Note: Descriptions are shown in the official language in which they were submitted.
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
TRISPECIFIC AND/OR TRIVALENT BINDING PROTEINS USING THE CROSS-OVER-DUAL-
VARIABLE
DOMAIN (CODV) FORMAT FOR TREATMENT OF HIV INFECTION
100011 This application claims priority to U.S. Provisional Application No.
62/831,415,
filed April 9, 2019, and EP Application No. EP19306312.0, filed October 8,
2019, the
disclosures of each of which are incorporated herein by reference in their
entirety.
SUBMISSION OF SEQUENCE LISTING ON ASCII TEXT FILE
[0002] The content of the following submission on ASCII text file is
incorporated
herein by reference in its entirety: a computer readable form (CRF) of the
Sequence Listing
(file name: 183952031940SEQLIST.TXT, date recorded: March 25, 2020, size: 580
KB).
FIELD
[0003] The disclosure relates to trispecific and/or trivalent binding
proteins comprising
four polypeptide chains that form three antigen binding sites that
specifically bind one or
more HIV target proteins, wherein a first pair of polypeptides forming the
binding protein
possess dual variable domains having a cross-over orientation. The disclosure
also relates
to methods for making trispecific and/or trivalent binding proteins and uses
of such binding
proteins for treating and/or preventing HIV/AIDS.
BACKGROUND
[0004] Anti-retroviral therapy (ART) has been the standard of care for
HIV/AIDS
patients in the past decades. ART drugs target internal proteins such as
reverse
transcriptase (RT), integrase (IN), and viral protease (P1) by inhibiting
reverse transcription
of HIV-1 genome, integration of HIV-1 genome, and proteolytic cleavage of
protein
precursors that are necessary for the production of infectious viral
particles. Treatment
using ART or combination of different classes of ART results in inhibition of
HIV-1
replication and subsequent reduction of viremia, often to undetectable level
(aviremic
status). Although ART greatly helps HIV patients in controlling their disease
progression,
and containing the global HIV epidemic, it does require patients taking daily
medicines
often following a strict regimen. About 10% patients fail therapy each year
due to drug
toxicity, suboptimal adherence and emerging drug resistance. As more HIV
patients can
live a normal life span (over 80 years), chronic complications are of
particular concern,
1
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
such as aging and drug-drug interaction, and cardiovascular/renal/bone
toxicities. The
economic burden treating HIV/AIDS has not subsided thus far.
[0005] HIV latently infects long-lived resting memory CD4+ T cells and
others as a
form of proviral DNA integrated into the host genome. The latently infected
cells survive
for decades and self-renew like stem cells via homeostatic proliferation,
which is regarded
as an HIV-1 reservoir. The HIV-1 reservoirs are neither affected by ART nor
the host
immune system as they do not express viral proteins. Yet, a small proportion
of cells among
the reservoirs are randomly reactivated by unknown mechanism(s), which are
responsible
for recurrence of viremia once ART is stopped.
[0006] Therefore, a need exists for developing HIV/AIDS treatments to
target the HIV-
1 reservoir(s), and ultimately eliminate them completely, achieving a cure, or
long term
remission of HIV without any further treatment. Any therapeutic strategy to
eliminate the
HIV-1 reservoir needs to activate the reservoir first, followed by elimination
of the
activated HIV-1 reservoir cells.
[0007] All references cited herein, including patent applications, patent
publications,
and UniProtKB/Swiss-Prot Accession numbers are herein incorporated by
reference in their
entirety, as if each individual reference were specifically and individually
indicated to be
incorporated by reference.
BRIEF SUMMARY
[0008] To meet these and other needs, provided herein are multispecific
binding
proteins (e.g., antibodies) that form three antigen binding sites. In some
embodiments, the
binding proteins bind one or more HIV target proteins and a CD3 polypeptide.
In some
embodiments, the binding proteins bind an HIV target protein, a CD28
polypeptide, and a
CD3 polypeptide. The trispecific anti-HIV/CD28xCD3 T cell engager (TCE)
concept
disclosed herein is thought to be an effective eliminator of the HIV-1
reservoir through
activation by anti-CD3, co-activation by anti-CD28, and subsequent killing of
activated
HIV-1 reservoir cells through anti-HIV/anti-CD28 by engaging activated CD8 T
cells,
providing a potential strategy for attacking the HIV-1 reservoir. In addition,
anti-CD3
binding sites are described with high affinity binding to human CD3
polypeptides and
potential manufacturing liabilities (e.g., deamidation sites) removed.
[0009] In some embodiments, provided herein are binding proteins comprising
four
polypeptide chains that form the three antigen binding sites that specifically
bind one or
2
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
more HIV target proteins, wherein a first polypeptide chain comprises a
structure
represented by the formula:
VL2-L 1 -VL 1 -L2- CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH 1-L3 -VH2 -L4 -CH -hinge -CH2 -CH3 RI]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3 -CH -hinge-CH2-Cm [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3 -CL [IV]
wherein:
VIA is a first immunoglobulin light chain variable domain;
V1,2 is a second immunoglobulin light chain variable domain;
V1,3 is a third immunoglobulin light chain variable domain;
VH1 is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CH1 is an immunoglobulin CH1 heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CH1 and CH2 domains;
and
L1, L2, L3 and L4 are amino acid linkers;
wherein the polypeptide of formula I and the polypeptide of formula II form a
cross-over
light chain-heavy chain pair;
wherein VH1 and VLi form a first antigen binding site;
wherein VH2 and VL2 form a second antigen binding site that binds a CD3
polypeptide,
wherein the VH2 domain comprises a CDR-H1 sequence comprising the amino acid
sequence of GFTFTKAW (SEQ ID NO:20), a CDR-H2 sequence comprising the amino
acid sequence of IKDKSNSYAT (SEQ ID NO:21), and a CDR-H3 sequence comprising
the amino acid sequence of RGVYYALSPFDY (SEQ ID NO:22), and the VL2 domain
comprises a CDR-L1 sequence comprising the amino acid sequence of
QSLVHX1NX2X3TY, wherein Xi is E or Q, X2 is A or L, and X3 is Q, R, or F (SEQ
ID
NO:293), a CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID
NO:29), and a CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT
(SEQ ID NO:30); and
3
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
wherein VH3 and VL3 form a third antigen binding site that binds an HIV target
protein.
[0010] In some embodiments, the first binding site binds a CD28 polypeptide
(e.g., a
human CD28 polypeptide). In some embodiments, the VH1 domain comprises a CDR-
H1
sequence comprising the amino acid sequence of GYTFTSYY (SEQ ID NO:31), a CDR-
H2 sequence comprising the amino acid sequence of IYPGNVNT (SEQ ID NO:32), and
a
CDR-H3 sequence comprising the amino acid sequence of TRSHYGLDWNFDV (SEQ ID
NO:33), and the VIA domain comprises a CDR-L1 sequence comprising the amino
acid
sequence of QNIYVW (SEQ ID NO:34), a CDR-L2 sequence comprising the amino acid
sequence of KAS (SEQ ID NO:35), and a CDR-L3 sequence comprising the amino
acid
sequence of QQGQTYPY (SEQ ID NO:36). In some embodiments, the VH1 domain
comprises the amino acid sequence of
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSTYPGN
VNTNYAQKFQGRATLTVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDV
WGKGTTVTVSS (SEQ ID NO:59), and/or the VIA domain comprises the amino acid
sequence of
DIQMTQSPSSLSASVGDRVTITCQASQNIYVWLNWYQQKPGKAPKWYKASNLHT
GVPSRF SGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIK (SEQ ID
NO:60).
[0011] In some embodiments, the CDR-L1 sequence of the VL2 domain comprises
an
amino acid sequence selected from the group consisting of QSLVHQNAQTY (SEQ ID
NO:24), QSLVHENLQTY (SEQ ID NO:25), QSLVHENLFTY (SEQ ID NO:26), and
QSLVHENLRTY (SEQ ID NO:27). In some embodiments, the VH2 domain comprises: an
antibody heavy chain variable (VH) domain comprising a CDR-H1 sequence
comprising
the amino acid sequence of GFTFTKAW (SEQ ID NO:20), a CDR-H2 sequence
comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:21), and a CDR-H3
sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID NO:22);
and
the VL2 domain comprises a CDR-L1 sequence comprising the amino acid sequence
of
QSLVHQNAQTY (SEQ ID NO:24), a CDR-L2 sequence comprising the amino acid
sequence of KVS (SEQ ID NO:29), and a CDR-L3 sequence comprising the amino
acid
sequence of GQGTQYPFT (SEQ ID NO:30). In some embodiments, the VH2 domain
comprises: a CDR-H1 sequence comprising the amino acid sequence of GFTFTKAW
(SEQ
ID NO:20), a CDR-H2 sequence comprising the amino acid sequence of IKDKSNSYAT
(SEQ ID NO:21), and a CDR-H3 sequence comprising the amino acid sequence of
4
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
RGVYYALSPFDY (SEQ ID NO:22); and the VL2 domain comprises a CDR-L1 sequence
comprising the amino acid sequence of QSLVHENLQTY (SEQ ID NO:25), a CDR-L2
sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:30). In
some
embodiments, the VH2 domain comprises: a CDR-H1 sequence comprising the amino
acid
sequence of GFTFTKAW (SEQ ID NO:20), a CDR-H2 sequence comprising the amino
acid sequence of IKDKSNSYAT (SEQ ID NO:21), and a CDR-H3 sequence comprising
the amino acid sequence of RGVYYALSPFDY (SEQ ID NO:22); and the VL2 domain
comprises a CDR-L1 sequence comprising the amino acid sequence of QSLVHENLFTY
(SEQ ID NO:26), a CDR-L2 sequence comprising the amino acid sequence of KVS
(SEQ
ID NO:29), and a CDR-L3 sequence comprising the amino acid sequence of
GQGTQYPFT
(SEQ ID NO:30). In some embodiments, the VH2 domain comprises: a CDR-H1
sequence
comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20), a CDR-H2
sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:21), and
a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:22); and the VL2 domain comprises a CDR-L1 sequence comprising the amino
acid
sequence of QSLVHENLRTY (SEQ ID NO:27), a CDR-L2 sequence comprising the
amino acid sequence of KVS (SEQ ID NO:29), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:30). In some embodiments, the VH2
domain comprises the amino acid sequence of
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKD
KSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPF
DYWGQGTLVTVSS (SEQ ID NO:52), and/or the VL2 domain comprises an amino acid
sequence selected from the group consisting of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:54),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:55),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:56), and
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVS
NRF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK
(SEQ ID NO:57). In some embodiments, a binding protein of the present
disclosure
comprises an antigen binding site comprising: an antibody heavy chain variable
(VH)
domain comprising the amino acid sequence of SEQ ID NO:52, and/or an antibody
light
chain variable (VL) domain comprising the amino acid sequence of SEQ ID NO:54.
In
some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising the
amino acid sequence of SEQ ID NO:52, and/or an antibody light chain variable
(VL)
domain comprising the amino acid sequence of SEQ ID NO:55. In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ
ID NO:52, and/or an antibody light chain variable (VL) domain comprising the
amino acid
sequence of SEQ ID NO:56. In some embodiments, a binding protein of the
present
disclosure comprises an antigen binding site comprising: an antibody heavy
chain variable
(VH) domain comprising the amino acid sequence of SEQ ID NO:52, and/or an
antibody
light chain variable (VL) domain comprising the amino acid sequence of SEQ ID
NO:57.
[0012] In
some embodiments, the third antigen binding site binds an HIV target protein
selected from the group consisting of glycoprotein 120, glycoprotein 41 and
glycoprotein
160. In some embodiments, the VH3 domain comprises a CDR-H1 sequence
comprising the
amino acid sequence of NCPIN (SEQ ID NO:1) a CDR-H2 sequence comprising the
amino
acid sequence of WMKPRHGAVSYARQLQG (SEQ ID NO:2), and a CDR-H3 sequence
comprising the amino acid sequence of GKYCTARDYYNWDFEH (SEQ ID NO:3), and
the VL3 domain comprises a CDR-L1 sequence comprising the amino acid sequence
of
RTSQYGSLA (SEQ ID NO:4), a CDR-L2 sequence comprising the amino acid sequence
of SGSTRAA (SEQ ID NO:5), and a CDR-L3 sequence comprising the amino acid
sequence of QQYEF (SEQ ID NO:6). In some embodiments, the VH3 domain comprises
a
CDR-H1 sequence comprising the amino acid sequence of GYTFTAHI (SEQ ID NO:7) a
CDR-H2 sequence comprising the amino acid sequence of IKPQYGAV (SEQ ID NO:8)
or
IKPQYGAT (SEQ ID NO:9), and a CDR-H3 sequence comprising the amino acid
sequence of DRSYGDSSWALDA (SEQ ID NO:10), and the VL3 domain comprises a
CDR-L1 sequence comprising the amino acid sequence of QGVGSD (SEQ ID NO:11), a
CDR-L2 sequence comprising the amino acid sequence of HTS (SEQ ID NO:12), and
a
6
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
CDR-L3 sequence comprising the amino acid sequence of CQVLQF (SEQ ID NO:13).
In
some embodiments, the VH3 domain comprises a CDR-H1 sequence comprising the
amino
acid sequence of DCTLN (SEQ ID NO:14) a CDR-H2 sequence comprising the amino
acid
sequence of WLKPRWGAVNYARPLQG (SEQ ID NO:15), and a CDR-H3 sequence
comprising the amino acid sequence of GKNCDYNWDFEH (SEQ ID NO:16), and the VL3
domain comprises a CDR-L1 sequence comprising the amino acid sequence of
RTSQYGSLA (SEQ ID NO:17), a CDR-L2 sequence comprising the amino acid sequence
of SGSTRAA (SEQ ID NO:18), and a CDR-L3 sequence comprising the amino acid
sequence of QQYEF (SEQ ID NO:19). In some embodiments, the VH3 domain
comprises
the amino acid sequence of
QVRLSQSGGQMKKPGDSMRISCRASGYEFINCPINWIRLAPGKRPEWMGWMKPRH
GAVSYARQLQGRVTMTRDMYSETAFLELRSLTSDDTAVYFCTRGKYCTARDYYN
WDFEHWGQGTPVTVSS (SEQ ID NO:43), and/or the VL3 domain comprises the amino
acid sequence of
SLTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRF
SGSRWGPDYNLTISNLESGDFGVYYCQQYEFFGQGTKVQVDIK (SEQ ID NO :45).
In some embodiments, the VH3 domain comprises the amino acid sequence of
QVRLSQSGGQMKKPGDSMRISCRASGYEFINCPINWIRLAPGKRPEWMGWMKPRH
GAVSYARQLQGRVTMTRQLSQDPDDPDWGTAFLELRSLTSDDTAVYFCTRGKYC
TARDYYNWDFEHWGQGTPVTVSS (SEQ ID NO:44), and/or the VL3 domain comprises
the amino acid sequence of
SLTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRF
SGSRWGPDYNLTISNLESGDFGVYYCQQYEFFGQGTKVQVDIK (SEQ ID NO :45).
In some embodiments, the VH3 domain comprises the amino acid sequence of
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQ
YGAVNFGGGFRDRVTLTRDVYREIAYMDIRGLKPDDTAVYYCARDRSYGDSSWA
LDAWGQGTTVVVSA (SEQ ID NO:46), and/or the VL3 domain comprises the amino acid
sequence of
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDG
VPSRFSGSGFHTSFNLTISDLQADDIATYYCQVLQFFGRGSRLHIK (SEQ ID NO :49).
In some embodiments, the VH3 domain comprises the amino acid sequence of
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQ
YGATNFGGGFRDRVTLTRDVYREIAYMDIRGLKPDDTAVYYCARDRSYGDSSWA
7
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
LDAWGQGTTVVVSA (SEQ ID NO:47), and/or the VL3 domain comprises the amino acid
sequence of
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDG
VPSRFSGSGFHTSFNLTISDLQADDIATYYCQVLQFFGRGSRLHIK (SEQ ID NO :49).
In some embodiments, the VH3 domain comprises the amino acid sequence of
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQ
YGAVNFGGGFRDRVTLTRQLSQDPDDPDWGIAYMDIRGLKPDDTAVYYCARDRS
YGDSSWALDAWGQGTTVVVSA (SEQ ID NO:48), and/or the VL3 domain comprises
the amino acid sequence of
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDG
VPSRFSGSGFHTSFNLTISDLQADDIATYYCQVLQFFGRGSRLHIK (SEQ ID NO :49).
In some embodiments, the VH3 domain comprises the amino acid sequence of
QVQLVQSGGQMKKPGESMRISCRASGYEFIDCTLNWIRLAPGKRPEWMGWLKPR
WGAVNYARPLQGRVTMTRQLSQDPDDPDWGTAFLELRSLTVDDTAVYFCTRGKN
CDYNWDFEHWGRGTPVIVSS (SEQ ID NO:50), and/or the VL3 domain comprises the
amino acid sequence of
LTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRFS
GSRWGPDYNLTISNLESGDFGVYYCQQYEFFGQGTKVQVDIK (SEQ ID NO:51).
[0013] In some embodiments that may be combined with any other embodiments
described herein, at least one of Li, L2, L3 or L4 is independently 0 amino
acids in length.
In some embodiments, Li, L2, L3 and L4 each independently are zero amino acids
in length
or comprise a sequence selected from the group consisting of GGGGSGGGGS (SEQ
ID
NO:40), GGGGSGGGGSGGGGS (SEQ ID NO: 41), S, RT, TKGPS (SEQ ID NO: 39),
GQPKAAP (SEQ ID NO: 38), and GGSGSSGSGG (SEQ ID NO: 42). In some
embodiments, Li, L2, L3 and L4 each independently comprise a sequence selected
from the
group consisting of GGGGSGGGGS (SEQ ID NO:40), GGGGSGGGGSGGGGS (SEQ ID
NO:41), S, RT, TKGPS (SEQ ID NO:39), GQPKAAP (SEQ ID NO: 38), and
GGSGSSGSGG (SEQ ID NO:42). In some embodiments, Li comprises the sequence
GQPKAAP (SEQ ID NO: 38), L2 comprises the sequence TKGPS (SEQ ID NO:39), L3
comprises the sequence S, and L4 comprises the sequence RT. In some
embodiments, at
least one of Li, L2, L3 or L4 comprises the sequence DKTHT (SEQ ID NO:37). In
some
embodiments, Li, L2, L3 and L4 comprise the sequence DKTHT (SEQ ID NO:37).
8
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
[0014] In some embodiments that may be combined with any other embodiments
described herein, the hinge-CH2-CH3 domains of the second and the third
polypeptide chains
are human IgG4 hinge-CH2-CH3 domains, and wherein the hinge-CH2-CH3 domains
each
comprise amino acid substitutions at positions corresponding to positions 234
and 235 of
human IgG4 according to EU Index, wherein the amino acid substitutions are
F234A and
L235A. In some embodiments, the hinge-CH2-CH3 domains of the second and the
third
polypeptide chains are human IgG4 hinge-CH2-CH3 domains, and wherein the hinge-
Cm-
CH3 domains each comprise amino acid substitutions at positions corresponding
to positions
233-236 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
E233P, F234V, L235A, and a deletion at 236. In some embodiments, the hinge-CH2-
CH3
domains of the second and the third polypeptide chains are human IgG4 hinge-
CH2-CH3
domains, and wherein the hinge-CH2-CH3 domains each comprise amino acid
substitutions at
positions corresponding to positions 228 and 409 of human IgG4 according to EU
Index,
wherein the amino acid substitutions are S228P and R409K. In some embodiments,
the
hinge-CH2-CH3 domains of the second and the third polypeptide chains are human
IgG1
hinge-CH2-CH3 domains, and wherein the hinge-CH2-CH3 domains each comprise
amino acid
substitutions at positions corresponding to positions 234, 235, and 329 of
human IgG1
according to EU Index, wherein the amino acid substitutions are L234A, L235A,
and
P329A. In some embodiments, the hinge-CH2-CH3 domains of the second and the
third
polypeptide chains are human IgG1 hinge-CH2-CH3 domains, and wherein the hinge-
Cm-
CH3 domains each comprise amino acid substitutions at positions corresponding
to positions
298, 299, and 300 of human IgG1 according to EU Index, wherein the amino acid
substitutions are S298N, T299A, and Y300S. In some embodiments, the hinge-CH2-
CH3
domain of the second polypeptide chain comprises amino acid substitutions at
positions
corresponding to positions 349, 366, 368, and 407 of human IgG1 or IgG4
according to EU
Index, wherein the amino acid substitutions are Y349C, T366S, L368A, and
Y407V; and
wherein the hinge-CH2-CH3 domain of the third polypeptide chain comprises
amino acid
substitutions at positions corresponding to positions 354 and 366 of human
IgG1 or IgG4
according to EU Index, wherein the amino acid substitutions are S354C and
T366W. In
some embodiments, the hinge-CH2-CH3 domain of the second polypeptide chain
comprises
amino acid substitutions at positions corresponding to positions 354 and 366
of human
IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions are
S354C and
T366W; and wherein the hinge-CH2-CH3 domain of the third polypeptide chain
comprises
9
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
amino acid substitutions at positions corresponding to positions 349, 366,
368, and 407 of
human IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions
are
Y349C, T366S, L368A, and Y407V.
[0015] In some embodiments, the first polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:61 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:61; the second polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:62 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:62; the third polypeptide chain comprises
the
amino acid sequence of SEQ ID NO:63 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:63; and the fourth
polypeptide chain
comprises the amino acid sequence of SEQ ID NO:64 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:64. In some
embodiments,
the first polypeptide chain comprises the amino acid sequence of SEQ ID NO:65
or an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:65; the second polypeptide chain comprises the amino acid sequence of SEQ
ID NO:66
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:66; the third polypeptide chain comprises the amino acid sequence of SEQ
ID
NO:67 or an amino acid sequence that is at least 95% identical to the amino
acid sequence
of SEQ ID NO:67; and the fourth polypeptide chain comprises the amino acid
sequence of
SEQ ID NO:68 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:68. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:69 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:69; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:70 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:70; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:71 or an amino acid
sequence that
is at least 95% identical to the amino acid sequence of SEQ ID NO:71; and the
fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:72 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:72. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:73 or an amino acid sequence that is at least 95% identical to the amino
acid
sequence of SEQ ID NO:73; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:74 or an amino acid sequence that is at least 95%
identical to the
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
amino acid sequence of SEQ ID NO:74; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:75 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:75; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:76 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:76. In some embodiments, the
first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:77 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:77; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:78 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:78; the third polypeptide chain comprises the amino acid sequence of SEQ ID
NO:79
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:79; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ ID
NO:80 or an amino acid sequence that is at least 95% identical to the amino
acid sequence
of SEQ ID NO:80. In some embodiments, the first polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:81 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:81; the second polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:82 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:82; the third polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:83 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:83; and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:84 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:84. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:85 or an amino acid sequence that is at least 95% identical to the amino
acid
sequence of SEQ ID NO:85; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:86 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:86; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:87 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:87; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:88 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:88. In some embodiments, the
first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:89 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:89; the
11
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:90 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:90; the third polypeptide chain comprises the amino acid sequence of SEQ ID
NO:91
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:91; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ ID
NO:92 or an amino acid sequence that is at least 95% identical to the amino
acid sequence
of SEQ ID NO:92. In some embodiments, the first polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:93 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:93; the second polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:94 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:94; the third polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:95 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:95; and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:96 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:96. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:97 or an amino acid sequence that is at least 95% identical to the amino
acid
sequence of SEQ ID NO:97; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:98 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:98; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:99 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:99; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:100 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:100. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:101 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:101; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:102 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:102; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:103
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:103; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:104 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:104. In some embodiments, the first polypeptide chain
comprises
12
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
the amino acid sequence of SEQ ID NO:105 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:105; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:106 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:106; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:107 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:107;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:108 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:108. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:109 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:109; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:110 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:110; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:111 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:111; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:112 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:112. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:113 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:113; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:114 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:114; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:115
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:115; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:116 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:116. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:117 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:117; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:118 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:118; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:119 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:119;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:120 or an
amino acid
13
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:120. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:121 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:121; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:122 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:122; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:123 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:123; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:124 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:124. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:129 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:129; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:130 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:130; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:131
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:131; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:132 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:132. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:133 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:133; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:134 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:134; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:135 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:135;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:136 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:136. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:137 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:137; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:138 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:138; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:139 or an amino acid sequence that is at least 95%
identical
14
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
to the amino acid sequence of SEQ ID NO:139; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:140 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:140. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:141 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:141; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:142 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:142; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:143
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:143; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:144 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:144. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:145 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:145; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:146 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:146; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:147 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:147;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:148 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:148. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:149 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:149; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:150 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:150; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:151 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:151; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:152 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:152. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:153 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:153; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:154 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
NO:154; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:155
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:155; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:156 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:156. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:157 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:157; the second polypeptide
chain
comprises the amino acid sequence of SEQ ID NO:158 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:158; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:159 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:159;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:160 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:160. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:161 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:161; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:162 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:162; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:163 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:163; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:164 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:164. In some embodiments,
the first
polypeptide chain comprises the amino acid sequence of SEQ ID NO:165 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:165; the
second polypeptide chain comprises the amino acid sequence of SEQ ID NO:166 or
an
amino acid sequence that is at least 95% identical to the amino acid sequence
of SEQ ID
NO:166; the third polypeptide chain comprises the amino acid sequence of SEQ
ID NO:167
or an amino acid sequence that is at least 95% identical to the amino acid
sequence of SEQ
ID NO:167; and the fourth polypeptide chain comprises the amino acid sequence
of SEQ
ID NO:168 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:168. In some embodiments, the first polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:169 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:169; the second polypeptide
chain
16
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprises the amino acid sequence of SEQ ID NO:170 or an amino acid sequence
that is at
least 95% identical to the amino acid sequence of SEQ ID NO:170; the third
polypeptide
chain comprises the amino acid sequence of SEQ ID NO:171 or an amino acid
sequence
that is at least 95% identical to the amino acid sequence of SEQ ID NO:171;
and the fourth
polypeptide chain comprises the amino acid sequence of SEQ ID NO:172 or an
amino acid
sequence that is at least 95% identical to the amino acid sequence of SEQ ID
NO:172. In
some embodiments, the first polypeptide chain comprises the amino acid
sequence of SEQ
ID NO:173 or an amino acid sequence that is at least 95% identical to the
amino acid
sequence of SEQ ID NO:173; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:174 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:174; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:175 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:175; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:176 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:176.
[0016] In some embodiments, provided herein are isolated nucleic acid
molecules
comprising a nucleotide sequence encoding the binding protein of any one of
the above
embodiments. In some embodiments, provided herein are expression vectors
comprising
the nucleic acid molecule of any one of the above embodiments. In some
embodiments,
provided herein are isolated host cells comprising the nucleic acid molecule
of any one of
the above embodiments or the expression vector of any one of the above
embodiments. In
some embodiments, the host cell is a mammalian or insect cell.
[0017] In some embodiments, provided herein are pharmaceutical compositions
comprising the binding protein of any one of the above embodiments and a
pharmaceutically acceptable carrier.
[0018] In some embodiments, provided herein are methods of preventing
and/or
treating HIV infection in a patient comprising administering to the patient a
therapeutically
effective amount of at least one binding protein of any one of the above
embodiments or the
pharmaceutical composition of any one of the above embodiments. In some
embodiments,
the binding protein is co-administered with standard anti-retroviral therapy.
In some
embodiments, administration of the at least one binding protein results in the
elimination of
one or more latently and/or chronically HIV-infected cells in the patient. In
some
embodiments, the patient is a human.
17
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
[0019] In some embodiments, the binding protein or pharmaceutical
composition of any
one of the above embodiments is provided for the prevention and/or treatment
of HIV
infection in a patient. In some embodiments, the binding protein is to be co-
administered
with standard anti-retroviral therapy. In some embodiments, the binding
protein causes the
elimination of one or more latently and/or chronically HIV-infected cells in
the patient. In
some embodiments, the patient is a human.
[0020] In some embodiments, the binding protein or pharmaceutical
composition of any
one of the above embodiments is provided for use in the manufacture of a
medicament for
the prevention and/or treatment of HIV infection in a patient. In some
embodiments, the
binding protein is to be co-administered with standard anti-retroviral
therapy. In some
embodiments, the binding protein causes the elimination of one or more
latently and/or
chronically HIV-infected cells in the patient. In some embodiments, the
patient is a human.
[0021] In some embodiments, provided herein is a vector system comprising
one or
more vectors encoding a first, second, third, and fourth polypeptide chain of
a binding
protein of any one of the above embodiments. In some embodiments, the vector
system
comprises a first vector encoding the first polypeptide chain of the binding
protein, a
second vector encoding the second polypeptide chain of the binding protein, a
third vector
encoding the third polypeptide chain of the binding protein, and a fourth
vector encoding
the fourth polypeptide chain of the binding protein.
[0022] In some embodiments, provided herein are kits comprising one, two,
three, or
four polypeptide chains of a binding protein according to any one of the above
embodiments. In some embodiments, the kits further comprise instructions for
using the
polypeptide chain or binding protein according to any of the methods or uses
described
herein, e.g., supra.
[0023] In some embodiments, provided herein are kits comprising one, two,
three, or
four polynucleotides according to any one of the above embodiments. In some
embodiments, provided herein are kits of polynucleotides comprising one, two,
three, or
four polynucleotides of a kit of polynucleotides comprising: (a) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:177, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:178, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:179, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:180; (b) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:181, a second
polynucleotide
18
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprising the polynucleotide sequence of SEQ ID NO:182, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:183, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:184; (c) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:185, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:186, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:187, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:188; (d) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:189, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:190, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:191, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:192; (e) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:193, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:194, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:195, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:196; (f) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:197, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:198, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:199, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:200; (g) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:201, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:202, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:203, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:204; (h) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:205, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:206, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:207, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:208; (i) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:209, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:210, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:211, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:212; (j) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:213, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:214, a third
polynucleotide
19
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprising the polynucleotide sequence of SEQ ID NO:215, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:216; (k) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:217, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:218, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:219, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:220; (1) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:221, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:222, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:223, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:224; (m) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:225, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:226, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:227, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:228; (n) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:229, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:230, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:231, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:232; (o) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:233, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:234, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:235, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:236; (p) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:237, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:238, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:239, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:240; (q) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:241, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:242, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:243, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:244; (r) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:245, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:246, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:247, and a fourth
polynucleotide
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprising the polynucleotide sequence of SEQ ID NO:248; (s) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:249, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:250, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:251, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:252; (t) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:253, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:254, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:255, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:256; (u) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:257, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:258, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:259, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:260; (v) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:261, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:262, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:263, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:264; (w) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:265, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:266, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:267, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:268; (x) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:269, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:270, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:271, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:272; (y) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:273, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:274, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:275, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:276; (z) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:277, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:278, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:279, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:280; (aa) a first
polynucleotide
21
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprising the polynucleotide sequence of SEQ ID NO:281, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:282, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:283, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:284; (bb) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:285, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:286, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:287, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:288; or (cc) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:289, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:290, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:291, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:292. In some embodiments,
the
first, second, third, and fourth polynucleotides are present on one or more
expression
vectors, e.g., one, two, three, or four expression vectors.
[0024] It is to be understood that one, some, or all of the properties of
the various
embodiments described herein may be combined to form other embodiments of the
present
invention. These and other aspects of the invention will become apparent to
one of skill in
the art.
BRIEF DESCRIPTION OF THE DRAWINGS
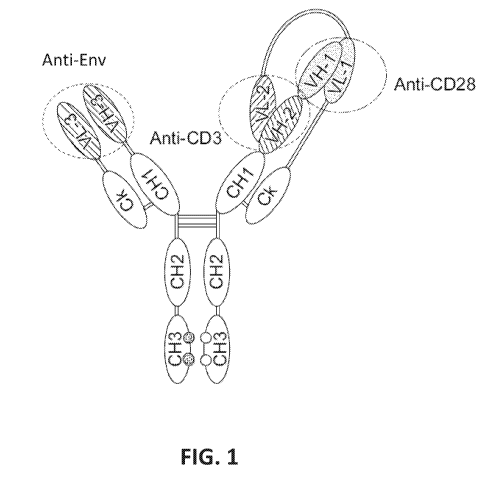
[0025] FIG. 1 provides a schematic representation of a trispecific binding
protein
comprising four polypeptide chains that form three antigen binding sites that
binds three
target proteins: CD28, CD3, and HIV Env. A first pair of polypeptides possess
dual
variable domains having a cross-over orientation (VH1-VH2 and VL2-VL1) forming
two
antigen binding sites (VH1 and VL1; VH2 and VL2) that recognize CD28 and CD3,
resepectively, and a second pair of polypeptides possess a single variable
domain (VH3 and
VL3) forming a single antigen binding site that recognizes HIV Env. The
trispecific
binding protein shown in FIG. 1 uses a constant region with a "knobs-into-
holes" mutation,
where the knob is on the second pair of polypeptides with a single variable
domain.
[0026] FIG. 2 shows a schematic representation of a trispecific T cell
Engager (TCE)
strategy for using the anti-HIV trispecific binding protein shown in FIG. 1 to
target and
eliminate the HIV reservoir.
22
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
DETAILED DESCRIPTION
[0027] The present disclosure provides trispecific and/or trivalent binding
proteins
comprising four polypeptide chains that form three antigen binding sites that
specifically
bind to one or more human immunodeficiency virus (HIV) target proteins and/or
one or
more T-cell receptor target proteins, wherein a first pair of polypeptides
forming the
binding protein possess dual variable domains having a cross-over orientation,
and wherein
a second pair of polypeptides possess a single variable domain.
[0028] The following description sets forth exemplary methods, parameters,
and the
like. It should be recognized, however, that such description is not intended
as a limitation
on the scope of the present disclosure but is instead provided as a
description of exemplary
embodiments.
Definitions
[0029] As utilized in accordance with the present disclosure, the following
terms,
unless otherwise indicated, shall be understood to have the following
meanings. Unless
otherwise required by context, singular terms shall include pluralities and
plural terms shall
include the singular.
[0030] It is understood that aspects and embodiments of the disclosure
described herein
include "comprising," "consisting," and "consisting essentially of' aspects
and
embodiments.
[0031] The term "polynucleotide" as used herein refers to single-stranded
or double-
stranded nucleic acid polymers of at least 10 nucleotides in length. In
certain embodiments,
the nucleotides comprising the polynucleotide can be ribonucleotides or
deoxyribonucleotides or a modified form of either type of nucleotide. Such
modifications
include base modifications such as bromuridine, ribose modifications such as
arabinoside
and 2',3'-dideoxyribose, and internucleotide linkage modifications such as
phosphorothioate, phosphorodithioate, phosphoroselenoate,
phosphorodiselenoate,
phosphoroanilothioate, phoshoraniladate and phosphoroamidate. The term
"polynucleotide" specifically includes single-stranded and double-stranded
forms of DNA.
[0032] An "isolated polynucleotide" is a polynucleotide of genomic, cDNA,
or
synthetic origin or some combination thereof, which: (1) is not associated
with all or a
portion of a polynucleotide in which the isolated polynucleotide is found in
nature, (2) is
linked to a polynucleotide to which it is not linked in nature, or (3) does
not occur in nature
as part of a larger sequence.
23
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
[0033] An "isolated polypeptide" is one that: (1) is free of at least some
other
polypeptides with which it would normally be found, (2) is essentially free of
other
polypeptides from the same source, e.g., from the same species, (3) is
expressed by a cell
from a different species, (4) has been separated from at least about 50
percent of
polynucleotides, lipids, carbohydrates, or other materials with which it is
associated in
nature, (5) is not associated (by covalent or noncovalent interaction) with
portions of a
polypeptide with which the "isolated polypeptide" is associated in nature, (6)
is operably
associated (by covalent or noncovalent interaction) with a polypeptide with
which it is not
associated in nature, or (7) does not occur in nature. Such an isolated
polypeptide can be
encoded by genomic DNA, cDNA, mRNA or other RNA, of synthetic origin, or any
combination thereof. Preferably, the isolated polypeptide is substantially
free from
polypeptides or other contaminants that are found in its natural environment
that would
interfere with its use (therapeutic, diagnostic, prophylactic, research or
otherwise).
[0034] Naturally occurring antibodies typically comprise a tetramer. Each
such
tetramer is typically composed of two identical pairs of polypeptide chains,
each pair
having one full-length "light" chain (typically having a molecular weight of
about 25 kDa)
and one full-length "heavy" chain (typically having a molecular weight of
about 50-70
kDa). The terms "heavy chain" and "light chain" as used herein refer to any
immunoglobulin polypeptide having sufficient variable domain sequence to
confer
specificity for a target antigen. The amino-terminal portion of each light and
heavy chain
typically includes a variable domain of about 100 to 110 or more amino acids
that typically
is responsible for antigen recognition. The carboxy-terminal portion of each
chain typically
defines a constant domain responsible for effector function. Thus, in a
naturally occurring
antibody, a full-length heavy chain immunoglobulin polypeptide includes a
variable domain
(VH) and three constant domains (CHi, CH2, and CH3), wherein the VH domain is
at the
amino-terminus of the polypeptide and the CH3 domain is at the carboxyl-
terminus, and a
full-length light chain immunoglobulin polypeptide includes a variable domain
(VL) and a
constant domain (CL), wherein the VL domain is at the amino-terminus of the
polypeptide
and the CL domain is at the carboxyl-terminus.
[0035] Human light chains are typically classified as kappa and lambda
light chains,
and human heavy chains are typically classified as mu, delta, gamma, alpha, or
epsilon, and
define the antibody's isotype as IgM, IgD, IgG, IgA, and IgE, respectively.
IgG has several
subclasses, including, but not limited to, IgGl, IgG2, IgG3, and IgG4. IgM has
subclasses
24
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
including, but not limited to, IgM1 and IgM2. IgA is similarly subdivided into
subclasses
including, but not limited to, IgAl and IgA2. Within full-length light and
heavy chains, the
variable and constant domains typically are joined by a "J" region of about 12
or more
amino acids, with the heavy chain also including a "D" region of about 10 more
amino
acids. See, e.g., FUNDAMENTAL IMMUNOLOGY (Paul, W., ed., Raven Press, 2nd ed.,
1989),
which is incorporated by reference in its entirety for all purposes. The
variable regions of
each light/heavy chain pair typically form an antigen binding site. The
variable domains of
naturally occurring antibodies typically exhibit the same general structure of
relatively
conserved framework regions (FR) joined by three hypervariable regions, also
called
complementarity determining regions or CDRs. The CDRs from the two chains of
each
pair typically are aligned by the framework regions, which may enable binding
to a specific
epitope. From the amino-terminus to the carboxyl-terminus, both light and
heavy chain
variable domains typically comprise the domains FR1, CDR1, FR2, CDR2, FR3,
CDR3,
and FR4.
[0036] The term "CDR set" refers to a group of three CDRs that occur in a
single
variable region capable of binding the antigen. The exact boundaries of these
CDRs have
been defined differently according to different systems. The system described
by Kabat
(Kabat et at., SEQUENCES OF PROTEINS OF IMMUNOLOGICAL INTEREST (National
Institutes of
Health, Bethesda, Md. (1987) and (1991)) not only provides an unambiguous
residue
numbering system applicable to any variable region of an antibody, but also
provides
precise residue boundaries defining the three CDRs. These CDRs may be referred
to as
Kabat CDRs. Chothia and coworkers (Chothia and Lesk, 1987, J. Mot. Biol. 196:
901-17;
Chothia et at., 1989, Nature 342: 877-83) found that certain sub-portions
within Kabat
CDRs adopt nearly identical peptide backbone conformations, despite having
great
diversity at the level of amino acid sequence. These sub-portions were
designated as Li,
L2, and L3 or H1, H2, and H3 where the "L" and the "H" designates the light
chain and the
heavy chain regions, respectively. These regions may be referred to as Chothia
CDRs,
which have boundaries that overlap with Kabat CDRs. Other boundaries defining
CDRs
overlapping with the Kabat CDRs have been described by Padlan, 1995, FASEB J
9: 133-
39; MacCallum, 1996, J. Mot. Biol. 262(5): 732-45; and Lefranc, 2003, Dev.
Comp.
Immunol. 27: 55-77. Still other CDR boundary definitions may not strictly
follow one of
the herein systems, but will nonetheless overlap with the Kabat CDRs, although
they may
be shortened or lengthened in light of prediction or experimental findings
that particular
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
residues or groups of residues or even entire CDRs do not significantly impact
antigen
binding. The methods used herein may utilize CDRs defined according to any of
these
systems, although certain embodiments use Kabat or Chothia defined CDRs.
Identification
of predicted CDRs using the amino acid sequence is well known in the field,
such as in
Martin, A.C. "Protein sequence and structure analysis of antibody variable
domains," In
Antibody Engineering, Vol. 2. Kontermann R., Dithel S., eds. Springer-Verlag,
Berlin, p.
33-51 (2010). The amino acid sequence of the heavy and/or light chain variable
domain
may be also inspected to identify the sequences of the CDRs by other
conventional
methods, e.g., by comparison to known amino acid sequences of other heavy and
light
chain variable regions to determine the regions of sequence hypervariability.
The
numbered sequences may be aligned by eye, or by employing an alignment program
such
as one of the CLUSTAL suite of programs, as described in Thompson, 1994,
Nucleic Acids
Res. 22: 4673-80. Molecular models are conventionally used to correctly
delineate
framework and CDR regions and thus correct the sequence-based assignments.
[0037] The term "Fc" as used herein refers to a molecule comprising the
sequence of a
non-antigen-binding fragment resulting from digestion of an antibody or
produced by other
means, whether in monomeric or multimeric form, and can contain the hinge
region. The
original immunoglobulin source of the native Fc is preferably of human origin
and can be
any of the immunoglobulins, although IgG1 and IgG2 are preferred. Fc molecules
are
made up of monomeric polypeptides that can be linked into dimeric or
multimeric forms by
covalent (i.e., disulfide bonds) and non-covalent association. The number of
intermolecular
disulfide bonds between monomeric subunits of native Fc molecules ranges from
1 to 4
depending on class (e.g., IgG, IgA, and IgE) or subclass (e.g., IgGl, IgG2,
IgG3, IgAl, and
IgGA2). One example of a Fc is a disulfide-bonded dimer resulting from papain
digestion
of an IgG. The term "Fc" as used herein is generic to the monomeric, dimeric,
and
multimeric forms.
[0038] A F(ab) fragment typically includes one light chain and the VH and
Cm domains
of one heavy chain, wherein the VH-Cm heavy chain portion of the F(ab)
fragment cannot
form a disulfide bond with another heavy chain polypeptide. As used herein, a
F(ab)
fragment can also include one light chain containing two variable domains
separated by an
amino acid linker and one heavy chain containing two variable domains
separated by an
amino acid linker and a Cm domain.
26
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
[0039] A F(ab') fragment typically includes one light chain and a portion
of one heavy
chain that contains more of the constant region (between the CFH and CH2
domains), such
that an interchain disulfide bond can be formed between two heavy chains to
form a F(al302
molecule.
[0040] The term "binding protein" as used herein refers to a non-naturally
occurring (or
recombinant or engineered) molecule that specifically binds to at least one
target antigen.
A trispecific binding protein of the present disclosure, unless otherwise
specified, typically
comprises four polypeptide chains that form at least three antigen binding
sites, wherein a
first polypeptide chain has a structure represented by the formula:
VL2- L1-VL1- L2-CL [I]
and a second polypeptide chain has a structure represented by the formula:
VH1-L3-VH2-L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain has a structure represented by the formula:
VH3-CH1 [M]
and a fourth polypeptide chain has a structure represented by the formula:
VL3-CL [IV]
wherein:
VLi is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VFH is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is the immunoglobulin CHi heavy chain constant domain; and
hinge is an immunoglobulin hinge region connecting the Cm and CH2 domains;
Li, L2, L3 and L4 are amino acid linkers;
and wherein the polypeptide of formula I and the polypeptide of formula II
form a cross-
over light chain-heavy chain pair.
[0041] A "recombinant" molecule is one that has been prepared, expressed,
created, or
isolated by recombinant means.
[0042] One embodiment of the disclosure provides binding proteins having
biological
and immunological specificity to between one and three target antigens.
Another
27
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
embodiment of the disclosure provides nucleic acid molecules comprising
nucleotide
sequences encoding polypeptide chains that form such binding proteins. Another
embodiment of the disclosure provides expression vectors comprising nucleic
acid
molecules comprising nucleotide sequences encoding polypeptide chains that
form such
binding proteins. Yet another embodiment of the disclosure provides host cells
that express
such binding proteins (i.e., comprising nucleic acid molecules or vectors
encoding
polypeptide chains that form such binding proteins).
[0043] The term "swapability" as used herein refers to the
interchangeability of variable
domains within the binding protein format and with retention of folding and
ultimate
binding affinity. "Full swapability" refers to the ability to swap the order
of both VH1 and
VH2 domains, and therefore the order of VIA and VL2 domains, in the
polypeptide chain of
formula I or the polypeptide chain of formula II (i.e., to reverse the order)
while
maintaining full functionality of the binding protein as evidenced by the
retention of
binding affinity. Furthermore, it should be noted that the designations VH and
VL refer only
to the domain's location on a particular protein chain in the final format.
For example, VH1
and VH2 could be derived from VIA and VL2 domains in parent antibodies and
placed into
the VH1 and VH2 positions in the binding protein. Likewise, VIA and VL2 could
be derived
from VH1 and VH2 domains in parent antibodies and placed in the VH1 and VH2
positions in
the binding protein. Thus, the VH and VL designations refer to the present
location and not
the original location in a parent antibody. VH and VL domains are therefore
"swappable."
[0044] The term "antigen" or "target antigen" or "antigen target" as used
herein refers to
a molecule or a portion of a molecule that is capable of being bound by a
binding protein,
and additionally is capable of being used in an animal to produce antibodies
capable of
binding to an epitope of that antigen. A target antigen may have one or more
epitopes.
With respect to each target antigen recognized by a binding protein, the
binding protein is
capable of competing with an intact antibody that recognizes the target
antigen.
[0045] The term "HIV" as used herein means Human Immunodeficiency Virus. As
used herein, the term "HIV infection" generally encompasses infection of a
host,
particularly a human host, by the human immunodeficiency virus (HIV) family of
retroviruses including, but not limited to, HIV I, HIV II, HIV III (also known
as HTLV-II,
LAV-1, LAV-2). HIV can be used herein to refer to any strains, forms,
subtypes, clades and
variations in the HIV family. Thus, treating HIV infection will encompass the
treatment of
a person who is a carrier of any of the HIV family of retroviruses or a person
who is
28
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
diagnosed with active AIDS, as well as the treatment or prophylaxis of the
AIDS-related
conditions in such persons.
[0046] The term "AIDS" as used herein means Acquired Immunodeficiency
Syndrome.
AIDS is caused by HIV.
[0047] The terms "CD4bs" or "CD4 binding site" refer to the binding site
for CD4
(cluster of differentiation 4), which is a glycoprotein found on the surface
of immune cells
such as T helper cells, monocytes, macrophages, and dendritic cells.
[0048] The term "CD3" is cluster of differentiation factor 3 polypeptide
and is a T-cell
surface protein that is typically part of the T cell receptor (TCR) complex.
[0049] "CD28" is cluster of differentiation 28 polypeptide and is a T-cell
surface
protein that provides co-stimulatory signals for T-cell activation and
survival.
[0050] The term "glycoprotein 160" or "gp160 protein" refers to the
envelope
glycoprotein complex of HIV and which is a homotrimer that is cleaved into
gp120 and
gp41 subunits.
[0051] The term "MPER" refers to the membrane-proximal external region of
glycoprotein 41 (gp41), which is a subunit of the envelope protein complex of
retroviruses,
including HIV.
[0052] The term "glycan" refers to the carbohydrate portion of a
glycoconjugate, such
as a glycoprotein, glycolipid, or a proteoglycan. In the disclosed binding
proteins, glycan
refers to the HIV-1 envelope glycoprotein gp120.
[0053] The term "T-cell engager" refers to binding proteins directed to a
host's immune
system, more specifically the T cells' cytotoxic activity as well as directed
to a HIV target
protein.
[0054] The term "trimer apex" refers to apex of HIV-1 envelope glycoprotein
gp120.
[0055] The term "monospecific binding protein" refers to a binding protein
that
specifically binds to one antigen target.
[0056] The term "monovalent binding protein" refers to a binding protein
that has one
antigen binding site.
[0057] The term "bispecific binding protein" refers to a binding protein
that specifically
binds to two different antigen targets.
[0058] The term "bivalent binding protein" refers to a binding protein that
has two
binding sites.
29
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
[0059] The term "trispecific binding protein" refers to a binding protein
that specifically
binds to three different antigen targets.
[0060] The term "trivalent binding protein" refers to a binding protein
that has three
binding sites. In particular embodiments the trivalent binding protein can
bind to one
antigen target. In other embodiments, the trivalent binding protein can bind
to two antigen
targets. In other embodiments, the trivalent binding protein can bind to three
antigen
targets.
[0061] An "isolated" binding protein is one that has been identified and
separated
and/or recovered from a component of its natural environment. Contaminant
components
of its natural environment are materials that would interfere with diagnostic
or therapeutic
uses for the binding protein, and may include enzymes, hormones, and other
proteinaceous
or non-proteinaceous solutes. In some embodiments, the binding protein will be
purified:
(1) to greater than 95% by weight of antibody as determined by the Lowry
method, and
most preferably more than 99% by weight, (2) to a degree sufficient to obtain
at least 15
residues of N-terminal or internal amino acid sequence by use of a spinning
cup sequenator,
or (3) to homogeneity by SDS-PAGE under reducing or nonreducing conditions
using
Coomassie blue or, preferably, silver stain. Isolated binding proteins include
the binding
protein in situ within recombinant cells since at least one component of the
binding
protein's natural environment will not be present.
[0062] The terms "substantially pure" or "substantially purified" as used
herein refer to
a compound or species that is the predominant species present (i.e., on a
molar basis it is
more abundant than any other individual species in the composition). In some
embodiments, a substantially purified fraction is a composition wherein the
species
comprises at least about 50% (on a molar basis) of all macromolecular species
present. In
other embodiments, a substantially pure composition will comprise more than
about 80%,
85%, 90%, 95%, or 99% of all macromolar species present in the composition. In
still
other embodiments, the species is purified to essential homogeneity
(contaminant species
cannot be detected in the composition by conventional detection methods)
wherein the
composition consists essentially of a single macromolecular species.
[0063] A "neutralizing" binding protein as used herein refers to a molecule
that is able
to block or substantially reduce an effector function of a target antigen to
which it binds.
As used herein, "substantially reduce" means at least about 60%, preferably at
least about
70%, more preferably at least about 75%, even more preferably at least about
80%, still
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
more preferably at least about 85%, most preferably at least about 90%
reduction of an
effector function of the target antigen.
[0064] The term "epitope" includes any determinant, preferably a
polypeptide
determinant, capable of specifically binding to an immunoglobulin or T-cell
receptor. In
certain embodiments, epitope determinants include chemically active surface
groupings of
molecules such as amino acids, sugar side chains, phosphoryl groups, or
sulfonyl groups,
and, in certain embodiments, may have specific three-dimensional structural
characteristics
and/or specific charge characteristics. An epitope is a region of an antigen
that is bound by
an antibody or binding protein. In certain embodiments, a binding protein is
said to
specifically bind an antigen when it preferentially recognizes its target
antigen in a complex
mixture of proteins and/or macromolecules. In some embodiments, a binding
protein is
said to specifically bind an antigen when the equilibrium dissociation
constant is < 10' M,
more preferably when the equilibrium dissociation constant is < 10-9M, and
most
preferably when the dissociation constant is < 10-10 M.
[0065] The dissociation constant (KD) of a binding protein can be
determined, for
example, by surface plasmon resonance. Generally, surface plasmon resonance
analysis
measures real-time binding interactions between ligand (a target antigen on a
biosensor
matrix) and analyte (a binding protein in solution) by surface plasmon
resonance (SPR)
using the BIAcore system (Pharmacia Biosensor; Piscataway, NJ). Surface
plasmon
analysis can also be performed by immobilizing the analyte (binding protein on
a biosensor
matrix) and presenting the ligand (target antigen). The term "KID," as used
herein refers to
the dissociation constant of the interaction between a particular binding
protein and a target
antigen.
[0066] The term "specifically binds" as used herein refers to the ability
of a binding
protein or an antigen-binding fragment thereof to bind to an antigen
containing an epitope
with an Kd of at least about lx 106M, lx 107M, lx 10-8M, lx 109M, lx 10' M, lx
10-11 M, 1 x 10-12 M, or more, and/or to bind to an epitope with an affinity
that is at least
two-fold greater than its affinity for a nonspecific antigen.
[0067] The term "linker" as used herein refers to one or more amino acid
residues
inserted between immunoglobulin domains to provide sufficient mobility for the
domains
of the light and heavy chains to fold into cross over dual variable region
immunoglobulins.
A linker is inserted at the transition between variable domains or between
variable and
constant domains, respectively, at the sequence level. The transition between
domains can
31
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
be identified because the approximate size of the immunoglobulin domains are
well
understood. The precise location of a domain transition can be determined by
locating
peptide stretches that do not form secondary structural elements such as beta-
sheets or
alpha-helices as demonstrated by experimental data or as can be assumed by
techniques of
modeling or secondary structure prediction. The linkers described herein are
referred to as
Li, which is located on the light chain between the C-terminus of the VL2 and
the N-
terminus of the VLi domain; and L2, which is located on the light chain
between the C-
terminus of the VLi and the N-terminus of the CL domain. The heavy chain
linkers are
known as L3, which is located between the C-terminus of the Vui and the N-
terminus of the
Vii2 domain; and L4, which is located between the C-terminus of the Vii2 and
the N-
terminus of the Cul domain.
[0068] The term "vector" as used herein refers to any molecule (e.g.,
nucleic acid,
plasmid, or virus) that is used to transfer coding information to a host cell.
The term
"vector" includes a nucleic acid molecule that is capable of transporting
another nucleic
acid to which it has been linked. One type of vector is a "plasmid," which
refers to a
circular double-stranded DNA molecule into which additional DNA segments may
be
inserted. Another type of vector is a viral vector, wherein additional DNA
segments may
be inserted into the viral genome. Certain vectors are capable of autonomous
replication in
a host cell into which they are introduced (e.g., bacterial vectors having a
bacterial origin of
replication and episomal mammalian vectors). Other vectors (e.g., non-episomal
mammalian vectors) can be integrated into the genome of a host cell upon
introduction into
the host cell and thereby are replicated along with the host genome. In
addition, certain
vectors are capable of directing the expression of genes to which they are
operatively
linked. Such vectors are referred to herein as "recombinant expression
vectors" (or simply,
"expression vectors"). In general, expression vectors of utility in
recombinant DNA
techniques are often in the form of plasmids. The terms "plasmid" and "vector"
may be
used interchangeably herein, as a plasmid is the most commonly used form of
vector.
However, the disclosure is intended to include other forms of expression
vectors, such as
viral vectors (e.g., replication defective retroviruses, adenoviruses, and
adeno-associated
viruses), which serve equivalent functions.
[0069] The phrase "recombinant host cell" (or "host cell") as used herein
refers to a cell
into which a recombinant expression vector has been introduced. A recombinant
host cell
or host cell is intended to refer not only to the particular subject cell, but
also to the progeny
32
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
of such a cell. Because certain modifications may occur in succeeding
generations due to
either mutation or environmental influences, such progeny may not, in fact, be
identical to
the parent cell, but such cells are still included within the scope of the
term "host cell" as
used herein. A wide variety of host cell expression systems can be used to
express the
binding proteins, including bacterial, yeast, baculoviral, and mammalian
expression
systems (as well as phage display expression systems). An example of a
suitable bacterial
expression vector is pUC19. To express a binding protein recombinantly, a host
cell is
transformed or transfected with one or more recombinant expression vectors
carrying DNA
fragments encoding the polypeptide chains of the binding protein such that the
polypeptide
chains are expressed in the host cell and, preferably, secreted into the
medium in which the
host cells are cultured, from which medium the binding protein can be
recovered.
[0070] The term "transformation" as used herein refers to a change in a
cell's genetic
characteristics, and a cell has been transformed when it has been modified to
contain a new
DNA. For example, a cell is transformed where it is genetically modified from
its native
state. Following transformation, the transforming DNA may recombine with that
of the cell
by physically integrating into a chromosome of the cell, or may be maintained
transiently as
an episomal element without being replicated, or may replicate independently
as a plasmid.
A cell is considered to have been stably transformed when the DNA is
replicated with the
division of the cell. The term "transfection" as used herein refers to the
uptake of foreign or
exogenous DNA by a cell, and a cell has been "transfected" when the exogenous
DNA has
been introduced inside the cell membrane. A number of transfection techniques
are well
known in the art. Such techniques can be used to introduce one or more
exogenous DNA
molecules into suitable host cells.
[0071] The term "naturally occurring" as used herein and applied to an
object refers to
the fact that the object can be found in nature and has not been manipulated
by man. For
example, a polynucleotide or polypeptide that is present in an organism
(including viruses)
that can be isolated from a source in nature and that has not been
intentionally modified by
man is naturally-occurring. Similarly, "non-naturally occurring" as used
herein refers to an
object that is not found in nature or that has been structurally modified or
synthesized by
man.
[0072] As used herein, the twenty conventional amino acids and their
abbreviations
follow conventional usage. Stereoisomers (e.g., D-amino acids) of the twenty
conventional
amino acids; unnatural amino acids and analogs such as a-, a-disubstituted
amino acids, N-
33
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
alkyl amino acids, lactic acid, and other unconventional amino acids may also
be suitable
components for the polypeptide chains of the binding proteins. Examples of
unconventional amino acids include: 4-hydroxyproline, y-carboxyglutamate, c-
N,N,N-
trimethyllysine, c-N-acetyllysine, 0-phosphoserine, N-acetylserine, N-
formylmethionine,
3-methylhistidine, 5-hydroxylysine, a-N-methylarginine, and other similar
amino acids and
imino acids (e.g., 4-hydroxyproline). In the polypeptide notation used herein,
the left-hand
direction is the amino terminal direction and the right-hand direction is the
carboxyl-
terminal direction, in accordance with standard usage and convention.
[0073] Naturally occurring residues may be divided into classes based on
common side
chain properties:
(1) hydrophobic: Met, Ala, Val, Leu, Ile, Phe, Trp, Tyr, Pro;
(2) polar hydrophilic: Arg, Asn, Asp, Gln, Glu, His, Lys, Ser, Thr ;
(3) aliphatic: Ala, Gly, Ile, Leu, Val, Pro;
(4) aliphatic hydrophobic: Ala, Ile, Leu, Val, Pro;
(5) neutral hydrophilic: Cys, Ser, Thr, Asn, Gln;
(6) acidic: Asp, Glu;
(7) basic: His, Lys, Arg;
(8) residues that influence chain orientation: Gly, Pro;
(9) aromatic: His, Trp, Tyr, Phe; and
(10) aromatic hydrophobic: Phe, Trp, Tyr.
[0074] Conservative amino acid substitutions may involve exchange of a
member of
one of these classes with another member of the same class. Non-conservative
substitutions
may involve the exchange of a member of one of these classes for a member from
another
class.
[0075] A skilled artisan will be able to determine suitable variants of the
polypeptide
chains of the binding proteins using well-known techniques. For example, one
skilled in
the art may identify suitable areas of a polypeptide chain that may be changed
without
destroying activity by targeting regions not believed to be important for
activity.
Alternatively, one skilled in the art can identify residues and portions of
the molecules that
are conserved among similar polypeptides. In addition, even areas that may be
important
for biological activity or for structure may be subject to conservative amino
acid
substitutions without destroying the biological activity or without adversely
affecting the
polypeptide structure.
34
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
[0076] The term "patient" as used herein includes human and animal
subjects.
[0077] The terms "treatment" or "treat" as used herein refer to both
therapeutic
treatment and prophylactic or preventative measures. Those in need of
treatment include
those having the disorder as well as those prone to have a disorder or those
in which the
disorder is to be prevented. In particular embodiments, binding proteins can
be used to
treat humans infected with HIV, or humans susceptible to HIV infection, or
ameliorate HIV
infection in a human subject infected with HIV. The binding proteins can also
be used to
prevent HIV in a human patient.
[0078] It should be understood as that treating humans infected with HIV
include those
subjects who are at any one of the several stages of HIV infection
progression, which, for
example, include acute primary infection syndrome (which can be asymptomatic
or
associated with an influenza-like illness with fevers, malaise, diarrhea and
neurologic
symptoms such as headache), asymptomatic infection (which is the long latent
period with
a gradual decline in the number of circulating CD4+T cells), and AIDS (which
is defined by
more serious AIDS-defining illnesses and/or a decline in the circulating CD4
cell count to
below a level that is compatible with effective immune function). In addition,
treating or
preventing HIV infection will also encompass treating suspected infection by
HIV after
suspected past exposure to HIV by e.g., contact with HIV-contaminated blood,
blood
transfusion, exchange of body fluids, "unsafe" sex with an infected person,
accidental
needle stick, receiving a tattoo or acupuncture with contaminated instruments,
or
transmission of the virus from a mother to a baby during pregnancy, delivery
or shortly
thereafter.
[0079] The terms "pharmaceutical composition" or "therapeutic composition"
as used
herein refer to a compound or composition capable of inducing a desired
therapeutic effect
when properly administered to a patient.
[0080] The term "pharmaceutically acceptable carrier" or "physiologically
acceptable
carrier" as used herein refers to one or more formulation materials suitable
for
accomplishing or enhancing the delivery of a binding protein.
[0081] The terms "effective amount" and "therapeutically effective amount"
when used
in reference to a pharmaceutical composition comprising one or more binding
proteins refer
to an amount or dosage sufficient to produce a desired therapeutic result.
More specifically,
a therapeutically effective amount is an amount of a binding protein
sufficient to inhibit, for
some period of time, one or more of the clinically defined pathological
processes associated
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
with the condition being treated. The effective amount may vary depending on
the specific
binding protein that is being used, and also depends on a variety of factors
and conditions
related to the patient being treated and the severity of the disorder. For
example, if the
binding protein is to be administered in vivo, factors such as the age,
weight, and health of
the patient as well as dose response curves and toxicity data obtained in
preclinical animal
work would be among those factors considered. The determination of an
effective amount
or therapeutically effective amount of a given pharmaceutical composition is
well within
the ability of those skilled in the art.
[0082] One embodiment of the disclosure provides a pharmaceutical
composition
comprising a pharmaceutically acceptable carrier and a therapeutically
effective amount of
a binding protein.
Trispecific and/or Trivalent Binding Proteins
[0083] Certain aspects of the present disclosure relate to trispecific
and/or trivalent
binding proteins. Any of the CDRs or variable domains of any of the antigen
binding
proteins described herein may find use in a trispecific binding protein of the
present
disclosure. Trispecific binding proteins of various formats are contemplated.
In some
embodiments, the binding protein of the disclosure is a trispecific and/or
trivalent binding
protein comprising four polypeptide chains that form three antigen binding
sites that
specifically bind one or more HIV target proteins, wherein a first polypeptide
chain
comprises a structure represented by the formula:
VL2- L2-CL [I]
and a second polypeptide chain comprises a structure represented by the
formula:
VH1- L3-VH2- L4-CH1-hinge-CH2-CH3 [II]
and a third polypeptide chain comprises a structure represented by the
formula:
VH3-CH1-hinge-CH2-CH3 [III]
and a fourth polypeptide chain comprises a structure represented by the
formula:
VL3-CL [IV]
wherein:
Vu is a first immunoglobulin light chain variable domain;
VL2 is a second immunoglobulin light chain variable domain;
VL3 is a third immunoglobulin light chain variable domain;
VFH is a first immunoglobulin heavy chain variable domain;
VH2 is a second immunoglobulin heavy chain variable domain;
36
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
VH3 is a third immunoglobulin heavy chain variable domain;
CL is an immunoglobulin light chain constant domain;
CHi is an immunoglobulin CHi heavy chain constant domain;
CH2 is an immunoglobulin CH2 heavy chain constant domain;
CH3 is an immunoglobulin CH3 heavy chain constant domain;
hinge is an immunoglobulin hinge region connecting the CHi and CH2 domains;
and
Li, L2, L3 and L4 are amino acid linkers;
and wherein the polypeptide of formula I and the polypeptide of formula II
form a cross-over
light chain-heavy chain pair.
[0084] In some embodiments, the first polypeptide chain and the second
polypeptide
chain have a cross-over orientation that forms two distinct antigen binding
sites. In some
embodiments, the VH1 and VLi form a binding pair and form the first antigen
binding site.
In some embodiments, the VH2 and VL2 form a binding pair and form the second
antigen
binding site. In some embodiments, the third polypeptide and the fourth
polypeptide form a
third antigen binding site. In some embodiments, the VH3 and VL3 form a
binding pair and
form the third antigen binding site.
[0085] In some embodiments, one or more of the antigen binding sites binds
an HIV
target protein. In some embodiments, VH3 and VL3 form a third antigen binding
site that
binds an HIV target protein. In some embodiments, VH1 and VLi form a first
antigen
binding site that binds a T cell target protein, VH2 and VL2 form a second
antigen binding
site that binds a T cell target protein, and VH3 and VL3 form a third antigen
binding site that
binds an HIV target protein. In some embodiments, VH1 and VLi form a first
antigen
binding site that binds a T cell target protein, VH2 and VL2 form a second
antigen binding
site that binds a CD3 polypeptide, and VH3 and VL3 form a third antigen
binding site that
binds an HIV target protein. In some embodiments, VH1 and VLi form a first
antigen
binding site that binds a CD28 polypeptide, VH2 and VL2 form a second antigen
binding site
that binds a CD3 polypeptide, and VH3 and VL3 form a third antigen binding
site that binds
an HIV target protein.
[0086] In some embodiments, the binding proteins specifically bind to one
or more HIV
target proteins (e.g., as described infra) and one or more target proteins on
a T-cell
including T cell receptor complex. These T-cell engager binding proteins are
capable of
recruiting T cells transiently to target cells and, at the same time,
activating the cytolytic
activity of the T cells. The T-cell engager trispecific antibodies can be used
to activate HIV-
37
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
1 reservoirs and redirect/activate T cells to lyse latently infected HIV-1+ T
cells. Examples
of target proteins on T cells include but are not limited to CD3 and CD28,
among others. In
some embodiments, the trispecific binding proteins may be generated by
combining the
antigen binding domains of two or more monospecific antibodies (parent
antibodies) into
one antibody. See International Publication Nos. WO 2011/038290 A2, WO
2013/086533
Al, WO 2013/070776 Al, WO 2012/154312 Al, and WO 2013/163427 Al. The binding
proteins of the disclosure may be prepared using domains or sequences obtained
or derived
from any human or non-human antibody, including, for example, human, murine,
or
humanized antibodies.
[0087] In some embodiments of the disclosure, the trivalent binding protein
is capable
of binding three different antigen targets. In one embodiment, the binding
protein is
trispecific and one light chain-heavy chain pair is capable of binding two
different antigen
targets or epitopes and one light chain-heavy chain pair is capable of binding
one antigen
target or epitope.
[0088] In some embodiments, a binding protein of the present disclosure
binds one or
more HIV target proteins and one or more T cell target proteins. In some
embodiments, the
binding protein is capable of specifically binding one HIV target protein and
two different
epitopes on a single T cell target protein. In some embodiments, the binding
protein is
capable of specifically binding one HIV target protein and two different T
cell target
proteins (e.g., CD28 and CD3). In some embodiments, the first and second
polypeptide
chains of the binding protein form two antigen binding sites that specifically
target two T
cell target proteins, and the third and fourth polypeptide chains of the
binding protein form
an antigen binding site that specifically binds an HIV target protein. In some
embodiments,
the one or more HIV target proteins are one or more of glycoprotein 120,
glycoprotein 41,
and glycoprotein 160. In some embodiments, the one or more T cell target
proteins are one
or more of CD3 and CD28.
[0089] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:61 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:61; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:62 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:62; the third polypeptide chain comprises the
amino
38
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
acid sequence of SEQ ID NO:63 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:63; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:64 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:64.
[0090] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:65 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:65; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:66 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:66; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:67 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:67; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:68 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:68.
[0091] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:69 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:69; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:70 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:70; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:71 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:71; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:72 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:72.
[0092] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:73 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:73; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:74 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:74; the third polypeptide chain comprises the
amino
39
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
acid sequence of SEQ ID NO:75 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:75; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:76 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:76.
[0093] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:77 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:77; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:78 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:78; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:79 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:79; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:80 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:80.
[0094] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:81 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:81; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:82 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:82; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:83 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:83; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:84 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:84.
[0095] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:85 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:85; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:86 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:86; the third polypeptide chain comprises the
amino
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
acid sequence of SEQ ID NO:87 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:87; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:88 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:88.
[0096] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:89 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:89; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:90 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:90; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:91 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:91; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:92 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:92.
[0097] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:93 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:93; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:94 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:94; the third polypeptide chain comprises the
amino
acid sequence of SEQ ID NO:95 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:95; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:96 or an amino acid sequence that is at least
95%
identical to the amino acid sequence of SEQ ID NO:96.
[0098] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:97 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:97; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:98 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:98; the third polypeptide chain comprises the
amino
41
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
acid sequence of SEQ ID NO:99 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:99; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:100 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:100.
[0099] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:101 or an amino acid sequence that is at least 95% identical to
the amino
acid sequence of SEQ ID NO:101; the second polypeptide chain comprises the
amino acid
sequence of SEQ ID NO:102 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:102; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:103 or an amino acid sequence that is at least 95%
identical
to the amino acid sequence of SEQ ID NO:103; and the fourth polypeptide chain
comprises
the amino acid sequence of SEQ ID NO:104 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:104.
[0100] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:105 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:105; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:106 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:106; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:107 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:107; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:108 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:108.
[0101] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:109 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:109; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:110 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:110; the third polypeptide chain comprises
the amino
42
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
acid sequence of SEQ ID NO:111 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:111; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:112 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:112.
[0102] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:113 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:113; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:114 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:114; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:115 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:115; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:116 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:116.
[0103] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:117 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:117; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:118 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:118; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:119 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:119; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:120 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:120.
[0104] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:121 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:121; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:122 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:122; the third polypeptide chain comprises
the amino
43
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
acid sequence of SEQ ID NO:123 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:123; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:124 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:124.
[0105] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:129 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:129; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:130 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:130; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:131 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:131; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:132 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:132.
[0106] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:133 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:133; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:134 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:134; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:135 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:135; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:136 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:136.
[0107] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:137 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:137; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:138 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:138; the third polypeptide chain comprises
the amino
44
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
acid sequence of SEQ ID NO:139 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:139; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:140 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:140.
[0108] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:141 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:141; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:142 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:142; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:143 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:143; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:144 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:144.
[0109] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:145 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:145; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:146 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:146; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:147 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:147; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:148 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:148.
[0110] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:149 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:149; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:150 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:150; the third polypeptide chain comprises
the amino
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
acid sequence of SEQ ID NO:151 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:151; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:152 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:152.
[0111] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:153 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:153; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:154 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:154; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:155 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:155; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:156 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:156.
[0112] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:157 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:157; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:158 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:158; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:159 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:159; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:160 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:160.
[0113] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:161 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:161; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:162 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:162; the third polypeptide chain comprises
the amino
46
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
acid sequence of SEQ ID NO:163 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:163; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:164 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:164.
[0114] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:165 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:165; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:166 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:166; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:167 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:167; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:168 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:168.
[0115] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:169 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:169; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:170 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:170; the third polypeptide chain comprises
the amino
acid sequence of SEQ ID NO:171 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:171; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:172 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:172.
[0116] In some embodiments, a binding protein of the present disclosure
comprises four
polypeptide chains that form three antigen binding sites that specifically
bind one or more
HIV target proteins, wherein the first polypeptide chain comprises the amino
acid sequence
of SEQ ID NO:173 or an amino acid sequence that is at least 95% identical to
the amino acid
sequence of SEQ ID NO:173; the second polypeptide chain comprises the amino
acid
sequence of SEQ ID NO:174 or an amino acid sequence that is at least 95%
identical to the
amino acid sequence of SEQ ID NO:174; the third polypeptide chain comprises
the amino
47
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
acid sequence of SEQ ID NO:175 or an amino acid sequence that is at least 95%
identical to
the amino acid sequence of SEQ ID NO:175; and the fourth polypeptide chain
comprises the
amino acid sequence of SEQ ID NO:176 or an amino acid sequence that is at
least 95%
identical to the amino acid sequence of SEQ ID NO:176.
Exemplary and non-limiting polypeptides that may find use in any of the
trispecific binding
proteins described herein are provided in Table 4.
Anti-HIV Binding Sites
[0117] Certain aspects of the present disclosure relate to binding proteins
that comprise
an antigen binding site that binds an HIV target protein or polypeptide.
[0118] In some embodiments, the HIV target protein is glycoprotein 120,
glycoprotein
41, or glycoprotein 160. In some embodiments, a binding protein binds one or
more of:
glycoprotein 120, glycoprotein 41, and glycoprotein 160. Exemplary HIV target
proteins
include, without limitation, MPER of the HIV-1 gp41 protein, a CD4 binding
site of the HIV-
1 gp120 protein, a glycan in the V3 loop of the HIV-1 gp120 protein, or a
trimer apex of the
HIV-1 gp120 protein or gp160. For example, in some embodiments, a binding
protein of the
present disclosure comprises an antigen binding site that binds a CD4 binding
site of the
HIV-1 gp120 protein. Exemplary antigen binding sites that bind HIV target
proteins
contemplated for use herein include, without limitation, those described in
International
Publication No. W02017/074878, such as those from antibodies CD4BS "a", CD4BS
"b",
MPER, MPER 100W, V1/V2 "a", V1/V2 "b", or V3.
[0119] In some embodiments, a binding protein comprising an antigen binding
site that
binds an HIV target protein is monospecific and/or monovalent, bispecific
and/or bivalent,
trispecific and/or trivalent, or multispecific and/or multivalent. In some
embodiments, a
binding protein that comprises an antigen binding site that binds an HIV
target protein is a
trispecific binding protein comprising four polypeptides that form three
antigen binding sites.
[0120] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of NCPIN (SEQ ID NO:1) a
CDR-
H2 sequence comprising the amino acid sequence of WMKPRHGAVSYARQLQG (SEQ ID
NO:2), and a CDR-H3 sequence comprising the amino acid sequence of
GKYCTARDYYNWDFEH (SEQ ID NO:3); and/or an antibody light chain variable (VL)
domain comprising a CDR-L1 sequence comprising the amino acid sequence of
RTSQYGSLA (SEQ ID NO:4), a CDR-L2 sequence comprising the amino acid sequence
of
48
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
SGSTRAA (SEQ ID NO:5), and a CDR-L3 sequence comprising the amino acid
sequence of
QQYEF (SEQ ID NO:6). In some embodiments, a binding protein of the present
disclosure
comprises an antigen binding site comprising: an antibody heavy chain variable
(VH) domain
comprising a CDR-H1 sequence comprising the amino acid sequence of NCPIN (SEQ
ID
NO:1) a CDR-H2 sequence comprising the amino acid sequence of
WMKPRHGAVSYARQLQG (SEQ ID NO:2), and a CDR-H3 sequence comprising the
amino acid sequence of GKYCTARDYYNWDFEH (SEQ ID NO:3); and an antibody light
chain variable (VL) domain comprising a CDR-L1 sequence comprising the amino
acid
sequence of RTSQYGSLA (SEQ ID NO:4), a CDR-L2 sequence comprising the amino
acid
sequence of SGSTRAA (SEQ ID NO:5), and a CDR-L3 sequence comprising the amino
acid
sequence of QQYEF (SEQ ID NO:6).
[0121] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GYTFTAHI (SEQ ID NO:7) a
CDR-H2 sequence comprising the amino acid sequence of IKPQYGAV (SEQ ID NO:8)
or
IKPQYGAT (SEQ ID NO:9); and/or an antibody light chain variable (VL) domain
comprising a CDR-L1 sequence comprising the amino acid sequence of QGVGSD (SEQ
ID
NO:11), a CDR-L2 sequence comprising the amino acid sequence of HTS (SEQ ID
NO:12),
and a CDR-L3 sequence comprising the amino acid sequence of CQVLQF (SEQ ID
NO:13).
In some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising a CDR-
H1 sequence comprising the amino acid sequence of GYTFTAHI (SEQ ID NO:7) a CDR-
H2
sequence comprising the amino acid sequence of IKPQYGAV (SEQ ID NO:8) or
IKPQYGAT (SEQ ID NO:9); and an antibody light chain variable (VL) domain
comprising a
CDR-L1 sequence comprising the amino acid sequence of QGVGSD (SEQ ID NO:11), a
CDR-L2 sequence comprising the amino acid sequence of HTS (SEQ ID NO:12), and
a
CDR-L3 sequence comprising the amino acid sequence of CQVLQF (SEQ ID NO:13).
In
some embodiments, a binding protein of the present disclosure comprises an
antigen binding
site comprising: an antibody heavy chain variable (VH) domain comprising a CDR-
H1
sequence comprising the amino acid sequence of GYTFTAHI (SEQ ID NO:7) a CDR-H2
sequence comprising the amino acid sequence of IKPQYGAV (SEQ ID NO:8); and/or
an
antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the
amino acid sequence of QGVGSD (SEQ ID NO: ii), a CDR-L2 sequence comprising
the
49
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
amino acid sequence of HTS (SEQ ID NO:12), and a CDR-L3 sequence comprising
the
amino acid sequence of CQVLQF (SEQ ID NO:13). In some embodiments, a binding
protein of the present disclosure comprises an antigen binding site
comprising: an antibody
heavy chain variable (VH) domain comprising a CDR-H1 sequence comprising the
amino
acid sequence of GYTFTAHI (SEQ ID NO:7) a CDR-H2 sequence comprising the amino
acid sequence of IKPQYGAV (SEQ ID NO:8); and an antibody light chain variable
(VL)
domain comprising a CDR-L1 sequence comprising the amino acid sequence of
QGVGSD
(SEQ ID NO:11), a CDR-L2 sequence comprising the amino acid sequence of HTS
(SEQ ID
NO:12), and a CDR-L3 sequence comprising the amino acid sequence of CQVLQF
(SEQ ID
NO:13). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GYTFTAHI (SEQ ID NO:7) a
CDR-H2 sequence comprising the amino acid sequence of IKPQYGAT (SEQ ID NO:9);
and/or an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QGVGSD (SEQ ID NO:11), a CDR-L2 sequence
comprising the amino acid sequence of HTS (SEQ ID NO:12), and a CDR-L3
sequence
comprising the amino acid sequence of CQVLQF (SEQ ID NO:13). In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising a CDR-H1 sequence
comprising the
amino acid sequence of GYTFTAHI (SEQ ID NO:7) a CDR-H2 sequence comprising the
amino acid sequence of IKPQYGAT (SEQ ID NO:9); and an antibody light chain
variable
(VL) domain comprising a CDR-L1 sequence comprising the amino acid sequence of
QGVGSD (SEQ ID NO:11), a CDR-L2 sequence comprising the amino acid sequence of
HTS (SEQ ID NO:12), and a CDR-L3 sequence comprising the amino acid sequence
of
CQVLQF (SEQ ID NO:13).
[0122] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of DCTLN (SEQ ID NO:14) a
CDR-
H2 sequence comprising the amino acid sequence of WLKPRWGAVNYARPLQG (SEQ ID
NO:15), and a CDR-H3 sequence comprising the amino acid sequence of
GKNCDYNWDFEH (SEQ ID NO:16); and/or an antibody light chain variable (VL)
domain
comprising a CDR-L1 sequence comprising the amino acid sequence of RTSQYGSLA
(SEQ
ID NO:17), a CDR-L2 sequence comprising the amino acid sequence of SGSTRAA
(SEQ ID
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
NO:18), and a CDR-L3 sequence comprising the amino acid sequence of QQYEF (SEQ
ID
NO:19). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of DCTLN (SEQ ID NO:14) a
CDR-
H2 sequence comprising the amino acid sequence of WLKPRWGAVNYARPLQG (SEQ ID
NO:15), and a CDR-H3 sequence comprising the amino acid sequence of
GKNCDYNWDFEH (SEQ ID NO:16); and an antibody light chain variable (VL) domain
comprising a CDR-L1 sequence comprising the amino acid sequence of RTSQYGSLA
(SEQ
ID NO:17), a CDR-L2 sequence comprising the amino acid sequence of SGSTRAA
(SEQ ID
NO:18), and a CDR-L3 sequence comprising the amino acid sequence of QQYEF (SEQ
ID
NO:19).
[0123] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site with a VH domain comprising an extended heavy chain FR3
loop of
antibody VRC03, e.g., as described in Liu, Q. et at. (2019) Nat. Commun.
10:721.
[0124] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
QVRLSQSGGQMKKPGDSMRISCRASGYEFINCPINWIRLAPGKRPEWMGWMKPRHG
AVSYARQLQGRVTMTRDMYSETAFLELRSLTSDDTAVYFCTRGKYCTARDYYNWD
FEHWGQGTPVTVSS (SEQ ID NO:43), and/or an antibody light chain variable (VL)
domain comprising an amino acid sequence that is at least 85%, at least 86%,
at least 87%, at
least 88%, at least 89%, at least 90%, at least 91%, at least 92%, at least
93%, at least 94%, at
least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100%
identical to the
amino acid sequence of
SLTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRFS
GSRWGPDYNLTISNLESGDFGVYYCQQYEFFGQGTKVQVDIK (SEQ ID NO:45). In
some embodiments, a binding protein of the present disclosure comprises an
antigen binding
site comprising: an antibody heavy chain variable (VH) domain comprising the
amino acid
sequence of SEQ ID NO:43, and/or an antibody light chain variable (VL) domain
comprising
the amino acid sequence of SEQ ID NO:45. In some embodiments, a binding
protein of the
51
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
present disclosure comprises an antigen binding site comprising: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:43, and
an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:45.
[0125] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
QVRLSQSGGQMKKPGDSMRISCRASGYEFINCPINWIRLAPGKRPEWMGWMKPRHG
AVSYARQLQGRVTMTRQLSQDPDDPDWGTAFLELRSLTSDDTAVYFCTRGKYCTA
RDYYNWDFEHWGQGTPVTVSS (SEQ ID NO:44), and/or an antibody light chain variable
(VL) domain comprising an amino acid sequence that is at least 85%, at least
86%, at least
87%, at least 88%, at least 89%, at least 90%, at least 91%, at least 92%, at
least 93%, at least
94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or
100% identical to
the amino acid sequence of
SLTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRFS
GSRWGPDYNLTISNLESGDFGVYYCQQYEFFGQGTKVQVDIK (SEQ ID NO:45). In
some embodiments, a binding protein of the present disclosure comprises an
antigen binding
site comprising: an antibody heavy chain variable (VH) domain comprising the
amino acid
sequence of SEQ ID NO:44, and/or an antibody light chain variable (VL) domain
comprising
the amino acid sequence of SEQ ID NO:45. In some embodiments, a binding
protein of the
present disclosure comprises an antigen binding site comprising: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:44, and
an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:45.
[0126] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
52
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQY
GAVNFGGGFRDRVTLTRDVYREIAYMDIRGLKPDDTAVYYCARDRSYGDSSWALD
AWGQGTTVVVSA (SEQ ID NO:46), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGV
PSRFSGSGFHTSFNLTISDLQADDIATYYCQVLQFFGRGSRLHIK (SEQ ID NO:49). In
some embodiments, a binding protein of the present disclosure comprises an
antigen binding
site comprising: an antibody heavy chain variable (VH) domain comprising the
amino acid
sequence of SEQ ID NO:46, and/or an antibody light chain variable (VL) domain
comprising
the amino acid sequence of SEQ ID NO:49. In some embodiments, a binding
protein of the
present disclosure comprises an antigen binding site comprising: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:46, and
an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:49.
[0127] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQY
GATNFGGGFRDRVTLTRDVYREIAYMDIRGLKPDDTAVYYCARDRSYGDSSWALD
AWGQGTTVVVSA (SEQ ID NO:47), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGV
PSRFSGSGFHTSFNLTISDLQADDIATYYCQVLQFFGRGSRLHIK (SEQ ID NO:49). In
some embodiments, a binding protein of the present disclosure comprises an
antigen binding
53
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
site comprising: an antibody heavy chain variable (VH) domain comprising the
amino acid
sequence of SEQ ID NO:47, and/or an antibody light chain variable (VL) domain
comprising
the amino acid sequence of SEQ ID NO:49. In some embodiments, a binding
protein of the
present disclosure comprises an antigen binding site comprising: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:47, and
an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:49.
[0128] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQY
GAVNFGGGFRDRVTLTRQLSQDPDDPDWGIAYMDIRGLKPDDTAVYYCARDRSYG
DSSWALDAWGQGTTVVVSA (SEQ ID NO:48), and/or an antibody light chain variable
(VL) domain comprising an amino acid sequence that is at least 85%, at least
86%, at least
87%, at least 88%, at least 89%, at least 90%, at least 91%, at least 92%, at
least 93%, at least
94%, at least 95%, at least 96%, at least 97%, at least 98%, at least 99%, or
100% identical to
the amino acid sequence of
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGV
PSRFSGSGFHTSFNLTISDLQADDIATYYCQVLQFFGRGSRLHIK (SEQ ID NO:49). In
some embodiments, a binding protein of the present disclosure comprises an
antigen binding
site comprising: an antibody heavy chain variable (VH) domain comprising the
amino acid
sequence of SEQ ID NO:48, and/or an antibody light chain variable (VL) domain
comprising
the amino acid sequence of SEQ ID NO:49. In some embodiments, a binding
protein of the
present disclosure comprises an antigen binding site comprising: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:48, and
an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:49.
[0129] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
54
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
89%, at least 90%, at least 91%, at least 92%, at least 930 o, at least 940 o,
at least 950 o, at least
960 , at least 970 o, at least 98%, at least 990 o, or 10000 identical to the
amino acid sequence
of
QVQLVQSGGQMKKPGESMRISCRASGYEFIDCTLNWIRLAPGKRPEWMGWLKPRW
GAVNYARPLQGRVTMTRQLSQDPDDPDWGTAFLELRSLTVDDTAVYFCTRGKNCD
YNWDFEHWGRGTPVIVSS (SEQ ID NO:50), and/or an antibody light chain variable
(VL)
domain comprising an amino acid sequence that is at least 85%, at least 86%,
at least 87%, at
least 88%, at least 89%, at least 90%, at least 91%, at least 92%, at least
930, at least 940, at
least 950, at least 96%, at least 970, at least 98%, at least 990, or 100%
identical to the
amino acid sequence of
LTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRF SG
SRWGPDYNLTISNLESGDFGVYYCQQYEFFGQGTKVQVDIK (SEQ ID NO:51). In
some embodiments, a binding protein of the present disclosure comprises an
antigen binding
site comprising: an antibody heavy chain variable (VH) domain comprising the
amino acid
sequence of SEQ ID NO:50, and/or an antibody light chain variable (VL) domain
comprising
the amino acid sequence of SEQ ID NO:51. In some embodiments, a binding
protein of the
present disclosure comprises an antigen binding site comprising: an antibody
heavy chain
variable (VH) domain comprising the amino acid sequence of SEQ ID NO:50, and
an
antibody light chain variable (VL) domain comprising the amino acid sequence
of SEQ ID
NO:51.
[0130] In
some embodiments of any of the above embodiments, the binding protein is a
trispecific binding protein. In some embodiments, the trispecific binding
protein comprising
an antigen binding site that binds an HIV target protein, an antigen binding
site that binds a
CD28 polypeptide, and an antigen binding site that binds a CD3 polypeptide. In
some
embodiments, the binding protein is a trispecific binding protein comprising
four
polypeptides comprising three antigen binding sites, wherein the polypeptide
of formula I and
the polypeptide of formula II form a cross-over light chain-heavy chain pair
(e.g., as
described herein). In some embodiments, the VH and VL domains of any of the
anti-CD38
antigen binding sites described above represent VH3 and VL3 and form a third
antigen binding
site that binds an HIV target protein. In some embodiments, VFH and VIA form a
first antigen
binding site that binds a CD28 polypeptide, VH2 and VL2 form a second antigen
binding site
that binds a CD3 polypeptide, and the VH and VL domains of any of the anti-HIV
antigen
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
binding sites described above and/or in Table 1 represent VH3 and VL3 and form
a third
antigen binding site that binds an HIV target protein.
[0131] Sequences of exemplary anti-HIV antigen binding sites are provided
in Table 1. In
some embodiments, a binding protein comprising an anti-HIV antigen binding
site of the
present disclosure comprises 1, 2, 3, 4, 5, or all 6 CDR sequences of an anti-
HIV antibody
described in Table 1. In some embodiments, a binding protein comprising an
anti-HIV
antigen binding site of the present disclosure comprises a VH domain sequence
and/or VL
domain sequence of an anti-HIV antibody described in Table 1.
56
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
Table 1. Anti-HIV binding protein sequences.
Sequence Molecule Description SEQ Sequence
Type ID NO
CDR VRCO7 523 CDR-H1 1 NCPIN
(anti-Env
gp120 CDR-H2 2 WMKPRHGAVSYARQLQG
CD4bs)
CDR-H3 3 GKYCTARDYYNWDFEH
CDR-L1 4 RTSQYGSLA
CDR-L2 5 SGSTRAA
CDR-L3 6 QQYEF
N6 CDR-H1 7 GYTFTAHI
(anti-Env
gp120 CDR-H2 8 IKPQYGAV
CD4bs) Original
CDR-H2 rw52 9 IKPQYGAT
CDR-H3 10 DRSYGDSSWALDA
CDR-L1 11 QGVGSD
CDR-L2 12 HTS
CDR-L3 13 CQVLQF
VRC01.23 CDR-H1 14 DCTLN
CDR-H2 15 WLKPRWGAVNYARPLQG
CDR-H3 16 GKNCDYNWDFEH
CDR-L1 17 RTSQYGSLA
CDR-L2 18 SGSTRAA
CDR-L3 19 QQYEF
Variable VRCO7 523 VH 43 QVRLSQSGGQMKKPGDSMRISCRASG
domain YEFINCPINWIRLAPGKRPEWMGWM
KPRHGAVSYARQLQGRVTMTRDMYS
ETAFLELRSLTSDDTAVYFCTRGKYC
TARDYYNWDFEHWGQGTPVTVSS
FR3-03 VH 44 QVRLSQSGGQMKKPGDSMRISCRASG
YEFINCPINWIRLAPGKRPEWMGWM
KPRHGAVSYARQLQGRVTMTRQLSQ
DPDDPDWGTAFLELRSLTSDDTAVYF
CTRGKYCTARDYYNWDFEHWGQGT
PVTVSS
57
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
VL 45 SLTQSPGTLSLSPGETAIISCRTSQYGS
LAWYQQRPGQAPRLVIYSGSTRAAGI
PDRFSGSRWGPDYNLTISNLESGDFG
VYYCQQYEFFGQGTKVQVDIK
N6 VH 46 RAHLVQSGTAMKKPGASVRVSCQTS
GYTFTAHILFWFRQAPGRGLEWVGWI
KPQYGAVNFGGGFRDRVTLTRDVYR
EIAYMDIRGLKPDDTAVYYCARDRSY
GDS SWALDAWGQGTTVVVSA
rw52 VH 47 RAHLVQSGTAMKKPGASVRVSCQTS
GYTFTAHILFWFRQAPGRGLEWVGWI
KPQYGATNFGGGFRDRVTLTRDVYR
EIAYMDIRGLKPDDTAVYYCARDRSY
GDS SWALDAWGQGTTVVVSA
FR3-03 VH 48 RAHLVQSGTAMKKPGASVRVSCQTS
GYTFTAHILFWFRQAPGRGLEWVGWI
KPQYGAVNFGGGFRDRVTLTRQLSQ
DPDDPDWGIAYMDIRGLKPDDTAVY
YCARDRSYGDSSWALDAWGQGTTV
VVSA
VL 49 YIHVTQSPSSLSVSIGDRVTINCQTSQG
VGSDLHWYQHKPGRAPKLLIHHTSSV
EDGVPSRFSGSGFHTSFNLTISDLQAD
DIATYYCQVLQFFGRGSRLHIK
VRC01.23 VH 50 QVQLVQSGGQMKKPGESMRISCRAS
GYEFIDCTLNWIRLAPGKRPEWMGW
LKPRWGAVNYARPLQGRVTMTRQLS
QDPDDPDWGTAFLELRSLTVDDTAV
YFCTRGKNCDYNWDFEHWGRGTPVI
VSS
VL 51 LTQSPGTLSLSPGETAIISCRTSQYGSL
AWYQQRPGQAPRLVIYSGSTRAAGIP
DRFSGSRWGPDYNLTISNLESGDFGV
YYCQQYEFFGQGTKVQVDIK
Anti-CD28 Binding Sites
[0132]
Certain aspects of the present disclosure relate to binding proteins that
comprise
an antigen binding site that binds a CD28 polypeptide. In some embodiments,
the CD28
polypeptide is a human CD28 polypeptide, also known as Tp44. Human CD28
polypeptides
are known in the art and include, without limitation, the polypeptides
represented by NCBI
Accession Numbers XP 011510499.1 XP 011510497.1, XP 011510496.1,
_
NP 001230007.1, NP 001230006.1, or NP 006130.1, or a polypeptide produced from
NCBI
Gene ID Number 940. In some embodiments, a binding protein comprising an
antigen
binding site that binds a CD28 polypeptide is monospecific and/or monovalent,
bispecific
and/or bivalent, trispecific and/or trivalent, or multispecific and/or
multivalent. In some
embodiments, a binding protein that comprises an antigen binding site that
binds a CD28
58
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
polypeptide is a trispecific binding protein comprising four polypeptides that
form three
antigen binding sites. In some embodiments, a binding protein that comprises
an antigen
binding site that binds a CD28 polypeptide is a trispecific binding protein
comprising four
polypeptides that form three antigen binding sites, one of which binds a CD28
polypeptide,
and one of which binds a CD3 polypeptide. In some embodiments, a binding
protein that
comprises an antigen binding site that binds a CD28 polypeptide is a
trispecific binding
protein comprising four polypeptides that form three antigen binding sites,
one of which
binds a CD28 polypeptide, one of which binds a CD3 polypeptide, and one of
which binds an
HIV target protein.
[0133] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GYTFTSYY (SEQ ID NO:31),
a
CDR-H2 sequence comprising the amino acid sequence of IYPGNVNT (SEQ ID NO:32),
and a CDR-H3 sequence comprising the amino acid sequence of TRSHYGLDWNFDV (SEQ
ID NO:33); and/or an antibody light chain variable (VL) domain comprising a
CDR-L1
sequence comprising the amino acid sequence of QNIYVW (SEQ ID NO:34), a CDR-L2
sequence comprising the amino acid sequence of KAS (SEQ ID NO:35), and a CDR-
L3
sequence comprising the amino acid sequence of QQGQTYPY (SEQ ID NO:36). In
some
embodiments, a binding protein of the present disclosure comprises an antigen
binding site
comprising: an antibody heavy chain variable (VH) domain comprising a CDR-H1
sequence
comprising the amino acid sequence of GYTFTSYY (SEQ ID NO:31), a CDR-H2
sequence
comprising the amino acid sequence of IYPGNVNT (SEQ ID NO:32), and a CDR-H3
sequence comprising the amino acid sequence of TRSHYGLDWNFDV (SEQ ID NO:33);
and an antibody light chain variable (VL) domain comprising a CDR-L1 sequence
comprising the amino acid sequence of QNIYVW (SEQ ID NO:34), a CDR-L2 sequence
comprising the amino acid sequence of KAS (SEQ ID NO:35), and a CDR-L3
sequence
comprising the amino acid sequence of QQGQTYPY (SEQ ID NO:36).
[0134] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
59
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIEWVRQAPGQGLEWIGSIYPGNV
NTNYAQKFQGRATLTVDTSISTAYMELSRLRSDDTAVYYCTRSHYGLDWNFDVWG
KGTTVTVSS (SEQ ID NO:59), and/or an antibody light chain variable (VL) domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIQMTQSPSSLSASVGDRVTITCQASQNIYVWLNWYQQKPGKAPKWYKASNLHTG
VPSRF SGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIK (SEQ ID
NO:60). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
the amino acid sequence of SEQ ID NO:59, and/or an antibody light chain
variable (VL)
domain comprising the amino acid sequence of SEQ ID NO:60. In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:59, and an antibody light chain variable (VL) domain comprising the amino
acid
sequence of SEQ ID NO:60.
[0135] In some embodiments of any of the above embodiments, the binding
protein is a
trispecific binding protein. In some embodiments, the trispecific binding
protein comprising
an antigen binding site that binds an HIV target protein, an antigen binding
site that binds a
CD28 polypeptide, and an antigen binding site that binds a CD3 polypeptide. In
some
embodiments, the binding protein is a trispecific binding protein comprising
four
polypeptides comprising three antigen binding sites, wherein the polypeptide
of formula I and
the polypeptide of formula II form a cross-over light chain-heavy chain pair
(e.g., as
described herein). In some embodiments, the VH and VL domains of any of the
anti-CD28
antigen binding sites described above represent VFH and Vu and form a first
antigen binding
site that binds a CD28 polypeptide. In some embodiments, the VH and VL domains
of any
of the anti-CD28 antigen binding sites described above represent Vm and VIA
and form a first
antigen binding site that binds a CD28 polypeptide, VH2 and VL2 form a second
antigen
binding site that binds a CD3 polypeptide, and VH3 and VL3 and form a third
antigen binding
site that binds an HIV target protein.
[0136] Sequences of exemplary anti-CD28 antigen binding sites are provided
in Table 2.
In some embodiments, a binding protein comprising an anti-CD28 antigen binding
site of the
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
present disclosure comprises 1, 2, 3, 4, 5, or all 6 CDR sequences of an anti-
CD28 antibody
described in Table 2. In some embodiments, a binding protein comprising an
anti-CD28
antigen binding site of the present disclosure comprises a VH domain sequence
and/or VL
domain sequence of an anti-CD28 antibody described in Table 2.
61
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
Table 2. Anti-CD28 binding protein sequences.
Sequence Molecule Description SEQ Sequence
Type ID NO
CDR Anti-CD28 CDR-H1 31 GYTFTSYY
(sup)
CDR-H2 32 IYPGNVNT
CDR-H3 33 TRSHYGLDWNFDV
CDR-L1 34 QNIYVW
CDR-L2 35 KAS
CDR-L3 36 QQGQTYPY
Variable Anti-CD28 VH 59 QVQLVQSGAEVVKPGASVKVSCKAS
Domain (sup) GYTFTSYYIHWVRQAPGQGLEWIGSI
YPGNVNTNYAQKFQGRATLTVDTSIS
TAYMELSRLRSDDTAVYYCTRSHYG
LDWNFDVWGKGTTVTVS S
VL 60 DIQMTQSPS SLSASVGDRVTITCQASQ
NIYVWLNWYQQKPGKAPKLLIYKAS
NLHTGVPSRFSGSGSGTDFTLTIS SLQP
EDIATYYCQQGQTYPYTFGQGTKLEI
Anti-CD3 Binding Sites
[0137]
Certain aspects of the present disclosure relate to binding proteins that
comprise
an antigen binding site that binds a CD3 polypeptide. In some embodiments, the
CD3
polypeptide is a human CD3 polypeptide, including CD3-delta (also known as
T3D, IMD19,
and CD3-DELTA), CD3-epsilon (also known as T3E, IMD18, and TCRE), and CD3-
gamma
(also known as T3G, IMD17, and CD3-GAMMA). Human CD3 polypeptides are known in
the art and include, without limitation, the polypeptides represented by NCBI
Accession
Numbers XP 006510029.1 or NP 031674.1, or a polypeptide produced from NCBI
Gene ID
Numbers 915, 916, or 917. In some embodiments, a binding protein comprising an
antigen
binding site that binds a CD3 polypeptide is monospecific and/or monovalent,
bispecific
and/or bivalent, trispecific and/or trivalent, or multispecific and/or
multivalent. In some
embodiments, a binding protein that comprises an antigen binding site that
binds a CD3
polypeptide is a trispecific binding protein comprising four polypeptides that
form three
antigen binding sites. In some embodiments, a binding protein that comprises
an antigen
binding site that binds a CD3 polypeptide is a trispecific binding protein
comprising four
polypeptides that form three antigen binding sites, one of which binds a CD28
polypeptide,
62
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
and one of which binds a CD3 polypeptide. In some embodiments, a binding
protein that
comprises an antigen binding site that binds a CD3 polypeptide is a
trispecific binding protein
comprising four polypeptides that form three antigen binding sites, one of
which binds a
CD28 polypeptide, one of which binds a CD3 polypeptide, and one of which binds
an HIV
target protein.
[0138] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20),
a
CDR-H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID
NO:21),
and a CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ
ID NO:22); and/or an antibody light chain variable (VL) domain comprising a
CDR-L1
sequence comprising the amino acid sequence of QSLVHX1NX2X3TY, wherein Xi is E
or Q,
X2 is A or L, and X3 is Q, R, or F (SEQ ID NO:293), a CDR-L2 sequence
comprising the
amino acid sequence of KVS (SEQ ID NO:29), and a CDR-L3 sequence comprising
the
amino acid sequence of GQGTQYPFT (SEQ ID NO:30). In some embodiments, the CDR-
Li sequence of the VL2 domain comprises an amino acid sequence selected from
the group
consisting of QSLVHQNAQTY (SEQ ID NO:24), QSLVHENLQTY (SEQ ID NO:25),
QSLVHENLFTY (SEQ ID NO:26), and QSLVHENLRTY (SEQ ID NO:27).
[0139] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20),
a
CDR-H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID
NO:21),
and a CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ
ID NO:22); and/or an antibody light chain variable (VL) domain comprising a
CDR-L1
sequence comprising the amino acid sequence of QSLVHQNAQTY (SEQ ID NO:24), a
CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and
a
CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID
NO:30).
In some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising a CDR-
H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20), a
CDR-
H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:21),
and a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:22); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
63
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprising the amino acid sequence of QSLVHQNAQTY (SEQ ID NO:24), a CDR-L2
sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:30).
[0140] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20),
a
CDR-H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID
NO:21),
and a CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ
ID NO:22); and/or an antibody light chain variable (VL) domain comprising a
CDR-L1
sequence comprising the amino acid sequence of QSLVHENLQTY (SEQ ID NO:25), a
CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and
a
CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID
NO:30).
In some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising a CDR-
H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20), a
CDR-
H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:21),
and a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:22); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QSLVHENLQTY (SEQ ID NO:25), a CDR-L2
sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:30).
[0141] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20),
a
CDR-H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID
NO:21),
and a CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ
ID NO:22); and/or an antibody light chain variable (VL) domain comprising a
CDR-L1
sequence comprising the amino acid sequence of QSLVHENLFTY (SEQ ID NO:26), a
CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and
a
CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID
NO:30).
In some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising a CDR-
H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20), a
CDR-
64
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:21),
and a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:22); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QSLVHENLFTY (SEQ ID NO:26), a CDR-L2
sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:30).
[0142] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising a
CDR-H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20),
a
CDR-H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID
NO:21),
and a CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ
ID NO:22); and/or an antibody light chain variable (VL) domain comprising a
CDR-L1
sequence comprising the amino acid sequence of QSLVHENLRTY (SEQ ID NO:27), a
CDR-L2 sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and
a
CDR-L3 sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID
NO:30).
In some embodiments, a binding protein of the present disclosure comprises an
antigen
binding site comprising: an antibody heavy chain variable (VH) domain
comprising a CDR-
H1 sequence comprising the amino acid sequence of GFTFTKAW (SEQ ID NO:20), a
CDR-
H2 sequence comprising the amino acid sequence of IKDKSNSYAT (SEQ ID NO:21),
and a
CDR-H3 sequence comprising the amino acid sequence of RGVYYALSPFDY (SEQ ID
NO:22); and an antibody light chain variable (VL) domain comprising a CDR-L1
sequence
comprising the amino acid sequence of QSLVHENLRTY (SEQ ID NO:27), a CDR-L2
sequence comprising the amino acid sequence of KVS (SEQ ID NO:29), and a CDR-
L3
sequence comprising the amino acid sequence of GQGTQYPFT (SEQ ID NO:30).
[0143] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:52), and/or an antibody light chain variable (VL)
domain
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to an amino
acid sequence selected from the group consisting of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK (SEQ
ID NO:54),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK (SEQ
ID NO:55),
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNR
F SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK (SEQ
ID NO:56), and
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK (SEQ
ID NO:57). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
the amino acid sequence of SEQ ID NO:52, and/or an antibody light chain
variable (VL)
domain comprising an amino acid sequence selected from the group consisting of
SEQ ID
NO:54, SEQ ID NO:55, SEQ ID NO:56, and SEQ ID NO:57. In some embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:52, and an antibody light chain variable (VL) domain comprising an amino
acid sequence
selected from the group consisting of SEQ ID NO:54, SEQ ID NO:55, SEQ ID
NO:56, and
SEQ ID NO:57.
[0144] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
QVQLVESGGGVVQPGRSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRETISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
66
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
WGQGTLVTVSS (SEQ ID NO:52), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:54). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
the amino acid sequence of SEQ ID NO:52, and/or an antibody light chain
variable (VL)
domain comprising the amino acid sequence of SEQ ID NO:54. In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:52, and an antibody light chain variable (VL) domain comprising the amino
acid
sequence of SEQ ID NO:54.
[0145] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:52), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:55). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
the amino acid sequence of SEQ ID NO:52, and/or an antibody light chain
variable (VL)
67
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
domain comprising the amino acid sequence of SEQ ID NO:55. In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:52, and an antibody light chain variable (VL) domain comprising the amino
acid
sequence of SEQ ID NO:55.
[0146] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
of
QVQLVESGGGVVQPGRSLRLSCAASGETFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRETISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:52), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNR
F SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTEGSGTKVEIK (SEQ
ID NO:56). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
the amino acid sequence of SEQ ID NO:52, and/or an antibody light chain
variable (VL)
domain comprising the amino acid sequence of SEQ ID NO:56. In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:52, and an antibody light chain variable (VL) domain comprising the amino
acid
sequence of SEQ ID NO:56.
[0147] In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
an amino acid sequence that is at least 85%, at least 86%, at least 87%, at
least 88%, at least
89%, at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at
least 95%, at least
96%, at least 97%, at least 98%, at least 99%, or 100% identical to the amino
acid sequence
68
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
of
QVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMHWVRQAPGKQLEWVAQIKDKS
NSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDY
WGQGTLVTVSS (SEQ ID NO:52), and/or an antibody light chain variable (VL)
domain
comprising an amino acid sequence that is at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% identical
to the amino
acid sequence of
DIVMTQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVSN
RF SGVPDRF SGSGSGTDFTLKISRVEAEDVGVYYCGQGTQYPFTFGSGTKVEIK (SEQ
ID NO:57). In some embodiments, a binding protein of the present disclosure
comprises an
antigen binding site comprising: an antibody heavy chain variable (VH) domain
comprising
the amino acid sequence of SEQ ID NO:52, and/or an antibody light chain
variable (VL)
domain comprising the amino acid sequence of SEQ ID NO:57. In some
embodiments, a
binding protein of the present disclosure comprises an antigen binding site
comprising: an
antibody heavy chain variable (VH) domain comprising the amino acid sequence
of SEQ ID
NO:52, and an antibody light chain variable (VL) domain comprising the amino
acid
sequence of SEQ ID NO:57.
[0148] Advantageously, anti-CD3 binding sites are described herein with
high affinity
binding to human CD3 polypeptides and potential manufacturing liabilities
(e.g., deamidation
sites) removed.
[0149] In some embodiments of any of the above embodiments, the binding
protein is a
trispecific binding protein. In some embodiments, the trispecific binding
protein comprising
an antigen binding site that binds an HIV target protein, an antigen binding
site that binds a
CD28 polypeptide, and an antigen binding site that binds a CD3 polypeptide. In
some
embodiments, the binding protein is a trispecific binding protein comprising
four
polypeptides comprising three antigen binding sites, wherein the polypeptide
of formula I and
the polypeptide of formula II form a cross-over light chain-heavy chain pair
(e.g., as
described herein). In some embodiments, the VH and VL domains of any of the
anti-CD3
antigen binding sites described above represent VH2 and VL2and form a second
antigen
binding site that binds a CD3 polypeptide. In some embodiments, VFH and VIA
form a first
antigen binding site that binds a CD28 polypeptide, the VH and VL domains of
any of the
anti-CD3 antigen binding sites described above and/or in Table 3 represent VH2
and VL2 and
69
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
form a second antigen binding site that binds a CD3 polypeptide, and VH3 and
VL3 form a
third antigen binding site that binds an HIV target protein.
[0150]
Sequences of exemplary anti-CD3 antigen binding sites are provided in Table 3.
In some embodiments, a binding protein comprising an anti-CD3 antigen binding
site of the
present disclosure comprises 1, 2, 3, 4, 5, or all 6 CDR sequences of an anti-
CD3 antibody
described in Table 3. In some embodiments, a binding protein comprising an
anti-CD3
antigen binding site of the present disclosure comprises a VH domain sequence
and/or VL
domain sequence of an anti-CD3 antibody described in Table 3.
Table 3. Anti-CD3 binding protein sequences.
Sequence Molecule Description SEQ Sequence
Type ID NO
CDR Anti-CD3 CDR-H1 20 GFTFTKAW
(mid) original
CDR-H2 21 IKDKSNSYAT
original
CDR-H3 22 RGVYYALSPFDY
original
CDR-L1 23 QSLVHNNANTY
original
CDR-L1 QQ 24 QSLVHQNAQTY
CDR-L1 ENLQ 25 QSLVHENLQTY
CDR-L1 ENLF 26 QSLVHENLFTY
CDR-L1 ENLR 27 QSLVHENLRTY
CDR-L1 28 QSLVHDNAQTY
DNAQ
CDR-L2 29 KVS
original
CDR-L3 30 GQGTQYPFT
Original
consensus 293 QSLVHX1NX2X3TY,
CDR-L1 wherein Xi is E or Q, X2 is A or L, and X3
is Q, R, or F
Variable Anti-CD3 VH 52 QVQLVESGGGVVQPGRSLRLSCAASG
domain (mid) FTFTKAWMHWVRQAPGKQLEWVAQ
IKDKSNSYATYYADSVKGRFTISRDDS
KNTLYLQMNSLRAEDTAVYYCRGVY
YALSPFDYWGQGTLVTVSS
VL 53 DIVMTQTPLSLSVTPGQPASISCKSSQS
Original LVHNNANTYLSWYLQKPGQSPQSLIY
KVSNRFSGVPDRFSGSGSGTDFTLKIS
RVEAEDVGVYYCGQGTQYPFTFGSG
TKVEIK
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
VL 54
DIVMTQTPLSLSVTPGQPASISCKSSQS
32/35 QQ
LVHQNAQTYLSWYLQKPGQSPQSLIY
KVSNRFSGVPDRFSGSGSGTDFTLKIS
RVEAEDVGVYYCGQGTQYPFTFGSG
TKVEIK
VL 55
DIVMTQTPLSLSVTPGQPASISCKSSQS
ENLQ
LVHENLQTYLSWYLQKPGQSPQSLIY
KVSNRFSGVPDRFSGSGSGTDFTLKIS
RVEAEDVGVYYCGQGTQYPFTFGSG
TKVEIK
VL 56
DIVMTQTPLSLSVTPGQPASISCKSSQS
ENLF LVHENLFTYLSWYLQKPGQSPQSLIY
KVSNRFSGVPDRFSGSGSGTDFTLKIS
RVEAEDVGVYYCGQGTQYPFTFGSG
TKVEIK
VL 57
DIVMTQTPLSLSVTPGQPASISCKSSQS
ENLR LVHENLRTYLSWYLQKPGQSPQSLIY
KVSNRFSGVPDRFSGSGSGTDFTLKIS
RVEAEDVGVYYCGQGTQYPFTFGSG
TKVEIK
VL 58
DIVMTQTPLSLSVTPGQPASISCKSSQS
DNAQ
LVHDNAQTYLSWYLQKPGQSPQSLIY
KVSNRFSGVPDRFSGSGSGTDFTLKIS
RVEAEDVGVYYCGQGTQYPFTFGSG
TKVEIK
Linkers
[0151] In some embodiments, the linkers Li, L2, L3, and L4 range from no
amino acids
(length=0) to about 100 amino acids long, or less than 100, 50, 40, 30, 20, or
15 amino acids
or less. The linkers can also be 10, 9, 8, 7, 6, 5, 4, 3, 2, or 1 amino acids
long. Li, L2, L3, and
L4 in one binding protein may all have the same amino acid sequence or may all
have
different amino acid sequences.
[0152] Examples of suitable linkers include, for example, GGGGSGGGGS (SEQ
ID
NO:40), GGGGSGGGGSGGGGS (SEQ ID NO: 41), S, RT, TKGPS (SEQ ID NO: 39),
GQPKAAP (SEQ ID NO: 38), GGSGSSGSGG (SEQ ID NO: 42), and DKTHT (SEQ ID
NO:37), as well as those disclosed in International Publication Nos.
W02017/074878 and
W02017/180913. The examples listed above are not intended to limit the scope
of the
disclosure in any way, and linkers comprising randomly selected amino acids
selected from
the group consisting of valine, leucine, isoleucine, serine, threonine,
lysine, arginine,
histidine, aspartate, glutamate, asparagine, glutamine, glycine, and proline
have been shown
to be suitable in the binding proteins.
71
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
[0153] The identity and sequence of amino acid residues in the linker may
vary
depending on the type of secondary structural element necessary to achieve in
the linker. For
example, glycine, serine, and alanine are best for linkers having maximum
flexibility. Some
combination of glycine, proline, threonine, and serine are useful if a more
rigid and extended
linker is necessary. Any amino acid residue may be considered as a linker in
combination
with other amino acid residues to construct larger peptide linkers as
necessary depending on
the desired properties.
[0154] In some embodiments, the length of Li is at least twice the length
of L3. In some
embodiments, the length of L2 is at least twice the length of L4. In some
embodiments, the
length of Li is at least twice the length of L3, and the length of L2 is at
least twice the length
of L4. In some embodiments, Li is 3 to 12 amino acid residues in length, L2 is
3 to 14 amino
acid residues in length, L3 is 1 to 8 amino acid residues in length, and L4 is
1 to 3 amino acid
residues in length. In some embodiments, Li is 5 to 10 amino acid residues in
length, L2 is 5
to 8 amino acid residues in length, L3 is 1 to 5 amino acid residues in
length, and L4 is 1 to 2
amino acid residues in length. In some embodiments, Li is 7 amino acid
residues in length, L2
is 5 amino acid residues in length, L3 is 1 amino acid residue in length, and
L4 is 2 amino acid
residues in length.
[0155] In some embodiments, Li, L2, L3 and L4 each independently are zero
amino acids
in length or comprise a sequence selected from the group consisting of
GGGGSGGGGS
(SEQ ID NO:40), GGGGSGGGGSGGGGS (SEQ ID NO: 41), S, RT, TKGPS (SEQ ID NO:
39), GQPKAAP (SEQ ID NO: 38), and GGSGSSGSGG (SEQ ID NO: 42). In some
embodiments, Li, L2, L3 and L4 each independently comprise a sequence selected
from the
group consisting of GGGGSGGGGS (SEQ ID NO:40), GGGGSGGGGSGGGGS (SEQ ID
NO:41), S, RT, TKGPS (SEQ ID NO:39), GQPKAAP (SEQ ID NO: 38), and
GGSGSSGSGG (SEQ ID NO:42). In some embodiments, Li comprises the sequence
GQPKAAP (SEQ ID NO: 38), L2 comprises the sequence TKGPS (SEQ ID NO:39), L3
comprises the sequence S, and L4 comprises the sequence RT.
[0156] In some embodiments, at least one of Li, L2, L3 or L4 comprises the
sequence
DKTHT (SEQ ID NO:37). In some embodiments, Li, L2, L3 and L4 comprise the
sequence
DKTHT (SEQ ID NO:37).
72
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
Fe regions and constant domains
[0157] In some embodiments, a binding protein of the present disclosure
comprises a
second polypeptide chain further comprising an Fe region linked to CHi, the Fe
region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains. In some embodiments, a binding protein of the present
disclosure
comprises a third polypeptide chain further comprising an Fe region linked to
CHi, the Fe
region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy
chain constant domains. In some embodiments, a binding protein of the present
disclosure
comprises a second polypeptide chain further comprising an Fe region linked to
CHi, the Fe
region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy
chain constant domains, and a third polypeptide chain further comprising an Fe
region linked
to CHi, the Fe region comprising an immunoglobulin hinge region and CH2 and
CH3
immunoglobulin heavy chain constant domains.
[0158] In some embodiments, a binding protein of the present disclosure
comprises a
full-length antibody heavy chain or a polypeptide chain comprising an Fe
region. In some
embodiments, the Fe region is a human Fe region, e.g., a human IgGl, IgG2,
IgG3, or IgG4
Fe region. In some embodiments, the Fe region includes an antibody hinge, CHi,
CH2, CH3,
and optionally CH4 domains. In some embodiments, the Fe region is a human IgG1
Fe
region. In some embodiments, the Fe region is a human IgG4 Fe region. In some
embodiments, the Fe region includes one or more of the mutations described
infra. In some
embodiments, the Fe region is an Fe region of one of the heavy chain
polypeptides (e.g.,
polypeptide 2 or 3) of a binding protein shown in Table 4. In some
embodiments, the heavy
chain constant region is a constant region of one of the heavy chain
polypeptides (e.g.,
polypeptide 2 or 3) of a binding protein shown in Table 4. In some
embodiments, the light
chain constant region is a constant region of one of the light chain
polypeptides (e.g.,
polypeptide 1 or 4) of a binding protein shown in Table 4.
[0159] In some embodiments, a binding protein of the present disclosure
includes one or
two Fe variants. The term "Fe variant" as used herein refers to a molecule or
sequence that is
modified from a native Fe but still comprises a binding site for the salvage
receptor, FcRn
(neonatal Fe receptor). Exemplary Fe variants, and their interaction with the
salvage receptor,
are known in the art. Thus, the term "Fe variant" can comprise a molecule or
sequence that is
humanized from a non-human native Fe. Furthermore, a native Fe comprises
regions that can
be removed because they provide structural features or biological activity
that are not
73
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
required for the antibody-like binding proteins of the invention. Thus, the
term "Fe variant"
comprises a molecule or sequence that lacks one or more native Fe sites or
residues, or in
which one or more Fe sites or residues has be modified, that affect or are
involved in: (1)
disulfide bond formation, (2) incompatibility with a selected host cell, (3) N-
terminal
heterogeneity upon expression in a selected host cell, (4) glycosylation, (5)
interaction with
complement, (6) binding to an Fe receptor other than a salvage receptor, or
(7) antibody-
dependent cellular cytotoxicity (ADCC).
[0160] In some embodiments, a binding protein of the present disclosure
(e.g., a
trispecific binding protein) comprises a "knob" mutation on the second
polypeptide chain and
a "hole" mutation on the third polypeptide chain. In some embodiments, a
binding protein of
the present disclosure comprises a "knob" mutation on the third polypeptide
chain and a
"hole" mutation on the second polypeptide chain. In some embodiments, the
"knob" mutation
comprises substitution(s) at positions corresponding to positions 354 and/or
366 of human
IgG1 or IgG4 according to EU Index. In some embodiments, the amino acid
substitutions are
S354C, T366W, T366Y, S354C and T366W, or S354C and T366Y. In some embodiments,
the "knob" mutation comprises substitutions at positions corresponding to
positions 354 and
366 of human IgG1 or IgG4 according to EU Index. In some embodiments, the
amino acid
substitutions are S354C and T366W. In some embodiments, the "hole" mutation
comprises
substitution(s) at positions corresponding to positions 407 and, optionally,
349, 366, and/or
368 and of human IgG1 or IgG4 according to EU Index. In some embodiments, the
amino
acid substitutions are Y407V or Y407T and optionally Y349C, T366S, and/or
L368A. In
some embodiments, the "hole" mutation comprises substitutions at positions
corresponding to
positions 349, 366, 368, and 407 of human IgG1 or IgG4 according to EU Index.
In some
embodiments, the amino acid substitutions are Y349C, T366S, L368A, and Y407V.
[0161] In some embodiments, the second polypeptide chain further comprises
a first Fe
region linked to CHL the first Fe region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first Fe
region
comprises amino acid substitution(s) at positions corresponding to positions
366 and
optionally 354 of human IgG1 or IgG4 according to EU Index, wherein the amino
acid
substitutions are T366W or T366Y and optionally S354C; and wherein the third
polypeptide
chain further comprises a second Fe region linked to CHL the second Fe region
comprising
an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the second Fe region comprises amino acid substitution(s) at
positions
74
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
corresponding to positions 407 and optionally 349, 366, and/or 368 and of
human IgG1 or
IgG4 according to EU Index, wherein the amino acid substitutions are Y407V or
Y407T and
optionally Y349C, T366S, and/or L368A.
[0162] In
some embodiments, the second polypeptide chain further comprises a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region
comprises amino acid substitution(s) at positions corresponding to positions
407 and
optionally 349, 366, and/or 368 and of human IgG1 or IgG4 according to EU
Index, wherein
the amino acid substitutions are Y407V or Y407T and optionally Y349C, T366S,
and/or
L368A; and wherein the third polypeptide chain further comprises a second Fc
region linked
to CH1, the second Fc region comprising an immunoglobulin hinge region and CH2
and CH3
immunoglobulin heavy chain constant domains, wherein the second Fc region
comprises
amino acid substitution(s) at positions corresponding to positions 366 and
optionally 354 of
human IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions
are
T366W or T366Y and optionally S354C.
[0163] In
some embodiments, the second polypeptide chain further comprises a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region
comprises amino acid substitution at position corresponding to position 366 of
human IgG1
or IgG4 according to EU Index, wherein the amino acid substitution is T366W;
and wherein
the third polypeptide chain further comprises a second Fc region linked to
CH1, the second
Fc region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin
heavy chain constant domains, wherein the second Fc region comprises amino
acid
substitution(s) at positions corresponding to positions 366, 368, and/or 407
and of human
IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions are
T366S,
L368A, and/or Y407V.
[0164] In
some embodiments, the second polypeptide chain further comprises a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region
comprises amino acid substitution(s) at positions corresponding to positions
366, 368, and/or
407 and of human IgG1 or IgG4 according to EU Index, wherein the amino acid
substitutions
are T366S, L368A, and/or Y407V; and wherein the third polypeptide chain
further comprises
a second Fc region linked to CH1, the second Fc region comprising an
immunoglobulin hinge
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
region and CH2 and CH3 immunoglobulin heavy chain constant domains, wherein
the
second Fc region comprises amino acid substitution at position corresponding
to position 366
of human IgG1 or IgG4 according to EU Index, wherein the amino acid
substitution is
T366W.
[0165] In
some embodiments, the second polypeptide chain further comprises a first Fc
region linked to CH1, the first Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region
comprises amino acid substitutions at positions corresponding to positions 354
and 366 of
human IgG1 or IgG4 according to EU Index, wherein the amino acid substitutions
are S354C
and T366W; and wherein the third polypeptide chain further comprises a second
Fc region
linked to CH1, the second Fc region comprising an immunoglobulin hinge region
and CH2
and CH3 immunoglobulin heavy chain constant domains, wherein the second Fc
region
comprises amino acid substitutions at positions corresponding to positions
349, 366, 368, and
407 of human IgG1 or IgG4 according to EU Index, wherein the amino acid
substitutions are
Y349C, T366S, L368A, and Y407V. In some embodiments, the second polypeptide
chain
further comprises a first Fc region linked to CH1, the first Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the first Fc region comprises amino acid substitutions at
positions
corresponding to positions 349, 366, 368, and 407 of human IgG1 or IgG4
according to EU
Index, wherein the amino acid substitutions are Y349C, T366S, L368A, and
Y407V; and
wherein the third polypeptide chain further comprises a second Fc region
linked to CH1, the
second Fc region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy chain constant domains, wherein the second Fc region
comprises
amino acid substitutions at positions corresponding to positions 354 and 366
of human IgG1
or IgG4 according to EU Index, wherein the amino acid substitutions are S354C
and T366W.
In some embodiments, the first and/or second Fc regions are human IgG1 Fc
regions. In
some embodiments, the first and/or second Fc regions are human IgG4 Fc
regions.
[0166] In
some embodiments, the second polypeptide chain further comprises a first Fc
region linked to CH1, wherein the first Fc region is a human IgG4 Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the first Fc region comprises amino acid substitutions at
positions
corresponding to positions 228, 354, 366, and 409 of human IgG4 according to
EU Index,
wherein the amino acid substitutions are S228P, S354C, T366W, and R409K; and
wherein
76
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
the third polypeptide chain further comprises a second Fc region linked to
CH1, wherein the
second Fc region is a human IgG4 Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the second Fc
region
comprises amino acid substitutions at positions corresponding to positions
228, 349, 366,
368, 407, and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are S228P, Y349C, T366S, L368A, Y407V, and R409K. In some
embodiments,
the second polypeptide chain further comprises a first Fc region linked to
CH1, wherein the
first Fc region is a human IgG4 Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region
comprises amino acid substitutions at positions corresponding to positions
228, 349, 366,
368, 407, and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are S228P, Y349C, T366S, L368A, Y407V, and R409K; and wherein
the third
polypeptide chain further comprises a second Fc region linked to CH1, wherein
the second Fc
region is a human IgG4 Fc region comprising an immunoglobulin hinge region and
CH2 and
CH3 immunoglobulin heavy chain constant domains, wherein the second Fc region
comprises amino acid substitutions at positions corresponding to positions
228, 354, 366, and
409 of human IgG4 according to EU Index, wherein the amino acid substitutions
are S228P,
S354C, T366W, and R409K.
[0167] In
some embodiments, the second polypeptide chain further comprises a first Fc
region linked to CH1, wherein the first Fc region is a human IgG4 Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the first Fc region comprises amino acid substitutions at
positions
corresponding to positions 234, 235, 354, and 366 of human IgG4 according to
EU Index,
wherein the amino acid substitutions are F234A, L235A, S354C, and T366W; and
wherein
the third polypeptide chain further comprises a second Fc region linked to
CH1, wherein the
second Fc region is a human IgG4 Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the second Fc
region
comprises amino acid substitutions at positions corresponding to positions
234, 235, 349,
366, 368, and 407 of human IgG4 according to EU Index, wherein the amino acid
substitutions are F234A, L235A, Y349C, T366S, L368A, and Y407V. In some
embodiments,
the second polypeptide chain further comprises a first Fc region linked to
CH1, wherein the
first Fc region is a human IgG4 Fc region comprising an immunoglobulin hinge
region and
CH2 and CH3 immunoglobulin heavy chain constant domains, wherein the first Fc
region
77
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprises amino acid substitutions at positions corresponding to positions
234, 235, 349,
366, 368, and 407 of human IgG4 according to EU Index, wherein the amino acid
substitutions are F234A, L235A, Y349C, T366S, L368A, and Y407V; and wherein
the third
polypeptide chain further comprises a second Fc region linked to CH1, wherein
the second Fc
region is a human IgG4 Fc region comprising an immunoglobulin hinge region and
CH2 and
CH3 immunoglobulin heavy chain constant domains, wherein the second Fc region
comprises amino acid substitutions at positions corresponding to positions
234, 235, 354, and
366 of human IgG4 according to EU Index, wherein the amino acid substitutions
are F234A,
L235A, S354C, and T366W.
[0168] In
some embodiments, a binding protein of the present disclosure comprises one
or more mutations to reduce effector function, e.g., Fc receptor-mediated
antibody-dependent
cellular phagocytosis (ADCP), complement-dependent cytotoxicity (CDC), and/or
antibody-
dependent cellular cytotoxicity (ADCC). In some embodiments, the second
polypeptide
chain further comprises a first Fc region linked to CHi, the first Fc region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains; wherein the third polypeptide chain further comprises a second Fc
region linked to
CHi, the second Fc region comprising an immunoglobulin hinge region and CH2
and CH3
immunoglobulin heavy chain constant domains; wherein the first and second Fc
regions are
human IgG1 Fc regions; and wherein the first and the second Fc regions each
comprise
amino acid substitutions at positions corresponding to positions 234 and 235
of human IgG1
according to EU Index, wherein the amino acid substitutions are L234A and
L235A. In some
embodiments, the Fc regions of the second and the third polypeptide chains are
human IgG1
Fc regions, and wherein the Fc regions each comprise amino acid substitutions
at positions
corresponding to positions 234 and 235 of human IgG1 according to EU Index,
wherein the
amino acid substitutions are L234A and L235A. In some embodiments, the second
polypeptide chain further comprises a first Fc region linked to CHi, the first
Fc region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains; wherein the third polypeptide chain further comprises a
second Fc region
linked to CHi, the second Fc region comprising an immunoglobulin hinge region
and CH2 and
CH3 immunoglobulin heavy chain constant domains; wherein the first and second
Fc regions
are human IgG1 Fc regions; and wherein the first and the second Fc regions
each comprise
amino acid substitutions at positions corresponding to positions 234, 235, and
329 of human
IgG1 according to EU Index, wherein the amino acid substitutions are L234A,
L235A, and
78
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
P329A. In some embodiments, the Fe regions of the second and the third
polypeptide chains
are human IgG1 Fe regions, and wherein the Fe regions each comprise amino acid
substitutions at positions corresponding to positions 234, 235, and 329 of
human IgG1
according to EU Index, wherein the amino acid substitutions are L234A, L235A,
and P329A.
In some embodiments, the Fe regions of the second and the third polypeptide
chains are
human IgG4 Fe regions, and the Fe regions each comprise amino acid
substitutions at
positions corresponding to positions 234 and 235 of human IgG4 according to EU
Index,
wherein the amino acid substitutions are F234A and L235A. In some embodiments,
the
binding protein comprises a second polypeptide chain further comprising a
first Fe region
linked to CH1, the first Fe region comprising an immunoglobulin hinge region
and CH2 and
CH3 immunoglobulin heavy chain constant domains, and a third polypeptide chain
further
comprising a second Fe region linked to CH1, the second Fe region comprising
an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains; and wherein the first and the second Fe regions each comprise amino
acid
substitutions at positions corresponding to positions 234 and 235 of human
IgG4 according to
EU Index, wherein the amino acid substitutions are F234A and L235A.
[0169] In
some embodiments, the second polypeptide chain further comprises a first Fe
region linked to CH1, wherein the first Fe region is a human IgG4 Fe region
comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the first Fe region comprises amino acid substitutions at
positions
corresponding to positions 228, 234, 235, 354, 366, and 409 of human IgG4
according to EU
Index, wherein the amino acid substitutions are S228P, F234A, L235A, S354C,
T366W, and
R409K; and wherein the third polypeptide chain further comprises a second Fe
region linked
to CH1, wherein the second Fe region is a human IgG4 Fe region comprising an
immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy chain
constant
domains, wherein the second Fe region comprises amino acid substitutions at
positions
corresponding to positions 228, 234, 235, 349, 366, 368, 407, and 409 of human
IgG4
according to EU Index, wherein the amino acid substitutions are S228P, F234A,
L235A,
Y349C, T366S, L368A, Y407V, and R409K. In some embodiments, the second
polypeptide
chain further comprises a first Fe region linked to CH1, wherein the first Fe
region is a
human IgG4 Fe region comprising an immunoglobulin hinge region and CH2 and CH3
immunoglobulin heavy chain constant domains, wherein the first Fe region
comprises amino
acid substitutions at positions corresponding to positions 228, 234, 235, 349,
366, 368, 407,
79
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
S228P, F234A, L235A, Y349C, T366S, L368A, Y407V, and R409K; and wherein the
third
polypeptide chain further comprises a second Fc region linked to CH1, wherein
the second Fc
region is a human IgG4 Fc region comprising an immunoglobulin hinge region and
CH2 and
CH3 immunoglobulin heavy chain constant domains, wherein the second Fc region
comprises amino acid substitutions at positions corresponding to positions
228, 234, 235,
354, 366, and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are S228P, F234A, L235A, S354C, T366W, and R409K.
[0170] In some embodiments, the Fc region is a human IgG4 Fc region
comprising one or
more mutations that reduce or eliminate FcyI and/or FcyII binding. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising one or more mutations that
reduce or
eliminate FcyI and/or FcyII binding but do not affect FcRn binding. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising amino acid substitutions at
positions
corresponding to positions 228 and/or 409 of human IgG4 according to EU Index.
In some
embodiments, the amino acid substitutions are S228P and /or R409K. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising amino acid substitutions at
positions
corresponding to positions 234 and/or 235 of human IgG4 according to EU Index.
In some
embodiments, the amino acid substitutions are F234A and/or L235A. In some
embodiments,
the Fc region is a human IgG4 Fc region comprising amino acid substitutions at
positions
corresponding to positions 228, 234, 235, and/or 409 of human IgG4 according
to EU Index.
In some embodiments, the amino acid substitutions are S228P, F234A, L235A, and
/or
R409K. In some embodiments, the Fc region is a human IgG4 Fc region comprising
amino
acid substitutions at positions corresponding to positions 233-236 of human
IgG4 according
to EU Index. In some embodiments, the amino acid substitutions are E233P,
F234V, L235A,
and a deletion at 236. In some embodiments, the Fc region is a human IgG4 Fc
region
comprising amino acid mutations at substitutions corresponding to positions
228, 233-236,
and/or 409 of human IgG4 according to EU Index. In some embodiments, the amino
acid
mutations are S228P; E233P, F234V, L235A, and a deletion at 236; and /or
R409K.
[0171] In some embodiments, the Fc region comprises one or more mutations
that reduce
or eliminate Fc receptor binding and/or effector function of the Fc region
(e.g., Fc receptor-
mediated antibody-dependent cellular phagocytosis (ADCP), complement-dependent
cytotoxicity (CDC), and/or antibody-dependent cellular cytotoxicity (ADCC)).
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
[0172] In some embodiments, the Fe region is a human IgG1 Fe region
comprising one or
more amino acid substitutions at positions corresponding to positions 234,
235, and/or 329 of
human IgG1 according to EU Index. In some embodiments, the amino acid
substitutions are
L234A, L235A, and/or P329A. In some embodiments, the Fe region is a human IgG1
Fe
region comprising amino acid substitutions at positions corresponding to
positions 298, 299,
and/or 300 of human IgG1 according to EU Index. In some embodiments, the amino
acid
substitutions are S298N, T299A, and/or Y300S.
[0173] In some embodiments, a binding protein of the present disclosure
comprises one
or more mutations to improve stability, e.g., of the hinge region and/or dimer
interface of
IgG4 (See e.g., Spiess, C. et al. (2013)1 Biol. Chem. 288:26583-26593). In
some
embodiments, the mutation comprises substitutions at positions corresponding
to positions
228 and 409 of human IgG4 according to EU Index, wherein the amino acid
substitutions are
S228P and R409K. In some embodiments, the binding protein comprises a second
polypeptide chain further comprising a first Fe region linked to CHi, the
first Fe region
comprising an immunoglobulin hinge region and CH2 and CH3 immunoglobulin heavy
chain
constant domains, and a third polypeptide chain further comprising a second Fe
region linked
to CHi, the second Fe region comprising an immunoglobulin hinge region and CH2
and CH3
immunoglobulin heavy chain constant domains; wherein the first and second Fe
regions are
human IgG4 Fe regions; and wherein the first and the second Fe regions each
comprise
amino acid substitutions at positions corresponding to positions 228 and 409
of human IgG4
according to EU Index, wherein the amino acid substitutions are S228P and
R409K. In some
embodiments, a binding protein of the present disclosure comprises knob and
hole mutations
and one or more mutations to improve stability. In some embodiments, the first
and/or second
Fe regions are human IgG4 Fe regions.
[0174] In some embodiments, the Fe region is a human IgG1 Fe region
comprising one or
more amino acid substitutions at positions corresponding to positions 234,
235, and/or 329 of
human IgG1 according to EU Index. In some embodiments, the amino acid
substitutions are
L234A, L235A, and/or P329A. In some embodiments, the Fe region is a human IgG1
Fe
region comprising amino acid substitutions at positions corresponding to
positions 298, 299,
and/or 300 of human IgG1 according to EU Index. In some embodiments, the amino
acid
substitutions are S298N, T299A, and/or Y300S.
81
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
Nucleic acids
[0175] Other aspects of the present disclosure relate to isolated nucleic
acid molecules
comprising a nucleotide sequence encoding any of the binding proteins
described herein.
Exemplary and non-limiting nucleic acid sequences are provided in Table 5.
[0176] Other aspects of the present disclosure relate to kits of
polynucleotides, e.g., that
encode one or more polypeptides of a binding protein as described herein. In
some
embodiments, a kit of polynucleotides of the present disclosure comprises one,
two, three, or
four polynucleotides of a kit of polynucleotides comprising: (a) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:177, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:178, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:179, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:180; (b) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:181, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:182, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:183, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:184; (c) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:185, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:186, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:187, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:188; (d) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:189, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:190, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:191, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:192; (e) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:193, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:194, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:195, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:196; (f) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:197, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:198, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:199, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:200; (g) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:201, a second
polynucleotide
82
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprising the polynucleotide sequence of SEQ ID NO:202, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:203, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:204; (h) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:205, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:206, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:207, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:208; (i) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:209, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:210, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:211, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:212; (j) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:213, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:214, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:215, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:216; (k) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:217, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:218, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:219, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:220; (1) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:221, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:222, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:223, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:224; (m) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:225, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:226, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:227, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:228; (n) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:229, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:230, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:231, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:232; (o) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:233, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:234, a third
polynucleotide
83
CA 03136147 2021-10-04
WO 2020/210386
PCT/US2020/027313
comprising the polynucleotide sequence of SEQ ID NO:235, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:236; (p) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:237, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:238, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:239, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:240; (q) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:241, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:242, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:243, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:244; (r) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:245, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:246, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:247, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:248; (s) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:249, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:250, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:251, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:252; (t) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:253, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:254, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:255, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:256; (u) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:257, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:258, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:259, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:260; (v) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:261, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:262, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:263, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:264; (w) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:265, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:266, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:267, and a fourth
polynucleotide
84
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
comprising the polynucleotide sequence of SEQ ID NO:268; (x) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:269, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:270, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:271, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:272; (y) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:273, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:274, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:275, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:276; (z) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:277, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:278, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:279, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:280; (aa) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:281, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:282, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:283, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:284; (bb) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:285, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:286, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:287, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:288; or (cc) a first
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:289, a second
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:290, a third
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:291, and a fourth
polynucleotide
comprising the polynucleotide sequence of SEQ ID NO:292.
[0177] Other aspects of the present disclosure relate to a vector system
comprising one or
more vectors encoding a first, second, third, and fourth polypeptide chain of
any of the
binding proteins described herein. In some embodiments, the vector system
comprises a first
vector encoding the first polypeptide chain of the binding protein, a second
vector encoding
the second polypeptide chain of the binding protein, a third vector encoding
the third
polypeptide chain of the binding protein, and a fourth vector encoding the
fourth polypeptide
chain of the binding protein, e.g., as shown in the polynucleotides of Table
5. In some
embodiments, the vector system comprises a first vector encoding the first and
second
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
polypeptide chains of the binding protein, and a second vector encoding the
third and fourth
polypeptide chains of the binding protein. In some embodiments, the vector
system comprises
a first vector encoding the first and third polypeptide chains of the binding
protein, and a
second vector encoding the second and fourth polypeptide chains of the binding
protein. In
some embodiments, the vector system comprises a first vector encoding the
first and fourth
polypeptide chains of the binding protein, and a second vector encoding the
second and third
polypeptide chains of the binding protein. In some embodiments, the vector
system comprises
a first vector encoding the first, second, third, and fourth polypeptide
chains of the binding
protein. The one or more vectors of the vector system may be any of the
vectors described
herein. In some embodiments, the one or more vectors are expression vectors.
In some
embodiments, the first, second, third, and fourth polynucleotides are present
on one or more
expression vectors, e.g., one, two, three, or four expression vectors.
[0178] Standard recombinant DNA methodologies are used to construct the
polynucleotides that encode the polypeptides which form the binding proteins,
incorporate
these polynucleotides into recombinant expression vectors, and introduce such
vectors into
host cells. See e.g., Sambrook et at., 2001, MOLECULAR CLONING: A LABORATORY
MANUAL
(Cold Spring Harbor Laboratory Press, 3rd ed.). Enzymatic reactions and
purification
techniques may be performed according to manufacturer's specifications, as
commonly
accomplished in the art, or as described herein. Unless specific definitions
are provided, the
nomenclature utilized in connection with, and the laboratory procedures and
techniques of,
analytical chemistry, synthetic organic chemistry, and medicinal and
pharmaceutical
chemistry described herein are those well-known and commonly used in the art.
Similarly,
conventional techniques may be used for chemical syntheses, chemical analyses,
pharmaceutical preparation, formulation, delivery, and treatment of patients.
[0179] In some embodiments, the isolated nucleic acid is operably linked to
a
heterologous promoter to direct transcription of the binding protein-coding
nucleic acid
sequence. A promoter may refer to nucleic acid control sequences which direct
transcription
of a nucleic acid. A first nucleic acid sequence is operably linked to a
second nucleic acid
sequence when the first nucleic acid sequence is placed in a functional
relationship with the
second nucleic acid sequence. For instance, a promoter is operably linked to a
coding
sequence of a binding protein if the promoter affects the transcription or
expression of the
coding sequence. Examples of promoters may include, but are not limited to,
promoters
obtained from the genomes of viruses (such as polyoma virus, fowlpox virus,
adenovirus
86
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
(such as Adenovirus 2), bovine papilloma virus, avian sarcoma virus,
cytomegalovirus, a
retrovirus, hepatitis-B virus, Simian Virus 40 (5V40), and the like), from
heterologous
eukaryotic promoters (such as the actin promoter, an immunoglobulin promoter,
from heat-
shock promoters, and the like), the CAG-promoter (Niwa et al., Gene 108(2):193-
9, 1991),
the phosphoglycerate kinase (PGK)-promoter, a tetracycline-inducible promoter
(Masui et al.,
Nucleic Acids Res. 33:e43, 2005), the lac system, the trp system, the tac
system, the trc
system, major operator and promoter regions of phage lambda, the promoter for
3-
phosphoglycerate kinase, the promoters of yeast acid phosphatase, and the
promoter of the
yeast alpha-mating factors. Polynucleotides encoding binding proteins of the
present
disclosure may be under the control of a constitutive promoter, an inducible
promoter, or any
other suitable promoter described herein or other suitable promoter that will
be readily
recognized by one skilled in the art.
[0180] In some embodiments, the isolated nucleic acid is incorporated into
a vector. In
some embodiments, the vector is an expression vector. Expression vectors may
include one
or more regulatory sequences operatively linked to the polynucleotide to be
expressed. The
term "regulatory sequence" includes promoters, enhancers and other expression
control
elements (e.g., polyadenylation signals). Examples of suitable enhancers may
include, but are
not limited to, enhancer sequences from mammalian genes (such as globin,
elastase, albumin,
a-fetoprotein, insulin and the like), and enhancer sequences from a eukaryotic
cell virus (such
as 5V40 enhancer on the late side of the replication origin (bp 100-270), the
cytomegalovirus
early promoter enhancer, the polyoma enhancer on the late side of the
replication origin,
adenovirus enhancers, and the like). Examples of suitable vectors may include,
for example,
plasmids, cosmids, episomes, transposons, and viral vectors (e.g., adenoviral,
vaccinia viral,
Sindbis-viral, measles, herpes viral, lentiviral, retroviral, adeno-associated
viral vectors, etc.).
Expression vectors can be used to transfect host cells, such as, for example,
bacterial cells,
yeast cells, insect cells, and mammalian cells. Biologically functional viral
and plasmid
DNA vectors capable of expression and replication in a host are known in the
art, and can be
used to transfect any cell of interest.
Host cells
[0181] Other aspects of the present disclosure relate to a host cell (e.g.,
an isolated host
cell) comprising one or more isolated polynucleotides, vectors, and/or vector
systems
described herein. In some embodiments, an isolated host cell of the present
disclosure is
87
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
cultured in vitro. In some embodiments, the host cell is a bacterial cell
(e.g., an E. coil cell).
In some embodiments, the host cell is a yeast cell (e.g., an S. cerevisiae
cell). In some
embodiments, the host cell is an insect cell. Examples of insect host cells
may include, for
example, Drosophila cells (e.g., S2 cells), Trichoplusia ni cells (e.g., High
FiveTM cells), and
Spodoptera frupperda cells (e.g., Sf21 or SP9 cells). In some embodiments, the
host cell is a
mammalian cell. Examples of mammalian host cells may include, for example,
human
embryonic kidney cells (e.g., 293 or 293 cells subcloned for growth in
suspension culture),
Expi293Tm cells, CHO cells, baby hamster kidney cells (e.g., BHK, ATCC CCL
10), mouse
sertoli cells (e.g., TM4 cells), monkey kidney cells (e.g., CV1 ATCC CCL 70),
African green
monkey kidney cells (e.g., VERO-76, ATCC CRL-1587), human cervical carcinoma
cells
(e.g., HELA, ATCC CCL 2), canine kidney cells (e.g., MDCK, ATCC CCL 34),
buffalo rat
liver cells (e.g., BRL 3A, ATCC CRL 1442), human lung cells (e.g., W138, ATCC
CCL 75),
human liver cells (e.g., Hep G2, HB 8065), mouse mammary tumor cells (e.g.,
MMT 060562,
ATCC CCL51), TM cells, MRC 5 cells, F54 cells, a human hepatoma line (e.g.,
Hep G2),
and myeloma cells (e.g., NSO and 5p2/0 cells).
[0182] Other aspects of the present disclosure relate to a method of
producing any of the
binding proteins described herein. In some embodiments, the method includes a)
culturing a
host cell (e.g., any of the host cells described herein) comprising an
isolated nucleic acid,
vector, and/or vector system (e.g., any of the isolated nucleic acids,
vectors, and/or vector
systems described herein) under conditions such that the host cell expresses
the binding
protein; and b) isolating the binding protein from the host cell. Methods of
culturing host
cells under conditions to express a protein are well known to one of ordinary
skill in the art.
Methods of isolating proteins from cultured host cells are well known to one
of ordinary skill
in the art, including, for example, by affinity chromatography (e.g., two step
affinity
chromatography comprising protein A affinity chromatography followed by size
exclusion
chromatography).
Pharmaceutical Compositions
[0183] Therapeutic or pharmaceutical compositions comprising binding
proteins are
within the scope of the disclosure. Such therapeutic or pharmaceutical
compositions can
comprise a therapeutically effective amount of a binding protein, or binding
protein-drug
conjugate, in admixture with a pharmaceutically or physiologically acceptable
formulation
agent selected for suitability with the mode of administration.
88
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
[0184] Acceptable formulation materials are nontoxic to recipients at the
dosages and
concentrations employed.
[0185] The pharmaceutical composition can contain formulation materials for
modifying,
maintaining, or preserving, for example, the pH, osmolarity, viscosity,
clarity, color,
isotonicity, odor, sterility, stability, rate of dissolution or release,
adsorption, or penetration of
the composition. Suitable formulation materials include, but are not limited
to, amino acids
(such as glycine, glutamine, asparagine, arginine, or lysine), antimicrobials,
antioxidants
(such as ascorbic acid, sodium sulfite, or sodium hydrogen-sulfite), buffers
(such as borate,
bicarbonate, Tris-HC1, citrates, phosphates, or other organic acids), bulking
agents (such as
mannitol or glycine), chelating agents (such as ethylenediamine tetraacetic
acid (EDTA)),
complexing agents (such as caffeine, polyvinylpyrrolidone, beta-cyclodextrin,
or
hydroxypropyl-beta-cyclodextrin), fillers, monosaccharides, disaccharides, and
other
carbohydrates (such as glucose, mannose, or dextrins), proteins (such as serum
albumin,
gelatin, or immunoglobulins), coloring, flavoring and diluting agents,
emulsifying agents,
hydrophilic polymers (such as polyvinylpyrrolidone), low molecular weight
polypeptides,
salt-forming counterions (such as sodium), preservatives (such as benzalkonium
chloride,
benzoic acid, salicylic acid, thimerosal, phenethyl alcohol, methylparaben,
propylparaben,
chlorhexidine, sorbic acid, or hydrogen peroxide), solvents (such as glycerin,
propylene
glycol, or polyethylene glycol), sugar alcohols (such as mannitol or
sorbitol), suspending
agents, surfactants or wetting agents (such as pluronics; PEG; sorbitan
esters; polysorbates
such as polysorbate 20 or polysorbate 80; triton; tromethamine; lecithin;
cholesterol or
tyloxapal), stability enhancing agents (such as sucrose or sorbitol), tonicity
enhancing agents
(such as alkali metal halides ¨ e.g., sodium or potassium chloride ¨ or
mannitol sorbitol),
delivery vehicles, diluents, excipients and/or pharmaceutical adjuvants (see,
e.g.,
REMINGTON'S PHARMACEUTICAL SCIENCES (18th Ed., A.R. Gennaro, ed., Mack
Publishing
Company 1990), and subsequent editions of the same, incorporated herein by
reference for
any purpose).
[0186] The optimal pharmaceutical composition will be determined by a
skilled artisan
depending upon, for example, the intended route of administration, delivery
format, and
desired dosage. Such compositions can influence the physical state, stability,
rate of in vivo
release, and rate of in vivo clearance of the binding protein.
[0187] The primary vehicle or carrier in a pharmaceutical composition can
be either
aqueous or non-aqueous in nature. For example, a suitable vehicle or carrier
for injection can
89
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
be water, physiological saline solution, or artificial cerebrospinal fluid,
possibly
supplemented with other materials common in compositions for parenteral
administration.
Neutral buffered saline or saline mixed with serum albumin are further
exemplary vehicles.
Other exemplary pharmaceutical compositions comprise Tris buffer of about pH
7.0-8.5, or
acetate buffer of about pH 4.0-5.5, which can further include sorbitol or a
suitable substitute.
In one embodiment of the disclosure, binding protein compositions can be
prepared for
storage by mixing the selected composition having the desired degree of purity
with optional
formulation agents in the form of a lyophilized cake or an aqueous solution.
Further, the
binding protein can be formulated as a lyophilizate using appropriate
excipients such as
sucrose.
[0188] The pharmaceutical compositions of the disclosure can be selected
for parenteral
delivery or subcutaneous. Alternatively, the compositions can be selected for
inhalation or
for delivery through the digestive tract, such as orally. The preparation of
such
pharmaceutically acceptable compositions is within the skill of the art.
[0189] The formulation components are present in concentrations that are
acceptable to
the site of administration. For example, buffers are used to maintain the
composition at
physiological pH or at a slightly lower pH, typically within a pH range of
from about 5 to
about 8.
[0190] When parenteral administration is contemplated, the therapeutic
compositions for
use can be in the form of a pyrogen-free, parenterally acceptable, aqueous
solution
comprising the desired binding protein in a pharmaceutically acceptable
vehicle. A
particularly suitable vehicle for parenteral injection is sterile distilled
water in which a
binding protein is formulated as a sterile, isotonic solution, properly
preserved. Yet another
preparation can involve the formulation of the desired molecule with an agent,
such as
injectable microspheres, bio-erodible particles, polymeric compounds (such as
polylactic acid
or polyglycolic acid), beads, or liposomes, that provides for the controlled
or sustained
release of the product which can then be delivered via a depot injection.
Hyaluronic acid can
also be used, and this can have the effect of promoting sustained duration in
the circulation.
Other suitable means for the introduction of the desired molecule include
implantable drug
delivery devices.
[0191] In one embodiment, a pharmaceutical composition can be formulated
for
inhalation. For example, a binding protein can be formulated as a dry powder
for inhalation.
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
Binding protein inhalation solutions can also be formulated with a propellant
for aerosol
delivery. In yet another embodiment, solutions can be nebulized.
[0192] It is also contemplated that certain formulations can be
administered orally. In
one embodiment of the disclosure, binding proteins that are administered in
this fashion can
be formulated with or without those carriers customarily used in the
compounding of solid
dosage forms such as tablets and capsules. For example, a capsule can be
designed to release
the active portion of the formulation at the point in the gastrointestinal
tract where
bioavailability is maximized and pre-systemic degradation is minimized.
Additional agents
can be included to facilitate absorption of the binding protein. Diluents,
flavorings, low
melting point waxes, vegetable oils, lubricants, suspending agents, tablet
disintegrating
agents, and binders can also be employed.
[0193] Another pharmaceutical composition can involve an effective quantity
of binding
proteins in a mixture with non-toxic excipients that are suitable for the
manufacture of
tablets. By dissolving the tablets in sterile water, or another appropriate
vehicle, solutions
can be prepared in unit-dose form. Suitable excipients include, but are not
limited to, inert
diluents, such as calcium carbonate, sodium carbonate or bicarbonate, lactose,
or calcium
phosphate; or binding agents, such as starch, gelatin, or acacia; or
lubricating agents such as
magnesium stearate, stearic acid, or talc.
[0194] Additional pharmaceutical compositions of the disclosure will be
evident to those
skilled in the art, including formulations involving binding proteins in
sustained- or
controlled-delivery formulations. Techniques for formulating a variety of
other sustained- or
controlled-delivery means, such as liposome carriers, bio-erodible
microparticles or porous
beads and depot injections, are also known to those skilled in the art.
Additional examples of
sustained-release preparations include semipermeable polymer matrices in the
form of shaped
articles, e.g. films, or microcapsules. Sustained release matrices can include
polyesters,
hydrogels, polylactides, copolymers of L-glutamic acid and gamma ethyl-L-
glutamate,
poly(2-hydroxyethyl-methacrylate), ethylene vinyl acetate, or poly-D(-)-3-
hydroxybutyric
acid. Sustained-release compositions can also include liposomes, which can be
prepared by
any of several methods known in the art.
[0195] Pharmaceutical compositions to be used for in vivo administration
typically must
be sterile. This can be accomplished by filtration through sterile filtration
membranes.
Where the composition is lyophilized, sterilization using this method can be
conducted either
prior to, or following, lyophilization and reconstitution. The composition for
parenteral
91
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
administration can be stored in lyophilized form or in a solution. In
addition, parenteral
compositions generally are placed into a container having a sterile access
port, for example,
an intravenous solution bag or vial having a stopper pierceable by a
hypodermic injection
needle.
[0196] Once the pharmaceutical composition has been formulated, it can be
stored in
sterile vials as a solution, suspension, gel, emulsion, solid, or as a
dehydrated or lyophilized
powder. Such formulations can be stored either in a ready-to-use form or in a
form (e.g.,
lyophilized) requiring reconstitution prior to administration.
[0197] The disclosure also encompasses kits for producing a single-dose
administration
unit. The kits can each contain both a first container having a dried protein
and a second
container having an aqueous formulation. Also included within the scope of
this disclosure
are kits containing single and multi-chambered pre-filled syringes (e.g.,
liquid syringes and
lyosyringes).
[0198] The effective amount of a binding protein pharmaceutical composition
to be
employed therapeutically will depend, for example, upon the therapeutic
context and
objectives. One skilled in the art will appreciate that the appropriate dosage
levels for
treatment will thus vary depending, in part, upon the molecule delivered, the
indication for
which the binding protein is being used, the route of administration, and the
size (body
weight, body surface, or organ size) and condition (the age and general
health) of the patient.
Accordingly, the clinician can titer the dosage and modify the route of
administration to
obtain the optimal therapeutic effect.
[0199] Dosing frequency will depend upon the pharmacokinetic parameters of
the
binding protein in the formulation being used. Typically, a clinician will
administer the
composition until a dosage is reached that achieves the desired effect. The
composition can
therefore be administered as a single dose, as two or more doses (which may or
may not
contain the same amount of the desired molecule) over time, or as a continuous
infusion via
an implantation device or catheter. Further refinement of the appropriate
dosage is routinely
made by those of ordinary skill in the art and is within the ambit of tasks
routinely performed
by them. Appropriate dosages can be ascertained through use of appropriate
dose-response
data.
[0200] The route of administration of the pharmaceutical composition is in
accord with
known methods, e.g., orally; through injection by intravenous,
intraperitoneal, intracerebral
(intraparenchymal), intracerebroventricular, intramuscular, intraocular,
intraarterial,
92
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
intraportal, or intralesional routes; by sustained release systems; or by
implantation devices.
Where desired, the compositions can be administered by bolus injection or
continuously by
infusion, or by implantation device.
[0201] The composition can also be administered locally via implantation of
a membrane,
sponge, or other appropriate material onto which the desired molecule has been
absorbed or
encapsulated. Where an implantation device is used, the device can be
implanted into any
suitable tissue or organ, and delivery of the desired molecule can be via
diffusion, timed-
release bolus, or continuous administration.
[0202] The pharmaceutical compositions can be used to prevent and/or treat
HIV
infection. The pharmaceutical compositions can be used as a standalone therapy
or in
combination with standard anti-retroviral therapy.
[0203] The disclosure also relates to a kit comprising a binding protein
and other reagents
useful for detecting target antigen levels in biological samples. Such
reagents can include a
detectable label, blocking serum, positive and negative control samples, and
detection
reagents. In some embodiments, the kit comprises a composition comprising any
binding
protein, polynucleotide, vector, vector system, and/or host cell described
herein. In some
embodiments, the kit comprises a container and a label or package insert on or
associated
with the container. Suitable containers include, for example, bottles, vials,
syringes, IV
solution bags, etc. The containers may be formed from a variety of materials
such as glass or
plastic. The container holds a composition which is by itself or combined with
another
composition effective for treating, preventing and/or diagnosing a condition
(e.g., HIV
infection) and may have a sterile access port (for example the container may
be an
intravenous solution bag or a vial having a stopper pierceable by a hypodermic
injection
needle). In some embodiments, the label or package insert indicates that the
composition is
used for preventing, diagnosing, and/or treating the condition of choice.
Alternatively, or
additionally, the article of manufacture or kit may further comprise a second
(or third)
container comprising a pharmaceutically-acceptable buffer, such as
bacteriostatic water for
injection (BWFI), phosphate-buffered saline, Ringer's solution and dextrose
solution. It may
further include other materials desirable from a commercial and user
standpoint, including
other buffers, diluents, filters, needles, and syringes.
Methods and Uses for Binding Proteins
[0204] Certain aspects of the present disclosure relate to methods of
preventing HIV
infection in a patient, treating HIV infection in a patient, preventing AIDS
in a patient, and
93
CA 03136147 2021-10-04
WO 2020/210386 PCT/US2020/027313
treating AIDS in a patient, using any of the binding proteins or
pharmaceutical compositions
disclosed herein. Any of the binding proteins or pharmaceutical compositions
disclosed
herein may find use in a method of the present disclosure, e.g., methods for
preventing HIV
infection in a patient, treating HIV infection in a patient, preventing AIDS
in a patient, and
treating AIDS in a patient.
[0205] FIG. 1 illustrates an exemplary and non-limiting format for a
trispecific binding
protein that may be employed in the methods and uses described herein. As
shown in FIG. 2,
a proposed mechanism by which the binding protein shown in FIG. 1 may result
in
elimination of the HIV reservoir cells in a patient involves: (1) activation
of latently infected
CD4+ T cells via the anti-CD28 and anti-CD3 arms of the trispecific binding
protein; (2)
recruitment of CD8+ T cells to activated, latently infected CD4+ T cells via
anti-Env and
anti-CD3 arms; (3) activation of engaged CD8+ T cells via the anti-CD28 and
anti-CD3
arms; and (4) killing of latently infected CD4+ T cells through a
Perforin/Granzyme
mechanism. Advantageously, this mechanism is thought to activate and
subsequently kill
HIV-1 reservoir cells, providing a novel strategy for attacking the HIV-1
reservoir in a
patient.
[0206] In some embodiments, the methods of the present disclosure comprise
administering to the patient a therapeutically effective amount of at least
one of the binding
proteins or pharmaceutical compositions described herein.
[0207] In some embodiments, the at least one binding protein is
administered in
combination with an anti-retroviral therapy (e.g., an anti-HIV therepy). In
some
embodiments, the at least one binding protein is administered before the anti-
retroviral
therapy. In some embodiments, the at least one binding protein is administered
concurrently
with the anti-retroviral therapy. In some embodiments, the at least one
binding protein is
administered after the anti-retroviral therapy. In some embodiments, the at
least one binding
protein is co-administered with any standard anti-retroviral therapy known in
the art.
[0208] In some embodiments, administration of the at least one binding
protein or
pharmaceutical composition results in elimination of one or more latently
and/or chronically
HIV-infected cells in the patient. In some embodiments, administration of the
at least one
binding protein results in neutralization of one or more HIV virions and
results in elimination
of one or more latently and/or chronically HIV-infected cells in the patient.
In some
embodiments, the patient is a human.
94
Table 4. Trispecific binding protein polypeptide sequences
0
Molecule Polypeptide SEQ Sequence
t..)
o
Number ID
r..)
o
(ace. to NO
t''J
1¨,
formula)
Trispecific 1 1 61
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHNNANTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR oe
cA
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKGQPKAAPDIQMTQSPS
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGK
VRC07_523 /
APKLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKTKGPSRTVAAPSVF
IFPPS
CD28sup x
DEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQG
CD3mid IgG1 LSSPVTKSFNRGEC
LALA/P329A
2 62
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYWG
QGTLVTVSSRTASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV
VT Q
VPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDV
SH
,..
,
EDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALAAPIEKTISKAKGQPREP
,..
,
QVCTLPPSRDELTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQGNVFSC
S .
...,
s:)
VMHEALHNHYTQKSLSLSPG
"
,D
3 63
QVRLSQSGGQMKKPGDSMRISCRASGYEFINCPINWIRLAPGKRPEWMGWMKPRHGAVSYARQLQGRVTMTRDMYSET
,
,
,
AFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVSSASTKGPSVFPLAPSSKSTSGGTAALGCLVKDY
F ,
,D
PEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPC
PA .
PEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLH
QDWLNGKEYKCKVSNKALAAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDIAVEWESNGQPEN
NYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPG
4 64
SLTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRFSGSRWGPDYNLTISNLESGDF
GVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQDS
K
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 2 1 65
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHNNANTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR IV
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKGQPKAAPDIQMTQSPS
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGK
n
VRC07_523 /
APKLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKTKGPSRTVAAPSVF
IFPPS g
CD28sup x
DEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQG
4
CD3mid IgG1 LSSPVTKSFNRGEC
2
NNAS
o
'a
2
66
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
r..)
--.1
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
c,.)
1¨,
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYWG
QGTLVTVS SRTASTKGPSVFPLAPS SKSTSGGTAALGCLVKDYFPEPVTVSWNS GALT S GVHTFPAVLQ S
SGLYSLS SVVT
VP S S SLGTQTYICNVNHKP SNTKVDKKVEPKS CDKTHTCPP CPAPELL GGP S VFLFPPKPKD TLMI
SRTPEVTCVVVD VSHE o
DPEVKFNWYVD GVEVHNAKTKPREEQYNNASRVVSVL TVLHQDWLNGKEYKCKVSNKALPAPIEKTI
SKAKGQPREPQ r..)
o
VCTLPPSRDELTKNQVSL SCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLD SD GSFFLVSKL TVDK
SRWQQGNVF S C SV r..)
o
MHEALHNHYTQKSL SL SP G
t''J
1¨,
3 67 QVRL SQSGGQMKKPGD SMRIS CRAS GYEFINCPINWIRL AP
GKRPEWMGWMKPRHGAV SYARQLQ GRVTMTRD MY SET e,
AFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVS S A S TKGP S VFPLAP
SSKSTSGGTAALGCLVKDYF c'cg
PEPVTVS WNS GALT S GVHTFPAVLQ S SGLYSLS SVVTVPS S SL GTQ TYICNVNHKP
SNTKVDKKVEPKS CDKTHT CPP CPA
PELL GGP SVFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNNASRVVSVLTVLHQ
DWLNGKEYKCKVSNKALPAPIEKTI SKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP
SDIAVEWESNGQPENNY
KTTPPVLD SD G SFFLY SKLTVDK SRWQQ GNVF S C S VMHEALHNHYTQK SL SL SP G
4 68 SLTQSPGTL SL SP GETAII S CRTS
QYGSLAWYQQRPGQAPRLVIYS GS TRAAGIPDRF S G SRWGPDYNL TI SNLE S GDFGVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS GTA S VVCLLNNFYPREAKVQWKVDNAL Q S GNS
QE S V 1EQD SK
D S TY SL S STLTLSKADYEKHKVYACEVTHQGL S SPVTKSFNRGEC
Trispecific 3 1 69 DIVNITQTPL SL S VTPGQPA SI S CK S SQ
SLVHNNANTYL SWYLQKPGQ SPQ SLIYKVSNRF S GVPDRF S GS GS GTDFTLKI SR
VEAEDVGVYYCGQGTQYPFTFGS GTKVEIKGQPKAAPDIQMTQ SP S
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGK
P
VRC07_523 / APKLLIYKASNLHTGVPSRF S GS GS GTDFTLTIS
SLQPEDIATYYCQQGQTYPYTFGQGTKLEIKTKGPSRTVAAPSVFIFPPS L.
,
L.
CD28sup x DEQLKS
GTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQD SKD STY SL S STLTL
SKADYEKHKVYACEVTHQG .
,
s:) CD3mid IgG4 L S SPVTKSFNRGEC
,
cs, FAL A/409K
2
,
2 70 QVQLVQ S GAEVVKPGASVKVS CKAS GYTFT
SYYIHWVRQAPGQGLEWIG SIYP GNVNTNYAQKFQGRATLTVDTSI STA ,
,
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS S SQVQLVESGGGVVQPGRSLRL S CAA S
GFTFTKAWMH '
WVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD SKNTLYLQMNSLRAEDTAVYYCRGVYYAL
SPFDYWG
QGTLVTVS SRTA S TKGP S VFPLAP C SR S T SE S TAAL GCL VKDYFPEPVTVS WNS GALT S
GVHTFPAVL Q S SGLYSLS SVVTV
PS S SL GTKTYT CNVDHKP SNTKVDKRVE SKYGPP CPP CPAPEAAGGP S VFLFPPKPKD TLMI
SRTPEVT CVVVD VS QEDPE
VQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS
SIEKTISKAKGQPREPQVCTL
PP S QEEMTKNQVSL S CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SD GSFFL VSKLTVDK
SRWQEGNVF S CSVMHEA
LHNHYTQKSLSL SLG
3 71 QVRL SQSGGQMKKPGD SMRIS CRAS GYEFINCPINWIRL AP
GKRPEWMGWMKPRHGAV SYARQLQ GRVTMTRD MY SET
AFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVS
SASTKGPSVFPLAPCSRSTSESTAALGCLVKDYF
Iv
PEPVTVSWNS GAL TS GVHTFPAVLQ S SGLYSL S SVVTVP S S
SLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEA
AGGP S VFLFPPKPKDTLMI SRTPEVTCVVVDVSQEDPEVQFNWYVD
GVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDW
LNGKEYKCKVSNKGLPS S IEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYP
SDIAVEWESNGQPENNYKT 4
TPPVLD SD GSFFLY SKL TVDK SRWQE GNVF S C S VMHEALHNHYTQK SL SL SLG
o
r..)
o
4 72 SLTQSPGTL SL SP GETAII S CRTS
QYGSLAWYQQRPGQAPRLVIYS GS TRAAGIPDRF S G SRWGPDYNL TI SNLE S GDFGVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS
GTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQD SK
D S TY SL S STLTLSKADYEKHKVYACEVTHQGL S SPVTKSFNRGEC
c,.)
Trispecific 4 1 73
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKGQPKAAPDIQMTQSPS
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGK
0
VRC07_523 /
APKLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKTKGPSRTVAAPSVF
IFPPS ao
CD28sup x
DEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQG
a'
CD3mid_QQ LSSPVTKSFNRGEC
1-,
IgG4
o
FALA/409K
oe
cr
2 74
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
WVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYAL
SPFDYWG
QGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV
VTV
PS
SSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPE
VQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVCTL
PP SQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLD
SDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEA
LHNHYTQKSLSLSLG
3 75
QVRLSQSGGQMKKPGDSMRISCRASGYEFINCPINWIRLAPGKRPEWMGWMKPRHGAVSYARQLQGRVTMTRDMYSET
P
AFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVSSASTKGPSVFPLAPCSRSTSESTAALGCLVKDY
F .
,
PEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAP
EA
,
s:)
AGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDW
.
,
---1
LNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYK
T
r.,
TPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLG
,
,
4 76
SLTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRFSGSRWGPDYNLTISNLESGDF
GVYY .
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQDS
K
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 6 1 77
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSL
SASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
VRC07_523 /
KLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIF
PPSD
CD28sup x
EQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL
CD3mid_QQ SSPVTKSFNRGEC
IgG4
IV
FALA/409K
n
1-3
DKTHT linker
2
78
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
cp
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
o
n.o
o
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYWG
'a
QGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV
VTV = t : 1
W
PS
SSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPE
VQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP
SSIEKTISKAKGQPREPQVCTL
PP S QEEMTKNQVS L S CAVKGFYP SD IAVEWE SNGQPENNYKTTPPVLD SD G SFFL VSKLTVDK
SRWQEGNVF S CSVMHEA
LHNHYTQKSLSL SLG
0
3 79 QVRL SQSGGQMKKPGD SMRISCRASGYEFIN CPINWIRL AP GKRPEWMGWMKPRH GAV
SYARQLQGRVTNITRD MY SET a'
AFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVSSASTKGPSVFPLAPCSRSTSESTAALGCLVKDY
F
PEPVTVSWNSGALTSGVHTFPAVLQSSGLYSL
SSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEA
AGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDW
e,
LNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYP
SDIAVEWESNGQPENNYKT c'cg
TPPVLD SD G SFFLY SKL TVDK SRWQE GNVF S C S VMHEALHNHYTQK S L SL SLG
4 80 SLTQSPGTL SL SP GETAII S CRTS QYGSLAWYQQRPGQAPRLVIYS GS TRAAGIPDRF
S G SRWGPDYNL TI SNLE S GDFGVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS GTA S VVCLLNNFYPREAKVQWKVDNAL Q S GNS
QE S V 1EQD SK
D S TY SL SSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC
Trispecific 8 1 81 DIVNITQTPLSLSVTPGQPASISCKS SQ SLVHQNAQTYL
SWYLQKP GQ SPQ SLIYKVSNRF SGVPDRF SGSGSGTDFTLKI SR
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQ SP S SL S A S VGDRVTITCQA S
QNIYVVVLNWYQQKP GKAP
VRC07_523 /
KLLIYKASNLHTGVP SRF S GS G SGTDFTLTI
S SLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAP SVFIFPP SD
CD28sup x
EQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKD STY SL
SSTLTLSKADYEKHKVYACEVTHQGL
CD3mid_QQ SSPVTKSFNRGEC
P
IgG1 NNA S/4
,
09K_DKTHT
.
,
s:) linker
,
oc
r.,
2 82 QVQLVQ S GAEVVKPGASVKVS CKAS GYTFT
SYYIHWVRQAPGQGLEWIG SIYP GNVNTNYAQKFQGRATLTVDTSI STA .
r.,
,
, YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA ,
, WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD
SKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD .
YWGQGTLVTVS SDKTHTASTKGP SVFPL AP S
SKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLY
SL S SVVTVPS S SL GTQTYICNVNHKP SNTKVDKKVEPKSCDKTHT CPP CPAPELLGGP SVFLFPPKPKD
TLMI SRTPEVT CV
VVDVSHEDPEVKFNWYVD GVEVHNAKTKPREEQYNNASRVVSVL TVLHQDWLNGKEYKCKVSNKALPAPIEKTI
SKAK
GQPREPQVCTLPPSRDELTKNQVSL S CAVKGFYP SD IAVEWE SNGQPENNYKTTPPVLD SD G
SFFLVSKLTVDK SRWQQ G
NVF SCSVMHEALHNHYTQKSLSL SP G
3 83 QVRL SQSGGQMKKPGD SMRISCRASGYEFIN CPINWIRL AP GKRPEWMGWMKPRH GAV
SYARQLQ GRVTMTRD MY SET
AFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVS S A S TKGP S VFPLAP
SSKSTSGGTAALGCLVKDYF
PEPVTVSWNSGALTSGVHTFPAVLQSSGLYSL S SVVTVP S S SL GTQTYICNVNHKP
SNTKVDKKVEPKSCDKTHTCPP CPA Iv
PELL GGP SVFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNNASRVVSVLTVLHQ
DWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDEL TKNQVSLWCLVKGFYP
SDIAVEWESNGQPENNY
KTTPPVLD SD G SFFLY SKL TVDK SRWQ Q GNVF S C S VMHEALHNHYTQK SL SL SP G
cp
n.)
4 84 SLTQ SP GTL SL SP GETAIIS CRT S QYG SLAWYQQRP GQAPRLVIY S G S
TRAAGIPDRF S G SRWGPDYNLTI SNLE S GDF GVYY E4
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS GTA S VVCLLNNFYPREAKVQWKVDNAL Q S GNS
QE S V 1EQD SK
n.)
D S TY SL SSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC
-4
Trispecific 9 1
85 DIVNITQTPL SL S VTPGQPA SI S CKS
SQ SLVHENLQTYL SWYLQKPGQ SPQ SLIYKVSNRF S GVPDRF S GS GS GTDFTLKI SRV
EAEDVGVYYCGQGTQYPFTFGS GTKVEIKDKTHTDIQMTQ SP S
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAPK
VRC07_523_F
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE
R3-03 /
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
o
CD28sup x SPVTKSFNRGEC
n.)
o
CD3mid_ENL
n.)
o
Q IgG4
1--,
FALA/409K
o
DKTHT linker
oe
cA
2 86
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYWG
QGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV
VTV
PS
SSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPE
VQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS
SIEKTISKAKGQPREPQVCTL
PPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHE
A
LHNHYTQKSLSLSLG
3 87
QVRLSQSGGQMKKPGDSMRISCRASGYEFINCPINWIRLAPGKRPEWMGWMKPRHGAVSYARQLQGRVTMTRQLSQDP
P
DDPDWGTAFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVSSASTKGPSVFPLAPCSRSTSESTAAL
G .
L.
,
CLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPP
CP L.
,
s:)
PCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVL
T .
,
s:)
VLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQ
r.,
PENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLG
,
,
4 88
SLTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRFSGSRWGPDYNLTISNLESGDF
GVYY .
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQDS
K
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 10 1 89
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K
VRC07_523_F
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE
R3-03 /
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
CD28sup x SPVTKSFNRGEC
CD3mid_ENL
IV
F IgG4
n
,-i
FALA/409K
DKTHT linker
cp
n.)
2
90
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
o
n.)
o
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
C-5
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYWG
n.)
--.1
QGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV
VTV c`i'44
PS
SSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPE
VQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP S
SIEKTISKAKGQPREPQVCTL
PP S QEEMTKNQVSL S CAVKGFYP SD IAVEWE SNGQPENNYKTTPPVLD SD GSFFL VSKL TVDK
SRWQEGNVF S CSVMHEA o
LHNHYTQKSLSL SLG
n.)
o
3
91 QVRL SQSGGQMKKPGD
SMRISCRASGYEFINCPINWIRL AP GKRPEWMGWMKPRHGAV SYARQLQ GRVTMTRQL SQDP n.)
o
DDPDWGTAFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVSSASTKGPSVFPLAPCSRSTSESTAAL
G
CL VKDYFPEPVTVS WNSGALTSGVH TFPAVLQS SGLYSLS SVVTVPS S SLGTKTYTCNVDHKP
SNTKVDKRVESKYGPP CP e,
PCPAPEAAGGPSVFLFPPKPKDTLM ISRTPEVTCVVVD VSQEDPEVQFNWYVD
GVEVHNAKTKPREEQFNSTYRVVSVLT c'cg
VLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQ
PENNYKTTPPVLD SD GSFFLY SKL TVDK SRWQEGNVF S CSVMHEALHNHYTQKSLSL SLG
4 92 SLTQSPGTL SL SP GETAII S CRTS
QYGSLAWYQQRPGQAPRLVIYS GS TRAAGIPDRF S G SRWGPDYNLTI SNLE S GDFGVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS GTA S VVCLLNNFYPREAKVQWKVDNAL Q S GNS
QES V 1EQD SK
D S TY SL SSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC
Trispecific 11 1 93 DIVNITQTPL SLSVTPGQPASISCKSSQSLVHENLQTYL
SWYLQKPGQSPQSLIYKVSNRFS GVPDRFSGSGSGTDFTLKISRV
EAEDVGVYYCGQGTQYPFTFGS GTKVEIKDKTHTDIQMTQ SP S
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAPK
VRC07_523_F
LLIYKASNLHTGVP SRF S GS GS GTDFTLTIS
SLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAP SVFIFPP SDE
R3-03 /
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQD SKD STY SL SSTLTL
SKADYEKHKVYACEVTHQGLS P
CD28sup x SPVTKSFNRGEC
,
CD3mid_ENL
.
,
Q
.
,
IgG1 NNAS_
o
r.,
,
, DKTHT linker
,
, 2 94 QVQLVQ S GAEVVKPGASVKVS CKAS GYTFT SYYIHWVRQAPGQGLEWIG SIYP
GNVNTNYAQKFQGRATLTVDTSI STA .
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD SKNTLYLQMNSLRAEDTAVYYCRGVYYAL
SPFD
YWGQGTLVTVS SDKTHTASTKGP SVFPL AP S
SKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLY
SL S SVVTVPS S SL GTQTYICNVNHKP SNTKVDKKVEPKSCDKTHT CPP CPAPELLGGP SVFLFPPKPKD
TLMI SRTPEVT CV
VVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNNASRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAK
GQPREPQVCTLPP SRDELTKNQVSL S CAVKGFYP SDIAVEWESNGQPENNYKTTPPVLD SD
GSFFLVSKLTVDKSRWQQG
NVFSCSVMHEALHNHYTQKSLSL SP G
3
95 QVRL SQSGGQMKKPGD
SMRISCRASGYEFINCPINWIRL AP GKRPEWMGWMKPRHGAV SYARQLQ GRVTMTRQL SQDP IV
DDPDWGTAFLELRSLTSDDTAVYFCTRGKYCTARDYYNWDFEHWGQGTPVTVS S A S TKGP S VFPL AP S
SKSTSGGTAAL n
GCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSLS SVVTVPS S
SLGTQTYICNVNHKPSNTKVDKKVEPKSCDKT
HTCPPCPAPELLGGP SVFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNNASRVV 4
SVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTI SKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP
SDIAVEWE S g
NGQPENNYKTTPPVLD SD GSFFLY SKLTVDK SRWQQ GNVF S C S VMHEALHNHYTQK SL SL SP G
-a 5
4 96 SLTQSPGTL SL SP GETAII S CRTS
QYGSLAWYQQRPGQAPRLVIYS GS TRAAGIPDRF S G SRWGPDYNLTI SNLE S GDFGVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDS
K
D S TY SL SSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC
Trispecific 12 1 97 DIVNITQTPL SL S VTPGQPA SI S CK S SQ SLVHENLFTYL
SWYLQKP GQ SPQ SLIYKVSNRF S GVPDRF S GS GS GTDFTLKI SRV
EAEDVGVYYCGQGTQYPFTFGS GTKVEIKDKTHTDIQMTQ SP S SL S
ASVGDRVTITCQASQNIYVVVLNWYQQKPGKAPK 0
VRC07_523_F LLIYKASNLHTGVP SRF S GS GS GTDFTLTIS
SLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAP SVFIFPP SDE n.)
o
R3-03 / QLKSGTASVVCLLNNFYPREAKVQWKVDNALQ SGNSQESVTEQD SKD
STY SL SSTLTL SKADYEKHKVYACEVTHQGLS a'
CD28sup x SPVTKSFNRGEC
1--,
CD3mid_ENL
o
F
oe
o
IgG1 NNAS_
DKTHT linker
2 98 QVQLVQ S GAEVVKPGASVKVS CKAS GYTFT
SYYIHWVRQAPGQGLEWIG SIYP GNVNTNYAQKFQGRATLTVDTSI STA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYAL SPFD
YWGQGTLVTVS SDKTHTASTKGP SVFPL AP S SKST SGGTAAL GCLVKDYFPEPVTVS WNSGALT
SGVHTFPAVLQ S SGLY
SL S SVVTVP S S SL GTQTYICNVNHKP SNTKVDKKVEPKSCDKTHT CPP CPAPELLGGP
SVFLFPPKPKD TLMI SRTPEVT CV
VVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNNASRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAK
GQPREPQVCTLPP SRDELTKNQVSL S CAVKGFYP SDIAVEWESNGQPENNYKTTPPVLD SD
GSFFLVSKLTVDKSRWQQG P
NVF SC SVMHEALHNHYTQKSL SL SP G
.
,
3 99 QVRL SQ SGGQMKKP GD SMRISCRASGYEFINCPINWIRL AP
GKRPEWMGWMKPRHGAVSYARQLQ GRVTMTRQL SQDP
,
DDPD W GTAFLELRSL T SDD TAVYF CTRGKY CTARDYYNWDFEHWGQ GTPVTVS S A S TKGP S
VFPL AP S SK S T S GGTAAL .
,
GCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSL S SVVTVP S S SLGTQTYICNVNHKP
SNTKVDKKVEPKSCDKT .
r.,
HTCPPCPAPELLGGP SVFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNNASRVV ,
,
,
SVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTI SKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP
SDIAVEWE S .
,
NGQPENNYKTTPP VLD SD GSFFLY SKLTVDK SRWQQ GNVF S CS VMHEALHNHYTQK SL SL SP G
.
4 100 SL TQ SP GTL SL SP GETAII S CRT S QYGSLAWYQQRP
GQAPRLVIY S G S TRAAGIPDRF S GSRWGPDYNLTI SNLE S GDF GVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS GTA S VVCLLNNFYPREAKVQWKVDNAL Q S GNS
QES V 1EQD SK
D S TY SL SSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC
Trispecific 13 1 101 DIVNITQTPL SLSVTPGQPASISCKSSQSLVHNNANTYL SWYL QKP GQ
SPQ SLIYKVSNRF S GVPDRF SGSGSGTDFTLKI SR
VEAEDVGVYYCGQGTQYPFTFGS GTKVEIKGQPKAAPDIQMTQ SP S
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGK
N6 / CD28sup APKLLIYKASNLHTGVPSRF
SGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKTKGPSRTVAAPSVFIFPPS
x CD3mid DEQLKS GTA S VVCLLNNFYPREAKVQWKVDNALQ S GNS QES
VTEQD SKD S TY SL SSTLTLSKADYEKHKVYACEVTHQG 00
IgG4 L SSPVTKSFNRGEC
n
1-3
2 102 QVQLVQSGAEVVKPGASVKVS CKAS GYTFTSYYIHWVRQAP GQ
GLEWIGSIYP GNVNTNYAQKFQGRATLTVDTSIS TA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRL S CAA S
GFTFTKAWMH cp
n.)
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYAL SPFDYWG
o
n.)
o
QGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSL
SSVVTV
PS S SL GTKTYT CNVDHKP SNTKVDKRVESKYGPP CPP CPAPPVAGP S VFLFPPKPKD TLMI
SRTPEVT CVVVD VS QEDPEV n.)
-4
QFNWYVD GVEVHNAKTKPREEQFNS TYRVVS VLTVLHQD WLNGKEYKCKVSNKGLP S SIEKTI
SKAKGQPREPQVCTLP '&44
PSQEEMTKNQVSL SCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLD
SDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEAL
HNHYTQKSLSLSLG
0
3 103
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
a'
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
n.)
o
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPPVA
GP
SVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGK
e,
EYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTPP
VL c'cg
DSDGSFFLYSKLTVDKSRWQEGNVFSCSVNIHEALHNHYTQKSLSL SLG
4 104
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 14 1 105
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHNNANTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKGQPKAAPDIQMTQSPS
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGK
N6 / CD28sup
APKLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKTKGPSRTVAAPSVF
IFPPS
x CD3mid
DEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQG
IgG4 LSSPVTKSFNRGEC
P
2 106
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
,
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
.
,
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYWG
,
t.)
QGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV
VTV 0
r.,
,
, PS
SSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPE
,
, VQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS
SIEKTISKAKGQPREPQVCTL .
PP SQEEMTKNQVSL
SCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEA
LHNHYTQKSLSLSLG
3 107
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G
GP
SVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSL SLG
IV
4 108
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
cp
n.)
o
Trispecific 15 1 109
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR
o
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKGQPKAAPDIQMTQSPS
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGK
'a
N6 / CD28sup
APKLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKTKGPSRTVAAPSVF
IFPPS
X CD3mid_QQ
DEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQG
LSSPVTKSFNRGEC
IgG4
FALA/409K
0
2 110
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
r..)
o
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSSQVQLVESGGGVVQPGRSLRLSCAASGFTFTKAWMH
r..)
o
WVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFDYWG
1-,
QGTLVTVSSRTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSV
VTV F,3
PS
SSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPE
oe
cA
VQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS
SIEKTISKAKGQPREPQVCTL
PP SQEEMTKNQVSL
SCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVFSCSVMHEA
LHNHYTQKSLSLSLG
3 111
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G
GP
SVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSL SLG
P
4 112
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI .
L.
,
L.
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D .
,
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
.
,
w Trispecific 16 1
113
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR .
r.,
,
, VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSL
SASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP ,
, N6 / CD28sup
KLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIF
PPSD .
x CD3mid_QQ
EQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL
IgG4 SSPVTKSFNRGEC
FALA/409K
DKTHT linker
2 114
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS Iv
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD n
,-i
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP
SSIEKTISKAKGQPR
EPQVCTLPPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVF
S cp
r..)
CSVMHEALHNHYTQKSLSLSLG
o
r..)
o
3 115
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
ti
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVF SCSVMHEALHNHYTQKSLSL SLG
0
4
116
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI a'
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKS GTASVVCLLNNFYPREAKVQWKVDNALQS
GNSQESVIEQD
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
1-,
Trispecific 17 1 117
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHNNANTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR o
oe
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSL
SASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
o
N6 / CD28sup
KLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIF
PPSD
x CD3mid
EQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL
IgG4 SSPVTKSFNRGEC
FALA/409K
DKTHT linker
2 118
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS P
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD
,
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPR
.
,
EPQVCTLPPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVF
S ,
-i. CSVMHEALHNHYTQKSLSLSLG
o
r.,
,
, 3
119
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
,
,
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
.
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVF SCSVMHEALHNHYTQKSLSL SLG
4 120
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKS
GTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVIEQD
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 19 1 121
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHQNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR IV
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSL
SASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
n
1-3
N6 / CD28sup
KLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIF
PPSD
x CD3mid_QQ
EQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL
4
=
IgG1 NNAS_ SSPVTKSFNRGEC
o
DKTHT linker
'a
n.)
2 122
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
-4
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
Y
SLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEV
TCV 0
VVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNNASRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAK
ao
GQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQ
G a'
NVFSCSVMHEALHNHYTQKSLSLSPG
1¨,
3
123
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
e,
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEP
c'cg
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAP
EL
LGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNNASRVVSVLTVLHQDW
LNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYK
T
TPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSL SLSPG
4 124
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 21 1 125
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHDNAQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISR
VEAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSL
SASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP P
N6 / CD28sup
KLLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIF
PPSD
,
x
EQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGL
.
,
CD3mid DNA SSPVTKSFNRGEC
,
r.,
Q IgG4
o
r.,
FALA/409K
,
,
,
, DKTHT linker
.
2 126
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPR
EPQVCTLPPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVF
S
CSVMHEALHNHYTQKSLSLSLG
IV
3
127
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
r'
1-3
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G cp
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
E. 0
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSL SLG
iµ.0
-4
1¨,
c,.)
4 128
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D 0
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
n.)
o
Trispecific 22 1 129
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K iZ.1
1¨,
N6 / CD28sup
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE o
x
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
c'cg
CD3mid_ENL SPVTKSFNRGEC
Q IgG4
FALA/409K
DKTHT linker
2 130
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD P
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPR
.
,
EPQVCTLPPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVF
S
,
CSVMHEALHNHYTQKSLSLSLG
.
,
r.,
cs, 3 131 RAHLVQ S GTAMKKP GA S VRVS CQ T S
GYTFTAHILFWFRQAP GRGLEWVGWIKPQYGAVNF GGGFRDRVTL TRD VYREIA o
r.,
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
,
,
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G .
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLG
4 132
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 23 1 133
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLRTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K IV
N6 / CD28sup
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE n
x
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
CD3mid_ENL SPVTKSFNRGEC
cp
n.)
o
R IgG4
r..)
o
FALA/409K
-a 5
DKTHT linker
n.)
-4
2 134
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD
SKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVS SDKTHTASTKGP SVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNS GALT S
GVHTFPAVLQ S SGLYS o
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD n.)
o
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP
SSIEKTISKAKGQPR a'
EPQVCTLPPSQEEMTKNQVSLS CAVKGFYP SD IAVEWE SNGQPENNYKTTPPVLD SD G SFFLVSKLTVDK
SRWQEGNVF S
1-,
CSVMHEALHNHYTQKSLSLSLG
o
3
135
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
c'cg
YMDIRGLKPDDTAVYYCARDRSYGD S S WALD AW GQ GTTVVVS AA S TKGP S VFPL AP C SR S T
SE S TAAL GCLVKDYFPEP
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPS
SIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTTP
PVLD SD G SFFLY SKL TVDK SRWQEGNVF SCSVMHEALHNHYTQKSLSL SLG
4 136 YIHVTQ SP S SL
SVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTS SVED GVP SRF S GS GFHT SFNLTI SDL
QADD I
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKS GTA S VVCLLNNFYPREAKVQWKVDNALQ S GN S
QES V 1EQD
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 24 1 137 DIVNITQTPL SL S VTP GQPAS IS CKS
SQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRF S GS GS GTDFTLKI SRV P
EAEDVGVYYCGQGTQYPFTFGS GTKVEIKDKTHTDIQMTQ SP S
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAPK
L.
,
L.
N6 / CD28sup LLIYKASNLHTGVP SRF S GS GS GTDFTLTIS
SLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAP SVFIFPP SDE .
,
x QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQD
SKD STY SL S STLTL SKADYEKHKVYACEVTHQGLS ,
r.,
--1 CD3mid_ENL SPVTKSFNRGEC
o
r.,
F IgG4
,
FALA/409K
,
DKTHT linker
2 138
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD SKNTLYLQMNSLRAEDTAVYYCRGVYYAL
SPFD
YWGQGTLVTVS SDKTHTASTKGP SVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNS GALT S
GVHTFPAVLQ S S GLYS
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP
SSIEKTISKAKGQPR
EPQVCTLPP SQEEMTKNQVSL S CAVKGFYP SDIAVEWE SNGQPENNYKTTPPVLD SD G SFFLVSKL
TVDKSRWQEGNVF S IV
CSVMHEALHNHYTQKSLSLSLG
n
1-3
3 139
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRDVYREIA
YMDIRGLKPDDTAVYYCARDRSYGD S S WALD AWGQ GTTVVV S AA S TK GP S VFPLAP C SR S T
SE S TAAL GCLVKDYFPEP 4
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G o
n.)
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLP S SIEKTISKAKGQPREP QVYTLPP CQEEMTKNQVSLW CL VKGFYP SD IAVEWE
SNGQPENNYKTTP n.)
--.1
PVLD SD G SFFLY SKL TVDK SRWQEGNVF SCSVMHEALHNHYTQKSLSL SLG
c,.)
4 140
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D 0
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
r..)
o
Trispecific 25 1 141
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K
1¨,
N6 rw52 /
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE o
CD28sup x
oe
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
o
CD3mid_ENL SPVTKSFNRGEC
Q IgG4
FALA/409K
DKTHT linker
2 142
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD P
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPR
.
L.
,
EPQVCTLPPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVF
S L.
,
CSVMHEALHNHYTQKSLSLSLG
.
,
r.,
oc 3 143
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGATNFGGGFRDRVTLTRDVYREIA
o
r.,
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
,
,
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G .
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLG
4 144
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSSEEGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 26 1 145
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K IV
N6 rw52 /
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE n
,-i
CD28sup x
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
CD3mid_ENL SPVTKSFNRGEC
cp
r..)
o
F IgG4
r..)
o
FALA/409K
C-5
DKTHT linker
r..)
--.1
2 146
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS o
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD n.)
o
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPR
a'
EPQVCTLPPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVF
S iZ.1
1-,
CSVMHEALHNHYTQKSLSLSLG
o
3
147
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGATNFGGGFRDRVTLTRDVYREIA
c'cg
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEP
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAA
G
GPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLN
GKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYKTT
P
PVLDSDGSFFLYSKLTVDKSRWQEGNVF SCSVMHEALHNHYTQKSLSL SLG
4 148
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSSEEGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 27 1 149
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV P
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K
,
N6 rw52 /
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE .
,
CD28sup x
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
.
,
r.,
s:) CD3mid_ENL SPVTKSFNRGEC
o
r.,
F
,
,
,
, IgG1 NNAS_
.
DKTHT linker
2 150
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
Y
SLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEV
TCV
VVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNNASRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAK
GQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQ
G IV
NVFSCSVMHEALHNHYTQKSLSLSPG
n
,-i
3 151
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGATNFGGGFRDRVTLTRDVYREIA
YMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEP
4
VTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAP
EL o
r..)
LGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNNASRVVSVLTVLHQDW
LNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDIAVEWESNGQPENNYK
T ti
TPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSL SLSPG
c,.)
4
152
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSSEEGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D 0
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
r..)
o
Trispecific 28 1 153
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K
1--,
N6 FR3-03 /
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE o
CD28sup x
oe
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
o
CD3mid_ENL SPVTKSFNRGEC
Q IgG4
FALA/409K
DKTHT linker
2 154
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD P
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPR
.
L.
,
EPQVCTLPPSQEEMTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQEGNVF
S L.
,
. CSVMHEALHNHYTQKSLSLSLG
.
,
3
155
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRQLSQDP
o
r.,
DDPDWGIAYMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPCSRSTSESTAALGCL
,
,
,
,
VKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCP
PC .
PAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTV
L
HQDWLNGKEYKCKVSNKGLPSSIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYPSDIAVEWESNGQPE
NNYKTTPPVLDSDGSFFLYSKLTVDKSRWQEGNVFSCSVMHEALHNHYTQKSLSLSLG
4 156
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 29 1
157
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K IV
N6 FR3-03 /
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE n
,-i
CD28sup x
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
CD3mid_ENL SPVTKSFNRGEC
cp
r..)
o
F IgG4
r..)
o
FALA/409K
C-5
r..)
DKTHT linker
--.1
2
158
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
1--,
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD
SKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVS SDKTHTASTKGP
SVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYS o
L SSVVTVP S S SL GTKTYT CNVDHKP SNTKVDKRVE SKYGPP CPP CP APEAAG GP SVFLFPPKPKD
TLMI SRTPEVTCVVVD n.)
o
VS QEDPEVQFNWYVD GVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS
SIEKTISKAKGQPR a'
EPQVCTLPPSQEEMTKNQVSLS CAVKGFYP SD IAVEWE SNGQPENNYKTTPP VLD SD G SFFLVSKLTVDK
SRWQEGNVF S iZ.1
1--,
CSVMHEALHNHYTQKSL SLSLG
o
3 159 RAHLVQ S GTAMKKP GA S VRV S CQ T S GYTFTAHILFWFRQAP
GRGLEWVGWIKPQYGAVNF GGGFRDRVTL TRQL S QDP oe
cr
DDPDWGIAYMDIRGLKPDDTAVYYCARDRSYGD S S WALD AW GQ GTTVVVS AA S TKGP S VFPL AP C
SR S T SE S TAAL G CL
VKDYFPEPVTVSWNS GALT SGVHTFPAVLQ SS GLYSL
SSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPC
PAPEAAGGP S VFLFPPKPKDTLMI SRTPEVTCVVVDVSQEDPEVQFNWYVD
GVEVHNAKTKPREEQFNSTYRVVSVLTVL
HQDWLNGKEYKCKVSNKGLP S SIEKTI SKAKGQPREPQVYTLPP CQEEMTKNQVSLW CLVKGFYP SD
IAVEWE SNGQPE
NNYKTTPPVLD SD G SFFLY SKL TVDK SRWQEGNVF S CSVMHEALHNHYTQKSLSLSL G
4 160 YIHVTQ SP S SL SVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTS
SVEDGVPSRFSGS GFHT SFNLTI SDL QADD I
ATYYCQVLQFF GRG SRLHIKRTVAAP S VFIFPP SD EQLK S GTA S VVCLLNNFYPREAKVQWKVDNAL
Q S GNS QE S V 1EQD
SKD STY SL SSTLTL SKADYEKHKVYACEVTHQGLS SP VTK SFNRGEC
Trispecific 30 1
161 DIVNITQTPL SLSVTPGQPASISCKS
SQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTLKISRV P
EAEDVGVYYCGQGTQYPFTFGS GTKVEIKDKTHTDIQMTQ SP S
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAPK
,
N6 FR3-03/ LLIYKASNLHTGVP SRF S GS GS GTDFTLTIS
SLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAP SVFIFPP SDE .
,
. CD28sup x QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQD
SKD STY SL SSTLTL SKADYEKHKVYACEVTHQGLS ,
.
r.,
. CD3mid ENL SPVTKSFNRGEC
o
r.,
,
, QIgG1 NNAS
,
, DKTHT
.
linker
2 162 QVQLVQSGAEVVKPGASVKVS CKAS GYTF TSYYIHWVRQAP GQ GLEWIGSIYP
GNVNTNYAQKFQGRATLTVDTSIS TA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD SKNTLYLQMNSLRAEDTAVYYCRGVYYAL
SPFD
YWGQGTLVTVS SDKTHTASTKGP SVFPLAPS SKSTSGGTAAL GCLVKDYFPEPVTVSWNS GAL T S
GVHTFPAVLQ S SGLY
SL S SVVTVPS S SL GTQTYICNVNHKP SNTKVDKKVEPKSCDKTHT CPP CPAPELL GGP
SVFLFPPKPKD TLMI SRTPEVT CV
VVDVSHEDPEVKFNWYVD GVEVHNAKTKPREEQYNNASRVVSVL TVLHQDWLNGKEYKCKVSNKALPAPIEKTI
SKAK
GQPREPQVCTLPP SRDEL TKNQVSL S CAVKGFYP SDIAVEWESNGQPENNYKTTPPVLD SD
GSFFLVSKLTVDKSRWQQG IV
NVFSCSVMHEALHNHYTQKSLSL SP G
n
,-i
3 163 RAHLVQ S GTAMKKP GA S VRV S CQ T S GYTFTAHILFWFRQAP
GRGLEWVGWIKPQYGAVNF GGGFRDRVTL TRQL S QDP
DDPDWGIAYMDIRGLKPDDTAVYYCARDRSYGD S S WALD AW GQ GTTVVVS AA S TKGP S VFPL AP S
SKSTSGGTAALGC cp
n.)
LVKDYFPEPVTVSWNSGALTSGVHTFPAVLQS SGLYSLS SVVTVPS S
SLGTQTYICNVNHKPSNTKVDKKVEPKS CDKTHT g
CPPCPAPELLGGP SVFLFPPKPKDTLMI SRTPEVTCVVVDVSHEDPEVKFNWYVD
GVEVHNAKTKPREEQYNNASRVVSV
L TVLHQDWLNGKEYKCKVSNKALPAPIEKTI SKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYP
SDIAVEWESNG ti
QPENNYKTTPPVLD SD G SFFLY SKL TVDK SRWQQ GNVF S C S VMHEALHNHYTQK SL SL SP G
1--,
c,.)
4 164
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 31 1 165
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K
N6 FR3-03 /
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE
CD28sup x
oe
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
CD3mid_ENL SPVTKSFNRGEC
IgG1 NNAS_
DKTHT linker
2 166
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVSSDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYADSVKGRFTISRDDSKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPSSKSTSGGTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
Y
SLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKTHTCPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEV
TCV
VVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNNASRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAK
GQPREPQVCTLPPSRDELTKNQVSLSCAVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLVSKLTVDKSRWQQ
G
NVFSCSVMHEALHNHYTQKSLSLSPG
3 167
RAHLVQSGTAMKKPGASVRVSCQTSGYTFTAHILFWFRQAPGRGLEWVGWIKPQYGAVNFGGGFRDRVTLTRQLSQDP
DDPDWGIAYMDIRGLKPDDTAVYYCARDRSYGDSSWALDAWGQGTTVVVSAASTKGPSVFPLAPSSKSTSGGTAALGC
LVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTQTYICNVNHKPSNTKVDKKVEPKSCDKT
HT
CPPCPAPELLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNNASRVVS
V
LTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPCRDELTKNQVSLWCLVKGFYPSDIAVEWESN
G
QPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPG
4 168
YIHVTQSPSSLSVSIGDRVTINCQTSQGVGSDLHWYQHKPGRAPKLLIHHTSSVEDGVPSRFSGSGFHTSFNLTISDLQ
ADDI
ATYYCQVLQFFGRGSRLHIKRTVAAPSVFIFPPSDEQLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQ
D
SKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 34 1 169
DIVNITQTPLSLSVTPGQPASISCKSSQSLVHENLQTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRFSGSGSGTDFTL
KISRV
EAEDVGVYYCGQGTQYPFTFGSGTKVEIKDKTHTDIQMTQSPSSLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAP
K
VRC01.23 /
LLIYKASNLHTGVPSRFSGSGSGTDFTLTISSLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAPSVFIFP
PSDE
CD28sup x
QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQDSKDSTYSLSSTLTLSKADYEKHKVYACEVTHQGLS
CD3mid_ENL SPVTKSFNRGEC
IgG4
FALA/409K
C-5
DKTHT linker
2 170
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD SKNTLYLQMNSLRAEDTAVYYCRGVYYAL
SPFD
YWGQGTLVTVSSDKTHTASTKGPSVFPLAPCSRSTSESTAALGCLVKDYFPEPVTVSWNSGALTSGVHTFPAVLQSSGL
YS
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD
VSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLP
SSIEKTISKAKGQPR
EPQVCTLPPSQEEMTKNQVSLS CAVKGFYP SD IAVEWE SNGQPENNYKTTPP VLD SD G SFFLVSKLTVDK
SRWQEGNVF S
CSVMHEALHNHYTQKSLSLSLG
3 171 QVQL VQ S GGQMKKP GE SMRI S CRA S GYEF ID CTLNWIRL AP
GKRPEWMGWLKPRWGAVNYARPLQ GRVTMTRQL S QD P c'cg
DDPDWGTAFLELRSLTVDDTAVYFCTRGKNCDYNWDFEHWGRGTPVIVSSASTKGPSVFPLAPCSRSTSESTAALGCLV
K
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPC
PA
PEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLH
Q
DWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYP SD IAVEWE
SNGQPENN
YKTTPPVLD SD G SFFLY SKL TVDK SRWQEGNVF SCSVMHEALHNHYTQKSLSL SLG
4 172 L TQ SP GTL SL SP GETAII S CRT S QYG SLAWYQQRP GQ APRLVIY S G S
TRAAGIPD RF SGSRWGPDYNLTISNLESGDFGVYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS GTA S VVCLLNNFYPREAKVQWKVDNAL Q S GNS
QES V 1EQD SK
DSTYSLSSTLTLSKADYEKHKVYACEVTHQGLSSPVTKSFNRGEC
Trispecific 35 1 173 DIVNITQTPL SL S VTP GQPAS IS CKS
SQSLVHENLFTYLSWYLQKPGQSPQSLIYKVSNRFSGVPDRF S GS GS GTDFTLKI SRV
EAEDVGVYYCGQGTQYPFTFGS GTKVEIKDKTHTDIQMTQ SP S
SLSASVGDRVTITCQASQNIYVVVLNWYQQKPGKAPK
VRC01.23 / LLIYKASNLHTGVP SRF S GS GS GTDFTL TI S
SLQPEDIATYYCQQGQTYPYTFGQGTKLEIKDKTHTRTVAAP SVFIFPP SDE
CD28sup x QLKSGTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESVTEQD SKD S
TY SL S STLTL SKADYEKHKVYACEVTHQGLS
CD3mid ENL SPVTKSFNRGEC
IgG4
FALA/409K
DKTHT linker
2 174
QVQLVQSGAEVVKPGASVKVSCKASGYTFTSYYIHWVRQAPGQGLEWIGSIYPGNVNTNYAQKFQGRATLTVDTSISTA
YMELSRLRSDDTAVYYCTRSHYGLDWNFDVVVGKGTTVTVS
SDKTHTQVQLVESGGGVVQPGRSLRLSCAASGFTFTKA
WMHWVRQAPGKQLEWVAQIKDKSNSYATYYAD SVKGRFTISRDD
SKNTLYLQMNSLRAEDTAVYYCRGVYYALSPFD
YWGQGTLVTVS SDKTHTASTKGP SVFPL APCSRSTSE STAAL GCLVKDYFPEPVTVS WN S GALT S
GVHTFPAVLQ S SGLYS
LSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPCPAPEAAGGPSVFLFPPKPKDTLMISRTPEVTCVV
VD
VS QEDPEVQFNWYVD GVEVHNAKTKPREEQFNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKGLPS
SIEKTISKAKGQPR
EPQVCTLPPSQEEMTKNQVSLS CAVKGFYP SD IAVEWE SNGQPENNYKTTPPVLD SD
GSFFLVSKLTVDKSRWQEGNVFS
CSVMHEALHNHYTQKSLSLSLG
3 175
QVQLVQSGGQMKKPGESMRISCRASGYEFIDCTLNWIRLAPGKRPEWMGWLKPRWGAVNYARPLQGRVTMTRQLSQDP
DDPDWGTAFLELRSLTVDDTAVYFCTRGKNCDYNWDFEHWGRGTPVIVSSASTKGPSVFPLAPCSRSTSESTAALGCLV
K
DYFPEPVTVSWNSGALTSGVHTFPAVLQSSGLYSLSSVVTVPSSSLGTKTYTCNVDHKPSNTKVDKRVESKYGPPCPPC
PA
PEAAGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSQEDPEVQFNWYVDGVEVHNAKTKPREEQFNSTYRVVSVLTVLH
Q
DWLNGKEYKCKVSNKGLPS SIEKTISKAKGQPREPQVYTLPPCQEEMTKNQVSLWCLVKGFYP SD IAVEWE
SNGQPENN
YKTTPPVLD SD G SFFLY SKL TVDK SRWQEGNVF SCSVMHEALHNHYTQKSLSL SLG
4 176
LTQSPGTLSLSPGETAIISCRTSQYGSLAWYQQRPGQAPRLVIYSGSTRAAGIPDRFSGSRWGPDYNLTISNLESGDFG
VYY
CQQYEFFGQGTKVQVDIKRTVAAPSVFIFPPSDEQLKS
GTASVVCLLNNFYPREAKVQWKVDNALQSGNSQESV1EQD SK o
DSTYSL SSTLTLSKADYEKHKVYACEVTHQGL SSPVTKSFNRGEC
n.)
o
n.)
o
Table 5. Trispecific binding protein polynucleotide sequences
oe
cr
Molecule Polypeptide SEQ Sequence
Number ID
(ace. to NO
formula)
Trispecific 1 1 177
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
VRC07_523 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD3mid IgG1
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGAGCCCCA
P
LALA/P329A
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAAC
.
TGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCA
,
GCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACC
,
.
-,
TACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGACCAAGGGCC
CCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCT
"
,
,
GTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCG
,
,
GCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAG
CAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAG
AGCTTCAACCGGGGCGAGTGT
2 178
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTGGTGGAATCT
GGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTCACCAAGGCCT
00
n
GGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAAGAGCAACAGCTA 1-
3
CGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAACACCCTGTACCTG
c7,
CAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCTGAGCCCCTTCGA
a'
TTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCCAGCGTGTTCCCTCTGG
a'
CCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAGTCCAGCGGCCTGTAC
d
AGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACATCTGCAACGTGAACCACAAGCC
i:4
CAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACACCTGTCCCCCTTGTCCTGCC
CCCGAAGCCGCCGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCC
o
CGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCAATTGGTACGTGGACGGCGTGG
ao
AAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGT
a'
GCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGCCCTGGCCGCCCCCATCGAG
AAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGCCCCCAAGCAGGGACGAGCTGA a
CCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATCGCCGTGGAATGGGAGAGCAAC
g
GGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCT
GACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCAC
TACACCCAGAAGTCCCTGAGCCTGAGCCCCGGCTAGTGA
3 179
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGGACATGTACAGCG
AGACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTCTGCACCCGGGGCAAGTACTGC
ACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTGTGACAGTGTCTAGCGCTTCGAC
CAAGGGCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCG
P
TGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCA
.
GCTGTGCTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGAC
.
CTACATCTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAG
.."
,
(..,
ACCCACACCTGTCCCCCTTGTCCTGCCCCCGAAGCCGCCGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAG
2
GACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGA
,
,
AGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAGCAC
0"
,
CTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCC
2
AACAAGGCCCTGGCCGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTACA
CACTGCCCCCATGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGTGGTGTCTGGTGAAAGGCTTCTACCCCTCC
GATATCGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCG
ACGGCTCATTCTTCCTGTACTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCC
GTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
4 180
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
00
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
n
1-3
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG -
--
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
cp
n.o
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
2
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
!II
ACCGGGGCGAGTGT
c,.)
c,.)
Trispecific 2 1 181
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
o
VRC07_523 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
ao
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
a'
CD3mid IgG1
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGAGCCCCA
NNAS
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAAC
a
TGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCA
oe
c:
GCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACC
TACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGACCAAGGGCC
CCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCT
GTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCG
GCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAG
CAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAG
AGCTTCAACCGGGGCGAGTGT
2 182
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
P
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
.
,
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
,
.
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTGGTGGAATCT
.
,
GGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTCACCAAGGCCT
2
GGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAAGAGCAACAGCTA
,
,
,
CGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAACACCCTGTACCTG
.
,
CAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCTGAGCCCCTTCGA
.
TTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCCAGCGTGTTCCCTCTGG
CCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTGCTGCAGTCCAGCGGCCTGTAC
AGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACATCTGCAACGTGAACCACAAGCC
CAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCACACCTGTCCCCCTTGTCCTGCC
CCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCC
CGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCAATTGGTACGTGGACGGCGTGG
,t
AAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCGGGTGGTGTCCGTGCTGACCGT
n
1-3
GCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGCCCTGCCTGCCCCCATCGAG
----
AAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGCCCCCAAGCAGGGACGAGCTGA 4
CCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATCGCCGTGGAATGGGAGAGCAAC
2
GGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCT
GACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCAC
iµ.0
--4
TACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
c,.)
c,.)
3
183
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
o
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGGACATGTACAGCG
ao
AGACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTCTGCACCCGGGGCAAGTACTGC
a'
ACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTGTGACAGTGTCTAGCGCTTCGAC
CAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGG
ca,
TCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCG
g
GCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGAC
CTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAA
ACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAA
GGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCA
AGTTCAACTGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAATGC
CTCCCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCA
ACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACAC
CCTGCCCCCATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGGTAAAAGGCTTCTATCCCAGCG
ACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGA
P
CGGCTCCTTCTTCCTCTATTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGT
0
,
GCTGCATGAGGCTCTGCACAGCCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGT
0
,
. 4
184
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
.
,
L---1
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
2
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
,
,
,
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
0
,
0
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
.
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
Trispecific 3 1 185
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
VRC07_523 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
00
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
n
1-3
CD3mid IgG4
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGAGCCCCA
---
FALA/409K
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAAC
4
TGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCA
o
GCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACC
,---,
TACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGACCAAGGGCC
tµ.0
-4
CCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCT
C4
W
GTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCG
GCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAG
CAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAG
o
AGCTTCAACCGGGGCGAGTGT
n.)
o
2
186
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
Ce,
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
j2-
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
F,3
oe
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
cA
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTGGTGGAATCT
GGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTCACCAAGGCCT
GGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAAGAGCAACAGCTA
CGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAACACCCTGTACCTG
CAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCTGAGCCCCTTCGA
TTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGG
CCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTA
CTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGC
P
CCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCT
2
GCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGAC
.
CTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACA
.."
,
cc,
ACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCA
2
GGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATC
,
AGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACC
Z
AGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCC
.
GAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGA
CAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAG
AAGTCCCTGTCTCTGTCCCTGGGC
3 187
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGGACATGTACAGCG
AGACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTCTGCACCCGGGGCAAGTACTGC
00
ACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTGTGACAGTGTCTAGCGCTTCGAC
n
1-3
CAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCG
---
TGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCA
cp
n.)
GCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGAC
2
=
CTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCC
TGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTG
!II
ATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTG
C4
GTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
`'"
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCC
TGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCT
o
TGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGT
GGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTC
a'
TTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGA
GGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
a
4
188
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
oe
c:
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
Trispecific 4 1
189
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
P
GCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
0
,
VRC07_523 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
0
,
,¨ CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
.
,
CD3mid_QQ
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGAGCCCCA
2
IgG4
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAAC
,
,
,
FALA/409K
TGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCA
0
,
0
GCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACC
.
TACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGACCAAGGGCC
CCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCT
GTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCG
GCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAG
CAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAG
AGCTTCAACCGGGGCGAGTGT
2
190
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
00
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
n
1-3
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
4
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTGGTGGAATCT
2
=
GGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTCACCAAGGCCT
'a
GGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAAGAGCAACAGCTA
!II
CGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAACACCCTGTACCTG
CAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCTGAGCCCCTTCGA
TTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGG
CCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
o
ACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTA
ao
CTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGC
a'
CCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCT
GCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGAC
a
CTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACA
crc'e
ACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCA
GGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATC
AGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACC
AGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCC
GAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGA
CAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAG
AAGTCCCTGTCTCTGTCCCTGGGC
3
191
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
P
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGGACATGTACAGCG
2
AGACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTCTGCACCCGGGGCAAGTACTGC
i.i
ACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTGTGACAGTGTCTAGCGCTTCGAC
.."
,
CAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCG
2
TGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCA
,
,
GCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGAC
,
CTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCC
2
TGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTG
ATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTG
GTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCC
TGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCT
TGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGT
GGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTC
,t
TTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGA
n
1-3
GGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4
192
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
4
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
2
=
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
----
o
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
!II
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
c,.)
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
w
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
o
ACCGGGGCGAGTGT
n.)
o
Trispecific 6 1 193
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
n.)
o
GCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
VRC07_523 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
F,3
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
g
CD3mid_QQ
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
IgG4
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
FALA/409K
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
DKTHT linker
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
P
ACCGGGGCGAGTGT
0
,
2 194
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
0
,
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
.
,
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
2
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
,
,
,
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
0
,
0
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
.
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
00
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
n
1-3
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
---
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
4
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC 2
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
n.)
--4
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
c,.)
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
w
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
3 195
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
C4,
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGGACATGTACAGCG
AGACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTCTGCACCCGGGGCAAGTACTGC
ACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTGTGACAGTGTCTAGCGCTTCGAC
CAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCG
TGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCA
GCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGAC
CTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCC
TGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTG
ATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTG
GTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTG
GTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCC
TGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCT
TGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGT
GGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTC
TTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGA
GGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 196
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
Trispecific 8 1
197
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
1-3
VRC07_523 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD3mid_QQ
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
IgG1 NNAS/4
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
09K_DKTHT
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
linker
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 198
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
oe
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTG
CTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACAT
CTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCA
CACCTGTCCCCCTTGTCCTGCCCCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACAC
CCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCA
ATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GCCCTGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGC
CCCCAAGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATC
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCT
CATTCTTCCTGGTGTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATG
CACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
3 199
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGGACATGTACAGCG
AGACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTCTGCACCCGGGGCAAGTACTGC
ACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTGTGACAGTGTCTAGCGCTTCGAC
CAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGG
TCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCG
GCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGAC
CTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAA
ACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAA
GGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCA
AGTTCAACTGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAATGC
o
CTCCCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCA
ao
ACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACAC
a'
CCTGCCCCCATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGGTAAAAGGCTTCTATCCCAGCG
ACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGA
a
CGGCTCCTTCTTCCTCTATTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGT
g
GCTGCATGAGGCTCTGCACAGCCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGT
4 200
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
P
ACCGGGGCGAGTGT
0
,
Trispecific 9 1
201
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
0
,
GCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
.
,
-1" VRC07 523 F
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
2
R3-03 /
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
,
,
,
CD28sup x
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
0
,
0
CD3mid_ENL
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
.
Q IgG4
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
FALA/409K
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
DKTHT linker
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
00
ACCGGGGCGAGTGT
n
1-3
2 202
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
2
=
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
,---,
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
iµ.0
--4
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
o
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
ao
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
.. a'
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
a
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
.. oe
cA
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
P
3 203
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
2
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
i.i
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGCAGCTGAGCCAGG
.. .."
,
(..,
ACCCTGATGATCCGGATTGGGGCACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTC
2
TGCACCCGGGGCAAGTACTGCACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTG
,
,
TGACAGTGTCTAGCGCTTCGACCAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCT
,
ACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGAC
2
AAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCA
GCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGT
GGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCC
CCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAA
GATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAAC
AGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAA
GTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAG
,t
CCTCAAGTGTATACCCTGCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGG
n
1-3
CTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
GTGCTGGACAGCGACGGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGT
4
GTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4
204
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
.. ---':'
o
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
!II
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
c,.)
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
w
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
Trispecific 10 1 205
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
VRC07_523_F
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
R3-03
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
CD28sup x
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
CD3mid_ENL
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
IgG4
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
FALA/409K
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
DKTHT linker
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
cs, 2 206
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
0
0
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
o
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
?,
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
n4
o
3 207
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
F,3
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGCAGCTGAGCCAGG
CR
ACCCTGATGATCCGGATTGGGGCACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTC
TGCACCCGGGGCAAGTACTGCACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTG
TGACAGTGTCTAGCGCTTCGACCAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCT
ACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGAC
AAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCA
GCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGT
GGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCC
CCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAA
GATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAAC
P
AGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAA
2
GTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAG
CCTCAAGTGTATACCCTGCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGG
.."
,
---1
CTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCT
2
GTGCTGGACAGCGACGGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGT
,
,
GTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0"
,
4 208
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
2
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
00
ACCGGGGCGAGTGT
n
1-3
Trispecific 11 1 209
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
cp
n4
VRC07_523_F
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
o
n4
R3-03 /
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD28sup x
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
!II
CD3mid_ENL
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
Q
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
IgG1 NNAS_
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
DKTHT linker
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
o
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
n.)
o
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
a'
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
a
ACCGGGGCGAGTGT
oe
cA
2 210
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
P
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
2
GCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTG
.."
,
oc
CTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACAT
2
CTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCA
,
,
CACCTGTCCCCCTTGTCCTGCCCCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACAC
,
CCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCA
2
ATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GCCCTGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGC
CCCCAAGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATC
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCT
CATTCTTCCTGGTGTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATG
CACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
00
3 211
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
n
1-3
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG -
--
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGCAGCTGAGCCAGG 4
ACCCTGATGATCCGGATTGGGGCACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTC
2
TGCACCCGGGGCAAGTACTGCACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTG
TGACAGTGTCTAGCGCTTCGACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGC
n.)
--.1
ACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGAC
C4
CAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTC
`'"
CAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTT
GAGCCCAAATCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTT
CCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGA
GCCACGAAGACCCTGAGGTCAAGTTCAACTGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCG
GGAGGAGCAGTACAACAATGCCTCCCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGG
AGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCC
o e
CCGAGAACCACAGGTGTACACCCTGCCCCCATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGG
TAAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCAC
GCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGG
GGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCG
GGT
4 212
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
Trispecific 12 1 213
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
0
0
VRC07_523_F
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
R3-03
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
CD28sup x
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
CD3mid_ENL
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
IgG1 NNAS_
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
DKTHT linker
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
1-3
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 214
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
C
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
o
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
ao
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
a'
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
a
GCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAG
g
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTG
CTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACAT
CTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCA
CACCTGTCCCCCTTGTCCTGCCCCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACAC
CCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCA
ATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GCCCTGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGC
CCCCAAGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATC
P
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCT
2
CATTCTTCCTGGTGTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATG
.
CACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
.."
,
w
3 215
CAAGTGCGGCTGTCTCAGTCTGGCGGCCAGATGAAGAAACCCGGCGACAGCATGCGGATCAGCTGCAGAGCCAGCG
2
GCTACGAGTTCATCAACTGCCCCATCAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGATG
,
,
AAGCCCAGACACGGGGCCGTGTCCTACGCCAGACAGCTGCAGGGCAGAGTGACCATGACCCGGCAGCTGAGCCAGG
0"
,
ACCCTGATGATCCGGATTGGGGCACAGCCTTCCTGGAACTGCGGAGCCTGACCAGCGACGATACCGCCGTGTACTTC
2
TGCACCCGGGGCAAGTACTGCACCGCCAGAGACTACTACAACTGGGACTTCGAGCACTGGGGCCAGGGCACACCTG
TGACAGTGTCTAGCGCTTCGACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGC
ACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGAC
CAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTC
CAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTT
GAGCCCAAATCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTT
CCTCTTCCCCCCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGA
,t
GCCACGAAGACCCTGAGGTCAAGTTCAACTGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCG
n
1-3
GGAGGAGCAGTACAACAATGCCTCCCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGG -
---
AGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCC
4
CCGAGAACCACAGGTGTACACCCTGCCCCCATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGG
2
TAAAAGGCTTCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCAC
GCCTCCCGTGCTGGACTCCGACGGCTCCTTCTTCCTCTACTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGG
!II
GGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCG
c,.)
GGT
4
216
AGCCTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGT
ACGGCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGC
CGCCGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGC
GGCGACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
Trispecific 13 1 217
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 / CD28sup
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
CD3mid
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
IgG4
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGAGCCCCA
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAAC
TGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCA
GCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACC
TACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGACCAAGGGCC
CCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCT
GTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCG
GCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAG
CAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAG
0
0
AGCTTCAACCGGGGCGAGTGT
2 218
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTGGTGGAATCT
GGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTCACCAAGGCCT
GGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAAGAGCAACAGCTA
CGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAACACCCTGTACCTG
1-3
CAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCTGAGCCCCTTCGA
TTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGG
CCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTA
CTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGC
CCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTCCTGTG
GCTGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTG
CGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAAC
GCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGG
o
ACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAG
ao
CAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACCAG
a'
GTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGA
GAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGACA
a
AGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAA
g
GTCCCTGTCTCTGTCCCTGGGC
3 219
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGAC
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
P
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
2
GCCCAGCCCCTCCTGTGGCTGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGG
0
.
ACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGG
.."
,
w
t.)
CGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTG
2
ACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCA
,
,
TCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGAAGA
0"
,
GATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGA
2
GCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTACTCC
AAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACA
ACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 220
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
00
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
n
1-3
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
---
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA 4
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
o
n4
TTCAACCGGGGCGAGTGT
o
'a
Trispecific 14 1
221
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
n4
d
GCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
N6 / CD28sup
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD3mid
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGAGCCCCA
IgG4
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAAC
t=-0
TGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCA
GCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACC
TACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGACCAAGGGCC
CCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCT
GTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCG
GCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAG
CAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAG
AGCTTCAACCGGGGCGAGTGT
2 222
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTGGTGGAATCT
GGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTCACCAAGGCCT
GGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAAGAGCAACAGCTA
CGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAACACCCTGTACCTG
CAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCTGAGCCCCTTCGA
TTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGG
CCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTA
CTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGC
CCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCT
GCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGAC
CTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACA
ACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCA
GGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATC
AGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACC
AGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCC
1-3
GAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGA
CAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAG
AAGTCCCTGTCTCTGTCCCTGGGC
3 223
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 224
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 15 1 225
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 / CD28sup
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
x CD3mid_QQ
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
IgG4
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGGCCAGCCCAAGGCCGCCCCCGACATCCAGATGACCCAGAGCCCCA
FALA/409K
GCAGCCTGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAAC
TGGTATCAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCA
GCAGATTTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACC
1-3
TACTACTGCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGACCAAGGGCC
CCAGCCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCT
GTCGTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCG
tµ.0
GCAACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAG
CAAGGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAG
AGCTTCAACCGGGGCGAGTGT
2 226
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
o
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
ao
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
a'
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCAGCCAGGTGCAGCTGGTGGAATCT
GGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCTTCACCAAGGCCT
a
GGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAAGAGCAACAGCTA
cr,c4
CGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAACACCCTGTACCTG
CAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCTGAGCCCCTTCGA
TTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTCGGACCGCCAGCACAAAGGGCCCATCGGTGTTCCCTCTGG
CCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTG
ACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTA
CTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGC
CCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCT
GCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGAC
CTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACA
P
ACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCA
2
GGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATC
.
AGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCAGGAAGAGATGACCAAGAACC
.."
,
w
(..,
AGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCC
0"
GAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGGTGTCCAAGCTGACCGTGGA
,
,
CAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAG
0"
,
AAGTCCCTGTCTCTGTCCCTGGGC
2
3 227
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
,t
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
2
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
c6
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
2
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
!II
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
c,.)
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
w
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 228
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 16 1 229
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 / CD28sup
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
x CD3mid_QQ
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
IgG4
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
FALA/409K
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
DKTHT linker
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
cs,
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 230
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
1-3
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
C
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
oe
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 231
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 232
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 17 1 233
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACAACAACGCCAACACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
0
N6 / CD28sup
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
t=-0
x CD3mid
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
IgG4
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
FALA/409K
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
DKTHT linker
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCAG
TCAGCCCAAGGCTGCCCCCTCGGTCACTCTGTTCCCGCCCTCGAGTGAGGAGCTTCAAGCCAACAAGGCCACACTGG
TGTGTCTCATAAGTGACTTCTACCCGGGAGCCGTGACAGTGGCCTGGAAGGCAGATAGCAGCCCCGTCAAGGCGGG
AGTGGAGACCACCACACCCTCCAAACAAAGCAACAACAAGTACGCGGCCAGCAGCTACCTGAGCCTGACGCCTGAG
CAGTGGAAGTCCCACAGAAGCTACAGCTGCCAGGTCACGCATGAAGGGAGCACCGTGGAGAAGACAGTGGCCCCTA
CAGAATGT
2 234
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
0
0
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 235
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
o
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
n.)
o
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
n.)
o
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
a
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
g
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
P
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
0
,
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
,
. 4 236
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
.
,
w
s:)
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
2
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
,
,
,
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
0
,
0
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
.
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 19 1 237
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACCAGAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 / CD28sup
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
00
x CD3mid_QQ
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
n
1-3
IgG1 NNAS_
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
---
DKTHT linker
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
4
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
2
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
!II
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
c,.)
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
w
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
o
ACCGGGGCGAGTGT
n.)
o
2
238
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
Ce,
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
j2-
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
F,3
oe
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
cA
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTG
CTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACAT
P
CTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCA
2
CACCTGTCCCCCTTGTCCTGCCCCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACAC
CCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCA
.."
,
ATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCG
2
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
,
,
GCCCTGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGC
0"
,
CCCCAAGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATC
2
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCT
CATTCTTCCTGGTGTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATG
CACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
3 239
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
00
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
n
1-3
CATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGAC
---
TACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCT
4
ACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCT
o
n.)
GCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACAC
ATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCC
!II
TCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAAC
C4
TGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAATGCCTCCCGTG
`'"
TGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCC
CTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGCCCC
CATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGGTAAAAGGCTTCTATCCCAGCGACATCGCC
GTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCT
TCTTCCTCTACTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCAT
GAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGT
4 oe
240
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 21 1 241
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGACAACGCCCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 / CD28sup
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD3mid_DNA
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
IgG4
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
FALA/409K
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
DKTHT linker
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCAG
TCAGCCCAAGGCTGCCCCCTCGGTCACTCTGTTCCCGCCCTCGAGTGAGGAGCTTCAAGCCAACAAGGCCACACTGG
TGTGTCTCATAAGTGACTTCTACCCGGGAGCCGTGACAGTGGCCTGGAAGGCAGATAGCAGCCCCGTCAAGGCGGG
AGTGGAGACCACCACACCCTCCAAACAAAGCAACAACAAGTACGCGGCCAGCAGCTACCTGAGCCTGACGCCTGAG
CAGTGGAAGTCCCACAGAAGCTACAGCTGCCAGGTCACGCATGAAGGGAGCACCGTGGAGAAGACAGTGGCCCCTA
CAGAATGT
2 242
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
1-3
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
o
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
ao
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
a'
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
t'-4
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
a
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
g
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 243
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
P
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
2
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
.."
,
t.)
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
2
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
,
,
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
0"
,
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
2
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
,t
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
n
1-3
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 244
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
4
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
o
n.o
o
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
,---,
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
!II
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
C4
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
`'"
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
o
TTCAACCGGGGCGAGTGT
n.)
o
Trispecific 22 1 245
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
n.)
o
GCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 / CD28sup
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
F,3
x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
g
CD3mid_ENL
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
Q IgG4
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
FALA/409K
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
DKTHT linker
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
P
ACCGGGGCGAGTGT
0
,
2 246
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
0
,
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
.
,
w
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
2
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
,
,
,
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
0
,
0
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
.
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
00
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
n
1-3
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
----
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
4
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC 2
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
n.)
--4
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
c,.)
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
w
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
3 247
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 248
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 23 1 249
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGCGTACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
1-3
N6 / CD28sup
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
CD3mid_ENL
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
IgG4
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
FALA/409K
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
DKTHT linker
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
o
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
a'
ACCGGGGCGAGTGT
2
250
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
F,3
oe
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
cA
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
.. P
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
2
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
.."
,
(..,
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
2
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
,
,
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
,
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
2
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3
251
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
00
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
n
1-3
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
---
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
4
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
2
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
!II
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
C4
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
`'"
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG o
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
ao
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
iµ.0
o
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
a
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
oe
c:
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 252
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
P
TTCAACCGGGGCGAGTGT
0
,
Trispecific 24 1 253
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
0
,
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
.
,
c' N6 / CD28sup
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
2
x
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
,
,
,
CD3mid_ENL
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
0
,
0
F IgG4
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
.
FALA/409K
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
DKTHT linker
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
00
ACCGGGGCGAGTGT
n
1-3
2 254
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
2
=
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
,---,
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
iµ.0
--4
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
wl-,
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
o
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
ao
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
a'
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
a
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
oe
cA
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
P
3 255
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
2
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
.."
,
---1
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
2
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
,
,
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTGGGCTGCCTCGTGAAGGAC
,
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
2
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
00
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
n
1-3
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC -
---
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
4
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4
256
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
n.o
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
`'"
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 25 1 257
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
oe
N6 rw52 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD3mid_ENL
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
IgG4
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
FALA/409K
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
DKTHT linker
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 258
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
0
0
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
tµ.0
3 259
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCACCAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
oe
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGAC
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
0
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
0
0
4 260
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCTCCGAAGAGGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
1-3
Trispecific 26 1 261
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
N6 rw52 /
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
CD28sup x
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
CD3mid_ENL
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
IgG4
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
FALA/409K
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
DKTHT linker
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
o
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
n.)
o
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
a'
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
a
ACCGGGGCGAGTGT
oe
cA
2 262
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
P
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
.
L.
,
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
L.
,
.
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
.
,
(..,
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
,
,
,
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
.
,
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
.
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
00
3 263
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
n
1-3
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
---
AAGCCCCAGTATGGCGCCACCAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
cp
n.)
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
o
n.)
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
CATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAGGAC
"II
TACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTGCTC
C4
CAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACACCTG
`'"
TAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCTCCTT
GCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATCAGC
CGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGTGGA
CGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCCGTG
CTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCAGCT
CCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATACCCTGCCCCCTTGCCAGGA
AGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAATGGG
AGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCTGTAC
TCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCCCTGC
ACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 264
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCTCCGAAGAGGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 27 1 265
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
N6 rw52 /
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
0
0
CD28sup x
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
CD3mid_ENL
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
IgG1 NNAS_
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
DKTHT linker
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 266
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA o
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ao
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
a'
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAG
a
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTG
g
CTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACAT
CTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCA
CACCTGTCCCCCTTGTCCTGCCCCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACAC
CCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCA
ATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GCCCTGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGC
CCCCAAGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATC
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCT
P
CATTCTTCCTGGTGTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATG
2
CACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
0
. 3 267
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
.."
,
t.)
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
2
AAGCCCCAGTATGGCGCCACCAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGGACGTGTACCGCG
,
,
AGATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTACTGCGCCAGAGACAGAAGCTA
0"
,
CGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGTCTGCCGCCTCTACAAAGGGCC
2
CATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGAC
TACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCT
ACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCT
GCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGACAAAACTCACAC
ATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCCCCAAAACCCAAGGACACCC
TCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAGACCCTGAGGTCAAGTTCAAC
TGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAACAATGCCTCCCGTG
,t
TGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCC
n
,-i
CTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGTGTACACCCTGCCCC
CATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGGTAAAAGGCTTCTATCCCAGCGACATCGCC
4
GTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGGACTCCGACGGCTCCT
2
TCTTCCTCTACTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCAT
GAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGT
n.o
--.1
4 268
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCTCCGAAGAGGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
oe
Trispecific 28 1 269
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 FR3-03 /
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD3mid_ENL
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
IgG4
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
FALA/409K
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
DKTHT linker
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 270
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
0
0
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
,4
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
`'"
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
0
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
n.)
o
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
a'
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
a
3 271
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
oe
cA
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGCAGCTGAGCCAGG
ACCCTGATGATCCGGATTGGGGCATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTAC
TGCGCCAGAGACAGAAGCTACGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGT
CTGCCGCCTCTACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCC
NTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGT
GCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAA
GTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCC
P
CAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAG
2
GTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACA
.
GCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGT
.."
,
(..,
-i.
GTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTG
2
TATACCCTGCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCC
,
,
CAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGAC
,
AGCGACGGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTG
2
CTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 272
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
00
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA n
1-3
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
---
TTCAACCGGGGCGAGTGT
cp
n.)
Trispecific 29 1 273
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
o
n.)
o
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
,---,
N6 FR3-03 /
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
!II
CD28sup x
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
wl-,
CD3mid_ENL
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
IgG4
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
FALA/409K
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
DKTHT linker
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 274
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
0
0
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
1-3
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 275
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGCAGCTGAGCCAGG
ACCCTGATGATCCGGATTGGGGCATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTAC
TGCGCCAGAGACAGAAGCTACGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGT
CTGCCGCCTCTACAAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCC
NTGGGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGT
GCACACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCC
TGGGCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAA
GTACGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCC
CAAGGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAG
GTGCAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACA
GCACCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGT
oe
GTCCAACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTG
TATACCCTGCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCC
CAGCGACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGAC
AGCGACGGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTG
CTCCGTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 276
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
cs,
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 30 1 277
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
0
0
GCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
N6 FR3-03/
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
CD28sup x
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD3mid ENL
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
QIgG1 NNAS
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
DKTHT
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
linker
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
1-3
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 278
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
o
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
ao
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
a'
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
a
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
g
GCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTG
CTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACAT
CTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCA
CACCTGTCCCCCTTGTCCTGCCCCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACAC
CCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCA
ATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCG
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
GCCCTGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGC
P
CCCCAAGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATC
2
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCT
.
CATTCTTCCTGGTGTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATG
.."
,
(..,
---1
CACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
3 279
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
,
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
,
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGCAGCTGAGCCAGG
2
ACCCTGATGATCCGGATTGGGGCATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTAC
TGCGCCAGAGACAGAAGCTACGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGT
CTGCCGCCTCTACAAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCC
CTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGT
GCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTT
GGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAA
TCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCC
,t
CCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAG
n
ACCCTGAGGTCAAGTTCAACTGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCA
..-1
GTACAACAATGCCTCCCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGT
4
GCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACC
2
ACAGGTGTACACCCTGCCCCCATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGGTAAAAGGCT
TCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGT
!II
GCTGGACTCCGACGGCTCCTTCTTCCTCTACTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCT
TCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGT
4
280
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
Trispecific 31 1 281
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
N6 FR3-03 /
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
CD28sup x
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
CD3mid_ENL
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
IgG1 NNAS_
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
DKTHT linker
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
oc
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
0
0
ACCGGGGCGAGTGT
2 282
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCCAGCGTGTTCCCTCTGGCCCCTAGCAGCAAGAGCACATCTGGCGGAACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAATTCTGGCGCCCTGACCAGCGGCGTGCACACCTTTCCAGCTGTG
CTGCAGTCCAGCGGCCTGTACAGCCTGAGCAGCGTCGTGACAGTGCCCAGCAGCTCTCTGGGCACCCAGACCTACAT
CTGCAACGTGAACCACAAGCCCAGCAACACCAAGGTGGACAAGAAGGTGGAACCCAAGAGCTGCGACAAGACCCA
c,4
CACCTGTCCCCCTTGTCCTGCCCCCGAACTGCTGGGAGGCCCTTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACAC
CCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCACGAGGACCCTGAAGTGAAGTTCA
ATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCAAGAGAGGAACAGTACAACAATGCCTCCCG o
GGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAG
ao
GCCCTGCCTGCCCCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAACCCCAGGTGTGCACACTGC
n4
o
CCCCAAGCAGGGACGAGCTGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCTCCGATATC
GCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCT a
CATTCTTCCTGGTGTCCAAGCTGACAGTGGACAAGTCCCGGTGGCAGCAGGGCAACGTGTTCAGCTGCTCCGTGATG
g
CACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGAGCCTGAGCCCCGGC
3
283
AGAGCCCACCTGGTGCAGTCTGGCACCGCCATGAAGAAACCAGGCGCCTCTGTGCGGGTGTCCTGTCAGACAAGCG
GCTACACCTTCACCGCCCACATCCTGTTCTGGTTCCGGCAGGCCCCTGGCAGAGGACTGGAATGGGTGGGATGGATC
AAGCCCCAGTATGGCGCCGTGAACTTCGGCGGAGGCTTCCGGGATAGAGTGACCCTGACCCGGCAGCTGAGCCAGG
ACCCTGATGATCCGGATTGGGGCATCGCCTACATGGACATCCGGGGCCTGAAGCCCGATGACACCGCCGTGTACTAC
TGCGCCAGAGACAGAAGCTACGGCGACAGCAGCTGGGCTCTGGATGCTTGGGGCCAGGGCACAACCGTGGTGGTGT
CTGCCGCCTCTACAAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCC
CTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGT
GCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTT
P
GGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAA
2
TCTTGTGACAAAACTCACACATGCCCACCGTGCCCAGCACCTGAACTCCTGGGGGGACCGTCAGTCTTCCTCTTCCCC
0
.
CCAAAACCCAAGGACACCCTCATGATCTCCCGGACCCCTGAGGTCACATGCGTGGTGGTGGACGTGAGCCACGAAG
.."
,
(..,
s:)
ACCCTGAGGTCAAGTTCAACTGGTATGTTGACGGCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCA
2
GTACAACAATGCCTCCCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTGAATGGCAAGGAGTACAAGT
,
,
GCAAGGTCTCCAACAAAGCCCTCCCAGCCCCCATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACC
0"
,
ACAGGTGTACACCCTGCCCCCATGCCGGGATGAGCTGACCAAGAATCAAGTCAGCCTGTGGTGCCTGGTAAAAGGCT
2
TCTATCCCAGCGACATCGCCGTGGAGTGGGAGAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGT
GCTGGACTCCGACGGCTCCTTCTTCCTCTACTCAAAACTCACCGTGGACAAGAGCAGGTGGCAGCAGGGGAACGTCT
TCTCATGCTCCGTGATGCATGAGGCTCTGCACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGT
4
284
TACATCCACGTGACCCAGAGCCCCAGCAGCCTGTCCGTGTCCATCGGCGACAGAGTGACCATCAACTGCCAGACCTC
TCAGGGCGTGGGCAGCGACCTGCACTGGTATCAGCACAAGCCTGGCAGAGCCCCCAAGCTGCTGATCCACCACACA
AGCAGCGTGGAAGATGGCGTGCCCAGCAGATTTTCCGGCAGCGGCTTCCACACCAGCTTCAACCTGACCATCAGCGA
TCTGCAGGCCGACGACATTGCCACCTACTATTGTCAGGTGCTGCAGTTCTTCGGCAGAGGCAGCAGACTGCACATCA
00
AGCGTACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTC
n
1-3
GTGTGCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCA
---
ACAGCCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAA 4
GGCCGACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGC
TTCAACCGGGGCGAGTGT
o
'a
Trispecific 34 1
285
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
n4
d
GCCAGAGCCTGGTGCACGAGAACCTGCAGACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTC
CCTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCA
VRC01.23 /
CCCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACC
CD28 sup x
TTTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCC
CD3mid_ENL
TGTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTAT
IgG4
CAGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGAT
FALA/409K
TTTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
DKTHT linker
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
oe
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 286
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
1-3
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
3 287
CAGGTGCAGCTGGTGCAGTCTGGCGGCCAGATGAAGAAACCCGGCGAGAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCGACTGCACCCTGAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGCTG
AAGCCTAGATGGGGAGCCGTGAACTACGCCAGACCTCTGCAGGGCAGAGTGACCATGACCCGGCAGCTGAGCCAGG
ACCCTGATGATCCGGATTGGGGCACCGCCTTCCTGGAACTGCGGAGCCTGACCGTGGATGATACCGCCGTGTACTTC
`'"
TGCACCCGGGGCAAGAACTGCGACTACAACTGGGACTTCGAGCACTGGGGCAGAGGCACCCCTGTGATCGTGTCAA
GCGCGTCGACCAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTG
GGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGNTNTGACAAGCGGCGTGCA
CACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGG
GCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTA
CGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAA
GGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTG
oe
CAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCA
CCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCC
AACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATA
CCCTGCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGC
GACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCG
ACGGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCC
GTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
4 288
CTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGTACG
GCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGCCGC
CGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGCGGC
GACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCGTAC
GGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGTGCCT
GCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAGCCAG
GAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCCGACT
ACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCAACCG
0
0
GGGCGAGTGT
Trispecific 35 1 289
GACATCGTGATGACCCAGACCCCCCTGAGCCTGAGCGTGACACCTGGACAGCCTGCCAGCATCAGCTGCAAGAGCA
GCCAGAGCCTGGTGCACGAGAACCTGTTCACCTACCTGAGCTGGTATCTGCAGAAGCCCGGCCAGAGCCCCCAGTCC
VRC01.23 /
CTGATCTACAAGGTGTCCAACAGATTCAGCGGCGTGCCCGACAGATTCTCCGGCAGCGGCTCTGGCACCGACTTCAC
CD28sup x
CCTGAAGATCAGCCGGGTGGAAGCCGAGGACGTGGGCGTGTACTATTGTGGCCAGGGCACCCAGTACCCCTTCACCT
CD3mid_ENL
TTGGCAGCGGCACCAAGGTGGAAATCAAGGACAAAACCCATACCGACATCCAGATGACCCAGAGCCCCAGCAGCCT
IgG4
GTCTGCCAGCGTGGGCGACAGAGTGACCATCACCTGTCAGGCCAGCCAGAACATCTACGTGTGGCTGAACTGGTATC
FALA/409K
AGCAGAAGCCCGGCAAGGCCCCCAAGCTGCTGATCTACAAGGCCAGCAACCTGCACACCGGCGTGCCCAGCAGATT
DKTHT linker
TTCTGGCAGCGGCTCCGGCACCGACTTCACCCTGACAATCAGCTCCCTGCAGCCCGAGGACATTGCCACCTACTACT
1-3
GCCAGCAGGGCCAGACCTACCCCTACACCTTTGGCCAGGGCACCAAGCTGGAAATCAAGGATAAGACCCACACCCG
TACGGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGT
GCCTGCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAG
CCAGGAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCC
GACTACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCA
ACCGGGGCGAGTGT
2 290
CAGGTGCAGCTGGTGCAGTCTGGCGCCGAGGTCGTGAAACCTGGCGCCTCTGTGAAGGTGTCCTGCAAGGCCAGCGG
CTACACCTTTACCAGCTACTACATCCACTGGGTGCGCCAGGCCCCTGGACAGGGACTGGAATGGATCGGCAGCATCT
o
ACCCCGGCAACGTGAACACCAACTACGCCCAGAAGTTCCAGGGCAGAGCCACCCTGACCGTGGACACCAGCATCAG
ao
CACCGCCTACATGGAACTGAGCCGGCTGAGAAGCGACGACACCGCCGTGTACTACTGCACCCGGTCCCACTACGGCC
a'
TGGATTGGAACTTCGACGTGTGGGGCAAGGGCACCACCGTGACAGTGTCTAGCGACAAAACCCATACCCAGGTGCA
GCTGGTGGAATCTGGCGGCGGAGTGGTGCAGCCTGGCAGAAGCCTGAGACTGAGCTGTGCCGCCAGCGGCTTCACCT
a
TCACCAAGGCCTGGATGCACTGGGTGCGCCAGGCCCCTGGAAAGCAGCTGGAATGGGTGGCCCAGATCAAGGACAA
g
GAGCAACAGCTACGCCACCTACTACGCCGACAGCGTGAAGGGCCGGTTCACCATCAGCCGGGACGACAGCAAGAAC
ACCCTGTACCTGCAGATGAACAGCCTGCGGGCCGAGGACACCGCCGTGTACTACTGTCGGGGCGTGTACTATGCCCT
GAGCCCCTTCGATTACTGGGGCCAGGGAACCCTCGTGACCGTGTCTAGTGATAAGACCCACACCGCCAGCACAAAGG
GCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCCTGGGCTGCCTCGTGAAG
GACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGCTCTGACAAGCGGCGTGCACACCTTTCCAGCCGTG
CTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGGGCACCAAGACCTACAC
CTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTACGGCCCTCCCTGCCCT
CCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAAGGACACCCTGATGATC
AGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTGCAGTTCAATTGGTACGT
P
GGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCACCTACCGGGTGGTGTCC
2
GTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCCAACAAGGGCCTGCCCA
GCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTGTACCCTGCCCCCTAGCCA
.."
,
t.)
GGAAGAGATGACCAAGAACCAGGTGTCCCTGAGCTGTGCCGTGAAAGGCTTCTACCCCAGCGACATTGCCGTGGAA
TGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCGACGGCTCATTCTTCCT
,
,
GGTGTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCCGTGATGCACGAGGCC
,
CTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
2
3 291
CAGGTGCAGCTGGTGCAGTCTGGCGGCCAGATGAAGAAACCCGGCGAGAGCATGCGGATCAGCTGCAGAGCCAGCG
GCTACGAGTTCATCGACTGCACCCTGAACTGGATCAGACTGGCCCCTGGCAAGCGGCCTGAGTGGATGGGATGGCTG
AAGCCTAGATGGGGAGCCGTGAACTACGCCAGACCTCTGCAGGGCAGAGTGACCATGACCCGGCAGCTGAGCCAGG
ACCCTGATGATCCGGATTGGGGCACCGCCTTCCTGGAACTGCGGAGCCTGACCGTGGATGATACCGCCGTGTACTTC
TGCACCCGGGGCAAGAACTGCGACTACAACTGGGACTTCGAGCACTGGGGCAGAGGCACCCCTGTGATCGTGTCAA
GCGCGTCGACCAAGGGCCCATCGGTGTTCCCTCTGGCCCCTTGCAGCAGAAGCACCAGCGAATCTACAGCCGCCNTG
GGCTGCCTCGTGAAGGACTACTTTCCCGAGCCCGTGACCGTGTCCTGGAACTCTGGCGNTNTGACAAGCGGCGTGCA
,t
CACCTTTCCAGCCGTGCTCCAGAGCAGCGGCCTGTACTCTCTGAGCAGCGTCGTGACAGTGCCCAGCAGCAGCCTGG
n
1-3
GCACCAAGACCTACACCTGTAACGTGGACCACAAGCCCAGCAACACCAAGGTGGACAAGCGGGTGGAATCTAAGTA --
-
CGGCCCTCCCTGCCCTCCTTGCCCAGCCCCTGAAGCTGCCGGCGGACCCTCCGTGTTCCTGTTCCCCCCAAAGCCCAA
4
GGACACCCTGATGATCAGCCGGACCCCCGAAGTGACCTGCGTGGTGGTGGATGTGTCCCAGGAAGATCCCGAGGTG
CAGTTCAATTGGTACGTGGACGGCGTGGAAGTGCACAACGCCAAGACCAAGCCCAGAGAGGAACAGTTCAACAGCA
CCTACCGGGTGGTGTCCGTGCTGACCGTGCTGCACCAGGACTGGCTGAACGGCAAAGAGTACAAGTGCAAGGTGTCC
t.1
AACAAGGGCCTGCCCAGCTCCATCGAGAAAACCATCAGCAAGGCCAAGGGCCAGCCCCGCGAGCCTCAAGTGTATA
C4,4
CCCTGCCCCCTTGCCAGGAAGAGATGACCAAGAACCAGGTGTCCCTGTGGTGTCTCGTGAAAGGCTTCTACCCCAGC
`'"
GACATTGCCGTGGAATGGGAGAGCAACGGCCAGCCCGAGAACAACTACAAGACCACCCCCCCTGTGCTGGACAGCG
ACGGCTCATTCTTCCTGTACTCCAAGCTGACCGTGGACAAGAGCCGGTGGCAGGAAGGCAACGTGTTCAGCTGCTCC
o
GTGATGCACGAGGCCCTGCACAACCACTACACCCAGAAGTCCCTGTCTCTGTCCCTGGGC
n.)
o
4 292
CTGACACAGAGCCCTGGCACCCTGTCACTGAGCCCAGGCGAGACAGCCATCATCAGCTGCCGGACAAGCCAGTACG
n.)
o
GCAGCCTGGCCTGGTATCAGCAGAGGCCTGGACAGGCCCCCAGACTCGTGATCTACAGCGGCAGCACAAGAGCCGC
j2-
CGGAATCCCCGATAGATTCAGCGGCTCCAGATGGGGACCCGACTACAACCTGACCATCAGCAACCTGGAAAGCGGC
F,3
GACTTCGGCGTGTACTACTGCCAGCAGTACGAGTTCTTCGGCCAGGGCACCAAGGTGCAGGTGGACATCAAGCGTAC
g
GGTGGCCGCTCCCAGCGTGTTCATCTTCCCACCTAGCGACGAGCAGCTGAAGTCCGGCACAGCCTCTGTCGTGTGCCT
GCTGAACAACTTCTACCCCCGCGAGGCCAAAGTGCAGTGGAAGGTGGACAACGCCCTGCAGAGCGGCAACAGCCAG
GAAAGCGTGACCGAGCAGGACAGCAAGGACTCCACCTACAGCCTGAGCAGCACCCTGACACTGAGCAAGGCCGACT
ACGAGAAGCACAAGGTGTACGCCTGCGAAGTGACCCACCAGGGCCTGTCTAGCCCCGTGACCAAGAGCTTCAACCG
GGGCGAGTGT
P
,,0
µ,"
.."
..,
,,..)
,,0
'7
'8
,
2.
00
n
,-i
cp
w
=
w
=
-a
w
-.,
,,,