide,
dimethyl
acetamide and dimethylformamide.
[178] For transmucosal administration, nasal sprays, rectal or vaginal
suppositories can
be used. The active compounds can be incorporated into any of the known
suppository
bases by methods known in the art. Examples of such bases include cocoa
butter,
polyethylene glycols (carbowaxes), polyethylene sorbitan monostearate, and
mixtures of
these with other compatible materials to modify the melting point or
dissolution rate.
[179] The present invention relates to a method for treating a disease or
disorder
caused by bacteria comprising administering a therapeutically amount of the
pharmaceutical or veterinary composition as disclosed herein to a subject
having such
disease or disorder and in need of treatment. It also relates to the
pharmaceutical or
veterinary composition as disclosed herein for use in the treatment of a
disease or
disorder caused by bacteria. It further relates to the use of a pharmaceutical
or veterinary
composition as disclosed herein for the manufacture of a medicament for
treating a
disease or disorder caused by bacteria.
[180] The diseases or disorders caused by bacteria may be selected from the
group
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
69
consisting of abdominal cramps, acne vulgaris, acute epiglottitis, arthritis,
bacteraemia,
bloody diarrhea, botulism, Brucellosis, brain abscess, chancroid venereal
disease,
Chlamydia, Crohn's disease, conjunctivitis, cholecystitis, colorectal cancer,
polyposis,
dysbiosis, Lyme disease, diarrhea, diphtheria, duodenal ulcers, endocarditis,
erysipelothricosis, enteric fever, fever, glomerulonephritis, gastroenteritis,
gastric ulcers,
Guillain-Barre syndrome tetanus, gonorrhoea, gingivitis, inflammatory bowel
diseases,
irritable bowel syndrome, leptospirosis, leprosy, listeriosis, tuberculosis,
Lady
Widermere syndrome, Legionaire's disease, meningitis, mucopurulent
conjunctivitis,
multi-drug resistant bacterial infections, multi-drug resistant bacterial
carriage,
myonecrosis-gas gangrene, mycobacterium avium complex, neonatal necrotizing
enterocolitis, nocardiosis, nosocomial infection, otitis, periodontitis,
phalyngitis,
pneumonia, peritonitis, purpuric fever, Rocky Mountain spotted fever,
shigellosis,
syphilis, sinusitis, sigmoiditis, septicaemia, subcutaneous abscesses,
tularaemia,
tracheobronchitis, tonsillitis, typhoid fever, ulcerative colitis, urinary
infection and
whooping cough.
[181] The disease or disorder caused by bacteria may be a bacterial infection
selected
from the group consisting of skin infections such as acne, intestinal
infections such as
esophagitis, gastritis, enteritis, colitis, sigmoiditis, rectitis, and
peritonitis, urinary tract
infections, vaginal infections, female upper genital tract infections such as
salpingitis,
endometritis, oophoritis, myometritis, parametritis and infection in the
pelvic
peritoneum, respiratory tract infections such as pneumonia, intra-amniotic
infections,
odontogenic infections, endodontic infections, fibrosis, meningitis,
bloodstream
infections, nosocomial infection such as catheter-related infections, hospital
acquired
pneumonia, post-partum infection, hospital acquired gastroenteritis, hospital
acquired
urinary tract infections, and a combination thereof. Preferably, the infection
according to
the disclosure is caused by a bacterium presenting an antibiotic resistance.
In a particular
embodiment, the infection is caused by a bacterium as listed above in the
targeted
bacteria.
[182] The disease or disorder caused by bacteria may also be a metabolic
disorder, for
example, obesity and diabetes. The disclosure thus also concerns a
pharmaceutical or
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
veterinary composition as disclosed herein for use in the treatment of
metabolic disorder
including, for example, obesity and diabetes. It further concerns a method for
treating a
metabolic disorder comprising administering a therapeutically amount of the
pharmaceutical or veterinary composition as disclosed herein, and the use of a
5 pharmaceutical or veterinary composition as disclosed herein for the
manufacture of a
medicament for treating a metabolic disorder.
[183] The disease or disorder caused by bacteria may also be a pathology
involving
bacteria of the human microbiome. Thus, in a particular embodiment, the
disclosure
concerns a pharmaceutical or veterinary composition as disclosed herein for
use in the
10 treatment of pathologies involving bacteria of the human microbiome, such
as
inflammatory and auto-immune diseases, cancers, infections or brain disorders.
It further
concerns a method for treating a pathology involving bacteria of the human
microbiome
comprising administering a therapeutically amount of the pharmaceutical or
veterinary
composition as disclosed herein, and the use of a pharmaceutical or veterinary
15 composition as disclosed herein for the manufacture of a medicament for
treating a
pathology involving bacteria of the human microbiome. Indeed, some bacteria of
the
microbiome, without triggering any infection, can secrete molecules that will
induce
and/or enhance inflammatory or auto-immune diseases or cancer development.
More
specifically, the present disclosure relates also to modulating microbiome
composition to
20 improve the efficacy of immunotherapies based, for example, on CAR-T
(Chimeric
Antigen Receptor T) cells, TIL (Tumor Infiltrating Lymphocytes) and Tregs
(Regulatory
T cells) also known as suppressor T cells. Modulation of the microbiome
composition to
improve the efficacy of immunotherapies may also include the use of immune
checkpoint inhibitors well known in the art such as, without limitation, PD-1
25 (programmed cell death protein 1) inhibitor, PD-Li (programmed death ligand
1)
inhibitor and CTLA-4 (cytotoxic T lymphocyte associated protein 4).
[184] Some bacteria of the microbiome can also secrete molecules that will
affect the
brain.
[185] Therefore, a further object of the disclosure is a method for
controlling the
30 microbiome of a subject, comprising administering an effective amount of
the
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
71
pharmaceutical composition as disclosed herein in said subject.
[186] In a particular embodiment, the disclosure also relates to a method for
personalized treatment for an individual in need of treatment for a bacterial
infection
comprising: i) obtaining a biological sample from the individual and
determining a
group of bacterial DNA sequences from the sample; ii) based on the determining
of the
sequences, identifying one or more pathogenic bacterial strains or species
that were in
the sample; and iii) administering to the individual a pharmaceutical
composition
according to the disclosure capable of recognizing each pathogenic bacterial
strain or
species identified in the sample and to deliver the packaged plasmid.
[187] Preferably, the biological sample comprises pathological and non-
pathological
bacterial species, and subsequent to administering the pharmaceutical or
veterinary
composition according to the disclosure to the individual, the amount of
pathogenic
bacteria on or in the individual are reduced, but the amount of non-pathogenic
bacteria is
not reduced.
[188] In another particular embodiment, the disclosure concerns a
pharmaceutical or
veterinary composition according to the disclosure for use in order to improve
the
effectiveness of drugs. Indeed, some bacteria of the microbiome, without being
pathogenic by themselves, are known to be able to metabolize drugs and to
modify them
in ineffective or harmful molecules.
[189] In another particular embodiment, the disclosure concerns a composition
according to the invention that may further comprise at least one additional
active
ingredient, for instance a prebiotic and/or a probiotic and/or an antibiotic,
and/or another
antibacterial or antibiofilm agent, and/or any agent enhancing the targeting
of the
bacterial delivery vehicle to a bacteria and/or the delivery of the payload
into a bacteria.
[190] In another particular embodiment, the disclosure concerns the in-situ
bacterial
production of any compound of interest, including therapeutic compound such as
prophylactic and therapeutic vaccine for mammals. The compound of interest can
be
produced inside the targeted bacteria, secreted from the targeted bacteria or
expressed on
the surface of the targeted bacteria. In a more particular embodiment, an
antigen is
expressed on the surface of the targeted bacteria for prophylactic and/or
therapeutic
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
72
vaccination.
[191] The present disclosure also relates to a non-therapeutic use of the
bacterial
delivery particles. For instance, the non-therapeutic use can be a cosmetic
use or a use
for improving the well-being of a subject, in particular a subject who does
not suffer
from a disease, and in particular from a disease or disorder caused by
bacteria.
Accordingly, the present disclosure also relates to a cosmetic composition or
a non-
therapeutic composition comprising the bacterial delivery particles of the
disclosure.
EXAMPLE 1
[192] The example below demonstrates that a significative portion of a lambda
receptor binding protein (RBP), e.g. the stf protein, can be exchanged with a
portion of a
different RBP. More particularly, specific fusion positions in the lambda RBP
have been
identified which allow one to obtain a functional chimeric RBP. Specifically,
the data
demonstrate in a non-limiting embodiment that in the case of packaged
phagemids
derived from bacteriophage lambda, modifying the side tail fiber protein
results in an
expanded host range. The addition of chimeric stf proteins to lambdoid
packaged
phagemids, is demonstrated to be a very powerful approach to modify and
increase their
host range, and in some cases is more efficient than the modification of the
gpJ gene. In
addition, modification of the side tail fiber protein to encode enzymatic
activity such as
depolymerase activities can dramatically increase the delivery efficiency. In
some cases,
the addition of this enzymatic activity allows for 100% delivery efficiency
while the
wild-type lambda packaged phagemid showed no entry at all. These two
approaches can
be combined to generate packaged phagemid variants with different
specificities and
delivery efficiencies to many strains of bacterial species.
Materials and Methods
[193] Tests were conducted to determine whether the modification of the tail
tip gene
(gpJ) would have an impact in the host range of lambda packaged phagemids. The
lambda tail tip was modified to include the mutations described in [11] to
generate
OMPF-lambda. This packaged phagemid should now use OmpF instead of LamB as a
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
73
primary receptor in the cell surface. Next, the delivery efficiency was tested
in a
collection of E. coli strains that spans a variety of 0 and K serotypes, as
shown in FIG.
1.
[194] As can be seen in FIG. 1, using packaged phagemids that recognize a
different
cell surface receptor has a minimal impact on efficiency delivery and host
range. Only 3
strains show a marginal improvement in the number of colonies after treatment
with the
modified packaged phagemid. This result may be due to the presence of a
capsule
around the majority of the cells that forms a physical barrier to the packaged
phagemids,
thus rendering this approach unsuccessful. In view of these results, the
lambda stf gene
was modified to include enzymatic activities against bacterial capsules.
[195] The sequence of lambda stf (SEQ ID NO:1) is:
MAVKISGVLKDGTGKPVQNCTIQLKARRNSTTVVVNTVGSENP DEAGRYSMDVE
YGQYSVILQVDGFPPSHAGTITVYEDSQPGTLNDFLCAMTEDDARPEVLRRLELM
VEEVARNASVVAQSTADAKKSAGDASASAAQVAALVTDATDSARAASTSAGQAA
SSAQEASSGAEAASAKATEAE KSAAAAE SSKNAAATSAGAAKTSE TNAAAS QQSA
ATSASTAATKASEAATSARDAVASKEAAKSSE TNASSSAGRAASSATAAE N SARA
AKTSE TNARSSE TAAE RSASAAADAKTAAAGSAS TASTKATEAAGSAVSAS QSKS
AAEAAAI RAKN SAKRAE D IASAVAL E DAD TTRKGIVQLSSATNSTSETLAATP KA
VKVVM D E TN RKAP L D SPALTGTP TAP TAL RGTN N TQIAN TAFVLAAIAD VI DASP
DAL N TL N E LAAAL GN D P DFATTMTNALAGKQP KNATLTALAGLSTAKNKLPYF
AE N DAASLTE LTQVGRD I LAKN SVADVLEYLGAGENSAFPAGAPIPWPSDIVPS
GYVLMQGQAFDKSAYPKLAVAYPSGVLPDMRGWTIKGKPASGRAVLSQEQD
GIKSHTHSASASGTDLGTKTTSSFDYGTKTTGSFDYGTKSTNNTGAHAHSLS
GSTGAAGAHAHTSGLRMNSSGWSQYGTATITGSLSTVKGTSTQGIAYLSKTD
SQGSHSHSLSGTAVSAGAHAHTVGIGAHQHPVVIGAHAHSFSIGSHGHTITVN
AAGNAENTVKNIAFNYIVRLA
[196] The bold and underlined sequence represents the part of the protein that
was
introduced in the T4 phage (Montag et al. J Bacteriol. 1989 Aug; 171(8): 4378-
4384).
Experiments were conducted to investigate if it was possible to exchange the C-
terminus
of the lambda stf with a tail fiber from a different phage to yield chimeric
side tail fibers
with an enzymatic activity against encapsulated E. coli. The tail fiber from
the KlF
phage which has been studied in depth and its structure solved [19], [20] was
chosen.
KlF encodes an enzyme with endosialidase activity, which is active against
polymer of
sialic acid secreted by K1-encapsulated E. coli. In fact, Kl+ strains are
immune to T7
infection because the capsule forms a physical barrier that prevents
attachment of the
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
74
phage, but if purified K 1F enzyme is added to the cells before infection, T7
is able to
lyse them [21], confirming that the presence of bacterial capsules is a
powerful
mechanism to avoid recognition by bacteriophages. Thus, by testing delivery of
modified lambda-stf-K1 packaged phagemids in Kl+ strains it was possible to
verify
whether the lambda-stf chimeric proteins retain its enzymatic activity.
[197] The sequence of KlF tail fiber (SEQ ID NO:121) is:
MSTITQFPSGNTQYRIEFDYLARTFVVVTLVNSSNPTLNRVLEVGRDYRF
LNPTMIEMLVDQSGFDIVRIHRQTGTDLVVDFRNGSVLTASDLTTAELQA
IHIAEEGRDQTVDLAKEYADAAGSSAGNAKDSEDEARRIAESIRAAGLIG
YMTRRSFEKGYNVTTWSEVLLWEEDGDYYRWDGTLPKNVPAGSTPETS
GGIGLGAWVSVGDAALRSQISNPEGAILYPELHRARWLDEKDARGWGA
KGDGVTDDTAALTSALNDTPVGOKINGNGKTYKVTSLPDISRFINTR
FVYERIPGCOPLYYASEEFVOGELFKITDTPYYNAWPODKAFVYENVI
YAPYMGSDRHGVSRLHVSWVKSGDDGOTWSTPEWLTDLHPDYPTV
NYHCMSMGVCRNRLFAMIETRTLAKNALTNCALWDRPMSRSLHLT
GGITKAANCORYATIHVPDHGLFVGDFVNFSNSAVTGVSGDMTVATVI
DKDNFTVLTPNOCITSDLNNAGKNWHMGTSFHKSPWRKTDLGLIPSV
TEVHSFATIDNNGFAMGYHOGDVAPREVGLFYFPDAFNSPSNYVRRO
IPSEYEPDASEPCIKYYDGVLYLITRGTRGDRLGSSLHRSRDIGOTWE
SLRFPHNVHHTTLPFAKVGDDLIMFGSERAENEWEAGAPDDRYKAS
YPRTFYARLNVNNWNADDIEWVNITDOIYOGGIVNSGVGVGSVVVK
DNYIYYMFGGEDHFNPWTYGDNSAKDPFKSDGHPSDLYCYKMKIGP
DNRVSRDFRYGAVPNRAVPVFFDTNGVRTVPAPMEFTGDLGLGHVT
IRASTSSNIRSEVLMEGEYGFIGKSIPTDNPAGORHFCGGEGTSSTTG
ACIITLYGANNTDSRRIVYNGDEHLFOSADVKPYNDNVTALGGPSNRF
TTAYLGSNPIVTSNGERKTEPVVFDDAFLDAWGDVHYIMYCOWLDAV
COLKGNDARIHFGVIACIOIRDVFIAHGLMDENSTNCRYAVLCYDKYPR
MTDTVFSHNEIVEHTDEEGNVTTTEEPVYTEVVIHEEGEEWGVRPD
GIFFAEAAYORRKLERIEARLSALECOK
[198] The bold and underlined sequence represents the part of the protein that
has been
crystalized and has been shown to retain its endosialidase activity. Since
there is no
identity between the lambda stf protein and the KlF tail fiber, the insertion
site was
made based on conclusions extracted from different sources of information,
including
literature and crystal structures.
[199] The stf gene was modified to include the KlF endosialidase at its C-
terminus
using a Cas9-mediated gene exchange protocol [22] and resulted in a lambda-K1F
chimeric stf of nucleotide sequence SEQ ID NO: 106 and aminoacid sequence SEQ
ID
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
NO: 46. Lambda-K1F packaged phagemids were produced as in [23] and titrated
against
some K1+ strains, specifically E. coli UTI89 and S88. The results were
striking; in
these strains, there is no delivery if lambda wild-type stf is used, but the
addition of the
lambda-K1F variant of SEQ ID NO: 46 to the packaged phagemid gives 100%
delivery
5 (FIG. 2).
[200] The same principle was followed to create a different variant of lambda-
stf, this
time with KS-capsule degrading activity (K5 lyase tail fiber from phage K5A).
As in the
case of K1F, there is no homology between lambda-stf and K5 lyase, but its
crystal
structure has been published [24]. Hence, the same approach as for K 1F was
used to
10 generate lambda-KS chimeric side tail fibers of nucleotide sequence SEQ ID
NO: 107
and aminoacid sequence SEQ ID NO: 47 and tested the produced packaged
phagemids
against a KS-encapsulated strain of E. coli (ECOR 55). In this case, however,
a delta-stf
lambda production strain was produced with the lambda-KS stf fusion gene
expressed in
trans under the control of an inducible promoter. As depicted in FIG. 3, there
was some
15 residual delivery using the wild-type lambda-stf, probably due to the
presence of some
cells with a thinner K5 capsule. However, the addition of lambda-stf-K5
chimeras
allows for an improvement in delivery of more than 106 fold.
[201] In some other cases, side tail fibers can be found that have some degree
of
homology to lambda stf, although no crystal structure is available. In these
cases, the
20 insertion site was designed as the last stretch of amino acids with
identity to lambda stf.
For example, in two in-house sequenced phages, the predicted side tail fiber
proteins are
as follows:
[202] Phage AG22 stf:
[203] MAIYRQGQASMDAQGYVTGYGTKWREQLTLIRPGATIFFLAQPLQAAVI
25 TEVISDTSIRAITTGGAVVQKTNYLILLHDSLTVDGLAQDVAETLRYYQGKESEF
AGFIEIIKDFDWDKLQKIQEDVKTNADAAAAS QQAAKTSENNAKTS ATNAANS
KKGADTAKAAAESARDAANTAKTGAEAAKSGAESARDAANTAKAGAESARD
QAEEYAKQAAEPYKDLLQPLPDVWIPFNDSLDMITGFSPSYKKIVIGDDEITMPG
DKIVKFKRASTATYINKSGVLTNAAIDEPRFEKDGLLIEGQRTNLLINSTNPSKW
30 NKSSNMILDRSGVDDFGFQYAKFTLKPEMVGQTSSINIVTVSGSRGFDVTGNEK
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
76
YVTIS CRAQS GTPNLRCRLRFENYD GS AYAS LGDAYVNLTDLS IEKTGGAANRI
TARAVKDEASKWIFFEATIKALDTENMIGAMVQYAPAKDGGGTGADDYIYIAT
PQVEGGVCASSFIITEATPVTRASDMVTIPIKNNLYNLPFTVLCEVHKNVVYITPN
AAPRVFDTGGHQS GAAIILAFGS AD GD ND GFPYCDIGKS NRRVNENAKLKKMII
GMRVKSDYNTCCVSNARISSETKTEWRYIVSTATIRIGGQTSTGERHLFGHVRNF
RIVVHKALTDHQLGEIV (SEQ ID NO:204) and corresponding nucleotide sequence of
SEQ ID NO: 213.
[204] Its alignment to lambda stf is as follows:
Lambda 156
STSAGQAASSAQEASSGAEAASARATEAEKSAAAAESSKNAAATSAGAAKTSETNAAASQ
AG22= 92
ETLRYYQGKESEFAGFIETIKDFDWDKLQKIQEDVKTNADAAAASQQAAKTSENNAKTSA
*** * ****** ** *
[205] The sequence of the stf of a second in-house phage is as follows:
[206] Phage SIEAll stf:
[207] MSTKFKTVITTAGAAKLAAATVPGGKKVTLSAMAVGDGNGKLPVPDAG
QTKLVHEVWRHALNKVSVDNKNKNYIVAELVVPPEVGGFWMRELGLYDDAG
TLIAVSNMAESYKPELAEGSGRAQTCRMVIIVSNVASVELSIDASTVMATQDYV
DDKIAEHEQS RRHPDATLTEKGFTQLS S ATNS TS ES LAATPKAVKAANDNANS R
LAKNQNGAD IQDKS AFLDNVGVTS LTFMKNNGEMPVDADLNTFGS VKAYS GI
WSKATSTNATLEKNFPEDNAVGVLEVFTGGNFAGTQRYTTRDGNLYIRKLIGT
WNGNDGPWGAWRHVQAVTRALSTTIDLNSLGGAEHLGLWRNSSSAIASFERH
YPEQGGDAQGILEIFEGGLYGRTQRYTTRNGTMYIRGLTAKWDAENPQWEDW
NQIGYQTSSTFYEDDLDDLMSPGIYSVTGKATHTPIQGQSGFLEVIRRKDGVYVL
QRYTTTGTSAATKDRLYERVFLGGSFNAWGEWRQIYNSNSLPLELGIGGAVAK
LTS LDWQTYD FVPGS LITVRLDNMTNIPD GMDWGVID GNLINIS V GPS DD S GS G
RSMHVWRSTVSKANYRFFMVRISGNPGSRTITTRRVPIIDEAQTWGAKQTFSAG
LS GELS GNAATATKLKTARKINNVS FD GTS D INLTPKNIGAFAS GKTGDTVAND
KAVGWNWSSGAYNATIGGASTLILHFNIGEGSCPAAQFRVNYKNGGIFYRSARD
GYGFEADWSEFYTTTRKPTAGDVGALPLSGGQLNGALGIGTSSALGGNSIVLGD
NDTGFKQNGD GNLDVYANS VHVMRFVS GS VQS NKTINITGRVNPS DYGNFD S R
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
77
YVRDVRLGTRVVQTMQKGVMYEKAGHVITGLGIVGEVDGDDPAVFRPIQKYIN
GTWYNVAQV (SEQ ID NO:205) and corresponding nucleotide sequence of SEQ ID
NO: 214
[208] Its alignment to lambda stf is as follows:
Lambda 367
SSATNSTSETLANITKAVICVVMDETNRKAPLDSPALTGTPTAPTALRGTNNIVIANTAFV
SIEA11 180 SSATNS TSESLAATPISAVKAANDNANSRL- - -AKNONGADIQDKSAF-
LDNVGVTSLTF14
********* ********* * * * * *
[209] In these two specific cases, it was unknown which antigen these side
tail fibers
were able to recognize, so lambda packaged phagemids with the chimeric side
tail fibers
lambda-AG22 and lambda-SIEAll enginereed based respectively on SEQ ID NO: 1
and
SEQ ID NO: 213, and SEQ ID NO: 1 and SEQ ID NO: 214 were produced and their
delivery efficiency was tested in a E. coli collection that contains a very
diverse group of
0 and K serotypes.
[210] As shown in FIG. 4, the addition of a chimeric stf allows the lambda-
based
packaged phagemid to show increased delivery efficiency in 25 out of 96
strains tested
(more than 25% of the collection). In some cases, the increase is modest; in
others, it
allows for very good delivery efficiency in strains that had no or very low
entry with
wild-type lambda packaged phagemids. It is also worth noting that AG22 belongs
to the
Siphovirus_family, like lambda, but SIEA1 1 is a P2-like phage. This
highlights the
significant observation that stf modules can be exchanged across bacteriophage
genera.
[211] Other side tail fiber genes have been analyzed as shown in FIG. 4 and
several
insertion sites into the lambda stf gene have been identified that give
chimeric variants
with differential entry in the E. coli collection as shown previously. These
insertion sites
are based on the results for the non-homologous tail fiber variants (such as
in the cases
for K 1F and K5 above) or on varying degrees of homology between lambda stf
and the
variant to be tested. This homology can be short, about 5-10 aminoacids, or
substantially
similar. The insertion sites tested are shown in bold and underlined below:
[212] lambda stf
[213] MAVKISGVLKDGTGKPVQNCTIQLKARRNSTTVVVNTVGSENPDEAGRYSMDVE
YGQYSVILQVDGEPPSHAGTITVYEDSQPGTLNDFLCAMTEDDARPEVLRRLELMVEEVA
RNASVVAQSTADAKKSAGDASASAAQVAALVTDATDSARAASTSAGQAASSAQEASSGA
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
78
EAASAKATEAE KSAAAAE SSKNAAATSAGAAKTSETNAAASQQSAATSASTAATKASEA
ATSARDAVASKEAAKSSETNASSSAGRAASSATAAE N SARAAKTSETNARSSETAAE RSA
SAAADAKTAAAGSASTASTKATEAAGSAVSASQSKSAAEAAAIRAKNSAKRAEDIASAVA
LEDADTTRKGIVQLSSATNSTSETLAATPKAVKVVMDETNRKAP L D SPALTGTP TAP TA
LRGTNNTQIANTAFVLAAIADVIDASP DAL NTL N ELAAALGNDP DFATTMTNALAGKQP
KNATLTALAGLSTAKN KL PYFAE N DAASLTE LTQVGRD I LAKN SVADVL EYL GAGENSAF
PAGAP I PWP SD IVP SGYVL M QG QAF D KSAYP KLAVAYP SGVL P D M RGWTI KGKPASGRA
VLSQEQDGIKSHTH SASASGTD L GTKTTSSF DYGTKTTGSF DYGTKSTN NTGAHAH SL SG
STGAAGAHAHTSGLRMNSSGWSQYGTATITGSLSTVKGTSTQGIAYLSKTDSQGSHSHSL
SGTAVSAGAHAHTVGIGAH QH PVVIGAHAH SF SI GSH GH TITVNAAGNAE NTVKN IAF NY
IVRLA (SEQ ID NO:1)
The lambda stf protein consists of 774 aminoacids. The insertion sites can be
found
closer to the N-terminus (amino acid 131, insertion point ADAKKS (SEQ ID
NO:191))
or closer to the C-terminus (amino acid 529, insertion point GAGENS (SEQ ID
NO:194)). Stf chimeras with aminoacid sequences of SEQ ID NO: 2-45 and 48-61
and
corresponding nucleotide sequences of SEQ ID NO: 62-105 and 108-120 were
engineered using these insertion sites. FIG. 5 depicts delivery of some
selected examples
of stf chimeras for the insertion sites ADAKKS (SEQ ID NO:191), SASAAA (SEQ ID
NO:193) and MDETNR (SEQ ID NO:192).The results described herein show that it
is
possible to build chimeric tail fibers that combine the part of one tail fiber
that attaches
to the capsid of one phage (usually the N-terminus of the protein) with the
part of
another fiber that interacts with the bacterium (usually the C-terminus of the
protein).
Stretches of homology between the sequences of different tail fibers can be
considered
as preferable recombination sites. In order to identify such sites for the stf
protein of
phage lambda a scan of the stf sequence was performed with a 50aa window and a
phmmer search [25] was performed on each window to identify homologous
sequences
in the representative proteome 75 database (FIG. 6).
EXAMPLE 2
[214] Many phages contain a single stf protein, which is a very important
factor
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
79
determining their host specificity. However, there are also several examples
of phages
encoding more than one stf gene, which is a beneficial trait since,
presumably, each of
them recognizes a different host. These phages have found different solutions
to achieve
this feature: some of them, like phi92, encode up to 6 stfs that bind to
different parts of
the baseplate/viral particle, and probably to other stfs [29]; others, like
CBA120 [30],
encode 4 stfs that form a tetrameric structure in which one of the stfs
attaches to the
phage particle while the other three attach to the first one through
interaction; and others,
like DT57C, contain an stf that binds the particle and a second one that
attaches to the
first through an interaction domain (branched stfs) [32] (FIG. 7A). In terms
of
engineering, having a particle that is able to recognize different hosts could
have a great
impact in terms of production costs and host range expansion.
[215] As a proof of concept, an engineered lambda stf was constructed based on
a
branched architecture. A phage, referred to as WWII, contains two stfs of SEQ
ID NO:
124 and 125 that follow the same order and contain homology to phage DT57C,
which
has been suggested to have branched stfs. The interaction domains of stf-1 and
stf-2 in
phage WWII have been identified and used as modules to attach to the lambda
stf. The
final construct contains the N-terminal part of the lambda stf of SEQ ID NO: 1
up to the
GAGENS insertion site SEQ ID NO: 194 fused to WWII stf-1 interaction domain
ID1
of SEQ ID NO: 280 and WW1 1 stf-1 proper; after this, a synthetic RBS was
inserted
and immediately after, the stf-2 interaction domain ID2 of SEQ ID NO: 281 was
fused
to the C-terminal part of the KlF tail fiber of SEQ ID NO: 121 (see bold
sequence of
section [198] (FIG. 7B). The GAGENS insertion site of SEQ ID NO: 194 was
chosen as
the insertion site. Both chimeric proteins were transcribed from a
polycistronic mRNA.
This construction resulted in a final branched stf WW11-K1F of aminoacid
sequence
SEQ ID NO: 282 and 283 and of nucleotide sequence of SEQ ID NO: 284.
[216] The original host range of WWII phage is 0157 strains, while that of KlF
phage
is K1 strains. As demonstrated in FIG. 8, when producing packaged phagemids
containing the branched chimeric stf of SEQ ID NO: 282 and 283, it can be seen
that the
host range of the branched stf is now the combination of each single stf, i.e.
a
combination of both single stf activities (0157 and K1). Although the KlF stf
in the
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
branched architecture seems to be less efficient than in the particle
containing only one
stf, further engineering such as, for example, (i) choosing a more efficient
RBS between
both stfs,(ii) increasing the length or (iii) introducing flexible linkers
between the
interaction domains and the fusion stfs and/or (iv) increasing the translation
rate of the
5 first stf in the polycistron (since this is known to affect the translation
rate of the second
CDS in a polycistronic message). An advantage of this approach is also that
the stfs are
present in the phagemid particle in a 1:1 ratio, assuming proper expression of
both
components, which may be important for regulatory purposes.
[217] Following a similar approach to the "dimeric" branched stf (Lambda-ID1-
WW11
10 ID2-K1F), a lambda particle whose stf carried 4 different activities was
designed.
CBA120 is a phage with 4 fibers (called stf; See, FIG. 9). Their sequences are
below:
5tf4 (also called TSP4) of FIG. 9 (orf213, protein ID YP 004957867.1) (SEQ ID
NO:127), stf3 (also called TSP3) of FIG. 9 (orf212, protein ID YP 004957866.1)
(SEQ ID NO:128), stf2 (also called TSP2) of FIG. 9
(orf211, protein ID
15 YP 004957865.1) (SEQ ID NO:129), and stfl(also called TSP1) of FIG. 9
(orf210,
protein ID YP 004957864.1) (SEQ ID NO: 130).
During analysis of these 4 TSPs, it was noticed that the N-termini have
homology to
phages G7C/WW11/DT57C, which is a strong indication that these stfs associate
to
form a tetrameric complex. The first stf to be translated, according to the
TSP operon in
20 the CBA120 genome, is TSP4, then TSP3, then TSP2, then TSP1. From this
information, it was assumed that the main stf is TSP4, since it's the one to
be expressed
first. It may encode an ID domain to which more than one TSP attaches. The N-
terminal
domains also share homology to one another, which helps in the identification
of the
presumable ID domains. The following interaction domains (ID) were found with
25 probable branching/association activities, making BLAST analysis to the
phages
mentioned above: TSP4 ID4 Of FIG. 9 (SEQ ID NO: 131) (ID4 seems to have an
extra
domain at the N-terminus that may be involved in capsid binding, so it will be
left out of
the final construct), TSP3 ID3 of FIG. 9 (SEQ ID NO: 132), TSP2 ID2 of FIG. 9
(SEQ
ID NO: 133), and TSP1 ID1 of FIG. 9 (SEQ ID NO:134).
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
81
The following describes the assembly process to be used to construct the
tetrameric
branched RBP. First, the domains were recoded and two gene blocks were
ordered.
Recoding is a very common process, and the objective is two-fold. First
recoding is done
to avoid codon bias based on the fact that the codons used for a given amino
acid vary
depending on the organism. Accordingly, to avoid expression problems, which
can lead
to truncated, mutated or misfolded proteins, sequences were recoded with the
codon
usage of the host organism (in this case E. coli). Second, removal of unknown
layers of
regulation may be advantageous.This is especially true for operons, phages and
in
general any other sequences with high "density information", like a phage
genome; they
have a limited amount of DNA that can be packaged, so several signals and
functions
may be encoded within a region, and this may impact the ability to use a
genetic part in
the desired way.
[218] Plasmid pPh1F-Tetra STF BsaI cloning (pSC101 37C, KanR) has been cloned,
containing a fusion to Lambda stf GAGENS of SEQ ID NO: 194 with TSP4
interaction
domain SEQ ID NO: 131 to 134. The other TSP interaction domains are preceded
by a
synthetic RBS to avoid including unknown layers of regulation from the phage.
This
plasmid allows the TypeIIS cloning (BSaI) of 4 stfs fused to each of the TSP
interaction
domains. Four stfs and 4 strains were identified for readout of each stf
activity: (i)- V10:
strain V10; (ii) - K1F: strain K1F; (iii)- K5: strain K5; and - stf48: strain
48. These four
stfs will be cloned as fusions with ID4 SEQ ID NO: 131, ID3 SEQ ID NO: 132,
ID2
SEQ ID NO: 133 and ID1 SEQ ID NO: 134, respectively. ID4 is fused to the N-
terminus
of Lambda stf at the GAGENS SEQ ID NO: 194 insertion site (FIG. 9A-B). The
final
architecture for expression of the tetrameric is depicted in FIG. 9B, although
such an
expression scheme may be modified, since the plasmid will be large (>16 kb)
and the
length of the transcribed mRNA is >9kb.
EXAMPLE 3
[219] T4-like phages are a very diverse family of bacteriophages that share a
common
long tail fiber architecture: a proximal tail fiber that attaches to the phage
particle and a
distal tail fiber (DTF) that encodes host specificity linked by proteins
acting as "hinge
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
82
connectors" (Desplats and Krisch, 2003, Res. Microbiol. 154:259-267; Bartual
et al.
2010, Proc. Natl. Acad. Sci. 107: 20287-20292). It is thought that the main
host range
determinants of the tail fiber reside in the distal part. Hence, it is very
important to
understand if it is possible to translate the host range of a given T4-like
phage, which are
known to be very broad, to any other phage or packaged phagemid of interest.
The distal
tail fiber (C-terminal domain of the T4-like long tail fiber) of several T4-
like phages
were screened for possible functional insertion sites, several fusions with
the Lambda stf
gene were generated and their host range screened.
[220] Possible insertion sites in the DTF that, when fused to a heterologous
tail fiber
(the lambda phage stf), will give a functional chimera were searched. The DTF
of the
phage (WW13) was used as a testbed. This phage possesses a classical T4-like
architecture, with a proximal and a distal tail fiber separated by hinge
connectors, a gp38
chaperone/adhesin (to assist folding of the tail fiber and recognition of the
host (Trojet et
al., 2011, Genome Biol. Evol. 3:674-686) and a gp57A chaperone known to be
needed
for proper folding of the tail fiber (Matsui et al., 1997, J. Bacteriol.
179:1846-1851).
Since the endogenous genomic regulation of T4-like phages is complex and may
include
unknown layers of regulation (Miller et al., 2003, Microbio. Mol. Biol. Rev.
67:86-156),
a synthetic linker encoding a RBS was designed to replace the natural DNA
linker
between the DTF gene and the adhesin; immediately downstream, another
synthetic
RBS preceding the chaperone gp57A was added, hence creating a polycistronic
mRNA
encoding for all the functions needed for the proper folding of the DTF (FIG.
10). This
construct was built in a plasmid under the control of an inducible promoter
and
complemented in trans in a strain producing lambda-based phagemids.
[221] FIG 10. depicts the architecture of an engineered lambda stf-T4-like DTF
chimera. The semicircles denote RBS sites; the T sign, a transcriptional
terminator; the
arrow, a promoter. Several parts of the C-terminus of the DTF were screened
and fused
to the lambda stf gene at the GAGENS (SEQ ID NO: 194) insertion site. Several
variants of the chimera lambda stf-WW13 were functional, as assessed by
production of
phagemid particles and transduction of a chloramphenicol marker in a
collection of E.
co/i strains. The functional chimeras shown in FIG. 11 were obtained with
fusion at the
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
83
IIQLED (SEQ ID NO:196) insertion site in WW13. Additional functional chimeras
were
obtained by fusion at the lambda stf MDETNR (SEQ ID NO:192) insertion site and
at
the WW13 DTF GNIIDL (SEQ ID NO:197), VDRAV (SEQ ID NO:203) and IIQLED
(SEQ ID NO:196) insertion sites (FIG. 13).
[222] Other T4-like phages, like PP-1, sharing sequence homology with WW13
were
also tested and verified to produce functional chimeras (FIG. 11). These
functional
chimeras show a IATRV (SEQ ID NO:198) insertion site at the beginning of PP-1
DTF
part.
[223] FIG. 11 depicts screening of phagemid particles with chimeric lambda stf-
T4-
like DTFs and in particular chimeric lambda stf-WW13 and chimeric lambda stf-
PP1 of
aminoacids sequences SEQ ID NO: 142 to 149 and nucleotide sequences of SEQ ID
NO: 166 to 173 including their respective chaperones proteins. A collection of
96
different wild type E. coli strains, encompassing different serotypes, was
transduced
with lambda-based packaged phagemids and plated on Cm LB agar. Left panel
represents wild-type lambda stf; the middle panel represents chimeric lambda-
stf-
WW13; and the right panel, represents chimeric lambda-stf-PP-1.
[224] The insertion sites found for WW13 do not always exist in a given T4-
like DTF,
thereby complicating the analysis. Another functional insertion site without
homology to
WW13 was discovered for a second phage (WW55, FIG. 12). The same TPGEL (SEQ
ID NO:199) insertion site could be found in a subset of T4-like phages and
proven to
yield functional chimeras with at least one of them, WW34 (FIG. 12), and at
MDETNR
(SEQ ID NO:192) insertion site in lambda stf.
[225] FIG. 12. shows screening of phagemid particles with chimeric lambda stf-
T4-like
DTFs and in particular chimeric lambda stf-WW55 and chimeric lambda stf-WW34
of
aminoacids sequences SEQ ID NO: 150 to 156 and nucleotide sequences of SEQ ID
NO: 174 to 180 including their respective chaperones proteins. A collection of
96
different wild type E. coli strains, encompassing different serotypes, was
transduced
with lambda-based phagemids and plated on Cm LB agar. The left panel
represents
wild-type lambda stf; the middle panel represents chimeric lambda-stf-WW55;
and the
right panel represents chimeric lambda-stf-WW34.
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
84
[226] Since T4-like DTF proteins may or may not share common sites for
insertion,
attempts were made to identify a universal insertion site that exists in all
T4-like DTFs.
When several T4-like DTFs are aligned, no homology along the whole DTF gene
present in all the sequences exists, except for the N-terminus which is well
conserved.
The N-terminus of the DTF is thought to interact with the hinge connectors for
attachment to the main phage particle.
[227] Although the classic view is that the host range determinants reside in
the C-
terminal part of the DTF, recent studies have proven that the N-terminus may
also be
involved in this process (Chen et al., 2017, Appl. Environ. Microbiol. Vl. 83
No. 23).
The N-terminus of the DTF was then scanned to look for an insertion site that
exists in
all T4-like phages and that is able to yield functional chimeras. Phage WW13
DTF and
insertion site MDETNR (SEQ ID NO:192) in lambda stf were used. While the
direct
fusion of the complete DTF gene (starting at amino acid 2) gives particles
with some
activity, a region from amino acid 1 to 90, with a preferred region from amino
acid 40 to
50 of the DTF, that recapitulates the behavior of the DTF fusion was
identified and is
shown in FIG. 13. Importantly, this region exists in all T4-like phages
screened and
could be very rapidly used to generate chimeras with a diverse set of DTFs,
including
WV/55 (FIG. 13), and in particular chimeric lambda stf-WW14, chimeric lambda
stf-
WW170 and chimeric lambda stf-202 of aminoacids sequences SEQ ID NO: 157 to
165
and nucleotide sequences of SEQ ID NO: 181 to 189 including their respective
chaperones proteins.
[228] Accordingly, the present disclosure is useful for the generation of
phage and
phagemid particles with altered host ranges, since it provides a practical
framework for
the construction of chimeras using the DTFs from any T4-like phage,
highlighting its
modularity and translatability.
EXAMPLE 4
[229] The human microbiome comprises different zones of the body, including
gut,
skin, vagina and mouth [29]. The microbiota in these areas is composed of
different
3Ctommunities of microorganisms, such as bacteria, archaea and fungi
[29]¨[31]. While
CA 03120160 2021-05-17
WO 2020/128108
PCT/EP2019/086990
numerous studies have been made that try to elucidate the specific composition
of these
communities, it is becoming clear that while there may exist a "core
microbiome", there
are many variations in the relative content of each microorganism depending on
several
factors, such as geographical location, diet or age [32]¨[35].
5 [230] Specifically, in the case of the human gut microbiota, it is not
possible to know a
priori what are the bacterial species that a given person possesses without
running a
diagnostic method. In the case of Escherichia coli, some studies have been
made that
point out to the prevalence of some serotypes and phylogenetic groups in the
majority of
humans; however, there are significant changes in the composition of the
samples
10 depending on the geographic distribution as well as the time of sampling:
for example,
samples isolated from Europe, Africa, Asia and South America in the 1980s show
a
prevalence for phylogroups A and B1 (55% and 21%, respectively); but samples
obtained in the 2000s in Europe, North America, Asia and Australia belong
mainly to
the B2 group (43%), followed by the A (24%), D (21%), and B1 (12%) [36]. It is
also
15 thought that phylogenetic groups B2 and D are usually more commonly
associated with
pathogenic strains than with commensal strains [37], but there are studies
showing a
number of human- and non-human-specific strains belonging to phylogenetic
group B2
that are commensals and belong to different serotypes [38].
[231] The intrinsic variability of the human microbiome, and specifically that
of
20 Escherichia coli subtypes, makes it difficult to design targeted
therapeutic approaches.
In the case of phage therapy aimed at killing a target bacterial population,
for instance,
two possible approaches are possible: first, the use of narrow host range
particles that are
able to recognize and target a specific E. coli serotype or second the use of
broad host
range phages that are able to recognize many different strains, sometimes even
from
25 different genera [39]. This difficulty is exacerbated if one takes into
account strategies
that do not aim to kill the target bacterial population, but that seek to add
a function to
them (i.e. delivery of a factor that will have an effect in the host and that
will be
expressed by the targeted microbiota). In this specific case, the use of
packaged
phagemids is of great interest, since they do not kill the host (unless their
payload carries
30 genes aimed at killing the host), payload does not replicate and expand and
does not
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
86
contain any endogenous phage genes. However, as in the case of phages, a
diagnostic
study would be needed to identify the specific serotypes/variants of bacteria
that exist in
the patient before the treatment in order to find or design a packaged
phagemid that
allows for delivery of a payload adding a function to the target bacteria
without killing
them.
[232] By combining these two approaches, it was proposed to use engineered
delivery
vehicles that are able to recognize a large number of strains belonging to
different
serotypes and phylogenetic groups (i.e., engineered particles having a "broad
host
range"), with a focus on Escherichia coli. As opposed to a killing-oriented
approach,
where the targeted bacterial population needs to be as close as possible to
100% to
reduce their numbers, a therapeutic delivery approach does not need a priori
to reach a
large percentage of bacteria; the delivery needs to be high enough for the
therapeutic
payload to be expressed at the correct levels, which may be highly variable
depending
on the application. Additionally, the payload can be expressed by different
serotypes or
phylogenetic groups. This approach increases the chance that the particle will
deliver a
payload expressed in vivo in the majority of patients.
[233] To achieve the delivery in bacterial communities composed of unknown
serotypes/variants of target strains, delivery vehicles were engineered to
contain
chimeric side tail fibers (stf) that have been selected due to their ability
to recognize a
large variety of target strains. There are many phages that have been
described as having
a broad host range in E. coli and many of these belong to the T4 family,
although in
general, phages against E. coli and related bacteria have a restricted host
range.
[234] However, according to [41], there is no consensus as to how many strains
need to
be targeted by a phage to be considered as a "broad host range".
[235] In the case of Escherichia coli, the ECOR collection is a set of strains
isolated
from different sources that is thought to represent the variability of this
bacterium in
Nature [42]. Some phage have been shown to have a broad host range against
this
collection (for instance, about 53% of the ECOR strains can be lysed with
phage AR1
[43] and about 60% with phage 5U16 [44]). As opposed to this, a single phage
is able to
infect 95% of Staphylococcus aureus strains [40].
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
87
[236] It was decided to use human strains of this collection to test
engineered delivery
vehicles with chimeric stf and assess their host range in an attempt to
identify variants
that are able to recognize as many hosts as possible, as has been described in
the
literature [45]. The difference is that the present assays measure delivery
efficiency as
opposed to lysis.
[237] Strains from an overnight culture were diluted 1:100 in 600 uL of LB
supplemented with 5mM CaCl2 in deep 96 well plates and grown for 2 hours at 37
C at
900 rpm. 10 lit of packaged phagemids, containing a DNA payload p7.3 of SEQ ID
NO: 277, produced at an average of 1064iL were then added to 90 uL of the
bacterial
cultures, incubated 30 minutes at 37 C and 10 lit of the mixtures plated on LB
agar
supplemented with 24 lig/mL chloramphenicol and incubated overnight at 37 C.
The
next day, the density of the dots was scored from 0 to 5, with 0 being no
transductants
and 5 being a spot with very high density [FIG. 14]. The density of the spots
is directly
related to the delivery efficiency of the packaged phagemids, since it
corresponds to the
number of bacteria that have received a payload containing a chloramphenicol
acetyltransferase gene.
[238] Several stf chimeras were tested and screened in 40 human strains of the
ECOR
collection. As a control, the delivery efficiency of the wild-type lambda stf
of SEQ ID
NO: 1 was tested. The packaged phagemid variant used for the delivery
experiments was
modified so that its tail tip gpJ now recognizes a receptor other than LamB
(gpJ 1A2
variant of aminoacid sequence SEQ ID NO: 278 and nucleotide sequence SEQ ID
NO:
279). In FIG. 15, the raw dot titrations for 18 chimeric stf of aminoacid
sequence of SEQ
ID NO: 215 to 242 and nucleotide sequences of SEQ ID NO: 243 to 270 including
their
respective chaperones proteins are shown and in FIG. 16 a bar-formatted table
is shown
with the delivery efficiencies scored by dot density as well as the delivery
statistics.
[239] Taking only into account dots with density scores of 3 and higher
(considered as
medium to high delivery efficiency), some stf s can be considered as broad
host range
because the delivery efficiency in the selected ECOR strains is significantly
higher than
when using the wild type stf. For example, for stf EB6 or stf 68B, about 50%
of the
strains show medium to high delivery efficiencies, as compared to 17.5% of the
strains
CA 03120160 2021-05-17
WO
2020/128108 PCT/EP2019/086990
88
with the wild type stf. These stf are good candidates for in vivo delivery,
since they are
able to deliver in different phylogenetic groups as well as serotypes. At the
bottom of the
Table in FIG. 16, a bar-formatted representation for density scores higher
than 3 is
shown, where the threshold for a broad host range stf is set at an increase of
at least 2X
compared to the basal line of the wild type stf; this is, stf that are able to
deliver with
scores of 3 and higher in at least 35% of the strains. Other stf also show an
increased
delivery as compared to the wild type stf, so a less stringent threshold was
set for stf able
to deliver with scores 3 or higher with at least a 50% increase compared to
the number
of strains delivered with the wild-type stf (this is, delivery with scores of
3 and higher in
at least 26.25% of the strains). As a comparison, data for stf K1 and stf 66D
is shown:
these stf seem to be delivering efficiently in a small number of strains (for
instance,
strains B and AB for stf Kl; and strains E and AF for stf 66D), which means
that they
probably have a narrow host range; this is to be expected, since in the case
of the K1 stf
the cognate receptor is the K1 capsule [46]. Additionally, data are shown for
a chimera
with a stf originating in a T4-like phage; as the literature suggests, this
chimera shows a
broad host range although it does not seem to be the best candidate.
[240] Taken together, these results suggest that the stf of a delivery vehicle
can be
engineered to recognize a wide number of target E. coli strains, hence
rendering it
"broad host range". This type of particles can be very useful to deliver
payloads adding a
function to the target bacteria without having to engineer a specific variant
that
recognizes a given bacterial strain.
List of References Cited
Each of the reference cited within the specification and those listed below
are hereby
incorporated by reference in their entirety.
[1] G. P. C. Salmond and P. C. Fineran, "A century of the phage: past,
present and
future," Nat. Rev. Microbiol., vol. 13, no. 12, pp. 777-786, Dec. 2015.
[2] P. Hyman and S. T. Abedon, "Bacteriophage host range and bacterial
resistance," Adv. Appl. Microbiol., vol. 70, pp. 217-248, 2010.
[3] S. Chatterjee and E. Rothenberg, "Interaction of Bacteriophage 2\.,
with Its E. coli
Receptor, LamB," Viruses, vol. 4, no. 11, pp. 3162-3178, Nov. 2012.
[4] Nobrega et al, Nat Rev, 2018 "Targeting mechanisms of tailed
bacteriophages"
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
89
[5] A. Flayhan, F. Wien, M. Paternostre, P. Boulanger, and C. Breyton, "New
insights into pb5, the receptor binding protein of bacteriophage T5, and its
interaction
with its Escherichia coli receptor FhuA," Biochimie, vol. 94, no. 9, pp. 1982-
1989, Sep.
2012.
[5] M. G. Rossmann, V. V. Mesyanzhinov, F. Arisaka, and P. G. Leiman, "The
bacteriophage T4 DNA injection machine," Curr. Opin. Struct. Biol., vol. 14,
no. 2, pp.
171-180, Apr. 2004.
[6] Y. Zivanovic et al., "Insights into Bacteriophage T5 Structure from
Analysis of
Its Morphogenesis Genes and Protein Components," J. Virol., vol. 88, no. 2,
pp. 1162-
1174, Jan. 2014.
[7] R. W. Hendrix and R. L. Duda, "Bacteriophage lambda PaPa: not the
mother of
all lambda phages," Science, vol. 258, no. 5085, pp. 1145-1148, Nov. 1992.
[8] M. A. Speed, T. Morshead, D. I. Wang, and J. King, "Conformation of P22
tailspike folding and aggregation intermediates probed by monoclonal
antibodies,"
Protein Sci. Publ. Protein Soc., vol. 6, no. 1, pp. 99-108, Jan. 1997.
[9] S. J. Labrie, J. E. Samson, and S. Moineau, "Bacteriophage resistance
mechanisms," Nat. Rev. Microbiol., vol. 8, no. 5, pp. 317-327, Mar. 2010.
[10] C. Whitfield, "Biosynthesis and assembly of capsular polysaccharides in
Escherichia coli," Annu. Rev. Biochem., vol. 75, pp. 39-68,2006.
[11] J. R. Meyer, D. T. Dobias, J. S. Weitz, J. E. Barrick, R. T. Quick,
and R. E.
Lenski, "Repeatability and contingency in the evolution of a key innovation in
phage
lambda," Science, vol. 335, no. 6067, pp. 428-432, Jan. 2012.
[12] D. S. Gupta et al., "Coliphage K5, specific for E. coli exhibiting the
capsular K5
antigen," FEMS Microbiol. Lett., vol. 14, no. 1, pp. 75-78, May 1982.
[13] R. J. Gross, T. Cheasty, and B. Rowe, "Isolation of bacteriophages
specific for
the K1 polysaccharide antigen of Escherichia coli," J. Clin. Microbiol., vol.
6, no. 6, pp.
548-550, Dec. 1977.
[14] D. Schwarzer et al., "A Multivalent Adsorption Apparatus Explains the
Broad
Host Range of Phage phi92: a Comprehensive Genomic and Structural Analysis,"
J.
Virol., vol. 86, no. 19, pp. 10384-10398, Oct. 2012.
[15] F. 'Mart, F. Repoila, C. Monod, and H. M. Krisch, "Bacteriophage T4 host
range
is expanded by duplications of a small domain of the tail fiber adhesin," J.
Mol. Biol.,
vol. 258, no. 5, pp. 726-731, May 1996.
[16] E. HaggArd-Ljungquist, C. Halling, and R. Calendar, "DNA sequences of the
tail
fiber genes of bacteriophage P2: evidence for horizontal transfer of tail
fiber genes
among unrelated bacteriophages.," J. Bacteriol., vol. 174, no. 5, pp. 1462-
1477, Mar.
1992.
[17] L.-T. Wu, S.-Y. Chang, M.-R. Yen, T.-C. Yang, and Y.-H. Tseng,
"Characterization of Extended-Host-Range Pseudo-T-Even Bacteriophage Kpp95
Isolated on Klebsiella pneumoniae," Appl. Environ. Microbiol., vol. 73, no. 8,
pp. 2532-
2540, Apr. 2007.
[18] D. Montag, H. Schwarz, and U. Henning, "A component of the side tail
fiber of
Escherichia coli bacteriophage lambda can functionally replace the receptor-
recognizing
part of a long tail fiber protein of the unrelated bacteriophage T4," J.
Bacteriol., vol.
171, no. 8, pp. 4378-4384, Aug. 1989.
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
[19] E. R. Vimr, R. D. McCoy, H. F. Vollger, N. C. Wilkison, and F. A. Troy,
"Use
of prokaryotic-derived probes to identify poly(sialic acid) in neonatal
neuronal
membranes," Proc. Natl. Acad. Sci., vol. 81, no. 7, pp. 1971-1975, Apr. 1984.
[20] K. Stummeyer, A. Dickmanns, M. Miihlenhoff, R. Gerardy-Schahn, and R.
5 Ficner, "Crystal structure of the polysialic acid-degrading endosialidase of
bacteriophage K1F," Nat. Struct. Mol. Biol., vol. 12, no. 1, pp. 90-96, Jan.
2005.
[21] D. Scholl, S. Adhya, and C. Merril, "Escherichia coli K1 's Capsule Is a
Barrier
to Bacteriophage T7," Appl. Environ. Microbiol., vol. 71, no. 8, pp. 4872-
4874, Aug.
2005.
10 [22] Y. Jiang, B. Chen, C. Duan, B. Sun, J. Yang, and S. Yang, "Multigene
Editing in
the Escherichia coli Genome via the CRISPR-Cas9 System," Appl. Environ.
Microbiol.,
vol. 81, no. 7, pp. 2506-2514, Apr. 2015.
[23] J. E. Cronan, "Improved Plasmid-Based System for Fully Regulated Off-To-
On
Gene Expression in Escherichia coli: Application to Production of Toxic
Proteins,"
15 Plasmid, vol. 69, no. 1, pp. 81-89, Jan. 2013.
[24] J. E. Thompson et al., "The K5 Lyase KflA Combines a Viral Tail Spike
Structure with a Bacterial Polysaccharide Lyase Mechanism," J. Biol. Chem.,
vol. 285,
no. 31, pp. 23963-23969, Jul. 2010.
[25] S. C. Potter, A. Luciani, S. R. Eddy, Y. Park, R. Lopez, and R. D. Finn,
20 "HMMER web server: 2018 update," Nucleic Acids Res., vol. 46, no. Wl, pp.
W200-
W204, Jul. 2018.
[26] E. I. Marusich, L. P. Kurochkina, and V. V. Mesyanzhinov, "Chaperones in
bacteriophage T4 assembly," Biochem. Biokhimiia, vol. 63, no. 4, pp. 399-406,
Apr.
1998.
25 [27] J. Xu, R. W. Hendrix, and R. L. Duda, "Chaperone-protein interactions
that
mediate assembly of the bacteriophage lambda tail to the correct length," J.
Mol. Biol.,
vol. 426, no. 5, pp. 1004-1018, Mar. 2014.
[28] D. Schwarzer et al., "Proteolytic Release of the Intramolecular Chaperone
Domain Confers Processivity to Endosialidase F," J. Biol. Chem., vol. 284, no.
14, pp.
30 9465-9474, Apr. 2009.
[29] D. Schwarzer et al., "A Multivalent Adsorption Apparatus Explains the
Broad Host
Range of Phage phi92: a Comprehensive Genomic and Structural Analysis," J.
Virol.,
vol. 86, no. 19, pp. 10384-10398, Oct. 2012.
[30] "Characterization of a VI-like phage specific to Escherichia coli
0157:H7. -
35 PubMed NCBI." [Online].
Available:
https ://www.ncbi.nlm.nih.gov/pubmed/21899740. [Accessed: 10-D ec-2018] .
[31] C. Chen, P. Bales, J. Greenfield, R. D. Heselpoth, D. C. Nelson, and 0.
Herzberg, "Crystal structure of 0RF210 from E. coli 0157:H1 phage CBA120
(TSP1), a
putative tailspike protein," PloS One, vol. 9, no. 3, p. e93156,2014.
40 [32] A. K. Golomidova et al., "Branched Lateral Tail Fiber Organization in
T5-Like
Bacteriophages DT57C and DT571/2 is Revealed by Genetic and Functional
Analysis,"
Viruses, vol. 8, no. 1, Jan. 2016.
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
91
SEQUENCES
,
SEQ ID Name Protein (PRT) Origin Insertion site
NO or DNA
i
1
+lambda stf PRT lambda bacteriophage
-i
2 STF-25 PRT t- Artificial sequence
ADAKKS
______________________________________ i
3 STF25-AP1 ' P= RT Artificial sequence
4 STF-27 PRT Artificial sequence
ADAKKS
t
STF27-AP1 PRT -1- Artificial sequence
______________________________________ i
6 STF27-AP2 PRT Artificial sequence
______________________________________ i
7 STF-28 PRT Artificial sequence
ADAKKS
8 +STF28-AP1 PRT +Artificial sequence
______________________________________ i
9 STF-15 ' PRT Artificial sequence
SASAAA
_____ i _____________________________ 1
STF15-AP1 PRT Artificial sequence
, t
11 STF15-AP2 PRT +Artificial sequence
12 STF-16 ' PRT Artificial sequence
SASAAA
13 STF16-AP1 PRT Artificial sequence
_____ i _____________________________ 1
14 STF16-AP2 PRT Artificial sequence
STF-17 ' PRT Artificial sequence SASAAA
16 STF17-AP1 PRT Artificial sequence
17 + STF 4-13 PRT Artificial sequence
SASAAA
_____ i ______________ , ___________ 1
18 STF13-AP1 PRT Artificial sequence
4 - _
19 STF13-AP2 PRT +Artificial sequence
STF-12 ' PRT Artificial sequence SASAAA
21 STF12-AP1 PRT Artificial sequence
22 STF12-AP2 PRT Artificial sequence
23 STF-63 PRT Artificial sequence
SASAAA
24 STF-62 ' PRT Artificial sequence
SASAAA
STF-71 ' P= RT Artificial sequence SASAAA
_____ i _____________________________ 1
26 STF71-AP1 PRT Artificial sequence
27 STF-20 PRT Artificial sequence
MDETNR
_____ i _____________________________ 1
28 STF20-AP1 ' PRT Artificial sequence
_____ i _____________________________ 1
29 STF-23 PRT Artificial sequence
MDETNR
4 -I
STF23-AP1 PRT +Artificial sequence
31 STF-24 PRT Artificial sequence
MDETNR
32 4 STF24-AP1 PRT t Artificial sequence
_____ 1 _____________________________ 1
33 0111-2.0 ' P= RT Artificial sequence
MDETNR
4- 4- - _
34 0111 2.0-API PRT Artificial sequence
, , .
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
92
[35 ' STF-74 ' PRT ' Artificial sequence i
MDETNR
I 36 STF74-AP1 PRT Artificial sequence
I 37 t
STF-86 PRT I-Artificial sequence
MDETNR
______ 1 _____________ , ___________ 1
I 38 STF86-AP1 PRT Artificial sequence
t
I 39 STF-84 PRT +Artificial sequence
MDETNR
______ 1 ____________________________ i
I 40 STF84-AP1 PRT Artificial sequence
+
41 STF-93 PRT +Artificial sequence
MDETNR
______ i ____________________________ 1
42 STF-95 PRT Artificial sequence
MDETNR 1
______ i ____________________________ 1
43 STF95-AP1 PRT Artificial sequence
______ 1 ____________________________ i
[ 44 STF-132 PRT Artificial sequence
MDETNR
______ i ____________________________ 1
I 45 STF132-AP1 ' P= RT Artificial sequence
__ i ! 46 KlF PRT __ iArtificial
sequence GAGENS
j 47 K5 PRT Artificial sequence
GAGENS
I 48 STF-37 ' PRT Artificial sequence
GAGENS
______ t ____________________________ i
1 49 1JL PRT Artificial sequence
GAGENS
t
I 50 STF-48 PRT 1- Artificial sequence
GAGENS
I 51 STF-49 PRT Artificial sequence
GAGENS
i 52 t
STF-52 PRT +Artificial sequence
GAGENS
______ i ____________________________ I
I 53 1 AR ' P= RT Artificial sequence
GAGENS
-4- 4- -
1 54 lAR-API PRT Artificial sequence
______ i ____________________________ }
i 55 1AR-AP2 ' P= RT Artificial sequence
' 56 13-13.0 PRT Artificial sequence
GAGENS
-I
I 57 -I- 13-13.0-AP1 PRT +Artificial sequence
______ i ____________________________ I
I 58 13-13.0-AP2 ' P= RT Artificial sequence
.1-
I 59 13-14.3 PRT +Artificial sequence
SAGDAS
______ i ____________________________ t
' 60 13-14.3-AP1 ' PRT Artificial sequence
______ 1 ____________________________ 1
61 13-14.3-AP2 PRT Artificial sequence
I ____________
______ 1 ____________________________ 1
62 STF-25 ' DNA Artificial sequence
______ 1 ____________________________ 1
63 STF25-AP1 DNA Artificial sequence
______ i ____________________________ I
64 STF-27 ' DNA Artificial sequence
______ 1 _____________ , ___________ 1
65 STF27-AP1 DNA Artificial sequence
66 STF27-AP2 ' DNA Artificial sequence
______ 1 _____________ , ___________ 1
67 STF-28 DNA Artificial sequence
t 1-
68 STF28-AP1 DNA Artificial sequence
______ i _____________ , ___________ 1
69 STF-15 DNA Artificial sequence
_ -
70 STF15-AP1 DNA +Artificial sequence
- ____________
71 STF15-AP2 DNA Artificial sequence
4-
+ _ -
72 STF-16 DNA Artificial sequence
' ______________________
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
93
[73 ' STF16-AP1 "-T DNA ' Artificial sequence
______________________________________ I
1 74 STF16-AP2 ___ ,
DNA Artificial sequence
175 t STF-17 DNA 1- Artificial sequence
______ 1 ____________ , ____________ 1
76 STF17-AP1 DNA Artificial sequence
t
77 STF-13 DNA +Artificial sequence
______ 1 ____________________________ i
78 STF13-AP1 DNA Artificial sequence
4-
79 STF13-AP2 DNA +Artificial sequence
______ i ____________________________ 1
80 STF-12 ' D= NA Artificial sequence
1
81 STF12-AP1 DNA Artificial sequence
______ 1 ____________________________ i
82 STF12-AP2 ' DNA Artificial sequence
______ i ____________________________ 1
83 STF-63 ' D= NA Artificial sequence
84 STF-62 DNA i ____________ Artificial sequence
I
85 STF-71 DNA Artificial sequence
86 STF71-AP1 ' DNA Artificial sequence
87 STF-20 DNA Artificial sequence
t ,
88 STF20-AP1 DNA 1- Artificial sequence
______ i ____________ 1 ____________ i
89 STF-23 DNA Artificial sequence
t
90 STF23-AP1 DNA +Artificial sequence
91 f 1 STF-24 ' D= NA Artificial sequence
92 STF24-API DNA Artificial sequence
______ i ____________________________ }
93 0111-2.0 ' D= NA Artificial sequence
94 +0111 2.0-AP1 DNA +Artificial sequence
95 STF-74 ' DNA Artificial sequence
______ f ____________________________ i
96 STF74-AP1 DNA Artificial sequence
97 STF-86 DNA Artificial sequence
98 STF86-AP1 DNA Artificial sequence
______ I ____________________________ 1
99 STF-84 ' DNA Artificial sequence
!
100 STF84-AP1 DNA Artificial sequence
--1- _________________________________ 1
101 STF-93 ' DNA Artificial sequence
102 STF-95 DNA Artificial sequence
103 4 STF95-AP1 DNA t- Artificial sequence
______________________ ,
104 STF-132 DNA Artificial sequence
105 STF132-API DNA +Artificial sequence
106 - K 1F ' D= NA Artificial sequence
+K5
107 DNA +Artificial sequence
108 STF-37 DNA Artificial sequence
--4 __________________________________ 1
109 1JL DNA Artificial sequence
¨t 1
[ 110 STF-48 : DNA Artificial sequence
i ,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
94
' 111 ' STF-49 "-T DNA ' Artificial sequence
112 STF-52 DNA Artificial sequence
i-
113 1AR DNA I- Artificial sequence
______ 1 ____________ , ____________ 1
114 1AR-AP1 DNA Artificial sequence
115 -i-1AR-AP2 DNA +Artificial sequence
....._+... ___________________________ i
116 13-13.0 DNA Artificial sequence
i-
117 13-13.0-AP1 DNA +Artificial sequence
______ i ____________________________ 1
118 13-13.0-AP2 ' D= NA Artificial sequence 1
119 13-14.3 DNA Artificial sequence
______ 1 ____________________________ i
120 13-14.3-AP1 ' DNA Artificial sequence
______ i ____________________________ 1
121 KlF ' P= RT KlF phage
122 13-14.3-AP2 DNA Artificial sequence
-I
123 +1 amb da stf PRT t" lambda phage
124 WWII stfl ' PRT WWII phage
i 125 WWII 5tf2 ' PRT i WWII phage
126 KlF ' PRT KlF phage
,..._1.
127 TSP4 Branched PRT CBA120 phage
______ i
128 TSP3 Branched PRT CBA120 phage
--t
129 TSP2 Branched PRT CBA120 phage
130 TSP1 Branched PRT CBA120 phage ,
131 ---1 ID4 Branched ' PRT CBA120 phage
132 ID3 Branched PRT CBA120 phage
,
133 ID2 Branched ' PRT CBA120 phage
134 ID1 branched PRT CBA120 phage
135 WW13 PRT -t-
Artificial sequence GNIIDL
_..i. ________________ ,
136 PP-1 PRT Artificial sequence I ATRV
. -I- -i- -
137 W W55 PRT Artificial sequence TPGEL
..._
138 WW34 ' P= RT Artificial sequence ! TPGEL
-I-W W14 139 PRT +Artificial sequence NQIID
-I-
140 WW170 PRT Artificial sequence GAIIN
--i-
141 W W202 PRT Artificial sequence GQIVN
142 ¨ WW13 13.0 ' PRT Artificial
sequence IIQLED
_____i_
143 WW13 10.0 PRT Artificial sequence VDRAV
___!....
144 WW13-G8 ' PRT Artificial sequence GNIIDL
..._ .
145 WW13 gp38 ' PRT ww13 phage
______ i
146 WW13 gp57A PRT ww13 phage
i _________________________________________________________
147 PP-1 ' P= RT Artificial sequence IATRV
, ____________________________________
148 PP-1 gp38 PRT pp-1 phage
149 PP-1 gp57A , PRT i pp-1 phage
-1-
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
150 ' WW55 3.0 ' PRT ' Artificial sequence TPGEL
¨t- _________________________ i
151 WW55-G8 ' PRT Artificial sequence GAIIN
152 WW55 gp38 ' PRT ww55 phage
i _________________________________________________________________________
153 WW55 gp57A ' P= RT ww55 phage
154 WW34 3.0 PRT Artificial sequence TPGEL
----i- i _________________________________
155 WW34 gp38 PRT ww34 phage
_____ 1 _____________ , i _________________________________
156 WW34 gp57A PRT ww34 phage
157 - WW14-G8 PRT Artificial sequence NQIID
1 I _________________________________
158 --I-WW14 gp38 PRT ww14 phage
______________________________________ i
159 WW 14 gp57A ' P= RT w\,14 pliagc
- _
160 +N\ NN, 170-G8 PRT +Artificial sequence GAI1N
4-
161 +1A, WI70 gp38 PRT ww170 phage
-I-- -t-
162 W W170 gp57A PRT ww170 phage
163 +WW202-G8 PRT +Artificial sequence GQIVN
1 _________________________________________________________________________
164 'WW202 gp38 ' PRT ww202 phage
1 _________________________________________________________________________
165 WW202 gp57A PRT ww202 phage
13.0
----1- i _________________________________
166 WW13 ' DNA Artificial sequence
167 WW13 10.0 ' D= NA Artificial sequence
_____ i _____________________________ 1 , ___________
168 WW13-G8 DNA Artificial sequence
169 WW13 GP38 DNA ww13 phage
¨t- _______________________________________________________________________ t
170 WW13 GP57A ' DNA ww13 phage
171 PP-1 DNA Artificial sequence
172 PP-1 GP38 DNA pp-1 phage
173 ' PP-1 GP57A DNA pp-1 phage
t
174 t WW55 3.0 DNA Artificial sequence
______________________________________ i
175 WW55-G8 ' D= NA Artificial sequence
-
176 +Vr W55 GP38 DNA + ww55 phage
177 - Vr W55 GP57A DNA i ww55 phage
.4- - _
178 +IN W34 3.0 DNA Artificial sequence
--f- ________________________ 1
179 WW34 GP38 DNA ww34 phage
¨i- _________________________ i
180 W W34 GP57A DNA ww34 phage
181 WW14-G8 ' DNA Artificial sequence
182 WW14 GP38 DNA ww14 phage
183 WW14 GP57A ' DNA ww14 phage
184 WW170-G8 DNA Artificial sequence
185 ThWW170 GP38 DNA +ww170 phage
186 - WW170 GP57A DNA ww170 phage
187 f-WW202-G8 DNA -i-
Artificial sequence
188 WW202 GP38 DNA ww202 phage
i
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
96
189 WW202 GP57A DNA ww202 phage
190 insertion sequence PRT Artificial
sequence
t-.
191 insertion sequence PRT t-' Artificial
sequence
_____ 1 ____________________________ 1
192 insertion sequence PRT Artificial
sequence
193 t insertion sequence PRT +Artificial
sequence
----f. ______________________ 1
194 insertion sequence PRT Artificial
sequence
t
195 insertion sequence PRT +Artificial
sequence
_____________________________________ I
196 insertion sequence PRT
Artificial sequence 1
197 insertion sequence PRT I Artificial
sequence
198 insertion sequence PRT Artificial
sequence
199 insertion sequence PRT Artificial
sequence
i
200 nsertion sequence PRT __ Artificial
sequence I
_____________________________________ i
201 insertion sequence PRT Artificial
sequence
202 insertion sequence PRT Artificial
sequence
_____ f ____________________________ i
203 insertion sequence PRT Artificial
sequence
,
204 f- AG22 PRT /- AG22 phage
205 SIEAll PRT SIEAll phage
206 t WW13 DNA +Artificial sequence
207 PP-1 ' D= NA Artificial sequence
4 -I- _
208 IA W DNA Artificial sequence
209 IN W34 ' D= NA Artificial sequence
_
+-W W14 _
210 DNA +Artificial sequence
_____ 1 ____________________________ I
211 WW170 ' DNA Artificial sequence
212 W W202 DNA Artificial sequence
_____________________________________ i
213 ¨I¨AG22 DNA AG22 phage
214 SIEAll DNA SIEAll phage
215 0111 ' PRT Artificial sequence
216 SIED6 PRT Artificial sequence
,
217 SIED6 AP1 ' PRT Artificial sequence
218 SIED6 AP2 PRT Artificial sequence
-1
219 SIEAll PRT t Artificial sequence
220 STEAll AP1 PRT Artificial sequence
_
221 DC1 PRT +Artificial sequence
_____________________________________ i _________________________________ 1
222 ' DC1 AP1 ' P= RT Artificial sequence
223 EB6 PRT +Artificial sequence
_____________________________________ i
224 EB6 AP1 ' P= RT Artificial sequence
225 AH11L PRT Artificial sequence
¨i- _________________________ 1
226 -AH11L AP1 : PRT Artificial sequence
I.- i
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
97
227 ' STF-94A ' PRT ' Artificial sequence
228 STF-94A AP1 PRT Artificial sequence
t
229 STF-69A PRT -1-- Artificial sequence
_______ 1 ____________ , ___________ 1
230 STF-69A AP1 PRT Artificial sequence
t
231 STF-69A AP2 PRT +Artificial sequence
----f. ______________________ 1
232 STF-68B PRT Artificial sequence
233 STF-68B AP1 PRT +Artificial sequence
¨i- _________________________ 1
234 STF-68B AP2 PRT Artificial sequence 1
-4 I
235 STF-118 PRT Artificial sequence
--i _________________________ I
236 STF-118 AP1 PRT Artificial sequence
-4 __________________________ 1
237 STF-90B ' P= RT Artificial sequence
238 STF-90B AP1 PRT l ____________ Artificial sequence I
-4 I
239 STF-117 PRT Artificial sequence
-I- _________________________ I
240 STF-117 AP1 ' PRT Artificial sequence
241 STF-66D PRT Artificial sequence
242 t STF-66D AP1 PRT 1- Artificial sequence
243 0111 DNA Artificial sequence
244 SIED6 DNA +Artificial sequence
245 SIED6 AP1 ' D= NA Artificial sequence
. -4 4- _
246 S I ED6 AP2 DNA Artificial sequence
--I _________________________ 1
247 SIEAll ' D= NA Artificial sequence
_
248 SIEAll AP1 DNA +Artificial sequence
--I _________________________ 1
249 DC1 ' DNA Artificial sequence
______________________________________ 1
250 DC1 AP1 DNA Artificial sequence
______________________________________ 1
251 ' EB6 DNA Artificial sequence
______________________________________ 1
252 EB6 AP1 DNA Artificial sequence
[ 253 ¨1 AH11L ' DNA _______ i
Artificial sequence
I
I 254 AH11L AP1 DNA Artificial sequence
--1- ________________________ I
I 255 STF-94A ' DNA Artificial sequence
I 256 STF-94A AP1 __ ,
DNA Artificial sequence
[ 257 4 STF-69A DNA t- Artificial
sequence
I 258 STF-69A AP1 DNA Artificial sequence
1 259 STF-69A AP2 DNA +Artificial sequence
,
I 260 STF-68B ' D= NA Artificial sequence
1 261 STF-68B AP1 DNA +Artificial sequence
1 262 STF-68B AP2 DNA Artificial sequence
-4 __________________________ I
I 263 STF-118 DNA Artificial sequence
¨t __________________________ 1
[264 STF-118 AP1 : DNA Artificial sequence
, ,
CA 03120160 2021-05-17
WO 2020/128108 PCT/EP2019/086990
98
I 265 ' STF-90B "-T DNA ' Artificial
sequence
266 STF-90B AP1 __ ,
DNA Artificial sequence
t
267 STF-117 DNA i- Artificial sequence
______ 1 ____________________________ 1
268 STF-117 AP1 __ ,
DNA Artificial sequence
t
269 STF-66D DNA + Artificial sequence
--I- _________________________________ I
270 STF-66D AP1 DNA Artificial sequence
271 + WW55 3.0 AP1 PRT +Artificial sequence
--f __________________________________ 1
272 WW55 3.0 AP1 DNA Artificial sequence 1
-4 ___________________________________ I
273 WW55 3.0 AP2 PRT Artificial sequence
274 WW55 3.0 AP2 ' DNA Artificial sequence
______ i ____________________________ 1
275 lambda stf AP1 PRT Artificial sequence
276 lambda stf AP1 DNA Artificial sequence I
277 payload p7.3 DNA Artificial sequence
______________________________________ i
278 1A2 gpJ variant ' PRT Artificial
sequence
279 1A2 gpJ variant DNA Artificial sequence
280 t WWII ID 1 DNA 1- Artificial sequence
281 WWII ID2 DNA Artificial sequence
282 WWII KlF PRT + Artificial sequence
chimeric stfa
¨I 4---
283 WWII KlF PRT Artificial sequence
chimeric stfb
-H --t-
284 WWII KlF DNA Artificial sequence
..i. chimeric stf i
_