Note: Descriptions are shown in the official language in which they were submitted.
WO 2021/168014
PCT/US2021/018422
KNOWLEDGE DISTILLATION AND GRADIENT PRUNING-BASED COMPRESSION
OF ARTIFICIAL INTELLIGENCE-BASED BASE CALLER
FIELD OF THE TECHNOLOGY DISCLOSED
100011 The technology disclosed relates to artificial intelligence
type computers and digital
data processing systems and corresponding data processing methods and products
for emulation
of intelligence (i.e., knowledge based systems, reasoning systems, and
knowledge acquisition
systems); and including systems for reasoning with uncertainty (e.g., fuzzy
logic systems),
adaptive systems, machine learning systems, and artificial neural networks. In
particular, the
technology disclosed relates to using deep neural networks such as deep
convolutional neural
networks for analyzing data.
PRIORITY APPLICATION
100021 This PCT application claims priority to and benefit of U.S.
Provisional Patent
Application No. 62/979,385, titled "KNOWLEDGE DISTILLATION-BASED COMPRESSION
OF ARTIFICIAL INTELLIGENCE-BASED BASE CALLER," filed 20 February 2020
(Attorney Docket No. ILLM 1017-1/IP-1859-PRV) and U.S. Patent Application No.
17/176,151,
titled "KNOWLEDGE DISTILLATION-BASED COMPRESSION OF ARTIFICIAL
INTELLIGENCE-BASED BASE CALLER," filed 15 February 2021 (Attorney Docket No.
ILLM 1017-2/IP-1859-US). The priority applications are hereby incorporated by
reference for all
purposes as if fully set forth herein.
INCORPORATIONS
100031 The following are incorporated by reference as if fully set
forth herein.
100041 U.S. Provisional Patent Application No. 62/979,384, titled
"ARTIFICIAL
INTELLIGENCE-BASED BASE CALLING OF INDEX SEQUENCES," filed 20 February
2020 (Attorney Docket No. ILLM 1015-1/IP-1857-PRV);
100051 U. S . Provisional Patent Application No. 62/979,414, titled
"ARTIFICIAL
INTELLIGENCE-BASED MANY-TO-MANY BASE CALLING," filed 20 February 2020
(Attorney Docket No. ILLM 1016-1/IP-1858-PRV);
100061 U.S. Provisional Patent Application No. 63/072,032, titled
"DETECTING AND
FILTERING CLUSTERS BASED ON ARTIFICIAL INTELLIGENCE-PREDICTED BASE
CALLS," filed 28 August 2020 (Attorney Docket No. ILLM 1018-1/IP-1860-PRV);
100071 U.S. Provisional Patent Application No. 62/979,412, titled
"MULTI-CYCLE
CLUSTER BASED REAL TIME ANALYSIS SYSTEM," filed 20 February 2020 (Attorney
Docket No. ILLM 1020-1/IP-1866-PRV);
1
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0008] U. S . Provisional Patent Application No. 62/979,411, titled
"DATA COMPRESSION
FOR ARTIFICIAL INTELLIGENCE-BASED BASE CALLING," filed 20 February 2020
(Attorney Docket No. ILLM 1029-1/IP-1964-PRV);
[0009] U. S . Provisional Patent Application No. 62/979,399, titled
"SQUEEZING LAYER
FOR ARTIFICIAL INTELLIGENCE-BASED BASE CALLING," filed 20 February 2020
(Attorney Docket No. ILLM 1030-1/IP-1982-PRV);
[0010] U. S . Nonprovisional Patent Application No. 16/825,987,
titled "TRAINING DATA
GENERATION FOR ARTIFICIAL INTELLIGENCE-BASED SEQUENCING," filed 20
March 2020 (Attorney Docket No. ILLM 1008-16/IP-1693-US);
[0011] U. S . Nonprovisional Patent Application No. 16/825,991
titled "ARTIFICIAL
INTELLIGENCE-BASED GENERATION OF SEQUENCING METADATA," filed 20 March
2020 (Attorney Docket No. ILLM 1008-17/IP-1741-US);
[0012] U. S . Nonprovisional Patent Application No. 16/826,126,
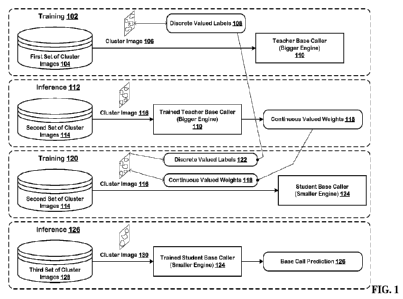
titled "ARTIFICIAL
INTELLIGENCE-BASED BASE CALLING," filed 20 March 2020 (Attorney Docket No.
ILLM 1008-18/IP-1744-US);
[0013] U. S . Nonprovisional Patent Application No. 16/826,134,
titled "ARTIFICIAL
INTELLIGENCE-BASED QUALITY SCORING," filed 20 March 2020 (Attorney Docket No.
ILLM 1008-19/IP-1747-US); and
[0014] U. S . Nonprovisional Patent Application No. 16/826,168,
titled "ARTIFICIAL
INTELLIGENCE-BASED SEQUENCING," filed 21 March 2020 (Attorney Docket No. ILLM
1008-20/IP-1752-PRV-US).
BACKGROUND
[0015] The subject matter discussed in this section should not be
assumed to be prior art
merely as a result of its mention in this section. Similarly, a problem
mentioned in this section or
associated with the subject matter provided as background should not be
assumed to have been
previously recognized in the prior art. The subject matter in this section
merely represents
different approaches, which in and of themselves can also correspond to
implementations of the
claimed technology.
[0016] In order to deploy efficient deep neural networks on mobile
devices, academia and
industry have put forward a number of model compression methods. The
compression methods
can be broadly classified into four categories: parameter sharing, network
pruning, low-rank
factorization, and knowledge distillation. In knowledge distillation, the
knowledge embedded in
the cumbersome model, known as the teacher model, is distilled to guide the
training of a smaller
model called the student model. The student model has a different architecture
and fewer
2
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
parameters but can achieve comparable performance by mimicking the behavior of
the
cumbersome model. Other compression methods like quantization and low-rank
factorization are
complementary to knowledge distillation and can also be used to further reduce
the size of
student models.
[0017] An opportunity arises to accelerate artificial intelligence-
based base calling using
knowledge distillation.
BRIEF DESCRIPTION OF THE DRAWINGS
[0018] In the drawings, like reference characters generally refer
to like parts throughout the
different views. Also, the drawings are not necessarily to scale, with an
emphasis instead
generally being placed upon illustrating the principles of the technology
disclosed. In the
following description, various implementations of the technology disclosed are
described with
reference to the following drawings, in which:
[0019] Figure 1 illustrates various aspects of using the disclosed
knowledge distillation for
artificial intelligence-based base calling
[0020] Figure 2A depicts one implementation of training a teacher
base caller by using a first
set of cluster images that are annotated with first ground truth data which
uses discrete valued
labels (one-hot encoding) to identify a correct base call.
[0021] Figure 2B depicts another implementation of training the
teacher base caller by using
the first set of cluster images that are annotated with the first ground truth
data which uses the
discrete valued labels (softened one-hot encoding) to identify the correct
base call.
[0022] Figure 3 shows one implementation of applying the trained
teacher base caller on a
second set of cluster images and generating base call predictions that are
represented by
continuous valued weights.
[0023] Figures 4A and 4B illustrate one implementation of so-called
"hybrid ground truth
data" generation using a combination of the discrete valued labels and the
continuous valued
weights.
[0024] Figure 5 is one implementation of training the student base
caller using the second set
of cluster images that are annotated with the hybrid ground truth data which
identifies a correct
base call based on the discrete valued labels and the continuous valued
weights.
[0025] Figure 6 shows one implementation of applying the trained
student base caller on a
third set of cluster images and generating base call predictions.
[0026] Figure 7 illustrates one implementation of data processing
by the teacher and student
base callers.
3
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0027] Figures 8A and 8B depict one implementation of a sequencer
that uses the student
base caller for base calling.
[0028] Figure 8C is a simplified block diagram of a system for
analysis of sensor data from
the sequencing system, such as base call sensor outputs.
[0029] Figure 8D is a simplified diagram showing aspects of the
base calling operation,
including functions of a runtime program executed by a host processor.
[0030] Figure 8E is a simplified diagram of a configuration of a
configurable processor 846
such as that of Figure 8C.
[0031] Figure 9 is a simplified block diagram of a computer system
that can be used to
implement the technology disclosed.
[0032] Figure 10A shows one implementation of training a first base
caller over cluster
intensity images and producing a first trained base caller. Figure 10B shows
one implementation
of the first trained base caller mapping the cluster intensity images to base
call predictions
[0033] Figures 11A and 11B show various aspects of a loop
implemented by the technology
disclosed to perform computationally efficient base calling.
[0034] Figure 12 illustrates one implementation of generating
contribution scores for the
cluster feature maps.
[0035] Figure 13 shows one implementation of an artificial
intelligence-based method of
performing computationally efficient base calling.
[0036] Figure 14 shows another implementation of an artificial
intelligence-based method of
performing computationally efficient base calling.
[0037] Figures 15A, 15B, 15C, 15D, 15E, and 15F are performance
results that demonstrate
that the technology disclosed implements computationally efficient base
calling.
[0038] Figure 16 shows one implementation of a larger, teacher base
caller with 251,220
total parameters.
[0039] Figure 17 shows one implementation of a smaller, student
base caller with 12,970
total parameters that is distilled from the larger, teacher base caller of
Figure 16 using the
technology disclosed.
[0040] Figure 18 shows the base calling performance of a smaller,
student base caller against
the base calling performance of a larger, teacher base caller.
[0041] Figure 19 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution weights for a distilled base
caller.
[0042] Figure 20 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution biases for a distilled base
caller.
4
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0043] Figure 21 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution weights for a distilled base
caller in which
regularization is applied to both the convolution weights and the convolution
biases.
[0044] Figure 22 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution biases for a distilled base caller
in which
regularization is applied to both the convolution weights and the convolution
biases.
DETAILED DESCRIPTION
[0045] The following discussion is presented to enable any person
skilled in the art to make
and use the technology disclosed and is provided in the context of a
particular application and its
requirements. Various modifications to the disclosed implementations will be
readily apparent to
those skilled in the art, and the general principles defined herein may be
applied to other
implementations and applications without departing from the spirit and scope
of the technology
disclosed. Thus, the technology disclosed is not intended to be limited to the
implementations
shown but is to be accorded the widest scope consistent with the principles
and features
disclosed herein.
Introduction
[0046] The technology disclosed compresses a larger, teacher base
caller into a smaller,
distilled student base caller. The student base caller has fewer processing
modules and
parameters than the teacher base caller. The larger, teacher base caller can
comprise an ensemble
of larger, teacher base callers. The teacher base caller is trained using hard
labels (e.g., one-hot
encodings). The trained teacher base caller is used to generate soft labels as
output probabilities
during the inference phase. The soft labels are used to train the student base
caller.
[0047] A hard label is a one-hot vector where all entries are set
to zero aside from a single
entry, the one corresponding to the correct class, which is set to one. In
contrast, the soft labels
form a probability distribution over the possible classes. The idea is that a
cluster image contains
information about more than one class (e.g., a cluster image of the base call
"A" looks a lot like
other cluster images of base call "A," but it also looks like some cluster
images of the base call
"C"). Using soft labels allows us to convey more information about the
associated cluster image,
which is particularly useful in detecting boundaries between clusters in a
cluster image.
[0048] This application refers to the teacher base caller as the
first base caller, the bigger
engine, and the bigger model. This application refers to the student base
caller as the second base
caller, the smaller engine, and the smaller model. This application refers to
the hard labels as
discrete valued labels. This application refers to the soft labels as
continuous valued weights.
The student base caller can be used for executing the sequencing run in an
online model where
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
base calls are generated in real-time on a cycle-by-cycle basis such as that
the student base caller
processes incoming images for a current sequencing cycle, generates base calls
for the current
sequencing cycle, processes incoming images for a next sequencing cycle,
generates base calls
for the next sequencing cycle, and so on.
Base Callers
[0049] The discussion begins with data processing by the teacher
base caller 110 and the
student base caller 124, which are trained to map sequencing images to base
calls. In Figure 7,
for purposes of illustration of the data processing, base caller 730 is
representative of both the
teacher base caller 110 and the student base caller 124; however, the student
base caller 124 has
fewer processing modules and parameters than the teacher base caller 110. In
one
implementation, one of the processing modules is neural network layers. In one
implementation,
one of the parameters is interconnections between the neural network layers.
In one
implementation, one of the processing modules is neural network filters. In
one implementation,
one of the processing modules is neural network kernels In one implementation,
one of the
parameters is multiplication and addition operations.
[0050] Base calling is the process of determining the nucleotide
composition of a sequence.
Base calling involves analyzing image data, i.e., sequencing images produced
during the
sequencing reaction carried out by a sequencing instrument such as Illumina's
i Seq, HiSeqX,
HiSeq 3000, HiSeq 4000, Hi Seq 2500, NovaSeq 6000, NextSeq, NextSeqDx, MiSeq,
and
MiSeqDx. The following discussion outlines how the sequencing images are
generated and what
they depict, in accordance with one implementation.
[0051] Base calling decodes the raw signal of the sequencing
instrument, i.e., intensity data
extracted from the sequencing images, into nucleotide sequences. In one
implementation, the
Illumina platforms employ cyclic reversible termination (CRT) chemistry for
base calling. The
process relies on growing nascent strands complementary to template strands
with fluorescently-
labeled nucleotides, while tracking the emitted signal of each newly added
nucleotide. The
fluorescently-labeled nucleotides have a 3' removable block that anchors a
fluorophore signal of
the nucleotide type.
[0052] Sequencing occurs in repetitive cycles, each comprising
three steps: (a) extension of a
nascent strand by adding the fluorescently-labeled nucleotide; (b) excitation
of the fluorophore
using one or more lasers of an optical system of the sequencing instrument and
imaging through
different filters of the optical system, yielding the sequencing images; and
(c) cleavage of the
fluorophore and removal of the 3' block in preparation for the next sequencing
cycle.
Incorporation and imaging cycles are repeated up to a designated number of
sequencing cycles,
6
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
defining the read length. Using this approach, each cycle interrogates a new
position along the
template strands.
[0053] The tremendous power of the Illumina platforms stems from
their ability to
simultaneously execute and sense millions or even billions of analytes (e.g.,
clusters) undergoing
CRT reactions. A cluster comprises approximately one thousand identical copies
of a template
strand, though clusters vary in size and shape. The clusters are grown from
the template strand,
prior to the sequencing run, by bridge amplification of the input library. The
purpose of the
amplification and cluster growth is to increase the intensity of the emitted
signal since the
imaging device cannot reliably sense fluorophore signal of a single strand.
However, the physical
distance of the strands within a cluster is small, so the imaging device
perceives the cluster of
strands as a single spot.
[0054] Sequencing occurs in a flow cell ¨ a small glass slide that
holds the input strands. The
flow cell is connected to the optical system, which comprises microscopic
imaging, excitation
lasers, and fluorescence filters. The flow cell comprises multiple chambers
called lanes. The
lanes are physically separated from each other and may contain different
tagged sequencing
libraries, distinguishable without sample cross contamination. The imaging
device of the
sequencing instrument (e.g., a solid-state imager such as a charge-coupled
device (CCD) or a
complementary metal¨oxide¨semiconductor (CMOS) sensor) takes snapshots at
multiple
locations along the lanes in a series of non-overlapping regions called tiles.
For example, there
are hundred tiles per lane in Illumina's Genome Analyzer II and sixty-eight
tiles per lane in
Illumina's HiSeq 2000. A tile holds hundreds of thousands to millions of
clusters.
[0055] The output of the sequencing is the sequencing images, each
depicting intensity
emissions of the clusters and their surrounding background. The sequencing
images depict
intensity emissions generated as a result of nucleotide incorporation in the
sequences during the
sequencing. The intensity emissions are from associated analytes and their
surrounding
background.
[0056] The following discussion is organized as follows. First, the
input to the base caller
730 is described, in accordance with one implementation. Then, examples of the
structure and
form of the base caller 730 are provided. Finally, the output of the base
caller 730 is described, in
accordance with one implementation.
[0057] Additional details about the base caller 730 can be found in
US Provisional Patent
Application No. 62/821,766, titled "ARTIFICIAL INTELLIGENCE-BASED SEQUENCING,"
(Attorney Docket No. ILLM 1008-9/IP-1752-PRV), filed on March 21, 2019, which
is
incorporated herein by reference.
7
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0058] In one implementation, image patches are extracted from the
sequencing images. The
extracted image patches are provided to the base caller 730 as "input image
data" 726 for base
calling. The image patches have dimensions 14) x h, where Iv (width) and h
(height) are any
numbers ranging from 1 and 10,000 (e.g., 3 x 3, 5 x 5, 7 x 7, 10 x 10, 15 x
15, 25 x 25). In some
implementations, 14) and h are the same. In other implementations, w and h are
different.
[0059] Sequencing produces m image(s) per sequencing cycle for
corresponding m image
channels. In one implementation, each image channel corresponds to one of a
plurality of filter
wavelength bands. In another implementation, each image channel corresponds to
one of a
plurality of imaging events at a sequencing cycle. In yet another
implementation, each image
channel corresponds to a combination of illumination with a specific laser and
imaging through a
specific optical filter.
[0060] An image patch is extracted from each of the m image(s) to
prepare the input image
data 726 for a particular sequencing cycle. In different implementations such
as 4-, 2-, and 1-
channel chemistries, in is 4 or 2. In other implementations, Fr/ is 1, 3, or
greater than 4. The input
image data 726 is in the optical, pixel domain in some implementations, and in
the upsampled,
subpixel domain in other implementations.
[0061] Consider, for example, that sequencing uses two different
image channels. a red
channel and a green channel. Then, at each sequencing cycle, sequencing
produces a red image
and a green image. This way, for a series of k sequencing cycle, a sequence
with k pairs of red
and green images is produced as output.
[0062] The input image data 726 comprises a sequence of per-cycle
image patches generated
for a series of k sequencing cycles of a sequencing run. The per-cycle image
patches contain
intensity data for associated analytes and their surrounding background in one
or more image
channels (e.g., a red channel and a green channel). In one implementation,
when a single target
analyte (e.g., cluster) is to be base called, the per-cycle image patches are
centered at a center
pixel that contains intensity data for a target associated analyte and non-
center pixels in the per-
cycle image patches contain intensity data for associated analytes adjacent to
the target
associated analyte.
[0063] The input image data 726 comprises data for multiple
sequencing cycles (e.g., a
current sequencing cycle, one or more preceding sequencing cycles, and one or
more successive
sequencing cycles). In one implementation, the input image data 726 comprises
data for three
sequencing cycles, such that data for a current (time t) sequencing cycle to
be base called is
accompanied with (i) data for a left flanking/context/previous/preceding/prior
(time t-1)
sequencing cycle and (ii) data for a right
flanking/context/next/successive/subsequent (time t+1)
sequencing cycle. In other implementations, the input image data 726 comprises
data for a single
8
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
sequencing cycle. In yet other implementations, the input image data 726
comprises data for 58,
75, 92, 130, 168, 175, 209, 225, 230, 275, 318, 325, 330, 525, or 625
sequencing cycles.
[0064] In one implementation, the base caller 730 is a multilayer
perceptron (MLP). In
another implementation, the base caller 730 is a feedforward neural network.
In yet another
implementation, the base caller 730 is a fully-connected neural network. In a
further
implementation, the base caller 730 is a fully convolutional neural network.
In yet further
implementation, the base caller 730 is a semantic segmentation neural network.
In yet another
further implementation, the base caller 730 is a generative adversarial
network (GAN).
[0065] In one implementation, the base caller 730 is a
convolutional neural network (CNN)
with a plurality of convolution layers. In another implementation, it is a
recurrent neural network
(RNN) such as a long short-term memory network (LSTM), hi-directional LSTM (Bi-
LSTM), or
a gated recurrent unit (GRU) In yet another implementation, it includes both a
CNN and a RNN
[0066] In yet other implementations, the base caller 730 can use 1D
convolutions, 2D
convolutions, 3D convolutions, 4D convolutions, 5D convolutions, dilated or
atrous
convolutions, transpose convolutions, depthwise separable convolutions,
pointwise convolutions,
1 x 1 convolutions, group convolutions, flattened convolutions, spatial and
cross-channel
convolutions, shuffled grouped convolutions, spatial separable convolutions,
and
deconvolutions. It can use one or more loss functions such as logistic
regression/log loss, multi-
class cross-entropy/softmax loss, binary cross-entropy loss, mean-squared
error loss, Li loss, L2
loss, smooth Li loss, and Huber loss. It can use any parallelism, efficiency,
and compression
schemes such TFRecords, compressed encoding (e.g, PNG), sharding, parallel
calls for map
transformation, batching, prefetching, model parallelism, data parallelism,
and
synchronous/asynchronous stochastic gradient descent (SGD). It can include
upsampling layers,
downsampling layers, recurrent connections, gates and gated memory units (like
an LSTM or
GRU), residual blocks, residual connections, highway connections, skip
connections, peephole
connections, activation functions (e.g., non-linear transformation functions
like rectifying linear
unit (ReLU), leaky ReLU, exponential liner unit (ELU), sigmoid and hyperbolic
tangent (tanh)),
batch normalization layers, regularization layers, dropout, pooling layers
(e.g., max or average
pooling), global average pooling layers, and attention mechanisms.
[0067] In one implementation, the base caller 730 outputs a base
call for a single target
analyte for a particular sequencing cycle. In another implementation, it
outputs a base call for
each target analyte in a plurality of target analytes for the particular
sequencing cycle. In yet
another implementation, it outputs a base call for each target analyte in a
plurality of target
analytes for each sequencing cycle in a plurality of sequencing cycles,
thereby producing a base
call sequence for each target analyte.
9
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0068] In one implementation, the sequencing images 704, 714 from
the current (time t)
sequencing cycle are accompanied with the sequencing images 702, 712 from the
preceding
(time t-1) sequencing cycle and the sequencing images 706, 716 from the
succeeding (time t+1)
sequencing cycle. The base caller 730 processes the sequencing images 702,
712, 704, 714, 706,
and 716 through its convolution layers and produces an alternative
representation, according to
one implementation. The alternative representation is then used by an output
layer (e.g., a
softmax layer) for generating a base call for either just the current (time t)
sequencing cycle or
each of the sequencing cycles, i.e., the current (time t) sequencing cycle,
the preceding (time t-1)
sequencing cycle, and the succeeding (time t+1) sequencing cycle. The
resulting base calls 732
form the sequencing reads.
[0069] In one implementation, a patch extraction process 724
extracts patches from the
sequencing images 702, 712, 704, 714, 706, and 716 and generates the input
image data 726
Then, the extracted images patches in the input image data 726 are provided to
the base caller
730 as input.
[0070] The teacher base caller 110 and the student base caller 124
are trained using
backpropagation-based gradient update techniques. Some types of gradient
descent techniques
that can be used for training the teacher base caller 110 and the student base
caller 124 are
stochastic gradient descent, batch gradient descent, and mini-batch gradient
descent. Some
examples of gradient descent optimization algorithms that can be used for
training the teacher
base caller 110 and the student base caller 124 are Momentum, Nesterov
accelerated gradient,
Adagrad, Adadelta, RMSprop, Adam, AdaMax, Nadam, and AMSGrad.
Knowledge Distillation
[0071] Figure 1 illustrates various aspects of using the disclosed
knowledge distillation for
artificial intelligence-based base calling. The disclosed knowledge
distillation comprises:
= Training the teacher base caller on training data using "hard labels"
= Generating "soft labels" by applying the trained teacher base caller on
inference data
= Training the student base caller on training data using a "combination"
of the
hard and soft labels, i.e., "hybrid- ground truth data
[0072] The student base caller 124 has fewer processing modules and
parameters than the
teacher base caller 110. In one implementation, one of the processing modules
is neural network
layers. In one implementation, one of the parameters is interconnections
between the neural
network layers. In one implementation, one of the processing modules is neural
network filters.
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
In one implementation, one of the processing modules is neural network
kernels. In one
implementation, one of the parameters is multiplication and addition
operations.
Training the Teacher Base Caller
[0073] During training 102, the teacher base caller 110 is trained
on training data comprising
a first set of cluster images 104. The first set of cluster images 104 are
annotated with ground
truth data that uses discrete valued labels 108.
[0074] In one implementation, a cluster image 106 is annotated with
the discrete valued
labels 108 that are one-hot encoded with a one-value for a correct base and
zero-values for
incorrect bases. The following is an example of one-hot encoding for the four
nucleotide bases:
A 1
0
0
0
[0075] Figure 2A depicts one implementation of training 200A the
teacher base caller 110 by
using the first set of cluster images 104 that are annotated with first ground
truth data 214 which
uses discrete valued labels 216 (one-hot encoding 218) to identify a correct
base call. During
forward propagation 206, the input to the teacher base caller 110 is a cluster
image 202 that
depicts intensities of clusters 204A, 204B, 204C, and 204D and their
surrounding background.
[0076] In one implementation, the cluster image 202 is accompanied
with supplemental data
224 such as a distance channel and a scaling channel. Additional details about
the supplemental
data 224 can be found in US Provisional Patent Application No. 62/821,766,
titled
"ARTIFICIAL INTELLIGENCE-BASED SEQUENCING," (Attorney Docket No. ILLM 1008-
9/IP-1752-PRV), filed on March 21, 2019, which is incorporated herein by
reference.
[0077] In response to processing the cluster image 202, the teacher
base caller 110 produces
an output 208. Based on the output 208, a base call prediction 210 is made
that identifies
confidence scores assigned by the teacher base caller 110 to each of the bases
A, C, T, and G.
[0078] Then, an error 212 is computed between the base call
prediction 210 and the discrete
valued labels 216, e.g., one-hot encoding 218, i.e., [1, 0, 0, 0]. Backward
propagation 220
updates weights and parameters of the teacher base caller 110 based on the
error 212.
[0079] This process is iterated until the teacher base caller 110
converges to a desired base
call accuracy on a validation dataset. The training is operationalized
(implemented) by a trainer
222 using backpropagation-based gradient update techniques (such as the ones
discussed above).
[0080] In another implementation, the cluster image 106 is
annotated with the discrete
valued labels 108 that have a near-one-value for the correct base and near-
zero-values for the
11
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
incorrect bases, referred to herein as "softened one-hot encoding." The
following is an example
of softened one-hot encoding for the four nucleotide bases:
A 0.95
0.02
0.017
0.013
[0081] Figure 2B depicts another implementation of training 200B
the teacher base caller
110 by using the first set of cluster images 104 that are annotated with the
first ground truth data
226 which uses the discrete valued labels 216 (softened one-hot encoding 228)
to identify the
correct base call. Here, the error 212 is computed between the base call
prediction 210 and the
softened one-hot encoding 228, i.e., [0.95, 0.02, 0.017, 0.013].
Generating Soft Labels
[0082] During inference 112, the trained teacher base caller 110 is
applied on inference data
comprising a second set of cluster images 114. The trained teacher base caller
110 processes the
second set of cluster images 114 and generates base call predictions as
output. The base call
predictions are represented by continuous valued weights 118 (soft labels)
that identify a
predicted base call. The continuous valued weights 118 are part of a
probability distribution for a
correct base being Adenine (A), Cytosine (C), Thymine (T), and Guanine (G). In
one
implementation, the continuous valued weights 118 are softmax scores, i.e.,
posterior
probabilities.
[0083] In one implementation, a cluster image 116 is fed as input
to the trained teacher base
caller 110. In response, the trained teacher base caller 110 generates
exponentially normalized
likelihood of a base incorporated in a cluster depicted by the cluster image
116 at a current
sequencing cycle being A, C, T, and G.
[0084] The following is an example of the continuous valued weights
118:
A 0.175
0.024
0.475
0.326
[0085] Figure 3 shows one implementation of applying 300 the
trained teacher base caller
110 on the second set of cluster images 114 and generating a base call
prediction 312 that is
represented by the continuous valued weights 310. During forward propagation
306, the input to
the trained teacher base caller 110 is a cluster image 302 that depicts
intensities of clusters 304A,
304B, 304C, and 304D and their surrounding background. In one implementation,
the cluster
12
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
image 302 is accompanied with supplemental data 316 such as the distance
channel and the
scaling channel.
[0086] In response to processing the cluster image 302, the trained
teacher base caller 110
produces an output 308. Based on the output 308, the base call prediction 310
is generated that
identifies confidence scores assigned by the trained teacher base caller 110
to each of the bases A
(0.175), C (0.024), T (0.475), and G (0.326). These confidence scores are
represented as
continuous values, i.e., the continuous valued weights 310.
[0087] This process is iterated over numerous images in the second
set of cluster images 114,
such that a set of continuous valued weights is generated for each evaluated
cluster image. The
evaluation is operationalized (implemented) by a tester 314.
Generating Hybrid Ground Truth Data
[0088] Figures 4A and 4B illustrate one implementation of so-called
"hybrid ground truth
data" generation 400A and 400B using a combination of the discrete valued
labels 216 and the
continuous valued weights 310
[0089] In one implementation, the discrete valued labels 216 and
the continuous valued
weights 310 are accessed for a same cluster image 302 and combined to generate
hybrid ground
truth data for the cluster image 302. In some implementations, the discrete
valued labels 216 are
summed 402 with the continuous valued weights 310. In other implementations,
the discrete
valued labels 216 are multiplied with the continuous valued weights 310. In
some other
implementations, the discrete valued labels 216 are concatenated with the
continuous valued
weights 310.
[0090] In one implementation, the discrete valued labels 216 and
the continuous valued
weights 310 are combined on class-wise basis. That is, discrete valued label
for base call A is
summed, multiplied, or concatenated with continuous valued weight for base
call A, discrete
valued label for base call C is summed, multiplied, or concatenated with
continuous valued
weight for base call C, discrete valued label for base call T is summed,
multiplied, or
concatenated with continuous valued weight for base call T, and discrete
valued label for base
call G is summed, multiplied, or concatenated with continuous valued weight
for base call G.
[0091] In some implementations, prior to being combined with the
discrete valued labels
216, the continuous valued weights 310 are modified using a modification
parameter()) 404. In
one implementation, the modification parameter (X) 404 is iteratively learned
based on
performance of the student base caller 124 over a validation dataset. After
the modification
parameter (X) 404 is applied on the continuous valued weights 310, what
results is modified
continuous valued weights 406.
13
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0092] In one implementation, the discrete valued labels 216 are
combined with the is
modified continuous valued weights 406 to produce unnormalized ground truth
data 408 for the
cluster image 302. The unnormalized ground truth data 408 is then normalized
to produce
noimalized ground truth data 412 for the cluster image 302. In some
implementations, an
exponential normalizer 410 (e.g., softmax) is used to produce the normalized
ground truth data
412.
[0093] In one implementation, the unnormalized ground truth data
408 is considered the
hybrid ground truth data 414 for the cluster image 302. In another
implementation, the
normalized ground truth data 412 is considered the hybrid ground truth data
416 for the cluster
image 302.
Training the Student Base Caller
[0094] During training 120, the student base caller 124 is trained
on training data comprising
the second set of cluster images 114. The second set of cluster images 114 are
annotated with
ground truth data 414/416 that identifies a correct base call based on (i) the
discrete valued labels
122 and (ii) the continuous valued weights 118
[0095] Figure 5 is one implementation of training the student base
caller 124 using the
second set of cluster images 114 that are annotated with the hybrid ground
truth data 414/416
which identifies a correct base call based on the discrete valued labels 216
and the continuous
valued weights 310. During forward propagation 506, the input to the student
base caller 124 is
the cluster image 302 that depicts intensities of clusters 304A, 304B, 304C,
and 304D and their
surrounding background. In one implementation, the cluster image 302 is
accompanied with
supplemental data 316 such as the distance channel and the scaling channel.
[0096] In response to processing the cluster image 302, the student
base caller 124 produces
an output 508. Based on the output 508, a base call prediction 510 is made
that identifies
confidence scores assigned by the student base caller 124 to each of the bases
A, C, T, and G.
[0097] Then, an error 512 is computed between the base call
prediction 510 and the hybrid
ground truth data 414/416. Backward propagation 514 updates weights and
parameters of the
student base caller 124 based on the error 512.
[0098] This process is iterated until the student base caller 124
converges to a desired base
call accuracy on a validation dataset. The training is operationalized
(implemented) by the trainer
222 using backpropagation-based gradient update techniques (such as the ones
discussed above).
14
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
Applying the Trained Student Base Caller
10991 During inference 126, the trained student base caller 124 is
applied on inference data
comprising a third set of cluster images 128. The trained student base caller
124 processes the
third set of cluster images 128 and generates base call predictions 126 as
output.
[0100] In one implementation, a cluster image 130 is fed as input
to the trained student base
caller 124. In response, the trained student base caller 124 generates
exponentially normalized
likelihood of a base incorporated in a cluster depicted by the cluster image
130 at a current
sequencing cycle being A, C, T, and G.
[0101] Figure 6 shows one implementation of applying 600 the
trained student base caller
124 on the third set of cluster images 128 and generating a base call
prediction 610. During
forward propagation 606, the input to the trained student base caller 124 is
the cluster image 602
that depicts intensities of clusters 604A, 604B, and 604C and their
surrounding background_ In
one implementation, the cluster image 602 is accompanied with supplemental
data 612 such as
the distance channel and the scaling channel.
[0102] In response to processing the cluster image 602, the trained
student base caller 124
produces an output 608. Based on the output 608, the base call prediction 610
is generated that
identifies confidence scores assigned by the trained student base caller 124
to each of the bases A
(0.1), C (0.1), T (0.2), and G (0.6).
[0103] This process is iterated over numerous images in the third
set of cluster images 128,
such that the base call prediction is generated for each evaluated cluster
image. The evaluation is
operationalized (implemented) by the tester 314.
Technical Effect/Advantage
[0104] The teacher and student approach to transporting an
intensely trained model from a
resource-rich platform to a compact platform confers substantial technical
benefits. The
technology disclosed effectively shrinks the model and, with it, the execution
time and resources
required to analyze a particular input.
[0105] The extent of shrinkage is substantial, in most every
proportion Figures 16-17 show a
reduction in filter depth from 64 filters to 14 filters. The shrunken model
(the smaller, student
base caller) has 21.9 percent as many filters as the large model (the larger,
teacher base caller)
for the resource-rich platform. The reduction in parameters is more dramatic,
approximately
quadratic with the reduction in filter depth. Figures 16-17 show a reduction
in trainable
parameter count form 250,060 to just 12,710. The shrunken model has 5.1
percent as many
trainable parameters as the large model. For execution resources, the core
calculations can be
performed at the same time with compute resources reduced by 20-fold:
computations scale
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
about linearly with parameter count, so 5.1 percent as many parameters
translates into 5.1
percent as many calculations required to process the same inputs. Memory usage
also is reduced.
[0106] Reducing resource requirements is enabling for application
of at least some
commercially available computation accelerators, such as Xilinx FPGAs. In
general, FPGAs
have limited on-board memory and programmable footprint. The model in Figure
16 will not run
on a commercial FPGA offering such as Xilinx Alveo U200, Xilinx Alveo U250,
Xilinx Alveo
U280, Intel/Altera Stratix GX2800, Intel/Altera Stratix GX2800, and Intel
Stratix GX10M, but
the model in Figure 17 will.
[0107] Reduced resource requirements and elapsed run time are
accomplished without
compromising accuracy of results. Figure 18 graphs results accomplished
running the shrunken
model against the large model. For all models, the error rate increases
measurably as errors
accumulate over multiple cycles. At 120 cycles, the error rate for the large
model has crept up to
0.2 percent (0.002 error rate).
[0108] A new class of compact machine results, at a lower cost than
one that with resources
sufficient to run the large model. Results become available in real time,
instead of being delayed
by server-based computations. The technical improvements are manifest.
[0109] Figure 16 shows one implementation of a larger, teacher base
caller with 251,220
total parameters. The larger, teacher base caller has convolution layers that
contain 64 filters per
convolution layer.
[0110] Figure 17 shows one implementation of a smaller, student
base caller with 12,970
total parameters that is distilled from the larger, teacher base caller of
Figure 16 using the
technology disclosed. The smaller, student base caller has convolution layers
that contain 14
filters per convolution layer.
[0111] As shown in Figure 17, the smaller, student base caller has
about 5.1% of the total
parameters as the larger, teacher base caller. In other implementations when
the larger, teacher
base caller comprises an ensemble of larger, teacher base callers, the
smaller, student base caller
has about 1% to 3% of the total parameters as the ensemble of larger, teacher
base callers in the
larger, teacher base caller. This significant reduction in the total number of
model parameters
makes the smaller, student base caller much more suitable for execution on on-
chip processors
like FPGAs, GPUs, ASICs, CGRAs.
[0112] Figure 18 shows the base calling performance of a smaller,
student base caller against
the base calling performance of a larger, teacher base caller. The y-axis
represents the base
calling error rate (Error %) and the x-axis represents the sequencing cycles
of a sequencing run.
The purple line represents the larger, teacher base caller comprising an
ensemble of four larger,
teacher base callers that have 64 convolution filters per convolution layer.
The cyan line
16
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
represents the smaller, student base caller comprising that has 14 convolution
filters per
convolution layer. The smaller, student base caller (cyan line) is distilled
from the larger, teacher
base caller (purple line) using the technology disclosed.
[0113] As shown, the smaller, student base caller (cyan line) has a
base calling error rate that
is close to the base calling error rate (purple line) of the larger, teacher
base caller comprising the
ensemble of four larger, teacher base callers. Therefore, the technical
advantage and technical
effect of the technology disclosed is that the smaller, student base caller
has much smaller
compute footprint than the larger, teacher base caller, but similar/comparable
base calling
accuracy. This enables efficient execution of the smaller, student base caller
during inference on
on-chip processors like FPGAs, GPUs, ASICs, and CGRAs. This also improves the
speed of
base calling and reduces latency. This also leads to conservation of compute
resources.
[0114] More importantly, the student model, as a distilled version
of the teacher model,
outperforms another model with the same architecture whose coefficients are
independently
learned and not derived from a teacher model.
Sequencing System
[0115] Figures 8A and 8B depict one implementation of a sequencing
system 800A. The
sequencing system 800A comprises a configurable processor 846. The
configurable processor
846 implements the base calling techniques disclosed herein. The sequencing
system is also
referred to as a "sequencer."
[0116] The sequencing system 800A can operate to obtain any
information or data that
relates to at least one of a biological or chemical substance. In some
implementations, the
sequencing system 800A is a workstation that may be similar to a bench-top
device or desktop
computer. For example, a majority (or all) of the systems and components for
conducting the
desired reactions can be within a common housing 802.
[0117] In particular implementations, the sequencing system 800A is
a nucleic acid
sequencing system configured for various applications, including but not
limited to de novo
sequencing, resequencing of whole genomes or target genomic regions, and
metagenomics. The
sequencer may also be used for DNA or RNA analysis. In some implementations,
the sequencing
system 800A may also be configured to generate reaction sites in a biosensor.
For example, the
sequencing system 800A may be configured to receive a sample and generate
surface attached
clusters of clonally amplified nucleic acids derived from the sample. Each
cluster may constitute
or be part of a reaction site in the biosensor.
[0118] The exemplary sequencing system 800A may include a system
receptacle or interface
810 that is configured to interact with a biosensor 812 to perform desired
reactions within the
17
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
biosensor 812. In the following description with respect to Figure 8A, the
biosensor 812 is
loaded into the system receptacle 810. However, it is understood that a
cartridge that includes the
biosensor 812 may be inserted into the system receptacle 810 and in some
states the cartridge
can be removed temporarily or permanently. As described above, the cartridge
may include,
among other things, fluidic control and fluidic storage components.
[0119] In particular implementations, the sequencing system 800A is
configured to perform
a large number of parallel reactions within the biosensor 812. The biosensor
812 includes one or
more reaction sites where desired reactions can occur. The reaction sites may
be, for example,
immobilized to a solid surface of the biosensor or immobilized to beads (or
other movable
substrates) that are located within corresponding reaction chambers of the
biosensor. The
reaction sites can include, for example, clusters of clonally amplified
nucleic acids. The
biosensor 812 may include a solid-state imaging device (e.g., CCD or CMOS
imager) and a flow
cell mounted thereto. The flow cell may include one or more flow channels that
receive a
solution from the sequencing system 800A and direct the solution toward the
reaction sites.
Optionally, the biosensor 812 can be configured to engage a thermal element
for transferring
thermal energy into or out of the flow channel.
[0120] The sequencing system 800A may include various components,
assemblies, and
systems (or sub-systems) that interact with each other to perform a
predetermined method or
assay protocol for biological or chemical analysis. For example, the
sequencing system 800A
includes a system controller 806 that may communicate with the various
components,
assemblies, and sub-systems of the sequencing system 800A and also the
biosensor 812. For
example, in addition to the system receptacle 810, the sequencing system 800A
may also include
a fluidic control system 808 to control the flow of fluid throughout a fluid
network of the
sequencing system 800A and the biosensor 812; a fluid storage system 814 that
is configured to
hold all fluids (e.g., gas or liquids) that may be used by the bioassay
system; a temperature
control system 804 that may regulate the temperature of the fluid in the fluid
network, the fluid
storage system 814, and/or the biosensor 812; and an illumination system 816
that is configured
to illuminate the biosensor 812. As described above, if a cartridge having the
biosensor 812 is
loaded into the system receptacle 810, the cartridge may also include fluidic
control and fluidic
storage components.
[0121] Also shown, the sequencing system 800A may include a user
interface 818 that
interacts with the user. For example, the user interface 818 may include a
display 820 to display
or request information from a user and a user input device 822 to receive user
inputs. In some
implementations, the display 820 and the user input device 822 are the same
device. For
example, the user interface 818 may include a touch-sensitive display
configured to detect the
18
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
presence of an individual's touch and also identify a location of the touch on
the display.
However, other user input devices 822 may be used, such as a mouse, touchpad,
keyboard,
keypad, handheld scanner, voice-recognition system, motion-recognition system,
and the like. As
will be discussed in greater detail below, the sequencing system 800A may
communicate with
various components, including the biosensor 812 (e.g., in the form of a
cartridge), to perform the
desired reactions. The sequencing system 800A may also be configured to
analyze data obtained
from the biosensor to provide a user with desired information.
[0122] The system controller 806 may include any processor-based or
microprocessor-based
system, including systems using microcontrollers, reduced instruction set
computers (RISC),
application specific integrated circuits (ASICs), field programmable gate
array (FPGAs), coarse-
grained reconfigurable architectures (CGRAs), logic circuits, and any other
circuit or processor
capable of executing functions described herein The above examples are
exemplary only and are
thus not intended to limit in any way the definition and/or meaning of the
term system controller.
In the exemplary implementation, the system controller 806 executes a set of
instructions that are
stored in one or more storage elements, memories, or modules in order to at
least one of obtain
and analyze detection data. Detection data can include a plurality of
sequences of pixel signals,
such that a sequence of pixel signals from each of the millions of sensors (or
pixels) can be
detected over many base calling cycles. Storage elements may be in the form of
information
sources or physical memory elements within the sequencing system 800A.
[0123] The set of instructions may include various commands that
instruct the sequencing
system 800A or biosensor 812 to perform specific operations such as the
methods and processes
of the various implementations described herein. The set of instructions may
be in the form of a
software program, which may form part of a tangible, non-transitory computer
readable medium
or media. As used herein, the terms "software" and "firmware" are
interchangeable and include
any computer program stored in memory for execution by a computer, including
RAM memory,
ROM memory, EPROM memory, EEPROM memory, and non-volatile RAM (NVRAM)
memory. The above memory types are exemplary only, and are thus not limiting
as to the types
of memory usable for storage of a computer program.
[0124] The software may be in various forms such as system software
or application
software. Further, the software may be in the form of a collection of separate
programs, or a
program module within a larger program or a portion of a program module. The
software also
may include modular programming in the form of object-oriented programming.
After obtaining
the detection data, the detection data may be automatically processed by the
sequencing system
800A, processed in response to user inputs, or processed in response to a
request made by
another processing machine (e.g., a remote request through a communication
link). In the
19
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
illustrated implementation, the system controller 806 includes an analysis
module 844. In other
implementations, system controller 806 does not include the analysis module
844 and instead has
access to the analysis module 844 (e.g., the analysis module 844 may be
separately hosted on
cloud).
[0125] The system controller 806 may be connected to the biosensor
812 and the other
components of the sequencing system 800A via communication links. The system
controller 806
may also be communicatively connected to off-site systems or servers. The
communication links
may be hardwired, corded, or wireless. The system controller 806 may receive
user inputs or
commands, from the user interface 818 and the user input device 822.
[0126] The fluidic control system 808 includes a fluid network and
is configured to direct
and regulate the flow of one or more fluids through the fluid network. The
fluid network may be
in fluid communication with the biosensor 812 and the fluid storage system 814
For example,
select fluids may be drawn from the fluid storage system 814 and directed to
the biosensor 812 in
a controlled manner, or the fluids may be drawn from the biosensor 812 and
directed toward, for
example, a waste reservoir in the fluid storage system 814. Although not
shown, the fluidic
control system 808 may include flow sensors that detect a flow rate or
pressure of the fluids
within the fluid network. The sensors may communicate with the system
controller 806.
[0127] The temperature control system 804 is configured to regulate
the temperature of
fluids at different regions of the fluid network, the fluid storage system
814, and/or the biosensor
812. For example, the temperature control system 804 may include a
thermocycler that interfaces
with the biosensor 812 and controls the temperature of the fluid that flows
along the reaction
sites in the biosensor 812. The temperature control system 804 may also
regulate the temperature
of solid elements or components of the sequencing system 800A or the biosensor
812. Although
not shown, the temperature control system 804 may include sensors to detect
the temperature of
the fluid or other components. The sensors may communicate with the system
controller 806.
[0128] The fluid storage system 814 is in fluid communication with
the biosensor 812 and
may store various reaction components or reactants that are used to conduct
the desired reactions
therein. The fluid storage system 814 may also store fluids for washing or
cleaning the fluid
network and biosensor 812 and for diluting the reactants. For example, the
fluid storage system
814 may include various reservoirs to store samples, reagents, enzymes, other
biomolecules,
buffer solutions, aqueous, and non-polar solutions, and the like. Furthermore,
the fluid storage
system 814 may also include waste reservoirs for receiving waste products from
the biosensor
812. In implementations that include a cartridge, the cartridge may include
one or more of a fluid
storage system, fluidic control system or temperature control system.
Accordingly, one or more
of the components set forth herein as relating to those systems can be
contained within a
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
cartridge housing. For example, a cartridge can have various reservoirs to
store samples,
reagents, enzymes, other biomolecules, buffer solutions, aqueous, and non-
polar solutions,
waste, and the like. As such, one or more of a fluid storage system, fluidic
control system or
temperature control system can be removably engaged with a bioassay system via
a cartridge or
other biosensor.
[0129] The illumination system 816 may include a light source
(e.g., one or more LEDs) and
a plurality of optical components to illuminate the biosensor. Examples of
light sources may
include lasers, arc lamps, LEDs, or laser diodes. The optical components may
be, for example,
reflectors, dichroics, beam splitters, collimators, lenses, filters, wedges,
prisms, mirrors,
detectors, and the like. In implementations that use an illumination system,
the illumination
system 816 may be configured to direct an excitation light to reaction sites.
As one example,
fluorophores may be excited by green wavelengths of light, as such the
wavelength of the
excitation light may be approximately 532 nm. In one implementation, the
illumination system
816 is configured to produce illumination that is parallel to a surface normal
of a surface of the
biosensor 812. In another implementation, the illumination system 816 is
configured to produce
illumination that is off-angle relative to the surface normal of the surface
of the biosensor 812. In
yet another implementation, the illumination system 816 is configured to
produce illumination
that has plural angles, including some parallel illumination and some off-
angle illumination.
[0130] The system receptacle or interface 810 is configured to
engage the biosensor 812 in at
least one of a mechanical, electrical, and fluidic manner. The system
receptacle 810 may hold the
biosensor 812 in a desired orientation to facilitate the flow of fluid through
the biosensor 812.
The system receptacle 810 may also include electrical contacts that are
configured to engage the
biosensor 812 so that the sequencing system 800A may communicate with the
biosensor 812
and/or provide power to the biosensor 812. Furthermore, the system receptacle
810 may include
fluidic ports (e.g., nozzles) that are configured to engage the biosensor 812.
In some
implementations, the biosensor 812 is removably coupled to the system
receptacle 810 in a
mechanical manner, in an electrical manner, and also in a fluidic manner.
[0131] In addition, the sequencing system 800A may communicate
remotely with other
systems or networks or with other bioassay systems 800A. Detection data
obtained by the
bioassay system(s) 800A may be stored in a remote database.
[0132] Figure 8B is a block diagram of a system controller 806 that
can be used in the
system of Figure 8A. In one implementation, the system controller 806 includes
one or more
processors or modules that can communicate with one another. Each of the
processors or
modules may include an algorithm (e.g., instructions stored on a tangible
and/or non-transitory
computer readable storage medium) or sub-algorithms to perform particular
processes. The
21
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
system controller 806 is illustrated conceptually as a collection of modules,
but may be
implemented utilizing any combination of dedicated hardware boards, DSPs,
processors, etc.
Alternatively, the system controller 806 may be implemented utilizing an off-
the-shelf PC with a
single processor or multiple processors, with the functional operations
distributed between the
processors. As a further option, the modules described below may be
implemented utilizing a
hybrid configuration in which certain modular functions are performed
utilizing dedicated
hardware, while the remaining modular functions are performed utilizing an off-
the-shelf PC and
the like. The modules also may be implemented as software modules within a
processing unit.
101331 During operation, a communication port 850 may transmit
information (e.g.,
commands) to or receive information (e.g., data) from the biosensor 812
(Figure 8A) and/or the
sub-systems 808, 814, 804 (Figure 8A). In implementations, the communication
port 850 may
output a plurality of sequences of pixel signals A communication link 834 may
receive user
input from the user interface 818 (Figure 8A) and transmit data or information
to the user
interface 818. Data from the biosensor 812 or sub-systems 808, 814, 804 may be
processed by
the system controller 806 in real-time during a bioassay session. Additionally
or alternatively,
data may be stored temporarily in a system memory during a bioassay session
and processed in
slower than real-time or off-line operation.
[0134] As shown in Figure 8B, the system controller 806 may include
a plurality of modules
828-844 that communicate with a main control module 824, along with a central
processing unit
(CPU) 852. The main control module 824 may communicate with the user interface
818 (Figure
8A). Although the modules 828-844 are shown as communicating directly with the
main control
module 824, the modules 828-844 may also communicate directly with each other,
the user
interface 818, and the biosensor 812. Also, the modules 828-844 may
communicate with the
main control module 824 through the other modules.
[0135] The plurality of modules 828-844 include system modules 828-
832, 826 that
communicate with the sub-systems 808, 814, 804, and 816, respectively. The
fluidic control
module 828 may communicate with the fluidic control system 808 to control the
valves and flow
sensors of the fluid network for controlling the flow of one or more fluids
through the fluid
network. The fluid storage module 830 may notify the user when fluids are low
or when the
waste reservoir is at or near capacity. The fluid storage module 830 may also
communicate with
the temperature control module 832 so that the fluids may be stored at a
desired temperature. The
illumination module 826 may communicate with the illumination system 816 to
illuminate the
reaction sites at designated times during a protocol, such as after the
desired reactions (e.g.,
binding events) have occurred. In some implementations, the illumination
module 826 may
22
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
communicate with the illumination system 816 to illuminate the reaction sites
at designated
angles.
101361 The plurality of modules 828-844 may also include a device
module 836 that
communicates with the biosensor 812 and an identification module 838 that
determines
identification information relating to the biosensor 812. The device module
836 may, for
example, communicate with the system receptacle 810 to confirm that the
biosensor has
established an electrical and fluidic connection with the sequencing system
800A. The
identification module 838 may receive signals that identify the biosensor 812.
The identification
module 838 may use the identity of the biosensor 812 to provide other
information to the user.
For example, the identification module 838 may determine and then display a
lot number, a date
of manufacture, or a protocol that is recommended to be run with the biosensor
812.
101371 The plurality of modules 828-844 also includes an analysis
module 844 (also called
signal processing module or signal processor) that receives and analyzes the
signal data (e.g.,
image data) from the biosensor 812. Analysis module 844 includes memory (e.g.,
RAM or
Flash) to store detection/image data. Detection data can include a plurality
of sequences of pixel
signals, such that a sequence of pixel signals from each of the millions of
sensors (or pixels) can
be detected over many base calling cycles. The signal data may be stored for
subsequent analysis
or may be transmitted to the user interface 818 to display desired information
to the user. In
some implementations, the signal data may be processed by the solid-state
imager (e.g-,., CMOS
image sensor) before the analysis module 844 receives the signal data.
[0138] The analysis module 844 is configured to obtain image data
from the light detectors
at each of a plurality of sequencing cycles. The image data is derived from
the emission signals
detected by the light detectors and process the image data for each of the
plurality of sequencing
cycles through the student base caller 124 and produce a base call for at
least some of the
analytes at each of the plurality of sequencing cycle. The light detectors can
be part of one or
more over-head cameras (e.g., Illumina's GAIIx's CCD camera taking images of
the clusters on
the biosensor 812 from the top), or can be part of the biosensor 812 itself
(e.g., Illumina's iSeq's
CMOS image sensors underlying the clusters on the biosensor 812 and taking
images of the
clusters from the bottom).
[0139] The output of the light detectors is the sequencing images,
each depicting intensity
emissions of the clusters and their surrounding background. The sequencing
images depict
intensity emissions generated as a result of nucleotide incorporation in the
sequences during the
sequencing. The intensity emissions are from associated analytes and their
surrounding
background. The sequencing images are stored in memory 848.
23
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0140] Protocol modules 840 and 842 communicate with the main
control module 824 to
control the operation of the sub-systems 808, 814, and 804 when conducting
predetermined
assay protocols. The protocol modules 840 and 842 may include sets of
instructions for
instructing the sequencing system 800A to perform specific operations pursuant
to
predetermined protocols. As shown, the protocol module may be a sequencing-by-
synthesis
(SBS) module 840 that is configured to issue various commands for performing
sequencing-by-
synthesis processes. In SBS, extension of a nucleic acid primer along a
nucleic acid template is
monitored to determine the sequence of nucleotides in the template. The
underlying chemical
process can be polymerization (e.g., as catalyzed by a polymerase enzyme) or
ligation (e.g.,
catalyzed by a ligase enzyme). In a particular polymerase-based SBS
implementation,
fluorescently labeled nucleotides are added to a primer (thereby extending the
primer) in a
template dependent fashion such that detection of the order and type of
nucleotides added to the
primer can be used to determine the sequence of the template. For example, to
initiate a first SBS
cycle, commands can be given to deliver one or more labeled nucleotides, DNA
polymerase,
etc., into/through a flow cell that houses an array of nucleic acid templates.
The nucleic acid
templates may be located at corresponding reaction sites. Those reaction sites
where primer
extension causes a labeled nucleotide to be incorporated can be detected
through an imaging
event. During an imaging event, the illumination system 816 may provide an
excitation light to
the reaction sites. Optionally, the nucleotides can further include a
reversible termination
property that terminates further primer extension once a nucleotide has been
added to a primer.
For example, a nucleotide analog having a reversible terminator moiety can be
added to a primer
such that subsequent extension cannot occur until a deblocking agent is
delivered to remove the
moiety. Thus, for implementations that use reversible termination a command
can be given to
deliver a deblocking reagent to the flow cell (before or after detection
occurs). One or more
commands can be given to effect wash(es) between the various delivery steps.
The cycle can
then be repeated n times to extend the primer by n nucleotides, thereby
detecting a sequence of
length n. Exemplary sequencing techniques are described, for example, in
Bentley et al., Nature
456:53-59 (2008); WO 04/018497; US 7,057,026; WO 91/06678; WO 07/123744; US
7,329,492; US 7,211,414; US 7,315,019; US 7,405,281, and US 2008/014708082,
each of which
is incorporated herein by reference.
[0141] For the nucleotide delivery step of an SBS cycle, either a
single type of nucleotide
can be delivered at a time, or multiple different nucleotide types (e.g., A,
C, T and G together)
can be delivered. For a nucleotide delivery configuration where only a single
type of nucleotide
is present at a time, the different nucleotides need not have distinct labels
since they can be
distinguished based on temporal separation inherent in the individualized
delivery. Accordingly,
24
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
a sequencing method or apparatus can use single color detection. For example,
an excitation
source need only provide excitation at a single wavelength or in a single
range of wavelengths.
For a nucleotide delivery configuration where delivery results in multiple
different nucleotides
being present in the flow cell at one time, sites that incorporate different
nucleotide types can be
distinguished based on different fluorescent labels that are attached to
respective nucleotide types
in the mixture. For example, four different nucleotides can be used, each
having one of four
different fluorophores. In one implementation, the four different fluorophores
can be
distinguished using excitation in four different regions of the spectrum. For
example, four
different excitation radiation sources can be used. Alternatively, fewer than
four different
excitation sources can be used, but optical filtration of the excitation
radiation from a single
source can be used to produce different ranges of excitation radiation at the
flow cell.
[0142] In some implementations, fewer than four different colors
can be detected in a
mixture having four different nucleotides. For example, pairs of nucleotides
can be detected at
the same wavelength, but distinguished based on a difference in intensity for
one member of the
pair compared to the other, or based on a change to one member of the pair
(e.g., via chemical
modification, photochemical modification or physical modification) that causes
apparent signal
to appear or disappear compared to the signal detected for the other member of
the pair.
Exemplary apparatus and methods for distinguishing four different nucleotides
using detection of
fewer than four colors are described for example in US Pat. App. Ser. Nos.
61/538,294 and
61/619,878, which are incorporated herein by reference in their entireties.
U.S. Application No.
13/624,200, which was filed on September 21, 2012, is also incorporated by
reference in its
entirety.
[0143] The plurality of protocol modules may also include a sample-
preparation (or
generation) module 842 that is configured to issue commands to the fluidic
control system 808
and the temperature control system 804 for amplifying a product within the
biosensor 812. For
example, the biosensor 812 may be engaged to the sequencing system 800A. The
amplification
module 842 may issue instructions to the fluidic control system 808 to deliver
necessary
amplification components to reaction chambers within the biosensor 812. In
other
implementations, the reaction sites may already contain some components for
amplification,
such as the template DNA and/or primers. After delivering the amplification
components to the
reaction chambers, the amplification module 842 may instruct the temperature
control system
804 to cycle through different temperature stages according to known
amplification protocols. In
some implementations, the amplification and/or nucleotide incorporation is
performed
isothermally.
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0144] The SBS module 840 may issue commands to perform bridge PCR
where clusters of
clonal amplicons are formed on localized areas within a channel of a flow
cell. After generating
the amplicons through bridge PCR, the amplicons may be "linearized" to make
single stranded
template DNA, or sstDNA, and a sequencing primer may be hybridized to a
universal sequence
that flanks a region of interest. For example, a reversible terminator-based
sequencing by
synthesis method can be used as set forth above or as follows.
[0145] Each base calling or sequencing cycle can extend an sstDNA
by a single base which
can be accomplished for example by using a modified DNA polymerase and a
mixture of four
types of nucleotides. The different types of nucleotides can have unique
fluorescent labels, and
each nucleotide can further have a reversible terminator that allows only a
single-base
incorporation to occur in each cycle. After a single base is added to the
sstDNA, excitation light
may be incident upon the reaction sites and fluorescent emissions may be
detected After
detection, the fluorescent label and the terminator may be chemically cleaved
from the sstDNA.
Another similar base calling or sequencing cycle may follow. In such a
sequencing protocol, the
SBS module 840 may instruct the fluidic control system 808 to direct a flow of
reagent and
enzyme solutions through the biosensor 812. Exemplary reversible terminator-
based SBS
methods which can be utilized with the apparatus and methods set forth herein
are described in
US Patent Application Publication No. 2007/0166705 Al, US Patent Application
Publication
No. 2006/0188901 Al, US Patent No. 7,057,026, US Patent Application
Publication No.
2006/0240439 Al, US Patent Application Publication No. 2006/02814714709 Al,
PCT
Publication No. WO 05/065814, US Patent Application Publication No.
2005/014700900 Al,
PCT Publication No. WO 06/08B199 and PCT Publication No. WO 07/01470251, each
of which
is incorporated herein by reference in its entirety. Exemplary reagents for
reversible terminator-
based SBS are described in US 7,541,444; US 7,057,026; US 7,414,14716; US
7,427,673; US
7,566,537; US 7,592,435 and WO 07/14835368, each of which is incorporated
herein by
reference in its entirety.
[0146] In some implementations, the amplification and SBS modules
may operate in a single
assay protocol where, for example, template nucleic acid is amplified and
subsequently
sequenced within the same cartridge.
[0147] The sequencing system 800A may also allow the user to
reconfigure an assay
protocol. For example, the sequencing system 800A may offer options to the
user through the
user interface 818 for modifying the determined protocol. For example, if it
is determined that
the biosensor 812 is to be used for amplification, the sequencing system 800A
may request a
temperature for the annealing cycle. Furthermore, the sequencing system 800A
may issue
26
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
warnings to a user if a user has provided user inputs that are generally not
acceptable for the
selected assay protocol.
[0148] In implementations, the biosensor 812 includes millions of
sensors (or pixels), each
of which generates a plurality of sequences of pixel signals over successive
base calling cycles.
The analysis module 844 detects the plurality of sequences of pixel signals
and attributes them to
corresponding sensors (or pixels) in accordance to the row-wise and/or column-
wise location of
the sensors on an array of sensors.
[0149] Figure 8C is a simplified block diagram of a system for
analysis of sensor data from
the sequencing system 800A, such as base call sensor outputs. In the example
of Figure 8C, the
system includes the configurable processor 846. The configurable processor 846
can execute a
base caller (e.g., the student base caller 124) in coordination with a runtime
program executed by
the central processing unit (CPU) 852 (i.e., a host processor) The sequencing
system 800A
comprises the biosensor 812 and flow cells. The flow cells can comprise one or
more tiles in
which clusters of genetic material are exposed to a sequence of analyte flows
used to cause
reactions in the clusters to identify the bases in the genetic material. The
sensors sense the
reactions for each cycle of the sequence in each tile of the flow cell to
provide tile data. Genetic
sequencing is a data intensive operation, which translates base call sensor
data into sequences of
base calls for each cluster of genetic material sensed in during a base call
operation.
[0150] The system in this example includes the CPU 852, which
executes a runtime program
to coordinate the base call operations, memory 848B to store sequences of
arrays of tile data,
base call reads produced by the base calling operation, and other information
used in the base
call operations. Also, in this illustration the system includes memory 848A to
store a
configuration file (or files), such as FPGA bit files, and model parameters
for the neural
networks used to configure and reconfigure the configurable processor 846, and
execute the
neural networks. The sequencing system 800A can include a program for
configuring a
configurable processor and in some embodiments a reconfigurable processor to
execute the
neural networks.
[0151] The sequencing system 800A is coupled by a bus 854 to the
configurable processor
846. The bus 854 can be implemented using a high throughput technology, such
as in one
example bus technology compatible with the PCIe standards (Peripheral
Component
Interconnect Express) currently maintained and developed by the PCI-SIG (PCI
Special Interest
Group). Also in this example, a memory 848A is coupled to the configurable
processor 846 by
bus 856. The memory 848A can be on-board memory, disposed on a circuit board
with the
configurable processor 846. The memory 848A is used for high speed access by
the configurable
processor 846 of working data used in the base call operation. The bus 856 can
also be
27
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
implemented using a high throughput technology, such as bus technology
compatible with the
PCIe standards.
[0152] Configurable processors, including field programmable gate
arrays FPGAs, coarse
grained reconfigurable arrays CGRAs, and other configurable and reconfigurable
devices, can be
configured to implement a variety of functions more efficiently or faster than
might be achieved
using a general purpose processor executing a computer program. Configuration
of configurable
processors involves compiling a functional description to produce a
configuration file, referred to
sometimes as a bitstream or bit file, and distributing the configuration file
to the configurable
elements on the processor. The configuration file defines the logic functions
to be executed by
the configurable processor, by configuring the circuit to set data flow
patterns, use of distributed
memory and other on-chip memory resources, lookup table contents, operations
of configurable
logic blocks and configurable execution units like multiply-and-accumulate
units, configurable
interconnects and other elements of the configurable array. A configurable
processor is
reconfigurable if the configuration file may be changed in the field, by
changing the loaded
configuration file. For example, the configuration file may be stored in
volatile SRAM elements,
in non-volatile read-write memory elements, and in combinations of the same,
distributed among
the array of configurable elements on the configurable or reconfigurable
processor. A variety of
commercially available configurable processors are suitable for use in a base
calling operation as
described herein. Examples include Google's Tensor Processing Unit (TPU)Tm,
rackmount
solutions like GX4 Rackmount SeriesTM, GX9 Rackmount SeriesTM, NVIDIA DGX-1Tm,
Microsoft' Stratix V FPGATM, Graphcore's Intelligent Processor Unit (IPU)TM,
Qualcomm's
Zeroth PlatformTM with Snapdragon processorsTM, NVIDIA' s VoltaTm, NVIDIA' s
DRIVE PXTM,
NV1DIA' s JETSON TX1/TX2 MODULETM, Intel's NirvanaTM, Movidius VPUTM, Fujitsu
DPITM, ARM' s DynamiclQTM, IBM TrueNorthTm, Lambda GPU Server with Testa
V100sT1,
Xilinx AlveoTM U200, Xilinx AlveoTM U250, Xilinx AlveoTM U280, Intel/Altera
StratixTM
GX2800, Intel/Altera StratixTM GX2800, and Intel StratixTM GX10M. In some
examples, a host
CPU can be implemented on the same integrated circuit as the configurable
processor.
[0153] Embodiments described herein implement the student base
caller 124 using the
configurable processor 846. The configuration file for the configurable
processor 846 can be
implemented by specifying the logic functions to be executed using a high
level description
language EIDL or a register transfer level RTL language specification. The
specification can be
compiled using the resources designed for the selected configurable processor
to generate the
configuration file. The same or similar specification can be compiled for the
purposes of
generating a design for an application-specific integrated circuit which may
not be a configurable
processor.
28
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0154] Alternatives for the configurable processor configurable
processor 846, in all
embodiments described herein, therefore include a configured processor
comprising an
application specific ASIC or special purpose integrated circuit or set of
integrated circuits, or a
system-on-a-chip SOC device, or a graphics processing unit (GPU) processor or
a coarse-grained
reconfigurable architecture (CGRA) processor, configured to execute a neural
network based
base call operation as described herein.
[0155] In general, configurable processors and configured
processors described herein, as
configured to execute runs of a neural network, are referred to herein as
neural network
processors.
[0156] The configurable processor 846 is configured in this example
by a configuration file
loaded using a program executed by the CPU 852, or by other sources, which
configures the
array of configurable elements 866 (e.g., configuration logic blocks (CLB)
such as look up tables
(LUTs), flip-flops, compute processing units (PMUs), and compute memory units
(CMUs),
configurable I/0 blocks, programmable interconnects), on the configurable
processor to execute
the base call function. In this example, the configuration includes data flow
logic 862 which is
coupled to the buses 854 and 856 and executes functions for distributing data
and control
parameters among the elements used in the base call operation.
[0157] Also, the configurable processor 846 is configured with base
call execution logic 862
to execute the student base caller 124. The logic 862 comprises multi-cycle
execution clusters
(e.g., 864) which, in this example, includes execution cluster 1 through
execution cluster X. The
number of multi-cycle execution clusters can be selected according to a trade-
off involving the
desired throughput of the operation, and the available resources on the
configurable processor
846.
[0158] The multi-cycle execution clusters are coupled to the data
flow logic 862 by data flow
paths 858 implemented using configurable interconnect and memory resources on
the
configurable processor 846. Also, the multi-cycle execution clusters are
coupled to the data flow
logic 862 by control paths 860 implemented using configurable interconnect and
memory
resources for example on the configurable processor 846, which provide control
signals
indicating available execution clusters, readiness to provide input units for
execution of a run of
the student base caller 124 to the available execution clusters, readiness to
provide trained
parameters for the student base caller 124, readiness to provide output
patches of base call
classification data, and other control data used for execution of the student
base caller 124.
[0159] The configurable processor 846 is configured to execute runs
of the student base
caller 124 using trained parameters to produce classification data for the
sensing cycles of the
base calling operation. A run of the student base caller 124 is executed to
produce classification
29
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
data for a subject sensing cycle of the base calling operation. A run of the
student base caller 124
operates on a sequence including a number N of arrays of tile data from
respective sensing cycles
of N sensing cycles, where the N sensing cycles provide sensor data for
different base call
operations for one base position per operation in time sequence in the
examples described herein.
Optionally, some of the N sensing cycles can be out of sequence if needed
according to a
particular neural network model being executed. The number N can be any number
greater than
one. In some examples described herein, sensing cycles of the N sensing cycles
represent a set of
sensing cycles for at least one sensing cycle preceding the subject sensing
cycle and at least one
sensing cycle following the subject cycle in time sequence. Examples are
described herein in
which the number N is an integer equal to or greater than five.
[0160] The data flow logic 862 is configured to move tile data and
at least some trained
parameters of the model parameters from the memory 848A to the configurable
processor 846
for runs of the student base caller 124, using input units for a given run
including tile data for
spatially aligned patches of the N arrays The input units can be moved by
direct memory access
operations in one DMA operation, or in smaller units moved during available
time slots in
coordination with the execution of the neural network deployed.
[0161] Tile data for a sensing cycle as described herein can
comprise an array of sensor data
having one or more features. For example, the sensor data can comprise two
images which are
analyzed to identify one of four bases at a base position in a genetic
sequence of DNA, RNA, or
other genetic material. The tile data can also include metadata about the
images and the sensors.
For example, in embodiments of the base calling operation, the tile data can
comprise
information about alignment of the images with the clusters such as distance
from center
information indicating the distance of each pixel in the array of sensor data
from the center of a
cluster of genetic material on the tile.
[0162] During execution of the student base caller 124 as described
below, tile data can also
include data produced during execution of the student base caller 124,
referred to as intermediate
data, which can be reused rather than recomputed during a run of the student
base caller 124. For
example, during execution of the student base caller 124, the data flow logic
862 can write
intermediate data to the memory 848A in place of the sensor data for a given
patch of an array of
tile data. Embodiments like this are described in more detail below.
[0163] As illustrated, a system is described for analysis of base
call sensor output,
comprising memory (e.g., 848A) accessible by the runtime program storing tile
data including
sensor data for a tile from sensing cycles of a base calling operation. Also,
the system includes a
neural network processor, such as configurable processor 846 having access to
the memory. The
neural network processor is configured to execute runs of a neural network
using trained
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
parameters to produce classification data for sensing cycles. As described
herein, a run of the
neural network is operating on a sequence of N arrays of tile data from
respective sensing cycles
of N sensing cycles, including a subject cycle, to produce the classification
data for the subject
cycle. The data flow logic 862 is provided to move tile data and the trained
parameters from the
memory to the neural network processor for runs of the neural network using
input units
including data for spatially aligned patches of the N arrays from respective
sensing cycles of N
sensing cycles.
[0164] Also, a system is described in which the neural network
processor has access to the
memory, and includes a plurality of execution clusters, the execution clusters
in the plurality of
execution clusters configured to execute a neural network. The data flow logic
862 has access to
the memory and to execution clusters in the plurality of execution clusters,
to provide input units
of tile data to available execution clusters in the plurality of execution
clusters, the input units
including a number N of spatially aligned patches of arrays of tile data from
respective sensing
cycles, including a subject sensing cycle, and to cause the execution clusters
to apply the N
spatially aligned patches to the neural network to produce output patches of
classification data
for the spatially aligned patch of the subject sensing cycle, where N is
greater than 1.
[0165] Figure 8D is a simplified diagram showing aspects of the
base calling operation,
including functions of a runtime program executed by a host processor. In this
diagram, the
output of image sensors from a flow cell are provided on lines 868 to image
processing threads
869, which can perform processes on images such as alignment and arrangement
in an array of
sensor data for the individual tiles and resampling of images, and can be used
by processes
which calculate a tile cluster mask for each tile in the flow cell, which
identifies pixels in the
array of sensor data that correspond to clusters of genetic material on the
corresponding tile of
the flow cell. The outputs of the image processing threads 869 are provided on
lines 870 to a
dispatch logic 877 in the CPU which routes the arrays of tile data to a data
cache 872 (e.g., SSD
storage) on a high-speed bus 871, or on high-speed bus 873 to the neural
network processor
hardware 874, such as the configurable processor 846 of Figure 8C, according
to the state of the
base calling operation. The processed and transformed images can be stored on
the data cache
872 for sensing cycles that were previously used. The hardware 874 returns
classification data
output by the neural network to the dispatch logic 877, which passes the
information to the data
cache 872, or on lines 875 to threads 870 that perform base call and quality
score computations
using the classification data, and can arrange the data in standard formats
for base call reads. The
outputs of the threads 870 that perform base calling and quality score
computations are provided
on lines 876 to threads 871 that aggregate the base call reads, perform other
operations such as
31
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
data compression, and write the resulting base call outputs to specified
destinations for
utilization by the customers.
[0166] In some embodiments, the host can include threads (not
shown) that perform final
processing of the output of the hardware 874 in support of the neural network.
For example, the
hardware 874 can provide outputs of classification data from a final layer of
the multi-cluster
neural network. The host processor can execute an output activation function,
such as a softmax
function, over the classification data to configure the data for use by the
base call and quality
score threads 870. Also, the host processor can execute input operations (not
shown), such as
batch normalization of the tile data prior to input to the hardware 874.
[0167] Figure 8E is a simplified diagram of a configuration of a
configurable processor 846
such as that of Figure 8C. In Figure 8E, the configurable processor 846
comprises an FPGA with
a plurality of high speed PCIe interfaces The FPGA is configured with a
wrapper 880 which
comprises the data flow logic 862 described with reference to Figure 8C. The
wrapper 880
manages the interface and coordination with a runtime program in the CPU
across the CPU
communication link 878 and manages communication with the on-board DRAM 879
(e.g.,
memory 848A) via DRAM communication link 881. The data flow logic 862 in the
wrapper 880
provides patch data retrieved by traversing the arrays of tile data on the on-
board DRAM 879 for
the number N cycles to a cluster 884, and retrieves process data 882 from the
cluster 884 for
delivery back to the on-board DRAM 879. The wrapper 880 also manages transfer
of data
between the on-board DRAM 879 and host memory, for both the input arrays of
tile data, and for
the output patches of classification data. The wrapper transfers patch data on
line 887 to the
allocated cluster 884. The wrapper provides trained parameters, such as
weights and biases on
line 886 to the cluster 884 retrieved from the on-board DRAM 302. The wrapper
provides
configuration and control data on line 885 to the cluster 884 provided from,
or generated in
response to, the runtime program on the host via the CPU communication link
878. The cluster
can also provide status signals on line 883 to the wrapper 880, which are used
in cooperation
with control signals from the host to manage traversal of the arrays of tile
data to provide
spatially aligned patch data, and to execute the multi-cycle neural network
over the patch data
using the resources of the cluster 884.
[0168] As mentioned above, there can be multiple clusters on a
single configurable processor
managed by the wrapper 880 configured for executing on corresponding ones of
multiple patches
of the tile data. Each cluster can be configured to provide classification
data for base calls in a
subject sensing cycle using the tile data of multiple sensing cycles described
herein.
[0169] In examples of the system, model data, including kernel data
like filter weights and
biases can be sent from the host CPU to the configurable processor, so that
the model can be
32
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
updated as a function of cycle number. A base calling operation can comprise,
for a
representative example, on the order of hundreds of sensing cycles. Base
calling operation can
include paired end reads in some embodiments. For example, the model trained
parameters may
be updated once every 20 cycles (or other number of cycles), or according to
update patterns
implemented for particular systems and neural network models. In some
embodiments including
paired end reads in which a sequence for a given string in a genetic cluster
on a tile includes a
first part extending from a first end down (or up) the string, and a second
part extending from a
second end up (or down) the string, the trained parameters can be updated on
the transition from
the first part to the second part.
[0170] In some examples, image data for multiple cycles of sensing
data for a tile can be sent
from the CPU to the wrapper 880. The wrapper 880 can optionally do some pre-
processing and
transformation of the sensing data and write the information to the on-board
DRAM 879 The
input tile data for each sensing cycle can include arrays of sensor data
including on the order of
4000 x 3000 pixels per sensing cycle per tile or more, with two features
representing colors of
two images of the tile, and one or two bytes per feature per pixel. For an
embodiment in which
the number N is three sensing cycles to be used in each run of the multi-cycle
neural network,
the array of tile data for each run of the multi-cycle neural network can
consume on the order of
hundreds of megabytes per tile. In some embodiments of the system, the tile
data also includes
an array of DFC data, stored once per tile, or other type of metadata about
the sensor data and the
tiles.
[0171] In operation, when a multi-cycle cluster is available, the
wrapper allocates a patch to
the cluster. The wrapper fetches a next patch of tile data in the traversal of
the tile and sends it to
the allocated cluster along with appropriate control and configuration
information. The cluster
can be configured with enough memory on the configurable processor to hold a
patch of data
including patches from multiple cycles in some systems, that is being worked
on in place, and a
patch of data that is to be worked on when the current patch of processing is
finished using a
ping-pong buffer technique or raster scanning technique in various
embodiments.
[0172] When an allocated cluster completes its run of the neural
network for the current
patch and produces an output patch, it will signal the wrapper. The wrapper
will read the output
patch from the allocated cluster, or alternatively the allocated cluster will
push the data out to the
wrapper. Then the wrapper will assemble output patches for the processed tile
in the DRAM 879.
When the processing of the entire tile has been completed, and the output
patches of data
transferred to the DRAM, the wrapper sends the processed output array for the
tile back to the
host/CPU in a specified format. In some embodiments, the on-board DRAM 879 is
managed by
memory management logic in the wrapper 880. The runtime program can control
the sequencing
33
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
operations to complete analysis of all the arrays of tile data for all the
cycles in the run in a
continuous flow to provide real time analysis.
[0173] "Logic" (e.g., data flow logic), as used herein, can be
implemented in the form of a
computer product including a non-transitory computer readable storage medium
with computer
usable program code for performing the method steps described herein. The -
logic- can be
implemented in the form of an apparatus including a memory and at least one
processor that is
coupled to the memory and operative to perform exemplary method steps. The
"logic" can be
implemented in the form of means for carrying out one or more of the method
steps described
herein; the means can include (i) hardware module(s), (ii) software module(s)
executing on one
or more hardware processors, or (iii) a combination of hardware and software
modules; any of
(i)-(iii) implement the specific techniques set forth herein, and the software
modules are stored in
a computer readable storage medium (or multiple such media) In one
implementation, the logic
implements a data processing function The logic can be a general purpose,
single core or
multicore, processor with a computer program specifying the function, a
digital signal processor
with a computer program, configurable logic such as an FPGA with a
configuration file, a
special purpose circuit such as a state machine, or any combination of these.
Also, a computer
program product can embody the computer program and configuration file
portions of the logic.
Computer System
[0174] Figure 9 is a computer system 900 that can be used by the
sequencing system 800A to
implement the base calling techniques disclosed herein. Computer system 900
includes at least
one central processing unit (CPU) 972 that communicates with a number of
peripheral devices
via bus subsystem 955. These peripheral devices can include a storage
subsystem 858 including,
for example, memory devices and a file storage subsystem 936, user interface
input devices 938,
user interface output devices 976, and a network interface subsystem 974. The
input and output
devices allow user interaction with computer system 900. Network interface
subsystem 974
provides an interface to outside networks, including an interface to
corresponding interface
devices in other computer systems.
[0175] In one implementation, the system controller 806 is
communicably linked to the
storage subsystem 858 and the user interface input devices 938.
[0176] User interface input devices 938 can include a keyboard;
pointing devices such as a
mouse, trackball, touchpad, or graphics tablet; a scanner; a touch screen
incorporated into the
display; audio input devices such as voice recognition systems and
microphones; and other types
of input devices. In general, use of the term -input device" is intended to
include all possible
types of devices and ways to input information into computer system 900.
34
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0177] User interface output devices 976 can include a display
subsystem, a printer, a fax
machine, or non-visual displays such as audio output devices. The display
subsystem can include
an LED display, a cathode ray tube (CRT), a flat-panel device such as a liquid
crystal display
(LCD), a projection device, or some other mechanism for creating a visible
image. The display
subsystem can also provide a non-visual display such as audio output devices.
In general, use of
the term "output device" is intended to include all possible types of devices
and ways to output
information from computer system 900 to the user or to another machine or
computer system.
[0178] Storage subsystem 858 stores programming and data constructs
that provide the
functionality of some or all of the modules and methods described herein.
These software
modules are generally executed by deep learning processors 978.
[0179] Deep learning processors 978 can be graphics processing
units (GPUs), field-
programmable gate arrays (FPGAs), application-specific integrated circuits
(ASICs), and/or
coarse-grained reconfigurable architectures (CGRAs). Deep learning processors
978 can be
hosted by a deep learning cloud platform such as Google Cloud PlatformTm,
XilinxTM, and
CirrascaleTM. Examples of deep learning processors 978 include Google's Tensor
Processing
Unit (TPU)Tm, rackmount solutions like GX4 Rackmount SeriesTM, GX9 Rackmount
SeriesTM,
NVIDIA DGX-1 TM, Microsoft' Stratix V FPGATM, Graphcore' s Intelligent
Processor Unit
opuvrivi
) , Qualcomm's Zeroth PlatformTM with Snapdragon processorsTM,
NVIDIA's VoltaTM,
NVIDIA' s DRIVE PXTM, NVIDIA' s JETSON TX1/TX2 MODULETM, Intel's NirvanaTM,
Movidius VPUTM, Fujitsu DPITM, ARM' s DynamiclQTM, IBM TrueNorthm, Lambda GPU
Server with Testa V100sT11, and others.
[0180] Memory subsystem 922 used in the storage subsystem 858 can
include a number of
memories including a main random access memory (RAM) 932 for storage of
instructions and
data during program execution and a read only memory (ROM) 934 in which fixed
instructions
are stored. A file storage subsystem 936 can provide persistent storage for
program and data
files, and can include a hard disk drive, a floppy disk drive along with
associated removable
media, a CD-ROM drive, an optical drive, or removable media cartridges. The
modules
implementing the functionality of certain implementations can be stored by
file storage
subsystem 936 in the storage subsystem 858, or in other machines accessible by
the processor.
[0181] Bus subsystem 955 provides a mechanism for letting the
various components and
subsystems of computer system 900 communicate with each other as intended.
Although bus
subsystem 955 is shown schematically as a single bus, alternative
implementations of the bus
subsystem can use multiple busses.
[0182] Computer system 900 itself can be of varying types including
a personal computer, a
portable computer, a workstation, a computer terminal, a network computer, a
television, a
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
mainframe, a server farm, a widely-distributed set of loosely networked
computers, or any other
data processing system or user device. Due to the ever-changing nature of
computers and
networks, the description of computer system 900 depicted in Figure 9 is
intended only as a
specific example for purposes of illustrating the preferred implementations of
the present
invention. Many other configurations of computer system 900 are possible
having more or less
components than the computer system depicted in Figure 9.
Pruning
[0183] We also disclose an artificial intelligence-based technology
of performing
computationally efficient base calling. Figure 10A shows one implementation of
training 1004 a
first base caller 1006 over cluster intensity images 1002 and producing a
first trained base caller
1006. Figure 10B shows one implementation of the first trained base caller
1006 mapping the
cluster intensity images 1002 (e.g., the cluster image 1008) to base call
predictions 1010.
[0184] Figures 11A and 11B show various aspects of a loop
implemented by the technology
disclosed to perform computationally efficient base calling
[0185] A controller 1148 begins with the first trained base caller
1006 and executes a loop
1102 in which each iteration uses a starting trained base caller 1112 as input
and produces a
pruned trained base caller 1142 as output. The pruned trained base caller 1142
has fewer
processing elements than the starting trained base caller 1112. In one
implementation, the first
trained base caller 1006 is a neural network, and the processing elements are
neurons of the
neural network. In another implementation, the first trained base caller 1006
is a convolutional
neural network, and the processing elements are convolutional filters of the
convolutional neural
network. In yet another implementation, the processing elements are
convolutional kernels of the
convolutional neural network. In yet further implementation, the processing
elements are weights
of the convolutional kernels of the convolutional neural network. In another
implementation, the
first trained base caller 1006 is a recurrent neural network, and the
processing elements are
weights of gates of the recurrent neural network.
[0186] In yet further implementation, the first trained base caller
1006 is a fully-connected
neural network.
[0187] In yet further implementation, the processing elements are
the cluster feature maps.
The cluster feature maps can be convolved features or convolved
representations when the first
trained base caller 1006 is a convolutional neural network. The cluster
feature maps can be
hidden state features or hidden state representations when the first trained
base caller 1006 is a
recurrent neural network.
36
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0188] When a convolution filter convolves over a cluster image (or
a cluster intensity
image), the resulting output is called the cluster feature map. Similarly,
when a convolution filter
convolves over a cluster feature map produced in an another convolution layer
(e.g., a preceding
convolution layer), the resulting output is also called the cluster feature
map. In one
implementation, the cluster feature map is produced by element-wise
multiplying the elements of
the convolution filter (neurons) with corresponding elements (e.g., intensity
values) of the cluster
intensity image, or of a cluster feature map produced in an another
convolution layer (e.g., a
preceding convolution layer), and summing the results of the element-wise
multiplication to
produce the cluster feature map.
[0189] A cluster feature maps generator 1150, in each iteration,
during forward propagation
1108, processes a subset 1106 of the clusters intensity images (e.g, cluster
image 1110) through
the processing elements of the starting trained base caller 1112, generates
one or more cluster
feature maps 1114 using each processing element, and produces the base call
predictions 1116
based on the cluster feature maps 1114. This is considered the cluster feature
maps generation
step 1104.
[0190] A gradient determiner 1152, in each iteration, during
backward propagation 1126,
determines gradients 1124 for the cluster feature maps 1114 based on error
1122 between the
base call predictions 1116 and ground truth base calls 1120. This is
considered the gradient
determination step 1118.
[0191] A contribution measurer 1154, in each iteration, applies the
gradients 1124 to
respective ones of the cluster feature maps 1114 and generates a contribution
score 1130 for each
of the cluster feature maps 1114 that identifies how much a cluster feature
map contributed to the
base call predictions 1116. This is considered the contribution measurement
step 1128.
[0192] Figure 12 illustrates one implementation of generating
contribution scores for the
cluster feature maps. In one implementation, the contribution score 1214 for a
cluster feature
map 1202 is generated by multiplying each of the feature values 1204 in the
cluster feature map
1202 with a respective one of the gradients 1206 and producing intermediate
feature values
1208, applying an absolute function 1210 to the intermediate feature values
1208 and generating
absolute intermediate feature values 1212, and summing the absolute
intermediate feature values
1212 and producing the contribution score 1214 for the cluster feature map
1202.
[0193] In another implementation, the contribution score 1214 for
the cluster feature map
1202 is generated without using the gradients 1206. This includes applying the
absolute function
1210 to the feature values (weights) 1204 in the cluster feature map 1202 and
generating
absolute feature values, and summing the absolute feature values to produce
the contribution
score 1214 for the cluster feature map 1202.
37
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0194] A pruner 1156, in each iteration, selects a subset 1134 of
the cluster feature maps
based on their contribution scores 1130 and produces the pruned trained base
caller 1142 by
removing, from the starting trained base caller 1112, those processing
elements 1138 that were
used to generate the selected subset 1134 of the cluster feature maps during
the forward
propagation 1108. This is considered the pruning step 1132.
[0195] A retrainer 1158, in each iteration, further trains the
pruned trained base caller 1142
over the cluster intensity images 1002 and makes the pruned trained base
caller 1142 available
for a successive iteration as the starting trained base caller 1112.
[0196] A terminator 1160 terminates the loop 1102 after n
iterations and uses the pruned
trained base caller 1142 produced by the nth iteration for further base
calling.
[0197] In one implementation, each iteration, during the forward
propagation, processes the
subset of the clusters intensity images through the processing elements of the
starting trained
base caller in batches. In such an implementation, the gradients for the
cluster feature map are
determined on a batch-by-batch basis, the absolute intermediate feature values
for the cluster
feature map are generated on the batch-by-batch basis, and the contribution
score for the cluster
feature map is generated by summing the absolute intermediate feature values
for each of the
batches.
[0198] In one implementation, Lp normalization is used for the
training of the first base
caller 1106. The Lp normalization can be L-1 normalization, L-2 normalization,
and L-infinity
normalization. In one implementation, for a first iteration, the Lp
normalization produces a
subset of the cluster feature maps whose contribution score is zero. For the
first iteration, the
pruning step 1132 first removes, from the first trained base caller 1006,
those processing
elements that were used generate the cluster feature maps whose contribution
score is zero due to
the Lp normalization, and then removes, from the first trained base caller
1006, the processing
elements that were used to generate the selected subset 1134 of the cluster
feature maps during
the forward propagation.
[0199] Other examples of normalization include L-0 normalization,
absolute-value
normalization, Euclidean normalization, Taxicab or Manhattan normalization, p-
normalization,
maximum normalization, infinity normalization, uniform normalization, supremum
normalization, and zero normalization. Additional information about and
examples of
normalization techniques can be found at Wikipedia
(https://en.wikipedia.org/wiki/Norm_(mathematics) - 1/15/2029, 9:54 AM).
[0200] In some implementations, each convolution filter can be
normalized by the count of
pixels in the convolution filter. The normalization can be along the spatial
and/or time
dimension. That is, the count of the pixels/feature
values/units/size/dimensionality of the
38
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
resulting feature map for an input cluster intensity image of size 115x115 at
the spatial layer 1
can be 113x113, whereas at the temporal layer 7 it can be 101x101. The
resulting sum of dot
product of absolute values can be normalized by the filter size. Also, the
temporal layers of the
base caller have a time component dimension that varies, which can be used for
the
normalization.
[0201] In one implementation, the subset 1106 of the cluster
feature maps are selected based
on a percentage of the cluster feature maps that have the lowest contribution
scores. In some
implementations, the percentage ranges from 2% to 5%.
[0202] In one implementation, the number of epochs used in the
retraining step 1144 to
further train the pruned trained base caller 1142 is less than the number of
epochs used in the
training 1004 of the first base caller 1006. For example, the number of epochs
used in the
retraining step 1144 to further train the pruned trained base caller 1142 is
fifteen and the number
of epochs used in the training 1004 of the first base caller 10006 is fifty.
[0203] In one implementation, the subset 1106 of the clusters
intensity images used in the
cluster feature maps generation step 1104 is 15% to 30% of the cluster
intensity images 1002
used for the training 1004 of the first base caller 1006, and is randomly
selected at each iteration.
In other implementations, the subset 1106 of the clusters intensity images
used in the cluster
feature maps generation step 1104 can be less than 15% and more than 30% of
the cluster
intensity images 1002, or between 15% and 30%.
[0204] Figure 13 shows one implementation of an artificial
intelligence-based method of
performing computationally efficient base calling.
[0205] At action 1302, the method incudes training a first base
caller over cluster intensity
images and producing a first trained base caller that maps the cluster
intensity images to base call
predictions.
[0206] At action 1312, the method includes beginning with the first
trained base caller,
executing a loop in which each iteration uses a starting trained base caller
as input and produces
a pruned trained base caller as output, wherein the pruned trained base caller
has fewer
processing elements than the starting trained base caller.
[0207] Each iteration comprises (i) a cluster feature maps
generation step, (ii) a gradient
determination step, (iii) a contribution measurement step, (iv) a pruning
step, and (v) a retraining
step.
[0208] At action 1322, the cluster feature maps generation step,
during forward propagation,
processes a subset of the clusters intensity images through the processing
elements of the starting
trained base caller, generates one or more cluster feature maps using each
processing element,
and produces the base call predictions based on the cluster feature maps.
39
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0209] At action 1332, the gradient determination step, during
backward propagation,
determines gradients for the cluster feature maps based on error between the
base call predictions
and ground truth base calls.
[0210] At action 1342, the contribution measurement step applies
the gradients to respective
ones of the cluster feature maps and generates a contribution score for each
of the cluster feature
maps that identifies how much a cluster feature map contributed to the base
call predictions.
[0211] At action 1352, the pruning step selects a subset of the
cluster feature maps based on
their contribution scores and produces the pruned trained base caller by
removing, from the
starting trained base caller, those processing elements that were used to
generate the selected
subset of the cluster feature maps during the forward propagation.
[0212] At action 1362, the retraining step further trains the
pruned trained base caller over
the cluster intensity images and makes the pruned trained base caller
available for a successive
iteration as the starting trained base caller.
[0213] At action 1372, the method includes terminating the loop
after, iterations and using
the pruned trained base caller produced by the nth iteration for further base
calling.
[0214] Other implementations of the method described in this
section can include a non-
transitory computer readable storage medium storing instructions executable by
a processor to
perform any of the methods described above. Yet another implementation of the
method
described in this section can include a system including memory and one or
more processors
operable to execute instructions, stored in the memory, to perform any of the
methods described
above.
[0215] Figure 14 shows another implementation of an artificial
intelligence-based method of
performing computationally efficient base calling.
[0216] At action 1402, the method incudes training a first base
caller over cluster intensity
images and producing a first trained base caller that maps the cluster
intensity images to base call
predictions.
[0217] At action 1412, the method includes beginning with the first
trained base caller,
executing a loop in which each iteration uses a starting trained base caller
as input and produces
a pruned trained base caller as output, wherein the pruned trained base caller
has fewer
processing elements than the starting trained base caller.
[0218] Each iteration comprises (i) a cluster feature maps
generation step, (ii) a contribution
measurement step, (iii) a pruning step, and (iv) a retraining step.
[0219] At action 1422, the cluster feature maps generation step,
during forward propagation,
processes a subset of the clusters intensity images through the processing
elements of the starting
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
trained base caller, generates one or more cluster feature maps using each
processing element,
and produces the base call predictions based on the cluster feature maps.
[0220] At action 1432, the contribution measurement step generates
a contribution score for
each of the cluster feature maps that identifies how much a cluster feature
map contributed to the
base call predictions.
[0221] At action 1442, the pruning step selects a subset of the
cluster feature maps based on
their contribution scores and produces the pruned trained base caller by
removing, from the
starting trained base caller, those processing elements that were used to
generate the selected
subset of the cluster feature maps during the forward propagation.
[0222] At action 1452, the retraining step further trains the
pruned trained base caller over
the cluster intensity images and makes the pruned trained base caller
available for a successive
iteration as the starting trained base caller.
[0223] At action 1462, the method includes terminating the loop
after n iterations and using
the pruned trained base caller produced by the nth iteration for further base
calling.
[0224] Other implementations of the method described in this
section can include a non-
transitory computer readable storage medium storing instructions executable by
a processor to
perform any of the methods described above. Yet another implementation of the
method
described in this section can include a system including memory and one or
more processors
operable to execute instructions, stored in the memory, to perform any of the
methods described
above.
[0225] Figures 15A, 15B, 15C, 15D, 15E, and 15F are performance
results that demonstrate
that the technology disclosed implements computationally efficient base
calling.
[0226] The following plots illustrate the iterative pruning process
output of the technology
disclosed. We start from our standard Multicluster algorithm which the first
trained base caller
1006 has seven spatial layers each with 48 filters, and 2 timewise
convolutional layers each with
96 filters. We add an Li norm regularization criterion to obtain a sparse set
of filters, this results
in an initial trained model that contains many of its convolutional kernels
set to all zeros (the
blue model in the first plot with suffix " tf2 human"). A higher Li
regularization parameter
leads to more filters being set to all zeros on this initially trained model.
[0227] Starting from this model we start a round of pruning
iterations, during which we
compute a pruning criterion on a random 15% of the training set. After ranking
the filters
according to this criterion, we eliminate the filters (typically we eliminate
2% of all filters across
the model at each iteration) that are deemed least important, and then retrain
the model for fine
tuning. Each model resulting from pruning and tine tuning at each iteration is
labelled in the
following plots according to the suffix "prunedxx human" where xx is a number
from 00 to 35.
41
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0228] Initially we see that at each new pruning iteration the
model performs better and
better until iteration 14 (model "pruned13 human"). This is likely due to
retraining the model
with our learning rate annealing approach (train with high learning rate and
progressively lower
our learning rate). The use of cyclical learning rate training schedules (high
to low learning rate
as we add more training epochs, followed by more training epochs of high to
low learning rate,
leads to better models according to literature on the subject).
[0229] Later on, after iteration 14 as the model is trimmed down
even further, we notice
progressive degradation of the model mismatch rate.
[0230] Training iteration 24 (model -_pruned23 human") shows to be
a good candidate,
from our pipeline output logs the model has the following filters:
= model fine-tuned from loss 0.029168058224022388 to
0.022608762811869382
= pruning iteration 24/35
= spatial correction convolutional stack
= Li keeping 14/14 filters
= L2 keeping 14/14 filters
= L3 keeping 11/12 filters
= L4 keeping 16/16 filters
= L5 keeping 15/15 filters
= L6 keeping 18/18 filters
= L7 keeping 6/6 filters
= timewise correction convolutional stack
= L8 keeping 12/13 filters
= L9 keeping 17/18 filters
[0231] Taking these filter counts and translating them into numbers
of operations per patch
we have 295813196 operations which is 8% less operations than our standard
K=14 model.
[0232] Interestingly, we also notice the squeezing of the last
filter in the spatial
convolutional stack down to 6 filters which corroborates our findings
(disclosed in U.S.
Provisional Patent Application No. 62/979,411) that data can be compressed
significantly
between the spatial correction layers and the timewise convolution layers.
[0233] Each of the following plots shows different models pruned
and fine-tuned iteratively.
A pruning session has been executed for each cycle along the x-axis, hence the
lines between
points at different cycles are here only to link independent models that are
at the same pruning
iteration.
42
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0234] The legend is partially hidden on these plots and the colors
are from top to bottom (in
the legend) blue, orange, green, red, black. Black fitted line represents
performance measured on
the same clusters as the disclosed deep learning model by Illumina's Real-Time
Analysis (RTA)
software (used herein as the baseline model).
[0235] Figure 19 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution weights for a distilled base
caller.
[0236] Figure 20 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution biases for a distilled base
caller.
[0237] Figure 21 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution weights for a distilled base
caller in which
regularization is applied to both the convolution weights and the convolution
biases.
[0238] Figure 22 depicts a box and whisker plot for one
implementation of the technology
disclosed that generates pruned convolution biases for a distilled base caller
in which
regularization is applied to both the convolution weights and the convolution
biases.
[0239] In some implementations, the technology disclosed uses an
alternative learning
scheduler, which starts from a higher learning rate and results in a better
distilled model. Figures
19 to 22 illustrate different regularization parameters and convergence on
12(0.00001) as both
kernel and bias regularizers when distilling models. By doing so, the accuracy
of the distilled
model is not affected but the weights and biases are reduced to the range
which can be
accommodated on FPGA.
Terminology and Additional Implementations
[0240] Base calling includes incorporation or attachment of a
fluorescently-labeled tag with
an analyte. The analyte can be a nucleotide or an oligonucleotide, and the tag
can be for a
particular nucleotide type (A, C, T, or G). Excitation light is directed
toward the analyte having
the tag, and the tag emits a detectable fluorescent signal or intensity
emission. The intensity
emission is indicative of photons emitted by the excited tag that is
chemically attached to the
analyte.
[0241] Throughout this application, including the claims, when
phrases such as or similar to
"images, image data, or image regions depicting intensity emissions of
analytes and their
surrounding background" are used, they refer to the intensity emissions of the
tags attached to
the analytes. A person skilled in the art will appreciate that the intensity
emissions of the
attached tags are representative of or equivalent to the intensity emissions
of the analytes to
which the tags are attached, and are therefore used interchangeably.
Similarly, properties of the
analytes refer to properties of the tags attached to the analytes or of the
intensity emissions from
43
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
the attached tags. For example, a center of an analyte refers to the center of
the intensity
emissions emitted by a tag attached to the analyte. In another example, the
surrounding
background of an analyte refers to the surrounding background of the intensity
emissions emitted
by a tag attached to the analyte.
[0242] All literature and similar material cited in this
application, including, but not limited
to, patents, patent applications, articles, books, treatises, and web pages,
regardless of the format
of such literature and similar materials, are expressly incorporated by
reference in their entirety.
In the event that one or more of the incorporated literature and similar
materials differs from or
contradicts this application, including but not limited to defined terms, term
usage, described
techniques, or the like, this application controls.
[0243] The technology disclosed uses neural networks to improve the
quality and quantity of
nucleic acid sequence information that can be obtained from a nucleic acid
sample such as a
nucleic acid template or its complement, for instance, a DNA or RNA
polynucleotide or other
nucleic acid sample. Accordingly, certain implementations of the technology
disclosed provide
higher throughput polynucleotide sequencing, for instance, higher rates of
collection of DNA or
RNA sequence data, greater efficiency in sequence data collection, and/or
lower costs of
obtaining such sequence data, relative to previously available methodologies.
[0244] The technology disclosed uses neural networks to identify
the center of a solid-phase
nucleic acid cluster and to analyze optical signals that are generated during
sequencing of such
clusters, to discriminate unambiguously between adjacent, abutting or
overlapping clusters in
order to assign a sequencing signal to a single, discrete source cluster.
These and related
implementations thus permit retrieval of meaningful information, such as
sequence data, from
regions of high-density cluster arrays where useful information could not
previously be obtained
from such regions due to confounding effects of overlapping or very closely
spaced adjacent
clusters, including the effects of overlapping signals (e.g., as used in
nucleic acid sequencing)
emanating therefrom.
[0245] As described in greater detail below, in certain
implementations there is provided a
composition that comprises a solid support having immobilized thereto one or a
plurality of
nucleic acid clusters as provided herein. Each cluster comprises a plurality
of immobilized
nucleic acids of the same sequence and has an identifiable center having a
detectable center label
as provided herein, by which the identifiable center is distinguishable from
immobilized nucleic
acids in a surrounding region in the cluster. Also described herein are
methods for making and
using such clusters that have identifiable centers.
[0246] The presently disclosed implementations will find uses in
numerous situations where
advantages are obtained from the ability to identify, determine, annotate,
record or otherwise
44
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
assign the position of a substantially central location within a cluster, such
as high-throughput
nucleic acid sequencing, development of image analysis algorithms for
assigning optical or other
signals to discrete source clusters, and other applications where recognition
of the center of an
immobilized nucleic acid cluster is desirable and beneficial.
[0247] In certain implementations, the present invention
contemplates methods that relate to
high-throughput nucleic acid analysis such as nucleic acid sequence
determination (e.g.,
"sequencing"). Exemplary high-throughput nucleic acid analyses include without
limitation de
novo sequencing, re-sequencing, whole genome sequencing, gene expression
analysis, gene
expression monitoring, epigenetic analysis, genome methylation analysis,
allele specific primer
extension (APSE), genetic diversity profiling, whole genome polymorphism
discovery and
analysis, single nucleotide polymorphism analysis, hybridization based
sequence determination
methods, and the like One skilled in the art will appreciate that a variety of
different nucleic
acids can be analyzed using the methods and compositions of the present
invention.
[0248] Although the implementations of the present invention are
described in relation to
nucleic acid sequencing, they are applicable in any field where image data
acquired at different
time points, spatial locations or other temporal or physical perspectives is
analyzed. For
example, the methods and systems described herein are useful in the fields of
molecular and cell
biology where image data from microarrays, biological specimens, cells,
organisms and the like
is acquired and at different time points or perspectives and analyzed. Images
can be obtained
using any number of techniques known in the art including, but not limited to,
fluorescence
microscopy, light microscopy, confocal microscopy, optical imaging, magnetic
resonance
imaging, tomography scanning or the like. As another example, the methods and
systems
described herein can be applied where image data obtained by surveillance,
aerial or satellite
imaging technologies and the like is acquired at different time points or
perspectives and
analyzed. The methods and systems are particularly useful for analyzing images
obtained for a
field of view in which the analytes being viewed remain in the same locations
relative to each
other in the field of view. The analytes may however have characteristics that
differ in separate
images, for example, the analytes may appear different in separate images of
the field of view.
For example, the analytes may appear different with regard to the color of a
given analyte
detected in different images, a change in the intensity of signal detected for
a given analyte in
different images, or even the appearance of a signal for a given analyte in
one image and
disappearance of the signal for the analyte in another image.
[0249] Examples described herein may be used in various biological
or chemical processes
and systems for academic or commercial analysis. More specifically, examples
described herein
may be used in various processes and systems where it is desired to detect an
event, property,
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
quality, or characteristic that is indicative of a designated reaction. For
example, examples
described herein include light detection devices, biosensors, and their
components, as well as
bioassay systems that operate with biosensors. In some examples, the devices,
biosensors and
systems may include a flow cell and one or more light sensors that are coupled
together
(removably or fixedly) in a substantially unitary structure.
[0250] The devices, biosensors and bioassay systems may be
configured to perform a
plurality of designated reactions that may be detected individually or
collectively. The devices,
biosensors and bioassay systems may be configured to perform numerous cycles
in which the
plurality of designated reactions occurs in parallel. For example, the
devices, biosensors and
bioassay systems may be used to sequence a dense array of DNA features through
iterative
cycles of enzymatic manipulation and light or image detection/acquisition. As
such, the devices,
biosensors and bioassay systems (e.g., via one or more cartridges) may include
one or more
microfluidic channel that delivers reagents or other reaction components in a
reaction solution to
a reaction site of the devices, biosensors and bioassay systems. In some
examples, the reaction
solution may be substantially acidic, such as comprising a pH of less than or
equal to about 5, or
less than or equal to about 4, or less than or equal to about 3. In some other
examples, the
reaction solution may be substantially alkaline/basic, such as comprising a pH
of greater than or
equal to about 8, or greater than or equal to about 9, or greater than or
equal to about 10. As used
herein, the term "acidity" and grammatical variants thereof refer to a pH
value of less than about
7, and the terms "basicity," "alkalinity" and grammatical variants thereof
refer to a pH value of
greater than about 7.
[0251] In some examples, the reaction sites are provided or spaced
apart in a predetermined
manner, such as in a uniform or repeating pattern. In some other examples, the
reaction sites are
randomly distributed. Each of the reaction sites may be associated with one or
more light guides
and one or more light sensors that detect light from the associated reaction
site. In some
examples, the reaction sites are located in reaction recesses or chambers,
which may at least
partially compartmentalize the designated reactions therein.
[0252] As used herein, a "designated reaction" includes a change in
at least one of a
chemical, electrical, physical, or optical property (or quality) of a chemical
or biological
substance of interest, such as an analyte-of-interest. In particular examples,
a designated reaction
is a positive binding event, such as incorporation of a fluorescently labeled
biomolecule with an
analyte-of-interest, for example. More generally, a designated reaction may be
a chemical
transformation, chemical change, or chemical interaction. A designated
reaction may also be a
change in electrical properties. In particular examples, a designated reaction
includes the
incorporation of a fluorescently-labeled molecule with an an alyte. The
analyte may be an
46
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
oligonucleotide and the fluorescently-labeled molecule may be a nucleotide. A
designated
reaction may be detected when an excitation light is directed toward the
oligonucleotide haying
the labeled nucleotide, and the fluorophore emits a detectable fluorescent
signal. In alternative
examples, the detected fluorescence is a result of chemiluminescence or
bioluminescence. A
designated reaction may also increase fluorescence (or Forster) resonance
energy transfer
(FRET), for example, by bringing a donor fluorophore in proximity to an
acceptor fluorophore,
decrease FRET by separating donor and acceptor fluorophores, increase
fluorescence by
separating a quencher from a fluorophore, or decrease fluorescence by co-
locating a quencher
and fluorophore.
[0253] As used herein, a "reaction solution," "reaction component"
or "reactant" includes
any substance that may be used to obtain at least one designated reaction. For
example, potential
reaction components include reagents, enzymes, samples, other biomolecules,
and buffer
solutions, for example. The reaction components may be delivered to a reaction
site in a solution
and/or immobilized at a reaction site. The reaction components may interact
directly or indirectly
with another substance, such as an analyte-of-interest immobilized at a
reaction site. As noted
above, the reaction solution may be substantially acidic (i.e., include a
relatively high acidity)
(e.g., comprising a pH of less than or equal to about 5, a pH less than or
equal to about 4, or a pH
less than or equal to about 3) or substantially alkaline/basic (i.e., include
a relatively high
alkalinity/basicity) (e ,g, , comprising a pH of greater than or equal to
about 8, a pH of greater
than or equal to about 9, or a pH of greater than or equal to about 10).
[0254] As used herein, the term "reaction site" is a localized
region where at least one
designated reaction may occur. A reaction site may include support surfaces of
a reaction
structure or substrate where a substance may be immobilized thereon. For
example, a reaction
site may include a surface of a reaction structure (which may be positioned in
a channel of a flow
cell) that has a reaction component thereon, such as a colony of nucleic acids
thereon. In some
such examples, the nucleic acids in the colony have the same sequence, being
for example,
clonal copies of a single stranded or double stranded template. However, in
some examples a
reaction site may contain only a single nucleic acid molecule, for example, in
a single stranded or
double stranded form.
[0255] A plurality of reaction sites may be randomly distributed
along the reaction structure
or arranged in a predetermined manner (e.g., side-by-side in a matrix, such as
in microarrays). A
reaction site can also include a reaction chamber or recess that at least
partially defines a spatial
region or volume configured to compartmentalize the designated reaction. As
used herein, the
term "reaction chamber" or "reaction recess" includes a defined spatial region
of the support
structure (which is often in fluid communication with a flow channel). A
reaction recess may be
47
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
at least partially separated from the surrounding environment other or spatial
regions. For
example, a plurality of reaction recesses may be separated from each other by
shared walls, such
as a detection surface. As a more specific example, the reaction recesses may
be nanowells
comprising an indent, pit, well, groove, cavity or depression defined by
interior surfaces of a
detection surface and have an opening or aperture (i.e., be open-sided) so
that the nanowells can
be in fluid communication with a flow channel.
[0256] In some examples, the reaction recesses of the reaction
structure are sized and shaped
relative to solids (including semi-solids) so that the solids may be inserted,
fully or partially,
therein. For example, the reaction recesses may be sized and shaped to
accommodate a capture
bead. The capture bead may have clonally amplified DNA or other substances
thereon.
Alternatively, the reaction recesses may be sized and shaped to receive an
approximate number
of beads or solid substrates As another example, the reaction recesses may be
filled with a
porous gel or substance that is configured to control diffusion or filter
fluids or solutions that
may flow into the reaction recesses.
[0257] In some examples, light sensors (e.g., photodiodes) are
associated with corresponding
reaction sites. A light sensor that is associated with a reaction site is
configured to detect light
emissions from the associated reaction site via at least one light guide when
a designated reaction
has occurred at the associated reaction site. In some cases, a plurality of
light sensors (e.g.
several pixels of a light detection or camera device) may be associated with a
single reaction site.
In other cases, a single light sensor (e.g. a single pixel) may be associated
with a single reaction
site or with a group of reaction sites. The light sensor, the reaction site,
and other features of the
biosensor may be configured so that at least some of the light is directly
detected by the light
sensor without being reflected.
[0258] As used herein, a "biological or chemical substance"
includes biomolecules, samples-
of-interest, analytes-of-interest, and other chemical compound(s). A
biological or chemical
substance may be used to detect, identify, or analyze other chemical
compound(s), or function as
intermediaries to study or analyze other chemical compound(s). In particular
examples, the
biological or chemical substances include a biomolecule. As used herein, a
"biomolecule"
includes at least one of a biopolymer, nucleoside, nucleic acid,
polynucleotide, oligonucleotide,
protein, enzyme, polypeptide, antibody, antigen, ligand, receptor,
polysaccharide, carbohydrate,
polyphosphate, cell, tissue, organism, or fragment thereof or any other
biologically active
chemical compound(s) such as analogs or mimetics of the aforementioned
species. In a further
example, a biological or chemical substance or a biomolecule includes an
enzyme or reagent
used in a coupled reaction to detect the product of another reaction such as
an enzyme or reagent,
such as an enzyme or reagent used to detect pyrophosphate in a pyrosequencing
reaction.
48
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
Enzymes and reagents useful for pyrophosphate detection are described, for
example, in U.S.
Patent Publication No. 2005/0244870 Al, which is incorporated by reference in
its entirety.
[0259] Biomolecules, samples, and biological or chemical substances
may be naturally
occurring or synthetic and may be suspended in a solution or mixture within a
reaction recess or
region. Biomolecules, samples, and biological or chemical substances may also
be bound to a
solid phase or gel material. Biomolecules, samples, and biological or chemical
substances may
also include a pharmaceutical composition. In some cases, biomolecules,
samples, and biological
or chemical substances of interest may be referred to as targets, probes, or
analytes.
[0260] As used herein, a -biosensor" includes a device that
includes a reaction structure with
a plurality of reaction sites that is configured to detect designated
reactions that occur at or
proximate to the reaction sites. A biosensor may include a solid-state light
detection or
"imaging" device (e.g., CCD or CMOS light detection device) and, optionally, a
flow cell
mounted thereto. The flow cell may include at least one flow channel that is
in fluid
communication with the reaction sites. As one specific example, the biosensor
is configured to
fluidically and electrically couple to a bioassay system. The bioassay system
may deliver a
reaction solution to the reaction sites according to a predetermined protocol
(e.g., sequencing-by-
synthesis) and perform a plurality of imaging events. For example, the
bioassay system may
direct reaction solutions to flow along the reaction sites. At least one of
the reaction solutions
may include four types of nucleotides having the same or different fluorescent
labels. The
nucleotides may bind to the reaction sites, such as to corresponding
oligonucleotides at the
reaction sites. The bioassay system may then illuminate the reaction sites
using an excitation
light source (e.g., solid-state light sources, such as light-emitting diodes
(LEDs)). The excitation
light may have a predetermined wavelength or wavelengths, including a range of
wavelengths.
The fluorescent labels excited by the incident excitation light may provide
emission signals (e.g.,
light of a wavelength or wavelengths that differ from the excitation light
and, potentially, each
other) that may be detected by the light sensors.
[0261] As used herein, the term "immobilized," when used with
respect to a biomolecule or
biological or chemical substance, includes substantially attaching the
biomolecule or biological
or chemical substance at a molecular level to a surface, such as to a
detection surface of a light
detection device or reaction structure. For example, a biomolecule or
biological or chemical
substance may be immobilized to a surface of the reaction structure using
adsorption techniques
including non-covalent interactions (e.g., electrostatic forces, van der
Waals, and dehydration of
hydrophobic interfaces) and covalent binding techniques where functional
groups or linkers
facilitate attaching the bi omol ecul es to the surface. Immobilizing bi om ol
ecul es or biological or
chemical substances to the surface may be based upon the properties of the
surface, the liquid
49
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
medium carrying the biomolecule or biological or chemical substance, and the
properties of the
biomolecules or biological or chemical substances themselves. In some cases,
the surface may be
functionalized (e.g., chemically or physically modified) to facilitate
immobilizing the
biomolecules (or biological or chemical substances) to the surface.
[0262] In some examples, nucleic acids can be immobilized to the
reaction structure, such as
to surfaces of reaction recesses thereof. In particular examples, the devices,
biosensors, bioassay
systems and methods described herein may include the use of natural
nucleotides and also
enzymes that are configured to interact with the natural nucleotides. Natural
nucleotides include,
for example, ribonucleotides or deoxyribonucleotides. Natural nucleotides can
be in the mono-,
di-, or tri-phosphate form and can have a base selected from adenine (A),
Thymine (T), uracil
(U), guanine (G) or cytosine (C). It will be understood, however, that non-
natural nucleotides,
modified nucleotides or analogs of the aforementioned nucleotides can be used
[0263] As noted above, a biomolecule or biological or chemical
substance may be
immobilized at a reaction site in a reaction recess of a reaction structure.
Such a biomolecule or
biological substance may be physically held or immobilized within the reaction
recesses through
an interference fit, adhesion, covalent bond, or entrapment. Examples of items
or solids that may
be disposed within the reaction recesses include polymer beads, pellets,
agarose gel, powders,
quantum dots, or other solids that may be compressed and/or held within the
reaction chamber.
In certain implementations, the reaction recesses may be coated or filled with
a hydrogel layer
capable of covalently binding DNA oligonucleotides. In particular examples, a
nucleic acid
superstructure, such as a DNA ball, can be disposed in or at a reaction
recess, for example, by
attachment to an interior surface of the reaction recess or by residence in a
liquid within the
reaction recess. A DNA ball or other nucleic acid superstructure can be
performed and then
disposed in or at a reaction recess. Alternatively, a DNA ball can be
synthesized in situ at a
reaction recess. A substance that is immobilized in a reaction recess can be
in a solid, liquid, or
gaseous state.
[0264] As used herein, the term "analyte" is intended to mean a
point or area in a pattern that
can be distinguished from other points or areas according to relative
location. An individual
analyte can include one or more molecules of a particular type. For example,
an analyte can
include a single target nucleic acid molecule having a particular sequence or
an analyte can
include several nucleic acid molecules having the same sequence (and/or
complementary
sequence, thereof). Different molecules that are at different analytes of a
pattern can be
differentiated from each other according to the locations of the analytes in
the pattern. Example
analytes include without limitation, wells in a substrate, beads (or other
particles) in or on a
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
substrate, projections from a substrate, ridges on a substrate, pads of gel
material on a substrate,
or channels in a substrate.
[0265] Any of a variety of target analytes that are to be detected,
characterized, or identified
can be used in an apparatus, system or method set forth herein. Exemplary
analytes include, but
are not limited to, nucleic acids (e.g., DNA, RNA or analogs thereof),
proteins, polysaccharides,
cells, antibodies, epitopes, receptors, ligands, enzymes (e.g. kinases,
phosphatases or
polymerases), small molecule drug candidates, cells, viruses, organisms, or
the like.
[0266] The terms -analyte", -nucleic acid", -nucleic acid
molecule", and -polynucleotide"
are used interchangeably herein. In various implementations, nucleic acids may
be used as
templates as provided herein (e.g., a nucleic acid template, or a nucleic acid
complement that is
complementary to a nucleic acid nucleic acid template) for particular types of
nucleic acid
analysis, including but not limited to nucleic acid amplification, nucleic
acid expression analysis,
and/or nucleic acid sequence determination or suitable combinations thereof.
Nucleic acids in
certain implementations include, for instance, linear polymers of
deoxyribonucleotides in 3'-,5'
phosphodiester or other linkages, such as deoxyribonucleic acids (DNA), for
example, single-
and double-stranded DNA, genomic DNA, copy DNA or complementary DNA (cDNA),
recombinant DNA, or any form of synthetic or modified DNA. In other
implementations, nucleic
acids include for instance, linear polymers of ribonucleotides in 3 '-5'
phosphodiester or other
linkages such as ribonucleic acids (RNA), for example, single- and double-
stranded RNA,
messenger (mRNA), copy RNA or complementary RNA (cRNA), alternatively spliced
mRNA,
ribosomal RNA, small nucleolar RNA (snoRNA), microRNAs (miRNA), small
interfering
RNAs (sRNA), piwi RNAs (piRNA), or any form of synthetic or modified RNA.
Nucleic acids
used in the compositions and methods of the present invention may vary in
length and may be
intact or full-length molecules or fragments or smaller parts of larger
nucleic acid molecules. In
particular implementations, a nucleic acid may have one or more detectable
labels, as described
elsewhere herein.
[0267] The terms "analyte", "cluster", "nucleic acid cluster",
"nucleic acid colony", and
"DNA cluster" are used interchangeably and refer to a plurality of copies of a
nucleic acid
template and/or complements thereof attached to a solid support. Typically and
in certain
preferred implementations, the nucleic acid cluster comprises a plurality of
copies of template
nucleic acid and/or complements thereof, attached via their 5' termini to the
solid support. The
copies of nucleic acid strands making up the nucleic acid clusters may be in a
single or double
stranded form. Copies of a nucleic acid template that are present in a cluster
can have nucleotides
at corresponding positions that differ from each other, for example, due to
presence of a label
moiety. The corresponding positions can also contain analog structures having
different chemical
51
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
structure but similar Watson-Crick base-pairing properties, such as is the
case for uracil and
thymine.
[0268] Colonies of nucleic acids can also be referred to as
"nucleic acid clusters". Nucleic
acid colonies can optionally be created by cluster amplification or bridge
amplification
techniques as set forth in further detail elsewhere herein. Multiple repeats
of a target sequence
can be present in a single nucleic acid molecule, such as a concatamer created
using a rolling
circle amplification procedure.
[0269] The nucleic acid clusters of the invention can have
different shapes, sizes and
densities depending on the conditions used. For example, clusters can have a
shape that is
substantially round, multi-sided, donut-shaped or ring-shaped. The diameter of
a nucleic acid
cluster can be designed to be from about 0.2 pm to about 6 p.m, about 0.3 pm
to about 4 pm,
about 0.4 p.m to about 3 pm, about 0.5 p.m to about 2 pm, about 0.75 p.m to
about 1.5 pm, or any
intervening diameter. In a particular implementation, the diameter of a
nucleic acid cluster is
about 0.5 pm, about 1 pm, about 1.5 pm, about 2 pm, about 2.5 pm, about 3 pm,
about 4 p.m,
about 5 pm, or about 6 p.m. The diameter of a nucleic acid cluster may be
influenced by a
number of parameters, including, but not limited to the number of
amplification cycles
performed in producing the cluster, the length of the nucleic acid template or
the density of
primers attached to the surface upon which clusters are formed. The density of
nucleic acid
clusters can be designed to typically be in the range of 0.1/mm2, 1/mm2,
10/mm2, 100/mm2,
1,000/mm2, 10,000/mm2 to 100,000/mm2. The present invention further
contemplates, in part,
higher density nucleic acid clusters, for example, 100,000/mm2 to
1,000,000/mm2 and
1,000,000/mm2 to 10,000,000/mm2.
[0270] As used herein, an "analyte" is an area of interest within a
specimen or field of view.
When used in connection with microarray devices or other molecular analytical
devices, an
analyte refers to the area occupied by similar or identical molecules. For
example, an analyte can
be an amplified oligonucleotide or any other group of a polynucleotide or
polypeptide with a
same or similar sequence. In other implementations, an analyte can be any
element or group of
elements that occupy a physical area on a specimen. For example, an analyte
could be a parcel of
land, a body of water or the like. When an analyte is imaged, each analyte
will have some area.
Thus, in many implementations, an analyte is not merely one pixel.
[0271] The distances between analytes can be described in any
number of ways. In some
implementations, the distances between analytes can be described from the
center of one analyte
to the center of another analyte. In other implementations, the distances can
be described from
the edge of one analyte to the edge of another analyte, or between the outer-
most identifiable
points of each analyte. The edge of an analyte can be described as the
theoretical or actual
52
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
physical boundary on a chip, or some point inside the boundary of the analyte.
In other
implementations, the distances can be described in relation to a fixed point
on the specimen or in
the image of the specimen.
[0272] Generally several implementations will be described herein
with respect to a method
of analysis. It will be understood that systems are also provided for carrying
out the methods in
an automated or semi-automated way. Accordingly, this disclosure provides
neural network-
based template generation and base calling systems, wherein the systems can
include a
processor; a storage device; and a program for image analysis, the program
including
instructions for carrying out one or more of the methods set forth herein.
Accordingly, the
methods set forth herein can be carried out on a computer, for example, having
components set
forth herein or otherwise known in the art.
[0273] The methods and systems set forth herein are useful for
analyzing any of a variety of
objects. Particularly useful objects are solid supports or solid-phase
surfaces with attached
analytes. The methods and systems set forth herein provide advantages when
used with objects
having a repeating pattern of analytes in an xy plane. An example is a
microarray having an
attached collection of cells, viruses, nucleic acids, proteins, antibodies,
carbohydrates, small
molecules (such as drug candidates), biologically active molecules or other
analytes of interest.
[0274] An increasing number of applications have been developed for
arrays with analytes
having biological molecules such as nucleic acids and polypeptides. Such
microarrays typically
include deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) probes. These
are specific for
nucleotide sequences present in humans and other organisms. In certain
applications, for
example, individual DNA or RNA probes can be attached at individual analytes
of an array. A
test sample, such as from a known person or organism, can be exposed to the
array, such that
target nucleic acids (e.g., gene fragments, mRNA, or amplicons thereof)
hybridize to
complementary probes at respective analytes in the array. The probes can be
labeled in a target
specific process (e.g., due to labels present on the target nucleic acids or
due to enzymatic
labeling of the probes or targets that are present in hybridized form at the
analytes). The array
can then be examined by scanning specific frequencies of light over the
analytes to identify
which target nucleic acids are present in the sample.
[0275] Biological microarrays may be used for genetic sequencing
and similar applications.
In general, genetic sequencing comprises determining the order of nucleotides
in a length of
target nucleic acid, such as a fragment of DNA or RNA. Relatively short
sequences are typically
sequenced at each analyte, and the resulting sequence information may be used
in various
bioinformatics methods to logically fit the sequence fragments together so as
to reliably
determine the sequence of much more extensive lengths of genetic material from
which the
53
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
fragments were derived. Automated, computer-based algorithms for
characteristic fragments
have been developed, and have been used more recently in genome mapping,
identification of
genes and their function, and so forth. Microarrays are particularly useful
for characterizing
genomic content because a large number of variants are present and this
supplants the alternative
of performing many experiments on individual probes and targets. The
microarray is an ideal
format for performing such investigations in a practical manner.
[0276] Any of a variety of analyte arrays (also referred to as
"microarrays") known in the art
can be used in a method or system set forth herein. A typical array contains
analytes, each having
an individual probe or a population of probes. In the latter case, the
population of probes at each
analyte is typically homogenous having a single species of probe. For example,
in the case of a
nucleic acid array, each analyte can have multiple nucleic acid molecules each
having a common
sequence However, in some implementations the populations at each analyte of
an array can be
heterogeneous. Similarly, protein arrays can have analytes with a single
protein or a population
of proteins typically, but not always, having the same amino acid sequence.
The probes can be
attached to the surface of an array for example, via covalent linkage of the
probes to the surface
or via non-covalent interaction(s) of the probes with the surface. In some
implementations,
probes, such as nucleic acid molecules, can be attached to a surface via a gel
layer as described,
for example, in U.S. patent application Ser. No. 13/784,368 and US Pat. App.
Pub. No.
2011/0059865 Al, each of which is incorporated herein by reference.
[0277] Example arrays include, without limitation, a BeadChip Array
available from
Illumina, Inc. (San Diego, Calif.) or others such as those where probes are
attached to beads that
are present on a surface (e.g. beads in wells on a surface) such as those
described in U.S. Pat. No.
6,266,459; 6,355,431; 6,770,441; 6,859,570; or 7,622,294; or PCT Publication
No. WO
00/63437, each of which is incorporated herein by reference. Further examples
of commercially
available microarrays that can be used include, for example, an Affymetrix
GeneChip
microarray or other microarray synthesized in accordance with techniques
sometimes referred to
as VLS1PST1 (Very Large Scale Immobilized Polymer Synthesis) technologies. A
spotted
microarray can also be used in a method or system according to some
implementations of the
present disclosure. An example spotted microarray is a CodeLinkTM Array
available from
Amersham Biosciences. Another microarray that is useful is one that is
manufactured using
inkjet printing methods such as SurePrintTM Technology available from Agilent
Technologies.
[0278] Other useful arrays include those that are used in nucleic
acid sequencing
applications. For example, arrays having amplicons of genomic fragments (often
referred to as
clusters) are particularly useful such as those described in Bentley et al.,
Nature 456:53-59
(2008), WO 04/018497; WO 91/06678; WO 07/123744; U.S. Pat. No. 7,329,492;
7,211,414;
54
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
7,315,019; 7,405,281, or 7,057,026; or US Pat. App. Pub. No. 2008/0108082 Al,
each of which
is incorporated herein by reference. Another type of array that is useful for
nucleic acid
sequencing is an array of particles produced from an emulsion PCR technique.
Examples are
described in Dressman et al., Proc. Natl. Acad. Sci. USA 100:8817-8822 (2003),
WO 05/010145,
US Pat. App. Pub. No. 2005/0130173 or US Pat. App. Pub. No. 2005/0064460, each
of which is
incorporated herein by reference in its entirety.
[0279] Arrays used for nucleic acid sequencing often have random
spatial patterns of nucleic
acid analytes. For example, Hi Seq or MiSeq sequencing platforms available
from Illumina Inc.
(San Diego, Calif) utilize flow cells upon which nucleic acid arrays are
formed by random
seeding followed by bridge amplification. However, patterned arrays can also
be used for nucleic
acid sequencing or other analytical applications. Example patterned arrays,
methods for their
manufacture and methods for their use are set forth in U.S. Ser. No.
13/787,396; U.S. Ser. No.
13/783,043; U.S. Ser. No. 13/784,368; US Pat. App. Pub. No. 2013/0116153 Al;
and US Pat.
App. Pub. No. 2012/0316086 Al, each of which is incorporated herein by
reference. The
analytes of such patterned arrays can be used to capture a single nucleic acid
template molecule
to seed subsequent formation of a homogenous colony, for example, via bridge
amplification.
Such patterned arrays are particularly useful for nucleic acid sequencing
applications.
[0280] The size of an analyte on an array (or other object used in
a method or system herein)
can be selected to suit a particular application. For example, in some
implementations, an analyte
of an array can have a size that accommodates only a single nucleic acid
molecule. A surface
having a plurality of analytes in this size range is useful for constructing
an array of molecules
for detection at single molecule resolution. Analytes in this size range are
also useful for use in
arrays having analytes that each contain a colony of nucleic acid molecules.
Thus, the analytes of
an array can each have an area that is no larger than about 1 mm2, no larger
than about 500 gm2,
no larger than about 100 tim2, no larger than about 10 [.t.m2, no larger than
about 1 IL.tm2, no larger
than about 500 nm2, or no larger than about 100 nm2, no larger than about 10
nm2, no larger than
about 5 nm2, or no larger than about 1 nm2. Alternatively or additionally, the
analytes of an array
will be no smaller than about 1 mm2, no smaller than about 500 im2, no smaller
than about 100
lam2, no smaller than about 10m2, no smaller than about 1 gm2, no smaller than
about 500 nm2,
no smaller than about 100 nm2, no smaller than about 10 nm2, no smaller than
about 5 nm2, or no
smaller than about 1 nm2. Indeed, an analyte can have a size that is in a
range between an upper
and lower limit selected from those exemplified above. Although several size
ranges for analytes
of a surface have been exemplified with respect to nucleic acids and on the
scale of nucleic acids,
it will be understood that analytes in these size ranges can be used for
applications that do not
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
include nucleic acids. It will be further understood that the size of the
analytes need not
necessarily be confined to a scale used for nucleic acid applications.
[0281] For implementations that include an object having a
plurality of analytes, such as an
array of analytes, the analytes can be discrete, being separated with spaces
between each other.
An array useful in the invention can have analytes that are separated by edge
to edge distance of
at most 100 pm, 50 gm, 10 pm, 5 pm, 1 pm, 0.5 pm, or less. Alternatively or
additionally, an
array can have analytes that are separated by an edge to edge distance of at
least 0.5 p.m, 1 pm, 5
pm, 10 pm, 50 pm, 100 pm, or more. These ranges can apply to the average edge
to edge spacing
for analytes as well as to the minimum or maximum spacing.
[0282] In some implementations the analytes of an array need not be
discrete and instead
neighboring analytes can abut each other. Whether or not the analytes are
discrete, the size of the
analytes and/or pitch of the analytes can vary such that arrays can have a
desired density. For
example, the average analyte pitch in a regular pattern can be at most 100 pm,
50 pm, 10 pm, 5
pm, 1 pm, 0.5 p.m, or less. Alternatively or additionally, the average analyte
pitch in a regular
pattern can be at least 0.5 pm, 1 pm, 5 pm, 10 pm, 50 pm, 100 pm, or more.
These ranges can
apply to the maximum or minimum pitch for a regular pattern as well. For
example, the
maximum analyte pitch for a regular pattern can be at most 100 m, 50 pm, 10
pm, 5 pm, 1 prn,
0.5 pm, or less; and/or the minimum analyte pitch in a regular pattern can be
at least 0.5 p.m, 1
pm, 5 pm, 10 pm, 50 pm, 100 pm, or more.
[0283] The density of analytes in an array can also be understood
in terms of the number of
analytes present per unit area. For example, the average density of analytes
for an array can be at
least about 1x103 analytes/mm2, 1x104 analytes/mm2, 1x105 analytes/mm2, 1x106
analytes/mm2,
1x107 analytes/mm2, 1x108 analytes/mm2, or 1x109 analytes/mm2, or higher.
Alternatively or
additionally the average density of analytes for an array can be at most about
1x109
analytes/mm2, lx108 analytes/mm2, lx107 analytes/mm2, lx106 analytes/mm2,
lx105
analytes/mm2, 1x104 analytes/mm2, or 1x103 analytes/mm2, or less.
[0284] The above ranges can apply to all or part of a regular
pattern including, for example,
all or part of an array of analytes.
[0285] The analytes in a pattern can have any of a variety of
shapes. For example, when
observed in a two dimensional plane, such as on the surface of an array, the
analytes can appear
rounded, circular, oval, rectangular, square, symmetric, asymmetric,
triangular, polygonal, or the
like. The analytes can be arranged in a regular repeating pattern including,
for example, a
hexagonal or rectilinear pattern. A pattern can be selected to achieve a
desired level of packing.
For example, round analytes are optimally packed in a hexagonal arrangement.
Of course other
packing arrangements can also be used for round analytes and vice versa.
56
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0286] A pattern can be characterized in terms of the number of
analytes that are present in a
subset that forms the smallest geometric unit of the pattern. The subset can
include, for example,
at least about 2, 3, 4, 5, 6, 10 or more analytes. Depending upon the size and
density of the
analytes the geometric unit can occupy an area of less than 1 mm2, 500 lam2,
100m2, 50 Ilm2,
wn2, 1 pm2, 500 nm2, 100 nm2, 50 nm2, 10 nm2, or less. Alternatively or
additionally, the
geometric unit can occupy an area of greater than 10 nm2, 50 nm2, 100 nm2, 500
nm2, 1 [Im2, 10
pm2, 50 u,m2, 100 um2, 500 wn2, 1 mm2, or more. Characteristics of the
analytes in a geometric
unit, such as shape, size, pitch and the like, can be selected from those set
forth herein more
generally with regard to analytes in an array or pattern.
[0287] An array having a regular pattern of analytes can be ordered
with respect to the
relative locations of the analytes but random with respect to one or more
other characteristic of
each analyte For example, in the case of a nucleic acid array, the nuclei acid
analytes can be
ordered with respect to their relative locations but random with respect to
one's knowledge of the
sequence for the nucleic acid species present at any particular analyte As a
more specific
example, nucleic acid arrays formed by seeding a repeating pattern of analytes
with template
nucleic acids and amplifying the template at each analyte to form copies of
the template at the
analyte (e.g., via cluster amplification or bridge amplification) will have a
regular pattern of
nucleic acid analytes but will be random with regard to the distribution of
sequences of the
nucleic acids across the array. Thus, detection of the presence of nucleic
acid material generally
on the array can yield a repeating pattern of analytes, whereas sequence
specific detection can
yield non-repeating distribution of signals across the array.
[0288] It will be understood that the description herein of
patterns, order, randomness and
the like pertain not only to analytes on objects, such as analytes on arrays,
but also to analytes in
images. As such, patterns, order, randomness and the like can be present in
any of a variety of
formats that are used to store, manipulate or communicate image data
including, but not limited
to, a computer readable medium or computer component such as a graphical user
interface or
other output device.
[0289] As used herein, the term "image" is intended to mean a
representation of all or part of
an object. The representation can be an optically detected reproduction. For
example, an image
can be obtained from fluorescent, luminescent, scatter, or absorption signals.
The part of the
object that is present in an image can be the surface or other xy plane of the
object. Typically, an
image is a 2 dimensional representation, but in some cases information in the
image can be
derived from 3 or more dimensions. An image need not include optically
detected signals. Non-
optical signals can be present instead. An image can be provided in a computer
readable format
or medium such as one or more of those set forth elsewhere herein.
57
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0290] As used herein, "image" refers to a reproduction or
representation of at least a portion
of a specimen or other object. In some implementations, the reproduction is an
optical
reproduction, for example, produced by a camera or other optical detector. The
reproduction can
be a non-optical reproduction, for example, a representation of electrical
signals obtained from
an array of nanopore analytes or a representation of electrical signals
obtained from an ion-
sensitive CMOS detector. In particular implementations non-optical
reproductions can be
excluded from a method or apparatus set forth herein. An image can have a
resolution capable of
distinguishing analytes of a specimen that are present at any of a variety of
spacings including,
for example, those that are separated by less than 100 ium, 50 ttm, 10 [un,
5ium, 1 ium or 0.5 ium.
[0291] As used herein, "acquiring", "acquisition" and like terms
refer to any part of the
process of obtaining an image file. In some implementations, data acquisition
can include
generating an image of a specimen, looking for a signal in a specimen,
instructing a detection
device to look for or generate an image of a signal, giving instructions for
further analysis or
transformation of an image file, and any number of transformations or
manipulations of an image
file.
[0292] As used herein, the term "template" refers to a
representation of the location or
relation between signals or analytes. Thus, in some implementations, a
template is a physical
grid with a representation of signals corresponding to analytes in a specimen.
In some
implementations, a template can be a chart, table, text file or other computer
file indicative of
locations corresponding to analytes. In implementations presented herein, a
template is generated
in order to track the location of analytes of a specimen across a set of
images of the specimen
captured at different reference points. For example, a template could be a set
of x,y coordinates
or a set of values that describe the direction and/or distance of one analyte
with respect to
another analyte.
[0293] As used herein, the term "specimen" can refer to an object
or area of an object of
which an image is captured. For example, in implementations where images are
taken of the
surface of the earth, a parcel of land can be a specimen. In other
implementations where the
analysis of biological molecules is performed in a flow cell, the flow cell
may be divided into
any number of subdivisions, each of which may be a specimen. For example, a
flow cell may be
divided into various flow channels or lanes, and each lane can be further
divided into 2, 3, 4, 5,
6,7, 8,9, 10, 20, 30, 40, 50, 60 70, 80, 90, 100, 110, 120, 140, 160, 180,
200, 400, 600, 800,
1000 or more separate regions that are imaged. One example of a flow cell has
8 lanes, with each
lane divided into 120 specimens or tiles. In another implementation, a
specimen may be made up
of a plurality of tiles or even an entire flow cell. Thus, the image of each
specimen can represent
a region of a larger surface that is imaged.
58
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0294] It will be appreciated that references to ranges and
sequential number lists described
herein include not only the enumerated number but all real numbers between the
enumerated
numbers.
[0295] As used herein, a "reference point" refers to any temporal
or physical distinction
between images. In a preferred implementation, a reference point is a time
point. In a more
preferred implementation, a reference point is a time point or cycle during a
sequencing reaction.
However, the term "reference point" can include other aspects that distinguish
or separate
images, such as angle, rotational, temporal, or other aspects that can
distinguish or separate
images.
[0296] As used herein, a "subset of images" refers to a group of
images within a set. For
example, a subset may contain 1,2, 3,4, 6, 8, 10, 12, 14, 16, 18, 20, 30, 40,
50, 60 or any
number of images selected from a set of images_ In particular implementations,
a subset may
contain no more than 1, 2, 3, 4, 6, 8, 10, 12, 14, 16, 18, 20, 30, 40, 50, 60
or any number of
images selected from a set of images. In a preferred implementation, images
are obtained from
one or more sequencing cycles with four images correlated to each cycle. Thus,
for example, a
subset could be a group of 16 images obtained through four cycles.
[0297] A base refers to a nucleotide base or nucleotide, A
(adenine), C (cytosine), T
(thymine), or G (guanine). This application uses "base(s)" and "nucleotide(s)"
interchangeably.
[0298] The term "chromosome" refers to the heredity-bearing gene
carrier of a living cell,
which is derived from chromatin strands comprising DNA and protein components
(especially
histones). The conventional internationally recognized individual human genome
chromosome
numbering system is employed herein.
[0299] The term "site" refers to a unique position (e.g.,
chromosome ID, chromosome
position and orientation) on a reference genome. In some implementations, a
site may be a
residue, a sequence tag, or a segment's position on a sequence. The term
"locus" may be used to
refer to the specific location of a nucleic acid sequence or polymorphism on a
reference
chromosome.
[0300] The term "sample" herein refers to a sample, typically
derived from a biological fluid,
cell, tissue, organ, or organism containing a nucleic acid or a mixture of
nucleic acids containing
at least one nucleic acid sequence that is to be sequenced and/or phased. Such
samples include,
but are not limited to sputum/oral fluid, amniotic fluid, blood, a blood
fraction, fine needle
biopsy samples (e.g., surgical biopsy, fine needle biopsy, etc.), urine,
peritoneal fluid, pleural
fluid, tissue explant, organ culture and any other tissue or cell preparation,
or fraction or
derivative thereof or isolated therefrom. Although the sample is often taken
from a human
subject (e.g., patient), samples can be taken from any organism having
chromosomes, including,
59
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
but not limited to dogs, cats, horses, goats, sheep, cattle, pigs, etc. The
sample may be used
directly as obtained from the biological source or following a pretreatment to
modify the
character of the sample. For example, such pretreatment may include preparing
plasma from
blood, diluting viscous fluids and so forth. Methods of pretreatment may also
involve, but are not
limited to, filtration, precipitation, dilution, distillation, mixing,
centrifugation, freezing,
lyophilization, concentration, amplification, nucleic acid fragmentation,
inactivation of
interfering components, the addition of reagents, lysing, etc.
[0301] The term -sequence" includes or represents a strand of
nucleotides coupled to each
other. The nucleotides may be based on DNA or RNA. It should be understood
that one
sequence may include multiple sub-sequences. For example, a single sequence
(e.g., of a PCR
amplicon) may have 350 nucleotides. The sample read may include multiple sub-
sequences
within these 350 nucleotides_ For instance, the sample read may include first
and second flanking
subsequences having, for example, 20-50 nucleotides. The first and second
flanking sub-
sequences may be located on either side of a repetitive segment haying a
corresponding sub-
sequence (e.g., 40-100 nucleotides). Each of the flanking sub-sequences may
include (or include
portions of) a primer sub-sequence (e.g., 10-30 nucleotides). For ease of
reading, the term "sub-
sequence" will be referred to as "sequence," but it is understood that two
sequences are not
necessarily separate from each other on a common strand. To differentiate the
various sequences
described herein, the sequences may be given different labels (e.g., target
sequence, primer
sequence, flanking sequence, reference sequence, and the like). Other terms,
such as "allele,"
may be given different labels to differentiate between like objects. The
application uses "read(s)"
and "sequence read(s)" interchangeably.
[0302] The term "paired-end sequencing" refers to sequencing
methods that sequence both
ends of a target fragment. Paired-end sequencing may facilitate detection of
genomic
rearrangements and repetitive segments, as well as gene fusions and novel
transcripts.
Methodology for paired-end sequencing are described in PCT publication
W007010252, PCT
application Serial No. PCTGB2007/003798 and US patent application publication
US
2009/0088327, each of which is incorporated by reference herein. In one
example, a series of
operations may be performed as follows; (a) generate clusters of nucleic
acids; (b) linearize the
nucleic acids; (c) hybridize a first sequencing primer and carry out repeated
cycles of extension,
scanning and deblocking, as set forth above; (d) "invert" the target nucleic
acids on the flow cell
surface by synthesizing a complimentary copy; (e) linearize the resynthesized
strand; and (f)
hybridize a second sequencing primer and carry out repeated cycles of
extension, scanning and
deblocking, as set forth above. The inversion operation can be carried out be
delivering reagents
as set forth above for a single cycle of bridge amplification.
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0303] The term "reference genome" or "reference sequence" refers
to any particular known
genome sequence, whether partial or complete, of any organism which may be
used to reference
identified sequences from a subject. For example, a reference genome used for
human subjects as
well as many other organisms is found at the National Center for Biotechnology
Information at
ncbi.nlm.nih.gov. A -genome- refers to the complete genetic information of an
organism or
virus, expressed in nucleic acid sequences. A genome includes both the genes
and the noncoding
sequences of the DNA. The reference sequence may be larger than the reads that
are aligned to
it. For example, it may be at least about 100 times larger, or at least about
1000 times larger, or
at least about 10,000 times larger, or at least about 105 times larger, or at
least about 106 times
larger, or at least about 107 times larger. In one example, the reference
genome sequence is that
of a full length human genome. In another example, the reference genome
sequence is limited to
a specific human chromosome such as chromosome 13 In some implementations, a
reference
chromosome is a chromosome sequence from human genome version hg19. Such
sequences may
be referred to as chromosome reference sequences, although the term reference
genome is
intended to cover such sequences. Other examples of reference sequences
include genomes of
other species, as well as chromosomes, sub-chromosomal regions (such as
strands), etc., of any
species. In various implementations, the reference genome is a consensus
sequence or other
combination derived from multiple individuals. However, in certain
applications, the reference
sequence may be taken from a particular individual. In other implementations,
the "genome" also
covers so-called "graph genomes", which use a particular storage format and
representation of
the genome sequence. In one implementation, graph genomes store data in a
linear file. In
another implementation, the graph genomes refer to a representation where
alternative sequences
(e.g., different copies of a chromosome with small differences) are stored as
different paths in a
graph. Additional information regarding graph genome implementations can be
found in
https://www.biorxiv.org/content/biorxiv/early/2018/03/20/194530.full.pdf, the
content of which
is hereby incorporated herein by reference in its entirety.
[0304] The term "read" refer to a collection of sequence data that
describes a fragment of a
nucleotide sample or reference. The term "read" may refer to a sample read
and/or a reference
read. Typically, though not necessarily, a read represents a short sequence of
contiguous base
pairs in the sample or reference. The read may be represented symbolically by
the base pair
sequence (in ACTG) of the sample or reference fragment. It may be stored in a
memory device
and processed as appropriate to determine whether the read matches a reference
sequence or
meets other criteria. A read may be obtained directly from a sequencing
apparatus or indirectly
from stored sequence information concerning the sample. In some cases, a read
is a DNA
sequence of sufficient length (e.g., at least about 25 bp) that can be used to
identify a larger
61
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
sequence or region, e.g., that can be aligned and specifically assigned to a
chromosome or
genomic region or gene.
[0305] Next-generation sequencing methods include, for example,
sequencing by synthesis
technology (IIlumina), pyrosequencing (454), ion semiconductor technology (Ion
Torrent
sequencing), single-molecule real-time sequencing and sequencing by ligation
(SOLiD
sequencing). Depending on the sequencing methods, the length of each read may
vary from
about 30 bp to more than 10,000 bp. For example, the DNA sequencing method
using SOLiD
sequencer generates nucleic acid reads of about 50 bp. For another example,
Ion Torrent
Sequencing generates nucleic acid reads of up to 400 bp and 454 pyrosequencing
generates
nucleic acid reads of about 700 bp. For yet another example, single-molecule
real-time
sequencing methods may generate reads of 10,000 bp to 15,000 bp. Therefore, in
certain
implementations, the nucleic acid sequence reads have a length of 30-100 bp,
50-200 bp, or 50-
400 bp.
[0306] The terms "sample read", "sample sequence" or "sample
fragment" refer to sequence
data for a genomic sequence of interest from a sample. For example, the sample
read comprises
sequence data from a PCR amplicon having a forward and reverse primer
sequence. The
sequence data can be obtained from any select sequence methodology. The sample
read can be,
for example, from a sequencing-by-synthesis (SBS) reaction, a sequencing-by-
ligation reaction,
or any other suitable sequencing methodology for which it is desired to
determine the length
and/or identity of a repetitive element. The sample read can be a consensus
(e.g., averaged or
weighted) sequence derived from multiple sample reads. In certain
implementations, providing a
reference sequence comprises identifying a locus-of-interest based upon the
primer sequence of
the PCR amplicon.
[0307] The term "raw fragment" refers to sequence data for a
portion of a genomic sequence
of interest that at least partially overlaps a designated position or
secondary position of interest
within a sample read or sample fragment. Non-limiting examples of raw
fragments include a
duplex stitched fragment, a simplex stitched fragment, a duplex un-stitched
fragment and a
simplex un-stitched fragment. The term "raw" is used to indicate that the raw
fragment includes
sequence data having some relation to the sequence data in a sample read,
regardless of whether
the raw fragment exhibits a supporting variant that corresponds to and
authenticates or confirms
a potential variant in a sample read. The term "raw fragment" does not
indicate that the fragment
necessarily includes a supporting variant that validates a variant call in a
sample read. For
example, when a sample read is determined by a variant call application to
exhibit a first variant,
the variant call application may determine that one or more raw fragments lack
a corresponding
62
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
type of "supporting" variant that may otherwise be expected to occur given the
variant in the
sample read.
[0308] The terms "mapping", "aligned," "alignment," or "aligning"
refer to the process of
comparing a read or tag to a reference sequence and thereby determining
whether the reference
sequence contains the read sequence. If the reference sequence contains the
read, the read may
be mapped to the reference sequence or, in certain implementations, to a
particular location in
the reference sequence. In some cases, alignment simply tells whether or not a
read is a member
of a particular reference sequence (i.e., whether the read is present or
absent in the reference
sequence). For example, the alignment of a read to the reference sequence for
human
chromosome 13 will tell whether the read is present in the reference sequence
for chromosome
13. A tool that provides this information may be called a set membership
tester. In some cases,
an alignment additionally indicates a location in the reference sequence where
the read or tag
maps to. For example, if the reference sequence is the whole human genome
sequence, an
alignment may indicate that a read is present on chromosome 13, and may
further indicate that
the read is on a particular strand and/or site of chromosome 13.
[0309] The term "indel" refers to the insertion and/or the deletion
of bases in the DNA of an
organism. A micro-indel represents an indel that results in a net change of 1
to 50 nucleotides. In
coding regions of the genome, unless the length of an indel is a multiple of
3, it will produce a
frameshift mutation. Indels can be contrasted with point mutations. An indel
inserts and deletes
nucleotides from a sequence, while a point mutation is a form of substitution
that replaces one of
the nucleotides without changing the overall number in the DNA. Indels can
also be contrasted
with a Tandem Base Mutation (TBM), which may be defined as substitution at
adjacent
nucleotides (primarily substitutions at two adjacent nucleotides, but
substitutions at three
adjacent nucleotides have been observed.
[0310] The term "variant" refers to a nucleic acid sequence that is
different from a nucleic
acid reference. Typical nucleic acid sequence variant includes without
limitation single
nucleotide polymorphism (SNP), short deletion and insertion polymorphisms
(Indel), copy
number variation (CNV), microsatellite markers or short tandem repeats and
structural variation.
Somatic variant calling is the effort to identify variants present at low
frequency in the DNA
sample. Somatic variant calling is of interest in the context of cancer
treatment. Cancer is caused
by an accumulation of mutations in DNA. A DNA sample from a tumor is generally
heterogeneous, including some normal cells, some cells at an early stage of
cancer progression
(with fewer mutations), and some late-stage cells (with more mutations).
Because of this
heterogeneity, when sequencing a tumor (e.g., from an FFPE sample), somatic
mutations will
often appear at a low frequency. For example, a SNV might be seen in only 10%
of the reads
63
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
covering a given base. A variant that is to be classified as somatic or
germline by the variant
classifier is also referred to herein as the "variant under test".
[0311] The term "noise" refers to a mistaken variant call resulting
from one or more errors in
the sequencing process and/or in the variant call application.
[0312] The term -variant frequency" represents the relative
frequency of an allele (variant of
a gene) at a particular locus in a population, expressed as a fraction or
percentage. For example,
the fraction or percentage may be the fraction of all chromosomes in the
population that carry
that allele. By way of example, sample variant frequency represents the
relative frequency of an
allele/variant at a particular locus/position along a genomic sequence of
interest over a
"population" corresponding to the number of reads and/or samples obtained for
the genomic
sequence of interest from an individual. As another example, a baseline
variant frequency
represents the relative frequency of an allele/variant at a particular
locus/position along one or
more baseline genomic sequences where the "population" corresponding to the
number of reads
and/or samples obtained for the one or more baseline genomic sequences from a
population of
normal individuals.
[0313] The term "variant allele frequency (VAF)" refers to the
percentage of sequenced
reads observed matching the variant divided by the overall coverage at the
target position. VAF
is a measure of the proportion of sequenced reads carrying the variant.
[0314] The terms "position", "designated position", and "locus"
refer to a location or
coordinate of one or more nucleotides within a sequence of nucleotides. The
terms "position",
"designated position", and "locus" also refer to a location or coordinate of
one or more base pairs
in a sequence of nucleotides.
[0315] The term "haplotype" refers to a combination of alleles at
adjacent sites on a
chromosome that are inherited together. A haplotype may be one locus, several
loci, or an entire
chromosome depending on the number of recombination events that have occurred
between a
given set of loci, if any occurred.
[0316] The term "threshold" herein refers to a numeric or non-
numeric value that is used as a
cutoff to characterize a sample, a nucleic acid, or portion thereof (e.g., a
read). A threshold may
be varied based upon empirical analysis. The threshold may be compared to a
measured or
calculated value to determine whether the source giving rise to such value
suggests should be
classified in a particular manner. Threshold values can be identified
empirically or analytically.
The choice of a threshold is dependent on the level of confidence that the
user wishes to have to
make the classification. The threshold may be chosen for a particular purpose
(e.g., to balance
sensitivity and selectivity). As used herein, the term "threshold" indicates a
point at which a
course of analysis may be changed and/or a point at which an action may be
triggered. A
64
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
threshold is not required to be a predetermined number. Instead, the threshold
may be, for
instance, a function that is based on a plurality of factors. The threshold
may be adaptive to the
circumstances. Moreover, a threshold may indicate an upper limit, a lower
limit, or a range
between limits.
[0317] In some implementations, a metric or score that is based on
sequencing data may be
compared to the threshold. As used herein, the terms "metric" or "score" may
include values or
results that were determined from the sequencing data or may include functions
that are based on
the values or results that were determined from the sequencing data. Like a
threshold, the metric
or score may be adaptive to the circumstances. For instance, the metric or
score may be a
normalized value. As an example of a score or metric, one or more
implementations may use
count scores when analyzing the data. A count score may be based on number of
sample reads.
The sample reads may have undergone one or more filtering stages such that the
sample reads
have at least one common characteristic or quality. For example, each of the
sample reads that
are used to determine a count score may have been aligned with a reference
sequence or may be
assigned as a potential allele. The number of sample reads having a common
characteristic may
be counted to determine a read count. Count scores may be based on the read
count. In some
implementations, the count score may be a value that is equal to the read
count. In other
implementations, the count score may be based on the read count and other
information. For
example, a count score may be based on the read count for a particular allele
of a genetic locus
and a total number of reads for the genetic locus. In some implementations,
the count score may
be based on the read count and previously-obtained data for the genetic locus.
In some
implementations, the count scores may be normalized scores between
predetermined values. The
count score may also be a function of read counts from other loci of a sample
or a function of
read counts from other samples that were concurrently run with the sample-of-
interest. For
instance, the count score may be a function of the read count of a particular
allele and the read
counts of other loci in the sample and/or the read counts from other samples.
As one example,
the read counts from other loci and/or the read counts from other samples may
be used to
normalize the count score for the particular allele.
[0318] The terms "coverage" or "fragment coverage" refer to a count
or other measure of a
number of sample reads for the same fragment of a sequence. A read count may
represent a
count of the number of reads that cover a corresponding fragment.
Alternatively, the coverage
may be determined by multiplying the read count by a designated factor that is
based on
historical knowledge, knowledge of the sample, knowledge of the locus, etc.
[0319] The term "read depth" (conventionally a number followed by
"x') refers to the
number of sequenced reads with overlapping alignment at the target position.
This is often
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
expressed as an average or percentage exceeding a cutoff over a set of
intervals (such as exons,
genes, or panels). For example, a clinical report might say that a panel
average coverage is
1,105x with 98% of targeted bases covered >100x.
[0320] The terms "base call quality score" or "Q score" refer to a
PHRED-scaled probability
ranging from 0-50 inversely proportional to the probability that a single
sequenced base is
correct. For example, a T base call with Q of 20 is considered likely correct
with a probability of
99.99%. Any base call with Q<20 should be considered low quality, and any
variant identified
where a substantial proportion of sequenced reads supporting the variant are
of low quality
should be considered potentially false positive.
[0321] The terms "variant reads" or "variant read number" refer to
the number of sequenced
reads supporting the presence of the variant.
[0322] Regarding "strandedness" (or DNA strandedness), the genetic
message in DNA can
be represented as a string of the letters A, G, C, and T. For example, 5' ¨
AGGACA ¨ 3'. Often,
the sequence is written in the direction shown here, i.e., with the 5' end to
the left and the 3' end
to the right. DNA may sometimes occur as single-stranded molecule (as in
certain viruses), but
normally we find DNA as a double-stranded unit. It has a double helical
structure with two
antiparallel strands. In this case, the word "antiparallel" means that the two
strands run in
parallel, but have opposite polarity. The double-stranded DNA is held together
by pairing
between bases and the pairing is always such that adenine (A) pairs with
thymine (T) and
cytosine (C) pairs with guanine (G). This pairing is referred to as
complementarity, and one
strand of DNA is said to be the complement of the other. The double-stranded
DNA may thus be
represented as two strings, like this: 5' ¨ AGGACA ¨ 3' and 3' ¨ TCCTGT ¨ 5'.
Note that the
two strands have opposite polarity. Accordingly, the strandedness of the two
DNA strands can be
referred to as the reference strand and its complement, forward and reverse
strands, top and
bottom strands, sense and antisense strands, or Watson and Crick strands.
[0323] The reads alignment (also called reads mapping) is the
process of figuring out where
in the genome a sequence is from. Once the alignment is performed, the
"mapping quality" or the
"mapping quality score (MAPQ)" of a given read quantifies the probability that
its position on
the genome is correct. The mapping quality is encoded in the phred scale where
P is the
probability that the alignment is not correct. The probability is calculated
as: P =1 010)
where MAPQ is the mapping quality. For example, a mapping quality of 40 = 10
to the power of
-4, meaning that there is a 0.01% chance that the read was aligned
incorrectly. The mapping
quality is therefore associated with several alignment factors, such as the
base quality of the read,
the complexity of the reference genome, and the paired-end information.
Regarding the first, if
the base quality of the read is low, it means that the observed sequence might
be wrong and thus
66
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
its alignment is wrong. Regarding the second, the mappability refers to the
complexity of the
genome. Repeated regions are more difficult to map and reads falling in these
regions usually get
low mapping quality. In this context, the MAPQ reflects the fact that the
reads are not uniquely
aligned and that their real origin cannot be determined. Regarding the third,
in case of paired-end
sequencing data, concordant pairs are more likely to be well aligned. The
higher is the mapping
quality, the better is the alignment. A read aligned with a good mapping
quality usually means
that the read sequence was good and was aligned with few mismatches in a high
mappability
region. The MAPQ value can be used as a quality control of the alignment
results. The
proportion of reads aligned with an MAPQ higher than 20 is usually for
downstream analysis.
[0324] As used herein, a "signal" refers to a detectable event such
as an emission, preferably
light emission, for example, in an image. Thus, in preferred implementations,
a signal can
represent any detectable light emission that is captured in an image (i.e., a
"spot") Thus, as used
herein, "signal" can refer to both an actual emission from an analyte of the
specimen, and can
refer to a spurious emission that does not correlate to an actual analyte.
Thus, a signal could arise
from noise and could be later discarded as not representative of an actual
analyte of a specimen.
[0325] As used herein, the term "clump" refers to a group of
signals. In particular
implementations, the signals are derived from different analytes. In a
preferred implementation, a
signal clump is a group of signals that cluster together. In a more preferred
implementation, a
signal clump represents a physical region covered by one amplified
oligonucleotide. Each signal
clump should be ideally observed as several signals (one per template cycle,
and possibly more
due to cross-talk). Accordingly, duplicate signals are detected where two (or
more) signals are
included in a template from the same clump of signals.
[0326] As used herein, terms such as "minimum," "maximum,"
"minimize," "maximize"
and grammatical variants thereof can include values that are not the absolute
maxima or minima.
In some implementations, the values include near maximum and near minimum
values. In other
implementations, the values can include local maximum and/or local minimum
values. In some
implementations, the values include only absolute maximum or minimum values.
[0327] As used herein, "cross-talk" refers to the detection of
signals in one image that are
also detected in a separate image. In a preferred implementation, cross-talk
can occur when an
emitted signal is detected in two separate detection channels. For example,
where an emitted
signal occurs in one color, the emission spectrum of that signal may overlap
with another emitted
signal in another color. In a preferred implementation, fluorescent molecules
used to indicate the
presence of nucleotide bases A, C, G and T are detected in separate channels.
However, because
the emission spectra of A and C overlap, some of the C color signal may be
detected during
detection using the A color channel. Accordingly, cross-talk between the A and
C signals allows
67
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
signals from one color image to appear in the other color image. In some
implementations, G and
T cross-talk. In some implementations, the amount of cross-talk between
channels is asymmetric.
It will be appreciated that the amount of cross-talk between channels can be
controlled by,
among other things, the selection of signal molecules having an appropriate
emission spectrum
as well as selection of the size and wavelength range of the detection
channel.
[0328] As used herein, "register", "registering", "registration"
and like terms refer to any
process to correlate signals in an image or data set from a first time point
or perspective with
signals in an image or data set from another time point or perspective. For
example, registration
can be used to align signals from a set of images to form a template. In
another example,
registration can be used to align signals from other images to a template. One
signal may be
directly or indirectly registered to another signal. For example, a signal
from image "S" may be
registered to image "G" directly_ As another example, a signal from image "N.'
may be directly
registered to image "G", or alternatively, the signal from image "N" may be
registered to image
"S", which has previously been registered to image "G". Thus, the signal from
image "N" is
indirectly registered to image "G".
[0329] As used herein, the term "fiducial" is intended to mean a
distinguishable point of
reference in or on an object. The point of reference can be, for example, a
mark, second object,
shape, edge, area, irregularity, channel, pit, post or the like. The point of
reference can be present
in an image of the object or in another data set derived from detecting the
object. The point of
reference can be specified by an x and/or y coordinate in a plane of the obj
ect. Alternatively or
additionally, the point of reference can be specified by a z coordinate that
is orthogonal to the xy
plane, for example, being defined by the relative locations of the object and
a detector. One or
more coordinates for a point of reference can be specified relative to one or
more other analytes
of an object or of an image or other data set derived from the object.
[0330] As used herein, the term "optical signal" is intended to
include, for example,
fluorescent, luminescent, scatter, or absorption signals. Optical signals can
be detected in the
ultraviolet (UV) range (about 200 to 390 nm), visible (VIS) range (about 391
to 770 nm),
infrared (IR) range (about 0.771 to 25 microns), or other range of the
electromagnetic spectrum.
Optical signals can be detected in a way that excludes all or part of one or
more of these ranges.
[0331] As used herein, the term "signal level" is intended to mean
an amount or quantity of
detected energy or coded information that has a desired or predefined
characteristic. For
example, an optical signal can be quantified by one or more of intensity,
wavelength, energy,
frequency, power, luminance or the like. Other signals can be quantified
according to
characteristics such as voltage, current, electric field strength, magnetic
field strength, frequency,
68
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
power, temperature, etc. Absence of signal is understood to be a signal level
of zero or a signal
level that is not meaningfully distinguished from noise.
[0332] As used herein, the term "simulate" is intended to mean
creating a representation or
model of a physical thing or action that predicts characteristics of the thing
or action. The
representation or model can in many cases be distinguishable from the thing or
action. For
example, the representation or model can be distinguishable from a thing with
respect to one or
more characteristic such as color, intensity of signals detected from all or
part of the thing, size,
or shape. In particular implementations, the representation or model can be
idealized,
exaggerated, muted, or incomplete when compared to the thing or action. Thus,
in some
implementations, a representation of model can be distinguishable from the
thing or action that it
represents, for example, with respect to at least one of the characteristics
set forth above. The
representation or model can be provided in a computer readable format or
medium such as one or
more of those set forth elsewhere herein
[0333] As used herein, the term "specific signal" is intended to
mean detected energy or
coded information that is selectively observed over other energy or
information such as
background energy or information. For example, a specific signal can be an
optical signal
detected at a particular intensity, wavelength or color, an electrical signal
detected at a particular
frequency, power or field strength; or other signals known in the art
pertaining to spectroscopy
and analytical detection.
[0334] As used herein, the term "swath" is intended to mean a
rectangular portion of an
object. The swath can be an elongated strip that is scanned by relative
movement between the
object and a detector in a direction that is parallel to the longest dimension
of the strip.
Generally, the width of the rectangular portion or strip will be constant
along its full length.
Multiple swaths of an object can be parallel to each other. Multiple swaths of
an object can be
adjacent to each other, overlapping with each other, abutting each other, or
separated from each
other by an interstitial area.
[0335] As used herein, the term "variance" is intended to mean a
difference between that
which is expected and that which is observed or a difference between two or
more observations.
For example, variance can be the discrepancy between an expected value and a
measured value.
Variance can be represented using statistical functions such as standard
deviation, the square of
standard deviation, coefficient of variation or the like.
[0336] As used herein, the term "xy coordinates" is intended to
mean information that
specifies location, size, shape, and/or orientation in an xy plane. The
information can be, for
example, numerical coordinates in a Cartesian system. The coordinates can be
provided relative
to one or both of the x and y axes or can be provided relative to another
location in the xy plane.
69
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
For example, coordinates of a analyte of an object can specify the location of
the analyte relative
to location of a fiducial or other analyte of the object.
[0337] As used herein, the term "xy plane" is intended to mean a 2
dimensional area defined
by straight line axes x and y. When used in reference to a detector and an
object observed by the
detector, the area can be further specified as being orthogonal to the
direction of observation
between the detector and object being detected.
[0338] As used herein, the term "z coordinate" is intended to mean
information that specifies
the location of a point, line or area along an axes that is orthogonal to an
xy plane. In particular
implementations, the z axis is orthogonal to an area of an object that is
observed by a detector.
For example, the direction of focus for an optical system may be specified
along the z axis.
[0339] In some implementations, acquired signal data is transformed
using an affine
transformation_ In some such implementations, template generation makes use of
the fact that the
affine transforms between color channels are consistent between runs Because
of this
consistency, a set of default offsets can be used when determining the
coordinates of the analytes
in a specimen. For example, a default offsets file can contain the relative
transformation (shift,
scale, skew) for the different channels relative to one channel, such as the A
channel. In other
implementations, however, the offsets between color channels drift during a
run and/or between
runs, making offset-driven template generation difficult. In such
implementations, the methods
and systems provided herein can utilize offset-less template generation, which
is described
further below.
[0340] In some implementations of the above implementations, the
system can comprise a
flow cell. In some implementations, the flow cell comprises lanes, or other
configurations, of
tiles, wherein at least some of the tiles comprise one or more arrays of
analytes. In some
implementations, the analytes comprise a plurality of molecules such as
nucleic acids. In certain
aspects, the flow cell is configured to deliver a labeled nucleotide base to
an array of nucleic
acids, thereby extending a primer hybridized to a nucleic acid within a
analyte so as to produce a
signal corresponding to a analyte comprising the nucleic acid. In preferred
implementations, the
nucleic acids within a analyte are identical or substantially identical to
each other.
[0341] In some of the systems for image analysis described herein,
each image in the set of
images includes color signals, wherein a different color corresponds to a
different nucleotide
base. In some implementations, each image of the set of images comprises
signals having a
single color selected from at least four different colors. In some
implementations, each image in
the set of images comprises signals having a single color selected from four
different colors. In
some of the systems described herein, nucleic acids can be sequenced by
providing four different
labeled nucleotide bases to the array of molecules so as to produce four
different images, each
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
image comprising signals having a single color, wherein the signal color is
different for each of
the four different images, thereby producing a cycle of four color images that
corresponds to the
four possible nucleotides present at a particular position in the nucleic
acid. In certain aspects,
the system comprises a flow cell that is configured to deliver additional
labeled nucleotide bases
to the array of molecules, thereby producing a plurality of cycles of color
images.
[0342] In preferred implementations, the methods provided herein
can include determining
whether a processor is actively acquiring data or whether the processor is in
a low activity state.
Acquiring and storing large numbers of high-quality images typically requires
massive amounts
of storage capacity. Additionally, once acquired and stored, the analysis of
image data can
become resource intensive and can interfere with processing capacity of other
functions, such as
ongoing acquisition and storage of additional image data. Accordingly, as used
herein, the term
low activity state refers to the processing capacity of a processor at a given
time In some
implementations, a low activity state occurs when a processor is not acquiring
and/or storing
data. In some implementations, a low activity state occurs when some data
acquisition and/or
storage is taking place, but additional processing capacity remains such that
image analysis can
occur at the same time without interfering with other functions.
[0343] As used herein, "identifying a conflict" refers to
identifying a situation where
multiple processes compete for resources. In some such implementations, one
process is given
priority over another process. In some implementations, a conflict may relate
to the need to give
priority for allocation of time, processing capacity, storage capacity or any
other resource for
which priority is given. Thus, in some implementations, where processing time
or capacity is to
be distributed between two processes such as either analyzing a data set and
acquiring and/or
storing the data set, a conflict between the two processes exists and can be
resolved by giving
priority to one of the processes.
[0344] Also provided herein are systems for performing image
analysis. The systems can
include a processor; a storage capacity; and a program for image analysis, the
program
comprising instructions for processing a first data set for storage and the
second data set for
analysis, wherein the processing comprises acquiring and/or storing the first
data set on the
storage device and analyzing the second data set when the processor is not
acquiring the first
data set. In certain aspects, the program includes instructions for
identifying at least one instance
of a conflict between acquiring and/or storing the first data set and
analyzing the second data set;
and resolving the conflict in favor of acquiring and/or storing image data
such that acquiring
and/or storing the first data set is given priority. In certain aspects, the
first data set comprises
image files obtained from an optical imaging device In certain aspects, the
system further
71
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
comprises an optical imaging device. In some implementations, the optical
imaging device
comprises a light source and a detection device.
[0345] As used herein, the term "program" refers to instructions or
commands to perform a
task or process. The term "program" can be used interchangeably with the term
module. In
certain implementations, a program can be a compilation of various
instructions executed under
the same set of commands. In other implementations, a program can refer to a
discrete batch or
file.
[0346] Set forth below are some of the surprising effects of
utilizing the methods and
systems for performing image analysis set forth herein. In some sequencing
implementations, an
important measure of a sequencing system's utility is its overall efficiency.
For example, the
amount of mappable data produced per day and the total cost of installing and
running the
instrument are important aspects of an economical sequencing solution To
reduce the time to
generate mappable data and to increase the efficiency of the system, real-time
base calling can be
enabled on an instrument computer and can run in parallel with sequencing
chemistry and
imaging. This allows much of the data processing and analysis to be completed
before the
sequencing chemistry finishes. Additionally, it can reduce the storage
required for intermediate
data and limit the amount of data that needs to travel across the network.
[0347] While sequence output has increased, the data per run
transferred from the systems
provided herein to the network and to secondary analysis processing hardware
has substantially
decreased. By transforming data on the instrument computer (acquiring
computer), network
loads are dramatically reduced. Without these on-instrument, off-network data
reduction
techniques, the image output of a fleet of DNA sequencing instruments would
cripple most
networks.
[0348] The widespread adoption of the high-throughput DNA
sequencing instruments has
been driven in part by ease of use, support for a range of applications, and
suitability for virtually
any lab environment. The highly efficient algorithms presented herein allow
significant analysis
functionality to be added to a simple workstation that can control sequencing
instruments. This
reduction in the requirements for computational hardware has several practical
benefits that will
become even more important as sequencing output levels continue to increase.
For example, by
performing image analysis and base calling on a simple tower, heat production,
laboratory
footprint, and power consumption are kept to a minimum. In contrast, other
commercial
sequencing technologies have recently ramped up their computing infrastructure
for primary
analysis, with up to five times more processing power, leading to commensurate
increases in
heat output and power consumption. Thus, in some implementations, the
computational
72
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
efficiency of the methods and systems provided herein enables customers to
increase their
sequencing throughput while keeping server hardware expenses to a minimum.
[0349] Accordingly, in some implementations, the methods and/or
systems presented herein
act as a state machine, keeping track of the individual state of each
specimen, and when it detects
that a specimen is ready to advance to the next state, it does the appropriate
processing and
advances the specimen to that state. A more detailed example of how the state
machine monitors
a file system to determine when a specimen is ready to advance to the next
state according to a
preferred implementation is set forth in Example 1 below.
[0350] In preferred implementations, the methods and systems
provided herein are multi-
threaded and can work with a configurable number of threads. Thus, for example
in the context
of nucleic acid sequencing, the methods and systems provided herein are
capable of working in
the background during a live sequencing run for real-time analysis, or it can
be run using a pre-
existing set of image data for off-line analysis. In certain preferred
implementations, the methods
and systems handle multi-threading by giving each thread its own subset of
specimen for which
it is responsible. This minimizes the possibility of thread contention.
[0351] A method of the present disclosure can include a step of
obtaining a target image of
an object using a detection apparatus, wherein the image includes a repeating
pattern of analytes
on the object. Detection apparatus that are capable of high resolution imaging
of surfaces are
particularly useful. In particular implementations, the detection apparatus
will have sufficient
resolution to distinguish analytes at the densities, pitches, and/or analyte
sizes set forth herein.
Particularly useful are detection apparatus capable of obtaining images or
image data from
surfaces. Example detectors are those that are configured to maintain an
object and detector in a
static relationship while obtaining an area image. Scanning apparatus can also
be used. For
example, an apparatus that obtains sequential area images (e.g., so called
'step and shoot'
detectors) can be used. Also useful are devices that continually scan a point
or line over the
surface of an obj ect to accumulate data to construct an image of the surface.
Point scanning
detectors can be configured to scan a point (i.e., a small detection area)
over the surface of an
object via a raster motion in the x-y plane of the surface. Line scanning
detectors can be
configured to scan a line along they dimension of the surface of an object,
the longest dimension
of the line occurring along the x dimension. It will be understood that the
detection device,
object or both can be moved to achieve scanning detection. Detection apparatus
that are
particularly useful, for example in nucleic acid sequencing applications, are
described in US Pat
App. Pub. Nos. 2012/0270305 Al; 2013/0023422 Al; and 2013/0260372 Al; and U.S.
Pat. Nos.
5,528,050; 5,719,391; 8,158,926 and 8,241,573, each of which is incorporated
herein by
reference.
73
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0352] The implementations disclosed herein may be implemented as a
method, apparatus,
system, or article of manufacture using programming or engineering techniques
to produce
software, firmware, hardware, or any combination thereof. The term "article of
manufacture" as
used herein refers to code or logic implemented in hardware or computer
readable media such as
optical storage devices, and volatile or non-volatile memory devices. Such
hardware may
include, but is not limited to, field programmable gate arrays (FPGAs), coarse
grained
reconfigurable architectures (CGRAs), application-specific integrated circuits
(ASICs), complex
programmable logic devices (CPLDs), programmable logic arrays (PLAs),
microprocessors, or
other similar processing devices. In particular implementations, information
or algorithms set
forth herein are present in non-transient storage media.
[0353] In particular implementations, a computer implemented method
set forth herein can
occur in real time while multiple images of an object are being obtained Such
real time analysis
is particularly useful for nucleic acid sequencing applications wherein an
array of nucleic acids is
subjected to repeated cycles of fluidic and detection steps. Analysis of the
sequencing data can
often be computationally intensive such that it can be beneficial to perform
the methods set forth
herein in real time or in the background while other data acquisition or
analysis algorithms are in
process. Example real time analysis methods that can be used with the present
methods are those
used for the MiSeq and HiSeq sequencing devices commercially available from
Illumina, Inc.
(San Diego, Calif.) and/or described in US Pat. App. Pub. No. 2012/0020537 Al,
which is
incorporated herein by reference.
[0354] An example data analysis system, formed by one or more
programmed computers,
with programming being stored on one or more machine readable media with code
executed to
carry out one or more steps of methods described herein. In one
implementation, for example,
the system includes an interface designed to permit networking of the system
to one or more
detection systems (e.g., optical imaging systems) that are configured to
acquire data from target
objects. The interface may receive and condition data, where appropriate. In
particular
implementations the detection system will output digital image data, for
example, image data
that is representative of individual picture elements or pixels that,
together, form an image of an
array or other object. A processor processes the received detection data in
accordance with a one
or more routines defined by processing code. The processing code may be stored
in various types
of memory circuitry.
[0355] In accordance with the presently contemplated
implementations, the processing code
executed on the detection data includes a data analysis routine designed to
analyze the detection
data to determine the locations and metadata of individual analytes visible or
encoded in the data,
as well as locations at which no analyte is detected (i.e., where there is no
analyte, or where no
74
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
meaningful signal was detected from an existing analyte). In particular
implementations, analyte
locations in an array will typically appear brighter than non-analyte
locations due to the presence
of fluorescing dyes attached to the imaged analytes. It will be understood
that the analytes need
not appear brighter than their surrounding area, for example, when a target
for the probe at the
analyte is not present in an array being detected. The color at which
individual analytes appear
may be a function of the dye employed as well as of the wavelength of the
light used by the
imaging system for imaging purposes. Analytes to which targets are not bound
or that are
otherwise devoid of a particular label can be identified according to other
characteristics, such as
their expected location in the microarray.
[0356] Once the data analysis routine has located individual
analytes in the data, a value
assignment may be carried out. In general, the value assignment will assign a
digital value to
each analyte based upon characteristics of the data represented by detector
components (e.g.,
pixels) at the corresponding location. That is, for example when imaging data
is processed, the
value assignment routine may be designed to recognize that a specific color or
wavelength of
light was detected at a specific location, as indicated by a group or cluster
of pixels at the
location. In a typical DNA imaging application, for example, the four common
nucleotides will
be represented by four separate and distinguishable colors. Each color, then,
may be assigned a
value corresponding to that nucleotide.
[0357] As used herein, the terms "module", "system," or "system
controller" may include a
hardware and/or software system and circuitry that operates to perform one or
more functions.
For example, a module, system, or system controller may include a computer
processor,
controller, or other logic-based device that performs operations based on
instructions stored on a
tangible and non-transitory computer readable storage medium, such as a
computer memory.
Alternatively, a module, system, or system controller may include a hard-wired
device that
performs operations based on hard-wired logic and circuitry. The module,
system, or system
controller shown in the attached figures may represent the hardware and
circuitry that operates
based on software or hardwired instructions, the software that directs
hardware to perform the
operations, or a combination thereof. The module, system, or system controller
can include or
represent hardware circuits or circuitry that include and/or are connected
with one or more
processors, such as one or computer microprocessors.
[0358] As used herein, the terms "software" and "firmware" are
interchangeable and include
any computer program stored in memory for execution by a computer, including
RAM memory,
ROM memory, EPROM memory, EEPROM memory, and non-volatile RAM (NVRAM)
memory. The above memory types are examples only and are thus not limiting as
to the types of
memory usable for storage of a computer program.
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0359] In the molecular biology field, one of the processes for
nucleic acid sequencing in use
is sequencing-by-synthesis. The technique can be applied to massively parallel
sequencing
projects. For example, by using an automated platform, it is possible to carry
out hundreds of
thousands of sequencing reactions simultaneously. Thus, one of the
implementations of the
present invention relates to instruments and methods for acquiring, storing,
and analyzing image
data generated during nucleic acid sequencing.
[0360] Enormous gains in the amount of data that can be acquired
and stored make
streamlined image analysis methods even more beneficial. For example, the
image analysis
methods described herein permit both designers and end users to make efficient
use of existing
computer hardware. Accordingly, presented herein are methods and systems which
reduce the
computational burden of processing data in the face of rapidly increasing data
output. For
example, in the field of DNA sequencing, yields have scaled 15-fold over the
course of a recent
year and can now reach hundreds of gigabases in a single run of a DNA
sequencing device. If
computational infrastructure requirements grew proportionately, large genome-
scale experiments
would remain out of reach to most researchers. Thus, the generation of more
raw sequence data
will increase the need for secondary analysis and data storage, making
optimization of data
transport and storage extremely valuable. Some implementations of the methods
and systems
presented herein can reduce the time, hardware, networking, and laboratory
infrastructure
requirements needed to produce usable sequence data.
[0361] The present disclosure describes various methods and systems
for carrying out the
methods. Examples of some of the methods are described as a series of steps.
However, it should
be understood that implementations are not limited to the particular steps
and/or order of steps
described herein. Steps may be omitted, steps may be modified, and/or other
steps may be added.
Moreover, steps described herein may be combined, steps may be performed
simultaneously,
steps may be performed concurrently, steps may be split into multiple sub-
steps, steps may be
performed in a different order, or steps (or a series of steps) may be re-
performed in an iterative
fashion. In addition, although different methods are set forth herein, it
should be understood that
the different methods (or steps of the different methods) may be combined in
other
implementations.
[0362] In some implementations, a processing unit, processor,
module, or computing system
that is "configured to" perform a task or operation may be understood as being
particularly
structured to perform the task or operation (e.g., having one or more programs
or instructions
stored thereon or used in conjunction therewith tailored or intended to
perform the task or
operation, and/or having an arrangement of processing circuitry tailored or
intended to perform
the task or operation). For the purposes of clarity and the avoidance of
doubt, a general purpose
76
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
computer (which may become "configured to" perform the task or operation if
appropriately
programmed) is not "configured to" perform a task or operation unless or until
specifically
programmed or structurally modified to perform the task or operation.
[0363] Moreover, the operations of the methods described herein can
be sufficiently complex
such that the operations cannot be mentally performed by an average human
being or a person of
ordinary skill in the art within a commercially reasonable time period. For
example, the methods
may rely on relatively complex computations such that such a person cannot
complete the
methods within a commercially reasonable time.
[0364] Throughout this application various publications, patents or
patent applications have
been referenced. The disclosures of these publications in their entireties are
hereby incorporated
by reference in this application in order to more fully describe the state of
the art to which this
invention pertains
[0365] The term "comprising" is intended herein to be open-ended,
including not only the
recited elements, but further encompassing any additional elements.
[0366] As used herein, the term "each", when used in reference to a
collection of items, is
intended to identify an individual item in the collection but does not
necessarily refer to every
item in the collection. Exceptions can occur if explicit disclosure or context
clearly dictates
otherwise.
[0367] Although the invention has been described with reference to
the examples provided
above, it should be understood that various modifications can be made without
departing from
the invention.
[0368] The modules in this application can be implemented in
hardware or software and
need not be divided up in precisely the same blocks as shown in the figures.
Some can also be
implemented on different processors or computers or spread among a number of
different
processors or computers. In addition, it will be appreciated that some of the
modules can be
combined, operated in parallel or in a different sequence than that shown in
the figures without
affecting the functions achieved. Also as used herein, the term "module" can
include "sub-
modules", which themselves can be considered herein to constitute modules. The
blocks in the
figures designated as modules can also be thought of as flowchart steps in a
method.
[0369] As used herein, the "identification" of an item of
information does not necessarily
require the direct specification of that item of information. Information can
be "identified" in a
field by simply referring to the actual information through one or more layers
of indirection, or
by identifying one or more items of different information which are together
sufficient to
determine the actual item of information. In addition, the term "specify" is
used herein to mean
the same as "identify".
77
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0370] As used herein, a given signal, event or value is "in
dependence upon" a predecessor
signal, event or value of the predecessor signal, event or value influenced by
the given signal,
event, or value. If there is an intervening processing element, step or time
period, the given
signal, event, or value can still be "in dependence upon" the predecessor
signal, event, or value.
If the intervening processing element or step combines more than one signal,
event or value, the
signal output of the processing element or step is considered "in dependence
upon" each of the
signal, event, or value inputs. If the given signal, event, or value is the
same as the predecessor
signal, event, or value, this is merely a degenerate case in which the given
signal, event or value
is still considered to be -in dependence upon" or -dependent on" or -based on"
the predecessor
signal, event, or value. "Responsiveness" of a given signal, event or value
upon another signal,
event or value is defined similarly.
[0371] As used herein, "concurrently" or "in parallel" does not
require exact simultaneity. It
is sufficient if the evaluation of one of the individuals begins before the
evaluation of another of
the individuals completes.
[0372] This application refers to "cluster images" and "cluster
intensity images"
interchangeably.
Particular Implementations
[0373] We describe various implementations of artificial
intelligence-based base calling
using knowledge distillation techniques. One or more features of an
implementation can be
combined with the base implementation. Implementations that are not mutually
exclusive are
taught to be combinable. One or more features of an implementation can be
combined with other
implementations. This disclosure periodically reminds the user of these
options. Omission from
some implementations of recitations that repeat these options should not be
taken as limiting the
combinations taught in the preceding sections - these recitations are hereby
incorporated forward
by reference into each of the following implementations.
[0374] We disclose an artificial intelligence-based method of base
calling. The method
includes training a teacher (first, bigger) base caller by using a first set
of cluster images as
training data. The first set of cluster images are annotated with first ground
truth data that uses
discrete valued labels to identify a correct base call. In one implementation,
the discrete valued
labels are one-hot encoded with a one-value for a correct base and zero-values
for incorrect
bases. In one implementation, the discrete valued labels have a near-one-value
for the correct
base and near-zero-values for the incorrect bases.
[0375] The method includes evaluating a second set of cluster
images as inference data by
applying the trained teacher (first, bigger) base caller on the second set of
cluster images and
78
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
generating base call predictions. The base call predictions are represented by
continuous valued
weights that identify a predicted base call. In one implementation, the
continuous valued weights
are part of a probability distribution for a correct base being Adenine (A),
Cytosine (C), Thymine
(T), and Guanine (G).
[0376] The method includes training a student (second, smaller)
base caller using the second
set of cluster images as training data. The second set of cluster images are
annotated with second
ground truth data that identifies a correct base call based on (i) the
discrete valued labels and (ii)
the continuous valued weights.
[0377] The student (second, smaller) base caller has fewer
processing modules and
parameters than the teacher (first, bigger) base caller. In one
implementation, one of the
processing modules is neural network layers. In one implementation, one of the
parameters is
interconnections between the neural network layers In one implementation, one
of the
processing modules is neural network filters. In one implementation, one of
the processing
modules is neural network kernels. In one implementation, one of the
parameters is
multiplication and addition operations.
[0378] The method includes evaluating a third set of cluster images
as inference data by
applying the trained student (second, smaller) base caller on the third set of
cluster images and
generating base call predictions.
[0379] The method described in this section and other sections of
the technology disclosed
can include one or more of the following features and/or features described in
connection with
additional methods disclosed. In the interest of conciseness, the combinations
of features
disclosed in this application are not individually enumerated and are not
repeated with each base
set of features. The reader will understand how features identified in these
implementations can
readily be combined with sets of base features identified in other
implementations.
[0380] In one implementation, the method includes training the
student (second, smaller)
base caller using the second set of cluster images as training data. The
second set of cluster
images are annotated with the second ground truth data that identifies the
correct base call based
on the continuous valued weights.
[0381] In one implementation, a cluster image depicts intensity
emissions of clusters. The
intensity emissions are captured during a sequencing cycle of a sequencing
run. In one
implementation, the cluster image further depicts intensity emissions of
background surrounding
the clusters.
[0382] In one implementation, the first, second, and third sets of
cluster images share one or
more common cluster images.
79
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
[0383] In one implementation, the method includes training an
ensemble of the teacher (first,
bigger) base caller by using the first set of cluster images as training data.
The first set of cluster
images are annotated with the first ground truth data that uses the discrete
valued labels to
identify the correct base call. The ensemble comprises two or more instances
of the teacher (first,
bigger) base caller.
[0384] The method includes evaluating the second set of cluster
images as inference data by
applying the trained teacher (first, bigger) base caller on the second set of
cluster images and
generating the base call predictions. The base call predictions are
represented by the continuous
valued weights that identify the predicted base call.
[0385] The method includes training the student (second, smaller)
base caller using the
second set of cluster images as training data. The second set of cluster
images are annotated with
the second ground truth data that identifies the correct base call based on
(i) the discrete valued
labels and (ii) the continuous valued weights. The student (second, smaller)
base caller has fewer
processing modules and parameters than the ensemble of the teacher (first,
bigger) base caller.
[0386] The method includes evaluating the third set of cluster
images as inference data by
applying the trained student (second, smaller) base caller on the third set of
cluster images and
generating the base call predictions.
[0387] In one implementation, the method includes implementing the
trained student
(second, smaller) base caller on one or more parallel processors of a
sequencing instrument for
real-time base calling.
[0388] Other implementations of the method described in this
section can include a non-
transitory computer readable storage medium storing instructions executable by
a processor to
perform any of the methods described above. Yet another implementation of the
method
described in this section can include a system including memory and one or
more processors
operable to execute instructions, stored in the memory, to perform any of the
methods described
above.
[0389] In another implementation, we disclose a system for
artificial intelligence-based base
calling. The system comprises a base caller (student, second, smaller base
caller/engine) trained
on cluster images that are annotated with ground truth data that identifies a
correct base call
based on (i) discrete valued labels of ground truth data used to train another
base caller and (ii)
continuous valued weights of base call predictions generated by the another
base caller (teacher,
first, bigger base caller/engine) for the cluster images during inference.
[0390] The base caller (student, second, smaller base
caller/engine) has fewer processing
modules and parameters than the another base caller (teacher, first, bigger
base caller/engine). In
one implementation, one of the processing modules is neural network layers. In
one
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
implementation, one of the parameters is interconnections between the neural
network layers. In
one implementation, one of the processing modules is neural network filters.
In one
implementation, one of the processing modules is neural network kernels. In
one
implementation, one of the parameters is multiplication and addition
operations.
[0391] The base caller (student, second, smaller base
caller/engine) is configured to evaluate
additional cluster images and generate, for the additional cluster images,
base call predictions.
[0392] The discrete valued labels are one-hot encoded with a one-
value for a correct base
and zero-values for incorrect bases. The continuous valued weights are part of
a probability
distribution for a correct base being Adenine (A), Cytosine (C), Thymine (T),
and Guanine (G).
[0393] In yet another implementation, we disclose a system for
artificial intelligence-based
base calling. The system comprises a teacher (first, bigger) base caller
trained on cluster images
that are annotated with ground truth data that identifies a correct base call
based on base call
predictions generated by a student (second, smaller) base caller.
[0394] In yet further implementation, we disclose an artificial
intelligence-based method of
base calling. The method includes training a teacher (first, bigger) base
caller by using a first set
of cluster images as training data. The first set of cluster images are
annotated with first ground
truth data that uses discrete valued labels to identify a correct base call.
In one implementation,
the discrete valued labels are one-hot encoded with a one-value for a correct
base and zero-
values for incorrect bases. In one implementation, the discrete valued labels
have a near-one-
value for the correct base and near-zero-values for the incorrect bases.
[0395] The method includes evaluating a second set of cluster
images as inference data by
applying the trained teacher (first, bigger) base caller on the second set of
cluster images and
generating base call predictions. The base call predictions are represented by
continuous valued
weights that identify a predicted base call. In one implementation, the
continuous valued weights
are part of a probability distribution for a correct base being Adenine (A),
Cytosine (C), Thymine
(T), and Guanine (G).
[0396] The method includes training a student (second, smaller)
base caller using the second
set of cluster images as training data. The second set of cluster images are
annotated with second
ground truth data that identifies a correct base call based on (i) the
discrete valued labels and (ii)
the continuous valued weights.
[0397] In some implementations, the teacher base caller (first,
bigger engine/model) is a
neural network-based base caller. In one implementation, the teacher base
caller (first, bigger
engine/model) is a convolutional neural network (CNN) with a plurality of
convolution layers. In
another implementation, it is a recurrent neural network (RNN) such as a long
short-term
81
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
memory network (LSTM), bi-directional LSTM (Bi-LSTM), or a gated recurrent
unit (GRU). In
yet another implementation, it includes both a CNN and an RNN.
[0398] In yet other implementations, the teacher base caller
(first, bigger engine/model) can
use 1D convolutions, 2D convolutions, 3D convolutions, 4D convolutions, 5D
convolutions,
dilated or atrous convolutions, transpose convolutions, depthwise separable
convolutions,
pointwise convolutions, 1 x 1 convolutions, group convolutions, flattened
convolutions, spatial
and cross-channel convolutions, shuffled grouped convolutions, spatial
separable convolutions,
and deconvolutions. It can use one or more loss functions such as logistic
regression/log loss,
multi-class cross-entropy/softmax loss, binary cross-entropy loss, mean-
squared error loss, Li
loss, L2 loss, smooth Li loss, and Huber loss. It can use any parallelism,
efficiency, and
compression schemes such TFRecords, compressed encoding (e.g, PNG), sharding,
parallel
calls for map transformation, batching, prefetching, model parallelism, data
parallelism, and
synchronous/asynchronous stochastic gradient descent (SGD). It can include
upsampling layers,
downsampling layers, recurrent connections, gates and gated memory units (like
an LSTM or
GRU), residual blocks, residual connections, highway connections, skip
connections, peephole
connections, activation functions (e.g., non-linear transformation functions
like rectifying linear
unit (ReLU), leaky ReLU, exponential liner unit (ELU), sigmoid and hyperbolic
tangent (tanh)),
batch normalization layers, regularization layers, dropout, pooling layers
(e.g., max or average
pooling), global average pooling layers, and attention mechanisms.
[0399] In some implementations, the student base caller (second,
smaller engine/model) is a
neural network-based base caller. In one implementation, the student base
caller (second, smaller
engine/model) is a convolutional neural network (CNN) with a plurality of
convolution layers. In
another implementation, it is a recurrent neural network (RNN) such as a long
short-term
memory network (LSTM), bi-directional LSTM (Bi-LSTM), or a gated recurrent
unit (GRU). In
yet another implementation, it includes both a CNN and a RNN.
[0400] In yet other implementations, the student base caller
(second, smaller engine/model)
can use 11) convolutions, 2D convolutions, 3D convolutions, 4D convolutions,
5D convolutions,
dilated or atrous convolutions, transpose convolutions, depthwise separable
convolutions,
pointwise convolutions, 1 x I convolutions, group convolutions, flattened
convolutions, spatial
and cross-channel convolutions, shuffled grouped convolutions, spatial
separable convolutions,
and deconvolutions. It can use one or more loss functions such as logistic
regression/log loss,
multi-class cross-entropy/softmax loss, binary cross-entropy loss, mean-
squared error loss, Li
loss, L2 loss, smooth Li loss, and Huber loss. It can use any parallelism,
efficiency, and
compression schemes such TFRecords, compressed encoding (e.g., PNG), sharding,
parallel
calls for map transformation, batching, prefetching, model parallelism, data
parallelism, and
82
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
synchronous/asynchronous stochastic gradient descent (SGD). It can include
upsampling layers,
downsampling layers, recurrent connections, gates and gated memory units (like
an LSTM or
GRU), residual blocks, residual connections, highway connections, skip
connections, peephole
connections, activation functions (e.g., non-linear transformation functions
like rectifying linear
unit (ReLU), leaky ReLU, exponential liner unit (ELU), sigmoid and hyperbolic
tangent (tanh)),
batch normalization layers, regularization layers, dropout, pooling layers
(e.g., max or average
pooling), global average pooling layers, and attention mechanisms.
Clauses
[0401] We disclose the following clauses:
36. An artificial intelligence-based method of performing computationally
efficient base calling,
the method including:
training a first base caller over cluster intensity images and producing a
first trained base
caller that maps the cluster intensity images to base call predictions;
beginning with the first trained base caller, executing a loop in which each
iteration uses a
starting trained base caller as input and produces a pruned trained base
caller as output, wherein
the pruned trained base caller has fewer processing elements than the starting
trained base caller;
wherein each iteration comprises (i) a base call prediction step, (ii) a
contribution
measurement step, (iii) a pruning step, and (iv) a retraining step;
wherein the base call prediction step, during forward propagation, processes a
subset of the
clusters intensity images through processing elements of the starting trained
base caller and
produces the base call predictions;
wherein the contribution measurement step generates a contribution score for
each of the
processing elements that identifies how much a processing element contributed
to the base call
predictions;
wherein the pruning step selects a subset of the processing elements based on
their
contribution scores and produces the pruned trained base caller by removing,
from the starting
trained base caller, the selected subset of the processing elements;
wherein the retraining step further trains the pruned trained base caller over
the cluster
intensity images and makes the pruned trained base caller available for a
successive iteration as
the starting trained base caller; and
terminating the loop after n iterations and using the pruned trained base
caller produced by
the nth iteration for further base calling.
83
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
37. The artificial intelligence-based method of clause 36, wherein the
contribution score for the
processing element is generated by:
applying an absolute function to weights of the processing element and
generating absolute
weight values; and
summing the absolute weight values and producing the contribution score for
the processing
element.
38. The artificial intelligence-based method of clause 36, implementing each
of the clauses
which ultimately depend from clauses 1 and 27.
39. An artificial intelligence-based method of performing computationally
efficient base
calling, the method including:
training a first base caller over cluster intensity images and producing a
first trained base
caller that maps the cluster intensity images to base call predictions;
beginning with the first trained base caller, executing a loop in which each
iteration uses a
starting trained base caller as input and produces a pruned trained base
caller as output, wherein
the pruned trained base caller has fewer processing elements than the starting
trained base caller;
wherein each iteration comprises (i) a cluster feature maps generation step,
(ii) a gradient
determination step, (iii) an intermediate feature value generation step, (iv)
a feature sum
generation step, (v) a subset output generation step, (vi) a subset selection
step, (vii) a pruning
step, (viii) a cluster feature map identification step, and (ix) a retraining
step;
wherein the cluster feature maps generation step, during forward propagation,
processes a
subset of the clusters intensity images through the processing elements of the
starting trained
base caller, generates one or more cluster feature maps using each processing
element, and
produces the base call predictions based on the cluster feature maps;
wherein the gradient determination step, during backward propagation,
determines gradients
for the cluster feature maps based on error between the base call predictions
and ground truth
base calls;
wherein the intermediate feature value generation step multiplies feature
values in the
cluster feature maps with respective ones of the gradients and produces a set
of intermediate
feature values for each of the cluster features maps;
wherein the feature sum generation step sums the intermediate feature values
in the set of
intermediate feature values and produces a feature sum for each of the cluster
features maps,
thereby producing a set of feature sums for the starting trained base caller;
wherein the subset output generation step processes subsets of feature sums in
the set of
feature sums and generates a subset output for each of the subsets;
wherein the subset selection step selects one or more of the subsets of
feature sums based on
84
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
evaluating their respective subset outputs against one or more of the feature
sums in the set of
feature sums;
wherein the cluster feature map identification step identifies those cluster
feature maps
whose feature sums are part of the selected subsets of feature sums;
wherein the pruning step produces the pruned trained base caller by removing,
from the
starting trained base caller, those processing elements that were used to
generate the identified
cluster feature maps during the forward propagation;
wherein the retraining step further trains the pruned trained base caller over
the cluster
intensity images and makes the pruned trained base caller available for a
successive iteration as
the starting trained base caller; and
terminating the loop after n iterations and using the pruned trained base
caller produced by
the nth iteration for further base calling
40. The artificial intelligence-based method of clause 39, wherein the subset
output for a subset
of feature sums is based on an additive sum of the feature sums in the subset
41. The artificial intelligence-based method of clause 40, wherein the subset
output for the subset
of feature sums is based on an average of the feature sums in the subset.
42. The artificial intelligence-based method of clause 41, wherein the subset
output for the subset
of feature sums is based on an exponential sum of the feature sums in the
subset.
43. The artificial intelligence-based method of clause 42, wherein the subset
output for the subset
of feature sums is based on a multiplicative interaction of the feature sums
in the subset.
44. The artificial intelligence-based method of clause 43, wherein the subset
selection step
selects the subsets of feature sums based on evaluating their respective
subset outputs against a
lowest one of the feature sums in the set of feature sums and selecting those
subsets of feature
sums whose subset outputs are lower than the lowest one of the feature sums in
the set of feature
sums.
45. The artificial intelligence-based method of clause 44, wherein the subset
selection step
selects the subsets of feature sums based on evaluating their respective
subset outputs against a
plurality of lowest ones of the feature sums in the set of feature sums and
selecting those subsets
of feature sums whose subset outputs are lower than the lowest ones of the
feature sums in the
plurality of lowest ones of the feature sums.
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
46. The artificial intelligence-based method of clause 45, wherein the subset
selection step
selects those subsets of feature sums whose subset outputs are zero.
47. The artificial intelligence-based method of clause 46, wherein the subset
selection step
selects those subsets of feature sums whose subset outputs are closest to
zero.
48. An artificial intelligence-based method of performing computationally
efficient base calling,
the method including:
training a first base caller over cluster intensity images and producing a
first trained base
caller that maps the cluster intensity images to base call predictions;
beginning with the first trained base caller, executing a loop in which an
iteration uses a
starting trained base caller as input and produces a pruned trained base
caller as output, wherein
the pruned trained base caller has fewer processing elements than the starting
trained base caller;
wherein the iteration comprises (i) a base call prediction step, (ii) a
contribution
measurement step, and (iii) a pruning step;
wherein the base call prediction step, during forward propagation, processes
one or more of
the clusters intensity images through processing elements of the starting
trained base caller and
produces the base call predictions;
wherein the contribution measurement step determines a contribution score for
each of the
processing elements that identifies how much a processing element contributed
to the base call
predictions; and
wherein the pruning step selects a subset of the processing elements based on
their
contribution scores and produces the pruned trained base caller by removing,
from the starting
trained base caller, the selected subset of the processing elements.
49. The artificial intelligence-based method of claim 48, wherein the
contribution score for each
of the processing elements is determined based on their corresponding feature
maps.
50. The artificial intelligence-based method of claim 48, wherein the loop
comprises one or more
iterations.
51. The artificial intelligence-based method of claim 48, wherein the
processing element is a
filter.
52. The artificial intelligence-based method of claim 51, wherein the
processing element is a
convolution filter.
86
CA 03168451 2022- 8- 18
WO 2021/168014
PCT/US2021/018422
53. The artificial intelligence-based method of claim 48, wherein the
processing element is a
kernel.
54. The artificial intelligence-based method of claim 53, wherein the
processing element is a
convolution kernel.
55. The artificial intelligence-based method of claim 48, wherein the
processing element is a
layer.
56. The artificial intelligence-based method of claim 55, wherein the
processing element is a
convolution layer.
[0402] Other implementations of the method described above can
include a non-transitory
computer readable storage medium storing instructions executable by a
processor to perform any
of the methods described above. Yet another implementation of the method
described in this
section can include a system including memory and one or more processors
operable to execute
instructions, stored in the memory, to perform any of the methods described
above.
[0403] What is claimed is:
87
CA 03168451 2022- 8- 18