Note: Descriptions are shown in the official language in which they were submitted.
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
CONTENANT LES PAGES 1 A 234
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 234
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME:
NOTE POUR LE TOME / VOLUME NOTE:
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
OPTIMIZED TETRAHYDROCANNABINOLIC ACID (THCA) SYNTHASE
POLYPEPTIDES
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application is a continuation of U.S. Patent Application
Serial No.
62/902,300, filed on September 18, 2019, the entire disclosure of each of
which is
incorporated herein by reference in its entirety.
SEQUENCE LISTING STATEMENT
[0002] A sequence listing text (.txt) file is submitted herewith under 37
CFR.
1.821(c) and is hereby incorporated by reference in its entirely. The details
of the file as
required under 37 CFR. 1.52(e)(5) and 37 CFR 1.77(b)(5) are as follows: Name
of file is
DEMT 007 01W0 SeqList ST25.txt; date of creation is September 17, 2020; size
is
697176 bytes. The content of the sequence listing information recorded in
computer readable
form is identical to the written sequence listing (if any) and identical to
the sequence
information provided with the original filed application and contains no new
matter. The
information recorded in electronic form (if any) submitted under Rule 13ter
with this
application is identical to the sequence listing as contained in the
application as filed.
BACKGROUND
[0003] Plants from the genus Cannabis have been used by humans for their
medicinal properties for thousands of years. In modern times, the bioactive
effects of
Cannabis are attributed to a class of compounds termed "cannabinoids," of
which there are
hundreds of structural analogs including tetrahydrocannabinol (THC) and
cannabidiol
(CBD). These molecules and preparations of Cannabis material have recently
found
application as therapeutics for chronic pain, multiple sclerosis, cancer-
associated nausea and
vomiting, weight loss, appetite loss, spasticity, seizures, and other
conditions.
1
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
HO HO
OH 0
Cannabidiol / CBD Tetrahydrocannabinol / THC / Dronabinol /
Marino!
[0004] The physiological effects of certain cannabinoids are thought to
be mediated
by their interaction with two cellular receptors found in humans and other
animals.
Cannabinoid receptor type 1 (CB1) is common in the brain, the reproductive
system, and the
eye. Cannabinoid receptor type 2 (CB2) is common in the immune system and
mediates
therapeutic effects related to inflammation in animal models. The discovery of
cannabinoid
receptors and their interactions with plant-derived cannabinoids predated the
identification
of endogenous ligands.
[0005] Besides THC and CBD, hundreds of other cannabinoids have been
identified
in Cannabis. However, many of these compounds exist at low levels and
alongside more
abundant cannabinoids, making it difficult to obtain pure samples from plants
to study their
therapeutic potential. Similarly, methods of chemically synthesizing these
types of products
have been cumbersome and costly, and tend to produce insufficient yield.
Accordingly,
additional methods of making pure cannabinoids or cannabinoid derivatives are
needed.
[0006] One possible method is production via fermentation of engineered
microbes,
such as yeast. By engineering production of the relevant plant enzymes in
microbes, it may
be possible to achieve conversion of various feedstocks into a range of
cannabinoids,
potentially at much lower cost and with much higher purity than what is
available from the
plant. A key challenge to this effort is the difficulty of expressing plant
enzymes in the
microbe, particularly secreted enzymes such as the cannabinoid synthases,
which must
successfully traverse the microbe's secretory pathway to fold and function
properly.
Engineered variants of cannabinoid synthases, modified host cells, and new
methods are
needed to address these challenges.
Summary
[0007] The present disclosure provides engineered variants of a
tetrahydrocannabinolic acid synthase (THCAS) polypeptide comprising an amino
acid
sequence of SEQ ID NO:44 with one or more amino acid substitutions, nucleic
acids
comprising nucleotide sequences encoding said engineered variants, methods of
making
2
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
modified host cells comprising said nucleic acids, modified host cells for
producing
cannabinoids or cannabinoid derivatives, methods of producing cannabinoids or
cannabinoid
derivatives, and methods of screening engineered variants of the
tetrahydrocannabinolic acid
synthase (THCAS) polypeptide. The engineered variants of the disclosure may be
useful for
producing cannabinoids or cannabinoid derivatives (e.g., non-naturally
occurring
cannabinoids). The modified host cells of the disclosure may be useful for
producing
cannabinoids or cannabinoid derivatives (e.g., non-naturally occurring
cannabinoids) and/or
for expressing engineered variants of the disclosure. The disclosure also
provides for
modified host cells for expressing the engineered variants of the disclosure.
Additionally, the
disclosure provides for preparation of engineered variants of the disclosure.
[0008] An aspect of the disclosure relates to an engineered variant of a
tetrahydrocannabinolic acid synthase (THCAS) polypeptide comprising an amino
acid
sequence of SEQ ID NO:44 with one or more amino acid substitutions. In some
embodiments, the engineered variant comprises an amino acid sequence with at
least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%,
at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, or at least
99% sequence identity to SEQ ID NO:44. In some embodiments, the engineered
variant
comprises at least one amino acid substitution in a signal polypeptide, a
flavin adenine
dinucleotide (FAD) binding domain, a berberine bridge enzyme (BBE) domain, or
a
combination of the foregoing. In some embodiments, the engineered variant
comprises
substitution of at least one surface exposed amino acid.
[0009] In some embodiments, the engineered variant comprises at least one
amino
acid substitution at an amino acid selected from the group consisting of R31,
P43, P49, K50,
L51, Q55, H56, L59, M61, M61, S62, L71, S100, V103, T109, Q124, V125, L132,
S137,
H143, V149, W161, K165, N168, E167, S170, F171, P172, Y175, G180, N196, H208,
G235, A250, 1257, K261, L269, G311, F317, L327, K390, T379, S429, N467, Y500,
N528,
P539, P542, H543, H544, and H545. In some embodiments, the engineered variant
comprises at least one amino acid substitution selected from the group
consisting of R31Q,
P43E, P49E, P49K, P49Q, K50T, L51I, Q55E, Q55P, H56E, L59E, M61W, M61H, M615,
562Q, L71A, S100A, V103F, T109V, Q124D, Q124E, Q124N, V125E, V125Q, L132M,
5137G, H143D, W161R, W161Y, W161K, K165A, N1685, E167P, Y175F, G180A,
N196Q, N196V, H208T, A250T, I257V, K261C, G311A, F317Y, L327I, K390E, T3795,
Y500M, Y500V, N528E, P542E, P542V, H543V, H544A, H545D, and H545E
3
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0010] In some embodiments, the engineered variant comprises an amino
acid
sequence selected from the group consisting of SEQ ID NO:50, SEQ ID NO:52, SEQ
ID
NO:54, SEQ ID NO:56, SEQ ID NO:58, SEQ ID NO:60, SEQ ID NO:62, SEQ ID NO:64,
SEQ ID NO:66, SEQ ID NO:68, SEQ ID NO:70, SEQ ID NO:72, SEQ ID NO:74, SEQ ID
NO:76, SEQ ID NO:78, SEQ ID NO:80 SEQ ID NO:82, SEQ ID NO:84, SEQ ID NO:86,
SEQ ID NO:88, SEQ ID NO:90, SEQ ID NO:92, SEQ ID NO:94, SEQ ID NO:96, SEQ ID
NO:98, SEQ ID NO:100, SEQ ID NO:102, SEQ ID NO:104, SEQ ID NO:106, SEQ ID
NO:108, SEQ ID NO:110, SEQ ID NO:112, SEQ ID NO:114, SEQ ID NO:116, SEQ ID
NO:118, SEQ ID NO:120, SEQ ID NO:122, SEQ ID NO:124, SEQ ID NO:126, SEQ ID
NO:128, SEQ ID NO:130, SEQ ID NO:132, SEQ ID NO: 134, SEQ ID NO:136, SEQ ID
NO:138, SEQ ID NO: 140, SEQ ID NO:142, SEQ ID NO:144, SEQ ID NO:146, SEQ ID
NO:148, SEQ ID NO:150, SEQ ID NO: 152, SEQ ID NO:154, SEQ ID NO:156, SEQ ID
NO:158, SEQ ID NO:160, SEQ ID NO:162, SEQ ID NO:164, SEQ ID NO:166, SEQ ID
NO:168, SEQ ID NO:170, SEQ ID NO:172, SEQ ID NO:174, SEQ ID NO:176, SEQ ID
NO:178, SEQ ID NO:180, SEQ ID NO:182, SEQ ID NO:184, and SEQ ID NO:186.
[0011] In some embodiments, the engineered variant comprises an amino
acid
sequence selected from the group consisting of SEQ ID NO:50, SEQ ID NO:52, SEQ
ID
NO:54, SEQ ID NO:56, SEQ ID NO:58, SEQ ID NO:60, SEQ ID NO:70, SEQ ID NO:72,
SEQ ID NO:74, SEQ ID NO:76, SEQ ID NO:78, SEQ ID NO:80 SEQ ID NO:82, SEQ ID
NO:84, SEQ ID NO:86, SEQ ID NO:88, SEQ ID NO:90, SEQ ID NO:92, SEQ ID NO:94,
SEQ ID NO:96, SEQ ID NO:98, SEQ ID NO:100, SEQ ID NO:102, SEQ ID NO:104, SEQ
ID NO:106, SEQ ID NO:108, SEQ ID NO:110, SEQ ID NO:112, SEQ ID NO:114, SEQ ID
NO:116, SEQ ID NO:118, SEQ ID NO:120, SEQ ID NO:124, SEQ ID NO:126, SEQ ID
NO:128, SEQ ID NO:130, SEQ ID NO:132, SEQ ID NO: 134, SEQ ID NO:138, SEQ ID
NO: 140, SEQ ID NO:142, SEQ ID NO:144, SEQ ID NO:146, SEQ ID NO:148, SEQ ID
NO: 152, SEQ ID NO:156, SEQ ID NO:158, SEQ ID NO:160, SEQ ID SEQ ID NO:166,
SEQ ID NO:168, SEQ ID NO:170, SEQ ID NO:172, SEQ ID NO:174, SEQ ID NO:176,
SEQ ID NO:178, SEQ ID NO:180, SEQ ID NO:182, SEQ ID NO:184, and SEQ ID NO:186.
[0012] In some embodiments, the engineered variant comprises an amino
acid
sequence of SEQ ID NO:44 with at least 1, at least 2, at least 3, at least 4,
at least 5, at least
6, at least 7, at least 8, at least 9, at least 10, at least 11, at least 12,
at least 13, at least 14, at
least 15, at least 16, at least 17, at least 18, at least 19, at least 20, at
least 21, at least 22, at
least 23, at least 24, at least 25, at least 26, at least 27, at least 28, at
least 29, or at least 30
4
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
amino acid substitutions. In some embodiments, the engineered variant
comprises an amino
acid sequence of SEQ ID NO:44 with 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13,
14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 amino acid
substitutions.
[0013] In some embodiments, the engineered variant comprises at least one
immutable amino acid in a flavin adenine dinucleotide (FAD) binding domain, a
berberine
bridge enzyme (BBE) domain, or a combination of the foregoing. In some
embodiments, the
engineered variant comprises at least 1, at least 2, at least 3, at least 4,
at least 5, at least 6, at
least 7, at least 8, at least 9, at least 10, at least 11, at least 12, at
least 13, at least 14, or at
least 15 immutable amino acids in the FAD binding domain. In some embodiments,
the
engineered variant comprises at least 1, at least 2, at least 3, at least 4,
at least 5, at least 6, at
least 7, at least 8, at least 9, at least 10, at least 11, at least 12, at
least 13, at least 14, or at
least 15 immutable amino acids in the BBE domain.
[0014] In some embodiments, the engineered variant comprises at least one
immutable amino acid selected from the group consisting of A28, F34, L35, C37,
L64, N70,
P87, 193, C99, R108, R110, G112, E117, G118, S120, P126, F127, D131, D141,
W148,
G152, A153, L155, G156, E157, Y159, Y160, N163, A173, G174, C176, P177, T178,
V179, G182, G183, H184, F185, G187, G188, G189, Y190, G191, P192, L193, R195,
A201, D202, 1205, D206, V210, G214, G223, D225, L226, F227, W228, R231, G234,
S237,
F238, G239, K245, 1246, L248, V251, V259, Q276, F312, S313, L323, C341, F352,
S354,
F380, K381, 1382, K383, D385, Y386, 1391, M412, L415, G419, M422, 1425, 1430,
P431,
P433, H434, R435, G437, Y440, W443, Y444, 1445, 1464, Y465, M468, T469, Y471,
V472,
P476, R484, N498, A502, N513, F514, K521, N528, F529, E533, Q534, and S535. In
some
embodiments, the engineered variant comprises at least one immutable amino
acid selected
from the group consisting of C37, N70, 193, C99, E117, S120, F127, D131, G156,
E157,
Y159, G174, C176, G182, G183, F185, G187, G188, G189, Y190, G191, P192, R195,
D202, D206, G214, W228, G234, F238, and L248.
[0015] In some embodiments, the engineered variant comprises at least 1,
at least 2,
at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at
least 9, at least 10, at least 11,
at least 12, at least 13, at least 14, at least 15, at least 16, at least 17,
at least 18, at least 19, at
least 20, at least 21, at least 22, at least 23, at least 24, or at least 25
immutable amino acids.
[0016] In some embodiments, the engineered variant produces
tetrahydrocannabinolic acid (THCA) from cannabigerolic acid (CBGA) in a
greater amount,
as measured in mg/L or mM, than an amount of THCA produced from CBGA by a
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
tetrahydrocannabinolic acid synthase (THCAS) polypeptide having an amino acid
sequence
of SEQ ID NO:44 under similar conditions for the same length of time. In some
embodiments, the engineered variant produces THCA from CBGA in an amount, as
measured in mg/L or mM, at least 5%, at least 10%, at least 15%, at least 20%,
at least 25%,
at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least
60%, at least
70%, at least 80%, at least 90%, at least 100%, at least 150% at least 200%,
at least 500%, or
at least 1000% greater than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time.
[0017] In some embodiments, the engineered variant produces
tetrahydrocannabinolic acid (THCA) from cannabigerolic acid (CBGA) in an
increased ratio
of THCA over another cannabinoid (e.g., cannabichromenic acid (CBCA)) compared
to that
produced by a tetrahydrocannabinolic acid synthase polypeptide having an amino
acid
sequence of SEQ ID NO:44 under similar conditions for the same length of time.
In some
embodiments, the engineered variant produces THCA from CBGA in a ratio of THCA
over
another cannabinoid (e.g., CBCA) of about 11:1, about 11.5:1, about 12:1,
about 12.5:1,
about 13:1, about 13.5:1, about 14:1, about 14.5:1, about 15:1, about 15.5:1,
about 16:1,
about 16.5:1, about 17:1, about 17.5:1, about 18:1, about 18.5:1, about 19:1,
about 19.5:1,
about 20:1, about 25:1, about 30:1, about 35:1, about 40:1, about 45:1, about
50:1, about
60:1, about 70:1, about 80:1, about 90:1, about 100:1, about 150:1, about
200:1, about 500:1,
or greater than about 500:1.
[0018] In some embodiments, the engineered variant comprises a truncation
at an N-
terminus, at a C-terminus, or at both the N- and C-termini. In some
embodiments, the
truncated engineered variant comprises a signal polypeptide or a membrane
anchor. In some
embodiments, the engineered variant lacks a native signal polypeptide. In some
embodiments, the engineered variant comprises a truncation of at least 1, at
least 2, at least
3, at least 4, at least 5, at least 6, at least 7, at least 8, at least 9, or
at least 10 amino acids at
the C-terminus. In some embodiments, the engineered variant comprises a
truncation of 1, 2,
3, 4, 5, 6, 7, 8, 9, or 10 amino acids at the C-terminus.
[0019] Another aspect of the disclosure relates to a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant of the disclosure. In some
embodiments,
the nucleic acid comprising a nucleotide sequence encoding an engineered
variant of the
disclosure comprises a nucleotide sequence selected from the group consisting
of SEQ ID
6
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NO:49, SEQ ID NO:51, SEQ ID NO:53, SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59,
SEQ ID NO:61, SEQ ID NO:63, SEQ ID NO:65, SEQ ID NO:67, SEQ ID NO:69, SEQ ID
NO:71, SEQ ID NO:73, SEQ ID NO:75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81,
SEQ ID NO:83, SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO:93, SEQ ID
NO:95, SEQ ID NO:97, SEQ ID NO:99, SEQ ID NO:101, SEQ ID NO:103, SEQ ID
NO:105, SEQ ID NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID
NO:115, SEQ ID NO:117, SEQ ID NO:119, SEQ ID NO:121, SEQ ID NO:123, SEQ ID
NO:125, SEQ ID NO:127, SEQ ID NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID
NO:135, SEQ ID NO:137, SEQ ID NO:139, SEQ ID NO:141, SEQ ID NO:143, SEQ ID
NO:145, SEQ ID NO:147, SEQ ID NO:149, SEQ ID NO:151, SEQ ID NO:153, SEQ ID
NO:155, SEQ ID NO:157, SEQ ID NO:159, SEQ ID NO:161, SEQ ID NO:163, SEQ ID
NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID NO:171, SEQ ID NO:173, SEQ ID
NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID NO:181, SEQ ID NO:183, and SEQ ID
NO:185. In some embodiments, the nucleic acid comprising a nucleotide sequence
encoding
an engineered variant of the disclosure comprises a nucleotide sequence
selected from the
group consisting of SEQ ID NO:49, SEQ ID NO:51, SEQ ID NO:53, SEQ ID NO:55,
SEQ
ID NO:57, SEQ ID NO:59, SEQ ID NO:69, SEQ ID NO:71, SEQ ID NO:73, SEQ ID
NO:75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81, SEQ ID NO:83, SEQ ID NO:85,
SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO:93, SEQ ID NO:95, SEQ ID NO:97, SEQ ID
NO:99, SEQ ID NO:101, SEQ ID NO:103, SEQ ID NO:105, SEQ ID NO:107, SEQ ID
NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID NO:115, SEQ ID NO:117, SEQ ID
NO:119, SEQ ID NO:123, SEQ ID NO:125, SEQ ID NO:127, SEQ ID NO:129, SEQ ID
NO:131, SEQ ID NO:133, SEQ ID NO:137, SEQ ID NO:139, SEQ ID NO:141, SEQ ID
NO:143, SEQ ID NO:145, SEQ ID NO:147, SEQ ID NO:151, SEQ ID NO:155, SEQ ID
NO:157, SEQ ID NO:159, SEQ ID NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID
NO:171, SEQ ID NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID
NO:181, SEQ ID NO:183, and SEQ ID NO:185. In some embodiments of the nucleic
acids
of the disclosure, the nucleotide sequence is codon-optimized.
[0020] An aspect of the disclosure relates to a method of making a
modified host cell
for producing a cannabinoid or a cannabinoid derivative, the method comprising
introducing
one or more nucleic acids comprising a nucleotide sequence encoding an
engineered variant
of the disclosure into a host cell.
7
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0021] Another aspect of the disclosure relates to a vector comprising
one or more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure.
[0022] An aspect of the disclosure relates to a method of making a
modified host cell
for producing a cannabinoid or a cannabinoid derivative, the method comprising
introducing
one or more vectors comprising one or more nucleic acids comprising a
nucleotide sequence
encoding an engineered variant of the disclosure into a host cell.
[0023] Another aspect of the disclosure relates to a modified host cell
for producing
a cannabinoid or a cannabinoid derivative, wherein the modified host cell
comprises one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
disclosure.
[0024] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
geranyl
pyrophosphate:olivetolic acid geranyltransferase (GOT) polypeptide. In certain
such
embodiments, the GOT polypeptide comprises an amino acid sequence having at
least 85%
sequence identity to SEQ ID NO:17. In some embodiments, the modified host cell
comprises
two or more heterologous nucleic acids comprising the nucleotide sequence
encoding the
GOT polypeptide.
[0025] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
NphB
polypeptide. In certain such embodiments, the NphB polypeptide comprises an
amino acid
sequence having at least 85% sequence identity to SEQ ID NO:188.
[0026] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
tetraketide
synthase (TKS) polypeptide and one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding an olivetolic acid cyclase (OAC) polypeptide. In
certain such
embodiments, the TKS polypeptide comprises an amino acid sequence having at
least 85%
sequence identity to SEQ ID NO:19. In some embodiments, the modified host cell
comprises
three or more heterologous nucleic acids comprising a nucleotide sequence
encoding a TKS
polypeptide. In some embodiments, the OAC polypeptide comprises an amino acid
sequence
having at least 85% sequence identity to SEQ ID NO:21 or SEQ ID NO:48. In some
embodiments, the modified host cell comprises three or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an OAC polypeptide.
8
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0027] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an acyl-
activating enzyme (AAE) polypeptide. In certain such embodiments, the AAE
polypeptide
comprises an amino acid sequence having at least 85% sequence identity to SEQ
ID NO:23.
In some embodiments, the modified host cell comprises two or more heterologous
nucleic
acids comprising a nucleotide sequence encoding an AAE polypeptide.
[0028] In some embodiments of the disclosure, the modified host cell
comprises one
or more of the following: a) one or more heterologous nucleic acids comprising
a nucleotide
sequence encoding a HMG-CoA synthase (HMGS) polypeptide; b) one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a
truncated 3-
hydroxy-3-methyl-glutaryl-CoA reductase (tHMGR) polypeptide; c) one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a
mevalonate kinase
(MK) polypeptide; d) one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding a phosphomevalonate kinase (PMK) polypeptide; e) one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a
mevalonate
pyrophosphate decarboxylase (MVD1) polypeptide; or f) one or more heterologous
nucleic
acids comprising a nucleotide sequence encoding a isopentenyl diphosphate
isomerase
(IDI1) polypeptide. In some embodiments, the IDI1 polypeptide comprises an
amino acid
sequence having at least 85% sequence identity to SEQ ID NO:25. In some
embodiments,
the tHMGR polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:27. In some embodiments, the HMGS polypeptide comprises
an
amino acid sequence having at least 85% sequence identity to SEQ ID NO:29. In
some
embodiments, the MK polypeptide comprises an amino acid sequence having at
least 85%
sequence identity to SEQ ID NO:39. In some embodiments, the PMK polypeptide
comprises
an amino acid sequence having at least 85% sequence identity to SEQ ID NO:37.
In some
embodiments, the MVD1 polypeptide comprises an amino acid sequence having at
least
85% sequence identity to SEQ ID NO:33.
[0029] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an
acetoacetyl-CoA thiolase polypeptide. In certain such embodiments, the
acetoacetyl-CoA
thiolase polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:31.
9
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0030] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
pyruvate
decarboxylase (PDC) polypeptide. In certain such embodiments, the PDC
polypeptide
comprises an amino acid sequence having at least 85% sequence identity to SEQ
ID NO:35.
[0031] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
geranyl
pyrophosphate synthetase (GPPS) polypeptide. In certain such embodiments, the
GPPS
polypeptide comprises an amino acid sequence having at least 85% sequence
identity to SEQ
ID NO:41.
[0032] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
KAR2
polypeptide. In certain such embodiments, the KAR2 polypeptide comprises an
amino acid
sequence having at least 85% sequence identity to SEQ ID NO:5. In some
embodiments, the
modified host cell comprises two or more heterologous nucleic acids comprising
a
nucleotide sequence encoding a KAR2 polypeptide.
[0033] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
PDI1
polypeptide. In certain such embodiments, the PDI1 polypeptide comprises an
amino acid
sequence having at least 85% sequence identity to SEQ ID NO:9.
[0034] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an IRE1
polypeptide. In certain such embodiments, the IRE1 polypeptide comprises an
amino acid
sequence having at least 85% sequence identity to SEQ ID NO:11 or SEQ ID
NO:190.
[0035] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an ER01
polypeptide. In certain such embodiments, the ER01 polypeptide comprises an
amino acid
sequence having at least 85% sequence identity to SEQ ID NO:7.
[0036] In some embodiments of the disclosure, the modified host cell
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
FAD1
polypeptide. In certain such embodiments, the FAD1 polypeptide comprises an
amino acid
sequence having at least 85% sequence identity to SEQ ID NO:192.
[0037] In some embodiments of the disclosure, the modified host cell
comprises a
deletion or downregulation of one or more genes encoding a PEP4 polypeptide.
In certain
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
such embodiments, the PEP4 polypeptide comprises an amino acid sequence having
at least
85% sequence identity to SEQ ID NO:15.
[0038] In some embodiments of the disclosure, the modified host cell
comprises a
deletion or downregulation of one or more genes encoding a ROT2 polypeptide.
In certain
such embodiments, the ROT2 polypeptide comprises an amino acid sequence having
at least
85% sequence identity to SEQ ID NO:13.
[0039] In some embodiments of the disclosure, the modified host cell is a
eukaryotic
cell. In certain such embodiments, the eukaryotic cell is a yeast cell. In
certain such
embodiments, the yeast cell is Saccharomyces cerevisiae. In certain such
embodiments, the
Saccharomyces cerevisiae is a protease-deficient strain of Saccharomyces
cerevisiae.
[0040] In some embodiments of the disclosure, at least one of the one or
more
nucleic acids are integrated into the chromosome of the modified host cell. In
some
embodiments of the disclosure, at least one of the one or more nucleic acids
are maintained
extrachromosomally. In some embodiments of the disclosure, at least one of the
one or more
nucleic acids are operably-linked to an inducible promoter. In some
embodiments of the
disclosure, at least one of the one or more nucleic acids are operably-linked
to a constitutive
promoter.
[0041] In some embodiments of the disclosure, the modified host cell
produces a
cannabinoid or a cannabinoid derivative in an amount, as measured in mg/L or
mM, greater
than an amount of the cannabinoid or the cannabinoid derivative produced by a
modified
host cell comprising one or more nucleic acids comprising a nucleotide
sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, wherein the modified host cell comprising one or more nucleic acids
comprising the
nucleotide sequence encoding the tetrahydrocannabinolic acid synthase
polypeptide having
the amino acid sequence of SEQ ID NO:44 lacks a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, grown under similar
culture
conditions for the same length of time.
[0042] In some embodiments of the disclosure, the modified host cell
produces a
cannabinoid or a cannabinoid derivative in an amount, as measured in mg/L or
mM, at least
5%, at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at
least 35%, at least
40%, at least 45%, at least 50%, at least 60%, at least 70%, at least 80%, at
least 90%, at
least 100%, at least 150% at least 200%, at least 500%, or at least 1000%
greater than an
amount of the cannabinoid or the cannabinoid derivative produced by a modified
host cell
11
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, wherein the modified host cell comprising one or more nucleic acids
comprising the
nucleotide sequence encoding the tetrahydrocannabinolic acid synthase
polypeptide having
the amino acid sequence of SEQ ID NO:44 lacks a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, grown under similar
culture
conditions for the same length of time.
[0043] In some embodiments of the disclosure, the modified host cell has
a faster
growth rate and/or higher biomass yield compared to a growth rate and/or
higher biomass
yield of a modified host cell comprising one or more nucleic acids comprising
a nucleotide
sequence encoding a tetrahydrocannabinolic acid synthase polypeptide having an
amino acid
sequence of SEQ ID NO:44, wherein the modified host cell comprising one or
more nucleic
acids comprising the nucleotide sequence encoding the tetrahydrocannabinolic
acid synthase
polypeptide having the amino acid sequence of SEQ ID NO:44 lacks a nucleic
acid
comprising a nucleotide sequence encoding an engineered variant of the
disclosure, grown
under similar culture conditions for the same length of time.
[0044] In some embodiments of the disclosure, the modified host cell has
a growth
rate and/or higher biomass yield at least 5%, at least 10%, at least 15%, at
least 20%, at least
25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at
least 60%, at
least 70%, at least 80%, at least 90%, at least 100%, at least 150% at least
200%, at least
500%, or at least 1000% faster than a growth rate and/or higher biomass yield
of a modified
host cell comprising one or more nucleic acids comprising a nucleotide
sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, wherein the modified host cell comprising one or more nucleic acids
comprising the
nucleotide sequence encoding the tetrahydrocannabinolic acid synthase
polypeptide having
the amino acid sequence of SEQ ID NO:44 lacks a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, grown under similar
culture
conditions for the same length of time.
[0045] In some embodiments of the disclosure, the modified host cell
produces
tetrahydrocannabinolic acid (THCA) from cannabigerolic acid (CBGA) in an
increased ratio
of THCA over another cannabinoid (e.g., cannabichromenic acid (CBCA)) compared
to that
produced by a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
12
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
amino acid sequence of SEQ ID NO:44, wherein the modified host cell comprising
one or
more nucleic acids comprising the nucleotide sequence encoding the
tetrahydrocannabinolic
acid synthase polypeptide having the amino acid sequence of SEQ ID NO:44 lacks
a nucleic
acid comprising a nucleotide sequence encoding an engineered variant of the
disclosure,
grown under similar culture conditions for the same length of time.
[0046] In some embodiments of the disclosure, the modified host cell
produces
THCA from CBGA in a ratio of THCA over another cannabinoid (e.g.,
cannabichromenic
acid (CBCA)) of about 11:1, about 11.5:1, about 12:1, about 12.5:1, about
13:1, about
13.5:1, about 14:1, about 14.5:1, about 15:1, about 15.5:1, about 16:1, about
16.5:1, about
17:1, about 17.5:1, about 18:1, about 18.5:1, about 19:1, about 19.5:1, about
20:1, about
25:1, about 30:1, about 35:1, about 40:1, about 45:1, about 50:1, about 60:1,
about 70:1,
about 80:1, about 90:1, about 100:1, about 150:1, about 200:1, about 500:1, or
greater than
about 500:1.
[0047] Another aspect of the disclosure relates to a method of producing
a
cannabinoid or a cannabinoid derivative, the method comprising: a) culturing a
modified
host cell of the disclosure in a culture medium. In certain such embodiments,
the method
comprises: b) recovering the produced cannabinoid or cannabinoid derivative.
In some
embodiments, the culture medium comprises a carboxylic acid. In certain such
embodiments, the carboxylic acid is an unsubstituted or substituted C3-C18
carboxylic acid.
In certain such embodiments, the unsubstituted or substituted C3-Ci8
carboxylic acid is an
unsubstituted or substituted hexanoic acid. In some embodiments, the culture
medium
comprises olivetolic acid or an olivetolic acid derivative. In some
embodiments, the
cannabinoid is cannabidiolic acid, cannabidiol, cannabidivarinic acid, or
cannabidivarin. In
some embodiments, the culture medium comprises a fermentable sugar. In some
embodiments, the culture medium comprises a pretreated cellulosic feedstock.
In some
embodiments, the culture medium comprises a non-fermentable carbon source. In
certain
such embodiments, the non-fermentable carbon source comprises ethanol. In some
embodiments, the cannabinoid or the cannabinoid derivative is produced in an
amount of
more than 100 mg/L culture medium.
[0048] In some embodiments of the methods of the disclosure, the
cannabinoid or the
cannabinoid derivative is produced in an amount, as measured in mg/L or mM,
greater than
an amount of the cannabinoid or the cannabinoid derivative produced in a
method
comprising culturing a modified host cell comprising one or more nucleic acids
comprising a
13
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44 instead of the modified host cell of the
disclosure,
wherein the modified host cell comprising one or more nucleic acids comprising
the
nucleotide sequence encoding the tetrahydrocannabinolic acid synthase
polypeptide having
the amino acid sequence of SEQ ID NO:44 lacks a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, and wherein the
modified host
cell of the disclosure and the modified host cell comprising one or more
nucleic acids
comprising the nucleotide sequence encoding the tetrahydrocannabinolic acid
synthase
polypeptide having the amino acid sequence of SEQ ID NO:44 but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant of the
disclosure, are
cultured under similar culture conditions for the same length of time.
[0049] In some embodiments of the methods of the disclosure, the
cannabinoid or the
cannabinoid derivative is produced in an amount, as measured in mg/L or mM, at
least 5%,
at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least
35%, at least
40%, at least 45%, at least 50%, at least 60%, at least 70%, at least 80%, at
least 90%, at
least 100%, at least 150% at least 200%, at least 500%, or at least 1000%
greater than an
amount of the cannabinoid or the cannabinoid derivative produced in a method
comprising
culturing a modified host cell comprising one or more nucleic acids comprising
a nucleotide
sequence encoding a tetrahydrocannabinolic acid synthase polypeptide having an
amino acid
sequence of SEQ ID NO:44 instead of the modified host cell of the disclosure,
wherein the
modified host cell comprising one or more nucleic acids comprising the
nucleotide sequence
encoding the tetrahydrocannabinolic acid synthase polypeptide having the amino
acid
sequence of SEQ ID NO:44 lacks a nucleic acid comprising a nucleotide sequence
encoding
an engineered variant of the disclosure, and wherein the modified host cell of
the disclosure
and the modified host cell comprising one or more nucleic acids comprising the
nucleotide
sequence encoding the tetrahydrocannabinolic acid synthase polypeptide having
the amino
acid sequence of SEQ ID NO:44, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, are cultured under
similar culture
conditions for the same length of time.
[0050] In some embodiments of the methods of the disclosure, the
cannabinoid is
tetrahydrocannabinolic acid (THCA), and wherein the method produces THCA in an
increased ratio of THCA over another cannabinoid (e.g., cannabichromenic acid
(CBCA))
compared to that produced in a method comprising culturing a modified host
cell comprising
14
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
one or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 instead of the modified host cell of the disclosure, wherein the
modified host cell
comprising one or more nucleic acids comprising the nucleotide sequence
encoding the
tetrahydrocannabinolic acid synthase polypeptide having the amino acid
sequence of SEQ
ID NO:44 lacks a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant of the disclosure, grown under similar culture conditions for the same
length of time.
[0051] An aspect of the disclosure relates to a method of producing a
cannabinoid or
a cannabinoid derivative, the method comprising use of an engineered variant
of the
disclosure. In certain such embodiments, the method comprises recovering the
produced
cannabinoid or cannabinoid derivative. In some embodiments of the methods of
the
disclosure, the cannabinoid is tetrahydrocannabinolic acid,
tetrahydrocannabivarinic acid, or
tetrahydrocannabivarin.
[0052] In some embodiments of the methods of the disclosure, the
cannabinoid or the
cannabinoid derivative is produced in an amount, as measured in mg/L or mM,
greater than
an amount of the cannabinoid or the cannabinoid derivative produced in a
method
comprising use of a tetrahydrocannabinolic acid synthase polypeptide having an
amino acid
sequence of SEQ ID NO:44 instead of the engineered variant of the disclosure,
wherein the
engineered variant of the disclosure and the tetrahydrocannabinolic acid
synthase
polypeptide having the amino acid sequence of SEQ ID NO:44 are used under
similar
conditions for the same length of time.
[0053] In some embodiments of the methods of the disclosure, the
cannabinoid or the
cannabinoid derivative is produced in an amount, as measured in mg/L or mM, at
least 5%,
at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least
35%, at least
40%, at least 45%, at least 50%, at least 60%, at least 70%, at least 80%, at
least 90%, at
least 100%, at least 150% at least 200%, at least 500%, or at least 1000%
greater than an
amount of the cannabinoid or the cannabinoid derivative produced in a method
comprising
use of a tetrahydrocannabinolic acid synthase polypeptide having an amino acid
sequence of
SEQ ID NO:44 instead of the engineered variant of the disclosure, wherein the
engineered
variant of the disclosure and the tetrahydrocannabinolic acid synthase
polypeptide having the
amino acid sequence of SEQ ID NO:44 are used under similar conditions for the
same length
of time.
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0054] In some embodiments of the methods of the disclosure, the
cannabinoid is
tetrahydrocannabinolic acid (THCA), and wherein the method produces THCA in an
increased ratio of THCA over another cannabinoid (e.g., cannabichromenic acid
(CBCA)
compared to that produced in a method comprising use of a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44 instread of
the
engineered variant of the disclosure, wherein the engineered variant of the
disclosure and the
tetrahydrocannabinolic acid synthase polypeptide having the amino acid
sequence of SEQ
ID NO:44 are used under similar conditions for the same length of time.
[0055] In some embodiments of the methods of the disclosure, the method
produces
THCA from CBGA in a ratio of THCA over another cannabinoid (e.g., CBCA) of
about
11:1, about 11.5:1, about 12:1, about 12.5:1, about 13:1, about 13.5:1, about
14:1, about
14.5:1, about 15:1, about 15.5:1, about 16:1, about 16.5:1, about 17:1, about
17.5:1, about
18:1, about 18.5:1, about 19:1, about 19.5:1, about 20:1, about 25:1, about
30:1, about 35:1,
about 40:1, about 45:1, about 50:1, about 60:1, about 70:1, about 80:1, about
90:1, about
100:1, about 150:1, about 200:1, about 500:1, or greater than about 500:1.
[0056] Another aspect of the disclosure relates to a method of screening
an
engineered variant of a tetrahydrocannabinolic acid synthase (THCAS)
polypeptide
comprising an amino acid sequence of SEQ ID NO:44 with one or more amino acid
substitutions, the method comprising: a) dividing a population of host cells
into a control
population and a test population; b) co-expressing in the control population a
THCAS
polypeptide having an amino acid sequence of SEQ ID NO:44 and a comparison
cannabinoid synthase polypeptide, wherein the THCAS polypeptide having an
amino acid
sequence of SEQ ID NO:44 can convert cannabigerolic acid (CBGA) to a first
cannabinoid,
tetrahydrocannabinolic acid (THCA), and the comparison cannabinoid synthase
polypeptide
can convert the same CBGA to a different second cannabinoid; c) co-expressing
in the test
population the engineered variant and the comparison tetrahydrocannabinolic
acid synthase
polypeptide, wherein the engineered variant may convert CBGA to the same first
cannabinoid, tetrahydrocannabinolic acid (THCA), as the THCAS polypeptide
having an
amino acid sequence of SEQ ID NO:44, and wherein the comparison cannabinoid
synthase
polypeptide can convert the same CBGA to the second cannabinoid and is
expressed at
similar levels in the test population and in the control population; d)
measuring a ratio of the
first cannabinoid, tetrahydrocannabinolic acid (THCA), over the second
cannabinoid
produced by both the test population and the control population; and e)
measuring an
16
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
amount, in mg/L or mM, of the first cannabinoid produced by both the test
population and
the control population. In certain such embodiments, the test population is
identified as
comprising an engineered variant having improved in vivo performance compared
to the
tetrahydrocannabinolic acid synthase (THCAS) polypeptide having an amino acid
sequence
of SEQ ID NO:44, wherein improved in vivo performance is demonstrated by an
increase in
the ratio of the first cannabinoid over the second cannabinoid produced by the
test
population compared to that produced by the control population under similar
culture
conditions for the same length of time. In some embodiments of the method of
screening the
engineered variant of a THCAS polypeptide, the test population is identified
as comprising
an engineered variant having improved in vivo performance compared to the
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:3 by producing the first cannabinoid in a greater amount, as measured in
mg/L or mM,
by the test population compared to the amount produced by the control
population under
similar culture conditions for the same length of time.
[0057] In some embodiments of the method of screening the engineered
variant of a
THCAS polypeptide, the cannabinoid synthase polypeptide is a cannabidiolic
acid synthase
polypeptide. In certain such embodiments, the cannabidiolic acid synthase
(CBDAS)
polypeptide comprises an amino acid sequence having at least 85% sequence
identity to SEQ
ID NO :3. In some embodiments of the method of screening the engineered
variant of a
THCAS polypeptide, the second cannabinoid is cannabidiolic acid (CBDA).
[0058] In some embodiments of the method of screening the engineered
variant of a
THCAS polypeptide, the engineered variant is an engineered variant of the
disclosure.
Brief Description Of The Drawings
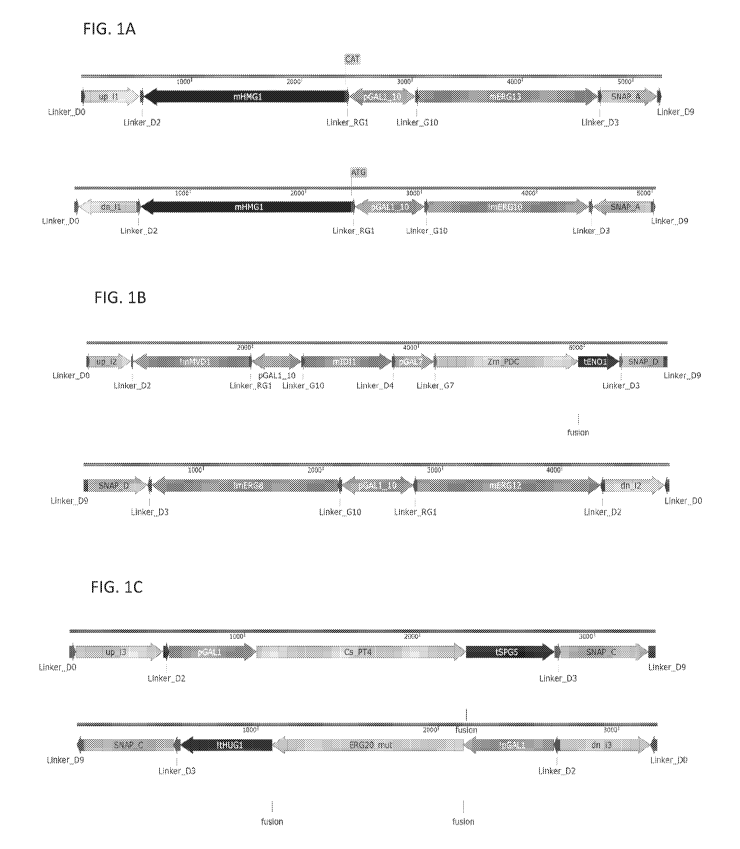
[0059] FIGS. 1A, 1B, and 1C depict expression constructs used in the
production of
the S29 strain. The expression constructs depicted in FIGS. 1A, 1B, and 1C
were also used
in the production of the following strains: S61, S122, S171, S181, S220, S241,
S270, S487,
S951, S1000-S1059, S1072-1079, and S1081. Throughout the figures, in addition
to the
specified coding sequences from Table 1, construct maps depict regulatory, non-
coding and
genomic cassette sequences described in Table 5. Construct maps also depict
genes denoted
with a preceding "m" (e.g. mERG13), which specify open reading frames from
Table 1 with
200-250 base pairs (bp) of downstream regulatory (terminator) sequence. Arrows
in
construct maps indicate the directionality of certain DNA parts. The "!"
preceding a part
17
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
name is an output of the DNA design software used, is redundant with the arrow
directionality, and can be ignored.
[0060] FIG. 2 depicts an expression construct used in the production of
the S181
strain. The expression construct depicted in FIG. 2 was also used in the
production of
following strains: S220, S241, S270, S487, S951, S1000-S1059, S1072-1079, and
S1081.
[0061] FIG. 3 depicts an expression construct used in the production of
the S220
strain. The expression construct depicted in FIG. 3 was also used in the
production of
following strains: S241, S270, S487, S951, S1000-S1059, S1072-1079, and S1081.
[0062] FIG. 4 depicts expression constructs used in the production of the
S241 strain.
The expression constructs depicted in FIG. 4 were also used in the production
of following
strains: S270, S487, S951, S1000-S1059, S1072-1079, and S1081.
[0063] FIG. 5 depicts a landing pad construct used in the production of
the S61
strain. The construct depicted in FIG. 5 was also used in the production of
the following
strains: S122, S171, S181, S220, S241, S270, S487, S951, S1000-S1059, S1072-
1079, and
S1081.
[0064] FIG. 6 depicts expression constructs used in the production of the
S122 strain.
The expression constructs depicted in FIG. 6 were also used in the production
of the
following strains: S171, S181, S220, S241, S270, S487, S951, S1000-S1059,
S1072-1079,
and S1081.
[0065] FIG. 7 depicts an expression construct used in the production of
the S171
strain. The expression construct depicted in FIG. 7 was also used in the
production of the
following strains: S181, S220, S241, S270, S487, S951, S1000-S1059, S1072-
1079, and
S1081.
[0066] FIG. 8 depicts expression constructs used in the production of the
S270 strain.
The expression constructs depicted in FIG. 8 were also used in the production
of the
following strains: S487, S951, S1000-S1059, S1072-1079, and S1081.
[0067] FIGS. 9A and 9B depict an expression construct used in the
production of the
S487 strain. The expression constructs depicted in FIGS. 9A and 9B were also
used in the
production of the following strains: S951 S1000-S1059, S1072-1079, and S1081.
[0068] FIG. 10 depicts an expression construct used in the production of
the S1042
strain.
[0069] FIG. 11 depicts an expression construct used in the production of
the
following strains: S951, S1000-S1041, S1043-S1059, S1072-1079, and S1081.
18
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Detailed Description
[0070] Synthetic biology allows for the engineering of industrial host
organisms ¨
e.g., microbes ¨ to convert simple sugar feedstocks into medicines. This
approach includes
identifying genes that produce the target molecules and optimizing their
activities in the
industrial host. Microbial production can be significantly cost-advantaged
over agriculture
and chemical synthesis, less variable, and allow tailoring of the target
molecule. However,
reconstituting or creating a pathway to produce a target molecule in an
industrial host
organism can require significant engineering of both the pathway genes and the
host. The
present disclosure provides engineered variants of a tetrahydrocannabinolic
acid synthase
(THCAS) polypeptide comprising an amino acid sequence of SEQ ID NO:44 with one
or
more amino acid substitutions, nucleic acids comprising nucleotide sequences
encoding said
engineered variants, methods of making modified host cells comprising said
nucleic acids,
modified host cells for producing cannabinoids or cannabinoid derivatives,
methods of
producing cannabinoids or cannabinoid derivatives, and methods of screening
engineered
variants of the THCAS polypeptide. The engineered variants of the disclosure
may be useful
for producing cannabinoids or cannabinoid derivatives (e.g., non-naturally
occurring
cannabinoids). The modified host cells of the disclosure may be useful for
producing
cannabinoids or cannabinoid derivatives (e.g., non-naturally occurring
cannabinoids) and/or
for expressing engineered variants of the disclosure. The disclosure also
provides for
modified host cells for expressing the engineered variants of the disclosure.
Additionally, the
disclosure provides for preparation of engineered variants of the disclosure.
[0071] Cannabinoid synthase polypeptides, such as tetrahydrocannabinolic
acid
synthase, cannabichromenic acid synthase, or cannabidiolic acid synthase
polypeptides, play
an important role in the biosynthesis of cannabinoids. However, reconstituting
their activity
in a modified host cell has proven challenging, hampering progress in the
production of
cannabinoids or cannabinoid derivatives. Cannabinoid synthases must
successfully traverse
the secretory pathway to fold and function properly. These secreted plant
enzymes have not
evolved to be expressed in a yeast cell, and as a result have poor activity,
with limited
conversion of their substrate cannabigerolic acid (CBGA) into cannabidiolic
acid (CBDA) or
tetrahydrocannabinolic acid (THCA). A simple method to increase activity of an
enzyme is
to increase its copy number (expression). However, expression of the CBDAS and
tetrahydrocannabinolic acid synthase (THCAS) genes in yeast is toxic (likely
owing to
19
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
misfolding of the protein), frustrating straightforward attempts to boost
activity by
integrating multiple copies of the genes. Product profile presents another
problem. While the
primary product of the natural CBDAS enzyme is CBDA, the enzyme also makes
significant
amounts of THCA, an undesired byproduct, which would require expensive
additional
downstream purification steps to separate in an industrial process.
[0072] For
these reasons, the natural CBDAS or THCAS enzymes are not optimal
for industrial purposes, and improved enzymes are required. Parameters of
interest include
catalytic activity, product profile, enzyme stability, and pH and temperature
optima. Enzyme
improvement is typically accomplished by coupling the generation of diversity
(a library of
engineered variants) to a screen or selection for the properties of interest.
DNA libraries
encoding engineered variants can be generated in a variety of ways. For
example, libraries
can be generated using error prone PCR using the wild type gene sequence as a
template.
The resulting library can be quite large, consisting of genes with variable
numbers of
mutations at random positions. Error prone PCR is inexpensive and convenient
but has
several drawbacks. First, instead of a precise number of mutations per
construct, a
distribution is obtained. This presents an unfortunate trade-off A
distribution centered
around a low number of mutations will include a significant amount of zero-
mutation wild-
type constructs that waste screening capacity. A distribution centered around
a higher
number of mutations is likely to generate constructs that have accumulated
loss of function
mutations that would prevent identification of the desired gain of function
mutations.
Second, error prone PCR introduces mutational bias (an intrinsic property of
the low fidelity
polymerases used) which means that the library underrepresents certain types
of mutation. A
powerful alternative to error prone PCR is saturation mutagenesis, which
involves synthesis
of a library containing every possible amino acid at every position in the
protein. Recent
advances in DNA synthesis technologies have improved the quality of these
libraries
significantly.
[0073] Once a
library encoding engineered variants is generated, it is necessary to
select or screen for engineered variants with the properties of interest. This
can be
accomplished by using a protein production host to express and purify the
engineered
variants, followed by testing in vitro. Such an approach allows careful
measurement of the
engineered variants' kinetic parameters and assessment of performance under
carefully
controlled conditions. However, for application in an engineered microbial
strain, in vitro
data can be highly misleading as no in vitro system can represent the cellular
milieu
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
accurately. In this case, the best option is to test the engineered variants
in the exact context
they must eventually perform¨inside an engineered production strain. In the
case of the
cannabinoid synthases, such a production strain would be engineered to produce
the
substrate CBGA in excess. One challenge with this in vivo system is that
variability is
higher. When testing a large library, this variability can make it difficult
to distinguish
clones with more subtle improvements over the wild type enzyme activity. By
calculating
the ratio of the library enzyme product titer and the invariant competition
enzyme titer, it is
possible to reduce the variability in data significantly. This is because
biological variables
tend to affect both of the enzymes in the same way, allowing normalization of
the effect.
Unlike a kinetic parameter, the competition ratio reports on both changes in
both enzyme
catalytic parameters such as Km and Kcat as well as changes in the steady
state levels of
functional engineered variant (expression and stability).
[0074] Through use of the above methods, the present disclosure provides
engineered variants of a tetrahydrocannabinolic acid synthase (THCAS)
polypeptide. Herein,
various engineered variants were screened. THCA titers were improved (outside
standard
deviation of wild type) in 58 distinct variants. These engineered variants of
the disclosure
may be useful for producing cannabinoids or cannabinoid derivatives (e.g., non-
naturally
occurring cannabinoids). The engineered variants of the disclosure may produce
tetrahydrocannabinolic acid (THCA) from cannabigerolic acid (CBGA) in a
greater amount,
as measured in mg/L or mM, than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time. Additionally, the
engineered
variants of the disclosure may produce THCA from CBGA in an increased ratio of
THCA
over another cannabinoid (e.g., CBCA) compared to that produced by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time. Similar conditions
may include
the same temperature, pH, buffer, and/or fermentation conditions and in the
same culture
medium and/or reaction solvent.
[0075] The methods of the disclosure may include using engineered
microorganisms
(e.g., modified host cells) or engineered variants of a THCAS polypeptide of
the disclosure
to produce naturally-occurring and non-naturally occurring cannabinoids.
Naturally-
occurring cannabinoids and non-naturally occurring cannabinoids (e.g.,
cannabinoid
derivatives) are challenging to produce using chemical synthesis due to their
complex
21
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
structures. The methods of the disclosure enable the construction of metabolic
pathways
inside living cells to produce bespoke cannabinoids or cannabinoid derivatives
from simple
precursors such as sugars and carboxylic acids. One or more nucleic acids
(e.g., heterologous
nucleic acids) disclosed herein comprising nucleotide sequences encoding one
or more
polypeptides or engineered variants disclosed herein can be introduced into
host
microorganisms allowing for the stepwise conversion of inexpensive feedstocks,
e.g., sugar,
into final products: cannabinoids or cannabinoid derivatives. These products
can be specified
by the choice and construction of expression constructs or vectors comprising
one or more
nucleic acids (e.g., heterologous nucleic acids) disclosed herein, allowing
for the efficient
bioproduction of chosen cannabinoids, such as THC and THCA and less common
cannabinoid species found at low levels in Cannabis; or cannabinoid
derivatives.
Bioproduction also enables synthesis of cannabinoids or cannabinoid
derivatives with
defined stereochemistries, which is challenging to do using chemical
synthesis. To produce
cannabinoids or cannabinoid derivatives and create biosynthetic pathways
within modified
host cells, modified host cells comprising one or more nucleic acids
comprising a nucleotide
sequence encoding an engineered variant of a THCAS polypeptide of the
disclosure may
express or overexpress combinations of heterologous nucleic acids comprising
nucleotide
sequences encoding one or more polypeptides involved in cannabinoid or
cannabinoid
precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates, olivetolic
acid, or
hexanoyl-CoA) biosynthesis. In some embodiments, the nucleotide sequences
encoding the
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis are
codon-
optimized.
[0076] The disclosure also provides for modification of the secretory
pathway of a
host cell modified with one or more nucleic acids (e.g., heterologous nucleic
acids)
comprising a nucleotide sequence encoding an engineered variant of a THCAS
polypeptide
of the disclosure. In some embodiments, the nucleotide sequence encoding the
engineered
variant of a THCAS polypeptide is codon-optimized. Modification of the
secretory pathway
in the host cell may improve expression and solubilization of the engineered
variants of the
disclosure, as these variants are processed through the secretory pathway.
Reconstituting the
activity of polypeptides processed through the secretory pathway, such as the
engineered
variants of the disclosure, in a modified host cell, such as a modified yeast
cell, can be
challenging and unreliable. Often the expressed engineered variants may be
misfolded or
22
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
mislocalized, resulting in low expression, expressed engineered variants
lacking activity,
engineered variant aggregation, reduced host cell viability, and/or cell
death. Additionally, a
backlog of misfolded or mislocalized expressed engineered variants can induce
metabolic
stress within the modified host cell, harming the modified host cell. The
expressed
engineered variants may lack necessary posttranslational modifications for
folding and
activity, such as disulfide bonds, glycosylation and trimming, and cofactors,
affording
inactive polypeptides or polypeptides with reduced enzymatic activity.
[0077] The modified host cell of the disclosure may be a modified yeast
cell. Yeast
cells may be cultured using known conditions, grow rapidly, and are generally
regarded as
safe. Yeast cells contain the secretory pathway common to all eukaryotes. As
disclosed
herein, manipulation of that secretory pathway in yeast host cells modified
with one or more
nucleic acids (e.g., heterologous nucleic acids) comprising a nucleotide
sequence encoding
an engineered variant of a THCAS polypeptide of the disclosure may improve
expression,
folding, and enzymatic activity of the engineered variant as well as viability
of the modified
yeast host cell, such as modified Saccharomyces cerevisiae. Further, use of
codon-
optimized nucleotide sequences encoding engineered variants of the disclosure,
may
improve expression and activity of the engineered variant and viability of
modified yeast
host cells, such as modified Saccharomyces cerevisiae.
[0078] Besides allowing for the production of desired cannabinoids or
cannabinoid
derivatives, the present disclosure provides a more reliable and economical
process than
agriculture-based production. Microbial fermentations can be completed in days
versus the
months necessary for an agricultural crop, are not affected by climate
variation or soil
contamination (e.g., by heavy metals), and can produce pure products at high
titer.
[0079] The present disclosure also provides a platform for the economical
production
of high-value cannabinoids, including THC, as well as derivatives thereof. It
also provides
for the production of different cannabinoids or cannabinoid derivatives for
which no viable
method of production exists. Using the engineered variants, methods, and
modified host cells
disclosed herein, cannabinoids and cannabinoid derivatives may be produced in
an amount
of over 100 mg per liter of culture medium, over 1 g per liter of culture
medium, over 10 g
per liter of culture medium, over 100 g per liter of culture medium.
[0080] Additionally, the disclosure provides engineered variants of a
THCAS
polypeptide, methods, modified host cells, and nucleic acids to produce
cannabinoids or
cannabinoid derivatives in vivo or in vitro from simple precursors. Nucleic
acids (e.g.,
23
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
heterologous nucleic acids) disclosed herein can be introduced into
microorganisms (e.g.,
modified host cells), resulting in expression or overexpression of one or more
polypeptides,
such as the engineered variants of the disclosure, which can then be utilized
in vitro or in
vivo for the production of cannabinoids or cannabinoid derivatives. In some
embodiments,
the in vitro methods are cell-free.
Cannabinoid Biosynthesis
[0081] In addition to one or more nucleic acids (e.g., heterologous
nucleic acids)
encoding an engineered variant of a THCAS polypeptide, one or more nucleic
acids (e.g.,
heterologous nucleic acids) encoding one or more polypeptides having at least
one activity
of a polypeptide present in the cannabinoid or cannabinoid precursor
biosynthetic pathway
may be useful in the methods and modified host cells for the synthesis of
cannabinoids or
cannabinoid derivatives. Cannabinoid precursors may include, for example,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA.
[0082] In Cannabis, cannabinoids are produced from the common metabolite
precursors geranylpyrophosphate (GPP) and hexanoyl-CoA by the action of three
polypeptides. Hexanoyl-CoA and malonyl-CoA are combined to afford a 12-carbon
tetraketide intermediate by a tetraketide synthase (TKS) polypeptide. This
tetraketide
intermediate is then cyclized by an olivetolic acid cyclase (OAC) polypeptide
to produce
olivetolic acid. Olivetolic acid is then prenylated with the common isoprenoid
precursor GPP
by a geranyl pyrophosphate:olivetolic acid geranyltransferase (GOT)
polypeptide (e.g., a
CsPT4 polypeptide) to produce CBGA, the cannabinoid also known as the "mother
cannabinoid." The engineered variants of a THCAS polypeptide of the disclosure
then
convert CBGA into other cannabinoids, e.g., THCA, etc. In the presence of heat
or light, the
acidic cannabinoids can undergo decarboxylation, e.g., THCA producing THC.
[0083] GPP and hexanoyl-CoA can be generated through several pathways.
One or
more nucleic acids (e.g., heterologous nucleic acids) encoding one or more
polypeptides
having at least one activity of a polypeptide present in these pathways can be
useful in the
methods and modified host cells for the synthesis of cannabinoids or
cannabinoid
derivatives.
[0084] Polypeptides that generate GPP or are part of a biosynthetic
pathway that
generates GPP may be one or more polypeptides having at least one activity of
a polypeptide
present in the mevalonate (MEV) pathway (e.g., one or more MEV pathway
polypeptides).
24
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
The term "mevalonate pathway" or "MEV pathway," as used herein, may refer to
the
biosynthetic pathway that converts acetyl-CoA to isopentenyl pyrophosphate
(IPP) and
dimethylallyl pyrophosphate (DMAPP). The mevalonate pathway comprises
polypeptides
that catalyze the following steps: (a) condensing two molecules of acetyl-CoA
to generate
acetoacetyl-CoA (e.g., by action of an acetoacetyl-CoA thiolase polypeptide);
(b) condensing
acetoacetyl-CoA with acetyl-CoA to form hydroxymethylglutaryl-CoA (HMG-CoA)
(e.g.,
by action of a HMG-CoA synthase (HMGS) polypeptide); (c) converting HMG-CoA to
mevalonate (e.g., by action of a HMG-CoA reductase (HMGR) polypeptide); (d)
phosphorylating mevalonate to mevalonate 5-phosphate (e.g., by action of a
mevalonate
kinase (MK) polypeptide); (e) converting mevalonate 5-phosphate to mevalonate
5-
pyrophosphate (e.g., by action of a phosphomevalonate kinase (PMK)
polypeptide); (f)
converting mevalonate 5-pyrophosphate to isopentenyl pyrophosphate (e.g., by
action of a
mevalonate pyrophosphate decarboxylase (MVD1) polypeptide); and (g) converting
isopentenyl pyrophosphate (IPP) to dimethylallyl pyrophosphate (DMAPP) (e.g.,
by action
of an isopentenyl pyrophosphate isomerase (IDI1) polypeptide). A geranyl
pyrophosphate
synthetase (GPPS) polypeptide then acts on IPP and/or DMAPP to generate GPP.
[0085] Polypeptides that generate hexanoyl-CoA may include polypeptides
that
generate acyl-CoA compounds or acyl-CoA compound derivatives (e.g., an acyl-
activating
enzyme polypeptide, a fatty acyl-CoA synthetase polypeptide, or a fatty acyl-
CoA ligase
polypeptide). Hexanoyl CoA derivatives, acyl-CoA compounds, or acyl-CoA
compound
derivatives may also be formed via such polypeptides.
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Sugar
/ 1 1
i'
Acetyl-CoA
/
ACC1 / 1
0
SCoA
Hexanoyl CoA
0 0
HO).)LSCoA Tetraketide
3x Malonyl CoA N,Synthase (TKS)
ENZ¨S
0 0 0 0
Olivetolic acid 1
cyclase (OAC)
OH 0
OH
HO
Olivetolic Acid
MEV -7.-
pathway )0PP-N Prenyltransferase
GPP I (GOT)
0 OH
HO
H
¨A-- Other cannabinoids
/ Other
OH synthases+heat
\
Cannabigerolic acid (CBGA)
C NTHCA synthase BDA synthasy
(CBDAS) (THCAS)
0 OH 0 OH
HO HO
H H
7
OH 0
H H
Cannabidiolic acid / CBDA .. Tetrahydrocannabinolic acid / THCA
1 Heat 1Heat
HO HO
H H
7
OH 0
H H
Cannabidiol / CBD Tetrahydrocannabinol / Dronabinol /
Marino! / THC
Biosynthetic pathways to cannabinoids
26
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0086] GPP and hexanoyl-CoA may also be generated through pathways
comprising
polypeptides that condense two molecules of acetyl-CoA to generate acetoacetyl-
CoA and
pyruvate decarboxylase polypeptides that generate acetyl-CoA from pyruvate via
acetaldehyde. Hexanoyl CoA derivatives, acyl-CoA compounds, or acyl-CoA
compound
derivatives may also be formed via such pathways.
General Information
[0087] In certain aspects, the practice of the present disclosure will
employ, unless
otherwise indicated, conventional techniques of molecular biology (including
recombinant
techniques), microbiology, cell biology, biochemistry, and immunology, which
are within
the skill of the art. Such techniques are explained fully in the literature:
"Molecular
Cloning: A Laboratory Manual," second edition (Sambrook et al., 1989);
"Oligonucleotide
Synthesis" (M. J. Gait, ed., 1984); "Animal Cell Culture" (R. I. Freshney,
ed., 1987);
"Methods in Enzymology" (Academic Press, Inc.); "Current Protocols in
Molecular
Biology" (F. M. Ausubel et al., eds., 1987, and periodic updates); "PCR: The
Polymerase
Chain Reaction," (Mullis et al., eds., 1994). Singleton et al., Dictionary of
Microbiology and
Molecular Biology 2nd ed., J. Wiley & Sons (New York, N.Y. 1994), and March,
Advanced
Organic Chemistry Reactions, Mechanisms and Structure 4th ed., John Wiley &
Sons (New
York, N.Y. 1992), provide one skilled in the art with a general guide to many
of the terms
used in the present application.
[0088] "Cannabinoid" or "cannabinoid compound" as used herein may refer
to a
member of a class of unique meroterpenoids found until now only in Cannabis
sativa.
Cannabinoids may include, but are not limited to, cannabichromene (CBC) type
(e.g.
cannabichromenic acid), cannabigerol (CBG) type (e.g. cannabigerolic acid),
cannabidiol
(CBD) type (e.g. cannabidiolic acid), A9-trans-tetrahydrocannabinol (A9 -THC)
type (e.g. A9-
tetrahydrocannabinolic acid), A8-trans-tetrahydrocannabinol (A8 -THC) type,
cannabicyclol
(CBL) type, cannabielsoin (CBE) type, cannabinol (CBN) type, cannabinodiol
(CBND)
type, cannabitriol (CBT) type, cannabigerolic acid (CBGA), cannabigerolic acid
monomethylether (CBGAM), cannabigerol (CBG), cannabigerol monomethylether
(CBGM), cannabigerovarinic acid (CBGVA), cannabigerovarin (CBGV),
cannabichromenic
acid (CBCA), cannabichromene (CBC), cannabichromevarinic acid (CBCVA),
cannabichromevarin (CBCV), cannabidiolic acid (CBDA), cannabidiol (CBD),
cannabidiol
monomethylether (CBDM), cannabidiol-C4 (CBD-C4), cannabidivarinic acid
(CBDVA),
27
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
cannabidivarin (CBDV), cannabidiorcol (CBD-C1), A9 ¨tetrahydrocannabinolic
acid A
(THCA-A), A9 ¨tetrahydrocannabinolic acid B (THCA-B), A9 ¨tetrahydrocannabinol
(THC),
A9 ¨tetrahydrocannabinolic acid-C4 (THCA-C4), A9 ¨tetrahydrocannabinol-C4 (THC-
C4), A9
¨tetrahydrocannabivarinic acid (THCVA), A9 ¨tetrahydrocannabivarin (THCV), A9
¨
tetrahydrocannabiorcolic acid (THCA-C1), A9 ¨tetrahydrocannabiorcol (THC-C1),
A7 ¨cis-
iso-tetrahydrocannabivarin, A8 ¨tetrahydrocannabinolic acid (A' ¨THCA), A8 ¨
tetrahydrocannabinol (A' ¨THC), cannabicyclolic acid (CBLA), cannabicyclol
(CBL),
cannabicyclovarin (CBLV), cannabielsoic acid A (CBEA-A), cannabielsoic acid B
(CBEA-
B), cannabielsoin (CBE), cannabielsoinic acid, cannabicitranic acid,
cannabinolic acid
(CBNA), cannabinol (CBN), cannabinol methylether (CBNM), cannabinol-C4, (CBN-
C4),
cannabivarin (CBV), cannabinol-C2 (CNB-C2), cannabiorcol (CBN-C1),
cannabinodiol
(CBND), cannabinodivarin (CBVD), cannabitriol (CB T), 10-ethyoxy-9-hydroxy-
delta-6a-
tetrahydrocannabinol, 8,9-dihydroxyl-delta-6a-tetrahydrocannabinol,
cannabitriolvarin
(CBTVE), dehydrocannabifuran (DCBF), cannabifuran (CBF), cannabichromanon
(CBCN),
cannabicitran (CBT), 10-oxo-delta-6a-tetrahydrocannabinol (OTHC), delta-9-cis-
tetrahydrocannabinol (cis-THC), 3,4,5,6-tetrahydro-7-hydroxy-alpha-alpha-2-
trimethy1-9-n-
propy1-2,6-methano-2H-1-benzoxocin-5-methanol (OH-iso-HHCV), cannabiripsol
(CBR),
and trihydroxy-delta-9-tetrahydrocannabinol (tri0H-THC).
[0089] An acyl-CoA compound as detailed herein may include compounds with
the
0
RõCoA
following structure: S , wherein R may be an unsubstituted fatty acid
side
chain or a fatty acid side chain substituted with or comprising one or more
functional and/or
reactive groups as disclosed herein (i.e., an acyl-CoA compound derivative).
[0090] As used herein, a hexanoyl CoA derivative, an acyl-CoA compound
derivative, a cannabinoid derivative, or an olivetolic acid derivative may
refer to hexanoyl
CoA, an acyl-CoA compound, a cannabinoid, or olivetolic acid substituted with
or
comprising one or more functional and/or reactive groups. Functional groups
may include,
but are not limited to, azido, halo (e.g., chloride, bromide, iodide,
fluorine), methyl, alkyl
(including branched and straight chain alkyl groups), alkynyl, alkenyl,
methoxy, alkoxy,
acetyl, amino, carboxyl, carbonyl, oxo, ester, hydroxyl, thio (e.g., thiol),
cyano, aryl,
heteroaryl, cycloalkyl, cycloalkenyl, cycloalkylalkenyl, cycloalkylalkynyl,
cycloalkenylalkyl, cycloalkenylalkenyl, cycloalkenylalkynyl,
heterocyclylalkenyl,
heterocyclylalkynyl, heteroarylalkenyl, heteroarylalkynyl, arylalkenyl,
arylalkynyl,
28
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
heterocyclyl, spirocyclyl, heterospirocyclyl, thioalkyl (or alkylthio),
arylthio, heteroarylthio,
sulfone, sulfonyl, sulfoxide, amido, alkylamino, dialkylamino, arylamino,
alkylarylamino,
diarylamino, N-oxide, imide, enamine, imine, oxime, hydrazone, nitrile,
aralkyl,
cycloalkylalkyl, haloalkyl, heterocyclylalkyl, heteroarylalkyl, nitro, thioxo,
and the like.
Suitable reactive groups may include, but are not necessarily limited to,
azide, carboxyl,
carbonyl, amine (e.g., alkyl amine (e.g., lower alkyl amine), aryl amine),
halide, ester (e.g.,
alkyl ester (e.g., lower alkyl ester, benzyl ester), aryl ester, substituted
aryl ester), cyano,
thioester, thioether, sulfonyl halide, alcohol, thiol, succinimidyl ester,
isothiocyanate,
iodoacetamide, maleimide, hydrazine, alkynyl, alkenyl, and the like. A
reactive group may
facilitate covalent attachment of a molecule of interest. Suitable molecules
of interest may
include, but are not limited to, a detectable label; imaging agents; a toxin
(including
cytotoxins); a linker; a peptide; a drug (e.g., small molecule drugs); a
member of a specific
binding pair; an epitope tag; ligands for binding by a target receptor; tags
to aid in
purification; molecules that increase solubility; molecules that enhance
bioavailability;
molecules that increase in vivo half-life; molecules that target to a
particular cell type;
molecules that target to a particular tissue; molecules that provide for
crossing the blood-
brain barrier; molecules to facilitate selective attachment to a surface; and
the like.
Functional and reactive groups may be unsubstituted or substituted with one or
more
functional or reactive groups.
[0091] A cannabinoid derivative or olivetolic acid derivative may also
refer to a
compound lacking one or more chemical moieties found in naturally-occurring
cannabinoids
or olivetolic acid. Such chemical moieties may include, but are not limited
to, methyl, alkyl,
alkenyl, methoxy, alkoxy, acetyl, carboxyl, carbonyl, oxo, ester, hydroxyl,
aryl, heteroaryl,
cycloalkyl, cycloalkenyl, cycloalkylalkenyl, cycloalkenylalkyl,
cycloalkenylalkenyl,
heterocyclylalkenyl, heteroarylalkenyl, arylalkenyl, heterocyclyl, aralkyl,
cycloalkylalkyl,
heterocyclylalkyl, heteroarylalkyl, and the like. In some embodiments, a
cannabinoid
derivative or olivetolic acid derivative may also comprise one or more of any
of the
functional and/or reactive groups described herein. Functional and reactive
groups may be
unsubstituted or substituted with one or more functional or reactive groups.
[0092] The term "nucleic acid" used herein, may refer to a polymeric form
of
nucleotides of any length, either ribonucleotides or deoxynucleotides. Thus,
this term may
include, but is not limited to, single-, double-, or multi-stranded DNA or
RNA, genomic
DNA, cDNA, genes, synthetic DNA or RNA, DNA-RNA hybrids, or a polymer
comprising
29
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
purine and pyrimidine bases or other naturally-occurring, chemically or
biochemically
modified, non- naturally-occurring, or derivatized nucleotide bases.
[0093] The terms "peptide," "polypeptide," and "protein" may be used
interchangeably herein, and may refer to a polymeric form of amino acids of
any length,
which can include coded and non-coded amino acids and chemically or
biochemically
modified or derivatized amino acids. The polypeptides disclosed herein may
include full-
length polypeptides, fragments of polypeptides, truncated polypeptides, fusion
polypeptides,
or polypeptides having modified peptide backbones. The polypeptides disclosed
herein may
also be variants differing from a specifically recited "reference" polypeptide
(e.g., a wild-
type polypeptide) by amino acid insertions, deletions, mutations, and/or
substitutions.
[0094] An "engineered variant of a tetrahydrocannabinolic acid synthase
polypeptide" or "engineered variant of the disclosure" may indicate a non-wild
type
polypeptide having tetrahydrocannabinolic acid synthase activity. One skilled
in the art can
measure the tetrahydrocannabinolic acid synthase activity of the engineered
variants using
known methods. For example, by GC-MS or LC-MS or as described in the examples
provided herein. Engineered variants may have amino acid substitutions
compared to a wild
type tetrahydrocannabinolic acid synthase sequence, such as the
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44. In
addition to
substitutions, engineered variants may comprise truncations, additions, and/or
deletions,
and/or other mutations compared to a wild type tetrahydrocannabinolic acid
synthase
sequence, such as the tetrahydrocannabinolic acid synthase polypeptide having
an amino
acid sequence of SEQ ID NO:44. Engineered variants may have substitutions
compared a
non-wild type tetrahydrocannabinolic acid synthase sequence. In addition to
substitutions,
engineered variants may comprise truncations, additions, and/or deletions
and/or other
mutations compared to a non-wild type tetrahydrocannabinolic acid synthase
sequence. The
engineered variants described herein contain at least one amino acid residue
substitution
from a parent tetrahydrocannabinolic acid synthase polypeptide. In some
embodiments, the
parent tetrahydrocannabinolic acid synthase polypeptide is a wild type
sequence. In some
embodiments, the parent tetrahydrocannabinolic acid synthase polypeptide is a
non-wild
type sequence.
[0095] As used herein, the term "heterologous" may refer to what is not
normally
found in nature. As such, a heterologous nucleotide sequence may be: (a)
foreign to its host
cell (i.e., is "exogenous" to the cell); (b) naturally found in the host cell
(i.e., "endogenous")
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
but present at an unnatural quantity in the cell (i.e., greater or lesser
quantity than naturally
found in the host cell); (c) be naturally found in the host cell but
positioned outside of its
natural locus; or (d) be naturally found in the host cell, but with introns
removed or added.
The term "heterologous nucleotide sequence" or the term "heterologous nucleic
acid" may
refer to a nucleic acid or nucleotide sequence not normally found in a given
cell in nature. A
codon-optimized nucleotide sequence may be an example of a heterologous
nucleotide
sequence. The term "heterologous enzyme" or "heterologous polypeptide" may
refer to an
enzyme or polypeptide that is not normally found in a given cell in nature.
The term
encompasses an enzyme or polypeptide that is: (a) exogenous to a given cell
(i.e., encoded
by a nucleic acid that is not naturally present in the host cell or not
naturally present in a
given context in the host cell); or (b) naturally found in the host cell
(e.g., the enzyme or
polypeptide is encoded by a nucleic acid that is endogenous to the cell) but
that is produced
in an unnatural amount (e.g., greater or lesser than that naturally found) in
the host cell. For
example, a heterologous polypeptide may include a mutated version of a
polypeptide
naturally occurring in a host cell. A heterologous nucleic acid may be: (a)
foreign to its host
cell (i.e., is "exogenous" to the cell); (b) naturally found in the host cell
(i.e., "endogenous")
but present at an unnatural quantity in the cell (i.e., greater or lesser
quantity than naturally
found in the host cell); or (c) be naturally found in the host cell but
positioned outside of its
natural locus. In some embodiments, a heterologous nucleic acid may comprise a
codon-
optimized nucleotide sequence.
[0096] As used herein, the term "one or more heterologous nucleic acids"
or "one or
more heterologous nucleotide sequences" may refer to heterologous nucleic
acids
comprising one or more nucleotide sequences encoding one or more polypeptides.
In some
embodiments, these one or more heterologous nucleic acids may comprise a
nucleotide
sequence encoding one polypeptide. In other embodiments, these one or more
heterologous
nucleic acids may comprise nucleotide sequences encoding more than one
polypeptide. In
some embodiments, these one or more heterologous nucleic acids may comprise
nucleotide
sequences encoding multiple copies of the same polypeptide. In some
embodiments, these
one or more heterologous nucleic acids may comprise nucleotide sequences
encoding
multiple copies of different polypeptides.
[0097] As used herein, "increased ratio" may refer to an increase in the
molar ratio,
an increase in the mass (or weight) ratio, an increase in the molarity ratio,
or an increase in
the mass concentration (e.g., mg/L or mg/mL) ratio between two products
produced by a
31
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide, engineered variant, method, and/or modified host cell disclosed
herein
compared to the molar ratio, mass (or weight) ratio, molarity ratio, or mass
concentration
ratio between the same two products produced by another polypeptide,
engineered variant,
method, and/or modified host cell disclosed herein (e.g., a comparative
polypeptide,
engineered variant, method, and/or modified host cell disclosed herein). For
example, a
100:1 ratio of THCA over CBCA produced by an engineered variant disclosed
herein would
be an increased ratio of THCA over CBCA compared to an 11:1 ratio of THCA over
CBCA
produced by a different engineered variant disclosed herein.
[0098] As used herein, a ratio of products produced by a polypeptide,
engineered
variant, method, and/or modified host cell disclosed herein, such as the ratio
of THCA over
CBCA, may refer to a molar ratio, a mass (or weight) ratio, molarity ratio, or
a mass
concentration (e.g., mg/L or mg/mL) ratio. For example, if a modified host
cell disclosed
herein produced 4 mM THCA and 1 mM CBCA, the ratio of THCA over CBCA would be
4:1.
[0099] "Operably linked" may refer to an arrangement of elements wherein
the
components so described are configured so as to perform their usual function.
Thus, control
sequences operably linked to a coding sequence are capable of effecting the
expression of
the coding sequence. The control sequences need not be contiguous with the
coding
sequence, so long as they function to direct the expression thereof. Thus, for
example,
intervening untranslated yet transcribed sequences can be present between a
promoter
sequence and the coding sequence and the promoter sequence can still be
considered
"operably linked" to the coding sequence.
[0100] "Isolated" may refer to polypeptides or nucleic acids that are
substantially or
essentially free from components that normally accompany them in their natural
state.
An isolated polypeptide or nucleic acid may be other than in the form or
setting in which it is
found in nature. Isolated polypeptides and nucleic acids therefore may be
distinguished
from the polypeptides and nucleic acids as they exist in natural cells. An
isolated nucleic
acid or polypeptide may be purified from one or more other components in a
mixture with
the isolated nucleic acid or polypeptide, if such components are present.
[0101] A "modified host cell" (also may be referred to as a "recombinant
host cell")
may refer to a host cell into which has been introduced a heterologous nucleic
acid, e.g., an
expression vector or construct. For example, a modified eukaryotic host cell
may be
32
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
produced through introduction into a suitable eukaryotic host cell of a
heterologous nucleic
acid.
[0102] As used herein, a "cell-free system" may refer to a cell lysate,
cell extract or
other preparation in which substantially all of the cells in the preparation
have been
disrupted or otherwise processed so that all or selected cellular components,
e.g., organelles,
proteins, nucleic acids, the cell membrane itself (or fragments or components
thereof), or the
like, are released from the cell or resuspended into an appropriate medium
and/or purified
from the cellular milieu. Cell-free systems can include reaction mixtures
prepared from
purified and/or isolated polypeptides and suitable reagents and buffers.
[0103] In some embodiments, conservative substitutions may be made in the
amino
acid sequence of a polypeptide without disrupting the three-dimensional
structure or function
of the polypeptide. Conservative substitutions may be accomplished by the
skilled artisan by
substituting amino acids with similar hydrophobicity, polarity, and R-chain
length for one
another. Additionally, by comparing aligned sequences of homologous proteins
from
different species, conservative substitutions may be identified by locating
amino acid
residues that have been mutated between species without altering the basic
functions of the
encoded proteins. The term "conservative amino acid substitution" may refer to
the
interchangeability in proteins of amino acid residues having similar side
chains. For
example, a group of amino acids having aliphatic side chains may consist of
glycine, alanine,
valine, leucine, and isoleucine; a group of amino acids having aliphatic-
hydroxyl side chains
may consist of serine and threonine; a group of amino acids having amide
containing side
chains may consist of asparagine and glutamine; a group of amino acids having
aromatic
side chains may consist of phenylalanine, tyrosine, and tryptophan; a group of
amino acids
having basic side chains may consist of lysine, arginine, and histidine; a
group of amino
acids having acidic side chains may consist of glutamate and aspartate; and a
group of amino
acids having sulfur containing side chains may consist of cysteine and
methionine.
Exemplary conservative amino acid substitution groups are: valine-leucine-
isoleucine,
phenylalanine-tyrosine, lysine-arginine, alanine-valine, and asparagine-
glutamine.
[0104] A polynucleotide or polypeptide has a certain percent "sequence
identity" to
another polynucleotide or polypeptide, meaning that, when aligned, that
percentage of bases
or amino acids are the same, and in the same relative position, when comparing
the two
sequences. Sequence identity can be determined in a number of different
manners. To
determine sequence identity, sequences can be aligned using various methods
and computer
33
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
programs (e.g., BLAST, T-COFFEE, MUSCLE, MAFFT, etc.), available over the
world
wide web at sites including
ncbi.nlm.nili.gov/BLAST,ebi.ac.uk/Tools/msa/tcoffee/ebi.ac.uk/
Tools/msa/muscle/mafft.cbrc.jp/alignment/software/. See, e.g., Altschul et al.
(1990), J. Mol.
Biol. 215:403-10.
[0105] Before the present disclosure is further described, it is to be
understood that
this disclosure is not limited to particular embodiments described, as such
may, of course,
vary. It is also to be understood that the terminology used herein is for the
purpose of
describing particular embodiments only, and is not intended to be limiting,
since the scope of
the present disclosure will be limited only by the appended claims.
[0106] Where a range of values is provided, it is understood that each
intervening
value, to the tenth of the unit of the lower limit unless the context clearly
dictates otherwise,
between the upper and lower limit of that range and any other stated or
intervening value in
that stated range, is encompassed within the disclosure. The upper and lower
limits of these
smaller ranges may independently be included in the smaller ranges, and are
also
encompassed within the disclosure, subject to any specifically excluded limit
in the stated
range. Where the stated range includes one or both of the limits, ranges
excluding either or
both of those included limits are also included in the disclosure.
[0107] Unless defined otherwise, all technical and scientific terms used
herein have
the same meaning as commonly understood by one of ordinary skill in the art to
which this
disclosure belongs. Although any methods and materials similar or equivalent
to those
described herein can also be used in the practice or testing of the present
disclosure, the
preferred methods and materials are now described. All publications mentioned
herein are
incorporated herein by reference to disclose and describe the methods and/or
materials in
connection with which the publications are cited.
[0108] It must be noted that as used herein and in the appended claims,
the singular
forms "a," "an," and "the" may include plural referents unless the context
clearly dictates
otherwise. Thus, for example, reference to "a cannabinoid compound" or
"cannabinoid" may
include a plurality of such compounds and reference to "the modified host
cell" may include
reference to one or more modified host cells and equivalents thereof known to
those skilled
in the art, and so forth. It is further noted that the claims may be drafted
to exclude any
optional element. As such, this statement is intended to serve as antecedent
basis for use of
such exclusive terminology as "solely," "only" and the like in connection with
the recitation
of claim elements, or use of a "negative" limitation.
34
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0109] It is appreciated that certain features of the disclosure, which
are, for clarity,
described in the context of separate embodiments, may also be provided in
combination in a
single embodiment. Conversely, various features of the disclosure, which are,
for brevity,
described in the context of a single embodiment, may also be provided
separately or in any
suitable sub-combination. All combinations of the embodiments pertaining to
the disclosure
are specifically embraced by the present disclosure and are disclosed herein
just as if each
and every combination was individually and explicitly disclosed. In addition,
all sub-
combinations of the various embodiments and elements thereof are also
specifically
embraced by the present disclosure and are disclosed herein just as if each
and every such
sub-combination was individually and explicitly disclosed herein.
Engineered Variants of the Tetrahydrocannabinolic Acid Synthase (THCAS)
Polypeptide
[0110] Disclosed herein are engineered variants of a
tetrahydrocannabinolic acid
synthase (THCAS) polypeptide comprising an amino acid sequence of SEQ ID NO:44
with
one or more amino acid substitutions. The inventors have identified amino acid
locations of
the THCAS polypeptide comprising an amino acid sequence of SEQ ID NO:44 that
when
substituted, may result in one or more improved properties of the engineered
variant. In one
aspect of the disclosure, the substitution is at a location corresponding to
the position in the
THCAS polypeptide of SEQ ID NO:44 from Cannabis sativa. The THCAS polypeptide
of
SEQ ID NO:44 from Cannabis sativa comprises the following domains:
1. Signal polypeptide: amino acids 1-28.
2. FAD binding domain: amino acids 77-251.
3. BBE domain: amino acids 480-538.
[0111] The THCAS polypeptide of SEQ ID NO:44 from Cannabis sativa also
comprises the following domains surface exposed amino acids: 28-33, 35, 36, 39-
45, 47-50,
52, 55-59, 61, 62, 65, 66, 69, 71-77, 79, 80, 82, 88, 89, 90, 94, 98, 101,
102, 104, 109, 114,
115, 124, 125, 126, 133, 134, 136-139, 141-145, 148, 150, 161, 164-168, 176,
183, 197, 202,
205, 208, 213, 215-221, 223, 224, 225, 231, 236, 245, 247, 250, 252, 253, 259,
261, 262-
268, 271, 274, 275, 278, 279, 281, 282, 284, 285, 286, 292, 294, 296-306, 312,
318, 321,
322, 323, 326, 327, 329, 330, 331, 333, 334, 336, 338-341, 343, 344, 349, 356,
358-368,
371-374, 377, 378, 389, 390, 391, 393, 394, 395, 399, 402, 403, 405, 406, 408,
409, 410,
413, 422, 424-430, 437, 438, 444, 446, 448, 450, 451-454, 456, 457, 460, 463,
464, 467,
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
468, 470, 471, 472, 475-478, 483, 484, 487, 488, 491, 493-502, 504, 505, 508,
509, 513,
516, 517, 520, 524, 525, 527, 528, 530, 532, and 540-545.
[0112] Residue positions in the engineered variants discussed herein are
identified
with respect to a reference amino acid sequence, the THCAS polypeptide of SEQ
ID NO:44
from Cannabis sativa (shown herein in Table 1; UniProtKB/Swiss-Prot: Q8GTB6).
Accordingly, a reference to "F317" identifies an amino acid that, in the THCAS
polypeptide
of SEQ ID NO:44 from Cannabis sativa, is the 317th amino acid from the N-
terminus,
wherein the methionine is the first amino acid. The 317th amino acid is a
phenylalanine (F)
in the THCAS polypeptide of SEQ ID NO:44 from Cannabis sativa. Those of skill
in the art
appreciate that the F317 amino acid may have a different position in the THCAS
polypeptides from different species or in different isoforms. These engineered
variants are
intended to be encompassed by this disclosure.
[0113] The polypeptide sequence position at which a particular amino acid
or amino
acid change ("residue difference") is present is sometimes described herein as
"Xn", or
"position n", where n refers to the amino acid position with respect to the
reference
sequence. Accordingly, a reference to "X317" identifies an amino acid that, in
the THCAS
polypeptide of SEQ ID NO:44 from Cannabis sativa, is the 317th amino acid from
the N-
terminus.
[0114] A specific substitution mutation, which is a replacement of the
specific amino
acid in a reference sequence with a different specified residue may be denoted
by the
conventional notation "X (number)Y", where X is the single letter identifier
of the amino in
the reference sequence, "number" is the amino acid position in the reference
sequence, and
Y is the single letter identifier of the amino acid substitution in the
engineered sequence.
Accordingly, a reference to "F317Y" identifies a substitution that, in the
THCAS
polypeptide of SEQ ID NO:44 from Cannabis sativa, is the 317th amino acid from
the N-
terminus, phenylalanine, being replaced by tyrosine.
[0115] Cannabinoid synthase polypeptides, secreted polypeptides, have
structural
features that may hinder expression in modified host cells, such as modified
yeast cells.
Cannabinoid synthase polypeptides comprise disulfide bonds, numerous
glycosylation sites,
including N-glycosylation sites, and a bicovalently attached flavin adenine
dinucleotide
(FAD) cofactor moiety. Accordingly, reconstituting the activity of or
expressing cannabinoid
synthase polypeptides in a modified host cell, such as a modified yeast cell,
can be
challenging and unreliable. Often these secreted polypeptides are misfolded or
mislocalized,
36
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
resulting in low expression, polypeptides lacking activity, reduced host cell
viability, and/or
cell death. As disclosed herein, engineered variants may have improved
expression, folding,
and enzymatic activity compared to the THCAS polypeptide comprising an amino
acid
sequence of SEQ ID NO:44. Additionally, expression of the engineered variants
of the
disclosure may enhance viability of the modified host cells disclosed herein
compared to
modified host cells expressing a THCAS polypeptide comprising an amino acid
sequence of
SEQ ID NO:44.
[0116] The disclosure provides for an engineered variant of a
tetrahydrocannabinolic
acid synthase (THCAS) polypeptide comprising an amino acid sequence of SEQ ID
NO:44
with one or more amino acid substitutions. In certain such embodiments, the
engineered
variant comprises an amino acid sequence with at least 85%, at least 86%, at
least 87%, at
least 88%, at least 89%, at least 90%, at least 91%, at least 92%, at least
93%, at least 94%,
at least 95%, at least 96%, at least 97%, at least 98%, or at least 99%
sequence identity to
SEQ ID NO:44 . In some embodiments, the engineered variant comprises an amino
acid
sequence with at least 75%, at least 76%, at least 77%, at least 78%, at least
79%, at least
80%, at least 81%, at least 82%, at least 83%, or at least 84% sequence
identity to SEQ ID
NO:44.
[0117] The disclosure provides for an engineered variant of a
tetrahydrocannabinolic
acid synthase (THCAS) polypeptide comprising an amino acid sequence of SEQ ID
NO:44
with one or more amino acid substitutions, wherein the engineered variant
comprises at least
one amino acid substitution in a signal polypeptide, a flavin adenine
dinucleotide (FAD)
binding domain, a berberine bridge enzyme (BBE) domain, or a combination of
the
foregoing. In some embodiments, at least one amino acid substitution is
present in the signal
polypeptide. In certain such embodiments, the engineered variant comprises at
least 1, at
least 2, at least 3, at least 4, at least 5, at least 6, at least 7, at least
8, at least 9, at least 10, at
least 11, at least 12, at least 13, at least 14, or at least 15 amino acid
substitutions in the
signal polypeptide. In some embodiments, the engineered variant comprises 1,
2, 3, 4, 5, 6,
7, 8, 9, 10, 11, 12, 13, 14, or 15 amino acid substitutions in the signal
polypeptide. In some
embodiments, at least one amino acid substitution is present in the FAD
binding domain. In
certain such embodiments, the engineered variant comprises at least 1, at
least 2, at least 3, at
least 4, at least 5, at least 6, at least 7, at least 8, at least 9, at least
10, at least 11, at least 12,
at least 13, at least 14, or at least 15 amino acid substitutions in the FAD
binding domain. In
some embodiments, the engineered variant comprises 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13,
37
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
14, or 15 amino acid substitutions in the FAD binding domain. In some
embodiments,
wherein at least one amino acid substitution is present in the FAD domain, the
engineered
variant comprises at least one amino acid substitution at an amino acid
selected from the
group consisting of X100, X103, X109, X124, X125, X132, X137, X143, X149,
X161,
X165, X167, X168, X170, X171, X172, X175, X180, X196, X208, X235, and X250. In
some embodiments, wherein at least one amino acid substitution is present in
the FAD
domain, the engineered variant comprises at least one amino acid substitution
at an amino
acid selected from the group consisting of S100, V103, T109, Q124, V125, L132,
S137,
H143, V149, W161, K165, E167, N168, S170, F171, P172, Y175, G180, N196, H208,
G235, and A250. In some embodiments, wherein at least one amino acid
substitution is
present in the FAD domain, the engineered variant comprises at least one amino
acid
substitution selected from the group consisting of L132M, S170T, F171I, N196T,
N196Q,
and N196V. In some embodiments, at least one amino acid substitution is
present in the BBE
domain. In certain such embodiments, the engineered variant comprises at least
1, at least 2,
at least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at
least 9, at least 10, at least 11,
at least 12, at least 13, at least 14, or at least 15 amino acid substitutions
in the BBE domain.
In some embodiments, the engineered variant comprises 1, 2, 3, 4, 5, 6, 7, 8,
9, 10, 11, 12,
13, 14, or 15 amino acid substitutions in the BBE domain. In some embodiments,
wherein at
least one amino acid substitution is present in the BBE domain, the engineered
variant
comprises at least one amino acid substitution at an amino acid selected from
the group
consisting of X500 and X528. BBE domain, the engineered variant comprises at
least one
amino acid substitution at an amino acid selected from the group consisting of
Y500 and
N528. In some embodiments, wherein at least one amino acid substitution is
present in the
BBE domain, the engineered variant comprises at least one amino acid
substitution selected
from the group consisting of Y500M, Y500V, and N528E.
[0118] The disclosure provides for an engineered variant of a
tetrahydrocannabinolic
acid synthase (THCAS) polypeptide comprising an amino acid sequence of SEQ ID
NO:44
with one or more amino acid substitutions, wherein the engineered variant
comprises
substitution of at least one surface exposed amino acid. In certain such
embodiments, at least
one hydrophobic surface exposed amino acid is substituted with a hydrophilic
amino acid. In
some embodiments, at least one hydrophilic surface exposed amino acid is
substituted with a
hydrophobic amino acid. In some embodiments, the engineered variant comprises
substitution of at least 1, at least 2, at least 3, at least 4, at least 5, at
least 6, at least 7, at least
38
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
8, at least 9, at least 10, at least 11, at least 12, at least 13, at least
14, or at least 15 surface
exposed amino acids. In some embodiments, the engineered variant comprises
substitution
of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, or 15 surface exposed amino
acids. In some
embodiments, wherein the engineered variant comprises substitution of at least
one surface
exposed amino acid, the engineered variant comprises at least one amino acid
substitution
selected from the group consisting of X132, X170, X171, X196, X261, X269,
X317, and
X539. In some embodiments, wherein the engineered variant comprises
substitution of at
least one surface exposed amino acid, the engineered variant comprises at
least one amino
acid substitution selected from the group consisting of L132, S170, F171,
N196, K261,
L269, F317, and P539. In some embodiments, wherein the engineered variant
comprises
substitution of at least one surface exposed amino acid, the engineered
variant comprises at
least one amino acid substitution selected from the group consisting of L132M,
S170T,
F171I, N196T, N196Q, N196V, K261C, L269I, F317Y, and P539T. Substitution of
hydrophobic surface exposed amino acids with hydrophilic amino acids may
increase the
hydrophilicity of solvent-exposed amino acids, which may improve solubility of
the
engineered variants of the disclosure in an aqueous (non-trichome)
environment.
[0119] The
disclosure provides for an engineered variant, wherein the engineered
variant comprises at least one amino acid substitution at an amino acid
selected from the
group consisting of X31, X43, X49, X50, X 51, X55, X56, X59, X61, X62, X71,
X100,
X103, X109, X124, X125, X132, X137, X143, X149, X161, X165, X168, X167, X170,
X171, X172, X175, X180, X196, X208, X235, X250, X257, X261, X269, X311, X317,
X327, X390, X379, X429, X467, X500, X528, X539, X542, X543, X544, and X545.
Such
engineered variants may produce THCA from CBGA in a greater amount, as
measured in
mg/L or mM, than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44 under
similar
conditions for the same length of time. In some embodiments, such engineered
variants may
produce THCA from CBGA in a greater amount, as measured in mg/L or mM, than an
amount of THCA produced from CBGA by a tetrahydrocannabinolic acid synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 under similar
conditions for
the same length of time and may produce THCA from CBGA in an increased ratio
of THCA
over another another cannabinoid (e.g., CBCA) compared to that produced by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time.
39
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
[0120] The
disclosure provides for an engineered variant, wherein the engineered
variant comprises at least one amino acid substitution at an amino acid
selected from the
group consisting of R31, P43, P49, K50, L51, Q55, H56, L59, M61, S62, L71,
S100, V103,
T109, Q124, V125, L132, S137, H143, W161, K165, N168, E167, Y175, G180, N196,
H208, A250, 1257, K261, G311, F317, L327, K390, T379, D429, Y500, N528, P542,
H543,
H544, and H545. Such engineered variants may produce THCA from CBGA in a
greater
amount, as measured in mg/L or mM, than an amount of THCA produced from CBGA
by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time. In some
embodiments, such
engineered variants may produce THCA from CBGA in a greater amount, as
measured in
mg/L or mM, than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44 under
similar
conditions for the same length of time and may produce THCA from CBGA in an
increased
ratio of THCA over another cannabinoid (e.g., CBCA) compared to that produced
by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time.
[0121] The
disclosure provides for an engineered variant, wherein the engineered
variant comprises at least one amino acid substitution selected from the group
consisting of
R31Q, P43E, P49E, P49K, P49Q, K50T, L51I, Q55E, Q55P, H56E, L59E, M61W, M61H,
M615, 562Q, L71A, S100A, V103F,T109V,Q124D, Q124E, Q124N, V125E, V125Q,
L132M, 5137G, H143D, W161R, W161Y, W161K, K165A, N1685, E167P, Y175F,
G180A, N196Q, N196V, H208T, A250T, I257V, K261C, G311A, F317Y, L327I, K390E,
T37975, Y500M, Y500V, N528E, P542E, P542V, H543V, H544A, H545D, and H545E.
Such engineered variants may produce THCA from CBGA in a greater amount, as
measured
in mg/L or mM, than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time. In some
embodiments, such
engineered variants may produce THCA from CBGA in a greater amount, as
measured in
mg/L or mM, than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44 under
similar
conditions for the same length of time and may produce THCA from CBGA in an
increased
ratio of THCA over another cannabinoid (e.g., CBCA) compared to that produced
by a
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time.
[0122] The
disclosure provides for an engineered variant, wherein the engineered
variant comprises an amino acid sequence selected from the group consisting of
SEQ ID
NO:50, SEQ ID NO:52, SEQ ID NO:54, SEQ ID NO:56, SEQ ID NO:58, SEQ ID NO:60,
SEQ ID NO:70, SEQ ID NO:72, SEQ ID NO: 74, SEQ ID NO: 76, SEQ ID NO: 78, SEQ
ID
NO:80 SEQ ID NO:82, SEQ ID NO:84, SEQ ID NO:86, SEQ ID NO: 88, SEQ ID NO:90,
SEQ ID NO:92, SEQ ID NO:94, SEQ ID NO:96, SEQ ID NO:98, SEQ ID NO:100, SEQ ID
NO:102, SEQ ID NO:104, SEQ ID NO:106, SEQ ID NO:108, SEQ ID NO:110, SEQ ID
NO:112, SEQ ID NO:114, SEQ ID NO:116, SEQ ID NO:118, SEQ ID NO:120, SEQ ID
NO:124, SEQ ID NO:126, SEQ ID NO:128, SEQ ID NO:130, SEQ ID NO:132, SEQ ID
NO: 134, SEQ ID NO:138, SEQ ID NO: 140, SEQ ID NO:142, SEQ ID NO:144, SEQ ID
NO:146, SEQ ID NO:148, SEQ ID NO: 152, SEQ ID NO:156, SEQ ID NO:158, SEQ ID
NO:160, SEQ ID NO:164, SEQ ID NO:166, SEQ ID NO:168, SEQ ID NO:170, SEQ ID
NO:172, SEQ ID NO:174, SEQ ID NO:176, SEQ ID NO: 178, SEQ ID NO:180, SEQ ID
NO: 182, SEQ ID NO: 184, and SEQ ID NO:186. Such engineered variants may
produce
THCA from CBGA in a greater amount, as measured in mg/L or mM, than an amount
of
THCA produced from CBGA by a tetrahydrocannabinolic acid synthase polypeptide
having
an amino acid sequence of SEQ ID NO:44 under similar conditions for the same
length of
time. In some embodiments, such engineered variants may produce THCA from CBGA
in a
greater amount, as measured in mg/L or mM, than an amount of THCA produced
from
CBGA by a tetrahydrocannabinolic acid synthase polypeptide having an amino
acid
sequence of SEQ ID NO:44 under similar conditions for the same length of time
and may
produce THCA from CBGA in an increased ratio of THCA over another cannabinoid
(e.g.
CBCA) compared to that produced by a tetrahydrocannabinolic acid synthase
polypeptide
having an amino acid sequence of SEQ ID NO:44 under similar conditions for the
same
length of time.
[0123] The
disclosure provides for an engineered variant, wherein the engineered
variant comprises an amino acid sequence of SEQ ID NO:44 with at least 1, at
least 2, at
least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least
9, at least 10, at least 11, at
least 12, at least 13, at least 14, at least 15, at least 16, at least 17, at
least 18, at least 19, at
least 20, at least 21, at least 22, at least 23, at least 24, at least 25, at
least 26, at least 27, at
least 28, at least 29, or at least 30 amino acid substitutions. The disclosure
provides for an
41
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
engineered variant, wherein the engineered variant comprises an amino acid
sequence of
SEQ ID NO:44 with 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 amino acid substitutions. Combinations of
the amino acid
substitutions described herein can be made and the resulting engineered
variants screened for
improved tetrahydrocannabinolic acid synthase (THCAS) properties. Engineered
variants
comprising combinations of all of the substitutions described herein are
intended to be
encompassed by this disclosure. In some embodiments, the engineered variant
comprises at
least 1, at least 2, at least 3, at least 4, at least 5, at least 6, at least
7, at least 8, at least 9, at
least 10, at least 11, at least 12, at least 13, at least 14, at least 15, at
least 16, at least 17, at
least 18, at least 19, at least 20, at least 21, at least 22, at least 23, at
least 24, at least 25, at
least 26, at least 27, at least 28, at least 29, or at least 30 of the amino
acid substitutions
described herein. In some embodiments, the engineered variant comprises 1, 2,
3, 4, 5, 6, 7,
8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, or 30 of the
amino acid substitutions described herein (e.g., 1-30 of the amino acid
substitutions
described herein). In some embodiments, the engineered variant comprises 1, 2,
3, 4, 5, 6, 7,
8,9, 10, 11, 12, 13, 14, or 15 of the amino acid substitutions described
herein (e.g., 1-15 of
the amino acid substitutions described herein). In some embodiments, the
engineered
variant comprises 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 of the amino acid
substitutions described
herein (e.g., 1-10 of the amino acid substitutions described herein). In some
embodiments,
the engineered variant comprises 1, 2, 3, 4, or 5 of the amino acid
substitutions described
herein (e.g., 1-5 of the amino acid substitutions described herein). In some
embodiments, the
engineered variant comprises 1, 2, 3, or 4 of the amino acid substitutions
described herein
(e.g., 1-4 of the amino acid substitutions described herein). In some
embodiments, the
engineered variant comprises 1, 2, or 3 of the amino acid substitutions
described herein (e.g.,
1-3 of the amino acid substitutions described herein). In some embodiments,
the engineered
variant comprises 1 or 2 of the amino acid substitutions described herein
(e.g., 1-2 of the
amino acid substitutions described herein). In some embodiments, the
engineered variant
comprises 1 of the amino acid substitutions described herein. In some
embodiments, the
engineered variant comprises 2 of the amino acid substitutions described
herein. In some
embodiments, the engineered variant comprises 3 of the amino acid
substitutions described
herein. In some embodiments, the engineered variant comprises 4 of the amino
acid
substitutions described herein. In some embodiments, the engineered variant
comprises 5 of
the amino acid substitutions described herein.
42
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0124] The disclosure provides for an engineered variant, wherein the
engineered
variant comprises at least one immutable amino acid. The disclosure provides
for an
engineered variant, wherein the engineered variant comprises at least one
immutable amino
acid in a flavin adenine dinucleotide (FAD) binding domain, a berberine bridge
enzyme
(BBE) domain, or a combination of the foregoing.
[0125] In some embodiments, the engineered variant comprises at least one
immutable amino acid in the FAD binding domain. In certain such embodiments,
the
engineered variant comprises at least 1, at least 2, at least 3, at least 4,
at least 5, at least 6, at
least 7, at least 8, at least 9, at least 10, at least 11, at least 12, at
least 13, at least 14, or at
least 15 immutable amino acids in the FAD binding domain. In some embodiments,
the
engineered variant comprises at least one immutable amino acid in the FAD
binding domain,
wherein the at least one immutable amino acid is selected from the group
consisting of X87,
X93, X99, X108, X110, X112, X117, X118, X120, X126, X127, X131, X141, X148,
X152,
X153, X155, X156, X157, X159, X160, X163, X170, X171, X172, X173, X174, X176,
X177, X178, X179, X182, X183, X184, X185, X187, X188, X189, X190, X191, X192,
X193, X195, X201, X202, X205, X206, X210, X214, X223, X225, X226, X227, X228,
X231, X234, X237, X238, X239, X245, X246, X248, and X251. In some embodiments,
mutation of one or more of these immutable amino acids reduces titer of one or
more
cannabinoids. In some embodiments, mutation of one or more of amino acids
X170, X171,
and/or X172 reduces titer of one or more cannabinoids. In some embodiments,
wherein the
engineered variant comprises at least one immutable amino acid in the FAD
binding domain,
the at least one immutable amino acid is selected from the group consisting of
P87, 193, C99,
R108, R110, G112, E117, G118, S120, P126, F127, D131, D141, W148, G152, A153,
L155,
G156, E157, Y159, Y160, N163, S170, F171, P172, G173, G174, C176, P177, T178,
V179,
G182, G183, H184, F185, G187, G188, G189, Y190, G191, A192, L193, R195, A201,
D202, 1205, D206, V210, G214, G223, D225, L226, F227, W228, R231, G234, N237,
F238,
G239, K245, 1246, L248, and V251. In some embodiments, mutation of one or more
of
amino acids S170, F171, and/or P172 reduces titer of one or more cannabinoids.
[0126] In some embodiments, the engineered variant comprises at least one
immutable amino acid in the BBE domain. In certain such embodiments, the
engineered
variant comprises at least 1, at least 2, at least 3, at least 4, at least 5,
at least 6, at least 7, at
least 8, at least 9, at least 10, at least 11, at least 12, at least 13, at
least 14, or at least 15
immutable amino acids in the BBE domain. In some embodiments, wherein the
engineered
43
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
variant comprises at least one immutable amino acid in the BBE domain, the at
least one
immutable amino acid is selected from the group consisting of X485, X499,
X503, X514,
X515, X522, X529, X530, X534, X535, and X536. In some embodiments, wherein the
engineered variant comprises at least one immutable amino acid in the BBE
domain, the at
least one immutable amino acid is selected from the group consisting of R485,
N499, A503,
N514, F515, K522, N529, F530, E534, Q535, and S536.
[0127] The
disclosure provides for an engineered variant, wherein the engineered
variant comprises at least one immutable amino acid selected from the group
consisting of
X28, X34, X35, X37, X64, X70, X87, X93, X99, X108, X110, X112, X117, X118,
X120,
X126, X127, X131, X141, X148, X152, X153, X155, X156, X157, X159, X160, X163,
X173, X174, X176, X177, X178, X179, X182, X183, X184, X185, X187, X188, X189,
X190, X191, X192, X193, X195, X201, X202, X205, X206, X210, X214, X223, X225,
X226, X227, X228, X231, X234, X237, X238, X239, X245, X246, X248, X251, X260,
X277, X313, X314, X324, X342, X353, X355, X381, X382, X383, X384, X386, X387,
X392, X413, X416, X420, X423, X426, X431, X432, X434, X435, X436, X438, X441,
X444, X445, X446, X465, X466, X469, X470, X472, X473, X477, X485, X499, X503,
X514, X515, X522, X529, X530, X534, X535, and X536. In certain such
embodiments, the
engineered variant comprises at least one immutable amino acid selected from
the group
consisting of X37, X70, X93, X99, X117, X120, X127, X131, X156, X157, X159,
X174,
X176, X182, X183, X185, X187, X188, X189, X190, X191, X192, X195, X202, X206,
X214, X228, X234, X238, X248, X277, X314, X324, X355, X382, X384, X386, X420,
X423, X436, X441, X444, X445, X472, X477, X514, X515, X529, and X535. In some
embodiments, the engineered variant comprises at least one immutable amino
acid selected
from the group consisting of A28, F34, L35, C37, L64, N70, P87, 193, C99,
R108, R110,
G112, E117, G118, S120, P126, F127, D131, D141, W148, G152, A153, L155, G156,
E157,
Y159, Y160, N163, A173, G174, C176, P177, T178, V179, G182, G183, H184, F185,
G187, G188, G189, Y190, G191, P192, L193, R195, A201, D202, 1205, D206, V210,
G214,
G223, D225, L226, F227, W228, R231, G234, S237, F238, G239, K245, 1246, L248,
V251,
V260, Q277, F313, S314, L324, C342, F353, S355, F381, K382, 1383, K384, D386,
Y387,
1392, M413, L416, G420, M423, 1426, 1431, P432, P434, H435, R436, G438, Y441,
W444,
Y445, 1446, 1465, Y466, M469, T470, Y472, V473, P477, R485, N499, A503, N514,
F515,
K522, N529, F530, E534, Q535, and S536. In certain such embodiments, the
engineered
variant comprises at least one immutable amino acid selected from the group
consisting of
44
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
C37, N70, 193, C99, E117, S120, F127, D131, G156, E157, Y159, G174, C176,
G182,
G183, F185, G187, G188, G189, Y190, G191, P192, R195, D202, D206, G214, W228,
G234, F238, L248, Q277, S314, L324, S355, K382, K384, D386, G420, M423, R436,
Y441,
W444, Y445, Y472, P477, N514, F515, N529, and Q535.
[0128] The disclosure provides for an engineered variant, wherein the
engineered
variant comprises at least 1, at least 2, at least 3, at least 4, at least 5,
at least 6, at least 7, at
least 8, at least 9, at least 10, at least 11, at least 12, at least 13, at
least 14, at least 15, at least
16, at least 17, at least 18, at least 19, at least 20, at least 21, at least
22, at least 23, at least
24, or at least 25 immutable amino acids, provided that the engineered variant
has at least
one amino acid substitution compared to SEQ ID NO:44. Engineered variants with
combinations of the immutable amino acids and substitiutions described herein
can be made
and the resulting engineered variants screened for improved
tetrahydrocannabinolic acid
synthase (THCAS) properties. Engineered variants comprising combinations of
all of the
substitutions and immutable amino acids described herein are intended to be
encompassed
by this disclosure.
[0129] The disclosure provides for an engineered variant, wherein the
engineered
variant comprises at least one amino acid substitution at the C-terminus. In
certain such
embodiments, a hydrophilic amino acid is replaced with a hydrophobic amino
acid. In some
embodiments, wherein the engineered variant comprises at least one amino acid
substitution
at the C-terminus, a hydrophobic amino acid is replaced with a hydrophilic
amino acid. Such
engineered variants may produce THCA from CBGA in a greater amount, as
measured in
mg/L or mM, than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44 under
similar
conditions for the same length of time.
[0130] The disclosure provides for an engineered variant, wherein the
engineered
variant comprises a truncation at the N-terminus, at the C-terminus, or at
both the N- and C-
termini. In some embodiments, the engineered variant comprises a truncation at
the N-
terminus. In some embodiments, the engineered variant comprises a truncation
at the C-
terminus. In some embodiments, the engineered variant comprises a truncation
at both the N-
and C-termini. In some embodiments, the engineered variant lacks a native
signal
polypeptide (i.e., amino acids 1-28 of SEQ ID NO:44).
[0131] In some embodiments, the engineered variant comprises a truncation
at the N-
terminus, at the C-terminus, or at both the N- and C-termini, and comprises an
amino acid
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
sequence with at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 9300, at least 9400, at least 9500,
at least 96%, at
least 9700, at least 98%, or at least 9900 sequence identity to SEQ ID NO:44.
In some
embodiments, the engineered variant comprises a truncation at the N-terminus,
at the C-
terminus, or at both the N- and C-termini, and comprises an amino acid
sequence with at
least '75%, at least '76%, at least '7'7%, at least '78%, at least '79%, at
least 80%, at least 81%,
at least 82%, at least 83%, or at least 84% sequence identity to SEQ ID NO:44.
[0132] In some
embodiments, the engineered variant comprises a truncation of at
least 1, at least 2, at least 3, at least 4, at least 5, at least 6, at least
7, at least 8, at least 9, or
at least 10 amino acids at the C-terminus. In some embodiments, the engineered
variant
comprises a truncation of at least 11, at least 12, at least 13, at least 14,
at least 15, at least
16, at least 17, at least 18, at least 19, or at least 20 amino acids at the C-
terminus. In some
embodiments, the engineered variant comprises a truncation of at least 21, at
least 22, at
least 23, at least 24, at least 25, at least 26, at least 27, at least 28, at
least 29, or at least 30
amino acids at the C-terminus. In some embodiments, the engineered variant
comprises a
truncation of 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 amino acids at the C-terminus
(e.g., 1-10 amino
acids at the C-terminus). In some embodiments, the engineered variant
comprises a
truncation of 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 amino acids at the C-
terminus (e.g., 11-
20 amino acids at the C-terminus). In some embodiments, the engineered variant
comprises
a truncation of 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 amino acids at the C-
terminus (e.g.,
21-30 amino acids at the C-terminus).
[0133] In some
embodiments, the engineered variant comprises a truncation of at
least 1, at least 2, at least 3, at least 4, at least 5, at least 6, at least
7, at least 8, at least 9, or
at least 10 amino acids at the N-terminus. In some embodiments, the engineered
variant
comprises a truncation of at least 11, at least 12, at least 13, at least 14,
at least 15, at least
16, at least 17, at least 18, at least 19, or at least 20 amino acids at the N-
terminus. In some
embodiments, the engineered variant comprises a truncation of at least 21, at
least 22, at
least 23, at least 24, at least 25, at least 26, at least 27, at least 28, at
least 29, or at least 30
amino acids at the N-terminus. In some embodiments, the engineered variant
comprises a
truncation of 1, 2, 3, 4, 5, 6, 7, 8, 9, or 10 amino acids at the N-terminus
(e.g., 1-10 amino
acids at the N-terminus). In some embodiments, the engineered variant
comprises a
truncation of 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 amino acids at the N-
terminus (e.g., 11-
20 amino acids at the N-terminus). In some embodiments, the engineered variant
comprises
46
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
a truncation of 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 amino acids at the N-
terminus (e.g.,
21-30 amino acids at the N-terminus).
[0134] In some embodiments, a truncated engineered variant of the
disclosure may
comprise a signal polypeptide. In certain such embodiments, the truncated
engineered
variant lacks a native signal polypeptide. In some embodiments, the signal
polypeptide is a
secretory signal polypeptide. In some embodiments, the secretory signal
polypeptide is a
native secretory signal polypeptide. In some embodiments, the secretory signal
polypeptide
is a synthetic secretory signal polypeptide. In some embodiments, the
secretory signal
polypeptide is an endoplasmic reticulum retention signal polypeptide. In
certain such
embodiments, the endoplasmic reticulum retention signal polypeptide is a HDEL
polypeptide or a KDEL polypeptide. In some embodiments, the secretory signal
polypeptide
is a mitochondrial targeting signal polypeptide. In some embodiments, the
secretory signal
polypeptide is a Golgi targeting signal polypeptide. In some embodiments, the
secretory
signal polypeptide is a vacuolar localization signal polypeptide. In certain
such
embodiments, the vacuolar localization signal polypeptide is a PEP4t
polypeptide or a
PRClt polypeptide. In certain such embodiments, the vacuolar localization
signal
polypeptide is a PEP4t polypeptide. In some embodiments, the secretory signal
polypeptide
is a plasma membrane localization signal polypeptide. In some embodiments, the
secretory
signal polypeptide is a peroxisome targeting signal polypeptide. In some
embodiments, the
peroxisome targeting signal polypeptide is a PEX8 polypeptide. In some
embodiments, the
secretory signal polypeptide is a mating factor secretory signal polypeptide
(e.g., a MF
polypeptide or an evolved MF polypeptide (MFev)). In some embodiments, the
signal
polypeptide is linked to the N-terminus of the engineered variant.
[0135] In some embodiments, a truncated engineered variant of the
disclosure may
comprise a membrane anchor. A membrane anchor may be a sequence that inserts
into a
membrane in the cell and anchor an attached polypeptide there. A membrane
anchor may be
present in a membrane external to the cell (e.g., GPI polypeptides) or
internal to the cell
(e.g., tail anchors, ER anchoring). Examples of membrane anchors include, but
are not
limited to, glycosylphosphatidylinositol membrane anchors (GPI polypeptides,
e.g. AGA1),
CAAX box polypeptides (get prenylated, e.g. RAS1), or tail anchored
polypeptides with a
hydrophobic C-terminus (e.g. phosphatidylinositol 4,5-bisphosphate 5-
phosphatase (INP54)
has a hydrophobic tail anchor in ER membrane or synaptobrevin 2 (VAMP2) has a
hydrophobic poly-I tail anchor in vesicle membranes).
47
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0136] The disclosure provides for an engineered variant, wherein the
engineered
variant comprises an addition and/or deletion of one or more amino acids.
[0137] Engineered variants of a THCAS polypeptide can be made and
screened for
improved properties, such as, production of THCA from CBGA in a greater
amount, as
measured in mg/L or mM, than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time. Additionally,
engineered
variants of a THCAS polypeptide can be made and screened for improved
properties, such
as, production of THCA from CBGA in an increased ratio of THCA over another
cannabinoid (e.g., CBCA) compared to that produced by a tetrahydrocannabinolic
acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44 under
similar
conditions for the same length of time. Similar conditions may refer to
reaction conditions at
the same temperature, pH, buffer, and/or fermentation conditions and in the
same culture
medium and/or reaction solvent.
[0138] In some embodiments of the disclosure, the engineered variant
produces
tetrahydrocannabinolic acid (THCA) from cannabigerolic acid (CBGA) in an
amount, as
measured in mg/L or mM, at least 5%, at least 10%, at least 15%, at least 20%,
at least 25%,
at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least
60%, at least
70%, at least 80%, at least 90%, at least 100%, at least 150% at least 200%,
at least 500%, or
at least 1000% greater than an amount of THCA produced from CBGA by a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 under similar conditions for the same length of time.
[0139] In some embodiments of the disclosure, the engineered variant
produces
THCA from CBGA in a ratio of THCA over another cannabinoid (e.g., CBCA) of
about
11:1, about 11.5:1, about 12:1, about 12.5:1, about 13:1, about 13.5:1, about
14:1, about
14.5:1, about 15:1, about 15.5:1, about 16:1, about 16.5:1, about 17:1, about
17.5:1, about
18:1, about 18.5:1, about 19:1, about 19.5:1, about 20:1, about 25:1, about
30:1, about 35:1,
about 40:1, about 45:1, about 50:1, about 60:1, about 70:1, about 80:1, about
90:1, about
100:1, about 150:1, about 200:1, about 500:1, or greater than about 500:1.
[0140] These improved properties may be assessed by the conversion of
CBGA to
THCA, or alternatively the conversion of another starting material to a
desired cannabinoid
or cannabinoid derivative, in vitro with isolated and/or purified engineered
variants of the
disclosure or in vivo in the context of a modified host cell expressing the
engineered variant.
48
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
In some embodiments, the modified host cell expresses polypeptides involved in
the MEV
pathway and/or polypeptides involved in cannabinoid biosynthesis and/or
comprises
modifications to the secretory pathway. It is contemplated that engineered
variants of the
disclosure having various degrees of stability, solubility, activity, and/or
expression level in
one or more of the test conditions will find use in the present disclosure for
the production of
cannabinoids or cannabinoid derivatives in a diversity of host cells.
[0141] Additionally, engineered variants of a THCAS polypeptide can be
made and
screened for improved properties, such as, production of cannabinoids or
cannabinoid
derivatives by modified host cells comprising one or more nucleic acids
comprising a
nucleotide sequence encoding the engineered variant in an amount, as measured
in mg/L or
mM, greater than an amount of the cannabinoid or the cannabinoid derivative
produced by
modified host cells comprising one or more nucleic acids comprising a
nucleotide sequence
encoding a tetrahydrocannabinolic acid synthase polypeptide having an amino
acid sequence
of SEQ ID NO:44, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, grown under similar culture conditions for the same length
of time.
[0142] Additionally, engineered variants of a THCAS polypeptide can be
made and
screened for improved properties, such as, modified host cells comprising one
or more
nucleic acids comprising a nucleotide sequence encoding the engineered variant
have a
faster growth rate and/or higher biomass yield compared to a growth rate
and/or higher
biomass yield of modified host cells comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant, grown under similar culture
conditions for the
same length of time. Additionally, engineered variants of a THCAS polypeptide
can be made
and screened for improved properties, such as, modified host cells comprising
one or more
nucleic acids comprising a nucleotide sequence encoding the engineered variant
produce
THCA from CBGA in an increased ratio of THCA over another cannabinoid (e.g.,
CBCA)
compared to that produced by modified host cells comprising one or more
nucleic acids
comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, grown under
similar
culture conditions for the same length of time. Similar culture conditions may
refer to host
49
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
cells grown in the same culture medium at the same temperature, pH, and/or
fermentation
conditions.
[0143] Moreover, engineered variants of a THCAS polypeptide can be made
and
screened for improved properties, such as, modified host cells comprising one
or more
nucleic acids comprising a nucleotide sequence encoding the engineered variant
do not have
significantly decreased growth or viability compared to modified host cells
comprising one
or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
similar culture conditions for the same length of time. Additionally,
engineered variants of a
THCAS polypeptide can be made and screened for improved properties, such as,
modified
host cells comprising one or more nucleic acids comprising a nucleotide
sequence encoding
the engineered variant do not have significantly decreased growth or viability
compared to
an unmodified host cell.
Nucleic Acids Comprising Nucleotide Sequences Encoding Engineered Variants of
the
Tetrahydrocannabinolic Acid Synthase (THCAS) Polypeptide and Expression
Vectors
and Constructs
[0144] The disclosure provides for nucleic acids comprising nucleotide
sequences
encoding engineered variants of the tetrahydrocannabinolic acid synthase
(THCAS)
polypeptide disclosed herein and expression vectors and constructs comprising
said nucleic
acids.
[0145] The disclosure provides nucleic acids comprising nucleotide
sequences
encoding engineered variants of the disclosure. Some embodiments of the
disclosure relate
to a nucleic acid comprising a nucleotide sequence encoding an engineered
variant of the
disclosure comprising an amino acid sequence set forth in SEQ ID NO:50, SEQ ID
NO:52,
SEQ ID NO:54, SEQ ID NO:56, SEQ ID NO:58, SEQ ID NO:60, SEQ ID NO:62, SEQ ID
NO:64, SEQ ID NO:66, SEQ ID NO:68, SEQ ID NO:70, SEQ ID NO:72, SEQ ID NO: 74,
SEQ ID NO: 76, SEQ ID NO: 78, SEQ ID NO:80 SEQ ID NO:82, SEQ ID NO:84, SEQ ID
NO:86, SEQ ID NO: 88, SEQ ID NO:90, SEQ ID NO:92, SEQ ID NO:94, SEQ ID NO:96,
SEQ ID NO:98, SEQ ID NO:100, SEQ ID NO:102, SEQ ID NO:104, SEQ ID NO:106, SEQ
ID NO:108, SEQ ID NO:110, SEQ ID NO:112, SEQ ID NO:114, SEQ ID NO:116, SEQ ID
NO:118, SEQ ID NO:120, SEQ ID NO:122, SEQ ID NO:124, SEQ ID NO:126, SEQ ID
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NO:128, SEQ ID NO:130, SEQ ID NO:132, SEQ ID NO: 134, SEQ ID NO:136, SEQ ID
NO:138, SEQ ID NO: 140, SEQ ID NO:142, SEQ ID NO:144, SEQ ID NO:146, SEQ ID
NO:148, SEQ ID NO:150, SEQ ID NO: 152, SEQ ID NO:154, SEQ ID NO:156, SEQ ID
NO:158, SEQ ID NO:160, SEQ ID NO:162, SEQ ID NO:164, SEQ ID NO:166, SEQ ID
NO:168, SEQ ID NO:170, SEQ ID NO:172, SEQ ID NO:174, SEQ ID NO:176, SEQ ID
NO:178, SEQ ID NO:180, SEQ ID NO:182, SEQ ID NO:184, and SEQ ID NO:186. In
some
embodiments, the nucleotide sequence is codon-optimized.
[0146] Some embodiments of the disclosure relate to a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant of the disclosure
comprising an amino
acid sequence set forth in SEQ ID NO:50, SEQ ID NO:52, SEQ ID NO:54, SEQ ID
NO:56,
SEQ ID NO:58, SEQ ID NO:60, SEQ ID NO:70, SEQ ID NO:72, SEQ ID NO: 74, SEQ ID
NO:76, SEQ ID NO:78, SEQ ID NO:80 SEQ ID NO:82, SEQ ID NO:84, SEQ ID NO:86,
SEQ ID NO: 88, SEQ ID NO:90, SEQ ID NO:92, SEQ ID NO:94, SEQ ID NO:96, SEQ ID
NO:98, SEQ ID NO:100, SEQ ID NO:102, SEQ ID NO:104, SEQ ID NO:106, SEQ ID
NO:108, SEQ ID NO:110, SEQ ID NO:112, SEQ ID NO:114, SEQ ID NO:116, SEQ ID
NO:118, SEQ ID NO:120, SEQ ID NO:124, SEQ ID NO:126, SEQ ID NO:128, SEQ ID
NO:130, SEQ ID NO:132, SEQ ID NO: 134, SEQ ID NO:138, SEQ ID NO: 140, SEQ ID
NO:142, SEQ ID NO:144, SEQ ID NO:146, SEQ ID NO:148, SEQ ID NO: 152, SEQ ID
NO:156, SEQ ID NO:158, SEQ ID NO:160, SEQ ID NO:166, SEQ ID NO:168, SEQ ID
NO:170, SEQ ID NO:172, SEQ ID NO:174, SEQ ID NO:176, SEQ ID NO: 178, SEQ ID
NO:180, SEQ ID NO: 182, SEQ ID NO: 184, or SEQ ID NO:186. In some embodiments,
the nucleotide sequence is codon-optimized.
[0147] The disclosure also provides a nucleic acid comprising a
nucleotide sequence
encoding an engineered variant, wherein the nucleotide sequence is that set
forth in SEQ ID
NO:49, SEQ ID NO:51, SEQ ID NO:53, SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59,
SEQ ID NO:61, SEQ ID NO:63, SEQ ID NO:65, SEQ ID NO:67, SEQ ID NO:69, SEQ ID
NO:71, SEQ ID NO:73, SEQ ID NO:75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81,
SEQ ID NO:83, SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO:93, SEQ ID
NO:95, SEQ ID NO:97, SEQ ID NO:99, SEQ ID NO:101, SEQ ID NO:103, SEQ ID
NO:105, SEQ ID NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID
NO:115, SEQ ID NO:117, SEQ ID NO:119, SEQ ID NO:121, SEQ ID NO:123, SEQ ID
NO:125, SEQ ID NO:127, SEQ ID NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID
NO:135, SEQ ID NO:137, SEQ ID NO:139, SEQ ID NO:141, SEQ ID NO:143, SEQ ID
51
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NO:145, SEQ ID NO:147, SEQ ID NO:149, SEQ ID NO:151, SEQ ID NO:153, SEQ ID
NO:155, SEQ ID NO:157, SEQ ID NO:159, SEQ ID NO:161, SEQ ID NO:163, SEQ ID
NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID NO:169, SEQ ID NO:171, SEQ ID
NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID NO:181, SEQ ID
NO:183, or SEQ ID NO:185. In some embodiments, the nucleotide sequence is
codon-
optimized.
[0148] The disclosure provides a nucleic acid comprising a nucleotide
sequence
encoding an engineered variant, wherein the nucleotide sequence is that set
forth in SEQ ID
NO:49, SEQ ID NO:51, SEQ ID NO:53, SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59,
SEQ ID NO:61, SEQ ID NO:63, SEQ ID NO:65, SEQ ID NO:67, SEQ ID NO:69, SEQ ID
NO:71, SEQ ID NO: 73, SEQ ID NO: 75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81,
SEQ ID NO:83, SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO: 93, SEQ ID
NO: 95, SEQ ID NO: 97, SEQ ID NO: 99, SEQ ID NO: 101, SEQ ID NO:103, SEQ ID
NO:105, SEQ ID NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID
NO:115, SEQ ID NO:117, SEQ ID NO:119, SEQ ID NO:121, SEQ ID NO:123, SEQ ID
NO:125, SEQ ID NO:127, SEQ ID NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID
NO:135, SEQ ID NO:137, SEQ ID NO:139, SEQ ID NO:141, SEQ ID NO:143, SEQ ID
NO:145, SEQ ID NO:147, SEQ ID NO:151, SEQ ID NO:155, SEQ ID NO:157, SEQ ID
NO:159, SEQ ID NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID NO:171, SEQ ID
NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID NO:181, SEQ ID
NO:183, or SEQ ID NO:185, or a codon degenerate sequence of any of the
foregoing. In
some embodiments, the nucleotide sequence is codon-optimized.
[0149] The disclosure provides a nucleic acid comprising a nucleotide
sequence
encoding an engineered variant, wherein the nucleotide sequence is that set
forth in SEQ ID
NO:49, SEQ ID NO:51, SEQ ID NO:53, SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59,
SEQ ID NO:69, SEQ ID NO:71, SEQ ID NO: 73, SEQ ID NO: 75, SEQ ID NO:77, SEQ ID
NO:79, SEQ ID NO:81, SEQ ID NO:83, SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91,
SEQ ID NO: 93, SEQ ID NO: 95, SEQ ID NO: 97, SEQ ID NO: 99, SEQ ID NO: 101,
SEQ
ID NO:103, SEQ ID NO:105, SEQ ID NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID
NO:113, SEQ ID NO:115, SEQ ID NO:117, SEQ ID NO:119, SEQ ID NO:123, SEQ ID
NO:125, SEQ ID NO:127, SEQ ID NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID
NO:137, SEQ ID NO:139, SEQ ID NO:141, SEQ ID NO:143, SEQ ID NO:145, SEQ ID
NO:147, SEQ ID NO:151, SEQ ID NO:155, SEQ ID NO:157, SEQ ID NO:159, SEQ ID
52
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID NO:171, SEQ ID NO:173, SEQ ID
NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID NO:181, SEQ ID NO:183, or SEQ ID
NO:185, or a codon degenerate sequence of any of the foregoing. In some
embodiments, the
nucleotide sequence is codon-optimized.
[0150] The disclosure provides a nucleic acid comprising a nucleotide
sequence
encoding an engineered variant, wherein the nucleotide sequence is that set
forth in SEQ ID
NO:49, SEQ ID NO:51, SEQ ID NO:53, SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59,
SEQ ID NO:61, SEQ ID NO:63, SEQ ID NO:65, SEQ ID NO:67, SEQ ID NO:69, SEQ ID
NO:71, SEQ ID NO: 73, SEQ ID NO: 75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81,
SEQ ID NO:83, SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO: 93, SEQ ID
NO: 95, SEQ ID NO: 97, SEQ ID NO: 99, SEQ ID NO: 101, SEQ ID NO:103, SEQ ID
NO:105, SEQ ID NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID
NO:115, SEQ ID NO:117, SEQ ID NO:119, SEQ ID NO:121, SEQ ID NO:123, SEQ ID
NO:125, SEQ ID NO:127, SEQ ID NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID
NO:135, SEQ ID NO:137, SEQ ID NO:139, SEQ ID NO:141, SEQ ID NO:143, SEQ ID
NO:145, SEQ ID NO:147, SEQ ID NO:151, SEQ ID NO:155, SEQ ID NO:157, SEQ ID
NO:159, SEQ ID NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID NO:171, SEQ ID
NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID NO:181, SEQ ID
NO:183, or SEQ ID NO:185. In some embodiments, the nucleotide sequence is
codon-
optimized.
[0151] Further included are nucleic acids that hybridize to the nucleic
acids disclosed
herein. Hybridization conditions may be stringent in that hybridization will
occur if there is
at least a 90%, at least a 95%, or at least a 97% sequence identity with the
nucleotide
sequence present in the nucleic acid encoding the polypeptides disclosed
herein. The
stringent conditions may include those used for known Southern hybridizations
such as, for
example, incubation overnight at 42 C in a solution having 50% formamide, 5x
SSC (150
mM NaCl, 15 mM trisodium citrate), 50 mM sodium phosphate (pH 7.6), 5
xDenhardt' s
solution, 10% dextran sulfate, and 20 micrograms/milliliter denatured, sheared
salmon sperm
DNA, following by washing the hybridization support in 0.1x SSC at about 65
C. Other
known hybridization conditions are well known and are described in Sambrook et
al.,
Molecular Cloning: A Laboratory Manual, Third Edition, Cold Spring Harbor,
N.Y. (2001).
[0152] The length of the nucleic acids disclosed herein may depend on the
intended
use. For example, if the intended use is as a primer or probe, for example for
PCR
53
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
amplification or for screening a library, the length of the nucleic acid will
be less than the
full length sequence, for example, 15-50 nucleotides. In certain such
embodiments, the
primers or probes may be substantially identical to a highly conserved region
of the
nucleotide sequence or may be substantially identical to either the 5' or 3'
end of the
nucleotide sequence. In some cases, these primers or probes may use universal
bases in some
positions so as to be "substantially identical" but still provide flexibility
in sequence
recognition. It is of note that suitable primer and probe hybridization
conditions are well
known in the art.
[0153] Some embodiments of the disclosure relate to a vector comprising
one or
more nucleic acids disclosed herein. Some embodiments of the disclosure relate
to an
expression construct comprising one or more nucleic acids disclosed herein.
Some
embodiments of the disclosure relate to nucleic acids comprising codon-
optimized
nucleotide sequences encoding the engineered variants of the disclosure. In
some
embodiments, the nucleic acids disclosed herein are heterologous.
Methods of Screening Engineered Variants of the Tetrahydrocannabinolic Acid
Synthase (THCAS) Polypeptide
[0154] The disclosure provides a method of screening an engineered
variant of a
tetrahydrocannabinolic acid synthase (THCAS) polypeptide comprising an amino
acid
sequence of SEQ ID NO:44 with one or more amino acid substitutions. In certain
such
embodiments, the method involves a competition assay wherein the engineered
variant of the
disclosure is expressed in a modified host cells alongside a related enzyme.
[0155] Some embodiments of the disclosure relate to a method of screening
an
engineered variant of a tetrahydrocannabinolic acid synthase (THCAS)
polypeptide
comprising an amino acid sequence of SEQ ID NO:44 with one or more amino acid
substitutions, the method comprising:
a) dividing a population of host cells into a control population and a test
population;
b) co-expressing in the control population a THCAS polypeptide having an amino
acid sequence of SEQ ID NO:44 and a comparison cannabinoid synthase
polypeptide,
wherein the THCAS polypeptide having an amino acid sequence of SEQ ID NO:44
can
convert CBGA to a first cannabinoid, THCA, and the comparison cannabinoid
synthase
polypeptide can convert the same CBGA to a different second cannabinoid;
54
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
c) co-expressing in the test population the engineered variant and the
comparison
cannabinoid synthase polypeptide, wherein the engineered variant may convert
CBGA to the
same first cannabinoid, THCA, as the THCAS polypeptide having an amino acid
sequence
of SEQ ID NO:44, and wherein the comparison cannabinoid synthase polypeptide
can
convert the same CBGA to the second cannabinoid and is expressed at similar
levels in the
test population and in the control population;
d) measuring a ratio of the first cannabinoid, THCA, over the second
cannabinoid
produced by both the test population and the control population; and
e) measuring an amount, in mg/L or mM, of the first cannabinoid produced by
both
the test population and the control population. In cetain such embodiments,
the engineered
variant is an engineered variant of the disclosure.
[0156] In some embodiments, the test population is identified as
comprising an
engineered variant having improved in vivo performance compared to the
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 by producing the first cannabinoid in a greater amount, as measured in
mg/L or mM,
by the test population compared to the amount produced by the control
population under
similar culture conditions for the same length of time. In some embodiments,
the test
population is identified as comprising an engineered variant having improved
in vivo
performance compared to the tetrahydrocannabinolic acid synthase polypeptide
having an
amino acid sequence of SEQ ID NO:44, wherein improved in vivo performance is
demonstrated by an increase in the ratio of the first cannabinoid over the
second cannabinoid
produced by the test population compared to that produced by the control
population under
similar culture conditions for the same length of time.
[0157] In some embodiments, the cannabinoid synthase polypeptide is a
cannabidiolic acid synthase (CBDAS) polypeptide. In certain such embodiments,
the
CBDAS polypeptide comprises an amino acid sequence having at least 85%, at
least 86%, at
least 87%, at least 88%, at least 89%, at least 90%, at least 91%, at least
92%, at least 93%,
at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least
99%, at least
99.5%, at least 99.6%, at least 99.7%, at least 99.8%, at least 99.9%, or 100%
sequence
identity to SEQ ID NO:3. In some embodiments, a nucleotide sequence encoding
the
CBDAS polypeptide is the nucleotide sequence set forth in SEQ ID NO:1 or SEQ
ID NO:2.
In some embodiments, a nucleotide sequence encoding the CBDAS polypeptide is
the
nucleotide sequence set forth in SEQ ID NO:1 or SEQ ID NO:2, or a codon
degenerate
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
nucleotide sequence thereof In some embodiments, a nucleotide sequence
encoding the
CBDAS polypeptide has at least 85%, at least 86%, at least 87%, at least 88%,
at least 89%,
at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least
95%, at least
96%, at least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%,
at least 99.7%,
at least 99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:1 or
SEQ ID NO:2.
In some embodiments, the second cannabinoid is CBDA.
Modified Host Cells for Expressing Engineered Variants of the
Tetrahydrocannabinolic Acid Synthase (THCAS) Polypeptide and for Producing
Cannabinoids and Cannabinoid Derivatives
[0158] The present disclosure provides modified host cells comprising one
or more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure. In certain such embodiments, the modified host cells of the
disclosure are for
expressing an engineered variant and/or for producing a cannabinoid or a
cannabinoid
derivative. In some embodiments, the nucleotide sequence encoding the
engineered variant
is codon-optimized.
[0159] The disclosure also provides nucleic acids (e.g., heterologous
nucleic acids),
which can be introduced into microorganisms (e.g., modified host cells),
resulting in
expression or overexpression of the engineered variants of the disclosure,
which can then be
utilized in vitro (e.g., cell-free) or in vivo for the production of
cannabinoids or cannabinoid
derivatives. In some embodiments, these nucleic acids comprise a codon-
optimized
nucleotide sequence encoding the engineered variant.
[0160] Cannabinoid synthase polypeptides, secreted polypeptides, such as
the
engineered variants of the disclosure, have structural features that may
hinder expression in
modified host cells, such as modified yeast cells. Cannabinoid synthase
polypeptides,
including the engineered variants of the disclosure, comprise disulfide bonds,
numerous
glycosylation sites, including N-glycosylation sites, and a bicovalently
attached flavin
adenine dinucleotide (FAD) cofactor moiety. Often these secreted polypeptides
are
misfolded or mislocalized, resulting in low expression, polypeptides lacking
activity,
reduced host cell viability, and/or cell death. As disclosed herein,
manipulation of secretory
pathway in host cells modified with one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure may improve
expression, folding,
and enzymatic activity of the engineered variant of the disclosure as well as
viability of the
56
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
modified host cell. In certain such embodiments, the nucleotide sequence
encoding the
engineered variant is codon-optimized.
[0161] To produce cannabinoids or cannabinoid derivatives and create
biosynthetic
pathways within modified host cells, modified host cells comprising one or
more nucleic
acids comprising a nucleotide sequence encoding an engineered variant of the
disclosure
may express or overexpress combinations of heterologous nucleic acids
comprising
nucleotide sequences encoding polypeptides involved in cannabinoid or
cannabinoid
precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates, olivetolic
acid, or
hexanoyl-CoA) biosynthesis. In some embodiments, the nucleotide sequences
encoding the
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis are
codon-
optimized. In some embodiments, the modified host cells of the disclosure for
producing
cannabinoid or cannabinoid derivatives comprising one or more nucleic acids
comprising a
nucleotide sequence encoding an engineered variant of the disclosure comprise
one or more
modifications to modulate the expression of one or more secretory pathway
polypeptides.
The one or more modifications to modulate the expression of one or more
secretory pathway
polypeptides may include introducing into a host cell one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more secretory pathway
polypeptides
and/or deletion or downregulation of one or more genes encoding one or more
secretory
pathway polypeptides in a host cell. In some embodiments, a modified host cell
of the
present disclosure for producing cannabinoids or cannabinoid derivatives
comprising one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
disclosure comprises one or more heterologous nucleic acids comprising
nucleotide
sequences encoding one or more secretory pathway polypeptides, resulting in
expression or
overexpression of the one or more secretory pathway polypeptides. In some
embodiments,
the nucleotide sequences encoding the one or more secretory pathway
polypeptides are
codon-optimized. In some embodiments, the modified host cell for producing
cannabinoids
or cannabinoid derivatives comprising one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure comprises a deletion
or
downregulation of one or more genes encoding one or more secretory pathway
polypeptides,
reducing or eliminating the expression of the one or more secretory pathway
polypeptides.
In certain such embodiments, the modified host cells comprise a deletion of
one or more
genes encoding one or more secretory pathway polypeptides. In some
embodiments, the
57
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
modified host cells comprise a downregulation of one or more genes encoding
one or more
secretory pathway polypeptides.
[0162] In some embodiments, culturing of a modified host cell for
producing
cannabinoids or cannabinoid derivatives in a culture medium provides for
synthesis of the
cannabinoid or the cannabinoid derivative.
[0163] To express an engineered variant of the disclosure, the modified
host cells
may express or overexpress one or more nucleic acids comprising a nucleotide
sequence
encoding the engineered variant. In some embodiments, the nucleotide sequences
encoding
the engineered variants are codon-optimized. In some embodiments, the modified
host cells
of the disclosure for expressing an engineered variant of the disclosure
comprising one or
more nucleic acids comprising a nucleotide sequence encoding the engineered
variant
comprise one or more modifications to modulate the expression of one or more
secretory
pathway polypeptides. The one or more modifications to modulate the expression
of one or
more secretory pathway polypeptides may include introducing into a host cell
one or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more secretory
pathway polypeptides and/or deletion or downregulation of one or more genes
encoding one
or more secretory pathway polypeptides in a host cell. In some embodiments, a
modified
host cell of the present disclosure for expressing an engineered variant of
the disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
the
engineered variant comprises one or more heterologous nucleic acids comprising
nucleotide
sequences encoding one or more secretory pathway polypeptides, resulting in
expression or
overexpression of the one or more secretory pathway polypeptides. In some
embodiments,
the nucleotide sequences encoding the one or more secretory pathway
polypeptides are
codon-optimized. In some embodiments, the modified host cell for expressing an
engineered
variant of the disclosure comprising one or more nucleic acids comprising a
nucleotide
sequence encoding the engineered variant comprises a deletion or
downregulation of one or
more genes encoding one or more secretory pathway polypeptides, reducing or
eliminating
the expression of the one or more secretory pathway polypeptides. In certain
such
embodiments, the modified host cells comprise a deletion of one or more genes
encoding
one or more secretory pathway polypeptides. In some embodiments, the modified
host cells
comprise a downregulation of one or more genes encoding one or more secretory
pathway
polypeptides.
58
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Secretory Pathway Modifications
[0164] Secretory pathway polypeptides with modulated expression in the
modified
host cells of the disclosure may include, but are not limited to: a KAR2
polypeptide, a ROT2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, FAD1 polypeptide, a PEP4
polypeptide, and an IRE1 polypeptide. Expression of secretory pathway
polypeptides may
be modulated by introducing into a host cell one or more heterologous nucleic
acids
comprising nucleotide sequences encoding one or more secretory pathway
polypeptides
and/or deletion or downregulation of one or more genes encoding one or more
secretory
pathway polypeptides in a host cell. In some embodiments, the nucleotide
sequences
encoding the one or more secretory pathway polypeptides are codon-optimized.
[0165] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of one or more of the following genes: a ROT2 gene
or a PEP4
gene. In some embodiments, the modified host cells of the disclosure comprise
a deletion of
one or more of the following genes: a ROT2 gene or a PEP4 gene. In some
embodiments,
the modified host cells of the disclosure comprise a downregulation of one or
more of the
following genes: a ROT2 gene or a PEP4 gene.
[0166] The secretory pathway polypeptides and heterologous nucleic acids
comprising nucleotide sequences encoding one or more secretory pathway
polypeptides may
be derived from any suitable source, for example, bacteria, yeast, fungi,
algae, human, plant,
or mouse. In some embodiments, the secretory pathway polypeptides and
heterologous
nucleic acids comprising nucleotide sequences encoding one or more secretory
pathway
polypeptides may be derived from Pichia pastoris (now known as Komagataella
phaffii),
Pichia finlandica, Pichia trehalophila, Pichia koclamae, Pichia
membranaefaciens, Pichia
opuntiae, Pichia thermotolerans, Pichia salictaria, Pichia guercuum, Pichia
pijperi, Pichia
stiptis, Pichia methanol/ca, Pichia sp., Saccharomyces cerevisiae,
Saccharomyces sp.,
Hansenula polymorpha (now known as Pichia angusta), Yarrowia hpolytica,
Kluyveromyces sp., Kluyveromyces lactis, Kluyveromyces marxianus,
Schizosaccharomyces
pombe, Scheffersomyces stipites, Dekkera bruxellensis, Blastobotrys
adeninivorans
(formerly Arxula adeninivorans), Candida alb/cans, Aspergillus nidulans,
Aspergillus niger, ,
Aspergillus oryzae, Trichoderma reesei, Chrysosporium lucknowense, Fusarium
sp.,
Fusarium gramineum, Fusarium venenatum, Neurospora crassa, and the like. In
some
embodiments, the disclosure also encompasses orthologous genes encoding the
secretory
59
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
pathway polypeptides disclosed herein. Exemplary secretory pathway
polypeptides disclosed
herein may also include a full-length secretory pathway polypeptide, a
fragment of a
secretory pathway polypeptide, a variant of a secretory pathway polypeptide, a
truncated
secretory pathway polypeptide, or a fusion polypeptide that has at least one
activity of a
secretory pathway polypeptide.
[0167] Exemplary KAR2 polypeptides disclosed herein may include a full-
length
KAR2 polypeptide, a fragment of a KAR2 polypeptide, a variant of a KAR2
polypeptide, a
truncated KAR2 polypeptide, or a fusion polypeptide that has at least one
activity of a KAR2
polypeptide.
[0168] Exemplary ROT2 polypeptides disclosed herein may include a full-
length
ROT2 polypeptide, a fragment of a ROT2 polypeptide, a variant of a ROT2
polypeptide, a
truncated ROT2 polypeptide, or a fusion polypeptide that has at least one
activity of a ROT2
polypeptide.
[0169] Exemplary PDI1 polypeptides disclosed herein may include a full-
length
PDI1 polypeptide, a fragment of a PDI1 polypeptide, a variant of a PDI1
polypeptide, a
truncated PDI1 polypeptide, or a fusion polypeptide that has at least one
activity of a PDI1
polypeptide.
[0170] Exemplary ER01 polypeptides disclosed herein may include a full-
length
ER01 polypeptide, a fragment of an ER01 polypeptide, a variant of an ER01
polypeptide, a
truncated ER01 polypeptide, or a fusion polypeptide that has at least one
activity of an
ER01 polypeptide.
[0171] Exemplary FAD1 polypeptides disclosed herein may include a full-
length
FAD1 polypeptide, a fragment of a FAD1 polypeptide, a variant of a FAD1
polypeptide, a
truncated FAD1 polypeptide, or a fusion polypeptide that has at least one
activity of a FAD1
polypeptide.
[0172] Exemplary PEP4 polypeptides disclosed herein may include a full-
length
PEP4 polypeptide, a fragment of a PEP4 polypeptide, a variant of a PEP4
polypeptide, a
truncated PEP1 polypeptide, or a fusion polypeptide that has at least one
activity of a PEP4
polypeptide.
[0173] Exemplary IRE1 polypeptides disclosed herein may include a full-
length
IRE1 polypeptide, a fragment of an IRE1 polypeptide (e.g., missing the first 7
amino acids),
a variant of an IRE1 polypeptide, a truncated IRE1 polypeptide, or a fusion
polypeptide that
has at least one activity of an IRE1 polypeptide.
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0174] Modified host cells of the disclosure may comprise one or more
modifications
to modulate the expression of one or more of a KAR2 polypeptide, a ROT2
polypeptide, a
PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide, a PEP4 polypeptide,
or an
IRE1 polypeptide. The one or more modifications to modulate the expression of
one or more
of a KAR2 polypeptide, a ROT2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, a PEP4 polypeptide, or an IRE1 polypeptide may include
introducing
into a host cell one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of the KAR2 polypeptide, the PDI1 polypeptide, the ER01
polypeptide, the FAD1 polypeptide, or the IRE1 polypeptide and/or deletion or
downregulation of one or more genes encoding one or more of the ROT2
polypeptide or the
PEP4 polypeptide in a host cell. In some embodiments, a modified host cell of
the present
disclosure comprises one or more heterologous nucleic acids comprising
nucleotide
sequences encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an
ER01
polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide resulting in
expression or
overexpression of the KAR2 polypeptide, the PDI1 polypeptide, the ER01
polypeptide, the
FAD1 polypeptide, or the IRE1 polypeptide. In some embodiments, the modified
host cells
of the disclosure comprise a deletion or downregulation of one or more genes
encoding one
or more of a ROT2 polypeptide or a PEP4 polypeptide, reducing or eliminating
the
expression of the ROT2 polypeptide or PEP4 polypeptide.
[0175] In some embodiments, the one or more modifications to modulate the
expression of one or more secretory pathway polypeptides may improve modified
host cell
viability. Improving modified host cell viability may improve the industrial
fermentation
process. The ER01 polypeptide may serve as a partner to the PDI1 polypeptide,
a protein
disulfide isomerase polypeptide. Modulating the expression of an IRE1
polypeptide may
prevent degradation of expressed engineered variants of the disclosure.
[0176] In some embodiments, the modified host cells of the disclosure
comprise one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1
polypeptide, or
an IRE1 polypeptide.
[0177] In some embodiments, the modified host cells of the disclosure
comprise one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
secretory pathway polypeptides comprising the amino acid sequences set forth
in SEQ ID
NO:5 (a KAR2 polypeptide), SEQ ID NO:9 (a PDI1 polypeptide), SEQ ID NO:7 (an
ER01
61
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide), SEQ ID NO:192 (a FAD1 polypeptide), SEQ ID NO:11 (an IRE1
polypeptide), or SEQ ID NO:190 (a fragment IRE1 polypeptide).
[0178] In some embodiments, the modified host cells of the disclosure
comprise one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
secretory pathway polypeptides comprising the amino acid sequences set forth
in SEQ ID
NO:5 (a KAR2 polypeptide), SEQ ID NO:9 (a PDI1 polypeptide), SEQ ID NO:7 (an
ER01
polypeptide), SEQ ID NO:192 (a FAD1 polypeptide), SEQ ID NO:11 (an IRE1
polypeptide), or SEQ ID NO:190 (a fragment IRE1 polypeptide), or a
conservatively
substituted amino acid sequence of any of the foregoing.
[0179] In some embodiments, the modified host cells of the disclosure
comprise one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
secretory pathway polypeptides comprising amino acid sequences having at least
50%, at
least 55%, at least 60%, at least 65%, at least 70%, at least 75%, at least
80%, at least 81%,
at least 82%, at least 83%, at least 84%, at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at
least 95%, at least 96%, at least 97%, at least 98%, at least 99%, at least
99.5%, at least
99.6%, at least 99.7%, at least 99.8%, at least 99.9%, or 100% amino acid
sequence identity
to SEQ ID NO:5 (a KAR2 polypeptide), SEQ ID NO:9 (a PDI1 polypeptide), SEQ ID
NO:7
(an ER01 polypeptide), SEQ ID NO:192 (a FAD1 polypeptide), SEQ ID NO:11 (an
IRE1
polypeptide), or SEQ ID NO:190 (a fragment IRE1 polypeptide).
[0180] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of one or more genes encoding encoding one or more
of a ROT2
polypeptide or a PEP4 polypeptide.
[0181] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of one or more genes encoding one or more secretory
pathway
polypeptides comprising the amino acid sequences set forth in SEQ ID NO:13 (a
ROT2
polypeptide) or SEQ ID NO:15 (a PEP4 polypeptide).
[0182] In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
an IRE1 polypeptide. In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding two or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
62
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
an IRE1 polypeptide. In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding three or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide,
or an IRE1 polypeptide. In some embodiments, a modified host cell of the
present
disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding a KAR2 polypeptide, one or more heterologous nucleic acids
comprising
a nucleotide sequence encoding a PDI1 polypeptide, one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an ER01 polypeptide, and one or more
heterologous nucleic acids comprising a nucleotide sequence encoding an IRE1
polypeptide.
[0183] In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
a FAD1 polypeptide. In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding two or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
a FAD1 polypeptide. In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding three or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide,
or a FAD1 polypeptide. In some embodiments, a modified host cell of the
present disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding a KAR2 polypeptide, one or more heterologous nucleic acids comprising
a
nucleotide sequence encoding a PDI1 polypeptide, one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an ER01 polypeptide, and one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a FAD1
polypeptide.
In some embodiments, the nucleotide sequences encoding the one or more of a
KAR2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or
an IRE1
polypeptide are codon-optimized.
[0184] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of one or more genes encoding one or more of a ROT2
polypeptide or a PEP4 polypeptide. In some embodiments, the modified host
cells of the
disclosure comprise a deletion or downregulation of genes encoding a ROT2
polypeptide
and a PEP4 polypeptide.
63
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0185] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a secretory pathway
polypeptide, such
as, a full-length secretory pathway polypeptide, a fragment of a secretory
pathway
polypeptide, a variant of a secretory pathway polypeptide, a truncated
secretory pathway
polypeptide, or a fusion polypeptide that has at least one activity of a
secretory pathway
polypeptide. In some embodiments, the nucleotide sequence is codon-optimized.
[0186] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a KAR2 polypeptide, such
as, a full-
length KAR2 polypeptide, a fragment of a KAR2 polypeptide, a variant of a KAR2
polypeptide, a truncated KAR2 polypeptide, or a fusion polypeptide that has at
least one
activity of a KAR2 polypeptide. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0187] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a ROT2 polypeptide, such
as, a full-
length ROT2 polypeptide, a fragment of a ROT2 polypeptide, a variant of a ROT2
polypeptide, a truncated ROT2 polypeptide, or a fusion polypeptide that has at
least one
activity of a ROT2 polypeptide. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0188] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a PDI1 polypeptide, such
as, a full-
length PDI1 polypeptide, a fragment of a PDI1 polypeptide, a variant of a PDI1
polypeptide,
a truncated PDI1 polypeptide, or a fusion polypeptide that has at least one
activity of a PDI1
polypeptide. In some embodiments, the nucleotide sequence is codon-optimized.
[0189] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes an ER01 polypeptide, such
as, a full-
length ER01 polypeptide, a fragment of an ER01 polypeptide, a variant of an
ER01
polypeptide, a truncated ER01 polypeptide, or a fusion polypeptide that has at
least one
activity of an ER01 polypeptide. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0190] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a FAD1 polypeptide, such
as, a full-
length FAD1 polypeptide, a fragment of a FAD1 polypeptide, a variant of a FAD1
polypeptide, a truncated FAD1 polypeptide, or a fusion polypeptide that has at
least one
64
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
activity of a FAD1 polypeptide. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0191] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a PEP4 polypeptide, such
as, a full-
length PEP4 polypeptide, a fragment of a PEP4 polypeptide, a variant of a PEP4
polypeptide, a truncated PEP1 polypeptide, or a fusion polypeptide that has at
least one
activity of a PEP4 polypeptide. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0192] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes an IRE1 polypeptide, such
as, a full-
length IRE1 polypeptide, a fragment of an IRE1 polypeptide (e.g., missing the
first 7 amino
acids), a variant of an IRE1 polypeptide, a truncated IRE1 polypeptide, or a
fusion
polypeptide that has at least one activity of an IRE1 polypeptide. In some
embodiments, the
nucleotide sequence is codon-optimized.
[0193] In some embodiments, one or more secretory pathway polypeptides,
such as a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide,
or an
IRE1 polypeptide, are overexpressed in the modified host cell. Overexpression
may be
achieved by increasing the copy number of the one or more heterologous nucleic
acids
comprising nucleotide sequences encoding one or more secretory pathway
polypeptides,
such as a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1
polypeptide, or an IRE1 polypeptide, e.g., through use of a high copy number
expression
vector (e.g., a plasmid that exists at 10-40 copies or about 100 copies per
cell) and/or by
operably linking the nucleotide sequences encoding one or more secretory
pathway
polypeptides, such as a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, or an IRE1 polypeptide, to a strong promoter. In some
embodiments, the
modified host cell has one copy of a heterologous nucleic acid comprising a
nucleotide
sequence encoding a secretory pathway polypeptide, such as a KAR2 polypeptide,
a PDI1
polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide.
In some
embodiments, the modified host cell has two copies of a heterologous nucleic
acid
comprising a nucleotide sequence encoding a secretory pathway polypeptide,
such as a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide,
or an
IRE1 polypeptide. In some embodiments, the modified host cell has three copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding a
secretory pathway
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide, such as a KAR2 polypeptide, a PDI1 polypeptide, a FAD1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide. In some embodiments, the modified
host cell
has four copies of a heterologous nucleic acid comprising a nucleotide
sequence encoding a
secretory pathway polypeptide, such as a KAR2 polypeptide, a PDI1 polypeptide,
an ER01
polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide. In some embodiments,
the
modified host cell has five copies of a heterologous nucleic acid comprising a
nucleotide
sequence encoding a secretory pathway polypeptide, such as a KAR2 polypeptide,
a PDI1
polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide.
In some
embodiments, the modified host cell has five or more copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding a secretory pathway polypeptide,
such as a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide,
or an
IRE1 polypeptide.
[0194] In some embodiments, the modified host cells of the disclosure
comprise one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
secretory pathway polypeptides selected from the group consisting of
nucleotide sequences
set forth in SEQ ID NO:4 (encodes a KAR2 polypeptide), SEQ ID NO:8 (encodes a
PDI1
polypeptide), SEQ ID NO:6 (encodes an ER01 polypeptide), SEQ ID NO:191
(encodes a
FAD1 polypeptide), SEQ ID NO:10 (encodes an IRE1 polypeptide), and SEQ ID
NO:189
(encodes a fragment IRE1 polypeptide).
[0195] In some embodiments, the modified host cells of the disclosure
comprise one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
secretory pathway polypeptides selected from the group consisting of
nucleotide sequences
set forth in SEQ ID NO:4 (encodes a KAR2 polypeptide), SEQ ID NO:8 (encodes a
PDI1
polypeptide), SEQ ID NO:6 (encodes an ER01 polypeptide), SEQ ID NO:191
(encodes a
FAD1 polypeptide), SEQ ID NO:10 (encodes an IRE1 polypeptide), and SEQ ID
NO:189
(encodes a fragment IRE1 polypeptide), or a codon degenerate nucleotide
sequence of any of
the foregoing.
[0196] In some embodiments, the modified host cells of the disclosure
comprise one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
secretory pathway polypeptides selected from the group consisting of
nucleotide sequences
having at least 80%, at least 81%, at least 82%, at least 83%, at least 84%,
at least 85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%,
at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least
66
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
99%, at least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%, at least
99.9%, or 100%
sequence identity to SEQ ID NO:4 (encodes a KAR2 polypeptide), SEQ ID NO:8
(encodes a
PDI1 polypeptide), SEQ ID NO:6 (encodes an ER01 polypeptide), SEQ ID NO:191
(encodes a FAD1 polypeptide), SEQ ID NO:10 (encodes an IRE1 polypeptide), and
SEQ ID
NO:189 (encodes a fragment IRE1 polypeptide).
[0197] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of one or more genes encoding one or more secretory
pathway
polypeptides encoded by nucleotide sequences selected from the group
consisting of
nucleotide sequences set forth in SEQ ID NO:12 (encodes a ROT2 polypeptide)
and SEQ ID
NO:14 (encodes a PEP4 polypeptide).
[0198] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of a ROT2 gene. In some embodiments, the modified
host cells
of the disclosure comprise a deletion of a ROT2 gene. In some embodiments, the
modified
host cells of the disclosure comprise a downregulation of a ROT2 gene.
[0199] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of a PEP4 gene. In some embodiments, the modified
host cells of
the disclosure comprise a deletion of a PEP4 gene. In some embodiments, the
modified host
cells of the disclosure comprise a downregulation of a PEP4 gene.
[0200] In some embodiments, the modified host cells of the disclosure
comprise a
deletion or downregulation of a PEP4 gene and a ROT2 gene. In some
embodiments, the
modified host cells of the disclosure comprise a deletion of a PEP4 gene and a
ROT2 gene.
In some embodiments, the modified host cells of the disclosure comprise a
downregulation
of a PEP4 gene and a ROT2 gene.
Cannabinoid and Cannabinoid Precursor Biosynthetic Pathway Modifications
[0201] A modified host cell of the present disclosure comprising one or
more nucleic
acids comprising a nucleotide sequence encoding an engineered variant of the
disclosure
may also comprise one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In addition to engineered variants of the disclosure, such
polypeptides may
include, but are not limited to: a geranyl pyrophosphate:olivetolic acid
geranyltransferase
(GOT) polypeptide, a tetraketide synthase (TKS) polypeptide, an olivetolic
acid cyclase
67
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
(OAC) polypeptide, one or more polypeptides having at least one activity of a
polypeptide
present in the mevalonate (MEV) pathway (e.g., one or more MEV pathway
polypeptides),
an acyl-activating enzyme (AAE) polypeptide, a polypeptide that generates GPP
(e.g., a
geranyl pyrophosphate synthetase (GPPS) polypeptide), a polypeptide that
condenses two
molecules of acetyl-CoA to generate acetoacetyl-CoA (e.g., an acetoacetyl-CoA
thiolase
polypeptide), and a pyruvate decarboxylase polypeptide. In some embodiments,
the
nucleotide sequences encoding one or more polypeptides involved in cannabinoid
or
cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates,
olivetolic acid,
or hexanoyl-CoA) biosynthesis are codon-optimized.
[0202] The polypeptides involved in cannabinoid or cannabinoid precursor
biosynthesis and heterologous nucleic acids comprising nucleotide sequences
encoding one
or more polypeptides involved in cannabinoid or cannabinoid precursor
biosynthesis may be
derived from any suitable source, for example, bacteria, yeast, fungi, algae,
human, plant
(e.g., Cannabis), or mouse. In some embodiments, the disclosure also
encompasses
orthologous genes encoding the polypeptides involved in cannabinoid or
cannabinoid
precursor biosynthesis disclosed herein.
Engineered Variants of the Tetrahydrocannabinolic Acid Synthase (THCAS)
Polypeptide
[0203] A modified host cell of the present disclosure may comprise one or
more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
tetrahydrocannabinolic acid synthase (THCAS) polypeptide disclosed herein. In
certain
such embodiments, the tetrahydrocannabinolic acid synthase polypeptide has an
amino acid
sequence of SEQ ID NO:44.
[0204] In some embodiments a modified host cell of the disclosure
comprises one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
disclosure, wherein the engineered variant comprises the amino acid sequence
set forth in
SEQ ID NO:50, SEQ ID NO:52, SEQ ID NO:54, SEQ ID NO:56, SEQ ID NO:58, SEQ ID
NO:60, SEQ ID NO:62, SEQ ID NO:64, SEQ ID NO:66, SEQ ID NO:68, SEQ ID NO:70,
SEQ ID NO:72, SEQ ID NO:74, SEQ ID NO:76, SEQ ID NO:78, SEQ ID NO:80 SEQ ID
NO:82, SEQ ID NO:84, SEQ ID NO:86, SEQ ID NO:88, SEQ ID NO:90, SEQ ID NO:92,
SEQ ID NO:94, SEQ ID NO:96, SEQ ID NO:98, SEQ ID NO:100, SEQ ID NO:102, SEQ
ID NO:104, SEQ ID NO:106, SEQ ID NO:108, SEQ ID NO:110, SEQ ID NO:112, SEQ ID
NO:114, SEQ ID NO:116, SEQ ID NO:118, SEQ ID NO:120, SEQ ID NO:122, SEQ ID
68
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NO:124, SEQ ID NO:126, SEQ ID NO:128, SEQ ID NO:130, SEQ ID NO:132, SEQ ID
NO:134, SEQ ID NO:136, SEQ ID NO:138, SEQ ID NO:140, SEQ ID NO:142, SEQ ID
NO:144, SEQ ID NO:146, SEQ ID NO:148, SEQ ID NO:150, SEQ ID NO:152, SEQ ID
NO:154, SEQ ID NO:156, SEQ ID NO:158, SEQ ID NO:160, SEQ ID NO:162, SEQ ID
NO:164, SEQ ID NO:166, SEQ ID NO:168, SEQ ID NO:170, SEQ ID NO:172, SEQ ID
NO:174, SEQ ID NO:176, SEQ ID NO:178, SEQ ID NO:180, SEQ ID NO:182, SEQ ID
NO:184, or SEQ ID NO:186. In some embodiments, the nucleotide sequence is
codon-
optimized.
[0205] In some embodiments, a modified host cell of the disclosure
comprises one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
disclosure, wherein the engineered variant comprises the amino acid sequence
set forth in
SEQ ID NO:50, SEQ ID NO:52, SEQ ID NO:54, SEQ ID NO:56, SEQ ID NO:58, SEQ ID
NO:60, SEQ ID NO:70, SEQ ID NO:72, SEQ ID NO:74, SEQ ID NO:76, SEQ ID NO:78,
SEQ ID NO:80 SEQ ID NO:82, SEQ ID NO:84, SEQ ID NO:86, SEQ ID NO:88, SEQ ID
NO:90, SEQ ID NO:92, SEQ ID NO:94, SEQ ID NO:96, SEQ ID NO:98, SEQ ID NO:100,
SEQ ID NO:102, SEQ ID NO:104, SEQ ID NO:106, SEQ ID NO:108, SEQ ID NO:110,
SEQ ID NO:112, SEQ ID NO:114, SEQ ID NO:116, SEQ ID NO:118, SEQ ID NO:120,
SEQ ID NO:124, SEQ ID NO:126, SEQ ID NO:128, SEQ ID NO:130, SEQ ID NO:132,
SEQ ID NO:134, SEQ ID NO:138, SEQ ID NO:140, SEQ ID NO:142, SEQ ID NO:144,
SEQ ID NO:146, SEQ ID NO:148, SEQ ID NO:152, SEQ ID NO:156, SEQ ID NO:158,
SEQ ID NO:160, SEQ ID NO:164, SEQ ID NO:166, SEQ ID NO:168, SEQ ID NO:170,
SEQ ID NO:172, SEQ ID NO:174, SEQ ID NO:176, SEQ ID NO:178, SEQ ID NO:180,
SEQ ID NO:182, SEQ ID NO:184, or SEQ ID NO:186. In some embodiments, the
nucleotide sequence is codon-optimized.
[0206] In some embodiments, the engineered variant of the disclosure is
overexpressed in the modified host cell. Overexpression may be achieved by
increasing the
copy number of the one or more nucleic acids comprising a nucleotide sequence
encoding
the engineered variant of the disclosure, e.g., through use of a high copy
number expression
vector (e.g., a plasmid that exists at 10-40 copies or about 100 copies per
cell) and/or by
operably linking the nucleotide sequence encoding the engineered variant of
the disclosure
to a strong promoter. In some embodiments, the modified host cell has one copy
of a nucleic
acid comprising a nucleotide sequence encoding the engineered variant of the
disclosure. In
some embodiments, the modified host cell has two copies of a nucleic acid
comprising a
69
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
nucleotide sequence encoding the engineered variant of the disclosure. In some
embodiments, the modified host cell has three copies of a nucleic acid
comprising a
nucleotide sequence encoding the engineered variant of the disclosure. In some
embodiments, the modified host cell has four copies of a nucleic acid
comprising a
nucleotide sequence encoding the engineered variant of the disclosure. In some
embodiments, the modified host cell has five copies of a nucleic acid
comprising a
nucleotide sequence encoding the engineered variant of the disclosure. In some
embodiments, the modified host cell has six copies of a nucleic acid
comprising a nucleotide
sequence encoding the engineered variant of the disclosure. In some
embodiments, the
modified host cell has seven copies of a nucleic acid comprising a nucleotide
sequence
encoding the engineered variant of the disclosure. In some embodiments, the
modified host
cell has eight copies of a nucleic acid comprising a nucleotide sequence
encoding the
engineered variant of the disclosure. In some embodiments, the modified host
cell has eight
or more copies of a nucleic acid comprising a nucleotide sequence encoding the
engineered
variant of the disclosure.
[0207] In some embodiments, a modified host cell of the disclosure
comprises one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
tetrahydrocannabinolic acid synthase (THCAS) polypeptide disclosed herein,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:49, SEQ ID NO:51, SEQ ID
NO:53,
SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59, SEQ ID NO:61, SEQ ID NO:63, SEQ ID
NO:65, SEQ ID NO:67, SEQ ID NO:69, SEQ ID NO:71, SEQ ID NO:73, SEQ ID NO:75,
SEQ ID NO:77, SEQ ID NO: 79, SEQ ID NO: 81, SEQ ID NO: 83, SEQ ID NO: 85, SEQ
ID NO: 87, SEQ ID NO:91, SEQ ID NO:93, SEQ ID NO:95, SEQ ID NO:97, SEQ ID
NO:99, SEQ ID NO:101, SEQ ID NO:103, SEQ ID NO:105, SEQ ID NO:107, SEQ ID
NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID NO:115, SEQ ID NO:117, SEQ ID
NO:119, SEQ ID NO:121, SEQ ID NO:123, SEQ ID NO:125, SEQ ID NO:127, SEQ ID
NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID NO:135, SEQ ID NO:137, SEQ ID
NO:139, SEQ ID NO:141, SEQ ID NO:143, SEQ ID NO:145, SEQ ID NO:147, SEQ ID
NO:149, SEQ ID NO:151, SEQ ID NO:153, SEQ ID NO:155, SEQ ID NO:157, SEQ ID
NO:159, SEQ ID NO:161, SEQ ID NO:163, SEQ ID NO:165, SEQ ID NO:167, SEQ ID
NO:169, SEQ ID NO:171, SEQ ID NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID
NO:179, SEQ ID NO:181, SEQ ID NO:183, or SEQ ID NO:185. In some embodiments,
the
nucleotide sequence is codon-optimized.
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0208] In some embodiments, a modified host cell of the disclosure
comprises one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
tetrahydrocannabinolic acid synthase (THCAS) polypeptide disclosed herein,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:49, SEQ ID NO:51, SEQ ID
NO:53,
SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59, SEQ ID NO:61, SEQ ID NO:63, SEQ ID
NO:65, SEQ ID NO:67, SEQ ID NO:69, SEQ ID NO:71, SEQ ID NO:73, SEQ ID NO:75,
SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81, SEQ ID NO:83, SEQ ID NO:85, SEQ ID
NO:87, SEQ ID NO:91, SEQ ID NO:93, SEQ ID NO:95, SEQ ID NO:97, SEQ ID NO:99,
SEQ ID NO:101, SEQ ID NO:103, SEQ ID NO:105, SEQ ID NO:107, SEQ ID NO:109,
SEQ ID NO:111, SEQ ID NO:113, SEQ ID NO:115, SEQ ID NO:117, SEQ ID NO:119,
SEQ ID NO:121, SEQ ID NO:123, SEQ ID NO:125, SEQ ID NO:127, SEQ ID NO:129,
SEQ ID NO:131, SEQ ID NO:133, SEQ ID NO:135, SEQ ID NO:137, SEQ ID NO:139,
SEQ ID NO:141, SEQ ID NO:143, SEQ ID NO:145, SEQ ID NO:147, SEQ ID NO:149,
SEQ ID NO:151, SEQ ID NO:153, SEQ ID NO:155, SEQ ID NO:157, SEQ ID NO:159,
SEQ ID NO:161, SEQ ID NO:163, SEQ ID NO:165, SEQ ID NO:167, SEQ ID NO:169,
SEQ ID NO:171, SEQ ID NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID NO:179,
SEQ ID NO:181, SEQ ID NO:183, or SEQ ID NO:185, or a codon degenerate
nucleotide
sequence of any of the foregoing. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0209] In some embodiments, a modified host cell of the disclosure
comprises one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
tetrahydrocannabinolic acid synthase (THCAS) polypeptide disclosed herein,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:49, SEQ ID NO:51, SEQ ID
NO:53,
SEQ ID NO:55, SEQ ID NO:57, SEQ ID NO:59, SEQ ID NO:69, SEQ ID NO:71, SEQ ID
NO: 73, SEQ ID NO: 75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81, SEQ ID NO:83,
SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO: 93, SEQ ID NO: 95, SEQ ID
NO: 97, SEQ ID NO: 99, SEQ ID NO: 101, SEQ ID NO:103, SEQ ID NO:105, SEQ ID
NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID NO:115, SEQ ID
NO:117, SEQ ID NO:119, SEQ ID NO:123, SEQ ID NO:125, SEQ ID NO:127, SEQ ID
NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID NO:137, SEQ ID NO:139, SEQ ID
NO:141, SEQ ID NO:143, SEQ ID NO:145, SEQ ID NO:147, SEQ ID NO:151, SEQ ID
NO:155, SEQ ID NO:157, SEQ ID NO:159, SEQ ID NO:165, SEQ ID NO:167, SEQ ID
NO:169, SEQ ID NO:171, SEQ ID NO:173, SEQ ID NO:175, SEQ ID NO:177, SEQ ID
71
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NO:179, SEQ ID NO:181, SEQ ID NO:183, or SEQ ID NO:185. In some embodiments,
the
nucleotide sequence is codon-optimized.
[0210] In some embodiments, a modified host cell of the disclosure
comprises one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
tetrahydrocannabinolic acid synthase (THCAS) polypeptide disclosed herein,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:49, SEQ ID NO:51, SEQ ID
NO:53,
SEQ ID NO:55, SEQ ID NO:57, or SEQ ID NO:59, SEQ ID NO:69, SEQ ID NO:71, SEQ
ID NO: 73, SEQ ID NO: 75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID NO:81, SEQ ID
NO:83, SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO: 93, SEQ ID NO: 95,
SEQ ID NO: 97, SEQ ID NO: 99, SEQ ID NO: 101, SEQ ID NO:103, SEQ ID NO:105,
SEQ ID NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID NO:115,
SEQ ID NO:117, SEQ ID NO:119, SEQ ID NO:123, SEQ ID NO:125, SEQ ID NO:127,
SEQ ID NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID NO:137, SEQ ID NO:139,
SEQ ID NO:141, SEQ ID NO:143, SEQ ID NO:145, SEQ ID NO:147, SEQ ID NO:151,
SEQ ID NO:155, SEQ ID NO:157, SEQ ID NO:159, SEQ ID NO:165, SEQ ID NO:167,
SEQ ID NO:169, SEQ ID NO:171, SEQ ID NO:173, SEQ ID NO:175, SEQ ID NO:177,
SEQ ID NO:179, SEQ ID NO:181, SEQ ID NO:183, or SEQ ID NO:185 or a codon
degenerate sequence of any of the foregoing. In some embodiments, the
nucleotide sequence
is codon-optimized.
[0211] In some embodiments, at least one of the one or more nucleic acids
comprising a nucleotide sequence encoding the engineered variant of the
disclosure is
operably linked to an inducible promoter. In some embodiments, at least one of
the one or
more nucleic acids comprising a nucleotide sequence encoding the engineered
variant of the
disclosure is operably linked to a constitutive promoter.
Geranyl Pyrophosphate. Olivetolic Acid Geranyhransferase (GOT) Polypeptides
[0212] A modified host cell of the present disclosure may comprise one or
more
heterologous nucleic acids comprising a nucleotide sequence encoding a geranyl
pyrophosphate:olivetolic acid geranyltransferase (GOT) polypeptide.
[0213] Exemplary GOT polypeptides disclosed herein may include a full-
length
GOT polypeptide, a fragment of a GOT polypeptide, a variant of a GOT
polypeptide, a
truncated GOT polypeptide, or a fusion polypeptide that has at least one
activity of a GOT
polypeptide. In some embodiments, the GOT polypeptide has aromatic
prenyltransferase
72
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
(PT) activity. In some embodiments, the GOT polypeptide modifies a cannabinoid
precursor
or a cannabinoid precursor derivative. In certain such embodiments, the GOT
polypeptide
modifies olivetolic acid or an olivetolic acid derivative.
[0214] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
GOT
polypeptide, wherein the GOT polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:17. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
GOT
polypeptide, wherein the GOT polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:17, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a GOT polypeptide,
wherein the
GOT polypeptide comprises an amino acid sequence having at least 65%, at least
70%, at
least 75%, at least 80%, at least 81%, at least 82%, at least 83%, at least
84%, at least 85%,
at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least
92%, at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at
least 98%, at
least 99%, at least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%, at
least 99.9%, or
100% amino acid sequence identity to SEQ ID NO:17.
[0215] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
GOT
polypeptide, wherein the GOT polypeptide comprises an amino acid sequence
having at least
65%, at least 70%, or at least 75% amino acid sequence identity to SEQ ID
NO:17. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a GOT polypeptide,
wherein the
GOT polypeptide comprises an amino acid sequence having at least 80%, at least
81%, at
least 82%, at least 83%, or at least 84% amino acid sequence identity to SEQ
ID NO:17. In
some embodiments, a modified host cell of the disclosure comprises one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a GOT
polypeptide,
wherein the GOT polypeptide comprises an amino acid sequence having at least
85%, at
least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least 92%,
at least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least
99%, at least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%, at least
99.9%, or 100%
amino acid sequence identity to SEQ ID NO:17.
73
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0216] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a GOT polypeptide, such
as, a full-
length GOT polypeptide, a fragment of a GOT polypeptide, a variant of a GOT
polypeptide,
a truncated GOT polypeptide, or a fusion polypeptide that has at least one
activity of a GOT
polypeptide. In some embodiments, the nucleotide sequence is codon-optimized.
[0217] In some embodiments, the GOT polypeptide is overexpressed in the
modified
host cell. Overexpression may be achieved by increasing the copy number of the
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
GOT
polypeptide, e.g., through use of a high copy number expression vector (e.g.,
a plasmid that
exists at 10-40 copies or about 100 copies per cell) and/or by operably
linking the nucleotide
sequence encoding the GOT polypeptide to a strong promoter. In some
embodiments, the
modified host cell has one copy of a heterologous nucleic acid comprising a
nucleotide
sequence encoding the GOT polypeptide. In some embodiments, the modified host
cell has
two copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
GOT polypeptide. In some embodiments, the modified host cell has three copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the GOT
polypeptide.
In some embodiments, the modified host cell has four copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding the GOT polypeptide. In some
embodiments,
the modified host cell has five copies of a heterologous nucleic acid
comprising a nucleotide
sequence encoding the GOT polypeptide. In some embodiments, the modified host
cell has
six copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
GOT polypeptide. In some embodiments, the modified host cell has seven copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the GOT
polypeptide.
In some embodiments, the modified host cell has eight copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding the GOT polypeptide. In some
embodiments, the
modified host cell has eight or more copies of a heterologous nucleic acid
comprising a
nucleotide sequence encoding the GOT polypeptide.
[0218] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
GOT
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:16. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a GOT polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:16, or a codon degenerate
nucleotide
74
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a GOT
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, at least 84%, at least 85%, at least 86%, at least 87%, at least
88%, at least 89%,
at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least
95%, at least
96%, at least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%,
at least 99.7%,
at least 99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:16.
[0219] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
GOT
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:16. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a GOT polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:16.
[0220] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
GOT
polypeptide, wherein the nucleotide sequence has at least 80% sequence
identity to SEQ ID
NO:16. In some embodiments, a modified host cell of the disclosure comprises
one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a GOT
polypeptide,
wherein the nucleotide sequence has at least 85% sequence identity to SEQ ID
NO:16. In
some embodiments, a modified host cell of the disclosure comprises one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a GOT
polypeptide,
wherein the nucleotide sequence has at least 90% sequence identity to SEQ ID
NO:16. In
some embodiments, a modified host cell of the disclosure comprises one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a GOT
polypeptide,
wherein the nucleotide sequence has at least 95% sequence identity to SEQ ID
NO:16.
NphB Polypeptides
[0221] In some embodiments, a NphB polypeptide is used instead of a GOT
polypeptide to generate cannabigerolic acid from GPP and olivetolic acid. A
modified host
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
cell of the present disclosure may comprise one or more heterologous nucleic
acids
comprising a nucleotide sequence encoding a NphB polypeptide.
[0222] Exemplary NphB polypeptides disclosed herein may include a full-
length
NphB polypeptide, a fragment of a NphB polypeptide, a variant of a NphB
polypeptide, a
truncated NphB polypeptide, or a fusion polypeptide that has at least one
activity of a NphB
polypeptide. In some embodiments, the NphB polypeptide has aromatic
prenyltransferase
(PT) activity. In some embodiments, the NphB polypeptide modifies a
cannabinoid
precursor or a cannabinoid precursor derivative. In certain such embodiments,
the NphB
polypeptide modifies olivetolic acid or an olivetolic acid derivative.
[0223] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
NphB
polypeptide, wherein the NphB polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:188. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a NphB
polypeptide, wherein the NphB polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:188, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a NphB polypeptide,
wherein the
NphB polypeptide comprises an amino acid sequence having at least 65%, at
least 70%, or at
least 75% amino acid sequence identity to SEQ ID NO:188. In some embodiments,
a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a NphB polypeptide, wherein the NphB
polypeptide comprises an amino acid sequence having at least 80%, at least
81%, at least
82%, at least 83%, or at least 84% amino acid sequence identity to SEQ ID
NO:188. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a NphB polypeptide,
wherein the
NphB polypeptide comprises an amino acid sequence having at least 85%, at
least 86%, at
least 87%, at least 88%, at least 89%, at least 90%, at least 91%, at least
92%, at least 93%,
at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at least
99%, at least
99.5%, at least 99.6%, at least 99.7%, at least 99.8%, at least 99.9%, or 100%
amino acid
sequence identity to SEQ ID NO:188.
[0224] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a NphB polypeptide, such
as, a full-
76
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
length NphB polypeptide, a fragment of a NphB polypeptide, a variant of a NphB
polypeptide, a truncated NphB polypeptide, or a fusion polypeptide that has at
least one
activity of a NphB polypeptide. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0225] In some embodiments, the NphB polypeptide is overexpressed in the
modified host cell. Overexpression may be achieved by increasing the copy
number of the
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding the
NphB polypeptide, e.g., through use of a high copy number expression vector
(e.g., a
plasmid that exists at 10-40 copies or about 100 copies per cell) and/or by
operably linking
the nucleotide sequence encoding the NphB polypeptide to a strong promoter. In
some
embodiments, the modified host cell has one copy of a heterologous nucleic
acid comprising
a nucleotide sequence encoding the NphB polypeptide. In some embodiments, the
modified
host cell has two copies of a heterologous nucleic acid comprising a
nucleotide sequence
encoding the NphB polypeptide. In some embodiments, the modified host cell has
three
copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the NphB
polypeptide. In some embodiments, the modified host cell has four copies of a
heterologous
nucleic acid comprising a nucleotide sequence encoding the NphB polypeptide.
In some
embodiments, the modified host cell has five copies of a heterologous nucleic
acid
comprising a nucleotide sequence encoding the NphB polypeptide. In some
embodiments,
the modified host cell has six copies of a heterologous nucleic acid
comprising a nucleotide
sequence encoding the NphB polypeptide. In some embodiments, the modified host
cell has
seven copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
NphB polypeptide. In some embodiments, the modified host cell has eight copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the NphB
polypeptide.
In some embodiments, the modified host cell has eight or more copies of a
heterologous
nucleic acid comprising a nucleotide sequence encoding the NphB polypeptide.
[0226] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
NphB
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:187. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a NphB polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:187, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
77
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a NphB
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:187. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a NphB polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:187.
[0227] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
NphB
polypeptide, wherein the nucleotide sequence has at least 80% sequence
identity to SEQ ID
NO:187. In some embodiments, a modified host cell of the disclosure comprises
one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a NphB
polypeptide,
wherein the nucleotide sequence has at least 85% sequence identity to SEQ ID
NO:187. In
some embodiments, a modified host cell of the disclosure comprises one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a NphB
polypeptide,
wherein the nucleotide sequence has at least 90% sequence identity to SEQ ID
NO:187. In
some embodiments, a modified host cell of the disclosure comprises one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a NphB
polypeptide,
wherein the nucleotide sequence has at least 95% sequence identity to SEQ ID
NO:187.
Polyp eptides that Generate Acyl-CoA Compounds or Acyl-CoA Compound
Derivatives
[0228] A modified host cell of the present disclosure may comprise one or
more
heterologous nucleic acids comprising a nucleotide sequence encoding a
polypeptide that
generates acyl-CoA compounds or acyl-CoA compound derivatives. Such
polypeptides may
include, but are not limited to, acyl-activating enzyme (AAE) polypeptides,
fatty acyl-CoA
synthetases (FAA) polypeptides, or fatty acyl-CoA ligase polypeptides. In some
embodiments, a modified host cell of the present disclosure comprises one or
more
heterologous nucleic acids comprising a nucleotide sequence encoding an AAE
polypeptide.
[0229] AAE polypeptides, FAA polypeptides, and fatty acyl-CoA ligase
polypeptides can convert carboxylic acids to their CoA forms and generate acyl-
CoA
compounds or acyl-CoA compound derivatives. Promiscuous acyl-activating enzyme
78
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptides, such as CsAAE1 and CsAAE3 polypeptides, FAA polypeptides, or
fatty acyl-
CoA ligase polypeptides, may permit generation of cannabinoid derivatives
(e.g.,
cannabigerolic acid derivatives), as well as cannabinoids (e.g.,
cannabigerolic acid). In some
embodiments, unsubstituted or substituted hexanoic acid or carboxylic acids
other than
unsubstituted or substituted hexanoic acid are fed to modified host cells
expressing an AAE
polypeptide, FAA polypeptide, or fatty acyl-CoA ligase polypeptide (e.g., are
present in the
culture medium in which the cells are grown) to generate hexanoyl-CoA, acyl-
CoA
compounds, derivatives of hexanoyl-CoA, or derivatives of acyl-CoA compounds.
The
hexanoyl-CoA, acyl-CoA compounds, derivatives of hexanoyl-CoA, or derivatives
of acyl-
CoA compounds can then be further utilized by a modified host cell to generate
cannabinoids or cannabinoid derivatives. In certain such embodiments, the cell
culture
medium comprising the modified host cells comprises unsubstituted or
substituted
hexanoate. In some embodiments, the cell culture medium comprising the
modified host
cells comprises a carboxylic acid other than unsubstituted or substituted
hexanoate.
[0230] Exemplary AAE, FAA, or fatty acyl-CoA ligase polypeptides
disclosed
herein may include a full-length AAE, FAA, or fatty acyl-CoA ligase
polypeptide; a
fragment of an AAE, FAA, or fatty acyl-CoA ligase polypeptide; a variant of an
AAE, FAA,
or fatty acyl-CoA ligase polypeptide; a truncated AAE, FAA, or fatty acyl-CoA
ligase
polypeptide; or a fusion polypeptide that has at least one activity of an AAE,
FAA, or fatty
acyl-CoA ligase polypeptide.
[0231] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
AAE
polypeptide, wherein the AAE polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:23. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an AAE
polypeptide, wherein the AAE polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:23, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding an AAE polypeptide,
wherein the
AAE polypeptide comprises an amino acid sequence having at least 50%, at least
55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:23. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
AAE
79
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide, wherein the AAE polypeptide comprises an amino acid sequence
having at least
80%, at least 81%, at least 82%, at least 83%, or at least 84% amino acid
sequence identity
to SEQ ID NO:23. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding an AAE
polypeptide, wherein the AAE polypeptide comprises an amino acid sequence
having at least
85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at
least 91%, at
least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least
97%, at least 98%,
at least 99%, at least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%,
at least 99.9%, or
100% amino acid sequence identity to SEQ ID NO:23.
[0232] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes an AAE, FAA, or fatty acyl-
CoA ligase
polypeptide, such as, a full-length AAE, FAA, or fatty acyl-CoA ligase
polypeptide; a
fragment of an AAE, FAA, or fatty acyl-CoA ligase polypeptide; a variant of an
AAE, FAA,
or fatty acyl-CoA ligase polypeptide; a truncated AAE, FAA, or fatty acyl-CoA
ligase
polypeptide; or a fusion polypeptide that has at least one activity of an AAE,
FAA, or fatty
acyl-CoA ligase polypeptide. In some embodiments, the nucleotide sequence is
codon-
optimized.
[0233] In some embodiments, one or more AAE, FAA, or fatty acyl-CoA
ligase
polypeptide are overexpressed in the modified host cell. Overexpression may be
achieved
by increasing the copy number of the one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the AAE, FAA, or fatty acyl-CoA ligase
polypeptide, e.g.,
through use of a high copy number expression vector (e.g., a plasmid that
exists at 10-40
copies or about 100 copies per cell) and/or by operably linking a nucleotide
sequence
encoding the AAE, FAA, or fatty acyl-CoA ligase polypeptide to a strong
promoter. In some
embodiments, the modified host cell has one copy of a heterologous nucleic
acid comprising
a nucleotide sequence encoding an AAE, FAA, or fatty acyl-CoA ligase
polypeptide. In
some embodiments, the modified host cell has two copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding an AAE, FAA, or fatty acyl-CoA
ligase
polypeptide. In some embodiments, the modified host cell has three copies of a
heterologous
nucleic acid comprising a nucleotide sequence encoding an AAE, FAA, or fatty
acyl-CoA
ligase polypeptide. In some embodiments, the modified host cell has four
copies of a
heterologous nucleic acid comprising a nucleotide sequence encoding an AAE,
FAA, or
fatty acyl-CoA ligase polypeptide. In some embodiments, the modified host cell
has five
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding an AAE,
FAA, or fatty acyl-CoA ligase polypeptide. In some embodiments, the modified
host cell has
six copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding an
AAE, FAA, or fatty acyl-CoA ligase polypeptide. In some embodiments, the
modified host
cell has seven copies of a heterologous nucleic acid comprising a nucleotide
sequence
encoding an AAE, FAA, or fatty acyl-CoA ligase polypeptide. In some
embodiments, the
modified host cell has eight copies of a heterologous nucleic acid comprising
a nucleotide
sequence encoding an AAE, FAA, or fatty acyl-CoA ligase polypeptide. In some
embodiments, the modified host cell has eight or more copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding an AAE, FAA, or fatty acyl-CoA
ligase
polypeptide.
[0234] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
AAE
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:22. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding an AAE polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:22, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding an AAE
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:22. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an AAE polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:22.
Polyp eptides that Condense an Acyl-CoA Compound or an Acyl-CoA Compound
Derivative
with Malonyl-CoA to Generate Olivetolic Acid or Derivatives of Olivetolic Acid
[0235] A modified host cell of the present disclosure may comprise one or
more
heterologous nucleic acids comprising a nucleotide sequence encoding one or
more
polypeptides that condense an acyl-CoA compound, such as hexanoyl-CoA, or an
acyl-CoA
81
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
compound derivative, such as a hexanoyl-CoA derivative, with malonyl-CoA to
generate
olivetolic acid, or a derivative of olivetolic acid. Polypeptides that react
an acyl-CoA
compound or an acyl-CoA compound derivative with malonyl-CoA to generate
olivetolic
acid, or a derivative of olivetolic acid, may include TKS and OAC
polypeptides. TKS and
OAC polypeptides have been found to have broad substrate specificity, enabling
production
of cannabinoid derivatives or cannabinoids. In some embodiments, a modified
host cell of
the present disclosure comprises one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a TKS polypeptide. In some embodiments, a
modified host
cell of the present disclosure comprises one or more heterologous nucleic
acids comprising a
nucleotide sequence encoding an OAC polypeptide.
[0236] Exemplary TKS or OAC polypeptides disclosed herein may include a
full-
length TKS or OAC polypeptide, a fragment of a TKS or OAC polypeptide, a
variant of a
TKS or OAC polypeptide, a truncated TKS or OAC polypeptide, or a fusion
polypeptide that
has at least one activity of a TKS or OAC polypeptide.
[0237] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
TKS
polypeptide, wherein the TKS polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:19. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
TKS
polypeptide, wherein the TKS polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:19, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a TKS polypeptide,
wherein the
TKS polypeptide comprises an amino acid sequence having at least 50%, at least
55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:19. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
TKS
polypeptide, wherein the TKS polypeptide comprises an amino acid sequence
having at least
80%, at least 81%, at least 82%, at least 83%, or at least 84% amino acid
sequence identity
to SEQ ID NO:19. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a TKS
polypeptide, wherein the TKS polypeptide comprises an amino acid sequence
having at least
85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at
least 91%, at
82
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
least 92%, at least 930 o, at least 940 o, at least 950 o, at least 96%, at
least 970 o, at least 98%,
at least 990 o, at least 99.50, at least 99.6%, at least 99.70, at least
99.8%, at least 99.90, or
10000 amino acid sequence identity to SEQ ID NO:19.
[0238] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
OAC
polypeptide, wherein the OAC polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:21 or SEQ ID NO:48. In some embodiments, a modified host cell of the
disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding an OAC polypeptide, wherein the OAC polypeptide comprises
the amino
acid sequence set forth in SEQ ID NO:21 or SEQ ID NO:48, or a conservatively
substituted
amino acid sequence thereof. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding an OAC polypeptide, wherein the OAC polypeptide comprises an amino
acid
sequence having at least 50%, at least 55%, at least 60%, at least 65%, at
least 70%, or at
least 750 amino acid sequence identity to SEQ ID NO:21 or SEQ ID NO:48. In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding an OAC polypeptide,
wherein the
OAC polypeptide comprises an amino acid sequence having at least 80%, at least
81%, at
least 82%, at least 83%, or at least 84% amino acid sequence identity to SEQ
ID NO:21 or
SEQ ID NO:48. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an OAC
polypeptide, wherein the OAC polypeptide comprises an amino acid sequence
having at
least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least
90%, at least 91%,
at least 92%, at least 930, at least 940, at least 950, at least 96%, at least
970, at least
98%, at least 990, at least 99.50, at least 99.6%, at least 99.70, at least
99.8%, at least
99.9%, or 100% amino acid sequence identity to SEQ ID NO:21 or SEQ ID NO:48.
[0239] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
OAC
polypeptide, wherein the OAC polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:21. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an OAC
polypeptide, wherein the OAC polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:21, or a conservatively substituted amino acid sequence thereof In
some
83
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding an OAC polypeptide,
wherein the
OAC polypeptide comprises an amino acid sequence having at least 50%, at least
55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:21. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
OAC
polypeptide, wherein the OAC polypeptide comprises an amino acid sequence
having at
least 80%, at least 81%, at least 82%, at least 83%, or at least 84% amino
acid sequence
identity to SEQ ID NO:21. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding an OAC polypeptide, wherein the OAC polypeptide comprises an amino
acid
sequence having at least 85%, at least 86%, at least 87%, at least 88%, at
least 89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% amino acid sequence identity to SEQ ID NO:21.
[0240] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
OAC
polypeptide, wherein the OAC polypeptide is a variant OAC (Y27F variant)
polypeptide
comprising the amino acid sequence set forth in SEQ ID NO:48. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an OAC polypeptide, wherein the OAC
polypeptide is a variant OAC (Y27F variant) polypeptide comprising the amino
acid
sequence set forth in SEQ ID NO:48, or a conservatively substituted amino acid
sequence
thereof. In some embodiments, a modified host cell of the disclosure comprises
one or more
heterologous nucleic acids comprising a nucleotide sequence encoding an OAC
polypeptide,
wherein the OAC polypeptide is a variant OAC (Y27F variant) polypeptide
comprising an
amino acid sequence having at least 50%, at least 55%, at least 60%, at least
65%, at least
70%, or at least 75% amino acid sequence identity to SEQ ID NO:48. In some
embodiments,
a modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an OAC polypeptide, wherein the OAC
polypeptide is a variant OAC (Y27F variant) polypeptide comprising an amino
acid
sequence having at least 80%, at least 81%, at least 82%, at least 83%, or at
least 84% amino
acid sequence identity to SEQ ID NO:48. In some embodiments, a modified host
cell of the
84
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding an OAC polypeptide, wherein the OAC polypeptide is a variant
OAC
(Y27F variant) polypeptide comprising an amino acid sequence having at least
85%, at least
86%, at least 87%, at least 88%, at least 89%, at least 90%, at least 91%, at
least 92%, at
least 93%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, at least 99%,
at least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%, at least
99.9%, or 100% amino
acid sequence identity to SEQ ID NO:48.
[0241] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a TKS or OAC polypeptide,
such as, a
full-length TKS or OAC polypeptide, a fragment of a TKS or OAC polypeptide, a
variant of
a TKS or OAC polypeptide, a truncated TKS or OAC polypeptide, or a fusion
polypeptide
that has at least one activity of a TKS or OAC polypeptide. In some
embodiments, the
nucleotide sequence is codon-optimized.
[0242] In some embodiments, the TKS polypeptide is overexpressed in the
modified
host cell. Overexpression may be achieved by increasing the copy number of the
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
TKS
polypeptide, e.g., through use of a high copy number expression vector (e.g.,
a plasmid that
exists at 10-40 copies or about 100 copies per cell) and/or by operably
linking the nucleotide
sequence encoding the TKS polypeptide to a strong promoter. In some
embodiments, the
modified host cell has one copy of a heterologous nucleic acid comprising a
nucleotide
sequence encoding the TKS polypeptide. In some embodiments, the modified host
cell has
two copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
TKS polypeptide. In some embodiments, the modified host cell has three copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the TKS
polypeptide.
In some embodiments, the modified host cell has four copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding the TKS polypeptide. In some
embodiments,
the modified host cell has five copies of a heterologous nucleic acid
comprising a nucleotide
sequence encoding the TKS polypeptide. In some embodiments, the modified host
cell has
six copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
TKS polypeptide. In some embodiments, the modified host cell has seven copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the TKS
polypeptide.
In some embodiments, the modified host cell has eight copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding the TKS polypeptide. In some
embodiments, the
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
modified host cell has nine copies of a heterologous nucleic acid comprising a
nucleotide
sequence encoding the TKS polypeptide. In some embodiments, the modified host
cell has
ten copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
TKS polypeptide. In some embodiments, the modified host cell has eleven copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the TKS
polypeptide.
In some embodiments, the modified host cell has twelve copies of a
heterologous nucleic
acid comprising a nucleotide sequence encoding the TKS polypeptide. In some
embodiments, the modified host cell has twelve or more copies of a
heterologous nucleic
acid comprising a nucleotide sequence encoding the TKS polypeptide.
[0243] In some embodiments, the OAC polypeptide is overexpressed in the
modified
host cell. Overexpression may be achieved by increasing the copy number of the
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
OAC
polypeptide, e.g., through use of a high copy number expression vector (e.g.,
a plasmid that
exists at 10-40 copies or about 100 copies per cell) and/or by operably
linking the nucleotide
sequence encoding the OAC polypeptide to a strong promoter. In some
embodiments, the
modified host cell has one copy of a heterologous nucleic acid comprising a
nucleotide
sequence encoding the OAC polypeptide. In some embodiments, the modified host
cell has
two copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
OAC polypeptide. In some embodiments, the modified host cell has three copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the OAC
polypeptide.
In some embodiments, the modified host cell has four copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding the OAC polypeptide. In some
embodiments,
the modified host cell has five copies of a heterologous nucleic acid
comprising a nucleotide
sequence encoding the OAC polypeptide. In some embodiments, the modified host
cell has
six copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
OAC polypeptide. In some embodiments, the modified host cell has seven copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the OAC
polypeptide.
In some embodiments, the modified host cell has eight copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding the OAC polypeptide. In some
embodiments, the
modified host cell has nine copies of a heterologous nucleic acid comprising a
nucleotide
sequence encoding the OAC polypeptide. In some embodiments, the modified host
cell has
ten copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
OAC polypeptide. In some embodiments, the modified host cell has eleven copies
of a
86
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
heterologous nucleic acid comprising a nucleotide sequence encoding the OAC
polypeptide.
In some embodiments, the modified host cell has twelve copies of a
heterologous nucleic
acid comprising a nucleotide sequence encoding the OAC polypeptide. In some
embodiments, the modified host cell has twelve or more copies of a
heterologous nucleic
acid comprising a nucleotide sequence encoding the OAC polypeptide.
[0244] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
TKS
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:18. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a TKS polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:18, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a TKS
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:18. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a TKS polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:18.
[0245] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
OAC
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID NO:20
or SEQ ID
NO:47. In some embodiments, a modified host cell of the disclosure comprises
one or more
heterologous nucleic acids comprising a nucleotide sequence encoding an OAC
polypeptide,
wherein the nucleotide sequence is that set forth in SEQ ID NO:20 or SEQ ID
NO:47, or a
codon degenerate nucleotide sequence of any of the foregoing. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an OAC polypeptide, wherein the
nucleotide
sequence has at least 80%, at least 81%, at least 82%, at least 83%, or at
least 84% sequence
identity to SEQ ID NO:20 or SEQ ID NO:47. In some embodiments, a modified host
cell of
the disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
87
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
sequence encoding an OAC polypeptide, wherein the nucleotide sequence has at
least 85%,
at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at least
91%, at least
92%, at least 930 o, at least 940 o, at least 950 o, at least 96%, at least
970 o, at least 98%, at
least 990, at least 99.50, at least 99.6%, at least 99.70, at least 99.8%, at
least 99.90, or
10000 sequence identity to SEQ ID NO:20 or SEQ ID NO:47.
[0246] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
OAC
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:20. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding an OAC polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:20, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding an OAC
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:20. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an OAC polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:20.
[0247] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
OAC
polypeptide, wherein the OAC polypeptide is a variant OAC (Y27F variant)
polypeptide,
wherein the nucleotide sequence is that set forth in SEQ ID NO:47. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an OAC polypeptide, wherein the OAC
polypeptide is a variant OAC (Y27F variant) polypeptide, wherein the
nucleotide sequence
is that set forth in SEQ ID NO:47, or a codon degenerate nucleotide sequence
thereof In
some embodiments, a modified host cell of the disclosure comprises one or more
heterologous nucleic acids comprising a nucleotide sequence encoding an OAC
polypeptide,
wherein the OAC polypeptide is a variant OAC (Y27F variant) polypeptide,
wherein the
nucleotide sequence has at least 80%, at least 81%, at least 82%, at least
83%, or at least
88
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
84% sequence identity to SEQ ID NO:47. In some embodiments, a modified host
cell of the
disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding an OAC polypeptide, wherein the OAC polypeptide is a variant
OAC
(Y27F variant) polypeptide, wherein the nucleotide sequence has at least 85%,
at least 86%,
at least 87%, at least 88%, at least 89%, at least 90%, at least 91%, at least
92%, at least
93%, at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, at
least 99%, at
least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%, at least 99.9%,
or 100% sequence
identity to SEQ ID NO:47.
Polyp eptides that Generate Geranyl Pyrophosphate
[0248] A modified host cell of the present disclosure may comprise one or
more
heterologous nucleic acids comprising a nucleotide sequence encoding a
polypeptide that
generates GPP. In some embodiments, the polypeptide that generates GPP is a
geranyl
pyrophosphate synthetase (GPPS) polypeptide. In some embodiments, the GPPS
polypeptide
also has farnesyl diphosphate synthase (FPPS) polypeptide activity. In some
embodiments,
the GPPS polypeptide is modified such that it has reduced FPPS polypeptide
activity (e.g., at
least 10%, at least 20%, at least 30%, at least 40%, at least 50%, at least
60%, at least 70%,
at least 80%, at least 90%, or more than at least 90%, less FPPS polypeptide
activity) than
the corresponding wild-type or parental GPPS polypeptide from which the
modified GPPS
polypeptide is derived. In some embodiments, the GPPS polypeptide is modified
such that it
has substantially no FPPS polypeptide activity. In some embodiments, a
modified host cell
of the present disclosure comprises one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a GPPS polypeptide.
[0249] Exemplary GPPS polypeptides disclosed herein may include a full-
length
GPPS polypeptide, a fragment of a GPPS polypeptide, a variant of a GPPS
polypeptide, a
truncated GPPS polypeptide, or a fusion polypeptide that has at least one
activity of a GPPS
polypeptide.
[0250] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
GPPS
polypeptide, wherein the GPPS polypeptide is a variant GPPS (ERG20mut, F96W,
N127W)
polypeptide comprising the amino acid sequence set forth in SEQ ID NO:41. In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a GPPS polypeptide,
wherein the
89
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
GPPS polypeptide is a variant GPPS (ERG20mut, F96W, N127W) polypeptide
comprising
the amino acid sequence set forth in SEQ ID NO:41, or a conservatively
substituted amino
acid sequence thereof. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding a GPPS polypeptide, wherein the GPPS polypeptide is a variant GPPS
(ERG20mut, F96W, N127W) polypeptide comprising an amino acid sequence having
at
least 50%, at least 55%, at least 60%, at least 65%, at least 70%, or at least
75% amino acid
sequence identity to SEQ ID NO:41. In some embodiments, a modified host cell
of the
disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding a GPPS polypeptide, wherein the GPPS polypeptide is a
variant GPPS
(ERG20mut, F96W, N127W) polypeptide comprising an amino acid sequence having
at
least 80%, at least 81%, at least 82%, at least 83%, or at least 84% amino
acid sequence
identity to SEQ ID NO:41. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding a GPPS polypeptide, wherein the GPPS polypeptide is a variant GPPS
(ERG20mut, F96W, N127W) polypeptide comprising an amino acid sequence having
at
least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least
90%, at least 91%,
at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least
97%, at least
98%, at least 99%, at least 99.5%, at least 99.6%, at least 99.7%, at least
99.8%, at least
99.9%, or 100% amino acid sequence identity to SEQ ID NO:41. The mutation in
this amino
acid sequence shifts the ratio of GPP to farnesyl diphosphate (FPP),
increasing the
production of the GPP required to produce CBDA or THCA.
[0251] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a GPPS polypeptide, such
as, a full-
length GPPS polypeptide, a fragment of a GPPS polypeptide, a variant of a GPPS
polypeptide, a truncated GPPS polypeptide, or a fusion polypeptide that has at
least one
activity of a GPPS polypeptide. In some embodiments, the nucleotide sequence
is codon-
optimized.
[0252] In some embodiments, the GPPS polypeptide is overexpressed in the
modified host cell. Overexpression may be achieved by increasing the copy
number of the
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding the
GPPS polypeptide, e.g., through use of a high copy number expression vector
(e.g., a
plasmid that exists at 10-40 copies or about 100 copies per cell) and/or by
operably linking
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
the nucleotide sequence encoding the GPPS polypeptide to a strong promoter. In
some
embodiments, the modified host cell has one copy of a heterologous nucleic
acid comprising
a nucleotide sequence encoding the GPPS polypeptide. In some embodiments, the
modified
host cell has two copies of a heterologous nucleic acid comprising a
nucleotide sequence
encoding the GPPS polypeptide. In some embodiments, the modified host cell has
three
copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the GPPS
polypeptide. In some embodiments, the modified host cell has four copies of a
heterologous
nucleic acid comprising a nucleotide sequence encoding the GPPS polypeptide.
In some
embodiments, the modified host cell has five copies of a heterologous nucleic
acid
comprising a nucleotide sequence encoding the GPPS polypeptide. In some
embodiments,
the modified host cell has six copies of a heterologous nucleic acid
comprising a nucleotide
sequence encoding the GPPS polypeptide. In some embodiments, the modified host
cell has
seven copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
GPPS polypeptide. In some embodiments, the modified host cell has eight copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the GPPS
polypeptide.
In some embodiments, the modified host cell has eight or more copies of a
heterologous
nucleic acid comprising a nucleotide sequence encoding the GPPS polypeptide.
[0253] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
GPPS
polypeptide, wherein the GPPS polypeptide is a variant GPPS (ERG20mut, F96W,
N127W)
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:40. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a GPPS polypeptide,
wherein the
GPPS polypeptide is a variant GPPS (ERG20mut, F96W, N127W) polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:40, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a GPPS
polypeptide, wherein the GPPS polypeptide is a variant GPPS (ERG20mut, F96W,
N127W)
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:40. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a GPPS polypeptide, wherein the GPPS
polypeptide is a variant GPPS (ERG20mut, F96W, N127W) polypeptide, wherein the
91
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
nucleotide sequence has at least 85%, at least 86%, at least 87%, at least
88%, at least 89%,
at least 90%, at least 91%, at least 92%, at least 930 o, at least 940 o, at
least 950 o, at least
960 o, at least 970 o, at least 98%, at least 990 o, at least 99.50 o, at
least 99.6%, at least 99.70
,
at least 99.8%, at least 99.90 o, or 1000o sequence identity to SEQ ID NO:40.
Polyp eptides that Generate Acetyl-CoA from Pyruvate
[0254] A modified host cell of the present disclosure may comprise one or
more
heterologous nucleic acids comprising a nucleotide sequence encoding a
polypeptide that
generates acetyl-CoA from pyruvate. Polypeptides that generate acetyl-CoA from
pyruvate
may include a pyruvate decarboxylase (PDC) polypeptide. In some embodiments, a
modified
host cell of the present disclosure comprises one or more heterologous nucleic
acids
comprising a nucleotide sequence encoding a PDC polypeptide.
[0255] Exemplary PDC polypeptides disclosed herein may include a full-
length PDC
polypeptide, a fragment of a PDC polypeptide, a variant of a PDC polypeptide,
a truncated
PDC polypeptide, or a fusion polypeptide that has at least one activity of a
PDC polypeptide.
[0256] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
PDC
polypeptide, wherein the PDC polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:35. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
PDC
polypeptide, wherein the PDC polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:35, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a PDC polypeptide,
wherein the
PDC polypeptide comprises an amino acid sequence having at least 50%, at least
55%, at
least 60%, at least 65%, at least 70%, or at least 750 amino acid sequence
identity to SEQ
ID NO:35. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
PDC
polypeptide, wherein the PDC polypeptide comprises an amino acid sequence
having at least
800o, at least 810o, at least 82%, at least 83%, or at least 84% amino acid
sequence identity
to SEQ ID NO:35. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a PDC
polypeptide, wherein the PDC polypeptide comprises an amino acid sequence
having at least
850o, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at
least 91%, at
92
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
least 92%, at least 9300, at least 9400, at least 9500, at least 96%, at least
9700, at least 98%,
at least 9900, at least 99.500, at least 99.6%, at least 99.700, at least
99.8%, at least 99.900, or
100% amino acid sequence identity to SEQ ID NO:35.
[0257] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a PDC polypeptide, such
as, a full-
length PDC polypeptide, a fragment of a PDC polypeptide, a variant of a PDC
polypeptide, a
truncated PDC polypeptide, or a fusion polypeptide that has at least one
activity of a PDC
polypeptide. In some embodiments, the nucleotide sequence is codon-optimized.
[0258] In some embodiments, the PDC polypeptide is overexpressed in the
modified
host cell. Overexpression may be achieved by increasing the copy number of the
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
PDC
polypeptide, e.g., through use of a high copy number expression vector (e.g.,
a plasmid that
exists at 10-40 copies or about 100 copies per cell) and/or by operably
linking the nucleotide
sequence encoding the PDC polypeptide to a strong promoter. In some
embodiments, the
modified host cell has one copy of a heterologous nucleic acid comprising a
nucleotide
sequence encoding the PDC polypeptide. In some embodiments, the modified host
cell has
two copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
PDC polypeptide. In some embodiments, the modified host cell has three copies
of a
heterologous nucleic acid comprising a nucleotide sequence encoding the PDC
polypeptide.
In some embodiments, the modified host cell has four copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding the PDC polypeptide. In some
embodiments,
the modified host cell has five copies of a heterologous nucleic acid
comprising a nucleotide
sequence encoding the PDC polypeptide. In some embodiments, the modified host
cell has
five or more copies of a heterologous nucleic acid comprising a nucleotide
sequence
encoding the PDC polypeptide.
[0259] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
PDC
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:34. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a PDC polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:34, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a PDC
93
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:34. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a PDC polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:34.
Polyp eptides that Condense Two Molecules of Acetyl-CoA to Generate
Acetoacetyl-CoA
[0260] A modified host cell of the disclosure may comprise one or more
heterologous nucleic acids comprising a nucleotide sequence encoding a
polypeptide that
condenses two molecules of acetyl-CoA to generate acetoacetyl-CoA. In some
embodiments,
the polypeptide that condenses two molecules of acetyl-CoA to generate
acetoacetyl-CoA is
an acetoacetyl-CoA thiolase polypeptide. In some embodiments, a modified host
cell of the
present disclosure comprises one or more heterologous nucleic acids comprising
a nucleotide
sequence encoding an acetoacetyl-CoA thiolase polypeptide.
[0261] Exemplary acetoacetyl-CoA thiolase polypeptides disclosed herein
may
include a full-length acetoacetyl-CoA thiolase polypeptide, a fragment of an
acetoacetyl-
CoA thiolase polypeptide, a variant of an acetoacetyl-CoA thiolase
polypeptide, a truncated
acetoacetyl-CoA thiolase polypeptide, or a fusion polypeptide that has at
least one activity of
an acetoacetyl-CoA thiolase polypeptide.
[0262] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
acetoacetyl-
CoA thiolase polypeptide, wherein the acetoacetyl-CoA thiolase polypeptide
comprises the
amino acid sequence set forth in SEQ ID NO:31. In some embodiments, a modified
host cell
of the disclosure comprises one or more heterologous nucleic acids comprising
a nucleotide
sequence encoding an acetoacetyl-CoA thiolase polypeptide, wherein the
acetoacetyl-CoA
thiolase polypeptide comprises the amino acid sequence set forth in SEQ ID
NO:31, or a
conservatively substituted amino acid sequence thereof. In some embodiments, a
modified
host cell of the disclosure comprises one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding an acetoacetyl-CoA thiolase polypeptide, wherein
the
acetoacetyl-CoA thiolase polypeptide comprises an amino acid sequence having
at least
50%, at least 55%, at least 60%, at least 65%, at least 70%, or at least 75%
amino acid
94
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
sequence identity to SEQ ID NO:31. In some embodiments, a modified host cell
of the
disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding an acetoacetyl-CoA thiolase polypeptide, wherein the
acetoacetyl-CoA
thiolase polypeptide comprises an amino acid sequence having at least 80%, at
least 81%, at
least 82%, at least 83%, or at least 84% amino acid sequence identity to SEQ
ID NO:31. In
some embodiments, a modified host cell of the disclosure comprises one or more
heterologous nucleic acids comprising a nucleotide sequence encoding an
acetoacetyl-CoA
thiolase polypeptide, wherein the acetoacetyl-CoA thiolase polypeptide
comprises an amino
acid sequence having at least 85%, at least 86%, at least 87%, at least 88%,
at least 89%, at
least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least
95%, at least 96%,
at least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at
least 99.8%, at least 99.9%, or 100% amino acid sequence identity to SEQ ID
NO:31.
[0263] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes an acetoacetyl-CoA
thiolase
polypeptide, such as, a full-length acetoacetyl-CoA thiolase polypeptide, a
fragment of an
acetoacetyl-CoA thiolase polypeptide, a variant of an acetoacetyl-CoA thiolase
polypeptide,
a truncated acetoacetyl-CoA thiolase polypeptide, or a fusion polypeptide that
has at least
one activity of an acetoacetyl-CoA thiolase polypeptide. In some embodiments,
the
nucleotide sequence is codon-optimized.
[0264] In some embodiments, the acetoacetyl-CoA thiolase polypeptide is
overexpressed in the modified host cell. Overexpression may be achieved by
increasing the
copy number of the one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding the acetoacetyl-CoA thiolase polypeptide, e.g., through use
of a high
copy number expression vector (e.g., a plasmid that exists at 10-40 copies or
about 100
copies per cell) and/or by operably linking the nucleotide sequence encoding
the acetoacetyl-
CoA thiolase polypeptide to a strong promoter. In some embodiments, the
modified host cell
has one copy of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
acetoacetyl-CoA thiolase polypeptide. In some embodiments, the modified host
cell has two
copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
acetoacetyl-CoA thiolase polypeptide. In some embodiments, the modified host
cell has
three copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
acetoacetyl-CoA thiolase polypeptide. In some embodiments, the modified host
cell has four
copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
acetoacetyl-CoA thiolase polypeptide. In some embodiments, the modified host
cell has five
copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding the
acetoacetyl-CoA thiolase polypeptide. In some embodiments, the modified host
cell has five
or more copies of a heterologous nucleic acid comprising a nucleotide sequence
encoding
the acetoacetyl-CoA thiolase polypeptide.
[0265] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
acetoacetyl-
CoA thiolase polypeptide, wherein the nucleotide sequence is that set forth in
SEQ ID
NO:30. In some embodiments, a modified host cell of the disclosure comprises
one or more
heterologous nucleic acids comprising a nucleotide sequence encoding an
acetoacetyl-CoA
thiolase polypeptide, wherein the nucleotide sequence is that set forth in SEQ
ID NO:30, or a
codon degenerate nucleotide sequence thereof In some embodiments, a modified
host cell
of the disclosure comprises one or more heterologous nucleic acids comprising
a nucleotide
sequence encoding an acetoacetyl-CoA thiolase polypeptide, wherein the
nucleotide
sequence has at least 80%, at least 81%, at least 82%, at least 83%, or at
least 84% sequence
identity to SEQ ID NO:30. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding an acetoacetyl-CoA thiolase polypeptide, wherein the nucleotide
sequence has at
least 85%, at least 86%, at least 87%, at least 88%, at least 89%, at least
90%, at least 91%,
at least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least
97%, at least
98%, at least 99%, at least 99.5%, at least 99.6%, at least 99.7%, at least
99.8%, at least
99.9%, or 100% sequence identity to SEQ ID NO:30.
Mevalonate Pathway Polyp eptides
[0266] A modified host cell of the present disclosure may comprise one or
more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides having at least one activity of a polypeptide present in the
mevalonate (MEV)
pathway. In certain such embodiments, the one or more polypeptides having at
least one
activity of a polypeptide present in the mevalonate (MEV) pathway comprise one
or more
MEV pathway polypeptides.
[0267] In some embodiments, the one or more polypeptides that are part of
a
biosynthetic pathway that generates GPP are one or more polypeptides having at
least one
activity of a polypeptide present in the mevalonate pathway. The mevalonate
pathway may
comprise polypeptides that catalyze the following steps: (a) condensing two
molecules of
96
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
acetyl-CoA to generate acetoacetyl-CoA (e.g., by action of an acetoacetyl-CoA
thiolase
polypeptide); (b) condensing acetoacetyl-CoA with acetyl-CoA to form
hydroxymethylglutaryl-CoA (HMG-CoA) (e.g., by action of a HMGS polypeptide);
(c)
converting HMG-CoA to mevalonate (e.g., by action of an HMGR polypeptide); (d)
phosphorylating mevalonate to mevalonate 5-phosphate (e.g., by action of a MK
polypeptide); (e) converting mevalonate 5-phosphate to mevalonate 5-
pyrophosphate (e.g.,
by action of a PMK polypeptide); (f) converting mevalonate 5-pyrophosphate to
isopentenyl
pyrophosphate (e.g., by action of a mevalonate pyrophosphate decarboxylase
(MPD or
MVD1) polypeptide); and (g) converting isopentenyl pyrophosphate to
dimethylallyl
pyrophosphate (e.g., by action of an isopentenyl pyrophosphate isomerase
(IDI1)
polypeptide).
[0268] In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding a MEV pathway polypeptide. In some embodiments, a modified host cell
of the
present disclosure comprises one or more heterologous nucleic acids comprising
nucleotide
sequences encoding more than one MEV pathway polypeptide. In some embodiments,
a
modified host cell of the present disclosure comprises one or more
heterologous nucleic
acids comprising nucleotide sequences encoding more than two MEV pathway
polypeptides.
In some embodiments, a modified host cell of the present disclosure comprises
one or more
heterologous nucleic acids comprising nucleotide sequences encoding more than
three MEV
pathway polypeptides. In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding more than four MEV pathway polypeptides. In some embodiments, a
modified
host cell of the present disclosure comprises one or more heterologous nucleic
acids
comprising nucleotide sequences encoding more than five MEV pathway
polypeptides. In
some embodiments, a modified host cell of the present disclosure comprises one
or more
heterologous nucleic acids comprising nucleotide sequences encoding more than
six MEV
pathway polypeptides. In some embodiments, a modified host cell of the present
disclosure
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding all MEV pathway polypeptides.
[0269] Exemplary MEV pathway polypeptides disclosed herein may include a
full-
length MEV pathway polypeptide, a fragment of a MEV pathway polypeptide, a
variant of a
MEV pathway polypeptide, a truncated MEV pathway polypeptide, or a fusion
polypeptide
97
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
that has at least one activity of a MEV pathway polypeptide. In some
embodiments, the one
or more MEV pathway polypeptides are selected from the group consisting of an
acetoacetyl-CoA thiolase polypeptide, a HMGS polypeptide, a HMGR polypeptide,
an MK
polypeptide, a PMK polypeptide, an MVD1 polypeptide, and an IDI1 polypeptide.
[0270] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
HMGS
polypeptide, wherein the HMGS polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:29. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
HMGS
polypeptide, wherein the HMGS polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:29, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a HMGS polypeptide,
wherein the
HMGS polypeptide comprises an amino acid sequence having at least 50%, at
least 55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:29. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
HMGS
polypeptide, wherein the HMGS polypeptide comprises an amino acid sequence
having at
least 80%, at least 81%, at least 82%, at least 83%, or at least 84% amino
acid sequence
identity to SEQ ID NO:29. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding a HMGS polypeptide, wherein the HMGS polypeptide comprises an amino
acid
sequence having at least 85%, at least 86%, at least 87%, at least 88%, at
least 89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% amino acid sequence identity to SEQ ID NO:29.
[0271] In some embodiments, the HMGR polypeptide is a truncated HMGR
(tHMGR) polypeptide. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding a tHMGR polypeptide, wherein the tHMGR polypeptide comprises the
amino acid
sequence set forth in SEQ ID NO:27. In some embodiments, a modified host cell
of the
disclosure comprises one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding a tHMGR polypeptide, wherein the tHMGR polypeptide comprises
the
98
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
amino acid sequence set forth in SEQ ID NO:27, or a conservatively substituted
amino acid
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a
tHMGR polypeptide, wherein the tHMGR polypeptide comprises an amino acid
sequence
having at least 50%, at least 55%, at least 60%, at least 65%, at least 70%,
or at least 75%
amino acid sequence identity to SEQ ID NO:27. In some embodiments, a modified
host cell
of the disclosure comprises one or more heterologous nucleic acids comprising
a nucleotide
sequence encoding a tHMGR polypeptide, wherein the tHMGR polypeptide comprises
an
amino acid sequence having at least 80%, at least 81%, at least 82%, at least
83%, or at least
84% amino acid sequence identity to SEQ ID NO:27. In some embodiments, a
modified host
cell of the disclosure comprises one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a tHMGR polypeptide, wherein the tHMGR
polypeptide
comprises an amino acid sequence having at least 85%, at least 86%, at least
87%, at least
88%, at least 89%, at least 90%, at least 91%, at least 92%, at least 93%, at
least 94%, at
least 95%, at least 96%, at least 97%, at least 98%, at least 99%, at least
99.5%, at least
99.6%, at least 99.7%, at least 99.8%, at least 99.9%, or 100% amino acid
sequence identity
to SEQ ID NO:27.
[0272] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a MK
polypeptide, wherein the MK polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:39. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
MK
polypeptide, wherein the MK polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:39, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a MK polypeptide,
wherein the
MK polypeptide comprises an amino acid sequence having at least 50%, at least
55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:39. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a MK
polypeptide, wherein the MK polypeptide comprises an amino acid sequence
having at least
80%, at least 81%, at least 82%, at least 83%, or at least 84% amino acid
sequence identity
to SEQ ID NO:39. In some embodiments, a modified host cell of the disclosure
comprises
99
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a MK
polypeptide, wherein the MK polypeptide comprises an amino acid sequence
having at least
85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at
least 91%, at
least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least
97%, at least 98%,
at least 99%, at least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%,
at least 99.9%, or
100% amino acid sequence identity to SEQ ID NO:39.
[0273] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
PMK
polypeptide, wherein the PMK polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:37. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
PMK
polypeptide, wherein the PMK polypeptide comprises the amino acid sequence set
forth in
SEQ ID NO:37, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a PMK polypeptide,
wherein the
PMK polypeptide comprises an amino acid sequence having at least 50%, at least
55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:37. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
PMK
polypeptide, wherein the PMK polypeptide comprises an amino acid sequence
having at
least 80%, at least 81%, at least 82%, at least 83%, or at least 84% amino
acid sequence
identity to SEQ ID NO:37. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding a PMK polypeptide, wherein the PMK polypeptide comprises an amino
acid
sequence having at least 85%, at least 86%, at least 87%, at least 88%, at
least 89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% amino acid sequence identity to SEQ ID NO:37.
[0274] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
MVD1
polypeptide, wherein the MVD1 polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:33. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding a
MVD1
100
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide, wherein the MVD1 polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:33, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a MVD1 polypeptide,
wherein the
MVD1 polypeptide comprises an amino acid sequence having at least 50%, at
least 55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:33. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
MVD1
polypeptide, wherein the MVD1 polypeptide comprises an amino acid sequence
having at
least 80%, at least 81%, at least 82%, at least 83%, or at least 84% amino
acid sequence
identity to SEQ ID NO:33. In some embodiments, a modified host cell of the
disclosure
comprises one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding a MVD1 polypeptide, wherein the MVD1 polypeptide comprises an amino
acid
sequence having at least 85%, at least 86%, at least 87%, at least 88%, at
least 89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% amino acid sequence identity to SEQ ID NO:33.
[0275] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
IDI1
polypeptide, wherein the IDI1 polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:25. In some embodiments, a modified host cell of the disclosure
comprises one
or more heterologous nucleic acids comprising a nucleotide sequence encoding
an IDI1
polypeptide, wherein the IDI1 polypeptide comprises the amino acid sequence
set forth in
SEQ ID NO:25, or a conservatively substituted amino acid sequence thereof In
some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding an IDI1 polypeptide,
wherein the
IDI1 polypeptide comprises an amino acid sequence having at least 50%, at
least 55%, at
least 60%, at least 65%, at least 70%, or at least 75% amino acid sequence
identity to SEQ
ID NO:25. In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
IDI1
polypeptide, wherein the IDI1 polypeptide comprises an amino acid sequence
having at least
80%, at least 81%, at least 82%, at least 83%, or at least 84% amino acid
sequence identity
to SEQ ID NO:25. In some embodiments, a modified host cell of the disclosure
comprises
101
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding an IDI1
polypeptide, wherein the IDI1 polypeptide comprises an amino acid sequence
having at least
85%, at least 86%, at least 87%, at least 88%, at least 89%, at least 90%, at
least 91%, at
least 92%, at least 93%, at least 94%, at least 95%, at least 96%, at least
97%, at least 98%,
at least 99%, at least 99.5%, at least 99.6%, at least 99.7%, at least 99.8%,
at least 99.9%, or
100% amino acid sequence identity to SEQ ID NO:25.
[0276] Exemplary heterologous nucleic acids disclosed herein may include
nucleic
acids comprising a nucleotide sequence that encodes a MEV pathway polypeptide,
such as, a
full-length MEV pathway polypeptide, a fragment of a MEV pathway polypeptide,
a variant
of a MEV pathway polypeptide, a truncated MEV pathway polypeptide, or a fusion
polypeptide that has at least one activity of a polypeptide that is part of
the MEV pathway.
In some embodiments, the nucleotide sequence is codon-optimized.
[0277] In some embodiments, one or more MEV pathway polypeptides are
overexpressed in the modified host cell. Overexpression may be achieved by
increasing the
copy number of the one or more heterologous nucleic acids comprising
nucleotide sequences
encoding a MEV pathway polypeptide, e.g., through use of a high copy number
expression
vector (e.g., a plasmid that exists at 10-40 copies or about 100 copies per
cell) and/or by
operably linking the nucleotide sequences encoding a MEV pathway polypeptide
to a strong
promoter. In some embodiments, the modified host cell has one copy of a
heterologous
nucleic acid comprising a nucleotide sequence encoding a MEV pathway
polypeptide. In
some embodiments, the modified host cell has two copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding a MEV pathway polypeptide. In some
embodiments, the modified host cell has three copies of a heterologous nucleic
acid
comprising a nucleotide sequence encoding a MEV pathway polypeptide. In some
embodiments, the modified host cell has four copies of a heterologous nucleic
acid
comprising a nucleotide sequence encoding a MEV pathway polypeptide. In some
embodiments, the modified host cell has five copies of a heterologous nucleic
acid
comprising a nucleotide sequence encoding a MEV pathway polypeptide. In some
embodiments, the modified host cell has five or more copies of a heterologous
nucleic acid
comprising a nucleotide sequence encoding a MEV pathway polypeptide.
[0278] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
HMGS
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:28. In some
102
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a HMGS polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:28, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a HMGS
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:28. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a HMGS polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:28.
[0279] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
tHMGR
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:26. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a tHMGR polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:26, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a
tHMGR polypeptide, wherein the nucleotide sequence has at least 80%, at least
81%, at least
82%, at least 83%, or at least 84% sequence identity to SEQ ID NO:26. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a tHMGR polypeptide,
wherein the
nucleotide sequence has at least 85%, at least 86%, at least 87%, at least
88%, at least 89%,
at least 90%, at least 91%, at least 92%, at least 93%, at least 94%, at least
95%, at least
96%, at least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%,
at least 99.7%,
at least 99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:26.
[0280] In some embodiments, a modified host cell of the present
disclosure
comprises two or more heterologous nucleic acids comprising a nucleotide
sequence that
encodes a tHMGR polypeptide. In some embodiments, a modified host cell of the
present
103
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
disclosure comprises two heterologous nucleic acids comprising a nucleotide
sequence that
encodes a tHMGR polypeptide.
[0281] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a MK
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:38. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a MK polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:38, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a MK
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:38. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a MK polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:38.
[0282] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
PMK
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:36. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a PMK polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:36, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a PMK
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:36. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a PMK polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
104
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:36.
[0283] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
MVD1
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:32. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a MVD1 polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:32, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a MVD1
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:32. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a MVD1 polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:32.
[0284] In some embodiments, a modified host cell of the disclosure
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding an
IDI1
polypeptide, wherein the nucleotide sequence is that set forth in SEQ ID
NO:24. In some
embodiments, a modified host cell of the disclosure comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding an IDI1 polypeptide,
wherein the
nucleotide sequence is that set forth in SEQ ID NO:24, or a codon degenerate
nucleotide
sequence thereof. In some embodiments, a modified host cell of the disclosure
comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding an IDI1
polypeptide, wherein the nucleotide sequence has at least 80%, at least 81%,
at least 82%, at
least 83%, or at least 84% sequence identity to SEQ ID NO:24. In some
embodiments, a
modified host cell of the disclosure comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an IDI1 polypeptide, wherein the
nucleotide
sequence has at least 85%, at least 86%, at least 87%, at least 88%, at least
89%, at least
90%, at least 91%, at least 92%, at least 93%, at least 94%, at least 95%, at
least 96%, at
105
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
least 97%, at least 98%, at least 99%, at least 99.5%, at least 99.6%, at
least 99.7%, at least
99.8%, at least 99.9%, or 100% sequence identity to SEQ ID NO:24.
Modified Host Cells to Produce Cannabinoids or Cannabinoid Derivatives and/or
Express
Engineered Variants of the Disclosure
[0285] The present disclosure provides modified host cells comprising one
or more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure. The modified host cells of the disclosure comprising one or more
nucleic acids
comprising a nucleotide sequence encoding an engineered variant of the
disclosure may be
for producing cannabinoids or cannabinoid derivatives and/or for expressing an
engineered
variant of the disclosure.
[0286] The disclosure provides for modified host cells for producing
cannabinoids or
cannabinoid derivatives. For producing cannabinoids or cannabinoid
derivatives, modified
host cells disclosed herein may be modified to express or overexpress one or
more nucleic
acids disclosed herein comprising nucleotide sequences encoding an engineered
variant of
the disclosure, one or more of a KAR2 polypeptide, a PDT] polypeptide, an ER01
polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide, and/or one or more
polypeptides
involved in cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate
(GPP), prenyl
phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis. A modified host
cell for
producing cannabinoids or cannabinoid derivatives may comprise a deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide. In certain such embodiments, the modified host cell for
producing
cannabinoids or cannabinoid derivatives may comprise a deletion of one or more
genes
encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide. In some
embodiments,
the modified host cell for producing cannabinoids or cannabinoid derivatives
may comprise
a downregulation of one or more genes encoding one or more of a ROT2
polypeptide or a
PEP4 polypeptide. In some embodiments, the modified host cell for producing a
cannabinoid
or a cannabinoid derivative comprises one or more nucleic acids comprising a
codon-
optimized nucleotide sequence encoding an engineered variant of the
disclosure. In some
embodiments, the nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDT] polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or an IRE1
polypeptide,
and/or one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
106
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis are codon-optimized.
[0287] The disclosure also provides modified host cells modified to
express or
overexpress one or more nucleic acids comprising a nucleotide sequence
encoding an
engineered variant of the disclosure. In some embodiments of the modified host
cell for
expressing an engineered variant of the disclosure, the modified host cell
comprises one or
more nucleic acids comprising a nucleotide sequence encoding the engineered
variant of the
disclosure and one or more heterologous nucleic acids disclosed herein
comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide. In some
embodiments of
the modified host cell for expressing an engineered variant of the disclosure,
the modified
host cell comprises one or more nucleic acids comprising a nucleotide sequence
encoding
the engineered variant of the disclosure and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide. In
certain such
embodiments, the modified host cell may comprise a deletion of one or more
genes encoding
one or more of a ROT2 polypeptide or a PEP4 polypeptide. In some embodiments,
the
modified host cell may comprise a downregulation of one or more genes encoding
one or
more of a ROT2 polypeptide or a PEP4 polypeptide. In some embodiments of the
modified
host cell for expressing an engineered variant of the disclosure, the
nucleotide sequence
encoding the engineered variant of the disclosure is a codon-optimized
nucleotide sequence.
In some embodiments, the nucleotide sequences encoding one or more of a KAR2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or
an IRE1
polypeptide are codon-optimized.
[0288] To produce cannabinoids or cannabinoid derivatives, expression or
overexpression of one or more nucleic acids comprising a nucleotide sequence
encoding an
engineered variant of the disclosure in a modified host cell may be done in
combination with
expression or overexpression by the modified host cell of one or more
heterologous nucleic
acids disclosed herein (e.g., one or more heterologous nucleic acids
comprising nucleotide
sequences encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an
ER01
polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide) and/or with deletion
or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide. In some embodiments, the nucleotide sequence encoding the
engineered
variant of the disclosure is a codon-optimized nucleotide sequence.
107
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0289] To express or overexpress an engineered variant of the disclosure,
expression
or overexpression of one or more nucleic acids comprising a nucleotide
sequence encoding
the engineered variant in a modified host cell may be done in combination with
expression
or overexpression by the modified host cell of one or more heterologous
nucleic acids
disclosed herein (e.g., one or more heterologous nucleic acids comprising
nucleotide
sequences encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an
ER01
polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide) and/or with deletion
or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide. In some embodiments, the nucleotide sequence encoding the
engineered
variant is a codon-optimized nucleotide sequence.
[0290] In some embodiments, a modified host cell of the disclosure for
producing
cannabinoids or cannabinoid derivatives produces a cannabinoid or a
cannabinoid derivative
in an amount, as measured in mg/L or mM, greater than an amount of the
cannabinoid or the
cannabinoid derivative produced by a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, grown under
similar
culture conditions for the same length of time. In some embodiments, the
modified host cell
for producing cannabinoids or cannabinoid derivatives produces a cannabinoid
or a
cannabinoid derivative in an amount, as measured in mg/L or mM, at least 5%,
at least 10%,
at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at least
40%, at least
45%, at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, at
least 100%, at
least 150% at least 200%, at least 500%, or at least 1000% greater than an
amount of the
cannabinoid or the cannabinoid derivative produced by a modified host cell
comprising one
or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
similar culture conditions for the same length of time.
[0291] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure produces a cannabinoid or a cannabinoid derivative in an
amount, as measured
in mg/L or mM, greater than an amount of the cannabinoid or the cannabinoid
derivative
produced by a modified host cell comprising one or more nucleic acids
comprising a
108
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant, grown under similar culture
conditions for the
same length of time. In some embodiments, the modified host cell comprising
one or more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure produces a cannabinoid or a cannabinoid derivative in an amount, as
measured in
mg/L or mM, at least 5%, at least 10%, at least 15%, at least 20%, at least
25%, at least 30%,
at least 35%, at least 40%, at least 45%, at least 50%, at least 60%, at least
70%, at least
80%, at least 90%, at least 100%, at least 150% at least 200%, at least 500%,
or at least
1000% greater than an amount of the cannabinoid or the cannabinoid derivative
produced by
a modified host cell comprising one or more nucleic acids comprising a
nucleotide sequence
encoding a tetrahydrocannabinolic acid synthase polypeptide having an amino
acid sequence
of SEQ ID NO:44, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, grown under similar culture conditions for the same length
of time. In
some embodiments of the modified host cell of the disclosure comprising one or
more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure, the modified host cell comprises one or more heterologous nucleic
acids
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis. In some embodiments, a
modified host cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, but lacking a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant, comprises one or more heterologous nucleic acids comprising
nucleotide sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis.
[0292] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure and one or more heterologous nucleic acids comprising
nucleotide sequences
encoding one or more of a KAR2 polypeptide, a PIM polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, or an IRE1 polypeptide produces a cannabinoid or a
cannabinoid
derivative in an amount, as measured in mg/L or mM, greater than an amount of
the
109
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
cannabinoid or the cannabinoid derivative produced by a modified host cell
comprising one
or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44 and
one or
more heterologous nucleic acids comprising nucleotide sequences encoding one
or more of a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide,
or an
IRE1 polypeptide, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, grown under similar culture conditions for the same length
of time. In
some embodiments, a modified host cell of the disclosure comprising one or
more nucleic
acids comprising a nucleotide sequence encoding an engineered variant of the
disclosure and
one or more heterologous nucleic acids comprising nucleotide sequences
encoding one or
more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1
polypeptide, or an IRE1 polypeptide produces a cannabinoid or a cannabinoid
derivative in
an amount, as measured in mg/L or mM, at least 5%, at least 10%, at least 15%,
at least
20%, at least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at
least 50%, at
least 60%, at least 70%, at least 80%, at least 90%, at least 100%, at least
150% at least
200%, at least 500%, or at least 1000% greater than an amount of the
cannabinoid or the
cannabinoid derivative produced by a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 and one or more
heterologous
nucleic acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or
an IRE1
polypeptide, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, grown under similar culture conditions for the same length
of time. In
some embodiments of the modified host cell of the disclosure comprising one or
more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure and one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, a FAD1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, the modified host cell comprises one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis. In
some
embodiments, a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
110
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
amino acid sequence of SEQ ID NO:44 and one or more heterologous nucleic acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDI1
polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide,
but
lacking a nucleic acid comprising a nucleotide sequence encoding an engineered
variant,
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis.
[0293] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure and a deletion or downregulation of one or more genes encoding
one or more
of a ROT2 polypeptide or a PEP4 polypeptide produces a cannabinoid or a
cannabinoid
derivative in an amount, as measured in mg/L or mM, greater than an amount of
the
cannabinoid or the cannabinoid derivative produced by a modified host cell
comprising one
or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44 and a
deletion
or downregulation of one or more genes encoding one or more of a ROT2
polypeptide or a
PEP4 polypeptide, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, grown under similar culture conditions for the same length
of time. In
some embodiments, a modified host cell of the disclosure comprising one or
more nucleic
acids comprising a nucleotide sequence encoding an engineered variant of the
disclosure and
a deletion or downregulation of one or more genes encoding one or more of a
ROT2
polypeptide or a PEP4 polypeptide produces a cannabinoid or a cannabinoid
derivative in an
amount, as measured in mg/L or mM, at least 5%, at least 10%, at least 15%, at
least 20%, at
least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least
50%, at least 60%,
at least 70%, at least 80%, at least 90%, at least 100%, at least 150% at
least 200%, at least
500%, or at least 1000% greater than an amount of the cannabinoid or the
cannabinoid
derivative produced by a modified host cell comprising one or more nucleic
acids
comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 and a deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, grown under similar culture conditions for the same length
of time. In
111
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
some embodiments of the modified host cell of the disclosure comprising one or
more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure and a deletion or downregulation of one or more genes encoding one
or more of a
ROT2 polypeptide or a PEP4 polypeptide, the modified host cell comprises one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis. In
some
embodiments, a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44 and a deletion or downregulation of one or
more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
comprises
one or more heterologous nucleic acids comprising nucleotide sequences
encoding one or
more polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis.
[0294] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure, one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
an IRE1 polypeptide, and a deletion or downregulation of one or more genes
encoding one
or more of a ROT2 polypeptide or a PEP4 polypeptide produces a cannabinoid or
a
cannabinoid derivative in an amount, as measured in mg/L or mM, greater than
an amount of
the cannabinoid or the cannabinoid derivative produced by a modified host cell
comprising
one or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, one or more heterologous nucleic acids comprising nucleotide sequences
encoding
one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or
an IRE1
polypeptide, and a deletion or downregulation of one or more genes encoding
one or more of
a ROT2 polypeptide or a PEP4 polypeptide, but lacking a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant, grown under similar
culture conditions
for the same length of time. In some embodiments, a modified host cell of the
disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
112
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
engineered variant of the disclosure, one or more heterologous nucleic acids
comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide
produces a
cannabinoid or a cannabinoid derivative in an amount, as measured in mg/L or
mM, at least
5%, at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at
least 35%, at least
40%, at least 45%, at least 50%, at least 60%, at least 70%, at least 80%, at
least 90%, at
least 100%, at least 150% at least 200%, at least 500%, or at least 1000%
greater than an
amount of the cannabinoid or the cannabinoid derivative produced by a modified
host cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, one or more heterologous nucleic acids comprising nucleotide sequences
encoding
one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or
an IRE1
polypeptide, and a deletion or downregulation of one or more genes encoding
one or more of
a ROT2 polypeptide or a PEP4 polypeptide, but lacking a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant, grown under similar
culture conditions
for the same length of time. In some embodiments of the modified host cell of
the disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure, one or more heterologous nucleic acids
comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide, the
modified
host cell comprises one or more heterologous nucleic acids comprising
nucleotide sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, one or more
heterologous
nucleic acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or an IRE1 polypeptide,
and a
deletion or downregulation of one or more genes encoding one or more of a ROT2
polypeptide or a PEP4 polypeptide, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant, comprises one or more heterologous
nucleic acids
113
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis.
[0295] In some embodiments, the modified host cell of the disclosure for
producing
cannabinoids or cannabinoid derivatives has a growth rate and/or biomass yield
similar to, or
lower than, a growth rate and/or biomass yield of a modified host cell
comprising one or
more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
similar culture conditions for the same length of time. In some embodiments,
the modified
host cell of the disclosure for producing cannabinoids or cannabinoid
derivatives has a
growth rate and/or biomass yield similar to, or lower than, a growth rate
and/or biomass
yield and an increased titer of THCA compared to a modified host cell
comprising one or
more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
similar culture conditions for the same length of time.
[0296] In some embodiments, the modified host cell of the disclosure for
producing
cannabinoids or cannabinoid derivatives has a faster growth rate and/or higher
biomass yield
compared to a growth rate and/or higher biomass yield of a modified host cell
comprising
one or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, but lacking a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant, grown under similar culture conditions for the same length of time.
In some
embodiments, the modified host cell of the disclosure for producing
cannabinoids or
cannabinoid derivatives has a growth rate and/or higher biomass yield at least
5%, at least
10%, at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at
least 40%, at
least 45%, at least 50%, at least 60%, at least 70%, at least 80%, at least
90%, at least 100%,
at least 150% at least 200%, at least 500%, or at least 1000% faster than a
growth rate and/or
higher biomass yield of a modified host cell comprising one or more nucleic
acids
comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
114
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
comprising a nucleotide sequence encoding an engineered variant, grown under
similar
culture conditions for the same length of time.
[0297] In some embodiments, the modified host cell of the disclosure for
expressing
an engineered variant of the disclosure has a growth rate and/or biomass yield
similar to, or
lower than, a growth rate and/or biomass yield of a modified host cell
comprising one or
more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
similar culture conditions for the same length of time. In some embodiments,
the modified
host cell of the disclosure expressing an engineered variant of the disclosure
has a growth
rate and/or biomass yield similar to, or lower than, a growth rate and/or
biomass yield and an
increased titer of THCA compared to a modified host cell comprising one or
more nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, grown under
similar
culture conditions for the same length of time.
[0298] In some embodiments, the modified host cell of the disclosure for
expressing
an engineered variant of the disclosure has a faster growth rate and/or higher
biomass yield
compared to a growth rate and/or higher biomass yield of a modified host cell
comprising
one or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, but lacking a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant, grown under similar culture conditions for the same length of time.
In some
embodiments, the modified host cell of the disclosure for expressing an
engineered variant
of the disclosure has a growth rate and/or higher biomass yield at least 5%,
at least 10%, at
least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at least
40%, at least 45%,
at least 50%, at least 60%, at least 70%, at least 80%, at least 90%, at least
100%, at least
150% at least 200%, at least 500%, or at least 1000% faster than a growth rate
and/or higher
biomass yield of a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant, grown under similar culture
conditions for the
same length of time.
115
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0299] In some embodiments, the modified host cell of the disclosure
comprising
one or more nucleic acids comprising a nucleotide sequence encoding an
engineered variant
of the disclosure has a growth rate and/or biomass yield similar to, or lower
than, a growth
rate and/or biomass yield of a modified host cell comprising one or more
nucleic acids
comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, grown under
similar
culture conditions for the same length of time. In some embodiments, the
modified host cell
of the disclosure comprising one or more nucleic acids comprising a nucleotide
sequence
encoding an engineered variant of the disclosure has a growth rate and/or
biomass yield
similar to, or lower than, a growth rate and/or biomass yield and an increased
titer of THCA
compared to a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant, grown under similar culture
conditions for the
same length of time.
[0300] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure has a faster growth rate and/or higher biomass yield compared
to a growth rate
and/or higher biomass yield of a modified host cell comprising one or more
nucleic acids
comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, grown under
similar
culture conditions for the same length of time. In some embodiments, the
modified host cell
of the disclosure comprising one or more nucleic acids comprising a nucleotide
sequence
encoding an engineered variant of the disclosure has a growth rate and/or
higher biomass
yield at least 5%, at least 10%, at least 15%, at least 20%, at least 25%, at
least 30%, at least
35%, at least 40%, at least 45%, at least 50%, at least 60%, at least 70%, at
least 80%, at
least 90%, at least 100%, at least 150% at least 200%, at least 500%, or at
least 1000% faster
than a growth rate and/or higher biomass yield of a modified host cell
comprising one or
more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
116
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
similar culture conditions for the same length of time. In some embodiments of
the modified
host cell of the disclosure comprising one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, the modified host
cell comprises
one or more heterologous nucleic acids comprising nucleotide sequences
encoding one or
more polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, comprises one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis.
[0301] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure and one or more heterologous nucleic acids comprising
nucleotide sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, or an IRE1 polypeptide has a faster growth rate and/or
higher biomass
yield compared to a growth rate and/or higher biomass yield of a modified host
cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 and one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, or an IRE1 polypeptide, but lacking a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant, grown under similar
culture conditions
for the same length of time. In some embodiments, a modified host cell of the
disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure and one or more heterologous nucleic
acids comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide has a growth rate
and/or
higher biomass yield at least 5%, at least 10%, at least 15%, at least 20%, at
least 25%, at
least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least
60%, at least 70%,
at least 80%, at least 90%, at least 100%, at least 150% at least 200%, at
least 500%, or at
117
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
least 1000% faster than a growth rate and/or higher biomass yield of a
modified host cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 and one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, or an IRE1 polypeptide, but lacking a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant, grown under similar
culture conditions
for the same length of time. In some embodiments of the modified host cell of
the disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure and one or more heterologous nucleic
acids comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide, the modified
host cells
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 and one or more
heterologous
nucleic acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or
an IRE1
polypeptide, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, comprises one or more heterologous nucleic acids
comprising nucleotide
sequences encoding one or more polypeptides involved in cannabinoid or
cannabinoid
precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates, olivetolic
acid, or
hexanoyl-CoA) biosynthesis.
[0302] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure and a deletion or downregulation of one or more genes encoding
one or more
of a ROT2 polypeptide or a PEP4 polypeptide has a faster growth rate and/or
higher biomass
yield compared to a growth rate and/or higher biomass yield of a modified host
cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 and a deletion or downregulation of one or more genes encoding one or
more of a
118
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
ROT2 polypeptide or a PEP4 polypeptide, but lacking a nucleic acid comprising
a nucleotide
sequence encoding an engineered variant, grown under similar culture
conditions for the
same length of time. In some embodiments, a modified host cell of the
disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure and a deletion or downregulation of one
or more genes
encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide has a growth
rate
and/or higher biomass yield at least 5%, at least 10%, at least 15%, at least
20%, at least
25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at
least 60%, at
least 70%, at least 80%, at least 90%, at least 100%, at least 150% at least
200%, at least
500%, or at least 1000% faster than a growth rate and/or higher biomass yield
of a modified
host cell comprising one or more nucleic acids comprising a nucleotide
sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 and a deletion or downregulation of one or more genes encoding one or
more of a
ROT2 polypeptide or a PEP4 polypeptide, but lacking a nucleic acid comprising
a nucleotide
sequence encoding an engineered variant, grown under similar culture
conditions for the
same length of time. In some embodiments of the modified host cell of the
disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure and a deletion or downregulation of one
or more genes
encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide, the modified
host cell
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 and a deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, comprises one or more heterologous nucleic acids
comprising nucleotide
sequences encoding one or more polypeptides involved in cannabinoid or
cannabinoid
precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates, olivetolic
acid, or
hexanoyl-CoA) biosynthesis.
[0303] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
119
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
the disclosure, one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
an IRE1 polypeptide, and a deletion or downregulation of one or more genes
encoding one
or more of a ROT2 polypeptide or a PEP4 polypeptide has a faster growth rate
and/or higher
biomass yield compared to a growth rate and/or higher biomass yield of a
modified host cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, one or more heterologous nucleic acids comprising nucleotide sequences
encoding
one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or
an IRE1
polypeptide, and a deletion or downregulation of one or more genes encoding
one or more of
a ROT2 polypeptide or a PEP4 polypeptide, but lacking a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant, grown under similar
culture conditions
for the same length of time. In some embodiments, a modified host cell of the
disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure, one or more heterologous nucleic acids
comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide has a
growth rate
and/or higher biomass yield at least 5%, at least 10%, at least 15%, at least
20%, at least
25%, at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at
least 60%, at
least 70%, at least 80%, at least 90%, at least 100%, at least 150% at least
200%, at least
500%, or at least 1000% faster than a growth rate and/or higher biomass yield
of a modified
host cell comprising one or more nucleic acids comprising a nucleotide
sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, one or more heterologous nucleic acids comprising nucleotide sequences
encoding
one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or
an IRE1
polypeptide, and a deletion or downregulation of one or more genes encoding
one or more of
a ROT2 polypeptide or a PEP4 polypeptide, but lacking a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant, grown under similar
culture conditions
for the same length of time. In some embodiments of the modified host cell of
the disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure, one or more heterologous nucleic acids
comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
120
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
ER01 polypeptide, or an IRE1 polypeptide, and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide, the
modified
host cell comprises one or more heterologous nucleic acids comprising
nucleotide sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, one or more
heterologous
nucleic acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or an IRE1 polypeptide,
and a
deletion or downregulation of one or more genes encoding one or more of a ROT2
polypeptide or a PEP4 polypeptide, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant, comprises one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis.
[0304] In some embodiments, a modified host cell of the disclosure for
producing
cannabinoids or cannabinoid derivatives produces THCA from CBGA in an
increased ratio
of THCA over another cannabinoid (e.g., CBCA) compared to that produced by a
modified
host cell comprising one or more nucleic acids comprising a nucleotide
sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, but lacking a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant, grown under similar culture conditions for the same length of time.
In some
embodiments, the modified host cell for producing cannabinoids or cannabinoid
derivatives
produces THCA from CBGA in a ratio of THCA over another cannabinoid (e.g.,
CBCA) of
about 11:1, about 11.5:1, about 12:1, about 12.5:1, about 13:1, about 13.5:1,
about 14:1,
about 14.5:1, about 15:1, about 15.5:1, about 16:1, about 16.5:1, about 17:1,
about 17.5:1,
about 18:1, about 18.5:1, about 19:1, about 19.5:1, about 20:1, about 25:1,
about 30:1, about
35:1, about 40:1, about 45:1, about 50:1, about 60:1, about 70:1, about 80:1,
about 90:1,
about 100:1, about 150:1, about 200:1, about 500:1, or greater than about
500:1.
[0305] In some embodiments, a modified host cell of the disclosure for
expressing an
engineered variant of the disclosure produces THCA from CBGA in an increased
ratio of
THCA over another cannabinoid (e.g., CBCA) compared to that produced by a
modified
121
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
host cell comprising one or more nucleic acids comprising a nucleotide
sequence encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, but lacking a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant, grown under similar culture conditions for the same length of time.
In some
embodiments, the modified host cell for expressing an engineered variant of
the disclosure
produces THCA from CBGA in a ratio of THCA over another cannabinoid (e.g.,
CBCA) of
about 11:1, about 11.5:1, about 12:1, about 12.5:1, about 13:1, about 13.5:1,
about 14:1,
about 14.5:1, about 15:1, about 15.5:1, about 16:1, about 16.5:1, about 17:1,
about 17.5:1,
about 18:1, about 18.5:1, about 19:1, about 19.5:1, about 20:1, about 25:1,
about 30:1, about
35:1, about 40:1, about 45:1, about 50:1, about 60:1, about 70:1, about 80:1,
about 90:1,
about 100:1, about 150:1, about 200:1, about 500:1, or greater than about
500:1.
[0306] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure produces THCA from CBGA in an increased ratio of THCA over
another
cannabinoid (e.g., CBCA) compared to that produced by a modified host cell
comprising one
or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
similar culture conditions for the same length of time. In some embodiments, a
modified
host cell of the disclosure comprising one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure produces THCA from
CBGA in a
ratio of THCA over another cannabinoid (e.g., CBCA) of about 11:1, about
11.5:1, about
12:1, about 12.5:1, about 13:1, about 13.5:1, about 14:1, about 14.5:1, about
15:1, about
15.5:1, about 16:1, about 16.5:1, about 17:1, about 17.5:1, about 18:1, about
18.5:1, about
19:1, about 19.5:1, about 20:1, about 25:1, about 30:1, about 35:1, about
40:1, about 45:1,
about 50:1, about 60:1, about 70:1, about 80:1, about 90:1, about 100:1, about
150:1, about
200:1, about 500:1, or greater than about 500:1. In some embodiments of the
modified host
cell of the disclosure comprising one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, the modified host
cell comprises
one or more heterologous nucleic acids comprising nucleotide sequences
encoding one or
more polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, a modified host cell comprising one or more
nucleic
122
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, comprises one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis.
[0307] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure and one or more heterologous nucleic acids comprising
nucleotide sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, or an IRE1 polypeptide produces THCA from CBGA in an
increased
ratio of THCA over another cannabinoid (e.g., CBCA) compared to that produced
by a
modified host cell comprising one or more nucleic acids comprising a
nucleotide sequence
encoding a tetrahydrocannabinolic acid synthase polypeptide having an amino
acid sequence
of SEQ ID NO:44 and one or more heterologous nucleic acids comprising
nucleotide
sequences encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an
ER01
polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide, but lacking a nucleic
acid
comprising a nucleotide sequence encoding an engineered variant, grown under
similar
culture conditions for the same length of time. In some embodiments, a
modified host cell
of the disclosure comprising one or more nucleic acids comprising a nucleotide
sequence
encoding an engineered variant of the disclosure and one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDI1
polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide
produces
THCA from CBGA in a ratio of THCA over another cannabinoid (e.g., CBCA) of
about
11:1, about 11.5:1, about 12:1, about 12.5:1, about 13:1, about 13.5:1, about
14:1, about
14.5:1, about 15:1, about 15.5:1, about 16:1, about 16.5:1, about 17:1, about
17.5:1, about
18:1, about 18.5:1, about 19:1, about 19.5:1, about 20:1, about 25:1, about
30:1, about 35:1,
about 40:1, about 45:1, about 50:1, about 60:1, about 70:1, about 80:1, about
90:1, about
100:1, about 150:1, about 200:1, about 500:1, or greater than about 500:1. In
some
embodiments of the modified host cell of the disclosure comprising one or more
nucleic
acids comprising a nucleotide sequence encoding an engineered variant of the
disclosure and
one or more heterologous nucleic acids comprising nucleotide sequences
encoding one or
more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1
123
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide, or an IRE1 polypeptide, the modified host cell comprises one or
more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis. In
some
embodiments, a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44 and one or more heterologous nucleic acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDI1
polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide,
but
lacking a nucleic acid comprising a nucleotide sequence encoding an engineered
variant,
comprises one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis.
[0308] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure and a deletion or downregulation of one or more genes encoding
one or more
of a ROT2 polypeptide or a PEP4 polypeptide produces THCA from CBGA in an
increased
ratio of THCA over another cannabinoid (e.g., CBCA) compared to that produced
by a
modified host cell comprising one or more nucleic acids comprising a
nucleotide sequence
encoding a tetrahydrocannabinolic acid synthase polypeptide having an amino
acid sequence
of SEQ ID NO :44 and a deletion or downregulation of one or more genes
encoding one or
more of a ROT2 polypeptide or a PEP4 polypeptide, but lacking a nucleic acid
comprising a
nucleotide sequence encoding an engineered variant, grown under similar
culture conditions
for the same length of time. In some embodiments, a modified host cell of the
disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure and a deletion or downregulation of one
or more genes
encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide produces THCA
from
CBGA in a ratio of THCA over another cannabinoid (e.g., CBCA) of about 11:1,
about
11.5:1, about 12:1, about 12.5:1, about 13:1, about 13.5:1, about 14:1, about
14.5:1, about
15:1, about 15.5:1, about 16:1, about 16.5:1, about 17:1, about 17.5:1, about
18:1, about
18.5:1, about 19:1, about 19.5:1, about 20:1, about 25:1, about 30:1, about
35:1, about 40:1,
about 45:1, about 50:1, about 60:1, about 70:1, about 80:1, about 90:1, about
100:1, about
124
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
150:1, about 200:1, about 500:1, or greater than about 500:1. In some
embodiments of the
modified host cell of the disclosure comprising one or more nucleic acids
comprising a
nucleotide sequence encoding an engineered variant of the disclosure and a
deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide, the modified host cell comprises one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis. In some embodiments, a
modified host cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a THCAS
polypeptide having an amino acid sequence of SEQ ID NO:44 and a deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, comprises one or more heterologous nucleic acids
comprising nucleotide
sequences encoding one or more polypeptides involved in cannabinoid or
cannabinoid
precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates, olivetolic
acid, or
hexanoyl-CoA) biosynthesis.
[0309] In some embodiments, a modified host cell of the disclosure
comprising one
or more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of
the disclosure, one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
an IRE1 polypeptide, and a deletion or downregulation of one or more genes
encoding one
or more of a ROT2 polypeptide or a PEP4 polypeptide produces THCA from CBGA in
an
increased ratio of THCA over another a cannabinoid (e.g., CBCA) compared to
that
produced by a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44, one or more heterologous nucleic acids
comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide, but
lacking a
nucleic acid comprising a nucleotide sequence encoding an engineered variant,
grown under
similar culture conditions for the same length of time. In some embodiments, a
modified
host cell of the disclosure comprising one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, one or more
heterologous nucleic
125
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDI1 polypeptide, an ER01 polypeptide, or an IRE1 polypeptide, and a deletion
or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide produces THCA from CBGA in a ratio of THCA over another
cannabinoid (e.g., CBCA) of about 11:1, about 11.5:1, about 12:1, about
12.5:1, about 13:1,
about 13.5:1, about 14:1, about 14.5:1, about 15:1, about 15.5:1, about 16:1,
about 16.5:1,
about 17:1, about 17.5:1, about 18:1, about 18.5:1, about 19:1, about 19.5:1,
about 20:1,
about 25:1, about 30:1, about 35:1, about 40:1, about 45:1, about 50:1, about
60:1, about
70:1, about 80:1, about 90:1, about 100:1, about 150:1, about 200:1, about
500:1, or greater
than about 500:1. In some embodiments of the modified host cell of the
disclosure
comprising one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure, one or more heterologous nucleic acids
comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide, the
modified
host cell comprises one or more heterologous nucleic acids comprising
nucleotide sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, a modified host cell comprising one or more
nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, one or more
heterologous
nucleic acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or an IRE1 polypeptide,
and a
deletion or downregulation of one or more genes encoding one or more of a ROT2
polypeptide or a PEP4 polypeptide, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant, comprises one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis.
[0310] In some embodiments, the growth and/or viability of modified host
cells of
the disclosure for producing cannabinoids or cannabinoid derivatives is not
significantly
decreased compared to the growth and/or viability of an unmodified host cell.
In some
embodiments, a culture of modified host cells of the disclosure for producing
cannabinoids
126
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
or cannabinoid derivatives has a cell density of at least 25% or more, at
least 30% or more,
at least 35% or more, at least 40% or more, at least 45% or more, at least 50%
or more, at
least 55% or more, at least 60% or more, at least 65% or more, at least 70% or
more, at least
75% or more, at least 80% or more, at least 85% or more at least 90% or more,
at least 95%
or more, at least 100% or more, at least 110% or more, at least 120% or more,
at least 130%
or more, at least 140% or more, or at least 150% or more compared to the cell
density of a
culture of unmodified control host cells grown for the same period, in the
same culture
medium, and under the same culture conditions.
[0311] In some embodiments, the growth and/or viability of modified host
cells of
the disclosure for expressing an engineered variant of the disclosure is not
significantly
decreased compared to the growth and/or viability of an unmodified host cell.
In some
embodiments, a culture of modified host cells of the disclosure for expressing
an engineered
variant of the disclosure has a cell density of at least 25% or more, at least
30% or more, at
least 35% or more, at least 40% or more, at least 45% or more, at least 50% or
more, at least
55% or more, at least 60% or more, at least 65% or more, at least 70% or more,
at least 75%
or more, at least 80% or more, at least 85% or more at least 90% or more, at
least 95% or
more, at least 100% or more, at least 110% or more, at least 120% or more, at
least 130% or
more, at least 140% or more, or at least 150% or more compared to the cell
density of a
culture of unmodified control host cells grown for the same period, in the
same culture
medium, and under the same culture conditions.
[0312] In some embodiments, the growth and/or viability of modified host
cells of
the disclosure comprising one or more nucleic acids comprising a nucleotide
sequence
encoding an engineered variant of the disclosure is not significantly
decreased compared to
the growth and/or viability of an unmodified host cell. In some embodiments, a
culture of
modified host cells of the disclosure comprising one or more nucleic acids
comprising a
nucleotide sequence encoding an engineered variant of the disclosure has a
cell density of at
least 25% or more, at least 30% or more, at least 35% or more, at least 40% or
more, at least
45% or more, at least 50% or more, at least 55% or more, at least 60% or more,
at least 65%
or more, at least 70% or more, at least 75% or more, at least 80% or more, at
least 85% or
more at least 90% or more, at least 95% or more, at least 100% or more, at
least 110% or
more, at least 120% or more, at least 130% or more, at least 140% or more, or
at least 150%
or more compared to the cell density of a culture of unmodified control host
cells grown for
the same period, in the same culture medium, and under the same culture
conditions. In
127
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
some embodiments of the modified host cell of the disclosure comprising one or
more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure, the modified host cell comprises one or more heterologous nucleic
acids
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis.
[0313] In some embodiments, the growth and/or viability of modified host
cells of
the disclosure comprising one or more nucleic acids comprising a nucleotide
sequence
encoding an engineered variant of the disclosure and one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDI1
polypeptide, an ER01 polypeptide, a FAD1 polypeptide, or an IRE1 polypeptide
is not
significantly decreased compared to the growth and/or viability of an
unmodified host cell.
In some embodiments, a culture of modified host cells of the disclosure
comprising one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
disclosure and one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, a
FAD1 polypeptide, or an IRE1 polypeptide has a cell density of at least 25% or
more, at
least 30% or more, at least 35% or more, at least 40% or more, at least 45% or
more, at least
50% or more, at least 55% or more, at least 60% or more, at least 65% or more,
at least 70%
or more, at least 75% or more, at least 80% or more, at least 85% or more at
least 90% or
more, at least 95% or more, at least 100% or more, at least 110% or more, at
least 120% or
more, at least 130% or more, at least 140% or more, or at least 150% or more
compared to
the cell density of a culture of unmodified control host cells grown for the
same period, in
the same culture medium, and under the same culture conditions. In some
embodiments of
the modified host cell of the disclosure comprising one or more nucleic acids
comprising a
nucleotide sequence encoding an engineered variant of the disclosure and one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more of a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, a FAD1 polypeptide,
or an
IRE1 polypeptide, the modified host cell comprises one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis.
128
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0314] In some embodiments, the growth and/or viability of modified host
cells of
the disclosure comprising one or more nucleic acids comprising a nucleotide
sequence
encoding an engineered variant of the disclosure and a deletion or
downregulation of one or
more genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide is
not
significantly decreased compared to the growth and/or viability of an
unmodified host cell.
In some embodiments, a culture of modified host cells of the disclosure
comprising one or
more nucleic acids comprising a nucleotide sequence encoding an engineered
variant of the
disclosure and a deletion or downregulation of one or more genes encoding one
or more of a
ROT2 polypeptide or a PEP4 polypeptide has a cell density of at least 25% or
more, at least
30% or more, at least 35% or more, at least 40% or more, at least 45% or more,
at least 50%
or more, at least 55% or more, at least 60% or more, at least 65% or more, at
least 70% or
more, at least 75% or more, at least 80% or more, at least 85% or more at
least 90% or more,
at least 95% or more, at least 100% or more, at least 110% or more, at least
120% or more,
at least 130% or more, at least 140% or more, or at least 150% or more
compared to the cell
density of a culture of unmodified control host cells grown for the same
period, in the same
culture medium, and under the same culture conditions. In some embodiments of
the
modified host cell of the disclosure comprising one or more nucleic acids
comprising a
nucleotide sequence encoding an engineered variant of the disclosure and a
deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide, the modified host cell comprises one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more polypeptides involved in
cannabinoid or cannabinoid precursor (e.g., geranylpyrophosphate (GPP), prenyl
phosphates,
olivetolic acid, or hexanoyl-CoA) biosynthesis.
[0315] In some embodiments, the growth and/or viability of modified host
cells of
the disclosure comprising one or more nucleic acids comprising a nucleotide
sequence
encoding an engineered variant of the disclosure, one or more heterologous
nucleic acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDI1
polypeptide, an ER01 polypeptide, or an IRE1 polypeptide, and a deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide is not significantly decreased compared to the growth and/or
viability of
an unmodified host cell. In some embodiments, a culture of modified host cells
of the
disclosure comprising one or more nucleic acids comprising a nucleotide
sequence encoding
an engineered variant of the disclosure, one or more heterologous nucleic
acids comprising
129
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and a deletion or downregulation of
one or more
genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide has a
cell density
of at least 25% or more, at least 30% or more, at least 35% or more, at least
40% or more, at
least 45% or more, at least 50% or more, at least 55% or more, at least 60% or
more, at least
65% or more, at least 70% or more, at least 75% or more, at least 80% or more,
at least 85%
or more at least 90% or more, at least 95% or more, at least 100% or more, at
least 110% or
more, at least 120% or more, at least 130% or more, at least 140% or more, or
at least 150%
or more compared to the cell density of a culture of unmodified control host
cells grown for
the same period, in the same culture medium, and under the same culture
conditions. In
some embodiments of the modified host cell of the disclosure comprising one or
more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure, one or more heterologous nucleic acids comprising nucleotide
sequences
encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01
polypeptide, or
an IRE1 polypeptide, and a deletion or downregulation of one or more genes
encoding one
or more of a ROT2 polypeptide or a PEP4 polypeptide, the modified host cell
comprises one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis.
Suitable Host Cells
[0316] Parent host cells that are suitable for use in generating a
modified host cell of
the present disclosure may include eukaryotic cells. In some embodiments, the
eukaryotic
cells are yeast cells.
[0317] Host cells (including parent host cells and modified host cells)
are in some
embodiments unicellular organisms, or are grown in culture as single cells. In
some
embodiments, the host cell is a eukaryotic cell. Suitable eukaryotic host
cells may include,
but are not limited to, yeast cells and fungal cells. Suitable eukaryotic host
cells may
include, but are not limited to, Pichia pastoris (now known as Komagataella
phaffii), Pichia
finlandica, Pichia trehalophila, Pichia koclamae, Pichia membranaefaciens,
Pichia
opuntiae, Pichia thermotolerans, Pichia salictaria, Pichia guercuum, Pichia
pijperi, Pichia
stiptis, Pichia methanol/ca, Pichia sp., Saccharomyces cerevisiae,
Saccharomyces sp.,
Hansenula polymorpha (now known as Pichia angusta), Yarrowia hpolytica,
130
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Kluyveromyces sp., Kluyveromyces lactis, Kluyveromyces marxianus,
Schizosaccharomyces
pombe, Scheffersomyces stipites, Dekkera bruxellensis, Blastobotrys
adeninivorans
(formerly Arxula adeninivorans), Candida albicans, Aspergillus nidulans,
Aspergillus niger,
Aspergillus oryzae, Trichoderma reesei, Chrysosporium lucknowense, Fusarium
sp.,
Fusarium gramineum, Fusarium venenatum, Neurospora crassa, and the like. In
some
embodiments, the modified host cell disclosed herein is cultured in vitro.
[0318] In some embodiments, the host cell of the disclosure is a yeast
cell. In some
embodiments, the host cell is a protease-deficient strain of Saccharomyces
cerevisiae.
Protease-deficient yeast strains may be effective in reducing the degradation
of expressed
heterologous proteins. Examples of proteases deleted in such strains may
include one or
more of the following: PEP4,PRB1, and KEX1.
[0319] In some embodiments, the host cell is Saccharomyces cerevisiae. In
some
embodiments, the host cell for use in generating a modified host cell of the
present
disclosure may be selected because of ease of culture; rapid growth;
availability of tools for
modification, such as promoters and vectors; and the host cell's safety
profile. In some
embodiments, the host cell for use in generating a modified host cell of the
present
disclosure may be selected because of its ability or inability to introduce
certain
posttranslational modifications onto expressed polypeptides, such as
engineered variants of
the disclosure. For instance, modified Komagataella phaffii host cells may
hyperglycosylate
engineered variants of the disclosure and hyperglycosylation may alter the
activity of the
resultant expressed polypeptide.
Genetic Modification of Host Cells and Exemplary Modified Host Cells of the
Disclosure
[0320] The present disclosure provides for modified host cells and
methods of
making modified host cells comprising one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure. In some
embodiments, the
method of making a modified host cell of the disclosure comprises introducing
into a host
cell one or more nucleic acids comprising a nucleotide sequence encoding an
engineered
variant of the disclosure. In some embodiments, the modified host cell of the
disclosure
comprises one or more nucleic acids comprising a nucleotide sequence encoding
an
engineered variant of the disclosure. In some embodiments, the nucleic acids
comprise
codon-optimized nucleotide sequences.
[0321] The present disclosure provides for modified host cells and
methods of
making modified host cells for producing a cannabinoid or a cannabinoid
derivative,
131
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
comprising introducing into a host cell one or more nucleic acids (e.g.,
heterologous)
disclosed herein. In some embodiments, the nucleic acids comprise codon-
optimized
nucleotide sequences.
[0322] The disclosure provides a method of making a modified host cell
for
producing a cannabinoid or a cannabinoid derivative, the method comprising a)
introducing
into a host cell one or more nucleic acids comprising a nucleotide sequence
encoding an
engineered variant of the disclosure. In certain such embodiments, the method
comprises b)
introducing into the host cell one or more heterologous nucleic acids
comprising nucleotide
sequences encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an
ER01
polypeptide, or an IRE1 polypeptide. In some embodiments, the method comprises
b)
introducing into the host cell one or more heterologous nucleic acids
comprising nucleotide
sequences encoding one or more of a KAR2 polypeptide, a PDI1 polypeptide, an
ER01
polypeptide, or a FAD1 polypeptide. In some embodiments, the modified host
cell for
producing a cannabinoid or a cannabinoid derivative comprises one or more
nucleic acids
comprising a codon-optimized nucleotide sequence encoding an engineered
variant of the
disclosure.
[0323] In some embodiments, the modified host cell for producing a
cannabinoid or
a cannabinoid derivative comprises one or more heterologous nucleic acids
comprising
nucleotide sequences encoding an engineered variant of the disclosure and one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more of a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or an IRE1
polypeptide. In
certain such embodiments, the modified host cell comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding the KAR2 polypeptide,
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
PDI1
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the ER01 polypeptide, and one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the IRE1 polypeptide. In some embodiments, the
modified
host cell for producing a cannabinoid or a cannabinoid derivative comprises
two or more
heterologous nucleic acids comprising a nucleotide sequence encoding a KAR2
polypeptide.
[0324] In some embodiments, the modified host cell for producing a
cannabinoid or
a cannabinoid derivative comprises one or more heterologous nucleic acids
comprising
nucleotide sequences encoding an engineered variant of the disclosure and one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more of a
132
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide or a FAD1
polypeptide. In
certain such embodiments, the modified host cell comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding the KAR2 polypeptide,
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
PDI1
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the ER01 polypeptide, and one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the FAD1 polypeptide. In some embodiments, the
modified
host cell for producing a cannabinoid or a cannabinoid derivative comprises
two or more
heterologous nucleic acids comprising a nucleotide sequence encoding a KAR2
polypeptide.
[0325] In some embodiments, the modified host cell for producing a
cannabinoid or
a cannabinoid derivative comprises one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure and a deletion or
downregulation
of one or more genes encoding one or more of ROT2 polypeptide or a PEP4
polypeptide. In
certain such embodiments, the modified host cell comprises a deletion or
downregulation of
one or more genes encoding the ROT2 polypeptide and the PEP4 polypeptide. The
disclosure provides a method of making a modified host cell for producing a
cannabinoid or
a cannabinoid derivative, the method comprising introducing into a host cell
one or more
nucleic acids comprising a nucleotide sequence encoding an engineered variant
of the
disclosure and a deletion or downregulation of one or more genes encoding one
or more of a
ROT2 polypeptide or a PEP4 polypeptide.
[0326] In some embodiments, the modified host cell for producing a
cannabinoid or
a cannabinoid derivative comprises one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, one or more
heterologous nucleic
acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDI1 polypeptide, an ER01 polypeptide, or an IRE1 polypeptide, and a deletion
or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide. In certain such embodiments, the modified host cell
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
KAR2
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the PDI1 polypeptide, one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the ER01 polypeptide, and one or more
heterologous nucleic
acids comprising a nucleotide sequence encoding the IRE1 polypeptide and a
deletion or
downregulation of one or more genes encoding the ROT2 polypeptide and the PEP4
133
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
polypeptide. In some embodiments, the modified host cell for producing a
cannabinoid or a
cannabinoid derivative comprises two or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a KAR2 polypeptide.
[0327] The disclosure provides a method of making a modified host cell
for
producing a cannabinoid or a cannabinoid derivative, the method comprising
introducing
into a host cell: a) one or more nucleic acids comprising a nucleotide
sequence encoding an
engineered variant of the disclosure, b) one or more heterologous nucleic
acids comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and c) a deletion or downregulation
of one or
more genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide.
[0328] The disclosure provides a method of making a modified host cell
for
producing a cannabinoid or a cannabinoid derivative, the method comprising
introducing
into a host cell: a) one or more nucleic acids comprising a nucleotide
sequence encoding an
engineered variant of the disclosure and b) one or more heterologous nucleic
acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDI1
polypeptide, an ER01 polypeptide, or a FAD1 polypeptide.
[0329] In some embodiments, the modified host cell for producing a
cannabinoid or
a cannabinoid derivative may comprise one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure and express or
overexpress
combinations of heterologous nucleic acids comprising nucleotide sequences
encoding one
or more polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, the methods of making a modified host cell
for
producing a cannabinoid or a cannabinoid derivative comprise introducing into
a host cell
one or more heterologous nucleic acids comprising nucleotide sequences
encoding one or
more polypeptides involved in cannabinoid or cannabinoid precursor
biosynthesis.
[0330] In some embodiments, the modified host cell for producing a
cannabinoid or
a cannabinoid derivative comprises one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, one or more
heterologous nucleic
acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDI1 polypeptide, an ER01 polypeptide, or an IRE1 polypeptide, a deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide, and one or more heterologous nucleic acids comprising
nucleotide
134
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
sequences encoding one or more polypeptides involved in cannabinoid or
cannabinoid
precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates, olivetolic
acid, or
hexanoyl-CoA) biosynthesis. In certain such embodiments, the modified host
cell comprises
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding the
KAR2 polypeptide, one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding the PDI1 polypeptide, one or more heterologous nucleic acids
comprising a nucleotide sequence encoding the ER01 polypeptide, one or more
heterologous nucleic acids comprising a nucleotide sequence encoding the IRE1
polypeptide
and a deletion or downregulation of the genes encoding the ROT2 polypeptide
and the PEP4
polypeptide. In some embodiments, the modified host cell for producing a
cannabinoid or a
cannabinoid derivative comprises two or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a KAR2 polypeptide.
[0331] In some embodiments, the modified host cell for producing a
cannabinoid or
a cannabinoid derivative comprises one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, one or more
heterologous nucleic
acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDI1 polypeptide, an ER01 polypeptide, or a FAD1 polypeptide, and one or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis. In
certain such
embodiments, the modified host cell comprises one or more heterologous nucleic
acids
comprising a nucleotide sequence encoding the KAR2 polypeptide, one or more
heterologous nucleic acids comprising a nucleotide sequence encoding the PDI1
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the ER01 polypeptide, and one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the FAD1 polypeptide. In some embodiments, the
modified
host cell for producing a cannabinoid or a cannabinoid derivative comprises
two or more
heterologous nucleic acids comprising a nucleotide sequence encoding a KAR2
polypeptide.
[0332] The present disclosure provides for a method of making a modified
host cell
for expressing an engineered variant of the disclosure, the method comprising
introducing
into a host cell one or more nucleic acids disclosed herein. The disclosure
provides a
method of making a modified host cell for expressing an engineered variant of
the
disclosure, the method comprising introducing into a host cell: a) one or more
nucleic acids
135
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
comprising a nucleotide sequence encoding an engineered variant of the
disclosure and b)
one or more heterologous nucleic acids comprising nucleotide sequences
encoding one or
more of a KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or an
IRE1
polypeptide. The disclosure provides a method of making a modified host cell
for
expressing an engineered variant of the disclosure, the method comprising
introducing into a
host cell: a) one or more nucleic acids comprising a nucleotide sequence
encoding an
engineered variant of the disclosure and b) one or more heterologous nucleic
acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDI1
polypeptide, an ER01 polypeptide, or a FAD1 polypeptide. In some embodiments,
the
modified host cell for expressing an engineered variant of the disclosure
comprises one or
more nucleic acids comprising a codon-optimized nucleotide sequence encoding
the
engineered variant of the disclosure.
[0333] In some
embodiments, the modified host cell for expressing an engineered
variant of the disclosure comprises one or more nucleic acids comprising a
nucleotide
sequence encoding the engineered variant of the disclosure and comprises one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more of a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or an IRE1
polypeptide. In
certain such embodiments, the modified host cell comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding the KAR2 polypeptide,
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
PDI1
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the ER01 polypeptide, and one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the IRE1 polypeptide. In some embodiments, the
modified
host cell for expressing an engineered variant of the disclosure comprises two
or more
heterologous nucleic acids comprising a nucleotide sequence encoding a KAR2
polypeptide.
[0334] In some
embodiments, the modified host cell for expressing an engineered
variant of the disclosure comprises one or more nucleic acids comprising a
nucleotide
sequence encoding the engineered variant of the disclosure and comprises one
or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more of a
KAR2 polypeptide, a PDI1 polypeptide, an ER01 polypeptide, or a FAD1
polypeptide. In
certain such embodiments, the modified host cell comprises one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding the KAR2 polypeptide,
one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
PDI1
136
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the ER01 polypeptide, and one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the FAD1 polypeptide. In some embodiments, the
modified
host cell for expressing an engineered variant of the disclosure comprises two
or more
heterologous nucleic acids comprising a nucleotide sequence encoding a KAR2
polypeptide.
[0335] In some
embodiments, the modified host cell for expressing an engineered
variant of the disclosure comprising one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure comprises a deletion
or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide. In certain such embodiments, the modified host cell
comprises a deletion
or downregulation of one or more genes encoding the ROT2 polypeptide and the
PEP4
polypeptide. The disclosure provides a method of making a modified host cell
for expressing
an engineered variant of the disclosure, the method comprising introducing
into a host cell:
a) one or more nucleic acids comprising a nucleotide sequence encoding an
engineered
variant of the disclosure and b) a deletion or downregulation of one or more
genes encoding
one or more of a ROT2 polypeptide or a PEP4 polypeptide.
[0336] In some
embodiments, the modified host cell for expressing an engineered
variant of the disclosure comprises one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, one or more
heterologous nucleic
acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDI1 polypeptide, an ER01 polypeptide, or an IRE1 polypeptide, and a deletion
or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide. In certain such embodiments, the modified host cell
comprises one or
more heterologous nucleic acids comprising a nucleotide sequence encoding the
KAR2
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the PDI1 polypeptide, one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the ER01 polypeptide, and one or more
heterologous nucleic
acids comprising a nucleotide sequence encoding the IRE1 polypeptide and a
deletion or
downregulation of one or more genes encoding the ROT2 polypeptide and the PEP4
polypeptide. In some embodiments, the modified host cell for expressing an
engineered
variant of the disclosure comprises two or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a KAR2 polypeptide.
137
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0337] The disclosure provides a method of making a modified host cell
for
expressing an engineered variant of the disclosure, the method comprising
introducing into a
host cell: a) one or more nucleic acids comprising a nucleotide sequence
encoding an
engineered variant of the disclosure, b) one or more heterologous nucleic
acids comprising
nucleotide sequences encoding one or more of a KAR2 polypeptide, a PDI1
polypeptide, an
ER01 polypeptide, or an IRE1 polypeptide, and c) a deletion or downregulation
of one or
more genes encoding one or more of a ROT2 polypeptide or a PEP4 polypeptide.
[0338] The disclosure provides a method of making a modified host cell
for
expressing an engineered variant of the disclosure, the method comprising
introducing into a
host cell: a) one or more nucleic acids comprising a nucleotide sequence
encoding an
engineered variant of the disclosure and b) one or more heterologous nucleic
acids
comprising nucleotide sequences encoding one or more of a KAR2 polypeptide, a
PDT]
polypeptide, an ER01 polypeptide, or a FAD1 polypeptide.
[0339] In some embodiments, the modified host cell for expressing an
engineered
variant of the disclosure may comprise one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure and express or
overexpress
combinations of heterologous nucleic acids comprising nucleotide sequences
encoding one
or more polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate (GPP), prenyl phosphates, olivetolic acid, or hexanoyl-
CoA)
biosynthesis. In some embodiments, the methods of making a modified host cell
for
expressing an engineered variant of the disclosure comprise introducing into a
host cell one
or more heterologous nucleic acids comprising nucleotide sequences encoding
one or more
polypeptides involved in cannabinoid or cannabinoid precursor biosynthesis.
[0340] In some embodiments, the modified host cell for expressing an
engineered
variant of the disclosure comprises one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, one or more
heterologous nucleic
acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDT] polypeptide, an ER01 polypeptide, or an IRE1 polypeptide, a deletion or
downregulation of one or more genes encoding one or more of a ROT2 polypeptide
or a
PEP4 polypeptide, and one or more heterologous nucleic acids comprising
nucleotide
sequences encoding one or more polypeptides involved in cannabinoid or
cannabinoid
precursor (e.g., geranylpyrophosphate (GPP), prenyl phosphates, olivetolic
acid, or
hexanoyl-CoA) biosynthesis. In certain such embodiments, the modified host
cell comprises
138
CA 03152803 2022-02-25
WO 2021/055597
PCT/US2020/051261
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding the
KAR2 polypeptide, one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding the PDI1 polypeptide, one or more heterologous nucleic acids
comprising a nucleotide sequence encoding the ER01 polypeptide, one or more
heterologous nucleic acids comprising a nucleotide sequence encoding the IRE1
polypeptide
and a deletion or downregulation of the genes encoding the ROT2 polypeptide
and the PEP4
polypeptide. In some embodiments, the modified host cell for expressing an
engineered
variant of the disclosure comprises two or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a KAR2 polypeptide.
[0341] In some
embodiments, the modified host cell for expressing an engineered
variant of the disclosure comprises one or more nucleic acids comprising a
nucleotide
sequence encoding an engineered variant of the disclosure, one or more
heterologous nucleic
acids comprising nucleotide sequences encoding one or more of a KAR2
polypeptide, a
PDI1 polypeptide, an ER01 polypeptide, or a FAD1 polypeptide and one or more
heterologous nucleic acids comprising nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor (e.g.,
geranylpyrophosphate
(GPP), prenyl phosphates, olivetolic acid, or hexanoyl-CoA) biosynthesis. In
certain such
embodiments, the modified host cell comprises one or more heterologous nucleic
acids
comprising a nucleotide sequence encoding the KAR2 polypeptide, one or more
heterologous nucleic acids comprising a nucleotide sequence encoding the PDI1
polypeptide, one or more heterologous nucleic acids comprising a nucleotide
sequence
encoding the ER01 polypeptide, one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding the FAD1 polypeptide. In some embodiments, the
modified
host cell for expressing an engineered variant of the disclosure comprises two
or more
heterologous nucleic acids comprising a nucleotide sequence encoding a KAR2
polypeptide.
[0342] To
modify a parent host cell to produce a modified host cell of the present
disclosure, one or more nucleic acids (e.g., heterologous) disclosed herein
may be introduced
stably or transiently into a host cell, using established techniques. Such
techniques may
include, but are not limited to, electroporation, calcium phosphate
precipitation, DEAE-
dextran mediated transfection, liposome-mediated transfection, the lithium
acetate method,
and the like. See Gietz, R.D. and R.A. Woods. (2002) TRANSFORMATION OF YEAST
BY THE Liac/SS CARRIER DNA/PEG METHOD. For stable transformation, a nucleic
acid
(e.g., heterologous) will generally include a selectable marker, e.g., any of
several well-
139
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
known selectable markers such as neomycin resistance, ampicillin resistance,
tetracycline
resistance, chloramphenicol resistance, kanamycin resistance, and the like. In
some
embodiments, a parent host cell is modified to produce a modified host cell of
the present
disclosure using a CRISPR/Cas9 system to modify a parent host cell with one or
more
nucleic acids (e.g., heterologous) disclosed herein.
[0343] In some embodiments, varying engineered variant expression level
and/or the
production of cannabinoids or cannabinoid derivatives in a modified host cell
may be done
by changing the gene copy number, promoter strength, and/or promoter
regulation.
[0344] One or more nucleic acids (e.g., heterologous) disclosed herein
can be present
in an expression vector or construct. Suitable expression vectors may include,
but are not
limited to, plasmids, yeast plasmids, yeast artificial chromosomes, and any
other vectors
specific for specific hosts of interest (such as yeast). Thus, for example,
one or more nucleic
acids (e.g., heterologous) comprising nucleotide sequences encoding a
mevalonate pathway
gene product(s) is included in any one of a variety of expression vectors for
expressing the
mevalonate pathway gene product(s). Such vectors may include chromosomal, non-
chromosomal, and synthetic DNA sequences.
[0345] The present disclosure provides for a method of making a modified
host cell
for producing a cannabinoid or a cannabinoid derivative, the method comprising
introducing
into a host cell one or more vectors disclosed herein. In certain such
embodiments, the one
or more vectors comprise one or more vectors comprising one or more nucleic
acids (e.g.,
heterologous) comprising a nucleotide sequence encoding an engineered variant
of the
disclosure. In certain such embodiments, the one or more vectors comprise one
or more
vectors comprising one or more nucleic acids (e.g., heterologous) comprising
nucleotide
sequences encoding one or more secretory pathway polypeptides. In some
embodiments, the
method comprises introducing into the host cell a deletion or downregulation
of one or more
genes encoding one or more secretory pathway polypeptides. In some
embodiments, the
nucleotide sequences encoding one or more secretory pathway polypeptides are
codon-
optimized. In some embodiments, the one or more vectors comprise one or more
vectors
comprising one or more nucleic acids (e.g., heterologous) comprising
nucleotide sequences
encoding one or more polypeptides involved in cannabinoid or cannabinoid
precursor
biosynthesis. In some embodiments, the nucleotide sequences encoding one or
more
polypeptides involved in cannabinoid or cannabinoid precursor biosynthesis are
codon-
optimized.
140
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0346] The present disclosure provides for a method of making a modified
host cell
for expressing a cannabinoid synthase polypeptide, the method comprising
introducing into a
host cell one or more vectors disclosed herein. In certain such embodiments,
the one or
more vectors comprise one or more vectors comprising one or more nucleic acids
(e.g.,
heterologous) comprising a nucleotide sequence encoding an engineered variant
of the
disclosure. In certain such embodiments, the one or more vectors comprise one
or more
vectors comprising one or more nucleic acids (e.g., heterologous) comprising
nucleotide
sequences encoding one or more secretory pathway polypeptides. In some
embodiments, the
nucleotide sequences encoding one or more secretory pathway polypeptides are
codon-
optimized. In some embodiments, the method comprises introducing into the host
cell a
deletion or downregulation of one or more genes encoding one or more secretory
pathway
polypeptides.
[0347] Numerous additional suitable expression vectors are known to those
of skill
in the art, and many are commercially available. The following vectors are
provided by way
of example; for yeast, the low copy CEN ARS and high copy 2 micron plasmids.
However,
any other plasmid or other vector may be used so long as it is compatible with
the host cell.
[0348] In some embodiments, one or more of the nucleic acids (e.g.,
heterologous)
disclosed herein are present in a single expression vector. In some
embodiments, two or
more of the nucleic acids (e.g., heterologous) disclosed herein are present in
a single
expression vector. In some embodiments, three or more of the nucleic acids
(e.g.,
heterologous) disclosed herein are present in a single expression vector. In
some
embodiments, four or more of the nucleic acids (e.g., heterologous) disclosed
herein are
present in a single expression vector. In some embodiments, five or more of
the nucleic
acids (e.g., heterologous) disclosed herein are present in a single expression
vector. In some
embodiments, six or more of the nucleic acids (e.g., heterologous) disclosed
herein are
present in a single expression vector. In some embodiments, seven or more of
the nucleic
acids (e.g., heterologous) disclosed herein are present in a single expression
vector.
[0349] In some embodiments, two or more nucleic acids (e.g.,
heterologous)
disclosed herein are in separate expression vectors. In some embodiments,
three or more
nucleic acids (e.g., heterologous) disclosed herein are in separate expression
vectors. In
some embodiments, four or more nucleic acids (e.g., heterologous) disclosed
herein are in
separate expression vectors. In some embodiments, five or more nucleic acids
(e.g.,
heterologous) disclosed herein are in separate expression vectors. In some
embodiments, six
141
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
or more nucleic acids (e.g., heterologous) disclosed herein are in separate
expression vectors.
In some embodiments, seven or more nucleic acids (e.g., heterologous)
disclosed herein are
in separate expression vectors. In some embodiments, eight or more nucleic
acids (e.g.,
heterologous) disclosed herein are in separate expression vectors. In some
embodiments,
nine or more nucleic acids (e.g., heterologous) disclosed herein are in
separate expression
vectors. In some embodiments, ten or more nucleic acids (e.g., heterologous)
disclosed
herein are in separate expression vectors.
[0350] In some embodiments, one or more of the nucleic acids (e.g.,
heterologous)
disclosed herein are present in a single expression construct. In some
embodiments, two or
more of the nucleic acids (e.g., heterologous) disclosed herein are present in
a single
expression construct. In some embodiments, three or more of the nucleic acids
(e.g.,
heterologous) disclosed herein are present in a single expression construct.
In some
embodiments, four or more of the nucleic acids (e.g., heterologous) disclosed
herein are
present in a single expression construct. In some embodiments, five or more of
the nucleic
acids (e.g., heterologous) disclosed herein are present in a single expression
construct. In
some embodiments, six or more of the nucleic acids (e.g., heterologous)
disclosed herein are
present in a single expression construct. In some embodiments, seven or more
of the nucleic
acids (e.g., heterologous) disclosed herein are present in a single expression
construct.
[0351] In some embodiments, two or more nucleic acids (e.g.,
heterologous)
disclosed herein are in separate expression constructs. In some embodiments,
three or more
nucleic acids (e.g., heterologous) disclosed herein are in separate expression
constructs. In
some embodiments, four or more nucleic acids (e.g., heterologous) disclosed
herein are in
separate expression constructs. In some embodiments, five or more nucleic
acids (e.g.,
heterologous) disclosed herein are in separate expression constructs. In some
embodiments,
six or more nucleic acids (e.g., heterologous) disclosed herein are in
separate expression
constructs. In some embodiments, seven or more nucleic acids (e.g.,
heterologous) disclosed
herein are in separate expression constructs. In some embodiments, eight or
more nucleic
acids (e.g., heterologous) disclosed herein are in separate expression
constructs. In some
embodiments, nine or more nucleic acids (e.g., heterologous) disclosed herein
are in separate
expression constructs. In some embodiments, ten or more nucleic acids (e.g.,
heterologous)
disclosed herein are in separate expression constructs.
[0352] In some embodiments, one or more of the nucleic acids (e.g.,
heterologous)
disclosed herein is present in a high copy number plasmid, e.g., a plasmid
that exists in about
142
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
10-50 copies per cell, or more than 50 copies per cell. In some embodiments,
one or more of
the nucleic acids (e.g., heterologous) disclosed herein is present in a low
copy number
plasmid. In some embodiments, one or more of the nucleic acids (e.g.,
heterologous)
disclosed herein is present in a medium copy number plasmid. The copy number
of the
plasmid may be selected to reduce expression of one or more polypeptides
disclosed herein,
such as an engineered variant of the disclosure. Reducing expression by
limiting the copy
number of the plasmid may prevent saturation of the secretory pathway leading
to possible
protein degradation and/or modified host cell death or a loss of modified host
cell viability.
[0353] In some embodiments, the modified host cell has one copy of a
nucleic acid
(e.g., heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed
herein. In some embodiments, the modified host cell has two copies of a
nucleic acid (e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has three copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has four copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has five copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has six copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has seven copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has eight copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has nine copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has ten copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has eleven copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
some embodiments, the modified host cell has twelve copies of a nucleic acid
(e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein. In
143
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
some embodiments, the modified host cell has twelve or more copies of a
nucleic acid (e.g.,
heterologous) comprising a nucleotide sequence encoding a polypeptide
disclosed herein.
[0354] Depending on the host/vector or host/construct system utilized,
any of a
number of suitable transcription and translation control elements, including
constitutive and
inducible promoters, transcription enhancer elements, transcription
terminators, etc. may be
used in the expression vector or construct (see e.g., Bitter et al.
(1987)Methods in
Enzymology, 153:516-544).
[0355] In some embodiments, the nucleic acids (e.g., heterologous)
disclosed herein
are operably linked to a promoter. In some embodiments, the promoter is a
constitutive
promoter. In some embodiments, the promoter is an inducible promoter. In some
embodiments, the promoter is functional in a eukaryotic cell. In some
embodiments, the
promoter can be a strong driver of expression. In some embodiments, the
promoter can be a
weak driver of expression. In some embodiments, the promoter can be a medium
driver of
expression. The promoter may be selected to reduce expression of one or more
polypeptides
disclosed herein, such as an engineered variant of the disclosure. Reducing
expression
through promoter selection may prevent saturation of the secretory pathway
leading to
possible protein degradation and/or modified host cell death or a loss of
modified host cell
viability. Examples of strong constitutive promoters include, but are not
limited to: pTDH3
and pFBAl. Examples of medium constitutive promoters include, but are not
limited to:
pACT1 and pCYCl. An example of a weak constitutive promoter includes, but is
not limited
to: pSLN1. Examples of strong inducible promoters include, but are not limited
to: pGAL1
and pGAL10. An example of a medium inducible promoter includes, but is not
limited to:
pGAL7. An example of a weak inducible promoter includes, but is not limited
to: pGAL3.
[0356] Non-limiting examples of suitable eukaryotic promoters may include
CMV
immediate early, HSV thymidine kinase, early and late SV40, LTRs from
retrovirus, and
mouse metallothionein-I. Selection of the appropriate vector, construct, and
promoter is well
within the level of ordinary skill in the art. The expression vector or
construct may also
contain a ribosome binding site for translation initiation and a transcription
terminator. The
expression vector or construct may also include appropriate sequences for
amplifying
expression.
[0357] In yeast, a number of vectors or constructs containing
constitutive or
inducible promoters may be used. For a review see, Current Protocols in
Molecular Biology,
Vol. 2, 1988, Ed. Ausubel, et al., Greene Publish. Assoc. & Wiley
Interscience, Ch. 13;
144
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Grant, et al., 1987, Expression and Secretion Vectors for Yeast, in Methods in
Enzymology,
Eds. Wu & Grossman, 31987, Acad. Press, N.Y., Vol. 153, pp.516-544; Glover,
1986, DNA
Cloning, Vol. II, IRL Press, Wash., D.C., Ch. 3; and Bitter, 1987,
Heterologous Gene
Expression in Yeast, Methods in Enzymology, Eds. Berger & Kimmel, Acad. Press,
N.Y.,
Vol. 152, pp. 673-684; and The Molecular Biology of the Yeast Saccharomyces,
1982, Eds.
Strathern et al., Cold Spring Harbor Press, Vols. I and II. A constitutive
yeast promoter such
as pADH, pTDH3, pFBA1, pACT1, pCYCL and pSLN1 or an inducible promoter such as
pGAL1, pGAL10, pGAL7, and pGAL3 may be used (Cloning in Yeast, Ch. 3, R.
Rothstein
In: DNA Cloning Vol. 11, A Practical Approach, Ed. DM Glover, 1986, IRL Press,
Wash.,
D.C.). Alternatively, vectors may be used which promote integration of foreign
DNA
sequences into the yeast chromosome.
[0358] Generally, recombinant expression vectors will include origins of
replication
and selectable markers permitting transformation of the host cell, e.g., the
S. cerevisiae
TRP1 gene or a gene cassette encoding resistance to an antibiotic, etc.; and a
promoter
derived from a highly-expressed gene to direct transcription of the coding
sequence. Such
promoters can be derived from genetic sequences encoding glycolytic enzymes
such as 3-
phosphoglycerate kinase (PGK), a-factor, acid phosphatase, or heat shock
proteins, among
others.
[0359] Inducible promoters are well known in the art. Suitable inducible
promoters
may include, but are not limited to, a tetracycline-inducible promoter; an
estradiol inducible
promoter, a sugar inducible promoter, e.g, pGall or pSUC2, an amino acid
inducible
promoter, e.g. pMet25; a metal inducible promoter, e.g. pCupl, a methanol-
inducible
promoter, e.g. pA0X1, and the like.
[0360] In addition, the expression vectors or constructs will in many
embodiments
contain one or more selectable marker genes to provide a phenotypic trait for
selection of
transformed host cells such as dihydrofolate reductase or neomycin resistance
for eukaryotic
cell culture.
[0361] In some embodiments, one or more nucleic acids (e.g.,
heterologous)
disclosed herein is integrated into the genome of the modified host cell
disclosed herein. In
some embodiments, one or more nucleic acids (e.g., heterologous) disclosed
herein is
integrated into a chromosome of the modified host cell disclosed herein. In
some
embodiments, one or more nucleic acids (e.g., heterologous) disclosed herein
remains
episomal (i.e., is not integrated into the genome or a chromosome of the
modified host cell).
145
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
In some embodiments, at least one of the one or more nucleic acids (e.g.,
heterologous)
disclosed herein is maintained extrachromosomally. The gene copy number of one
or more
genes encoding one or more polypeptides disclosed herein, such as an
engineered variant of
the disclosure, may be selected to reduce expression of the one or more
polypeptides
disclosed herein, such as an engineered variant of the disclosure. Reducing
expression by
limiting the gene copy number may prevent saturation of the secretory pathway
leading to
possible protein degradation and/or modified host cell death or a loss of
modified host cell
viability.
[0362] As will be appreciated by the skilled artisan, slight changes in
nucleotide
sequence do not necessarily alter the amino acid sequence of the encoded
polypeptide. It will
be appreciated by persons skilled in the art that changes in the identities of
nucleotides in a
specific gene sequence that change the amino acid sequence of the encoded
polypeptide may
result in reduced or enhanced effectiveness of the genes and that, in some
applications (e.g.,
anti-sense, co-suppression, or RNAi), partial sequences often work as
effectively as full
length versions. The ways in which the nucleotide sequence can be varied or
shortened are
well known to persons skilled in the art, as are ways of testing the
effectiveness of the
altered genes. In certain embodiments, effectiveness may easily be tested by,
for example,
conventional gas chromatography. All such variations of the genes are
therefore included as
part of the present disclosure.
[0363] Genomic deletion of the open reading frame encoding the protein
may abolish
all expression of a gene. Downregulation of a gene can be accomplished in
several ways at
the DNA, RNA, or protein level, with the result being a reduction in the
amount of active
protein in the cell. Truncations of the open reading frame or the introduction
of mutations
that destabilize the protein or reduce catalytic activity achieve a similar
goal, as does fusing
a "degron" polypeptide that destabilizes the protein. Engineering of the
regulatory regions of
the gene can also be used to change gene expression. Alteration of the
promoter sequence or
replacement with a different promoter is one method. Truncation of the
terminator, known as
decreased abundance of mRNA perturbation (DAmP) is also known to reduce gene
expression. Other methods that reduce the stability of the mRNA include the
use of cis- or
trans-acting ribozymes, e.g., self-cleaving ribozymes, or RNA elements that
recruite an
exonuclease, or antisense DNA. RNAi may be used to silence genes in budding
yeast strains
via import of the required protein factors from other species, e.g., Drosha or
Dice
(Drinnenberg et al 2009). Gene expression may also be silenced in S.
cerevisiae via
146
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
recruitment of native or heterologous silencing factors or repressors, which
may be
accomplished at arbitrary loci using the D-Cas9 CRISPR system (Qi et al 2013).
Protein
level can also be reduced by engineering the amino acid sequence of the target
protein. A
variety of degron sequences may be used to target the protein for rapid
degradation,
including, but not limited to, ubiquitin fusions and N-end rule residues at
the amino
terminus. These methods may be implemented in a constitutive or conditional
fashion.
Induction Systems
[0364] To adapt to a constantly changing environment, microbes such as
yeast have
evolved a wide range of natural inducible promoter systems. Any promoter that
is regulated
by a small molecule or change in environment (temperature, pH, oxygen level,
osmolarity,
oxidative damage) can in principle be converted into an inducible system for
the expression
of heterologous genes. The best known system in S. cerevisiae is the galactose
regulon,
which is strongly repressed by glucose and activated by galactose.
Heterologous genetic
pathways under the control of galactose-inducible promoters are regulated in
the same way,
and thus an engineered strain can be grown in glucose media to build biomass,
and then
switched to galactose to induce pathway expression. A range of expression
levels can be
achieved, from very strong pGAL1 to relatively weak pGAL3. However, galactose
may be
expensive and a poor carbon source for S. cerevisiae. Therefore, for
industrial applications, it
may be advantageous to re-engineer the regulon such that the cells can be
induced in a non-
galactose media. The galactose regulon can be modified for this purpose in
many ways,
including:
= Overexpressing the negative regulator of GAL80, GAL3, from an inducible
promoter, e.g. pSUC2-GAL3, such that switching from glucose to sucrose
relieves GAL80 expression and activates the pathway.
= Deleting the repressor GAL80 and replacing the native GAL4 cassette with
a
version under the control of a sucrose inducible promoter, e.g. pSUC2-GAL4,
such that expression is induced by a switch from glucose to sucrose.
= Replacing the native GAL80 gene with an inducible version, e.g. pSUC2-
GAL80, such that expression is induced by a switch from sucrose to glucose.
[0365] These strategies often require fine-tuning of the activator and
repressor levels
to achieve the proper dynamics (very low or no expression in the off state,
and desired
expression level in the on state). There are a variety of ways to fine tune
protein expression,
147
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
including use of protein stabilization or degradation tags (e.g. degrons) or
use of temperature
sensitive mutants of the activators or regulators. In the examples above, the
pSUC2 promoter
is used to induce the galactose regulon in sucrose media. However, any
inducible promoter
can be used for this purpose, or for control of individual genes outside of
the context of the
galactose regulon. The list below provides some examples:
= Phosphate regulated promoters, e.g. pPHO5
= Carbon source regulated promoters, e.g. pADH2
= Amino acid regulated promoters, e.g. pMET25
= Metal ion induced promoters, e.g. pCUP1
= Temperature regulated promoters, e.g. pHSP12, pHSP26
= pH regulated promoters, e.g. pHSP12, pHSP26
= Oxygen level regulated promoters, e.g. pDAN1
= Oxidative stress regulated promoters, e.g. promoters from AHP1, TRR1,
TRX2, TSAI, GPX2, GSH1, GSH2, GLR1, SOD1, or SOD2 genes.
= ER stress regulated promoters, e.g. unfolded protein response element
promoters.
[0366] In addition to these natural examples, there are a variety of
synthetic
inducible promoter systems. These are generally based on re-arrangement of
native or
foreign transcriptional elements into a basal promoter scaffold and/or fusions
of activator
domains and DNA binding domains to create novel transcription factors. Two
examples are
provided below:
= Estradiol-inducible systems involving fusion of the estradiol receptor to
DNA-binding and transcriptional activation domain, paired with synthetic or
native promoters with binding sites.
= tet Trans Activator (tTA) or reverse tet Trans Activator (rtTA) systems
paired
with tet0-containing promoters.
[0367] In some embodiments, one of the above inducible promoter systems
is used in
a modified host cell of the disclosure. In some embodiments, the inducible
promoter system
is a natural inducible promoter system. In some embodiments, the inducible
promoter system
is a synthetic inducible promoter system. In some embodiments, a suitable
media for
culturing modified host cells of the disclosure comprises one or more of the
inducers
disclosed herein. Possible inducers include:
= Phosphate regulated promoters, e.g. pPHO5
148
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
o KH2PO4
= Carbon source regulated promoters, e.g. pADH2
o Galactose (e.g. pGAL1)
o Glucose (e.g. pADH2)
o Sucrose (e.g. pSUC2, pGPH1, pMAL12)
o Maltose (e.g. pMAL12, pMAL32)
= Amino acid regulated promoters, e.g. pMET25
o Methionine (e.g. pMET25)
o Lysine (e.g. pLYS9)
o Other amino acids
= Metal ion induced promoters, e.g. pCUP1
o CuSO4
= Temperature regulated promoters, e.g. pHSP12, pHSP26
o Change in temperature, e.g. 30 C to 37 C
= pH regulated promoters, e.g. pHSP12, pHSP26
o Change in pH, e.g. pH 6 to pH 4
= Oxygen level regulated promoters, e.g. pDAN1
o Change in oxygen level, e.g. 20% to 1% dissolved oxygen levels
= Oxidative stress regulated promoters, e.g. pSOD1
o Addition of hydrogen peroxide or superoxide-generating drug menadione
= ER stress regulated promoters, e.g. unfolded protein response element
promoters.
o Tunicamycin, or expression of proteins prone to misfolding (e.g.,
cannabinoid
synthases)
= Estradiol-inducible systems involving fusion of the estradiol receptor to
DNA-
binding and transcriptional activation domain, paired with synthetic or native
promoters with binding sites.
o estradiol
= tet Trans Activator (tTA) or reverse tet Trans Activator (rtTA) systems
paired with
tet0-containing promoters.
o doxycyclin
149
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Codon Usage
[0368] As is well known to those of skill in the art, it is possible to
improve the
expression of a heterologous nucleic acid in a host organism by replacing the
nucleotide
sequences coding for a particular amino acid (i.e., a codon) with another
codon which is
better expressed in the host organism (i.e., codon-optimization). One reason
that this effect
arises is due to the fact that different organisms show preferences for
different codons. In
some embodiments, a nucleic acid disclosed herein is modified or optimized
such that the
nucleotide sequence reflects the codon preference for the particular host
cell. For example,
the nucleotide sequence will in some embodiments be modified or optimized for
yeast codon
preference. In some embodiments, a nucleotide sequence disclosed herein is
codon-
optimized. See, e.g., Bennetzen and Hall (1982)1 Biol. Chem. 257(6): 3026-
3031.
[0369] Statistical methods have been generated to analyze codon usage
bias in
various organisms and many computer algorithms have been developed to
implement these
statistical analyses in the design of codon optimized gene sequences (Lithwick
G, Margalit
H (2003) Hierarchy of sequence-dependent features associated with prokaryotic
translation.
Genome Research 13: 2665-73). Other modifications in codon usage to increase
protein
expression that are not dependent on codon bias have also been described
(Welch et al.
(2009).
[0370] In some embodiments, the codon usage of a nucleotide sequence is
modified
or optimized such that the level of translation of the encoded mRNA is
decreased. In some
embodiments, a codon-optimized nucleotide sequence may be optimized such that
the level
of translation of the encoded mRNA is decreased. Reducing the level of
translation of an
mRNA by modifying codon usage may be achieved by modifying the nucleotide
sequence to
include codons that are rare or not commonly used by the host cell. Codon
usage tables for
many organisms are available that summarize the percentage of time a specific
organism
uses a specific codon to encode for an amino acid. Certain codons are used
more often than
other, "rare" codons. The use of "rare" codons in a nucleotide sequence
generally decreases
its rate of translation. Thus, e.g., the nucleotide sequence is modified by
introducing one or
more rare codons, which affect the rate of translation, but not the amino acid
sequence of the
polypeptide translated. For example, there are six codons that encode for
arginine: CGT,
CGC, CGA, CGG, AGA, and AGG. In E. coil the codons CGT and CGC are used far
more
often (encoding approximately 40% of the arginines in E. coil each) than the
codon AGG
150
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
(encoding approximately 2% of the arginines in E. coil). Modifying a CGT codon
within the
sequence of a gene to an AGG codon would not change the sequence of the
polypeptide, but
would likely decrease the gene's rate of translation.
[0371] In some embodiments, a codon-optimized nucleotide sequence may be
optimized for expression in a yeast cell. In certain such embodiments, the
yeast cell is
Saccharomyces cerevisiae.
[0372] Further, it will be appreciated that this disclosure embraces the
degeneracy of
codon usage as would be understood by one of ordinary skill in the art and
illustrated in the
following table.
Codon Degeneracies
Amino Acid Codons
AlalA GCT, GCC, GCA, GCG
.Arg/R CGT, CGC, CGA, COG, AGA, AGG
A.sn/N AAT, AAC
.Asp/D OAT, GAC
Cys/C TGT, 717GC
Gni() CAA, CAG-
GlulE GAA, GAG
Gly/G CiGT, CiGC, GGA, CiGG
CAT, CAC
Ile/I ATT, ATC, ATA
LeulL TTA, TTG, CIT, CTC, CTA, CTG
Lys/K. AAA, AAG
Met/M ATG
Ph.e/F ]ITC
Pro/P ccr, ccA, ccG-
Ser/S TCT, ICC, ICAõ TCG, AGT, AGC
Thr/T ACT, ACC, ACA, ACG
Trp/W TGG
TyrlY TAT, TAC
Val/V Gil, GTC, GIA, GIG
START ATG
STOP TAG, TGA, TAA
Methods of Producing a Cannabinoid or a Cannabinoid Derivative or of
Expressing
and/or Preparing Engineered Variants of the Tetrahydrocannabinolic Acid
Synthase
(THCAS) Polypeptide
[0373] The disclosure provides methods for expressing an engineered
variant of the
tetrahydrocannabinolic acid synthase (THCAS) polypeptide of the disclosure. In
certain such
embodiments, the methods comprise culturing a modified host cell of the
disclosure in a
151
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
culture medium. The disclosure also provides methods for preparing an
engineered variant of
the tetrahydrocannabinolic acid synthase (THCAS) polypeptide of the
disclosure. The
disclosure also provides methods of producing a cannabinoid or a cannabinoid
derivative,
the method comprising use of an engineered variant of the disclosure.
[0374] The present disclosure also provides methods of producing a
cannabinoid or a
cannabinoid derivative. The methods of the present disclosure may involve
production of
cannabinoids or cannabinoid derivatives using an engineered variant disclosed
herein. The
methods may involve culturing a modified host cell of the present disclosure
in a culture
medium and recovering the produced cannabinoid or cannabinoid derivative. The
methods
may also involve cell-free production of cannabinoids or cannabinoid
derivatives using one
or more polypeptides disclosed herein, such as an engineered variant of the
disclosure,
expressed or overexpressed by a modified host cell of the disclosure. The
methods may also
involve cell-free production of cannabinoids or cannabinoid derivatives using
an engineered
variant disclosed herein.
[0375] Cannabinoids or cannabinoid derivatives that can be produced with
the
engineered variants, methods, or modified host cells of the present disclosure
may include,
but are not limited to, cannabichromene (CBC) type (e.g., cannabichromenic
acid),
cannabidiol (CBD) type (e.g. cannabidiolic acid), A9-trans-
tetrahydrocannabinol (A9 -THC)
type (e.g. A9-tetrahydrocannabinolic acid), A8-trans-tetrahydrocannabinol (A8 -
THC) type,
cannabicyclol (CBL) type, cannabielsoin (CBE) type, cannabinol (CBN) type,
cannabinodiol
(CBND) type, cannabitriol (CBT) type, derivatives of any of the foregoing, and
others as
listed in Elsohly M.A. and Slade D., Life Sci. 2005 Dec 22;78(5):539-48. Epub
2005 Sep 30.
In some embodiments, the cannabinoid or cannabinoid derivative is produced in
an amount
of more than 100 mg/L culture medium. In some embodiments, the cannabinoid or
cannabinoid derivative is produced in an amount of more than 50 mg/L culture
medium.
[0376] Cannabinoids or cannabinoid derivatives that can be produced with
the
engineered variants, methods, or modified host cells of the present disclosure
may also
include, but are not limited to, cannabichromenic acid (CBCA), cannabichromene
(CBC),
cannabichromevarinic acid (CBCVA), cannabichromevarin (CBCV), CBDA,
cannabidiol
(CBD), cannabidiol monomethylether (CBDM), cannabidiol-C4 (CBD-C4),
cannabidivarinic
acid (CBDVA), cannabidivarin (CBDV), cannabidiorcol (CBD-C1), A9 ¨
tetrahydrocannabinolic acid A (THCA-A), A9 ¨tetrahydrocannabinolic acid B
(THCA-B), A9
¨tetrahydrocannabinol (THC), A9 ¨tetrahydrocannabinolic acid-C4 (THCA-C4), A9
¨
152
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
tetrahydrocannabinol-C4 (THC-C4), A9 ¨tetrahydrocannabivarinic acid (THCVA),
A9 ¨
tetrahydrocannabivarin (THCV), A9 ¨tetrahydrocannabiorcolic acid (THCA-C1), A9
¨
tetrahydrocannabiorcol (THC-C1), A7 ¨cis-iso-tetrahydrocannabivarin, A8 ¨
tetrahydrocannabinolic acid (A8 ¨THCA), A8 ¨tetrahydrocannabinol (A8 ¨THC),
cannabicyclolic acid (CBLA), cannabicyclol (CBL), cannabicyclovarin (CBLV),
cannabielsoic acid A (CBEA-A), cannabielsoic acid B (CBEA-B), cannabielsoin
(CBE),
cannabielsoinic acid, cannabicitranic acid, cannabinolic acid (CBNA),
cannabinol (CBN),
cannabinol methylether (CBNM), cannabinol-C4, (CBN-C4), cannabivarin (CBV),
cannabinol-C2 (CNB-C2), cannabiorcol (CBN-C1), cannabinodiol (CBND),
cannabinodivarin
(CBVD), cannabitriol (CB T), 10-ethyoxy-9-hydroxy-delta-6a-
tetrahydrocannabinol, 8,9-
dihydroxyl-delta-6a-tetrahydrocannabinol, cannabitriolvarin (CBTVE),
dehydrocannabifuran
(DCBF), cannabifuran (CBF), cannabichromanon (CBCN), cannabicitran (CBT), 10-
oxo-
delta-6a-tetrahydrocannabinol (OTHC), delta-9-cis-tetrahydrocannabinol (cis-
THC), 3,4,5,6-
tetrahydro-7-hydroxy-alpha-alpha-2-trimethy1-9-n-propy1-2,6-methano-2H-1-
benzoxocin-5-
methanol (OH-iso-HHCV), cannabiripsol (CBR), trihydroxy-delta-9-
tetrahydrocannabinol
(tri0H-THC), CBGA-hydrocinnamic acid (3-[(2E)-3,7-dimethylocta-2,6-dien-l-y1]-
2,4-
dihydroxy-6-(2-phenylethyl)benzoic acid), CBG-hydrocinnamic acid (2-[(2E)-3,7-
dimethylocta-2,6-dien-l-y1]-5-(2-phenylethyl)benzene-1,3-diol), CBDA-
hydrocinnamic acid
(2,4-dihydroxy-3 -[3 -methyl-6-(prop-1-en-2-y1)cycl ohex-2-en-l-yl] -6-(2-
phenylethyl)benzoic acid), CBD-hydrocinnamic acid (2-[3-methy1-6-(prop-1-en-2-
y1)cyclohex-2-en-1-y1]-5-(2-phenylethyl)benzene-1,3-diol), THCA-hydrocinnamic
acid (1-
hydroxy-6,6,9-trimethy1-3 -(2-phenylethyl)-6H,6aH,7H,8H,10aH-benzo[c]i
sochromene-2-
carb oxyli c acid), THC-hydrocinnamic acid (6,6,9-trimethy1-3-(2-phenylethyl)-
6H,6aH,7H,8H,10aH-benzo[c]isochromen-1-ol, perrottetinene), and derivatives of
any of the
foregoing. In some embodiments, the cannabinoid or cannabinoid derivative is
produced in
an amount of more than 100 mg/L culture medium. In some embodiments, the
cannabinoid
or cannabinoid derivative is produced in an amount of more than 50 mg/L
culture medium.
[0377] In some embodiments, the cannabinoid produced with the engineered
variants, methods, or modified host cells of the present disclosure is A9-
tetrahydrocannabinolic acid, A9-tetrahydrocannabinol, A8-
tetrahydrocannabinolic acid, A8-
tetrahydrocannabinol, cannabidiolic acid, cannabidiol, cannabichromenic acid,
cannabichromene, cannabinolic acid, cannabinol, cannabidivarinic acid,
cannabidivarin,
tetrahydrocannabivarinic acid, tetrahydrocannabivarin, cannabichromevarinic
acid,
153
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
cannabichromevarin, cannabigerovarinic acid, cannabigerovarin, cannabicyclolic
acid,
cannabicyclol, cannabielsoinic acid, cannabielsoin, cannabicitranic acid, or
cannabicitran. In
some embodiments, the cannabinoid is produced in an amount of more than 100
mg/L
culture medium. In some embodiments, the cannabinoid is produced in an amount
of more
than 50 mg/L culture medium.
[00100] In some embodiments, the cannabinoid produced with the engineered
variants, methods, or modified host cells of the present disclosure is
tetrahydrocannabinolic
acid, tetrahydrocannabivarinic acid, or tetrahydrocannabivarin. In some
embodiments, the
cannabinoid is produced in an amount of more than 100 mg/L culture medium. In
some
embodiments, the cannabinoid is produced in an amount of more than 50 mg/L
culture
medium.
[0378] Additional cannabinoids and cannabinoid derivatives that can be
produced
with the engineered variants, methods, or modified host cells of the present
disclosure may
also include, but are not limited to, CBDA, CBD, CBGA, THC, THCA, THCVA,
CBDVA,
(6aR,10aR)-1-hydroxy-6,6,9-trimethy1-3 -butyl-6a,7,8,10a-tetrahydro-6H-dib
enzo [b,d]pyran-
2-carboxylic acid, (6aR,10aR)-1-hydroxy-6,6,9-trimethy1-3 -(3 -methylpenty1)-
6a,7,8,10a-
tetrahydro-6H-dib enzo [b,d]pyran-2-carb oxyli c acid, (6aR,10aR)-1-hydroxy-
6,6,9-trimethyl-
3 -(4-penteny1)-6 a,7,8,10a-tetrahydro-6H-dib enzo [b, d]pyran-2-carboxylic
acid, (6aR,10aR)-
1-hydroxy-6,6,9-trimethy1-3-hexy1-6a,7,8,10a-tetrahydro-6H-dibenzo[b,d]pyran-2-
carboxylic acid, (6aR,10aR)-1-hydroxy-6,6,9-trimethy1-3-(5-hexyny1)-6a,7,8,10a-
tetrahydro-
6H-dibenzo[b,d]pyran-2-carboxylic acid, and others as listed in Bow, E. W. and
Rimoldi, J.
M., "The Structure¨Function Relationships of Classical Cannabinoids: CB1/CB2
Modulation," Perspectives in Medicinal Chemistry 2016:8 17-39 doi:
10.4137/PMC.S32171, incorporated by reference herein. In some embodiments, the
cannabinoid or cannabinoid derivative is produced in an amount of more than
100 mg/L
culture medium. In some embodiments, the cannabinoid or cannabinoid derivative
is
produced in an amount of more than 50 mg/L culture medium.
[0379] Additional cannabinoids and cannabinoid derivatives that can be
produced
with the engineered variants, methods, or modified host cells of the present
disclosure may
also include, but are not limited to, (1'R,2'R)-4-(hexan-2-y1)-5'-methyl-2'-
(prop-1-en-2-y1)-
1',2',3',4'-tetrahydro-[1,1'-bipheny1]-2,6-diol, (1'R,2'R)-4-hexy1-5'-methy1-
2'-(prop-1-en-2-
y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-(3-
methylpenty1)-2'-
(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-4-
(4-chlorobuty1)-5'-
154
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
methyl-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-bipheny1]-2,6-diol,
(1'R,2'R)-5'-methy1-
4-(4-methylpenty1)-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-
2,6-diol,
(1'R,2'R)-5'-methy1-4-(4-(methylthio)butyl)-2'-(prop-1-en-2-y1)-1',2',3',4'-
tetrahydro-[1,1'-
biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-((E)-pent-1-en-l-y1)-2'-(prop-1-en-2-
y1)-1',2',3',4'-
tetrahydro-[1,1'-biphenyl] -2,6-di ol, (1'R,2'R)-5'-methy1-4-((E)-pent-3-en-1-
y1)-2'-(prop-1-en-
2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-
((E)-pent-2-en-1-
y1)-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol,
(1'R,2'R)-4-(but-3-yn-1-
y1)-5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-
diol, (1'R,2'R)-4-
((E)-but-l-en-l-y1)-5'-methyl-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-
biphenyl]-2,6-
diol, (1'R,2'R)-5'-methy1-4-(pent-4-yn-l-y1)-2'-(prop-1-en-2-y1)-1',2',3',4'-
tetrahydro-[1,1'-
biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-2'-(prop-1-en-2-y1)-4-undecyl-
1',2',3',4'-tetrahydro-
[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-4-(hex-5-yn-l-y1)-5'-methy1-2'-(prop-1-en-
2-y1)-1',2',3',4'-
tetrahydro-[1,1'-biphenyl] -2,6-di ol, (1'R,2'R)-4-((E)-hept-l-en-1-y1)-5'-
methyl-2'-(prop-1-en-
2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-
octy1-2'-(prop-1-en-
2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-
((E)-oct-1-en-1-y1)-
2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-bipheny1]-2,6-diol, (1'R,2'R)-
5'-methy1-4-
nony1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-bipheny1]-2,6-diol,
(1'R,2'R)-5'-methy1-
4-(3-phenylpropy1)-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-
2,6-diol,
(1'R,2'R)-5'-methy1-4-(4-phenylbuty1)-2'-(prop-1-en-2-y1)-1',2',3',4'-
tetrahydro-[1,1'-
biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-(5-phenylpenty1)-2'-(prop-1-en-2-y1)-
1',2',3',4'-
tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-(6-phenylhexyl)-2'-
(prop-1-en-2-
y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-5'-methy1-4-(2-
methylpenty1)-2'-
(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, (1'R,2'R)-4-
isopropy1-5'-
methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol,
(1'R,2'R)-4-decy1-
5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-bipheny1]-2,6-diol,
(1'R,2'R)-5'-
methy1-2'-(prop-1-en-2-y1)-4-tridecyl-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-
2,6-diol, (E)-3-
((1'R,2'R)-2,6-dihydroxy-5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-
[1,1'-bipheny1]-
4-y1)acrylic acid, (Z)-341'R,2'R)-2,6-dihydroxy-5'-methy1-2'-(prop-1-en-2-y1)-
1',2',3',4'-
tetrahydro-[1,1'-biphenyl]-4-y1)acrylic acid, 7-((l'R,2'R)-2,6-dihydroxy-5'-
methy1-2'-(prop-
1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-4-y1)heptanoic acid,
841'R,2'R)-2,6-
dihydroxy-5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-
4-y1)octanoic
acid, 9-((l'R,2'R)-2,6-dihydroxy-5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-
tetrahydro-[1,1'-
biphenyl]-4-y1)nonanoic acid, 11-((1'R,2'R)-2,6-dihydroxy-5'-methy1-2'-(prop-1-
en-2-y1)-
155
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
1',2',3',4'-tetrahydro-[1,1'-bipheny1]-4-yl)undecanoic acid, (1"R,2"R)-3',5'-
dihydroxy-5"-
methy1-2"-(prop-1-en-2-y1)-1",2",3",4"-tetrahydro-[1,1':4',1"-terphenyl]-2-
carboxylic acid,
(1"R,2"R)-3',5'-dihydroxy-5"-methy1-2"-(prop-1-en-2-y1)-1",2",3",4"-tetrahydro-
[1,1':4',1"-
terpheny1]-3-carboxylic acid, (1"R,2"R)-3',5'-dihydroxy-5"-methy1-2"-(prop-1-
en-2-y1)-
1",2",3",4"-tetrahydro-[1,1':4',1"-terphenyl]-4-carboxylic acid, (1"R,2"R)-
3',5'-dihydroxy-5"-
methy1-2"-(prop-1-en-2-y1)-1",2",3",4"-tetrahydro-[1,1':4',1"-terpheny1]-3,5-
dicarboxylic
acid, (1'R,2'R)-4-(4-hydroxybuty1)-5'-methyl-2'-(prop-1-en-2-y1)-1',2',3',4'-
tetrahydro-[1,1'-
biphenyl]-2,6-diol, (1'R,2'R)-4-(4-aminobuty1)-5'-methy1-2'-(prop-1-en-2-y1)-
1',2',3',4'-
tetrahydro-[1,1'-biphenyl]-2,6-diol, 5-((1'R,2'R)-2,6-dihydroxy-5'-methy1-2'-
(prop-1-en-2-
y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-4-y1)pentanenitrile, (1'R,2'R)-5'-
methy1-4-(3-
methylhexan-2-y1)-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-
2,6-diol,
(1'R,2'R)-5'-methy1-2'-(prop-1-en-2-y1)-4-propy1-1',2',3',4'-tetrahydro-[1,1'-
biphenyl]-2,6-
diol, (1'R,2'R)-4-buty1-5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-
[1,1'-bipheny1]-
2,6-diol, (1'R,2'R)-5'-methy1-4-penty1-2'-(prop-1-en-2-y1)-1',2',3',4'-
tetrahydro-[1,1'-
bipheny1]-2,6-diol, (1'R,2'R)-4-hepty1-5'-methy1-2'-(prop-1-en-2-y1)-
1',2',3',4'-tetrahydro-
[1,1'-bipheny1]-2,6-diol, (1'R,2'R)-5'-methy1-4-(pent-4-en-1-y1)-2'-(prop-1-en-
2-y1)-1',2',3',4'-
tetrahydro-[1,1'-bipheny1]-2,6-diol, 3-((1'R,2'R)-2,6-dihydroxy-5'-methy1-2'-
(prop-1-en-2-
y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-4-y1)propanoic acid, (1'R,2'R)-4,5'-
dimethy1-2'-
(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-2,6-diol, 24(1'R,2'R)-
2,6-dihydroxy-5'-
methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-4-y1)acetic
acid, 4-
((1'R,2'R)-2,6-dihydroxy-5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-
[1,1'-biphenyl]-
4-yl)butanoic acid, (1'R,2'R)-2,6-dihydroxy-5'-methy1-2'-(prop-1-en-2-y1)-
1',2',3',4'-
tetrahydro-[1,1'-bipheny1]-4-carboxylic acid, 5-((1'R,2'R)-2,6-dihydroxy-5'-
methy1-2'-(prop-
1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-4-y1)pentanoic acid, and 6-
((l'R,2'R)-2,6-
dihydroxy-5'-methy1-2'-(prop-1-en-2-y1)-1',2',3',4'-tetrahydro-[1,1'-biphenyl]-
4-y1)hexanoic
acid. In some embodiments, the cannabinoid or cannabinoid derivative is
produced in an
amount of more than 100 mg/L culture medium. In some embodiments, the
cannabinoid or
cannabinoid derivative is produced in an amount of more than 50 mg/L culture
medium.
[0380] A cannabinoid derivative may lack one or more chemical moieties
found in a
naturally-occurring cannabinoid. Such chemical moieties may include, but are
not limited
to, methyl, alkyl, alkenyl, methoxy, alkoxy, acetyl, carboxyl, carbonyl, oxo,
ester, hydroxyl,
aryl, heteroaryl, cycloalkyl, cycloalkenyl, cycloalkylalkenyl,
cycloalkenylalkyl,
cycloalkenylalkenyl, heterocyclylalkenyl, heteroarylalkenyl, arylalkenyl,
heterocyclyl,
156
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
aralkyl, cycloalkylalkyl, heterocyclylalkyl, heteroarylalkyl, and the like. In
some
embodiments, a cannabinoid derivative lacking one or more chemical moieties
found in a
naturally-occurring cannabinoid may also comprise one or more of any of the
functional
and/or reactive groups described herein. Functional and reactive groups may be
unsubstituted or substituted with one or more functional or reactive groups.
[0381] A cannabinoid derivative may be a cannabinoid substituted with or
comprising one or more functional and/or reactive groups. Functional groups
may include,
but are not limited to, azido, halo (e.g., chloride, bromide, iodide,
fluorine), methyl, alkyl,
alkynyl, alkenyl, methoxy, alkoxy, acetyl, amino, carboxyl, carbonyl, oxo,
ester, hydroxyl,
thio (e.g., thiol), cyano, aryl, heteroaryl, cycloalkyl, cycloalkenyl,
cycloalkylalkenyl,
cycloalkylalkynyl, cycloalkenylalkyl, cycloalkenylalkenyl,
cycloalkenylalkynyl,
heterocyclylalkenyl, heterocyclylalkynyl, heteroarylalkenyl,
heteroarylalkynyl, arylalkenyl,
arylalkynyl, spirocyclyl, heterospirocyclyl, heterocyclyl, thioalkyl (or
alkylthio), arylthio,
heteroarylthio, sulfone, sulfonyl, sulfoxide, amido, alkylamino, dialkylamino,
arylamino,
alkylarylamino, diarylamino, N-oxide, imide, enamine, imine, oxime, hydrazone,
nitrile,
aralkyl, cycloalkylalkyl, haloalkyl, heterocyclylalkyl, heteroarylalkyl,
nitro, thioxo, and the
like. Suitable reactive groups may include, but are not necessarily limited
to, azide, carboxyl,
carbonyl, amine (e.g., alkyl amine (e.g., lower alkyl amine), aryl amine),
halide, ester (e.g.,
alkyl ester (e.g., lower alkyl ester, benzyl ester), aryl ester, substituted
aryl ester), cyano,
thioester, thioether, sulfonyl halide, alcohol, thiol, succinimidyl ester,
isothiocyanate,
iodoacetamide, maleimide, hydrazine, alkynyl, alkenyl, acetyl, and the like.
In some
embodiments, the reactive group is selected from a carboxyl, a carbonyl, an
amine, an ester,
a thioester, a thioether, a sulfonyl halide, an alcohol, a thiol, an alkyne,
alkene, an azide, a
succinimidyl ester, an isothiocyanate, an iodoacetamide, a maleimide, and a
hydrazine.
Functional and reactive groups may be unsubstituted or substituted with one or
more
functional or reactive groups.
[0382] "Alkyl" may refer to a straight or branched chain saturated
hydrocarbon. For
example, C1-C6alkyl groups contain 1 to 6 carbon atoms. Examples of a C1-
C6alkyl group
include, but are not limited to, methyl, ethyl, propyl, butyl, pentyl,
isopropyl, isobutyl, sec-
butyl and tert-butyl, isopentyl, and neopentyl.
[0383] "Alkenyl" may include an unbranched (i.e., straight) or branched
hydrocarbon chain containing 2-12 carbon atoms. The "alkenyl" group contains
at least one
double bond. The double bond of an alkenyl group can be unconjugated or
conjugated to
157
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
another unsaturated group. Examples of alkenyl groups may include, but are not
limited to,
ethylenyl, vinyl, allyl, butenyl, pentenyl, hexenyl, butadienyl, pentadienyl,
hexadienyl, 2-
ethylhexenyl, 2-propy1-2-butenyl, 4-(2-methyl-3-butene)-pentenyl and the like.
[0384] Compounds disclosed herein, such as cannabinoids and cannabinoid
derivatives, may be substituted with one or more substituents, such as those
illustrated
generally herein, or as exemplified by particular classes, subclasses, and
species of the
present disclosure. In general, the term "substituted" refers to the
replacement of a hydrogen
atom in a given structure with a specified sub stituent. Combinations of
substituents
envisioned by the present disclosure are typically those that result in the
formation of stable
or chemically feasible compounds.
[0385] As used herein, the term "unsubstituted" may mean that the
specified group
bears no substituents beyond the moiety recited (e.g., where valency satisfied
by hydrogen).
[0386] A reactive group may facilitate covalent attachment of a molecule
of interest.
Suitable molecules of interest may include, but are not limited to, a
detectable label; imaging
agents; a toxin (including cytotoxins); a linker; a peptide; a drug (e.g.,
small molecule
drugs); a member of a specific binding pair; an epitope tag; ligands for
binding by a target
receptor; tags to aid in purification; molecules that increase solubility; and
the like. A linker
may be a peptide linker or a non-peptide linker.
[0387] In some embodiments, a cannabinoid derivative substituted with an
azide may
be reacted with a compound comprising an alkyne group via "click chemistry" to
generate a
product comprising a heterocycle, also known as an azide-alkyne cycloaddition.
In some
embodiments, a cannabinoid derivative substituted with an alkyne may be
reacted with a
compound comprising an azide group via click chemistry to generate a product
comprising a
heterocycle.
[0388] Additional molecules of interest that may be desirable for
attachment to a
cannabinoid derivative may include, but are not necessarily limited to,
detectable labels (e.g.,
spin labels, fluorescence resonance energy transfer (FRET)-type dyes, e.g.,
for studying
structure of biomolecules in vivo); small molecule drugs; cytotoxic molecules
(e.g., drugs);
imaging agents; ligands for binding by a target receptor; tags to aid in
purification by, for
example, affinity chromatography (e.g., attachment of a FLAG epitope);
molecules that
increase solubility (e.g., poly(ethylene glycol); molecules that enhance
bioavailability;
molecules that increase in vivo half-life; molecules that target to a
particular cell type (e.g.,
an antibody specific for an epitope on a target cell); molecules that target
to a particular
158
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
tissue; molecules that provide for crossing the blood-brain barrier; and
molecules to facilitate
selective attachment to a surface, and the like.
[0389] In some embodiments, a molecule of interest comprises an imaging
agent.
Suitable imaging agents may include positive contrast agents and negative
contrast agents.
Suitable positive contrast agents may include, but are not limited to,
gadolinium
tetraazacyclododecanetetraacetic acid (Gd-DOTA); gadolinium-
diethylenetriaminepentaacetic acid (Gd-DTPA); gadolinium-1,4,7-
tris(carbonylmethyl)-10-
(2'-hydroxypropy1)-1,4,7,10-tetraazacyclododecane (Gd-HP-D03A); Manganese(II)-
dipyridoxal diphosphate (Mn-DPDP); Gd-diethylenetriaminepentaacetate-
bis(methylamide)
(Gd-DTPA-BMA); and the like. Suitable negative contrast agents may include,
but are not
limited to, a superparamagnetic iron oxide (SPIO) imaging agent; and a
perfluorocarbon,
where suitable perfluorocarbons may include, but are not limited to,
fluoroheptanes,
fluorocycloheptanes, fluoromethylcycloheptanes, fluorohexanes,
fluorocyclohexanes,
fluoropentanes, fluorocyclopentanes, fluoromethylcyclopentanes,
fluorodimethylcyclopentanes, fluoromethylcyclobutanes,
fluorodimethylcyclobutanes,
fluorotrimethylcyclobutanes, fluorobutanes, fluorocyclobutanse,
fluoropropanes,
fluoroethers, fluoropolyethers, fluorotriethylamines, perfluorohexanes,
perfluoropentanes,
perfluorobutanes, perfluoropropanes, sulfur hexafluoride, and the like.
[0390] Additional cannabinoid derivatives that can be produced with an
engineered
variant, method, or modified host cell of the present disclosure may include
derivatives that
have been modified via organic synthesis or an enzymatic route to modify drug
metabolism
and pharmacokinetics (e.g. solubility, bioavailability, absorption,
distribution, plasma half-
life and metabolic clearance). Modification examples may include, but are not
limited to,
halogenation, acetylation, and methylation.
[0391] The cannabinoids or cannabinoid derivatives described herein
further include
all pharmaceutically acceptable isotopically labeled cannabinoids or
cannabinoid derivatives.
An "isotopically-" or "radio-labeled" compound is a compound where one or more
atoms are
replaced or substituted by an atom having an atomic mass or mass number
different from the
atomic mass or mass number typically found in nature (i.e., naturally
occurring). For
example, in some embodiments, in the cannabinoids or cannabinoid derivatives
described
herein, hydrogen atoms are replaced or substituted by one or more deuterium or
tritium.
Certain isotopically labeled cannabinoids or cannabinoid derivatives of this
disclosure, for
example, those incorporating a radioactive isotope, are useful in drug and/or
substrate tissue
159
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
distribution studies. The radioactive isotopes tritium, i.e., 3H, and carbon
14, i.e., 14C, are
particularly useful for this purpose in view of their ease of incorporation
and ready means of
detection. Substitution with heavier isotopes such as deuterium, i.e., 2H, may
afford certain
therapeutic advantages resulting from greater metabolic stability, for
example, increased in
vivo half-life or reduced dosage requirements, and hence may be preferred in
some
circumstances. Suitable isotopes that may be incorporated in cannabinoids or
cannabinoid
derivatives described herein include but are not limited to 2H (also written
as D for
deuterium), 3H (also written as T for tritium), HC, 13C, 14C, 13N, 15N, 150,
170, 180, 18F, 35s,
36C1 , 82Br, 75Br, 76Br, 77Br, 1231, 1241, 1251, and 1311. Substitution with
positron emitting
isotopes, such as HC, 18F, 15,,,
and 13N, can be useful in Positron Emission Topography
(PET) studies.
[0392] The methods of bioproduction, modified host cells, and engineered
variants
disclosed herein enable synthesis of cannabinoids or cannabinoid derivatives
with defined
stereochemistries, which is challenging to do using chemical synthesis.
Cannabinoids or
cannabinoid derivatives disclosed herein may be enantiomers or disastereomers.
The term
"enantiomers" may refer to a pair of stereoisomers which are non-
superimposable mirror
images of one another. In some embodiments the cannabinoids or cannabinoid
derivatives
may be the (S)-enantiomer. In some embodiments the cannabinoids or cannabinoid
derivatives may be the (R)-enantiomer. In some embodiments, the cannabinoids
or
cannabinoid derivatives may be the (+) or (-) enantiomers. The term
"diastereomers" may
refer to the set of stereoisomers which cannot be made superimposable by
rotation around
single bonds. For example, cis- and trans- double bonds, endo- and exo-
substitution on
bicyclic ring systems, and compounds containing multiple stereogenic centers
with different
relative configurations may be considered to be diastereomers. The term
"diastereomer"
may refer to any member of this set of compounds. Cannabinoids or cannabinoid
derivatives
disclosed herein may include a double bond or a fused ring. In certain such
embodiments,
the double bond or fused ring may be cis or trans, unless the configuration is
specifically
defined. If the cannabinoid or cannabinoid derivative contains a double bond,
the substituent
may be in the E or Z configuration, unless the configuration is specifically
defined.
[0393] In some embodiments when the cannabinoid or cannabinoid derivative
is
recovered from a cell lysate; from a culture medium; from a modified host
cell; from both
the cell lysate and the culture medium; from both the modified host cell and
the culture
medium; from the cell lysate, the modified host cell, and the culture medium;
or from a cell-
160
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
free reaction mixture comprising one or more polypeptides and/or engineered
variants
disclosed herein, the recovered cannabinoid or cannabinoid derivative is in
the form of a salt.
In certain such embodiments, the salt is a pharmaceutically acceptable salt.
In some
embodiments, the salt of the recovered cannabinoid or cannabinoid derivative
is then
purified as disclosed herein.
[0394] The disclosure includes pharmaceutically acceptable salts of the
cannabinoids
or cannabinoid derivatives described herein. "Pharmaceutically acceptable
salts" may refer
to those salts which retain the biological effectiveness and properties of the
free bases, which
are not biologically or otherwise undesirable. Representative pharmaceutically
acceptable
salts include, but are not limited to, e.g., water-soluble and water-insoluble
salts, such as the
acetate, amsonate (4,4-diaminostilbene-2,2-disulfonate), benzenesulfonate,
benzonate,
bicarbonate, bisulfate, bitartrate, borate, bromide, butyrate, calcium,
calcium edetate,
camsylate, carbonate, chloride, citrate, clavulariate, dihydrochloride,
edetate, edisylate,
estolate, esylate, fiunarate, gluceptate, gluconate, glutamate,
glycollylarsanilate,
hexafluorophosphate, hexylresorcinate, hydrabamine, hydrobromide,
hydrochloride,
hydroxynaphthoate, iodide, sethionate, lactate, lactobionate, laurate,
magnesium, malate,
maleate, mandelate, mesylate, methylbromide, methylnitrate, methyl sulfate,
mucate,
napsylate, nitrate, N-methylglucamine ammonium salt, 3-hydroxy-2-naphthoate,
oleate,
oxalate, palmitate, pamoate (1,1-methene-bis-2-hydroxy-3-naphthoate,
einbonate),
pantothenate, phosphate/diphosphate, picrate, polygalacturonate, propionate, p-
toluenesulfonate, salicylate, stearate, subacetate, succinate, sulfate,
sulfosalicylate, suramate,
tannate, tartrate, teoclate, tosylate, triethiodide, and valerate salts.
[0395] "Pharmaceutically acceptable salt" also includes both acid and
base addition
salts. "Pharmaceutically acceptable acid addition salt" may refer to those
salts which retain
the biological effectiveness and properties of the free bases, which are not
biologically or
otherwise undesirable, and which are formed with inorganic acids such as, but
are not
limited to, hydrochloric acid, hydrobromic acid, sulfuric acid, nitric acid,
phosphoric acid
and the like, and organic acids such as, but not limited to, acetic acid, 2,2-
dichloroacetic
acid, adipic acid, alginic acid, ascorbic acid, aspartic acid, benzenesulfonic
acid, benzoic
acid, 4-acetamidobenzoic acid, camphoric acid, camphor-10-sulfonic acid,
capric acid,
caproic acid, caprylic acid, carbonic acid, cinnamic acid, citric acid,
cyclamic acid,
dodecylsulfuric acid, ethane-1,2-disulfonic acid, ethanesulfonic acid, 2-
hydroxyethanesulfonic acid, formic acid, fumaric acid, galactaric acid,
gentisic acid,
161
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
glucoheptonic acid, gluconic acid, glucuronic acid, glutamic acid, glutaric
acid, 2-oxo-
glutaric acid, glycerophosphoric acid, glycolic acid, hippuric acid,
isobutyric acid, lactic
acid, lactobionic acid, lauric acid, maleic acid, malic acid, malonic acid,
mandelic acid,
methanesulfonic acid, mucic acid, naphthalene-1,5-disulfonic acid, naphthalene-
2-sulfonic
acid, 1-hydroxy-2-naphthoic acid, nicotinic acid, oleic acid, orotic acid,
oxalic acid, palmitic
acid, pamoic acid, propionic acid, pyroglutamic acid, pyruvic acid, salicylic
acid, 4-
aminosalicylic acid, sebacic acid, stearic acid, succinic acid, tartaric acid,
thiocyanic acid, p-
toluenesulfonic acid, trifluoroacetic acid, undecylenic acid, and the like.
[0396] "Pharmaceutically acceptable base addition salt" may refer to
those salts
which retain the biological effectiveness and properties of the free acids,
which are not
biologically or otherwise undesirable. These salts are prepared from addition
of an inorganic
base or an organic base to the free acid. Salts derived from inorganic bases
include, but are
not limited to, the sodium, potassium, lithium, ammonium, calcium, magnesium,
iron, zinc,
copper, manganese, aluminum salts and the like. For example, inorganic salts
include, but
are not limited to, ammonium, sodium, potassium, calcium, and magnesium salts.
Salts
derived from organic bases include, but are not limited to, salts of primary,
secondary, and
tertiary amines, substituted amines including naturally occurring substituted
amines, cyclic
amines and basic ion exchange resins, such as ammonia, isopropylamine,
trimethylamine,
diethylamine, triethylamine, tripropylamine, diethanolamine, ethanolamine,
deanol, 2-
dimethylaminoethanol, 2-diethylaminoethanol, dicyclohexylamine, lysine,
arginine,
histidine, caffeine, procaine, hydrabamine, choline, betaine, benethamine,
benzathine,
ethylenediamine, glucosamine, methylglucamine, theobromine, triethanolamine,
tromethamine, purines, piperazine, piperidine, N-ethylpiperidine, polyamine
resins and the
like.
[0397] The disclosure provides a method of producing a cannabinoid or a
cannabinoid derivative, the method comprising use of an engineered variant of
of the
disclosure. In certain such embodiments, the cannabinoid or the cannabinoid
derivative is
produced in an amount, as measured in mg/L or mM, greater than an amount of
the
cannabinoid or the cannabinoid derivative produced in a method comprising use
of a
THCAS polypeptide having an amino acid sequence of SEQ ID NO:44 instead of the
engineered variant of the disclosure. In certain such embodiments, the
engineered variant of
the disclosure and the tetrahydrocannabinolic acid synthase polypeptide having
the amino
acid sequence of SEQ ID NO:44 are used under similar conditions for the same
length of
162
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
time. In some embodiments of the methods of producing a cannabinoid or a
cannabinoid
derivative of the disclosure, the cannabinoid or the cannabinoid derivative is
produced in an
amount, as measured in mg/L or mM, at least 5%, at least 10%, at least 15%, at
least 20%, at
least 25%, at least 30%, at least 35%, at least 40%, at least 45%, at least
50%, at least 60%,
at least 70%, at least 80%, at least 90%, at least 100%, at least 150% at
least 200%, at least
500%, or at least 1000% greater than an amount of the cannabinoid or the
cannabinoid
derivative produced in a method comprising use of a tetrahydrocannabinolic
acid synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 instead of the
engineered
variant of the disclosure. In certain such embodiments, the engineered variant
of the
disclosure and the tetrahydrocannabinolic acid synthase polypeptide having the
amino acid
sequence of SEQ ID NO:44 are used under similar conditions for the same length
of time.
[0398] In some embodiments of the methods of producing a cannabinoid or a
cannabinoid derivative of the disclosure, the cannabinoid is THCA and the
method produces
THCA in an increased ratio of THCA over another cannabinoid (e.g., CBCA)
compared to
that produced in a method comprising use of a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 instead of the
engineered
variant of the disclosure. In certain such embodiments, the engineered variant
of the
disclosure and the tetrahydrocannabinolic acid synthase polypeptide having the
amino acid
sequence of SEQ ID NO:44 are used under similar conditions for the same length
of time. In
some embodiments of the methods of producing a cannabinoid or a cannabinoid
derivative
of the disclosure, the cannabinoid is THCA and the method produces THCA from
CBGA in
a ratio of THCA over another cannabinoid (e.g., CBCA) of about 11:1, about
11.5:1, about
12:1, about 12.5:1, about 13:1, about 13.5:1, about 14:1, about 14.5:1, about
15:1, about
15.5:1, about 16:1, about 16.5:1, about 17:1, about 17.5:1, about 18:1, about
18.5:1, about
19:1, about 19.5:1, about 20:1, about 25:1, about 30:1, about 35:1, about
40:1, about 45:1,
about 50:1, about 60:1, about 70:1, about 80:1, about 90:1, about 100:1, about
150:1, about
200:1, about 500:1, or greater than about 500:1.
Methods of Using Host Cells to Generate Cannabinoids or Cannabinoid
Derivatives
[0399] The disclosure provides methods of producing a cannabinoid or a
cannabinoid derivative, such as those described herein, the method comprising:
culturing a
modified host cell of the disclosure in a culture medium. In certain such
embodiments, the
method comprises recovering the produced cannabinoid or cannabinoid
derivative. In
163
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
certain such embodiments, the produced cannabinoid or cannabinoid derivative
is then
purified as disclosed herein.
[0400] In some embodiments, culturing of the modified host cells of the
disclosure in
a culture medium provides for synthesis of a cannabinoid or a cannabinoid
derivative, such
as those described herein, in an increased amount compared to an unmodified
host cell
cultured under similar conditions.
[0401] The disclosure provides methods of producing a cannabinoid or a
cannabinoid derivative, such as those described herein, the method comprising:
culturing a
modified host cell of the disclosure in a culture medium comprising a
carboxylic acid. In
certain such embodiments, the method comprises recovering the produced
cannabinoid or
cannabinoid derivative. In certain such embodiments, the produced cannabinoid
or
cannabinoid derivative is then purified as disclosed herein.
[0402] In some embodiments, the cannabinoid or cannabinoid derivative is
recovered
from a cell lysate; from a culture medium; from a modified host cell; from
both the cell
lysate and the culture medium; from both the modified host cell and the
culture medium;
from the cell lysate, the modified host cell, and the culture medium. In
certain such
embodiments, the recovered cannabinoid or cannabinoid derivative is then
purified as
disclosed herein. In some embodiments when the cannabinoid or cannabinoid
derivative is
recovered from the cell lysate; from the culture medium; from the modified
host cell; from
both the cell lysate and the culture medium; from both the modified host cell
and the culture
medium; from the cell lysate, the modified host cell, and the culture medium;
or from the
cell-free reaction mixture comprising one or more polypeptides disclosed
herein, the
recovered cannabinoid or cannabinoid derivative is in the form of a salt. In
certain such
embodiments, the salt is a pharmaceutically acceptable salt. In some
embodiments, the salt
of the recovered cannabinoid or cannabinoid derivative is then purified as
disclosed herein.
[0403] In some embodiments, the modified host cell of the present
disclosure is
cultured in a culture medium comprising a carboxylic acid. In some
embodiments, the
carboxylic acid may be substituted with or comprise one or more functional
and/or reactive
groups. Functional groups may include, but are not limited to, azido, halo
(e.g., chloride,
bromide, iodide, fluorine), methyl, alkyl, alkynyl, alkenyl, methoxy, alkoxy,
acetyl, amino,
carboxyl, carbonyl, oxo, ester, hydroxyl, thio (e.g., thiol), cyano, aryl,
heteroaryl, cycloalkyl,
cycloalkenyl, cycloalkylalkenyl, cycloalkylalkynyl, cycloalkenylalkyl,
cycloalkenylalkenyl,
cycloalkenylalkynyl, heterocyclylalkenyl, heterocyclylalkynyl,
heteroarylalkenyl,
164
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
heteroarylalkynyl, arylalkenyl, arylalkynyl, spirocyclyl, heterospirocyclyl,
heterocyclyl,
thioalkyl (or alkylthio), arylthio, heteroarylthio, sulfone, sulfonyl,
sulfoxide, amido,
alkylamino, dialkylamino, arylamino, alkylarylamino, diarylamino, N-oxide,
imide,
enamine, imine, oxime, hydrazone, nitrile, aralkyl, cycloalkylalkyl,
haloalkyl,
heterocyclylalkyl, heteroarylalkyl, nitro, thioxo, and the like. Reactive
groups may include,
but are not necessarily limited to, azide, halogen, carboxyl, carbonyl, amine
(e.g., alkyl
amine (e.g., lower alkyl amine), aryl amine), ester (e.g., alkyl ester (e.g.,
lower alkyl ester,
benzyl ester), aryl ester, substituted aryl ester), cyano, thioester,
thioether, sulfonyl halide,
alcohol, thiol, succinimidyl ester, isothiocyanate, iodoacetamide, maleimide,
hydrazine,
alkynyl, alkenyl, and the like. In some embodiments, the reactive group is
selected from a
carboxyl, a carbonyl, an amine, an ester, thioester, thioether, a sulfonyl
halide, an alcohol, a
thiol, a succinimidyl ester, an isothiocyanate, an iodoacetamide, a maleimide,
an azide, an
alkyne, an alkene, and a hydrazine. Functional and reactive groups may be
unsubstituted or
substituted with one or more functional or reactive groups.
[0404] In some embodiments, the carboxylic acid is isotopically- or radio-
labeled. In
some embodiments, the carboxylic acid may be an enantiomer or disastereomer.
In some
embodiments the carboxylic acid may be the (S)-enantiomer. In some embodiments
the
carboxylic acid may be the (R)-enantiomer. In some embodiments, the carboxylic
acid may
be the (+) or (-) enantiomer. In some embodiments, the carboxylic acid may
include a
double bond or a fused ring. In certain such embodiments, the double bond or
fused ring
may be cis or trans, unless the configuration is specifically defined. If the
carboxylic acid
contains a double bond, the substituent may be in the E or Z configuration,
unless the
configuration is specifically defined.
[0405] In some embodiments, the carboxylic acid comprises a C=C group. In
some
embodiments, the carboxylic acid comprises an alkyne group. In some
embodiments, the
carboxylic acid comprises an N3 group. In some embodiments, the carboxylic
acid comprises
a halogen. In some embodiments, the carboxylic acid comprises a CN group. In
some
embodiments, the carboxylic acid comprises iodo. In some embodiments, the
carboxylic acid
comprises bromo. In some embodiments, the carboxylic acid comprises chloro. In
some
embodiments, the carboxylic acid comprises fluoro. In some embodiments, the
carboxylic
acid comprises a carbonyl. In some embodiments, the carboxylic acid comprises
an acetyl.
In some embodiments, the carboxylic acid comprises an alkyl group. In some
embodiments,
the carboxylic acid comprises an aryl group.
165
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0406] Carboxylic acids may include, but are not limited to,
unsubstituted or
substituted C3-Ci8 fatty acids, C3-Ci8 carboxylic acids, Ci-Cis carboxylic
acids, butyric
acid, isobutyric acid, valeric acid, hexanoic acid, heptanoic acid, octanoic
acid, nonanoic
acid, decanoic acid, undecanoic acid, lauric acid, myristic acid, Cis-CB fatty
acids, Cis-CB
carboxylic acids, fumaric acid, itaconic acid, malic acid, succinic acid,
maleic acid,
malonic acid, glutaric acid, glucaric acid, oxalic acid, adipic acid, pimelic
acid, suberic acid,
azelaic acid, sebacic acid, dodecanedioic acid, glutaconic acid, ortho-
phthalic acid,
isophthalic acid, terephthalic acid, citric acid, isocitric acid, aconitic
acid, tricarballylic acid,
and trimesic acid. Carboxylic acids may include unsubstituted or substituted
Ci-Cis
carboxylic acids. Carboxylic acids may include unsubstituted or substituted C3-
Ci8
carboxylic acids. Carboxylic acids may include unsubstituted or substituted C3-
Ci2
carboxylic acids. Carboxylic acids may include unsubstituted or substituted C4-
Cm
carboxylic acids. In some embodiments, the carboxylic acid is an unsubstituted
or
substituted C4 carboxylic acid. In some embodiments, the carboxylic acid is an
unsubstituted
or substituted Cs carboxylic acid. In some embodiments, the carboxylic acid is
an
unsubstituted or substituted C6 carboxylic acid. In some embodiments, the
carboxylic acid is
an unsubstituted or substituted C7 carboxylic acid. In some embodiments, the
carboxylic acid
is an unsubstituted or substituted Cs carboxylic acid. In some embodiments,
the carboxylic
acid is an unsubstituted or substituted C9 carboxylic acid. In some
embodiments, the
carboxylic acid is an unsubstituted or substituted Cio carboxylic acid. In
some embodiments,
the carboxylic acid is unsubstituted or substituted butyric acid. In some
embodiments,
carboxylic acid is unsubstituted or substituted valeric acid. In some
embodiments, the
carboxylic acid is unsubstituted or substituted hexanoic acid. In some
embodiments, the
carboxylic acid is unsubstituted or substituted heptanoic acid. In some
embodiments, the
carboxylic acid is unsubstituted or substituted octanoic acid. In some
embodiments, the
carboxylic acid is unsubstituted or substituted nonanoic acid. In some
embodiments, the
carboxylic acid is unsubstituted or substituted decanoic acid.
[0407] Carboxylic acids may include, but are not limited to, 2-
methylhexanoic acid,
3-methylhexanoic acid, 4-methylhexanoic acid, 5-methylhexanoic acid, 2-
hexenoic acid, 3-
hexenoic acid, 4-hexenoic acid, 5-hexenoic acid, 5-chlorovaleric acid, 5-
aminovaleric acid,
5-cyanovaleric acid, 5-(methylsulfanyl)valeric acid, 5-hydroxyvaleric acid, 5-
phenylvaleric
acid, 2,3-dimethylhexanoic acid, d3-hexanoic acid, 4-pentynoic acid, trans-2-
pentenoic acid,
5-hexynoic acid, trans-2-hexenoic acid, 6-heptynoic acid, trans-2-octenoic
acid, trans-2-
166
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
nonenoic acid, 4-phenylbutyric acid, 6-phenylhexanoic acid, 7-phenylyheptanoic
acid, and
the like. In some embodiments, the carboxylic acid is 2-methylhexanoic acid.
In some
embodiments, the carboxylic acid is 3-methylhexanoic acid. In some
embodiments, the
carboxylic acid is 4-methylhexanoic acid. In some embodiments, the carboxylic
acid is 5-
methylhexanoic acid. In some embodiments, the carboxylic acid is 2-hexenoic
acid. In
some embodiments, the carboxylic acid is 3-hexenoic acid. In some embodiments,
the
carboxylic acid is 4-hexenoic acid. In some embodiments, the carboxylic acid
is 5-hexenoic
acid. In some embodiments, the carboxylic acid is 5-chlorovaleric acid. In
some
embodiments, the carboxylic acid is 5-aminovaleric acid. In some embodiments,
the
carboxylic acid is 5-cyanovaleric acid. In some embodiments, the carboxylic
acid is 5-
(methylsulfanyl)valeric acid. In some embodiments, the carboxylic acid is 5-
hydroxyvaleric
acid. In some embodiments, the carboxylic acid is 5-phenylvaleric acid. In
some
embodiments, the carboxylic acid is 2,3-dimethylhexanoic acid. In some
embodiments, the
carboxylic acid is d3-hexanoic acid. In some embodiments, the carboxylic acid
is 4-
pentynoic acid. In some embodiments, the carboxylic acid is trans-2-pentenoic
acid. In some
embodiments, the carboxylic acid is 5-hexynoic acid. In some embodiments, the
carboxylic
acid is trans-2-hexenoic acid. In some embodiments, the carboxylic acid is 6-
heptynoic acid.
In some embodiments, the carboxylic acid is trans-2-octenoic acid. In some
embodiments,
the carboxylic acid is trans-2-nonenoic acid. In some embodiments, the
carboxylic acid is 4-
phenylbutyric acid. In some embodiments, the carboxylic acid is 6-
phenylhexanoic acid. In
some embodiments, the carboxylic acid is 7-phenylheptanoic acid.
[0408] In some embodiments wherein the modified host cell of the present
disclosure
is cultured in a culture medium comprising a carboxylic acid, the carboxylic
acid is an
unsubstituted or substituted C3-Ci8 carboxylic acid. In certain such
embodiments, the
unsubstituted or substituted C3-C18 carboxylic acid is an unsubstituted or
substituted
hexanoic acid. In some embodiments, the cannabinoid or cannabinoid derivative
is produced
in an amount of more than 100 mg/L culture medium. In some embodiments, the
cannabinoid or cannabinoid derivative is produced in an amount of more than 50
mg/L
culture medium.
[0409] In some embodiments wherein the modified host cell of the present
disclosure
is cultured in a culture medium comprising a carboxylic acid, the carboxylic
acid is butyric
acid, valeric acid, hexanoic acid, octanoic acid, 2-methylhexanoic acid, 3-
methylhexanoic
acid, 4-methylhexanoic acid, 5-methylhexanoic acid, 2-hexenoic acid, 3-
hexenoic acid, 4-
167
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
hexenoic acid, 5-hexenoic acid, heptanoic acid, 5-chlorovaleric acid, 5-
(methylsulfanyl)valeric acid, 4-pentynoic acid, trans-2-pentenoic acid, 5-
hexynoic acid,
trans-2-hexenoic acid, 6-heptynoic acid, trans-2-octenoic acid, nonanoic acid,
trans-2-
nonenoic acid, decanoic acid, undecanoic acid, dodecanoic acid, myristic acid,
4-
phenylbutyric acid, 5-phenylvaleric acid, 6-phenylhexanoic acid, 7-
phenylheptanoic acid,
isobutyric acid, fumaric acid, itaconic acid, malic acid, succinic acid,
maleic acid, malonic
acid, glutaric acid, glucaric acid, oxalic acid, adipic acid, pimelic acid,
suberic acid, azelaic
acid, sebacic acid, dodecandioic acid, glutaconic acid, ortho-phthalic acid,
isophthalic acid,
terephthalic acid, citric acid, isocitric acid, aconitic acid, tricarballylic
acid, trimesic acid, 5-
aminovaleric acid, 5-cyanovaleric acid, 5-hydroxyvaleric acid, or 2,3-
dimethylhexanoic acid.
In some embodiments, the cannabinoid or cannabinoid derivative is produced in
an amount
of more than 100 mg/L culture medium. In some embodiments, the cannabinoid or
cannabinoid derivative is produced in an amount of more than 50 mg/L culture
medium.
[0410] In some embodiments wherein the modified host cell of the present
disclosure
is cultured in a culture medium comprising a carboxylic acid, the carboxylic
acid is butyric
acid, valeric acid, hexanoic acid, octanoic acid, 2-methylhexanoic acid, 3-
methylhexanoic
acid, 4-methylhexanoic acid, 5-methylhexanoic acid, 2-hexenoic acid, 3-
hexenoic acid, 4-
hexenoic acid, 5-hexenoic acid, heptanoic acid, 5-chlorovaleric acid, 5-
(methylsulfanyl)valeric acid, 4-pentynoic acid, trans-2-pentenoic acid, 5-
hexynoic acid,
trans-2-hexenoic acid, 6-heptynoic acid, trans-2-octenoic acid, nonanoic acid,
trans-2-
nonenoic acid, decanoic acid, undecanoic acid, dodecanoic acid, myristic acid,
4-
phenylbutyric acid, 5-phenylvaleric acid, 6-phenylhexanoic acid, 7-
phenylheptanoic acid,
isobutyric acid, fumaric acid, succinic acid, maleic acid, malonic acid,
glutaric acid, oxalic
acid, adipic acid, pimelic acid, suberic acid, azelaic acid, sebacic acid,
dodecandioic acid,
ortho-phthalic acid, isophthalic acid, terephthalic acid, trimesic acid, 5-
aminovaleric acid, 5-
cyanovaleric acid, 5-hydroxyvaleric acid, or 2,3-dimethylhexanoic acid. In
some
embodiments, the cannabinoid or cannabinoid derivative is produced in an
amount of more
than 100 mg/L culture medium. In some embodiments, the cannabinoid or
cannabinoid
derivative is produced in an amount of more than 50 mg/L culture medium.
[0411] In some embodiments wherein the modified host cell of the present
disclosure
is cultured in a culture medium comprising a carboxylic acid, the carboxylic
acid is 2-
methylhexanoic acid, 3-methylhexanoic acid, 4-methylhexanoic acid, 5-
methylhexanoic
acid, 2-hexenoic acid, 3-hexenoic acid, 4-hexenoic acid, heptanoic acid, 5-
chlorovaleric
168
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
acid, 5-(methylsulfanyl)valeric acid, 4-pentynoic acid, trans-2-pentenoic
acid, 5-hexynoic
acid, trans-2-hexenoic acid, 6-heptynoic acid, trans-2-octenoic acid, nonanoic
acid, trans-2-
nonenoic acid, decanoic acid, undecanoic acid, dodecanoic acid, myristic acid,
4-
phenylbutyric acid, 5-phenylvaleric acid, 6-phenylhexanoic acid, 7-
phenylheptanoic acid,
isobutyric acid, fumaric acid, itaconic acid, malic acid, maleic acid,
glucaric acid, suberic
acid, azelaic acid, sebacic acid, dodecandioic acid, glutaconic acid, ortho-
phthalic acid,
isophthalic acid, terephthalic acid, citric acid, isocitric acid, aconitic
acid, tricarballylic acid,
trimesic acid, 5-aminovaleric acid, 5-cyanovaleric acid, 5-hydroxyvaleric
acid, or 2,3-
dimethylhexanoic acid. In some embodiments, the cannabinoid or cannabinoid
derivative is
produced in an amount of more than 100 mg/L culture medium. In some
embodiments, the
cannabinoid or cannabinoid derivative is produced in an amount of more than 50
mg/L
culture medium.
[0412] In some embodiments wherein the modified host cell of the present
disclosure
is cultured in a culture medium comprising a carboxylic acid, the carboxylic
acid is 2-
methylhexanoic acid, 3-methylhexanoic acid, 4-methylhexanoic acid, 5-
methylhexanoic
acid, 2-hexenoic acid, 3-hexenoic acid, 4-hexenoic acid, heptanoic acid, 5-
chlorovaleric
acid, 5-(methylsulfanyl)valeric acid, 4-pentynoic acid, trans-2-pentenoic
acid, 5-hexynoic
acid, trans-2-hexenoic acid, 6-heptynoic acid, trans-2-octenoic acid, nonanoic
acid, trans-2-
nonenoic acid, decanoic acid, undecanoic acid, dodecanoic acid, myristic acid,
4-
phenylbutyric acid, 5-phenylvaleric acid, 6-phenylhexanoic acid, 7-
phenylheptanoic acid,
isobutyric acid, fumaric acid, maleic acid, suberic acid, azelaic acid,
sebacic acid,
dodecandioic acid, ortho-phthalic acid, isophthalic acid, terephthalic acid,
trimesic acid, 5-
aminovaleric acid, 5-cyanovaleric acid, 5-hydroxyvaleric acid, or 2,3-
dimethylhexanoic acid.
In some embodiments, the cannabinoid or cannabinoid derivative is produced in
an amount
of more than 100 mg/L culture medium. In some embodiments, the cannabinoid or
cannabinoid derivative is produced in an amount of more than 50 mg/L culture
medium.
[0413] In some embodiments wherein the modified host cell of the present
disclosure
is cultured in a culture medium comprising a carboxylic acid, the carboxylic
acid is 4-
pentynoic acid, trans-2-pentenoic acid, 5-hexynoic acid, trans-2-hexenoic
acid, 6-heptynoic
acid, trans-2-octenoic acid, nonanoic acid, trans-2-nonenoic acid, decanoic
acid, undecanoic
acid, dodecanoic acid, 4-phenylbutyric acid, 5-phenylvaleric acid, 6-
phenylhexanoic acid, or
7-phenylheptanoic acid. In some embodiments, the cannabinoid or cannabinoid
derivative is
produced in an amount of more than 100 mg/L culture medium. In some
embodiments, the
169
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
cannabinoid or cannabinoid derivative is produced in an amount of more than 50
mg/L
culture medium.
[0414] In some embodiments wherein the modified host cell of the present
disclosure
is cultured in a culture medium comprising a carboxylic acid, the carboxylic
acid is 2-
methylhexanoic acid, 4-methylhexanoic acid, 5-methylhexanoic acid, 2-hexenoic
acid, 3-
hexenoic acid, 4-hexenoic acid, heptanoic acid, 5-chlorovaleric acid, or 5-
(methylsulfanyl)valeric acid. In some embodiments, the cannabinoid or
cannabinoid
derivative is produced in an amount of more than 100 mg/L culture medium. In
some
embodiments, the cannabinoid or cannabinoid derivative is produced in an
amount of more
than 50 mg/L culture medium.
[0415] The disclosure also provides methods of producing a cannabinoid or
a
cannabinoid derivative, such as those described herein, the method comprising:
culturing a
modified host cell of the disclosure in a culture medium comprising olivetolic
acid or an
olivetolic acid derivative. In certain such embodiments, the method comprises
recovering the
produced cannabinoid or cannabinoid derivative. In certain such embodiments,
the produced
cannabinoid or cannabinoid derivative is then purified as disclosed herein.
[0416] Olivetolic acid derivatives used herein may be substituted with or
comprise
one or more reactive and/or functional groups as disclosed herein. In some
embodiments, an
olivetolic acid derivative may lack one or more chemical moieties found in
olivetolic acid. In
some embodiments when the culture medium comprises an olivetolic acid
derivative, the
olivetolic acid derivative is orsellinic acid. In some embodiments when the
culture medium
comprises an olivetolic acid derivative, the olivetolic acid derivative is
divarinic acid. In
some embodiments, the cannabinoid or cannabinoid derivative is produced in an
amount of
more than 100 mg/L culture medium. In some embodiments, the cannabinoid or
cannabinoid
derivative is produced in an amount of more than 50 mg/L culture medium.
[0417] The disclosure provides methods of using a modified host cell of
the
disclosure for producing a cannabinoid or cannabinoid derivative. In some
embodiments of
the methods of using a modified host cell of the disclosure for producing a
cannabinoid or
cannabinoid derivative, the cannabinoid or the cannabinoid derivative is
produced in an
amount, as measured in mg/L or mM, greater than an amount of the cannabinoid
or the
cannabinoid derivative produced in a method instead comprising culturing a
modified host
cell comprising one or more nucleic acids comprising a nucleotide sequence
encoding a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
170
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NO:44, but lacking a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant. In certain such embodiments, the modified host cell of the disclosure
and the
modified host cell comprising one or more nucleic acids comprising the
nucleotide sequence
encoding the tetrahydrocannabinolic acid synthase polypeptide having the amino
acid
sequence of SEQ ID NO:44, but lacking a nucleic acid comprising a nucleotide
sequence
encoding an engineered variant, are cultured under similar culture conditions
for the same
length of time.
[0418] In some embodiments of the methods of using a modified host cell
of the
disclosure for producing a cannabinoid or cannabinoid derivative, the
cannabinoid or the
cannabinoid derivative is produced in an amount, as measured in mg/L or mM, at
least 5%,
at least 10%, at least 15%, at least 20%, at least 25%, at least 30%, at least
35%, at least
40%, at least 45%, at least 50%, at least 60%, at least 70%, at least 80%, at
least 90%, at
least 100%, at least 150% at least 200%, at least 500%, or at least 1000%
greater than an
amount of the cannabinoid or the cannabinoid derivative produced in a method
instead
comprising culturing a modified host cell comprising one or more nucleic acids
comprising a
nucleotide sequence encoding a tetrahydrocannabinolic acid synthase
polypeptide having an
amino acid sequence of SEQ ID NO:44, but lacking a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant. In certain such embodiments, the
modified host
cell of the disclosure and the modified host cell comprising one or more
nucleic acids
comprising the nucleotide sequence encoding the tetrahydrocannabinolic acid
synthase
polypeptide having the amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant, are cultured
under similar
culture conditions for the same length of time.
[0419] In some embodiments of the methods of using a modified host cell
of the
disclosure for producing a cannabinoid or cannabinoid derivative, the
cannabinoid is THCA
and the method produces THCA in an increased ratio of THCA over another
cannabinoid
(e.g., CBCA) compared to that produced in a method instead comprising
culturing a
modified host cell comprising one or more nucleic acids comprising a
nucleotide sequence
encoding a tetrahydrocannabinolic acid synthase polypeptide having an amino
acid sequence
of SEQ ID NO:44, but lacking a nucleic acid comprising a nucleotide sequence
encoding an
engineered variant, grown under similar culture conditions for the same length
of time. In
some embodiments of the methods of using a modified host cell of the
disclosure for
producing a cannabinoid or cannabinoid derivative, the cannabinoid is THCA and
the
171
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
method produces THCA from CBGA in a ratio of THCA over another cannabinoid
(e.g.,
CBCA) of about 11:1, about 11.5:1, about 12:1, about 12.5:1, about 13:1, about
13.5:1,
about 14:1, about 14.5:1, about 15:1, about 15.5:1, about 16:1, about 16.5:1,
about 17:1,
about 17.5:1, about 18:1, about 18.5:1, about 19:1, about 19.5:1, about 20:1,
about 25:1,
about 30:1, about 35:1, about 40:1, about 45:1, about 50:1, about 60:1, about
70:1, about
80:1, about 90:1, about 100:1, about 150:1, about 200:1, about 500:1, or
greater than about
500:1.
Exemplary Cell Culture Conditions
[0420] Suitable media for culturing modified host cells of the disclosure
may include
standard culture media (e.g., Luria-Bertani broth, optionally supplemented
with one or more
additional agents, such as an inducer (e.g., where nucleic acids disclosed
herein are under the
control of an inducible promoter, etc.); standard yeast culture media; and the
like). In some
embodiments, the culture medium can be supplemented with a fermentable sugar
(e.g., a
hexose sugar, e.g., glucose, xylose, and the like). Sugars fermentable by
yeast may include,
but are not limited to, sucrose, dextrose, glucose, fructose, mannose,
galactose, and maltose.
[0421] In some embodiments, the culture medium can be supplemented with
unsubstituted or substituted hexanoate, carboxylic acids other than
unsubstituted or
substituted hexanoate, olivetolic acid, or olivetolic acid derivatives. In
some embodiments,
the culture medium can be supplemented with pretreated cellulosic feedstock
(e.g., wheat
grass, wheat straw, barley straw, sorghum, rice grass, sugarcane straw,
bagasse, switchgrass,
corn stover, corn fiber, grains, or any combination thereof). In some
embodiments, the
culture medium can be supplemented with oleic acid. In some embodiments, the
culture
medium comprises a non-fermentable carbon source. In certain such embodiments,
the non-
fermentable carbon source comprises ethanol. In some embodiments, the suitable
media
comprises an inducer. In certain such embodiments, the inducer comprises
galactose. In
some embodiments, the inducer comprises KH2PO4, galactose, glucose, sucrose,
maltose, an
amino acid (e.g., methionine, lysine), CuSO4, a change in temperature (e.g.,
30 C to 37 C),
a change in pH (e.g., pH 6 to pH 4), a change in oxygen level (e.g., 20% to 1%
dissolved
oxygen levels), addition of hydrogen peroxide or superoxide-generating drug
menadione,
tunicamycin, expression of proteins prone to misfolding (e.g. cannabinoid
synthases),
estradiol, or doxycycline. Additional induction systems are detailed herein.
172
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0422] The carbon source in the suitable media can vary significantly,
from simple
sugars like glucose to more complex hydrolysates of other biomass, such as
yeast extract.
The addition of salts generally provide essential elements such as magnesium,
nitrogen,
phosphorus, and sulfur to allow the cells to synthesize polypeptides and
nucleic acids. The
suitable media can also be supplemented with selective agents, such as
antibiotics, to select
for the maintenance of certain plasmids and the like. For example, if a
microorganism is
resistant to a certain antibiotic, such as ampicillin or tetracycline, then
that antibiotic can be
added to the medium in order to prevent cells lacking the resistance from
growing. The
suitable media can be supplemented with other compounds as necessary to select
for desired
physiological or biochemical characteristics, such as particular amino acids
and the like.
[0423] In some embodiments, modified host cells disclosed herein are
grown in
minimal medium. As used herein, the terms "minimal medium" may refer to growth
medium containing the minimum nutrients possible for cell growth, generally,
but not
always, without the presence of one or more amino acids (e.g., 1, 2, 3, 4, 5,
6, 7, 8, 9, 10, or
more amino acids). Minimal medium typically contains: (1) a carbon source for
cellular (e.g.
bacterial or yeast) growth; (2) various salts, which can vary among cellular
(e.g. bacterial or
yeast) species and growing conditions; and (3) water.
[0424] In some embodiments, modified host cells disclosed herein are
grown in rich
medium. In certain such embodiments, the rich medium or rich media comprises
yeast
extract peptone dextrose (YPD) media comprising water, 10 g/L yeast extract,
20 g/L Bacto
peptone, and 20 g/L dextrose (glucose). In some embodiments, the rich medium
or rich
media comprises YP + 20 g/L galactose and 1 g/L glucose. In some embodiments,
the rich
medium or rich media comprises a carboxylic acid (e.g., 1 mM olivetolic acid,
1 mM
olivetolic acid derivative, 2 mM unsubstituted or substituted hexanoic acid,
or 2 mM of a
carboxylic acid other than unsubstituted or substituted hexanoic acid). In
some
embodiments, rich medium or rich media affords more rapid cell growth compared
to
minimal media or minimal medium.
[0425] Materials and methods suitable for the maintenance and growth of
the
recombinant cells of the disclosure are described herein, e.g., in the
Examples section. Other
materials and methods suitable for the maintenance and growth of cell (e.g.
bacterial or
yeast) cultures are well known in the art. Exemplary techniques can be found
in International
Publication No. W02009/076676, U.S. Patent Application No. 12/335,071 (U.S.
Publ. No.
2009/0203102), WO 2010/003007, US Publ. No. 2010/0048964, WO 2009/132220, US
173
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Pub!. No. 2010/0003716, Manual of Methods for General Bacteriology Gerhardt et
al, eds),
American Society for Microbiology, Washington, D.C. (1994) or Brock in
Biotechnology: A
Textbook of Industrial Microbiology, Second Edition (1989) Sinauer Associates,
Inc.,
Sunderland, MA.
[0426] Standard cell culture conditions can be used to culture the
modified host cells
disclosed herein (see, for example, WO 2004/033646 and references cited
therein). In some
embodiments, cells are grown and maintained at an appropriate temperature, gas
mixture,
and pH (such as at about 20 C to about 37 C, at about 0.04% to about 84%
CO2, at about
0% to about 100% dissolved oxygen, and at a pH between about 2 to about 9). In
some
embodiments, modified host cells disclosed herein are grown at about 34 C in
a suitable cell
culture medium. In some embodiments, modified host cells disclosed herein are
grown at
about 20 C to about 37 C in a suitable cell culture medium. While the growth
optimum for
S. cerevisiae is about 30 C, culturing cells at a higher temperature, e.g. 34
C may be
advantageous by reducing the costs to cool industrial fermentation tanks. In
some
embodiments, modified host cells disclosed herein are grown at about 20 C,
about 21 C,
about 22 C, about 23 C, about 24 C, about 25 C, about 26 C, about 27 C,
about 28 C,
about 29 C, about 30 C, about 31 C, about 32 C, about 33 C, about 34 C,
about 35 C,
about 36 C, or about 37 C in a suitable cell culture medium. In some
embodiments, the pH
ranges for fermentation are between about pH 3.0 to about pH 9.0 (such as
about pH 3.0,
about pH 3.5, about pH 4.0, about pH 4.5, about pH 5.0, about pH 5.5, about pH
6.0, about
pH 6.5, about pH 7.0, about pH 7.5, about pH 8.0, about pH 8.5, about pH 6.0
to about pH
8.0 or about 6.5 to about 7.0). In some embodiments, the pH ranges for
fermentation are
between about pH 4.5 to about pH 5.5. In some embodiments, the pH ranges for
fermentation are between about pH 4.0 to about pH 6Ø In some embodiments,
the pH
ranges for fermentation are between about pH 3.0 to about pH 6Ø In some
embodiments,
the pH ranges for fermentation are between about pH 3.0 to about pH 5.5. In
some
embodiments, the pH ranges for fermentation are between about pH 3.0 to about
pH 5Ø In
some embodiments, the dissolved oxygen is between about 0% to about 10%, about
0% to
about 20%, about 0% to about 30%, about 0% to about 40%, about 0% to about
50%, about
0% to about 60%, about 0% to about 70%, about 0% to about 80%, about 0% to
about 90%,
about 5% to about 10%, about 5% to about 20%, about 5% to about 30%, about 5%
to about
40%, about 5% to about 50%, about 5% to about 60%, about 5% to about 70%,
about 5% to
about 80%, about 5% to about 90%, about 10% to about 20%, about 10% to about
30%,
174
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
about 10% to about 40% or about 10% to about 500o. In some embodiments, the
CO2 level
is between about 0.04% to about 0.1% CO2, about 0.04% to about 1% CO2, about
0.04% to
about 500 CO2, about 0.04 A to about 10% CO2, about 0.04 A to about 20 A CO2,
about
0.04 A to about 30 A CO2, about 0.04 A to about 40 A CO2, about 0.04 A to
about 5000 CO2,
about 0.04% to about 60 A CO2, about 0.04% to about 70 A CO2, about 0.1% to
about 5%
CO2, about 0.1% to about 10% CO2, about 0.1% to about 20 A CO2, about 0.1% to
about
30 A CO2, about 0.1 A to about 40 A CO2, about 0.1 A to about 5000 CO2, about
1 A to about
50 CO2, about 1% to about 10% CO2, about 1% to about 20 A CO2, about 1% to
about 30 A
CO2, about 1% to about 40 A CO2, about 1% to about 50% CO2, about 5% to about
10%
CO2, about 10% to about 20 A CO2, about 10% to about 30 A CO2, about 10% to
about 40 A
CO2, about 10% to about 50% CO2, about 10% to about 60 A CO2, about 10% to
about 70 A
CO2, about 10% to about 80 A CO2, about 50% to about 60 A CO2, about 50% to
about 70 A
CO2, or about 50% to about 80 A CO2. Modified host cells disclosed herein
disclosed herein
can be grown under aerobic, anoxic, microaerobic, or anaerobic conditions
based on the
requirements of the cells.
[0427] Standard culture conditions and modes of fermentation, such as
batch, fed-
batch, or continuous fermentation that can be used are described in
International Publication
No. WO 2009/076676, U.S. Patent Application No. 12/335,071 (U.S. Publ. No.
2009/0203102), WO 2010/003007, US Publ. No. 2010/0048964, WO 2009/132220, US
Publ. No. 2010/0003716, the contents of each of which are incorporated by
reference herein
in their entireties. Batch and Fed-Batch fermentations are common and well
known in the art
and examples can be found in Brock, Biotechnology: A Textbook of Industrial
Microbiology, Second Edition (1989) Sinauer Associates, Inc.
Production and Recovery of Produced Cannabinoids or Cannabinoid Derivatives
[0428] The present disclosure provides for production of a cannabinoid or
a
cannabinoid derivative in an amount. In some embodiments, a method of the
present
disclosure provides for production of a cannabinoid or a cannabinoid
derivative, such as
those disclosed herein, by modified host cells of the disclosure in an amount
of from about 1
mg/L culture medium to about 1 g/L culture medium. In some embodiments, a
method of the
present disclosure provides for production of a cannabinoid or a cannabinoid
derivative in an
amount of from about 1 mg/L culture medium to about 500 mg/L culture medium.
In some
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
175
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
a cannabinoid derivative in an amount of from about 1 mg/L culture medium to
about 100
mg/L culture medium. For example, in some embodiments, a method of the present
disclosure provides for production of a cannabinoid or a cannabinoid
derivative in an amount
of from about 1 mg/L culture medium to about 5 mg/L culture medium, from about
5 mg/L
culture medium to about 10 mg/L culture medium, from about 10 mg/L culture
medium to
about 25 mg/L culture medium, from about 25 mg/L culture medium to about 50
mg/L
culture medium, from about 50 mg/L culture medium to about 75 mg/L culture
medium, or
from about 75 mg/L culture medium to about 100 mg/L culture medium. In some
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
a cannabinoid derivative in an amount of from about 100 mg/L culture medium to
about 150
mg/L culture medium, from about 150 mg/L culture medium to about 200 mg/L
culture
medium, from about 200 mg/L culture medium to about 250 mg/L culture medium,
from
about 250 mg/L culture medium to about 500 mg/L culture medium, from about 500
mg/L
culture medium to about 750 mg/L culture medium, or from about 750 mg/L
culture medium
to about 1 g/L culture medium. In some embodiments, a method of the present
disclosure
provides for production of a cannabinoid or a cannabinoid derivative in an
amount of from
about from about 50 mg/L culture medium to about 100 mg/L culture medium, 50
mg/L
culture medium to about 150 mg/L culture medium, from about 50 mg/L culture
medium to
about 200 mg/L culture medium, from about 50 mg/L culture medium to about 250
mg/L
culture medium, from about 50 mg/L culture medium to about 500 mg/L culture
medium, or
from about 50 mg/L culture medium to about 750 mg/L culture medium.
[0429] In some embodiments, a method of the present disclosure provides
for
production of a cannabinoid or a cannabinoid derivative, such as those
disclosed herein, in
an amount of from about 50 mg/L culture medium to about 100 g/L culture
medium, or more
than 100 g/L culture medium. In some embodiments, a method of the present
disclosure
provides for production of a cannabinoid or a cannabinoid derivative, such as
those disclosed
herein, in an amount of from about 50 mg/L culture medium to about 100 mg/L
culture
medium, or more than 100 mg/L culture medium. In some embodiments, a method of
the
present disclosure provides for production of a cannabinoid or a cannabinoid
derivative, such
as those disclosed herein, in an amount of more than 50 mg/L culture medium.
In some
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
a cannabinoid derivative, such as those disclosed herein, in an amount of more
than 100
mg/L culture medium. In some embodiments, a method of the present disclosure
provides
176
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
for production of a cannabinoid or a cannabinoid derivative in an amount of
from about 100
mg/L culture medium to about 500 mg/L culture medium, or more than 500 mg/L
culture
medium. In some embodiments, a method of the present disclosure provides for
production
of a cannabinoid or a cannabinoid derivative in an amount of from about 500
mg/L culture
medium to about 1 g/L culture medium, or more than 1 g/L culture medium. In
some
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
a cannabinoid derivative in an amount of from about 1 g/L culture medium to
about 10 g/L
culture medium, or more than 10 g/L culture medium. In some embodiments, a
method of
the present disclosure provides for production of a cannabinoid or a
cannabinoid derivative
in an amount of from about 10 g/L culture medium to about 100 g/L culture
medium, or
more than 100 g/L culture medium. In some embodiments, a method of the present
disclosure provides for production of a cannabinoid or a cannabinoid
derivative in an amount
of from about 1 g/L culture medium to about 20 g/L culture medium, or more
than 20 g/L
culture medium. In some embodiments, a method of the present disclosure
provides for
production of a cannabinoid or a cannabinoid derivative in an amount of from
about 1 g/L
culture medium to about 30 g/L culture medium, or more than 30 g/L culture
medium. In
some embodiments, a method of the present disclosure provides for production
of a
cannabinoid or a cannabinoid derivative in an amount of from about 1 g/L
culture medium to
about 40 g/L culture medium, or more than 40 g/L culture medium. In some
embodiments, a
method of the present disclosure provides for production of a cannabinoid or a
cannabinoid
derivative in an amount of from about 1 g/L culture medium to about 50 g/L
culture
medium, or more than 50 g/L culture medium. In some embodiments, a method of
the
present disclosure provides for production of a cannabinoid or a cannabinoid
derivative in an
amount of from about 1 g/L culture medium to about 60 g/L culture medium, or
more than
60 g/L culture medium. In some embodiments, a method of the present disclosure
provides
for production of a cannabinoid or a cannabinoid derivative in an amount of
from about 1
g/L culture medium to about 70 g/L culture medium, or more than 70 g/L culture
medium.
In some embodiments, a method of the present disclosure provides for
production of a
cannabinoid or a cannabinoid derivative in an amount of from about 1 g/L
culture medium to
about 80 g/L culture medium, or more than 80 g/L culture medium. In some
embodiments, a
method of the present disclosure provides for production of a cannabinoid or a
cannabinoid
derivative in an amount of from about 1 g/L culture medium to about 90 g/L
culture
medium, or more than 90 g/L culture medium. In some embodiments, a method of
the
177
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
present disclosure provides for production of a cannabinoid or a cannabinoid
derivative in an
amount of from about 10 g/L culture medium to about 20 g/L culture medium, or
more than
20 g/L culture medium. In some embodiments, a method of the present disclosure
provides
for production of a cannabinoid or a cannabinoid derivative in an amount of
from about 10
g/L culture medium to about 30 g/L culture medium, or more than 30 g/L culture
medium. In
some embodiments, a method of the present disclosure provides for production
of a
cannabinoid or a cannabinoid derivative in an amount of from about 10 g/L
culture medium
to about 40 g/L culture medium, or more than 40 g/L culture medium. In some
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
a cannabinoid derivative in an amount of from about 10 g/L culture medium to
about 50 g/L
culture medium, or more than 50 g/L culture medium. In some embodiments, a
method of
the present disclosure provides for production of a cannabinoid or a
cannabinoid derivative
in an amount of from about 10 g/L culture medium to about 60 g/L culture
medium, or more
than 60 g/L culture medium. In some embodiments, a method of the present
disclosure
provides for production of a cannabinoid or a cannabinoid derivative in an
amount of from
about 10 g/L culture medium to about 70 g/L culture medium, or more than 70
g/L culture
medium. In some embodiments, a method of the present disclosure provides for
production
of a cannabinoid or a cannabinoid derivative in an amount of from about 10 g/L
culture
medium to about 80 g/L culture medium, or more than 80 g/L culture medium. In
some
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
a cannabinoid derivative in an amount of from about 10 g/L culture medium to
about 90 g/L
culture medium, or more than 90 g/L culture medium. In some embodiments, a
method of
the present disclosure provides for production of a cannabinoid or a
cannabinoid derivative
in an amount of from about 50 g/L culture medium to about 100 g/L culture
medium, or
more than 100 g/L culture medium. In some embodiments, a method of the present
disclosure provides for production of a cannabinoid or a cannabinoid
derivative in an amount
of from about 50 g/L culture medium to about 60 g/L culture medium, or more
than 60 g/L
culture medium. In some embodiments, a method of the present disclosure
provides for
production of a cannabinoid or a cannabinoid derivative in an amount of from
about 50 g/L
culture medium to about 70 g/L culture medium, or more than 70 g/L culture
medium. In
some embodiments, a method of the present disclosure provides for production
of a
cannabinoid or a cannabinoid derivative in an amount of from about 50 g/L
culture medium
to about 80 g/L culture medium, or more than 80 g/L culture medium. In some
178
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
a cannabinoid derivative in an amount of from about 50 g/L culture medium to
about 90 g/L
culture medium, or more than 90 g/L culture medium. In some embodiments, a
method of
the present disclosure provides for production of a cannabinoid or a
cannabinoid derivative
in an amount of from about 20 g/L culture medium to about 100 g/L culture
medium, or
more than 100 g/L culture medium. In some embodiments, a method of the present
disclosure provides for production of a cannabinoid or a cannabinoid
derivative in an amount
of from about 20 g/L culture medium to about 30 g/L culture medium, or more
than 30 g/L
culture medium. In some embodiments, a method of the present disclosure
provides for
production of a cannabinoid or a cannabinoid derivative in an amount of from
about 20 g/L
culture medium to about 40 g/L culture medium, or more than 40 g/L culture
medium. In
some embodiments, a method of the present disclosure provides for production
of a
cannabinoid or a cannabinoid derivative in an amount of from about 20 g/L
culture medium
to about 50 g/L culture medium, or more than 50 g/L culture medium. In some
embodiments, a method of the present disclosure provides for production of a
cannabinoid or
a cannabinoid derivative in an amount of from about 20 g/L culture medium to
about 60 g/L
culture medium, or more than 60 g/L culture medium. In some embodiments, a
method of
the present disclosure provides for production of a cannabinoid or a
cannabinoid derivative
in an amount of from about 20 g/L culture medium to about 70 g/L culture
medium, or more
than 70 g/L culture medium. In some embodiments, a method of the present
disclosure
provides for production of a cannabinoid or a cannabinoid derivative in an
amount of from
about 20 g/L culture medium to about 80 g/L culture medium, or more than 80
g/L culture
medium. In some embodiments, a method of the present disclosure provides for
production
of a cannabinoid or a cannabinoid derivative in an amount of from about 20 g/L
culture
medium to about 90 g/L culture medium, or more than 90 g/L culture medium.
[0430] In some embodiments, the modified host cell disclosed herein is
cultured in a
liquid medium comprising a carboxylic acid, olivetolic acid, or an olivetolic
acid derivative.
[0431] In some embodiments, a method of producing a cannabinoid or a
cannabinoid
derivative, such as those disclosed herein, may involve culturing a modified
yeast cell of the
present disclosure under conditions that favor production of a cannabinoid or
a cannabinoid
derivative; wherein the cannabinoid or the cannabinoid derivative is produced
by the
modified yeast cell and is present in the culture medium (e.g., a liquid
culture medium) in
which the modified yeast cell is cultured. In some embodiments, the culture
medium in
179
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
which the modified yeast cell is cultured comprises a cannabinoid or a
cannabinoid
derivative in an amount of from 1 ng/L to 1 g/L (e.g., from 1 ng/L to 50 ng/L,
from 50 ng/L
to 100 ng/L, from 100 ng/L to 500 ng/L, from 500 ng/L to 1 [tg/L, from 1 [tg/L
to 50 [tg/L,
from 50 [tg/L to 100 [tg/L, from 100 [tg/L to 500 [tg/L, from 500 [tg/L to 1
mg/L, from 1
mg/L to 50 mg/L, from 50 mg/L to 100 mg/L, from 100 mg/L to 500 mg/L, or from
500
mg/L to 1 g/L). In certain such embodiments, the modified yeast cell is a
modified S.
cerevisiae. In some embodiments, the culture medium in which the modified
yeast cell is
cultured comprises a cannabinoid or a cannabinoid derivative in an amount from
50 mg/L to
100 mg/L. In certain such embodiments, the modified yeast cell is a modified
S. cerevisiae.
In some embodiments, the culture medium in which the modified yeast cell is
cultured
comprises a cannabinoid or a cannabinoid derivative in an amount from 100 mg/L
to 500
mg/L. In certain such embodiments, the modified yeast cell is a modified S.
cerevisiae. In
some embodiments, the culture medium in which the modified yeast cell is
cultured
comprises a cannabinoid or a cannabinoid derivative in an amount from 500 mg/L
to 1 g/L.
In certain such embodiments, the modified yeast cell is a modified S.
cerevisiae. In some
embodiments, the culture medium in which the modified yeast cell is cultured
comprises a
cannabinoid or a cannabinoid derivative in an amount more than 1 g/L. In
certain such
embodiments, the modified yeast cell is a modified S. cerevisiae.
[0432] In some embodiments, a method of producing a cannabinoid or a
cannabinoid
derivative, such as those disclosed herein, may involve culturing a modified
yeast cell of the
present disclosure under conditions that favor fermentation of a sugar, and
under conditions
that favor production of a cannabinoid or a cannabinoid derivative; wherein
the cannabinoid
or the cannabinoid derivative is produced by the modified yeast cell and is
present in alcohol
produced by the modified yeast cell. The present disclosure provides an
alcoholic beverage
produced by the modified yeast cell, where the alcoholic beverage comprises
the
cannabinoid or cannabinoid derivative produced by the modified yeast cell.
Alcoholic
beverages may include beer, wine, and distilled alcoholic beverages. In some
embodiments,
an alcoholic beverage of the present disclosure comprises a cannabinoid or a
cannabinoid
derivative in an amount of from 1 ng/L to 1 g/L (e.g., from 1 ng/L to 50 ng/L,
from 50 ng/L
to 100 ng/L, from 100 ng/L to 500 ng/L, from 500 ng/L to 1 [tg/L, from 1 [tg/L
to 50 [tg/L,
from 50 [tg/L to 100 [tg/L, from 100 [tg/L to 500 [tg/L, from 500 [tg/L to 1
mg/L, from 1
mg/L to 50 mg/L, from 50 mg/L to 100 mg/L, from 100 mg/L to 500 mg/L, or from
500
180
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
mg/L to 1 g/L). In some embodiments, an alcoholic beverage of the present
disclosure
comprises a cannabinoid or a cannabinoid derivative in an amount more than 1
g/L.
[0433] The present disclosure provides a beverage produced by the
modified yeast
cell, where the beverage comprises the cannabinoid or cannabinoid derivative,
such as those
disclosed herein, produced by the modified yeast cell. In some embodiments, a
beverage of
the present disclosure comprises a cannabinoid or a cannabinoid derivative in
an amount of
from 1 ng/L to 1 g/L (e.g., from 1 ng/L to 50 ng/L, from 50 ng/L to 100 ng/L,
from 100 ng/L
to 500 ng/L, from 500 ng/L to 1 [tg/L, from 1 [tg/L to 50 [tg/L, from 50 [tg/L
to 100 [tg/L,
from 100 [tg/L to 500 [tg/L, from 500 [tg/L to 1 mg/L, from 1 mg/L to 50 mg/L,
from 50
mg/L to 100 mg/L, from 100 mg/L to 500 mg/L, or from 500 mg/L to 1 g/L). In
some
embodiments, a beverage of the present disclosure comprises a cannabinoid or a
cannabinoid
derivative in an amount more than 1 g/L. In some embodiments, a beverage of
the present
disclosure is non-alcoholic.
[0434] In some embodiments, a method of the present disclosure provides
for
increased production of a cannabinoid or a cannabinoid derivative, such as
those disclosed
herein. In certain such embodiments, culturing of the modified host cell
disclosed herein in
a culture medium provides for synthesis of a cannabinoid or a cannabinoid
derivative in an
increased amount compared to an unmodified host cell cultured under similar
conditions.
The production of a cannabinoid or a cannabinoid derivative by the modified
host cells
disclosed herein may be increased by about 5% to about 1,000,000 folds
compared to an
unmodified host cell cultured under similar conditions. The production of a
cannabinoid or a
cannabinoid derivative by the modified host cells disclosed herein may be
increased by
about 10% to about 1,000,000 folds (e.g., about 50% to about 1,000,000 folds,
about 1 to
about 500,000 folds, about 1 to about 50,000 folds, about 1 to about 5,000
folds, about 1 to
about 1,000 folds, about 1 to about 500 folds, about 1 to about 100 folds,
about 1 to about 50
folds, about 5 to about 100,000 folds, about 5 to about 10,000 folds, about 5
to about 1,000
folds, about 5 to about 500 folds, about 5 to about 100 folds, about 10 to
about 50,000 folds,
about 50 to about 10,000 folds, about 100 to about 5,000 folds, about 200 to
about 1,000
folds, about 50 to about 500 folds, or about 50 to about 200 folds) compared
to the
production of a cannabinoid or a cannabinoid derivative by unmodified host
cells cultured
under similar conditions. The production of a cannabinoid or a cannabinoid
derivative by
modified host cells disclosed herein may also be increased by at least about
any of 10%,
20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 1 fold, 2 folds, 5 folds, 10 folds, 20
folds, 50
181
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
folds, 100 folds, 200 folds, 500 folds, 1000 folds, 2000 folds, 5000 folds,
10,000 folds,
20,000 folds, 50,000 folds, 100,000 folds, 200,000 folds, 500,000 folds, or
1,000,000 folds
compared to the production of a cannabinoid or a cannabinoid derivative by
unmodified host
cells cultured under similar conditions.
[0435] In some embodiments, the production of a cannabinoid or a
cannabinoid
derivative, such as those disclosed herein, by modified host cells of the
disclosure may also
be increased by at least about any of 10%, 20%, 30%, 40%, 50%, 60%, 70%, 80%,
or 90%
compared to the production of a cannabinoid or a cannabinoid derivative by
unmodified host
cells cultured under similar conditions. In some embodiments, the production
of a
cannabinoid or a cannabinoid derivative by modified host cells disclosed
herein may also be
increased by at least about any of 1-20%, 2-20%, 5-20%, 10-20%, 15-20%, 1-15%,
1-10%,
2-15%, 2-10%, 5-15%, 10-15%, 1-50%, 10-50%, 20-50%, 30-50%, 40-50%, 50-100%,
50-
60%, 50-70%, 50-80%, or 50-90% compared to the production of a cannabinoid or
a
cannabinoid derivative by unmodified host cells cultured under similar
conditions.
[0436] In some embodiments, production of a cannabinoid or a cannabinoid
derivative by modified host cells of the disclosure is determined by LC-MS
analysis. In
certain such embodiments, each cannabinoid or cannabinoid derivative is
identified by
retention time, determined from an authentic standard, and multiple reaction
monitoring
(MRM) transition.
[0437] In some embodiments, the modified host cell of the disclosure is a
yeast cell.
In certain such embodiments, the modified host cell disclosed herein is
cultured in a
bioreactor. In some embodiments, the modified host cell is cultured in a
culture medium
supplemented with unsubstituted or substituted hexanoic acid, a carboxylic
acid other than
unsubstituted or substituted hexanoic acid, olivetolic acid, or an olivetolic
acid derivative. In
some embodiments, the modified yeast cell is a modified S. cerevisiae.
[0438] In some embodiments, the cannabinoid or cannabinoid derivative,
such as
those disclosed herein, is recovered from a cell lysate, e.g., by lysing the
modified host cell
disclosed herein and recovering the cannabinoid or cannabinoid derivative
derivative from
the lysate. In other cases, the cannabinoid or cannabinoid derivative is
recovered from the
culture medium in which the modified host cell disclosed herein is cultured.
In other cases,
the cannabinoid or cannabinoid derivative is recovered from both the cell
lysate and the
culture medium. In other cases, the cannabinoid or cannabinoid derivative is
recovered from
a modified host cell. In other cases, the cannabinoid or cannabinoid
derivative is recovered
182
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
from both the modified host cell and the culture medium. In other cases, the
cannabinoid or
cannabinoid derivative is recovered from the cell lysate, the modified host
cell, and the
culture medium. In some embodiments when the cannabinoid or cannabinoid
derivative is
recovered from a cell lysate; from a culture medium; from a modified host
cell; from both
the cell lysate and the culture medium; from both the modified host cell and
the culture
medium; from the cell lysate, the modified host cell, and the culture medium;
or from a cell-
free reaction mixture comprising one or more polypeptides disclosed herein,
the recovered
cannabinoid or cannabinoid derivative is in the form of a salt. In certain
such embodiments,
the salt is a pharmaceutically acceptable salt. In some embodiments, the salt
of the recovered
cannabinoid or cannabinoid derivative is then purified as disclosed herein.
[0439] In some embodiments, the recovered cannabinoid or cannabinoid
derivative,
such as those disclosed herein, is then purified. In some embodiments, whole-
cell broth from
cultures comprising modified host cells of the disclosure may be extracted
with a suitable
organic solvent to afford cannabinoids or cannabinoid derivatives. Suitable
organic solvents
include, but are not limited to, hexane, heptane, ethyl acetate, petroleum
ether, and di-ethyl
ether, chloroform, and ethyl acetate. In some embodiments, the suitable
organic solvent
comprises hexane. In some embodiments, the suitable organic solvent may be
added to the
whole-cell broth from fermentations comprising modified host cells of the
disclosure at a
10:1 ratio (10 parts whole-cell broth ¨ 1 part organic solvent) and stirred
for 30 minutes. In
certain such embodiments, the organic fraction may be separated and extracted
twice with an
equal volume of acidic water (pH 2.5). The organic layer may then be separated
and dried in
a concentrator (rotary evaporator or thin film evaporator under reduced
pressure) to obtain
crude cannabinoid or cannabinoid derivative crystals. In certain such
embodiments, the
crude crystals may be heated or exposed to light to decarboxylate the crude
cannabinoid or
cannabinoid derivative. In certain such embodiments, the crude crystals may be
heated to
105 C for 15 minutes followed by 145 C for 55 minutes to decarboxylate the
crude
cannabinoid or cannabinoid derivative. In certain such embodiments, the crude
crystalline
product may be re-dissolved and recrystallized in a suitable solvent (e.g., n-
pentane) and
filtered to remove any insoluble material. In certain such embodiments, the
solvent may then
be removed e.g. by rotary evaporation, to produce pure crystalline product.
[0440] In some embodiments, the cannabinoid or cannabinoid derivative is
pure, e.g.,
at least about 40% pure, at least about 50% pure, at least about 60% pure, at
least about 70%
pure, at least about 80% pure, at least about 90% pure, at least about 95%
pure, at least about
183
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
98%, or more than 98% pure, where "pure" in the context of a cannabinoid or a
cannabinoid
derivative may refer to a cannabinoid or a cannabinoid derivative that is free
from other
cannabinoids or cannabinoid derivatives, macromolecules, contaminants, etc.
Methods of Preparing Engineered Variants of a Tetrahydrocannabinolic Acid
Synthase
(THCAS) Polypeptide
[0441] In an aspect, the present disclosure provides methods for
preparing
engineered variants of a tetrahydrocannabinolic acid synthase (THCAS)
polypeptide. In
certain such embodiments, the methods may comprise culturing a modified host
cell of the
disclosure in a culture medium. In some embodiments, the modified host cell of
the
disclosure is a Pichia sp. The method can comprise isolating and/or purifying
the expressed
engineered variants, as described herein.
[0442] In some embodiments, the method for preparing engineered variants
comprises the step of isolating or purifying the engineered variants. The
engineered variants
of the disclosure can be expressed in modified host cells, as described
herein, and isolated
from the modified host cells and/or culture medium using any one or more of
the well
known techniques used for protein purification, including, among others,
lysozyme
treatment, sonication, filtration, salting-out, ultra-centrifugation, and
chromatography.
Chromatographic techniques for isolation of the engineered variants of the
disclosure may
include, among others, reverse phase chromatography high performance liquid
chromatography, ion exchange chromatography, gel electrophoresis, and affinity
chromatography. In some embodiments, affinity chromatography is used.
[0443] In some embodiments, the engineered variants of the disclosure
expressed in
the modified host cells of the disclosure can be prepared and used in various
forms including
but not limited to crude extracts (e.g., cell-free lysates), powders (e.g.,
shake-flask powders),
lyophilizates, frozen stocks made with glycerol or another cryoprotectant, and
substantially
pure preparations (e.g., D SP powders).
[0444] In some embodiments, the engineered variants of the disclosure
expressed in
the modified host cells of the disclosure can be prepared and used in purified
form.
Generally, conditions for purifying a particular engineered variant will
depend, in part, on
factors such as net charge, hydrophobicity, hydrophilicity, molecular weight,
molecular
shape, etc., and will be apparent to those having skill in the art.
184
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Cell-Free Methods of Producing Cannabinoids or Cannabinoid Derivatives
[0445] The methods of the disclosure may involve cell-free production of
cannabinoids or cannabinoid derivatives, such as those disclosed herein, using
engineered
variants disclosed herein expressed or overexpressed by a modified host cell
of the
disclosure. In some embodiments, an engineered variant disclosed herein is
used in a cell-
free system for the production of cannabinoids or cannabinoid derivatives. In
certain such
embodiments, the engineered variant of the disclosure is isolated and/or
purified. In some
embodiments, appropriate starting materials for use in producing cannabinoids
or
cannabinoid derivatives may be mixed together with engineered variants
disclosed herein in
a suitable reaction vessel to effect the reaction. The engineered variants
disclosed herein
may be used in combination to effect a complete synthesis of a cannabinoid or
cannabinoid
derivative from the appropriate starting materials. In some embodiments, the
cannabinoid or
cannabinoid derivative is recovered from a cell-free reaction mixture
comprising engineered
disclosed herein.
[0446] In some embodiments, the recovered cannabinoids or cannabinoid
derivatives, such as those disclosed herein, are then purified. In certain
such embodiments, a
cell-free reaction mixture comprising an engineered variant disclosed herein
may be
extracted with a suitable organic solvent to afford cannabinoids or
cannabinoid derivatives.
Suitable organic solvents include, but are not limited to, hexane, heptane,
ethyl acetate,
petroleum ether, and di-ethyl ether, chloroform, and ethyl acetate. In some
embodiments,
the suitable organic solvent comprises hexane. In some embodiments, the
suitable organic
solvent may be added to the cell-free reaction mixture comprising one or more
of the
polypeptides disclosed herein at a 10:1 ratio (10 parts reaction mixture ¨ 1
part organic
solvent) and stirred for 30 minutes. In certain such embodiments, the organic
fraction may
be separated and extracted twice with an equal volume of acidic water (pH
2.5). The organic
layer may then be separated and dried in a concentrator (rotary evaporator or
thin film
evaporator under reduced pressure) to obtain crude cannabinoid or cannabinoid
derivative
crystals. In certain such embodiments, the crude crystals may be heated or
exposed to light
to decarboxylate the crude cannabinoid or cannabinoid derivative. In certain
such
embodiments, the crude crystals may be heated to 105 C for 15 minutes
followed by 145 C
for 55 minutes to decarboxylate the crude cannabinoid or cannabinoid
derivative. In certain
such embodiments, the crude crystalline product may be re-dissolved and
recrystallized in a
185
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
suitable solvent (e.g., n-pentane) and filtered to remove any insoluble
material. In certain
such embodiments, the solvent may then be removed e.g. by rotary evaporation,
to produce
pure crystalline product.
[0447] In some embodiments when the cannabinoid or cannabinoid derivative
is
recovered from a cell-free reaction mixture comprising one or more engineered
variants
disclosed herein, the recovered cannabinoid or cannabinoid derivative is in
the form of a salt.
In certain such embodiments, the salt is a pharmaceutically acceptable salt.
In some
embodiments, the salt of the recovered cannabinoid or cannabinoid derivative
is then
purified as disclosed herein.
[0448] In some embodiments, cell-free production of a cannabinoid or a
cannabinoid
derivative by engineered variants disclosed herein is determined by LC-MS
analysis. In
certain such embodiments, each cannabinoid or cannabinoid derivative is
identified by
retention time, determined from an authentic standard, and multiple reaction
monitoring
(MRM) transition.
EXAMPLES OF NON-LIMITING EMBODIMENTS OF THE DISCLOSURE
[0449] Embodiments of the present subject matter disclosed herein may be
beneficial
alone or in combination with one or more other embodiments. Without limiting
the
foregoing description, certain non-limiting embodiments of the disclosure,
numbered I-1 to
1-121 are provided below. As will be apparent to those of skill in the art
upon reading this
disclosure, each of the individually numbered embodiments may be used or
combined with
any of the preceding or following individually numbered embodiments. This is
intended to
provide support for all such combinations of embodiments and is not limited to
combinations of embodiments explicitly provided below.
[0450] Some embodiments of the disclosure are of Embodiment I:
[0451] Embodiment I-1. An engineered variant of a
tetrahydrocannabinolic acid
synthase (THCAS) polypeptide comprising an amino acid sequence of SEQ ID NO:44
with
one or more amino acid substitutions.
[0452] Embodiment 1-2. The engineered variant of Embodiment I-1,
wherein
the engineered variant comprises an amino acid sequence with at least 85%, at
least 86%, at
least 87%, at least 88%, at least 89%, at least 90%, at least 91%, at least
92%, at least 93%,
at least 94%, at least 95%, at least 96%, at least 97%, at least 98%, or at
least 99% sequence
identity to SEQ ID NO:44.
186
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0453] Embodiment 1-3. The engineered variant of Embodiment I-1 or 1-
2,
wherein the engineered variant comprises at least one amino acid substitution
in a signal
polypeptide, a flavin adenine dinucleotide (FAD) binding domain, a berberine
bridge
enzyme (BBE) domain, or a combination of the foregoing.
[0454] Embodiment 1-4. The engineered variant of Embodiment 1-3,
wherein
the engineered variant comprises at least one amino acid substitution in the
signal
polypeptide.
[0455] Embodiment 1-5. The engineered variant of Embodiment 1-3 or 1-
4,
wherein the engineered variant comprises at least one amino acid substitution
in the FAD
binding domain.
[0456] Embodiment 1-6. The engineered variant of any one of
Embodiments 1-3
to 1-5, wherein the engineered variant comprises at least one amino acid
substitution in the
BBE domain.
[0457] Embodiment 1-7. The engineered variant of any one of
Embodiments 1-3
to 1-6, wherein the engineered variant comprises substitution of at least one
surface exposed
amino acid.
[0458] Embodiment 1-8. The engineered variant of Embodiment I-1 or 1-
2,
wherein the engineered variant comprises at least one amino acid substitution
at an amino
acid selected from the group consisting of R31, P43, P49, K50, L51, Q55, H56,
L59, M61,
S62, L71, S100, V103, T109, Q124, V125, L132, S137, H143, V149, W161, K165,
N168,
E167, S170, F171, P172, Y175, G180, N196, H208, G235, A250, 1257, K261, L269,
G311,
F317, L327, K390, T379, S429, N467, Y500, N528, P539, P542, H543, H544, and
H545
[0459] Embodiment 1-9. The engineered variant of Embodiment 1-8,
wherein
the engineered variant comprises at least one amino acid substitution selected
from the group
consisting of R31Q, P43E, P49E, P49K, P49Q, K50T, L51I, Q55E, Q55P, H56E,
L59E,
M61W, M61H, M61S, S62Q, L71A, S100A, V103F,T109V,Q124D, Q124E, Q124N,
V125E, V125Q, L132M, S137G, H143D, V1491, W161R, W161Y, W161K, K165A,
N168S, E167P, S170T, F1711, P172V, Y175F, G180A, N196Q, N196V, H208T, G235P,
A250T, I257V, K261W, K261C, L269I, G311A, G311C, F317Y, L327I, K390E, T379S,
S429L, N467D, Y500M, Y500V, N528E, P539T, P542E, P542V, H543V, H544A, H545D,
and H545E.
[0460] Embodiment I-10. The engineered variant of Embodiment I-1 or 1-
2,
wherein the engineered variant comprises an amino acid sequence selected from
the group
187
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
consisting of SEQ ID NO:50, SEQ ID NO:52, SEQ ID NO:54, SEQ ID NO:56, SEQ ID
NO:58, SEQ ID NO:60, SEQ ID NO:62, SEQ ID NO:64, SEQ ID NO:66, SEQ ID NO:68,
SEQ ID NO:70, SEQ ID NO:72, SEQ ID NO:74, SEQ ID NO:76, SEQ ID NO:78, SEQ ID
NO:80 SEQ ID NO:82, SEQ ID NO:84, SEQ ID NO:86, SEQ ID NO:88, SEQ ID NO:90,
SEQ ID NO:92, SEQ ID NO:94, SEQ ID NO:96, SEQ ID NO:98, SEQ ID NO:100, SEQ ID
NO:102, SEQ ID NO:104, SEQ ID NO:106, SEQ ID NO:108, SEQ ID NO:110, SEQ ID
NO:112, SEQ ID NO:114, SEQ ID NO:116, SEQ ID NO:118, SEQ ID NO:120, SEQ ID
NO:122, SEQ ID NO:124, SEQ ID NO:126, SEQ ID NO:128, SEQ ID NO:130, SEQ ID
NO:132, SEQ ID NO:134, SEQ ID NO:136, SEQ ID NO:138, SEQ ID NO:140, SEQ ID
NO:142, SEQ ID NO:144, SEQ ID NO:146, SEQ ID NO:148, SEQ ID NO:150, SEQ ID
NO:152, SEQ ID NO:154, SEQ ID NO:156, SEQ ID NO:158, SEQ ID NO:160, SEQ ID
NO:162, SEQ ID NO:164, SEQ ID NO:166, SEQ ID NO:168, SEQ ID NO:170, SEQ ID
NO:172, SEQ ID NO:174, SEQ ID NO:176, SEQ ID NO:178, SEQ ID NO:180, SEQ ID
NO:182, SEQ ID NO:184, and SEQ ID NO:186.
[0461] Embodiment I-11. The engineered variant of any one of
Embodiments I-1
to 1-9, wherein the engineered variant comprises an amino acid sequence of SEQ
ID NO:44
with at least 1, at least 2, at least 3, at least 4, at least 5, at least 6,
at least 7, at least 8, at least
9, at least 10, at least 11, at least 12, at least 13, at least 14, at least
15, at least 16, at least 17,
at least 18, at least 19, at least 20, at least 21, at least 22, at least 23,
at least 24, at least 25, at
least 26, at least 27, at least 28, at least 29, or at least 30 amino acid
substitutions.
[0462] Embodiment 1-12. The engineered variant of any one of
Embodiments I-1
to 1-9, wherein the engineered variant comprises an amino acid sequence of SEQ
ID NO:44
with 1, 2, 3, 4, 5, 6, 7, 8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26,
27, 28, 29, or 30 amino acid substitutions.
[0463] Embodiment 1-13. The engineered variant of any one of
Embodiments I-1
to 1-12, wherein the engineered variant comprises at least one immutable amino
acid in a
flavin adenine dinucleotide (FAD) binding domain, a berberine bridge enzyme
(BBE)
domain, or a combination of the foregoing.
[0464] Embodiment 1-14. The engineered variant of Embodiment 1-13,
wherein
the engineered variant comprises at least one immutable amino acid in the FAD
binding
domain.
[0465] Embodiment 1-15. The engineered variant of Embodiment 1-14,
wherein
the engineered variant comprises at least 1, at least 2, at least 3, at least
4, at least 5, at least
188
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
6, at least 7, at least 8, at least 9, at least 10, at least 11, at least 12,
at least 13, at least 14, or
at least 15 immutable amino acids in the FAD binding domain.
[0466] Embodiment 1-16. The engineered variant of any one of
Embodiments I-
13 to 1-15, wherein the engineered variant comprises at least one immutable
amino acid in
the BBE domain.
[0467] Embodiment 1-17. The engineered variant of Embodiment 1-16,
wherein
the engineered variant comprises at least 1, at least 2, at least 3, at least
4, at least 5, at least
6, at least 7, at least 8, at least 9, at least 10, at least 11, at least 12,
at least 13, at least 14, or
at least 15 immutable amino acids in the BBE domain.
[0468] Embodiment 1-18. The engineered variant of any one of
Embodiments I-1
to 1-17, wherein the engineered variant comprises at least one immutable amino
acid selected
from the group consisting of A28, F34, L35, C37, L64, N70, P87, 193, C99,
R108, R110,
G112, E117, G118, S120, P126, F127, D131, D141, W148, G152, A153, L155, G156,
E157,
Y159, Y160, N163, A173, G174, C176, P177, T178, V179, G182, G183, H184, F185,
G187, G188, G189, Y190, G191, P192, L193, R195, A201, D202, 1205, D206, V210,
G214,
G223, D225, L226, F227, W228, R231, G234, S237, F238, G239, K245, 1246, L248,
V251,
V259, Q276, F312, S313, L323, C341, F352, S354, F380, K381, 1382, K383, D385,
Y386,
1391, M412, L415, G419, M422, 1425, 1430, P431, P433, H434, R435, G437, Y440,
W443,
Y444, 1445, 1464, Y465, M468, T469, Y471, V472, P476, R484, N498, A502, N513,
F514,
K521, N528, F529, E533, Q534, and S535.
[0469] Embodiment 1-19. The engineered variant of Embodiment 1-18,
wherein
the engineered variant comprises at least one immutable amino acid selected
from the group
consisting of C37, N70, 193, C99, E117, S120, F127, D131, G156, E157, Y159,
G174,
C176, G182, G183, F185, G187, G188, G189, Y190, G191, P192, R195, D202, D206,
G214, W228, G234, F238, L248, Q277, S314, L324, S355, K382, K384, D386, G420,
M423, R436, Y441, W444, Y445, Y472, P477, N514, F515, N529, and Q535.
[0470] Embodiment 1-20. The engineered variant of any one of
Embodiments I-1
to 1-19, wherein the engineered variant comprises at least 1, at least 2, at
least 3, at least 4, at
least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at least
11, at least 12, at least 13,
at least 14, at least 15, at least 16, at least 17, at least 18, at least 19,
at least 20, at least 21, at
least 22, at least 23, at least 24, or at least 25 immutable amino acids.
[0471] Embodiment 1-21. The engineered variant of any one of
Embodiments I-1
to 1-20, wherein the engineered variant produces tetrahydrocannabinolic acid
(THCA) from
189
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
cannabigerolic acid (CBGA) in a greater amount, as measured in mg/L or mM,
than an
amount of THCA produced from CBGA by a tetrahydrocannabinolic acid synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 under similar
conditions for
the same length of time.
[0472] Embodiment 1-22 The engineered variant of any one of
Embodiments I-1
to 1-21, wherein the engineered variant produces tetrahydrocannabinolic acid
(THCA) from
cannabigerolic acid (CBGA) in an amount, as measured in mg/L or mM, at least
5%, at least
10%, at least 15%, at least 20%, at least 25%, at least 30%, at least 35%, at
least 40%, at
least 45%, at least 50%, at least 60%, at least 70%, at least 80%, at least
90%, at least 100%,
at least 150% at least 200%, at least 500%, or at least 1000% greater than an
amount of
THCA produced from CBGA by a tetrahydrocannabinolic acid synthase polypeptide
having
an amino acid sequence of SEQ ID NO:44 under similar conditions for the same
length of
time.
[0473] Embodiment 1-23. The engineered variant of any one of
Embodiments I-1
to 1-22, wherein the engineered variant produces tetrahydrocannabinolic acid
(THCA) from
cannabigerolic acid (CBGA) in an increased ratio of THCA over another
cannabinoid (e.g.
cannabichromenic acid (CBCA)) compared to that produced by a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44 under
similar
conditions for the same length of time.
[0474] Embodiment 1-24. The engineered variant of any one of
Embodiments I-1
to 1-23, wherein the engineered variant produces THCA from CBGA in a ratio of
THCA
over another cannabinoid (e.g., CBCA) of about 11:1, about 11.5:1, about 12:1,
about
12.5:1, about 13:1, about 13.5:1, about 14:1, about 14.5:1, about 15:1, about
15.5:1, about
16:1, about 16.5:1, about 17:1, about 17.5:1, about 18:1, about 18.5:1, about
19:1, about
19.5:1, about 20:1, about 25:1, about 30:1, about 35:1, about 40:1, about
45:1, about 50:1,
about 60:1, about 70:1, about 80:1, about 90:1, about 100:1, about 150:1,
about 200:1, about
500:1, or greater than about 500:1.
[0475] Embodiment 1-25. The engineered variant of any one of
Embodiments I-1
to 1-24, wherein the engineered variant comprises a truncation at an N-
terminus, at a C-
terminus, or at both the N- and C-termini.
[0476] Embodiment 1-26. The engineered variant of Embodiment 1-25,
wherein
the truncated engineered variant comprises a signal polypeptide or a membrane
anchor.
190
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0477] Embodiment 1-27. The engineered variant of Embodiment 1-25 or 1-
26,
wherein the engineered variant lacks a native signal polypeptide.
[0478] Embodiment 1-28. The engineered variant of any one of
Embodiments I-
25 to 1-27, wherein the engineered variant comprises a truncation of at least
1, at least 2, at
least 3, at least 4, at least 5, at least 6, at least 7, at least 8, at least
9, or at least 10 amino
acids at the C-terminus.
[0479] Embodiment 1-29. The engineered variant of any one of
Embodiments I-
25 to 1-27, wherein the engineered variant comprises a truncation of 1, 2, 3,
4, 5, 6, 7, 8, 9, or
amino acids at the C-terminus.
[0480] Embodiment 1-30. A nucleic acid comprising a nucleotide
sequence
encoding an engineered variant of any one of Embodiments I-1 to 1-29.
[0481] Embodiment 1-31. A nucleic acid comprising a nucleotide
sequence
encoding an engineered variant of a tetrahydrocannabinolic acid synthase
(THCAS)
polypeptide comprising an amino acid sequence of SEQ ID NO:44 with one or more
amino
acid substitutions, wherein the nucleotide sequence is selected from the group
consisting of
SEQ ID NO:49, SEQ ID NO:51, SEQ ID NO:53, SEQ ID NO:55, SEQ ID NO:57, SEQ ID
NO:59, SEQ ID NO:61, SEQ ID NO:63, SEQ ID NO:65, SEQ ID NO:67, SEQ ID NO:69,
SEQ ID NO:71, SEQ ID NO:73, SEQ ID NO:75, SEQ ID NO:77, SEQ ID NO:79, SEQ ID
NO:81, SEQ ID NO:83, SEQ ID NO:85, SEQ ID NO:87, SEQ ID NO:91, SEQ ID NO:93,
SEQ ID NO:95, SEQ ID NO:97, SEQ ID NO:99, SEQ ID NO:101, SEQ ID NO:103, SEQ
ID NO:105, SEQ ID NO:107, SEQ ID NO:109, SEQ ID NO:111, SEQ ID NO:113, SEQ ID
NO:115, SEQ ID NO:117, SEQ ID NO:119, SEQ ID NO:121, SEQ ID NO:123, SEQ ID
NO:125, SEQ ID NO:127, SEQ ID NO:129, SEQ ID NO:131, SEQ ID NO:133, SEQ ID
NO:135, SEQ ID NO:137, SEQ ID NO:139, SEQ ID NO:141, SEQ ID NO:143, SEQ ID
NO:145, SEQ ID NO:147, SEQ ID NO:149, SEQ ID NO:151, SEQ ID NO:153, SEQ ID
NO:155, SEQ ID NO:157, SEQ ID NO:159, SEQ ID NO:161, SEQ ID NO:163, SEQ ID
NO:165, SEQ ID NO:167, SEQ ID NO:169, SEQ ID NO:171, SEQ ID NO:173, SEQ ID
NO:175, SEQ ID NO:177, SEQ ID NO:179, SEQ ID NO:181, SEQ ID NO:183, and SEQ ID
NO:185.
[0482] Embodiment 1-32. The nucleic acid of Embodiment 1-30 or 1-31,
wherein
the nucleotide sequence is codon-optimized.
191
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0483] Embodiment 1-33. A method of making a modified host cell for
producing
a cannabinoid or a cannabinoid derivative, the method comprising introducing
one or more
nucleic acids of any one of Embodiments 1-30 to 1-32 into a host cell.
[0484] Embodiment 1-34. A vector comprising one or more nucleic acids
of any
one of Embodiments 1-30 to 1-32.
[0485] Embodiment 1-35. A method of making a modified host cell for
producing
a cannabinoid or a cannabinoid derivative, the method comprising introducing
one or more
vectors of Embodiment 1-34 into a host cell.
[0486] Embodiment 1-36. A modified host cell for producing a
cannabinoid or a
cannabinoid derivative, wherein the modified host cell comprises one or more
nucleic acids
of any one of Embodiments 1-30 to 1-32.
[0487] Embodiment 1-37. The modified host cell of Embodiment 1-36,
wherein
the modified host cell comprises one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a geranyl pyrophosphate:olivetolic acid
geranyltransferase
(GOT) polypeptide.
[0488] Embodiment 1-38. The modified host cell of Embodiment 1-37,
wherein
the GOT polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:17.
[0489] Embodiment 1-39. The modified host cell of Embodiment 1-37 or 1-
38,
wherein the modified host cell comprises two or more heterologous nucleic
acids comprising
the nucleotide sequence encoding the GOT polypeptide.
[0490] Embodiment 1-40. The modified host cell of Embodiment 1-36,
wherein
the modified host cell comprises one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a NphB polypeptide.
[0491] Embodiment 1-41. The modified host cell of Embodiment 1-40,
wherein
the NphB polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:188.
[0492] Embodiment 1-42. The modified host cell of any one of
Embodiments I-
36 to 1-41, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a tetraketide synthase (TKS)
polypeptide and
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding an
olivetolic acid cyclase (OAC) polypeptide.
192
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0493] Embodiment 1-43. The modified host cell of Embodiment 1-42,
wherein
the TKS polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:19.
[0494] Embodiment 1-44. The modified host cell of Embodiment 1-42 or 1-
43,
wherein the modified host cell comprises three or more heterologous nucleic
acids
comprising a nucleotide sequence encoding a TKS polypeptide.
[0495] Embodiment 1-45. The modified host cell of any one of
Embodiments I-
42 to 1-44, wherein the OAC polypeptide comprises an amino acid sequence
having at least
85% sequence identity to SEQ ID NO:21 or SEQ ID NO:48.
[0496] Embodiment 1-46. The modified host cell of any one of
Embodiments I-
42 to 1-45, wherein the modified host cell comprises three or more
heterologous nucleic
acids comprising a nucleotide sequence encoding an OAC polypeptide.
[0497] Embodiment 1-47. The modified host cell of any one of
Embodiments I-
36 to 1-46, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an acyl-activating enzyme (AAE)
polypeptide.
[0498] Embodiment 1-48. The modified host cell of Embodiment 1-47,
wherein
the AAE polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:23.
[0499] Embodiment 1-49. The modified host cell of Embodiment 1-47 or 1-
48,
wherein the modified host cell comprises two or more heterologous nucleic
acids comprising
a nucleotide sequence encoding an AAE polypeptide.
[0500] Embodiment I-50. The modified host cell of any one of
Embodiments I-
36 to 1-49, wherein the modified host cell comprises one or more of the
following: a) one or
more heterologous nucleic acids comprising a nucleotide sequence encoding a
HMG-CoA
synthase (HMGS) polypeptide; b) one or more heterologous nucleic acids
comprising a
nucleotide sequence encoding a truncated 3-hydroxy-3-methyl-glutaryl-CoA
reductase
(tHMGR) polypeptide; c) one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding a mevalonate kinase (MK) polypeptide; d) one or more
heterologous
nucleic acids comprising a nucleotide sequence encoding a phosphomevalonate
kinase
(PMK) polypeptide; e) one or more heterologous nucleic acids comprising a
nucleotide
sequence encoding a mevalonate pyrophosphate decarboxylase (MVD1) polypeptide;
or f)
one or more heterologous nucleic acids comprising a nucleotide sequence
encoding a
isopentenyl diphosphate isomerase polypeptide.
193
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0501] Embodiment 1-51. The modified host cell of Embodiment 1-50,
wherein
the IDI1 polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:25.
[0502] Embodiment 1-52. The modified host cell of Embodiment I-50 or 1-
51,
wherein the tHMGR polypeptide comprises an amino acid sequence having at least
85%
sequence identity to SEQ ID NO:27.
[0503] Embodiment 1-53. The modified host cell of any one of
Embodiments I-
50 to 1-52, wherein the HMGS polypeptide comprises an amino acid sequence
having at
least 85% sequence identity to SEQ ID NO:29.
[0504] Embodiment 1-54. The modified host cell of any one of
Embodiments I-
50 to 1-53, wherein the MK polypeptide comprises an amino acid sequence having
at least
85% sequence identity to SEQ ID NO:39.
[0505] Embodiment 1-55. The modified host cell of any one of
Embodiments I-
50 to 1-54, wherein the PMK polypeptide comprises an amino acid sequence
having at least
85% sequence identity to SEQ ID NO:37.
[0506] Embodiment 1-56. The modified host cell of any one of
Embodiments I-
50 to I-55, wherein the MVD1 polypeptide comprises an amino acid sequence
having at least
85% sequence identity to SEQ ID NO:33.
[0507] Embodiment 1-57. The modified host cell of any one of
Embodiments I-
36 to I-56, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an acetoacetyl-CoA thiolase
polypeptide.
[0508] Embodiment 1-58. The modified host cell of Embodiment 1-57,
wherein
the acetoacetyl-CoA thiolase polypeptide comprises an amino acid sequence
having at least
85% sequence identity to SEQ ID NO:31.
[0509] Embodiment 1-59. The modified host cell of any one of
Embodiments I-
36 to I-58, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a pyruvate decarboxylase (PDC)
polypeptide.
[0510] Embodiment 1-60. The modified host cell of Embodiment 1-59,
wherein
the PDC polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:35.
[0511] Embodiment 1-61. The modified host cell of any one of
Embodiments I-
36 to 1-60, wherein the modified host cell comprises one or more heterologous
nucleic acids
194
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
comprising a nucleotide sequence encoding a geranyl pyrophosphate synthetase
(GPPS)
polypeptide.
[0512] Embodiment 1-62. The modified host cell of Embodiment 1-61,
wherein
the GPPS polypeptide comprises an amino acid sequence haying at least 85%
sequence
identity to SEQ ID NO:41.
[0513] Embodiment 1-63. The modified host cell of any one of
Embodiments I-
36 to 1-62, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a KAR2 polypeptide.
[0514] Embodiment 1-64. The modified host cell of Embodiment 1-63,
wherein
the KAR2 polypeptide comprises an amino acid sequence haying at least 85%
sequence
identity to SEQ ID NO:5.
[0515] Embodiment 1-65. The modified host cell of Embodiment 1-63 or 1-
64,
wherein the modified host cell comprises two or more heterologous nucleic
acids comprising
a nucleotide sequence encoding a KAR2 polypeptide.
[0516] Embodiment 1-66. The modified host cell of any one of
Embodiments I-
36 to 1-65, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a PDI1 polypeptide.
[0517] Embodiment 1-67. The modified host cell of Embodiment 1-66,
wherein
the PDI1 polypeptide comprises an amino acid sequence haying at least 85%
sequence
identity to SEQ ID NO:9.
[0518] Embodiment 1-68. The modified host cell of any one of
Embodiments I-
36 to 1-67, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an IRE1 polypeptide.
[0519] Embodiment 1-69. The modified host cell of Embodiment 1-68,
wherein
the IRE1 polypeptide comprises an amino acid sequence haying at least 85%
sequence
identity to SEQ ID NO:11 or SEQ ID NO:190.
[0520] Embodiment 1-70. The modified host cell of any one of
Embodiments I-
36 to 1-69, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding an ER01 polypeptide.
[0521] Embodiment 1-71. The modified host cell of Embodiment 1-70,
wherein
the ER01 polypeptide comprises an amino acid sequence haying at least 85%
sequence
identity to SEQ ID NO:7.
195
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0522] Embodiment 1-72. The modified host cell of any one of
Embodiments I-
36 to 1-71, wherein the modified host cell comprises one or more heterologous
nucleic acids
comprising a nucleotide sequence encoding a FAD1 polypeptide.
[0523] Embodiment 1-73. The modified host cell of Embodiment 1-72,
wherein
the FAD1 polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:192.
[0524] Embodiment 1-74. The modified host cell of any one of
Embodiments I-
36 to 1-73, wherein the modified host cell comprises a deletion or
downregulation of one or
more genes encoding a PEP4 polypeptide.
[0525] Embodiment 1-75. The modified host cell of Embodiment 1-74,
wherein
the PEP4 polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:15.
[0526] Embodiment 1-76. The modified host cell of any one of
Embodiments I-
36 to 1-75, wherein the modified host cell comprises a deletion or
downregulation of one or
more genes encoding a ROT2 polypeptide.
[0527] Embodiment 1-77. The modified host cell of Embodiment 1-76,
wherein
the ROT2 polypeptide comprises an amino acid sequence having at least 85%
sequence
identity to SEQ ID NO:13.
[0528] Embodiment 1-78. The modified host cell of any one of
Embodiments I-
36 to 1-77, wherein the modified host cell is a eukaryotic cell.
[0529] Embodiment 1-79. The modified host cell of Embodiment 1-78,
wherein
the eukaryotic cell is a yeast cell.
[0530] Embodiment 1-80. The modified host cell of Embodiment 1-79,
wherein
the yeast cell is Saccharomyces cerevisiae.
[0531] Embodiment 1-81. The modified host cell of Embodiment 1-80,
wherein
the Saccharomyces cerevisiae is a protease-deficient strain of Saccharomyces
cerevisiae.
[0532] Embodiment 1-82. The modified host cell of any one of
Embodiments I-
36 to 1-81, wherein at least one of the one or more nucleic acids are
integrated into the
chromosome of the modified host cell.
[0533] Embodiment 1-83. The modified host cell of any one of
Embodiments I-
36 to 1-81, wherein at least one of the one or more nucleic acids are
maintained
extrachromosomally.
196
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0534] Embodiment 1-84. The modified host cell of any one of
Embodiments I-
36 to 1-83, wherein at least one of the one or more nucleic acids are operably-
linked to an
inducible promoter.
[0535] Embodiment 1-85. The modified host cell of any one of
Embodiments I-
36 to 1-83, wherein at least one of the one or more nucleic acids are operably-
linked to a
constitutive promoter.
[0536] Embodiment 1-86. The modified host cell of any one of
Embodiments I-
36 to 1-85, wherein the modified host cell produces a cannabinoid or a
cannabinoid
derivative in an amount, as measured in mg/L or mM, greater than an amount of
the
cannabinoid or the cannabinoid derivative produced by a modified host cell
comprising one
or more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic
acid synthase polypeptide having an amino acid sequence of SEQ ID NO:44,
wherein the
modified host cell comprising one or more nucleic acids comprising the
nucleotide sequence
encoding the tetrahydrocannabinolic acid synthase polypeptide having the amino
acid
sequence of SEQ ID NO:44 lacks a nucleic acid comprising a nucleotide sequence
encoding
an engineered variant of any one of Embodiments I-1 to 1-29, grown under
similar culture
conditions for the same length of time.
[0537] Embodiment 1-87. The modified host cell of any one of
Embodiments I-
36 to 1-86, wherein the modified host cell produces a cannabinoid or a
cannabinoid
derivative in an amount, as measured in mg/L or mM, at least 5%, at least 10%,
at least 15%,
at least 20%, at least 25%, at least 30%, at least 35%, at least 40%, at least
45%, at least
50%, at least 60%, at least 70%, at least 80%, at least 90%, at least 100%, at
least 150% at
least 200%, at least 500%, or at least 1000% greater than an amount of the
cannabinoid or
the cannabinoid derivative produced by a modified host cell comprising one or
more nucleic
acids comprising a nucleotide sequence encoding a tetrahydrocannabinolic acid
synthase
polypeptide having an amino acid sequence of SEQ ID NO:44, wherein the
modified host
cell comprising one or more nucleic acids comprising the nucleotide sequence
encoding the
tetrahydrocannabinolic acid synthase polypeptide having the amino acid
sequence of SEQ
ID NO:44 lacks a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant of any one of Embodiments I-1 to 1-29, grown under similar culture
conditions for
the same length of time.
[0538] Embodiment 1-88. The modified host cell of any one of
Embodiments I-
36 to 1-87, wherein the modified host cell has a faster growth rate and/or
higher biomass
197
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
yield compared to a growth rate and/or higher biomass yield of a modified host
cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, wherein the modified host cell comprising one or more nucleic acids
comprising the
nucleotide sequence encoding the tetrahydrocannabinolic acid synthase
polypeptide having
the amino acid sequence of SEQ ID NO:44 lacks a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant of any one of Embodiments I-1 to 1-29,
grown
under similar culture conditions for the same length of time.
[0539] Embodiment 1-89. The modified host cell of any one of
Embodiments I-
36 to 1-88, wherein the modified host cell has a growth rate and/or higher
biomass yield at
least 5%, at least 10%, at least 15%, at least 20%, at least 25%, at least
30%, at least 35%, at
least 40%, at least 45%, at least 50%, at least 60%, at least 70%, at least
80%, at least 90%,
at least 100%, at least 150% at least 200%, at least 500%, or at least 1000%
faster than a
growth rate and/or higher biomass yield of a modified host cell comprising one
or more
nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44, wherein
the
modified host cell comprising one or more nucleic acids comprising the
nucleotide sequence
encoding the tetrahydrocannabinolic acid synthase polypeptide having the amino
acid
sequence of SEQ ID NO:44 lacks a nucleic acid comprising a nucleotide sequence
encoding
an engineered variant of any one of Embodiments I-1 to 1-29, grown under
similar culture
conditions for the same length of time.
[0540] Embodiment 1-90. The modified host cell of any one of
Embodiments I-
36 to 1-89, wherein the modified host cell produces tetrahydrocannabinolic
acid (THCA)
from cannabigerolic acid (CBGA) in an increased ratio of THCA over another
cannabinoid
(e.g., cannabichromenic acid (CBCA)) compared to that produced by a modified
host cell
comprising one or more nucleic acids comprising a nucleotide sequence encoding
a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44, wherein the modified host cell comprising one or more nucleic acids
comprising the
nucleotide sequence encoding the tetrahydrocannabinolic acid synthase
polypeptide having
the amino acid sequence of SEQ ID NO:44 lacks a nucleic acid comprising a
nucleotide
sequence encoding an engineered variant of any one of Embodiments I-1 to 1-29,
grown
under similar culture conditions for the same length of time.
198
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0541] Embodiment 1-91. The modified host cell of any one of
Embodiments I-
36 to 1-90, wherein the modified host cell produces THCA from CBGA in a ratio
of THCA
over another cannabinoid (e.g., CBCA) of about 11:1, about 11.5:1, about 12:1,
about
12.5:1, about 13:1, about 13.5:1, about 14:1, about 14.5:1, about 15:1, about
15.5:1, about
16:1, about 16.5:1, about 17:1, about 17.5:1, about 18:1, about 18.5:1, about
19:1, about
19.5:1, about 20:1, about 25:1, about 30:1, about 35:1, about 40:1, about
45:1, about 50:1,
about 60:1, about 70:1, about 80:1, about 90:1, about 100:1, about 150:1,
about 200:1, about
500:1, or greater than about 500:1.
[0542] Embodiment 1-92. A method of producing a cannabinoid or a
cannabinoid
derivative comprising: a) culturing a modified host cell of any one of
Embodiments 1-36 to I-
91 in a culture medium.
[0543] Embodiment 1-93. The method of Embodiment 1-92, wherein the
method
comprises: b) recovering the produced cannabinoid or cannabinoid derivative.
[0544] Embodiment 1-94. The method of Embodiment 1-92 or 1-93, wherein
the
culture medium comprises a carboxylic acid.
[0545] Embodiment 1-95. The method of Embodiment 1-94, wherein the
carboxylic acid is an unsubstituted or substituted C3-C18 carboxylic acid.
[0546] Embodiment 1-96. The method of Embodiment 1-95, wherein the
unsubstituted or substituted C3-C18 carboxylic acid is an unsubstituted or
substituted
hexanoic acid.
[0547] Embodiment 1-97. The method of Embodiment 1-92 or 1-93, wherein
the
culture medium comprises olivetolic acid or an olivetolic acid derivative.
[00101] Embodiment 1-98. The method of Embodiment 1-92 or 1-93, wherein
the
cannabinoid is tetrahydrocannabinolic acid, tetrahydrocannabivarinic acid, or
tetrahydrocannabivarin.
[0548] Embodiment 1-99. The method of any one of Embodiments 1-92 to 1-
98,
wherein the culture medium comprises a fermentable sugar.
[0549] Embodiment I-100. The method of any one of Embodiments 1-92 to 1-
98,
wherein the culture medium comprises a pretreated cellulosic feedstock.
[0550] Embodiment I-101. The method of any one of Embodiments 1-92 to 1-
98,
wherein the culture medium comprises a non-fermentable carbon source.
[0551] Embodiment 1-102. The method of Embodiment I-101, wherein the non-
fermentable carbon source comprises ethanol.
199
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0552] Embodiment 1-103. The method of any one of Embodiments 1-92 to 1-
102,
wherein the cannabinoid or the cannabinoid derivative is produced in an amount
of more
than 100 mg/L culture medium.
[0553] Embodiment 1-104. The method of any one of Embodiments 1-92 to 1-
103,
wherein the cannabinoid or the cannabinoid derivative is produced in an
amount, as
measured in mg/L or mM, greater than an amount of the cannabinoid or the
cannabinoid
derivative produced in a method comprising culturing a modified host cell
comprising one or
more nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44 instead of
the
modified host cell of any one of Embodiments 1-34 to 1-89, wherein the
modified host cell
comprising one or more nucleic acids comprising the nucleotide sequence
encoding the
tetrahydrocannabinolic acid synthase polypeptide having the amino acid
sequence of SEQ
ID NO:44 lacks a nucleic acid comprising a nucleotide sequence encoding an
engineered
variant of any one of Embodiments I-1 to 1-29, and wherein the modified host
cell of any
one of Embodiments 1-36 to 1-91 and the modified host cell comprising one or
more nucleic
acids comprising the nucleotide sequence encoding the tetrahydrocannabinolic
acid synthase
polypeptide having the amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant of any one of
Embodiments I-1 to 1-29, are cultured under similar culture conditions for the
same length of
time.
[0554] Embodiment 1-105. The method of any one of Embodiments 1-92 to 1-
104,
wherein the cannabinoid or the cannabinoid derivative is produced in an
amount, as
measured in mg/L or mM, at least 5%, at least 10%, at least 15%, at least 20%,
at least 25%,
at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least
60%, at least
70%, at least 80%, at least 90%, at least 100%, at least 150% at least 200%,
at least 500%, or
at least 1000% greater than an amount of the cannabinoid or the cannabinoid
derivative
produced in a method comprising culturing a modified host cell comprising one
or more
nucleic acids comprising a nucleotide sequence encoding a
tetrahydrocannabinolic acid
synthase polypeptide having an amino acid sequence of SEQ ID NO:44 instead of
the
modified host cell of any one of Embodiments 1-36 to 1-91, wherein the
modified host cell
comprising one or more nucleic acids comprising the nucleotide sequence
encoding the
tetrahydrocannabinolic acid synthase polypeptide having the amino acid
sequence of SEQ
ID NO:44 lacks a nucleic acid comprising a nucleotide sequence encoding an
engineered
200
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
variant of any one of Embodiments I-1 to 1-29, and wherein the modified host
cell of any
one of Embodiments 1-36 to 1-91 and the modified host cell comprising one or
more nucleic
acids comprising the nucleotide sequence encoding the tetrahydrocannabinolic
acid synthase
polypeptide having the amino acid sequence of SEQ ID NO:44, but lacking a
nucleic acid
comprising a nucleotide sequence encoding an engineered variant of any one of
Embodiments I-1 to 1-29, are cultured under similar culture conditions for the
same length of
time.
[0555] Embodiment 1-106. The method of any one of Embodiments 1-92 to 1-
105,
wherein the cannabinoid is tetrahydrocannabinolic acid (THCA), and wherein the
method
produces THCA in an increased ratio of THCA over another cannabinoid (e.g.,
cannabichromenic acid (CBCA)) compared to that produced in a method comprising
culturing a modified host cell comprising one or more nucleic acids comprising
a nucleotide
sequence encoding a tetrahydrocannabinolic acid synthase polypeptide having an
amino acid
sequence of SEQ ID NO:44 instead of the modified host cell of any one of
Embodiments I-
36 to 1-91, wherein the modified host cell comprising one or more nucleic
acids comprising
the nucleotide sequence encoding the tetrahydrocannabinolic acid synthase
polypeptide
having the amino acid sequence of SEQ ID NO:44 lacks a nucleic acid comprising
a
nucleotide sequence encoding an engineered variant of any one of Embodiments I-
1 to 1-29,
grown under similar culture conditions for the same length of time.
[0556] Embodiment 1-107. The method of any one of Embodiments 1-92 to 1-
106,
wherein the method produces THCA from CBGA in a ratio of THCA over another
cannabinoid (e.g., CBCA) of about 11:1, about 11.5:1, about 12:1, about
12.5:1, about 13:1,
about 13.5:1, about 14:1, about 14.5:1, about 15:1, about 15.5:1, about 16:1,
about 16.5:1,
about 17:1, about 17.5:1, about 18:1, about 18.5:1, about 19:1, about 19.5:1,
about 20:1,
about 25:1, about 30:1, about 35:1, about 40:1, about 45:1, about 50:1, about
60:1, about
70:1, about 80:1, about 90:1, about 100:1, about 150:1, about 200:1, about
500:1, or greater
than about 500:1.
[0557] Embodiment 1-108. A method of producing a cannabinoid or a
cannabinoid
derivative, the method comprising use of an engineered variant of any one of
Embodiments
I-1 to 1-29.
[0558] Embodiment 1-109. The method of Embodiment 1-108, wherein the
method
comprises recovering the produced cannabinoid or cannabinoid derivative.
201
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0559] Embodiment I-110. The method of Embodiment 1-108 or 1-109, wherein
the cannabinoid is tetrahydrocannabinolic acid, tetrahydrocannabivarinic acid,
or
tetrahydrocannabivarin.
[0560] Embodiment I-111. The method of any one of Embodiments 1-108 to I-
110,
wherein the cannabinoid or the cannabinoid derivative is produced in an
amount, as
measured in mg/L or mM, greater than an amount of the cannabinoid or the
cannabinoid
derivative produced in a method comprising use of a tetrahydrocannabinolic
acid synthase
polypeptide having an amino acid sequence of SEQ ID NO:44 instead of the
engineered
variant of any one of Embodiments I-1 to 1-29, wherein the engineered variant
of any one of
Embodiments I-1 to 1-29, and the tetrahydrocannabinolic acid synthase
polypeptide having
the amino acid sequence of SEQ ID NO:44 are used under similar conditions for
the same
length of time.
[0561] Embodiment 1-112. The method of any one of Embodiments 1-108 to I-
111,
wherein the cannabinoid or the cannabinoid derivative is produced in an
amount, as
measured in mg/L or mM, at least 5%, at least 10%, at least 15%, at least 20%,
at least 25%,
at least 30%, at least 35%, at least 40%, at least 45%, at least 50%, at least
60%, at least
70%, at least 80%, at least 90%, at least 100%, at least 150% at least 200%,
at least 500%, or
at least 1000% greater than an amount of the cannabinoid or the cannabinoid
derivative
produced in a method comprising use of a tetrahydrocannabinolic acid synthase
polypeptide
having an amino acid sequence of SEQ ID NO:44 instead of the engineered
variant of any
one of Embodiments I-1 to 1-29, wherein the engineered variant of any one of
Embodiments
I-1 to 1-29 and the tetrahydrocannabinolic acid synthase polypeptide having
the amino acid
sequence of SEQ ID NO:44 are used under similar conditions for the same length
of time.
[0562] Embodiment 1-113. The method of any one of Embodiments 1-108 to 1-
112,
wherein the cannabinoid is tetrahydrocannabinolic acid (THCA), and wherein the
method
produces THCA in an increased ratio of THCA over another cannabinoid (e.g.,
cannabichromenic acid (CBCA)) compared to that produced in a method comprising
use of a
tetrahydrocannabinolic acid synthase polypeptide having an amino acid sequence
of SEQ ID
NO:44 instread of the engineered variant of any one of Embodiments I-1 to 1-
29, wherein the
engineered variant of any one of Embodiments I-1 to 1-29 and the
tetrahydrocannabinolic
acid synthase polypeptide having the amino acid sequence of SEQ ID NO:44 are
used under
similar conditions for the same length of time.
202
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
[0563] Embodiment 1-114. The method of any one of Embodiments 1-108 to 1-
113,
wherein the method produces THCA from CBGA in a ratio of THCA over another
cannabinoid (e.g., CBCA) of about 11:1, about 11.5:1, about 12:1, about
12.5:1, about 13:1,
about 13.5:1, about 14:1, about 14.5:1, about 15:1, about 15.5:1, about 16:1,
about 16.5:1,
about 17:1, about 17.5:1, about 18:1, about 18.5:1, about 19:1, about 19.5:1,
about 20:1,
about 25:1, about 30:1, about 35:1, about 40:1, about 45:1, about 50:1, about
60:1, about
70:1, about 80:1, about 90:1, about 100:1, about 150:1, about 200:1, about
500:1, or greater
than about 500:1.
[0564] Embodiment 1-115. A method of screening an engineered variant of a
tetrahydrocannabinolic acid synthase (THCAS) polypeptide comprising an amino
acid
sequence of SEQ ID NO:44 with one or more amino acid substitutions, the method
comprising: a) dividing a population of host cells into a control population
and a test
population; b) co-expressing in the control population a THCAS polypeptide
having an
amino acid sequence of SEQ ID NO:44 and a comparison tetrahydrocannabinolic
synthase
polypeptide, wherein the THCAS polypeptide having an amino acid sequence of
SEQ ID
NO:44 can convert cannabigerolic acid (CBGA) to a first cannabinoid,
tetrahydrocannabinolic acid (THCA), and the comparison cannabinoid synthase
polypeptide
can convert the same CBGA to a different second cannabinoid; c) co-expressing
in the test
population the engineered variant and the comparison cannabinoid synthase
polypeptide,
wherein the engineered variant may convert CBGA to the same first cannabinoid,
tetrahydrocannabinolic acid (THCA), as the THCAS polypeptide having an amino
acid
sequence of SEQ ID NO:44, and wherein the comparison tetrahydrocannabinolic
synthase
polypeptide can convert the same CBGA to the second cannabinoid and is
expressed at
similar levels in the test population and in the control population; d)
measuring a ratio of the
first cannabinoid, tetrahydrocannabinolic acid (THCA), over the second
cannabinoid
produced by both the test population and the control population; and e)
measuring an
amount, in mg/L or mM, of the first cannabinoid produced by both the test
population andthe
control population.
[0565] Embodiment 1-116. The method of Embodiment 1-115, wherein the test
population is identified as comprising an engineered variant having improved
in vivo
performance compared to the tetrahydrocannabinolic acid synthase polypeptide
having an
amino acid sequence of SEQ ID NO:44, wherein improved in vivo performance is
demonstrated by an increase in the ratio of the first cannabinoid over the
second cannabinoid
203
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
produced by the test population compared to that produced by the control
population under
similar culture conditions for the same length of time.
[0566] Embodiment 1-117. The method of Embodiment 1-115 or 1-116, wherein
the test population is identified as comprising an engineered variant having
improved in vivo
performance compared to the tetrahydrocannabinolic acid synthase polypeptide
having an
amino acid sequence of SEQ ID NO:44 by producing the first cannabinoid in a
greater
amount, as measured in mg/L or mM, by the test population compared to the
amount
produced by the control population under similar culture conditions for the
same length of
time.
[0567] Embodiment 1-118. The method of any one of Embodiments 1-115 to 1-
117,
wherein the cannabinoid synthase polypeptide is a cannabidiolic acid synthase
polypeptide.
[0568] Embodiment 1-119. The method of Embodiment 1-118, wherein the
cannabidiolic acid synthase polypeptide comprises an amino acid sequence
having at least
85% sequence identity to SEQ ID NO:3.
[0569] Embodiment 1-120. The method of any one of Embodiments 1-115 to 1-
119,
wherein the second cannabinoid is cannabidiolic acid (CBDA).
[0570] Embodiment 1-121. The method of any one of Embodiments 1-115 to 1-
119,
wherein the engineered variant is an engineered variant of any one of
Embodiments I-1 to I-
29.
[0571] Provided in Table 1 are amino acid and nucleotide sequences
disclosed
herein. Where a genus and/or species is noted, the sequence should not be
construed to be
limited only to the specified genus and/or species, but also includes other
genera and/or
species expressing said sequence. Orthologs of the sequences disclosed in
Table 1 may also
be encompassed by this disclosure. Nucleotide sequences indicated as codon
optimized in
Table 1 are codon optimized for expression in S. cerevisiae. In Table 1, "*"
used as the end
of a sequence denotes a stop codon. In reference to OAC*, "*" denotes a
mutation is present
in the sequence.
Table 1: Amino acid and nucleotide sequences of the disclosure
SEQ ID NO:1 ATGAAATGCTCTACCTTTTCTTTCTGGTTCGTTTGTAAGATTATCTTCTTCTT
CTTCTCCTTCAACATCCAAACCTCTATCGCTAACCCTCGTGAAAACTTTTTG
Cannabidiolic Acid
AAATGTTTTTCCCAATACATCCCAAATAACGCTACTAATTTGAAGTTGGTTT
(CBDA) Synthase
ACACCCAAAACAACCCATTGTATATGTCCGTTTTAAACTCTACTATTCACA
Codon opt 2 ATTTGCGTTTTACCTCTGATACTACCCCTAAACCATTGGTCATTGTTACCCC
Artificial sequence ATCCCATGTTTCTCATATCCAAGGTACTATCTTGTGTTCTAAAAAGGTTGGT
TTGCAAATTAGAACTCGTTCCGGTGGTCACGATTCTGAAGGTATGTCTTAC
204
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Codon optimized ATTTCTCAAGTTCCTTTCGTCATTGTCGACTTGAGAAACATGAGATCCATCA
AAATTGATGTTCACTCTCAAACTGCTTGGGTCGAAGCCGGTGCCACTTTAG
GTGAGGTCTACTATTGGGTTAACGAGAAGAACGAAAACTTGTCTTTGGCTG
CCGGTTACTGTCCAACTGTCTGTGCTGGTGGTCATTTTGGTGGTGGTGGTTA
CGGTCCATTGATGAGAAACTACGGTTTGGCTGCTGATAACATTATTGATGC
TCACTTAGTTAACGTCCACGGTAAAGTCTTGGATAGAAAGTCCATGGGTGA
AGACTTGTTCTGGGCTTTAAGAGGTGGTGGTGCTGAATCCTTCGGTATTAT
TGTTGCTTGGAAAATCAGATTGGTCGCTGTTCCAAAATCCACCATGTTTTCT
GTCAAGAAAATCATGGAAATTCATGAATTAGTTAAGTTGGTCAACAAATG
GCAAAACATTGCCTATAAATACGACAAGGATTTGTTGTTGATGACTCATTT
CATCACTCGTAACATCACTGATAATCAAGGTAAGAACAAGACTGCTATCCA
TACTTACTTCTCTTCCGTCTTCTTGGGTGGTGTTGACTCTTTGGTCGATTTGA
TGAACAAATCCTTTCCAGAGTTAGGTATTAAGAAGACTGACTGTAGACAAT
TATCTTGGATTGACACTATTATCTTCTACTCTGGTGTTGTCAATTACGATAC
TGATAACTTTAACAAGGAAATTTTGTTGGACCGTTCTGCTGGTCAAAACGG
TGCCTTCAAGATTAAGTTAGATTACGTTAAGAAGCCAATCCCAGAATCTGT
CTTCGTCCAAATTTTGGAGAAATTGTATGAAGAGGACATTGGTGCTGGTAT
GTACGCCTTGTATCCTTACGGTGGTATCATGGACGAGATCTCCGAATCTGC
CATCCCTTTTCCTCATCGTGCTGGTATCTTGTACGAGTTGTGGTACATCTGT
TCCTGGGAGAAGCAAGAAGATAATGAAAAGCACTTGAACTGGATTAGAAA
TATTTATAATTTCATGACTCCATACGTTTCTAAGAACCCACGTTTGGCTTAC
TTAAATTACAGAGATTTGGATATTGGTATCAACGACCCTAAGAACCCTAAC
AACTACACTCAAGCTAGAATTTGGGGTGAGAAATATTTCGGTAAGAACTTC
GATAGATTGGTCAAGGTTAAAACTTTAGTTGATCCAAATAACTTTTTTAGA
AACGAACAATCTATTCCACCATTGCCAAGACACAGACACTAG
SEQ ID NO :2 ATGAAATGCTCCACTTTCTCTTTCTGGTTCGTTTGTAAGATTATCTTCTTCTT
C bid l A d CTTTTCTTTCAACATCCAAACTTCCATTGCCAACCCTCGTGAGAACTTCTTG
annaioic ci
(CBDA AAATGTTTTTCTCAATATATCCCAAATAACGCTACTAACTTGAAGTTAGTCT
Synthase )
ATACTCAAAACAACCCATTATATATGTCTGTCTTAAACTCTACCATTCACA
Codon opt 5 ACTTACGTTTCACTTCTGATACTACTCCAAAACCTTTGGTCATCGTCACCCC
Artificial se quence ATCCCACGTTTCTCACATCCAAGGTACCATCTTGTGTTCCAAAAAGGTTGG
TTTACAAATCCGTACTAGATCCGGTGGTCATGACTCCGAAGGTATGTCTTA
Codon optimized CATTTCCCAAGTCCCTTTCGTCATCGTCGACTTAAGAAATATGCGTTCCATC
AAGATTGATGTCCATTCCCAAACTGCTTGGGTTGAAGCCGGTGCCACTTTA
GGTGAAGTCTATTACTGGGTTAACGAGAAGAATGAGAACTTATCTTTGGCT
GCCGGTTACTGTCCAACTGTTTGTGCTGGTGGTCATTTCGGTGGTGGTGGTT
ACGGTCCATTAATGCGTAACTACGGTTTGGCTGCCGATAACATCATTGATG
CCCACTTAGTCAACGTTCATGGTAAGGTCTTGGACCGTAAGTCTATGGGTG
AGGATTTATTCTGGGCTTTGAGAGGTGGTGGTGCTGAATCTTTCGGTATTA
TCGTCGCTTGGAAGATTAGATTAGTTGCTGTTCCAAAGTCTACTATGTTCTC
TGTTAAGAAGATCATGGAAATTCACGAGTTGGTTAAATTAGTTAACAAATG
GCAAAACATTGCCTACAAGTACGATAAAGATTTGTTATTAATGACTCACTT
TATCACTAGAAACATTACTGATAACCAAGGTAAGAATAAGACTGCCATTC
ACACTTACTTCTCTTCTGTTTTCTTGGGTGGTGTTGATTCCTTGGTCGATTTG
ATGAACAAGTCTTTTCCAGAATTAGGTATTAAGAAGACCGATTGTCGTCAA
TTATCTTGGATTGATACCATTATTTTTTACTCCGGTGTTGTCAACTACGACA
CTGATAATTTTAATAAGGAGATTTTGTTAGATAGATCTGCTGGTCAAAATG
GTGCCTTTAAAATCAAATTGGACTACGTTAAGAAGCCTATTCCAGAATCCG
TCTTTGTTCAAATTTTGGAGAAGTTATACGAAGAAGATATTGGTGCTGGTA
TGTACGCCTTGTATCCATATGGTGGTATTATGGATGAAATTTCTGAATCCG
CCATCCCTTTCCCTCATCGTGCTGGTATCTTATACGAGTTGTGGTACATCTG
TTCTTGGGAAAAGCAAGAAGATAATGAAAAGCATTTGAACTGGATCCGTA
ACATCTATAACTTCATGACTCCATACGTTTCCAAAAACCCTAGATTGGCTT
ACTTAAATTACAGAGACTTAGATATTGGTATTAACGACCCTAAGAACCCAA
ACAATTACACTCAAGCTAGAATCTGGGGTGAAAAGTACTTCGGTAAGAAT
TTCGACAGATTAGTTAAGGTCAAGACTTTAGTTGACCCAAATAACTTCTTC
AGAAACGAACAATCTATCCCACCATTGCCTAGACATAGACACTAG
SEQ ID NO :3 MKCSTFSFWFVCKIIFFFF SFNIQTSIANPRENFLKCFSQYIPNNATNLKLVYTQ
NNPLYMS VLNSTIHNLRFTSDTTPKPLVIVTP SHVSHIQGTILCSKKVGLQIRTR
205
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Cannabidiolic Acid SGGHD SEGMSYISQVPFVIVDLRNMRSIKIDVHSQTAWVEAGATL GEVYYWV
(CBDA) Synthase NEKNENL SL AAGYCPTVCAGGHFGGGGYGPLMRNYGLAADNIIDAHLVNVH
GKVLDRKSMGEDLFWALRGGGAESFGIIVAWKIRLVAVPKS TMF SVKKIMEIH
Polypeptide from codon
ELVKLVNKWQNIAYKYDKDLLLMTHFITRNITDNQGKNKTAIHTYFS SVFLGG
opts 2 and 5
VD SLVDLMNKSFPEL GIKKTDCRQL SWIDTIIFYSGVVNYDTDNFNKEILLDRS
Cannabis sativa AGQNGAFKIKLDYVKKPIPESVFVQILEKLYEEDIGAGMYALYPYGGIMDEISE
SAIPFPHRAGILYELWYICSWEKQEDNEKHLNWIRNIYNFMTPYVSKNPRLAY
LNYRDLD IGINDPKNPNNYTQARIWGEKYFGKNFDRLVKVKTLVDPNNFFRN
EQSIPPLPRHRH*
SEQ ID NO :4 ATGTTTTTCAACAGACTAAGCGCTGGCAAGCTGCTGGTACCACTCTCCGTG
KAR2 GTCCTGTACGCCCTTTTCGTGGTAATATTACCTTTACAGAATTCTTTCCACT
CCTCCAATGTTTTAGTTAGAGGTGCCGATGATGTAGAAAACTACGGAACTG
Saccharomyces sp. TTATCGGTATTGACTTAGGTACTACTTATTCCTGTGTTGCTGTGATGAAAAA
TGGTAAGACTGAAATTCTTGCTAATGAGCAAGGTAACAGAATCACCCCATC
TTACGTGGCATTCACCGATGATGAAAGATTGATTGGTGATGCTGCAAAGAA
CCAAGTTGCTGCCAATCCTCAAAACACCATCTTCGACATTAAGAGATTGAT
CGGTTTGAAATATAACGACAGATCTGTTCAGAAGGATATCAAGCACTTGCC
ATTTAATGTGGTTAATAAAGATGGGAAGCCCGCTGTAGAAGTAAGTGTCA
AAGGAGAAAAGAAGGTTTTTACTCCAGAAGAAATTTCTGGTATGATCTTGG
GTAAGATGAAACAAATTGCCGAAGATTATTTAGGCACTAAGGTTACCCAT
GCTGTCGTTACTGTTCCTGCTTATTTCAATGACGCGCAAAGACAAGCCACC
AAGGATGCTGGTACCATCGCTGGTTTGAACGTTTTGAGAATTGTTAATGAA
CCAACCGCAGCCGCCATTGCCTACGGTTTGGATAAATCTGATAAGGAACAT
CAAATTATTGTTTATGATTTGGGTGGTGGTACTTTCGATGTCTCTCTATTGT
CTATTGAAAACGGTGTTTTCGAAGTCCAAGCCACTTCTGGTGATACTCATT
TAGGTGGTGAAGATTTTGACTATAAGATCGTTCGTCAATTGATAAAAGCTT
TCAAGAAGAAGCATGGTATTGATGTGTCTGACAACAACAAGGCCCTAGCT
AAATTGAAGAGAGAAGCTGAAAAGGCTAAACGTGCCTTGTCCAGCCAAAT
GTCCACCCGTATTGAAATTGACTCCTTCGTTGATGGTATCGACTTAAGTGA
AACCTTGACCAGAGCTAAGTTTGAGGAATTAAACCTAGATCTATTCAAGAA
GACCTTGAAGCCTGTCGAGAAGGTTTTGCAAGATTCTGGTTTGGAAAAGAA
GGATGTTGATGATATCGTTTTGGTTGGTGGTTCTACTAGAATTCCAAAGGT
CCAACAATTGTTAGAATCATACTTTGATGGTAAGAAGGCCTCCAAGGGTAT
TAACCCAGATGAAGCTGTTGCATACGGTGCAGCCGTTCAAGCTGGTGTCTT
ATCCGGTGAAGAAGGTGTCGAAGATATTGTTTTATTGGATGTCAACGCTTT
GACTCTTGGTATTGAAACCACTGGTGGTGTCATGACTCCATTAATTAAGAG
AAATACTGCTATTCCTACAAAGAAATCCCAAATTTTCTCTACTGCCGTTGA
CAACCAACCAACCGTTATGATCAAGGTATACGAGGGTGAAAGAGCCATGT
CTAAGGACAACAATCTATTAGGTAAGTTTGAATTAACCGGCATTCCACCAG
CACCAAGAGGTGTACCTCAAATTGAAGTCACATTTGCACTTGACGCTAATG
GTATTCTGAAGGTGTCTGCCACAGATAAGGGAACTGGTAAATCCGAATCTA
TCACCATCACTAACGATAAAGGTAGATTAACCCAAGAAGAGATTGATAGA
ATGGTTGAAGAGGCTGAAAAATTCGCTTCTGAAGACGCTTCTATCAAGGCC
AAGGTTGAATCTAGAAACAAATTAGAAAACTACGCTCACTCTTTGAAAAA
CCAAGTTAATGGTGACCTAGGTGAAAAATTGGAAGAAGAAGACAAGGAA
ACCTTATTAGATGCTGCTAACGATGTTTTAGAATGGTTAGATGATAACTTT
GAAACCGCCATTGCTGAAGACTTTGATGAAAAGTTCGAATCTTTGTCCAAG
GTCGCTTATCCAATTACTTCTAAGTTGTACGGAGGTGCTGATGGTTCTGGT
GCCGCTGATTATGACGACGAAGATGAAGATGACGATGGTGATTATTTCGA
ACACGACGAATTGTAG
SEQ ID NO :5 MFFNRL SAGKLLVPLSVVLYALFVVILPLQNSFHS SNVLVRGADDVENYGTVI
KAR2 GIDLGTTYSCVAVIVIKNGK IEILANEQGNRITPSYVAFTDDERLIGDAAKNQVA
ANPQNTIFDIKRLIGLKYNDRSVQKDIKHLPFNVVNKD GKPAVEVSVKGEKKV
Saccharomyces sp. FTPEEISGMILGKMKQIAEDYLGTKVTHAVVTVPAYFNDAQRQATKDAGTIA
GLNVLRIVNEPTAAAIAYGLDKSDKEHQIIVYDLGGGTFDVSLL SIENGVFEVQ
ATSGDTHLGGEDFDYKIVRQLIKAFKKKHGIDVSDNNKALAKLKREAEKAKR
ALS SQMSTRIEID SFVDGIDL SETLTRAKFEELNLDLFKKTLKPVEKVLQD SGLE
KKDVDDIVLVGGSTRIPKVQQLLESYFDGKKASKGINPDEAVAYGAAVQAGV
LSGEEGVEDIVLLDVNALTLGIETTGGVIVITPLIKRNTAIPTKKSQIFSTAVDNQP
206
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TVMIKVYEGERAMSKDNNLLGKFELTGIPPAPRGVPQIEVTFALDANGILKVS
ATDKGTGKSESITITNDKGRLTQEEIDRIVIVEEAEKFASEDASIKAKVESRNKLE
NYAHSLKNQVNGDLGEKLEEEDKETLLDAANDVLEWLDDNFETAIAEDFDEK
FESLSKVAYPITSKLYGGADGSGAADYDDEDEDDDGDYFEHDEL*
SEQ ID NO :6 ATGAGATTAAGAACCGCCATTGCCACACTGTGCCTCACGGCTTTTACATCT
ER01 GCAACTTCAAACAATAGCTACATCGCCACCGACCAAACACAAAATGCCTTT
AATGACACTCACTTTTGTAAGGTCGACAGGAATGATCACGTTAGTCCCAGT
Saccharomyces sp. TGTAACGTAACATTCAATGAATTAAATGCCATAAATGAAAACATTAGAGA
TGATCTTTCGGCGTTATTAAAATCTGATTTCTTCAAATACTTTCGGCTGGAT
TTATACAAGCAATGTTCATTTTGGGACGCCAACGATGGTCTGTGCTTAAAC
CGCGCTTGCTCTGTTGATGTCGTAGAGGACTGGGATACACTGCCTGAGTAC
TGGCAGCCTGAGATCTTGGGTAGTTTCAATAATGATACAATGAAGGAAGC
GGATGATAGCGATGACGAATGTAAGTTCTTAGATCAACTATGTCAAACCA
GTAAAAAACCTGTAGATATCGAAGACACCATCAACTACTGTGATGTAAAT
GACTTTAACGGTAAAAACGCCGTTCTGATTGATTTAACAGCAAATCCGGAA
CGATTTACAGGTTATGGTGGTAAGCAAGCTGGTCAAATTTGGTCTACTATC
TACCAAGACAACTGTTTTACAATTGGCGAAACTGGTGAATCATTGGCCAAA
GATGCATTTTATAGACTTGTATCCGGTTTCCATGCCTCTATCGGTACTCACT
TATCAAAGGAATATTTGAACACGAAAACTGGTAAATGGGAGCCCAATCTG
GATTTGTTTATGGCAAGAATCGGGAACTTTCCTGATAGAGTGACAAACATG
TATTTCAATTATGCTGTTGTAGCTAAGGCTCTCTGGAAAATTCAACCATATT
TACCAGAATTTTCATTCTGTGATCTAGTCAATAAAGAAATCAAAAACAAAA
TGGATAACGTTATTTCCCAGCTGGACACAAAAATTTTTAACGAAGACTTAG
TTTTTGCCAACGACCTAAGTTTGACTTTGAAGGACGAATTCAGATCTCGCT
TCAAGAATGTCACGAAGATTATGGATTGTGTGCAATGTGATAGATGTAGAT
TGTGGGGCAAAATTCAAACTACCGGTTACGCAACTGCCTTGAAAATTTTGT
TTGAAATCAACGACGCTGATGAATTCACCAAACAACATATTGTTGGTAAGT
TAACCAAATATGAGTTGATTGCACTATTACAAACTTTCGGTAGATTATCTG
AATCTATTGAATCTGTTAACATGTTCGAAAAAATGTACGGGAAAAGGTTAA
ACGGTTCTGAAAACAGGTTAAGCTCATTCTTCCAAAATAACTTCTTCAACA
TTTTGAAGGAGGCAGGCAAGTCGATTCGTTACACCATAGAGAACATCAATT
CCACTAAAGAAGGAAAGAAAAAGACTAACAATTCTCAATCACATGTATTT
GATGATTTAAAAATGCCCAAAGCAGAAATAGTTCCAAGGCCCTCTAACGG
TACAGTAAATAAATGGAAGAAAGCTTGGAATACTGAAGTTAACAACGTTT
TAGAAGCATTCAGATTTATTTATAGAAGCTATTTGGATTTACCCAGGAACA
TCTGGGAATTATCTTTGATGAAGGTATACAAATTTTGGAATAAATTCATCG
GTGTTGCTGATTACGTTAGTGAGGAGACACGAGAGCCTATTTCCTATAAGC
TAGATATACAATAA
SEQ ID NO:? MRLRTAIATL CLTAFTSAT SNNSYIATDQTQNAFNDTHFCKVDRNDHVSP S CN
ER01 VTFNELNAINENIRDDL SAUK SDFFKYFRLDLYKQC SFWDAND GL CLNRAC S
VDVVEDWDTLPEYWQPEIL GSFNNDTMKEADD SDDECKFLDQLCQTSKKPV
Saccharomyces sp. DIEDTINYCDVNDFNGKNAVLIDLTANPERFTGYGGKQAGQIWSTIYQDNCFTI
GETGESLAKDAFYRLVSGFHASIGTHLSKEYLNTKTGKWEPNLDLFMARIGNF
PDRVTNMYFNYAVVAKALWKIQPYLPEF SFCDLVNKEIKNKMDNVISQLDTK
IFNEDLVFANDL SLTLKDEFRSRFKNVTKIMDCVQCDRCRLWGKIQTTGYATA
LKILFEINDADEFTKQHIVGKLTKYELIALLQTFGRL SESIESVNMFEKMYGKR
LNGSENRL S SFFQNNFFNILKEAGKSIRYTIENINSTKEGKKKTNNSQ SHVFDDL
KMPKAEIVPRP SNGTVNKWKKAWN 1EVNNVLEAFRFIYRSYLDLPRNIWEL S
LMKVYKFWNKFIGVADYVSEETREPISYKLDIQ*
SEQ ID NO :8 ATGAAGTTTTCTGCTGGTGCCGTCCTGTCATGGTCCTCCCTGCTGCTCGCCT
PDI1 CCTCTGTTTTCGCCCAACAAGAGGCTGTGGCCCCTGAAGACTCCGCTGTCG
TTAAGTTGGCCACCGACTCCTTCAATGAGTACATTCAGTCGCACGACTTGG
Saccharomyces sp. TGCTTGCGGAGTTTTTTGCTCCATGGTGTGGCCACTGTAAGAACATGGCTC
CTGAATACGTTAAAGCCGCCGAGACTTTAGTTGAGAAAAACATTACCTTGG
CCCAGATCGACTGTACTGAAAACCAGGATCTGTGTATGGAACACAACATTC
CAGGGTTCCCAAGCTTGAAGATTTTCAAAAACAGCGATGTTAACAACTCGA
TCGATTACGAGGGACCTAGAACTGCCGAGGCCATTGTCCAATTCATGATCA
AGCAAAGCCAACCGGCTGTCGCCGTTGTTGCTGATCTACCAGCTTACCTTG
CTAACGAGACTTTTGTCACTCCAGTTATCGTCCAATCCGGTAAGATTGACG
207
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
CCGACTTCAACGCCACCTTTTACTCCATGGCCAACAAACACTTCAACGACT
ACGACTTTGTCTCCGCTGAAAACGCAGACGATGATTTCAAGCTTTCTATTT
ACTTGCCCTCCGCCATGGACGAGCCTGTAGTATACAACGGTAAGAAAGCC
GATATCGCTGACGCTGATGTTTTTGAAAAATGGTTGCAAGTGGAAGCCTTG
CCCTACTTTGGTGAAATCGACGGTTCCGTTTTCGCCCAATACGTCGAAAGC
GGTTTGCCTTTGGGTTACTTATTCTACAATGACGAGGAAGAATTGGAAGAA
TACAAGCCTCTCTTTACCGAGTTGGCCAAAAAGAACAGAGGTCTAATGAA
CTTTGTTAGCATCGATGCCAGAAAATTCGGCAGACACGCCGGCAACTTGAA
CATGAAGGAACAATTCCCTCTATTTGCCATCCACGACATGACTGAAGACTT
GAAGTACGGTTTGCCTCAACTCTCTGAAGAGGCGTTTGACGAATTGAGCGA
CAAGATCGTGTTGGAGTCTAAGGCTATTGAATCTTTGGTTAAGGACTTCTT
GAAAGGTGATGCCTCCCCAATCGTGAAGTCCCAAGAGATCTTCGAGAACC
AAGATTCCTCTGTCTTCCAATTGGTCGGTAAGAACCATGACGAAATCGTCA
ACGACCCAAAGAAGGACGTTCTTGTTTTGTACTATGCCCCATGGTGTGGTC
ACTGTAAGAGATTGGCCCCAACTTACCAAGAACTAGCTGATACCTACGCCA
ACGCCACATCCGACGTTTTGATTGCTAAACTAGACCACACTGAAAACGATG
TCAGAGGCGTCGTAATTGAAGGTTACCCAACAATCGTCTTATACCCAGGTG
GTAAGAAGTCCGAATCTGTTGTGTACCAAGGTTCAAGATCCTTGGACTCTT
TATTCGACTTCATCAAGGAAAACGGTCACTTCGACGTCGACGGTAAGGCCT
TGTACGAAGAAGCCCAGGAAAAAGCTGCTGAGGAAGCCGATGCTGACGCT
GAATTGGCTGACGAAGAAGATGCCATTCACGATGAATTGTAA
SEQ ID NO :9 MKFSAGAVL SWS SLLL AS SVFAQQEAVAPED SAVVKLATD SFNEYIQSHDLVL
PDI1 AEFFAPWCGHCKNMAPEYVKAAETLVEKNITLAQIDC 1ENQDL CMEHNIPGF
PSLKIFKNSDVNNSIDYEGPRTAEAIVQFMIKQ SQPAVAVVADLPAYLANETFV
Saccharomyces sp. TPVIVQSGKIDADFNATFYSMANKHFNDYDFVSAENADDDFKL SIYLPSAMDE
PVVYNGKKADIADADVFEKWLQVEALPYFGEID GSVFAQYVE S GLPLGYLFY
NDEEELEEYKPLF 1ELAKKNRGLMNFVSIDARKFGRHAGNLNMKEQFPLFAIH
DM 1EDLKYGLPQL SEEAFDELSDKIVLESKAIESLVKDFLKGDASPIVKSQEIFE
NQD S SVFQLVGKNHDEIVNDPKKDVLVLYYAPWCGHCKRLAPTYQELADTY
ANATSDVLIAKLDH 1ENDVRGVVIEGYPTIVLYPGGKKSESVVYQGSRSLDSL
FDFIKENGHFDVDGKALYEEAQEKAAEEADADAELADEEDAIHDEL*
SEQ ID NO:10 ATGCGTCTACTTCGAAGAAACATGTTAGTATTGACACTGCTCGTTTGTGTG
IRE 1 TTTTCATCCATCATTTCATGCTCAATCCCATTGTCGTCTCGCACCTCAAGGC
GGCAGATAGTGGAAGATGAAGTTGCCTCCACTAAAAAGCTCAATTTCAAC
Saccharomyces sp. TATGGTGTGGATAAAAATATAAACTCGCCCATTCCTGCTCCAAGAACCACT
GAAGGTTTACCAAATATGAAACTCAGCTCATATCCAACTCCTAACTTATTG
AATACTGCTGATAATCGACGTGCTAACAAAAAAGGACGTAGGGCTGCCAA
TTCTATAAGTGTACCCTATTTGGAGAATCGTTCCTTGAACGAACTGAGTTT
ATCAGATATACTAATCGCAGCCGACGTTGAGGGTGGACTTCATGCTGTAGA
TAGAAGAAATGGTCATATCATATGGTCAATCGAACCAGAAAATTTTCAACC
TCTGATAGAAATACAAGAACCTTCGAGGTTAGAAACATATGAAACGTTGA
TTATAGAACCTTTCGGTGATGGGAACATTTACTACTTTAACGCCCATCAAG
GGTTACAAAAACTGCCTTTATCCATACGACAACTTGTATCAACTTCCCCGC
TGCACTTGAAAACAAATATTGTGGTTAATGACTCTGGAAAAATTGTTGAAG
ATGAAAAGGTCTACACTGGATCGATGAGAACTATAATGTATACTATAAAC
ATGTTGAATGGTGAAATTATATCAGCGTTCGGACCTGGTTCAAAAAACGGG
TATTTCGGGAGCCAGAGTGTGGATTGCTCACCTGAGGAGAAGATAAAACT
TCAGGAATGTGAAAATATGATTGTAATAGGCAAAACTATTTTTGAGCTGGG
AATTCACTCTTATGATGGAGCAAGCTACAATGTCACTTACTCTACATGGCA
GCAAAATGTTTTAGATGTTCCCCTAGCGCTTCAGAATACATTTTCAAAGGA
CGGCATGTGCATAGCGCCTTTCCGTGATAAATCATTGCTAGCAAGCGATTT
AGATTTTAGAATTGCTAGATGGGTTTCTCCGACATTCCCCGGAATTATTGTT
GGGCTTTTCGATGTGTTTAATGATCTCCGCACCAATGAAAATATACTGGTA
CCGCATCCCTTTAATCCTGGTGATCATGAAAGTATATCGAGTAACAAAGTT
TACTTGGATCAGACTTCGAACCTCTCCTGGTTTGCATTATCTAGTCAGAATT
TTCCATCTTTAGTCGAATCAGCTCCCATATCAAGATACGCTTCCAGTGACC
GTTGGAGGGTGTCTTCAATTTTTGAAGATGAGACTTTATTCAAGAACGCAA
TCATGGGTGTTCATCAGATATATAATAATGAATATGATCACCTTTATGAAA
ACTATGAAAAAACGAATAGTTTGGACACTACGCACAAATATCCACCTCTG
208
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
ATGATTGATTCGTCCGTTGATACAACCGATTTACATCAGAATAACGAGATG
AATTCACTAAAGGAATACATGTCACCAGAAGACCTTGAGGCATATAGAAA
AAAGATACACGAGCAAATATCGAGAGAATTAGATGAAAAGAACCAAAATT
CTTTGCTACTGAAGTTTGGAAGTCTAGTATATCGAATTATAGAGACTGGAG
TATTTCTGTTGTTATTTCTCATTTTTTGTGCAATACTACAAAGATTCAAAAT
TTTGCCGCCACTATATGTATTATTATCCAAAATTGGATTTATGCCTGAAAA
GGAAATCCCCATAGTTGAGTCGAAATCGCTAAATTGTCCCTCTTCATCGGA
AAATGTAACCAAGCCATTCGATATGAAATCAGGGAAGCAAGTTGTTTTTGA
AGGTGCTGTGAACGATGGAAGTCTAAAATCTGAAAAAGATAACGATGATG
CTGATGAAGATGATGAAAAATCACTAGATTTAACCACAGAAAAGAAGAAG
AGGAAAAGAGGTTCGAGAGGAGGCAAAAAGGGCCGAAAATCACGCATTG
CAAATATACCAAACTTTGAGCAATCTTTAAAAAATTTGGTAGTATCCGAAA
AAATTTTAGGTTACGGTTCATCAGGAACAGTAGTTTTTCAGGGAAGTTTTC
AAGGAAGACCTGTTGCGGTAAAGAGAATGTTAATTGATTTTTGTGACATAG
CTTTAATGGAAATAAAACTTTTGACTGAAAGCGATGATCACCCTAACGTCA
TACGATACTACTGTTCAGAAACAACAGACAGATTTTTGTATATTGCTTTAG
AGCTCTGCAATTTGAACCTTCAAGATTTGGTGGAGTCTAAGAATGTATCAG
ATGAAAACCTGAAATTACAGAAAGAGTATAATCCAATTTCGTTATTGAGAC
AAATAGCGTCCGGGGTAGCACATTTACATTCTTTAAAGATTATCCATCGAG
ATTTAAAGCCTCAAAATATTCTCGTTTCTACTTCGAGTAGGTTTACTGCCGA
TCAGCAAACAGGAGCAGAAAATCTTCGAATTTTGATATCAGACTTTGGTCT
TTGCAAAAAACTAGACTCTGGTCAGTCTTCATTTAGAACAAATTTGAATAA
CCCTTCTGGCACAAGTGGTTGGAGGGCCCCAGAGCTGCTTGAAGAATCAA
ACAATTTGCAGTGCCAAGTCGAAACGGAACACTCTTCTAGTAGGCATACA
GTAGTTTCATCTGATTCTTTTTATGATCCGTTCACCAAGAGGAGGCTAACA
AGATCTATTGATATTTTTTCTATGGGATGTGTATTCTATTATATCCTATCCA
AAGGGAAGCATCCATTTGGAGATAAATATTCACGTGAAAGCAATATCATA
AGAGGAATATTCAGTCTTGATGAAATGAAATGTCTACATGATAGATCCTTA
ATTGCAGAAGCTACAGATCTGATCTCCCAAATGATTGATCACGATCCGTTA
AAAAGACCTACTGCTATGAAAGTTCTAAGGCATCCGTTGTTTTGGCCAAAG
TCGAAAAAATTGGAGTTCCTTTTAAAAGTTAGTGATAGGCTTGAAATTGAA
AACAGAGACCCTCCAAGTGCCCTGTTAATGAAATTTGACGCCGGTTCTGAC
TTTGTAATACCCAGTGGAGATTGGACTGTCAAGTTTGATAAAACATTCATG
GACAACCTTGAAAGGTACAGAAAATACCATTCATCAAAGTTAATGGATCT
ATTAAGAGCACTTAGGAATAAATATCATCATTTTATGGATTTACCTGAAGA
TATAGCAGAACTAATGGGGCCGGTACCCGATGGATTTTACGATTACTTCAC
CAAGCGTTTTCCAAACCTATTAATAGGTGTTTATATGATTGTCAAGGAAAA
TTTAAGTGACGATCAAATTTTACGTGAATTTTTGTATTCATAA
SEQ ID NO:11 MRLLRRNMLVLTLLVCVFS S II S C S IPL S SRT
SRRQIVEDEVASTKKLNFNYGVD
IRE1 KNINSPIPAPRTIEGLPNMKLSSYPTPNLLNTADNRRANKKGRRAANSISVPYL
ENRSLNEL SL SDILIAADVEGGLHAVDRRNGHIIWSIEPENFQPLIEIQEP SRLET
Saccharomyces sp. YETLIIEPFGDGNIYYFNAHQGLQKLPL SIRQLVST SPLHLKTNIVVND
SGKIVE
DEKVYTGSMRTIMYTINMLNGEIISAFGPGSKNGYFGSQ SVD CSPEEKIKLQEC
ENMIVIGKTIFELGIHSYDGASYNVTYSTWQQNVLDVPLALQNTF SKDGMCIA
PFRDKSLLASDLDFRIARWVSPTFPGIIVGLFDVFNDLRTNENILVPHPFNPGDH
ESISSNKVYLDQTSNL SWFAL SSQNFPSLVESAPISRYASSDRWRVSSIFEDETL
FKNAIMGVHQIYNNEYDHLYENYEKTNSLDTTHKYPPLMIDSSVDTTDLHQN
NEMNSLKEYMSPEDLEAYRKKIHEQISRELDEKNQNSLLLKFGSLVYRIIETGV
FLLLFL IF CAIL QRFKILPPLYVLL SKI GFMPEKEIPIVE SK SLN CP S S SENVTKPFD
MKSGKQVVFEGAVNDGSLKSEKDNDDADEDDEKSLDLTTEKKKRKRGSRGG
KKGRKSRIANIPNFEQSLKNLVVSEKILGYGSSGTVVFQGSFQGRPVAVKRMLI
DFCDIALMEIKLL 1ESDDHPNVIRYYCSETTDRFLYIALELCNLNLQDLVESKN
VSDENLKLQKEYNPISLLRQIASGVAHLH SLKIIHRDLKPQNILVSTS SRFTADQ
QTGAENLRILISDFGLCKKLD SGQ SSFRTNLNNP S GT SGWRAPELLEESNNLQC
QVETEHSS SRHTVVSSDSFYDPFTKRRL TRSIDIF SMGCVFYYIL SKGKHPFGDK
YSRESNIIRGIF SLDEMKCLHDRSLIAEATDLISQMIDHDPLKRPTAMKVLRHPL
FWPKSKKLEFLLKVSDRLEIENRDPP SALLMKFDAGSDFVIPSGDWTVKFDKT
FMDNLERYRKYH S SKLMDLLRALRNKYHHFMDLPEDIAELMGPVPDGFYDY
FTKRFPNLLIGVYMIVKENLSDDQILREFLYS*
209
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
SEQ ID NO:12 ATGGTCCTTTTGAAATGGCTCGTATGCCAATTGGTCTTCTTTACCGCTTTTT
CGCATGCGTTTACCGACTATCTATTAAAGAAGTGTGCGCAATCTGGGTTTT
rot2
GCCATAGAAACAGGGTTTATGCAGAAAATATTGCCAAATCTCATCACTGCT
Saccharomyces sp. ATTACAAAGTGGACGCCGAGTCTATTGCACACGATCCTTTAGAGAATGTGC
TTCATGCTACCATAATTAAAACTATACCAAGATTGGAGGGCGATGATATAG
CCGTTCAGTTCCCATTCTCTCTCTCTTTTTTACAGGATCACTCAGTAAGGTT
CACTATAAATGAGAAAGAGAGAATGCCAACCAACAGCAGCGGTTTGTTGA
TCTCTTCACAACGGTTCAATGAGACCTGGAAGTACGCATTCGACAAGAAAT
TTCAAGAGGAGGCGAACAGGACCAGTATTCCACAATTCCACTTCCTTAAGC
AAAAACAAACTGTGAACTCATTCTGGTCGAAAATATCTTCATTTTTGTCAC
TTTCAAACTCCACTGCAGACACATTTCATCTTCGAAACGGTGATGTATCCG
TAGAAATCTTTGCTGAACCTTTTCAATTGAAAGTTTACTGGCAAAATGCGC
TGAAACTTATTGTAAACGAGCAAAATTTCCTGAACATTGAACATCATAGAA
CTAAGCAGGAAAACTTCGCACACGTGCTGCCAGAAGAAACAACTTTCAAC
ATGTTTAAGGACAATTTCTTGTATTCAAAGCATGACTCTATGCCTTTGGGG
CCTGAATCGGTTGCGCTAGATTTCTCTTTCATGGGTTCTACTAATGTCTACG
GTATACCGGAACATGCGACGTCGCTAAGGCTGATGGACACTTCAGGTGGA
AAGGAACCCTACAGGCTTTTCAACGTTGATGTCTTTGAGTACAACATCGGT
ACCAGCCAACCAATGTACGGTTCGATCCCATTCATGTTTTCATCTTCGTCCA
CATCTATCTTTTGGGTCAATGCAGCTGACACTTGGGTAGACATAAAGTATG
ACACCAGTAAAAATAAAACGATGACTCATTGGATCTCCGAAAATGGTGTC
ATAGATGTAGTCATGTCCCTGGGGCCAGATATTCCAACTATCATTGACAAA
TTTACCGATTTGACTGGTAGACCCTTTTTACCGCCCATTTCCTCTATAGGGT
ACCATCAATGTAGATGGAATTATAATGATGAGATGGACGTTCTCACAGTGG
ACTCTCAGATGGATGCTCATATGATTCCTTACGATTTTATTTGGTTGGACTT
GGAGTATACGAACGACAAAAAATATTTTACTTGGAAGCAGCACTCCTTTCC
CAATCCAAAAAGGCTGTTATCCAAATTAAAAAAGTTGGGTAGAAATCTTGT
CGTACTAATCGATCCTCATTTAAAGAAAGATTATGAAATCAGTGACAGGGT
AATTAATGAAAATGTAGCAGTCAAGGATCACAATGGAAATGACTATGTAG
GTCATTGCTGGCCAGGTAATTCTATATGGATTGATACCATAAGCAAATATG
GCCAAAAGATTTGGAAGTCCTTTTTCGAACGGTTTATGGATCTGCCGGCTG
ATTTAACTAATTTATTCATTTGGAATGATATGAACGAGCCTTCGATTTTCGA
TGGCCCAGAGACCACAGCTCCAAAAGATTTGATTCACGACAATTACATTGA
GGAAAGATCCGTCCATAACATATATGGTCTATCAGTGCATGAAGCTACTTA
CGACGCAATAAAATCGATTTATTCACCATCCGATAAGCGTCCTTTCCTTCT
AACAAGGGCTTTTTTTGCCGGCTCTCAACGTACTGCTGCCACATGGACTGG
TGACAATGTGGCCAATTGGGATTACTTAAAGATTTCCATTCCTATGGTTCT
GTCAAACAACATTGCTGGTATGCCATTTATAGGAGCCGACATAGCTGGCTT
TGCTGAGGATCCTACACCTGAATTGATTGCACGTTGGTACCAAGCGGGCTT
ATGGTACCCATTTTTTAGAGCACACGCCCATATAGACACCAAGAGAAGAG
AACCATACTTATTCAATGAACCTTTGAAGTCGATAGTACGTGATATTATCC
AATTGAGATATTTCCTGCTACCTACCTTATACACCATGTTTCATAAATCAAG
TGTCACTGGATTTCCGATAATGAATCCAATGTTTATTGAACACCCTGAATTT
GCTGAATTGTATCATATCGATAACCAATTTTACTGGAGTAATTCAGGTCTA
TTAGTCAAACCTGTCACGGAGCCTGGTCAATCAGAAACGGAAATGGTTTTC
CCACCCGGTATATTCTATGAATTCGCATCTTTACACTCTTTTATAAACAATG
GTACTGATTTGATAGAAAAGAATATTTCTGCACCATTGGATAAAATTCCAT
TATTTATTGAAGGCGGTCACATTATCACTATGAAAGATAAGTATAGAAGAT
CTTCAATGTTAATGAAAAACGATCCATATGTAATAGTTATAGCCCCTGATA
CCGAGGGACGAGCCGTTGGAGATCTTTATGTTGATGATGGAGAAACTTTTG
GCTACCAAAGAGGTGAGTACGTAGAAACTCAGTTCATTTTCGAAAACAAT
ACCTTAAAAAATGTTCGAAGTCATATTCCCGAGAATTTGACAGGCATTCAC
CACAATACTTTGAGGAATACCAATATTGAAAAAATCATTATCGCAAAGAA
TAATTTACAACACAACATAACGTTGAAAGACAGTATTAAAGTCAAAAAAA
ATGGCGAAGAAAGTTCATTGCCGACTAGATCGTCATATGAGAATGATAAT
AAGATCACCATTCTTAACCTATCGCTTGACATAACTGAAGATTGGGAAGTT
ATTTTTTGA
SEQ ID NO:13 MVLLKWLVCQLVFFTAF SHAFTDYLLKKCAQ SGFCHRNRVYAENIAKSHHCY
rot2 YKVDAESIAHDPLENVLHATIIKTIPRLEGDDIAVQFPFSL SFLQDHS VRFTINEK
210
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Saccharomyces sp. ERMPTNS SGLLIS SQRFNETWKYAFDKKFQEEANRTSIPQFHFLKQKQTVNSF
W SKI S SFL SL SN STADTFHLRNGD VS VEIF AEPFQLKVYWQNALKLIVNEQNFL
NIEHHRTKQENFAHVLPEETTFNMFKDNFLYSKHD SMPLGPESVALDF SFMGS
TNVYGIPEHATSLRLMDTSGGKEPYRLFNVDVFEYNIGTSQPMYGSIPFMFS SS
STSIFWVNAADTWVDIKYDTSKNKTMTHWISENGVIDVVMSLGPDIPTIIDKFT
DLTGRPFLPPIS SIGYHQCRWNYNDEMDVLTVD SQMDAHMIPYDFIWLDLEYT
NDKKYFTWKQHSFPNPKRLL SKLKKL GRNLVVLIDPHLKKDYEISDRVINENV
AVKDHNGNDYVGHCWPGNSIWIDTISKYGQKIWKSFFERFMDLPADLTNLFI
WNDMNEP SIFDGPETTAPKDLIHDNYIEERSVHNIYGL SVHEATYDAIKSIYSP S
DKRPFLLTRAFFAGSQRTAATWTGDNVANWDYLKISIPMVL SNNIAGMPFIGA
DIAGFAEDPTPELIARWYQAGLWYPFFRAHAHIDTKRREPYLFNEPLKSIVRDII
QLRYFLLPTLYTMFHKS SVTGFPIMNPMFIEHPEFAELYHIDNQFYWSNSGLLV
KPVTEPGQ SE lEMVFPPGIFYEFASLHSFINNGTDLIEKNISAPLDKIPLFIEGGHI
ITMKDKYRRS SMLMKNDPYVIVIAPD lEGRAVGDLYVDD GETFGYQRGEYVE
TQFIFENNTLKNVRSHIPENLTGIHHNTLRNTNIEKIIIAKNNLQHNITLKDSIKV
KKNGEES SLPTRS SYENDNKITILNL SLDITEDWEVIF*
SEQ ID NO:14 ATGTTCAGCTTGAAAGCATTATTGCCATTGGCCTTGTTGTTGGTCAGCGCC
AACCAAGTTGCTGCAAAAGTCCACAAGGCTAAAATTTATAAACACGAGTT
pep4
GTCCGATGAGATGAAAGAAGTCACTTTCGAGCAACATTTAGCTCATTTAGG
Saccharomyces sp. CCAAAAGTACTTGACTCAATTTGAGAAAGCTAACCCCGAAGTTGTTTTTTC
TAGGGAGCATCCTTTCTTCACTGAAGGTGGTCACGATGTTCCATTGACAAA
TTACTTGAACGCACAATATTACACTGACATTACTTTGGGTACTCCACCTCA
AAACTTCAAGGTTATTTTGGATACTGGTTCTTCAAACCTTTGGGTTCCAAGT
AACGAATGTGGTTCCTTGGCTTGTTTCCTACATTCTAAATACGATCATGAA
GCTTCATCAAGCTACAAAGCTAATGGTACTGAATTTGCCATTCAATATGGT
ACTGGTTCTTTGGAAGGTTACATTTCTCAAGACACTTTGTCCATCGGGGATT
TGACCATTCCAAAACAAGACTTCGCTGAGGCTACCAGCGAGCCGGGCTTA
ACATTTGCATTTGGCAAGTTCGATGGTATTTTGGGTTTGGGTTACGATACC
ATTTCTGTTGATAAGGTGGTCCCTCCATTTTACAACGCCATTCAACAAGATT
TGTTGGACGAAAAGAGATTTGCCTTTTATTTGGGAGACACTTCAAAGGATA
CTGAAAATGGCGGTGAAGCCACCTTTGGTGGTATTGACGAGTCTAAGTTCA
AGGGCGATATCACTTGGTTACCTGTTCGTCGTAAGGCTTACTGGGAAGTCA
AGTTTGAAGGTATCGGTTTAGGCGACGAGTACGCCGAATTGGAGAGCCAT
GGTGCCGCCATCGATACTGGTACTTCTTTGATTACCTTGCCATCAGGATTA
GCTGAAATGATTAATGCTGAAATTGGGGCCAAGAAGGGTTGGACCGGTCA
ATATACTCTAGACTGTAACACCAGAGACAATCTACCTGATCTAATTTTCAA
CTTCAATGGCTACAACTTCACTATTGGGCCATACGATTACACGCTTGAAGT
TTCAGGCTCCTGTATCTCTGCAATTACACCAATGGATTTCCCAGAACCTGTT
GGCCCACTGGCCATCGTTGGTGATGCCTTCTTGCGTAAATACTATTCTATTT
ACGATTTGGGCAACAATGCGGTTGGTTTGGCCAAAGCAATTTGA
SEQ ID NO:15 MFSLKALLPLALLLVSANQVAAKVHKAKIYKHEL SDEMKEVTFEQHLAHLGQ
KYLTQFEKANPEVVFSREHPFF lEGGHDVPLTNYLNAQYYTDITLGTPPQNFK
pep4
VILDTGS SNLWVP SNECGSLACFLHSKYDHEAS S SYKANGTEFAIQYGTGSLE
Saccharomyces sp. GYISQDTL SIGDLTIPKQDFAEATSEPGLTFAFGKFDGILGLGYDTISVDKVVPP
FYNAIQQDLLDEKRFAFYLGDTSKDTENGGEATF GGIDESKFKGDITWLPVRR
KAYWEVKFEGIGLGDEYAELESHGAAIDTGTSLITLP SGLAEMINAEIGAKKG
WTGQYTLDCNTRDNLPDLIFNFNGYNFTIGPYDYTLEVSGSCISAITPMDFPEP
VGPLAIVGDAFLRKYYSIYDLGNNAVGLAKAI*
SEQ ID NO:16 ATGGGTTTATCTTTGGTCTGCACCTTCTCCTTTCAAACTAACTACCACACTT
TATTGAATCCACATAATAAGAATCCTAAGAACTCTTTATTGTCCTACCAAC
Geranyl pyrophosphate
ACCCAAAGACTCCTATTATCAAGTCCTCTTACGATAACTTCCCATCTAAGT
olivetolic acid
ACTGTTTGACTAAGAATTTCCATTTGTTGGGTTTGAATTCTCACAACAGAAT
geranyltransferase
TTCCTCCCAATCCCGTTCTATTAGAGCCGGTTCTGATCAAATCGAAGGTTCC
CsPT4 nucleotide
CCTCATCATGAGTCCGATAACTCCATTGCTACTAAAATTTTAAATTTCGGTC
sequence
ATACTTGTTGGAAGTTGCAACGTCCTTACGTTGTCAAGGGTATGATCTCTA
(GOT) TTGCTTGTGGTTTGTTCGGTAGAGAATTGTTTAACAACAGACACTTGTTCTC
Artificial se quence TTGGGGTTTGATGTGGAAAGCTTTCTTCGCTTTGGTCCCAATTTTGTCTTTC
AATTTCTTCGCCGCCATCATGAACCAAATCTACGATGTTGATATCGACCGT
Codon optimized ATCAACAAGCCAGACTTACCTTTAGTTTCCGGTGAAATGTCCATTGAAACT
211
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
GCTTGGATCTTGTCTATCATTGTTGCCTTGACTGGTTTAATTGTTACTATTA
AGTTGAAGTCCGCTCCATTGTTTGTCTTCATCTACATCTTCGGTATCTTCGC
TGGTTTCGCTTACTCCGTCCCACCTATTAGATGGAAACAATATCCTTTTACC
AATTTCTTGATCACTATTTCCTCTCATGTTGGTTTGGCTTTCACTTCTTACTC
TGCCACCACTTCTGCTTTAGGTTTGCCTTTCGTTTGGCGTCCTGCCTTCTCTT
TCATTATTGCTTTCATGACTGTCATGGGTATGACTATTGCCTTTGCTAAAGA
CATTTCTGATATCGAAGGTGATGCTAAGTACGGTGTCTCTACCGTTGCTAC
CAAGTTAGGTGCTAGAAATATGACTTTTGTTGTTTCTGGTGTCTTATTGTTG
AACTACTTGGTTTCTATCTCTATTGGTATCATTTGGCCACAAGTTTTCAAGT
CTAACATTATGATCTTGTCTCATGCTATTTTGGCTTTCTGTTTGATCTTTCAA
ACTCGTGAATTAGCCTTAGCCAATTATGCCTCTGCCCCATCCCGTCAATTTT
TCGAATTCATCTGGTTGTTATACTATGCCGAATACTTCGTTTACGTCTTCAT
TTAA
SEQ ID NO:17 MGLSLVCTFSFQTNYHTLLNPHNKNPKNSLLSYQHPKTPIIKSSYDNFPSKYCL
TKNFHLLGLNSHNRISSQSRSIRAGSDQIEGSPHHESDNSIATKILNFGHTCWKL
Geranyl pyrophosphate
QRPYVVKGMISIACGLFGRELFNNRHLFSWGLMWKAFFALVPILSFNFFAAIM
olivetolic acid
NQIYDVDIDRINKPDLPLVSGEMSIETAWIL SIIVALTGLIVTIKLKSAPLFVFIYI
geranyltransferase
FGIFAGFAYSVPPIRWKQYPFTNFLITISSHVGLAFTSYSATTSALGLPFVWRPA
CsPT4
FSFIIAFMTVMGMTIAFAKDISDIEGDAKYGVSTVATKLGARNMTFVVSGVLL
(GOT) LNYLVSISIGIIWPQVFKSNIMIL SHAILAFCLIFQTRELALANYASAPSRQFFEFI
Cannib is sativa WLLYYAEYFVYVFI*
SEQ ID NO:18 ATGAACCATTTAAGAGCTGAGGGTCCAGCTTCCGTCTTGGCTATCGGTACT
Tetraketid syntha GCTAATCCAGAGAACATTTTATTACAAGATGAGTTTCCAGATTACTATTTC
(TKS e
CGTGTTACTAAGTCCGAGCATATGACCCAATTGAAAGAAAAGTTCCGTAA
) n ucleotide se
AATCTGTGATAAATCTATGATTAGAAAAAGAAACTGCTTTTTAAACGAAGA
sequence
ACACTTGAAGCAAAACCCAAGATTAGTTGAACACGAGATGCAAACCTTGG
Artificial sequence ACGCTAGACAAGATATGTTGGTTGTCGAGGTTCCTAAATTGGGTAAAGACG
Codon optimized CCTGTGCTAAAGCTATCAAAGAGTGGGGTCAACCTAAGTCCAAGATCACTC
ACTTAATCTTCACTTCCGCTTCCACCACTGACATGCCTGGTGCTGATTACCA
CTGTGCCAAGTTGTTGGGTTTGTCTCCTTCTGTCAAGAGAGTTATGATGTAC
CAATTAGGTTGTTACGGTGGTGGTACTGTCTTAAGAATTGCTAAGGACATC
GCTGAAAACAACAAAGGTGCTAGAGTTTTAGCCGTTTGTTGTGACATCATG
GCTTGTTTATTTCGTGGTCCATCTGAATCTGACTTGGAGTTGTTGGTTGGTC
AAGCTATTTTTGGTGATGGTGCCGCTGCCGTCATCGTTGGTGCTGAGCCAG
ATGAATCCGTTGGTGAAAGACCAATTTTCGAATTAGTCTCTACTGGTCAAA
CTATTTTGCCAAACTCCGAGGGTACTATCGGTGGTCATATTCGTGAAGCCG
GTTTAATCTTTGATTTGCACAAAGACGTTCCAATGTTGATCTCTAACAACAT
CGAAAAGTGTTTAATTGAGGCTTTTACTCCAATTGGTATCTCTGACTGGAA
CTCTATCTTCTGGATCACTCATCCAGGTGGTAAGGCTATCTTGGACAAGGT
TGAAGAAAAATTACATTTAAAGTCCGATAAATTCGTCGATTCTCGTCATGT
TTTGTCTGAACACGGTAACATGTCTTCCTCCACTGTCTTGTTTGTTATGGAT
GAATTACGTAAGAGATCTTTGGAGGAGGGTAAGTCTACTACTGGTGATGGT
TTCGAATGGGGTGTTTTGTTCGGTTTCGGTCCTGGTTTGACTGTTGAACGTG
TTGTTGTTAGATCTGTTCCAATTAAGTACTAG
SEQ ID NO:19 MNHLRAEGPASVLAIGTANPENILLQDEFPDYYFRVTKSEHMTQLKEKFRKIC
Tetraketid DKSMIRKRNCFLNEEHLKQNPRLVEHEMQTLDARQDMLVVEVPKLGKDACA
e synthase
KAIKEWGQPKSKITHLIFTSASTTDMPGADYHCAKLLGL SPSVKRVMMYQLG
(TKS)
CYGGGTVLRIAKDIAENNKGARVLAVCCDIMACLFRGPSESDLELLVGQAIFG
GenBank B1Q2B6 DGAAAVIVGAEPDESVGERPIFELVSTGQTILPNSEGTIGGHIREAGLIFDLHKD
Cannabis sativa VPMLISNNIEKCLIEAFTPIGISDWNSIFWITHPGGKAILDKVEEKLHLKSDKFV
DSRHVLSEHGNMSSSTVLFVMDELRKRSLEEGKSTTGDGFEWGVLFGFGPGL
TVERVVVRSVPIKY*
SEQ ID NO :20 ATGGCCGTCAAACACTTGATCGTCTTAAAATTCAAGGATGAAATTACTGAA
Ol tol GCTCAAAAAGAAGAGTTCTTCAAAACCTATGTCAATTTAGTCAACATTATT
ic =acid cyclase ive
CCTGCTATGAAGGACGTTTACTGGGGTAAGGATGTCACCCAAAAGAACAA
(OAC) nucleotide
GGAAGAAGGTTACACTCACATTGTTGAAGTCACTTTCGAATCTGTTGAAAC
sequence
TATCCAAGATTATATTATCCACCCAGCTCATGTCGGTTTTGGTGATGTTTAC
Artificial sequence AGATCTTTTTGGGAAAAATTGTTGATCTTTGACTATACTCCAAGAAAATAA
212
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Codon optimized
SEQ ID NO :21 MAVKHLIVLKFKDEI lEAQKEEFFKTYVNLVNIIPAMKDVYWGKDVTQKNKE
EGYTHIVEVTFESVETIQDYIIHPAHVGFGDVYRSFWEKLLIFDYTPRK*
Olivetolic acid cyclase
(OAC)
GenBank AFN42527
Cannabis sativa
SEQ ID NO :22 ATGGGTAAGAATTACAAGTCCTTAGACTCTGTTGTTGCTTCTGACTTTATTG
CTTTAGGTATTACTTCCGAAGTTGCTGAAACCTTACACGGTAGATTGGCTG
Acyl-activating enzyme
AAATTGTTTGCAACTACGGTGCTGCTACCCCTCAAACTTGGATTAACATTG
Cs AAE1 nucleotide
CTAATCATATTTTGTCTCCAGATTTGCCATTTTCTTTACACCAAATGTTGTT
sequence
CTACGGTTGTTACAAGGATTTCGGTCCTGCTCCTCCAGCTTGGATTCCTGAT
Artificial sequence CCAGAAAAAGTCAAATCTACTAACTTGGGTGCTTTGTTGGAAAAGAGAGG
Codon optimized TAAGGAGTTTTTGGGTGTTAAGTACAAGGACCCAATTTCTTCTTTCTCTCAC
TTCCAAGAATTCTCTGTTAGAAACCCTGAAGTTTACTGGAGAACTGTTTTG
ATGGATGAGATGAAGATTTCTTTTTCTAAGGACCCAGAGTGTATCTTAAGA
AGAGACGACATTAACAATCCAGGTGGTTCTGAGTGGTTACCAGGTGGTTAC
TTGAACTCTGCCAAAAATTGCTTGAACGTTAACTCTAACAAGAAATTGAAT
GACACTATGATTGTCTGGAGAGATGAGGGTAACGATGATTTGCCTTTGAAT
AAATTGACTTTGGATCAATTGAGAAAAAGAGTCTGGTTGGTTGGTTACGCT
TTGGAAGAAATGGGTTTAGAAAAAGGTTGTGCTATCGCCATCGATATGCCT
ATGCACGTTGATGCTGTTGTTATTTATTTGGCTATTGTTTTAGCTGGTTATG
TTGTTGTTTCCATCGCCGACTCCTTCTCTGCTCCAGAAATCTCCACCAGATT
GAGATTGTCTAAAGCCAAAGCCATTTTCACCCAAGACCACATCATTAGAGG
TAAGAAGCGTATTCCATTGTATTCTCGTGTTGTTGAAGCTAAATCTCCTATG
GCTATCGTCATCCCATGCTCTGGTTCTAACATCGGTGCTGAATTAAGAGAC
GGTGATATTTCTTGGGACTACTTTTTAGAAAGAGCTAAAGAATTCAAAAAC
TGCGAGTTTACTGCTAGAGAACAACCTGTCGACGCTTATACTAATATTTTA
TTCTCTTCTGGTACTACTGGTGAACCTAAGGCTATTCCATGGACCCAAGCT
ACTCCTTTGAAAGCCGCTGCTGATGGTTGGTCCCATTTAGACATCAGAAAA
GGTGATGTCATCGTCTGGCCAACTAACTTAGGTTGGATGATGGGTCCATGG
TTAGTCTACGCTTCTTTGTTGAATGGTGCCTCTATCGCCTTATATAATGGTT
CCCCTTTAGTCTCTGGTTTTGCTAAATTCGTTCAAGATGCTAAGGTTACCAT
GTTAGGTGTTGTCCCTTCTATCGTTAGATCTTGGAAATCTACTAACTGTGTT
TCTGGTTACGACTGGTCCACTATTCGTTGTTTCTCTTCTTCTGGTGAAGCTT
CCAATGTCGATGAGTACTTATGGTTAATGGGTCGTGCTAACTACAAGCCAG
TCATCGAAATGTGCGGTGGTACTGAAATTGGTGGTGCTTTTTCCGCTGGTT
CTTTTTTACAAGCCCAATCCTTGTCTTCCTTCTCCTCTCAATGTATGGGTTGT
ACTTTATATATCTTAGATAAGAATGGTTACCCTATGCCTAAAAACAAGCCA
GGTATTGGTGAATTAGCTTTGGGTCCTGTTATGTTTGGTGCTTCTAAAACCT
TGTTAAATGGTAATCATCACGACGTTTACTTCAAAGGTATGCCTACTTTGA
ACGGTGAGGTTTTGAGACGTCATGGTGATATTTTCGAATTAACTTCCAACG
GTTATTATCACGCTCACGGTAGAGCTGATGATACTATGAACATTGGTGGTA
TTAAGATCTCTTCCATCGAAATTGAGAGAGTTTGTAACGAGGTTGACGATC
GTGTTTTCGAAACTACTGCTATTGGTGTCCCTCCTTTAGGTGGTGGTCCAGA
ACAATTGGTTATCTTTTTCGTCTTGAAGGACTCCAACGACACCACTATCGA
CTTAAACCAATTAAGATTGTCTTTCAACTTGGGTTTGCAAAAGAAGTTGAA
TCCATTATTTAAGGTTACTCGTGTCGTTCCATTGTCCTCCTTGCCAAGAACT
GCTACCAACAAGATTATGCGTAGAGTCTTGAGACAACAATTCTCTCACTTT
GAGTAA
SEQ ID NO :23 MGKNYKSLD SVVASDFIAL GITSEVAETLHGRLAEIVCNYGAATPQTWINIAN
HIL SPDLPF SLHQMLFYGCYKDFGPAPPAWIPDPEKVKSTNLGALLEKRGKEFL
Acyl-activating enzyme
GVKYKDPIS SF SHFQEF SVRNPEVYWRTVLMDEMKI SF SKDPECILRRDDINNP
(CsAAE1)
GGSEWLPGGYLNSAKNCLNVNSNKKLNDTMIVWRDEGNDDLPLNKLTLDQL
Cannabis sativa RKRVWLVGYALEEMGLEKGCAIAIDMPMHVDAVVIYL AIVLAGYVVVSIAD S
FSAPEISTRLRL SKAKAIFTQDHIIRGKKRIPLYSRVVEAKSPMAIVIPCSGSNIG
AELRD GD IS WDYFLERAKEFKNCEFTAREQPVDAYTNILF S SGTTGEPKAIPWT
QATPLKAAADGWSHLDIRKGDVIVWPTNL GWM MGPWLVYASLLNGASIALY
213
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
NGSPLVSGFAKFVQDAKVTML GVVPSIVRSWKSTNCVS GYDWSTIRCFS S S GE
ASNVDEYLWLMGRANYKPVIEMCGG lEIGGAFSAGSFLQAQSLS SFS SQCMG
CTLYILDKNGYPMPKNKPGIGELALGPVMFGASKTLLNGNHHDVYFKGMPTL
NGEVLRRHGDIFELTSNGYYHAHGRADDTMNIGGIKIS SIEIERVCNEVDDRVF
ETTAIGVPPLGGGPEQLVIFFVLKD SNDTTIDLNQLRL SFNL GLQKKLNPLFKV
TRVVPL S SLPRTATNKIMRRVLRQQFSHFE*
SEQ ID NO :24 ATGACTGCCGACAACAATAGTATGCCCCATGGTGCAGTATCTAGTTACGCC
AAATTAGTGCAAAACCAAACACCTGAAGACATTTTGGAAGAGTTTCCTGA
Isopentenyl
AATTATTCCATTACAACAAAGACCTAATACCCGATCTAGTGAGACGTCAAA
pyrophosphate
TGACGAAAGCGGAGAAACATGTTTTTCTGGTCATGATGAGGAGCAAATTA
isomerase (Sc IDI1)
¨ AGTTAATGAATGAAAATTGTATTGTTTTGGATTGGGACGATAATGCTATTG
Saccharomyces sp. GTGCCGGTACCAAGAAAGTTTGTCATTTAATGGAAAATATTGAAAAGGGTT
TACTACATCGTGCATTCTCCGTCTTTATTTTCAATGAACAAGGTGAATTACT
TTTACAACAAAGAGCCACTGAAAAAATAACTTTCCCTGATCTTTGGACTAA
CACATGCTGCTCTCATCCACTATGTATTGATGACGAATTAGGTTTGAAGGG
TAAGCTAGACGATAAGATTAAGGGCGCTATTACTGCGGCGGTGAGAAAAC
TAGATCATGAATTAGGTATTCCAGAAGATGAAACTAAGACAAGGGGTAAG
TTTCACTTTTTAAACAGAATCCATTACATGGCACCAAGCAATGAACCATGG
GGTGAACATGAAATTGATTACATCCTATTTTATAAGATCAACGCTAAAGAA
AACTTGACTGTCAACCCAAACGTCAATGAAGTTAGAGACTTCAAATGGGTT
TCACCAAATGATTTGAAAACTATGTTTGCTGACCCAAGTTACAAGTTTACG
CCTTGGTTTAAGATTATTTGCGAGAATTACTTATTCAACTGGTGGGAGCAA
TTAGATGACCTTTCTGAAGTGGAAAATGACAGGCAAATTCATAGAATGCTA
TAA
SEQ ID NO :25 MTADNNSMPHGAVS SYAKLVQNQTPEDILEEFPEIIPLQQRPNTRS SETSNDES
GETCF S GHDEEQIKLMNENCIVLDWDDNAIGAGTKKVCHLMENIEKGLLHRA
Isopentenyl
F SVFIFNEQGELLLQQRA 1EKITFPDLWTNTCC SHPL CIDDELGLKGKLDDKIK
pyrophosphate
GAITAAVRKLDHEL GIPEDETKTRGKFHFLNRIHYMAPSNEPWGEHEIDYILFY
isomerase (Sc_IDI1)
KINAKENLTVNPNVNEVRDFKWVSPNDLKTMFADPSYKFTPWFKIICENYLFN
Saccharomyces sp. WWEQLDDL SEVENDRQIHRML*
SEQ ID NO :26 ATGCAATTGGTGAAGACTGAAGTCACCAAGAAGTCTTTTACTGCTCCTGTA
T CAAAAGGCTTCTACACCAGTTTTAACCAATAAAACAGTCATTTCTGGATCG
runcated 3-hydroxy-3-
AAAGTCAAAAGTTTATCATCTGCGCAATCGAGCTCATCAGGACCTTCATCA
methyl-glutaryl-CoA
TCTAGTGAGGAAGATGATTCCCGCGATATTGAAAGCTTGGATAAGAAAAT
reductase (tHMG1,
ACGTCCTTTAGAAGAATTAGAAGCATTATTAAGTAGTGGAAATACAAAAC
tHMGR)
AATTGAAGAACAAAGAGGTCGCTGCCTTGGTTATTCACGGTAAGTTACCTT
Artificial sequence TGTACGCTTTGGAGAAAAAATTAGGTGATACTACGAGAGCGGTTGCGGTA
CGTAGGAAGGCTCTTTCAATTTTGGCAGAAGCTCCTGTATTAGCATCTGAT
CGTTTACCATATAAAAATTATGACTACGACCGCGTATTTGGCGCTTGTTGT
GAAAATGTTATAGGTTACATGCCTTTGCCCGTTGGTGTTATAGGCCCCTTG
GTTATCGATGGTACATCTTATCATATACCAATGGCAACTACAGAGGGTTGT
TTGGTAGCTTCTGCCATGCGTGGCTGTAAGGCAATCAATGCTGGCGGTGGT
GCAACAACTGTTTTAACTAAGGATGGTATGACAAGAGGCCCAGTAGTCCG
TTTCCCAACTTTGAAAAGATCTGGTGCCTGTAAGATATGGTTAGACTCAGA
AGAGGGACAAAACGCAATTAAAAAAGCTTTTAACTCTACATCAAGATTTG
CACGTCTGCAACATATTCAAACTTGTCTAGCAGGAGATTTACTCTTCATGA
GATTTAGAACAACTACTGGTGACGCAATGGGTATGAATATGATTTCTAAGG
GTGTCGAATACTCATTAAAGCAAATGGTAGAAGAGTATGGCTGGGAAGAT
ATGGAGGTTGTCTCCGTTTCTGGTAACTACTGTACCGACAAAAAACCAGCT
GCCATCAACTGGATCGAAGGTCGTGGTAAGAGTGTCGTCGCAGAAGCTAC
TATTCCTGGTGATGTTGTCAGAAAAGTGTTAAAAAGTGATGTTTCCGCATT
GGTTGAGTTGAACATTGCTAAGAATTTGGTTGGATCTGCAATGGCTGGGTC
TGTTGGTGGATTTAACGCACATGCAGCTAATTTAGTGACAGCTGTTTTCTTG
GCATTAGGACAAGATCCTGCACAAAATGTCGAAAGTTCCAACTGTATAAC
ATTGATGAAAGAAGTGGACGGTGATTTGAGAATTTCCGTATCCATGCCATC
CATCGAAGTAGGTACCATCGGTGGTGGTACTGTTCTAGAACCACAAGGTGC
CATGTTGGACTTATTAGGTGTAAGAGGCCCACATGCTACCGCTCCTGGTAC
CAACGCACGTCAATTAGCAAGAATAGTTGCCTGTGCCGTCTTGGCAGGTGA
ATTATCCTTATGTGCTGCCCTAGCAGCCGGCCATTTGGTTCAAAGTCATAT
214
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
GACCCACAACAGGAAACCTGCTGAACCAACAAAACCTAACAATTTGGACG
CCACTGATATAAATCGTTTGAAAGATGGGTCCGTCACCTGCATTAAATCCT
AA
SEQ ID NO :27 MQLVK 1EVTKKSFTAPVQKASTPVLTNKTVISGSKVKSL SSAQ SSSSGPSS SSE
T EDD SRDIESLDKKIRPLEELEALL S SGNTKQLKNKEVAALVIHGKLPLYALEKK
runcated 3-hydroxy-3-
LGDTTRAVAVRRKALSILAEAPVLASDRLPYKNYDYDRVFGACCENVIGYMP
me thy 1-glutaryl-Co A
(Sc_tHMG1 LPVGVIGPLVIDGTSYHIPMATTEGCLVASAMRGCKAINAGGGATTVLTKDG
H
reductase ,
MTRGPVVRFPTLKRSGACKIWLDSEEGQNAIKKAFNSTSRFARLQHIQTCLAG
tMGR)
DLLFMRFRTTTGDAMGMNMISKGVEYSLKQMVEEYGWEDMEVVSVSGNYC
Artificial sequence TDKKPAAINWIEGRGKSVVAEATIPGDVVRKVLKSD VSALVELNIAKNLVGSA
MAGSVGGFNAHAANLVTAVFLALGQDPAQNVES SNCITLMKEVDGDLRISVS
MP SIEVGTIGGGTVLEPQGAMLDLL GVRGPHATAPGTNARQLARIVACAVL A
GEL SLCAALAAGHLVQSHMTHNRKPAEPTKPNNLDATDINRLKD GSVTCIKS*
SEQ ID NO :28 ATGACTGAACTAAAAAAACAAAAGACCGCTGAACAAAAAACCAGACCTCA
HMG-CoA
AAATGTCGGTATTAAAGGTATCCAAATTTACATCCCAACTCAATGTGTCAA
synthase
IS ERG13HMGS CCAATCTGAGCTAGAGAAATTTGATGGCGTTTCTCAAGGTAAATACACAAT
c ) _,
TGGTCTGGGCCAAACCAACATGTCTTTTGTCAATGACAGAGAAGATATCTA
Saccharomyces sp. CTCGATGTCCCTAACTGTTTTGTCTAAGTTGATCAAGAGTTACAACATCGA
CACCAACAAAATTGGTAGATTAGAAGTCGGTACTGAAACTCTGATTGACA
AGTCCAAGTCTGTCAAGTCTGTCTTGATGCAATTGTTTGGTGAAAACACTG
ACGTCGAAGGTATTGACACGCTTAATGCCTGTTACGGTGGTACCAACGCGT
TGTTCAACTCTTTGAACTGGATTGAATCTAACGCATGGGATGGTAGAGACG
CCATTGTAGTTTGCGGTGATATTGCCATCTACGATAAGGGTGCCGCAAGAC
CAACCGGTGGTGCCGGTACTGTTGCTATGTGGATCGGTCCTGATGCTCCAA
TTGTATTTGACTCTGTAAGAGCTTCTTACATGGAACACGCCTACGATTTTTA
CAAGCCAGATTTCACCAGCGAATATCCTTACGTCGATGGTCATTTTTCATT
AACTTGTTACGTCAAGGCTCTTGATCAAGTTTACAAGAGTTATTCCAAGAA
GGCTATTTCTAAAGGGTTGGTTAGCGATCCCGCTGGTTCGGATGCTTTGAA
CGTTTTGAAATATTTCGACTACAACGTTTTCCATGTTCCAACCTGTAAATTG
GTCACAAAATCATACGGTAGATTACTATATAACGATTTCAGAGCCAATCCT
CAATTGTTCCCAGAAGTTGACGCCGAATTAGCTACTCGCGATTATGACGAA
TCTTTAACCGATAAGAACATTGAAAAAACTTTTGTTAATGTTGCTAAGCCA
TTCCACAAAGAGAGAGTTGCCCAATCTTTGATTGTTCCAACAAACACAGGT
AACATGTACACCGCATCTGTTTATGCCGCCTTTGCATCTCTATTAAACTATG
TTGGATCTGACGACTTACAAGGCAAGCGTGTTGGTTTATTTTCTTACGGTTC
CGGTTTAGCTGCATCTCTATATTCTTGCAAAATTGTTGGTGACGTCCAACAT
ATTATCAAGGAATTAGATATTACTAACAAATTAGCCAAGAGAATCACCGA
AACTCCAAAGGATTACGAAGCTGCCATCGAATTGAGAGAAAATGCCCATT
TGAAGAAGAACTTCAAACCTCAAGGTTCCATTGAGCATTTGCAAAGTGGTG
TTTACTACTTGACCAACATCGATGACAAATTTAGAAGATCTTACGATGTTA
AAAAATAAT
SEQ ID NO :29 M1ELKKQKTAEQKTRPQNVGIKGIQIYIPTQCVNQSELEKFDGVSQGKYTIGL
HMG-CoA synthase GQTNMSFVNDREDIYSMSLTVL SKLIKSYNIDTNKIGRLEVGTETLIDKSKSVK
(Sc ERG13 HMGS SVLMQLFGENTDVEGIDTLNACYGGTNALFNSLNWIESNAWDGRDAIVVCGD
_, )
IAIYDKGAARPTGGAGTVAMWIGPDAPIVFDSVRASYMEHAYDFYKPDFTSE
Saccharomyces sp. YPYVDGHF SLTCYVKALDQVYKSYSKKAISKGLVSDPAGSDALNVLKYFDYN
VFHVPTCKLVTKSYGRLLYNDFRANPQLFPEVDAELATRDYDESLTDKNIEKT
FVNVAKPFHKERVAQ SLIVPTNTGNMYTASVYAAFASLLNYVGSDDLQGKRV
GLFSYGSGLAASLYSCKIVGDVQHIIKELDITNKLAKRI1ETPKDYEAAIELREN
AHLKKNFKPQGSIEHLQSGVYYLTNIDDKFRRSYDVKK*
SEQ ID NO :30 ATGTCTCAGAACGTTTACATTGTATCGACTGCCAGAACCCCAATTGGTTCA
A t l CoA TTCCAGGGTTCTCTATCCTCCAAGACAGCAGTGGAATTGGGTGCTGTTGCT
ceoacety
TTAAAAGGCGCCTTGGCTAAGGTTCCAGAATTGGATGCATCCAAGGATTTT
thiolase (ERG10)
GACGAAATTATTTTTGGTAACGTTCTTTCTGCCAATTTGGGCCAAGCTCCG
Saccharomyces GCCAGACAAGTTGCTTTGGCTGCCGGTTTGAGTAATCATATCGTTGCAAGC
cerevisiae ACAGTTAACAAGGTCTGTGCATCCGCTATGAAGGCAATCATTTTGGGTGCT
CAATCCATCAAATGTGGTAATGCTGATGTTGTCGTAGCTGGTGGTTGTGAA
TCTATGACTAACGCACCATACTACATGCCAGCAGCCCGTGCGGGTGCCAAA
TTTGGCCAAACTGTTCTTGTTGATGGTGTCGAAAGAGATGGGTTGAACGAT
215
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
GCGTACGATGGTCTAGCCATGGGTGTACACGCAGAAAAGTGTGCCCGTGA
TTGGGATATTACTAGAGAACAACAAGACAATTTTGCCATCGAATCCTACCA
AAAATCTCAAAAATCTCAAAAGGAAGGTAAATTCGACAATGAAATTGTAC
CTGTTACCATTAAGGGATTTAGAGGTAAGCCTGATACTCAAGTCACGAAGG
ACGAGGAACCTGCTAGATTACACGTTGAAAAATTGAGATCTGCAAGGACT
GTTTTCCAAAAAGAAAACGGTACTGTTACTGCCGCTAACGCTTCTCCAATC
AACGATGGTGCTGCAGCCGTCATCTTGGTTTCCGAAAAAGTTTTGAAGGAA
AAGAATTTGAAGCCTTTGGCTATTATCAAAGGTTGGGGTGAGGCCGCTCAT
CAACCAGCTGATTTTACATGGGCTCCATCTCTTGCAGTTCCAAAGGCTTTG
AAACATGCTGGCATCGAAGACATCAATTCTGTTGATTACTTTGAATTCAAT
GAAGCCTTTTCGGTTGTCGGTTTGGTGAACACTAAGATTTTGAAGCTAGAC
CCATCTAAGGTTAATGTATATGGTGGTGCTGTTGCTCTAGGTCACCCATTG
GGTTGTTCTGGTGCTAGAGTGGTTGTTACACTGCTATCCATCTTACAGCAA
GAAGGAGGTAAGATCGGTGTTGCCGCCATTTGTAATGGTGGTGGTGGTGCT
TCCTCTATTGTCATTGAAAAGATATGA
SEQ ID NO :31 MSQNVYIVSTARTPIGSFQGSL S SKTAVELGAVALKGALAKVPELDASKDFDEI
Acetoace l CoA IFGNVL SANL GQAPARQVALAAGL SNHIVASTVNKVCASAMKAIILGAQ SIKC
ty
thiolase (ERG10 GNADVVVAGGCESMTNAPYYMPAARAGAKFGQTVLVDGVERDGLNDAYD
)
GLAMGVHAEKCARDWDITREQQDNFAIESYQKSQKSQKEGKFDNEIVPVTIK
Saccharomyces GFRGKPDTQVTKDEEPARLHVEKLRSARTVFQKENGTVTAANASPINDGAAA
cerevisiae VILVSEKVLKEKNLKPLAIIKGWGEAAHQPADFTWAP SLAVPKALKHAGIEDI
NSVDYFEFNEAFSVVGLVNTKILKLDP SKVNVYGGAVALGHPL GCS GARVVV
TLL SILQQEGGKIGVAAICNGGGGAS SIVIEKI*
SEQ ID NO :32 ATGACCGTTTACACAGCATCCGTTACCGCACCCGTCAACATCGCAACCCTT
M l AAGTATTGGGGGAAAAGGGACACGAAGTTGAATCTGCCCACCAATTCGTC
evaonate
CATATCAGTGACTTTATCGCAAGATGACCTCAGAACGTTGACCTCTGCGGC
pyrophosphate
TACTGCACCTGAGTTTGAACGCGACACTTTGTGGTTAAATGGAGAACCACA
decarboxylase
CAGCATCGACAATGAAAGAACTCAAAATTGTCTGCGCGACCTACGCCAAT
(Sc_ERG19, MVD1)
TAAGAAAGGAAATGGAATCGAAGGACGCCTCATTGCCCACATTATCTCAA
Saccharomyces sp. TGGAAACTCCACATTGTCTCCGAAAATAACTTTCCTACAGCAGCTGGTTTA
GCTTCCTCCGCTGCTGGCTTTGCTGCATTGGTCTCTGCAATTGCTAAGTTAT
ACCAATTACCACAGTCAACTTCAGAAATATCTAGAATAGCAAGAAAGGGG
TCTGGTTCAGCTTGTAGATCGTTGTTTGGCGGATACGTGGCCTGGGAAATG
GGAAAAGCTGAAGATGGTCATGATTCCATGGCAGTACAAATCGCAGACAG
CTCTGACTGGCCTCAGATGAAAGCTTGTGTCCTAGTTGTCAGCGATATTAA
AAAGGATGTGAGTTCCACTCAGGGTATGCAATTGACCGTGGCAACCTCCG
AACTATTTAAAGAAAGAATTGAACATGTCGTACCAAAGAGATTTGAAGTC
ATGCGTAAAGCCATTGTTGAAAAAGATTTCGCCACCTTTGCAAAGGAAAC
AATGATGGATTCCAACTCTTTCCATGCCACATGTTTGGACTCTTTCCCTCCA
ATATTCTACATGAATGACACTTCCAAGCGTATCATCAGTTGGTGCCACACC
ATTAATCAGTTTTACGGAGAAACAATCGTTGCATACACGTTTGATGCAGGT
CCAAATGCTGTGTTGTACTACTTAGCTGAAAATGAGTCGAAACTCTTTGCA
TTTATCTATAAATTGTTTGGCTCTGTTCCTGGATGGGACAAGAAATTTACTA
CTGAGCAGCTTGAGGCTTTCAACCATCAATTTGAATCATCTAACTTTACTG
CACGTGAATTGGATCTTGAGTTGCAAAAGGATGTTGCCAGAGTGATTTTAA
CTCAAGTCGGTTCAGGCCCACAAGAAACAAACGAATCTTTGATTGACGCA
AAGACTGGTCTACCAAAGGAATAA
SEQ ID NO :33 MTVYTASVTAPVNIATLKYWGKRDTKLNLPTNS SI SVTL SQDDLRTLTSAATA
M l PEFERDTL WLNGEPH SIDNERTQNCLRDLRQLRKEMESKDASLPTL SQWKLHI
evaonate
VSENNFPTAAGLAS SAAGF AALVSAIAKLYQLPQ ST SEI SRIARKGS G SACRSLF
pyrophosphate
boxylase GGYVAWEMGKAEDGHD SMAVQIAD S SDWPQMKACVLVVSDIKKDVS STQG
decar
(Sc ERG19 MVD1 MQLTVATSELFKERIEHVVPKRFEVMRKAIVEKDFATFAKETMMD SNSFHAT
_, )
CLD SFPPIFYMNDTSKRIISWCHTINQFYGETIVAYTFDAGPNAVLYYLAENES
Saccharomyces sp. KLFAFIYKLFGSVPGWDKKFT1EQLEAFNHQFESSNFTARELDLELQKDVARV
ILTQVGSGPQETNESLIDAKTGLPKE*
SEQ ID NO :34 ATGTCCTACACCGTTGGTACCTACTTAGCTGAGCGTTTGGTCCAAATCGGT
TTGAAGCACCATTTCGCCGTTGCTGGTGATTACAACTTGGTCTTGTTAGATA
Pyruvate decarboxylase
ATTTATTATTGAACAAGAACATGGAACAAGTCTACTGCTGTAATGAATTGA
(Zin_PDC)
ACTGTGGTTTCTCTGCTGAAGGTTATGCTAGAGCTAAAGGTGCCGCTGCCG
216
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Artificial sequence CTGTTGTCACTTACTCTGTTGGTGCTTTGTCTGCCTTCGACGCTATTGGTGG
C odon TGCTTACGCCGAGAATTTACCTGTTATTTTAATTTCTGGTGCCCCTAACAAT
optimized
AACGATCATGCTGCTGGTCATGTTTTACACCACGCTTTGGGTAAAACTGAC
TACCATTATCAATTAGAGATGGCCAAAAACATCACCGCCGCTGCCGAGGC
CATTTACACTCCAGAAGAAGCCCCAGCCAAAATTGATCACGTCATCAAAA
CCGCCTTGAGAGAGAAAAAACCTGTTTACTTGGAAATCGCCTGTAATATCG
CCTCTATGCCTTGCGCCGCTCCTGGTCCTGCTTCCGCCTTATTCAACGATGA
GGCTTCTGATGAAGCTTCCTTAAACGCTGCTGTTGAGGAGACTTTAAAGTT
CATCGCTAATAGAGATAAGGTCGCTGTTTTAGTCGGTTCTAAGTTGCGTGC
TGCCGGTGCCGAGGAAGCTGCTGTTAAATTCGCCGATGCTTTAGGTGGTGC
TGTCGCCACCATGGCCGCCGCCAAATCCTTTTTCCCTGAAGAAAACCCACA
CTACATCGGTACTTCTTGGGGTGAAGTCTCTTACCCAGGTGTCGAAAAGAC
TATGAAGGAAGCCGATGCCGTCATCGCCTTGGCCCCAGTTTTTAATGATTA
TTCCACCACTGGTTGGACTGATATCCCAGATCCTAAAAAGTTAGTTTTAGC
CGAGCCTAGATCCGTTGTTGTTAACGGTATTAGATTCCCTTCCGTTCACTTG
AAGGATTACTTAACTAGATTGGCTCAAAAGGTTTCCAAGAAGACCGGTGCT
TTGGACTTTTTCAAATCTTTGAACGCCGGTGAGTTAAAGAAGGCCGCCCCT
GCTGACCCATCTGCTCCATTGGTTAACGCTGAGATTGCTAGACAAGTCGAA
GCTTTATTGACCCCAAACACTACCGTTATCGCCGAAACTGGTGACTCTTGG
TTTAATGCTCAAAGAATGAAGTTACCAAATGGTGCCAGAGTTGAGTACGA
AATGCAATGGGGTCATATCGGTTGGTCTGTCCCAGCTGCTTTTGGTTATGCT
GTTGGTGCCCCTGAGAGAAGAAACATCTTGATGGTTGGTGACGGTTCCTTC
CAATTGACTGCTCAAGAAGTCGCTCAAATGGTTAGATTAAAATTACCAGTC
ATCATCTTCTTGATCAATAACTACGGTTACACTATCGAAGTCATGATTCAC
GATGGTCCTTACAATAATATTAAGAACTGGGACTATGCTGGTTTGATGGAA
GTCTTTAATGGTAACGGTGGTTACGATTCCGGTGCTGGTAAGGGTTTAAAG
GCTAAGACTGGTGGTGAATTAGCTGAAGCCATTAAGGTTGCCTTGGCTAAC
ACCGACGGTCCTACTTTAATCGAATGTTTCATTGGTAGAGAGGATTGTACC
GAAGAGTTAGTTAAGTGGGGTAAGAGAGTTGCCGCTGCTAATTCCCGTAA
GCCTGTCAATAAATTGTTATAA
SEQ ID NO :35 MSYTVGTYLAERLVQIGLKHHFAVAGDYNLVLLDNLLLNKNMEQVYCCNEL
NCGFSAEGYARAKGAAAAVVTYSVGAL SAFDAIGGAYAENLPVILISGAPNN
Pymvate decarbovlase
NDHAAGHVLHHALGKTDYHYQLEMAKNITAAAEAlYTPEEAPAKIDHVIKTA
(Zm_PDC)
LREKKPVYLEIACNIASMPCAAPGPA SALFNDEASDEA SLNAAVEETLKFIANR
Zymomonas mobilis DKVAVLVGSKLRAAGAEEAAVKFADAL GGAVATMAAAKSFFPEENPHYIGT
SWGEVSYPGVEKTMKEAD AVIALAP VFNDYSTTGWTDIPDPKKL VLAEPRSV
VVNGIRFP SVHLKDYLTRLAQKVSKKTGALDFFKSLNAGELKKAAPADP SAPL
VNAEIARQVEALLTPNTTVIAETGD SWFNAQRMKLPNGARVEYEMQWGHIG
WSVPAAF GYAVGAPERRNILMVGDGSFQLTAQEVAQMVRLKLPVIIFLINNYG
YTIEVMIHDGPYNNIKNWDYAGLMEVFNGNGGYD S GAGKGLKAKTGGELAE
AIKVALANTDGPTLIECFIGREDC lEELVKWGKRVAAANSRKPVNKLL*
SEQ ID NO :36 ATGTCAGAGTTGAGAGCCTTCAGTGCCCCAGGGAAAGCGTTACTAGCTGGT
Ph osphomevalonate GGATATTTAGTTTTAGATCCGAAATATGAAGCATTTGTAGTCGGATTATCG
GCAAGAATGCATGCTGTAGCCCATCCTTACGGTTCATTGCAAGAGTCTGAT
kinase
AAGTTTGAAGTGCGTGTGAAAAGTAAACAATTTAAAGATGGGGAGTGGCT
(Sc_ERG8, PMK) GTACCATATAAGTCCTAAAACTGGCTTCATTCCTGTTTCGATAGGCGGATC
Saccharomyces TAAGAACCCTTTCATTGAAAAAGTTATCGCTAACGTATTTAGCTACTTTAA
cerevisiae GCCTAACATGGACGACTACTGCAATAGAAACTTGTTCGTTATTGATATTTT
CTCTGATGATGCCTACCATTCTCAGGAGGACAGCGTTACCGAACATCGTGG
CAACAGAAGATTGAGTTTTCATTCGCACAGAATTGAAGAAGTTCCCAAAA
CAGGGCTGGGCTCCTCGGCAGGTTTAGTCACAGTTTTAACTACAGCTTTGG
CCTCCTTTTTTGTATCGGACCTGGAAAATAATGTAGACAAATATAGAGAAG
TTATTCATAATTTATCACAAGTTGCTCATTGTCAAGCTCAGGGTAAAATTG
GAAGCGGGTTTGATGTAGCGGCGGCAGCATATGGATCTATCAGATATAGA
AGATTCCCACCCGCATTAATCTCTAATTTGCCAGATATTGGAAGTGCTACT
TACGGCAGTAAACTGGCGCATTTGGTTAATGAAGAAGACTGGAATATAAC
GATTAAAAGTAACCATTTACCTTCGGGATTAACTTTATGGATGGGCGATAT
TAAGAATGGTTCAGAAACAGTAAAACTGGTCCAGAAGGTAAAAAATTGGT
ATGATTCGCATATGCCGGAAAGCTTGAAAATATATACAGAACTCGATCATG
217
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
CAAATTCTAGATTTATGGATGGACTATCTAAACTAGATCGCTTACACGAGA
CTCATGACGATTACAGCGATCAGATATTTGAGTCTCTTGAGAGGAATGACT
GTACCTGTCAAAAGTATCCTGAGATCACAGAAGTTAGAGATGCAGTTGCC
ACAATTAGACGTTCCTTTAGAAAAATAACTAAAGAATCTGGTGCCGATATC
GAACCTCCCGTACAAACTAGCTTATTGGATGATTGCCAGACCTTAAAAGGA
GTTCTTACTTGCTTAATACCTGGTGCTGGTGGTTATGACGCCATTGCAGTGA
TTGCTAAGCAAGATGTTGATCTTAGGGCTCAAACCGCTGATGACAAAAGAT
TTTCTAAGGTTCAATGGCTGGATGTAACTCAGGCTGACTGGGGTGTTAGGA
AAGAAAAAGATCCGGAAACTTATCTTGATAAATAA
SEQ ID NO :37 MSELRAFSAPGKALLAGGYLVLDPKYEAFVVGL SARMHAVAHPYGSLQESD
Ph osphomevalonate KFEVRVKSKQFKDGEWLYHISPKTGFIPVSIGGSKNPFIEKVIANVF SYFKPNM
DDYCNRNLFVIDIFSDDAYHSQED SVTEHRGNRRL SFH SHRIEEVPKTGL GS SA
kinase
GLVTVLTTALASFFVSDLENNVDKYREVIHNL SQVAHCQAQGKIGSGFDVAA
(Sc_ERG8, PMK) AAYGSIRYRRFPPALISNLPDIGSATYGSKLAHLVNEEDWNITIKSNHLP SGLTL
Saccharomyces WMGDIKNGSETVKLVQKVKNWYD SHMPESLKIYTELDHANSRFMDGL SKLD
cerevisiae RLHETHDDYSDQIFESLERNDCTCQKYPEI lEVRDAVATIRRSFRKITKESGADI
EPPVQTSLLDDCQTLKGVLTCLIPGAGGYDAIAVIAKQDVDLRAQTADDKRFS
KVQWLDVTQADWGVRKEKDPETYLDK*
SEQ ID NO :38 ATGTCATTACCGTTCTTAACTTCTGCACCGGGAAAGGTTATTATTTTTGGTG
M evalonate kinase AACACTCTGCTGTGTACAACAAGCCTGCCGTCGCTGCTAGTGTGTCTGCGT
TGAGAACCTACCTGCTAATAAGCGAGTCATCTGCACCAGATACTATTGAAT
(ERG12, MK) TGGACTTCCCGGACATTAGCTTTAATCATAAGTGGTCCATCAATGATTTCA
ATGCCATCACCGAGGATCAAGTAAACTCCCAAAAATTGGCCAAGGCTCAA
Saccharomyces sp.
CAAGCCACCGATGGCTTGTCTCAGGAACTCGTTAGTCTTTTGGATCCGTTG
TTAGCTCAACTATCCGAATCCTTCCACTACCATGCAGCGTTTTGTTTCCTGT
ATATGTTTGTTTGCCTATGCCCCCATGCCAAGAATATTAAGTTTTCTTTAAA
GTCTACTTTACCCATCGGTGCTGGGTTGGGCTCAAGCGCCTCTATTTCTGTA
TCACTGGCCTTAGCTATGGCCTACTTGGGGGGGTTAATAGGATCTAATGAC
TTGGAAAAGCTGTCAGAAAACGATAAGCATATAGTGAATCAATGGGCCTT
CATAGGTGAAAAGTGTATTCACGGTACCCCTTCAGGAATAGATAACGCTGT
GGCCACTTATGGTAATGCCCTGCTATTTGAAAAAGACTCACATAATGGAAC
AATAAACACAAACAATTTTAAGTTCTTAGATGATTTCCCAGCCATTCCAAT
GATCCTAACCTATACTAGAATTCCAAGGTCTACAAAAGATCTTGTTGCTCG
CGTTCGTGTGTTGGTCACCGAGAAATTTCCTGAAGTTATGAAGCCAATTCT
AGATGCCATGGGTGAATGTGCCCTACAAGGCTTAGAGATCATGACTAAGTT
AAGTAAATGTAAAGGCACCGATGACGAGGCTGTAGAAACTAATAATGAAC
TGTATGAACAACTATTGGAATTGATAAGAATAAATCATGGACTGCTTGTCT
CAATCGGTGTTTCTCATCCTGGATTAGAACTTATTAAAAATCTGAGCGATG
ATTTGAGAATTGGCTCCACAAAACTTACCGGTGCTGGTGGCGGCGGTTGCT
CTTTGACTTTGTTACGAAGAGACATTACTCAAGAGCAAATTGACAGTTTCA
AAAAGAAATTGCAAGATGATTTTAGTTACGAGACATTTGAAACAGACTTG
GGTGGGACTGGCTGCTGTTTGTTAAGCGCAAAAAATTTGAATAAAGATCTT
AAAATCAAATCCCTAGTATTCCAATTATTTGAAAATAAAACTACCACAAAG
CAACAAATTGACGATCTATTATTGCCAGGAAACACGAATTTACCATGGACT
TCATAA
SEQ ID NO :39 MSLPFLTSAPGKVIIFGEH SAVYNKPAVAASVSALRTYLLI SE S SAPDTIELDFP
M evalonate kinase DI SFNHKWSINDFNAI lEDQVNSQKLAKAQQATDGL SQELVSLLDPLLAQL
SE
SFHYHAAFCFLYMFVCLCPHAKNIKF SLK STLPIGAGL GS SASISVSLALAMAY
(ERG12, MK) LGGLIGSNDLEKL SENDKHIVNQWAFIGEKCIHGTP SGIDNAVATYGNALLFEK
D SHNGTINTNNFKFLDDFPAIPMILTYTRIPRSTKDLVARVRVL VTEKFPEVMK
Saccharomyces sp.
PILDAMGECALQGLEIMTKLSKCKGTDDEAVETNNELYEQLLELIRINHGLLVS
IGVSHPGLELIKNL SDDLRIGSTKLTGAGGGGCSLTLLRRDITQEQID SFKKKLQ
DDFSYETFETDLGGTGCCLL SAKNLNKDLKIKSLVFQLFENKTTTKQQIDDLLL
PGNTNLPWTS*
SEQ ID NO :40 ATGGCTTCTGAGAAGGAGATTCGTCGTGAGAGATTCTTGAATGTTTTTCCT
V t f AAATTAGTCGAGGAATTGAACGCTTCTTTGTTGGCTTATGGTATGCCTAAG
arnesyl arian
GAAGCTTGTGATTGGTATGCTCACTCCTTGAATTATAATACTCCAGGTGGT
pyrophosphate synthase
AAATTGAACCGTGGTTTGTCTGTTGTTGACACTTACGCTATTTTATCTAACA
AGACCGTCGAGCAATTGGGTCAAGAAGAGTATGAAAAGGTCGCTATTTTA
218
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
(ERG20mut, F96W, GGTTGGTGTATTGAATTGTTGCAAGCTTACTGGTTGGTTGCCGATGACATG
N127W; GPPS) ATGGACAAGTCTATTACTCGTCGTGGTCAACCTTGCTGGTATAAGGTCCCA
GAGGTTGGTGAAATTGCTATCTGGGACGCTTTCATGTTGGAAGCTGCTATC
Artificial sequence
TATAAATTGTTGAAATCCCACTTCAGAAACGAGAAATACTACATTGACATC
Codon optimized ACCGAGTTGTTCCACGAAGTCACTTTCCAAACTGAGTTAGGTCAATTAATG
GACTTGATCACCGCTCCAGAAGACAAAGTTGACTTGTCCAAGTTTTCCTTG
AAAAAGCACTCTTTCATCGTTACTTTCAAGACTGCTTATTACTCTTTCTACT
TACCAGTTGCCTTGGCTATGTACGTCGCCGGTATCACTGACGAAAAGGACT
TGAAGCAAGCTCGTGACGTTTTGATTCCATTAGGTGAATATTTCCAAATCC
AAGATGACTACTTAGACTGTTTTGGTACCCCTGAACAAATCGGTAAGATCG
GTACTGATATTCAAGATAACAAGTGCTCTTGGGTTATCAACAAGGCTTTAG
AGTTAGCCTCCGCCGAACAACGTAAAACTTTAGATGAAAACTACGGTAAA
AAAGACTCTGTTGCTGAGGCCAAGTGTAAGAAGATTTTTAACGATTTAAAA
ATCGAACAATTGTATCACGAATATGAAGAGTCCATTGCTAAGGATTTGAAG
GCTAAAATTTCTCAAGTTGACGAATCCCGTGGTTTCAAAGCTGACGTTTTG
ACTGCTTTTTTAAACAAGGTTTACAAGCGTTCCAAATAA
SEQ ID NO :41 MASEKEIRRERFLNVFPKLVEELNASLLAYGMPKEACDWYAHSLNYNTPGGK
Variant farnesyl LNRGLSVVDTYAIL SNKTVEQLGQEEYEKVAILGWCIELLQAYWLVADDMM
rophosphate synthase DKSITRRGQPCWYKVPEVGEIAIWDAFMLEAAIYKLLKSHFRNEKYYIDI1ELF
py
HEVTFQ1ELGQLMDLITAPEDKVDLSKFSLKKHSFIVTFKTAYYSFYLPVALA
(ERG20mut, F96W'
N127W GPPS MYVAGITDEKDLKQARDVLIPLGEYFQIQDDYLDCFGTPEQIGKIGTDIQDNKC
; )
SWVINKALELASAEQRKTLDENYGKKD SVAEAKCKKIFNDLKIEQLYHEYEES
Artificial sequence IAKDLKAKISQVDESRGFKADVLTAFLNKVYKRSK*
SEQ ID NO :42 ATGTCTACCGCACTAACAGAAGGAGCTAAACTATTCGAAAAGGAGATTCC
GFP TTACATTACAGAATTAGAGGGTGATGTCGAAGGAATGAAATTCATTATCAA
GGGCGAGGGTACTGGTGACGCTACTACCGGTACGATTAAAGCAAAGTACA
Artificial sequence TCTGTACAACAGGTGACCTTCCTGTTCCGTGGGCTACTCTGGTGAGCACTTT
GTCTTATGGAGTTCAATGTTTTGCTAAATACCCTTCGCACATTAAAGACTTT
TTCAAAAGTGCAATGCCTGAGGGCTATACTCAGGAGAGAACAATATCTTTC
GAAGGAGATGGTGTGTATAAGACTAGGGCTATGGTCACGTATGAAAGAGG
ATCCATCTACAATAGAGTAACTTTAACTGGTGAAAACTTCAAAAAGGACG
GTCACATCCTTAGAAAGAATGTTGCCTTTCAATGCCCACCATCCATCTTGT
ACATTTTGCCAGACACAGTTAACAATGGTATCAGAGTTGAGTTTAACCAAG
CTTATGACATAGAGGGTGTCACCGAAAAGTTGGTTACAAAATGTTCACAG
ATGAATCGTCCCCTGGCAGGATCAGCTGCCGTCCATATCCCACGTTACCAT
CATATCACTTATCATACCAAGCTGTCCAAAGATCGTGATGAGAGAAGGGA
TCACATGTGTTTGGTTGAAGTGGTAAAGGCCGTGGATTTGGATACTTACCA
AGGTTGA
SEQ ID NO :43 MSTAL lEGAKLFEKEIPYI1ELEGDVEGMKFIIKGEGTGDATTGTIKAKYICTTG
GFP DLPVPWATLVSTLSYGVQCFAKYPSHIKDFFKSAMPEGYTQERTISFEGDGVY
KTRAMVTYERGSIYNRVTLTGENFKKDGHILRKNVAFQCPPSILYILPDTVNN
Artificial sequence GIRVEFNQAYDIEGVTEKLVTKCSQMNRPLAGSAAVHIPRYHHITYHTKLSKD
RDERRDHMCLVEVVKAVDLDTYQG*
SEQ ID NO :44 MNCSAFSFWFVCKIIFFFLSFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCA
DQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATILCSKKVGLQIRTRSG
synthase
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Cannabis sativa KNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVDGK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VDSLVDLMNKSFPELGIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAFSIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEIS
ESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO :45 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCA
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
synthase
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
219
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
Codon optimized CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
sequence 1 GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :46 TGTTGTGGAAATGTAAAGAGCCCCATTATCTTAGCCTAAAAAAACCTTCTC
GALl tTDH1 TTTGGAACTTTCAGTAATACGCTTAACTGCTCATTGCTATATTGAAGTACG
p_
GATTAGAAGCCGCCGAGCGGGCGACAGCCCTCCGACGGAAGACTCTCCTC
Saccharomyces sp. CGTGCGTCCTGGTCTTCACCGGTCGCGTTCCTGAAACGCAGATGTGCCTCG
CGCCGCACTGCTCCGAACAATAAAGATTCTACAATACTAGCTTTTATGGTT
ATGAAGAGGAAAAATTGGCAGTAACCTGGCCCCACAAACCTTCAAATCAA
CGAATCAAATTAACAACCATAGGATAATAATGCGATTAGTTTTTTAGCCTT
ATTTCTGGGGTAATTAATCAGCGAAGCGATGATTTTTGATCTATTAACAGA
TATATAAATGCAAAAGCTGCATAACCACTTTAACTAATACTTTCAACATTT
TCGGTTTGTATTACTTCTTATTCAAATGTCATAAAAGTATCAACAAAAAAT
TGTTAATATACCTCTATACTTTAACGTCAAGGAGAAAAAACTATACTCTTA
TTACCCTATCCTATGGATAAAGCAATCTTGATGAGGATAATGATTTTTTTTT
GAATATACATAAATACTACCGTTTTTCTGCTAGATTTTGTGAAGACGTAAA
TAAGTACATATTACTTTTTAAGCCAAGACAAGATTAAGCATTAACTTTACC
CTTTTCTCTTCTAAGTTTCAATACTAGTTATCACTGTTTAAAAGTTATGGCG
AGAACGTCGGCGGTTAAAATATATTACCCTGAACGTGGTGAATTGAAGTTC
TAGGATGGTTTAAAGATTTTTCCTTTTTGGGAAATAAGTAAACAATATATT
GCTGCCTTTGC
SEQ ID NO :47 ATGGCCGTCAAACACTTGATCGTCTTAAAATTCAAGGATGAAATTACTGAA
OAC Y27F
GCTCAAAAAGAAGAGTTCTTCAAAACCTTCGTCAATTTAGTCAACATTATT
(OAC*) variant
CCTGCTATGAAGGACGTTTACTGGGGTAAGGATGTCACCCAAAAGAACAA
GGAAGAAGGTTACACTCACATTGTTGAAGTCACTTTCGAATCTGTTGAAAC
Artificial sequence TATCCAAGATTATATTATCCACCCAGCTCATGTCGGTTTTGGTGATGTTTAC
Codon optimized AGATCTTTTTGGGAAAAATTGTTGATCTTTGACTATACTCCAAGAAAATAA
SEQ ID NO :48 MAVKHLIVLKFKDEI lEAQKEEFFKTFVNLVNIIPAMKDVYWGKDVTQKNKE
OAC Y27F variant EGYTHIVEVTFESVETIQDYIIHPAHVGFGDVYRSFWEKLLIFDYTPRK*
(OAC*)
Artificial sequence
220
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
SEQ ID NO: 49 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS F31 CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant 7Y
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon Optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTATCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :50 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS F31 variant D LYMSILNSTIQNLRFISDTTPKPLVIVTP SNNSHIQATIL CSKKVGLQIRTRSG
7Y
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATL GEVYYWINE
Artificial sequence KNENL SFPGGYCPTVGVGGHF S GGGYGALMRNYGLAADNIIDAHL VNVD
GK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVP SKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIYHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTAS WEKQEDNEKHINWVRS VYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO :51 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS 196T
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAACCTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
221
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :52 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS 196T variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTP SNNSHIQATIL CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHFSGGGYGALMRTYGLAADNIIDAHLVNVDGK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVP SKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTAS WEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO :53 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS 196 CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
Q variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGACAATACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
222
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :54 MNCSAFSFWFVCKIIFFFLSFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS 196Q variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATILCSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHF S GGGYGALMRQYGLAADNIIDAHL VNVD
GK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO :55 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS K261C
C TATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCTGCAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :56 MNCSAFSFWFVCKIIFFFLSFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS K261C variant D LYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATILCSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHF S GGGYGALMRNYGLAADNIIDAHL VNVD
GK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVCKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
223
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
SEQ ID NO :57 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS N196V
C TATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
valiant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAGTCTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :58 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS N196V variant D LYMSILNSTIQNLRFISDTTPKPLVIVTP SNNSHIQATIL CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHF S GGGYGALMRVYGLAADNIIDAHL VNVD
GK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVP SKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTAS WEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO :59 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS Li 32M
C TATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
valiant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATATGAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
224
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :60 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS Li 32M variant D LYMSILNSTIQNLRFISDTTPKPLVIVTP SNNSHIQATIL
CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDMRNMHSIKIDVH SQTAWVEAGATLGEVYYWIN
Artificial Sequence EKNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVDG
KVLDRKSMGEDLFWAIRGGGGENF GIIAAWKIKLVAVP SKSTIF SVKKNMEIH
GLVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHG
GVD SLVDLMNKSFPEL GIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLDR
SAGKKTAFSIKLDYVKKPIPETAMVKILEKLYEED VGAGMYVLYPYGGIMEEI
SESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO:61 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS S170T
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTAACCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
225
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :62 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS S170T variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENLTFPGGYCPTVGVGGHF SGGGYGALMRNYGLAADNIIDAHLVNVDGK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAFSIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEIS
ESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO :63 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS P53 9T
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCAACTTTACCACCACACCACCATT
AG
SEQ ID NO :64 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS P53 9T variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHF S GGGYGALMRNYGLAADNIIDAHL VNVD
GK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAFSIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEIS
ESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPTLPPHHH*
226
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
SEQ ID NO :65 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS L269I
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTATCGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :66 MNCSAFSFWFVCKIIFFFLSFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS L269I variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATL GEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVDGK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEIHGI
IVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYF S SIFHGGV
D SLVDLMNKSFPELGIKKTDCKEF SWIDTTIFYS GVVNFNTANFKKEILLDRSA
GKKTAFSIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEISE
SAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRLA
YLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNFF
RNEQSIPPLPPHHH*
SEQ ID NO :67 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS F171I
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCATCC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
227
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :68 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTQH
THCAS F171I variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTP SNNSHIQATIL CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATL GEVYYWINE
Artificial Sequence KNENL SIPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVDGK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVP SKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTAS WEKQEDNEKHINWVRS VYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQ SIPPLPPHHH*
SEQ ID NO :69 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS R3 1Q
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCAAGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
228
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :70 MNCSAFSFWFVCKIIFFFL SFHIQISIANPQENFLKCFSKHIPNNVANPKLVYTQ
THCAS R31 variant HDQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRS
Q
GGHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWI
Artificial Sequence NEKNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVD
GKVLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEI
HGLVKLFNKWQNIAYKYDKDL VLMTHFITKNITDNHGKNKTTVHGYF S SIFH
GGVD SLVDLMNKSFPELGIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLD
RSAGKKTAF SIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEE
ISESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPR
LAYLNYRDLDL GKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNN
FFRNEQSIPPLPPHHH*
SEQ ID NO :71 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS P43E
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCGAGAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :72 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIENNVANPKLVYTQ
THCAS P43E variant HDQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRS
GGHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWI
Artificial Sequence NEKNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVD
GKVLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEI
HGLVKLFNKWQNIAYKYDKDL VLMTHFITKNITDNHGKNKTTVHGYF S SIFH
GGVD SLVDLMNKSFPELGIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLD
RSAGKKTAF SIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEE
ISESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPR
LAYLNYRDLDL GKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNN
FFRNEQSIPPLPPHHH*
229
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
SEQ ID NO :73 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS P49E
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACGAGAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :74 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCF SKHIPNNVANEKLVYTQ
THCAS P49E variant HDQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRS
GGHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWI
Artificial Sequence NEKNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVD
GKVLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEI
HGLVKLFNKWQNIAYKYDKDL VLMTHFITKNITDNHGKNKTTVHGYF S SIFH
GGVD SLVDLMNKSFPELGIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLD
RSAGKKTAF SIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEE
I SESAIPFPHRAGIMYEL WYTA SWEKQEDNEKHINWVRSVYNFTTPYVSQNPR
LAYLNYRDLDL GKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNN
FFRNEQSIPPLPPHHH*
SEQ ID NO :75 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS P49K
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACAAAAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
230
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :76 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANKKLVYTQ
THCAS P49K variant HDQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRS
GGHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWI
Artificial Sequence NEKNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVD
GKVLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEI
HGLVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFH
GGVD SLVDLMNKSFPELGIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLD
RSAGKKTAF SIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEE
I SESAIPFPHRAGIMYELWYTA SWEKQEDNEKHINWVRSVYNFTTPYVSQNPR
LAYLNYRDLDL GKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNN
FFRNEQSIPPLPPHHH*
SEQ ID NO:?? ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS P49 CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant Q
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCAAAAGTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
231
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :78 MNCSAFSFWFVCKIIFFFLSFHIQISIANPRENFLKCFSKHIPNNVANQKLVYTQ
THCAS P49Q variant HDQLYMS ILNSTIQNLRFI SDTTPKPLVIVTP SNN SHIQATIL
CSKKVGLQIRTRS
GGHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWI
Artificial Sequence NEKNENLSFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVD
GKVLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEI
HGLVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFH
GGVD SLVDLMNKSFPELGIKKTDCKEFSWIDTTIFYSGVVNFNTANFKKEILLD
RSAGKKTAF SIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEE
ISESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPR
LAYLNYRDLDL GKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNN
FFRNEQSIPPLPPHHH*
SEQ ID NO :79 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS K5OT
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTACCTTAGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :80 MNCSAFSFWFVCKIIFFFLSFHIQISIANPRENFLKCFSKHIPNNVANPTLVYTQH
THCAS K50T variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATILCSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHFSGGGYGALMRNYGLAADNIIDAHLVNVDGK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPELGIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAFSIKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEIS
ESAIPFPHRAGIMYELWYTASWEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
232
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
SEQ ID NO :81 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS L51I
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGATCGTT
Artificial Sequence TACACTCAACATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :82 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKIVYTQH
THCAS L51I variant DQLYMSILNSTIQNLRFISDTTPKPLVIVTP SNNSHIQATIL
CSKKVGLQIRTRSG
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATL GEVYYWINE
Artificial Sequence KNENL SFP GGYCPTVGVGGHF S GGGYGALMRNYGLAADNIIDAHL VNVD
GK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVP SKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTAS WEKQEDNEKHINWVRS VYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQ SIPPLPPHHH*
SEQ ID NO :83 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS 55E
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant Q
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTGAGCATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAA
Codon optimized AACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACC
CCATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTC
GGTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCT
TACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTA
TCAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTT
TAGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTC
CAGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTG
GTTATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCG
ACGCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGG
GTGAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTA
TCATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTAT
233
CA 03152803 2022-02-25
WO 2021/055597 PCT/US2020/051261
TTTCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAA
CAAGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGAC
CCACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTAC
TGTTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTC
GATTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGT
AAAGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACT
TCAACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTA
AAAAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAG
AAACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGT
GCCGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCT
GAATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGG
TACACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTG
GGTTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAG
ATTAGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGC
TTCCCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGG
TAAGAACTTCAACCGTTTAGTCAAGGTCAAGACTAAAGTTGATCCAAACA
ATTTTTTCAGAAACGAACAATCTATCCCACCTTTACCACCACACCACCATT
AG
SEQ ID NO :84 MNCSAFSFWFVCKIIFFFL SFHIQISIANPRENFLKCFSKHIPNNVANPKLVYTEH
THCAS 55E variant
DQLYMSILNSTIQNLRFISDTTPKPLVIVTPSNNSHIQATIL CSKKVGLQIRTRSG
Q
GHDAEGMSYISQVPFVVVDLRNMHSIKIDVHSQTAWVEAGATLGEVYYWINE
Artificial Sequence KNENL SFPGGYCPTVGVGGHF S GGGYGALMRNYGLAADNIIDAHL VNVD
GK
VLDRKSMGEDLFWAIRGGGGENFGIIAAWKIKLVAVPSKSTIFSVKKNMEIHG
LVKLFNKWQNIAYKYDKDLVLMTHFITKNITDNHGKNKTTVHGYFS SIFHGG
VD SLVDLMNKSFPEL GIKKTD CKEFSWIDTTIFYSGVVNFNTANFKKEILLDRS
AGKKTAF S IKLDYVKKPIPETAMVKILEKLYEEDVGAGMYVLYPYGGIMEEI S
ESAIPFPHRAGIMYELWYTAS WEKQEDNEKHINWVRSVYNFTTPYVSQNPRL
AYLNYRDLDLGKTNHASPNNYTQARIWGEKYFGKNFNRLVKVKTKVDPNNF
FRNEQSIPPLPPHHH*
SEQ ID NO :85 ATGAATTGTTCTGCTTTCTCTTTCTGGTTCGTTTGTAAGATCATCTTTTTCTT
THCAS 55P
CTTATCTTTCCATATTCAAATCTCTATCGCTAACCCTCGTGAGAACTTCTTG
variant Q
AAATGTTTCTCCAAACATATCCCAAACAATGTCGCTAACCCTAAGTTAGTT
Artificial Sequence TACACTCCTCATGATCAATTATATATGTCTATCTTGAACTCTACCATCCAAA
Codon optimized ACTTGAGATTCATCTCCGATACCACCCCAAAACCATTGGTTATTGTTACCC
CATCCAACAATTCTCATATTCAAGCTACCATTTTGTGCTCCAAAAAGGTCG
GTTTGCAAATCCGTACTAGATCTGGTGGTCACGATGCTGAAGGTATGTCTT
ACATTTCCCAAGTCCCATTCGTTGTTGTCGATTTAAGAAATATGCACTCTAT
CAAAATCGACGTTCACTCTCAAACTGCTTGGGTTGAAGCCGGTGCCACTTT
AGGTGAGGTTTACTACTGGATTAACGAAAAGAATGAAAACTTATCCTTTCC
AGGTGGTTACTGTCCAACTGTTGGTGTTGGTGGTCACTTCTCTGGTGGTGGT
TATGGTGCCTTGATGAGAAACTACGGTTTAGCTGCTGATAATATTATCGAC
GCTCACTTGGTTAATGTCGACGGTAAGGTTTTGGACAGAAAATCCATGGGT
GAAGATTTATTCTGGGCCATTAGAGGTGGTGGTGGTGAAAACTTCGGTATC
ATTGCTGCTTGGAAAATTAAATTGGTCGCTGTCCCATCCAAGTCTACTATTT
TCTCCGTCAAGAAAAACATGGAAATTCATGGTTTGGTTAAATTATTCAACA
AGTGGCAAAACATTGCTTACAAATACGACAAAGACTTAGTTTTGATGACCC
ACTTCATTACTAAAAACATTACCGACAACCATGGTAAAAATAAAACTACTG
TTCACGGTTACTTCTCTTCCATTTTTCATGGTGGTGTCGACTCCTTGGTCGA
TTTAATGAACAAATCTTTCCCTGAGTTGGGTATCAAGAAGACCGACTGTAA
AGAATTCTCTTGGATCGACACTACTATTTTCTACTCTGGTGTCGTTAACTTC
AACACCGCTAATTTCAAGAAGGAAATTTTATTAGATAGATCCGCTGGTAAA
AAGACCGCTTTCTCTATCAAATTAGACTACGTTAAAAAACCAATCCCAGAA
ACCGCTATGGTCAAAATCTTGGAAAAATTATATGAAGAAGACGTTGGTGC
CGGTATGTACGTCTTATATCCATATGGTGGTATTATGGAAGAGATCTCTGA
ATCCGCTATCCCTTTTCCACACAGAGCCGGTATTATGTACGAATTATGGTA
CACTGCTTCCTGGGAGAAACAAGAAGATAATGAAAAGCACATTAACTGGG
TTAGATCTGTTTACAACTTCACTACTCCATACGTCTCTCAAAACCCAAGATT
AGCCTACTTAAACTACCGTGATTTGGATTTAGGTAAAACTAATCACGCTTC
CCCAAACAACTACACCCAAGCTAGAATTTGGGGTGAGAAGTACTTTGGTA
234
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
CONTENANT LES PAGES 1 A 234
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 234
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME:
NOTE POUR LE TOME / VOLUME NOTE: