Note: Descriptions are shown in the official language in which they were submitted.
WO 2021/239758
PCT/EP2021/063954
MULTIPLEX CRISPR/CAS SYSTEM FOR MODIFYING CELL GENOMES
TECHNICAL FIELD
[0001] The invention relates to methods of modifying cell genomes using
multiple CRISPR/Cas
systems. The invention also relates to compositions, crRNAs, Cas and vectors
for carrying out such
methods as disclosed herein.
BACKGROUND
[0002] The state of the art describes vectors and uses of these that employ
CRISPR/Cas systems. For
example, reference is made to W02020078893, W02019185551, W02019105821,
W02017118598,
US20180140698, US20170246221, US20180273940, US20160115488, US20180179547,
US20170175142, US20160024510, US20150064138, US20170022499, US20160345578,
US20180155729, US20180200342, W02017112620, W02018081502, PCT/EP2018/066954,
PCT/EP2018/066980, PCT/EP2018/071454, EP3356533, EP3307872, W02020072253,
W02020072254, W02020072250, W02020072248, W02019236566, W02019144061,
W02017112620, W02015066119, EP16804164, W02019227080, EP3362571, EP3356533,
W02016205623, EP3307872, US2016/0324938 and US2019/0160120 and equivalent
publications by
the US Patent and Trademark Office (USPTO) or WIPO, the disclosures of which
are incorporated
herein by reference.
SUMMARY OF THE INVENTION
[0003] The invention provides the following configurations.
[0004] In a First Configuration
A method of modifying the genome of a cell, the method comprising
(a) using a first CRISPR/Cas system to modify a first protospacer of the
genome; and
(b) using a second CRISPR/Cas system to modify a second protospacer of the
genome, wherein
the second protospacer is different to the first protospacer;
wherein the systems comprise different Cas and are provided simultaneously in
the cell.
100051 In a Second Configuration
A method of modifying the genome of one or more cells, the method comprising
introducing into
each cell components (a), (b), (c) or (d):-
(a) first and second crRNAs;
(b) nucleic acid encoding first and second crRNAs, wherein the nucleic acid is
expressed in the
cell for producing the crRNAs;
1
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
(c) a first nucleic acid encoding a first crRNA, and a second nucleic acid
encoding a second
crRNA, wherein the nucleic acids are expressed in the cell for producing the
crRNAs; or
(d) a first crRNA and a nucleic acid encoding a second crRNA, wherein the
nucleic acid is
expressed in the cell for producing the second crRNA;
wherein for each cell
(e) the first crRNA (crRNA1) is capable of guiding a first Cas (C1) to a
protospacer sequence
(PS1) comprised by the cell genome to modify PS1; and
(f) the second crRNA (crRNA2) is capable of guiding a second Cas (C2) to a
protospacer
sequence (PS2) comprised by the cell genome to modify PS2;
(g) Cl and C2 are different;
(h) PS1 and PS2 are different; and
(i) crRNA1, crRNA2. Cl and C2 are provided in the cell, whereby the genome
of the cell is
subjected to Cas modification.
In one aspect the invention uses the method to kill target cells. In another
aspect the invention uses
the method to edit the genomes of cells.
[0006] In a third Configuration
A composition for use in a method treating or preventing a disease or
condition in a human or animal
subject that is mediated by target cells, the composition comprising
components (a), (b) or (d):-
(a) first and second crRNAs;
(b) nucleic acid encoding first and second crRNAs, wherein the nucleic acid is
expressible in a
target cell for producing the crRNAs;
(c) a first nucleic acid encoding a first crRNA, and a second nucleic acid
encoding a second
crRNA, wherein the nucleic acids are expressible in a target cell for
producing the crRNAs; or
(d) a first crRNA and a nucleic acid encoding a second crRNA, wherein the
nucleic acid is
expressible in a target cell for producing the second crRNA;
wherein
(e) the first crRNA (crRNA1) is capable of guiding a first Cas (Cl) to a
protospacer sequence
(PS1) comprised by a target cell genome to modify PS1; and
(f) the second crRNA (crRNA2) is capable of guiding a second Cas (C2) to a
protospacer
sequence (PS2) comprised by the target cell genome to modify PS2;
(g) Cl and C2 are different;
(h) PS1 and PS2 are different; and
wherein the method comprises administering the composition to the subject
whereby said components
of the composition are introduced into target cells wherein crRNA1, crRNA2, Cl
and C2 are provided
in each cell and the gcnomc of each cell is subjected to Cas modification and
the disease or condition
is treated or prevented.
2
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[0007] In a fourth Configuration
A composition comprising components (a), (b) or (d):-
(a) first and second crRNAs;
(b) nucleic acid encoding first and second crRNAs, wherein the nucleic acid is
expressible in a
target cell for producing the crRNAs;
(c) a first nucleic acid encoding a first crRNA, and a second nucleic acid
encoding a second
crRNA, wherein the nucleic acids are expressible in a target cell for
producing the crRNAs; or
(d) a first crRNA and a nucleic acid encoding a second crRNA, wherein the
nucleic acid is
expressible in a target cell for producing the second crRNA;
wherein
(e) the first crRNA (crRNA1) is capable of guiding a first Cas (C1) to a
protospacer sequence
(PS1) comprised by a target cell genome to modify PS1; and
(f) the second crRNA (crRNA2) is capable of guiding a second Cas (C2) to a
protospacer
sequence (PS2) comprised by the target cell genome to modify PS2;
(g) Cl and C2 are different;
(h) PS1 and PS2 are different; and
wherein when said components of the composition are introduced into a target
cell whereby crRNA1,
crRNA2, Cl and C2 are provided in the cell, the genome of the cell is
subjected to Cas modification.
[0008] Aspects provide:-
The method of the invention for
(a) producing synergistic Cas nuclease cutting of a cell genome;
(b) reducing a population of cells of a first species or strain by at least
100,000, 1,000,000 or
10,000,000-fold;
(c) killing at least at least 99%. 99.9%. 99.99%, 99.999%, 99.9999% or
99.99999% cells of a
first species or strain comprised by a microbiome;
(d) producing synergistic Class 1 Cas modification of a cell genome; or
(e) reducing bacterial cells of a first species or strain (eg, E coli
cells) in a cell population by at
least 105, 106 or 107 -fold, wherein the population comprises at least
100,000; 1,000,000; or
10,000,000 cells respectively.
100091 Aspects also provide pharmaceutical compositions, methods of making
such compositions
and medical methods using such compositions.
BRIEF DESCRIPTION OF THE DRAWINGS
[00010] Figure 1. Type T CRISPR-Cas system of E. coli and C. difficile
targeting E. coli MG1655.
Layout of the CRISPR Guided VectorTm, CGVTm. (A) E. coli CRISPR-Cas CGV: ColE1
on, cas3 and
3
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
cascade of E. coil, CRISPR array. (B) C. difficlle CRISPR Cas CGVs. Plasmid 1:
pSC101 on, cas3
and cascade of C. difficile. Plasmid 2: pC1oDF13 on, CRISPR array of C.
difficile.
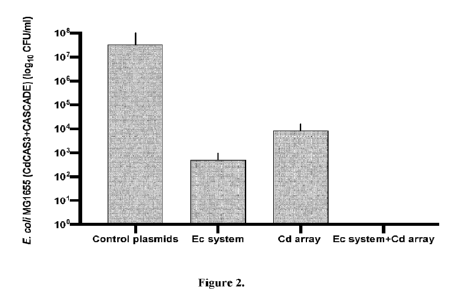
[00011] Figure 2. Killing of E. coli MG1655 with type IE CRISPR-Cas system of
E. coli and type I-B
CRISPR-Cas system of C. difficile. E. coil MG1655 harboring cas genes of C.
difficile was
transformed with cognate CRISPR array and E. coli CRISPR-Cas CGV. Both CRISPR
systems
together surprisingly synergistically killed 7 logio E. coli MG1655, compared
to empty vectors.
Additionally, single transformations with the CGVs were tested. E. coli CRISPR-
Cas system resulted
in ¨4-log to reductions; C. diffiede CRISPR-Cas system resulted ¨3-log to
(n=3).
DETAILED DESCRIPTION
[00012] The invention relates to methods of modifying cell genomes using
multiple CRISPR/Cas
systems. The invention also relates to compositions, crRNAs, Cas and vectors
for carrying out such
methods as disclosed herein.
1000131 The invention is useful to provide one or more of the following
advantages:-
(a) producing synergistic Cas nuclease cutting of a cell genome,
(b) reducing a population of cells of a first species or strain by at least
100,000, 1,000,000 or
10,000,000-fold;
(c) killing at least at least 99%. 99.9%. 99.99%, 99.999%, 99.9999% or
99.99999% cells of a
first species or strain comprised by a microbiome;
(d) producing synergistic Class 1 Cas modification of a cell genome; or
(e) reducing bacterial cells of a first species or strain in a cell population
by at least 105, 106 or
107-fold, wherein the population comprises at least 100,000; 1,000,000; or
10,000,000 cells
respectively.
[00014] Advantageously, by reducing the number of target cells in a cell
population, this may be
beneficaial where the cells are undesirable (eg, detrimental to the health of
a subject to which the
method is applied, or detrimental to an ex vivo environment or in vitro cell
sample to which the
method is applied or the composition is administered). For example, the cells
are cancer cells
comprised by a patient and the multiple Cas cutting of the invention
synergistically kills a very high
number (cg, at least 99.999% or 105-fold) of the cells. By reducing the cells
in this way, the number
of seeder cells to re-grow the cancer is reduced. In another example, the
cells may be bacterial or
archaeal cells and by reducing the cells in this way, the number of seeder
cells to re-grow an
undesirable cell population will be reduced.
[00015] In a configuration, the invention provides in one aspect:-
A method of modifying the genome of a cell, the method comprising
(a) using a first CRISPR/Cas system to modify a first protospacer
of the genome; and
4
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
(b)
using a second CRISPR/Cas system to modify a second protospacer of the
genome, wherein
the second protospacer is different to the first protospacer;
wherein the systems comprise different Cas and are used simultaneously.
[00016] Another aspect of the configuration provides:-
A method of modifying the genome of one or more cells, the method comprising
introducing into
each cell components (a), (b), (c) or (d):-
(a) first and second crRNAs;
(b) nucleic acid encoding first and second crRNAs, wherein the nucleic acid is
expressed in the
cell for producing the crRNAs;
(c) a first nucleic acid encoding a first crRNA, and a second nucleic acid
encoding a second
crRNA, wherein the nucleic acids are expressed in the cell for producing the
crRNAs; or
(d) a first crRNA and a nucleic acid encoding a second crRNA, wherein the
nucleic acid is
expressed in the cell for producing the second crRNA;
wherein for each cell
(e) the first crRNA (crRNA1) is capable of guiding a first Cas (Cl) to a
protospacer sequence
(PS1) comprised by the cell genome to modify PS1; and
(f) the second crRNA (crRNA2) is capable of guiding a second Cas (C2) to a
protospacer
sequence (PS2) comprised by the cell genome to modify PS2;
(g) Cl and C2 are different;
(h) PS1 and PS2 are different; and
(i) crRNA1, crRNA2, Cl and C2 are provided in the cell, whereby the genome
of the cell is
subjected to Cas modification.
[00017] Optionally, Cl and/or C2 is a Cas nuclease, eg, Cl and C2 each is a
Cas3. Optionally, Cl
and/or C2 is a Cascade Cas, eg, CasA, CasB, CasC, CasD or CasE. Optionally, Cl
is a Cas3 that
operates in the cell with Cascade Cas, eg, one, more or all of CasA, B, C, D
and E.
[00018] Optionally, the crRNAs of component (a) are introduced simultaneously
or sequentially.
Optionally, the nucleic acid and crRNA of component (c) are introduced
simultaneously or
sequentially. Optionally, the nucleic acids of component (d) arc introduced
simultaneously or
sequentially. The method, however, includes the presence of crRNA1, crRNA2, Cl
and C2 at the
same time in each cell, whereby multiple CRISPR/Cas systems are used to modify
the genome.
crRNA1, crRNA2, CI and C2 are provided in the cell, whereby PSI and PS2 are
subjected to Cas
nuclease modification wherein the genome of the cell is modified.
[00019] Preferably, the first and second protospacers are different or
comprised by different genes or
intergenic sequences of the genome.
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00020] Modification of the genome may be cutting of nucleic acid of the
genome, repression of
transcription or translation of a gene comprised by the genome, upregulation
of transcription or
translation of a gene comprised by the genome, or editing of the genome (eg,
to insert and/or delete
one or more nucleic acid sequences). The invention may advantageously be
useful for synergistically
or efficiently cutting, modifying or editing the genome of each cell. In an
example, DNA comprised
by the genome is cut, modified or edited and/or RNA comprised by the genome is
cut, modified or
edited. In an example, DNA comprised by the genome is degraded (eg, in a
process comprising Cos
exo- or endonuclease activity) and/or RNA comprised by the genome is cut,
modified or edited (eg, in
a process comprising Cas exo- or endonuclease activity).
[00021] In an example, the component is comprised by a nucleic acid vector. In
an example the
component (a), (b), (c) or (d) is introduced into each cell by transfection,
electroporation, transduction
or conjugative transfer. For example, the vector is a virus or phage and the
component is introduced
by transduction. For example, the vector is a plasmid and the component is
introduced by
conjugation, transfection or electroporation. For example, the vector is a
phagemid (optionally a
phagemid comprised by a virus or phage) and the component is introduced by
conjugation,
transduction, transfection or elcctroporation. For example, the nucleic
acid(s) of the component
is(are) introduced by electroporation thereof. For example, a phage herein is
a tailed phage. For
example a phage herein is a lytic phage. For example, a phage herein is a non-
lytic phage.
[00022] Optionally, the method is a recombineering method carried out in
vitro, and for example the
cell is an E coli cell.
[00023] Optionally, each said crRNA is encoded by a CRISPR array comprising
first and second
repeat sequences and a spacer sequence joining the repeat sequences.
Optionally, the nucleic acid of
(b) comprises a CRISPR array encoding crRNA1 and crRNA2. Optionally, the first
nucleic acid of
(c) comprises a first CRISPR array encoding crRNA1 and the second nucleic acid
comprises a second
CRISPR array encoding crRNA2. Optionally, the nucleic acid of (b) comprises a
CRISPR array
encoding crRNA2.
[00024] In an example each repeat sequence is GAGTTCCCCGCGCCAGCGGGGATAAACCG or
GTTTTATATTAACTAAGTGGTATGTAAAT. In an example, each protospacer or spacer
sequence
consists of from 15 to 70, 20 to 50, 17 to 45, 18 to 40, 18 to 35 or 20 to 40
contiguous nucleotides.
[00025] Optionally, Cosi and/or Cas2 are not introduced into each cell.
Optionally, each nucleic
acid is devoid of nucleic acid sequence encoding Cosi and/or Cas2. Optionally,
additionally Cas4 is
not introduced into each cell, or each nucleic acid is devoid of a Cas4-
encoding nucleic acid sequence.
Optionally, said introducing comprises (i) introducing into each cell an
operon comprising nucleotide sequences
encoding a type I Cas3 (wherein the Cas3 is Cl) and Cascade proteins under the
control of a common
constitutive promoter and/or introducing into each cell an operon comprising
nucleotide sequences encoding a
type I Cas3 (wherein the Cas3 is C2) and Cascade proteins under the control of
a common constitutive
promoter. In an example, Cl is a Type-IB Cas3 and/or C2 is a Type-IE Cas3.
Examples of suitable operons are
6
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
disclosed in W02020078893 or US20200115716, the disclosures of which are
expressly incorporated herein by
reference for possible use in the present invention. The term -operon- is
known to the skilled person such
as relating to a functioning unit of DNA containing at least expressible 2
nucleotide sequences
respectively encoding for an expression product (eg, a respective translatable
mRNA), wherein the
sequences are under common promoter control.
[00026] In an example, Cl is a Cas disclosed in W02019002218 and optionally
the first crRNA is
encoded by a CRISPR array comprising cognate repeat sequences, such as when Cl
is a Cas (eg,
Cas3) disclosed in W02019002218 the repeat sequences are the cognate repeat
sequence disclosed in
W02019002218. Additionally or alternatively, In an example, C2 is a Cas
disclosed in
W02019002218 and optionally the first crRNA is encoded by a CRISPR array
comprising cognate
repeat sequences, such as when C2 is a Cas (eg, Cas3) disclosed in
W02019002218 the repeat
sequences are the cognate repeat sequence disclosed in W02019002218. In an
example, the first
crRNA is encoded by an array comprising a repeat sequence disclosed in
W02019002218 and/or the
second crRNA is encoded by an array comprising a repeat sequence disclosed in
W02019002218.
For example, one or more nucleotide sequences encoding one or more Cascade Cas
(eg, which are
cognate to C 1 or C2) are introduced into the cell, wherein the Cascade Cas
are Cascade Cos disclosed
in W02019002218. All of these disclosures in W02019002218 are expressly
incorporated herein by
reference for possible use in the present invention.
[00027] Optionally,
(a) Cl is a Class 1 Cas and C2 is a Class 1 Cas;
(b) Cl is a Class 1 Cas and C2 is a Class 2 Cas;
(c) Cl is a Class 2 Cas and C2 is a Class 2 Cas;
(d) Cl is a Type I Cas (optionally Type I-A, B, C, D, E, F or U) and C2 is a
Type I Cas
(optionally Type I-A, B, C, D, E, F or U);
(e) Cl is a Type I (optionally Type I-A, B, C, D. E, F or U) or II Cas and C2
is a Type II Cas;
(f) Cl is a Type I (optionally Type I-A, B, C, D, E, F or U) or II Cas and C2
is a Type III Cas
(optionally Type I-A or B);
(g) Cl is a Type I (optionally Type I-A, B, C, D, E, F or U) or II Cas and C2
is a Type IV Cas;
(h) Cl is a Type 1 (optionally Type I-A, B, C, D, E, F or U) or 11 Cas and C2
is a Type V Cas; or
(i) Cl is a Type I or II Cas and C2 is a Type VI Cas.
[00028] Optionally. Cl and C2 are different Class 1 Cas selected from the Cas
disclosed in Table 2.
Optionally, Cl is an E coli Cas (eg, Cas3) and C2 is a Cas selected from the
Cas disclosed in Table 2.
Optionally, Cl is an C dificile Cas (eg, Cas3) and C2 is a Cas selected from
the Cas disclosed in
Table 2.
[00029] Optionally, Cl is a Type I-A, B, C, D, E, F or U Cas. Optionally, C2
is a Type I-A, B, C, D,
E, F or U Cas.
7
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00030] Optionally, Cl is a Type I-A Cas and C2 is a Type I-B, C, E, F or U
Cas. Optionally, Cl is a
Type I-B Cas and C2 is a Type I-B, C, E, F or U Cas. Optionally, CI is a Type
I-C Cas and C2 is a
Type I-B, C, E, F or U Cas. Optionally, Cl is a Type I-D Cas and C2 is a Type
I-B, C, E, F or U Cas.
Optionally, Cl is a Type I-E Cas and C2 is a Type I-B, C, E, F or U Cos.
Optionally, Cl is a Type I-F
Cas and C2 is a Type I-B, C, E, F or U Cas. Optionally, CI is a Type I-U Cas
and C2 is a Type I-B,
C, E, F or U Cas.
[00031] Optionally,
(a) Cl is a Type TB or C Cos and C2 is a Type I-E or F Cas (optionally Cl is a
Type TB Cas3 and
C2 is a Type IE Cas);
(b) Cl is a Type IC or C Cas and C2 is a Type I-E or F Cas (optionally Cl is a
Type IC Cas3 and
C2 is a Type IE Cas3); or
(c) Cl is a Type II Cas9 and C2 is a Type I Cas3 (optionally C2 is an E coil
Type IE or F Cas3;
or a C difficile Cas TB).
[00032] Optionally_
(a) Cl is a Cas3 (optionally a Type 1-A, B, C, D, E, F or U Cas3) and C2 is a
Cas3 (optionally a
Type I-A, B, C, D, E, F or U Cas3);
(b) Cl is a Cas9 and C2 is a Cas3 (optionally a Type I-A, B, C, D, E, F or U
Cas3);
(c) CI is a Cas3 (optionally a Type I-A, B, C, D, E, F or U Cas3) and C2 is a
Cas10 (optionally
Cas10 subtype A, B, C or D);
(d) Cl is a Cas9 and C2 is a Cas10 (optionally Cas10 subtype A, B, C or D);
(e) Cl is a Cas9 and C2 is a Cas12 (optionally Cas12a);
(f) Cl is a Cas3 (optionally a Type 1-A, B, C, D, E, F or U Cas3) and C2 is a
Cas12 (optionally
Cas12a);
(g) Cl is a Cas9 and C2 is a Cas13 (optionally Cas13a, Cas13b, Cas13c or
Cas13d); or
(h) Cl is a Cas3 (optionally a Type I-A, B, C, D, E, F or U Cas3) and C2 is a
Cas13 (optionally
Cas13a, Cas13b, Cas13c or Cas13d).
[00033] Optionally, PS1 and PS2 are protospacers comprised by
(a) RNA and RNA respectively;
(b) DNA and RNA respectively;
(c) RNA and DNA respectively; or
(d) DNA and DNA respectively.
[00034] Optionally, Cl is a Clostridiaceae Cas3 (optionally a C difficile
Cas3, such as a Type T-B
Cas3) and C2 is an Enterobacteriaceae Cas3 (optionally an E coli Cas3, such as
a Type I-E Cas3).
8
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00035] In an alternative, Cl and C2 are the same. In an alternative, Cl and
C2 are the same type of
Cas, eg, each is a Cas9, or each is a Cas3, or each is a Cas 1 2, or each is a
Cas 1 3, or each is the same
type of Cascade Cas.
[00036] Optionally, Cl is a Biostraticola, Buttiauxella, Cedecea, Citrobacter,
Cronobacter,
Enterobacillus, Enterobacter, Escherichia, Franconibacter, Gibbsiella,
Izhakiella, Klebsiella,
Kluyvera, Kosakonia, Leclercia, Lelliottia, Limnobaculum, Mangrovibacter,
Metakosakonia,
Pluralibacter, Pseudescherichia, Pseudocitrobacter, Raoultella or
Rosenbergiella Cas (eg, Cas3 or
Cascade Cas).
[00037] Optionally, Cl is a spCas9 (S pyogenes Cas9) or saCas9 (S aureus Cas9)
and C2 is a Type I
Cas3 (optionally C2 is an E coli Type I-E or F Cas3).
[00038] Optionally, the modification is cutting of the genome, eg, cutting DNA
(eg, ssDNA or
dsDNA) of the genome, RNA (eg, mRNA, crRNA, tracrRNA, tRNA, snRNA or rRNA,
preferably
mRNA), endonuclease cutting or exonuclease cutting, or cutting of one, but not
both strands of
dsDNA (double-stranded DNA) of the genome, or nicking of dsDNA of the genome.
[00039] Optionally_ PS 1 is a chromosomal sequence of the cell. Optionally,
PS1 is an episomal (eg,
plasmid) sequence of the cell.
[00040] Optionally, PS 1 is a chromosomal sequence of the cell and PS2 is a
chromosomal sequence of
the cell. Optionally, PS1 is a chromosomal sequence of the cell and PS2 is an
episomal (eg, plasmid)
sequence of the cell.
[00041] Optionally, each cell is a human, animal (ie, non-human animal),
plant, yeast, fungus,
amoeba, insect, mammalian, vertebrate, bird, fish, reptile, rodent, mouse,
rat, livestock animal, cow,
pig, sheep, goat, rabbit, frog, toad, protozoan, invertebrate, mollusc, fly,
grass, tree, flowering plant,
fruiting plant, crop plant, wheat, corn, maize, barley, potato, carrot or
lichen cell. Optionally, each
cell is a prokaryotic cell or eukaryotic cell. For example, each cell is a
bacterial or archaeal cell,
optionally an E coli cell or C difficile cell. In an embodiment, the cell or
the cells are of a genus or
species disclosed in Table 1. In an embodiment, the cell or the cells are gram
positive cells. In an
embodiment, the cell or the cells are gram negative cells.
[00042] Optionally, the step of introducing comprises infecting the cell with
a virus (optionally a
bacteriophage wherein the cell is a bacterial cell) or introducing a plasmid
(optionally a conjugative
plasmid) or introducing a phagcmid into thc cell, wherein the virus, plasmid
or phagcmid encodes the
crRNAs. Optionally, the virus, plasmid or phagemid encodes Cl and/or C2.
Optionally, the virus,
plasmid or phagemid encodes one of said Cl and C2, and the other Cas is an
endogenous Cas encoded
by the genome of the cell. Optionally, the each of C 1 and C2 is an endogenous
Cas encoded by the
genome of the cell. In an example, the Cas is encoded by a chromosome of the
cell.
[00043] Optionally Cl is a Cas3 and the virus or plasmid encodes a Cas5, Cas6,
Cas7 and Cas8 (and
optionally a Cas 11) that arc cognate to the Cas3. Additionally or
alternatively, optionally C2 is a
9
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
Cas3 and the virus or plasmid encodes a Cas5, Cas6, Cas7 and Cas8 (and
optionally a Cash) that are
cognate to the Cas3.
[00044] The Cas are simultaneously present in the cell and the Cas may cut the
genome
simultaneously or sequentially.
[00045] Optionally, the method comprises introducing into each cell or
expressing in each cell at least
3, 4 or 5 different types of crRNAs wherein the types target different
protospacer sequences
comprised by the cell genome (e,g different chromosomal sequences). In an
example, the cell is a
bacterial or archaeal cell and the protospacers are comprised by the cell
chromosome. For example, at
least one or two of said crRNA types targets a respective chromosomal sequence
and at least one or
more of the crRNA types targets a sequence comprised by an episome (eg, a
plasmid) of the cell,
wherein the cell is a bacterial or archaeal cell. For example, the cell (eg, a
human or mammalian cell)
comprises a plurality of chromosomes and the crRNAs target protospacer
sequences comprised by
two or more of said chromosomes (eg, wherein the chromosomes are not members
of the same diploid
chromosomal pair).
[00046] For example, the method comprised introducing a nucleic acid into each
cell, wherein the
nucleic acid comprises, in 5' to 3' direction a nucleotide sequence encoding a
Cas nuclease (eg, a
cas3) and one or more sequences encoding one or more Cascade Cas (eg, cas8e,
cas 11, cas7, cas5,
and cas6; or cas6, cas8b, cas7, and cas5) that are operable with the Cas
nuclease to modify a cognate
protospacer sequence.
[00047] The nucleic acid(s) is(are) preferably devoid of an adaptation module.
Optionally, the module
encodes a Cas 1 and a Cas2; or a Cas 1, a Cas2 and a Cas4
[00048] In an embodiment, a said nucleic acid comprises a CRISPR array
encoding crRNAs, such as
an array comprising at least 3, 4 or 5 spacer sequences targeting at least 3,
4 or 5 sequences of the cell
respectively. For example, a plurality of chromosomal intergenic regions are
targeted. Optionally,
each spacer sequence consists of from 20 to 50, 20 to 40, 22 to 40, 25 to 40
or 30 to 35 consecutive
nucleotides, eg, 32 or 37 nucleotides.
[00049] In an example, the array comprises the following spacer sequences
(Spacers 1-3):
TGATTGACGGCTACGGTAAACCGGCAACGTTC;
GCTGTTAACGTACGTACCGCGCCGCATCCGGC; and
CGGACTTAGTGCCAAAACATGGCATCGAAATT
separated by repeat sequence (ie, Spacer 1 ¨ repeat ¨ Spacer 2 ¨ repeat ¨
Spacer 3).
[00050] In another example, the array comprises 3, 4 or 5 of the following
spacer sequences (Spacers
4-8):
GCCATAATCTGGATCAGGAAGTCTTCCTTATCCATAT;
GGCTTTACGCCAGCGACGTATTGCCACAGGAATAACT;
GGGGATAGCGCGCCTGGAGCGTGCGATAGAGACTTTG;
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00051] GGCATTTACCGACCAGCCCATCAGCAGTACAGCAAAC; and
TCCTGAATCAAATCCGCCTGTGGCAGGCCATAGCCCG
separated by repeat sequence (ie, Spacer 4 - repeat - Spacer 5 - repeat -
Spacer 6- repeat - Spacer 7
- repeat - Spacer 8).
[00052] Optionally, each repeat sequence consists of from 20 to 50, 20 to 40,
22 to 40, 25 to 40 or 30
to 35 consecutive nucleotides, eg, 29 nucleotides. For example, each repeat
sequence consists of:
GAGTTCCCCGCGCCAGCGGGGATAAACCG (and optionally the Cas is/are E coli Cos). In
another example, each repeat sequence consists of: (and optionally the Cas
is/are C dificile Cas).
[00053] Optionally, each crRNA is expressed from the nucleic acid(s) under the
control of a common
or respective constitutive promoter.
[00054] Optionally, each Cas is expressed from the nucleic acid(s) under the
control of a common or
respective constitutive promoter. In an embodiment, the first crRNA and Cl are
expressed under the
control of a common constitutive promoter and/or the second crRNA and C2 are
expressed under the
control of a common constitutive promoter. For example, the promoters are the
same promoter or
they are different promoters. In an example, one, more of all of said
promoters is a strong promoter.
A promoter may be any promoter disclosed in W02020078893 or U S20200115716,
the disclosures of
such promoters (and nucleic acids, operons and vectors comprising one or more
such promoters)
being expressly incorporated herein by reference for possible use in the
present invention.
[00055] In an embodiment, a first plurality of different crRNAs are expressed
in one or more of the in
each cell, wherein each crRNA is operable with CS1 to guide modification of
the genome and the
plurality targets at least 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16,
17, 18, 19 or 20 (preferably, at
least 2, 3, 4 or 5; or exactly 2, 3, 4 or 5) different protospacers comprised
by the genome of the cell;
and/or a second plurality of different crRNAs are expressed in each cell
wherein each crRNA is
operable with CS2 and the second plurality targets at least 2, 3, 4, 5, 6, 7,
8, 9, 10, 11, 12, 13, 14, 15,
16, 17, 18, 19 or 20 (preferably, at least 2, 3, 4 or 5; or exactly 2, 3, 4 or
5) different comprised by the
genome of the cell. For example, the first plurality comprises from 2 to 10,
eg, from 2 to 7, different
crRNAs. For example, the second plurality comprises from 2 to 10, eg, from 2
to 7, different
crRNAs.
[00056] Optionally, one or more or all of said cells are killed by the method.
Optionally, the growth
or proliferation of one or more or all of said cells is reduced by the method.
Usefully, when the cell is
a prokaryotic cell (eg, a bacterial or archaeal cell) the chromosome of the
cell is cut by Cas. For
example, a bacterial cell chromosome is cut by Cl and C2 and the cell is
killed.
[00057] Optionally, the first crRNA (or each crRNA of said first plurality) is
comprised by a guide
RNA wherein the guide RNA further comprises a tracrRNA and/or the second crRNA
(or each
crRNA of said second plurality) is comprised by a guide RNA wherein the guide
RNA further
comprises a tracrRNA. Optionally, the first crRNA (or each crRNA of said first
plurality) is
11
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
comprised by a chimaeric guide RNA and/or the second crRNA (or each crRNA of
said second
plurality) is comprised by a chimaeric guide RNA.
[00058] For example, the genome modification of a plurality of cells is
cutting of genomic nucleic
acid (eg, chromosomal DNA) and the cells are killed, wherein said killing of
the plurality of cells is
synergistic compared to killing using CI or C2 alone.
[00059] An aspect provides:-
A method of killing or reducing the growth or proliferation of a plurality of
cells (optionally
prokaryotic cells, such as bacterial cells) of a first species or strain, the
method comprising carrying
out the method of the invention using the cells, wherein Cl and/or C2 is a Cas
nuclease and the
genomes of the cells are cut by Cas nuclease cutting and the cells are killed
or the growth or
proliferation of the cells is reduced.
1000601 Optionally, as exemplified herein, the method reduces the number of
cells of said plurality at
least 105, 106 or 107-fo1d, eg, between 105 and 107-fold, or between 105 and
10s-fold or between 105
and 109-fold. The skilled person will be familiar with determining fold-
killing or reduction in cells,
eg, using a cell sample that is representative of a microbiome or cell
population. An illustrative
example is given in the Examples below. For example, the extent of killing or
reduction in growth or
proliferation is determined using a cell sample, eg, a sample obtained from a
subject to which the
composition of the invention has been administered, or an environmental sample
(eg, aqueous, water
or soil sample) obtained from an environment (eg, a water source, waterway or
field) that has been
contacted with the composition of the invention.
1000611 For example, the method reduces the number of cells of said plurality
at least 105, 106 or 107¨
fold and optionally the plurality comprises at least 100,000; 1,000,000; or
10,000,000 cells
respectively.
[00062] Optionally, the plurality of cells is comprised by a cell population,
wherein at least 5, 6 or 7
log10 of cells of the population are killed by the method, and optionally the
plurality comprises at
least 100,000; 1,000,000; or 10,000,000 cells respectively.
[00063] When a cell herein is a bacterial cell, it may be of a first species
or genus selected from Table
1. Similarly, a plurality of cells herein may be cells which arc of a species
or genus selected from
Table 1.
[00064] Optionally, as exemplified herein, the method kills at least 99%.
99.9%. 99.99%, 99.999%,
99.9999% or 99.99999% cells of said plurality.
[00065] In an example, the method is carried out on a population (or said
plurality) of said cells and
the method kills, modifies or edits all (or essentially all) of the cells of
said population (or said
plurality). In an example, the method is carried out on a population (or said
plurality) of said cells and
the method kills, modifies or edits 100% (or about 100%) of the cells of said
population (or plurality).
12
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00066] Optionally, the species is E coil or C difficile.
[00067] An aspect of the invention provides:-
A method of editing the genome of one or more cells, the method comprising
(a) modifying the genome of each cell by carrying out the method of the
invention, wherein the
genome is subjected to Cas cutting; and
(b) inserting a nucleic acid at or adjacent to a Cos cut site in the genome
and/or deleting a nucleic
acid sequence from the genome at or adjacent to a Cas cut site in the genome,
wherein a cell
with an edited genome is produced; and
(c) optionally isolating from the cell a nucleic acid comprising the insertion
or the deletion; or
sequencing a nucleic acid sequence of the cell wherein the nucleic acid
sequence comprises
the insertion or the deletion.
1000681 In an example, the method is carried out on a population of said
cells, wherein the population
comprises at least 100 of said cells and at least 90 or 99% of said cells are
edited.
1000691 In an embodiment, the method is a method of recombineering, cg, in one
or more E coil cells.
[00070] The insertion may be immediately adjacent to, or overlapping the cut
site, or the insertion
may be within lkb, 2kb or 200, 150, 100, 50, 25, 10 or 5 nucleotides of the
cut site. For example, the
nucleic acid is inserted by homologous recombination. In an embodiment, the
nucleic acid is inserted
by homologous recombination and replaces (the sequence is inserted in the
place of genome sequence
that is deleted) genome sequence of 1 to 100, 90, 80, 70, 60, 50, 40, 30, 20,
10 or 5kb, or 200, 150,
100, 50, 25, 10 or 5 nucleotides of the genome. For example, the deleted
genome sequence flanks
either side of the cut site, or is at the 5'- or 3'-side of the cut site. In
an embodiment, the nucleic acid
is inserted by homologous recombination and does not replace any genomic
sequence.
[00071] The deletion may be immediately adjacent to, or overlapping the cut
site, or the deletion may
be within lkb, 2kb or 200, 150, 100, 50, 25, 10 or 5 nucleotides of the cut
site. For example, deletion
is a deletion of 1 to 100, 90, 80, 70, 60, 50, 40, 30, 20, 10, 5, 2 or lkb, or
200, 150, 100, 50, 25, 10 or
nucleotides of the genome. For example, the deleted genome sequence flanks
either side of the cut
site, or is at the 5'- or 3'-side of the cut site.
[00072] For example, the inserted nucleic acid is DNA. For example, the
deleted nucleic acid is
DNA, eg, chromosomal or episomal DNA).
[00073] For example, the inserted nucleic acid is at least (or no more than)
100, 90, 80, 70, 60, 50, 40,
30, 20, 10, 5, 2 or Ikb; or 200, 150, 100, 50, 25, 10 or 5 consecutive
nucleotides in length. For
example, the deleted genomic nucleic acid is at least (or no more than) 100,
90, 80, 70, 60, 50, 40, 30,
20, 10, 5, 2 or lkb; or 200, 150, 100, 50, 25, 10 or 5 consecutive nucleotides
in length.
13
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00074] For example, the genomic sequence is DNA. For example, genomic DNA is
deleted or
replaced. For example, genomic DNA is deleted or replaced and the editing
inserts DNA sequence
into the genome (eg, at or flanking the cut site).
[00075] For example, the genomic sequence is RNA. For example, genomic RNA is
deleted or
replaced. For example, genomic RNA is deleted or replaced and the editing
inserts RNA sequence
into the genome (eg, at or flanking the cut site).
[00076] Optionally, the method further comprises
(a) culturing the modified cell(s) to produce progeny thereof; and optionally
isolating the
progeny cells; or
(b) inserting a sequence obtained from a cell in step (c) into a recipient
cell and growing a cell
line therefrom.
[00077] Optionally, the progeny cells or cell line expresses a protein,
wherein the protein is encoded
(all or in part) by a nucleotide sequence that comprises the inserted nucleic
acid sequence, the method
further comprising obtaining the expressed protein or isolating the expressed
protein from the cells or
cell line.
[00078] Optionally, the method further comprises combining the progeny cells,
cell line or protein
with a pharmaceutically acceptable carrier, diluent or excipient, thereby
producing a pharmaceutical
composition.
[00079] In an embodiment, the inserted nucleic acid comprises a transcription
and/or translation
regulatory element for controlling expression of one or more nucleic acid
sequences of the edited
genome that are adjacent to the insertion. For example, the inserted nucleic
acid comprises a
promoter, eg, a constitutive or strong promoter. In another example, the
element is a transcription or
translation terminator, eg, the inserted sequence comprises a stop codon. In
this way, transcription of
a gene (or a part of a gene) that is adjacent to the inserted sequence in the
edited genome is terminated
or prevented or reduced.
[00080] In an example, the deleted genomic sequence is a RNA (eg, mRNA)
sequence. For example,
the deletion of the RNA sequence reduces or prevents expression of an amino
acid sequence in the
cell, wherein the amino acid sequence is encoded by the deleted RNA sequence.
This may be useful
for reducing or preventing expression in the cell of a protein comprising the
amino acid sequence,
such as where the protein is not desirable or required or detrimental to the
cell or is a subject or
environment that comprises the cell.
[00081] An aspect provides:-
A method of treating or preventing a disease or condition in a human or animal
subject, the method
comprising (i) administering to the subject a pharmaceutical composition
according to the invention
wherein the composition comprises said protein, wherein the protein mediates
treatment or prevention
of the disease or condition; or (ii) administering to the subject a
pharmaceutical composition
14
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
according to the invention, wherein when the composition comprises said
progeny cells or cell line,
the cells or cell line expresses a protein or RNA in the subject, and wherein
the protein or RNA
mediates treatment or prevention of the disease or condition.
[00082] Example diseases and conditions are disclosed below.
[00083] For example, the RNA encodes a therapeutic or prophylactic protein
that is expressed in the
subject. For example, the protein is a therapeutic or prophylactic protein.
The protein may exert a
therapeutic or prophylactic cell by interacting with a further protein (eg, an
endogenously-encoded
protein) in the subject, or by interacting with a further cell of the subject.
[00084] Optionally, the protein is an antibiotic, antibacterial agent, enzyme,
growth factor, antigen-
binding protein (eg, an antibody or fragment thereof), hormone, blood
component, cytokine, immune
checkpoint modulator (eg, inhibitor or upregulator), analgesic,
neurotransmitter, anti-inflammatory
agent or anti-neoplastic agent.
1000851 Optionally, the plurality of cells is comprised by a microbiome
sample, wherein the method is
carried out in vitro and produces a modified cell sample in which cells of the
first species or strain
have been killed, the method further comprising combining the modified sample
with a
pharmaceutically acceptable carrier, diluent or excipient, thereby producing a
pharmaceutical
composition comprising a cell transplant. For example, the transplant may be
administered to the
gastrointestinal (GI) tract or gut of a human or animal subject, eg, by oral
administration, or by rectal
administration. For example the transsplant may be administered by vaginal
administration.
[00086] Optionally, a microbiome herein is a gut, lung, kidney, urethral,
bladder, blood, vaginal, eye,
ear, nose, penile, bowel, liver, heart, tongue, hair or skin microbiome.
[00087] An aspect provides:-
A method of treating or preventing a disease or condition in a human or animal
subject, the method
comprising administering to the subject a pharmaceutical composition of the
invention.
[00088] An aspect provides:-
An ex vivo or in vitro method of treating an environment or cell sample, the
method comprising
exposing the environment or sample to a composition of the invention, wherein
cells comprised by the
environment or sample arc modified, edited or killed, or the growth or
proliferation of cells of the
environment or sample is reduced.
For example, the cells are killed. For example, the cells are edited by the
editing method of the
invention. Optionally, the treated sample is administered to a human or animal
subject or is contacted
with an environment.
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00089] Optionally, the plurality of cells is comprised by an environmental
sample (eg, an aqueous,
water, oil, petroleum, soil or fluid (such as an air or liquid) sample). A
suitable environment may be
contents of an industrial or laboratory apparatus or container, eg, a
fermentation vessel.
[00090] Optionally, the method of the invention is carried out in vitro.
Optionally, the method of the
invention is carried out ex vivo.
[00091] An aspect provides:-
A composition for use in a method treating or preventing a disease or
condition in a human or animal
subject that is mediated by target cells, the composition comprising
components (a), (b) or (d):-
(a) first and second crRNAs;
(b) nucleic acid encoding first and second crRNAs, wherein the nucleic acid is
expressible in a
target cell for producing the crRNAs;
(c) a first nucleic acid encoding a first crRNA, and a second nucleic acid
encoding a second
crRNA, wherein the nucleic acids are expressible in a target cell for
producing the crRNAs;
or
(d) a first crRNA and a nucleic acid encoding a second crRNA, wherein the
nucleic acid is
expressible in a target cell for producing the second crRNA;
wherein
(e) the first crRNA (crRNA1) is capable of guiding a first Cas (Cl) to a
protospacer sequence
(PS1) comprised by a target cell genome to modify PS1; and
(f) the second crRNA (crRNA2) is capable of guiding a second Cas (C2) to a
protospacer
sequence (PS2) comprised by the target cell genome to modify PS2;
(g) Cl and C2 are different;
(h) PS1 and PS2 are different; and
wherein the method comprises administering the composition to the subject
whereby said components
of the composition are introduced into target cells wherein crRNA1, crRNA2, Cl
and C2 are provided
in each cell and the genome of each cell is subjected to Cas modification and
the disease or condition
is treated or prevented.
[00092] Optionally, the treating or preventing compriscs carrying out the
method of thc invention.
1000931 Optionally, the method is for reducing an infection of the subject by
target cells (optionally
wherein the target cells are pathogenic cells, such as pathogenic prokaryotic
cells, such as pathogenic
bacterial cells).
[00094] Optionally, the components are comprised by one (or one or more)
nucleic acid vectors. In an
example, each vector is a virus, phage, plasmid (eg, a conjugative plasmid),
cosmid, phagemid or
nanoparticic (cg, a liposomc).
16
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[00095] In an example, any method herein is carried out on a population (or
said plurality) of said
cells, wherein the population comprises at least 100 of said cells and the
genome of at least 90, 99,
99.9, 99.99, 99.999, 99.9999, 99.99999, 99.999999, 99.9999999, 99.99999999 or
99.999999999% of
said cells are modified, eg, subjected to Cas nuclease cutting. In an
embodiment, the population (or
said plurality) comprises at least 1000 of said cells. In an embodiment, the
population (or said
plurality) comprises at least 10,000 of said cells. In an embodiment, the
population (or said plurality)
comprises at least 100,000 of said cells. In an embodiment, the population (or
said plurality)
comprises at least 1,000,000 of said cells. In an embodiment, the population
(or said plurality)
comprises at least 10,000,000 of said cells. In an embodiment, the population
(or said plurality)
comprises at least 100,000,000 of said cells. In an embodiment, the population
(or said plurality)
comprises at least 1000,000,000 of said cells. In an embodiment, the
population (or said plurality)
comprises at least 10,000,000,000 of said cells.
[00096] In an example, the population or said plurality is comprised by a
microbiome of a human,
animal (eg, a livestock animal or companion pet), plant or environment (eg, a
waterway, soil, fluid
microbiome).
[00097] An aspect provides:-
A composition comprising components (a), (b) or (d):-
(a) first and second crRNAs;
(b) nucleic acid encoding first and second crRNAs, wherein the nucleic acid is
expressible in a
target cell for producing the crRNAs;
(c) a first nucleic acid encoding a first crRNA, and a second nucleic acid
encoding a second
crRNA, wherein the nucleic acids are expressible in a target cell for
producing the crRNAs; or
(d) a first crRNA and a nucleic acid encoding a second crRNA, wherein the
nucleic acid is
expressible in a target cell for producing the second crRNA;
wherein
(e) the first crRNA (crRNA1) is capable of guiding a first Cas (C1) to a
protospacer sequence
(PS1) comprised by a target cell genome to modify PS1; and
(f) the second crRNA (crRNA2) is capable of guiding a second Cas (C2) to a
protospacer
sequence (PS2) comprised by the target cell genome to modify PS2;
(g) CI and C2 are different;
(h) PS1 and PS2 are different; and
wherein when said components of the composition are introduced into a target
cell whereby crRNA1,
crRNA2, Cl and C2 are provided in the cell, the genome of the cell is
subjected to Cas modification.
[00098] Optionally, the genome of each cell is edited or the cell is killed.
1000991 Optionally, each cell is a prokaryotic cell (optionally bacterial or
archaeal cell).
17
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[000100] Optionally, said nucleic acid(s) is(are) comprised by a
virus (eg, an AAV, or
cytomegalovirus, optionally wherein each cell is a mammalian cell, such as a
human cell), phage (eg,
wherein each cell is a bacterial cell), plasmid (optionally a conjugative
plasmid, eg, wherein each cell
is a bacterial cell), nanoparticle (eg, a liposome or gold particle) or
phagemid (eg, wherein each cell
is a bacterial cell).
[000101] When the nucleic acid is comprised by a virus, the cell
may be a mammalian (eg,
human or rodent, mouse or rat) cell, a bacterial cell, an archaeal cell or an
amoeba cell. When the
nucleic acid is comprised by a phage, the cell may be a bacterial cell.
[000102] Optionally, said nucleic acid(s) encode Cl and/or C2.
[000103] Optionally, Cl is a Type I Cas and said nucleic acid(s)
encode one or more Cascade
Cas that are operable with Cl and/or wherein C2 is a Type I Cas and said
nucleic acid(s) encode one
or more Cascade Cas that are operable with C2.
10001041 An aspect provides:-
A pharmaceutical composition which is a composition according to the
invention, wherein the
composition comprises a pharmaceutically acceptable excipient, diluent or
carrier.
[000105] The composition may be an aqueous composition. The
composition may be a
lyophilised or freeze-dried composition, eg, in a formulation that is suitable
for inhaled delivery to the
patient.
[000106] Optionally, the composition is comprised by a sterile
medicament administration
device, optionally a syringe, IV bag, intranasal delivery device, inhaler,
nebuliser or rectal
administration device). Optionally, the composition is comprised by a cosmetic
product, dental
hygiene product, personal hygiene product, laundry product, oil or petroleum
additive, water additive,
shampoo, hair conditioner, skin moisturizer, soap, hand detergent, clothes
detergent, cleaning agent,
environmental remediation agent, cooling agent (eg, an air cooling agent) or
air treatment agent.
[000107] In an example the composition is comprised by a device
for delivering the
composition as a liquid or dry powder spray. This may be useful for
administration topically to
patients or for administration to large environmental areas, such as fields or
waterways.
[000108] Optionally, the cells arc comprise by a gut, lung,
kidney, urethral, bladder, blood,
vaginal or skin microbiome of the subject.
[000109] Optionally, the method is carried out on a human or
animal subject, wherein the cells
are killed by the method and the killing upregulates or downregulates immune
cells (optionally (i)
upregulating CD8+, CD4+,TH1, TH2, TH17, NK cells, TILS, T regulatory or T
effector cells; or (ii)
downregulating CDg+, CD4,'TH1, 'TH2, 'TH17, T regulatory or T effector cells)
in the subject,
thereby treating or preventing a disease or condition in the subject. In one
preferred embodiment,
CD8+, NK or TILS cells are upregulated, eg, wherein the disease or condition
is a cancer. In one
18
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
preferred embodiment, CD8+ or NK cells are upregulated, eg, wherein the
disease or condition is a
viral infection. In one preferred embodiment, TH1, TH2 or TH17 cells are
downregulated, eg,
wherein the disease or condition is an autoimmune or inflammatory disease or
condition. For
example, the disease or condition is a cancer or an autoimmune disease or
condition. For example,
the disease or condition is a cancer and CD8+ or T effector cells are
upregulated in the subject and/or
T regulatory cells are downregulated in the subject. For example, the disease
or condition is an
autoimmune disease or condition and CD 8+ or T effector cells are
downregulated in the subject and/or
T regulatory cells are upregulated in the subject.
[000110] Optionally, the method comprises introducing into each
cell or expressing in each cell
at least 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 or 20
(preferably, at least 2, 3, 4 or 5;
or exactly 2, 3, 4 or 5, or exactly 8, or at least 8) different types of
crRNAs wherein the different types
target different protospacer sequences comprised by the cell genome; and
optionally wherein Cl and
C2 are Class 1 Cas nucleases, eg, Cas 3 nucleases.
10001111 Optionally, the method comprises introducing into each
cell a nucleic acid encoding a
Cas3, Cas8e, Casll, Cas7, Cas5, and Cas6 (optionally, the Cas are E coli Cas)
and/or a nucleic acid
encoding a Cas3, Cas6, Cas8b, Cas7, and Cas5 (optionally, the Cas arc C
dijIcile Cas).
[000112] In another example, the method comprises introducing into
each cell a nucleic acid
encoding a Cas3, Cas8e, Casll, Cas7, Cas5, and a nucleic acid encoding a Cas9.
In another example,
the method comprises introducing into each cell a nucleic acid encoding a
Cas3, Cas6, Cas8b, Cas7,
and Cas5 and a nucleic acid encoding a Cas9.
[000113] An aspect provides:
A method of modifying the genome of a cell, the method comprising
(a) using a first CRISPR/Cas system to modify a first protospacer of the
genome; and
(b) using a second CRISPR/Cas system to modify a second protospacer of the
genome, wherein
the second protospacer is different to the first protospacer;
wherein the systems comprise different Cas and are provided simultaneously in
the cell.
[000114] Optionally, the method comprises using a third CRISPR/Cas
system to modify a third
protospaccr of the genome, wherein the third protospacer is different to the
first and second
protospacers. For example, 3 different Cas3 are used; 3 different Cas9 are
used; a Cas3 and two
different Cas9 are used; or two different Cas3 and a Cas9 are used.
[000115] The method of claim 51, wherein the method is according
to any one of claims 1 to
35.
[000116] In certain aspects:-
The method of the invention is a method of
19
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
(a) producing synergistic Cas nuclease cutting of a cell genome;
(b) reducing a population of cells of a first species or strain by at least
100,000, 1,000,000 or
10,000,000-fold;
(c) killing at least at least 99%. 99.9%. 99.99%, 99.999%, 99.9999% or
99.99999% cells of a
first species or strain comprised by a microbiome;
(d) producing synergistic Class 1 Cas modification of a cell genome; or
(e) reducing bacterial cells of a first species or strain (eg, E coli cells)
in a cell population by at
least 105, 106 or 107-fold, wherein the population comprises at least 100,000;
1,000,000; or
10,000,000 cells respectively.
10001171 DISEASES AND CONDITIONS
Optionally, the disease or condition is selected from
(a) A neurodegeneratiye disease or condition;
(b) A brain disease or condition;
(c) A CNS disease or condition-
,
(d) Memory loss or impairment;
(e) A heart or cardiovascular disease or condition, eg, heart attack,
stroke or atrial fibrillation;
A liver disease or condition;
(g) A kidney disease or condition, eg, chronic kidney disease (CKD);
(h) A pancreas disease or condition;
(1) A lung disease or condition, eg, cystic fibrosis or COPD;
(1) A gastrointestinal disease or condition;
(k) A throat or oral cavity disease or condition;
(1) An ocular disease or condition;
(m) A genital disease or condition, eg, a vaginal, labial, penile or
scrotal disease or condition;
(n) A sexually-transmissible disease or condition, eg, gonorrhea, HIV
infection, syphilis or
Chlamydia infection;
(o) An ear disease or condition;
(13) A skin disease or condition;
(c1) A heart disease or condition;
(r) A nasal disease or condition
(s) A haematological disease or condition, eg, anaemia, eg, anaemia of
chronic disease or cancer;
(t) A viral infection;
(u) A pathogenic bacterial infection;
(v) A cancer;
(w) An autoimmunc disease or condition, cg, SLE;
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
(X) An inflammatory disease or condition, eg, rheumatoid arthritis,
psoriasis, eczema, asthma,
ulcerative colitis, colitis, Crohn's disease or IBD;
(y) Autism;
(z) ADHD;
(an) Bipolar disorder;
(bb) ALS [Amyotrophic Lateral Sclerosis];
(cc) Osteoarthritis;
(dd) A congenital or development defect or condition;
(ee) Miscarriage;
(if) A blood clotting condition;
(gg) Bronchitis;
(hh) Dry or wet AMD;
(ii) Neovascularisation (eg, of a tumour or in the eye);
(j1) Common cold;
(kk) Epilepsy;
(11) Fibrosis, eg, liver or lung fibrosis;
(mm) A fungal disease or condition, eg, thrush;
(nn) A metabolic disease or condition, eg, obesity, anorexia,
diabetes, Type I or Type II diabetes.
(oo) Ulcer(s), eg, gastric ulceration or skin ulceration;
(1313) Dry skin;
(WI) Sjogren's syndrome;
(1-1) Cytokine storm;
(ss) Deafness, hearing loss or impairment;
(-11) Slow or fast metabolism (ie, slower or faster than average for
the weight, sex and age of the
subject);
(uu) Conception disorder, eg, infertility or low fertility;
(vv) Jaundice;
(ww) Skin rash;
(xx) Kawasaki Disease;
(yy) Lyme Disease;
(zz) An allergy, eg, a nut, grass, pollen, dust mite, cat or dog fur
or dander allergy;
(aaa) Malaria, typhoid fever, tuberculosis or cholera;
(bbb) Depression;
(ccc) Mental retardation;
(ddd) Microcephaly;
(coo) Malnutrition;
(fff) Conjunctivitis;
21
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
(ggg) Pneumonia;
(hhh) Pulmonary embolism;
(iii) Pulmonary hypertension;
(jjj) A bone disorder;
(kkk) Sepsis or septic shock;
(111) Sinusitus;
(nimm) Stress (eg, occupational stress);
(nnn) Thalassaemia, anaemia, von Willebrand Disease, or haemophilia;
(000) Shingles or cold sore;
(ppp) Menstruation;
(qqq) Low sperm count.
NEURODEGENERATIVE OR CNS DISEASES OR CONDITIONS FOR TREATMENT OR
PREVENTION
[00113] In an example, a neurodegenerative or CNS disease or condition is
selected from the group
consisting of Alzheimer disease , geriopsychosis, Down syndrome, Parkinson's
disease, Crcutzfeldt-
jakob disease, diabetic neuropathy, Parkinson syndrome, Huntington's disease,
Machado-Joseph
disease, amyotrophic lateral sclerosis, diabetic neuropathy, and Creutzfeldt
Creutzfeldt- Jakob
disease. For example, the disease is Alzheimer disease. For example, the
disease is Parkinson
syndrome.
[00114] In an example, wherein the method of the invention is practised on a
human or animal subject
for treating a CNS or neurodegenerative disease or condition, the method
causes downregulation of
Treg cells in the subject, thereby promoting entry of systemic monocyte-
derived macrophages and/or
Treg cells across the choroid plexus into the brain of the subject, whereby
the disease or condition (eg,
Alzheimer's disease) is treated, prevented or progression thereof is reduced.
In an embodiment the
method causes an increase of IFN-gamma in the CNS system (eg, in the brain
and/or CSF) of the
subject. In an example, the method restores nerve fibre and//or reduces the
progression of nerve fibre
damage. In an example, the method restores nerve myelin and//or reduces the
progression of nerve
myelin damage. In an example, the method of the invention treats or prevents a
disease or condition
disclosed in W02015136541 and/or the method can be used with any method
disclosed in
W02015136541 (the disclosure of this document is incorporated by reference
herein in its entirety,
eg, for providing disclosure of such methods, diseases, conditions and
potential therapeutic agents that
can be administered to the subject for effecting treatement and/or prevention
of CNS and
neurodegenerative diseases and conditions, eg, agents such as immune
checkpoint inhibitors, eg, anti-
PD-1, anti-PD-L1, anti-TIM3 or other antibodies disclosed therein).
22
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
CANCERS FOR TREATMENT OR PREVENTION
[00115] Cancers that may be treated include tumours that are not vascularized,
or not substantially
vascularized, as well as vascularized tumours. The cancers may comprise non-
solid tumours (such as
haematological tumours, for example, leukaemias and lymphomas) or may comprise
solid tumours.
Types of cancers to be treated with the invention include, but are not limited
to, carcinoma, blastoma,
and sarcoma, and certain leukaemia or lymphoid malignancies, benign and
malignant tumours, and
malignancies e.g., sarcomas, carcinomas, and melanomas. Adult tumours/cancers
and paediatric
tumours/cancers are also included.
[00116] Haematologic cancers are cancers of the blood or bone marrow. Examples
of haematological
(or haematogenous) cancers include leukaemias, including acute leukaemias
(such as acute
lymphocytic leukaemia, acute myelocytic leukaemia, acute myelogenous leukaemia
and myeloblasts,
promyeiocytic, myelomonocytic, monocytic and erythroleukaemia), chronic
leukaemias (such as
chronic myelocytic (granulocytic) leukaemia, chronic myelogenous leukaemia,
and chronic
lymphocytic leukaemia), polycythemia vera, lymphoma, Hodgkin's disease, non-
Hodgkin's lymphoma
(indolent and high grade forms), multiple myeloma, Waldenstrom's
macroglobulinemia, heavy chain
disease, myeiodysplastic syndrome, hairy cell leukaemia and myelodysplasia.
[00117] Solid tumours are abnormal masses of tissue that usually do not
contain cysts or liquid areas.
Solid tumours can be benign or malignant. Different types of solid tumours are
named for the type of
cells that form them (such as sarcomas, carcinomas, and lymphomas). Examples
of solid tumours,
such as sarcomas and carcinomas, include fibrosarcoma, myxosarcoma,
liposarcoma,
chondrosarcoma, osteosarcoma, and other sarcomas, synovioma, mesothelioma,
Ewing's tumour,
leiomyosarcoma, rhabdomyosarcoma, colon carcinoma, lymphoid malignancy,
pancreatic cancer,
breast cancer, lung cancers, ovarian cancer, prostate cancer, hepatocellular
carcinoma, squamous eel!
carcinoma, basal cell carcinoma, adenocarcinoma, sweat gland carcinoma,
medullary thyroid
carcinoma, papillary thyroid carcinoma, pheochromocytomas sebaceous gland
carcinoma, papillary
carcinoma, papillary adenocarcinomas, medullary carcinoma, bronchogenic
carcinoma, renal cell
carcinoma, hepatoma, bile duct carcinoma, choriocarcinoma, Wilms' tumour,
cervical cancer,
testicular tumour, seminoma, bladder carcinoma, melanoma, and CNS tumours
(such as a glioma
(such as brainstem glioma and mixed gliomas), glioblastoma (also known as
glioblastoma
multiformc) astrocytoma, CNS lymphoma, gcrminoma, mcdu!loblastoma, Schwannoma
craniopharyogioma, ependymoma, pineaioma, hemangioblastoma, acoustic neuroma,
oligodendroglioma, menangioma, neuroblastoma, retinoblastoma and brain
metastases).
[00118] AUTOIMMUNE DISEASES FOR TREATMENT OR PREVENTION
= Acute Disseminated Encephalomyelitis (ADEM)
= Acute necrotizing hemorrhagic leukoencephalitis
= Addison's disease
23
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
= Agammaglobulinemia
= Alopecia areata
= Amyloidosis
= Ankylosing spondylitis
= Anti-GBIVI/Anti-TBM nephritis
= Antiphospholipid syndrome (APS)
= Autoimmune angioedema
= Autoimmune aplastic anemia
= Autoimmune dysautonomia
= Autoimmune hepatitis
= Autoimmune hyperlipidemia
= Autoimmune immunodeficiency
= Autoimmune inner ear disease (AIED)
= Autoimmune myocarditis
= Autoimmune oophoritis
= Autoimmune pancreatitis
= Autoimmune retinopathy
= Autoimmune thrombocytopenic purpura (ATP)
= Autoimmune thyroid disease
= Autoimmune urticaria
= Axonal & neuronal neuropathies
= Balo disease
= Behcet's disease
= Buttons pemphigoid
= Cardiomyopathy
= Castleman disease
= Celiac disease
= Chagas disease
= Chronic fatigue syndrome
= Chronic inflammatory dcmyclinating polyncuropathy (CIDP)
= Chronic recurrent multifocal ostomyelitis (CRMO)
= Churg-Strauss syndrome
= Cicatricial pemphigoid/benign mucosal pemphigoid
= Crohn's disease
= Cogans syndrome
= Cold agglutinin disease
= Congenital heart block
24
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
= Coxsackie myocarditis
= CREST disease
= Essential mixed cryoglobulinemia
= Demyelinating neuropathies
= Dermatitis herpetiformis
= Dermatomyositis
= Devic's disease (neuromyelitis optica)
= Discoid lupus
= Dressler's syndrome
= Endometriosis
= Eosinophilic esophagitis
= Eosinophilic fasciitis
= Erythema nodosum
= Experimental allergic encephalomyelitis
= Evans syndrome
= Fibromyalgia
= Fibrosing alveolitis
= Giant cell arteritis (temporal arteritis)
= Giant cell myocarditis
= Glomerulonephritis
= Goodpasture's syndrome
= Granulomatosis with Polyangiitis (GPA) (formerly called Wegener's
Granulomatosis)
= Graves' disease
= Guillain-Barre syndrome
= Hashimoto's encephalitis
= Hashimoto's thyroiditis
= Hemolytic anemia
= Henoch-Schonlein puipura
= Herpes gestationis
= Hypogammaglobulinemia
= Idiopathic thrombocytopenic purpura (ITP)
= IgA nephropathy
= IgG4-related sclerosing disease
= Immunoregulatory lipoproteins
= Inclusion body myosins
= Interstitial cystitis
= Juvenile arthritis
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
= Juvenile diabetes (Type 1 diabetes)
= Juvenile myositis
= Kawasaki syndrome
= Lambert-Eaton syndrome
= Leukocytoclastic vasculitis
= Lichen planus
= Lichen sclerosus
= Ligneous conjunctivitis
= Linear IgA disease (LAD)
= Lupus (SLE)
= Lyme disease, chronic
= Meniere's disease
= Microscopic polyangiitis
= Mixed connective tissue disease (MCTD)
= Mooren's ulcer
= Mucha-Habermann disease
= Multiple sclerosis
= Myasthenia gravis
= Myositis
= Narcolepsy
= Neuromyelitis optica (Devic's)
= Neutropenia
= Ocular cicatricial pemphigoid
= Optic neuritis
= Palindromic rheumatism
= PANDAS (Pediatric Autoimmune Neuropsychiatric Disorders Associated with
Streptococcus)
= Paraneoplastic cerebellar degeneration
= Paroxysmal nocturnal hemoglobinuria (PNH)
= Parry Romberg syndrome
= Parsonnage-Turner syndrome
= Pars planitis (peripheral uveitis)
= Pemphigus
= Peripheral neuropathy
= Perivenous encephalomyelitis
= Pernicious anemia
= POEMS syndrome
26
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
= Polyarteritis nodosa
= Type I, II, & III autoimmune polyglandular syndromes
= Polymyalgia rheumatica
= Polymyositis
= Postmyocardial infarction syndrome
= Postpericardiotomy syndrome
= Progesterone dermatitis
= Primary biliary cirrhosis
= Primary sclerosing cholangitis
= Psoriasis
= Psoriatic arthritis
= Idiopathic pulmonary fibrosis
= Pyoderma gangrenosum
= Pure red cell aplasia
= Raynauds phenomenon
= Reactive Arthritis
= Reflex sympathetic dystrophy
= Reiter's syndrome
= Relapsing polychondritis
= Restless legs syndrome
= Retroperitoneal fibrosis
= Rheumatic fever
= Rheumatoid arthritis
= Sarcoidosis
= Schmidt syndrome
= Scleritis
= Scleroderma
= Sjogren's syndrome
= Sperm & testicular autoimmunity
= Stiff person syndrome
= Subacute bacterial endocarditis (SBE)
= Susac's syndrome
= Sympathetic ophthalmia
= Takayasu's arteritis
= Temporal arteritis/Giant cell arteritis
= Thrombocytopcnic purpura (TTP)
= Tolosa-Hunt syndrome
27
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
= Transverse myelitis
= Type I diabetes
= Ulcerative colitis
= Undifferentiated connective tissue disease (UCTD)
= Uveitis
= Vasculitis
= Vesiculobullous dennatosis
= Vitiligo
= Wegener's granulomatosis (now termed Granulomatosis with Polyangiitis
(GPA).
[00119] INFLAMMATORY DISEASES FOR TREATMENT OR PREVENTION
= Alzheimer's
= ankylosing spondylitis
= arthritis (osteoarthritis, rheumatoid arthritis (RA), psoriatic
arthritis)
= asthma
= atherosclerosis
= Crohn's disease
= colitis
= dermatitis
= diverticulitis
= fibromyalgia
= hepatitis
= irritable bowel syndrome (IBS)
= systemic lupus erythematous (SLE)
= nephritis
= Parkinson's disease
= ulcerative colitis.
[000120] Optionally, the cells are C dificile, P aeruginosa, K
pneumoniae (eg, carbapenem-
resistant Klebsiella pneumoniae or Extended-Spectrum Beta-Lactamase (ESBL)-
producing K
pneumonicte), E coli (eg, ESBL-producing E. coli, or E. coli ST131-025b:H4), H
pylori, S
pneumoniae or S aureus cells.
[000121] A vector herein may be a high copy number plasmid or
phagemid comprising a
constitutive promoter for controlling the expression of crRNAs and optionally
one or more Cas
proteins
28
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[000122] In an example, promoter is a medium strength promoter. In
another example, the
promoter is a repressible promoter or an inducible promoter cell. Examples of
suitable repressible
promoters are Ptac (repressed by lad) and the Leftward promoter (pL) of phage
lambda (which
repressed by the ),,cI repressor). In an example, the promoter comprises a
repressible operator (eg,
tet0 or lac0) fused to a promoter sequence. Optionally, the promoter has an
Anderson Score (AS) of
0.5>AS >0.1.
GENERALLY APPLICABLE FEATURES:
[000123] Any cell herein may be a bacterial cell, archaeal cell,
algal cell, fungal cell, protozoan
cell, invertebrate cell, vertebrate cell, fish cell, bird cell, mammal cell,
companion animal cell, dog
cell, cat cell, horse cell, mouse cell, rat cell, rabbit cell, eukaryotic
cell, prokaryotic cell, human cell,
animal cell, rodent cell, insect cell or plant cell. Preferably, the cell is a
bacterial cell. Alternatively,
the cell is a human cell.
10001241 Optionally, Cl and C2 is any Cas (eg, a Cas2, 3, 4, 5, or
6) of a Type I system. In
this example, in an embodiment_ the Cas may be fused or conjugated to a moiety
that is operable to
increase or reduce transcription of a gene comprising the target protospacer
sequence. For example
the nucleic acid encoding the Cas that is introduced into a cell may comprise
a nucleotide sequence
encoding the moiety, wherein the Cas and moiety are expressed in the host cell
as a fusion protein. In
one embodiment, the Cas is N-terminal of the moiety; in another embodiment it
is C-terminal to the
moiety.
[000125] In an example, a vector herein is a DNA vector, eg, ssDNA
vector or dsDNA vector.
Optionally, the vector comprises a second nucleotide sequence encoding one or
more Cascade
proteins. For example, the Cascade protein(s) are cognate with the Cl or C2,
which is a Cas3.
[000126] In an example, Casl or Cas2 is a Cas3 that is cognate
with Cascade proteins encoded
by the cell.
[000127] Optionally, the Cas3 is a Cas3 encoded by a CRISPR/Cas
locus of a first bacterial or
archaeal species, wherein in the locus the Cas3-encoding sequence is 3' of
Cascade protein-encoding
sequences (ie, the latter are between the Cas3 and the 5'-most promoter of the
locus). Optionally, the
Cas3 is a ygcB protein.
[000128] Optionally, the Cascade proteins comprise or consist of
cas5 (casD, csy2), cas6
(cas6f, cse3, casE), cas7 (csc2, csy3, cse4, casC) and cas8 (casA, cas8al,
cas8b1, cas8c, caslOd,
cas8e, csel, cas8f, csy 1).
[000129] Optionally herein the promoter and the Cas3-encoding or
crRNA-encoding sequence
are spaced no more than 150, 100, 50, 40, 30, 20 or 10bp apart, eg, from 30-
45, or 30-40, or 39 or
around 39bp apart. Optionally herein a ribosome binding site and the Cas3-
encoding or crRNA-
encoding sequence are spaced no more than 20, 15, 14, 13, 12, 11, 10, 9, 8, 7,
6, 4 or 3bp apart, cg,
from 10-5, 6 or around 6bp apart.
29
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[000130] In an example, a promoter herein is in combination with a
Shine-Dalgarno sequence
comprising the sequence 5'- anagaggagaaa 3' (SEQ ID NO: 5) or a ribosome
binding site homologue
thereof. Optionally the promoter has an Anderson Score (AS) of AS >0.5; or an
Anderson Score (AS)
of 0.5>AS >0.1; or an Anderson Score (AS) of <0.1.
[000131] Optionally, the first crRNA-encoding nucleic acid
sequence, the second crRNA-
encoding nucleic acid sequence or operon is comprised by a mobile genetic
element. Suitable mobile
genetic elements, eg, transposons, are disclosed in W02016177682 and
US20170246221, the
disclosures of which are explicitly incorporated herein for possible use in
the invention and for
providing one or more features for the claims herein.
[000132] Optionally, the vector is devoid of nucleotide sequence
encoding one, more or all of a
Casl, Cas2, Cas4, Cas6 (optionally Cas6f), Cas7 and Cas 8 (optionaly Cas8f).
Optionally, the vector
is devoid of a sequence encoding a Cas6 (optionally a Cas6f). Optionally, tlhe
vector comprises
(optionally in 5' to 3- direction) nucleotide sequence encoding one, more or
all of Cas 11, Cas7 and
Cas8a1. Optionally, the vector comprises nucleotide sequence encoding Cas3'
and/or Cas3". In one
embodiment, the vector comprises nucleotide sequences (in 5' to 3' direction)
that encode a Cas3 (eg,
Cas3' and/or Cas3"), Cash, Cas7 and Cas8a1.
[000133] Optionally, a nucleotide sequence encoding Cas6 is
between the Cas3 sequence(s)
and the Cas 11 sequence. Optionally, the vector comprises a Type IA CRISPR
array or one or more
nucleotide sequences encoding single guide RNA(s) (gRNA(s)), wherein the array
and each gRNA
comprises repeat sequence that is cognate with the Cas3. Thus, the array is
operable in a host cell
when the vector has been introduced into the cell for production of guide
RNAs, wherein the guide
RNAs are operable with the Cas and Cascade proteins to target and modify (eg,
cut) a target
nucleotide sequence in the host cell, optionally thereby killing the host
cell. Similarly, the single
guide RNAs encoded by the vector in one embodiment are operable with the Cas
and Cascade
proteins to target and modify (eg, cut) a target nucleotide sequence in the
host cell, optionally thereby
killing the host cell.
[000134] Optionally, each cell comprises a Type IA CRISPR array
that is cognate with the
Cas3 (Cl or C2). Optionally, each cell comprises an endogenous Type IB, C, U,
D, E or F
CRISPR/Cas system. Optionally, the vector comprises (optionally in 5' to 3'
direction) nucleotide
sequence encoding one, more or all of Cas8b1, Cas7 and Cas5. In one
embodiment, the vector
comprises nucleotide sequences (in 5' to 3' direction) that encode a Cas3,
Cas8b1, Cas7 and Cas5.
Optionally, a nucleotide sequence encoding Cas6 is between the Cas3
sequence(s) and the Cas8b1
sequence. Optionally, the vector comprises a Type TB CRISPR array or one or
more nucleotide
sequences encoding single guide RNA(s) (gRNA(s)), wherein the array and each
gRNA comprises
repeat sequence that is cognate with the Cas3. Thus, the array is operable in
a host cell when the
vector has been introduced into the cell for production of guide RNAs, wherein
the guide RNAs are
operable with the Cas and Cascade proteins to target and modify (eg, cut) a
target nucleotide sequence
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
in the host cell, optionally thereby killing the host cell. Similarly, the
single guide RNAs encoded by
the vector in one embodiment are operable with the Cos and Cascade proteins to
target and modify
(eg, cut) a target nucleotide sequence in the host cell, optionally thereby
killing the host cell.
[000135] Optionally, the cell comprises a Type TB CRISPR array
that is cognate with the Cas3.
Optionally, the cell comprises an endogenous Type IA, C, U, D, E or F
CRISPR/Cas system.
Optionally, the vector comprises (optionally in 5' to 3' direction) nucleotide
sequence encoding one,
more or all of Cas5, Cas8c and Cas7. In one embodiment, the vector comprises
nucleotide sequences
(in 5' to 3' direction) that encode a Cas3, Cas5, Cas8c and Cas7. Optionally,
a nucleotide sequence
encoding Cas6 is between the Cas3 sequence(s) and the Cas5 sequence.
Optionally, the vector
comprises a Type IC CRISPR array or one or more nucleotide sequences encoding
single guide
RNA(s) (gRNA(s)), wherein the array and each gRNA comprises repeat sequence
that is cognate with
the Cas3. Thus, the array is operable in a host cell when the vector has been
introduced into the cell
for production of guide RNAs, wherein the guide RNAs are operable with the Cos
and Cascade
proteins to target and modify (eg, cut) a target nucleotide sequence in the
host cell, optionally thereby
killing the host cell. Similarly, the single guide RNAs encoded by the vector
in one embodiment are
operable with the Cas and Cascade proteins to target and modify (cg, cut) a
target nucleotide sequence
in the host cell, optionally thereby killing the host cell.
[000136] Optionally, the host cell comprises a Type IC CRISPR
array that is cognate with the
Cas3. Optionally, the host cell comprises an endogenous Type IA, B, U, D, E or
F CRISPR/Cas
system. Optionally, the vector comprises (optionally in 5' to 3' direction)
nucleotide sequence
encoding one, more or all of Cas8U2, Cas7, Cas5 and Cas6. In one embodiment,
the vector
comprises nucleotide sequences (in 5' to 3' direction) that encode a Cas3,
Cas8U2, Cas7, Cas5 and
Cas6. Optionally, a nucleotide sequence encoding Cas6 is between the Cas3
sequence(s) and the
Cas8U2 sequence.
[000137] Optionally, the vector comprises a Type IU CRISPR array
or one or more nucleotide
sequences encoding single guide RNA(s) (gRNA(s)), wherein the array and each
gRNA comprises
repeat sequence that is cognate with the Cas3. Thus, the array is operable in
a host cell when the
vector has been introduced into the cell for production of guide RNAs, wherein
the guide RNAs are
operable with the Cas and Cascade proteins to target and modify (eg, cut) a
target nucleotide sequence
in the host cell, optionally thereby killing the host cell. Similarly, the
single guide RNAs encoded by
the vector in one embodiment are operable with the Cos and Cascade proteins to
target and modify
(eg, cut) a target nucleotide sequence in the host cell, optionally thereby
killing the host cell.
[000138] Optionally, the host cell comprises a Type IU CRISPR
array that is cognate with the
Cas3. Optionally, the host cell comprises an endogenous Type IA, B, C, D, E or
F CRISPR/Cas
system. Optionally, the vector comprises (optionally in 5' to 3' direction)
nucleotide sequence
encoding one, more or all of Cas lOd, Cas7 and Cas5. Optionally, the vector
comprises a nucleotide
sequence encoding Cas3' and/or Cas3¨. In one embodiment, the vector comprises
nucleotide
31
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
sequences (in 5' to 3' direction) that encode a Cas3, CaslOd, Cas7 and Cas5.
Optionally, a nucleotide
sequence encoding Cas6 is between the Cas3 sequence(s) and the CaslOd
sequence. Optionally, the
vector comprises a Type ID CRISPR array or one or more nucleotide sequences
encoding single guide
RNA(s) (gRNA(s)), wherein the array and each gRNA comprises repeat sequence
that is cognate with
the Cas3. Thus, the array is operable in a host cell when the vector has been
introduced into the cell
for production of guide RNAs, wherein the guide RNAs are operable with the Cos
and Cascade
proteins to target and modify (eg, cut) a target nucleotide sequence in the
host cell, optionally thereby
killing the host cell. Similarly, the single guide RNAs encoded by the vector
in one embodiment are
operable with the Cas and Cascade proteins to target and modify (eg, cut) a
target nucleotide sequence
in the host cell, optionally thereby killing the host cell.
[000139] Optionally, the host cell comprises a Type ID CRISPR
array that is cognate with the
Cas3.
[000140] Optionally, the host cell comprises an endogenous Type
IA, B, C, U, E or F
CRISPR/Cas system.
[000141] Optionally, the vector comprises (optionally in 5' to 3'
direction) nucleotide sequence
encoding one, more or all of Cas8e, Os' I, Cas7, Cas5 and Cas6. In one
embodiment, the vector
comprises nucleotide sequences (in 5' to 3' direction) that encode a Cas3,
Cas8e, Casll, Cas7, Cas5
and Cas6. Optionally, a nucleotide sequence encoding Cas6 is between the Cas3
sequence(s) and the
Casll sequence. Optionally, the vector comprises a Type IE CRISPR array or one
or more nucleotide
sequences encoding single guide RNA(s) (gRNA(s)), wherein the array and each
gRNA comprises
repeat sequence that is cognate with the Cas3. Thus, the array is operable in
a host cell when the
vector has been introduced into the cell for production of guide RNAs, wherein
the guide RNAs are
operable with the Cas and Cascade proteins to target and modify (eg, cut) a
target nucleotide sequence
in the host cell, optionally thereby killing the host cell. Similarly, the
single guide RNAs encoded by
the vector in one embodiment are operable with the Cas and Cascade proteins to
target and modify
(eg, cut) a target nucleotide sequence in the host cell, optionally thereby
killing the host cell.
[000142] Optionally, the host cell comprises a Type IE CRISPR
array that is cognate with the
Cas3.
[000143] Optionally, the host cell comprises an endogenous Type
IA, B, C, D, U or F
CRISPR/Cas system.
10001441 Optionally, the vector comprises (optionally in 5' to 3'
direction) nucleotide sequence
encoding one, more or all of Cas8f, Cas5, Cas7 and Cas6f. In one embodiment,
the vector comprises
nucleotide sequences (in 5' to 3' direction) that encode a Cas3, Cas8f, Cas5,
Cas7 and Cas6f.
Optionally, a nucleotide sequence encoding Cas6 is between the Cas3
sequence(s) and the Cas8f
sequence. Optionally, the vector comprises a Type IF CRISPR array or one or
more nucleotide
sequences encoding single guide RNA(s) (gRNA(s)), wherein the array and each
gRNA comprises
repeat sequence that is cognate with the Cas3. Thus, the array is operable in
a host cell when the
32
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
vector has been introduced into the cell for production of guide RNAs, wherein
the guide RNAs are
operable with the Cas and Cascade proteins to target and modify (eg, cut) a
target nucleotide sequence
in the host cell, optionally thereby killing the host cell. Similarly, the
single guide RNAs encoded by
the vector in one embodiment are operable with the Cos and Cascade proteins to
target and modify
(eg, cut) a target nucleotide sequence in the host cell, optionally thereby
killing the host cell.
[000145] Optionally, the host cell comprises a Type IF CRISPR
array that is cognate with the
Cas3.
[000146] Optionally, the host cell comprises an endogenous Type
IA, B, C, D, U or E
CRISPR/Cas system.
[000147] Optionally, the Cas and Cascade are Type IA Cas and
Cascade proteins.
[000148] Optionally, the Cas and Cascade are Type TB Cas and
Cascade proteins.
[000149] Optionally, the Cas and Cascade are Type IC Cas and
Cascade proteins.
[000150] Optionally, the Cas and Cascade are Type ID Cas and
Cascade proteins.
10001511 Optionally, the Cas and Cascade are Type IE Cas and
Cascade proteins.
[000152] Optionally, the Cas and Cascade are Type IF Cas and
Cascade proteins.
10001531 Optionally, the Cas and Cascade are Type 1U Cas and
Cascade proteins.
[000154] Optionally, the Cas and Cascade are E coil (optionally
Type IE or IF) Cas and
Cascade proteins, optionally wherein the E coil is ESBL-producing E. coil or E
coil ST13 1-025b:H4.
[000155] Optionally, the Cas and Cascade are Clostridium (eg, C
dificile) Cas and Cascade
proteins, optionally C dificile resistant to one or more antibiotics selected
from aminoglycosides,
lincomycin, tetracyclines, erythromycin, clindamycin, penicillins,
cephalosporins and
fluoroquinolones.
10001561 Optionally, the Cas and Cascade are Pseudomonas
aeruginosa Cas and Cascade
proteins, optionally P aeruginosa resistant to one or more antibiotics
selected from carbapenems,
aminoglycosides, cefepime, ceftazidime, fluoroquinolones, piperacillin and
tazobactam.
[000157] Optionally, the Cas and Cascade are Klebsiella pneumoniae
(eg, carbapenem-resistant
Klebsiella pneumoniae or Extended-Spectrum Beta-Lactamase (ESBL)-producing K
pneumoniae)
Cas and Cascade proteins.
[000158] Optionally, the Cas and Cascade are E coli, C difficile,
P aeruginosa, K pneumoniae,
P furiosus or B halodurans Cos and Cascade proteins.
10001591 Optionally, each crRNAs or gRNAs comprises a spacer
sequence that is capable of
hybridising to a protospacer nucleotide sequence of the cell, wherein the
protospacer sequence is
adjacent a PAM, the PAM being cognate to the CI or C2, wherein CI or C2 is a
Cas nuclease, eg, a
Cas3. Thus, the spacer hybridises to the protospacer to guide the Cas3 to the
protospacer. Optionally,
the Cas3 cuts the protospacer, eg, using exo- and/or endonuclease activity of
the Cas3. Optionally,
the Cas3 removes a plurality (cg, at least 2, 3,4, 5, 6, 7, 8, 9 or 10)
nucleotides from the protospacer.
33
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
[000160] Optionally, the vector is a phage or non-replicative
transduction particle. The phage
or particles comprise phage coat proteins encapsidating DNA, wherein the DNA
comprises the vector.
Suitable examples of phage and particles are disclosed in U52019/0160120 the
disclosures of which
are incorporated herein by reference for possible use in the invention and for
providing one or more
features that may be included in gthe claims herein. Phage or particle is
capable of infecting the cell,
thereby introducing the vector into the cell.
[000161] It will be understood that particular embodiments
described herein are shown by way
of illustration and not as limitations of the invention. The principal
features of this invention can be
employed in various embodiments without departing from the scope of the
invention. Those skilled in
the art will recognize, or be able to ascertain using no more than routine
study, numerous equivalents
to the specific procedures described herein. Such equivalents are considered
to be within the scope of
this invention and are covered by the claims. All publications and patent
applications mentioned in the
specification are indicative of the level of skill of those skilled in the art
to which this invention
pertains. All publications and patent applications and all US equivalent
patent applications and patents
are herein incorporated by reference to the same extent as if each individual
publication or patent
application was specifically and individually indicated to be incorporated by
reference. Reference is
made to the publications mentioned herein and equivalent publications by the
US Patent and
Trademark Office (USPTO) or WIPO, the disclosures of which are incorporated
herein by reference
for providing disclosure that may be used in the present invention and/or to
provide one or more
features (eg, of a vector) that may be included in one or more claims herein.
[000162] The use of the word "a" or "an" when used in conjunction
with the term "comprising"
in the claims and/or the specification may mean "one," but it is also
consistent with the meaning of
"one or more," "at least one," and "one or more than one." The use of the term
"or" in the claims is
used to mean "and/or" unless explicitly indicated to refer to alternatives
only or the alternatives are
mutually exclusive, although the disclosure supports a definition that refers
to only alternatives and
"and/or." Throughout this application, the term "about" is used to indicate
that a value includes the
inherent variation of error for the device, the method being employed to
determine the value, or the
variation that exists among the study subjects.
[000163] As used in this specification and claim(s), the words
"comprising" (and any form of
comprising, such as "comprise" and "comprises"), "having" (and any form of
having, such as "have"
and "has"), "including" (and any form of including, such as "includes" and
"include") or "containing"
(and any form of containing, such as "contains" and "contain") are inclusive
or open-ended and do not
exclude additional, unrecited elements or method steps.
[000164] The term "or combinations thereof' or similar as used
herein refers to all
permutations and combinations of the listed items preceding the term. For
example, "A, B, C, or
combinations thereof is intended to include at least one of: A, B, C, AB, AC,
BC, or ABC, and if
order is important in a particular context, also BA, CA, CB, CBA, BCA, ACB,
BAC, or CAB.
34
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
Continuing with this example, expressly included are combinations that contain
repeats of one or
more item or term, such as BB, AAA, MB, BBC, AAABCCCC, CBBAAA, CABABB, and so
forth.
The skilled artisan will understand that typically there is no limit on the
number of items or terms in
any combination, unless otherwise apparent from the context.
[000165] Any part of this disclosure may be read in combination
with any other part of the
disclosure, unless otherwise apparent from the context.
[000166] All of the compositions and/or methods disclosed and
claimed herein can be made
and executed without undue experimentation in light of the present disclosure.
While the
compositions and methods of this invention have been described in terms of
preferred embodiments, it
will be apparent to those of skill in the art that variations may be applied
to the compositions and/or
methods and in the steps or in the sequence of steps of the method described
herein without departing
from the concept, spirit and scope of the invention. All such similar
substitutes and modifications
apparent to those skilled in the art are deemed to be within the spirit, scope
and concept of the
invention as defined by the appended claims.
[000167] The present invention is described in more detail in the
following non-limiting
Examples.
EXAMPLES
EXAMPLE 1. Combination of Type I CRISPR-Cas systems to synergistically target
multiple
genomic protospacers
[000168] A plasmid (which we call a CRISPR Guided Vector, CGV) was
constructed
comprising an operon with nucleotide sequences encoding a type 1 Cas3 and
Cascade proteins under
the control of a constitutive promoter. E. coli type IE Cas3 and Cascade was
used. A cognate CRISPR
array comprising E. coli direct repeat sequences and spacers for targeting an
E. coli host cell
chromosome was also cloned in the vector. An adaptation module containing Casl
and Cas2 was
omitted in the vector (see Figure 1A).
[000169] A plasmid was constructed comprising an operon with
nucleotide sequences encoding
a type I Cas3 and Cascade proteins under the control of constitutive
promoters. C. difficile type IB
Cas3 and Cascade was used. An adaptation module containing Casl, Cas2 and Cas4
was omitted in
the vector (see Figure 1B). A cognate CRISPR array comprising C. difficile
repeat sequences and
spacers for targeting an E. coli host cell chromosome was cloned in a second
vector, under the control
of constitutive promoters (see Figure 1B).
[000170] The CGV encoding C. difficile type TB Cas3 and Cascade
was transformed into E.
coli MG1655. Subsequently, the vector encoding C. difficile array and the CGV
encoding E. coli type
IE CRISPR-Cas system were transformed into the cells. CFU assays arc shown in
Figure 2. In Figure
2 there is shown CRISPR killing of -target strain E. coli MG1655 by C.
difficile CRISPR-Cas system
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
in combination with E. coli CRISPR-Cas system. Killing of essentially 100% of
the population was
achieved when combining both systems (a bit more than 7- logioreduction in
viable cells of E. coli
MG1655). However, transformation of E. coli CRISPR-Cas system alone resulted
in ¨4-logio
reduction in the bacterial population, and transformation of C. difficile
array in a bit less than 3-logio
reduction. These results indicate that E. coil CRISPR-Cas system and C.
difficile CRISPR-Cas system
are compatible, and their combination synergistically improves the killing
efficiency of target strain
greatly, hampering the growth of escapers.
Materials and methods
[000171] E. coli MG1655 was grown in lysogeny broth (LB) with
shaking (250 rpm) at 37 C.
When necessary, cultures were supplemented with tetracycline (10 m/mL),
kanamycin (50 Kg/mL),
and spectinomycin (100 lig/mL).
[000172] To construct a plasmid containing E. coli CRISPR-Cas
system under a constitutive
promoter, cas3, cas8e, cash, cas7, cas5, and cas6 genes from E. coli were
amplified and cloned in a
ColEl-type plasmid, pZE21 (Lutz and Bujard, 1997. Nucleic Acids Research, 25,
1203-1210) under
the control of a promoter. cas3 was located in the beginning of the operon
followed by cas8e, cash,
cas7, cas5, and cas6. The adaptation module (consisting of earl and cas2) was
omitted in the vector.
Additionally, a 3-spacer array targeting 3 chromosomal intergenic regions in
E. coli MG1655 was
included in the CGV under the control of a promoter. It contained 32
nucleotides from the genome of
E. coli MG1655 per target locus (TGATTGACGGCTACGGTAAACCGGCAACGTTC;
GCTGTTAACGTACGTACCGCGCCGCATCCGGC; and
CGGACTTAGTGCCAAAACATGGCATCGAAATT) separated by 29 bp direct repeats (each
repeat
was GAGTTCCCCGCGCCAGCGGGGATAAACCG). Additionally, the 3'-AAG protospacer
adjacent motif (PAM) is located adjacent to the selected target sequences in
the genome of E. coli
MG1655 (Figure 1A).
[000173] C. difficile CRISPR-Cas system was constructed in a two-
plasmid system. To
construct a plasmid containing C. difficile cas genes, cas3, cas6, cas8b,
cas7, and cas5 genes from C.
difficile were amplified and cloned in a pSC101 backbonep under the control of
a promoter. The
cas3 was located in the beginning of the operon followed by cas6, cas8b, cas7,
and cas5. The
adaptation module (consisting of casl, cas2, and cas4) was omitted in the
vector (Figure 1B). A
second plasmid containing a 5-spacer array was cloned in a CloDF13 on backbone
under the control
of a promoter J23100. It contained 37 nucleotides from the genome of E. coli
MG1655 per target
locus (GCCATAATCTGGATCAGGAAGTCTTCCTTATCCATAT;
GGCTTTACGCCAGCGACGTATTGCCACAGGAATAACT;
GGGGATAGCGCGCCTGGAGCGTGCGATAGAGACTTTG;
GGCATTTACCGACCAGCCCATCAGCAGTACAGCAAAC; and
36
CA 03171200 2022- 9-9
WO 2021/239758
PCT/EP2021/063954
TCCTGAATCAAATCCGCCTGTGGCAGGCCATAGCCCG) separated by 29 bp direct repeats
(each repeat was GTTTTATATTAACTAAGTGGTATGTAAAT). Additionally, the 3'-CCT
protospacer adjacent motif (PAM) is located adjacent to the selected target
sequences in the genome
of E. coil MG1655 (Figure 1B).
[000174] To perform killing assays, the plasmid harboring cas3 and
cascade genes of C.
difficile was transformed into E. coli MG1655 by electroporation.
Transformants were grown in liquid
LB with the antibiotic to mid-log phase, and further electroporated with a
plasmid harboring C.
difficile array and a plasmid with E. coil CRISPR-Cas system. Controls with
empty vectors, and with
each CGV separately were performed. Killing efficiency was determined by
plating the
transformations onto LB with antibiotics. Viability was calculated by counting
colony forming units
(CFUs) on the plates and data were calculated as viable cell concentration
(CFU/ml).
37
CA 03171200 2022- 9-9
9
a
-4
.
00
8
'.'
P
. TABLE 1: Example Bacteria
Optionally, the cell or cells are cell(s) of a genus or species selected from
this Table. 0
t..)
Abiotrophia Acidocella Actinomyces
Alkalilimnicola Aquaspirillum is.)
,-,
,
k..)
Abiotrophia defectiva Acidocella aminolytica
Actinomyces bovis Alkalilimnicola ehrlichii Aquaspirillum polyntorphum
-.1
Acaricomes Acidocella facilis Actinomyces denticolens
Alkaliphilus Aquaspirillum x
Acaricomes phytoseiuli Acidomonas Actinomyces europaetts
Alkaliphilus oremlandii putridiconchyliwn
Acetitomaculum Acidomonas methanolica
Actinomyces georgiae Alkaliphilus transvaalensis Aquaspirillum serpens
Acetitomaculum ruminis Acidothermus Actinomyces gerencseriae
Allochromatium Aquimarina
Acetivibrio Acidothermus cellulolyticus Actinomyces
Allochromatium vinosum Aquimarina latercula
Acetivibrio cellulolyficus Acidovorax horcleovulneris
Alloiococcus Arcanobacterium
Acetivibrio ethanolgignens Acidovorax anthurii Actinomyces howellii
Alloiococcus otitis Arcanobacterium
Acetivibrio multivorans Acidovorax caeni Actinomyces hyovaginalis
Allokutzneria haemolyticum ci.)
oe
Acetoanaerobium Acidovorax cattle yae Actinomyces israelii
Allokutzneria albata Arcanobacterittm pyo genes
Acetoanaerobium note rae Acidovorax citrulli Actinomyces johnsonii
Altererythrobacter Archangium
Acetobacter Acidovorax defluvii Actinomyces meyeri
Altererythrobacter Archangium gephyra
Acetobacter aceti Acidovorax delafieldii
Actinomyces naeslundii ishigakiensis Arcobacter
Acetobacter cerevisiae Acidovorax facilis Actinomyces neuii
Altermonas Arcobacter butzleri
Acetobacter cibinongensis Acidovorax konjaci Actinomyces odontolyticus
Altennonas haloplanktis Arcobacter cryaerophilus
Acetobacter estunensis Acidovorax temperans Actinomyces oris
Altennonas macleodii Arcobacter halophilus od
r)
Acetobacter fabarum Acidovorax valerianellae
Actinomyces radingae Alysiella Arcobacter nitrofigilis
....1
t=i
ot
Acetobacter ghanensis Acinetobacter Actinomyces slackii
Alysiella crassa Arcobacter skirrowii t..)
o
w
1-,
Acetobacter indonesiensis Acinetobacter baumannii
Actinomyces turicensis Alysiella filifortnis Arhodomonas -6-
c,
w
Acetobacter lovaniensis Acinetobacter baylyi Actinomyces viscosus
Arhodomonas aquaeolei
ul
.6.
9
a
-4
.
.
8
,..,
P
Lo
Acetobacter motor= Acinetobacter bouvetii
Actinoplanes Aminobacter Arsenophonus
Acetobacter nitrogenifigens Acinetobacter calcoaceficus Actinoplanes
auranticolor Aminobacter aganoensis Arsenophonus nasoniae 0
t..)
o
Acetobacter oeni Acinetobacter gemeri Actinoplanes
brasiliensis Aminobacter aminovorans
ts.)
1¨
,
k..)
Acetobacter orientalis Acinetobacter haemolyticus Actinoplanes
consettensis Aminobacter niigataensis Arthrobacter w
-.1
Acetobacter orleanensis Acinetobacter johnsonii
Actinoplanes deccanensis Aminobacterium Arthrobacter
agilis w
Acetobacter pasteuri anus Acinetobacter junii Actinoplanes denventensis
Aminobacterium mobile Arthrobacter albus
Acetobacter pomorum Acinetobacter lwoffi Actinoplanes digitatis
Aminomonas Arthrobacter aurescens
Acetobacter senegalensis Acinetobacter parvus Actinoplanes durhamensis
Aminomonas paucivorans Arthrobacter
Acetobacter xylinus Acinetobacter radioresistens Actinoplanes
ferrugineus Ammoniphilus chloroplienolicus
Acetobacterium Acinetobacter schindleri
Actinoplanes globisporus Ammoniphilus oxalaticus Arthrobacter citreus
Acetobacterium bakii Acinetobacter soli Actinoplanes humidus
Ammoniphilus oxalivorans Arthrobacter crystallopoietes
Acetobacterium carbinolicum Acinetobacter tancloii
Actinoplanes italicus Amphibacillus Arthrobacter cumminsii w
v:
Acetobacterium dehalogenans Acinetobacter tjembergiae
Actinoplanes liguriensis Arnphibacillus xylanus Arthrobacter globiformis
Acetobacterium fimetarium Acinetobacter towneri Actinoplanes lobatus
Amphritea Arthrobacter
Acetobacterium malicum Acinetobacter ursingii
Actinoplanes missouriensis Amphritea balenae
histidinolovorans
Acetobacterium paludosum Acinetobacter veneti anus
Actinoplanes palleronii Amphritea japonica Arthrobacter ilicis
Acetobacterium tundrae Acrocarpospora Actinoplanes philippinensis
Amycolatopsis Arthrobacter luteus
Acetobacterium wieringae Acrocarpospora corrugata
Actinoplanes rectilineatus Amycolatopsis alba Arthrobacter
methylotrophus
Acetobacterium woodii Acrocarpospora Actinoplanes regularis
Amycolatopsis albidoflavus Arthrobacter mysorens od
r)
Acetofilamentum macrocephala Actinoplanes
Amycolatopsis azurea Arthrobacter nicotianae ....1
t=i
ot
Acetofitamentum rigidum Acrocarpospora teichomyceticus
Amycolatopsis coloradensis Arthrobacter nicotinovorans
t..)
o
w
1-,
Acetohalobium pleiomorpha Actinoplanes utahensis
Amycolatopsis lurida Arthrobacter oxydans -O-
w
Acetohalobium arabaticum
Amycolatopsis mediterranei Arthrobacter pascens v:
ul
.6.
9
a
-4
.
.
8
,..,
P
Lo
Acetomicrobium Actibacter Actinopolyspora
Amycolatopsis rifamycinica Arthrobacter
Acetoinicrobium faecale Actibacter sediminis Actinopolyspora
halophila Amycolatopsis rubido phenanthrenivorans 0
t..)
o
Acetomicrobium flovidum Actinoalloteichus Actinopolyspora
Amycolatopsis sulphurea Arthrobacter ts.)
1.-
,
k..)
Acetonema Actinoalloteichus 171Ortivallis
Amycolatopsis tolypomycina polychromogenes w
-.1
Acetonema longum cyanogriseus Actinosynnema
Anabaena Atrhrobacter protophormiae x
Acetothermus Actinoalloteichus Actinosynnema mirum
Anctbaena cylindrica Arthrobacter
Acetothermtts paticivorans hymeniacidonis Actinotalea
Anctbaena flos -aquae psychrolactophilus
Acholeplasma Actinoalloteichus spitiensis
Actinotalea fermentans Anabctena variabilis Arthrobacter ramosus
Acholeplasma axanthum Actinobaccillus Aerococcus
Anaeroarcus Arthrobacter sulfonivorans
Acholeplasma brassicae Actinobacillus capsulatus
Aerococcus sanguinicola Anaeroarcus burkinensis Arthrobacter sulfureus
Acholeplasma cavigenitalium Actinobacillus delphinicola Aerococcus urinae
Anaerobaculum Arthrobacter uratoxydans
Acholeplasma equiletale Actinobacillus hoininis
Aerococcus urinaeequi Ancterobaculum mobile Arthrobacter ureafaciens
4,
o
Acholeplasma granulartun Actinobacillus indolicus
Aerococcus urinaehominis Anaerobiospirillum Arthrobacter
viscosus
Acholeplasma hippikon Actinobacillus lignieresii
Aerococcus viridans Ancterobiospirillum Arthrobacter woluwensis
Acholeplasma laidlawii Actinobacillus minor Aeromicrobium
succiniciproducens Asaia
Acholeplasma modicum Actinobacillus muris Aeromicrobium erythrewn
Ancterobiospirillum thomasii Asaia bogorensis
Acholeplasma morum Actinobacillus Aeromonas
Anaerococcus Asanoa
Acholeplasma multilocale pleuropneumoniae Aeromonas
Anaerococcus hydrogenalis Asanoa ferruginea
Acholeplasma oculi Actinobacillus porcinus
allosaccharophila Anaerococcus lactolyticus Asticcacaulis It
r)
Acholeplasma palmae Actinobacillus rossii Aeromonas bestiarum
Anaerococcus prevotii Asticcacaulis biprosthecium ....1
t=i
ot
t..)
Acholeplasma parvum Actinobacillus scotiae
Aeromonas caviae Anaerococcus tetradius Asticcacaulis excentricus
o
w
1-,
Acholeplasma pleciae Actinobacillus seminis
Aeromonas encheleia Anaerococcus vaginalis Atopobacter -6-
c,
w
Acholeplasma vituli Actinobacillus succinogenes Aeromonas
Atopobacter phocae
ul
4-
9
a
-4
.
.
8
,..,
P
. Achromobacter Actinobaccillus suis enteropelo genes
Anaerofustis Atopobium
Achromobacter denitrift cans Actinobacillus ureae
Aeromonas eucrenophila Ancterofustis stercorihomints Atopobiwn fossor
0
t..)
o
Achromobacter insolitus Actinobaculum Aeromonas ichthiosmia
Anaeromusa Atopobium minutum ts.)
1.-
,
k..)
w
Achroznobacter piechaudii Actinobaculum massiliense
Aeromonas jandaei Ancteromusa acidaminophila Atopobiwn parvulwn
-.1
Achromobacter ruhlandii Actinobaculum schaalii
Aeromonas media Anaeromyxobacter Atopobium rimae
Achromobacter spanius Actinobaculum suis Aeromonas pop offii
Ancteromyxobacter Atopobiwn vaginae
Acidaminobacter Actinonzyces urinate Aeromonas sobria
dehalogenans Aureobacterium
Acidaminobacter Actinocatenispora Aeromonas veronii
Anaerorhabdus Aureobacterium barkeri
hydrogenoformans Actinocatenispora rupis
Agrobacterium Ancterorhabdus furcosa Aurobacterium
Acidaminococcus Actinocatenispora Agrobacterium
Anaerosinus Aurobacterium liquefaciens
Acidaminococcus fermen tans thailandica gelatinovo rum
Ancterosinus glycerini Avibacterium
Acidaminococcus intestini Actinocatenispora sera
Agrococcus Anaerovirgula Avibacterium avium 4,
1-,
Acidicaldus Actinocorallia Agrococcus citreus
Anaerovirgula multivorans Avibacterium gallinarum
Acidicaldus organivorans Actinocorallia aurantiaca
Agrococcus jenensis Ancalomicrobium Avibacterium paragallinarum
Acidimicrobium Actinocorallia aurea Agromonas
Ancalomicrobium adetum Avibacterium volantium
Acidimicrobium ferrooxidans Actinocorallia cavernae
Agromonas oligotrophica Ancylobacter Azoarcus
Acidiphilium Actinocorallia glomerata
Agromyces Ancylobacter aquaticus Azoarcus indigens
Acidiphilium acidophilum Actinocorallia herbida
Agromyces fucosus Aneurinibacillus Azoarcus tolulyticus
Acidiphilium angustum Actinocorallia libanotica
Agromyces hippuratus Aneurinibacillus Azoarcus toluvorans od
r)
Acidiphilium crypt= Actinocorallia longicatena
Agromyces luteolus aneurinilyticus Azohydromonas ....1
t=i
ot
Acidiphilium multivorum Actinomadura Agromyces mediolanus
Aneurinibacillus migulanus Azohydromonas attstralica t..)
o
w
1-,
Acidiphilium organovorwn Actinomadttra alba Agromyces ramosus
Aneurinibacillus Azohydromonas lata -O-
w
Acidiphilium rubrum Actinomadura atramentaria Agromyces rhizospherae
thermoaerophilus v:
ul
4-
9
a
-4
.
.
8
,..,
P
. Acidisoma Actinomadura Akkermansia
Angiococcus Azomonas
Acidisoma sibiricum bangladeshensis Akkermansia nzuciniphila
Angiococcus discifonnis Azomonas agilis 0
t..)
Acidisoma tundrae Actinomadura catellafispora Albidiferax
Angulomicrobium Azomonas insignis ts.)
1.-
,
k..)
Acidisphaera Actinomadura chibensis
Albidiferax ferriredttcens Angtdomicrobium tetraedrale
Azomonas nzacrocytogenes w
-.1
Acidisphaera rttbrifaciens Actinomadura chokoriensis Albidovulum
Anoxybacillus Azorhizobium x
Acidithiobacillus Actinomadura citrea Albidovulum inexpectatum
Anoxybctcillus pushchinoensis Azorhizobium caulinodans
Acidithiobacillus albertensis Actinomadura coendea
Alcaligenes Aquabacterium Azorhizophilus
Acidithiobacillus caldus Actinomadura echinospora
Alculigenes denitrificans Aquabacterium commune Azorhizophilus paspuli
Acidithiobacillus ferrooxidans Actinomadura fibrosa Alcaligenes faecalis
Aquabacterium parvum Azospirillum
Acidithiobacillus thiooxidans Ac finomadura formosensis
Alcanivorax Azospirdlum brasdense
Acidobacterium Actinomadura hibisca Alcanivorax borkumensis
Azospirdlum halopraeferens
Acidobacterium capsulatum Actinomadura kijaniata
Alcanivorax jadensis Azospirdlum irakense 4,
t.)
Actinomadura lafina Algicola
Azotobacter
Actinomadura livida Algicola bacteriolytica
Azotobacter beijerinckii
Actinomadura Alicyclobacillus
Azotobacter chroococcum
luteofluorescens Alicyclobacillus
Azotobacter nigri cans
Actinomadura macra disulfidooxidans
Azotobacter salinestris
Actinomadura madurae Alicyclobacillus
Azotobacter vinelandii
Actinomadura oligospora sendaiensis
od
r)
Actinomadura pelletieri Alicyclobacillus vulcanalis
....1
t=i
ot
Actinomadura rubrobrunea Alishewanella
t..)
o
w
1-,
Actinomadura rugatobispora Alishewanella fetalis
-6-
w
Actinomadura umbrina
v:
ul
4-
.0
Actinomadara Alkalibacillus
verrucosospora Alkalibacillus
Actinomadttra vinacea haloalkaliphilus
ts.)
Actinonzadara viridilutea
rJI
Actinomadttra viridis
Actinomadttra yumaensis
Bacillus Bacteroides Bibersteinia
Borrelia Brevinema
[see below] Bacteroides caccae Bibersteinia trehalosi
Borrelia afzelii Brevinema andersonii
Bacteroides coagulans Bifidobacterium
Borrelia americana Brevundimonas
Bacteriovorax Bacteroides eggerthii Bifidobacterium
adolescentis Borrelia burgdorferi Brevundimonas alba
Bactenovorax stolpii Bacteroides fragilis Bifidobacterium angulatum
Borrelia carolinensis Brevundimonas aurantiaca
Bacteroides galacturonicus Bifidobacterium animalis
Borrelia coriaceae Brevundimonas diminuta
Bacteroides helco genes Bifidobacteriutn asteroides
Borrelia garinii Brevundimonas intermedia
Bacteroides ovatus Bifidobacterium bifidum
Borrelia japonica Brevundimonas subvibrio ides
Bacteroides pectinophilus Bifidobacterium bourn
Bosea Brevundimonas vancanneytii
Bacteroides pyo genes Bifidobacterium breve
Bosea minatitlanensis Brevundimonas variabilis
Bacteroides salyersiae Bifidobacteritun catenulatum
Bosea thiooxidans Brevundimonas vesicularis
Bacteroides stercoris Bifidobacterium choerinum
Brachybacterium Brochothrix
Bacteroides suis Bifidobacterium corynefonne
Brachybacterium Brochothrix campestris
Bacteroides tectus Bifidobacteritun cuniculi
alimentarium Brochothrix thermosphacta
Bacteroides thetaiotaomicron Bifidobacterium dentium
Brachybacteriwn faeclum
9
a
-4
.
00
8
'.'
P
Lo
Bacteroides uniformis Bifidobacteriutn gallicutn
Brachybacterium Brucella
Bacteroides ureolyticus Bifidobacterium gallinartun
paraconglomeratum Brucella cards 0
t..)
o
Bacteroides vulgatus Bifidobacterium indicum
Brachybacterium rhamnosum Brucella
neotomae ts.)
1..,
-,
k..)
Balnearium Bifidobacterium longum
Brachybacterium Bryobacter w
-.1
Balneariufn lithotrophicum Bifidobacterium
tyrofermentans Bryobacter aggregatus x
Balneatrix magnumBifidobacterium
Brachyspira Burkholderia
Balneatrix alp ica rnerycicum
Brachyspira alvinipulli Burkholderia ambifaria
Balneola Bifidobucterium minimum
Brachyspira hyodysenteriae Burkholderia andropogonis
Balneola vulgaris Bifidobacterium
Brachyspira innocens Burkholderia anthina
Barnesiella pseualocatentdatunt
Brachyspira murdochii Burkholderia adedonica
Barnesiella viscericola Bifidobacterium
Brachyspira pilosicoli Burkholderia caryophylli
Bartonella pseudolongum
Burkholderia cenocepacia 4,
4,
Bartonella alsatica Bifidobacterhtm pullorum
Bradyrhizobium Burkholderia cepacia
Bartonella bacillifonnis Bifidobacterium ruminantium
Bradyrhizobium canariense Burkholderia cocovenenans
Bartonella clarridgeiae Bifidobacterium saeculare
Bradyrhizobium elkanii Burkholderia dolosa
Bartonella doshiae Bifidobacterium subtile
Bradyrhizobium japonicum Burkholderia fungortun
Bartonella elizabethae Bifidobacterium
Bradyrhizobium liaoningense Burkholderia glathei
Bartonella grahamii thermophilum
Brenneria Burkholderia glumae
Bartonella henselae Bilophila
Brenneria alni Burkholderia graminis od
r)
Bartonella rochalimae Bilophila wadsworthia
Brenneria nigrifluens Burkholderia kururiensis ....1
t=i
ot
Bartonella vinsonii Biostraticola
Brenneria quercina Burkholderia multivorans t..)
o
1..,
Bavariicoccus Biostraticola tofi
Brenneria quercina Burkholderia phenazinium -O-
w
Bavariicoccus seileri
Brenneria salicis Burkholderia plantarii
fil
4.
9
a
-4
.
' '6-
.
8
,..,
P
Lo
Bdellovibrio Bizionia
Brevibacillus Burkholderia pyrrocinia
Bdellovibrio bacteriovorus Bizionia argentinensis
Brevibacillus agri Burkholderia silvatlantica 0
t..)
o
Bdellovibrio exovorus Blastobacter
Brevibacillus borstelensis Burkholderia stabilis ts.)
0-
,
k..)
Beggiatoa Blastobacter capsulatus
Brevibacillus brevis Burkholderia thailandensis w
-.1
Beggiatoa alba Blastobacter denitrificans
Brevibacillus centrosportts Burkholderia tropica oo
Beijerinckia Blastococcus
Brevibacillus choshinensis Burkholderia unamae
Beijerinckia derxii Blastococcus aggregatus
Brevibacillus invocatus Burkholderia vietnamiensis
Beijerinckia fluminensis Blastococcus suxobsidens
Brevibucillus luterosporus Buttiauxella
Beijerinckia indica Blastochloris
Brevibacillus parabrevis Buttiauxella agrestis
Beijerinckia mobilis Blastochloris viridis
Brevibacillus reuszeri Buttiauxella brennerae
Belliella Blastomonas
Brevibacterium Buttiauxella ferragutiae
Belliella baltica Blastornonas natatoria
Brevibacterium abidum Buttiauxella gaviniae 4,
Bellilinea Blastopirellula
Brevibacterium album Buttiauxella izardii
Bellilinea caldifistulae Blastopirellula marina
Brevibacterium aurantiacum Buttiauxella noackiae
Belnapia Blautia
Brevibacterium celere Buttiauxella wannboldiae
Belnapia moabensis Blautia coccoides
Brevibacterium epidermidis Butyrivibrio
Bergeriella Blautia hansenii
Brevibacterium Butyrivibrio fibrisolvens
Bergeriella denitrificans Blautia producta
frigoritolerans Butyrivibrio hungatei
Beutenbergia Blautia wexlerae
Brevibacterium halotolerans Butyrivibrio proteoclasticus od
r)
Beutenbergia cavernae Bogoriella
Brevibacterium iodinum ....1
t=i
It
Bogoriella caseilytica
Brevibacterium linens t..)
o
w
1-,
Bordetella
Brevibacterium lyticum -O-
w
Bordetella avi urn
Brevibacterium mcbrellneri
ul
4-
9
a
,-.
6-''
.
8
,..,
P
Lo
Bordetella bronchiseptica
Brevibacterium otitidis
Bordetella hinzii
Brevibacterium oxydans 0
t..)
o
Bordetella holmesii
Brevibacterium paucivorans ts.)
1.-
,
k..)
w
Bordetella parapertussis
Brevibacterium stationis
-.1
Bordetella pertussis
oo
Bordetella petrii
Bordetella trematum
Bacillus
B. acidiceler B. cuninovorans B. glucanolyticus
B. taeanensis B. lautus
B. acidicola B. cunylolyticus B. gordonae
B. tequilensis B. lehensis
4,
o
B. acidiproducens B. andreesenii B. gottheilii
B. the nnantarcticus B. lentimorbus
B. acidocaldarius B. aneurinilyticus B. graminis
B. thennoaerophilus B. lentus
B. acidoterrestris B. anthracis B. halmapalus
B. the nnoamylovorans B. lichen iformis
B. aeolius B. aquimaris B. haloalkaliphilus
B. thennocatenulatus B. ligniniphilus
B. aerius B. arenosi B. halochares
B. thennocloacae B. litoralis
B. aerophilus B. arseniciselenatis B. halodenitrifi cans
B. thennocopriae B. locisalis
B. agaradhaerens B. arsenicus B. halodurctns
B. thennodenitrificans B. luciferensis
od
B. agri B. aurantiacus B. halophilus
B. thennoglucosidasius B. luteolus r)
....1
t=i
B. aidingensis B. arvi B. halosaccharovorans
B. thennolactis B. luteus ot
t..)
o
B. akibai B. cuyabhattai B. hemicellulosilyticus
B. the nnoleovorans B. macauensis w
1-,
-O-
B. alcalophilus B. asahii B. hemicentroti
B. thennophilus B. macerans
w
ul
4-
9
a
,-.
6-''
.
8
,..,
P
Lo
B. algicola B. atrophaeus B. herbersteinensis
B. thennontber B. rnacquariensis
B. alginolyticus B. axarquiensis B. horikoshii
B. thennosphaericus B. rnacyae 0
t..)
o
B. alkalidiazotrophicus B. azotofixans B. horneckiae
B. thiaminolyticus B. malachensis ts.)
1.-
,
k..)
B. alkalinitrilicus B. azotofonnans B. horti
B. thioparans B. mannanilyticus w
-.1
B. alkalisediminis B. badius B. huizhouensis
B. thuringiensis B. marisflavi
B. alkalitelluris B. barbaricus B. humi
B. tianshenii B. marismortui
B. altitudinis B. bataviensis B. hwajinpoensis
B. trypoxylicola B. mannarensis
B. alveayuensis B. beijingensis B. idriensis
B. tusciae B. massiliensis
B. alvei B. benzoevorans B. indicus
B. validus B. megateriuin
B. amvloliquefaciens B. beringensis B. infantis
B. vallismortis B. mesonae
. B. B. berkeleyi
B. infernus B. vedderi B. methanolicus
a. subsp. amyloliquefaciens B. beveridgei B. insolitus
B. velezensis B. methylotrophicus 4,
-4
= B. a. subsp. plantanun B. bogoriensis
B. invictae B. vietnamensis B. migulanus
B. boroniphilus B. iranensis
B. vireti B. mojavensis
B. dipsosauri B. borstelensis B. isabeliae
B. vulcani B. mucilaginosus
B. drentensis B. brevis Migula B. isronensis
B. wakoensis B. =rails
B. edaphicus B. butanolivorans B. jeotg,ali
B. weihenstephanensis B. murimartini
B. ehimensis B. canaverahus B. kaustophilus
B. xiamenensis B. mvco ides
B. eiseniae B. carboniphilus B. kobensis
B. xiaoxiensis B. naganoensis od
r)
B. enclensis B. cecembensis B. kochii
B. zhanjiangensis B. nanhaiensis ....1
t=i
ot
B. endophyticus B. celhdosilyticus B. kokeshiiformis
B. peoriae B. nanhaiisediminis t..)
o
w
1-,
B. endoradicis B. centrosporus B. koreensis
B. persepolensis B. nealsonii -O-
w
B. farraginis B. cereus B. korlensis
B. persicus B. neidei
ul
4-
9
a
8
8
'.'
P
Lo
B. fastidiosus B. chagannorensis B. kribbensis
B. pervagus B. neizhouensis
B. fengqiuensis B. chitinolyticus B. krulwichiae
B. plakortidis B. niabensis 0
r..)
o
B. firmus B. chondroitinus B. laevolacticus
B. pocheonensis B. niacini ts.)
1.-
,
k..)
B. flexus B. choshinensis B. larvae
B. polvgoni B. novalis w
-.1
B. foraminis B. chungangensis B. laterosporus
B. polvmyxa B. oceanisediminis
B. fordii B. cibi B. salexigens
B. popilliae B. odysseyi
B. formosus B. cirettlans B. saliphilus
B. pseudalealophilus B. okhensis
B. fortis B. clarkii B. schlegelii
B. pseudofinnus B. okuhidensis
B. fumarioli B. clausii B. sediminis
B. pseudomycoides B. oleronius
B. fun iculus B. coagulans B. selenatarsenatis
B. psvchrodurans B. oryzaecorticis
B. fusiformis B. coahuilensis B. selenitireducens
B. psychrophilus B. oshimensis
B. galactophilus B. cohnii B. seohaeanensis
B. psvchrosaccharolyticus B. pabuli 4,
oe
B. galactosidilyticus B. composti B. shacheensis
B. psychrotolerans B. pakistctnensis
B. galliciensis B. curdlanolyticus B. shackletonii
B. pulvifaciens B. pallidus
B. gelatini B. cycloheptanicus B. siamensis
B. pumilus B. pallidus
B. gibsonii B. cytotoxicus B. silvestris
B. purgationiresistens B. panacisoli
B. ginsengi B. daliensis B. simplex
B. pycnus B. panaciterrae
B. ginsengihumi B. decisifrondis B. siralis
B. qingdaonensis B. pantothenticus
B. ginsengisoli B. decolorationis B. smithii
B. qingshengii B. parctbrevis od
r)
B. globisporus (eg, B. B. deserti B. soli
B. reuszeri B. paraflexus ....1
t=i
ot
g. subsp. Globisportts; or B. B. solimangrovi
B. rhizosphaerae B. pasteurii r..)
o
w
1-,
g. subsp. Marinus) B. solisalsi
B. rigui B. patagoniensis -O-
w
B. songklensis
B. runs
ul
4-
9
B. sonorensis
B. safensis
B. sphaericus
B. salarius
B. sporothermodurans
ts.)
B. stearothermophdus
B. stratosphericus
B. subterraneus
B. subtilis (eg, B.
s. subsp. Inaquosorum, or B.
s. subsp. Spizizeni, or B.
s. subsp. Subtilis)
Caenimonas Campylobacter Cardiobacterium
Catenuloplanes Curtobacterium
Caenhnonas koreensis Campylobacter coli Cardiobacterium hominis
Catenuloplanes atrovinosus Curtobacterium
Caldalkalibacillus Campylobacter concisus Carnimonas
Catenuloplanes castoneus albidum
Caldalkalibacillus uzonensis Campylobacter curvus Carnimonas
nigrificans Catenuloplanes crispus Curtobacterium citretts
Caldanaerobacter Campylobacter fetus Carnobacterium
Catenuloplanes indicus
Caldanaerobacter subterraneus Campylobacter gracilis Carnobacterium
Catenuloplanes japonicus
Caldanaerobius Campylobacter helveticus
alterfunditum Catenuloplanes nepalensis
Caldaraterobius fijiensis Campylobacter hominis Carnobacte riurn
dive rgetts Catenuloplanes niger
Caldanaerobius Campylobacter hyointestinatis Carnobacterium
funditum Chryseobacterium
polysaccharolyticus Campylobacter jejttni Carnobacterium
gallinarum Chtyseobacterium
Caldanaerobius zeae Campylobacter (art Carnobacterium
balustinum
Campylobacter mucosalis maltaromaticum
Caldanaerovirga Campylobacter rectus Carnobacterium mobile
Citrobacter
Caldanaerovirga acetigignens Campylobacter showae
Carnobacterium viridans C. amalonaticus
Caldicellulosiruptor Campylobacter sputorum Caryophanon C.
braakii
Caldicellulosiruptor bescii Campylobacter upsaliensis
Caryophanon latum C. diversus
Caldicellulosiruptor kristjanssonii Capnocytophaga
Caryophanon tenue C. fartneri
Caldicellulosiruptor owensensis Capnocytophaga canimorsus
Catellatospora C. freundii
Capnocytophaga cynodegmi Catellatospora citrea C.
gillenii
Capnocytophaga gingivalis Catellatospora C. koseri
Capnocytophaga granulosa methionotrophica C.
murliniae
Capnocytophaga haemolytica Catenococcus C.
pasteurii[11
Capnocytophaga ochracea Catenococcus thiocycli .. C.
rodentium
Capnocytophaga sputigena C.
sedlakii
C. werkmanii
C. youngae
Clostridium
(see below)
Coccochloris
Coccochloris elabens
Corynebacterium
Cmynebacterium flavescens
Cotynebacterium variabile
Clostridium
9
.0
Clostridium absonum, Clostridium aceticum, Clostridium acetireducens,
Clostridium ace tobutylicum, Clostridium acidisoli, Clostridium aciditolerans,
Clostridium acidurici, Clostridium aerotolerans, Clostridium aestuarii,
Clostridium akagii, Clostridium aldenense, Clostridium aldrichii, Clostridium
algidicami, Clostridium algidixylanolyticum, Clostridium algifaecis,
Clostridium algoriphilum, Clostridium alkalicellulosi, Clostridium
arninophilum, ts.)
Clostridium aminovalericum, Clostridium amygdalinum, Clostridium amylolyticum,
Clostridium arbusti, Clostridium arcticum, Clostridium argentinense,
Clostridium asparagifonne, Clostridium aurantibutyricum, Clostridium
autoethanogenum, Clostridium baratii, Clostridium barkeri, Clostridium
bartlettii,
Clostridium beijerinckii, Clostridium bifermentans, Clostridium bolteae,
Clostridium bornimense, Clostridium botulinttm, Clostridium bowmanii,
Clostridium
bryantii, Clostridium butyricum, Clostridium cadaveris, Clostridium caenicola,
Clostridium caminithermale, Clostridium carboxidivorans, Clostridium carnis,
Clostridium cavendishii, Clostridium celatum, Clostridium celerecrescens,
Clostridium cellobioparum, Clostridium cellulofermentans, Clostridium
cellulolyticum, Clostridium cellulosi, Clostridium cellulovorans, Clostridium
chartatabidum, Clostridium chauvoei, Clostridium chromiireducens, Clostridium
citron iae, Clostridium clariflavum, Clostridium clostridioforme, Clostridium
cocco ides, Clostridium cochlearium, Clostridium colletant, Clostridium
colicanis,
Clostridium colinum, Clostridium collagenovorans, Clostridium cylindrospo rum,
Clostridium difficile, Clostridium diolis, Clostridium disporicum,
Clostridium drakei, Clostridium durum, Clostridium estertheticum, Clostridium
estertheticum estertheticum, Clostridium estertheticum laramiense,
Clostridium fallax, Clostridium felsineum, Clostridium fervidum, Clostridium
fimetarium, Clostridium fonnicaceticurn, Clostridium fi-igidicarnis,
Clostridium
frigoris, Clostridium ganghwense, Clostridium gasigenes, Clostridium ghonii,
Clostridium glycolicum, Clostridium glycyrrhizinilyticum, Clostridium grantii,
Clostridium haemolyticum, Clostridium halophilum, Clostridium hastiforme,
Clostridium hathewayi, Clostridium herbivorans, Clostridium hiranonis,
Clostridium histolyticum, Clostridium homopropionicum, Clostridium huakuii,
Clostridium hungatei, Clostridium hydrogeniformans, Clostridium
hydroxybenzoicwn, Clostridium hylemonae, Clostridium jejuense, Clostridium
indolis, Clostridium innocuum, Clostridium intestinale, Clostridium
irregulare,
Clostridium isatidis, Clostridium josui, Clostridium kluyveri, Clostridium
lactatifermentans, Clostridium lacusflyxellense, Clostridium laramiense,
Clostridium
lavalense, Clostridium lentocelluin, Clostridium lentoputrescens, Clostridium
leptum, Clostridium limosum, Clostridium litorale, Clostridium lituseburense,
Clostridium ljungdahlii, Clostridium lortetii, Clostridium lundense,
Clostridium magnum, Clostridium malenominatum, Clostridium man genotii,
Clostridium
mayombei, Clostridium methoxybenzovorans, Clostridium methylpentosum,
Clostridium neopropionicum, Clostridium flexile, Clostridium nitrophenolicum,
Clostridium novyi, Clostridium oceanicwn, Clostridium orbiscindens,
Clostridium oroticum, Clostridium oxalicum, Clostridium papyrosolvens,
Clostridium
paradoxum, Clostridium paraperfringens (Alias: C. welchii), Clostridium
paraputrificum, Clostridium pascui, Clostridium pasteurianum, Clostridium
9
.0
peptidivorans, Clostridium perenne, Clostridium perfringens, Clostridium
pfennigii, Clostridium phytofermentans, Clostridium pilifonne, Clostridium
polysaccharolyticum, Clostridium populeti, Clostridium propionicum,
Clostridium proteoclasticum, Clostridium proteolyticum, Clostridium
psychrophilum,
Clostridium puniceum, Clostridium purinilyticum, Clostridium putrefaciens,
Clostridium putrificum, Clostridium quercicolum, Clostridium quinii,
Clostridium ,t2,
ramosum, Clostridium rectum, Clostridium roseum, Clostridium
saccharobutylicum, Clostridium saccharogumia, Clostridium sacchctrolvticum,
Clostridium
saccharoperbutylacetonicum, Clostridium sardiniense, Clostridium sartagoforme,
Clostridium scatolo genes, Clostridium schirmacherense, Clostridium
scindens, Clostridium septicum, Clostridium sordellii, Clostridium sphenoides,
Clostridium spiroforme, Clostridium sporo genes, Clostridium
sporosphaeroides, Clostridium stercorarium, Clostridium stercorarium
leptospartum, Clostridium stercorarium stercorarium, Clostridium stercorarium
thermolacticum, Clostridium sticklandii, Clostridium stratninisolvens,
Clostridium subtenninale, Clostridium sufflavum, Clostridium sttlfidigenes,
Clostridium
symbiosum, Clostridium tagluense, Clostridium tepidiprofundi, Clostridium
tennitidis, Clostridium tertium, Clostridium tetctni, Clostridium tetanomorp
hum,
Clostridium thermaceticum, Clostridium thennautotrophicum, Clostridium
thermoalcaliphilum, Clostridium thermobutyricum, Clostridium thermocellum,
Clostridium thermocopriae, Clostridittin thennohydrosulfuricum, Clostridium
thermolacticum, Clostridium thermopalinarium, Clostridium
thennopapyrolyticum, Clostridium thermosaccharolyticum, Clostridium
thermosuccinogenes, Clostridium thermosulfurigenes, Clostridium
thiosulfatireducens, Clostridium tyrobutyricum, Clostridium uliginosum,
Clostridium ultunense, Clostridium villosum, Clostridium vincentii,
Clostridium
viride, Clostridiwn xylanolyticwn, Clostridium xylanovorans
Dactylosporangium Deinococcus Delftia
Echinicola
Dactvlosporangium aurantiacum Deinococcus aeritts Delftia acidovorans
Echinicola pacifica
Dactvlosporangium fulvum Deinococcus apachensis Desulfovibrio
Echinicola vietnamensis
Dactvlosporangium matsuzakiense Deinococcus aqua ticus Desulfovibrio
desulfuricans
Dactvlosporangium roseum Deinococcus aquatilis Diplococcus
Dactvlosporangium thailandense Deinococcus caeni Diplococcus pneumoniae
Dactvlosporangium vinaceum Deinococcus radiodurans
Deinococcus radiophilus
Enterobacter Enterobacter kobei Faecalibacterium
Flavobacterium
E. aerogenes E. ludwigil Faecalibacterittm prausnitzii
Flavobacterium antarcticum
E. amnigenus E. mori Fangia
Flavobacterium aquatile ts.)
E. agglomerans E. nimipressuralis Fangio hongkongensis
Flavobacterium
E. arachidis E. olyzae Fastidiosipila
aquidurense
E. asburiae E. pulveris Fastidiosipila sanguinis
Flavobacterium balustinum
E. cancerogenous E. pyrinus Fusobacterium
Flavobacterium croceum
E. cloacae E. radicincitans Fttsobacterium nucleatum
Flavobacterium cucumis
E. cowanii E. taylorae
Flavobacterium
E. dissolvens E. turicensis daej eon
ense
E. gergoviae E. sakazakii Enterobacter soli
Flavobacteriwn defluvii
E. helveticus Enterococcus
Flavobacterium degerlachei
E. hormaechei Enterococcus durans
Flavobacterium
E. intermedius Enterococcus faecalis
denitrificans
Enterococcus faecium
Flavobacterium filum
Erwinia
Flavobacterium flevense
Erwinia hapontici
Flavobacterium frigidarium
Escherichia
Flavobacterium mizutaii
Escherichia coli
Flavobacterium
okeanokoites
9
a
-4
.
.
8
,..,
P
. Gaetbulibacter Haemophilus Ideonella
Janibacter
Gaetbulibacter saemankumensis Haemophilus aegyptius Ideonella
azotifigens Janibacter anophelis 0
t..)
o
Gallibacterium Haemophilus aphrophilus
Idiomarina Janibacter corallicola ts.)
1.-
,
k..)
Gallibacterium anatis Haemophilus felts Idiomarina abyssalis
Janibacter limosus w
-.1
Gallicola Haemophilus gallinarum Idiontarina baltica
Janibacter melonis
Gallicola barnesae Haemophilus haemolyticus
Idiomarina fontislapidosi Janibacter terrae
Garciella Haemophilus influenzae Idiomarina
loihiensis Jannaschia
Garciella nitratireducens Haemophilus paracuniculus
Idiomarina ramblicola Jannaschia cystaugens
Geobacillus Haemophilus parahaemolyticus Idiomarina
seosinensis Jannaschia helgolandensis
Geobacillus the rmoglucosidasius Haemophilus parainfluenzae
Idiomarina zobellii Jannaschia pohangensis
Geobacillus stearothermophilus Haemophilus Ignatzschineria
Jannaschia rubra
Geobacter paraphrohaemolytictts Ignatzschineria
larvae vi
4,
Geobacter bemidjiensis Haemophilus parasuis
Janthinobacterium
Geobacter bremensis Haemophilus pittmaniae Ignavigranum
Janthinobacterium
Geobacter chapellei Hafnia Ignavigranum ruoffiae
agaricidamnosum
Geobacter grbiciae Hafitia alvei llumatobacter
Janthinobacterium lividum
Geobacter hydrogenophilus Hahella Ilumatobacter fluminis
Jejuia
Geobacter lovleyi Hahella ganghwensis Ilyobacter
Jejuia pallidilutea
Geobacter metallireducens Halalkalibacillus Ilyobacter delafieldii
Jeotgalibacillus od
r)
Geobacter pelophilus Halalkalibacillus hcdophilus
Ilyobacter insuetus Jeotgalibacillus ....1
t=i
ot
Geobacter pickeringii Helicobacter Ilyobacter polytropus
alimentarius t..)
o
w
1-,
Geobacter sulfurreducens Helicobacter pylori Ilyobacter tartaricus
Jeotgalicoccus -6-
w
Jeotgalicoccus halotolerans
v:
ul
4-
Geodermatophilus
Geodermatophilus obscurus
Gluconacetobacter
Gluconacetobacter xylinus
Gordonia
Gordonia rubripertincta
Kaistia Labedella Listeria ivanovii
Micrococcus Nesterenkonia
Kaistia adipata Labedella gwakjiensis L. marthii
Micrococcus luteus Nesterenkonia ho labia
Kaistia soli Labrenzia L. monocyto genes
Micrococcus lylae Nocardia
Kangiella Labrenzia aggregata L. newyorkensis Moraxella
Nocardia argentinensis
Kangiella aquimarina Labrenzia alba L. riparia Moraxella
bovis Nocardia corallina
Kangiella koreensis Labrenzia alexandrii L. rocourtiae Moraxella
nonliquefaciens Nocardia
Labrenzia marina L. seeligeri Moraxella
osloensis otitidiscaviarum
Kerstersia Labrys L. weihenstephanensis
Nakamurella
Kerstersia gviorum Labrys methylaminiphilus L.
welshimeri Nakamurella multipartita
Kiloniella Labrys miyagiensis Listonella
Nannocystis
Kiloniella laminariae Labrys monachus Listonella anguillarum
Nannocystis pusilla
Klebsiella Labrys okinawensis Macrococcus
Natranaerobius
K. granulomatis Labrys portucalensis Macrococcus bovicus
Natranaerobius
K. oxytoca Marinobacter the
rmophilus
K. pneumoniae Lactobacillus Marinobacter algicola
Natranaerobius trueperi
9
K. terrigena [see below] Marinobacter brvozoorum
Naxibacter
K. variicola Laceyella
Marinobacter flavimaris Naxibacter alkalitolerans
Kluyver a Laceyella putido Meiothermus
Neisseria ts.)
Kluyvera ascorbata Lechevalieria Meiothennus ruber
Neisseria cinerea
Kocuria Lechevalieria aerocolonigenes Methylophilus
Neisseria denitrificans
Kocuria roasea Legionella Methylophilus
Neisseria gonorrhoeae
Kocuria varians [see below] methylotrophus
Neisseria lactamica
Kurthia Listeria Microbacterium
Neisseria mucosa
Kurthia zopfii L. aquatica Microbacterium
Neisseria sicca
L. booriae ammoniaphilum
Neisseria subflava
L. cornellensis Microbacterium
arborescens Neptunomonas
L. fleischmannii Microbacterium
liquefaciens Neptunomonas japonica
L. floridensis Microbacterium oxydans
L. grandensis
L. grayi
L. innocua
Lactobacillus
L. acetotolerans L. catenaformis
L. mall L. parakefiri L. sakei
L. acidifarinae L. ceti L. rnanihotivorans
L. paralimentarius L. sativarius
L. acidipiscis L. coleohominis L. mindensis
L. paraplantarum L. sanfranciscensis
L. acidophilus L. collinoides L. mucosae
L. pentosus L. satsumensis
9
a
-4
.
' '6-
.
8
,..,
P
. Lactobacillus agilis L. composti L. murinus
L. perolens L. secaliphilus
L. algidus L. conatvus L. nagelii
L. plantarum L. sharpeae 0
t..)
o
L. alimentarins L. coryniformis L. namurensis
L. pontis L. siliginis ts.)
1¨
,
k..)
L. amylolyticus L. crispatus L. nantensis
L. protectus L. spicheri w
-.1
L. amylophilus L. crustorum L. oligofennentans
L. psittaci L. suebicus
L. amylotrophicus L. curvatus L. oris
L. rennini L. thailandensis
L. amylovorus L. delbrueckii subsp. L. panis
L. reuteri L. ultunensis
L. animalis bulgaricus L. pantheris
L. rhamnosus L. vaccinostercus
L. antri L. delbrueckii subsp. L. parabrevis
L. rimae L. vaginalis
L. apodeini delbrueckii L. parabuchneri
L. rogosae L. versmoldensis
L. aviarius L. delbrueckii subsp. lactis
L. paracasei L. rossiae L. vini
L. bifennentans L. dextrinicus L. paracollino ides
L. ruminis L. vitulinus un
-4
L. brevis L. diolivorans L. parafarraginis
L. saerimneri L. zeae
L. buchneri L. equi L. homohiochii
L. jensenii L. zymae
L. camelliae L. equigenerosi L. iners
L. johnsonii L. gastricus
L. casei L. farraginis L. ingluviei
L. kalixensis L. ghanensis
L. kitasatonis L. farciminis L. intestinalis
L. kefiranofaciens L. graminis
L. kunkeei L. fermentum L. fuchuensis
L. kefiri L. hammesii
L. leichmannii L. fomicalis L. gallinarum
L. kimchli L. hamsteri od
r)
L. lindneri L. fructivorans L. gasseri
L. helvetictts L. harbinensis ....1
t=i
ot
L. malefermentans L. frumenti
L. hilgardii L. hayakitensis t..)
o
w
1-,
-O-
w
ul
.6.
.0
Legionella
Legionella adelaidensis Legionella drancourtit Candidatus
Legionella jconii Legionella quinlivanii
Legionella anisa Legionella dresdenensis
Legionella jordanis Legionella rowbothamii ts.)
Legionella beliardensis Legionella drozanskii Legionella
lansingensis Legionella rubrilucens
Legionella binninghamensis Legionella dumoffii Legionella
londiniensis Legionella sainthelensi
Legionella bozemanae Legionella erythra Legionella Ion gbeachae
Legionella santicrttcis
Legionella brunensis Legionella fairfieldensis
Legionella lytica Legionella shakespearei
Legionella busanensis Legionella fallonii Legionella maceachenni
Legionella spiritensis
Legionella cardiaca Legionella feeleii Legionella massiliensis
Legionella steelei
Legionella cherrii Legionella geestiana Legionella micdadei
Legionella steigenvalth
Legionella cincinnatiensis Legionella gcnomospccics
Legionella monrovica Legionella taurinensis
Legionella clemsonensis Legionella gonnanii Legionella moravica
Legionella tucsonensis
oe
Legionella donaldconti Legionella gratiana Legionella
nctga,sakiensis Legionella tunisiensis
Legionella gresilensis Legionella nautarum
Legionella wadsworthii
Legionella hackeliae Legionella norrlandica
Legionella waltersii
Legionella impletisoli Legionella oakridgensis
.. Legionella worsleiensis
Legionella israelensis Legionella parisiensis
Legionella yabuuchiae
Legionella jamestowniensis Legionella
pittsburghensis
Legionella pneumophila
Legionella quateirensis
9
a
-4
.
.
8
,..,
P
. Oceanibulbus Paenibacillus Prevotella
Quadrisphaera
Oceanibulbus inclolifex Paenibacillus thiaminolvticus
Prevotella albensis Quadrisphaera granulorum 0
t..)
o
Oceanicaulis Pantoea Prevotella amnii
Quatrionicoccus ts.)
0.
,
k..)
Oceanicaulis alexandrii Pantom agglomerans Prevotella bergensis
Quatrionicoccus w
-.1
Oceanicola Prevotella bivia
australiensis
Oceanicola batsensis Paracoccus Prevotella brevis
Oceanicola granulosus Paracoccus alcaliphilus
Prevotella buantii Quinella
Oceanicola nanhaiensis Paucimonas Prevotella buccae
Quinella ovalis
Oceanimonas Pauchnonas lemoignei Prevotella buccalis
Ocean imonas baumannh Pectobacterium Prevotella copri
Ralstonia
Oceaniserpentilla Pectobacterium aroidearum
Prevotella dentalis Ralstonia eutropha
Oceaniserpentilla haliotis Pectobacterium atrosepticum
Prevotella denticola Ralstonia insidiosa vi
v:
Oceanisphaera Pectobacterium Prevotella disienc
Ralstonia mannitolllytica
Oceanisphaera don ghaensis betavasculorum Prevotella histicola
Ralstonia pickettii
Oceanisphaera litoralis Pectobacterium cacticida
Prevotella intermedia Ralstonia
Oceanithermus Pectobacterium carnegieana
Prevotella maculosa pseudosolanacearum
Oceanithermus desulfurans Pectobacterium carotovorum
Prevotella marshii Ralstonia syzygii
Oceanithermus profundus Pectobacterium chrysanthemi Prevotella
tnelaninogenica Ralstonia solanacearum
Oceanobacillus Pectobacterium cypripedu
Prevotella micans Ramlibacter od
r)
Oceanobacillus caeni Pectobacterium rhapontici
Prevotella multifonnis Ramlibacter henchirensis ....1
t=i
ot
Oceanospirillum Pectobacterium wasabiae
Prevotella nigrescens Ramlibacter tataouinensis t..)
o
w
1-,
Oceanospirilluin linum Planococcus Prevotella oralis
-O-
w
Planococcus citreus Prevotella oris
v:
ul
.6.
Planomicrobium Prevotella oulorum Raoultella
Planomicrobium okeanokoites Prevotella pallens Raoultella ornithinolvtica
Plesiomonas Prevotella salivae Raoultella planticola
ts.)
Plesiomonas shigelloides Prevotella stercorea Raoultella
terrigena
Proteus Prevotella tannerae Rathayibacter
Proteus vulgaris Prevotella timonensis Rathavibacter caricis
Prevotella veroralis Rathayibacter festucae
Providencia Rathavibacter iranicus
Providencia stuartii Rathavibacter rathayi
Pseudomonas Rathavibacter toxicus
Pseudomonas aeruginosa Rathavibacter tritici
Pseudomonas alcaligenes Rhodobacter
Pseudomonas anguillispetica Rhodohacter sphaero ides
Pseudomonas fluorescens Ruegeria
Pseudoalteromonas Rue geria gelatinovorans
haloplanktis
Pseudomonas mendocina
Pseudomonas
pseudoalcaligenes
Pseudomonas putida
Pseudomonas tutzeri
Pseudomonas syringae
9
a
-4
.
.0
8
'.'
P
. Psychrobacter
Psychrobacter faecalis
0
t..)
o
Psychrobacter
ts.)
1¨
,
k..)
phenylpyrttvicus
w
¨1
cii
oo
Saccharococcus Sagittula Sanguibacter
Stenotrophomonas Tatlockia
Saccharococcus thermophilus Sagittala stellata Sanguibacter keddieii
Stenotrophomonas Tatlockia maceachemii
Saccharomonospora Salegentibacter Sanguibacter
sttarezii maltophilia Tatlockia micdadei
Saccharomonospora azurea Salegentibacter salegens
Saprospira Streptococcus Tenacibaculum
Saccharomonospora cyanea Salimicrobium Saprospira grandis
Tenacibaculum
Saccharomonospora viridis Salimicrobium album Sarcina
[also see below] amylolyticum o
1¨,
Saccharophagus Salinibacter Sarcina maxima
Streptomyces Tenacibaculum discolor
Saccharophagus degradans Salinibacter ruber Sarcina ventriculi
Streptomyces Tenacibaculum
Saccharopolyspora Salinicoccus Sebaldella
achromogenes gallaicum
Saccharopolyspora erythraeu Salinicoccus alkaliphilus
Sebaldella termitidis Streptomyces cesalbus Tenacibaculum
Saccharopolyspora gregorii Salinicoccus hispanicus
Streptomyces cescaepitosus lutimaris
Saccharopolyspora hirsuta Salinicoccus roseus Serratia
Streptomyces cesdiastaticus Tenacibaculum
Saccharopolyspora hordei Salinispora Serratia fonticola
Streptomyces cesexfoliatus mesophilum od
r)
Saccharopolyspora rectivirgula Salinispora arenicola Serratia marcescens
Streptomyces fitnbriatus Tenacibaculum ....1
t=i
ot
Saccharopolyspora spin osa Salinispora tropica Sphaerotilus
Streptomyces fradiae skagerrakense t.)
o
w
Saccharopolyspora taberi Salinivibrio Sphaerotilus natans
Streptontyces fulvissimus
-O-
w
Salinivibrio costicola
Streptomyces griseorttber
ul
.6.
Saccharothrix Salmonella Sphingobacterium
Streptomyces griseus Tepidanaerobacter
Saccharothrix australiensis Salmonella bongori
Sphingobacterium multivorttm Streptomyces lavendulae
Tepidanaerobacter
Saccharothrix coeruleofitsca Salmonella enterica
Staphylococcus Streptornyces syntrophicus ts.)
Saccharothrix espanaensis Salmonella subterranea [see
below] phae ochromo genes Tepidibacter
Saccharothrix longispora Salmonella typhi
Streptomyces Tepidibacter
Saccharothrix mutabilis the
rmodiastaticus fomticigenes
Saccharothrix syringae
Streptomyces tube rcidicus Tepidibacter
Saccharothrix tangerinus
thalassicus
Saccharothrix texasensis
Thermus
Thermus aquaticus
Themws filiformis
Therms thermophilus
Staphylococcus
S. arlettae S. equorurn S. micron S.
schleiferi
S. agnetis S. fells S. muscae S.
sciuri
S. aureus S. fleurettii S. nepalensis S.
sintiae
S. auricularis S. gallinarum S. pasteuri S.
simulans
S. cap itis S. haemolyticus S. petrasii S.
stepanovicii
S. caprae S. hominis S. pettenkoferi S.
succinus
S. carnosus S. hyicus S. pisciferrnentans S.
vitulinus
S. caseolyticus S. intermedius S. pseudintermedius S.
warneri
S. chromo genes S. kloosii S. pseudolugdunensis S.
xylosus
.0
S. cohnii S. leei S. pulvereri
S. condimenti S. lentus S. rostri
S. delphini S. lugdunensis S. saccharolyticus
S. devriesei S. lutrae S. saprophyticus
S. epideMlidis S. lyticans
S. massiliensis
Streptococcus
Streptococcus agalactiae Streptococcus infantarius
Streptococcus orisratti Streptococcus thermophilus
Streptococcus anginosus Streptococcus in/ac Streptococcus
parasanguinis Streptococcus sanguinis
Streptococcus bovis Streptococcus intennedius
Streptococcus peroris Streptococcus sobrinus
Streptococcus canis Streptococcus lactarius Streptococcus
pneumoniae Streptococcus suis
Streptococcus constellatus Streptococcus milleri Streptococcus
Streptococcus uberis
Streptococcus downei Streptococcus mitis pseudopneumoniae
Streptococcus vestibularis
Streptococcus dysgalactiae Streptococcus mutans Streptococcus pyo
genes Streptococcus viridans
Streptococcus equines Streptococcus rails Streptococcus ratti
Streptococcus
Streptococcus faecalis Streptococcus tigurinus Streptococcus
salivariu zooepidemicus
Streptococcus ferus
Uliginosibacterium Vagococcus Vibrio
Virgibacillus Xanthobacter
Vagococcus carniphdus Vibrio aerogenes
Virgibacillus Xanthobacter agilis
Uliginosibacterium gangwonense Vagococcus elongatus Vibrio aestuarianus
halodenitrificans Xanthobacter
9
a
-4
.
.0
8
'.'
P
. Ulvibacter Vagococcus fessus Vibrio albensis
Virgibacillus aminoxidans
Ulvibacter litoralis Vagococcus fluvialis Vibrio alginolyticus
pantothenticus Xanthobacter 0
t..)
o
Umezawaea Vagococcus lutrae Vibrio cainpbellii
Weissella autotrophicus ts.)
0-
,
k..)
Umezawaea tan gerina Vagococcus salmoninarum Vibrio cholerae
Weissella cibaria Xanthobacter flavus w
-.1
Undibacterium Variovorax Vibrio
cincinnatiensis Weissella confusa Xanthobacter tagetidis
Undibacterium pigrum Variovorax boronicumulans
Vibrio coralliilvticus Weissella halotolerans Xanthobacter viscosus
Ureaplasma Variovorax dokdonensis Vibrio
cyclitrophicus Weissella hellenica Xanthomonas
Ureaplasma urealyticum Variovorax paradoxus Vibrio
diazotrophicus Weissella kandleri Xanthomonas
Variovorax soli Vibrio fhivialis
Weissella koreensis albilinecins
Ureibacillus Veillonella Vibrio furnissii
Weissella minor Xanthomonas alfalfae
Ureibacillus composti Veillonella atypica Vibrio gazo genes
Weissella Xanthomonas
Ureibacillus stnvonensis Veillonella caviae Vibrio halioticoli
paramesenteroides arboricola o
4,
Ureibacillus terrenus Veillonella criceti Vibrio harveyi
Weissella soli Xanthomonas
Ureibacillus thermophilus Veillonella dispar Vibrio ichthyoenteri
Weissella thailandensis axonopodis
Ureibacillus thermosphaericus Veillonella montpellierensis
Vibrio mediterranei Weissella yin' descens Xanthomonas
Veillonella parvula Vibrio metschnikovii
Williamsia cainpestris
Veillonella ratti Vibrio mytili
Williamsia marianensis Xanthomonas citri
Veillonella rodentium Vibrio natriegens
Williamsia mans Xanthomonas codiaei
Venenivibrio Vibrio navarrensis
Williamsia serinedens Xanthomonas od
r)
Venenivibrio stagnispumantis Vibrio nereis
Winogradskyella cucurbitae ....1
t=i
It
Vibrio nigripulchritudo
Wino gradskyella Xanthomonas t..)
o
w
1-,
Verminephrobacter Vibrio ordalii
thalassocola euvesicatoria -O-
w
Verminephrobacter eiseniae Vibrio orientalis
Xanthomonas fragariae
4-
Vibrio parahaemolyticus
Wolbachia Xanthomonas fuscans
Verrucomicrobium Vibrio pectenicida
Wolbachia persica Xanthomonas gardneri
Verrucomicrobium spinosurn Vibrio penaeicida
Xanthornonas hortorum
Vibrio proteolyticus
Wolinella Xanthomonas hyacinthi ;=,s'
Vibrio shilonii
Wolinella succino genes Xanthomonas perforans
Vibrio splendidus
Xanthomonas phaseoli
Vibrio tubiashii Zobellia
Xanthomonas pisi
Vibrio vulmficus Zobellia
galactanivorans Xanthomonas populi
Zobellia uliginosa
Xanthomonas theicola
Zoogloea
Xanthomonas
Zoo gioea ramigera
translucens
Zoogloea resiniphila
Xanthomonas
vesicatoria
Xylella
Xylella fastidiosa
Xylophilus
Xylophilus ampelinus
Xenophilus Yangia Yersinia mollaretii
Zooshikella Zobellella
Xenophilus azovorans Yangia pacifica Yersinia philomiragia
Zooshikella ganghwensis Zobellella denitrificans
Yersinia pestis
Zobellella taiwanensis ,õ`s"
Xenorhabdus Yaniella Yersinia pseudotuberculosis
Zunongwangia
Xenorhabdus bedding ii Yaniellct flava Yersinia rohdei
Zunongwangia pro funda Zeaxanthinibacter
Xenorhabdus bovienii Yaniella halototerans Yersinia ruckeri
Zymobacter Zeaxanthinibacter ts.)
Xenorhabdus cabanillash Yeosuana Yokenella
Zvmobacter palmae enoshimensis
Xenorhabdus doucetiae Yeosuana aromativorans Yokenella regensburgei
Zymomonas Zhihengliuella
Xenorhabdus grfffiniae Yersinia Yonghaparkia
Zvmomonas mobilis Zhihengliuella
Xenorhabdus hominickh Yersinia aldovae Yonghaparkia alkaliphila
Zymophilus halotolerans
Xenorhabdus koppenhoeferi Yersinia bercovieri Zavarzinia
Zvmophilus paucivorans Xylanibacterium
Xenorhabdus nematophila Yersinia enterocolitica
Zavarzinia compransoris Zvmophilus raffinosivorans Xvlanibacterium ulmi
Xenorhabdus poinarii Yersinia entomophaga
Xylanibacter Yersinia frederiksenh
Xylanibacter oryzae Yersinia intermedia
Yersinia kristensenii
WO 2021/239758
PCT/EP2021/063954
Table 2: Example Cos
Cl may be a Cas (eg, a Cas3 or a Cascade Cos) selected from the following
types. Additionally or
alternatively, C2 may be a Cas (eg, a Cas3 or a Cascade Cas) selected from the
following types.
Cascade Cas may be selected from the following types.
Bacteria / Phage CRISPR-Cas Type
Clostridium botulinum Type I-B
Clostridium tetani Type I-B
Eggerthella lenta Type I-C
Moraxella bovoculi Type I-C
Streptococcus mutans Type I-C
Streptococcus mutans Type I-C
Streptococcus pyogenes Type I-C
Bacillus halodurans Type I-C
Prevotella enoeca Type I-C
Bacteroides fragilis Type I-C
Pseudomonas aeniginosa Type I-C
Nostoc sp. CENA543 Type I-D
Escherichia coli Type I-E
Vibrio cholerae Type I-E
Citrobacter freundii Type I-E
Salmonella enterica Type I-E
Klebsiella pneumoniae Type I-E
Streptococcus mutans Type I-E
Pseudomonas aeruginosa Type I-F
67
CA 03171200 2022- 9-9
WO 2021/239758 PCT/EP2021/063954
68
Yersinia pestis Type 1-F
Serralia marcescens Type I-F
Geobacter sulfurreducens Type I-U
Salinispora arenicola Type I-U
Vibrio phage ICP1 Type I-F
Table 3: Example Cas, Types and Classes
Type Cas Nuclease Target
Cas3 DNA
Class 1 III Cas 10 DNA or RNA
IV
II Cas9 DNA
Class 2 V Cas 12 DNA
VI Cas 13 RNA
CA 03171200 2022- 9-9