Note: Descriptions are shown in the official language in which they were submitted.
WO 2021/113772
PCT/US2020/063488
PARTICLE DELIVERY SYSTEMS
CROSS REFERENCE TO RELATED APPLICATIONS
100011 This application claims priority to U.S. provisional patent application
numbers
62/944,982, filed on December 6, 2019, 62/968,915, filed on January 31, 2020,
62/983,460, filed
on February 28, 2020, 63/035,576, filed on June 5, 2020 and 63/120,864, filed
on December 3,
2020, the contents of each of which are incorporated herein by reference in
their entireties.
INCORPORATION BY REFERENCE OF SEQUENCE LISTING
100021 This application contains a Sequence Listing which has been submitted
in ASCII
format via EFS-WEB and is hereby incorporated by reference in its entirety.
Said ASCII copy,
created on December 4, 2020 is named SCRB 024 05W0 SeqList ST25.txt and is 114
MB in
size.
BACKGROUND
100031 The delivery of protein or nucleic acid therapeutics to particular
cells or organs of the
body generally requires complex systems in which a targeting modality or
vehicle is linked to or
contains the therapeutic. Even with highly selective targeting modalities,
such as monoclonal
antibodies, the selectivity of the system for the target cells or organs is
not absolute, and off-
target toxicity can be a consequence.
100041 The Retroviridae family of viruses encompass several genera of viruses
that cause
chronic and deadly diseases characterized by long incubation periods, in
humans and other
mammalian species. The Retroviridae family includes Othoretrovirinae
(Lentivirus,
Alpharetrovirus, Betaretrovirus, Deltaretrovirus, Epsilonretrovirus,
Gammaretrovirus), and
Spurnaretrovirinae . The best known lentivirus is the Human Immunodeficiency
Virus (HIV),
which causes acquired immune deficiency syndrome (AIDS). As with all
retroviruses,
lentiviruses have gag, pot and env genes, coding for viral proteins in the
order: 5"-gag-pol-env-
3'. The lentivirus system has been adapted to introduce gene editing systems
into human or
animal cells by the creation of virus-like particles (VLP) containing the gene
editing systems.
Retroviral systems have advantages over other gene-therapy methods, including
high-efficiency
infection of dividing and non-dividing cells, long-term stable expression of a
transgene, and low
immunogenicity. Lentiviruses have been successfully used for transduction of
diabetic mice with
1
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
the gene encoding PDGF (platelet-derived growth factor), a therapy being
considered for use in
humans (Lee JA, et at. Lentiviral transfection with the PDGF-B gene improves
diabetic wound
healing. Plast. Reconstr. Surg. 116 (2): 532 (2005)). However, one major
difficulty with use of
certain therapeutics, like CRISPR nucleases, in VLP is off-target effects,
particularly with long-
term expression of the nuclease when traditional expression methods such as
via plasmid/viral
vectors are used. Accordingly, there remains a need for improved systems for
delivery of gene
editing systems using particles derived from viral vectors.
SUMMARY
100051 The present disclosure provides delivery particle (XDP) systems for the
delivery of
therapeutic payloads, including proteins, nucleic acids, small molecules and
the like to target
cells and tissues.
100061 In some embodiments, the XDP system comprises
nucleic acids encoding components
selected from all or a portion of a retroviral gag polyprotein, a therapeutic
payload, and a tropism
factor, wherein the tropism factor is selected from the group consisting of a
glycoprotein, an
antibody fragment, a receptor, and a ligand to a target cell marker. In one
embodiment of the
foregoing, the tropism factor is a glycoprotein having a sequence selected
from the group of
sequences consisting of SEQ ID NOS: 438, 440, 442, 444, 446, 448, 450, 452,
454, 456, 458,
460, 462, 464, 466, 468, 470, 472, 474, 476, 478, 480, 482, 484, 486, 488,
490, 492, 494, 496,
498, 500, 502, 504, 506, 508, 510, 512, 514, 516, 518, 520, 522, 524, 526,
528, 530, 532, 534,
536, 538, 540, 542, 544, 546, 548, 550, 552, 554, 556, 558, 560, 562, 564,
566, 568, 570, 572,
574, 576, 578, 580, 582, 584, 586, 588, 590, 592, 594 and 596 as set forth in
Table 4, or a
sequence having at least about 85%, at least about 90%, at least about 91%, at
least about 92%,
at least about 93%, at least about 94%, at least about 95%, at least about
96%, at least about
97%, at least about 98%, or at least about 99% sequence identity thereto. In a
particular
embodiment, the glycoprotein is VSV-G. In a particular embodiment, the
glycoprotein
comprises a sequence of SEQ ID NO: 438.
100071 The therapeutic payload can be a protein, a nucleic acid, or both a
protein and a nucleic
acid. In some embodiments of the XDP system, the protein payload is selected
from the group
consisting of a cytokine, an interleukin, an enzyme, a receptor, a
microprotein, a hormone,
erythropoietin, a ribonuclease (RNAse), a deoxyribonuclease (DNAse), a blood
clotting factor,
an anticoagulant, a bone morphogenetic protein, an engineered protein
scaffold, a thrombolytic
protein, a CRISPR protein, and an anti-cancer modality. In one embodiment, the
therapeutic
2
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
payload is a Class 1 or Class 2 CRISPR protein, wherein the Class 2 CRISPR
protein selected
from the group consisting of a Type II, Type V, or Type VI protein. In one
embodiment, the
Class 2 CRISPR Type V protein is selected from the group consisting of Cas12a,
Cas12b,
Cas12c, Cas12d (CasY), Cas12j and CasX, wherein the CasX comprises a sequence
of SEQ ID
NOS: 21-233, 343-345, 350-353, 355-367 or 388-397 as set forth in Tables 1, 7,
8, 9, or 11, or a
sequence having at least about 85%, at least about 90%, at least about 91%, at
least about 92%,
at least about 93%, at least about 94%, at least about 95%, at least about
96%, at least about
97%, at least about 98%, or at least about 99% sequence identity thereto. In
some embodiments,
the CasX comprises a sequence of SEQ ID NOS: 21-233, 343-345, 350-353, 355-367
or 388-
397. In some embodiments, the therapeutic payload is a nucleic acid selected
from the group
consisting of a single-stranded anti sense oligonucleotide (AS0s), a double-
stranded RNA
interference (RNAi) molecule, a DNA aptamer, and a CRISPR guide nucleic acid,
wherein the
CRISPR guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and
a targeting sequence, wherein the targeting sequence comprises between 14 and
30 nucleotides
and is complementary to a target nucleic acid sequence, and wherein the
scaffold sequence
comprises a sequence of SEQ ID NOS: 597-781 as set forth in Table 3, or a
sequence having at
least about 85%, at least about 90%, at least about 91%, at least about 92%,
at least about 93%,
at least about 94%, at least about 95%, at least about 96%, at least about
97%, at least about
98%, or at least about 99% sequence identity thereto. In some embodiments, the
scaffold
sequence comprises a sequence of SEQ ID NOS: 597-781.
[0008] In some embodiments, the XDP system further comprises nucleic acids
encoding one
or more components selected from one or more protease cleavage sites, a gag-
transframe region-
poi protease polyprotein (gag-TFR-PR), a retroviral gag-pol polyprotein, and a
non-retroviral
protease capable of cleaving the protease cleavage sites. In some embodiments,
the retroviral
components of the XDP system are derived from a Orthoretrovirthae virus or a
Spumaretrovirinae virus wherein the Orthoretrovirinae virus is selected from
the group
consisting of Alpharetrovirus, Befaretrovirus, Deltaretrovirus,
Epsilonretrovirus,
Gammaretrovirus, and Lentivirus, and the Spumaretrovirinae virus is selected
from the group
consisting of Bovispurnavirus, Equispumavirus, Felispumavirus,
Prosimiispumavirus,
Simiispumavirus, and Spumavirus.
100091 In some embodiments, the components of the XDP system are encoded on a
single
nucleic acid, on two nucleic acids, on three nucleic acids, on four nucleic
acids, or on five
3
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
nucleic acids, and the nucleic acids are configured according to any one of
FIGS. 36-68. In some
embodiments, the components of the XDP system are encoded by nucleic acids
selected from
the group of sequences of SEQ ID NOS: 192, 193, 195, 196, 198-201, 782, and
234-339 as set
forth in Table 5.
[0010] In some embodiments, the components of the XDP system are capable of
self-
assembling into an XDP when the one or more nucleic acids are introduced into
a eukaryotic
host cell and are expressed. In the foregoing embodiment, the therapeutic
payload is
encapsidated within the XDP upon self-assembly of the XDP. In a particular
embodiment,
wherein the therapeutic payload comprises a CasX and a guide RNA, the CasX and
guide RNA
are complexed as a ribonucleoprotein complex (RNP) and, optionally, a donor
template is also
encapsidated in the XDP. In another particular embodiment, the tropism factor
is incorporated
on the XDP surface upon self-assembly of the XDP.
[0011] In some embodiments of the XDP system, the nucleic acids encoding the
retroviral
components are all or a portion of an Alpharetrovirus gag polyprotein, wherein
the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a P2A peptide, a P2B peptide, a P10 peptide, a capsid
polypeptide (CA), and
a nucleocapsid polypeptide (NC). In some embodiments of the XDP system, the
nucleic acids
further comprise sequences encoding one or more components selected from an
HIV p1 peptide,
an HIV p6 peptide, a Gag-Pol polyprotein, one or more protease cleavage sites,
a non-retroviral,
heterologous protease capable of cleaving the cleavage sites, and a gag-
transframe region-pol
protease polyprotein.
[0012] In some embodiments of the XDP system, the nucleic acids encoding the
retroviral
components are all or a portion of an Betcweirovirus gag polyprotein, wherein
the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a PP21/24 peptide, a P12/P3/P8 peptide, a capsid polypeptide
(CA), and a
nucleocapsid polypeptide (NC). In some embodiments of the XDP system, the
nucleic acids
further comprise sequences encoding one or more components selected from an
HIV pl peptide,
an I-11V p6 peptide, a Gag-Pol polyprotein, one or more protease cleavage
sites, a non-retroviral,
heterologous protease capable of cleaving the cleavage sites, and a gag-
transframe region-pol
protease polyprotein.
[0013] In some embodiments of the XDP system, the nucleic acids encoding the
retroviral
components are all or a portion of a Deharetrovirus gag polyprotein, wherein
the gag
4
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a capsid polypeptide (CA), and a nucleocapsid polypeptide
(NC). In some
embodiments of the XDP system, the nucleic acids further comprise sequences
encoding one or
more components selected from an HIV p1 peptide, an HW p6 peptide, a Gag-Pol
polyprotein,
one or more protease cleavage sites, a non-retroviral, heterologous protease
capable of cleaving
the cleavage sites, and a gag-transframe region-pol protease polyprotein.
100141 In some embodiments of the XDP system, the nucleic acids encoding the
retroviral
components are all or a portion of a Epsdonretrovirus gag polyprotein, wherein
the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a p20 peptide, a capsid polypeptide (CA), and a nucleocapsid
polypeptide
(NC). In some embodiments of the XDP system, the nucleic acids further
comprise sequences
encoding one or more components selected from an HIV p1 peptide, an HIV p6
peptide, a Gag-
Pol polyprotein, one or more protease cleavage sites, a non-retroviral,
heterologous protease
capable of cleaving the cleavage sites, and a gag-transframe region-pol
protease polyprotein.
100151 In some embodiments of the XDP system, the nucleic acids encoding the
retroviral
components are all or a portion of a Ganunattreirovirus gag polyprotein,
wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a p12 peptide, a capsid polypeptide (CA), and a nucleocapsid
polypeptide
(NC). In some embodiments of the XDP system, the nucleic acids further
comprise sequences
encoding one or more components selected from an HIV p1 peptide, an HIV p6
peptide, a Gag-
Pol polyprotein, one or more protease cleavage sites, a non-retroviral,
heterologous protease
capable of cleaving the cleavage sites, and a gag-transfranie region-pol
protease polyprotein.
100161 In some embodiments of the XDP system, the nucleic acids encoding the
retroviral
components are all or a portion of a Lentivirus gag polyprotein, wherein the
gag polyprotein
comprises one or more components selected from the group consisting of a
matrix polypeptide
(MA), a capsid polypeptide (CA), a p2 peptide, a nucleocapsid polypeptide
(NC), a pl peptide,
and a p6 peptide. In some embodiments of the XDP system, the nucleic acids
further comprise
sequences encoding one or more components selected from a Gag-Pol polyprotein,
one or more
protease cleavage sites, a non-retroviral, heterologous protease capable of
cleaving the cleavage
sites, and a gag-transframe region-pol protease polyprotein.
100171 In some embodiments of the XDP system, the nucleic acids encoding the
retroviral
components are all or a portion of a Sputnaretrovirinae gag polyprotein,
wherein the gag
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
polyprotein comprises one or more components selected from the group
consisting of a p68 Gag
polypeptide and a p3 Gag polypeptide. In some embodiments of the XDP system,
the nucleic
acids further comprise sequences encoding one or more components selected from
an HIV pl
peptide, an p6 peptide, a Gag-Pol polyprotein, one or
more protease cleavage sites, a non-
retroviral, heterologous protease capable of cleaving the cleavage sites, and
a gag-transframe
region-pot protease polyprotein.
100181 In some embodiments of the CasX system, the CasX further comprises one
or more
NLS selected from the group of sequences consisting of PICKICRKV (SEQ ID NO:
130),
KRPAATKKAGQAICICKK (SEQ ID NO: 131), PAAKRVKLD (SEQ ID NO: 132),
RQRRNELICRSP (SEQ ID NO: 133),
NQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGY (SEQ ID NO: 134),
RMRIZFKNKGKDTAELRRRRVEVSVELRKAICKDEQILICRRNV (SEQ ID NO: 135),
VSRKRPRP (SEQ ID NO: 136), PPKKARED (SEQ ID NO: 137), PQPICKKPL (SEQ ID NO:
138), SALIKKICKKMAP (SEQ ID NO: 139), DRLRR. (SEQ ID NO: 140), PKQKKRK (SEQ
ID NO: 141), RICL,KICKIKKL (SEQ ID NO: 142), REICICKFLICRR (SEQ ID NO: 143),
KRKGDEVDGVDEVAKKKSKK (SEQ ID NO: 144), RKCLQAGMNLEARKTICK (SEQ ID
NO: 145), PRPRKIPR (SEQ ID NO: 146), PPRKKRTVV (SEQ ID NO: 147),
NLSKICKKRKREK (SEQ ID NO: 148), RRPSRPFRKP (SEQ ID NO: 149), KRPRSPSS (SEQ
ID NO: 150), KRGINDRNFWRGENERKTR (SEQ ID NO: 151), PRPPKMARYDN (SEQ ID
NO: 152), KRSFSKAF (SEQ ID NO: 153), KLICIKRPVK (SEQ ID NO: 154),
PKTRRRPRRSQRKRPPT (SEQ ID NO: 156), RRICKRRPRRKKRR (SEQ ID NO: 159),
PICICKSRKPKKKSRK (SEQ ID NO: 160), HICKKITPDASVNFSEFSK (SEQ ID NO: 161),
QRPGPYDRPQRPGPYDRP (SEQ ID NO: 162), LSPSLSPLLSPSLSPL (SEQ ID NO: 163),
RGKGGKGLGKGGAKR.HRIC (SEQ ID NO: 164), PKRGRGRPKR.GRGR (SEQ ID NO: 165),
MSRRRKANPTKLSENAKKLAICEVEN (SEQ ID NO: 157), PICKKRICVPPPPAAKRVKLD
(SEQ ID NO: 155), and PICKICRKVPPPPICKICRKV (SEQ ID NO: 166), wherein the NLS
are
located at or near the N-terminus and/or the C-terminus.
100191 In some embodiments of the XDP system, the non-retroviral, heterologous
protease is
selected from the group consisting of tobacco etch virus protease (TEV),
potyvirus HC protease,
potyvirus P1 protease, PreScission (HRV3C protease), b virus Ma protease, B
virus RNA-2-
encoded protease, aphthovirus L protease, enterovirus 2A protease, rhinovirus
2A protease,
picoma 3C protease, comovirus 24K protease, nepovirus 24K protease, RTSV (rice
tungro
6
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
spherical virus) 3C4ike protease, parsnip yellow fleck virus protease, 3C-like
protease, heparin,
cathepsin, thrombin, factor Xa, metalloproteinase, and enterokinase.
[0020] In other aspects, the present disclosure provides eukaryotic cells
comprising the XDP
system of any one of the foregoing embodiments, wherein the cell is a
packaging cell. In some
embodiments, the eukaryotic cell is selected from the group consisting of 1-
IEK293 cells, Lenti-X
293T cells, BHK cells, HepG2, Saos-2, HuH7, NSO cells, SP2/0 cells, YO myeloma
cells, A549
cells, P3X63 mouse myeloma cells, PER cells, PER.C6 cells, hybridoma cells,
VERO, NIH3T3
cells, COS, WI38, MRCS, A549, HeLa cells, CHO cells, and HT1080 cells.
[0021] In other aspects, the present disclosure provides methods of making an
XDP
comprising a therapeutic payload, the method comprising propagating the
packaging cell of any
of the embodiments under conditions such that XDPs are produced, and
harvesting the XDPs
produced by the packaging cell. The present disclosure further provides an XDP
produced by
the foregoing methods. In a particular embodiment, the XDP comprises a
therapeutic payload of
an RNP of a CasX and guide RNA and, optionally, a donor template of any of the
embodiments
disclosed herein.
[0022] In other aspects, the present disclosure provides methods of modifying
a target nucleic
acid sequence in a cell, the methods comprising contacting the cell with the
XDP comprising an
RNP of any of the embodiments disclosed herein, wherein said contacting
comprises introducing
into the cell the RNP comprising the CasX protein, the guide RNA, and,
optionally, the donor
template nucleic acid sequence, resulting in modification of the target
nucleic acid sequence. In
some cases, the modification comprises introducing one or more single-stranded
breaks in the
target nucleic acid sequence In other cases, the modification comprises
introducing one or more
double-stranded breaks in the target nucleic acid sequence. In still other
cases, the modification
comprises insertion of the donor template into the target nucleic acid
sequence. In one
embodiment, the cell is modified in vitro or ex viva In another embodiment,
the cell is modified
in vivo, In the foregoing embodiment, the XDP is administered to a subject at
a therapeutically
effective dose, wherein the subject is the subject is selected from the group
consisting of mouse,
rat, pig, non-human primate, and human. In some embodiments, the XDP is
administered by a
route of administration selected from the group consisting of subcutaneous,
intradermal,
intraneural, intranodal, intramedullary, intramuscular, intravenous,
intracerebroventricular,
intracistemal, intrathecal, intracranial, intralumbar, intratracheal,
intraosseous, inhalatory,
intracontralateral striatum, intraocular, intravitreal, intralymphatical,
intraperitoneal routes and
7
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
sub-retinal routes. In some embodiments, the therapeutically effective dose is
at least about 1 x
105 particles/kg, or at least about 1 x 106 particles/kg, or at least about 1
x 107 particles/kg, or at
least about 1 x 103 particles/kg, or at least about 1 x 109 particles/kg, or
at least about 1 x 1010
particles/kg, or at least about 1 x 10" particles/kg, or at least about 1 x
1012 particles/kg,, or at
least about 1 x 10" particles/kg, or at least about 1 x 10" particles/kg, or
at least about 1 x 1015
particles/kg, or at least about 1 x 1016 particles/kg. In some embodiments,
the XDP is
administered to the subject according to a treatment regimen comprising one or
more
consecutive doses using a therapeutically effective dose of the XDP. In some
embodiments, the
therapeutically effective dose is administered to the subject as two or more
doses over a period
of at least two weeks, or at least one month, or at least two months, or at
least three months, or at
least four months, or at least five months, or at least six months, or once a
year, or every 2 or 3
years.
[0023] In another aspect, provided herein are XDP particles, and XDP systems,
for use as a
medicament for the treatment of a subject having a disease.
INCORPORATION BY REFERENCE
[0024] All publications, patents, and patent applications mentioned in this
specification are
herein incorporated by reference to the same extent as if each individual
publication, patent, or
patent application was specifically and individually indicated to be
incorporated by reference.
The contents of PCT/U52020/036505, filed on June 5, 2020, and a U.S.
provisional application
entitled "Engineered CasX Systems", filed on December 3, 2020, both
applications which
disclose CasX variants and gNA variants, are hereby incorporated by reference
in their
entireties.
BRIEF DESCRIPTION OF THE DRAWINGS
100251 The novel features of the invention are set forth with particularity in
the appended
claims. A better understanding of the features and advantages of the present
invention will be
obtained by reference to the following detailed description that sets forth
illustrative
embodiments, in which the principles of the invention are utilized, and the
accompanying
drawings of which:
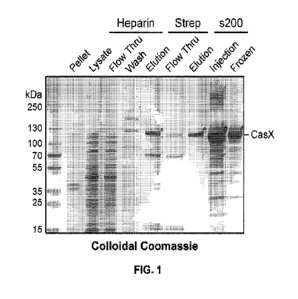
[0026] FIG. 1 shows an SDS-PAGE gel of StX2 purification fractions visualized
by colloidal
Coomassie staining, as described in Example 1.
8
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[0027] FIG. 2 shows the chromatogram from a size exclusion chromatography
assay of the
StX2, using of Superdex 200 16/600 pg Gel Filtration, as described in Example
1.
[0028] FIG. 3 shows an SDS-PAGE gel of StX2 purification fractions visualized
by colloidal
Coomassie staining, as described in Example 1.
[0029] FIG. 4 is a schematic showing the organization of the components in the
p5TX34
plasmid used to assemble the CasX constructs, as described in Example 2.
[0030] FIG. 5 is a schematic showing the steps of generating the CasX 119
variant, as
described in Example 2.
[0031] FIG. 6 shows an SDS-PAGE gel of purification samples, visualized on a
Bio-Rad
Stain-FreeTm gel, as described in Example 2.
[0032] FIG. 7 shows the chromatogram of Superdex 200 16/600 pg Gel Filtration,
as
described in Example 2.
[0033] FIG. 8 shows an SDS-PAGE gel of gel filtration samples, stained with
colloidal
Coomassie, as described in Example 2.
[0034] FIG. 9 shows an SDS-PAGE gel of purification samples of CasX 438,
visualized on a
Bio-Rad StainFreeTM gel, as described in Example 2.
[0035] FIG. 10 shows the chromatogram from a size exclusion chromatography
assay of the
CasX 438, using of Superdex 200 16/600 pg gel filtration, as described in
Example 2.
[0036] FIG. 11 shows an SDS-PAGE gel of CasX 438 purification fractions
visualized by
colloidal Coomassie staining, as described in Example, as described in Example
2.
[0037] FIG. 12 shows an SDS-PAGE gel of purification samples of CasX 457,
visualized on a
Bio-Rad StainFreeTM gel, as described in Example 2.
[0038] FIG. 13 shows the chromatogram from a size exclusion chromatography
assay of the
CasX 457, using of Superdex 200 16/600 pg gel filtration, as described in
Example 2.
[0039] FIG. 14 shows an SDS-PAGE gel of CasX 457 purification fractions
visualized by
colloidal Coomassie staining, as described in Example 2.
[0040] FIG. 15 is a graph of the results of an assay for the quantification of
active fractions of
RNP formed by sgRNA174 and the CasX variants, as described in Example 9,
Equimolar
amounts of RNP and target were co-incubated and the amount of cleaved target
was determined
at the indicated timepoints. Mean and standard deviation of three independent
replicates are
shown for each timepoint. The biphasic fit of the combined replicates is
shown. "2" refers to the
reference CasX protein of SEQ ID Na2,
9
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[0041] FIG. 16 shows the quantification of active fractions of RNP formed by
CasX2
(reference CasX protein of SEQ ID NO:2) and the modified sgRNAs, as described
in Example 9.
Equimolar amounts of RNP and target were co-incubated and the amount of
cleaved target was
determined at the indicated timepoints. Mean and standard deviation of three
independent
replicates are shown for each timepoint. The biphasic fit of the combined
replicates is shown.
[0042] FIG. 17 shows the quantification of active fractions of RNP formed by
CasX 491 and
the modified sgR_NAs under guide-limiting conditions, as described in Example
9. Equimolar
amounts of RNP and target were co-incubated and the amount of cleaved target
was determined
at the indicated timepoints. The biphasic fit of the data is shown.
[0043] FIG. 18 shows the quantification of cleavage rates of RNP formed by
sgRNA174 and
the CasX variants, as described in Example 9. Target DNA was incubated with a
20-fold excess
of the indicated RNP and the amount of cleaved target was determined at the
indicated time
points. Mean and standard deviation of three independent replicates are shown
for each
timepoint, except for 488 and 491 where a single replicate is shown. The
monophasic fit of the
combined replicates is shown.
[0044] FIG. 19 shows the quantification of cleavage rates of RNP formed by
CasX2 and the
sgRNA variants, as described in Example 9. Target DNA was incubated with a 20-
fold excess of
the indicated RNP and the amount of cleaved target was determined at the
indicated time points.
Mean and standard deviation of three independent replicates are shown for each
timepoint. The
monophasic fit of the combined replicates is shown.
[0045] FIG. 20 shows the quantification of initial velocities of RNP formed by
CasX2 and the
sgRNA variants, as described in Example 9. The first two time-points of the
previous cleavage
experiment were fit with a linear model to determine the initial cleavage
velocity.
[0046] FIG. 21 shows the quantification of cleavage rates of RNP formed by
CasX491 and the
sgRNA variants, as described in Example 9. Target DNA was incubated with a 20-
fold excess of
the indicated RNP at 10 C and the amount of cleaved target was determined at
the indicated time
points. The monophasic fit of the timepoints is shown.
[0047] FIGS. 22A-D shows the quantification of cleavage rates of CasX variants
on NTC
PAMs, as described in Example 10. Target DNA substrates with identical spacers
and the
indicated PAM sequence were incubated with a 20-fold excess of the indicated
RNP at 37 C and
the amount of cleaved target was determined at the indicated time points.
Monophasic fit of a
single replicate is shown. FIG. 22A shows the results for sequences having a
TTC PAM. FIG.
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
228 shows the results for sequences having a CTC PAM. FIG. 22C shows the
results for
sequences having a GTC PAM. FIG. 22D shows the results for sequences having a
ATC PAM.
[0048] FIG. 23 depicts the plasmids utilized in the creation of XDP comprising
CasX, gNA_,
and pseudotyping proteins, as described in Example 13.
[0049] FIG. 24 is a schematic of the steps using in the creation of XDP, as
described in
Example 13.
[0050] FIG. 25 is a graph of the results of the editing of the dtTomato assay,
as described in
Example 16.
[0051] FIG. 26A shows the results of percentage editing in mouse tdTomato
neural progenitor
cells (NPCs) with XDPs pseudotyped with serial concentrations of VSV-G, as
described in
Example 17.
[0052] FIG. 26B shows the XDP titers determined by a commercially available
Lenti-X p24
ELISA kit, as described in Example 17.
[0053] FIG. 27 shows the percentage of editing in mouse tdTomato NPCs with
XDPs
pseudotyped with different glycoproteins, as described in Example 17.
[0054] FIG. 28A shows the results of size distributions and viral titer
comparisons of VSV-G
pseudotyped XDP (both lx and 10X concentrated), rabies pseudotyped XDP and
lentivirus
(LV), as described in Example 17.
[0055] FIG. 28B shows the size comparisons between VSV-G XDP, LV and Rabies
XDP, as
described in Example 17.
[0056] FIG. 29 shows the results of percentage editing in mouse tdTomato NPCs
with VSV-G
pseudotyped XDPs carrying different RNPs, as described in Example 18.
[0057] FIG. 30 shows the percentage editing in mouse tdTomato NPCs with VSV-G
pseudotyped XDPs with titrated amounts of Gag-Pol vs Gag-Stx (Stx construct),
as described in
Example 19.
[0058] FIG. 31 shows the titers for these different XDPs with varying amounts
of Gag-Pol vs
Gag-Stx constructs, as described in Example 19.
[0059] FIG. 32 shows the amount of guide RNA per XDP titer for different
constructs as
assessed by QPCR, as described in Example 19.
[0060] FIG. 33 shows the results of the ref alive knockout rates of B2M by
XDPs containing
two different B2M targeting spacers and one non targeting spacer, as described
in Example 20.
11
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[0061] FIG. 34 shows representative SDS-PAGE and Western blot images of
samples taken
from throughout the centrifugation purification process for XDP particles, as
described in
Example 14.
[0062] FIG. 35 shows the results of an editing assay for XDP configured as
version 7, version
122 and version 123, as described in Example 21.
[0063] FIG. 36A shows the schematic for the configuration of the components
for version 1
XDP and the four plasmids used in the transfection to create the XDP.
[0064] FIG. 36B shows the schematic for the configuration of the components
for version 2
XDP and the four plasmids used in the transfection to create the XDP.
[0065] FIG. 37A shows the schematic for the configuration of the components
for version 3
XDP and the four plasmids used in the transfection to create the XDP.
[0066] FIG. 37B shows the schematic for the configuration of the components
for version 4
XDP and the three plasmids used in the transfection to create the XDP.
[0067] FIG. 38A shows the schematic for the configuration of the components
for version 5
XDP and the three plasmids used in the transfection to create the XDP.
[0068] FIG. 38B shows the schematic for the configuration of the components
for version 6
XDP and the four plasmids used in the transfection to create the XDP.
[0069] FIG. 39A shows the schematic for the configuration of the components
for version 7
XDP and the three plasmids used in the transfection to create the XDP.
[0070] FIG. 39B shows the schematic for the configuration of the components
for version 8
XDP and the four plasmids used in the transfection to create the XDP.
[0071] FIG. 40A shows the schematic for the configuration of the components
for version 9
XDP and the three plasmids used in the transfection to create the XDP.
[0072] FIG. 40B shows the schematic for the configuration of the components
for version 10
XDP and the three plasmids used in the transfection to create the XDP.
[0073] FIG. 41A shows the schematic for the configuration of the components
for version 11
XDP and the three plasmids used in the transfection to create the XDP.
[0074] FIG. 41B shows the schematic for the configuration of the components
for version 12
XDP and the three plasmids used in the transfection to create the XDP.
[0075] FIG. 42A shows the schematic for the configuration of the components
for version 13
XDP and the three plasmids used in the transfection to create the XDP.
12
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[0076] FIG. 42B shows the schematic for the configuration of the components
for version 14
XDP and the three plasmids used in the transfection to create the XDP.
[0077] FIG 43A shows the schematic for the configuration of the components for
version 15
XDP and the three plasmids used in the transfection to create the XDP.
[0078] FIG. 43B shows the schematic for the configuration of the components
for version 16
XDP and the three plasmids used in the transfection to create the XDP.
[0079] FIG. 44A shows the schematic for the configuration of the components
for version 24
XDP and the four plasmids used in the transfection to create the XDP.
[0080] FIG. 44B shows the schematic for the configuration of the components
for version 25
XDP and the four plasmids used in the transfection to create the XDP.
[0081] FIG. 45A shows the schematic for the configuration of the components
for version 26
XDP and the four plasmids used in the transfection to create the XDP.
[0082] FIG. 45B shows the schematic for the configuration of the components
for version 27
XDP and the four plasmids used in the transfection to create the XDP.
[0083] FIG. 46A shows the schematic for the configuration of the components
for version 31
XDP and the four plasmids used in the transfection to create the XDP.
[0084] FIG. 46B shows the schematic for the configuration of the components
for version 32
XDP and the four plasmids used in the transfection to create the XDP.
[0085] FIG. 47A shows the schematic for the configuration of the components
for version 33
XDP and the four plasmids used in the transfection to create the XDP.
[0086] FIG. 47B shows the schematic for the configuration of the components
for version 34
XDP and the four plasmids used in the transfection to create the XDP
[0087] FIG. 48A shows the schematic for the configuration of the components
for version 35
XDP and the four plasmids used in the transfection to create the XDP.
[0088] FIG. 48B shows the schematic for the configuration of the components
for version 36
XDP and the four plasmids used in the transfection to create the XDP.
[0089] FIG. 49A shows the schematic for the configuration of the components
for version 37
XDP and the four plasmids used in the transfection to create the XDP.
[0090] FIG. 49B shows the schematic for the configuration of the components
for version 38
XDP and the four plasmids used in the transfection to create the XDP.
[0091] FIG. 50A shows the schematic for the configuration of the components
for version 39
XDP and the four plasmids used in the transfection to create the XDP.
13
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
100921 FIG. 50B shows the schematic for the configuration of the components
for version 40
XDP and the four plasmids used in the transfection to create the XDP.
[0093] FIG. 51A shows the schematic for the configuration of the components
for version 17
XDP and the three plasmids used in the transfection to create the XDP.
[0094] FIG. 51B shows the schematic for the configuration of the components
for version 18
XDP and the three plasmids used in the transfection to create the XDP.
[0095] FIG. 52A shows the schematic for the configuration of the components
for versions 44
and 45 XDP and the three plasmids used in the transfection to create the XDP.
[0096] FIG. 52B shows the schematic for the configuration of the components
for versions 46,
47, 62, and 90 XDP and the three plasmids used in the transfection to create
the XDP.
[0097] FIG. 53A shows the schematic for the configuration of the components
for versions 48,
49, and 63 XDP and the three plasmids used in the transfection to create the
XDP.
[0098] FIG. 53B shows the schematic for the configuration of the components
for version 50
XDP and the three plasmids used in the transfection to create the XDP.
[0099] FIG. 54A shows the schematic for the configuration of the components
for versions 51
and 52 XDP and the three plasmids used in the transfection to create the XDP.
[00100] FIG. 54B shows the schematic for the configuration of the components
for versions 53,
54, 55 and 91 XDP and the three plasmids used in the transfection to create
the XDP.
[00101] FIG. 55A shows the schematic for the configuration of the components
for versions 56-
61 and 92 XDP and the three plasmids used in the transfection to create the
XDP.
[00102] FIG. 55B shows the schematic for the configuration of the components
for versions
66a and 67a XDP and the three plasmids used in the transfection to create the
XDP.
[00103] FIG. 56A shows the schematic for the configuration of the components
for versions
66b and 67b XDP and the four plasmids used in the transfection to create the
)(DP_
[00104] FIG. 56B shows the schematic for the configuration of the components
for versions
68a, 69a, 70a and 87a XDP and the three plasmids used in the transfection to
create the XDP.
[00105] FIG. 57A shows the schematic for the configuration of the components
for versions
68b, 6%, 70b and 87b XDP and the four plasmids used in the transfection to
create the XDP,
[00106] FIG. 57B shows the schematic for the configuration of the components
for versions
71a, 72a and 88a XDP and the three plasmids used in the transfection to create
the XDP.
[00107] FIG. 58A shows the schematic for the configuration of the components
for versions
71b, 72b and 88b XDP and the four plasmids used in the transfection to create
the XDP.
14
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1001081 FIG. 58B shows the schematic for the configuration of the components
for versions
73a XDP and the three plasmids used in the transfection to create the XDP.
[00109] FIG. 59A shows the schematic for the configuration of the components
for version 736
XDP and the four plasmids used in the transfection to create the XDP.
[00110] FIG. 59B shows the schematic for the configuration of the components
for versions
74a and 75a XDP and the three plasmids used in the transfection to create the
XDP.
[00111] FIG. 60A shows the schematic for the configuration of the components
for versions
74b and 751, XDP and the four plasmids used in the transfection to create the
XDP.
[00112] FIG. 60B shows the schematic for the configuration of the components
for versions
76a, 77a, 78a, and 79a XDP and the three plasmids used in the transfection to
create the XDP.
[00113] FIG. 61A shows the schematic for the configuration of the components
for versions
76b, 77b, 78b, and 79b XDP and the four plasmids used in the transfection to
create the XDP.
1001141 FIG. 61B shows the schematic for the configuration of the components
for versions
80a, 81a, 82a, 83a, 84a, 85a and 86a XDP and the three plasmids used in the
transfection to
create the XDP.
[00115] FIG. 62A shows the schematic for the configuration of the components
for versions
80b, 81b, 82b, 83b, 84b, 85b, and 86b XDP and the four plasmids used in the
transfection to
create the XDP.
[00116] FIG. 62B shows the schematic for the configuration of the components
for versions
102 and 114 XDP and the three plasmids used in the transfection to create the
XDP.
[00117] FIG. 63A shows the schematic for the configuration of the components
for versions
103, 108, and 109 XDP and the three plasmids used in the transfection to
create the XDP.
[00118] FIG. 63B shows the schematic for the configuration of the components
for versions
104, 105, 115, 116 and 117 XDP and the three plasmids used in the transfection
to create the
XDP.
[00119] FIG. 64A shows the schematic for the configuration of the components
for versions
106, 111, 112, 83b and 113 XDP and the three plasmids used in the transfection
to create the
XDP.
[00120] FIG. 64B shows the schematic for the configuration of the components
for versions
107 and 110 XDP and the three plasmids used in the transfection to create the
XDP.
[00121] FIG. 65 shows the schematic for the configuration of the components
for version 118
XDP and the three plasmids used in the transfection to create the XDP.
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00122] FIG. 66A shows the schematic for the configuration of the components
for version 122
XDP and the three plasmids used in the transfection to create the XDP.
[00123] FIG. 66B shows the schematic for the configuration of the components
for version 103
XDP and the three plasmids used in the transfection to create the XDP.
[00124] FIG. 67A shows the schematic for the configuration of the components
for version 124
XDP and the three plasmids used in the transfection to create the XDP.
[00125] FIG. 67B shows the schematic for the configuration of the components
for version 126
XDP and the three plasmids used in the transfection to create the XDP.
[00126] FIG. 68 shows the schematic for the configuration of the components
for versions 128
XDP and the three plasmids used in the transfection to create the XDP.
[00127] FIGS. 69A and 698 show the results of editing assays of the various
XDP versions, as
described in Example 22.
[00128] FIG. 70 shows the results of editing assays of the various XDP
versions, as described
in Example 22.
[00129] FIGS. 71A and 71B shows the results of editing assays of the various
XDP versions, as
described in Example 23.
[00130] FIG. 72 shows the results of editing assays of the various XDP
versions, as described
in Example 23.
[00131] FIGS. 73A and 73B shows the results of editing assays of the various
XDP versions, as
described in Example 23.
[00132] FIG. 74 shows the results of editing assays of the various XDP
versions, as described
in Example 23.
[00133] FIGS. 75A and 75B shows the results of editing assays of the various
XDP versions, as
described in Example 25.
[00134] FIG. 76 shows the results of editing assays of the various XDP
versions, as described
in Example 25.
[00135] FIG. 77 shows the results of editing assays of the various XDP
versions, as described
in Example 26.
[00136] FIG. 78 shows the results of editing assays of the various XDP
versions, as described
in Example 26.
16
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DETAILED DESCRIPTION
[00137] While preferred embodiments of the present invention have been shown
and described
herein, it will be obvious to those skilled in the art that such embodiments
are provided by way
of example only. Numerous variations, changes, and substitutions will now
occur to those
skilled in the art without departing from the invention. It should be
understood that various
alternatives to the embodiments of the invention described herein may be
employed in practicing
the invention. It is intended that the following claims define the scope of
the invention and that
methods and structures within the scope of these claims and their equivalents
be covered
thereby.
[00138] Unless otherwise defined, all technical and scientific terms used
herein have the same
meaning as commonly understood by one of ordinary skill in the art to which
this invention
belongs. Although methods and materials similar or equivalent to those
described herein can be
used in the practice or testing of the present embodiments, suitable methods
and materials are
described below. In case of conflict, the patent specification, including
definitions, will control.
In addition, the materials, methods, and examples are illustrative only and
not intended to be
limiting. Numerous variations, changes, and substitutions will now occur to
those skilled in the
art without departing from the invention.
DEFINITIONS
[00139] The terms "polynucleotide" and "nucleic acid," used interchangeably
herein, refer to a
polymeric form of nucleotides of any length, either ribonucleotides or
deoxyribonucleotides.
Thus, terms "polynucleotide" and "nucleic acid" encompass single-stranded DNA;
double-
stranded DNA; multi-stranded DNA; single-stranded RNA; double-stranded RNA;
multi-
stranded RNA; genomic DNA; cDNA; DNA-RNA hybrids; and a polymer comprising
purine
and pyrimidine bases or other natural, chemically or biochemically modified,
non-natural, or
derivatized nucleotide bases.
[00140] "Hybridizable" or "complementary" are used interchangeably to mean
that a nucleic
acid (e.g., RNA, DNA) comprises a sequence of nucleotides that enables it to
non-c,ovalently
bind, i.e., form Watson-Crick base pairs and/or G/U base pairs, "anneal", or
"hybridize," to
another nucleic acid in a sequence-specific, antiparallel, manner (i.e., a
nucleic acid specifically
binds to a complementary nucleic acid) under the appropriate in vitro and/or
in vivo conditions
of temperature and solution ionic strength. It is understood that the sequence
of a polynucleotide
need not be 100% complementary to that of its target nucleic acid to be
specifically
17
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
hybridizable; it can have at least about 70%, at least about 80%, or at least
about 90%, or at least
about 95% sequence identity and still hybridize to the target nucleic acid.
Moreover, a
polynucleotide may hybridize over one or more segments such that intervening
or adjacent
segments are not involved in the hybridization event (e.g., a loop structure
or hairpin structure, a
'bulge', 'bubble' and the like).
[00141] A "gene," for the purposes of the present disclosure, includes a DNA
region encoding a
gene product (e.g., a protein, RNA), as well as all DNA regions which regulate
the production of
the gene product, whether or not such regulatory sequences are adjacent to
coding and/or
transcribed sequences. Accordingly, a gene may include regulatory element
sequences including,
but not necessarily limited to, promoter sequences, terminators, translational
regulatory
sequences such as ribosome binding sites and internal ribosome entry sites,
enhancers, silencers,
insulators, boundary elements, replication origins, matrix attachment sites
and locus control
regions. Coding sequences encode a gene product upon transcription or
transcription and
translation; the coding sequences of the disclosure may comprise fragments and
need not contain
a full-length open reading frame. A gene can include both the strand that is
transcribed as well
as the complementary strand containing the anticodons.
[00142] The term "downstream" refers to a nucleotide sequence that is located
3' to a reference
nucleotide sequence. In certain embodiments, downstream nucleotide sequences
relate to
sequences that follow the starting point of transcription. For example, the
translation initiation
codon of a gene is located downstream of the start site of transcription.
[00143] The term "upstream" refers to a nucleotide sequence that is located 5'
to a reference
nucleotide sequence In certain embodiments, upstream nucleotide sequences
relate to
sequences that are located on the 5' side of a coding region or starting point
of transcription. For
example, most promoters are located upstream of the start site of
transcription.
[00144] The term "regulatory element" is used interchangeably herein with the
term "regulatory
sequence," and is intended to include promoters, enhancers, and other
expression regulatory
elements (e.g. transcription termination signals, such as polyadenylation
signals and poly-U
sequences). Exemplary regulatory elements include a transcription promoter
such as, but not
limited to, CMV, CMV-Fintron A, SV40, RSV, HIV-Ltr, elongation factor 1 alpha
(EF la),
MMLV-ltr, internal ribosome entry site (lRES) or P2A peptide to permit
translation of multiple
genes from a single transcript, metallothionein, a transcription enhancer
element, a transcription
termination signal, polyadenylation sequences, sequences for optimization of
initiation of
18
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
translation, and translation termination sequences. In the case of systems
utilized for exon
skipping, regulatory elements include exonic splicing enhancers. It will be
understood that the
choice of the appropriate regulatory element will depend on the encoded
component to be
expressed (e.g., protein or RNA) or whether the nucleic acid comprises
multiple components that
require different polymerases or are not intended to be expressed as a fusion
protein.
[00145] The term "promoter" refers to a DNA sequence that contains an RNA
polymerase
binding site, transcription start site, TATA box, and/or B recognition element
and assists or
promotes the transcription and expression of an associated transcribable
polynucleotide sequence
and/or gene (or transgene). A promoter can be synthetically produced or can be
derived from a
known or naturally occurring promoter sequence or another promoter sequence. A
promoter can
be proximal or distal to the gene to be transcribed. A promoter can also
include a chimeric
promoter comprising a combination of two or more heterologous sequences to
confer certain
properties. A promoter of the present disclosure can include variants of
promoter sequences that
are similar in composition, but not identical to, other promoter sequence(s)
known or provided
herein. A promoter can be classified according to criteria relating to the
pattern of expression of
an associated coding or transcribable sequence or gene operably linked to the
promoter, such as
constitutive, developmental, tissue-specific, inducible, etc.
[00146] The term "enhancer" refers to regulatory DNA sequences that, when
bound by specific
proteins Sled transcription factors, regulate the expression of an associated
gene. Enhancers
may be located in the intron of the gene, or 5' or 3' of the coding sequence
of the gene.
Enhancers may be proximal to the gene (i.e., within a few tens or hundreds of
base pairs (bp) of
the promoter), or may be located distal to the gene (i.e., thousands of bp,
hundreds of thousands
of bp, or even millions of bp away from the promoter). A single gene may be
regulated by more
than one enhancer, all of which are envisaged as within the scope of the
instant disclosure.
[00147] "Recombinant," as used herein, means that a particular nucleic acid
(DNA or RNA) is
the product of various combinations of cloning, restriction, and/or ligation
steps resulting in a
construct having a structural coding or non-coding sequence distinguishable
from endogenous
nucleic acids found in natural systems. Generally, DNA sequences encoding the
structural
coding sequence can be assembled from cDNA fragments and short oligonucleotide
linkers, or
from a series of synthetic oligonucleotides, to provide a synthetic nucleic
acid which is capable
of being expressed from a recombinant transcriptional unit contained in a cell
or in a cell-free
transcription and translation system Such sequences can be provided in the
form of an open
19
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
reading frame uninterrupted by internal non-translated sequences, or introns,
which are typically
present in eukaryotic genes. Genomic DNA comprising the relevant sequences can
also be used
in the formation of a recombinant gene or transcriptional unit. Sequences of
non-translated DNA
may be present 5' or 3' from the open reading frame, where such sequences do
not interfere with
manipulation or expression of the coding regions, and may indeed act to
modulate production of
a desired product by various mechanisms (see "enhancers" and "promoters",
above).
[00148] The term "recombinant polynucleotide" or "recombinant nucleic acid"
refers to one
which is not naturally occurring, e.g., is made by the artificial combination
of two otherwise
separated segments of sequence through human intervention. This artificial
combination is often
accomplished by either chemical synthesis means, or by the artificial
manipulation of isolated
segments of nucleic acids, e.g., by genetic engineering techniques. Such is
usually done to
replace a codon with a redundant codon encoding the same or a conservative
amino acid, while
typically introducing or removing a sequence recognition site. Alternatively,
it is performed to
join together nucleic acid segments of desired functions to generate a desired
combination of
functions. This artificial combination is often accomplished by either
chemical synthesis means,
or by the artificial manipulation of isolated segments of nucleic acids, e.g.,
by genetic
engineering techniques.
[00149] Similarly, the term "recombinant polypeptide" or "recombinant protein"
refers to a
polypeptide or protein which is not naturally occurring, e.g., is made by the
artificial
combination of two otherwise separated segments of amino sequence through
human
intervention. Thus, e.g., a protein that comprises a heterologous amino acid
sequence is
recombinant.
[00150] As used herein, the term "contacting" means establishing a physical
connection
between two or more entities. For example, contacting a target nucleic acid
with a guide nucleic
acid means that the target nucleic acid and the guide nucleic acid are made to
share a physical
connection; e.g., can hybridize if the sequences share sequence similarity.
[00151] "Dissociation constant", or "Kd", are used interchangeably and mean
the affinity
between a ligand "L" and a protein "P"; i.e., how tightly a ligand binds to a
particular protein. It
can be calculated using the formula Ku=[L] [P]/[LP], where [P], [L] and [LP]
represent molar
concentrations of the protein, ligand and complex, respectively.
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1001521 The disclosure provides compositions and methods useful for modifying
a target
nucleic acid. As used herein "modifying" includes but is not limited to
cleaving, nicking,
editing, deleting, knocking in, knocking out, and the like.
[00153] The term "knock-out" refers to the elimination of a gene or the
expression of a gene.
For example, a gene can be knocked out by either a deletion or an addition of
a nucleotide
sequence that leads to a disruption of the reading frame. As another example,
a gene may be
knocked out by replacing a part of the gene with an irrelevant sequence. The
term "knock-down"
as used herein refers to reduction in the expression of a gene or its gene
product(s). As a result of
a gene knock-down, the protein activity or function may be attenuated or the
protein levels may
be reduced or eliminated.
[00154] As used herein, "homology-directed repair" (HDR) refers to the form of
DNA repair
that takes place during repair of double-strand breaks in cells. This process
requires nucleotide
sequence homology, and uses a donor template to repair or knock-out a target
DNA, and leads to
the transfer of genetic information from the donor to the target. Homology-
directed repair can
result in an alteration of the sequence of the target sequence by insertion,
deletion, or mutation if
the donor template differs from the target DNA sequence and part or all of the
sequence of the
donor template is incorporated into the target DNA.
[00155] As used herein, "non-homologous end joining" (NHEJ) refers to the
repair of double-
strand breaks in DNA by direct ligation of the break ends to one another
without the need for a
homologous template (in contrast to homology-directed repair, which requires a
homologous
sequence to guide repair). NHEJ often results in the loss (deletion) of
nucleotide sequence near
the site of the double- strand break.
[00156] As used herein "micro-homology mediated end joining" (MMEJ) refers to
a mutagenic
DSB repair mechanism, which always associates with deletions flanking the
break sites without
the need for a homologous template (in contrast to homology-directed repair,
which requires a
homologous sequence to guide repair). MMEJ often results in the loss
(deletion) of nucleotide
sequence near the site of the double- strand break. A polynucleotide or
polypeptide has a certain
percent "sequence similarity" or "sequence identity" to another polynucleotide
or polypeptide,
meaning that, when aligned, that percentage of bases or amino acids are the
same, and in the
same relative position, when comparing the two sequences. Sequence similarity
(sometimes
referred to as percent similarity, percent identity, or homology) can be
determined in a number
of different manners. To determine sequence similarity, sequences can be
aligned using the
21
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
methods and computer programs that are known in the art, including BLAST,
available over the
world wide web at ncbi.nlm.nih.gov/BLAST. Percent complementarity between
particular
stretches of nucleic acid sequences within nucleic acids can be determined
using any convenient
method. Example methods include BLAST programs (basic local alignment search
tools) and
PowerBLAST programs (Altschul et al., J. Mol. Biol., 1990, 215, 403-410; Zhang
and Madden,
Genome Res., 1997, 7, 649-656) or by using the Gap program (Wisconsin Sequence
Analysis
Package, Version 8 for Unix, Genetics Computer Group, University Research
Park, Madison
Wis.), e.g., using default settings, which uses the algorithm of Smith and
Waterman (Adv. Appl.
Math., 1981, 2, 482-489).
1001571 The terms "polypeptide," and "protein" are used interchangeably
herein, and refer to a
polymeric form of amino acids of any length, which can include coded and non-
coded amino
acids, chemically or biochemically modified or derivatized amino acids, and
polypeptides
having modified peptide backbones. The term includes fusion proteins,
including, but not limited
to, fusion proteins with a heterologous amino acid sequence.
[00158] A "vector" or "expression vector" is a replicon, such as plasmid,
phage, virus, or
cosmid, to which another DNA segment, i.e., an "insert", may be attached so as
to bring about
the replication or expression of the attached segment in a cell.
[00159] The term "naturally-occurring" or "unmodified" or "wild type" as used
herein as
applied to a nucleic acid, a polypeptide, a cell, or an organism, refers to a
nucleic acid,
polypeptide, cell, or organism that is found in nature.
[00160] As used herein, a "mutation" refers to an insertion, deletion,
substitution, duplication,
or inversion of one or more amino acids or nucleotides as compared to a wild-
type or reference
amino acid sequence or to a wild-type or reference nucleotide sequence.
[00161] As used herein the term "isolated" is meant to describe a
polynucleotide, a polypeptide,
or a cell that is in an environment different from that in which the
polynucleotide, the
polypeptide, or the cell naturally occurs. An isolated genetically modified
host cell may be
present in a mixed population of genetically modified host cells.
[00162] A "host cell," as used herein, denotes a eukaryotic cell, a
prokaryotic cell, or a cell
from a multicellular organism (e.g., a cell line) cultured as a unicellular
entity, which eukaryotic
or prokaryotic cells are used as recipients for a nucleic acid (e.g., an
expression vector), and
include the progeny of the original cell which has been genetically modified
by the nucleic acid.
It is understood that the progeny of a single cell may not necessarily be
completely identical in
22
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
morphology or in genomic or total DNA complement as the original parent, due
to natural,
accidental, or deliberate mutation. A "recombinant host cell" (also referred
to as a "genetically
modified host cell") is a host cell into which has been introduced a
heterologous nucleic acid,
e.g., an expression vector.
1001631 The term "tropism" as used herein refers to preferential entry of the
XDP into certain
cell or tissue type(s) and/or preferential interaction with the cell surface
that facilitates entry into
certain cell or tissue types, optionally and preferably followed by expression
(e.g., transcription
and, optionally, translation) of sequences carried by the XDP into the cell.
1001641 The terms "pseudotype" or "pseudotyping" as used herein, refers to
viral envelope
proteins that have been substituted with those of another virus possessing
preferable
characteristics. For example, HIV can be pseudotyped with vesicular stomatitis
virus G-protein
(VSV-G) envelope proteins (amongst others, described herein, below), which
allows HIV to
infect a wider range of cells because HIV envelope proteins target the virus
mainly to CD4+
presenting cells.
[00165] The term "tropism factor" as used herein refers to components
integrated into the
surface of an XDP that provides tropism for a certain cell or tissue type. Non-
limiting examples
of tropism factors include glycoproteins, antibody fragments (e.g., scFv,
nanobodies, linear
antibodies, etc.), receptors and ligands to target cell markers.
[00166] A "target cell marker" refers to a molecule expressed by a target cell
including but not
limited to cell-surface receptors, cytokine receptors, antigens, tumor-
associated antigens,
glycoproteins, oligonucleotides, enzymatic substrates, antigenic determinants,
or binding sites
that may be present in the on the surface of a target tissue or cell that may
serve as ligands for a
tropism factor.
[00167] An "antibody fragment" refers to a molecule other than an intact
antibody that
comprises a portion of an intact antibody and that binds the antigen to which
the intact antibody
binds. Examples of antibody fragments include but are not limited to Fv, Fab,
Fab', Fab'-SH,
F(abs)2, diabodies, single chain diabodies, linear antibodies, a single domain
antibody, a single
domain camelid antibody, single-chain variable fragment (scFv) antibody
molecules, and
multispecific antibodies formed from antibody fragments.
[00168] The term "conservative amino acid substitution" refers to the
interchangeability in
proteins of amino acid residues having similar side chains. For example, a
group of amino acids
having aliphatic side chains consists of glycine, alanine, valine, leucine,
and isoleucine; a group
23
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
of amino acids having aliphatic-hydroxyl side chains consists of serine and
threonine; a group of
amino acids having amide-containing side chains consists of asparagine and
glutamine; a group
of amino acids having aromatic side chains consists of phenylalanine,
tyrosine, and tryptophan; a
group of amino acids having basic side chains consists of lysine, arginine,
and histidine, and a
group of amino acids having sulfur-containing side chains consists of cysteine
and methionine.
Exemplary conservative amino acid substitution groups are: valine-leucine-
isoleucine,
phenylalanine-tyrosine, lysine-arginine, alanine-valine, and asparagine-
glutamine.
[00169] As used herein, "treatment" or "treating," are used interchangeably
herein and refer to
an approach for obtaining beneficial or desired results, including but not
limited to a therapeutic
benefit and/or a prophylactic benefit. By therapeutic benefit is meant
eradication or amelioration
of the underlying disorder or disease being treated. A therapeutic benefit can
also be achieved
with the eradication or amelioration of one or more of the symptoms or an
improvement in one
or more clinical parameters associated with the underlying disease such that
an improvement is
observed in the subject, notwithstanding that the subject may still be
afflicted with the
underlying disorder.
[00170] The terms "therapeutically effective amount" and "therapeutically
effective dose", as
used herein, refer to an amount of a drug or a biologic, alone or as a part of
a composition, that is
capable of having any detectable, beneficial effect on any symptom, aspect,
measured parameter
or characteristics of a disease state or condition when administered in one or
repeated doses to a
subject such as a human or an experimental animal. Such effect need not be
absolute to be
beneficial.
[00171] As used herein, "administering" means a method of giving a dosage of a
compound
(e.g., a composition of the disclosure) or a composition (e.g., a
pharmaceutical composition) to a
subject.
[00172] A "subject" is a mammal. Mammals include, but are not limited to,
domesticated
animals, non-human primates, humans, dogs, rabbits, mice, rats and other
rodents.
I. General Methods
1001731 The practice of the present invention employs, unless otherwise
indicated, conventional
techniques of immunology, biochemistry, chemistry, molecular biology,
microbiology, cell
biology, genomics and recombinant DNA, which can be found in such standard
textbooks as
Molecular Cloning: A Laboratory Manual, 3rd Ed (Sambrook et al., Cold Spring
Harbor
24
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Laboratory Press 2001); Short Protocols in Molecular Biology, 4th Ed. (Ausubel
et al. eds., John
Wiley & Sons 1999); Protein Methods (Bollag et al., John Wiley & Sons 1996);
Nonviral
Vectors for Gene Therapy (Wagner et al_ eds., Academic Press 1999); Viral
Vectors (Kaplift &
Loewy eds., Academic Press 1995); Immunology Methods Manual (I. Lefkovits ed.,
Academic
Press 1997); and Cell and Tissue Culture: Laboratory Procedures in
Biotechnology (Doyle &
Griffiths, John Wiley & Sons 1998), the disclosures of which are incorporated
herein by
reference.
[00174] Where a range of values is provided, it is understood that endpoints
are included and
that each intervening value, to the tenth of the unit of the lower limit
unless the context clearly
dictates otherwise, between the upper and lower limit of that range and any
other stated or
intervening value in that stated range, is encompassed. The upper and lower
limits of these
smaller ranges may independently be included in the smaller ranges, and are
also encompassed,
subject to any specifically excluded limit in the stated range. Where the
stated range includes
one or both of the limits, ranges excluding either or both of those included
limits are also
included.
[00175] Unless defined otherwise, all technical and scientific terms used
herein have the same
meaning as commonly understood by one of ordinary skill in the art to which
this invention
belongs. All publications mentioned herein are incorporated herein by
reference to disclose and
describe the methods and/or materials in connection with which the
publications are cited.
[00176] It must be noted that as used herein and in the appended claims, the
singular forms "a,"
"an," and "the" include plural referents unless the context clearly dictates
otherwise.
[00177] It will be appreciated that certain features of the disclosure, which
are, for clarity,
described in the context of separate embodiments, may also be provided in
combination in a
single embodiment. In other cases, various features of the disclosure, which
are, for brevity,
described in the context of a single embodiment, may also be provided
separately or in any
suitable sub-combination. It is intended that all combinations of the
embodiments pertaining to
the disclosure are specifically embraced by the present disclosure and are
disclosed herein just as
if each and every combination was individually and explicitly disclosed. In
addition, all sub-
combinations of the various embodiments and elements thereof are also
specifically embraced
by the present disclosure and are disclosed herein just as if each and every
such sub-combination
was individually and explicitly disclosed herein.
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Particle Delivery Systems for Use in Targeting Cells
[00178] In a first aspect, the present disclosure relates to particle delivery
systems (XDP)
designed to self-assemble particles comprising therapeutic payloads wherein
the particles are
designed for selective delivery to targeted cells. As used herein, the term
"XDP" refers to a non-
replicating, self-assembling, non-naturally occurring multicomponent structure
composed of one
or more viral proteins, polyproteins, virally-derived peptides or
polypeptides, such as, but not
limited to, capsid, coat, shell, as well as tropism factors such as envelope
glycoproteins derived
from viruses, antibody fragments, receptors or ligand utilized for tropism to
direct the XDP to
target cells or tissues, with a lipid layer (derived from the host cell),
wherein the XDP are
capable of self-assembly in a host cell and encapsidating or encompassing a
therapeutic payload.
The XDP of present disclosure can be utilized to specifically and selectively
deliver therapeutic
payloads to target cells or tissues. The XDP of the disclosure have utility in
a variety of methods,
including, but not limited to, use in delivering a therapeutic in a selective
fashion to a target cell
or organ for the treatment of a disease.
[00179] In some embodiments, the present disclosure provides XDP systems
comprising one or
more nucleic acids comprising sequences encoding the components of the XDP,
the therapeutic
payload, and tropism factors that, that, when introduced into an appropriate
eukaryotic host cell,
result in the expression of the individual XDP structural components,
processing proteins,
therapeutic payloads, and tropism factors that self-assemble into XDP
particles that encapsidate
the therapeutic payload, and that can be collected and purified for the
methods and uses
described herein.
[00180] In some embodiments, the therapeutic payloads packaged within the XDP
comprise
therapeutic proteins, described more frilly below. In other embodiments, the
therapeutic payloads
packaged within XDP comprise therapeutic nucleic acids or nucleic acids that
encode
therapeutic proteins. In still other embodiments, the XDP comprise therapeutic
proteins and
nucleic acids. In some cases, the therapeutic payloads include gene editing
systems such as
CRISPR nucleases and guide RNA or zinc finger proteins useful for the editing
of nucleic acids
in target cells. In some embodiments, the therapeutic payloads include Class 2
CRISPR-Cas
systems. Class 2 systems are distinguished from Class 1 systems in that they
have a single multi-
domain effector protein and are further divided into a Type II, Type V. or
Type VI system,
described in Makarova, et al. Evolutionary classification of CRISPR-Cas
systems: a burst of
class 2 and derived variants Nature Rev Microbiol. 18:67 (2020), incorporated
herein by
26
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
reference. In some embodiments, the nucleases include Class 2, Type 11
CRISPR/Cas effector
polypeptides such as Cas9. In other cases, the nucleases include Class 2, Type
V CRISPR/Cas
effector polypeptides such as a Cas12a, Cas12b, Cas12c, Cas12d (CasY), Cas12J,
and CasX
wherein the CRISPR nuclease and guide system can do one or more of the
following: (i) modify
(e.g., edit) a target ssDNA, dsDNA or RNA (e.g., cleave, nick, or methylate);
(ii) modulate
transcription of the target nucleic acid; (iii) bind the target nucleic acid
(e.g., for purposes of
isolation, blocking transcription, labeling, or imaging, etc.); or (v) modify
a polypeptide
associated with a target nucleic acid. In a particular embodiment, the present
disclosure provides
XDP compositions, and methods to make the XDP compositions, designed to
package
ribonucleic acid particles (RNP) comprising CasX and guide RNA systems
(CasX:gNA system)
useful for the editing of nucleic acids in target cells, described more fully,
below. Accordingly,
the present disclosure provides XDP compositions, nucleic acids that encode
the components of
the XDP (both structural as well as gene-editing components), as well as
methods of making and
using the XDP. The nucleic acids, the components of the compositions, and the
methods of
making and using them, are described herein, below.
a. XDP Components
[00181] XDP can be created in multiple forms and configurations (see, e.g.,
FIGS. 36-68)
utilizing components derived from various sources and in different
combinations.
[00182] The structural components of the XDP of the present disclosure are
derived from
members of the Retroviridae family of viruses, described more fully, below.
The major
structural component of retroviruses is the polyprotein Gag, which also
typically contain
protease cleavage sites that, upon action by the viral protease, processes the
Gag into
subcomponents that, in the case of the replication of the source virus, then
self-assemble in the
host cell to make the core inner shell of the virus. The expression of Gag
alone is sufficient to
mediate the assembly and release of virus-like particles (VLPs) from host
cells. Gag proteins
from all retroviruses contain an N-terminal membrane-binding matrix (MA)
domain, a capsid
(CA) domain (with two subdomthns), and a nucleocapsid (NC) domain that are
structurally
similar across retroviral genera but differ greatly in sequence. Outside these
core domains, Gag
proteins vary among retroviruses, and other linkers and domains may be present
(Shur, F., et al.
The Structure of Immature Virus-Like Rous Sarcoma Virus Gag Particles Reveals
a Structural
Role for the p10 Domain in Assembly. J Virol. 89(20):10294 (2015)). The
assembly pathway of
Gag into immature particles in the host cell is mediated by interactions
between MA (which is
27
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
responsible for targeting Gag polyprotein to the plasma membrane), between NC
and RNA, and
between CA domains (which, in the context of the present disclosure, assemble
into the XDP
capsid). For most retrovirus genera, assembly takes place on the plasma
membrane, but for
betaretroviruses the particles are assembled in the cytoplasm and then
transported to the plasma
membrane. In the context of the retroviruses, concomitant with, or shortly
after, particle release,
cleavage of Gag by the viral protease (PR) gives rise to separate MA, CA, and
NC proteins,
inducing a rearrangement of the internal viral structure, with CA forming the
shell of the mature
viral core. Full proteolytic cleavage of Gag into its individual domains is
necessary for virus
infectivity for the native viruses. However, it has been discovered that for
self-assembly of XDP
within a host cell comprising retroviral components that are then capable of
being taken up by
target cells and delivering the active therapeutic payload, the XDP does not
require, in some
configuration embodiments, cleavage of Gag; hence the omission of a protease
and cleavage
sites is dispensable in some embodiments, described more fully, below,
including the Examples.
[00183] In some embodiments, the present disclosure provides XDP comprising
one or more
structural components derived from a Retroviridae virus, a therapeutic payload
(described more
fully, below), and a tropism factor (described more fully, below). In some
embodiments, the
virus structural components are derived from a Orthoretrovirinae virus_ In
some embodiments,
the Orthoretrovirinae virus is an Alpharetrovirus, a Betaretrovints, a
Deltaretrovirus, an
Epsilonretrovirus, a Gammaretrovirus or a Lentivirus. In other embodiments,
the virus
structural components are derived from a Spumaretrovirinae virus. In some
embodiments, the
Spumaretrovirinae virus is aBovaspumcrvirus, an Equispumavirus, aFel/spun/a-
virus, a
Prosimiispumavirus or a Sitniispumavirus.
b. Retroviral Components
[00184] The Retroviridae family of viruses have different subfamilies,
including
Orthoretrovirinae, Sputnaretrovirinae, and unclassified Retroviridae. Many
retroviruses cause
serious diseases in humans, other mammals, and birds. Human retroviruses
include Human
Immunodeficiency Virus 1 (HIV-1) and HIV-2, the cause of the disease AIDS, and
human T-
lymphotropic virus (HTLV) also cause disease in humans. The subfamily
Orthoretrovirinae
include the genera Alpharetrovirus, Betaretrovirus, Deltaretrovirus,
Epsilonretrovirus,
Gammaretrovirus, and Lentivirus. Members of ahareirovirus, including Avian
leukosis virus
and Rous sarcoma virus, can cause sarcomas, tumors, and anemia of wild and
domestic birds.
Examples of Betaretrovirus include mouse mammary tumor virus, Mason-Pfizer
monkey virus,
28
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
and enzootic nasal tumor virus. Examples of Deltareirovirus include the bovine
leukemia virus
and the human T-lymphotropic viruses. Members of Epsilonretrovirus include
Walleye dermal
sarcoma virus, and Walleye epidermal hyperplasia virus 1 and 2. Members of
Gcrmmaretrovints
include murine leukemia virus, Maloney murine leukemia virus, and feline
leukemia virus, as
well as viruses that infect other animal species. Lent/virus is a genus of
retroviruses that cause
chronic and deadly diseases, including 1-1IV-1 and HIV-2, the cause of the
disease AIDS, and
also includes Simian immunodeficiency virus. The subfamily Spurnaretrovirinae
include the
genera Bovispuntavirus, Equispuntavirus, Felispumavirus, Prositniispurnavirus,
Sitniispumavirus, and Spumavirus. Members of the Retroviridae have provided
valuable research
tools in molecular biology, and, in the context of the present disclosure,
have been used in the
generation of XDP for delivery systems. It has been discovered that the
retroviral-derived
structural components of XDP can be derived from each of the genera of
Retroviridae, and that
the resulting XDP are capable self-assembly in a host cell and encapsidating
(or encompassing)
therapeutic payloads that have utility in the targeted and selective delivery
of the therapeutic
payloads to target cells and tissues.
[00185] In some embodiments, the XDP retroviral components are derived from
Alpharetrovirus, including but not limited to avian leukosis virus (ALV) and
Rous sarcoma virus
(RSV). In such embodiments, the present disclosure provides XDP wherein the
XDP comprises
components selected from the group consisting of: a matrix polypeptide (MA); a
p2A spacer
peptide; ap2B spacer peptide; a p10 spacer peptide; a capsid polypeptide (CA);
a nueleocapsid
polypeptide (NC), a Gag polyprotein comprising a matrix polypeptide (MA), a
capsid
polypeptide (CA), p2A, p214, p10, a nucleocapsid polypeptide (NC); a
therapeutic payload; a
tropism factor; a Gag-Pol polyprotein; a Gag-transframe region-Pal protease
polyprotein; a
cleavage site(s); and a non-retroviral, heterologous protease capable of
cleaving the protease
cleavage sites. In the forgoing embodiment, Gag components (e.g., MA, CA, p2A,
p2B, p10,
and NC), and optionally the cleavage site and protease, are derived from an
Alpharetrovirus,
including but not limited to Avian leukosis virus and Rous sarcoma virus. The
encoding
sequences for these components are provided in Table 5, and the methods to
create the encoding
plasmids and produce the XDP in host cells are described herein, below. In
some embodiments,
the XDP comprises one or more Alpharetrovirus structural components encoded by
the
sequences selected from the group consisting SEQ ID NOS: 192, 193, 195, 196,
198-201, 782,
and 234 as set forth in Table 5, or a sequence having at least 80%, at least
90%, at least 95%, at
29
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
least 95%, at least 97%, at least 98%, or at least 99% identity thereto. In
some embodiments, the
XDP comprises one or more Alphcrretrovirus structural components encoded by
the sequences
selected from the group consisting SEQ ID NOS: 192, 193, 195, 196, 198-201,
782, 234 as set
forth in Table 5. The XDP having Alpharetrovirus components can be designed in
various
configurations, including the configurations of FIGS. 36-68, and may be
encoded by one, two,
three or four nucleic acids, described more fully, below. In some embodiments,
the XDP
comprise a subset of the components listed supra, such as depicted in FIGS. 36-
68, which depict
CasX and gNA as the therapeutic payloads. These alternative configurations are
described more
fully, below, as well as in the Examples. In a particular embodiment, the
therapeutic payload is
an RNP of a complexed CasX and gNA embodiment described herein, while the
tropism factor
is a viral glycoprotein embodiment described herein.
1001861 In some embodiments, the XDP viral components are derived from
Betaretrovirus,
including but not limited to mouse mammary tumor virus (MMTV), Mason-Pfizer
monkey virus
(MPMV), and enzootic nasal tumor virus (ENTV). In such embodiments, the
present disclosure
provides XDP wherein the XDP comprises components selected from the group
consisting of: a
matrix polypeptide (MA); a pp21/24 spacer peptide; a p3-p8/p12 spacer peptide;
a capsid
polypeptide (CA); a nucleocapsid polypeptide (NC); a Gag polyprotein
comprising a matrix
polypeptide (MA), a capsid polypeptide (CA), pp21/24, p3-p8/p12, a
nucleocapsid polypeptide
(NC); a therapeutic payload; a tropism factor; a Gag-Po1 polyprotein; a Gag-
transframe region-
Pol protease polyprotein; a cleavage site(s); and a non-retroviral,
heterologous protease capable
of cleaving the protease cleavage sites. In the forgoing embodiment, Gag
components (e.g., MA,
CA, pp2124 spacer, p3-p8/p12 spacer, and NC), and optionally the cleavage site
and protease,
are derived from an Betaretrovirus, including but not limited to mouse mammary
tumor virus,
Mason-Pfizer monkey virus, and enzootic nasal tumor virus. The encoding
sequences for these
components are provided in Table 5, and the methods to create the encoding
plasmids and
produce the XDP in host cells are described herein, below. In some
embodiments, the XDP
comprises one or more Betaretrovirus structural components encoded by the
sequences selected
from the group consisting SEQ ID NOS: 235-257 as set forth in Table 5, or a
sequence having at
least 80%, at least 90%, at least 95%, at least 95%, at least 97%, at least
98%, or at least 99%
identity thereto. In some embodiments, the XDP comprises one or more
Betaretrovirus
structural components encoded by the sequences selected from the group
consisting SEQ ID
NOS: 235-257 as set forth in Table 5, The XDP having Betaretrovirus components
can be
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
designed in various configurations, including the configurations of FIGS. 36-
68, and may be
encoded by one, two, three or four nucleic acids, described more fully, below.
In some
embodiments, the XDP comprise a subset of the components listed in the
paragraph, such as
depicted in FIGS. 36-68, which depict CasX and gNA as the therapeutic
payloads. These
alternative configurations are described more fully, below, as well as in the
Examples. In a
particular embodiment, the therapeutic payload is an RNP of a complexed CasX
and gNA
embodiment described herein, while the tropism factor is a viral glycoprotein
embodiment
described herein.
1001871 In some embodiments, the XDP viral components are derived from
Deltaretrovirus,
including but not limited to bovine leukemia virus (BLV) and the human T-
Iymphotropie viruses
(HTLV1). In such embodiments, the present disclosure provides XDP wherein the
XDP
comprises components selected from the group consisting of: a matrix
polypeptide (MA); a
capsid polypeptide (CA); a nucleocapsid polypeptide (NC); a Gag polyprotein
comprising a
matrix polypeptide (MA), a capsid polypeptide (CA), and a nucleocapsid
polypeptide (NC); a
therapeutic payload; a tropism factor; a Gag-Pol polyprotein; a Gag-transframe
region-Pol
protease polyprotein; a cleavage site(s); and a non-retroviral, heterologous
protease capable of
cleaving the protease cleavage sites. In the forgoing embodiment, Gag
components (e.g., MA,
CA, and NC), and optionally the cleavage site and protease, are derived from
an
Deftaretrovirus, including but not limited to bovine leukemia virus and the
human T-
lymphotropic viruses. The encoding sequences for these components are provided
in Table 5,
and the methods to create the encoding plasmids and produce the XDP in host
cells are described
herein, below. In some embodiments, the XDP comprises one or more
Deftaretrovirus
structural components encoded by the sequences selected from the group
consisting SEQ ID
NOS: 258-272 as set forth in Table 5, or a sequence having at least 80%, at
least 90%, at least
95%, at least 95%, at least 97%, at least 98%, or at least 99% identity
thereto. In some
embodiments, the XDP comprises one or more Deftaretrovirus structural
components encoded
by the sequences selected from the group consisting SEQ ID NOS: 258-272 as set
forth in Table
5. The XDP having Deltaretrovirus components can be designed in various
configurations,
including the configurations of FIGS. 36-68, and may be encoded by one, two,
three or four
nucleic acids, described more fully, below. In some embodiments, the XDP
comprise a subset of
the components listed in the paragraph, such as depicted in FIGS. 36-68, which
depict CasX and
gNA as the therapeutic payloads. These alternative configurations are
described more fully,
31
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
below, as well as in the Examples. In a particular embodiment, the therapeutic
payload is an
RNP of a complexed CasX and gNA embodiment described herein, while the tropism
factor is a
viral giycoprotein embodiment described herein.
1001881 In some embodiments, the XDP viral components are derived from
Epsilonretrovirus,
including but not limited to Walleye dermal sarcoma virus (WDSV), and Walleye
epidermal
hyperplasia virus 1 and 2. In such embodiments, the present disclosure
provides XDP wherein
the XDP comprises components selected from the group consisting of: a matrix
polypeptide
(MA); a p20 spacer peptide; a capsid polypeptide (CA); a nucleocapsid
polypeptide (NC); a Gag
polyprotein comprising a matrix polypeptide (MA), a capsid polypeptide (CA),
p20, a
nucleocapsid polypeptide (NC); a therapeutic payload; a tropism factor; a Gag-
Pot polyprotein; a
Gag-transframe region-Pol protease polyprotein; a cleavage site(s); and a non-
retroviral,
heterologous protease capable of cleaving the protease cleavage sites. In the
forgoing
embodiment, Gag components (e.g., MA, CA, p20, and NC), and optionally the
cleavage site
and protease, are derived from an Epsilonretrovirus, including but not limited
to Walleye dermal
sarcoma virus, and Walleye epidermal hyperplasia virus 1 and 2. The encoding
sequences for
these components are provided in Table 5, and the methods to create the
encoding plasmids and
produce the XDP in host cells are described herein, below. In some
embodiments, the XDP
comprises one or more Epsilonretrovirus structural components encoded by the
sequences
selected from the group consisting SEQ ID NOS: 273-277 as set forth in Table
5, or a sequence
having at least 80%, at least 90%, at least 95%, at least 95%, at least 97%,
at least 98%, or at
least 99% identity thereto. In some embodiments, the XDP comprises one or more
Epsilonretrovirus structural components encoded by the sequences selected from
the group
consisting SEQ ID NOS: 273-277 as set forth in Table 5. The XDP having
Epsilonretrovirus
components can be designed in various configurations, including the
configurations of FIGS. 36-
68, and may be encoded by one, two, three or four nucleic acids, described
more fully, below. In
some embodiments, the XDP comprise a subset of the components listed in the
paragraph, such
as depicted in FIGS. 36-68, which depict CasX and gNA as the therapeutic
payloads. These
alternative configurations are described more fully, below, as well as in the
Examples. In a
particular embodiment, the therapeutic payload is an RNP of a complexed CasX
and gNA
embodiment described herein, while the tropism factor is a viral glycoprotein
embodiment
described herein.
32
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00189] In some embodiments, the XDP viral components are derived from
Gammaretrovirus,
including but not limited to murine leukemia virus (MLV), Maloney murine
leukemia virus
(MMLV), and feline leukemia virus (FLV). In such embodiments, the present
disclosure
provides XDP wherein the XDP comprises components selected from the group
consisting of: a
matrix polypeptide (MA); a pp12 spacer peptide; a capsid polypeptide (CA); a
nucleocapsid
polypeptide (NC); a Gag polyprotein comprising a matrix polypeptide (MA), a
pp12 spacer, a
capsid polypeptide (CA), a nucleocapsid polypeptide (NC); a therapeutic
payload; a tropism
factor; a Gag-Pol polyprotein; a Gag-transframe region-Pol protease
polyprotein; a cleavage
site(s); and a non-retroviral, heterologous protease capable of cleaving the
protease cleavage
sites. In the forgoing embodiment, Gag components (e.g., MA, pp12, CA, and
NC), and
optionally the cleavage site and protease, are derived from an
Gammaretrovirus, including but
not limited to Walleye dermal sarcoma virus, and Walleye epidermal hyperplasia
virus 1 and 2.
The encoding sequences for these components are provided in Table 5, and the
methods to create
the encoding plasmids and produce the XDP in host cells are described herein,
below. In some
embodiments, the XDP comprises one or more Gamtnaretrovirus structural
components encoded
by the sequences selected from the group consisting SEQ ID NOS: 278-287 as set
forth in Table
5, or a sequence having at least 80%, at least 90%, at least 95%, at least
95%, at least 97%, at
least 98%, or at least 99% identity thereto. In some embodiments, the XDP
comprises one or
more Gammaretrovirus structural components encoded by the sequences selected
from the
group consisting SEQ ID NOS: 278-287 as set forth in Table 5. The XDP having
Gammaretrovirus components can be designed in various configurations,
including the
configurations of FIGS. 36-68, and may be encoded by one, two, three or four
nucleic acids,
described more fully, below. In some embodiments, the XDP comprise a subset of
the
components listed in the paragraph, such as depicted in FIGS. 36-68, which
depict CasX and
gNA as the therapeutic payloads. These alternative configurations are
described more fully,
below, as well as in the Examples. In a particular embodiment, the therapeutic
payload is an
RNP of a complexed CasX and gNA embodiment described herein, while the tropism
factor is a
viral glycoprotein embodiment described herein.
1001901 In some embodiments, the XDP viral components are derived from
Lentivirus,
including but not limited to HIV-1 and HIV-2, and Simian immunodeficiency
virus (SIV). In
such embodiments, the present disclosure provides XDP wherein the XDP
comprises
components selected from the group consisting of: a matrix polypeptide (MA); a
capsid (CA), a
33
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
p2 spacer peptide, a nucleocapsid (NC), a pi/p6 spacer peptide; ); a Gag
polyprotein comprising
a matrix polypeptide (MA), CA, p2, NC, and pl/p6; a therapeutic payload; a
tropism factor; a
Gag-Pot polyprotein; a Gag-transframe region-Pol protease polyprotein; a
cleavage site(s); and a
non-retroviral, heterologous protease capable of cleaving the protease
cleavage sites_ In the
forgoing embodiment, Gag components (e.g., MA, CA, NC, and pl/p6), and
optionally the
cleavage site and protease, are derived from an Lent/virus, including but not
limited to HIV-1,
HIV-2, and Simian immunodeficiency virus (SW). The encoding sequences for
these
components are provided in Table 5, and the methods to create the encoding
plasmids and
produce the XDP in host cells are described herein, below. In some
embodiments, the XDP
comprises one or more Lent/virus structural components encoded by the
sequences selected from
the group consisting SEQ ID NOS: 288-312 and 334-339 as set forth in Table 5,
or a sequence
having at least 80%, at least 90%, at least 95%, at least 95%, at least 97%,
at least 98%, or at
least 99% identity thereto. In some embodiments, the XDP comprises one or more
Lent/virus
structural components encoded by the sequences selected from the group
consisting SEQ ID
NOS: 288-312 and 334-339 as set forth in Table 5. The XDP having Lentivirus
components can
be designed in various configurations, including the configurations of FIGS.
36-68, and may be
encoded by one, two, three or four or more nucleic acids, described more
fully, below. In some
embodiments, the XDP comprise a subset of the components listed in the
paragraph, such as
depicted in FIGS. 36-68, which depict CasX and gNA as the therapeutic
payloads. These
alternative configurations are described more fully, below, as well as in the
Examples. In a
particular embodiment, the therapeutic payload is an RNP of a complexed CasX
and g,NA
embodiment described herein, while the tropism factor is a viral glycoprotein
embodiment
described herein.
[00191] In some embodiments, the XDP viral components are derived from
Spumaretrovirinae,
including but not limited to Bovisputnavirus, Equisputnavirtts,
Felispuntavirus,
Prosimiispumavirus, Simiispumavirus, and Spumavirus. In such cases, the
present disclosure
provides XDP wherein the XDP comprises components selected from the group
consisting of:
p68 Gag; a p3 Gag; a Gag polyprotein comprising of p68 Gag and p3 gag; a
therapeutic payload;
a tropism factor; a Gag-Pol polyprotein; a Gag-transframe region-Pol protease
polyprotein; a
cleavage site(s); and a non-retroviral, heterologous protease capable of
cleaving the protease
cleavage sites. In the forgoing embodiment, Gag components (e.g., p68 AND
p3p20), and
optionally the cleavage site and protease, are derived from an
Sputnaretrovirinae including but
34
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
not limited to Bovispumavirus, Equispumavirus, Felispuma-virus,
Prosimiisputnavirus,
Simiispumavirus, and Spumavirus. The encoding sequences for these components
are provided
in Table 5, and the methods to create the encoding plasmids and produce the
XDP in host cells
are described herein, below. In some embodiments, the XDP comprises one or
more
Spurnaretrovirinae structural components encoded by the sequences selected
from the group
consisting SEQ ID NOS: 313-333 as set forth in Table 5, or a sequence having
at least 80%, at
least 90%, at least 95%, at least 95%, at least 97%, at least 98%, or at least
99% identity thereto.
In some embodiments, the XDP comprises one or more Sputnareirovirinae
structural
components encoded by the sequences selected from the group consisting SEQ ID
NOS: 313-
333 as set forth in Table 5. The XDP having Spumaretrovirus components can be
designed in
various configurations, including the configurations of FIGS. 36-68, and may
be encoded by
one, two, three or four nucleic acids, described more fully, below. In some
embodiments, the
XDP comprise a subset of the components listed in the paragraph, such as
depicted in FIGS. 36-
68, which depict CasX and gNA as the therapeutic payloads. These alternative
configurations
are described more fully, below, as welt as in the Examples. In a particular
embodiment, the
therapeutic payload is an RNP of a complexed CasX and gNA embodiment described
herein,
while the tropism factor is a viral glycoprotein embodiment described herein.
1001921 In other embodiments, the present disclosure provides XDP wherein the
retroviral
components of the XDP are selected from different genera of the Retroviridae.
Thus the XDP
can comprise two or more components selected from a matrix polypeptide (MA), a
p2A spacer
peptide, a p2B spacer peptide; a p10 spacer peptide, a capsid polypeptide
(CA), a nucleocapsid
polypeptide (NC), a pp21/24 spacer peptide, a p3-P8 spacer peptide, a pp12
spacer peptide, a
p20 spacer peptide, a pi/p6 spacer peptide, a p68 Gag, a p3 Gag, a cleavage
site(s), a Gag-Pol
polyprotein; a Gag-transframe region-Pot protease polyprotein; and a non-
retroviral,
heterologous protease capable of cleaving the protease cleavage sites wherein
the components
are derived from Alpharetrovirus, Betaretrovirus, Deharetrovirus,
Epsdonretrovirus,
Gattunaretrovirus, Lent/virus, Bovisputnavirus, Equispumavirus,
Felisputnavirus,
Prositniisputnavirus, Simlispumavirus, or Spumavirus..
1001931 In retroviral components derived from HIV-1, the accessory protein
integrase (or its
encoding nucleic acid) can be omitted from the XDP systems, as well as the HIV
functional
accessory genes vpr, vpx (HIV-2), which are dispensable for viral replication
in vitro.
Additionally, the nucleic acids of the XDP system do not require reverse
transcriptase for the
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
creation of the XDP compositions of the embodiments. Thus, in one embodiment,
the HIV-1
Gag-Pot component of the XDP can be truncated to Gag linked to the transframe
region (TFR)
composed of the transframe octapeptide (TFP) and 48 amino acids of the p6pol,
separated by a
protease cleavage site, hereinafter referred to as Gag-TFR-PR, described more
fully, below.
c. Proteases
[00194] In some embodiments of the XDP systems, the protease capable of
cleaving the
protease cleavage sites is selected from a retroviral protease, including any
of the genera of the
Retroviridae. For example, the protease can be encoded by a sequence selected
from the group
consisting of SEQ ID NOS: 198, 234, 239, 245, 251, 257, 261, 266, 271, 276,
282, 287, 291,
296, 301, and 306 as set forth in Table 5, or a sequence having at least 80%,
at least 90%, at least
95%, at least 95%, at least 97%, at least 98%, or at least 99% identity
thereto. In other
embodiments, the protease capable of cleaving the protease cleavage sites is a
non-retroviral,
heterologous protease selected from the group of proteases consisting of
tobacco etch virus
protease (TEV), potyvirus HC protease, potyvirus Plprotease, PreScission
(HRV3C protease), b
virus NIa protease, B virus RNA-2-encoded protease, aphthovims L protease,
enterovirus 2A
protease, rhinovirus 2A protease, picoma 3C protease, comovirus 24K protease,
nepovirus 24K
protease, RTSV (rice tungro spherical virus) 3C-like protease, PYVF (parsnip
yellow fleck
virus) 3C-like protease, heparin, cathepsin, thrombin, factor Xa,
metalloproteinases (including
MMP-2, -3, -7, -9, -10, and -11), and enterokinasa In a particular embodiment,
the protease
capable of cleaving the protease cleavage sites is PreScission Protease; a
fusion protein of
human rhinovirus (HRV) 3C protease and glutathione S-transferase (GST). In
another particular
embodiment, the protease capable of cleaving the protease cleavage sites is
tobacco etch virus
protease (TEV), In another particular embodiment, the protease capable of
cleaving the protease
cleavage sites is HIV-1 protease. In the case of HIV-1 protease, the 99-amino
acid protease (PR)
of the precursor Gag--Pol polyprotein (which are encoded by overlapping open
reading frames
such that the synthesis of the of the Gag--Pol precursor results from a -1
frameshifting event) is
flanked at its N-terminus by a transframe region (TFR) composed of the
transframe octapeptide
(TFP) and 48 amino acids of the p6po1, separated by a protease cleavage site.
Cleavage at the
p6po1-PR site to release a free N-terminus of protease is concomitant with the
appearance of
enzymatic activity and formation of a stable tertiary structure that is
characteristic of the mature
protease (Louis, .11µ4. Et at. Autoprocessing of HIV-1 protease is tightly
coupled to protein
folding. Nat Struct Mol Biol 6, 868-875 (1999)). In some embodiments of the
XDP systems,
36
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
wherein the nucleic acid encodes all or a portion of the HIV-1 Gag-Pol
polyprotein, the Gag-Pol
sequence comprises the encoded TFR-PR to facilitate the-1 frameshifting event.
In some cases,
wherein the XDP system utilizes a component comprised of the Gag polyprotein
and a portion of
the pol polyprotein comprising the TER and the protease, the component is
referred to herein as
"Gag-TFR-PR", wherein the capability to facilitate the -1 frameshifting event
is retained, along
with the capability to produce the encoded protease. In non-limiting examples
of nucleic acids
encoding a Retroviral protease the can be incorporated into an encoding
plasmid of the XDP
system embodiments, representative sequences are provided in Table 5.
1001951 In a corresponding fashion, wherein pro-tease cleavage sites are
incorporated in the
XDP systems, the protease cleavage sites utilized in the encoded proteins of
the XDPs and their
encoding sequences in the nucleic acids will correlate with the protease that
is incorporated into
the XDP system. In some embodiments, the protease cleavage site of the XDP
component
comprising all or a portion of a Gag polyprotein is located between the Gag
polyprotein and the
therapeutic payload such that upon maturation of the XDP particle, the
therapeutic payload is not
tethered to any component of the Gag polyprotein. In other embodiments, the
protease cleavage
site is incorporated between the individual components of the Gag polyprotein
as well as
between the Gag polyprotein and the therapeutic payload. In a representative
embodiment,
wherein the protease capable of cleaving the protease cleavage sites is TEV,
the encoded TEV
protease cleavage sites can have the sequences EXXYXQ(G/S) (SEQ ID NO: 17),
ENLYFQG
(SEQ ID NO: 18) or ENLYFQS (SEQ ID NO: 19), wherein X represents any amino
acid and
cleavage by TEV occurs between Q and G or Q and S. In another embodiment,
wherein the
protease is protease, the encoded mV-1 cleavage sites
can have the sequence
SQNYPIVQ (SEQ ID NO: 20). In another embodiment, wherein the protease is
PreScission, the
protease cleavage sites include the core amino acid sequence Leu-Phe-Gln/Gly-
Pro (SEQ ID
NO: 1010), cleaving between the Gln and Gly residues. In one embodiment, the
XDP
comprising cleavage sites have protease cleavage sites that are identical. In
another
embodiment, the XDP comprising cleavage sites have protease cleavage sites
that are different
and are substrates for different proteases. In another embodiment, the XDP
system can comprise
a cleavage sequence that is susceptible to cleavage by two different
proteases; e.g., 11IV-1 and
PreScission protease. In such cases, the nucleic acids encoding the XDP would
include
encoding sequences for both proteases.
37
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00196] Additional protease cleavage sites are envisaged as within the scope
of the XDP of the
instant invention, and include, inter alia, SEQ ID NOS: 874-897, and 934-946.
d. Protein and Nucleic Acid Therapeutic Payloads of the XDP Systems
[00197] Protein therapeutic payloads suitable for inclusion in the XDP of the
present disclosure
include a diversity of categories of protein-based therapeutics, including,
but not limited to
cytokines (e.g., 1FNs a, 13, and 7, TNF-ct, G-CSF, GM-CSF)), interleukins
(e.g., IL-1 to IL-40),
growth factors (e.g., VEGF, PDGF, IGF-1, EGF, and TGF-I3), enzymes, receptors,
microproteins, hormones (e.g., growth hormone, insulin), erythropoietin,
RNAse, DNAse, blood
clotting factors (e.g. FVII, FVIII, FIX, FX), anticoagulants, bone
morphogenetic proteins,
engineered protein scaffolds, thrombolytics (e.g., streptokinase, tissue
plasminogen activator,
plasminogen, and plasmid), CRISPR proteins (Class 1 and Class 2 Type II, Type
V, or Type VI)
as well as engineered proteins such as anti-cancer modalities or biologics
intended to treat
diseases such as neurologic, metabolic, cardiovascular, liver, renal, or
endocrine diseases and
disorders. Nucleic acid payloads suitable for inclusion in the XDP of the
present disclosure
include a diversity of categories, including sequences encoding the foregoing
protein therapeutic
payloads, as well as single-stranded antisense oligonucleotides (AS0s), double-
stranded RNA
interference (RNAi) molecules, DNA aptamers, nucleic acids utilized in gene
therapy (e.g.,
guide RNAs utilized in CRISPR systems and donor templates), micro RNAs,
ribozymes, RNA
decoys and circular RNAs. In a particular embodiment, the protein payload of
the XDP
comprises a CasX variant protein of any of the embodiments described herein,
including the
CasX variants of SEQ 1D NOS: 21-233, 343-345, 350-353, 355-367 and 388-397 as
set forth in
Tables 1, 7, 8, 9 and 11, while the nucleic acid payload comprises one or more
guide RNAs of
any of the embodiments described herein, including the gNA variants with a
scaffold sequence
of SEQ ID NOS: 597-781 as set forth in Table 3 and, optionally, a donor
template.
e. CRISPR Proteins of the XDP Systems
[00198] In some embodiments, the present disclosure provides XDP compositions
and systems
comprising a CRISPR nuclease and one or more guide nucleic acids engineered to
bind target
nucleic acid that have utility in genome editing of eukaryotic cells. In some
embodiments, the
CRISPR nuclease employed in the XDP systems is a Class 2 nuclease. In other
embodiments,
the CRISPR nuclease is a Class 2, Type V nuclease. Although members of Class
2, Type V
CRISPR-Cas systems have differences, they share some common characteristics
that distinguish
them from the Cas9 systems. Firstly, the Type V nucleases possess a single RNA-
guided RuvC
38
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
domain-containing effector but no 1-INH domain, and they recognize T-rich PAM
5' upstream to
the target region on the non-targeted strand, which is different from Cas9
systems which rely on
G-rich PAM at 3' side of target sequences. Type V nucleases generate staggered
double-stranded
breaks distal to the PAM sequence, unlike Cas9, which generates a blunt end in
the proximal site
close to the PAM. In addition, Type V nucleases degrade ssDNA in trans when
activated by
target dsDNA or ssDNA binding in cis. In some embodiments, the Type V
nucleases utilized in
the XDP embodiments recognize a 5' TC PAM motif and produce staggered ends
cleaved solely
by the RuvC domain. In some embodiments, the XDP comprise a Class 2, Type V
nuclease
selected from the group consisting of Cas12a, Cas12b, Cas12c, Cas12d (CasY),
Cas12j and
CasX. In a particular embodiment, the present disclosure provides XDP
comprising a
ribonucleoprotein (RNP) of a complexed CasX protein and one or more guide
nucleic acids
(gNA) that are specifically designed to modify a target nucleic acid sequence
in eukaryotic cells.
1001991 The term "CasX protein", as used herein, refers to a family of
proteins, and
encompasses all naturally occurring CasX proteins (also referred to herein as
a "wild-type" or
"reference' CasX), as well as CasX variants with one or more modifications in
at least one
domain relative to a naturally-occurring reference CasX protein. Reference
CasX proteins
include, but are not limited to those isolated or derived from
Deltaproteobacter, , Planctornycetes,
or Candidatus (as described in US20180346927A1 and W02018064371A1,
incorporated herein
by reference). Exemplary embodiments of CasX variants envisaged as being
within the scope of
the disclosure are described herein, below.
1002001 In some cases, a Type V reference CasX protein is isolated or derived
from
Deltaproteobacteria. In some embodiments, a CasX protein comprises a sequence
at least 50%
identical, at least 60% identical, at least 65% identical, at least 70%
identical, at least 75%
identical, at least 80% identical, at least 81% identical, at least 82%
identical, at least 83%
identical, at least 84% identical, at least 85% identical, at least 86%
identical, at least 86%
identical, at least 87% identical, at least 88% identical, at least 89%
identical, at least 89%
identical, at least 90% identical, at least 91% identical, at least 92%
identical, at least 93%
identical, at least 94% identical, at least 95% identical, at least 96%
identical, at least 97%
identical, at least 98% identical, at least 99% identical, at least 99.5%
identical or 100% identical
to a sequence of:
1 MEKRINKIRK KLSADNATKP VSRSGPMKTL LVRVMTDDLK KRLEKRRKKP
EVMPQVISNN
39
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
61 AANNLRMLLD DYTEMKEAIL QVYWQEFKDD HVGLMCKFAQ PASKKIDQNK
LKPEMDEKGN
121 LT TAGFACSQ CGQPLFVYKL EQVSEKGKAY TNYFGRCNVA EHEKL I LLAQ
LKPEKDS DEA
181 VTYSLGKFGQ RALDFYS IHV TKESTHPVKP LAQIAGNRYA SGPVGKALSD
ACMGT IAS FL
241 SKYQDI I IEH QKVVKGNQKR LESLRELAGK ENLEYPSVTL PPQPHTKEGV
DAYNEVIARV
301 RMWVNLNLWQ KLKLSRDDAK PLLRLKG FPS FPVVERRENE VDWWNT INEV
KKL I DAKRDM
361 GRVFWSGVTA EKRNT I LEGY NYLPNENDHK KREGSLENPK KPAKRQFGDL
LLYLEKKYAG
421 DWGKVFDEAW ER I DKKIAGL T SH I EREEAR NAEDAQSKAV LT DWLRAKAS
FVLERLKEMD
481 EKE FYACE IQ LQKWYGDLRG NPFAVEAENR VVDI SGFS I G SDGHS I QYRN
LLAWKYLENG
541 KREFYLLMNY GKKGRIRFTD GTD I KKSGKW QGLLYGGGKA KVI DLT FDPD
DEQL I I L PLA
601 FGTRQGREFI WNDLLSLETG LIKLANGRVI EKT IYNKK G RDE PAL FVAL
T FE RREVVDP
661 SNIKPVNL I G VDRGENI PAV IALTDPEGCP LPEFKDS SGG PTDILRIGEG
Y KE KORA I QA
721 AKEVEQRRAG GYSRKFASKS RNLADDMVRN SARDLFYHAV THDAVLVFEN
LSRGFGRQGK
781 RTFMTERQYT KMEDWLTAKL AYEGLTSKTY LSKTLAQYTS KTCSNCGFT I
TTADYDGMLV
841 RLKKTSDGWA T TLNNKELKA EGQ I TYYNRY KRQTVEKELS AELDRLSEES
GNND I SKWTK
901 GRRDEALFLL KKRFSHRPVQ EQFVCLDCGH EVHADEQAAL NIARSWL FLN
SNSTEFKSYK
961 SGKQPFVGAW QAFYKRRLKE VWKPNA (SEQ ID NO: 1).
1002011 In some cases, a Type V reference CasX protein is isolated or derived
from
Planctomycetes. In some embodiments, a CasX protein comprises a sequence at
least 50%
identical, at least 60% identical, at least 65% identical, at least 70%
identical, at least 75%
identical, at least 80% identical, at least 81% identical, at least 82%
identical, at least 83%
identical, at least 84% identical, at least 85% identical, at least 86%
identical, at least 86%
identical, at least 87% identical, at least 88% identical, at least 89%
identical, at least 89%
identical, at least 90% identical, at least 91% identical, at least 92%
identical, at least 93%
identical, at least 94% identical, at least 95% identical, at least 96%
identical, at least 97%
identical, at least 98% identical, at least 99% identical, at least 99.5%
identical or 100% identical
to a sequence of:
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1 MQEIKRINKI RRRLVKDSNT KKAGKTGPMK TLLVRVMTPD LRERLENLRK
KPENIPQPIS
61 NTSRANLNKL LTDYTEMKKA ILHVYWEEFQ KDPVGLMSRV AQPAPKNIDQ
RKL I PVKDGN
121 ERL TS SGFAC SQCCQPLYVY KLEQVNDKGK PHTNYFGRCH VSEHERL ILL
SPHKPEANDE
181 LVTYSLGKFG QRALDFYSIH VTRESNHPVK PLEQ I GGNSC ASGPVGKALS
DACMGAVAS F
241 LTKYQDI ILE HQKVIKKNEK RLANLKDIAS .ANGLAFPKIT LPPQPHTKEG
I EAYNNVVAQ
301 IVIWVNLNLW QKLKIGRDEA KPLQRLKGFP SFPLVERQAN EVDWWDMVCN
VKKL I NEKKE
361 DGKVFWQNLA GYKRQEALLP YLSSEEDRKK GKKFARYQFG DLLLHLEKKH
GE DWGKVYDE
421 AWERI DKKVE GLSKHI KLEE ERRSEDAQSK AALTDWLRAK AS FV IEGLKE
ADKDE FORCE
481 LKLQKWYGDL RGKPFAIEAE NS ILDISGFS KQYNCAFIWQ KDGVKKLNLY
L I I NY FKGGK
541 LRFKKIKPEA FEANRFYTVI NKKS GE IVPM EVNENFDDPN LI IL PLA FGK
RQGRE F I WND
601 LLSLETGSLK LANGRVIEKT LYNRRTRQDE PALFVALT FE RREVLDSSNI
KPMNL I G I DR
661 GENIPAVIAL TDPEGCPLSR FKDSLGNPTH ILRIGESYKE KQRT IQAAKE
VEQRRAGGYS
721 RKYASKAKNL ADDMVRNTAR DLLYYAVTQD AML I FENL SR GFGRQGKRT F
MAE RQYTRME
781 DWLTAKLAYE GLPSKTYLSK TLAQYTSKTC SNCG FT I T SA DYDRVLEKLK
KTATGWMTT I
841 NGKELKVEGQ I TYYNRYKRQ NVVKDLSVEL DRLSEESVNN DI S SWTKGRS
GEALSLLKKR
901 FSHRPVQEKF VCLNCGFETH ADEQ.AALNIA RSWLFLRSQE YKKYQTNKTT
GNTDKRAFVE
961 TWQSFYRKKL KEVWKPAV ( SEQ ID NO: 2 ) .
1002021 In some cases, a Type V reference CasX protein is isolated or derived
from Candidaius
Sungbacteria. In some embodiments, a CasX protein comprises a sequence at
least 50%
identical, at least 60% identical, at least 65% identical, at least 70%
identical, at least 75%
identical, at least 80% identical, at least 81% identical, at least 82%
identical, at least 83%
identical, at least 84% identical, at least 85% identical, at least 86%
identical, at least 86%
identical, at least 87% identical, at least 88% identical, at least 89%
identical, at least 89%
identical, at least 90% identical, at least 91% identical, at least 92%
identical, at least 93%
identical, at least 94% identical, at least 95% identical, at least 96%
identical, at least 97%
identical, at least 98% identical, at least 99% identical, at least 99.5%
identical or 100% identical
to a sequence of
41
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1 MDNANKPSTK SLVNTTRISD HFGVTPGQVT RVFSFGIIPT KRQYAIIERW
FAAVEAARER
61 LYGMLYAHFQ ENPPAYLKEK FSYETFFKGR PVLNGLRDID PTIMTSAVFT
ALRHKAEGAM
121 AAFHTNHRRL FEEARKKMRE YAECLKANEA LLRGAADIDW DKIVNALRTR
LNTCLAPEYD
181 AVIADFGALC AFRALIAETN ALKGAYNHAL NQMLPALVKV DEPEEAEESP
RLRFFNGRIN
241 DLPKFPVAER ETPPDTETII RQLEDMARVI PDTAEILGYI HRIRHKAARR
KPGSAVPLPQ
301 RVALYCAIRM ERNPEEDPST VAGHFLGEID RVCEKRRQGL VRTPFDSQIR
ARYMDIISFR
361 ATLAHPDRWT EIQFLRSNAA SRRVRAETIS APFEGFSWTS NRTNPAPQYG
MALAKDANAP
421 ADAPELCICL SPSSAAFSVR EKGGDLIYMR PTGGRRGKDN PGKEITWVPG
SFDEYPASGV
481 ALKLRLYFGR SQARRMLTNK TWGLLSDNPR VFAANAELVG KKRNPQDRWK
LFFHMVISGP
541 PPVEYLDFSS DVRSRARTVI GINRGEVNPL AYAVVSVEDG QVLEEGLLGK
KEYIDQLIET
601 RRRISEYQSR EQTPPRDLRQ RVRHLQDTVL GSARAKIHSL LAFWKGILAI
ERLDDQFHGR
661 EQKIIPKKTY LANKTGFMNA LSFSGAVRVD KKGNPWGGMI EIYPGGISRT
CTQCGTVWLA
721 RRPKNPGHRD AMVVIPDIVD DAAATGFDNV DCDAGTVDYG ELFTLSREWV
RLTPRYSRVM
781 RGTLGDLERA IRQGDDRKSR QMLELALEPQ PQWGQFFCHR CGFNGQSDVL
AATNLARRAI
841 SLIRRLPDTD TPPTP (SEQ ID NO: 3).
1002031 In some embodiments of the XDP systems, the disclosure provides CasX
variant
proteins for use in the XDP comprising a sequence that has at least 1, at
least 2, at least 3, at least
4, at least 5, at least 6, at least 7, at least 8, at least 9, at least 10, at
least 20, at least 30, at least
40 or at least 50 or more individual or sequential mutations relative to the
sequence of a
reference CasX protein of SEQ ID NO: 1, SEQ ID NO:2, or SEQ ID NO:3. These
mutations can
be insertions, deletions, amino acid substitutions, or any combinations
thereof In some
embodiments, in addition to the aforementioned mutations, a CasX variant can
further comprise
a substitution of a portion or all of a domain from a heterologous reference
CasX, and the
substituted domain can further comprise one or more mutations. Suitable
mutagenesis methods
for generating CasX variant proteins of the disclosure may include, for
example, Deep
Mutational Evolution (DME), deep mutational scanning (DMS), error prone PCR,
cassette
mutagenesis, random mutagenesis, staggered extension PCR, gene shuffling, or
domain
42
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
swapping. In some embodiments, the CasX variants are designed, for example by
selecting one
or more desired mutations in a reference CasX. Any amino acid can be
substituted for any other
amino acid in the substitutions described herein. The substitution can be a
conservative
substitution (e.g., a basic amino acid is substituted for another basic amino
acid). The
substitution can be a non-conservative substitution (e.g., a basic amino acid
is substituted for an
acidic amino acid or vice versa). For example, a proline in a reference CasX
protein can be
substituted for any of arginine, histidine, lysine, aspartic acid, glutarnic
acid, serine, threonine,
asparagine, glutamine, cysteine, glycine, alanine, isoleucine, leucine,
methionine, phenylalanine,
tryptophan, tyrosine or valine to generate a CasX variant protein of the
disclosure. In certain
embodiments, the activity of a reference CasX protein is used as a benchmark
against which the
activity of one or more CasX variants are compared, thereby measuring
improvements in
function of the CasX variants.
[00204] In some embodiments, a CasX variant protein comprises at least one
amino acid
deletion relative to a reference CasX protein. In some embodiments, a CasX
variant protein
comprises a deletion of 1-4 amino acids, 1-10 amino acids, 1-20 amino acids, 1-
30 amino acids,
1-40 amino acids, 1-50 amino acids, 1-60 amino acids, 1-70 amino acids, 1-80
amino acids, 1-90
amino acids, 1-100 amino acids, 2-10 amino acids, 2-20 amino acids, 2-30 amino
acids, 3-10
amino acids, 3-20 amino acids, 3-30 amino acids, 4-10 amino acids, 4-20 amino
acids, 3-300
amino acids, 5-10 amino acids, 5-20 amino acids, 5-30 amino acids, 10-50 amino
acids or 20-50
amino acids relative to a reference CasX protein. In some embodiments, a CasX
protein
comprises a deletion of at least about 100 consecutive amino acids relative to
a reference CasX
protein. In some embodiments, a CasX variant protein comprises a deletion of
at least 1, 2, 3, 4,
5, 6, 7, 8, 9, 10, 20, 30, 40, 50 or 100 consecutive amino acids relative to a
reference CasX
protein. In some embodiments, a CasX variant protein comprises a deletion of
1, 2, 3, 4, 5, 6, 7,
8, 9 or 10 consecutive amino acids.
[00205] In some embodiments, a CasX variant protein comprises two or more
deletions relative
to a reference CasX protein, and the two or more deletions are not consecutive
amino acids. For
example, a first deletion may be in a first domain of the reference CasX
protein, and a second
deletion may be in a second domain of the reference CasX protein! In some
embodiments, a
CasX variant protein comprises 2, 3,4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19 or 20
non-consecutive deletions relative to a reference CasX protein. In some
embodiments, a CasX
variant protein comprises at least 20 non-consecutive deletions relative to a
reference CasX
43
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
protein. Each non-consecutive deletion may be of any length of amino acids
described herein,
e.g., 1-4 amino acids, 1-10 amino acids, and the like.
[00206] In some embodiments, the CasX variant protein comprises one or more
amino acid
insertions relative to the sequence of SEQ ID NOS:1, 2, or 3. In some
embodiments, a CasX
variant protein comprises an insertion of 1 amino acid, an insertion of 2-3
consecutive or non-
consecutive amino acids, 2-4 consecutive or non-consecutive amino acids, 2-5
consecutive or
non-consecutive amino acids, 2-6 consecutive or non-consecutive amino acids, 2-
7 consecutive
or non-consecutive amino acids, 2-8 consecutive or non-consecutive amino
acids, 2-9
consecutive or non-consecutive amino acids, 2-10 consecutive or non-
consecutive amino acids,
2-20 consecutive or non-consecutive amino acids, 2-30 consecutive or non-
consecutive amino
acids, 2-40 consecutive or non-consecutive amino acids, 2-50 consecutive or
non-consecutive
amino acids, 2-60 consecutive or non-consecutive amino acids, 2-70 consecutive
or non-
consecutive amino acids, 2-80 consecutive or non-consecutive amino acids, 2-90
consecutive or
non-consecutive amino acids, 2-100 consecutive or non-consecutive amino acids,
3-10
consecutive or non-consecutive amino acids, 3-20 consecutive or non-
consecutive amino acids,
3-30 consecutive or non-consecutive amino acids, 4-10 consecutive or non-
consecutive amino
acids, 4-20 consecutive or non-consecutive amino acids, 3-300 consecutive or
non-consecutive
amino acids, 5-10 consecutive or non-consecutive amino acids, 5-20 consecutive
or non-
consecutive amino acids, 5-30 consecutive or non-consecutive amino acids, 10-
50 consecutive
or non-consecutive amino acids or 20-50 consecutive or non-consecutive amino
acids relative to
a reference CasX protein. In some embodiments, the CasX variant protein
comprises an insertion
of 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 01 20
consecutive or non-consecutive
amino acids. In some embodiments, a CasX variant protein comprises an
insertion of at least
about 100 consecutive or non-consecutive amino acids. Any amino acid, or
combination of
amino acids, can be inserted in the insertions described herein to generate a
CasX variant
protein.
[00207] Any permutation of the substitution, insertion and deletion
embodiments described
herein can be combined to generate a CasX variant protein of the disclosure.
For example, a
CasX variant protein can comprise at least one substitution and at least one
deletion relative to a
reference CasX protein sequence, at least one substitution and at least one
insertion relative to a
reference CasX protein sequence, at least one insertion and at least one
deletion relative to a
44
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
reference CasX protein sequence, or at least one substitution, one insertion
and one deletion
relative to a reference CasX protein sequence.
1002081 A CasX variant comprises some or all of the following domains: a non-
target strand
binding (NTSB) domain, a target strand loading (TSL) domain, a helical I
domain, a helical 11
domain, an oligonucleotide binding domain (OBD), and a RuvC DNA cleavage
domain (the
latter which may be deleted in a catalytically dead CasX variant), described
more fully, below.
In some embodiments, the at least one modification of the CasX variant protein
comprises a
deletion of at least a portion of one domain of the reference CasX protein,
including the
sequences of SEQ ID NOS:1-3. In some embodiments, the deletion is in the
NTSBD, TSLD,
Helical I domain, Helical II domain, ODD, or RuvC DNA cleavage domain. In some
embodiments, the CasX variant comprises at least one modification in the NTSB
domain. In
some embodiments, the CasX variant comprises at least one modification in the
TSL domain. In
some embodiments, the at least one modification in the TSL domain comprises an
amino acid
substitution of one or more of amino acids Y857, 5890, or 5932 of SEQ ID NO:2.
In some
embodiments, the CasX variant comprises at least one modification in the
helical I domain. In
some embodiments, the at least one modification in the helical I domain
comprises an amino
acid substitution of one or more of amino acids S219, L249, E259, Q252, E292,
L307, or D318
of SEQ ID NO:2. In some embodiments, the CasX variant comprises at least one
modification in
the helical II domain. In some embodiments, the at least one modification in
the helical II
domain comprises an amino acid substitution of one or more of amino acids
D361, L379, E385,
E386, D387, F399, L404, R458, C477, or D489 of SEQ 1D NO:2. In some
embodiments, the
CasX variant comprises at least one modification in the ODD domain. In some
embodiments, the
at least one modification in the ODD comprises an amino acid substitution of
one or more of
amino acids F536, E552, T620, or 1658 of SEQ ID NO:2. In some embodiments, the
CasX
variant comprises at least one modification in the RuvC DNA cleavage domain.
In some
embodiments, the at least one modification in the RuvC DNA cleavage domain
comprises an
amino acid substitution of one or more of amino acids K682, 6695, A708, V711,
D732, A739,
D733, L742, V747, F755, M771, M779, W782, A788, G791, L792, P793, Y797, M799,
Q804,
S819, or Y857 or a deletion of amino acid P793 of SEQ ID NO:2.
[00209] In some embodiments, the CasX variant comprises at least one
modification compared
to the reference CasX sequence of SEQ ID NO:2 is selected from one or more of:
(a) an amino
acid substitution of L379R; (b) an amino acid substitution of A7081C; (c) an
amino acid
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
substitution of T620P; (d) an amino acid substitution of E385P; (e) an amino
acid substitution of
Y857R; (f) an amino acid substitution of I658V; (g) an amino acid substitution
of F399L; (h) an
amino acid substitution of Q252K; (i) an amino acid substitution of L404K; and
(j) an amino
acid deletion of P793.
[00210] The CasX variant proteins of the disclosure have an enhanced ability
to efficiently edit
and/or bind target DNA, when complexed with a gNA as an RNP, utilizing PAM TC
motif,
including PAM sequences selected from TTC, ATC, GTC, or CTC, compared to an
RNP of a
reference CasX protein and reference gNA. In the foregoing, the PAM sequence
is located at
least 1 nucleotide 5' to the non-target strand of the protospacer having
identity with the targeting
sequence of the gNA in a assay system compared to the editing efficiency
and/or binding of an
RNP comprising a reference CasX protein and reference gNA in a comparable
assay system. In
one embodiment, an RNP of a CasX variant and gNA variant exhibits greater
editing efficiency
and/or binding of a target sequence in the target DNA compared to an RNP
comprising a
reference CasX protein and a reference gNA in a comparable assay system,
wherein the PAM
sequence of the target DNA is TTC. In another embodiment, an RNP of a CasX
variant and
gNA variant exhibits greater editing efficiency and/or binding of a target
sequence in the target
DNA compared to an RNP comprising a reference CasX protein and a reference gNA
in a
comparable assay system, wherein the PAM sequence of the target DNA is ATC. In
another
embodiment, an RNP of a CasX variant and gNA variant exhibits greater Siting
efficiency
and/or binding of a target sequence in the target DNA compared to an RNP
comprising a
reference CasX protein and a reference gNA in a comparable assay system,
wherein the PAM
sequence of the target DNA is CTC. In another embodiment, an RNP of a CasX
variant and
gNA variant exhibits greater editing efficiency and/or binding of a target
sequence in the target
DNA compared to an RNP comprising a reference CasX protein and a reference gNA
in a
comparable assay system, wherein the PAM sequence of the target DNA is GTC. In
the
foregoing embodiments, the increased editing efficiency and/or binding
affinity for the one or
more PAM sequences is at least 1.5-fold greater or more compared to the
editing efficiency
and/or binding affinity of an RNP of any one of the CasX proteins of SEQ ID
NOS:1-3 and the
gNA of Table 2 for the PAM sequences.
[00211] All variants that improve one or more functions or characteristics of
the CasX variant
protein when compared to a reference CasX protein described herein are
envisaged as being
within the scope of the disclosure. Exemplary improved characteristics of the
CasX variant
46
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
embodiments include, but are not limited to improved folding of the variant,
improved binding
affinity to the gNA, improved binding affinity to the target nucleic acid,
improved ability to
utilize a greater spectrum of PAM sequences in the editing and/or binding of
target DNA,
improved unwinding of the target DNA, increased editing activity, improved
editing efficiency,
improved editing specificity, increased percentage of a eukaryotic genome that
can be efficiently
edited, increased activity of the nuclease, increased target strand loading
for double strand
cleavage, decreased target strand loading for single strand nicking, decreased
off-target cleavage,
improved binding of the non-target strand of DNA, improved protein stability,
improved
protein:gNA (RNP) complex stability, improved protein solubility, improved
protein:gNA
(RNP) complex solubility, improved protein yield, improved protein expression,
and improved
fusion characteristics, as described more fully, below. In some embodiments,
the RNP of the
CasX variant and the gNA variant exhibit one or more of the improved
characteristics that are at
least about 1.1 to about 100,000-fold improved relative to an RNP of the
reference CasX protein
of SEQ ID NO:1, SEQ ID NO:2, or SEQ ID NO:3 and the gNA of Table 2, when
assayed in a
comparable fashion. In other cases, the one or more improved characteristics
of an RNP of the
CasX variant and the gNA variant are at least about 1.1, at least about 10, at
least about 100, at
least about 1000, at least about 10,000, at least about 100,000-fold or more
improved relative to
an RNP of the reference CasX protein of SEQ ID NO:1, SEQ ID NO:2, or SEQ ID
NO:3 and the
gNA of Table 2. In other cases, the one or more of the improved
characteristics of an RNP of the
CasX variant and the gNA variant are about 1.1 to 100,00-fold, about 1.1 to
10,00-fold, about
1.1 to 1,000-fold, about 1.1 to 500-fold, about 1.1 to 100-fold, about 1.1 to
50-fold, about 1.1 to
20-fold, about 10 to 100,00-fold, about 10 to 10,00-fold, about 10 to 1,000-
fold, about 10 to 500-
fold, about 10 to 100-fold, about 10 to 50-fold, about 10 to 20-fold, about 2
to 70-fold, about 2
to 50-fold, about 2 to 30-fold, about 2 to 20-fold, about 2 to 10-fold, about
5 to 50-fold, about 5
to 30-fold, about 5 to 10-fold, about 100 to 100,00-fold, about 100 to 10,00-
fold, about 100 to
1,000-fold, about 100 to 500-fold, about 500 to 100,00-fold, about 500 to
10,00-fold, about 500
to 1,000-fold, about 500 to 750-fold, about 1,000 to 100,00-fold, about 10,000
to 100,00-fold,
about 20 to 500-fold, about 20 to 250-fold, about 20 to 200-fold, about 20 to
100-fold, about 20
to 50-fold, about 50 to 10,000-fold, about 50 to 1,000-fold, about 50 to 500-
fold, about 50 to
200-fold, or about 50 to 100-fold, improved relative to an RNP of the
reference CasX protein of
SEQ ID NO:1, SEQ 1D NO:2, or SEQ ID NO:3 and the reference gNA of SEQ ID NOS:
4-16 as
set forth in Table 2, when assayed in a comparable fashion. In other cases,
the one or more
47
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
improved characteristics of an RNP of the CasX variant and the gNA variant are
about 1.1-fold,
1.2-fold, E3-fold, 1.4-fold, 1.5-fold, 1.6-fold, 1.7-fold, 1.8-fold, 1.9-fold,
2-fold, 3-fold, 4-fold,
5-fold, 6-fold, 7-fold, 8-fold, 9-fold, 10-fold, 11-fold, 12-fold, 13-fold, 14-
fold, 15-fold, 16-fold,
17-fold, 18-fold, 19-fold, 20-fold, 25-fold, 30-fold, 40-fold, 45-fold, 50-
fold, 55-fold, 60-fold,
70-fold, 80-fold, 90-fold, 100-fold, 110-fold, 120-fold, 130-fold, 140-fold,
150-fold, 160-fold,
170-fold, 180-fold, 190-fold, 200-fold, 210-fold, 220-fold, 230-fold, 240-
fold, 250-fold, 260-
fold, 270-fold, 280-fold, 290-fold, 300-fold, 310-fold, 320-fold, 330-fold,
340-fold, 350-fold,
360-fold, 370-fold, 380-fold, 390-fold, 400-fold, 425-fold, 450-fold, 475-
fold, or 500-fold
improved relative to an RNP of the reference CasX protein of SEQ ID NO:!, SEQ
ID NO:2, or
SEQ ID NO:3 and the gNA SEQ ID NOS: 4-16 as set forth in Table 2, when assayed
in a
comparable fashion. An exemplary improved characteristic includes improved
editing
efficiency. In some embodiments, an RNP comprising a CasX variant protein and
a gNA of the
disclosure, at a concentration of 20 pM or less, is capable of cleaving a
double stranded DNA
target with an efficiency of at least 80%. In some embodiments, the RNP at a
concentration of
20 pM or less, is capable of cleaving a double stranded DNA target with an
efficiency of at least
40%, at least 50%, at least 60%, at least 70%, at least 80%, at least 85%, at
least 90% or at least
95%. In some embodiments, the RNP at a concentration of 50 pM or less, 40 pM
or less, 30 pM
or less, 20 pM or less, 10 pM or less, or 5 pM or less, is capable of cleaving
a double stranded
DNA target with an efficiency of at least 40%, at least 50%, at least 60%, at
least 700/s, at least
80%, at least 85%, at least 90% or at least 95%. The improved editing
efficiency of the CasX
variant, in combination with the gNA of the disclosure, make them well-suited
for inclusion in
the XDP of the disclosure
[00212] The term "CasX variant" is inclusive of variants that are fusion
proteins; i.e., the CasX
is "fused to" a heterologous sequence. This includes CasX variants comprising
CasX variant
sequences and N-terminal, C-terminal, or internal fusions of the CasX to a
heterologous protein
or domain thereof
[00213] In some embodiments, the CasX variant protein comprises between 400
and 2000
amino acids, between 500 and 1500 amino acids, between 700 and 1200 amino
acids, between
800 and 1100 amino acids or between 900 and 1000 amino acids.
[00214] In some embodiments, the CasX variant protein comprises one or more
modifications
comprising a region of non-contiguous residues that form a channel in which
gNA:target DNA
complexing occurs. In some embodiments, the CasX variant protein comprises one
or more
48
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
modifications comprising a region of non-contiguous residues that form an
interface which binds
with the gNA. For example, in some embodiments of a reference CasX protein,
the helical I,
helical II and OBD domains all contact or are in proximity to the gNA:target
DNA complex, and
one or more modifications to non-contiguous residues within any of these
domains may improve
function of the CasX variant protein.
[00215] In some embodiments, the CasX variant protein comprises one or more
modifications
comprising a region of non-contiguous residues that form a channel which binds
with the non-
target strand DNA. For example, a CasX variant protein can comprise one or
more modifications
to non-contiguous residues of the NTSBD. In some embodiments, the CasX variant
protein
comprises one or more modifications comprising a region of non-contiguous
residues that form
an interface which binds with the PAM. For example, a CasX variant protein can
comprise one
or more modifications to non-contiguous residues of the helical I domain or
OBD. In some
embodiments, the CasX variant protein comprises one or more modifications
comprising a
region of non-contiguous surface-exposed residues. As used herein, "surface-
exposed residues"
refers to amino acids on the surface of the CasX protein, or amino acids in
which at least a
portion of the amino acid, such as the backbone or a part of the side chain is
on the surface of the
protein. Surface exposed residues of cellular proteins such as CasX, which are
exposed to an
aqueous intracellular environment, are frequently selected from positively
charged hydrophilic
amino acids, for example arginine, asparagine, aspartate, glutamine,
glutamate, histidine, lysine,
serine, and threonine. Thus, for example, in some embodiments of the variants
provided herein,
a region of surface exposed residues comprises one or more insertions,
deletions, or substitutions
compared to a reference CasX protein. In some embodiments, one or more
positively charged
residues are substituted for one or more other positively charged residues, or
negatively charged
residues, or uncharged residues, or any combinations thereof In some
embodiments, one or
more amino acids residues for substitution are near bound nucleic acid, for
example residues in
the RuvC domain or helical I domain that contact target DNA, or residues in
the OBD or helical
II domain that bind the gNA, can be substituted for one or more positively
charged or polar
amino acids.
1002161 In some embodiments, the CasX variant protein comprises one or more
modifications
comprising a region of non-contiguous residues that form a core through
hydrophobic packing in
a domain of the reference CasX protein. Without wishing to be bound by any
theory, regions that
form cores through hydrophobic packing are rich in hydrophobic amino acids
such as valine,
49
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
isoleucine, leucine, methionine, phenylalanine, tryptophan, and cysteine. For
example, in some
reference CasX proteins, RuvC domains comprise a hydrophobic pocket adjacent
to the active
site. In some embodiments, between 2 to 15 residues of the region are charged,
polar, or base-
stacking. Charged amino acids (sometimes referred to herein as residues) may
include, for
example, arginine, lysine, aspartic acid, and glutarnic acid, and the side
chains of these amino
acids may form salt bridges provided a bridge partner is also present Polar
amino acids may
include, for example, glutamine, asparagine, histidine, serine, threonine,
tyrosine, and cysteine.
Polar amino acids can, in some embodiments, form hydrogen bonds as proton
donors or
acceptors, depending on the identity of their side chains. As used herein,
"base-stacking"
includes the interaction of aromatic side chains of an amino acid residue
(such as tryptophan,
tyrosine, phenylalanine, or histidine) with stacked nucleotide bases in a
nucleic acid. Any
modification to a region of non-contiguous amino acids that are in close
spatial proximity to
form a functional part of the CasX variant protein is envisaged as within the
scope of the
disclosure.
I CasX Variant Proteins with Domains from Multiple Source Proteins
[00217] Also contemplated within the scope of the disclosure are XDP
comprising chimeric
CasX proteins comprising protein domains from two or more different CasX
proteins, such as
two or more naturally occurring CasX proteins, or two or more CasX variant
protein sequences
as described herein. As used herein, a "chimeric CasX protein" refers to a
CasX containing at
least two domains isolated or derived from different sources, such as two
naturally occurring
proteins, which may, in some embodiments, be isolated from different species.
For example, in
some embodiments, a chimeric CasX protein comprises a first domain from a
first CasX protein
and a second domain from a second, different CasX protein. In some
embodiments, the first
domain can be selected from the group consisting of the NTSB, TSL, helical I,
helical H, OBD
and RuvC domains. In some embodiments, the second domain is selected from the
group
consisting of the NTSB, TSL, helical I, helical H, OBD and RuvC domains with
the second
domain being different from the foregoing first domain. For example, a
chimeric CasX protein
may comprise an NTSB, TSL, helical I, helical II, OBD domains from a CasX
protein of SEQ
ID NO: 2, and a RuvC domain from a CasX protein of SEQ ID NO: 1, or vice
versa. As a further
example, a chimeric CasX protein may comprise an NTSB, TSL, helical II, OBD
and RuvC
domain from CasX protein of SEQ ID NO: 2, and a helical I domain from a CasX
protein of
SEQ ID NO: 1, or vice versa. Thus, in certain embodiments, a chimeric CasX
protein may
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
comprise an NTSB, TSL, helical II, OBD and RuvC domain from a first CasX
protein, and a
helical I domain from a second CasX protein. In some embodiments of the
chimeric CasX
proteins, the domains of the first CasX protein are derived from the sequences
of SEQ ID NO: 1,
SEQ ID NO: 2 or SEQ ID NO: 3, and the domains of the second CasX protein are
derived from
the sequences of SEQ ID NO: 1, SEQ ID NO: 2 or SEQ ID NO: 3, and the first and
second
CasX proteins are not the same. In some embodiments, domains of the first CasX
protein
comprise sequences derived from SEQ ID NO: 1 and domains of the second CasX
protein
comprise sequences derived from SEQ ID NO: 2. In some embodiments, domains of
the first
CasX protein comprise sequences derived from SEQ ID NO: 1 and domains of the
second CasX
protein comprise sequences derived from SEQ ID NO: 3. In some embodiments,
domains of the
first CasX protein comprise sequences derived from SEQ ID NO: 2 and domains of
the second
CasX protein comprise sequences derived from SEQ ID NO: 3. In some
embodiments, the CasX
variant is selected of group consisting of CasX variants with sequences of SEQ
II) NO: 102,
113, 114, 115, 103, 104, 105, 106, 107, 108, 109, and 110, as described in
Table 1.
[00218] In some embodiments of the XDP systems, a CasX variant protein
comprises at least
one chimeric domain comprising a first part from a first CasX protein and a
second part from a
second, different CasX protein. As used herein, a "chimeric domain" refers to
a domain
containing at least two parts isolated or derived from different sources, such
as two naturally
occurring proteins or portions of domains from two reference CasX proteins.
The at least one
chimeric domain can be any of the NTSB, TSL, helical I, helical II, OBD or
RuvC domains as
described herein. In some embodiments, the first portion of a CasX domain
comprises a
sequence of SEQ ID NO: 1 and the second portion of a CasX domain comprises a
sequence of
SEQ ID NO: 2. In some embodiments, the first portion of the CasX domain
comprises a
sequence of SEQ ID NO: 1 and the second portion of the CasX domain comprises a
sequence of
SEQ ID NO: 3. In some embodiments, the first portion of the CasX domain
comprises a
sequence of SEQ ID NO: 2 and the second portion of the CasX domain comprises a
sequence of
SEQ ID NO: 3. In some embodiments, the at least one chimeric domain comprises
a chimeric
RuvC domain. As an example of the foregoing, the chimeric RuvC domain
comprises amino
acids 661 to 824 of SEQ ID NO: 1 and amino acids 922 to 978 of SEQ ID NO: 2.
As an
alternative example of the foregoing, a chimeric RuvC domain comprises amino
acids 648 to
812 of SEQ ID NO: 2 and amino acids 935 to 986 of SEQ ID NO: 1. In some
embodiments, a
CasX protein comprises a first domain from a first CasX protein and a second
domain from a
51
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
second CasX protein, and at least one chimeric domain comprising at least two
parts isolated
from different CasX proteins using the approach of the embodiments described
in this paragraph.
In the foregoing embodiments, the chimeric CasX proteins having domains or
portions of
domains derived from SEQ ID NOS: 1, 2 and 3, can further comprise amino acid
insertions,
deletions, or substitutions of any of the embodiments disclosed herein.
[00219] In some embodiments of the XDP systems, a CasX variant protein
comprises a
sequence of SEQ ID NOS: 21-233, 343-345, 350-353, 355-367 or 388-397 as set
forth in Tables
1, 7, 8, 9 or 11. In some embodiments, a CasX variant protein consists of a
sequence of SEQ ID
NOS: 21-233 as set forth in Table 1. In other embodiments, a CasX variant
protein comprises a
sequence at least 60% identical, at least 65% identical, at least 70%
identical, at least 75%
identical, at least 80% identical, at least 81% identical, at least 82%
identical, at least 83%
identical, at least 84% identical, at least 85% identical, at least 86%
identical, at least 86%
identical, at least 87% identical, at least 88% identical, at least 89%
identical, at least 89%
identical, at least 90% identical, at least 91% identical, at least 92%
identical, at least 93%
identical, at least 94% identical, at least 95% identical, at least 96%
identical, at least 97%
identical, at least 98% identical, at least 99% identical, at least 99.5%
identical to a sequence of
SEQ ID NOS: 21-233, 343-345, 350-353, 355-367 or 388-397 as set forth in
Tables 1, 7, 8, 9 or
11. In other embodiments, a CasX variant protein comprises a sequence set
forth in Table 1, and
further comprises one or more NLS disclosed herein at or near either the N-
terminus, the C-
terminus, or both. It will be understood that in some cases, the N-terminal
methionine of the
CasX variants of the Tables is removed from the expressed CasX variant during
post-
translational modification.
Table 1: CasX Variant Sequences
Descriptio Amino Acid
Sequence
n*
TSL, MQEIKRINKI
RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
Helical I, PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
Helical II, QPASKKIDQNKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGK
OBD and AYTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIH
RuvC
VTRESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIILE
domains
HQKVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVI
from SEQ VVVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERIDANEVDVVVVDMVCNV
ID NO:2 KKLINEKKEDGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQFG
and an
DLLLHLEKKHGEDWGIWYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSK
52
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
NTSB
AALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQIONYGDLRGKPFAIEAE
domain
NSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
from SEQ NRFYTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLE
ID NO: 1 TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN
IKPMNLI
GI DRGEN IPAVIALTDPEGC PLS RFKDSLGNPTHILRIGESYKEKQ RTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQ'YTRMEDVVLTAKLAYEGLSKTYLSKTLAQ'YTS
KTCSNCGFTITSADYDRVLEKLKKTATG1NMTTINGKELKVEGQITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVWKPAV (SEQ ID NO: 21)
NTSB, MQE
IKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
Helical I, PEN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYWE EFQ KDPVGLMS RVA
Helical II, Q PAP KN IDQ RKLIPVKDGNERLTSSGFACSO CCQ PLYVYKLEQVN DKGKP
OBD and HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RuvC
RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHQ
domains
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
from SEQ NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVCNVKKL
ID NO:2 INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
and a TSL LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
domain TDVVLRAKAS FVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAI
EAENSIL
from SEQ DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
ID NO: 1 . YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIE KTLYNRRTRQDEPALFVALTFERREVLDSSNIKPM NLIGIDR
GEN IPAVIALTDP EGC P LSRFKDSLGNPTH ILRIGESYKEKQ RTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGROGKRTFMAERQ'YTRMED1NLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITTADYDGMLVRLKKTSDGWATTLNNKELKAEGQITYYNRYKRQT
VEKELSAELDRLSEESGNNDISKVVI-KGRRDEALFLLKKRFSHRPVQEQFV
CLDCGHEVHADEQAALNIARS1NLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 22)
TSL,
MEKRINKIRKKLSADNATKPVSRSGPMKTUVRVMTDDLKKRLEKRRKKP
Helical I, EVMPQVISNNAANNLRMLLDDYTKMKEAILQVYWQEFKDDHVGLMCKFA
Helical II, Q PAP KN IDQ RKLIPVKDGNERLTSSGFAC SQ CCQ PLYVYKLEQVN DKGKP
OBD and HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLOKFGQRALDFYSIHVT
RuvC KESTHPVKPLAQ
IAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQKV
domains VKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
from SEQ WVNLNLWQKLKLSRDDAKPLLRLKGFPSFPVVERRENEVDWVVNTIN EVK
lD NO:1
KLIDAKRDMGRVFVVSGVTAEKRNTILEGYNYLPNENDHKKREGSLENPICK
and an
PAKRQFGDLLLYLEKKYAGDWGKVFDEAVVERIDKKIAGLTSHIEREEARN
NTSB
AEDAQSKAVLTDWLRAKASFVLERLKEMDEKEFYACEIQLQKVVYGDLRG
domain
NPFAVEAENRVVDISGFSIGSDGHSIQYRNLLA1NKYLENGKREFYLLMNY
from SEQ GKKGR IRFTDGTDIKKSGKWQGLLYGGGKAKVIDLTFDP DDE QV ILP LAF
ID NO:2
GTRQGREFIVVNDLLSLETGLIKLANGRVIEKTIYNKKIGRDEPALFVALTFE
RREVVDPSN IKPVNLIGVDRGEN IPAVIALTDPEGCP LP EFKDSSGGPTDI L
53
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
RIGEGYKEKQRAIQAAKEVEQRRAGGYSRKFASKSRNLADDMVRNSARD
LFYHAVTHDAVLVFENLSRGFGRQGKRTFMTERQYTKMED1NLTAKLAYE
GLTSKTYLSKTLAQYTSKTCSNCGFTITTADYDGMLVRLKKTSDGWATTL
NNKELKAEGQ ITYYNRYKRQTVEKELSAELDRLSEESGNNDIS KVVTKGRR
DEALF LLKKRFS H RPVQ EQ FVC LDCG H EVHADEQAALN IARSWLFL NS NS
TEFKSYKSGKQPFVGAWQAFYKRRLKEV1NKPNA (SEQ ID NO: 23)
NTSB,
MEKRINKIRKKLSADNATKPVSRSGPMKTLLVRVMTDDLKKRLEKRRKKP
Helical I, EVMPQVISNNAAN NLRMLLDDYTKMKEAILQVYWQ EFKDDHVGLMCKFA
Helical II, QPASKKIDQNKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGK
ODD and AYTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIH
RuvC VTKESTHPVKPLAQ IAG N RYASGPVG KALSDACMGTIASFLS
KYQ D I IIEHQ
domains KWKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVR
from SEQ MWVN L N LWQ KLKLSRD DAKP L LRLKG F PS FPVVE RR EN EVDWVVNTIN EV
ID NO:! KKLIDAKR DMGRVFWSGVTAE KR NTILEGYNYLP N E N
DHKKREGS LE N PK
and an TSL KPAKRQFGDLLLYLEKKYAGDWGKVFDEAVVERIDKKIAGLTSH IE RE EAR
domain NAEDAQS KAVLTDWLRAKASFVLE RLKEM DE KEFYACE IQ LQ
KWYG D LR
from SEQ GN PFAVEAENRVVD ISG FS I GS DGHS IQYRNLLAWKYLENGKREFYLLMN
ID NO:2. YOKKG RIRFTDGTDIKKSGKVVQ GLLYGGGKAKVIDLTFDP DD EQ LI I LPLAF
GTRQGREFIVVNDLLSLETGLIKLANGRVIEKTIYNKKIGRDEPALFVALTFE
RREVVDPS N IKPVN L IGVD RGE N IPAVIALTDP EGC PLP EF KDS SGG PTD IL
RIGEGYKEKQRAIQAAKEVEQ RRAGGYSR KFAS KS RN LADDMVRNSARD
LFYHAVTHDAVLVFENLSRGFGRQGKRTFMTERQYTKMEDVVLTAKLAYE
GLTSKTYLSKTLAQYTSKTCSNCGFTITSADYDRVLEKLKKTATGWMTTIN
GKELKVEGQITYYN RYKRQNVVKDLSVELDRLSEESVNNDISSWTKGRSG
EALSLLKKRFS HRPVQEKFVCLNCGFETHADEQAALN IARSWLFLN S N ST
EFKSYKSGKQPFVGAVVQAFYKRRLKEVWKPNA (SEQ ID NO: 24)
NTSB, MQE IKRI N KI RRRLVKDS NTKKAGKTG PM
KTLLVRVMTPDLRE RLE NLRKK
TSL, PEN I PQ P ISNTS RAN LN KLLTDYTEM KKAILHVYVVE EFQ
KD PVG LMS RVA
Helical I, QPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCQPLYVYKLEQVNDKGKP
Helical II HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
and OBD RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVAS FLTKYQD I I LEHQ
domains KVIKKNEKRLANLKDIASANGLAFPKITLPPQ PHTKEG I EAYN
NVVAQ IVIVVV
SEQ ID NLNLWQ KLKI GRDEAKP LQ RLKGFPS F PLVE ROAN
EVDVVWDMVCNVKKL
NO:2 and INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
an LH LE KKHG E DWG KVYDEAWERIDKKVEG LS KH I KLEE
ERRS EDAQ S KAAL
exogenous TDWLRAKASFVI EGLKEADKDEFCRCELKLQ KVVYG D LRGKPFA I EAENSI L
RuvC DISGFSKOYNCAFIWQKDGVKKLNLYLI I NYFKGG KLRFKKI KP
EAFEAN RF
domain or a YTV I NKKSG E IVPMEVN FN FDD P N LIILPLAFGKRQG RE FIVVN D LLS
LETGS
portion LKLANGRVIEKTLYNRRTRODEPALEVALTFERREVLDSSN I KPVN
LIGVDR
thereof GEN IPAVIALTD PEGG P LP E FKDSS GG PTDILR IG
EGYKEKQ RAI QAAKEVE
from a OR RAGGYSRKFAS KS RN LADDMVRNSAR DLFYHAVTH
DAVLVFE N LSRG
second
FGRQGKRTFMTERQYTKMEDVVLTAKLAYEGLTSKTYLSKTLAQYTSKTC
CasX SNCGFTITSADYD RVLEKLKKTATGVVMTTI NC KELKVEGQ
ITYYNRYKRQ
protein.
54
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
NVVKDLSVELD RLS EESVN N D IS SVVTKGRSGEALS L LKKRFSH R PVQ E KF
VCLNCGFETHA (SEQ ID NO: 25)
MQEIKRINKI RRR LVKDSNTKKAG KTGPM KTLLVRVMTPD LRE R LEN LRKK
PEN I PQ P ISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
Q PAPKN I DQRKL IPVKDGN ERLTSSGFACS QCCQ P LYVYKL EQVN DKGKP
HTNYFG RC NVS EH ERL I LLSP H KP EAN D ELVTYSLGKFGQ RALDFYSI HVT
RES N HP VK P LEQIGG N SCASG PVG KALS DAC MGAVAS FLTKYQ D I ILEHQ
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIINV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVCNVKKL
I NE KKEDG KVFWQN LAGYKRQ EALRPYLSSEE DRKKGKKFARYQ FGDLL
LH LE KKH GEDWGIWYD EAWE RI D KKVE GLSKH IKL EE ERRS E DAQS KAAL
TDWLRAKASFVIEGLKEADKDEFC RCELKLQ KVVYGDLRG KP FAI EAE N S IL
DISGFSKQYNCAFIWQKDGVKKLN LYL II NYFKGGKLRFKKIKP EAFEAN R F
YTVI N KKSGEIVP MEVN FN FDDP N LI I LP LAFG KRQ GRE F IWN DLLSLETGS
LKLANGRVI E KTLYNRRTRQ D EPALFVALTFE RREVLDS SN I KP M N L IG IDR
G EN I PAVIALTD PEGCP LS RFKDSLGN PTH I LR IG ESYKE KQ RTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDINLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGINMTTINGKELKVEGQ ITYYNRYKRQ N
VVKDLSVE L DR LSE ESVN N D I SSVVTKGRSGEALSLLKKRF SH RPVQ EKFV
CLNCGFETHA (SEQ ID NO: 26)
NTSB,
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
TSL, PEN I PQ P ISN NAANN LRM LLD DYTKM KEAILQVYINQ
EFKDDHVGLMCKFA
Helical II, Q PAPKN I DQRKL IPVKDGN ERLTSSGFACS QCC Q P LYVYKL EQVN DKGKP
OBD and HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RuvC KESTHPVKPLAQ IAGN RYASGPVG KALS DACM GTIAS F LS
KYQ D II I EH Q KV
domains
VKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
from SEQ INVNLNLWQ KLKLS RDDAKP LL RLKGFPS FP LVER QAN EVDWINDMVCNV
ID NO:2 KKL I N EKKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQFG
and a D LLLH LEKKHGEDWG KVYD EAVVE RID KKVEG LSKH IKL
E E ERRSEDAQS K
Helical I
AALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVY'GDLRGKPFAIEAE
domain N S ILD ISG FSKQYN CAF IWQ KDGVKKLN LYL I INYF
KGG KLRF KKI KPEAFEA
from SEQ N RFYTVI N KKSGE IVP M EVN FN FDD PN L I I LPLAFGKRQG REFIVVN D L LS
L E
ID NO: 1 TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN I
KPM N L I
G IDRGEN IPAV IALTD PEGCP LS RFKDSLGN PTH I LR I GESYKEKQRTIQAK
KEVEQ RRAGGYSRKYASKAKN LAD DMVR NTARD L LYYAVTQ DAM LIFE N
LSRGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ ITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSWLFLRSQ EYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVWKPAV (SEQ ID NO: 27)
NTSB,
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
TSL, PEN I PQ P
ISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
Helical I, QPAPKNIDORKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
OBD and HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RuvC RESNH PVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQ
DIILEHQ
domains
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
from SEQ NLNLWQKLKIGRDEAKPLQRLKGFPSFPVVERRENEVDWVVNTINEVKKLI
ID NO:2 DAKRDMGRVFWSGVTAEKRNTILEGYNYLPNENDHKKREGSLENPKKPA
and a
KRQFGDLLLYLEKKYAGDWGKVFDEAVVERIDKKIAGLTSHIEREEARNAE
Helical II DAQSKAVLTDVVLRAKASFVLE RLKEM D EKE FYAC E I Q LQ KVVYGDLRG N P
domain
FAVEAENSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIK
from SEQ P EAFEANRFYTVINKKSGE IVP MEVNFNFDDPNLI I LPLAFGKRQGREFIWN
ID NO:
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIK
PMNLIGIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQR
TIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAM
LIFENLSROFGROGKRTFMAERQYTRMED1NLTAKLAYEGLSKTYLSKTLA
QYTSKTCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYY
NRYKRQNWKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHR
PVQEKFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNT
DKRAFVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 28)
NTSB,
MISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQPAPKN
TSL,
IDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKPHTNYFG
Helical I, RC NVSEH ERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVTRESNH P
Helical II VKPLEQ IGG N SCASG PVGKALS DACMGAVASFLTKYQ D I ILEHQ KVI KKNE
and RuvC KRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVI1NVNLNLWQ
domains
KLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVWVDMVCNVKKLINEKKE
from a first DGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLLLHLEKK
CasX
HGEDWGIWYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAALTDWLR
protein and AKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENRVVDISGF
an
SIGSDGHSIQYRNLLAVVICYLENGKREFYLLMNYGICKGRIRFTDGTDIKKS
exogenous GKINQGLLYGGGKAKVIDLTFDPDDEQUILPLAFGTROGREFIWNDLLSLE
OBD or a TGLIKLANGRVIEKTIYNKKIGRDEPALFVALTFERREVVDPSNIKPMNLIGI
part thereof DRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKE
from a
VEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLS
second
RGFGRQGKRTFMAERQ'YTRMEDWLTAKLAYEGLSKTYLSKTLAQ'YTSKT
CasX
CSNCGFTITSADYDRVLEKLKICATGWMTTINGKELKVEGQITYYNRYKR
protein
QNWKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFV
ETINQSFYRKKLKEVVVKPAV (SEQ ID NO: 29)
MEKRINKIRKKLSADNATKPVSRSGPMKTLLVRVMTDDLKKRLEKRRKKP
EVMPQVISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
PAPKNIDORKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKPH
TNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVTR
ESNHPVKP LEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQ DI ILEHQK
VI KKN EKRLAN LKD IASANGLAF PKITLP PQ P HTKE G I EAYN NVVAQ IVIWV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWVVDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LH LE KKHGE DWG KVYDEAVVE RI D KKVEG LS KH I KLE E E RRS EDAQ S KAAL
56
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
TDWLRAKASFVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GEN IPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYAS KAKN LADDMVRNTARDLLYYAVTQ DAM LI FE N LSRG
FGRQGKRTFMAERQ'YTRMEDVVLTAKLAYEGLSICYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGINMTTINGKELKVEGQITYYNRYKRQN
VVKDLSVE LDR LSE ESVNN D IS SWTKG RSG EALS LLKKRFSH RPVQ EKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WOSFYRKKLKEVVVKPAV (SEQ ID NO: 30)
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PEN IPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
QPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVAS FLTKYQDI I LEK/
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDWLRAKAS FVI EG LKEADKD E FCRC E LKLQ KWYG D LRGKPFA I EAEN RV
VDISGFSIGSDGHSIQYRNLLAVVKYLENGKREFYLLMNYGKKGRIRFTDGT
DIKKSGKWQGLLYGGGKAKVIDLTFDP DDEQ LI ILP LAFGTRQGREFIWN D
LLSLETGLIKLANGRVIEKTIYNKKIGRDEPALFVALTFERREVVDPSNIKPM
NLIGIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQ
AKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIF
ENLSRGFGROGKRTFMAERQ'YTRMEDWLTAKLAYEGLSKTYLSKTLAQY
TS KTCS NCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ ITYYNR
YKRQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPV
QEKFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDK
RAFVETWQSFYRKKLKEV1A/KPAV (SEQ ID NO: 31)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of L379R, PEN IPQP ISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
a
QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGORALDFYSIHVT
of C477K, RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVAS FLTKYQDI I LEHQ
a KVIKKNEKRLANLKDIASANGLAFPKITLPPQ PHTKEGIEAYNNVVAQ
IVIVVV
substitution NLNLWQ KLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVCNVKKL
of A708K, INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
a deletion LH LEKKHGEDWGIWYDEAWERIDKKVEGLS KH IKLEEERRSEDAQSKAAL
of P at
TDWLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGDLRGKPFAIEAENSIL
position
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
793 and a YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
substitution LKLANGRVIEKPLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
of T62013 GEN IPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
57
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
of SEQ ID QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
NO:2
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NC GFT ITSADYD RVLEKLKKTATGINMTTI NG KELKVE GQ ITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALN IARSWLFLRSQ EYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 32)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of M771A PE N I PQ P ISNTS RAN L N KLLTDYTEM KKAI LHVYVVE EF Q KDPVGLMSRVA
of SEQ ID QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
NO:2.
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVAS FLTICYQDI I LEHQ
KVI KKN EKRLAN LKDIASANGLAF PKITLP PQ PHTKEG I EAYN NVVAQ IVIWV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDINVVDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDWGIWYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDWLRAKASFVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GEN IPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKQRTIQAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFAAERQ'YTRMEDWLTAKLAYEGLPSKTYLSKTLAQ'YTSKTC
SNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQ ITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPA (SEQ ID NO: 33)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of L379R, PEN I PQ P ISNTS RAN LN KLLTDYTEM KKAI LHVYVVE EFQ KD PVG LMS RVA
a
QPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCQPLYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of A708K, RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVAS FLTICYQD11 LEHQ
a deletion KVI KKN EKRLAN LKDIASANGLAF PKITLP PQ PHTKEG I EAYN NVVAQ IVIVVV
of P at
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDININDMVCNVKKL
position
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
793 and a LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
substitution TDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
of D732N DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
of SEQ ID YTV I NKKSGE IVPMEVNFN FDD PN LIILPLAFGKRQG RE FIWN D LLS LETGS
NO:2. LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN
IKPMNLIGIDR
GEN IPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLANDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQ'YTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQN
VVKDLSVELDRLSEESVNNDISSVITTKGRSGEALSLLKKRFSHRPVQEKFV
58
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
CLNCGFETHADEQAALNIARSWLFLRSOEYKKYOTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 34)
substitution MQ E I KRI N KI RRRLVKDSNTKKAG KTGPM laLLVRVMTPD LRE RLENLRKK
of W782Q PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYINEEFOKDPVGLMSRVA
of SEQ ID OPAPKNIDORKLIPVKDGNERLTSSGFACSOCCePLYVYKLEQVNDKGKP
NO:2.
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGORALDFYSIHVT
RESNHPVKPLEGIGGNSCASGPVGKALSDACMGAVASFLTKYQD1ILEHO
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDININDMVCNVKKL
INEKKEDGKVFWONLAGYKRQEALLPYLSSEEDRKKGKKFARYOFGDLLL
HLEKKHGEDWGIWYDEAVVERIDKKVEGLSKHIKLEEERRSEDAGSKAAL
TDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIINNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKORTIOAAKEVE
ORRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTODAMLIFENLSRG
FGRQGKRTFMAEROYTRMEDQLTAKLAYEGLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGINMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHR PVQ EKF
VCLNCGFETHADEQAALNIARSINLFLRSOEYKKYOTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 35)
substitution MOEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of M77 1 Q PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYINEEFQKDPVGLMSRVA
of SEQ ID QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
NO:2
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYODIILEHO
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQ IVIWV
NLNLWOKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVIANDMVCNVKKL
INEKKEDGKVFVVQNLAGYKROEALLPYLSSEEDRKKGKKFARYOFGDLLL
HLEKKHGEDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDINLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
DISGFSKOYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIINNDLLSLETGS
LKLANGRVIEKTLYNRRTRODEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIOAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTODAMLIFENLSRG
FGRQGKRTFQAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGINMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHR PVe EKF
VCLNCGFETHADEGAALNIARSINLFLRSQEYKKYOTNKTTGNTDKRAFVE
TWOSFYRKKLKEVVVKPAV (SEQ ID NO: 36)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of R4581 PENIPOP ISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
59
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
and a Q PAP KN IDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVN
DKGKP
substitution HTNYFG RCNVS EH E RLI LLSP H KP EAN D ELVTYS LG KFGQ RALDFYSI HVT
of A739V RESNH PVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQ DIILEHQ
of SEQ ID KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQ IVIWV
NO:2. NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWVVDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDWGIWYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDINLIAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
D ISGFSKQYNCAFIWQ KDGVKKLN LYLI I NYFKGGKLR FKKIKPEAFEANR F
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIE KTLYNRRTRQDEPALFVALTFERREVLDSSNIKPM NLIGIDR
GEN IPAVIALTDP EGC P LSRFKDSLGNPTH I LR IGESYKEKQ RTIQAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNIVRDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVILTAKLAYEGLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQ EKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 37)
L379R, a MQE IKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
substitution PEN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYVVE EFQKDPVGLMS RVA
of A708K, Q PAP KN IDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVN DKGKP
a deletion HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of P at
RESNHPVKPLEIDIGGNSCASGPVGKALSDACMGAVASFLTKYQD1ILEHQ
position KVI KKN EKRLAN LKD IASAN GLAFP KITLPP Q PHTKEG I
EAYN NVVAQ IVIWV
793 and a NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
substitution INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
of M771N LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
of SEQ ID TDWLRAKAS FVIEGLKEADKDE FCRCE LKLQKVVYGDLRGKPFAI EAE NS I L
NO:2
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIE KTLYNRRTRQDEPALFVALTFERREVLDSSNIKPM NLIGIDR
G EN I PAVIALTD P EGC P LSRFKDS LG N PTH I LRIG ESYKE KQ RTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFNAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQN
VVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
VVQSFYRKKLKEVVVKPAV (SEQ ID NO: 38)
substitution MOE IKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of L379R, PEN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYVVE EFQKDPVGLMS RVA
a Q PAP KN IDQ R KLIPVKDGNERLTSSGFAC S QCCQ
PLYVYKLEQVN DKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of A708K, RESNH PVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQ DIILEHQ
a deletion KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQ IVIWV
of P at NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWVVDMVCNVKKL
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
position
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
793 and a LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
substitution TDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
of A739T DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
of SEQ ID YWINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
NO:2
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GEN I PAVIALTD PEGCP LS RFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTTRDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 39)
substitution MQE I KR I N KI RRR LVKDSNTKKAGKTGPMKTLLVRVMTPD LR ER LE N LRKK
of L379R, PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFOKDPVGLMSRVA
a 0
PAPKNIDQRKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of C477K, RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYODIILEHO
a KVIKKN EKRLAN LKD IASANG LAFP KITLPPQ PHTKEG I
EAYN NVVAQ IVIWV
substitution NLNLWQKLKIGRDEAKPLQRLKGF PSFPLVEROANEVDWVVDMVCNVKKL
of A708K, INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
a deletion LH LE KKHGE DWGIWYD EAWE R I DKKVEGLS KH I KLE EE RRS EDAQS KAAL
of P at
TDV'VLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGSLRGKPFAIEAENSIL
position
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
793 and a YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
substitution LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
of D4895 GEN I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
of SEQ ID Q RRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
NO: 2.
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 40)
substitution MQE I KR I N KI RRRLVKDSNTKKAGKTGPMKTLLVRVMTPD LRERLE N LRKK
of L379R, PENIPQPISNTSRANLNKLLTD'YTEMKKAILHVYWEEFQKDPVGLMSRVA
a
CIPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCQPLYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of C477K, RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHQ
a KVIKKN EKRLAN LKD IASANG LAFP KITLPPQ PHTKEG I
EAYN NVVAQ IVIWV
substitution NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVEROANEVDWWDMVCNVKKL
of A708K, INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
a deletion LHLE KKHGEDWGIONDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
of P at
TDVVLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGDLRGKPFAIEAENSIL
position
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
793 and a 'YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
61
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
substitution LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
of D732N GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
of SEQ ID QRRAGGYSRKYASKAKNLANDMVRNTARDLLYYAVTQDAMLIFENLSRG
NO:2. FGRQ
GKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NG KE L KVE GQ ITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLN CG FE THAD EQAALN IARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 41)
substitution MQ E I KRI N KIRRRLVKDS NTKKAGKTG P M KTLLVRVMTP DLRERL EN LRKK
of V71 1K PEN IP Q P I SNTS RANL N KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMSRVA
of SEQ ID QPAPKN I DQ RKL I PVKDG N ER LTSSGFACSQC CQ P LYVYKLE QVN DKGKP
NO:2.
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHQ
KVI KKN E KR LANL KD IASANG LAF P KITLP PQ PHTKEG I EAYNNVVAQ IVIWV
N LN LWQ KLKI GRD EAKP LQ R L KGF PS FP LVE RQAN EVDVVVVDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
H LE KKHG EDWG KVY DEAVVERI DKKVEG LS KH I KLE EERRS EDAQS KAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDD P N LI I L PLAFGKRQ GRE FIWNDL LSL ETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEIWRTIQAAKEKE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLPSKTYLSKTLAQYTS KTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTI NGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNN DISSWTKG RSG EALS LL KKR FS H RPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNICTGNTDKRAFVE
TVVQSFYRKKLKEV'VVKPAV (SEQ ID NO: 42)
substitution MQ E I KRI N KIRRRLVKDS NTKKAGKTG P M KTLLVRVMTP DLRERL EN LRKK
of L379R, PEN IP Q P I SNTS RANL N KLLTDYTEM KKAI LHVYWEE FQ KDPVG LMS RVA
a QPAPKN I DQ RKL I PVKDG N ER LTSSGFACSQC C Q P
LYVYKLE QVN DKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of C477K, RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQD1ILEHQ
a KVIKKNEKRLANLKD IASANG LAF P KITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
substitution NLN LWQ KLKI GRD EAKP LQ R L KGF PS FP LVE ROAN EVDVVVVDMVCNVKKL
of A708K, INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
a deletion LH L E KKHGE DWGINY DEAWER I DKKVEGLSKH I KLEE ERRS EDAQS KAAL
of P at
TDVVLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGDLRGKPFAIEAE NS IL
position DISGFSKQYNCAFIWQKDGVKKLNLYLI
INYFKGGKLRFKKIKPEAFEANRF
793 and a YIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
substitution LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
of Y797L GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
of SEQ ID QRRAGGYSRICY'ASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
NO: 2. FGRQ
GKRTFMAERQYTRMEDVVLTAKLAYEGLSKTLLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NC KE L KVE GO ITYYNRYKRQ N
62
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 43)
119:
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
substitution PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
of L379R, Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
a
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
substitution RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
of A708K KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
and a N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDVVVVDMVC NVKKL
deletion of INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
P at LHLEKKHGE DWGKVYDEAVVER I DKKVE GLSKH IKLEE
ERRSEDAQSKAAL
position TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ
KVVYGDLRGKPFAIEAE NS I L
793 of SEQ DISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAFEANR F
ID NO:2. YTV INKKSGEIVPMEVNFN FDD P NLI I LPLAFGKRQ GRE FIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQD EPALFVALTFE RR EVLDSSN IKPMNLIGID R
GENIPAVIALTDPEGC P LSRFKDSLGNPTH ILR IGESYKEKQRTIQAKKEVE
ORRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQ'YTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NC KE LKVE GO ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WOSFYRKKLKEVVVKPAV (SEQ ID NO: 44)
substitution MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLE N LRKK
of L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
a Q PAPKN IDQRKL I PVKDGNER LTSSGFACSQCCQP
LYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEH ERL ILLSP HKP EAN D E LVTYSLGKFGQRALD FYS I HVT
of C477K, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
a KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIVVV
substitution NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
of A708K, INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
a deletion LHLEKKHGE DWGIWYDEAWER I DKKVE GLSKH IKLEE ERRSEDAOSKAAL
of P at TDWLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGDLRGKPFAIEAE
NS IL
position
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
793 and a YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIWNDLLSLETGS
substitution LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
of M77 1N GENIPAVIALTDPEGCP LSRFKDSLGNPTH ILR IGESYKEKQRTIQAKKEVE
of SEQ ID QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
NO: 2.
FGROGKRTFNAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CO FTITSADYD RVLE KLKKTATGVVMTTI NO KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
C LN CG FETHAD EQAALN IARSWLFLRSQ EYKKYQTN KTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 45)
63
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
substitution MQ E I KR I N KIRRRLVKDS NTKKAGKTG P M KTLLVRVMTP DLRERL EN LRKK
of A708K, PEN IPQ P I SNTS RANL N KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
a deletion Q PAPKN I DQRKL I PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
of P at
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
position RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
793 and a KVIKKNEKRLANLKD IASANGLAFPKITLPPQPHTKEGIEAYNNVVAQ IVIWV
substitution N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN EVDWVVDMVC NVKKL
of E386S INEKKEDGKVFWQNLAGYKRIDEALLPYLSSESDRKKGKKFARYQFGDLLL
of SEQ ID H LE KKHG EDVVG KVYDEAVVER I DKKVEG LS KH I KLE EER RS EDAQS KAAL
NO:2. TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ
KVVYGDLRGKPFAIEAENS I L
DISGFSKOYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVI N KKSG E IVP MEVNFN FDD P N LI I L PLAFGKR Q GR E FIWNDL LSL ETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RREVLDSS N IKPM NLIG I D R
GEN I PAVIALTDPEGCP LS RFKDSLGN PTH ILR I GESYKE KQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CC FTITSADYD RVLE KLKKTATGVVMTTI NC KE LKVE GQ ITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLN CGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 46)
substitution MQ E I KR I N KIRRRLVKDS NTKKAGKTG P M KTLLVRVMTP DLRERL EN LRKK
of L379R, PE N IPQ P I SNTS RANL N KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
a Q PAPKN I DQRKL I PVKDG N ER LTSSGFACSQCCQ P
LYVYKLEQVN DKGKP
substitution HTNYFGRCNVSEHERL ILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of C477K, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
a KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIWV
substitution N LN LWQ KLKI GRD EAKP LORLKGFPS FP LVE ROAN EVDWNDMVC NVKKL
of A708K INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
and a LH L E KKHGE DWGKVYDEAWER I DKKVE GLS KH IKLEE
ERRS EDAQS KAAL
deletion of TDVVLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGDLRGKPFAIEAE NS IL
P at
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
position YTVI N KKSG E IVP MEVNFN FDD P N LI I L PLAFGKRQ
GRE FIWNDL LSL ETGS
793 of SEQ LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RREVLDSS N IKPM NLIG I D R
ID NO:2, GEN I PAVIALTDPEGCP LS RFKDSLGN PTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGROGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NC KE LKVE GO ITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLN CG FETHAD EQAALN IARSVVLFL RSQ EYKKYQTN KTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 47)
substitution MQ E I KR I N KIRRRLVKDS NTKKAGKTG P M KTLLVRVMTP DLRERL EN LRKK
of L792D PE N IPQ P I SNTS RANL N KLLTDYTEM KKAI LHVYWEE FQ KDPVG LMSRVA
of SEQ Q PAPKN I DQRKL I PVKDG N ER LTSSGFACSQCCQ P
LYVYKLEQVN DKGKP
NO:2.
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNH PVKP LEG IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H
64
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
NLNLVVQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDWGIWYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQICNYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGDPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 48)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of G791F PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
of SEQ ID OPAPKNIDORKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKOKP
NO:2.
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHQ
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
NLNLVVQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWNDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDVVGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEIWRTIQAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEFLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARS1NLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 49)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of A708K, PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
a deletion QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
of P at
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKEGORALDFYSIHVT
position
RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHQ
793 and a KVIKKNEKRLANLKD IASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
substitution NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVEROANEVDWNDMVCNVKKL
of A739V INEKKEDGKVFWQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDVVGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
of SEQ ID DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
NO:2. YTVINKKSGEIVPMEVNFN FDDPNLI I
LPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTVRDLLYYAVTQDAMLIFENLSRG
FGROGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGVVMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 50)
substitution MQ E IKR I N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
of L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
a
QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of A708K, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
a deletion KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIVVV
of P at NLNLWQKLKIGRDEAKP LQRLKGFPS FP LVE ROAN
EVDWVVDMVC NVKKL
position
INEKKEDGKVFVVONLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
793 and a LHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
substitution TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAENSIL
of A739V DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
of SEQ ID 'YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIWNDLLSLETGS
NO:2. LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN
IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
ORRAGGYSRKYASKAKNLADDMVRNIVRDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGVVMTTI NC KE LKVE GO ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 51)
substitution MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
of C477K, PEN IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
a Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSOCCCIPLYV'YKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of A708K RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQDI1LEHQ
and a KVI KKN E KR LANLKD !MANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
deletion of NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
P at
INEKKEDGKVFVVQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
position HLEKKHG
EDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
793 of SEQ TDVVLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGDLRGKPFAIEAE NS IL
ID NO:2. DISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
66
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYYNRYKRQN
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 52)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of L249I PEN !POP I SNTS RANL N KLLTDYTEM KKAI LHVYWEE FQ
KDPVG LMS RVA
and a
QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of M77 IN RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIIIEHQK
of SEQ ID VIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIVVV
NO:2.
NLNLVVQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWWDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDVVGIONDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFNAERQYTRMEDVVLTAKLAYEGLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOS FYRKKLKEVVVKPAV (SEQ ID NO: 53)
substitution MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
of V747K PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
of SEQ ID QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
NO:2.
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQIGGNSCASGPVGKALSDACIVIGAVASFLTKYQD1ILEHQ
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDWWDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDVVGIONDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDWLRAKASFVIEGLKEADKDEFCRCELKLQIONYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKORTIQAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAKTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
67
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWQSFYRKKLKEVVVKPAV (SEQ ID NO: 54)
substitution MQ E I KR I N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
of L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
a
OPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCIDPLYVYKLEQVNDKGKP
substitution HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
of C477K, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
a KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIVVV
substitution NLNLVVQKLKIGRDEAKPLQRLKGFPSFPLVEROANEVDWNDMVCNVKKL
of A708K, INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
a deletion LH LE KKHGE DWGKVYDEAWER I DKKVE GLSKH IKLEE ER RS EDAQSKAAL
of P at
TDVVLRAKASFVIEGLKEADKDEFKRCELKLQKVVYGDLRGKPFAIEAE NS IL
position
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
793 and a YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIWNDLLSLETGS
substitution LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
of M779N GEN I PAVIALTDPEGCP LS RFKDSLGN PTH ILR I GESYKE KORTIQAKKEVE
of SEQ ID QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
NO: 2.
FGROGKRTFMAERQYTRNEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NC KE LKVE GQ ITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 55)
MQ EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
F755M Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIVVV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVEROANEVDWVVDMVCNVKKL
INEKKEDGIWFVVONLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDVVGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWY'GD LRG KP FAI EAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAAKEVE
ORRAGGYSRICY'ASKAKNLADDMVRNTARDLLYYAVTODAMLIMENLSRG
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGOITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOSFYRKKLKEVVVICPAV (SEQ ID NO: 56)
429: MQ
EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
68
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN EVDVIANDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ K1NYGD LRG KP FAIEAENS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
ORRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGINMTTI NG KE LKVE GQ ITYYN RRKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVINKPAV (SEQ ID NO: 57)
430: MQ E IKRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V KVI KKN E KR LANLKD !MANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAOSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RR EVLDSS N IKPM NLIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGN PTH ILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDINLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVIVITTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSVITIKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWQSFYRKKLKEVWKPAV (SEQ ID NO: 58)
431: MQ E IKRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
KVI KKN E KRLANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERGANEVDVVVVDMVCNVKKL
69
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSENDRKKGKKFARYQFGDLL
E386N LHLEKKHGE DWGKVYDEAWER I DKKVE GLSKH IKLEE ER
RSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTV INKKSGE IVPMEVNFN FDD P NLI I LPLAFGKRQ GRE FIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQD EPALFVALTFE RREVLDSSN IKPMNLIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEV
EQRRAGGYS RKYASKAKNLADDMVRNTARDLLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDVILTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTI N GKELKVEGQITYYN RRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOSFYRKKLKEVWKPAV (SEQ ID NO: 59)
432: MQ EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I PVKDGNER LTSSGFACSQCCQP
LYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H 0
I658V, KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNWAQ IVIWV
L404K N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDWNDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
KHLEKKH GE DWGKVYD EAVVE R IDKKVEGLSKH IKLEEERRSEDAQSKAA
LTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSI
LD I SG FS KQYNCAFIWQ KDGVKKLNLYL I I NYF KGGKLRF KKI KPEAFEAN R
FYIVINKKSGEIVPMEVNFNFDDPNLI ILP LAFGKRQ GRE Fl WN DLLSLETG
SLKLANGRVIEKTLYNRRTRQ DE PALFVALTFER REVLDSSN I KPMNLIGV
DRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKE
VEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLS
RGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKT
C SNC GFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGQ ITYYNRRKR
QNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 60)
433: MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQVRALDFYSIHV
Y857R, TRESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQD I
ILE H
I658V, Q KVIKKN E KRLAN LKD IASANGLAFPKITLPPQ P HTKEG I
EAYN NVVAQ IVI
AV192
1NVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWVVDMVCNV
KKL I N EKKEDGKVFWQ N LAGYKRQ EALRPYLSS E E DRKKG KKFARYQFG
DLLLHLEKKH GE DWGKVYD EAWER IDKKVEGLSKH IKLEEERRSEDAQSK
AALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGE IVPMEVNFNFD DP NLI ILPLAFGKRQGRE FIWND LLSLE
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTC S N CGFTITSADYD RVLEKLKKTATGVVMTTI NG KE LKVEGQ ITYYN RRK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVVVKPAV (SEQ ID NO: 61)
434: MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERL EN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIWV
L404K,
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
E386N
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSENDRKKGKKFARYQFGDLL
KHLEKKHGEDWGIWYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAA
LTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSI
LD I SG FS KQYNCAFIWQ KDGVKKLNLYL I I NYF KGGKLRF KKIKPEAFEAN R
FYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLETG
SLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN I KPMNLIGV
DRGEN IPAVIALTDPEGGPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKE
VEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLS
RGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKT
CSNCGFTITSADYD RVLEKLKKTATGVVMTTI NG KELKVEGQ ITYYN RRKR
QNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFV
ETWQSFYRKKLKEVWKPAV (SEQ ID NO: 62)
435: MQEIKRINKIRRRLVKDSNTKKAGICTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ
IGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHQ
I658V, KVI KKN E KRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
F399L
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVWDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLGDLL
LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFGRCELKLQ KVVYGDLRG KP FAIEAE NS IL
DISGFSKOYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGN PTH ILRIGESYKEKORTIOAKKEV
EQ R RAGGYS RKYASKAKN LADDMVR NTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
71
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVWKPAV (SEQ ID NO: 63)
436: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PEN IP Q P I SNTS RANL N KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
F399L, N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDVVWDMVC NVKKL
E386N
INEKKEDGKVFWQNLAGYKRQEALRPYLSSENDRKKGKKFARYQLGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RR EVLDSS N IKPM NLIGVD
RGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVWKPAV (SEQ ID NO: 64)
437: MQ E IKRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLE N LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIVVV
F399L,
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
C477S
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLGDLL
LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAOSKAAL
TDWLRAKASFVIEGLKEADKDEFSRCELKLQKVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKROGREFIVVNDLLSLETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RR EVLDSS N IKPM NLIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGN PTH ILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAEROYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWQSFYRKKLKEVWKPAV (SEQ ID NO: 65)
72
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
438: MQ EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
F399L, N LN LVVQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDWWDMVC NVKKL
L404K INEKKEDGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
LGDLL
KHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKH IKLEEERRSEDAQSKAA
LTDWLRAKAS FVI E GLKEADKDE FC RC E LKLQ KVVYGDLRG KP FAI EAE N S I
LD I SG FS KOYNCAFIWQ KDGVKKLNLYL I I NYF KGGKLRF KKI KPEAFEAN R
FYTVINKKSGEIVPMEVNFNFDDPNLI ILP LAFGKRQ GREFI WN DLLSLETG
SLKLANGRVIEKTLYNRRTRQ DEPALFVALTFERREVLDSSN I KPMNLIGV
DRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKE
VEQ RRAGGYSRKYASKAKN LAD DMVRNTARDLLYYAVTQ DAMLI FE NLS
RGFGRQGKRTFMAE RQYTRM E DVVLTAKLAYE GLSKTYLSKTLAQYTS KT
C SNC GFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGQ ITYYN RRKR
QNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVINKPAV (SEQ ID NO: 66)
439: MQ EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIWV
F399L, N LN LWQ KLKI GRD EAKP LORLKGFPS FP LVE ROAN
EVDWWDMVC NVKKL
E386N,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSENDRKKGKKFARYQLGDLL
C477S, KHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKH
IKLEEERRSEDAQSKAA
L404K
LTDWLRAKASFVIEGLKEADKDEFSRCELKLQKVVYGDLRGKPFAIEAENSI
LD I SG FS KQYNCAFIWQ KDGVKKLNLYL I I NYF KGGKLRF KKI KPEAFEAN R
FYTVINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKROGREFIWN DLLSLETG
SLKLANGRVIEKTLYNRRTRQ DEPALFVALTFERREVLDSSN I KPMNLIGV
DRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKE
VEQ RRAGGYSRKYASKAKN LAD DMVRNTARDLLYYAVTQ DAMLI FEN LS
RGFGRQGKRTFMAE RQYTRM EDWLTAKLAYEGLSKTYLSKTLAQYTS KT
C SNC GFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGO ITYYN RRKR
QNVVKDLSVELDRLSEESVNNDISSWIKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 67)
440: MQ EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG WS RVA
A708K, Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
73
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
I658V, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIWV
F399L,
NLNLWOKLKIGRDEAKPLORLKGFPSFPLVERDANEVDWWDMVCNVKKL
Y797L
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLGDLL
LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ KlNYGD LRG KP FAI EAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGVD
RGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTLLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 68)
441: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A7081C,
OPAPKNIDORKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKOKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIVVV
F399L, N LN LVVQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDWWDMVC NVKKL
Y797L,
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSENDRKKGKKFARYQLGDLL
E386N
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGD LRG KP FAI EAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGN PTH ILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDVILTAKLAYEGLSKTLLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVINKPAV (SEQ ID NO: 69)
442: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGORALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIVVV
F399L, N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDWNDMVC NVKKL
Y797L,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSENDRKKGKKFARYQLGDLL
E386N,
KHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAA
LTDVVLRAKASFVIEGLKEADKDEFSRCELKLQKVVYGDLRGKPFAIEAENSI
74
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
C477S, LD I SG FS KQYNCAFIWQ KDGVKKLNLYL I I NYF
KGGKLRF KKI KPEAFEAN R
L4041( FYTVINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKRQGREFIWN
DLLS LETG
SLKLANGRVIEKTLYNRRTRQ DE PALFVALTFERREVLDSSN I KPMNLIGV
DRG EN IPAVIALTDPEGCPLS RF KDS LG NPTH ILR IG ESYKEKQ RTI QAKKE
VEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLS
RGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTLLSKTLAQYTSKT
C SNC GFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGQ ITYYNRRKR
QNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVVVISPAV (SEQ ID NO: 70)
443: MQ EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIVVV
Y797L N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDWNDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LH LE KKHGE DWGKVYDEAWER I DKKVE GLSKH IKLEE ERRS EDAQSKAAL
TDWLRAKAS FVI EG LKEAD KD E FC RC ELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSG E IVPMEVNFN FDD P NLI I LPLAFGKRQ GRE FIVVNDLLSLETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RR EVLDSS N IKPM NLIGVD
RGEN IPAVIALTD P EGCP LS R FKDS LG N PTH ILRIGESYKEKQRTIQAKKEV
EQRRAGGYS RKYASKAKNLADDMVRNTARDLLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTLLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTI N GKELKVEGQITYYN RRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOSFYRKKLKENNVKPAV (SEQ ID NO: 71)
444: MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMSRVA
A708K, Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H O
I658V, KVI KKN E KR LANLKD !MANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
Y797L,
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVIDVVVVDMVCNVKKL
L404K
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
KH LE KKH GE DWGKVYD EAVVE R IDKKVEGLSKH IKLEEERRSEDAQSKAA
LTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSI
LD I SG FS KQYNCAFIWQ KDGVKKLNLYL I I NYF KGGKLRF KKI KPEAFEAN R
FYIVINKKSGEIVPMEVNENFDDPNLI ILPLAFGKRQGREFIVVN DLLS LETG
SLKLANGRVIEKTLYNRRTRQ DE PALFVALTFER REVLDSSN I KPMNLIGV
DRG EN IPAVIALTDPEGCPLS RF KDS LG NPTH ILRIG ESYKEKQ RTI QAKKE
VEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLS
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
RGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTLLSKTLAQYTSKT
CSNCGFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGQ ITYYN R R KR
QNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 72)
445: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IN) P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQKDPVG LMS RVA
A708K, QPAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQP
LYVYKLEQVNDKGKP
P793,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIWV
Y797L, N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDWWDMVC NVKKL
E386N
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSENDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGVD
RGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTLLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOSFYRKKLKEVWKPAV (SEQ ID NO: 73)
446: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQP
LYVYKLEQVNDKGKP
P793,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
I658V, KVI KKN E KRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIVVV
Y797L, N LN LWQ KLKI GRD EAKP LORLKGFPS FP LVE ROAN
EVDWWDMVC NVKKL
E386N,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSENDRKKGKKFARYQFGDLL
C477S,
KHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAA
L404K
LTDVVLRAKASFVIEGLKEADKDEFSRCELKLQKWYGDLRGKPFAIEAENSI
LD I SG FS KeYNCAFIWQ KDGVKKLNLYL I I NYF KGGKLRF KKI KPEAFEAN R
FYWINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKROGREFIWN DLLSLETG
SLKLANGRVIEKTLYNRRTRODEPALFVALTFERREVLDSSN I KPMNLIGV
DRGENIPAVIALTDPEGCPLSRAWSLGNPTHILRIGESYKEKORTIOAKKE
VEQ RRAGGYSRKYASKAKN LAD DMVRNTARDLLYYAVTO DAMLI FE NLS
RGFGROGKRTFMAERQYTRMEDWLTAKLAYEGLSKTLLSKTLAWTSKT
C SNC GFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGO ITYYN RRKR
Q NVVKDLSVELDRLSEESVN NDISSWTKGRSGEALSLLKKR FSHR PVC)EK
76
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVWKPAV (SEQ ID NO: 74)
447: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K,
OPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
E386N KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
NLNLVVQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWWDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSENDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGROGKRTFMAERQYTRMEDWLTAKLAYEGLSICTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGVVMTTI NG KE LKVE GQ ITYYN RRKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 75)
448: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K,
QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Y857R, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
E386N, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIWV
L404K NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVEROANEVDWVVDMVC
NVKKL
INEKKEDGKVFWONLAGYKRQEALRPYLSSENDRKKGKKFARYQFGDLL
KHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAA
LTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAENSI
LDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANR
FYTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVN DLLSLETG
SLKLANGRVIEKTLYNRRTRQ DEPALFVALTFERREVLDSSN I KP MNLIG ID
RGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEV
EQRRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAM LI FENLSR
GFGRQGKRTFIVIAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGOITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOS FYRKKLKEVVVKPAV (SEQ ID NO: 76)
449: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMSRVA
77
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
A708K,
QPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
D732N,
RESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHQ
E385P, KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVIWV
Y857R N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDVIANDMVC NVKKL
INEKKEDGKVFWQ NLAGYKRQEALRPYLSSP EDRKKGKKFARYQ FGDLL
LHLEKKHGEDWGKVYDEAWER I DKKVE GLSKH IKLEE ERRSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ K1NYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCP LSRFKDSLGNPTH ILR IGESYKEKQRTIQAKKEVE
0 RRAGGYSRKYAS KAKN LAN DMVRNTARDLLYYAVTQ DAM LI FEN LS RG
FGRQ GKRTFMAE RQYTRME DVVLTAKLAYEG LS KTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGINMTTI NG KE LKVE GQ ITYYNRRKRQN
VVKD LSVE LDRLS E ESVN ND IS SVVTKGRSGEALS LLKKRFSH RPVQE KFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVINKPAV (SEQ ID NO: 77)
450: MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I PVKDGNER LTSSGFACSQCCQP
LYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
D732N, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
E385P, KVI KKN E KR LANLKD !MANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
Y857R, N LN LWQ KLKI GRD EAKP LORLKGFPS FP LVE ROAN
EVDVINVDMVC NVKKL
I658V
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWER I DKKVE GLSKH IKLEE ERRSEDAOSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIVVNDLLSLETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RR EVLDSS N I KPM NLIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEV
EQRRAGGYS RKYASKAKN LAN DMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDINLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVIVITTINGKELKVEGQITYYNRRKRQ
NVVKDLSVE LDRLS E ESVNN DISSWTKG RSG EALS LLKKRFS H R PVQ E KF
VC LNC GFETHAD EQAALNIARSVVLFLRSQ EYKKYQTN KTTG NTD KRAFVE
TWOS FYRKKLKEVWKPAV (SEQ ID NO: 78)
451: MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I PVKDGNER LTSSGFACSQC C Q P
LYVYKLEQVNDKGKP
P793_, HTNYFGRCNVSEHERL ILLSP HKP
EANDELVTYSLGKFGQRALDFYSIHVT
D732N, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
E385P, KVI KKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
Y857R, N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE RQAN
EVDVVVVDMVC NVKKL
78
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQLGDLL
F399L
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGVD
RGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LAN DMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOSFYRKKLKEVWKPAV (SEQ ID NO: 79)
452: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
D732N, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
E385P, KVI KKN E KRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNWAQ IVIWV
Y857R, N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE RQAN
EVDWA/DMVC NVKKL
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPNDRKKGKKFARYQFGDLL
E386N
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKROGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGN PTH ILRIGESYKEKORTIOAKKEV
EQ RRAGGYS RKYASKAKN LAN DMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWOS FYRKKLKEVWKPAV (SEQ ID NO: 80)
453: MQ E IKRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMSRVA
A708K, Q PAPKN IDQRKL I PVKDGNER LTSSGFACSQCCQ P
LYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
D73N, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
E385P, KVI KKN E KRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIVVV
Y857R,
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVWDMVCNVKKL
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQFGDLL
L404K KHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKH
IKLEEERRSEDAQSKAA
LTDWLRAKAS FVI E GLKEADKDE FC RCE LKLQ KWYGDLRG KP FAI EAE N S I
LO I SG FS KQYNCAFIWQ KDGVKKLNLYL I I NYF KGGKLRF KKI KPEAFEAN R
FYTVINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKRQGREFIWN DLLSLETG
79
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
SLKLANGRVIEKTLYNRRTRQ DEPALFVALTFERREVLDSSN I KP MNLIGV
DRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKE
VEQRRAGGYSRKYASKAKNLANDMVRNTARDLLYYAVTQDAMLIFENLS
RGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKT
C SNC GFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGQ ITYYN RRKR
ONVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 81)
454: MCIEIKRINKIRRRLVKDSNTKKAGKTGPMKTUNRVMTPDLRERLENLRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQ P
LYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
T620P, RESNHPVKPLEQ
IGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHK
E385P, KVI KKN E KR LANLKD
IASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
Y857R,
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVCNVKKL
Q252K
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIVVNDLLSLETGS
LKLANGRVIEKPLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEIWRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGVVMTTI NG KE LKVE GQ ITYYN RRKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 82)
455: MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQ P
LYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGORALDFYSIHVT
T620P, RESNHPVKPLEQ
IGGNSCASGPVGKALSDACMGAVASFLTKYQDIILEHK
E385P, KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
Y857R,
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVWDMVCNVKKL
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQFGDLL
Q252K
LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKOYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIVVNDLLSLETGS
LKLANG RVIE KPLYN RRTRQ D E PALFVALTFE RR EVLDSS N IKP MN LIGVD
RGENIPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKORTIOAKKEV
EQRRAGGYS RKYASKAKN LADDMVR NTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVWKPAV (SEQ ID NO: 83)
456: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R, PEN IP Q P I SNTS RANL N KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
T620P, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H K
E385P, KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIWV
Y857R, N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDVVWDMVC NVKKL
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPNDRKKGKKFARYQFGDLL
E386N,
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
Q252K TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ
KWYGDLRGKPFAIEAE NS I L
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIVVNDLLSLETGS
LKLANG RVIE KPLYN RRTRQ D E PALFVALTFE RR EVLDSS N IKP MN LIGVD
RGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVWKPAV (SEQ ID NO: 84)
457: MQ E IKRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP DLRERLEN LRKK
L379R, PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
A708K, QPAPKN IDQRKLI
PVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
T620P, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H K
E385P, KVI KKN E KRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVIVVV
Y857R,
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQLGDLL
F399L,
LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAOSKAAL
Q252K TDWLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE
NS I L
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKROGREFIVVNDLLSLETGS
LKLANG RVIE KPLYN RRTRQ D E PALFVALTFE RR EVLDSS N IKP MN LIGVD
RGEN IPAVIALTDPEGCPLSRFKDSLGN PTH ILRIGESYKEKQRTIQAKKEV
EQ RRAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSR
GFGRQGKRTFMAEROYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TWQSFYRKKLKEVWKPAV (SEQ ID NO: 85)
81
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
458: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R,
PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
A708K,
OPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
P793
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
T620P, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H K
E385P, KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I
EAYNNVVAQ IVI WV
Y857R, N LN LVVQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN
EVDWVVDMVC NVKKL
I658V,
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQFGDLL
L404K, KH L E KKH GE DWGKVYD EAVVE R IDKKVEGLSKH
IKLEEERRSEDAQSKAA
Q252K
LTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSI
LDISGFSKOYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANR
FYN! N KKSGEIVPMEVN F N F DDP N LI I LP LAFG KRQ GRE Fl WN DLLS LETG
SLKLANGRVIEKPLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGV
DRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKE
VEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLS
RGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKT
CSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYYNRRKR
QNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVINKPAV (SEQ ID NO: 86)
459: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R,
PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
A708K,
QPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
T620P, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H 0
Y857R, KVI KKN E KR LANL KD IASANG LAF PKITLP PQ PHTKEG
I EAYNNVVAQ IVI WV
I658V, N LN LWQ KLKI GRD EAKP LORLKGFPS FP LVE ROAN
EVDWWDMVC NVKKL
E386N
INEKKEDGKVFWQNLAGYKRQEALRPYLSSENDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKD E FC RC ELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIVVNDLLSLETGS
LKLANGRVIEKPLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGVD
RGEN IPAVIALTD P EGCP LS R FKDS LG N PTH ILR I GESYKE KORTIQAKKEV
EQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSR
GFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRKRQ
NVVKDLSVELDRLSEESVNNDISSVITTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 87)
460: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R,
PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVA
A708K,
QPAPKNIDQRKLIPVKDGNERLTSSGFACSOCCQPLYVYKLEQVNDKGKP
P793_,
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
T620P, RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H K
82
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
E385P, KVIKKNEKRLANLKD
IASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
Q252K N LN LWQ KLKI GRD EAKP LQR LKGFPS FP LVE ROAN
EVDWWDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSPEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWER I DKKVE GLSKH IKLEE ER RSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ KlNYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIWNDLLSLETGS
LKLANGRVIEKPLYN RRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKORTIQAKKEVE
Q RRAGGYSRKYAS KAKN LAD DMVRNTARDLLYYAVTQ DAM LI FEN LS RG
FGRQ GKRTFMAE RQYTRME DVVLTAKLAYEG LS KTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGWMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 88)
278 QEIKRINKI RR R LVKDSNTKKAGKTGPM KTLLVRVMTPDLRER
LENLRKKP
EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYWE E FQ KDPVGLM SRVAQ
PAPKNIDORKLIPVKDONERLTSSGFACSOCCOPLYVYKLEQVNDKGKPH
TNYFGRC NVS EH E RL ILLSP H KP EAN D E LVTY8 LGKFGQ RALD FYSIHVTR
ESN HPVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQD I ILEHQK
VI KKN EKR LAN LKD IASAN GLAFP KITLPPQ PHTKEG I EAYN NVVAQ IVIVIN
N LN LVVQ KLKI GRD EAKP LQRLKGFP8 FP LVE ROAN EVDWWDMVC NVKKL
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWER I DKKVE GLSKH IKLEE ER RSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAFEANR F
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
Q RRAGGYSRKYAS KAKN LAD DMVRNTARDLLYYAVTQ DAM LI FEN LS RG
FGRQ GKRTFMAE RQYTRME DVVLTAKLAYEG LS KTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKD LSVE LDRLS E ESVN ND IS SVVTKGRSGEALS LLKKRFSH RPVQE KFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 89)
279 MQ
EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE FQ KDPVG LMS RVA
Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGORALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN EVDWNDMVC NVKKL
IN EKKEDGKVFWQ N LAGYKRQEALR PYLS SE E D R KKGKKFARYQ FG DLL
LHLEKKHGEDWGKVYDEAWER I DKKVE GLSKH IKLEE ERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
83
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVI N KKSG E IVP MEVNFN FDD P N LI I L PLAFGKR Q GR E FIWNDL LSL ETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RREVLDSS N IKPM NLIG I D R
GEN I PAVIALTDPEGCP LS RFKDSLGN PTH ILR I GESYKE KQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGROGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NC KE L KVE GQ ITYYNRYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 90)
280 MQ E IKR I N KIRRRLVKDS NTKKAGKTG P M KTLLVRVMTP
DLRERL EN LRKK
PE N IP Q P I SNTS RANL N KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
Q PAPKN I DQRKL I PVKDG N ER LTSSGFACSQCCQ P LYVYKLEQVN DKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANL KD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIVVV
N LN LWQ KLKI GRD EAKP LQRL KGF PS FP LVE ROAN EVDWNDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LH L E KKHGE DINGKVYDEAWER I DKKVE GLS KH IKLEE ERRS EDAQS KAAL
TDINLRAKAS FVI EG LKEADKD E FC RC ELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVI N KKSG E IVP MEVNFN FDD P N LI I L PLAFGKRQ GRE FIVVNDL LSL ETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RR EVLDSS N IKPM NLIG I D R
GEN I PAVIALTDPEGCP LS RFKDSLGN PTH ILR I GESYKE KQRTIQAKKEVE
ORRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTODAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTI NC KE L KVE GO ITYYNRYKRQN
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 91)
285 MQ E I KRI N KIRRRLVKDS NTKKAGKTG P M KTLLVRVMTP
DLRERL EN LRKK
PE N IP Q P I SNTS RANL N KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
Q PAPKN I DQRKL I PVKDG N ER LTSSGFACSQCCQ P LYVYKLEQVN DKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANL KD !MANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVIDVINVDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LH L E KKHGE DWGKVYDEAWER I DKKVE GLS KH IKLEE ERRS EDAQS KAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVI N KKSG E IVP MEVNFN FDD P N LI I L PLAFGKRQ GRE FIWNDL LSL ETGS
LKLANG RVIE KTLYNRRTRQD EPALFVALTFE RR EVLDSS N IKPM NLIG I D R
GEN I PAVIALTDPEGC P LS RFKDSLGN PTH ILR I GESYKE KQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
84
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGWMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 92)
286
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQ P LYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
NLNLVVQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWWDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CC FTITSADYD RVLE KLKKTATGVVMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 93)
287
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE FQ KDPVG LMS RVA
Q PAPKN IDQRKL I PVKDGNER LTSSGFACSQCCQ P LYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDWWDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAENSIL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKROGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKORTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGVVMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVWKPAV (SEQ ID NO: 94)
288
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
OPAPKNIDQRKLIPVKDGNERLTMSSGFACSOCCOPLYVYKLEQVNDKG
KPHTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIH
VTRESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIILE
HQKVIKKNEKRLANLKDIASANGLAFPKITLPPQ P HTKEG I EAYN NVVAQ IVI
VVVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVWDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFG
DLL LH L E KKH GE DWGKVYD EAWER IDKKVEGLSKH IKLEE ERRS EDAQS K
AALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAE
N S I LD ISGF SKQYN CAF IVVQ KDGVKKL N LYLI I NYFKGG KLRFKKIKP EAF EA
NRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSROFGROGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALN IARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVVVKPAV (SEQ ID NO: 95)
290
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANL KD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIVVV
N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN EVDWVVDMVC NVKKL
INEKKEDGKVFWONLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKAS FVI EG LKEAD KD E FC RC ELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
ORRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTODAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
NCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQN
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 96)
291
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFOKDPVGLMSRVA
86
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQ P LYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN EVDVIANDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ K1NYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
ORRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGINMTTI NC KE LKVE GO ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 97)
293 MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP
DLRERLEN LRKK
PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQP LYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANLKD !MANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVINVDMVCNVKKL
INEKKEDGKVFVVQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAOSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CC FTITSADYD RVLE KLKKTATGWMTTI NC KE LKVE GO ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
C LN CG FETHAD EQAALN IARSWLFLRSQ EYKKYQTN KTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 98)
300 MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP
DLRERLEN LRKK
PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQ P LYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERGANEVDVVVVDMVCNVKKL
87
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CO FTITSADYD RVLE KLKKTATGVVMTTI NO KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
CLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 99)
492
MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQP LYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNWAQ IVIWV
N LN LWQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN EVDWA/DMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKROGREFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKORTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGVVMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
C LN CG FETHAD EQAALN IARSWLFLRSQ EYKKYQTN KTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 100)
493 MQ E I KRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP
DLRERLEN LRKK
PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE FQ KDPVG LMS RVA
Q PAPKN IDQRKL I PVKDGNERLTSSGFACSQCCQ P LYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVIKKNEKRLANLKD IASANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIVVV
NLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWVVDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDWLRAKASFVIEGLKEADKDEFCRCELKLQ KWYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKROGREFIVVNDLLSLETGS
88
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCP LSRFKDSLGNPTH ILRIGESYKEKQRTIQAKKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
FGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSKTCS
N CG FTITSADYD RVLE KLKKTATGVVMTTI NG KE LKVE GQ ITYYN RYKRQ N
VVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKFV
C LN CG FETHAD EQAALN IARSVVLFLRSQ EYKKYQTN KTTGNTDKRAFVET
WQSFYRKKLKEVVVKPAV (SEQ ID NO: 101)
387:
QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
NTSB EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E
FQ KDPVGLM SRVAQ
swa from PASKKIDQ NKLKPEM DE KGNLTTAGFACSQCGQP LFVYKLEQVSEKGKA
p
SEQ ID YTNYFGRCNVAE
HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
NO:1 TRESNHPVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQD I
ILEH
Q KVIKKN E KR LAN LKD IASANGLAFPKITLPPQ P HTKEG I EAYN NVVAQ IVI
WVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDINVVDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSI LDISGFSKQYNCAF IWQKDGVKKLN LYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGE IVPMEVNFNFDDP NLI ILPLAFGKRQGREFIINNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPMNL I
GIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTCS N CGFTITSADYD RVLEKLKKTATGVVMTTI NG KE LKVEGQ ITYYN RYK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYOTNKTTGNTDKRAF
VETWQSFYRKKLKEVWKPAV (SEQ ID NO: 102)
395:
QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
Helical 1B EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM SRVAQ
swa from PAPKN IDQRKL IPVKDGN ERLTSSGFACSQCCQP LYVYKLEQVNDKGKPH
p
SEQ ID TNYFGRC NVS EH E
RLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVTK
NO:1 ESTH PVKP LAQIAGN RYASGPVG KALSDACM GTIASFLS KYQ
DI I IEHQKVV
KGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRMW
VN LN LWQ KLKLSRDDAKPLLRLKG FPSF PLVE ROAN EVDWVVDMVC NVK
KLI N EKKEDGKVFWQ N LAGYKRQ EALR PYLS SE E D RKKGKKFARYQFGD
LLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDAQSKA
ALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAEN
S I LD I SG FSKQYNCAF IWQ KDGVKKLN LYL I I NYFKGG KLRFKKIKP EAFEAN
RFYTVINKKSGE IVPMEVNFNFDDPNLI ILPLAFGKRQGREFIWNDLLSLET
GSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN I KPMNLIGI
DRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKE
VEQ RRAGGYSR KYASKAKN LAD DMVRNTAR DLLYYAVTQ DAMLI FE NLS
RGFGRQGKRTFMAE RQYTRM EDWLTAKLAYEGLSKTYLSKTLAQYTS KT
CSNCGFTITSADYD RVLEKLKKTATGWMTTI NG KELKVEGQ ITYYN RYKR
89
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
QNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEV1NKPAV (SEQ ID NO: 103)
485: QEIKRINKIRRRLVKOSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
Helical 1B EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM SRVAQ
swa from PAPKN IDQRKL IPVKDGN ERLTSS GFACSQCCQ P LYVYKLEQVNDKGKPH
p
SEQ ID TNYFGRC NVS EH E RL ILLSP H KP EAN D E LVTYS
LGKFGQ RALD FYSIHVTK
NO:1
ESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQKVV
KGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRMW
VNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVWVDMVCNVK
KLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLGD
LLLHLEKKHG EDWG KVYDEAVVE RID KKVEG LS KH IKLE EERRSE DAQSKA
ALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAEN
SILDISGFSKQYNCAF IWQ KDGVKKLNLYL I INYFKGGKLRFKKIKP EAFEAN
RFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLET
GSLKLAN GRVIEKTLYNR RTRQ D EPALFVALTFERREVLDSSN I KPMNLIG
VDRGENIPAVIALTDPE GCPLSRFKDSLGN PTH ILR IGESYKEKQRTIQAKK
EVEQR RAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENL
SRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSK
TCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQ ITYYNRRK
RQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALN IARSVVLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 104)
486: QEIKRINKI RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
Helical 1B EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM SRVAQ
swa from PAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKPH
p
SEQ ID TNYFGRCNVS EH E RL ILLSP H KP EAN D E LVTYS
LGKFGQ RALD FYSIHVTK
NO:1
ESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQKVV
KGNQKRLESLRELAGKENLEYPSVTLPPCIPHTKEGVDAYNEVIARVRMW
VNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVEROANEVDVINVDMVCNVK
KLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLGD
LLKHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKA
ALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAEN
S I LD I SG FSKQYNCAF IWQ KDGVKKLN LYL I I NYFKGG KLRFKKIKP EAFEAN
RFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLET
GSLKLAN GRVIEKTLYNR RTRQ D EPALFVALTFERREVLDSSN I KPMNLIG
VDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKORTIQAKK
EVEQR RAGGYS RKYASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENL
SRG FGRQ GKRTFMAERQYTRM E DVVLTAKLAYEGLS KTYLS KTLAQYTS K
TCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQ ITYYNRRK
RQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALN IARSVVLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 105)
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
487: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
Helical 1B ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRVAQ
swa from PAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKPH
p
SEQ ID
TNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVIK
Nal
ESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQKVV
KGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRMW
VNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDINVVDMVCNVK
KLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLGD
LLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKA
ALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAEN
SILDISGFSKOYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEAN
RFYTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLET
GSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIG
VDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKElaRTIQAKK
EVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENL
SRGFGRQGKRTFMAERQYTRMEDINLTAKLAYEGLSKTYLSKTLAQYTSK
TCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKR
QNVVKDLSVELDRLSEESVNNDISSINTKGRSGEALSLLKKRFSHRPVQEK
FVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAFV
ETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 106)
488: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
NTSB and ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
Helical 1B PASKKIDQ NKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKA
swa from YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
p
SEQ ID
TKESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQK
NO:1
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVVVDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFG
DLLLHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEIWRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 107)
489: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
NTSB and ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
Helical 1B PASKKIDQ NKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKA
swa from YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
p
TKESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQK
91
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
SEQ ID
VVKGNOKRLESLRELAGKENLEYPSVTLPPOPHTKEGVDAYNEVIARVRM
NO:1
WVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVVVDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIVVQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRK
RQNVVKDLSVELDRLSEESVNNDISSINTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVVVKPAV (SEQ ID NO: 108)
490: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
NTSB and ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
Helical 1B PASKKIDQNKLKPEMDEKGNLTTAGFACSQCGOPLFVYKLEQVSEKGKA
swa from YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
p
SEQ ID
TKESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQK
NO:1
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDWVVDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLKHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQS
KAALTDINLRAKASFVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEA
ENSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFE
ANRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSL
ETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRRK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVVVKPAV (SEQ ID NO: 109)
491: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
NTSB and ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
Helical 1B PASKKIDQNKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKA
swa from YTNYFGRCNVAEHEKLILLAOLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
p
SEQ ID
TKESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQK
NO:1
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
1NVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDWVVDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
92
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
NSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTS
KTC S N CGFTITSADYD RVLEKLKKTATGVVMTTI NG KE LKVEGQ ITYYN RYK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVVVKPAV (SEQ ID NO: 110)
494:
QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
NTSB EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E
FQ KDPVGLM SRVAQ
swa from PASKKIDQ NKLKPEM DE KGNLTTAGFACSQCGQPLFVYKLEQVSEKGKA
p
SEQ ID YTNYFGRCNVAE
HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
NO:1 TRESN HPVKPLEQIGGNSCASGPVGKALS DACMGAVASFLTKYQD
I ILEH
Q KVIKKN E KR LAN LKD IASANGLAFPKITLPPQ P HTKEG I EAYN NVVAQ IVI
1NVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVCNV
KKLINEKKEDGKVFWONLAGYKRQEALRPYLSSEEDRKKOKKFARYQLG
DLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDAQSK
AALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTC S N CGFTITSADYD RVLEKLKKTATGVVMTTI NG KE LKVEGQ ITYYN RYK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 111)
328: MQ E IKRI N KIRRRLVKDS NTKKAGKTG PM KTLLVRVMTP
DLRERLEN LRKK
S867G PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE
FQ KDPVG LMS RVA
QPAPKN IDQRKLI PVKDGNERLTSSGFACSOCCOPLYVYKLEQVNDKGKP
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
KVI KKN E KR LANLKD !MANG LAF PKITLP PQ PHTKEG I EAYNNVVAQ IVIWV
NLNLWQKLKIGRDEAKPLORLKGFPSFPLVEROANEVDVVVVDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALLPYLSSEEDRKKGKKFARYQFGDLLL
HLEKKHGEDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKASFVIEGLKEADKDEFCRCELKLQ KVVYGD LRG KP FAI EAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDPNLI I LPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAAKEVE
QRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRG
93
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
FGRQGKRTFMAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGINMTTINGKELKVEGQITYYNRYKRQ
NVVKDLGVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVINKPAV (SEQ ID NO: 112)
388: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R+A7 PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVA
08K+
QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
[P793] + HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
Xi RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ
D I I LE H Q
Helical2 KVIKKNEKRLANLKD
IASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
swap
NLNLVVQKLKIGRDEAKPLQRLKGFPSFPVVERRENEVDWINNTINEVKKLI
DAKRDMGRVFVVSGVTAEKRNTILEGYNYLPNENDHKKREGSLENPKKPA
KRQFGDLLLYLEKKYAGDWGKVFDEAWERIDKKIAGLTSHIEREEARNAE
DAQSKAVLTDVVLRAKASFVLERLKEMDEKEFYACEIQLQKVVYGDLRGNP
FAVEAENSILDISGFSKQYNCAFIVVQKDGVKKLNLYLIINYFKGGKLRFKKIK
PEAFEANRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIK
PMNLIGIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQR
TIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAM
LIFENLSRGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLA
QYTSKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ ITYY
NRYKRQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHR
PVQEKFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNT
DKRAFVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 113)
389: MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R+A7 PENIPQPISNTSRANLNKLLTDYTEMKKAILHVYINEEFQKDPVGLMSRVA
08K+
QPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCOPLYVYKLEQVNDKGKP
[P793] +
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
X1 RuvC1 RESNHPVKPLEQ IGG N SCASGPVGKALSDACMGAVAS FLTKYQ D I I LE H Q
swap
KVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIVVV
NLNLVVQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDIMNDMVCNVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPVNLIGVDR
GENIPAVIALTDPEGCPLPEFKDSSGGPTDILRIGEGYKEKQRAIQAAKEVE
ORRAGGYSRKFASKSRNLADDMVRNSARDLFYHAVTHDAVLVFENLSRG
FGRQGKRTFMTERQYTKIVIEDINLTAKLAYEGLTSKTYLSKTLAQYTSKTC
SNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYKRQ
NVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQEKF
VCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRAFVE
TVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 114)
94
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
390: MQ
EIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKK
L379R+A7 PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYVVEE FQ KDPVG LMS RVA
08K+ Q PAPKN IDQRKL I
PVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKGKP
[P793] +
HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVT
X1 RuvC2 RESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQ DI ILEH Q
swap KVIKKNEKRLANLKD
IASANGLAFPKITLPPQPHTKEGIEAYNNVVAQIVIWV
N LN LVVQ KLKI GRD EAKP LQRLKGFPS FP LVE ROAN EVDWWDMVC NVKKL
INEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQFGDLL
LHLEKKHGEDWGKVYDEAWER I DKKVE GLSKH IKLEE ER RSEDAQSKAAL
TDVVLRAKAS FVI EG LKEADKDEFC RC ELKLQ KVVYGDLRGKPFAIEAE NS IL
DISGFSKOYNCAFIWQKDGVKKLNLYLIINYFKGOKLRFKKIKPEAFEANRF
YTVINKKSGEIVPMEVNFN FDDP NLI I LPLAFGKRQ GR EFIVVNDLLSLETGS
LKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN IKPMNLIGIDR
GENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAKKEVE
Q RRAGGYSRKYAS KAKN LAD DMVRNTARDLLYYAVTQ DAM LI FEN LS RG
FGRQ GKRTFMAE RQ YTRME DWLTAKLAYEG LS KTYLSKTLAQYTSKTCS
N CC FTITSADYD RVLE KLKKTATGVVMTTI NC KE LKVE GQ ITYYN RYKRQ N
VVKD LSVE LDRLS E ESVN ND IS SVVTKGRSGEALS LLKKRFSH RPVQE KFV
CLNCGFETHADEQAALNIARSWLFLNSNSTEFKSYKSGKQPFVGAWQAF
YKRRLKEVVVKPNA (SEQ ID NO: 115)
514: QEIKRINKI RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
AH817 in EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM SRVAQ
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQC GQ P
LFVYKLEQVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
WVNLN LWQKLKLS RD DAKPLLRLKG FPSFP LVE RQANEVDVVWDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKH IKLEEERRSEDAQSK
AALTDVVLRAKASFVI EGLKEADKDE FC RC E LKLQ KVVYGDLRG KPFAI EAE
NSI LDISGFSKQYNCAF I VVQ KDGVKKLN LYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPMNL I
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKCIRTIQAK
KEVEQRRAGGYSR KYASKAKN LAD DMVRNTAR DLLYYAVTQDAM LI FEN
LS RG FGRQGKRTFMAERQYTRM EDVVLTAKLAYEGLSKTYLS KTLAQYTS
KTCSNCGFTIHTSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYYNRY
KRQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVID
EKFVC LN CC FETHADEQAALN IARSWLF LRSQ EYKKYQTNKTTGNTDKRA
FVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 217)
515: QEIKRINKI RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
AP793 in EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E
FQ KDPVGLM SRVAQ
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQC GQ P
LFVYKLEQVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Desc riptio Amino Acid
Sequence
n*
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVWDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIVVQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYTVIN KKSGE IVPMEVNFNFDDPNLI ILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRY
KRONVVKDLSVELDRLSEESVNNDISSWTKORSGEALSLLKKRFSHRPVQ
EKFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRA
FVETVVQSFYRKKLKEVVVKPAV (SEQ lin NO: 218)
516: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
L307H in ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
491
PASKKIDQNKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKA
YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNHNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVVVDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIONDEAWERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIVVQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYTVIN KKSGE IVPMEVNFNFDDPNLI ILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVVVKPAV (SEQ 1:13 NO: 219)
517: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
AA224 in ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
491
PASKKIDQNKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKA
YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTHPVKPLAQIAGNRYASGAPVGKALSDACMGTIASFLSKYQDIIIEHQ
KVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVR
MVINNLNLVVQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVWVDMVGN
VKKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQL
GDLLLHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQ
SKAALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIE
96
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
AE N S I LD ISGFSKQYNCAF IWQ KDGVKKLN LYL I INYF KGG KLRF KKI KP EAF
EAN RFYTVINKKSGE IVPMEVNFNFDDPNL I ILPLAFGKRQGREF IWNDLLS
LETGSLKLANGRVIE KTLYN RRTRQ DE PALFVALTFE RREVLDSSN IKPM N
LIGVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQ
AKKEVEQ RRAGGYS RKYASKAKN LAD DMVRNTARDLLYYAVTQ DAM LIF
EN LS RGFGRQGKRTFMAE RQYTRM EDWLTAKLAYE GLS KTYLSKTLAQY
TS KTC S N CC FTITSADYDRVLE KLKKTATGWMTTI NC KE LKVEGQ ITYYNR
YKRQ NVVKDLSVELDRLS E ESVN ND I SSVVTKGRSGEALSLLKKRFSH RPV
QEKFVCLNCGFETHADEQAALN IARSVVLFLRSQEYKKYQTNKTTGNTDK
RAFVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 220)
518: RQ E I KRIN KI RRRLVKDSNTKKAG KTG PMKTLLVRVMTPDLRE RLEN LRKK
AR! in 491 PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE FQ KDPVG LMS RVA
Q PASKKI DON KLKP EMDE KG N LTTAG FAC SQC GQ P LFVYKLEQVSE KGK
AYTNYFG RC NVAE H E KLI LLAQ LKPE KDS DEAVTYSLGKFGQ RALD FYS I H
VTKESTHPVKPLAQ IAGN RYASGAPVGKALSDACMGTIASFLSKYQ D I II E H
Q KVVKGNQ KRLES LRE LAGKE N LEYPSVTLPPQ P HTKEGVDAYN EVIARV
RMWVN LNLWQ KLKLSRD DAKP LLRLKGFPS FP LVERQAN EVDVVVVDMVC
NVKKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
LGDLLLHLEKKHGEDWGKVYDEAVVERIDKKVEGLSKHIKLEEERRSEDAQ
SKAALTDWLRAKASFVI EGLKEAD KDE FC RC E LKLQ KVVYGDLRGKP FAIE
AE N S I LD ISGFSKQYNCAF IWQ KDGVKKLN LYL I INYF KGG KLRF KKI KP EAF
EAN RFYTVINKKSGE IVPMEVNFNFDDPNL I ILPLAFGKRQGREF IVVNDLLS
LETGSLKLANGRVIE KTLYN RRTRQ DE PALFVALTFE RREVLDSSN IKPM N
LIGVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQ
AKKEVEQ RRAGGYS RKYASKAKN LAD DMVRNTAR DLLYYAVTQ DAM LIF
EN LS RGFGRQGKRTFMAE RQYTRM EDWLTAKLAYE GLS KTYLSKTLAQY
TS KTCS N CG FTITSADYDRVLE KLKKTATGWMTTI NC KE LKVEGQ ITYYNR
YKRQ NVVKDLSVELDRLS E ESVN ND I SSVVTKGRSGEALSLLKKRFSH RPV
QEKFVCLNCGFETHADEQAALN IARSWLFLRSQEYKKYQTNKTTGNTDK
RAFVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 221)
519: QEIKRINKI RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
AQ692 i EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM
SRVAQ
n
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQC GOP
LEVYKLEOVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLN LWQKLKLS RD DAKPLLRLKG FPSFP LVE RQANEVDVVWDMVCNV
KKL IN EKKEDGKVFWQ N LAGYKRQ EALRPYLSS E E DRKKG KKFARYQLG
DLLLHLEKKHGEDWGKVYDEAINERIDKKVEGLSKH IKLEEERRSEDAQSK
AALTDWLRAKASFVI EGLKEADKDE FC RC E LKLQ KVVYGDLRG KPFAI EAE
NSI LDISGFSKQYNCAF I VVQ KDGVKKLN LYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGE IVPMEVNFNFDDP NLI ILPLAFGKRQGREFIWNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPMNL I
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH IQ LRIGESYKEKQRTIQA
KKEVEQRRAGGYSRKYAS KAKN LADDMVRNTARDLLYYAVTQ DAM L IFE
97
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
NLSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRY
KRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQ
EKFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRA
FVETVVQSFYRKKLKEVWKPAV (SEQ ID NO: 222)
520:
QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
1705T in EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E
FQ KDPVGLM SRVAQ
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQCGQ P
LFVYKLEQVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLN LWQKLKLS RD DAKPLLRLKG FPSFP LVE RQANEVDVVVVDMVC NV
KKL IN EKKEDGKVFWQ N LAGYKRQ EALRPYLSS E E DRKKG KKFARYOLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKH IKLEEERRSEDAQSK
AALTDWLRAKASFVI EGLKEADKDE FC RC E LKLQ KVVYGDLRG KPFAI EAE
NSI LDISGFSKQYNCAF I VVQ KDGVKKLN LYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGE IVPMEVNFNFDDP NLI ILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPMNL I
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTTQA
KKEVEQRRAGGYSRKYAS KAKN LADDMVRNTARDLLYYAVTQ DAM L IFE
N LS RGFGRQGKRTFMAERQYTRME DWLTAKLAYEG LS KTYLS KTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ ITYYNRY
KRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQ
EKFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDKRA
FVETWQSFYRKKLKEVWKPAV (SEQ ID NO: 223)
522: QEIKRINKI
RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
D683R in EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM SRVAQ
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQC GQ P
LFVYKLEQVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLN LWQKLKLS RD DAKPLLRLKG FPSFP LVE RQANEVDVVWDMVCNV
KKL IN EKKEDGKVFWQ N LAGYKRQ EALRPYLSS E E DRKKG KKFARYOLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKH IKLEEERRSEDAQSK
AALTDVVLRAKASFVI EGLKEADKDE FC RC E LKLQ KVVYGDLRG KPFAI EAE
NSILDISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPMNL I
GVDRGEN IPAVIALTDPEGCPLSRFKRSLGNPTH ILRIGESYKEKQRTIQAK
KEVEQRRAGGYSR KYASKAKN LAD DMVRNTAR DLLYYAVTQDAM LI FEN
LS RG FGRQGKRTFMAERQYTRM EDWLTAKLAYEGLSKTYLS KTLAQYTS
KTCS N CGFTITSADYD RVLEKLKKTATGVVMTTI NG KE LKVEGQ ITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALN IARSVVLFLRSQEYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVVVKPAV (SEQ ID NO: 224)
98
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
523: Q E I KRI NKI RR R LVKDSNTKKAGKTYP MKTLLVRVMTPDLR E R LE NLR KKP
G26Y in EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E
FQ KDPVGLM SRVAQ
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQCGQ P
LFVYKLEQVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQ KRLESLRELAGKEN LEYPSVTLP PO PHTKEGVDAYN EVIARVRM
VVVNLN LWQKLKLS RD DAKPLLRLKG FPSFP LVE RQANEVDVVVVDMVC NV
KKL IN EKKEDGKVFWQ N LAGYKRQ EALRPYLSS E E DRKKG KKFARYQLG
DLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDAQSK
AALTDWLRAKASFVI EGLKEADKDE FC RC E LKLQ KVVYGDLRG KPFAI EAE
NSILDISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGE IVPMEVNFNFDDP NLI ILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPIVINL I
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILR IGESYKEKQ RTIQAK
KEVEQRRAGGYSRKYASKAKN LAD DMVRNTARDLLYYAVTQDAM LI FEN
LS RG FGRQGKRTFMAERQYTRM EDVVLTAKLAYEGLSKTYLS KTLAQYTS
KTC S N CGFTITSADYD RVLEKLKKTATGVVMTTI NG KE LKVEGQ ITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALN IARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVVVKPAV (SEQ 1:13 NO: 225)
524: QEIKRINKI RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
T817H in EN I PQ PI S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM SRVAQ
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQC GQ P
LFVYKLEQVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
WVNLN LWQKLKLS RD DAKPLLRLKG FPSFP LVE RQANEVDVVWDMVCNV
KKL IN EKKEDGKVFWQ N LAGYKRQ EALRPYLSS E E DRKKG KKFARYQLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKH IKLEEERRSEDAQSK
AALTDVVLRAKASFVI EGLKEADKDE FC RC E LKLQ KVVYGDLRG KPFAI EAE
NSI LDISGFSKQYNCAF I VVQ KDGVKKLN LYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSGE IVPMEVNFNFDDP NLI ILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPMNL I
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRIGESYKEKQRTIQAK
KEVEQRRAGGYSR KYASKAKN LAD DMVRNTAR DLLYYAVTQDAM LI FEN
LS RG FGRQGKRTFMAERQYTRM EDVVLTAKLAYEGLSKTYLS KTLAQYTS
KTCSNCGFTIHSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYN RY
KRQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVICI
EKFVC LN CC FETHADEQAALN IARSWLF LRSQ EYKKYQTNKTTGNTDKRA
FVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 226)
525; QEIKRINKI
RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
V746A in EN I PQ PIS NTS RAN LN KLLTDYTEMKKAILHVYVVEEFQ KDPVGLMSRVAQ
491 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQC GQ P
LFVYKLEQVSEKGKA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
99
CA 03159320 2022-5-24
WO 2021/113772
PCTMS2020/063488
Desc riptio Amino Acid
Sequence
n*
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VWNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVWDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDVVLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIVVQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYTVIN KKSGE IVPMEVNFNFDDPNLI ILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNIKPMNLI
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAATQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETWQSFYRKKLKEVWKPAV (SEQ ID NO: 227)
526: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
K708A in ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
491
PASKKIDQNKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKA
YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVVVDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIONDEAWERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
NSILDISGFSKQYNCAFIVVQKDGVKKLNLYLIINYFKGGKLRFKKIKPEAFEA
NRFYTVIN KKSGE IVPMEVNFNFDDPNLI ILPLAFGKRQGREFIVVNDLLSLE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSS NIKPM N L I
GVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQAA
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYYNRYK
RQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQE
KFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRAF
VETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 228)
527: QEIKRINKIRRRLVKDSNTKKAGKTRGPMKTLLVRVMTPDLRERLENLRKK
AR26 in PE N IPQ P I SNTS RANLN KLLTDYTEM KKAI LHVYWEE
FQ KDPVG LMS RVA
491
QPASKKIDQNKLKPEMDEKGNLTTAGFACSQCGQPLFVYKLEQVSEKGK
AYTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIH
VTKESTHPVKPLAQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQ
KVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVR
MVINNLNLVVQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVWVDMVGN
VKKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQL
GDLLLHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDAQ
SKAALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIE
100
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
AE N S I LD ISGFSKQYNCAF IWQ KDGVKKLN LYL I INYF KGG KLRF KKI KP EAF
EAN RFYTVINKKSGE IVPMEVNFNFD DPNL I ILPLAFGKRQG REF IWND LLS
LETGSLKLANGRVIE KTLYN RRTRQ DE PALFVALTFE RREVLDSSN IKPM N
LIGVDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQRTIQ
AKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIF
ENLSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQY
TSKTCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQITYYNR
YKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPV
QEKFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTDK
RAFVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 229)
528: QEIKRINKI RRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
G223Y
ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
in
515 PASKKIDO NKLKPEM DE KGNLTTAGFACSQC GQ P
LEVYKLEOVS EKGKA
YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTHPVKPLAQIAGNRYASYPVGKALSDACMGTIASFLSKYQDIIIEHQK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
WVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVWDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDVVLRAKASFVI EGLKEADKDE FC RC E LKLQ KVVYGDLRG KPFAI EAE
N S I LD ISGFSKQYN CAF I VVQ KDGVKKLN LYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSG E IVPMEVNFNFD DP NLI ILPLAFGKRQGRE FIVVND LLS LE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSS N I KPM N L I
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILR I GE SYKE KQ RTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDVVLTAKLAYEGLPSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQITYYNRY
KRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQ
EKFVCLNCGFETHADEQAALNIARSWLFLRSQEYKKYQTNKTTGNTDKRA
FVETVVQSFYRKKLKEVVVKPAV (SEQ ID NO: 230)
529: QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKP
G223N in ENIPQPISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQKDPVGLMSRVAQ
515 PASKKIDQ NKLKPEM DE KGNLTTAGFACSQC GO P
LEVYKLEOVS EKGKA
YTNYFGRCNVAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYAS N PVGKALS DACMGTIAS F LS KYQ D III E H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVWDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQLG
DLLLHLEKKHGEDWGIWYDEAINERIDKKVEGLSKHIKLEEERRSEDAQSK
AALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAE
N S I LD ISGFSKQYN CAF I VVQ KDGVKKLN LYLI INYFKGGKLRFKKIKPEAF EA
NRFYTVIN KKSG E IVPMEVNFNFD DP NLI ILPLAFGKRQGRE FIWND LLS LE
TGSLKLANGRVIEKTLYNRRTRQ DEPALFVALTF ERREVLDSSN IKPMNL I
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILRI GE SYKE KQ RTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
101
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Descriptio Amino Acid
Sequence
n*
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ ITYYNRY
KRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKRFSHRPVQ
EKFVCLNCGFETHADEQAALN IARSVVLFLRSQEYKKYQTNKTTGNTDKRA
FVETVVQSFYRKKLKEVWKPAV (SEQ ID NO: 231)
530: Q E I KRI NKI RRRLVKDSNTKKAGKTGPM KTLLVRVMTPDLRERLENLRKKP
Aw539 in EN I PQ P I S NTS RAN LN KLLTDYTEM KKAILHVYWE E FQ KDPVGLM SRVAQ
515 PASKKIDQ NKLKPEM DE KGN LTTAGFACSQCGQ P
LFVYKLEQVS EKG KA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VsA/NLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVWDMVCNV
KKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYOLG
DLL LH L E KKH GE DWGKVYD EA1NER IDKKVEGLSKH IKLEE ERRS EDAQS K
AALTDWLRAKASFVIEGLKEADKDEFC RC E LKLQ KVVYGDLRG KP FA I EAE
N S I LD ISGF SKQYN CAF I VVQ KDGVKKL N LYLI I NYFKGWG KLRF KKI KPEAF
EAN RFYTVINKKSGE IVPMEVN FN FD DPN L I I LP LAFG KRQG REF IVVN D LLS
LETGSLKLANGRVIE KTLYN RRTRQ DE PAL FVALTFE RREVLDSSN IKPM N
LIGVD R GEN I PAVIALTDP EGC PLS RF KDS LG N PTH I L RIG ESYKE KQ RTIQ
AKKEVEQ RRAGGYS RKYASKAKN LAD DMVRNTARDLLYYAVTQ DAM LIF
ENLSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQ
YTS KTCSNC G FTITSADY DRVL EKLKKTATGVVMTTI N GKELKVEGQ ITYYN
RYKRQNVVKDLSVE LDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRP
VQEKFVC LNCGFETHADEQAALNIARSVVLFLRSQEYKKYQTNKTTGNTD
KRAFVETWQSFYRKKLKEVWKPAV (SEQ ID NO: 232)
531: Q E I KRI NKI RRRLVKDSNTKKAGKTGPM KTLLVRVMTPDLRERLENLRKKP
Ay539 in EN I PQ P I S NTS RAN LN KLLTDYTEM KKAILHVYVVE E FQ KDPVGLM SRVAQ
515 PASKKIDQ NKLKPEM DE KGN LTTAGFACSQC GQ P
LFVYKLEQVS EKG KA
YTNYFGRCNVAE HEKLILLAQLKPEKDSDEAVTYSLGKFGQRALDFYSIHV
TKESTH PVKPLAQ IAG N RYASGPVGKALS DACMGTIAS FLS KYQ D I IIE H QK
VVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVIARVRM
VVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQANEVDVVWDMVCNV
KKLINEKKEDGKVFMNLAGYKRQEALRPYLSSEEDRKKGKKFARYOLG
DLL LH L E KKH GE DWGKVYD EA1NER IDKKVEGLSKH IKLEE ERRS EDAQS K
AALTDVVLRAKASFVIEGLKEADKDEFC RC E LKLQ KVVYGDLRG KP FA I EAE
N S I LD ISGF SKQYN CAF IWQ KDGVKKL N LYLI I NYFKGYGKLRFKKIKP EAF E
ANRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQGREFIWNDLLSL
ETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSN I KPMNLI
GVDRGEN IPAVIALTDPEGCPLSRFKDSLGNPTH ILR I GE SYKE KQ RTIQAK
KEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFEN
LSRGFGRQGKRTFMAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ ITYYNRY
KRQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKRFSHRPVQ
EKFVC LN CC F ETHADEQAAL N IARSWLFLRSQEYKKYQTNKTTGNTDKRA
FVETVVQSFYRKKLKEVWKPAV (SEQ ID NO: 233)
102
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
* Where a number is indicated in the left column, it designates the CasX
variant numerically;
changes, where indicated, are relative to SEQ ID NO2
g. CasX Fusion Proteins
1002201 Also contemplated within the scope of the disclosure are XDP
comprising CasX
variant proteins comprising a heterologous protein fused to the CasX. In some
embodiments,
the CasX variant protein is fused to one or more proteins or domains thereof
that has a different
activity of interest, resulting in a fusion protein. For example, in some
embodiments, the CasX
variant protein is fused to a protein (or domain thereof) that inhibits
transcription, modifies a
target nucleic acid, or modifies a polypeptide associated with a nucleic acid
(e.g., histone
modification).
1002211 In some embodiments, a heterologous polypeptide (or heterologous amino
acid such as
a cysteine residue or a non-natural amino acid) can be inserted at one or more
positions within a
CasX protein to generate a CasX fusion protein utilized in the XDP systems. In
other
embodiments, a cysteine residue can be inserted at one or more positions
within a CasX protein
followed by conjugation of a heterologous polypeptide described below. In some
alternative
embodiments, a heterologous polypeptide or heterologous amino acid can be
added at the N- or
C-terminus of the CasX variant protein. In other embodiments, a heterologous
polypeptide or
heterologous amino acid can be inserted internally within the sequence of the
CasX protein.
1002221 A variety of heterologous polypeptides are suitable for inclusion in a
CasX variant
fusion protein utilized in the XDP systems of the disclosure. In some cases,
the fusion partner
can modulate transcription (e.g., inhibit transcription, increase
transcription) of a target DNA.
For example, in some cases the fusion partner is a protein (or a domain from a
protein) that
inhibits transcription (e.g., a transcriptional repressor, a protein that
functions via recruitment of
transcription inhibitor proteins, modification of target DNA such as
methylation, recruitment of
a DNA modifier, modulation of histones associated with target DNA, recruitment
of a histone
modifier such as those that modify acetylation and/or methylation of histones,
and the like). In
some cases the fusion partner is a protein (or a domain from a protein) that
increases
transcription (e.g., a transcription activator, a protein that acts via
recruitment of transcription
activator proteins, modification of target DNA such as demethylation,
recruitment of a DNA
modifier, modulation of histones associated with target DNA, recruitment of a
histone modifier
such as those that modify acetylation and/or methylation of histones, and the
like).
103
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00223] In some cases, a CasX fusion partner utilized in the XDP systems has
enzymatic
activity that modifies a target nucleic acid (e.g., nuclease activity,
methyltransferase activity,
demethylase activity, DNA repair activity, DNA damage activity, deamination
activity,
dismutase activity, alkylation activity, depurination activity, oxidation
activity, pyrimidine dimer
forming activity, integrase activity, transposase activity, recombinase
activity, polymerase
activity, ligase activity, helicase activity, photolyase activity or
glycosylase activity).
[00224] In some cases, a CasX fusion partner utilized in the XDP systems has
enzymatic
activity that modifies a polypeptide (e.g., a histone) associated with a
target nucleic acid (e.g.,
methyltransferase activity, demethylase activity, acetyltransferase activity,
deacetylase activity,
kinase activity, phosphatase activity, ubiquitin ligase activity,
deubiquitinating activity,
adenylation activity, deadenylation activity, SUIVIOylating activity,
deSUMOylating activity,
ribosylation activity, deribosylation activity, myristoylation activity or
demyristoylation
activity).
[00225] Examples of proteins (or fragments thereof) that can be used as a CasX
fusion partner
utilized in the XDP systems to increase transcription include but are not
limited to.
transcriptional activators such as VP16, VP64, VP48, VP160, p65 subdomain
(e.g., from NFkB),
and activation domain of EDLL and/or TAL activation domain (e.g., for activity
in plants);
histone lysine methyltransferases such as SET1A, SET1B, MILLI to 5, ASH1,
SYMD2, NSD1,
and the like; histone lysine demethylases such as JIIDM2a/b, UTX, IMID3, and
the like; histone
acetyltransferases such as GCN5, PCAF, CBP, p300, TAF1, TIP60/PLIP, MOZ/MYST3,
MORF/MYST4, SRC1, ACTR, P160, CLOCK, and the like; and DNA demethylases such
as
Ten-Eleven Translocation (TET) dioxygenase 1 (TETICD), TETI, DME, DML I, DML2,
ROS I, and the like.
[00226] Examples of proteins (or fragments thereof) that can be used as a CasX
fusion partner
in an XDP to decrease transcription include but are not limited to:
transcriptional repressors such
as the Kruppel associated box (KRAB or 51(D); KOX1 repression domain; the Mad
mSIN3
interaction domain (SID), the ERF repressor domain (ERD), the SRDX repression
domain (e.g.,
for repression in plants), and the like; histone lysine methyltransferases
such as Pr-SET7/8,
SUV4- 20111, RIZ1, and the like; histone lysine demethylases such as
JNIJD2A/JHDM3A,
JN1JD2B, JMJD2C/GASC1, JMJD2D, JARID1A/RBP2, JARID1B/PLU-1, JAME) 1C/SMCX,
JARID1D/SMCY, and the like; histone lysine deacetylases such as HDAC1, HDAC2,
HDAC3,
HDAC8, HDAC4, FIDAC5, HDAC7, HDAC9, SIRTI, SIRT2, HDAC 11, and the like; DNA
104
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
methylases such as HhaI DNA m5c-methyltransferase (M.HhaI), DNA
methyltransferase 1
(DNMT1), DNA methyltransferase 3a (DNMT3a), DNA methyltransferase 3b (DNMT3b),
HETI, DRM3 (plants), ZMET2, CMT1, CMT2 (plants), and the like; and periphery
recruitment
elements such as Lamin A, Lamin B, and the like.
[00227] In some cases, the CasX fusion partner utilized in the XDP systems has
enzymatic
activity that modifies the target nucleic acid (e.g., ssRNA, dsRNA, ssDNA,
dsDNA). Examples
of enzymatic activity that can be provided by the fusion partner include but
are not limited to:
nuclease activity such as that provided by a restriction enzyme (e.g., Fold
nuclease),
methyltransferase activity such as that provided by a methyltransferase (e.g.,
Hhal DNA m5c-
methyltransferase (M.Hhal), DNA methyltransferase 1 (DNMT1), DNA
methyltransferase 3a
(DNMT3a), DNA methyltransferase 3b (DNMT3b), METI, DR1VI3 (plants), ZMET2,
CMT1,
CMT2 (plants), and the like); demethylase activity such as that provided by a
demethylase (e.g.,
Ten-Eleven Translocation (TET) dioxygenase 1 (TET 1 CD), TETI, DME, DML1,
DML2,
ROS1, and the like), DNA repair activity, DNA damage activity, deamination
activity such as
that provided by a deaminase (e.g., a cytosine deaminase enzyme, e.g., an
APOBEC protein such
as rat APOBEC1), dismutase activity, alkylation activity, depurination
activity, oxidation
activity, pyrimidine dimer forming activity, integrase activity such as that
provided by an
integrase and/or resolvase (e.g., Gin invertase such as the hyperactive mutant
of the Gin
invertase, GinH106Y; human immunodeficiency virus type 1 integrase (IN); Tn3
resolvase; and
the like), transposase activity, recombinase activity such as that provided by
a recombinase (e.g.,
catalytic domain of Gin recombinase), polymerase activity, ligase activity,
helicase activity,
photolyase activity, and g,lycosylase activity).
[00228] In other cases, CasX variant protein of the present disclosure
utilized in the XDP
systems is fused to a polypeptide selected from: a domain for increasing
transcription (e.g., a
VP16 domain, a VP64 domain), a domain for decreasing transcription (e.g., a
KRAB domain,
e.g., from the Koxl protein), a core catalytic domain of a histone
acetyltransferase (e.g., histone
acetyltransferase p300), a protein/domain that provides a detectable signal
(e.g., a fluorescent
protein such as GFP), a nuclease domain (e.g., a Fold nuclease), and a base
editor (e.g., cytidine
deaminase such as APOBEC1).
[00229] In still other cases, the CasX fusion partner utilized in the XDP
systems has enzymatic
activity that modifies a protein associated with the target nucleic acid
(e.g., ssRNA, dsRNA,
ssDNA, dsDNA) (e.g., a histone, an RNA binding protein, a DNA binding protein,
and the like).
105
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Examples of enzymatic activity (that modifies a protein associated with a
target nucleic acid)
that can be provided by the fusion partner include but are not limited to:
methyltransferase
activity such as that provided by a histone methyltransferase (FINIT) (e.g.,
suppressor of
variegation 3-9 homolog 1 (SUV39H1, also known as KMT1A), euchromatic histone
lysine
methyltransferase 2 (G9A, also known as KMT1C and EHMT2), SUV39H2, ESET/SETDB
1,
and the like, SET1A, SET1B, MILLI to 5, ASH1, SYMD2, NSD1, DOT IL, Pr-SET7/8,
SUV4-
20H1, EZH2, R1Z1), demethylase activity such as that provided by a histone
demethylase (e.g.,
Lysine Demethylase lA (KDM1A also known as LSD1), JHDM2a/b, JMJD2A/JHDM3A,
JIVLID2B, JMJD2C/GASC1, JMJD2D, JARID1A/RBP2, JARID1B/PLU-1, JARID1C/SMCX,
JAR1D1D/SMCY, lUTX, JMJD3, and the like), acetyltransferase activity such as
that provided
by a histone acetylase transferase (e.g., catalytic core/fragment of the human
acetyltransferase
p300, GCN5, PCAF, CBP, TAF1, TIP60/PLIP, MOZ/MYST3, MORF/MYST4, FIB01/MYST2,
HMOF/MYST1, SRC1, ACTR, P160, CLOCK, and the like), deacetylase activity such
as that
provided by a histone deacetylase (e.g., FIDAC1, HDAC2, HDAC3, MACS, HDAC4,
HDAC5,
HDAC7, HDAC9, SIRT1, S1RT2, HDAC11, and the like), kinase activity,
phosphatase activity,
ubiquitin ligase activity, deubiquitinating activity, adenylation activity,
deadenylation activity,
SUMOylating activity, deSUMOylating activity, ribosylation activity,
deribosylation activity,
myristoylation activity, and demyristoylation activity.
1002301 Suitable chloroplast transit peptides include, but are not limited to:
MASMISSSAVTTVSRASRGQSAANIAPFGGLKSMTGFPVRICVNTDITSITSNGGR
VKCMQVWPPIGKICKFETLSYLPPLTRDSRA (SEQ ID NO: 116);
MASMISSSAVTTVSRASRGQSAA1VIAPFGGLKSMTGFPVRKVNTDITSITSNGGRVKS
(SEQ ID NO: 117);
MASSMILSSATMVASPAQATMVAPFNGLKSSAAFPATRKANNDITSITSNGGRVNCMQV
WPPIEKKKFETLSYLPDLTDSGGRVNC (SEQ ID NO: 118;
MAQVSR1CNGVQNPSLISNLSKSSQRKSPLSVSLKTQQHPRAYPISSSWGLKKSGMTLIG
SELRPLKVMSSVSTAC (SEQ ID NO: 119);
MAQVSRICNGVWNPSLISNLSKSSQRKSPLSVSLKTQQIIPRAYPISSSWGLICKSGMTLIG
SELRPLKVMSSVSTAC (SEQ ID NO: 120);
MAQINNMAQGIQTLNPNSNFHKPQVPKSSSFLVFGSKICLKNSANSMiLVLICKDSIFMQLF
CSFR1SASVATAC (SEQ ID NO: 121);
MAALVTSQLATSGTVLSVTDRFRRPGFQGLRPRNPADAALG1VIRTVGASAAPKQSRKPH
106
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
RFDRRCLSMVV (SEQ ID NO: 122);
MAALTTSQLATSATGFGIADRSAPSSLLRHGFQGLICPRSPAGGDATSLSVTTSARATPICQ
QRSVQRGSRRFPSVVVC (SEQ ID NO: 123);
MASSVLSSAAVATRSNVAQANMVAPFTGLKSAASFPVSRKQNLDITSIASNGGRVQC
(SEQ ID NO: 124);
MESLAATSVFAPSRVAVPAARALVRAGTVVPTRRTSSTSGTSGVKCSAAVTPQASPVIS
RSAAAA (SEQ ID NO: 125); and
MGAAATSMQSLICFSNRLVPPSRRLSPVPNNVTCNNLPKSAAPVRTVKCCASSWNSTING
AAATTNGASAASS (SEQ ID NO: 126).
[00231] In some cases, a CasX variant polypeptide of the present disclosure
can include an
endosomal escape peptide. In some cases, an endosomal escape polypeptide
comprises the
amino acid sequence GLFXALLXLLXSLWXLLLXA (SEQ ID NO: 127), wherein each X is
independently selected from lysine, histidine, and arginine. In some cases, an
endosomal escape
polypeptide comprises the amino acid sequence GLFHALLHLLEISLWHILLLHA (SEQ ID
NO:
128), or FILIHRHIII-IFIH (SEQ ID NO: 129).
[00232] Non-limiting examples of CasX fusion partners for use when targeting
ssRNA target
nucleic acids include (but are not limited to): splicing factors (e.g., RS
domains); protein
translation components (e.g., translation initiation, elongation, and/or
release factors; e.g.,
elF4G); RNA methylases; RNA editing enzymes (e.g., RNA deaminases, e.g.,
adenosine
deaminase acting on RNA (ADAR), including A to I and/or C to U editing
enzymes); helicases;
RNA-binding proteins; and the like. It is understood that a heterologous
polypeptide can include
the entire protein or in some cases can include a fragment of the protein (e
g., a functional
domain).
[00233] A fusion partner can be any domain capable of interacting with ssRNA
(which, for the
purposes of this disclosure, includes intramolecular and/or intermolecular
secondary structures,
e.g., double-stranded RNA duplexes such as hairpins, stem-loops, etc.),
whether transiently or
irreversibly, directly or indirectly, including but not limited to an effector
domain selected from
the group comprising; endonucleases (for example RNase III, the CRR22 DYW
domain, Dicer,
and PIN (PilT N-terminus) domains from proteins such as SMG5 and SMG6);
proteins and
protein domains responsible for stimulating RNA cleavage (for example CPSF,
CstF, CFIm and
CFIIm); exonucleases (for example XRN-1 or Exonuclease T); deadenylases (for
example
IINT3); proteins and protein domains responsible for nonsense mediated RNA
decay (for
107
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
example UPF1, UPF2, UPF3, UPF3b, RNP SI, Y14, DEK, REF2, and SRm160); proteins
and
protein domains responsible for stabilizing RNA (for example PABP); proteins
and protein
domains responsible for repressing translation (for example Ago2 and Ago4);
proteins and
protein domains responsible for stimulating translation (for example Staufen);
proteins and
protein domains responsible for (e.g., capable of) modulating translation
(e.g., translation factors
such as initiation factors, elongation factors, release factors, etc., e.g.,
e1F4G); proteins and
protein domains responsible for polyadenylation of RNA (for example PAP1, GLD-
2, and Star-
PAP); proteins and protein domains responsible for polyuridinylation of RNA
(for example CI
DI and terminal uridylate transferase); proteins and protein domains
responsible for RNA
localization (for example from 111/1P1, ZBP1, She2p, She3p, and Bicaudal-D);
proteins and
protein domains responsible for nuclear retention of RNA (for example Rrp6);
proteins and
protein domains responsible for nuclear export of RNA (for example TAP, NXF1,
THO, TREX,
REF, and My); proteins and protein domains responsible for repression of RNA
splicing (for
example PTB, Sam68, and hnRNP Al); proteins and protein domains responsible
for stimulation
of RNA splicing (for example serine/arginine-rich (SR) domains); proteins and
protein domains
responsible for reducing the efficiency of transcription (for example FUS
(TLS)); and proteins
and protein domains responsible for stimulating transcription (for example
CDK7 and HIV Tat).
Alternatively, the effector domain may be selected from the group comprising
endonucleases;
proteins and protein domains capable of stimulating RNA cleavage;
exonucleases; deadenylases;
proteins and protein domains having nonsense mediated RNA decay activity;
proteins and
protein domains capable of stabilizing RNA; proteins and protein domains
capable of repressing
translation; proteins and protein domains capable of stimulating translation;
proteins and protein
domains capable of modulating translation (e.g., translation factors such as
initiation factors,
elongation factors, release factors, etc., e.g., elF4G); proteins and protein
domains capable of
polyadenylation of RNA; proteins and protein domains capable of
polyuridinylation of RNA;
proteins and protein domains having RNA localization activity; proteins and
protein domains
capable of nuclear retention of RNA, proteins and protein domains having RNA
nuclear export
activity; proteins and protein domains capable of repression of RNA splicing,
proteins and
protein domains capable of stimulation of RNA splicing; proteins and protein
domains capable
of reducing the efficiency of transcription; and proteins and protein domains
capable of
stimulating transcription. Another suitable heterologous polypeptide is a PUF
RNA-binding
108
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
domain, which is described in more detail in W02012068627, which is hereby
incorporated by
reference in its entirety.
1002341 Some RNA splicing factors that can be used (in whole or as fragments
thereof) as a
CasX fusion partners in the XDP systems have modular organization, with
separate sequence-
specific RNA binding modules and splicing effector domains. For example,
members of the
serine/arginine-rich (SR) protein family contain N-terminal RNA recognition
motifs (RRMs)
that bind to exonic splicing enhancers (ESEs) in pre-mRNAs and C-terminal RS
domains that
promote exon inclusion. As another example, the hnRNP protein hnRNP Al binds
to exonic
splicing silencers (ESSs) through its RRM domains and inhibits exon inclusion
through a C-
terminal glycine-rich domain. Some splicing factors can regulate alternative
use of splice site
(ss) by binding to regulatory sequences between the two alternative sites. For
example, ASF/SF2
can recognize ESEs and promote the use of intron proximal sites, whereas hnRNP
Al can bind
to ESSs and shift splicing towards the use of intron distal sites. One
application for such factors
is to generate ESFs that modulate alternative splicing of endogenous genes,
particularly disease
associated genes. For example, Bcl-x pre-mRNA produces two splicing isoforms
with two
alternative 5' splice sites to encode proteins of opposite functions. The long
splicing isoform Bcl-
xL is a potent apoptosis inhibitor expressed in long-lived post mitotic cells
and is up-regulated in
many cancer cells, protecting cells against apoptotic signals. The short
isoform Bc1-xS is a pro-
apoptotic isoform and expressed at high levels in cells with a high turnover
rate (e.g., developing
lymphocytes). The ratio of the two Bcl-x splicing isoforms is regulated by
multiple cis -elements
that are located in either the core exon region or the exon extension region
(i.e., between the two
alternative 5' splice sites) For more examples, see W02010075303, which is
hereby
incorporated by reference in its entirety.
1002351 Further suitable CasX fusion partners utilized in the XDP systems
include, but are not
limited to, proteins (or fragments thereof) that are boundary elements (e.g.,
CTCF), proteins and
fragments thereof that provide periphery recruitment (e.g., Lamin A, Lamin B,
etc.), and protein
docking elements (e.g., FKBP/FRB, Pill/Abyl, etc.).
1002361 In some cases, a heterologous polypeptide (a fusion partner) provides
for subcellular
localization of the CasX to which it is fused, i.e., the heterologous
polypeptide contains a
subcellular localization sequence (e.g., a nuclear localization signal (NLS)
for targeting to the
nucleus, a sequence to keep the fusion protein out of the nucleus, e.g., a
nuclear export sequence
(NES), a sequence to keep the fusion protein retained in the cytoplasm, a
mitochondrial
109
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
localization signal for targeting to the mitochondria, a chloroplast
localization signal for
targeting to a chloroplast, an ER retention signal, and the like). In some
embodiments, a subject
RNA-guided polypeptide does not include a NLS so that the protein is not
targeted to the
nucleus (which can be advantageous, e.g., when the target nucleic acid is an
RNA that is present
in the cytosol). In some embodiments, a fusion partner can provide a tag
(i.e., the heterologous
polypeptide is a detectable label) for ease of tracking and/or purification
(e.g., a fluorescent
protein, e.g., green fluorescent protein (GFP), yellow fluorescent protein
(YFP), red fluorescent
protein (RFP), cyan fluorescent protein (CFP), mCherry, tdTomato, and the
like; a histidine tag,
e.g., a 6XHis tag; a hemagglutinin (HA) tag; a FLAG tag; a Myc tag; and the
like).
1002371 In some cases, a CasX variant protein for use in the XDP systems
includes (is fused to)
a nuclear localization signal (NLS). In some cases, a CasX variant protein is
fused to 2 or more,
3 or more, 4 or more, or 5 or more 6 or more, 7 or more, 8 or more NLSs. In
some cases, one or
more NLSs (2 or more, 3 or more, 4 or more, or 5 or more NLSs) are positioned
at or near (e.g.,
within 50 amino acids of) the N-terminus and/or the C-terminus. In some cases,
one or more
NLSs (2 or more, 3 or more, 4 or more, or 5 or more NLSs) are positioned at or
near (e.g., within
50 amino acids of) the N-terminus. In some cases, one or more NLSs (2 or more,
3 or more, 4 or
more, or 5 or more NLSs) are positioned at or near (e.g., within 50 amino
acids of) the C-
terminus. In some cases, one or more NLSs (3 or more, 4 or more, or 5 or more
NLSs) are
positioned at or near (e.g., within 50 amino acids of) both the N-terminus and
the C-terminus. In
some cases, an NLS is positioned at the N-terminus and an NLS is positioned at
the C-terminus.
In some cases, a CasX variant protein includes (is fused to) between 1 and 10
NLSs (e.g., 1-9, 1-
8, 1-7, 1-6, 1-5, 2-10, 2-9, 2-8, 2-7, 2- 6, or 2-5 NLSs) In some cases, a
CasX variant protein
includes (is fused to) between 2 and 5 NLSs (e.g., 2-4, or 2-3 NLSs).
1002381 Non-limiting examples of NLSs include sequences derived from: the NLS
of the SV40
virus large T-antigen, having the amino acid sequence PKICICRICV (SEQ ID NO:
130); the NLS
from nucleoplasmin (e.g., the nucleoplasmin bipartite NLS with the sequence
KRPAATKKAGQAICKKK (SEQ ID NO: 131); the c-myc NLS having the amino acid
sequence
PAAKRVKLD (SEQ ID NO: 132) or RQRRNELICRSP (SEQ ID NO, 133); the hRNPA1 M9
NLS having the sequence NQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGY (SEQ
ID NO: 134); the sequence
RMRIZFICNKGKDTAELRRRRVEVSVELRKAKKDEQ1LICRRNV (SEQ ID NO: 135) of the
IBB domain from importin-alpha; the sequences VSRKRPRP (SEQ ID NO: 136) and
110
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
PP1CKARED (SEQ ID NO: 137) of the myoma T protein; the sequence PQPKICICPL
(SEQ ID
NO: 138) of human p53; the sequence SALIKKICKKMAP (SEQ ID NO: 139) of mouse c-
abl
IV; the sequences DRLRR (SEQ ID NO: 140) and PKQICKRK (SEQ ID NO: 141) of the
influenza virus NS1; the sequence RICLICICKIKKL (SEQ ID NO: 142) of the
Hepatitis virus
delta antigen; the sequence REKKKFLICRR (SEQ ID NO: 143) of the mouse Mxl
protein; the
sequence ICRKGDEVDGVDEVAKICKSKK (SEQ ID NO: 144) of the human poly(ADP-ribose)
polymerase; the sequence RKCLQAGMNLEARKT1CK (SEQ ID NO: 145) of the steroid
hormone receptors (human) glucocorticoid; the sequence PRPRICIPR (SEQ ID NO:
146) of
Boma disease virus P protein (BDV-P1); the sequence PPRKICRTVV (SEQ ID NO:
147) of
hepatitis C virus nonstructural protein (HCV-NS5A); the sequence NLSKKKKRKREK
(SEQ
ID NO: 148) of LEF1; the sequence RRPSRPFRKP (SEQ ID NO: 149) of ORF57
simirae; the
sequence KRPRSPSS (SEQ ID NO: 150) of EBV LANA; the sequence
KRGINDRNFWRGENERKTR (SEQ ID NO: 151) of Influenza A protein; the sequence
PRPPKMARYDN (SEQ ID NO: 152) of human RNA helicase A (RI-IA); the sequence
KRSFSKAF (SEQ ID NO: 153) of nucleolar RNA helicase II; the sequence
KLKIKRPVIC (SEQ
ID NO: 154) of TUS-protein; the sequence PKKICRKVPPPPAAICRVICLD (SEQ ID NO:
155)
associated with importin-alpha; the sequence PKTRRRPRRSQRICRPPT (SEQ ID NO:
156)
from the Rex protein in HTLV-I; the sequence MSRRRKANPTKLSENAKKLAKEVEN (SEQ
ID NO: 157) from the EGL-13 protein of Caenorhabditis elegans; and the
sequences
KTRRRPRRSQRICRPPT (SEQ ID NO: 158), RRICKRRPRRKKRR (SEQ ID NO: 159),
PKKKSRKPKKKSRK (SEQ ID NO: 160), TIKKKHPDASVNFSEFSK (SEQ ID NO: 161),
QRPGPYDRPQRPGPYDRP (SEQ ID NO: 162), LSPSLSPLLSPSLSPL (SEQ ID NO: 163),
RGKGGKGLGKGGAKRIIRIC (SEQ ID NO: 164), PKRGRGRPKRGRGR (SEQ ID NO: 165),
and PKKKRKVPPPPICKICRKV (SEQ ID NO: 166). In general, NLS (or multiple NLSs)
are of
sufficient strength to drive accumulation of a reference or CasX variant
fusion protein in the
nucleus of a eukaryotic cell. Detection of accumulation in the nucleus may be
performed by any
suitable technique. For example, a detectable marker may be fused to a
reference or CasX
variant fusion protein such that location within a cell may be visualized.
Cell nuclei may also be
isolated from cells, the contents of which may then be analyzed by any
suitable process for
detecting protein, such as immunohistochemistry, Western blot, or enzyme
activity assay.
Accumulation in the nucleus may also be determined.
111
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
002391 In some cases, a reference or CasX variant fusion protein includes a
"Protein
Transduction Domain" or PTD (also known as a CPP - cell penetrating peptide),
which refers to
a protein, polynucleotide, carbohydrate, or organic or inorganic compound that
facilitates
traversing a lipid bilayer, micelle, cell membrane, organelle membrane, or
vesicle membrane. A
PTD attached to another molecule, which can range from a small polar molecule
to a large
macromolecule and/or a nanoparticle, facilitates the molecule traversing a
membrane, for
example going from an extracellular space to an intracellular space, or from
the cytosol to within
an organelle. In some embodiments, a PTD is covalently linked to the amino
terminus of a
reference or CasX variant fusion protein. In some embodiments, a PTD is
covalently linked to
the carboxyl terminus of a reference or CasX variant fusion protein. In some
cases, the PTD is
inserted internally in the sequence of a reference or CasX variant fusion
protein at a suitable
insertion site. In some cases, a reference or CasX variant fusion protein
includes (is conjugated
to, is fused to) one or more PTDs (e.g., two or more, three or more, four or
more PTDs). In some
cases, a PTD includes one or more nuclear localization signals (NLS). Examples
of PTDs
include but are not limited to peptide transduction domain of HIV TAT
comprising
YGRKKRRQRRR (SEQ ID NO: 167), RICKRRQRR (SEQ ID NO: 168); YARAAARQARA
(SEQ ID NO: 169); THRLPRRRRRR (SEQ ID NO: 170); and GGRRARRRRRR (SEQ ID NO:
171); a polyarginine sequence comprising a number of arginines sufficient to
direct entry into a
cell (e.g., 3, 4, 5, 6, 7, 8, 9, 10, or 10-50 arginines (SEQ ID NO: 172)); a
VP22 domain (Zender
et al. (2002) Cancer Gene Ther. 9(6):489-96); an Drosophila Antennapedia
protein transduction
domain (Noguchi et al. (2003) Diabetes 52(7): 1732-1737); a truncated human
calcitonin peptide
(Trehin et at. (2004) Pharm. Research 21 :1248-1256); polylysine (Wender et
al. (2000) Proc.
Natl. Acad. Sci. USA 97: 13003-13008); RRQRRTSICLM1CR (SEQ ID NO: 173);
Transportan
GWTLNSAGYLLGKINLKALAALAKKIL (SEQ ID NO: 174);
KALAWEAICLAKALAICALAKHLAKALAKALKCEA (SEQ ID NO: 175); and
RQIICIWFQNRRMKWIC_K (SEQ ID NO: 176). In some embodiments, the PTD is an
activatable
CPP (ACPP) (Aguilera et al. (2009) Integr Biol (Camb) June; 1(5-6): 371-381).
ACPPs
comprise a polycationic CPP (e.g,, Arg9 or "R9") connected via a cleavable
linker to a matching
polyanion (e.g., Glu9 or "E9"), which reduces the net charge to nearly zero
and thereby inhibits
adhesion and uptake into cells. Upon cleavage of the linker, the polyanion is
released, locally
unmasking the polyarginine and its inherent adhesiveness, thus "activating"
the ACPP to
traverse the membrane.
112
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00240] In some embodiments, a reference or CasX variant fusion protein can
include a CasX
protein that is linked to an internally inserted heterologous amino acid or
heterologous
polypeptide (a heterologous amino acid sequence) via a linker polypeptide
(e.g., one or more
linker polypeptides). In some embodiments, a reference or CasX variant fusion
protein can be
linked at the C-terminal and/or N-terminal end to a heterologous polypeptide
(fusion partner) via
a linker polypeptide (e.g., one or more linker polypeptides) The linker
polypeptide may have
any of a variety of amino acid sequences. Proteins can be joined by a spacer
peptide, generally
of a flexible nature, although other chemical linkages are not excluded.
Suitable linkers include
polypeptides of between 4 amino acids and 40 amino acids in length, or between
4 amino acids
and 25 amino acids in length. These linkers are generally produced by using
synthetic, linker-
encoding oligonucleotides to couple the proteins. Peptide linkers with a
degree of flexibility can
be used. The linking peptides may have virtually any amino acid sequence,
bearing in mind that
the preferred linkers will have a sequence that results in a generally
flexible peptide. The use of
small amino acids, such as glycine and alanine, are of use in creating a
flexible peptide. The
creation of such sequences is routine to those of skill in the art. A variety
of different linkers are
commercially available and are considered suitable for use. Example linker
polypeptides include
glycine polymers (G)n, glycine-serine polymer (including, for example, (GS)n,
GSGGSn (SEQ
ID NO: 177), GGSGGSn (SEQ ID NO: 178), and GGGSn (SEQ ID NO: 179), where n is
an
integer of at least one), glycine-alanine polymers, alanine-serine polymers,
glycine-proline
polymers, proline polymers and proline-alanine polymers. Example linkers can
comprise amino
acid sequences including, but not limited to, GGSG (SEQ ID NO: 180), GGSGG
(SEQ ID NO:
181), GSGSG (SEQ ID NO: 182), GSGGG (SEQ ID NO: 183), GGGSG (SEQ ID Na 184),
GSSSG (SEQ ID NO: 185),GPGP (SEQ ID NO: 186), GOP, PPP, PPAPPA (SEQ ID NO:
187),
PPPGPPP (SEQ ID NO: 188) and the like. The ordinarily skilled artisan will
recognize that
design of a peptide conjugated to any elements described above can include
linkers that are all or
partially flexible, such that the linker can include a flexible linker as well
as one or more
portions that confer less flexible structure.
h. Guide Nucleic Acids of XDP Systems
[00241] In another aspect, the disclosure relates to XDP system components
that encode or
incorporate guide nucleic acids (gNA) of the CasX:gNA systems wherein the gNA
comprises a
targeting sequence engineered to be complementary to a target nucleic acid
sequence to be
edited. In some embodiments, the gNA is capable of forming a complex with a
CRISPR protein
113
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
that has specificity to a protospacer adjacent motif (PAM) sequence comprising
a TC motif in
the complementary non-target strand, and wherein the PAM sequence is located 1
nucleotide 5'
of the sequence in the non-target strand that is complementary to the target
nucleic acid
sequence in the target strand of the target nucleic acid. In some embodiments,
the gNA is
capable of forming a complex with a Class 2, Type V CRISPR nuclease. In a
particular
embodiment, the gNA is capable of forming a complex with a CasX nuclease.
[00242] Reference, or naturally-occurring gNA include, but are not limited to
those isolated or
derived from Deltaproteobacter, Matteson:ewe/es, or Candidatus (as described
in
US20180346927A1 and W02018064371A1, incorporated herein by reference),
including the
sequences of Table 2. In some embodiments of the XDP systems, the disclosure
provides gNA
variants having one or more modifications relative to a naturally-occurring
gNA, the modified
gNA hereinafter referred to as a "gNA variant". hi some cases, the encoded gNA
variant
comprises or consists of a sequence that has at least 1, at least 2, at least
3, at least 4, at least 5, at
least 6, at least 7, at least 8, at least 9, at least 10, or at least 20, or
at least 21, or at least 22, or at
least 23, or at least 24, or at least 25 mutations relative to the sequence of
a reference gNA.
These mutations can be insertions, deletions, nucleotide substitutions, or any
combinations
thereof In some embodiments, the gNA variant is a ribonucleic acid molecule
("gRNA"). In
other embodiments, the gNA variant is a deoxyribonucleic acid molecule
("gDNA") in which
uridine nucleotides have been replaced with thymidina In some embodiments, the
gNA is a
chimera, and comprises both DNA and RNA.
[00243] It is envisioned that in some embodiments of the XDP system, multiple
gNAs (e.g.,
two, three, four or more gNA) are delivered to the target cells or tissues in
the XDP particles for
the modification of a target nucleic acid. For example, when a deletion of a
protein-encoding
gene and/or regulatory element is desired, a pair of gNAs with targeting
sequences to different
regions of the target nucleic acid can be used in order to bind and cleave at
two different sites
within the gene or regulatory element, which is then edited by non-homologous
end joining
(NHEJ), homology-directed repair (HDR), homology-independent targeted
integration (HITI),
micro-homology mediated end joining (MMEJ), single strand annealing (SSA) or
base excision
repair (BER). For example, when an editing event designed to delete one or
more mutant exons
or a sequence of the target nucleic acid having two or more mutations that are
distal to one
another, a pair of gNAs can be incorporated into the XDP such that the CRISPR
nuclease can
bind and cleave at two different sites 5' and 3' of the exon(s) bearing the
mutation(s) within the
114
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
gene. In the context of nucleic acids, cleavage refers to the breakage of the
covalent backbone of
a nucleic acid molecule; either DNA or RNA, by the nuclease. Both single-
stranded cleavage
and double-stranded cleavage are possible, and double-stranded cleavage can
occur as a result of
two distinct single-stranded cleavage events. In some embodiments, small
indels introduced by
the CasX:gNA systems of the embodiments described herein and cellular repair
systems can
restore the protein reading frame of the mutant gene ("reframing" strategy).
When the reframing
strategy is used, the cells may be contacted with a single gNA. In the case of
deleting a long
segment of the gene, the disclosure contemplates use of targeting sequences
that flank the
segment 5' and 3' such that it can be deleted or replaced with a donor
template having the
correct sequence. In other cases, when a deletion or a knock-down/knock-out of
the HTT gene is
desired, a pair of gNAs with targeting sequences to different or overlapping
regions of the target
nucleic acid sequence can be used in order to bind and the CasX to cleave at
two different or
overlapping sites within or proximal to the exon or regulatory element of the
gene, which is then
edited by non-homologous end joining (NHEJ), homology-directed repair (HDR,
which can
include, for example, insertion of a donor template to replace all or a
portion of an HTT exon),
homology-independent targeted integration (HITI), micro-homology mediated end
joining
(MMEJ), single strand annealing (SSA) or base excision repair (BER).
1002441 The gNA variants of the disclosure can be designed and created by a
number of
mutagenesis methods, which may include Deep Mutational Evolution (DME) (as
described in
U.S. patent application serial number PCT/US20/36506, incorporated by
reference, herein), deep
mutational scanning (DMS), error prone PCR, cassette mutagenesis, random
mutagenesis,
staggered extension PCR, gene shuffling, or domain swapping, in order to
generate one or more
8NA variants with enhanced or varied properties relative to the reference gNA.
The activity of
reference gNAs may be used as a benchmark against which the activity of gNA
variants are
compared, thereby measuring improvements in function or other characteristics
of the gNA
variants. In other embodiments, a reference gNA may be subjected to one or
more deliberate,
targeted mutations in order to produce a gNA variant, for example a rationally
designed variant.
1002451 The gNAs of the disclosure comprise two segments: a targeting sequence
and a
protein-binding segment. The targeting segment of a gNA includes a nucleotide
sequence
(referred to interchangeably as a guide sequence, a spacer, a targeter, or a
targeting sequence)
that is complementary to (and therefore hybridizes with) a specific sequence
(a target site) within
the target nucleic acid sequence (e.g., a target ssRNA, a target ssDNA, a
strand of a double
115
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
stranded target DNA, etc.), described more fully below. The targeting sequence
of a gNA is
capable of binding to a target nucleic acid sequence, including a coding
sequence, a complement
of a coding sequence, a non-coding sequence, and to regulatory elements. The
protein-binding
segment (or "activator" or "protein-binding sequence") interacts with (e.g.,
binds to) a CasX
protein as a complex, forming an RNP (described more fully, below). The
protein-binding
segment is alternatively referred to herein as a "scaffold", which is
comprised of several regions,
described more fully, below.
[00246] In the case of a dual guide RNA (dgRNA), the targeter and the
activator portions each
have a duplex-forming segment, where the duplex forming segment of the
targeter and the
duplex-forming segment of the activator have complementarity with one another
and hybridize
to one another to form a double stranded duplex (dsRNA duplex for a gRNA) When
the gNA is
a gRNA, the term "targeter" or "targeter RNA" is used herein to refer to a
crRNA-like molecule
(crRNA: "CRISPR RNA") of a CasX dual guide RNA (and therefore of a CasX single
guide
RNA when the "activator" and the "targeter" are linked together; e.g., by
intervening
nucleotides). The crRNA has a 5' region that anneals with the tracrRNA
followed by the
nucleotides of the targeting sequence. Thus, for example, a guide RNA (dgRNA
or sgRNA)
comprises a guide sequence and a duplex-forming segment of a crRNA, which can
also be
referred to as a crRNA repeat. A corresponding tracrRNA-like molecule
(activator) also
comprises a duplex-forming stretch of nucleotides that forms the other half of
the dsRNA duplex
of the protein-binding segment of the guide RNA. Thus, a targeter and an
activator, as a
corresponding pair, hybridize to form a dual guide NA, referred to herein as a
"dual guide NA",
a "dual-molecule gNA", a "dgNA", a "double-molecule guide NA", or a "two-
molecule guide
NA". Site-specific binding and/or cleavage of a target nucleic acid sequence
(e.g., genomic
DNA) by the CasX protein can occur at one or more locations (e.g., a sequence
of a target
nucleic acid) determined by base-pairing complementarity between the targeting
sequence of the
gNA and the target nucleic acid sequence. Thus, for example, the gNA of the
disclosure have
sequences complementarity to and therefore can hybridize with the target
nucleic acid that is
adjacent to a sequence complementary to a TC PAM motif or a PAM sequence, such
as ATC,
CTC, GTC, or TTC. Because the targeting sequence of a guide sequence
hybridizes with a
sequence of a target nucleic acid sequence, a targeter can be modified by a
user to hybridize with
a specific target nucleic acid sequence, so long as the location of the PAM
sequence is
considered. Thus, in some cases, the sequence of a targeter may be a non-
naturally occurring
116
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
sequence. In other cases, the sequence of a targeter may be a naturally-
occurring sequence,
derived from the gene to be edited. In other embodiments, the activator and
targeter of the gNA
are covalently linked to one another (rather than hybridizing to one another)
and comprise a
single molecule, referred to herein as a "single-molecule gNA," "one-molecule
guide NA,"
"single guide NA", "single guide RNA", a "single-molecule guide RNA," a "one-
molecule
guide RNA", a "single guide DNA", a "single-molecule DNA", or a "one-molecule
guide
DNA", ("sgNA", "sgRNA", or a "sgDNA"). In some embodiments, the sgNA includes
an
"activator" or a "targeter" and thus can be an "activator-RNA" and a "targeter-
RNA,"
respectively.
[00247] Collectively, the assembled gNAs of the disclosure comprise four
distinct regions, or
domains: the RNA triplex, the scaffold stem, the extended stem, and the
targeting sequence that,
in the embodiments of the disclosure is specific for a target nucleic acid and
is located on the
3'end of the gNA. The RNA triplex, the scaffold stem, and the extended stem,
together, are
referred to as the "scaffold" of the gNA.
RNA Triplex
[00248] In some embodiments of the guide NAs provided herein (including
reference sgNAs),
there is a RNA-triplex, and the RNA triplex comprises the sequence of a UUU--
nX(---4-15)--
UUU stem loop (SEQ ID NO: 189) that ends with an AAAG after 2 intervening stem
loops (the
scaffold stem loop and the extended stem loop), forming a pseudoknot that may
also extend past
the triplex into a duplex pseudoknot. The UU-UUU-AAA sequence of the triplex
forms as a
nexus between the spacer, scaffold stem, and extended stem. In exemplary
reference CasX
sgNAs, the UUU-loop-UUU region is coded for first, then the scaffold stem
loop, and then the
extended stem loop, which is linked by the tetraloop, and then an AAAG closes
off the triplex
before becoming the spacer.
f. Scaffold Stem Loop
[00249] In some embodiments of CasX sgNAs of the disclosure, the triplex
region is followed
by the scaffold stem loop. The scaffold stem loop is a region of the gNA that
is bound by CasX
protein (such as a reference or CasX variant protein). In some embodiments,
the scaffold stem
loop is a fairly short and stable stem loop. In some cases, the scaffold stem
loop does not tolerate
many changes, and requires some form of an RNA bubble. In some embodiments,
the scaffold
stem is necessary for CasX sgNA function. While it is perhaps analogous to the
nexus stem of
Cas9 as being a critical stem loop, the scaffold stem of a CasX sgNA, in some
embodiments, has
117
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
a necessary bulge (RNA bubble) that is different from many other stem loops
found in
CRISPR/Cas systems. In some embodiments, the presence of this bulge is
conserved across
sgNA that interact with different CasX proteins. An exemplary sequence
encoding a scaffold
stem loop sequence of a gNA comprises the sequence CCAGCGACTATGTCGTATGG (SEQ
ID NO: 190). In other embodiments, the disclosure provides gNA variants
wherein the scaffold
stem loop is replaced with an RNA stem loop sequence from a heterologous RNA
source with
proximal 5' and 3' ends, such as, but not limited to stem loop sequences
designated as MS2, Q13,
Ul hairpin II, Uvsx, or PP7 stem loops, which can be used, in some cases, to
facilitate transport
out of the host cell nucleus. In some cases, the heterologous RNA stem loop of
the gNA is
capable of binding a protein, an RNA structure, a DNA sequence, or a small
molecule, which
can facilitate the binding of gNA to CasX.
It. Extended Stem Loop
1002501 In some embodiments of the sgNAs of the disclosure, the scaffold stem
loop is
followed by the extended stem loop. In some embodiments, the extended stem
comprises a
synthetic tracr and crRNA fusion that is largely unbound by the CasX protein.
In some
embodiments, the extended stem loop can be highly malleable. In some
embodiments, a single
guide gRNA is made with a GAAA tetraloop linker or a GAGAAA linker between the
tracr and
crRNA in the extended stem loop. In some cases, the targeter and activator of
a CasX sgNA are
linked to one another by intervening nucleotides and the linker can have a
length of from 3 to 20
nucleotides. In some embodiments of the CasX sgNAs of the disclosure, the
extended stem is a
large 32-bp loop that sits outside of the CasX protein in the
ribonucleoprotein complex. An
exemplary sequence encoding an extended stem loop sequence of a sgNA comprises
GCGCTTATTTATCGGAGAGAAATCCGATAAATAAGAAGC (SEQ ID NO: 191). In some
embodiments, the extended stem loop comprises a GAGAAA spacer sequence. In
some
embodiments, the disclosure provides gNA variants wherein the extended stem
loop is replaced
with an RNA stem loop sequence from a heterologous RNA source with proximal 5'
and 3'
ends, such as, but not limited to stem loop sequences designated MS2, Q13, Ul
hairpin II, Uvsx,
or PP7 stem loops. In such cases, the heterologous RNA stem loop increases the
stability of the
gNA. In other embodiments, the disclosure provides gNA variants having an
extended stem
loop region comprising at least 10, at least 100, at least 500, at least 1000,
or at least 10,000
nucleotides, or at least 10-10,000, at least 10-1000, or at least 10-100
nucleotides. In some
embodiments, the extended stem loop comprises a GAGAAA spacer sequence.
118
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
L Targeting Sequence (a.k.a. Spacer)
1002511 In some embodiments of the gNAs of the disclosure utilized in the XDP
systems, the
extended stem loop is followed by a region that forms part of the triplex, and
then the targeting
sequence (or "spacer) at the 3' end of the gNA. The targeting sequence targets
the CasX
ribonucleoprotein hobo complex to a specific region of the target nucleic acid
sequence of the
gene to be modified. Thus, for example, gNA targeting sequences of the
disclosure have
sequences complementarity to, and therefore can hybridize to, a portion of the
HTT gene in a
nucleic acid in a eukaryotic cell (e.g., a eukaryotic chromosome, chromosomal
sequence, a
eukaryotic RNA, etc.) as a component of the RNP when the TC PAM motif or any
one of the
PAM sequences TTC, ATC, GTC, or CTC is located 1 nucleotide 5' to the non-
target strand
sequence complementary to the target sequence. The targeting sequence of a gNA
can be
modified so that the gNA can target a desired sequence of any desired target
nucleic acid
sequence, so long as the PAM sequence location is taken into consideration. In
some
embodiments, the gNA scaffold is 5' of the targeting sequence, with the
targeting sequence on
the 3' end of the gNA. In some embodiments, the PAM motif sequence recognized
by the
nuclease of the RNP is TC. In other embodiments, the PAM sequence recognized
by the
nuclease of the RNP is NTC.
1002521 In some embodiments, the gNA of the XDP systems comprises a targeting
sequence (a)
complementary to a nucleic acid sequence encoding i) a target protein, which
may be a wild-type
sequence or may comprise one or more mutations or ii) the regulatory element
of the protein,
which may be a wild-type sequence; or (b) complementary to a complement of a
nucleic acid
sequence encoding a protein or its regulatory element, which may comprise one
or more
mutations. In some embodiments, the targeting sequence of the gNA is specific
for a portion of
a gene encoding a target protein comprising one or more mutations. In some
embodiments, the
targeting sequence of a gNA is specific for a target gene exon. In some
embodiments, the
targeting sequence of a gNA is specific for a target gene intron. In some
embodiments, the
targeting sequence of the gNA is specific for a target gene intron-exon
junction. In some
embodiments, the targeting sequence of the gNA is complementary to a sequence
comprising
one or more single nucleotide polymorphisms (SNPs) of the target gene or its
complement. In
other embodiments, the targeting sequence of the gNA is complementary to a
sequence of an
intergenic region of the target gene or a sequence complementary to an
intergenic region of the
target gene.
119
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00253] In some embodiments, the targeting sequence of a gNA is specific for a
regulatory
element that regulates expression of a target gene. Such regulatory elements
include, but are not
limited to promoter regions, enhancer regions, intergenic regions, 5'
untranslated regions (5'
UTR), 3' untranslated regions (3' UTR), intergenic regions, gene enhancer
elements, conserved
elements, and regions comprising cis-regulatory elements. The promoter region
is intended to
encompass nucleotides within 5 kb of the target gene initiation point or, in
the case of gene
enhancer elements or conserved elements, can be 1 Mb or more distal to the
target gene. In some
embodiments, the disclosure provides a gNA with a targeting sequence that
hybridizes with
target gene regulatory element. In the foregoing, the targets are those in
which the encoding
gene of the target is intended to be knocked out or knocked down such that the
target protein
comprising mutations is not expressed or is expressed at a lower level in a
cell. In some
embodiments, the disclosure provides a CasX:gNA system wherein the targeting
sequence (or
spacer) of the gNA is complementary to a nucleic acid sequence encoding the
target protein, a
portion of the target protein, a portion of a regulatory element, or the
complement of a portion of
a gene or a regulatory element for the target gene. In some embodiments, the
targeting sequence
has between 14 and 35 consecutive nucleotides. In some embodiments, the
targeting sequence
has 14, 15, 16, 18, 18, 19, 20, 21, 22, 23 24, 25, 26, 27, 28, 29, 30, 31, 32,
33, 34 or 35
consecutive nucleotides. In some embodiments, the targeting sequence consists
of 20
consecutive nucleotides. In some embodiments, the targeting sequence consists
of 19
consecutive nucleotides. In some embodiments, the targeting sequence consists
of 18
consecutive nucleotides. In some embodiments, the targeting sequence consists
of 17
consecutive nucleotides. In some embodiments, the targeting sequence consists
of 16
nucleotides. In some embodiments, the targeting sequence consists of 15
nucleotides. In some
embodiments, the targeting sequence has 14, 15, 16, 17, 18, 19, 20, 21, 22,
23, 24, 25, 26, 27,
28, 29, 30, 31, 32, 33, 34 or 35 consecutive nucleotides and the targeting
sequence can comprise
0 to 5, 0 to 4, 0 to 3, or 0 to 2 mismatches relative to the target nucleic
acid sequence and retain
sufficient binding specificity such that the RNP comprising the gNA comprising
the targeting
sequence can form a complementary bond with respect to the target nucleic
acid.
[00254] In some embodiments, the CasX:gNA of the XDP system comprises a first
gNA and
further comprises a second (and optionally a third, fourth or fifth) gNA,
wherein the second gNA
has a targeting sequence complementary a different portion of the target
nucleic acid or its
complement compared to the targeting sequence of the first gNA. By selection
of the targeting
120
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
sequences of the gNA, defined regions of the target nucleic acid can be
modified or edited using
the CasX:gNA systems described herein.
gNA scaffolds
1002551 With the exception of the targeting sequence region, the remaining
regions of the gNA
are referred to herein as the scaffold. In some embodiments, the gNA scaffolds
are derived from
naturally-occurring sequences, described below as reference gNA. In other
embodiments, the
gNA scaffolds are variants of reference gNA wherein mutations, insertions,
deletions or domain
substitutions are introduced to confer desirable properties on the gNA
variant.
1002561 In some embodiments, a reference gRNA comprises a sequence isolated or
derived
from Deltaproteobacteria. In some embodiments, the sequence is a CasX tracrRNA
sequence.
Exemplary CasX reference tracrRNA sequences isolated or derived from
Deltaproteobacteria
may include:
ACAUCUGGCGCGUUUAUUCCAUUAC1UUUGGAGCCAGUCCCAGCGACUAUGUCGU
AUGGACGAAGCGCUUAUUUAUCGGAGA (SEQ ID NO: 6) and
ACAUCUGGCGCGUUUAUUCCAUUACUIJUGGAGCCAGUCCCAGCGACUAUGUCGU
AUGGACGAAGCGCIUUAUUUAUCGG (SEQ ID NO: 7). Exemplary crRNA sequences
isolated or derived from Deltaproteobacter may comprise a sequence of
CCGAUAAGUAAAACGCAUCAAAG (SEQ ID NO: 194). In some embodiments, a CasX
reference gNA comprises a sequence at least 60% identical, at least 65%
identical, at least 70%
identical, at least 75% identical, at least 80% identical, at least 81%
identical, at least 82%
identical, at least 83% identical, at least 84% identical, at least 85%
identical, at least 86%
identical, at least 86% identical, at least 87% identical, at least 88%
identical, at least 89%
identical, at least 89% identical, at least 90% identical, at least 91%
identical, at least 92%
identical, at least 93% identical, at least 94% identical, at least 95%
identical, at least 96%
identical, at least 97% identical, at least 98% identical, at least 99%
identical, at least 99.5%
identical or 100% identical to a sequence isolated or derived from
Deltaproteobacter. In some
embodiments, a reference guide RNA comprises a sequence isolated or derived
from
Planctomycetes. In some embodiments, the sequence is a CasX tracrRNA sequence.
Exemplary
reference tracrRNA sequences isolated or derived from Planctomycetes may
include:
UACUGGCGCULTUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGACUAUGUCGUA
UGGGUAAAGCGCUUAUUUAUCGGAGA (SEQ ID NO: 8) and
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGACUAUGUCGUA
121
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
UGGGUAAAGCGCUUAUUUAUCGG (SEQ ID NO: 9). Exemplary crRNA sequences
isolated or derived from Planctotnycetes may comprise a sequence of
UCUCCGAUAAAUAAGAAGCAUCAAAG (SEQ ID NO: 197)_ In some embodiments, a
CasX reference gNA comprises a sequence at least 60% identical, at least 65%
identical, at least
70% identical, at least 75% identical, at least 80% identical, at least 81%
identical, at least 82%
identical, at least 83% identical, at least 84% identical, at least 85%
identical, at least 86%
identical, at least 86% identical, at least 87% identical, at least 88%
identical, at least 89%
identical, at least 89% identical, at least 90% identical, at least 91%
identical, at least 92%
identical, at least 93% identical, at least 94% identical, at least 95%
identical, at least 96%
identical, at least 97% identical, at least 98% identical, at least 99%
identical, at least 99.5%
identical or 100% identical to a sequence isolated or derived from
Planctomycetes.
1002571 In some embodiments, a reference gNA comprises a sequence isolated or
derived from
Candidatus Sungbacteria. In some embodiments, the sequence is a CasX tracrRNA
sequence.
Exemplary CasX reference tracrRNA sequences isolated or derived from
Candidatus
Sungbacteria may comprise sequences of: GUULTACACACUCCCUCUCAUAGGGU (SEQ ID
NO: 10), GUUUACACACUCCCUCUCAUGAGGU (SEQ ID NO: 11),
UUUUACAUACCCCCUCUCAUGGGAU (SEQ ID NO: 12) and
GUUUACACACUCCCUCUCAUGGGGG (SEQ ID NO: 13). In some embodiments, a CasX
reference guide RNA comprises a sequence at least 60% identical, at least 65%
identical, at least
70% identical, at least 75% identical, at least 80% identical, at least 81%
identical, at least 82%
identical, at least 83% identical, at least 84% identical, at least 85%
identical, at least 86%
identical, at least 86% identical, at least 87% identical, at least 88%
identical, at least 89%
identical, at least 89% identical, at least 90% identical, at least 91%
identical, at least 92%
identical, at least 93% identical, at least 94% identical, at least 95%
identical, at least 96%
identical, at least 97% identical, at least 98% identical, at least 99%
identical, at least 99.5%
identical or 100% identical to a sequence isolated or derived from Candidatus
Sungbacteria.
[00258] Table 2 provides the sequences of reference gRNAs tracr, cr and
scaffold sequences.
In some embodiments, the disclosure provides gNA sequences wherein the gNA has
a scaffold
comprising a sequence having at least one nucleotide modification relative to
a reference gNA
sequence having a sequence of any one of SEQ ID NOS: 4-16 of Table 2. It will
be understood
that in those embodiments wherein a vector comprises a DNA encoding sequence
for a gNA, or
where a gNA is a gDNA or a chimera of RNA and DNA, that thymine (T) bases can
be
122
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
substituted for the uracil (U) bases of any of the gNA sequence embodiments
described herein,
including the sequences of Table 2 and Table 3.
Table 2. Reference gRNA tracr and scaffold sequences
SEQ ID NO. Nucleotide Sequence
4
ACAUCUGGCGCGUUUAUUCCAUUACUUUGGAGCCAGUCCCAGCG
ACUAUGUCGUAUGGACGAAGCGCUUAUUUAUCGGAGAGAAACCG
AUAAGUAAAACGCAUCAAAG
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGA
CUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAGAGAAAUCCG
AUAAAUAAGAAGCAUCAAAG
6
ACAUCUGGCGCGUUUAUUCCAUUACUUUGGAGCCAGUCCCAGCG
ACUAUGUCGUAUGGACGAAGCGCUUAUUUAUCGGAGA
7
ACAUCUGGCGCGUUUAUUCCAUUACUUUGGAGCCAGUCCCAGCG
ACUAUGUCGUAUGGACGAAGCGCUUAUUUAUCGG
8
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGA
CUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAGA
9
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGA
CUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGG
GUUUACACACUCCCUCUCAUAGGGU
11 GUUUACACACUCCCUCUCAUGAGGU
12 UUUUACAUACCCCCUCUCAUGGGAU
13 GUUUACACACUCCCUCUCAUGGGGG
14 CCAGCGACUAUGUCGUAUGG
GCGCUUAUUUAUCGGAGAGAAAUCCGAUAAAUAAGAAGC
16
GGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGACUAU
GUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
n. gNA Variants
[00259] In another aspect, the disclosure relates to guide nucleic acid
variants (referred to
herein alternatively as "gNA variant" or "gRNA variant" when the nucleic acid
variant
comprises RNA), which comprise one or more modifications relative to a
reference gRNA
scaffold. As used herein, "scaffold" refers to all parts to the gNA necessary
for gNA function
with the exception of the spacer sequence
[00260] In some embodiments, a gNA variant comprises one or more nucleotide
substitutions,
insertions, deletions, or swapped or replaced regions relative to a reference
gRNA sequence of
the disclosure. In some embodiments, a mutation can occur in any region of a
reference gRNA to
produce a gNA variant. In some embodiments, the scaffold of the gNA variant
sequence has at
123
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
least 20%, at least 30%, at least 40%, at least 50%, at least 60%, or at least
70%, at least 80%, at
least 85%, at least about 90%, at least about 95%, at least about 96%, at
least about 97%, at least
about 98%, or at least about 99% identity to the sequence of SEQ ID NO: 4 or
SEQ ID NO: 5.
[00261] In some embodiments, a gNA variant comprises one or more nucleotide
changes within
one or more regions of the reference gRNA that improve a characteristic
relative to the reference
gRNA. Exemplary regions include the RNA triplex, the pseudoknot, the scaffold
stem loop, and
the extended stem loop. In some cases, the variant scaffold stem further
comprises a bubble. In
other cases, the variant scaffold further comprises a triplex loop region. In
still other cases, the
variant scaffold further comprises a 5' unstructured region. In one
embodiment, the gNA variant
scaffold comprises a scaffold stem loop having at least 60% sequence identity
to SEQ ID NO:
14. In another embodiment, the gNA variant comprises a scaffold stem loop
having the
sequence of CCAGCGACUAUGUCGUAGUGG (SEQ ID NO: 202). In another embodiment,
the disclosure provides a gNA scaffold comprising, relative to SEQ ID NO:5, a
C I8G
substitution, a G55 insertion, a U1 deletion, and a modified extended stem
loop in which the
original 6 nt loop and 13 most-loop-proximal base pairs (32 nucleotides total)
are replaced by a
Uvsx hairpin (4 nt loop and 5 loop-proximal base pairs; 14 nucleotides total)
and the loop-distal
base of the extended stem was converted to a fully base-paired stem contiguous
with the new
Uvsx hairpin by deletion of the A99 and substitution of 664U. In the foregoing
embodiment,
the gNA scaffold comprises the sequence
ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCAGCGACUAUGUCGUAG
UGGGUAAAGCUCCCUCUUCGGAGGGAGCAUCAAAG (SEQ ID NO: 734).
[00262] All gNA variants that have one or more improved functions or
characteristics, or add
one or more new functions when the variant gNA is compared to a reference gRNA
described
herein, are envisaged as within the scope of the disclosure. A representative
example of such a
gNA variant is guide 174 (SEQ ID NO: 734). In some embodiments, the gNA
variant adds a
new function to the RNP comprising the gNA variant. In some embodiments, the
gNA variant
has an improved characteristic selected from: improved stability; improved
solubility; improved
transcription of the gNA; improved resistance to nuclease activity; increased
folding rate of the
gNA; decreased side product formation during folding; increased productive
folding; improved
binding affinity to a CasX protein; improved binding affinity to a target DNA
when complexed
with a CasX protein; improved gene editing when complexed with a CasX protein;
improved
specificity of editing when complexed with a CasX protein; and improved
ability to utilize a
124
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
greater spectrum of one or more PAM sequences, including ATC, CTC, GTC, or
TTC, in the
editing of target DNA when complexed with a CasX protein, or any combination
thereof In
some cases, the one or more of the improved characteristics of the gNA variant
is at least about
1.1 to about 100,000-fold improved relative to the reference gNA of SEQ ID NO:
4 or SEQ ID
NO: 5. In other cases, the one or more improved characteristics of the gNA
variant is at least
about 1.1, at least about 10, at least about 100, at least about 1000, at
least about 10,000, at least
about 100,000-fold or more improved relative to the reference gNA of SEQ ID
NO: 4 or SEQ ID
NO: 5. In other cases, the one or more of the improved characteristics of the
gNA variant is
about 1.1 to 100,00-fold, about 1.1 to 10,00-fold, about 1.1 to 1,000-fold,
about 1.1 to 500-fold,
about 1.1 to 100-fold, about 1.1 to 50-fold, about 1.1 to 20-fold, about 10 to
100,00-fold, about
to 10,00-fold, about 10 to 1,000-fold, about 10 to 500-fold, about 10 to 100-
fold, about 10 to
50-fold, about 10 to 20-fold, about 2 to 70-fold, about 2 to 50-fold, about 2
to 30-fold, about 2 to
20-fold, about 2 to 10-fold, about 5 to 50-fold, about 5 to 30-fold, about 5
to 10-fold, about 100
to 100,00-fold, about 100 to 10,00-fold, about 100 to 1,000-fold, about 100 to
500-fold, about
500 to 100,00-fold, about 500 to 10,00-fold, about 500 to 1,000-fold, about
500 to 750-fold,
about 1,000 to 100,00-fold, about 10,000 to 100,00-fold, about 20 to 500-fold,
about 20 to 250-
fold, about 20 to 200-fold, about 20 to 100-fold, about 20 to 50-fold, about
50 to 10,000-fold,
about 50 to 1,000-fold, about 50 to 500-fold, about 50 to 200-fold, or about
50 to 100-fold,
improved relative to the reference gNA of SEQ ID NO: 4 or SEQ ID NO: 5. In
other cases, the
one or more improved characteristics of the gNA variant is about 1.1-fold, 1.2-
fold, 13-fold,
1.4-fold, 1.5-fold, 1.6-fold, 1.7-fold, 1.8-fold, 1.9-fold, 2-fold, 3-fold, 4-
fold, 5-fold, 6-fold, 7-
fold, 8-fold, 9-fold, 10-fold, 11-fold, 12-fold, 13-fold, 14-fold, 15-fold, 16-
fold, 17-fold, 18-fold,
19-fold, 20-fold, 25-fold, 30-fold, 40-fold, 45-fold, 50-fold, 55-fold, 60-
fold, 70-fold, 80-fold,
90-fold, 100-fold, 110-fold, 120-fold, 130-fold, 140-fold, 150-fold, 160-fold,
170-fold, 180-fold,
190-fold, 200-fold, 210-fold, 220-fold, 230-fold, 240-fold, 250-fold, 260-
fold, 270-fold, 280-
fold, 290-fold, 300-fold, 310-fold, 320-fold, 330-fold, 340-fold, 350-fold,
360-fold, 370-fold,
380-fold, 390-fold, 400-fold, 425-fold, 450-fold, 475-fold, or 500-fold
improved relative to the
reference gNA of SEQ ID NO: 4 or SEQ ID NO; 5.
1002631 In some embodiments, a gNA variant can be created by subjecting a
reference gRNA
to a one or more mutagenesis methods, such as the mutagenesis methods
described herein,
below, which may include Deep Mutational Evolution (DME), deep mutational
scanning
(DMS), error prone PCR, cassette mutagenesis, random mutagenesis, staggered
extension PCR,
125
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
gene shuffling, or domain swapping, in order to generate the gNA variants of
the disclosure. The
activity of reference gRNAs may be used as a benchmark against which the
activity of gNA
variants are compared, thereby measuring improvements in function of gNA
variants. In other
embodiments, a reference gRNA may be subjected to one or more deliberate,
targeted mutations,
substitutions, or domain swaps in order to produce a gNA variant, for example
a rationally
designed variant. Exemplary gRNA variants produced by such methods are
described in the
Examples and representative sequences of gNA scaffolds are presented in Table
3.
[00264] In some embodiments, the gNA variant comprises one or more
modifications
compared to a reference guide nucleic acid scaffold sequence, wherein the one
or more
modification is selected from: at least one nucleotide substitution in a
region of the gNA variant;
at least one nucleotide deletion in a region of the gNA variant; at least one
nucleotide insertion
in a region of the gNA variant; a substitution of all or a portion of a region
of the gNA variant; a
deletion of all or a portion of a region of the gNA variant; or any
combination of the foregoing.
In some cases, the modification is a substitution of 1 to 15 consecutive or
non-consecutive
nucleotides in the gNA variant in one or more regions. In other cases, the
modification is a
deletion of 1 to 10 consecutive or non-consecutive nucleotides in the gNA
variant in one or more
regions. In other cases, the modification is an insertion of 1 to 10
consecutive or non-consecutive
nucleotides in the gNA variant in one or more regions. In other cases, the
modification is a
substitution of the scaffold stem loop or the extended stem loop with an RNA
stem loop
sequence from a heterologous RNA source with proximal 5' and 3' ends. In some
cases, a gNA
variant of the disclosure comprises two or more modifications in one region.
In other cases, a
gNA variant of the disclosure comprises modifications in two or more regions_
In other cases, a
gNA variant comprises any combination of the foregoing modifications described
in this
paragraph.
[00265] In some embodiments, a 5' G is added to a gNA variant sequence for
expression in
vivo, as transcription from a U6 promoter is more efficient and more
consistent with regard to
the start site when the +1 nucleotide is a G. In other embodiments, two 5' Gs
are added to a gNA
variant sequence for in vitro transcription to increase production efficiency,
as T7 polymerase
strongly prefers a G in the +1 position and a purine in the +2 position. In
some cases, the 5' G
bases are added to the reference scaffolds of Table 2. In other cases, the 5'
G bases are added to
the variant scaffolds of Table 3.
126
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1002661 Table 3 provides exemplary gNA variant scaffold sequences of the
disclosure. In
Table 3, (-) indicates a deletion at the specified position(s) relative to the
reference sequence of
SEQ ID NO: 5, (+) indicates an insertion of the specified base(s) at the
position indicated
relative to SEQ ID NO: 5, (:) indicates the range of bases at the specified
start: stop coordinates
of a deletion or substitution relative to SEQ ID NO: 5, and multiple
insertions, deletions or
substitutions are separated by commas; e.g., A14C, T17G. In some embodiments,
the gNA
variant scaffold comprises any one of the sequences listed in Table 3, or SEQ
ID NOS: 597-781,
or a sequence having at least about 50%, at least about 60%, at least about
70%, at least about
80%, at least about 90%, at least about 95%, at least about 95%, at least
about 96%, at least
about 97%, at least about 98%, at least about 99% sequence identity thereto.
It will be
understood that in those embodiments wherein a vector comprises a DNA encoding
sequence for
a gNA, or where a gNA is a gDNA or a chimera of RNA and DNA, that thymine (T)
bases can
be substituted for the uracil (U) bases of any of the gNA sequence embodiments
described
herein.
Table 3. Exemplary gNA Variant Scaffold Sequences
SEQ
NUCLEOTIDE SEQUENCE
NAME or
ID,
NO: Modification
597 phage
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
replication
AGCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGACGA
stable CCUCUCGGUCGUCCUAUCUGAAGCAUCAAAG
598 Kissing
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop_bl
AGCGACUAUGUCGUAUGGGUAAAGCGCUGCUCGACGCG
UCCUCGAGCAGAAGCAUCAAAG
599 Kissing
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop_a
AGCGACUAUGUCGUAUGGGUAAAGCGCUGCUCGCUCCG
UUCGAGCAGAAGCAUCAAAG
600 32: uvsX
GUACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCAC
hairpin
CAGCGACUAUGUCGUAUGGGUAAAGCOCCCUCUUCGGA
GGGAAGCAUCAAAG
601 PP7
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCAGGAGUUUCUAU
GGAAACCCUGAAGGAUCAAAG
602 64: trip mut, GUACUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCAC
extended stem CAGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUC
truncation GGUCCGUAAGAAGCAUCAAAG
127
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
603 hyperstable UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
tetraloop
AGCGACUAUGUCGUAUGGGUAAAGCGCUGCGCUUGGGC
AGAAGCAUCAAAG
604 C18G
UACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
605 U17G
UACUGGCGCUUUUAUCGCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
606 CUUCGG UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GACUUCGGUCCGAUAAAUAAGAAGCAUCAAAG
607 MS2
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCACAUGAGGAUUA
CCCAUGUGAAGCAUCAAAG
608
-1, A2G, -78,
GCUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
G77U
GCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGUGA
GAAAUCCGAUAAAUAAGAAGCAUCAAAG
609 QB
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUGCAUGUCUAAG
ACAGCAGAAGCAUCAAAG
610
45,44 hairpin
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCAGGGCUUCGGC
CGAAGCAUCAAAG
611 U 1 A
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCAAUCCAUUGCAC
UCCGGAUUGAAGCAUCAAAG
612
A14C, U17G
UACUGGCGCUUUUCUCGCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
613 CUUCGG UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop modified AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
CUUCGGUCCGAUAAAUAAGAAGCAUCAAAG
614 Kissing
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop ¨b2
AGCGACUAUGUCGUAUGGGUAAAGCGCUGCUCGUUUGC
GGCUACGAGCAGAAGCAUCAAAG
615
-76:78, -83:87
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGAG
AGAUAAAUAAGAAGCAUCAAAG
128
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
in NAME or
NO: Modification
616 -4
UACGGCGCU
UUUAUCUCAUUACUUUGAGAGCCAUCACCA
GCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
617
extended stem
UACUGGCGCCUUUUAUCUCAUUACUUUGAGAGCCAUCAC
truncation
CAGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACU UC
GGUCCGUAAGAAGCAUCAAAG
618 C55
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUCGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
619 trip mut
UAC
UGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
CUUCGGUCCGAUAAAUAAGAAGCAUCAAAG
620 -76:78
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGAG
AAAU CC GAUAAAUAAGAAGCAUCAAAG
621 -1:5
GCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGA
CUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAGAGAA
AUCCGAUAAAUAAGAAGCAUCAAAG
622 -83:87
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAGAUAAAUAAGAAGCAUCAAAG
623 =+G28,
UACUGGCGCUUUUAUCUCAUUACUUUGGAGAGCCAUCAC
A82U, -84, CAGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGG
AGAGUAUCCGAUAAAUAAGAAGCAUCAAAG
624 =+51U
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUUCGUAUGGGUAAAGCGCUUAUUUAUCGG
AGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
625
-1:4, +GSA,
AGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCG
+G86,
ACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAGAGA
AAUGCCGAUAAAUAAGAAGCAUCAAAG
626 =+A94
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAAUAAGAAGCAUCAAAG
627 =+G72
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUGUAUCG
GAGAGAAAU CC GAUAAAUAAGAAG CAU CAAAG
628
shorten front,
GCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGA
CLTUCGG
CUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGACUUCG
loop modified. GUCCGAUAAAUAAGCGCAUCAAAG
129
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
in NAME or
NO: Modification
extend
extended
629 A14C
UACUGGCGCUUUUCUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
630 -1:3, +63
GUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAG
CGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAU CGGAGA
GAAAUCCGAUAAAUAAGAAGCAUCAAAG
631 =+C45, +U46 UAC UGGCGC UUUUAUCUCAUUAC U U UGAGAGC CAU
CAC C
AGC GAC C UUAUG U CGUAU GGG UAAAGC GC U UAUU UAUCG
GAGAGAAAU CC GAUAAAUAAGAAG CAU CAAAG
632 CUUCGG GAUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
loop modified, GCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAC
fun start UUCGGUCCGAUAAAUAAGAAGCAUCAAAG
633 -93:94 UAC UGGCGC U U U UAUCU CAU UAC U
U UGAGAGCCAU CAC C
AGC GAC UAU GUCGUAUGGGUAAAGC GC U UAUU UAUCGGA
GAGAAAUCC GAUAAAAGAAGCAUCAAAG
634 =+U45 UAC UGGCGC UUUUAUCUCAUUAC U U
UGAGAGC CAU CAC C
AGCGAUCUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGG
AGA GAAA U CCGAUAAAUAAGAAG CAU CAAAG
635 -69, -94 UAC UGGCGC U U U UAUCU CAU UAC U
U UGAGAGCCAU CAC C
AGCGACUAUGUCGUAUGGGUAAAGGCUUAUUUAUCGGAG
AGAAAU C C GAUAAAAAGAAGCAU CAAAG
636 -94 UAC UGGCGC U U U UAUCU CAU UAC U
U UGAGAGCCAU CAC C
AGC GAC UAU GUCGUAUGGGUAAAGC GC U UAUU UAUCGGA
GAGAAAUCC GAUAAAAAGAAGCAU CAAAG
637 modified
UACUGGCGCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
CUUCGG, GCOACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUGGGAC
minus U in 1st UUCGGUCCGAUAAAUAAGAAGCAUCAAAG
triplex
638 -1:4, +C4, CGGCGCU UUUC
UCGCAUUACUUUGAGAGCCAUCACCAGC
A14C, U17G, GACUAUGUCGUAUGGGUAAAGCGCUUAUUGUAUCGAGAG
+G72, -76:78, AUAAAUAAGAAGCAUCAAAG
-83:87
639 U1C, -73 CAC UGGCGC U U U UAUCU CAU UAC U
U UGAGAGCCAU CAC C
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
640 Scaffold
UACUGGCGCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
uuCG, stem GCGACUUCGGUCGUAUGGGUAAAGCGCUUAUGUAUCGG
uuCG. Stem CUUCGGCCGAUACAUAAGAAGCAUCAAAG
130
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
swap, t
shorten
641 Scaffold
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
uuCG, stem AGCGACUUCGGUCGUAUGGGUAAAGCGCUUAUGUAUCG
uuCG. Stem GCUUCGGCCGAUACAUAAGAAGCAUCAAAG
swap
642 =+G60
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUGAAAGCGCUUAUUUAUCG
GAGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
643 no stem
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
Scaffold AGCGACUUCGGUCGUAUGGGUAAAG
uuCG
644 no stem
GAUGGGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAG
Scaffold CGACUUCGGUCGUAUGGGUAAAG
uuCG, fun
start
645 Scaffold
GAUGGGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAG
uuCG, stem CGACUUCGGUCGUAUGGGUAAAGCGCUUAUUUAUCGGC
uuCG, fun UUCGGCCGAUAAAUAAGAAGCAUCAAAG
start
646 Pseudoknots UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUACACUGGGAUC
GCUGAAUUAGAGAUCGGCGUCCUUUCAUUCUAUAUACUU
UGGAGUUUUAAAAUGUCUCUAAGUACAGAAGCAUCAAAG
647 Scaffold
GGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCG
uuCG, stem ACUUCGGUCGUAUGGGUAAAGCGCUUAUUUAUCGGCUU
uuCG CGGCCGAUAAAUAAGAAGCAUCAAAG
648 Scaffold
GCUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
uuCG, stem GCGACUUCGGUCGUAUGGGUAAAGCGCUUAUUUAUCGG
uuCG, no start CUUCGGCCGAUAAAUAAGAAGCAUCAAAG
649 Scaffold
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
uuCG
AGCGACUUCGGUCGUAUGGGUAAAGCGCUUAUUUAUCG
GAGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
650 =+GCUC36 UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUGCUC
CACCAGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAU
CGGAGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
651 G quadriplex UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
te1omere
AGCGACUAUGUCGUAUGGGUAAAGCGGGGUUAGGGUUA
basket+ ends GGGUUAGGGAAGCAUCAAAG
131
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
652 G quadriplex UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
M3q
AGCGACUAUGUCGUAUGGGUAAAGCGGAGGGAGGGAGG
GAGAGGGAAAGCAUCAAAG
653 G quadriplex UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
telomere
AGCGACUAUGUCGUAUGGGUAAAGCGUUGGGUUAGGGU
basket no ends UAGGGUUAGGGAAAAGCAUCAAAG
654 45,44 hairpin UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
(old version) AGCGACUAUGUCGUAUGGGUAAAGCGC-----
AGGGCUUCGGCCG----GAAGCAUCAAAG
655 Sarcin-ricin UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop
AGCGACUAUGUCGUAUGGGUAAAGCGCCUGCUCAGUAC
GAGAGGAACCGCAGGAAGCAUCAAAG
656 uvsX, C18G UACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCC CUCUUCGGAG
GGAAGCAUCAAAG
657 truncated stem UACUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACC
loop, C18G, AGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUC
trip mut GGUCCGUAAGAAGCAUCAAAG
(U10C)
658 short phage UACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACC
rep, C18G
AGCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUCU
CGGUCGUCCGAAGCAUCAAAG
659 phage rep
UACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACC
loop, C18G AGCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGAGGA
CCUCUCGGUCGUCCUAUCUGAAGCAUCAAAG
660 =+G18,
UACUGGCGCCUUUAUCUGCAUUACUUUGAGAGCCAUCAC
stacked onto CAGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUC
64 GGUCCGUAAGAAGCAUCAAAG
661 truncated stem GCUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, C18G, - GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
1 MG GUCCGUAAGAAGCAUCAAAG
662 phage rep
UACUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACC
loop, C18G, AGCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGACGA
trip mut CC UCUCGGUCGUCCUAUC UGAAGCAUCAAAG
(U10C)
663 short phage UACUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACC
rep, C18G,
AGCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUCU
nip mut CGGUCGUCCGAAGCAUCAAAG
(U10C)
132
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
664 uvsX, trip mut UACUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACC
(U1 OC)
AGCGACUAUGUCGUAUGGGUAAAGCGCCCUCUUCGGAG
GGAAGCAUCAAAG
665 truncated stem UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop
AGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUC
GGUCCGUAAGAAGCAUCAAAG
666 =+A17,
UACUGGCGCCUUUAUCAUCAUUACUUUGAGAGCCAUCAC
stacked onto CAGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUC
64 GGUCCGUAAGAAGCAUCAAAG
667 3' HDV
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
genomic
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
ribozyme
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGGGCCGGCAU
GGUCCCAGCCUCCUCGCUGGCGCCGGCUGGGCAACAUU
CCGAGGGGACCGUCCCCUCGGUAAUGGCGAAUGGGACC
C
668 phage rep
UACUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACC
loop, trip mut AGCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGAGGA
(U10C) CCUCUCGGUCGUCCUAUCUGAAGCAUCAAAG
669 -79:80
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAAAUCCGAUAAAUAAGAAGCAUCAAAG
670 short phage UACUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACC
rep, trip mut AGCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUCU
(U1 OC) CGGUCGUCCGAAGCAUCAAAG
671 extra
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
truncated stem AGCGACUAUGUCGUAUGGGUAAAGCGCCGGACUUCGGU
loop CCGGAAGCAUCAAAG
672 U17G, C18G UAC UGGCGC UUUUAUCGGAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
673 short phage UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
rep
AGCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUC U
CGGUCGUCCGAAGCAUCAAAG
674 uvsX, C 18G, - GCUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
1 A2G
GCGACUAUGUCGUAUGGGUAAAGCGCCCUCUUCGGAGG
GAAGCAUCAAAG
675 uvsX, Cl8G, GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
trip mut
GCGACUAUGUCGUAUGGGUAAAGCUCCCUCUUCGGAGG
(U10C), -1 GAGCAUCAAAG
133
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
A2G, FIDV -
99 G65U
676 3' HDV
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
antigenomic AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
ribozyme
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGGGGUCGGCA
UGGCAUCUCCACCUCCUCGCGGUCCGACCUGGGCAUCC
GAAGGAGGACGCACGUCCACUCGGAUGGCUAAGGGAGA
GCCA
677 uvsX, Cl8G, GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
trip mut
GCGACUAUGUCGUAUGGGUAAAGCGCCCUCUUCGGAGG
(U10C), -1 GCGCAUCAAAG
A2G, HDV
AA(98:99)C
678 3' HDV
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
ribozyme AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
(Lior Nissim, GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGUUUUGGCCG
Timothy Lu) GCAUGGUCCCAGCCUCCUCGCUGGCGCCGGCUGGGCAA
CAUGCUUCGGCAUGGCGAAUGGGACCCCGGG
679 TAC(1:3)GA, GAUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
stacked onto GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
64 GUCCGUAAGAAGCAUCAAAG
680 uvsX, -1 A2G GCUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
GCGACUAUGUCGUAUGGGUAAAGCGCCCUCUUCGGAGG
GAAGCAUCAAAG
681 truncated stem GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, C18G, GCGACUAUGUCGUAUGGGUAAAGCUCUUACGGACUUCG
trip mut GUCCGUAAGAGCAUCAAAG
(U10C), -1
AUG, HDV -
99 G65U
682 short phage GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
rep, Cl 8G,
GCGACUAUGUCGUAUGGGUAAAGCUCGGACGACCUCUC
trip mut GGUCGUCCGAGCAUCAAAG
(U10C), -1
A2G, HDV -
99 G65U
683 3' sTRSV WT UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
viral
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
Hammerhead GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGCCUGUCACC
ribozyme
GGAUGUGCUUUCCGGUCUGAUGAGUCCGUGAGGACGAA
ACAGG
134
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
684 short phage GCUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
rep, C18G, -1 GCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUCUC
A2G GGUCGUCCGAAGCAUCAAAG
685 short phage GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
rep, C18G,
GCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUCUC
trip mut GGUCGUCCGAAGCAUCAAAG
(-CHOC), -1
A2G, 3'
genomic HDV
686 phage rep
GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, C18G, GCGACUAUGUCGUAUGGGUAAAGCUCAGGUGGGACGAC
trip mut CUCUCGGUCGUCCUAUCUGAGCAUCAAAG
(U10C), -1
A2G, HDV -
99 G65U
687 3' HDV
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
ribozyme AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
(Owen Ryan, GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGGAUGGCCOG
Jamie Cate) CAUGGUCCCAGCCUCCUCGCUGGCGCCGGCUGGGCAAC
ACC UUCGGGUGGCGAAUGGGAC
688 phage rep
GCUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, C18G, - GCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGACGAC
1 A2G CUCUCGGUCGUCCUAUCUGAAGCAUCAAAG
689 0.14
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUACUGGGUAAAGCGCUUAUUUAUCGG
AGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
690 -78, G77U
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGUG
AGAAAUCCGAUAAAUAAGAAGCAUCAAAG
691
GUACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCAC
CAGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGG
AGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
692 short phage GCUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
rep, -1 A2G GCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUCUC
GGUCGUCCGAAGCAUCAAAG
693 truncated stem GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, C18G, GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
trip mut GUCCGUAAGAAGCAUCAAAG
(U10C), -1
A2G
135
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
694 -1, A2G
GCUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
GCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
695 truncated stem GCUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
loop, trip mut GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
(U10C), -1 GUCCGUAAGAAGCAUCAAAG
A2G
696 uvsX, C 18G, GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
trip mut
GCGACUAUGUCGUAUGGGUAAAGCGCCCUCUUCGGAGG
(U10C), -1 GAAGCAUCAAAG
A2G
697 phage rep
GCUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
loop, -1 MG GCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGACGAC
CUCUCGGUCGUCCUAUCUGAAGCAUCAAAG
798 phage rep
GCUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
loop, nip mut GCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGACGAC
(U10C), -1 CUCUCGGUCGUCCUAUCUGAAGCAUCAAAG
MG
699 phage rep
GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, C18G, GCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGACGAC
hip mut CUCUCGGUCGUCCUAUCUGAAGCAUCAAAG
(U10C), -1
MG
700 truncated stem UACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACC
loop, C 18G AGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUC
GGUCCGUAAGAAGCAUCAAAG
701 uysX, trip mut GCUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
(U10C), -1
GCGACUAUGUCGUAUGGGUAAAGCGCCCUCUUCGGAGG
MG GAAGCAUCAAAG
702 truncated stem GCUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCA
loop, -1 MG GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
GUCCGUAAGAAGCAUCAAAG
703 short phage GCUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
rep, trip mut GCGACUAUGUCGUAUGGGUAAAGCGCGGACGACCUCUC
(U10C), -1 GGUCGUCCGAAGCAUCAAAG
MG
704 511-IDV
GAUGGCCGGCAUGGUCCCAGCCUCCUCGCUGGCGCCGG
ribozyme
CUGGGCAACACCUUCGGGUGGCGAAUGGGACUAC UGGC
(Owen Ryan, GCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGCGACU
Jamie Cate)
136
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
AUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAGAGAAAU
CCGAUAAAUAAGAAGCAUCAAAG
705 5111DV
GGCCGGCAUGGUCCCAGCCUCCUCGCUGGCGCCGGCUG
genomic
GGCAACAUUCCGAGGGGACCGUCCCCUCGGUAAUGGCG
ribozyme
AAUGGGACCCUACUGGCGCUUUUAUCUCAUUACUU UGAG
AGCCAUCACCAGCGACUAUGUCGUAUGGGUAAAGCGCUU
AU U UAU CGGAGAGAAAU CC GAUAAAUAAGAAGCAU CAAAG
706 truncated stem GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, C18G, GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
trip mut GUCCGUAAGCGCAUCAAAG
(U10C), -1
A2G, HDV
AA(98:99)C
707 5'env25 pistol CGUGGUUAGGGCCACGUUAAAUAGUUGCUUAAGCCCUAA
ribozyme GCOUUGAUC
UUCGGAUCAGGUGCAAUACUGGCGCUUUU
(with an added AUCUCAUUACUUUGAGAGCCAUCACCAGCGACUAUGUCG
CUUCGG
UAUGGGUAAAGCGCUUAUUUAUCGGAGAGAAAUCCGAUA
loop) AAUAAGAAGCAUCAAAG
708 5111DV
GGGUCGGCAUGGCAUCUCCACCUCCUCGCGGUCCGACC
antigenomic UGGGCAUCCGAAGGAGGACGCACGUCCACUCGGAUGGC
ribozyme
UAAGGGAGAGCCAUACUGGCGCUUUUAUCUCAUUACUUU
GAGAGCCAUCACCAGCGACUAUGUCGUAUGGGUAAAGCG
CUUAUUUAUCGGAGAGAAAUCCGAUAAAUAAGAAGCAUC
AAAG
709 3
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
Hammerhead AGC GAC UAU GUCGUAUGGGUAAAGC GC U UAUU UAUCGGA
ribozyme
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGCCAGUACUGA
(Lior Nissim, UGAGUCCGUGAGGACGAAACGAGUAAGCUCGUCUACUG
Timothy Lu) GCGCUUUUAUCUCAU
guide scaffold
scar
710 =+A27,
UACUGGCGCCUUUAUCUCAUUACUUUAGAGAGCCAUCAC
stacked onto CAGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACU UC
64 GGUCCGUAAGAAGGAUCAAAG
711 5'Hammerhea CGACUACUGAUGAGUCCGUGAGGACGAAACGAGUAAGCU
d ribozyme CGUCUAGUCGUACUGGCGCUUUUAUCUCAUUACUUUGAG
(Liar Nissim, AGCCAUCACCAGCGACUAUGUCGUAUGGGUAAAGCGCUU
Timothy Lu) AU U UAU C GGAGAGAAAU CC GAUAAAUAAGAAGCAU CAAAG
smaller scar
137
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
in NAME or
NO: Modification
712 phage rep
GCUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
loop, Cl8G, GCGACUAUGUCGUAUGGGUAAAGCGCAGGUGGGACGAC
nip mut CUCUCGGUCGUCCUAUCUGCGCAUCAAAG
(U10C), -1
A2G, 1-113V
AA(98:99)C
713 -27, stacked UACUGGCGCCUUUAUCUCAUUACUUUAGAGCCAUCACCA
onto 64
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
GUCCGUAAGAAGCAUCAAAG
714 3' Hatchet
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGCAUUCCUCAG
AAAAUGACAAACCUGUGGGGCGUAAGUAGAUCUUCGGAU
CUAUGAUCGUGCAGACGUUAAAAUCAGGU
715 3'
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
Hammerhead AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
ribozyme
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGCGACUACUGA
(Lior Nissim, UGAGUCCGUGAGGACGAAACGAGUAAGCUCGUCUAGUC
Timothy Lu) GCGUGUAGCGAAGCA
716 5 Hatchet
CAUUCCUCAGAAAAUGACAAACCUGUGGGGCGUAAGUAG
AUCUUCGGAUCUAUGAUCGUGCAGACGUUAAAAUCAGGU
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
717 5' HDV
UUUUGGCCGGCAUGGUCCCAGCCUCCUCGCUGGCGCCG
ribozyme GCUGGGCAACAUGCUUCGGCAUGGCGAAUGGGACCCCG
(Lior Nissim, GGUAGUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCA
Timothy Lu) CCAGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCG
GAGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
718 5'
CGACUACUGAUGAGUCCGUGAGGACGAAACGAGUAAGCU
Hammerhead CGUCUAGUCGCGUGUAGCGAAGCAUACUGGCGCUUUUA
ribozyme
UCUCAUUACUUUGAGAGCCAUCACCAGCGACUAUGUCGU
(Lior Nissim, AUGGGUAAAGCGCUUAUUUAUCGGAGAGAAAUCCGAUAA
Timothy Lu) AUAAGAAGCAUCAAAG
719 3' 111115
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
Minimal
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
Hammerhead GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGGGGAGCCCC
ribozyme
GCUGAUGAGGUCGGGGAGACCGAAAGGGACUUCGGUCC
CUACGGGGCUCCC
138
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
in NAME or
NO: Modification
720 5' RB1VIX
CCACCCCCACCACCACCCCCACCCCCACCACCACCCUAC
recruiting
UGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAGC
motif
GACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAGAG
AAAUCCGAUAAAUAAGAAGCAUCAAAG
721 3'
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
Hammerhead AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
ribozyme
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGCGACUACUGA
(Lior Nissim, UGAGUCCGUGAGGACGAAACGAGUAAGCUCGUCUAGUC
Timothy Lu) G
smaller scar
722 3' env25 pistol
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
ribozyme
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
(with an added GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGCGUGGUUAG
CUUCGG
GGCCACGUUAAAUAGUUGCUUAAGCCCUAAGCGUUGAUC
loop) UUCGGAUCAGGUGCAA
723 3' Env-9
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
Twister
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGGGCAAUAAAG
CGGUUACAAGCCCGCAAAAAUAGCAGAGUAAUGUCGCGA
UAGCGCGGCAUUAAUGCAGCUUUAUUG
724 =-FAUUAUC UACUGGCGCUUUUAUCUCAUUACUAUUAUCUCAUUACUU
UCAUUACU UGAGAGCCAUCACCAGCGACUAUGUCGUAUGGGUAAAGC
25
GCUUAUUUAUCGGAGAGAAAUCCGAUAAAUAAGAAGCAU
CAAAG
725 5 Env-9
GGCAAUAAAGCGGUUACAAGCCCGCAAAAAUAGCAGAGU
Twister
AAUGUCGCGAUAGCGCGGCAUUAAUGCAGCUUUAUUGUA
CUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACCAG
CGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGAGA
GAAAUCCGAUAAAUAAGAAGCAUCAAAG
726 3' Twisted
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
Sister 1
AGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGGA
GAGAAAUCCGAUAAAUAAGAAGCAUCAAAGACCCGCAAG
GCCGACGGCAUCCGCCGCCGCUGGUGCAAGUCCAGCCG
CCCCUUCGGGGGCGGGCGCUCAUGGGUAAC
727 no stem
UACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCACC
AGCGACUAUGUCGUAUGGGUAAAG
728 5' HH15
GGGAGCCCCGCUGAUGAGGUCGGGGAGACCGAAAGGGA
Minimal
CUUCGGUCCCUACGGGGCUCCCUACUGGCGCUUUUAUC
Hammerhead UCAUUACUUUGAGAGCCAUCACCAGCGACUAUGUCGUAU
ribozyme
GGGUAAAGCGCUUAUUUAUCGGAGAGAAAUCCGAUAAAU
AAGAAGCAUCAAAG
139
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
729 5'
CCAGUACUGAUGAGUCCGUGAGGACGAAACGAGUAAGCU
Hammerhead CGUCUACUGGCGCUUUUAUCUCAUUACUGGCGCUUUUAU
ribozyme
CUCAUUACUUUGAGAGCCAUCACCAGCGACUAUGUCGUA
(Lior Nissim, UGGGUAAAGCGCUUAUUUAUCGGAGAGAAAUCCGAUAAA
Timothy Lu) UAAGAAGCAUCAAAG
guide scaffold
scar
730 5' Twisted
ACCCGCAAGGCCGACGGCAUCCGCCGCCGCUGGUGCAA
Sister 1
GUCCAGCCGCCCCUUCGGGGGCGGGCGCUCAUGGGUAA
CUACUGGCGCUUUUAUCUCAUUACUUUGAGAGCCAUCAC
CAGCGACUAUGUCGUAUGGGUAAAGCGCUUAUUUAUCGG
AGAGAAAUCCGAUAAAUAAGAAGCAUCAAAG
731 5' sTRSV WT CCUGUCACCGGAUGUGCUUUCCGGUCUGAUGAGUCCGU
viral
GAGGACGAAACAGGUACUGGCGCUUUUAUCUCAUUACUU
Hammerhead UGAGAGCCAUCACCAGCGACUAUGUCGUAUGGGUAAAGC
ribozyme
GCUUAUUUAUCGGAGAGAAAUCCGAUAAAUAAGAAGGAU
CAAAG
732 148: =+G55, GUACUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCAC
stacked onto CAGCGACUAUGUCGUAGUGGGUAAAGCGCUUACGGACU
64 UCGGUCCGUAAGAAGCAUCAAAG
733 158:
GUACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCAC
103+148(+65 CAGCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGG
5) -99, G65U AGGGAGCAUCAAAG
734 174: Uvsx
ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
Extended stem GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
with [A99] GGAGCAUCAAAG
G65U),
Cl 8G,AG55,
[GU-1]
735 175: extended ACUGGCGCCUUUAUCUCAUUACUUUGAGAGCCAUCACCA
stem
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
truncation, GUCCGUAAGAAGCAUCAAAG
U10C, [GU-1]
736 176: 174 with GCUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
AlG
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
substitution GGAGCAUCAAAG
for T7
transcription
737 177: 174 with ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
bubble (+G55) GCGACUAUGUCGUAUGGGUAAAGCUCCCUCUUCGGAGG
removed GAGCAUCAAAG
140
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
in NAME or
NO: Modification
738 181: stem 42 ACUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
(truncated
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
stem loop); GUCCGUAAGAAGCAUCAAAG
U10C,C18G,[
GU-1]
(95+[GU-1])
739 182: stem 42 ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
(truncated
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
stem loop); GUCCGUAAGAAGCAUCAAAG
C18G,[GU-1]
740 183: stem 42 ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
(truncated
GCGACUAUGUCGUAGUGGGUAAAGCGCUUACGGACUUC
stem loop); GGUCCGUAAGAAGCAUCAAAG
C186,1\6-551
GU-1]
741 184: stem 48 ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
(uvsx, -99
GCGACUAUGUCGUAUUGGGUAAAGCUCCCUCUUCGGAG
g650; GGAGCAUCAAAG
Cl 86,AT55,[
GU-1]
742 185: stem 42 ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
(truncated
GCGACUAUGUCGUAUUGGGUAAAGCGCUUACGGACUUC
stem loop); GGUCCGUAAGAAGCAUCAAAG
Cl8G,AU55,[
GU-1]
743 186: stem 42 ACUGGCGCCUUUAUCAUCAUUACUUUGAGAGCCAUCACC
(truncated
AGCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUC
stem loop); GGUCCGUAAGAAGCAUCAAAG
U10C,AA17,[
GU-1]
744 187: stem 46 ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
(uvsx);
GCGACUAUGUCGUAGUGGGUAAAGCGCCCUCUUCGGAG
Cl8G,AG55,[ GGAAGCAUCAAAG
GU-1]
745 188: stem 50 ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
(ms2 Ul 5C, - GCGACUAUGUCGUAGUGGGUAAAGCUCACAUGAGGAUCA
99, g65t); CCCAUGUGAGCAUCAAAG
Cl8G,AG55,[
GU-1]
141
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
in NAME or
NO: Modification
746 189: 174 +
ACUGGCACUUUUACCUGAUUACUUUGAGAGCCAACACCA
G8A;U15C;U GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
35A GGAGCAUCAAAG
747
ACUGGCACUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
190: 174+
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
GSA G GAG CAU CAAAG
748
ACUGGCCCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
191: 174
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
G8C G GAG CAU CAAAG
749 ACUGGCGCU
UUUACCUGAUUACUUUGAGAGCCAUCACCA
192: 174+
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
U15C G GAG CAU CAAAG
750 ACUGGCGCU
UUUAUCUGAUUACUUUGAGAGCCAACACCA
193, 174+
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
U35A G GAG CAU CAAAG
751 195: 175+ AC U GGCACCU UUACC UGAUUAC U
UUGAGAGCCAACAC CA
C 8G
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
G8A;U15C;U GUCCGUAAGAAGCAUCAAAG
35A
752
ACUGGCACCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
196: 175 +
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
C18G + G8A GUCCGUAAGAAGCAUCAAAG
753 AC U GGCCCC U UUAUCUGAU UAC U U
UGAGAGCCAUCACCA
197: 175 +
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
C1 8G + G8C GUCCGUAAGAAGCAUCAAAG
754
ACUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAACACCA
198: 175 +
GCGACUAUGUCGUAUGGGUAAAGCGCUUACGGACUUCG
C18G + U35A GUCCGUAAGAAGCAUCAAAG
755 199: 174+
GCUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
A2G (test G GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
transcription GGAGCAUCAAAG
at start;
ccGCT...)
756 200: 174 + GACUGGCGCUUUUAUCUGAUUACUU
UGAGAGCCAUCACC
AG1
AGCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGA
(ccGACU...) GGGAGCAUCAAAG
757
ACUGGCGCCUUUAUCUGAUUACUUUGGAGAGCCAUCACC
201: 174+
AGCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGA
U1OC;AG28 GGGAGCAUCAAAG
142
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
758
ACUGGCGCAUUUAUCUGAUUACUUUGUGAGCCAUCACCA
202: 174 +
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
U10A;A28U GGAGCAUCAAAG
759
ACUGGCGCCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
203: 174+
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
UlOC GGAGCAUCAAAG
760
ACUGGCGCUUUUAUCUGAUUACUUUGGAGAGCCAUCACC
204: 174
AGCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGA
AG28 GGGAGCAUCAAAG
761
ACUGGCGCAUUUAUCUGAUUACUUUGAGAGCCAUCACCA
205: 174+
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
UlOA GGAGCAUCAAAG
762
ACUGGCGCUUUUAUCUGAUUACUUUGUGAGCCAUCACCA
206, 174+
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
A28U GGAGCAUCAAAG
763 AC U GGC GC U UUUAUUC UGAUUAC
UUUGAGAGCCAUCACC
207: 174 +
AGCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGA
AU15 GGGAGCAUCAAAG
764
ACGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCAG
208: 174 +
CGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAGG
[U4] GAGCAUCAAAG
765
ACUGGCGCUUUUAUAUGAUUACUUUGAGAGCCAUCACCA
209: 174 +
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
C16A GGAGCAUCAAAG
766 AC U GGC GC U UUUAUCUUGAUUAC
UUUGAGAGCCAUCACC
210: 174+
AGCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGA
AU17 GGGAGCAUCAAAG
767 211: 174+
ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAGCACCA
U35G
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
(compare with GGAGCAUCAAAG
174 + U35A
above)
768 212: 174
ACUGGCGCUGUUAUCUGAUUACUUCGAGAGCCAUCACCA
+U11G,
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
A105G GGAGCAUCGAAG
(A86G),
U26C
769 213: 174
ACUGGCGCUCUUAUCUGAUUACUUCGAGAGCCAUCACCA
+1111C,
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
A105G GGAGCAUCGAAG
143
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
ID NAME or
NO: Modification
(A86G),
U26C
770 214:
ACUGGCGCUUGUAUCUGAUUACUCUGAGAGCCAUCACCA
174+Ul2G; GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
A106G GGAGCAUCAGAG
(AS 7G),
U25C
771 215:
ACUGGCGCUUCUAUCUGAUUACUCUGAGAGCCAUCACCA
174+Ul2C; GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
A106G GGAGCAUCAGAG
(AS 7G),
U25C
772 216:
ACUGGCGCUUUGAUCUGAUUACCUUGAGAGCCAUCACCA
174_tx_11.G, GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
87.G,22.0 GGAGCAUCAAGG
773 217:
ACUGGCGCUUUCAUCUGAUUACCUUGAGAGCCAUCACCA
174 tx_11.C,8 GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
7.G,22.0 GGAGCAUCAAGG
774
ACUGGCGCUGUUAUCUGAUUACUUUGAGAGCCAUCACCA
218: 174
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
+U11G GGAGCAUCAAAG
775 219: 174
ACUGGCGCUUUUAUCUGAUUACUUUGAGAGCCAUCACCA
+A105G
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
(A86G) GGAGCAUCGAAG
776
ACUGGCGCUUUUAUCUGAUUACUUCGAGAGCCAUCACCA
220: 174
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
+U26C GGAGCAUCAAAG
777 221: 182+
ACUGGCACUUCUAUCUGAUUACUCUGAGAGCCAUCACCA
GSA (196)
GCGACUAUGUCGUAUGGGUAAAGCCGCUUACGGACUUC
+215 GGUCCGUAAGAGGCAUCAGAG
mutations +
AC63, A88G
778 222: 174 +
ACUGGCACUUCUAUCUGAUUACUCUGAGAGCCAUCACCA
GSA (196)
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
+215 GGAGCAUCAGAG
mutations
779 223: 181 +
ACUGGCACCUUUAUCUGAUUACUUUGAGAGCCAUCACCA
GSA (196) + GCGACUAUGUCGUAUGGGUAAAGCCGCUUACGGACUUC
AC63, A88G GGUCCGUAAGAGGCAUCAAAG
144
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ
NUCLEOTIDE SEQUENCE
1D NAME or
NO: Modification
780 224: 182 +
ACUGGCACUUGUAUCUGAUUACUCUGAGAGCCAUCACCA
G8A (196)
GCGACUAUGUCGUAUGGGUAAAGCCGCUUACGGACUUC
+214 GGUCCGUAAGAGGCAUCAGAG
mutations +
AC63, A88G
781 225: 174+
ACUGGCACUUGUAUCUGAUUACUCUGAGAGCCAUCACCA
G8A (196)
GCGACUAUGUCGUAGUGGGUAAAGCUCCCUCUUCGGAG
+214 GGAGCAUCAGAG
mutations
[00267] In some embodiments, the gNA variant comprises a tracrRNA stem loop
comprising
the sequence -UUU-N4-25UUU- (SEQ ID NO: 203). For example, the gNA variant
comprises
a scaffold stem loop or a replacement thereof, flanked by two triplet U motifs
that contribute to
the triplex region. In some embodiments, the scaffold stem loop or replacement
there of
comprises at least 4 nucleotides, at least 5 nucleotides, at least 6
nucleotides, at least 7
nucleotides, at least 7 nucleotides, at least 8 nucleotides, at least 9
nucleotides, at least 10
nucleotides, at least 11 nucleotides, at least 12 nucleotides, at least 13
nucleotides, at least 14
nucleotides, at least 15 nucleotides, at least 16 nucleotides, at least 17
nucleotides, at least 18
nucleotides, at least 19 nucleotides, at least 20 nucleotides, at least 21
nucleotides, at least 22
nucleotides, at least 23 nucleotides, at least 24 nucleotides, or at least 25
nucleotides.
[00268] In some embodiments, the gNA variant comprises a crRNA sequence with
-AAAG- in a location 5' to the spacer region. In some embodiments, the -AAAG-
sequence is
immediately 5' to the spacer region.
[00269] In some embodiments, the at least one nucleotide modification
comprises at least one
nucleotide deletion in the CasX variant gNA relative to the reference gRNA. In
some
embodiments, a gNA variant comprises a deletion of 1, 2, 3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14,
15, 16, 17, 18, 19 or 20 consecutive or non-consecutive nucleotides relative
to a reference
gRNA. In some embodiments, the at least one deletion comprises a deletion of
1, 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 01 20 or more consecutive
nucleotides relative to a
reference gRNA. In some embodiments, the gNA variant comprises 2, 3, 4, 5, 6,
7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18, 19 or 20 or more nucleotide deletions relative to
the reference gRNA,
and the deletions are not in consecutive nucleotides. In those embodiments
where there are two
or more non-consecutive deletions in the gNA variant relative to the reference
gRNA, any length
145
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
of deletions, and any combination of lengths of deletions, as described
herein, are contemplated
as within the scope of the disclosure. For example, in some embodiments, a gNA
variant may
comprise a first deletion of one nucleotide, and a second deletion of two
nucleotides and the two
deletions are not consecutive. In some embodiments, a gNA variant comprises at
least two
deletions in different regions of the reference gRNA. In some embodiments, a
gNA variant
comprises at least two deletions in the same region of the reference gRNA. For
example, the
regions may be the extended stem loop, scaffold stem loop, scaffold stem
bubble, triplex loop,
pseudoknot, triplex, or a 5' end of the gNA variant. Any deletion of any
nucleotide in a reference
gRNA is contemplated as within the scope of the disclosure.
[00270] In some embodiments, the at least one nucleotide modification
comprises at least one
nucleotide insertion. In some embodiments, a gNA variant comprises an
insertion of 1, 2, 3,4, 5,
6, 7, 8, 9 or 10 consecutive or non-consecutive nucleotides relative to a
reference gRNA. In
some embodiments, the at least one nucleotide insertion comprises an insertion
of 1, 2, 3,4, 5, 6,
7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20 or more consecutive
nucleotides relative to a
reference gRNA. In some embodiments, the gNA variant comprises 2 or more
insertions relative
to the reference gRNA, and the insertions are not consecutive. In those
embodiments where there
are two or more non-consecutive insertions in the gNA variant relative to the
reference gRNA,
any length of insertions, and any combination of lengths of insertions, as
described herein, are
contemplated as within the scope of the disclosure. For example, in some
embodiments, a gNA
variant may comprise a first insertion of one nucleotide, and a second
insertion of two
nucleotides and the two insertions are not consecutive. In some embodiments, a
gNA variant
comprises at least two insertions in different regions of the reference gRNA.
In some
embodiments, a gNA variant comprises at least two insertions in the same
region of the
reference gRNA. For example, the regions may be the extended stem loop,
scaffold stem loop,
scaffold stem bubble, triplex loop, pseudoknot, triplex, or a 5' end of the
gNA variant. Any
insertion of A, G, C, U (or T, in the corresponding DNA) or combinations
thereof at any location
in the reference gRNA is contemplated as within the scope of the disclosure.
1002711 In some embodiments, the at least one nucleotide modification
comprises at least one
nucleic acid substitution. In some embodiments, a gNA variant comprises 1, 2,
3, 4, 5, 6, 7, 8, 9,
10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20 or more consecutive or non-
consecutive substituted
nucleotides relative to a reference gRNA. In some embodiments, a gNA variant
comprises 1-4
nucleotide substitutions relative to a reference gRNA. In some embodiments,
the at least one
146
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
substitution comprises a substitution of 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11,
12, 13, 14, 15, 16, 17, 18,
19 or 20 or more consecutive nucleotides relative to a reference gRNA. In some
embodiments,
the gNA variant comprises 2 or more substitutions relative to the reference
gRNA, and the
substitutions are not consecutive. In those embodiments where there are two or
more non-
consecutive substitutions in the gNA variant relative to the reference gRNA,
any length of
substituted nucleotides, and any combination of lengths of substituted
nucleotides, as described
herein, are contemplated as within the scope of the disclosure. For example,
in some
embodiments, a gNA variant may comprise a first substitution of one
nucleotide, and a second
substitution of two nucleotides and the two substitutions are not consecutive.
In some
embodiments, a gNA variant comprises at least two substitutions in different
regions of the
reference gRNA. In some embodiments, a gNA variant comprises at least two
substitutions in
the same region of the reference gRNA. For example, the regions may be the
triplex, the
extended stem loop, scaffold stem loop, scaffold stem bubble, triplex loop,
pseudoknot, triplex,
or a 5' end of the gNA variant. Any substitution of A, G, C, U (or T, in the
corresponding DNA)
or combinations thereof at any location in the reference gRNA is contemplated
as within the
scope of the disclosure.
1002721 Any of the substitutions, insertions and deletions described herein
can be combined to
generate a gNA variant of the disclosure. For example, a gNA variant can
comprise at least one
substitution and at least one deletion relative to a reference gRNA, at least
one substitution and
at least one insertion relative to a reference gRNA, at least one insertion
and at least one deletion
relative to a reference gRNA, or at least one substitution, one insertion and
one deletion relative
to a reference gRNA.
1002731 In some embodiments, the gNA variant comprises a scaffold region at
least 20%
identical, at least 30% identical, at least 40% identical, at least 50%
identical, at least 60%
identical, at least 65% identical, at least 70% identical, at least 75%
identical, at least 80%
identical, at least 85% identical, at least 90% identical, at least 91%
identical, at least 92%
identical, at least 93% identical, at least 94% identical, at least 95%
identical, at least 96%
identical, at least 97% identical, at least 98% identical, or at least 99%
identical to any one of
SEQ ID NOS: 4-16. In some embodiments, the gNA variant comprises a scaffold
region at least
60% homologous (or identical) to any one of SEQ ID NOS: 4-16.
1002741 In some embodiments, the gNA variant comprises a tracr stem loop at
least 60%
identical, at least 65% identical, at least 70% identical, at least 75%
identical, at least 80%
147
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
identical, at least 85% identical, at least 90% identical, at least 91%
identical, at least 92%
identical, at least 93% identical, at least 94% identical, at least 95%
identical, at least 96%
identical, at least 97% identical, at least 98% identical, or at least 99%
identical to SEQ ID NO:
14. In some embodiments, the gNA variant comprises a tracr stem loop at least
60%
homologous (or identical) to SEQ ID NO: 14.
1002751 In some embodiments, the gNA variant comprises an extended stem loop
at least 60%
identical, at least 65% identical, at least 70% identical, at least 75%
identical, at least 80%
identical, at least 85% identical, at least 90% identical, at least 91%
identical, at least 92%
identical, at least 93% identical, at least 94% identical, at least 95%
identical, at least 96%
identical, at least 97% identical, at least 98% identical, or at least 99%
identical to SEQ ID NO:
15. In some embodiments, the gNA variant comprises an extended stem loop at
least 60%
homologous (or identical) to SEQ ID NO: 15.
[00276] In some embodiments, the gNA variant comprises an exogenous extended
stem loop,
with such differences from a reference gNA described as follows. In some
embodiments, an
exogenous extended stem loop has little or no identity to the reference stem
loop regions
disclosed herein (e.g., SEQ ID NO: 15). In some embodiments, an exogenous stem
loop is at
least 10 bp, at least 20 bp, at least 30 bp, at least 40 bp, at least 50 bp,
at least 60 bp, at least 70
bp, at least 80 bp, at least 90 bp, at least 100 bp, at least 200 bp, at least
300 bp, at least 400 bp,
at least 500 bp, at least 600 bp, at least 700 bp, at least 800 bp, at least
900 bp, at least 1,000 bp,
at least 2,000 bp, at least 3,000 bp, at least 4,000 bp, at least 5,000 bp, at
least 6,000 bp, at least
7,000 bp, at least 8,000 bp, at least 9,000 bp, at least 10,000 bp, at least
12,000 bp, at least
15,000 bp or at least 20,000 bp. In some embodiments, the gNA variant
comprises an extended
stem loop region comprising at least 10, at least 100, at least 500, at least
1000, or at least 10,000
nucleotides. In some embodiments, the heterologous stem loop increases the
stability of the
gNA. In some embodiments, the heterologous RNA stem loop is capable of binding
a protein, an
RNA structure, a DNA sequence, or a small molecule. In some embodiments, an
exogenous
stem loop region comprises an RNA stem loop or hairpin, for example a
thermostable RNA such
as MS2 (ACAUGAGGAUUACCCAUGU (SEQ ID NO: 204)), QI3
(UGCAUGUCUAAGACAGCA (SEQ ID NO: 205)), Ul hairpin II
(AAUCCAUUGCACUCCGGAUU (SEQ ID NO: 206)), Uvsx (CCUCUUCGGAGG (SEQ ID
NO: 207)), PP7 (AGGAGUUUCUAUGGAAACCCU (SEQ ID NO: 208)), Phage replication
loop (AGGUGGGACGACCUCUCGGUCGUCCUAUCU (SEQ ID NO: 209)), Kissing loop_a
148
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(UGCUCGCUCCGUUCGAGCA (SEQ ID NO: 210)), Kissing loop_b1
(UGCUCGACGCGUCCUCGAGCA (SEQ ID NO: 211)), Kissing loop b2
(UGCUCGUUUGCGGCUACGAGCA (SEQ ID NO: 212)), G quadriplex M3q
(AGGGAGGGAGGGAGAGG (SEQ ID NO: 213)), G quadriplex telomere basket
(GGUUAGGGUUAGGGUUAGG (SEQ ID NO: 214)), Sarcin-ricin loop
(CUGCUCAGUACGAGAGGAACCGCAG (SEQ ID NO: 215)) or Pseudoknots
(UACACUGGGAUCGCUGAAUUAGAGAUCGGCGUCCUUUCAUUCUAUAUACUUUGG
AGUUUUAAAAUGUCUCUAAGUACA (SEQ lID NO: 216)). In some embodiments, an
exogenous stem loop comprises a long non-coding RNA (lncRNA). As used herein,
a lncRNA
refers to a non-coding RNA that is longer than approximately 200 bp in length.
In some
embodiments, the 5' and 3' ends of the exogenous stem loop are base paired;
i.e., interact to
form a region of duplex RNA. In some embodiments, the 5' and 3' ends of the
exogenous stem
loop are base paired, and one or more regions between the 5' and 3' ends of
the exogenous stem
loop are not base paired. In some embodiments, the at least one nucleotide
modification
comprises: (a) substitution of 1 to 15 consecutive or non-consecutive
nucleotides in the gNA
variant in one or more regions; (b) a deletion of 1 to 10 consecutive or non-
consecutive
nucleotides in the gNA variant in one or more regions; (c) an insertion of 1
to 10 consecutive or
non-consecutive nucleotides in the gNA variant in one or more regions; (d) a
substitution of the
scaffold stem loop or the extended stem loop with an RNA stem loop sequence
from a
heterologous RNA source with proximal 5' and 3' ends; or any combination of
(a)-(d),
[00277] In some embodiments, the gNA variant comprises a scaffold stem loop
sequence of
CCAGCGACUAUGUCGUAGUGG (SEQ ID NO: 202). In some embodiments, the gNA
variant comprises a scaffold stem loop sequence of CCAGCGACUAUGUCGUAGUGG (SEQ
ID NO: 202) and at least 1, 2, 3, 4, or 5 mismatches thereto.
[00278] In some embodiments, the gNA variant comprises an extended stem loop
region
comprising less than 32 nucleotides, less than 31 nucleotides, less than 30
nucleotides, less than
29 nucleotides, less than 28 nucleotides, less than 27 nucleotides, less than
26 nucleotides, less
than 25 nucleotides, less than 24 nucleotides, less than 23 nucleotides, less
than 22 nucleotides,
less than 21 nucleotides, or less than 20 nucleotides. In some embodiments,
the gNA variant
comprises an extended stem loop region comprising less than 32 nucleotides. In
some
embodiments, the gNA variant further comprises a thermostable stem loop.
149
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00279] In some embodiments, the gNA comprises an RNA binding domain. The RNA
binding
domain can be a retroviral Psi packaging element inserted into the gNA or is a
stem loop with
affinity to a protein selected from the group consisting of MS2, PP7, Qbeta,
U1A, or phage R-
loop, which can facilitate the binding of gNA to CasX. Similar RNA components
with affinity to
protein structures incorporated into the CasX include kissing loop_a, kissing
loop_bl, kissing
loop_b2, G quadriplex M3q, G quadriplex telomere basket, sarcin-ricin loop,
and pseudoknots. It
has been discovered that the incorporation of the Psi packaging element
inserted into the guide
RNA facilitates the packaging of the XDP particle due, in part, to the high
affinity binding of Psi
sequences for the Gag NC protein. Further, due to the affinity of the CasX for
the gNA,
resulting in an RNP, the incorporation of the RNP into the >CDP is further
facilitated.
1002801 In some embodiments, an sgRNA variant comprises a sequence of SEQ ID
NOS: 597-
781 or a sequence having having at least about 80%, at least about 90%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, at least about 99%
identity thereto. In
some embodiments, an sgRNA variant comprises a sequence of SEQ ID NOS: 597-
781. In some
embodiments, an sgRNA variant comprises a sequence of SEQ ID NOS: 597-781 and
a targeting
sequence.
1002811 In some embodiments, a sgRNA variant comprises a sequence of SEQ ID
NO: 600,
SEQ ID NO: 602, SEQ ID NO: 659, SEQ ID NO: 603, SEQ ID NO: 660, SEQ ID NO:
661,
SEQ ID NO: 662, SEQ ID NO: 599, SEQ ID NO: 663, SEQ ID NO: 601, SEQ ID NO:
604,
SEQ ID NO: 608, SEQ ID NO: 656, SEQ ID NO: 666, SEQ ID NO: 610, SEQ ID NO:
667,
SEQ ID NO: 608, SEQ ID NO: 669, SEQ ID NO: 598, SEQ ID NO: 670, SEQ ID NO:
671,
SEQ ID NO: 605, SEQ ID NO: 672, SEQ ID NO: 734, SEQ ID NO: 735, SEQ ID NO:
736,
SEQ ID NO: 737, SEQ 1D NO: 770, SEQ ID NO:771, SEQ ID NO: 775, or SEQ ID NO:
781.
1002821 In some embodiments, the gNA variant comprises one or more additional
changes to a
sequence of any one of SEQ ID NOS: 732, 733, 734, 737, 740, 744, 745, or 755-
781, or having
at least about 80%, at least about 90%, at least about 95%, at least about
96%, at least about
97%, at least about 98%, at least about 99% identity thereto. In some
embodiments, the gNA
variant comprises one or more additional changes to a sequence of any one of
SEQ ID NOs:
597-781. In some embodiments, the gNA variant comprises the sequence of any
one of SEQ ID
NOS:732, 733, 734, 737, 740, 744, 745, or 755-781. In some embodiments, the
gNA variant
scaffold consists of the sequence of any one of SEQ ID NOS:732, 733, 734, 737,
740, 744, 745,
150
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
or 755-781, and further comprises a targeting sequence of any of the
embodiments described
herein.
[00283] In some embodiments, a sgRNA variant comprises one or more additional
changes to a
sequence of SEQ ID NO: 600, SEQ ID NO: 659, SEQ ID NO: 603, SEQ ID NO: 660,
SEQ ID
NO: 661, SEQ 1D NO: 662, SEQ ID NO: 599, SEQ ID NO: 663, SEQ ID NO: 601, SEQ
ID NO:
604, SEQ ID NO: 608, SEQ 1D NO: 656, SEQ ID NO: 666, SEQ 1D NO: 610, SEQ ID
NO: 667,
SEQ ID NO: 608, SEQ ID NO: 669, SEQ ID NO: 598, SEQ ID NO: 670, SEQ ID NO:
671,
SEQ ID NO: 605, SEQ ID NO: 672, SEQ ID NO: 734, SEQ ID NO: 735, SEQ ID NO:
736,
SEQ ID NO: 737, SEQ ID NO:770, SEQ ID NO:771, SEQ ID NO: 775, or SEQ ID NO:
781.
[00284] In some embodiments of the gNA variants of the disclosure, the gNA
variant
comprises at least one modification, wherein the at least one modification
compared to the
reference guide scaffold of SEQ ID NO: 5 is selected from one or more of: (a)
a Cl8G
substitution in the triplex loop; (b) a G55 insertion in the stem bubble; (c)
a Ul deletion; (d) a
modification of the extended stem loop wherein (i) a 6 nt loop and 13 loop-
proximal base pain
are replaced by a Uvsx hairpin; and (ii) a deletion of A99 and a substitution
of G65U that results
in a loop-distal base that is fully base-paired. In some embodiments, the gNA
variant comprises
the sequence of any one of SEQ ID NOS: 732, 733, 734, 737, 740, 744, 745, or
755-781.
[00285] The gNA variants utilized in the XDP systems further comprises a
spacer (or targeting
sequence) region located at the 3' end of the gNA, described more fully,
supra, wherein the
spacer is designed with a sequence that is complementary to a target nucleic
acid to be edited. In
some embodiments, the gNA variant comprises a targeting sequence of at least
14 to 30
nucleotides, wherein the sequence is complementary to the target nucleic acid
to be edited. In
some embodiments, the targeting sequence has 14, 15, 16, 17, 18, 19, 20, 21,
22, 23, 24, 25, 26,
27, 28, 29, 30, 31, 32, 33, 34 or 35 nucleotides. In some embodiments, the gNA
variant
comprises a targeting sequence having 20 nucleotides. In some embodiments, the
targeting
sequence has 25 nucleotides. In some embodiments, the targeting sequence has
24 nucleotides.
In some embodiments, the targeting sequence has 23 nucleotides. In some
embodiments, the
targeting sequence has 22 nucleotides. In some embodiments, the targeting
sequence has 21
nucleotides. In some embodiments, the targeting sequence has 20 nucleotides.
In some
embodiments, the targeting sequence has 19 nucleotides. In some embodiments,
the targeting
sequence has 18 nucleotides. In some embodiments, the targeting sequence has
17 nucleotides.
In some embodiments, the targeting sequence has 16 nucleotides. In some
embodiments, the
151
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
targeting sequence has 15 nucleotides. In some embodiments, the targeting
sequence has 14
nucleotides. In some embodiments, the target nucleic acid comprises a PAM
sequence located 5'
of the targeting sequence with at least a single nucleotide separating the PAM
from the first
nucleotide of the targeting sequence. In some embodiments, the PAM is located
on the non-
targeted strand of the target region, i.e. the strand that is complementary to
the target nucleic
acid. In some embodiments, the PAM sequence is a TC motif In some embodiments,
the PAM
sequence is a ATC. In other embodiments, the PAM sequence is a TTC. In other
embodiments,
the PAM sequence is a GTC. In other embodiments, the PAM sequence is a CTC.
[00286] In some embodiments, the scaffold of the gNA variant is a variant
comprising one or
more additional changes to a sequence of a reference gRNA that comprises SEQ
ID NO: 4 or
SEQ ID NO: 5. In those embodiments where the scaffold of the reference gRNA is
derived from
SEQ ID NO: 4 or SEQ ID NO: 5, the one or more improved or added
characteristics of the gNA
variant are improved compared to the same characteristic in SEQ ID NO: 4 or
SEQ ID NO: 5.
[00287] In some embodiments of the XDP system, the scaffold of the gNA variant
is part of an
RNP with a CasX variant protein comprising any one of the sequences of SEQ ID
NOS: 21-
233, 343-345, 350-353, 355-367 or 388-397, or a sequence having at least about
50%, at least
about 60%, at least about 70%, at least about 80%, at least about 85%, at
least about 90%, at
least about 91%, at least about 92%, at least about 93%, at least about 94%,
at least about 95%,
at least about 96%, at least about 97%, at least about 98%, or at least about
99% identity thereto.
In the foregoing embodiments, the gNA further comprises a targeting sequence.
a Chemically Modified gNA
[00288] In some embodiments, the disclosure relates to chemically-modified
gNA. In some
embodiments, the present disclosure provides a chemically-modified gNA that
has guide RNA
functionality and has reduced susceptibility to cleavage by a nuclease. A gNA
that comprises
any nucleotide other than the four canonical ribonucleotides A, C, G, and U,
or a
deoxynucleotide, is a chemically modified gNA. In some cases, a chemically-
modified gNA
comprises any backbone or intemucleotide linkage other than a natural
phosphodiester
intemucleotide linkage. In certain embodiments, the retained functionality
includes the ability of
the modified gNA to bind to a CasX of any of the embodiments described herein.
In certain
embodiments, the retained functionality includes the ability of the modified
gNA to bind to a
target nucleic acid sequence. In certain embodiments, the retained
functionality includes
targeting a CasX protein or the ability of a pre-complexed CasX protein-gNA to
bind to a target
152
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
nucleic acid sequence. In certain embodiments, the retained functionality
includes the ability to
nick a target polynucleotide by a CasX-gNA. In certain embodiments, the
retained functionality
includes the ability to cleave a target nucleic acid sequence by a CasX-gNA In
certain
embodiments, the retained functionality is any other known function of a gNA
in a CasX system
with a CasX protein of the embodiments of the disclosure.
[00289] In some embodiments, the disclosure provides a chemically-modified gNA
in which a
nucleotide sugar modification is incorporated into the gNA selected from the
group consisting of
2'43 ______________ C1-4a1ky1 such as 2'-0-methyl (2'-0Me), T-deoxy (T-H), 2'-
0 __________________ C1-3alky1-0 ____ C1-
3a1ky1 such as 2'-methoxyethyl ("2'-MOE"), 2'-fluoro ("2'-F"), 2`-amino ("2`-
NH2"), 2'-
arabinosyl ("2'-arabino") nucleotide, 2'-F-arabinosyl ("2'-F-arabino")
nucleotide, 2'-locked
nucleic acid ("LNA") nucleotide, 2'-unlocked nucleic acid ("ULNA") nucleotide,
a sugar in L
form ("L-sugar"), and 4'-thioribosyl nucleotide. In other embodiments, an
internucleotide
linkage modification incorporated into the guide RNA is selected from the
group consisting of:
phosphorothioate "P(SI (P(S)), phosphonocarboxylate (P(CH2)nCOOR) such as
phosphonoacetate "PACE" (P(CH2C00-)), thiophosphonocarboxyl ate
((S)P(CH2)nCOOR)
such as thiophosphonoacetate "thioPACE" ((S)P(CH2)nC00-)), alkylphosphonate
(P(C1-
3alkyl) such as methylphosphonate -P(CH3), boranophosphonate (P(BH3)), and
phosphorodithioate (P(S)2).
[00290] In certain embodiments, the disclosure provides a chemically-modified
gNA in which a
nucleobase ("base") modification is incorporated into the gNA selected from
the group
consisting of: 2-thiouracil ("2-thioU"), 2-thiocytosine ("2-thioC"), 4-
thiouracil ("4-thioU"), 6-
thioguanine ("6-thioG"), 2-aminoadenine ("2-aminoA"), 2-aminopurine,
pseudouracil,
hypoxanthine, 7-deazaguanine, 7-deaza-8-azaguanine, 7-deazaadenine, 7-deaza-8-
azaadenine, 5-
methylcytosine ("5-methylC"), 5-methyluracil ("5-methylU"), 5-
hydroxymethylcytosine, 5-
hydroxymethyluracil, 5,6-dehydrouracil, 5-propynylcytosine, 5-propynyluracil,
5-
ethynylcytosine, 5-ethynyluracil, 5-allyluracil ("5-ally1U"), 5-allylcytosine
("5-ally1C"), 5-
aminoallyluracil ("5-aminoallyIU"), 5-aminoallyl-cytosine ("5-aminoallylC"),
an abasic
nucleotide, Z base, P base, Unstructured Nucleic Acid ("UNA"), isoguanine
("isoG"),
isocytosine ("isoC"), 5-methyl-2-pytimidine, x(A_,G,C,T) and y(A,G,C,T).
[00291] In other embodiments, the disclosure provides a chemically-modified
gNA in which
one or more isotopic modifications are introduced on the nucleotide sugar, the
nucleobase, the
phosphodiester linkage and/or the nucleotide phosphates, including nucleotides
comprising one
153
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
or more 15N, 13C, MC, deuterium, 31-1, 32P, 1251, 1311 atoms or other atoms or
elements used
as tracers.
[00292] In some embodiments, an "end" modification incorporated into the gNA
is selected
from the group consisting of: PEG (polyethyleneglycol), hydrocarbon linkers
(including:
heteroatom (0,S,N)-substituted hydrocarbon spacers; halo-substituted
hydrocarbon spacers;
keto-, carboxyl-, amido-, thionyl-, carbamoyl-, thionocarbamaoyl-containing
hydrocarbon
spacers), spermine linkers, dyes including fluorescent dyes (for example
fluoresceins,
rhodamines, cyanines) attached to linkers such as for example 6-fluorescein-
hexyl, quenchers
(for example dabcyl, BHQ) and other labels (for example biotin, digoxigenin,
acridine,
streptavidin, avidin, peptides and/or proteins). In some embodiments, an "end"
modification
comprises a conjugation (or ligation) of the gNA to another molecule
comprising an
oligonucleotide of deoxynucleotides and/or ribonucleotides, a peptide, a
protein, a sugar, an
oligosaccharide, a steroid, a lipid, a folic acid, a vitamin and/or other
molecule. In certain
embodiments, the disclosure provides a chemically-modified gNA in which an
"end"
modification (described above) is located internally in the gNA sequence via a
linker such as, for
example, a 2-(4-butylamidofluorescein)propane-1,3-diol bis(phosphodiester)
linker, which is
incorporated as a phosphodiester linkage and can be incorporated anywhere
between two
nucleotides in the gNA.
[00293] In some embodiments, the disclosure provides a chemically-modified gNA
having an
end modification comprising a terminal functional group such as an amine, a
thiol (or
sulfhydryl), a hydroxyl, a carboxyl, carbonyl, thionyl, thiocarbonyl, a
carbamoyl, a
thiocarbamoyl, a phoshoryl, an alkene, an alkyne, an halogen or a functional
group-terminated
linker that can be subsequently conjugated to a desired moiety selected from
the group
consisting of a fluorescent dye, a non-fluorescent label, a tag (for 14C,
example biotin, avidin,
streptavidin, or moiety containing an isotopic label such as 15N, 13C,
deuterium, 3H, 32P, 1251 and
the like), an oligonucleotide (comprising deoxynucleotides and/or
ribonucleotides, including an
aptamer), an amino acid, a peptide, a protein, a sugar, an oligosaccharide, a
steroid, a lipid, a
folic acid, and a vitamin. The conjugation employs standard chemistry well-
known in the art,
including but not limited to coupling via N-hydroxysuccinimide,
isothiocyanate, DCC (or DCI),
and/or any other standard method as described in "Bioconjugate Techniques" by
Greg T.
Hermanson, Publisher Elsevier Science, S. (2013), the contents of which are
incorporated
herein by reference in its entirety.
154
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Tropism Factors and Pseudotyping of XDP Systems
1002941 In another aspect, the disclosure relates to the incorporation of
tropism factors in the
XDP to increase tropism and selectivity for target cells or tissues intended
for gene editing.
Tropism factors of the XDP embodiments include, but are not limited to,
envelope glycoproteins
derived from viruses, antibody fragments, and receptors or ligands that have
binding affinity to
target cell markers. The inclusion of such tropism factors on the surface of
XDP particles
enhances the ability of the XDP to selectively bind to and fuse with the cell
membrane of a
target cell bearing such target cell markers, increasing the therapeutic index
and reducing
unintended side effects of the therapeutic payload incorporated into the )(DP,
1002951 In some embodiments, the XDP comprises one or more glycoproteins (GP)
on the
surface of the particle wherein the GP provides for enhanced or selective
binding and fusion of
the XDP to a target cell. In other embodiments, the XDP comprises one or more
antibody
fragments on the surface of the particle wherein the antibody fragments
provides for enhanced or
selective binding and fusion of the XDP to a target cell. In other
embodiments, the XDP
comprises one or more cell surface receptors, including G-protein-linked
receptors, and enzyme-
linked receptors, on the surface of the particle wherein the receptor provides
for enhanced or
selective binding and fusion of the XDP to a target cell. In some embodiments,
the XDP
comprises one or more ligands on the surface of the particle wherein the
ligand provides for
enhanced or selective binding and fusion of the XDP to a target cell bearing a
receptor to the
ligand on the cell surface. In still other embodiments, the XDP comprises a
combinations of one
or more glycoproteins, antibody fragments, cell receptors, or ligands on the
surface of the
particle to provide for enhanced or selective binding and fusion of the XDP to
a target cell.
1002961 For enveloped viruses, membrane fusion for viral entry is mediated by
membrane
glycoprotein complexes. Two basic mechanistic principles of membrane fusion
have emerged as
conserved among enveloped viruses; target membrane engagement and refolding
into hairpin-
like structures (Plemper, RK. Cell Entry of Enveloped Viruses. Curt Opin
Virol. 1:92 (2011)).
The envelope glycoproteins are typically observed as characteristic protein
"spikes" on the
surface of purified vitions in electron microscopic images. The underlying
mechanism of viral
entry by enveloped viruses can be utilized to preferentially direct XDP to
target particular cells
or organs in a process known as pseudotyping. In some embodiments, the XDP of
the disclosure
are pseudotyped by incorporation of a glycoprotein derived from an enveloped
virus that has a
155
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
demonstrated tropism for a particular organ or cell. Representative
glycoproteins within the
scope of the instant disclosure are listed in Table 4 and in the Examples. In
some embodiments,
the viruses used to provide the glycoprotein include, but are not limited to
Argentine
hemorrhagic fever virus, Australian bat virus, Autographa califomica multiple
nucleopolyhedrovirus, Avian leukosis virus, baboon endogenous virus, Bolivian
hemorrhagic
fever virus, Borna disease virus, Breda virus, Bunyamwera virus, Chandipura
virus,
Chikungunya virus, Crimean-Congo hemorrhagic fever virus, Dengue fever virus,
Duvenhage
virus, Eastern equine encephalitis virus, Ebola hemorrhagic fever virus, Ebola
Zaire virus,
enteric adenovirus, Ephemerovirus, Epstein-Bar virus (EBV), European bat virus
1, European
bat virus 2, Fug Synthetic gP Fusion, Gibbon ape leukemia virus, Hantavirus,
Hendra virus,
hepatitis A virus, hepatitis B virus, hepatitis C virus, hepatitis D virus,
hepatitis E virus, hepatitis
G Virus (GB virus C), herpes simplex virus type 1, herpes simplex virus type
2, human
cytomegalovirus (HEWS), human foamy virus, human herpesvirus (HHV), human
Herpesvirus
7, human herpesvirus type 6, human herpesvirus type 8, human immunodeficiency
virus 1 (MV-
O, human metapneumovirus, human T-lymphotro pic virus 1, influenza A,
influenza B,
influenza C virus, Japanese encephalitis virus, Kaposi's sarcoma-associated
herpesvirus (HHV8),
Kaysanur Forest disease virus, La Crosse virus, Lagos bat virus, Lassa fever
virus, lymphocytic
choriomeningitis virus (LCMV), Machupo virus, Marburg hemorrhagic fever virus,
measles
virus, Middle eastern respiratory syndrome-related coronavirus, Mokola virus,
Moloney murine
leukemia virus, monkey pox, mouse mammary tumor virus, mumps virus, murine
gammaherpesvirus, Newcastle disease virus, Nipah virus, Nipah virus, Norwalk
virus, Omsk
hemorrhagic fever virus, papilloma virus, parvovirus, pseudorabies virus,
Quaranfil virus, rabies
virus, RD114 Endogenous Feline Retrovirus, respiratory syncytial virus (RSV),
Rift Valley
fever virus, Ross River virus, rRotavirus, Rous sarcoma virus, rubella virus,
Sabia-associated
hemorrhagic fever virus, SARS-associated coronavirus (SARS-CoV), Sendai virus,
Tacaribe
virus, Thogotovirus, tick-borne encephalitis causing virus, varicella zoster
virus (HHV3),
varicella zoster virus (IHIV3), variola major virus, variola minor virus,
Venezuelan equine
encephalitis virus, Venezuelan hemorrhagic fever virus, vesicular stomatitis
virus (VSV),
glycoprotein G from vesicular stomatitis virus (VSV-G), Vesiculovirus, West
Nile virus,
western equine encephalitis virus, and Zika Virus. Non-limiting examples of
glycoprotein
sequences are provided in Table 4. In some embodiments, the XDP comprises one
or more
glycoprotein sequences of Table 4, or a sequence having at least 80%, at least
90%, at least 95%,
156
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
at least 96%, at least 97%, at least 98%, at least 99% sequence identity
thereto, wherein the
glycoproteins are incorporated into the particle and exposed on the surface,
providing tropism
and enhanced selectivity for the XDP to the target cell to be edited.
Table 4: Glycoproteins for XDP
SEQ ID NO
SEQ ID NO
Virus Glycoprotein
(DNA)
(Protein)
Vesicular Stomatitis Virus pGP2
437 438
Human Immunodeficiency
pGP3 439 440
Virus
Avian leukosis virus pGP4
441 442
Rous Sarcoma virus pGP5
443 444
Mouse mammary tumor virus pGP6
445 446
Human T-lymphotropic virus 1 pGP7
447 448
RD114 Endogenous Feline
pGP8 449 450
Retrovirus
Gibbon ape leukemia virus pGP9
451 452
Moloney Murine leukemia virus pGP10
453 454
Baboon Endogenous Virus pGP11
455 456
Human Foamy Virus pGP12
457 458
pGP13.1
459 460
Pseudorabies virus pGP13.2
461 462
pGP13.3
463 464
pGP13.4
465 466
pGP14.1
467 468
pGP14.2
469 470
Herpes simplex virus 1 (HHV1)
pGP14.3
471 472
pGP14.4
473 474
Hepatitis C Virus pGP23
475 476
Rabies Virus pGP29
477 478
Mokola Virus pGP30
479 480
157
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ ID NO
SEQ ID NO
Virus Glycoprotein
(DNA)
(Protein)
pGP32.1
481 482
Measles Virus
pGP32.2
483 484
Ebola Zaire Virus pGP41 485 486
Dengue Virus pGP25
487 488
Zika virus pGP26
489 490
West Nile Virus pGP27
491 492
Japanese Encephalitis Virus pGP28
493 494
Hepatitis G Virus pGP24 495 496
Mumps Virus F pGP31.1
497 498
Mumps Virus I-IN pGP31.2 499 500
Sendai Virus F pGP33.1
501 502
Sendai Virus FIN pGP33.2 503 504
AcMNPV gp64 pGP59
505 506
Ross River Virus pGP54 507 508
Codon optimized rabies virus pGP29.2
509 510
Rabies virus (strain Nishigahara
pGP29.3
511 512
RCEH) (RABV)
Rabies virus (strain India)
pGP29.4
513 514
(RABV)
Rabies virus (strain CVS-11)
pGP29.5
515 516
(RABV)
Rabies virus (strain ERA)
pGP29.6
517 518
(RABV)
Rabies virus (strain SAD B19)
pGP29.7
519 520
(RABV)
Rabies virus (strain Vnukovo-
pGP29.8
521 522
32) (RABV)
Rabies virus (strain Pasteur
pGP29.9
523 524
vaccins / PV) (RABV)
158
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ ID NO
SEQ ID NO
Virus Glycoprotein
(DNA)
(Protein)
Rabies virus (strain
pGP29.1
525 526
PM1503/AVO1) (RABV)
Rabies virus (strain China/DRY)
pGP29.11 527 528
(BABY)
Rabies virus (strain
pGP29.12 529 530
China/MRV) (RABV)
Rabies virus (isolate
pGP29.13 531 532
Human/Algeria/1991) (RABV)
Rabies virus (strain HEP-Flury)
pGP29.14 533 534
(BABY)
Rabies virus (strain silver-haired
bat-associated) (RABV) pGP29.15
535 536
(SHBRV)
HSV2 gB pGP15.1
537 538
HSV2 gD pGP15.2
539 540
HSV2 gH pGP15.3
541 542
HSV2 gL pGP15.4
543 544
Varicella gB pGP16.1
545 546
Varicella gK pGP16.2
547 548
Varicella gH pGP16.3
549 550
Varicella gL pGP16.4
551 552
Hepatitis B gL pGP22.1
553 554
Hepatitis B gM pGP22.2
555 556
Hepatitis B gS pGP22.3
557 558
Eastern equine encephalitis
pGP65
559 560
virus (EEEV)
Venezuelan equine encephalitis
pGP66
561 562
viruses (VEEV)
159
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
SEQ ID NO
SEQ ID NO
Virus Glycoprotein
(DNA)
(Protein)
Western equine encephalitis
pGP67
563 564
virus (WEEV)
Semliki Forest virus pGP68
565 566
Sindbis virus pGP69
567 568
Chikungunya virus (CHIKV) pGP70
569 570
Bomavirus BoDV-I pGP58
571 572
Tick-borne encephalitis virus
pGP71
573 574
(TBEV)
Usutu virus pGP72
575 576
St. Louis encephalitis virus pGP73
577 578
Yellow fever virus pGP74
579 580
Dengue virus 2 pGP75
581 582
Dengue virus 3 pGP76
583 584
Dengue virus 4 pGP77
585 586
Murray Valley encephalitis
pGP78
587 588
virus (MVEV)
Powassan virus pGP79
589 590
Influenza A virus H5N1 pGP80
591 592
Influenza A virus H7N9 pGP81
593 594
Canine Distemper Virus pGP82
595 596
[00297] In some embodiments, the glycoprotein has a sequence selected from the
group
consisting of SEQ ID NOS: 438, 440, 442, 444, 446, 448, 450, 452, 454, 456,
458, 460, 462,
464, 466, 468, 470, 472, 474, 476, 478, 480, 482, 484, 486, 488, 490, 492,
494, 496, 498, 500,
502, 504, 506, 508, 510, 512, 514, 516, 518, 520, 522, 524, 526, 528, 530,
532, 534, 536, 538,
540, 542, 544, 546, 548, 550, 552, 554, 556, 558, 560, 562, 564, 566, 568,
570, 572, 574, 576,
578, 580, 582, 584, 586, 588, 590, 592, 594 and 596 as set forth in Table 4,
or a sequence having
at least 80%, at least 85%, at least 90%, at least 95%, at least 97%, at least
98%, at least 99% or
100% identity thereto. In some embodiments, the glycoprotein has a sequence
selected from the
group consisting of SEQ ID NOS: 438, 440, 442, 444, 446, 448, 450, 452, 454,
456, 458, 460,
160
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
462, 464, 466, 468, 470, 472, 474, 476, 478, 480, 482, 484, 486, 488, 490,
492, 494, 496, 498,
500, 502, 504, 506, 508, 510, 512, 514, 516, 518, 520, 522, 524, 526, 528,
530, 532, 534, 536,
538, 540, 542, 544, 546, 548, 550, 552, 554, 556, 558, 560, 562, 564, 566,
568, 570, 572, 574,
576, 578, 580, 582, 584, 586, 588, 590, 592, 594 and 596 as set forth in Table
4.
1002981 In some embodiments, the glycoprotein is incorporated into the XDP
system by
inclusion of a nucleic acid encoding the glycoprotein in a plasmid vector of
the XDP system,
described below. In some embodiments, the glycoprotein is encoded by a
sequence selected
from the group consisting of SEQ ID NOS: 437, 439, 441, 443, 445, 447, 449,
451, 453, 455,
457, 459, 461, 463, 465, 467, 469, 471, 473, 475, 477, 479, 481, 483, 485,
487, 489, 491, 493,
495, 497, 499, 501õ 503, 505, 507, 509, 511, 513, 515, 517, 519, 521, 523,
525, 527, 529, 531,
533, 535, 537, 539, 541, 543, 545, 547, 549, 551, 553, 555, 557, 559, 561,
563, 565, 567, 569,
571, 573, 575, 577, 579, 581, 583, 585, 587, 589, 591, 593 and 595 as set
forth in Table 4, or a
sequence having at least 80%, at least 85%, at least 90%, at least 95%, at
least 97%, at least
98%, at least 99% or 100% identity thereto. In some embodiments, the
,glycoprotein is encoded
by a sequence selected from the group consisting of SEQ ID NOS: 437, 439, 441,
443, 445, 447,
449, 451, 453, 455, 457, 459, 461, 463, 465, 467, 469, 471, 473, 475, 477,
479, 481, 483, 485,
487, 489, 491, 493, 495, 497, 499, 501õ 503, 505, 507, 509, 511, 513, 515,
517, 519, 521, 523,
525, 527, 529, 531, 533, 535, 537, 539, 541, 543, 545, 547, 549, 551, 553,
555, 557, 559, 561,
563, 565, 567, 569, 571, 573, 575, 577, 579, 581, 583, 585, 587, 589, 591, 593
and 595 as set
forth in Table 4,
[00299] In some embodiments, a XDP comprising a glycoprotein derived from an
enveloped
virus in a capsid of a XDP of the embodiments exhibits at least a 2-fold, or
at least a 3-fold, or at
least a 4-fold, or at least a 5-fold, or at least a 10-fold increase in
binding of the XDP to a target
cell compared to a XDP that does not have the glycoprotein. Representative
examples
demonstrating enhanced binding and uptake of XDP bearing glycoproteins to
target cells leading
to, in this case, enhance gene editing of target nucleic acid, are provided in
the Examples, below.
[00300] In some embodiments, the present disclosure provides XDP comprising an
antibody
fragment linked to the exterior of the particle wherein the antibody fragment
has specific binding
affinity to a target cell marker or receptor on a target cell, tissue or
organ, providing tropism for
the XDP for the target cell. In one embodiment, the antibody fragment is
selected from the
group consisting of an Fv, Fab, Fab', Fab'-SH, F(ab')2, diabody, single chain
diabody, linear
antibody, a single domain antibody, a single domain camelid antibody, and a
single-chain
161
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
variable fragment (scFv) antibody. Exemplary target cells include T cells, B
cells, macrophages,
liquid cancer cells (such as leukemia or myeloma cells), solid tumor cells,
muscle cells,
epithelial cells, endothelial cells, stem cells, dendritic cells, retinal
cells, hepatic cells, cardiac
cells, thyroid cells, neurons, glial cells, oligodendrocytes, Schwann cells,
and pancreatic cells.
Exemplary target organs include the brain, heart, liver, pancreas, lung, eye,
stomach, small
intestine, colon, and kidney. Exemplary tissues include skin, muscle, bone,
epithelial, and
connective tissue. The target cell marker or ligand can include cell receptors
or surface proteins
known to be expressed preferentially on a target cell for which nucleic acid
editing is desired. In
such cases, a XDP comprising an antibody fragment in a capsid of a XDP of the
embodiments
exhibits at least a 2-fold, or at least a 3-fold, or at least a 4-fold, or at
least a 5-fold, or at least a
10-fold increase in binding to a target cell bearing the target cell marker or
receptor compared to
a XDP that does not have the antibody fragment. In the case of antibody
fragments with affinity
to cancer cell markers or receptors, the cancer cell markers or receptors can
include, but not be
limited to cluster of differentiation 19 (CD19), cluster of differentiation 3
(CD3), CD3d
molecule (CD3D), CD3g molecule (CD3G), CD3e molecule (CD3E), CD247 molecule
(CD247, or CD3Z), CD8a molecule (CD8), CD7 molecule (CD7), membrane
metalloendopeptidase (CD10), membrane spanning 4-domains Al (CD20), CD22
molecule
(CD22), TNF receptor superfamily member 8 (CD30), C-type lectin domain family
12 member
A (CLL1), CD33 molecule (CD33), CD34 molecule (CD34), CD38 molecule (CD38),
integrin
subunit alpha 2b (CD41), CD44 molecule (Indian blood group) (CD44), CD47
molecule
(CD47), integrin alpha 6 (CD49f), neural cell adhesion molecule 1 (CD56), CD70
molecule
(CD70), CD74 molecule (CD74), CD99 molecule (Xg blood group) (CD99),
interleukin 3
receptor subunit alpha (CD123), prominin 1 (CD133), syndecan 1 (CD138),
carbonix anhydrase
IX (CAIX), CC chemokine receptor 4 (CCR4), ADAM metallopeptidase domain 12
(ADAM12), adhesion G protein-coupled receptor E2 (ADGRE2), alkaline
phosphatase
placental-like 2 (ALPPL2), alpha 4 Integrin, angiopoietin-2 (ANG2), B-cell
maturation antigen
(BCMA), CD44V6, carcinoembryonic antigen (CEA), CEAC, CEA cell adhesion
molecule 5
(CEACAM5), Claudin 6 (CLDN6), CLDN18, C-type lectin domain family 12 member A
(CLEC12A), mesenchymal-epithelial transition factor (cMET), cytotoxic T-
lymphocyte-
associated protein 4 (CTLA4), epidermal growth factor receptor 1 (EGF1R),
epidermal growth
factor receptor variant III (EGFRvIII), epithelial glycoprotein 2 (EGP-2),
epithelial cell adhesion
molecule (EGP-40 or EpCAM), EPH receptor A2 (EphA2), ectonucleotide
162
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
pyrophosphatase/phosphodiesterase 3 (ENPP3), erb-b2 receptor tyrosine kinase 2
(ERBB2),
erb-b2 receptor tyrosine kinase 3 (ERBB3), erb-b2 receptor tyrosine kinase 4
(ERBB4), folate
binding protein (FBP), fetal nicotinic acetylcholine receptor (AChR), folate
receptor alpha
(Fralpha or FOLR1), G protein-coupled receptor 143 (GPR143), glutamate
metabotropic
receptor 8 (GRM8), glypican-3 (GPC3), ganglioside GD2, ganglioside GD3, human
epidermal
growth factor receptor I (HERI), human epidermal growth factor receptor 2
(HER2), human
epidermal growth factor receptor 3 (HER3)õ Integrin B7, intercellular cell-
adhesion molecule-1
(ICAM-1), human telomerase reverse transcriptase (hTERT), Interleukin-13
receptor a2 (IL-
13R-a2), K-light chain, Kinase insert domain receptor (KDR), Lewis-Y (LeY),
chondromodulin-
1 (LECT1), Ll cell adhesion molecule (L1CAM), Lysophosphatidic acid receptor 3
(LPAR3),
melanoma-associated antigen 1 (MAGE-A1), mesothelin (MSLN), mucin 1 (MUC1),
mucin 16,
cell surface associated (MUC16), melanoma-associated antigen 3 (MAGEA3), tumor
protein
p53 (p53), Melanoma Antigen Recognized by T cells 1 (MART!), glycoprotein 100
(GPI00),
Proteinase3 (PRO, ephrin-A receptor 2 (EphA2), Natural killer group 2D ligand
(NKG2D
ligand), New York esophageal squamous cell carcinoma 1 (NY-ESO-1), oncofetal
antigen
(h5T4), prostate-specific membrane antigen (PSMA), programmed death ligand 1
(PDL-1),
receptor tyrosine kinase-like orphan receptor 1 (ROR1), trophoblast
glycoprotein (TPBG),
tumor-associated glycoprotein 72 (TAG-72), tumor-associated calcium signal
transducer 2
(TROP-2), tyrosinase, survivin, vascular endothelial growth factor receptor 2
(VEGF- R2),
Wilms tumor-1 (WT-1), leukocyte immunoglobulin-like receptor B2 (LILRB2),
Preferentially
Expressed Antigen In Melanoma (PRAME), T cell receptor beta constant l(TRBC1),
TRBC2,
and (T-cell immunoglobulin mucin-3) TTM-3. In the case of antibody fragments
with affinity to
neuron receptors, the cell markers or receptors can include, but not be
limited to Adrenergic
(e.g., alA, alb, alc, ald, a2a, a2b, a2c, a2d, 131, 132, I33), Dopaminergic
(e.g., D1, D2, D3, D4,
D5), GABAergic (e.g., GABAA, GABA131a, GABAB15, GABAB2, GABAC), Glutaminergic
(e.g., NMDA, AMPA, kainate, inGluR1, mGluR2, mGluR3, mGluR4, mGluR5, mGluR6,
mGluR7), Histaminergic (e.g., HI, H2, H3), Cholinergic (e.g., Muscarinic
(e.g., MI, M2, M3,
M4, M5; Nicotinic (e.g., muscle, neuronal (a-bungarotoxin-insensitive),
neuronal (a-
bungarotoxin-sensitive)), Opioid (e.g., i, 51, 52, ic), and Serotonergic
(e.g., 5-HT1A, 5-HT1B, 5-
HT1D, 5-HT1E, 5-HT IF, 5-HT2A, 5-HT2B, 5-HT2C, 5-HT3, 5-HT4, 5-HT5, 5-HT6, 5-
HT7).
1003011 In one embodiment, the antibody fragment is conjugated to the XDP
after its
production and isolation from the producing host cell. In another embodiment,
the antibody
163
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
fragment is produced as a part of the XDP capsid expressed by the producing
host cell of the
XDP system. In some cases, the present disclosure provides a nucleic acid
comprising a
sequence encoding the antibody fragment operably linked to the nucleic acid
encoding the XDP
capsid or other XDP components
IV. Nucleic Acids Encoding XDP Systems
[00302] In another aspect, the present disclosure relates to nucleic acids
encoding components
of the XDP system and the incorporated therapeutic payloads, and the vectors
that comprise the
nucleic acids, as well as methods to make the nucleic acids and vectors.
[00303] In some embodiments, the present disclosure provides one or more
nucleic acids
encoding components including retroviral-derived XDP structural and processing
components,
therapeutic payloads, and tropism factors. The nucleic acids and vectors
utilized for the key
structural components and for processing and the assembly of XDP particles of
the embodiments
can be derived from a variety of viruses, such as retroviruses, including but
not limited to
Retroviridae family members Alpharetroviruses, Betaretroviruses,
Gammaretroviruses,
Deltaretroviruses, Epsdonreiroviruses, Spumaretrovirinae, or lentiviruses such
as human
immunodeficiency-1 (HIV-1), human immunodeficiency-2 (HIV-2), simian
immunodeficiency
virus (Sly), feline immunodeficiency virus (Hy), bovine immunodeficiency virus
(BIV),
Jembrana Disease Virus (JDV), equine infectious anemia virus (EIAV), caprine
arthritis
encephalitis virus (CASEY) and the like.
[00304] In some embodiments, the nucleic acids encoding the XDP retroviral
components are
derived from Alpharetrovirus, including but not limited to avian leukosis
virus (ALV) and Rous
sarcoma virus (RSV). In some embodiments, the present disclosure provides
nucleic acids
encoding components selected from the group consisting of: a matrix
polypeptide (MA); a p2A
spacer peptide; ap2B spacer peptide; a p10 spacer peptide; a capsid
polypeptide (CA); a
nucleocapsid polypeptide (NC); a Gag polyprotein comprising a matrix
polypeptide (MA), a
capsid polypeptide (CA), p2A, p2B, p10, a nucleocapsid polypeptide (NC); a
therapeutic
payload; a tropism factor; a Gag-transframe region-Pol protease polyprotein; a
protease cleavage
site(s); and a protease capable of cleaving the protease cleavage sites. In
the forgoing
embodiment, Gag components (e.g., MA, CA, p24, p2B, p10, and NC), and
optionally the
protease cleavage site and protease, are derived from an Alpharetrovirus,
including but not
limited to Avian leukosis virus and Rous sarcoma virus. In some embodiments,
the encoding
164
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
sequences for the Alphareirovirus-derived components are selected from the
group consisting of
SEQ ID NOS: 192, 193, 195, 196, 198-201, 782, and 234 as set forth in Table 5,
or a sequence
having at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at
least about 98%, or at least about 99% identity thereto. In some embodiments,
the nucleic acids
encode a subset of the components listed in the paragraph, such as depicted in
FIGS. 36-68,
which depict CasX and gNA as the therapeutic payloads. In some embodiments of
the foregoing,
encoding nucleotides for protease cleavage sites are located between each of
the individual
components. In other cases, the protease cleavage sites are omitted. In a
particular embodiment,
an encoding sequence for a single protease cleavage site is located between
the sequence
encoding the nuclease and the linked retroviral component, which may be a
retroviral sequence
or a non-viral sequence, such as one that can be cleaved by TENT, PreScission
Protease, or any of
the other proteases disclosed herein. Representative configurations and
sequences are presented
in the Examples. In a particular embodiment, the encoded therapeutic payload
is a CasX and
gNA embodiment described herein, while the encoded tropism factor is a viral
glycoprotein
embodiment described herein.
1003051 In some embodiments, the nucleic acids encoding the XDP viral
components are
derived from Betaretrovirus, including but not limited to mouse mammary tumor
virus
(MMTV), Mason-Pfizer monkey virus (MPMV), and enzootic nasal tumor virus
(ENTV). In
such embodiments, the present disclosure provides nucleic acids encoding the
XDP wherein the
XDP comprises components selected from the group consisting of: a matrix
polypeptide (MA); a
pp21/24 spacer peptide; a p3-P8/p12 spacer peptide; a capsid polypeptide (CA);
a nucleocapsid
polypeptide (NC); a Gag polyprotein comprising a matrix polypeptide (MA), a
capsid
polypeptide (CA), pp21/24, p3-81p12, a nucleocapsid polypeptide (NC); a
therapeutic payload; a
tropism factor; a Gag-transframe region-Pol protease polyprotein; a protease
cleavage site(s);
and a protease capable of cleaving the protease cleavage sites. In the
forgoing embodiment, Gag
components (e.g., MA, CA, pp21/24 spacer, p3-p8/p12 spacer, and NC), and
optionally the
protease cleavage site and protease, are derived from an Betareirovirus,
including but not limited
to mouse mammary tumor virus, Mason-Pfizer monkey virus, and enzootic nasal
tumor virus. In
some embodiments, the encoding sequences for the Betaretrovirus-derived
components are
selected from the group consisting of SEQ ID NOS: 235-257 as set forth in
Table 5, or a
sequence having at least about 85%, at least about 90%, at least about 91%, at
least about 92%,
165
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
at least about 93%, at least about 94%, at least about 95%, at least about
96%, at least about
97%, at least about 98%, or at least about 99% identity thereto. In some
embodiments, the
nucleic acids encode a subset of the components listed in the paragraph, such
as depicted in
FIGS. 36-68, which depict CasX and gNA as the therapeutic payloads. In some
embodiments of
the foregoing, encoding nucleotides for protease cleavage sites are located
between each of the
individual components. In other cases, the protease cleavage sites are
omitted. In a particular
embodiment, an encoding sequence for a single protease cleavage site is
located between the
sequence encoding the nuclease and the linked retroviral component, which may
be a retroviral
sequence or a non-viral sequence, such as one that can be cleaved by TEV,
PreScission Protease,
or any of the other proteases disclosed herein. Representative configurations
and sequences are
presented in the Examples. In a particular embodiment, the encoded therapeutic
payload is a
CasX and gNA embodiment described herein, while the encoded tropism factor is
a viral
glycoprotein embodiment described herein.
1003061 In some embodiments, the nucleic acids encoding the XDP viral
components are
derived from Deltaretrovirus, including but not limited to bovine leukemia
virus (BLV) and the
human T-lymphotropic viruses (HTLV1). In such embodiments, the present
disclosure provides
nucleic acids encoding the XDP wherein the XDP comprises components selected
from the
group consisting of: a matrix polypeptide (MA); a capsid polypeptide (CA); a
nucleocapsid
polypeptide (NC); a Gag polyprotein comprising a matrix polypeptide (MA), a
capsid
polypeptide (CA)õ a nucleocapsid polypeptide (NC); a therapeutic payload; a
tropism factor; a
Gag-transframe region-Pol protease polyprotein; a protease cleavage site(s);
and a protease
capable of cleaving the protease cleavage sites. In the forgoing embodiment,
Gag components
(e.g., MA, CA, and NC), and optionally the protease cleavage site and
protease, are derived
from an Deltaretrovirus, including but not limited to bovine leukemia virus
and the human T-
lymphotropic viruses. In some embodiments, the encoding sequences for the
Deltaretrovirus-
derived components are selected from the group consisting of the sequences SEQ
ID NOS: 258-
272 as set forth in Table 5, or a sequence having at least about 85%, at least
about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, or at least about 99%
identity thereto. In
some embodiments, the nucleic acids encode a subset of the components listed
in the paragraph,
such as depicted in FIGS. 36-68, which depict CasX and gNA as the therapeutic
payloads. In
some embodiments of the foregoing, encoding nucleotides for protease cleavage
sites are located
166
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
between each of the individual components. In other cases, the protease
cleavage sites are
omitted. In a particular embodiment, an encoding sequence for a single
protease cleavage site is
located between the sequence encoding the nuclease and the linked retroviral
component, which
may be a retroviral sequence or a non-viral sequence, such as one that can be
cleaved by TEV,
PreScission Protease, or any of the other proteases disclosed herein.
Representative
configurations and sequences are presented in the Examples. In a particular
embodiment, the
encoded therapeutic payload is a CasX and gNA embodiment described herein,
while the
encoded tropism factor is a viral glycoprotein embodiment described herein.
1003071 In some embodiments, the nucleic acids encoding the XDP viral
components are
derived from Epsilonretrovirus, including but not limited to Walleye dermal
sarcoma virus
(WDSV), and Walleye epidermal hyperplasia virus 1 and 2. In such embodiments,
the present
disclosure provides nucleic acids encoding the XDP wherein the XDP comprises
components
selected from the group consisting of: a matrix polypeptide (MA); a p20 spacer
peptide; a capsid
polypeptide (CA); a nucleocapsid polypeptide (NC); a Gag polyprotein
comprising a matrix
polypeptide (MA), a capsid polypeptide (CA), p20, a nucleocapsid polypeptide
(NC); a
therapeutic payload; a tropism factor; a Gag-transframe region-Pol protease
polyprotein; a
protease cleavage site(s); and a protease capable of cleaving the protease
cleavage sites. In the
forgoing embodiment, Gag components (e.g., MA, CA, p20, and NC), and
optionally the
protease cleavage site and protease, are derived from an Epsilonreirovirus,
including but not
limited to Walleye dermal sarcoma virus, and Walleye epidermal hyperplasia
virus 1 and 2. In
some embodiments, the encoding sequences for the Epsilonretrovirus-derived
components are
selected from the group consisting of the sequences of SEQ ID NOS: 273-277 as
set forth in
Table 5, or a sequence having at least about 85%, at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at
least about 97%, at least about 98%, or at least about 99% identity thereto.
In some
embodiments, the nucleic acids encode a subset of the components listed in the
paragraph, such
as depicted in FIGS. 36-68, which depict CasX and gNA as the therapeutic
payloads. In some
embodiments of the foregoing, encoding nucleotides for protease cleavage sites
are located
between each of the individual components. In other cases, the protease
cleavage sites are
omitted. In a particular embodiment, an encoding sequence for a single
protease cleavage site is
located between the sequence encoding the nuclease and the linked retroviral
component, which
may be a retroviral sequence or a non-viral sequence, such as one that can be
cleaved by TEV,
167
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
PreScission Protease, or any of the other proteases disclosed herein.
Representative
configurations and sequences are presented in the Examples. In a particular
embodiment, the
encoded therapeutic payload is a CasX and gNA embodiment described herein,
while the
encoded tropism factor is a viral glycoprotein embodiment described herein.
1003081 In some embodiments, the nucleic acids encoding the XDP viral
components are
derived from Garnmaretrovirus, including but not limited to murine leukemia
virus (IVILV),
Maloney murine leukemia virus (MMLV), and feline leukemia virus (FLV). In such
embodiments, the nucleic acids encoding the present disclosure provides XDP
wherein the XDP
comprises components selected from the group consisting of: a matrix
polypeptide (MA); a pp12
spacer peptide; a capsid polypeptide (CA); a nucleocapsid polypeptide (NC); a
Gag polyprotein
comprising a matrix polypeptide (MA), a pp12 spacer, a capsid polypeptide
(CA), a
nucleocapsid polypeptide (NC); a therapeutic payload; a tropism factor; a Gag-
transframe
region-Pol protease polyprotein; a protease cleavage site(s); and a protease
capable of cleaving
the protease cleavage sites. In the forgoing embodiment, Gag components (e.g.,
MA, pp12, CA,
and NC), and optionally the protease cleavage site and protease, are derived
from an
Gammareirovirus, including but not limited to Walleye dermal sarcoma virus,
and Walleye
epidermal hyperplasia virus 1 and 2. In some embodiments, the encoding
sequences for the
Gannnaretrovirus-derived components are selected from the group consisting of
the sequences
of SEQ ID NOS: 278-287 as set forth in Table 5, or a sequence having at least
about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%,
at least about 95%, at least about 96%, at least about 97%, at least about
98%, or at least about
99% identity thereto. In some embodiments, the nucleic acids encode a subset
of the components
listed in the paragraph, such as depicted in FIGS. 36-68, which depict CasX
and gNA as the
therapeutic payloads. In some embodiments of the foregoing, encoding
nucleotides for protease
cleavage sites are located between each of the individual components. In other
cases, the
protease cleavage sites are omitted_ In a particular embodiment, an encoding
sequence for a
single protease cleavage site is located between the sequence encoding the
nuclease and the
linked retroviral component, which may be a retroviral sequence or a non-viral
sequence, such as
one that can be cleaved by TEV, PreScission Protease, or any of the other
proteases disclosed
herein. Representative configurations and sequences are presented in the
Examples. In a
particular embodiment, the encoded therapeutic payload is a CasX and gNA
embodiment
168
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
described herein, while the encoded tropism factor is a viral glycoprotein
embodiment described
herein.
1003091 In some embodiments, the nucleic acids encoding the XDP viral
components are
derived from Lentivirus, including but not limited to HIV-1 and MV-2, and
Simian
immunodeficiency virus (SW). In such embodiments, the present disclosure
provides nucleic
acids encoding the XDP wherein the XDP comprises components selected from the
group
consisting of: a matrix polypeptide (MA); a capsid (CA), a p2 spacer peptide,
a nucleocapsid
(NC), a p1/p6 spacer peptide; ); a Gag polyprotein comprising a matrix
polypeptide (MA), CA,
P2, NC, and pl/p6; a therapeutic payload; a tropism factor; a Gag-transframe
region-Pol
protease polyprotein; a protease cleavage site(s); and a protease capable of
cleaving the protease
cleavage sites. In the forgoing embodiment, Gag components (e.g., MA, CA, NC,
and pl/p6),
and optionally the protease cleavage site and protease, are derived from an
Lent/virus, including
but not limited to HIV-1, HIV-2, and Simian immunodeficiency virus (SIV). In
some
embodiments, the encoding sequences for the Lent/virus-derived components are
selected from
the group consisting of the sequences of SEQ ID NOS: 288-312 and 334-339 as
set forth in
Table 5, or a sequence having at least about 85%, at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at
least about 97%, at least about 98%, or at least about 99% identity thereto.
In some
embodiments, the nucleic acids encode a subset of the components listed in the
paragraph, such
as depicted in FIGS. 36-68, which depict CasX and gNA as the therapeutic
payloads. In some
embodiments of the foregoing, encoding nucleotides for protease cleavage sites
are located
between each of the individual components. In other cases, the protease
cleavage sites are
omitted. In a particular embodiment, an encoding sequence for a single
protease cleavage site is
located between the sequence encoding the nuclease and the linked retroviral
component, which
may be a retroviral sequence or a non-viral sequence, such as one that can be
cleaved by TEV,
PreScission Protease, or any of the other proteases disclosed herein.
Representative
configurations and sequences are presented in the Examples. In a particular
embodiment, the
encoded therapeutic payload is a CasX and gNA embodiment described herein,
while the
encoded tropism factor is a viral glycoprotein embodiment described herein.
1003101 In some embodiments, the nucleic acids encoding the XDP viral
components are
derived from Spumaretrovirinae, including but not limited to Bovispumavirus,
Equispumavirus,
Felispumavirus, Prosimiispumavirus, Similspumavirus, and Spumavirus. In such
cases, the
169
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
present disclosure provides nucleic acids encoding the XDP wherein the XDP
comprises
components selected from the group consisting of. P68 Gag; a p3 Gag; a Gag
polyprotein
comprising of P68 Gag and p3 gag; a therapeutic payload; a tropism factor; a
Gag-transframe
region-Pol protease polyprotein; a protease cleavage site(s); and a protease
capable of cleaving
the protease cleavage sites. In the forgoing embodiment, Gag components (e.g.,
MA, CA, p20,
and NC), and optionally the protease cleavage site and protease, are derived
from an
Spumaretrovirthae including but not limited to Bovispuntavnws,
Equispuntavirus,
Felisputnavirus, Prosintlisputnavirus, Simiisputnavirus, and Sputnavirus. In
some embodiments,
the encoding sequences for the Sumaretrovirinae-derived components are
selected from the
group consisting of the sequences of SEQ ID NOS: 313-333 as set forth in Table
5, or a
sequence having at least about 85%, at least about 90%, at least about 91%, at
least about 92%,
at least about 93%, at least about 94%, at least about 95%, at least about
96%, at least about
97%, at least about 98%, or at least about 99% identity thereto. In some
embodiments, the
nucleic acids encode a subset of the components listed in the paragraph, such
as depicted in
FIGS. 36-68, which depict CasX and gNA as the therapeutic payloads. In some
embodiments of
the foregoing, encoding nucleotides for protease cleavage sites are located
between each of the
individual components. In other cases, the protease cleavage sites are
omitted. In a particular
embodiment, an encoding sequence for a single protease cleavage site is
located between the
sequence encoding the nuclease and the linked retroviral component, which may
be a retroviral
sequence or a non-viral sequence, such as one that can be cleaved by TEV,
PreScission Protease,
or any of the other proteases disclosed herein. Representative configurations
and sequences are
presented in the Examples. In a particular embodiment, the encoded therapeutic
payload is a
CasX and gNA embodiment described herein, while the encoded tropism factor is
a viral
glycoprotein embodiment described herein.
1003111 In other embodiments, the present disclosure provides nucleic acids
encoding the XDP
wherein the retroviral components of the XDP are selected from different
genera of the
Retroviridae. Thus the nucleic acids encoding the XDP can comprise two or more
components
selected from a matrix polypeptide (MA), a p2A spacer peptide, a p211 spacer
peptide; a p10
spacer peptide, a capsid polypeptide (CA), a nucleocapsid polypeptide (NC), a
pp21/24 spacer
peptide, a p3-p8 spacer peptide, a pp12 spacer peptide, a p20 spacer peptide,
a pl /p6 spacer
peptide, a p68 Gag, a p3 Gag, a cleave site(s), and a protease capable of
cleaving the protease
cleavage sites wherein the components are derived from two or more of
*hare/row/its,
170
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus,
Lent/virus,
Bovispumavirns, Equispumcrvirus, Felispumavirus, Prosimiispumcrvirus,
Simiispumavirus, or
Spumavirns.
1003121 In retroviral components derived from FITV-1, the accessory protein
integrase (or its
encoding nucleic acid) can be omitted from the XDP systems, as well as the HIV
functional
accessory genes vpr, vpx (HIV-2), which are dispensable for viral replication
in vitro.
Additionally, the nucleic acids of the XDP system do not require reverse
transcriptase for the
creation of the XDP compositions of the embodiments. Thus, in one embodiment,
the HIV-1
Gag-Pol component of the XDP can be truncated to Gag linked to the transframe
region (TFR)
composed of the transframe octapeptide (TFP) and 48 amino acids of the p6pol,
separated by a
protease cleavage site, hereinafter referred to as Gag-TFR-PR, described more
fully, below.
Table 5: Retroviral structural component encoding DNA sequences
DNA Sequences Encoding Components
Virus MATRIX P2A-P2B- CAPSID NUCLE
PROTEASE*
P1O-PP24
OCAPSI
D
ALV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 192) 193) NO: 195)
NO: 198)
196)
RSV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ (SEQ ID NO:
NO: 199) 200) NO: 782)
ID NO: 234)
201)
Virus MATRIX PP21/24 P3-P8/P12 CAPSID
NUCLEOCA Protease
PSID
ENTV (SEQ ID (SEQ ID NO:
(SEQ ID (SEQ ID NO: (SEQ ID
NO: 235) 236)
NO: 238) NO: 239)
237)
MMT (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO: (SEQ ID
V NO: 240) 241) NO: 242)
NO: 244) NO: 245)
243)
MPM (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO: SEQ ID
V NO: 246) 247) NO: 248)
NO: 250) NO: 251)
249)
MPM (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO: (SEQ ID
V NO: 252) 253) NO: 254)
NO: 256) NO: 257)
255)
171
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DNA Sequences Encoding Components
Virus MATRIX P2A-P2B- CAPSID NUCLE
PROTEASE*
P1O-PP24
OCAPSI
D
Nativ
e
Virus MATRIX CAPSID NUCLEOC PROTE MA-
CA-
APSID ASE* CLEAVE
SITE*
BLV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 258) 259) NO: 260)
NO: 262)
261)
HTLV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
1 NO: 263) 264) NO: 265)
NO: 267)
266)
HTLV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
1 NO: 268) 269) NO: 270)
NO: 272)
Nativ
271)
e
Virus MATRIX P20 CAPSID NUCLE
PROTEASE*
OCAPSI
D
WDS (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
V NO: 273) 274) NO: 275)
NO: 277)
276)
Virus MATRIX P12 CAPSID NUCLE
PROTEASE*
OCAPSI
D
FLV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 278) 279) NO: 280)
NO: 282)
281)
MML (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
V NO: 283) 284) NO: 285)
NO: 287)
286)
Virus MATRIX CAPSID NUCLEOC PROTE MA-
CA-
APSID ASE' CLEAVE
SITE*
CAEV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 288) 289) NO: 290)
NO: 292)
291)
172
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DNA Sequences Encoding Components
Virus MATRIX P2A-P2B- CAPSID NUCLE
PROTEASE*
P1O-PP24
OCAPSI
D
EIAV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 293) 294) NO: 295)
NO: 297)
296)
Sly (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 298) 299) NO: 300)
NO: 302)
301)
Sly (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
Nativ NO: 303) 304) NO: 305)
NO: 307)
e
306)
VMV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 308) 309) NO: 310)
NO: 312)
311)
Virus GAG PROTEASE* CLEAVAG
E SITE*
BFV (SEQ ID (SEQ ID NO: (SEQ ID
NO: 313) 314) NO: 315)
BGPF (SEQ ID (SEQ ID NO: (SEQ ID
V NO: 316) 317) NO: 318)
CCFV (SEQ ID (SEQ ID NO: (SEQ ID
NO: 319) 320) NO: 321)
EFV (SEQ ID (SEQ ID NO: (SEQ ID
NO: 322) 323) NO: 324)
FFV (SEQ ID (SEQ ID NO: (SEQ ID
NO: 325) 326) NO: 327)
RHSF (SEQ ID (SEQ ID NO: (SEQ ID
V NO: 328) 329) NO: 330)
SFV (SEQ ID (SEQ ID NO: (SEQ ID
NO: 331) 332) NO: 333)
Virus MATRIX CAPSID P2
NC P1/P6
HIV (SEQ ID (SEQ ID NO: (SEQ ID
(SEQ ID (SEQ ID NO:
NO: 334) 335) NO: 336)
NO: 338)
337)
Protease*
(SEQ ID NO:
339)
* Wild-type sequence (optionally incorporated, depending on configuration)
173
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1003131 In some embodiments, the present disclosure provides nucleic acids
encoding
sequences for the tropism factors that are incorporated in, and displayed on
the surface of the
XDP, wherein the tropism factor confers an increased ability of the XDP to
bind and fuse with
the membrane of a target cell or tissue. In one embodiment, the tropism factor
is a glycoprotein,
wherein the encoding nucleic acid is selected from the group consisting of the
sequences of
Table 4, or a sequence having at least about 85%, at least about 90%, at least
about 91%, at least
about 92%, at least about 93%, at least about 94%, at least about 95%, at
least about 96%, at
least about 97%, at least about 98%, or at least about 99% identity thereto.
In another
embodiment, the disclosure provides a nucleic acids encoding an antibody
fragment, wherein the
antibody fragment has specific binding affinity for a target cell marker or
receptor on a target
cell or tissue. In another embodiment, the disclosure provides nucleic acids
encoding a cell
receptor, wherein the cell receptor has specific binding affinity for a target
cell marker on a
target cell or tissue. In another embodiment, the disclosure provides nucleic
acids encoding a
ligand, wherein the ligand has specific binding affinity for a target cell
marker or receptor on a
target cell or tissue. By inclusion of the nucleic acids encoding for the
tropism factors, it will be
understood that the resulting XDP will have increased selectivity for the
target cell or tissue,
resulting in an increased therapeutic index and reduced off-target effects.
1003141 The present disclosure further provides nucleic acids encoding or
comprising the
therapeutic payloads incorporated into the XDP. Exemplary therapeutic payloads
have been
described herein, supra. In some embodiments, the therapeutic payload of the
XDP is a CRISPR
nuclease and one or more guide RNAs In a particular embodiment of the
foregoing, the
disclosure provides nucleic acids encoding the CasX nucleases of Table 1, or a
sequence having
at least about 85%, at least about 90%, at least about 91%, at least about
92%, at least about
93%, at least about 94%, at least about 95%, at least about 96%, at least
about 97%, at least
about 98%, or at least about 99% identity thereto. Representative examples of
such nucleic acids
are presented in Tables 6-8, 11 and 16 of the Examples, which disclose nucleic
acids of SEQ ID
NOS: 354, 340-342, 346-349, 378-387 and 426-431. In another particular
embodiment of the
foregoing, the disclosure provides nucleic acids encoding the gNA variants of
SEQ ID NO: 597-
781 set forth in Table 3, or a sequence having at least about 85%, at least
about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, or at least about 99%
identity thereto,
174
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
and wherein the gNA further comprises a targeting sequence complementary to a
target nucleic
acid.
1003151 In some embodiments of the disclosure, the components of the XDP
systems are
encoded by one, two, three, four, five or more nucleic acids (see FIGS. 36-68,
which are
schematics of the representative plasmids and XDP configurations), which can
encode single
components or multiple components that are operably linked to (under the
control of) regulatory
elements operable in a eukaryotic cell and appropriate for the component to be
expressed. It will
be understood that in the descriptions of the XDP system configurations, the
absolute order of
the components encoded within a nucleic acid may be varied in order to take
advantage of the
placement of the regulatory elements, cleavage sequences, etc., such that each
component can be
expressed and/or utilized in the assembly of the XDP in an optimal fashion, as
would be
understood by one of ordinary skill in the art. For example, where a nucleic
acid encodes the
Gag polyprotein, the therapeutic payload, and a protease cleavage site, the
order (5' to 3') may
be Gag-cleavage site-therapeutic payload or it may be therapeutic payload-
cleavage site-gag, and
it is intended that the same would apply for any combination of components
encoded in a single
nucleic acid. Representative regulatory elements are described herein.
1003161 In some embodiments, the disclosure provides nucleic acids comprising
sequences
encoding components of the XDP system selected from two or more of a
retroviral Gag
polyprotein (all or portions thereof), a protease cleavage site, a therapeutic
payload, a Gag-Pol
polyprotein, and a tropism factor, wherein the components are encoded on one,
two, three, or
four individual nucleic acids. In some embodiments of the foregoing, the
components are
encoded on a single nucleic acid In some embodiments of the foregoing, a first
nucleic acid
encodes the Gag polyprotein (or portions thereof) and the CasX protein as the
therapeutic
payload with, optionally, an intervening protease cleavage site between the
two components, and
a second nucleic acid encodes the Gag-Pol polyprotein (or portions thereof),
the tropism factor
and the gNA. In another embodiment of the foregoing, a first nucleic acid
encodes the Gag
polyprotein (or portions thereof) and the CasX protein as the therapeutic
payload with,
optionally, and intervening protease cleavage site separating the two
components, a second
nucleic acid encodes the Gag-Pol polyprotein, and a third nucleic acid encodes
the tropism factor
and the gNA. In another embodiment, a first nucleic acid encodes the Gag
polyprotein (or
portions thereof) and the CasX protein as the therapeutic payload with,
optionally, an
intervening protease cleavage site separating the two components, a second
nucleic acid encodes
175
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
the tropism factor, a third nucleic acid encodes the Gag-Pol polyprotein (or
portions thereof),
and a fourth nucleic acid encodes the gNA. In some cases, the protease
cleavage sites are
omitted In other cases, protease cleavage sites are located between each
component of the Gag
polyprotein and, optionally, the therapeutic payload. Representative examples
of the encoding
nucleic acids of the foregoing embodiments are presented in the Examples.
1003171 In other embodiments, the disclosure provides nucleic acids comprising
sequences
encoding components of the XDP system comprising the Gag-TFR-PR polyprotein
(or portions
thereof), the protease cleavage site, the CasX protein as the therapeutic
payload, the gNA, and
the tropism factor, wherein the components are encoded on one, two, or three
individual nucleic
acids. In some embodiments of the foregoing, the components are encoded on a
single nucleic
acid. In another embodiment of the foregoing, a first nucleic acid encodes the
Gag-TFR-PR
polyprotein and the CasX protein as the therapeutic payload with an
intervening protease
cleavage site separating the two components, and a second nucleic acid encodes
the tropism
factor and the gNA. In another embodiment, a first nucleic acid encodes the
Gag-TFR-PR
polyprotein and the CasX protein as the therapeutic payload with an
intervening protease
cleavage site separating the two components, a second nucleic acid encodes the
tropism factor,
and a third nucleic acid encodes the gNA. In some embodiments of the
foregoing, protease
cleavage sites are located between each component of the Gag polyprotein and,
optionally, the
CasX protein. Representative examples of the encoding nucleic acids of the
foregoing
embodiments are presented in the Examples (see Tables 16, 17, 19, 20, 22, 23,
24, 27, 30, 33 and
36 and the sequences contained therein).
1003181 In other embodiments, the disclosure provides nucleic acids comprising
sequences
encoding components of the XDP system comprising the Gag polyprotein (or
portions thereof),
the protease cleavage site, the protease, the CasX protein, the gNA and the
tropism factor
wherein the components are encoded on one, two, or three individual nucleic
acids. In some
embodiments of the foregoing, the components are encoded on a single nucleic
acid. In another
embodiment of the foregoing, a first nucleic acid encodes the Gag polyprotein,
the protease, the
CasX protein, and intervening protease cleavage sites located between the
components, and a
second nucleic acid encodes the pseudotyping viral envelope glycoprotein or
antibody fragment
and the gNA. In another embodiment of the foregoing, a first nucleic acid
encodes the Gag
polyprotein, the protease, the CasX protein and intervening protease cleavage
sites between the
176
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
components, a second nucleic acid encodes the tropism factor; and a third
nucleic acid encodes
one or more gNA.
1003191 In other embodiments, the disclosure provides nucleic acids comprising
sequences
encoding components of the XDP system comprising the Gag-Pol polyprotein, the
CasX protein,
the protease cleavage site, the gNA, the RNA binding domain, and the tropism
factor, wherein
the components are encoded on one, two, or three individual nucleic acids. In
some
embodiments of the foregoing, the components are encoded on a single nucleic
acid. In another
case of the foregoing, a first nucleic acid encodes the Gag-Pol polyprotein
and the CasX with an
intervening protease cleavage site between the two components, and a second
nucleic acid
encodes the tropism factor, the gNA and the RNA binding domain. In another
case of the
foregoing, a first nucleic acid encodes the Gag-Pol polyprotein and the CasX
with an intervening
protease cleavage site between the two components, and a second nucleic acid
encodes the
tropism factor, and a third nucleic acid encodes the gNA and the RNA binding
domain.
1003201 In some embodiments, the disclosure provides nucleic acids comprising
sequences
encoding components of the )(DP system comprising the Gag-Pol polyprotein, the
CasX protein,
the protease cleavage site, the tropism factor, and the gNA, wherein the
components are encoded
on one, two, or three individual nucleic acids. In some embodiments of the
foregoing, the
components are encoded on a single nucleic acid. In another case of the
foregoing, a first
nucleic acid encodes the first nucleic acid encodes the Gag-Pol polyprotein
and the CasX with
an intervening protease cleavage site between the two components, and a second
nucleic acid
encodes the tropism factor and the gNA. In another case, a first nucleic acid
encodes the Gag-Pol
polyprotein and the CasX with an intervening protease cleavage site between
the two
components, a second nucleic acid encodes the tropism factor, and a third
nucleic acid encodes
the gNA.
1003211 In other embodiments, the disclosure provides nucleic acids comprising
sequences
encoding components of the XDP system comprising the MA, the CasX protein, the
protease,
the protease cleavage site, the gNA, and the tropism factor, wherein the
components are encoded
on one, two, three, or four individual nucleic acids. In some embodiments of
the foregoing, the
components are encoded on a single nucleic acid. In other cases of the
foregoing, a first nucleic
acid encodes the first nucleic acid encodes the MA, the CasX protein, the
protease, and
intervening protease cleavage sites between the three components, and a second
nucleic acid
encodes the tropism factor and the gNA. In other cases, a first nucleic acid
encodes the MA, the
177
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
CasX protein the protease, and intervening protease cleavage sites between the
three
components, a second nucleic acid encodes the tropism factor; and a third
nucleic acid encodes
the gNA. In other cases, a first nucleic acid encodes the MA and the CasX
protein with an
intervening protease cleavage site between the two components, a second
nucleic acid encodes
the tropism factor, a third nucleic acid encodes the ,gNA, and a fourth
nucleic acid encodes the
protease. In the foregoing embodiments, the first nucleic acid can further
encode a CA
component linked to the MA by an additional intervening protease cleavage
site. In some
embodiments of the foregoing, the protease and protease cleavage sites are
omitted.
1003221 In some embodiments, the disclosure provides nucleic acids comprising
sequences
encoding components of the XDP system comprising the Gag polyprotein (all or
portions
thereof), the CasX protein, the protease, the protease cleavage site, the gNA,
the tropism factor,
and the Gag-Pol polyprotein (all or portions thereof), wherein the components
are encoded on
two, three, or four individual nucleic acids. In some embodiments of the
foregoing, a first
nucleic acid encodes the Gag polyprotein, the CasX protein, the protease, and
intervening
protease cleavage sites between the three components, and a second nucleic
acid encodes the
Gag-Pol polyprotein, the tropism factor, and the gNA. In other embodiments, a
first nucleic acid
encodes the Gag polyprotein and the CasX protein with an intervening protease
cleavage site
between the two components, a second nucleic acid encodes the protease, and a
third nucleic
acid encodes the tropism factor, the gNA, and the Gag-Pol polyprotein. In
other embodiments, a
first nucleic acid encodes the Gag polyprotein, and the CasX protein with an
intervening
protease cleavage site between the two components, a second nucleic acid
encodes the protease,
a third nucleic acid encodes the tropism factor, and a fourth nucleic acid
encodes the gNA and
the Gag-Pol polyprotein. In some embodiments of the foregoing, the protease
and protease
cleavage sites are omitted.
1003231 In other embodiments, the XDP system is encoded by a portion or all of
a sequence
selected from the group consisting of the nucleic acid sequences of SEQ ID
NOs: 426-436, 784-
823, 828-873, 880-933, 947-1009 as set forth in Tables 16, 17, 19, 20, 22, 23,
24, 27, 30, 33, or
36, or a sequence having at least about 80%, at least about 90%, at least
about 95%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, at
least about 99%
sequence identity thereto_
1003241 In some embodiments, the nucleic acids encoding the XDP system of any
of the
embodiments described herein further comprises a donor template nucleic acid
wherein the
178
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
donor template comprises a sequence to be inserted into a target nucleic acid
to either correct a
mutation or to knock-down or knock-out a gene. In some embodiments, the donor
template
sequence comprises a non-homologous sequence flanked by two regions of
homology 5' and 3'
to the break sites of the target nucleic acid (i.e., homologous arms),
facilitating insertion of the
non-homologous sequence at the target region which can be mediated by HDR or
HITI. The
exogenous donor template inserted by HITI can be any length, for example, a
relatively short
sequence of between 1 and 50 nucleotides in length, or a longer sequence of
about 50-1000
nucleotides in length. The lack of homology can be, for example, having no
more than 20-50%
sequence identity and/or lacking in specific hybridization at low stringency.
In other cases, the
lack of homology can further include a criterion of having no more than 5, 6,
7, 8, or 9 bp
identity. In such cases, the use of homologous arms facilitates the insertion
of the non-
homologous sequence at the break site(s) introduced by the nuclease. In some
embodiments, the
donor template polynucleotide comprises at least about 10, at least about 50,
at least about 100,
or at least about 200, or at least about 300, or at least about 400, or at
least about 500, or at least
about 600, or at least about 700, or at least about 800, or at least about
900, or at least about
1000, or at least about 10,000, or at least about 15,000 nucleotides. In other
embodiments, the
donor template comprises at least about 10 to about 15,000 nucleotides, or at
least about 100 to
about 10,000 nucleotides, or at least about 400 to about 8,000 nucleotides, or
at least about 600
to about 5000 nucleotides, or at least about 1000 to about 2000 nucleotides.
The donor template
sequence may comprise certain sequence differences as compared to the genomic
sequence; e.g.,
restriction sites, nucleotide polymorphisms, selectable markers (e.g., drug
resistance genes,
fluorescent proteins, enzymes etc.), etc., which may be used to assess for
successful insertion of
the donor nucleic acid at the cleavage site or in some cases may be used for
other purposes (e.g.,
to signify expression at the targeted genomic locus). Alternatively, these
sequence differences
may include flanking recombination sequences such as FLPs, loxP sequences, or
the like, that
can be activated at a later time for removal of the marker sequence. In
another embodiment, the
donor template comprises a nucleic acid encoding at least a portion of a
target gene wherein the
donor template nucleic acid comprises all or a portion of the wild-type
sequence compared to the
target gene comprising a mutation, wherein the donor template is inserted into
the target nucleic
acid of the cell by HDR during the gene editing process. In such cases, upon
insertion into the
target nucleic acid, the target gene is corrected such that the functional
gene product can be
expressed. In some embodiments, the donor template ranges in size from 10-
10,000 nucleotides.
179
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
In other embodiments, the donor template ranges in size from 100-1,000
nucleotides. In some
embodiments, the donor template is a single-stranded DNA template or a single
stranded RNA
template. In other embodiments, the donor template is a double-stranded DNA
template. In
another embodiment of the XDP system, the donor template nucleic acid is
incorporated in the
first nucleic acid of the XDP system. In another embodiment of the XDP system,
the donor
template nucleic acid is incorporated in the second nucleic acid. In another
embodiment of the
XDP system, the donor template nucleic acid is incorporated in the third
nucleic acid. In another
embodiment of the XDP system, the donor template nucleic acid is incorporated
in the fourth or
a fifth nucleic acid.
[00325] In some embodiments, each of the individual nucleic acids are
incorporated into
plasmid vectors appropriate for transfection into a eukaryotic packaging cell,
examples of which
are detailed more fully, below, such that the XDP system will involve one,
two, three, four, or
five plasmids, as depicted in FIGS. 36-68. In each case, the nucleotide
sequence encoding the
components of the XDP system are operably linked to (under the control of)
regulatory elements
operable in a eukaryotic cell and appropriate for the component to be
expressed. Exemplary
regulatory elements include a transcription promoter (e.g., CMV, CMV+intron A,
SV40, RSV,
HIV-Ltr, MMLV-ltr, and metallothionein), a transcription enhancer element, a
transcription
termination signal, internal ribosome entry site (1RES) or p2A peptide to
permit translation of
multiple genes from a single transcript, polyadenylation sequences to promote
downstream
transcriptional termination, sequences for optimization of initiation of
translation, and translation
termination sequences. In some cases the promoter is a constitutive promoter,
such as a CMV
promoter, CAGG, PGK, U6 (for RNA pot III, which synthesizes shRNAs),
elongation factor 1
alpha (EF1-alpha), or Ill. In one embodiment, a constitutive promoter, such as
the human
cytomegalovirus immediate early (HCMV-1E) enhancer/promoter is used to
compensate for the
regulation of transcription normally provided by tat. In other cases, the
promoter can be an
inducible promoter such as, but are not limited to, T7 RNA polymerase
promoter, T3 RNA
polymerase promoter, isopropyl-beta-D-thiogalactopyranoside (IPTG)-regulated
promoter, heat
shock promoter, or tetracycline-regulated promoter (TRE), or a negative
inducible pLac
promoter. Any strong promoter known to those skilled in the art can be used
for driving the
expression of the nucleic acid. In the case of the nucleic acid encoding the
lentiviral packaging
components, the vector can be a psPax2 (detailed in the Examples, SEQ ID NO:
430) or
180
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
pMDLg/pRRE plasmid. In the case of the nucleic acid encoding the VSV-G
pseudotyping viral
envelope glycoprotein, the vector can be a pMD2.G plasmid.
1003261 The vectors of the embodiments may also comprise a polyadenylation
signal, which
may be downstream, for example, of the therapeutic payload, such as the CasX
sequence. The
polyadenylation signal may be a SV40 polyadenylation signal, LTR
polyadenylation signal,
bovine growth hormone (bGH) polyadenylation signal, human growth hormone (hGH)
polyadenylation signal, or human globin polyadenylation signal. The SV40
polyadenylation
signal may be a polyadenylation signal from a pCEP4 vector (Invitrogen, San
Diego, CA).
1003271 The vectors of the embodiments may also comprise an enhancer upstream
of the
therapeutic payload, such as the CasX sequence or gNA sequence. The enhancer
may be
necessary for DNA expression. The enhancer may be human actin, human myosin,
human
hemoglobin, human muscle creatine or a viral enhancer such as one from CMV,
HA, RSV, or
EBV. Polynucleotide function enhancers are described in U.S. Patent Nos.
5,593,972, 5,962,428,
and W094/016737, the contents of each are fully incorporated by reference. The
vector may also
comprise a mammalian origin of replication in order to maintain the vector
extrachromosomally
and produce multiple copies of the vector in a cell. The vector may also
comprise a regulatory
element, which may be well suited for gene expression in a mammalian or human
cell into
which the vector is administered. The vector may also comprise a reporter
gene, such as green
fluorescent protein ("GFP") and/or a selectable marker, such as hygromycin
("Hygro").
1003281 In embodiments involving the use of HIV-based vectors, the vectors can
include
additional sequences encoding factors or accessory proteins that assist in the
replication of viral
proteins. In one embodiment, the HIV-based vector comprises a sequence
encoding tat, a protein
involved in the activation of RNA Polymerase II, and that stimulates
transcription and
translation (Das, A., et at. The HIV-1 Tat Protein Has a Versatile Role in
Activating Viral
Transcription. J Virol. 85(18): 9506 (2011)). In another embodiment, the HIV-
based vector
comprises a sequence encoding Rev, an RNA binding protein that is critical in
the nuclear export
of intron-containing HIV-1 RNA (Pollard, V., et al. The HIV-1 Rev protein. Ann
Rev Microbiol.
52A91 (1998)). In another embodiment, the HIV-based vector comprises a
sequence encoding
viral infectivity factor (Vii), an accessory proteins essential for viral
replication that disrupts the
antiviral activity of the mammalian enzyme APOBEC by targeting it for
ubiquitination and
cellular degradation (Yang, G., et al. Viral infectivity factor: a novel
therapeutic strategy to
block HIV-1 replication. Minireviw Med Chem 13(7):1047 (2013)). In another
embodiment, the
181
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
HIV-based vector comprises a sequence encoding Viral protein U (Vpu), an
accessory protein
essential for suppressing the antiviral activity of host cell restriction
factors as well as the
efficient release of viral particles from infected cells (Gonzalez, M. Vpu
Protein: The Viroporin
Encoded by HIV-1. Viruses 7:4352 (2015). In another embodiment, the HIV-based
vector
comprises a sequence encoding Negative Factor (Nee, an accessory protein
essential for both
evading host adaptive cell-mediated immunity as well as enhancing infectivity
in the target cell
(Basmaciogullari, S., et at. The activity of Nef on HIV-1 infectivity.
Frontiers Microbiol 5:232
(2014). In another embodiment, the HIV-based vector comprises a sequence
encoding Viral
protein R (VpR), an accessory protein important for its interactions with a
number of cellular
proteins that impact viral replication in addition to a potential role in
restricting host anti-viral
pathways (Zhao, Richard Y, and Michael I Bukrinsky. HIV-1 accessory proteins:
VpR. Methods
Mol Biol 1087:125 (2014). In some embodiments, the HIV-based vector comprises
a sequence
encoding any combination of tat, Vii', Rev, Vpu, Nef, and VpR.
1003291 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding a
matrix polypeptide (MA), a capsid polypeptide (CA), a nucleocapsid polypeptide
(NC), a p1/p6
polypeptide and a CasX polypeptide. In some embodiments, the second nucleic
acid comprises,
from 5' to 3', MA, CA, pl/p6 operably linked, for example by a ribosomal
frameshift, to a
protease (PRO), a reverse transcriptase (RT) and an integrase (INT). In some
embodiments, the
third nucleic acid comprises a sequence encoding a glycoprotein, for example
VSV-G. In some
embodiments, the fourth nucleic acid comprises a sequence encoding a gNA.
1003301 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, a NC, pl/p6 and CasX. In some embodiments, the second nucleic acid
comprises, from 5'
to 3', MA, CA, NC, pl/p6, CasX and PRO. In some embodiments, the third nucleic
acid
comprises a sequence encoding a glycoprotein, for example VSV-G. In some
embodiments, the
fourth nucleic acid comprises a sequence encoding a gNA.
1003311 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding a
matrix polypeptide (MA), a capsid polypeptide (CA), a nucleocapsid polypeptide
(NC), a pl./p6
polypeptide and a CasX polypeptide. In some embodiments, the second nucleic
acid comprises,
from 5' to 3', MA, CA, NC, pl/p6 operably linked, for example by a ribosomal
frameshift, to a
182
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
CasX polypeptide, and PRO. In some embodiments, the third nucleic acid
comprises a sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the fourth
nucleic acid
comprises a sequence encoding a gNA.
1003321 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6 operably linked, for example by a ribosomal frameshift, to PRO,
and CasX. In
some embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence encoding a
gNA.
1003331 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6, CasX and PRO. In some embodiments, the second nucleic acid
comprises a
sequence encoding a glycoprotein, for example VSV-G. In some embodiments, the
third nucleic
acid comprises a sequence encoding a gNA.
1003341 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6, and CasX. In some embodiments, the second nucleic acid
comprises, from 5' to
3', sequences encoding MA, CA, NC, pl/p6, CasX and PRO. In some embodiments,
the third
nucleic acid comprises, from 5' to 3', sequence encoding MA, CA, NC and pl/p6.
In some
embodiments, the fourth nucleic acid comprises a sequence encoding a gNA.
[00335] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, pl/p6, and CasX. In some embodiments, the second nucleic acid
comprises a sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the third
nucleic acid
comprises a sequence encoding a gNAµ
[00336] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6, and CasX In some embodiments, the second nucleic acid
comprises, from 5' to
3', sequences encoding MA, CA, NC, and p1/p6. In some embodiments, the third
nucleic acid
comprises a sequence encoding a glycoprotein, for example VSV-G. In some
embodiments, the
fourth nucleic acid comprises a sequence encoding a gNA.
183
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1003371 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, and CasX. In some embodiments, the second nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the third
nucleic acid
comprises a sequence encoding a gNA.
1003381 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, pl and CasX. In some embodiments, the second nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the third
nucleic acid
comprises a sequence encoding a gNA,
1003391 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, CasX, and pl/p6 operably linked, for example by a ribosomal
frameshift, to PRO. In
some embodiments, the second nucleic acid comprises a sequence encoding a
,glycoprotein, for
example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence encoding a
gNA.
1003401 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, CasX, and pi/p6 operably linked, for example by a ribosomal frameshift, to
PRO. In some
embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence encoding a
gNA.
1003411 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CasX, and p1/p6 operably linked, for example by a ribosomal frameshift, to
PRO. In some
embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G In some embodiments, the third nucleic acid comprises a sequence
encoding a
gNA.
1003421 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CasX, and PRO. In some embodiments, the second nucleic acid comprises a
sequence encoding
184
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
a glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNAµ
1003431 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, CasX, and PRO. In some embodiments, the second nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the third
nucleic acid
comprises a sequence encoding a gNA.
[00344] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6, tev cleavage sequence (TCS), and CasX. In some embodiments, the
second
nucleic acid comprises, from 5' to 3', MA, CA, NC, p1/p6, TCS and a TEV
protease (TEV). In
some embodiments, the third nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the fourth nucleic acid comprises a
sequence encoding
a gNA.
1003451 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6, TCS, and CasX. In some embodiments, the second nucleic acid
comprises, from
5' to 3', MA, CA, NC, pl/p6, PreScission cleavage sequence (PCS) and a
PreScission protease
(PSP). In some embodiments, the third nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the fourth nucleic acid
comprises a
sequence encoding a gNA.
1003461 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6, TCS, and CasX. In some embodiments, the second nucleic acid
comprises, from
5' to 3', MA, CA, NC, pl/p6, PCS and a PreScission protease (PSP). In some
embodiments, the
third nucleic acid comprises a sequence encoding a glycoprotein, for example
VSV-G. In some
embodiments, the fourth nucleic acid comprises a sequence encoding a gNA.
[00347] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, pl/p6, PCS, and CasX. In some embodiments, the second nucleic acid
comprises, from
5' to 3', MA, CA, NC, p1/p6, PCS and PSP. In some embodiments, the third
nucleic acid
185
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
comprises a sequence encoding a glycoprotein, for example VSV-G. In some
embodiments, the
fourth nucleic acid comprises a sequence encoding a gNA.
1003481 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, p1/p6, PCS, and CasX. In some embodiments, the second nucleic acid
comprises, from
5' to 3', MA, CA, NC, pl/p6, PCS and TEV. In some embodiments, the third
nucleic acid
comprises a sequence encoding a glycoprotein, for example VSV-G. In some
embodiments, the
fourth nucleic acid comprises a sequence encoding a gNA.
1003491 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, and CasX. In some embodiments, the second nucleic acid comprises, from
5' to 3',
MA, CA, NC, and p1/p6. In some embodiments, the third nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the fourth
nucleic acid
comprises a sequence encoding a gNA.
1003501 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, P1 and CasX. In some embodiments, the second nucleic acid comprises,
from 5' to 3',
MA, CA, NC, and pl/p6. In some embodiments, the third nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the fourth
nucleic acid
comprises a sequence encoding a gNA.
1003511 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, CasX and P1/p6 operably linked, for example by a ribosomal frameshift,
to PRO. In
some embodiments, the second nucleic acid comprises, from 5' to 3', MA, CA,
NC, and p1/p6.
In some embodiments, the third nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the fourth nucleic acid comprises a
sequence encoding
a gNA.
1003521 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, CasX and Pl/p6 operably linked, for example by a ribosomal frameshift, to
PRO. In some
embodiments, the second nucleic acid comprises, from 5' to 3', MA, CA, NC, and
p1/p6. In
some embodiments, the third nucleic acid comprises a sequence encoding a
glycoprotein, for
186
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
example VSV-G. In some embodiments, the fourth nucleic acid comprises a
sequence encoding
a gNA.
1003531 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CasX, NC, and P1/p6 operably linked, for example by a ribosomal frameshift, to
PRO. In some
embodiments, the second nucleic acid comprises, from 5' to 3', MA, CA, NC, and
pl/p6. In
some embodiments, the third nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the fourth nucleic acid comprises a
sequence encoding
a gNA.
1003541 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CasX and P1/p6 operably linked, for example by a ribosomal frameshift, to PRO.
In some
embodiments, the second nucleic acid comprises, from 5' to 3', MA, CA, NC, and
pl/p6. In
some embodiments, the third nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the fourth nucleic acid comprises a
sequence encoding
a gNA.
1003551 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
NC, CasX and PRO. In some embodiments, the second nucleic acid comprises, from
5' to 3',
MA, CA, NC, and pl/p6. In some embodiments, the third nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the fourth
nucleic acid
comprises a sequence encoding a gNA.
1003561 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, CasX and PRO. In some embodiments, the second nucleic acid comprises, from
5' to 3',
MA, CA, NC, and pl/p6. In some embodiments, the third nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G In some embodiments, the fourth
nucleic acid
comprises a sequence encoding a gNA
1003571 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA
and CasX. In some embodiments, the second nucleic acid comprises, from 5' to
3', MA, CA,
NC, and p1/p6. In some embodiments, the third nucleic acid comprises a
sequence encoding a
187
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
glycoprotein, for example VSV-G. In some embodiments, the fourth nucleic acid
comprises a
sequence encoding a gNAµ
[00358] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, and CasX. In some embodiments, the second nucleic acid comprises, from 5'
to 3', MA,
CA, NC, and pl/p6. In some embodiments, the third nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the fourth
nucleic acid
comprises a sequence encoding a gNA.
[00359] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA
and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00360] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00361] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
the Alpharetrovirus gag polyprotein components P2A, P2B, and P10, as well as
CA, NC, PRO
and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00362] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
pp21/24, P12/P3/P8, CA, NC operably linked, for example by a ribosomal
frameshift, to PRO,
and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00363] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
188
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
NC operably linked, for example by a ribosomal frameshift, to PRO, and CasX.
In some
embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence encoding a
gNA.
1003641 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
p20, CA, NC, PRO, and CasX. In some embodiments, the second nucleic acid
comprises a
sequence encoding a glycoprotein, for example VSV-G. In some embodiments, the
third nucleic
acid comprises a sequence encoding a gNA.
1003651 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
pp12, CA, NC, PRO, and CasX. In some embodiments, the second nucleic acid
comprises a
sequence encoding a glycoprotein, for example VSV-G. In some embodiments, the
third nucleic
acid comprises a sequence encoding a gNA.
1003661 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA, NC, P6 operably linked, for example by a ribosomal frameshift, to PRO, and
CasX. In some
embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence encoding a
gNA.
[00367] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding p68-
Gag operably linked, for example by a ribosomal frameshift, to PRO, and CasX.
In some
embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence encoding a
gNA.
[00368] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
P2A, P2B, P10, CA and CasX. In some embodiments, the second nucleic acid
comprises a
sequence encoding a glycoprotein, for example VSV-G. In some embodiments, the
third nucleic
acid comprises a sequence encoding a gNA.
189
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00369] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
P2A, P213, P10, CA and CasX. In some embodiments the second nucleic acid
comprises, from 5'
to 3', MA, P2A, P2B, P10, CA, NC, PRO and CasX. In some embodiments, the third
nucleic
acid comprises a sequence encoding a glycoprotein, for example VSV-G. In some
embodiments,
the fourth nucleic acid comprises a sequence encoding a gNA.
[00370] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
pp21/24, P12/P3/P8, CA and CasX. In some embodiments, the second nucleic acid
comprises a
sequence encoding a glycoprotein, for example VSV-G. In some embodiments, the
third nucleic
acid comprises a sequence encoding a gNA.
[00371] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
pp21/24, P12/P3/P8, CA and CasX. In some embodiments the second nucleic acid
comprises,
from 5' to 3', MA, pp21/24, P12/P3/P8, CA, NC operably linked, for example by
a ribosomal
frameshift, to PRO and CasX. In some embodiments, the third nucleic acid
comprises a
sequence encoding a glycoprotein, for example VSV-G. In some embodiments, the
fourth
nucleic acid comprises a sequence encoding a gNA.
[00372] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00373] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA and CasX. In some embodiments, the second nucleic acid comprises, from 5'
to 3',
sequences encoding MA, CA, NC operably linked, for example by a ribosomal
frameshift, to
PRO and CasX. In some embodiments, the third nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the fourth nucleic acid
comprises a
sequence encoding a gNA.
[00374] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
190
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
p20, CA and CasX. In some embodiments, the second nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the third
nucleic acid
comprises a sequence encoding a gNA.
1003751 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
p20, CA and CasX. In some embodiments, the second nucleic acid comprises, from
5' to 3',
sequences encoding MA, p20, CA, NC operably linked, for example by a ribosomal
frameshift,
to PRO and CasX. in some embodiments, the third nucleic acid comprises a
sequence encoding
a glycoprotein, for example VSV-G. In some embodiments, the fourth nucleic
acid comprises a
sequence encoding a gNA.
1003761 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
pp12, CA and CasX, In some embodiments, the second nucleic acid comprises a
sequence
encoding a glycoprotein, for example VSV-G. In some embodiments, the third
nucleic acid
comprises a sequence encoding a gNA.
1003771 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
pp12, CA and CasX. In some embodiments, the second nucleic acid comprises,
from 5' to 3',
sequences encoding MA, pp12, CA, NC, PRO and CasX. In some embodiments, the
third
nucleic acid comprises a sequence encoding a glycoprotein, for example VSV-G.
In some
embodiments, the fourth nucleic acid comprises a sequence encoding a gNA.
1003781 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
1003791 In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding MA,
CA and CasX. In some embodiments, the second nucleic acid comprises, from 5'
to 3',
sequences encoding MA, CA, NC, P6 operably linked, for example by a ribosomal
frameshift, to
PRO and CasX. In some embodiments, the third nucleic acid comprises a sequence
encoding a
191
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
glycoprotein, for example VSV-G. In some embodiments, the fourth nucleic acid
comprises a
sequence encoding a gNAµ
[00380] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', p68-Gag,
p3-Gag and
CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00381] In some embodiments, the XDP system of the disclosure comprises four
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3',
sequences encoding p68-
Gag, p3-Gag and CasX. In some embodiments, the second nucleic acid comprises,
from 5' to 3',
sequences encoding p68-Gag, p3-Gag operably linked, for example by a ribosomal
frameshift, to
PRO and CasX. In some embodiments, the third nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the fourth nucleic acid
comprises a
sequence encoding a gNA.
[00382] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, P2A,
P2B, P10, CA,
NC and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00383] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, CA,
NC and CasX. In
some embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein, for
example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence encoding a
gNA.
[00384] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, CA,
NC, p6 and
CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00385] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA,
pp21/24, P12/P3/P8,
CA, NC and CasX. In some embodiments, the second nucleic acid comprises a
sequence
192
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
encoding a glycoprotein, for example VSV-G. In some embodiments, the third
nucleic acid
comprises a sequence encoding a gNAµ
[00386] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA,
pp12, CA, NC and
CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00387] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, p20,
CA, NC and
CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00388] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, CA,
pl/p6 and CasX.
In some embodiments, the second nucleic acid comprises a sequence encoding a
glycoprotein,
for example VSV-G. In some embodiments, the third nucleic acid comprises a
sequence
encoding a gNA.
[00389] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, CA,
NC, pl/p6, p1/p6
and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
[00390] In some embodiments, the XDP system of the disclosure comprises three
nucleic acids.
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, CA,
NC, CasX and
pl/p6. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G. In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA
1003911 In some embodiments, the XDP system of the disclosure comprises three
nucleic acids
In some embodiments, the first nucleic acid comprises, from 5' to 3', MA, CA,
NC, P2, pl/p6
and CasX. In some embodiments, the second nucleic acid comprises a sequence
encoding a
glycoprotein, for example VSV-G In some embodiments, the third nucleic acid
comprises a
sequence encoding a gNA.
193
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00392] In any of the foregoing, any of the components may be separated by
sequences
encoding protease cleavage sites, self-cleaving polypeptides, or internal
ribosome entry sites, or
any combination thereof
V. XDP Packaging Cells
1003931 In another aspect, the present disclosure relates to packaging cells
utilized in the
production of XDP. As used herein, the term "packaging cell" is used in
reference to cell lines
that do not contain a packaging signal, but do stably or transiently express
viral structural
proteins and replication enzymes (e.g., Gag, pot, etc.) which are necessary or
useful for the
correct packaging of XDP particles. In the embodiments, the cell line can be
any cell line
suitable for the production of XDP, including primary ex vivo cultured cells
(from an individual
organism) as well as established cell lines. Cell types may include bacterial
cells, yeast cells,
and mammalian cells. Exemplary bacterial cell types may include E co/i.
Exemplary yeast cell
types may include Saccharomyces cerevisiae. Also suitable for use as packaging
cells are insect
cell lines, such as Spodoptera ft-up-perdu sf9 cells. Exemplary mammalian cell
types may
include mouse, hamster, and human primary cells, as we as cell lines such as
human embryonic
kidney 293 (HEK293) cells, Lenti-X 293T cells, baby hamster kidney (BHK)
cells, HepG2 cells,
Saos-2 cells, HuH7 cells, NSO cells, SP2/0 cells, YO myeloma cells, A549
cells, P3X63 mouse
myeloma cells, PER cells, PER.C6 cells, hybridoma cells, VERO cells, N1H3T3
cells, COS
cells, WI38 cells, MRCS cells, A549 cells, HeLa cells, Chinese hamster ovary
(CHO) cells, or
HT1080 cells. The choice of the appropriate vector for the cell type will be
readily apparent to
the person of ordinary skill in the art. In some embodiments, the eukaryotic
cell is modified by
one or more mutations one or more mutations to reduce expression of a cell
surface marker that
could be incorporated into the XDP. Such markers can include receptors or
proteins capable of
being bound by MEC receptors or that would otherwise trigger an immune
response in a subject.
1003941 In the embodiments of the XDP system, vectors are introduced into the
packaging cell
that encode the particular therapeutic payload (e.g., a CasKgNA designed for
editing target
nucleic acid), as well as the other viral-derived structural components,
detailed above, (e.g., the
Gag polyprotein, the pot polyprotein, the tropism factor, and, optionally, the
donor template
nucleic acid sequence). The vectors can remain as extra-chromosomal elements
or some or all
can be integrated into the host cell chromosomal DNA to create a stably-
transformed packaging
cell.
194
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00395] In some embodiments, the vectors comprising the nucleic acids of the
XDP system are
introduced into the cell via transfection, transduction, lipofection or
electroporation to generate a
packaging cell line. The introduction of the vectors can use one or more of
the commercially
available TransMessenger reagents from Qiagen, Stemfect RNA Transfection Kit
from
Stemgent, and TransIT-mRNA Transfection Kit from Minis ho LLC, Lanza
nucleofection,
Maxagen electroporation and the like. Methods for transfection, transduction
or infection are
well known to those of skill in the art.
[00396] In some cases, the packaging vectors are introduced into the cells
together with a
dominant selectable marker, such as neo, DHFR, Gin synthetase or ADA, followed
by selection
in the presence of the appropriate drug and isolation of clones. A selectable
marker gene can be
linked physically to genes encoding by the packaging vector.
[00397] Assembly and release of XDP with the encapsidated therapeutic payload
from the
transfected host cell can be mediated by the viral structural protein, Gag.
Human
immunodeficiency virus type 1 (HIV-1) Gag is synthesized as a precursor
polyprotein, Pr55gag.
This polyprotein is comprised of four major structural domains, which are
cleaved by the viral
protease into p17 matrix (MA), p24 capsid (CA), p7 nucleocapsid (NC), and p6,
during or
immediately after the budding process (Adamson CS., and Freed EO. Human
immunodeficiency
virus type 1 assembly, release, and maturation. Adv. Phannacol. 55:347
(2007)). Utilizing an
HIV-1 system, it is sufficient to express the p55 Gag protein to allow the
efficient production of
XDPs from cells (Gheysen et al., Assembly and release of HTV-1 precursor
Pr55Gag virus-like
particles from recombinant baculovirus-infected insect cells. Cell. 59(1):103
(1989)). In the
context of the uncleaved Pr55Gag, MA constitutes the N-terminal domain of the
Gag protein and
is essential for membrane binding and localization of the Gag precursor to the
plasma
membrane. CA and NC domains promote Gag multimerization through direct protein-
protein
interactions and indirect RNA-mediated interactions, respectively. Inclusion
of the late domain
motif within p6 can promote release of XDP particles from the cell surface.
Upon expression,
the Gag polypeptide is targeted to the cell membrane and incorporated in the
XDP during
membrane budding. During or shortly after virus budding from the host cell,
the 11IV-1 protease
cleaves Pr5 stag into the mature Gag proteins p17 matrix (MA), p24 capsid
(CA), p7
nucleocapsid (NC), and p6. The proteolytic processing of Gag results in a
major transformation
in XDP structure: MA remains associated with the inner face of the viral
membrane, whereas
CA condenses to form a shell around the NC complex (if incorporated). This
rearrangement
195
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
produces a morphological transition to a particle with a conical core
characteristic similar to an
infectious virion.
[00398] It has been discovered that components derived, in part, from
retroviruses can be
utilized to create XDP within packaging cells for delivery of the therapeutic
payload to the target
cells. In one embodiment, the packaging cell transformed with the XDP system
plasmids
produce XDP that facilitate delivery of the encapsidated RNP of a CasX:gNA
system to cells to
effect editing of target nucleic acid.
VI. XDP Expression Systems and Methods of Producing XDP
[00399] In another aspect, the present disclosure provides a recombinant
expression system for
use in the production of XDP in a selected host packaging cell, comprising an
expression
cassette comprising the nucleic acids of the XDP system described herein
operably linked to
regulatory elements compatible with expression in the selected host cell. The
expression
cassettes may be included on one or more vectors as described herein and in
the Examples, and
may use the same or different promoters. Exemplary regulatory elements include
a transcription
promoter such as, but not limited to, CMV, CMV+intron A, SV40, RSV, HIV-Ltr,
elongation
factor 1 alpha (EF1a), MMLV-Itr, internal ribosome entry site ORES) or p2A
peptide to permit
translation of multiple genes from a single transcript, metallothionein, a
transcription enhancer
element, a transcription termination signal, polyadenylation sequences,
sequences for
optimization of initiation of translation, and translation termination
sequences. It will be
understood that the choice of the appropriate control element will depend on
the encoded
component to be expressed (e.g., protein or RNA) or whether the nucleic acid
comprises
multiple components that require different polymerases or are not intended to
be expressed as a
fusion protein.
[00400] In some embodiments, the present disclosure provides methods of making
an XDP
comprising a therapeutic payload (e.g., an RNP of a CasX protein and a gNA),
the method
comprising propagating the packaging cell of the embodiments described herein
comprising the
expression cassettes or the integrated nucleic acids encoding the XDP systems
of any one of the
embodiments described herein under conditions such that XDPs are produced with
the
encapsidated therapeutic payload, followed by harvesting the XDPs produced by
the packaging
cell, as described below or in the Examples. In some embodiments, the
packaging cell produces
196
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
XDP comprising RNP of a CasX and gNA and, optionally, a donor template for the
editing of
the target nucleic acid by HDR.
1004011 The packaging cell can be, for example, a mammalian cell (e.g., HEK293
cells, Lenti-
X 293T cells, 131-IK cells, HepG2 cells, Saos-2 cells, HuH7 cells, NSO cells,
SP2/0 cells, YO
myeloma cells, A549 cells, P3X63 mouse myeloma cells, PER cells, PER.C6 cells,
hybridoma
cells, VERO cells, NIH3T3 cells, COS cells, WI38 cells, MRCS cells, A549
cells, HeLa cells,
CHO cells, and HT1080 cells), an insect cell (e.g., Trichoplusia ni (Tn5) or
Sf9), a bacterial cell,
a plant cell, a yeast cell, an antigen presenting cell (e.g., primary,
immortalized or tumor-derived
lymphoid cells such as macrophages, monocytes, dendritic cells, B-cells, T-
cells, stem cells, and
progenitor cells thereof). Packaging cells can be transfected by conventional
methods, including
electroporation, use of cationic polymers, calcium phosphate, virus-mediated
transfection,
transduction, or lipofection. In some embodiments, the packaging cell can be
modified to reduce
or eliminate cell surface markers or receptors that would otherwise be
incorporated into the
XDP, thereby reducing an immune response to the cell surface markers or
receptors by the
subject receiving an administration of the XDP.
1004021 The introduction of the vectors into the packaging cell can use one or
more of the
commercially available TransMessenger reagents from Qiagen, Stemfect RNA
Transfection Kit
from Stemgent, and TransIT-mRNA Transfection Kit from Minis Bio LLC, Lonza
nucleofection, Maxagen electroporation and the like. Methods for transfection,
transduction or
infection are well known to those of skill in the art.
[00403] In one embodiment, XDP are produced by the incubation of the
transfected packaging
cells in appropriate growth medium for 48 to 96 hours and are collected by
filtration of the
growth medium through a 0.45 micron filter. In some cases, the XDP can be
further
concentrated by centrifugation in a 10% or a 10-30% density gradient sucrose
buffer. In other
cases, the XDP can be concentrated by column chromatography, such as by use of
an ion-
exchange resin or a size exclusion resin.
Applications
[00404] The XDP systems comprising CasX proteins and guides provided herein
are useful in
methods for modifying target nucleic acids in cells. In the XDP systems of
modifying a target
nucleic acid, the method utilizes any of the embodiments of the CasX:gNA
systems described
herein, and optionally includes a donor template embodiment described herein.
In some cases,
197
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
the method knocks-down the expression of a mutant protein in cells comprising
the target
nucleic. In other cases, the method knocks-out the expression of the mutant
protein. In still
other cases, the method results in the correction of the mutation in the
target nucleic acid,
resulting in the expression of fiinctional protein.
1004051 In some embodiments, the method comprises contacting the cells
comprising the target
nucleic acid with an effective dose of XDPs comprising RNPs of CasX protein
and a guide
nucleic acid (gNA) comprising a targeting sequence complementary to the target
nucleic acid,
wherein said contacting results in modification of the target nucleic acid by
the CasX protein. In
another embodiment, the XDP further comprises a donor template wherein the
contacting of the
cell with the XDP results in insertion of the donor template into the target
nucleic acid sequence.
In some cases the donor template is used in conjunction with the RNP to
correct a mutation in
the target nucleic acid gene, while in other cases the donor template is used
to insert a mutation
to knock-down or knock-out expression of the expression product of the target
nucleic acid gene.
1004061 In some embodiments, the method of modifying a target nucleic acid in
a cell
comprises contacting the cells comprising the target nucleic acid with an
effective dose of XDPs
wherein the cell is modified in vitro or ex vivo.
1004071 In other embodiments of the method of modifying a target nucleic acid
in a cell, the
cells are modified in vivo, wherein a therapeutically-effective dose of the
XDP is administered to
a subject. The method has the advantage over viral delivery systems in that
the RNP are
comparatively short-lived relative to the nucleic acids delivered in viral
systems such as AAV. A
further advantage of the XDP system is the ability to match the system to
specific cell types by
manipulating the tropism of the XDP. In some embodiments, the half-life of the
delivered RNP
is about 24h, or about 48h, or about 72h, or about 96h, or about 120h, or
about 1 week. By the
methods of treatment, the administration of the XDP results in the improvement
of one, two, or
more symptoms, clinical parameters or endpoints associated with the disease in
the subject.
1004081 In some embodiments, the subject administered the XDP is selected from
the group
consisting of mouse, rat, pig, non-human primate, and human. In a particular
embodiment, the
subject is a human. In one embodiment of the method, the XDP is administered
to the subject at
a dose of at least about 1 x 105 XDP particles/kg, or at least about 1 x 106
particles/kg, or at least
about 1 x 107 particles/kg, or at least about 1 x 108 particles/kg, or at
least about 1 x 109
particles/kg, or at least about 1 x 1010 particles/kg, or at least about 1 x
1011 particles/kg, or at
least about 1 x 1012 particles/kg, or at least about 1 x 10" particles/kg, or
at least about 1 x 10"
198
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
particles/kg, or at least about 1 x 1015 particles/kg, or at least about 1 x
1016 particles/kg. In other
embodiments, the VLP is administered to the subject at a dose of at least
about 1 x 105
particles/kg to at least about 1 x 10' particles/kg. In another embodiment,
the VLP is
administered to the subject at a dose of at least about 1 x 105 particles/kg
to about 1 x 1016
particles/kg, or at least about 1 x 106 particles/kg to about 1 x 1015
particles/kg, or at least about
1 x 107 particles/kg to about 1 x 1014 particles/kg. In other embodiments, the
VLP is
administered to the subject at a dose of at least about 1 x 105 particles/kg
to at least about 1 x
1016 particles/kg. In one embodiment, the XDP is administered by a route of
administration
selected from the group consisting of subcutaneous, intradermal, intraneural,
intranodal,
intramedullary, intramuscular, intravenous, intra-arterial,
intracerebroventricular, intracisternal,
intrathecal, intracra.nial, intralumbar, intratracheal, intraosseous,
inhalatory, intracontralateral
striatum, intraocular, intravitreal, intralymphatical, intraperitoneal routes
and sub-retinal routes.
[00409] In another embodiment, the disclosure provides a method of treatment
of a subject
having a disease according to a treatment regimen comprising one or more
consecutive doses
using a therapeutically effective dose of an XDP of any of the embodiments
described herein. In
one embodiment of the treatment regimen, the therapeutically effective dose is
administered as a
single dose. In another embodiment of the treatment regimen, the
therapeutically effective dose
is administered to the subject as two or more doses over a period of at least
two weeks, or at
least one month, or at least two months, or at least three months, or at least
four months, or at
least five months, or at least six months, or once a year, or every 2 or 3
years.
VEIL Kits and Articles of Manufacture
[00410] In another aspect, provided herein are kits comprising the
compositions of the
embodiments described herein. In some embodiments, the kit comprises an XDP
comprising a
therapeutic payload of any of the embodiment described herein, an excipient
and a suitable
container (for example a tube, vial or plate). In a particular embodiment, the
therapeutic payload
is an RNP of a CasX and a gNA.
[00411] In some embodiments, the kit further comprises a buffer, a nuclease
inhibitor, a
protease inhibitor, a liposome, a therapeutic agent, a label, a label
visualization reagent, or any
combination of the foregoing. In some embodiments, the kit further comprises a
pharmaceutically acceptable carrier, diluent or excipient. In some
embodiments, the kit further
comprises instructions for use.
199
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
IX. Exemplary Embodiments
1004121 The following exemplary embodiments, are provided by way of example
only.
1004131 In some embodiments, the XDP system comprises an editing efficiency of
at least 75%,
at least 80%, at least 85%, at least 87%, at least 90% or at least 91% as per
the editing assay
dilution in Table 25, or at least 70%, at least 75%, at least 80% or at least
85% as per the editing
assay dilution of Table 26_ In some embodiments, the XDP system comprises
version 44,
encoded by plasmid pXDP40 (SEQ ID NO: 882) as described in Table 24. In some
embodiments, the XDP system comprises a VSV glycoprotein as encoded by pGP2,
and an
sgRNA.
1004141 In some embodiments, the XDP system comprises an editing efficiency of
at least 25%,
at least 30%, at least 35% or at least 37% as per the editing assay dilution
in Table 25 or at least
5%, at least 10% or at least 13% as per the editing assay dilution of Table
26. In some
embodiments, the XDP system comprises version 63, encoded by plasmid pXDP62
(SEQ ID
NO: 904) as described in Table 24. In some embodiments, the XDP system
comprises a VSV
glycoprotein as encoded by pGP2, and an sgRNA.
1004151 In some embodiments, the XDP system comprises an editing efficiency of
at least 60%,
at least 65%, at least 70%, at least 75% or at least 77% as per the editing
assay dilution in Table
28, or at least 20%, at least 25%, at least 30% or at least 32% as per the
editing assay dilution of
Table 29. In some embodiments, the XDP system comprises version 74a, encoded
by plasmid
pXDP72 (SEQ ID NO:917) as described in Table 27. In some embodiments, the XDP
system
comprises a VSV glycoprotein as encoded by pGP2, and an sgRNA
1004161 In some embodiments, the XDP system comprises an editing efficiency of
at least at
least 50%, at least 55%, at least 60%, at least 65% or at least 67% as per the
editing assay
dilution in Table 28, or at least 25%, at least 30%, at least 35% or at least
38% as per the editing
assay dilution of Table 29_ In some embodiments, the XDP system comprises
version 75a,
encoded by plasmid pXDP73 (SEQ ID NO=918) as described in Table 27. In some
embodiments, the XDP system comprises a VSV glycoprotein as encoded by pGP2,
and an
sgRNA.
1004171 In some embodiments, the XDP system comprises an editing efficiency of
at least 75%,
at least 80%, at least 85%, at least 87%, at least 90% or at least 91% as per
the editing assay
dilution in Table 31, or at least 70%, at least 75%, at least 80% or at least
85% as per the editing
200
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
assay dilution of Table 32. In some embodiments, the XDP system comprises
version 44,
encoded by plasmid pXDP40 (SEQ ID NO: 949) as described in Table 30. In some
embodiments, the XDP system comprises a VSV glycoprotein as encoded by pGP2,
and an
sgRNA.
1004181 In some embodiments, the XDP system comprises an editing efficiency of
at least 25%,
at least 30%, at least 35% or at least 37% as per the editing assay dilution
in Table 31 or at least
5%, at least 10% or at least 13% as per the editing assay dilution of Table
32. In some
embodiments, the XDP system comprises version 63, encoded by plasmid pXDP62
(SEQ ID
NO: 971) as described in Table 30. In some embodiments, the XDP system
comprises a VSV
glycoprotein as encoded by pGP2, and an sgRNA.
1004191 In some embodiments, the XDP system comprises an editing efficiency of
at least 75%,
at least 80%, at least 85%, at least 87%, at least 90% or at least 94% as per
the editing assay
dilution in Table 34 or at least 75%, at least 80%, at least 85%, at least
87%, at least 90% or at
least 95% as per the editing assay dilution of Table 35. In some embodiments,
the XDP system
comprises version 102, encoded by plasmid pXDP127 (SEQ ID NO: 976) as
described in Table
33. In some embodiments, the XDP system comprises a VSV glycoprotein as
encoded by pGP2,
and an sgRNA.
[00420] In some embodiments, the XDP system comprises an editing efficiency of
at least 70%,
at least 75%, at least 80% or at least 84% as per the editing assay dilution
in Table 34 or at least
70%, at least 75%, or at least 800/u as per the editing assay dilution of
Table 35. In some
embodiments, the XDP system comprises version 7, encoded by plasmid pXDP0017.
In some
embodiments, the XDP system comprises a VSV glycoprotein as encoded by pGP2,
and an
sgRNA.
[00421] In some embodiments, the XDP system comprises an editing efficiency of
at least at
least 25%, at least 25%, at least 30% or at least 33% as per the editing assay
dilution in Table 37
or at least 1.8 % as per the editing assay dilution of Table 38. In some
embodiments, the XDP
system comprises version 6613, encoded by plasmid pXDP78 + pXDP54. In some
embodiments,
the XDP system comprises a VSV glycoprotein as encoded by pGP2, and an sgRNA
[00422] In some embodiments, the XDP system comprises an editing efficiency of
at least 10%,
at least 15%, at least 20% or at least 21% as per the editing assay dilution
in Table 37 or at least
5%, at least 7% or at least 9% as per the editing assay dilution of Table 38.
In some
embodiments, the XDP system comprises version 87B, encoded by plasmids pXDP83
+
201
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
pXDP59, In some embodiments, the XDP system comprises a VSV glycoprotein as
encoded by
pGP2, and an sgRNA.
[00423] Editing efficiency may be measured by any known method or assay in the
art. A person
of skill in the art would know how to identify and use such assays. In some
embodiments, the
editing efficiency may be measured as %TDT positive cells, for example as
shown in FIG. 69-
70.
[00424] In some embodiments, an XDP system comprises one or more plasmids or
elements in
an arrangement resulting in an increased editing efficiency compared an XDP
system not
comprising said arrangement. In some embodiments, the XDP system may have an
increased
editing efficiency of at least 1%, 3%, 5%, 7%, 10%, 15%, 20%, 25%, 30%, 35%,
40%, 45%,
50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 99% or 100% compared to an
XDP
system not comprising the same elements and/or arrangement.
[00425] In some embodiments, an XDP system may be derived from
Alpharetroviruses (avian
leukosis virus (ALV) and rous sarcoma virus (RSV)), and encoded by the three
plasmids
encoding the Gag-protease-CasX, the glycoprotein (VSV-G), and the guide RNA
(sgRNA). The
elements of the structural plasmid may be arranged as: MA, P2A, P28, P10, CA,
NC, Pro and
CasX (FIG. 52A). In an exemplary embodiment, the XDP system version 44
comprises elements
of a structural plasmid arranged as: MA, P2A, P2B, P10, CA, NC, Pro and CasX
(FIG 52A),
wherein version 44 has an increased editing efficiency of at least 1%, 3%, 5%,
7%, 10%, 15%,
20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%,
95%,
99% or 100% compared to an XDP not comprising the same elements and/or
arrangement.
[00426] In some embodiments, an XDP system may be encoded by the three
plasmids as shown
in FIG. 53A. The elements of the structural plasmid may be arranged as: MA,
CA, NC, Pro and
CasX. In an exemplary embodiment, the XDP system version 63 comprises elements
of a
structural plasmid arranged as: MA, CA, NC, Pro and CasX, wherein version 63
has an
increased editing efficiency of at least 1%, 3%, 5%, 7%, 10%, 15%, 20%, 25%,
30%, 35%, 40%,
45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 99% or 100% compared to
an
XDP not comprising the same elements and/or arrangement.
[00427] In some embodiments, an XDP system may be derived from
Gammaretroviruses (FLV
and MMLV), and encoded by the three plasmids as shown in FIG. 5911. The
elements of the
structural plasmid may be arranged as: MA, pp12, CA, and CasX. In an exemplary
embodiment,
the XDP system version 74a comprises elements of a structural plasmid arranged
as: MA, pp12,
202
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
CA, and CasX, wherein version 74a has an increased editing efficiency of at
least 1%, 3%, 5%,
7%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%,
85%, 90%, 95%, 99% or 100% compared to an XDP not comprising the same elements
and/or
arrangement.
[00428] In some embodiments, an XDP system may be derived from
Alpharetroviruses (avian
leukosis virus (ALV) and rous sarcoma virus (RSV) and encoded by the three
plasmids as shown
in FIG. 62B. The elements of the structural plasmid may be arranged as: MA,
P2A, P2B, P10,
CA, NC, and CasX. In an exemplary embodiment, the XDP system version 102
comprises
elements of a structural plasmid arranged as: MA, P2A, P2B, P10, CA, NC, and
CasX, wherein
version 102 has an increased editing efficiency of at least 1%, 3%, 5%, 7%,
10%, 15%, 20%,
25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 99%
or
100% compared to an XDP not comprising the same elements and/or arrangement.
[00429] In some embodiments, an XDP system may be encoded by three plasmids as
shown in
FIG. 39A. The elements of the structural plasmid may be arranged as: MA, CA,
NC, p1/p6, and
CasX. In an exemplary embodiment, the XDP system version 7 comprises elements
of a
structural plasmid arranged as: MA, CA, NC, pl/p6, and CasX, wherein version 7
has an
increased editing efficiency of at least 1%, 3%, 5%, 7%, 10%, 15%, 20%, 25%,
30%, 35%, 40%,
45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 99% or 100% compared to
an
XDP not comprising the same elements and/or arrangement.
[00430] In some embodiments, an XDP system may be encoded by the four plasmids
as shown
in FIG. 56A. The elements of structural plasmid 1 may be arranged as: MA, P2A,
P2B, P10, CA,
and CasX, and elements of structural plasmid 2 may be arranged as: MA, P2A,
P28, P10, CA,
NC, Pro, and CasX. In an exemplary embodiment, the XDP system version 668
comprises
elements of a structural plasmid 1 arranged as: MA, P2A, P2B, P10, CA, and
CasX, and
elements of structural plasmid 2 arranged as: MA, P2A, P2B, P10, CA, NC, Pro,
and CasX,
wherein version 66B has an increased editing efficiency of at least 1%, 3%,
5%, 7%, 10%, 15%,
20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%,
95%,
99% or 100% compared to an XDP not comprising the same elements and/or
arrangement.
[00431] In some embodiments, an XDP system may be encoded by the four plasmids
as shown
in FIG. 57A. The elements of structural plasmid 1 may be arranged as: MA,
pp21/24,
P12/P3/P8, CA, and CasX, and elements of structural plasmid 2 may be arranged
as: MA,
pp21/24, P12/P3/P8, CA, NC, Pro, and CasX. In an exemplary embodiment, the XDP
system
203
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
version 87B comprises elements of a structural plasmid larranged as: MA,
pp21/24, P12/P3/P8,
CA, and CasX, and elements of structural plasmid 2 arranged as: MA, pp21/24,
P12/P3/P8, CA,
NC, Pro, and CasX, wherein version 87B has an increased editing efficiency of
at least 1%, 3%,
5%, 7%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%,
80%,
85%, 90%, 95%, 99% or 100% compared to an XDP not comprising the same elements
and/or
arrangement.
[00432] The XDP systems disclosed herein may be derived from the Retroviridae
virus family,
including Oihoretrovirinae (Lentivirus, Alpharetrovirus, Betaretrovirus,
Deltaretrovirus,
Epsilonretrovirus, Gammaretrovirus), and Spurnaretrovirinae. Exemplary XDP
system versions
and their corresponding virus are shown in Tables 25, 26, 28, 29, 31, 32, 34,
35, 37 and 38.
X. Enumerated Embodiments
[00433] The invention may be defined by reference to the following sets of
enumerated,
illustrative embodiments:
Set I
[00434] Embodiment I-1. A CasX delivery particle (CasX XDP) system comprising:
a. a first nucleic acid comprising a sequence encoding a
fusion polypeptide that comprises:
i) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC);
ii) a CasX protein; and
iii) a protease cleavage site between the gag polyprotein and the CasX
protein;
b. a second nucleic acid comprising a sequence encoding a
guide RNA;
c. a third nucleic acid comprising a sequence encoding a
fusion polypeptide that comprises:
i) a gag polyprotein; and
ii) a pol polyprotein comprising at least a protease capable of cleaving
the protease
cleavage site between the CasX protein and the gag polyprotein; and
d. a fourth nucleic acid, comprising a sequence encoding a
pseudotyping viral envelope
glycoprotein or an antibody fragment that provides for binding and fusion of
the XDP to a target
cell.
[00435] Embodiment 1-2. A CasX delivery particle (CasX XDP) system comprising:
a. a first nucleic acid comprising a sequence encoding a
fusion polypeptide that comprises:
204
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
i) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC);
ii) a CasX protein;
iii) a protease cleavage site between the gag polyprotein and the CasX
protein; and
iv) a protease capable of cleaving the protease cleavage site between the
CasX
protein and the gag polyprotein;
b. a second nucleic acid comprising a sequence encoding a guide RNA; and
c. a third nucleic acid, comprising a sequence encoding a pseudotyping
viral envelope
glycoprotein or antibody fragment that provides for binding and fusion of the
XDP to a target
cell.
1004361 Embodiment 1-3. A CasX delivery particle (CasX XDP) system comprising:
a. a first nucleic acid comprising a sequence encoding a
fusion polypeptide that comprises:
i) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC);
ii) a CasX protein; and
iii) a protease cleavage site between the gag polyprotein and the CasX
protein;
b. a second nucleic acid comprising a sequence encoding a
guide RNA;
c. a third nucleic acid comprising a sequence encoding a
protease capable of cleaving the
protease cleavage site between the CasX protein and the gag polyprotein; and
d. a fourth nucleic acid, comprising a sequence encoding a
pseudotyping viral envelope
glycoprotein or antibody fragment that provides for binding and fusion of the
XDP to a target
cell.
1004371 Embodiment 1-4. A CasX delivery particle (CasX XDP) system comprising:
a. a first nucleic acid comprising a sequence encoding
i) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC); and
ii) a chimeric RNA comprising a guide RNA and a retroviral Psi packaging
element
inserted into the guide RNA;
b. a second nucleic acid comprising a sequence encoding a Cas X protein;
and
c. a third nucleic acid, comprising a sequence encoding a pseudotyping
viral
envelope glycoprotein or antibody fragment that provides for binding and
fusion of the XDP to a
target cell.
205
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1004381 Embodiment 1-5. A CasX delivery particle (CasX XDP) system comprising:
a. a first nucleic acid comprising a sequence encoding:
i) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC);
ii) an RNA binding domain protein; and
iii) an optional protease cleavage site between the gag polyprotein and the
RNA
binding domain protein;
b. a second nucleic acid comprising a sequence encoding a
guide RNA and a CasX protein;
c. a third nucleic acid comprising a sequence encoding a
protease capable of cleaving the
protease cleavage site between the gag polyprotein and the RNA binding domain
protein; and
d. a fourth nucleic acid, comprising a sequence encoding a
pseudotyping viral envelope
glycoprotein or antibody fragment that provides for binding and fusion of the
XDP to a target
cell.
[00439] Embodiment 1-6. The XDP system of embodiment 5, wherein the RNA
binding
domain protein is selected from the group consisting of MS2, PP7 or Qbeta, U1
A, phage
replication loop, kissing loop_a, kissing loop b1, kissing loop_b2, G
quadriplex M3q, G
quadriplex telomere basket, sarcin-ricin loop, and pseudoknots.
[00440] Embodiment 1-7. The XDP system of any one of embodiments 1-3,
comprising all
or a portion of any one of the nucleic acid sequences of Table 8 or Table 9.
1004411 Embodiment 1-8. The XDP system of any one of the preceding embodiments
of Set
I, wherein the gag polypeptide comprises one or more protease cleavage sites
between the matrix
polypeptide (MA) and the capsid polypeptide (CA) and/or between the capsid
polypeptide (CA)
and the nucleocapsid polypeptide (NC), wherein the one or more protease cleave
sites are
capable of being cleaved by the protease.
[00442] Embodiment 1-9. The XDP system of any one of the preceding embodiments
of Set
I, wherein the protease is selected from the group of proteases consisting of
HIV-1 protease,
tobacco etch virus protease (TEV), potyvirus HC protease, potyvirus P1
protease, PreScission, b
virus Ma protease, B virus RNA-2-encoded protease, aphthovirus L protease,
enterovirus 2A
protease, rhinovirus 2A protease, picorna 3C protease, comovirus 24K protease,
nepovirus 24K
protease, RTSV (rice tungro spherical virus) 3C-like protease, PYVF (parsnip
yellow fleck
virus) 3C-like protease, cathepsin, thrombin, factor Xa, metalloproteinases
MMP-2, -3, -7, -9, -
10, and -11, and enterokinase.
206
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00443] Embodiment I-10. The XDP system of embodiment 1, wherein the poi
polyprotein is
a retroviral polyprotein.
[00444] Embodiment I-11. The XDP system of embodiment 10, wherein the
retrovirus is an
alpharetrovirus, a betaretrovirus, a gammaretrovirus, a deltaretrovirus, a
epsilonretrovirus, or a
lentivirus.
[00445] Embodiment 1-12. The XDP system of embodiment 11, wherein the
lentivirus is a
human immunodeficiency virus (HIV).
[00446] Embodiment 1-13. The XDP system of any one of the preceding
embodiments of Set
I, wherein the gag polyprotein is a retroviral polyprotein.
1004471 Embodiment I-14, The XDP system of embodiment 13, wherein the gag
polyprotein
is derived from a alpharetrovirus, a betaretrovirus, a gammaretrovirus, a
deltaretrovirus, a
epsilonretrovirus, or a lentivirus.
[00448] Embodiment 1-15. The XDP system of embodiment 14, wherein the gag
polyprotein
is a lentiviral polyprotein.
[00449] Embodiment 1-16. The XDP system of embodiment 15, wherein the
lentiviral gag
polypeptide is an HIV-1 gag polyprotein_
[00450] Embodiment 1-17. The XDP system of any one of embodiments 13-16,
wherein the
gag polypeptide further comprises a p6 polypeptide.
[00451] Embodiment 1-18. The XDP system of embodiment 16 or embodiment 17,
wherein
the HIV-1 gag polypeptide comprises a MA polypeptide, a CA polypeptide, a p2
polypeptide, an
NC polypeptide, a pl polypeptide, and a p6 polypeptide, and wherein the HIV
gag polyprotein
comprises one or more protease cleavage sites located between one or more of:
a. the MA polypeptide and the CA polypeptide;
b. the CA polypeptide and the p2 polypeptide;
c. the p2 polypeptide and the NC polypeptide;
d. the NC polypeptide and the pl polypeptide; and
e. the pl polypeptide and the p6 polypeptide.
[00452] Embodiment 1-19. The XDP system of embodiment 18, wherein the protease
capable
of cleaving the protease cleavage site is selected from the group of proteases
consisting of HIV-1
protease, tobacco etch virus protease (TEV), potyvirus HC protease, potyvirus
PI protease,
PreScission, b virus Ma protease, B virus RNA-2-encoded protease, aphthovirus
L protease,
enterovirus 2A protease, rhinovirus 2A protease, picorna 3C protease,
comovirus 24K protease,
207
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
nepovirus 24K protease, RTSV (rice tungro spherical virus) 3C-like protease,
PYVF (parsnip
yellow fleck virus) 3C-like protease, cathepsin, thrombin, factor Xa,
metalloproteinases MMP-2,
-3, -7, -9, -10, and -11, and enterokinase.
[00453] Embodiment 1-20. The XDP system of embodiment 19, wherein the protease
capable
of cleaving the protease cleavage site is MV-1 protease.
[00454] Embodiment 1-21. The XDP system of any one of the preceding
embodiments of Set
I, further comprising a nucleic acid encoding a retroviral packaging signal
and further
comprising a donor template nucleic acid complementary to a target nucleic
acid.
[00455] Embodiment 1-22. The XDP system of embodiment 21, wherein the donor
template
nucleic acid sequence comprises at least a portion of a target nucleic acid
gene or a regulatory
element of the target nucleic acid gene.
[00456] Embodiment 1-23. The XDP system of embodiment 21 or embodiment 22,
wherein
the donor template nucleic acid sequence comprises a corrective sequence for a
mutation in the
target nucleic acid gene or regulatory element of the target nucleic acid
gene.
[00457] Embodiment 1-24. The XDP system of embodiment 21 or embodiment 22,
wherein
the donor template nucleic acid sequence comprises a mutation compared to the
target nucleic
acid gene or regulatory element of the target nucleic acid gene.
[00458] Embodiment 1-25. The XDP system of embodiment 24, where the mutation
is an
insertion, a deletion, or a substitution of one or more nucleotides in the
donor template nucleic
acid sequence.
1004591 Embodiment 1-26. The XDP system of any one of the preceding
embodiments of Set
1, wherein the guide RNA is a single-molecule guide RNA comprising a scaffold
sequence and a
targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
[00460] Embodiment 1-27. The XDP system of embodiment 26, wherein the guide
RNA
scaffold sequence has at least 80%, at least 90%, at least 95%, at least 96%,
at least 97%, at least
98%, at least 99%, or 100% sequence identity to a sequence selected from the
group of
sequences consisting of SEQ ID NOS: 4, 5, and 597-781.
[00461] Embodiment 1-28. The XDP system of embodiment 26 or embodiment 27,
wherein
the targeting sequence of the guide RNA consists of 14, 15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25,
26, 27, 28, 29, or 30 consecutive nucleotides.
208
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00462] Embodiment 1-29. The XDP system of embodiment 28, wherein the
targeting
sequence of the guide RNA consists of 20 nucleotides.
[00463] Embodiment 1-30. The XDP system of embodiment 28, wherein the
targeting
sequence of the guide RNA consists of 19 nucleotides.
[00464] Embodiment 1-31. The XDP system of embodiment 28, wherein the
targeting
sequence of the guide RNA consists of 18 nucleotides.
[00465] Embodiment 1-32. The XDP system of embodiment 28, wherein the
targeting
sequence of the guide RNA consists of 17 nucleotides.
[00466] Embodiment 1-33. The XDP system of embodiment 28, wherein the
targeting
sequence of the guide RNA consists of 16 nucleotides.
[00467] Embodiment 1-34, The XDP system of embodiment 28, wherein the
targeting
sequence of the guide RNA consists of 15 nucleotides.
[00468] Embodiment 1-35. The XDP system of any one of the preceding
embodiments of Set
I, wherein the guide RNA further comprises one or more ribozymes.
[00469] Embodiment 1-36. The XDP system of embodiment 35, wherein the one or
more
ribozymes are independently fused to a terminus of the guide RNA.
[00470] Embodiment 1-37. The XDP system of embodiment 35 or embodiment 36,
wherein at
least one of the one or more ribozymes are a hepatitis delta virus (HDV)
ribozyme, hammerhead
ribozyme, pistol ribozyme, hatchet ribozyme, or tobacco ringspot virus (TRSV)
ribozyme.
[00471] Embodiment 1-38. The XDP system of any one of the preceding
embodiments of Set
I, wherein the guide RNA is chemically modified.
[00472] Embodiment 1-39. The XDP system of any one of the preceding
embodiments of Set
I, wherein the CasX protein comprises a sequence having at least about 90%, or
at least about
91%, or at least about 92%, or at least about 93%, or at least about 94%, or
at least about 95%,
or at least about 96%, or at least about 97%, or at least about 98%, or at
least about 99%, or at
least 100% sequence identity to a sequence selected from the group consisting
of the sequences
set forth in Table 1.
[00473] Embodiment 1-40. The XDP system of any one of the preceding
embodiments of Set
I, wherein the CasX protein has binding affinity for a protospacer adjacent
motif (PAM)
sequence selected from the group consisting of TTC, ATC, GTC, and CTC.
[00474] Embodiment 1-41. The XDP system of any one of the preceding
embodiments of Set
I, wherein the CasX protein further comprises one or more nuclear localization
signals (NLS).
209
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1004751 Embodiment 1-42. The XDP system of embodiment 41, wherein the one or
more NLS
are selected from the group of sequences consisting of SEQ ID NOS: 130-166.
[00476] Embodiment 1-43. The CasX variant of embodiment 41 or embodiment 42,
wherein
the one or more NLS are expressed at the C-terminus of the CasX protein.
[00477] Embodiment 1-44. The CasX variant of embodiment 41 or embodiment 42,
wherein
the one or more NLS are expressed at the N-terminus of the CasX protein.
[00478] Embodiment 1-45. The CasX variant of embodiment 41 or embodiment 42,
wherein
the one or more NLS are expressed at the N-terminus and C-terminus of the CasX
protein.
[00479] Embodiment 1-46. The XDP system of any one of the preceding
embodiments of Set
I, wherein the CasX protein comprises a nuclease domain having nickase
activity,
[00480] Embodiment 1-47, The XDP system of any one of embodiments 1-45,
wherein the
CasX protein comprises a nuclease domain having double-stranded cleavage
activity.
[00481] Embodiment 1-48. The XDP system of any one of embodiments 1-45,
wherein the
CasX protein is a catalytically inactive CasX (dCasX) protein, and wherein the
dCasX and the
guide RNA retain the ability to bind to the target nucleic acid.
[00482] Embodiment 1-49. The XDP system of embodiment 48, wherein the dCasX
comprises a mutation at residues:
a. D672, E769, and/or D935 corresponding to the CasX protein of SEQ ID NO:
1; or
b. D659, E756 and/or D922 corresponding to the CasX protein of SEQ ID NO:
2.
[00483] Embodiment 1-50. The XDP system of embodiment 49, wherein the mutation
is a
substitution of alanine for the residue.
[00484] Embodiment 1-51. The XDP system of any one of the preceding
embodiments of Set
1, wherein the envelope glycoprotein is derived from an enveloped virus
selected from the group
consisting of influenza A, influenza B, influenza C virus, hepatitis A virus,
hepatitis B virus,
hepatitis C virus, hepatitis D virus, hepatitis E virus, rotavirus, Norwalk
virus, enteric
adenovirus, parvovirus, Dengue fever virus, monkey pox, Mononegavirales,
rabies virus, Lagos
bat virus, Mokola virus, Duvenhage virus, European bat virus 1, European bat
virus 2,
Australian bat virus, Ephemerovirus, Vesiculovirus, vesicular stomatitis virus
(VSV), herpes
simplex virus type 1, herpes simplex virus type 2, varicella zoster,
cytomegalovirus, Epstein-Bar
virus (EBV), human herpesvirus (I HIV), human herpesvirus type 6, human
herpesvirus type 8,
human immunodeficiency virus (HIV), papilloma virus, murine gammaherpesvirus,
Argentine
hemorrhagic fever virus, Bolivian hemorrhagic fever virus, Sabia-associated
hemorrhagic fever
210
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
virus, Venezuelan hemorrhagic fever virus, Lassa fever virus, Machupo virus,
lymphocytic
choriomeningitis virus (LCMV), Crimean-Congo hemorrhagic fever virus,
Hantavirus, Rift
Valley fever virus, Ebola hemorrhagic fever virus, Marburg hemorrhagic fever
virus, Kaysanur
Forest disease virus, Omsk hemorrhagic fever virus, tick-borne encephalitis
causing virus,
Hendra virus, Nipah virus, variola major virus, variola minor virus,
Venezuelan equine
encephalitis virus, eastern equine encephalitis virus, western equine
encephalitis virus, SARS-
associated coronavirus (SARS-Coy), and West Nile virus.
[00485] Embodiment 1-52. The XDP system of embodiment 51, wherein the envelope
glycoprotein is derived from vesicular stomatitis virus (VSV).
[00486] Embodiment 1-53, The XDP system of any one of embodiments 1-50,
wherein the
antibody fragment has binding affinity for a cell surface marker or receptor
of a target cell.
[00487] Embodiment 1-54. The XDP system of embodiment 53, wherein the antibody
fragment is a scFv.
[00488] Embodiment 1-55. A eukaryotic cell comprising the XDP system of any
one of the
preceding embodiments of Set I.
[00489] Embodiment 1-56. The eukaryotic cell of embodiment 54, wherein the
cell is a
packaging cell.
[00490] Embodiment 1-57. The eukaryotic cell of embodiment 55 or embodiment
56, wherein
the eukaryotic cell is selected from the group consisting of HEK293 cells,
Lenti-X 293T cells,
BHK cells, HepG2, Saos-2, HuH7, NSO cells, SP2/0 cells, YO myeloma cells, A549
cells,
P3X63 mouse myeloma cells, PER cells, PER.C6 cells, hybridoma cells, VERO, N11-
13T3 cells,
COS, W138, MRCS, A549, HeLa cells, CHO cells, or HT1080 cells.
[00491] Embodiment 1-58. The eukaryotic cell of embodiment 56 or embodiment
57, wherein
the packaging cell comprises one or more mutations to reduce expression of a
cell surface
marker.
[00492] Embodiment 1-59. A method of making an XDP comprising a CasX protein,
the
method comprising:
a. introducing the XDP system of any one of embodiments 1-54 into the
packaging cell of
any one of embodiments 56-58;
b. propagating the packaging cell under conditions such that XDPs are
produced; and
c. harvesting the XDPs produced by the packaging cell.
[00493] Embodiment 1-60. An XDP produced by the method of embodiment 59.
211
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00494] Embodiment 1-61. An XDP comprising:
a. a retroviral capsid (CA), matrix, (MA), and nucleocapsid (NC)
polypeptides
b. a pseudotyping viral envelope glycoprotein or an antibody fragment that
provides for
binding and fusion to a target cell; and
c. a CasX protein and a guide RNA associated together in a ribonuclear
protein complex
(RNP) within the XDP.
[00495] Embodiment 1-62. The XDP of embodiment 61, comprising the CasX of any
one of
embodiments 39-50 and the guide RNA of any one of embodiments 26-38.
[00496] Embodiment 1-63. The XDP of embodiment 61, wherein the pseudotyping
viral
envelope glycoprotein is derived from the packaging cell of embodiment 57 or
embodiment 58
or a nucleic acid encoding the glycoprotein introduced into the packaging
cell.
[00497] Embodiment 1-64. The XDP of embodiment 60-63, further comprising a
donor
template nucleic acid sequence of any one of embodiments 21-25.
[00498] Embodiment 1-65. A method of method of modifying a target nucleic acid
sequence
in a cell, the method comprising contacting the cell with the XDP of any one
of embodiments
60-64, wherein said contacting comprises introducing into the cell the CasX,
the guide RNA,
and, optionally, the donor template nucleic acid sequence, resulting in
modification of the target
nucleic acid sequence.
[00499] Embodiment 1-66. The method of embodiment 65, wherein the modification
comprises introducing one or more single-stranded breaks in the target nucleic
acid sequence_
[00500] Embodiment 1-67. The method of embodiment 65, wherein the modification
comprises introducing a double-stranded break in the target nucleic acid
sequence.
[00501] Embodiment 1-68. The method of any one of embodiments 65-67, wherein
the
modification comprises insertion of the donor template into the target nucleic
acid sequence.
[00502] Embodiment 1-69. The method of any one of embodiments 65-68, wherein
the cell is
modified in vitro.
[00503] Embodiment 1-70. The method of any one of embodiments 65-68, wherein
the cell is
modified in viva
[00504] Embodiment 1-71. The method of embodiment 70, wherein the XDP is
administered
to a subject.
[00505] Embodiment 1-72. The method of embodiment 71, wherein the subject is
the subject
is selected from the group consisting of mouse, rat, pig, non-human primate,
and human.
212
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00506] Embodiment 1-73. The method of embodiment 71 or embodiment 72, wherein
the
XDP is administered by a route of administration selected from the group
consisting of
intravenous, intracerebroyentricular, intracistemal, intrathecal,
intracranial, lumbar,
intratracheal, intraosseous, inhalatory, intracontralateral striatum,
intraocular, intravitreal, and
sub-retinal routes.
[00507] Embodiment 1-74. The method of any one of embodiments 71-73, wherein
the XDP
is administered to the subject using a therapeutically effective dose.
[00508] Embodiment 1-75. The method of embodiment 74, wherein the XDP is
administered
at a dose of at least about 1 x 105 particles, or at least about 1 x 106
particles, or at least about 1 x
107 particles, or at least about 1 x 108 panicles, or at least about 1 x 109
particles, or at least
about 1 x 1010 particles, or at least about 1 x 10" particles, or at least
about 1 x 1012 particles, or
at least about 1 x 10" particles, or at least about 1 x 10' particles, or at
least about 1 x 10"
particles, or at least about 1 x 1016 particles.
Set II
1005091 Embodiment
A CasX delivery particle (XDP)
system comprising one or more
nucleic acids comprising sequences encoding components selected from:
a. a matrix polypeptide (MA);
b. a capsid polypeptide (CA);
c. a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide (CA), and
a nucleocapsid polypeptide (NC);
d. a CasX protein;
e. a guide nucleic acid (gNA);
f. a pseudotyping viral envelope glycoprotein or antibody fragment that
provides for
binding and fusion of the XDP to a target cell;
g. an RNA binding domain;
h. a protease cleavage site;
i. a gag-transframe region-pol protease polyprotein (gag-TFR-PR);
j. a gag-pol polyprotein; and
k. a protease capable of cleaving the protease cleavage sites.
[00510] Embodiment 11-2. The XDP system of Embodiment II-1, wherein the
encoded
components comprise the gag polyprotein, the protease cleavage site, the CasX
protein, the gag-
213
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
pol polyprotein, the gNA, and the pseudotyping viral envelope glycoprotein or
antibody
fragment, wherein the components are encoded on two, three, or four individual
nucleic acids.
1005111 Embodiment 11-3. The XDP system of Embodiment 11-2, wherein
a. a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an intervening
protease cleavage site between the components; and a second nucleic acid
encodes the gag-pol
polyprotein, the pseudotyping viral envelope glycoprotein or antibody fragment
and the gNA;
b. a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an intervening
protease cleavage site between the components; a second nucleic acid encodes
the gag-pol
polyprotein; and a third nucleic acid encodes the pseudotyping viral envelope
glycoprotein or
antibody fragment and the gNA, or
c. a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an intervening
protease cleavage site between the components; a second nucleic acid encodes
the pseudotyping
viral envelope glycoprotein or antibody fragment; a third nucleic acid encodes
the gag-pol
polyprotein; and a fourth nucleic acid encodes the gNA.
1005121 Embodiment 11-4. The XDP system of Embodiment II-1, wherein the
encoded
components are selected from the gag-TFR-PR polyprotein, the protease cleavage
site, the CasX
protein, the gNA, and the pseudo-typing viral envelope glycoprotein or
antibody fragment,
wherein the components are encoded on one, two, or three individual nucleic
acids.
[00513] Embodiment 11-5. The XDP system of Embodiment 11-4, wherein
a. the components are encoded on a single nucleic acid;
b. a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and an
intervening protease cleavage site between the components; and a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment and the gNA;
c. a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA.
[00514] Embodiment 11-6. The XDP system of Embodiment II-1, wherein the
encoded
components are selected from the gag polyprotein, the protease cleavage site,
the protease, the
CasX protein, the gNA and the pseudotyping viral envelope glycoprotein or
antibody fragment,
wherein the components are encoded on one, two, or three individual nucleic
acids.
[00515] Embodiment 11-7. The XDP system of Embodiment 11-6, wherein
214
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
a. the components are encoded on a single nucleic acid;
ii a first nucleic acid encodes the gag polyprotein, the
protease, the CasX protein, and
intervening protease cleavage sites between the components; and a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment and the gNA;
c. a first nucleic acid encodes the gag polyprotein, the protease, the CasX
protein and
intervening protease cleavage sites between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA.
1005161 Embodiment 11-8. The XDP system of Embodiment II-1, wherein the
encoded
components are selected from the gag-pal polyprotein, the CasX protein, the
protease cleavage
site, the gNA, the RNA binding domain, and the pseudotyping viral envelope
glycoprotein or
antibody fragment, wherein the components are encoded on one, two, or three
individual nucleic
acids.
1005171 Embodiment 11-9. The XDP system of Embodiment 11-8, wherein
a. the components are encoded on a single nucleic acid;
b. a first nucleic acid encodes the gag-pd polyproteinõ the CasX protein,
and intervening
protease cleavage sites between the components; and a second nucleic acid
encodes the
pseudotyping viral envelope glycoprotein or antibody fragment, the gNA and the
RNA binding
domain; or
c. a first nucleic acid encodes the gag-pol polyprotein, the CasX protein,
and an intervening
protease cleavage site between the components; a second nucleic acid encodes
the pseudotyping
viral envelope glycoprotein or antibody fragment; and a third nucleic acid
encodes the gNA and
the RNA binding domain.
1005181 Embodiment II-10. The XDP system of Embodiment I1-1, wherein the
encoded
components are selected from the gag-TFR-PR polyprotein, the CasX protein, the
protease
cleavage site, the gNA, the RNA binding domain, and the pseudotyping viral
envelope
glycoprotein or antibody fragment, wherein the components are encoded on one,
two, or three
individual nucleic acids.
[00519] Embodiment II-11. The XDP system of Embodiment II-10, wherein
a. the components are encoded on a single nucleic acid;
b. a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and an
intervening protease cleavage site between the components; and a second
nucleic acid encodes
215
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
the pseudotyping viral envelope glycoprotein or antibody fragment, the gNA and
the RNA
binding domain; or
c. a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA and the RNA binding domain.
[00520] Embodiment 11-12. The XDP system of any one of Embodiments 11-8-11,
wherein the
RNA binding domain is a retroviral Psi packaging element inserted into the gNA
or is a protein
selected from the group consisting of MS2, PP7 or Qbeta, U1 A, phage
replication loop, kissing
loop_a, kissing loop_b1, kissing loop_b2, G quadriplex M3q, G quadriplex
telomere basket,
sarcin-ricin loop, and pseudoknots.
[00521] Embodiment 11-13. The XDP system of Embodiment II-1, wherein the
encoded
components are selected from the gag-pol polyprotein, the CasX protein, the
protease cleavage
site, the pseudotyping viral envelope glycoprotein or antibody fragment, and
the gNA, wherein
the components are encoded on one, two, or three individual nucleic acids.
[00522] Embodiment 11-14. The XDP system of Embodiment 11-13, wherein
a. the components are encoded on a single nucleic acid;
b. a first nucleic acid encodes the gag-pot polyprotein, an intervening
protease cleavage
site, the CasX protein; and a second nucleic acid encodes the pseudotyping
viral envelope
glycoprotein or antibody fragment and the gNA; or
c. a first nucleic acid encodes the gag-pot polyprotein, an intervening
protease cleavage
site, and the CasX protein; a second nucleic acid encodes the pseudotyping
viral envelope
glycoprotein or antibody fragment; and a third nucleic acid encodes the gNA.
[00523] Embodiment 11-15. The XDP system of Embodiment I1-1, wherein the
encoded
components are selected from the MA, the CasX protein, the protease, the
protease cleavage site,
the 8NA, and the pseudotyping viral envelope glycoprotein or antibody
fragment, wherein the
components are encoded on one, two, three, or four individual nucleic acids.
[00524] Embodiment 11-16 The XDP system of Embodiment 11-15, wherein
a. the components are encoded on a single nucleic acid;
b. a first nucleic acid encodes the MA, the CasX protein, the protease, and
intervening
protease cleavage sites between the components; and a second nucleic acid
encodes the
pseudotyping viral envelope glycoprotein or antibody fragment and the gNA;
216
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
c. a first nucleic acid encodes the MA, the CasX protein the protease, and
intervening
protease cleavage sites between the components; a second nucleic acid encodes
the pseudotyping
viral envelope glycoprotein or antibody fragment; and a third nucleic acid
encodes the gNA; or
d. a first nucleic acid encodes the MA, an intervening protease cleavage
site, and the CasX
protein; a second nucleic acid encodes the pseudotyping viral envelope
glycoprotein or antibody
fragment; a third nucleic acid encodes the gNA; and a fourth nucleic acid
encodes the protease.
[00525] Embodiment 11-17. The XDP system of Embodiment 11-15 or Embodiment 11-
16,
further comprising the CA component linked between the MA and the CasX protein
components
with intervening protease cleavage sites.
[00526] Embodiment II-18, The XDP system of Embodiment II-1, wherein the
encoded
components are selected from the gag polyprotein, the CasX protein, the
protease, the protease
cleavage site, the gNA, the pseudotyping viral envelope glycoprotein or
antibody fragment, and
the gag-pol polyprotein, wherein the components are encoded on two, three, or
four individual
nucleic acids.
1005271 Embodiment 11-19. The XDP system of Embodiment 11-18, wherein
a. a first nucleic acid encodes the gag polyprotein, the CasX protein, the
protease, and
intervening protease cleavage sites between the components; and a second
nucleic acid encodes
the gag-pol polyprotein, the pseudotyping viral envelope glycoprotein or
antibody fragment, and
the gNA; or
b. a first nucleic acid encodes the gag polyprotein, the intervening
protease cleavage site,
and the CasX protein; a second nucleic acid encodes the protease; and a third
nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment, the
gNA and the
gag-pol polyprotein; or
c. a first nucleic acid encodes the gag polyprotein, the intervening
protease cleavage site,
and the CasX protein; a second nucleic acid encodes the protease; a third
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment; and a
fourth nucleic acid
encodes the gNA and the gag-pot polyprotein
[00528] Embodiment 11-20 The XDP system of Embodiment 11-2 or Embodiment 11-3,
comprising all or a portion of any one of the nucleic acid sequences of Table
6.
[00529] Embodiment 11-21. The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the MA, the CA, the gag-TER-PR polyprotein, the gag
polyprotein, and the
gag-pol polyprotein are derived from a retrovirus.
217
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1005301 Embodiment 11-22. The XDP system of Embodiment 11-21, wherein the
retrovirus is
selected from the group consisting of an alpharetrovirus, a betaretrovirus, a
gammaretrovirus, a
deltaretrovirus, an epsilonretrovirus, and a lentivirus.
[00531] Embodiment 11-23. The XDP system of Embodiment 11-22, wherein the
lentivirus is
selected from the group consisting of human immunodeficiency-1 (TITV-1), human
immunodeficiency-2 (HIV-2), simian immunodeficiency virus (Sly), feline
immunodeficiency
virus (FIV), and bovine immunodeficiency virus (B1V).
[00532] Embodiment 11-24. The XDP system of Embodiment 11-23, wherein the
lentivirus is
REV-1 or S1V.
1005331 Embodiment 11-25, The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the gag polypeptide further comprises a p6 polypeptide.
[00534] Embodiment 11-26. The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the gag polypeptide comprises a MA polypeptide, a CA
polypeptide, a p2
polypeptide, an NC polypeptide, a p1 polypeptide, and a p6 polypeptide, and
wherein the gag
polyprotein comprises one or more protease cleavage sites located between one
or more of:
a. the MA polypeptide and the CA polypeptide,
b. the CA polypeptide and the p2 polypeptide;
c. the p2 polypeptide and the NC polypeptide;
d. the NC polypeptide and the p1 polypeptide; and
e. the pl polypeptide and the p6 polypeptide.
1005351 Embodiment 11-27. The XDP system of any one of the preceding
embodiments of Set
1 of Set IT, wherein the protease capable of cleaving the protease cleavage
site is selected from
the group of proteases consisting of HIV-1 protease, tobacco etch virus
protease (TEV),
potyvirus HC protease, potyvirus P1 protease, PreScission, b virus Ma
protease, B virus RNA-2-
encoded protease, aphthovirus L protease, enterovirus 2A protease, rhinovirus
2A protease,
picorna 3C protease, comovirus 24K protease, nepovirus 24K protease, RTSV
(rice tungro
spherical virus) 3C-like protease, PYVF (parsnip yellow fleck virus) 3C-like
protease, cathepsin,
thrombin, factor Xa, metalloproteinase-2 (MMP-2), NIMP -3, MMP-7,
MMP-10,
MMP-11, and enterokinase.
[00536] Embodiment 11-28. The XDP system of Embodiment 11-27, wherein the
protease
capable of cleaving the protease cleavage site is HIV-1 protease.
218
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1005371 Embodiment 11-29. The XDP system of any one of the preceding
embodiments of Set
I of Set 11, wherein the pseudotyping viral envelope glycoprotein is derived
from an enveloped
virus selected from the group consisting of Argentine hemorrhagic fever virus,
Australian bat
virus, Autographa californica multiple nucleopolyhedrovirus, Avian leukosis
virus, baboon
endogenous virus, Bolivian hemorrhagic fever virus, Borna disease virus, Breda
virus,
Bunyamwera virus, Chandipura virus, Chikungunya virus, Crimean-Congo
hemorrhagic fever
virus, Dengue fever virus, Duvenhage virus, Eastern equine encephalitis virus,
Ebola
hemorrhagic fever virus, Ebola Zaire virus, enteric adenovirus, Ephemerovirus,
Epstein-Bar
virus (EBV), European bat virus 1, European bat virus 2, Gibbon ape leukemia
virus,
Hantavirus, Hendra virus, hepatitis A virus, hepatitis B virus, hepatitis C
virus, hepatitis D virus,
hepatitis E virus, hepatitis G Virus (GB virus C), herpes simplex virus type
1, herpes simplex
virus type 2, human cytomegalovirus (HHV5), human foamy virus, human
herpesvirus (HHV),
human Herpesvirus 7, human herpesvirus type 6, human herpesvirus type 8, human
immunodeficiency virus 1 (111V-1), human metapneumovirus, human T-Iymphotro
pic virus 1,
influenza A, influenza B, influenza C virus, Japanese encephalitis virus,
Kaposirs sarcoma-
associated herpesvirus (REM), Kaysanur Forest disease virus, La Crosse virus,
Lagos bat virus,
Lassa fever virus, lymphocytic choriomeningitis virus (LCMV), Machupo virus,
Marburg
hemorrhagic fever virus, measles virus, Middle eastern respiratory syndrome-
related
coronavirus, Mokola virus, Moloney murine leukemia virus, monkey pox, mouse
mammary
tumor virus, mumps virus, murine gammaherpesvirus, Newcastle disease virus,
Nipah virus,
Nipah virus, Norwalk virus, Omsk hemorrhagic fever virus, papilloma virus,
parvovirus,
pseudorabies virus, Quaranfil virus, rabies virus, RD114 endogenous feline
retrovirus,
respiratory syncytial virus (RSV), Rift Valley fever virus, Ross River virus,
rotavirus, Rous
sarcoma virus, rubella virus, Sabia-associated hemorrhagic fever virus, SARS-
associated
coronavirus (SARS-CoV), Sendai virus, Tacaribe virus, Thogotovirus, tick-borne
encephalitis
causing virus, varicella zoster virus (HHV3), varicella zoster virus (1111V3),
variola major virus,
variola minor virus, Venezuelan equine encephalitis virus, Venezuelan
hemorrhagic fever virus,
vesicular stomatitis virus (VSV), Vesiculovirus, West Nile virus, western
equine encephalitis
virus, and Zika Virus.
[00538] Embodiment 11-30. The XDP system of Embodiment 11-29, wherein the
pseudotyping
viral envelope glycoprotein is derived from vesicular stomatitis virus (VSV).
219
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00539] Embodiment 11-31. The XDP system of any one of Embodiments 11-1-29,
wherein the
pseudotyping viral envelope glyc,oprotein comprises a sequence having at least
90%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence
identity to a
sequence selected from the group consisting of the sequences set forth in
Table 4.
1005401 Embodiment 11-32. The XDP system of any one of Embodiments 11-1-28,
wherein the
antibody fragment has binding affinity for a cell surface marker or receptor
of a target cell.
1005411 Embodiment 11-33. The XDP system of Embodiment 11-32, wherein the
antibody
fragment is a scFv.
1005421 Embodiment 11-34. The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the gNA is a single-molecule guide RNA comprising a
scaffold sequence and
a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
1005431 Embodiment 11-35. The XDP system of Embodiment 11-29, wherein the
guide RNA
scaffold sequence has at least 80%, at least 90%, at least 95%, at least 96%,
at least 97%, at least
98%, at least 99%, or 100% sequence identity to a sequence selected from the
group of
sequences consisting of SEQ ID NOS: 4, 5, and 2101-2241.
1005441 Embodiment 11-36. The XDP system of Embodiment 11-29 or Embodiment II-
Embodiment 11-35, wherein the targeting sequence of the guide RNA consists of
14, 15, 16, 17,
18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides.
[00545] Embodiment 11-37. The XDP system of Embodiment 11-36, wherein the
targeting
sequence of the guide RNA consists of 20 nucleotides.
[00546] Embodiment 11-38. The XDP system of Embodiment 11-36, wherein the
targeting
sequence of the guide RNA consists of 19 nucleotides.
[00547] Embodiment 11-39. The XDP system of Embodiment 11-36, wherein the
targeting
sequence of the guide RNA consists of 18 nucleotides.
[00548] Embodiment 11-40. The XDP system of Embodiment 11-36, wherein the
targeting
sequence of the guide RNA consists of 17 nucleotides.
[00549] Embodiment 11-41. The XDP system of Embodiment 11-36, wherein the
targeting
sequence of the guide RNA consists of 16 nucleotides.
[00550] Embodiment 11-42. The XDP system of Embodiment 11-36, wherein the
targeting
sequence of the guide RNA consists of 15 nucleotides.
220
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00551] Embodiment 11-43. The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the guide RNA further comprises one or more ribozymes.
[00552] Embodiment 11-44. The XDP system of Embodiment 11-43, wherein the one
or more
ribozymes are independently fused to a terminus of the guide RNA.
[00553] Embodiment 11-45. The XDP system of Embodiment 11-43 or Embodiment 11-
44,
wherein at least one of the one or more ribozymes is a hepatitis delta virus
(HDV) ribozyme,
hammerhead ribozyme, pistol ribozyme, hatchet ribozyme, or tobacco ringspot
virus (TRSV)
ribozyme.
[00554] Embodiment 11-46. The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the guide RNA is chemically modified.
[00555] Embodiment 11-47, The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the CasX protein comprises a sequence having at least
about 90%, or at least
about 91%, or at least about 92%, or at least about 93%, or at least about
94%, or at least about
95%, or at least about 96%, or at least about 97%, or at least about 98%, or
at least about 99%,
or at least 100% sequence identity to a sequence selected from the group
consisting of the
sequences set forth in Table 1.
[00556] Embodiment 11-48. The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the CasX protein has binding affinity for a protospacer
adjacent motif (PAM)
sequence selected from the group consisting of TTC, ATC, GTC, and CTC.
[00557] Embodiment 11-49. The XDP system of Embodiment 11-48, wherein the
binding
affinity of the CasX protein for the PAM sequence is at least 1.5-fold greater
compared to the
binding affinity of any one of the CasX proteins of SEQ ID NOS: 1-3 for the
PAM sequences.
[00558] Embodiment 11-50. The XDP system of any one of the preceding
embodiments of Set
of Set IT, wherein the CasX protein further comprises one or more nuclear
localization signals
(NLS).
[00559] Embodiment 11-51. The XDP system of Embodiment 11-50, wherein the one
or more
NLS are selected from the group of sequences consisting of PKKKRKV,
KRPAATKKAGQAKICKK, PAAKRVKLD, RQRRNELKRSP,
NQSSNEGPMKGGNEGGRSSGPYGGGGQYFAKPRNQGGY,
RMRIZEKNKGKDTAELRRRRVEVSVELRKAKKDEQILKRRNV, VSRICRPRP,
PPICICARED, PQPKK1CPL, SALIKKKKKMAP, DRLRR, PKQKKRK, RICLICKKIKKL,
REKKICFLICRR, KRICGDEVDGVDEVAKICKSKK, RICCLQAGMNLEARKTICK,
221
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
PRPRKIPR, PPRKICRTVV, NLSKICICKRKREK, RRPSRPFRKP, KRPRSPSS,
KRGINDRNFWRGENERKTR, PRPPICMARYDN, KRSFSKAF, KLICIKRPVK,
PKTRRRPRRSQRKRPPT, RRKKRRPRRICKRR, PKKKSRKPKICKSRK,
THCICKHPDASVNFSEFSK, QRPGPYDRPQRPGPYDRP, LSPSLSPLLSPSLSPL,
RGKGGKGLGKGGAKRFIRK, PICRGRGRPKRGRGR, and
MSRRRKANPTICL,SENAICKLAICEVEN.
[00560] Embodiment 11-52. The CasX variant of Embodiment II-50 or Embodiment
11-51,
wherein the one or more NLS are fused to the C-terminus of the CasX protein.
[00561] Embodiment 11-53. The CasX variant of Embodiment 11-50 or Embodiment
11-51,
wherein the one or more NLS are fused to the N-terminus of the CasX protein.
[00562] Embodiment II-54, The CasX variant of Embodiment II-50 or Embodiment
11-51,
wherein the one or more NLS are fused to the N-terminus and C-terminus of the
CasX protein.
[00563] Embodiment 11-55. The XDP system of any one of the preceding
embodiments of Set
I of Set II, wherein the CasX protein comprises a nuclease domain having
nickase activity.
[00564] Embodiment 11-56. The XDP system of any one of Embodiments 11-1-54,
wherein the
CasX protein comprises a nuclease domain having double-stranded cleavage
activity.
[00565] Embodiment 11-57. The XDP system of any one of the preceding
embodiments of Set
I of Set II, further comprising a nucleic acid encoding a retroviral packaging
signal.
[00566] Embodiment 11-58. The XDP system of any one of the preceding
embodiments of Set
I of Set II, further comprising a donor template nucleic acid complementary to
a target nucleic
acid.
[00567] Embodiment 11-59. The XDP system of Embodiment 11-58, wherein the
donor
template comprises two homologous arms complementary to sequences flanking a
cleavage site
in the target nucleic acid.
[00568] Embodiment 11-60. The XDP system of Embodiment 11-58 or Embodiment 11-
59,
wherein the donor template nucleic acid sequence comprises a corrective
sequence for a
mutation in the target nucleic acid
[00569] Embodiment 11-61 The XDP system of Embodiment 11-58 or Embodiment 11-
59,
wherein the donor template nucleic acid sequence comprises a mutation compared
to the target
nucleic acid.
222
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00570] Embodiment 11-62. The XDP system of Embodiment 11-61, where the
mutation is an
insertion, a deletion, or a substitution of one or more nucleotides in the
donor template nucleic
acid sequence.
[00571] Embodiment 11-63. The XDP system of any one of Embodiments 11-1-54,
wherein the
CasX protein is a catalytically inactive CasX (dCasX) protein, and wherein the
dCasX and the
guide RNA retain the ability to bind to the target nucleic acid.
[00572] Embodiment 11-64. The XDP system of Embodiment 11-63, wherein the
dCasX
comprises a mutation at residues:
a. D672, E769, and/or D935 corresponding to the CasX protein of SEQ ID NO:
1; or
b. D659, E756 and/or D922 corresponding to the CasX protein of SEQ ID NO:
2.
[00573] Embodiment 11-65, The XDP system of Embodiment 11-64, wherein the
mutation is a
substitution of alanine for the residue.
[00574] Embodiment 11-66. A eukaryotic cell comprising the XDP system of any
one of the
preceding embodiments of Set I of Set II.
[00575] Embodiment 11-67. The eukaryotic cell of Embodiment 11-66, wherein the
cell is a
packaging cell.
[00576] Embodiment 11-68. The eukaryotic cell of any one of Embodiments 11-66
or
Embodiment 11-67, wherein the eukaryotic cell is selected from the group
consisting of HEK293
cells, Lenti-X 293T cells, BHK cells, HepG2, Saos-2, HuH7, NSO cells, SP2/0
cells, YO
myeloma cells, A549 cells, P3X63 mouse myeloma cells, PER cells, PER.C6 cells,
hybridoma
cells, VERO, NIH3T3 cells, COS, W138, MRCS, A549, HeLa cells, CHO cells, and
HT1080
cells.
[00577] Embodiment 11-69. The eukaryotic cell of Embodiment 11-67 or
Embodiment 11-68,
wherein the packaging cell comprises one or more mutations to reduce
expression of a cell
surface marker.
[00578] Embodiment 11-70. The eukaryotic cell of any one of Embodiments 11-66-
69, wherein
all or a portion of the nucleic acids encoding the XDP system of any one of
Embodiments 11-1-
56 are integrated into the genotne of the eukaryotic cell.
[00579] Embodiment 11-71. A method of making an XDP comprising a CasX protein
and a
gNA, the method comprising:
a. propagating the packaging cell of any one of Embodiments
11-67-70 under conditions
such that XDPs are produced; and
223
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
b. harvesting the XDPs produced by the packaging cell.
[00580] Embodiment 11-72. An XDP produced by the method of Embodiment H-71.
[00581] Embodiment 11-73. An XDP comprising one or more components selected
from:
a. a matrix polypeptide (MA);
b. a capsid polypeptide (CA);
c. a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide (CA), and
a nucleocapsid polypeptide (NC);
d. a CasX protein;
e. a guide nucleic acid (gNA);
f. a pseudotyping viral envelope glycoprotein or antibody fragment that
provides for
binding and fusion of the XDP to a target cell; and
g. an RNA binding domain;
[00582] Embodiment 11-74. The XDP of Embodiment 11-73, wherein the XDP
comprises
a. the matrix polypeptide (MA);
b. the pseudotyping viral envelope glycoprotein or antibody fragment; and
c. the CasX and the gNA contained within the XDP.
[00583] Embodiment 11-75. The XDP of Embodiment 11-74, further comprising the
capsid
polypeptide (CA).
[00584] Embodiment 11-76. The XDP of Embodiment 11-74 or Embodiment 11-75,
further
comprising the nucleocapsid polypeptide (NC).
1005851 Embodiment 11-77. The XDP of any one of Embodiments 11-74-76, further
comprising
an RNA binding domain.
[00586] Embodiment 11-78. The XDP of Embodiment 11-77, wherein the RNA binding
domain
is a retroviral Psi packaging element inserted into the gNA or is a protein
selected from the
group consisting of MS2, PP7 or Qbeta, U1A, phage replication loop, kissing
loop_a, kissing
loop_bl, kissing loop_b2, G quadriplex M3q, G quadriplex telomere basket,
sarcin-ricin loop,
and pseudoknots.
[00587] Embodiment 11-79, The XDP of any one of Embodiments 11-74-78, wherein
the CasX
and the gNA are associated together in a ribonuclear protein complex (RNP)
within the XDP.
[00588] Embodiment 11-80. The XDP of any one of Embodiments 11-74-79,
comprising the
CasX of any one of Embodiments 11-47-65 and the guide RNA of any one of
Embodiments II-
34-46.
224
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00589] Embodiment 11-81. The XDP of any one of Embodiments 11-74-80, wherein
the
pseudotyping viral envelope glycoprotein comprises a sequence having at least
90%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence
identity to a
sequence selected from the group consisting of the sequences set forth in
Table 4.
1005901 Embodiment 11-82. The XDP of any one of Embodiments 11-73-80, wherein
the
pseudotyping viral envelope glycoprotein is derived from an enveloped virus
selected from the
group consisting of influenza A, influenza B, influenza C virus, hepatitis A
virus, hepatitis B
virus, hepatitis C virus, hepatitis D virus, hepatitis E virus, rotavirus,
Norwalk virus, enteric
adenovirus, parvovirus, Dengue fever virus, monkey pox, Mononegavirales,
rabies virus, Lagos
bat virus, Mokola virus, Duvenhage virus, European bat virus 1, European bat
virus 2,
Australian bat virus, Ephemerovirus, Vesiculovirus, vesicular stomatitis virus
(VSV), herpes
simplex virus type 1, herpes simplex virus type 2, varicella zoster,
cytomegalovirus, Epstein-Bar
virus (EBV), human herpesvirus (HHV), human herpesvirus type 6, human
herpesvirus type 8,
human immunodeficiency virus (HIV), papilloma virus, murine gammaherpesvirus,
Argentine
hemorrhagic fever virus, Bolivian hemorrhagic fever virus, Sabia-associated
hemorrhagic fever
virus, Venezuelan hemorrhagic fever virus, Lassa fever virus, Machupo virus,
lymphocytic
choriomeningitis virus (LCMV), Crimean-Congo hemorrhagic fever virus,
Hantavirus, Rift
Valley fever virus, Ebola hemorrhagic fever virus, Marburg hemorrhagic fever
virus, Kaysanur
Forest disease virus, Omsk hemorrhagic fever virus, tick-borne encephalitis
causing virus,
Hendra virus, Nipah virus, variola major virus, variola minor virus,
Venezuelan equine
encephalitis virus, eastern equine encephalitis virus, western equine
encephalitis virus, SARS-
associated coronavirus (SARS-CoV), and West Nile virus.
1005911 Embodiment 11-83. The XDP of any one of Embodiments 11-73-82, further
comprising
the donor template nucleic acid sequence of any one of Embodiments 1I-58-62.
1005921 Embodiment 11-84. A method of method of modifying a target nucleic
acid sequence
in a cell, the method comprising contacting the cell with the XDP of any one
of Embodiments II-
73-83, wherein said contacting comprises introducing into the cell the CasX
protein, the guide
RNA, and, optionally, the donor template nucleic acid sequence, resulting in
modification of the
target nucleic acid sequence.
1005931 Embodiment 11-85. The method of Embodiment 1I-84, wherein the
modification
comprises introducing one or more single-stranded breaks in the target nucleic
acid sequence.
225
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00594] Embodiment 11-86. The method of Embodiment 11-84, wherein the
modification
comprises introducing one or more double-stranded breaks in the target nucleic
acid sequence.
[00595] Embodiment 11-87 The method of any one of Embodiments 11-84-86,
wherein the
modification comprises insertion of the donor template into the target nucleic
acid sequence.
[00596] Embodiment 11-88. The method of any one of Embodiments 11-84-87,
wherein the cell
is modified in vitro.
[00597] Embodiment 11-89. The method of any one of Embodiments 11-84-87,
wherein the cell
is modified in vivo.
[00598] Embodiment 11-90. The method of Embodiment 11-89, wherein the XDP is
administered to a subject.
[00599] Embodiment 11-91, The method of Embodiment 11-90, wherein the subject
is the
subject is selected from the group consisting of mouse, rat, pig, non-human
primate, and human,
[00600] Embodiment 11-92. The method of Embodiment 11-90 or Embodiment 11-91,
wherein
the XDP is administered by a route of administration selected from the group
consisting of
subcutaneous, intradermal, intraneural, intranodal, intrarnedullary,
intramuscular, intravenous,
intracerebroventricular, intracistemal, intrathecal, intracranial,
intralumbar, intratracheal,
intraosseous, inhalatoty, intraeontralateral striatum, intraocular,
intravitreal, intralymphatical,
intraperitoneal routes and sub-retinal routes.
[00601] Embodiment 11-93. The method of any one of Embodiments 11-90-92,
wherein the
XDP is administered to the subject using a therapeutically effective dose.
[00602] Embodiment 11-94. The method of Embodiment 11-93, wherein the XDP is
administered at a dose of at least about 1 x 105 particles, or at least about
1 x 106 particles, or at
least about 1 x 107 particles, or at least about 1 x 108particles, or at least
about 1 x 109 particles,
oral least about 1 x 1010 particles, or at least about 1 x 10" particles, or
at least about 1 x 1012
particles, or at least about 1 x 10E3 particles, or at least about 1 x 1014
particles, or at least about 1
x 1015 particles, or at least about 1 x 10' particles.
[00603] Embodiment 11-95 A method for introducing a CasX and gNA RNP into a
cell having
a target nucleic acid, comprising contacting the cell with the XDP of any one
of Embodiments
11-79-83, such that the RNP enters the cell.
[00604] Embodiment 11-96. The method of Embodiment 11-95, wherein the RNP
binds to the
target nucleic acid.
226
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00605] Embodiment 11-97. The method of Embodiment 11-96, wherein the target
nucleic acid
is cleaved by the CasX.
[00606] Embodiment 11-98. The method of any one of Embodiments 11-95-97,
wherein the cell
is modified in vitro.
[00607] Embodiment 11-99. The method of any one of Embodiments 11-95-97,
wherein the cell
is modified in viva
[00608] Embodiment 11-100. The method of Embodiment 11-99, wherein the XDP is
administered to a subject.
[00609] Embodiment H-101.The method of Embodiment 11-100, wherein the subject
is the
subject is selected from the group consisting of mouse, rat, pig, non-human
primate, and human.
[00610] Embodiment II-102.The method of any one of Embodiments 11-99-101,
wherein the
XDP is administered to the subject using a therapeutically effective dose.
[00611] Embodiment II-103.The method of Embodiment 11-102, wherein the XDP is
administered at a dose of at least about 1 x 105 particles, or at least about
1 x 106 particles, or at
least about 1 x 107 particles, or at least about 1 x iO3 particles, or at
least about 1 x 109 particles,
or at least about 1 x 1010 particles, or at least about 1 x 1011 particles, or
at least about 1 x 1012
particles, or at least about 1 x 1013 particles, or at least about 1 x 1014
particles, or at least about 1
x 1015 particles, or at least about 1 x 10' particles.
Set In
[00612] Embodiment I11-1. A CasX delivery particle (XDP) system comprising one
or more
nucleic acids comprising sequences encoding components selected from:
(a) a matrix polypeptide (MA);
(b) a capsid polypeptide (CA);
(c) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC);
(d) a CasX protein;
(e) a guide nucleic acid (gNA);
(f) a pseudotyping viral envelope glycoprotein or antibody fragment that
provides
for binding and fusion of the XDP to a target cell;
(g) an RNA binding domain;
(h) a protease cleavage site;
227
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(i) a gag-transframe region-pol protease polyprotein
(gag-TFR-PR);
a gag-pol polyprotein; and
(k) a protease capable of cleaving the protease
cleavage sites.
[00613] Embodiment The XDP system of Embodiment
wherein the encoded
components comprise the gag polyprotein, the protease cleavage site, the CasX
protein, the gag-
pol polyprotein, the gNA_, and the pseudotyping viral envelope glycoprotein or
antibody
fragment, wherein the components are encoded on two, three, or four individual
nucleic acids.
Embodiment 111-3. The XDP system of Embodiment
wherein
(a) a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an
intervening protease cleavage site between the components; and a second
nucleic acid encodes
the gag-pol polyprotein, the pseudotyping viral envelope glycoprotein or
antibody fragment and
the gNA;
(b) a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
gag-poi polyprotein; and a third nucleic acid encodes the pseudotyping viral
envelope
glycoprotein or antibody fragment and the gNA; or
(c) a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; a third nucleic
acid encodes the
gag-pol polyprotein; and a fourth nucleic acid encodes the gNA.
[00614] Embodiment III-4. The XDP system of Embodiment
wherein the encoded
components are selected from the gag-TFR-PR polyprotein, the protease cleavage
site, the CasX
protein, the gNA, and the pseudo-typing viral envelope glycoprotein or
antibody fragment,
wherein the components are encoded on one, two, or three individual nucleic
acids.
[00615] Embodiment The XDP system of Embodiment
wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; and a second
nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment and
the gNA;
(c) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; a second nucleic
acid encodes the
228
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA.
[00616] Embodiment 111-6. The XDP system of Embodiment III- 1, wherein the
encoded
components are selected from the gag polyprotein, the protease cleavage site,
the protease, the
CasX protein, the gNA and the pseudotyping viral envelope glycoprotein or
antibody fragment,
wherein the components are encoded on one, two, or three individual nucleic
acids.
[00617] Embodiment 111-7. The XDP system of Embodiment
wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag polyprotein, the protease, the
CasX protein,
and intervening protease cleavage sites between the components; and a second
nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment and
the gNA,
(c) a first nucleic acid encodes the gag polyprotein, the protease, the
CasX protein
and intervening protease cleavage sites between the components; a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid
encodes the gNA.
[00618] Embodiment 111-8. The XDP system of Embodiment In-1, wherein the
encoded
components are selected from the gag-pol polyprotein, the CasX protein, the
protease cleavage
site, the gNA, the RNA binding domain, and the pseudotyping viral envelope
glycoprotein or
antibody fragment, wherein the components are encoded on one, two, or three
individual nucleic
acids.
1006191 Embodiment 111-9. The XDP system of Embodiment 111-8, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-pol polyprotein, the CasX protein,
and
intervening protease cleavage sites between the components; and a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment, the gNA and
the RNA
binding domain; or
(c) a first nucleic acid encodes the gag-pol polyprotein, the CasX protein,
and an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA and the RNA binding domain.
[00620] Embodiment II1-10. The XDP system of Embodiment III-1, wherein the
encoded
components are selected from the gag-TFR-PR polyprotein, the CasX protein, the
protease
229
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
cleavage site, the gNA, the RNA binding domain, and the pseudotyping viral
envelope
glycoprotein or antibody fragment, wherein the components are encoded on one,
two, or three
individual nucleic acids.
[00621] Embodiment III-11. The XDP system of Embodiment I11-10, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; and a second
nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment, the
8NA and the
RNA binding domain; or
(c) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid
encodes the gNA and the RNA binding domain.
[00622] Embodiment 111-12. The XDP system of any one of Embodiments In- 8-11,
wherein
the RNA binding domain is a retroviral Psi packaging element inserted into the
gNA or is a
protein selected from the group consisting of MS2, PP7 or Qbeta, U1A, phage
replication loop,
kissing loop_a, kissing loop_bl, kissing loop_b2, G quadriplex M3q, G
quadriplex telomere
basket, sarcin-ricin loop, and pseudoknots.
[00623] Embodiment 111-13. The XDP system of Embodiment
wherein the encoded
components are selected from the gag-pol polyprotein, the CasX protein, the
protease cleavage
site, the pseudotyping viral envelope glycoprotein or antibody fragment, and
the gNA, wherein
the components are encoded on one, two, or three individual nucleic acids.
[00624] Embodiment 111-14. The XDP system of Embodiment 111-13, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-pol polyprotein, an intervening
protease
cleavage site, the CasX protein; and a second nucleic acid encodes the
pseudotyping viral
envelope glycoprotein or antibody fragment and the gNA; or
(c) a first nucleic acid encodes the gag-pol polyprotein, an intervening
protease
cleavage site, and the CasX protein; a second nucleic acid encodes the
pseudotyping viral
envelope glycoprotein or antibody fragment; and a third nucleic acid encodes
the gNA.
[00625] Embodiment 111-15. The XDP system of Embodiment III-1, wherein the
encoded
components are selected from the MA, the CasX protein, the protease, the
protease cleavage site,
230
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
the gNA, and the pseudotyping viral envelope glycoprotein or antibody
fragment, wherein the
components are encoded on one, two, three, or four individual nucleic acids.
1006261 Embodiment 111-16 The XDP system of Embodiment 111-15, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the MA, the CasX protein, the protease,
and
intervening protease cleavage sites between the components; and a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment and the gNA;
(c) a first nucleic acid encodes the MA, the CasX protein the protease, and
intervening protease cleavage sites between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA; or
(d) a first nucleic acid encodes the MA, an intervening protease cleavage
site, and the
CasX protein; a second nucleic acid encodes the pseudotyping viral envelope
glycoprotein or
antibody fragment; a third nucleic acid encodes the gNA; and a fourth nucleic
acid encodes the
protease.
[00627] Embodiment III-17. The XDP system of Embodiment I11-15 or Embodiment
11I-16,
further comprising the CA component linked between the MA and the CasX protein
components
with intervening protease cleavage sites.
[00628] Embodiment 111-18. The XDP system of Embodiment
wherein the encoded
components are selected from the gag polyprotein, the CasX protein, the
protease, the protease
cleavage site, the gNA, the pseudotyping viral envelope glycoprotein or
antibody fragment, and
the gag-pol polyprotein, wherein the components are encoded on two, three, or
four individual
nucleic acids.
[00629] Embodiment 111-19. The XDP system of Embodiment 111-18, wherein
(a) a first nucleic acid encodes the gag polyprotein, the CasX protein, the
protease,
and intervening protease cleavage sites between the components; and a second
nucleic acid
encodes the gag-pol polyprotein, the pseudotyping viral envelope glycoprotein
or antibody
fragment, and the gNA; or
(b) a first nucleic acid encodes the gag polyprotein, the intervening
protease cleavage
site, and the CasX protein; a second nucleic acid encodes the protease; and a
third nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment, the
gNA and the
gag-pol polyprotein; or
231
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(c) a first nucleic acid encodes the gag
polyprotein, the intervening protease cleavage
site, and the CasX protein; a second nucleic acid encodes the protease; a
third nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment; and
a fourth
nucleic acid encodes the gNA and the gag-pol polyprotein.
[00630] Embodiment III-20. The XDP system of Embodiment III-2 or Embodiment
111-3,
comprising all or a portion of any one of the nucleic acid sequences of Table
6.
[00631] Embodiment III-21. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the MA, the CA, the gag-TFR-PR polyprotein, the gag
polyprotein, and the
gag-pol polyprotein are derived from a retrovirus.
1006321 Embodiment III-22, The XDP system of Embodiment III-21, wherein the
retrovirus is
selected from the group consisting of an alpharetrovirus, a betaretrovirus, a
gammaretrovirus, a
deltaretrovirus, an epsilonretrovirus, and a lentivirus.
[00633] Embodiment III-23. The XDP system of Embodiment III-22, wherein the
lentivirus is
selected from the group consisting of human immunodeficiency-1 (HIV-1), human
immunodeficiency-2 (HIV-2), simian immunodeficiency virus (SIV), feline
immunodeficiency
virus (FIV), and bovine immunodeficiency virus (BIV).
[00634] Embodiment III-24. The XDP system of Embodiment 111-23, wherein the
lentivirus is
HIV-1 or SIV.
[00635] Embodiment III-25. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the gag polypeptide further comprises a p6 polypeptide.
1006361 Embodiment III-26. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the gag polypeptide comprises a MA polypeptide, a CA
polypeptide, a p2
polypeptide, an NC polypeptide, a p1 polypeptide, and a p6 polypeptide, and
wherein the gag
polyprotein comprises one or more protease cleavage sites located between one
or more of:
(a) the MA polypeptide and the CA polypeptide;
(b) the CA polypeptide and the p2 polypeptide;
(c) the p2 polypeptide and the NC polypeptide;
(d) the NC polypeptide and the p1 polypeptide; and
(e) the p1 polypeptide and the p6 polypeptide.
[00637] Embodiment III-27. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the protease capable of cleaving the protease cleavage
site is selected from
the group of proteases consisting of HIV-1 protease, tobacco etch virus
protease (TEV),
232
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
potyvirus HC protease, potyvirus P1 protease, PreScission, b virus NIa
protease, B virus RNA-2-
encoded protease, aphthovirus L protease, enterovirus 2A protease, rhinovirus
2A protease,
picorna 3C protease, comovirus 24K protease, nepovirus 24K protease, RTSV
(rice tungro
spherical virus) 3C-like protease, PYVF (parsnip yellow fleck virus) 3C-like
protease, cathepsin,
thrombin, factor Xa, metalloproteinase-2 (MMP-2), WIMP -3, MMP-7, MMP-9, WIMP-
b,
MMP-11, and enterokinase.
1006381 Embodiment III-28. The XDP system of Embodiment III-27, wherein the
protease
capable of cleaving the protease cleavage site is HIV-1 protease.
1006391 Embodiment III-29. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the pseudotyping viral envelope glycoprotein is derived
from an enveloped
virus selected from the group consisting of Argentine hemorrhagic fever virus,
Australian bat
virus, Autographa califorrtica multiple nucleopolyhedrovirus, Avian leukosis
virus, baboon
endogenous virus, Bolivian hemorrhagic fever virus, Bona disease virus, Breda
virus,
Bunyamwera virus, Chandipura virus, Chikungunya virus, Crimean-Congo
hemorrhagic fever
virus, Dengue fever virus, Duvenhage virus, Eastern equine encephalitis virus,
Ebola
hemorrhagic fever virus, Ebola Zaire virus, enteric adenovirus, Ephemerovirus,
Epstein-Bar
virus (EBV), European bat virus 1, European bat virus 2, Gibbon ape leukemia
virus,
Hantavirus, Hendra virus, hepatitis A virus, hepatitis B virus, hepatitis C
virus, hepatitis D virus,
hepatitis E virus, hepatitis G Virus (GB virus C), herpes simplex virus type
1, herpes simplex
virus type 2, human cytomegalovirus (FIFIV5), human foamy virus, human
herpesvirus (HHV),
human Herpesvirus 7, human herpesvirus type 6, human herpesvirus type 8, human
immunodeficiency virus 1 (mV-1), human metapneumovirus, human T-lymphotro pic
virus 1,
influenza A, influenza B, influenza C virus, Japanese encephalitis virus,
Kaposi's sarcoma-
associated herpesvirus (HEIV8), Kaysanur Forest disease virus, La Crosse
virus, Lagos bat virus,
Lassa fever virus, lymphocytic choriomeningitis virus (LCMV), Machupo virus,
Marburg
hemorrhagic fever virus, measles virus, Middle eastern respiratory syndrome-
related
coronavirus, Mokola virus, Moloney murine leukemia virus, monkey pox, mouse
mammary
tumor virus, mumps virus, murine gammaherpesvirus, Newcastle disease virus,
Nipah virus,
Nipah virus, Norwalk virus, Omsk hemorrhagic fever virus, papilloma virus,
parvovirus,
pseudorabies virus, Quaranfil virus, rabies virus, RD114 endogenous feline
retrovirus,
respiratory syncytial virus (RSV), Rift Valley fever virus, Ross River virus,
rotavirus, Rous
sarcoma virus, rubella virus, Sabia-associated hemorrhagic fever virus, SARS-
associated
233
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
coronavirus (SARS-CoV), Sendai virus, Tacaribe virus, Thogotovirus, tick-borne
encephalitis
causing virus, varicella zoster virus (HHV3), varicella zoster virus (HEIV3),
variola major virus,
variola minor virus, Venezuelan equine encephalitis virus, Venezuelan
hemorrhagic fever virus,
vesicular stomatitis virus (VSV), Vesiculovirus, West Nile virus, western
equine encephalitis
virus, and Zika Virus.
[00640] Embodiment III-30. The XDP system of Embodiment 111-29, wherein the
pseudotyping
viral envelope glycoprotein is derived from vesicular stomatitis virus (VSV).
[00641] Embodiment III-31. The XDP system of any one of Embodiments III-1-29,
wherein the
pseudotyping viral envelope glycoprotein comprises a sequence having at least
90%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence
identity to a
sequence selected from the group consisting of the sequences set forth in
Table 4.
[00642] Embodiment III-32. The XDP system of any one of Embodiments III-
Embodiments
III-1-28, wherein the antibody fragment has binding affinity for a cell
surface marker or receptor
of a target cell.
[00643] Embodiment I11-33. The XDP system of Embodiment I11-32, wherein the
antibody
fragment is a scFv.
[00644] Embodiment 1I-34. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the gNA is a single-molecule guide RNA comprising a
scaffold sequence
and a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic
acid sequence.
[00645] Embodiment III-35. The XDP system of Embodiment III-29, wherein the
guide RNA
scaffold sequence has at least 80%, at least 90%, at least 95%, at least 96%,
at least 97%, at least
98%, at least 99%, or 100% sequence identity to a sequence selected from the
group of
sequences consisting of SEQ ID NOS: 4, 5, and 2101-2241.
[00646] Embodiment 111-36. The XDP system of Embodiment 111-29 or Embodiment
I11-35,
wherein the targeting sequence of the guide RNA consists of 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides,
[00647] Embodiment III-37 The XDP system of Embodiment 111-36, wherein the
targeting
sequence of the guide RNA consists of 20 nucleotides.
[00648] Embodiment III-38. The XDP system of Embodiment 111-36, wherein the
targeting
sequence of the guide RNA consists of 19 nucleotides.
234
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00649] Embodiment III-39. The XDP system of Embodiment III-36, wherein the
targeting
sequence of the guide RNA consists of 18 nucleotides.
[00650] Embodiment III-40. The XDP system of Embodiment III-36, wherein the
targeting
sequence of the guide RNA consists of 17 nucleotides.
[00651] Embodiment III-41. The XDP system of Embodiment III-36, wherein the
targeting
sequence of the guide RNA consists of 16 nucleotides.
[00652] Embodiment 111-42. The XDP system of Embodiment III-36, wherein the
targeting
sequence of the guide RNA consists of 15 nucleotides.
[00653] Embodiment III-43. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the guide RNA further comprises one or more ribozymes.
[00654] Embodiment III-44, The XDP system of Embodiment III-43, wherein the
one or more
ribozymes are independently fused to a terminus of the guide RNA.
[00655] Embodiment III-45. The XDP system of Embodiment III-43 or Embodiment
III-44,
wherein at least one of the one or more ribozymes is a hepatitis delta virus
(HDV) ribozyme,
hammerhead ribozyme, pistol ribozyme, hatchet ribozyme, or tobacco ringspot
virus (TRSV)
ribozyme.
[00656] Embodiment 111-46. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the guide RNA is chemically modified.
[00657] Embodiment III-47. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the CasX protein comprises a sequence having at least
about 90%, or at least
about 91%, or at least about 92%, or at least about 93%, or at least about
94%, or at least about
95%, or at least about 96%, or at least about 97%, or at least about 98%, or
at least about 99%,
oral least 100% sequence identity to a sequence selected from the group
consisting of the
sequences set forth in Table 1.
[00658] Embodiment III-48. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the CasX protein has binding affinity for a protospacer
adjacent motif
(PAM) sequence selected from the group consisting of TTC, ATC, GTC, and CTC.
[00659] Embodiment III-49. The XDP system of Embodiment III-48, wherein the
binding
affinity of the CasX protein for the PAM sequence is at least 1.5-fold greater
compared to the
binding affinity of any one of the CasX proteins of SEQ ID NOS: 1-3 for the
PAM sequences.
235
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00660] Embodiment III-50. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the CasX protein further comprises one or more nuclear
localization signals
(NLS).
[00661] Embodiment Ill-Si. The XDP system of Embodiment III-50, wherein the
one or more
NLS are selected from the group of sequences consisting of PKKKRKV,
KRPAATKKAGQAKKKK, PAAKRVKLD, RQRRNELKRSP,
NQSSNEGPMKGGNEGGRSSGPYGGGGQYFAKPRNQGGY,
RMRIZFKNKGKDTAELRRRRVEVSVELRKAKKDEQILICRRNV, VSRICRPRP,
PPKKARED, PQPICICKPL, SALIKKKKKMAP, DRLRR, PKQICICRIC, RICLICKKIICKL,
REKKICFLICRR, ICRKGDEVDGVDEVAKKKSKK, RKCLQAGMNLEARKTICK,
PRPRKIPR, PPRKICRTVV, NLSKICKKRKREK, RRPSRPFRKP, KRPRSPSS,
KRGINDRNEWRGENERKTR, PRPPICMARYDN, KRSFSKAF, KLKIKRPVK,
PKTRRRPRRSQRICRPPT, RRKICRRPRRICKRR, PKKKSRKPKICKSRK,
HICICICHPDASVNESEFSK, QRPGPYDRPQRPGPYDRP, LSPSLSPLLSPSLSPL,
RGKGGKGLGKGGAKRHRK, PKRGRGRPKRGRGR, and
MSRRRKANPTKLSENAICKLAICEVEN.
[00662] Embodiment 1I-52. The CasX variant of Embodiment III-50 or Embodiment
1,
wherein the one or more NLS are fused to the C-terminus of the CasX protein.
[00663] Embodiment III-53. The CasX variant of Embodiment III-50 or Embodiment
III-51,
wherein the one or more NLS are fused to the N-terminus of the CasX protein.
[00664] Embodiment 111-54. The CasX variant of Embodiment III-50 or Embodiment
II1-5 1,
wherein the one or more NLS are fused to the N-terminus and C-terminus of the
CasX protein.
[00665] Embodiment III-55. The XDP system of any one of the preceding
embodiments of Set
I of Set III, wherein the CasX protein comprises a nuclease domain having
nickase activity.
[00666] Embodiment III-56. The XDP system of any one of Embodiments III-
Embodiments
III-1-54, wherein the CasX protein comprises a nuclease domain having double-
stranded
cleavage activity.
[00667] Embodiment III-57. The XDP system of any one of the preceding
embodiments of Set
I of Set III, further comprising a nucleic acid encoding a retroviral
packaging signal.
[00668] Embodiment III-58. The XDP system of any one of the preceding
embodiments of Set
I of Set III, further comprising a donor template nucleic acid complementary
to a target nucleic
acid.
236
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00669] Embodiment III-59. The XDP system of Embodiment 111-58, wherein the
donor
template comprises two homologous arms complementary to sequences flanking a
cleavage site
in the target nucleic acid.
[00670] Embodiment III-60. The XDP system of Embodiment III-58 or Embodiment
111-59,
wherein the donor template nucleic acid sequence comprises a corrective
sequence for a
mutation in the target nucleic acid.
[00671] Embodiment III-61. The XDP system of Embodiment III-58 or Embodiment
11I-59,
wherein the donor template nucleic acid sequence comprises a mutation compared
to the target
nucleic acid.
[00672] Embodiment 111-62, The XDP system of Embodiment 111-61, where the
mutation is an
insertion, a deletion, or a substitution of one or more nucleotides in the
donor template nucleic
acid sequence.
[00673] Embodiment III-63. The XDP system of any one of Embodiments III-
Embodiments
III-1-54, wherein the CasX protein is a catalytically inactive CasX (dCasX)
protein, and wherein
the dCasX and the guide RNA retain the ability to bind to the target nucleic
acid.
[00674] Embodiment 1I-64. The XDP system of Embodiment 111-63, wherein the
dCasX
comprises a mutation at residues:
(a) D672, E769, and/or D935 corresponding to the CasX protein of SEQ ID NO:
1;
or
(b) D659, E756 and/or D922 corresponding to the CasX protein of SEQ ID NO:
2.
[00675] Embodiment 111-65. The XDP system of Embodiment 11I-64, wherein the
mutation is a
substitution of alanine for the residue.
[00676] Embodiment III-66. A eukaryotic cell comprising the XDP system of any
one of the
preceding embodiments of Set I of Set III.
[00677] Embodiment III-67. The eukaryotic cell of Embodiment 111-66, wherein
the cell is a
packaging cell.
[00678] Embodiment 11I-68, The eukaryotic cell of any one of Embodiments III-
Embodiments
III-66 or Embodiment III-67, wherein the eukaryotic cell is selected from the
group consisting of
ITEK293 cells, Lenti-X 293T cells, BHK cells, 11epG2, Saos-2, flu117, NSO
cells, SP2/0 cells,
YO myeloma cells, A549 cells, P3X63 mouse myeloma cells, PER cells, PER.C6
cells,
hybridoma cells, VERO, NIH3T3 cells, COS, WI38, MRCS, A549, HeLa cells, CHO
cells, and
HT1080 cells.
237
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00679] Embodiment 11I-69. The eukaryotic cell of Embodiment 111-67 or
Embodiment 111-68,
wherein the packaging cell comprises one or more mutations to reduce
expression of a cell
surface marker.
[00680] Embodiment 11I-70. The eukaryotic cell of any one of Embodiments III-
Embodiments
11I-66-69, wherein all or a portion of the nucleic acids encoding the XDP
system of any one of
Embodiments 111-1-56 are integrated into the genome of the eukaryotic cell.
[00681] Embodiment 11I-7 1. A method of making an XDP comprising a CasX
protein and a
gNA, the method comprising:
(a) propagating the packaging cell of any one of Embodiments 111-67-70
under
conditions such that XDPs are produced; and
(b) harvesting the XDPs produced by the packaging cell.
[00682] Embodiment 11I-72. An XDP produced by the method of Embodiment HI-71.
[00683] Embodiment 11I-73. An XDP comprising one or more components selected
from:
(a) a matrix polypeptide (MA);
(b) a capsid polypeptide (CA),
(c) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC);
(d) a CasX protein;
(e) a guide nucleic acid (gNA);
(t) a pseudotyping viral envelope glycoprotein or
antibody fragment that provides
for binding and fusion of the XDP to a target cell; and
(g) an RNA binding domain;
[00684] Embodiment 11I-74. The XDP of Embodiment 11I-73, wherein the XDP
comprises
(a) the matrix polypeptide (MA);
(b) the pseudotyping viral envelope glycoprotein or antibody fragment; and
(c) the CasX and the gNA contained within the XDP.
[00685] Embodiment 11I-75. The XDP of Embodiment 11I-74, further comprising
the capsid
polypeptide (CA).
[00686] Embodiment 11I-76. The XDP of Embodiment 111-74 or Embodiment
further
comprising the nucleocapsid polypeptide (NC).
[00687] Embodiment 11I-77. The XDP of any one of Embodiments 11I-74-76,
further
comprising an RNA binding domain.
238
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00688] Embodiment 111-78. The XDP of Embodiment III-77, wherein the RNA
binding
domain is a retroviral Psi packaging element inserted into the gNA or is a
protein selected from
the group consisting of MS2, PP7 or Qbeta, U1 A, phage replication loop,
kissing loop_a, kissing
loop bl, kissing loop b2, G quadriplex M3q, G quadriplex telomere basket,
sarcin-ricin loop,
and pseudoknots.
[00689] Embodiment III-79. The XDP of any one of Embodiments 111-74-78,
wherein the CasX
and the gNA are associated together in a ribonuclear protein complex (RNP)
within the XDP.
[00690] Embodiment III-80, The XDP of any one of Embodiments 111-74-79,
comprising the
CasX of any one of Embodiments 111-47-65 and the guide RNA of any one of
Embodiments III-
34-46,
[00691] Embodiment 111-81, The XDP of any one of Embodiments 111-74-80,
wherein the
pseudotyping viral envelope glycoprotein comprises a sequence having at least
90%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence
identity to a
sequence selected from the group consisting of the sequences set forth in
Table 4.
1006921 Embodiment III-82. The XDP of any one of Embodiments 111-73-80,
wherein the
pseudotyping viral envelope glycoprotein is derived from an enveloped virus
selected from the
group consisting of Argentine hemorrhagic fever virus, Australian bat virus,
Autographa
californica multiple nucleopolyhedrovirus, Avian leukosis virus, baboon
endogenous virus,
Bolivian hemorrhagic fever virus, Boma disease virus, Breda virus, Bunyamwera
virus,
Chandipura virus, Chikungunya virus, Crimean-Congo hemorrhagic fever virus,
Dengue fever
virus, Duvenhage virus, Eastern equine encephalitis virus, Ebola hemorrhagic
fever virus, Ebola
Zaire virus, enteric adenovirus, Ephemerovirus, Epstein-Bar virus (EBV),
European bat virus 1,
European bat virus 2, Gibbon ape leukemia virus, Hantavirus, Hendra virus,
hepatitis A virus,
hepatitis B virus, hepatitis C virus, hepatitis D virus, hepatitis E virus,
hepatitis G Virus (GB
virus C), herpes simplex virus type 1, herpes simplex virus type 2, human
cytomegalovirus
(HEWS), human foamy virus, human herpesvirus (HMV), human Herpesvirus 7, human
herpesvirus type 6, human herpesvirus type 8, human immunodeficiency virus 1
(I-11V-1), human
metapneumovirus, human T-lymphotro pic virus 1, influenza A, influenza B,
influenza C virus,
Japanese encephalitis virus, Kaposi's sarcoma-associated herpesvirus (FI11V8),
Kaysanur Forest
disease virus, La Crosse virus, Lagos bat virus, Lassa fever virus,
lymphocytic choriomeningitis
virus (LCMV), Machupo virus, Marburg hemorrhagic fever virus, measles virus,
Middle eastern
respiratory syndrome-related coronavirus, Mokola virus, Moloney murine
leukemia virus,
239
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
monkey pox, mouse mammary tumor virus, mumps virus, murine gammaherpesvirus,
Newcastle disease virus, Nipah virus, Nipah virus, Norwalk virus, Omsk
hemorrhagic fever
virus, papilloma virus, parvovirus, pseudorabies virus, Quaranfil virus,
rabies virus, RD114
endogenous feline retrovirus, respiratory syncytial virus (RSV), Rift Valley
fever virus, Ross
River virus, rotavirus, Rous sarcoma virus, rubella virus, Sabia-associated
hemorrhagic fever
virus, SARS-associated coronavirus (SARS-CoV), Sendai virus, Tacaribe virus,
Thogotovirus,
tick-borne encephalitis causing virus, varicella zoster virus (HEIV3),
varicella zoster virus
(HHV3), variola major virus, variola minor virus, Venezuelan equine
encephalitis virus,
Venezuelan hemorrhagic fever virus, vesicular stomatitis virus (VSV),
Vesiculovirus, West Nile
virus, western equine encephalitis virus, and Zika Virus.
[00693] Embodiment 111-83, The XDP of any one of Embodiments 111-73-82,
further
comprising the donor template nucleic acid sequence of any one of Embodiments
111-58-62.
[00694] Embodiment 111-84. A method of method of modifying a target nucleic
acid sequence
in a cell, the method comprising contacting the cell with the XDP of any one
of Embodiments
111-73-83, wherein said contacting comprises introducing into the cell the
CasX protein, the
guide RNA, and, optionally, the donor template nucleic acid sequence,
resulting in modification
of the target nucleic acid sequence.
[00695] Embodiment 111-85. The method of Embodiment 111-84, wherein the
modification
comprises introducing one or more single-stranded breaks in the target nucleic
acid sequence_
1006961 Embodiment 111-86. The method of Embodiment 111-84, wherein the
modification
comprises introducing one or more double-stranded breaks in the target nucleic
acid sequence.
[00697] Embodiment 111-87 The method of any one of Embodiments 111-84-86,
wherein the
modification comprises insertion of the donor template into the target nucleic
acid sequence.
[00698] Embodiment 111-88. The method of any one of Embodiments 111-84-87,
wherein the
cell is modified in vitro
[00699] Embodiment 111-89. The method of any one of Embodiments 111-84-87,
wherein the
cell is modified in vivo
[00700] Embodiment 111-90 The method of Embodiment 111-89, wherein the XDP is
administered to a subject.
[00701] Embodiment 111-91. The method of Embodiment 111-90, wherein the
subject is the
subject is selected from the group consisting of mouse, rat, pig, non-human
primate, and human.
240
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1007021 Embodiment 111-92. The method of Embodiment 111-90 or Embodiment 111-
91, wherein
the XDP is administered by a route of administration selected from the group
consisting of
subcutaneous, intradermal, intraneural, intranodal, intramedullary,
intramuscular, intravenous,
intracerebroventricular, intracisternal, intrathecal, intracranial,
intralumbar, intratracheal,
intraosseous, inhalatory, intracontralateral striatum, intraocular,
intravitreal, intralymphatical,
intraperitoneal routes and sub-retinal routes.
[00703] Embodiment III-93. The method of any one of Embodiments 111-90-92,
wherein the
XDP is administered to the subject using a therapeutically effective dose.
[00704] Embodiment III-94. The method of Embodiment III-93, wherein the XDP is
administered at a dose of at least about 1 x 105 particles, or at least about
1 x 106 panicles, or at
least about 1 x 107 particles, or at least about 1 x 108 particles, or at
least about 1 x 109 particles,
or at least about 1 x 1010 particles, or at least about 1 x 1011 particles, or
at least about 1 x 1012
particles, or at least about 1 x 1013 particles, or at least about 1 x 1014
particles, or at least about 1
x 1015 particles, or at least about 1 x 1016 particles.
1007051 Embodiment I11-95. A method for introducing a CasX and gNA RNP into a
cell having
a target nucleic acid, comprising contacting the cell with the XDP of any one
of Embodiments
111-79-83, such that the RNP enters the cell.
[00706] Embodiment III-96. The method of Embodiment 111-95, wherein the RNP
binds to the
target nucleic acid.
1007071 Embodiment 111-97. The method of Embodiment 111-96, wherein the target
nucleic acid
is cleaved by the CasX
[00708] Embodiment 111-98. The method of any one of Embodiments 111-95-97,
wherein the
cell is modified in vitro
[00709] Embodiment 111-99. The method of any one of Embodiments 111-95-97,
wherein the
cell is modified in viva
[00710] Embodiment III-100. The method of
Embodiment 111-99, wherein the XDP is
administered to a subject.
[00711] Embodiment III-101. The method of
Embodiment III-100, wherein the subject is
the subject is selected from the group consisting of mouse, rat, pig, non-
human primate, and
human.
[00712] Embodiment III-102. The method of any one
of Embodiments III-99-101,
wherein the XDP is administered to the subject using a therapeutically
effective dose.
241
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00713] Embodiment II1-103.
The method of Embodiment III-102,
wherein the XDP is
administered at a dose of at least about 1 x 105 particles, or at least about
1 x 106 particles, or at
least about 1 x 107 particles, or at least about 1 x 103 particles, or at
least about 1 x 109 particles,
oral least about 1 x 1010 particles, or at least about 1 x 1011 particles, or
at least about 1 x 1012
particles, or at least about 1 x 10" particles, or at least about 1 x 1014
particles, or at least about 1
x 1015 particles, or at least about 1 x 1016 particles.
Set IV
[00714] Embodiment IV-1. A delivery particle (XDP) system for CasX and one or
more
nucleic acids comprising sequences encoding one or more components selected
from (a) to (o) or
encoding one or more portions of the components selected from (a) to (o):
(a) a matrix polypeptide (MA);
(b) a capsid polypeptide (CA);
(c) a nucelocapsid polypeptide (NC);
(d) a pl spacer peptide;
(e) a p2 spacer peptide,
(f) p6 spacer peptide;
(g) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), a nucleocapsid polypeptide (NC), a p1 spacer, and a p6 spacer;
(h) a CasX protein;
(i) a guide nucleic acid (gNA);
(i) a pseudotyping viral envelope glycoprotein or
antibody fragment that provides
for binding and fusion of the XDP to a target cell;
(k) an RNA binding domain;
(I) a protease cleavage site;
(m) a gag-transframe region-pol protease polyprotein (gag-TFR-PR);
(n) a gag-pal polyprotein; and
(o) a protease capable of cleaving the protease cleavage sites.
[00715] Embodiment IV-2. The XDP system of Embodiment IV-1, wherein the
encoded
components comprise the gag polyprotein, the protease cleavage site, the CasX
protein, the gag-
pol polyprotein, the gNA, and the pseudotyping viral envelope glycoprotein or
antibody
fragment, wherein the components are encoded on two, three, or four individual
nucleic acids.
242
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1007161 Embodiment IV-3, The XDP system of Embodiment IV-2, wherein
(a) a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an
intervening protease cleavage site between the components; and a second
nucleic acid encodes
the gag-pol polyprotein, the pseudotyping viral envelope glycoprotein or
antibody fragment and
the gNA;
(b) a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
gag-pol polyprotein; and a third nucleic acid encodes the pseudotyping viral
envelope
glycoprotein or antibody fragment and the gNA; or
(c) a first nucleic acid encodes the gag polyprotein, the CasX protein, and
an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; a third nucleic
acid encodes the
gag-pol polyprotein; and a fourth nucleic acid encodes the gNA.
1007171 Embodiment IV-4. The XDP system of Embodiment IV-1, wherein the
encoded
components are selected from the gag-TFR-PR polyprotein, the protease cleavage
site, the CasX
protein, the gNA, and the pseudotyping viral envelope glycoprotein or antibody
fragment,
wherein the components are encoded on one, two, or three individual nucleic
acids.
1007181 Embodiment IV-5. The XDP system of Embodiment IV-4, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; and a second
nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment and
the gNA;
(c) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA.
1007191 Embodiment IV-6. The XDP system of Embodiment IV-I, wherein the
encoded
components are selected from the gag polyprotein, the protease cleavage site,
the protease, the
CasX protein, the 8NA and the pseudotyping viral envelope glycoprotein or
antibody fragment,
wherein the components are encoded on one, two, or three individual nucleic
acids.
1007201 Embodiment IV-7. The XDP system of Embodiment IV-6, wherein
(a) the components are encoded on a single nucleic
acid;
243
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(b) a first nucleic acid encodes the gag polyprotein, the protease, the
CasX protein,
and intervening protease cleavage sites between the components; and a second
nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment and
the gNA;
(c) a first nucleic acid encodes the gag polyprotein, the protease, the
CasX protein
and intervening protease cleavage sites between the components; a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid
encodes the gNA.
[00721] Embodiment IV-8. The XDP system of Embodiment IV-1, wherein the
encoded
components are selected from the gag-pol polyprotein, the CasX protein, the
protease cleavage
site, the gNA, the RNA binding domain, and the pseudotyping viral envelope
glycoprotein or
antibody fragment, wherein the components are encoded on one, two, or three
individual nucleic
acids.
[00722] Embodiment IV-9. The XDP system of Embodiment IV-8, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-poi polyprotein, the CasX protein,
and
intervening protease cleavage sites between the components; and a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment, the gNA and
the RNA
binding domain; or
(c) a first nucleic acid encodes the gag-pol polyprotein, the CasX protein,
and an
intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA and the RNA binding domain.
[00723] Embodiment IV-10. The XDP system of Embodiment IV-1, wherein the
encoded
components are selected from the gag-TFR-PR polyprotein, the CasX protein, the
protease
cleavage site, the gNA, the RNA binding domain, and the pseudotyping viral
envelope
glycoprotein or antibody fragment, wherein the components are encoded on one,
two, or three
individual nucleic acids.
[00724] Embodiment IV-11 The XDP system of Embodiment IV-10, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; and a second
nucleic acid
244
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
encodes the pseudotyping viral envelope glycoprotein or antibody fragment, the
gNA and the
RNA binding domain; or
(c) a first nucleic acid encodes the gag-TFR-PR polyprotein, the CasX
protein, and
an intervening protease cleavage site between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA and the RNA binding domain.
[00725] Embodiment IV-12. The XDP system of any one of Embodiments IV-8-11,
wherein the
RNA binding domain is a retroviral Psi packaging element inserted into the gNA
or is a protein
selected from the group consisting of MS2, PP7 or Qbeta, U1A, phage
replication loop, kissing
loop_a, kissing loop_bl, kissing loop_b2, G quadriplex M3q, G quadriplex
telomere basket,
sarcin-ricin loop, and pseudoknots.
[00726] Embodiment IV-13. The XDP system of Embodiment IV-1, wherein the
encoded
components are selected from the gag-pol polyprotein, the CasX protein, the
protease cleavage
site, the pseudotyping viral envelope glycoprotein or antibody fragment, and
the gNA, wherein
the components are encoded on one, two, or three individual nucleic acids.
[00727] Embodiment IV-14. The XDP system of Embodiment IV-13, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the gag-pol polyprotein, an intervening
protease
cleavage site, the CasX protein; and a second nucleic acid encodes the
pseudotyping viral
envelope glycoprotein or antibody fragment and the gNA; or
(c) a first nucleic acid encodes the gag-pol polyprotein, an intervening
protease
cleavage site, and the CasX protein; a second nucleic acid encodes the
pseudotyping viral
envelope glycoprotein or antibody fragment; and a third nucleic acid encodes
the gNA.
[00728] Embodiment IV-15. The XDP system of Embodiment IV-1, wherein the
encoded
components are selected from the MA, the CasX protein, the protease, the
protease cleavage site,
the gNA, and the pseudotyping viral envelope glycoprotein or antibody
fragment, wherein the
components are encoded on one, two, three, or four individual nucleic acids.
[00729] Embodiment IV-16 The XDP system of Embodiment IV-15, wherein
(a) the components are encoded on a single nucleic acid;
(b) a first nucleic acid encodes the MA, the CasX protein, the protease,
and
intervening protease cleavage sites between the components; and a second
nucleic acid encodes
the pseudotyping viral envelope glycoprotein or antibody fragment and the gNA;
245
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(c) a first nucleic acid encodes the MA, the CasX protein the protease, and
intervening protease cleavage sites between the components; a second nucleic
acid encodes the
pseudotyping viral envelope glycoprotein or antibody fragment; and a third
nucleic acid encodes
the gNA; or
(d) a first nucleic acid encodes the MA, an intervening protease cleavage
site, and the
CasX protein; a second nucleic acid encodes the pseudotyping viral envelope
g,lycoprotein or
antibody fragment; a third nucleic acid encodes the gNA; and a fourth nucleic
acid encodes the
protease.
[00730] Embodiment IV-17. The XDP system of Embodiment IV-15 or Embodiment IV-
16,
further comprising the CA component linked between the MA and the CasX protein
components
with intervening protease cleavage sites.
[00731] Embodiment IV-18. The XDP system of Embodiment IV-1, wherein the
encoded
components are selected from the gag polyprotein, the CasX protein, the
protease, the protease
cleavage site, the gNA, the pseudotyping viral envelope glycoprotein or
antibody fragment, and
the gag-poi polyprotein, wherein the components are encoded on two, three, or
four individual
nucleic acids.
[00732] Embodiment IV-19. The XDP system of Embodiment IV-18, wherein
(a) a first nucleic acid encodes the gag polyprotein, the CasX protein, the
protease,
and intervening protease cleavage sites between the components; and a second
nucleic acid
encodes the gag-pol polyprotein, the pseudotyping viral envelope glycoprotein
or antibody
fragment, and the gNA; or
(b) a first nucleic acid encodes the gag polyprotein, the intervening
protease cleavage
site, and the CasX protein; a second nucleic acid encodes the protease; and a
third nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment, the
gNA and the
gag-pol polyprotein; or
(c) a first nucleic acid encodes the gag polyprotein, the intervening
protease cleavage
site, and the CasX protein; a second nucleic acid encodes the protease; a
third nucleic acid
encodes the pseudotyping viral envelope glycoprotein or antibody fragment; and
a fourth nucleic
acid encodes the gNA and the gag-pol polyprotein.
[00733] Embodiment I1v-20. The XDP system of Embodiment IV-2 or Embodiment IV-
3,
comprising all or a portion of any one of the nucleic acid sequences of Tables
6-8.
246
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1007341 Embodiment IV-21. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the MA, the CA, the gag-TFR-PR polyprotein, the gag
polyprotein, and the
gag-pol polyprotein are derived from a retrovirus.
1007351 Embodiment IV-22. The XDP system of Embodiment IV-21, wherein the
retrovirus is
selected from the group consisting of an alpharetrovirus, a betaretrovirus, a
gammaretrovirus, a
deltaretrovirus, an epsilonretrovirus, and a lentivirus.
1007361 Embodiment IV-23. The XDP system of Embodiment IV-22, wherein the
lentivirus is
selected from the group consisting of human immunodeficiency-1 (H1V-1), human
immunodeficiency-2 (HIV-2), simian immunodeficiency virus (SIV), feline
immunodeficiency
virus (FLY), and bovine immunodeficiency virus (BIV).
1007371 Embodiment IV-24, The XDP system of Embodiment IV-23, wherein the
lentivirus is
HEY-1 or SIN.
1007381 Embodiment IV-25. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the gag polypeptide further comprises a p6 polypeptide.
1007391 Embodiment IV-26. The XDP system of any one Embodiments IV-1 to 25,
wherein the
gag polypeptide comprises a MA polypeptide, a CA polypeptide, a p2
polypeptide, an NC
polypeptide, a p1 polypeptide, and a p6 polypeptide, and wherein the gag
polyprotein comprises
one or more protease cleavage sites located between one or more of:
(a) the MA polypeptide and the CA polypeptide;
(b) the CA polypeptide and the p2 polypeptide;
(c) the p2 polypeptide and the NC polypeptide;
(d) the NC polypeptide and the p1 polypeptide; and
(e) the pl polypeptide and the p6 polypeptide.
1007401 Embodiment IV-27. The XDP system of any one Embodiments IV-1 to 26,
wherein the
protease capable of cleaving the protease cleavage site is selected from the
group of proteases
consisting of HIV-1 protease, tobacco etch virus protease (TEV), potyvirus HC
protease,
potyvirus P1 protease, PreScission, b virus NIa protease, B virus RNA-2-
encoded protease,
aphthovirus L protease, enterovirus 2A protease, rhinovirus 2A protease,
picoma 3C protease,
comovirus 24K protease, nepovirus 24K protease, RTSV (rice tungro spherical
virus) 3C-like
protease, PYVF (parsnip yellow fleck virus) 3C-like protease, cathepsin,
thrombin, factor Xa,
metalloproteinase-2 (MMP-2), MMP -3, MMP-7, MMP-9, MMP-10, MMP-11, and
enterokinase.
247
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1007411 Embodiment 1V-28. The XDP system of Embodiment 1V-27, wherein the
protease
capable of cleaving the protease cleavage site is HIV-1 protease.
1007421 Embodiment IV-29 The XDP system of any one of Embodiments IV-1 to 28,
wherein
the pseudotyping viral envelope glycoprotein is derived from an enveloped
virus selected from
the group consisting of Argentine hemorrhagic fever virus, Australian bat
virus, Autographa
californica multiple nucleopolyhedrovirus, Avian leukosis virus, baboon
endogenous virus,
Bolivian hemorrhagic fever virus, Boma disease virus, Breda virus, Bunyamwera
virus,
Chandipura virus, Chikungunya virus, Crimean-Congo hemorrhagic fever virus,
Dengue fever
virus, Duvenhage virus, Eastern equine encephalitis virus, Ebola hemorrhagic
fever virus, Ebola
Zaire virus, enteric adenovirus, Ephemerovirus, Epstein-Bar virus (EBV),
European bat virus 1,
European bat virus 2, Gibbon ape leukemia virus, Hantavirus, Hendra virus,
hepatitis A virus,
hepatitis B virus, hepatitis C virus, hepatitis D virus, hepatitis E virus,
hepatitis G Virus (GB
virus C), herpes simplex virus type 1, herpes simplex virus type 2, human
cytomegalovirus
(HEWS), human foamy virus, human herpesvirus (HMV), human Herpesvirus 7, human
herpesvirus type 6, human herpesvirus -type 8, human immunodeficiency virus 1
(HIV-1), human
metapneumovirus, human T-lymphotro pie virus 1, influenza A, influenza B,
influenza C virus,
Japanese encephalitis virus, Kaposi's sarcoma-associated herpesvirus (HHV8),
Kaysanur Forest
disease virus, La Crosse virus, Lagos bat virus, Lassa fever virus,
lymphocytic choriomeningitis
virus (LCMV), Machupo virus, Marburg hemorrhagic fever virus, measles virus,
Middle eastern
respiratory syndrome-related coronavirus, Mokola virus, Moloney murine
leukemia virus,
monkey pox, mouse mammary tumor virus, mumps virus, murine gammaherpesvirus,
Newcastle disease virus, Nipah virus, Nipah virus, Norwalk virus, Omsk
hemorrhagic fever
virus, papilloma virus, parvovirus, pseudorabies virus, Quaranfil virus,
rabies virus, RD114
endogenous feline retrovirus, respiratory syncytial virus (RSV), Rift Valley
fever virus, Ross
River virus, rotavirus, Rous sarcoma virus, rubella virus, Sabia-associated
hemorrhagic fever
virus, SARS-associated coronavirus (SARS-CoV), Sendai virus, Tacaribe virus,
Thogotovirus,
tick-borne encephalitis causing virus, varicella zoster virus (1111V3),
varicella zoster virus
(HEIV3), variola major virus, variola minor virus, Venezuelan equine
encephalitis virus,
Venezuelan hemorrhagic fever virus, vesicular stomatitis virus (VSV),
Vesiculovirus, West Nile
virus, western equine encephalitis virus, and Zika Virus.
1007431 Embodiment IV-30. The XDP system of Embodiment IV-29, wherein the
pseudotyping
viral envelope glycoprotein is derived from vesicular stomatitis virus (VSV).
248
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00744] Embodiment IV-31. The XDP system of any one of Embodiments IV-1-29,
wherein the
pseudotyping viral envelope glyc,oprotein comprises a sequence having at least
90%, at least
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence
identity to a
sequence selected from the group consisting of the sequences set forth in
Table 4.
[00745] Embodiment IV-32. The XDP system of any one of Embodiments IV-1-28,
wherein the
antibody fragment has binding affinity for a cell surface marker or receptor
of a target cell.
[00746] Embodiment IV-33. The XDP system of Embodiment IV-32, wherein the
antibody
fragment is a scFv.
[00747] Embodiment IV-34. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the gNA is a single-molecule guide RNA comprising a
scaffold sequence
and a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic
acid sequence.
[00748] Embodiment IV-35. The XDP system of Embodiment IV-29, wherein the
guide RNA
scaffold sequence has at least 80%, at least 90%, at least 95%, at least 96%,
at least 97%, at least
98%, at least 99%, or 100% sequence identity to a sequence selected from the
group of
sequences consisting of SEQ ID NOS: 4, 5, and 2101-2241.
[00749] Embodiment IV-36. The XDP system of Embodiment IV-29 or Embodiment D/-
35,
wherein the targeting sequence of the guide RNA consists of 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides.
[00750] Embodiment IV-37. The XDP system of Embodiment IV-36, wherein the
targeting
sequence of the guide RNA consists of 20 nucleotides.
[00751] Embodiment IV-38. The XDP system of Embodiment IV-36, wherein the
targeting
sequence of the guide RNA consists of 19 nucleotides.
[00752] Embodiment IV-39. The XDP system of Embodiment IV-36, wherein the
targeting
sequence of the guide RNA consists of 18 nucleotides.
[00753] Embodiment D/-40. The XDP system of Embodiment IV-36, wherein the
targeting
sequence of the guide RNA consists of 17 nucleotides.
[00754] Embodiment IV-41, The XDP system of Embodiment IV-36, wherein the
targeting
sequence of the guide RNA consists of 16 nucleotides.
[00755] Embodiment D/-42. The XDP system of Embodiment IV-36, wherein the
targeting
sequence of the guide RNA consists of 15 nucleotides.
249
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00756] Embodiment IV-43. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the guide RNA further comprises one or more ribozymes.
[00757] Embodiment IV-44. The XDP system of Embodiment IV-43, wherein the one
or more
ribozymes are independently fused to a terminus of the guide RNA.
[00758] Embodiment IV-45. The XDP system of Embodiment IV-43 or Embodiment IV-
44,
wherein at least one of the one or more ribozymes is a hepatitis delta virus
(HDV) ribozyme,
hammerhead ribozyme, pistol ribozyme, hatchet ribozyme, or tobacco ringspot
virus (TRSV)
ribozyme.
[00759] Embodiment IV-46. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the guide RNA is chemically modified.
[00760] Embodiment IV-47. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the CasX protein comprises a sequence having at least
about 90%, or at least
about 91%, or at least about 92%, or at least about 93%, or at least about
94%, or at least about
95%, or at least about 96%, or at least about 97%, or at least about 98%, or
at least about 99%,
or at least 100% sequence identity to a sequence selected from the group
consisting of the
sequences set forth in Table 1.
[00761] Embodiment IV-48. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the CasX protein has binding affinity for a protospacer
adjacent motif
(PAM) sequence selected from the group consisting of TTC, ATC, GTC, and CTC.
[00762] Embodiment IV-49. The XDP system of Embodiment IV-48, wherein the
binding
affinity of the CasX protein for the PAM sequence is at least 1.5-fold greater
compared to the
binding affinity of any one of the CasX proteins of SEQ ID NOS: 1-3 for the
PAM sequences.
[00763] Embodiment IV-50. The XDP system of any one Embodiments IV-1 to 49,
wherein,
wherein the CasX protein further comprises one or more nuclear localization
signals (NLS).
[00764] Embodiment IV-51. The XDP system of Embodiment IV-50, wherein the one
or more
NLS are selected from the group of sequences consisting of PKKKRKV,
KRPAATKKAG-QAKICKK, PAAKRVKLD, RQRRNELKRSP,
NQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGY,
RMRIZFICNKGKDTAELRRRRVEVSVELRKAKICDEQILKRRNV, VSRICRPRP,
PPIUCARED, PQPKKICPL, SALLKKKKKMAP, DRLRR, PKQKKRK, RICLICKKIKKL,
REKKKFLICRR, ICRKGDEVDGVDEVAKICKSKK, RKCLQAGMNLEARKTICK,
PRPRKIPR, PPRK1CRTVV, NLSKICICKRKREK, RRPSRPFRKP, KRPRSPSS,
250
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
KRGINDRNFWRGENERKTR, PRPPICMARYDN, KRSFSKAF, KLKIKRPVK,
PKTRRRPRRSQRICRPPT, RRKKRRPRRKKRR, PKKKSRKPKICKSRK,
HICICKUPDASVNFSEFSK, QRPGPYDRPQRPGPYDRP, LSPSLSPLLSPSLSPL,
RGKGGKGLGKGGAKRHRK, PICRGRGRPKRGRGR, and
MSRRRKANPTICLSENAICKLAKEVEN.
[00765] Embodiment IV-52. The CasX variant of Embodiment IV-50 or Embodiment
IV-51,
wherein the one or more NLS are fused to the C-terminus of the CasX protein.
[00766] Embodiment IV-53. The CasX variant of Embodiment IV-50 or Embodiment
IV-51,
wherein the one or more NLS are fused to the N-terminus of the CasX protein.
[00767] Embodiment IV-54, The CasX variant of Embodiment IV-50 or Embodiment
IV-51,
wherein the one or more NLS are fused to the N-terminus and C-terminus of the
CasX protein.
[00768] Embodiment IV-55. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, wherein the CasX protein comprises a nuclease domain having
nickase activity.
1007691 Embodiment IV-56. The XDP system of any one of Embodiments IV-I-54,
wherein the
CasX protein comprises a nuclease domain having double-stranded cleavage
activity.
[00770] Embodiment IV-57. The XDP system of any one Embodiments IV-1 to 56,
further
comprising a nucleic acid encoding a retroviral packaging signal.
[00771] Embodiment IV-58. The XDP system of any one of the preceding
embodiments of Set
I of Set IV, further comprising a donor template nucleic acid complementary to
a target nucleic
acid.
[00772] Embodiment IV-59. The XDP system of Embodiment IV-58, wherein the
donor
template comprises two homologous arms complementary to sequences flanking a
cleavage site
in the target nucleic acid.
[00773] Embodiment IV-60. The XDP system of Embodiment IV-58 or Embodiment IV-
59,
wherein the donor template nucleic acid sequence comprises a corrective
sequence for a
mutation in the target nucleic acid.
[00774] Embodiment IV-61 The XDP system of Embodiment IV-58 or Embodiment W-
59,
wherein the donor template nucleic acid sequence comprises a mutation compared
to the target
nucleic acid.
[00775] Embodiment IV-62. The XDP system of Embodiment IV-61, where the
mutation is an
insertion, a deletion, or a substitution of one or more nucleotides in the
donor template nucleic
acid sequence.
251
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00776] Embodiment IV-63. The XDP system of any one of Embodiments IV-1-54,
wherein the
CasX protein is a catalytically inactive CasX (dCasX) protein, and wherein the
dCasX and the
guide RNA retain the ability to bind to the target nucleic acid.
[00777] Embodiment IV-64. The XDP system of Embodiment IV-63, wherein the
dCasX
comprises a mutation at residues:
(a) D672, E769, and/or D935 corresponding to the CasX protein of SEQ ID NO:
1;
or
(b) D659, E756 and/or D922 corresponding to the CasX protein of SEQ ID NO:
2.
[00778] Embodiment IV-65. The XDP system of Embodiment IV-64, wherein the
mutation is a
substitution of alanine for the residue.
[00779] Embodiment IV-66. A eukaryotic cell comprising the XDP system of any
one of the
preceding embodiments of Set I of Set IV.
[00780] Embodiment IV-67. The eukaryotic cell of Embodiment IV-66, wherein the
cell is a
packaging cell.
[00781] Embodiment IV-68. The eukaryotic cell of any one of Embodiments IV-66
or
Embodiment IV-67, wherein the eukaryotic cell is selected from the group
consisting of
HEK293 cells, Lenti-X 293T cells, BHK cells, HepG2, Saos-2, HuH7, NSO cells,
SP2/0 cells,
YO myeloma cells, A549 cells, P3X63 mouse myeloma cells, PER cells, PER.C6
cells,
hybridoma cells, VERO, NIH3T3 cells, COS, WI38, MRCS, A549, HeLa cells, CHO
cells, and
HT1080 cells.
[00782] Embodiment IV-69. The eukaryotic cell of Embodiment IV-67 or
Embodiment IV-68,
wherein the packaging cell comprises one or more mutations to reduce
expression of a cell
surface marker.
[00783] Embodiment IV-70. The eukaryotic cell of any one of Embodiments IV-66-
69, wherein
all or a portion of the nucleic acids encoding the XDP system of any one of
Embodiments IV-1-
56 are integrated into the genome of the eukaryotic cell.
[00784] Embodiment IV-71, A method of making an XDP comprising a CasX protein
and a
gNA, the method comprising:
(a) propagating the packaging cell of any one of Embodiments IV-67-70 under
conditions such that XDPs are produced; and
(b) harvesting the XDPs produced by the packaging cell.
[00785] Embodiment IV-72. An XDP produced by the method of Embodiment IV-71.
252
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00786] Embodiment IV-73. An XDP comprising one or more components selected
from:
(a) a matrix polypeptide (MA);
(b) a capsid polypeptide (CA);
(c) a gag polyprotein comprising a matrix polypeptide (MA), a capsid
polypeptide
(CA), and a nucleocapsid polypeptide (NC);
(d) a CasX protein;
(e) a guide nucleic acid (gNA);
(f) a pseudotyping viral envelope glycoprotein or antibody fragment that
provides
for binding and fusion of the XDP to a target cell; and
(g) an RNA binding domain;
[00787] Embodiment IV-74, The XDP of Embodiment IV-73, wherein the XDP
comprises
(a) the matrix polypeptide (MA);
(b) the pseudotyping viral envelope glycoprotein or antibody fragment; and
(c) the CasX and the gNA contained within the XDP.
[00788] Embodiment IV-75. The XDP of Embodiment IV-74, further comprising the
capsid
polypeptide (CA).
[00789] Embodiment IV-76. The XDP of Embodiment IV-74 or Embodiment IV-75,
further
comprising the nucleocapsid polypeptide (NC).
[00790] Embodiment IV-77. The XDP of any one of Embodiments IV-74-76, further
comprising an RNA binding domain.
[00791] Embodiment IV-78. The XDP of Embodiment IV-77, wherein the RNA binding
domain is a retroviral Psi packaging element inserted into the gNA or is a
protein selected from
the group consisting of MS2, PP7 or Qbeta, U1 A, phage replication loop,
kissing loop a, kissing
loop_b I, kissing loop_b2, G quadriplex M3q, G quadriplex telomere basket,
sarcin-ricin loop,
and pseudoknots.
[00792] Embodiment IV-79. The XDP of any one of Embodiments IV-74-78, wherein
the CasX
and the gNA are associated together in a ribonuclear protein complex (RNP)
within the XDP.
[00793] Embodiment IV-80 The XDP of any one of Embodiments IV-74-79,
comprising the
CasX of any one of Embodiments IV-47-65 and the guide RNA of any one of
Embodiments IV-
34-46.
[00794] Embodiment IV-81. The XDP of any one of Embodiments IV-74-80, wherein
the
pseudotyping viral envelope glycoprotein comprises a sequence having at least
90%, at least
253
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
95%, at least 96%, at least 97%, at least 98%, at least 99%, or 100% sequence
identity to a
sequence selected from the group consisting of the sequences set forth in
Table 4.
1007951 Embodiment IV-82 The XDP of any one of Embodiments IV-73-80, wherein
the
pseudotyping viral envelope glycoprotein is derived from an enveloped virus
selected from the
group consisting of Argentine hemorrhagic fever virus, Australian bat virus,
Autographa
californica multiple nucleopolyhedrovirus, Avian leukosis virus, baboon
endogenous virus,
Bolivian hemorrhagic fever virus, Boma disease virus, Breda virus, Bunyamwera
virus,
Chandipura virus, Chikungunya virus, Crimean-Congo hemorrhagic fever virus,
Dengue fever
virus, Duvenhage virus, Eastern equine encephalitis virus, Ebola hemorrhagic
fever virus, Ebola
Zaire virus, enteric adenovirus, Ephemerovirus, Epstein-Bar virus (EBV),
European bat virus 1,
European bat virus 2, Gibbon ape leukemia virus, Hantavirus, Hendra virus,
hepatitis A virus,
hepatitis B virus, hepatitis C virus, hepatitis D virus, hepatitis E virus,
hepatitis G Virus (GB
virus C), herpes simplex virus type 1, herpes simplex virus type 2, human
cytomegalovirus
(HEWS), human foamy virus, human herpesvirus (HMV), human Herpesvirus 7, human
herpesvirus type 6, human herpesvirus -type 8, human immunodeficiency virus 1
(HIV-1), human
metapneumovirus, human T-Iymphotro pie virus 1, influenza A, influenza B,
influenza C virus,
Japanese encephalitis virus, Kaposi's sarcoma-associated herpesvirus (HHV8),
Kaysanur Forest
disease virus, La Crosse virus, Lagos bat virus, Lassa fever virus,
lymphocytic choriomeningitis
virus (LCMV), Machupo virus, Marburg hemorrhagic fever virus, measles virus,
Middle eastern
respiratory syndrome-related coronavirus, Mokola virus, Moloney murine
leukemia virus,
monkey pox, mouse mammary tumor virus, mumps virus, murine gammaherpesvirus,
Newcastle disease virus, Nipah virus, Nipah virus, Norwalk virus, Omsk
hemorrhagic fever
virus, papilloma virus, parvovirus, pseudorabies virus, Quaranfil virus,
rabies virus, RD114
endogenous feline retrovirus, respiratory syncytial virus (RSV), Rift Valley
fever virus, Ross
River virus, rotavirus, Rous sarcoma virus, rubella virus, Sabia-associated
hemorrhagic fever
virus, SARS-associated coronavirus (SARS-CoV), Sendai virus, Tacaribe virus,
Thogotovirus,
tick-borne encephalitis causing virus, varicella zoster virus (HEIV3),
varicella zoster virus
(HIIV3), variola major virus, variola minor virus, Venezuelan equine
encephalitis virus,
Venezuelan hemorrhagic fever virus, vesicular stomatitis virus (VSV),
Vesiculovirus, West Nile
virus, western equine encephalitis virus, and Zika Virus.
1007961 Embodiment IV-83. The XDP of any one of Embodiments IV-73-82, further
comprising the donor template nucleic acid sequence of any one of Embodiments
IV-58-62.
254
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00797] Embodiment IV-84. A method of method of modifying a target nucleic
acid sequence
in a cell, the method comprising contacting the cell with the XDP of any one
of Embodiments
IV-73-83, wherein said contacting comprises introducing into the cell the CasX
protein, the
guide RNA, and, optionally, the donor template nucleic acid sequence,
resulting in modification
of the target nucleic acid sequence.
[00798] Embodiment IV-85. The method of Embodiment IV-84, wherein the
modification
comprises introducing one or more single-stranded breaks in the target nucleic
acid sequence_
[00799] Embodiment IV-86. The method of Embodiment IV-84, wherein the
modification
comprises introducing one or more double-stranded breaks in the target nucleic
acid sequence.
[00800] Embodiment IV-87, The method of any one of Embodiments W-84-86,
wherein the
modification comprises insertion of the donor template into the target nucleic
acid sequence.
[00801] Embodiment IV-88. The method of any one of Embodiments W-84-87,
wherein the
cell is modified in vitro.
[00802] Embodiment IV-89. The method of any one of Embodiments IV-84-87,
wherein the
cell is modified in viva
[00803] Embodiment IV-90. The method of Embodiment D/-89, wherein the XDP is
administered to a subject.
[00804] Embodiment IV-91. The method of Embodiment IV-90, wherein the subject
is the
subject is selected from the group consisting of mouse, rat, pig, non-human
primate, and human.
[00805] Embodiment IV-92. The method of Embodiment IV-90 or Embodiment W-91,
wherein
the XDP is administered by a route of administration selected from the group
consisting of
subcutaneous, intradermal, intraneural, intranodal, intramedullary,
intramuscular, intravenous,
intracerebroventricular, intracisternal, intrathecal, intracranial,
intralumbar, intratracheal,
intraosseous, inhalatory, intracontralateral striatum, intraocular,
intravitreal, intralymphatical,
intraperitoneal routes and sub-retinal routes.
[00806] Embodiment IV-93. The method of any one of Embodiments W-90-92,
wherein the
XDP is administered to the subject using a therapeutically effective dose
[00807] Embodiment IV-94 The method of Embodiment W-93, wherein the XDP is
administered at a dose of at least about 1 x 105 particles, or at least about
1 x 106 particles, or at
least about 1 x 10 particles, or at least about 1 x 108 particles, or at least
about 1 x 109 particles,
or at least about 1 x 1010 particles, or at least about 1 x 10" particles, or
at least about 1 x 1012
255
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
particles, or at least about 1 x 10" particles, or at least about 1 x 1014
particles, or at least about 1
x 1015 particles, or at least about 1 x 1016 particles.
[00808] Embodiment IV-95. A method for introducing a CasX and gNA RNP into a
cell having
a target nucleic acid, comprising contacting the cell with the XDP of any one
of Embodiments
IV-79-83, such that the RNP enters the cell.
[00809] Embodiment IV-96. The method of Embodiment IV-95, wherein the RNP
binds to the
target nucleic acid.
[00810] Embodiment IV-97. The method of Embodiment IV-96, wherein the target
nucleic acid
is cleaved by the CasX.
[00811] Embodiment IV-98, The method of any one of Embodiments B1-95-97,
wherein the
cell is modified in vitro.
[00812] Embodiment IV-99. The method of any one of Embodiments IV-95-97,
wherein the
cell is modified in viva
[00813] Embodiment IV-100. The method of
Embodiment IV-99, wherein the XDP is
administered to a subject.
[00814] Embodiment IV-101. The method of
Embodiment IV-100, wherein the subject is
the subject is selected from the group consisting of mouse, rat, pig, non-
human primate, and
human.
[00815] Embodiment IV-102. The method of any one
of Embodiments IV-99-101,
wherein the XDP is administered to the subject using a therapeutically
effective dose.
[00816] Embodiment IV-103. The method of
Embodiment IV-102, wherein the XDP is
administered at a dose of at least about 1 x 105 particles, or at least about
1 x 106 particles, or at
least about 1 x 107 particles, or at least about 1 x 108 particles, or at
least about 1 x 109 particles,
oral least about 1 x 1010 particles, or at least about 1 x 1011 particles, or
at least about 1 x 1012
particles, or at least about 1 x 10E3 particles, or at least about 1 x 1014
particles, or at least about 1
x 1015 particles, or at least about 1 x 10' particles.
Set V
[00817] Embodiment V-1. A delivery particle (XDP) system comprising one or
more nucleic
acids encoding:
(a) one or more retroviral components;
(b) a therapeutic payload; and
256
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(c) a tropism factor
[00818] Embodiment V-2. The XDP system of Embodiment V-1, wherein the tropism
factor
is selected from the group consisting of a glycoprotein, an antibody fragment,
a receptor, and a
ligand to a target cell marker.
[00819] Embodiment V-3. The XDP system of Embodiment V-2, wherein the tropism
factor
is a glycoprotein having an encoding sequence selected from the group
consisting of SEQ 1D
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501,
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595 as set forth in Table 4, or a sequence having at
least about 85%, at
least about 90%, at least about 91%, at least about 92%, at least about 93%,
at least about 94%,
at least about 95%, at least about 96%, at least about 97%, at least about
98%, or at least about
99% sequence identity thereto.
[00820] Embodiment V-4. The XDP system of Embodiment V-2, wherein the tropism
factor
is a glycoprotein having an encoding sequence selected from the group
consisting of SEQ ID
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595.
[00821] Embodiment V-5. The XDP system of any one of the preceding embodiments
of Set
V, wherein the therapeutic payload comprises a protein, a nucleic acid, or
comprises both a
protein and a nucleic acid.
[00822] Embodiment V-6. The XDP system of Embodiment V-5, wherein the protein
payload
is selected from the group consisting of a cytokine, an interleukin, an
enzyme, a receptor, a
microprotein, a hormone, erythropoietin, ribonuclease (RNAse),
deoxyribonuclease (DNAse), a
blood clotting factor, an anticoagulant, a bone morphogenetic protein, an
engineered protein
scaffold, a thrombolytic protein, a CRISPR protein, and an anti-cancer
modality.
[00823] Embodiment V-7. The XDP system of Embodiment V-6, wherein the CR1SPR
protein is a Class 1 or Class 2 CRISPR protein.
257
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1008241 Embodiment V-8, The XDP system of Embodiment V-7, wherein the CRISPR
protein is a Class 2 CRISPR protein selected from the group consisting of a
Type II, a Type V,
or a Type VI protein.
[00825] Embodiment V-9. The XDP system of Embodiment V-8, wherein the CRISPR
protein is a Type V protein selected from the group consisting of Cas12a,
Cas12b, Cas12c,
Cas12d (CasY), Cas12j and CasX.
[00826] Embodiment V-10. The XDP system of Embodiment V-9, wherein the CRISPR
protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345, 350-
353, 355-367
or 388-397, or a sequence having at least about 85%, at least about 90%, at
least about 91%, at
least about 92%, at least about 93%, at least about 94%, at least about 95%,
at least about 96%,
at least about 97%, at least about 98%, or at least about 99% sequence
identity thereto.
[00827] Embodiment V-11. The XDP system of Embodiment V-5, wherein the
therapeutic
payload comprises a nucleic acid selected from the group consisting of a
single-stranded
antisense oligonucleotide (AS0s), a double-stranded RNA interference (RNAi)
molecule, a
DNA aptamer, and a CRISPR guide nucleic acid.
[00828] Embodiment V-12. The XDP system of Embodiment V-11, wherein the CRISPR
guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and a
targeting sequence, wherein the targeting sequence comprises between 14 and 30
nucleotides
and is complementary to a target nucleic acid sequence.
[00829] Embodiment V-13. The XDP system of Embodiment V-12, wherein the
scaffold
sequence comprises a sequence of SEQ ID NOS: 597-781 as set forth in Table 3,
or a sequence
having at least about 85%, at least about 90%, at least about 91%, at least
about 92%, at least
about 93%, at least about 94%, at least about 95%, at least about 96%, at
least about 97%, at
least about 98%, or at least about 99% sequence identity thereto.
[00830] Embodiment V-14. The XDP system of Embodiment V-13, wherein the
scaffold
sequence comprises a sequence of SEQ ID NOS: 597-781.
[00831] Embodiment V-15. The XDP system of any one of the preceding
embodiments of Set
V, wherein the nucleic acids further encode one or more components selected
from:
(a) all or a portion of a retroviral gag polyprotein;
(b) one or more protease cleavage sites;
(c) a gag-transframe region-pol protease polyprotein (gag-TFR-PR);
(d) a retroviral gag-pot polyprotein; and
258
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(e) a non-retroviral protease capable of cleaving the
protease cleavage sites.
[00832] Embodiment V-16. The XDP system of any one of the preceding
embodiments of Set
V, wherein one or more of the retroviral components are derived from an
Orthoretrovirinae virus
or a Spumaretrovirinae virus.
[00833] Embodiment V-17. The XDP system of Embodiment V-16, wherein the
Orthoretrovirinae virus is selected from the group consisting of an
Alpharetrovirus,
Betaretrovirus, Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus, and
Lentivirus.
[00834] Embodiment V-18. The XDP system of Embodiment V-16, wherein the
Spumaretrovirinae virus is selected from the group consisting of
Bovispumavirus,
Equispumavirus, Felispumavirus, Prosimiispumavirus, Simiispumavirus, or
Spumavirus.
[00835] Embodiment V-19. The XDP system of any one of the preceding
embodiments of Set
V. wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoded on two nucleic acids;
(c) the components are encoded on three nucleic acids;
(d) the components are encoded on four nucleic acids; or
(e) the components are encoded on five nucleic acids.
[00836] Embodiment V-20. The XDP system of Embodiment V-19, wherein the one or
more
of the components encoded by the nucleic acids are configured according to any
one of FIGS.
36-68.
[00837] Embodiment V-21. The XDP system of Embodiment V-19 or Embodiment V-20,
wherein the one or more of the retroviral components are encoded by a nucleic
acid selected
from the group of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-
201, 782, and
234-339 as set forth in Table 5.
[00838] Embodiment V-22. The XDP system of any one of the preceding
embodiments of Set
V, wherein the components are capable of self-assembling into an XDP when the
one or more
nucleic acids are introduced into a eukaryotic host cell and are expressed.
[00839] Embodiment V-23, The XDP of Embodiment V-22, wherein the therapeutic
payload is
encapsidated within the XDP upon self-assembly of the XDP.
[00840] Embodiment V-24. The XDP system of Embodiment V-23, wherein the
therapeutic
payload comprises a CasX and a guide RNA complexed as a ribonucleoprotein
complex (RNP)
and, optionally, a donor template.
259
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00841] Embodiment V-25. The XDP of Embodiment V-22, wherein the tropism
factor is
incorporated on the XDP surface upon self-assembly of the XDP.
[00842] Embodiment V-26. The XDP system of Embodiment V-25, wherein the
tropism factor
confers preferential interaction of the XDP with the cell surface of a target
cell and facilitates
entry of the XDP into the target cell
[00843] Embodiment V-27. An XDP system comprising one or more nucleic acids
encoding
components:
(a) all or a portion of an Alpharetrovirus gag polyprotein;
(b) a therapeutic payload; and
(c) a tropism factor.
[00844] Embodiment V-28. The XDP system of Embodiment V-27, wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a P2A peptide, a P2B peptide, a P10 peptide, a capsid
polypeptide (CA), and
a nucleocapsid polypeptide (NC).
[00845] Embodiment V-29. The XDP system of Embodiment V-28, wherein the gag
polyprotein comprises, from N-terminus to C-terminus, a matrix polypeptide
(MA), a P2A
peptide, a P28 peptide, a P10 peptide, a capsid polypeptide (CA), and a
nucleocapsid
polypeptide (NC).
[00846] Embodiment V-30. The XDP system of any one of Embodiments V-27-29,
wherein
the one or more nucleic acids encode one or more components selected from
(a) an HIV pl peptide;
(b) an HIV p6 peptide;
(c) a Gag-Pol polyprotein;
(d) one or more protease cleavage sites;
(e) a non-retroviral, heterologous protease capable of cleaving the
cleavage sites; and
(f) a gag-transframe region-pol protease polyprotein.
[00847] Embodiment V-31. The XDP system of any one of Embodiments V-27-30,
wherein
the tropism factor is selected from the group consisting of a glycoprotein, an
antibody fragment,
a receptor, and a ligand to a target cell marker.
[00848] Embodiment V-32. The XDP system of Embodiment V-31, wherein the
tropism factor
is a glycoprotein having an encoding sequence selected from the group
consisting of SEQ ID
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
260
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595, or a sequence having at least about 85%, at least
about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[00849] Embodiment V-33. The XDP system of Embodiment V-31, wherein the
tropism factor
is a glycoprotein having an encoding sequence selected from the group of
sequences consisting
of SEQ ID NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459,
461, 463, 465, 467,
469, 471, 473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497,
499, 501, 503, 505,
507, 509, 511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535,
537, 539, 541, 543,
545, 547, 549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573,
575, 577, 579, 581,
583, 585, 587, 589, 591, 593 and 595 as set forth in Table 4.
[00850] Embodiment V-34. The XDP system of Embodiment V-33, wherein the
tropism factor
is glycoprotein G from vesicular stomatitis virus (VSV-G), optionally wherein
the VSV-G
glycoprotein comprises a sequence of SEQ ID NO: 438.
[00851] Embodiment V-35. The XDP system of any one of Embodiments V-27-34,
wherein
the therapeutic payload comprises a protein, a nucleic acid, or comprises both
a protein and a
nucleic acid.
[00852] Embodiment V-36. The XDP system of Embodiment V-35, wherein the
protein
payload is selected from the group consisting of a cytokine, an interleukin,
an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
thrombolytic
protein, a CRISPR protein, and an anti-cancer modality.
[00853] Embodiment V-37. The XDP system of Embodiment V-36, wherein the CRISPR
protein is a Class 1 or Class 2 CRISPR protein.
[00854] Embodiment V-38, The XDP system of Embodiment V-37, wherein the CRISPR
protein is a Class 2 CRISPR protein selected from the group consisting of Type
II, Type V, or
Type VI protein.
261
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00855] Embodiment V-39. The XDP system of Embodiment V-38, wherein the CRISPR
protein is a Type V protein selected from the group consisting of Cas12a,
Cas12b, Cas12c,
Caslal (CasY), Cas12j and CasX.
1008561 Embodiment V-40. The XDP system of Embodiment V-39, wherein the CRISPR
protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345, 350-
353, 355-367
or 388-397, or a sequence having at least about 85%, at least about 90%, at
least about 91%, at
least about 92%, at least about 93%, at least about 94%, at least about 95%,
at least about 96%,
at least about 97%, at least about 98%, or at least about 99% sequence
identity thereto.
1008571 Embodiment V-41. The XDP system of Embodiment V-39, wherein the CRISPR
protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345, 350-
353, 355-367
or 388-397.
[00858] Embodiment V-42. The XDP system of any one of Embodiments V-39-41,
wherein
the CasX further comprises one or more NLS selected from the group of
sequences consisting of
PKKKRKV (SEQ ID NO: 130), KRPAATKKAGQAKKKK (SEQ ID NO: 131),
PAAKRVKLD (SEQ ID NO: 132), RQRRNELKRSP (SEQ ID NO: 133),
NQSSNFGPMKGGNFGGRSSGPYGGGGQYFAKPRNQGGY (SEQ ID NO: 134),
RMRIZFKNKGKDTAELRRRRVEVSVELRKAKICDEQILKRRNV (SEQ ID NO: 135),
VSR1CRPRP (SEQ ID NO: 136), PPICKARED (SEQ ID NO: 137), PQPICKICPL (SEQ ID NO:
138), SALIICKICICKMAP (SEQ ID NO: 139), DRLRR (SEQ ID NO: 140), PKQKKRK (SEQ
ID NO: 141), RICLKKKIKKL (SEQ ID NO: 142), REKKICFLKRR (SEQ ID NO: 143),
KRKGDEVDGVDEVAICKKSICIC (SEQ ID NO: 144), RKCLQAGMNLEARKTKK (SEQ ID
NO: 145), PRPRK1PR (SEQ ID NO: 146), PPRKICRTVV (SEQ ID NO: 147),
NLSKICKICRICREK (SEQ ID NO: 148), RRPSRPFRKP (SEQ ID NO: 149), KRPRSPSS (SEQ
ID NO: 150), ICR.GINDRNFWRGENERKTR (SEQ ID NO: 151), PRPPICMARYDN (SEQ ID
NO: 152), KRSFSKAF (SEQ ID NO: 153), KLKIKRPVK (SEQ ID NO: 154),
PKTRRRPRRSQRICRPPT (SEQ ID NO: 156), RRKKRRPRRICICRR (SEQ ID NO: 159),
P1CKKSRKPKICKSRK (SEQ ID NO: 160), HICICKHPDASVNFSEFSK (SEQ ID NO: 161),
QRPGPYDRPQRPGPYDRP (SEQ ID NO: 162), LSPSLSPLLSPSLSPL (SEQ ID NO: 163),
RGKGGKGLGKGGAKRHRK (SEQ ID NO: 164), PKRGRGRPKRGRGR (SEQ ID NO: 165),
MSRRRKANPTICLSENAICKLAKEVEN (SEQ ID NO: 157), PKKICRKVPPPPAA1CRVKLD
(SEQ ID NO: 155), and PICKKRKVPPPPICKKRKV (SEQ ID NO: 166), wherein the NLS
are
located at or near the N-terminus and/or the C-terminus.
262
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00859] Embodiment V-43. The XDP system of Embodiment V-35, wherein the
therapeutic
payload is a nucleic acid selected from the group consisting of a single-
stranded antisense
oligonucleotide (AS0s), a double-stranded RNA interference (RNAi) molecule, a
DNA aptamer,
and a CRISPR guide nucleic acid.
[00860] Embodiment V-44. The XDP system of Embodiment V-43, wherein the CRISPR
guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and a
targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
[00861] Embodiment V-45. The XDP system of Embodiment V-44, wherein the
scaffold
sequence comprises a sequence of SEQ ID NOS: 597-781, or a sequence having at
least about
85%, at least about 90%, at least about 91%, at least about 92%, at least
about 93%, at least
about 94%, at least about 95%, at least about 96%, at least about 97%, at
least about 98%, or at
least about 99% sequence identity thereto.
[00862] Embodiment V-46. The XDP system of Embodiment V-45, wherein the
scaffold
sequence of the guide RNA comprises a sequence of SEQ ID NOS: 597-781.
[00863] Embodiment V-47. The XDP system of any one of Embodiments V-44-46,
wherein
the targeting sequence of the guide RNA consists of 14, 15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25,
26, 27, 28, 29, or 30 consecutive nucleotides.
[00864] Embodiment V-48. The XDP system of any one of Embodiments V-27-47,
wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoding on two nucleic acids;
(c) the components are encoding on three nucleic acids;
(d) the components are encoding on four nucleic acids; or
(e) the components are encoding on five nucleic acids.
1008651 Embodiment V-49. The XDP system of Embodiment V-48, wherein the one or
more
of the components encoded by the nucleic acids are configured according to any
one of FIGS.
36-68.
[00866] Embodiment V-50. The XDP system of Embodiment V-48 or Embodiment V-49,
wherein the one or more of the components are encoded by nucleic acids
selected from the group
of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-201, 782, 234-
339, 880-933,
and 947-1000 as set forth in Tables 5, 24, 27, 30, and 33, or sequences having
at least about
85%, at least about 90%, at least about 91%, at least about 92%, at least
about 93%, at least
263
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
about 94%, at least about 95%, at least about 96%, at least about 97%, at
least about 98%, or at
least about 99% sequence identity thereto.
[00867] Embodiment V-51. The XDP system of any one of Embodiments V-27-50,
wherein
the components are capable of self-assembling into an XDP when the one or more
nucleic acids
are introduced into a eukaryotic host cell and are expressed.
[00868] Embodiment V-52. The XDP of Embodiment V-51, wherein the therapeutic
payload is
encapsidated within the XDP upon self-assembly of the XDP.
[00869] Embodiment V-53. The XDP system of Embodiment V-52, wherein the
therapeutic
payload comprises a CasX and a guide RNA complexed as a ribonucleoprotein
complex (RNP)
and, optionally, a donor template.
[00870] Embodiment V-54. The XDP of Embodiment V-51, wherein the tropism
factor is
incorporated on the XDP surface upon self-assembly of the XDP.
[00871] Embodiment V-55. The XDP system of Embodiment V-54, wherein the
tropism factor
confers preferential interaction with the cell surface of a target cell and
facilitates entry of the
XDP into the target cell.
[00872] Embodiment V-56. An XDP system comprising one or more nucleic acids
encoding
components:
(a) all or a portion of an Betaretrovirus gag polyprotein;
(b) a therapeutic payload; and
(c) a tropism factor.
[00873] Embodiment V-57. The XDP system of Embodiment V-56, wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a PP21/24 peptide, a P12/P3/P8 peptide, a capsid polypeptide
(CA), and a
nucleocapsid polypeptide (NC).
[00874] Embodiment V-58. The XDP system of Embodiment V-56, wherein the gag
polyprotein comprises, from N-terminus to C-terminus, a matrix polypeptide
(MA), a PP21/24
peptide, a P12/P3/P8 peptide, a capsid polypeptide (CA), and a nucleocapsid
polypeptide (NC).
[00875] Embodiment V-59, The XDP system of any one of Embodiments V-56-58,
wherein
the nucleic acids further encode one or more components selected from
(a) an HIV pi peptide;
(b) an HIV p6 peptide;
(c) a Gag-Pol polyprotein;
264
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(d) one or more protease cleavage sites;
(e) a non-retroviral, heterologous protease capable of cleaving the
cleavage sites; and
(f) a gag-transframe region-pol protease polyprotein.
[00876] Embodiment V-60. The XDP system of any one of Embodiments V-56-59,
wherein
the tropism factor is selected from the group consisting of a glycoprotein, an
antibody fragment,
a receptor, and a ligand to a target cell marker.
[00877] Embodiment V-61. The XDP system of Embodiment V-60, wherein the
tropism factor
is a glycoprotein having an encoding sequence selected from the group
consisting of SEQ ID
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595, or a sequence having at least about 85%, at least
about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[00878] Embodiment V-62. The XDP system of Embodiment V-61, wherein the
tropism factor
is a glycoprotein having a sequence selected from the group consisting of SEQ
ID NOS: 437,
439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463, 465, 467,
469, 471, 473, 475,
477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ 503, 505,
507, 509, 511, 513,
515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539, 541, 543,
545, 547, 549, 551,
553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577, 579, 581,
583, 585, 587, 589,
591, 593 and 595.
[00879] Embodiment V-63. The XDP system of Embodiment V-62, wherein the
tropism factor
is glycoprotein G from vesicular stomatitis virus (VSV-G).
1008801 Embodiment V-64. The XDP system of any one of Embodiments V-56-63,
wherein
the therapeutic payload comprises a protein, a nucleic acid, or comprises both
a protein and a
nucleic acid.
1008811 Embodiment V-65. The XDP system of Embodiment V-64, wherein the
protein
payload is selected from the group consisting of a cytokine, an interleukin,
an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
265
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
thrombolytic
protein, a CRISPR protein, and an anti-cancer modality.
[00882] Embodiment V-66. The XDP system of Embodiment V-65, wherein the CRISPR
protein is a Class 1 or Class 2 CRISPR protein.
[00883] Embodiment V-67. The XDP system of Embodiment V-66, wherein the CRISPR
protein is a Class 2 CRISPR protein selected from the group consisting of Type
II, Type V, or
Type VI protein.
[00884] Embodiment V-68. The XDP system of Embodiment V-67, wherein the CRISPR
protein is a Type V protein selected from the group consisting of Cas12a,
Cas12b, Cas12c,
Cas12d (CasY), Cas12j and CasX.
[00885] Embodiment V-69. The XDP system of Embodiment V-68, wherein the CRISPR
protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345, 350-
353, 355-367
or 388-397, or 11, or a sequence having at least about 85%, at least about
90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[00886] Embodiment V-70. The XDP system of Embodiment V-68, wherein the CRISPR
protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345, 350-
353, 355-367
or 388-397.
[00887] Embodiment V-71. The XDP system of any one of Embodiments V-68-70,
wherein
the CasX further comprises one or more NLS selected from the group of
sequences consisting of
SEQ ID NOS: 130-166, wherein the NLS are located at or near the N-terminus
and/or the C-
terminus.
[00888] Embodiment V-72. The XDP system of Embodiment V-64, wherein the
therapeutic
payload is a nucleic acid selected from the group consisting of a single-
stranded antisense
oligonucleotide (AS0s), a double-stranded RNA interference (RNA molecule, a
DNA aptamer,
and a CRISPR guide nucleic acid.
[00889] Embodiment V-73, The XDP system of Embodiment V-72, wherein the CRISPR
guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and a
targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
266
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00890] Embodiment V-74. The XDP system of Embodiment V-73, wherein the
scaffold
sequence comprises a sequence of SEQ ID NOS: 597-781, or a sequence having at
least about
85%, at least about 90%, at least about 91%, at least about 92%, at least
about 93%, at least
about 94%, at least about 95%, at least about 96%, at least about 97%, at
least about 98%, or at
least about 99% sequence identity thereto.
[00891] Embodiment V-75. The XDP system of Embodiment V-73, wherein the
scaffold
sequence of the guide RNA comprises a sequence of SEQ ID NOS: 597-781.
[00892] Embodiment V-76. The XDP system of any one of Embodiments V-73-75,
wherein
the targeting sequence of the guide RNA consists of 14, 15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25,
26, 27, 28, 29, or 30 consecutive nucleotides.
[00893] Embodiment V-77. The XDP system of any one of Embodiments V-56-76,
wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoding on two nucleic acids;
(c) the components are encoding on three nucleic acids;
(d) the components are encoding on four nucleic acids; or
(e) the components are encoding on five nucleic acids.
[00894] Embodiment V-78. The XDP system of Embodiment V-77, wherein the one or
more
of the components encoded by the nucleic acids are configured according to any
one of FIGS.
36-68.
[00895] Embodiment V-79. The XDP system of Embodiment V-77 or Embodiment V-78,
wherein the one or more of the components are encoded by nucleic acids
selected from the group
of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-201, 782, 234-
339, 880-933,
and 947-1000 as set forth in Tables 5, 24, 27, 30, and 33, or sequences having
at least about
85%, at least about 90%, at least about 91%, at least about 92%, at least
about 93%, at least
about 94%, at least about 95%, at least about 96%, at least about 97%, at
least about 98%, or at
least about 99% sequence identity thereto.
[00896] Embodiment V-80. The XDP system of any one of Embodiments V-56-79,
wherein
the components are capable of self-assembling into an XDP when the one or more
nucleic acids
are introduced into a eukaryotic host cell and are expressed.
[00897] Embodiment V-81. The XDP of Embodiment V-80, wherein the therapeutic
payload is
encapsidated within the XDP upon self-assembly of the XDP.
267
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00898] Embodiment V-82. The XDP system of Embodiment V-81, wherein the
therapeutic
payload comprises a CasX and a guide RNA complexed as a ribonucleoprotein
complex (RNP)
and, optionally, a donor template.
[00899] Embodiment V-83. The XDP of Embodiment V-80, wherein the tropism
factor is
incorporated on the XDP surface upon self-assembly of the XDP.
[00900] Embodiment V-84. The XDP system of Embodiment V-83, wherein the
tropism factor
confers preferential interaction with the cell surface of a target cell and
facilitates entry of the
XDP into the target cell.
[00901] Embodiment V-85. An XDP system comprising one or more nucleic acid
encoding
components:
(a) all or a portion of an Deltaretrovirus gag polyprotein;
(b) a therapeutic payload; and
(c) a tropism factor.
[00902] Embodiment V-86. The XDP system of Embodiment V-85, wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a capsid polypeptide (CA), and a nucleocapsid polypeptide
(NC).
[00903] Embodiment V-87. The XDP system of Embodiment V-86, wherein the gag
polyprotein comprises, from N-terminus to C-terminus, matrix polypeptide (MA),
a capsid
polypeptide (CA), and a nucleocapsid polypeptide (NC).
[00904] Embodiment V-88. The XDP system of any one of Embodiments V-85-87,
wherein
the nucleic acids encode one or more components selected from
(a) an p1 peptide;
(b) an Inv p6 peptide;
a Gag-Pol polyprotein;
(d) one or more protease cleavage sites;
(e) a non-retroviral, heterologous protease capable of
cleaving the cleavage sites; and
(0 a gag-transframe region-pol protease polyprotein.
[00905] Embodiment V-89 The XDP system of any one of Embodiments V-85-88,
wherein
the tropism factor is selected from the group consisting of a glycoprotein, an
antibody fragment,
a receptor, and a ligand to a target cell marker.
[00906] Embodiment V-90. The XDP system of Embodiment V-89, wherein the
tropism factor
is a glycoprotein having an encoding sequence selected from the group
consisting of SEQ ID
268
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595, or a sequence having at least about 85%, at least
about 90%, at least
about 91%, at least about 92%, at least about 93%, at least about 94%, at
least about 95%, at
least about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[00907] Embodiment V-91. The XDP system of Embodiment V-89, wherein the
tropism factor
is a glycoprotein having a sequence selected from the group consisting of SEQ
ID NOS: 437,
439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463, 465, 467,
469, 471, 473, 475,
477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ 503, 505,
507, 509, 511, 513,
515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539, 541, 543,
545, 547, 549, 551,
553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577, 579, 581,
583, 585, 587, 589,
591, 593 and 595.
[00908] Embodiment V-92. The XDP system of Embodiment V-91, wherein the
tropism factor
is glycoprotein G from vesicular stomatitis virus (VSV-G).
[00909] Embodiment V-93. The XDP system of any one of Embodiments V-85-92,
wherein
the therapeutic payload comprises a protein, a nucleic acid, or comprises both
a protein and a
nucleic acid.
[00910] Embodiment V-94. The XDP system of Embodiment V-93, wherein the
protein
payload is selected from the group consisting of a cytokine, an interleukin,
an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
thrombolytic
protein, a CRISPR protein, and an anti-cancer modality.
[00911] Embodiment V-95. The XDP system of Embodiment V-94, wherein the CRISPR
protein is a Class 1 or Class 2 CRISPR protein.
[00912] Embodiment V-96, The XDP system of Embodiment V-95, wherein the CRISPR
protein is a Class 2 CRISPR protein selected from the group consisting of Type
II, Type V, or
Type VI protein.
269
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
1009131 Embodiment V-97. The XDP system of Embodiment V-96, wherein the CRISPR
protein is a Type V protein selected from the group consisting of Cas12a,
Cas12b, Cas12c,
Caslal (CasY), Cas12j and CasX.
[00914] Embodiment V-98. The XDP system of Embodiment V-97, wherein the CRISPR
protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345, 350-
353, 355-367
or 388-397, or a sequence having at least about 85%, at least about 90%, at
least about 91%, at
least about 92%, at least about 93%, at least about 94%, at least about 95%,
at least about 96%,
at least about 97%, at least about 98%, or at least about 99% sequence
identity thereto.
[00915] Embodiment V-99. The XDP system of Embodiment V-97, wherein the CRISPR
protein is a CasX comprising a sequence of SEQ ID NOS; 21-233, 343-345, 350-
353, 355-367
or 388-397.
[00916] Embodiment V-100. The XDP system of any
one of Embodiments V-97-99,
wherein the CasX further comprises one or more NLS selected from the group of
sequences
consisting of SEQ ID NOS: 130-166, wherein the NLS are located at or near the
N-terminus
and/or the C-terminus.
[00917] Embodiment V-101. The XDP system of
Embodiment V-93, wherein the
therapeutic payload is a nucleic acid selected from the group consisting of a
single-stranded
antisense oligonucleotide (AS0s), a double-stranded RNA interference (RNAi)
molecule, a
DNA aptamer, and a CRISPR guide nucleic acid.
[00918] Embodiment V-102. The XDP system of
Embodiment V-101, wherein the
CRISPR guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and
a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
[00919] Embodiment V-103. The XDP system of
Embodiment V-102, wherein the
scaffold sequence comprises a sequence of SEQ ID NOS: 597-781, or a sequence
having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
[00920] Embodiment V-104. The XDP system of
Embodiment V-102, wherein the
scaffold sequence of the guide RNA comprises a sequence of SEQ ID NOS: 597-
781_
270
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00921] Embodiment V-105. The XDP system of any
one of Embodiments V-102-104,
wherein the targeting sequence of the guide RNA consists of 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides.
[00922] Embodiment V-106. The XDP system of any
one of Embodiments V-85-105,
wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoding on two nucleic acids;
(c) the components are encoding on three nucleic acids;
(d) the components are encoding on four nucleic acids; or
(e) the components are encoding on five nucleic acids.
[00923] Embodiment V-107. The XDP system of
Embodiment V-106, wherein the one
or more of the components encoded by the nucleic acids are configured
according to any one of
FIGS. 36-68.
[00924] Embodiment V-108. The XDP system of
Embodiment V-106 or Embodiment
V-107, wherein the one or more of the components are encoded by nucleic acids
selected from
the group of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-201,
782, 234-339,
880-933, and 947-1000 as set forth in Tables 5, 24, 27, 30, and 33, or
sequences having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
[00925] Embodiment V-109. The XDP system of any
one of Embodiments V-85-108,
wherein the components are capable of self-assembling into an XDP when the one
or more
nucleic acids are introduced into a eukaryotic host cell and are expressed.
[00926] Embodiment V-110. The XDP of Embodiment
V-109, wherein the therapeutic
payload is encapsidated within the XDP upon self-assembly of the XDP.
[00927] Embodiment V-111. The XDP system of
Embodiment V-110, wherein the
therapeutic payload comprises a CasX and a guide RNA complexed as a
ribonucleoprotein
complex (RNP) and, optionally, a donor template.
[00928] Embodiment V-112. The XDP of Embodiment
V-109, wherein the tropism
factor is incorporated on the XDP surface upon self-assembly of the XDP.
271
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00929] Embodiment V-113. The XDP system of
Embodiment V-112, wherein the
tropism factor confers preferential interaction with the cell surface of a
target cell and facilitates
entry of the XDP into the target cell
[00930] Embodiment V-114. An XDP system
comprising one or more nucleic acid
encoding components:
(a) all or a portion of an Epsilonretrovirus gag polyprotein;
(b) a therapeutic payload; and
(c) a tropism factor.
[00931] Embodiment V-115. The XDP system of
Embodiment V-114, wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a p20 peptide, a capsid polypeptide (CA), and a nucleocapsid
polypeptide
(NC).
[00932] Embodiment V-116. The XDP system of
Embodiment V-114, wherein the gag
polyprotein comprises, from N-terminus to C-terminus, matrix polypeptide (MA),
a p20 peptide,
a capsid polypeptide (CA), and a nucleocapsid polypeptide (NC).
[00933] Embodiment V-117. The XDP system of any
one of Embodiments V-114-116,
wherein the nucleic acids encode one or more components selected from
(a) an HIV pl peptide;
(b) an HIV p6 peptide;
(c) a Gag-Pal polyprotein;
(d) one or more protease cleavage sites;
(e) a non-retroviral, heterologous protease capable of cleaving the
cleavage sites; and
(0 a gag-transframe region-pol protease polyprotein.
[00934] Embodiment V-118. The XDP system of any
one of Embodiments V-114-117,
wherein the tropism factor is selected from the group consisting of a
glycoprotein, an antibody
fragment, a receptor, and a ligand to a target cell marker.
1009351 Embodiment V-119. The XDP system of
Embodiment V-118, wherein the
tropism factor is a g,lycoprotein having an encoding sequence selected from
the group consisting
of SEQ ID NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459,
461, 463, 465, 467,
469, 471, 473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497,
499, 501õ 503, 505,
507,509,511,513,515,517,519,521,523,525,527,529,531,533,535,537,539,541,543,
545,547,549,551,553,555,557,559,561,563,565,567,569,571,573,575,577,579,581,
272
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
583, 585, 587, 589, 591, 593 and 595, or a sequence having at least about 85%,
at least about
90%, at least about 91%, at least about 92%, at least about 93%, at least
about 94%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
sequence identity thereto.
1009361 Embodiment V-120. The XDP system of
Embodiment V-118, wherein the
tropism factor is a g,lycoprotein having a sequence selected from the group
consisting of SEQ ID
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595.
1009371 Embodiment V-121. The XDP system of
Embodiment V-120, wherein the
tropism factor is glycoprotein G from vesicular stomatitis virus (VSV-G).
1009381 Embodiment V-122. The XDP system of any
one of Embodiments V-114-121,
wherein the therapeutic payload comprises a protein, a nucleic acid, or
comprises both a protein
and a nucleic acid.
1009391 Embodiment V-123. The XDP system of
Embodiment V-122, wherein the
protein payload is selected from the group consisting of a cytokine, an
interleukin, an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
thrombolytic
protein, a CRISPR protein, and an anti-cancer modality.
[00940] Embodiment V-124. The XDP system of
Embodiment V-123, wherein the
CRISPR protein is a Class 1 or Class 2 CRISPR protein.
[00941] Embodiment V-125. The XDP system of
Embodiment V-124, wherein the
CRISPR protein is a Class 2 CRISPR protein selected from the group consisting
of Type II,
Type V, or Type VI protein.
[00942] Embodiment V-126. The XDP system of
Embodiment V-125, wherein the
CRISPR protein is a Type V protein selected from the group consisting of
Cas12a, Cas12b,
Cas12c, Cas12d (CasY), Cas12j and CasX.
[00943] Embodiment V-127. The XDP system of
Embodiment V-126, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397, or a sequence having at least about 85%, at least about
90%, at least about
273
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[00944] Embodiment V128. The XDP system of
Embodiment V-126, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397.
[00945] Embodiment V-129. The XDP system of any
one of Embodiments V-126-128,
wherein the CasX further comprises one or more NLS selected from the group of
sequences
consisting of SEQ ID NOS: 130-166, wherein the NLS are located at or near the
N-terminus
and/or the C-terminus.
[00946] Embodiment V-130. The XDP system of
Embodiment V-122, wherein the
therapeutic payload is a nucleic acid selected from the group consisting of a
single-stranded
antisense oligonucleotide (AS0s), a double-stranded RNA interference (RNAi)
molecule, a
DNA aptamer, and a CRISPR guide nucleic acid.
[00947] Embodiment V-131. The XDP system of
Embodiment V-130, wherein the
CRISPR guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and
a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
[00948] Embodiment V-132. The XDP system of
Embodiment V-131, wherein the
scaffold sequence comprises a sequence of SEQ ID NOS: 597-78 lot a sequence
having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
[00949] Embodiment V-133. The XDP system of
Embodiment V-131, wherein the
scaffold sequence of the guide RNA comprises a sequence of SEQ ID NOS: 597-
781.
[00950] Embodiment V-134. The XDP system of any
one of Embodiments V-131-133,
wherein the targeting sequence of the guide RNA consists of 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides.
[00951] Embodiment V-135. The XDP system of any
one of Embodiments V-114-134,
wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoding on two nucleic acids;
274
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(c) the components are encoding on three nucleic acids;
(d) the components are encoding on four nucleic acids; or
(e) the components are encoding on five nucleic acids.
1009521 Embodiment V136. The XDP system of
Embodiment V-135, wherein the one
or more of the components encoded by the nucleic acids are configured
according to any one of
FIGS. 36-68.
1009531 Embodiment V-137. The XDP system of
Embodiment V-135 or Embodiment
V-136, wherein the one or more of the components are encoded by nucleic acids
selected from
the group of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-201,
782, 234-339,
880-933, and 947-1000 as set forth in Tables 5, 24, 27, 30, and 33, or
sequences having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
1009541 Embodiment V-138. The XDP system of any
one of Embodiments V-114-137,
wherein the components are capable of self-assembling into an XDP when the one
or more
nucleic acids are introduced into a eukaryotic host cell and are expressed.
1009551 Embodiment V-139. The XDP of Embodiment
V-138, wherein the therapeutic
payload is encapsidated within the XDP upon self-assembly of the XDP.
[00956] Embodiment V-140. The XDP system of
Embodiment V-139, wherein the
therapeutic payload comprises a CasX and a guide RNA complexed as a
ribonucleoprotein
complex (RNP) and, optionally, a donor template.
1009571 Embodiment V-141. The XDP of Embodiment
V-139, wherein the tropism
factor is incorporated on the XDP surface upon self-assembly of the XDP.
1009581 Embodiment V-142. The XDP system of
Embodiment V-141, wherein the
tropism factor confers preferential interaction with the cell surface of a
target cell and facilitates
entry of the XDP into the target cell.
[00959] Embodiment V-143. An XDP system
comprising one or more nucleic acid
encoding components:
(a) all or a portion of an Gammaretrovirus gag polyprotein;
(b) a therapeutic payload; and
(c) a tropism factor.
275
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00960] Embodiment V-144. The XDP system of
Embodiment V-143, wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a p12 peptide, a capsid polypeptide (CA), and a nucleocapsid
polypeptide
(NC).
[00961] Embodiment V-145. The XDP system of
Embodiment V-144, wherein the gag
polyprotein comprises, from N-terminus to C-terminus, matrix polypeptide (MA),
a p12 peptide,
a capsid polypeptide (CA), and a nucleocapsid polypeptide (NC).
[00962] Embodiment V-146. The XDP system of any
one of Embodiments V-143-145,
wherein the nucleic acids encode one or more components selected from
(a) an HIV p 1 peptide;
(b) an HIV p6 peptide;
(c) a Gag-Pol polyprotein;
(d) one or more protease cleavage sites;
(e) a non-retroviral, heterologous protease capable of cleaving the
cleavage sites; and
(0 a gag-transframe region-pol protease polyprotein.
[00963] Embodiment V-147. The XDP system of any
one of Embodiments V-143-146,
wherein the tropism factor is selected from the group consisting of a
glycoprotein, an antibody
fragment, a receptor, and a ligand to a target cell marker.
[00964] Embodiment V-148. The XDP system of
Embodiment V-147, wherein the
tropism factor is a g,lycoprotein having an encoding sequence selected from
the group consisting
of SEQ ID NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459,
461, 463, 465, 467,
469, 471, 473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497,
499, 501õ 503, 505,
507, 509, 511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535,
537, 539, 541, 543,
545, 547, 549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573,
575, 577, 579, 581,
583, 585, 587, 589, 591, 593 and 595, or a sequence having at least about 85%,
at least about
90%, at least about 91%, at least about 92%, at least about 93%, at least
about 94%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
sequence identity thereto.
[00965] Embodiment V-149. The XDP system of
Embodiment V-147, wherein the
tropism factor is a g,lycoprotein having a sequence selected from the group
consisting of SEQ lD
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501,
503, 505, 507, 509,
276
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595.
[00966] Embodiment V-150. The XDP system of
Embodiment V-149, wherein the
tropism factor is glycoprotein G from vesicular stomatitis virus (VSV-G).
[00967] Embodiment V-151. The XDP system of any
one of Embodiments V-143-150,
wherein the therapeutic payload comprises a protein, a nucleic acid, or
comprises both a protein
and a nucleic acid.
[00968] Embodiment V-152. The XDP system of
Embodiment V-151, wherein the
protein payload is selected from the group consisting of a cytokine, an
interleukin, an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
thrombolytic
protein, a CRISPR protein, and an anti-cancer modality.
[00969] Embodiment V-153. The XDP system of
Embodiment V-152, wherein the
CRISPR protein is a Class 1 or Class 2 CRISPR protein.
[00970] Embodiment V-154. The XDP system of
Embodiment V-153, wherein the
CRISPR protein is a Class 2 CRISPR protein selected from the group consisting
of Type II,
Type V, or Type VI protein.
[00971] Embodiment V-155. The XDP system of
Embodiment V-154, wherein the
CRISPR protein is a Type V protein selected from the group consisting of
Cas12a, Cas12b,
Cas12c, Cas12d (CasY), Cas12j and CasX.
[00972] Embodiment V-156. The XDP system of
Embodiment V-155, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397, or a sequence having at least about 85%, at least about
90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[00973] Embodiment V-157. The XDP system of
Embodiment V-155, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397.
[00974] Embodiment V-158. The XDP system of any
one of Embodiments V-155-157,
wherein the CasX further comprises one or more NLS selected from the group of
sequences
277
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
consisting of SEQ ID NOS: 130-166, wherein the NLS are located at or near the
N-terminus
and/or the C-terminus.
[00975] Embodiment V-159. The XDP system of
Embodiment V-151, wherein the
therapeutic payload is a nucleic acid selected from the group consisting of a
single-stranded
antisense oligonucleotide (AS0s), a double-stranded RNA interference (RNAi)
molecule, a
DNA aptamer, and a CRISPR guide nucleic acid.
[00976] Embodiment V-160. The XDP system of
Embodiment V-159, wherein the
CRISPR guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and
a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
[00977] Embodiment V-161. The XDP system of
Embodiment V-160, wherein the
scaffold sequence comprises a sequence of SEQ ID NOS: 597-781, or a sequence
having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
[00978] Embodiment V-162. The XDP system of
Embodiment V-160, wherein the
scaffold sequence of the guide RNA comprises a sequence of SEQ ID NOS: 597-
781.
[00979] Embodiment V-163. The XDP system of any
one of Embodiments V-160-162,
wherein the targeting sequence of the guide RNA consists of 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides.
[00980] Embodiment V-164. The XDP system of any
one of Embodiments V-143-163,
wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoding on two nucleic acids;
(c) the components are encoding on three nucleic acids;
(d) the components are encoding on four nucleic acids; or
(e) the components are encoding on five nucleic acids.
[00981] Embodiment V-165. The XDP system of
Embodiment V-164, wherein the one
or more of the components encoded by the nucleic acids are configured
according to any one of
FIGS. 36-68.
[00982] Embodiment V-166. The XDP system of
Embodiment V-164 or Embodiment
V-165, wherein the one or more of the components are encoded by nucleic acids
selected from
278
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
the group of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-201,
782, 234-339,
880-933, and 947-1000 as set forth in Tables 5, 24, 27, 30, and 33, or
sequences having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
[00983] Embodiment V-167. The XDP system of any
one of Embodiments V-164-166,
wherein the components are capable of self-assembling into an XDP when the one
or more
nucleic acids are introduced into a eukaryotic host cell and are expressed.
[00984] Embodiment V-168. The XDP of Embodiment
V-167, wherein the therapeutic
payload is encapsidated within the XDP upon self-assembly of the XDP.
[00985] Embodiment V-169. The XDP system of
Embodiment V-168, wherein the
therapeutic payload comprises a CasX and a guide RNA complexed as a
ribonucleoprotein
complex (RNP) and, optionally, a donor template.
[00986] Embodiment V-170. The XDP of Embodiment
V-167, wherein the tropism
factor is incorporated on the XDP surface upon self-assembly of the XDP.
[00987] Embodiment V-171. The XDP system of
Embodiment V-170, wherein the
tropism factor confers preferential interaction with the cell surface of a
target cell and facilitates
entry of the XDP into the target cell.
[00988] Embodiment V-172. An XDP system
comprising one or more nucleic acid
encoding components:
(a) all or a portion of an Lentivirus gag polyprotein;
(b) a therapeutic payload; and
(c) a tropism factor.
[00989] Embodiment V-173. The XDP system of
Embodiment V-172, wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a matrix
polypeptide (MA), a capsid polypeptide (CA), a p2 peptide, a nucleocapsid
polypeptide (NC), a
p1 peptide, and a p6 peptide.
[00990] Embodiment V-174. The XDP system of
Embodiment V-173, wherein the gag
polyprotein comprises, from N-terminus to C-terminus, matrix polypeptide (MA),
a capsid
polypeptide (CA), a p2 peptide, a nucleocapsid polypeptide (NC), a pl peptide,
and a p6 peptide.
1009911 Embodiment V-175. The XDP system of any
one of Embodiments V-172-173,
wherein the nucleic acids encode one or more components selected from
279
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(a) a Gag-Pal polyprotein;
(b) one or more protease cleavage sites;
(c) a non-retroviral, heterologous protease capable of cleaving the
cleavage sites; and
(d) a gag-transframe region-pol protease polyprotein.
1009921 Embodiment V-176. The XDP system of any
one of Embodiments V-172-175,
wherein the lentivirus is selected from the group consisting of human
immunodeficiency-1
(HIV-1), human immunodeficiency-2 (111V-2), simian immunodeficiency virus
(Sty), feline
immunodeficiency virus (Hy), and bovine immunodeficiency virus (BIV).
1009931 Embodiment V-177. The XDP system of
Embodiment V-176, wherein the
lentivirus is HIV-1
1009941 Embodiment V-178. The XDP system of any
one of Embodiments V-172-177,
wherein the tropism factor is selected from the group consisting of a
glycoprotein, an antibody
fragment, a receptor, and a ligand to a target cell marker.
1009951 Embodiment V-179. The XDP system of
Embodiment V-178, wherein the
tropism factor is a glycoprotein having an encoding sequence selected from the
group consisting
of SEQ ID NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459,
461, 463, 465, 467,
469, 471, 473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497,
499, 501õ 503, 505,
507, 509, 511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535,
537, 539, 541, 543,
545, 547, 549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573,
575, 577, 579, 581,
583, 585, 587, 589, 591, 593 and 595, or a sequence having at least about 85%,
at least about
90%, at least about 91%, at least about 92%, at least about 93%, at least
about 94%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
sequence identity thereto.
1009961 Embodiment V-180. The XDP system of
Embodiment V-178, wherein the
tropism factor is a glycoprotein having a sequence selected from the group
consisting of SEQ ID
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501,
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595.
1009971 Embodiment V-181. The XDP system of
Embodiment V-180, wherein the
tropism factor is glycoprotein G from vesicular stomatitis virus (VSV-G).
280
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[00998] Embodiment V-182. The XDP system of any
one of Embodiments V-172-181,
wherein the therapeutic payload comprises a protein, a nucleic acid, or
comprises both a protein
and a nucleic acid.
1009991 Embodiment V-183. The XDP system of
Embodiment V-182, wherein the
protein payload is selected from the group consisting of a cytolcine, an
interleulcin, an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
thrombolytic
protein, a CRISPR protein, and an anti-cancer modality.
[001000] Embodiment V-184. The XDP system of
Embodiment V-183, wherein the
CRISPR protein is a Class 1 or Class 2 CRISPR protein.
[0010011Embodiment V-185. The XDP system of
Embodiment V-184, wherein the
CRISPR protein is a Class 2 CRISPR protein selected from the group consisting
of Type II,
Type V. or Type VI protein.
[001002] Embodiment V-186. The XDP system of
Embodiment V-185, wherein the
CRISPR protein is a Type V protein selected from the group consisting of
Cas12a, Cas12b,
Cas12c, Cas12d (CasY), Cas12j and CasX.
[001003] Embodiment V-187. The XDP system of
Embodiment V-186, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397, or a sequence having at least about 85%, at least about
90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[001004] Embodiment V-188. The XDP system of
Embodiment V-186, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397.
[001005] Embodiment V-189. The XDP system of any
one of Embodiments V-186-188,
wherein the CasX further comprises one or more NLS selected from the group of
sequences
consisting of SEQ ID NOS: 130-166, wherein the NLS are located at or near the
N-terminus
and/or the C-terminus.
[001006] Embodiment V-190. The XDP system of
Embodiment V-182, wherein the
therapeutic payload is a nucleic acid selected from the group consisting of a
single-stranded
281
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
antisense oligonucleotide (AS0s), a double-stranded RNA interference (RNAi)
molecule, a
DNA aptamer, and a CRISPR guide nucleic acid.
[0010071Embodiment V-191. The XDP system of
Embodiment V-190, wherein the
CRISPR guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and
a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
10010081 Embodiment V-192. The XDP system of
Embodiment V-191, wherein the
scaffold sequence comprises a sequence of SEQ ID NOS: 597-781or a sequence
having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
10010091Embodiment V-193. The XDP system of
Embodiment V-191, wherein the
scaffold sequence of the guide RNA comprises a sequence of SEQ ID NOS: 597-
781.
10010101Embodiment V-194. The XDP system of any
one of Embodiments V-191-193,
wherein the targeting sequence of the guide RNA consists of 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides.
[0010111Embodiment V-195. The XDP system of any
one of Embodiments V-172-194,
wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoding on two nucleic acids;
(c) the components are encoding on three nucleic acids;
(d) the components are encoding on four nucleic acids; or
(e) the components are encoding on five nucleic acids.
10010121 Embodiment V-196. The XDP system of
Embodiment V-195, wherein the one
or more of the components encoded by the nucleic acids are configured
according to any one of
FIGS. 36-68.
10010131Embodiment V-197. The XDP system of
Embodiment V-195 or Embodiment
V-196, wherein the one or more of the components are encoded by nucleic acids
selected from
the group of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-201,
782, 234-339,
880-933, and 947-1000 as set forth in Tables 5, 24, 27, 30, and 33, or
sequences having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
282
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
[0010141Embodiment V-198. The XDP system of any
one of Embodiments V-195-197,
wherein the components are capable of self-assembling into an XDP when the one
or more
nucleic acids are introduced into a eukaryotic host cell and are expressed.
10010151Embodiment V-199. The XDP of Embodiment
V-198, wherein the therapeutic
payload is encapsidated within the XDP upon self-assembly of the XDP.
10010161Embodiment V-200. The XDP system of
Embodiment V-198, wherein the
therapeutic payload comprises a CasX and a guide RNA complexed as a
ribonucleoprotein
complex (RNP) and, optionally, a donor template.
10010171Embodiment V-201. The XDP of Embodiment
V-198, wherein the tropism
factor is incorporated on the XDP surface upon self-assembly of the XDP.
10010181Embodiment V-202. The XDP system of
Embodiment V-201, wherein the
tropism factor confers preferential interaction with the cell surface of a
target cell and facilitates
entry of the XDP into the target cell.
100101.91Embodiment V-203. An XDP system
comprising one or more nucleic acid
encoding components:
(a) all or a portion of an Spumaretrovirinae gag polyprotein;
(b) a therapeutic payload; and
(c) a tropism factor.
10010201Embodiment V-204. The XDP system of
Embodiment V-203, wherein the gag
polyprotein comprises one or more components selected from the group
consisting of a p68 Gag
polypeptide and a p3 Gag polypeptide.
10010211Embodiment V-205. The XDP system of
Embodiment V-204, wherein the gag
polyprotein comprises, from N-terminus to C-terminus, p68 Gag polypeptide and
a p3 Gag
polypeptide.
10010221Embodiment V-206. The XDP system of any
one of Embodiments V-203-205,
wherein the nucleic acids encode one or more components selected from
(a) an HIV pl peptide;
(b) an HIV p6 peptide;
(c) a Gag-Pol polyprotein;
(d) one or more protease cleavage sites;
283
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(e) a non-retroviral, heterologous protease capable of
cleaving the cleavage sites; and
(1) a gag-transframe region-pol protease polyprotein.
10010231Embodiment V-207. The XDP system of any
one of Embodiments V-203-206,
wherein the tropism factor is selected from the group consisting of a
glycoprotein, an antibody
fragment, a receptor, and a ligand to a target cell marker.
10010241Embodiment V-208. The XDP system of
Embodiment V-207, wherein the
tropism factor is a g,lycoprotein having an encoding sequence selected from
the group consisting
of SEQ ID NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459,
461, 463, 465, 467,
469, 471, 473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497,
499, 501õ 503, 505,
507, 509, 511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535,
537, 539, 541, 543,
545, 547, 549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573,
575, 577, 579, 581,
583, 585, 587, 589, 591, 593 and 595, or a sequence having at least about 85%,
at least about
90%, at least about 91%, at least about 92%, at least about 93%, at least
about 94%, at least
about 95%, at least about 96%, at least about 97%, at least about 98%, or at
least about 99%
sequence identity thereto.
10010251Embodiment V-209. The XDP system of
Embodiment V-207, wherein the
tropism factor is a glycoprotein having a sequence selected from the group
consisting of SEQ ID
NOS: 437, 439, 441, 443, 445, 447, 449, 451, 453, 455, 457, 459, 461, 463,
465, 467, 469, 471,
473, 475, 477, 479, 481, 483, 485, 487, 489, 491, 493, 495, 497, 499, 501õ
503, 505, 507, 509,
511, 513, 515, 517, 519, 521, 523, 525, 527, 529, 531, 533, 535, 537, 539,
541, 543, 545, 547,
549, 551, 553, 555, 557, 559, 561, 563, 565, 567, 569, 571, 573, 575, 577,
579, 581, 583, 585,
587, 589, 591, 593 and 595.
10010261 Embodiment V-210. The XDP system of
Embodiment V-209, wherein the
tropism factor is glycoprotein G from vesicular stomatitis virus (VSV-G).
10010271Embodiment V-211. The XDP system of any
one of Embodiments V-203-210,
wherein the therapeutic payload comprises a protein, a nucleic acid, or
comprises both a protein
and a nucleic acid.
[001028] Embodiment V-212. The XDP system of
Embodiment V-211, wherein the
protein payload is selected from the group consisting of a cytokine, an
interleukin, an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
thrombolytic
protein, a CRISPR protein, and an anti-cancer modality.
284
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
[0010291Embodiment V-213. The XDP system of
Embodiment V-212, wherein the
CRISPR protein is a Class 1 or Class 2 CRISPR protein.
[0010301Embodiment V-214. The XDP system of
Embodiment V-213, wherein the
CRISPR protein is a Class 2 CRISPR protein selected from the group consisting
of Type II,
Type V, or Type VI protein.
[0010311Embodiment V-215. The XDP system of
Embodiment V-214, wherein the
CRISPR protein is a Type V protein selected from the group consisting of
Cas12a, Cas12b,
Cas12c, Cas12d (CasY), Cas12j and CasX.
[0010321Embodiment V-216. The XDP system of
Embodiment V-215, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397, or a sequence having at least about 85%, at least about
90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
[0010331Embodiment V-217. The XDP system of
Embodiment V-216, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397.
[0010341Embodiment V-218. The XDP system of any
one of Embodiments V-203-217,
wherein the CasX further comprises one or more NLS selected from the group of
sequences
consisting of SEQ ID NOS: 130-166, wherein the NLS are located at or near the
N-terminus
and/or the C-terminus.
[0010351Embodiment V-219. The XDP system of
Embodiment V-211, wherein the
therapeutic payload is a nucleic acid selected from the group consisting of a
single-stranded
antisense oligonucleotide (AS0s), a double-stranded RNA interference (RNAi)
molecule, a
DNA aptamer, and a CRISPR guide nucleic acid.
[0010361Embodiment V-220. The XDP system of
Embodiment V-219, wherein the
CRISPR guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and
a targeting sequence, wherein the targeting sequence is complementary to a
target nucleic acid
sequence.
[0010371Embodiment V-221. The XDP system of
Embodiment V-220, wherein the
scaffold sequence comprises a sequence of SEQ ID NOS: 597-781, or a sequence
having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
285
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
10010381 Embodiment V-222. The XDP system of
Embodiment V-221, wherein the
scaffold sequence of the guide RNA comprises a sequence of SEQ ID NOS: 597-
781.
10010391 Embodiment V-223. The XDP system of any
one of Embodiments V-220-222,
wherein the targeting sequence of the guide RNA consists of 14, 15, 16, 17,
18, 19, 20, 21, 22,
23, 24, 25, 26, 27, 28, 29, or 30 consecutive nucleotides.
10010401 Embodiment V-224. The XDP system of any
one of Embodiments V-203-223,
wherein
(a) the components are encoded on a single nucleic acid;
(b) the components are encoding on two nucleic acids;
(c) the components are encoding on three nucleic acids;
(d) the components are encoding on four nucleic acids; or
(e) the components are encoding on five nucleic acids.
10010411Embodiment V-225. The XDP system of
Embodiment V-224, wherein the one
or more of the components encoded by the nucleic acids are configured
according to any one of
FIGS. 36-68.
10010421 Embodiment V-226. The XDP system of
Embodiment V-224 or Embodiment
V-225, wherein the one or more of the components are encoded by nucleic acids
selected from
the group of sequences consisting of SEQ ID NOS: 192, 193, 195, 196, 198-201,
782, 234-339,
880-933, and 947-1000 as set forth in Tables 5, 24, 27, 30, and 33, or
sequences having at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
10010431Embodiment V-227. The XDP system of any
one of Embodiments V-224-226,
wherein the components are capable of self-assembling into an XDP when the one
or more
nucleic acids are introduced into a eukaryotic host cell and are expressed.
10010441Embodiment V-228. The XDP of Embodiment
V-227, wherein the therapeutic
payload is encapsidated within the XDP upon self-assembly of the XDP.
10010451 Embodiment V-229. The XDP system of
Embodiment V-228, wherein the
therapeutic payload comprises a CasX and a guide RNA complexed as a
ribonucleoprotein
complex (RNP) and, optionally, a donor template.
286
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10010461Embodiment V-230. The XDP of Embodiment
V-227, wherein the tropism
factor is incorporated on the XDP surface upon self-assembly of the XDP.
10010471Embodiment V-231. The XDP system of
Embodiment V-230, wherein the
tropism factor confers preferential interaction with the cell surface of a
target cell and facilitates
entry of the XDP into the target cell
10010481Embodiment V-232. The XDP system of any
one of the preceding embodiments
of Set V. wherein the gag polyprotein and the therapeutic payload is expressed
as a fusion
protein.
10010491Embodiment V-233. The XDP system of
Embodiment V-232, wherein the
fusion protein does not comprise a protease cleavage site between the gag
polyprotein and the
therapeutic payload.
10010501Embodiment V-234. The XDP system of
Embodiment V-232, wherein the
fusion protein comprises a protease cleavage site between the gag polyprotein
and the
therapeutic payload.
10010511Embodiment V-235. The XDP system of any
one of Embodiments V-232-234,
wherein the fusion protein comprises protease cleavage sites between the
components of the gag
polyprotein.
10010521Embodiment V-236. The XDP system of
Embodiment V-234 and/or
Embodiment V-235, wherein the cleavage sites are capable of being cleaved by
the protease of
the Gag-Pal polyprotein, the protease of the gag-transframe region-pal
protease polyprotein, or
the non-retroviral, heterologous protease.
10010531Embodiment V-237. The XDP system of
Embodiment V-236, wherein the
cleavage sites are capable of being cleaved by the protease of the gag-
transframe region-pol
protease polyprotein.
10010541 Embodiment V-238. The XDP system of
Embodiment V-236, wherein the
cleavage sites are capable of being cleaved by the protease of the Gag-Pot
polyprotein
0010551 Embodiment V-239. The XDP system of
Embodiment V-236, wherein the non-
retroviral, heterologous protease is selected from the group consisting of
tobacco etch virus
protease (TEV), potyvirus HC protease, potyvirus P1 protease, PreScission
(HRV3C protease), b
virus Ma protease, B virus RNA-2-encoded protease, aphthovirus L protease,
enterovirus 2A
protease, rhinovirus 2A protease, picoma 3C protease, comovirus 24K protease,
nepovirus 24K
protease, RTSV (rice tungro spherical virus) 3C-like protease, parsnip yellow
fleck virus
287
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
protease, 3C-like protease, heparin, cathepsin, thrombin, factor Xa,
metalloproteinase, and
enterokinase.
10010561Embodiment V-240. The XDP system of
Embodiment V-239, wherein the non-
retroviral, heterologous protease is PreScission (IIRV3C protease).
10010571Embodiment V-241. The XDP system of
Embodiment V-239, wherein the non-
retroviral, heterologous protease is tobacco etch virus protease (TEV).
10010581Embodiment V-242. The XDP system of any
one of Embodiments V-12-13, 44-
47, 73-76, 96-99, 103-106, 132-135, 161-164, 192-195 or 221-224, wherein the
guide RNA
further comprises one or more ribozymes.
10010591Embodiment V-243. The XDP system of
Embodiment V-242, wherein the one
or more ribozymes are independently fused to a terminus of the guide RNA.
10010601Embodiment V-244. The XDP system of
Embodiment V-242 or Embodiment
V-243, wherein at least one of the one or more ribozymes is a hepatitis delta
virus (HDV)
ribozyme, hammerhead ribozyme, pistol ribozyme, hatchet ribozyme, or tobacco
ringspot virus
(TRSV) ribozyme.
10010611Embodiment V-245. The XDP system of any
one of Embodiments V-12-13, 44-
47, 73-76, 96-99, 103-106, 132-135, 161-164, 192-195 or 221-224, wherein the
guide RNA is
chemically modified.
10010621Embodiment V-246. The XDP system of any
one of Embodiments V-12-13, 44-
47, 73-76, 96-99, 103-106, 132-135, 161-164, 192-195 or 221-224, wherein the
guide RNA
comprises an element selected from the group consisting of a Psi packaging
element, kissing
loop_a, kissing loop_bl, kissing loop_b2, G quadriplex M3q, G quadriplex
telomere basket,
sarcin-ricin loop, or pseudoknot, wherein the element has affinity to a
protein incorporated into
the CasX selected from the group consisting of MS2, PP7, Qbeta, Ul A, and
phage I&-loop.
10010631Embodiment V-247. A eukaryotic cell
comprising the XDP system of any one
of the preceding embodiments of Set V.
10010641Embodiment V-248. The eukaryotic cell
of Embodiment V-247, wherein the
cell is a packaging cell
10010651Embodiment V-249. The eukaryotic cell
of Embodiment V-247 or Embodiment
V-248, wherein the eukaryotic cell is selected from the group consisting of
11EIC293 cells, Lenti-
X 293T cells, MIK cells, HepG2, Saos-2, HuH7, NSO cells, SP2/0 cells, YO
myeloma cells,
288
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
A549 cells, P3X63 mouse myeloma cells, PER cells, PER.C6 cells, hybridoma
cells, VERO,
NIFI3T3 cells, COS, W138, MRCS, A549, HeLa cells, CHO cells, and HT1080 cells.
10010661 Embodiment V-250. The eukaryotic cell
of Embodiment V-248 or Embodiment
V-249, wherein the packaging cell comprises one or more mutations to reduce
expression of a
cell surface marker.
10010671 Embodiment V-251. The eukaryotic cell
of any one of Embodiments V-247-
250, wherein all or a portion of the nucleic acids encoding the XDP system are
integrated into
the genome of the eukaryotic cell.
10010681Embodiment V-252. A method of making an
XDP comprising a therapeutic
payload, the method comprising:
(a) propagating the packaging cell of any one of Embodiments V-248-251
under conditions
such that XDPs are produced; and
(b) harvesting the XDPs produced by the packaging cell.
10010691Embodiment V-253. An XDP produced by
the method of Embodiment V-252.
10010701 Embodiment V-254. The XDP of Embodiment
V-253, comprising a therapeutic
payload of an RNP of a CasX and guide RNA and, optionally, a donor template.
[0010711Embodiment V-255. A method of method of
modifying a target nucleic acid
sequence in a cell, the method comprising contacting the cell with the XDP of
Embodiment V-
254, wherein said contacting comprises introducing into the cell the RNP and,
optionally, the
donor template nucleic acid sequence, wherein the target nucleic acid targeted
by the guide RNA
is modified by the CasX.
10010721 Embodiment V-256. The method of
Embodiment V-255, wherein the
modification comprises introducing one or more single-stranded breaks in the
target nucleic acid
sequence.
10010731Embodiment V-257. The method of
Embodiment V-255, wherein the
modification comprises introducing one or more double-stranded breaks in the
target nucleic
acid sequence.
10010741Embodiment V-258. The method of any one
of Embodiments V-255-257,
wherein the modification comprises insertion of the donor template into the
target nucleic acid
sequence.
10010751 Embodiment V-259. The method of any one
of Embodiments V-255-258,
wherein the cell is modified in vitro or ex vivo.
289
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10010761Embodiment V-260. The method of any one
of Embodiments V-255-258,
wherein the cell is modified in viva
10010771 Embodiment V-261. The method of
Embodiment V-260, wherein the XDP is
administered to a subject.
10010781Embodiment V-262. The method of
Embodiment V-261, wherein the subject is
the subject is selected from the group consisting of mouse, rat, pig, non-
human primate, and
human.
10010791 Embodiment V-263. The method of
Embodiment V-261 or Embodiment V-262,
wherein the XDP is administered by a route of administration selected from the
group consisting
of subcutaneous, intradermal, intraneural, intranodal, intramedullary,
intramuscular, intravenous,
intracerebroventricular, intracisternal, intrathecal, intracranial,
intralumbar, intratracheal,
intraosseous, inhalatory, intracontralateral striatum, intraocular,
intravitreal, intralymphatical,
intraperitoneal routes and sub-retinal routes.
10010801 Embodiment V-264. The method of any one
of Embodiments V-261-263,
wherein the XDP is administered to the subject using a therapeutically
effective dose.
10010811 Embodiment V-265. The method of
Embodiment V-264, wherein the XDP is
administered at a dose of at least about 1 x 10A5 particles/kg, or at least
about 1 x 10^6
particles/kg, or at least about 1 x 101\7 particles/kg, or at least about 1 x
HY'S particles/kg, or at
least about 1 x 10.'9 particles/kg, or at least about 1 x 1090 particles/kg,
or at least about 1 x
1091 particles/kg, or at least about 1 x 1092 particles/kg, or at least about
1 x 1093
particles/kg, or at least about 1 x 1094 particles/kg, or at least about 1 x
101\15 particles/kg, or
at least about 1 x 10^16 particles/kg.
10010821Embodiment V-266. The method of any one
of Embodiments V-261-265,
wherein the XDP is administered to the subject according to a treatment
regimen comprising one
or more consecutive doses using a therapeutically effective dose of the XDP.
10010831 Embodiment V-267. The method of
Embodiment V-266, wherein the
therapeutically effective dose is administered to the subject as two or more
doses over a period
of at least two weeks, or at least one month, or at least two months, or at
least three months, or at
least four months, or at least five months, or at least six months, or once a
year, or every 2 or 3
years.
290
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10010841Embodiment V-268. A method for
introducing a CasX and gNA RNP into a cell
having a target nucleic acid, comprising contacting the cell with the XDP of
Embodiment V-253
or Embodiment V-254, such that the RNP enters the cell.
10010851Embodiment V-269. The method of
Embodiment V-268, wherein the RNP
binds to the target nucleic acid.
10010861 Embodiment V-270. The method of
Embodiment V-269, wherein the target
nucleic acid is cleaved by the CasX.
10010871 Embodiment V-271. The method of any one
of Embodiments V-268-270,
wherein the cell is modified in vitro.
10010881 Embodiment V-272. The method of any one
of Embodiments V-268-270,
wherein the cell is modified in vivo.
10010891Embodiment V-273. The method of
Embodiment V-272, wherein the XDP is
administered to a subject.
10010901Embodiment V-274. The method of
Embodiment V-273, wherein the subject is
the subject is selected from the group consisting of mouse, rat, pig, non-
human primate, and
human.
[0010911Embodiment V-275. The method of any one
of Embodiments V-272-274,
wherein the XDP is administered to the subject using a therapeutically
effective dose.
10010921Embodiment V-276. The method of
Embodiment V-275, wherein the XDP is
administered at a dose of at least about 1 x 10^5 particles/kg, or at least
about 1 x 101\6
particles/kg, or at least about 1 x 10'7 particles/kg, or at least about 1 x
10^8 particles/kg, or at
least about 1 x 10^9 particles/kg, or at least about 1 x 101\10 particles/kg,
or at least about 1 x
10^11 particles/kg, or at least about 1 x 1092 particles/kg, or at least about
1 x 1093
particles/kg, or at least about 1 x 1094 particles/kg, or at least about 1 x
101\15 particles/kg, or
at least about 1 x 1096 particles/kg.
10010931 Embodiment V-277. A XDP particle
comprising:
(a) a retroviral matrix (MA) polypeptide;
(b) a therapeutic payload encapsidated within the XDP; and
(c) a tropism factor incorporated on the XDP surface.
10010941Embodiment V-278. The XDP particle of Embodiment V-277, further
comprising one
or more retroviral components selected from:
(a) a capsid polypeptide (CA);
291
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(b) a nucleocapsid polypeptide (NC);
(c) a P2A peptide, a P2B peptide;
(d) a P10 peptide;
(e) a p12 peptide
(f) a PP21/24 peptide;
(8) a P12/P3/P8 peptide;
(h) a P20 peptide;
(1) A pl peptide; and
(i) a p6 peptide.
10010951 Embodiment V-279. The XDP particle of
Embodiment V-277 or Embodiment
V-278, wherein the tropism factor is selected from the group consisting of a
glycoprotein, an
antibody fragment, a receptor, and a ligand to a target cell marker.
10010961 Embodiment V-280. The XDP particle of
Embodiment V-279, wherein the
tropism factor is a ,glycoprotein having an sequence selected from the group
consisting of SEQ
ID NOS: 438, 440, 442, 444, 446, 448, 450, 452, 454, 456, 458, 460, 462, 464,
466, 468, 470,
472, 474, 476, 478, 480, 482, 484, 486, 488, 490, 492, 494, 496, 498, 500,
502, 504, 506, 508,
510, 512, 514, 516, 518, 520, 522, 524, 526, 528, 530, 532, 534, 536, 538,
540, 542, 544, 546,
548, 550, 552, 554, 556, 558, 560, 562, 564, 566, 568, 570, 572, 574, 576,
578, 580, 582, 584,
586, 588, 590, 592, 594 and 596, or a sequence having at least about 85%, at
least about 90%, at
least about 91%, at least about 92%, at least about 93%, at least about 94%,
at least about 95%,
at least about 96%, at least about 97A, at least about 98%, or at least about
99% sequence
identity thereto.
10010971Embodiment V-281. The XDP particle of
Embodiment V-279, wherein the
tropism factor is a glycoprotein having an encoding sequence selected from the
group consisting
of SEQ ID NOS: 438, 440, 442, 444, 446, 448, 450, 452, 454, 456, 458, 460,
462, 464, 466,
468, 470, 472, 474, 476, 478, 480, 482, 484, 486, 488, 490, 492, 494, 496,
498, 500, 502, 504,
506, 508, 510, 512, 514, 516, 518, 520, 522, 524, 526, 528, 530, 532, 534,
536, 538, 540, 542,
544, 546, 548, 550, 552, 554, 556, 558, 560, 562, 564, 566, 568, 570, 572,
574, 576, 578, 580,
582, 584, 586, 588, 590, 592, 594 and 596_
10010981Embodiment V-282. The XDP particle of
any one of Embodiments V-277-281,
wherein the therapeutic payload comprises a protein, a nucleic acid, or
comprises both a protein
and a nucleic acid.
292
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10010991 Embodiment V-283. The XDP particle of
Embodiment V-282, wherein the
protein payload is selected from the group consisting of a cytokine, an
interleukin, an enzyme, a
receptor, a microprotein, a hormone, erythropoietin, RNAse, DNAse, a blood
clotting factor, an
anticoagulant, a bone morphogenetic protein, an engineered protein scaffold, a
tlu-ombolytic
protein, a CRISPR protein, and an anti-cancer modality.
10011001 Embodiment V-284. The XDP particle of
Embodiment V-283, wherein the
CRISPR protein is a Class 1 or Class 2 CRISPR protein.
[001101jEmbodiment V-285. The XDP particle of
Embodiment V-284, wherein the
CRISPR protein is a Class 2 CRISPR protein selected from the group consisting
of Type II,
Type V. or Type VI protein.
10011021 Embodiment V-286. The XDP particle of
Embodiment V-285, wherein the
CRISPR protein is a Type V protein selected from the group consisting of
Cas12a, Cas12b,
Cas12c, Cas12d (CasY), Cas12j and CasX.
10011031Embodiment V-287. The XDP particle of
Embodiment V-286, wherein the
CRISPR protein is a CasX comprising a sequence of SEQ ID NOS: 21-233, 343-345,
350-353,
355-367 or 388-397, or a sequence having at least about 85%, at least about
90%, at least about
91%, at least about 92%, at least about 93%, at least about 94%, at least
about 95%, at least
about 96%, at least about 97%, at least about 98%, or at least about 99%
sequence identity
thereto.
10011041 Embodiment V-288. The XDP particle of
Embodiment V-282, wherein the
therapeutic payload is a nucleic acid selected from the group consisting of a
single-stranded
antisense oligonucleotide (AS0s), a double-stranded RNA interference (RNAi)
molecule, a
DNA aptamer, and a CRISPR guide nucleic acid.
10011051 Embodiment V-289. The XDP particle of
Embodiment V-288, wherein the
CRISPR guide nucleic acid is a single-molecule guide RNA comprising a scaffold
sequence and
a targeting sequence, wherein the targeting sequence comprises between 14 and
30 nucleotides
and is complementary to a target nucleic acid sequence.
10011061Embodiment V-290. The XDP particle of
Embodiment V-289, wherein the
scaffold sequence comprises a sequence of SEQ ID NOS: 597-781, or a sequence
haying at least
about 85%, at least about 90%, at least about 91%, at least about 92%, at
least about 93%, at
least about 94%, at least about 95%, at least about 96%, at least about 97%,
at least about 98%,
or at least about 99% sequence identity thereto.
293
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10011071Embodiment V-291. The XDP particle of
Embodiment V-290, wherein the
scaffold sequence comprises a sequence of SEQ ID NOS: 597-781.
10011081Embodiment V-292. The XDP particle of
any one of Embodiments V-286-291,
wherein the therapeutic payload comprises a CasX and a guide RNA complexed as
a
ribonucleoprotein complex (RNP) and, optionally, a donor template.
10011091Embodiment V-293. The XDP particle of
any one of Embodiments V-277-292,
wherein the retroviral components are derived from a Orthoretrovirinae virus
or a
Spumaretrovirinae virus.
10011101Embodiment V-294. The XDP particle of
Embodiment V-293, wherein the
Orthoretrovirinae virus is selected from the group consisting of
Alpharetrovirus, Betaretrovirus,
Deltaretrovirus, Epsilonretrovirus, Gammaretrovirus, and Lentivirus,
[0011111Embodiment V-295. The XDP particle of
Embodiment V-293, wherein the
Spumaretrovirinae virus is selected from the group consisting of
Bovispumavirus,
Equispumavirus, Felispumavirus, Prosimiispumavirus, Simiispumavirus, and
Spumavirus.
10011121Embodiment V-296. The XDP particles, or
the XDP systems of any one of the
preceding embodiments, for use as a medicament for the treatment of a subject
having a disease.
10011131The present description sets forth numerous exemplary configurations,
methods,
parameters, and the like. It should be recognized, however, that such
description is not intended
as a limitation on the scope of the present disclosure, but is instead
provided as a description of
exemplary embodiments. Embodiments of the present subject matter described
above may be
beneficial alone or in combination, with one or more other aspects or
embodiments. Without
limiting the foregoing description, certain non-limiting embodiments of the
disclosure are
provided below. As will be apparent to those of skill in the art upon reading
this disclosure, each
of the individually numbered embodiments may be used or combined with any of
the preceding
or following individually numbered embodiments. This is intended to provide
support for all
such combinations of embodiments and is not limited to combinations of
embodiments explicitly
provided below.
EXAMPLES
Example 1: Creation, Expression and Purification of CasX Constructs
1. Growth and Expression
10011141An expression construct for CasX Stx2 (also referred to herein as
CasX2), derived
from Planctomycetes (having the amino acid sequence of SEQ ID NO: 2 and
encoded by the
294
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
sequence of the Table 6, below), was constructed from gene fragments (Twist
Biosciences) that
were codon optimized for E.coll. The assembled construct contains a TEV-
cleavable, C-
terminal, TwinStrep tag and was cloned into a pBR322-derivative plasmid
backbone containing
an ampicillin resistance gene. The expression construct was transformed into
chemically
competent BL21* (DE3)E. coil and a starter culture was grown overnight in LB
broth
supplemented with carbenicillin at 37 C, 200 RPM, in UltraYield Flasks
(Thomson Instrument
Company). The following day, this culture was used to seed expression cultures
at a 1:100 ratio
(starter culture:expression culture). Expression cultures were Terrific Broth
(Novagen)
supplemented with carbenicillin and grown in UltraYield flasks at 37 C, 200
RPM. Once the
cultures reached an OD of 2, they were chilled to 16 C and IPTG (isopropyl 13-
D-1-
thiogalactopyranoside) was added to a final concentration of 1 mM, from a 1 M
stock. The
cultures were induced at 16 C, 200 RPM for 20 hours before being harvested by
centrifugation
at 4,000xg for 15 minutes, 4 C. The cell paste was weighed and resuspended in
lysis buffer (50
mM REPES-NaOH, 250 m1VI NaC1, 5 mM MgCl2, 1 mM TCEP, 1 mM benzamidine-HCL, 1
mM PMSF, 0.5% CHAPS, 10% glycerol, pH 8) at a ratio of 5 mL of lysis buffer
per gram of
cell paste. Once resuspended, the sample was frozen at -80 C until
purification.
Table 6: DNA sequence of CasX Stx2 construct
Construct
DNA Sequence
SV40 NLS-CasX-SV40 NLS-TEV cleavage site ¨
(SEQ ID NO: 354)
TwinStrep tag
2. Purification
10011151Frozen samples were thawed overnight at 4 C with magnetic stirring.
The viscosity of
the resulting lysate was reduced by sonication and lysis was completed by
homogenization in
three passes at 17k PSI using an Emulsiflex C3 (Avestin). Lysate was clarified
by centrifugation
at 50,000x g, 4 C, for 30 minutes and the supernatant was collected. The
clarified supernatant
was applied to a Heparin 6 Fast Flow column (GE Life Sciences) by gravity
flow. The column
was washed with 5 CV of Heparin Buffer A (50 m/vl HEPES-NaOH, 250 mM NaCl, 5
mM
MgCl2, 1 mM TCEP, 10% glycerol, pH 8), then with 5 CV of Heparin Buffer B
(Buffer A with
the NaC1 concentration adjusted to 500 mM). Protein was eluted with 5 CV of
Heparin Buffer C
(Buffer A with the NaCl concentration adjusted to 1 M), collected in
fractions. Fractions were
assayed for protein by Bradford Assay and protein-containing fractions were
pooled. The pooled
295
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
heparin eluate was applied to a Strep-Tactin XT Superflow column (IBA Life
Sciences) by
gravity flow. The column was washed with 5 CV of Strep Buffer (50 mM HEPES-
NaOH, 500
m114 NaC1, 5 mM MgCl2, 1 mM TCEP, 10% glycerol, pH 8). Protein was eluted from
the
column using 5 CV of Strep Buffer with 50 mM D-Biotin added and collected in
fractions.
CasX-containing fractions were pooled, concentrated at 4 C using a 30 kDa cut-
off spin
concentrator, and purified by size exclusion chromatography on a Superdex 200
pg column (GE
Life Sciences). The column was equilibrated with SEC Buffer (25 mM sodium
phosphate, 300
mM NaC1, 1 mM TCEP, 10% glycerol, pH 7.25) operated by an AKTA Pure FPLC
system (GE
Life Sciences). CasX-containing fractions that eluted at the appropriate
molecular weight were
pooled, concentrated at 4 C using a 30 kDa cut-off spin concentrator,
aliquoted, and snap-frozen
in liquid nitrogen before being stored at -80 C.
3. Results
10011161 Samples from throughout the purification were resolved by SDS-PAGE
and visualized
by colloidal Coomassie staining, as shown in FIG. 1 and FIG. 3. In FIG. 1, the
lanes, from left to
right, are: molecular weight standards, Pellet: insoluble portion following
cell lysis, Lysate.
soluble portion following cell lysis, Flow Thai: protein that did not bind the
Heparin column,
Wash: protein that eluted from the column in wash buffer, Elution: protein
eluted from the
heparin column with elution buffer, Flow Thai: Protein that did not bind the
StrepTactinXT
column, Elution: protein eluted from the StrepTactin XT column with elution
buffer, Injection:
concentrated protein injected onto the s200 gel filtration column, Frozen:
pooled fractions from
the s200 elution that have been concentrated and frozen. In FIG. 3, the lanes
from right to left,
are the injection (sample of protein injected onto the gel filtration column)
molecular weight
markers, lanes 3 -9 are samples from the indicated elution volumes. Results
from the gel
filtration are shown in FIG. 2. The 68.36 mL peak corresponds to the apparent
molecular weight
of CasX and contained the majority of CasX protein. The average yield was 0.75
mg of purified
CasX protein per liter of culture, with 75% purity, as evaluated by colloidal
Coomassie staining.
Example 2: CasX construct CasX 119, 438 and 457
10011171 In order to generate the CasX 119, 438, and 457 constructs (sequences
in Table 7), the
codon-optimized CasX 37 construct (based on the CasX Stx2 construct of Example
1, encoding
Planctomyeetes CasX SEQ Wi NO: 2, with a A708K substitution and a [P793]
deletion with
fused NLS, and linked guide and non-targeting sequences) was cloned into a
mammalian
296
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
expression plasmid (pStX; see FIG. 4) using standard cloning methods. To build
CasX 119, the
CasX 37 construct DNA was PCR amplified in two reactions using Q5 DNA
polymerase (New
England BioLabs Cat# M0491L) according to the manufacturer's protocol, using
primers
oIC539 and oIC88 as well as oIC87 and oIC540 respectively (see FIG. 5). To
build CasX 457,
the CasX 365 construct DNA was PCR amplified in four reactions using Q5 DNA
polymerase
(New England BioLabs Cat# M0491L) according to the manufacturer's protocol,
using primers
oIC539 and oIC212, oIC211 and oIC376, oIC375 and oIC551, and oIC550 and oIC540
respectively. To build CasX 438, the CasX 119 construct DNA was PCR amplified
in four
reactions using Q5 DNA polymerase according to the manufacturer's protocol,
using primers
oIC539 and oIC689, oIC688 and oIC376, oIC375 and oIC551, and oIC550 and oIC540
respectively. The resulting PCR amplification products were then purified
using Zymoclean
DNA clean and concentrator (Zymo Research Cat# 4014) according to the
manufacturer's
protocol. The pStX backbone was digested using XbaI and SpeI in order to
remove the 2931
base pair fragment of DNA between the two sites in plasmid pStx34. The
digested backbone
fragment was purified by gel extraction from a 1% agarose gel (Gold Bio Cat# A-
201-500) using
Zymoclean Gel DNA Recovery Kit (Zymo Research Cat#D4002) according to the
manufacturer's protocol. The three fragments were then pieced together using
Gibson assembly
(New England BioLabs Cat* E2621S) following the manufacturer's protocol.
Assembled
products in the pStx34 were transformed into chemically-competent or electro-
competent Turbo
Competent E coil bacterial cells, plated on LB-Agar plates (LB: Teknova Cat#
L9315, Agar:
Quartzy Cat* 214510) containing carbenicillin. Individual colonies were picked
and
miniprepped using Qiagen spin Miniprep Kit (Qiagen Cat# 27104) following the
manufacturer's
protocol. The resultant plasmids were sequenced using Sanger sequencing to
ensure correct
assembly. pStX34 includes an EF-la promoter for the protein as well as a
selection marker for
both puromycin and carbenicillin. Sequences encoding the targeting sequences
that target the
gene of interest were designed based on CasX PAM locations. Targeting sequence
DNA was
ordered as single-stranded DNA (ssDNA) oligos (Integrated DNA Technologies)
consisting of
the targeting sequence and the reverse complement of this sequence. These two
oligos were
annealed together and cloned into pStX individually or in bulk by Golden Gate
assembly using
T4 DNA Ligase (New England BioLabs Cat# M0202L) and an appropriate restriction
enzyme
for the plasmid. Golden Gate products were transformed into chemically or
electro-competent
cells such as NEB Turbo competent E. coh (NEB Cat #C2984I), plated on LB-Agar
plates
297
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
containing carbenicillin. Individual colonies were picked and miniprepped
using Qiagen spin
Miniprep Kit and following the manufacturer's protocol. The resultant plasmids
were sequenced
using Sanger sequencing to ensure correct ligation. SaCas9 and SpyCas9 control
plasmids were
prepared similarly to pStX plasmids described above, with the protein and
guide regions of pStX
exchanged for the respective protein and guide. Targeting sequences for SaCas9
and SpyCas9
were either obtained from the literature or were rationally designed according
to established
methods. The expression and recovery of the CasX 119, 438 and 457 proteins was
performed
using the general methodologies of Example 1 (however the DNA sequences were
codon
optimized for expression in E. coli).
10011181 CasX Variant 119: following the same expression and purification
scheme for WT
CasX, the following results were obtained for CasX variant 119. Samples from
throughout the
purification procedure were resolved by SDS-PAGE and visualized by colloidal
Coomassie
staining, as shown in FIG. 6 and FIG. 8. Results from the gel filtration are
shown in FIG. 7. The
average yield was 11.7 mg of purified CasX protein per liter of culture at 95%
purity, as
evaluated by colloidal Coomassie staining.
10011191CasX Variant 438: Following the same expression and purification
scheme for WT
CasX, the following results were obtained for CasX variant 438. Samples from
throughout the
purification procedure were resolved by SDS-PAGE and visualized by colloidal
Coomassie
staining, as shown in FIGS. 9 and 11. Results from the gel filtration are
shown in FIG. 10. The
average yield was 13.1 mg of purified CasX protein per liter of culture at
97.5% purity, as
evaluated by colloidal Coomassie staining.
10011201 CasX Variant 457: Following the same expression and purification
scheme for WT
CasX, the following results were obtained for CasX variant 457. Samples from
throughout the
purification procedure were resolved by SDS-PAGE and visualized by colloidal
Coomassie
staining, as shown in FIGS. 12 and 14 and gel filtration, as shown in FIG. 13.
The average yield
was 9.76 mg of purified CasX protein per liter of culture at 91.6% purity, as
evaluated by
colloidal Coomassie staining.
10011211 Overall, the results support that CasX variants can be produced and
recovered at high
levels of purity sufficient for experimental assays and evaluation.
Table 7: Sequences of CasX 119,438 and 457
298
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA Amino
Acid Sequence
Sequence
CasX 119 (SEQ ID
QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRKKPE
NO: 340) NI PQPI SNTSRAN LN KLLTDYTEM KKAI LHVYWEEFQKDPVG LMSRVAQP
APKN I DQR KLI PVKDG N ER LTSSG FACSQCCQPLYVYKLEQVN DKGKPHTN
YFGRCNVSEHERLI LLSPH KPEAN D ELVTYSLG KFGQRALD FYSI HVTR ESN H
PVKPLECEIGGNSCASGPVGKALSDACMGAVASFLTKYQD1ILEHQKVI KKNE
KRLANLKDIASANG LAFPKITLPPQPHTKEGIEAYNNVVAQIVIWVNLNLW
QKLKIG RDEAKPLQRLKGEPSFPLVERQAN EVDWWDMVCNVKKLINEKKE
DGKVFWQN LAGYKRQEALRPYLSSE EDRKKG KKFARYQFGDLLLH LE KKH
G EDWG KVYDEAWERI DKKVEGLSKH I KLEEE RRSEDAQSKAALTDWLRAK
ASFVIEG LKEADKDEFCRCELKLQKWYG DLRG KPFAI EAENSI LDISG FSKQY
NCAFI WQKDGVKKLN LYLI I NYFKGG KLRFKKI KPEAFEAN RFYTVI N KKSG El
VPMEVNINFDDPNLIILPLAFGKRO.GREFIWNDLLSLETGSLKLANGRVIEK
TLYN RRTRQDEPALEVALTFE RREVLDSSN I KPM N LI GI DRGENI PAVIALTD
PEGCPLSRFKDSLG N PTH I LRI GESYKEKQRTI QAKKEVEQRRAGGYSRKYAS
KAKN LADDMVRNTARDLLYYAVTQDAM LI FEN LSRGFG RQG KRTFMAER
QYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCSNCGITITSADYDRVLE
KLKKTATGWMTTINGKELKVEGQITYYNRYKRQNVVKDLSVELDRLSEESV
NN DI SSWTKGRSGEALSLLKKR FSH RPVQEKFVCLNCGFETHADEQAALN I
ARSWLFLRSQEYKKYQTN KTTG NTDKRAFVETWQSFYRKKLKEVWKPAV
(SEQ ID NO: 343)
CasX 457 (SEQ ID
QEIKRINKIRRRLVKDSNTKKAGICGPMKTLLVRVMTPDLRERLENLRKKPE
NO: 341) NI PQPI SNTSRAN LN KLLTDYTEM KKAI LHVYWEEFQKDPVG LMSRVAQP
APKN I DQR KLI PVKDG N ER LTSSG FACSQCCOPLYVYKLEQVN DKGKPHTN
YFGRCNVSEHERLI LLSPH KPEAN D ELVTYSLG KFGQRALD FYSI HVTR ESN H
PVKPLECII GGNSCASG PVGKALSDACMGAVASFLTKYQDII LEHKKVIKKNE
KRLANLKDIASANG LAFPKITLPPQPHTKEGI EAYN NVVAQI VI WVNLNLW
QKLKIG RDEAKPLQRLKGEPSFPLVERQAN EVDWWDMVCNVKKLINEKKE
DGKVFWQN LAGYKRQEALRPYLSSPEDRKKGKKFARYQLGDLLLHLEKKH
G EDWG KVYDEAWERI DKKVEGLSKH I KLEEE RRSEDAQSKAALTDWLRAK
ASEVIEGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAENSILDISGESKQY
NCAFI WQKDGVKKLN LYLI I NYFKGG KLRFKKI KPEAFEAN RFYTVI N KKSG El
VPMEVNENFDDPNLIILPLAFGKRQGREFIWNDLLSLETGSLKLANGRVIEK
PLYNRRTRQDEPALEVALTFERREVLDSSN I KPM N LIGVDRG EN I PAVIALT
DPEGCPLSRFKDSLG N PTH I LRIG ESYKEKQRTIQAKKEVEQRRAGGYSRKY
ASKAKN LAD DMVRNTAR DLLYYAVTQDAM LI FEN LSRG FG RQG KRTFMA
E RQYTR ME DW LTAKLAYEGLSKTYLSKTLAQYTSKTCSNCG FTITSA DYD RV
LEKLKKTATGWMTTINGKELKVEGQITYYNRRKRQNVVKDLSVELDRLSEE
SVN ND ISSWTKGRSGEALSLLKKRFSH RPVQEKFVCLN CG FETHADEQAAL
NIARSWLFLRSQEYKKYQTN KTTGNTDKRAFVENVQSFYRKKLKEVWKPA
V (SEQ ID NO: 344)
CasX 438 (SEQ ID ..
QEIKRINKIRRRLVKDSNIKKAGKTGPMKTLLVRVMTPDLRERLENLRKKPE
NO: 342) NI PQPI SNTSRAN LN KLLTDYTEM KKAI LHVYWEEFQKDPVG LMSRVAQP
APKN I DQR KLI PVKDG N ER LTSSG FACSOCCOPLYVYKLEQVN DKGKPHTN
299
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
YFGRCNVSEHERLI LLSPHKPEANDELVTYSLGKFGQRALDFYSI HVTR ESN H
PVKPLECEIGGNSCASGPVGKALSDACMGAVASFLTKYQD1ILEHQKVI KKNE
KRLANLKDIASANG LAFPKITLPPQPHTKEGI EAYN NVVAQI VI WVNLNLW
QKLKIGRDEAKPLQRLKGFPSFPLVERQAN EVDWWDMVCNVKKLINEKKE
DGKVFWQN LAGYKRQEALRPYLSSEEDRKKG KKFARYQLGDLLKHLEKKH
GEDWGKVYDEAWERI DKKVEGLSKH I KLEEE RRSEDAQSKAALTDW LRAK
ASFVI EGLKEADKDEFCRCELKLQKWYGDLRGKPFAIEAENSI LDISGFSKQY
NCAF I WQKDGVKKLN LYLI I NYFKGG KLRFKKI KPEAFEAN RFYTVI N KKSG El
VP M EVN FN FD DPN LI I LPLAFG KRQG REF IWN DLLSLETGSLKLA NG RVI EK
TLYN RRTRQDEPALFVALTFE RREVLDSSN I KPM N LI GVDRGENI PAVIALTD
PEGCPLSRFKDSLG N PTH I LRIGESYKEKORTIQAKKEVEQRRAGGYSRKYAS
KAKN LADDMVRNTARDLLYYAVTQDAM LI F EN LSRGFG RQG KRTFMAER
QYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTSKTCSNCGFTITSADYDRVLE
KLKKTATGWMTTI NGKELKVEGQITYYNRRKRQNVVKDLSVELDRLSEESV
N N DI SSWTKGRSGEALSLLKKR FSH RPVQEKFVCLNCGF ETHADEQAALN I
ARSWLFLRSQEYKKYQTNKTTG NTDKRAFVETVVQSFYRKKLKEVWKPAV
(SEQ ID NO: 345)
Example 3: CasX construct 438, 491, 515 and 527
10011221 In order to generate the CasX 488 construct (sequences in Table 8),
the codon-
optimized CasX 119 construct (based on the CasX Stx2 construct of Example 1,
encoding
Planctomycetes CasX SEQ II NO: 2, with a A708K substitution, a L379R
substitution, and a
[P793] deletion with fused NLS, and linked guide and non-targeting sequences)
was cloned into
a destination plasmid (pStX; see FIG. 4) using standard cloning methods. In
order to generate
the CasX 491 construct (sequences in Table 8), the codon-optimized CasX 484
construct (based
on the CasX Stx2 construct of Example 1, encoding Planctomycetes CasX SEQ lD
NO: 2, with
a A708K substitution, a L379R substitution, a [P793] deletion, a I658V
substitution, and a
F399L substitution with fused NLS, and linked guide and non-targeting
sequences) was cloned
into a destination plasmid (pStX; see FIG. 4) using standard cloning methods.
Construct CasX 1
(CasX SEQ ID NO: 1) was cloned into a destination vector using standard
cloning methods. To
build CasX 488, the CasX 119 construct DNA was PCR amplified using Q5 DNA
polymerase
according to the manufacturer's protocol, using primers oIC765 and oIC762 (see
FIG. 5). To
build CasX 491, the codon optimized CasX 484 construct DNA was PCR amplified
using Q5
DNA polymerase according to the manufacturer's protocol, using primers oIC765
and oIC762
(see FIG. 5). The CasX 1 construct was PCR amplified using Q5 DNA polymerase
according to
the manufacturer's protocol, using primers oIC766 and oIC784. Each of the PCR
products were
purified by gel extraction from a 1% agarose gel (Gold Rio Cat# A-201-500)
using Zymoclean
300
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Gel DNA Recovery Kit according to the manufacturer's protocol. The
corresponding fragments
were then pieced together using Gibson assembly (New England BioLabs Cat#
E2621S)
following the manufacturer's protocol. Assembled products in pStx1 were
transformed into
chemically-competent Turbo Competent E. coli bacterial cells, plated on LB-
Agar plates
containing kanamycin. Individual colonies were picked and miniprepped using
Qiagen spin
Miniprep Kit following the manufacturer's protocol. The resultant plasmids
were sequenced
using Sanger sequencing to ensure correct assembly. The correct clones were
then subcloned
into the mammalian expression vector pStx34 using restriction enzyme cloning.
The pStx34
backbone and the CasX 488 and 491 clones in pStx1 were digested with XbaI and
BamHIE
respectively. The digested backbone and respective insert fragments were
purified by gel
extraction from a 1% agarose gel (Gold Bio Cat# A-201-500) using Zymoclean Gel
DNA
Recovery Kit according to the manufacturer's protocol. The clean backbone and
insert were then
ligated together using T4 Ligase (New England Biolabs Cat# M0202L) according
to the
manufacturer's protocol. The ligated products were transformed into chemically-
competent
Turbo Competent E. coli bacterial cells, plated on LB-Agar plates containing
carbenicillin.
Individual colonies were picked and miniprepped using Qiagen spin Miniprep Kit
following the
manufacturer's protocol. The resultant plasmids were sequenced using Sanger
sequencing to
ensure correct assembly.
10011231To build CasX 515 (sequences in Table 8), the CasX 491 construct DNA
was PCR
amplified in two reactions using Q5 DNA polymerase according to the
manufacturer's protocol,
using primers oIC539 and oSH556 as well as oSH555 and oIC540 respectively (see
FIG. 5). To
build CasX 527 (sequences in Table 8), the CasX 491 construct DNA was PCR
amplified in two
reactions using Q5 DNA polymerase according to the manufacturer's protocol,
using primers
oIC539 and oSH584 as well as oSH583 and oIC540 respectively. The PCR products
were
purified by gel extraction from a 1% agarose gel using Zymoclean Gel DNA
Recovery Kit
according to the manufacturer's protocol. The pStX backbone was digested using
XbaI and SpeI
in order to remove the 2931 base pair fragment of DNA between the two sites in
plasmid
pStx56. The digested backbone fragment was purified by gel extraction from a
1% agarose gel
using Zymoclean Gel DNA Recovery Kit according to the manufacturer's protocol.
The insert
and backbone fragments were then pieced together using Gibson assembly (New
England
BioLabs Cat# E2621S) following the manufacturer's protocol. Assembled products
in the
pStx56 were transformed into chemically-competent Turbo Competent E. coli
bacterial cells,
301
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
plated on LB-Agar plates containing kanamycin. Individual colonies were picked
and
miniprepped using Qiagen spin Miniprep Kit following the manufacturer's
protocol. The
resultant plasmids were sequenced using Sanger sequencing to ensure correct
assembly. pStX34
includes an EF-1ot promoter for the protein as well as a selection marker for
both puromycin and
carbenicillin. pStX56 includes an EF-1ot promoter for the protein as well as a
selection marker
for both puromycin and kanamycin Sequences encoding the targeting sequences
that target the
gene of interest were designed based on CasX PAM locations. Targeting sequence
DNA was
ordered as single-stranded DNA (ssDNA) oligos (Integrated DNA Technologies)
consisting of
the targeting sequence and the reverse complement of this sequence. These two
oligos were
annealed together and cloned into pStX individually or in bulk by Golden Gate
assembly using
T4 DNA Ligase and an appropriate restriction enzyme for the plasmid. Golden
Gate products
were transformed into chemically or electro-competent cells such as NEB Turbo
competent E.
coli (NEB Cat #C2984I), plated on LB-Agar plates containing the appropriate
antibiotic.
Individual colonies were picked and miniprepped using Qiaprep spin Miniprep
Kit and
following the manufacturer's protocol. The resultant plasmids were sequenced
using Sanger
sequencing to ensure correct ligation. SaCas9 and SpyCas9 control plasmids
were prepared
similarly to pStX plasmids described above, with the protein and guide regions
of pStX
exchanged for the respective protein and guide. Targeting sequences for SaCas9
and SpyCas9
were either obtained from the literature or were rationally designed according
to established
methods. The expression and recovery of the CasX constructs was performed
using the general
methodologies of Example 1 and are summarized as follows:
10011241CasX variant 488: following the same expression and purification
scheme for WT
CasX SEQ ID NO: 2, the following results were obtained for CasX variant 488.
Samples from
throughout the purification procedure were resolved by SDS-PAGE and visualized
by colloidal
Coomassie staining, as well as resolved by gel filtration. The average yield
was 2.7 mg of
purified CasX protein per liter of culture at 98.8% purity, as evaluated by
colloidal Coomassie
staining.
0011251 CasX Variant 491: following the same expression and purification
scheme for WT
CasX SEQ ID NO: 2, the following results were obtained for CasX variant 488.
Samples from
throughout the purification procedure were resolved by SDS-PAGE and visualized
by colloidal
Coomassie staining, as well as resolved by gel filtration. The average yield
was 12.4 mg of
302
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
purified CasX protein per liter of culture at 99.4% purity, as evaluated by
colloidal Coomassie
staining.
10011261CasX variant 515: following the same expression and purification
scheme for WT
CasX SEQ 113 NO: 2, the following results were obtained for CasX variant 488.
Samples from
throughout the purification procedure were resolved by SDS-PAGE and visualized
by colloidal
Coomassie staining, as well as resolved by gel filtration. The average yield
was 7.8 mg of
purified CasX protein per liter of culture at 87.2% purity, as evaluated by
colloidal Coomassie
staining.
Table 8: Sequences of CasX 488, 491, 515 and 527
Construct DNA Amino
Acid Sequence
Segue
nce
CasX 488 (SEQ QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRER
ID NO: LENLRKKPENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWE
346) EFQKDPVGLMSRVAQPASKKIDQNKLKPEMDEKGNLTTAGF
ACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCNVAEHEKLILL
AOLKPEKDSDEAVTYSLGKFGORALDFYSIHVTKESTHPVKPL
AQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQKVVK
GNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEVI
ARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQA
NEVDVVVVDMVCNVKKLINEKKEDGKVFWQNLAGYKRQEALR
PYLSSEEDRKKGKKFARYQFGDLLLHLEKKHGEDWGKVYDE
AWERIDKKVEGLSKHIKLEEERRSEDAQSKAALTDVVLRAKAS
FVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSILDI
SGFSKQYNCAFIVVQKDGVKKLNLYLIINYFKGGKLRFKKIKPE
AFEANRFYTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQG
REFIVVNDLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVA
LTFERREVLDSSNIKPMNLIGIDRGENIPAVIALTDPEGCPLSR
FKDSLGNPTHILRIGESYKEKQRTIQAKKEVEQRRAGGYSRKY
ASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFGRQ
GKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSK
TCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQI
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSWTKGRSGE
ALSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSWL
FLRSQEYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVVVK
PAV (SEQ ID NO: 350)
CasX 491 (SEQ QEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRER
ID NO: LENLRKKPENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWE
347) EFQKDPVGLMSRVAQPASKKIDQNKLKPEMDEKGNLTTAGF
ACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCNVAEHEKLILL
AQLKPEKDSDEAVTYSLGKFGQRALDFYSIHVTKESTHPVKPL
AQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQKVVK
303
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
GNQ KR LESLR ELAGKENLEYPSVTLPPQ PHTKEGVDAYNEVI
ARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQA
NEVDVVVVDMVCNVKKLINEKKEDGKVFWQNLAGYKRQEALR
PYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWGIONDE
AWER I DKKVEGLSKH IKLEE ERRSE DAQSKAALTDVVLRAKAS
FVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSILDI
SGFS KQYNCAF IWQ KDGVKKLN LYL I INYF KGG KLRFKKI KPE
AFEANRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQG
REF IWNDLLSLETGS LKLANG RVI EKTLYN RRTRQ DE PALFVA
LTFERREVLDSSN IKPM NLIGVDRGEN I PAVIALTDP EGC PLSR
F KDSLGNPTHILR IGESYKEKQ RTIQAKKEVEQR RAGGYSRKY
ASKAKN LADDMVRNTARD LLYYAVTQ DAML I FENLSRGFGRQ
GKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSK
TCS N CGFTITSADYD RVLEKLKKTATGVVMTTI NGKE LKVEGQ I
TYYN RYKRQ NVVKD LSVELDRLS E ESVN ND I SSWTKGRSGE
ALS LLKKRFS H RPVQ EKFVC LNCGFETHADEQAALN IARSWL
FLRSQEYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVVVK
PAV (SEQ ID NO: 351)
CasX 515 (SEQ QEIKRIN KIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRER
ID NO: LENLRKKPENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWE
348) EFQKDPVGLMSRVAQPASKKIDQNKLKPEMDEKGNLTTAGF
ACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCNVAEHEKLILL
AOLKP EKDSDEAVTYSLGKFGQ RALD FYS I HVTKESTH PVKPL
AQIAGNRYASGPVGKALSDACMGTIASFLSKYQDIIIEHQKVVK
GNQKRLESLRELAGKENLEYPSVTLPPQ PHTKEGVDAYNEVI
ARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQA
NEVDVVVVDMVCNVKKLINEKKEDGKVFWQNLAGYKRQEALR
PYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWGKVYDE
AWER I DKKVEGLSKH IKLEE ERRSE DAQSKAALTDVVLRAKAS
FVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSILDI
SGFS KQYNCAF IVVQ KDGVKKLN LYL I INYF KGG KLRFKKI KPE
AFEANRFYTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQG
REF IVVNDLLSLETGS LKLANG RVI EKTLYN RRTRQ DE PALFVA
LTFERREVLDSSN IKPM NLIGVDRGEN I PAVIALTDP EGGPLSR
F KDSLGNPTHILR IGESYKEKQ RTIQAKKEVEQR RAGGYSRKY
AS KAKN LADDMVRNTARD LLYYAVTQ DAM L I FE N LS RGFG RQ
GKRTFMAERQYTRMEDWLTAKLAYEGLPSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEG
IDITYYNRYKRONVVKDLSVELDRLSEESVNNDISSVVTKGRSG
EALSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALN IARSW
LFLRSQEYKKYQTNKTTGNTDKRAFVETWQSFYRKKLKEVW
KPAV (SEQ ID NO: 352)
CasX 527 (SEQ Q EIKR IN KIR RR LVKDSNTKKAGKTRGPM KTLLVRVMTP DLR E
ID NO: RLEN LRKKPEN IPQP ISNTSRANLNKLLTDYTEMKKAILHVYW
349) EEFQ KD PVGLMSRVAQPASKKIDQNKLKPEMDEKGNLTTAG
FACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCNVAEHEKLIL
LAQ LKP E KDS DEAVTYSLGKFGQ RALD FYS I HVTKE STH PVKP
LAO IAGN RYASG PVGKALS DACMGTIAS FLS KYQ DI II EH QKVV
304
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
KGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGVDAYNEV
IARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSFPLVERQA
NEVDVVVVDMVCNVKKLINEKKEDGKVFWQNLAGYKRQEALR
PYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWGIONDE
AWERIDKKVEGLSKHIKLEEERRSEDAQSKAALTDVVLRAKAS
FVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENSILDI
SGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKPE
AFEANRFYTVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKRQG
REF IWNDLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVA
LTFERREVLDSSNIKPMNLIGVDRGENIPAVIALTDPEGCPLSR
FKDSLGNPTHILRIGESYKEKQRTIQAKKEVEQRRAGGYSRKY
ASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFGRQ
GKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTSK
TCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQI
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVIKGRSGE
ALSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSWL
FLRSQEYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVVVK
PAV (SEQ ID NO: 353)
Example 4: Design and Generation of CasX Constructs 278-280, 285-288, 290,
291, 293,
300, 492, and 493
10011271ln order to generate the CasX 278-280, 285-288, 290, 291, 293, 300,
492, and 493
constructs (sequences in Table 9), the N- and C-termini of the codon-optimized
CasX 119
construct (based on the CasX Stx37 construct of Example 2, encoding
Planctomycetes CasX
SEQ ID NO: 2, with a A708K substitution and a [P793] deletion with fused NLS,
and linked
guide and non-targeting sequences) in a mammalian expression vector were
manipulated to
delete or add NLS sequences (sequences in Table 10). Constructs 278, 279, and
280 were
manipulations of the N- and C-termini using only an SV40 NLS sequence.
Construct 280 had no
NLS on the N-terminus and added two SV40 NLS' on the C-terminus with a triple
proline linker
in between the two SV40 NLS sequences. Constructs 278, 279, and 280 were made
by
amplifying pStx34.119.174.NT with Q5 DNA polymerase according to the
manufacturer's
protocol, using primers oIC527 and oIC528, oIC730 and oIC522, and oIC730 and
oIC530 for
the first fragments each and using oIC529 and oIC520, oIC519 and oIC731, and
oIC529 and
oIC731 to create the second fragments each. These fragments were purified by
gel extraction
from a 1% agarose gel using Zymoclean Gel DNA Recovery Kit according to the
manufacturer's
protocol. The respective fragments were cloned together using Gibson assembly
(New England
BioLabs Cat# E2621S) following the manufacturer's protocol. Assembled products
in the
pStx.34 were transformed into chemically-competent Turbo Competent E. colt
bacterial cells,
305
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
plated on LB-Agar plates containing carbenicillin and incubated at 37 C.
Individual colonies
were picked and miniprepped using Qiagen spin Miniprep Kit following the
manufacturer's
protocol. The resultant plasmids were sequenced using Sanger sequencing to
ensure correct
assembly. Sequences encoding the targeting sequences that target the gene of
interest were
designed based on CasX PAM locations. Targeting sequence DNA was ordered as
single-
stranded DNA (ssDNA) oligos (Integrated DNA Technologies) consisting of the
targeting
sequence and the reverse complement of this sequence. These two oligos were
annealed together
and cloned into pStX individually or in bulk by Golden Gate assembly using T4
DNA Ligase
(New England BioLabs Cat# M0202L) and an appropriate restriction enzyme for
the plasmid.
Golden Gate products were transformed into chemically- or electro-competent
cells such as NEB
Turbo competent E. cob (NEB Cat #C2984I), plated on LB-Agar plates containing
carbenicillin
and incubated at 37 C. Individual colonies were picked and miniprepped using
Qiagen spin
Miniprep Kit and following the manufacturer's protocol. The resultant plasmids
were sequenced
using Sanger sequencing to ensure correct ligation.
10011281 In order to generate constructs 285-288, 290, 291, 293, and 300, a
nested PCR method
was used for cloning. The backbone vector and PCR template used was construct
pStx34
279.119.174.NT, having the CasX 119, guide 174, and non-targeting spacer (see
Examples 8 and
9 and Tables therein for sequences). Construct 278 has the configuration
SV4ONLS-CasX119.
Construct 279 has the configuration CasX119-SV4ONLS. Construct 280 has the
configuration
CasX119-SV4ONLS-PPP linker-SV4ONLS. Construct 285 has the configuration
CasX119-
SV4ONLS-PPP linker-SynthNLS3. Construct 286 has the configuration CasX119-
SV4ONLS-
PPP linker-SynthNLS4. Construct 287 has the configuration CasX119-SV4ONLS-PPP
linker-
SynthNLS5. Construct 288 has the configuration CasX119-SV4ONLS-PPP linker-
SynthNLS6.
Constrict 290 has the configuration CasX119-SV4ONLS-PPP linker-EGL-13 NLS.
Construct
291 has the configuration CasX119-SV4ONLS-PPP linker-c-Myc NLS. Construct 293
has the
configuration CasX119-SV4ONLS-PPP linker-Nucleolar RNA Helicase II NLS.
Construct 300
has the configuration CasX119-SV4ONLS-PPP linker-Influenza A protein NLS.
Construct 492
has the configuration SV4ONLS-CasX119- SV4ONLS-PPP linker-SV4ONLS. Construct
493 has
the configuration SV4ONLS-CasX119- SV4ONLS-PPP linker-c-Myc NLS. Each variant
had a
set of three PCRs; two of which were nested, were purified by gel extraction,
digested, and then
ligated into the digested and purified backbone. Assembled products in the
pStx34 were
transformed into chemically-competent Turbo Competent E. coil bacterial cells,
plated on LB-
306
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Agar plates containing carbenicillin and incubated at 37 C. Individual
colonies were picked and
miniprepped using Qiagen spin Miniprep Kit following the manufacturer's
protocol. The
resultant plasmids were sequenced using Sanger sequencing to ensure correct
assembly.
Sequences encoding the targeting sequences that target the gene of interest
were designed based
on CasX PAM locations. Targeting sequence DNA was ordered as single-stranded
DNA
(ssDNA) oligos (Integrated DNA Technologies) consisting of the targeting
sequence and the
reverse complement of this sequence. These two oligos were annealed together
and cloned into
the resulting pStX individually or in bulk by Golden Gate assembly using T4
DNA Ligase (New
England BioLabs Cat# M0202L) and an appropriate restriction enzyme for the
plasmid. Golden
Gate products were transformed into chemically- or electro-competent cells
such as NEB Turbo
competent E. cold (NEB Cat #C2984I), plated on LB-Agar plates containing
carbenicillin and
incubated at 37 C. Individual colonies were picked and miniprepped using
Qiagen spin Miniprep
Kit and following the manufacturer's protocol. The resultant plasmids were
sequenced using
Sanger sequencing to ensure correct ligation.
10011291 In order to generate constructs 492 and 493, constructs 280 and 291
were digested
using XbaI and Banaill (NEB# R01455 and NEB# R31365) according to the
manufacturer's
protocol. Next, they were purified by gel extraction from a 1% agarose gel
using Zymoclean Gel
DNA Recovery Kit according to the manufacturer's protocol. Finally, they were
ligated using T4
DNA ligase (NEB# M02025) according to the manufacturer's protocol into the
digested and
purified pSbc34,119.174,NT using XbaI and BamHE and Zymoclean Gel DNA Recovery
Kit.
Assembled products in the pStx34 were transformed into chemically-competent
Turbo
Competent E call bacterial cells, plated on LB-Agar plates containing
carbenicillin and
incubated at 37 C. Individual colonies were picked and miniprepped using
Qiagen spin Miniprep
Kit following the manufacturer's protocol. The resultant plasmids were
sequenced using Sanger
sequencing to ensure correct assembly. Sequences encoding the targeting spacer
sequences that
target the gene of interest were designed based on CasX PAM locations.
Targeting sequence
DNA was ordered as single-stranded DNA (ssDNA) oligos (Integrated DNA
Technologies)
consisting of the targeting spacer sequence and the reverse complement of this
sequence. These
two oligos were annealed together and cloned into each pStX individually or in
bulk by Golden
Gate assembly using T4 DNA Ligase (New England BioLabs Cat# M0202L) and an
appropriate
restriction enzyme for the respective plasmids. Golden Gate products were
transformed into
chemically- or electro-competent cells such as NEB Turbo competent E cold (NEB
Cat
307
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
#C2984I), plated on LB-Agar plates containing carbenicillin and incubated at
37 C. Individual
colonies were picked and miniprepped using Qiagen spin Miniprep Kit and
following the
manufacturer's protocol. The resultant plasmids were sequenced using Sanger
sequencing to
ensure correct ligation. The plasmids would be used to produce and recover
CasX protein
utilizing the general methodologies of Examples 1 and 2.
Table 9: CasX 278-280, 285-288, 290, 291, 293, 300, 492, and 493 sequences
Construct Amino Acid
Sequence
278 MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTP
DLRERLENLRKKPENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEF
QKDPVGLMSRVAQPAPKNIDORKLIPVKDGNERLTSSGFACSOCCOPLY
VYKLEQVNDKGKPHTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLG
KFGQRALDFYSIHVTRESNHPVKPLEQIGGNSCASGPVGKALSDACMGA
VASFLTKYQD1ILEHQKVIKKNEKRLANLKDIASANGLAFPKITLPPOPHTK
EGIEAYNNVVAQIVIVVVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQ
ANEVDWVVDMVCNVKKLINEKKEDGKVFWONLAGYKRQEALRPYLSSE
EDRKKGKKFARYQFGDLLLHLEKKHGEDWGIWYDEAWERIDKKVEGLS
KHIKLEEERRSEDAQSKAALTDVVLRAKASFVIEGLKEADKDEFCRCELKL
QKWYGDLRGKPFAIEAENSILDISGFSKQYNCAFIWQKDGVKKLNLYLIIN
YFKGGKLRFKKIKPEAFEANRFYIVINKKSGEIVPMEVNFNFDDPNLIILPL
AFGKROGREFIVVNDLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVA
LTFERREVLDSSNIKPMNLIGIDRGENIPAVIALTDPEGCPLSRFKDSLGN
PTHILRIGESYKEKQRTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVR
NTARDLLYYAVTQDAMLIFENLSRGFGRQGKRTFMAERQYTRMEDWLT
AKLAYEGLSKTYLSKTLAQYTSKTCSNCGFTITSADYDRVLEKLKKTATG
WMTTINGKELKVEGQITYYNRYKRQNVVKDLSVELDRLSEESVNNDISS
WTKGRSGEALSLLKKRFSHRPVQEKFVGLNCGFETHADEQAALNIARS
WLFLRSQEYKKYQTNKTTGNTDKRAFVETWQSFYRKKLKEVVVKPAV
(SEQ ID NO: 355)
279 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KPENIPQPISNTSRANLNKLLTDYTEMKKAILHVYWEEFQKDPVGLMSRV
AQPAPKNIDQRKLIPVKDGNERLTSSGFACSQCCQPLYVYKLEQVNDKG
KPHTNYFGRCNVSEHERLILLSPHKPEANDELVTY'SLGKFGQRALDFYSI
HVTRESNHPVKPLEGIGGNSCASGPVGKALSDACMGAVASFLTKYQD1IL
EHQKVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVAQI
VIWVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWVVDMVC
NVKKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FGDLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKHIKLEEERRSEDA
OSKAALTDV'VLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKP FA
lEAENSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRF'YTVINKKSGEIVPMEVNENFDDPNLIILPLAFGKROGREFIVVN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNI
KPMNLIGIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
308
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct] Amino Acid
Sequence
M L I FE N LS RGFGRQ GKRTFMAERQYTRM EDVVLTAKLAYEG LS KTYLS KT
LAQYTSKTCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVC LN CGFETHADEQAALN IARSVVLF LRSQ EYKKYQTN KT
TGNTDKRAFVETVVQSFYRKKLKEVWKPAVTSPKKKRKV (SEQ ID NO:
356)
280 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN IPQ P IS NTSRANLNKLLTDYTEMKKAILHVYWE EFQ KDPVGLMSRV
AQ PAP KN IDQ RKLIPVKDGNE RLTSSGFACSQ CCQ PLYVYKLEQVNDKG
KPHTNYFGRCNVSEH ERLI LLS PH KPEAN DELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQDIIL
EHQKVI KKN E KRLANLKD IASANGLAFPKITLP PQ PHTKEG I EAYNNVVAQ I
VIVVVNLNLWOKLKIGRDEAKPLQRLKGFPSFPLVEROANEVDVVVVDMVC
NVKKL I N E KKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FG DLLLH LEKKHGEDWG KVYD EAWE RIDKKVEG LSKH IKLEE ERRSE DA
QS KAALTDVVLRAKAS FVI E GLKEAD KD E FCRC E LKLQ KVVYGDLRGKP FA
I EAENS ILD ISG FSKQYN CAF IWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRFYTVINKKSGE IVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVN
D LLS LETGSLKLAN G RVI E KTLYN R RTRQ D E PALFVALTFE RREVLDS SN I
KPM NLIG ID RGE N I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAERQ'YTRM EDVVLTAKLAYEG LS KTYLS KT
LAQYTSKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSWTKGRSGEALSLLKKR
FS H RPVQ E KFVC LN CGFETHADEQAALN IARSVVLF LRSQ EYKKYQTN KT
TGNTDKRAFVETWQSFYRKKLKEVWKPAVTSPKKKRKVPPPPKKKRKV
(SEQ ID NO: 357)
285 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYWE EFQ KDPVGLMSRV
AQ PAP KN IDQ RKLIPVKDGNE RLTSSGFAC SQ CCQ PLYVYKLEQVNDKG
KPHTNYFGRCNVSEH ERLI LLS PH KPEAN DELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKPLEQ IGGNSCASGPVGKALS DACMGAVASF LTKYQD It
EHQKVI KKN E KRLANLKD IASANGLAFPKITLP PQ PHTKEG I EAYN NVVAQ I
VIVVVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVWDMVC
NVKKL I N E KKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FG DLLLH LEKKHGEDWGKVYD EAWE RIDKKVEG LSKH IKLEE ERRSE DA
QS KAALTDVVLRAKAS FVI E GLKEAD KD E FCRC E LKLQ KVVYGDLRGKP FA
I EAENS ILD ISG FSKQYN CAF IWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRFYTVINKKSGE IVPMEVNFNFDDPNLIILPLAFGKRQGREFIWN
D LLS LETGSLKLAN G RVI E KTLYN RRTRQ D E PALFVALTFE RREVLDS SN I
KPM NLIG ID RGE N I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAERQ'YTRM EDWLTAKLAYEG LS KTYLS KT
LAQYTSKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVCLN CGFETHADEQAALN IARSVVLF LRSQ EYKKYQTN KT
309
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct] Amino Acid
Sequence
TGNTDKRAFVETVVQSFYRKKLKEVVVKPAVISPKKKRKVPPPHKKKHPD
ASVNFSEFSK (SEQ ID NO: 358)
286 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYVVE EFQ KDPVGLMSRV
AQ PAP KNIDQRKLIPVKDGNERLTSSGFACSQCCQ PLYVYKLEQVNDKG
KPHTNYFGRCNVSEH ER LILLSPHKPEANDELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIIL
EHQKVI KKN E KR LANLKD IASANGLAFPKITLP PQ PHTKEG I EAYNNVVAQ I
VIVVVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVWDMVC
NVKKLINEKKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FGDLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDA
QS KAALTDWLRAKAS FVI E GLKEAD KD E FC RC E LKLQ KVVYGDLRGKP FA
I EAENS ILD ISGFSKQYNCAF IWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRF'YTVINKKSGE IVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNI
KPMNLIGIDRGEN I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAEROYTRM EDVVLTAKLAYEG LS KTYLS KT
LAQYTS KTCSNCGFTITSADYD RVLEKLKKTATGVVMTTI NGKELKVEGQ I
TYYNRYKRONVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVCLN CGFETHADEQAALN IARSWLF LRSQ EYKKYQTN KT
TGNTDKRAFVETWQSFYRKKLKEVINKPAVISPKKKRKVPPPQRPGPYD
RPQRPGPYDRP (SEQ ID NO: 359)
287 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYWE EFQ KDPVGLMSRV
AQ PAP KNIDQRKLIPVKDGNERLTSSGFACSQCCQ PLYVYKLEQVNDKG
KPHTNYFGRCNVSEH ER LILLSPHKPEANDELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIIL
EHQKVI KKN E KRLANLKD IASANGLAFPKITLP PQ PHTKEG I EAYNNVVAQ I
VIWVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVWDMVC
NVKKLINEKKE DGKVFWQ N LAGYKRO EALRPYLSS EE D RKKGKKFARYQ
FGDLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDA
QS KAALTDVVLRAKAS FVI E GLKEAD KD E FC RC E LKLQ KVVYGDLRGKP FA
lEAENSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRFYTVINKKSGE IVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNI
KPMNLIGIDRGEN I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAERQYTRM EDVVLTAKLAYEG LS KTYLS KT
LAQYTS KTCSNCGFTITSADYD RVLEKLKKTATGVVMTTI NGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVCLN CGFETHADEQAALN IARSWLF LRSQ EYKKYQTN KT
TGNTDKRAFVETINQSFYRKKLKEVVVKPAVTSPKKKRKVPPPLSPSLSPL
LSPSLSPL (SEQ ID NO: 360)
288 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYVVE EFQ KDPVGLMSRV
310
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct] Amino Acid
Sequence
AQ PAP KN IDQ RKLIPVKDGNERLTMSSGFACS Q CC Q PLYVYKLEQVNDK
GKPHTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFY
S IHVTR ESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVAS FLTKYQ DI
ILEHQKVIKKNEKRLANLKDIASANGLAFPKITLPPQPHTKEGIEAYNNVVA
QIVIVVVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWWDMV
C NVKKL I N E KKE DGKVFWQ N LAGYKRQEALRPYLSS EE DRKKG KKFARY
QFGDLLLHLEKKHGEDWGIWYDEAVVERIDKKVEGLSKH IKLEEERRSED
AQSKAALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPF
AlEAENSILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIK
PEAFEANRFYTVI NKKSGEIVPMEVN FN FDDP N LI ILPLAFGKRQGREFIW
NDLLSLETGSLKLANGRVI EKTLYNRRTRQDEPALFVALTFERREVLDSS
NIKPMNLIGIDRGENIPAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKE
KQRTIQAKKEVEQ RRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVT
Q DAM L I F EN LSRGFG RQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTY
LSKTLAQYTSKTCSNCOFTITSADYDRVLEKLKKTATGVVMTTINGKELKV
EGQITYYN RYKRQ NVVKD LSVE LD R LS E ESVN N D ISSWTKG RSG EALS L
LKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVLFLRSQEYKKYQ
TN KTTGNTD KRAFVETWQ SFYRKKLKEV1NKPAVTS PKKKRKVPP PRG K
GGKGLGKGGAKRHRK (SEQ ID NO: 361)
290 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN IPQ P ISNTSRANLNKLLTDYTEMKKAILHVYWEEFQ KDPVGLMSRV
AC:213AP KN IDQ RKLIPVKDGNERLTSSGFAC SQCCQ PLYVYKLEQVNDKG
KPHTNYFGRCNVSEH ERLI LLS PH KPEAN DELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKP LEO IGGNSCASGPVGKALSDACMGAVASF LTKYQDIIL
EHQKVI KKN E KRLANLKD IASANGLAFPKITLP PQ PHTKEG I EAYNNVVAQ I
VIWVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWVVDMVC
NVKKLINEKKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FGDLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDA
QS KAALTDVVLRAKAS FVI E GLKEAD KD E FCRC E LKLQ KVVYGDLRGKP FA
lEAENSILDISGFSKQYNCAF IWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRFYTVINKKSGE IVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNI
KPMNLIGIDRGEN I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAERQYTRM ED1NLTAKLAYEG LS KTYLS KT
LAQYTS KTCSNCGFTITSADYD RVLEKLKKTATGVVMTTI NGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVC LN CGFETHADEQAALN IARSVVLF LRSQ EYKKYQTN KT
TGNTDKRAFVERNQSFYRKKLKEVVVKPAVISPKKKRKVPPPSRRRKAN
PTKLSENAKKLAKEVEN (SEQ ID NO: 362)
291 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN IPQ P ISNTSRANLNKLLTDYTEMKKAILHVYWEEFQ KDPVGLMSRV
AQ PAP KN IDQ RKLIPVKDGNERLTSSGFAC SOCCQ PLYVYKLEQVNDKG
KPHTNYFGRCNVSEH ER LILLSPHKPEANDELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTICYQD1IL
EHQKVI KKN E KRLANLKD IASANGLAFPKITLP PQ PHTKEG I EAYNNVVAQ I
311
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct] Amino Acid
Sequence
VIVVVNLNLWQKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDVVVVDMVC
NVKKLINEKKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FGDLLLHLEKKHGEDWGIWYDEAWERIDKKVEGLSKHIKLEEERRSEDA
QS KAALTDVVLRAKAS FVI E GLKEAD KD E FC RC E LKLQ KVVYGDLRGKP FA
I EAENS ILD ISGFSKQYNCAF IWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRFYTVINKKSGE IVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNI
KPMNLIGIDRGEN I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAERQYTRM EDVVLTAKLAYEG LS KTYLS KT
LAQYTS KTCSNCGFTITSADYD RVLEKLKKTATGVVMTTI NGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVCLN CGFETHADEQAALN IARSVVLF LRSQ EYKKYQTN KT
TGNTDKRAFVETWQSFYRKKLKEVVVKPAVISPKKKRKVPPPPAAKRVK
LD (SEQ ID NO: 363)
293 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYVVE EFQ KDPVGLMSRV
AQ PAP KNIDQRKLIPVKDGNERLTSSGFACSOCCQ PLYVYKLEQVNDKG
KPHTNYFGRCNVSEH ER LILLSPHKPEANDELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIIL
EHQKVI KKN E KR LANLKD IASANGLAFPKITLP PQ PHTKEG I EAYNNVVAQ I
VIWVNLNLWOKLKIGRDEAKPLORLKGFPSFPLVEROANEVDWWDMVC
NVKKLINEKKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FGDLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDA
QS KAALTDVVLRAKAS FVI E GLKEAD KD E FC RC E LKLQ KVVYGDLRGKP FA
I EAENS ILD ISGFSKQYNCAF IWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRF'YTVINKKSGE IVPMEVNFNFDDPNLIILPLAFGKRQGREFIVVN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNI
KPMNLIGIDRGEN I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAERQYTRM EDVVLTAKLAYEG LS KTYLS KT
LAQYTS KTCSNCGFTITSADYD RVLEKLKKTATGVVMTTI NGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVCLN CGFETHADEQAALN IARSWLF LRSQ EYKKYQTN KT
TG NTD KRAFVETVVQ S FYRKKLKEVVVKPAVTS PKKKRKVPPP KRS FS KA
F (SEQ ID NO: 364)
300 MQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLLVRVMTPDLRERLENLRK
KP EN I PQ P IS NTSRAN LN KLLTDYTEM KKAI LHVYVVE EFQ KDPVGLMSRV
AQ PAP KNIDQRKLIPVKDGNERLTSSGFACSQCCQ PLYVYKLEQVNDKG
KPHTNYFGRC NVSEH ERLI LLS PH KPEAN DELVTYSLGKFGQ RALDFYS I
HVTRESNH PVKPLEQIGGNSCASGPVGKALSDACMGAVASFLTKYQDIIL
EHQKVI KKN E KR LANLKD IASANGLAFPKITLP PQ PHTKEG I EAYNNVVAQ I
VIWVNLNLWOKLKIGRDEAKPLQRLKGFPSFPLVERQANEVDWWDMVC
NVKKLINEKKE DGKVFWQ NLAGYKRQEALRPYLSSEEDRKKGKKFARYQ
FGDLLLHLEKKHGEDWGKVYDEAWERIDKKVEGLSKH IKLEEERRSEDA
QS KAALTDWLRAKAS FVI E GLKEAD KD E FCRC E LKLQ KVVYGDLRGKP FA
312
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct] Amino Acid
Sequence
I EAENS ILD ISG FSKQYN CAF IWQKDGVKKLNLYLIINYFKGGKLRFKKIKP
EAFEANRF'YTVINKKSGE IVPM EVN F NFDD P N LI ILPLAFGKRQ GRE F IVVN
DLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFERREVLDSSNI
KPM N LI G ID RGE N I PAVIALTDPEGCPLSRFKDSLGNPTHILRIGESYKEKQ
RTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARDLLYYAVTQDA
M L I FE N LS RGFGRQ GKRTFMAERQYTRM EDVVLTAKLAYEG LSKTYLS la
LAQYTSKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ I
TYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRSGEALSLLKKR
FS H RPVQ E KFVC LN CGFETHADEQAALN IARSVVLF LRSQ EYKKYQTN KT
TGNTDKRAFVETVVQSFYRKKLKEV'VVKPAVTSPKKKRKVPPPKRGINDR
NFVVRGENERKTR (SEQ ID NO: 365)
492 MAP KKKRKVS RMQ E I KRI N KI RRRLVKDS NTKKAG KTGPM
KTLLVRVMTP
DLRERLENLRKKP EN I PQ PISNTSRANLNKLLTDYTEMKKAILHVYVVEEFQ
KD PVG LMSRVAQ PAP KN I DQ RKLIPVKDGN E RLTSSGFACSQ CCQ PLYV
YKLEOVNDKGKP HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKF
GQRALDFYS I HVTRESN H PVKPLEQ I GGNSCASG PVG KALS DACM CAVA
SFLTKYQDI I LEHQ KVI KKN EKRLAN LKD IASAN GLAFP KITLPPQPHTKE G I
EAYNNVVAQ IVIWVN LN LWQ KLKI GR DEAKP LQ RLKG FPSF PLVE ROAN E
VDVVVVDMVCNVKKLINEKKEDGKVFWQNLAGYKRQEALRPYLSSEEDRK
KGKKFARYQ FGDLLLH LE KKHG EDWG KVYDEAVVERI DKKVEG LS KH I KL
EEERRSEDAQSKAALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVY
G DLRGKP FAIEAE NS ILD ISGFSKQYNCAFIWOKDGVKKLN LYLIINYFKG
GKLRFKKIKPEAFEANRFYTVI NKKSG EIVPM EVNFNF D DP NL I ILPLAFGK
RQGREFIWNDLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFE
RREVLDS SNIKPM N LIGIDRG EN IPAVIALTDPE GC P LSRFKDS LG NPTH IL
RIGESYKEKQRTIQAKKEVEQRRAGGYSRKYASKAKNLADDMVRNTARD
LLYYAVTQ DAML I FENLSRGFGRQGKRTFMAERQYTRMEDVVLTAKLAYE
GLSKTYLS KTLAQYTS KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTIN
GKELKVEGQ ITYYNRYKRQNVVKDLSVELDRLSEESVNNDISSWTKGRS
GEALSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVLFLRSQ
EYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVVVKPAVTSPKKKRKV
PPPPKKKRKV (SEQ ID NO: 366)
493 MAP KKKRKVS RMQ E I KRI N KI RRRLVKDS NTKKAG KTGPM
KTLLVRVMTP
DLRERLENLRKKP EN I PQ PISNTSRANLNKLLTDYTEMKKAILHVYWEEFQ
KD PVG LMSRVAQ PAP KN I DQ R KLIPVKDGN E RLTSSGFACSQ CCQ PLYV
YKLEQVNDKGKP HTNYFGRCNVSEHERLILLSPHKPEANDELVTYSLGKF
GQRALDFYS I HVTRESN H PVKPLEQ I GGNSCASG PVG KALS DACM CAVA
SFLTKYQDIILEHQKVIKKN EKRLAN LKD IASAN GLAFP KITLPPQPHTKE G I
EAYNNVVAQ IVIVVVNLNLWQKLKIGRDEAKPLQ RLKG FPSF PLVE ROAN E
VDVVVVDMVCNVKKLINEKKEDGKVFWONLAGYKRQEALRPYLSSEEDRK
KGKKFARYQ FGDLLLH LE KKHG EDWG KVYDEAVVERI DKKVEG LS KH I KL
EEERRSEDAQSKAALTDWLRAKASFVIEGLKEADKDEFCRCELKLQKVVY
G DLRGKP FAIEAE NS ILD ISGFSKOYNCAFIWQKDGVKKLN LYLIINYFKG
GKLRFKKIKPEAFEANRFYTVI NKKSG EIVPM EVNFNF D DP NL I ILPLAFGK
ROGREFIVVNDLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALFVALTFE
RREVLDS SNIKPM N LIGIDRG EN IPAVIALTDPE GC P LSRFKDS LG NPTH IL
313
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct] Amino Acid
Sequence
RIGESYKEKQRTIOAKKEVEORRAGGYSRKYASKAKNLADDMVRNTARD
LLYYAVTODAMLIFENLSRGFGRQGKRTFIVIAERQYTRMEDVVLTAKLAYE
GLSKTYLSKTLAQYTSKTCSNCGFTITSADYDRVLEKLKKTATGVVMTTIN
GKELKVEGQITYYNRYKRQNVVKDLSVELDRLSEESVNNDISSVVTKGRS
GEALSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSWLFLRSQ
EYKKYQTNKTTGNTDKRAFVETVVOSFYRKKLKEVVVKPAVTSPKKKRKV
PPPPAAKRVKLD (SEQ ID NO: 367)
Table 10: Nuclear localization sequence list
CasX NLS DNA Sequence
Amino Acid Sequence
278, 279, SV40 CCAAAGAAGAAGCGGAAGG
PKKKRKV (SEQ ID
280, 492, TC (SEQ ID NO: 368)
NO: 130)
493
285 SynthNL S3 CACAAGAAGAAACATCCAGA
HICKICHPDASVNFSE
CGCATCAGTCAACTTTAGCG
FSK (SEQ ID NO:
AGTTCAGTAAA (SEQ ID NO:
369)
376)
286 SynthNLS4 CAGCGCCCTGGGCCTTACGA
QRPGPYDRPQRPGP
TAGGCCGCAAAGACCCGGAC
YDRP (SEQ ID NO:
CGTATGATCGCCCT (SEQ ID
NO: 370)
162)
287 SynthNLS5 CTCAGCCCGAGTCTTAGTCC LSPSLSPLLSPSLSPL
ACTGCTTTCCCCGTCCCTGTC
(SEQ ID NO: 163)
TCCACTG (SEQ ID NO: 371)
288 SynthNLS6 CGGGGCAAGGGTGGCAAGG RGKGGKGLGKGGA
GGCTTGGCAA
A
KRIIRK (SEQ ID NO:
AAGAGGCACAGGAAG (SEQ
ID NO: 372)
164)
290 EGL-13 AGCCGCCGCAGAAAAGCCAA
SRRRKANPTKLSEN
TCCTACAAAACTGTCAGAAA
AICICLAKEVEN (SEQ
ATGCGAAAAAACTTGCTAAG
GAGGTGGAAAAC (SEQ ID
ID NO: 157)
NO: 373)
291 c-Myc CCTGCCGCAAAGCGAGTGAA
PAAICRVKLD (SEQ
ATTGGAC (SEQ ID NO: 374)
ID NO: 132)
314
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
CasX NLS DNA Sequence
Amino Acid Sequence
293 Nucleolar RNA AAGCGGTCCTTCAGTAAGGC KRSFSKAF
(SEQ ID
Helicase II CTTT (SEQ ID NO: 375)
NO: 153)
300 Influenza A AAACGGGGAATAAACGACC
KRGINDRNFIATRGEN
protein GGAACTTCTGGCGCGGGGAA
ERKTR (SEQ ID NO:
AACGAGCGCAAAACCCGA
(SEQ ID NO: 376)
151)
Example 5: Design and Generation of CasX Constructs 387, 395, 485-491, and 494
10011301ln order to generate CasX 395, CasX 485, CasX 486, CasX 487, the codon
optimized
CasX 119 (based on the CasX 37 construct of Example 2, encoding Planctomycetes
CasX SEQ
ID NO: 2, with a A708K substitution and a [P793] deletion with fused NLS, and
linked guide
and non-targeting sequences), CasX 435, CasX 438, and CasX 484 (each based on
CasX 119
construct of Example 2 encoding Planctomycetes CasX SEQ ID NO: 2, with a L379R
substitution, a A708K substitution, and a [P793] deletion with fused NLS, and
linked guide and
non-targeting sequences) were cloned respectively into a 4kb staging vector
comprising a KanR
marker, colE1 ori, and CasX with fused NLS (pStx1) using standard cloning
methods. Gibson
primers were designed to amplify the CasX SEQ ID NO: 1 Helical I domain from
amino acid
192-331 in its own vector to replace this corresponding region (aa 193-332) on
CasX 119, CasX
435, CasX 438, and CasX 484 in pStx1 respectively. The Helical I domain from
CasX SEQ ID
NO: 1 was amplified with primers oIC768 and oIC784 using Q5 DNA polymerase
according to
the manufacturer's protocol. The destination vector containing the desired
CasX variant was
amplified with primers oIC765 and oIC764 using Q5 DNA polymerase according to
the
manufacturer's protocol. The two fragments were purified by gel extraction
from a 1% agarose
gel using Zymoclean Gel DNA Recovery Kit according to the manufacturer's
protocol. The
insert and backbone fragments were then pieced together using Gibson assembly
(New England
BioLabs Cat# E2621S) following the manufacturer's protocol. Assembled products
in the pStx1
staging vector were transformed into chemically-competent Turbo Competent E:
coil bacterial
cells, plated on LB-Agar plates (LB: Teknova Cat# L9315, Agar: Quartzy Cat#
214510)
containing kanamycin and incubated at 37 C. Individual colonies were picked
and miniprepped
using Qiagen spin Miniprep Kit following the manufacturer's protocol. The
resultant plasmids
were sequenced using Sanger sequencing to ensure correct assembly. Correct
clones were then
315
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
cut and pasted into a mammalian expression plasmid (see FIG. 5) using standard
cloning
methods. The resultant plasmids were sequenced using Sanger sequencing to
ensure correct
assembly.
10011311 Sequences encoding the targeting spacer sequences that target the
gene of interest were
designed based on CasX PAM locations. Targeting spacer sequence DNA was
ordered as single-
stranded DNA (ssDNA) oligos (Integrated DNA Technologies) consisting of the
targeting
sequence and the reverse complement of this sequence. These two oligos were
annealed together
and cloned into pStX individually or in bulk by Golden Gate assembly using T4
DNA Ligase
(New England BioLabs Cat# M0202L) and an appropriate restriction enzyme for
the plasmid.
Golden Gate products were transformed into chemically or electro-competent
cells such as NEB
Turbo competent E. coli (NEB Cat #C2984I), plated on LB-Agar plates (LB:
Teknova Cat#
L9315, Agar: Quartzy Cat# 214510) containing carbenicillin and incubated at
37oC. Individual
colonies were picked and miniprepped using Qiagen spin Miniprep Kit following
the
manufacturer's protocol. The resultant plasmids were sequenced using Sanger
sequencing to
ensure correct ligation.
10011321 In order to generate CasX 488, CasX 489, CasX 490, and CasX 491
(sequences in
Table 11), the codon optimized CasX 119) CasX 435, CasX 438, and CasX 484
(each based on
CasX119 construct of Example 2) were cloned respectively into a 4kb staging
vector that was
made up of a KanR marker, colE1 ori, and STX with fused NLS (pStx1) using
standard cloning
methods. Gibson primers were designed to amplify the CasX Stx1 NTSB domain
from amino
acid 101-191 and Helical I domain from amino acid 192-331 in its own vector to
replace this
similar region (aa 103-332) on CasX 119, CasX 435, CasX 438, and CasX 484 in
pStx1
respectively. The NTSB and Helical I domain from CasX SEQ ID NO: I were
amplified with
primers oIC766 and oIC784 using Q5 DNA polymerase according to the
manufacturer's
protocol. The destination vector containing the desired CasX variant was
amplified with primers
oIC762 and oIC765 using Q5 DNA polymerase according to the manufacturer's
protocol. The
two fragments were purified by gel extraction from a 1% agarose gel using
Zymoclean Gel DNA
Recovery Kit according to the manufacturer's protocol. The insert and backbone
fragments were
then pieced together using Gibson assembly (New England BioLabs Cat# E2621S)
following the
manufacturer's protocol. Assembled products in the pStx1 staging vector were
transformed into
chemically-competent Turbo Competent E. coli bacterial cells, plated on LB-
Agar plates (LB:
Teknova Cat# L9315, Agar: Quartzy Cat# 214510) containing kanamycin and
incubated at
316
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
37oC, Individual colonies were picked and miniprepped using Qiagen spin
Miniprep Kit
following the manufacturer's protocol. The resultant plasmids were sequenced
using Sanger
sequencing to ensure correct assembly. Correct clones were then cut and pasted
into a
mammalian expression plasmid (see FIG. 5) using standard cloning methods. The
resultant
plasmids were sequenced using Sanger sequencing to ensure correct assembly.
Sequences
encoding the targeting spacer sequences that target the gene of interest were
designed based on
CasX PAM locations. Targeting spacer sequence DNA was ordered as single-
stranded DNA
(ssDNA) oligos (Integrated DNA Technologies) consisting of the targeting
sequence and the
reverse complement of this sequence. These two oligos were annealed together
and cloned into
pStX individually or in bulk by Golden Gate assembly using T4 DNA Ligase (New
England
BioLabs Cat# M0202L) and an appropriate restriction enzyme for the plasmid.
Golden Gate
products were transformed into chemically or electro-competent cells such as
NEB Turbo
competent E. coil (NEB Cat #C2984I), plated on LB-Agar plates (LB: Teknova
Cat# L9315,
Agar: Quartzy Cat# 214510) containing carbenicillin and incubated at 37oC.
Individual colonies
were picked and miniprepped using Qiagen spin Miniprep Kit and following the
manufacturer's
protocol. The resultant plasmids were sequenced using Sanger sequencing to
ensure correct
ligation.
10011331ln order to generate CasX 387 and CasX 494 (sequences in Table 11),
the codon
optimized CasX 119 and CasX 484 were cloned respectively into a 4kb staging
vector that was
made up of a KanR marker, colE1 ori, and STX with fused NLS (pStx1) using
standard cloning
methods. Gibson primers were designed to amplify the CasX Stx1 NTSB domain
from amino
acid 101-191 in its own vector to replace this similar region (aa 103-192) on
CasX 119 and
CasX 484 in pStx1 respectively. The NTSB domain from CasX Stx1 was amplified
with primers
oIC766 and oIC767 using Q5 DNA polymerase according to the manufacturer's
protocol. The
destination vector containing the desired CasX variant was amplified with
primers oIC763 and
o1C762 using Q5 DNA polymerase according to the manufacturer's protocol. The
two fragments
were purified by gel extraction from a 1% agarose gel using Zymoclean Gel DNA
Recovery Kit
according to the manufacturer's protocol. The insert and backbone fragments
were then pieced
together using Gibson assembly (New England BioLabs Cat# E2621S) following the
manufacturer's protocol. Assembled products in the pStx1 staging vector were
transformed into
chemically-competent Turbo Competent E. coli bacterial cells, plated on LB-
Agar plates (LB:
Teknova Cat# L9315, Agar: Quartzy Cat# 214510) containing kanamycin and
incubated at
317
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
37oC, Individual colonies were picked and miniprepped using Qiagen spin
Miniprep Kit
following the manufacturer's protocol. The resultant plasmids were sequenced
using Sanger
sequencing to ensure correct assembly. Correct clones were then cut and pasted
into a
mammalian expression plasmid ( see FIG. 5) using standard cloning methods. The
resultant
plasmids were sequenced using Sanger sequencing to ensure correct assembly.
Sequences
encoding the targeting sequences that target the gene of interest were
designed based on CasX
PAM locations. Targeting sequence DNA was ordered as single-stranded DNA
(ssDNA) oligos
(Integrated DNA Technologies) consisting of the targeting sequence and the
reverse complement
of this sequence. These two oligos were annealed together and cloned into pStX
individually or
in bulk by Golden Gate assembly using T4 DNA Ligase (New England BioLabs Cat#
M0202L)
and an appropriate restriction enzyme for the plasmid. Golden Gate products
were transformed
into chemically or electro-competent cells such as NEB Turbo competent E. coli
(NEB Cat
#C2984I), plated on LB-Agar plates (LW Teknova Cat# L9315, Agar: Quartzy Cat#
214510)
containing carbenicillin and incubated at 37oC. Individual colonies were
picked and
miniprepped using Qiagen spin Miniprep Kit and following the manufacturer's
protocol. The
resultant plasmids were sequenced using Sanger sequencing to ensure correct
ligation.
Sequences of the resulting constructs are listed in Table 11.
Table 11: Sequences of CasX 395 and 485-491
DNA
Construct Amino Acid Sequence
Sequence
CasX 387 (SEQ ID MAP KKKRKVSRQ E IKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRERLEN LRKKP EN IP QP
ISNTSRAN LN KLLTDYTE
378) M KKAILHVYVVEEFQKDPVGLMSRVAQ
PASKKIDQ NKLKPEMD
EKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCN
VAE H E KLI LLAQ LKP E KDS DEAVTYSLGKFGQ RALD FYS I HVTR
ESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQ
DI I LEHQ KV IKKNEKRLANLKDIASANGLAFP KITLP PQ PHTKEGI
EAYNNVVAQ IVIWVN LN LWQ KLKI GR D EAKP LQ RLKG FPS FPL
VERQANEVDVWVDMVCNVKKL IN E KKEDG KVFWQ N LAGYKR
QEALRPYLSSEEDRKKGKKFARYQFGDLLLHLEKKHGED WOK
VYDEAINERIDKKVEGLSKH IKLEEERRSEDAQSKAALTDVVLRA
KASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENS
ILDISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKIK
PEAFEANRFYIVINKKSGEIVPMEVNFNFDDPNLIILPLAFGKR
QGREFIINNDLLSLETGSLKLANGRVIEKTLYNRRTRQDEPALF
VALTFERREVLDSSNIKPMNLIGIDRGENIPAVIALTDPEGCPLS
RFKDSLGNPTHILRIGESYKEKQRTIQAKKEVEQRRAGGYSRK
318
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DNA
Construct Amino Acid Sequence
Sequence
YASKAKN LADDMVR NTARD LLYYAVTQ DAM L I FE NLSRG FG R
QGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ
ITYYN RYKRQ NVVKD LSVE LDRLS E ESVN N D I SSWTKGRSG EA
LSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVLFL
RSQEYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVVVKPA
VTSPKKKRKV (SEQ ID NO: 388)
CasX 395 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRE RLEN LRKKP E N IP QP
ISNTSRAN LN KLLTDYTE
379) M KKAILHVYVVEEFQKDPVGLMSRVAQ PAPKN I DQ RKL I PVKDG
NERLTSSGFACSQCCQPLYVYKLEQVNDKGKPHTNYFGRCN
VSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVTKE
STHPVKPLAQ IAGNRYASGPVGKALSDACMGTIASFLSKYQD1I
IEHQKVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGV
DAYNEVIARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSFP
LVERQAN EVDVVVVDMVC NVKKL I N EKKE DG KVFWQ N LAGYKR
QEALRPYLSSEEDRKKGKKFARYQFGDLLLHLEKKHGEDWGK
VYDEAWER I D KKVE GLSKH I KLEE E R R SE DAQ SKAA LTDINLRA
KASFVIEGLKEADKDEFCRCELKLQKVVY'GDLRGKPFAIEAENS
ILDISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIK
PEAFEAN RFYTVINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKR
QGREFIVVNDLLSLETGSLKLANGRVIEKTLYNRRTRQ DEPALF
VALTFERREVLDSSNIKPMNLIGIDRGENIPAVIALTDPEGCPLS
RFKDSLGNPTH I LRIG ESYKE KQ RTIQAKKEVEQ RRAGGYS RK
YASKAKN LADDMVR NTARD LLYYAVTQ DAM L I FE NLSRG FG R
QGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ
ITYYN RYKRQ NVVKD LSVE LDRLS E ESVN N D I SSVVTKGRSG EA
LSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVLFL
RSOEYKKYQTNKTTGNTDKRAFVETWQSFYRKKLKEVVIMPA
VTSPKKKRKVTSPKKKRKV (SEQ ID NO: 389)
CasX 485 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRE RLEN LRKKP E N IP QP
ISNTSRAN LN KLLTDYTE
380) M KKAILHVYVVEEFQKDPVGLMSRVAQ PAPKN I DQ RKL I PVKDG
NERLTSSGFACSOCCOPLYVYKLEQVNDKGKPHTNYFGRCN
VSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVTKE
STHPVKPLAQ IAGNRYASGPVGKALSDACMGTIASFLSKYQD11
IEHQKVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGV
DAYNEVIARVRMVVVNLNLVVQKLKLSRDDAKPLLRLKGFPSFP
LVERQAN EVDWVVDMVC NVKKL I N EKKE DG KVFWQ N LAGYKR
QEALRPYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWGK
VYDEAINE R I DKKVEGLSKH I KLEE ERRSE DAQ SKAALTDVVLRA
KAS FV I EGLKEAD KDE FC RCE LKLQ KVVYGDLRG KP FAIEAE N S
ILDISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIK
PEAFEAN RF'YTVINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKR
Q GR E F IINN DLLS LETGS LKLANG RVI E KTLYN RRTRQ DEPALF
319
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DNA
Construct Amino Acid Sequence
Sequence
VALTFERREVLDSSNIKPMNLIGVDRGEN IPAVIALTDPEGCPL
SRFKDSLGN PTH I LR IGESYKEKQ RTI QAKKEVEQ R RAGGYSR
KYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFG
RQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGVVIVITTINGKELKVEG
0ITYYNRRKRQNVVKDLSVELDRLSEESVNNDISSWTKGRSG
EALS LLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVL
FLRSQ EYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVVVKP
AVTSPKKKRKV (SEQ ID NO: 390)
CasX 486 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRER LEN LRKKP EN IP QP
ISNTSRAN LN KLLTDYTE
381) M KKAILHVYVVEEFQKDPVGLMSRVAQ PAPKN I DQ RKL I PVKDG
NERLTSSGFACSQCCQPLYVYKLEQVNDKGKPHTNYFGRCN
VSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVTKE
STHPVKPLAQ IAGN RYASG PVG KALSDACMGTIASFLSKYQDI I
IEHQKVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGV
DAYNEVIARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSFP
LVERQANEVDIANVDMVCNVKKLINEKKEDGKVFWQNLAGYKR
QEALRPYLSSEEDRKKGKKFARYQLGDLLKHLEKKHGEDWGK
VYDEAWER I DKKVEGLSKH I KLEE ER RSE DAQ SKAALTDVVLRA
KASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENS
ILDISGFSKQYNCAFIVVQKDGVKKLNLYLI INYFKGGKLRFKKIK
PEAFEAN RFYTVINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKR
QGREFIVVNDLLSLETGSLKLANGRVIEKTLYNRRTRQ DEPALF
VALTFERREVLDSSNIKPMNLIGVDRGEN IPAVIALTDPEGCPL
SRFKDSLGN PTH I LR IGESYKEKQ RTI QAKKEVEQ RRAGGYSR
KYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFG
RQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEG
Q ITYYNRRKRQNVVKDLSVELDRLSEESVNNDISSWTKGRSG
EALS LLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVL
FLRSQ EYKKYQTNKTTGNTD KRAFVETWQ S FYRKKLKEVWKP
AVTSPKKKRKV (SEQ ID NO: 391)
CasX 487 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRERLEN LRKKP EN IP QP
ISNTSRAN LN KLLTDYTE
382) M KKAILHVYWEEFQKDPVGLMSRVAQ PAPKN I DQ RKL I PVKDG
NERLTSSGFACSQCCQPLYVYKLEQVNDKGKPHTNYFGRCN
VSEHERLILLSPHKPEANDELVTYSLGKFGQRALDFYSIHVTKE
5TH PVKP LAO IAGN RYASG PVG KALSDACMGTIASFLSKYQDI I
IEHQKVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEGV
DAYNEVIARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSFP
LVEROAN EVDVVVVDMVC NVKKL I N EKKE DG KVFWQ N LAGYKR
QEALRPYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWGK
VYDEAINE R I DKKVEGLSKH I KLEE ERRSE DAQ SKAALTDINLRA
KASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAENS
ILDISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIK
320
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DNA
Construct Amino Acid Sequence
Sequence
PEAFEAN RFYIVINKKSGEIVPMEVNENFDDPNLI ILPLAFGKR
Q GR E F IVVN DLLS LETGS LKLANG RVI E KTLYN RRTRQ DEPALF
VALTFERREVLDSSNIKPMNLIGVDRGEN IPAVIALTDPEGCPL
SRFKDSLGN PTH I LR IGESYKEKQ RTI QAKKEVEQ RRAGGYSR
KYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFG
RQGKRTFMAERQ'YTRMEDVVLTAKLAYEGLSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGINMTTINGKELKVEG
Q ITYYN RYKRQ NVVKDLSVE LD RLS EESVN N D I SSVVTKGRSG
EALS LLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVL
FLRSQ EYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVWKP
AVTSPKKKRKV (SEQ ID NO: 392)
CasX 488 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGICTGPMKTLL
NO: VRVMTPDLRE R LEN LRKKP E N IP QP
ISNTSRAN LN KLLTDYTE
383) M KKAILHVYWEEFQKDPVGLMSRVAQ PASKKIDQ NKLKPEMD
EKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKAYTNYFGRGN
VAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQ RALD FYS I HVTK
ESTHPVKPLAQ IAGNRYASGPVGKALSDACMGTIASFLSKYQD
III EHQ KVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEG
VDAYNEVIARVRMVVVNLNLWQKLKLSRDDAKPLLRLKGFPSF
PLVE ROAN EVDVVVVDMVCNVKKLI NE KKE DGKVFWQ N LAGYK
RQEALRPYLSSEEDRKKGKKFARYQFGDLLLHLEKKHGEDWG
KVYDEAVVE RI DKKVEG LS KH I KLE EE RRS EDAQS KAALTDVVLR
AKASFVIEGLKEADKDEFCRCELKLQKVVYGDLRGKPFAIEAEN
SILD ISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKI
KPEAFEANRFYTVIN KKSGEIVPMEVNFNFDDPNLIILPLAFGKR
Q GRE F IVVN DLLS LETGS LKLANG RVI E KTLYN RRTRQ DEPALF
VALTFERREVLDSSNIKPMNLIGIDRGENIPAVIALTDPEGCPLS
RFKDSLGNPTH I LR IG ESYKEKQ RTIQAKKEVEQ RRAGGYSR K
YASKAKN LADDMVR NTARD LLYYAVTQ DAM L I FE NLSRG FG R
QGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYTS
KTCSNCGFTITSADYDRVLEKLKKTATGVVMTTINGKELKVEGQ
ITYYN RYKRQ NVVKD LSVE LDRLS E ESVN N D I SSWTKGRSG EA
LSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVLFL
RSQEYKKYQTNKTTGNTDKRAFVETWQSFYRKKLKEVVVKPA
VTSPKKKRKV (SEQ ID NO: 393)
CasX 489 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRE RLEN LRKKP E N IP QP
ISNTSRAN LN KLLTDYTE
384) M KKAILHVYVVEEFQKDPVGLMSRVAQ PASKKIDQ NKLKPEMD
EKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCN
VAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQ RALD FYS I HVTK
ESTHPVKPLAQ IAGNRYASGPVGKALSDACMGTIASFLSICYQD
III EHQ KVVKGNQKRLESLRELAGKENLEYPSVTLPPQ PHTKEG
VDAYNEVIARVRMWVNLNLWQKLKLSRDDAKPLLRLKGFPSF
PLVE RQAN EVDWVVDMVC NVKKLI NE KKE DGKVFWQ N LAGYK
RQEALRPYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWG
KVYDEAWE R I DKKVEG LS KH I KLE EE RRS EDAQS KAALTDWLR
321
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DNA
Construct Amino Acid Sequence
Sequence
AKASFVIEG LKEADKDEFC RC ELKLQ KVVYG DLRGKP FAI EAE N
SILD ISGFSKQYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKI
KPEAFEANRFYTVIN KKSGEIVPMEVNFNFDDPNLIILPLAFGKR
QGREFIINNDLLSLETGSLKLANGRVIEKTLYNRRTRQ DEPALF
VALTFERREVLDSSNIKPMNLIGVDRGENIPAVIALTDPEGCPL
SRFKDSLGN PTH I LR IGESYKEKQ RTI QAKKEVEQ RRAGGYSR
KYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFG
RQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEG
Q ITYYN RRKRQNVVKDLSVE LD RLS EESVN N D ISSVVTKG RS G
EALS LLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARSVVL
FLRSQ EYKKYQTNKTTGNTD KRAFVETWQ S FYRKKLKEVWKP
AVTSPKKKRKV (SEQ ID NO: 394)
CasX 490 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRERLEN LRKKP EN IP QP
ISNTSRAN LN KLLTDYTE
385) M KKAILHVYWEEFQKDPVGLMSRVAQ PASKKIDQ NKLKPEMD
EKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCN
VAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQ RALD FYS I HVTK
ESTHPVKPLAQ IAGNRYASGPVGKALSDACMGTIASFLSKYQD
III EHQ KVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEG
VDAYNEVIARVRMWVNLNLWQKLKLSRDDAKPLLRLKGFPSF
PLVE RQAN EVDVVVVDMVCNVKKLI NE KKE DGKVFWQ N LAGYK
RQEALRPYLSSEEDRKKGKKFARYQLGDLLKHLEKKHGEDW
GKVYDEAVVE RI D KKVEG LSKH IKLE E E RRS E DAQSKAALTDW
LRAKAS FVI EG LKEADKD EFCRCE LKLQ KVVYG DLRGKPFAI EA
ENSI LD ISG FSKQYN CAF IVVQ KDGVKKLN LYLI I NYFKGGKLRFK
KIKP EAFEANRFYTVI NKKS GE IVP MEVNF NFDDP NLIILP LAFG
KROGREFIWNDLLSLETGSLKLANGRVIEKTLYNRRTRQDEPA
LFVALTFERREVLDSSNIKPMNLIGVDRGENIPAVIALTDPEGC
PLSR FKDSLGNPTH I LR I GESYKE KQ RTIQAKKEVEQ R RAGGY
SRKYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGF
GRQGKRTFMAERQYTRMEDWLTAKLAYEGLSKTYLSKTLAQY
TS KTCS N CGFTITSADYD RVLE KLKKTATGINMTTI NC KE LKVE
GQ ITYYNRRKRQ NVVKDLSVELDRLSEESVNND I SSVVTKGRS
GEALSLLKKRFSHRPVQEKFVCLNCGFETHADEQAALNIARS
VVLFLRSQ EYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEV
VVKPAVTSPKKKRKV (SEQ ID NO: 395)
CasX 491 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRER LEN LRKKP EN IP QP
ISNTSRAN LN KLLTDYTE
386) M KKAILHVYWEEFQKDPVGLMSRVAQ PASKKIDQ NKLKPEMD
EKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKAYTNYFGRCN
VAEHEKLILLAQLKPEKDSDEAVTYSLGKFGQ RALD FYS I HVTK
ESTHPVKPLAQ IAGNRYASGPVGKALSDACMGTIASFLSKYQD
III EHQ KVVKGNQKRLESLRELAGKENLEYPSVTLPPQPHTKEG
VDAYNEVIARVRMWVNLNLWQKLKLSRDDAKPLLRLKGFPSF
PLVE RQAN EVDVVVVDMVCNVKKLI NE KKE DGKVFWQ N LAGYK
322
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
DNA
Construct Amino
Acid Sequence
Sequence
RQEALRPYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWG
KVYDEAWE R I DKKVEG LS KH I KLE EE RRS EDAQS KAALTDWLR
AKASFVIEGLKEADKDEFC RC ELKLQ KVVYG DLRGKP FAI EAE N
SILD ISGFSKIDYNCAFIWQKDGVKKLNLYLIINYFKGGKLRFKKI
KPEAFEANRFYTVIN KKSGE IVP M EVN FN F DDP NLI I LP LAFGKR
Q GRE F IINN DLLS LETGS LKLANG RVI E KTLYN RRTRQ DEPALF
VALTFERREVLDSSNIKPMNLIGVDRGEN IPAVIALTDPEGCPL
SRFKDSLGN PTH I LR IGESYKEKQ RTI QAKKEVEQ RRAGGYSR
KYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFG
RQGKRTFMAERQYTRMEDVVLTAKLAYEGLSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGWMTTINGKELKVEG
Q ITYYN RYKRQ NVVKDLSVE LD R LS EESVN N D I SSWTKGRSG
EALS LLKKRFSH RPVQEKFVCLNCGFETHADEQAALNIARSVVL
FLRSQ EYKKYQTNKTTGNTDKRAFVETVVQSFYRKKLKEVVVKP
AVTSPKKKRKV (SEQ ID NO: 396)
CasX 494 (SEQ ID MAPKKKRKVSRQEIKRINKIRRRLVKDSNTKKAGKTGPMKTLL
NO: VRVMTPDLRE RL EN L RKKP E N IP QP I
SNTS RAN LN KLLTDYTE
387) M KKAILHVYINEEFQKDPVGLMSRVAQ
PASKKIDQ NKLKPEMD
EKGNLTTAGFACSQCGQPLFVYKLEQVSEKGKAYTNYFGRC N
VAE H E KL I LLAQ LKP E KDS DEAVTYSLGKFGQ RAL D FYS I HVTR
ESNHPVKPLEQ IGGNSCASGPVGKALSDACMGAVASFLTKYQ
D I I LEHQ KV IKKN E KRLAN LKD IASANG LAFP KITLP PQ P HTKEGI
EAYNNVVAQ IVIVVVN LN LWQ KLKI GR D EAKP LQ RLKG FPS FPL
VERQANEVDVWVDMVCNVKKLINEKKEDGKVFVVQNLAGYKR
QEALRPYLSSEEDRKKGKKFARYQLGDLLLHLEKKHGEDWGK
VYDEAVVE R I DKKVEGLSKH I KLEE ERRSE DAQ SKAALTDINLRA
KAS FV I EGLKEAD KDE FC RCE LKLQ KVVYGDLRGKPFAIEAENS
ILDISGFSKQYNCAFIWQKDGVKKLNLYLI INYFKGGKLRFKKIK
PEAFEAN RFYTVINKKSGEIVPMEVNFNFDDPNLI ILPLAFGKR
Q GR E F IINN DLLS LETGS LKLANG RVI E KTLYN RRTRQ DEPALF
VALTFERREVLDSSNIKPMNLIGVDRGENIPAVIALTDPEGCPL
SRFKDSLGN PTH I LR IGESYKEKQ RTI QAKKEVEQ RRAGGYSR
KYASKAKNLADDMVRNTARDLLYYAVTQDAMLIFENLSRGFG
RQGKRTFMAERQ'YTRMEDVVLTAKLAYEGLSKTYLSKTLAQYT
SKTCSNCGFTITSADYDRVLEKLKKTATGINMTTINGKELKVEG
Q ITYYN RYKRQ NVVKDLSVE LD RLS EESVN N D I SSVVTKGRSG
EALS LLKKRFSH RPVQEKFVCLNCGFETHADEQAALNIARSVVL
FLRSQ EYKKYQTNKTTGNTD KRAFVETWQ S FYRKKLKEVVVKP
AVTSPKKKRKV (SEQ ID NO: 397)
Example 6: Generation of RNA guides
10011341For the generation of RNA single guides and spacers, templates for in
vitro
transcription were generated by performing PCR with Q5 polymerase (NEB M0491)
according
to the recommended protocol, with template oligos for each backbone and
amplification primers
323
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
with the T7 promoter and the spacer sequence. The DNA primer sequences for the
T7 promoter,
guide and spacer for guides and spacers are presented in Table 12, below. The
template oligos,
labeled "backbone fwd" and "backbone rev" for each scaffold, were included at
a final
concentration of 20 rtM each, and the amplification primers (T7 promoter and
the unique spacer
primer) were included at a final concentration of 1 gM each. The sg2, sg32,
sg64, and sg174
guides correspond to SEQ ID NOS. 5, 600, 602, and 734, respectively, with the
exception that
sg2, sg32, and sg64 were modified with an additional 5' G to increase
transcription efficiency
(compare sequences in Table 12 to Table 2). The 7.37 spacer targets beta2-
microglobulin
(B2M). Following PCR amplification, templates were cleaned and isolated by
phenol-
chloroform-isoamyl alcohol extraction followed by ethanol precipitation.
10011351ln vitro transcriptions were carried out in buffer containing 50 mM
Ttis pH 8.0, 30 mM
MgCl2, 0.01% Triton X-100, 2 mM spermidine, 20 mM DTT, 5 mM N'TPs, 0.5 ttM
template,
and 100 gg/mL T7 RNA polymerase. Reactions were incubated at 37 C overnight.
20 units of
DNase I (Promega #M6101)) were added per 1 mL of transcription volume and
incubated for
one hour. RNA products were purified via denaturing PAGE, ethanol
precipitated, and
resuspended in lx phosphate buffered saline. To fold the sgRNAs, samples were
heated to 700
C for 5 min and then cooled to room temperature. The reactions were
supplemented to 1 mM
final MgCl2 concentration, heated to 50 C for 5 min and then cooled to room
temperature_ Final
RNA guide products were stored at -80 C.
Table 12: Sequences for generation of guide RNA
Primer Primer sequence
RNA product
T7 promoter GAAATTAATACGACTCACTATA (SEQ ID NO: Used for all
primer 398)
sg2 backbone GAAATTAATACGACTCACTATAGGTACTGG GGUACUGGCGCU
fwd CGCTTTTATCTCATTACTTTGAGAGCCATC UUUAUCUCAUUAC
ACCAGCGACTATGTCGTATGGGTAAAG
UUUGAGAGCCAU
(SEQ ID NO: 399)
CACCAGCGACUAU
sg2 backbone CTTTGATGCTTCTTATTTATCGGATTTCTCT GUCGUAUGGGUA
rev CCGATAAATAAGCGCTTTACCCATACGACA AAGCGCUUAUUUA
TAGTCGCTGGTGATGGC (SEQ ID NO: 400) UCGGAGAGAAAU
sg2.7.37 CGGAGCGAGACATCTCGGCCCTTTGATGC CCGAUAAAUAAGA
spacer primer TTCTTATTTATCGGATTTCTCTCCG (SEQ ID AGCAUCAAAGGG
NO: 401)
CCGAGAUGUCUC
GCUCCG (SEQ ID
NO: 411)
324
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Primer Primer sequence
RNA product
sg32 GAAATTAATACGACTCACTATAGGTACTGG GGUACUGGCGCU
backbone fwd CGCTTTTATCTCATTACTTTGAGAGCCATC UUUAUCUCAUUAC
ACCAGCGACTATGTCGTATGGGTAAAGCG UUUGAGAGCCAU
C (SEQ ID NO: 402)
CACCAGCGACUAU
sg32 CTTTGATGCTTCCCTCCGAAGAGGGCGCT GUCGUAUGGGUA
backbone rev TTACCCATACGACATAG (SEQ ID NO: 403) AAGCGCCCUCUU
CGGAGGGAAGCA
sg32.7.37 CGGAGCGAGACATCTCGGCCCTTTGATGC UCAAAGGGCCGA
spacer primer TTCCCTCCGAAGAG (SEQ ID NO: 404)
GAUGUCUCG
(SEQ ID NO: 412)
sg64 GAAATTAATACGACTCACTATAGGTACTGG GGUACUGGCGCC
backbone fwd CGCCTTTATCTCATTACTTTGAGAGCCATC UUUAUCUCAUUAC
ACCAGCGACTATGTCGTATGGGTAAAGCG UUUGAGAGCCAU
C (SEQ ID NO: 405)
CACCAGCGACUAU
sg64 CTTTGATGCTTCTTACGGACCGAAGTCCGT GUCGUAUGGGUA
backbone rev AAGCGCTTTACCCATACGACATAG (SEQ ID AAGCGCUUACGG
NO: 406)
ACUUCGGUCCGU
sg64.7.37 CGGAGCGAGACATCTCGGCCCTTTGATGC AAGAAGCAUCAAA
spacer primer TTCTTACGGACCGAAG (SEQ ID NO: 407)
GGGCCGAGAUGU
CUCGCUCCG
(SEQ ID NO: 413)
sg174 GAAATTAATACGACTCACTATAACTGGCGC ACUGGCGCUUUU
backbone fwd TTTTATCTGATTACTTTGAGAGCCATCACCA AUCUgAUUACUUU
GCGACTATGTCGTAGTGGGTAAAGCT
GAGAGCCAUCAC
(SEQ ID NO: 408)
CAGCGACUAUGU
sg174 CTTT
TT T GATGCTCCCTCCGAAGAGGGAGC
CGUAgUGGGUAAA
backbone rev ACCCACTACGACATAGTCGC (SEQ ID NO: GCUCCCUCUUCG
409)
GAGGGAGCAUCA
sg174.7,37 CGGAGCGAGACATCTCGGCCCTTTGATGC AAGGGCCGAGAU
spacer primer TCCCTCC (SEQ ID NO: 410)
GUCUCGCUCCG
(SEQ ID NO: 414)
Example 7: Assessing binding affinity to the guide RNA
10011361Purified wild-type and improved CasX will be incubated with synthetic
single-guide
RNA containing a 3' Cy7.5 moiety in low-salt buffer containing magnesium
chloride as well as
heparin to prevent non-specific binding and aggregation. The sgRNA will be
maintained at a
concentration of 10 pM, while the protein will be titrated from 1 pM to 100
ELM in separate
binding reactions. After allowing the reaction to come to equilibrium, the
samples will be run
through a vacuum manifold filter-binding assay with a nitrocellulose membrane
and a positively
charged nylon membrane, which bind protein and nucleic acid, respectively. The
membranes
will be imaged to identify guide RNA, and the fraction of bound vs unbound RNA
will be
determined by the amount of fluorescence on the nitrocellulose vs nylon
membrane for each
325
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
protein concentration to calculate the dissociation constant of the protein-
sgRNA complex. The
experiment will also be carried out with improved variants of the sgRNA to
determine if these
mutations also affect the affinity of the guide for the wild-type and mutant
proteins. We will also
perform electromobility shift assays to qualitatively compare to the filter-
binding assay and
confirm that soluble binding, rather than aggregation, is the primary
contributor to protein-RNA
association.
Example 8: Assessing binding affinity to the target DNA
10011371Purified wild-type and improved CasX will be complexed with single-
guide RNA
bearing a targeting sequence complementary to the target nucleic acid. The RNP
complex will
be incubated with double-stranded target DNA containing a PAM and the
appropriate target
nucleic acid sequence with a 5' Cy7.5 label on the target strand in low-salt
buffer containing
magnesium chloride as well as heparin to prevent non-specific binding and
aggregation. The
target DNA will be maintained at a concentration of 1 n.M, while the RNP will
be titrated from 1
pM to 100 M in separate binding reactions. After allowing the reaction to
come to equilibrium,
the samples will be run on a native 5% polyacrylamide gel to separate bound
and unbound target
DNA. The gel will be imaged to identify mobility shifts of the target DNA, and
the fraction of
bound vs unbound DNA will be calculated for each protein concentration to
determine the
dissociation constant of the RNP-target DNA ternary complex.
Example 9: CasX:gNA In Vitro Cleavage Assays
1. Determining cleavage-competent fractions for protein variants compared to
wild-type
reference CasX
10011381The ability of CasX variants to form active RNP compared to reference
CasX was
determined using an in vitro cleavage assay. The beta-2 microglobulin (B2M)
7.37 target for the
cleavage assay was created as follows. DNA oligos with the sequence
TGAAGCTGACAGCATTCGGGCCGAGATGTCTCGCTCCGTGGCCTTAGCTGTGCTCGC
GCT (non-target strand, NTS (SEQ ID NO: 415)) and
TGAACCTGACAGCATTCGGGCCGAGATGTCTCGCTCCGTCGCCTTAGCTGTGCTOGC
GCT (target strand, TS (SEQ ED NO: 416)) were purchased with 5' fluorescent
labels (LI-COR
IRDye 700 and 800, respectively). dsDNA targets were formed by mixing the
oligos in a 1:1
ratio in lx cleavage buffer (20 mM Tris HC1 pH 7.5, 150 mM NaC1, 1 mM TCEP, 5%
glycerol,
326
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
mM MgC12), heating to 95 C for 10 minutes, and allowing the solution to cool
to room
temperature.
10011391 CasX RNPs were reconstituted with the indicated CasX and guides (see
graphs) at a
final concentration of 1 rtM with 1.5-fold excess of the indicated guide
unless otherwise
specified in lx cleavage buffer (20 mM Tris HCl pH 7.5, 150 mM NaCl, 1 mM
TCEP, 5%
glycerol, 10 mM MgCl2) at 37' C for 10 min before being moved to ice until
ready to use_ The
7.37 target was used, along with sgRNAs having spacers complementary to the
7.37 target.
10011401 Cleavage reactions were prepared with final RNP concentrations of 100
nM and a final
target concentration of 100 nM. Reactions were carried out at 370 C and
initiated by the addition
of the 7.37 target DNA. Aliquots were taken at 5, 10, 30, 60, and 120 minutes
and quenched by
adding to 95% formamide, 20 mM EDTA. Samples were denatured by heating at 950
C for 10
minutes and run on a 10% urea-PAGE gel. The gels were either imaged with a LI-
COR Odyssey
CLx and quantified using the LI-COR Image Studio software or imaged with a
Cytiva Typhoon
and quantified using the Cytiva 1Q11. software. The resulting data were
plotted and analyzed
using Prism. We assumed that CasX acts essentially as a single-turnover enzyme
under the
assayed conditions, as indicated by the observation that sub-stoichiometric
amounts of enzyme
fail to cleave a greater-than-stoichiometric amount of target even under
extended time-scales and
instead approach a plateau that scales with the amount of enzyme present.
Thus, the fraction of
target cleaved over long time-scales by an equimolar amount of RNP is
indicative of what
fraction of the RNP is properly formed and active for cleavage. The cleavage
traces were fit with
a biphasic rate model, as the cleavage reaction clearly deviates from
monophasic under this
concentration regime, and the plateau was determined for each of three
independent replicates.
The mean and standard deviation were calculated to determine the active
fraction (Table 13).
The graph is shown in FIG. 15.
10011411 Apparent active (competent) fractions were determined for RNPs formed
for CasX2 +
guide 174 + 7.37 spacer, CasX119 + guide 174 + 7.37 spacer, CasX457 + guide
174 +7.37
spacer, CasX488 + guide 174 + 7.37 spacer, and CasX491 + guide 174 + 7.37
spacer. The
determined active fractions are shown in Table 13. All CasX variants had
higher active fractions
than the wild-type CasX2, indicating that the engineered CasX variants form
significantly more
active and stable RNP with the identical guide under tested conditions
compared to wild-type
CasX. This may be due to an increased affinity for the sgRNA, increased
stability or solubility in
the presence of sgRNA, or greater stability of a cleavage-competent
conformation of the
327
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
engineered CasX:sgRNA complex_ An increase in solubility of the RNP was
indicated by a
notable decrease in the observed precipitate fanned when CasX457, CasX488, or
CasX491 was
added to the sgRNA compared to CasX2.
2. In vitro Cleavage Assays ¨ Determining kcieave for CasX variants compared
to wild-type
reference CasX
10011421Cleavage-competent fractions were also determined using the same
protocol for
CasX2.2.7.37, CasX2.32.7.37, CasX2.64.7.37, and CasX2.174.7.37 to be 16 3%,
13 3%, 5
2%, and 22 5%, as shown in FIG. 16 and Table 13.
10011431 A second set of guides were tested under different conditions to
better isolate the
contribution of the guide to RNP formation. 174, 175, 185, 186, 196, 214, and
215 guides with
7.37 spacer were mixed with CasX491 at final concentrations of 1 M for the
guide and 1,5 jiM
for the protein, rather than with excess guide as before. Results are shown in
FIG. 17 and Table
13. Many of these guides exhibited additional improvement over 174, with 185
and 196
achieving 91 4% and 91 1% competent fractions, respectively, compared with
80 9% for
174 under these guide-limiting conditions.
10011441 The data indicate that both CasX variants and sgRNA variants are able
to form a higher
degree of active RNP with guide RNA compare to wild-type CasX and wild-type
sgRNA.
10011451 The apparent cleavage rates of CasX variants 119, 457, 488, and 491
compared to
wild-type reference CasX were determined using an in vitro fluorescent assay
for cleavage of the
target 7,37.
10011461 CasX RNPs were reconstituted with the indicated CasX (see FIG. 18) at
a final
concentration of 1 uM with 1.5-fold excess of the indicated guide in lx
cleavage buffer (20 tnivl
Tris HC1 pH 7.5, 150 in.M NaC1, 1 mM TCEP, 5% glycerol, 10 mM MgCl2) at 37 C
for 10 min
before being moved to ice until ready to use. Cleavage reactions were set up
with a final RNP
concentration of 200 nIvl and a final target concentration of 10 WO. Reactions
were carried out at
37 C except where otherwise noted and initiated by the addition of the target
DNA. Aliquots
were taken at 025, 0.5, 1, 2, 5, and 10 minutes and quenched by adding to 95%
formamide, 20
mM EDTA. Samples were denatured by heating at 950 C for 10 minutes and run on
a 10% urea-
PAGE gel. The gels were imaged with a LI-COR Odyssey CLx and quantified using
the LI-
COR Image Studio software or imaged with a Cytiva Typhoon and quantified using
the Cytiva
IQTL software. The resulting data were plotted and analyzed using Prism, and
the apparent first-
order rate constant of non-target strand cleavage (lccieave) was determined
for each CasX:sgRNA
328
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
combination replicate individually. The mean and standard deviation of three
replicates with
independent fits are presented in Table 13, and the cleavage traces are shown
in FIG 18.
10011471 Apparent cleavage rate constants were determined for wild-type CasX2,
and CasX
variants 119, 457, 488, and 491 with guide 174 and spacer 7.37 utilized in
each assay (see Table
13 and FIG. 18). All CasX variants had improved cleavage rates relative to the
wild-type
CasX2. CasX457 cleaved more slowly than 119, despite having a higher competent
fraction as
determined above. CasX488 and CasX491 had the highest cleavage rates by a
large margin; as
the target was almost entirely cleaved in the first timepoint, the true
cleavage rate exceeds the
resolution of this assay, and the reported kthave should be taken as a lower
bound.
10011481 The data indicate that the CasX variants have a higher level of
activity, with ndeave rates
reaching at least 30-fold higher compared to wild-type CasX2.
3. In vitro Cleavage Assays: Comparison of guide variants to wild-type guides
10011491 Cleavage assays were also performed with wild-type reference CasX2
and reference
guide 2 compared to guide variants 32, 64, and 174 to determine whether the
variants improved
cleavage. The experiments were performed as described above. As many of the
resulting RNPs
did not approach full cleavage of the target in the time tested, we determined
initial reaction
velocities (Vo) rather than first-order rate constants. The first two
timepoints (15 and 30 seconds)
were fit with a line for each CasX:sgRNA combination and replicate. The mean
and standard
deviation of the slope for three replicates were determined.
10011501 Under the assayed conditions, the Vo for CasX2 with guides 2, 32, 64,
and 174 were
20.4 1.4 nM/min, 18.4 2.4 n.M/min, 7.8 1.8 aM/min, and 49.3 1.4
n.M/min (see Table 13
and FIG. 19 and FIG. 20). Guide 174 showed substantial improvement in the
cleavage rate of the
resulting RNP (-2.5-fold relative to 2, see FIG. 20), while guides 32 and 64
performed similar to
or worse than guide 2. Notably, guide 64 supports a cleavage rate lower than
that of guide 2 but
performs much better in vivo (data not shown). Some of the sequence
alterations to generate
guide 64 likely improve in vivo transcription at the cost of a nucleotide
involved in triplex
formation. Improved expression of guide 64 likely explains its improved
activity in vivo, while
its reduced stability may lead to improper folding in vitro.
10011511 Additional experiments were carried out with guides 174, 175, 185,
186, 196, 214, and
215 with spacer 7.37 and CasX491 to determine relative cleavage rates. To
reduce cleavage
kinetics to a range measurable with our assay, the cleavage reactions were
incubated at 100 C.
Results are in FIG. 21 and Table 13. Under these conditions, 215 was the only
guide that
329
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
supported a faster cleavage rate than 174. 196, which exhibited the highest
active fraction of
RNP under guide-limiting conditions, had kinetics essentially the same as 174,
again
highlighting that different variants result in improvements of distinct
characteristics.
10011521The data support that, under the conditions of the assay, use of the
majority of the
guide variants with CasX results in RNP with a higher level of activity than
one with the wild-
type guide, with improvements in initial cleavage velocity ranging from ¨2-
fold to >6-fold.
Numbers in Table 13 indicate, from left to right, CasX variant, sgRNA
scaffold, and spacer
sequence of the RNP construct. In the RNP construct names in the table below,
CasX protein
variant, guide scaffold and spacer are indicated from left to right.
Table 13: Results of cleavage and RNP formation assays
RNP Construct likleave*
Initial velocity* Competent fraction
2.2.7.37 20.4 1.4 nM/min 16
3%
2.32.7.37 18.4 2.4 nM/min
13 3%
1641.37 7.8 1.8 nM/min
5 2%
2.174.7.37 0.51 0.01 min" 49.3
1.4 nM/min 22 5%
119.174.7.37 6.29 1 2.11 min"
35 6%
457.174.7.37 3.01 0.90 min"
53 7%
488.1747.37 15.19 min-1
67%
16.59 min" / 0.447
83% / 79 9%
491.174.7.37
0.031 min" (10 C)
(guide-limited)
491.175.7.37 0.089 min" (10 C)
5% (guide-limited)
491 185737 0.2828 0.014 min"
91 4% (guide-
. ..
(10 C)
limited)
491 186.7.37 0.118 0.010 min"
32 2% (guide-
.
(10 C)
limited)
491 196737 0.439 0.067 min"
91 1% (guide-
. ..
C)
limited)
491 214.7.37 0.413 0.059 min"
73 8% (guide-
.
(10 C)
limited)
491.215.7.37
0.668 0.165 min"
81 6% (guide-
(10
limited)
*Mean and standard deviation
330
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Example 10: Assessing differential PAM recognition in vitro
10011531ln vitro cleavage assays were performed essentially as described in
Example 9, using
CasX2, CasX119, and CasX438 complexed with sg174.7.37. Fluorescently labeled
dsDNA
targets with a 7.37 spacer and either a TIC, CTC, GTC, or ATC PAM were used
(sequences are
in Table 14). Time points were taken at 0.25, 0.5, 1,2, 5, 10, 30, and 60
minutes. Gels were
imaged with an Cytiva Typhoon and quantified using the IQTL 8.2 software.
Apparent first-
order rate constants for non-target strand cleavage (kcleave) were determined
for each
Casx:sgRNA complex on each target. Rate constants for targets with non-TTC PAM
were
compared to the TTC PAM target to determine whether the relative preference
for each PAM
was altered in a given protein variant.
10011541For all variants, the TTC target supported the highest cleavage rate,
followed by the
ATC, then the CTC, and finally the GTC target (FIG. 22A-D, Table 15). For each
combination
of CasX variant and NTC PAM, the cleavage rate keleave is shown. For all non-
NTC PAMs, the
relative cleavage rate as compared to the ITC rate for that variant is shown
in parentheses. All
non-TTC PAMs exhibited substantially decreased cleavage rates (>10-fold for
all). The ratio
between the cleavage rate of a given non-TTC PAM and the TTC PAM for a
specific variant
remained generally consistent across all variants. The CTC target supported
cleavage 3.5-4.3%
as fast as the TTC target; the GTC target supported cleavage 1.0-1.4% as fast;
and the ATC
target supported cleavage 6.5-8.3% as fast. The exception is for 491, where
the kinetics of
cleavage at nic PAMs are too fast to allow accurate measurement, which
artificially decreases
the apparent difference between TTC and non-TTC PAMs. Comparing the relative
rates of 491
on GTC, CTC, and ATC PAMs, which fall within the measurable range, results in
ratios
comparable to those for other variants when comparing across non-TTC PAMs,
consistent with
the rates increasing in tandem. Overall, differences between the variants are
not substantial
enough to suggest that the relative preference for the various NTC PAMs have
been altered.
However, the higher basal cleavage rates of the variants allow targets with
ATC or CTC PAMs
to be cleaved nearly completely within 10 minutes, and the apparent lc,
¨leaves are comparable to or
greater than the kcleave of CasX2 on a TTC PAM (Table 14). This increased
cleavage rate may
cross the threshold necessary for effective genome editing in a human cell,
explaining the
apparent increase in PAM flexibility for these variants.
331
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Table 14. Sequences of DNA substrates used in in vitro PAM cleavage assay.
Guide* DNA Sequence
7.37 TIC AGCGCGAGCACAGCTAAGGCCACGGAGCGAGACATCTCGGCCCGAA
PAM TS TGCTGTCAGCTTCA (SEQ ID NO: 417)
7.37 TIC TGAAGCTGACAGCATTCGGGCCGAGATGTCTCGCTCCGTGGCCTTAG
PAM NTS CTGTGCTCGCGCT (SEQ ID NO: 418)
7.37 CTC AGCGCGAGCACAGCTAAGGCCAGGGAGCGAGACATCTCGGCCCGAG
PAM IS TGCTGTCAGCTTCA (SEQ ID NO: 419)
7.37 CTC TGAAGCTGACAGCACTCGGGCCGAGATGTCTCGCTCCGTGGCCTTAG
PAM NTS CTGTGCTCGCGCT (SEQ ID NO: 420)
7.37 GTC AGCGCGAGCACAGCTAAGGCCACGGAGCGAGACATCTCGGCCCGAC
PAM IS TGCTGTCAGCTTCA (SEQ ID NO: 421)
7.37 GTC TGAAGCTGACAGCAGTCGGGCCGAGATGTCTCGCTCCGTGGCCTTAG
PAM NTS CTGTGCTCGCGCT (SEC) ID NO: 422)
7.37 ATC AGCGCGAGCACAGCTAAGGCCAGGGAGCGAGACATCTCGGCCCGAT
PAM IS TGCTGTCAGCTTCA (SEQ ID NO: 423)
7.37 ATC TGAAGCTGACAGCAATCGGGCCGAGATGTCTCGCTCCGTGGCCTTAG
PAM NTS CTGTGCTCGCGCT (SEQ ID NO: 424)
*The PAM sequences for each are bolded. TS - target strand. NTS -Non-target
strand.
Table 15. Apparent cleavage rates of CasX variants against NTC PAMs.
Variant TTC CTC
GTC ATC
2 0.267 min-1 9.29E-3 min-1
3.75E-3 min-1 1.87E-2 min-1
(0.035) (0.014) (0.070)
119 8.33 min-1 0.303 min-1
8.64E-2 min-1 0.540 m1n-1
(0.036) (0.010) (0.065)
438 4.94 min-1 0.212 m1n-1
1.31E-2 min-1 0.408 m1n-1
(0.043) (0.013) (0.083)
491 16.42 m1n-1 8.605 m1n-1
2.447 min4 11.33 min-1
(0.524) (0.149) (0.690)
Example 11: Identification of nicking variants
10011551Purified modified CasX variants will be complexed with single-guide
RNA bearing a
fixed targeting sequence. The RNP complexes will be added to buffer containing
MgCl2 at a
final concentration of 100 n1V1 and incubated with double-stranded target DNA
with a 5'
fluorescein label on the target strand and a 5' Cy5 label on the non-target
strand at a
concentration of 10 tiM. Aliquots of the reactions will be taken at fixed time
points and
quenched by the addition of an equal volume of 50 mM EDTA and 95% formamide.
The
samples will be run on a denaturing polyacrylamide gel to separate cleaved and
uncleaved DNA
332
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
substrates. Efficient cleavage of one strand but not the other would be
indicative that the variant
possessed single-strand nickase activity.
Example 12: Assessing improved expression and solubility characteristics of
CasX variants
for RNP production
10011561Wild-type and modified CasX variants will be expressed in BL21 (DE3)
E. coli under
identical conditions. All proteins will be under the control of an IPTG-
inducible T7 promoter_
Cells will be grown to an OD of 0.6 in TB media at 37 C, at which point the
growth temperature
will be reduced to 16 C and expression will be induced by the addition of 0.5
mM IPTG. Cells
will be harvested following 18 hours of expression. Soluble protein fractions
will be extracted
and analyzed on an SDS-PAGE gel. The relative levels of soluble CasX
expression will be
identified by Coomassie staining. The proteins will be purified in parallel
according to the
protocol above, and final yields of pure protein will be compared. To
determine the solubility of
the purified protein, the constructs will be concentrated in storage buffer
until the protein begins
to precipitate. Precipitated protein will be removed by centrifugation and the
final concentration
of soluble protein will be measured to determine the maximum solubility for
each variant.
Finally, the CasX variants will be complexed with single guide RNA and
concentrated until
precipitation begins. Precipitated RNP will be removed by centrifugation and
the final
concentration of soluble RNP will be measured to determine the maximum
solubility of each
variant when bound to guide RNA.
Example 13: XDP construct, transfection and recovery.
Plasmids and Cell lines
10011571CasX delivery particles (XDPs) containing RNP of CasX, CasX 119, CasX
438, or
CasX 457 protein and single guide RNA 174 with spacer sequence 12/ (encoded by
CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825) targeting tdTomato were produced by
transient transfection of LentiX HEK293T cells (Takara Biosciences) using the
four plasmids
portrayed in FIG. 23 and listed in Table 16 (with different plasmids depending
on which CasX
was utilized). The pStx43 plasmid contains the Gag polyprotein sequence
followed by a CasX
protein fused at the C-terminus (pXID10 encodes CasX 119; pXD11 encodes CasX
438; pXD12
encodes CasX 457). A SQNYPIVQ (SEQ ID NO: 20) HIV-1 cleavage site separated
the Gag
protein and CasX protein sequences to mediate separation of the editing
molecules during XDP
333
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
maturation. The pStx42.174.12.7 plasmid was created with a human U6 promoter
upstream of a
CasX guide cassette having scaffold and spacer components (targeted to
tdTomato) in a single-
guide format. Another pStx42 plasmid was utilized to make a CasX guide
cassette having
scaffold and non-targeting spacer components, used as a control in the editing
assay. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP and Gag-Pot (psPax2) proteins
were also
used. MI plasmids contained either an ampicillin or kanamycin resistance gene.
The sequences
incorporated into the plasmids are presented in Table 16.
Table 16: Plasmid Encoding Sequences
Construct DNA
SEQUENCE
pStx42.174. ACTGGCGCTTTTATCTGATTACTTTGAGAGCCATCACCAGCGACTAT
12.7 GTCGTAGTGGGTAAAGCTCCCTCTTCGGAGGGAGCATCAAAGCTGC
ATTCTAGTTGTGGTTT (SEQ ID NO: 426)
pXDP10 ATGGGTGCGAGAGCGTCAGTATTAAGCGGGGGAGAATTAGATCGAT
GGGAAAAAATTCGGTTAAGGCCAGGGGGAAAGAAAAAATATAAATTA
AAACATATAGTATGGGCAAGCAGGGAGCTAGAACGATTCGCAGTTA
ATCCTGGCCTGTTAGAAACATCAGAAGGCTGTAGACAAATACTGGG
ACAGCTACAACCATCCCTTCAGACAGGATCAGAAGAACTTAGATCAT
TATATAATACAGTAGCAACCCTCTATTGTGTGCATCAAAGGATAGAG
ATAAAAGACACCAAGGAAGCTTTAGACAAGATAGAGGAAGAGCAAA
ACAAAAGTAAGAAAAAAGCACAGCAAGCAGCAGCTGACACAGGACA
CAGCAATCAGGTCAGCCAAAATTACCCTATAGTGCAGAACATCCAG
GGGCAAATGGTACATCAGGCCATATCACCTAGAACTTTAAATGCATG
GGTAAAAGTAGTAGAAGAGAAGGCTTTCAGCCCAGAAGTGATACCC
ATGTTTTCAGCATTATCAGAAGGAGCCACCCCACAAGATTTAAACAC
CATGCTAAACACAGTGGGGGGACATCAAGCAGCCATGCAAATGTTA
AAAGAGACCATCAATGAGGAAGCTGCAGAATGGGATAGAGTGCATC
CAGTGCATGCAGGGCCTATTGCACCAGGCCAGATGAGAGAACCAA
GGGGAAGTGACATAGCAGGAACTACTAGTACCCTTCAGGAACAAAT
AGGATGGATGACACATAATCCACCTATCCCAGTAGGAGAAATCTATA
AAAGATGGATAATCCTGGGATTAAATAAAATAGTAAGAATGTATAGC
CCTACCAGCATTCTGGACATAAGACAAGGACCAAAGGAACCCTTTA
GAGACTATGTAGACCGATTCTATAAAACTCTAAGAGCCGAGCAAGCT
TCACAAGAGGTAAAAAATTGGATGACAGAAACCTTGTTGGTCCAAAA
TGCGAACCCAGATTGTAAGACTATTTTAAAAGCATTGGGACCAGGAG
CGACACTAGAAGAAATGATGACAGCATGTCAGGGAGTGGGGGGAC
CCGGCCATAAAGCAAGAGTTTTGGCTGAAGCAATGAGCCAAGTAAC
AAATCCAGCTACCATAATGATACAGAAAGGCAATTTTAGGAACCAAA
GAAAGACTGTTAAGTGTTTCAATTGTGGCAAAGAAGGGCACATAGC
CAAAAATTGCAGGGCCCCTAGGAAAAAGGGCTGTTGGAAATGTGGA
334
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
AAGGAAGGAGACCAAATGAAAGATTGTACTGAGAGACAGGCTAATTT
TTTAG GGAAGATCTGGCCTTCC CACAAGGGAAG GC CAGG GAATTTT
CTTCAGAGCAGACCAGAGCCAACAGC CCCACCAGAAGAGAGCTTCA
GGTTTG GGGAAGAGACAACAACTCCCTCTCAGAAG GAG GAGC C GAT
AGACAAGGAACTGTATC CTTTAGCTTCC CTCAGATCACTCTTTGGCA
GC GAC CC CTCGTCACAAAAC TTTAGC CAGAACTATC CGATTGTG CA
GACCG GTG C CC CAAAGAAGAAG CGGAAG GTCTCTAGACAAGAGAT
CAAGAGAATCAACAAGATCAGAAGGAGACTGGTCAAGGACAGCAAC
ACAAAGAAG GC C GG CAAGACAG GCCC CATGAAAACCCTGCTC GTCA
GAGTGATGACCCCTGACCTGAGAGAGCGGCTGGAAAACCTGAGAA
AGAAG CC CGAGAACATCCCTCAGC CTATCAG CAACACCAG CAGGGC
CAACCTGAACAAGCTGCTGACCGACTACACCGAGATGAAGAAAGCC
ATC CTG CAC GTGTACTG GGAAGAGTTC CAGAAAGACCC CGTGGGCC
TGATGAG CAGAGTTG CTCAG C CC G CTCCTAAGAACATCGACCAGAG
AAAGCTGATCC CCGTGAAGGACGGCAACGAGAGACTGACCTCTAGC
GGCTTTGCCTGCAGCCAGTGTTGCCAGCCTCTGTAC GTGTACAAGC
TGGAACAAGTGAACGACAAGG GCAAGC CC CACAC CAACTACTTC GG
CAGATGCAAC GTGTCC GAG CAC GAGAG GCTGATC CTG CTGTCTC CT
CACAAGC CC GAGGCCAACGATGAGCTG GTCACATACAGC CTGG GC
AAGTTCGGACAGAGAG CC CTG GACTTCTACAGCATC CACGTGAC CA
GG GAGAGCAATCAC CCTGTGAAGC C CCTG GAACAGATCG GC GG CA
ATAGCTGTGCCTCTGGACCTGTGGGAAAAGCCCTGAGCGACGCCT
GTATGGGAGCCGTGGCATC CTTCCTGACCAAGTAC CAC GACATCAT
CCTG GAACACCAGAAAGTGATCAAGAAGAACGAGAAAAGACTGG CC
AACCTCAAGGATATC GC CAGC GCTAAC GGCCTGGC CTTTCCTAAGA
TCACCCTGCCTCCACAGCCTCACAC CAAAGAGGGCATCGAGGCCTA
CAACAACGTGGTGGCCCAGATCGTGATTTGGGTCAACCTGAATCTG
TGGCAGAAGCTGAAGATCGGCAGGGACGAAGC CAAGC CACTGCAG
AGACTGAAG G G CTTCC CTAGCTTCC CTCTGGTGGAAAGACAG GC CA
ATGAAGTGGATTGGTGGGACATG GTCTGCAACGTGAAGAAGCTGAT
CAACGAGAAGAAAGAGGATGGCAAGGTTTTCTGGCAGAACCTGGCC
GG CTACAAGAGACAAGAAG CC CTGAGGCCTTACCTGAGCAGCGAA
GAG GAC C GGAAGAAG GGCAAGAAGTTCG C CAGATAC CAGTTCGGC
GACCTGCTG CTG CAC CTG GAAAAGAAGCACGG C GAG GACTG G GG C
AAAGTGTACGATGAGGCCTGGGAGAGAATCGACAAGAAGGTGGAA
GGC CTGAGCAAGCACATTAAG CTG GAAGAG GAAAGAAG GAG C GAG
GACGCCCAATCTAAAGCCGCTCTGACCGATTGGCTGAGAGCCAAGG
CCAGCTTTGTGATC GAG G GC CTGAAAGAG GCC GACAAGGAC GAGT
TCTGCAGATGCGAGCTGAAGCTGCAGAAGTG GTACG GC GATCTGA
GAG G CAAG C CCTTC GC CATTGAG G CCGAGAACAGCATCCTGGACAT
CAGCGGCTTCAGCAAGCAGTACAACTGCGC CTTCATTTGGCAGAAA
GACG GC GTCAAGAAACTGAAC CTGTACCTGATCATCAATTACTTCAA
AG GC GG CAAGCTGC G GTTCAAGAAGATCAAAC CC GAG G C CTTC GA
GG CTAACAGATTCTACACCGTGATCAACAAAAAGTCCG GCGAGATC
GTG CC CATG GAAGTGAACTTGAACTTC GAC GAC CC CAACCTGATTAT
335
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
CCTGCCTCTGGC CTTCGGCAAGAGACAGGGCAGAGAGTTCATCTG
GAAC GATCTGCTGAGC CTGGAAAC C G G CTCTCTGAAG CTG GC CAAT
GGCAGAGTGATCGAGAAAACC CTGTACAACAGGAGAACCAGACAG
GACGAGGCTGCTCTGTTTGTGGCCCTGACCTTCGAGAGAAGAGAGG
TGCTGGACAGCAGCAACATCAAGCCCATGAACCTGATC GGCATC GA
CC GGGGCGAGAATATCC CTGCTGTGATCGCCCTGACAGACCCTGAA
GGATGCCCACTGAGCAGATTCAAGGACTCCCTGGGCAACCCTACAC
ACATC CTGAGAATC GGC GAGAGCTACAAAGAGAAGCAGAGGACAAT
CCAGGCCAAGAAAGAGGTGGAAGAGAGAAGAGCCGGCGGATACTC
TAGGAAGTAC GC CAG CAAG GC CAAGAATCTG G CC GAC GACATGGT
CC GAAACAC C G CCAGAGATCTGCTGTACTAC GC CGTGACACAGGAC
GC CATGCTGATCTTCGAGAATCTGAG CAGAGG CTTCG GC C GG CAG
GGCAAGAGAACCTTTATGGC C GAGAGGCAGTACAC CAGAATGGAAG
ATTGGCTCACAGCTAAACTGGC CTACGAGGGACTGAGCAAGACCTA
CCTGTCCAAAACACTGGC CCAGTATAC C TCCAAGACCTGCAGCAAT
TGCGGCTTCACCATCAC CAGC GC C GACTACGACAGAGTGCTGGAAA
AGCTCAAGAAAACCGCCACCGGCTGGATGACCACCATCAACGGCAA
AGAGCTGAAGGTTGAGGGC CAGATCAC C TACTACAACAGGTACAAG
AG G CAGAAC GTCGTGAAGGATCTGAGC GTGGAACTGGACAGACTG
AG C GAAGAGAG C GTGAACAAC GACATCAG CAG CTG GACAAAG G G C
AGATCAG GC GAGGCTCTGAGCCTGCTGAAGAAGAGGTTTAGCCACA
GACCTGTGCAAGAGAAGTTCGTGTGCCTGAACTGCGGCTTCGAGAC
ACAC GCC GATGAACAGGCTGCC CTGAACATTGCCAGAAGCTGGCTG
TTCCTGAGAAGCCAAGAGTACAAGAAGTACCAGACCAACAAGACCA
CC GG CAACAC C GACAAGAG G GC CTTTGTG GAAAC CTG G CAGAG CT
TCTACAGAAAAAAGCTGAAAGAAGTCTGGAAGCC C GC C GTGACTAG
TCCAAAAAAGAAGAGAAAGGTAGCC CTCGAGTACC CATATGATGTC
CCTGACTACGCT (SEQ ID NO: 427)
pXDP 11 ATGGGTGC GAGAGC GTCAGTATTAAG CGG G
GGAGAATTAGATC GAT
GGGAAAAAATTC GGTTAAGGCCAGGGGGAAAGAAAAAATATAAATTA
AAACATATAGTATGGGCAAGCAGGGAGCTAGAACGATTCGCAGTTA
ATCCTGGCCTGTTAGAAACATCAGAAGGCTGTAGACAAATACTGGG
ACAGCTACAACCATCCCTTCAGACAGGATCAGAAGAACTTAGATCAT
TATATAATACAGTAGCAACC CTCTATTGTGTGCATCAAAGGATAGAG
ATAAAAGACACCAAGGAAGCTTTAGACAAGATAGAGGAAGAGCAAA
ACAAAAGTAAGAAAAAAG CACAG CAAG CAG CAG CTGACACAG GACA
CAGCAATCAGGTCAGC CAAAATTACCCTATAGTGCAGAACATC CAG
GGGCAAATGGTACATCAGGCCATATCACCTAGAACTTTAAATGCATG
GGTAAAAGTAGTAGAAGAGAAGGCTTTCAGCC CAGAAGTGATACC C
ATGTTTTCAG CATTATCAGAAG GAG C CACC CCACAAGATTTAAACAC
CATGCTAAACACAGTGGGGGGACATCAAGCAGCCATGCAAATGTTA
AAAGAGACCATCAATGAGGAAGCTGCAGAATGGGATAGAGTGCATC
CAGTGCATGCAGGG CCTATTG CAC CAGG C CAGATGAGAGAAC CAA
GGGGAAGTGACATAGCAGGAACTACTAGTAC CCTTCAGGAACAAAT
336
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
AGGATGGATGACACATAATCCACCTATCCCAGTAGGAGAAATCTATA
AAAGATGGATAATCCTGGGATTAAATAAAATAGTAAGAATGTATAGC
CCTACCAGCATTCTGGACATAAGACAAGGACCAAAGGAACCCTTTA
GAGACTATGTAGACCGATTCTATAAAACTCTAAGAGCCGAGCAAGCT
TCACAAGAGGTAAAAAATTGGATGACAGAAACCTTGTTGGTCCAAAA
TGCGAACCCAGATTGTAAGACTATTTTAAAAGCATTGGGACCAGGAG
CGACACTAGAAGAAATGATGACAGCATGTCAGGGAGTGGGGGGAC
CCGGCCATAAAGCAAGAGTTTTGGCTGAAGCAATGAGCCAAGTAAC
AAATC CAGCTAC CATAATGATACAGAAAGGCAATTTTAGGAAC CAAA
GAAAGACTGTTAAGTGTTTCAATTGTGGCAAAGAAGGGCACATAGC
CAAAAATTGCAGGGCC C CTAG GAAAAAG GG CTGTTG GAAATGTG GA
AAGGAAGGACACCAAATGAAAGATTGTACTGAGAGACAGGCTAATTT
TTTAGGGAAGATCTGGCCTTCC CACAAGGGAAG GC CAGG GAATTTT
CTTCAGAGCAGACCAGAGCCAACAGCCCCACCAGAAGAGAGCTTCA
GGTTTGGGGAAGAGACAACAACTC CCTCTCAGAAGCAGGAGCC GAT
AGACAAGGAACTGTATCCTTTAGCTTCCCTCAGATCACTCTTTGGCA
GCGACCCCTCGTCACAAAACTTTAGCCAGAACTATCCGATTGTGCA
GAC CGGTGCCCCAAAGAAGAAGC GGAAGGTCTCTAGACAAGAGAT
CAAGAGAATCAACAAGATCAGAAGGAGACTGGTCAAGGACAGCAAC
ACAAAGAAGGCCGGCAAGACAGGCCCCATGAAAACCCTGCTCGTCA
GAGTGATGACCCCTGACCTGAGAGAGCGGCTGGAAAACCTGAGAA
AGAAGCCCGAGAACATCCCTCAGCCTATCAGCAACACCAGCAGGGC
CAACCTGAACAAGCTGCTGAC CGACTACACC GAGATGAAGAAAGCC
ATCCTG CAC GTGTACTG GGAAGAGTTC CAGAAAGACC CCGTGGGCC
TGATGAGCAGAGTTGCTCAGCCCGCTCCTAAGAACATCGACCAGAG
AAAGCTGATCC CCGTGAAGGACGGCAACGAGAGACTGACCTCTAGC
GGCTTTGCCTGCAGCCAGTGTTGCCAGCCTCTGTACGTGTACAAGC
TGGAACAAGTGAACGACAAGGGCAAGC C CCACAC CAACTACTTC GG
CAGATGCAACGTGTCC GAG CACGAGAG G CTGATC CTG CTGTCTC CT
CACAAGCCCGAGGCCAACGATGAGCTGGTCACATACAGCCTGGGC
AAGTTCGGACAGAGAGCCCTGGACTTCTACAGCATCCACGTGACCA
GGGAGAGCAATCACCCTGTGAAGCCCCTGGAACAGATCGG C GG CA
ATAGCTGTGCCTCTGGACCTGTGGGAAAAGC CCTGAGCGACGC CT
GTATG GGAG CC GTG GCATC CTTCCTGACCAAGTAC CAGGACATCAT
CCTGGAACACCAGAAAGTGATCAAGAAGAACGAGAAAAGACTGGCC
AACCTCAAGGATATC GC CAGC GCTAAC GGCCTGGC CTTTCCTAAGA
TCACCCTGC CTCCACAGCCTCACAC CAAAGAGGGCATC GAG G CCTA
CAACAAC GTG GTG GC CCAGATC GTGATTTGGGTCAACCTGAATCTG
TGGCAGAAGCTGAAGATC G GCAG G CAC GAAG C CAAGC CACTGCAG
AGACTGAAGGGCTTCC CTAGCTTCC CTCTG GTG GAAAGACAG GC CA
ATGAAGTGGATTGGTGGGACATGGTCTGCAACGTGAAGAAGCTGAT
CAACGAGAAGAAAGAGGATGGCAAGGTTTTCTGGCAGAACCTGGCC
GG CTACAAGAGACAAGAAG CC CTGAGGCCTTACCTGAGCAGCGAA
GAGGACCGGAAGAAGGGCAAGAAGTTCGCCAGATACCAGCTGGGC
GACCTGCTGAAGCACCTGGAAAAGAAGCACGGCGAGGACTGGGGC
337
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
AAAGTGTACGATGAGGCCTGGGAGAGAATCGACAAGAAGGTGGAA
GG C CTGAG CAAG CACATTAAG CTG GAAGAG GAAAGAAG GAG C GAG
GACGC CCAATCTAAAGCCGCTCTGACCGATTGGCTGAGAGCCAAGG
CCAGCTTTGTGATC GAG G GC CTGAAAGAG G CC GACAAGGAC GAGT
TCTGCAGATGCGAGCTGAAGCTGCAGAAGTGGTACGGCGATCTGA
GAG G CAAG C CCTTC GC CATTGAG G C CGAGAACAGCATC CTGGACAT
CAGCGGCTTC AGCAAGCAGTACAACTGCGCCTTCATTTGGCAGAAA
GAC G GC GTCAAGAAACTGAAC CTGTAC CTGATCATCAATTACTTCAA
AG G C GG CAAGCTGC G GTTCAAGAAGATCAAAC CC GAG G C CTTC GA
GGCTAACAGATTCTACACCGTGATCAACAAAAAGTCCGGCGAGATC
GTGC CCATGGAAGTGAACTTCAACTTC GAC GAC CC CAACCTGATTAT
CCTGCCTCTGGC CTTCGGCAAGAGACAGGGCAGAGAGTTCATCTG
GAAC GATCTGCTGAGC CTGGAAAC C G G CTCTCTGAAG CTG GC CAAT
GGCAGAGTGATCGAGAAAACC CTGTACAACAGGAGAACCAGACAG
GAC GAG C CTGCTCTGTTTGTG GC C CTGAC CTTC GAGAGAAGAGAGG
TGCTGGACAGCAGCAACATCAAGCCCATGAACCTGATC GGC GTG GA
CCGGGGCGAGAATATCCCTGCTGTGATCGCCCTGACAGACCCTGAA
GGATGC C CACTGAGCAGATTCAAGGACTCCCTGGGCAAC C CTACAC
ACATCCTGAGAATC GGCGAGAGCTACAAAGAGAAGCAGAGGACAAT
CCAGGCCAAGAAAGAGGTGGAACAGAGAAGAGCCGGCGGATACTC
TAGGAAGTAC GC CAG CAAG GC CAAGAATCTG G CC GAC GACATGGT
CCGAAACACCGCCAGAGATCTGCTGTACTACGCCGTGACACAGGAC
GC CATGCTGATCTTC GAGAATCTGAG CAGAGG CTTC G GC C GG CAG
GGCAAGAGAACCTTTATGGCCGAGAGGCAGTACACCAGAATGGAAG
ATTGGCTCACAGCTAAACTGGC CTACGAGGGACTGAGCAAGAC CTA
CCTGTCCAAAACACTGGC CCAGTATAC C TCCAAGACCTGCAGCAAT
TGCGGCTTCACCATCACCAGCGCCGACTACGACAGAGTGCTGGAAA
AG CTCAAGAAAAC CGC CAC C G GCTGGATGAC CAC CATCAAC GGCAA
AGAGCTGAAGGTTGAGGGCCAGATCAC CTACTACAACAG GAG GAAG
AG G CAGAAC GTCGTGAAGGATCTGAGC GTGGAACTGGACAGACTG
AG C GAAGAGAG C GTGAACAAC GACATCAG GAG CTG GACAAAG G G C
AGATCAG GC GAGGCTCTGAGCCTGCTGAAGAAGAGGTTTAGCCACA
GAC CTGTGCAAGAGAAGTTC GTGTGC CTGAACTGC GGCTTC GAGAC
ACAC G CC GATGAACAGGCTGCC CTGAACATTGCCAGAAGCTGGCTG
TTCCTG AGAAGCCAAGAGTACAAGAAGTACCAGAC CAACAAGACCA
CC GG CAACAC C GACAAGAG G GC C TTTGTG GAAAC CTG G CAGAG CT
TCTACAGAAAAAAGCTGAAAGAAGTCTGGAAGCC C GC CGTGACTAG
TC CAAAAAAGAAGAGAAAGGTAGCC CTC GAGTACC CATATGATGTC
CCTGACTACGCT (SEQ ID NO: 428)
pXDP1 2 ATGG GTG C GAGAG C GTCAGTATTAAG CG G G GGAGAATTAGATCGAT
GGGAAAAAATTC GGTTAAGGC CAGGGGGAAAGAAAAAATATAAATTA
AAACATATAGTATGGGCAAGCAGGGAGCTAGAACGATTCGCAGTTA
ATCCTGGCCTGTTAGAAACATCAGAAGGCTGTAGACAAATACTGGG
ACAGCTACAACCATC CCTTCAGACAGGATCAGAAGAACTTAGATCAT
338
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
TATATAATACAGTAGCAACCCTCTATTGTGTGCATCAAAGGATAGAG
ATAAAAGACACCAAGGAAGCTTTAGACAAGATAGAGGAAGAGCAAA
ACAAAAGTAAGAAAAAAGCACAGCAAGCAGCAGCTGACACAGGACA
CAGCAATCAGGTCAGCCAAAATTACCCTATAGTGCAGAACATCCAG
GG GCAAATG GTACATCAGG C CATATCAC CTAGAACTTTAAATGCATG
GGTAAAAGTAGTAGAAGAGAAGGCTTTCAGGCCAGAAGTGATACCC
ATGTTTTCAG CATTATCAGAAG GAG CCACC CCACAAGATTTAAACAC
CATGCTAAACACAGTGGGGGGACATCAAGCAGCCATGCAAATGTTA
AAAGAGACCATCAATGAGGAAGCTGCAGAATGGGATAGAGTGCATC
CAGTGCATG CAC GG CCTATTG CAC CAGG C CAGATGAGAGAAC CAA
GGGGAAGTGACATAGCAGGAACTACTAGTACCCTTCAGGAACAAAT
AGGATGGATGACACATAATCCACCTATCCCAGTAGGAGAAATCTATA
AAAGATGGATAATCCTGGGATTAAATAAAATAGTAAGAATGTATAGC
CCTACCAGCATTCTGGACATAAGACAAGGACCAAAGGAACCCTTTA
GAGACTATGTAGACC GATTCTATAAAACTCTAAGAGC C GAG CAAGCT
TCACAAGAGGTAAAAAATTGGATGACAGAAACCTTGTTGGTCCAAAA
TGCGAAC CCAGATTGTAAGACTATTTTAAAAGCATTGGG AC CAGGAG
CGACACTAGAAGAAATGATGACAGCATGTCAGGGAGTGGGGGGAC
CC GGCCATAAAG CAAGAGTTTTGGCTGAAG CAATGAG CCAAGTAAC
AAATC CAGCTAC CATAATGATACAGAAAGG CAATTTTAGGAAC CAAA
GAAAGACTGTTAAGTGTTTCAATTGTGGCAAAGAAGGGCACATAGC
CAAAAATTG CAG GGCC C CTAGGAAAAAGGGCTGTTGGAAATGTG GA
AAGGAAGGACACCAAATGAAAGATTGTACTGAGAGACAGGCTAATTT
TTTAG GGAAGATCTGGCCTTCC CACAAGGGAAG GC CAGG GAATTTT
CTTCAGAGCAGACCAGAGC CAACAGC C C CAC CAGAAGAGAG CTTCA
GGTTTG GGGAAGAGACAACAACTC C CTCTCAGAAG CAG GAGC C GAT
AGACAAG GAACTGTATC CTTTAGCTTCC CTCAGATCACTCTTTGGCA
GC GAC C C CTC GTCACAAAACTTTAGCCAGAACTATC C GATTGTG CA
GACCG GTG C CC CAAAGAAGAAG CGGAAG GTCTCTAGACAAGAGAT
CAAGAGAATCAACAAGATCAGAAGGAGACTGGTCAAGGACAGCAAC
ACAAAGAAGGCCGGCAAGACAGGCCCCATGAAAACCCTGCTCGTCA
GAGTGATGACC C CTGACCTGAGAGAG CGOCTOGAAAACCTGAGAA
AGAAGCC C GAGAACATC CCTCAGC CTATCAGCAACACCAG CAGGGC
CAAC CTGAACAAGCTGCTGACCGACTACAC CGAGATGAAGAAAG C C
ATCCTG CAC GTGTACTG GGAAGAGTTC CAGAAAGACC CCGTG GGCC
TGATGAGCAGAGTTGCTCAGC CCGCTCCTAAGAACATC GAC CAGAG
AAAG CTGATCCCCGTGAAGGACGG CAACGAGAGACTGACCTCTAGC
GG CTTTGCCTGCAG CCAGTGTTG CCAGCCTCTGTAC GTGTACAAG C
TGGAACAAGTGAACGACAAGG GCAAGC C C CACAC CAACTACTTC GG
CAGATGCAACGTGTCC GAG CACGAGAG GCTGATC CTG CTGTCTC CT
CACAAGC C C GAGGC CAAC GATGAGCTG GTCACATACAGC CTGG GC
AAGTTCGGACAGAGAG CC CTG GACTTCTACAGCATC CACGTGAC CA
GG GAGAGCAATCAC CCTGTGAAGC CCCTG GAACAGATCG GC GG CA
ATAGCTGTGCCTCTGGACCTGTGGGAAAAGCCCTGAGCGACG C CT
GTATGGGAG CC GTG GCATC CTTCCTGACCAAGTAC GAG GACATCAT
339
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
CCTG GAACACAAGAAAGTGATCAAGAAGAACGAGAAAAGACTGGC C
AACCTCAAGGATATCGC CAGCGCTAAC G GC CTG G C CTTTC CTAAGA
TCACCCTGC CTCCACAGCCTCACAC CAAAGAG GGCATC GAG G CCTA
CAACAACGTGGTGGCCCAGATCGTGATTTGGGTCAACCTGAATCTG
TGGCAGAAGCTGAAGATCGGCAGGGACGAAGC CAAGC CACTGCAG
AGACTGAAG G G CFTC C CTAGCTTCC CTCTG GTGGAAAGACAG GC CA
ATGAAGTGGATTGGTGGGACATG GTCTGCAACGTGAAGAAGCTGAT
CAACGAGAAGAAAGAGGATGGCAAGGTTTTCTGGCAGAAC C TGGCC
GG CTACAAGAGACAAGAAG CC CTGAGG GOTTA CCTGAGCAGC CC C
GAG GAC CG GAAGAAG GGCAAGAAGTTCGCCAGATACCAGCTGG GC
GAC CTGCTG CTG CAC CTG GAAAAGAAGCACGG C GAG GACTG G GG C
AAAGTGTACGATGAGGCCTGGGAGAGAATCGACAAGAAGGTGGAA
GGC CTGAG CAAG CACATTAAG CTG GAAGAG GAAAGAAG GAG C GAG
GACGCCCAATCTAAAGCCGCTCTGACCGATTGGCTGAGAGCCAAGG
CCAGCTTTGTGATC GAG G GC CTGAAAGAG GCC GACAAGGAC GAGT
TCTGCAGATGCGAGCTGAAGCTGCAGAAGTGGTACGGCGATCTGA
GAG G CAAG C CCTTC GC CATTGAG G C CGAGAACAGCATC CTGGACAT
CAGCGGCTTCAGCAAGCAGTACAACTGCGC CTTCATTTGGCAGAAA
GACG GC GTCAAGAAACTGAAC CTGTACCTGATCATCAATTACTTCAA
AG GC GG CAAGCTGC G GTTCAAGAAGATCAAAC CC GAG G C CTTC GA
GGCTAACAGATTCTACACCGTGATCAACAAAAAGTCCG GCGAGATC
GTGC CCATG GAAGTGAACTTCAACTTC GAC GAC CC CAACCTGATTAT
CCTGC CTCTGGCC TTCGGCAAGAGACAGGGCAGAGAGTTCATCTG
GAAC GATCTGCTGAGC CTGGAAACCG G CTCTCTGAAG CTG GC CAAT
GGCAGAGTGATCGAGAAAC CCCTGTACAACAGGAGAACCAGACAG
GAC GAG C CTGCTCTGTTTGTG GC C CTGAC CTTCGAGAGAAGAGAGG
TG CTG GACAG CAG CAACATCAAG CC CATGAACCTGATC GG C GTG GA
CC GGGGC GAGAATATCC CTGCTGTGATCGC CCTGACAGAC CCTGAA
GGATGCCCACTGAGCAGATTCAAGGACTCCCTGGGCAACCCTACAC
ACATCCTGAGAATC GGCGAGAGCTACAAAGAGAAGCAGAGGACAAT
CCAGGCCAAGAAAGAGGTGGAACAGAGAAGAGCCGGCGGATACTC
TAGGAAGTAC GC CAGCAAG GC CAAGAATCTG G CC GAC GACATGGT
CC GAAACAC C GC CAGAGATCTGCTGTACTAC GC C GTGACACAGGAC
GC CATGCTGATCTTCGAGAATCTGAGCAGAGGCTTCG GC C GGCAG
GGCAAGAGAACCTTTATGGCCGAGAG GCAGTACACCAGAATGGAAG
ATTGGCTCACAGCTAAACTGGC CTACGAGGGACTGAGCAAGACCTA
CCTGTCCAAAACACTG G CC CAGTATAC CTCCAAGACCTGCAGCAAT
TGCGGCTTCACCATCACCAGC GC C GACTACGACAGAGTGCTGGAAA
AG CTCAAGAAAAC CGC CAC C G GCTGGATGAC CAC CATCAAC GGCAA
AGAGCTGAAGGTTGAG GGCCAGATCAC CTACTACAACAG GAG GAAG
AG GCAGAAC GTCGTGAAGGATCTGAGC GTGGAACTGGACAGAC TG
AG C GAAGAGAG C GTGAACAAC GACATCAG CAG CTG GACAAAG GGC
AGATCAG GC GAGGCTCTGAGCCTGCTGAAGAAGAGGTTTAGCCACA
GACCTGTGCAAGAGAAGTTCGTGTGCCTGAACTGC GGCTTCGAGAC
ACACGCCGATGAACAGGCTGCCCTGAACATTGCCAGAAGCTGGCTG
340
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
TTCCTGAGAAGCCAAGAGTACAAGAAGTACCAGAC CAACAAGACCA
CCGGCAACACCGACAAGAGGGCCTTTGTGGAAACCTGGCAGAGCT
TCTACAGAAAAAAG CT GAAAGAAGTCTG GAAGCC C GC C GT GACTA G
TCCAAAAAAGAAGAGAAAG GTAGCC CTCGAGTACC CATATGATGTC
CCTGACTACGCT (SEG ID NO: 429)
psPax2 AT GG GTG C GAGA G C GTCAGTATTAAG CG G
GGGAGAATTAGATCGAT
GO GAAAAAATTC GGTTAAGG CCAGGG GGAAAGAAAAAATATAAATTA
AAACATATAGTATGGG CAA GC A GG GA GC TAGAACGATTCG CAGTTA
AT C CTG G C C TGTTA GAAAC AT CAGAAG GC TGTAGACAAATAC T GG G
ACAG CTACAAC CAT CC CTTCAGACAG GAT CAGAA GAACTTAGATCAT
TATATAATACAGTAG CAACC CTCTATTGTGTGCATC AAAGGATAGAG
ATAAAAGACAC CAAGGAAG CTTTAGACAAGATAGAGGAAGAGCAAA
AC AAAA GTAAGAAAAAAG CAC AG CAAG C A G CAG CT GACAC AG GAC A
CAGCAATCAGGTCAGC CAAAATTAC CCTATAGTGCAGAACATC C AG
GO GCAAATG GTACATCAGG CC ATATCAC CTA GAAC TTTAAATG CATG
GGTAAAAGTAGTAGAAGAGAAGG CTTT CA G CC CAGAAGTGATACC C
AT GTTTTCAG CATTATCAGAAG GAG CCACC CCACAAGATTTAAACAC
CATG CTAAACACAGTG G G GG GAC ATCAA GC A G CC AT G CAAATGTTA
AAAGAGACCATCAATGAGGAAGCTG CAGAATG GGATAGAGTG CATC
CAGTGCATG CAC GO CC TATTG CAC CA GG C CAGATGAGAGAAC CAA
GG GGAAGTGACATAGCAGGAACTACTAGTAC C C TTC AG GAACAAAT
AG GATGGATGACACATAATC CAC CTATC C CAGTAGGAGAAATC TATA
AAAGATGGATAATC CTG GGATTAAATAAAATAGTAAGAATGTATAG C
CCTAC CAGCATTCTGGACATAAGACAAGGACCAAAGGAAC CCTTTA
GAGACTATGTAGAC C GATT CTATAAAACTCTAAGA G C C GAG CAA GC T
TCACAA GAG GTAAAAAATTGGATGACAGAAACCTTGTTGGTCCAAAA
TGCGAAC C CA GATTGTAAGAC TATTTTAAAAG CATTG GGAC CAGGAG
CGACACTAGAAGAAATGAT GACAG C ATGT CAC GGAGTGGGGG GAG
CCGGCCATAAAG CAAGAGTTTTGGCTGAAG CAATGAG C CAAGTAAC
AAATC CAGCTAC CATAATGATACAGAAAGG CAATTTTAGGAAC CAAA
GAAAGACTGTTAAGTGTTTCAATTGTGG CAAAGAAGGG C AC ATAG C
CAAAAATTG CAG GGCCCC TAG GAAAAAG GG C TGTTGGAAATGTG GA
AA GGAAG GACACC AAATGAAA GATTGTACTGA GA GAC A GG CTAATTT
TTTAG GGAAGATCTGGC CTTCC CACAAGGGAAG GC C AGG GAATTTT
CTTCAGAGCAGACCAGAGC CAACAGC CCCACCAGAAGAGAG CTTCA
GGTTTG GG GAA GAGAC AACAACTC C CTC TC A GAA G CAG GAGC C GAT
AGACAAG GAACTGTATC CTTTAGCTTCC CT CAGATCACTCTTTG GCA
GCGACCCCTCGTCACAATAAAGATAGGGGGGCAATTAAAGGAAGCT
CTATTAGATACAGGAGCAGATGATACAGTATTAGAAGAAATGAATTT
GC CAG GAAGATGGAAAC CAAAAATGATAGGGGGAATTG GAGGTTTT
AT CAAAGTAAGAC AGTATGATCAGATAC TC ATAGAAATC TG CC GACA
TAAAGCTATAGGTACAGTATTAGTAGGACCTACAC CT GTC AACATAA
TTGGAAGAAATCTGTTGACTCAGATTGG CTGCACTTTAAATTTTCC C
ATTAGTC CTATTGAGACTGTACCAGTAAAATTAAAGCCAGGAATGGA
341
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
TGGCCCAAAAGTTAAACAATGGCCATTGACAGAAGAAAAAATAAAAG
CATTAGTAGAAATTTGTACAGAAATGGAAAAGGAAGGAAAAATTTCA
AAAATTG GGCCTGAAAATC CATACAATACTCCAGTATTTGC CATAAA
GAAAAAAGACAGTAGTAAATGGAGAAAATTAGTAGATTTCAGAGAAC
TTAATAAGAGAACTCAAGATTTCTGGGAAGTTCAATTAGGAATACCA
CATCCTGCAGGGTTAAAACAGAAAAAATCAGTAACAGTACTGGATGT
GG GCGATGCATATTTTTCAGTTC CCTTAGATAAAGACTTCAGGAAGT
ATAC TGCATTTAC CATAC CTAGTATAAACAATGAGACAC CAGG GATT
AGATATCAGTACAATGTGCTTCCACAGG GATGGAAAGGATCA C CAG
CAATATTCCAGTG TAG CATGACAAAAATCTTAGAGC CTTTTAGAAAA
CAAAATCCAGACATAGTCATCTATCAATACATGGATGATTTGTATGTA
GGATCTGACTTAGAAATAGGGCAGCATAGAACAAAAATAGAGGAAC
TGAGACAACATCTGTTGAG GTG GGGATTTACCACACCAGACAAAAA
ACATCAGAAAGAACCTCCATTCCTTTGGATGGGTTATGAACTCCATC
CTGATAAATG GACAGTACAGCCTATAGTGCTGCCAGAAAAG GACAG
CTGGACTGTCAATGACATACAGAAATTAGTGGGAAAATTGAATTGGG
CAAGTCAGATTTATGCAGGGATTAAAGTAAGGCAATTATGTAAACTT
CTTAGGG GAAC CAAAG CACTAACAGAAGTAGTAC CACTAACAGAAG
AAGCAGAGCTAGAACTGGCAGAAAACAGGGAGATTCTAAAAGAACC
GGTACATGGAGTGTATTATGACCCATCAAAAGACTTAATAGCAGAAA
TACAGAAGCAGGGGCAAGGCCAATGGACATATCAAATTTATCAAGA
GCCATTTAAAAATCTGAAAACAGGAAAGTATGCAAGAATGAAGGGTG
CCCACACTAATGATGTGAAACAATTAACAGAGGCAGTACAAAAAATA
GCCACAGAAAGCATAGTAATATGGGGAAAGACTCCTAAATTTAAATT
ACC CATACAAAAGGAAACATGG GAAGCATGGTGGACAGAGTATTGG
CAAG C CAC CTG GATTCCTGAGTGGGAGTTTGTCAATAC C C CTC C CT
TAGTGAAGTTATGGTACCAGTTAGAGAAAGAACCCATAATAGGAGCA
GAAACTTTCTATGTAGATGGGGCAGCCAATAGGGAAACTAAATTAGG
AAAAGCAGGATATGTAACTGACAGAGGAAGACAAAAAGTTGTCCCC
CTAACGGACACAACAAATCAGAAGACTGAGTTACAAGCAATTCATCT
AGCTTTGCAGGATTCGGGATTAGAAGTAAACATAGTGACAGACTCAC
AATATGCATTGGGAATCATTCAAGCACAACCAGATAAGAGTGAATCA
GAGTTAGTCAGTCAAATAATAGAGCAGTTAATAAAAAAGGAAAAAGT
CTACCTGGCATGG GTAC CAGCACACAAAGGAATTG GAG GAAATGAA
CAAGTAGATAAATTGGTCAGTGCTGGAATCAGGAAAGTACTATTTTT
AGATGGAATAGATAAGGCCCAAGAAGAACATGAGAAATATCACAGTA
ATTGGAGAGCAATGGCTAGTGATTTTAACCTACCACCTGTAGTAGCA
AAAGAAATAGTAGCCAGCTGTGATAAATGTCAGCTAAAAGGGGAAG
CCATGCATG GACAAGTAGACTGTAGCC CAGGAATATG GCAGCTAGA
TTGTACACATTTAGAAGGAAAAGTTATCTTGGTAGCAGTTCATGTAG
CCAGTGGATATATAGAAGCAGAAGTAATTCCAGCAGAGACAGGGCA
AGAAACAGCATACTTCCTCTTAAAATTAGCAGGAAGATGGCCAGTAA
AAACAGTACATACAGACAATGGCAGCAATTTCACCAGTACTACAGTT
AAGGCCGCCTGTTGGTGGGCGGGGATCAAGCAGGAATTTGGCATT
CCCTACAATCCCCAAAGTCAAGGAGTAATAGAATCTATGAATAAAGA
342
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
ATTAAAGAAAATTATAG GACAG GTAAGAGATCAGG CTGAACATCTTA
AGA C A GC A GTAC AAATG G CAGTATTC ATC CA CAATTTTAAAAGAAAA
GG GGGGATTGG GGGGTACAGTGCAGGGGAAAGAATAGTAGACATA
ATAGCAAGAGACATACAAACTAAAGAATTACAAAAACAAATTACAAAA
ATTCAAAATTTTC GG GTTTATTACAG GGACAGCAGAGATC CAGTTTG
GAAAGGACCAGCAAAG CTC CT CT GGAAA GGTGAA GGG GCAGTAGT
AATA C AA GATAATA GTGA C ATAAAAGTA GTG C CAAGAA GAAAAG CAA
AGATCATCAGGGATTATGGAAAACAGATGGCAGGTGATGATTGTGT
GG CAA GTA GA CAG GAT GAG GATTAACACATGGAATTCTGCAACAAC
TG CTGTTTATC CATTTC A GAATTG G GT GTC GACATAG CAGAATA GG C
GTTACTC GACAGAGGAGAG CAAGAAATG GAG CCAGTAGATC CTA GA
CTAGA GC C CT G GAAGCATC C A GGAA GTC A G CC TAAAAC TG CTTGTA
C CAATT G C TATTGTAAAAAGTGTT GC TTTCATTG C CAAGTTT GTTT CA
TGACAAAAG C CTTAGG CAT CT CC TATGG CAG GAAGAAG CG GAGAGA
GC GAC GAAGAG CTC AT CAGAA CAGTC AGAC T CATCAAG C TTCTC TAT
CAAAGCAGTAAG (SEQ ID NO: 430)
pGP2 AT GAAGTGC CTTTTGTACTTAG CC TTTTTATT CATTG GG
GTGAATTGC
AA GTTC A CC ATA GTTTTT C CA CAC AA C CAAAAA G GAAA CTG GAAAAA
TGTTC C TT C TAATTAC C ATTATTGCC C GTCAAG CTCAGATTTAAATTG
GCATAATGACTTAATAGGCACAGC CTTACAAGTCAAAATGC CCAAGA
GT CAC AA GG C TATTCAAGCAGAC GGTTGGATGTGTCATGCTTC CAA
AT GG GTCAC TAC TT GTGATTT C C GCTGGTATGGACC GAAGTATATAA
CAC ATT C CATCC GATC CTTC A C TCC ATCTGTA GAA CAATG C AAG GAA
AG CATTGAACAAAC GAAACAAGGAACTTGGCTGAATCCAGGCTTC C
CT CC TCAAAGTTGT GGATATG CAA CTGTGA C GGATG CCGAAGCAGT
GATTGTCCAGGTGACTCCTCAC CATGT GC TG GTT GATGAATA C A CA
GGAGAATGG GTTGATTCACAGTTCATCAACGGAAAATG CAGCAATTA
CATATG C CC CA CTGTC CATAACTCTAC AA C CTG GCATTCTGACTATA
AG GTCAAAG GG C TATGT GATTC TAAC C TC ATTTC C AT G GAC AT CAC C
TTCTTCTCAGAG GACG GAGAG CTATCATC CC TG G GAAAGGAG G G CA
CAGG GTTCAGAAGTAACTACTTTGCTTATGAAACTGGAGG CAAGG C
CTGCAAAATGCAATAC TG C AA GCATT GG G GAGTCAGACTC C CATC A
GGTGTCTGGTTCGAGATG GC TGATAA GGATCTCTTT G C TG CA GC CA
GATTC C CTGAATGC C CAGAAGGGTCAAGTATC TCTGC TC CATCTCA
GAC CT CA GTG GATGTAAGTCTAATTCAG GACGTTGAGAGGATCTTG
GATTATTC CC TCTGC CAAGAAAC CTG GAG CAAAATCAGA G CG GGTC
TTC CAATC TC TC CAGTG GATCTCAG C TATCTT GC TC CTAAAAAC CCA
GGAACC GGTCC TG C TTT CA C CATAATCAATG GTA CC CTAAAATAC TT
TGAGACCAGATACATCAGAGTC GATATTG C T GC TC CAATC CTCTCAA
GAATG GT CG GAATGATCAGTGGAACTACCACAGAAAG GGAACTGTG
GGATGACTG G G CAC CATATGAAGACGTGGAAATTG GAC C CAATG GA
GTTCTGAGGACCAGTTCAGGATATAAGTTTC CTTTATACATGATTG G
AC ATG GTAT GTTG GAC TC C GATC TTC AT C TTA GC TC AAA GG C TC A GG
TGTTC GAACATC C T CAC ATTC AAGAC GC TGC TTC GCAACTTC C TGAT
343
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Construct DNA
SEQUENCE
GATGAGAGTTTA I I 1 1 1 1GGTGATACTGGGCTATCCAAAAATCCAAT
CGAGCTTGTAGAAGGTTGGTTCAGTAGTTGGAAAAGCTCTATTGCCT
CTTTTTTCTTTATCATAGGGTTAATCATTGGACTATTCTTGGTTCTCC
GAGTTGGTATCCATCTTTGCATTAAATTAAAGCACACCAAGAAAAGA
CAGATTTATACAGACATAGAGATGAACCGACTTGGAAAGTAA (SEQ
ID NO: 431)
Transfection
10011581 The steps for creation of the XDP are depicted graphically in FIG.
24. HEK293T
Lenti-X cells were maintained in 10% FBS supplemented DMEM with HEPES,
penicillin/streptomycin (Pen/Step), sodium pyruvate, and 2-mercaptoethanol.
Cells were seeded
in 10 cm dishes at 8th cells per dish in 10 mL of media. Cells were allowed to
settle and grow
for 24 hours before transfection. At the time of transfection cells were 70-
90% confluent. For
transfection, the following plasmid amounts were used: 19.8 jig of pXDP10,
pXDP11, or
pXDP12.5 pig of pStx42.174.12.7, 3.3 jig of psPax2, and 1 pg of pGP2 in 680 gl
of Opti-MEM
media. 87.5 gl of 1 mg/ml linear polyethylenimine (PEI, MW=25,000 Da) was then
added to the
plasmid mixture, mixed, and allowed to incubate at room temperature before
being added to the
cell culture.
Collection and concentration
10011591Media was changed on cells 24 hours post-transfection. XDP-containing
media was
collected 72 hours post-transfection and filtered through a 0.45 i.tM filter
using a 10 mL syringe.
1 ml of the approximately 8 mL remaining after filtration was stored at 4 C
for titering and
subsequent assays. The remaining filtered supernatant was used directly for
cell editing or was
concentrated by centrifugation at 10,000 x g at 4 C for 4h using a 10% sucrose
buffer in N'TE, as
described below.
Example 14: Purification of XDP
10011601 As described in the various Examples for production of XDP,
production cells were
maintained in DMEM supplemented with 10% fetal bovine system at 37 C in a
humidified 5%
CO2 atmosphere. Cells were plated in 15 cm plates 24 hours before
transfection. Transfections
were carried out using PEI with the appropriate plasmids. The media was
removed and replaced
with Opti-MEM containing 6.25 U/mL of Benzonase 24 hours after transfection.
XDP-
344
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
containing supernatant was collected 72 hours after transfection and filtered
through 0_45 jaM
PES filters before being stored at 4 C until purification.
Centrifugation Protocol
[0011611Filtered supernatant was divided evenly into an appropriate number of
centrifuge tubes
or bottles and 115th of the supernatant volume of Sucrose Buffer (50mM Tris-
HCL, 100mM
NaCl, 10% Sucrose, pH 7.4) was underlaid using serological pipettes. The
samples were
centrifuged at 10,000xg, 4 C, in a swinging-bucket rotor for 4 hours with no
brake. The
supernatant was carefully removed and the pellet briefly dried by inverting
the centrifuge
vessels. Pellets were then resuspended in Storage Buffer (PBS + 113 mM NaC1,
15% Trehalose
dihydrate, pH 8) or an appropriate media by gentle trituration and vortexing.
Column Protocol
10011621Filtered supernatant was purified by anion exchange chromatography
(AEX) using an
FPLC instrument, at 4 C. The AEX column was equilibrated with buffer A, the
supernatant was
applied, and the column was washed with 10 CV of Buffer A (100 mM Tris-HCI, pH
7.5).
Bound material was eluted using a gradient elution from 0% - 100% Buffer B
(100 mM Tris-
HCI, 1M NaCI, pH 7.5) over 40 column volumes. XDP-containing fractions were
pooled and
further purified using a CaptoCore 700 column (Cytiva), equilibrated with
buffer C (100 mlkil
Tris-HC1, 300 mM NaCl, pH 7.5). The XDP-containing flow-through was then
concentrated
using 100 kDa cutoff spin concentrators at room temperature. The resulting
concentrated sample
was diafiltered into Storage buffer, aliquoted, and snap-frozen in liquid
nitrogen before being
stored at -80 C.
Quantification
10011631Samples were quickly thawed at 37 C in a heat bath, vortexed, and
diluted in 2xPBS
supplemented with 0.1% Tween 20. Particle titer and size was evaluated using
the qNano Gold
TRPS system (Izon Science) on an NP150 nanopore.
10011641FIG. 34 shows representative SDS-PAGE and Western blot images of
samples taken
from throughout the centrifugation purification process. Lanes from left to
right Cells: producer
cells, Pre: Supernatant pre-filtration, Post: 0.45 pM filtered supernatant,
Supe: Supernatant
remaining after centrifugation, Pellet: resuspended XDP pellet. Total protein
was visualized with
StainFree technology (BioRad), Western blotting was performed with the
indicated antibodies.
These figures show that XDPs can be purified and concentrated from mammalian
producer cell
supernatant either by centrifugation or by the column chromatography. In FIG.
34, the total
345
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
protein staining shows that certain proteins are concentrated in the
supernatant, that are not over-
represented in the whole cell lysate (Cells lane). The pre, post, and supe
lanes are
indistinguishable, demonstrating that the bulk proteins are not being
concentrated into the XDP
pellet. This is further shown by the change in makeup of the pellet lane,
which has unique bands
consistent with the molecular weight of gag-CasX-HA, VSV-G, and gag. Western
blotting
confirmed these results, showing that, despite the same amount of protein
being loaded in each
lane, the most significant staining is in the pellet lane. The second darkest
staining can be seen in
the input lane, showing that the particles are concentrated by this process.
The lack of staining in
the other lanes indicates that only an insignificant amount of particles are
lost at each step.
10011651On average, this purifications process yields 4.13 x 1012 particles
per liter of filtered
supernatant, at a concentration of 2,48 x 1011 particles per milliliter,
averaging 113 nm in
diameter, as measured by TRPS. The average activity of particles purified in
this way was 4,27 x
107 editing units (EU) per mL, once purified. This works out to 1.42 x 107
EU/L of culture,
which is a feasible yield for production of vectors for therapeutic use.
Example 15: XDP construct, transfection and recovery
10011661 Alternative configuration versions of the CasX delivery particles
(XDPs) named
Versions 1-24 (see Table 17) were designed to contain RNP of four different
CasX variants
proteins; CasX119, CasX438, CasX 457, or CasX 491, complexed with single guide
RNA
variant 174 having spacer sequence 12.7 targeted to tdTomato (encoded by
CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825). The XDP were produced by transient
transfection of LentiX FIEK293T cells (Takara Biosciences) using one or more
structural
plasmids (derived from one or more components of the Gag-Pol
system, a plasmid
encoding a pseudotyping glycoprotein, and a plasmid encoding a single guide
RNA (see FIG.
17, representing Version 1), using the methods described below. Table 17,
grouped by version
number, lists the plasmids (and their sequences) that were used to produce
each version of the
XDP containing the components indicated in the column "Design", and FIG. 24
shows
schematics of the organization of the various plasmids in the versions The
plasmids were
constructed utilizing the methods outlined in Example 13. For the plasmid
encoding the guide
RNA, the pStx42 plasmid was created with a human U6 promoter upstream of a
guide RNA
cassette having scaffold and spacer components targeted to tdTomato in a
single-guide format,
as described in Example 13. Another pStx42 plasmid was utilized to make a
guide RNA cassette
346
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
having scaffold and non-targeting spacer components, used as a control in the
editing assay.
Plasmids encoding VSV-G (pGP2) for pseudotyping the XDP and Gag-Pol (psPax2)
proteins
was also used (representative sequence in Table 16). All plasmids contained
either an ampicillin
or kanamycin resistance gene.
Table 17: Plasmid Encoding Sequences
Versio Design XDP plasmid
DNA SEQUENCE
n& number
CasX
V1 MA-CA-NC-Pl/P6-X pXDP17
(SEQ ID NO: 432)
MA-CA-NC-PI/P6-(-1)- 491 POL
pXDP4 (SEQ ID NO: 433)
V2 MA-CA-NC-P1/P6-X pXDP17
(SEQ ID NO: 434)
491 MA-CA-NC-P1/P6-X-PR pXDP22
(SEQ ID NO: 435)
V3 MA-CA-NC-P1/P6-X pXDP17
(SEQ ID NO: 436)
491 MA-CA-NC-P1/P6-(-1)-
pXDP13
(SEQ ID NO: 783)
X-PR
V4
MA-CA-NC-Pl/P6-(-1)-
pX0P13
(SEQ ID NO: 784)
X-PR
119
V5
MA-CA-NC-P1/P6-X-PR pXDP22
(SEQ ID NO: 785)
491
MA-CA-NC-P1/P6-X-PR pXDP22
(SEQ ID NO: 786)
V6
MA-CA-NC-P1/P6-X pXDP17
(SEQ ID NO:787)
491
MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 788)
V7
MA-CA-NC-P1/P6-X pXDP17
SEQ ID NO: 789)
491
V8 MA-CA-NC-P1/P6-X pXDP17
(SEQ ID NO: 790)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 791)
V9
MA-CA-NC-X (no p1/p6) pXDP0023
(SEQ ID NO: 792)
491
V10
MA-CA-NC-P1-X pXDP0024
(SEQ ID NO: 793)
491
V11 MA-CA-NC-X-(-1)-PR pXDP0025
(SEQ ID NO: 794)
347
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Versio Design XDP plasm id
DNA SEQUENCE
n& number
CasX
491
V12
MA-CA-X-(-1)-PR
pXDP0026 (SEQ ID NO: 795)
491
V13
MA-X-NC-(-1)-PR
pXDP0027 (SEQ ID NO: 796)
491
V14
MA-X-(-1)-PR
pXDP0028 (SEQ ID NO: 797)
491
V15
MA-X-PR
pXDP0029 (SEQ ID NO: 798)
491
V16
MA-CA-X-PR
pXDP0030 (SEQ ID NO: 799)
491
V17
MA-X
pXDP0031 (SEQ ID NO: 800)
491
V18
MA-CA-X
pXDP0032 (SEQ ID NO: 801)
491
V19
MA-X-X-(-1)-PR
pXDP0033 (SEQ ID NO: 802)
491
V20
MA-CA-X-X-(-1)-PR
pXDP0034 (SEQ ID NO: 803)
491
V21
MA-CA-NC-X-X-(-1)-PR pXDP0035 (SEQ ID NO: 804)
491
V22 Gag-1%TCS-STx-HA
pXDP0036 (SEQ ID NO: 805)
491 Gag-PCS-HRV3c
pXDP0039 (SEQ ID NO: 806)
V23 Gag-TCS-STx-HA
pXDP0037 (SEQ ID NO: 807)
491 Gag-PCS-HRV3c
pXDP0039 (SEQ ID NO: 808)
V24 Gag-PCS-STx-HA
pXDP0038 (SEQ ID NO: 809)
491 Gag-PCS-IIRV3c
pXDP0039 (SEQ ID NO: 810)
348
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Transfection
10011671 The steps for creation of the XDP are depicted graphically in FIG.
24. HEK293T
Lenti-X cells were maintained in 10% FBS supplemented DMEM with HEPES,
penicillin/streptomycin (Pen/Step), sodium pyruvate, and 2-mercaptoethanol.
Cells were seeded
in 10 cm dishes at 8e6 cells per dish in 10 mL of media. Cells were allowed to
settle and grow
for 24 hours before transfection. At the time of transfection cells were 70-
90% confluent. For
transfection, the plasmids of Table 17, together with the 5 tug of the guide
plasmid, and 0.1 Kg of
pMD2.G in 680 Eil of Opti-MEM media. 87.5 gl of 1 mg/ml linear
polyethylenimine (PEI,
MW=40,000 Da) was then added to the plasmid mixture, mixed, and allowed to
incubate at
room temperature before being added to the cell culture.
Collection and concentration
10011681 Media was changed on cells 24 hours post-transfection. XDP-containing
media was
collected 72 hours post-transfection and filtered through a 0.45 KM filter
using a 10 mL syringe.
1 ml of the approximately 8 mL remaining after filtration was stored at 4 C
for titering and
subsequent assays. The remaining filtered supernatant was used directly for
cell editing or was
concentrated by centrifugation at 10,000 x g at 4 C for 4h using a 10% sucrose
buffer in NTE.
Example 16: Editing of tdTomato neural progenitor cells using XDP
10011691tdTomato neural progenitor cells (tdT NPCs) were grown in DMEM F12
supplemented
with glutamax, HEPES, non-essential amino acids, Pen/Strep, 2-mercaptoethanol,
B-27 without
vitamin A, and N2. Cells were harvested using a Takara Biosciences Neuron
Dissociation Kit
and seeded on PLF coated 96 well plates. Cells were allowed to grow at 37 C
for 48 hours
before being treated with targeting XDPs (having spacer 12.7 for tdTomato) and
non-targeting
XDPs (having a non-targeting spacer) as a 10x concentrate from the sucrose
buffer concentrates
using half-log dilutions, as well as a Opti-MEM negative control. NPCs were
grown for 96 hours
before analysis of fluorescence as a marker of editing of tdTomato.
[0011701Results: The results of the editing assay are shown in FIG, 25 and in
Table 18, below,
FIG. 25 shows results of a single experiment (Targeting XLP is XDP CasX119
with VSV-g;
Bald VLP is XDP CasX119 with no GP; and Negative Control is Buffer Control as
labelled in
Table 18, while the table represents the mean results of 3 experiments showing
20% editing of
the dtTomato target sequence was achieved with the XDP comprising the CasX 119
construct.
349
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Table 18: Results of Editing Assay
Group % TDT+ cells (% editing)
VSV-G XDP 20%
CasX119
No Glycoprotein <0.5%
XDP CasX119
Buffer control <0.5%
Example 17: Construction of XDP with incorporated glycoproteins to evaluate
tropism and
editing capabilities
10011711 Viral vectors including lentiviral and retroviral vectors are most
often pseudotyped
with the envelope protein of vesicular stomatitis virus (VSV-G), a
glycoprotein that endows both
a broad host cell range and high vector particle stability. Experiments were
performed in which
XDPs with incorporated RNP of CasX and gNA specific for editing tdTomato in
mouse neural
progenitor cells (tdT NPCs) were created with varying concentrations of
incorporated VSV-G to
determine the corresponding effects on editing in tdT NPCs via the enhanced
delivery of the
editing moiety by the VSV-G.
10011721Experiments shown in FIGS. 26-28 follow the XDP production methods
(for the CasX
119 and single guide RNA 174 with spacer sequence 12.7 targeted to tdT) and,
where applicable,
testing procedures detailed in Examples 13 and 15. Sequences are shown in
Table 19. For the
experiments resulting in the data in FIGS. 26A and 26B, the effects of varying
concentrations of
the pseudotyping (VSV-G) plasmid incorporated into the XDP were evaluated as
follows: 1 fig
of the VSV-G plasmid was used for the 100% VSV-G group, 0.3 mg was used for
the 30% VSV-
G group, 0.1 pig was used for the 10% VSV-G group, 0.03 lig was used for the
3% VSV-G
group, 0.01 gg was used for the 1% VSV-G group, and 0.003 gg was used for the
0.3% VSV-G
group. Titering of the XDPs produced was done using the Takara p24 rapid titer
kit. Editing was
assessed in the tdTomato NPC cells as detailed in Example 16.
10011731 The results for the 10% and 30% VSV-G groups trend towards a better
editing
outcome as compared to the 100% VSV-G group, as shown in FIG. 26A, without
affecting viral
titer or stability, as shown in FIG. 2613
10011741 As the results indicate that one can achieve, under the experimental
conditions, the
same, if not higher editing with 10-30% VSV-G compared to the 100% VSV-G
group, this
350
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
opens up the possibility of pseudotyping the XDP particle with other encoded
glycoproteins,
either with or without VSV-G, to confer differential or enhanced cellular
tropism to the resulting
XDP, including the viral glycoproteins disclosed herein, examples of which
were produced and
evaluated as follows. Utilizing the XDP production and editing methods of
Example 13 and 15,
each XDP transfection used 3.3 pg (0.467 pM) of psPax2 plasmid, 19.8 lig (3.24
pM) of
pStx43.119 plasmid, 5 pg (3.13 pM) of pStx42 plasmid (with guide 174)
targeting the tdTomato
locus using spacer 12.7 and 0.262 pM of the respective g,lycoprotein(s)
plasmid which varied in
molecular weight. Glycoprotein plasmids contained the same backbone pGP2 and
only varied by
expressing different viral envelope proteins which they expressed. The
following plasmids were
used for transfections: rabies used 0.94 pg of pGP29; FUG E used 0.95 pg of
pGP60; HSV-1
used 028 pg of pGP14.1, 022 pg of pGP14.2, 0.27 pg of pGP14.3, and 020 pg of
pGP14.4;
RD114 used 0.96 jig of pGP8; HCV used 0.97 ug of pGP23; EBOV used 1.02 pg of
pGP41;
Mokola used 1.02 jig of pGP30. Canonical HSV-1 pseudotyping requires four
glycoproteins
which were used in equimolar amounts in this assay (Polpitiya Arachchige, S.,
Henke, W.,
Kalamvoki, M. et al. Analysis of herpes simplex type 1 gB, gD, and gH/gL on
production of
infectious HIV-1: HSV-1 gD restricts HIV-1 by exclusion of HIV-1 Env from
maturing viral
particles. Retrovirology 16:9 (2019)). Glycoprotein amino acid sequences come
from wild type
viral sequences. Nucleic acid sequences also came from wild type viral
sequences though some
were codon optimized for synthesis and expression in human cell lines.
10011751The editing efficiencies in mouse tdTomato NPCs were tested with an
initial panel of
pseudotyped XDPs having glycoproteins from VSV-G, rabies, FUG E, HSV-1, RD114,
hepatitis
C virus (HCV), and Ebola virus (EBOV), produced as described above. The
results are shown in
FIG. 27. While constructs with FUG E, Mokola and herpes simplex virus-1 (HSV-
1)
incorporated glycoproteins were expected to achieve some level of cell entry
in NPCs, rabies
was the only glycoprotein other than VSV-G resulting in an observable level of
editing under the
conditions of the assay, which is a readout for cell entry into mouse neural
progenitor cells.
Conversely, XDPs pseudotyped with HCV, EBOV and RD114 did not achieve any
editing in
mouse NPCs, which indicates the potential cell specificity requirements for
this cell type.
10011761We also assessed whether pseudotyping with different viral
glycoproteins could have
an impact on overall size distributions, which could have an impact on in vivo
editing
efficiencies in different tissues of interest. For this experiment, the rabies
pseudotyped XDP 10X
and VSV-G pseudotyped XDP lx were produced using the protocol described above
scaled to a
351
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
6 well format and using pGP29 in place of the pGP2 plasmid. All plasmid
quantities and cells
used were scaled down 8-fold. The VSV-G pseudotyped XDP lx were generated as
described
above. These preparations were then concentrated at 20,000 x g at 4 C for 90
minutes without a
sucrose buffer. LV was transfected with the following plasmid weights: 5.4 jig
of psPax2, 1.8 itg
of pGP2, and 7.2 mg of pStx34.119.174.12.7, generating lentivirus designed to
induce production
and incorporation of RNP with the same enzymatic capabilities as VSV-G
pseudotyped XDP
1X. Samples were diluted appropriately for analysis. The size and number of
particles were
assessed using a Tunable Resistive Pulse Sensor (Izon Biosciences qNano Gold).
While both
rabies and VSV-G XDPs ranged in size from 75-140 nm, lentiviruses (LVs) tend
to be a bit
larger, ranging in size from 85-160 nm, as shown in FIG. 28A. FIG. 28B shows
that rabies
pseudotyped XDPs trend towards a smaller mode as compared to VSV-G pseudotyped
XDPs.
Table 19. Plasmid encoding sequences for glycoproteins.
Glycoprotein
SEQUENCE
pGP2 (VSV-G)
(SEQ ID NO: 811)
pGP29 (Rabies)
(SEQ ID NO: 812)
pGP60 (FUG E)
(SEQ ID NO: 813)
pGP14.1 (HSV-1 gB)
(SEQ ID NO: 814)
pGP14.2 (HSV-1 gD)
(SEQ ID NO: 815)
pGP14.3 (HSV-1 gH)
(SEQ ID NO: 816)
pGP14.4 (HSV-1 gL)
(SEQ ID NO: 817)
pGP8 (RD114)
(SEQ ID NO: 818)
pGP23 (HCV)
(SEQ ID NO: 819)
pGP41 (EBOV)
(SEQ ID NO: 820)
pGP30 (Mokola)
(SEQ ID NO: 821)
352
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Example 18: Construction and evaluation of XDP with RNP comprising CasX with
enhanced editing capabilities
10011771ln addition to improving the targeting capability and specificity
within the XDP
platform, the ability to concurrently improve the editing capability of XDPs
incorporating
improved RNP variants having CasX 438 and CasX 457 (compared to CasX 119) was
examined
(with guide 174 and spacer 12.7). The RNP variants were constructed by
exchanging the CasX
encoding sequences within the pStx43 plasmid. RNP 457 was transfected using
19_8 jig of
pStx43.119, RNP 438 was transfected using 19.8 pig of pStx43.438, and RNP 119
was
transfected using 19.8 jig of pStx43.119 (sequences in Table 20). Percent
editing in mouse NPCs
was assessed using the tdTomato assay described above and read-out was
performed using an
Attune NxT Flow Cytometer. Titers were assessed using a Takara p24 Rapid Titer
Kit. The
results, shown in FIG. 29, demonstrate enhanced editing of the tdTomato NPCs
by the XDP with
RNP comprising the CasX 438 and CasX 457 compared to RNP comprising CasX 119.
Example 19: Construction and evaluation of XDP with non-essential lentiviral
components
removed
10011781 The ability to improve XDP editing by optimizing RNP packaging into
the viral
vectors was evaluated by stripping away non-essential components such as the
viral genome
(Gag-Pol) from the Gag-CasX construct. Moreover, the removal of these
components would
alleviate some of the safety concerns with these platforms by taking away the
reverse
transcriptase (RT), integrase (IN) components that have been a source of
concern for their use in
humans. Furthermore, it offers the possibility of increased packaging of the
RNP complex into
an XDP molecule, as every Gag molecule packaged would have a CasX molecule
attached to it.
10011791 The XDP were created using the same approach as described above
(i.e., 8 x 106
LentiX cells were plated in a 10 cm dish, 24 hours later cells were
transfected with DNA, media
was changed 16 hours after transfeetion, XDPs were collected 72 hours post-
transfection and
concentrated). Here, we introduced a new plasmid having the components Gag,
CasX, and
protease, referred to as Gag-CasX-PR (or pMRG103; sequence in Table 20). This
plasmid
contains a gag polyprotein followed by a CasX molecule linked by a SQNYPIVQ
(SEQ ID NO:
20)11IV-1 cleavage site. The CasX molecule is followed by an HA tag and
another SQNYPIVQ
(SEQ ID NO: 20) HIV-1 cleavage site linked to a component of the Pol protein
from HIV-1.
This component contains the H1V-1 protease (PR) and lacks the HIV-1 reverse
transcriptase
353
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
(RT), p15, and integrase (INT) components. Upon budding of the XDP from the
cell membrane,
the protease functions identically to the protease found in the native Gag-Pot
complex; it
dimerizes and facilitates cleavage of the SQNYPIVQ (SEQ ID NO: 20) HIV-1
cleavage sites,
freeing CasX from Gag and PR To generate XDPs with this new construct, the
following
plasmid amounts were used: 0.3 pug of pGP2, 5 pg of pStx42 (guide 174) with
spacer 12.7, and
19.8 pig of pStx43.119 (CasX 119). Additional constructs used the following
plasmid amounts:
100% Gag-Pol used 3.3 mg of psPax2; the 50% Gag-Pol + 50% Gag-CasX construct
used 1.65
1.tg of psPax2 and 1.48 pg of Gag-CasX-PR; the 30% Gag-Pot + 70% Gag-CasX
construct used
0.99 pg of psPax2 and 1.47 pg of Gag-CasX-PR; the 15% Gag-Pol + 85% Gag-CasX
construct
used 0.50 pg of psPax2 and 2.51 tug of Gag-CasX-PR, and the 100% Gag-CasX
construct used
3.00 lug of Gag-CasX-PR. Sequences are provided in Table 20.
10011801Editing of tdTomato NPCs was assessed as described above, and the
titer of the XDP
preparations was assessed using the Takara p24 Rapid Titer Kit. The results,
shown in FIG. 30,
demonstrated that XDP created with Gag-CasX-PR and no inclusion of Gag-Pol
were able to
achieve the same amount of editing at ¨106 particles as compared to ¨108
particles with XDPs
that have 100% Gag-Pd. The other constructs showed editing in proportion to
the titer of the
particles. The titer data for the various constructs that were produced is
shown in FIG. 31. We
believe that this observed enhancement in editing efficiency is due to
enhanced packaging of
RNP molecules per XDP, as shown by guide RNA quantification for the different
XDP
constructs as depicted in FIG. 32.
Table 20: Plasmid encoding sequences
Construct Sequence
pMRG103 (Gag-CasX119-
PR) (SEQ ID NO:
822)
pM1RG103 (Gag-CasX438-
PR) (SEQ ID NO:
823)
Example 20: Construction and evaluation of XDP targeting human cells
10011811 The tdTomato mouse neural progenitor cell model is a powerful tool to
assess the
potency of XDPs. However, in view of the intended clinical application of XDP,
the potency of
354
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
these particles must be assessed on human cells using easily accessible,
quantifiable and
therapeutically relevant cell lines. As the human HLA locus for MHC I beta 2
microglobulin
(B2M) fits these criteria, XDP were generated using the methodology described
in Examples 13
and 15 above, with RNP comprising CasX 119 and gNA 174 with spacer sequences
targeting
B2M for assessment in Jurkat cells, a human T- cell line. The spacers 7.9
(GTGTAGTACAAGAGATAGAA, SEQ ID NO: 824) and 7.37
(GGCCGAGATGTCTCGCTCCG, SEQ ID NO: 826) target the human B2M locus and spacer
12.7 (CTG-CATTCTAGTTGTGGTTT, SEQ ID NO: 825), which targets the artificial
tdTomato
locus in mice, was used as the non-targeting spacer. Jurkat cells were seeded
in a 96 well plate in
RPM1 media supplemented with 10% FBS, sodium pyruvate, and GlutalvIAX. XDPs,
resuspended in Opti-MEM, were diluted in half-log serial dilutions in RPMI
media before being
put on Jurkat cells and were spinfected at 1000 x g for 15 minutes. Cells were
incubated at 37 C
for 120 hours before analysis. To stain HLA, we used DAPI to mark dead cells,
and the PE-Cy7
Mouse Anti-Human HLA-ABC staining kit (BD Pharmingen) was used to stain major
histocompatibility complex, class I. Expression of this complex at the cell
surface is blocked by
B2M knockout.
Results:
10011821The results, shown in FIG. 33, depicts the relative HLA negative
(edited) populations
in Jurkat cells, after being treated with XDPs containing CasX molecules with
spacer 7.9, spacer
7.37, or a non-targeting spacer. The results indicate that under the
experimental conditions, the
XDPs with spacer 7.9 are capable of knocking out B2M in ¨10% of Jurkat cells.
Example 21: The generation and assessment of potency of HIV-1 XDPs with
alternative
structures of HIV-1 Gag in various configurations.
10011831The purpose of these experiments was to make various configurations of
XDP
constructs comprising CasX and guide RNA as RNP to demonstrate their utility
in the editing of
eukaryotic cells; either by in vitro or by in vivo delivery. To generate the
most efficient and
minimal HRT-1 capsid designed specifically for RNP delivery, we created thirty-
five different
versions of HINT-1 based XDPs with CasX 491 and guide RNA 174 and spacer 12.7
to tdTomato
to 1) determine which components of HIV-1 were and were not necessary for the
successful
delivery of RNP to cells capable of editing target nucleic acid; and 2)
demonstrate that multiple
355
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
configurations of XDP were able to successfully delivery RNP to cells and edit
target nucleic
acid. Methods
Method for the generation of XDPs
10011841 Alternative configuration versions of the XDPs, referred to as
versions 1, 4, 5, 7-27,
32-40, and 122-124, 126 and 128 (see FIGS. 36-68), were designed to contain
RNP of CasX
491 complexed with a single guide RNA variant having spacer sequence 12.7
targeted to
tdTomato (encoded by CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825). Utilizing methods
described in the sections below, the XDP versions were produced by transient
transfection of
LentiX HEK293T cells (Takara Biosciences) with one or more structural plasmids
encoding
components of the gag-pol HIV-1 system, a plasmid encoding a pseudotyping
glycoprotein, and
a plasmid encoding a single guide RNA (see FIGS. 36-68 for schematics of each
version, the
plasmids employed and the components the plasmid encoded)). Table 21, grouped
by version
number, lists the plasmids and their sequences that were used to produce each
version of the
XDP containing the components indicated in the Table and the corresponding
version of the
Figures. For the plasmid encoding the guide RNA, the pStx42 plasmid was
created with a human
U6 promoter upstream of a guide RNA cassette having scaffold and spacer
components targeted
to tdTomato in a single-guide format (p42.174.12.7). Another pStx42 plasmid
was utilized to
make a guide RNA cassette having scaffold and non-targeting spacer components
(Stx42.174.NT), used as a control in the editing assays. A plasmid encoding
VSV-G (pGP2) for
pseudotyping the XDP was also used (Table 22). All plasmids contained either
an ampicillin or
kanamycin resistance gene.
Structural plasmid cloning
10011851 In order to generate pXDP3, pXDP17, pXDP23-32, pXDP98-100, pXDP102
and
pXDP103, pXDP1 (UC Berkeley) was digested using EcoRI to remove the gag-pol
sequence.
Between one and three fragments containing CasX and HIV-1 components were
amplified using
In Fusion primers with 15-20 base pair overlaps and Kapa HiFi DNA polymerase
according to
the manufacturer's protocols. The fragments were purified by gel extraction
from a 1% agarose
gel using Zymoclean Gel DNA Recovery Kit according to the manufacturer's
protocol. These
fragments were cloned into plasmid backbones using In-Fusion UD Cloning Kit
from Takara
(Cat# 639650) according to the manufacturer's protocols. Assembled products
were transformed
into chemically-competent Turbo Competent E. tole bacterial cells, plated on
LB-Agar plates
(LB: Teknova Cat# L9315, Agar: Quartzy Cat# 214510) containing ampicillin and
incubated at
356
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
37 C. Individual colonies were picked and miniprepped using Qiagen spin
Miniprep Kit
following the manufacturer's protocol. The resultant plasmids were sequenced
using Sanger
sequencing to ensure correct assembly. The encoding sequences are presented in
Table 23. The
first column of the table describes the version number and CasX molecule
included. The second
is the configuration of the HIV components and CasX molecules. The plasmid
number for those
design plasmids are in the third column. The fourth column contains SEQ IDS
for only the
encoding sequences for HIV-1 gag, HIV-1 pol, and CasX molecules, as
applicable.
Guide plasmid cloning
10011861The p42.174.NT (NT sequence CGAGACGTAATTACGTCTCG, SEQ ID NO: 827)
plasmid encoding the guide RNA 174 and the non-targeting spacer and the
p42.174.12.7
targeting tdTomato were cloned using standard cloning methods. The mammalian
expression
backbone contained a cPPT, ampicillin resistance, and a colEI replication site
and was amplified
using primers with appropriate overlaps to accept the U6 promoter and guide
RNA scaffold
cassette. These fragments were amplified using Kapa HiFi DNA polymerase
according to the
manufacturer's protocols and primers appropriate for In-Fusion cloning. The
fragments were
purified by gel extraction from a 1% agarose gel using Zymoclean Gel DNA
Recovery Kit
according to the manufacturer's protocol. These fragments were cloned into
plasmid backbones
using In-Fusion HD Cloning Kit from Tak.ara (Cat 639650) according to
manufacturer
protocols. Assembled products were transformed into chemically-competent Turbo
Competent
E. coil bacterial cells, plated on LB-Agar plates (LB: Teknova Cat# L9315,
Agar: Quartzy Cat's/
214510) containing ampicillin and incubated at 37 C. Individual colonies were
picked and
miniprepped using Qiagen spin Miniprep Kit following the manufacturer's
protocol. The
resultant plasmids were sequenced using Sanger sequencing to ensure correct
assembly.
Cloning tdTomato spacer 12.7 into p42.174.NT
10011871 The targeting spacer sequence DNA for the tdTomato targeting spacer
12.7 was
ordered as single-stranded DNA (ssDNA) oligos (Integrated DNA Technologies)
consisting of
the targeting sequence (CTGCATTCTAGTTGTGGITT, SEQ ID NO: 825) and the reverse
complement of this sequence. These two oligos were annealed together and
cloned into p42.174
NT or a p42 plasmids with an alternate scaffold. This was done by Golden Gate
assembly using
T4 DNA Ligase (New England BioLabs Cat# M0202L) and Esp3I restriction enzyme
from NEB
(New England BioLabs Cat# R0734L), Golden Gate products were transformed into
chemically
competent NEB Turbo competent E. call (NEB Cat #C2984I), plated on LB-Agar
plates (LB:
357
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Teknova Cat #L9315, Agar: Quartzy Cat# 214510) containing carbenicillin and
incubated at
37 C. Individual colonies were picked and miniprepped using Qiagen spin
Miniprep Kit
following the manufacturer's protocol. The resultant plasmids were sequenced
using Sanger
sequencing to ensure correct ligation.
pGP2 Glycoprotein plasmid cloning
10011881 Sequences encoding the VSV-G glycoprotein and the CMV promoter were
amplified
from pMD2.G (UC Berkeley) using Kapa HiFi DNA polymerase according to the
manufacturer's protocols and primers appropriate for In-Fusion cloning. The
backbone was
taken from a kanamycin resistant plasmid and amplified using the same methods.
These were
purified by gel extraction from a 1% agarose gel using Zymoclean Gel DNA
Recovery Kit
according to the manufacturer's protocol. These fragments were cloned into
plasmid backbones
using In-Fusion HD Cloning Kit from Takara (Cat 639650) according to
manufacturer
protocols. Assembled products were transformed into chemically-competent Turbo
Competent
E coil bacterial cells, plated on LB-Agar plates containing kanarnycin and
incubated at 37t.
Individual colonies were picked and miniprepped using Qiagen spin Miniprep Kit
following the
manufacturer's protocol. The resultant plasmids were sequenced using Sanger
sequencing to
ensure correct assembly.
Cell culture and transfection
10011891HEK293T Lenti-X cells were maintained in 10% FBS supplemented DMEM
with
HEPES and Glutamax (Thermo Fisher). Cells were seeded in 15 cm dishes at 20 x
106 cells per
dish in 20 mL of media. Cells were allowed to settle and grow for 24 hours
before transfection.
At the time of transfection, cells were 70-90% confluent. For transfection,
the XDP structural
plasmids (also encoding the CasX variants) of Table 21 were used in amounts
ranging from 13
to 80.0 pig. Each transfection also received 13 pig of p42.174.12.7 and 0.25
rig of pGP2.
Polyethylenimine (PEI Max, Polyplus) was then added to the plasmid mixture,
mixed, and
allowed to incubate at room temperature before being added to the cell
culture.
Collection and concentration
10011901 Media was aspirated from the plates 24 hours post-transfection and
replaced with Opti-
MEM (Thermo Fisher). XDP-containing media was collected 72 hours post-
transfection and
filtered through a 0.45 pm PES filter. The supernatant was concentrated and
purified via
centrifugation at 10,000 x g at 4 C for 4h using a 10% sucrose buffer in NTE
(50mM Tris-HCL,
100mM NaCl, 10% Sucrose, pH 7.4). XDPs were resuspended in 300 Ed, of DMEM/
F12
358
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
supplemented with glutamax, HEPES, non-essential amino acids, Pen/Strep, 2-
mercaptoethanol,
B-27 without vitamin A, and N2.
Resuspension and transduction
[0011911tdTomato neural progenitor cells (tdT NPCs) were grown in DMEM/ F12
supplemented with glutamax, HEPES, non-essential amino acids, Pen/Strep, 2-
mercaptoethanol,
B-27 without
vitamin A, and N2. Cells were harvested using StemPro Accutase Cell
Dissociation Reagent and
seeded on PLF coated 96 well plates. Cells were allowed to grow for 48 hours
before being
treated for targeting XDPs (having a spacer for tdTomato) starting with neat
resuspended virus
and proceeding through 5 half-log dilutions. Cells were then centrifuged for
15 minutes at 1000
g. NPCs were grown for 96 hours before analysis of fluorescence as a marker of
editing of
tdTomato. The assays were run 2-3 times for each sample with similar results.
Editing results for
a single assay are shown in Table 21.
Results
10011921 The editing results confirmed that, under the conditions of the
assay, the majority of
the 35 alternative configurations were able to edit the NPCs with at least 10%
or greater editing,
with 7 versions showing >80% editing. Additionally, it was confirmed that some
of the HIV
structural components of Gag were dispensable, with editing seen in one
configuration in which
only the matrix (MA) component was linked to the CasX. The pl/p6 component,
which
promotes budding from the host cell, was associated in all versions with high
levels of editing
(>= 70%, VI, V7, V8, V33, V34, V40, V123, V124) suggesting that this component
is important
for potency. Particles without NC, such as versions 34, 40 and 123, were able
to achieve high
levels of editing whereas particles without CA (such as version 17) had lower
levels of editing
(37%). The results also demonstrated that the protease component is not
necessary for the XDP
to retain high levels of editing potency, as demonstrated by versions 7, 8,40,
123, and 124.
Furthermore, p2, a component of NC, was also detrimental to potency as seen
when comparing
versions 122 and 128 on table X_X where 122 (MA-CA-pl/p6) has no p2 and
achieves 444%
editing and versions 128 (MA-CA-p2-pl/p6) includes p2 and archives only 29.2%
editing. In
addition, constructs with multiple p 1 /p6 may contribute to enhance editing,
as seen in FIG. 35
(version 122 versus 123), however, this did not prove to be the case for other
configurations;
e.g., version 7 (MA-CA-NC-pl/p6-X) versus version 124 (MA-CA-NC-pl/p6-pl/p6),
where
version 7 achieved 92.2% editing and version 124 achieved only 72.3% editing.
359
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10011931Overall, the results support that, under the conditions of the assays,
multiple
configurations of XDP are able to successfully assemble particles able to
deliver the CasX and
guide RNA therapeutic payloads to eukaryotic cells, resulting in editing of
the target nucleic
acid.
Table 21: Editing of NPCs by XDP constructs, by version configuration.
XDP Structural Structural Structural Structural
Version Plasmid 1 Plasmid 2 Plasmid 1 plasmid 2
CasX % Editing
1 Gag-stx gag-pol pXDP17
pXDP1 491 95.4
gag(-1)
gag(-1)-PR-
4 Stx pXDP88
491 2.78
7 Gag-Stx pXDP17
491 92.2
8 Gag-Stx gag pXDP17
pXDP3 491 87.5
Ma-CA-NC-
9 X pXDP23
491 14.20
MA-CA-
NC-P1-X pXDP24 491
7.19
MA-CA-
NC-X(-1)-
11 Pro pXDP25
491 35.8
MA-CA-X(-
12 1)-Pro pXDP26
491 28.5
MA-Stx-
13 NC-(-1)-PR pXDP27
491 17.3
MA-X-(-1)-
14 Pro pXDP28
491 32.6
MA-X-Pro pXDP29 491
0
MA-CA-
16 Stx-Pro OCDP30
491 1.86
17 MA-X pXDP31
491 37.9
18 MA-CA-X pXDP32
491 18.3
31 MA-CA- Gag pXDP23 pXDP3 491 17.0
360
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
XDP Structural Structural Structural Structural
Version Plasmid 1 Plasmid 2 Plasmid 1 plasmid 2
CasX % Editing
NC-X
MA-CA-
32 NC-P1-X Gag pXDP24 pXDP3 491 13.40
MA-CA-
NC-X(-1)-
33 PR Gag pXDP25 pXDP3 491 90.1
MA-CA-X(-
34 1)-PR Gag pXDP26 pXDP3 491 95.3
MA-X-NC-
pl/p6-(-1)-
35 Pro Gag pXDP27
pXDP3 491 11.6
MA-X-(-1)-
36 Pro Gag pXDP28
pXDP3 491 25.10
MA-CA-
38 STx-Pro Gag pXDP30 pXDP3 491 8.5
39 MA-X Gag pXDP31 pXDP3 491 30.7
40 MA-CA-X Gag pXDP32 pXDP3 491 843
MA-CA-
122 p1/p6-X pXDP98
491 44.4
MA-CA-
p 1 /p6-pl/p6-
123 X pXDP99
491 91.5
MA-CA-
NC-p1/136-
124 p1/p6-X pXDP100
491 73.2
MA-CA-
126 NC-X-pl/p6 pXDP102
491 44.8
MA-CA-p2-
128 pl/p6-x pXDP104
491 29.2
*% Editing was calculated by taking the maximum editing percentage of the 5
dilutions'
averaged replicates.
361
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Table 22: Encoding sequences for guides and glycoproteins
Plasmid DNA
sequence
p42.174,12.7 A CTG G C G CTTTTATC TgATTA CTTT G A GAG C CATC AC CAC C GA CT
ATGTC GTAgTG GGTAAAG CT C C CTC TTC G GAG G GAG CAT CAAAG
CTGCATTCTAGTTGTGGTTT (SEQ ID NO: 828)
A C TG G C GC TTTTATC TgATTA C TTT G A GAG C C ATC AC C AG C GA C T
ATGTC GTAgTG GGTAAAG CTC CCTCTTC G GAG G GAG CATCAAAG
p42.174,NT CGAGACGTAATTACGTCTCG (SEQ ID NO: 829)
pGP2 ATG AA GTG CCTTTTGTACTTAGCCTTTTTATTCATTG G G
GT GAATT
GCAAGTTCAC CATAGTTTTTC CACACAAC CAAAAAG GAAAC TG GA
AAAATGTTCCTTCTAATTAC CATTATTG C CC GTC AAG CTCAGATTT
AAATTGGCATAATGACTTAATAGG C AC AG C CTTACAAGTCAAAAT
GCCCAAGAGTCACAAGGCTATTCAAGCAGACG GTTGGATGTGTC
ATG CTTCCAAATG G GTCA CTA C TTGTG A TTT C C G CTGGTATGGAC
C GAAGTATATAACACATTC CATCC GATCCTTCACTC CATCTGTAG
AAC AATGC AAG GAAAG CATTG AA CAAAC GAAACAAGGAACTTG G
C TG AATCC AG GCTTCCCTCCTCAAAGTTGTGGATATG CAA CT GTG
ACG GATGCCGAAGCAGTGATTGTCCAGGTGACTCCTCACCATGT
GCTGGTTGATGAATACACAGGAGAATGGGTTGATTCACAGTTCAT
CAACG GAAAATG GAG CAATTACATATGC CC C AC TG TC CATAACTC
TACAACCTGG CATTCTGACTATAAGGTCAAAG GGCTATGTGATTC
TAAC CTCATTTC CATG GACATC AC CTTCTTCTCAGAGGAC G GAGA
GCTATCATCCCTG G GAAAG GAG GGCACAGGGTTCAGAAGTAACT
A CTTTG CTTATGAAACTG GA G G CAAG GCCTG CAAAATG CAATA CT
GCAAGCATTG GGGAGTCAGACTC C CATC AG GTGTCTGGTTC GAG
ATG G CT GATAA G GATCTCTTTGCTG CAG CCAGATTC C CTG AATG C
C CAGAAGGGTCAAGTATCTCTGCTCCATCTCAGACCTCAGTGGA
TGTAAGTCTAATTCAG GACGTTGAGAGGATCTTGGATTATTC C CT
CTG C CAA GAAAC CT GGAGC AAAATCAGAG C GGGTCTTCCAATCT
CTC C AG TG GATCTCAGCTATCTTGCTCCTAAAAAC C C AG GAAC C G
GTC CTG CTTTCACCATAATCAATG GTACC CTAAAATACTTT GAGA
C CAGATACATCAGAGTC GATATTGCTGCTC CAATC CTCTCAAGAA
TGGTC GGAATGATCAGTGGAACTAC CA CAGAAAG GGAACTGTGG
GATGACTG G G CAC CATATG AA GAC GTG G AAATTG GACCCAATG G
AGTTCTGAGGAC CAGTTCAGGATATAAGTTTC CTTTATACATGATT
GGACATGGTATGTTGGACTCCGATCTTCATCTTAGCTCAAAG G CT
C A G GTGTTCGAACATCCTCACATTCAAGACG CT G CTTC G CAAC TT
CCTGATGATGAGAGTTTA i 1111 i GGTGATACTGGGCTATCCAAA
AATCCAATCGAGCTTGTAGAAGGTTGGTTCAGTAGTTGGAAAAG C
TCTATTGC CTCTTTTTTCTTTATCATAGGGTTAATCATTGGACTATT
CTTGGTTCTCCGAGTTGGTATCCATCTTTGCATTAAATTAAAGCAC
AC CAAGAAAAGACAGATTTATACAGACATAGAGATGAAC C GACTT
GGAAAGTAA (SEQ ID NO: 830)
Table 23: XDP Versions and Component Encoding Sequences
362
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Versions & XDP
plasmid
Design
DNA Sequence
CasX
number
V1 MA-CA-NC-P1/P6-X pXDP17
(SEQ ID NO: 831)
491 MA-CA-NC-P1/P6-(-1)-POL pXDP1
(SEQ ID NO: 832)
V4 MA-CA-NC-P1/P6-(-1)-X-PR pXDP88
(SEQ ID NO: 833)
491
VS MA-CA-NC-P1/P6-X-PR pXDP22
(SEQ ID NO: 834)
491
V7 MA-CA-NC-P1/P6-X pXDP17
(SEQ ID NO: 835)
491
V8 MA-CA-NC-P1/P6-X pXDP17
(SEQ ID NO: 836)
MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 837)
491
j V9 MA-CA-NC-X (no p1/p6) pXDP23
(SEQ ID NO: 838)
491
V10 MA-CA-NC-P 1-X pXDP24
(SEQ ID NO: 839)
491
j V11 MA-CA-NC-X-(-1)-PR p3CDP25
(SEQ ID NO: 840)
491
V12 MA-CA-X-(-1)-PR p3CDP26
(SEQ ID NO: 841)
491
V13 MA-X-NC-(-1)-PR p3CDP27
(SEQ ID NO: 842)
363
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Versions & XDP
plasmid
Design
DNA Sequence
CasX
number
491
j V14 MA-X-(-1)-PR pXDP28
(SEQ ID NO: 843)
491
j V15 MA-X-PR p3CDP29
(SEC) ID NO: 844)
491
j V16 MA-CA-X-PR pXDP30
(SEQ ID NO: 845)
491
j V17 MA-X pXDP31
(SEQ ID NO: 846)
491
V18 MA-CA-X pXDP32
(SEQ ID NO: 847)
491
V31 MA-CA-NC-X 0CDP23
(SEQ ID NO: 848)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 849)
j V32 MA-CA-NC-P 1-X pXDP24
(SEQ ID NO: 850)
491 MA-CA-NC-PI/P6 pXDP3
(SEQ ID NO: 851)
j V33 MA-CA-NC-X-(-1)-PR p3CDP25
(SEQ ID NO: 852)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 853)
j V34 MA-CA-X-(-1)-PR p3CDP26
(SEQ ID NO: 854)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 855)
j V35 MA-X-NC-(-1)-PR p3CDP27
(SEQ ID NO: 856)
364
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Versions & XDP
plasmid
Design
DNA Sequence
CasX
number
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 857)
V36 MA-X-(-1)-PR pXDP28
(SEQ ID NO: 858)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 859)
V37 MA-X-PR pXDP29
(SEQ ID NO: 860)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 861)
V38 MA-CA-X-PR pXDP30
(SEQ ID NO: 862)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 863)
V39 MA-X pXDP31
(SEQ ID NO: 864)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 865)
V40 MA-CA-X pXDP32
(SEQ ID NO: 866)
491 MA-CA-NC-P1/P6 pXDP3
(SEQ ID NO: 867)
V122 MA-CA-P1/P6-X pXDP98
(SEC) ID NO: 868)
491
V123 MA-CA-P1/P6-Pl/P6-X pXDP99
(SEQ ID NO: 869)
491
V124 MA-CA-NC-P1/P6-P1/P6-X pXDP100
(SEQ ID NO: 870)
V125 MA-CA-X-P1/P6 pXDP101
(SEQ ID NO: 871)
V126 MA-CA-NC-X-P1/P6 pXDP102
(SEQ ID NO: 872)
V128 MA-CA-P2-P1/P6-X pXDP104
(SEQ ID NO: 873)
Example 22: Transfection and recovery of XDP constructs in the Gag-(4)-
protease-CasX
configuration derived from Retroviruses.
10011941Editing efficiency and specificity can be altered and enhanced with
the method of
CasX delivery that is employed. A wide variety of viral vector families,
including those of
retroviral origin, can be engineered for the transient delivery of CasX RNPs.
In addition to
potentially enhancing editing with altered cell and tissue tropism, use of
RNPs also offers the
365
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
unique advantage of negating the potential risks of insertional mutagenesis
and long-term
transgene expression. The purpose of the following experiment was to create
and identify unique
CasX delivery particles derived from different genera of the Retroviridae
family. The genera
investigated in the following experiments include Alpharetroviruses,
Betaretroviruses,
Gammaretroviruses, Deltaretroviruses, Epsilonretroviruses, Non-primate
lentiviruses and
Spumaretroviruses.
Method for the generation of XDPs
10011951 XDPs derived from Alpharetrovinises (avian leukosis virus (ALV) and
rous sarcoma
virus (RSV)) in the Gag-protease-CasX variation (Version 44 and 45; see FIG.
52A) were
produced by transient transfection of LentiX HEK293T cells (Takara
Biosciences) using the
three plasmids encoding the Gag-protease-CasX, the glycoprotein, and the guide
RNA,
respectively, and listed in Table 24. The pXDP40 and pXDP41 plasmid contains
the Gag
polyprotein sequence followed by a protease and a CasX 491 protein fused at
the C-terminus. A
TSCYHCGT (SEQ ID NO: 944) cleavage site separated the Protease protein and
CasX protein
sequences to mediate separation of the editing molecules during XDP
maturation. The
pStx.42.174.12.7 plasmid was created with a human U6 promoter upstream of a
CasX guide
RNA cassette having scaffold 174 and spacer components (targeted to tdTomato:
CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825) in a single-guide format. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP were also used. MI plasmids
contained
either an ampicillin or kanamycin resistance gene. The sequences incorporated
into the plasmids
are presented in Table 24.
10011961 XDPs derived from Betaretroviruses (Enzootic Nasal Tumor Virus
(ENTV), mouse
mammary tumor virus (IVINITV) and Mason-Pfizer monkey virus (MPMV)) in the Gag-
(-1)-
protease-CasX variation (Version 46, 47, 62 and 90; see FIG. 52B) were
produced by transient
transfection of LentiX HEIC293T cells using the three plasmids encoding the
Gag-(-1)-protease-
CasX, the glycoprotein, and the guide RNA, respectively, and listed in Table
24. The pXDP42,
pXDP43, pXDP44 and pXDP61 plasmid contains the Gag polyprotein sequence
followed by
ribosomal frameshift, a protease and a CasX protein fused at the C-terminus. A
DCLDFDND
(SEQ ID NO: 934), DLVLLSAE (SEQ ID NO: 935), PQVMAAVA (SEQ ID NO: 936) and
PQVMAAVA (SEQ ID NO: 936) cleavage site separated the Protease protein and
CasX protein
sequences to mediate separation of the editing molecules during XDP maturation
in the
pXDP42, pXDP43, pXDP44 and pXDP61 plasmids, respectively. The pStx42.174.12.7
plasmid
366
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
was created with a human U6 promoter upstream of a CasX guide cassette having
scaffold 174
and spacer components (targeted to tdTomato) in a single-guide format.
Plasmids containing
VSV-G (pGP2) for pseudotyping the XDP were also used. All plasmids contained
either an
ampicillin or kanamycin resistance gene. The sequences incorporated into the
plasmids are
presented in Table 24.
10011971 XDPs derived from Deltaretroviruses (bovine leukemia virus (BLV) and
human T
lymphotropic virus (HTLV1)) in the Gag-(-1)-protease-CasX variation (Version
48, 49 and 63)
were produced by transient transfection of LentiX HEIC293T cells using three
plasmids encoding
the Gag-(-1)-protease-CasX, the glycoprotein, and the guide RNA, respectively,
and listed in
Table 24. The pXDP45, pXDP46, and pXDP62 plasmid contains the Gag polyprotein
sequence
followed by ribosomal frameshift, a protease and a CasX protein fused at the C-
terminus. A
PAILPHS (SEQ ID NO: 945), PQVLPVMH (SEQ ID NO: 946) and PQVLPVMH (SEQ ID NO:
946) cleavage site separated the Protease protein and CasX protein sequences
to mediate
separation of the editing molecules during XDP maturation in the pXDP45,
pXDP46, and
pXDP62 plasmid respectively. The pStx42.174.12.7 plasmid was created with a
human U6
promoter upstream of a CasX guide cassette having scaffold 174 and spacer
components
(targeted to tdTomato) in a single-guide format. Plasmids containing VSV-G
(pGP2) for
pseudotyping the XDP were also used. All plasmids contained either an
ampicillin or kanamycin
resistance gene. The sequences incorporated into the plasmids are presented in
Table 24.
10011981XDPs derived from Epsilonretroviruses (walleye dermal sarcoma virus
(WDSV)) in
the Gag-protease-CasX variation (Version 50) were produced by transient
transfection of LentiX
HEK293T cells using three plasmids encoding the Gag-protease-CasX, the
glycoprotein, and the
guide RNA, respectively, and listed in Table 24. The pXDP47 plasmid contains
the Gag
polyprotein sequence followed by a protease and a CasX protein fused at the C-
terminus. A
ARQMTAHT (SEQ ID NO: 937) cleavage site separated the Protease protein and
CasX protein
sequences to mediate separation of the editing molecules during XDP maturation
in the pXDP47
plasmid. The pStx42.174.12.7 plasmid was created with a human U6 promoter
upstream of a
CasX guide cassette having scaffold 174 and spacer components (targeted to
tdTomato) in a
single-guide format. Plasmids containing VSV-G (pGP2) for pseudotyping the XDP
were also
used. All plasmids contained either an ampicillin or kanamycin resistance
gene. The sequences
incorporated into the plasmids are presented in Table 24.
367
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10011991 XDPs derived from Gammaretroviruses (feline leukemia virus (FLY) and
murine
leukemia virus (MMLV)) in the Gag-protease-CasX variation (Version 51 and 52)
were
produced by transient transfection of LentiX HEK293T cells using the three
plasmids portrayed
in FIG. 54A and listed in Table 24. The pXDP48, and p3CDP49 plasmid contains
the Gag
polyprotein sequence followed by a protease and a CasX protein fused at the C-
terminus. A
SSLYPVLP (SEQ ID NO: 938), and SSLYPALT (SEQ ID NO: 939) cleavage site
separated the
Protease protein and CasX protein sequences to mediate separation of the
editing molecules
during XDP maturation in the pXDP48, and pXDP49 plasmid respectively. The
pStx42.174.12.7
plasmid was created with a human U6 promoter upstream of a CasX guide cassette
having
scaffold 174 and spacer components (targeted to tdTomato) in a single-guide
format. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP were also used. All plasmids
contained
either an ampicillin or kanamycin resistance gene. The sequences incorporated
into the plasmids
are presented in Table 24.
10012001 XDPs derived from Non-primate Lentiviruses (caprine arthritis
encephalitis (CAEV),
equine infectious anaemia virus (EIAV), simian immunodeficiency virus (SIV)
and visna maedi
virus (VMV)) in the Gag-(-1)-protease-CasX variation (Version 53, 54, 55 and
91) were
produced by transient transfection of LentiX HEK293T cells using three
plasmids encoding the
Gag-(-1)-protease-CasX, the g,lycoprotein, and the guide RNA, respectively,
and listed in Table
24. The pXDP50, pXDP51, pXDP52, p3CDP53 plasmid contains the Gag polyprotein
sequence
followed by a ribosomal frameshift, a protease and a CasX protein fused at the
C-terminus_ A
AGGRSWKA (SEQ ID NO: 940), SEEYPIMT (SEQ ID NO: 941), G-GNYPVQQ (SEQ ID NO:
942) and REVYPIVN (SEQ 1D NO: 943) cleavage site separated the Protease
protein and CasX
protein sequences to mediate separation of the editing molecules during XDP
maturation in the
pXDP50, pXDP51, pXDP52, p3CDP53 plasmid respectively. The pStx42.174.12.7
plasmid was
created with a human U6 promoter upstream of a CasX guide cassette having
scaffold 174 and
spacer components (targeted to tdTomato) in a single-guide format. Plasmids
containing VSV-G
(pGP2) for pseudo-typing the XDP were also used. All plasmids contained either
an ampicillin
or kanamycin resistance gene. The sequences incorporated into the plasmids are
presented in
Table 24.
10012011 XDPs derived Spumaretrovirinae family (bovine foamy virus (BFV),
equine foamy
virus (EFV), feline foamy virus (FFV), Brown greater galago prosimian foamy
virus (BGPFV),
Rhesus macaque simian foamy virus (RHSFY) and Simian foamy virus (SFV)) in the
Gag-(-1)-
368
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
protease-CasX variation (Version 56, 57, 58, 59, 60, 61 and 92) were produced
by transient
transfection of LentiX HEK293T cells using the three plasmids encoding the Gag-
(-1)-protease-
CasX, the glycoprotein, and the guide RNA, respectively, and listed in Table
24. The pXDP54,
pXDP55, p3CDP56, p3CDP57, p3CDP58, pXDP59 and p3CDP60 plasmid contains the Gag
polyprotein sequence followed by a ribosomal frameshift, a protease and a CasX
protein fused at
the C-terminus. A SAVHSVRL (SEQ ID NO: 784), RTVNTVRV (SEQ 1D NO: 785),
NTVHTVRQVES (SEQ 1D NO: 786), AAVHTVKA (SEQ ID NO: 787), RTVNTVTT (SEQ ID
NO: 788) and RSVNTVTA (SEQ ID NO: 789) cleavage site separated the Protease
protein and
CasX protein sequences to mediate separation of the editing molecules during
XDP maturation
in the pXDP54, pXDP55, pXDP56, pXDP57, pXDP58, pXDP59 and pXDP60 plasmid
respectively. The pSix42.174.12.7 plasmid was created with a human U6 promoter
upstream of a
CasX guide cassette having scaffold 174 and spacer components (targeted to
tdTomato) in a
single-guide format. Plasmids containing VSV-G (pGP2) for pseudotyping the XDP
were also
used. All plasmids contained either an ampicillin or kanamycin resistance
gene. The sequences
incorporated into the plasmids are presented in Table 24.
Table 24: Plasmid Encoding Sequences for XDP Versions
Version XDP plasmid SEQ ID NO of DNA
number Sequence
pStx42.174.12. 880
pGP2 881
44 pX D P40 882
45 pXDP41 883
46 pX D P42 884
90 pX D P43 885
47 pX D P44 886
48 pX D P45 887
49 pXDP46 888
369
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version XDP plasmid SEO ID NO of DNA
number Sequence
50 pXDP47 889
51 pXDP48 890
52 pXDP49 891
91 pXDP50 892
53 pXDP51 893
54 pXDP52 894
55 pXDP53 895
56 pXDP54 896
57 pXDP55 897
58 pXDP56 898
59 pXDP57 899
92 pXDP58 900
60 pXDP59 901
61 pXDP60 902
62 pXDP61 903
63 pDP62 904
64 pXDP63 905
V29 pXDP88 906
Transfection
10012021The steps for creation of the XDP are depicted graphically in FIG. 24.
HEK293T
Lenti-X cells were maintained in 10% FBS supplemented DMEM with HEPES,
penicillin/streptomycin (Pen/Step), sodium pyruvate, and 2-mercaptoethanol.
Cells were seeded
in TWO 15 cm dishes at 8e6 cells per dish in 10 mL of media. Cells were
allowed to settle and
370
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
grow for 24 hours before transfection. At the time of transfection cells were
70-90% confluent.
For transfection, the following plasmid amounts were used for the structural
plasmid
individually: pXDP40 (151 pg), pXDP41(151 pg), pXDP42 (157 pg), pXDP43 (157
pig),
pXDP44 (159 pg), pXDP45 (145 rig), pXDP46 (149 pg), pXDP47 (152 pg), pXDP48
(148 p.g),
p3CDP49 (149 pg), pXDP50 (145 rig), pXDP51 (146 pg), p3CDP52 (147 pg), pXDP53
(144 p.g),
pXDP54 (149 pg), pXDP55 (153 rig), pXDP56 (154 pig), pXDP57 (150 pg), pXDP58
(146 pg),
pXDP59 (154 pg), pXDP60 (154 rig), pXDP61 (159 pig), pXDP62 (149 pg), pXDP63
(147 pg),
pXDP88 (146 pg). Along with the structural plasmid, each transfection also
received 26.3 pig of
pStx42.174.12.7, and the 5 pg of pGP2 in 3800 RE of Opti-MEM media. 1 mg/m1
linear
polyethylenimine (PEI, MW-25,000 Da) was then added to the plasmid mixture at
1:3 DNA:PEI
concentration, mixed, and allowed to incubate at room temperature before being
added to the
cell culture.
Collection and concentration
10012031 Media was changed on cells 24 hours post-transfection. XDP-containing
media was
collected 72 hours post-transfection and filtered through a 0.45 LIM filter
using a 60 mL syringe.
The filtered supernatant was concentrated by centrifugation at 17,000 x g at 4
C for 4h using a
10% sucrose buffer in NTE. The concentrated XDPs were held at -20 C until use.
Editing of tdTomato neural progenitor cells using XDP
10012041 tdTomato neural progenitor cells (tdT NPCs) were grown in DMEM F12
supplemented
with glutamax, HEPES, non-essential amino acids, Pen/Strep, 2-mercaptoethanol,
B-27 without
vitamin A, and N2. Cells were harvested using a Takara Biosciences Neuron
Dissociation Kit
and seeded on PLF coated 96 well plates. Cells were allowed to grow at 37 C
for 48 hours
before being treated with targeting XDPs (having spacer 12.7 for tdTomato) as
a 10x concentrate
from the sucrose buffer concentrates using half-log dilutions. NPCs were grown
for 96 hours
before analysis of fluorescence as a marker of editing of tdTomato. Version 29
XDP made with
pXDP88 is the HIV lentivirus control for these experiments testing out Gag-Pro-
Stx versions of
the various retroviruses.
[001205] Results: The results of the editing assay are shown in FIGS. 69A and
B, FIG. 70 and in
Table 25 and Table 26 below. FIGS. 69A and B show the percentage editing
efficacy for
specific amounts of the various XDP versions in tdTomato NPCs. FIG. 70 shows
specifically the
editing efficacy of the various XDP versions when 16.6 p.1 of the concentrated
XDP prep is used
to treat tdTomato NPCs. Tables 25 and 26 represent the results showing %
editing of the
371
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
dtTomato target sequence when 50W and 16.6 1 of the concentrated XDP prep
were used to
treat NPCs. The results indicate that, under the conditions of the assay, XDPs
constructed using
members of the Retroviridae in several different configurations of the XDP,
were able, for the
majority of the genera, to result in significant editing of the target nucleic
acid in the NPC cells,
with several editing above 10%.
Table 25: Results of Editing Assay for the first dilution (50 pl)
Version Genus/order
Virus XDP plasmid number
Editing %
number
44 Alpharetrovirus ALV
pXDP40 91.5
45 Alpharetrovirus RSV
pXDP41 4.3
46 Betaretrovirus
ENTV 0CDP42 9.1
90 Betaretrovirus MMTV
p3CDP43 7.3
47 Betaretrovirus MPMV
p3CDP44 30.5
62 Betaretrovirus MPMV Native
p3CDP61 30,8
48 Deltaretrovirus BLV
p3CDP45 194
49 Deltaretrovirus HTLV1
pXDP46 20.1
63 Deltaretrovirus HTLV1 Native
pXDP62 37.0
50 Epsilonretrovirus WDSV
pXDP47 10.9
51 Gammaretrovirus FLV
p3CDP48 6.7
52 Gammaretrovirus MMLV
p3CDP49 12.4
91 Non-primate
CAEV 0CDP50 8.2
lentivirus
53 Non-primate
EIAV p3CDP51 5.3
lentivirus
54 Non-primate SW
pXDP52 11.7
lentivirus
64 Non-primate SW Native
pXDP63 13.5
lentivirus
55 Non-primate VMV
p3CDP53 8.7
lentivirus
56 Spumaretrovirinae BFV
pXDP54 3.6
372
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
XDP plasmid number Editing %
number
57 Spumaretrovirinae BGPFV
pXDP55 8.9
58 Spumaretrovirinae CCFV
pXDP56 5.5
59 Spumaretrovirinae EFV
pXDP57 4.4
92 Spumaretrovirinae FFV
pXDP58 7.3
60 Spumaretrovirinae RHSFV
pXDP59 4.2
61 Spumaretrovirinae SFV
pXDP60 4.5
29 Lentivirus HIV
p3CDP88 7.4
Table 26: Results of Editing Assay for the second dilution (16.6u0
Version Genus/order Virus
XDP plasmid number Editing %
number
44 Al pharetrovi rus ALV
p3CDP40 85.7
45 Al pharetrovi rus RSV
p3CDP41 2_9
46 Betaretrovirus ENTV
pXDP42 2.3
90 Betaretrovirus MMTV
pXDP43 7.6
47 Betaretrovirus MPMV
pXDP44 2.6
62 Betaretrovirus MPMV Native
pXDP61 8.5
48 Deltaretrovirus BLV
pXDP45 15.2
49 Deltaretrovirus HTLV1
p3CDP46 1.8
63 Deltaretrovirus HTLV1 Native
pXDP62 13.0
50 Epsi1onretrovirus WDSV
pXDP47 1.1
51 Gammaretrovirus FLY
pXDP48 7.8
52 Gammaretrovirus MMLV
pXDP49 6.3
91 Non-primate CAEV
pXDP50 3.1
1entivirus
53 Non-primate EIAV
pXDP51 3.8
lentivirus
54 Non-primate SW
pXDP52 1.3
1entivirus
373
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
XDP plasm id number Editing "4
number
64 Non-primate SIV Native
pXDP63 1.0
lentivirus
55 Non-primate VMV
pXDP53 7.4
lentivirus
56 Spumaretrovirinae BFV
pXDP54 1.9
57 Spumaretrovirinae BGPFV
pXDP55 4.5
58 Spumaretrovirinae CCFV
pXDP56 3.7
59 Spumaretrovirinae EFV
pXDP57 2.7
92 Spumaretrovirinae FFV
pXDP58 1_7
60 Spumaretrovirinae RHSFV
pXDP59 3_4
61 Spumaretrovirinae SFV
pXDP60 1.8
29 Lentivirus HIV
pXDP88 53
Example 23: Transfection and recovery of XDP constructs in a MA-CA-CasX
configuration derived from Retroviruses
10012061Editing efficiency and specificity can be altered and enhanced with
the method of
CasX delivery that is employed. A wide variety of viral vector families,
including those of
retroviral origin, can be engineered for the transient delivery of CasX RNPs.
In addition to
potentially enhancing editing with altered cell and tissue tropism, use of
RNPs packaged within
these viral vectors also offers the unique advantage of negating the potential
risks of insertional
mutagenesis and long-term transgene expression. The purpose of the following
experiment was
to build upon the previous example and to create and identify unique CasX
delivery particles
derived from different genera of the Retroviridae family using different
architectures. The
genera investigated in the following experiments include Alpharetroviruses,
Betaretroviruses,
Gammaretroviruses, Deltaretroviruses, Epsilonretroviruses and Non-primate
lentiviruses in a
MA-CA-CasX configuration, thereby eliminating the NC and protease domains.
Methods
Method for the generation of XDPs
10012071XDPs derived from Alpharetroviruses (ALV and RSV) in the MA-CA-CasX
variation
(Version 66a and 67a; see FIG. 55B) were produced by transient transfection of
LentiX
374
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
HEK293T cells (Takara Biosciences) using the three plasmids encoding the MA-CA-
CasX, the
glycoprotein, and the guide RNA, respectively, and listed in Table 27. The
pXDP64 and
pXDP65 plasmid contains the Matrix sequence followed by the Capsid sequence
and a CasX
491 protein fused at the C-terminus. The cleavage site between the Capsid and
the Nucleocapsid
protein was kept intact for each virus and immediately preceded the CasX
protein sequences to
mediate separation of the editing molecules during XDP maturation, when
coupled with a
plasmid that contained the respective viral protease. The pS1x42.174.12.7
plasmid was created
with a human U6 promoter upstream of a CasX guide RNA cassette having scaffold
174 and
spacer components (targeted to tdTomato: CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825)
in a single-guide format Plasmids containing VSV-G (pGP2) for pseudotyping the
XDP were
also used. All plasmids contained either an ampicillin or kanamycin resistance
gene. The
sequences incorporated into the plasmids are presented in Table 27.
10012081 XDPs derived from Betaretroviruses (ENTV, MMTV and MPMV) in the MA-CA-
CasX variation (Version 68A, 69A, 70A and 87A, FIG. 56B) were produced by
transient
transfection of LentiX HEK293T cells using three plasmids encoding the MA-CA-
CasX, the
glycoprotein, and the guide RNA, respectively, and listed in Table 27. The
pXDP66, pXDP67,
pXDP68 and pXDP85 plasmid contains the Matrix sequence followed by the Capsid
sequence
and a CasX protein fused at the C-terminus. The cleavage site between the
Capsid and the
Nucleocapsid protein was kept intact for each virus and immediately preceded
the CasX protein
sequences to mediate separation of the editing molecules during XDP
maturation, when coupled
with a plasmid that contained the respective viral protease. The
pStx42.174.12.7 plasmid was
created with a human U6 promoter upstream of a CasX guide cassette having
scaffold 174 and
spacer components (targeted to tdTomato) in a single-guide format. Plasmids
containing VSV-G
(pGP2) for pseudotyping the XDP were also used. All plasmids contained either
an ampicillin or
kanamycin resistance gene. The sequences incorporated into the plasmids are
presented in Table
27.
10012091 XDPs derived from Deltaretroviruses (ELY and ITTLV1) in the MA-CA-
CasX
variation (Version 71A, 72A and 88A, FIG. 5713) were produced by transient
transfection of
LentiX HEK293T cells using the three plasmids portrayed in FIG. 5711 and
listed in Table 27.
The pXDP69, pXDP70, and pXDP86 plasmid contains the Matrix sequence followed
by the
Capsid sequence and a CasX protein fused at the C-terminus. The cleavage site
between the
Capsid and the Nucleocapsid protein was kept intact for each virus and
immediately preceded
375
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
the CasX protein sequences to mediate separation of the editing molecules
during XDP
maturation, when coupled with a plasmid that contained the respective viral
protease. The
pStx42.174.12/ plasmid was created with a human U6 promoter upstream of a CasX
guide
cassette having scaffold 174 and spacer components (targeted to tdTomato) in a
single-guide
format. Plasmids containing VSV-G (pGP2) for pseudotyping the XDP were also
used. All
plasmids contained either an ampicillin or kanamycin resistance gene. The
sequences
incorporated into the plasmids are presented in Table 27.
10012101 XDPs derived from Epsilonretroviruses (WDSV) in the MA-CA-CasX
variation
(Version 73A, FIG. 58B) were produced by transient transfection of LentiX
HEK293T cells
using the three plasmids portrayed in FIG. 58B and listed in Table 27. The
pXDP71 plasmid
contains the Matrix sequence followed by the Capsid sequence and a CasX
protein fused at the
C-terminus. The cleavage site between the Capsid and the Nucleocapsid protein
was kept intact
for each virus and immediately preceded the CasX protein sequences to mediate
separation of
the editing molecules during XDP maturation, when coupled with a plasmid that
contained the
respective viral protease. The pStx42.174.12.7 plasmid was created with a
human U6 promoter
upstream of a CasX guide cassette having scaffold 174 and spacer components
(targeted to
tdTomato) in a single-guide format. Plasmids containing VSV-G (pGP2) for
pseudotyping the
XDP were also used. All plasmids contained either an ampicillin or kanamycin
resistance gene.
The sequences incorporated into the plasmids are presented in Table 27.
10012111 XDPs derived from Gammaretroviruses (FLV and !ONLY) in the MA-CA-CasX
variation (Version 74A and 75A, FIG. 59B) were produced by transient
transfection of LentiX
IfEK293T cells using the three plasmids portrayed in FIG. 5913 and listed in
Table 27. The
pXDP72, and pXDP73 plasmid contains the Matrix sequence followed by the Capsid
sequence
and a CasX protein fused at the C-terminus. The cleavage site between the
Capsid and the
Nucleocapsid protein was kept intact for each virus and immediately preceded
the CasX protein
sequences to mediate separation of the editing molecules during XDP
maturation, when coupled
with a plasmid that contained the respective viral protease. The
pStx42.174.12.7 plasmid was
created with a human U6 promoter upstream of a CasX guide cassette having
scaffold 174 and
spacer components (targeted to tdTomato) in a single-guide format. Plasmids
containing VSV-G
(pGP2) for pseudotyping the XDP were also used. All plasmids contained either
an ampicillin or
kanamycin resistance gene. The sequences incorporated into the plasmids are
presented in Table
27.
376
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10012121XDPs derived from Non-primate Lentiviruses (CAEV, ELAN, SIV and VMV)
irk the
MA-CA-CasX variation (Version 76A, 77A, 78A, 79A and 89A, FIG. 60B) were
produced by
transient transfection of LentiX BEK293T cells using the three plasmids
portrayed in FIG. 6011
and listed in Table 27. The pXDP74, pXDP75, p3CDP76, pXDP77 and pXDP87 plasmid
contains the Matrix sequence followed by the Capsid sequence and a CasX
protein fused at the
C-terminus. The cleavage site between the Capsid and the Nucleocapsid protein
was kept intact
for each virus and immediately preceded the CasX protein sequences to mediate
separation of
the editing molecules during XDP maturation, when coupled with a plasmid that
contained the
respective viral protease. The pStx42.174.12.7 plasmid was created with a
human U6 promoter
upstream of a CasX guide cassette having scaffold 174 and spacer components
(targeted to
tdTomato) in a single-guide format. Plasmids containing VSV-G (pGP2) for
pseudo-typing the
XDP were also used. All plasmids contained either an ampicillin or kanamycin
resistance gene.
The sequences incorporated into the plasmids are presented in Table 27.
Table 27: Plasmid Encoding Sequences for XDP Versions
Version
XDP plasmid SEQ
ID NO of DNASequence
number
N/A pStx42.I74.12.7 907
pGP2
908
66a pXDP64 909
67a pXDP65 910
68a pXDP66 911
69a pXDP67 912
70a p3CDP68 913
71a pXDP69 914
72a 0CDP70 915
73a pXDP7I 916
74a pXDP72 917
377
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version
XDP plasmid SEQ
ID NO of DNASequence
number
75a pXDP73
918
76a pXDP74
919
77a pXDP75
920
78a pXDP76
921
79a pXDP77
922
87a pXDP85
923
88a pXDP86
924
89a pXDP87
925
59 pXDP57
926
92 pXDP58
927
60 pXDP59
928
61 pXDP60
929
62 pXDP61
930
63 pXDP62
931
64 pXDP63
932
V29 pXDP88
933
Transfection
10012131 The steps for creation of the XDP are depicted graphically in FIG.
24. HEK293T
Lenti-X cells were maintained in 10% FBS supplemented DMEM with HEPES,
penicillin/streptomycin (Pen/Step), sodium pyruvate, and 2-mercaptoethanol.
Cells were seeded
in TWO 15 cm dishes at 8th cells per dish in 10 mL of media. Cells were
allowed to settle and
grow for 24 hours before transfection. At the time of transfection cells were
70-90% confluent
For transfection, the following plasmid amounts were used for the structural
plasmid
individually: pXDP64 (143 jig), pXDP65 (143 jig), pXDP66 (142 jig), pXDP67
(143 jig),
378
CA 03159320 2022-5-24
WO 2021/113772
PCVUS2020/063488
pXDP68 (144 jig), pXDP69 (136 jig), pXDP70 (137 g), pXDP71 (141 jig), pXDP72
(140 lig),
pXDP73 (142 jig), pXDP74 (134 g), pXDP75 (134 jig), pXDP76 (134 jig), pXDP85
(144 lig),
pXDP86 (137 jig), pXDP87 (138 jig), pXDP32 (114 g). Along with the structural
plasmid,
each transfection also received 26.3 fig of pStx42.174.12.7, and the 5 jig of
pGP2 in 3800 pl of
Opti-MEM media. 1 mg/ml linear polyethylenimine (PEI, MW=25,000 Da) was then
added to
the plasmid mixture at 1:3 DNA:PEI concentration, mixed, and allowed to
incubate at room
temperature before being added to the cell culture.
Collection and concentration
10012141Media was changed on cells 24 hours post-transfection. XDP-containing
media was
collected 72 hours post-transfection and filtered through a 0,45 LAM filter
using a 60 mL syringe.
The filtered supernatant was concentrated by centrifugation at 17,000 x g at
4oC for 4h using a
10% sucrose buffer in NTE. The concentrated XDPs were held at -20 C until use.
Editing of tdTomato neural progenitor cells using XDP
10012151 tdTomato neural progenitor cells (tdT NPCs) were grown in DMEM F12
supplemented
with glutamax, HEPES, non-essential amino acids, Pen/Strep, 2-mereaptoethanol,
B-27 without
vitamin A, and N2. Cells were harvested using a Takara Bioscienc,es Neuron
Dissociation Kit
and seeded on PLF coated 96 well plates. Cells were allowed to grow at 37 C
for 48 hours
before being treated with targeting XDPs (having spacer 12.7 for tdTomato) as
a 10x concentrate
from the sucrose buffer concentrates using half-log dilutions. NPCs were grown
for 96 hours
before analysis of fluorescence as a marker of editing of tdTomato. Version 18
with pXDP32
serves as the control for these experiments.
[0012161Results: The results of the editing assay are shown in FIGS. 71A and
B, FIG. 72 and in
Tables 28 and 29 below. FIGS. 73A and B shows the percentage editing efficacy
for specific
amounts of the various XDP versions in tdTomato NPCs. FIG. 72 shows
specifically the editing
efficacy of the various XDP versions when 16.6 I of the concentrated XDP prep
is used to treat
tdTomato NPCs. Tables 28 and 29 represent the results showing % editing of the
dtTomato
target sequence when 50 pi and 16.6 I of the concentrated XDP prep were used
to treat NPCs,
The results indicate that, under the conditions of the assay, XDPs constructed
using members of
the Retroviridae in MA-CA-X configuration of the XDP, were able, for the
majority of the
genera, to result in significant editing of the target nucleic acid in the NPC
cells, with several
editing above 10%.
379
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Table 28: Results of Editing Assay for the first dilution (50 ul)
i
Version Genus/order
Virus XDP plasmid number
Editing V.
number
66a Alpharetrovirus ALV
pXDP64 47.0
67a Alpharetrovirus RSV
pXDP65 42.5
68a Betaretrovirus ENTV
pXDP66 18.3
69a Betaretrovirus
MMTV pXDP67 11.7
70a Betaretrovirus
MPMV pXDP68 28.3
87a Betaretrovirus MPMV Native
pXDP85 30.8
71a Deltaretrovirus BLV
pXDP69 31.1
72a Deltaretrovirus
HTLV1 pXDP70 22.4
88a Deltaretrovirus HTLV1 Native
pXDP86 37.0
73a Epsilonretrovirus
WDSV pXDP71 14.2
74a Gammaretrovirus FLY
pXDP72 77.5
75a Gammaretrovirus
MMLV p3CDP73 67.3
76a Non-primate
CAEV p3CDP74 18.5
lentivirus
77a Non-primate EIAV
p3CDP75 46.2
lentivirus
78a Non-primate SIV
pXDP76 17.6
tentivirus
89a Non-primate SIV Native
p3CDP87 13.5
lentivirus
18 Lentivirus HIV
pXDP32 21.3
Table 29: Results of Editing Assay for the second dilution (16.6 I)
Version Genusiorder
Virus XDP plasmid number
Editing %
number
66a Alpharetrovirus ALV
0CDP64 11.2
67a Alpharetrovirus RSV
pXDP65 14.3
68a Betaretrovirus ENTV
pXDP66 2.3
380
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
XDP plasmid number Editing %
number
69a Betaretrovirus
MMTV pXDP67 1.7
70a Betaretrovirus
MPMV pXDP68 6.0
87a Betaretrovirus MPMV Native
pXDP85 8.5
71a Deltaretrovirus BLV
pXDP69 3.2
72a Deltaretrovirus
HTLV1 pXDP70 2.9
88a Deltaretrovirus HTLV1 Native
pXDP86 13.0
73a Epsilonretrovirus
WDSV pXDP71 1.9
74a Gammaretrovirus FLV
pXDP72 32.0
75a Gammaretrovirus
MMLV pXDP73 38.0
76a Non-primate
CAEV pXDP74 5.6
lentivirus
77a Non-primate EIAV
pXDP75 29.1
lentivirus
78a Non-primate SIV
pXDP76 7.0
lentivirus
89a Non-primate SIV Native
pXDP87 1.0
lentivirus
18 Lentivirus 1-11V
pXDP32 9.3
Example 24: Transfection and recovery of XDP constructs in the Gag-(-1)-
protease-CasX
configuration derived from Retroviruses.
10012171Editing efficiency and specificity can be altered and enhanced with
the method of
CasX delivery that is employed. A wide variety of viral vector families,
including those of
retroviral origin, can be engineered for the transient delivery of CasX RNPs.
In addition to
potentially enhancing editing with altered cell and tissue tropism, use of
RNPs also offers the
unique advantage of negating the potential risks of insertional mutagenesis
and long-term
transgene expression. The purpose of the following experiment was to create
and identify unique
CasX delivery particles derived from different genera of the Retroviridae
family. The genera
investigated in the following experiments include Alphareiroviruse,
Beiareiroviruse,
381
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Gammaretroviruse, Deltaretroviruse, Epsilonretroviruse, Non-primate
lentiviruses and
Spumaretroviruse
Method for the generation of XDPs
10012181 XDPs derived from Alpharetroviruses (avian leukosis virus (ALV) and
rous sarcoma
virus (RSV)) in the Gag-protease-CasX variation (Version 44 and 45; see FIG.
52A) were
produced by transient transfection of LentiX HEK293T cells (Takara
Biosciences) using the
three plasmids portrayed in FIG. 52A and listed in Table 30. The pXDP40 and
pXDP41 plasmid
contains the Gag polyprotein sequence followed by a protease and a CasX 491
protein fused at
the C-terminus. A TSCYHCGT (SEQ ID NO: 944) cleavage site separated the
Protease protein
and CasX protein sequences to mediate separation of the editing molecules
during XDP
maturation. The pStx42.174.12.7 plasmid was created with a human U6 promoter
upstream of a
CasX guide RNA cassette having scaffold 174 and spacer components (targeted to
tdTomato:
CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825) in a single-guide format. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP were also used. All plasmids
contained
either an arnpicillin or kanamycin resistance gene. The sequences incorporated
into the plasmids
are presented in Table 30.
10012191 XDPs derived from Betaretroviruses (Enzootic Nasal Tumor Virus
(ENTV), mouse
mammary tumor virus (MMTV) and Mason-Pfizer monkey virus (MPMV)) in the Gag-(-
1)-
protease-CasX variation (Version 46, 47, 62 and 90; see FIG. 52B) were
produced by transient
transfection of LentiX HEK293T cells using the three plasmids portrayed in
FIG. 52B and listed
in Table 30. The pXDP42, pXDP43, pXDP44 and pXDP61 plasmid contains the Gag
polyprotein sequence followed by ribosomal frameshift, a protease and a CasX
protein fused at
the C-terminus. A DCLDFDND (SEQ ID NO: 934), DLVLLSAE (SEQ ID NO: 935),
PQVMAAVA (SEQ ID NO: 936) and PQVMAAVA (SEQ ID NO: 936) cleavage site
separated
the Protease protein and CasX protein sequences to mediate separation of the
editing molecules
during XDP maturation in the pXDP42, pXDP43, pXDP44 and pXDP61 plasmids,
respectively.
The pStx42.174.12.7 plasmid was created with a human U6 promoter upstream of a
CasX guide
cassette having scaffold 174 and spacer components (targeted to tdTomato) in a
single-guide
format. Plasmids containing VSV-G (pGP2) for pseudotyping the XDP were also
used. All
plasmids contained either an ampicillin or kanamycin resistance gene. The
sequences
incorporated into the plasmids are presented in Table 30.
382
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
10012201 XDPs derived from Deltaretroviruses (bovine leukemia virus (BLV) and
human T
lymphotropic virus (HTI.V1)) in the Gag-(-1)-protease-CasX variation (Version
48, 49 and 63;
see FIG. 53A) were produced by transient transfection of LentiX 11EK293T cells
using the three
plasmids portrayed in FIG. 53A and listed in Table 30. The pXDP45, pXDP46, and
pXDP62
plasmid contains the Gag polyprotein sequence followed by ribosomal
frameshift, a protease and
a CasX protein fused at the C-terminus_ A PAILHIS (SEQ ID NO: 945),
PQVLPVIVITI (SEQ ID
NO: 946) and PQVLPVMEI (SEQ ID NO: 946) cleavage site separated the Protease
protein and
CasX protein sequences to mediate separation of the editing molecules during
XDP maturation
in the pXDP45, pXDP46, and pXDP62 plasmid respectively. The pStx42.174.12.7
plasmid was
created with a human U6 promoter upstream of a CasX guide cassette having
scaffold 174 and
spacer components (targeted to tdTomato) in a single-guide format. Plasmids
containing VSV-G
(pGP2) for pseudotyping the XDP were also used. All plasmids contained either
an ampicillin or
kanamycin resistance gene. The sequences incorporated into the plasmids are
presented in Table
30.
10012211XDPs derived from Epsilonretroviruses (walleye dermal sarcoma virus
(WDSV)) in
the Gag-protease-CasX variation (Version 50; see FIG. 53B) were produced by
transient
transfection of LentiX HEK293T cells using the three plasmids portrayed in
FIG. 538 and listed
in Table 30. The pXDP47 plasmid contains the Gag polyprotein sequence followed
by a
protease and a CasX protein fused at the C-terminus. A ARQMTAHT (SEQ ID NO:
937)
cleavage site separated the Protease protein and CasX protein sequences to
mediate separation of
the editing molecules during XDP maturation in the pXDP47 plasmid. The
pStx42.174.12.7
plasmid was created with a human U6 promoter upstream of a CasX guide cassette
having
scaffold 174 and spacer components (targeted to tdTomato) in a single-guide
format. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP were also used. MI plasmids
contained
either an ampicillin or kanamycin resistance gene. The sequences incorporated
into the plasmids
are presented in Table 30.
10012221 XDPs derived from Gammaretroviruses (feline leukemia virus (FLV) and
murine
leukemia virus (MA/11,V)) in the Gag-protease-CasX variation (Version 51 and
52; see FIG. 54A)
were produced by transient transfection of LentiX ITEK293T cells using the
three plasmids
portrayed in FIG. 54A and listed in Table 30. The pXDP48, and pXDP49 plasmid
contains the
Gag polyprotein sequence followed by a protease and a CasX protein fused at
the C-terminus. A
SSLYPVLP (SEQ ID NO: 938), and SSLYPALT (SEQ ID NO: 939) cleavage site
separated the
383
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Protease protein and CasX protein sequences to mediate separation of the
editing molecules
during XDP maturation in the pXDP48, and pXDP49 plasmid respectively. The
pStx42.174.12.7
plasmid was created with a human U6 promoter upstream of a CasX guide cassette
having
scaffold 174 and spacer components (targeted to tdTomato) in a single-guide
format. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP were also used. MI plasmids
contained
either an ampicillin or kanamycin resistance gene. The sequences incorporated
into the plasmids
are presented in Table 30.
10012231 3CDPs derived from Non-primate Lentiviruses (caprine arthritis
encephalitis (CAEV),
equine infectious anaemia virus (EIAV), simian immunodeficiency virus (SIV)
and visna maedi
virus (VMV)) in the Gag-(-1)-protease-CasX variation (Version 53, 54, 55 and
91; see FIG.
54B) were produced by transient transfection of LentiX HEK293T cells using the
three plasmids
portrayed in FIG. 54B and listed in Table 30. The pXDP50, pXDP51, pXDP52,
0CDP53
plasmid contains the Gag polyprotein sequence followed by a ribosomal
frameshift, a protease
and a CasX protein fused at the C-terminus. A AGGRSWKA (SEQ ID NO: 940),
SEEYPIMI
(SEQ ID NO: 941), GGNYPVQQ (SEQ ID NO: 942) and REVYPIVN (SEQ ID NO: 943)
cleavage site separated the Protease protein and CasX protein sequences to
mediate separation of
the editing molecules during XDP maturation in the pXDP50, p3CDP51, pXDP52,
p3CDP53
plasmid respectively. The pStx42.174.12.7 plasmid was created with a human U6
promoter
upstream of a CasX guide cassette having scaffold 174 and spacer components
(targeted to
tdTomato) in a single-guide format. Plasmids containing VSV-G (pGP2) for
pseudo-typing the
XDP were also used. All plasmids contained either an ampicillin or kanamycin
resistance gene.
The sequences incorporated into the plasmids are presented in Table 30.
10012241 XDPs derived Spumaretrovifinae family (bovine foamy virus (BFV),
equine foamy
virus (EFV), feline foamy virus (FFV), Brown greater galago prosimian foamy
virus (BGPFV),
Rhesus macaque simian foamy virus (RHSFV) and Simian foamy virus (SFV)) in the
Gag-(-1)-
protease-CasX variation (Version 56, 57, 58, 59, 60, 61 and 92; see FIG. 55A)
were produced by
transient transfection of LentiX HEK293T cells using the three plasmids
portrayed in FIG, 55A
and listed in Table 30. The p3CDP54, p3CDP55, p3CDP56, pXDP57, pXDP58, pXDP59
and
pXDP60 plasmid contains the Gag polyprotein sequence followed by a ribosomal
frameshift, a
protease and a CasX protein fused at the C-terminus. A SAVHSVRL (SEQ ID NO:
784),
RTVNTVRV (SEQ ID NO: 785), NTVHTVRQVES (SEQ ID NO: 786), AAVHTVKA (SEQ
ID NO: 787), RTVNTVTT (SEQ ID NO: 788) and RSVNTVTA (SEQ ID NO: 789) cleavage
384
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
site separated the Protease protein and CasX protein sequences to mediate
separation of the
editing molecules during XDP maturation in the pXDP54, pXDP55, p3CDP56,
pXDP57,
pXDP58, pXDP59 and pXDP60 plasmid respectively. The pSix42.174.12.7 plasmid
was created
with a human U6 promoter upstream of a CasX guide cassette having scaffold 174
and spacer
components (targeted to tdTomato) in a single-guide format. Plasmids
containing VSV-G
(pGP2) for pseudotyping the XDP were also used. All plasmids contained either
an ampicillin or
kanamycin resistance gene. The sequences incorporated into the plasmids are
presented in Table
30.
Table 30: Plasmid and XDP Encoding Sequences
Version
XDP plasmid
SEQ ID NO of DNA Sequence
number
pStx42.174.12.7
947
pGP2
948
44 pXDP40
949
45 pXDP41
950
46 pXDP42
951
90 pXDP43
952
47 pXDP44
953
48 pXDP45
954
49 pXDP46
955
50 pXDP47
956
51 pXDP48
957
52 pXDP49
958
91 pXDP50
959
53 pXDP51
960
54 pXDP52
961
385
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version
XDP plasmid
SEQ ID NO of DNA Sequence
number
55 pXDP53
962
56 pXDP54
963
57 pXDP55
964
58 pXDP56
965
59 pXDP57
966
92 pXDP58
967
60 pXDP59
968
61 pXDP60
969
62 pXDP61
970
63 pXDP62
971
64 pXDP63
972
V29 pXDP88
973
Transfection
10012251 The steps for creation of the XDP are depicted graphically in FIG.
24. HEK293T
Lenti-X cells were maintained in 10% FBS supplemented DMEM with HEPES,
penicillin/streptomycin (Pen/Step), sodium pyruvate, and 2-mercaptoethanol.
Cells were seeded
in TWO 15 cm dishes at 8e6 cells per dish in 10 mL of media. Cells were
allowed to settle and
grow for 24 hours before transfection. At the time of transfection cells were
70-90% confluent
For transfection, the following plasmid amounts were used for the structural
plasmid
individually: pXDP40 (151 pg), pXDP41(151 jig), pXDP42 (157 pg), pXDP43 (157
pg),
pXDP44 (159 pg), pXDP45 (145 i's). pXDP46 (149 pg), pXDP47 (152 pg), p3CDP48
(148 pg),
pXDP49 (149 pg), pXDP50 (145 i's) , pXDP51 (146 pg), pXDP52 (147 pg), pXDP53
(144 pg),
pXDP54 (149 pg), pXDP55 (153 pg), pXDP56 (154 pg), p>CDP57 (150 pig), p>CD1P58
(146 lig),
pXDP59 (154 pg), pXDP60 (154 pg), pXDP61 (159 pg), p3CDP62 (149 pg), p3CDP63
(147 lig),
pXDP88 (146 pg). Along with the structural plasmid, each transfection also
received 26.3 pg of
386
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
pStx42.174,12.7, and the 5 jig of pGP2 in 3800 pl of Opti-MEM media. 1 mg/ml
linear
polyethylenimine (PEI, MW=25,000 Da) was then added to the plasmid mixture at
1:3 DNA:PEI
concentration, mixed, and allowed to incubate at room temperature before being
added to the
cell culture.
Collection and concentration
10012261 Media was changed on cells 24 hours post-transfection. XDP-containing
media was
collected 72 hours post-transfection and filtered through a 0.45 p.114 filter
using a 60 mL syringe.
The filtered supernatant was concentrated by centrifugation at 17,000 x g at 4
C for 4h using a
10% sucrose buffer in NTE. The concentrated XDPs were held at -20 C until use.
Editing of tdTomato neural progenitor cells using XDP
10012271tdTomato neural progenitor cells (tdT NPCs) were grown in DMEM F12
supplemented
with glutamax, HEPES, non-essential amino acids, Pen/Strep, 2-mercaptoethanol,
B-27 without
vitamin A, and N2. Cells were harvested using a Takara Biosciences Neuron
Dissociation Kit
and seeded on PLF coated 96 well plates. Cells were allowed to grow at 37 C
for 48 hours
before being treated with targeting XDPs (having spacer 12.7 for tdTomato) as
a 10x concentrate
from the sucrose buffer concentrates using half-log dilutions. NPCs were grown
for 96 hours
before analysis of fluorescence as a marker of editing of tdTomato. Version 29
XDP made with
pXDP88 is the HIV lentivirus control for these experiments testing out Gag-Pro-
Stx versions of
the various retroviruses.
10012281Results: The results of the editing assay are shown in FIGS. 69A and B
and in Table
31 and Table 32 below. FIGS. 69A and B show the percentage editing efficacy
for specific
amounts of the various XDP versions in tdTomato NPCs. Tables 31 and 32
represent the results
showing % editing of the dtTomato target sequence when 50 gl and 16.6 gl of
the concentrated
XDP prep were used to treat NPCs. The results indicate that, under the
conditions of the assay,
XDPs constructed using members of the Retroviridae in several different
configurations of the
XDP, were able, for the majority of the genera, to result in significant
editing of the target
nucleic acid in the NPC cells, with several editing above 10%.
Table 31: Results of Editing Assay for the first dilution (50 pd)
Version Genus/order
Virus XDP plasmid number
Editing %
number
44 Alpharetrovirus ALV
pXDP40 91.5
387
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
XDP plasmid number Editing %
number
45 Al pharetrovirus RSV
pXDP41 4.3
46 Betaretrovirus ENTV
pXDP42 9.1
90 Betaretrovirus MMTV
pXDP43 7.3
47 Betaretrovirus MPMV
pXDP44 30.5
62 Betaretrovirus MPMV Native
pXDP61 30.8
48 Deltaretrovirus BLV
pXDP45 19.4
49 Deltaretrovirus HTLV1
pXDP46 20.1
63 Deltaretrovirus HTLV1 Native
pXDP62 37.0
50 Epsi I onretrovirus WDSV
pXDP47 10.9
51 Gammaretrovirus FLV
pXDP48 6.7
52 Gammaretrovirus MMLV
pXDP49 12.4
91 Non-primate CAEV
pXDP50 8.2
lentivirus
53 Non-primate EIAV
pXDP51 5.3
lentivirus
54 Non-primate SW
pXDP52 11.7
lentivirus
64 Non-primate SW Native
pXDP63 13.5
lentivirus
55 Non-primate VMV
pXDP53 8.7
lentivirus
56 Spumaretrovirinae BFV
pXDP54 3.6
57 Spumaretrovirinae BGPFV
pXDP55 8.9
58 Spumaretrovirinae CCFV
pXDP56 5.5
59 Spumaretrovirinae EFV
pXDP57 4.4
92 Spumaretrovirinae FFV
pXDP58 7.3
60 Spumaretrovirinae RHSFV
pXDP59 4.2
61 Spumaretrovirinae SFV
pXDP60 4.5
29 Lentivirus HIV
pXDP88 7.4
388
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Table 32: Results of Editing Assay for the second dilution (16.6 I)
Version Genus/order
Virus XDP plasmid number
Editing %
number
44 Al pharetrovirus ALV
pXDP40 85.7
45 Al pharetrovirus RSV
pXDP41 2.9
46 Betaretrovirus
ENTV pXDP42 2.3
90 Betaretrovirus MMTV
p3CDP43 7.6
47 Betaretrovirus MPMV
pXDP44 2.6
62 Betaretrovirus MPMV Native
pXDP61 8.5
48 Deltaretrovirus BLV
pXDP45 15.2
49 Deltaretrovirus HTLV1
pXDP46 1.8
63 Deltaretrovirus HTLV1 Native
pXDP62 13.0
50 Epsi I onretrovirus WDSV
pXDP47 1.1
51 Gammaretrovirus FLY
pXDP48 7.8
52 Gammaretrovirus MMLV
p3CDP49 6.3
91 Non-primate
CAEV p3CDP50 3.1
lentivirus
53 Non-primate
EIAV pXDP51 3.8
lentivirus
54 Non-primate SW
pXDP52 1.3
lentivirus
64 Non-primate SW Native
pXDP63 1.0
lentivirus
55 Non-primate VMV
pXDP53 7.4
lentivirus
56 Spumaretrovirinae BFV
p3CDP54 1.9
57 Spumaretrovirinae BGPFV
p3CDP55 4.5
58 Spumaretrovirinae
CCFV p3CDP56 3.7
59 Spumaretrovirinae EFV
p3CDP57 2.7
92 Spumaretrovirinae FFV
pXDP58 1.7
60 Spumaretrovirinae RHSFV
pXDP59 3.4
389
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
XDP plasmid number Editing %
number
61 Spumaretrovirinae SFV
pXDP60 1.8
29 Lentivirus HIV
pXDP88 5.3
Example 25: Transfection and recovery of XDP constructs in the Gag-CasX
configuration
derived from Retroviruses.
10012291Editing efficiency and specificity can be altered and enhanced with
the method of
CasX delivery that is employed. A wide variety of viral vector families,
including those of
retroviral origin, can be engineered for the transient delivery of CasX RNPs.
In addition to
potentially enhancing editing with altered cell and tissue tropism, use of
RNPs packaged within
these viral vectors also offers the unique advantage of negating the potential
risks of insertional
mutagenesis and long-term transgene expression. The purpose of the following
experiment was
to build upon the previous example and to create and identify unique CasX
delivery particles
derived from different genera of the Retroviridae family using different
architectures. The
genera investigated in the following experiments include Alpharetroviruses,
Betaretroviruses,
Garnmaretroviruses, Deltaretroviruses, Epsilonretroviruses and Non-primate
lentiviruses in a
Gag-CasX configuration. The experiments were meant to be a direct comparison
with the HIV
Lentivirus based V7 construct, with the Gag component being replaced with the
corresponding
Gag components of Alpharetroviruses, Betaretroviruses, Gammaretroviruses,
Deltaretroviruses,
Epsilonretroviruses, Non-primate lentiviruses and Spumaretroviruses, with the
protease domains
eliminated in all constructs to test whether XDP capable of editing required
active release from
Gag.
Methods for the generation of XDPs
10012301XDPs derived from Alpharetroviruses (avian leukosis virus (ALV) and
rous sarcoma
virus (RSV)) in the Gag-CasX variation (V102 and V114; see FIG. 62B) were
produced by
transient transfection of LentiX ITEIC293T cells (Takara Biosciences) using
the three plasmids
portrayed in FIG. 62B and listed in Table 33. The pXDP127 and pXDP139 plasmid
contains the
Gag polyprotein sequence followed by the CasX 491 protein fused at the C-
terminus. The
pStx42.174.12.7 plasmid was created with a human U6 promoter upstream of a
CasX guide
RNA cassette having scaffold 174 and spacer components (targeted to tdTomato:
CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825) in a single-guide format. Plasmids
390
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
containing VSV-G (pGP2) for pseudotyping the XDP were also incorporated into
the constructs.
MI plasmids contained either an ampicillin or kanamycin resistance gene. The
sequences
incorporated into the plasmids are presented in Table 33.
10012311XDPs derived from Betaretroviruses (Enzootic Nasal Tumor Virus (ENTV),
mouse
mammary tumor virus (MMTV) and Mason-Pfizer monkey virus (MPMV)) in the Gag-
CasX
variation (V106, V111, V112 and V113, FIG. 64A) were produced by transient
transfection of
LentiX HEK293T cells using the three plasmids portrayed in FIG. 64A and listed
in Table 33.
The pXDP131, pXDP136, pXDP137 and pXDP138 plasmid contains the Gag polyprotein
sequence followed by the CasX 491 protein fused at the C-terminus. The
pStx42.174.12.7
plasmid was created with a human U6 promoter upstream of a CasX guide cassette
having
scaffold 174 and spacer components (targeted to tdTomato) in a single-guide
format. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP were also incorporated into
the constructs.
MI plasmids contained either an ampicillin or kanamycin resistance gene. The
sequences
incorporated into the plasmids are presented in Table 33 .
10012321XDPs derived from Deltaretroviruses (bovine leukemia virus (BLV) and
human T
lymphotropic virus (HTLV1)) in the Gag-CasX variation (Version V103, V108 and
V109, FIG.
63A) were produced by transient transfection of LentiX HEK293T cells using the
three
plasmids portrayed in FIG. 63A and listed in Table 33 . The pXDP128, pXDP133
and pXDP134
plasmid contains the Gag polyprotein sequence followed by the CasX 491 protein
fused at the
C-terminus. The pStx42.174.12.7 plasmid was created with a human U6 promoter
upstream of a
CasX guide cassette having scaffold 174 and spacer components (targeted to
tdTomato) in a
single-guide format. Plasmids containing VSV-G (pGP2) for pseudotyping the XDP
were also
incorporated into the constructs. All plasmids contained either an ampicillin
or kanamycin
resistance gene. The sequences incorporated into the plasmids are presented in
Table 33 .
10012331XDPs derived from Epsilonretroviruses (walleye dermal sarcoma virus
(WDSV)) in
the Gag-CasX variation (Version 73A, FIG. 58B) were produced by transient
transfection of
LentiX HEK293T cells using the three plasmids portrayed in FIG. 58B and listed
in Table 33.
The pXDP127 and pXDP139 plasmid contains the Gag polyprotein sequence followed
by the
CasX 491 protein fused at the C-terminus. The pStx42.174.12.7 plasmid was
created with a
human U6 promoter upstream of a CasX guide cassette having scaffold 174 and
spacer
components (targeted to tdTomato) in a single-guide format. Plasmids
containing VSV-G
(pGP2) for pseudotyping the XDP were also incorporated into the constructs.
All plasmids
391
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
contained either an ampicillin or kanamycin resistance gene. The sequences
incorporated into
the plasmids are presented in Table 33.
1001231 XDPs derived from Gammaretroviruses (feline leukemia virus (FLV) and
murine
leukemia virus (MMLV)) in the Gag-CasX variation (V107 and V110, FIG. 64B)
were produced
by transient transfection of LentiX HEK293T cells using the three plasmids
portrayed in FIG.
MB and listed in Table 33. The p3CDP132, and pXDP135 plasmid contains the Gag
polyprotein
sequence followed by the CasX 491 protein fused at the C-terminus. The
pStx42.174.12.7
plasmid was created with a human U6 promoter upstream of a CasX guide cassette
having
scaffold 174 and spacer components (targeted to tdTomato) in a single-guide
format. Plasmids
containing VSV-G (pGP2) for pseudotyping the XDP were also incorporated into
the constructs.
All plasmids contained either an ampicillin or kanamycin resistance gene. The
sequences
incorporated into the plasmids are presented in Table 33.
10012351 XDPs derived from Non-primate Lentiviruses (caprine arthritis
encephalitis (CAEV),
equine infectious anaemia virus (EIAV), simian immunodeficiency virus (SIV)
and visna maedi
virus (VMV)) in the Gag-CasX variation (V104, V105, V115, V116 and V117, FIG.
63B) were
produced by transient transfection of LentiX HEK293T cells using the three
plasmids portrayed
in FIG. 63B and listed in Table 33. The p3CDP129, p3CDP130, pXDP140, p3CDP141
and
pXDP142 plasmid contains the Gag polyprotein sequence followed by the CasX 491
protein
fused at the C-terminus. The pStx42.174.12.7 plasmid was created with a human
U6 promoter
upstream of a CasX guide cassette having scaffold 174 and spacer components
(targeted to
tdTomato) in a single-guide format. Plasmids containing VSV-G (pGP2) for
pseudo-typing the
3CDP were also incorporated into the constructs. All plasmids contained either
an ampicillin or
kanamycin resistance gene. The sequences incorporated into the plasmids are
presented in Table
33.
10012361 XDPs derived Spumaretrovirinae family (bovine foamy virus (BFV),
equine foamy
virus (EFV), feline foamy virus (FFV), Brown greater galas prosimian foamy
virus (BGPFV),
Rhesus macaque simian foamy virus (RHSFV) and Simian foamy virus (SFV)) in the
Gag-CasX
variation (V80a, V81a, V82a, V83a, V84a, V85a and V86a; see FIG. 62A) were
produced by
transient transfection of LentiX HEK293T cells using the three plasmids
portrayed in FIG. 62A
and listed in Table 33. The pXDP78, pXDP79, pXDP80, pXDP81, pXDP82, pXDP83 and
pXDP84 plasmid contains the Gag polyprotein sequence followed by the CasX
protein fused at
the C-terminus. The pStx42.174.12.7 plasmid was created with a human U6
promoter upstream
392
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
of a CasX guide cassette having scaffold 174 and spacer components (targeted
to tdTomato) in a
single-guide format. Plasmids containing VSV-G (pGP2) for pseudotyping the XDP
were also
incorporated into the constructs. All plasmids contained either an ampicillin
or kanamycin
resistance gene. The sequences incorporated into the plasmids are presented in
Table 33.
Table 33: XDP Plasmid and Encoding Sequences
Version XDP
number plasmid SEQ ID NO
of DNA Sequence
pStx42.1
N/A
974
74.12.7
pGP2
975
102 pXDP127
976
103 pXDP128
977
104 pXDP129
978
105 pXDP130
979
106 pXDP131
980
107 pXDP132
981
108 pXDP133
982
109 pXDP134
983
110 pXDP135
984
111 pXDP136
985
112 pXDP137
986
113 pXDP138
987
114 pXDP139
988
115 pXDP140
989
116 pXDP141
990
117 pXDP142
991
118 pXDP143
992
80a pXDP78
993
393
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version XDP
number plasmid SEQ ID NO of DNA Sequence
81a pXDP79 994
82a pXDP80 995
83a pXDP81 996
84a pXDP82 997
85a pXDP83 998
86a pXDP84 999
V29 pXDP88 1000
Transfection
10012371 The steps for creation of the XDP are depicted graphically in FIG.
24. HEK293T
Lenti-X cells were maintained in 10% FBS supplemented DMEM with HEPES,
penicillin/streptomycin (Pen/Step), sodium pyruvate, and 2-mercaptoethanol.
Cells were seeded
in TWO 15 cm dishes at 8e6 cells per dish in 10 mL of media. Cells were
allowed to settle and
grow for 24 hours before transfection. At the time of transfection cells were
70-90% confluent.
For transfection, the following plasmid amounts were used for the structural
plasmid
individually: pXDP127 (146 pg), pXDP129 (141 pg), pXDP130 (143 jig), pXDP131
(145 pg),
pXDP132 (143 pg), pXDP135 (145 jig), pXDP136 (152 pg), pXDP138 (149 gg),
pXDP139
(146 pg), pXDP140 (143 jig), pXDP141 (143 jig), pXDP142 (141 pg), pXDP143 (146
pg),
pXDP78 (145 pg), pXDP81 (141 pg), pXDP82 (139 pg), pXDP83 (145 pg), pXDP0017
(122
jig). Along with the structural plasmid, each transfection also received 26.3
jig of
pStx.42.174.12.7, and the 5 pig of pGP2 in 3800 pl of Opti-MEM media. 1 mg/m1
linear
polyethylenimine (PEI, MVV=25,000 Da) was then added to the plasmid mixture at
1:3 DNA:PEI
concentration, mixed, and allowed to incubate at room temperature before being
added to the
cell culture.
Collection and concentration
10012381 Media was changed on cells 24 hours post-transfection. XDP-containing
media was
collected 72 hours post-transfection and filtered through a 0.45 p.114 filter
using a 60 mL syringe.
394
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
The filtered supernatant was concentrated by centrifugation at 17,000 x g at 4
C for 4h using a
10% sucrose buffer in NTE. The concentrated XDPs were held at -20 C until use.
Editing of tdTomato neural progenitor cells using XDP
10012391 tdTomato neural progenitor cells (tdT NPCs) were grown in DMEM F12
supplemented
with glutamax, HEPES, non-essential amino acids, Pen/Strep, 2-mercaptoethanol,
B-27 without
vitamin A, and N2. Cells were harvested using a Takara Biosciences Neuron
Dissociation Kit
and seeded on PLF coated 96 well plates. Cells were allowed to grow at 37 C
for 48 hours
before being treated with targeting XDPs (having spacer 12.7 for tdTomato) as
a 10x concentrate
from the sucrose buffer concentrates using half-log dilutions. NPCs were grown
for 96 hours
before analysis of fluorescence as a marker of editing of tdTomato. Version 18
with pXDP32
serves as the control for these experiments.
[0012401Results: The results of the editing assay are shown in FIGS. 75A and
B, FIG. 76 and in
Table 34 and Table 35 below. FIGS. 75 A and B shows the percentage editing
efficacy for
specific amounts of the various XDP versions in tdTomato NPCs. Tables 34 and
35 represent the
results showing % editing of the tdTomato target sequence when 50 ul and 16.6
ul of the
concentrated XDP prep were used to treat NPCs. The results indicate that,
under the conditions
of the assay, XDPs constructed using members of the Retroviridae in Gag-CasX
configuration of
the XDP, were able, for the majority of the genera, to result in significant
editing of the target
nucleic acid in the NPC cells, with several editing above 4%.
Table 34: Results of Editing Assay for the first dilution (50u1)
Version Genus/order Virus
XDP plasmid number Editing %
102 Alpharetrovirus ALV
pXDP127 94.2
114 Alpharetrovirus RSV
pXDP139 43.4
106 Betaretrovirus ENTV
pXDP131 29.1
111 Betaretrovirus MIVITV
pXDP136 11.1
113 Betaretrovirus MPMV Native
pXDP138 19.2
118 Epsilonretrovinis WDSV
pXDP143 2,5
107 Gammaretrovirus FLY
pXDP132 6.8
110 Gammaretrovirus MMLV
pXDP135 45.2
395
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
XDP plasmid number Editing %
Non-primate
104 CAEV pXDP129 14.6
lenti virus
Non-primate
105 EIAV pXDP130 44.2
1enti virus
Non-primate
115 SIV pXDP140 43.1
lenti virus
Non-primate
116 S IV Native pXDP141 48.1
lenti virus
Non-primate
117 VIv1V pXDP142 9.6
lenti virus
7 Lentivirus 1-DV
pXDP0017 84.5
80a Spumaretrovirus BFV
pXDP78 29.2
83a Spumaretrovirus EFV
pXDP81 4.7
84a Spumaretrovirus FFV
pXDP82 4.9
85a Spumaretrovirus
RHSFV pXDP83 4.1
Table 35: Results of Editing Assay for the second dilution (16.6u1)
Version Genus/order Virus
XDP plasmid number Editing %
102 Alpharetrovirus
ALV pXDP127 95.8
114 Alpharetrovirus RSV
pXDP139 20.9
106 Betaretrovirus ENTV
pXDP131 10.5
111 Betaretrovirus MMTV
pXDP136 1.0
113 Betaretrovirus MPMV Native
pXDP138 2.7
118 Epsilonretrovirus
WDSV pXDP143 2.6
107 Gammaretrovirus FLV
pXDP132 6.4
110 Gammaretrovirus MMLV
pXDP135 12.8
Non-primate
104 CAEV pXDP129 1.3
lenti virus
Non-primate
105 EIAV pXDP130 21.7
lendvirus
Non-primate
115 SIV pXDP140 2.7
lendvirus
396
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
XDP plasmid number Editing %
Non-primate
116 SIV Native pXDP141 27.3
lentivirus
Non-primate
117 VMV pXDP142 1.0
lentivirus
7 Lentivirus HIV
pXDP0017 80.0
80a Spumaretrovirus BFV
pXDP78 11.4
83a Spumaretrovirus EFV
pXDP81 0.4
84a Spumaretrovirus FFV
pXDP82 0.7
85a Spumaretrovirus RHSFV
pXDP83 0.7
Example 26: Transfection and recovery of XDP constructs derived from
Spumaretrovirinae.
10012411Editing efficiency and specificity can be altered and enhanced with
the method of
CasX delivery that is employed. A wide variety of viral vector families,
including those of
retroviral origin, can be engineered for the transient delivery of CasX RNPs.
In addition to
potentially enhancing editing with altered cell and tissue tropism, use of
RNPs packaged within
these viral vectors also offers the unique advantage of negating the potential
risks of insertional
mutagenesis and long-term transgene expression. The purpose of the following
experiment was
to build upon the previous example and to create and identify unique CasX
delivery particles
derived from different genera of the Retroviridae family using different
architectures. The
genera investigated in the following experiments include Spumaretroviruses in
a Gag-CasX +
Gag-(-1)-Protease-CasX configuration. Here we hypothesized that by adding in
different
amounts of the protease with the Gag-Protease-CasX polyprotein along with the
Gag-CasX
polyproteins, we could potentially improve XDP particle formation and
maturation, mediated by
proteolytic cleavage.
Methods
Method for the generation of XDPs
10012421XDPs derived from Spumaretrovirinae family (BFV, EFV, FFV, BGPFV,
RHSFV and
SFV) in the 90% Gag-CasX + 10% Gag-(-1)-Protease-CasX variation (V80b, V81b,
V82b,
V83b, V84b, V85b and V86b; see FIG. 62A) were produced by transient
transfection of LentiX
HEK293T cells (Talcara Biosciences) using the plasmids portrayed in FIG. 62A
and listed in
397
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Table 36. The plasmids pXDP54, pXDP55, pXDP56, pXDP57, pXDP58, pXDP59 and
pXDP60
have been described in previous examples. The pStx42.174.12.7 plasmid was
created with a
human U6 promoter upstream of a CasX guide RNA cassette having scaffold 174
and spacer
components (targeted to tdTomato: CTGCATTCTAGTTGTGGTTT, SEQ ID NO: 825) in a
single-guide format. Plasmids containing VSV-G (pGP2) for pseudotyping the XDP
were also
used. MI plasmids contained either an ampicillin or kanamycin resistance gene.
The sequences
incorporated into the plasmids are presented in Table 36 and A .
Table 36: Plasmid Sequences
Version
XDP plasmid
SEQ ID NO of DNA Sequence
number
N/A p5tx42.174.12.7
1001
pGP2
1002
80a pXDP78
1003
81a pXDP79
1004
82a pXDP80
1005
83a pXDP81
1006
84a pXDP82
1007
85a pXDP83
1008
86a pXDP84
1009
Transfection
0012431 The steps for creation of the XDP are depicted graphically in FIG. 24.
HEK293T
Lenti-X cells were maintained in 10% FBS supplemented DMEM with HEPES,
penicillin/streptomycin (Pen/Step), sodium pyruvate, and 2-mercaptoethanol.
Cells were seeded
in two 15 cm dishes at 8e6 cells per dish in 10 inL of media. Cells were
allowed to settle and
grow for 24 hours before transfection. At the time of transfection cells were
70-90% confluent.
For transfection, the following plasmid amounts were used for the structural
plasmid
individually: pXDP78 + pXDP54 (146 pig + 15 pig), pXDP81 + pXDP57 (150 pig +
15 jig),
pXDP82 + pXDP58 (146 pig + 15 pig), pXDP83 + pXDP59 (154 pig + 15.4 pg). Along
with the
structural plasmid, each transfection also received 26.3 pig of
pStx42.174.12.7, and the 5 itg of
398
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
pGP2 in 3800 I of Opti-MEM media. 1 mg/ml linear polyethylenimine (PEI,
MW=25,000 Da)
was then added to the plasmid mixture at 1:3 DNA:PEI concentration, mixed, and
allowed to
incubate at room temperature before being added to the cell culture.
Collection and concentration
[0012441Media was changed on cells 24 hours post-transfection. XDP-containing
media was
collected 72 hours post-transfection and filtered through a 0.45 M filter
using a 60 mL syringe.
The filtered supernatant was concentrated by centrifugation at 17,000 x g at 4
C for 4h using a
10% sucrose buffer in NTE. The concentrated XDPs were held at -20 C until use.
Editing of tdTomato neural progenitor cells using XDP
10012451tdTomato neural progenitor cells (MT NPCs) were grown in DMEM F12
supplemented
with glutamax, I-IEPES, non-essential amino acids, Pen/Strep, 2-
mercaptoethanol, B-27 without
vitamin A, and N2. Cells were harvested using a Takara Biosciences Neuron
Dissociation Kit
and seeded on PLF coated 96 well plates. Cells were allowed to grow at 37 C
for 48 hours
before being treated with targeting XDPs (having a spacer for tdTomato) as a
10x concentrate
from the sucrose buffer concentrates using half-log dilutions. NPCs were grown
for 96 hours
before analysis of fluorescence as a marker of editing of tdTomato. Version 18
with pXDP32
serves as the control for these experiments.
[0012461Results: The results of the editing assay are shown in FIGS. 73A and
B, FIG. 74 and in
Table 37 and Table 38 below. FIGS. 73 A and B shows the percentage editing
efficacy for
specific amounts of the various XDP versions in tdTomato NPCs. Fig 74 shows
specifically the
editing efficacy of the various XDP versions when 16.6 I of the concentrated
XDP prep is used
to treat tdTomato NPCs. Tables 37 and 38 represent the results showing %
editing of the
dtTomato target sequence when 50 1 and 16.6 .I of the concentrated XDP prep
were used to
treat NPCs. The results indicate that, under the conditions of the assay, XDPs
constructed using
members of the Retroviridae in 90% Gag-CasX + 10% Gag-protease-CasX
configuration of the
XDP, were able, for the majority of the genera, to result in significant
editing of the target
nucleic acid in the NPC cells, with several editing above 10%.
Table 37: Results of Editing Assay for the first dilution (50u1)
Version Genus/order Virus
Plasmid Editing %
668 Spumaretrovirus BFV
pXDP78 + pXDP54 33,5
698 Spumaretrovirus EFV
pXDP81 + pXDP57 3.3
399
CA 03159320 2022-5-24
WO 2021/113772
PCT/US2020/063488
Version Genus/order Virus
Plasm id Editing %
70B Spumaretrovirus FFV
OCDP82 + OCDP58 3.5
87B Spumaretrovirus RHSFV
OCDP83 + OCDP59 213
Table 38: Results of Editing Assay for the second dilution (16.6u1)
1
1 1
Version Genus/order Virus
Plasm id Editing %
66B Spumaretrovirus BFV
pXDP78 + pXDP54 1.8
69B Spumaretrovirus EFV
pXDP81 + pXDP57 0.7
708 Spumaretrovirus FFV
pXDP82 + pXDP58 0.6
878 Spumaretrovirus RHSFV
pXDP83 + pXDP59 9.3
400
CA 03159320 2022-5-24