Note: Descriptions are shown in the official language in which they were submitted.
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
ENZYMES WITH RUVC DOMAINS
CROSS-REFERENCE
[0001] This application claims the benefit of U.S. Provisional Application No.
62/805,868, filed
February 14, 2019 and titled "MG1 ENZYMES WITH RUVC DOMAINS", U.S. Provisional
Application No. 62/874,414, filed July 15, 2019 and titled "MG1 ENZYMES WITH
RUVC
DOMAINS", U.S. Provisional Application No. 62/805,878, filed February 14, 2019
and titled
"MG2 ENZYMES CONTAINING RUVC DOMAINS", and U.S. Provisional Application No.
62/805,899, filed February 14, 2019 and titled "MG3 ENZYMES WITH RUVC
DOMAINS",
each of which is entirely incorporated herein by reference.
BACKGROUND
[0002] Cas enzymes along with their associated Clustered Regularly Interspaced
Short
Palindromic Repeats (CRISPR) guide ribonucleic acids (RNAs) appear to be a
pervasive (-45%
of bacteria, ¨84% of archaea) component of prokaryotic immune systems, serving
to protect such
microorganisms against non-self nucleic acids, such as infectious viruses and
plasmids by
CRISPR-RNA guided nucleic acid cleavage. While the deoxyribonucleic acid (DNA)
elements
encoding CRISPR RNA elements may be relatively conserved in structure and
length, their
CRISPR-associated (Cas) proteins are highly diverse, containing a wide variety
of nucleic acid-
interacting domains. While CRISPR DNA elements have been observed as early as
1987, the
programmable endonuclease cleavage ability of CRISPR/Cas complexes has only
been
recognized relatively recently, leading to the use of recombinant CRISPR/Cas
systems in diverse
DNA manipulation and gene editing applications.
SEQUENCE LISTING
[0003] The instant application contains a Sequence Listing which has been
submitted
electronically in ASCII format and is hereby incorporated by reference in its
entirety. Said ASCII
copy, created on February 13, 2020, is named 55921-703 601 SL.txt and is
23,363,113 bytes in
size.
SUMMARY
[0004] In some aspects, the present disclosure provides for an engineered
nuclease system,
comprising: (a) an endonuclease comprising a RuvC III domain and an HNH
domain, wherein
the endonuclease is derived from an uncultivated microorganism, wherein the
endonuclease is a
class 2, type II Cas endonuclease; and (b) an engineered guide ribonucleic
acid structure
configured to form a complex with the endonuclease comprising: (i) a guide
ribonucleic acid
sequence configured to hybridize to a target deoxyribonucleic acid sequence;
and (ii) a tracr
- 1 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
ribonucleic acid sequence configured to bind to the endonuclease. In some
embodiments, the
RuvC III domain comprises a sequence with at least 70%, at least 75%, at least
80% or at least
90% sequence identity to any one of SEQ ID NOs: 1827-3637.
[0005] In some aspects, the present disclosure provides for an engineered
nuclease system
comprising: (a) an endonuclease comprising a RuvC III domain having at least
75% sequence
identity to any one of SEQ ID NOs: 1827-3637; and (b) an engineered guide
ribonucleic acid
structure configured to form a complex with the endonuclease comprising: (i) a
guide ribonucleic
acid sequence configured to hybridize to a target deoxyribonucleic acid
sequence; and (ii) a tracr
ribonucleic acid sequence configured to bind to the endonuclease.
[0006] In some aspects, the present disclosure provides for an engineered
nuclease system
comprising: (a) an endonuclease configured to bind to a protospacer adjacent
motif (PAM)
sequence comprising SEQ ID NOs: 5512-5537, wherein the endonuclease is a class
2, type II Cas
endonuclease; and (b) an engineered guide ribonucleic acid structure
configured to form a
complex with the endonuclease comprising: (i) a guide ribonucleic acid
sequence configured to
hybridize to a target deoxyribonucleic acid sequence; and (ii) a tracr
ribonucleic acid sequence
configured to bind to the endonuclease.
[0007] In some embodiments, the endonuclease is derived from an uncultivated
microorganism.
In some embodiments, the endonuclease has not been engineered to bind to a
different PAM
sequence. In some embodiments, the endonuclease is not a Cas9 endonuclease, a
Cas14
endonuclease, a Cas12a endonuclease, a Cas12b endonuclease, a Cas 12c
endonuclease, a
Cas12d endonuclease, a Cas12e endonuclease, a Cas13a endonuclease, a Cas13b
endonuclease,
a Cas13c endonuclease, or a Cas 13d endonuclease. In some embodiments, the
endonuclease has
less than 80% identity to a Cas9 endonuclease. In some embodiments, the
endonuclease further
comprises an HNH domain. In some embodiments, the tracr ribonucleic acid
sequence
comprises a sequence with at least 80% sequence identity to about 60 to 90
consecutive
nucleotides selected from any one of SEQ ID NOs: 5476-5511 and SEQ ID NO:
5538.
[0008] In some aspects, the present disclosure provides for an engineered
nuclease system
comprising, (a) an engineered guide ribonucleic acid structure comprising: (i)
a guide ribonucleic
acid sequence configured to hybridize to a target deoxyribonucleic acid
sequence; and (ii) a tracr
ribonucleic acid sequence configured to bind to an endonuclease, wherein the
tracr ribonucleic
acid sequence comprises a sequence with at least 80% sequence identity to
about 60 to 90
consecutive nucleotides selected from any one of SEQ ID NOs: 5476-5511 and SEQ
ID NO:
5538; and (b) a class 2, type II Cas endonuclease configured to bind to the
engineered guide
ribonucleic acid. In some embodiments, the endonuclease is configured to bind
to a protospacer
adjacent motif (PAM) sequence selected from the group comprising SEQ ID NOs:
5512-5537.
- 2 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[0009] In some embodiments, the engineered guide ribonucleic acid structure
comprises at least
two ribonucleic acid polynucleotides. In some embodiments, the engineered
guide ribonucleic
acid structure comprises one ribonucleic acid polynucleotide comprising the
guide ribonucleic
acid sequence and the tracr ribonucleic acid sequence.
[0010] In some embodiments, the guide ribonucleic acid sequence is
complementary to a
prokaryotic, bacterial, archaeal, eukaryotic, fungal, plant, mammalian, or
human genomic
sequence. In some embodiments, the guide ribonucleic acid sequence is 15-24
nucleotides in
length. In some embodiments, the endonuclease comprises one or more nuclear
localization
sequences (NLSs) proximal to an N- or C-terminus of the endonuclease. In some
embodiments,
the NLS comprises a sequence selected from SEQ ID NOs: 5597-5612.
[0011] In some embodiments, the engineered nuclease system further comprises a
single- or
double-stranded DNA repair template comprising from 5' to 3': a first homology
arm comprising
a sequence of at least 20 nucleotides 5' to the target deoxyribonucleic acid
sequence, a synthetic
DNA sequence of at least 10 nucleotides, and a second homology arm comprising
a sequence of
at least 20 nucleotides 3' to the target sequence. In some embodiments, the
first or second
homology arm comprises a sequence of at least 40, 80, 120, 150, 200, 300, 500,
or 1,000
nucleotides.
[0012] In some embodiments, the system further comprises a source of Mg2+-
[0013] In some embodiments, the endonuclease and the tracr ribonucleic acid
sequence are
derived from distinct bacterial species within a same phylum. In some
embodiments, the
endonuclease is derived from a bacterium belonging to a genus Dermabacter. In
some
embodiments, the endonuclease is derived from a bacterium belonging to Phylum
Verrucomicrobia, Phylum Candidatus Peregrinibacteria, or Phylum Candidatus
Melainabacteria.
In some embodiments, the endonuclease is derived from a bacterium comprising a
16S rRNA
gene having at least 90% identity to any one of SEQ ID NOs: 5592-5595 .
[0014] In some embodiments, the HNH domain comprises a sequence with at least
70% or at
least 80% identity to any one of SEQ ID NOs: 5638-5460. In some embodiments,
the
endonuclease comprises SEQ ID NOs: 1-1826 or a variant thereof having at least
55% identity
thereto. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NOs:
1827-1830 or
SEQ ID NOs: 1827-2140.
[0015] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NOs: 3638-
3641 or SEQ ID
NOs: 3638-3954. In some embodiments, the endonuclease comprises at least 1, at
least 2, at least
3, at least 4, or at least 5 peptide motifs selected from the group consisting
of SEQ ID NOs:
- 3 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
5615-5632. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NOs:
1-4 or SEQ ID
NOs: 1-319.
[0016] In some embodiments, the guide RNA structure comprises a sequence at
least 70%, 80%,
or 90% identical to a sequence selected from the group consisting of SEQ ID
NOs: 5461-5464,
SEQ ID NOs: 5476-5479, or SEQ ID NOs: 5476-5489. In some embodiments, the
guide RNA
structure comprises an RNA sequence predicted to comprise a hairpin consisting
of a stem and a
loop, wherein the stem comprises at least 10, at least 12 or at least 14 base-
paired
ribonucleotides, and an asymmetric bulge within 4 base pairs of the loop.
[0017] In some embodiments, the endonuclease is configured to bind to a PAM
comprising a
sequence selected from the group consisting of SEQ ID NOs: 5512-5515 or SEQ ID
NOs: 5527-
5530.
[0018] In some embodiments: (a) the endonuclease comprises a sequence at least
70%, at least
80%, or at least 90% identical to SEQ ID NO: 1827; (b) the guide RNA structure
comprises a
sequence at least 70%, at least 80%, or at least 90% identical to at least one
of SEQ ID NO: 5461
or SEQ ID NO: 5476; and (c) the endonuclease is configured to bind to a PAM
comprising SEQ
ID NO: 5512 or SEQ ID NO: 5527. In some embodiments: (a) the endonuclease
comprises a
sequence at least 70%, at least 80%, or at least 90% identical to SEQ ID NO:
1828; (b) the guide
RNA structure comprises a sequence at least 70%, at least 80%, or at least 90%
identical to at
least one of SEQ ID NO: 5462 or SEQ ID NO: 5477; and (c) the endonuclease is
configured to
bind to a PAM comprising SEQ ID NO: 5513 or SEQ ID NO: 5528. In some
embodiments: (a)
the endonuclease comprises a sequence at least 70%, at least 80%, or at least
90% identical to
SEQ ID NO: 1829; (b) the guide RNA structure comprises a sequence at least
70%, at least 80%,
or at least 90% identical to at least one of SEQ ID NO: 5463 or SEQ ID NO:
5478; and (c) the
endonuclease is configured to bind to a PAM comprising SEQ ID NO: 5514 or SEQ
ID NO:
5529. In some embodiments: (a) the endonuclease comprises a sequence at least
70%, at least
80%, or at least 90% identical to SEQ ID NO: 1830; (b) the guide RNA structure
comprises a
sequence at least 70%, at least 80%, or at least 90% identical to at least one
of SEQ ID NO: 5464
or SEQ ID NO: 5479; and (c) the endonuclease is configured to bind to a PAM
comprising SEQ
ID NO: 5515 or SEQ ID NO: 5530.
[0019] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NOs: 2141-
2142 or SEQ ID
NOs: 2141-2241. In some embodiments, the endonuclease comprises a sequence at
least 70%,
80%, or 90% identical to a sequence selected from the group consisting of SEQ
ID NOs: 3955-
3956 or SEQ ID NOs: 3955-4055. In some embodiments, the endonuclease comprises
at least 1,
- 4 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
at least 2, at least 3, at least 4, or at least 5 peptide motifs selected from
the group consisting of
SEQ ID NOs: 5632-5638. In some embodiments, the endonuclease comprises a
sequence at least
70%, 80%, or 90% identical to a sequence selected from the group consisting of
SEQ ID NOs:
320-321 or SEQ ID NOs: 320-420. In some embodiments, the guide RNA structure
comprises a
sequence at least 70%, 80%, or 90% identical to a sequence selected from the
group consisting of
SEQ ID NO: 5465, SEQ ID NOs: 5490-5491 or SEQ ID NOs: 5490-5494. In some
embodiments, the guide RNA structure comprises a tracr ribonucleic acid
sequence comprising a
hairpin comprising at least 8, at least 10, or at least 12 base-paired
ribonucleotides. In some
embodiments, the endonuclease is configured to bind to a PAM comprising a
sequence selected
from the group consisting of SEQ ID NOs: 5516 and SEQ ID NOs: 5531. In some
embodiments:
(a) the endonuclease comprises a sequence at least 70%, 80%, or 90% identical
to SEQ ID NO:
2141; (b) the guide RNA structure comprises a sequence at least 70%, 80%, or
90% identical to
SEQ ID NO: 5490; and (c) the endonuclease is configured to binding to a PAM
comprising SEQ
ID NO: 5531. In some embodiments: (a) the endonuclease comprises a sequence at
least 70%,
80%, or 90% identical to SEQ ID NO: 2142; (b) the guide RNA structure
comprises a sequence
at least 70%, 80%, or 90% identical to SEQ ID NO: 5465 or SEQ ID NO: 5491; and
(c) the
endonuclease is configured to binding to a PAM comprising SEQ ID NO: 5516.
[0020] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NOs: 2245-
2246. In some
embodiments, the endonuclease comprises a sequence at least 70%, 80%, or 90%
identical to a
sequence selected from the group consisting of SEQ ID NOs: 4059-4060. In some
embodiments,
the endonuclease comprises at least 1, at least 2, at least 3, at least 4, or
at least 5 peptide motifs
selected from the group consisting of SEQ ID NOs: 5639-5648. In some
embodiments, the
endonuclease comprises a sequence at least 70%, 80%, or 90% identical to a
sequence selected
from the group consisting of SEQ ID NOs: 424-425. In some embodiments, the
guide RNA
structure comprises a sequence at least 70%, 80%, or 90% identical to a
sequence selected from
the group consisting of SEQ ID NOs: 5498-5499 and SEQ ID NO: 5539. In some
embodiments,
the guide RNA structure comprises a guide ribonucleic acid sequence predicted
to comprise a
hairpin with an uninterrupted base-paired region comprising at least 8
nucleotides of a guide
ribonucleic acid sequence and at least 8 nucleotides of a tracr ribonucleic
acid sequence, and
wherein the tracr ribonucleic acid sequence comprises, from 5' to 3', a first
hairpin and a second
hairpin, wherein the first hairpin has a longer stem than the second hairpin.
[0021] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NOs: 2242-
2244 or SEQ ID
NOs: 2247-2249. In some embodiments, the endonuclease comprises a sequence at
least 70%,
- 5 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
80%, or 90% identical to a sequence selected from the group consisting of SEQ
ID NOs: 4056-
4058 and SEQ ID NOs 4061-4063. In some embodiments, the endonuclease comprises
at least 1,
at least 2, at least 3, at least 4, or at least 5 peptide motifs selected from
the group consisting of
SEQ ID NOs: 5639-5648. In some embodiments, the endonuclease comprises a
sequence at least
70%, 80%, or 90% identical to a sequence selected from the group consisting of
SEQ ID NOs:
421-423 or SEQ ID NOs: 426-428. In some embodiments, the guide RNA structure
comprises a
sequence at least 70%, 80%, or 90% identical to a sequence selected from the
group consisting of
SEQ ID NOs: 5466-5467, SEQ ID NOs: 5495-5497, SEQ ID NO: 5500-5502, and SEQ ID
NO:
5539. In some embodiments, the guide RNA structure comprises a guide
ribonucleic acid
sequence predicted to comprise a hairpin with an uninterrupted base-paired
region comprising at
least 8 nucleotides of a guide ribonucleic acid sequence and at least 8
nucleotides of a tracr
ribonucleic acid sequence, and wherein the tracr ribonucleic acid sequence
comprises, from 5' to
3', a first hairpin and a second hairpin, wherein the first hairpin has a
longer stem than the second
hairpin. In some embodiments, the endonuclease is configured to binding to a
PAM comprising
a sequence selected from the group consisting of SEQ ID NOs: 5517-5518 or SEQ
ID NOs:
5532-5534. In some embodiments: (a) the endonuclease comprises a sequence at
least 70%,
80%, or 90% identical to SEQ ID NO: 2247; (b) the guide RNA structure
comprises a sequence
at least 70%, 80%, or 90% identical to SEQ ID NO: 5500; and (c) the
endonuclease is configured
to binding to a PAM comprising SEQ ID NO: 5517 or SEQ ID NO: 5532. In some
embodiments: (a) the endonuclease comprises a sequence at least 70%, 80%, or
90% identical to
SEQ ID NO: 2248; (b) the guide RNA structure comprises a sequence at least
70%, 80%, or 90%
identical to SEQ ID NO: 5501; and (c) the endonuclease is configured to
binding to a PAM
comprising SEQ ID NO: 5518 or SEQ ID NOs: 5533. In some embodiments: (a) the
endonuclease comprises a sequence at least 70%, 80%, or 90% identical to SEQ
ID NO: 2249;
(b) the guide RNA structure comprises a sequence at least 70%, 80%, or 90%
identical to SEQ
ID NO: 5502; and (c) the endonuclease is configured to binding to a PAM
comprising SEQ ID
NO: 5534.
[0022] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 2253
or SEQ ID NOs:
2253-2481. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
4067 or SEQ ID
NOs: 4067-4295. In some embodiments, the endonuclease comprises a peptide
motif according
to SEQ ID NO: 5649. In some embodiments, the endonuclease comprises a sequence
at least
70%, 80%, or 90% identical to a sequence selected from the group consisting of
SEQ ID NO:
432 or SEQ ID NOs: 432-660. In some embodiments, the guide RNA structure
comprises a
- 6 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
sequence at least 70%, 80%, or 90% identical to a sequence selected from the
group consisting of
SEQ ID NO: 5468 or SEQ ID NO: 5503. In some embodiments, the endonuclease is
configured
to binding to a PAM comprising a sequence selected from the group consisting
of SEQ ID NOs:
5519. In some embodiments: (a) the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to SEQ ID NO: 2253; (b) the guide RNA structure comprises a
sequence at least
70%, 80%, or 90% identical to SEQ ID NO: 5468 or SEQ ID NO: 5503; and (c) the
endonuclease is configured to binding to a PAM comprising SEQ ID NO: 5519.
[0023] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NOs: 2482-
2489. In some
embodiments, the endonuclease comprises a sequence at least 70%, 80%, or 90%
identical to a
sequence selected from the group consisting of SEQ ID NOs: 4296-4303. In some
embodiments,
the endonuclease comprises a sequence at least 70%, 80%, or 90% identical to a
sequence
selected from the group consisting of or SEQ ID NOs: 661-668. In some
embodiments, the
endonuclease comprises a sequence at least 70%, 80%, or 90% identical to a
sequence selected
from the group consisting of or SEQ ID NOs: 2490-2498. In some embodiments,
the
endonuclease comprises a sequence at least 70%, 80%, or 90% identical to a
sequence selected
from the group consisting of SEQ ID NOs: 4304-4312. In some embodiments, the
endonuclease
comprises a sequence at least 70%, 80%, or 90% identical to a sequence
selected from the group
consisting of SEQ ID NOs: 669-677. In some embodiments, the guide RNA
structure comprises
a sequence at least 70%, 80%, or 90% identical to a sequence selected from the
group consisting
of SEQ ID NO: 5504.
[0024] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 2499
or SEQ ID NOs:
2499-2750. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
4313 or SEQ ID
NOs: 4313-4564. In some embodiments, the endonuclease comprises at least 1, at
least 2, at least
3, at least 4, or at least 5 peptide motifs selected from the group consisting
of SEQ ID NOs:
5650-5667. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
678 or SEQ ID
NOs: 678-929. In some embodiments, the guide RNA structure comprises a
sequence at least
70%, 80%, or 90% identical to SEQ ID NO: 5469 or SEQ ID NO: 5505. In some
embodiments,
the endonuclease is configured to binding to a PAM comprising SEQ ID NOs: 5520
or SEQ ID
NOs: 5535. In some embodiments: (a) the endonuclease comprises a sequence at
least 70%,
80%, or 90% identical to SEQ ID NO: 2499; (b) the guide RNA structure
comprises a sequence
at least 70%, 80%, or 90% identical to SEQ ID NO: 5469 or SEQ ID NO: 5505; and
(c) the
- 7 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
endonuclease is configured to binding to a PAM comprising SEQ ID NO: 5520 or
SEQ ID NO:
5535.
[0025] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 2751
or SEQ ID NOs:
2751-2913. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
4565 or SEQ ID
NOs: 4565-4727. In some embodiments, the endonuclease comprises at least 1, at
least 2, at least
3, at least 4, or at least 5 peptide motifs selected from the group consisting
of SEQ ID NOs:
5668-5678. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
930 or SEQ ID
NOs: 930-1092. In some embodiments, the guide RNA structure comprises a
sequence at least
70%, 80%, or 90% identical to SEQ ID NO: 5470 or SEQ ID NOs: 5506. In some
embodiments,
the endonuclease is configured to binding to a PAM comprising a sequence
selected from the
group consisting of SEQ ID NOs: 5521 or SEQ ID NOs: 5536. In some embodiments:
(a) the
endonuclease comprises a sequence at least 70%, 80%, or 90% identical to SEQ
ID NO: 2751;
(b) the guide RNA structure comprises a sequence at least 70%, 80%, or 90%
identical to SEQ
ID NO: 5470 or SEQ ID NO: 5506; and (c) the endonuclease is configured to
binding to a PAM
comprising SEQ ID NO: 5521 or SEQ ID NO: 5536.
[0026] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 2914
or SEQ ID NOs:
2914-3174. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
4728 or SEQ ID
NOs: 4728-4988. In some embodiments, the endonuclease comprises at least 1, at
least 2, or at
least 3 peptide motifs selected from the group consisting of SEQ ID NOs: 5676-
5678. In some
embodiments, the endonuclease comprises a sequence at least 70%, 80%, or 90%
identical to a
sequence selected from the group consisting of SEQ ID NO: 1093 or SEQ ID NOs:
1093-1353.
In some embodiments, the guide RNA structure comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 5471,
SEQ ID NO:
5507, and SEQ ID NOs: 5540-5542. In some embodiments, the guide RNA structure
comprises
a tracr ribonucleic acid sequence predicted to comprise at least two hairpins
comprising less than
base-paired ribonucleotides. In some embodiments, the endonuclease is
configured to binding
to a PAM comprising SEQ ID NO: 5522. In some embodiments: (a) the endonuclease
comprises
a sequence at least 70%, 80%, or 90% identical to SEQ ID NO: 2914; (b) the
guide RNA
structure comprises a sequence at least 70%, 80%, or 90% identical to SEQ ID
NO: 5471 or SEQ
- 8 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
ID NO: 5507; and (c) the endonuclease is configured to binding to a PAM
comprising SEQ ID
NO: 5522.
[0027] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 3175
or SEQ ID NOs:
3175-3330. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
4989 or SEQ ID
NOs: 4989-5146. In some embodiments, the endonuclease comprises at least 1, at
least 2, at least
3, at least 4, or at least 5 peptide motifs selected from the group consisting
of SEQ ID NOs:
5679-5686. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
1354 or SEQ ID
NOs: 1354-1511. In some embodiments, the guide RNA structure comprises a
sequence at least
70%, 80%, or 90% identical to a sequence selected from the group consisting of
SEQ ID NOs:
5472 or SEQ ID NOs: 5508. In some embodiments, the endonuclease is configured
to binding to
a PAM comprising a sequence selected from the group consisting of SEQ ID NO:
5523 or SEQ
ID NO: 5537. In some embodiments: (a) the endonuclease comprises a sequence at
least 70%,
80%, or 90% identical to SEQ ID NO: 3175; (b) the guide RNA structure
comprises a sequence
at least 70%, 80%, or 90% identical to SEQ ID NO: 5472 or SEQ ID NO: 5508; and
(c) the
endonuclease is configured to binding to a PAM comprising SEQ ID NO: 5523 or
SEQ ID NO:
5537.
[0028] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NOs: 3331
or SEQ ID
NOs: 3331-3474. In some embodiments, the endonuclease comprises a sequence at
least 70%,
80%, or 90% identical to a sequence selected from the group consisting of SEQ
ID NOs: 5147 or
SEQ ID NOs: 5147-5290. In some embodiments, the endonuclease comprises at
least 1, at least
2, at least 3, at least 4, or at least 5 peptide motifs selected from the
group consisting of SEQ ID
NOs: 5674-5675 and SEQ ID NOs: 5687-5693. In some embodiments, the
endonuclease
comprises a sequence at least 70%, 80%, or 90% identical to a sequence
selected from the group
consisting of SEQ ID NO: 1512 or SEQ ID NOs: 1512-1655. In some embodiments,
the guide
RNA structure comprises a sequence at least 70%, 80%, or 90% identical to a
sequence selected
from the group consisting of SEQ ID NO: 5473 or SEQ ID NO: 5509. In some
embodiments, the
endonuclease is configured to binding to a PAM comprising SEQ ID NO: 5524. In
some
embodiments: (a) the endonuclease comprises a sequence at least 70%, 80%, or
90% identical to
SEQ ID NO: 3331; (b) the guide RNA structure comprises a sequence at least
70%, 80%, or 90%
identical to SEQ ID NO: 5473 or SEQ ID NO: 5509; and (c) the endonuclease is
configured to
binding to a PAM comprising SEQ ID NO: 5524.
- 9 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[0029] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 3475
or SEQ ID NOs:
3475-3568. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
5291 or SEQ ID
NOs: 5291-5389. In some embodiments, the endonuclease comprises at least 1, at
least 2, at least
3, at least 4, or at least 5 peptide motifs selected from the group consisting
of SEQ ID NOs:
5694-5699. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
1656 or SEQ ID
NOs: 1656-1755. In some embodiments, the guide RNA structure comprises a
sequence at least
70%, 80%, or 90% identical to SEQ ID NO: 5474 or SEQ ID NO: 5510. In some
embodiments,
the endonuclease is configured to binding to a PAM comprising SEQ ID NOs:
5525. In some
embodiments: (a) the endonuclease comprises a sequence at least 70%, 80%, or
90% identical to
SEQ ID NO: 3475; (b) the guide RNA structure comprises a sequence at least
70%, 80%, or 90%
identical to SEQ ID NO: 5474 or SEQ ID NO: 5510; and (c) the endonuclease is
configured to
binding to a PAM comprising SEQ ID NO: 5525.
[0030] In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or 90%
identical to a sequence selected from the group consisting of SEQ ID NO: 3569
or SEQ ID NOs:
3569-3637. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
5390 or SEQ ID
NOs: 5390-5460. In some embodiments, the endonuclease comprises at least 1, at
least 2, at least
3, at least 4, or at least 5 peptide motifs selected from the group consisting
of SEQ ID NOs:
5700-5717. In some embodiments, the endonuclease comprises a sequence at least
70%, 80%, or
90% identical to a sequence selected from the group consisting of SEQ ID NO:
1756 or SEQ ID
NOs: 1756-1826. In some embodiments, the guide RNA structure comprises a
sequence at least
70%, 80%, or 90% identical to SEQ ID NO: 5475 or SEQ ID NOs: 5511. In some
embodiments,
the endonuclease is configured to binding to a PAM comprising SEQ ID NO: 5526.
In some
embodiments: (a) the endonuclease comprises a sequence at least 70%, 80%, or
90% identical to
SEQ ID NO: 3569; (b) the guide RNA structure comprises a sequence at least
70%, 80%, or 90%
identical to SEQ ID NO: 5475 or SEQ ID NO: 5511; and (c) the endonuclease is
configured to
binding to a PAM comprising SEQ ID NO: 5526. In some embodiments, the sequence
identity
is determined by a BLASTP, CLUSTALW, MUSCLE, MAFFT, or Smith-Waterman homology
search algorithm. In some embodiments, the sequence identity is determined by
the BLASTP
homology search algorithm using parameters of a wordlength (W) of 3, an
expectation (E) of 10,
and a BLOSUM62 scoring matrix setting gap costs at existence of 11, extension
of 1, and using a
conditional compositional score matrix adjustment.
- 10 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[0031] In some aspects, the present disclosure provides for an engineered
guide ribonucleic acid
polynucleotide comprising: (a) a DNA-targeting segment comprising a nucleotide
sequence that
is complementary to a target sequence in a target DNA molecule; and (b) a
protein-binding
segment comprising two complementary stretches of nucleotides that hybridize
to form a double-
stranded RNA (dsRNA) duplex, wherein the two complementary stretches of
nucleotides are
covalently linked to one another with intervening nucleotides, and wherein the
engineered guide
ribonucleic acid polynucleotide is configured to forming a complex with an
endonuclease
comprising a RuvC III domain having at least 75% sequence identity to any one
of SEQ ID
NOs: 1827-3637 and targeting the complex to the target sequence of the target
DNA molecule.
In some embodiments, the DNA-targeting segment is positioned 5' of both of the
two
complementary stretches of nucleotides.
[0032] In some embodiments: (a) the protein binding segment comprises a
sequence having at
least 70%, at least 80%, or at least 90% identity to a sequence selected from
the group consisting
of SEQ ID NOs: 5476-5479 or SEQ ID NOs: 5476-5489; (b) the protein binding
segment
comprises a sequence having at least 70%, at least 80%, or at least 90%
identity to a sequence
selected from the group consisting of (SEQ ID NOs: 5490-5491 or SEQ ID NOs:
5490-5494) and
SEQ ID NO: 5538; (c) the protein binding segment comprises a sequence having
at least 70%, at
least 80%, or at least 90% identity to a sequence selected from the group
consisting of SEQ ID
NOs: 5498-5499; (d) the protein binding segment comprises a sequence having at
least 70%, at
least 80%, or at least 90% identity to a sequence selected from the group
consisting of SEQ ID
NOs: 5495-5497 and SEQ ID NOs: 5500-5502; (e) the protein binding segment
comprises a
sequence having at least 70%, at least 80%, or at least 90% identity to SEQ ID
NO: 5503; (f) the
protein binding segment comprises a sequence having at least 70%, at least
80%, or at least 90%
identity to SEQ ID NO: 5504; (g) the protein binding segment comprises a
sequence having at
least 70%, at least 80%, or at least 90% identity to SEQ ID NOs: 5505; (h)
protein binding
segment comprises a sequence having at least 70%, at least 80%, or at least
90% identity to SEQ
ID NO: 5506; (i) protein binding segment comprises a sequence having at least
70%, at least
80%, or at least 90% identity to SEQ ID NO: 5507; (j) the protein binding
segment comprises a
sequence having at least 70%, at least 80%, or at least 90% identity to SEQ ID
NO: 5508; (k) the
protein binding segment comprises a sequence having at least 70%, at least
80%, or at least 90%
identity to SEQ ID NO: 5509; (1) the protein binding segment comprises a
sequence having at
least 70%, at least 80%, or at least 90% identity to SEQ ID NO: 5510; or (m)
the protein binding
segment comprises a sequence having at least 70%, at least 80%, or at least
90% identity to SEQ
ID NO: 5511.
-11-
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[0033] In some embodiments: (a) the guide ribonucleic acid polynucleotide
comprises an RNA
sequence comprising a hairpin comprising a stem and a loop, wherein the stem
comprises at least
10, at least 12, or at least 14 base-paired ribonucleotides, and an asymmetric
bulge within 4 base
pairs of the loop; (b) the guide ribonucleic acid polynucleotide comprises a
tracr ribonucleic acid
sequence predicted to comprise a hairpin comprising at least 8, at least 10,
or at least 12 base-
paired ribonucleotides; (c) the guide ribonucleic acid polynucleotide
comprises a guide
ribonucleic acid sequence predicted to comprise a hairpin with an
uninterrupted base-paired
region comprising at least 8 nucleotides of a guide ribonucleic acid sequence
and at least 8
nucleotides of a tracr ribonucleic acid sequence, and wherein the tracr
ribonucleic acid sequence
comprises, from 5' to 3', a first hairpin and a second hairpin, wherein the
first hairpin has a
longer stem than the second hairpin; or (d) the guide ribonucleic acid
polynucleotide comprises a
tracr ribonucleic acid sequence predicted to comprise at least two hairpins
comprising less than 5
base-paired ribonucleotides.
[0034] In some aspects, the present disclosure provides for a deoxyribonucleic
acid
polynucleotide encoding any of the engineered guide ribonucleic acid
polynucleotides described
herein.
[0035] In some aspects, the present disclosure provides for a nucleic acid
comprising an
engineered nucleic acid sequence optimized for expression in an organism,
wherein the nucleic
acid encodes a class 2, type II Cas endonuclease comprising a RuvC III domain
and an HNH
domain, and wherein the endonuclease is derived from an uncultivated
microorganism.
[0036] In some aspects, the present disclosure provides for a nucleic acid
comprising an
engineered nucleic acid sequence optimized for expression in an organism,
wherein the nucleic
acid encodes an endonuclease comprising a RuvC III domain having at least 70%
sequence
identity to any one of SEQ ID NOs: 1827-3637. In some embodiments, the
endonuclease
comprises an HNH domain having at least 70% or at least 80% sequence identity
to any one of
SEQ ID NOs: 3638-5460. In some embodiments, the endonuclease comprises SEQ ID
NOs:
5572-5591 or a variant thereof having at least 70% sequence identity thereto.
In some
embodiments, the endonuclease comprises a sequence encoding one or more
nuclear localization
sequences (NLSs) proximal to an N- or C-terminus of the endonuclease. In some
embodiments,
the NLS comprises a sequence selected from SEQ ID NOs: 5597-5612.
[0037] In some embodiments, the organism is prokaryotic, bacterial,
eukaryotic, fungal, plant,
mammalian, rodent, or human. In some embodiments, the organism is E. coil,
and: (a) the
nucleic acid sequence has at least 70%, 80%, or 90% identity to a sequence
selected from the
group consisting of SEQ ID NOs: 5572-5575; (b) the nucleic acid sequence has
at least 70%,
80%, or 90% identity to a sequence selected from the group consisting of SEQ
ID NOs: 5576-
- 12 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
5577; (c) the nucleic acid sequence has at least 70%, 80%, or 90% identity to
a sequence selected
from the group consisting of SEQ ID NOs: 5578-5580; (d) the nucleic acid
sequence has at least
70%, 80%, or 90% identity to SEQ ID NO: 5581; (e) the nucleic acid sequence
has at least 70%,
80%, or 90% identity to SEQ ID NO: 5582; (f) the nucleic acid sequence has at
least 70%, 80%,
or 90% identity to SEQ ID NO: 5583; (g) the nucleic acid sequence has at least
70%, 80%, or
90% identity to SEQ ID NO: 5584; (h) the nucleic acid sequence has at least
70%, 80%, or 90%
identity to SEQ ID NO: 5585; (i) the nucleic acid sequence has at least 70%,
80%, or 90%
identity to SEQ ID NO: 5586; or (j) the nucleic acid sequence has at least
70%, 80%, or 90%
identity to SEQ ID NO: 5587. In some embodiments, the organism is human, and:
(a) the nucleic
acid sequence has at least 70%, 80%, or 90% identity to SEQ ID NO: 5588 or SEQ
ID NO: 5589;
or (b) the nucleic acid sequence has at least 70%, 80%, or 90% identity to SEQ
ID NO: 5590 or
SEQ ID NO: 5591.
[0038] In some aspects, the present disclosure provides for a vector
comprising a nucleic acid
sequence encoding a class 2, type II Cas endonuclease comprising a RuvC III
domain and an
HNH domain, wherein the endonuclease is derived from an uncultivated
microorganism.
[0039] In some aspects, the present disclosure provides for a vector
comprising the any of the
nucleic acids described herein. In some embodiments, the vector further
comprises a nucleic acid
encoding an engineered guide ribonucleic acid structure configured to form a
complex with the
endonuclease comprising: (a) a guide ribonucleic acid sequence configured to
hybridize to a
target deoxyribonucleic acid sequence; and (b) a tracr ribonucleic acid
sequence configured to
binding to the endonuclease. In some embodiments, the vector is a plasmid, a
minicircle, a
CELiD, an adeno-associated virus (AAV) derived virion, or a lentivirus.
[0040] In some aspects, the present disclosure provides for a cell comprising
any of the vectors
described herein.
[0041] In some aspects, the present disclosure provides for a method of
manufacturing an
endonuclease, comprising cultivating any of the cells described herein.
[0042] In some aspects, the present disclosure provides for a method for
binding, cleaving,
marking, or modifying a double-stranded deoxyribonucleic acid polynucleotide,
comprising: (a)
contacting the double-stranded deoxyribonucleic acid polynucleotide with a
class 2, type II Cas
endonuclease in complex with an engineered guide ribonucleic acid structure
configured to bind
to the endonuclease and the double-stranded deoxyribonucleic acid
polynucleotide; (b) wherein
the double-stranded deoxyribonucleic acid polynucleotide comprises a
protospacer adjacent
motif (PAM); and (c) wherein the PAM comprises a sequence selected from the
group consisting
of SEQ ID NOs: 5512-5526 or SEQ ID NOs: 5527-5537. In some embodiments, the
double-
stranded deoxyribonucleic acid polynucleotide comprises a first strand
comprising a sequence
- 13 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
complementary to a sequence of the engineered guide ribonucleic acid structure
and a second
strand comprising the PAM. In some embodiments, the PAM is directly adjacent
to the 3' end of
the sequence complementary to the sequence of the engineered guide ribonucleic
acid structure.
[0043] In some embodiments, the class 2, type II Cas endonuclease is not a
Cas9 endonuclease, a
Cas14 endonuclease, a Cas12a endonuclease, a Cas12b endonuclease, a Cas 12c
endonuclease, a
Cas12d endonuclease, a Cas12e endonuclease, a Cas13a endonuclease, a Cas13b
endonuclease,
a Cas13c endonuclease, or a Cas 13d endonuclease. In some embodiments, the
class 2, type II
Cas endonuclease is derived from an uncultivated microorganism. In some
embodiments, the
double-stranded deoxyribonucleic acid polynucleotide is a eukaryotic, plant,
fungal, mammalian,
rodent, or human double-stranded deoxyribonucleic acid polynucleotide.
[0044] In some embodiments: (a) the PAM comprises a sequence selected from the
group
consisting of SEQ ID NOs: 5512-5515 and SEQ ID NOs: 5527-5530; (b) the PAM
comprises
SEQ ID NO: 5516 or SEQ ID NO: 5531; (c) the PAM comprises SEQ ID NO: 5539; (d)
the
PAM comprises SEQ ID NO: 5517 or SEQ ID NO: 5518; (e) the PAM comprises SEQ ID
NO:
5519; (f) the PAM comprises SEQ ID NO: 5520 or SEQ ID NO: 5535; (g) the PAM
comprises
SEQ ID NO: 5521 or SEQ ID NO: 5536; (h) the PAM comprises SEQ ID NO: 5522; (i)
the PAM
comprises SEQ ID NO: 5523 or SEQ ID NO: 5537; (j) the PAM comprises SEQ ID NO:
5524;
(k) the PAM comprises SEQ ID NO: 5525; or (1) the PAM comprises SEQ ID NO:
5526.
[0045] In some aspects, the present disclosure provides for a method of
modifying a target
nucleic acid locus, the method comprising delivering to the target nucleic
acid locus any of the
engineered nuclease systems described herein, wherein the endonuclease is
configured to form a
complex with the engineered guide ribonucleic acid structure, and wherein the
complex is
configured such that upon binding of the complex to the target nucleic acid
locus, the complex
modifies the target nucleic locus. In some embodiments, modifying the target
nucleic acid locus
comprises binding, nicking, cleaving, or marking the target nucleic acid
locus. In some
embodiments, the target nucleic acid locus comprises deoxyribonucleic acid
(DNA) or
ribonucleic acid (RNA). In some embodiments, the target nucleic acid comprises
genomic DNA,
viral DNA, viral RNA, or bacterial DNA. In some embodiments, the target
nucleic acid locus is
in vitro. In some embodiments, the target nucleic acid locus is within a cell.
In some
embodiments, the cell is a prokaryotic cell, a bacterial cell, a eukaryotic
cell, a fungal cell, a plant
cell, an animal cell, a mammalian cell, a rodent cell, a primate cell, or a
human cell.
[0046] In some embodiments, delivering the engineered nuclease system to the
target nucleic
acid locus comprises delivering the nucleic acid of any of claims 135-140 or
the vector of any of
claims 142-146. In some embodiments, delivering the engineered nuclease system
to the target
nucleic acid locus comprises delivering a nucleic acid comprising an open
reading frame
- 14 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
encoding the endonuclease. In some embodiments, the nucleic acid comprises a
promoter to
which the open reading frame encoding the endonuclease is operably linked. In
some
embodiments, the engineered nuclease system to the target nucleic acid locus
comprises
delivering a capped mRNA containing the open reading frame encoding the
endonuclease. In
some embodiments, the engineered nuclease system to the target nucleic acid
locus comprises
delivering a translated polypeptide. In some embodiments, the engineered
nuclease system to the
target nucleic acid locus comprises delivering a deoxyribonucleic acid (DNA)
encoding the
engineered guide ribonucleic acid structure operably linked to a ribonucleic
acid (RNA) pol III
promoter. In some embodiments, the endonuclease induces a single-stranded
break or a double-
stranded break at or proximal to the target locus.
[0047] Additional aspects and advantages of the present disclosure will become
readily apparent
to those skilled in this art from the following detailed description, wherein
only illustrative
embodiments of the present disclosure are shown and described. As will be
realized, the present
disclosure is capable of other and different embodiments, and its several
details are capable of
modifications in various obvious respects, all without departing from the
disclosure.
Accordingly, the drawings and description are to be regarded as illustrative
in nature, and not as
restrictive.
INCORPORATION BY REFERENCE
[0048] All publications, patents, and patent applications mentioned in this
specification are
herein incorporated by reference to the same extent as if each individual
publication, patent, or
patent application was specifically and individually indicated to be
incorporated by reference.
BRIEF DESCRIPTION OF THE DRAWINGS
[0049] The novel features of the invention are set forth with particularity in
the appended claims.
A better understanding of the features and advantages of the present invention
will be obtained
by reference to the following detailed description that sets forth
illustrative embodiments, in
which the principles of the invention are utilized, and the accompanying
drawings (also "Figure"
and "FIG." herein), of which:
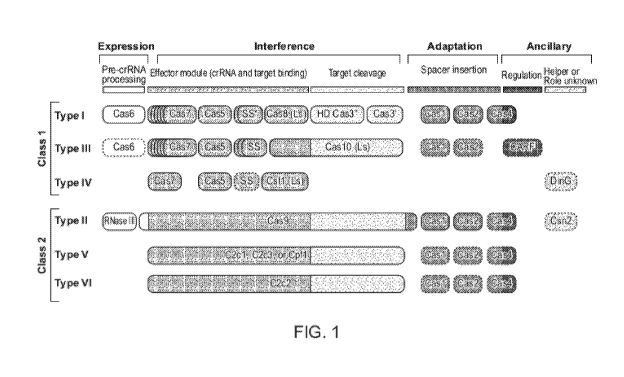
[0050] FIGURE 1 depicts typical organizations of CRISPR/Cas loci of different
classes and
types.
[0051] FIGURE 2 depicts the architecture of a natural Class2/Type II
crRNA/tracrRNA pair,
compared to a hybrid sgRNA wherein both are joined.
[0052] FIGURE 3 depicts schematics showing organization of CRISPR loci
encoding enzymes
from the MG1 family.
- 15 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[0053] FIGURE 4 depicts schematics showing organization of CRISPR loci
encoding enzymes
from the MG2 family.
[0054] FIGURE 5 depicts schematics showing organization of CRISPR loci
encoding enzymes
from the MG3 family.
[0055] FIGURE 6 depicts a structure-based alignment of an enzyme of the
present disclosure
(MG1-1) versus Cas9 from Staphylococcus aureus (SEQ ID NO:5613).
[0056] FIGURE 7 depicts a structure-based alignment of an enzyme of the
present disclosure
(MG2-1) versus Cas9 from Staphylococcus aureus (SEQ ID NO:5613).
[0057] FIGURE 8 depicts a structure-based alignment of an enzyme of the
present disclosure
(MG3-1) versus Cas9 from Actinomyces naeslundii (SEQ ID NO: 5614).
[0058] FIGURES 9A, 9B, 9C, 9D, 9E, 9F, 9G, and 9H depicts a structure-based
alignment of
MG1 family enzymes MG1-1 through MG1-6 (SEQ ID NOs: 5, 6, 9, 1, 2, and 3).
[0059] FIGURE 10 depicts in vitro cleavage of DNA by MG1-4 in complex with its
corresponding sgRNA containing targeting sequences of varying lengths.
[0060] FIGURE 11 depicts in cell cleavage of E. coli genomic DNA using MG1-4
along with its
corresponding sgRNA. Shown are dilution series of cells transformed with MG1-4
along with
target or non-target spacer (top); bottom panel shows the data quantitated,
where the left bar
represents non-target sgRNA and the right bar represents target sgRNA.
[0061] FIGURE 12 depicts in cell indel formation generated by transfection of
HEK cells with
MG1-4 or MG1-6 constructs described in Example 11 alongside their
corresponding sgRNAs
containing various different targeting sequences targeting various locations
in the human
genome.
[0062] FIGURE 13 depicts vitro cleavage of DNA by MG3-6 in complex with its
corresponding
sgRNA containing targeting sequences of varying lengths.
[0063] FIGURE 14 depicts in cell cleavage of E. coil genomic DNA using MG3-7
along with its
corresponding sgRNA. Shown are dilution series of cells transformed with MG3-7
along with
target or non-target spacer (top); bottom panel shows the data quantitated,
where the left bar
represents non-target sgRNA and the right bar represents target sgRNA.
[0064] FIGURE 15 depicts in cell indel formation generated by transfection of
HEK cells with
MG3-7 constructs described in Example 13 alongside their corresponding sgRNAs
containing
various different targeting sequences targeting various locations in the human
genome.
[0065] FIGURE 16 depicts in vitro cleavage of DNA by MG15-1 in complex with
its
corresponding sgRNA containing targeting sequences of varying lengths.
[0066] FIGURES 17, 18, 19, and 20 depict agarose gels showing the results of
PAM vector
library cleavage in the presence of TXTL extracts containing various MG family
nucleases and
- 16 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
their corresponding tracrRNAs or sgRNAs.
[0067] FIGURES 21, 22, 23, 24, 25 and 26 depict predicted structures
(predicted e.g., as in
Example 7) of corresponding sgRNAs of MG enzymes described herein.
[0068] FIGURES 27, 28, 29, 30, 31, 32 and 33 depict seqLogo representations of
PAM
sequences derived via NGS as described herein (e.g. as described in Example
6).
[0069] FIGURE 34 depicts in cell cleavage of E. coil genomic DNA using MG2-7
along with its
corresponding sgRNA. Shown are dilution series of cells transformed with MG2-7
along with
target or non-target spacer (top); bottom panel shows the data quantitated,
where the right bar
represents non-target sgRNA and the left bar represents target sgRNA.
[0070] FIGURE 35 depicts in cell cleavage of E. coil genomic DNA using MG14-1
along with
its corresponding sgRNA. Shown are dilution series of cells transformed with
MG14-1 along
with target or non-target spacer (top); bottom panel shows the data
quantitated, where the right
bar represents non-target sgRNA and the left bar represents target sgRNA.
[0071] FIGURE 36 depicts in cell cleavage of E. coil genomic DNA using MG15-1
along with
its corresponding sgRNA. Shown are dilution series of cells transformed with
MG15-1 along
with target or non-target spacer (top); bottom panel shows the data
quantitated, where the right
bar represents non-target sgRNA and the left bar represents target sgRNA.
BRIEF DESCRIPTION OF THE SEQUENCE LISTING
[0072] The Sequence Listing filed herewith provides exemplary polynucleotide
and polypeptide
sequences for use in methods, compositions and systems according to the
disclosure. Below are
exemplary descriptions of sequences therein.
[0073] MG1
[0074] SEQ ID NOs: 1-319 show the full-length peptide sequences of MG1
nucleases.
[0075] SEQ ID NOs: 1827-2140 show the peptide sequences of RuvC III domains of
MG1
nucleases above.
[0076] SEQ ID NOs: 3638-3955 show the peptide of HNH domains of MG1 nucleases
above.
[0077] SEQ ID NOs: 5476-5479 show the nucleotide sequences of MG1 tracrRNAs
derived from
the same loci as MG1 nucleases above (e.g., same loci as SEQ ID NO:1-4,
respectively).
[0078] SEQ ID NOs: 5461-5464 show the nucleotide sequences of sgRNAs
engineered to
function with an MG1 nuclease (e.g., SEQ ID NO:1-4, respectively), where Ns
denote
nucleotides of a targeting sequence.
[0079] SEQ ID NOs: 5572-5575 show nucleotide sequences for E. coil codon-
optimized coding
sequences for MG1 family enzymes (SEQ ID NOs: 1-4).
- 17 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[0080] SEQ ID NOs: 5588-5589 show nucleotide sequences for human codon-
optimized coding
sequences for MG1 family enzymes (SEQ ID NOs: 1 and 3).
[0081] SEQ ID NOs: 5616-5632 show peptide motifs characteristic of MG1 family
enzymes.
[0082] MG2
[0083] SEQ ID NOs: 320-420 show the full-length peptide sequences of MG2
nucleases.
[0084] SEQ ID NOs: 2141-2241 show the peptide sequences of RuvC III domains of
MG2
nucleases above.
[0085] SEQ ID NOs: 3955-4055 show the peptide of HNH domains of MG2 nucleases
above.
[0086] SEQ ID NOs: 5490-5494 show the nucleotide sequences of MG2 tracrRNAs
derived from
the same loci as MG2 nucleases above (e.g., same loci as SEQ ID NOs: 320, 321,
323, 325, and
326, respectively).
[0087] SEQ ID NO: 5465 shows the nucleotide sequence of an sgRNA engineered to
function
with an MG2 nuclease (e.g., SEQ ID NO: 321 above).
[0088] SEQ ID NOs: 5572-5575 show nucleotide sequences for E. coil codon-
optimized coding
sequences for MG2 family enzymes.
[0089] SEQ ID NOs: 5631-5638 show peptide sequences characteristic of MG2
family enzymes.
[0090] MG3
[0091] SEQ ID NOs: 421-431 show the full-length peptide sequences of MG3
nucleases.
[0092] SEQ ID NOs: 2242-2251 show the peptide sequences of RuvC III domains of
MG3
nucleases above.
[0093] SEQ ID NOs: 4056-4066 show the peptide of HNH domains of MG3 nucleases
above.
[0094] SEQ ID NOs: 5495-5502 show the nucleotide sequences of MG3 tracrRNAs
derived from
the same loci as MG3 nucleases above (e.g., same loci as SEQ ID NOs: 421-428,
respectively).
[0095] SEQ ID NOs: 5466-5467 show the nucleotide sequence of sgRNAs engineered
to
function with an MG3 nuclease (e.g., SEQ ID NOs: 421 - 423).
[0096] SEQ ID NOs: 5578-5580 show nucleotide sequences for E. coil codon-
optimized coding
sequences for MG3 family enzymes.
[0097] SEQ ID NOs: 5639-5648 show peptide sequences characteristic of MG3
family enzymes.
[0098] MG4
[0099] SEQ ID NOs: 432-660 show the full-length peptide sequences of MG4
nucleases.
[00100] SEQ ID NOs: 2253-2481 show the peptide sequences of RuvC III domains
of MG4
nucleases above.
[00101] SEQ ID NOs: 4067-4295 show the peptide of HNH domains of MG4 nucleases
above.
[00102] SEQ ID NO: 5503 shows the nucleotide sequences of an MG4 tracrRNA
derived from
the same loci as MG4 nucleases above.
- 18 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00103] SEQ ID NO: 5468 shows the nucleotide sequence of sgRNAs engineered to
function
with an MG4 nuclease.
[00104] SEQ ID NO: 5649 shows a peptide sequence characteristic of MG4 family
enzymes.
[00105] MG6
[00106] SEQ ID NOs: 661-668 show the full-length peptide sequences of MG6
nucleases.
[00107] SEQ ID NOs: 2482-2489 show the peptide sequences of RuvC III domains
of MG6
nucleases above.
[00108] SEQ ID NOs: 4296-4303 show the peptide of HNH domains of MG3 nucleases
above.
[00109] MG7
[00110] SEQ ID NOs: 669-677 show the full-length peptide sequences of MG7
nucleases.
[00111] SEQ ID NOs: 2490-2498 show the peptide sequences of RuvC III domains
of MG7
nucleases above.
[00112] SEQ ID NOs: 4304-4312 show the peptide of HNH domains of MG3 nucleases
above.
[00113] SEQ ID NO: 5504 shows the nucleotide sequence of an MG7 tracrRNA
derived from
the same loci as MG7 nucleases above.
[00114] MG14
[00115] SEQ ID NOs: 678-929 show the full-length peptide sequences of MG14
nucleases.
[00116] SEQ ID NOs: 2499-2750 show the peptide sequences of RuvC III domains
of MG14
nucleases above.
[00117] SEQ ID NOs: 4313-4564 show the peptide of HNH domains of MG14
nucleases above.
[00118] SEQ ID NO: 5505 shows the nucleotide sequences of MG14 tracrRNA
derived from the
same loci as MG14 nucleases above.
[00119] SEQ ID NO: 5581 shows a nucleotide sequence for an E. coil codon-
optimized coding
sequences for an MG14 family enzyme.
[00120] SEQ ID NOs: 5650-5667 show peptide sequences characteristic of MG14
family
enzymes.
[00121] MG15
[00122] SEQ ID NOs: 930-1092 show the full-length peptide sequences of MG15
nucleases.
[00123] SEQ ID NOs: 2751-2913 show the peptide sequences of RuvC III domains
of MG15
nucleases above.
[00124] SEQ ID NOs: 4565-4727 show the peptide of HNH domains of MG15
nucleases above.
[00125] SEQ ID NO: 5506 shows the nucleotide sequences of MG15 tracrRNA
derived from the
same loci as MG15 nucleases above.
[00126] SEQ ID NOs: 5470 shows the nucleotide sequence of an sgRNA engineered
to function
with an MG15 nuclease.
- 19 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00127] SEQ ID NO: 5582 shows a nucleotide sequence for an E. coil codon-
optimized coding
sequences for an MG15 family enzyme.
[00128] SEQ ID NOs: 5668-5675 show peptide sequences characteristic of MG15
family
enzymes.
[00129] MG16
[00130] SEQ ID NOs: 1093-1353 show the full-length peptide sequences of MG16
nucleases.
[00131] SEQ ID NOs: 2914-3174 show the peptide sequences of RuvC III domains
of MG16
nucleases above.
[00132] SEQ ID NOs: 4728-4988 show the peptide of HNH domains of MG16
nucleases above.
[00133] SEQ ID NOs: 5507 show the nucleotide sequences of an MG16 tracrRNA
derived from
the same loci as MG3 nucleases above.
[00134] SEQ ID NOs: 5471 shows the nucleotide sequence of sgRNAs engineered to
function
with an MG16 nuclease.
[00135] SEQ ID NO: 5583 shows a nucleotide sequence for an E. coil codon-
optimized coding
sequences for an MG16 family enzyme.
[00136] SEQ ID NOs: 5676-5678 show peptide sequences characteristic of MG16
family
enzymes.
[00137] MG18
[00138] SEQ ID NOs: 1354-1511 show the full-length peptide sequences of MG18
nucleases.
[00139] SEQ ID NOs: 3175-3330 show the peptide sequences of RuvC III domains
of MG18
nucleases above.
[00140] SEQ ID NOs: 4989-5146 show the peptide of HNH domains of MG18
nucleases above.
[00141] SEQ ID NO: 5508 shows the nucleotide sequences of MG18 tracrRNA
derived from the
same loci as MG18 nucleases above.
[00142] SEQ ID NOs: 5472 shows the nucleotide sequence of an sgRNA engineered
to function
with an MG18 nuclease.
[00143] SEQ ID NO: 5584 shows a nucleotide sequence for an E. coil codon-
optimized coding
sequences for an MG18 family enzyme.
[00144] SEQ ID NOs: 5679-5686 show peptide sequences characteristic of MG18
family
enzymes.
[00145] MG21
[00146] SEQ ID NOs: 1512-1655 show the full-length peptide sequences of MG21
nucleases.
[00147] SEQ ID NOs: 3331-3474 show the peptide sequences of RuvC III domains
of MG21
nucleases above.
[00148] SEQ ID NOs: 5147-5290 show the peptide of HNH domains of MG21
nucleases above.
- 20 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00149] SEQ ID NOs: 5509 show the nucleotide sequence of an MG21 tracrRNA
derived from
the same loci as MG21 nucleases above.
[00150] SEQ ID NOs: 5473 shows the nucleotide sequence of an sgRNA engineered
to function
with an MG21 nuclease.
[00151] SEQ ID NO: 5585 shows a nucleotide sequence for an E. coil codon-
optimized coding
sequences for an MG21 family enzyme.
[00152] SEQ ID NOs: 5687-5692 and 5674-5675 show peptide sequences
characteristic of
MG21 family enzymes.
[00153] MG22
[00154] SEQ ID NOs: 1656-1755 show the full-length peptide sequences of MG22
nucleases.
[00155] SEQ ID NOs: 3475-3568 show the peptide sequences of RuvC III domains
of MG22
nucleases above.
[00156] SEQ ID NOs: 5291-5389 show the peptide of HNH domains of MG22
nucleases above.
[00157] SEQ ID NO: 5510 show the nucleotide sequence of an MG22 tracrRNA
derived from
the same loci as MG22 nucleases above.
[00158] SEQ ID NOs: 5474 shows the nucleotide sequence of an sgRNAs engineered
to function
with an MG22 nuclease.
[00159] SEQ ID NO: 5586 shows a nucleotide sequence for an E. coil codon-
optimized coding
sequences for an MG22 family enzyme.
[00160] SEQ ID NOs: 5694-5699 show peptide sequences characteristic of MG22
family
enzymes.
[00161] MG23
[00162] SEQ ID NOs: 1756-1826 show the full-length peptide sequences of MG23
nucleases.
[00163] SEQ ID NOs: 3569-3637 show the peptide sequences of RuvC III domains
of MG23
nucleases above.
[00164] SEQ ID NOs: 5390-5460 show the peptide of HNH domains of MG23
nucleases above.
[00165] SEQ ID NO: 5511 shows the nucleotide sequences of an MG23 tracrRNA
derived from
the same loci as MG23 nucleases above.
[00166] SEQ ID NOs: 5475 shows the nucleotide sequence of an sgRNA engineered
to function
with an MG23 nuclease.
[00167] SEQ ID NO: 5587 shows a nucleotide sequence for an E. coil codon-
optimized coding
sequences for an MG23 family enzyme.
[00168] SEQ ID NOs: 5700-5717 show peptide sequences characteristic of MG23
family
enzymes.
-21 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
DETAILED DESCRIPTION
[00169] While various embodiments of the invention have been shown and
described herein, it
will be obvious to those skilled in the art that such embodiments are provided
by way of example
only. Numerous variations, changes, and substitutions may occur to those
skilled in the art
without departing from the invention. It should be understood that various
alternatives to the
embodiments of the invention described herein may be employed.
[00170] The practice of some methods disclosed herein employ, unless otherwise
indicated,
techniques of immunology, biochemistry, chemistry, molecular biology,
microbiology, cell
biology, genomics and recombinant DNA. See for example Sambrook and Green,
Molecular
Cloning: A Laboratory Manual, 4th Edition (2012); the series Current Protocols
in Molecular
Biology (F. M. Ausubel, et al. eds.); the series Methods In Enzymology
(Academic Press, Inc.),
PCR 2: A Practical Approach (M.J. MacPherson, B.D. Hames and G.R. Taylor eds.
(1995)),
Harlow and Lane, eds. (1988) Antibodies, A Laboratory Manual, and Culture of
Animal Cells: A
Manual of Basic Technique and Specialized Applications, 6th Edition (R.I.
Freshney, ed. (2010))
(which is entirely incorporated by reference herein).
[00171] As used herein, the singular forms "a", "an" and "the" are intended to
include the plural
forms as well, unless the context clearly indicates otherwise. Furthermore, to
the extent that the
terms "including", "includes", "having", "has", "with", or variants thereof
are used in either the
detailed description and/or the claims, such terms are intended to be
inclusive in a manner similar
to the term "comprising".
[00172] The term "about" or "approximately" means within an acceptable error
range for the
particular value as determined by one of ordinary skill in the art, which will
depend in part on
how the value is measured or determined, i.e., the limitations of the
measurement system. For
example, "about" can mean within one or more than one standard deviation, per
the practice in
the art. Alternatively, "about" can mean a range of up to 20%, up to 15%, up
to 10%, up to 5%,
or up to 1% of a given value.
[00173] As used herein, a "cell" generally refers to a biological cell. A cell
may be the basic
structural, functional and/or biological unit of a living organism. A cell may
originate from any
organism having one or more cells. Some non-limiting examples include: a
prokaryotic cell,
eukaryotic cell, a bacterial cell, an archaeal cell, a cell of a single-cell
eukaryotic organism, a
protozoa cell, a cell from a plant (e.g., cells from plant crops, fruits,
vegetables, grains, soy bean,
corn, maize, wheat, seeds, tomatoes, rice, cassava, sugarcane, pumpkin, hay,
potatoes, cotton,
cannabis, tobacco, flowering plants, conifers, gymnosperms, ferns, clubmosses,
hornworts,
liverworts, mosses), an algal cell, (e.g.õ Botryococcus braunii, Chlamydomonas
reinhardtii,
Nannochloropsis gaditana, Chlorella pyrenoidosa, Sargassum patens C. Agardh,
and the like),
- 22 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
seaweeds (e.g., kelp), a fungal cell (e.g.õ a yeast cell, a cell from a
mushroom), an animal cell, a
cell from an invertebrate animal (e.g., fruit fly, cnidarian, echinoderm,
nematode, etc.), a cell
from a vertebrate animal (e.g., fish, amphibian, reptile, bird, mammal), a
cell from a mammal
(e.g., a pig, a cow, a goat, a sheep, a rodent, a rat, a mouse, a non-human
primate, a human, etc.),
and etcetera. Sometimes a cell is not originating from a natural organism
(e.g., a cell can be a
synthetically made, sometimes termed an artificial cell).
[00174] The term "nucleotide," as used herein, generally refers to a base-
sugar-phosphate
combination. A nucleotide may comprise a synthetic nucleotide. A nucleotide
may comprise a
synthetic nucleotide analog. Nucleotides may be monomeric units of a nucleic
acid sequence
(e.g., deoxyribonucleic acid (DNA) and ribonucleic acid (RNA)). The term
nucleotide may
include ribonucleoside triphosphates adenosine triphosphate (ATP), uridine
triphosphate (UTP),
cytosine triphosphate (CTP), guanosine triphosphate (GTP) and
deoxyribonucleoside
triphosphates such as dATP, dCTP, dITP, dUTP, dGTP, dTTP, or derivatives
thereof. Such
derivatives may include, for example, [aS]dATP, 7-deaza-dGTP and 7-deaza-dATP,
and
nucleotide derivatives that confer nuclease resistance on the nucleic acid
molecule containing
them. The term nucleotide as used herein may refer to dideoxyribonucleoside
triphosphates
(ddNTPs) and their derivatives. Illustrative examples of dideoxyribonucleoside
triphosphates
may include, but are not limited to, ddATP, ddCTP, ddGTP, ddITP, and ddTTP. A
nucleotide
may be unlabeled or detectably labeled, such as using moieties comprising
optically detectable
moieties (e.g., fluorophores). Labeling may also be carried out with quantum
dots. Detectable
labels may include, for example, radioactive isotopes, fluorescent labels,
chemiluminescent
labels, bioluminescent labels and enzyme labels. Fluorescent labels of
nucleotides may include
but are not limited fluorescein, 5-carboxyfluorescein (FAM), 217'-dimethoxy-
4'5-dichloro-6-
carboxyfluorescein (JOE), rhodamine, 6-carboxyrhodamine (R6G), N,N,N,N1-
tetramethy1-6-
carboxyrhodamine (TAMRA), 6-carboxy-X-rhodamine (ROX), 4-
(4'dimethylaminophenylazo)
benzoic acid (DABCYL), Cascade Blue, Oregon Green, Texas Red, Cyanine and 5-
(2'-
aminoethyl)aminonaphthalene-1-sulfonic acid (EDANS). Specific examples of
fluorescently
labeled nucleotides can include [R6G]dUTP, [TAMRA]dUTP, [R110]dCTP, [R6G]dCTP,
[TAMRA]dCTP, [JOE]ddATP, [R6G]ddATP, [FAM]ddCTP, [R110]ddCTP, [TAMRA]ddGTP,
[ROX]ddTTP, [dR6G]ddATP, [dR110]ddCTP, [dTAMRA]ddGTP, and [dROX]ddTTP
available
from Perkin Elmer, Foster City, Calif; FluoroLink DeoxyNucleotides, FluoroLink
Cy3-dCTP,
FluoroLink Cy5-dCTP, FluoroLink Fluor X-dCTP, FluoroLink Cy3-dUTP, and
FluoroLink Cy5-
dUTP available from Amersham, Arlington Heights, Ill.; Fluorescein-15-dATP,
Fluorescein-12-
dUTP, Tetramethyl-rodamine-6-dUTP, IR770-9-dATP, Fluorescein-12-ddUTP,
Fluorescein-12-
UTP, and Fluorescein-15-2'-dATP available from Boehringer Mannheim,
Indianapolis, Ind.; and
- 23 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
Chromosome Labeled Nucleotides, BODIPY-FL-14-UTP, BODIPY-FL-4-UTP, BODIPY-TMR-
14-UTP, BODIPY-TMR-14-dUTP, BODIPY-TR-14-UTP, BODIPY-TR-14-dUTP, Cascade
Blue-7-UTP, Cascade Blue-7-dUTP, fluorescein-12-UTP, fluorescein-12-dUTP,
Oregon Green
488-5-dUTP, Rhodamine Green-5-UTP, Rhodamine Green-5-dUTP,
tetramethylrhodamine-6-
UTP, tetramethylrhodamine-6-dUTP, Texas Red-5-UTP, Texas Red-5-dUTP, and Texas
Red-12-
dUTP available from Molecular Probes, Eugene, Oreg. Nucleotides can also be
labeled or
marked by chemical modification. A chemically-modified single nucleotide can
be biotin-dNTP.
Some non-limiting examples of biotinylated dNTPs can include, biotin-dATP
(e.g., bio-N6-
ddATP, biotin-14-dATP), biotin-dCTP (e.g., biotin-11-dCTP, biotin-14-dCTP),
and biotin-dUTP
(e.g., biotin-11-dUTP, biotin-16-dUTP, biotin-20-dUTP).
[00175] The terms "polynucleotide," "oligonucleotide," and "nucleic acid" are
used
interchangeably to generally refer to a polymeric form of nucleotides of any
length, either
deoxyribonucleotides or ribonucleotides, or analogs thereof, either in single-
, double-, or multi-
stranded form. A polynucleotide may be exogenous or endogenous to a cell. A
polynucleotide
may exist in a cell-free environment. A polynucleotide may be a gene or
fragment thereof. A
polynucleotide may be DNA. A polynucleotide may be RNA. A polynucleotide may
have any
three-dimensional structure and may perform any function. A polynucleotide may
comprise one
or more analogs (e.g., altered backbone, sugar, or nucleobase). If present,
modifications to the
nucleotide structure may be imparted before or after assembly of the polymer.
Some non-
limiting examples of analogs include: 5-bromouracil, peptide nucleic acid,
xeno nucleic acid,
morpholinos, locked nucleic acids, glycol nucleic acids, threose nucleic
acids,
dideoxynucleotides, cordycepin, 7-deaza-GTP, fluorophores (e.g., rhodamine or
fluorescein
linked to the sugar), thiol containing nucleotides, biotin linked nucleotides,
fluorescent base
analogs, CpG islands, methyl-7-guanosine, methylated nucleotides, inosine,
thiouridine,
pseudourdine, dihydrouridine, queuosine, and wyosine. Non-limiting examples of
polynucleotides include coding or non-coding regions of a gene or gene
fragment, loci (locus)
defined from linkage analysis, exons, introns, messenger RNA (mRNA), transfer
RNA (tRNA),
ribosomal RNA (rRNA), short interfering RNA (siRNA), short-hairpin RNA
(shRNA), micro-
RNA (miRNA), ribozymes, cDNA, recombinant polynucleotides, branched
polynucleotides,
plasmids, vectors, isolated DNA of any sequence, isolated RNA of any sequence,
cell-free
polynucleotides including cell-free DNA (cfDNA) and cell-free RNA (cfRNA),
nucleic acid
probes, and primers. The sequence of nucleotides may be interrupted by non-
nucleotide
components.
[00176] The terms "transfection" or "transfected" generally refer to
introduction of a nucleic acid
into a cell by non-viral or viral-based methods. The nucleic acid molecules
may be gene
- 24 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
sequences encoding complete proteins or functional portions thereof. See,
e.g., Sambrook et al.,
1989, Molecular Cloning: A Laboratory Manual, 18.1-18.88.
[00177] The terms "peptide," "polypeptide," and "protein" are used
interchangeably herein to
generally refer to a polymer of at least two amino acid residues joined by
peptide bond(s). This
term does not connote a specific length of polymer, nor is it intended to
imply or distinguish
whether the peptide is produced using recombinant techniques, chemical or
enzymatic synthesis,
or is naturally occurring. The terms apply to naturally occurring amino acid
polymers as well as
amino acid polymers comprising at least one modified amino acid. In some
cases, the polymer
may be interrupted by non-amino acids. The terms include amino acid chains of
any length,
including full length proteins, and proteins with or without secondary and/or
tertiary structure
(e.g., domains). The terms also encompass an amino acid polymer that has been
modified, for
example, by disulfide bond formation, glycosylation, lipidation, acetylation,
phosphorylation,
oxidation, and any other manipulation such as conjugation with a labeling
component. The terms
"amino acid" and "amino acids," as used herein, generally refer to natural and
non-natural amino
acids, including, but not limited to, modified amino acids and amino acid
analogues. Modified
amino acids may include natural amino acids and non-natural amino acids, which
have been
chemically modified to include a group or a chemical moiety not naturally
present on the amino
acid. Amino acid analogues may refer to amino acid derivatives. The term
"amino acid" includes
both D-amino acids and L-amino acids.
[00178] As used herein, the "non-native" can generally refer to a nucleic acid
or polypeptide
sequence that is not found in a native nucleic acid or protein. Non-native may
refer to affinity
tags. Non-native may refer to fusions. Non-native may refer to a naturally
occurring nucleic acid
or polypeptide sequence that comprises mutations, insertions and/or deletions.
A non-native
sequence may exhibit and/or encode for an activity (e.g., enzymatic activity,
methyltransferase
activity, acetyltransferase activity, kinase activity, ubiquitinating
activity, etc.) that may also be
exhibited by the nucleic acid and/or polypeptide sequence to which the non-
native sequence is
fused. A non-native nucleic acid or polypeptide sequence may be linked to a
naturally-
occurring nucleic acid or polypeptide sequence (or a variant thereof) by
genetic engineering to
generate a chimeric nucleic acid and/or polypeptide sequence encoding a
chimeric nucleic acid
and/or polypeptide.
[00179] The term "promoter", as used herein, generally refers to the
regulatory DNA region
which controls transcription or expression of a gene and which may be located
adjacent to or
overlapping a nucleotide or region of nucleotides at which RNA transcription
is initiated. A
promoter may contain specific DNA sequences which bind protein factors, often
referred to as
transcription factors, which facilitate binding of RNA polymerase to the DNA
leading to gene
- 25 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
transcription. A 'basal promoter', also referred to as a 'core promoter', may
generally refer to a
promoter that contains all the basic necessary elements to promote
transcriptional expression of
an operably linked polynucleotide. Eukaryotic basal promoters typically,
though not necessarily,
contain a TATA-box and/or a CAAT box.
[00180] The term "expression", as used herein, generally refers to the process
by which a nucleic
acid sequence or a polynucleotide is transcribed from a DNA template (such as
into mRNA or
other RNA transcript) and/or the process by which a transcribed mRNA is
subsequently
translated into peptides, polypeptides, or proteins. Transcripts and encoded
polypeptides may be
collectively referred to as "gene product." If the polynucleotide is derived
from genomic DNA,
expression may include splicing of the mRNA in a eukaryotic cell.
[00181] As used herein, "operably linked", "operable linkage", "operatively
linked", or
grammatical equivalents thereof generally refer to juxtaposition of genetic
elements, e.g., a
promoter, an enhancer, a polyadenylation sequence, etc., wherein the elements
are in a
relationship permitting them to operate in the expected manner. For instance,
a regulatory
element, which may comprise promoter and/or enhancer sequences, is operatively
linked to a
coding region if the regulatory element helps initiate transcription of the
coding sequence. There
may be intervening residues between the regulatory element and coding region
so long as this
functional relationship is maintained.
[00182] A "vector" as used herein, generally refers to a macromolecule or
association of
macromolecules that comprises or associates with a polynucleotide and which
may be used to
mediate delivery of the polynucleotide to a cell. Examples of vectors include
plasmids, viral
vectors, liposomes, and other gene delivery vehicles. The vector generally
comprises genetic
elements, e.g., regulatory elements, operatively linked to a gene to
facilitate expression of the
gene in a target.
[00183] As used herein, "an expression cassette" and "a nucleic acid cassette"
are used
interchangeably generally to refer to a combination of nucleic acid sequences
or elements that are
expressed together or are operably linked for expression. In some cases, an
expression cassette
refers to the combination of regulatory elements and a gene or genes to which
they are operably
linked for expression.
[00184] A "functional fragment" of a DNA or protein sequence generally refers
to a fragment
that retains a biological activity (either functional or structural) that is
substantially similar to a
biological activity of the full-length DNA or protein sequence. A biological
activity of a DNA
sequence may be its ability to influence expression in a manner known to be
attributed to the full-
length sequence.
[00185] As used herein, an "engineered" object generally indicates that the
object has been
- 26 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
modified by human intervention. According to non-limiting examples: a nucleic
acid may be
modified by changing its sequence to a sequence that does not occur in nature;
a nucleic acid
may be modified by ligating it to a nucleic acid that it does not associate
with in nature such that
the ligated product possesses a function not present in the original nucleic
acid; an engineered
nucleic acid may synthesized in vitro with a sequence that does not exist in
nature; a protein may
be modified by changing its amino acid sequence to a sequence that does not
exist in nature; an
engineered protein may acquire a new function or property. An "engineered"
system comprises at
least one engineered component.
[00186] As used herein, "synthetic" and "artificial" are used interchangeably
to refer to a protein
or a domain thereof that has low sequence identity (e.g., less than 50%
sequence identity, less
than 25% sequence identity, less than 10% sequence identity, less than 5%
sequence identity, less
than 1% sequence identity) to a naturally occurring human protein. For
example, VPR and VP64
domains are synthetic transactivation domains.
[00187] The term "tracrRNA" or "tracr sequence", as used herein, can generally
refer to a
nucleic acid with at least about 5%, 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%,
90%, 95%, or
100% sequence identity and/or sequence similarity to a wild type exemplary
tracrRNA sequence
(e.g., a tracrRNA from S. pyogenes S. aureus, etc or SEQ ID NOs: 5476-5511).
tracrRNA can
refer to a nucleic acid with at most about 5%, 10%, 20%, 30%, 40%, 50%, 60%,
70%, 80%,
90%, or 100% sequence identity and/or sequence similarity to a wild type
exemplary tracrRNA
sequence (e.g., a tracrRNA from S. pyogenes S. aureus, etc). tracrRNA may
refer to a modified
form of a tracrRNA that can comprise a nucleotide change such as a deletion,
insertion, or
substitution, variant, mutation, or chimera. A tracrRNA may refer to a nucleic
acid that can be at
least about 60% identical to a wild type exemplary tracrRNA (e.g., a tracrRNA
from S. pyogenes
S. aureus, etc) sequence over a stretch of at least 6 contiguous nucleotides.
For example, a
tracrRNA sequence can be at least about 60% identical, at least about 65%
identical, at least
about 70% identical, at least about 75% identical, at least about 80%
identical, at least about 85%
identical, at least about 90% identical, at least about 95% identical, at
least about 98% identical,
at least about 99% identical, or 100 % identical to a wild type exemplary
tracrRNA (e.g., a
tracrRNA from S. pyogenes S. aureus, etc) sequence over a stretch of at least
6 contiguous
nucleotides. Type II tracrRNA sequences can be predicted on a genome sequence
by identifying
regions with complementarity to part of the repeat sequence in an adjacent
CRISPR array.
[00188] As used herein, a "guide nucleic acid" can generally refer to a
nucleic acid that may
hybridize to another nucleic acid. A guide nucleic acid may be RNA. A guide
nucleic acid may
be DNA. The guide nucleic acid may be programmed to bind to a sequence of
nucleic acid site-
specifically. The nucleic acid to be targeted, or the target nucleic acid, may
comprise nucleotides.
- 27 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
The guide nucleic acid may comprise nucleotides. A portion of the target
nucleic acid may be
complementary to a portion of the guide nucleic acid. The strand of a double-
stranded target
polynucleotide that is complementary to and hybridizes with the guide nucleic
acid may be called
the complementary strand. The strand of the double-stranded target
polynucleotide that is
complementary to the complementary strand, and therefore may not be
complementary to the
guide nucleic acid may be called noncomplementary strand. A guide nucleic acid
may comprise a
polynucleotide chain and can be called a "single guide nucleic acid." A guide
nucleic acid may
comprise two polynucleotide chains and may be called a "double guide nucleic
acid." If not
otherwise specified, the term "guide nucleic acid" may be inclusive, referring
to both single
guide nucleic acids and double guide nucleic acids. A guide nucleic acid may
comprise a
segment that can be referred to as a "nucleic acid-targeting segment" or a
"nucleic acid-targeting
sequence." A nucleic acid-targeting segment may comprise a sub-segment that
may be referred to
as a "protein binding segment" or "protein binding sequence" or "Cas protein
binding segment".
[00189] The term "sequence identity" or "percent identity" in the context of
two or more nucleic
acids or polypeptide sequences, generally refers to two (e.g., in a pairwise
alignment) or more
(e.g., in a multiple sequence alignment) sequences that are the same or have a
specified
percentage of amino acid residues or nucleotides that are the same, when
compared and aligned
for maximum correspondence over a local or global comparison window, as
measured using a
sequence comparison algorithm. Suitable sequence comparison algorithms for
polypeptide
sequences include, e.g., BLASTP using parameters of a wordlength (W) of 3, an
expectation (E)
of 10, and the BLOSUM62 scoring matrix setting gap costs at existence of 11,
extension of 1,
and using a conditional compositional score matrix adjustment for polypeptide
sequences longer
than 30 residues; BLASTP using parameters of a wordlength (W) of 2, an
expectation (E) of
1000000, and the PAM30 scoring matrix setting gap costs at 9 to open gaps and
1 to extend gaps
for sequences of less than 30 residues (these are the default parameters for
BLASTP in the
BLAST suite available at https://blast.ncbi.nlm.nih.gov); CLUSTALW with
parameters of; the
Smith-Waterman homology search algorithm with parameters of a match of 2, a
mismatch of -1,
and a gap of -1; MUSCLE with default parameters; MAFFT with parameters retree
of 2 and
maxiterations of 1000; Novafold with default parameters; HMMER hmmalign with
default
parameters.
[00190] As used herein, the term "RuvC III domain" generally refers to a third
discontinuous
segment of a RuvC endonuclease domain (the RuvC nuclease domain being
comprised of three
discontiguous segments, RuvC I, RuvC II, and RuvC III). A RuvC domain or
segments thereof
can generally be identified by alignment to known domain sequences, structural
alignment to
proteins with annotated domains, or by comparison to Hidden Markov Models
(HMIMs) built
- 28 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
based on known domain sequences (e.g., Pfam HMM PF18541 for RuvC III).
[00191] As used herein, the term "HNH domain" generally refers to an
endonuclease domain
having characteristic histidine and asparagine residues. An HNH domain can
generally be
identified by alignment to known domain sequences, structural alignment to
proteins with
annotated domains, or by comparison to Hidden Markov Models (HMMs) built based
on known
domain sequences (e.g., Pfam HMIM PF01844 for domain HNH).
[00192] Overview
[00193] The discovery of new Cas enzymes with unique functionality and
structure may offer the
potential to further disrupt deoxyribonucleic acid (DNA) editing technologies,
improving speed,
specificity, functionality, and ease of use. Relative to the predicted
prevalence of Clustered
Regularly Interspaced Short Palindromic Repeats (CRISPR) systems in microbes
and the sheer
diversity of microbial species, relatively few functionally characterized
CRISPR/Cas enzymes
exist in the literature. This is partly because a huge number of microbial
species may not be
readily cultivated in laboratory conditions. Metagenomic sequencing from
natural
environmental niches that represent large numbers of microbial species may
offer the potential to
drastically increase the number of new CRISPR/Cas systems known and speed the
discovery of
new oligonucleotide editing functionalities. A recent example of the
fruitfulness of such an
approach is demonstrated by the 2016 discovery of CasX/CasY CRISPR systems
from
metagenomic analysis of natural microbial communities.
[00194] CRISPR/Cas systems are RNA-directed nuclease complexes that have been
described to
function as an adaptive immune system in microbes. In their natural context,
CRISPR/Cas
systems occur in CRISPR (clustered regularly interspaced short palindromic
repeats) operons or
loci, which generally comprise two parts: (i) an array of short repetitive
sequences (30-40bp)
separated by equally short spacer sequences, which encode the RNA-based
targeting element;
and (ii) ORFs encoding the Cas encoding the nuclease polypeptide directed by
the RNA-based
targeting element alongside accessory proteins/enzymes. Efficient nuclease
targeting of a
particular target nucleic acid sequence generally requires both (i)
complementary hybridization
between the first 6-8 nucleic acids of the target (the target seed) and the
crRNA guide; and (ii)
the presence of a protospacer-adjacent motif (PAM) sequence within a defined
vicinity of the
target seed (the PAM usually being a sequence not commonly represented within
the host
genome). Depending on the exact function and organization of the system,
CRISPR-Cas systems
are commonly organized into 2 classes, 5 types and 16 subtypes based on shared
functional
characteristics and evolutionary similarity.
[00195] Class I CRISPR-Cas systems have large, multisubunit effector
complexes, and comprise
Types I, III, and IV.
- 29 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00196] Type I CRISPR-Cas systems are considered of moderate complexity in
terms of
components. In Type I CRISPR-Cas systems, the array of RNA-targeting elements
is transcribed
as a long precursor crRNA (pre-crRNA) that is processed at repeat elements to
liberate short,
mature crRNAs that direct the nuclease complex to nucleic acid targets when
they are followed
by a suitable short consensus sequence called a protospacer-adjacent motif
(PAM). This
processing occurs via an endoribonuclease subunit (Cas6) of a large
endonuclease complex
called Cascade, which also comprises a nuclease (Cas3) protein component of
the crRNA-
directed nuclease complex. Cas I nucleases function primarily as DNA
nucleases.
[00197] Type III CRISPR systems may be characterized by the presence of a
central nuclease,
known as Cas10, alongside a repeat-associated mysterious protein (RAMP) that
comprises Csm
or Cmr protein subunits. Like in Type I systems, the mature crRNA is processed
from a pre-
crRNA using a Cas6-like enzyme. Unlike type I and II systems, type III systems
appear to target
and cleave DNA-RNA duplexes (such as DNA strands being used as templates for
an RNA
polymerase).
[00198] Type IV CRISPR-Cas systems possess an effector complex that consists
of a highly
reduced large subunit nuclease (csfl), two genes for RAMP proteins of the Cas5
(csf3) and Cas7
(csf2) groups, and, in some cases, a gene for a predicted small subunit; such
systems are
commonly found on endogenous plasmids.
[00199] Class II CRISPR-Cas systems generally have single-polypeptide
multidomain nuclease
effectors, and comprise Types II, V and VI.
[00200] Type II CRISPR-Cas systems are considered the simplest in terms of
components. In
Type II CRISPR-Cas systems, the processing of the CRISPR array into mature
crRNAs does not
require the presence of a special endonuclease subunit, but rather a small
trans-encoded crRNA
(tracrRNA) with a region complementary to the array repeat sequence; the
tracrRNA interacts
with both its corresponding effector nuclease (e.g. Cas9) and the repeat
sequence to form a
precursor dsRNA structure, which is cleaved by endogenous RNAse III to
generate a mature
effector enzyme loaded with both tracrRNA and crRNA. Cas II nucleases are
known as DNA
nucleases. Type 2 effectors generally exhibit a structure consisting of a RuvC-
like endonuclease
domain that adopts the RNase H fold with an unrelated HNH nuclease domain
inserted within the
folds of the RuvC-like nuclease domain. The RuvC-like domain is responsible
for the cleavage
of the target (e.g., crRNA complementary) DNA strand, while the HNH domain is
responsible
for cleavage of the displaced DNA strand.
[00201] Type V CRISPR-Cas systems are characterized by a nuclease effector
(e.g. Cas12)
structure similar to that of Type II effectors, comprising a RuvC-like domain.
Similar to Type II,
most (but not all) Type V CRISPR systems use a tracrRNA to process pre-crRNAs
into mature
- 30 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
crRNAs; however, unlike Type II systems which requires RNAse III to cleave the
pre-crRNA
into multiple crRNAs, type V systems are capable of using the effector
nuclease itself to cleave
pre-crRNAs. Like Type-II CRISPR-Cas systems, Type V CRISPR-Cas systems are
again known
as DNA nucleases. Unlike Type II CRISPR-Cas systems, some Type V enzymes
(e.g., Cas12a)
appear to have a robust single-stranded nonspecific deoxyribonuclease activity
that is activated
by the first crRNA directed cleavage of a double-stranded target sequence.
[00202] Type VI CRIPSR-Cas systems have RNA-guided RNA endonucleases. Instead
of
RuvC-like domains, the single polypeptide effector of Type VI systems (e.g.
Cas13) comprises
two HEPN ribonuclease domains. Differing from both Type II and V systems, Type
VI systems
also appear to not need a tracrRNA for processing of pre-crRNA into crRNA.
Similar to type V
systems, however, some Type VI systems (e.g., C2C2) appear to possess robust
single-stranded
nonspecific nuclease (ribonuclease) activity activated by the first crRNA
directed cleavage of a
target RNA.
[00203] Because of their simpler architecture, Class II CRISPR-Cas have been
most widely
adopted for engineering and development as designer nuclease/genome editing
applications.
[00204] One of the early adaptations of such a system for in vitro use can be
found in Jinek et al.
(Science. 2012 Aug 17;337(6096):816-21, which is entirely incorporated herein
by reference).
The Jinek study first described a system that involved (i) recombinantly-
expressed, purified full-
length Cas9 (e.g., a Class II, Type II Cas enzyme) isolated from S. pyogenes
SF370, (ii) purified
mature ¨42 nt crRNA bearing a ¨20 nt 5' sequence complementary to the target
DNA sequence
desired to be cleaved followed by a 3' tracr-binding sequence (the whole crRNA
being in vitro
transcribed from a synthetic DNA template carrying a T7 promoter sequence);
(iii) purified
tracrRNA in vitro transcribed from a synthetic DNA template carrying a T7
promoter sequence,
and (iv) Mg2+. Jinek later described an improved, engineered system wherein
the crRNA of (ii)
is joined to the 5' end of (iii) by a linker (e.g., GAAA) to form a single
fused synthetic guide
RNA (sgRNA) capable of directing Cas9 to a target by itself (compare top and
bottom panel of
FIGURE 2).
[00205] Mali et al. (Science. 2013 Feb 15; 339(6121): 823-826.), which is
entirely incorporated
herein by reference, later adapted this system for use in mammalian cells by
providing DNA
vectors encoding (i) an ORF encoding codon-optimized Cas9 (e.g., a Class II,
Type II Cas
enzyme) under a suitable mammalian promoter with a C-terminal nuclear
localization sequence
(e.g., 5V40 NLS) and a suitable polyadenylation signal (e.g., TK pA signal);
and (ii) an ORF
encoding an sgRNA (having a 5' sequence beginning with G followed by 20 nt of
a
complementary targeting nucleic acid sequence joined to a 3' tracr-binding
sequence, a linker,
and the tracrRNA sequence) under a suitable Polymerase III promoter (e.g., the
U6 promoter) .
-31-
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00206] MG1 Enzymes
[00207] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 1827-2140. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 1827-2140. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 1827-2140. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 1827-1831. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 1827-
1831. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 1827-1831. In some cases, the endonuclease may comprise a RuvC III
domain
having at least about 70%, at least about 75%, at least about 80%, at least
about 85%, at least
about 90%, at least about 91%, at least about 92%, at least about 93%, at
least about 94%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, at
least about 99% identity
to SEQ ID NO: 1827. In some cases, the endonuclease may comprise a RuvC III
domain having
at least about 70%, at least about 75%, at least about 80%, at least about
85%, at least about 90%,
at least about 91%, at least about 92%, at least about 93%, at least about
94%, at least about 95%,
at least about 96%, at least about 97%, at least about 98%, at least about 99%
identity to SEQ ID
NO: 1828. In some cases, the endonuclease may comprise a RuvC III domain
having at least
about 70%, at least about 75%, at least about 80%, at least about 85%, at
least about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, at least about 99% identity
to SEQ ID NO:
1829. In some cases, the endonuclease may comprise a RuvC III domain having at
least about
- 32 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
70%, at least about 7500, at least about 800 o, at least about 85%, at least
about 900 o, at least about
91%, at least about 92%, at least about 930, at least about 940, at least
about 950, at least about
96%, at least about 9'7%, at least about 98%, at least about 99 A identity to
SEQ ID NO: 1830. In
some cases, the endonuclease may comprise a RuvC III domain having at least
about 70%, at
least about 750, at least about 80%, at least about 85%, at least about 90%,
at least about 91%, at
least about 92%, at least about 930, at least about 940, at least about 950,
at least about 96%, at
least about 9'7%, at least about 98%, at least about 99 A identity to SEQ ID
NO: 1831.
[00208] The endonuclease may comprise an HNH domain having at least about 70 A
identity to
any one of SEQ ID NOs: 3638-3955. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 750, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 930,
at least about 940, at
least about 950, at least about 96%, at least about 970, at least about 98%,
or at least about 99 A
identical to any one of SEQ ID NOs: 3638-3955. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 3638-3955. The
endonuclease may
comprise an HNH domain having at least about 70 A identity to any one of SEQ
ID NOs: 3638-
3955. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 750, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 930, at least about 940, at least
about 950, at least about
96%, at least about 97%, at least about 98%, or at least about 99 A identical
to any one of SEQ
ID NOs: 3638-3955. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 3638-3955. The endonuclease may comprise an HNH domain
having
at least about 70 A identity to any one of SEQ ID NOs: 3638-3641. In some
cases, the
endonuclease may comprise an HNH domain having at least about 70%, at least
about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 930, at least about 940, at least about 950, at least about 96%,
at least about 970, at
least about 98%, or at least about 99 A identical to any one of SEQ ID NOs:
3638-3641. The
endonuclease may comprise an HNH domain substantially identical to any one of
SEQ ID NOs:
3638-3641. The endonuclease may comprise an HNH domain having at least about
70 A identity
to any one of SEQ ID NOs: 3638. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 750, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 930,
at least about 940, at
least about 950, at least about 96%, at least about 970, at least about 98%,
or at least about 990
identical to any one of SEQ ID NOs: 3638. The endonuclease may comprise an HNH
domain
substantially identical to any one of SEQ ID NOs: 3638. The endonuclease may
comprise an
HNH domain having at least about 70 A identity to any one of SEQ ID NOs: 3639.
In some
- 33 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
cases, the endonuclease may comprise an HNH domain having at least about 70%,
at least about
75%, at least about 80%, at least about 85%, at least about 90%, at least
about 91%, at least about
92%, at least about 93%, at least about 94%, at least about 95%, at least
about 96%, at least about
97%, at least about 98%, or at least about 99% identical to any one of SEQ ID
NOs: 3639. The
endonuclease may comprise an HNH domain substantially identical to any one of
SEQ ID NOs:
3639. The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 3640. In some cases, the endonuclease may comprise an
HNH domain
having at least about 70%, at least about 75%, at least about 80%, at least
about 85%, at least
about 90%, at least about 91%, at least about 92%, at least about 93%, at
least about 94%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
identical to any one of SEQ ID NOs: 3640. The endonuclease may comprise an HNH
domain
substantially identical to any one of SEQ ID NOs: 3640. The endonuclease may
comprise an
HNH domain having at least about 70% identity to any one of SEQ ID NOs: 3641.
In some
cases, the endonuclease may comprise an HNH domain having at least about 70%,
at least about
75%, at least about 80%, at least about 85%, at least about 90%, at least
about 91%, at least about
92%, at least about 93%, at least about 94%, at least about 95%, at least
about 96%, at least about
97%, at least about 98%, or at least about 99% identical to any one of SEQ ID
NOs: 3641. The
endonuclease may comprise an HNH domain substantially identical to any one of
SEQ ID NOs:
3641.
[00209] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 1-6
or 9-319. In
some cases, the endonuclease may be substantially identical to any one of SEQ
ID NOs: 1-6 or 9-
319. In some cases, the endonuclease may comprise a variant having at least
about 30%, at least
about 35%, at least about 40%, at least about 45%, at least about 50%, at
least about 55%, at least
about 60%, at least about 65%, at least about 70%, at least about 75%, at
least about 80%, at least
about 85%, at least about 90%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to any one of SEQ ID NOs:1-4. In
some cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 1-4. In
some cases, the
endonuclease may comprise a peptide motif substantially identical to any one
of SEQ ID NOs:
5615, 5616, or 5617.
[00210] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
- 34 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
1-6 or 9-319, or to a variant having at least about 30%, at least about 35%,
at least about 40%, at
least about 45%, at least about 50%, at least about 55%, at least about 60%,
at least about 65%, at
least about 70%, at least about 75%, at least about 80%, at least about 85%,
at least about 90%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identity to any one of SEQ ID NOs: 1-319. The NLS may be an 5V40 large T
antigen NLS. The
NLS may be a c-myc NLS. The NLS can comprise a sequence with at least about
80%, at least
about 85%, at least about 90%, at least about 95%, at least about 99% identity
to any one of SEQ
ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical to
any one of
SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
below, or a
combination thereof:
[00211] Table 1: Example NLS Sequences that can be used with Cas Effectors
According to
the Disclosure
Source NLS amino acid sequence
SEQ ID NO:
SV40 PKKKRKV 5593
nucleoplasmin bipartite NLS KRPAATKKAGQAKKKK 5594
c-myc NLS PAAKRVKLD 5595
c-iny-c NLS RQRRNELKRSP 5596
hRNPA1 M9 NLS
NQSSNFGPMKGGNFGGRSSGPYGGGGQYFA 5597
KPRNQGCN
.Importin-alpha IBB domain RMRIZFKNKGKDTAELRRRRVEVSVELRKAK :5598
KDEQILKRRNV
Myoma T protein VSRKRPRP 5599
Myoma T protein PPKK.ARED 5600
P53 PQPKKKPL 5601
mouse c-abl IV SALIKKKKKMAP 5602
influenza virus NS1 DRLRR 5603
influenza virus NS1 PKQKKRK 5604
Hepatitis virus delta antigen RKLKKKIKKL 5605
mouse Mx l protein REKKKFLKRR 5606
human poly(ADP-ribose) KRKGDEVDGVDEVAKKKSKK 5607
polymerase
steroid hormone receptor RK.CLQAGMNLEARKTKK 5608
(human) glucocorticoid
[00212] In some cases, the endonuclease may be recombinant (e.g., cloned,
expressed, and
purified by a suitable method such as expression in E. coli followed by
epitope-tag purification).
In some cases, the endonuclease may be derived from a bacterium with a 16S
rRNA gene having
at least about 90% identity to any one of SEQ ID NOs: 5592-5595. The
endonuclease may be
- 35 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
derived from a species having a 16S rRNA gene at least about 80%, at least
about 82%, at least
about 83%, at least about 84%, at least about 85%, at least about 86%, at
least about 87%, at least
about 88%, at least about 89%, at least about 90%, at least about 91%, at
least about 92%, at least
about 9300, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 990 identity to any one of SEQ ID NOs: 5592-5595.
The
endonuclease may be derived from a species having a 16S rRNA gene
substantially identical to
any one of SEQ ID NOs: 5592-5595. The endonuclease may be derived from a
bacterium
belonging to the Phylum Verrucomicrobia or the Phylum Candidatus
Peregrinibacteria.
[00213] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00214] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00215] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80 A to at least about 60-100 (e.g., at least about
60, at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of any one of SEQ ID NOs: 5476-5489. In some cases,
the tracrRNA
may have at least about 80%, at least about 85%, at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, or at least about 99 A identity to at least
about 60-90 (e.g., at least
- 36 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
about 60, at least about 65, at least about 70, at least about 75, at least
about 80, at least about 85,
or at least about 90) consecutive nucleotides of any one of SEQ ID NOs: 5476-
5489. In some
cases, the tracrRNA may be substantially identical to at least about 60-100
(e.g., at least about
60, at least about 65, at least about 70, at least about 75, at least about
80, at least about 85, or at
least about 90) consecutive nucleotides of any one of SEQ ID NOs: 5476-5489.
The tracrRNA
may comprise any of SEQ ID NOs: 5476-5489.
[00216] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to any one of SEQ ID NOs: 5461-5464. The sgRNA may comprise
a
sequence having at least about 80%, at least about 85%, at least about 90%, at
least about 91%, at
least about 92%, at least about 93%, at least about 94%, at least about 95%,
at least about 96%, at
least about 97%, at least about 98%, or at least about 99% identity to any one
of SEQ ID NOs:
5461-5464. The sgRNA may comprise a sequence substantially identical to any
one of SEQ ID
NOs: 5461-5464.
[00217] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00218] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus. The method may comprise delivering to the target nucleic
acid locus any of
the non-natural systems disclosed herein, including an enzyme and at least one
synthetic guide
RNA (sgRNA) disclosed herein. The enzyme may form a complex with the at least
one sgRNA,
and upon binding of the complex to the target nucleic acid locus, may modify
the target nucleic
acid locus. Delivering the enzyme to said locus may comprise transfecting a
cell with the system
or nucleic acids encoding the system. Delivering the nuclease to said locus
may comprise
electroporating a cell with the system or nucleic acids encoding the system.
Delivering the
nuclease to said locus may comprise incubating the system in a buffer with a
nucleic acid
comprising the locus of interest. In some cases, the target nucleic acid locus
comprises
deoxyribonucleic acid (DNA) or ribonucleic acid (RNA). The target nucleic acid
locus may
comprise genomic DNA, viral DNA, viral RNA, or bacterial DNA. The target
nucleic acid locus
may be within a cell. The target nucleic acid locus may be in vitro. The
target nucleic acid locus
- 37 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
may be within a eukaryotic cell or a prokaryotic cell. The cell may be an
animal cell, a human
cell, bacterial cell, archaeal cell, or a plant cell. The enzyme may induce a
single or double-
stranded break at or proximal to the target locus of interest.
[00219] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 1827-
2140. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to any of SEQ ID
NOs: 5572-5575
or at variant having at least about 30%, at least about 35%, at least about
40%, at least about
45%, at least about 50%, at least about 55%, at least about 60%, at least
about 65%, at least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
95%, at least about 96%, at least about 97%, at least about 98%, or at least
about 99% identity to
any one of SEQ ID NOs: 5572-5575. In some cases, the nucleic acid comprises a
promoter to
which the open reading frame encoding the endonuclease is operably linked. The
promoter may
be a CMV, EFla, 5V40, PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa
promoter. The
endonuclease may be supplied as a capped mRNA containing said open reading
frame encoding
said endonuclease. The endonuclease may be supplied as a translated
polypeptide. The at least
one engineered sgRNA may be supplied as deoxyribonucleic acid (DNA) containing
a gene
sequence encoding said at least one engineered sgRNA operably linked to a
ribonucleic acid
(RNA) pol III promoter. In some cases, the organism may be eukaryotic. In some
cases, the
organism may be fungal. In some cases, the organism may be human.
[00220] In some cases, the present disclosure may provide for an expression
cassette comprising
the system disclosed herein, or the nucleic acid described herein. In some
cases, the expression
cassette or nucleic acid may be supplied as a vector. In some cases, the
expression cassette,
nucleic acid, or vector may be supplied in a cell. In some cases, the cell is
a cell of a bacterium
with a 16S rRNA gene having at least about 90% (e.g., at least about 99%)
identity to any one of
SEQ ID NOs: 5592-5595.
[00221] MG2 Enzymes
[00222] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2141-2241. In some cases, the endonuclease may comprise
a RuvC III
- 38 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
domain, wherein the RuvC III domain has at least about 20%, at least about 250
o, at least about
30%, at least about 3500, at least about 40%, at least about 45%, at least
about 50%, at least about
550, at least about 60%, at least about 65%, at least about 70%, at least
about 750, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
930, at least about 940, at least about 950, at least about 96%, at least
about 970, at least about
98%, at least about 990 identity to any one of SEQ ID NOs: 2141-2241. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2141-2142. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 2141-2142. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99 A identity to any one of SEQ ID NOs: 2141-
2142. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 2141-2142.
[00223] The endonuclease may comprise an HNH domain having at least about 70 A
identity to
any one of SEQ ID NOs: 3955-4055. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99 A
identical to any one of SEQ ID NOs: 3955-4055. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 3955-4055. The
endonuclease may
comprise an HNH domain having at least about 70 A identity to any one of SEQ
ID NOs: 3955-
3956. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99 A identical
to any one of SEQ
ID NOs: 3955-3956. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 3955-3956.
[00224] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
- 39 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 320-
420. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 320-420. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs:320-321. In some
cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 320-321.
[00225] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
320-420, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at least
about 45%, at least about 50%, at least about 55%, at least about 60%, at
least about 65%, at least
about 70%, at least about 75%, at least about 80%, at least about 85%, at
least about 90%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
identity to any one of SEQ ID NOs: 320-420. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00226] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00227] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
- 40 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00228] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of any one of SEQ ID NOs: 5490-5494. In some cases,
the tracrRNA
may have at least about 80%, at least about 85%, at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, or at least about 99% identity to at least
about 60-90 (e.g., at least
about 60, at least about 65, at least about 70, at least about 75, at least
about 80, at least about 85,
or at least about 90) consecutive nucleotides of any one of SEQ ID NOs: 5490-
5494. In some
cases, the tracrRNA may be substantially identical to at least about 60-100
(e.g., at least about
60, at least about 65, at least about 70, at least about 75, at least about
80, at least about 85, or at
least about 90) consecutive nucleotides of any one of SEQ ID NOs: 5490-5494.
The tracrRNA
may comprise any of SEQ ID NOs: 5490-5494.
[00229] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5465. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5465. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5465.
[00230] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
-41 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00231] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00232] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2141-
2241. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to any of SEQ ID
NOs: 5576-5577
or at variant having at least about 30%, at least about 35%, at least about
40%, at least about
45%, at least about 50%, at least about 55%, at least about 60%, at least
about 65%, at least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
95%, at least about 96%, at least about 97%, at least about 98%, or at least
about 99% identity to
any one of SEQ ID NOs: 5576-5577. In some cases, the nucleic acid comprises a
promoter to
which the open reading frame encoding the endonuclease is operably linked. The
promoter may
be a CMV, EFla, 5V40, PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa
promoter. The
endonuclease may be supplied as a capped mRNA containing said open reading
frame encoding
said endonuclease. The endonuclease may be supplied as a translated
polypeptide. The at least
one engineered sgRNA may be supplied as deoxyribonucleic acid (DNA) containing
a gene
sequence encoding said at least one engineered sgRNA operably linked to a
ribonucleic acid
- 42 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
(RNA) pol III promoter. In some cases, the organism may be eukaryotic. In some
cases, the
organism may be fungal. In some cases, the organism may be human.
[00233] MG3 Enzymes
[00234] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2242-2251. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 2242-2251. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2242-2251. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 2242-2244. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 2242-
2244. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 2242-2244.
[00235] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 4056-4066. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 4056-4066. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4056-4066. The
endonuclease may
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 4056-
4058. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
- 43 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
ID NOs: 4056-4058. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 4056-4058.
[00236] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 421-
431. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 421-431. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs:421-423. In some
cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 421-423.
[00237] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
421-431, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at least
about 45%, at least about 50%, at least about 55%, at least about 60%, at
least about 65%, at least
about 70%, at least about 75%, at least about 80%, at least about 85%, at
least about 90%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
identity to any one of SEQ ID NOs: 421-431. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00238] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
- 44 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00239] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00240] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of any one of SEQ ID NOs: 5495-5502. In some cases,
the tracrRNA
may have at least about 80%, at least about 85%, at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, or at least about 99% identity to at least
about 60-90 (e.g., at least
about 60, at least about 65, at least about 70, at least about 75, at least
about 80, at least about 85,
or at least about 90) consecutive nucleotides of any one of SEQ ID NOs: 5495-
5502. In some
cases, the tracrRNA may be substantially identical to at least about 60-100
(e.g., at least about
60, at least about 65, at least about 70, at least about 75, at least about
80, at least about 85, or at
least about 90) consecutive nucleotides of any one of SEQ ID NOs: 5495-5502.
The tracrRNA
may comprise any of SEQ ID NOs: 5495-5502.
[00241] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to any one of SEQ ID NOs: 5466-5467. The sgRNA may comprise
a
sequence having at least about 80%, at least about 85%, at least about 90%, at
least about 91%, at
least about 92%, at least about 93%, at least about 94%, at least about 95%,
at least about 96%, at
least about 97%, at least about 98%, or at least about 99% identity to any one
of SEQ ID NOs:
- 45 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
5466-5467. The sgRNA may comprise a sequence substantially identical to any
one of SEQ ID
NOs: 5466-5467.
[00242] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00243] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00244] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2242-
2251. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to any of SEQ ID
NOs: 5578-5580
or at variant having at least about 30%, at least about 35%, at least about
40%, at least about
45%, at least about 50%, at least about 55%, at least about 60%, at least
about 65%, at least about
- 46 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
95%, at least about 96%, at least about 97%, at least about 98%, or at least
about 99% identity to
any one of SEQ ID NOs: 5578-5580. In some cases, the nucleic acid comprises a
promoter to
which the open reading frame encoding the endonuclease is operably linked. The
promoter may
be a CMV, EFla, 5V40, PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa
promoter. The
endonuclease may be supplied as a capped mRNA containing said open reading
frame encoding
said endonuclease. The endonuclease may be supplied as a translated
polypeptide. The at least
one engineered sgRNA may be supplied as deoxyribonucleic acid (DNA) containing
a gene
sequence encoding said at least one engineered sgRNA operably linked to a
ribonucleic acid
(RNA) pol III promoter. In some cases, the organism may be eukaryotic. In some
cases, the
organism may be fungal. In some cases, the organism may be human.
[00245] MG4 Enzymes
[00246] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2253-2481. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 2253-2481. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2253-2481. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 2253-2481. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 2253-
2481. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 2253-2481.
- 47 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00247] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 4067-4295. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 4067-4295. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4067-4295. The
endonuclease may
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 4067-
4295. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
ID NOs: 4067-4295. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 4067-4295.
[00248] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 432-
660. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 432-660. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs: 432-660. In some
cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 432-660.
[00249] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
432-660, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at least
about 45%, at least about 50%, at least about 55%, at least about 60%, at
least about 65%, at least
about 70%, at least about 75%, at least about 80%, at least about 85%, at
least about 90%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
identity to any one of SEQ ID NOs: 432-660. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
- 48 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00250] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00251] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00252] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5503. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5503. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
- 49 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5503. The tracrRNA may comprise SEQ ID NO: 5503.
[00253] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5468. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5468. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5468.
[00254] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00255] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00256] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
- 50 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2253-
2481. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00257] MG6 Enzymes
[00258] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2482-2489. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 2482-2489. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2482-2489.
[00259] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 4296-4303. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 4296-4303. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4056-4066.
[00260] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
-51 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
least about 60%, at least about 65%, at least about 700 o, at least about
7500, at least about 800 o, at
least about 85%, at least about 90%, at least about 950, at least about 96%,
at least about 970, at
least about 98%, or at least about 990 identity to any one of SEQ ID NOs: 661-
668. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 661-668.
[00261] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
661-668, or to a variant having at least about 30%, at least about 350, at
least about 40%, at least
about 450, at least about 50%, at least about 550, at least about 60%, at
least about 65%, at least
about 70%, at least about 750, at least about 80%, at least about 85%, at
least about 90%, at least
about 950, at least about 96%, at least about 970, at least about 98%, or at
least about 99 A
identity to any one of SEQ ID NOs: 661-668. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 950, at least about 99 A
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00262] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00263] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
- 52 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00264] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence.
[00265] In some cases, the system above may comprise two different guide RNAs
targeting a
first region and a second region for cleavage in a target DNA locus, wherein
the second region is
3' to the first region. In some cases, the system above may comprise a single-
or double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00266] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00267] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2482-
2489. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
- 53 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00268] MG7 Enzymes
[00269] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2490-2498. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 2490-2498. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2490-2498. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 2490-2498. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 2490-
2498. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 2490-2498.
[00270] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 4304-4312. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
- 54 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
identical to any one of SEQ ID NOs: 4304-4312. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4304-4312. The
endonuclease may
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 4304-
4312. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
ID NOs: 4304-4312. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 4304-4312.
[00271] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 669-
677. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 669-677. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs: 669-677. In some
cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 669-677.
[00272] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
669-677, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at least
about 45%, at least about 50%, at least about 55%, at least about 60%, at
least about 65%, at least
about 70%, at least about 75%, at least about 80%, at least about 85%, at
least about 90%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
identity to any one of SEQ ID NOs: 669-677. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
- 55 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00273] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00274] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00275] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5504. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5504. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5504. The tracrRNA may comprise SEQ ID NO: 5504.
[00276] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
- 56 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00277] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00278] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2490-
2498. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
- 57 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00279] MG14 Enzymes
[00280] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2499-2750. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 2499-2750. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2499-2750. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 2499-2750. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 2499-
2750. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 2499-2750.
[00281] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 4313-4564. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 4313-4564. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4313-4564. The
endonuclease may
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 4313-
4564. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
- 58 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
ID NOs: 4067-4295. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 4313-4564.
[00282] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 678-
929. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 678-929. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs: 678-929. In some
cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 678-929.
[00283] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
678-929, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at least
about 45%, at least about 50%, at least about 55%, at least about 60%, at
least about 65%, at least
about 70%, at least about 75%, at least about 80%, at least about 85%, at
least about 90%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
identity to any one of SEQ ID NOs: 678-929. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00284] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00285] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
- 59 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00286] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5505. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5505. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5505. The tracrRNA may comprise SEQ ID NO: 5505.
[00287] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5469. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5469. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5469.
[00288] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
- 60 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00289] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00290] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2499-
2750. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to SEQ ID NO:
5581 or at variant
having at least about 30%, at least about 35%, at least about 40%, at least
about 45%, at least
about 50%, at least about 55%, at least about 60%, at least about 65%, at
least about 70%, at least
about 75%, at least about 80%, at least about 85%, at least about 90%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
identity to SEQ ID NO:
5581. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
- 61 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00291] MG15 Enzymes
[00292] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2751-2913. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 2751-2913. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2751-2913. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 2751-2913. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 2751-
2913. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 2751-2913.
[00293] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 4565-4727. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 4565-4727. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4565-4727. The
endonuclease may
- 62 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 4565-
4727. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
ID NOs: 4565-4727. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 4565-4727.
[00294] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 930-
1092. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 930-1092. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs: 930-1092. In
some cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 930-
1092.
[00295] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
930-1092, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at
least about 45%, at least about 50%, at least about 55%, at least about 60%,
at least about 65%, at
least about 70%, at least about 75%, at least about 80%, at least about 85%,
at least about 90%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identity to any one of SEQ ID NOs: 930-1092. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00296] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
- 63 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00297] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00298] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5506. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5506. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5506. The tracrRNA may comprise SEQ ID NO: 5506.
[00299] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5470. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
- 64 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
about 98%, or at least about 99% identity to SEQ ID NO: 5470. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5470.
[00300] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00301] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00302] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2751-
2913. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to SEQ ID NO:
5582 or at variant
having at least about 30%, at least about 35%, at least about 40%, at least
about 45%, at least
about 50%, at least about 55%, at least about 60%, at least about 65%, at
least about 70%, at least
- 65 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
about 75%, at least about 80%, at least about 85%, at least about 90%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
identity to SEQ ID NO:
5582. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00303] MG16 Enzymes
[00304] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 2914-3174. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 2914-3174. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 2914-3174. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 2914-3174. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 2914-
3174. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 2914-3174.
- 66 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00305] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 4728-4988. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 4728-4988. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4728-4988. The
endonuclease may
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 4728-
4988. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
ID NOs: 4728-4988. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 4728-4988.
[00306] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 1093-
1353. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 1093-1353. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs: 1093-1353. In
some cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 1093-
1353.
[00307] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
1093-1353, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at
least about 45%, at least about 50%, at least about 55%, at least about 60%,
at least about 65%, at
least about 70%, at least about 75%, at least about 80%, at least about 85%,
at least about 90%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identity to any one of SEQ ID NOs: 1093-1353. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
- 67 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00308] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00309] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00310] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5507. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5507. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
- 68 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5507. The tracrRNA may comprise SEQ ID NO: 5507.
[00311] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5471. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5471. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5471.
[00312] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00313] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00314] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
- 69 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 2914-
3174. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to SEQ ID NO:
5583 or at variant
having at least about 30%, at least about 35%, at least about 40%, at least
about 45%, at least
about 50%, at least about 55%, at least about 60%, at least about 65%, at
least about 70%, at least
about 75%, at least about 80%, at least about 85%, at least about 90%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
identity to SEQ ID NO:
5583. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00315] MG18 Enzymes
[00316] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 3175-3300. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 3175-3300. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 3175-3300. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 3175-3300. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
- 70 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
least about 5500, at least about 600 o, at least about 65%, at least about 700
o, at least about 750, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 930, at least about 9400, at least about 9500, at least about 96%,
at least about 970, at
least about 98%, at least about 990 identity to any one of SEQ ID NOs: 3175-
3300. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 3175-3300.
[00317] The endonuclease may comprise an HNH domain having at least about 70 A
identity to
any one of SEQ ID NOs: 4989-5146. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 750, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 930,
at least about 940, at
least about 950, at least about 96%, at least about 970, at least about 98%,
or at least about 99 A
identical to any one of SEQ ID NOs: 4989-5146. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 4989-5146. The
endonuclease may
comprise an HNH domain having at least about 70 A identity to any one of SEQ
ID NOs: 4989-
5146. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 750, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 930, at least about 940, at least
about 950, at least about
96%, at least about 97%, at least about 98%, or at least about 99 A identical
to any one of SEQ
ID NOs: 4989-5146. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 4989-5146.
[00318] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 350, at least about 40%, at least about 450, at least about 50%,
at least about 550, at
least about 60%, at least about 65%, at least about 70%, at least about 750,
at least about 80%, at
least about 85%, at least about 90%, at least about 950, at least about 96%,
at least about 970, at
least about 98%, or at least about 99 A identity to any one of SEQ ID NOs:
1354-1511. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 1354-1511. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
350, at least about 40%, at least about 450, at least about 50%, at least
about 550, at least about
60%, at least about 65%, at least about 70%, at least about 750, at least
about 80%, at least about
85%, at least about 90%, at least about 950, at least about 96%, at least
about 970, at least about
or at least about 99 A identity to any one of SEQ ID NOs: 1354-1511. In some
cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 1354-
1511.
[00319] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
- 71 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
1354-1511, or to a variant having at least about 30%, at least about 3500, at
least about 400 o, at
least about 450, at least about 5000, at least about 550, at least about 60%,
at least about 65%, at
least about 70%, at least about 750, at least about 80%, at least about 85%,
at least about 90%, at
least about 950, at least about 96%, at least about 970, at least about 98%,
or at least about 990
identity to any one of SEQ ID NOs: 1354-1511. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 950, at least about 99 A
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00320] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00321] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00322] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80 A to at least about 60-100 (e.g., at least about
60, at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5508. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
- 72 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5508. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5508. The tracrRNA may comprise SEQ ID NO: 5508.
[00323] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5472. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5472. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5472.
[00324] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00325] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
- 73 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00326] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 3175-
3300. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to SEQ ID NOs:
5584 or at variant
having at least about 30%, at least about 35%, at least about 40%, at least
about 45%, at least
about 50%, at least about 55%, at least about 60%, at least about 65%, at
least about 70%, at least
about 75%, at least about 80%, at least about 85%, at least about 90%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
identity to SEQ ID NOs:
5584. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00327] MG21 Enzymes
[00328] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 3331-3474. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 3331-3474. In some
cases, the
- 74 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 3331-3474. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 3331-3474. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 3331-
3474. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 3331-3474.
[00329] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 5147-5290. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 5147-5290. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 5147-5290. The
endonuclease may
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 5147-
5290. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
ID NOs: 5147-5290. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 5147-5290.
[00330] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 1512-
1655. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 1512-1655. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
- 75 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
98%, or at least about 99% identity to any one of SEQ ID NOs: 1512-1655. In
some cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 1512-
1655.
[00331] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
1512-1655, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at
least about 45%, at least about 50%, at least about 55%, at least about 60%,
at least about 65%, at
least about 70%, at least about 75%, at least about 80%, at least about 85%,
at least about 90%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identity to any one of SEQ ID NOs: 1512-1655. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00332] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00333] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00334] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
- 76 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5509. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5509. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5509. The tracrRNA may comprise SEQ ID NO: 5509.
[00335] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5473. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5473. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5473.
[00336] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00337] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
- 77 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00338] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 3331-
3474. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to SEQ ID NOs:
5585 or at variant
having at least about 30%, at least about 35%, at least about 40%, at least
about 45%, at least
about 50%, at least about 55%, at least about 60%, at least about 65%, at
least about 70%, at least
about 75%, at least about 80%, at least about 85%, at least about 90%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
identity to SEQ ID NOs:
5585. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00339] MG22 Enzymes
[00340] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 3475-3568. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
- 78 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
30%, at least about 3500, at least about 400 o, at least about 450, at least
about 500 o, at least about
550, at least about 60%, at least about 65%, at least about 70%, at least
about 750, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
930, at least about 940, at least about 950, at least about 96%, at least
about 970, at least about
98%, at least about 990 identity to any one of SEQ ID NOs: 3475-3568. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 3475-3568. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 3475-3568. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 350, at least about 40%, at least about 450,
at least about 50%, at
least about 550, at least about 60%, at least about 65%, at least about 70%,
at least about 750, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 930, at least about 940, at least about 950, at least about 96%,
at least about 970, at
least about 98%, at least about 99 A identity to any one of SEQ ID NOs: 3475-
3568. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 3475-3568.
[00341] The endonuclease may comprise an HNH domain having at least about 70 A
identity to
any one of SEQ ID NOs: 5291-5389. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 750, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 930,
at least about 940, at
least about 950, at least about 96%, at least about 970, at least about 98%,
or at least about 99 A
identical to any one of SEQ ID NOs: 5291-5389. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 5291-5389. The
endonuclease may
comprise an HNH domain having at least about 70 A identity to any one of SEQ
ID NOs: 5291-
5389. In some cases, the endonuclease may comprise an HNH domain having at
least about
700o, at least about 750, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 930, at least about 940, at least
about 950, at least about
96%, at least about 97%, at least about 98%, or at least about 99 A identical
to any one of SEQ
ID NOs: 5291-5389. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 5291-5389.
[00342] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 350, at least about 40%, at least about 450, at least about 50%,
at least about 550, at
least about 60%, at least about 65%, at least about 70%, at least about 750,
at least about 80%, at
least about 85%, at least about 90%, at least about 950, at least about 96%,
at least about 970, at
least about 98%, or at least about 99 A identity to any one of SEQ ID NOs:
1656-1755. In some
- 79 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 1656-1755. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs: 1656-1755. In
some cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 1656-
1755.
[00343] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
432-660, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at least
about 45%, at least about 50%, at least about 55%, at least about 60%, at
least about 65%, at least
about 70%, at least about 75%, at least about 80%, at least about 85%, at
least about 90%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
identity to any one of SEQ ID NOs: 1656-1755. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00344] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00345] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
- 80 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00346] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5510. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5510. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5510. The tracrRNA may comprise SEQ ID NO: 5510.
[00347] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5474. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5474. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5474.
[00348] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00349] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
- 81 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00350] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 3475-
3568. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to SEQ ID NOs:
5586 or at variant
having at least about 30%, at least about 35%, at least about 40%, at least
about 45%, at least
about 50%, at least about 55%, at least about 60%, at least about 65%, at
least about 70%, at least
about 75%, at least about 80%, at least about 85%, at least about 90%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
identity to SEQ ID NOs:
5586. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
[00351] MG23 Enzymes
- 82 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00352] In one aspect, the present disclosure provides for an engineered
nuclease system
comprising (a) an endonuclease. In some cases, the endonuclease is a Cas
endonuclease. In some
cases, the endonuclease is a Type II, Class II Cas endonuclease. The
endonuclease may comprise
a RuvC III domain, wherein said RuvC III domain has at least about 70%
sequence identity to
any one of SEQ ID NOs: 3569-3637. In some cases, the endonuclease may comprise
a RuvC III
domain, wherein the RuvC III domain has at least about 20%, at least about
25%, at least about
30%, at least about 35%, at least about 40%, at least about 45%, at least
about 50%, at least about
55%, at least about 60%, at least about 65%, at least about 70%, at least
about 75%, at least about
80%, at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, at least about 99% identity to any one of SEQ ID NOs: 3569-3637. In some
cases, the
endonuclease may comprise a RuvC III domain, wherein the substantially
identical to any one
of SEQ ID NOs: 3569-3637. The endonuclease may comprise a RuvC III domain
having at least
about 70% sequence identity to any one of SEQ ID NOs: 3569-3637. In some
cases, the
endonuclease may comprise a RuvC III domain having at least about 20%, at
least about 25%, at
least about 30%, at least about 35%, at least about 40%, at least about 45%,
at least about 50%, at
least about 55%, at least about 60%, at least about 65%, at least about 70%,
at least about 75%, at
least about 80%, at least about 85%, at least about 90%, at least about 91%,
at least about 92%, at
least about 93%, at least about 94%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, at least about 99% identity to any one of SEQ ID NOs: 3569-
3637. In some
cases, the endonuclease may comprise a RuvC III domain substantially identical
to any one of
SEQ ID NOs: 3569-3637.
[00353] The endonuclease may comprise an HNH domain having at least about 70%
identity to
any one of SEQ ID NOs: 5390-5460. In some cases, the endonuclease may comprise
an HNH
domain having at least about 70%, at least about 75%, at least about 80%, at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identical to any one of SEQ ID NOs: 5390-5460. The endonuclease may comprise
an HNH
domain substantially identical to any one of SEQ ID NOs: 5390-5460. The
endonuclease may
comprise an HNH domain having at least about 70% identity to any one of SEQ ID
NOs: 5390-
5460. In some cases, the endonuclease may comprise an HNH domain having at
least about
70%, at least about 75%, at least about 80%, at least about 85%, at least
about 90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least about
96%, at least about 97%, at least about 98%, or at least about 99% identical
to any one of SEQ
- 83 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
ID NOs: 5390-5460. The endonuclease may comprise an HNH domain substantially
identical to
any one of SEQ ID NOs: 5390-5460.
[00354] In some cases, the endonuclease may comprise a variant having at least
about 30%, at
least about 35%, at least about 40%, at least about 45%, at least about 50%,
at least about 55%, at
least about 60%, at least about 65%, at least about 70%, at least about 75%,
at least about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 96%,
at least about 97%, at
least about 98%, or at least about 99% identity to any one of SEQ ID NOs: 1756-
1826. In some
cases, the endonuclease may be substantially identical to any one of SEQ ID
NOs: 1756-1826. In
some cases, the endonuclease may comprise a variant having at least about 30%,
at least about
35%, at least about 40%, at least about 45%, at least about 50%, at least
about 55%, at least about
60%, at least about 65%, at least about 70%, at least about 75%, at least
about 80%, at least about
85%, at least about 90%, at least about 95%, at least about 96%, at least
about 97%, at least about
98%, or at least about 99% identity to any one of SEQ ID NOs: 1756-1826. In
some cases, the
endonuclease may be substantially identical to any one of SEQ ID NOs: 1756-
1826.
[00355] In some cases, the endonuclease may comprise a variant having one or
more nuclear
localization sequences (NLSs). The NLS may be proximal to the N- or C-terminus
of said
endonuclease. The NLS may be appended N-terminal or C-terminal to any one of
SEQ ID NOs:
1756-1826, or to a variant having at least about 30%, at least about 35%, at
least about 40%, at
least about 45%, at least about 50%, at least about 55%, at least about 60%,
at least about 65%, at
least about 70%, at least about 75%, at least about 80%, at least about 85%,
at least about 90%, at
least about 95%, at least about 96%, at least about 97%, at least about 98%,
or at least about 99%
identity to any one of SEQ ID NOs: 1756-1826. The NLS may be an 5V40 large T
antigen NLS.
The NLS may be a c-myc NLS. The NLS can comprise a sequence with at least
about 80%, at
least about 85%, at least about 90%, at least about 95%, at least about 99%
identity to any one of
SEQ ID NOs: 5593-5608. The NLS can comprise a sequence substantially identical
to any one
of SEQ ID NOs: 5593-5608. The NLS can comprise any of the sequences in Table 1
or a
combination thereof:
[00356] In some cases, sequence identity may be determined by the BLASTP,
CLUSTALW,
MUSCLE, MAFFT, Novafold, or Smith-Waterman homology search algorithm. The
sequence
identity may be determined by the BLASTP algorithm using parameters of a
wordlength (W) of
3, an expectation (E) of 10, and using a BLOSUM62 scoring matrix setting gap
costs at existence
of 11, extension of 1, and using a conditional compositional score matrix
adjustment.
[00357] In some cases, the system above may comprise (b) at least one
engineered synthetic
guide ribonucleic acid (sgRNA) capable of forming a complex with the
endonuclease bearing a
5' targeting region complementary to a desired cleavage sequence. In some
cases, the 5'
- 84 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
targeting region may comprises a PAM sequence compatible with the
endonuclease. In some
cases, the 5' most nucleotide of the targeting region may be G. In some cases,
the 5' targeting
region may be 15-23 nucleotides in length. The guide sequence and the tracr
sequence may be
supplied as separate ribonucleic acids (RNAs) or a single ribonucleic acid
(RNA). The guide
RNA may comprise a crRNA tracrRNA binding sequence 3' to the targeting region.
The guide
RNA may comprise a tracrRNA sequence preceded by a 4-nucleotide linker 3' to
the crRNA
tracrRNA binding region. The sgRNA may comprise, from 5' to 3': a non-natural
guide nucleic
acid sequence capable of hybridizing to a target sequence in a cell; and a
tracr sequence. In some
cases, the non-natural guide nucleic acid sequence and the tracr sequence are
covalently linked.
[00358] In some cases, the tracr sequence may have a particular sequence. The
tracr sequence
may have at least about 80% to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of a natural tracrRNA sequence. The tracr sequence may
have at least
about 80% sequence identity to at least about 60-100 (e.g., at least about 60,
at least about 65, at
least about 70, at least about 75, at least about 80, at least about 85, or at
least about 90)
consecutive nucleotides of SEQ ID NO: 5511. In some cases, the tracrRNA may
have at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to at least about 60-90 (e.g., at
least about 60, at least
about 65, at least about 70, at least about 75, at least about 80, at least
about 85, or at least about
90) consecutive nucleotides of SEQ ID NO: 5511. In some cases, the tracrRNA
may be
substantially identical to at least about 60-100 (e.g., at least about 60, at
least about 65, at least
about 70, at least about 75, at least about 80, at least about 85, or at least
about 90) consecutive
nucleotides of SEQ ID NO: 5511. The tracrRNA may comprise SEQ ID NO: 5511.
[00359] In some cases, the at least one engineered synthetic guide ribonucleic
acid (sgRNA)
capable of forming a complex with the endonuclease may comprise a sequence
having at least
about 80% identity to SEQ ID NO: 5475. The sgRNA may comprise a sequence
having at least
about 80%, at least about 85%, at least about 90%, at least about 91%, at
least about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to SEQ ID NO: 5475. The sgRNA may
comprise a
sequence substantially identical to SEQ ID NO: 5475.
[00360] In some cases, the system above may comprise two different sgRNAs
targeting a first
region and a second region for cleavage in a target DNA locus, wherein the
second region is 3' to
the first region. In some cases, the system above may comprise a single- or
double-stranded
DNA repair template comprising from 5' to 3': a first homology arm comprising
a sequence of at
- 85 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
least about 20 (e.g., at least about 40, 80, 120, 150, 200, 300, 500, or lkb)
nucleotides 5' to the
first region, a synthetic DNA sequence of at least about 10 nucleotides, and a
second homology
arm comprising a sequence of at least about 20 (e.g., at least about 40, 80,
120, 150, 200, 300,
500, or lkb) nucleotides 3' to the second region.
[00361] In another aspect, the present disclosure provides a method for
modifying a target
nucleic acid locus of interest. The method may comprise delivering to the
target nucleic acid
locus any of the non-natural systems disclosed herein, including an enzyme and
at least one
synthetic guide RNA (sgRNA) disclosed herein. The enzyme may form a complex
with the at
least one sgRNA, and upon binding of the complex to the target nucleic acid
locus of interest,
may modify the target nucleic acid locus of interest. Delivering the enzyme to
said locus may
comprise transfecting a cell with the system or nucleic acids encoding the
system. Delivering the
nuclease to said locus may comprise electroporating a cell with the system or
nucleic acids
encoding the system. Delivering the nuclease to said locus may comprise
incubating the system
in a buffer with a nucleic acid comprising the locus of interest. In some
cases, the target nucleic
acid locus comprises deoxyribonucleic acid (DNA) or ribonucleic acid (RNA).
The target
nucleic acid locus may comprise genomic DNA, viral DNA, viral RNA, or
bacterial DNA. The
target nucleic acid locus may be within a cell. The target nucleic acid locus
may be in vitro. The
target nucleic acid locus may be within a eukaryotic cell or a prokaryotic
cell. The cell may be
an animal cell, a human cell, bacterial cell, archaeal cell, or a plant cell.
The enzyme may induce
a single or double-stranded break at or proximal to the target locus of
interest.
[00362] In cases where the target nucleic acid locus may be within a cell, the
enzyme may be
supplied as a nucleic acid containing an open reading frame encoding the
enzyme having a
RuvC III domain having at least about 75% (e.g., at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at least
about 97%, at least about 98%, at least about 99%) identity to any one of SEQ
ID NOs: 3569-
3637. The deoxyribonucleic acid (DNA) containing an open reading frame
encoding said
endonuclease may comprise a sequence substantially identical to SEQ ID NOs:
5587 or at variant
having at least about 30%, at least about 35%, at least about 40%, at least
about 45%, at least
about 50%, at least about 55%, at least about 60%, at least about 65%, at
least about 70%, at least
about 75%, at least about 80%, at least about 85%, at least about 90%, at
least about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
identity to SEQ ID NOs:
5587. In some cases, the nucleic acid comprises a promoter to which the open
reading frame
encoding the endonuclease is operably linked. The promoter may be a CMV, EFla,
5V40,
PGK1, Ubc, human beta actin, CAG, TRE, or CaMKIIa promoter. The endonuclease
may be
supplied as a capped mRNA containing said open reading frame encoding said
endonuclease.
- 86 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
The endonuclease may be supplied as a translated polypeptide. The at least one
engineered
sgRNA may be supplied as deoxyribonucleic acid (DNA) containing a gene
sequence encoding
said at least one engineered sgRNA operably linked to a ribonucleic acid (RNA)
pol III promoter.
In some cases, the organism may be eukaryotic. In some cases, the organism may
be fungal. In
some cases, the organism may be human.
EXAMPLES
Example 1. ¨Metagenomic analysis for new proteins
[00363] Metagenomic samples were collected from sediment, soil and animal.
Deoxyribonucleic
acid (DNA) was extracted with a Zymobiomics DNA mini-prep kit and sequenced on
an Illumina
HiSeqg 2500. Samples were collected with consent of property owners.
Additional raw sequence
data from public sources included animal microbiomes, sediment, soil, hot
springs, hydrothermal
vents, marine, peat bogs, permafrost, and sewage sequences. Metagenomic
sequence data was
searched using Hidden Markov Models generated based on known Cas protein
sequences
including type II Cas effector proteins. Novel effector proteins identified by
the search were
aligned to known proteins to identify potential active sites. This metagenomic
workflow resulted
in delineation of the MG1, MG2, MG3, MG4, MG6, MG14, MG15, MG16, MG18, MG21,
MG22, and MG23 families of class II, type II CRISPR endonucleases described
herein.
Example 2A. ¨ Discovery of an MG! Family of CRISPR systems
[00364] Analysis of the data from the metagenomic analysis of Example 1
revealed a new cluster
of previously undescribed putative CRISPR systems initially comprising six
members (MG1-1,
MG1-2, MG1-3, MG1-4, MG1-5, and MG1-6 recorded as SEQ ID NOs: 5, 6, 1, 2, and
3
respectively). This family is characterized by an enzyme bearing HNH and RuvC
domains. The
RuvC domains of this family have a RuvC III portion having low homology to
previously
described Cas9 family members. Although the initial family members have a
maximum of
56.8% identity among them, all 6 enzymes exhibit a divergent RuvC III portion
of the RuvC
domain and bear the common motif of RHHALDAMV (SEQ ID NO:5615), KHHALDAMC
(SEQ ID NO:5616), or KHHALDAIC (SEQ ID NO:5617) . These motifs are not found
in other
described Cas9-like enzymes. The corresponding protein and nucleic acid
sequences for these
new enzymes and their relevant subdomains are presented in the sequence
listing. Putative
tracrRNA sequences were identified based on their location relative to the
other genes and are
presented as SEQ ID NOs: 5476-5479. The enzyme systems appear to derive from
the Phylum
Verrucomicrobia, the Phylum Candidatus Peregrinibacteria, or the Phylum
Candidatus
Melainabacteria based on the sequences of 16S rRNAs from genome bins
containing the
- 87 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
CRISPR systems. The 16S rRNA sequences are presented as SEQ ID NOs: 5592-
5596). A
detailed domain-level alignment of the CRISPR system sequences together
calling out the
features described by Shmakov et al. (Mol Cell. 2015 Nov 5;60(3):385-97),
which is entirely
incorporated by reference) is depicted in FIGURES 9A, 9B, 9C, 9D, 9E, 9F, 9G,
and 9H.A
comparison of MG1-1, 1-2, and 1-3 versus additional proprietary protein
datasets revealed
additional protein sequences with similar architecture, presented as SEQ NOs:
7-319. These
MG1 protein sequences led to the discovery of additional MG1 motifs as shown
in SEQ ID NOs:
5618-5632.
Example 2B. ¨ Discovery of an MG2 Family of CRISPR systems
[00365] Analysis of data from the metagenomic analysis of Example 1 revealed a
new cluster of
previously undescribed putative CRISPR systems comprising six members (MG2-1,
MG2-2,
MG2-3, MG2-5, and MG2-6). The corresponding protein and nucleic acid sequences
for these
new enzymes and exemplary subdomains are presented as SEQ ID NOs: 320, 322-
325. Based on
their location relative to the other genes, putative tracrRNA sequences were
identified in the
operon and are presented as SEQ ID NOs: 5490, 5492-5494, and 5538. A detailed
domain-level
alignment of these sequences versus Cas9 as outlined in Shmakov et al. (Mol
Cell. 2015 Nov
5;60(3):385-97.), is depicted in FIGURE 7.
[00366] A comparison of MG2-1, MG2-2, MG2-3, MG2-5, and MG2-6 versus
additional
proprietary protein datasets revealed additional protein sequences with
similar architecture,
presented as SEQ NOs: 321 and 326-420. Motifs commonly found in MG2 family
members are
presented as SEQ ID NOs: 5631-5638.
Example 2C. ¨ Discovery of an MG3 Family of CRISPR systems
[00367] Analysis of the data from the metagenomic analysis of Example 1
revealed a new
previously undescribed putative CRISPR system: MG3-1. The corresponding amino
acid
sequences for this new enzyme and its exemplary subdomains are presented as
SEQ ID NOs:
424, 2245, and 4059. Based on proximity to the other elements in the operon, a
putative
tracrRNA containing sequence was identified and is included as SEQ ID NO:
5498. A detailed
domain-level alignment of the sequence versus Cas9 from Actinomyces naeslundii
is depicted in
FIGURE 8.
[00368] A comparison of MG3-1 versus additional proprietary protein datasets
revealed
additional protein sequences with similar architecture, presented as SEQ NOs:
421-423, 425-431.
Example 2D. ¨ Discovery of MG4, 7, 14, 15, 16, 18, 21, 22, 23 Families of
CRISPR systems
- 88 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00369] Analysis of the data from the metagenomic analysis of Example 1
revealed new clusters
of previously undescribed putative CRISPR systems comprising 9 families of one
member each
(MG 4-5, MG7-2, MG14-1, MG15-1, MG16-2, MG18-1, MG21-1, MG22-1, MG23-1). The
corresponding protein and nucleic acid sequences for these new enzymes and
their exemplary
subdomains are presented as SEQ ID NOs: 432, 669, 678, 930, 1093, 1354, 1512,
1656, 1756.
Based on proximity to the other elements in the operon, a putative tracr
containing sequence was
identified for each family. These sequences are presented in the sequence
listing as SEQ ID NOs:
5503-5511, respectively.
[00370] A comparison of MG 4-5, MG7-2, MG14-1, MG15-1, MG16-2, MG18-1, MG21-1,
MG22-1, MG23-1 versus additional proprietary protein datasets revealed
additional protein
sequences with similar architecture, presented as SEQ NOs: 433-660, 670-677,
679-929, 931-
1092, 1094-1353, 1355-1511, 1513-1655, 1657-1755, and 1757-1826. Motifs common
to the
nucleases of these sets of CRISPR systems are presented as SEQ ID NO: 5649 for
MG4; SEQ ID
NOs: 5650-5667 for MG14; 5668-5675 for MG15; SEQ ID NOs: 5676-5678 for MG16;
SEQ ID
NOs: 5679-5686 for MG18; SEQ ID NOs: 5687-5693 and SEQ ID NOs: 5674-5675 for
MG21;
SEQ ID NOs: 5694-5699 for MG22; and SEQ ID NOs: 5700-5717 for MG23.
Example 3.-Prophetic--Determination of Protospacer-Adjacent Motif.
[00371] Experiments are performed as in any of the examples in Karvelis et al.
Methods. 2017
May 15;121-122:3-8, which is entirely incorporated by reference herein, to
identify the
protospacer adjacent motif (PAM) sequence specificity for the novel enzymes
described herein to
allow for optimal synthetic sequence targeting.
[00372] In one example (in-vivo screen), cells bearing plasmids encoding any
of the enzymes
described herein and protospacer-targeting guide RNA are co-transformed with a
plasmid library
containing an antibiotic resistance gene, and a protospacer sequence flanked
by a randomized
PAM sequence. Plasmids containing functional PAMs are cleaved by the enzyme,
leading to cell
death. Deep-sequencing of the enzyme cleavage-resistant plasmid pool isolated
from the
surviving cells displays a set of depleted plasmids that contain functional
cleavage-permitting
PAMs.
[00373] In another example (in vitro screen), PAM library in the form of DNA
plasmid or
concatemeric repeats is subjected to cleavage by the RNP complex (e.g.,
including the enzyme,
tracrRNA and crRNA or the enzyme and hybrid sgRNA) assembled in vitro or in
cell lysates.
Resulting free DNA ends from successful cleavage events are captured by
adapter ligation,
followed by the PCR amplification of the PAM-sided products. Amplified library
of functional
PAMs is subjected to deep sequencing and PAMs licensing DNA cleavage are
identified.
- 89 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
Example 4.-Prophetic¨Use of synthetic CRISPR system as described herein in a
mammalian cell for genome editing
[00374] DNA/RNA sequences encoding (i) an ORF encoding codon-optimized enzyme
under a
cell-compatible promoter with a cell-compatible C-terminal nuclear
localization sequence (e.g.,
SV40 NLS in the case of human cells) and a suitable polyadenylation signal
(e.g., TK pA signal
in the case of human cells); and (ii) an ORF encoding an sgRNA (having a 5'
sequence
beginning with G followed by 20 nt of a complementary targeting nucleic acid
sequence
targeting genomic DNA followed by a corresponding compatible PAM identified
via Example 3
and a 3' tracr-binding sequence, a linker, and the tracrRNA sequence) under a
suitable
Polymerase III promoter (e.g., the U6 promoter in mammalian cells) are
prepared. In some
embodiments, these sequences are prepared on the same or separate plasmid
vectors, which are
transfected via a suitable technique into eukaryotic cells. In some
embodiments, these sequences
are prepared as separate DNA sequences, which are transfected or microinjected
into cells. In
some embodiments, these sequences are prepared as synthesized RNAs or in-vitro
transcribed
RNAs which are transfected or microinjected into cells. In some embodiments,
these sequences
are translated into proteins and transfected or microinjected into cells.
[00375] Whichever transfection method is selected, (i) and (ii) are introduced
into cells. A
period of incubation is allowed to pass so that the enzyme and/or sgRNA can be
transcribed
and/or translated into active form. After the incubation period, genomic DNA
in the vicinity of
the targeting sequence is analyzed (e.g., by sequencing). An indel is
introduced into the genomic
DNA in the vicinity of the targeting sequence as a result of enzyme-mediated
cleavage and non-
homologous end joining.
[00376] In some embodiments, (i) and (ii) are introduced into cells with a
third repair nucleotide
that encodes regions of the genome flanking the cleavage site of sizes 25 bp
or larger, which will
facilitate homology directed repair. Containing within these flanking
sequences may be a single
base pair mutation, a functional gene fragment, a foreign or native gene for
expression, or
several genes composing a biochemical pathway.
Example 5.-Prophetic¨Use of synthetic CRISPR system as described herein in
vitro
[00377] Any of the enzymes described herein are cloned into a suitable E. coli
expression
plasmid containing a purification tag and are recombinantly expressed in E.
coli and purified
using the recombinant tag. RNAs comprising a 5' G followed by a 20 nt
targeting sequence and
PAM sequence, a tracrRNA binding region of a compatible crRNA, a GAAA linker,
and a
compatible tracrRNA are synthesized by suitable solid-phase RNA synthesis
methods.
- 90 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
Recombinant enzymes and sgRNA are combined in a suitable cleavage buffer
containing Mg2+
(e.g., 20 mM HEPES pH 7.5, 100 mM KC1, 5 mM MgCl2, 1 mM DTT, 5% glycerol) and
the
reaction is initiated by introducing a target DNA including a sequence
complementary to the
targeting sequence and PAM sequence. Cleavage of the DNA is monitored by a
suitable assay
(e.g., agarose gel electrophoresis followed by ethidium bromide staining (or
similarly acting
DNA-intercalating agent) and UV visualization).
Example 6.-(General protocol) PAM Sequence identification/confirmation for the
endonucleases described herein
[00378] PAM sequences were determined by sequencing plasmids containing
randomly-
generated PAM sequences that could be cleaved by putative endonucleases
expressed in an E.
coil lysate-based expression system (myTXTL, Arbor Biosciences). In this
system, an E. coil
codon optimized nucleotide sequence was transcribed and translated from a PCR
fragment under
control of a T7 promoter. A second PCR fragment with a tracr sequence under a
T7 promoter and
a minimal CRISPR array composed of a T7 promoter followed by a repeat-spacer-
repeat
sequence was transcribed in the same reaction. Successful expression of the
endonuclease and
tracr sequence in the TXTL system followed by CRISPR array processing provided
active in
vitro CRISPR nuclease complexes.
[00379] A library of target plasmids containing a spacer sequence matching
that in the minimal
array followed by 8N mixed bases (putative PAM sequences) was incubated with
the output of
the TXTL reaction. After 1-3 hr, the reaction was stopped and the DNA was
recovered via a
DNA clean-up kit, e.g., Zymo DCC, AMPure XP beads, QiaQuick etc. Adapter
sequences were
blunt-end ligated to DNA with active PAM sequences that had been cleaved by
the
endonuclease, whereas DNA that had not been cleaved was inaccessible for
ligation. DNA
segments comprising active PAM sequences were then amplified by PCR with
primers specific
to the library and the adapter sequence. The PCR amplification products were
resolved on a gel
to identify amplicons that corresponded to cleavage events. The amplified
segments of the
cleavage reaction were also used as template for preparation of an NGS
library. Sequencing this
resulting library, which was a subset of the starting 8N library, revealed the
sequences which
contain the correct PAM for the active CRISPR complex. For PAM testing with a
single RNA
construct, the same procedure was repeated except that an in vitro transcribed
RNA was added
along with the plasmid library and the tracr/minimal CRISPR array template was
omitted. For
endonucleases where NGS libraries were prepared, seqLogo (see e.g., Huber et
al. Nat Methods.
2015 Feb;12(2):115-21) representations were constructed and are presented in
Figures 27, 38, 29,
30, 31, 32, 33, 34, and 35. The seqLogo module used to construct these
representations takes the
- 91 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
position weight matrix of a DNA sequence motif (e.g. a PAM sequence) and plots
the
corresponding sequence logo as introduced by Schneider and Stephens (see e.g.
Schneider et al.
Nucleic Acids Res. 1990 Oct 25;18(20):6097-100. The characters representing
the sequence in
the seqLogo representations have been stacked on top of each other for each
position in the
aligned sequences (e.g. PAM sequences). The height of each letter is
proportional to its
frequency, and the letters have been sorted so the most common one is on top.
Example 7.-(General protocol) RNA Folding of tracrRNA and sgRNA structures
[00380] Folded structures of guide RNA sequences at 37 C were computed using
the method of
Andronescu et al. Bioinformatics. 2007 Jul 1;23(13):i19-28, which is
incorporated by reference
herein in its entirety. Predicted structures of exemplary sgRNAs described
herein are presented
in Figures 21, 22, 23, 24, 25, and 26.
Example 8.-(General protocol) In vitro cleavage efficiency of MG CRISPR
Complexes
[00381] Endonucleases were expressed as His-tagged fusion proteins from an
inducible T7
promoter in a protease deficient E. coil B strain. Cells expressing the His-
tagged proteins were
lysed by sonication and the His-tagged proteins were purified by Ni-NTA
affinity
chromatography on a HisTrap FF column (GE Lifescience) on an AKTA Avant FPLC
(GE
Lifescience). The eluate was resolved by SDS-PAGE on acrylamide gels (Bio-Rad)
and stained
with InstantBlue Ultrafast coomassie (Sigma-Aldrich). Purity was determined
using densitometry
of the protein band with ImageLab software (Bio-Rad). Purified endonucleases
were dialyzed
into a storage buffer composed of 50 mM Tris-HC1, 300 mM NaCl, 1 mM TCEP, 5%
glycerol;
pH 7.5 and stored at -80 C.
[00382] Target DNAs containing spacer sequences and PAM sequences (determined
e.g., as in
Example 6) were constructed by DNA synthesis. A single representative PAM was
chosen for
testing when the PAM had degenerate bases. The target DNAs comprised 2200 bp
of linear
DNA derived from a plasmid via PCR amplification with a PAM and spacer located
700 bp from
one end. Successful cleavage resulted in fragments of 700 and 1500 bp. The
target DNA, in vitro
transcribed single RNA, and purified recombinant protein were combined in
cleavage buffer (10
mM Tris, 100 mM NaCl, 10 mM MgCl2) with an excess of protein and RNA and
incubated for 5
minutes to 3 hours, usually 1 hr. The reaction was stopped via addition of
RNAse A and
incubation at 60 minutes. The reaction was then resolved on a 1.2% TAE agarose
gel and the
fraction of cleaved target DNA is quantified in ImageLab software.
- 92 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
Example 9.-(General protocol) Testing of Genome Cleavage Activity of MG CRISPR
Complexes in E. coil
[00383] E. coil lacks the capacity to efficiently repair double-stranded DNA
breaks. Thus,
cleavage of genomic DNA can be a lethal event. Exploiting this phenomenon,
endonuclease
activity was tested in E. coil by recombinantly expressing an endonuclease and
a tracrRNA in a
target strain with spacer/target and PAM sequences integrated into its genomic
DNA.
[00384] In this assay, the PAM sequence is specific for the endonuclease being
tested as
determined by the methods described in Example 6. sgRNA sequences were
determined based
upon the sequence and predicted structure of the tracrRNA. Repeat-anti-repeat
pairings of 8-12
bp (generally 10bp) were chosen, starting from the 5' end of the repeat. The
remaining 3' end of
the repeat and 5' end of the tracrRNA were replaced with a tetraloop.
Generally, the tetraloop
was GAAA, but other tetraloops can be used, particularly if the GAAA sequence
is predicted to
interfere with folding. In these cases, a TTCG tetraloop was used.
[00385] Engineered strains with PAM sequences integrated into their genomic
DNA were
transformed with DNA encoding the endonuclease. Transformants were then made
chemocompetent and transformed with 50 ng of single guide RNAs either specific
to the target
sequence ("on target"), or non-specific to the target ("non target"). After
heat shock,
transformations were recovered in SOC for 2 hrs at 37 C. Nuclease efficiency
was then
determined by a 5-fold dilution series grown on induction media. Colonies were
quantified from
the dilution series in triplicate.
Example 10.-(General protocol) Testing of Genome Cleavage Activity of MG
CRISPR
Complexes in Mammalian Cells
[00386] To show targeting and cleavage activity in mammalian cells, the MG Cas
effector
protein sequences were tested in two mammalian expression vectors: (a) one
with a C-terminal
5V40 NLS and a 2A-GFP tag, and (b) one with no GFP tag and two 5V40 NLS
sequences, one
on the N-terminus and one on the C-terminus. In some instances, nucleotide
sequences encoding
the endonucleases were codon-optimized for expression in mammalian cells.
[00387] The corresponding single guide RNA sequence (sgRNA) with targeting
sequence
attached is cloned into a second mammalian expression vector. The two plasmids
are
cotransfected into HEK293T cells. 72 hr after co-transfection of the
expression plasmid and a
sgRNA targeting plasmid into HEK293T cells, the DNA is extracted and used for
the
preparation of an NGS-library. Percent NHEJ is measured via indels in the
sequencing of the
target site to demonstrate the targeting efficiency of the enzyme in mammalian
cells. At least 10
different target sites were chosen to test each protein's activity.
- 93 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
Example 11.- Characterization of MG1 family members
[00388] PAM Specificity, tracrRNA/sgRNA Validation
[00389] The targeted endonuclease activity of MG1 family endonuclease systems
was confirmed
using the myTXTL system described in Example 6. In this assay, PCR
amplification of cleaved
target plasmids yields a product that migrates at approximately 170 bp in the
gel, as shown in
Figures 17-20. Amplification products were observed for MG1-4 (dual guide: see
gel 1, lane 3,
single guide: see gel 6 lane 2), MG1-5 (gel 2 lane 10), MG1-6 (dual guide: see
gel 5 lane 6,
single guide see: gel 6 lane 5), and MG1-7 (dual guide: see gel 3 lane 13,
single guide: see gel 3
lane 2) (protein SEQ ID NOs: 1-4, respectively). Sequencing the PCR products
revealed active
PAM sequences for these enzymes as shown in Table 2.
[00390] Table 2: PAM sequence specificities and related data for MG! enzymes
Enzyme Enzyme Native PAM tracrRNA sgRNA Synthetic Synthetic
protein (dual SEQ SEQ ID SEQ ID (single (single
SEQ ID guide) ID NO: NO: NO: guide) guide)
NO PAM PAM PAM
SEQ ID
NO:
MG1-4 1 nRRRAA 5527 5476 5461 nRRR 5512
MG1-5 2 nnnnCC 5528 5477 5462 nnnnYY 5513
MG1-6 3 nnRRWC 5529 5478 5463 nnRRAY 5514
MG1-7 4 nRRRAA 5530 5479 5464 nRRRAAG 5515
[00391] Synthetic single guide RNAs (sgRNAs) were designed based on the
sequences and
predicted structures of the tracrRNAs and are presented as SEQ ID NOs: 5461-
5464. The PAM
sequence screen of Example 6 was repeated with the sgRNAs. The results of this
experiment are
also presented in Table 2, which reveals that PAM specificity changed slightly
when using
sgRNAs.
[00392] Targeted endonuclease activity In Vitro
[00393] In vitro activity of the MG1-4 endonuclease system (protein SEQ ID NO:
1 with sgRNA
SEQ ID NO: 5461) on a target DNA with a PAM sequence CAGGAAGG was verified
using the
method of Example 8. The single guide sequence reported above (SEQ ID NO:
5461) was used,
with varying spacer/targeting sequence lengths from 18-24 nt replacing the Ns
of the sequence.
The results are shown in Figure 10, wherein the left panel shows a gel
demonstrating DNA
cleavage by MG1-4 in combination with corresponding single guide sgRNAs having
different
targeting sequence lengths (18-24nt), and the right panel shows the same data
quantified as a bar
- 94 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
graph. The data demonstrated that targeting sequences from 18-24 nucleotides
were functional
with the MG1-4/sgRNA system.
[00394] Targeted endonuclease activity in bacterial cells
[00395] In vivo activity of the MG1-4 endonuclease system (protein SEQ ID NO:
1, sgRNA SEQ
ID NO: 5461) was tested with the PAM sequence CAGGAAGG as in Example 9.
Transformed
E. coli were plated in serial dilution, and the results (showing E. coli
serial dilutions in the left
panel and quantitated growth in the right panel) are presented in Figure 11. A
substantial
reduction in the growth of E. coli expressing on target sgRNA compared to E.
coli expressing
non-target sgRNA indicates that genomic DNA was specifically cleaved by the
endonuclease in
E. coli cells.
[00396] Targeted endonuclease activity in mammalian cells
[00397] The method of Example 10 was used to demonstrate targeting and
cleavage activity in
mammalian cells. Open reading frames encoding the MG1-4 (protein SEQ ID NO:
5527) and
MG1-6 (protein SEQ ID NO: 5529) sequences were cloned into 2 mammalian
expression
vectors, one with a C-terminal 5V40 NLS and a 2A-GFP tag (E. coli MG-BB) and
one with no
GFP tag and 2 NLS sequences, one on the N-terminus and one on the C-terminus
(E. coli pMG5-
BB). For MG1-6, the open reading frame was additionally codon-optimized for
mammalian
expression (SEQ ID NO: 5589) and cloned into the 2-NLS plasmid backbone (MG-
16hs). The
results of this experiment are shown in Figure 12. The endonuclease expression
vectors were
cotransfected into HEK293T cells with a second vector for expressing a sgRNA
(e.g., SEQ ID
NOs: 5512 or 5515) with a tracr sequence specific for the endonuclease and a
guide sequence
selected from Tables 3-4. 72 hr after co-transfection the DNA was extracted
and used for the
preparation of an NGS-library. Cleavage activity was detected by the
appearance of internal
deletions (NHEJ remnants) proximal to the sequence of the target site. Percent
NHEJ was
measured via indels in the sequencing of the target site to demonstrate the
targeting efficiency of
the enzyme in mammalian cells and is presented in Figure 12.
[00398] Table 3: MG1-4 mammalian targeting sequences
MG1-4 MG1-4 Targeting Sequence Targeting sequence Targeted Gene
Target ID SEQ ID NO:
1 aatatgtagctgtttgggaggt 5543 VEGFA
2 ctagggggcgctcggccaccac 5544 VEGFA
3 tggctaaagagggaatgggctt 5545 VEGFA
4 cacaccccggctctggctaaag 5546 VEGFA
tcggaggagccgtggtccgcgc 5547 VEGFA
- 95 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
6 geggaccacggctectccgaag 5548 VEGFA
7 gtacaaacggcagaagctggag 5549 EMX1
8 gaggaagggcctgagtccgagca 5550 EMX1
9 aaggcaaacatcctgataatgg 5551 Apolipoprotein
[00399] Table 4: MG1-6 mammalian targeting sequences
MG1-6 MG1-6 Targeting Sequence Targeting sequence Targeted Gene
Target ID SEQ ID NO:
1 tctttagccagagccggggtgt 5552 VEGF A
2 tggaccccctatttctgacctc 5553 VEGF A
3 atgggagcccttcttcttctgc 5554 EMX1
4 tgccacgaagcaggccaatggg 5555 EMX1
tggtgtctgtttgaggttgcta 5556 HBB-R01
6 gggcaggttggtatcaaggtta 5557 HBB-R01
7 aggtgctgacgtaggtagtgct 5558 FANCF
8 gccctacttccgctttcacctt 5559 FANCF
9 aatgtatgctggcttttaaggg 5560 IVS40
gctcctttggctagggaagtgt 5561 IVS40
Example 12.- Characterization of MG2 family members
[00400] PAM Specificity, tracrRNA/sgRNA Validation
[00401] The targeted endonuclease activity of MG2 family members was confirmed
in the
myTXTL system as described in Example 6. Results of this assay are shown in
Figures 17-20. In
the assay shown in Figures 17-20, active proteins that successfully cleave the
library result in a
band around 170 bp in the gel. Amplification products were observed for MG2-1
(see gel 2 lane
11 and gel 4 lane 6) and MG2-7 (see gel 11 lane 10) (SEQ ID NOs: 320 and 321,
respectively).
Sequencing the PCR products revealed active PAM sequences in Table 5 below:
[00402] Table 5: PAM sequence specificities and related data for MG2 enzymes
Enzyme Enzyme Native PAM tracrRNA sgRNA Synthetic Syntheti
protein (dual SEQ SEQ ID SEQ ID (single c (single
SEQ ID guide) ID NO: NO: guide) guide)
NO PAM NO: PAM PAM
SEQ ID
NO:
MG2-1 320 nRCGT 5531 5490 N/A N/A N/A
A
MG2-7 321 N/A N/A 5491 5465 NNNRTA 5516
[00403] Targeted endonuclease activity in bacterial cells
- 96 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00404] In vivo activity of the MG2-7 endonuclease system with a sgRNA
(endonuclease SEQ
ID NO: 321; sgRNA SEQ ID NO: 5465) and an AGCGTAAG PAM sequence was confirmed
using the method described in Example 9. Transformed E. coil were plated in
serial dilution,
and the results (showing E. coil serial dilutions in the left panel and
quantitated growth in the
right panel) are presented in Figure 34. A substantial reduction in the growth
of E. coil
expressing on target sgRNA compared to E. coil expressing non-target sgRNA
indicates that
genomic DNA was specifically cleaved by the MG1-4 endonuclease in E. coil
cells.
[00405] Example 13.- Characterization of MG3 family members
[00406] PAM Specifici02, tracrRNA/sgRNA Validation
[00407] The targeted endonuclease activity of MG3 family members was confirmed
using the
myTXTL system as described in Example 6 using tracr sequences and CRISPR
arrays. In this
assay, PCR amplification of cleaved target plasmids yields a product that
migrates at
approximately 170 bp in the gel, as shown in Figures 17-20. Amplification
products were
observed for MG3-6 (dual guide: see gel 2 lane 8; single guide: see gel 3 lane
3), MG3-7 (dual
guide: see gel 2 lane 3, single guide: see gel 3 lane 4), and MG3-8 (dual
guide: see gel 9 lane 5)
(SEQ ID NOs: 421, 422, and 423, respectively). Sequencing the PCR products
revealed active
PAM sequences in Table 6 below:
[00408] Table 6: PAM sequence specificities and related data for MG3 enzymes
Enzyme Enzyme Native PAM tracrRN sgRNA Synthetic Synthetic
protein (dual SEQ A SEQ SEQ ID (single (single
SEQ ID guide) ID NO: ID NO: NO: guide) guide)
NO PAM PAM PAM
SEQ ID
NO:
MG3-6 421 nnRGGTT 5532 5500 5466 nnGGG 5517
MG3-7 422 nnRnYAY 5533 5501 5467 nnGnTnT 5518
MG3-8 423 nnRGGTT 5534 5502 N/A N/A N/A
[00409] Synthetic single guide RNAs (sgRNAs) were designed based on the
sequences and
predicted structures of the tracrRNAs and are presented as SEQ ID NOs: 5466-
5467. The PAM
sequence screen of Example 6 was repeated with the sgRNAs. The results of this
experiment are
also presented in Table 6, which reveals that PAM specificity changed slightly
when using
sgRNAs.
[00410] Targeted endonuclease activity In Vitro
[00411] In vitro activity of the MG3-6 (endonuclease SEQ ID NO: 421) was
verified with the
PAM sequence GTGGGTTA using the method of Example 8. The single guide sequence
- 97 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
reported above (SEQ ID NO: 5466) was used, with varying spacer/targeting
sequence lengths
from 18-24 nt replacing the Ns of the sequence. The results are shown in
Figure 13, wherein the
top panel shows a gel demonstrating DNA cleavage by MG3-6 in combination with
different
sgRNAs having different targeting sequence lengths (18-24nt), and the bottom
panel shows the
same data quantified as a bar graph. The data demonstrated that targeting
sequences from 18-24
nucleotides were functional with the MG3-6/sgRNA system.
[00412] Targeted endonuclease activity in bacterial cells
[00413] In vivo activity of the MG3-7 endonuclease system (protein SEQ ID NO:
422; sgRNA
SEQ ID NO: 5467) was tested with the PAM sequence TGGACCTG using the method of
Example 9. Transformed E. coli were plated in serial dilution, and the results
(showing E. coli
serial dilutions in the top panel and quantitated growth in the bottom panel)
are presented in
Figure 14. A substantial reduction in the growth of E. coli expressing on
target sgRNA
compared to E. coli expressing non-target sgRNA indicates that genomic DNA was
being
specifically cleaved by the MG3-7 endonuclease system.
[00414] Targeted endonuclease activity in mammalian cells
[00415] The method of Example 10 was used to demonstrate targeting and
cleavage activity in
mammalian cells. Open reading frames encoding MG3-7 (protein SEQ ID NO: 422)
was cloned
into 2 mammalian expression vectors, one with a C-terminal 5V40 NLS and a 2A-
GFP tag (E.
coli MG-BB) and one with no GFP tag and 2 NLS sequences, one on the N-terminus
and one on
the C-terminus (E. coli pMG5-BB). The endonuclease expression vectors were
cotransfected
into HEK293T cells with a second vector for expressing the sgRNA above with a
guide sequence
selected from Table 7. The results of this experiment are shown in Figure 12 .
72 hr after co-
transfection DNA was extracted and used for the preparation of an NGS-library.
Cleavage
activity was detected by the appearance of internal deletions (NHEJ remnants)
in the vicinity of
the target site. Results are presented in Figure 15.
[00416] The target site which were encoded on the sgRNA plasmids are shown in
Table 7 below.
[00417] Table 7: MG3-7 mammalian targeting sequences
MG3-7 MG3-7 Targeting Sequence Targeting sequence Targeted Gene
Target ID SEQ ID NO:
1 cccctatttctgacctcccaaa 5563 VEGF A
2 tgtggttccagaaccggaggac 5564 EMX1
3 ggccctgggcaggttggtatca 5565 HBB-R01
4 tecttaaacctgtettgtaacc 5566 HBB-R01
ctgactcctgaggagaagtctg 5567 HBB-R01
6 tccgagcttctggcggtctcaa 5568 FANCF
7 tatcatttcgcggatgttccaa 5569 FANCF
8 tcgggcagagggtgcatcacct 5570 Apolipoprotein
- 98 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
9 ataataagcagaacttttagtg 5571 Fibrinogen
gttttctttagttattaatttc 5572 Fibrinogen
Example 13.- Characterization of MG4 family members
[00418] PAM Specificity, tracrRNA/sgRNA Validation
[00419] The targeted endonuclease activity of MG4 family endonuclease systems
was confirmed
using the myTXTL system as described in Example 6. In this assay, PCR
amplification of
cleaved target plasmids yields a product that migrates at approximately 170 bp
in the gel, as
shown in Figures 17-20. Amplification products were observed for, MG4-2 (dual
guide: see ge12
lane 9, single guide: see gel 10 lane 7) (SEQ ID NO: 432). Sequencing the PCR
products
revealed active PAM sequences shown in Table 8 below.
Table 8: PAM sequence specificities and related data for MG4 enzymes
Enzyme Enzyme tracrRN sgRNA Syntheti Syntheti
protein A SEQ SEQ ID c (single c (single
SEQ ID ID NO: NO: guide) guide)
NO PAM PAM
SEQ ID
NO:
MG4-5 432 5503 5468 nnCCR 5519
Example 14.- Characterization of MG14 family members
[00420] PAM Specificity, tracrRNA/sgRNA Validation
[00421] The targeted endonuclease activity of MG14 family members (was
confirmed using the
myTXTL system as described in Example 6. In this assay, PCR amplification of
cleaved target
plasmids yields a product that migrates at approximately 170 bp in the gel, as
shown in Figures
17-20. Amplification products were observed for MG14-1 (dual guide: see gel 1
lane 4, single
guide: see gel 3 lane 8) (SEQ ID NO: 678). Sequencing the PCR products
revealed active PAM
sequence specificities shown in Table 9 below.
Enzyme Enzyme Native PAM tracrRN sgRNA Syntheti Syntheti
protein (dual SEQ ID A SEQ SEQ ID c (single c (single
SEQ ID guide) NO: ID NO: NO: guide) guide)
NO PAM PAM PAM
determined determi SEQ ID
ned NO:
MG14-1 678 NNNNGGT 5535 5505 5469 NNNGG 5520
A RTA
[00422] Targeted endonuclease activity in bacterial cells
- 99 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00423] In vivo activity of the MG14-1 endonuclease system with a sgRNA
(endonuclease SEQ
ID NO: 678; sgRNA SEQ ID NO: 5469) and a GGCGGGGA PAM sequence was confirmed
using the method described in Example 9. Transformed E. coil were plated in
serial dilution,
and the results (showing E. coil serial dilutions in the left panel and
quantitated growth in the
right panel) are presented in Figure 35. A substantial reduction in the growth
of E. coil
expressing on target sgRNA compared to E. coil expressing non-target sgRNA
indicates that
genomic DNA was specifically cleaved by the MG1-4 endonuclease in E. coil
cells.
Example 15.- Characterization of MG15 family members
[00424] PAM Specificity, tracrRNA/sgRNA Validation
[00425] The targeted endonuclease activity of MG15 family members was
confirmed using the
myTXTL system as described in Example 6. In this assay, PCR amplification of
cleaved target
plasmids yields a product that migrates at approximately 170 bp in the gel, as
shown in Figures
17-20. Amplification products were observed for MG15-1 (dual guide: see gel 7
lane 7, single
guide: see gel 3 lane 9) (SEQ ID NO: 930). Sequencing the PCR products
revealed active PAM
sequence specificities detailed in Table 10 below.
[00426] Table 10:
Enzyme Enzyme Native PAM tracrRN sgRNA Synthetic Synthetic
protein (dual SEQ ID A SEQ SEQ ID (single
(single
SEQ ID guide) NO: ID NO: NO: guide)
guide)
NO PAM PAM PAM
SEQ ID
NO:
MG15-1 930 nnnnC 5536 5506 5470 nnnnC 5521
[00427] In Vitro Activity
[00428] In vitro activity of the MG15-1 endonuclease system (protein SEQ ID
NO: 930; sgRNA
SEQ ID NO:5470) was tested with the PAM sequence GGGTCAAA using the method of
Example 8. The single guide sequence reported above (SEQ ID NO: 5470) was
used, with
varying spacer/targeting sequence lengths from 18-24 nt (replacing the Ns of
the sequence). The
results are shown in Figure 16, wherein the top panel shows a gel
demonstrating DNA cleavage
by MG15-1 in combination with different sgRNAs having different targeting
sequence lengths
(18-24nt), and the bottom panel shows the same data quantified as a bar graph.
The data
demonstrated that targeting sequences from 18-24 nucleotides were functional
with the MG15-
1/sgRNA system.
[00429] Targeted endonuclease activity in bacterial cells
- 100 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00430] In vivo activity of the MG15-1 endonuclease system with a sgRNA
(endonuclease SEQ
ID NO: 930; sgRNA SEQ ID NO: 5470) and a GGGTCAAA PAM sequence was confirmed
using the method described in Example 9. Transformed E. coil were plated in
serial dilution,
and the results (showing E. coil serial dilutions in the left panel and
quantitated growth in the
right panel) are presented in Figure 35. A substantial reduction in the growth
of E. coil
expressing on target sgRNA compared to E. coil expressing non-target sgRNA
indicates that
genomic DNA was specifically cleaved by the MG1-4 endonuclease in E. coil
cells.
Example 16.- Characterization of MG16 family members
[00431] PAM Specificity, tracrRNA/sgRNA Validation
[00432] The targeted endonuclease activity of MG16 family members was
confirmed using the
myTXTL system as described in Example 6. In this assay, PCR amplification of
cleaved target
plasmids yields a product that migrates at approximately 170 bp in the gel, as
shown in Figures
17-20. Amplification products were observed for MG16-2 (see gel 11, lane 17)
(SEQ ID NO:
1093). Sequencing the PCR products revealed active PAM sequence specificities
detailed in
Table 11 below.
[00433] Table 11:
Enzyme Enzyme sgRNA Synthetic Synthetic
protein SEQ ID (single guide) (single
SEQ ID NO: PAM guide) PAM
NO SEQ ID NO:
MG16-2 1093 5471 nRTnCC 5522
Example 17.- Characterization of MG18 family members
[00434] PAM Specificity, tracrRNA/sgRNA Validation
[00435] The targeted endonuclease activity of MG18 family members was
confirmed using the
myTXTL system as described in Example 6. In this assay, PCR amplification of
cleaved target
plasmids yields a product that migrates at approximately 170 bp in the gel, as
shown in Figures
17-20. Amplification products were observed for MG18-1 (dual guide: see gel 9
lane 9, single
guide: see gel 11 lane 12) (SEQ ID NO: 1354). Sequencing the PCR products
revealed active
PAM sequence specificities detailed in Table 12 below.
[00436] Table 12:
- 101 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
Enzyme Enzyme Native PAM tracrRN sgRNA Synthetic Synthetic
protein (dual SEQ ID A SEQ SEQ ID (single (single
SEQ ID guide) NO: ID NO: NO: guide) guide)
NO PAM PAM PAM
SEQ ID
NO:
MG18-1 1354 nRWART 5537 5508 5472 nnnRRT 5523
Example 18.- Characterization of MG21 family members
[00437] PAM Specificity, tracrRNA/sgRNA Validation
[00438] The targeted endonuclease activity of MG21 family was confirmed using
the myTXTL
system as described in Example 6. In this assay, PCR amplification of cleaved
target plasmids
yields a product that migrates at approximately 170 bp in the gel, as shown in
Figures 17-20.
Amplification products were observed for MG21-1 (see gel 11 lane 2) (SEQ ID
NO: 1512).
Sequencing the PCR products revealed active PAM sequence specificities
detailed in Table 13
below.
[00439] Table 13:
Enzyme Enzyme sgRNA Synthetic Synthetic
protein SEQ ID (single (single
SEQ ID NO: guide) PAM guide) PAM
NO SEQ ID NO:
MG21-1 1512 5473 nnRnR 5524
Example 19.- Characterization of MG22 family members
[00440] PAM Specificity, tracrRNA/sgRNA Validation
[00441] The targeted endonuclease activity of MG22 family members was
confirmed using the
myTXTL system as described in Example 6. In this assay, PCR amplification of
cleaved target
plasmids yields a product that migrates at approximately 170 bp in the gel, as
shown in Figures
17-20. In the assay shown Figures 17-20, active proteins that successfully
cleave the library
result in a band around 170 bp in the gel. Amplification products were
observed for MG22-1
(see gel 11 lane 3) (protein SEQ ID NO: 1656). Sequencing the PCR products
revealed active
PAM sequence specificities detailed in Table 14 below.
[00442] Table 14:
- 102 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
Enzyme Enzyme Native PAM tracrRN sgRNA Syntheti Syntheti
protein (dual SEQ ID A SEQ SEQ ID c (single c (single
SEQ ID guide) NO: ID NO: NO: guide) guide)
NO PAM PAM PAM
determi determi SEQ ID
ned ned NO:
MG22-1 1656 N/A N/A 5510 5474 nnRCnT 5525
Example 20.- Characterization of MG23 family members
[00443] PAM Specificity, tracrRNA/sgRNA Validation
[00444] The targeted endonuclease activity of MG23 family members was
confirmed using the
myTXTL system as described in Example 6. In this assay, PCR amplification of
cleaved target
plasmids yields a product that migrates at approximately 170 bp in the gel, as
shown in Figures
17-20. Amplification products were observed for MG23-1 (see gel 11 lane 4)
(SEQ ID NO:
1756). Sequencing the PCR products revealed active PAM sequences specificities
for these
enzymes detailed Table 15 below.
[00445] Table 15:
Enzyme Enzyme tracrRNA sgRNA Synthetic Synthetic
protein SEQ ID SEQ ID (single (single guide)
SEQ ID NO NO: NO: guide) PAM
PAM SEQ ID NO:
MG23-1 1756 5511 5475 nRRA 5526
[00446] Systems of the present disclosure may be used for various
applications, such as, for
example, nucleic acid editing (e.g., gene editing), binding to a nucleic acid
molecule (e.g.,
sequence-specific binding). Such systems may be used, for example, for
addressing (e.g.,
removing or replacing) a genetically inherited mutation that may cause a
disease in a subject,
inactivating a gene in order to ascertain its function in a cell, as a
diagnostic tool to detect
disease-causing genetic elements (e.g. via cleavage of reverse-transcribed
viral RNA or an
amplified DNA sequence encoding a disease-causing mutation), as deactivated
enzymes in
combination with a probe to target and detect a specific nucleotide sequence
(e.g. sequence
encoding antibiotic resistance int bacteria), to render viruses inactive or
incapable of infecting
host cells by targeting viral genomes, to add genes or amend metabolic
pathways to engineer
organisms to produce valuable small molecules, macromolecules, or secondary
metabolites, to
establish a gene drive element for evolutionary selection, to detect cell
perturbations by foreign
small molecules and nucleotides as a biosensor.
- 103 -
CA 03130135 2021-08-12
WO 2020/168291 PCT/US2020/018432
[00447] While preferred embodiments of the present invention have been shown
and described
herein, it will be obvious to those skilled in the art that such embodiments
are provided by way of
example only. It is not intended that the invention be limited by the specific
examples provided
within the specification. While the invention has been described with
reference to the
aforementioned specification, the descriptions and illustrations of the
embodiments herein are not
meant to be construed in a limiting sense. Numerous variations, changes, and
substitutions will
now occur to those skilled in the art without departing from the invention.
Furthermore, it shall
be understood that all aspects of the invention are not limited to the
specific depictions,
configurations or relative proportions set forth herein which depend upon a
variety of conditions
and variables. It should be understood that various alternatives to the
embodiments of the
invention described herein may be employed in practicing the invention. It is
therefore
contemplated that the invention shall also cover any such alternatives,
modifications, variations
or equivalents. It is intended that the following claims define the scope of
the invention and that
methods and structures within the scope of these claims and their equivalents
be covered thereby.
- 104 -