Note: Descriptions are shown in the official language in which they were submitted.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
PHAGE AND TRANSDUCTION PARTICLES
TECHNICAL FIELD
[0001] The invention relates to the production of phage and transduction
particles comprising phage
proteins using DNAs (eg, plasmids and helper phage, or plasmids with
chromosomally integrated
helper phage genes), as well as the phage, helper phage, kits, compositions
and methods involving
these. The particles are particularly useful for delivering toxic payloads
into target bacteria for
antibacterial action. Embodiments enable production of highly pure
compositions of such particles
for medical or environmental use and for containment of the particles, which
may be useful for
containing antibacterial action, controlling dosing or reducing the risk of
acquisition of undesirable
foreign genes.
BACKGROUND
[0002] The use of helper phage to package phagemid DNA into phage virus
particles is known. An
example is the M13K07 helper phage, a derivative of M13, used in E coli host
cells. Other examples
are R408 and CM13.
SUMMARY OF THE INVENTION
[0003] The invention relates to tbe production of phage and transduction
particles and provides:-
[0004] In a First Configuration
A kit comprising
a) A first DNA; and
b) One or more second DNAs;
Wherein
(i) the DNAs together comprise all phage structural protein genes required
to produce a
packaged phage particle comprising a copy of the first DNA;
(ii) the first DNA comprises none or at least one, but not all, of the
genes; and wherein the one or
more second DNAs comprise the remainder of the genes;
(iii) the first DNA comprises a phage packaging signal for producing the
packaged phage particle;
and
(iv) the second DNA is devoid of a nucleotide sequence (eg, a packaging
signal) required for
packaging the second DNA into phage particles;
wherein the DNAs are operable when co-existing in a host bacterium for
producing packaged phage
that comprise the first DNA, wherein the phage require the second DNA for
replicaton thereof to
produce further phage particles.
1
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
There is also provided
A method of producing phage, the method comprising expressing in a cell
comprising the DNAs the
phage protein genes, wherein packaged phage are produced that comprise the
first DNA, wherein the
phage require the second DNA for replicaton thereof to produce further phage
particles.
[0005] In a Seond Configuration
A population of helper phage, wherein the helper phage are capable of
packaging first phage, wherein
the first phage are different from the helper phage and the helper phage are
incapable of self-
replication.
[0006] In a third Configuration
A composition comprising a population of first phage, wherein the first phage
require helper phage
according to the First Configuration for replication; and wherein less than
[20%] of total phage
comprised by the composition are such helper phage.
[0007] In a Fourth Configuration
[0008] A method of producing first phage, wherein the first phage require
helper phage to replicate,
the method comprising
(a) Providing DNA comprising a packaging signal;
(b) Introducing the DNA into a host bacterial cell;
(c) Wherein the host bacterial cell comprises helper phage or wherein
helper phage are
introduced into the bacterial cell simultaneously or sequentially with step
(b);
(d) Wherein the helper phage are according to the invention;
(e) Causing or allowing the helper phage to produce phage proteins, wherein
the
packaging signal is recognised in the host cell, whereby first phage are
produced using the proteins,
the first phage packaging the DNA;
(f) Wherein helper phage replication in the host cell is inhibited or
reduced, thereby
limiting the availability of helper phage;
(g) Optionally lysing the host cell and obtaining the first phage;
(h) Thereby producing a composition comprising first phage which require
the helper
phage for replication, wherein propagation of first phage is prevented or
reduced by the limitation of
helper phage availability.
[0009] In a Fifth Configuration
A phage production system, for producing phage (eg, the first phage of any
preceding claim)
comprising a nucleotide sequence of interest (NSI-phage), the system
comprising components (i) to
(iii):-
(i) A first DNA;
2
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
(ii) A second DNA; and
(iii) a NSI-phage production factor (NPF) or an expressible nucleotide
sequence that encodes a
NPF;
Wherein
a) The first DNA encodes a helper phage (eg, said first helper phage
recited in any preceding
claim);
b) The second DNA comprises the nucleotide sequence of interest (NSI);
c) When the system is comprised by a bacterial host cell, helper phage
proteins are expressed
from the first DNA to form phage that package the second DNA in the presence
of the NPF, thereby
producing NSI-phage;
d) The system is devoid of a helper phage production factor (HPF) that is
required for forming
phage that package the first DNA, or is devoid of an expressible nucleotide
sequence that encodes a
functional HPF; or the system comprises a nucleotide sequence that comprises
or encodes a functional
HPF, the system further comprising means for targeted inactivation in the host
cell of the HPF
sequence to eliminate or minimise production of helper phage comprising the
first DNA; and
Whereby the system is capable of producing a product comprising a population
of NSI-phage,
wherein each NSI-phage requires a said helper phage for propagation, wherein
the NSI-phage in the
product are not mixed with helper phage or less than [20%] of total phage
comprised by the product
are said helper phage.
The invention also provides:_
[0010] A composition for use in antibacterial treatment of bacteria, the
composition comprising an
engineered mobile genetic element (MGE) that is capable of being mobilised in
a first bacterial host
cell of a first species or strain, the cell comprising a first phage genome,
wherein in the cell the MGE
is mobilised using proteins encoded by the phage and replication of first is
inhibited, wherein the
MGE encodes an antibacterial agent or encodes a component of such an agent.
[0011] A nucleic acid vector comprising the MGE integrated therein, wherein
the vector is capable of
transferring the MGE or a copy thereof into a host bacterial cell.
[0012] A non-self replicative transduction particle comprising said MGE or
vector of the invention.
[0013] A composition comprising a plurality of transduction particles, wherein
each particle
comprises a MGE or vector according to the invention, wherein the transduction
particles are capable
3
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
of transferring the MGEs, or nucleic acid encoding the agent or component, or
copies thereof into
target bacterial cells, wherein
(i)target cells are killed by the antibacterial agent;
(ii) growth or proliferation of target cells is reduced; or
(iii)target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
[0014] A composition comprising a plurality of non-self replicative
transduction particles, wherein
each particle comprises a MGE or plasmid according to the invention, wherein
the transduction
particles are capable of transferring the MGEs, or nucleic acid encoding the
agent or component, or
copies thereof into target bacterial cells, wherein the agent is a CRISPR/Cas
system and the
component comprises a nucleic acid encoding a crRNA or a guide RNA that is
operable with a Cas in
a target bacterial cell to guide the Cas to a target nucleic acid sequence of
the cell to modify the
sequence, whereby
(i)target cells are killed by the antibacterial agent;
(ii)growth or proliferation of target cells is reduced; or
(iii)target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
[0015] A method of producing a plurality of transduction particles, the method
comprising
combining the composition of the invention with host bacterial cells of said
first species, wherein the
cells comprise the first phage, allowing a plurality of said MGEs to be
introduced into host cells and
culturing the host cells under conditions in which first phage-encoded
proteins are expressed and
MGE copies are packaged by first phage proteins to produce a plurality of
transduction particles, and
optionally separating the transduction particles from cells and obtaining a
plurality of transduction
particles separated from cells.
[0016] A bacterial host cell comprising a first phage and a MGE, vector or
particle of the invention,
wherein the agent is toxic to cells of the same species as the host cell, and
wherein the host cell has
been engineered so that the agent is not toxic to the host cell.
[0017] A bacterial host cell comprising a first phage, wherein the cell is
comprised by a kit, the kit
further comprising a composition of the invention, wherein the agent is toxic
to cells of the same
species as the host cell, and wherein the host cell has been engineered so
that the agent is not toxic to
the host cell.
4
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[0018] A bacterial host cell comprising a first phage and a MGE, vector or
particle of the invention,
wherein the agent is not toxic to the host cell, but the agent is toxic to
second cells of a species or
strain that is different from the species or strain of the host cell, wherein
the MGE is mobilizable in
transduction particles producible by the host cell that are capable of
transferring the MGE or a copy
thereof into a said second cell, whereby the second cell is exposed to the
antibacterial agent.
[0019] A bacterial host cell comprising a first phage, wherein the cell is
comprised by a kit, the kit
further comprising a composition of the invention, wherein the agent is not
toxic to the host cell, but
the agent is toxic to second cells of a species or strain that is different
from the species or strain of the
host cell, wherein the MGE is mobilizable in transduction particles producible
by the host cell that are
capable of transferring the MGE or a copy thereof into a said second cell,
whereby the second cell is
exposed to the antibacterial agent.
[0020] A bacterial host cell comprising a MGE, vector or particle of the
invention and nucleic acid
under the control of one or more inducible promoters, wherein the nucleic acid
encodes all structural
proteins necessary to produce a transduction particle that packages a copy of
the MGE or plasmid,
wherein the agent is not toxic to the host cell, but the agent is toxic to
second cells of a species or
strain that is different from the species or strain of the host cell, wherein
the MGE is mobilizable in
transduction particles producible by the host cell that are capable of
transferring the MGE or a copy
thereof into a said second cell, whereby the second cell is exposed to the
antibacterial agent.
[0021] A plasmid comprising
(a) A nucleotide sequence encoding an antibacterial agent or component
thereof for expression in
target bacterial cells;
(b) A constitutive promoter for controlling the expression of the agent or
component;
(c) An optional terS nucleotide sequence;
(d) An origin of replication (on); and
(e) A phage packaging sequence (optionally pac, cos or a homologue
thereof); and
the plasmid being devoid of
(f) All nucleotide sequences encoding phage structural proteins necessary
for the production of a
transduction particle (optionally a phage), or the plasmid being devoid of at
least one of such
sequences; and
(g) Optionally terL.
5
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[0022] A bacterial host cell comprising the genome of a helper phage that is
incapable of self-
replication, optionally wherein the genome is present as a prophage, and a
plasmid according to the
invention, wherein the helper phage is operable to package copies of the
plasmid in transduction
particles, wherein the particles are capable of infecting bacterial target
cells to which the antibacterial
agent is toxic.
[0023] A method of making a plurality of transduction particles, the method
comprising culturing a
plurality of host cells according to the invention, optionally inducing a
lytic cycle of the helper phage,
and incubating the cells under conditions wherein transducing particles
comprising packaged copies
of the plasmid are created, and optionally separating the particles from the
cells to obtain a plurality of
transduction particles.
[0024] A plurality of transduction particles obtainable by the method of the
invention for use in
medicine, eg, for treating or preventing an infection of a human or animal
subject by target bacterial
.. cells, wherein transducing particles are administered to the subject for
infecting target cells and killing
the cells using the antibacterial agent.
[0025] A method of making a plurality of transduction particles, the method
comprising
(a) Producing host cells whose genomes comprise nucleic acid encoding
structural proteins
necessary to produce transduction particles that can package first DNA,
wherein the genomes are
devoid of a phage packaging signal, wherein the expression of the proteins is
under the control of
inducible promoter(s);
(b) Producing first DNA encoding an antibacterial agent or a component
thereof, wherein the
DNA comprises a phage packaging signal;
(c) Introducing the DNA into the host cells;
(d) Inducing production of the structural proteins in host cells, whereby
transduction particles are
produced that package the DNA;
(e) Optionally isolating a plurality of the transduction particles; and
(0 Optionally formulating the particles into a pharmaceutical
composition for administration to a
.. human or animal for medical use.
[0026] A plurality of transduction particles obtainable by the method.
[0027] A host bacterial cell comprising
6
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
a) A first DNA; and
b) One or more second DNAs;
wherein
(i) the DNAs together comprise all genes required to produce a transduction
particle comprising
a copy of the first DNA packaged by phage structural proteins;
(ii) the first DNA is devoid of at least one functional essential gene (eg,
encoding a phage
structural protein) required to produce the particle; and wherein the one or
more second DNAs
comprises said functional essential gene(s);
(iii) the first DNA comprises a phage packaging signal for producing the
particle; and
(iv) the second DNA is devoid of a nucleotide sequence required for
packaging the second DNA
into transduction particles;
wherein the second DNA is required for packaging first DNA to produce
particles, wherein the DNAs
are operable in the cell for producing transduction particles comprising phage
structural proteins that
package copies of the first DNA.
BRIEF DESCRIPTION OF THE DRAWINGS
[0028] Figure 1: The genetic map of P2 genome with non-essential genes boxed;
[0029] Figure 2: Schematic of SaPIbov1;
[0030] Figure 3: (A) Maps of P2 and P4¨ genomic architecture and regulatory
network of phage P2
and satellite phage P4; the grey boxed region in P2 was deleted and replaced
with a a kanamycin
marker sequence, and the box shown in dashed lines shows the region of P4
including the cos site that
was included in the CGV; (B) p94 genomic map;
[0031] Figure 4: Schematic view of the SA100 production srain ¨ the defective
helper phage will
only turn on phage production following induction; since the phage packaging
DNA sequence has
.. been removed from the helper and placed on the CGVTM, only CGVTM molecules
will be packaged
into the synthetic phage particles;
[0032] Figure 5: Efficiency of infection ¨ percent infected EMG-2 cells with
increasing ratio (MOT)
of SA100 particles to EMG-2;
[0033] Figure 6: CGVTM delivery by SA100 ¨ total CFU and CGVTm-containing CFU
after infection
of MG1655_pks cells (A) with SA100 containing p114 or p94 or X1-blue_pks cells
(B) containing
p114;
[0034] Figure 7: Killing of target cells by SA100-delivered CGVTM - killing
efficiency of the two
target strains by SA100 delivered CGV5TM (2nd, 4th and 6th bars) using wild-
type and optimised
copy number CGVTM compared to non-infected controls;
7
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[0035] Figure 8: Plotted is the abundance as expressed in colony forming units
(CFU) per 5 milliliter
faecal sample (n=15). Two groups, CRISPR induced and non-induced are shown. No
difference was
observed between the groups when the CFU was deteremined on agar plates
containing streptomycin;
and
[0036] Figure 9: Plotted is the abundance as expressed in colony forming units
(CFU) per 5 milliliter
faecal sample (n=15). Two groups, CRISPR induced and non-induced are shown.
The statistical
difference is calculated using the Student t-test, showing a statistically
significant increase in
abundance after 48 hours in the CRISPR non-induced group (p-value=0.011) on
agar plates
supplemented with spectinomycin and streptomycin. This shows that the CGV
(containing a
spectonomycin marker) can be delivered by non-self-replicative particles
SA100.
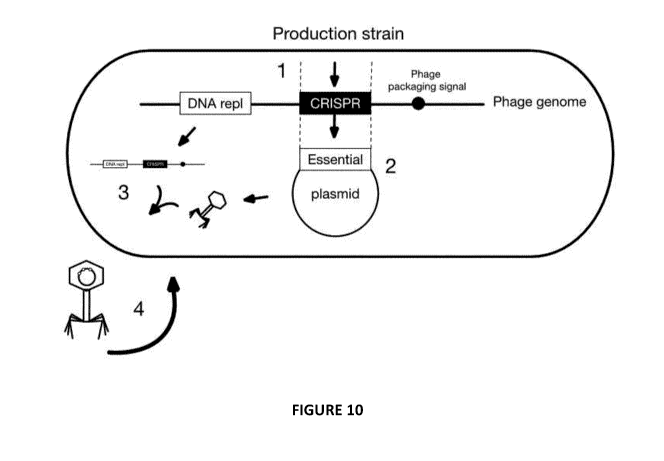
[0037] Figure 10:
1. CRISPR system is inserted into a phage genome containing at least
packaging signal and
DNA replication module thereby generating a vector, such as a plasmid (we call
a CRISPR Guided
VectorTM, CGV); the CRISPR system may comprise nucleic acid encoding a Cas
(eg, Cas3 or Cas9)
and/or one or more crRNAs or gRNAs, optionally also Cascade proteins when the
Cas is Cas3;
2. Essential gene(s) are removed or mutated in phage and the function
provided by expression in
trans from a plasmid (or this may instead be from essential genes integrated
in the chromosome of a
host bacterial cell (aka the production strain));
3. CGV is replicated and packaged in phage-like transduction particles
encoded by functions
encoded on CGV, but at least one essential function being expressed in trans
from plasmid or
chromosome; and
4. Production via propagation cycles on production strain. Propagation on
other bacterial strains
is advantageously prevented.
DETAILED DESCRIPTION
[0038] The invention relates to tbe production of phage using DNAs (eg,
plasmids with helper
phage), as well as the phage, helper phage, compositions and methods involving
these. The invention
finds utility, for example, for containing phage in environments ex vivo and
in vivo, reducing the risk
of acquisition of antibiotic resistance or other genes by phage, as well as
controlling dosing of phage
in an environment. The contamination of useful phage populations by helper
phage may in examples
also be restricted or eliminated, thereby controlling phage propagation and
enhancing the proportion
of desired phage in phage compositions, such as medicaments, herbicides and
other agents where
phage may usefully be used. Thus, the invention provides the following
embodiments.
[0039] A kit comprising
8
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
a) A first DNA; and
b) One or more second DNAs;
Wherein
(i) the DNAs together comprise all phage structural protein genes required
to produce a
packaged phage particle or a transduction particle comprising a copy of the
first DNA;
(ii) the first DNA comprises none or at least one, but not all, of the
genes; and wherein the one or
more second DNAs comprise the remainder of the genes;
(iii) the first DNA comprises a phage packaging signal for producing the
packaged phage particle;
and
(iv) the second DNA is devoid of a nucleotide sequence required for
packaging the second DNA
into phage particles;
wherein the DNAs are operable when co-existing in a host bacterium for
producing packaged phage
that comprise the first DNA, wherein the phage require the second DNA for
replicaton thereof to
produce further phage particles.
[0040] For example the second DNA is devoid of a packaging signal for
packaging second DNA.
Additionally or alternatively, the second DNA is devoid of a nucleotide
sequence required for
replication of helper phage. Optionally, the nucleotide sequence enodes a
sigma factor or comprises a
sigma factor recognition site, a DNA polymerisation recognition site, or a
promoter of a gene required
for helper phage DNA replication when the second DNA is comprised by a helper
prophage.
[0041] In an example, the second DNA is comprised by an M13 or M13-based
helper phage. M13
encodes the following proteins required for phage packaging:-
a) pill: host recognition
b) pV: coat protein
c) pVII, pVIII, pIX: membrane proteins
d) pI, pIV, pXI: Channel for translocating the phage to the extracellular
space.
[0042] In this example, the second DNA is devoid of one or more of the genes
coding for these
proteins, eg, is devoid of a gene endoding pill, a gene encoding pV, a gene
endoding pVII, a gene
endoding pVIII, a gene endoding pIX, a gene endoding pI, a gene endoding pIV
and/or a gene
endoding XI.
[0043] In an embodiment, the phage particle of (i) is capable of infecting a
target bacterium, the
phage comprising a nucleotide sequence of interest (NSI) that is capable of
expressing a protein or
RNA in the target bacterium, or wherein the NSI comprises a regulatory element
that is operable in
the target bacterium. In an example, the NSI is capable of recombination with
the target cell
chromosome or an episome comprised by the target cell to modify the chromosome
or episome.
Optionally, this is carried out in a method wherein the chromosome or episome
is cut (eg, at a
9
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
predetermined site using a guided nuclease, such as a Cas, TALEN, zinc finger
or meganuclease; or a
restriction endonuclease) and simultaneously or sequentially the cell is
infected by a phage particle
that comprises the first DNA, wherein the DNA is introduced into the cell and
the NSI or a sequence
thereof is introduced into the chromosome or episome at or adjacent the cut
site. In an example the
first DNA comprises one or more components of a CRISPR/Cas system operable to
perform the
cutting (eg, comprising at least a nucleotide sequence encoding a guide RNA or
crRNA for targeting
the site to be cut) and further comprising the NSI.
[0044] In an embodiment, the presence in the target bacterium of the NSI or
its encoded protein or
RNA mediates target cell killing, or downregulation of growth or propagation
of target cells, or
mediates switching off of expression of one or more RNA or proteins encoded by
the target cell
genome, or downregulation thereof.
[0045] In an embodiment, the presence in the target bacterium of the NSI or
its encoded protein or
RNA mediates upregulation of growth or propagation of the target cell, or
mediates switching on of
expression of one or more RNA or proteins encoded by the target cell genome,
or upregulation
thereof.
[0046] In an embodiment, the NSI encodes a component of a CRISPR/Cas system
that is toxic to the
target bacterium.
[0047] In an embodiment, the DNA is a first DNA as defined in any preceding
paragraph.
[0048] In an embodiment, the first DNA is comprised by a vector (eg, a plasmid
or shuttle vector).
[0049] In an embodiment, the second DNA is comprised by a vector (eg, a
plasmid or shuttle vector),
helper phage (eg, a helper phagemid) or is integrated in the genome of a host
bacterial cell.
[0050] An embodiment provides a bacterial cell comprising the first and second
DNAs. Optionally,
the cell is devoid of a functional CRISPR/Cas system before transfer therein
of a first DNA, eg, a first
DNA comprising a component of a CRISPR/Cas system that is toxic to the target
bacterium. An
embodiment provides an antibacterial composition comprising a plurality of
cells, wherein each cell is
optionally according to this paragraph, for administration to a human or
animal subject for medical
use.
[0051] A method of producing phage is provided, the method comprising
expressing in a host
bacterial cell the phage protein genes, wherein packaged phage are produced
that comprise the first
DNA, wherein the phage require the second DNA for replicaton thereof to
produce further phage
particles. Optionally, the method comprises isolating the phage particles.
[0052] A composition comprising a population of phage particles obtainable by
the method is
provided for administration to a human or animal subject for treating an
infection of target bacterial
cells, wherein the phage are capable of infecting and killing the target
cells.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[0053] A method of treating an environment ex vivo, the method comprising
exposing the
environment to a population of phage particles obtainable by the method is
provided, wherein the
environment comprises target bacteria and the phage infect and kill the target
bacteria. In an example
thje subject is further administered an agent simultaneously or sequentially
with the phage
administration. In an example, the agent is a herbicide, pesticide,
insecticide, plant fertilizer or
cleaning agent.
[0054] Optionally, the method is for containing the treatment in the
environment.
[0055] Optionally, the method is for controlling the dosing of the phage
treatment in the
environment.
[0056] Optionally, the method is for reducing the risk of acquisition of
foreign gene sequence(s) by
the phage in the environment.
[0057] A method of treating an infection of target bacteria in a human or
animal subject is provided,
the method comprising exposing the bacteria to a population of phage particles
obtainable by the
production method, wherein the phage infect and kill the target bacteria.
[0058] Optionally, the method for treating is for containing the treatment in
the subject.
[0059] Optionally, the method for treating is for containing the treatment in
the environment in
which the subject exists.
[0060] Optionally, the method for treating is for controlling the dosing of
the phage treatment in the
subject.
[0061] Optionally, the method for treating is for reducing the risk of
acquisition of foreign gene
sequence(s) by the phage in the subject.
[0062] Optionally, the method for treating is for reducing the risk of
acquisition of foreign gene
sequence(s) by the phage in the environment in which the subject exists.
[0063] Optionally, target bacteria herein are comprised by a microbiome of the
subject, eg, a gut
microbiome. Altertnatively, the microbiome is a skin, scalp, hair, eye, ear,
oral, throat, lung, blood,
rectal, anal, vaginal, scrotal, penile, nasal or tongue microbiome.
[0064] In an example thje subject is further administered a medicament
simultaneously or
sequentially with the phage administration. In an example, the medicament is
an antibiotic, antibody,
immune checkpoint inhibitor (eg, an anti-PD-1, anti-PD-Li or anti-CTLA4
antibody), adoptive cell
therapy (eg, CAR-T therapy) or a vaccine.
[0065] In an example, the invention employs helper phage for packaging the
phage nucleic acid of
interest. Thus, the invention provides the following illustrative Aspects:-
11
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
1. A population of helper phage, wherein the helper phage are capable
of packaging first phage
nucleic acid to produce first phage particles, wherein the first phage are
different from the helper
phage and the helper phage are incapable themselves of producing helper phage
particles.
2. A composition comprising a population of first phage, wherein the first
phage require helper
phage according to Aspect 1 for replication of first phage particles; and
optionally wherein less than
20, 15, 10, 5, 4, 3,2, 1, 0.5, 0.4, 0.2 or 0.1% of total phage particles
comprised by the composition are
particles of such helper phage.
In an example the composition comprises helper phage and less than 1% of total
phage particles
comprised by the composition are particles of such helper phage. In an example
the composition
comprises helper phage and less than 0.5% of total phage particles comprised
by the composition are
particles of such helper phage. In an example the composition comprises helper
phage and less than
0.1% of total phage particles comprised by the composition are particles of
such helper phage. In an
example the composition comprises helper phage and less than 0.01% of total
phage particles
comprised by the composition are particles of such helper phage. In an example
the composition
comprises helper phage and less than 0.001% of total phage particles comprised
by the composition
are particles of such helper phage. In an example the composition comprises
helper phage and less
than 0.0001% of total phage particles comprised by the composition are
particles of such helper
phage. In an example the composition comprises helper phage and less than
0.00001% of total phage
particles comprised by the composition are particles of such helper phage. In
an example the
composition comprises helper phage and less than 0.000001% of total phage
particles comprised by
the composition are particles of such helper phage. In an example the
composition comprises helper
phage and less than 0.0000001% of total phage particles comprised by the
composition are particles
of such helper phage. In an example the composition comprises helper phage and
less than
0.00000001% of total phage particles comprised by the composition are
particles of such helper
phage.
In an example, the population comprises at least 10,104' 105 or 106 phage
particles, as indicated a
transduction assay, for example. In an example, the population comprises at
least 10 phage particles
and eg, no more than 1014 particles. In an example, the population comprises
at least 104 phage
particles and eg, no more than 1014 particles. In an example, the population
comprises at least 105
phage particles and eg, no more than 1014 particles. In an example, the
population comprises at least
106 phage particles and eg, no more than 1014 particles. To have a measure of
the first phage
concentration, for example, one can perform a standard transduction assay when
the first phage
12
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
genome contains an antibiotic marker. Thus, in this case the first phage are
capable of infecting target
bacteria and in a sample of lml the population comprises at least 103,104' 105
or 106 transducing
particles, which can be determined by infecting susceptible bacteria at a
multiplicity of infection <0.1
and determining the number of infected cells by plating on a selective agar
plate corresponding to the
antibiotic marker in vitro at 20 to 37 degrees centigrade, eg, at 20 or 37
degrees centrigrade.
Optionally at least 99.9, 99.8, 99.7, 99.6, 99.5, 99.4, 99.3, 99.2, 99.1, 90,
85, 80, 70, 60, 50 or 40% of
total phage particles comprised by the composition are particles of first
phage.
In an example, the first phage genome comprises an fl origin of replication.
In an example, the helper phage are E coli phage. In an example, the first
phage are E co/i, C dificile,
Streptococcus, Klebsiella, Pseudomonas, Acitenobacter, Enterobacteracea,
Firmicutes or
Bacteroidetes phage. In an example, the helper phage are engineered M13 phage.
In an example, the first phage genome comprises a phagemid, wherein the
phagemid comprises a
packaging signal for packaging first phage particles in the presence of the
helper phage.
The first phage particles may contain a nucleotide sequence of interest (NSI),
eg, as defined herein,
such as a NSI that encodes a component of a CRISPR/Cas system operable in
target bacteria that can
be infected by the first phage particles. Once inside the target bacteria, the
first phage DNA is
incapable of being packaged to form first phage particles in the absence of
the helper phage. This
usefully contains the activity of the first phage genome and its encoded
products (protieins and/or
nucleic acid), as well as limits or controls dosing of the NSI and its encoded
products in an
environment comprising the target bacteria that have been exposed to the first
phage. This is useful,
for example to control the medical treatment of an environment comprised by a
human or animal
subject, plant or other environment (eg, soil or a foodstiff or food
ingredient).
3. The helper phage or composition of any preceding Aspect, wherein the
genome of each first
phage is devoid of genes encoding first phage structural proteins.
4. The composition of Aspect 2 or 3, wherein the composition comprises
helper phage DNA.
5. The composition of Aspect 4, wherein the DNA comprises helper DNA
fragments.
6. The helper phage or composition of any one preceding Aspect, wherein the
helper phage are
in the form of prophage.
13
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Thus, the prophage is integrated in the chromosome of a host cell.
Examples of phage structural proteins are phage coat proteins, collar proteins
and phage tail fibre
proteins.
7. The composition of any one of Aspects 2 or 3, wherein the
composition comprises no helper
phage DNA comprising a sequence of 20 contiguous nucleotides or more (eg, from
20 to 25, 30, 35,
40, 50 or 100 nucleotides), eg, no helper phage DNA.
This can be determined, for example, using DNA probes (designed on the basis
of the known heper
phge genome sequence) with PCR, as is conventional. In an example, the
composition may comprise
residual helper prophage DNA, but essentially otherwise is devoid of helper
DNA.
8. The composition of any one of Aspects 2 to 5 and 7, wherein the helper
phage are capable of
infecting host bacteria and the composition does not comprise host bacteria.
9. The composition of any one of Aspects 2 to 8, wherein the composition is
a lysate of host
bacterial cells, wherein the lysate comprises helper prophage DNA, eg, such
DNA comprises 20
contiguous nucleotides or more (eg, from 20 to 25, 30, 35, 40, 50 or 100
nucleotides) of helper phage
DNA.
10. The composition of any one of Aspects 2 to 8, wherein the composition
is a lysate of host
bacterial cells, wherein the lysate has been processed (eg, filtered) to
remove all or some helper phage
DNA; or the composition is a lysate of host bacterial cells that is devoid of
cellular material.
11. The composition of any one of Aspects 2 to 10, wherein the composition
does not comprise
helper phage particles.
12. The composition of any one of Aspects 2 to 11, wherein at least 95%
(eg, 100%) of phage
particles comprised by the composition are first phage particles.
In another embodiment, the composition comprises second phage particles,
wherein the second phage
.. are different from the first phage and are not helper phage.
13. The composition of any one of Aspects 2 to 12, wherein the population
comprises at least 103,
104, 105 or 106 phage particles, as indicated in a transduction assay.
14. The helper phage or composition of any preceding Aspect, wherein the
first phage are capable
of replicating in host bacteria in the presence of the helper phage (eg,
helper prophage), wherein the
14
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
first phage comprise antibacterial means for killing target bacteria of a
first strain or species, wherein
the target bacteria are of a different strain or species and the antibacterial
means is not operable to kill
the target bacteria.
15. A composition comprising a population of phage, the population
comprising
(a) A first sub-population of first phage that require a helper phage for
packaging the first phage;
(b) A second sub-population of phage comprising the helper phage,
wherein the helper phage are
as recited in any preceding Aspect.
16. The helper phage or composition of any preceding Aspect, wherein the
helper phage are
phagemids.
17. A composition comprising
(a) A population of helper phage as recited in any preceding Aspect; and
(b) A population of nucleic acid vectors comprising vector DNA that
comprises a first phage
packaging signal;
(c) wherein the helper phage are capable of packaging the vector DNA to
produce first phage.
18. The composition of Aspect 17, wherein the vectors are phage.
19. The composition of Aspect 17, wherein the vectors are plasmids or
phagemids.
20. The composition of Aspect 19, the vectors are shuttle vectors (eg, pUC
vectors) that can be
replicated in first bacteria, wherein the vectors can further be replicated
and packaged into first phage
in second bacteria (host bacteria) in the presence of the helper phage,
wherein the first bacteria are of
a strain or species that is different to the strain or species of the host
bacteria.
21. The composition of Aspect 20, wherein the first phage are capable of
infecting third bacteria
.. of a strain or species that is different to the second (and optionally also
the first) bacteria.
22. The composition of any one of Aspects 17 to 21, wherein the first phage
are capable of
replicating in host bacteria in the presence of the helper phage (eg, helper
prophage), wherein the first
phage comprise antibacterial means for killing target bacteria of a first
strain or species, wherein the
host bacteria are of a different strain or species and the antibacterial means
is not operable to kill the
host bacteria.
23. The helper phage or composition of any preceding Aspect, wherein the
genome is devoid of a
packaging signal (eg, SEQ ID NO: 2 below), wherein the helper phage are
incapable of self-
replication.
24. The helper phage or composition of Aspect 23, wherein the signal is a
pac or cos sequence.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
25. The helper phage or composition of any preceding Aspect, wherein the
helper phage genome
is capable of replication in a host cell.
Thus, the genome is capable of nucleic acid replication but not packaging of
helper phage.
26. The helper phage or composition of any one of Aspects 1 to 24, wherein
the genome is devoid
of a nucleotide sequence required for production of helper phage particles.
27. The helper phage or composition of Aspect 26, wherein the nucleotide
sequence enodes a
sigma factor (eg, sigma-70) or comprises a sigma factor recognition site, a
DNA polymerisation
recognition site, or a promoter of a gene required for helper phage DNA
replication.
28. The helper phage or composition of any preceding Aspect, wherein the
helper phage are
temperate phage.
29. The helper phage or composition of any one of Aspects 1 to 27, wherein
the helper phage are
lytic phage.
30. The helper phage or composition of any preceding Aspect, wherein the
first phage are capable
of infecting target bacteria, the first phage comprising a nucleotide sequence
of interest (NSI) that is
capable of expressing a protein or RNA (eg, gRNA or crRNA) in target bacteria,
or wherein the NSI
comprises a regulatory element that is operable in target bacteria.
31. The helper phage or composition of Aspect 30, wherein the presence in
target bacteria of the
NSI or its encoded protein or RNA mediates target cell killing, or
downregulation of growth or
propagation of target cells, or mediates switching off of expression of one or
more RNA or proteins
encoded by the target cell genomes, or downregulation thereof.
32. The helper phage or composition of Aspect 30, wherein the presence in
target bacteria of the
NSI or its encoded protein or RNA mediates upregulation of growth or
propagation of target cells, or
mediates switching on of expression of one or more RNA or proteins encoded by
the target cell
genomes, or upregulation thereof.
33. An antibacterial composition according to any one of Aspects 2 to 32,
wherein the first phage
are capable of infecting target bacteria and each first phage comprises
engineered antibacterial means
for killing target bacteria.
By use of the term "engineered" it will be readily apparent to the skilled
addressee that the relevant
means has been introduced and is not naturally-occurring in the phage. For
example, the means is
recombinant, artificial or synthetic.
16
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
34. The composition of Aspect 14, 22 or 33, wherein the antibacterial means
comprises one or
more components of a CRISPR/Cas system.
35. The composition of caim 34, wherein the component(s) comprise (i) a DNA
sequence
encoding a guide RNA (eg, a single guide RNA) or comprising a CRISPR array for
producing guide
RNA, wherein the guide RNA is capable of targeting the genome of target
bacteria; (ii) a Cas
nuclease-encoding DNA sequence; and/or (iii) a DNA sequence encoding one or
more components of
Cascade.
In an example, a Cas herein is a Cas9. In an example, a Cas herein is a Cas3.
The Cas may be
identical to a Cas encoded by the target bacteria.
36. The composition of any one of Aspects 14, 22 or 33 to 35, wherein the
antibacterial means
comprises a nucleic acid encoding a guided nuclease, such as a Cas nuclease,
TALEN, zinc finger
nuclease or meganuclease.
37. The helper phage or composition of any preceding Aspect, wherein the
helper phage is for use
in medicine practised on a human or animal subject, or the composition is a
pharmaceutical
composition for use in medicine practised on a human or animal subject.
In an example, the animak is a livestock or companion pet animal (eg, a cow,
pig, goat, sheep, horse,
dog, cat or rabbit). In an example, the animal is an insect (an insect at any
stage of its lifecycle, eg,
egg, larva or pupa). In an example, the animal is a protozoan. In an example,
the animal is a
cephalopod.
38. The composition of any one of Aspects 2 to 36, wherein the composition
is a herbicide,
pesticide, food or beverage processing agent, food or beverage additive,
petrochemical or fuel
processing agent, water purifying agent, cosmetic additive, detergent additive
or environmental (eg,
soil) additive or cleaning agent.
39. The helper phage or composition of any one of Aspects 1 to 37 for use
in a contained method
of treating a disease or condition of a human or animal subject, wherein the
disease or condition is
mediated by the target bacteria and the target bacteria are comprised by the
subject, the method
comprising administering the composition to the subject, whereby the target
bacteria are exposed to
the antibacterial means and killed and propagation of the first phage is
contained.
The inability of the first phage to self-replicate and to require helper phage
or second DNA to do this
usefully provides containment in the location (eg, gut) of action of the
composition and/or in the
17
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
environment of the subject, eg, when exposed to secretions such as urine and
faeces of the subject that
otherwise may contain replicated first phage. Inability of the helper phage or
second DNA to self-
package limits availability of factors required by the first phage to form
packaged particles, hence
providing containment by limiting first phage propagation. This may be useful,
for example, to
contain an antibacterial acitivity provided by the first phage, such as a
CRISPR/Cas killing principle.
40. A bacterial cell or a plurality of bacterial cells comprising the
helper phage or composition of
any preceding Aspect, wherein the first phage are capable of replication in
the presence of the helper
phage in the cell.
The cell may, for example, act as a carrier for the genome of the first phage,
wherein the first phage
DNA is capable of horizontal transfer from the carrier to the target bacteria
once the carrier bacteria
have been administered to an environment to be treated, eg, a soil or a human
gut or other
environment described herein. In an example, the environment is comprised by a
human or animal
subject and the carrier are commensal or probiotic in the subject. For example
the carrier bacteria are
Lactobacillus (eg, L reuteri or L lactis), E coli or Streptococcus (eg, S
thermophilus) bacteria. The
horizontal transfer can be transfer of a plasmid (such as a conjugative
plasmid) to the target bacteria
or first phage infection of the target bacteria, wherein the first phage have
been prior packaged in the
carrier. The use of a carrier is useful too for oral administration or other
routes where the carrier can
provide protection for the phage, helper or composition from the acid stomach
or other harsh
environments in the subject. Furthermore, the carrier can be formulated into a
beverage, for example,
a probiotic drink, eg, an adapted Yakult (trademark), Actimel (trademark),
Kevita (trademark),
Activia (trademark), Jarrow (trademark) or similar drink for human
consumption.
41. The cell(s) of Aspect 40 for administration to a human or animal
subject for medical use,
comprising killing target bacteria using first phage, wherein the target
bacteria mediate as disease or
condition in the subject.
In an example, when the subject is a human, the subject is not an embryo.
42. The cell(s) of Aspect 41, wherein the cell(s) comprises helper phage
and is symbiotic or
probiotic in the subject.
18
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
43. A method of killing target bacteria in an environment, optionally
wherein the method is not
practised on a human or animal body, wherein the method comprises exposing the
environment to the
cell(s) according to Aspect 42, or a composition obtained or obtainable by the
method of any one of
Aspects 57 to 65, wherein the environment is or has been exposed to first
phage or said vectors to
produce first phage in the presence of the helper phage, wherein the first
phage are capable of
replication in the environment and kill target bacteria.
44. The cell(s) or method of any one of Aspects 40 to 43, wherein the cell
is an E coli,
Lactobacillus (eg, L lactis or retueri) or Streptococcus (eg, thermophilus)
cell.
45. The cell(s) or method of Aspects 40 to 44 wherein the subject is
administered or has been
administered a cell comprising first phage.
46. The composition of any one of Aspects 2 to 45 in combination with a
target bacterial cell
wherein the first phage are capable of infecting the target bacterial cell.
47. Use of the helper phage, composition or cell(s) of any one of Aspects 1
to 42 and 44 to 46, or
a composition obtained or obtainable by the method of any one of Aspects 57 to
65, in the
manufacture of an antibacterial agent that kills target bacteria, for
containment of the antibacterial in
an environment, eg, containment ex vivo; or containment in a human or animal
subject comprising the
environment.
48. Use of the helper phage, composition or cell(s) of any one of Aspects 1
to 42 and 44 to 46, or
a composition obtained or obtainable by the method of any one of Aspects 57 to
65, in the
manufacture of an antibacterial agent that kills the target bacteria, for
reducing the risk of acquisition
by the first phage of foreign genes.
For example, this is useful for reducing the risk of antibiotic resistance
genes by the phage, such as
when the phage are in the presence of other phage or plasmids in the
environment.
49. Use of the helper phage, composition or cell(s) of any one of Aspects 1
to 42 and 44 to 46, or
a composition obtained or obtainable by the method of any one of Aspects 57 to
65, in the
manufacture of an antibacterial agent that kills the target bacteria, for
reducing the risk of acquisition
by the first phage of one or more antibiotic resistance genes.
50. A method of reducing the risk of acquisition by first phage of foreign
genes, the method
comprising
(a) Providing the composition of any one of Aspects 2 to 42 and 44 to 46,
or a composition
obtained or obtainable by the method of any one of Aspects 57 to 65; and
(b) Exposing target bacteria to the composition, wherein the first phage
infect the target bacteria;
19
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
(c) wherein the helper phage are incapable of self-replication and
propagation of first phage is
thereby limited, wherein propagation of first phage is prevented or reduced,
thereby reducing the risk
of acquisition of first phage of foreign genes (eg, antibiotic resistance
genes).
51. A method of containing an antibacterial activity in an environment
(e.g., ex vivo), the method
comprising
(a) Providing an antibacterial composition according to any one of Aspects
2 to 42 and 44 to 46,
or a composition obtained or obtainable by the method of any one of Aspects 57
to 65; and
(b) Exposing target bacteria in the environment to the composition, wherein
the bacteria are
exposed to the first phage and antibacterial means and are killed;
(c) wherein the helper phage are incapable of self-replication and
propagation of first phage is
thereby limited, wherein propagation of first phage is prevented or reduced,
thereby containing the
antibacterial activity.
52. A method of controlling the dosing of first phage in an environment
(e.g., ex vivo), the
method comprising
(a) Providing an antibacterial composition according to any one of
Aspects 2 to 42 and 44 to 46,
or a composition obtained or obtainable by the method of any one of Aspects 57
to 65; and
(b) Exposing target bacteria in the environment to the composition, wherein
the bacteria are
infected by first phage;
(c) wherein the helper phage are incapable of self-replication and
propagation of first phage is
thereby limited, wherein propagation of first phage is prevented or reduced,
thereby controlling
dosing of first phage in the environment.
53. The method of any one of Aspects 43 to 45, 51 and 52, or the use of
Aspect 47, wherein the
environment is a human or animal microbiome, e.g., a gut microbiome.
54. The method of any one of Aspects 43 to 45, 51 and 52, or the use of
Aspect 47, wherein the
environment is a microbiome of soil; a plant, part of a part (e.g., a leaf,
fruit, vegetable or flower) or
plant product (e.g., pulp); water; a waterway; a fluid; a foodstuff or
ingredient thereof; a beverage or
ingredient thereof; a medical device; a cosmetic; a detergent; blood; a bodily
fluid; a medical
apparatus; an industrial apparatus; an oil rig; a petrochemical processing,
storage or transport
apparatus; a vehicle or a container.
55. The method of any one of Aspects 43 to 45, 51 and 52, or the use of
Aspect 47, wherein the
environment is an ex vivo bodily fluid (e.g., urine, blood, blood product,
sweat, tears, sputum or spit),
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
bodily solid (e.g., faeces) or tissue of a human or animal subject that has
been administered the
composition.
56. The method of any one of Aspects 43 to 45, 51 and 52, or the use of
Aspect 47, wherein the
environment is an in vivo bodily fluid (e.g., urine, blood, blood product,
sweat, tears, sputum or spit),
bodily solid (e.g., faeces) or tissue of a human or animal subject that has
been administered the
composition.
57. A method of producing first phage, wherein the first phage require
helper phage to replicate,
the method comprising
(a) Providing DNA comprising a packaging signal;
(b) Introducing the DNA into a host bacterial cell;
(c) Wherein the host bacterial cell comprises helper phage or wherein
helper phage are
introduced into the bacterial cell simultaneously or sequentially with step
(b);
(d) Wherein the helper phage are according to any preceding Aspect;
(e) Causing or allowing the helper phage to produce phage coat proteins,
wherein the packaging
signal is recognised in the host cell, whereby first phage are produced using
the proteins, the first
phage packaging the DNA;
(0 Wherein helper phage particle production in the host cell is
inhibited or reduced, thereby
limiting the availability of helper phage particles;
(g) Optionally lysing the host cell and obtaining the first phage;
(h) Thereby producing a composition comprising first phage which require
the helper phage for
replication, wherein further production of first phage particles is prevented
or reduced by the
limitation of helper phage availability in the composition.
In an embodiment, the DNA is comprised by a phagemid or cloning vector (eg, a
shuttle vector, eg, a
pUC vector).
There may be a modest amount of helper phage DNA replication to enable first
phage protein
production efficiently, or should replication of helper phage DNA may be
eliminated totally
eliminated.
58. The method of Aspect 57, wherein in (c) the helper phage are
prophage integrated in the
bacterial cell chromosome.
59. The method of Aspect 59, wherein (e) comprises inducing replication
of helper phage DNA
and/or expression of the proteins, eg, using UV, mitomycin.
21
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
60. The method of any one of Aspects 57 to 59, wherein (g) comprises
further separating the first
phage from cellular material or helper phage DNA.
61. The method of any one of Aspects 57 to 60, wherein the composition
comprises a population
of first phage particles, wherein the composition does not comprise helper
phage DNA and/or
particles.
62. The method of any one of Aspects 57 to 61, wherein the DNA of (a)
comprises engineered
antibacterial means for killing target bacteria.
63. The method of Aspect 62, wherein the antibacterial means comprises one
or more
components of a CRISPR/Cas system.
64. The method of Aspect 63, wherein the component(s) comprise (i) a DNA
sequence encoding
a guide RNA (eg, a single guide RNA) or comprising a CRISPR array for
producing guide RNA,
wherein the guide RNA is capable of targeting the genome of target bacteria;
(ii) a Cas (eg, Cas9,
Cas3, Cpfl, CasX or CasY) nuclease-encoding DNA sequence; and/or (iii) a DNA
sequence encoding
one or more components of Cascade (eg, CasA).
65. The method of any one of Aspects 62 to 64, wherein the antibacterial
means comprises a
nucleic acid encoding a guided nuclease, such as a Cas nuclease, TALEN, zinc
finger nuclease or
meganuclease.
66. The helper phage, composition or cell(s) of any one of Aspects 1 to 42
and 44 to 46, or a
composition obtained or obtainable by the method of any one of Aspects 57 to
65, for antibacterial
treatment of target bacteria in a human or animal subject whereby the
antibacterial treatment is
contained in the subject.
67. The helper phage, composition or cell(s) of any one of Aspects 1 to 42
and 44 to 46, or a
composition obtained or obtainable by the method of any one of Aspects 57 to
65, for antibacterial
treatment of target bacteria in a gut of a human or animal subject whereby the
antibacterial activity in
one or more bodily excretions of the subject is reduced.
This is useful as a safety measure to reduce or eliminate first phage activity
outside the subject.
68. The helper phage, composition or cell(s) of Aspect 67, wherein the
antibacterial activity in
one or more bodily excretions of the subject is eliminated.
69. The helper phage, composition or cell(s) of any one of Aspects 1 to 42
and 44 to 46, or a
composition obtained or obtainable by the method of any one of Aspects 57 to
65, for controlling the
dosing of antibacterial treatment of target bacteria in a human or animal
subject, eg, in the gut of the
subject.
22
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Usefully, propagation of the first phage is restricted or eliminated, so
dosing in the subject can be
controlled, or even pre-determined within a narrow expected range. This is
useful, for example, for
medicaments comprising the first phage or composition, and may be aid approval
of such medicines
before FDA and simiar authorities.
Alternatively, the dosing is dosing of an environment, such as soil etc
disclosed herein, wherein
limitation of the first phage or composition activity is also desirable to
limit spread of activities in
natural and other terrains.
70. The helper phage, composition or cell(s) of any one of Aspects 1 to 42
and 44 to 46, or a
composition obtained or obtainable by the method of any one of Aspects 57 to
65, for fixing the
dosing of antibacterial treatment of target bacteria in a human or animal
subject, eg, in the gut of the
subject.
71. A phage production system, for producing phage (eg, the first phage of
any preceding Aspect)
comprising a nucleotide sequence of interest (NSI-phage), the system
comprising components (i) to
(iii):-
(a) A first DNA;
(b) A second DNA; and
(c) a NSI-phage production factor (NPF) or an expressible nucleotide
sequence that encodes a
NPF;
Wherein
(d) The first DNA encodes a helper phage (eg, said first helper phage
recited in any preceding
Aspect);
(e) The second DNA comprises the nucleotide sequence of interest (NSI);
(0 When the system is comprised by a bacterial host cell, helper phage
proteins are expressed
from the first DNA to form phage that package the second DNA in the presence
of the NPF, thereby
producing NSI-phage; and
(g) The system is devoid of a helper phage production factor (HPF) that
is required for forming
helper phage particles that package the first DNA, or is devoid of an
expressible nucleotide sequence
that encodes a functional HPF; or the system comprises a nucleotide sequence
that comprises or
encodes a functional HPF, the system further comprising means for targeted
inactivation in the host
23
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
cell of the HPF sequence to eliminate or minimise production of helper phage
comprising the first
DNA;
Whereby the system is capable of producing a product comprising a population
of NSI-phage,
wherein each NSI-phage requires a said helper phage for propagation,
optionally wherein the NSI-
phage in the product are not mixed with helper phage or less than 20% of total
phage comprised by
the product are said helper phage.
The invention includes within its concept relatively low level of helper phage
particle production if
there is a residual capability of helper phage to replicate to produce
particles, such as for example in
the case that a helper phage packaging signal or other HPF nucleotide sequence
in the helper phage
genome is mutated (eg, by deletion, substitution or addition of nucleotides
therein) to knock down the
ability to form phage particles. Preferably, there is no production of helper
phage particles, such as by
deleting all or part of the sequence from the helper phage genome or
inactivating the sequence.
72. A method of producing first phage, wherein the first phage require
helper phage to replicate,
the method comprising
(a) Providing in host cells the system of Aspect 71;
(b) Causing or allowing the helper phage proteins to be produced, whereby
the second DNA is
packaged to produce first phage; and
(c) Optionally lysing the host cells and obtaining a composition comprising
first phage.
73. The method of Aspect 72, wherein step (c) comprises separating the
first phage from cellular
material.
74. The method of Aspect 72 or 73, wherein the composition comprises a
population of first
phage, wherein less than 20, 10, 5, 4, 3, 2, 1, 0.5 or 0.1% of total phage
comprised by the composition
are helper phage.
75. The method of any one of Aspects 72 to 74, wherein the second DNA
comprises engineered
antibacterial means for killing target bacteria.
76. The method of Aspect 75, wherein the antibacterial means comprises
one or more
components of a CRISPR/Cas system.
77. The method of Aspect 76 wherein the component(s) comprise (i) a DNA
sequence encoding a
guide RNA (eg, a single guide RNA) or comprising a CRISPR array for producing
guide RNA,
wherein the guide RNA is capable of targeting the genome of target bacteria;
(ii) a Cas nuclease-
encoding DNA sequence; and/or (iii) a DNA sequence encoding one or more
components of Cascade.
24
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
78. The method of any one of Aspects 75 to 77, wherein the antibacterial
means comprises a
nucleic acid encoding a guided nuclease, such as a Cas nuclease, TALEN, zinc
finger nuclease or
meganuclease
79. The system of method of any one of Aspects 71 to 78, wherein the first
phage are capable of
infecting target bacteria, the NSI being capable of expressing a protein or
RNA in target bacteria, or
wherein the NSI comprises a regulatory element that is operable in target
bacteria.
80. The system or method of Aspect 79, wherein the presence in target
bacteria of the NSI or its
encoded protein or RNA mediates target cell killing, or downregulation of
growth or propagation of
target cells, or mediates switching off of expression of one or more RNA or
proteins encoded by the
target cell genomes, or downregulation thereof.
81. The system or method of Aspect 79, wherein the presence in target
bacteria of the NSI or its
encoded protein or RNA mediates upregulation of growth or propagation of
target cells, or mediates
switching on of expression of one or more RNA or proteins encoded by the
target cell genomes, or
upregulation thereof.
82. The system of method of any one of Aspects 71 to 81, wherein each of
the NPF and HPF is a
packaging signal, eg, SEQ ID NO: 2 or a sequence that is at least 70, 80, 90,
95, 96, 97, 98 or 99%
identical thereto, or is a homologue from a different species.
83. The system of method of Aspect 82, wherein each signal is a pac or
cos sequence, or is a
homologue.
84. The system of method of any one of Aspects 71 to 81, wherein the HPF is
a nucleotide
sequence required for replication of helper phage.
85. The system of method of any one of Aspects 71 to 81, wherein the HPF
enodes a sigma factor
(eg, sigma-70) or comprises a sigma factor recognition site, a DNA
polymerisation recognition site, or
a promoter of a gene required for helper phage DNA replication, a helper phage
integrase, a helper
phage excissionase or a helper phage origin of replication,
86. A composition comprising a population of first phage obtainable by the
method of any one of
Aspects 72 to 85, wherein the genome of each first phage is devoid of genes
encoding phage proteins.
87. The composition of Aspect 86, wherein the first phage comprise
antibacterial means as
recited in any one of Aspects 75 to 78.
88. The composition of Aspect 87, comprising DNA identical to the first DNA
or fragments
thereof.
89. The composition of Aspect 88, wherein the DNA of the composition is
identical to the first
DNA and is devoid of a helper phage packaging signal.
90. The composition of any one of Aspects 86 to 89 for antibacterial
treatment of target bacteria
in a human or animal subject whereby the antibacterial treatment is contained
in the subject.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
91. The composition of any one of Aspects 86 to 89 for antibacterial
treatment of target bacteria
in a gut of a human or animal subject whereby the antibacterial activity in
one or more bodily
excretions of the subject is reduced.
92. The composition of Aspect 91, wherein the antibacterial activity in one
or more bodily
excretions of the subject is eliminated.
93. The composition of any one of Aspects 86 to 89 for controlling the
dosing of antibacterial
treatment of target bacteria in a human or animal subject, eg, in the gut of
the subject.
94. The composition of any one of Aspects 86 to 89 for fixing the dosing of
antibacterial
treatment of target bacteria in a human or animal subject, eg, in the gut of
the subject.
95. An isolated DNA comprising all structural protein genes of a helper
phage genome that are
required for producing phage particles, wherein the DNA is devoid of a helper
phage production
factor (HPF) that is required for producing packaged helper phage, optionally
wherein the DNA
comprises one or more promoters for expression of the genes when the DNA is
integrated in the
genone of a host bacterial cell.
96. The DNA of Aspect 95, wherein the DNA is devoid of any phage packaging
signals.
97. The DNA of Aspect 95 or 96, wherein the HPF is a sigma factor-encoding
nucleotide
sequence or comprises a sigma factor recognition site, a DNA polymerisation
recognition site, a
promoter of a gene required for helper phage DNA replication, a helper phage
integrase-encoding
nucleotide sequence, a helper phage excissionase-encoding nucleotide sequence
or a helper phage
origin of replication.
98. The DNA of any one of Aspects 95 to 97, wherein the DNA comprises a
nucleotide sequence
encoding a CRISPR/Cas system repressor.
99. The DNA of any one of Aspects 95 to 98, wherein the DNA is integrated
in the chromosome
of a host bacterial cell, wherein the genes are expressible in the host cell.
100. The DNA of Aspect 99, wherein the cell is devoid of an active CRISPR/Cas
system.
101. The DNA of any one of Aspects 95 to 100 in combination with a second DNA,
wherein the
second DNA comprises the HPF.
102. The DNA of any one of Aspects 95 to 100 in combination with a second DNA,
wherein the
second DNA comprises a phage packaging signal and optionally the first DNA is
devoid of a phage
packaging signal.
103. The DNA of Aspect 101 or 102, wherein the second DNA is comprised by a
phagemid or a
plasmid (eg, a shuttle vector).
[0066] In an example, the kit, DNA(s), first phage, helper phage or
composition is comprised by a
medical container, eg, a syringe, vial, IV bag, inhaler, eye dropper or
nebulizer. In an example, the
26
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
kit, DNA(s), first phage, helper phage or composition is comprised by a
sterile container. In an
example, the kit, DNA(s), first phage, helper phage or composition is
comprised by a medically-
compatible container. In an example, the kit, DNA(s), first first phage,
helper phage or composition is
comprised by a fermentation vessel, eg, a metal, glass or plastic vessel.
[0067] In an example, the kit, DNA(s), first phage, helper phage or
composition is comprised by a
medicament, e,g in combination with instructions or a packaging label with
directions to administer
the medicament by oral, IV, subcutaneous, intranasal, intraocular, vaginal,
topical, rectal or inhaled
administration to a human or animal subject. In an example, the kit, DNA(s),
first phage, helper
phage or composition is comprised by an oral medicament formulation. In an
example, the kit,
DNA(s), first phage, helper phage or composition is comprised by an intranasal
or ocular medicament
formulation. In an example, the kit, DNA(s), first phage, helper phage or
composition is comprised
by a personal hygiene composition (eg, shampoo, soap or deodorant) or cosmetic
formulation. In an
example, the kit, DNA(s), first phage, helper phage or composition is
comprised by a detergent
formulation. In an example, the kit, DNA(s), first phage, helper phage or
composition is comprised
by a cleaning formulation, eg, for cleaning a medical or industrial device or
apparatatus. In an
example, the kit, DNA(s), first phage, helper phage or composition is
comprised by foodstuff,
foodstuff ingredient or foodstuff processing agent. In an example, the kit,
DNA(s), first phage, helper
phage or composition is comprised by beverage, beverage ingredient or beverage
processing agent. In
an example, the kit, DNA(s), first phage, helper phage or composition is
comprised by a medical
bandage, fabric, plaster or swab. In an example, the kit, DNA(s), first phage,
helper phage or
composition is comprised by a herbicide or pesticide. In an example, the kit,
DNA(s), first phage,
helper phage or composition is comprised by an insecticide.
[0068] In an example, the first phage is a is a Corticoviridae, Cystoviridae,
Inoviridae, Leviviridae,
Microviridae, Myoviridae, Podoviridae, Siphoviridae, or Tectiviridae virus. In
an example, the helper
phage is a is a Corticoviridae, Cystoviridae, Inoviridae, Leviviridae,
Microviridae, Myoviridae,
Podoviridae, Siphoviridae, or Tectiviridae virus. In an example, the helper
phage is a filamentous
M13, a Noviridae, a tailed phage (eg, a Myoviridae, Siphoviridae or
Podoviridae), or a non-tailed
phage (eg, a Tectiviridae).
[0069] In an example, both the first and helper phage are Corticoviridae. In
an example, both the
first and helper phage are Cystoviridae. In an example, both the first and
helper phage are Inoviridae.
In an example, both the first and helper phage are Leviviridae. In an example,
both the first and
helper phage are Microviridae. In an example, both the first and helper phage
are Podoviridae. In an
example, both the first and helper phage are Siphoviridae. In an example, both
the first and helper
phage are Tectiviridae.
27
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[0070] In an example, the CRISPR/Cas component(s) are component(s) of a Type I
CRISPR/Cas
system. In an example, the CRISPR/Cas component(s) are component(s) of a Type
II CRISPR/Cas
system. In an example, the CRISPR/Cas component(s) are component(s) of a Type
III CRISPR/Cas
system. In an example, the CRISPR/Cas component(s) are component(s) of a Type
IV CRISPR/Cas
system. In an example, the CRISPR/Cas component(s) are component(s) of a Type
V CRISPR/Cas
system. In an example, the CRISPR/Cas component(s) comprise a Cas9-encoding
nucleotide
sequence (eg, S pyogenes Cas9, S aureus Cas9 or S thermophilus Cas9). In an
example, the
CRISPR/Cas component(s) comprise a Cas3-encoding nucleotide sequence (eg, E
coli Cas3, C
difficile Cas3 or Salmonella Cas3). In an example, the CRISPR/Cas component(s)
comprise a Cpf-
encoding nucleotide sequence. In an example, the CRISPR/Cas component(s)
comprise a CasX-
encoding nucleotide sequence. In an example, the CRISPR/Cas component(s)
comprise a CasY-
encoding nucleotide sequence.
[0071] In an example, the first DNA, first phage or vector encode a CRISPR/Cas
component or
protein of interest from a nucleotide sequence comprising a promoter that is
operable in the target
bacteria.
[0072] In an example, the host bacteria and/or target bacteria are E coli. In
an example, the host
bacteria and/or target bacteria are C dificile (eg, the vector is a shuttle
vector operable in E coli and
the host bacteria are C dificile). In an example, the host bacteria and/or
target bacteria are
Streptococcus, such as S the rmophilus (eg, the vector is a shuttle vector
operable in E coli and the host
bacteria are Streptococcus). In an example, the host bacteria and/or target
bacteria are Pseudomonas,
such as P aeruginosa (eg, the vector is a shuttle vector operable in E coli
and the host bacteria are P
aeruginosa). In an example, the host bacteria and/or target bacteria are
Klebsiella (eg, the vector is a
shuttle vector operable in E coli and the host bacteria are Klebsiella). In an
example, the host bacteria
and/or target bacteria are Salmonella, eg, S typhimurium (eg, the vector is a
shuttle vector operable in
E coli and the host bacteria are Salmonella).
[0073] Optionally, host and/or target bacteria is a gram negative bacterium
(eg, a spirilla or vibrio).
Optionally, host and/or target bacteria is a gram positive bacterium.
Optionally, host and/or target
bacteria is a mycoplasma, chlamydiae, spirochete or mycobacterium. Optionally,
host and/or target
bacteria is a Streptococcus (eg, pyo genes or thermophilus). Optionally, host
and/or target bacteria is
a Staphylococcus (eg, aureus, eg, MRSA). Optionally, host and/or target
bacteria is an E. coli (eg,
0157: H7) host, eg, wherein the Cas is encoded by the vecor or an endogenous
host Cas nuclease
activity is de-repressed. Optionally, host and/or target bacteria is a
Pseudomonas (eg, aeruginosa).
Optionally, host and/or target bacteria is a Vibro (eg, cholerae (eg, 0139) or
vulnificus). Optionally,
host and/or target bacteria is a Neisseria (eg, gonnorrhoeae or meningitidis).
Optionally, host and/or
target bacteria is a Bordetella (eg, pertussis). Optionally, host and/or
target bacteria is a Haemophilus
28
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
(eg, influenzae). Optionally, host and/or target bacteria is a Shigella (eg,
dysenteriae). Optionally,
host and/or target bacteria is a Brucella (eg, abortus). Optionally, host
and/or target bacteria is a
Francisella host. Optionally, host and/or target bacteria is a Xanthomonas
host. Optionally, host
and/or target bacteria is a Agro bacterium host. Optionally, host and/or
target bacteria is a Erwinia
host. Optionally, host and/or target bacteria is a Legionella (eg,
pneumophila). Optionally, host
and/or target bacteria is a Listeria (eg, monocytogenes). Optionally, host
and/or target bacteria is a
Campylobacter (eg, jejuni). Optionally, host and/or target bacteria is a
Yersinia (eg, pestis).
Optionally, host and/or target bacteria is a Borelia (eg, burgdorferi).
Optionally, host and/or target
bacteria is a Helicobacter (eg, pylori). Optionally, host and/or target
bacteria is a Clostridium (eg,
dificile or botulinum). Optionally, host and/or target bacteria is a Erlichia
(eg, chaffeensis).
Optionally, host and/or target bacteria is a Salmonella (eg, typhi or
enterica, eg, serotype
typhimurium, eg, DT 104). Optionally, host and/or target bacteria is a
Chlamydia (eg, pneumoniae).
Optionally, host and/or target bacteria is a Parachlamydia host. Optionally,
host and/or target
bacteria is a Cotynebacterium (eg, amycolatum). Optionally, host and/or target
bacteria is a Klebsiella
(eg, pneumoniae). Optionally, host and/or target bacteria is an Enterococcus
(eg, faecalis or faecim,
eg, linezolid-resistant). Optionally, host and/or target bacteria is an
Acinetobacter (eg, baumannii, eg,
multiple drug resistant).
Further examples of target cells and targeting of antibiotic resistance in
such cells using the present
invention are as follows:-
1. Optionally the target bacteria are Staphylococcus aureus cells, eg,
resistant to an antibiotic
selected from methicillin, vancomycin, linezolid, daptomycin, quinupristin,
dalfopristin and
teicoplanin.
2. Optionally the target bacteria are Pseudomonas aeuroginosa cells, eg,
resistant to an
antibiotic selected from cephalosporins (eg, ceftazidime), carbapenems (eg,
imipenem or
meropenem), fluoroquinolones, aminoglycosides (eg, gentamicin or tobramycin)
and colistin.
3. Optionally the target bacteria are Klebsiella (eg, pneumoniae) cells,
eg, resistant to
carbapenem.
4. Optionally the target bacteria are Streptoccocus (eg, thermophilus,
pneumoniae or pyogenes)
cells, eg, resistant to an antibiotic selected from erythromycin, clindamycin,
beta-lactam, macrolide,
amoxicillin, azithromycin and penicillin.
5. Optionally the target bacteria are Salmonella (eg, serotype Typhi)
cells, eg, resistant to an
antibiotic selected from ceftriaxone, azithromycin and ciprofloxacin.
6. Optionally the target bacteria are Shigella cells, eg, resistant to an
antibiotic selected from
ciprofloxacin and azithromycin.
29
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
7. Optionally the target bacteria are mycobacterium tuberculosis cells, eg,
resistant to an
antibiotic selected from Resistance to isoniazid (INH), rifampicin (RMP),
fluoroquinolone, amikacin,
kanamycin and capreomycin and azithromycin.
8. Optionally the target bacteria are Enterococcus cells, eg, resistant to
vancomycin.
9. Optionally the target bacteria are Enterobacteriaceae cells, eg,
resistant to an antibiotic
selected from a cephalosporin and carbapenem.
10. Optionally the target bacteria are E. coli cells, eg, resistant to an
antibiotic selected from
trimethoprim, itrofurantoin, cefalexin and amoxicillin.
11. Optionally the target bacteria are Clostridium (eg, dificile) cells,
eg, resistant to an antibiotic
selected from fluoroquinolone antibiotic and carbapenem.
12. Optionally the target bacteria are Neisseria gonnorrhoea cells, eg,
resistant to an antibiotic
selected from cefixime (eg, an oral cephalosporin), ceftriaxone (an injectable
cephalosporin),
azithromycin and tetracycline.
13. Optionally the target bacteria are Acinetoebacter baumannii cells, eg,
resistant to an antibiotic
selected from beta-lactam, meropenem and a carbapenem.
14. Optionally the target bacteria are Campylobacter cells, eg, resistant
to an antibiotic selected
from ciprofloxacin and azithromycin.
15. Optionally, the target cell(s) produce Beta (13)-lactamase.
16. Optionally, the target cell(s) are bacterial cells that are resistant
to an antibiotic recited in any
one of examples 1 to 14.
Mobile Genetic Elements, Genomic Islands, Pathogenicity Islands etc.
[0072] Genetic variation of bacteria and archaea can be achieved through
mutations, rearrangements
and horizontal gene transfers and recombinations. Increasing genome sequence
data have
demonstrated that, besides the core genes encoding house-keeping functions
such as essential
metabolic activities, information processing, and bacterial structural and
regulatory components, a
vast number of accessory genes encoding antimicrobial resistance, toxins, and
enzymes that
contribute to adaptation and survival under certain environmental conditions
are acquired by
horizontal gene transfer of mobile genetic elements (MGEs). Mobile genetic
elements are a
heterogeneous group of molecules that include plasmids, bacteriophages,
genomic islands,
chromosomal cassettes, pathogenicity islands, and integrative and conjugative
elements. Genomic
islands are relatively large segments of DNA ranging from 10 to 200 kb often
integrated into tRNA
gene clusters flanked by 16-20 bp direct repeats. They are recognized as
discrete DNA segments
acquired by horizontal gene transfer since they can differ from the rest of
the chromosome in terms of
GC content (%G+C) and codon usage.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[0073] Pathogenicity islands (PTIs) are a subset of horizontally transferred
genetic elements known
as genomic islands. There exists a particular family of highly mobile PTIs in
Staphylococcus
aureus that are induced to excise and replicate by certain resident prophages.
These PTIs are
packaged into small headed phage-like particles and are transferred at
frequencies commensurate with
the plaque-forming titer of the phage. This process is referred to as the SaPI
excision replication-
packaging (ERP) cycle, and the high-frequency SaPI transfer is referred to as
SaPI-specific transfer
(SPST) to distinguish it from classical generalized transduction (CGT). The
SaPIs have a highly
conserved genetic organization that parallels that of bacteriophages and
clearly distinguishes them
from all other horizontally acquired genomic islands. The SaPThencoded and
SaPIbov2-encoded
integrases are used for both excision and integration of the corresponding
elements, and it is assumed
that the same is true for the other SaPIs. Phage 80a can induce several
different SaPIs, including
SaPI1, SaPI2, and SaPIbovl, whereas 911 can induce SaPIbovl but neither of the
other two SaPIs.
[0074] Reference is made to "Staphylococcal pathogenicity island DNA packaging
system involving
cos-site packaging and phage-encoded HNH endonucleases", Quiles-Puchalt et al,
PNAS April 22,
2014. 111(16) 6016-6021. Staphylococcal pathogenicity islands (SaPIs) are
highly mobile and carry
and disseminate superantigen and other virulence genes. It was reported that
SaPIs hijack the
packaging machinery of the phages they victimise, using two unrelated and
complementary
mechanisms. Phage packaging starts with the recognition in the phage DNA of a
specific sequence,
termed "pac" or "cos" depending on the phage type. The SaPI strategies involve
carriage of the helper
phage pac- or cos-like sequences in the SaPI genome, which ensures SaPI
packaging in full-sized
phage particles, depending on the helper phage machinery. These strategies
interfere with phage
reproduction, which ultimately is a critical advantage for the bacterial
population by reducing the
number of phage particles.
[0075] Staphylococcal pathogenicity islands (SaPIs) are the prototypical
members of a widespread
family of chromosomally located mobile genetic elements that contribute
substantially to intra- and
interspecies gene transfer, host adaptation, and virulence. The key feature of
their mobility is the
induction of SaPI excision and replication by certain helper phages and their
efficient encapsidation
into phage-like infectious particles. Most SaPIs use the headful packaging
mechanism and encode
small terminase subunit (TerS) homologs that recognize the SaPI-specific pac
site and determine SaPI
packaging specificity. Several of the known SaPIs do not encode a recognizable
TerS homolog but are
nevertheless packaged efficiently by helper phages and transferred at high
frequencies. Quiles-Puchalt
et al report that one of the non¨terS-coding SaPIs, SaPIbov5, and found that
it uses two different,
undescribed packaging strategies. SaPIbov5 is packaged in full-sized phage-
like particles either by
typical pac-type helper phages, or by cos-type phages¨i.e., it has both pac
and cossites and uses the
two different phage-coded TerSs. This is an example of SaPI packaging by a cos
phage, and in this, it
31
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
resembles the P4 plasmid of Escherichia coll. Cos-site packaging in
Staphylococcus aureus is
additionally unique in that it requires the HNH nuclease, carried only by cos
phages, in addition to the
large terminase subunit, for cos-site cleavage and melting.
[0076] Characterization of several of the phage-inducible SaPIs and their
helper phages has
established that the pac (or headful) mechanism is used for encapsidation. In
keeping with this
concept, some SaPIs encode a homolog of TerS, which complexes with the phage-
coded large
terminase subunit TerL to enable packaging of the SaPI DNA in infectious
particles composed of
phage proteins. These also contain a morphogenesis (cpm) module that causes
the formation of small
capsids commensurate with the small SaPI genomes. Among the SaPI sequences
first characterized,
there were several that did not include either a TerS homolog or a cpm
homolog, and the same is true
of several subsequently identified SaPIs from bovine sources and for many
phage-inducible
chromosomal islands from other species. It was assumed, for these several
islands, either that they
were defective derivatives of elements that originally possessed these genes,
or that terS
and cpm genes were present but not recognized by homology.
[0077] Quiles-Puchalt et al observed that an important feature of
4SLT/SaPIbov5 packaging is the
requirement for an HNH nuclease, which is encoded next to the OSLT terminase
module. Proteins
carrying HNH domains are widespread in nature, being present in organisms of
all kingdoms. The
HNH motif is a degenerate small nucleic acid-binding and cleavage module of
about 30-40 aa
residues and is bound by a single divalent metal ion. The HNH motif has been
found in a variety of
enzymes playing important roles in many different cellular processes,
including bacterial killing;
DNA repair, replication, and recombination; and processes related to RNA. HNH
endonucleases are
present in a number of cos-site bacteriophages of Gram-positive and -negative
bacteria, always
adjacent to the genes encoding the terminases and other morphogenetic
proteins. Quiles-Puchalt et al
have demonstrated that the HNH nucleases encoded by 4)12 and the closely
related OSLT have
nonspecific nuclease activity and are required for the packaging of these
phages and of SaPIbov5.
Quiles-Puchalt et al have shown that HNH and TerL are jointly required for cos-
site cleavage.
Quiles-Puchalt et al have also observed that only cos phages of Gram-negative
as well as of Gram-
positive bacteria encode HNH nucleases, consistent with a special requirement
for cos-site cleavage
as opposed to pac-site cleavage, which generates flush-ended products. The
demonstration that HNH
nuclease activity is required for some but not other cos phages suggests that
there is a difference
between the TerL proteins of the two types of phages¨one able to cut both
strands and the other
needing a second protein to enable the generation of a double-stranded cut.
[0078] The invention, also involves, in certain configurations the use of
mobile genetic elements
(MGEs). Thus, there are provided the following Clauses. Any of the other
configurations, Aspects,
32
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Examples or description of the invention above or elsewhere herein are
combinable mutatis mutandis
with any of these Clauses:-
1. A composition for use in antibacterial treatment of bacteria, the
composition comprising an
engineered mobile genetic element (MGE) that is capable of being mobilised in
a first bacterial host
cell of a first species or strain, the cell comprising a first phage genome,
wherein in the cell the MGE
is mobilised using proteins encoded by the phage and replication of first is
inhibited, wherein the
MGE encodes an antibacterial agent or encodes a component of such an agent.
In the alternative, instead of a bacteria, the host cell is a archaeal cell
and instead of a phage there is a
virus that is capable of infecting the archaeal cell.
In an example, the MGE is capable of integration into the genome of the host
cell comprising the
genome of a first phage, for example integration in the chromosome of the host
cell and/or an episome
thereof.
Optionally, the MGE inhibits first phage replication.
In an example, first phage replication is totally inhibited. In an example, it
is reduced by at least 50,
60, 70, 80 or 90% compared to replication in the absence of the MGE in host
cells. This can be
assessed by a standard in vitro plaque assay to determine the relative amount
of first phage plaque
formation.
Optionally, in the presence of the agent,
(i)host cells are killed by the antibacterial agent;
(ii) growth or proliferation of host cells is reduced; and/or
(iii)host cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
2. The composition of Clause 1, wherein the agent is toxic to cells of the
same species or strain
as the host cell.
3. The composition of Clause 1 or 2, wherein the agent is toxic to cells of
a species or strain that
is different from the strain or species of the host cell.
4. The composition of Clause 1, wherein the agent is toxic to cells of the
same species as the
host cell, and wherein the host cell has been engineered so that the agent is
not toxic to the host cell.
5. The composition of Clause 4, wherein the agent is a guided nuclease
system (optionally a
CRISPR/Cas system) and cells of the same species as the host cell comprise a
target sequence that is
cut by the nuclease, wherein the target sequence has been removed or altered
in the host cell whereby
the nuclease is not capable of cutting the target sequence.
33
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Viruses undergo lysogenic and lytic cycles in a host cell. If the lysogenic
cycle is adopted, the phage
chromosome can be integrated into the bacterial chromosome, or it can
establish itself as a stable
plasmid in the host, where it can remain dormant for long periods of time. If
the lysogen is induced,
the phage genome is excised from the bacterial chromosome and initiates the
lytic cycle, which
culminates in lysis of the cell and the release of phage particles. The lytic
cycle leads to the
production of new phage particles which are released by lysis of the host.
6. The composition of any preceding Clause, wherein the first phage is a
temperate phage.
7. The composition of any preceding Clause, wherein the first cell
comprises the first phage as a
prophage.
8. The composition of any one of Clauses 1 to 5, wherein the first phage is
a lytic phage.
9. The composition of any preceding Clause, wherein in the presence of a
first phage the
mobilisation of the MGE causes host cell lysis.
10. The composition of any preceding Clause, wherein the MGE is capable of
being packaged in
transduction particles that comprise some, but not all, structural proteins of
the first phage.
"Transduction particles" may be phage or smaller than phage and are particles
that are capable of
transducing nucleic acid encoding the antibiotic or component thereof or other
DNA into target
bacterial cells.
In some examples, instead of a nucleic acid encoding an antibiotic or
component thereof, there is used
a nucleic acid sequence of interest (eg, any NSI disclosed herein) that
encodes a protein or RNA of
interest for expression in the cell into which transduction from a
transduction particle can take place.
Examples of structural proteins are phage proteins selected from one, more or
all of the major head
and tail proteins, the portal protein, tail fibre proteins, and minor tail
proteins.
The MGE comprises a packaging signal sequence operable with proteins encoded
by the first phage
to package the MGE (or at least nucleic acid thereof encoding the agent or one
or more components
thereof) into transduction particles that are capable of infecting host cells
of the same species or strain
as the first host cell.
11. The composition of any preceding Clause, wherein mobilisation of the
MGE comprises
packaging of copies of the MGE or nucleic acid encoding the agent or component
into transduction
particles that are capable of transferring the copies into target bacterial
cells for antibacterial treatment
of the target cells.
34
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
12. The composition of Clause 10 or 11, wherein the transduction particles
are particles of second
phage that are capable of infecting cells of said first species or strain.
13. The composition of any one of Clauses 10 to 12, wherein the
transduction particles are non-
self replicative particles.
A "non-self replicative transduction particle" refers to a particle, (eg, a
phage or phage-like particle;
or a particle produced from a genomic island (eg, a SaPI) or a modified
version thereof) capable of
delivering a nucleic acid molecule encoding an antibacterial agent or
component (or any protein or
RNA) into a bacterial cell, but does not package its own replicated genome
into the transduction
particle. In an alternative herein, instead of a phage, there is used or
packaged a virus that infects an
animal, human, plant or yeast cell. For example, an adenovirus when the cell
is a human cell.
14. The composition of any preceding Clause, wherein the MGE is devoid of
genes encoding
phage structural proteins.
Optionally, the MGE is devoid of one or more phage genes rinA, terS and terL.
In an example, in a host cell a protein complex comprising the small terminase
(encoded by terS) and
large terminase (encoded by terL) proteins is able to recognise and cleave a
double-stranded DNA
molecule of the MGE at or near the pac site (cos site or other packaging
signal sequence comprised
by the MGE), and this allows the MGE or plasmid DNA molecule to be packaged
into a phage capsid.
When first phage as prophage in the host cell is induced, the lytic cycle of
the phage produces the
phage's structural proteins and the phage's large terminase protein. The MGE
or plasmid is replicated,
and the small terminase protein encoded by the MGE or plasmid is expressed.
The replicated MGE or
plasmid DNA containing the terS (and the nucleotide sequence encoding the
antibacterial agent or
component) are packaged into phage capsids, resulting in non-self replicative
transduction particles
carrying only MGE or plasmid DNA.
15. The composition of any one of Clauses 1 to 13, wherein the MGE
comprises phage structural
genes and a packaging signal sequence and the first phage is devoid of a
packaging signal sequence.
16. The composition of any preceding Clause, wherein the MGE is a modified
version of a MGE
that is naturally found in bacterial cells of the first species or strain.
17. The composition of any preceding Clause, wherein the MGE comprises a
modified genomic
island.
Optionally, the genomic island is an island that is naturally found in
bacterial cells of the first species
or strain. In an example, the genomic island is selected from the group
consisting of a SaPI, a SaPI1,
a SaPI2, a SaPIbov 1 and a SaPibov2 genomic island.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
18. The composition of any preceding Clause, wherein the MGE comprises a
modified
pathogenicity island.
Optionally, the pathogenicity island is an island that is naturally found in
bacterial cells of the first
species or strain, eg, a Staphylococcus SaPI or a Vibro PLE or a P. aeruginosa
pathogenicity island
(eg, a PAPI or a PAGI, eg, PAPI-1, PAGI-5, PAGI-6, PAGI-7, PAGI-8, PAGI-9,
PAGI-10, or PAGI-
19. The composition of Clause 18, wherein the pathogenicity island is a
SaPI (S aureus
pathogenicity island).
20. The composition of Clause 19, wherein the first phage is 011, 80a, 012
or 0SLT.
Staphylococcus phage 80a appears to mobilise all known SaPIs. Thus, in an
example, the MGE
comprises a modified SaPI and the first phage is a 80a.
21. The composition of Clause 18, wherein the pathogenicity island is a V.
cholerae PLE (phage-
inducible chromosomal island-like element) and optionally the first phage is
ICP1.
22. The composition of Clause 18, wherein the pathogenicity island is a E
coli PLE.
23. The composition of any one of Clauses 1 to 16, wherein the MGE
comprises P4 DNA, eg, a
P4 packaging signal sequence.
24. The composition of Clause 23, wherein the first phage are P2 phage or a
modified P2 phage
that is self-replicative defective; optionally present as a prophage.
25. The composition of any preceding Clause, wherein the MGE comprises a
pacA gene of the
Enterobacteriaceae bacteriophage Pl.
26. The composition of any preceding Clause, wherein the MGE comprises a
packaging initiation
site sequence, optionally a packaging initiation site sequence of Pl.
27. The composition of any preceding Clause, wherein the MGE comprises a
nucleotide sequence
that is beneficial to cells of the first species or strain, optionally
encoding a protein that is beneficial to
cells of the first species or strain.
This is useful where, not only does the presence of the MGE reduce first phage
replication in the host
cell, but also the MGE is taken up and may provide a survival, growth or other
benefit to the host cell,
promoting uptake and/or retention of MGEs by host cells. In an example,
expression of the
antibacterial agent in the host cell is under the control of an inducible
promoter or weak promoter to
allow for a period where uptake of MGEs into host cells may be favoured owing
to the presence of the
nucleotide sequence that is beneficial to cells of the first species or
strain.
28. The composition of any preceding Clause, wherein the MGE is devoid of
rinA.
29. The composition of any preceding Clause, wherein the MGE is is devoid
of terL.
36
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
30. The composition of any preceding Clause, wherein the MGE comprises a
terS or a homologue
thereof, and optionally is devoid of any other terminase gene.
The terS homologues are sequences which, like terS, recognise the SaPI-
specific pac site (or other
packaging sequence) comprised by the MGE or plasmid and determine packaging
specificity for
packaging the MGE.
Examples of terminase genes are pacA, pacB, terA, terB and terL.
31. The composition of any preceding Clause, wherein the first phage is a
pac-type phage (eg,
011 or 80a) operable with a pac comprised by the MGE.
32. The composition of any one of Clauses 1 to 30, wherein the first phage
is a cos-type phage
(eg, 012 or 0SLT) operable with a cos comprised by the MGE.
Optionally, the phage is P2. Optionally, the first phage is a T7 or T7-like
phage that recognises direct
repeat sequences comprised by the MGE for packaging.
33. The composition of any preceding Clause, wherein the plasmid or MGE
comprises a pac
and/or cos sequence or a homologue thereof.
34. The composition of any preceding Clause, wherein the plasmid or MGE
comprises a terS or a
homologue thereof and optionally devoid of terL.
The terS homologues are sequences which, like terS, recognise the SaPI-
specific pac site (or other
packaging sequence) comprised by the MGE or plasmid and determine packaging
specificity for
packaging the MGE.
In an example, the terS comprises the sequence of SEQ ID NO: 1.
35. The composition of Clause 34, wherein the terS is a S aureus
bacteriophage 980a terS or a
bacteriophage 911 terS.
36. The composition of any preceding Clause, wherein the MGE is a modified
SaPIbov 1 or
SaPIbov5 and is devoid of a terS.
37. The composition of any preceding Clause, wherein the first phage is
devoid of a functional
packaging signal sequence and the MGE comprises a packaging signal sequence
operable with
proteins encoded by the first phage for producing transduction particles that
package copies of the
MGE or copies of a nucleic acid encoding the agent or component.
38. The composition of any preceding Clause, wherein the MGE or plasmid
comprises a Ppi or
homologue, which is capable of complexing with first phage TerS, thereby
blocking function of the
TerS.
37
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
39. The composition of any preceding Clause, wherein the MGE comprises a
morphogenesis
(cpm) module.
40. The composition of any preceding Clause, wherein the MGE comprises cpmA
and/or cpmB.
Optionally the cpmA and B are from any SaPI disclosed herein. In an example
any SaPI is a SaPI
disclosed in Table 2 and optionally the host cell or target cell is any
corresponding Staphylococcus
disclosed in the table.
41. The composition of any preceding Clause, wherein the MGE or first phage
comprises one,
more or all genes cpl, cp2, and cp3.
In an example, the MGE comprises a modified SaPI and comprises one, more or
all genes cpl, cp2,
and cp3.
42. The composition of any preceding Clause, wherein the MGE or first phage
encodes a HNH
nuclease.
43. The composition of any preceding Clause, wherein the MGE or first phage
comprises an
integrase gene that encodes an integrase for excising the MGE and integrating
the MGE into a
bacterial cell genome.
44. The composition of any preceding Clause, wherein the MGE is devoid of a
functional
integrase gene, and the first phage or host cell genome (eg, bacterial
chromosome or a bacterial
episome) comprises a functional integrase gene.
45. The composition of any preceding Clause, wherein the transcription of
MGE nucleic acid is
under the control of a constitutive promoter, for transcription of copies of
the agent or component in a
host cell.
Optionally, Constitutive transcription and production of the agent in target
cells may be used where
the target cells should be killed, eg, in medical settings.
Optionally, the transcription of MGE nucleic acid is under the control of an
inducible promoter, for
transcription of copies of the agent or component in a host cell. This may be
useful, for example, to
control switching on of the antibacterial activity against target bacterial
cells, such as in an
environment (eg, soil or water) or in an industrial culture or fermentation
container containing the
target cells. For example, the target cells may be useful in an industrial
process (eg, for fermentation,
eg, in the brewing or dairy industry) and the induction enables the process to
be controlled (eg,
stopped or reduced) by using the antibacterial agent against the target
bacteria.
38
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
46. The composition of Clause 45, wherein the promoter is foreign to the
host cell.
47. The composition of Clause 45 or 46, wherein the promoter comprises a
nucleotide sequence
that is at least 80% identical to an endogenous promoter sequence of the host
cell.
48. The composition of any preceding Clause comprising a nucleic acid that
is separate from the
MGE, wherein the nucleic acid comprises all genes necessary for producing
first phage particles.
49. The composition of any one of Clauses 1 to 47 comprising a nucleic acid
that is separate from
the MGE, wherein the nucleic acid comprises less than, all genes necessary for
producing first phage
particles, but comprises genes encoding structural proteins for production of
transduction particles
that package MGE nucleic acid encoding the antibacterial agent or one or more
components thereof.
When the agent comprises a plurality of components, eg, wherein the agent is a
CRISPR/Cas system,
or is a CRISPR array encoding crRNA or a nucleic acid encoding a guide RNA
(eg, single guide
RNA) operable with a Cas in host cells, wherein the crRNA or gRNA guides the
Cas to a target
sequence in the host cell to modify the target (eg, cut it or repress
transcription from it).
50. The composition of Clause 48 or 49, wherein the genes are comprised by
the host cell
chromosome and/or one or more host cell episome(s).
51. The composition of Clause 50, wherein the genes are comprised by a
chromosomally-
integrated prophage of the first phage.
52. The composition of any preceding Clause, wherein the agent is a guided
nuclease system or a
component thereof, wherein the agent is capable of recognising and cutting
host cell DNA (eg,
chromosomal DNA).
In examples, such cutting causes one or more of the following:-
(i)The host cell is killed by the antibacterial agent;
(ii) growth or proliferation of the host cell is reduced; and/or
(iii)The host cell is sensitised to an antibiotic, whereby the antibiotic is
toxic to the cell.
53. The composition of Clause 52, wherein the guided nuclease system is
selected from a
CRISPR/Cas system, TALEN system, meganuclease system or zinc finger system.
54. The composition of Clause 52, wherein the system is a CRISPR/Cas system
and each MGE
encodes a (a) CRISPR array encoding crRNA or (b) a nucleic acid encoding a
guide RNA (gRNA, eg,
single guide RNA), wherein the crRNA or gRNA is operable with a Cas in target
bacterial cells,
wherein the crRNA or gRNA guides the Cas to a target nucleic acid sequence in
the host cell to
modify the target sequence (eg, cut it or repress transcription from it).
39
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Optionally, the Cas is a Cas encoded by a functional endogenous nucleic acid
of a host cell. For
example, the target is comprised by a DNA or RNA of the host cell.
55. The composition of Clause 52, wherein the system is a CRISPR/Cas system
and each MGE
encodes a Cas (eg, a Cas nuclease) that is operable in a target bacterial
cells to modify a target nucleic
acid sequence comprised by the target cell.
56. The composition of Clause 53, 54 or 55, wherein the Cas is a Cas3,
Cas9, Cas13, CasX, CasY
or Cpfl.
57. The composition of any one of Clauses 52 to 56, wherein the system is a
CRISPR/Cas system
and each MGE encodes one or more Cascade Cas (eg, Cas, A, B, C, D and E).
58. The composition of any one of Clauses 52 to 57, wherein each MGE
further encodes a Cas3
that is operable in a target bacterial cell with the Cascade Cas.
59. The composition of any preceding Clause, wherein the first species or
strain is a gram
positive species or strain.
60. The composition of any one of Clauses 1 to 58, wherein the first
species or strain is a gram
negative species or strain.
61. The composition of any preceding Clause, wherein the first species or
strain is selected from
Table 1.
In an example, the first species of strain is a Staphylococcus (eg, S aureus)
species or strain and
optionally the MGE is a modified SaPI; and optionally the first phage is a
980a or 911. In an
example, the first species of strain is a Vibrio (eg, V cholerae) species or
strain and optionally the
MGE is Vibrio (eg, V cholerae) PLE.
62. The composition of any preceding Clause, wherein the first species or
strain is selected from
Shigella, E coli, Salmonella, Serratia, Klebsiella, Yersinia, Pseudomonas and
Enterobacter.
These are species that P2 phage can infect. Thus, in an embodiment, the MGE
comprises one or more
P4 sequences (eg, a P4 packaging sequence) and the first phage is P2. Thus,
the MGE is packaged by
P2 structural proteins and the resultant transduction particles can infect a
broad spectrum of species,
ie, two or more of Shigella, E coli, Salmonella, Serratia, Klebsiella,
Yersinia, Pseudomonas and
Enterobacter.
63. A nucleic acid vector comprising a MGE integrated therein, wherein the
MGE is according to
any preceding Clause and the vector is capable of transferring the MGE or a
copy thereof into a host
bacterial cell.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Suitable vectors are plasmids (eg, conjugative plasmids) or viruses (eg, phage
or packaged
phagemids).
64. The vector of Clause 63, wherein the vector is a shuttle vector.
A shuttle vector is a vector (usually a plasmid) constructed so that it can
propagate in two different
host species. Therefore, DNA inserted into a shuttle vector can be tested or
manipulated in two
different cell types.
65. The vector of Clause 63, wherein the vector is a plasmid, wherein
the plasmid is capable of
being transformed into a host bacterial cell comprising a first phage.
66. A non-self replicative transduction particle comprising said MGE or
vector of any preceding
.. Clause.
By "non-replicative" it is meant that the MGE is not capable by itself of self-
replicating. For
example, the MGE is devoid of one or more nucleotide sequences encoding a
protein (eg, a structural
protein) that is necessary to produce a transduction particle comprising a
copy of the MGE.
67. A composition comprising a plurality of transduction particles,
wherein each particle
comprises a MGE or vector according to any one of Clauses 1 to 65, wherein the
transduction
particles are capable of transferring the MGEs, or nucleic acid encoding the
agent or component, or
copies thereof into target bacterial cells, wherein
a. target cells are killed by the antibacterial agent;
b. growth or proliferation of target cells is reduced; or
c. target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
In an example, the reduction in growth or proliferation of host cells is at
least 50, 60, 70, 80, 90 or
95%. The antibiotic can be any antibiotic disclosed herein.
68. The composition of Clause 67, wherein the agent is a guided nuclease
system or a component
thereof, wherein the agent is capable of recognising and cutting host cell DNA
(eg, chromosomal
DNA) whereby
a. target cells are killed by the antibacterial agent;
b. growth or proliferation of target cells is reduced; or
c. target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
69. A composition comprising a plurality of non-self replicative
transduction particles, wherein
each particle comprises a MGE or plasmid according to any one of Clauses 1 to
65, wherein the
transduction particles are capable of transferring the MGEs, or nucleic acid
encoding the agent or
41
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
component, or copies thereof into target bacterial cells, wherein the agent is
a CRISPR/Cas system
and the component comprises a nucleic acid encoding a crRNA or a guide RNA
that is operable with
a Cas in a target bacterial cell to guide the Cas to a target nucleic acid
sequence of the cell to modify
the sequence, whereby
a. target cells are killed by the antibacterial agent;
b. growth or proliferation of target cells is reduced; or
c. target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
In an example, the reduction in growth or proliferation of host cells is at
least 50, 60, 70, 80, 90 or
95%. The antibiotic can be any antibiotic disclosed herein.
70. A kit comprising the composition of Clause 69 and said antibiotic.
71. The composition of Clause 69, wherein the composition comprises said
antibiotic.
72. The composition of any one of Clauses 67 to 69, wherein less than 10%
of transduction
particles comprise by the composition are first phage particles.
73. The composition of any one of Clauses 67 to 69, wherein no first phage
particles are present
in the composition.
74. The MGE, vector, particle, composition or kit of any preceding
Clause for medical use in a
human or animal patient.
75. The MGE, vector, particle, composition or kit of any preceding
Clause for treating or
preventing an infection by target bacterial cells in a human or animal
patient, wherein the antibacterial
agent is toxic to the target cells.
76. The MGE, vector, particle, composition or kit of any preceding
Clause for treating or
preventing an infection by target bacterial cells in a human or animal
patient, wherein in the presence
of the antibacterial agent
a. target cells are killed by the antibacterial agent;
b. growth or proliferation of target cells is reduced; and/or
c. target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
77. A method of producing a plurality of transduction particles, the
method comprising
combining the composition of any one of Clauses 1 to 62, 67 to 69 and 71 to 76
with host bacterial
cells of said first species, wherein the cells comprise the first phage,
allowing a plurality of said
MGEs to be introduced into host cells and culturing the host cells under
conditions in which first
phage-encoded proteins are expressed and MGE copies are packaged by first
phage proteins to
produce a plurality of transduction particles, and optionally separating the
transduction particles from
cells and obtaining a plurality of transduction particles separated from
cells.
42
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
78. The method of Clause 77, comprising separating the transduction
particles from any first
phage, optionally by filtering or centrifugation, thereby obtaining a
plurality of transduction particles
in the absence of first phage.
79. The method of Clause 77 or 78, wherein the particles encode a guided
nuclease system
(optionally a CRISPR/Cas system) or component thereof for cutting a target
nucleic acid sequence
comprised by target bacterial cells.
80. The method of Clause 79, wherein the sequence is comprised by an
antibiotic resistance gene
and the method comprises combining the plurality of particles with said
antibiotic in a kit or a
mixture.
81. The method of any one of Clauses 77 to 80, wherein said conditions
comprise induction of a
lytic cycle of the first phage.
82. A bacterial host cell comprising a first phage and a MGE, vector or
particle as recited in any
one of Clauses 1 to 66, wherein the agent is toxic to cells of the same
species as the host cell, and
wherein the host cell has been engineered so that the agent is not toxic to
the host cell.
83. A bacterial host cell comprising a first phage, wherein the cell is
comprised by a kit, the kit
further comprising a composition as recited in any one of Clauses 1 to 62, 67
to 69 and 71 to 76,
wherein the agent is toxic to cells of the same species as the host cell, and
wherein the host cell has
been engineered so that the agent is not toxic to the host cell.
84. The cell of Clause 83, wherein the agent is a guided nuclease system
(optionally a
CRISPR/Cas system) and cells of the same species as the host cell comprise a
target sequence that is
cut by the nuclease, wherein the target sequence has been removed or altered
in the host cell whereby
the nuclease is not capable of cutting the target sequence.
85. A bacterial host cell comprising a first phage and a MGE, vector or
particle as recited in any
one of Clauses 1 to 66, wherein the agent is not toxic to the host cell, but
the agent is toxic to second
cells of a species or strain that is different from the species or strain of
the host cell, wherein the MGE
is mobilizable in transduction particles producible by the host cell that are
capable of transferring the
MGE or a copy thereof into a said second cell, whereby the second cell is
exposed to the antibacterial
agent.
86. A bacterial host cell comprising a first phage, wherein the cell is
comprised by a kit, the kit
further comprising a composition as recited in any one of Clauses 1 to 62, 67
to 69 and 71 to 76,
wherein the agent is not toxic to the host cell, but the agent is toxic to
second cells of a species or
strain that is different from the species or strain of the host cell, wherein
the MGE is mobilizable in
transduction particles producible by the host cell that are capable of
transferring the MGE or a copy
thereof into a said second cell, whereby the second cell is exposed to the
antibacterial agent.
87. The cell of Clause 86, wherein the first phage is a prophage.
43
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
88. A bacterial host cell comprising a MGE, vector or particle as recited
in any one of Clauses 1
to 66 and nucleic acid under the control of one or more inducible promoters,
wherein the nucleic acid
encodes all structural proteins necessary to produce a transduction particle
that packages a copy of the
MGE or plasmid, wherein the agent is not toxic to the host cell, but the agent
is toxic to second cells
of a species or strain that is different from the species or strain of the
host cell, wherein the MGE is
mobilizable in transduction particles producible by the host cell that are
capable of transferring the
MGE or a copy thereof into a said second cell, whereby the second cell is
exposed to the antibacterial
agent.
89. The cell of Clause 88, wherein the structural proteins are structural
proteins of a lytic phage.
90. The cell of Clause 88 or 89, wherein the nucleic acid comprises terS
and/or terL.
91. The cell of any one of Clauses 88 to 90, wherein the host and second
cells are of the same
species and the host cell has been engineered so that the antibiotic is not
toxic to the host cell.
92. The cell of any one of Clauses 88 to 91, wherein the nucleic acid is
comprised by a plasmid.
93. The cell of any one of Clauses 88 to 92, wherein the agent is a
guided nuclease system
(optionally a CRISPR/Cas system) and the second cells comprise a target
sequence that is cut by the
nuclease, wherein the target sequence is absent in the genome of the host cell
whereby the nuclease is
not capable of cutting the host cell genome.
94. The composition, vector, particle, kit or method of any preceding
Clause, wherein the cell,
host cell or target cell is selected from a Staphylococcal, Vibrio,
Pseudomonas, Clostridium, E coli,
Helicobacter, Klebsiella and Salmonella cell.
95. A plasmid comprising
a. A nucleotide sequence encoding an antibacterial agent or component
thereof for expression in
target bacterial cells;
b. A constitutive promoter for controlling the expression of the agent or
component;
c. An optional terS nucleotide sequence;
d. An origin of replication (on); and
e. A phage packaging sequence (optionally pac, cos or a homologue thereof);
and
f. the plasmid being devoid of
g. All nucleotide sequences encoding phage structural proteins necessary
for the production of a
transduction particle (optionally a phage), or the plasmid being devoid of at
least one of such
sequences; and
h. Optionally terL.
96. The plasmid of Clause 95, wherein the antibacterial agent is a
CRISPR/Cas system and the
plasmid encodes a crRNa or guide RNA (eg, single gRNA) that is operable with a
Cas in the target
cells to guide the Cas to a target nucleotide sequence to modify (eg, cut) the
sequence, whereby
44
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
a. target cells are killed by the antibacterial agent;
b. growth or proliferation of target cells is reduced; or
c. target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
97. The plasmid of Clause 95 or 96, wherein the antibacterial agent is a
CRISPR/Cas system and
the plasmid encodes a Cas that is operable with a crRNa or guide RNA (eg,
single gRNA) in the
target cells to guide the Cas to a target nucleotide sequence to modify (eg,
cut) the sequence, whereby
a. target cells are killed by the antibacterial agent;
b. growth or proliferation of target cells is reduced; or
c. target cells are sensitised to an antibiotic, whereby the antibiotic is
toxic to the cells.
98. The plasmid of Clause 97, wherein the plasmid further encodes said
crRNA or gRNA.
99. A host cell comprising the plasmid of any one of Clauses 95 to 98,
wherein the host cell does
not comprise the target nucleotide sequence.
100. The host cell of Clause 99, wherein the cell is capable of replicating
the plasmid and
packaging the replicated plasmid in transduction particles that are capable of
infecting target bacterial
cells.
101. The host cell of Clause 99 or 100, wherein the host cell comprises,
integrated in the cell
chromosome and/or one or more episomes of the cell,
a. A terL;
b. An optional terS; and
c. Expressible nucleotide sequences encoding all structural proteins
necessary for the production
of transduction particles that package copies of the plasmid;
d. wherein the chromosome and episomes of the cell (other than said
plasmid) are devoid of a
phage packaging sequence, wherein the phage packaging sequence comprised by
the plasmid is
operable together with the product of said terS and terL in the production of
packaged plasmid.
102. The cell of Clause 101, wherein the terL, optional terS and nucleotide
sequences encoding the
structural proteins are comprised by a phage (optionally a prophage) genome in
the host cell.
103. A bacterial host cell comprising the genome of a helper phage that is
incapable of self-
replication, optionally wherein the genome is present as a prophage, and a
plasmid according to any
one of Clauses 95 to 98, wherein the helper phage is operable to package
copies of the plasmid in
transduction particles, wherein the particles are capable of infecting
bacterial target cells to which the
antibacterial agent is toxic.
104. The cell of Clause 103, wherein the host cell is a cell of first
species or strain and the target
cells are of the same species or strain, and optionally wherein the hosts cell
is an engineered cell that
to which the antibacterial agent is not toxic.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
105. The cell of Clause 103, wherein the host cell is a cell of first
species or strain and the target
cells are of a different species or strain, wherein the antibacterial agent is
not toxic to the host cell.
106. A method of making a plurality of transduction particles, the method
comprising culturing a
plurality of host cells according to any one of Clauses 103 to 105, optionally
inducing a lytic cycle of
the helper phage, and incubating the cells under conditions wherein
transducing particles comprising
packaged copies of the plasmid are created, and optionally separating the
particles from the cells to
obtain a plurality of transduction particles.
107. A plurality of transduction particles obtainable by the method of
Clause 106 for use in
medicine, eg, for treating or preventing an infection of a human or animal
subject by target bacterial
cells, wherein transducing particles are administered to the subject for
infecting target cells and killing
the cells using the antibacterial agent.
108. A method of making a plurality of transduction particles, the method
comprising
i.Producing host cells whose genomes comprise nucleic acid encoding structural
proteins necessary
to produce transduction particles that can package first DNA, wherein the
genomes are devoid of
a phage packaging signal, wherein the expression of the proteins is under the
control of inducible
promoter(s);
ii.Producing first DNA encoding an antibacterial agent or a component thereof
(eg, as defined in
any preceding Clause), wherein the DNA comprises a phage packaging signal;
iii.Introducing the DNA into the host cells;
iv.Inducing production of the structural proteins in host cells, whereby
transduction particles are
produced that package the DNA;
v.Optionally isolating a plurality of the transduction particles; and
vi.Optionally formulating the particles into a pharmaceutical composition for
administration to a
human or animal for medical use.
109. The method of Clause 108, wherein the DNA comprises a MGE as defined in
any preceding
Clause.
110. The method of Clause 108 or 109, wherein the structural proteins are
P2 phage proteins and
optionally the packaging signal is a P4 phage packaging signal.
111. The method of Clause 108 or 109, wherein the DNA comprises a modified
SaPI or a genomic
island DNA.
112. The method of any one of Clauses 108 to 111, wherein the cells in step
(iv) comprise a gene
encoding a helper phage activator, optionally wherein the activator is a P4
phage delta or ogr protein
when the structural proteins are P2 proteins; or the activator is a SaPI rinA,
ptiA, ptiB or ptiM when
the MGE comprises a modified SaPI; and optionally the expression of the
activator(s) is controlled by
an inducible promoter, eg, a T7 promoter.
46
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
113. The method of any one of Clauses 108 to 112, wherein the packaging
signal is P4 phage Sid
and/or psu; or the signal is SaPI cpmA and/or cpmB.
This is useful for packaging DNAs into smaller capsids.
114. The method of any one of Clauses 108 to 113, wherein the cell genomes
comprise prophages,
wherein each prophage comprises said nucleic acid encoding structural
proteins.
115. The method of Clause 114, wherein the prophages are P2 prophages devoid
of cos and
optionally one, more or all genes selected from int, cox orf78, B, oif80,
oif81, oif82, orf83, A, oif91,
tin, old, orf30 andfun(Z); and optionally the packaging signal of (ii) is a
cos or P4 packaging signal.
116. The method of Clause 114 or 115, wherein the prophages are P2
prophages devoid of cos and
comprising genes from Q to S, V to G and Fr to ogr.
117. The method of Clause 114, wherein the prophages are phill prophages
devoid of a packaging
signal and comprising gene 29 (terS) to gene 53 (lysin); and optionally the
packaging signal of (ii) is a
phill packaging signal.
118. A plurality of transduction particles obtainable by the method of any
one of Clauses 108 to
117.
119. The particles of Clause 118 for administration to a human or animal
for medical use.
Further Concepts of the invention are as follows:-
[0079] The present invention is optionally for an industrial or domestic use,
or is used in a method
for such use. For example, it is for or used in agriculture, oil or petroleum
industry, food or drink
industry, clothing industry, packaging industry, electronics industry,
computer industry,
environmental industry, chemical industry, aeorspace industry, automotive
industry, biotechnology
industry, medical industry, healthcare industry, dentistry industry, energy
industry, consumer products
industry, pharmaceutical industry, mining industry, cleaning industry,
forestry industry, fishing
.. industry, leisure industry, recycling industry, cosmetics industry,
plastics industry, pulp or paper
industry, textile industry, clothing industry, leather or suede or animal hide
industry, tobacco industry
or steel industry.
[0080] The present invention is optionally for use in an industry or the
environment is an industrial
environment, wherein the industry is an industry of a field selected from the
group consisting of the
medical and healthcare; pharmaceutical; human food; animal food; plant
fertilizers; beverage; dairy;
meat processing; agriculture; livestock farming; poultry farming; fish and
shellfish farming;
veterinary; oil; gas; petrochemical; water treatment; sewage treatment;
packaging; electronics and
computer; personal healthcare and toiletries; cosmetics; dental; non-medical
dental; ophthalmic; non-
medical ophthalmic; mineral mining and processing; metals mining and
processing; quarrying;
47
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
aviation; automotive; rail; shipping; space; environmental; soil treatment;
pulp and paper; clothing
manufacture; dyes; printing; adhesives; air treatment; solvents; biodefence;
vitamin supplements; cold
storage; fibre retting and production; biotechnology; chemical; industrial
cleaning products; domestic
cleaning products; soaps and detergents; consumer products; forestry; fishing;
leisure; recycling;
plastics; hide, leather and suede; waste management; funeral and undertaking;
fuel; building; energy;
steel; and tobacco industry fields.
[0081] In an example, the ifirst DNA, first phage or vector comprises a CRISPR
array that targets
target bacteria, wherein the array comprises one, or two or more spacers (eg,
2, 3, 4, 5, 6, 7, 8, 9 ,10,
20, 30, 40, 50 or more spacers) for targeting the genome of target bacteria.
[0082] In an example, the target bacteria are comprised by an environment as
follows. In an
example, the environment is a microbiome of a human, eg, the oral cavity
microbiome or gut
microbiome or the bloodstream. In an example, the environment is not an
environment in or on a
human. In an example, the environment is not an environment in or on a non-
human animal. In an
embodiment, the environment is an air environment. In an embodiment, the
environment is an
agricultural environment. In an embodiment, the environment is an oil or
petroleum recovery
environment, eg, an oil or petroleum field or well. In an example, the
environment is an environment
in or on a foodstuff or beverage for human or non-human animal consumption.
[0083] In an example, the environment is a a human or animal microbiome (eg,
gut, vaginal, scalp,
armpit, skin or oral cavity microbiome). In an example, the target bacteria
are comprised by a human
or animal microbiome (eg, gut, vaginal, scalp, armpit, skin or oral cavity
microbiome).
[0084] In an example, the DNAs, phage or composition of the invention are
administered
intranasally, topically or orally to a human or non-human animal, or is for
such administration. The
skilled person aiming to treat a microbiome of the human or animal will be
able to determine the best
route of administration, depending upon the microbiome of interest. For
example, when the
microbiome is a gut microbiome, administration can be intranasally or orally.
When the microbiome
is a scalp or armpit microbiome, administration can be topically. When the
microbiome is in the
mouth or throat, the administration can be orally.
[0085] In any use or method herein, in an embodiment the first phage are
contacted with the target
bacteria at a multiplicity of infection (MOI) of at least 0.5, 1, 2, 3, 4, 5,
6, 7, 8,9, 10, 15, 20, 25, 30,
35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100, 110, 120, 130, 140,
150, 160, 170, 180, 190,
200, 300, 400, 500, 600 or 700. For example, the MOI is from 20 to 200, from
20 to 100, fro 50 to
200, from 50 to 100, from 75 to 150, 100 or about 100, or 200 or about 200. In
an example, this may
be determined by obtaining a sample of the microbiome containing the target
bacteria (eg, a sample of
a waterway or gut microbiome of a subject) and determining the number of
CFU/ml or mg in the
48
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
sample and using this to titrate the phage dose at the desired MOT to be
exposed to the microbiome or
administered to the environment or subject to be treated.
[0086] In an example, the environment is harboured by a beverage or water (eg,
a waterway or
drinking water for human consumption) or soil. The water is optionally in a
heating, cooling or
industrial system, or in a drinking water storage container.
[0087] In an example, the host and/or target bacteraia are Finnicutes selected
from Anaerotruncus,
Acetanaerobacterium, Acetitomaculum, Acetivibrio, Anaerococcus, Anaerofilum,
Anaerosinus,
Anaerostipes, Anaerovorax, Butyrivibrio, Clostridium, Capracoccus,
Dehalobacter, Dialister, Dorea,
Enterococcus, Ethanoligenens, Faecalibacterium, Fusobacterium, Gracilibacter,
Guggenheimella,
Hespellia, Lachnobacterium, Lachnospira, Lactobacillus, Leuconostoc,
Megamonas,
Mitsuokella, Oribacterium, Oxobacter, Papillibacter,
Proprionispira,Pseudobutyrivibrio,
Pseudoramibacter, Roseburia, Ruminococcus, Sarcina, Seinonella,
Shuttleworthia, Sporobacter,
Sporobacterium, Streptococcus, Subdoligranulum, Syntrophococcus,
Thermobacillus, Turibacter and
Weisella.
[0088] In an example, the kit, DNA(s), first phage, helper phage, composition,
use or method is for
reducing pathogenic infections or for re-balancing gut or oral microbiota eg,
for treating or preventing
obesity or disease in a human or animal. For example, the first phage, helper
phage, composition, use
or method is for knocking-down Clostridium dificile or E coli bacteria in a
gut microbiota of a human
or animal.
[0089] In an example, the packaging signal, NPF and/or HPF consists or
comprises SEQ ID NO: 2 or
a structural or functional homologue thereof.
[0090] In an example, the packaging signal, NPF and/or HPF consists or
comprises SEQ ID NO: 2 or
a nucleotide sequence that is at least 70, 75, 80, 85, 90, 91, 92, 93, 94, 95,
96, 97, 98 or 99% identical
thereto.
[0091] In an example, the disease or condition is a cancer, inflammatory or
autoimmune disease or
condition, eg, obesity, diabetes IBD, a GI tract condition or an oral cavity
condition.
[0092] Optionally, the environment is comprised by, or the target bacteria are
comprised by, a gut
microbiota, skin microbiota, oral cavity microbiota, throat microbiota, hair
microbiota, armpit
microbiota, vaginal microbiota, rectal microbiota, anal microbiota, ocular
microbiota, nasal
microbiota, tongue microbiota, lung microbiota, liver microbiota, kidney
microbiota, genital
microbiota, penile microbiota, scrotal microbiota, mammary gland microbiota,
ear microbiota, urethra
microbiota, labial microbiota, organ microbiota or dental microbiota.
Optionally, the environment is
comprised by, or the target bacteria are comprised by, a plant (eg, a tobacco,
crop plant, fruit plant,
vegetable plant or tobacco, eg on the surface of a plant or contained in a
plant) or by an environment
(eg, soil or water or a waterway or acqueous liquid).
49
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[0093] Optionally, the disease or condition of a human or animal subject is
selected from
(a) A neurodegenerative disease or condition;
(b) A brain disease or condition;
(c) A CNS disease or condition;
(d) Memory loss or impairment;
(e) A heart or cardiovascular disease or condition, eg, heart attack,
stroke or atrial fibrillation;
(0 A liver disease or condition;
(g) A kidney disease or condition, eg, chronic kidney disease (CKD);
(h) A pancreas disease or condition;
(i) A lung disease or condition, eg, cystic fibrosis or COPD;
(i) A gastrointestinal disease or condition;
(k) A throat or oral cavity disease or condition;
(1) An ocular disease or condition;
(m) A genital disease or condition, eg, a vaginal, labial, penile or
scrotal disease or condition;
(n) A sexually-transmissible disease or condition, eg, gonorrhea, HIV
infection, syphilis or
Chlamydia infection;
(o) An ear disease or condition;
(13) A skin disease or condition;
(q) A heart disease or condition;
(r) A nasal disease or condition
(s) A haematological disease or condition, eg, anaemia, eg, anaemia of
chronic disease or cancer;
(t) A viral infection;
(u) A pathogenic bacterial infection;
(v) A cancer;
(w) An autoimmune disease or condition, eg, SLE;
(x) An inflammatory disease or condition, eg, rheumatoid arthritis,
psoriasis, eczema, asthma,
ulcerative colitis, colitis, Crohn's disease or IBD;
(y) Autism;
(z) ADHD;
(aa) Bipolar disorder;
(bb) ALS [Amyotrophic Lateral Sclerosis];
(cc) Osteoarthritis;
(dd) A congenital or development defect or condition;
(cc) Miscarriage;
(ft) A blood clotting condition;
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
(gg) Bronchitis;
(hh) Dry or wet AMD;
(ii) Neovascularisation (eg, of a tumour or in the eye);
Common cold;
(kk) Epilepsy;
(11) Fibrosis, eg, liver or lung fibrosis;
(mm) A fungal disease or condition, eg, thrush;
(nn) A metabolic disease or condition, eg, obesity, anorexia, diabetes,
Type I or Type II diabetes.
(oo) Ulcer(s), eg, gastric ulceration or skin ulceration;
(pp) Dry skin;
(qq) Sjogren's syndrome;
(a) Cytokine storm;
(ss) Deafness, hearing loss or impairment;
(tt) Slow or fast metabolism (ie, slower or faster than average for the
weight, sex and age of the
subject);
(uu) Conception disorder, eg, infertility or low fertility;
(vv) Jaundice;
(ww) Skin rash;
(xx) Kawasaki Disease;
(yy) Lyme Disease;
(zz) An allergy, eg, a nut, grass, pollen, dust mite, cat or dog fur or
dander allergy;
(aaa) Malaria, typhoid fever, tuberculosis or cholera;
(bbb) Depression;
(ccc) Mental retardation;
(ddd) Microcephaly;
(eee) Malnutrition;
(fff) Conjunctivitis;
(ggg) Pneumonia;
(hhh) Pulmonary embolism;
(iii) Pulmonary hypertension;
A bone disorder;
(kkk) Sepsis or septic shock;
(111) Sinusitus;
(mmm) Stress (eg, occupational stress);
(nnn) Thalassaemia, anaemia, von Willebrand Disease, or haemophilia;
51
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
(000) Shingles or cold sore;
(ppp) Menstruation;
(qqq) Low sperm count.
[0094] NEURODEGENERATIVE OR CNS DISEASES OR CONDITIONS FOR
TREATMENT OR PREVENTION BY THE INVENTION
[0095] In an example, the neurodegenerative or CNS disease or condition is
selected from the group
consisting of Alzheimer disease , geriopsychosis, Down syndrome, Parkinson's
disease, Creutzfeldt-
j akob disease, diabetic neuropathy, Parkinson syndrome, Huntington's disease,
Machado-Joseph
disease, amyotrophic lateral sclerosis, diabetic neuropathy, and Creutzfeldt
Creutzfeldt- Jakob
disease. For example, the disease is Alzheimer disease. For example, the
disease is Parkinson
syndrome.
[0096] In an example, wherein the method of the invention is practised on a
human or animal subject
for treating a CNS or neurodegenerative disease or condition, the method
causes downregulation of
Treg cells in the subject, thereby promoting entry of systemic monocyte-
derived macrophages and/or
Treg cells across the choroid plexus into the brain of the subject, whereby
the disease or condition (eg,
Alzheimer's disease) is treated, prevented or progression thereof is reduced.
In an embodiment the
method causes an increase of IFN-gamma in the CNS system (eg, in the brain
and/or CSF) of the
subject. In an example, the method restores nerve fibre and//or reduces the
progression of nerve fibre
damage. In an example, the method restores nerve myelin and//or reduces the
progression of nerve
myelin damage. In an example, the method of the invention treats or prevents a
disease or condition
disclosed in W02015136541 and/or the method can be used with any method
disclosed in
W02015136541 (the disclosure of this document is incorporated by reference
herein in its entirety,
eg, for providing disclosure of such methods, diseases, conditions and
potential therapeutic agents that
can be administered to the subject for effecting treatement and/or prevention
of CNS and
neurodegenerative diseases and conditions, eg, agents such as immune
checkpoint inhibitors, eg, anti-
PD-1, anti-PD-L1, anti-TIM3 or other antibodies disclosed therein).
[0097] CANCERS FOR TREATMENT OR PREVENTION BY THE METHOD
[0098] Cancers that may be treated include tumours that are not vascularized,
or not substantially
vascularized, as well as vascularized tumours. The cancers may comprise non-
solid tumours (such as
haematological tumours, for example, leukaemias and lymphomas) or may comprise
solid tumours.
Types of cancers to be treated with the invention include, but are not limited
to, carcinoma, blastoma,
and sarcoma, and certain leukaemia or lymphoid malignancies, benign and
malignant tumours, and
52
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
malignancies e.g., sarcomas, carcinomas, and melanomas. Adult tumours/cancers
and paediatric
tumours/cancers are also included.
[0099] Haematologic cancers are cancers of the blood or bone marrow. Examples
of haematological
(or haematogenous) cancers include leukaemias, including acute leukaemias
(such as acute
lymphocytic leukaemia, acute myelocytic leukaemia, acute myelogenous leukaemia
and myeloblasts,
promyeiocytic, myelomonocytic, monocytic and erythroleukaemia), chronic
leukaemias (such as
chronic myelocytic (granulocytic) leukaemia, chronic myelogenous leukaemia,
and chronic
lymphocytic leukaemia), polycythemia vera, lymphoma, Hodgkin's disease, non-
Hodgkin's lymphoma
(indolent and high grade forms), multiple myeloma, Waldenstrom's
macroglobulinemia, heavy chain
disease, myeiodysplastic syndrome, hairy cell leukaemia and myelodysplasia.
[00100] Solid tumours are abnormal masses of tissue that usually do not
contain cysts or liquid areas.
Solid tumours can be benign or malignant. Different types of solid tumours are
named for the type of
cells that form them (such as sarcomas, carcinomas, and lymphomas). Examples
of solid tumours,
such as sarcomas and carcinomas, include fibrosarcoma, myxosarcoma,
liposarcoma,
chondrosarcoma, osteosarcoma, and other sarcomas, synovioma, mesothelioma,
Ewing's tumour,
leiomyosarcoma, rhabdomyosarcoma, colon carcinoma, lymphoid malignancy,
pancreatic cancer,
breast cancer, lung cancers, ovarian cancer, prostate cancer, hepatocellular
carcinoma, squamous eel!
carcinoma, basal cell carcinoma, adenocarcinoma, sweat gland carcinoma,
medullary thyroid
carcinoma, papillary thyroid carcinoma, pheochromocytomas sebaceous gland
carcinoma, papillary
carcinoma, papillary adenocarcinomas, medullary carcinoma, bronchogenic
carcinoma, renal cell
carcinoma, hepatoma, bile duct carcinoma, choriocarcinoma, Wilms' tumour,
cervical cancer,
testicular tumour, seminoma, bladder carcinoma, melanoma, and CNS tumours
(such as a glioma
(such as brainstem glioma and mixed gliomas), glioblastoma (also known as
glioblastoma
multiforme) astrocytoma, CNS lymphoma, germinoma, medu!loblastoma, Schwannoma
craniopharyogioma, ependymoma, pineaioma, hemangioblastoma, acoustic neuroma,
oligodendroglioma, menangioma, neuroblastoma, retinoblastoma and brain
metastases).
[00101] AUTOIMMUNE DISEASES FOR TREATMENT OR PREVENTION BY THE
METHOD
1. Acute Disseminated Encephalomyelitis (ADEM)
2. Acute necrotizing hemorrhagic leukoencephalitis
3. Addison's disease
4. Agammaglobulinemia
5. Alopecia areata
6. Amyloidosis
53
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
7. Ankylosing spondylitis
8. Anti-GBM/Anti-TBM nephritis
9. Antiphospholipid syndrome (APS)
10. Autoimmune angioedema
11. Autoimmune aplastic anemia
12. Autoimmune dysautonomia
13. Autoimmune hepatitis
14. Autoimmune hyperlipidemia
15. Autoimmune immunodeficiency
16. Autoimmune inner ear disease (AIED)
17. Autoimmune myocarditis
18. Autoimmune oophoritis
19. Autoimmune pancreatitis
20. Autoimmune retinopathy
21. Autoimmune thrombocytopenic putpura (ATP)
22. Autoimmune thyroid disease
23. Autoimmune urticaria
24. Axonal & neuronal neuropathies
25. Balo disease
26. Behcet's disease
27. Bullous pemphigoid
28. Cardiomyopathy
29. Castleman disease
30. Celiac disease
31. Chagas disease
32. Chronic fatigue syndrome
33. Chronic inflammatory demvelinating polyneuropathv (CIDP)
34. Chronic recurrent multifocal ostomvelitis (CRMO)
35. Churg-Strauss syndrome
36. Cicatricial pemphigoid/benign mucosal pemphigoid
37. Crohn's disease
38. Cogans syndrome
39. Cold agglutinin disease
40. Congenital heart block
41. Coxsackie mvocarditis
54
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
42. CREST disease
43. Essential mixed cryoglobulincm i a
44. Demyelinating neuropathics
45. Dermatitis herpetiformis
46. Dermatomyositis
47. Devic's disease (neuromyelitis optica)
48. Discoid lupus
49. Dressler's syndrome
50. Endometriosis
51. Eosinophilic esophagitis
52. Eosinophilic fasciitis
53. Erythema nodosum
54. Experimental allergic encephalomyelitis
55. Evans syndrome
56. Fibromyalgia
57. Fibrosing alveolitis
58. Giant cell arteritis (temporal artcritis)
59. Giant cell myocarditis
60. Glomerulonephritis
61. Goodpasture's syndrome
62. Granulomatosis with Polyangiitis (GPM (formerly called Wegener's
Granulomatosis)
63. Graves' disease
64. Guillain-Barre syndrome
65. Hashimoto's encephalitis
66. Hashimoto's thyroiditis
67. Hemolytic anemia
68. Hcnoch-Schonlein putpura
69. Herpes gestationis
70. Hvpogammaglobulinemia
71. Idiopathic thrombocvtopenic purpura (ITP)
72. IgA nephropathy
73. IgG4-related sclerosing disease
74. Immunoregulatoty lipoproteins
75. Inclusion body myositis
76. Interstitial cystitis
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
77. Juvenile arthritis
78. Juvenile diabetes (Type 1 diabetes)
79. Juvenile myositis
80. Kawasaki syndrome
81. Lambert-Eaton syndrome
82. Leukocvtoclastic vasculitis
83. Lichen planus
84. Lichen sclerosus
85. Ligneous conjunctivitis
86. Linear IgA disease (LAD)
87. Lupus (SLE)
88. Lyme disease, chronic
89. Meniere's disease
90. Microscopic polyangiitis
91. Mixed connective tissue disease (MCTD)
92. Mooren's ulcer
93. Mucha-Habermann disease
94. Multiple sclerosis
95. Myasthenia gravis
96. Myositis
97. Narcolepsy
98. Neuromvelitis optica (Devic's)
99. Neutropenia
100. Ocular cicatricial pemphigoid
101. Optic neuritis
102. Palindromic rheumatism
103. PANDAS (Pediatric Autoimmune Neuropsychiatric Disorders Associated with
Streptococcus)
104. Paraneoplastic cerebellar degeneration
105. Paroxysmal nocturnal hemoglobinuria (PNH)
106. Parry Romberg syndrome
107. Parsonnage-Turner syndrome
108. Pars planitis (peripheral uveitis)
109. Pemphigus
110. Peripheral neuropathy
56
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
111. Perivenous encephalomyelitis
112. Pernicious anemia
113. POEMS syndrome
114. Polyarteritis nodosa
115. Type I. H. & III autoimmune polvglandular syndromes
116. Polvinyalgia rheumatica
117. Polymyositis
118. Postmvocardial infarction syndrome
119. Postpericardiotomv syndrome
120. Progesterone dermatitis
121. Primary biliary cirrhosis
122. Primary sclerosing cholangitis
123. Psoriasis
124. Psoriatic arthritis
125. Idiopathic pulmonary fibrosis
126. Pyoderma gangrenosum
127. Pure red cell aplasia
128. Raynauds phenomenon
129. Reactive Arthritis
130. Reflex sympathetic dystrophy
131. Reiter's syndrome
132. Relapsing polvchondritis
133. Restless legs syndrome
134. Retroperitoneal fibrosis
135. Rheumatic fever
136. Rheumatoid arthritis
137. Sarcoidosis
138. Schmidt syndrome
139. Scleritis
140. Scleroderma
141. Sjogren's syndrome
142. Sperm & testicular autoimmunity
143. Stiff person syndrome
144. Subacute bacterial endocarditis (SBE)
145. Susac's syndrome
57
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
146. Sympathetic ophthalmia
147. Takayasu's arteritis
148. Temporal arteritis/Giant cell arteritis
149. Thrombocytopenic purpura (TTP)
150. Tolosa-Hunt syndrome
151. Transverse myelitis
152. Type 1 diabetes
153. Ulcerative colitis
154. Undifferentiated connective tissue disease (UCTD)
155. Uveitis
156. Vasculitis
157. Vesiculobullous dermatosis
158. Vitiligo
159. Wegener's granulomatosis (now termed Granulomatosis with Polyangiitis
(GPA).
[00102] INFLAMMATORY DISEASES FOR TREATMENT OR PREVENTION BY THE
METHOD
1. Alzheimer
2. ankylosing spondylitis
3. arthritis (osteoarthritis, rheumatoid arthritis (RA), psoriatic
arthritis)
4. asthma
5. atherosclerosis
6. Crohn's disease
7. colitis
8. dermatitis
9. diverticulitis
10. fibromyalgia
11. hepatitis
12. irritable bowel syndrome (IBS)
13. systemic lupus erythematous (SLE)
14. nephritis
15. Parkinson's disease
16. ulcerative colitis.
58
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[00103] PARAGRAPHS:
By way of example, the invention provides the following Paragraphs (which may
optionally be
combined with any of the disclosure above):-
1. An antibacterial composition comprising a population of first phage,
wherein the first phage
require helper phage for replication of first phage particles, wherein the
helper phage are capable of
packaging first phage nucleic acid to produce first phage particles, wherein
the first phage are
different from the helper phage and the helper phage are incapable themselves
of producing helper
phage particles, wherein the first phage are capable of infecting target
bacteria and each first phage
comprises antibacterial means for killing bacteria.
2. The composition of Paragraph 1, wherein at least 95% of phage particles
comprised by the
composition are first phage particles.
Optionally, the composition comprises helper phage. Optionally, the
composition comprises no more
than 1 helper phage particle per 1 x 106 or more of phage particles.
Optionally, the composition
comprises no more than 1 helper phage particle per 1 x 10' or more of phage
particles. Optionally,
the composition comprises no more than 1 helper phage particle per 1 x 109 or
more of phage
particles. Optionally, the composition comprises no more than 1 helper phage
particle per 1 x 1010 or
more of phage particles.
3. The composition of Paragraph 1 or 2, wherein each first phage comprises
one or more
components of a CRISPR/Cas system, wherein the component(s) comprise a DNA
sequence encoding
a guide RNA (optionally a single guide RNA) or comprising a CRISPR array for
producing guide
RNA, wherein the guide RNA is capable of targeting the genome of target
bacteria.
4. The composition of any preceding Paragraph, wherein each first phage
genome is devoid of
genes encoding phage proteins.
5. A composition according to any preceding Paragraph for use in
antibacterial treatment of
bacteria, the composition comprising an engineered mobile genetic element
(MGE) that is capable of
being mobilised in a first bacterial host cell of a first species or strain,
the cell comprising a helper
phage genome, wherein in the cell the MGE is mobilised using proteins encoded
by the helper phage
and replication of helper phage is inhibited, wherein the MGE encodes an
antibacterial agent or
encodes a component of such an agent, wherein the MGE comprises a modified
genomic island,
59
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
modified pathogenicity island, SaPI (S aureus Pathogenicity Island), V
cholerae PLE (Phage-Like
Inducible Chromosomal Island-Like Element) or E coli PLE.
6. A kit comprising
(a) A first DNA; and
(b) One or more second DNAs;
Wherein
(i) the DNAs together comprise all phage structural protein genes
required to produce a
packaged phage particle comprising a copy of the first DNA;
(ii) the first DNA comprises none or at least one, but not all, of the
genes; and wherein the one or
more second DNAs comprise the remainder of the genes;
(iii) the first DNA comprises a phage packaging signal for producing the
packaged phage particle;
and
(iv) the second DNA is devoid of a nucleotide sequence required for
packaging the second DNA
into phage particles;
wherein the DNAs are operable when co-existing in a host bacterium for
producing packaged phage
(first phage) that comprise the first DNA, wherein the first phage require the
second DNA for
replication thereof to produce further first phage particles.
7. The kit of Paragraph 6, wherein the phage particle of (i) is capable of
infecting a target
bacterium, the phage comprising a nucleotide sequence of interest (NSI) that
is capable of expressing
a protein or RNA in the target bacterium, wherein the presence in the target
bacterium of the NSI-
encoded protein or RNA mediates target cell killing, or downregulation of
target cell growth or
propagation.
8. The kit of Paragraph 6, wherein the phage particle of (i) is capable of
infecting a target
bacterium, the phage comprising a nucleotide sequence of interest (NSI) that
is capable of expressing
a protein or RNA in the target bacterium, or wherein the NSI comprises a
regulatory element that is
operable in the target bacterium.
9. The kit of Paragraph 8, wherein the presence in the target bacterium of
the NSI or its encoded
protein or RNA mediates target cell killing, or downregulation of target cell
growth or propagation, or
mediates switching off of expression of one or more RNA or proteins encoded by
the target cell
genome, or downregulation thereof.
60
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
10. The kit of Paragraph 8, wherein the presence in the target bacterium of
the NSI or its encoded
protein or RNA mediates upregulation of growth or propagation of the target
cell, or mediates
switching on of expression of one or more RNA or proteins encoded by the
target cell genome, or
upregulation thereofy.
11. The kit of any one of Paragraphs 7, 8 or 9, wherein the NSI encodes a
component of a
CRISPR/Cas system that is toxic to the target bacterium.
12. The kit of any one of Paragraphs 7, 8 or 9, wherein the NSI comprises
engineered
antibacterial means for killing target bacteria.
13. The kit of any one of Paragraphs 6 to 12, wherein the packaged phage
particle genome is
devoid of genes encoding phage proteins.
14. The kit of any one of Paragraphs 6 to 13, wherein the first DNA
comprises none of the
structural protein genes.
15. The kit of any one of Paragraphs 6 to 14, wherein the second DNA is
devoid of a phage
packaging signal.
16. The kit of any one of Paragraphs 6 to 15, wherein each signal is a pac
or cos sequence, or is a
homologue thereof.
17. The kit of any one of Paragraphs 6 to 16, wherein each signal comprises
SEQ ID NO: 2 or a
sequence that is at least 70, 80, 90, 95, 96, 97, 98 or 99% identical thereto,
or is a homologue from a
different species.
18. An isolated DNA, wherein the DNA is a first DNA as defined in any one
of Paragraphs 6 to
17.
19. The kit or DNA of any one of Paragraphs 6 to 18, wherein the first DNA
is comprised by a
vector (optionally, a plasmid, phagemid or shuttle vector).
20. The kit or DNA of any one of Paragraphs 6 to 19, wherein the second DNA
is comprised by a
vector (optionally a plasmid, phagemid or shuttle vector), helper phage
(optionally a helper phagemid)
61
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
or is integrated in the genome of a host bacterial cell.
21. A host bacterial cell comprising the first and second DNAs as defined
in any one of
Paragraphs 6 to 17, 19 and 20; or comprising the DNA of Paragraph 18, 19 or
20.
22. A method of producing a phage composition, the method comprising
expressing in a cell of
Paragraph 21 the phage proteins, wherein packaged first phage particles are
produced that comprise
the first DNA, wherein the first phage require the second DNA for replication
thereof to produce
further first phage; and optionally separating an amount of first phage from
cellular material wherein
an amount of purified phage is obtained.
In an example, the purified phage are mixed with a pharmaceutically-acceptable
excipient, carrier or
diluent (eg, an aqueous liquid or water) to produce a pharmaceutical
composition.
In an example any composition or kit of the invention is in combination with a
label or instructions
for use to treat and/or prevent a disease or condition in a human; optionally
wherein the label or
instructions comprise a marketing authorisation number (eg, an FDA or EMA
authorisation number);
optionally wherein the kit comprises an injection pen or IV container that
comprises the first DNA or
first phage.
23. The method of Paragraph 22, comprising isolating the first phage
particles.
24. A composition (eg, antibacterial composition, eg, for medcial use)
comprising a population of
first phage particles obtainable by the method of Paragraph 22 or 23.
In an example, the first phage particles are obtained by the method.
In an example, any composition of the invention comprises at least 1 x 103
first phage per ml or mg,
such as when the composition is comprised by a fluid (eg, a liquid) or solid.
In an example, any
composition of the invention comprises at least 1 x 104 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 105 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 106 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 107 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 10' first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 109 first phage per ml or
mg. In an example, any
62
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
composition of the invention comprises at least 1 x 1010 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 1011 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 1012 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 1013 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 1014 first phage per ml or
mg.
In an example, any composition of the invention comprises up to 1 x 1014 first
phage per ml or mg,
such as when the composition is comprised by a fluid (eg, a liquid) or solid.
In an example, any
composition of the invention comprises up to 1 x 1013 first phage per ml or
mg. In an example, any
composition of the invention comprises up to 1 x 1012 first phage per ml or
mg. In an example, any
composition of the invention comprises up to 1 x 1011 first phage per ml or
mg. In an example, any
composition of the invention comprises up to 1 x 1010 first phage per ml or
mg. In an example, any
composition of the invention comprises up to 1 x 109 first phage per ml or mg.
In an example, any composition of the invention comprises at least 1 x 103 to
1 x 1010, 1 x 1011, 1 x
1012, 1 x 10" or 1 x 1014 first phage per ml or mg, such as when the
composition is comprised by a
fluid (eg, a liquid) or solid. In an example, any composition of the invention
comprises at least 1 x
104 to 1 x 1010, 1 x 1011, 1 x 1012, 1 x 10" or 1 x 1014 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 105 to 1 x 1010, 1 x 1011,
1 x 1012, 1 x 10" or 1 x
1014 first phage per ml or mg. In an example, any composition of the invention
comprises at least 1 x
106 to 1 x 1010, 1 x 1011, 1 x 1012, 1 x 10" or 1 x 1014 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 107 to 1 x 1010, 1 x 1011,
1 x 1012, 1 x 10" or 1 x
1014 first phage per ml or mg. In an example, any composition of the invention
comprises at least 1 x
108 to 1 x 1010, 1 x 1011, 1 x 1012, 1 x 10" or 1 x 1014 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 109 to 1 x 1010, 1 x 1011,
1 x 1012, 1 x 10" or 1 x
1014 first phage per ml or mg. In an example, any composition of the invention
comprises at least 1 x
1010 to 1 x le, 1 x 1011, 1 x 1012, 1 x 10" or 1 x 1014 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 1011 to 1 x le, 1 x 1011,
1 x 1012, 1 x 1013 or 1 x
1014 first phage per ml or mg. In an example, any composition of the invention
comprises at least 1 x
1012 to 1 x le, 1 x 1011, 1 x 1012, 1 x 10" or 1 x 1014 first phage per ml or
mg. In an example, any
composition of the invention comprises at least 1 x 1013 to 1 x le, 1 x 1011,
1 x 1012, 1 x 1013 or 1 x
1014 first phage per ml or mg. In an example, any composition of the invention
comprises at least 1 x
1014 to 1 x le, 1 x 1011, 1 x 1012, 1 x 10" or 1 x 1014 first phage per ml or
mg.
63
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
In an example, the composition comprises one or more doses of the first phage
for administration to a
subject for medical use, eg, to treat or prevent a disease or condition in the
subject. In an example, the
composition comprises a single dose. In an example, the composition comprises
(or comprises at
least) 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26, 27, 28, 29 or
30 doses. In an example, each dose is (or is at least) a 0.5, 1, 2, 3, 4, 5,
10, 20, 25, 30, 40, 50, 75, 100,
125, 200 or 250mg or ml dose comprising said phage (ie, the dose is said
amount and comprises
phage and an excipient, diluent or carrier for example).
In an example, the composition comprises one or more doses of the first phage
for administration to a
subject for non-medical use, eg, for agricultural use. In an example, the
composition comprises a
single dose. In an example, the composition comprises (or comprises at least)
2, 3, 4, 5, 6, 7, 8, 9, 10,
11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 or
30 doses. In an example,
each dose is (or is at least) a 0.5, 1, 2, 3, 4, 5, 10, 20, 25, 30, 40, 50,
75, 100, 125, 200, 250, 500, 750,
1000, 2000, 3000, 4000, 5000, 10000, 50000, 100000 mg or ml dose comprising
said phage (ie, the
dose is said amount and comprises phage and an excipient, diluent or carrier
for example). The dose
may be dissolved or diluted in a solvent (eg, an aqueous solvent or water)
before use for contacting
with target bacteria. In an example 1 imperial gallon comprises one dose of
the first phage, eg, for
agricultural use, such as crop spraying, or for animal or livestock use, such
as use as a beverage.
25. The method of Paragraph 22 or 23 or the composition of Paragraph 24,
wherein the second
DNA is comprised by helper phage DNA and less than 5% of total phage particles
comprised by the
composition are helper phage particles.
26.
The method or composition of any one of Paragraphs 22 to 24, wherein the
second DNA is
comprised by helper phage DNA and the composition comprises no more than 1
helper phage particle
per 1 x 106 or more of phage particles.
This has been demonstrated in Example 3 and shown to be efficacious for target
bacteria killing in
Example 4. In an embodiment, the composition comprises no more than 1 helper
phage particle per 1
x 107 or more of phage particles. In an embodiment, the composition comprises
no more than 1
helper phage particle per 1 x 10' or more of phage particles. In an
embodiment, the composition
comprises no more than 1 helper phage particle per 1 x 109 or more of phage
particles. In an
embodiment, the composition comprises no more than 1 helper phage particle per
1 x 1010 or more of
phage particles.
64
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
In an embodiment, the composition comprises no more than 1 helper phage
particle per 1 x 106 of
phage particles. In an embodiment, the composition comprises no more than 1
helper phage particle
per 1 x 107 of phage particles. In an embodiment, the composition comprises no
more than 1 helper
phage particle per 1 x 108 of phage particles. In an embodiment, the
composition comprises no more
than 1 helper phage particle per 1 x 109 of phage particles.
In an embodiment, the composition comprises no more than 1 helper phage
particle per 1 x 106 to 1 x
109 of phage particles. In an embodiment, the composition comprises no more
than 1 helper phage
particle per 1 x 106 to 1 x 108 of phage particles. In an embodiment, the
composition comprises no
.. more than 1 helper phage particle per 1 x 106 to 1 x 107 of phage
particles. In an embodiment, the
composition comprises no more than 1 helper phage particle per 1 x 107 to 1 x
109 of phage particles.
In an embodiment, the composition comprises no more than 1 helper phage
particle per 1 x 107 to 1 x
108 of phage particles.
In an example, the proportion of helper phage is determined as plaque forming
units (PFU), eg,
PFU/ml of the phage composition comprising said number of phage particles. In
example, the
proportion of non-helper phage (ie, first phage) is determined as number of
transduced units of target
bacteria (TFU), eg, TFU/ml of the phage composition. PFU is determined on
lawns of a susceptible
indicator bacterium while TFU is determined in a transduction assay in which a
culture of susceptible
.. indicator bacteria is infected with the phage composition ensuring surplus
of indicator cells (i.e. at a
low multiplicity of infection (MOI < 0.01) and number of transduced cells are
determined by plating
on selective plates.
Thus, the composition may comprise one or more helper phage particles. The
level of helper particles
is, however, extremely low. This is beneficial as the composition is
relatively pure (and useful, for
example, therefore as a medicament). It is also useful as the chances of the
first phage being
replicated is extremely low, providing the advantages of dosing control of
phage, containment of
phage (eg, in a human or animal body or an environment), and the lack of phage
replication reduces
the chances of acquiring undesirable genes (eg, antibiotic resistance genes)
by the phage.
27. The composition of any one of Paragraphs 1 to 5, the method of
Paragraph 22, 23, 25 or 26 or
the composition of Paragraph 24, 25 or 26, wherein each first phage particle
comprises a nucleotide
sequence of interest (NSI) that is capable of expressing a protein or RNA in a
target bacterium,
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
wherein the presence in the target bacterium of the NSI-encoded protein or RNA
mediates target cell
killing, or downregulation of target cell growth or propagation.
28. The method or composition of Paragraph 27, wherein the NSI encodes a
component of a
CRISPR/Cas system that is toxic to target cells.
29. The method or composition of Paragraph 28, wherein the component
comprises (i) a DNA
sequence encoding a guide RNA (eg, a single guide RNA) or comprising a CRISPR
array for
producing guide RNA, wherein the guide RNA is capable of targeting the genome
of target bacteria;
(ii) a Cas nuclease-encoding DNA sequence; and/or (iii) a DNA sequence
encoding one or more
components of Cascade.
30. The method or composition of Paragraph 29, wherein the NSI encodes a
Cas nuclease and a
guide RNA of the system (or wherein the NSI encodes a Cas nuclease and a
CRISPR array for
producing guide RNA), wherein the guide RNA is capable of targeting the genome
of target bacteria,
wherein the guide RNA is capable of guiding the Cas in target cells to mediate
target cell killing, or
downregulation of target cell growth or propagation.
Optionally, the NSI encodes a Cas9, and a tracrRNA and a CRISPR array for
producing guide RNA.
31. The method or composition of any one of Paragraphs 1 to 5 and 22 to 30,
wherein the first
phage particles comprise no phage structural protein genes.
32. The kit, DNA, method or composition of any preceding Paragraph, wherein
the first DNA or
first phage DNA is comprised by a high copy number plasmid.
Optionally, the first DNA or first phage DNA is comprised by a medium copy
number plasmid.
The meaning of low, medium and high copy number on and plasmids is known to
the skilled
addressee and these are terms of art. As is known by the skilled person, copy
number denotes the
average number of plasmid copies per cell. For example, a low copy number
plasmid is a plasmid
that exists in from 1 to 10 copies per bacterial cell in which the plasmid is
harboured; a medium copy
number plasmid exists in from 11 to 50 (eg, 11 to 40 or 20 to 30 or 40) copies
per cell; and a high
copy number is >50 (eg, up to 100, 200, 250, 300, 400, 500, 600 or 700) copies
per cell. In an
example, the plasmid or vector comprising first DNA is a medium copy number
plasmid or vector. In
66
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
an example, the plasmid or vector comprising first DNA is a high copy number
plasmid or vector. An
example of common on and plasmids is shown in Table 7.
33. The method or composition of Paragraph 27 or any one of Paragraphs 28
to 32 when
dependent from Paragraph 27, wherein the NSI comprises engineered
antibacterial means for killing
target bacteria.
34. The method of composition of Paragraph 33, wherein the antibacterial
means comprises a
nucleic acid encoding a guided nuclease, such as a Cas nuclease, TALEN, zinc
finger nuclease or
meganuclease
35. The composition of any one of Paragraphs 1 to 5 and 24 to 34 for
administration to a human
or animal subject for medical use.
36. The composition of any one of Paragraphs 1 to 5 and 24 to 34 for
administration to a human
or animal subject for treating an infection of target bacterial cells, wherein
the first phage are capable
of infecting and killing the target cells, optionally wherein the infection is
a gut microbiome infection.
Optionally, the target cells are E coli cells the first DNA is comprised by a
high copy number plasmid.
In an example, the gut microbiome is an upper GI tract microbiome. In an
example the target cells are
comprised by the upper GI tract of the subject. In an example, the first phage
are delivered to the
upper GI tract of the subject.
In an example, the gut microbiome is a stomach or small intestine microbiome.
In an example the
target cells are comprised by the stomach or small intestine of the subject.
In an example, the first
phage are delivered to the stomach or small intestine of the subject.
37. The composition of Paragraph 33 or 34 for use in a contained method
of treating a disease or
condition of a human or animal subject, wherein the disease or condition is
mediated by target
bacteria and the target bacteria are comprised by the subject (optionally
comprised by a gut
microbiome), the method comprising administering the composition to the
subject, whereby the target
bacteria are exposed to the antibacterial means and killed and propagation of
the first phage is
contained.
67
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
38. A method of treating an environment ex vivo, the method comprising
exposing the
environment to a composition comprising a population of first phage particles,
wherein the
composition is obtainable by the method of any one of Paragraphs 22, 23 and 25
to 34, or the
composition is according to any one of Paragraphs 1 to 5 and 24 to 34, wherein
the environment
comprises target bacteria and the first phage infect and kill the target
bacteria.
39. The method of Paragraph 38, wherein the method is for containing the
treatment in the
environment.
40. The composition of any one of Paragraphs 1 to 5 and 24 to 34 for
controlling the dosing of the
first phage treatment in the subject; or the method of Paragraph 38 or 39 for
controlling the dosing of
the first phage treatment in the environment.
41. The composition of any one of Paragraphs 1 to 5 and 24 to 34 for
reducing the risk of
acquisition of foreign gene sequence(s) by the first phage in the subject; or
the method of Paragraph
38 or 39 for reducing the risk of acquisition of foreign gene sequence(s) by
the first phage in the
environment.
CONCEPTS:
[00104] The invention provides the following Concepts, as exemplified by
Example 6.
1. A host bacterial cell comprising
c) A first DNA; and
d) One or more second DNAs;
wherein
(v) the DNAs together comprise all genes required to produce a transduction
particle comprising
a copy of the first DNA packaged by phage structural proteins;
(vi) the first DNA is devoid of at least one functional essential gene (eg,
encoding a phage
structural protein) required to produce the particle; and wherein the one or
more second DNAs
comprises said functional essential gene(s);
(vii) the first DNA comprises a phage packaging signal for producing the
particle; and
(viii) the second DNA is devoid of a nucleotide sequence required for
packaging the second DNA
into transduction particles;
68
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
wherein the second DNA is required for packaging first DNA to produce
particles, wherein the DNAs
are operable in the cell for producing transduction particles comprising phage
structural proteins that
package copies of the first DNA.
2. The cell of Concept 1, wherein the host bacterial cell comprises
a) said first DNA; and
b) said one or more second DNAs;
wherein
(i) the DNAs together comprise encode all phage structural proteins
required to produce a
packaged transduction particle comprising a copy of the first DNA;
(ii) the first DNA encodes none or at least one, but not all, of the
structural proteins; and wherein
the one or more second DNAs encode the remainder of the structural proteins;
(iii) the first DNA comprises a phage packaging signal for producing the
particle; and
(iv) the second DNA is devoid of a nucleotide sequence required for
packaging the second DNA
into transduction particles;
wherein the second DNA is required for packaging first DNA to produce
particles, wherein the DNAs
are operable in the cell for producing transduction particles comprising phage
structural proteins that
package copies of the first DNA.
3. The cell of Concept 1, wherein (i) the first DNA is comprised by an
episome (eg, a plasmid)
that is devoid of said essential or structural protein gene(s) and/or the
second DNA is comprised by an
episome (eg, a plasmid) or a chromosome of the cell.
4. The cell of Concept 1, 2 or 3, wherein all of said essential genes or
phage structural protein
genes are comprised by the second DNA and the first DNA is devoid of said
genes.
5. The cell of any preceding Concept, wherein the first DNA encodes a
guided nuclease or a
component of a CRISPR/Cas system (optionally, a crRNA or a guide RNA).
6. The cell of any preceding Concept, wherein the first DNA comprises a
phage origin of
replication and/or the first DNA comprises phage replication genes and/or
phage lysis genes; and/or
(ii) the first DNA comprises a phage origin of replication but no bacterial
origin of replication.
69
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
7. The cell of any preceding Concept, wherein each transduction particle is
a non-self replicative
transduction particle.
8. The cell of any preceding Concept, wherein each particle comprises a
phage tail fibre.
9. The cell of any preceding Concept, wherein the essential genes or
structural protein genes;
and packaging signal are genes and a packaging signal of a tailed phage, eg, a
P2, T4, T7, Phi92,
lambda, K1-5 or 933w phage.
10. The cell of any preceding Concept, wherein the first DNA is comprised
by a phage genome,
wherein the phage genome is integrated in a plasmid; optionally wherein each
particle is capable of
infecting a target bacterium, the first DNA comprising a nucleotide sequence
of interest (NSI) that is
capable of expressing a protein or RNA in the target bacterium, wherein the
NSI replaces the essential
gene(s) or structural protein gene(s) of the phage.
11. The cell of any preceding Concept, wherein each particle is capable
of infecting a target
bacterium, the first DNA comprising a nucleotide sequence of interest (NSI)
that is capable of
expressing a protein or RNA in the target bacterium, wherein the presence in
the target bacterium of
the NSI-encoded protein or RNA mediates target cell killing, or downregulation
of target cell growth
or propagation; or (ii) wherein each particle is capable of infecting a target
bacterium, the first DNA
comprising a nucleotide sequence of interest (NSI) that is capable of
expressing a protein or RNA in
the target bacterium, or wherein the NSI comprises a regulatory element that
is operable in the target
bacterium.
12. The cell of Concept 11, wherein the presence in the target bacterium of
the NSI or its encoded
protein or RNA mediates target cell killing, or downregulation of target cell
growth or propagation, or
mediates switching off of expression of one or more RNA or proteins encoded by
the target cell
genome, or downregulation thereof.
13. The kit of Concept 11, wherein the presence in the target bacterium of
the NSI or its encoded
protein or RNA mediates upregulation of growth or propagation of the target
cell, or mediates
switching on of expression of one or more RNA or proteins encoded by the
target cell genome, or
upregulation thereof.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
14. The cell of any preceding Concept, wherein the packaging signal is a
pac or cos sequence, or
is a homologue thereof; or is a direct terminal repeat (DTR).
15. The cell of any preceding Concept, wherein the packaging signal
comprises SEQ ID NO: 2 or
a sequence that is at least 70, 80, 90, 95, 96, 97, 98 or 99% identical
thereto, or is a homologue from a
different phage.
16. An isolated DNA, comprising a first DNA as defined in any preceding
Concept; or
comprising a second DNA as defined in any preceding Concept.
17. The first DNA or the second DNA of any one of Concepts 1 to 15, wherein
the DNA is
comprised by a plasmid.
18. The second DNA of Concept 16 or 17, wherein the DNA is comprised by a
cell, wherein the
cell does not comprise the first DNA.
19. A kit comprising a cell as recited in Concept 18, wherein the kit
comprises a vector
(optionally a plasmid) comprising the first DNA, wherein the vector is not
comprised by the cell.
20. The kit of Concept 18 or 19, wherein the cell is a bacterial or
archaeal cell.
21. A method of producing a transduction particle composition, the method
comprising
expressing in a cell of any one of Concepts 1 to 15 phage structural proteins
and replicating the first
DNA, wherein transduction particles are produced that comprise packaged first
DNA; and optionally
separating an amount of transduction particles from cellular material wherein
an amount of purified
transduction particles is obtained.
22. The method of Concept 21, comprising isolating transduction particles.
23. A composition comprising a population of transduction particles
obtainable by the method of
Concept 21 or 22.
24. The method of Concept 21 or 22 or the composition of Concept 23,
wherein the second DNA
is comprised by plasmid DNA and less than 5% of total DNA comprised by the
composition is DNA
of said plasmid.
71
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
25. The method or composition of any one of Concepts 21 to 24, wherein each
transduction
particle comprises a nucleotide sequence of interest (NSI) that is capable of
expressing a protein or
RNA in a target bacterium, wherein the presence in the target bacterium of the
NSI-encoded protein
or RNA mediates target cell killing, or downregulation of target cell growth
or propagation.
26. The method or composition of Concept 25, wherein the NSI encodes a
guided nuclease or a
component of a CRISPR/Cas system that is toxic to target cells.
27. The method or composition of Concept 26, wherein the component
comprises (i) a DNA
sequence encoding a guide RNA (eg, a single guide RNA) or comprising a CRISPR
array for
producing guide RNA, wherein the guide RNA is capable of targeting the genome
of target bacteria;
(ii) a Cas nuclease-encoding DNA sequence; and/or (iii) a DNA sequence
encoding one or more
components of Cascade.
28. The method or composition of Concept 26, wherein the NSI encodes a Cas
nuclease and a
guide RNA of the system (or wherein the NSI encodes a Cas nuclease and a
CRISPR array for
producing guide RNA), wherein the guide RNA is capable of targeting the genome
of target bacteria,
wherein the guide RNA is capable of guiding the Cas in target cells to mediate
target cell killing, or
downregulation of target cell growth or propagation.
29. The method or composition of Concept 25, wherein the NSI comprises
engineered
antibacterial means for killing target bacteria.
30. The method or composition of Concept 29, wherein the antibacterial
means comprises a
nucleic acid encoding a guided nuclease, such as a Cas nuclease, TALEN, zinc
finger nuclease or
meganuclease
31. The cell of any one of Concepts 1 to 15, the DNA of any one of Concepts
16 to 18, or the
composition of any one of Concepts 23 to 30 for administration to a human or
animal subject for
medical use.
32. The cell of any one of Concepts 1 to 15, the DNA of any one of Concepts
16 to 18, or the
composition of any one of Concepts 23 to 30 for administration to a human or
animal subject for
treating an infection of target bacterial cells, wherein the particles are
capable of infecting and killing
72
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
the target cells, optionally wherein the infection is a gut, blood, lung or
uterine tract microbiome
infection.
33. The composition of Concept 31 or 32 for use in a contained method of
treating a disease or
condition of a human or animal subject, wherein the disease or condition is
mediated by target
bacteria and the target bacteria are comprised by the subject (optionally
comprised by a gut, blood,
lung or uterine tract microbiome), the method comprising administering the
composition to the
subject, whereby the target bacteria are exposed to antibacterial means
encoded by the first DNA and
killed, and propagation of the transduction particles is contained.
34. A method of treating an environment ex vivo, the method comprising
exposing the
environment to a composition comprising a population of particles, wherein the
particles are capable
of transducing first DNA into the target cells comprised by the environment,
the first DNA encoding
antibacterial means that is toxic to taraget cells whereby target cells are
killed, wherein the
composition is obtainable by the method of any one of Concepts 21, 22 and 24
to 30, or the
composition is according to any one of Concepts 23 to 30.
35. The method of Concept 34, wherein the method is for containing the
treatment in the
environment.
36. The composition of any one of Concepts 23 to 30 for controlling in a
human or animal subject
the dosing of transduction particle treatment of a target bacterial cell
infection in the subject, wherein
the particles are capable of transducing first DNA into the target cells, the
first DNA encoding
antibacterial means that is toxic to taraget cells whereby target cells are
killed; or the method of
Concept 34 or 35 for controlling the dosing of the particle treatment in the
environment.
37. The composition of any one of Concepts 23 to 30 for reducing in a human
or animal subject
the risk of acquisition of foreign gene sequence(s) by the particles in the
subject; or the method of
Concept 34 or 35 for reducing the risk of acquisition of foreign gene
sequence(s) by the particles in
the environment.
[00105] Optionally, each particle is capable of infecting a target bacterium,
the first DNA comprising
a nucleotide sequence of interest (NSI) that is capable of expressing a
protein or RNA in the target
bacterium. For example, the NSI encodes an antimibacterial agent. For example,
the NSI encodes a
guided nuclease (eg, a Cas, TALEN, zinc finger nuclease or meganuclease). For
example, the NSI
73
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
encodes a component of a CRISPR/Cas system. For example, the nuclease or
system is operable in
the target cell to cut a target nucleic acid (DNA or RNA) sequence comprised
by the target cell (eg,
comprised by a chromosome or episome thereof).
[00106] In an embodiment, an essential gene may be omitted from the first DNA,
whereby the first
.. DNA is devoid of the essential gene. In another embodiment, the first DNA
may comprise a mutant
of the essential gene which does not provide the essential gene function (ie,
this is not a functional
essential gene). In another embodiment, first DNA may comprise a non-
expressible form of the
essential gene, eg, wherein a regulatory element of the gene has been deleted
or mutated so that the
gene does not function, eg, does not express its encoded protein.
.. [00107] Optionally, the essential genes are not phage terminase genes.
Optionally, the first DNA is
not devoid of all phage terminase genes. Optionally, the first DNA is not
devoid of phage structural
protein genes. Optionally, the essential genes are not phage structural
protein genes. Optionally, the
essential genes are not phage terminase genes and not phage structural protein
genes. Optionally, the
first DNA does not comprise an origin of replication (on) operable in a
bacterial host cell for
.. replication of the first DNA (and optionally the first DNA further
comprises a pahge orgin of
replication).
[00108] Optionally, each particle comprises a tail fibre (eg, a tail fibre
comprising one or more tail
fibre domains of a wild-type phage).
[00109] Optionally, each particle comprises phage capsid proteins, a packaging
signal (comprised by
the first DNA) and optionally phage replication gene(s), wherein all of these
components are proteins,
packaging signal and gene(s) of the same phage (eg, a wild-type phage).
[00110] Optionally, each particle comprises phage capsid proteins, a packaging
signal (comprised by
the first DNA) and optionally phage replication gene(s), wherein the packaging
signal and gene(s) are
components of the same phage (eg, a wild-type phage) and the capsid proteins
are proteins of a
different phage (eg, a wild-type phage).
[00111] Optionally, the first DNA is devoid of all phage structural protein
genes and the second DNA
comprises all phage structural protein genes required for packaging first DNA
to produce particles,
and for example the packaging signal is a packaging signal of a phage selected
from a Caudovirales,
Myovirideae, Podovirideae or Siphovirideae phage. Optionally, the first DNA is
devoid of all phage
structural protein genes and the second DNA comprises all phage structural
protein genes required for
packaging first DNA to produce particles, and for example the packaging signal
is a packaging signal
of a phage selected from a P2, Phi92, T7, lambda, 933w, K1-5 and T4 phage.
[00112] Optionally, the first DNA is devoid of all phage structural protein
genes and the second DNA
comprises all phage structural protein genes required for packaging first DNA
to produce particles, and for
example the phage structural protein genes of the second DNA ae genes of a
phage selected from a
74
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Caudovirales, Myovirideae, Podovirideae or Siphovirideae phage. Optionally,
the first DNA is devoid of all
phage structural protein genes and the second DNA comprises all phage
structural protein genes required for
packaging first DNA to produce particles, and for example the phage structural
protein genes of the second
DNA ae genes of a phage selected from a P2, Phi92, T7, lambda, 933w, K1-5 and
T4 phage.
[00113] The essential or structural protein gene(s) and packaging signal may
be structural protein genes and a
packaging signal of a tailed phage. Alternatively, the essential or structural
protein gene(s) and packaging
signal may be essential or structural protein gene(s) and a packaging signal
of a temperate phage. Alternatively,
the essential or structural protein gene(s) and packaging signal may be
essential or structural protein gene(s) and
a packaging signal of a lytic phage.
[00114] Optionally, the essential or structural protein gene(s) and packaging
signal may be essential or
structural protein gene(s) and packaging signal of a P2, T4, T7, Phi92,
lambda, K1-5 or 933w phage.
Optionally, the essential or structural protein genes and packaging signal may
be essential or structural protein
genes and a packaging signal of a tailed phage. Optionally, the essential or
structural protein genes and
packaging signal may be essential or structural protein genes and a packaging
signal of a Caudovirales phage.
Optionally, the essential or structural protein genes and packaging signal may
be essential or structural protein
genes and a packaging signal of a Myovirideae, Podovirideae or Siphovirideae
phage. Optionally, the essential
or structural protein genes and packaging signal may be essential or
structural protein genes and a packaging
signal of a P2 phage. Optionally, the essential or structural protein genes
and packaging signal may be essential
or structural protein genes and a packaging signal of a T7 phage. Optionally,
the essential or structural protein
genes and packaging signal may be essential or structural protein genes and a
packaging signal of a Phi92
phage. Optionally, the essential or structural protein genes and packaging
signal may be essential or structural
protein genes and a packaging signal of a lambda phage. Optionally, the
essential or structural protein genes
and packaging signal may be essential or structural protein genes and a
packaging signal of a 933w phage.
Optionally, the essential or structural protein genes and packaging signal may
be essential or structural protein
.. genes and a packaging signal of a K1-5 phage. Optionally, the essential or
structural protein genes and
packaging signal may be essential or structural protein genes and a packaging
signal of a T4 phage.
[00115] Optionally, any phage herein may be a tailed phage. Optionally, any
phage herein may be a
Caudovirales phage. Optionally, any phage herein may be a Myovirideae,
Podovirideae or Siphovirideae phage.
Optionally, any phage herein may be a P2, T4, T7, Phi92, lambda, K1-5 or 933w
phage. Optionally, any phage
herein may be a P2 phage. Optionally, any phage herein may be a T7 phage.
Optionally, any phage herein may
be a Phi92 phage. Optionally, any phage herein may be a lambda phage.
Optionally, any phage herein may be a
933w phage. Optionally, any phage herein may be a K1-5 phage. Optionally, any
phage herein may be a T4
phage.
[00116] An essential gene as per the invention may be any nucleic acid
sequence (not necessarily encoding a
protein) that is required to produce the particle or a phage. In an example,
an essential gene encodes a protein.
In an example, the essential gene(s) are not a packaging signal or phage
origin of replication.
[00117] For example, each essential gene is selected from
= a phage gene encoding a phage structural protein;
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
= a phage gene encoding a gene expression activator (eg, a RNA polymerase,
such as a RNA
polymerase of coliphage T4, T3 or K1-5); gene Q (eg, gene Q of coliphage
lambda); gene Rin (eg,
Rin of staphylococcal phage NM1), gene ogr (eg, ogr of coliphage P2) or gene
delta (eg, delta of coli
satellite phage P4));
= a phage RNA metabolism gene (ie, encoding a protein that is comprised by
RNA metabolism
system of a phage);
= a phage DNA metabolism gene ie, encoding a protein that is comprised by
DNA metabolism
system of a phage);
= a phage DNA packaging gene (ie, encoding a protein that is comprised by
DNA packaging
.. system of a phage);
= a phage gene encoding a protein necessary for bacterial cell lysis.
[00118]For example, each essential gene is selected from the following P2
genes (function is given in
brackets):
= Gene B (DNA replication);
= Gene Q (portal protein);
= Genes P and M (terminase);
= Genes 0, L and N (capsid);
= Genes X, R, S, v, W, J, I, H, G, Fl, FIT, E, E', T, U, D (tail and tail
fiber);
= Genes X, Y, lysA, lysB, lysC (lysis cassette); and
= Ogr (activator of late gene expression).
[00119]For example, each essential gene is selected from the following T7
genes (function is given in
brackets):
= Gene 1 (RNA polymerase);
= Genes 1.3, 2.5, 3, 3.5, 4a, 5, 6 (DNA and RNA metabolism);
= Genes 6.7, 7.3, 8, 9, 10A, 10b, 14, 15, 16 (capsid and internal core);
= Genes 11, 12, 17 (tail);
= Genes 17.5, 18.5, 18.6, 18.6 (lysis); and
= Genes 18 and 19 (terminase).
[00120] The phage is a virus that is capable of infecting a target bacterial
cell or the transduction
particle is capable of transducing a target bacterial cell, ie, is capable of
introducing first DNA or a
portion thereof into the target cell.
76
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[00121] In an embodiment, the kit comprises a cell (eg, a bacterial cell)
comprising the first and
second DNAs. For example, the cell is a bacterial cell, the first DNA is
comprised by an episome (eg,
plasmid) that is devoid of said essential or structural protein gene(s) and
the second DNA is
comprised by a chromosome of the cell; and optionally all essential or phage
structural protein genes
are comprised by the second DNA and the first DNA and episome is devoid of
said genes.
Optionally, the first DNA (or portion thereof) encodes a guided nuclease or a
component of a
CRISPR/Cas system (eg, a Cas, Cascade protein, crRNA, guide RNA or tracrRNA).
Optionally, the
first DNA (or portion thereof) encodes a crRNA or a guide RNA.
[00122] Optionally, the first DNA comprises a phage origin of replication.
[00123] Optionally, the transduction particle is a non-self replicative
transduction particle.
[00124] Optionally, the first DNA comprises phage replication genes and/or
phage lysis genes.
[00125] Optionally, the phage or particle comprises a tail fibre, eg, a tail
fibre (or domain thereof) of a
wild-type phage that comprises the structural proteins.
[00126] In an alternative to a bacterial cell, the cell is an archaeal cell
and the phage is a virus that is
capable of infecting archaea or the transduction particle is capable of
transducing archaea.
[00127] Optionally, a packaging signal herein is a pac or cos sequence, or is
a homologue thereof; or a
direct terminal repeat (DTR).
[00128] It will be understood that particular embodiments described herein are
shown by way of
illustration and not as limitations of the invention. The principal features
of this invention can be
employed in various embodiments without departing from the scope of the
invention. Those skilled in
the art will be able to ascertain using no more than routine study, numerous
equivalents to the specific
procedures described herein. Such equivalents are considered to be within the
scope of this invention
and are covered by the claims. All publications and patent applications
mentioned in the specification
are indicative of the level of skill of those skilled in the art to which this
invention pertains. US Patent
Application number 15/985,658, PCT/EP2018/082053, all publications and patent
applications and all
US equivalent patent applications and patents are herein incorporated by
reference to the same extent
as if each individual publication or patent application was specifically and
individually indicated to be
incorporated by reference. The use of the word "a" or "an" when used in
conjunction with the term
"comprising" in the claims and/or the specification may mean "one," but it is
also consistent with the
meaning of "one or more," "at least one," and "one or more than one." The use
of the term "or" in the
claims is used to mean "and/or" unless explicitly indicated to refer to
alternatives only or the
alternatives are mutually exclusive, although the disclosure supports a
definition that refers to only
alternatives and "and/or." Throughout this application, the term "about" is
used to indicate that a
77
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
value includes the inherent variation of error for the device, the method
being employed to determine
the value, or the variation that exists among the study subjects.
[00105] As used in this specification and claim(s), the words "comprising"
(and any form of
comprising, such as "comprise" and "comprises"), "having" (and any form of
having, such as "have"
and "has"), "including" (and any form of including, such as "includes" and
"include") or "containing"
(and any form of containing, such as "contains" and "contain") are inclusive
or open-ended and do not
exclude additional, unrecited elements or method steps.
[00106] The term "or combinations thereof" or similar as used herein refers to
all permutations and
combinations of the listed items preceding the term. For example, "A, B, C, or
combinations thereof is
intended to include at least one of: A, B, C, AB, AC, BC, or ABC, and if order
is important in a
particular context, also BA, CA, CB, CBA, BCA, ACB, BAC, or CAB. Continuing
with this example,
expressly included are combinations that contain repeats of one or more item
or term, such as BB,
AAA, MB, BBC, AAABCCCC, CBBAAA, CABABB, and so forth. The skilled artisan will
understand that typically there is no limit on the number of items or terms in
any combination, unless
otherwise apparent from the context.
[00107] Any part of this disclosure may be read in combination with any other
part of the disclosure,
unless otherwise apparent from the context.
[00108] All of the compositions and/or methods disclosed and claimed herein
can be made and
executed without undue experimentation in light of the present disclosure.
While the compositions
and methods of this invention have been described in terms of preferred
embodiments, it will be
apparent to those of skill in the art that variations may be applied to the
compositions and/or methods
and in the steps or in the sequence of steps of the method described herein
without departing from the
concept, spirit and scope of the invention. All such similar substitutes and
modifications apparent to
those skilled in the art are deemed to be within the spirit, scope and concept
of the invention as
defined by the appended claims.
EXAMPLES
Example 1: Efficient phage CRISPR delivery vehicle production
[00109] Background
We designed a strategy for efficient production of phage particles comprising
components of a
CRISPR/Cas system for killing target E coli Nissle strain bacteria. So our
phage composition will
consist of a lysate primarily containing CRISPR/Cas system components packaged
in phage particles
which will be devoid of phage protein-encoding sequences and which will have
no or a very low
78
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
proportion of helper phage. Also the strategy will work alternatively in less
well 79amster7979ius79
phage/bacterial strain combinations. Worked exemplification is provided in
Examples 3 and 4.
[00110] Outline of strategy for CRISPR/Cas component packaging in hitherto
unknown phages
(a) Identify a high or medium copy number cloning/lshuttle vector (capable
of cloning
and propagation in a first E coli strain (cloning production strain) and then
transfer to a second
bacterial host strain of interest (target host strain)), the vector containing
an E coli on for replication
in the E coli cloning production strain;
(b) Isolate temperate phage against the target host (second) bacterium;
(c) To produce the cloning production strain, identify or engineer a phage
production
strain of the host target bacteria (or other bacteria) that has an inactive
CRISPR/Cas system (eg, a
repressed Cas3 or other nuclease) and/or lacks the target protospacer found in
the second strain, and
which can be infected and lysogenized with the temperate phage; or repress or
inactivate the system in
the production strain;
(d) In that production strain make a lysogen using the temperate phage
(helper phage)
.. and test that it can be induced;
(e) Identify the packaging sequence (pac or cos) using PhageTerm
(https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5557969/) on whole genome
sequenced phage;
Delete the pac/cos packaging signal sequence in the helper phage in the
production
strain bacteria;
(g) Incorporate the packaging signal in the shuttle vector along with a
CRISPR-array or
single gRNA-encoding sequence (and optionally other components of the
CRISPR/Cas system, such
as a Cas9-encoding nucleotide sequence and optionally tracrRNA-encoding
sequence, or Cas3 and/or
Cascade-encoding sequence);
(h) Transform the vector into production host strain;
(i) Induce (eg, UV or mitomycin C induce) and harvest phage comprising the
CRISPR/Cas component(s). Alternatively, use a system with inducible RecA in
trans to simulate SOS
(needs to be activated RecA).
[00111] Example of the above specifically for E coli Nissle using phage P2:
[00112] Nissle is useful due to its GRAS (Generally Regarded as Safe) status
and P2 has a relatively
broad host range (most E coli, Shigella, Klebsiella, Salmonella in 79amster79
to DNA delivery into
e.g. Pseudomonas; Kahn et al 1991, "Bacteriophage P2 and P4", Methods in
Enzymology, vol 204,
pp264-280).
79
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[00113]We will use pUC19 or other high or medium copy number cloning vector.
Temperate phage
P2 can lysogenize Nissle. Most E coli K strains have an inactive CRISPR/Cas
system and can be
infected by P2 and thus all regular cloning hosts can be used (here
exemplified by E coli TOP10).
[00114]P2 is introduced into TOP10 to produce a lysogen. P2 cannot be induced
with mitomycin C or
UV but we will use the epsilon anti-repressor from the parasite phage P4 that
derepresses P2 and
makes it go into lytic phase. We will express this gene from an inducible
promoter in the production
host strain.
[00115] The 325 bp packaging signal sequence of SEQ ID NO: 2 will be used.
[00116] The packaging sequence will be deleted in the P2 prophage of the
lysogenic production
TOP10 strain.
[00117] A pUC19 shuttle vector encoding a guide RNA that targets the genome of
the target Nissle
strain (or alternatively comprising a CRISPR array for producing such a guide
RNA) will be
constructed and the packaging signal SEQ ID NO: 2 will be added. If the target
Nissle harbours its
own endogenous CRISPR/Cas system, we will use an activation strategy to
activate the endogenous
Cas3 by including Cas activating genes in the vector. If not, we will include
an exogenous Cas3-
encoding nucleotide sequence (and optionally one or more nucleotide sequences
encoding one or
more required Cascade components) in the vector for expression in the target
Nissle. We will
transform the vector into the TOP10 production strain, induce the P4 anti-
repressor and harvest phage
comprising the CRISPR/Cas component(s).
[00118] Since the induced (helper) phage DNA does not contain a packaging
signal we will be able to
isolate particles with only the vector DNA packaged. Thus, we will obtain a
composition comprising
such phage which can be used to infect target Nissle E coli bacteria and
introduce the CRISPR/Cas
component(s) therein for killing the target bacteria.
Example 2: MGEs, Genomics Islands etc
[00119] Overview of possible different MGE packaging strategies follow.
[00120] Applicable to different types of phages:
= Identify packaging signal and structural genes in the helper phage
= Delete packaging signal in helper phage and place on plasmid comprising
MGE
= Place both helper and plasmid in production strain
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
= Induce structural gene transcription of helper to get production of
helper-phage-packaged
MGEs
[00121]For using parasitic mobile elements (P4 phage or SaPI etc) activation
of helper phage
structural genes is done by induction of a helper phage activator obtained
from the parasitic element
Delta in P4 or one, more or al of ptiA/B/M in SaPI.
[00122]If one wants smaller size particles one can choose to package in a
parasite-size capsid
(typically 10-20 kb) by including in the MGE or vector P4 Sid and psu or
cpmA/B from a SaPI.
[00123] One can use defective helper phages where at least the packaging
signal has been removed
and structural genes are either on a plasmid or integrated as a cryptic
prophage in the production host.
If for some reason one cannot use this approach and need to use functional
helper phages, one will
include in the MGE or vector the genes on the parasite that hijack the phage
packaging machinery to
preferentially package parasite DNA (in our case CGVTM) over phage DNA.
[00124]List of the minimal genes one could include on a plasmid vector from
P4.
[00125]P4 sequence: see https://www.ncbi.nlm.nih.gov/nuccore/x51522
[00126]Cos packaging site: SEQ ID NO: 3
[00127] The homologous sequence between P2 and P4; this may be used as an
alternative packaging
signal in the MGE or vector: SQ ID NO: 4
[00128]For small capsid size (packages 11.4kb instead of 33.5 kb) Sid and/or
Psu can be included in
the MGE or vector:-
Sid: SEQ ID NO: 5
Psu: SEQ ID NO: 6
[00129] To activate helper phage P2, Delta from P4 can be included in a host
cell genome (provided
separately in a host cell, not on the MGE or vector to be packaged)
Delta: SEQ ID NO: 7
[00130] Minimum genes to include in the host chromosome/episome from P2.
P2 sequence (acc.number: NC_001895)
[00131]Figure 1 shows the genetic map of P2 genome with non-essential genes
boxed ¨ one, more or
all of these can be excluded (but Cos is always deleted). Cos is deleted and
preferably the whole
81
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
region from int through Cos. This region may, for example, be swapped with a
resistance marker
while the orf30 and fun(Z) genes are left intact or may be deleted.
[00132] Thus in an embodiment of any aspect herein of the invention, int
through to Cos (ie, including
int and Cos) are omitted from the helper phage DNA or second DNA or a
homologous region is
omitted when different phage are used as the basis for the helper phage DNA or
second DNA.
The sequences of various stretches of the P2 genome are as follows:-
[00133]"Q" through "S": SEQ ID NO: 8
[00134]"V" through "G": SEQ ID NO: 9
[00135] "Fl" through "ogr": SEQ ID NO: 10
[00136]Minimal genes to include from a SaPI on a vector or MGE.
Several different SaPI systems exist. Figure 2 shows one of the well
characterized SaPIs (SaPIbov1),
which exploits phages phill or phi80a1pha as helper phage. SaPIbov 1 sequence
(acc.number:
AF217235.1)
Packaging signal
[00137]If one uses a defective helper phage with deleted packaging signal one
can use that signal
from the helper phage and include it in the DNA to be packaged. An example
from S. aureus phill
(acc. Number: AF424781) is SEQ ID NO: 11.
[00138]For small capsid size (packages 15.8 kb instead of 43.6 kb), one can
include cpmA and/or
cpmB in the MGE or vector (SEQ ID Nos: 12 and 13).
[00139] To activate helper phage phill one can include one, more or all of
ptiA, B and M (provided
separately in the production host cell and not on the MGE or vector to be
packaged) (SEQ ID Nos:
14-16).
[00140]Minimum genes to include in the host chromosome/episome from phill.
Phill sequence (acc.number: AF424781)
[00141]gene #29 (terS) through gene #53 (lysin): SEQ ID NO: 17
[00142]A list of phage that work with SaPIs
82
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[00143] Different SaPIs are linked to different helper phages (see Table 2
below)
[00144] One can mutate the helper phage to only contain structural genes to
direct the phage to
package in smaller capsids. If only looking at the genes responsible for small
capsid packaging (cpmA
and cpmB) these are highly conserved among staphylococci indicating that they
will function to
redirect packaging in a variety of phages broader than the list below (Table
2).
Example 3: Design, construction and in vitro efficacy of a synthetic
transduction particle
delivery platform
Executive summary
[00145] SA100 is a synthetic DNA delivery vehicle comprising non-self-
replicative transduction
particles comprising phage coat proteins packaging DNA plasmids that comprise
nucleic acid
sequences endoding CRISPR/Cas components, wherein the particles are capable of
infecting target E
coli host cells to introduce the DNAs therein. Introduced DNAs are then able
to be used in the host
cells to produce the components. We call such DNAs CRISPR-Guided Vectors
(CGVsTm). As
shown in this example, we made such particles using a defective helper phage.
Upon production of
the synthetic particles we obtain a pure synthetic lysate devoid of any phage
nucleic acid encoding
phage proteins. We show that this lysate is capable of very efficient delivery
of a CGVTM to E. coli in
vitro. The particles are non-self-replicative and require the helper phage for
replication; thus usefully
the lysate is pure and removes the possibility of replication of the particles
containing the CGVTM.
This is useful to contain the action of the antibacterial, to allow for more
controlled (eg,
predetermined) dosing for administration to a human or animal subject or an
environement for
example, and the elimination of replication in the subject or environment
reduces the chances of
phage acquiring undesirable genes or traits from surrounding phage or bacteria
(eg, undesirable
antibiotic resistance genes) ¨ and even then the limitation of particle
replication of the invention
affords a way of reducing or eliminating spread of such genes or traits. This
may be of interest to
authorities such as the US FDA or USDA who consider such aspects when
approving medicines or
antibacterials for use in the environment.
Study objectives
[00146] Objective 1: Design and engineer the fully synthetic phage-based CGV
delivery platform
SA100.
83
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[00147] Objective 2: Use SA100 to demonstrate CGV delivery to E.coli.
Materials and methods
Bacterial strains and growth conditions
[00148] We made a bacterial production strain for producing the synthetic
SAE100 transduction
partiles. E coli strain C-2323 harboring phage P2 was used as starting
material for engineering our
SA100/CGV production strain (gift from Gail E. Christie at the Virginia
Commonwealth University).
Other strains used for testing SA100 and infectivity were EMG-2, C-la and C-
1792, (purchased from
the Coli Genetic Stock Center http://cgsc2.bio1ogy.ya1e.edu/).
[00149] All bacterial cultures were grown in LB medium supplemented with 2mM
MgCl2. When
appropriate, cultures were supplemented with 50 g/mL kanamycin or 50 g/mL
spectinomycin for
selection of defective prophage and CGV, respectively. In SA100 infection
studies, the medium was
supplemented with 2.5 mM CaCl2 to aid adsorption of the SA100 particles to
bacterial cells. For
induction of the CRISPR/Cas system, cells were exposed to 0.5mM IPTG and 2mM
theophylline.
Construction of defective helper prophage
[00150] The pTK-red recombineering plasmid (p'72) was transformed in strain C-
2323.
A KamR kanamycin marker gene was amplified using oligos with 50 bp homology to
the regions of
the P2 prophage immediately up- and downstream of the approximately 10 kbp
region of P2 that we
wanted to delete (see Figure 3). Following successful replacement of the
desired region with the
KanR marker gene, the thermosensitive pTK-red recombineering plasmid was
cured.
Engineering CGV (p94) for packaging in the SA100 particles
[00151] CGV p77 was constructed using plasmid p53 as a template. Plasmid p53
is a plasmid
encoding components of a CRISPR/Cas system, the plasmid comprising nucleic
acid sequences
endoding a tracrRNA and the Cas9 protein from Streptococcus pyogenes (SpCas).
Nucleic acid
sequence for produing crRNAs was obtained, expression driven by a plac
promoter. The two
fragments were assembled using Gibson assembly (NEB E55 10S) and transformed
into E. coli
competent cells. Assembled p77 plasmid was verified by full sequencing.
[00152] p77 harboring the inducible CRISPR/Cas system was used as the backbone
for insertion of a
fragment comprising an arabinose inducible promoter transcribing the region
sid through cos from the
satellite phage P4 (Figure 3).
[00153] Both fragments were PCR amplified and cloned by restriction enzyme
cloning. The resulting
SA100 packable CGV (p94) was sequence verified.
84
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Production of SA100 packaged CGVs
[00154] CGV p94 was transformed into the previously constructed strain
harboring the defective
prophage thereby generating production strain #189.
[00155] The production strain was grown exponentially for approximately 10
generations in the
presence of kanamycin and spectinomycin, ensuring balanced growth of the
population. At optical
density (0D600) of 1 the culture was induced with 1% arabinose and incubation
was continued until
optical density measurements were low and stable. After spinning away cell
debris the lysate was
filtered (0.45 tim) and stored at +4C until further use.
Transduction protocol to determine SA100 titers.
[00156] Determination of titers was done by mixing the SA100 lysate with a
suitable bacterial strain
ensuring at least 100-fold surplus of bacterial cells in presence of 2.5 mM
CaCl2. Following 30 min
incubation, the bacteria were diluted and plated on LB containing
spectinomycin for enumeration of
bacteria that had received the p94 CGV by transduction. To verify purity of
the SA100 lysate,
undiluted lysate was spotted on a lawn of bacteria (C-1a) known to support P2
proliferation using
standard soft-agar overlay. Absence of plaque formation was used to verify
purity.
DNA delivery assay
[00157] A transduction protocol was used in which the SA100 lysate was diluted
before infecting the
bacterial culture to obtain multiplicities of infection (MOT) ranging from
0.01 to 100 effectively
spanning a 4 log-ratio of SA100 to bacterial cells.
[00158] Following 30 min infection in the presence of 2.5 mM CaCl2 the
bacteria were enumerated on
LB plates with and without spectinomycin selection for delivery of the p94
CGV. The obtained
numbers were used to calculate the percentage of the population infected at a
given MOT.
Results
Design of the SA100 vehicle
[00159] The overall design of the SA100 E. coli production strain harboring
CGV and defective
prophage is outlined schematically in Figure 4. We used the native E. coli
phage P2 integrated as a
prophage on the chromosome of the production strain as our starting material.
In the prophage, we
deleted genes that are essential for phage to initiate and carry out its own
proliferation thus generating
a defective helper phage.
[00160] We then placed expression of the structural genes needed to produce
SA100 transduction
particles under the control of an inducible promoter allowing us to turn on
production of particles.
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Finally, we removed the packaging sequence normally used by the phage to
package its DNA into the
phage particles from the phage DNA and placed it on our CGV (see cos sites
shown in Figure 3).
Upon induction of the phage structural genes we got packaging of CGV DNA into
the particles that
were subsequently released by lysis of the production cells.
Engineering defective helper phage
[00161] E. coli strain C-2323 harboring phage P2 on the chromosome was used as
starting material for
engineering the production strain with the defective helper phage. A region on
the P2 phage
comprising integrase, promoters for initiation of phage proliferation, origin
of replication, DNA
replication genes and the cos site (DNA sequence 86amster8686i when packaging
DNA into phage
particles) (see boxed P2 DNA shown in Figure 3), was replaced by a kanamycin
marker using
recombineering. The site-specific integration of the marker and deletion of
approximately 10 kbp of
phage DNA was verified by sequencing.
Engineering CGV to enable SA100 packaging
[00162] We used a single vector (p77) with the CRIPSR/Cas system expressed
from inducible
promoters for engineering our SA100 packable CGV.
[00163] To enable activation of the defective P2 helper phage and CGV
packaging into synthetic
transduction particles, we cloned the genetic region comprising sid-delta-psu-
cos (cos packaging site)
from the satellite phage P4 into the p77 plasmid. The cloned P4 region (SEQ ID
NO: 18), which is
known to be able to activate the P2 phage was already cloned in another vector
p93 where it was
expressed from an arabinose inducible promoter. The resulting CGV (p94),
containing all elements
necessary for P2 activation and SA100 packaging, was verified by complete
sequencing. P4 genomic
architecture and the region cloned in p94 is shown in the lower box in Figure
3A and the p94 genomic
map is shown in Figure 3B.
Production of SA100 packaged CGVs
[00164] The p94 CGV was electroporated into the strain harboring the defective
prophage resulting in
production strain #189.
[00165] SA100 packaged CGVs were produced as described above. Titers of the
SA100 packaged
CGV was approximately 1010 TFU/mL determined by transduction of SA100 packaged
p94 into 3
strains of E. coli. As expected, no contamination of the native P2 phage could
be observed (Table 3
below). Accordingly, we can assume a native phage contamination of less than 1
in 1e9 per synthetic
particle.
86
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
CGV delivery to E. coli using SA100
[00166] To determine the number of phages required for 100% infection of an E.
coli population, we
performed an infection experiment varying the ratio of SA100 particles to
bacterial cells, the so-called
multiplicity-of-infection (MOT) from 0.01 to 100.
[00167]We observed that a MOT of 1 meaning one SA100 particle per E. coli
cell, was sufficient for
100% infection of the bacterial population (Figure 4).
Discussion and conclusions
[00168] SA100, a fully synthetic phage-based CGV delivery vehicle was designed
and constructed.
Upon expression in a production strain also harboring a compatible CGV, a high
titer lysate of SA100
packaged CGVs devoid of native phage contamination was produced. Finally, the
obtained SA100
packaged CGVs were efficiently delivered into E. coli.
Example 4: CGV in vitro killing of E. coli following delivery by the SA100
synthetic
transduction particle delivery platform
Executive summary
[00169]We used the SA100 synthetic DNA delivery vehicle to deliver
87amster8787 CRISPR guided
vectors (CGVTM) to two E. coli target strains. Upon induction of the system we
could demonstrate up
to 4 logs killing of the target population.
Introduction
[00170]We have previously engineered the phage-based synthetic DNA delivery
vehicle SA100
(Example 3) for delivery of CGVs to E. coli target cells. Here, we
87amster8787 a CGV for delivery
by the SA100 vehicle and subsequently tested the efficacy in killing E. coli
target cells.
Study objectives
[00171] Objective 1: Optimize CGV for delivery by SA100.
[00172] Objective 2: kill E.coli by SA100 delivered CGVs
87
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Materials and methods
Bacterial strains and culture conditions
[00173] All bacterial cultures were grown in LB medium supplemented with 2mM
MgCl2. When
appropriate, cultures were supplemented with 50 g/mL kanamycin or 50 g/mL
spectinomycin for
.. selection of defective prophage and CGV, respectively. In SA100 infection
studies, the medium was
supplemented with 2.5 mM CaCl2 to aid adsorption of the SA100 particles to
bacterial cells. For
induction of the CRISPR/Cas system, cells were exposed to 0.5mM IPTG and 2mM
theophylline.
Target strains MG1655_pks and XL1-blue_pks were constructed by recombineering
of a 20-
nucleotide sequence complementary to the guide RNA spacer sequence in our CGVs
into the lacZ
gene of MG1655 and XL1-blue thereby generating target strains MG1655_pks and
XL1-blue_pks,
respectively.
Production of SA100 packaged CGVs
[00174] CGVs (p94 and p114) were separately transformed into a respective
strain harboring the
defective prophage thereby generating production strain #189 and #226,
respectively.
[00175] The production strains were grown exponentially for approximately 10
generations in the
presence of kanamycin and spectinomycin ensuring balanced growth of the
population. At optical
density (0D600) of 1 cultures were concentrated x10 and induced with 1%
arabinose and incubation
was continued until optical density measurements were low and stable. After
spinning away cell
debris the lysate was filtered (0.45 tim), further concentrated approximately
x4 using Amicon Ultra-
15 centrifugal filters and stored at +4C until further use.
CGV delivery and killing
[00176] Exponentially growing cultures of the E. coli target strains were
infected with SA100
packaged CGVs p94 or p114 at a MOI of 20 in the presence of 2.5mM CaCl2.
[00177] Following 30 min incubation at 37 degreees centigrade, the bacterial
cultures were serially
diluted and plated on LB plates containing IPTG and theophylline (for inducing
the expression of
CRISPR/Cas components) to investigate CGV mediated killing or on
LB+spectinomycin to
investigate CGV delivery to the target populations.
88
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Results
Optimising CGV DNA by increasing copy number
[00178] CGV p94 was previously shown to be efficiently delivered to E. coli
(Example 3). This CGV
contains the CloDF13 origin of replication, in which a single SNP will
increase copy number from 7
to 80 according to Stuitje et al. 1981, "Identification of mutations affecting
replication control of
plasmid Clo DF13", Nature, vol 290, pp264-267. By incorporating the SNP into
an oligo used for
PCR amplification of the whole CGV, we created p114 containing the mutated
CloDF13 on (SEQ ID
NO: 23). Sequence verification was carried out.
CGV delivery by SA100
[00179] We previously showed efficient CGV delivery using p94 packaged in
SA100 (Example 3).
Here, we compared delivery of CGVs p94 and p114 to MG1655_pks (Fig 6A and
additionally p114
delivery to XL1-blue_pks (Fig 6B. Using a multiplicity of infection (MOI) of
20 we demonstrate
100% delivery to the target populations.
Killing efficacy of SA100 delivered CGV
[00180] Using the same experimental setup as above (target strains, SA100
packaged CGVs and MOI
of 20) we investigated the ability of the delivered CGVs to kill the target
cell upon induction of the
expression of the CRISPR/Cas system.
[00181] The p94 CGV targeting the pks DNA sequence in MG1655_pks was able to
reduce the target
strain approximately 2 logs while p114 targeting the same sequence reduced the
target population
approximately 4 logs (Fig 7). The same magnitude of killing was observed when
p114 was targeting
pks in the XL1-blue_pks target population (Fig 7).
Discussion and conclusions
Following SA100 delivery of CGVs to target cells we were able to increase
killing of the target
population from approximately 2 logs using p94 to 4 logs using p114 probably
due to a high copy
number CGV.
89
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Example 5: In vivo antibacterial delivery to gut microbiome using non-self-
replicative particles
& target cell killing in microbiome setting
Executive summary
[00182]E. coli AMG1655-pks was used to colonize the murine gut. We show
statistically significant
delivery of CGVTM pSNP114 into MG1655-PKS by SA100 delivery vehicle (synthetic
non-self-
replicative phage particles, see Examples 3 and 4). We observered a reduction
in survival upon
activation of the CGVTM targeting E. coli MG1655-pks.
Introduction
[00183] We have previously shown that E. coli ATCC43888 colonizes the a murine
gut of NMRI
mice model (data not shown) and that activating of a CRISPR array in E. coli
ATCC43888 in the
mouse gut reduces survival, where guide RNA produced using the array target
the E coli genome for
Cas cutting (data not shown). In this Example we set out to assess the
survival of a CRISPR/Cas-
based antibacterial (a CGVTM) delivered by a Synthetic NanoboioticTM, namely
SA100, into MG1655-
pks.
Study objective:
[00184] Establish survival reduction of E. coli MG1655-pks in the murine gut
microbiome by delivery
of CGVTM pSNP114 with Synthetic Nanoboiotic TM SA100.
Materials and methods
[00185]E coli was E coli MG1655 with pks fragment inserted, with a strep
resistance marker, phylo
group A.
[00186] Cells were grown overnight in LB media supplemented with 50 g/mL
spectinomycin. Next
day the OD was measured and the cultures were diluted down to 108 CFU/mL.
Vehicle, induced and
uninduced groups are shown in Table 4.
= 15 female NMRI mice, 26-30 gram (Taconic)
= Inducer solution containing theophylline and L-arabinose
Laboratory animal facilities and housing of mice
[00187] The temperature was 21 C +/- 2 C and could be regulated by heating and
cooling, and
light/dark period was in 12-hours intervals of 6 a.m.- 6 p.m./6 p.m ¨ 6 a.m.
Mice had free access to
food and to domestic quality drinking water until 3 days prior to the study.
Mice were housed 6-7
mice /cage in the standard facility with bedding from Aspen Wood from Tapvei
and paper strands
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
from Sizzle-nest as nesting material until 1 day prior to the study and there
after moved into the GMO
facility 1 mouse per cage. The From day 0 the wooden bedding was changed to
white paper bedding
to facilitate collection of faeces from the cages
Preparation of streptomycin drinking water
[00188] 6 liters of 5 g/L of streptomycin was prepared by dissolving 6.94 g
streptomycin sulphate / L
of sterile water. Mice were given streptomycin water starting day -3 and
throughout the study.
Inoculation of mice
[00189] Inocula was formulated prior to inoculation as ready to use
suspensions. Mice were
inoculated with 0.25 ml by oral gavage at 8 am day 0. A titre of approximately
10" TFU/ml was used.
Treatment of mice
[00190] Mice were dosed with inducers and 10" TFU/mL SA100 by oral gavage at 7
am and 1 pm
day 1 and again at 7 am day 2. Inducers were mixed immediately prior to each
treatment time point
prior to the treatment mice were gavaged with antacid.
Collection of faecal samples from cages
[00191] Immediately prior to inoculation 0.5 ml faecal samples were collected
from cages into three
15 ml Nunc tubes. Day 1 and 2 after inoculation, 0.5 ml faecal samples were
collected prior to
treatment at 7 am and mice were moved to a new cage. Day 2 after inoculation
faecal samples were
also collected 4 hours after treatment
CFU determination
[0019210.5 ml faecal matter was dissolved in 5 ml sterile saline by vortexing
multiple times during 1-
4 hours. Colony counts were determined by 10 fold serially dilution of the
faecal samples in sterile
0.9% NaCl and 20 111_, spots were applied to agar plates. The lower detection
limit of faecal samples
was 50 CFU/ml. All agar plates were incubated 18-22 hrs at 35 C in ambient
air.
[00193] Delivery of the CGV was enumerated by plating on LB + streptomycin +
spectinomycin
plates plates. Abudance of the target strain was enumerated by plating on LB
with streptomycin only.
91
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Clinical monitoring of mice
[00194] The body weight of the mice were monitored throughout the study and
the mice were scored
0 ¨ 6 based on their behavior and clinical signs (Table 5). Mice were
euthanized if reaching score 3 or
20% weight loss.
Results
[00195]Microbiome analysis of mice prior to inoculation
No bacteria was detected in 2 out of 3 of the pooled samples collected prior
to inoculation. One of the
pooled samples had 6.54 10g10 CFU/ml of small white colonies. MALDI analysis
indicated
Paenibacillus odorifer with a score value of 1.71 and as second most likely
Staphylococcus xylosus
with a score value of 1.43 both with a low rank quality. Also in the 2 other
pooled samples 3-4 10g10
CFU/ml of small white colonies appeared after 48 hours of incubation.
[00196] At 24 h after inoculation the CFU levels in the faecal samples ranged
from 6.1 ¨ 8.6 10g10
CFU/sample as determined on the LB+ strep agar plates. Similar levels were
observed day 2 after
inoculation. No significant differences were observed with in each of the 2
inoculation groups.
[00197] On the LB+ strep + spec agar plates, no colonies were observed day 1
in in any of the groups.
Day 2 CFU counts ranging from 1 to 5 log10 CFU/sample was observed in the
inducer treated groups
but not in the vehicle treated groups.
Layout of animal study
[00198] Twenty four mice were used in each murine intervention study.
Clinical mice score
[00199]Mice did not show any clinical signs of infection or discomfort during
the entire study period.
Induction killing of MG1655-pks
[00200]Faecal matter extracts were plated out on on plates containing
streptomycin to determine the
CFU of the E. coli MG1655-PKS strain (Fig 8). Streptomycin CFU counts show no
difference
between the various treatment groups.
[00201]When plating the faecal samples on spectomycin and streptomycin to
determine delivery of
CGV pSNP114 (because this CGV carries a spectomycin resistance marker) we
observe a significant
increase in CFU, and thus CGV delivery, after 48 hours (Fig 9). When the
CRISPR system was
induced we observed a significant decrease in the mean CFU counts.
92
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Discussion and conclusions
[00202]We successfully demonstrated guided nuclease antibacterial (CGVTM)
delivery using
Synthetic NanobioticTM platform, SA100, into E. coli MG1655-pks in the murine
gut model. Thus,
delivery of an antibacterial using non-self-replicative particles to a
microbiome in vivo was
.. established. A statistically significant reduction in CFU counts was
observed using the activated
CRISPR system on the CGVTM.
Example 6: Production of Transduction Particles By Essential Functions in
Trans
[00203]Native phage are engineered to convert them into DNA carrier vehicles
(transduction
particles) for our CRISPR-Guided VectorsTM (CGVs). The particles can transduce
target bacterial to
introduce the vectors, from which CRISPR/Cas systems or one or more components
thereof are
expressed.We make use of lytic propagation in which essential helper functions
are provided only by
the host cell (SA800 & SA900).
SA800 & SA900
[00204] Production of SA800 & SA900 particles is based on lytic propagation on
a production host
bacterial strain. We engineer native (wild-type) phage by inactivating
essential phage functions (e.g.
by deleting structural genes) and adding nucleotide sequences encoding the
CRISPR-Cas systems.
[00205]By providing the essential genes on a separate helper plasmid, we can
make the CGV-phage
hybrid propagate on the production strain. However, CGV propagation is not
possible on any other
bacterial strains due to the lack of the helper plasmid.
[00206]We refer to Figure 10. To construct a non-self replicative particle
comprising a CGV
comprising nucleotide sequences encoding components of a CRISPR-Cas system, we
engineer a
native phage to contain a phage packaging signal and genes needed for phage
DNA replication. The
CGV may contain multiple phage genes as long as one or more essential genes
are deleted or mutated
to make the CGV-phage hybrid non-self replicative on strains not expressing
the essential function(s).
In the production strain, this function(s) is provided in trans expressed from
a plasmid or the bacterial
cell chromosome.
[00207] In one example, the production cell will contain a plasmid with a gene
encoding a structural
.. phage protein, that is expressed during CGV propagation and packaging. A
vector is constructed
comprising a native phage genome in which this gene is mutated and into which
are inserted
nucleotide sequences encoding components of a CRISPR-Cas system, forming a CGV-
phage hybrid.
Thus, the CGV-phage hybrid is non-functional to package the CGV DNA into
transduction particles
in the absence of the helper function provided in trans.
93
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
[00208] In a minimal version, all essential functions (ie, essential for
packaging) are carried by the
production host cell (eg, on a chromosome or one or more plasmids) except for
the phage packaging
signal and a phage DNA replication machinery (including phage origin of
replication) which are
instead carried on the CGV. Importantly, the DNA replication machinery may not
need originate from
the same phage as the essential genes on the helper plasmid or chromosome. For
example, the DNA
replication machinery from phage lambda may be included on a CGV (and used to
replicate this)
carrying a phage P2 packaging signal and packaged into P2 phage capsids (eg,
wherein the capsid
proteins are encoded by a helper plasmid or chromosome of the production
strain cell).
[00209] The CGV is capable of replicating as a lytic phage (i.e. propagation
by infection-lysis-
reinfection cycles) on a suitable production host strain carrying the helper
function(s). However, in
non-production cells such as clinical target bacteria, the phage cannot
propagate by the lytic cycle but
merely functions as a DNA delivery vehicle, thereby delivering sequences
encoding one or more
components encoding a CRISPR/Cas component(s) (or alternatively another
antibacterial agent or
other protein or RNA of interest).
94
Table 1: Example Bacteria
0
t..)
Optionally, the host cells are selected from this Table and/or the target
cells are selected from this Table (eg, wherein the host and target cells are
of a o
t..)
o
different species; or of the same species but are a different strain or the
host cells are engineered but the target cells are wild-type or vice versa).
For example
.6.
.6.
the host cells are E coli cells and the target cells are C dificile, E coli,
Akkermansia, Enterobacteriacea, Ruminococcus, Faecalibacterium, Firmicutes,
n.)
oe
Bacteroidetes, Salmonella, Klebsiella, Pseudomonas, Acintenobacter or
Streptococcus cells.
Abiotrophia Acidocella Actinomyces
Alkalilimnicola Aquaspirillum
Abiotrophia defectiva Acidocella aminolytica Actinomyces bovis
Alkalilimnicola ehrlichii Aquaspirillum polymorphum
Acidocella facilis Actinomyces denticolens
Aquaspirillum
Acaricomes
Alkaliphilus
Actinomyces europaeus
putridiconchylium
P
Acaricomes phytoseiuli Acidomonas
Alkaliphilus oremlandii
Actinomyces georgiae
Aquaspirillum serpens 2
Acidomonas methanolica
Alkapus ransvaaenss
o
Actinomyces gerencseriae ;3'lihil t l i
vi
?.
Acetitomaculum .
Actinomyces
Aquimarina
N)
Acetitomaculum ruminis Acidothermus
Allochromatium ,
hordeovulneris
Aquimarina late rcula
7
Acidothermus cellulolyticus
Allochromatium vinosum r.,
N)
Acetivibrio Actinomyces howellii
Arcanobacterium
li i h Actinomyces yovagnas
Acetivibrio cellulolyticus Acidovorax Ac
Alloiococcus Arcanobacterium
lii i Actinomyces srae
Acetivibrio ethanolgignens Acidovorax anthurii Act
Alloiococcus otitis
haemolyticum
Acetivibrio multivorans Acidovorax caeni Actinomyces johnsonii
Arcanobacterium pyo genes
Actinomyces meyeri
Allokutzneria
Acidovorax cattleyae
Iv
n
Acetoanaerobium Actinomyces naeslundii
Allokutzneria albata 1-3
Acidovorax citrulli
Archangium t=1
1-d
Acetoanaerobium noterae Actinomyces neuii
t..)
Acidovorax defluvii
Archangium gephyra o
t..)
o
Acidovorax delafieldii Actinomyces odontolyticus
'a
o
.6.
t..)
t..)
un
Acidovorax facilis Actinomyces oris
o
t..)
Acetobacter
Altererythrobacter Arcobacter o
Acidovorax konjaci Actinomyces radingae
t..)
o
Acetobacter aceti
Altereiythrobacter Arcobacter butzleri
Acidovorax temperans Actinomyces slackii
.6.
Acetobacter cerevisiae
ishigakiensis Arcobacter ciyaerophilus .6.
t...)
Acidovorax valerianellae Actinomyces turicensis
oe
Acetobacter cibinongensis
Arcobacter halophilus
Actinomyces viscosus
Altermonas
Acetobacter estunensis Acinetobacter
Arcobacter nitrofigilis
Altermonas haloplanktis
Acetobacter fabarum Acinetobacter baumannii
Actinoplanes Arcobacter ski rrowii
Altermonas macleodii
Acetobacter ghanensis Acinetobacter baylyi Actinoplanes
auranticolor
Arhodomonas
Acetobacter indonesiensis Acinetobacter bouvetii Actinoplanes
brasiliensis
Alysiella
Arhodomonas aquaeolei
P
Acetobacter lovaniensis Acinetobacter calcoaceticus Actinoplanes
consettensis
Alysiella crassa
2
Acetobacter malo rum Acinetobacter gerneri Actinoplanes
deccanensis ;3'
o
Alysiella filifonnis Arsenophonus
o,
..
Acetobacter nitrogenifigens Acinetobacter haemolyticus Actinoplanes
derwentensis
Arsenophonus nasoniae
2'
,
Acetobacter oeni Acinetobacter johnsonii
Actinoplanes digitatis Aminobacter
,
""
Acetobacter orientalis Acinetobacter junii Actinoplanes durhamensis
Aminobacter aganoensis
Acetobacter orleanensis Acinetobacter lwoffi Actinoplanes ferrugineus
Aminobacter aminovorans
Acetobacter pasteurianus Acinetobacter parvus Actinoplanes globisporus
Arthrobacter
Aminobacter niigataensis
Acetobacter pornorurn Acinetobacter radioresistens Actinoplanes
humidus Arthrobacter agilis
Acetobacter senegalensis Acinetobacter schindleri
Actinoplanes italicus Aminobacterium Arthrobacter albus
Iv
Acetobacter xylinus Acinetobacter soli Actinoplanes liguriensis
Aminobacterium mobile Arthrobacter aurescens n
,-i
m
Acinetobacter tandoii Actinoplanes lobatus
Arthrobacter Iv
t..)
o
Acetobacterium
Aminomonas r..)
Acinetobacter tjernbergiae Actinoplanes missouriensis
chlorophenolicus o
'a
Acetobacterium bakii
Aminomonas paucivorans o
.6.
t..)
t..)
un
Acetobacterium carbinolicum Acinetobacter towneri Actinoplanes
palleronii Arthrobacter citreus o
t..)
Ammoniphilus
Acetobacterium dehalogenans Acinetobacter ursingii Actinoplanes
philippinensis Arthrobacter ciystallopoietes 2
=
Ammoniphilus oxalaticus
Acetobacterium fimetarium Acinetobacter venetianus
Actinoplanes rectilineatus Arthrobacter cumminsii
c,.)
.6.
.6.
hilus oxalivorans
r..)
Acetobacterium malicum Actinoplanes regularis
Ammonip Arthrobacter globiformis oe
oraosp
Acetobacterium paludosum Acrocarp Actinoplanes
Arthrobacter
Amphibacillus
Acrocarpospora corrugata
Acetobacterium tundrae teichomyceticus
histidinolovorans
Amphibacillus xylanus
Acetobacterium wieringae Acrocarpospora Actinoplanes utahensis
Arthrobacter ilicis
l haa
Acetobacterium woodii macrocep
Arthrobacter luteus
Amphritea
Acrocarpospora Actinopolyspora
Arthrobacter methylotrophus
Amphritea balenae
P
Acetofilamentum pleiomorpha Actinopolyspora halophila
Arthrobacter mysorens
2
Amphritea japonica
Acetofilamentum rigidum Actinopolyspora
o
Arthrobacter nicotianae 03-
'
--.1 Actibacter
.
mortivallis
r.,
Amycolatopsis
Arthrobacter nicotinovorans r.,0
Acetohalobium Actibacter sediminis
,
Amycolatopsis alba
Arthrobacter oxydans
Acetohalobium arabaticum Actinosynnema
r.,7
r.,
Amycolatopsis albidoflavus
Arthrobacter pascens
Actinoalloteichus Actinosynnema mirum
Acetomicrobium Actinoalloteichus
Amycolatopsis azurea Arthrobacter
Acetomicrobium faecale cyanogriseus Actinotalea
Amycolatopsis coloradensis phenanthrenivorans
Amycolatopsis lurida
Arthrobacter
Acetomicrobium flavidum Actinoalloteichus Actinotalea fennentans
Amycolatopsis mediterranei
polychromo genes
hymeniacidonis
Iv
n
Acetonema Aerococcus
Amycolatopsis rifamycinica Atrhrobacter protophormiae
1-3
Actinoalloteichus spitiensis
t=1
Iv
Acetonema longum Aerococcus sanguinicola
Amycolatopsis rubida Arthrobacter w
o
t..)
o
Aerococcus urinae
psychrolactophilus 'a
o
.6.
r..)
t..)
un
Aerococcus urinaeequi
Amycolatopsis sulphurea Arthrobacter ramosus o
t..)
Acetothermus Actinobaccillus
o
Aerococcus urinaehominis Amycolatopsis tolypomycina
Arthrobacter sulfonivorans t..)
o
Acetothermus paucivorans Actinobacillus capsulatus
Aerococcus viridans
Arthrobacter sulfureus c,.)
.6.
.6.
Actinobacillus delphinicola
Anabaena t..)
Arthrobacter uratoxydans
oe
Acholeplasma Actinobacillus hominis Aeromicrobium
Anabaena cylindrica
Arthrobacter ureafaciens
Acholeplasma axanthum
Actinobacillus indolicus Aeromicrobium mythreum
Anabaena flos-aquae
Arthrobacter viscosus
Acholeplasma brassicae
Actinobacillus lignieresii
Anabaena variabilis
Arthrobacter woluwensis
Acholeplasma cavigenitalium Aeromonas
Actinobacillus minor
Acholeplasma equifetale Aeromonas
Anaeroarcus
Actinobacillus muris
Asaia
Acholeplasma granularum allosaccharophila
Anaeroarcus burkinensis P
Actinobacillus
Asaia bogorensis
2
Acholeplasma hippikon Aeromonas bestiarum
pleuropneumoniae
;3'
o
Anaerobaculum .?.
oe Acholeplasma laidlawii Aeromonas caviae
Asanoa
Actinobacillus porcinus
r.,
Anaerobaculum mobile
,30
,
Acholeplasma modicum Aeromonas encheleia
Asanoa ferruginea
Actinobacillus rossii
N)
Acholeplasma morum Aeromonas
r.,
Actinobacillus scotiae
Anaerobiospirillum Asticcacaulis
Acholeplasma multilocale enteropelo genes
Actinobacillus seminis Anaerobiospirillum
Asticcacaulis biprosthecium
Acholeplasma oculi Aeromonas eucrenophila
Actinobacillus succinogenes
succiniciproducens
Asticcacaulis excentricus
Acholeplasma palmae Aeromonas ichthiosmia
Actinobaccillus suis
Anaerobiospirillum thomasii
Acholeplasma parvum Aeromonas jandaei
Actinobacillus ureae
Atopobacter 1-d
Acholeplasma pleciae Aeromonas media
n
Anaerococcus
Atopobacter phocae 1-3
Acholeplasma vituli Actinobaculum Aeromonas popoffii
t=1
Anaerococcus hydrogenalis
Iv
t..)
o
Actinobaculum massiliense Aeromonas sobria
t..)
Anaerococcus lactolyticus
o
'a
Actinobaculum schaalii Aeromonas veronii
o
.6.
t..)
t..)
un
Actinobaculum suis
Anaerococcus prevotii o
t..)
Achromobacter Agrobacterium
Atopobium o
Actinomyces urinale
Anaerococcus tetradius t..)
o
Achromobacter denitrificans Agrobacterium
Atopobium fossor
Anaerococcus vaginalis
.6.
.6.
Achromobacter insolitus Actinocatenispora gelatinovorum
Atopobium minutum r..)
oe
Achromobacter piechaudii Actinocatenispora rupis
Anaerofustis Atopobium parvulum
Achromobacter ruhlandii Actinocatenispora Agrococcus
Anaerofustis stercorihominis Atopobium rimae
Agrococcus citreus
Achromobacter spanius thailandica
Atopobium vaginae
Agrococcus jenensis
Anaeromusa
Actinocatenispora sera
Acidaminobacter
Anaeromusa acidaminophila Aureobacterium
romonas
Acidaminobacter Actinocorallia Ag
Aureobacterium barkeri P
Agromonas oligotrophica Anaeromyxobacter
2
hydrogenoformans Actinocorallia aurantiaca
;'3
o
Anaeromyxobacter Aurobacterium ..
o
Actinocorallia aurea r.,
Acidaminococcus Agromyces
ens r.
dehalogenans
Aurobacterium liquefaci3O
,
Actinocorallia cavernae
Agromyces fucosus
0
Acidaminococcus fermentans
Actinocorallia glomerata
r.,^'
Acidaminococcus intestini Agromyces hippuratus
Anaerorhabdus Avibacterium
Actinocorallia herbida
Agromyces luteolus
Anaerorhabdus furcosa Avibacterium avium
Actinocorallia libanotica
Acidicaldus Agromyces mediolanus
Avibacterium gallinarum
Actinocorallia longicatena
Anaerosinus
Acidicaldus organivorans Agromyces ramosus
Avibacterium paragallinarum
Anaerosinus glycerini
Agromyces rhizospherae
Avibacterium volantium
Actinomadura
1-d
Acidimicrobium
n
,-i
Actinomadura alba
Anaerovirgula m
Acidimicrobium ferrooxidans Akkermansia
Azoarcus 1-d
t..)
Actinomadura atramentaria
o
i Anaerovrgula multivorans
t..)
Akkermansia muciniphila
Azoarcus indigens o
Actinomadura
'a
o
.6.
t..)
t..)
un
ban gladeshensis
Azoarcus tolulyticus 0
Acidiphilium Albidiferax
Ancalomicrobium t..)
o
Actinomadura catellatispora
Azoarcus toluvorans t..)
o
Acidiphilium acidophilum Actinomadura chibensis Albidiferax
ferrireducens Ancalomicrobium adetum
.6.
.6.
Acidiphilium angustum Actinomadura chokoriensis
Azohydromonas t..)
oe
Albidovulum
Ancylobacter
Acidiphilium ciy Azohydromonas australica ptum
Actinomadura citrea
Albidovulum inexpectatum Ancylobacter aquaticus
Azohydromonas lata Acidiphilium multivorum
Actinomadura coerulea
Acidiphilium organovorum
Actinomadura echinospora Alcaligenes
Aneurinibacillus Azomonas
Acidiphilium rubrum
Actinomadura fibrosa Alcaligenes denitrificans
Aneurinibacillus
Azomonas agilis
Actinomadura formosensis Alcaligenes faecalis
aneurinilyticus
Acidisoma
Azomonas insignis P
2
Actinomadura hibisca
i
i Aneurinibacillus mgulanus
Acidisoma sibiricum
Azomonas macrocyto genes
1-, Actinomadura ktjaniata Alcanivorax
;3'
o
Aneurinibacillus .?.
o
Acidisoma tundrae r.,
Actinomadura latina Alcanivorax borkumensis
the rmoaerophilus
Azorhizobium ,30
,
Actinomadura livida Alcanivorax jadensis
o
Acidisphaera
Azorhizobium caulinodans r,
N)
Actinomadura
Angiococcus
Acidisphaera rubrifaciens
Algicola
luteofluorescens
Angiococcus discifonnis Azorhizophilus
Algicola bacteriolytica
Acidithiobacillus Actinomadura macra
Azorhizophilus paspali
Angulomicrobium
Acidithiobacillus albertensis Actinomadura madurae
Alicyclobacillus
Angulomicrobium tetraedrale Azospirillum
Acidithiobacillus caldus Actinomadura oligospora
Iv
Alicyclobacillus
n
Azospirillum brasdense
Acidithiobacillus ferrooxidans Actinomadura pelletieri
1-3
disulfidooxidans
Anoxybacillus t=1
Azospirillum halopraeferens
Iv
Acidithiobacillus thiooxidans Actinomadura rubrobrunea
t..)
Alicyclobacillus
Anoxybacillus pushchinoensis o
t..)
Azospirillum irakense
o
Actinomadura rugatobispora
'a
o
.6.
t..)
t..)
un
Actinomadura umbrina sendaiensis
o
t..)
Acidobacterium
Aquabacterium Azotobacter o
Actinomadura Alicyclobacillus
vulcanalis t..)
o
Acidobacterium capsulatum
Aquabacterium commune Azotobacter beijerinckii
verrucosospora
.6.
.6.
Alishewanella
Aquabacterium parvum Azotobacter chroococcum t...)
Actinomadura vinacea
oe
Alishewanella fetalis
Azotobacter nigricans
Actinomadura viridilutea
Azotobacter salinestris
Actinomadura viridis
Alkalibacillus
Azotobacter vinelandii
Actinomadura yumaensis
Alkalibacillus
haloalkaliphilus
P
2
,
1¨ Bacillus Bacteroides Bibersteinia
Borrelia Brevinema
o 2
1-,
r.,
[see below] Bacteroides caccae Bibersteinia trehalosi
Borrelia afzelii Brevinema andersonii 2
'7
Bacteroides coagulans
Borrelia americana 0"
N)
Bifidobacterium
Brevundimonas "
Bacteroides eggerthii
Borrelia burgdorferi
Bifidobacterium ado lescentis
Brevundimonas alba
Bacteroides fragilis
Borrelia carolinensis
Bacteriovorax
Bifidobacterium angulatum
Brevundimonas aurantiaca
Bacteroides galacturonicus
Borrelia coriaceae
Bacteriovorax stolpii
Bifidobacterium animalis
Brevundimonas diminuta
Bacteroides helco genes
Borrelia garinii
Bifidobacterium asteroides
Brevundimonas intermedia
Bacteroides ovatus
Borrelia japonica Iv
Bifidobacterium bifidum
Brevundimonas subvibrioides n
Bacteroides pectinophilus
1-3
t=1
Bifidobacterium boum
Brevundimonas vancanneytii Iv
Bacteroides pyo genes
t..)
o
Bifidobacterium breve
t..)
o
Bacteroides salyersiae
'a
o
.6.
t..)
t..)
un
Bacteroides stercoris Bifidobacterium catenulatum
Brevundimonas variabilis 0
t..)
Bosea
o
Bacteroides suis Bifidobacterium choerinum
Brevundimonas vesicularis t..)
o
Bosea minatitlanensis
Bacteroides tectus Bifidobacterium coiyneforme
c...)
.6.
.6.
Bosea thiooxidans
Brochothrix t..)
Bacteroides thetaiotaomicron Bifidobacterium cuniculi
oe
Bacteroides uniformis Bifidobacterium dentium
Brochothrix campestris
Brachybacterium
Brochothrix thermosphacta
Bacteroides ureolyticus Bifidobacterium gallicum
Brachybacterium
Bacteroides vulgatus Bifidobacterium gallinarum
alimentarium
BruceIla
Bifidobacterium indicum
Balnearium
Brachybacterium faecium BruceIla canis
Bifidobacterium longum
P
Balnearium lithotrophicum
Bifi
Brachybacterium BruceIla neotomaedobacterium 2
paraconglomeratum
1-, magnumBifidobacterium
o
.? Bryobacter .
w Balneatrix
Brachybacterium rhamnosum r.,
meiycicum
2
atusre obacter aggg
,
Balneatrix alpica
Brachybacterium Biy ,
Bifidobacterium minimum
0"
,
tyrofermentans
r.,"
Balneola Bifidobacterium
Burkholderia
pseudocatenulatum
Burkholderia ambifaria
Balneola vulgaris
Brachyspira
Bifidobacterium
Brachyspira alvinipulli Burkholderia andropogonis
Barnesiella pseudolongum
Burkholderia anthina
Brachyspira hyodysenteriae
Barnesiella viscericola Bifidobacterium pullorum
Brachyspira innocens Burkholderia caledonica
Iv
n
Bifidobacterium ruminantium
Burkholderia caiyophylli 1-3
Brachyspira murdochii
t=1
Bartonella
1-d
Bifidobacterium saeculare
Brachyspira pilosicoli Burkholderia cenocepacia t..)
o
Bartonella alsatica
t..)
o
Bifidobacterium subtile
Burkholderia cepacia 'a
o
.6.
t..)
t..)
un
Bartonella bacilliformis Bifidobacterium
Burkholderia cocovenenans o
t..)
o
Bartonella clarridgeiae the rmophilum
Burkholderia dolosa t..)
o
Bartonella doshiae
Burkholderia fungorum c,.)
.6.
Bradyrhizobium
Bilophila
.6.
t..)
Bartonella elizabethae
Burkholderia glathei oe
Bilophila wadsworthia
Bradyrhizobium canariense
Bartonella grahamii
Burkholderia glumae
Bradyrhizobium elkanii
Bartonella henselae
Burkholderia graminis
Biostraticola
Bradyrhizobium japonicum
Bartonella rochalimae
Burkholderia kururiensis
Biostraticola tofi
Bradyrhizobium liaoningense
Bartonella vinsonii
Burkholderia multivorans
Bizionia
Brenneria Burkholderia phenazinium
P
Bavariicoccus
Burkholderia plantarii 2
Bizionia argentinensis
Brenneria alni
Bavariicoccus seileri
1-,
Burkholderia pyrrocinia ;3'
o
Brenneria nigrifluens .2
r.,
Blastobacter Burkholderia silvatlantica 2'
Brenneria quercina
Bdellovibrio
,
Blastobacter capsulatus
Burkholderia stabilis
Brenneria quercina
Bdellovibrio bacteriovorus
r.3^'
Blastobacter denitrificans
Burkholderia thailandensis
Brenneria salicis
Bdellovibrio exovorus
Burkholderia tropica
Blastococcus
Brevibacillus Burkholderia unamae
Beggiatoa
Blastococcus aggregatus
Brevibacillus agri
Burkholderia vietnamiensis
Beggiatoa alba
Blastococcus saxobsidens
Brevibacillus borstelensis
Iv
n
Beijerinckia
Brevibacillus brevis Buttiauxella
Blastochloris
t=1
Buttiauxella agrestis
Iv
Betjerinckia derxii
Brevibacillus centrosporus t..)
Blastochloris viridis
o
t..)
b ll iauxea rennerae
o
Betjerinckia fluminensis
Brevibacillus choshinensis Butt 'a
o
.6.
t..)
t..)
un
Betjerinckia indica
Brevibacillus invocatus Buttiauxella ferragutiae o
t..)
Blastomonas
o
Betjerinckia mobilis
Brevibacillus late rosporus Buttiauxella gaviniae t..)
o
Blastomonas natatoria
Brevibacillus parabrevis
Buttiauxella izardii c,.)
.6.
Belliella
.6.
t..)
Brevibacillus reuszeri
Buttiauxella noackiae oe
Belliella baltica Blastopirellula
Buttiauxella warmboldiae
Blastopirellula marina
Brevibacterium
Bellilinea
Brevibacterium abidum
Butyrivibrio
Blautia
Bellilinea caldifistulae
Brevibacterium album
Butyrivibrio fibrisolvens
Blautia coccoides
Brevibacterium aurantiacum Butyrivibrio hungatei
Belnapia Blautia hansenii
P
Brevibacterium celere
Butyrivibrio proteoclasticus
Belnapia moabensis Blautia producta
2
Brevibacterium epidermidis
1-,
;33'
o
Blautia wexlerae .2
.6.
Brevibacterium
Bergeriella
2
,
Bergeriella denitrificans Bogoriella
frigoritolerans
N)
Brevibacterium halotolerans
r.,
Bogoriella caseilytica
Beutenbergia
Brevibacterium iodinum
Beutenbergia cavernae Bordetella
Brevibacterium linens
Bordetella avium
Brevibacterium lyticum
Bordetella bronchiseptica
Brevibacterium mcbrellneri
Iv
Bordetella hinzii
Brevibacterium otitidis n
,-i
m
Bordetella holmesii
Brevibacterium oxydans Iv
t..)
o
Bordetella parapertussis
t..)
o
'a
o
.6.
t..)
t..)
un
Bordetella pertussis
Brevibacterium paucivorans o
tµ.)
o
Bordetella petrii
Brevibacterium stationis tµ.)
o
Bordetella trematum
c,.)
.6.
.6.
n.)
oe
Bacillus
B. acidiceler B. aminovorans B. glucanolyticus
B. taeanensis B. lautus
B. acidicola B. amylolyticus B. gordonae
B. tequilensis B. lehensis
B. acidiproducens B. andreesenii B. gottheilii
B. the rmantarcticus B. lentimorbus
P
B. acidocaldarius B. aneurindyncus B. graminis
B. the rmoaerophilus B. lentus
,
...]
o h li B l th B l l h B
i th B. acidoterrestris B. anracs B. amapaus B.
ermoamyovorans B. ceniformis
..
un
r.,
B. aeolius B. aquimaris B. haloalkaliphilus
B. the rmocatenulatus B. ligniniphilus
,
,
,
B. aerius B. arenosi B. halochares
B. the rmocloacae B. litoralis '
r.,
r.,
B. aerophilus B. arseniciselenatis B. halodenitrificans
B. the rmocopriae B. locisalis
B. agaradhaerens B. arsenicus B. halodurans
B. thermodenitrificans B. luciferensis
B. agri B. aurantiacus B. halophilus
B. the rmoglucosidasius B. luteolus
B. aidingensis B. arvi B. halosaccharovorans
B. the rmolactis B. luteus
B. akibai B. cnyabhattai B.
hemicellulosilyticus B. the rmoleovorans B. macauensis Iv
n
,-i
B. alcalophilus B. asahii B. hemicentroti
B. the rmophilus B. macerans t=1
Iv
B. algicola B. atrophaeus B. herbersteinensis
B. the rmoruber B. macquariensis tµ.)
o
tµ.)
o
B. alginolyticus B. axarquiensis B. horikoshii
B. the rmosphaericus B. macyae 'a
o
.6.
tµ.)
tµ.)
un
B. alkalidiazotrophicus B. azotofixans B. horneckiae
B. thiaminolyticus B. malacitensis 0
n.)
B. alkalinitrilicus B. azotoformans B. horti
B. thioparans B. mannandyticus o
t..)
o
B. alkalisediminis B. badius B. huizhouensis
B. thuringiensis B. marisflavi c,.)
.6.
.6.
B. alkalitelluris B. barbaricus B. humi
B. tianshenii B. marismortui t.)
oe
B. altitudinis B. bataviensis B. hwajinpoensis
B. bypoxylicola B. marmarensis
B. alveayuensis B. betjingensis B. idriensis
B. tusciae B. massiliensis
B. alvei B. benzoevorans B. indicus
B. validus B. megaterium
B. amyloliquefaciens B. beringensis B. infantis
B. vallismortis B. mesonae
B. berkeleyi B. infernus
B. vedderi B. methanolicus
P
= B.
B. beveridgei B. insolitus
B. velezensis B. methylotrophicus
,
a. subsp. Amyloliquefaciens
...]
1-, b i i B ii
Bi i Bil B. ogorenss B. nvctae B. vetnamenss
B. mguanus 0
o ..
o
=
B. a. subsp. Plantarum r.,
B. boroniphilus B. iranensis
B. vireti B. mojavensis
'7
B. borstelensis B. isabeliae
B. vulcani B. mucilaginosus 0"
r.,
B. brevis Migula B. isronensis
B. wakoensis B. muralis
B. dipsosauri
B. butanolivorans B. jeotgali
B. weihenstephanensis B. murimartini
B. drentensis
B. canaveralius B. kaustophilus
B. xiamenensis B. myco ides
B. edaphicus
B. carboniphilus B. kobensis
B. xiaoxiensis B. naganoensis
B. ehimensis
B. cecembensis B. kochii
B. zhanjiangensis B. nanhaiensis Iv
B. eiseniae
n
B. cellulosilyticus B. kokeshiiformis
B. nanhaiisediminis 1-3
B. enclensis
t=1
B. peoriae
Iv
B. centrosporus B. koreensis
B. nealsonii t..)
o
B. endophyticus
n.)
B. persepolensis
o
B. cereus B. korlensis
B. neidei 'a
B. endoradicis
o
.6.
t..)
t..)
un
B. farraginis B. chagannorensis B. kribbensis
B. persicus B. neizhouensis o
tµ.)
o
B. fastidiosus B. chitinolyticus B. krulwichiae
B. pervagus B. niabensis tµ.)
o
B. fengqiuensis B. chondroitinus B. laevolacticus
B. plakortidis B. niacini c,.)
.6.
.6.
tµ.)
B. firmus B. choshinensis B. larvae
B. pocheonensis B. novalis oe
B. flexus B. chungangensis B. laterosporus
B. polygoni B. oceanisediminis
B. foraminis B. cibi B. salexigens
B. polymyxa B. odysseyi
B. fordii B. circulans B. saliphilus
B. popilliae B. okhensis
B. formosus B. clarkii B. schlegelii
B. pseudalcalophilus B. okuhidensis
B. fortis B. clausii B. sediminis
B. pseudofirmus B. oleronius
P
B. fiimarioli B. coagulans B. selenatarsenatis
B. pseudomycoides B. oiyzaecorticis ,
1-, B. fiiniculus B. coahuilensis B. selenitireducens
B. psychrodurans B. oshimensis
o ..
B. fiisiformis B. cohnii B. seohaeanensis
B. psychrophilus B. pabuli
'7
,
B. galactophilus B. compost! B. shacheensis
B. psychrosaccharolyticus B. pakistanensis
r.,
B. galactosidilyticus B. curdlanolyticus B. shackletonii
B. psychrotolerans B. pallidus
B. galliciensis B. cycloheptanicus B. siamensis
B. pulvifaciens B. pallidus
B. gelatini B. cytotoxicus B. silvestris
B. pumilus B. panacisoli
B. gibsonii B. daliensis B. simplex
B. purgationiresistens B. panaciterrae
B. ginsengi B. decisifrondis B. siralis
B. pycnus B. pantothenticus Iv
n
B. ginsengihumi B. decolorationis B. smithii
B. qingdaonensis B. parabrevis 1-3
t=1
Iv
B. ginsengisoli B. deserti B. soli
B. qingshengii B. paraflexus tµ.)
o
tµ.)
B. globisporus (eg, B. B. solimangrovi
B. reuszeri B. pasteurii 'a
o
.6.
tµ.)
tµ.)
un
g. subsp. Globisporus; or B. B. solisalsi
B. rhizosphaerae B. patagoniensis c;;)
t..)
g. subsp. Marinus) B. songklensis
B. rigui o
t..)
o
B. sonorensis
B. runs c,.)
.6.
.6.
n.)
B. sphaericus
B. safensis oe
B. sporothermodurans
B. salarius
B. stearothermophilus
B. stratosphericus
B. subterraneus
B. subtilis (eg, B.
P
s. subsp. Inaquosorum; or B.
2
1-, s. subsp. Spizizeni;
or B.
o 2
oe
N,
s. subsp. Subtilis)
2
'7
,
Caenimonas Campylobacter Cardiobacterium
Catenuloplanes Curtobacterium
Caenimonas koreensis Campylobacter coli Cardiobacterium
hominis Catenuloplanes atrovinosus Curtobacterium
Campylobacter concisus
Catenuloplanes castaneus .. albidum
Caldalkalibacillus Carnimonas
Campylobacter curvus
Catenuloplanes crispus Curtobacterium citreus
Caldalkalibacillus uzonensis Carnimonas
nigrificans
Campylobacter fetus
Catenuloplanes indicus
Iv
Campylobacter gracilis
Catenuloplanes japonicus n
Caldanaerobacter Carnobacterium
Campylobacter helveticus
Catenuloplanes nepalensis t=1
Iv
Caldanaerobacter subterraneus Carnobacterium
t..)
Campylobacter hominis
Catenuloplanes niger o
t..)
alterfunditum
o
'a
Campylobacter hyointestinalis
o
.6.
t..)
t..)
un
Campylobacter jejuni Carnobacterium
divergens o
t..)
Caldanaerobius
Chryseobacterium o
Campylobacter lari Carnobacterium
funditum t...)
o
Caldanaerobius fijiensis Campylo
Chiyseobacterium
bacter mucosalis Carnobacterium
gallinarum c...)
.6.
.6.
Caldanaerobius
balustinum t..)
Campylobacter rectus Carnobacterium
oe
polysaccharolyticus
Campylobacter showae maltaromaticum
Citrobacter
Caldanaerobius zeae
Campylobacter sputorum Carnobacterium mobile
C. amalonaticus
Campylobacter upsaliensis Carnobacterium
viridans
Caldanaerovirga
C. braakii
Caldanaerovirga acetigignens
C. diversus
Capnocytophaga Caryophanon
P
Capnocytophaga canimorsus Caiyopha
C. farmeri
non latum
Caldicellulosiruptor
2
C. freundii
Capnocytophaga cynodegmi Caiyophanon tenue
1-,
;3'
o Caldicellulosiruptor bii
o
C. gillenii .?.
rup esc
Capnocytophaga gingivalis
r.,
2
Caldicellulosiruptor kristjanssonii
Catellatospora
Catellat
C. koseri ,
Capnocytophaga granulosa
0"1
Caldicellulosiruptor owensensis
Catellatospora citrea
C. murliniae ,3"1
Capnocytophaga haemolytica
Catellatospora
C. pasteuriim
Capnocytophaga ochracea
methionotrophica
C. rodentium
Capnocytophaga sputigena
C. sedlakii
Catenococcus
C. werkmanii
Catenococcus thiocycli
Iv
C. youngae
n
,-i
m
,-o
Clostridium
t..)
o
t..)
o
(see below)
'a
cA
.6.
t..)
t..)
un
C
t..)
Coccochloris
o
t..)
o
Coccochloris elabens
.6.
.6.
n.)
oe
Corynebacterium
Coiynebacterium flavescens
Coiynebacterium variabile
Clostridium
Clostridium absonum, Clostridium aceticum, Clostridium ace tireducens,
Clostridium acetobutylicum, Clostridium acidisoli, Clostridium aciditolerans,
P
Clostridium acidurici, Clostridium aerotolerans, Clostridium aestuarii,
Clostridium akagii, Clostridium aldenense, Clostridium aldrichii, Clostridium
2
1-, algidicami, Clostridium algidixylanolyticum, Clostridium algifaecis,
Clostridium algoriphilum, Clostridium alkalicellulosi, Clostridium
aminophilum,
1-,
.
o r.,
Clostridium aminovalericum, Clostridium amygdalinum, Clostridium amylolyticum,
Clostridium arbusti, Clostridium arcticum, Clostridium argentinense, 2
'7
Clostridium asparagiforme, Clostridium aurantibutyricum, Clostridium
autoethanogenum, Clostridium baratii, Clostridium barkeri, Clostridium
bartlettii,
r.,
Clostridium betjerinckii, Clostridium bifermentans, Clostridium bolteae,
Clostridium bomimense, Clostridium botulinum, Clostridium bowmanii,
Clostridium
biyantii, Clostridium butyricum, Clostridium cadaveris, Clostridium caenicola,
Clostridium cam inithermale, Clostridium carboxidivorans, Clostridium camis,
Clostridium cavendishii, Clostridium celatum, Clostridium celerecrescens,
Clostridium cellobioparum, Clostridium cellulofermentans, Clostridium
cellulolyticum, Clostridium cellulosi, Clostridium cellulovorans, Clostridium
chartatabidum, Clostridium chauvoei, Clostridium chromiireducens, Clostridium
citroniae, Clostridium clariflavum, Clostridium clostridioforme, Clostridium
cocco ides, Clostridium cochlearium, Clostridium colletant, Clostridium
colicanis, .0
n
Clostridium colinum, Clostridium collagenovorans, Clostridium cylindrosporum,
Clostridium difficile, Clostridium diolis, Clostridium disporicum, 1-3
t=1
Clostridium drakei, Clostridium durum, Clostridium estertheticum, Clostridium
estertheticum estertheticum, Clostridium estertheticum laramiense, Iv
t..)
o
t..)
Clostridium fallax, Clostridium felsineum, Clostridium fervidum, Clostridium
fimetarium, Clostridium formicaceticum, Clostridium frigidicamis, Clostridium
o
'a
o
.6.
t..)
t..)
un
frigoris, Clostridium ganghwense, Clostridium gasigenes, Clostridium ghonii,
Clostridium glycolicum, Clostridium glycyrrhizinilyticum, Clostridium grantii,
0
n.)
o
Clostridium haemolyticum, Clostridium halophilum, Clostridium hastiforme,
Clostridium hathewayi, Clostridium herbivorous, Clostridium hiranonis, tµ.)
o
Clostridium histolyticum, Clostridium homopropionicum, Clostridium huakuii,
Clostridium hungatei, Clostridium hydrogeniformans, Clostridium w
.6.
.6.
tµ.)
hydroxybenzoicum, Clostridium hylemonae, Clostridium jejuense, Clostridium
indolis, Clostridium innocuum, Clostridium intestinale, Clostridium
irregulare, oe
Clostridium isatidis, Clostridium josui, Clostridium kluyveri, Clostridium
lactatifermentans, Clostridium lacusfiyxellense, Clostridium laramiense,
Clostridium
lavalense, Clostridium lentocellum, Clostridium lentoputrescens, Clostridium
leptum, Clostridium limosum, Clostridium litorale, Clostridium lituseburense,
Clostridium ljungdahlii, Clostridium lortetii, Clostridium lundense,
Clostridium magnum, Clostridium malenominatum, Clostridium mangenotii,
Clostridium
mayombei, Clostridium methoxybenzovorans, Clostridium methylpentosum,
Clostridium neopropionicum, Clostridium nexile, Clostridium nitrophenolicum,
Clostridium novyi, Clostridium oceanicum, Clostridium orbiscindens,
Clostridium oroticum, Clostridium oxalicum, Clostridium papyrosolvens,
Clostridium
P
paradoxum, Clostridium paraperfringens (Alias: C. welchii), Clostridium
paraputrificum, Clostridium pascui, Clostridium pasteurianum, Clostridium 2
...]
1-, peptidivorans, Clostridium perenne, Clostridium perfringens,
Clostridium pfennigii, Clostridium phytofermentans, Clostridium piliforme,
Clostridium
1-,
..
polysaccharolyticum, Clostridium populeti, Clostridium propionicum,
Clostridium proteoclasticum, Clostridium proteolyticum, Clostridium
psychrophilum, 2
'7
,
Clostridium puniceum, Clostridium purinilyticum, Clostridium putrefaciens,
Clostridium putrificum, Clostridium quercicolum, Clostridium quinii,
Clostridium o
r.,
ramosum, Clostridium rectum, Clostridium roseum, Clostridium
saccharobutylicum, Clostridium saccharogumia, Clostridium saccharolyticum,
Clostridium
saccharoperbutylacetonicum, Clostridium sardiniense, Clostridium sartagoforme,
Clostridium scatolo genes, Clostridium schirmacherense, Clostridium
scindens, Clostridium septicum, Clostridium sordellii, Clostridium sphenoides,
Clostridium spiroforme, Clostridium sporo genes, Clostridium
sporosphaeroides, Clostridium stercorarium, Clostridium stercorarium
leptospartum, Clostridium stercorarium stercorarium, Clostridium stercorarium
the rmolacticum, Clostridium sticklandii, Clostridium straminisolvens,
Clostridium subterminale, Clostridium sufflavum, Clostridium sulfidigenes,
Clostridium ,t
n
symbiosum, Clostridium tagluense, Clostridium tepidiprofundi, Clostridium
termitidis, Clostridium tertium, Clostridium tetani, Clostridium
tetanomorphum, 1-3
t=1
Iv
Clostridium thermaceticum, Clostridium thermautotrophicum, Clostridium
thermoalcaliphilum, Clostridium thermobutyricum, Clostridium the rmocellum,
t.)
o
tµ.)
o
Clostridium thermocopriae, Clostridium thermohydrosulfuricum, Clostridium
thermolacticum, Clostridium the rmopalmarium, Clostridium 'a
o
.6.
tµ.)
tµ.)
un
thermopapyrolyticum, Clostridium thermosaccharolyticum, Clostridium
thermosuccinogenes, Clostridium thermosulfurigenes, Clostridium 0
t..)
o
thiosulfatireducens, Clostridium tyrobutyricum, Clostridium uliginosum,
Clostridium ultunense, Clostridium villosum, Clostridium vincentii,
Clostridium t...)
o
viride, Clostridium xylanolyticum, Clostridium xylanovorans
c...)
.6.
.6.
n.)
oe
Dactylosporangium Deinococcus Delftia
Echinicola
Dactylosporangium aurantiacum Deinococcus aerius Delftia acidovorans
Echinicola pacifica
Dactylosporangium fulvum Deinococcus apachensis Desulfovibrio
Echinicola vietnamensis
Dactylosporangium matsuzakiense Deinococcus aquaticus Desulfovibrio
desulfuricans
Dactylosporangium roseum Deinococcus aquatilis Diplococcus
Dactylosporangium thailandense Deinococcus caeni Diplococcus
pneumoniae P
2
Dactylosporangium vinaceum Deinococcus radiodurans
1-,
t..) Deinococcus radiophilus
..
0"
r.,
'7
Enterobacter Enterobacter kobei Faecalibacterium
Flavobacterium
"
E. aero genes E. ludwigii Faecalibacterium
prausnitzii Flavobacterium antarctic urn
E. amnigenus E. mori Fangia
Flavobacterium aquatile
E. agglomerans E. nimipressuralis Fangia hongkongensis
Flavobacterium
E. arachidis E. oiyzae Fastidiosipila
aquidurense
E. asburiae E. pulveris Fastidiosipila
sanguinis Flavobacterium balustinum
Iv
n
E. cancerogenous E. pyrinus Fusobacterium
Flavobacterium croceum 1-3
t=1
E. cloacae E. radicincitans Fusobacterium
nucleatum Flavobacterium cucumis Iv
t..)
o
t..)
E. cowanii E. taylorae
Flavobacterium =
'a
o
.6.
t..)
t..)
un
E. dissolvens E. turicensis
daejeonense 0
n.)
o
E. gergoviae E. sakazakii Enterobacter soli
Flavobacterium defluvii r..)
o
E. helveticus Enterococcus
Flavobacterium degerlachei c...)
.6.
.6.
t..)
E. honnaechei Enterococcus durans
Flavobacterium oe
E. intermedius Enterococcus faecalis
denitrificans
Enterococcus faecium
Flavobacterium filum
Erwinia
Flavobacterium flevense
Erwinia hapontici
Flavobacterium frigidarium
Escherichia
Flavobacterium mizutaii
P
Escherichia coli
Flavobacterium 2
-,
1-,
okeanokoites
1-,
..
r.,
N)
'7
'8
N)
N)
Gaetbulibacter Haemophilus Ideonella
Janibacter
Gaetbulibacter saemankumensis Haemophilus aegyptius Ideonella
azotifigens Janibacter anophelis
Gallibacterium Haemophilus aphrophilus Idiomarina
Janibacter corallicola
Gallibacterium anatis Haemophilus felis Idiomarina abyssalis
Janibacter limosus
Gallicola Haemophilus gallinarum Idiomarina baltica
Janibacter melonis Iv
n
,-i
Gallicola barnesae Haemophilus haemolyticus Idiomarina
fontislapidosi Janibacter terrae t=1
Iv
Garciella Haemophilus influenzae Idiomarina
loihiensis Jannaschia t..)
o
t..)
o
Garciella nitratireducens Haemophilus paracuniculus Idiomarina
ramblicola Jannaschia cystaugens 'a
o
.6.
t..)
t..)
un
Geobacillus Haemophilus parahaemolyticus Idiomarina
seosinensis Jannaschia helgolandensis
0
r..)
o
Geobacillus the rmoglucosidasius Haemophilus parainfluenzae
Idiomarina zobellii Jannaschia pohangensis r..)
o
Geobacillus stearothermophilus Haemophilus Ignatzschineria
Jannaschia rubra c...)
.6.
.6.
t..)
Geobacter paraphrohaemolyticus Ignatzschineria
larvae oe
Geobacter bemidjiensis Haemophilus parasuis
Janthinobacterium
Geobacter bremensis Haemophilus pittmaniae Ignavigranum
Janthinobacterium
Geobacter chapellei Hafnia Ignavigranum ruoffiae
agaric idamno sum
Geobacter grbiciae Hafnia alvei Ilumatobacter
Janthinobacterium lividum
Geobacter hydrogenophilus Hahella Ilumatobacter
fluminis Jejuia
P
Geobacter lovleyi Hahella ganghwensis Ilyobacter
Jejuia pallidilutea 2
,
1-, Geobacter metallireducens Halalkalibacillus Ilyobacter
delafieldii Jeotgalibacillus .
,-,
..
Geobacter pelophilus Halalkalibacillus halophilus
Ilyobacter insuetus Jeotgalibacillus 2
'7
Geobacter pickeringii Helicobacter Ilyobacter polytropus
alimentarius 0"
r.,
Geobacter sulfurreducens Helicobacter pylori Ilyobacter tartaricus
Jeotgalicoccus
Geodermatophilus
Jeotgalicoccus halotolerans
Geodermatophilus obscurus
Gluconacetobacter
Gluconacetobacter xylinus
Iv
n
Gordonia
m
1-d
Gordonia rubripertincta
w
o
t..)
o
'a
o
.6.
t..)
t..)
un
Kaistia Labedella Listeria ivanovii
Micrococcus Nesterenkonia o
t..)
Kaistia adipata Labedella gwakjiensis L. marthii
Micrococcus luteus Nesterenkonia holobia o
t...)
o
Kaistia soli Labrenzia L. monocyto genes
Micrococcus lylae Nocardia c,.)
.6.
.6.
Kangiella Labrenzia aggregata L. newyorkensis
Moraxella Nocardia argentinensis t..)
oe
Kangiella aquimarina Labrenzia alba L. riparia
Moraxella bovis Nocardia corallina
Kangiella koreensis Labrenzia alexandrii L. rocourtiae
Moraxella nonliquefaciens Nocardia
Labrenzia marina L. seeligeri
Moraxella osloensis otitidiscaviarum
Kerstersia Labrys L. weihenstephanensis
Nakamurella
Kerstersia gyiorum Labiys methylaminiphilus L. welshimeri
Nakamurella multipartita
P
Kiloniella Labiys miyagiensis Listonella
Nannocystis 2
1-, Kiloniella laminariae Labiys monachus Listonella
anguillarum Nannocystis pusilla
1-,
..
Klebsiella Labiys okinawensis Macrococcus
Natranaerobius 2
'7
,
K granulomatis Labiys portucalensis Macrococcus bovicus
Natranaerobius 0
r.,
K oxytoca Marinobacter
the rmophilus
K pneumoniae Lactobacillus Marinobacter algicola
Natranaerobius trueperi
K terrigena [see below] Marinobacter
biyozoorum Naxibacter
K variicola Laceyella Marinobacter
flavimaris Naxibacter alkalitolerans
Kluyvera Laceyella putida Meiothermus
Neisseria 1-d
n
Kluyvera ascorbata Lechevalieria Meiothermus ruber
Neisseria cinerea 1-3
t=1
Iv
Lechevalieria aerocolonigenes
Neisseria denitrificans w
o
t..)
Legionella
Neisseria gonorrhoeae o
'a
o
.6.
t..)
t..)
un
Kocuria [see below] Methylophilus
Neisseria lactamica 0
t..)
Kocuria roasea Listeria Methylophilus
Neisseria mucosa o
r..)
o
Kocuria varians L. aquatica methylotrophus
Neisseria sicca c...)
.6.
.6.
Kurthia L. booriae Microbacterium
Neisseria subflava t..)
oe
Kurthia zopfii L. cornellensis Microbacterium
Neptunomonas
L. fleischmannii ammoniaphilum
Neptunomonas japonica
L. floridensis Microbacterium
arborescens
L. grandensis Microbacterium
liquefaciens
L. grayi Microbacterium
oxydans
P
L. innocua
,
-,
o r.,
Lactobacillus
,
,
,
,D
L. acetotolerans L. catenaformis L. mali
L. parakefiri L. sakei
r.,
L. acidifarinae L. ceti L. manihotivorans
L. paralimentarius L. salivarius
L. acidipiscis L. coleohominis L. mindensis
L. paraplantarum L. sanfranciscensis
L. acidophilus L. collinoides L. mucosae
L. pentosus L. satsumensis
Lactobacillus agilis L. composti L. murinus
L. perolens L. secaliphilus
Iv
L. algidus L. concavus L. nagelii
L. plantarum L. sharpeae n
,-i
L. 116amster116116ius L. coryniformis L. namurensis
L. pontis L. siliginis t=1
Iv
t..)
L. amylolyticus L. crispatus L. nantensis
L. protectus L. spicheri o
t..)
o
'a
L. amylophilus L. crustorum L. oligofermentans
L. psittaci L. suebicus o
.6.
t..)
t..)
un
L. amylotrophicus L. curvatus L. oris
L. rennini L. thailandensis o
r..)
L. amylovorus L. delbrueckii subsp. L. panis
L. reuteri L. ultunensis o
t..)
o
L. animalis Bulgaricus L. pantheris
L. rhamnosus L. vaccinostercus c...)
.6.
.6.
L. antri L. delbrueckii subsp. L. parabrevis
L. rimae L. vaginalis t..)
oe
L. 117amster Delbrueckii L. parabuchneri
L. rogosae L. versmoldensis
L. 117amster117 L. delbrueckii subsp. Lactis
L. paracasei L. rossiae L. vini
L. bifermentans L. dextrinicus L. paracollinoides
L. ruminis L. vitulinus
L. brevis L. diolivorans L. parafarraginis
L. saerimneri L. zeae
L. buchneri L. equi L. homohiochii
L. jensenii L. zymae
P
L. camelliae L. equigenerosi L. iners
L. johnsonii L. gastricus 2
1-, L. casei L. farraginis L. ingluviei
L. kalixensis L. ghanensis
1-,
..
L. kitasatonis L. farciminis L. intestinalis
L. kefiranofaciens L. graminis 2
'7
L. kunkeei L. fermentum L. fuchuensis
L. kefiri L. hammesii ,
0
r.,
L. leichmannii L. fornicalis L. gallinarum
L. 117amste L. 117amster
L. lindneri L. fructivorans L. gasseri
L. helveticus L. harbinensis
L. malefermentans L. frumenti
L. hilgardii L. hayakitensis
Iv
n
,-i
Legionella
t=1
1-d
r..)
Legionella adelaidensis Legionella drancourtii Candidatus
Legionella jeonii Legionella quinlivanii o
t..)
o
'a
Legionella anisa Legionella dresdenensis Legionella jordanis
Legionella rowbothamii o
.6.
t..)
t..)
un
Legionella beliardensis Legionella drozanskii Legionella
lansingensis Legionella rubrilucens 0
t..)
o
Legionella birminghamensis Legionella dumoffii Legionella
londiniensis Legionella sainthelensi t..)
o
Legionella bozemanae Legionella eiythra Legionella
longbeachae Legionella santicrucis c,.)
.6.
.6.
t..)
Legionella brunensis Legionella fairfieldensis Legionella lytica
Legionella shakespearei oe
Legionella busanensis Legionella fallonii Legionella
maceachernii Legionella spiritensis
Legionella cardiaca Legionella feeleii Legionella
massiliensis Legionella steelei
Legionella cherrii Legionella geestiana Legionella micdadei
Legionella steigerwalth
Legionella cincinnatiensis Legionella genomospecies Legionella
monrovica Legionella taurinensis
Legionella clemsonensis Legionella gormanii Legionella moravica
Legionella tucsonensis
P
Legionella donaldsonii Legionella gratiana Legionella
nagasakiensis Legionella tunisiensis 2
1-, Legionella gresilensis Legionella nautarum
Legionella wadsworthii
1-,
.
Legionella hackeliae Legionella
norrlandica Legionella waltersii 2
'7
Legionella impletisoli Legionella
oakridgensis Legionella worsleiensis 0"
r.,
Legionella israelensis Legionella
parisiensis Legionella yabuuchiae
Legionella jamestowniensis Legionella
pittsburghensis
Legionella pneumophila
Legionella quateirensis
Iv
n
,-i
Oceanibulbus Paenibacillus Prevotella
Quadrisphaera m
1-d
t..)
Oceanibulbus indolifex Paenibacillus thiaminolyticus
Prevotella albensis Quadrisphaera granulorum o
t..)
o
'a
Prevotella amnii
o
.6.
t..)
t..)
un
Oceanicaulis Pantoea Prevotella bergensis
Quatrionicoccus 0
n.)
o
Oceanicaulis alexandrii Pantoea agglomerans Prevotella bivia
Quatrionicoccus r..)
o
Oceanicola Prevotella Prevotella
brevis australiensis c,.)
.6.
.6.
t..)
Oceanicola batsensis Paracoccus Prevotella biyantii
oe
Oceanicola granulosus Paracoccus alcaliphilus Prevotella buccae
Quinella
Oceanicola nanhaiensis Paucimonas Prevotella buccalis
Quinella ovalis
Oceanimonas Paucimonas lemoignei Prevotella copri
Oceanimonas baumannii Pectobacterium Prevotella dentalis
Ralstonia
Oceaniserpentilla Pectobacterium aroidearum Prevotella
denticola Ralstonia eutropha
P
Oceaniserpentilla haliotis Pectobacterium atrosepticum
Prevotella disiens Ralstonia insidiosa 2
,-, Oceanisphaera Pectobacterium Prevotella histicola
Ralstonia mannitolilytica
1-,
2
o r.,
Oceanisphaera don ghaensis betavasculorum Prevotella intermedia
Ralstonia pickettii 2
'7
,
Oceanisphaera litoralis Pectobacterium cacticida Prevotella
maculosa Ralstonia 0 ,
Oceanithermus Pectobacterium carnegieana
Prevotella marshii pseudosolanacearum
Oceanithermus desulfurans Pectobacterium carotovorum
Prevotella melaninogenica Ralstonia syzygii
Oceanithermus profundus Pectobacterium chiysanthemi Prevotella micans
Ralstonia solanacearum
Oceanobacillus Pectobacterium cypripedii Prevotella
multiformis Ramlibacter
Oceanobacillus caeni Pectobacterium rhapontici Prevotella
nigrescens Ramlibacter henchirensis
Iv
n
Oceanospirillum Pectobacterium wasabiae Prevotella oralis
Ramlibacter tataouinensis 1-3
t=1
Iv
Oceanospirillum linum Planococcus Prevotella oris
w
o
t..)
o
Planococcus citreus Prevotella oulorum
'a
o
.6.
t..)
t..)
un
Planomicrobium Prevotella pallens
Raoultella 0
n.)
o
Planomicrobium okeanokoites Prevotella salivae
Raoultella ornithinolytica t...)
o
Plesiomonas Prevotella stercorea
Raoultella planticola c...)
.6.
.6.
t..)
Plesiomonas shigelloides Prevotella tannerae
Raoultella terrigena oe
Proteus Prevotella timonensis
Rathayibacter
Proteus vulgaris Prevotella veroralis
Rathayibacter caricis
Providencia
Rathayibacter festucae
Providencia stuartii
Rathayibacter iranicus
Pseudomonas
Rathayibacter rathayi
P
Pseudomonas aeruginosa
Rathayibacter toxicus 2
,
1-, Pseudomonas
alcaligenes Rathayibacter tritici
t..)
.
o r.,
Pseudomonas anguillispetica Rhodobacter
2
'7
,
Pseudomonas fluorescens
Rhodobacter sphaero ides o
r.,
Pseudoalteromonas
Ruegeria
haloplanktis
Ruegeria gelatinovorans
Pseudomonas mendocina
Pseudomonas
pseudoalcaligenes
Iv
n
Pseudomonas putida
1-3
t=1
Iv
Pseudomonas tutzeri
w
o
t..)
o
Pseudomonas syringae
'a
o
.6.
t..)
t..)
un
Psychrobacter
o
t..)
o
Psychrobacter faecalis
t...)
o
Psychrobacter
c...)
.6.
.6.
t..)
phenylpyruvicus
oe
Saccharococcus Sagittula Sanguibacter
Stenotrophomonas Tatlockia
Saccharococcus the rmophilus Sagittula stellata Sanguibacter
keddieii Steno trophomonas Tatlockia maceachernii
Sanguibacter suarezii
maltophilia Tatlockia micdadei
Saccharomonospora Salegentibacter
P
Streptococcus
Tenacibaculum 2
Saccharomonospora azurea Salegentibacter sale gens Saprospira
1-,
Tenacibaculum
n . )
-J 2
1-, Saccharomonospora cyanea Saprospira grandis
r.,
[also see below]
amylolyticum r.,0
Salimicrobium
,
Saccharomonospora viridis
'
Tenacibaculum discolor
'8
Salimicrobium album Sarcina
Streptomyces
r.,
Tenacibaculum
Saccharophagus Sarcina maxima
Streptomyces
Salinibacter
gallaicum
Saccharophagus degradans Sarcina ventriculi
achromo genes
Tenacibaculum
Salinibacter ruber
Streptomyces cesalbus
Saccharopolyspora Sebaldella
lutimaris
Salinicoccus
Streptomyces cescaepitosus
Tenacibaculum
Saccharopolyspora eiythraea Sebaldella term
itidis
Iv
Streptomyces cesdiastaticus
n
Salinicoccus alkaliphilus
mesophilum 1-3
Saccharopolyspora gregorii
t=1
Streptomyces cesexfoliatus
Iv
Salinicoccus hispanicus
Tenacibaculum
Saccharopolyspora hirsuta
t..)
o
Streptomyces fimbriatus
t..)
Salinicoccus roseus
skagerrakense =
Saccharopolyspora hordei
'a
Streptomyces fradiae
o
.6.
t..)
t..)
un
Saccharopolyspora rectivirgula
Streptomyces fulvissimus Tepidanaerobacter o
t..)
Salinispora Serratia
o
Saccharopolyspora spinosa
Streptomyces griseoruber Tepidanaerobacter t...)
o
Salinispora arenicola Serratia fonticola
Saccharopolyspora taberi
Streptomyces griseus syntrophicus c...)
.6.
.6.
Salinispora tropica Serratia marcescens
t..)
Streptomyces lavendulae
Tepidibacter oe
Saccharothrix
Streptomyces
Tepidibacter
Salinivibrio Sphaerotilus
Saccharothrix australiensis
phaeochromo genes
formicigenes
Salinivibrio costicola Sphaerotilus natans
Saccharothrix coeruleofusca
Streptomyces
Tepidibacter
Saccharothrix espanaensis
the rmodiastaticus
thalassicus
Salmonella Sphingobacterium
Saccharothrix longispora
Streptomyces tube rcidicus Thermus
Salmonella bongori Sphingobacterium
multivorum
P
Saccharothrix mutabilis
Thermus aquaticus
Salmonella enterica
2
Saccharothrix syringae
-J,-, ermus fiiform
Staphylococcus
Thlis
t..) Salmonella subterranea
.i.
t..)
Saccharothrix tangerinus
r.,
[see below]
The rmus the
Salmonella typhi
h hil r.,0
,
Saccharothrix texasensis
0"
N)
N)
Staphylococcus
S. arlettae S. microti
S. equorum
S. schleiferi
S. agnetis S. muscae
S.
S. sciuri
felis S. nepalensis S.
aureus
S. fleurettii
S. simiae
S. auricularis S. pasteuri
Iv
n
S. gallinarum
S. simulans 1-3
S. capitis S. petrasii
t=1
S. haemolyticus
S. stepanovicii Iv
t..)
S. caprae S. pettenkoferi
o
t..)
S. hominis
S. succinus o
S. carnosus S. piscifermentans
'a
o
.6.
t..)
t..)
un
S. caseolyticus S. hyicus S. pseudintermedius
S. vitulinus 0
t..)
o
S. chromo genes S. intermedius S. pseudolugdunensis
S. warneri t...)
o
S. cohnii S. kloosii S. pulvereri
S. xylosus c...)
.6.
.6.
S. condimenti S. leei S. rostri
t..)
oe
S. delphini S. lentus S. saccharolyticus
S. devriesei S. lugdunensis S. saprophyticus
S. epidermidis S. lutrae
S. lyticans
S. massiliensis
P
Streptococcus
,
,
t..)
.
Streptococcus agalactiae Streptococcus infantarius Streptococcus
orisratti Streptococcus the rmophilus
..
N,
2
Streptococcus anginosus Streptococcus iniae Streptococcus
parasanguinis Streptococcus sanguinis ,
,
,
,D
,
Streptococcus bovis Streptococcus intermedius Streptococcus
peroris Streptococcus sobrinus "
N,
Streptococcus canis Streptococcus lactarius Streptococcus
pneumoniae Streptococcus suis
Streptococcus constellatus Streptococcus milleri Streptococcus
Streptococcus uberis
Streptococcus downei Streptococcus mitis pseudopneumoniae
Streptococcus vestibularis
Streptococcus dysgalactiae Streptococcus mutans Streptococcus pyo
genes Streptococcus viridans
Iv
Streptococcus equines Streptococcus oralis Streptococcus ratti
Streptococcus n
,-i
Streptococcus faecalis Streptococcus tigurinus Streptococcus
salivariu zooepidemicus t=1
Iv
t..)
o
Streptococcus ferus
t..)
o
'a
o
.6.
t..)
t..)
un
C
t..)
o
t..)
o
Uliginosibacterium Vagococcus Vibrio
Virgibacillus Xanthobacter c,.)
.6.
.6.
t..)
Vagococcus carniphilus Vibrio aero genes
Virgibacillus Xanthobacter agilis oe
Uliginosibacterium gangwonense Vagococcus elongatus Vibrio aestuarianus
halodenitrificans Xanthobacter
Ulvibacter Vagococcus fessus Vibrio albensis
Virgibacillus aminoxidans
Ulvibacter litoralis Vagococcus fluvialis Vibrio alginolyticus
pantothenticus Xanthobacter
Umezawaea Vagococcus lutrae Vibrio campbellii
autotrophicus
Weissella
Umezawaea tan gerina Vagococcus salmoninarum Vibrio cholerae
Xanthobacter flavus
P
Weissella cibaria
Undibacterium Vibrio
cincinnatiensis Xanthobacter tagetidis 2
,-, Variovorax
Weissella confusa ;3'
t..) Undibacterium pigrum Vibrio
coralliilyticus Xanthobacter viscosus 2
.6.
r.,
Variovorax boronicumulans
Weissella halotolerans
Ureaplasma Vibrio
cyclitrophicus Xanthomonas 2'
' 7
Variovorax dokdonensis
Weissella hellenica 0"
Ureaplasma urealyticum Vibrio
diazotrophicus Xanthomonas ,
,3"
Variovorax paradoxus
Vibrio fluvialis
Weissella kandleri
albilineans
Variovorax soli
Weissella koreensis
Ureibacillus Vibrio furnissii
Xanthomonas alfalfae
Weissella minor
Ureibacillus composti Vibrio gazo genes
Xanthomonas
Veillonella
Weissella
Ureibacillus suwonensis Vibrio halioticoli
arboricola
Veillonella atypica
aramesenteroides
Ureibacillus terrenus Vibrio harveyi
p Xanthomonas Iv
Veillonella caviae
n
Weissella soli
1-3
Ureibacillus the rmophilus Vibrio ichthyoenteri
axonopodis t=1
Veillonella criceti
Iv
Weissella thailandensis
t..)
Ureibacillus the rmosphaericus Vibrio mediterranei
Xanthomonas o
Veillonella dispar
t..)
Weissella viridescens
o
Vibrio metschnikovii
campestris 'a
Veillonella montpellierensis
o
.6.
t..)
t..)
un
Veillonella parvula Vibrio mytili
Xanthomonas citri o
Williamsia
t..)
o
Veillonella ratti Vibrio natriegens
Xanthomonas codiaei r..)
o
Williamsia marianensis
Veillonella rodentium Vibrio navarrensis
Xanthomonas c...)
.6.
.6.
Williamsia mans
t...)
Vibrio nereis
cucurbitae oe
Venenivibrio
Williamsia serinedens
Vibrio nigripulchritudo
Xanthomonas
Venenivibrio stagnispumantis
Vibrio ordalii
euvesicatoria
Winogradskyella
Vibrio orientalis Xanthomonas fragariae
Wino gradskyella
Vibrio parahaemolyticus Xanthomonas fuscans
thalassocola
Verminephrobacter Vibrio pectenicida
Xanthomonas gardneri
P
Vibrio penaeicida
Xanthomonas hortorum
Verminephrobacter eiseniae
Wolbachia 2
1-, Vibrio proteolyticus
Xanthomonas hyacinthi
? ;3'
t..)
Wolbachia persica ..
un
r.,
Vibrio shilonii
Xanthomonas perforans 2
'7
,
Vibrio splendidus
Xanthomonas phaseoli 0
N)
Verrucomicrobium
r.,
Vibrio tubiashii
Xanthomonas pisi
Verrucomicrobium spinosum
Wolinella
Vibrio vulnificus
Xanthomonas populi
Wolinella succino genes
Xanthomonas theicola
Xanthomonas
translucens
Iv
n
,-i
Zobellia
m
1-d
Zobellia galactanivorans
Xanthomonas w
o
t..)
o
Zobellia uliginosa
vesicatoria 'a
o
.6.
t..)
t..)
un
C
Zoogloea
Xylella t..)
o
t..)
o
Zoo gloea ramigera
Xylella fastidiosa
.6.
Zoo gloea resiniphila
Xylophilus .6.
t...)
oe
Xylophilus ampelinus
Xenophilus Yangia Yersinia mollaretii
Zooshikella Zobellella
Xenophilus azovorans Yangia pacifica Yersinia
philomiragia Zooshikella ganghwensis Zobellella denitrificans
Xenorhabdus Yersinia pestis
Zobellella taiwanensis
P
Yaniella
Zunongwangia
Xenorhabdus beddingii Yersinia
pseudotuberculosis 2
,
1-, Yaniella flava
Zunongwangia profunda .3
t..) Xenorhabdus bovienii Yersinia rohdei
.2
o r.,
Yaniella halotolerans
Xenorhabdus cabanillasii Yersinia ruckeri
2'
,
Zymobacter
Zeaxanthinibacter
Xenorhabdus doucetiae
,
Yeosuana
Yokenella
Zymobacter palmae Zeaxanthinibacter r.,"
Xenorhabdus griffiniae
Yeosuana aromativorans
enoshimensis
eiensburg
Xenorhabdus hominickii Yokenella reg
Zymomonas
Xenorhabdus koppenhoeferi Yersinia
Zymomonas mobilis Zhihengliuella
Yonghaparkia
Xenorhabdus nematophila Yersinia aldovae
Zhihengliuella
Yonghaparkia alkaliphila
Xenorhabdus poinarii
Zymophilus 1-d
Yersinia bercovieri
halotolerans n
,-i
Xylanibacter
Zymophilus paucivorans
Yersinia enterocolitica Zavarzinia
Xylanibacterium t=1
1-d
Xylanibacter oiyzae
Zymophilus raffinosivorans t..)
Yersinia entomophaga Zavarzinia
compransoris Xylanibacterium ulmi o
t..)
o
'a
Yersinia frederiksenii
o
.6.
t..)
t..)
un
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
.,..,
'.-"E3 e)
,'12.; = ,t2
4-=
= ,..4 ,-.=
= 4t = 4t
= ,4 = ,4
4.1 4.1
}... }...
127
CA 03137804 2021-10-22
WO 2020/234428 PCT/EP2020/064225
TABLE 2: SaPIs
Element Staphylococcal genome Baba* Lindsay and Size Inducing
Holden' (kb) phages
SaP14 S. aureus str. MRSA252 NA SaP14 15.1 Endogenous
prophage
SaP11028 S. aureus str. NY940 NA NA 15.6 Endogenous
prophage
SaPlbovl S. aureus str. RF122 vSa2 NA 15.8 91 1 and 80a
SaPlbov2 S. aureus str. V329 NA NA 27 80a
SaP1m4 S. aureus str. mu50 v5a3 NA 14.4 Endogenous
type I prophage
SaPlmw2 S. aureus str. mw2 v5a3 SaP13 14.4 Endogenous
type II prophage
SeP11 S. aureus str. FRI909 NA NA 9.9 Not known
ShP12 S. haemolyticus v5h2 NA 16.6 Not known
SaP11 S. aureus str. RN4282 vSal NA 15.2 80a and 913
SaP13 S. (WIWI'S' str. COL vSal SaP11 15.6 Not known
SaP15 S. aureus str. USA300 NA Na 14.0 Not known
SaP1n1 and S. aureus str. n315 and S. v5a4 SaP12 15 80a
SaP1m1 aureus str. mu50, respectively type 1
SaP12 S. aureus str. RN3984 NA NA 14.7 80 and 80a
SaRlfusB S. aureus European fusidic NA NA 20.7 Not
known
acid-resistant impetigo clone
CS6
SaP1122 S. aureus str. RF122 NA NA 17.9 Endogenous
prophage
SaP16.L S. aureus strains 8325, COL, v5a4 NA 3.14 Not
known
USA300, M55A476, type II
Newman and mw2
SsP115305 S. saprophyticus str. 15305 vSs15305 NA 16.7 Not
known
[N/A, not applicable; *Nomenclature proposed by Baba et al; # Nomenclature
proposed by Lindsey &
Holden et all
Table 3: Titers of SA100 packaged CGVs after production
SA100 Native phage
strain TFU/mL PFU/mL
EMG2 1.4E+10 n.d
C-la 2.2E+10 <20
C-1792 5.2E+09 <20
128
SUBSTITUTE SHEET (RULE 26)
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
Table 4: Groups (Example 5)
Group Preparation (per 500 I)
vehicle 139 ul 9% Sod.carb + 361 I H20
SA100 139 ul 9% Sod.carb + 330 I p114 + 31 I H20
SA100 + induction of CRISPR system 170 I Inducers+Sod.carb + 330 I p114 +
31 I H20
Table 5: Clinical Scores
Score 0 healthy
Score 1 Minor clinical signs of infection (slower movements, light
piloerection in the
skin)
Score 2 Moderate signs of infection (slower movements, slightly pinched
eyes, lack of
curiosity or changed activity)
Score 3 Severe signs of infection (reduced movements, pinched eyes,
changed body
posture)
Score 4 Severe signs of infection (stiff movements, forced ventilation,
pinched eyes)
Sacrificed.
Score 5 The mouse does not move, is cold, lying on the side. Sacrificed
Score 6 The mouse is dead
129
Table 6: Sequences
0
w
o
SEQ ID NO: DESCRIPTION SEQUENCE
w
o
i-J
4,.
1 terS nucleotide
AATTGGCAGTAAAGTGGCAGTTTTTGATACCTAAAATGAGATATTATGATAGTGTAGGATAT
w
oe
sequence
TGACTATCTTACTGCGTTTCCCTTATCGCAATTAGGAATAAAGGATCTATGTGGGTTGGCTG
ATTATAGCCAATCCTTTTTTAATTTTAAAAAGCGTATAGCGCGAGAGTTGGTGGTAAATGAA
ATGAACGAAAAACAAAAGAGATTCGCAGATGAATATATAATGAATGGATGTAATGGTAAAAA
AG CAG CAATTTCAG CAG GTTATAGTAAGAAAACAG CAGAGTCTTTAG CAAGTCGATTGTTAA
P
GAAATGTTAATGTTTCGGAATATATTAAAGAACGATTAGAACAGATACAAGAAGAGCGTTTA
,
,
1¨
.3
=
ATGAGCATTACAGAAGCTTTAGCGTTATCTGCTTCTATTGCTAGAGGAGAACCTCAAGAGGC
,
,
TTACAGTAAGAAATATGACCATTTAAACGATGAAGTGGAAAAAGAGGTTACTTACACAATCA
,
,
,,
,,
CACCAACTTTTGAAGAGCGTCAGAGATCTATTGACCACATACTAAAAGTTCATGGTGCGTAT
ATCGACAAAAAAGAAATTACTCAGAAGAATATTGAGATTAATATTGGTGAGTACGATGACGA
AAGTTAAATTAAACTTTAACAAACCATCTAATGTTTTCAACAG
2 Packaging signal
GCATGCGTTTTCCTGCCTCATTTTCTGCAAACCGCGCCATTCCCGGCGCGGTCTGAGCGTGTCAGTGCAACTGCATTAA
AACCG 1-d
n
sequence
CCCCGCAAAGCGGGCGGGCGAGGCGGGGAAAGCACCGCGCGCAAACCCAGAAGTTAGTTAATTATTTGTGTAGTCAAAG
TGC 1-3
t=1
CTTGACTACATACCTCGTTAATACATTG GAG CATAATGAAGAAAATCTATG G CCTATG
GTCCAAAACTGTCTTTTTTGATG G CAC 1-d
w
o
TATCCTGAAAAATATGCAAAAAATAGATTGATGTAAGGTGGTTCTTGTCAGTGTCGCAAGATCCTTAAGAATTC
w
o
'a
o
4,.
w
w
vi
3 Cos packaging signal
GCATGCGTTTTCCTGCCTCATTTTCTGCAAACCGCGCCATTCCCGGCGCGGTCTGAGCGTGTCAGTGCAACTGCATTAA
AACCG
0
sequence
CCCCGCAAAGCGGGCGGGCGAGGCGGGGAAAGCACCGCGCGCAAACCGACAAGTTAGTTAATTATTTGTGTAGTCAAAG
TGC w
o
w
CTTCAGTACATACCTCGTTAATACATTGGAGCATAATGAAGAAAATCTATGGCCTATGGTC
i-J
4,.
4,.
4 P2/P4 packaging
TGCATTAAAACCGCCCCGCAAAGCGGGCGGGCGAGGCGGGGAAAGCACCGCGCGC
w
oe
signal sequence
sid
ATGTCTGACCACACTATCCCTGAATATCTGCAACCCGCACTGGCACAACTGGAAAAGGCCAGAGCCGCCCATCTTGAGA
ACGC
CCGCCTGATGGATGAGACCGTCACGGCCATTGAACGGGCAGAGCAGGAAAAAAATGCGCTGGCGCAGGCCGACGGAAAC
GA
CGCTGACGACTGGCGCACGGCCTTTCGTGCAGCCGGTGGTGTCCTGAGCGACGAGCTGAAACAGCGCCACATTGAGCGC
GTG
GCACGCCGGGAGCTGGTACAGGAATATGACAATCTGGCCGTGGTGCTGAATTTCGAACGTGAACGCCTGAAAGGGGCGT
GTG
ACAGCACGGCCACCGCCTACCGGAAGGCACATCATCACCTTCTGAGTCTGTATGCAGAGCATGAGCTGGAACACGCCCT
GAAT P
GAAACCTGTGAGGCGCTTGTCCGGGCAATGCATCTGAGCATTCTGGTACAGGAAAATCCGCTCGCCAACACCACCGGCC
ATCA ,
1¨
GGGCTACGTCGCACCGGAAAAGGCTGTCATGCAGCAGGTGAAATCATCGCTGGAACAGAAAATTAAACAGATGCAAATC
AGC ,
0
1¨
CTCACCGGCGAGCCGGTTCTCCGGCTGACCGGACTGTCAGCGGCAACACTCCCGCACATGGATTATGAGGTGGCAGGCA
CACC
0
GGCACAGCGCAAGGTGTGGCAGGACAAAATAGACCAGCAGGGAGCAGAGCTTAAGGCCAGAGGGCTGCTGTCATGA
,
,
,
0
,
,,
6 psu
ATGGAAAGCACAGCCTTACAGCAGGCCTTTGACACCTGTCAGAATAACAAAGCAGCATGGCTGCAACGCAAAAATGAGC
TGG ,,
CAGCGGCCGAACAGGAATATCTGCGGCTTCTGTCAGGAGAAGGCAGAAACGTCAGTCGCCTGGACGAATTACGCAATAT
TATC
GAAGTCAGAAAATGGCAGGTGAATCAGGCCGCCGGTCGTTATATTCGTTCGCATGAAGCCGTTCAGCACATCAGCATCC
GCGA
CCGGCTGAATGATTTTATGCAGCAGCACGGCACAGCACTGGCGGCCGCACTGGCACCGGAGCTGATGGGCTACAGTGAG
CTG
ACGGCCATTGCCCGAAACTGTGCCATACAGCGTGCCACAGATGCCCTGCGTGAAGCCCTTCTGTCCTGGCTTGCGAAGG
GTGA
AAAAATTAATTATTCCGCACAGGATAGCGACATTTTAACGACCATCGGATTCAGGCCTGACGTGGCTTCGGTGGATGAC
AGCC 1-d
GTGAAAAATTCACCCCTGCGCAGAACATGATTTTTTCGCGTAAAAGTGCGCAACTGGCATCACGTCAGTCAGTGTAA
n
,-i
m
,-o
7 Delta
ATGATTTACTGTCCGTCGTGTGGACATGTTGCTCACACCCGTCGCGCACATTTCATGGACGATGGCACCAAGATAATGA
TTGCA w
o
w
CAGTGCCGGAATATTTATTGCTCTGCGACATTTGAAGCGAGTGAAAGCTTTTTCTCTGACAGTAAAGATTCAGGAATGG
AATAC
'a
o
ATTTCAGGCAAACAGAGATACCGCGATTCACTGACGTCAGCCTCCTGCGGTATGAAACGCCCGAAAAGAATGCTTGTTA
CCGG
w
w
vi
ATATTGTTGTCGGAGATGTAAAGGCCTTGCACTGTCAAGAACATCGCGGCGTCTGTCTCAGGAAGTCACCGAGCG 1
1 1 1 1 ATGT
0
GTGCACGGATCCGGGCTGTGGTCTGGTGTTTAAAACGCTTCAGACCATCAACCGCTTCATTGTCCGCCCGGTCACGCCG
GACG r..)
o
r..)
AACTGGCAGAACGCCTGCATGAAAAACAGGAACTGCCGCCAGTACGGTTAAAAACACAATCATATTCGCTGCGTCTGGA
ATGA
i-J
.6.
.6.
8 Q through S of P2
TCAGTCGTTGTCAGTGTCCAGTGAGTAGTTTTTAAAGCGGATGACCTCCTGACCGAGCCAGCCGTTTATCTCGCGGATC
CTGTC r..)
oe
CTGTAACGGGATAAGCTCATTGCGGACAAAGACCTTTGCCACTTTCTCAATATCACCCAGCGACCCGACGTTCTCCGGC
TTGCC
ACCCATCAACTGAAAGGGGATGCGGTGCGCGTCCAGCAGGTCAGCGGCGCTGGCTTTTTTGATATTAAAAAAATCGTCC
TTCG
TCGCCACTTCACTGAGGGGGATAATTTTAATGCCGTCGGCTTTCCCCTGTGGGGCATAGAGAAACAGGTTTTTAAAGTT
GTTGC
GGCCTTTCGACTTGACCATGTTTTCGCGAAGCATTTCGATATCGTTGCGATCCTGCACGGCATCGGTGACATACATGAT
GTATCC
GGCATGTGCGCCATTTTCGTAATACTTGCGGCGGAACAACGTGGCCGACTCATTCAGCCAGGCAGAGTTAAGGGCGCTG
AGAT
ATTCCGGCAGGCCGTACAGCTCCTGATTAATATCCGGCTCCAGCAGGTGAAACACGGAGCCGGGCGCGAAGGCTGTCGG
CTC
GTTGAAGGACGGCACCCACCAGTAAACATCCTCTTCCACGCCACGGCGGGTATATTTTGCCGGTGAGGTTTCCAGTCTG
ATGAC P
CTTACCGGTGGTGCTGTAACGCTTTTCCAGAAACGCATTACCGAACACCAGAAAATCCAGCACAAAGCGGCTGAAATCC
TGCT L.
,
L.
,
1¨
GGGAAAGCCATGGATGCGGGATAAATGTCGAGGCCAGAATATTGCGTTTGACGTAAATCGGCGAGCTGTGATGCACGGC
AGC .3
.
r..)
CCGCAGGCTTTTTGCCAGACCGGTAAAGCTGACCGGTGGCTCATACCATCTGCCGTTACTGATGCACTCGACGTAATCC
AGAAT
2
,
GTCACGGCGGTCGAGTACCGGCACCGGCTCACCAAAGGTGAATGCCTCCATTTTCGGGCCGCTGGCGGTCATTGTTTTT
GCCG ,
,
CAGGTTGCGGTGTTTTCCCTTTTTTCTTGCTCATCAGTAAAACTCCAGAATGGTGGATGTCAGCGGGGTGCTGATACCG
GCGGT
GAGTGGCTCATTTAACAGGGCGTGCATGGTCGCCCAGGCGAGGTCGGCGTGGCTGGCTTCCTCGCTGCGGCTGGCCTCA
TAG
GTGGCGCTGCGTCCGCTGCTGGTCATGGTCTTGCGGATAGCCATAAACGAGCTGGTGATGTCGGTGGCGCTGACGTCGT
ATTC
CAGACAGCCACGGCGGATAACGTCTTTTGCCTTGAGCACCATTGCGGTTTTCATTTCCGGCGTGTAGCGGATATCACGC
GCGGC
GGGATAGAACGAGCGCACGAGCTGGAACACGCCGACACCGAGGCCGGTGGCATCAATACCGATGTATTCGACGTTGTAT
TTT
TCGGTGAGTTTGCGGATGGATTCCGCCTGGGTGGCAAAGTCCATGCCTTTCCACTGGTGACGCTCAAGTATTCTGAATT
TGCCA
1-d
CCGGCCACCACCGGCGGTGCCAGTACCACGCATCCGGCGCTGTCGCCACGGTGTGACGGGTCGTAACCAATCCATACCG
GGC n
1-3
GGGAGCCGAACGGATTGGCGGCAAACGGCGCATAGTCTTCCCATTCTTCCAGCGTGTCGACCATGCAGCGTTGCAGCTC
CTCG t=1
1-d
AACGGGAACACCGATGCCTTGTCGTCAACAAATTCACACATGAACAGGTTTTTAAAATCGTCGGCGCTGTTTTCGCGTT
TGAGC r..)
o
r..)
TGCTCAATGTCGAACAGCGTGCAGCCGCCTTTCAGGGCGTCCTCAATGGTGACAATCTGTCGCCACTGGCCGTCCGCAC
AGAG =
'a
AAGCCCACCGGCAAGGGCGTTATGACTGACGTCGATTTCCACGCGTTCGGCGGCGCTGGCGCGTCCCCGGTTAAACAGT
TCAC o
.6.
r..)
r..)
un
CCGACCAGAACGGGTAGGCGTCGTGCGCCAGCGTGGACGGGGTGGAGAAATAGGTCGAGCGCAGGTGACTCTGTGAGGC
CA
0
TACCTGATGCCACCTTACGCAGTACCTGAAAATTCGGGATCCAGAAAATCTCATCGACGTACAGGTCGCCGTTATGACT
CTGCG n.)
o
n.)
CGGTGTTGGAGTTGGTGCCGAGAAAAATCAGTTTTGCGCCGTTATTGCCCAGGACAATCGGGTCACCGGTCAGGTCAAC
GTCA
i-J
ACCAGACGGGCAAAGGCGATGATGTATTCGCGGAACACATACGCCTGTGTTTTACTGGCCGACAGAAAAATCTGGTTAT
GACC .6.
.6.
n.)
GGTTTTCAGGGCACGCAGCAGCGCCTCGCGGGAAAAATAAAACGTCGCGCCAATCTGGCGGGATTTCAGGATATCGCGG
ATG oe
CGGTGCTCAAGCCCGGCACGATACCAGTGCAGCTGATAGTCGAAAGACTGCTCAAAGAAAATCTGCTCCAGCTTTTCGA
TGGC
CTCGTCACTGAAAAAATTCTTTTTCGGTTTGCGCCGTCCGCCTTTGTTACGGTTAGCGACGTTCGGATTAAGGTCTGCC
TCGTTG
CCGGTCTGGCTGTAGCGGTTGACCCGTGCCAGTCGTTCAATCTGGCGTCCCAGCAGGTCAATTTCCTTGAAGTCACCGC
CGGTT
TTCTGTGGTTTGATGATGAGCTGGGTCAGCCGCGCTTCCAGACTCATTTCGACACGGCTGATGGGGGCAACGCTGTCCC
AGCC
GTCGCGCTGTTTCCAGCTCTGCACTGTCGGGCGTTTCATCTGCAACATGGCGGCAATCTGCGGCACGGAAAACCCCTGC
CAGTA
CAGCAGCGCCGCCTGACGACGCGGGTCGTGTAAAAGAGTGGTGTCTGTGGTGATGGTCATGAATACCTCGCCGTGATGA
ATA
P
CACGGCAAGGCTACTGAGTCGCGCCCCGCGATTCGCTAAGGTGCTGTTGTGTCAGTGATAAGCCATCCGGGACTGATGG
CGG
2
AGGATGCGCATCGTCGGGAAACTGATGCCGACATGTGACTCCTCTAATCACTATTCAGGACTCCTGACAATGGCAAAAA
AAGT
CTCAAAATTCTTTCGTATCGGCGTTGAGGGTGACACCTGTGACGGGCGTGTCATCAGTGCGCAGGATATTCAGGAAATG
GCCG 2
.3
AAACCTTTGACCCGCGTGTCTATGGTTGCCGCATTAACCTGGAACATCTGCGCGGCATCCTGCCTGACGGTATTTTTAA
GCGTT 2
1-
ATGGCGATGTGGCCGAACTGAAGGCCGAAAAGATTGACGATGATTCGGCGCTGAAAGGCAAATGGGCGCTGTTTGCGAA
AAT
.31
.3
CACCCCGACCGATGACCTTATCGCGATGAACAAGGCCGCGCAGAAGGTCTACACCTCAATGGAAATTCAGCCGAACTTT
GCCA
ACACCGGCAAATGTTATCTGGTGGGTCTGGCCGTCACCGATGACCCGGCAAGCCTCGGCACGGAATACCTGGAATTCTG
CCGC
ACGGCAAAACACAACCCCCTGAACCGCTTCAAATTAAGCCCTGAAAACCTGATTTCAGTGGCAACGCCTGTTGAGCTGG
AATTT
GAAGACCTGCCTGAAACCGTGTTCACCGCCCTGACCGAAAAGGTGAAGTCCATTTTTGGCCGCAAACAGGCCAGCGATG
ATGC
CCGTCTGAATGACGTGCATGAAGCGGTGACCGCTGTTGCTGAACATGTGCAGGAAAAACTGAGCGCCACTGAGCAGCGC
CTC
GCTGAGATGGAAACCGCCTTTTCTGCACTTAAGCAGGAGGTGACTGACAGGGCGGATGAAACCAGCCAGGCATTCACCC
GCC
IV
TGAAAAACAGTCTCGACCACACCGAAAGTCTGACCCAGCAGCGCCGCAGCAAAGCCACCGGCGGTGGCGGTGACGCCCT
GAT n
1-3
GACGAACTGCTGACCGGCGTCAGTCAGTCCGGGAAAACCTTCACGATTAACCCTTAATTTCAGGAAAAACTATGCGCCA
GGAA t=1
tl
ACCCGCTTTAAATTTAATGCCTACCTGTCCCGTGTTGCCGAACTGAACGGCATCGACGCCGGTGATGTGTCGAAAAAAT
TCACC 2
o
GTTGAACCGTCGGTCACCCAGACCCTGATGAACACCATGCAGGAGTCCTCTGACTTTCTGACCCGCATCAATATTGTGC
CGGTC 'a
o
AGCGAAATGAAAGGGGAAAAAATTGGTATCGGTGTCACCGGCTCCATCGCCAGCACTACCGACACTGCCGGTGGTACCG
AGC tt
n.)
un
GTCAGCCGAAGGACTTCTCGAAGCTGGCGTCAAACAAGTACGAATGCGACCAGATTAACTTCGATTTTTATATCCGCTA
CAAAA
0
CGCTGGACCTGTGGGCGCGTTATCAGGATTTCCAGCTCCGTATCCGTAACGCCATTATCAAACGCCAGTCCCTTGATTT
CATCAT n.)
o
n.)
GGCCGGTTTTAACGGCGTGAAGCGTGCCGAAACCTCTGACCGCAGCAGCAATCCGATGTTGCAGGATGTGGCGGTCGGC
TGG
i-J
CTGCAGAAATACCGCAATGAAGCACCGGCGCGCGTGATGAGCAAGGTCACTGACGAGGAAGGCCGCACCACCTCTGAGG
TTA .6.
.6.
n.)
TCCGCGTGGGTAAGGGCGGTGATTATGCCAGCCTTGATGCACTGGTGATGGATGCGACCAACAACCTGATTGAACCGTG
GTAT oe
CAGGAAGACCCTGACCTTGTGGTGATTGTGGGGCGTCAGCTACTGGCGGACAAGTATTTCCCCATCGTCAACAAGGAGC
AGG
ACAACAGCGAAATGCTGGCCGCTGACGTCATCATCAGCCAGAAACGCATCGGTAACCTACCAGCGGTACGCGTCCCGTA
CTTC
CCGGCGGATGCGATGCTCATCACGAAGCTGGAAAACCTGTCCATCTACTACATGGATGACAGCCATCGCCGCGTGATTG
AGGA
AAACCCGAAACTCGACCGCGTGGAGAACTACGAGTCAATGAACATTGATTACGTGGTGGAAGACTACGCCGCCGGTTGT
CTG
GTGGAAAAAATCAAGGTCGGTGACTTCTCCACACCGGCTAAGGCGACCGCAGAGCCGGGAGCGTAACCGATGACGAGTC
CCG
CACAGCGCCACATGATGCGGGTCTCGGCAGCGATGACCGCGCAGCGGGAAGCCGCCCCGCTGCGACATGCAACTGTCTA
TGA
P
GCAGATGCTGGTTAAGCTCGCCGCAGACCAGCGCACACTGAAAGCGATTTACTCAAAAGAGCTGAAGGCCGCAAAAAAA
CGC
2
GAACTGCTGCCGTTCTGGTTGCCGTGGGTGAACGGCGTGCTGGAGCTGGGCAAAGGTGCACAGGATGACATTCTGATGA
CGG
TCATGCTGTGGCGTCTGGATACCGGCGATATTGCCGGTGCGCTGGAGATTGCCCGTTATGCCCTGAAGTACGGTCTGAC
CATG 2
.6.
.3
CCGGGTAAACACCGCCGTACCCCGCCGTACATGTTCACCGAGGAGGTAGCGCTTGCGGCCATGCGCGCTCACGCTGCCG
GTGA 2
1-
GTCTGTGGATACCCGCCTGCTGACGGAGACCCTTGAACTGACCGCCACGGCTGACATGCCTGATGAAGTGCGCGCAAAG
CTGC
.31
.3
ACAAAATCACCGGTCTGTTTCTGCGTGACGGTGGTGATGCCGCCGGTGCGCTGGCGCACCTGCAACGTGCGACACAGCT
CGAC
TGTCAGGCAGGCGTCAAAAAAGAGATTGAACGACTGGAGCGGGAGCTGAAACCGAAGCCGGAGCCGCAGCCCAAAGCGG
CC
ACCCGCGCCCCGCGTAAGACCCGGAGCGTGACACCGGCAAAACGTGGACGCCCGAAAAAGAAAGCCAGTTAACAACCGA
ATG
CGCCCCGCGCCAGGGCGGCACGCCGGTCAGTGACGGTGAATCACCTGACACTGCACCGGCGTCCACCGCCCGACTTTTC
AGAG
GTAGTCATGATGACGCTGATTATTCCGCGAAAGGAGGCTCCCGTGTCCGGTGAGGGTACGGTGGTCATCCCGCAACCGG
CAG
GCGACGAGCCGGTGATTAAAAACACGTTCTTTTTTCCCGATATCGACCCGAAGCGCGTCCGGGAACGTATGCGCCTTGA
GCAG
IV
ACCGTCGCCCCCGCCCGTCTGCGTGAGGCCATCAAGTCAGGCATGGCTGAAACGAATGCGGAGCTGTACGAGTACCGCG
AAC n
1-3
AGAAAATTGCCGCCGGTTTTACGCGTCTGGCTGACGTCCCGGCGGACGATATCGACGGTGAAAGCATCAAGGTTTTTTA
CTAC t=1
tl
GAGCGCGCCGTGTGTGCGATGGCGACCGCGTCGCTTTATGAGCGTTATCGCGGTGTGGATGCCAGTGCGAAAGGCGACA
AGA 2
o
AGGCTGACAGCATTGACAGCACCATTGATGAGCTGTGGCGGGATATGCGCTGGGCGGTGGCGCGCATCCAGGGCAAGCC
GC 'a
o
GCTGCATCGTGAGTCAAATCTGATGAAGACCTTTGCGCTACAGGGCGACACGCTCGACGCCATTTGTGTCCGCTATTAC
GGGC tt
n.)
un
GCACTGAGGGCGTGGTTGAGACCGTGCTCGCCGCAAATCCGGGACTGGCTGAACTGGGGGCGGTGCTGCCACACGGCAC
CG
0
CCGTCGAACTGCCCGACGTTCAGACCGCGCCCGTGGCTGAAACTGTCAATCTGTGGGAGTAACGCATGACAGCAGAAGA
AAA n.)
o
n.)
AAGCGTCCTGTCGCTTTTCATGATTGGGGTGCTGATTGTTGTCGGCAAGGTGCTTGCCGGTGGTGAACCTATCACCCCG
CGTCT
i-J
GTTTATCGGGCGCATGTTGCTCGGTGGTTTTGTCTCGATGGTTGCCGGTGTTGTTCTGGTGCAGTTTCCTGACCTGTCA
CTGCCA .6.
.6.
n.)
GCGGTGTGCGGCATCGGCTCCATGCTGGGTATCGCCGGTTATCAGGTGATTGAGATTGCCATTCAGCGCCGCTTTAAGG
GCAG oe
GGGGAAACAGTAATGCCGGTAATTAACACGCATCAGAATATCGCCGCCTTTCTCGACATGCTGGCCGTGTCCGAAGGGA
CGGC
GAATCATCCACTGACGAAAAACCGGGGCTATGACGTGATAGTCACCGGACTGGACGGGAAGCCGGAAATTTTCACCGAC
TAC
AGTGACCACCCGTTCGCACATGGCCGACCGGCGAAGGTGTTTAACCGTCGCGGTGAAAAATCCACGGCCTCCGGTCGCT
ATCA
GCAGCTTTACCTGTTCTGGCCGCATTACCGCAAACAGCTTGCCCTGCCGGATTTCAGTCCGTTGTCACAGGACAGACTC
GCCAT
TCAGTTGATCCGCGAACGCGGAGCACTGGATGACATCCGGGCGGGACGCATTGAGCGCGCCATTTCACGCTGTCGCAAT
ATCT
GGGCGTCCCTGCCGGGTGCCGGTTACGGTCAGCGTGAGCATTCACTGGAAAAACTGGTCACCGTCTGGCGTACCGCTGG
CGG
P
CGTACCGGCTTAAACGGAGTAAATACCATGAAGAAATTATCCCTTTCACTGATGCTGAACGTGTCGCTGGCGCTGATGC
TGGC
2
ACTGTCCCTGATTTACCCGCAGAGCGTGGCCGTCAATTTTGTCGCTGCCTGGGCGATTCTGGCGACGGTTATCTGTGTG
GTTGC
CGGTGGTGTGGGCGTGTATGCCACTGAGTATGTGCTGGAACGCTACGGGCGGGAGCTGCCGCCGGAATCGCTGGCCGTG
AA 2
un
.3
GATTGTCACGTCGCTGTTTTTGCAGCCGGTGCCGTGGCGCAGACGGGCGGCGGCTCTGGTAGTGGTGGTGGCGACGTTT
ATCT 2
1-
CGCTGGTCGCTGCCGGGTGGATTTTTACCGCGCTGATTTATCTTGTGGTGTCGCTGTTTTTCCGGCTGATACGTAAAGC
CTGTC
.31
.3
GTCAGCGTCTTGAGGGGCGGGAACCATGTCAAGGCTGATGATTGTGCTGGTCGTGTTGTTATCGCTGGCGGTGGCCGGT
CTGT
TTCTGGTGAAACACAAAAATGCCAGCCTGCGCGCCTCGCTGGACAGGGCGAACAACGTCGCCAGCGGTCAGCAGACGAC
CAT
CACCATGCTGAAAAATCAGCTTCATGTTGCGCTCACCAGGGCAGATAAAAACGAGCTGGCGCAGGTGGCACTGCGTCAG
GAA
CTGGAGAACGCCGCGAAACGTGAAGCACAGCGCGAGAAAACCATCACGAGGTTACTTAATGAGAACGAAGATTTTCGCC
GCT
GGTACGGTGCTGACCTGCCTGATGCTGTGCGCCGGTTGCACCAGCGCCCCGCCTGCACCGACGCCAGTGATTGTCCCCA
ACGC
ATGCCCGAAAGTGAGCCTTTGCCCGATGCCGGGCAGTGACCCGCAGACGAACGGCGATTTAAGTGCCGATATCCGGCAG
CTT
IV
GAGAACGCGCTGGCACGCTGTGCCAGCCAGGTAAAAATGATTAAACACTGTCAGGACGAAAACGATGCTCAAACCCGAC
AGC n
1-3
CTGCGCAGGGCGCTGACTGATGCCGTCACGGTGCTGAAAACTAACCCCGATATGCTGCGGATATTCGTGGATAACGGGA
GTAT t=1
tl
TGCCTCCACACTGGCGGCGTCGCTGTCATTCGAAAAGCGTTACACGCTCAATGTGATTGTGACCGACTTTACCGGTGAT
TTTGA 2
o
CCTGCTCATTGTGCCGGTGCTGGCGTGGCTGCGGGAAAATCAGCCCGACATCATGACCACCGACGAAGGCCAGAAAAAG
GGC 'a
o
TTCACGTTTTATGCAGACATCAACAATGACAGCAGCTTTGATATCAGTATCAGCCTGATGCTGACCGAGCGCACGCTGG
TCAGT tt
n.)
un
GAGGTGGACGGCGCACTGCATGTGAAGAATATCTCGGAACCCCCGCCGCCGGAGCCGGTCACCCGCCCGATGGAGCTGT
ATA
0
TCAATGGCGAACTGGTGAGTAAGTGGGATGAATGAGTTTAAGCGTTTTGAAGACCGGCTGACCGGACTGATTGAATCGC
TGTC n.)
o
n.)
ACCGTCAGGGCGTCGGCGACTGAGTGCCGAACTGGCGAAACGTCTGCGGCAGAGTCAGCAGCGTCGGGTGATGGCACAG
AA
AGCCCCGGACGGCACACCCTACGCGCCACGCCAGCAGCAGAGCGTCAGAAAAAAGACCGGTCGCGTTAAGCGAAAAATG
TTT .6.
.6.
n.)
GCGAAACTTATTACCAGTCGTTTTTTGCATATCCGTGCCAGCCCGGAGCAGGCATCAATGGAATTTTACGGCGGGAAGT
CGCC oe
GAAAATCGCCAGTGTGCATCAGTTTGGTCTGTCGGAAGAAAACCGGAAAGACGGTAAGAAAATTGATTATCCGGCGCGT
CCCC
TGCTCGGCTTTACCGGTGAGGATGTGCAGATGATTGAAGAGATTATCCTGGCTCACCTTGAGCGTTAG
9 V through G of P2
ATGAACACTCTCGCAAATATTCAGGAACTCGCGCGCGCACTGCGCAACATGATTCGCACTGGCATTATCGTCGAAACCG
ACCTT
AACGCCGGTCGCTGCCGCGTGCAGACCGGCGGCATGTGCACCGACTGGCTTCAGTGGCTGACCCATCGCGCAGGACGTT
CGC
GCACATGGTGGGCACCTTCCGTGGGGGAACAGGTGCTGATTCTGGCCGTGGGTGGTGAACTCGACACGGCGTTCGTTCT
GCC
GGGGATTTATTCCGGCGATAACCCCTCGCCGTCTGTGTCGGCGGATGCCCTGCATATCCGTTTCCCTGACGGGGCGGTG
ATTG P
AATATGAACCCGAAACCAGTGCACTCACGGTAAGCGGAATTAAAACGGCCAGCGTGACGGCTTCCGGTTCTGTTACTGC
CACG L.
,
L.
,
1-,
GTGCCGGTGGTCATGGTGAAAGCATCAACCCGCGTCACCCTGGACACCCCGGAGGTGGTCTGCACCAACAGGCTGATTA
CCG .3
.
eA
GCACGCTGGAAGTGCAGAAAGGCGGGACGATGCGCGGCAACATTGAACACACCGGCGGTGAACTCTCATCAAACGGTAA
GG
2
,
TACTGCATACCCATAAACACCCCGGCGACAGCGGCGGCACAACCGGGAGTCCTTTATGACAGCGCGTTATCTCGGAATG
AATC ,
,
GCAGTGATGGCCTGACTGTCACTGACCTTGAGCATATCAGCCAGAGTATCGGCGATATCCTGCGCACACCGGTCGGCTC
ACGG
GTGATGCGTCGTGATTACGGCTCGTTGCTGGCGTCAATGATTGACCAGCCGCAGACCCCGGCGCTTGAGTTGCAGATTA
AAGT
CGCCTGTTACATGGCAGTGCTGAAATGGGAACCCCGCGTCACCCTGTCATCCGTCACCACGGCGCGCAGTTTTGACGGG
CGAA
TGACGGTCACGTTAACCGGCCAGCACAACGACACCGGCCAGCCACTTTCATTAACCATCCCTGTGAGTTGAAACCATGC
CGATT
ATCGACCTGAACCAGCTACCCGCACCGGATGTGGTCGAGGAGCTGGACTTTGAAAGCATTCTCGCTGAACGCAAGGCGA
CACT
GATTTCCCTTTACCCGGAAGATCAGCAGGAGGCGGTCGCCCGTACCCTGACACTGGAATCTGAGCCTCTCGTCAAACTG
CTGG
Iv
AAGAAAATGCTTATCGTGAGCTTATCTGGCGTCAGCGTGTGAATGAGGCCGCACGGGCGGTGATGCTGGCCTGTGCCGC
CGG n
1-3
TAATGACCTTGATGTGATTGGTGCCAATTACAACACCACGCGCCTGACTATCACCCCGGCAGATGATTCGACCATCCCG
CCGAC t=1
Iv
ACCGGCAGTGATGGAATCTGACACCGATTATCGTCTGCGTATTCAGCAGGCTTTTGAGGGCTTAAGCGTCGCCGGGTCA
GTGG n.)
o
n.)
GAGCCTATCAGTATCATGGTCGCAGTGCTGACGGGCGTGTCGCGGATATTTCTGTCACCAGTCCGTCTCCGGCCTGTGT
CACCA
'a
TCTCTGTGCTGTCACGTGAAAATAACGGCGTCGCATCCGAAGACCTGCTGGCTGTGGTGCGTAACGCCCTTAATGGCGA
GGAC eA
.6.
n.)
n.)
un
GTCAGGCCGGTGGCCGACCGCGTGACCGTGCAGTCTGCCGCCATCGTTGAATACCAGATAAACGCCACGCTTTACCTTT
ACCCT
0
GGTCCCGAAAGCGAACCCATCCGCGCTGCCGCTGTGAAAAAGCTGGAAGCGTATATCACGGCACAGCACCGGCTGGGGC
GCG n.)
o
n.)
ACATCCGTCTGTCTGCCATTTATGCCGCTTTGCATGTGGAAGGTGTGCAGCGTGTCGAACTGGCTGCACCACTGGCCGA
CATCG
i-J
TGCTCAACAGTACGCAGGCGTCTTTCTGTACCGAATACCGCGTCGTGACCGGAGGCTCGGATGAGTGATTCGCGACTGC
TGCC .6.
.6.
n.)
GACCGGCTCATCACCGCTTGAGGTCGCCGCCGCAAAAGCCTGTGCGGAAATTGAAAAAACGCCGGTCAGTATTCGTGAA
CTGT oe
GGAACCCGGACACCTGTCCGGCAAATCTGCTGCCGTGGCTGGCGTGGGCGTTTTCGGTCGACAGGTGGGATGAAAAGTG
GCC
GGAAGCGACAAAACGCGCCGTTATCCGCGATGCCTATTTCATCCACTGTCATAAGGGCACGATAGGTGCAATCCGGCGT
GTGG
TGGAGCCGCTCGGCTATCTCATCAACGTGACGGAGTGGTGGGAAAACAGTGACCCGCCCGGCACCTTCCGGCTTGATAT
TGGT
GTACTGGAAAGCGGTATCACAGAGGCAATGTATCAGGAAATGGAACGGCTGATTGCTGATGCCAAACCTGCAAGCCGTC
ACC
TTATTGGCCTGAACATTACCCGGGACATTCCCGGCTATCTGTTCGCCGGTGGTGTGGCTTACGACGGCGATGTAATTAC
GGTTT
ACCCCGGATAAGTGAGGAATAATGAGCATAAAATTCAGAACCGTTATCACCACTGCCGGTGCAGCAAAGCTGGCAGCGG
CAA
P
CCGCGCCGGGAAGGCGGAAGGTCGGCATTACCACGATGGCCGTCGGGGATGGCGGTGGTAAATTGCCTGTCCCGGATGC
CG
2
GACAGACCGGGCTTATCCATGAAGTCTGGCGACATGCGCTGAACAAAATCAGCCAGGACAAACGAAACAGTAATTATAT
TATC
GCCGAGCTGGTTATTCCGCCGGAGGTGGGCGGTTTCTGGATGCGTGAGCTTGGCCTGTACGATGATGCGGGAACGTTAA
TTG 2
--.1
.3
CCGTGGCGAACATGGCCGAAAGCTATAAGCCAGCCCTTGCCGAAGGCTCAGGACGTTGGCAGACCTGTCGCATGGTCAT
CATC 2
1-
GTCAGCAGTGTGGCCTCAGTGGAGCTGACCATTGACACCACAACGGTGATGGCGACGCAGGATTACGTTGATGACAAAA
TTG
.31
.3
CAGAGCACGAACAGTCACGACGTCACCCGGACGCCTCGCTGACAGCAAAAGGTTTTACTCAGTTAAGCAGTGCGACCAA
CAGC
ACGTCTGAAACACTGGCCGCAACGCCGAAAGCGGTAAAGGCCGCGTATGACCTGGCTAACGGGAAATATACCGCACAGG
ACG
CCACCACAGCGCGAAAAGGCCTTGTCCAGCTTAGTAGCGCCACCAACAGCACGTCTGAAACGCTCGCCGCAACACCAAA
AGCC
GTTAAGACGGTAATGGATGAAACGAACAAAAAAGCGCCATTAAACAGCCCTGCACTGACCGGAACGCCAACGACGCCAA
CTG
CGCGACAGGGAACGAATAATACTCAGATCGCAAACACGGCTTTCGTTATGGCCGCGATTGCCGCCCTTGTAGACTCGTC
GCCT
GACGCACTGAATACGCTGAACGAGCTGGCGGCGGCGCTGGGCAATGACCCGAATTTTGCTACCACCATGACTAATGCGC
TTGC
IV
GGGTAAGCAACCGAAAGATGCTACCCTGACGGCGCTGGCGGGGCTTGCTACTGCGGCAGACAGGTTTCCGTATTTTACG
GGG n
1-3
AATGATGTTGCCAGCCTGGCGACCCTGACAAAAGTCGGGCGGGATATTCTGGCTAAATCGACCGTTGCCGCCGTTATCG
AATA t=1
tl
TCTCGGTTTACAGGAAACGGTAAACCGAGCCGGGAACGCCGTGCAAAAAAATGGCGATACCTTGTCCGGTGGACTTACT
TTTG 2
o
AAAACGACTCAATCCTTGCCTGGATTCGAAATACTGACTGGGCGAAGATTGGATTTAAAAATGATGCCGATGGTGACAC
TGAT 'a
o
TCATACATGTGGTTTGAAACGGGGGATAACGGCAATGAATATTTCAAATGGAGAAGCCGCCAGAGTACCACAACAAAAG
ACC tt
n.)
un
TGATGACGTTGAAATGGGATGCACTAAATATTCTTGTTAATGCCGTCATTAATGGCTGTTTTGGAGTTGGTACGACGAA
TGCAC
0
TAGGTGGTAGCTCTATTGTTCTTGGTGATAATGATACCGGATTTAAACAGAATGGAGACGGTATTCTTGATGTTTATGC
TAACA tµ.)
o
tµ.)
GTCAGCGTGTATTCCGTTTTCAGAATGGAGTGGCTATTGCTTTTAAAAATATTCAGGCAGGTGATAGTAAAAAGTTCTC
GCTAT
i-J
CCAGCTCTAATACATCCACGAAGAATATTACCTTTAATTTATGGGGTGCTTCCACCCGTCCAGTGGTTGCAGAGTTAGG
CGATG .6.
.6.
tµ.)
AGGCCGGATGGCATTTCTATAGCCAGCGAAATACAGATAACTCGGTAATATTTGCTGTTAACGGTCAGATGCAACCCAG
CAAC oe
TGGGGAAATTTTGATTCCCGCTATGTGAAAGATGTTCGCCTGGGTACGCGAGTTGTTCAATTGATGGCGCGAGGTGGTC
GTTA
TGAAAAAGCCGGACACACGATTACCGGATTAAGAATCATTGGTGAAGTAGATGGCGATGATGAAGCCATCTTCAGGCCG
ATA
CAAAAATACATCAATGGCACATGGTATAACGTTGCGCAGGTGTAAGTTATGCAGCATTTAAAGAACATTAAGTCAGGTA
ATCC
AAAAACAAAAGAGCAATATCAGCTAACAAAGAATTTTGATGTTATCTGGTTATGGTCCGAAGACGGAAAAAACTGGTAT
GAGG
AAGTGAAGAACTTTCAGCCAGACACAATAAAGATTGTTTACGATGAAAATAATATTATTGTCGCTATCACCAGAGATGC
TTCAA
CGCTTAATCCTGAAGGTTTTAGCGTTGTTGAGGTTCCTGATATTACCTCCAACCGACGTGCTGACGACTCAGGTAAATG
GATGT
P
TTAAGGATGGTGCTGTGGTTAAACGGATTTATACGGCAGATGAACAGCAACAACAGGCAGAATCACAAAAGGCCGCGTT
ACTT .
,
TCCGAAGCGGAAAACGTTATTCAGCCACTGGAACGCGCTGTCAGGCTGAATATGGCGACGGATGAGGAACGTGCACGAC
TGG
...]
1¨
.3
AGTCATGGGAACGTTACAGCGTTCTGGTCAGCCGTGTGGATCCTGCAAATCCTGAATGGCCGGAAATGCCGCAATAA
.
oe
,
,
Fl through ogr of P2
ATGAGTGACTATCATCACGGCGTGCAGGTGCTGGAGATTAACGAGGGCACCCGCGTCATTTCCACCGTATCCACGGCCA
TTGT ,
,
CGGCATGGTCTGCACGGCCAGCGATGCAGATGCGGAAACCTTCCCCCTCAATAAACCTGTGCTGATTACCAATGTGCAG
AGCG
CAATTTCAAAGGCCGGTAAAAAAGGCACGCTGGCGGCATCGTTGCAGGCCATCGCTGACCAGTCAAAACCGGTCACCGT
TGTC
ATGCGCGTGGAAGACGGCACCGGTGATGACGAGGAAACGAAACTCGCGCAGACCGTTTCCAATATCATCGGCACCACCG
ATG
AAAACGGTCAGTACACCGGACTAAAAGCCATGCTGGCGGCGGAGTCGGTAACCGGTGTTAAACCGCGTATTCTCGGCGT
GCC
GGGACTGGATACCAAAGAGGTGGCTGTTGCACTGGCATCAGTCTGTCAGAAGCTGCGTGCTTTCGGGTATATCAGCGCA
TGG
GGCTGTAAAACCATTTCCGAGGTGAAAGCCTATCGTCAGAATTTCAGCCAGCGTGAGCTGATGGTCATCTGGCCGGATT
TCCTC
1-d
GCATGGGATACGGTCACCAGTACCACCGCCACCGCGTATGCCACCGCCCGTGCGCTGGGGCTGCGCGCTAAAATCGACC
AGG n
,-i
AGCAGGGCTGGCATAAAACGCTGTCCAATGTCGGGGTGAACGGTGTTACCGGCATCAGCGCATCTGTATTCTGGGATTT
GCAG t=1
1-d
GAGTCCGGCACCGATGCTGACCTGCTTAACGAGTCAGGCGTCACTACGCTGATTCGCCGCGACGGTTTCCGCTTCTGGG
GTAA tµ.)
o
tµ.)
CCGTACCTGCTCTGATGACCCGCTGTTCCTCTTTGAAAACTACACCCGCACCGCGCAGGTCGTGGCCGACACGATGGCT
GAGGC =
'a
GCACATGTGGGCGGTGGACAAGCCCATCACTGCAACGCTGATTCGCGACATCGTTGACGGCATCAATGCCAAATTCCGT
GAGC
.6.
tµ.)
tµ.)
un
TGAAAACAAACGGCTATATCGTGGATGCGACCTGCTGGTTCAGCGAAGAATCCAACGATGCGGAAACCCTCAAGGCCGG
AAA
0
ACTGTATATCGACTACGACTATACACCGGTGCCTCCTCTCGAAAACCTGACCCTGCGCCAGCGTATTACCGATAAATAC
CTGGC n.)
o
n.)
AAATCTGGTCACCTCGGTTAACAGCAATTAAGGAGCCTGACCGATGGCAATGCCGCGCAAACTCAAGTTAATGAACGTC
TTTCT
i-J
GAACGGCTACAGCTATCAGGGCGTTGCAAAGTCCGTCACGCTGCCAAAACTGACCCGTAAGCTCGAAAACTATCGCGGT
GCGG .6.
.6.
n.)
GGATGAACGGCAGCGCACCGGTAGACCTCGGCCTTGATGACGATGCGCTGTCAATGGAGTGGTCGCTCGGTGGCTTCCC
GGA oe
TTCGGTTATCTGGGAGCTTTACGCCGCAACCGGTGTGGATGCCGTGCCGATTCGTTTTGCAGGCTCTTACCAGCGCGAC
GATAC
CGGCGAAACGGTGGCCGTCGAAGTGGTCATGCGTGGACGTCAGAAAGAAATCGACACCGGCGAGGGTAAACAGGGAGAA
G
ACACTGAGTCGAAAATCTCCGTGGTCTGCACCTATTTCCGGCTGACGATGGACGGTAAGGAGCTGGTCGAAATTGACAC
CATC
AACATGATTGAGAAGGTGAACGGCGTCGATCGGCTGGAGCAACACCGCCGCAATATCGGCCTGTGATTTTCATCCGGTC
AGCC
TGGCTGGCCGGTTAACCCTGATTCAGAAGTGAGAAAACCATGAACAAAGAAAATGTCATTACCCTGGACAATCCGGTCA
AACG
TGGTGAGCAGGTTATCGAACAGGTCACGCTGATGAAACCCAGTGCCGGGACGCTACGCGGTGTCAGTCTGGCTGCGGTT
GCA
P
AACTCCGAAGTCGATGCACTGATTAAGGTGCTGCCGCGCATGACGGCACCGATGCTGACCGAGCAGGAAGTCGCCGCGC
TGG
2
AACTGCCTGACCTTGTGGCGCTGGCCGGTAAGGTGGTCGGTTTTTTGTCGCCGAACTCGGTGCAGTGACGTTTCCGAAA
AATC
TCTCGGTCGATGACCTGATGGCGGATGTGGCAGTGATATTTCACTGGCCGCCATCAGAACTGTATCCCATGAGCCTGAC
CGAA 2
o .3
CTCATCACATGGCGCGAAAAGGCGCTCCGGCGAAGCGGAAACACGAATGAGTAACAATGTAAAATTACAGGTATTGCTC
AGG 2
1-
GCTGTTGACCAGGCATCCCGCCCGTTTAAATCCATCCGCACAGCGAGCAAGTCGCTGTCGGGGGATATCCGGGAAACAC
AAAA
.31
.3
ATCACTGCGCGAGCTGAACGGTCACGCATCCCGTATTGAGGGATTCCGCAAGACCAGTGCACAGCTCGCCGTGACTGGT
CATG
CACTTGAAAAGGCACGGCAGGAGGCCGAAGCCCTTGCCACACAGTTTAAAAACACCGAACGTCCGACCCGTGCTCAGGC
GAA
AGTCCTGGAATCCGCAAAGCGTGCGGCGGAGGACTTACAGGCGAAATATAACCGCCTGACAGATTCCGTTAAACGCCAG
CAG
CGGGAACTGGCCGCTGTGGGAATTAATACCCGCAATCTTGCACATGATGAGCAGGGACTGAAAAACCGTATCAGTGAAA
CCA
CCGCACAGCTTAACCGTCAGCGTGATGCGCTGGTGCGTGTCAGTGCGCAACAGGCAAAACTTAACGCAGTAAAACAGCG
TTAT
CAGGCCGGAAAGGAACTGGCCGGAAATATGGCCTCAGTGGGCGCTGCCGGTGTGGGGATTGCGGCGGCGGGAACGATGG
C
IV
CGGTGTTAAGCTACTGATGCCCGGTTATGAGTTTGCGCAGAAAAACTCAGAATTACAGGCTGTGATCGGAGTGGCAAAA
GACT n
1-3
CCGCCGAAATGGCCGCACTCCGCAAGCAGGCGCGCCAGCTCGGCGACAATACCGCCGCCTCGGCAGATGATGCAGCCGG
TGC t=1
tl
GCAGATTATTATTGCGAAAGCCGGTGGGGATGTTGATGCCATTCAGGCGGCAACGCCGGTCACGCTGAACATGGCGCTG
GCG 2
o
AACCGTCGCACAATGGAAGAAAACGCCGCCCTGCTGATGGGGATGAAATCCGCCTTTCAGCTTTCAAACGATAAGGTCG
CTCA 'a
o
TATCGGGGATGTTCTCTCCATGACGATGAACAAAACCGCCGCCGATTTTGACGGCATGAGCGATGCGCTGACCTATGCC
GCAC tt
n.)
un
CTGTGGCAAAAAATGCCGGTGTCAGCATTGAAGAAACCGCCGCAATGGTCGGGGCGCTGCATGATGCAAAAATCACAGG
CTC
0
AATGGCGGGGACGGGAAGCCGTGCCGTGTTAAGCCGCCTGCAGGCACCGACGGGAAAAGCATGGGATGCACTCAAAGAG
CT n.)
o
n.)
TGGAGTGAAAACCTCAGACAGCAAAGGAAACACCCGGCCAATATTTACCATTCTGAAAGAAATGCAGGCCAGTTTTGAG
AAAA
i-J
ACCGGCTCGGTACTGCCCAGCAGGCTGAATACATGAAAACTATTTTCGGGGAGGAGGCCAGCTCAGCCGCTGCCGTGCT
GAT .6.
.6.
n.)
GACTGCCGCCTCAACCGGAAAGCTGGACAAACTGACCGCTGCGTTTAAAGCCTCAGACGGGAAGACCGCCGAGCTGGTA
AAT oe
ATCATGCAGGACAACCTAGGCGGTGACTTTAAAGCGTTTCAGTCCGCTTATGAGGCGGTGGGGACTGACCTGTTTGACC
AGCA
GGAAGGCGCGCTGCGTAAGCTCACGCAGACGGCCACAAAGTATGTGTTAAAACTCGACGGCTGGATACAGAAAAACAAA
TCA
CTGGCGTCAACCATCGGCATCATTGCCGGCGGTGCACTGGCGCTTACTGGCATCATCGGTGCCATTGGCCTCGTAGCCT
GGCC
GGTTATCACCGGCATCAATGCCATCATCGCGGCAGCAGGCGCAATGGGGGCAGTCTTCACGACGGTTGGCAGTGCTGTT
ATGA
CCGCCATCGGGGCTATTAGCTGGCCGGTTGTGGCCGTGGTGGCTGCCATTGTCGCCGGTGCGTTGCTTATCCGTAAATA
CTGG
GAGCCTGTCAGCGCATTCTTTGGTGGTGTGGTTGAAGGGCTGAAAGCGGCATTTGCGCCGGTGGGGGAACTGTTCACGC
CAC
P
TTAAACCGGTTTTTGACTGGCTGGGCGAAAAGTTACAGGCCGCGTGGCAGTGGTTTAAAAACCTGATTGCCCCGGTCAA
AGCC
2
ACCCAGGACACCCTGAACCGTTGCCGTGACACGGGCGTCATGTTCGGGCAGGCACTGGCTGACGCGTTGATGCTGCCGC
TTAA
.6.
TGCGTTCAACAAACTGCGCAGTGGTATTGACTGGGTACTGGAAAAACTCGGTGTTATCAACAAAGAGTCAGACACACTT
GACC 2
o .3
AGACCGCCGCCAGAACTCATACCGCCACGTATGGTACCGGTGACTATATTCCGGCGACCAGCTCTTATGCAGGCTATCA
GGCTT 2
1-
ATCAGCCGGTCACGGCACCGGCTGGCCGCTCTTATGTAGACCAGAGTAAAAACGAATATCACATCAGCCTGACGGGGGG
GAC
.31
.3
TGCGCCGGGGACACAGCTTGACCGCCAGTTACAGGATGCGCTCGAAAAATACGAGCGGGATAAACGTGCGCGCGCCCGT
GCC
AGCATGATGCATGACGGTTAAGGAGGTGACGAAAAATGATGCTCGCGTTAGGTATGTTTGTTTTTATGCGCCAGACGCT
GCCA
CACCAGACCATGCAGCGTGAATCAGATTATCGCTGGCCGTCAAATTCCCGTATCGGTAAACGGGATGCCTTTCAGTTTC
TCGGT
GTGGGTGAGGAAAACATCACGCTGGCCGGTGTGCTTTATCCCGAACTGACCGGCGGCAAGCTGACGATGACCACGCTCA
GGC
TGATGGCAGAGGAGGGGCGGGCGTGGCCGTTGCTGGATGGCACCGGCATGATTTACGGCATGTATGTCATCAGCAGGGT
GA
GTGAAACAGGGAGTATTTTCTTTGCAGACGGCACACCCCGGAAAATTGATTTTACGCTGTCACTCACCCGCGTTGATGA
ATCAC
IV
TGGCCGCGCTTTATGGCGATATCGGTAAACAGGCGGAATCGCTCATCGGTAAGGCCGGCAGTATGGCGACCAGATTCAC
AGG n
1-3
TATGACGGGGGCGGGATAATGCTGGATGCGCTGACATTTGATGCAGGCAGTACGCTGACGCCGGATTACATGCTGATGC
TCG t=1
tl
ACAGCAGGGATATTACCGGCAATATCAGCGACCGTCTGATGAGCATGACCCTGACGGATAACCGGGGCTTTGAGGCTGA
CCA 2
o
GCTTGATATTGAACTGAACGATGCCGACGGGCAGGTCGGGCTGCCGGTTCGTGGCGCTGTCCTGACGGTGTATATCGGC
TGG 'a
o
AAAGGTTTTGCCCTGGTATGCAAAGGGAAATTTACCGTTGATGAGGTTGAACACCGGGGCGCACCGGATGTAGTCACCA
TCCG tt
n.)
un
CGCCCGGAGTGCAGATTTTCGCGGGACGCTCAATTCCCGCCGGGAAGGCTCCTGGCATGACACCACGCTCGGTGCGATT
GTTA
0
AGGCGATAGCCACCCGTAACAGGCTGGAAGCCAGTGTCGCTCCGTCACTGGCCGGAATAAAAATTCCACACATCGACCA
GTCG t,.)
o
CAGGAGTCTGATGCGAAATTCCTGACCCGTCTTGCAGAACGCAACGGCGGTGAGGTGTCGGTAAAAATGGGAAAACTGT
TGT
i-J
TTCTCAAAGCGGGGCAGGGAGTGACGGCCAGCGGTAAAAAAATCCCGCAGGTCACCATAACCCGCAGCGACGGCGACCG
CCA
4,.
TCATTTTGCGATTGCTGACCGTGGAGCCTACACCGGTGTAACGGCAAAATGGCTACACACTAAAGACCCGAAGCCGCAA
AAGC oe
AGAAGGTAAAACTGAAACGCAAAAAGAAAGAGAAACACCTGCGCGCACTGGAGCACCCGAAAGCGAAACCGGTCAGGCA
GA
AGAAAGCGCCTAAAGTACCGGAAGCGCGTGAAGGTGAATACATGGCCGGTGAGGCTGACAACGTTTTTGCCCTGACCAC
GGT
ATATGCCACGAAAGCGCAGGCCATGCGCGCCGCTCAGGCGAAGTGGGATAAACTGCAACGGGGCGTTGCGGAGTTCTCT
ATC
AGCCTGGCTACCGGTCGGGCAGATATTTACACGGAAACACCGGTCAAAGTGTCTGGCTTTAAGCGCGTCATAGACGAGC
AGG
ACTGGACAATCACTAAGGTGACACATTTTCTGAATAATAGCGGCTTCACGACGTCCTTAGAGCTTGAGGTCAGGCTTTC
TGATG
TGGAGTACGAAACAGAAGATGATGAGTGATGTTTTTGTTTTATCTGTTTGTTTTGTAAGGATAAATTAACTAAAATGGC
ACCAT
P
CAACAAAACCGGAAGAGGTGCTCGCGATGTTTCATTGTCCTTTATGCCAGCATGCCGCACATGCGCGTACAAGTCGCTA
TATCA .
,
CTGACACGACAAAAGAGCGTTATCATCAGTGCCAGAACGTGAATTGCAGCGCCACGTTCATCACTTATGAGTCGGTACA
GCGA
_.]
1¨
.3
4,.
TACATCGTGAAGCCGGGAGAAGTCCACGCCGTAAGGCCGCACCCGTTGCCATCAGGGCAGCAAATTATGTGGATGTAA
.
..
1¨
,
,
11 S. aureus phi11 ANGATTTANTCC
,
,
packaging signal
12 cpmA M KTESYF KEYNQFVLDQH KAI QE LEQE R NALESKI
KLDKSTYKQLIM DGQDD KADN LYQATDAD E KKLKALN KR LETKKSVSKEVKY
QKTI E LLKH QS ELSSLYESE KQSAI EKLKKAVDAYN EIIDEIEDIND RYE D EHQQYASVYSQEQLYDD
KEARKALNG H FKE N I FTSFINGN
DLPYEHNNKLFLKC
13 cpmB M KTKYELN NTKKVANAFCLN EE DTN LLI NAVD LD I
KN N M QE ISS E LQQAEQSKQKQYGTTLQN LAKQN RIIK 1-d
n
1-3
14 ptiA
MDKQQIKDFVCDYHERTRSDVLIDDDINTDEFFSIADENSNEWMADDNIDDHIVKNHLEMIVDRVANDKEFYIFDSLIQ
GRSYQDIS t=1
1-d
GVLDCSEQSVRFWYETLLD KIVEVI E
t,.)
o
o
'a
o
4,.
vi
15 ptiB
MESIAEKETYHLPTEHLQVFNVIKNTSNKYITKTKILNQLGYEYNSSNERWLRRVINSLVYDYGYPIGCSYKPSERGYV
IITTEQEKQQA
0
MRSIKKLADGSMKRYEALKRIEV
w
o
w
o
16 ptiM
MIAYPIRVGSVYRGEQMKLLKTKNCLYYRNGDNKLSEYQLLTQFNPTFINKKIRMCEFQIESMYHMSASTTTCDEMMGV
VSVSYPIE
.6.
.6.
KLVIKIIETKARLQNYKNRSISNMVLLKTVLNHYTEKEQKKVVKYMRSNGRYKPYNVIERLQVDLYQASIKQRSERQKQ
RNIAIENSKIA w
oe
RVNAYHQSSYVKVV
17 gene #29 (terS) [00119]
through gene #53
atgaacgaaaaacaaaagagattcgcagatgaatatataatgaatggatgtaatggtaaaaaagcagcaattacagcag
gttatagtaagaaaacagc
(lysin) of phi11
agagtctttagcaagtcgattgttaagaaatgttaatgtttcggaatatattaaagaacgattagaacagatacaagaa
gagcgtttaatgagtattacagaagcttta
gcgttatctgcttctattgctagaggagaacctcaagaggcttacagtaagaaatatgaccatttaaacgatgaagtgg
aaaaagaggttacttacacaatcacacca
acttttgaagagcgtcagagatctattgaccacatactaaaagtacatggtgcgtatatcgataaaaaagaaattactc
agaagaatattgagattaatattggtgagt P
acgatgacgaaagttaaattaaactttaacaaaccgtctaatgttttcaatagaaacatattcgaaatactaaccaatt
acgataacttcactgaagtacattacggtg .
,
1¨
gaggttcgagcggtaagtctcacggcgttatacaaaaagttgtactcaaagcattgcaagattggaaatatcctaggcg
tatactgtggcttagaaaagtacaatcaa ,
.3
.6.
.
w
caattaaagatagtttgttcgaagatgttaaagattgtttgataaactttggtatttgggacatgtgcctttggaataa
gactgataacaaagttgaattgccaaacggc
r.,
gcagtttttttgtttaaaggattagataacccagagaaaataaagtcgataaaaggcatatcagacatagtcatggaag
aagcgtctgaattcacactaaatgattac ,
,
,
,
acgcaattaacgttgcgtttgagggagcgtaaacacgtgaataagcaaatatttttgatgtttaacccagtatctaaac
tgaattgggtttataagtatttctttgaacat
r.,
ggtgaaccaatggaaaatgtcatgattagacaatctagttatcgagataataagtttcttgatgaaatgacacgacaaa
acttagagttgttagcaaatcgtaatcca
gcatattacaaaatttatgcgttaggtgaatttgctacactagacaaattggttttccctaagtatgaaaaacgtttaa
taaataaagatgagttaagacatttaccttct
tattttggattggactttggctacgttaatgatcctagtgcttttatacattctaaaatagatgtaaagaaaaagaagt
tatacatcattgaagagtatgttaaacaaggt
atgctgaatgatgaaatagctaatgtcataaagcaacttggttatgctaaagaagaaattacagcagatagtgcagaac
aaaaaagtatagctgaattaaggaatct
agggcttaaaaggattttaccaaccaaaaaagggaagggctcggttgtacaagggttacaattcttaatgcaatttgaa
atcattgttgatgaacgttgtttcaagact
attgaagagtttgacaactacacatggcaaaaggacaaagatacaggtgaatataccaatgaaccagtagatacataca
atcattgtatcgattcgttgcgttattca 1-d
n
gtggaacgattctacagaccggttagaaaacgcacaaatctcagttcgaaagttgacacaataaaatctctaggattat
aggagggaacaaatgttaaaagtaaacg 1-3
t=1
aatttgaaacagatacagatctacggggaaacataaattacttatttaatgatgaagccaatgttgtttacacatatga
cgggacggaatccgatttattacaaaacgt 1-d
w
o
taatgaagtaagtaaatacattgaacatcacatggattaccaacgacctagattgaaagtgttaagtgattattacgaa
ggtaaaactaagaacttagttgagttaac w
o
-a-,
acgacgcaaagaagagtacatggcagataaccgtgtagcgcatgattacgcatcttatattagcgattttatcaacggc
tatttcttgggtaatccaattcaatatcaa o
.6.
w
w
vi
gatgatgacaaagatgtattagaagttattgaggcgttcaatgatttaaatgatgttgagtcacacaatagatctttag
gattagatttgtcaatttatggcaaagcttat
0
gagttaatgattagaaaccaagatgatgaaacgcgtttatacaagagtgatgcaatgagtacttttgtcatatacgaca
atacaattgaacgtaatagtatcgcaggc n.)
o
n.)
gttagatatttaagaactaaaccaatagacaagactgacgaagatgaagtgtttacagttgatttattcacttcacacg
gtgtttatagatatcttaccagtagaacaa o
atggattgaagctcacaccacgtgaaaacggttttgaatcacactctttcgaacgtatgcctattacagaatttagcaa
caacgaaagaagaaaaggggattatgag .6.
.6.
n.)
aaagtaatcactttaattgatttgtatgataatgctgaatcagatactgctaactatatgagtgatttaaatgacgcta
tgttacttattaaaggtaatttaaatttagatc oe
ctgtagaagttagaaaacaaaaggaagctaacgtgttgtttttagaaccgactgtttatgctgatagcgaaggtagaga
aacagaaggctctgttgatggtggttatat
ttataagcaatacgatgtacaaggtaccgaagcttataaagaccgtttaaacagtgatatacacatgtttaccaacacg
cctaacatgaaagatgataactttagcgg
cactcaatcgggcgaggcaatgaaatacaaattatttggattggaacaacgtactaaaactaaagaaggattgtttact
aaagggttaagacgtcgtgctaagttgtt
agagacaatacttaaaaatacatggtcgattgacgctaacaaagatttcaatactgttagatacgtatacaacagaaac
ttacctaaatcattgattgaagaattaaa
agcttatattgattctggtgggaagattagccaaacaactttaatgtctctattctcgttcttccaagaccctgaatta
gaagttaagaaaatcgaagaagatgagaaa
gaatctattaaaaaagctcaaaaaggtatttataaagaccctagagacatcaatgatgacgaacaagatgatgatacaa
aagatactgttgataaaaaggaatgatt
P
gtaattgcctaacaaaaacactcaagaatattgggaagaacgcggacgcaaagcaatcgagaatgagttgaagcgtgat
aaaactaaagctgaagaaatagaacg .
,
tatattgaatatgatgattaagcgcattgaaaaagagatcaatgcgtttattgtcaagtacggagattttgcaggcgtt
acattacaagaagcacaaaagattattgat ,,
,
1¨
.3
.6.
.
c,.)
gagttcgatgtaaaagcgtttcaagaagaagcaaaaagattggtcgaaaacaaggagtttagcgatagagcaaatgaag
aattaaagaagtataacacgaaaatg .
r.,
tatgtatctagagaacagatgttaaagattcaaatagaattcttaattgcttatgcaacagctcaaacagaattatcga
tgagggaatatttcgaatcaacagcttatc "
,
,
,
gtgtgttcagtgatcaagcgggtattttaggtgaaggtgtacaagtagctaaagaagttatagatacaatcgttgatac
acaatttcatggtgtcgtttggtcagagcga 0 ,
N,
N,
ttatggactaataccgaagcaatgaaacaagaagtagaagaaataattgctaatgtagttattagaggtcgacatccta
atgaatatgttaaagatatgcgcaagca
cttaaataaattcgaaggcacagcacgacaaaagaccgcagcaattaaatcattgctttatacggaatcggcacgtgtt
cacgcacaatcaagcattgacagcatga
aagaaatttcaccggaaggatattatatgtatattgcaaaaatcgataatagaacaactaaagtatgcaaagggcttaa
tggagaaatattcaaagttaaagacgct
aaaattggtgttaatttctatcctatgcatatcaattgtcgttcagattgcgctttactacctaaatctatgtggccga
aaaaaccaagcaagaaacgaaaaacaaaat
acttcggagggaaagtgaaaagcggtgattgatttaaaagtgaagttttttaaaggcaagttagttttgtatgacagta
aattaaatgtttggaggatactaatatgag
taatactgacaaataccttagagacatagcaagagaattaaaaggtatacgtaaagagttacaaaagcgaaacgaaaca
gttattattgatgcaaacttagacagtt 1-0
n
taaggtcggcagtattagccgataaagaaaaatcgaaatataatgaacctctcttttaatagctagcacttaattgtgt
tggctattttttatgtccaaaacgtgctgatg 1-3
acataaaaagcacgcatggaaaaacagtcgacagactataaatggaggtatatctcatggaagaaaataaacttaagtt
taatttgcaattttttgcagaccaatcag t=1
1-0
n.)
atgatccggacgaaccaggcggagatggtaaaaaaggaaatcctgataagaaagaaaatgacgaaggtactgaaataac
tttcacgccagagcaacaaaagaaa =
n.)
o
gttgatgaaatacttgaacgtcgtgtagcccacgaaaagaaaaaagctgatgagtatgcaaaagaaaaagcagcagaag
ctgctaaagaagctgctaaattagcg -a-,
c,
.6.
aaaatgaacaaggatcaaaaagatgaatatgaacgcgaacaaatggaaaaagaactggaacaattacgttcagaaaaac
aattaaacgaaatgcgttcagaagc n.)
n.)
vi
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
OP OA , (0 OA r0 0 r0 0 OP OA
OD OD ,sr, ,(;)n 0 CLO (0 2 OA tj t,' tc; -u M MI
-uM-our04-,0,0(00(0c0cOutj-- 0 r0 03
(0 -I-' (0 (0 co 4-, (.0 " (13 OD U FO,
.iL-' (0 (0 OA 03 co 4_, 4_, (i, 4_,
rOtjr0r0rotj4-,(0cou,,?;oroOOrOrOrauSu3r0BOrOut,'
-1,- (0 U (0 (13 4-, (0 (0 co 2 U a-, co (0 t; (0 co (0 , -1e,., (0 co op (10
4., (13
4 (0 (0 (0 (0 4-, r0 (0 03 0 0,0 4-, 4-, (0 (.0 (0 CO CO (0s4.4., 4-, r0 03
0 r0 0 (0 4- +., 4-' -' (0 (0 4-, co (0
,(45, OP co 0
(13Z-r,E; 2 oo (-.
co 0 .(,-9, 2, r ( = 0 -' o(0- 4-,' 2
(.0 (0 4(2 0 4_, 0 3 n, 0, (-0 , 0 00 (.0 2
0,0^ tv ro +, ro 0 u ro r (0 OA 2 (ao (0 a ttotE; u (0
CO 0 U 4-' (0 r.: "M 4- 2 õro' tE; CO
ere CIO -'rir (0 CLO ti' 6) U
,.., .... (xi op 0.0 _ -_ .1-, 4_,-uu-o03c0
=L''(0 w:' 010 co CIO
-u -o (J4- co OP (0 co co -o M OLO M 0-0 co 0 t
, ^ .1- ; 00 op 0 (0 ,_,,., .1-; 541 (0 4_, (13 ,(7.),., OP n3 0 0
(0 0 0
OP (.0 co 03 co r0 n3 4.-; OA -3 OD 4-, 4_, ro 0 OD '4. .1-, 4-1 CO 4-, 4-' U
(0
03 0 (0 -1-J .1-; OD 0 (13 hn c0 ,...,(13 U c0
4- M M, (0 ro 0 t,,o ro .0 rr, H r (0 b' OA OA c.71 (0 61 M OD 2 Vo (0 ST MI
0,0
cOr0(0(00.04-,c0-u- m 0,13-aro4-, Ms (0-u
.E1 2 : I 2 t Lou .t , BO - u( . 0 c or 0 3 Lo a . 5 , ( . 0 .. , r 0 (. or 0
.. t : , , 4-J( . 0 mu 0 , 00 , 0 . 0( 0 (Z, 0 cjr BO t:), 0 r 0 t :,(13 (31
3
4-, 4-, 4-, U U CO _ 4_, 03
03 op (i3 u !,7, 4.., n3
CD U 4-1 a-, (0 0,0 0,13 CD 4-',..,0.0op op U 4-, (10 00 03 03
03 co 4_, 03 4_, 4_, _ n3 sam 00 .1-; 4-, OD (xi 4_, 4_, (0 ,,,, 4-, ttO .1_=
(0 4-'õ CO 0 4 (0
CO 013 CO 4-, 4-, U CO '' CO (0 (-0 (-0 OD `(.6" r0 03 03 0 , -..:, 03
.1-; 0 (13 U U 0 0 ,., U .1-; .1-; (0 ,r,
U 4-' (0 0 t' `:,' co co co 0 iii "(13 ((3.30 ((.73) (% 03 (a, õ(13 BO
(0 _, (0 4-, (.0 ,,,:, 4_, l:113 c0 U 4-' 0 u
CD CO +-, U OP
Ur0C0c0(0(13.t,' M r0+,(0r0a-'n32 2 t,' tE;
(00c0030.0(00(0+-,(04-,c0(0ZrOopur00-uut0'23,,r0(00
0 ---, co 03 03 (a0 4-, op -I-' 0 -u a, 4-, 0 -.It r0 --Ir-:: OW (13 a (0
(0 (13 -u (0
1
Ms 4-
t 0 o
mro Ms 0 0 mro 0 4- -, CD ro ...,- CD +., 0 -, ro 0.
CO CD
(13 (13 U ' ' (13 CO .1_= -I-, 0 U 0,0 4-, :i CO
.1-, 0 .1_= 0 0,0 0 j3 if U 4-' CO
0 CO CO 0 0 op op 0
j (0 0
CD (0 (13 4-, (13 (13 (0 0 03 0 Op õ,, 0 (0 co 03 co 4_, t5 03 ti r0 0 r0
r0 OA 0,0 tnn (:(0 OP OLO -o u-' (0 MI co -o co 0 h,-,(13 ,T, co
OP 0 s's 4-
(-0 ' (0 4--, Op 4-, 03 co 03
03 ..õ.,-, 0- OP 03 0
r0 Ms OA co 0,0 +., (0 MI r0 -u M M 0.0 4t1 0.0 OD ro 0-0 - r0 OP co OO
0 ^ -o co , ss-s" OP -o OP m co 03 0 0 op BO -,O,, co +., +_, (0 co 0.0 op T.0-
' , 0 (0 ts,
õ, (0 _ .... 4_, hn _ -
' OP µ6. MI U (70- ((.,, SIP, (a) 2 0.0 2 "(-0 0,0 0 0.0 (,,, ro (0 4-,
t.,, 0 -,t,' s,$).0 t,' co
03 r0 03 (0 -o tr, 4-, (0 (0 0
co (0 0 u .2 co (0 03 +., OP (0
op OP 0 co OP 0 , 0 4_, co , OP , r0 r0 0 op -.1,:,' OW 2 (.0 .1-, 1.T3' (0 (-
0 447.,, (:113
CO 0,13 CO t: 0.0 CO (13 OD OD (0 U (a U ,(513 2 ro -(0, op a 0 OA to op OP
(0 0 M
0 -u Ms 0,0 -r; -_,t; co -o 0 0 co S U -^-' , c0
03 OP U op uUr0r0r0
00,0UCCI -(13(13(130,00,0 00(00,0U
(00,00a-,+,+.,+,(13c0UUWCOM cot ''U Ur0r00.0+' (0 U 0
j3 U 0 j3 (X1 (0 CO 03 (0 U .o .o .1.-= 4_, (13 (.0 0 4_, , . 2
0,0 0 4-,
+., Ms MI (0 co (0 Ms r0
OA co 'um h n 4- ^ U -' -' OP hn Ms m u
-o OP 0-0 0 -o OP (0 4-, }I
0 4- '' OP -o 03 - - r0 OP (0 r0OD 0
OP a-, OP op -Jif (0 (0 a-,
03 OP t.r3 2
ro ,
(0O 4-, 4-, ,., 0 0 4-, U
,.,
'' MI MI ' M M (a CD (0 (a0 ta' 2 oo on 0.0 0.0 0
(0 0 .E19 4t: (),., .t,' U U
.F., U (13 (.0 CO (a CO +-= 4=- "jõ.., CO .F. , CO 4- 0 tp r o 4 -J 0 co co
co r 0 õ . ,,, s , t; - oõ.., Ms
-o 0 03 - r0 co 0 4-, OP s sss 0 03 03 -1--, op ,T, 4-
, CO (0 .1-, (0 (-0 CO " s . 4-,
0coo-uhnromo (0- Op4-, 4-,
+.,opc0+,0c0
CD 0 4-, (0 co "u 0,0 Op co tf, hn- 03 .':, MI +., hnc 0 MI 2 (ao 2 OP co ro -
u (0 0.0
U 4-, +.' (0 (0 2 (-0 (-0
417 +.1 op all u 4,4, 03 OP 4-, OP 4-, 03 a-, CD 8 oo -,
4-, 03 co 03
OP (0 Si? 29 Op 03 1_2 (AO (30 .1-, S19, S. 2 _it, 22:1, (0 t' -(1;r3 (If
a
t : U ( a 0 ( 0 6' rj (al -r. : µ( . ki 4 - ' B 2 0 (0 (0 2, t,'u 4u ro Lao 0
,r,(ao -_,t
,,0 n3 eto (0 , ..".õ0 ro (.0 0 (.0 4_, 0 (.0 4-, (0 -, ro - 4-,,
(.0
0 A OLO r 0 CD 1 0 O t 0 Z - ,u 0 i jp' r 0 op C ti;
%CI
(0 (0 OA (13 (0 U 4-, (13 0 (13 (13 ,., (13 (0 (13 (13 U OD
(13OP OP 0 op aT,"
4-',..,õ (.0 (.0 0 00 s sa -10, (.0 0 U 0 4-' 03
.1-; a-,
(0 03 M tp CtO -u
M 0 m OA CO 4-' -' (0 W3
(0 ^ M MI (0 W3 (a r (0 (a CO M
OD U MI (a M U OD (a CO MI r MI OD (0
OD U CO 0 CO rocOopMour0+,03-u-r%0t3u03(003-ur00-000
(0 r0 _,_,4-' OA co OA M r0 (a -u co (a MI co 2 (0 (ao 4-, r 4-, .-t--,' (0
r (ao r "ro
(0
0 2 -a -0.13-rorotitirotto0+-,4-Jurou'ro+-' CO 4-, OP
00 4_, 4_, 03 -
4-, OP (0 (0
0 0 03 op (0 01:1 4-' 01:1 4-, (0 OP 00 0,0 op .I.J U 4_, (0 r0 (0
r0 hi3 OP 4-, CO 4-j (13 4-, op 4_, 0 OP (0 OP co OP OP 0 , op -1--
, OP op (0 4-, 0
0,..õ,(04-,courOuro,,+-,00,0c04-,4-,c00,0000,,c0c0+,00A-u
CO CO ro (0 4--' (.0 op op -.1-,-;õ if a, co dA S U (0 4..; u a, all U -,-
',.., (0 4.7,j,, 3
'(-0 rom 4-'r 4-'r .).) t --Fi I?, 2 B 4-' co mr(E', Wt.,' tE; (0 r (-0 co
2 (a13 b' 3 (,0-- ro
Bo 1-. 4.7_, co (0 su; 4-, 4- 0.0 0 t; 3 03 op 4- op 0 U ST r-: 2 0 (70) OD OD
0.0 MI T^ 010 .1.'.1 ti; (0 õi, it?... t..2 4.5, (0 (0 õ .1-; OP (0 03 (.0 03
0 03 OP 0 r0 4-' OP t;
op op 4-, (.0 op µ(.6-' ',,-u 6uzi (0 .(i2, (0 -,-, (0 (0 ro 0 ttO (.0 4-, 4-'
.1-, CO (:)J3 (0 (13 4-,
CO U (13 0 U 4_, ,...," _,_, rr, 4.,, (0013 0 4-',,,, õ., t', (13 03 OP 0 co
OP
03 OP (0 03 r0 4-, ,..,.," Tip iii ' . +-, OLO co
(0 r0 Op _Ls.,, õ..,- õ..,- -F-0 co 0 4-, op r0 n3 -
i.i3
OP r0 (10 op ,47', OP ,__,,,s, 0.0 (.0 2 233 3 li 17;j3 -ILI co op srifi O.
(.0
OP r0 (0 4-, s4A, OP L) 0 4-, 03 03
0 0,0 (0 0 (0 op hi3 M 13,0' -(1-15 0 co -13k, 44 (0 2 OD co 1.T3 40 co OP 03
r0
0.04-' CO U CO +-, , (0 0 (.0 0 (13 ,, (.0 ....i 0,0 r tj (.0 (0 0,1:1(0 0
(.0 (0 (.0 0 .1-; .1-, 4_, (0 ,.., õ ,., (0 0 j:, (.0 u , CO _ ^ ro if
(.0 (0 03 n3 OP 0 n3 -14 .1-,' 44, U 03 0 (.0 µ,4,u 0,13 4(9 !!! 0 tu3 ,,,, BO
CO (13 0,0 CO CO
(0 C's M OLO Ms 4-' OA 3 ro (0 +, 6'3 -r.,' 0 gup r0 co µ0µ2,1 co ti M u --t--
: M
4-'
(.0 1::: Bo (.0 0 4_,
03 ,,,,OO 'LI OP Fo-
OP op (0 4-, 4-, OA 4_, t,j3 4-, 4,7',., 0,13 CO CO n3 if (13 a-, (xi (0 n3 4-
, (0 0 .iL-s 4_, co (.0
Ms 0 OP co co op u tx, U 6A+ +4 U u tx, , 03 OA 0 U 03 op 07 (0 0 j:, I.
4.5,
BO 0,0 CO ,4,7',., CO CO U (0 OD (.0 (0 (0 (0 .1-, 0 (0 OD (0 (0 .1-; Op OP 4-
, 4_, (.0 4_, .ifj
4- r 0 'Ilt.? ;(.7,)n MI MI (0 MI CO MOD CO CO 0.0 (.0 .t,' (0 (0 CO CO CO (0
()
- 4-, (13 0 (.0 4-,
03 op co OP OP r0 (0 CO 4-, (0 'I.'
(13 0 (13 (13 (13
013 CO ,., 013 r0 co O, (0 OA op
0.0 CO (0 +., CO Op 4-'
CW (0 (0 0 4' U t' U M
CD n3 (0 so 4_, 0 4-, 4-, 0 4_, 0 03 0 (0 OP 0,0 0
rOcOrourOcorour00cOurOc0(04-,00,0c04-,00,0+-,(08+-,
CD n3 n3 (.0 n3 (.0 co r0 03 03 n3 ta. OP 03 +, co (.0 4-, (-0 U 4- U (-0 CO
+., (-0 .,.,
CO .1-, OD (.0 (0 ro co co co co 03 T.-,,, ril u a-, CD 03 (0 co -2-,:,' ro,
r0 0,0 r0
0,00,0,(.0(00.00,0c003003'(.6'r4rocOrou.p.,03opT.0-c000,0c0003
0 (0 04_, 4_, 0 (13 (13 u Op 03 4., 4-, (13 (0 opn34_, 03,, (0 OPOPOPOPM
CD 0 OP a-, U .1-, CO CO CO all a-, a-, all U all all a-, .1-, U OP .1-, CO
013 CO 013 OD U
144
CA 03137804 2021-10-22
WO 2020/234428 PCT/EP2020/064225
-' CD+-, , op az) 4_, 0 0 CO .1-; " (0
4-, 0 (:10 0 0
0 ro (0 4_, 0 co 4--;, 4_, 0 0.0 0.0 op 03 0.0 ro ro 0 - +-, CO +., 4- 013 013
-X 4-, 0 0 44f (:10 0 (0 (0 (00 (0 (0 4-, (.0 (0 (0 0 (0 013 01:1 4(9 0 (13 00
013 4-' 4-'
0 -X 0 4- CD 00 00 rj -M, (0 4-' 4-' (0 U 00 CO 0' (0 (0 CO 0 (0 .M, 00 (0
00 W3 r
w 4- U 4-, (0 00 4-' 4-, (0 j:( (.0 4., 4_, (00 00 (0 0 (0 .1-, U 4-, .1-, U
(.0 00 ro ro ro
ro ro 0 op 0,0 0,0 0 ro 0 co OA 4- ro r0 ro OA Ms 0 00 r,' Ms an 0 co 0 -o OA
-r; 03 r; 0 03 , 0 (0 .t,'
03 -õ,-, r0 op ro 0.0 -a' -(-
ro (.0 4(3,ji co ro 2 0 L, , 4.,,,, UJJ (0 0 (00 4.7,4, .1-; -13'D (0 (.0
µØ/ 0 (0 (3j3 4,-! .,., µ2.,_
-4t !.(1,1 0-0 On (a0 (13 0 (a (0 t.,-( r0 (6 0-0 MI '' `rt3 tiE; 0 0 OA .2
tE; Ms ro '6j3 0 ..V.Y
+., 0 0D (,, õ (.0
co ro _03 (.0 (0 co 0.0 t1,0 On +., MI (0 4-, OD -'
tl op OA ro 0.0 T.0-' 03 3 -(õ; op (.7) 03 ro 03 w , 03 03 0 op +-, 0 4-, OA
OA 0,0 +-',.,
t1,0 -' OD (0 (a +., OA ro +., 03 +., ro (0
OA 0 0.0 MI OA ro OA -
4-, 0 -o co 0 0 ,
0 0.0 +., 2 co co 0 co 4-,õ ,,,,-, 0.0 .t,' 4- (.0 t.,, 3
(-õ, 2 2 .81, (.0 ro -6-3, 03 0 03 4-, a-, 2 CO 0 ro ro -(1.T3, 4(2, ,,,,ji
f,...; 1:E; ro 2 4t, (.0 (0
2 0.0 (.0 (.0 (-0 2 (0 (0 "r -4t, 83 -(7, Ms (rao (0 ,(7,0 ((70) co -o,T, ro
La0 Ms tE; op ro ro r0
On Ms OA
..., -(y3 CD r ri- (0 0-0 r (0 Ms 2 (0 (0 ro i:5 0 .t,' oh n (a0 (a ti t.) -
4t
4_, 0 (0 (00 (.0 .44 (0 r0 00 .4:,,, 4.-,. CD0 (0 is, 0 .1-; (0 4_, CD,,n ¨ 4-
, CD(1,3 4_, CD0 4_, 0 4-, 0 "-. U CO 4-, 03 tIA -6- y." 0 tIA 03 U CO
ro 0,0 op (-0 0,0 CO .1-; -14 m 41.1 (0 CO CO 0,0 f,;:', r13 tIA 0 03 03 0,0
.4' (13 r 0 (.0
4-,4-,t1,0-4-,Zt 0 m-t: (10r0+-,+-
,0r0-ro Ms roLaOtAro u-o op
U (0 co ro ro .1-, 100 (0 iii-:, 0 2 (-0 -,-, 00 4-' CO 0 ((II!, ro ro ob 4-=,
,-,-,-, 0 rts 0 4-'
00 CO U r 00 0,0 00 U op ro 0 ro ro 4-' 8 44:: OA 03 03
'' U U C'.0 CO 4- U
CO õ (00 ro 0 4(1; , (.0 44
õ 03 ro ',I-4 U 4--
(13 "Id Bo ro , , 4-, -1--' 013 41.1 0 L'
= (0 4¨' s ' 013 .1-; (0 .1_, (0 4-t, (a0 4-'
co ro ,Y,,, If -14:: co op t:ID 2 -.If 3
CD U (0 -',, CO CO (0 r 0.0 0 00 4-' CO MI MI MI 0 4' 4-' CO -(-% (0 (0 to co
4-' 4-t:
= -X CO ¶"
44 (.0 .1-; op .(2 (.0 4-, c13. 0.0 OA 4.7,_ .1-; tIA 03 õ.,
.1-; u. 00 13JS :3,2 to
w
OA (0 4-, 0 .(,?, 00 (a MI 0 OA 4+7,. ' (II t u OD 0_ 0
:if; tIA 4(3b
CO ro ro , (0 4_, 0 0 (ap co õ (.0 00 4_, ro 4-, 00 03
00 4-, _ op 00 03 4-,
2 Bc, to
.,,,,, mu -5, (0,0 ,,, mu t,n(-0 00.0 .,-4- (yo (6, 1.1 {05 5 1.,,,,,w3 B .
.,,,, 0 t , zr 0 , ow 2 . . -, , ) - + . ,- ( . 0( = , ,
, 0 - , ro (.0 ., 0 co '''' 0 00 03 4-
, , ro 03
03 tIA m õ, 0 0 03 U 2 4-if -X 4,7.: CO 4.if 2 4-, n,,
,,-, tto aD -
+t,' , (0 ro
co 0 ro 'or ro to to (a 4-' 0 MI (a 76 010 MI (-1'3 -Ft; (":' co ro 4-,
0 .1-, op 4-, 0 ro 0 4(2, 1-', ro ro 0 4_, ro ro _ co 0 ro ro 0
.1-; (:10 4-, 0
0 013 OA
tIA 4-, a-, ro ro ro +-, (.0 -F.E; ro tIA ro 4-, .0 ro ,õ, (0 ro 0 0,0 ro ,:vi
_ OA Eo_ z
op tIA 4-, 0,0 op 4-' ro (.0 4_, a-, CD OA t:IA ro ro .1.; 0 0-13 (13 ro 44,
ow 0 0,0 00
4'6'ro,(0rOrourOoprou,O,Or0(.00(0-(000,13-r:
0 4-, CO -X (.0 0 (-0 +.1 04-, 0 03 co 4-, 4-, (.0 0,0 130 8 (.0 , 0,02 0,:,
ro ro .t, ro
O m ro ro u 0,0 OA - r, 00 0D 0 Ms ro -o 0 (0 -o ro (13 co an
-'
roo.OoD-'0 ro mu 0 (-0 4-, co OD ty3 0 03
co .0 ty:, co 0 0 0,0 ro
..... 4_, t.ui op , 00 t,u3 4_, 1_3
03 0 , 03 .2.,,, r0 0 r0 -1-, 4-j 00 U ti CO ro .0 CO
w U U co OD ro ro
CD ro (3-13 ES ro F ) (L3 2 ¨ (-0 CS 010 (0 ro 4-' (0
c FoL (a 44:: r0 4-'(6 V.0
- U -it: (0 00 4-, ro 0,0 0(12 ro tf, 1-.6 4-, 00 03 op õ,(13 , 0 a-, _it:
m .,.,
= CO U (-0
(-0 (0 t: U 0 (a 00 CO r0 0.0 r r r -' uM 4-' (0 (a ti W tiO 4,jj
ro co tIA -o
.tjr 13 4 - 'MI ( OM t coo ( OM 4 - J+ ' ( 0 , 19 - M ,+ ' UM + '( = 0 ( ,30 (
0 or III mall g tr 71 c 0(4 : gj t 2 ti 3 rii OD 03 op 03
4-, ro ro tIA .1-; (0 174 4-, ro ro ro , , ro
0.0 ("3 (0 op an u
ro -u ro ro
CD oproro
ro(.0-(; ro 0.0ro.0roro.,(02t50.00 ()rpm 04-,0-c%
^ 0 a-, (0 00 4-, (.0 00 0 op
.1-, (0 0 0 OA 0,0 Op 00 4-, 4-, 03 23:, ro 4_, ,
CO tu3 tu3 CO , 0.0 (0 CO .1-, -1-, (.0 4-' 00 (.0 ro
4., CO tll3 CO -1-, (:11:1 0
4-, m .1-, .1-; tn
CO ,_,4-' (13 4-' CO 4-, ,+, 4.if ro ,T, 0 co 0 0 0 co ro (a.,3 0
ro ^ an 0.0 0 0 0.0 +-, ¨ tIA T-, +-' OA ro 03 - 4-, -' - 03 , ro 0 03 ro 03 4-
, 03
03 op 4_, 44f (.0 - 00 4_, (.0 (.0 ro 03 4õ-_, 3 , 4_, 4_, tõ:, 4_, 0 4-, ro
4_,
,-.U., ^ 0 'm `rEr on ro (3-13 an +-, (13 co ro ro ro ("3 ro '''' ro 0 4--,
a-, all tIA 4-' ro 4-,
- MI CO
0 W3 (a 00 U MI OD 4- U U r OA a (4 OD (a0 0 OD CO 4-' t; 0 0
'6'0,000(.0oproZurOtE;r0r0(00roum¨FErroro.20,0op.7)3.0zr0
0 0.0 0 (0 CLO OD +., 0 OD (0 ro r0 0 to 0 4- 0 (.0 s (0 +, tj-Shn mu (.0 -
'(.0 to roM
4_, 03 03 4-' 4-' tIA .1-, ro 0.0 0 ro (13 (13 ro ro
r^ oc 04-,rorow3couummr04-,-.,tror02,--0.0(10---(0(3(3-0-uu
a.0(04-,r04-,uroco-u..00ror0(10+-,(113+-, uõwrOdA,,wro.2030(0
, U CO
ro +-' U (13 (13 , ,., tIA 00 OA op -o ro ro +-, +-, ,444 ro 4-, .T-_, 0 .,,
ro co ro
ro ro ananu ro-u a.13-' co+, (000-0(04-'(00Øt:r00-04-'0:10.000(0
ro-or04-,(1300,0MrprOm õ., ro (.0 U (13 ojo 0 4-1 Op U (0 4-, 0 (0
4-, ro 4-, 4-, ro ,...., = - 4_, r0 - s '-' 4-, (-0
(0 4- 4-, OA 0 0 0 ro ro
ro '.4" co an ro (3-0 an co -' 0 (0 4-, .0
..__,('3 (1' 4-'.s,õ 4-,_ õ
co a. 4-, c(3 an 4- (a ro ro (a -' 4c; ro op ro op 0 -i.i, ,...., 41J UM -
I¨' 0.1 3 MI
4-, tIA co
4_, (.0 .1-; ro (.0 op (0 (.0 õ,ji 0 co 0 ro (0 4-, (00 (00 (.0 4(2, 03 r0
03 o
ro op OD ro co 00 4-,õ 03 03 tx0_, r0 co IT CO ,..,(13 ro ro co 0 4-'m +A 2 4-
, 03 1-3-(0 -1-' 03
03 03 3 -(i.T3, (.0 0 --- (õ:, 0 6, tu, co co ttO Cr CO (13 4-1 -1--' (.6 445
00 U CO 4-, To 4-,
U (13
CO U az, ro u (0 0 ro 0.0 0 1-3 ro a. .t,' co co eto 4,..-.: ,(2, , (0 , eto
44 0.0 ro (0
" r 4uroro--roror0r0""vmuroonr 4-,rouro
ro -1-, ro (.0 ro 0 03 4_, 03 op ,
, U 0 U ro ro 4_, 00 4-, (00 0 (0,0
OD MI 00 0 '' co (0 (00 MI co U CO 0 U U (0 (0 Zy
4-, tIA 03 ro 03 ro op Ms ro r0 _ ro 4-,. t1,13 _ 4F,0
ro ro (.0 (.0
, , ro -u 0 4-, (0 ro co ro ro ro
OA 0 0 ro co ;43' OA ro zo, (12 (-0 4-, b'
-c% (."op tIA U CO 2 t,,,:, (30 (.0 1=i 00 0 µ6, CO (.32
(0 .0 r, on on a F, .E(,-(3 ro ro 2 MI ((3),0 (rao mr 4- t.) 4- CO 00 (0 (0
CO 3 ',Lk?, -(-% ,t-:
ro (0 ro (3113 (L3 ro ro 0 to c(3 co rLo) ro ro ro (a ..'i 0 '' a4-(L3 '
OA 03 4-' (a
U tIA 03 ro tIA 4_, 03 0 ro on ro co n3 .i .14:
0,0 -1-' (.0 CO -1-, ozi .0 ojo op OD 4_, CO -I-'
ro --4 0 op 0 4-' 4--' t.E.; tl
(00 0 on 0 U (.0 õ 4-' 0 (.0 OA 03 ro CO ojo ro
+, ., .. co .0 (0 Y.
0 õ.,
õ., co .0 CO CO 0 (a ''' (13 ',4
(^ .0 co (.0 4_, ro ,(45, (.0 op tyzi (.0 co 44 if
(.0 4_, 0 .,.. t.,ji 0 ¨ (ap (.0 00 co 00 U .iL.,
CD 2, op 4-, 0 a-, CO -r% ro CO fo OA ro CO to 2 (%0 ro if (Y0D 0.0 r13 OA 03
00 op ro
4.2 4.2 ro r0 03 U On co 03 op 4-, 4-, 00 ro 4_, 03 co 03 03 _, 0 co 4-, 0
4.7.., (.0 (00
4-, (0 0 '' U U (13 0 U3 OD +'/-11 rill (13 4-' (13 '' ro 4-, µ,44 -
47.; U (13 co ' ,...,
(0 U OD r (0 MI -' '' ".. MI (0
MI 0,0 OD 2 4¨, 0.0 sc,. on -' 0-0 (..) ¨
4_, (.0 1¨,. ro (.0 ro 0.0 (.0 (.0 t, , 3 0 ro 4,74, (.0 (.0 4_, U
ro õ., .0 ro 4-,_ ro ro 4-'
(0 U 44 MI 0 4-' 00 (.
MI (13 (0 ". m .1-; (.0 4-, (.0 w (0,0 4-, W .1-; 4-, (00 ro
co ., .0 tIA op ro MI ro µ(.6 0
00 Ms (0 ro , 0 Ms 0 -tj op Ms
" (.0 03 0 4_, 0 r0 4-, 4_, ttO 47,,, 03 0,0
4_, ttO 4_, 4.7',., 03 0 On O4-.4:, 4,7.,' 00 0 r0
a , op op t-,. 0 pp m ro ro ,..L., 0 0,0 OA 0 47; +., ;:,..,,-, , ro 0 s,.-õ,
0 .,,,L, ., ro ro
CD 0 -t; 0 r o -(73 -4._-; Tr, tu 3 0 r o µr 64 4(3 co 0 c 0 2, tll3 ,(6., 4-
,, a3 .(i2, ,(6., on 0 0,0 0,0 op
4-, (0
CL:Ihn (13m 2 2 bo 0 0 (-0 To (.0 ?bm 2 (.0 (=:,,' 2 +.,,,,.. (.0 -,,,
0.00 15, 0,0 0D
0 a-a 4--' 0/3 (10 (13 CD, 4-' ro ro ro ro -1-
' 053 - 03 OA op
_ , 03 00 03 03 -L-_:, 00 0 4-, _ , t,u3 co 4F3, tie., co 1_3 _
03 co ;=.,": 4-, OA OA w op 03 03 op 0 Ilu.d. 4-, +' co "' 4-, ,,-,
2 (00 4_, 0
.1-, OD UJJ (13 CO co U ojo CO CO 4-, .1-; (0 CO (0
(13 (13 ro co co 0 .1-; 0 4-' U 4-, .1-; ro ro on co ro .1-, CO U (13 U co CO
0 +-,_ On
CO CO .1../ (0 (0 CO .I-, a-, CDU CDU CO .I-, CO a-, all OA OA all all OA a-,
U 4-, 0.0 a3
145
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
4¨, ro U 4_,
4_, (13 '81 4-, (0 õ., 0 ro OD 4-, CO U 44f ro -..17: OD 4-; ro
0 co ro op 4-, ro
CO (13 op co ro tie., ro co t,' 00 OP 0 '0'5 co ro tax) .2 U -.it s LO, .1!::
3 (.0 Fe
(.0 4-, (0 OP (.0 OP 4-, U 03 ro OP 4-, 4-' U 4F0' oo r 0 -1-, on
co ro 4_,
.,_, u 0 0 4-' (-)(04-'t:(13 0(10(02--
(113.t.,' OD
(23. .L.-! -(.10 4-, y (.0 ro -2-,:: 2 (.0 t) j3 c13 4_,
(3.13 4_, U 4-, (0
.Lt: '-' +.1 (13 1?, +-, U on , 4-' (0 U "a (0 (0 0D CO Z3' CO r U (0 to (13
ro ro ro , (13 4.-= 4-, , U ro ,.. 0 CO (.0 03 tu3 03
co 4-, OP rp OP 03 8
on 0 an 4-, (0 (0 c'D an ("0 ro 613 ro ("3 ro t 2, a (II 0=an ro (0 an ro4-'-
'-'roro-O.0 um opa.004-,mro op(094-'0-ti+-,,,
-' U+., U 4-'
r (113 4-' (I' (I' tj (113 4-' utl 4-' 447.: OD (0 t)
cati if 1:6' 2 ,,nnro u (0 H 0 ¨,,., nr93 õ., CD (.0 4_,
0 4_, _ 03 4_, _ 0D 4_, 4_, 0.0 ,,,,., , , 4-, (.0 0,
4,7 44=,' co ....,,,, 4_, ¨ ¨ .... 0
4_, CO 03 U "J , ,
Zuocororom¨urot'ro%4,74?6-' r.; 3 0D 0 (.0 2 (-0 2 g2 3 2 2 0
0,0 oz, 0.0 ., 0D (-0 .:, u (-0 on ,4 ij .1,- - .E.,4 r o
t:: (0 - bp OP co +., U co +,, (13
op CO -' 4-, OP OD U (0 U U (73 +,, op tj 03 00 00 CO all ., (0 co 00 03 +.,
(.0
(0 CO (0 CO r (0 (0 +.; -' CIO (-0 CO 4-= CO :.,' CI -' C1.0 U CO
6
- '
= CD (100.000U(10+-'03(0U(130,0oU(.0(.0(00,04-' 3 T.0-, u (-0 .õ 0,0
,,,op-03(.0r003 ,00303ro-ro co co 0
4, CD 4., CP CD MI CO (0 (II
0.0 rn m 0.13 ED
a-, co 4_, CD CD U op oz, .(i2, op 03 0 4t', OP ro 4--' (0 CO OP az 6 ro .03
OP U 4-, +-' (13 OP op ozi CO +-, Op Op 2 r0 2, OP .F., t,' U +a OP 03
ro
op op OP ro 'Jr:: -I-' 4_,
-ti (.0 (.0 (.0 ox, 0 co , ro ti BO U -ti 03 03 (I,' 0 00 U co
r/.9(10t5W30.13COUU U +., 4-' U t,' OD (-0 ,,9 CO U U OD 0 0 OA tj 0.0 0 OA
¨'' ro (134-'
tin CO MI (0 '' MI OD OA OD 4-' s ' co ro (0 u co 0 ro opOOro OA
0 ro 03 ro co , 03 03 a-, OP -,,.7,' co 03 4-, OD 2, 03 op OP 00 4_, CO co 4-;
OD 1.74 CO
(.0 co 4_, 0 CO 0 s . (-0 co ril OP 4-; (.0 OP ro
a-, +-' CO fo op ro g (0
co co 4-, (.0 ro ro 2, 0 (.0 (.0 , : 4
4_, ro ro ro ro (.0 (.0 ro - ro
OA ro 44f ro ttO LI 0 -'-', , u roc to 0 4-, 0.,n3 u 4-',., (10 4-'0, oro -1-
'0J3 (30 44(13 4-'0
(0 (-0 (0 U (2i3 .`=,':,. lf, :IL CO }:, -I.' yro õro iii ro 4:,
+.1 U
OP 03 U Op
+., .2 tj .(e, -0 :.; +-' SIP CO W3 Tz, ki ?LI 0 t, n ti OP -,..,,õ iii +., (0
03 CO
CO 03 t',õ OP co 03 00 4-; 4,74 (.0 2 (.0 4r2( BO (:,j3 0 (.0 (13 (1.3.. oz, -
ti (yo. (.0 ro ro m u
(0 0 4,"" u (0 ro 4-= (0 'r6 ro an 4-=
4-' MI CLOt,' (13t,4-' 0.13(0-(3 MI mr (Clj
U (a0 0.0 44 -14 0.0 CD 4,-', 0 (53) 44 ro _if 2 q 3 0 (.0 (.0
13 OP OA (0 Tx) 6 CD
r 0 OA OA ro OA (13
CO t:, +., (0 0.0 +., OA 83 2 0 Fe 3 on on .-2, t; ((:0) Fe (,.-0, 0 (-0 (,.-
0, 2 -t, 4-, 0 (0
22? (.0 s (.0 ,,,:, 25, 2.9 2 ,:,õ (-
õ, -(.6' (0Ø0 t.rõ 4_, 0 , 0 a 3 rp 0D CO 2 ro ".t'
4_, ro c13. CD " op OP 03 oz, 03 3 -,(T,,, ,:, (.0 (.0 u 44 (.0 if 4., 3 2
(0 4_, 44:, CO a, 4-, CD 4-,
030,,COBOopurO3UC044.t,S2UU
CO -' CO CO 010 4-' .F., MI -' ti' r MI
.F., ..t:: MI .F., CIO CO CO 2., r 4_, op ro 4_, ro 4-1 4_, fp .fzi 4_, tu3
ro 0 4-, 0 CO 03 2 03 oz, 1_3 03 03 0 ozi ,-0.1 CO
(0 (0 U OD all tp OD (1 4-, (13 OP 4-' if rill CO (1 (13 cp CO U 0 4-' OP co
4-; al/ 4-'
(0 .t: (13 (0 4-' 0 CIO OD CO U r
co .F,L 0 t,' oLO .,., CO 4-' CO -',-, ,õ('':'
00 44 +., 0 03 õ, 0.0 4-, OP OP .V.," `ii
,,õ,õ0113 OP OD CO (113 4-',D if ro a mr (0 ro OA Lo tj ,(1:1 ro
u LLO (. t: 2 8 mcio ¨ (-0 (-T3 ¨ (-0 0 .(,,, , 0 õ0 ¨ .- (-0 (.0 (.0
0 2 9 op co op - ,9 ro op
ro co 0 op 4-, 03 op 0,0 OA 4-' U 0 U 4_, ro ro 0.0 CO õ
,.,-- op ,
u CD 4-' CD 4-, CD 4-; 4t, ' . CO a-a 0 to 0 (13 44 CO CDco CD CD'4' 0
CD u '' CD W3 -t: r U U (0 ro op (i3 (:,j3 0 tu3 op 223, 4-, 03 tyn ti3
0.-0' OP 2 (.0 0õ
0 0 ¨ 0 0 4- r o
õnOPU 4_, 0j34-, 4_, CO 0 mu 4(1.00j303 4,,-,,' (\,!, tu34(101::: 83 .2,
,,.,,, ro (0 U 4-
' (0 "" ,000CO `(.',0.0(13+,,UrOCO'co -U+.,4-'+arOCOL4ZrOUO.0
OPcor0(.0Ut,õ.,COUUcoC00003+-,2õc0+,,COOPOPCO+aCOCOCO-
CO U 0 , U 03 , ,-, 03 CO 03 a-, U ro 4-' op ,aaa 0 03 03 4-, 4-' 4-, ro 03
U CO on 0,0 4t: 0 (13 U OP op U co , ra 00 4t, (.0 (0 4-, _it:OP OP OP
AM O Pt: ' ' I 3 + ar L3 + 'C 0 + '(..) + ar L3 't r (.- ,f, - '0 3 UM
+ ar L3 (tF, ), 0 + '(-) + -'r 0 c 0U te. )13 = 'M r:t I: I (V + -'(' 0 C
OM L'i 3 i 4_,(-0 to
2 on (.0oz, 0D (-0 -(,0J (.0 ., t: -u ., OPCO+,, CO2t,' ,,, MI co+a, 03 (0
4_, ¨ ro 4-, 4-; .F., 4_, 0 j3 (.0 U (.0 SIP OD U +' OP CO 03 0 613 2, tIZI
T.E; ro .p,
ro OA 4-' CO 4-' OD U (0
0 ro 4_, a-a all all ro u U U (V 4-, :lc:: OD (.0 4_, 2, (41_2,
ro ms L, -6, a (.0
03 OP 4-, CO U CO op OP ro_ ,9 u 3 0 3 oz, tf, 3 0 õ..(U 4-, CO -I-',õ op CO U
a-, 4t,
(.0 4-, 0,13 (:)J3 4-, 4-, 0,0 U OIJJ `-' (.0
oz, co 00 03 op 0,0 OP s . 0.0 ro .i.; 0 t 4-' tn ro
0.0 co -14:' OA (42, 4(9 4-, -1,7',., -1,-',, ((.11.), 4_, _ co (.0 (.0 4_, u
4_, ro 4.-,. co 4_, co .iL.. ro -44
" ' ., 0 '(.'6
-' -' MI +-, 4-'
4- CD (13 OP
4-, 4_, 4-; (.0 OP 4- U oo ro co 0 -1-, co ro 0 4_, co CO U õ, CO 0 (0
.0U a-,(0 -I-'u to õor 4-'(.0 (pro ro 9 4-'M 4-'CLII 4-,+' -1-9 tO ep out:4
.oror oroM ,r0 -1-'(.0(13 oroM 03ur sg3:0 a - ,(r 11 roc roc
ro co on 0.0 ro ro .,., (0 ro +, (.0
co , 03 op (.0 +., 4-; 1-3 (.0 ro 0 co OP 4.5, u -1-' (.0 0D 4.5, ozi 4., ro
õr., cp cp CO .(iap
U fp -I-, 4-' rp OP OP ro U U (SD all (al u op .2 -, oD op ro ro 0 0.,¨ ro OA
ro ,
um roro.,r0O.0¨ roro
rorouopopro-OproW
-4-'0Øt,'-'-'''''''0. O.Or00-0,9roo.Ororoaro um oorl:/+-'ro opm -
'0o.o+-,rororot10+-,uroroM.,
2,r0+-,uurot,'BOroroo.Oromcor 26µ53 .2ut;',0w3u 0
m 0 ro 0 u 4-, 4_, (-0 2, OP op õro oo 4(3 03 op 4-, 2
ro ro ro OP 4-, 4-' 03 44=,' OP ¨ op 4-, 4-; 4-, on co 0 6" 0 ti ro op OA op
Orouti.O(0+-,-0.00Øt,'(04-'opmroo.Oromro
00 OP 0 4(2, OP 4-' CO 44:' 0.0 4-; OP 44:' 2 OP u co OP 03 00 ro 4_, OA co
4-; OP 03 03 (0 (13 CO rp U OP OP rp rp t' 44 CO 03 +-, +-, -' +-, CO (.0 0
U 4-' 03 4_, CO (13 CO 00 (13 4-, CO (0 4-, 4-, OD U 4-' OD OD MI -1-' CO " op
.1_= U
MI (13 4-, OA (0 u CO +,, OD 0 co 0 U u 00 4-' t,' CO CO t,' 2
CD 0 r 0 r 0 4- , r o 0 r 0 r 0 o r 0 4 - (- 0 r o r 0 r 0
0.0-+t + - , r 0 .._,- (..) ,,9
0 ro ro 0 -1--, ti-'3 4-, ro ro U U ro (.0 3
(-0 u -,-, (-0 4-, ro -'-' U "rill OP n,
4-, U u OD 4-' .,,, co 4-; U 1.7',.. CO 0 U i. 03 _ 4_, 4_, CO 4-' 0 (13 4-;
ro OA co CO ¨
COCO0,04-,(0`"'OPCOctur0 -
co`(.00.0t,'( 0,04-'CO(0-04-'(''
ti' (0 U r r
COMUMMUCOCOMRICOM
4_, ro , 0 " Op 0 j3 4-1 2 4,,, 4(3 U 4., (.0 ozi
0.0 rp 4-1 , , U ,., n'w
CO 0 4-, OP (13 4-, ro
0.0 ro ro ro ro ro ro co 4-' .,, 4-, ro ,-; op ro 0,0 t a ro a, co (.0 u op -'
co
on +., 0 +, u t,' ro co u 4-, co ozi op ,
CO 4.5, 0D 4.5, H
0 ro OP 4-',õ 4-, 4-; op 0 CO 4-, .F., (.0
(:)J3 4-; 4-, tk., 4-, 4-; ,..¶., _ ro ro u 4_, ro ro4-' ,,, ro 4_, 4-, co.
4,7.: 2, r0 m 4-, c
ro ro -' an on ro '4" ,U ro ro an (0 __co tto (0 Fa' 0.0 an ro 70' 61, 4u an
ro L'D ro U 4-; (:113 U U 4-, 4-, 4-; CO OP 4--' op U -,:,-, CO t.-.,:, ,,3 0
4-; (13
4-' 4-' ro 4-, ro
4-, 4-' U U 013 tIZI U 4-, 4-, OD 4-' "" 4-' `-µ,, CO U 4-, U 0.04-,
(.0 4-, 0,0 0,0 0,0 4-, 4_, OP 0 4-, CD U OP OP 03 ro 4-, ro 0 4-, ro ro ro ro
ro ro 0 4-, OP 4-, a-, a-. CO a-, CO a-, CO CO tll3 CO CO a-, a-. CO tll3 U U
4-, CD CD 0
146
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
4-,a-,ouur000
(0 ro 4-, oo .1-; 4_,
03 (0 m r0 r0 oo u
, u
ro (0 CO 00 ro o co "" co -'' 00 ro co 0.0 ro 0,0 (''' i.3 co co
ro ro an
(49 3 (0 - t' 0 (0 tin (0 (0 U 4-, u ro 4-' ro a-,
MI '' 0 ro -' on ro 2 ro 4-, r 0 u if r 0 .o U (0 4.1: on (0 4-, 4-,
( ) On 'I.- U -' '' (-0 0.0 (0
0.0 (10 (0 (0
CO (11:10.13(13 MI 0.0(0 -' CO 0 MI - (..õ., 4-,
U on ro 4-, U On
W3 C0 (0.2 te:PrO (0 (13"+-, mtl 76 0,0 r0 te ro 0 00 (0,,, õ.,0 ,õ,(0
r0 B ro (0 2 A On hcM". (0 r U5 4-' (0 U 4-= (0 CO r ". ". "' 4-' ro on (0
u (0 4-, (.0 (0 u u 0 i.i:, ,,.(- (.0 c,- (33 44-, 4_, 4-, (0 CD 03
(:(j3 (:(. On 1.T; U
CO OA 4-' (0 ro u ljz, (0 r0 4F0' oto Fo" ro on r0 ro ro ((.1', o 1.-0' 4-'õ.,
ro ,4:',.. 01:1 1F .1-; 2
OA ri3 4-'
co 00 u on r0 ro ro +, -' on -u an r.,' ro 2 ro µr,Y., r, a t, 0 r 0 +-;
4_, 4_, .1-; (10 ere .1-, 4-, CO u OA m co oo u rou4-,
(0 0,00,04-,,,-(0 ooro u (0 ro 4-' 4-' on o o ro - (0
(0 4-, õ., 4-, (0 0 U+'
4- W3r0 (0 U W3(0 0.1:1CW.,,
0M 0%0 p. 0 0 , 0U t : ( MI UM .. t-J+ ' 00.0 3 ro +.,-- -ro Bo (0 ,,,,,r0
,.0 on (-,, tE; u
U 4-,
ro .1-; ro o 03 4_, ro u ro 44f,
03 ro
a004-, ro monro co, r04-,ro (pro on.F,uro mroc,0-(3'.2("3-(%.t,'ro.F,
r0 B 00 I'S u -c; (0 u an (0 0.0 (0 co u (0 4,74 (0 2, t-_,, en , (:to 2 +, 4-
= .C,:i U
(0 (10 0.1:1 (0 U "MI -(0' t,en 3 ro on .s,. 2, ro +, 0 u 00 m CO ro m u
4-' r0 4-, .1-=
MI (13 0 on 3 õ.,(13 4-5' 2 (134--10 hrio,orl3U-4-,(00.0(00-13c0 co OA , U
ro op 4-1 ======= ro ro u r0 OA u 0 u 4-, u 4-' oo
a,
t 41(00.0u6',0 2 3 0 0 0 + , r 0 r 0 r o r o o n + , r o r o ,,,0 r o t: .t,'
, . . , r 0 CD,
on (0 u (0 on oo t-,, rot, u+,+, onro um .,, 0,13-42-0, 0
on FO. (0 (.0 0 (3,3 o (0 ro u 4-' OA (0 tX, ro 4-' 4-, 4-,
0
(0 u rj 00 4-' +' 4-' MI ro 4_, oo OA ro 4-, hn 4-, U ''
CD -,-, m ro (0 m .0 .0 03 (34 CO 03 on o on 4-, ro (0 (0 (I,- (0 , u
u ro CO
I. 0 ' s' U U .1-; (0 (0 on (0 t, (13 (13 (0
OA (0 00 ro 4-; ,,., OA (0 :-., ro ro (0 u 4_, 4-, 4-,
-u0.0rorouror0robow:,(0(.0ro00`(.'o'roro4Z+,rororo
u op ro 4-, U (0 013 (-0 (0
4.5, u .,, 0 j0 (.0 .i., U (-0 t.: CO U On 4-, u ro -' 4-, , (0 2 on on on +-,
u ro
on ro OA m (0 U 2, ro µ(.6-= t, OA r0 44f, r0 (34 00 on 03
ri3 on -' 4-, OA on 4-, -(% +,4-= CO 4-, (0 on t, -
(% ro Z ro a onro
U ro 00 -u (0 on 4--,
romBO,,u0Orom4-,0,0-ourorOo,ror0 rOro co
oprou
on u 4_, ro ro 0.0 ro -u -u r0 4-, 4-' (0 MI MI -' 0,0 an
4_, (.0 m op (:,,:, 4_, ,:,,3 õo 4_, ro
oo (0 OA ro on 4., "ro 0 2 tr.,
(0 on o on 4_, u (.0 s_. on 4_, 4_, u u (.0 ro 4_, -
+, u (0 on u 0.0 ( 0 co r 0 r o r 0 oo u Ms (0
4-1 on co m 4.5, u 03 -(13js -8J-3, 4-, 03 ro 03 4-, 4.,., ri3 ((.3.. 4-' ri3
ro (0 OA 4- (0 CO
OA
u
MI -' ur 4- CO CIO on 00 U OA MU 0 t) CO (73) (0 On (0 a .t,' U CO
r
CO CO 4-' CO (0 - CO .1-, .1-; - ro ro O ,õ 4-,
4.,O ro 03 r0 .1-; CO on
4- .F., ., CO ,z, (.0 .,., (0 +
+., ro ro u 0 .,A OA ro 81, CD+, ro 03 CD CD CD CDCD ro 4-, (0 (.0 u
U -' (0 .1- = .1-; U
+., ro r0 +, on on co m (0 CDan (0 co (0 OA OA op u ro (0 4-, CDCD(0 (.0
4_, 4_, 4_, õ= 4-, CD(.0 (0 OA co 0 .1-; 4-; 01:1
CO U (0 .t: an -
(0 }-!, (0
co 0 (10 00 e, r, 00 +., ro hn 4-, u (0 u " 2 co õr -"r0 BC) ro
(0 ''' (1 t,' ' t.') (0 r r to Cr to
4-, OA (0 (4
(0 s ' ro OA m- OA 4-, 4-'
.1-; CO co , 0 co on CO 0 4_, 0 on u on
oo on 4-, on (0 4_, u , -,_ u ro 00
Ms r0 Ms romr 0.0 'o ,,,,(1 0 (I 0-0 0 2 Ms a, 4-, (0 0 (0 ro ro , .
rOrOror00.0r0-u --rororOuu 4-' on r0 CO 4-,
ro an on ro u ro p.,r,r0 u ' 0 -' (0 - -u u u (3-13 u
ro on ro u ro ro on OA (0 u u OA 0 on 0 ro 4_, 0 ro ro onro ononro -u hn _ 00
(13 On }I ,,., (13 .II .., 0 (13 0 0j0
On (0 4-' CO (0 On mr r ..6 mW (0 0 (0 tig3 s(1.; tj (0 (1 Z:54
(1301_30(.00 (0
co -I-, OA 4-, (13 (-0 ... ro 03 - 4_, ro 0 03 (34 on ro .1-; CO -1-' (0
,,,(10 oo .1-; CO (-0 4-, (0 (0 U U 0 (0 u ro 4-, on 4-, -1-' _,_,4-'
' w 0,0 .1-; 0 -L) U On U On (0 on U CO on ',,,.. 4-' OA +., a o 4, :0- r 0 r
0 r o r 0 : -, :, 4- , (0 + , r 0 oo +a r, r, ro +, 4-' u on -' on 4-, +,
+, (0
4-, , OS 4-, (-0 (.0 ,,, co ,:u3 (.0 ,z, s. .1-; U (0 on on n3 on 4-= 4-,
,z, r,,(:u3 4-, 0 4-, - -' on (..) 4-, U 4-,
(13 4., a-, U co ro ro CO 4., 0 CO (0 4_, 0.0 0.0 ro .(i2 (.0 ro on on 4 d 2 ,
i i ,- op
0,0 on u ro (0 0 on OA OA 4-, -81 4d 2 233 .-t: ,, (0130 ( 0 .t, v 2 CO (0 (T,
u u on ro
-' -' ro .,, , ro 4- CO
4- U (0 (0 0 +., on U '' (13 W3 (11:1 U '' 0 0 0 4..-...' (.0 U t u 1-,, OA
,(7,, rill. (4
.t,' CO 00 00 oo ro (0 0.0 (0 4-, (0 Ms U (0 (0 4-' 2 - ro (0 r 0 tE; z (0
(.'; -..-5 z.v
4- ,,, ro ro 0 r 0 4- - t; -J, õ,
y. ,, +, on +, -8;3 u , (0 E (0 OA r0 _ 4_, (0 oo 4-, 4-, - m 174' if on OA -
653 op - ,,,-, 03 3 (0 on ro 4-, 4-, .1-, (.0 oo oto
row mr0 mu 0.0 -' (0 (0 ro 4-,
õ., 4-, (0 .1-, OA 4-, 4--' õro -ti ri3 OA m (0 00 (0 r0 4-, ro 1-.,E,' -6' r0
4-, CO 4-'
4-, ".. ".. ,,,u onn3 (0 u- on,rorou 4-, 4_,
on ro
to +.,ro wro ,(5.),0 -0,0 (29 uu 3 mu mro +.,m, (.0 uro (pro 1_3(0 4-(0 +.,m,
mon mu .t' wro '3 .t.6, -ro
--' t'
4,õ -(i.T,; 4(9 (0
.,_= (0 (0 u on 3 (0 .(,2 õc, on (.0 u (0 2 OA ro ,:,j3 -(1.T3' _ET (.0
4 = ' + , 70 t . i -I, ro r o On 2 r 0 t; tj -5' ro u 0 4_, on
4_, 4_, u
(0 u ,
(34 ri3 (.0 4, .tian, a-, ro (.0 03 on CO OA on on u r0 0 co u OA m 4-,
CO 4-' 'I.'
On CO 4-, On 4., ro ro CO On 4., hn OA 44f hn U co (10
CO (0 (0 U co
U 0.0 +-' op (0 00 ro :C.," Ms ro -' -+t,' ro ..04 Ms m 4,"' (0 ro +., 4-,
,,..,,r0 u +., on r0 4-,
(0 co On õ OA 4-, (0 4-, on (0 03 r 0 u u (3,3 r 0 2, 4 - ' -4t f, 3 - -r% ti;
u (0 ano
an r o r o 0,0 co -.,t,' r 0 0.0 4-, 00 mum 0 mumroro,r0(.0(.0u0.0
4-, ro .,., ro 4-' 00 ro an (1 co ("3 u ro 00 -u ro 00 ("3 on ro (1' +,
0.0
u on 4_, (.0 44 ro u (.0 OA (0 r0 m m 4-, (13 ,...., (0 4r2( On u 4_, U on OA
4_, on ro
(0 on 44 u on OA ro BO co u ro .(,-9 'rt.,'t; OA r0 ro u
Ms 0,0 4-,
(0 CO on u ..,u on
ro ro (.0 ao ro ro (0 ro .2 t,' 4-, on 00 B ro 4-, 1-21 (0 U ,,,, +.; on ro
on +, +, 4-,
(0 u an t,' 2 ..), a 2 -,,,' hnr0 a BO ko) r0 mr0 t:,' (0 BO t C to' r0 8 4-m
Bo (0 .2
CO
(0 -' co (10 ili
(al 2 ro t, ro u (0 +, Ec, ro (0 u
an u -u 4- u u 00 4-, CO 4-, r0 u , ro 4-, ro
OS ro r0 u 4-, u 4-,
+.1 on On u ro ro ro (0 4-1 (0 (13 (0 0 4-, (0 (0 ro
CO OS OA
OA ro on ro 0 OA (0 .(,2 ro 0 ,:,z, co oo (0 4-, .1-
; 0,0 U 4-' OS (0 (0 OS
OA -I-'
(0 (0 -' u ro U (0 223, 2 1::: 233 (0 r0 -6, co t4 2 ro ro if
U u ro um ro ro ro ro 4_, 0.0w 4-, ro On 0 (13 (3
on 0 on 4-, ro 2 ro ro (0 OA u ro
co on OA 4-, (0 u -' .,., +, ro r0 ro 03 , on 4-, gjo , +, U (0 (.0
4(.T:,'
(0 .1-; a-, u On 4-1 4-' 4t1 4-, (13 (0 CO CO (.0 (0 (0 CO oo 4-, r0 On 00 u
ro ri3 ro
ro 0,0(0-u
ro+, co u 0.0+-= onro o 0+,4-, u ro-u o t,'1-2, on r,(113 (13 U -' 4_, on n3
ro (.0 4-, u õ., n3 u ,,., 4_, on .... 4-, u OA . n (13 CO ,,. .1_= u .iL-' CO
co (13
on ro t,' (0 ro (0 (0 ro ro
' an, , r, .L'2, on ro ro '-'a õflon ro µc, (0 (.0 (.0 on on '10
ro ro on 2 00 BO E.0, 0 2 (ojo -E, 0 4.,
rO co u (0 6.. 44 (.0 ,z, , 3
oo (0 4-, ro ,õ0 2 a 3 1-3 ro (0 oo u 0-0
, u (0 on (,:, ro op +., oo 00 '-"' r0 ' w _ (.0 (.0 õ,.,
4.5, (.0 U (-0 U
UtnUr0r0(000(1303(13MU4-'M CO 4- '4,' U CO MI -'
(0 .i.7 (0 (0 (0 u ro 4-'_ 4_, u "(13 (0 o 4-, ro ro_ 4-, on u ro 4-, 4-',.r,
õ(13 4-'õ.,
ro (.0 ,z, (0 (0 ,z, .,. 41.1 (0 On .iLl, .,. U
(13 (13 l 1.1 (13 ro on on (0 - - -
U 1
.,-==,
U u co u u 4-, (-0 .iti 4-, 4-, ; 4 .1--, ro ro CD 4-'
4' .-; u (0 4- , (0 ro ro ro
CD.,., +, OA r0 r0 +., +., op u +, co op , on CD CD CD CDu 4- on (0 u i.0 4-
CO OA rill OA OA OA rill U U rill rill U U U 4-, U U OA U 4-, OA rill 4-, CO 4-
, U ro
147
atatcaaaacgaaagaattgttatttaaacgtcgcatcgatataggcggtgtgaataacaactttaaaggagatttcca
agaggctgagggtctagatatgtattacg
0
atctagaaacaggacgtaaagcacttctaatcggggtaactattggacctggtaacaacagacatcattcaatttattc
tatcggtcaaagaggtgtaaaccaattctt n.)
o
n.)
gaaaaacatcgcacctcaagtatcaatgactgattcaggcggacgtgttaaaccgttaccaatacagaacccagcatat
ctaagtgatattacggaagttggtcatta o
ctatatctatacgcaagacacacaaaatgcattagatttcccgttaccgaaagcgtttagagatgcagggtggttcttg
gatgtactgcctggacactataatggtgctc .6.
.6.
n.)
taagacaagtacttaccagaaacagcacaggtagaaatatgcttaaattcgaacgtgtcattgacattttcaataagaa
aaacaacggagcatggaatttctgcccgc oe
aaaacgccggttattgggaacatatccctaagagtattacaaaattatcagatttaaaaatcgttggtttagatttcta
tatcactactgaagaatcaaaccgatttact
gattttcctaaagactttaaaggtattgcaggttggatattagaagtaaaatcgaatacaccaggtaatacaacacaag
tattaagacgtaataacttcccgtctgcac
atcaatttttagttagaaactttggtactggtggcgttggtaaatggagtttattcgaaggaaaggtggttgaataatg
gtagtagataatttttcgaaagatgataactt
aatcgagttacaaacaacatcacaatataatccggttattgacacaaacatcagtttctatgaatcagatagaggaact
ggtgttttaaattttgcagtaactaagaat
aacagacccttatctataagttctgaacatgttaaaacatctatcgtgttaaaaaccgatgattataacgtagatagag
gcgcttatatttcagacgaattaacgatagt
agacgcaattaatgggcgtttgcagtatgtgataccgaatgaatttttaaaacattcaggcaaggtgcatgctcaggca
ttctttacacaaaacgggagtaataatgtt
P
gttgttgaacgtcaatttagcttcaatattgaaaatgatttagttagtgggtttgatggtataacaaagcttgtttata
tcaaatctattcaagatactatcgaagctgtcg .
,
gtaaagattttaaccaattaaagcaaaatatggctgatacacaaacgttaatagcaaaagtgaatgatagtgcgacaaa
aggcattcaacaaatcgaaatcaagca ,,
,
1¨
.3
.6.
.
oe
aaacgaagctatacaagctattactgcgacgcaaactagtgcaacacaagctgttacagctgaattcgataaaatagtt
gaaaaagagcaagcgatttttgaacgtg .
r.,
ttaacgaagttgaacaacaaatcaatggcgctgaccttgttaaaggtaattcaacaacgaattggcaaaagtctaaact
tacagatgattacggtaaagcaattgaat "
,
,
,
cgtatgagcagtccatagatagcgttttaagcgcagttaacacatctaggattattcatattactaatgcaacagatgc
gccagaaaagacggatataggcacgttag 0
,
N,
N,
agaagcctggacaagatggtgttgatgacggttcttcgttcgatgaatcaacttatacatcaagcaaatctggtgtgtt
agttgtttatgttgttgataataatactgctc
gtgcaacatggtacccagacgattcaaacgatgagtacacaaaatacaaaatctacggcacatggtacccgttttataa
aaagaatgatggaaacttaactaagcaa
tttgttgaagaaacgtctaacaacgctttaaatcaagctaagcagtatgtagatgataaattcggaacaacgagctggc
aacaacataagatgacagaggcgaatgg
tcaatcaattcaagttaacttaaataatgcgcaaggcgatttgggatatttaactgctggtaattactatgcaacaaga
gtgccggatttaccaggtagcgttgaaagtt
atgagggttatttatcggtattcgttaaagatgatacaaacaagctatttaacttcacaccttataactctaaaaagat
ttacacacgatcaatcacaaacggcagactt
gagcaacagtggacagttcctaatgaacataaatcaacggtattgttcgacggtggcgcaaatggtgtaggtacaacaa
tcaatctaactgaaccgtacacaaactat 1-0
n
tctattttgttggtaagtggaacttatccaggtggcgttattgagggattcggactaaccgcattacctaacgcgattc
aattgagtaaagcgaatgtagttgactcaga 1-3
cggcaacggtggcggtatttatgagtgcttactatccaaaacaagtagcactactttaagaatagataacgatgtgtac
tttgatttaggtaaaacatcaggttctgga t=1
1-0
n.)
gcgaatgccaacaaagttactataactaaaattatggggtggaaataatgaaaatcacagtaaacgataaaaacgaagt
tatcggattcgttaatactggcggtttac =
n.)
o
gcaatagtttagatgtagatgataacaatgtgcctattaaatttaaagaagagttcgaacctagaaagtttgttttcac
taacggcgaaattaaatacaatagcaatttc -a-,
c,
.6.
gaaaaagaagacgtaccgaatgcatcaaaccaacaaagtgcgtcagatttaagtgatgaggaacttcgcggaatggttg
cgagtatgcaaatgcaggtggcacaag n.)
n.)
vi
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
-' u ro 4-, u tw_mtwro (0, m 2 n , n (.0 4-,
0 u m m 4-, 2 op oz, 0
^ `-µ,' op 03 ro 4-, (0 OP 0 ,,-, au u (II
(0 ' ro o ,õ u
r or o 2 . o4 - , or 0 + -=,r 0 t :Thu mop wrill t: 2 .,(0 60 (00A .,co
wro -r.103 0D ad + . ,r 0 roro coop row roro co- .,03
0-13 to op U 03 ozi op 03 co -1-' OA oo OA 4-, -' OP OP
r0 co
co 4-, OP 03 03 4_, ro CO U CO (-0 CO oo oz,
ro ro 4-, U co OP 4-, OP OP 0 OP -'
4-, -' -I-' 4., (-0 M CO CO (-0 4-,
CO
Mr C00.00.00.000CCIM'w4-'(-0--.F., ""r 4-'
0,13+,0(13- (0
CO ro ro ro ro ao u co on ,snu r u tto
,..,(1' BO ro ((r o) ,_,(1' õnu -u u ro ao ro 1-3 ro
, oz, op ro 03 co ro 4-_,w (1-3.- 3 ( (,0, 4-; 4-, co 44 co u a 3 (0 (0 0 4_,
co U
no M co 4-, OD 4-; r0 OP co
u .2 :0 m ro tto 0,0 3 oo o ro to V.1 ro ro 0 tE; ro ro t'-', +-, ro ro u -u u
(0 u
+.,- 0 t OA u Bo t,' co -(0' ro -0 õnnõ4.-
iirourorow,u
4-, 233 ',nal, op,c0'-
wro,uttOuurr,030
^ CO co op - õ, 4_, 0D ro co
ro u ro 03
(1 r0 tp OP ro ez, op -I-' OP OP op ro '', 4-, u ro (13
OP ro op ro ro OP 4-, ro u ro a, m, (0 u ao
4- on 0,0 +, (.0 ro ao ao
õ, (0 +-, OA otO -um oto m 0 u
co co 4_, ro on u 4_,urowro- 4_, (0 4-
, - ro - 4-, 0
OP o^ p 4_, CO ro u ro 4_, oz, U OA U ro 4-' OA ro ro
OA OD OD CIA all U a-, 0 a-, '4.. ro ro co U ro ,.., U U
op ,õ CO op 0 ( 0 op 4.,
(00,1:1(0rOUCOCCICOCCI.2(10(00.00+.1 -(3"drob'-'sr6uroa.o0.0(0-6'
OP ro 03 co U _it, CO a-a (13 tx, t:ll3 4-' u U ty0 0 ro .(i2, ro (0 ro ta ro
ro 4.5, co tw
4-,
U (0 MI (1 (1 CO MI (113 +' -' MI MI 1-.,) r l:IA CO d.13 CO MI CO
co U 4-' (13 ro ,,c, . 4,74 4(2, 4-' ,,,.,(13 2 (ao n, on n, n, 0 , ro tax)
4., on u 4-, ro m
, ro (0 +' 44=,' s . no 111 4`-_,'.. co .,(2,
4_, U u co 4-, OJO b) -1-' 4-, CO 4-' CO CD s '
4-' U (0 CO nn ro ro
0.0 otO (II (II r 0-0 ro L,:v3 ro (%ro' ro ro ro (0 Lto ro ro ((i,' Oo ro u
(0 , u rr,- u ro ror000mmooBorprorouromromm+-,coro+-,0.0,corom-ro,
(0 u co 0 u co
- õõ Op
ro ro on 4-, 4- ro ro ro ro ra m - ,
a, 0 m u ro ,,,, 4-, 0 CO 4-, 4_, CO OA P. 4_, r CO hnr 'WM 03 4-' U
(0 ro co ro ro (0 4 ao u oo +-, ro 0.13-a CO 0.04-' ..."' ro ''' ao ro -e, u
,r +'ro +-'op
ra 0 ra ,õ +., OP OP co co t 4-, U (-0 CO 4_, 4-, CO CO
co ro (13 u oz, 223, 03 co a-,
,,-, OP 4-, u ro ro 4-, co '' +., (113 u U (.
r, U CO
CD on 4., 4-1 co CO 1::: (0 ^ Op 0 0 0 op 0D r0 OP r0 ozi op op 4-, ro
OP - +., op
U 4., 4--' -I-, 4-, 4_,
co OP co (13 ro ro opOPrOc0 4-'rOur0O.PrOcOu
Ms op t r 0 2 r, 4-, co+-' MU co+, co CO ClIlt,' TIE; all U+,4-' 2 , + - , r
0 + -, r 0
CO all 0,0 ro rõ, co 0 ri, 2 on CO 4_, on 0 ro ro 0 ro ro CO 4-, OD 4. op 0 op
0D
-'(00.0Mco4-'CLCiourOMIcOc0(00-0r0(0+-,(0
0 +., ro OP OP 0 CO op -1-' -' +-' +, U 0 r
CD-CD
co ro .i.-; ozi all 4_,z, U CO 1T3' tu3 (.0 4_, no
,.., co 4-, co 4_, U 4_, no 4_,
MI 03 4- +, (13 4-' (0 (0 0,0 ca
0 ro ro 0 .,,,,, (.0 ao ro ro OP ro co (.0 ,
03 03 u a-a (13 4t: U CO BO 4-' ro
0,0 a, , all ro op OP 4-1 CO ojo (-0
MU (11:1-j CO+, Mr 0,13(0+' (113(0 OD+, CO CO+, COBOU all (044
, op 4-, OP op op OP 4-' op OP ro ro 0 co mu ro (0 co op co OP
" ro ro IT tp co U
U ro ro t 4-, 4-, U U +., OD 4-' +./ MI '
. OD co OP OP t:'- - ro u u -I-, 4--' U
CO 0,0 CO , CO U .t, (10 ro +., CO (-0 co (-0 01:1 44 ,,,, 4_, ojo
co co 4_, ,,, oo u
CO 0 tn U OD r 0 1-3 CO CA t: (10 (0 co
'cLY., U U OD CO OD (0 2 '44 4-,õ u
ao ro =z.,.- ro ro (0 tto ro ro ,-.4, u -' -r; ro t', , õ, OP co +., u op co u
OP _,_,=-= OP
-' Ms 0,0 OP ca OD 4-, +., 0,0 tj 0 MI (0 r0 ' OP 'v7
u u ra -' u u OP OP 'El Ms
0 (0 (10 co OA MI OP Ms OA ro ro '10 -0 oo 4-' CO +./ MI (1 -(Lo' MI CO OD U
CO 0 (10
CO ro (0 ro ro ro ro 4-, oz, tx, u ro ro op OP 03 4_, ro ro 2 rõ, (-õ, 03
4_, a-a -!--; U CO ozi op u all a-a (0 CO 4-, ro ro ro 0,0 (0
r, C00.0,...,''4.7,',tf,+4-'01:1C111+,4-JUUCOU0
0c000(0000
0,0 Ms 0 OP ttO .L'f, '-'-'4 op Ms +., (10 r0 MS OP ca -' 0,0 Ms Ms MI OA
OP co (13 CO
ro co
ro+,+,4-,ro.,,r0 0 u4-,r0 uttoro-u8r04-,ro , ro
u u 0,0 co co 4-, OP oz, co OP co U ro U 2 ,õ ro
ro õ OP WI
roc003cOuur00,003-60,0cOur0+,:,;0,0rOurocourorProrou
(0 4t, (-0 CO CO OA CO ,..,., OD 0 DOACOUCCI(13(00.t,'+,U(OUUCCIrat'
U OD CO MI (10 (0 -,t: 4j CO 0D (0 (13 -(0' -(0j OP 0,0 r0 op (0 OA (o co u U -
' co fo
03 oz, 03 0,0 U -I-' oz, (0 4-, _ u 1--,, 4_, 4_, .(1.3, oo 4.2 co u 4_, co
4_, 4_, u co tw I.
U op 0,0 (0 K1 3 , 0 (- 0 t: : 4 i - = , 0 z ; + -=, , 0 0 OP w 4-, Ms ca O
00U 2 -'
,T, 4-, ro
ro OP 4-' ro - +., u OP OP 0 - ro ro 4-, 4-, ro
OP rill -r, u õ..,"' co a-,
(13 4-; CO Cll:1 OD OD (13 4-, OJO co U µ s
(13 co ro ro t,o ro CIA (:113 CO /1
a-, 4-' OA ro co 4-, ro a-, op - ^ 4_,
03 4_, 4_, 4_, 4_, (.0 u - õõ co co
4., 4-, co 4_, (-0 4., u OP 4-, OA OD (-0 ro (0 4-, ro m (0 co ''' 4-' u
OP OP 03 4-, ro ro 03 OP -4 r0 0,00,0
co 4-, (0 OA, u co - OA U r0 MI OA
on +.1
ClAttOroUoproCCIroro+.1 Uotnr0 op 1-3 U3 OA (10
+.,(0 .000 ror0 (6,0 +-,(0 rou ror %.1:1 Zm ro(10 +-,(0 co 0DM 0,000 _our
uU cora (00-0 tn,..,, (00.0 al hj, .6,' ro õ9 ti
u44 u 0,0(0,4-,õ,(04:5 um u n, ro ro 4-, OP CO
co tx,
(13 u OP -5' ,õ(0 op 1-2, co co 0,0 0,0 mr0 ,T,U ro
LI 4t r or 13 r ou 0 3( 1 0 3( 1 4j LW mall (al,(13 ro to .t, Lto u 45.
u , ro -u ro ro - -
roun,rorouro
03 OP 03 co ,,,, 0 0
OP co -' +., co OP ,44-' rõR co CO u 4-, ro
ro (13 on , 0j3 4-, ro u - ro en (61 (.0 u (.0 ro (0 -0
3
co 4., ,_,U (0 ro OP 03 ll, -1-',_ 8 4-, 4-',,, 4-,
CIIIMU-+.,UU 0,0 0j3 co .1=, tto op u 4_, oz, , . 4_, s
. ro
U r0 u (10 4-' -' (õ,-,1 op u 0 op co 0 ^ op , (.0 r0 co ro (.0 co (.0 u
CD 4_, 4_, U 4-, CO µ...4 co 4_, co ojo u 4_, co 4_, co 4_, 4_, u no hn on
õ, co U a-,
ro 4-'ro 2 6a0 rpm 4-'u .I.-,t' U U 4-',., 4-'µ , CO 4_,4-' OD 4õ7,' OD OA U
CO CO C13--. CD 0 -0 6' t S 4 - 'a 13
0 u no 4_, co (a rub 3 4C2, 4t,. õ.,`-' tiO co 4-' ,_,"-' CO (:113 -I-' ro U
ro ,..õ r0 0.0 4_, 4_, 4-,
4-, ".. ro ro - 4-'
u ro a-, co a-, 'w CO 4., (-0 U 4-'
U (0 CO '' OD (1 OD U 4-' CO -' r U
OD oto ro m ro c'D t:,' ro 8 ( , :, ,, um - 6 ,:,õ` 1 2 2 3 s r
orc' ,4 - , 3( = 0 s g2 - , .0', ,
( . 0 n , n , 0 . 0 4-; OP U ...,, (13
,,,, , (-0 ro 4-, , , r0 tto µ..,_ u ro 4_, (.0 4_,
0 4_, _
4-,r0rOWZro+attOurO`' 4-, ,,-,
-' MI op 3 0 . 0 t E ; u r 0 , 0 , 0 4 -,c) .2 2 "( = 0 , ow or 0 mu 0 .
om ot, 0 -t:', z s . õ- ' r or 0 + - ',. 0 ,` '
CO a-,
CO a-, (0 co .2 0.0 W:Th r ti'o OD U Fcr to 4-' fa (0 CO r ti U (0 CO CO
-6' ro V , ro ro (.0 , TIE; 4., U u (53' ojo ro (-0 oo
Op 0 L,^ u OP (13
4-, U 2 U 3 4-, r (13 U OP to op R? F33 2 U - (1 T1; BO 83 If a-, 2
2 0.0 -, u u u oto 0 ro 0 0 on u
ro ro ro CO CO (:113 (10 -1--' U
CO U U U U ro oz, CO 4., 4-' m CO (13
u 4-, r CO CO op ro 0j3 (0 CO OP OP u
CD 0 c0-(0'corOc uu 4-
'CLOMIro
U u 4-, CD CD4-, 4-; ...,--
(-0 4., (0 l:ll3 Cll:1 U U w CO CO BO u 0 CD CD co co u u 03 0,0 op 0,0 OA u -
+t,' u
cor0c0u4-,4-,+-,c0OPc0(0c0(00,0000,0urPrOcouut,'-ur0
.,., +., , co r0 -' 4-' r CO OA
(1 on On CO ''" U '' CO r 0 m CO MI
4_, ro 4.5, ,S, ,y 0 OD CO ()J3 (0 ((ro) 4-, op (cil , ro ro z., 4-, 2 - i -=,
u .,, ro u r 4-' a (You .,,:i t, -+t: (:Lo -r.: (0 ro co u -m (.2 t 5 a t2, +
- , ( .' 73' "r 0 u r, u
+., ca 0.0 ro -.t:,' on 4-, on ti 4- m- u -',õ on Lao t ro - ro (ao (ao
oz, co (0 4-' CO (-0 4-' dA 4-, 4-, 4-' 4.D
(-0 , . 4-, - 4-; r (-0 4-, OA
co CO co ro ro 4-, a-, 4., CO -1-' 4-, 4-' -1-' (-0 (-0 ro , a, u 4-, OA 2 0
ro rõ,
co OD +./ t,' CO CO CO U CO CO U t: CO (0 +, OP OP .,., OP OP OP .,., u ca +.,
OP CO
03 rill rill a-, CO CO toiO U U OA CO 03 OP a-, CO CO 4-, (0 t1J3 OA CO a-, a-
, co ro a-, U
a-, CO CO CO CO CO OA CO CO CO OA a-, CO CO CO OA CO a-, U OA CO CO CO U 4-,
CO CO
149
CA 03137804 2021-10-22
WO 2020/234428 PCT/EP2020/064225
0 rn ro (.0
, ro ro u oo oo oo ho ro u 0.0 +, oto B ((:, )3 tj U CO r U CO
2 (II (a ro ro ro OA -' '' OLO -' U CO u (0 r MI r +., U 0 0 I.
0.0 on (0 (0 (.0 oz,
moo 2 (0 on u (0 0,3 (0 (ao (ao (ao 0 l5Aro 0+, Fo-ar0 44
0.0(0(000 inn U ¨ ¨ 0 ro bn CO MI ''( ) (0 OA OA +-, CO (0 rn m cO
4-, ty3 oo 4-, -' ¨ hn tto OD a, ro 0 u ,,,¨ m 0 .4¨ ro 4-, u u ro
oo ro +, ro u-' - (13 +,
CO OA U U it., 0.13 Z.:w (73 (13 Zj
co ro Op .1-, ro 0 (0 CO (0 _ 4_, U u , ,
OA
+-,(00.0(0 -Ot: 1 ( O.O.O.00.0Mr w3 4-'c'0' 3 ?.., (-L0) ti
(.0 , (.0 (4 u , 0 (.0 4., 03 0,0 0,0 CO .it', CO 4-, (.0 ,,,, (.0 co
(0 (0 ro (0 (0 (0 0.0 (-0 a. 0 (0 (0_ r 0 tE; ti ro 'µ,' 4-, 0 ro oo oo oo u
ro co
co U MI - La0 OD OD (5.0 (0 U " (0 (5,0 U (0 (5.0 U (0 (0
13;3 ((.11.), 03 co 4(:), 4(:), T.0 ro ro cc, 44 4(2( 0 (.0 (.0 (00 0.0
ro (.0 (.0 0 ro 0 OP
U OP 0 tE; U U U U CO CO t-.,' to +-, (a op (0 4-, (0 CO OP 4t,' 83 OP (0 U 0
CO
.1-; 03 CID oz, 4-, (13 4-, r0 P OA O CO
4-, CO U 03 u 4-, if U u oyo ozi 03 .!..! U 0D co
CD m to U co CO
CO U co co 4-, tto ttO , (0 OA 4_, OA .1-; rn _ 4_, -r,r, co .1-; 0
4-, = "' 0,13r CO 0 CO (10(0 +-,.F., tnr0 (0
U U 0.00,13(0 CO U '',....4-' f, -r, +-, ,
ro sc, op ro ro (-0 4-, oo -u ro u 4, op 1-', U 4-' -' oyo +-' co +-, " CO
CO " 4-, ro u m ro ro .--,:, u ro (13 4-, `-' Op CO OA 03 OA op CO tj OP co
oz, 4-' U
U%-",U-to-0003opc0 +.,Opouto ODopOP CO4-, OA
4-,(000.0+.,( C0+.,C0t0LWrOrOopCOOPrOUouUroto(a U
CO . CO m r OD 4-' 4-' U OD (0 r CO 4=- (0 r "..: (-13 CO .2 U CO p.09
hriCd3 OD
t) t.,' (0 '' OD U MI OP -' MI OA CO (0 U CO (13 '' r CO OP 0 U '' (0
L. co OA -r.r, U .,(a 4_,(13 tj 4-= 4-' 2., U 2 - ro
t,,o ro 4_, , ro 0.0 ro ¨ ro 4-, = ,-, 0.0 4_, , 0,3 ¨ 4-, õ., .1_=
.1-; l:113 U ,W ro
4t, 1-,. op 4t1 ri3 2 ro tri (13 0 1-', ri3 4-, (13 C1:1 +11 .1., 4-, OD CO U
U 76 di3 CO U(F0) 2 -(,-,-3, co OP (F0) 4-, op OP 4-, 0 -6' 03 CD
2 ao c'ci ro -'((:0' (.0 ti, 0 ro a , as Bo 3 2 (y, w3 (.0 2 ;0.0
murorOm
ro so U u C1:1 (.5 t' 4(3 C1:1 T.E; (V .(i2, 44:: 03 (13 (%/3 4-, TIE; 4-' U
C1:1 1553' u ro a 4-, 1.T3'
U CO MI (a 2 (:(.0 (:(.0 -' -813 CO OA OP (0 U (V co 2 ao 3 g2 cd:' (:(.0 Bo -
(;E; .0 (V (.0
(o:d3 0.0 mr Bo (.0 co -(0' ac9 4-' co -(t:; u -(0' t' u (0 (.0 s.,,o ro .0
(To (0 co (0
u 4-, 0 2 -(.6' (.0 .2 (4 (0 "(0 3 ro+,N32.2+,,,,r0 ro (0, u,
0.0^ 0.0 0.0 4-, ,,,, 0.0 (0 (0 (0 (0 -,,õ _51,0 .L,OO tj 8.0" 0 2 SG (-V
L'; 4r2' 4(2 (0 C(E)1 1-.,)
.1-, U OD (13 , C1:1 4_, (0j0
ro r0 rn ro +, -r, 4-' 0 (0 (0 .L'_I -63 '' CO
ro 0 .-t-', OP OA co to OA CO OA OP OA
U CO 03 ro OP (.0 1.T3' 0 CO CO oyo op (.0 2 ,(:),.,
(0 , -!-,, oz, (.0 õ,:, (0 2 (0 .i.,:,, ti
2 -,,,, (.0 (0 (0 (.0 ,,,, (.0 t 2
(u40.2sz-i2(04-,-.t:: oti_4-,(0
CO OP r0 t, 4_, (-0 -r.,, 0 (6,3, ,,, 0 (.0 0 (.0 .0 (.0 CO 0
ro ro 4(2, co u 2 (.0 ro (.0 (.0 (4 4_, (.0
(13 CO
rOCOOLOCOUM.2 4-'-'+.,d.00ON4-'+-'(-"130.0,,MIZ (-000mU.F.,-rnUt,'
CO CO OPOPoprOor00.0000h¨n4-'0OMO.0`"COOprOiirOii
(.0 4-, ro 4-, , 4-, ¨ co ro 0 0 .0 OA _
4-, u +-,OD
CO U +-, 4-' r CO OD OD OD 4-' (0 ". u 3 tn iii3) I CO ti
(.0(aci +.,4-' (.0(. .(,..i ro 3 rou u ro ro oo e., õas:, ,
,O.0 ro rn to ,,,,O.0 t:,' 1-2, +., cr 0D mCO õ.0 0
(0 (.0 (ao co (0 co co tto 013 (0 .1-; ...A., -.. U co ro -..., 0 3 (4 (.0
4_,
a. (0 u (.0 (0 u ri3 (0 CIA CO MI co (0 ..r.: -' 4-, U-',+./.,,..0
(0 W3 OD 0 -' CO U r '' (0 MI ro MI CO
.:,) '' OD `5 '= 0 I. 'thj.' 0 4-' .F., O .0 . = 3
03(-0,,,,r003(.0roro+,ro.0ro.,,(00(04-,,u 4-
,ror000,+, = co
OD (5.1:1 (11:1 COM 0,0 (0 CO t,'COCCI(5,000tt or 03 (0 03 !TRIMS U fl, 4-,
OA co
(0 U OA co ro .1-; ,T, n5 .1-; CO OP to
ro OP 4-'._ ,., U OA ro
co (0 CO U r CO sCj '' " +., CO CO .A.' (0 U
(13 .1-; OP 4-, ¨4-, CO OP -' CO CO
CO
MI 4-' ro CO r0 U Z.,. r0 (0 4-' r0 -u OD OD MI u OD ,(.7?r, u '' op MI u
4-1 +, a-, ro ro op 1:,, 03 U 4-1 U CO Op u ,õ--4 ro op , 0 .0 u oz, OA
CD
CD +., +., op +-, ro _ 0 .,, 0 (13
.t, ro ro u op -F0 U +, OD (13 U C1:1 op ' . CO
op U .1-, co OD (13 "-'.,., 4-, OA 0 co (0 4., 0 ro ro OP .1.., ro 4-' 4-, ro
oo 0 ro (13 OA
03 Op to , op 0,0 um co Op +., 0j0 u co +, u rn 4_, .,., 4-, ro ro ao +, 4-, 0
OA op
op op oyo 4., OP 4..1 OD (-0 4_, 4-' 4., OD .1-; OD (.0 CO u a-a (1:1
4-' ^ ro CO 4-, (-0 u ro ro ro u 0j3 (13 a-, CO OP 4-, op op co 4-' U 4-, (-0
CO (,.,-. 4-, (13
, OA 4-, U co u OA -I-' co .it', op TIE; .it', CO 03 4.,
(13 ro u 0.0 OA OA s w ro u
(:113 CO oyo OP 44f 44 co u 03 _ 4_, u CO `,.. 0j0 4-, 00 U U (15
0.0 MI CO 4-' +., U +.; CO +., U 0 "m 4-2, C 0 MI, , MI q p t U .,(11 Z 2.,
, Op ro OD .1-; OD 4-, ,T, 4-, (.., .1_= ¨ ¨ ro =-= to' 4_, 0,0 u 4_, 4-,
U C1:1 ri3 CIA (513 U C1:1 +11 C1:1 .;:r (0 (0 OD ro U õ., (13 4., (13 co CO
CO op to 03 0 = 0
OA 4-' op U 4-' OP OP ro ro s- 4_, ro 0 u u "¨ u (.0 4_, 0j0 (:(o ro
CO (a .1-; ro ro ro ro 0D 4-,, ro ¨ 4_, (.0 ro õ., 4_, co (.0
co OP co co CO44=,' ro ttO 0.0 ¨, ro OP (13 ro u 4-, .iL' ro -,-, u (13 (13
W3
CO (0 r r CO Z:r ro r rt:' r 2 -' 4-= Cd3 U +-' (a 4-' 4- U rnM 11(7)n r
(.7 CO
OP 03 op ro U op (o 0,0 +., CO CO CO +., .t, U CO +., +.1 4-, +., t' U ¨ ¨ -'
a..! ,.,., 03 03 op .1-; u ,.õ( 03 OD (0 U .1-; 03 C1:1 C1:1 2 a-, (a) U
ro U CO -1--,
U
(µ5'i3 '0'5 03 .1- i B) 0 j3 ( . 0 '0'5 1 f BO (13 BO 4t.1 - = 4- (%) 0 j3 a 4
_, C1:1 4, co (i13,.0 3
ro ti-3' (6,13) (13 0 a, op 0 3 (13 (r-)0 OP OA "t1-8 r (V
w3r0+,t,'4-'0.04-,w3tiouu000.oumu
.2 co co 010 u r t co -' co r ro r u OA 4-' ro u
ro ro .0 0 +, ro u tto OP o U CO +-, tt, (.0 (0 0,0 (.0 +-, 0,0 t,' (%.0
00 ^ 0,0 t 03 U (13 (5j3 CO (13 C1:1 U 0.0 'a t:: OP -(. a a CO OP (01 CO U
co to CO u ro U 0 03 op
(0 ro Li (017'0 gi2 44:: (a 2
29, õc= o 2 0 (.0 0,0 r 0 ti õ0 -(0' w:, (0 (-0 a -(.0' ("3 c'D .0 u r u u
co ro ro 4t,' (0
¨ ,,, 4-, tIA .1-, C1:1 ¨ .1-; 4-, 0.0-rr,+.,(1:1(0MCLOCLOUCOCO
t.1 r U OLO r 4- 0 2 U (0 -' MI -2 a to w3 613 U CO -,..,r, (a0 r0 u ro m
(loopt::(.0Urp.00(.00.0(0,0,00,0S,COok7,COUCOroC06-pt)
COmr0U-mtor131.2,4-'rou'C'04-'`wr rOrOUO.04-'CO
CD co 03 u OP co op 4-, CO ._,., OP oi3 1T3' 4-, .1-; 0,0
4-, OD U ro u op CO ro 4-,
µ6, (0 u (.0 Bo co oo u ro 1,...,.. tto 00 OP ((.1),
2 0 4-, 0 1.T3' CO (0 (0 oto u ro u
(0 03 , 03 4_, 03 4t, OA co 41.1 -I-, . 2 (-0 4_, , OA OP CO 0 4-, (4 u 03 ty3
U ro
rn u 0.0 oo ro 0 ro r (0 u r op ro .1t .' (a B .(.% r op td3 CO r0 u u ro
OD
(.0 op t (0 .1..1 ro oi3 fO, CO op 2 (.0 (.0 4_, u ro 4., (.0 ,(rõ) 4., co (13
-1--' 4-, OP U CO
U CO sowp OP -6,-.0 "(0' CO Z.; 1-2, "(0' +., U Op OP 2, CO OP 0 `(.'6 S co CO
2 2 O. BO CO
-(1-3-3' to ro OP 03 oi3 U U co 4_, CO 4-, .1-,4_, 0 (.0 (0 4_, 0 j3 op 4_, co
+., +-, CO MI OA op MI to' '' (0 MI .2 tE; 0 0D ro an tE; U RI r (13 M
.I-; U n3 C1:1 U
UU
4., u CO 0 CO .1.-= U (.0 u (.0 UroM U WCOCOOD
OA dA 4-, CDu (0 0 4-1 1.T3' On (13 03 u CDCD03 CDu 1-3 4-, OA t t.,1 4-, U CD
CDop 4-, (0 (-0 U OA oz, co .1-; 4t,' 03 co 03 4., dA dA 0 4., 4- u o a-
o A U U D p U 4-, U CO
co 1-3 op co CO ri3 4t', 4-, 03 fl5 OP 0 j3 op u 0 oi3 4-1 03 OD OA OD 03 0,0
OD CD-(1.T0' 0
rornroror04-,(0Z,,ro(.0+,roo(.01-3(0b) tzl t:: U OA 2 3 (- 0 Bo
CO ri, s ' .1_, 4-' 4-' 4-,
(0 ,...., CO O CO op o co 0
ro ., CDoo rn CD ii (0., +, 0.0 013 (0 CO, 0+,
''' C1:1 O C1:1 4-, 4-, (13 (13 CO , CO .1-; d A
U d A 4-, ro o CD ro h CDao
CD 4-,
^ a-, CD CO OA CD OA CD a-, a-, CD CO CD U U CD OA tll3 CO OA tll3 CO CD
tll3 a-, a-. OA
150
CA 03137804 2021-10-22
WO 2020/234428
PCT/EP2020/064225
on w3 co an - co 2, +, an Bo 2 0 (.0 .p, 0 < 0 H u
< 0 0 0 u <
ro OA co OA 1-T; -1-' co < 0 < H H H
0.0 -04-'0Opla 03(Or C000Ur 4-'
O op MI
OA - '' OAot La 0 < 0 0 (D L9 < H
4-, (0 r on ro
ro on 1_3 3 .-,t, (0 4t, (V (0
ro CD m co rill
,õ '=..... U 0 U , õI- (-9
CD
OD OA CO OA -I-' 4-, 0 ro rm. - L7 < U H U L7 L7 H
4_, (0 (-0 tn M U .1-,
µ(.6 CO ro .1.t,' CO CIA 65 0 ro -,1.74 on ro ro no H < U < <
01:1 U U CDU CO CD C0 1-3 `6'
u
g2 < W 0 u W W H < 0 < 0
(0 ro ro 2 4-, ^ 0 OA 4-; tE; ps,-,C4 .1-, tl CO mall (-90uUL-9(-9U<U
4-, , , on rou corn .t., 2 .2 .,-, to on to, .ro. UL7Ho<<L71-uL75
(.1:'
O
(0 (10 U r
U o ro - 4-' CD H U H < (-
9 5
CD CD0,0 co ro 4-, ro co U L7 U 0 U y +., < L9 (-9 <
<UULDH 8 <
(-0 OA OD CO ro 4-, all all U ro ro 4-, all op 1(2 8 * u 0
-',..., op õ., ,-, OA co 4-,
^ +-' ro ,,, 0 ,,, - U Q
H < U < W W
2 2 2 .,r 0 'r 0 r ou
.t,'
flII. t4 0 ro
(-0 tton3 4(2, 076, 0.0 2 n, H (D EH < UU LDIL2
(D
(0 CO CO 2 U CO roOA +-, 4-, -
0.0 CO OA CO .F., -õ.., U r, 0 , u 44 44 .-.1 < < < (-90,õ(-90<(DE
u op ro U CDs == CO 4-, all OA (13 co a-, co
0 U C-) 0 (-9 U W U
CD co op (13 4_, 4-, CD-I-, OD 4-1 OA co HI-0 00<0
001- 0
U '
(0 0.0 ' ,-rs -
0 CO OA tf, r -',õ, u u u < HI-
< H 0 CD4-, 4-, 4-, , ,4-, CO 4-; . s'
U 4-' (0 4-' ro (13 U M U
4-, ''' co ro tto ro on 0 H
- 4= 4- < U U
4-, M 4-, U 4-' 4-; u CD0 tto < co 0 0 U U 0 OH <- 0 4; -; (13
0.13 on 0 ttO ro U (13
CO CO U (0 CO CD4-, CO 0 (-0 on 4-, CDCO
OD OD (0 OD -' U U M 4-, U 4- +-'rn OD M U 0 u U U U (-9 - U
ro .1-, M co 4t, (00 OD all a-, On ro ,.õ.,--õH U L7 U u < (-7 I-
- 4-; M on M M CO 0 CO CO (0
0U 0 < H W
,_ - U .1= (-0 -, 4-; 4-'õ
O
o (0 pow ro 4-, 4_, t u on ro 4-, 0
-' Op ro L -' ro on u 4= +-, CO -' +-, m
(.0 s ''',,.. (D U
00000001-
OA 4_, .1= CO i.-, .1-, ,-,.. 0 op 4-, (.0 co tu 0
co 0 0 m co j0 .' 0 ro 0
ro OD (.0 +-, ro OD 0 4-, OA
O (%0 2 õc, -(iT,; 4(3 12 T ,,:, if (a tiej a op 1-0, 2 H t)UL7õ<01-
UH<Uuu ,1-
<U`"Uerl-
ro 4-, ro 4-, ro ro o 4-, co 4-, 4_, 4_, 0 (0
H U 0 , r, , ,.., H u - u -.
0 j0 ro 0 0 all U ro 0 0 0 dA U ro
MI tiO MI
(13 4-, 4-, 4-; 4_,
L9 0 U U U L9
U 4-, U (-0 .F., (0 (0 ,47',., (0 M
4-; 4-, co co U 0 4-, I-! C0 tto U
u ono roror,ro70$194-,(04-,0 ro f ri Uf)I-
L9QHQU U
ro on u u
=-.., < --- (.5 H , .-,
(0 (0 U RI OD p , < 5 u 0 H u H u < H
mu 42 2 2 0 0 t i , 2 2 - u
u < u < (.9 '9
0
o < u 0 u
0 < 0 H <
1.,) (.0 _(-0 u tE;
4 - ., (13 ro
CO
-(0'CCIa0O.t,'.t:COCO(.000rod.0 Ur0 < ILD(D<L7L9uUUHU<
oHU(D(DUu(D 0. U an4-,+-' 2 0 (0
_ _ . ,
u u u u , < c,--:, < =-= ,, <
ro (13 (13 ro U all
U , U ro H
((7, )3 :,ti CO tf, = U U 4- ' U (10 8 (0 0 u u (49 0 u u < H
CD< op dA 4-, CO
RI r
CD ro ro CD
4^ F ' co co co co OA 4-, L. 4-,
u (.0 4_, co (0 u t.,-?,
ro ro u , 0 u u ro t ro 0 U <
2 4- U U U < (11 U 0 < U U
3
(.0 , - (.0
ro ro ro U u on OA 0 <000 Lc:c)Q0QL<_S(D<
Qui 4-' U rj U (13 .1-, CO 4-'
CO 4-, ro 0 j0 CO co u co CIII U 4-'
(:Lo 0 0 -.,,, ,L0
4-; (0 (.0
ro 0 ro U < < L9 "L9 U < <
OD M CO W3 OD (113 +., 2 co op Bo
4t, .1f, 44:: ro
õ, -Id 44 .f2 r(.1k8r,.õ,(5,(2u0(.1
(0 .0 u CDu tt0_,_ õ1:10 u ro ttO u 1::: co 4-, 4_,
U ro ro u ro tal., .i...-, ro op C0 4.., 4-, ro (13 D -1-' < 0 < < < H 0
U H < U
U U CO O 4_, (0 u 4-
0 j0 ; (0 U M 4_, u CD
< I- < L7 < ( D < ( D U ( D <
CO U U CO 013 CO .1-, .1-, CO U CO M a-, U CO
CU
rq u
.(0 w
.+7 u
,1-
0_ ( = 0 0_ 0
..._... 4_,
-0 0
013 (11
U CO SD- CO
00
,-I
151
ACAGTAAAGATTCAGGAATGGAATACATTTCAGGCAAACAGAGATACCGCGATTCACTGACGTCAGCCTCCTGCGGTAT
GAAA
0
CGCCCGAAAAGAATGCTTGTTACCGGATATTGTTGTCGGAGATGTAAAGGCCTTGCACTGTCAAGAACATCGCGGCGTC
TGTC w
o
w
TCAGGAAGTCACCGAGCGTTTTTATGTGTGCACGGATCCGGGCTGTGGTCTGGTGTTTAAAACGCTTCAGACCATCAAC
CGCTT
i-J
CATTGTCCGCCCGGTCACGCCGGACGAACTGGCAGAACGCCTGCATGAAAAACAGGAACTGCCGCCAGTACGGTTAAAA
ACA
4,.
w
CAATCATATTCGCTGCGTCTGGAATGAGGGCTGCCGGTTAACACCGGCCGTCGCCGCACACCGTATTTTTATTCTTCAG
CATGA oe
TGAGAAAGAGATAACGATGGAAAGCACAGCCTTACAGCAGGCCTTTGACACCTGTCAGAATAACAAAGCAGCATGGCTG
CAA
CGCAAAAATGAGCTGGCAGCGGCCGAACAGGAATATCTGCGGCTTCTGTCAGGAGAAGGCAGAAACGTCAGTCGCCTGG
ACG
AATTACGCAATATTATCGAAGTCAGAAAATGGCAGGTGAATCAGGCCGCCGGTCGTTATATTCGTTCGCATGAAGCCGT
TCAG
CACATCAGCATCCGCGACCGGCTGAATGATTTTATGCAGCAGCACGGCACAGCACTGGCGGCCGCACTGGCACCGGAGC
TGA
TGGGCTACAGTGAGCTGACGGCCATTGCCCGAAACTGTGCCATACAGCGTGCCACAGATGCCCTGCGTGAAGCCCTTCT
GTCC
TGGCTTGCGAAGGGTGAAAAAATTAATTATTCCGCACAGGATAGCGACATTTTAACGACCATCGGATTCAGGCCTGACG
TGGC
P
TTCGGTGGATGACAGCCGTGAAAAATTCACCCCTGCGCAGAACATGATTTTTTCGCGTAAAAGTGCGCAACTGGCATCA
CGTC .
,
AGTCAGTGTAAAATTCCCCGAAAATCCGCCCGTTTTTACTGAAAAAAGCCATGCATCGATAAGGTGCATGGCTTTGCAT
GCGTT
,
1¨
.3
vi
TTCCTGCCTCATTTTCTGCAAACCGCGCCATTCCCGGCGCGGTCTGAGCGTGTCAGTGCAACTGCATTAAAACCGCCCC
GCAAA .
w
GCGGGCGGGCGAGGCGGGGAAAGCACCGCGCGCAAACCGACAAGTTAGTTAATTATTTGTGTAGTCAAAGTGCCTTCAG
TAC "
,
,
,
ATACCTCGTTAATACATTGGAGCATAATGAAGAAAATCTATGGCCTATGGTCCAAAACTGTCTTTTTTGATGGCACTAT
CCTGAA 0
,
N,
N,
AAATATGCAAAAAATAGATTGATGTAAGGTGGTTCTTGTCAGTGTCGCAAGATC
19 Native P2 (acc nr:
GGCGAGGCGGGGAAAGCACTGCGCGCTGACGGTGGTGCTGATTGTATTTTTTCAGCGTCTCAGCGCGTCGTGACGGCAC
TTA
NC_001895)
GTCTGCCCGTTGAGGCGTTGTGTGTCTGCGGGGTGTTTTGTGCGGTGGTGAGCGTGTGAGGGGGGATGACGGGGTGTAA
AA
AAGCCGCCCGCAGGCGGCGATGTTCAGTCGTTGTCAGTGTCCAGTGAGTAGTTTTTAAAGCGGATGACCTCCTGACCGA
GCCA
GCCGTTTATCTCGCGGATCCTGTCCTGTAACGGGATAAGCTCATTGCGGACAAAGACCTTTGCCACTTTCTCAATATCA
CCCAGC
1-d
GACCCGACGTTCTCCGGCTTGCCACCCATCAACTGAAAGGGGATGCGGTGCGCGTCCAGCAGGTCAGCGGCGCTGGCTT
TTTT n
1-3
GATATTAAAAAAATCGTCCTTCGTCGCCACTTCACTGAGGGGGATAATTTTAATGCCGTCGGCTTTCCCCTGTGGGGCA
TAGAG t=1
1-d
AAACAGGTTTTTAAAGTTGTTGCGGCCTTTCGACTTGACCATGTTTTCGCGAAGCATTTCGATATCGTTGCGATCCTGC
ACGGCA w
o
w
TCGGTGACATACATGATGTATCCGGCATGTGCGCCATTTTCGTAATACTTGCGGCGGAACAACGTGGCCGACTCATTCA
GCCAG =
'a
GCAGAGTTAAGGGCGCTGAGATATTCCGGCAGGCCGTACAGCTCCTGATTAATATCCGGCTCCAGCAGGTGAAACACGG
AGC o
4,.
w
w
vi
CGGGCGCGAAGGCTGTCGGCTCGTTGAAGGACGGCACCCACCAGTAAACATCCTCTTCCACGCCACGGCGGGTATATTT
TGCC
0
GGTGAGGTTTCCAGTCTGATGACCTTACCGGTGGTGCTGTAACGCTTTTCCAGAAACGCATTACCGAACACCAGAAAAT
CCAGC n.)
o
n.)
ACAAAGCGGCTGAAATCCTGCTGGGAAAGCCATGGATGCGGGATAAATGTCGAGGCCAGAATATTGCGTTTGACGTAAA
TCG
i-J
GCGAGCTGTGATGCACGGCAGCCCGCAGGCTTTTTGCCAGACCGGTAAAGCTGACCGGTGGCTCATACCATCTGCCGTT
ACTG .6.
.6.
n.)
ATGCACTCGACGTAATCCAGAATGTCACGGCGGTCGAGTACCGGCACCGGCTCACCAAAGGTGAATGCCTCCATTTTCG
GGCC oe
GCTGGCGGTCATTGTTTTTGCCGCAGGTTGCGGTGTTTTCCCTTTTTTCTTGCTCATCAGTAAAACTCCAGAATGGTGG
ATGTCA
GCGGGGTGCTGATACCGGCGGTGAGTGGCTCATTTAACAGGGCGTGCATGGTCGCCCAGGCGAGGTCGGCGTGGCTGGC
TTC
CTCGCTGCGGCTGGCCTCATAGGTGGCGCTGCGTCCGCTGCTGGTCATGGTCTTGCGGATAGCCATAAACGAGCTGGTG
ATGT
CGGTGGCGCTGACGTCGTATTCCAGACAGCCACGGCGGATAACGTCTTTTGCCTTGAGCACCATTGCGGTTTTCATTTC
CGGCG
TGTAGCGGATATCACGCGCGGCGGGATAGAACGAGCGCACGAGCTGGAACACGCCGACACCGAGGCCGGTGGCATCAAT
AC
CGATGTATTCGACGTTGTATTTTTCGGTGAGTTTGCGGATGGATTCCGCCTGGGTGGCAAAGTCCATGCCTTTCCACTG
GTGAC
P
GCTCAAGTATTCTGAATTTGCCACCGGCCACCACCGGCGGTGCCAGTACCACGCATCCGGCGCTGTCGCCACGGTGTGA
CGGG
2
TCGTAACCAATCCATACCGGGCGGGAGCCGAACGGATTGGCGGCAAACGGCGCATAGTCTTCCCATTCTTCCAGCGTGT
CGAC
un
CATGCAGCGTTGCAGCTCCTCGAACGGGAACACCGATGCCTTGTCGTCAACAAATTCACACATGAACAGGTTTTTAAAA
TCGTC 2
.3
GGCGCTGTTTTCGCGTTTGAGCTGCTCAATGTCGAACAGCGTGCAGCCGCCTTTCAGGGCGTCCTCAATGGTGACAATC
TGTCG 2
1-
CCACTGGCCGTCCGCACAGAGAAGCCCACCGGCAAGGGCGTTATGACTGACGTCGATTTCCACGCGTTCGGCGGCGCTG
GCG
.31
.3
CGTCCCCGGTTAAACAGTTCACCCGACCAGAACGGGTAGGCGTCGTGCGCCAGCGTGGACGGGGTGGAGAAATAGGTCG
AG
CGCAGGTGACTCTGTGAGGCCATACCTGATGCCACCTTACGCAGTACCTGAAAATTCGGGATCCAGAAAATCTCATCGA
CGTA
CAGGTCGCCGTTATGACTCTGCGCGGTGTTGGAGTTGGTGCCGAGAAAAATCAGTTTTGCGCCGTTATTGCCCAGGACA
ATCG
GGTCACCGGTCAGGTCAACGTCAACCAGACGGGCAAAGGCGATGATGTATTCGCGGAACACATACGCCTGTGTTTTACT
GGCC
GACAGAAAAATCTGGTTATGACCGGTTTTCAGGGCACGCAGCAGCGCCTCGCGGGAAAAATAAAACGTCGCGCCAATCT
GGC
GGGATTTCAGGATATCGCGGATGCGGTGCTCAAGCCCGGCACGATACCAGTGCAGCTGATAGTCGAAAGACTGCTCAAA
GAA
IV
AATCTGCTCCAGCTTTTCGATGGCCTCGTCACTGAAAAAATTCTTTTTCGGTTTGCGCCGTCCGCCTTTGTTACGGTTA
GCGACG n
1-3
TTCGGATTAAGGTCTGCCTCGTTGCCGGTCTGGCTGTAGCGGTTGACCCGTGCCAGTCGTTCAATCTGGCGTCCCAGCA
GGTCA t=1
tl
ATTTCCTTGAAGTCACCGCCGGTTTTCTGTGGTTTGATGATGAGCTGGGTCAGCCGCGCTTCCAGACTCATTTCGACAC
GGCTG 2
o
ATGGGGGCAACGCTGTCCCAGCCGTCGCGCTGTTTCCAGCTCTGCACTGTCGGGCGTTTCATCTGCAACATGGCGGCAA
TCTGC 'a
o
GGCACGGAAAACCCCTGCCAGTACAGCAGCGCCGCCTGACGACGCGGGTCGTGTAAAAGAGTGGTGTCTGTGGTGATGG
TCA tt
n.)
un
TGAATACCTCGCCGTGATGAATACACGGCAAGGCTACTGAGTCGCGCCCCGCGATTCGCTAAGGTGCTGTTGTGTCAGT
GATA
0
AGCCATCCGGGACTGATGGCGGAGGATGCGCATCGTCGGGAAACTGATGCCGACATGTGACTCCTCTAATCACTATTCA
GGAC n.)
o
n.)
TCCTGACAATGGCAAAAAAAGTCTCAAAATTCTTTCGTATCGGCGTTGAGGGTGACACCTGTGACGGGCGTGTCATCAG
TGCG
i-J
CAGGATATTCAGGAAATGGCCGAAACCTTTGACCCGCGTGTCTATGGTTGCCGCATTAACCTGGAACATCTGCGCGGCA
TCCT .6.
.6.
n.)
GCCTGACGGTATTTTTAAGCGTTATGGCGATGTGGCCGAACTGAAGGCCGAAAAGATTGACGATGATTCGGCGCTGAAA
GGC oe
AAATGGGCGCTGTTTGCGAAAATCACCCCGACCGATGACCTTATCGCGATGAACAAGGCCGCGCAGAAGGTCTACACCT
CAAT
GGAAATTCAGCCGAACTTTGCCAACACCGGCAAATGTTATCTGGTGGGTCTGGCCGTCACCGATGACCCGGCAAGCCTC
GGCA
CGGAATACCTGGAATTCTGCCGCACGGCAAAACACAACCCCCTGAACCGCTTCAAATTAAGCCCTGAAAACCTGATTTC
AGTGG
CAACGCCTGTTGAGCTGGAATTTGAAGACCTGCCTGAAACCGTGTTCACCGCCCTGACCGAAAAGGTGAAGTCCATTTT
TGGCC
GCAAACAGGCCAGCGATGATGCCCGTCTGAATGACGTGCATGAAGCGGTGACCGCTGTTGCTGAACATGTGCAGGAAAA
ACT
GAGCGCCACTGAGCAGCGCCTCGCTGAGATGGAAACCGCCTTTTCTGCACTTAAGCAGGAGGTGACTGACAGGGCGGAT
GAA
P
ACCAGCCAGGCATTCACCCGCCTGAAAAACAGTCTCGACCACACCGAAAGTCTGACCCAGCAGCGCCGCAGCAAAGCCA
CCGG
2
CGGTGGCGGTGACGCCCTGATGACGAACTGCTGACCGGCGTCAGTCAGTCCGGGAAAACCTTCACGATTAACCCTTAAT
TTCA
un
GGAAAAACTATGCGCCAGGAAACCCGCTTTAAATTTAATGCCTACCTGTCCCGTGTTGCCGAACTGAACGGCATCGACG
CCGG 2
.6.
.3
TGATGTGTCGAAAAAATTCACCGTTGAACCGTCGGTCACCCAGACCCTGATGAACACCATGCAGGAGTCCTCTGACTTT
CTGAC 2
1-
CCGCATCAATATTGTGCCGGTCAGCGAAATGAAAGGGGAAAAAATTGGTATCGGTGTCACCGGCTCCATCGCCAGCACT
ACCG
.31
.3
ACACTGCCGGTGGTACCGAGCGTCAGCCGAAGGACTTCTCGAAGCTGGCGTCAAACAAGTACGAATGCGACCAGATTAA
CTTC
GATTTTTATATCCGCTACAAAACGCTGGACCTGTGGGCGCGTTATCAGGATTTCCAGCTCCGTATCCGTAACGCCATTA
TCAAAC
GCCAGTCCCTTGATTTCATCATGGCCGGTTTTAACGGCGTGAAGCGTGCCGAAACCTCTGACCGCAGCAGCAATCCGAT
GTTGC
AGGATGTGGCGGTCGGCTGGCTGCAGAAATACCGCAATGAAGCACCGGCGCGCGTGATGAGCAAGGTCACTGACGAGGA
AG
GCCGCACCACCTCTGAGGTTATCCGCGTGGGTAAGGGCGGTGATTATGCCAGCCTTGATGCACTGGTGATGGATGCGAC
CAAC
AACCTGATTGAACCGTGGTATCAGGAAGACCCTGACCTTGTGGTGATTGTGGGGCGTCAGCTACTGGCGGACAAGTATT
TCCC
IV
CATCGTCAACAAGGAGCAGGACAACAGCGAAATGCTGGCCGCTGACGTCATCATCAGCCAGAAACGCATCGGTAACCTA
CCA n
1-3
GCGGTACGCGTCCCGTACTTCCCGGCGGATGCGATGCTCATCACGAAGCTGGAAAACCTGTCCATCTACTACATGGATG
ACAG t=1
tl
CCATCGCCGCGTGATTGAGGAAAACCCGAAACTCGACCGCGTGGAGAACTACGAGTCAATGAACATTGATTACGTGGTG
GAA 2
o
GACTACGCCGCCGGTTGTCTGGTGGAAAAAATCAAGGTCGGTGACTTCTCCACACCGGCTAAGGCGACCGCAGAGCCGG
GAG 'a
o
CGTAACCGATGACGAGTCCCGCACAGCGCCACATGATGCGGGTCTCGGCAGCGATGACCGCGCAGCGGGAAGCCGCCCC
GCT tt
n.)
un
GCGACATGCAACTGTCTATGAGCAGATGCTGGTTAAGCTCGCCGCAGACCAGCGCACACTGAAAGCGATTTACTCAAAA
GAGC
0
TGAAGGCCGCAAAAAAACGCGAACTGCTGCCGTTCTGGTTGCCGTGGGTGAACGGCGTGCTGGAGCTGGGCAAAGGTGC
AC n.)
o
n.)
AGGATGACATTCTGATGACGGTCATGCTGTGGCGTCTGGATACCGGCGATATTGCCGGTGCGCTGGAGATTGCCCGTTA
TGCC
i-J
CTGAAGTACGGTCTGACCATGCCGGGTAAACACCGCCGTACCCCGCCGTACATGTTCACCGAGGAGGTAGCGCTTGCGG
CCAT .6.
.6.
n.)
GCGCGCTCACGCTGCCGGTGAGTCTGTGGATACCCGCCTGCTGACGGAGACCCTTGAACTGACCGCCACGGCTGACATG
CCTG oe
ATGAAGTGCGCGCAAAGCTGCACAAAATCACCGGTCTGTTTCTGCGTGACGGTGGTGATGCCGCCGGTGCGCTGGCGCA
CCT
GCAACGTGCGACACAGCTCGACTGTCAGGCAGGCGTCAAAAAAGAGATTGAACGACTGGAGCGGGAGCTGAAACCGAAG
CC
GGAGCCGCAGCCCAAAGCGGCCACCCGCGCCCCGCGTAAGACCCGGAGCGTGACACCGGCAAAACGTGGACGCCCGAAA
AA
GAAAGCCAGTTAACAACCGAATGCGCCCCGCGCCAGGGCGGCACGCCGGTCAGTGACGGTGAATCACCTGACACTGCAC
CGG
CGTCCACCGCCCGACTTTTCAGAGGTAGTCATGATGACGCTGATTATTCCGCGAAAGGAGGCTCCCGTGTCCGGTGAGG
GTAC
GGTGGTCATCCCGCAACCGGCAGGCGACGAGCCGGTGATTAAAAACACGTTCTTTTTTCCCGATATCGACCCGAAGCGC
GTCC
P
GGGAACGTATGCGCCTTGAGCAGACCGTCGCCCCCGCCCGTCTGCGTGAGGCCATCAAGTCAGGCATGGCTGAAACGAA
TGC
2
GGAGCTGTACGAGTACCGCGAACAGAAAATTGCCGCCGGTTTTACGCGTCTGGCTGACGTCCCGGCGGACGATATCGAC
GGT
un
GAAAGCATCAAGGTTTTTTACTACGAGCGCGCCGTGTGTGCGATGGCGACCGCGTCGCTTTATGAGCGTTATCGCGGTG
TGGA 2
un
.3
TGCCAGTGCGAAAGGCGACAAGAAGGCTGACAGCATTGACAGCACCATTGATGAGCTGTGGCGGGATATGCGCTGGGCG
GT 2
1-
GGCGCGCATCCAGGGCAAGCCGCGCTGCATCGTGAGTCAAATCTGATGAAGACCTTTGCGCTACAGGGCGACACGCTCG
ACG
.31
.3
CCATTTGTGTCCGCTATTACGGGCGCACTGAGGGCGTGGTTGAGACCGTGCTCGCCGCAAATCCGGGACTGGCTGAACT
GGG
GGCGGTGCTGCCACACGGCACCGCCGTCGAACTGCCCGACGTTCAGACCGCGCCCGTGGCTGAAACTGTCAATCTGTGG
GAG
TAACGCATGACAGCAGAAGAAAAAAGCGTCCTGTCGCTTTTCATGATTGGGGTGCTGATTGTTGTCGGCAAGGTGCTTG
CCGG
TGGTGAACCTATCACCCCGCGTCTGTTTATCGGGCGCATGTTGCTCGGTGGTTTTGTCTCGATGGTTGCCGGTGTTGTT
CTGGT
GCAGTTTCCTGACCTGTCACTGCCAGCGGTGTGCGGCATCGGCTCCATGCTGGGTATCGCCGGTTATCAGGTGATTGAG
ATTG
CCATTCAGCGCCGCTTTAAGGGCAGGGGGAAACAGTAATGCCGGTAATTAACACGCATCAGAATATCGCCGCCTTTCTC
GACA
IV
TGCTGGCCGTGTCCGAAGGGACGGCGAATCATCCACTGACGAAAAACCGGGGCTATGACGTGATAGTCACCGGACTGGA
CGG n
1-3
GAAGCCGGAAATTTTCACCGACTACAGTGACCACCCGTTCGCACATGGCCGACCGGCGAAGGTGTTTAACCGTCGCGGT
GAAA t=1
tl
AATCCACGGCCTCCGGTCGCTATCAGCAGCTTTACCTGTTCTGGCCGCATTACCGCAAACAGCTTGCCCTGCCGGATTT
CAGTCC 2
o
GTTGTCACAGGACAGACTCGCCATTCAGTTGATCCGCGAACGCGGAGCACTGGATGACATCCGGGCGGGACGCATTGAG
CGC 'a
o
GCCATTTCACGCTGTCGCAATATCTGGGCGTCCCTGCCGGGTGCCGGTTACGGTCAGCGTGAGCATTCACTGGAAAAAC
TGGT tt
n.)
un
CACCGTCTGGCGTACCGCTGGCGGCGTACCGGCTTAAACGGAGTAAATACCATGAAGAAATTATCCCTTTCACTGATGC
TGAA
0
CGTGTCGCTGGCGCTGATGCTGGCACTGTCCCTGATTTACCCGCAGAGCGTGGCCGTCAATTTTGTCGCTGCCTGGGCG
ATTCT n.)
o
n.)
GGCGACGGTTATCTGTGTGGTTGCCGGTGGTGTGGGCGTGTATGCCACTGAGTATGTGCTGGAACGCTACGGGCGGGAG
CTG
i-J
CCGCCGGAATCGCTGGCCGTGAAGATTGTCACGTCGCTGTTTTTGCAGCCGGTGCCGTGGCGCAGACGGGCGGCGGCTC
TGG .6.
.6.
n.)
TAGTGGTGGTGGCGACGTTTATCTCGCTGGTCGCTGCCGGGTGGATTTTTACCGCGCTGATTTATCTTGTGGTGTCGCT
GTTTTT oe
CCGGCTGATACGTAAAGCCTGTCGTCAGCGTCTTGAGGGGCGGGAACCATGTCAAGGCTGATGATTGTGCTGGTCGTGT
TGTT
ATCGCTGGCGGTGGCCGGTCTGTTTCTGGTGAAACACAAAAATGCCAGCCTGCGCGCCTCGCTGGACAGGGCGAACAAC
GTC
GCCAGCGGTCAGCAGACGACCATCACCATGCTGAAAAATCAGCTTCATGTTGCGCTCACCAGGGCAGATAAAAACGAGC
TGGC
GCAGGTGGCACTGCGTCAGGAACTGGAGAACGCCGCGAAACGTGAAGCACAGCGCGAGAAAACCATCACGAGGTTACTT
AAT
GAGAACGAAGATTTTCGCCGCTGGTACGGTGCTGACCTGCCTGATGCTGTGCGCCGGTTGCACCAGCGCCCCGCCTGCA
CCGA
CGCCAGTGATTGTCCCCAACGCATGCCCGAAAGTGAGCCTTTGCCCGATGCCGGGCAGTGACCCGCAGACGAACGGCGA
TTTA
P
AGTGCCGATATCCGGCAGCTTGAGAACGCGCTGGCACGCTGTGCCAGCCAGGTAAAAATGATTAAACACTGTCAGGACG
AAA
2
ACGATGCTCAAACCCGACAGCCTGCGCAGGGCGCTGACTGATGCCGTCACGGTGCTGAAAACTAACCCCGATATGCTGC
GGAT
un
ATTCGTGGATAACGGGAGTATTGCCTCCACACTGGCGGCGTCGCTGTCATTCGAAAAGCGTTACACGCTCAATGTGATT
GTGA 2
o .3
CCGACTTTACCGGTGATTTTGACCTGCTCATTGTGCCGGTGCTGGCGTGGCTGCGGGAAAATCAGCCCGACATCATGAC
CACC 2
1-
GACGAAGGCCAGAAAAAGGGCTTCACGTTTTATGCAGACATCAACAATGACAGCAGCTTTGATATCAGTATCAGCCTGA
TGCT
.31
.3
GACCGAGCGCACGCTGGTCAGTGAGGTGGACGGCGCACTGCATGTGAAGAATATCTCGGAACCCCCGCCGCCGGAGCCG
GTC
ACCCGCCCGATGGAGCTGTATATCAATGGCGAACTGGTGAGTAAGTGGGATGAATGAGTTTAAGCGTTTTGAAGACCGG
CTG
ACCGGACTGATTGAATCGCTGTCACCGTCAGGGCGTCGGCGACTGAGTGCCGAACTGGCGAAACGTCTGCGGCAGAGTC
AGC
AGCGTCGGGTGATGGCACAGAAAGCCCCGGACGGCACACCCTACGCGCCACGCCAGCAGCAGAGCGTCAGAAAAAAGAC
CG
GTCGCGTTAAGCGAAAAATGTTTGCGAAACTTATTACCAGTCGTTTTTTGCATATCCGTGCCAGCCCGGAGCAGGCATC
AATGG
AATTTTACGGCGGGAAGTCGCCGAAAATCGCCAGTGTGCATCAGTTTGGTCTGTCGGAAGAAAACCGGAAAGACGGTAA
GAA
IV
AATTGATTATCCGGCGCGTCCCCTGCTCGGCTTTACCGGTGAGGATGTGCAGATGATTGAAGAGATTATCCTGGCTCAC
CTTGA n
1-3
GCGTTAGTTTTATCCAGGCAGAGGCTGATGCGCAATTAAACATTGAGCGGCCATGCTGGTCGCTCAATGTTTAGAGGTT
TATG t=1
tl
AGTGATTTTTATTTGATGCTTTGTATTCTAAAACCTTCTTATTGGCGTAAAAGAATTTTGTATATGACAGGAATATAAC
CAGACC 2
o
TGAAGTGAAATAGACGAGGGATAGTATTAATAATGCTTTTTTGTGACTGTTATTATCTTTAATCTCCTGGCTTAACCAT
TCGGAG 'a
o
TCCTCCTCGTTTAGCTGTAAGAGCTTATTGCAGGCGATCTCAGGAAGTGTGTCTTTTATAAATACGTTTTGCAGTCTCT
TGCAAT tt
n.)
un
CGGCAAGGCTATAAGTTTTATTAAATTCAACTGCTTTATTTTTGAAGGATAAAAGAACTTTGTCACTATAAACATAGTA
CATCAT
0
GTTTTTATATGGTATGCCTGTGGCATCCCTTACTATAACGGATTGTTCGTTGTGTATGTAACATGCGAGGAGAATGTAA
AAAAT n.)
o
n.)
ACTGGCCAGAATTACAATTATTGTTTTAATTATGTGTGGTGGTTTTGTTATGTCACCCCAGATACGAGTAAGGAAAAAA
TACGA
i-J
TGTTTTTAGTTTTCCATCAATCAGTCCCTGCTGTATCATTCTCACATTTTCAATGCCTGATACATTGATTCCGTTAATT
ATTTTAAA .6.
.6.
n.)
TAGTTGAATGTCGCGCCACTCGCGGTCAAGTCTTTTTAATTTTTTGTCTGAATATCCAAAATTGAAATAATGTGCAATA
AGCCTC oe
ATAAGGTTACTTTTACCAAAGCTAAAAAATGCTAATACTGCAAAGCTACAAAGGAAAAAAACGATTAGCCCCCACACAT
TAGTC
ACATTATAGCTGACCATTACGCTCTCCTTGAATGTTGTCTGGTAGTTCTACAAATGAATCCAGATAGCATAACTTTTAT
ATATTGT
GCAATCTCACATGCATGAACACTCTCGCAAATATTCAGGAACTCGCGCGCGCACTGCGCAACATGATTCGCACTGGCAT
TATCG
TCGAAACCGACCTTAACGCCGGTCGCTGCCGCGTGCAGACCGGCGGCATGTGCACCGACTGGCTTCAGTGGCTGACCCA
TCGC
GCAGGACGTTCGCGCACATGGTGGGCACCTTCCGTGGGGGAACAGGTGCTGATTCTGGCCGTGGGTGGTGAACTCGACA
CGG
CGTTCGTTCTGCCGGGGATTTATTCCGGCGATAACCCCTCGCCGTCTGTGTCGGCGGATGCCCTGCATATCCGTTTCCC
TGACG
P
GGGCGGTGATTGAATATGAACCCGAAACCAGTGCACTCACGGTAAGCGGAATTAAAACGGCCAGCGTGACGGCTTCCGG
TTC
2
TGTTACTGCCACGGTGCCGGTGGTCATGGTGAAAGCATCAACCCGCGTCACCCTGGACACCCCGGAGGTGGTCTGCACC
AACA
un
GGCTGATTACCGGCACGCTGGAAGTGCAGAAAGGCGGGACGATGCGCGGCAACATTGAACACACCGGCGGTGAACTCTC
ATC 2
--.1
.3
AAACGGTAAGGTACTGCATACCCATAAACACCCCGGCGACAGCGGCGGCACAACCGGGAGTCCTTTATGACAGCGCGTT
ATCT 2
1-
CGGAATGAATCGCAGTGATGGCCTGACTGTCACTGACCTTGAGCATATCAGCCAGAGTATCGGCGATATCCTGCGCACA
CCGG
.31
.3
TCGGCTCACGGGTGATGCGTCGTGATTACGGCTCGTTGCTGGCGTCAATGATTGACCAGCCGCAGACCCCGGCGCTTGA
GTTG
CAGATTAAAGTCGCCTGTTACATGGCAGTGCTGAAATGGGAACCCCGCGTCACCCTGTCATCCGTCACCACGGCGCGCA
GTTTT
GACGGGCGAATGACGGTCACGTTAACCGGCCAGCACAACGACACCGGCCAGCCACTTTCATTAACCATCCCTGTGAGTT
GAAA
CCATGCCGATTATCGACCTGAACCAGCTACCCGCACCGGATGTGGTCGAGGAGCTGGACTTTGAAAGCATTCTCGCTGA
ACGC
AAGGCGACACTGATTTCCCTTTACCCGGAAGATCAGCAGGAGGCGGTCGCCCGTACCCTGACACTGGAATCTGAGCCTC
TCGT
CAAACTGCTGGAAGAAAATGCTTATCGTGAGCTTATCTGGCGTCAGCGTGTGAATGAGGCCGCACGGGCGGTGATGCTG
GCC
IV
TGTGCCGCCGGTAATGACCTTGATGTGATTGGTGCCAATTACAACACCACGCGCCTGACTATCACCCCGGCAGATGATT
CGACC n
1-3
ATCCCGCCGACACCGGCAGTGATGGAATCTGACACCGATTATCGTCTGCGTATTCAGCAGGCTTTTGAGGGCTTAAGCG
TCGC t=1
tl
CGGGTCAGTGGGAGCCTATCAGTATCATGGTCGCAGTGCTGACGGGCGTGTCGCGGATATTTCTGTCACCAGTCCGTCT
CCGG 2
o
CCTGTGTCACCATCTCTGTGCTGTCACGTGAAAATAACGGCGTCGCATCCGAAGACCTGCTGGCTGTGGTGCGTAACGC
CCTTA 'a
o
ATGGCGAGGACGTCAGGCCGGTGGCCGACCGCGTGACCGTGCAGTCTGCCGCCATCGTTGAATACCAGATAAACGCCAC
GCT tt
n.)
un
TTACCTTTACCCTGGTCCCGAAAGCGAACCCATCCGCGCTGCCGCTGTGAAAAAGCTGGAAGCGTATATCACGGCACAG
CACC
0
GGCTGGGGCGCGACATCCGTCTGTCTGCCATTTATGCCGCTTTGCATGTGGAAGGTGTGCAGCGTGTCGAACTGGCTGC
ACCA n.)
o
n.)
CTGGCCGACATCGTGCTCAACAGTACGCAGGCGTCTTTCTGTACCGAATACCGCGTCGTGACCGGAGGCTCGGATGAGT
GATT
i-J
CGCGACTGCTGCCGACCGGCTCATCACCGCTTGAGGTCGCCGCCGCAAAAGCCTGTGCGGAAATTGAAAAAACGCCGGT
CAG .6.
.6.
n.)
TATTCGTGAACTGTGGAACCCGGACACCTGTCCGGCAAATCTGCTGCCGTGGCTGGCGTGGGCGTTTTCGGTCGACAGG
TGGG oe
ATGAAAAGTGGCCGGAAGCGACAAAACGCGCCGTTATCCGCGATGCCTATTTCATCCACTGTCATAAGGGCACGATAGG
TGCA
ATCCGGCGTGTGGTGGAGCCGCTCGGCTATCTCATCAACGTGACGGAGTGGTGGGAAAACAGTGACCCGCCCGGCACCT
TCC
GGCTTGATATTGGTGTACTGGAAAGCGGTATCACAGAGGCAATGTATCAGGAAATGGAACGGCTGATTGCTGATGCCAA
ACC
TGCAAGCCGTCACCTTATTGGCCTGAACATTACCCGGGACATTCCCGGCTATCTGTTCGCCGGTGGTGTGGCTTACGAC
GGCGA
TGTAATTACGGTTTACCCCGGATAAGTGAGGAATAATGAGCATAAAATTCAGAACCGTTATCACCACTGCCGGTGCAGC
AAAG
CTGGCAGCGGCAACCGCGCCGGGAAGGCGGAAGGTCGGCATTACCACGATGGCCGTCGGGGATGGCGGTGGTAAATTGC
CT
P
GTCCCGGATGCCGGACAGACCGGGCTTATCCATGAAGTCTGGCGACATGCGCTGAACAAAATCAGCCAGGACAAACGAA
ACA
2
GTAATTATATTATCGCCGAGCTGGTTATTCCGCCGGAGGTGGGCGGTTTCTGGATGCGTGAGCTTGGCCTGTACGATGA
TGCG
un
GGAACGTTAATTGCCGTGGCGAACATGGCCGAAAGCTATAAGCCAGCCCTTGCCGAAGGCTCAGGACGTTGGCAGACCT
GTC 2
oe
GCATGGTCATCATCGTCAGCAGTGTGGCCTCAGTGGAGCTGACCATTGACACCACAACGGTGATGGCGACGCAGGATTA
CGTT 2
1-
GATGACAAAATTGCAGAGCACGAACAGTCACGACGTCACCCGGACGCCTCGCTGACAGCAAAAGGTTTTACTCAGTTAA
GCAG
.31
.3
TGCGACCAACAGCACGTCTGAAACACTGGCCGCAACGCCGAAAGCGGTAAAGGCCGCGTATGACCTGGCTAACGGGAAA
TAT
ACCGCACAGGACGCCACCACAGCGCGAAAAGGCCTTGTCCAGCTTAGTAGCGCCACCAACAGCACGTCTGAAACGCTCG
CCGC
AACACCAAAAGCCGTTAAGACGGTAATGGATGAAACGAACAAAAAAGCGCCATTAAACAGCCCTGCACTGACCGGAACG
CCA
ACGACGCCAACTGCGCGACAGGGAACGAATAATACTCAGATCGCAAACACGGCTTTCGTTATGGCCGCGATTGCCGCCC
TTGT
AGACTCGTCGCCTGACGCACTGAATACGCTGAACGAGCTGGCGGCGGCGCTGGGCAATGACCCGAATTTTGCTACCACC
ATGA
CTAATGCGCTTGCGGGTAAGCAACCGAAAGATGCTACCCTGACGGCGCTGGCGGGGCTTGCTACTGCGGCAGACAGGTT
TCC
IV
GTATTTTACGGGGAATGATGTTGCCAGCCTGGCGACCCTGACAAAAGTCGGGCGGGATATTCTGGCTAAATCGACCGTT
GCCG n
1-3
CCGTTATCGAATATCTCGGTTTACAGGAAACGGTAAACCGAGCCGGGAACGCCGTGCAAAAAAATGGCGATACCTTGTC
CGGT t=1
tl
GGACTTACTTTTGAAAACGACTCAATCCTTGCCTGGATTCGAAATACTGACTGGGCGAAGATTGGATTTAAAAATGATG
CCGAT 2
o
GGTGACACTGATTCATACATGTGGTTTGAAACGGGGGATAACGGCAATGAATATTTCAAATGGAGAAGCCGCCAGAGTA
CCA 'a
o
CAACAAAAGACCTGATGACGTTGAAATGGGATGCACTAAATATTCTTGTTAATGCCGTCATTAATGGCTGTTTTGGAGT
TGGTA tt
n.)
un
CGACGAATGCACTAGGTGGTAGCTCTATTGTTCTTGGTGATAATGATACCGGATTTAAACAGAATGGAGACGGTATTCT
TGAT
0
GTTTATGCTAACAGTCAGCGTGTATTCCGTTTTCAGAATGGAGTGGCTATTGCTTTTAAAAATATTCAGGCAGGTGATA
GTAAA n.)
o
n.)
AAGTTCTCGCTATCCAGCTCTAATACATCCACGAAGAATATTACCTTTAATTTATGGGGTGCTTCCACCCGTCCAGTGG
TTGCAG
i-J
AGTTAGGCGATGAGGCCGGATGGCATTTCTATAGCCAGCGAAATACAGATAACTCGGTAATATTTGCTGTTAACGGTCA
GATG .6.
.6.
n.)
CAACCCAGCAACTGGGGAAATTTTGATTCCCGCTATGTGAAAGATGTTCGCCTGGGTACGCGAGTTGTTCAATTGATGG
CGCG oe
AGGTGGTCGTTATGAAAAAGCCGGACACACGATTACCGGATTAAGAATCATTGGTGAAGTAGATGGCGATGATGAAGCC
ATC
TTCAGGCCGATACAAAAATACATCAATGGCACATGGTATAACGTTGCGCAGGTGTAAGTTATGCAGCATTTAAAGAACA
TTAA
GTCAGGTAATCCAAAAACAAAAGAGCAATATCAGCTAACAAAGAATTTTGATGTTATCTGGTTATGGTCCGAAGACGGA
AAAA
ACTGGTATGAGGAAGTGAAGAACTTTCAGCCAGACACAATAAAGATTGTTTACGATGAAAATAATATTATTGTCGCTAT
CACCA
GAGATGCTTCAACGCTTAATCCTGAAGGTTTTAGCGTTGTTGAGGTTCCTGATATTACCTCCAACCGACGTGCTGACGA
CTCAG
GTAAATGGATGTTTAAGGATGGTGCTGTGGTTAAACGGATTTATACGGCAGATGAACAGCAACAACAGGCAGAATCACA
AAA
P
GGCCGCGTTACTTTCCGAAGCGGAAAACGTTATTCAGCCACTGGAACGCGCTGTCAGGCTGAATATGGCGACGGATGAG
GAA
2
CGTGCACGACTGGAGTCATGGGAACGTTACAGCGTTCTGGTCAGCCGTGTGGATCCTGCAAATCCTGAATGGCCGGAAA
TGCC
un
GCAATAAGTTGTATGACCTCTGTTGTGAACTTACATATCTATGGCACAGAGTAAAGCCTAATCTGAAGTCCACTCTGTG
CCAAA 2
o .3
AGCGGACCTTATAAACAAGAACAATGGTCATATCAGTGTGTTTTGATTGCATAGCTAACGTGCGTCTTCCTGTACAGAA
TCATA 2
1-
AGATGATAGGGCATAGGAGATGATTATTATCGCGTGTTTTAAAAATAGCTTTCTGCATCAAGTGTCACTTGCGAAGAGG
TATTG
.31
.3
GCGCTATGATTGGCATCACTCAGTTCAGATAATATTTTAAATATTTGGTAAATGTTCAGATTACACAATTCGGTCTGCG
TCCCGT
TAAAATTATCTACTATTAGTTATTACTATGAGGTGAATGGCAAGTGTTTTCCACTCGCCATTTTTGTTTTTTATTTTTT
TAAAGCCT
TTCCGACGTTTAAGGATTTTGCCAAGCCATAAAATATTTGATATTTCATTCCTGTTTTTATTTTTGGTGAGATATTTGC
GTAACTT
CTTTCTCTAAAACACCAACGTATAAACCTATTATTATCTTCGGTTGTTTCTATATAGCATATTACATTGAGACCAAAAA
GAAACTG
AAGGAAGTCATTTGCAGTTGTCATGAATTTTGGTTTTGTTGTGGAAATGGAACTAATATGAGTTTCAAGTTGTTTATAT
GCTAA
GAGGTATTCAGCGTAATCAAAAGTATCTTTTCCAGAGAGGAATTCAAAGAACTTTAAAAAAATCTCATAATGTTCACTT
CCATA
IV
ATAAAATGATAAATGATCTTTTATTTCTCCAAGTAAATAGTTAGAGTAATCTCTTTGCATGCTAGGACTTTCAAAGTCA
TCTAGA n
,-i
CTAAATTGATTTTTCCCATCATCACCGCTGGCGTGTGATTTGATAATTTCGAGCATCCGTAATATATCTCTTGGGCGAT
AATATG t=1
tl
ACCAGCGAAGGAAATTAATAAATGATGTGGGGTGGTTATAATTATCATGGACGTTGGGTGAATCCCAAGGGAAATAATG
GTC 2
o
CCATGCCTGTCCCTTTTTAAGGGCCATGTCTTGCTGAGTCCTAAGCAGATGATCTAATACCTCAAATAATTGAGAGCTA
CGGTG 'a
o
GTTAACATATTCTGTCCGCCAGTCCAAGAATACTGAATTATCTCTGATTTTTGCGTTTTGATTTTGTAATCCTATGGAT
TCAAATA tt
n.)
un
TATCTGGCCTGATCAAGAGAACGGCCTTCATTCTCCCTTTTCCTCCCTTTATACTTGGGAAGAAATCGTTGTTAATCTC
CCAGACT
0
GCGTGAGCTAATCCTTTTATACATTCTAGATAATCATCGTATGGAATTGAGGAAGGGCGGATATCAATACCATCGATAA
ATAAA n.)
o
n.)
ATATGATTTTGAGATAAACGTATTTGGCTAAAGGCCTCTTCAAATTTTTTTTGTATATAAAATAAATTTATCTGAAATC
TGCTCTC
i-J
AGAAAAAGATAAAGACTCTCTTTCTTCACCTTTTAAGGTTGCATGCTTATGAAGTAGTTCGGCTGCAACTTTTGACTCT
TGAACA .6.
.6.
n.)
AAACTGAGGGCTTGAATTATTTCTGGTGAAAATGCCTTAAGATAATACTCATCTATTGCATCATTAAGTGATGAGAATT
TTGAAT oe
AATTTAGTAACTTTGAAATTTTCCCTTCTTTATCTAATACTTGTTTTGATATTAATAAATATAGAATAACTTTCCATAT
GCCAGAGA
AGTCAGACAAATTTAAATGCTTTTCAGATTTAAGAGTGATAAATTTTTGATATTCTGTCTCTCTTATATATTTAGTTGT
AGCTAGC
GTATTGTTAATATTGTTATTTGAGAGATAGATAGAGTATGCGGTTTTACCAGTACCTTTCTCTCCTACAAGAAACGATA
TATTTG
GTTCACATAATCTAGATAGGTGATTATCACGTATGAACACTTTGTTAAGTAATTCTTTATTCTCCCTTCTCTTATAATT
TTCAGCAT
CAGCATAGCCAAGAGTTAGATCTTTGATTGGTATCATTATTACCTCAACTAAAGCAATTGAAAAACATATGGTTATAAA
AACAT
AGCGCTGCTCTGTCGCATATTGTGGATATCAACATATGAAATGTAAAGGAACTATTTGTGGATGAATGAACCGAATCGC
TGTA
P
GCTTCGGTAGTCATCTTTCAGACTTGTATAAATGAACAACTTCCGCTTCTCGCTCAAAGGAAACTGTCAGATTTGATAG
CTTTTG
2
GGCTATGTAAACTGTCAGTCGGAAAATGAGTGAGTACAAATCAGGGCAGGTGAGCGAATTGCCCGCCTTTTCTTTACCG
GTGG
o
TTGTGCTGTCGATTAGCCAACCGGGACAAATAGCCTGACATCTCCGGCGCAACTGAAAATACCACTCACCCATTAACCA
CGGAG 2
o .3
TTAAACGGATGAGTGACTATCATCACGGCGTGCAGGTGCTGGAGATTAACGAGGGCACCCGCGTCATTTCCACCGTATC
CACG 2
1-
GCCATTGTCGGCATGGTCTGCACGGCCAGCGATGCAGATGCGGAAACCTTCCCCCTCAATAAACCTGTGCTGATTACCA
ATGTG
.31
.3
CAGAGCGCAATTTCAAAGGCCGGTAAAAAAGGCACGCTGGCGGCATCGTTGCAGGCCATCGCTGACCAGTCAAAACCGG
TCA
CCGTTGTCATGCGCGTGGAAGACGGCACCGGTGATGACGAGGAAACGAAACTCGCGCAGACCGTTTCCAATATCATCGG
CAC
CACCGATGAAAACGGTCAGTACACCGGACTAAAAGCCATGCTGGCGGCGGAGTCGGTAACCGGTGTTAAACCGCGTATT
CTC
GGCGTGCCGGGACTGGATACCAAAGAGGTGGCTGTTGCACTGGCATCAGTCTGTCAGAAGCTGCGTGCTTTCGGGTATA
TCA
GCGCATGGGGCTGTAAAACCATTTCCGAGGTGAAAGCCTATCGTCAGAATTTCAGCCAGCGTGAGCTGATGGTCATCTG
GCCG
GATTTCCTCGCATGGGATACGGTCACCAGTACCACCGCCACCGCGTATGCCACCGCCCGTGCGCTGGGGCTGCGCGCTA
AAAT
IV
CGACCAGGAGCAGGGCTGGCATAAAACGCTGTCCAATGTCGGGGTGAACGGTGTTACCGGCATCAGCGCATCTGTATTC
TGG n
1-3
GATTTGCAGGAGTCCGGCACCGATGCTGACCTGCTTAACGAGTCAGGCGTCACTACGCTGATTCGCCGCGACGGTTTCC
GCTTC t=1
tl
TGGGGTAACCGTACCTGCTCTGATGACCCGCTGTTCCTCTTTGAAAACTACACCCGCACCGCGCAGGTCGTGGCCGACA
CGATG 2
o
GCTGAGGCGCACATGTGGGCGGTGGACAAGCCCATCACTGCAACGCTGATTCGCGACATCGTTGACGGCATCAATGCCA
AATT 'a
o
CCGTGAGCTGAAAACAAACGGCTATATCGTGGATGCGACCTGCTGGTTCAGCGAAGAATCCAACGATGCGGAAACCCTC
AAG tt
n.)
un
GCCGGAAAACTGTATATCGACTACGACTATACACCGGTGCCTCCTCTCGAAAACCTGACCCTGCGCCAGCGTATTACCG
ATAAA
0
TACCTGGCAAATCTGGTCACCTCGGTTAACAGCAATTAAGGAGCCTGACCGATGGCAATGCCGCGCAAACTCAAGTTAA
TGAA n.)
o
n.)
CGTCTTTCTGAACGGCTACAGCTATCAGGGCGTTGCAAAGTCCGTCACGCTGCCAAAACTGACCCGTAAGCTCGAAAAC
TATCG
i-J
CGGTGCGGGGATGAACGGCAGCGCACCGGTAGACCTCGGCCTTGATGACGATGCGCTGTCAATGGAGTGGTCGCTCGGT
GGC .6.
.6.
n.)
TTCCCGGATTCGGTTATCTGGGAGCTTTACGCCGCAACCGGTGTGGATGCCGTGCCGATTCGTTTTGCAGGCTCTTACC
AGCGC oe
GACGATACCGGCGAAACGGTGGCCGTCGAAGTGGTCATGCGTGGACGTCAGAAAGAAATCGACACCGGCGAGGGTAAAC
AG
GGAGAAGACACTGAGTCGAAAATCTCCGTGGTCTGCACCTATTTCCGGCTGACGATGGACGGTAAGGAGCTGGTCGAAA
TTG
ACACCATCAACATGATTGAGAAGGTGAACGGCGTCGATCGGCTGGAGCAACACCGCCGCAATATCGGCCTGTGATTTTC
ATCC
GGTCAGCCTGGCTGGCCGGTTAACCCTGATTCAGAAGTGAGAAAACCATGAACAAAGAAAATGTCATTACCCTGGACAA
TCCG
GTCAAACGTGGTGAGCAGGTTATCGAACAGGTCACGCTGATGAAACCCAGTGCCGGGACGCTACGCGGTGTCAGTCTGG
CTG
CGGTTGCAAACTCCGAAGTCGATGCACTGATTAAGGTGCTGCCGCGCATGACGGCACCGATGCTGACCGAGCAGGAAGT
CGC
P
CGCGCTGGAACTGCCTGACCTTGTGGCGCTGGCCGGTAAGGTGGTCGGTTTTTTGTCGCCGAACTCGGTGCAGTGACGT
TTCC
2
GAAAAATCTCTCGGTCGATGACCTGATGGCGGATGTGGCAGTGATATTTCACTGGCCGCCATCAGAACTGTATCCCATG
AGCC
o
TGACCGAACTCATCACATGGCGCGAAAAGGCGCTCCGGCGAAGCGGAAACACGAATGAGTAACAATGTAAAATTACAGG
TAT 2
1-,
.3
TGCTCAGGGCTGTTGACCAGGCATCCCGCCCGTTTAAATCCATCCGCACAGCGAGCAAGTCGCTGTCGGGGGATATCCG
GGAA 2
1-
ACACAAAAATCACTGCGCGAGCTGAACGGTCACGCATCCCGTATTGAGGGATTCCGCAAGACCAGTGCACAGCTCGCCG
TGAC
.31
.3
TGGTCATGCACTTGAAAAGGCACGGCAGGAGGCCGAAGCCCTTGCCACACAGTTTAAAAACACCGAACGTCCGACCCGT
GCTC
AGGCGAAAGTCCTGGAATCCGCAAAGCGTGCGGCGGAGGACTTACAGGCGAAATATAACCGCCTGACAGATTCCGTTAA
ACG
CCAGCAGCGGGAACTGGCCGCTGTGGGAATTAATACCCGCAATCTTGCACATGATGAGCAGGGACTGAAAAACCGTATC
AGT
GAAACCACCGCACAGCTTAACCGTCAGCGTGATGCGCTGGTGCGTGTCAGTGCGCAACAGGCAAAACTTAACGCAGTAA
AAC
AGCGTTATCAGGCCGGAAAGGAACTGGCCGGAAATATGGCCTCAGTGGGCGCTGCCGGTGTGGGGATTGCGGCGGCGGG
AA
CGATGGCCGGTGTTAAGCTACTGATGCCCGGTTATGAGTTTGCGCAGAAAAACTCAGAATTACAGGCTGTGATCGGAGT
GGCA
IV
AAAGACTCCGCCGAAATGGCCGCACTCCGCAAGCAGGCGCGCCAGCTCGGCGACAATACCGCCGCCTCGGCAGATGATG
CAG n
1-3
CCGGTGCGCAGATTATTATTGCGAAAGCCGGTGGGGATGTTGATGCCATTCAGGCGGCAACGCCGGTCACGCTGAACAT
GGC t=1
tl
GCTGGCGAACCGTCGCACAATGGAAGAAAACGCCGCCCTGCTGATGGGGATGAAATCCGCCTTTCAGCTTTCAAACGAT
AAGG 2
o
TCGCTCATATCGGGGATGTTCTCTCCATGACGATGAACAAAACCGCCGCCGATTTTGACGGCATGAGCGATGCGCTGAC
CTAT 'a
o
GCCGCACCTGTGGCAAAAAATGCCGGTGTCAGCATTGAAGAAACCGCCGCAATGGTCGGGGCGCTGCATGATGCAAAAA
TCA tt
n.)
un
CAGGCTCAATGGCGGGGACGGGAAGCCGTGCCGTGTTAAGCCGCCTGCAGGCACCGACGGGAAAAGCATGGGATGCACT
CA
0
AAGAGCTTGGAGTGAAAACCTCAGACAGCAAAGGAAACACCCGGCCAATATTTACCATTCTGAAAGAAATGCAGGCCAG
TTTT n.)
o
n.)
GAGAAAAACCGGCTCGGTACTGCCCAGCAGGCTGAATACATGAAAACTATTTTCGGGGAGGAGGCCAGCTCAGCCGCTG
CCG
i-J
TGCTGATGACTGCCGCCTCAACCGGAAAGCTGGACAAACTGACCGCTGCGTTTAAAGCCTCAGACGGGAAGACCGCCGA
GCT .6.
.6.
n.)
GGTAAATATCATGCAGGACAACCTAGGCGGTGACTTTAAAGCGTTTCAGTCCGCTTATGAGGCGGTGGGGACTGACCTG
TTTG oe
ACCAGCAGGAAGGCGCGCTGCGTAAGCTCACGCAGACGGCCACAAAGTATGTGTTAAAACTCGACGGCTGGATACAGAA
AAA
CAAATCACTGGCGTCAACCATCGGCATCATTGCCGGCGGTGCACTGGCGCTTACTGGCATCATCGGTGCCATTGGCCTC
GTAGC
CTGGCCGGTTATCACCGGCATCAATGCCATCATCGCGGCAGCAGGCGCAATGGGGGCAGTCTTCACGACGGTTGGCAGT
GCT
GTTATGACCGCCATCGGGGCTATTAGCTGGCCGGTTGTGGCCGTGGTGGCTGCCATTGTCGCCGGTGCGTTGCTTATCC
GTAA
ATACTGGGAGCCTGTCAGCGCATTCTTTGGTGGTGTGGTTGAAGGGCTGAAAGCGGCATTTGCGCCGGTGGGGGAACTG
TTC
ACGCCACTTAAACCGGTTTTTGACTGGCTGGGCGAAAAGTTACAGGCCGCGTGGCAGTGGTTTAAAAACCTGATTGCCC
CGGT
P
CAAAGCCACCCAGGACACCCTGAACCGTTGCCGTGACACGGGCGTCATGTTCGGGCAGGCACTGGCTGACGCGTTGATG
CTG
2
CCGCTTAATGCGTTCAACAAACTGCGCAGTGGTATTGACTGGGTACTGGAAAAACTCGGTGTTATCAACAAAGAGTCAG
ACAC
o
ACTTGACCAGACCGCCGCCAGAACTCATACCGCCACGTATGGTACCGGTGACTATATTCCGGCGACCAGCTCTTATGCA
GGCTA 2
n.)
.3
TCAGGCTTATCAGCCGGTCACGGCACCGGCTGGCCGCTCTTATGTAGACCAGAGTAAAAACGAATATCACATCAGCCTG
ACGG 2
1-
GGGGGACTGCGCCGGGGACACAGCTTGACCGCCAGTTACAGGATGCGCTCGAAAAATACGAGCGGGATAAACGTGCGCG
CG
.31
.3
CCCGTGCCAGCATGATGCATGACGGTTAAGGAGGTGACGAAAAATGATGCTCGCGTTAGGTATGTTTGTTTTTATGCGC
CAGA
CGCTGCCACACCAGACCATGCAGCGTGAATCAGATTATCGCTGGCCGTCAAATTCCCGTATCGGTAAACGGGATGCCTT
TCAGT
TTCTCGGTGTGGGTGAGGAAAACATCACGCTGGCCGGTGTGCTTTATCCCGAACTGACCGGCGGCAAGCTGACGATGAC
CAC
GCTCAGGCTGATGGCAGAGGAGGGGCGGGCGTGGCCGTTGCTGGATGGCACCGGCATGATTTACGGCATGTATGTCATC
AGC
AGGGTGAGTGAAACAGGGAGTATTTTCTTTGCAGACGGCACACCCCGGAAAATTGATTTTACGCTGTCACTCACCCGCG
TTGA
TGAATCACTGGCCGCGCTTTATGGCGATATCGGTAAACAGGCGGAATCGCTCATCGGTAAGGCCGGCAGTATGGCGACC
AGA
IV
TTCACAGGTATGACGGGGGCGGGATAATGCTGGATGCGCTGACATTTGATGCAGGCAGTACGCTGACGCCGGATTACAT
GCT n
1-3
GATGCTCGACAGCAGGGATATTACCGGCAATATCAGCGACCGTCTGATGAGCATGACCCTGACGGATAACCGGGGCTTT
GAG t=1
tl
GCTGACCAGCTTGATATTGAACTGAACGATGCCGACGGGCAGGTCGGGCTGCCGGTTCGTGGCGCTGTCCTGACGGTGT
ATAT 2
o
CGGCTGGAAAGGTTTTGCCCTGGTATGCAAAGGGAAATTTACCGTTGATGAGGTTGAACACCGGGGCGCACCGGATGTA
GTC 'a
o
ACCATCCGCGCCCGGAGTGCAGATTTTCGCGGGACGCTCAATTCCCGCCGGGAAGGCTCCTGGCATGACACCACGCTCG
GTGC tt
n.)
un
GATTGTTAAGGCGATAGCCACCCGTAACAGGCTGGAAGCCAGTGTCGCTCCGTCACTGGCCGGAATAAAAATTCCACAC
ATCG
0
ACCAGTCGCAGGAGTCTGATGCGAAATTCCTGACCCGTCTTGCAGAACGCAACGGCGGTGAGGTGTCGGTAAAAATGGG
AAA n.)
o
n.)
ACTGTTGTTTCTCAAAGCGGGGCAGGGAGTGACGGCCAGCGGTAAAAAAATCCCGCAGGTCACCATAACCCGCAGCGAC
GGC
i-J
GACCGCCATCATTTTGCGATTGCTGACCGTGGAGCCTACACCGGTGTAACGGCAAAATGGCTACACACTAAAGACCCGA
AGCC .6.
.6.
n.)
GCAAAAGCAGAAGGTAAAACTGAAACGCAAAAAGAAAGAGAAACACCTGCGCGCACTGGAGCACCCGAAAGCGAAACCG
GT oe
CAGGCAGAAGAAAGCGCCTAAAGTACCGGAAGCGCGTGAAGGTGAATACATGGCCGGTGAGGCTGACAACGTTTTTGCC
CTG
ACCACGGTATATGCCACGAAAGCGCAGGCCATGCGCGCCGCTCAGGCGAAGTGGGATAAACTGCAACGGGGCGTTGCGG
AG
TTCTCTATCAGCCTGGCTACCGGTCGGGCAGATATTTACACGGAAACACCGGTCAAAGTGTCTGGCTTTAAGCGCGTCA
TAGAC
GAGCAGGACTGGACAATCACTAAGGTGACACATTTTCTGAATAATAGCGGCTTCACGACGTCCTTAGAGCTTGAGGTCA
GGCT
TTCTGATGTGGAGTACGAAACAGAAGATGATGAGTGATGTTTTTGTTTTATCTGTTTGTTTTGTAAGGATAAATTAACT
AAAAT
GGCACCATCAACAAAACCGGAAGAGGTGCTCGCGATGTTTCATTGTCCTTTATGCCAGCATGCCGCACATGCGCGTACA
AGTC
P
GCTATATCACTGACACGACAAAAGAGCGTTATCATCAGTGCCAGAACGTGAATTGCAGCGCCACGTTCATCACTTATGA
GTCG
2
GTACAGCGATACATCGTGAAGCCGGGAGAAGTCCACGCCGTAAGGCCGCACCCGTTGCCATCAGGGCAGCAAATTATGT
GGA
o
TGTAATTACAAACAGAAAGCCCCTCAGTCGAGGGGCTTTTTTGTCGATGTGGTCAATGTGTGGACGTGACCAGAAATAA
ATCC 2
.3
TTTTATTTCAATTTATTGTACGTAAAAAATAAGCCCGTGTAAGGGAGATTTAGGGTGTCACCAGTAGGGGCTTTCAACG
GTACA 2
1-
ATGCGGGTTTGAGCGGCATAAATTACCACTGAAAGCCCTTAAACGTTACTCTACTGTGGACACTGTGTGGACACTCTCG
GCCTC
.31
.3
AGTACCACCTCTTAGCGGATTAAGAGAAATGGCGTCCTGAAGGTACTCTGGCGCAAAATGAGCGTAAACCATAGTTTGC
TCAA
TCCGCGTGTGACCTAGTATCCGTTGTAGCGTGATAATACTTCCTCCATTAATCATGAAATGAGTGGCAAAGCTGTGCCT
TAGTG
CATGTGTGGCTTGCCCCATTGGCAAATCCGGTTTTATTGCTTTCATTGTTCGTCTGAAGCGAGGGTAATCAGCATCAGG
GAATA
AAAAACCTCGTTTGTTATCCGCGATCATTTTGGCAACAGCCTCTGAGATCGGGACGGTGCGTGGTTTGTTTGTTTTCGT
TTTAAC
AAACGTGACGCGGTTATGGATGATATTTTCTGCTTTCAAACGAGCTGCTTCTCCCCAACGTGCTCCTGTACTCAGGCAA
AGAAT
CGCAATCTTTTTATTGTCGCCGTCAAGTGCTGCAAGCAGTAAGGCAATTTCTTCCTGTGTTAGATAGCCTGTTTCTGGT
TTTTCCT
IV
CCTTAAGCCTTTTTGTCCCTCTGATAGGGTGCTCACCAAAGAATAACTCCGCTTCAATCAGGGCTGTAAACATGCCGCT
AATACA n
1-3
TGTTAAATCACGATTGATACTCGAAGGTTTAATACCCTGACTTCTTCGGGTGGCGCAGTACTGGCTGATAAGCGATTTC
GTAAT t=1
tl
TTGAAATGCGCATGGGTCATTCGTTATTTTTGTGAAGATTTCAATTTTTCCAAGATTAGATTTCCCATGCTCTTCGTGT
TTACCCT 2
o
TTAAATCCCACCAGATCTGTGTCAGCTCCGACAGACGTCGCTTGTCTGTTGGTTTTGATAGCCATTCTTTATTGTGGTG
GTTGTA 'a
o
CAACGTGTATTTCTCGAAAGCGACAGCTTCGCTTTTCTTATCAAACTTCCTACGGATGCGTTTTCCATTACGTCCAGTA
GGGCGG tt
n.)
un
ATGTCCACTTCATATCGACCATCATCGAGTTTTTTGATTGCCATCAGAAAACCCTCCGAGTGGTGTGTTTTTTTGCGAC
TACTAAT
0
CGCTTTTTTCGTGGTGGCTGAAATTTAGCCACCAATAGTAGGCACTTGTGATGAATATATTCACGATGAATTGTTAACC
AGTCTT n.)
o
n.)
TTGACCGGAGTGGGGCGACGTTGTTTCGTTTTGCCCAAAGTGTGCGAGAGCGGGCGCAATTTGCCCGGACTCAGGAGCG
ATC
i-J
TGATTGGTCATGAACCATAAAGTGTATTTGGTGAATTGTGGGGTCTGCAGGATGTTCATCATGACATCTGTTGGAGGTG
TTGA .6.
.6.
n.)
ACGACCACTTTCATAGTAACTCAGCGTGCCATACGGAACCCCTGTTAAATCAGCAAGTTGTTGTCTGCTCAAATACTCT
GATTTT oe
CGCATTAAGACTATCTTCTCGCTTATCGTGTTTGACATGGTGTTTAGATCTCAATAGTATTTAGTTTAGATGTAGATTG
TTTAGTG
CTTGGATGTGGGCACTAAAAGGCATTATAAGACATTAAACGCAATTCATGAGGGCTAGAGGACGACATGAGCAAGCAAG
TAA
CACTCATGACTGATGCGATTCCTTATCAGGAGTTCGCAAAACTAATAGGAAAATCGACAGGAGCGGTTCGTCGGATGAT
CGAT
AAAGGAAAGCTGCCTGTAATTGATATGACCGATCCACAATCAGCTTCAGGTCGTGCAGGTGAATATTGGGTATACCTTC
CGGC
ATGGAATAACGGACTAAAACTGGCTTATGAAAGCCGCCCTAAAGAGATTCGTGACGGCTGGTTGATGTGGTTAGGTCTC
GGTG
AACCACGTTAAGGAGAACCGTATGAATGAGCCTCGTTGTATTGCTCAGTTACTGCGTAACGAAAGCCCCAGGGCGATTG
ACTT
P
CACCATCACCCACGGTAAGGGGCGTAAGGGAATCATTATCCGCACCAAAAAACAGAGTCCGTTAAAAAAGGCTCTGACC
TTTC
2
TGAAAAGCCGGAGGGTATGGAAATGACAGTGATGACGCTCAATCTCGTTGAAAAACAGCCAGCAGCTATGCGCCGGATA
ATT
o
GGTAAGCATCTTGCCGTTCCTCGCTGGCAGGATACATGTGATTATTATAATCAGATGATGGAGCGCGAACGGTTAACGG
TTTG 2
.6.
.3
CTTCCATGCTCAGTTAAAACAGCGTCACGCAACGATGTGTTTTGAAGAAATGAACGACGTCGAACGTGAACGACTGGTA
TGTG 2
1-
CAATTGATGAATTGCGTGGTGCATTCTCAAAACGCCGTCAGGTTGGCGCAAGTGAGTATGCATATATTAGTTTTTTAAC
AGTCA
.31
.3
GTCAGCGTCGTACTTTATTTATGCATGCCGGATTGACTGAAAAAGAATTCAACCAGCCATACTGGCGAATTAATGAAGA
GTCAT
GTTACTGGCGTGATGCTTTATTCCGTGCATTACGTGAATTATTCAGTCTGTTTGAGTATGCACCGACAATTCTGACGTC
GGTAAA
ACCGGAGCAATATCTGCATTAAGTAATTAACCAGAGTTTTTAACGCACTTAATTGTGCGGGGCTTCTTTTTGCCTGGAG
AAAGT
TATGCATACAGTTTCTGAAAATCAGTGCGGTATATACGCATTACTGCTGCAACAGGCCAGAACCGAAGCACAGGCCGAC
GCTG
CGACGCGCTTTTCTTCTCATCTTGATGCCATGATTCGCCACATCACAAAGGCGGAGTTATCCCGCGTGGAGATAGTCGA
GCTGC
TCAGTCAGGAGTCGGAAAAATTTCACAATATCGGATTGTCTCGCGGGGAGGTGCTTTGATGTTCTGTTCTCGTGCAGTT
GTATT
IV
ACTGAATAACGCCTTAAAAATCGCCGTTATGAAAAATGGCGATTTGTCTCTTATTCAACTTGGTCTTGATAAAGAAAAA
CGCGA n
,-i
AATAACTGAATCTGTTATCGCGATTTATCAGAGTGAATTAAACCTCCTGTCTGATGTGGTCAATTTACTTGTTAAACGC
GCTGTA t=1
tl
TTTCACAAGCAAATTTCCTCCGTGGATGAACTGACGAAATTAACGACAGAAATCGCCAGCTATTGCGCTGATGAATTTA
AAAAA 2
o
CTGAACGACAAAAGGAACTGGTAATGCCGGACAACGTAGATTTTATTCAGGAACAACAGGCTGAATTACTGGAGCGCCA
GATT 'a
o
AACGCGGCAAGGGTAAAACATTGCGGTGCTTCTGCGCTGGTTTGCGAAGAGTGTGACGCGCCAATACCTGCTGCCCGTC
GTGC tt
n.)
un
GGCTTACCCGTCAGCCACGCGTTGTGTTTCCTGTCAGTCAGTCTTTGAAGCAAAAAACAAACATTACCGGAGAACGGCA
TGAG
0
TATTCGTATTGAAATTGGCGAACGTTATGTCGTTACCAGTGACAGCTTTCAGTTTATTCTCCACGAGAAAAAGAGAGCG
GAAAG n.)
o
n.)
CGGTAAAAACGCCGGTCAGGAATGGCTGGCGGTGGTTGGTTATTACCCGAAATTAAGCCAGCTCGTTTCAGGCCTGATG
CATC
i-J
ACGATATTCTGACCGGAAGCGCAAAGTCTTTTGCTGATTTAAACGTGCAGGTTGAGCAACTCAGCAAGCGTTGTTCAGA
GGCT .6.
.6.
n.)
TTTGGCTCATATGGCCGTTAAAGCCTCCGGGCGTTTTGTCCCTCCGTCAGCATTTGCCGCAGGCACCGGTAAGATGTTT
ACCGG oe
TGCTTATGCATGGAACGCGCCACGCGAGGCCGTCGGGCGCGAAAGACCCCTTACACGTGACGAGATGCGTCAGATGCAA
GGT
GTTTTATCCACGATTAACCGCCTGCCTTACTTTTTGCGCTCGCTGTTTACTTCACGCTATGACTACATCCGGCGCAATA
AAAGCCC
GGTGCACGGGTTTTATTTCCTCACATCCACTTTTCAGCGTCGTTTATGGCCGCGCATTGAGCGTGTGAATCAGCGCCAT
GAAAT
GAACACCGACGCGTCGTTGCTGTTTCTGGCAGAGCGTGACCACTATGCGCGCCTGCCGGGAATGAATGACAAGGAGCTG
AAA
AAGTTTGCCGCCCGTATCTCATCGCAGCTTTTCATGATGTATGAGGAACTCAGCGATGCCTGGGTGGATGCACATGGCG
AAAA
AGAATCGCTGTTTACGGATGAGGCGCAGGCTCACCTCTATGGTCATGTTGCTGGCGCTGCACGTGCTTTCAATATTTCC
CCGCT
P
TTACTGGAAAAAATACCGTAAAGGACAGATGACCACGAGGCAGGCATATTCTGCCATTGCCCGTCTGTTTAACGATGAG
TGGT
2
GGACTCATCAGCTCAAAGGCCAGCGTATGCGCTGGCATGAGACGTTACTGATTGCTGTCGGGGAGGTGAATAAAGACCG
TTCT
o
CCTTATGCCAGTAAACATGCCATTCGTGATGTGCGTGCACGCCGCCAAGCAAATCTGGAATTTCTTAAATCGTGTGACC
TTGAA 2
un
.3
AACAGGGAAACCGGCGAGCGCATCGACCTTATCAGTAAGGTGATGGGCAGTATTTCTAATCCTGAAATTCGCCGGATGG
AGCT 2
1-
GATGAACACCATTGCCGGTATTGAGCGTTACGCCGCCGCAGAGGGTGATGTGGGGATGTTTATCACGCTTACCGCGCCT
TCAA
.31
.3
AGTATCACCCGACACGTCAGGTCGGAAAAGGCGAAAGTAAAACCGTCCAGCTAAATCACGGCTGGAACGATGAGGCATT
TAA
TCCAAAGGATGCGCAGCGTTATCTCTGCCATATCTGGAGCCTGATGCGCACGGCATTCAAAGATAATGATTTACAGGTC
TACG
GTTTGCGTGTCGTCGAGCCACACCACGACGGAACGCCGCACTGGCATATGATGCTTTTTTGTAATCCACGCCAGCGTAA
CCAGA
TTATCGAAATCATGCGTCGCTATGCGCTCAAAGAGGATGGCGACGAAAGAGGAGCCGCGCGAAACCGTTTTCAGGCAAA
ACA
CCTTAACCAGGGCGGTGCTGCGGGGTATATCGCGAAATACATCTCAAAAAACATCGATGGCTATGCACTGGATGGTCAG
CTCG
ATAACGATACCGGCAGACCGCTGAAAGACACTGCTGCGGCTGTTACCGCATGGGCGTCAACGTGGCGCATCCCACAATT
TAAA
IV
ACGGTTGGTCTGCCGACAATGGGGGCTTACCGTGAACTACGCAAATTGCCTCGCGGCGTCAGCATTGCTGATGAGTTTG
ACGA n
1-3
GCGCGTCGAGGCTGCACGCGCCGCCGCAGACAGTGGTGATTTTGCGTTGTATATCAGCGCGCAGGGTGGGGCAAATGTC
CCG t=1
tl
CGCGATTGTCAGACTGTCAGGGTCGCCCGTAGTCCGTCGGATGAGGTTAACGAGTACGAGGAAGAAGTCGAGAGAGTGG
TC 2
o
GGCATTTACGCGCCGCATCTCGGCGCGCGTCATATTCATATCACCAGAACGACGGACTGGCGCATTGTGCCGAAAGTTC
CGGT 'a
o
CGTTGAGCCTCTGACTTTAAAAAGCGGCATCGCCGCGCCTCGGAGTCCTGTCAATAACTGTGGAAAGCTCACCGGTGGT
GATA tt
n.)
un
CTTCGTTACCGGCTCCCACACCTTCTGAGCACGCCGCAGCAGTGCTTAATCTGGTTGATGACGGTGTTATTGAATGGAA
TGAAC
0
CGGAGGTCGTGAGGGCGCTCAGGGGCGCATTAAAATACGACATGAGAACGCCAAACCGTCAGCAAAGAAACGGAAGCCC
GT n.)
o
n.)
TAAAACCGCATGAAATTGCACCATCTGCCAGACTGACCAGGTCTGAACGATTGCAGATCACCCGTATCCGCGTTGACCT
TGCTC
i-J
AGAACGGTATCAGGCCTCAGCGATGGGAACTTGAGGCGCTGGCGCGTGGAGCAACCGTAAATTATGACGGGAAAAAATT
CAC .6.
.6.
n.)
GTATCCGGTCGCTGATGAGTGGCCGGGATTCTCAACAGTAATGGAGTGGACATGATGGCAAAAATTCACGAGGTAAAGC
TGC oe
ACGCAAAATATTTCGACCTTGTACTGGAAGGAAAGAAACGCGCAGAGTTTCGGAAAAAAGATCGTAATTATGAGCGCGG
GGA
CACGTTGATTTTGCATGAATGGGTGCAGGGTGTGTTTACGGGGCGAAAGGTTGAAGCCCGGATAACAGATGTTACTGAC
CTGT
CAGACTGGCTGGAAGATTATGTCTTGCTGCACAGAATACTAGGTGAGATACTGCCGCTGACTGTCAAGCTGGCAGTATC
CTCC
ACATTTTTTATAAATCTAAGGCGAAGAACGGTGGCAATATATTTGCTTTAAATTGTAAATAAGCAAATTATTCTAAGCC
ACAAAT
GATATCAATTGAGTATCGATTTTCGTTCGAATATAACCATAACGCCAAGCGTCAAATGTAAGTCATCTATATGTGATTC
GTATTC
ATATTGGTCAGCATCAGGAGATAGTAAATGCTCTGCTGCTGTAAAAACATCAATTGTAACAGATATATCGTCTGGAATT
TCTTC
P
ATTTAATTCGGATATGTATTTTTTATATTTGTTTTTTAATTTCCTTATCATGTCTTCCTTATGGTGGATTGTAGTGCAG
TAATTTTTT
2
TTAGTTGAATCATTGTATGAATGGAATGAACAGTTTAATGCTAGGCCAAGACTTGAATTTCCTGTAGGGCCTGTGTGTT
TAAAG
o
TATTCAAGATTTCTTCTTATGTCAGAAAATAAAAGACTTTTACAGGGGGCATTTCCTTTAACTTCTATTGCACATATTG
GAATGTC 2
o .3
GAAAAATGGTCTGCTGTCCAATATGGCAATATCAATTCTGCCTGATCTTGTTGTGTTTGTGTTTTTTCTGACAGTTTTT
TTTGAGA 2
1-
ATATTTTTTGGGGGTCAGACCGTTTCGATAAAGGAACTGTTGCATTAATGAATTTATTTGTATGGTACTCAAATATAAC
TTTGTA
.31
.3
ATCACCATGGCGAAAAGATTTTATTTCTAACAACGATAGACCTATAGATACGGTAGCTACATATTCAGTGTTTATTTTT
GCACCA
GACCTGAAATCCATTAAATAAGTTGCGTCGGCTGACTTTTTCATTCCATCTTTTACAGCTTTCACAATATCCATGTTAT
TCATATC
AACACCTTTTATATTCCAGAATTATAAATCTTAATGATTGATTTGTTTGTGTGTTTAGGTTACTAGATTGACGTACTTA
TAGCACA
TTCTGATTTTGAAATCACCATCCTTGGGTAAAGCACAGCCTATCACTATTGGTTTAGTACTGTGCTGTAATAGTATCAG
ATTAAG
TTAAATCCATTTTATGAAATCTTCCATTTCATCATATTTTTTTATTACTGCTCTAACATCTTTGCTTTCATCACATAAA
CAATCTAAA
AGACTATCCCATTCAGTTTGTTTTTTACCAAATCCATAAACAGCCTCTATATCACCGGATTTCCATATATAGATTCCGT
TATCTTTC
IV
AATTTTTGATGTATTTCATGTATATGCTTTTGTGTTTTTTCATGGTTACATATTTTGATGAAGTCTTTGCTTGAAATGC
TTTTGAAT n
,-i
GACTCAAAAGTAGTAACTTTTGTATTTAAACTAAGCTCTCCTGAGTTGATTGATTCAATCAAAGCAGTCAATAGATTAT
CACATT t=1
tl
CAGTACTAAGTAAGTCTTTATGCTCTGTTAATAGAATATTTGATAAGAAATCACAATCAGCTAGAATCCTTGTTTTTAT
GCCGAT 2
o
GGCATTGATGATTTGTGACATCTTAAATAAACTACCCTTACCGTCAACGGCAACAATGCAGATTTTACTCGGGTTGAGT
TCATGT 'a
o
CCGTTAATTTTTTTATAAAGTGCATATAGAACGTTTGTCTCTGTTTTCCCTTCAACAAGCAAAACTTCTTCAGAAAATA
AAAGGTA tt
n.)
un
TGATGAATTAGAAAGCGTGAATGCTGAGTGCAATTGCGGTGATGAAGATTTATATAATTCTTCGATTTTTTCAGATATA
GTCTTC
0
CTTGCTATGGTTCCATTAGAATCCTTACAAACCTGAATCGCATTTGCTGCATGCTTTGCAGAAAGCATACTGGCTGAGT
GAGTT w
o
w
GATATTATAACCTGATACCCTGATTCACTTAATGTGACAAGTGATTCTCTGACAGAATTAATG
GCTGAAGGGTGTAAATATAAC
i-J
TCAG GTTCATCAATG AAAATCAAAGTGTTTG ATTTTTTTG ATTC G CTGTTTTCTTTTTTTATTTC G GC
CAG GTATTG AATTAATG C
4,.
w
CATTTG AATG G AAC GTTGTGTTC CGTG ACCAAATC G G CTG ATATCTCTCATTACCG GTTCATCTTCC
CG AG ACTCAAAAACTTTC oe
AG AGTG C CG G ATTTAAATATCTCATCTAATGTCG GTGTG G G AAAGTGTAACTTTACACTTAC GTCAGG
AAAAAATTG GTTTACT
TTTTTATTTACACCTG AGTCTATTTTATTAAG G CTTTCTAATCTGTTCTCACC GTTGTG AG AAAG
ATATTTAC CTATTTCTGATATG
TTTTTTG AAAATTTTTCTTCGTGTTCTTGTTTTATTTCAG AAACAATTG C G G AAAGTATCTTTC
CTATTGTG GTCGTGTTTTTG CAT
TTTGTTG AGTCTTCG ACAG CGTCAG ACATTG CAG G G ATATG AATTG GTTC CG G AAATATATTG G
AG ATTG CAC CATCTATG C CG
CCAGGGTTTTTCTTCCACGTGGTACCGTCATATACATCCAGACTTTTTTTGGCTTTTCCTGTTTCCTTATTAAATTCCT
GTCTTCTT
G CAAAG GTAAG AGTCC CGTCAATTATAAAC G G AG CTATTTTTTGTTG ATTCTCTTCTGTTAACAAAG
ACAG AGTATCATCTGTTA
P
TACCTTG AATAACG C CTTCAACG G ATACG G G GTGTGTAG G ATCGTACACATCTG ATTCTG
AAATCAAAG AG C CATCTAACAG C C .
,
ACTTAATTGCTAAGATAATATTTGATTTTCCTGCGTTATTATAACCAACTAAAGCAGTGAAGGGGCGCAAAATAGCCGA
TGTTG
,
1¨
.3
o
ACTTACAAG AACG AAAGTTG CTAATTG
AAACTG AAG CAAG AC GTACAGTCATTATTTTTC CTTAAATGTG CTATTTGTATG CAAT .
--4
G AGTTCATACG AAACG CTTTTTTACATTTTATAGTCGTTGCATTCAAG G GTG CATG AG ATTG CATTAAG
G G AAACTGTG ATATG "
,
,
,
GCTTGGCTTTTGACTGGAAATACTGATGGCTCATTAGTTTTATTAAGGTGCATTAAAACCG CC CC GTG AAGCG
G G CGG GCG AG 0
,
N,
N,
GCGGGGAAAGCAC
20 Native P4 (acc nr:
TTTGCCGCTGCCGCCGGGACCGGTCACCTCCAGAAAGAGCTGCCAGTCGTAGCGGTTTGCCAGCACCATAAACAGTGCA
GCCA
X51522)
GAATCACGTCGCGTTTTTCCGCACGGCCACCGGCGGCACGGTCAAGCCAGCGCCAGAAGGCGGGGGCGTGGGTTTCCAG
CGT
TTCACCGTCCACCGGCGGGGTGAAATCCACATCGCACAGGGTGCGCATCCAGTGTGACGGACTGTGCGGGTGGAACGTG
CCG
TTCTGCGTGTCGAGCACGCCGTTACGAAAGCCAATCAGGCGGCGGGAGGGGGCTTCCTGCTGCGGAATAATCAGCTTCA
GGG
1-d
TATCCACCACGGAGGCCACCTTCCCGGAGGAGAACGGCGCACGCAGACGCTGAAACAGCCCGGCCACATCCCGGGCAAA
GTC n
1-3
CTGTGGCGGCAGCACCTTCCAGACACCATTTTCATAGCGGGACAGAAGCTGGCCGTTGGCATCGACCGCGAGCGCCTCG
CCGT t=1
1-d
AATGCTCATAGATACGCATGGCCTTTTCGCTGGTACTCATGGCGGAAAACTCCGCTTCGCTCATGGTGTCGAACGGGCT
TTCAG w
o
w
CCGGTGGCCGGATGGCATCGTAAATGGCCTTACGGGTGGCCTCCCCGCCGTACTGCGTGAAGGCATCATTCCAGTCACC
GAAG =
'a
ACCGGCGGCAGGGCAACAACACCTTCACACGCATCTGCGGCTGCGGCGGCTTTTTTCTGGCCGTCACCACTGAGGTCAC
GGTC o
4,.
w
w
vi
AGCGGCAAGGACAATCTGACAGGCGGGGTGCTTCTGCCGGGCAAGGCTGGCCAGAGAAAGGAGGTTCACGGAAGAAAGC
G
0
CCACCATCACCGTTTCACCGGTCAGGTGATGTACGGTAAGTGCGGTCGCGTATCCCTCCGCTATCCACAGACGTTTTCC
GGCCT n.)
o
n.)
GATTCTGTCCTTCAAGGGTGTGACAGGTGCCCCTGACCTGTCCGCCTTTCAGGGTGCGCTTACGGCCGTCAGCACTGAT
TAACT
i-J
GAAGGTTAACCAGTTCGCCGCTGTCGTCATACAGTGGCACCACAAGGTCACCGGCGCGCCAGCTCACGCCACCGGCTCT
GTGT .6.
.6.
n.)
GTGCCGGTCAGCATCCGGCATTCCCGGCCGGGAAAGCCCTTGCGGGTCAGGTAGGCGTTACCGGTTCCGGTACGGGTTT
TCGC oe
CATCAGGGTTTGTGCCAGTGCGGCGGCGTTCTTCCGGGCAGCGTCTGTTTCATCAACGGCGGCGGTCGTCACTGCCGGG
TCAG
CCGGTGGCAGGCTGCCGGTCACGGCAGCCACCTTTGCGGCCGCGTCGGACGGGGAAACACCAAAAACCTTTTCAACCAG
TTTC
AGGCCGTCACCGGCACCACACTGATTGCAGTACCAGGTGCCGCGCCCCTCCCTGTCATCAAAACGGAAGCGGTCACTCC
CGCC
ACAGACCGGACAGGGCTGATGACGGTTCTTCAGCACCTGAATCCCCAGCGCCGGGAGAATACGCGGCCAGTGGCCGAGC
GCA
TGGCTGACGGTGGCGGTTACGTTCATTTTCATGGTGTTGTTCTCCTTCAGTGCAGTACCGGCGCTTTTATGTGACGGGC
ACAGA
GTTCATCCATCACAACCAGCCCGAGAAAGGACAGCGACGGCGCGGCCTTCAGGGGGCCGGATTCCATTAAATCTTCCAG
CAGG
P
GCACAGGCTATCTGACGCCCTTTTTCCTCACCGTGCTGGCGCAGATAAAAGCCTTCCAGCTCAGCGGCGATGGCCGCCT
CCAGT
2
GACTCAAGGGTGAGATGCGGGTAGCGGTGCTGACGTTCGCACACGGTCAGCCAGGCACAGGCGACAGCGCGACGGTAAA
GG
o
GCAGCGCGTAAGACGGGCGGTAAGGGTGTTTTCATTTGCTTTTCTCCCTGTGACAGATGACTGCATTCCGTGCCGGTTG
CATTA 2
oe
ACTGATAAGGCATATCTGCGTCTCCTGAAGACGTGCGTATCCCTGCGCGAATACGCACATTTAATTTTTCGGGGGTCGT
TTTTTA 2
1-
ATTACAGATAATTGCGGTAACTGTTATCCGGGGTGGTTTCCGGGTCAGGCTCCGTGCGGGGAATTTCCCGCCATTCCCG
CGCCA
.31
.3
CCGGTGCTGCCCGGCTGACCGGAACAGTGTCCTGCGGGTAAATATCCAGATATTTTTCCCGCCATTTCTGTAATTCCGG
GTCTC
CGGCCATTTCTTTCAGTACCGCATGCCGGTTTACGGGGCTGCGTTTAAACAGGTCAGGACGGTCACAGGTAAATTCCCG
CAGA
AAACGCCCCAGCGGGATGTCTGTGGTGCGTCCGTCAGCGAGGATACGCACAAGGATACTGAATTTACGGCGGTACGGGT
TCC
AGACAATGTCCGGGCAGCGGTACGGCATTTCCCACGGAATACCGTCTTCCAGAATGCCGACCACGGCCACATCGGGAAA
ACCG
GCAGAACGGTAAATCTCACCGGGCTGGGGAAAATCAAACATGCGTCCTGTCTCCCCGGTCTTTCTGCTGGGCGAGAAAA
TCGC
GGCACAGGCCTTTGGCTTTCAGCTCATTCAGCACAAAATCAATATCTTCATTCAGGTAGCTGAAAATATGTGGAATGTA
GAGCT
IV
GATGCAGGCCGGAGAGTTCACGGTGAATCAAATCACCCCCAACAAACCGGGATACGGCGCTGGCGCGGTTGAGCTTATG
GTA n
1-3
AGCCTCAATGCTGAGGTGTTCACGGGCGTCATGACGCGCTGAGACGGTCTGAGGGGCTTTTTTATTACGCACGGGACAC
CTCC t=1
tl
ACCACCGGCAGACGGGCAGCAAGGGAGAGCACATAGTCACGGACAAGGGAACGGCGGGCACTGCGTTCATCACCGGCGA
CG 2
o
GTGCGAAGCATACAGATACGGGGATGACGGTCTGCGCGACGGACAGCCGCAAACACAAAGACAAATTCAGGGTGTGAGG
GG 'a
o
GTAAGGGTTGTAGCCATGATGGCAGCCTCCTGTGAATAGCAAATAACGCTATCGCCGGAGTTCTCACGCTCGATGGCGA
TAGC tt
n.)
un
CCAGACGGGGGTGAGAATACCGGCTTCACAGGATACCGGCCAGCCCGGAGGCTGCCCCGCCTGAGCTACCATTGACTCT
GCG
0
GCATAATGAGCGGACGCGGGCAGGATGCACGGAATGCCATCTGCACGACTGACCACACACCACACCATAATCTGGCGCT
CTGT n.)
o
n.)
GGCATTGATTGCGACACAAAAAAAGACGCGTGGCGCGTCATATGTCGCCTGTGAATTGCTCGGGTTCTCACGCCCGGCT
GCCG
i-J
ATTTTGCGGCAGGCGAAAAACTATATCCGCAAATGCCGGAAAAAGGCAAGCCAGAAAAAGGGAGTTTTTGCAGAGCGGG
CAT .6.
.6.
n.)
CATCATGCGTCGTACCCCCGTTTGCGTCCGGCAATGCGTCCGGCCATCCATGCGGTGACTTCAGAGTGCAGCCAGGCCA
CATTT oe
TTACCGCCAAGACTCACCTGCGGCGGAAATTCCCCCTTACGGATGAGTTCGTAGATGGTCGAGCGTGACAGGCCGCACA
GGTG
CATCACTTCCGGCACACGTAAAAAACGCTCCTGCGTGATGTCCGGCAGCGGCATCAGTGGCGTCACTGGGGCGGGAGAC
GGG
GAAGAAAAAACAGCTTGCATCGGGCTACCTCGTTAATGTCCATACAGCACCGGATAAGTCCGTCCGGCTTCGGGTAGCG
CTTT
ATTTTGTGAATATTTTCAGCAGACGCAACAGGGGGGATTTGTTCAGGCTGTCTTACAATGGCTGTGTGTTTTTTGTTCA
TCTCCA
CTTAAAGTCATTTAAAGCCACTTAAAGCAATTTGTAATTTTTATAGTGAAATACAAATCGTTTCTTCTTATTCATTCCC
GGCGAAT
TAATAAAAACAAACAGTAGTAAACAGCACAAAAAGCCCATCAACGGGTGAACAGTGGTGAACAGACGGTGAACAGTCAT
TAC
P
TGCGATTGTTCACCCTTTAACTTACTGTATTACTTATCTTTTTTATTAAGGTGAACAGAGGTGAACAGTAAAATATAAA
AAAACA
2
AACAGTAAGCCGGTTTTTCCTGCGACCTTTTCCTGGCTTGCCGGTCTGAGGATGAGTCTCCTGTGTCAGGGCTGGCACA
TCTGC
o
AATGCGTCGTGTTGTTGTCCGGTGTACGTCACAATTTTCTTAACCTGAAGTGACGAGGAGCCGGAAAATGTCTGACCAC
ACTAT 2
o .3
CCCTGAATATCTGCAACCCGCACTGGCACAACTGGAAAAGGCCAGAGCCGCCCATCTTGAGAACGCCCGCCTGATGGAT
GAGA 2
1-
CCGTCACGGCCATTGAACGGGCAGAGCAGGAAAAAAATGCGCTGGCGCAGGCCGACGGAAACGACGCTGACGACTGGCG
CA
.31
.3
CGGCCTTTCGTGCAGCCGGTGGTGTCCTGAGCGACGAGCTGAAACAGCGCCACATTGAGCGCGTGGCACGCCGGGAGCT
GGT
ACAGGAATATGACAATCTGGCCGTGGTGCTGAATTTCGAACGTGAACGCCTGAAAGGGGCGTGTGACAGCACGGCCACC
GCC
TACCGGAAGGCACATCATCACCTTCTGAGTCTGTATGCAGAGCATGAGCTGGAACACGCCCTGAATGAAACCTGTGAGG
CGCT
TGTCCGGGCAATGCATCTGAGCATTCTGGTACAGGAAAATCCGCTCGCCAACACCACCGGCCATCAGGGCTACGTCGCA
CCGG
AAAAGGCTGTCATGCAGCAGGTGAAATCATCGCTGGAACAGAAAATTAAACAGATGCAAATCAGCCTCACCGGCGAGCC
GGT
TCTCCGGCTGACCGGACTGTCAGCGGCAACACTCCCGCACATGGATTATGAGGTGGCAGGCACACCGGCACAGCGCAAG
GTG
IV
TGGCAGGACAAAATAGACCAGCAGGGAGCAGAGCTTAAGGCCAGAGGGCTGCTGTCATGATTTACTGTCCGTCGTGTGG
ACA n
1-3
TGTTGCTCACACCCGTCGCGCACATTTCATGGACGATGGCACCAAGATAATGATTGCACAGTGCCGGAATATTTATTGC
TCTGC t=1
tl
GACATTTGAAGCGAGTGAAAGCTTTTTCTCTGACAGTAAAGATTCAGGAATGGAATACATTTCAGGCAAACAGAGATAC
CGCG 2
o
ATTCACTGACGTCAGCCTCCTGCGGTATGAAACGCCCGAAAAGAATGCTTGTTACCGGATATTGTTGTCGGAGATGTAA
AGGC 'a
o
CTTGCACTGTCAAGAACATCGCGGCGTCTGTCTCAGGAAGTCACCGAGCGTTTTTATGTGTGCACGGATCCGGGCTGTG
GTCT tt
n.)
un
GGTGTTTAAAACGCTTCAGACCATCAACCGCTTCATTGTCCGCCCGGTCACGCCGGACGAACTGGCAGAACGCCTGCAT
GAAA
0
AACAGGAACTGCCGCCAGTACGGTTAAAAACACAATCATATTCGCTGCGTCTGGAATGAGGGCTGCCGGTTAACACCGG
CCGT n.)
o
n.)
CGCCGCACACCGTATTTTTATTCTTCAGCATGATGAGAAAGAGATAACGATGGAAAGCACAGCCTTACAGCAGGCCTTT
GACAC
i-J
CTGTCAGAATAACAAAGCAGCATGGCTGCAACGCAAAAATGAGCTGGCAGCGGCCGAACAGGAATATCTGCGGCTTCTG
TCA .6.
.6.
n.)
GGAGAAGGCAGAAACGTCAGTCGCCTGGACGAATTACGCAATATTATCGAAGTCAGAAAATGGCAGGTGAATCAGGCCG
CCG oe
GTCGTTATATTCGTTCGCATGAAGCCGTTCAGCACATCAGCATCCGCGACCGGCTGAATGATTTTATGCAGCAGCACGG
CACAG
CACTGGCGGCCGCACTGGCACCGGAGCTGATGGGCTACAGTGAGCTGACGGCCATTGCCCGAAACTGTGCCATACAGCG
TGC
CACAGATGCCCTGCGTGAAGCCCTTCTGTCCTGGCTTGCGAAGGGTGAAAAAATTAATTATTCCGCACAGGATAGCGAC
ATTTT
AACGACCATCGGATTCAGGCCTGACGTGGCTTCGGTGGATGACAGCCGTGAAAAATTCACCCCTGCGCAGAACATGATT
TTTT
CGCGTAAAAGTGCGCAACTGGCATCACGTCAGTCAGTGTAAAATTCCCCGAAAATCCGCCCGTTTTTACTGAAAAAAGC
CATGC
ATCGATAAGGTGCATGGCTTTGCATGCGTTTTCCTGCCTCATTTTCTGCAAACCGCGCCATTCCCGGCGCGGTCTGAGC
GTGTC
P
AGTGCAACTGCATTAAAACCGCCCCGCAAAGCGGGCGGGCGAGGCGGGGAAAGCACCGCGCGCAAACCGACAAGTTAGT
TA
2
ATTATTTGTGTAGTCAAAGTGCCTTCAGTACATACCTCGTTAATACATTGGAGCATAATGAAGAAAATCTATGGCCTAT
GGTCC
--.1
AAAACTGTCTTTTTTGATGGCACTATCCTGAAAAATATGCAAAAAATAGATTGATGTAAGGTGGTTCTTGTCAGTGTCG
CAAGA 2
o .3
TCCTTAAGAATTCGTGGCATGAGAGAGTTAAAGGATGCTGAATCATGTATATGGATTAATTGGCGTTGTTGGGACCATA
ATCA 2
1-
CTATTGCTACTAGCTTCATCCGCAACCAAGATATTTCACAATGGTTGCTCGTTTGTTCTGGATGGTTAGCTGCTTTACT
TATTGGA
.31
.3
TGGTTTACACACAGAACGATTAAAGCAATTAGCAACAACCACACGAATGTTATAAAAAGTAATATGGAGGTAATCAACA
GTCA
TAATGAATCCAATCAAAACTTGATGGCTGAAAACAAAGAATTGATTAAAGAACTTGCAAGAACTTCAGAACAGAAAGAG
AAGA
TGGAAAGTATAGCTGCTTATCTTGCTACTCAGAACCCCCAAATTAACGCTATGCCAAGAACAGCAAGCCGACCAGAAAA
TATCG
ATTCGGAGGCTAATTAAATATGAAAGTATATTTTGAAAATTACTCATACTATCCAGCCCTAAGAACACGATCAGCAGAA
ATGAC
CGGACTGAATAATTTATCATATGAGAATAAGAAAAAAATATTACCTTTAATTTCTTTGGGGAAATGGCCTCGCTCGGAA
GAAAT
ACAGGTATCGTTAGATAAAAGTTTAGAAGTGATGAGTAATCTCCCATTTATTTTGGATGTCACAAAGGATAATTCTCAT
CATTGC
IV
GCGTCAAGCTTCGAACTACTCTCTCCTGAGAATGGTTTTAAAAACTGGATAGAGTTTTGTTCCAGAAATGATAATATTA
TTCCAG n
,-i
TAGTCCAGATGCCGGATTCAGCAAAACTTAGAGATATATCTATCCAAGCCAGGGTTTTAGAGGAACTTAAAGGTTCAAT
TGCAT t=1
tl
TTAGAATTAGAAATCTAAATACAGACATTAATAAAACCCTGACGTCACTAGTATCGATGAACTCTCCAGAAAATGCCAT
AGTTTT 2
o
CATAGATTTGGGCTACATTAGAGGGAATGTATCTGCTATAACAGCTGCAGCAATAAATTCGATTAATCAGATTAGAACT
GAGAT 'a
o
TCCTGAAGCCATAATTAGTGTGCTAGCAACAAGCTTCCCTAGTTCAGTTACAAATTTTTGCCGTGAAAATGGACAGTCC
GGATA tt
n.)
un
TATTGATGTTATTGAGAGAGAATTGCATCAAAATATTGGTGGAAGTGATGTCGCTATTTATGGCGATCATGGTTCAATT
CATTC
0
AGTAGTGTATGACAACATAATTGGGCGATATGTTCCTAGAATTGATATAGCGTTAAATGATTCATGGTATTTCGAGCGC
CGCCC n.)
o
n.)
TGGGATGAATAAAGAAGGGTTCATTGAAGCTGCGAAATCAATTTTGGCAGAGTATCCACATTATCAAAGGGAGGACTCT
TGG
i-J
GGAGCTGCAATGATAAGAAATGCTGCTATAGGGGATATAGCAGGTATGGGAAGTCCAGCTAAATGGATTGCTGTTCGAG
TGA .6.
.6.
n.)
ATCTACACTTAAATAAGCAAATTGAGCTATCTGAAGCTCTACAGTATGGCTTTGATCATGATGAGGAAGATCTTATCTA
ATAAA oe
AAACTAGGGAAGGTTAGCCTTCCCTAGCCCACAACCCTTTTAAGTACTTTTTCCGGAATATAGGAAACATCATCGCCAT
ATTTCC
TAACTATAGAGTCAACATCAACCTTATGGTATTCATTTAAAGATTTCTCCGAACAAGCCTTAAACTCAAACTTAACCAA
AGAATT
ATTCCTTCTTTTAGCTTTGCTCAATAAGGCAAGATCGTTTATAGATATTTCTTGATTACCATGGAGTCTAGATAAAAAA
TCATTAG
ATGTCGCATGATACCTATTTTTAATAGATGTAATGACAAATTCTCTAATCTTGTTTAAAGATAACTTAGATGCCAAAAC
CTCAAG
ACAATCCCTATGTAGTTCATTTGAAAATGTAATTCCATACTGATTAAGCAGATCTTTAAGCTCAGACTTAAGCAAAAAA
CCCAAA
TAAACAAATTTATTTAAGCGAGAGCAAACCCTTCCTTTTTGAATTATTTTAAAGCGTACGCTACTATTATTAGATTGAA
ATTCTAC
P
AACGCCAACATCTGATGAAACCCGTTTAGATACTTCACTGGTAAATTTTGGCGCGCACACAATTGTAACTTTTTCGAAG
CTAGAT
2
TTAAAAATTTTTAACTGACCATCCAAGCGCTTAAGAGAGTCAAAATCGCTTTTTATTTCAAAAGCTTGTAGGCTTCCAT
TGGCAA
--.1
TAGCCAAATCAGCTCTTCTGCTCCAGTTAGCAACAACCATTTCATTAATGATCGTTGCATCACTAAGCATGCCTTTTTT
ATATAAC 2
1-,
.3
CATTCAATTAGGGCTATTTTAACTTCCTGCTCTCGCAATAACTCAGCCATGTTAATCACACTCACTTCACGTTGATTTT
TATTATA 2
1-
GCAACTAACAATTAAAAACTCCAGATTATGTTACTTAACACTCATTTTAAAAATCAACCAATAACTATTTTTATTTAAG
TATTTCTT
.31
.3
TGTATTTTGAATAATCATAAGGCTAATATATTTATTACGGTTCATATCCAAATAATCAGCCCACCACTGCATCATTAAA
CGCCGC
TCATCCAAATGCTCGGAGGTATGGATATATGCCGCACGTACATTATTACGCTCTGAGTGGCTCAGTTGCCTCTCTATCG
CGTCAT
CACTCCATAACCCCGACTCCCCCAACGCACCACGCGCCATAGTCCTAAACCCATGCCCACACACCTCGGTTTTAGTATC
ATAGCC
CATCGCACGCAATGCGCTGTTTACCGTGTTTTCACTCATAACCTTAGTTGCGTCATGATCCCCCGGAAAAAGCAGCTCT
TTATCA
CCACTAATCTGCTTTAACTGGTTTAATAAAATCATCGCCTGTCGACTAAGCGGAACGATATGTTCCTCTTTCATCTTCA
TGCCTCG
GTACGAATAACGCACACCTTTAATTTCTTCTCGCTTTGCAGGTATACGCCAAAGAGATTTATCGAAGTCGAATTCATCC
CAACGT
IV
GCGAAACGTAATTCACTGGAACGCACAAAAGTTAGCAAGGAAAGCTTGACCGCAATCCGTGTCATTACACGGCCACGAT
ATGC n
,-i
AGCAAGACGTGCAAGAAACTCAGGGAATCGGCTAGAAGGTAAAGCAGGGTAATGTCGCGCTTTGGTTGTCGATAGCGCA
CCA t=1
tl
GCCATATCGCTGGCTGGATTTGAGTCGATGTAATCGTTCTGTACGGCATAACGCATAATGGCTGTGACGCGCTGTTGCA
GGCG 2
o
CTGAGCGACATCGTGTTTGCCACTGGCATCAACTTTTTTAATCGGGGCTAACAGGTGGCTGGTTTTAAGCTGCCGAATG
TCGGA 'a
o
CGAACCGATATGAGGGAATATATAAAGCTCAAGATAGCGAAGAACGCGCGATCGATGGTCTTCACTCCAGCGCTTGTTA
CTGG tt
n.)
un
CATGCCATTCTCGGGCGATAGCTTCAAAAGTATATGCCCCCGAATTCTCGGCCCGAGCTTCTTTTTGTACGACTTTTGG
GTCTAT
0
GCCCTGTACTAAAAGCTTTTTAGCTTCATCGCGTTTTGCTCTTGCCTGAGCAAGCGTCACAGTAGGCCAAACACCAAAC
GCAAG w
o
w
ACGATCCTCTTTTTTGTCAGAGGGACGTCTGTATTTCATGCGCCAGTATTTGGAACCTTTGGCCGAAACCTCAAGATAC
AAACCA
i-J
CCACCATCGGCCATTTTGTAGGTTTTGTCTTTTGGCTTTGCGGTCTCGACCTGTCTGGCGTTGAGCTTCATTTGGGGGC
ACATTT
4,.
w
CTAATCGAAGTTAAGATGCCCCCAATTATGCCCCCAATGATATCCGGATTTCAACGGACAACCTCGGAAGACGCAGGAC
GAAA oe
AAATCGCTGCAAGCATTGATTTTAAAGGGATATTTGGACTTTCTCGGATGGTCTTGGAAGTAAGAATGGTGCCGAAGGC
CGGA
CTCAAACATCAAAATAAGTTAATGATAAAAAACAAATAATAAAACACAACAATGAAATATGCCCCCTTTTGTGCCCCCA
CTGTTT
TTCTGACCAATCTATTTTCAGCCCATCAATAAATCGGAAAGTTAAATCATTTTTAATCAGTAAGTTTGGATCCGTAGCT
CGGATC
CAAACCAGTGCATCTTTTATCCACATAAAAAATTTTTTTTCGAAAGAACTGTTCACACTGTTCACCTTTCTGTTTTCTC
CTTTTATT
TCAGAGTGATAGGTGGTGAATAATGGGTGAAGGGTGAACATTCGATTCTTCACCTCCGGCATTCTGCCGATGTGACTCA
TACC
GGTGATTAATCCTCCGCACTGAAATCACTCAGGAAGAAAAAAGTTTTTTTTGATTTGATTGTTCACACTGTTCACCTTT
CGTTTTT
P
CTCTTTTAATTTCAGTGTGATAACGGGTGAATATACGGTGAAGGGTGAACAGTGGATTGTTCACCTTCGGGGGATATCG
GGAT .
,
AAAAAAAGACCGGCAGATGCCGGTCAGGTGGGTCAGGCTGTTGTAGGGTCGTCACATTTTGG CAG
CCAGTCGCCGTAGCTTTC
,
1¨
.3
--4
CTCTTTCAGCGTCAGGTTGGTCTGTATCCCCTGTTTGGTATGGCGTTTCTCGTAATTCAGTCCGTATTCCTTCAGCATC
ACCGGCA .
w
GCCCCAGCCCGAACATTTTCAGACTGAGTACATTCCGGTAGCCGTTTGCCTCCATGTAGGCCAGATAGGCGTGATAGAG
GTATT "
,
,
,
TACGGTAATTGCGCGGGATGATACTGGCGTTCCCCATATACATGCCGCTGGTCTGCGGCAGGGTTTCCAGATAGCCGAT
AAAA 0
,
N,
N,
TCAAACGTCGGGTCGGCATCCCGTTTGATGTTCAGTGCCTCGTCTGAGTTCTGCTGGGACTGAAGCAGTGACCGGGCGA
GCAT
CGGGTCGCTGAACTTCTGCATCAGGTGACGCACGATGACCGCCAGCTCGCGGGTGATTTTGTCCTTAAGCTGCGGGTCG
CGCT
CCTGCGGGGCTATCTGTTCCGGGAAGTGAATAATCACCCGTCGGCGTGACACGCCGCCGCTGCGGTCGGTGAAGCGCAT
CGG
GTTATTGTTCACGGCCAGAATCACCGCCGGGATGTGCGTGGAGTACGCATCCCGGTATTTCGGGTCAACGGACACCGCA
TCGC
CGCCGGTGATGGCCTTGAGTCCGGCACCGTCGCCGCTCCATTTTTCCTGGTCCGGCAGGCGTATCAGTGAGAAGCCAGT
TAAC
GCGGCACGTTCACGCGGGGATTCCAGCGTCTCGATGGTGGCCGACGTGGCGTTATCCTCCCCGGCCAGCAGGGTGGCTA
TTTC 1-d
GGCCATGATACT
n
,-i
m
,-o
21 Sequence removed
TTTGAGCGGCATAAATTACCACTGAAAGCCCTTAAACGTTACTCTACTGTGGACACTGTGTGGACACTCTCGG
CCTCAGTACCA w
o
w
from P2 (replaced
CCTCTTAGCGGATTAAGAGAAATGGCGTCCTGAAGGTACTCTGGCGCAAAATGAGCGTAAACCATAGTTTGCTCAATCC
GCGT =
'a
GTGACCTAGTATCCGTTGTAGCGTGATAATACTTCCTCCATTAATCATGAAATGAGTGGCAAAGCTGTGCCTTAGTGCA
TGTGT o
4,.
w
w
vi
with kanamycin
GGCTTGCCCCATTGGCAAATCCGGTTTTATTGCTTTCATTGTTCGTCTGAAGCGAGGGTAATCAGCATCAGGGAATAAA
AAACC
0
marker; Example 3)
TCGTTTGTTATCCGCGATCATTTTGGCAACAGCCTCTGAGATCGGGACGGTGCGTGGTTTGTTTGTTTTCGTTTTAACA
AACGTG w
o
w
ACGCGGTTATGGATGATATTTTCTGCTTTCAAACGAGCTGCTTCTCCCCAACGTGCTCCTGTACTCAGGCAAAGAATCG
CAATCT
i-J
TTTTATTGTCGCCGTCAAGTGCTGCAAGCAGTAAGGCAATTTCTTCCTGTGTTAGATAGCCTGTTTCTGGTTTTTCCTC
CTTAAGC
4,.
w
CTTTTTGTCCCTCTGATAGGGTGCTCACCAAAGAATAACTCCGCTTCAATCAGGGCTGTAAACATGCCGCTAATACATG
TTAAAT oe
CACGATTGATACTCGAAGGTTTAATACCCTGACTTCTTCGGGTGGCGCAGTACTGGCTGATAAGCGATTTCGTAATTTG
AAATG
CGCATGGGTCATTCGTTATTTTTGTGAAGATTTCAA 11111
CCAAGATTAGATTTCCCATGCTCTTCGTGTTTACCCTTTAAATCCC
ACCAGATCTGTGTCAGCTCCGACAGACGTCGCTTGTCTGTTGGTTTTGATAGCCATTCTTTATTGTGGTGGTTGTACAA
CGTGTA
TTTCTCGAAAGCGACAGCTTCGCTTTTCTTATCAAACTTCCTACGGATGCGTTTTCCATTACGTCCAGTAGGGCGGATG
TCCACT
TCATATCGACCATCATCGAGTTTTTTGATTGCCATCAGAAAACCCTCCGAGTGGTGTGTTTTTTTGCGACTACTAATCG
CTTTTTT
CGTGGTGGCTGAAATTTAGCCACCAATAGTAGGCACTTGTGATGAATATATTCACGATGAATTGTTAACCAGTCTTTTG
ACCGG
P
AGTGGGGCGACGTTGTTTCGTTTTGCCCAAAGTGTGCGAGAGCGGGCGCAATTTGCCCGGACTCAGGAGCGATCTGATT
GGTC .
,
ATGAACCATAAAGTGTATTTG GTGAATTGTG G G GTCTG CAG GATGTTCATCATGACATCTGTTG G AG
GTGTTG AACGACCACTT
,
1¨
.3
--4
TCATAGTAACTCAGCGTGCCATACGGAACCCCTGTTAAATCAGCAAGTTGTTGTCTGCTCAAATACTCTGATTTTCGCA
TTAAGA .
CTATCTTCTCGCTTATCGTGTTTGACATGGTGTTTAGATCTCAATAGTATTTAGTTTAGATGTAGATTGTTTAGTGCTT
GGATGTG "
,
,
,
GGCACTAAAAGGCATTATAAGACATTAAACGCAATTCATGAGGGCTAGAGGACGACATGAGCAAGCAAGTAACACTCAT
GAC 0
,
N,
N,
TGATGCGATTCCTTATCAGGAGTTCGCAAAACTAATAGGAAAATCGACAGGAGCGGTTCGTCGGATGATCGATAAAGGA
AAG
CTGCCTGTAATTGATATGACCGATCCACAATCAGCTTCAGGTCGTGCAGGTGAATATTGGGTATACCTTCCGGCATGGA
ATAAC
GGACTAAAACTGGCTTATGAAAGCCGCCCTAAAGAGATTCGTGACGGCTGGTTGATGTGGTTAGGTCTCGGTGAACCAC
GTTA
AGGAGAACCGTATGAATGAGCCTCGTTGTATTGCTCAGTTACTGCGTAACGAAAGCCCCAGGGCGATTGACTTCACCAT
CACC
CACGGTAAGGGGCGTAAGGGAATCATTATCCGCACCAAAAAACAGAGTCCGTTAAAAAAGGCTCTGACCTTTCTGAAAA
GCCG
GAGGGTATGGAAATGACAGTGATGACGCTCAATCTCGTTGAAAAACAGCCAGCAGCTATGCGCCGGATAATTGGTAAGC
ATCT 1-d
TGCCGTTCCTCGCTGGCAGGATACATGTGATTATTATAATCAGATGATGGAGCGCGAACGGTTAACGGTTTGCTTCCAT
GCTCA n
,-i
GTTAAAACAGCGTCACGCAACGATGTGTTTTGAAGAAATGAACGACGTCGAACGTGAACGACTGGTATGTGCAATTGAT
GAAT t=1
1-d
w
TGCGTGGTGCATTCTCAAAACGCCGTCAGGTTGGCGCAAGTGAGTATGCATATATTAGTTTTTTAACAGTCAGTCAGCG
TCGTA =
w
o
CTTTATTTATGCATGCCGGATTGACTGAAAAAGAATTCAACCAGCCATACTGGCGAATTAATGAAGAGTCATGTTACTG
GCGTG 'a
o
ATG CTTTATTCCGTG CATTACGTGAATTATTCAGTCTGTTTGAGTATG
CACCGACAATTCTGACGTCGGTAAAACCG GAG CAAT
w
w
vi
ATCTGCATTAAGTAATTAACCAGAGTTTTTAACGCACTTAATTGTGCGGGGCTTCTTTTTGCCTGGAGAAAGTTATGCA
TACAGT
0
TTCTGAAAATCAGTGCGGTATATACGCATTACTGCTGCAACAGGCCAGAACCGAAGCACAGGCCGACGCTGCGACGCGC
TTTT n.)
o
n.)
CTTCTCATCTTGATGCCATGATTCGCCACATCACAAAGGCGGAGTTATCCCGCGTGGAGATAGTCGAGCTGCTCAGTCA
GGAGT
i-J
CGGAAAAATTTCACAATATCGGATTGTCTCGCGGGGAGGTGCTTTGATGTTCTGTTCTCGTGCAGTTGTATTACTGAAT
AACGC .6.
.6.
n.)
CTTAAAAATCGCCGTTATGAAAAATGGCGATTTGTCTCTTATTCAACTTGGTCTTGATAAAGAAAAACGCGAAATAACT
GAATC oe
TGTTATCGCGATTTATCAGAGTGAATTAAACCTCCTGTCTGATGTGGTCAATTTACTTGTTAAACGCGCTGTATTTCAC
AAGCAA
ATTTCCTCCGTGGATGAACTGACGAAATTAACGACAGAAATCGCCAGCTATTGCGCTGATGAATTTAAAAAACTGAACG
ACAA
AAGGAACTGGTAATGCCGGACAACGTAGATTTTATTCAGGAACAACAGGCTGAATTACTGGAGCGCCAGATTAACGCGG
CAA
GGGTAAAACATTGCGGTGCTTCTGCGCTGGTTTGCGAAGAGTGTGACGCGCCAATACCTGCTGCCCGTCGTGCGGCTTA
CCCG
TCAGCCACGCGTTGTGTTTCCTGTCAGTCAGTCTTTGAAGCAAAAAACAAACATTACCGGAGAACGGCATGAGTATTCG
TATTG
AAATTGGCGAACGTTATGTCGTTACCAGTGACAGCTTTCAGTTTATTCTCCACGAGAAAAAGAGAGCGGAAAGCGGTAA
AAAC
P
GCCGGTCAGGAATGGCTGGCGGTGGTTGGTTATTACCCGAAATTAAGCCAGCTCGTTTCAGGCCTGATGCATCACGATA
TTCT
2
GACCGGAAGCGCAAAGTCTTTTGCTGATTTAAACGTGCAGGTTGAGCAACTCAGCAAGCGTTGTTCAGAGGCTTTTGGC
TCAT
--.1
ATGGCCGTTAAAGCCTCCGGGCGTTTTGTCCCTCCGTCAGCATTTGCCGCAGGCACCGGTAAGATGTTTACCGGTGCTT
ATGCA 2
.6.
.3
TGGAACGCGCCACGCGAGGCCGTCGGGCGCGAAAGACCCCTTACACGTGACGAGATGCGTCAGATGCAAGGTGTTTTAT
CCA 2
1-
CGATTAACCGCCTGCCTTACTTTTTGCGCTCGCTGTTTACTTCACGCTATGACTACATCCGGCGCAATAAAAGCCCGGT
GCACGG
.31
.3
GTTTTATTTCCTCACATCCACTTTTCAGCGTCGTTTATGGCCGCGCATTGAGCGTGTGAATCAGCGCCATGAAATGAAC
ACCGAC
GCGTCGTTGCTGTTTCTGGCAGAGCGTGACCACTATGCGCGCCTGCCGGGAATGAATGACAAGGAGCTGAAAAAGTTTG
CCG
CCCGTATCTCATCGCAGCTTTTCATGATGTATGAGGAACTCAGCGATGCCTGGGTGGATGCACATGGCGAAAAAGAATC
GCTG
TTTACGGATGAGGCGCAGGCTCACCTCTATGGTCATGTTGCTGGCGCTGCACGTGCTTTCAATATTTCCCCGCTTTACT
GGAAA
AAATACCGTAAAGGACAGATGACCACGAGGCAGGCATATTCTGCCATTGCCCGTCTGTTTAACGATGAGTGGTGGACTC
ATCA
GCTCAAAGGCCAGCGTATGCGCTGGCATGAGACGTTACTGATTGCTGTCGGGGAGGTGAATAAAGACCGTTCTCCTTAT
GCCA
IV
GTAAACATGCCATTCGTGATGTGCGTGCACGCCGCCAAGCAAATCTGGAATTTCTTAAATCGTGTGACCTTGAAAACAG
GGAA n
1-3
ACCGGCGAGCGCATCGACCTTATCAGTAAGGTGATGGGCAGTATTTCTAATCCTGAAATTCGCCGGATGGAGCTGATGA
ACAC t=1
tl
CATTGCCGGTATTGAGCGTTACGCCGCCGCAGAGGGTGATGTGGGGATGTTTATCACGCTTACCGCGCCTTCAAAGTAT
CACC 2
o
CGACACGTCAGGTCGGAAAAGGCGAAAGTAAAACCGTCCAGCTAAATCACGGCTGGAACGATGAGGCATTTAATCCAAA
GGA 'a
o
TGCGCAGCGTTATCTCTGCCATATCTGGAGCCTGATGCGCACGGCATTCAAAGATAATGATTTACAGGTCTACGGTTTG
CGTGT tt
n.)
un
CGTCGAGCCACACCACGACGGAACGCCGCACTGGCATATGATGCTTTTTTGTAATCCACGCCAGCGTAACCAGATTATC
GAAAT
0
CATGCGTCGCTATGCGCTCAAAGAGGATGGCGACGAAAGAGGAGCCGCGCGAAACCGTTTTCAGGCAAAACACCTTAAC
CAG n.)
o
n.)
GGCGGTGCTGCGGGGTATATCGCGAAATACATCTCAAAAAACATCGATGGCTATGCACTGGATGGTCAGCTCGATAACG
ATAC
i-J
CGGCAGACCGCTGAAAGACACTGCTGCGGCTGTTACCGCATGGGCGTCAACGTGGCGCATCCCACAATTTAAAACGGTT
GGTC .6.
.6.
n.)
TGCCGACAATGGGGGCTTACCGTGAACTACGCAAATTGCCTCGCGGCGTCAGCATTGCTGATGAGTTTGACGAGCGCGT
CGAG oe
GCTGCACGCGCCGCCGCAGACAGTGGTGATTTTGCGTTGTATATCAGCGCGCAGGGTGGGGCAAATGTCCCGCGCGATT
GTC
AGACTGTCAGGGTCGCCCGTAGTCCGTCGGATGAGGTTAACGAGTACGAGGAAGAAGTCGAGAGAGTGGTCGGCATTTA
CGC
GCCGCATCTCGGCGCGCGTCATATTCATATCACCAGAACGACGGACTGGCGCATTGTGCCGAAAGTTCCGGTCGTTGAG
CCTC
TGACTTTAAAAAGCGGCATCGCCGCGCCTCGGAGTCCTGTCAATAACTGTGGAAAGCTCACCGGTGGTGATACTTCGTT
ACCG
GCTCCCACACCTTCTGAGCACGCCGCAGCAGTGCTTAATCTGGTTGATGACGGTGTTATTGAATGGAATGAACCGGAGG
TCGT
GAGGGCGCTCAGGGGCGCATTAAAATACGACATGAGAACGCCAAACCGTCAGCAAAGAAACGGAAGCCCGTTAAAACCG
CAT
P
GAAATTGCACCATCTGCCAGACTGACCAGGTCTGAACGATTGCAGATCACCCGTATCCGCGTTGACCTTGCTCAGAACG
GTATC
2
AGGCCTCAGCGATGGGAACTTGAGGCGCTGGCGCGTGGAGCAACCGTAAATTATGACGGGAAAAAATTCACGTATCCGG
TCG
--.1
CTGATGAGTGGCCGGGATTCTCAACAGTAATGGAGTGGACATGATGGCAAAAATTCACGAGGTAAAGCTGCACGCAAAA
TAT 2
un
.3
TTCGACCTTGTACTGGAAGGAAAGAAACGCGCAGAGTTTCGGAAAAAAGATCGTAATTATGAGCGCGGGGACACGTTGA
TTT 2
1-
TGCATGAATGGGTGCAGGGTGTGTTTACGGGGCGAAAGGTTGAAGCCCGGATAACAGATGTTACTGACCTGTCAGACTG
GCT
.31
.3
GGAAGATTATGTCTTGCTGCACAGAATACTAGGTGAGATACTGCCGCTGACTGTCAAGCTGGCAGTATCCTCCACATTT
TTTAT
AAATCTAAGGCGAAGAACGGTGGCAATATATTTGCTTTAAATTGTAAATAAGCAAATTATTCTAAGCCACAAATGATAT
CAATT
GAGTATCGATTTTCGTTCGAATATAACCATAACGCCAAGCGTCAAATGTAAGTCATCTATATGTGATTCGTATTCATAT
TGGTCA
GCATCAGGAGATAGTAAATGCTCTGCTGCTGTAAAAACATCAATTGTAACAGATATATCGTCTGGAATTTCTTCATTTA
ATTCGG
ATATGTATTTTTTATATTTGTTTTTTAATTTCCTTATCATGTCTTCCTTATGGTGGATTGTAGTGCAGTAA 1 1 1
1 1 TTTAGTTGAATC
ATTGTATGAATGGAATGAACAGTTTAATGCTAGGCCAAGACTTGAATTTCCTGTAGGGCCTGTGTGTTTAAAGTATTCA
AGATT
IV
TCTTCTTATGTCAGAAAATAAAAGACTTTTACAGGGGGCATTTCCTTTAACTTCTATTGCACATATTGGAATGTCGAAA
AATGGT n
,-i
CTGCTGTCCAATATGGCAATATCAATTCTGCCTGATCTTGTTGTGTTTGTGTTTTTTCTGACAGTTTTTTTTGAGAATA
TTTTTTGG t=1
tl
GGGTCAGACCGTTTCGATAAAGGAACTGTTGCATTAATGAATTTATTTGTATGGTACTCAAATATAACTTTGTAATCAC
CATGGC 2
o
GAAAAGATTTTATTTCTAACAACGATAGACCTATAGATACGGTAGCTACATATTCAGTGTTTATTTTTGCACCAGACCT
GAAATC 'a
o
CATTAAATAAGTTGCGTCGGCTGACTTTTTCATTCCATCTTTTACAGCTTTCACAATATCCATGTTATTCATATCAACA
CCTTTTAT tt
n.)
un
ATTCCAGAATTATAAATCTTAATGATTGATTTGTTTGTGTGTTTAGGTTACTAGATTGACGTACTTATAGCACATTCTG
ATTTTGA
0
AATC AC CATC CTT G G GTAAAG C AC AG CCTATCACTATTG G TTTA G TACTG T G CTG
TAATAG TAT CA G ATTAA G TTAAATC C ATTT n.)
o
n.)
TATGAAATCTTCCATTTCATCATATT 1 1 1 1 1
ATTACTGCTCTAACATCTTTGCTTTCATCACATAAACAATCTAAAAGACTATCCCA
i-J
TTCAGTTTGTTTTTTACCAAATCCATAAACAGCCTCTATATCACCGGATTTCCATATATAGATTCCGTTATCTTTCAAT
TTTTGATG .6.
.6.
n.)
TATTTCATGTATATG CTTTT GT G TTTTTTCATG GTTACATATTTTGATGAAGTCTTTG CTTGAAATG
CTTTTG AATG A CTCAAAA G oe
TA G TAACTTTTG TATTTAAACTAAG CTCTC CTG A GTTG ATTG ATT CAATC AAAG C AG T CAATA
G ATTATC AC ATT CA GTA CTAA G
TAAGTCTTTATG CTCTGTTAATAGAATATTTGATAAGAAATCACAATCAG CTAGAATCCTTGTTTTTATG C C G
AT G G CATTGATG
ATTTGTG AC AT CTTAAATAAA CTAC CCTTACC GTC AACG G CAA CAATG CAGATTTTACTCG G
GTTGAGTTCATGTCCGTTAATTT
TTTTATAAAGTG C ATATA G AAC G TTT GT CT CTG TTTTC C CTT CAAC AA G
CAAAACTTCTTCAGAAAATAAAAG GTATGATGAATT
AG AAAG CGTGAATG CTG AG TG CAATTG CG GTGATG AAGATTTATATAATTCTTC G ATTTTTTC AG
ATATAGTCTTCCTTG CTATG
GTTCCATTAGAATCCTTACAAACCTGAATCG CATTTG CTGCATG CTTTG CAGAAAG CATACTG G CTG
AGTG AG TTG ATATTATA
P
AC CTG ATAC CCTG ATTCA CTTAATGTG AC AA GTG ATTCT CTG ACAG AATTAATG G CTGAAG G
GTGTAAATATAA CT CAG GTTC A
2
TCAATGAAAATCAAAGTGTTTGATTTTTTTG ATTCG CTGTTTTCTTTTTTTATTTCG G C CA G G TATTG
AATTAATG CCATTTGAAT L."
--.1
GGAACGTTGTGTTCCGTGACCAAATCGGCTGATATCTCTCATTACCGGTTCATCTTCCCGAGACTCAAAAACTTTCAGA
GTGCC 2
o .3
G GATTTAAATATCTCATCTAATGTCG GT G TG G G AAAG T G TAACTTTA CA CTTAC GT CA G G
AAAAAATTG GTTTACTTTTTTATTT 2
1-
AC AC CT G AG T CTATTTTATTAA G G CTTT CTAATCTG TT CTC AC C G TT GT G A G AAAG
ATATTTA C CTATTTCTG ATAT G TTTTTTG A
.31
.3
AAATTTTTCTTC GTG TTCTTG TTTTATTTC AG AAA CAATTG C G G AAAG TAT CTTTC CTATTGTG
GTCGTGTTTTTG CATTTTG TTG A
GTCTTCGACAG CGTCAGACATTG CAG G GATATGAATTGGTTCCG GAAATATATTG G AG ATTG CAC
CATCTATG C CG CCAG G GT
TTTTCTTCCACGTGGTACCGTCATATACATCCAGACTTTTTTTGGCTTTTCCTGTTTCCTTATTAAATTCCTGTCTTCT
TGCAAAGG
TAAG AGTC CC GT CAATTATAAACG GAG CTATTTTTTGTTG ATTCTCTTCTG TTAA CAAAGACAG AG
TAT CATCTGTTATAC CTTG
AATAACGCCTTCAACGGATACGGGGTGTGTAGGATCGTACACATCTGATTCTGAAATCAAAGAGCCATCTAACAGCCAC
TTAAT
TG CTAAGATAATATTTGATTTTCCTGCGTTATTATAACCAACTAAAG CAGTGAAG G G G CG CAAAATAG C
CG ATG TTG ACTTA CA
IV
AG AAC G AAAG TTG CTAATTG AAA CT G AA G CAAG A C G TACA G TC ATTATTTTT C
CTTAAAT G TG CTATTTG TAT G CAATG AG TTC n
,-i
ATACGAAACGCTTTTTTACATTTTATAGTCGTTGCATTCAAGGGTGCATGAGATTGCATTAAGGGAAACTGTGATATGG
CTTGG t=1
tl
CTTTTGACTG GAAATACTGATG G CTCATTAGTTTTATTAAG GTG CATTAAAACCG C CC CGTG AAG
CGGG CG G G CG AG G CG G G G 2
o
'a
o
.6.
n.)
n.)
un
AAAGCACGGCGAGGCGGGGAAAGCACTGCGCGCTGACGGTGGTGCTGATTGTATTTTTTCAGCGTCTCAGCGCGTCGTG
ACG
0
GCACTTAGTCTGCCCGTTGAGGCGTTGTGTGTCTGCGGGGTGTTTTGTGCGGTGGT
w
o
w
o
22 WilcktypecloDF13
GCGCTGCGGACACATACAAAGTTACCCACAGATTCCGTGGATAAGCAGGGGACTAACATGTGAGGCAAAACAGCAGGGC
CGC
w
4.
4.
on
GCCGGTGGCGTTTTTCCATAGGCTCCGCCCTCCTGCCAGAGTTCACATAAACAGACGCTTTTCCGGTGCATCTGTGGGA
GCCGT w
m
GAGGCTCAACCATGAATCTGACAGTACGGGCGAAACCCGACAGGACTTAAAGATCCCCACCGTTTCCGGCGGGTCGCTC
CCTC
TTGCGCTCTCCTGTTCCGACCCTGCCGTTTACCGGATACCTGTTCCGCCTTTCTCCCTTACGGGAAGTGTGGCGCTTTC
TCATAGC
TCACACACTGGTATCTCGGCTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTAAGCAAGAACTCCCCGTTCAGCCCGA
CTGCT
GCGCCTTATCCGGTAACTGTTCACTTGAGTCCAACCCGGAAAAGCACGGTAAAACGCCACTGGCAGCAGCCATTGGTAA
CTGG
GAGTTCGCAGAGGATTTGTTTAGCTAAACACGCGGTTGCTCTTGAAGTGTGCGCCAAAGTCCGGCTACACTGGAAGGAC
AGAT
TTGGTTGCTGTGCTCTGCGAAAGCCAGTTACCACGGTTAAGCAGTTCCCCAACTGACTTAACCTTCGATCAAACCACCT
CCCCAG
GTGGTTTTTTCGTTTACAGGGCAAAAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATC
P
.
,
1-, 23 MutantcloDF13ori
GCGCTGCGGACACATACAAAGTTACCCACAGATTCCGTGGATAAGCAGGGGACTAACATGTGAGGCAAAACAGCAGGGC
CGC ,
--.1 (higher copy number
GCCGGTGGCGTTTTTCCATAGGCTCCGCCCTCCTGCCAGAGTTCACATAAACAGACGCTTTTCCGGTGCATCTGTGGGA
GCCGT
on cloD13_cop3;
GAGGCTCAACCATGAATCTGACAGTACGGGCGAAACCCGACAGGACTTAAAGATCCCCACCGTTTCCGGCGGGTCGCTC
CCTC ,
,
,
,
single mutation
TTGCGCTCTCCTGTTCCGACCCTGCCGTTTACCGGATACCTGTTCCGCCTTTCTCCCTTACGGGAAGTGTGGCGCTTTC
TCATAGC
versusmMcktypeis
TCACACACTGGTATCTCGGCTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTAAGCAAGAACTCCCCGTTCAGCCCGA
CTGCT
shown underlined
GCGCCTTATCCGGTAACTGTTCACTTGAGTCCAACCCGGAAAAGCACGGTAAAACGCCACTGGCAGCAGCCATTGGTAA
CTGG
andinbold
GAGTTCGCAGAGGATTTGTTTAGCTAAACACGCGGTTGCTCTTGAAGTGTGCGCCAAAGTCCGGCTACACTGGAAGGAC
AGAT
TTGGTTGCTGTGCTCTGCGAAATCCAGTTACCACGGTTAAGCAGTTCCCCAACTGACTTAACCTTCGATCAAACCACCT
CCCCAG
GTGGTTTTTTCGTTTACAGGGCAAAAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATC
Iv
n
,-i
m
,-o
t..)
=
t..)
=
'a
c.,
4,.
w
w
u,
Table 7: Example Plasmids and Copy Numbers
0
w
1
_______________________________________________________________________________
________________________________________ <
w
i Common Vectors Copy Number OF11
Incompatibliity Group Control
4-
4-
t.)
OC
pUC -500-700 0,481 (derivative)
A Relaxed
pBR322 -15-20 pME31
A Relaxed
PET -15-20 pBR322
A Relaxed
pG EX -15-20 pBR322
A Relaxed
pColE1 -15-20 ColE1
A Relaxed 0
0
1-
IpR6K -15-20 R6 K '
C Stringent .
0
1
.
oe
,pACYC -10 p15A
B Relaxed .
0 ..
i
..
i
IpSC101 -5 pSC 101
C Stringent
ipB1Liescript -300-500 0 1E1 (derivative) and Fl"
A Relaxed
pGENI -300-500 pUC and Fl -
A Relaxed
.0
n
,-3
ril
.0
w
<
F,
4-
t.)
t.)
!A