Note: Descriptions are shown in the official language in which they were submitted.
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
TITLE OF THE INVENTION
DIAGNOSIS AND TREATMENT OF COVID-19
FIELD OF THE INVENTION
This invention relates to the diagnosis and treatment of COVID-19.
BACKGROUND OF THE INVENTION
Throughout this application, various references are cited in brackets to
describe more fully
the state of the art to which this invention pertains. The disclosure of these
references is hereby
incorporated by reference into the present disclosure.
The Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) is the virus
that
causes COVID-19 (or COVID19). COVID-19 primarily affects lungs, and in the
most severe
cases results in acute respiratory distress syndrome (ARDS) associated with or
without multiorgan
dysfunction (1-4). Once COVID-19 patients are admitted to the intensive care
unit (ICU), the
mortality rate is reported at 31% with a median of 9 days to ICU death (5).
There are no specific
therapies for COVID19, and patients are provided only supportive care.
Identification of
pathophysiological mediators, as well as prognostic biomarkers and/or
therapeutic targets, is
essential for improving COVID19 patient outcomes.
Recent reports and commentaries have suggested that the severity of COVID19
may be
due to a "cytokine storm", (6) which is the excessive or uncontrolled release
of cytokines in
response to a pathologic event, such as a viral infection. (7) These
suggestions are due to increased
inflammatory cytokine levels, such as interleukin 6 (1L6), as well as fever,
cytopenia and
hyperferritinemia. (8, 9)
Moreover, these commentaries have been accompanied by calls for the use of
broad
immunosuppression with steroids, intravenous immunoglobulin, and/or selective
cytokine
blockade as a therapeutic approach for COVID19. (7,10) While patient mortality
could be
improved with immunosuppressive therapies, the evidence for changes in
specific cytokines is
incomplete, and often observed at a single timepoint with limited comparison
to control groups.
(8, 9)
1
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Additionally, as described in recent commentaries and reviews, the use of
immunosuppressive therapies to treat critically ill patients, including those
with ARDS, has often
been challenging due to the potential to cause harm highlighting the need for
rigorous data to
support any proposed trials. (11,12)
It would be beneficial to have a small number of blood biomarkers measured
that could
serve not just to obtain an accurate diagnosis of COVID-19, but also serve to
prognosticate the
outcome and recovery of the patients. In addition, these biomarkers may
identify and/or indicate
potential therapeutic targets for COVID-19 infection.
SUMMARY OF THE INVENTION
In one embodiment, the present invention is a method of diagnosing COVID-19 in
a
subject, the method comprising measuring levels of one or more metabolites
listed in Table 13 in
a sample taken from the subject, wherein a diagnosis of COVID-19 positive is
indicated when the
levels of said one or more metabolites are statistically different from known
normal levels of said
one or more metabolites.
In one embodiment of the method of diagnosing COVID-19, the one or more
metabolites
is one or more of kynurenine, arginine, sarcosine, lysoPC18:1, lysoPC20:4,
lysoPC14:0,
lysoPC17:0, lysoPC18:2, creatinine, creatine, C3OH, PC40:6AA, C5, C6:1, C3:1
and
methylmalonic acid.
In another embodiment of the method of diagnosing COVID-19, the one or more
metabolites is one more of kynurenine, arginine, sarcosine, lysoPC18:1,
lysoPC20:4, lysoPC14:0,
lysoPC17:0, and lysoPC18:2.
In another embodiment of the method of diagnosing COVID-19, the one or more
metabolites is one more of creatinine, creatine, C3OH, PC40:6AA, C5, C6:1,
C3:1 and
methylmalonic acid.
In another embodiment of the method of diagnosing COVID-19, the one or more
metabolites are kynurenine and arginine, and wherein the diagnosis of COVID-19
positive is
indicated when a ratio of arginine/kynurenine levels is statistically
decreased from known normal
ratio levels of arginine/kynurenine.
2
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
In another embodiment of the method of diagnosing COVID-19, the one or more
metabolite is kynurenine, and wherein the diagnosis of COVID-19 positive is
indicated when the
levels of kynurenine in the sample is statistically elevated from the known
normal levels of
kynurenine.
In another embodiment of the method of diagnosing COVID-19, the one or more
metabolites is arginine, and wherein the diagnosis of COVID-19 positive is
indicated when the
levels of arginine in the sample is statistically decreased from the known
normal levels of arginine.
In another embodiment of the method of diagnosing COVID-19, when the subject
is
indicated as being COVID-19 positive, the subject is treated with tryptophan,
arginine, sarcosine
and/or LysoPCs or any combinations thereof
In another embodiment of the method of diagnosing COVID-19 in the subject,
only when
the subject is COVID-19 positive, the method further comprising treating the
subject for COVID-
19.
In another embodiment, the present invention relates to a method of
determining COVID-
19 disease severity for a COVID-19 patient, the method comprising, (a)
measuring levels of one
or more metabolites selected from Table 13 in a sample from the patient, (b)
comparing the levels
of the one or more metabolites to the known normal levels of said one or more
metabolites, and
based on the comparison, determining the severity of the disease.
In one embodiment of the method determining disease severity, the one or more
metabolites is one or more of kynurenine, arginine, sarcosine, lysoPC18:1,
lysoPC20:4,
lysoPC14:0, lysoPC17:0, lysoPC18:2, creatinine, creatine, C3OH, PC40:6AA, C5,
C6:1, C3:1 and
methylmalonic acid.
In another embodiment of the method determining disease severity, the one or
more
metabolites is one more of kynurenine, arginine, sarcosine, lysoPC18:1,
lysoPC20:4, lysoPC14:0,
lysoPC17:0, and lysoPC18:2.
In another embodiment of the method determining disease severity, the one or
more
metabolites is one more of creatinine, creatine, C3OH, PC40:6AA, C5, C6:1,
C3:1 and
methylmalonic acid.
In another embodiment of the method determining disease severity, the one or
more
3
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
metabolites is creatinine and arginine.
In another embodiment of the method determining disease severity, the one or
more
metabolites is arginine.
In another embodiment of the method determining disease severity, the one or
more
metabolites is creatinine.
In another embodiment of the method determining disease severity, disease
severity
includes mortality risk.
In another embodiment of the method determining disease severity, the method
further
comprises treating the subject for COVID-19.
In another embodiment of the method determining disease severity, the method
further
comprises administering to the subject tryptophan, arginine, sarcosine and/or
LysoPCs or any
combinations thereof
In another embodiment, the present invention is a method for the diagnosis of
COVID-19
in a subject based on metabolomics analysis, said method comprising: (a)
obtaining a
metabolomics profile of biological samples collected from known COVID-19
positive patients and
metabolomics profile of biological samples collected from known COVID-19
negative subjects to
train a classification model and establish a COVID-19 positive class
membership and a control
class membership, and (b) analyzing an unknown biological sample collected
from the subject to
be diagnosed for COVID-19 and assigning a class membership for the unknown
biological sample
on the basis of the classification model established in step (a), wherein a
diagnosis of COVID-19
positive is indicated when the unknown biological sample is assigned to the
COVID-19 positive
class membership.
In one embodiment of the method for the diagnosis of COVID-19 based on
metabolomics
analysis, the metabolomics profile includes the metabolites listed in table
13.
In another embodiment of the method for the diagnosis of COVID-19 based on
metabolomics analysis, the metabolomics profile includes kynurenine, arginine,
sarcosine,
lysoPC18:1, lysoPC20:4, lysoPC14:0, lysoPC17:0 and lysoPC18:2.
In another embodiment of the method for the diagnosis of COVID-19 based on
metabolomics analysis, the metabolomics profile includes kynurenine, arginine,
sarcosine,
4
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
lysoPC18:1, lysoPC20:4, lysoPC14:0, lysoPC17:0, lysoPC18:2, creatinine,
creatine, C3OH,
PC40:6AA, C5, C6:1, C3:1 and methylmalonic acid.
In another embodiment of the method for the diagnosis of COVID-19 based on
metabolomics analysis, the metabolomics profile includes kynurenine and
arginine.
In another embodiment of the method for the diagnosis of COVID-19 based on
metabolomics analysis, the metabolomics profile include kynurenine.
In another embodiment of the method for the diagnosis of COVID-19 based on
metabolomics analysis, only when the subject is assigned in the COVID-19
positive class
membership, the method further comprising treating the subject for COVID-19.
In another embodiment of the method for the diagnosis of COVID-19 based on
metabolomics analysis, only when the subject is assigned in the COVID-19
positive class
membership, the subject is treated with tryptophan, arginine, sarcosine and/or
LysoPCs or any
combination thereof
In another embodiment, the present invention provides for a COVID-19
diagnostic
apparatus, the COVID-19 diagnostic apparatus including a computer readable
storage medium and
a computer program mechanism embedded therein, the computer program mechanism
comprising
executable instructions for performing a method of diagnosing for a COVID-19,
said executable
instructions comprising: (a) measuring levels of one or more metabolites
listed in Table 13 in a
sample taken from the subject, (b) providing a diagnosis of COVID-19 positive
when the levels of
said one or more metabolites are statistically different from known normal
levels of said one or
more metabolites.
In one embodiment of the COVID-19 diagnostic apparatus, the one or more
metabolites is
one or more of kynurenine, arginine, sarcosine, lysoPC18:1, lysoPC20:4,
lysoPC14:0, lysoPC17:0,
lysoPC18:2, creatinine, creatine, C3OH, PC40:6AA, C5, C6:1, C3:1 and
methylmalonic acid.
In another embodiment of the COVID-19 diagnostic apparatus, the one or more
metabolites
is one more of kynurenine, arginine, sarcosine, lysoPC18:1, lysoPC20:4,
lysoPC14:0, lysoPC17:0,
and lysoPC18:2.
In another embodiment of the COVID-19 diagnostic apparatus, the one or more
metabolites
is one more of creatinine, creatine, C3OH, PC40:6AA, C5, C6:1, C3:1 and
methylmalonic acid.
5
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
In another embodiment of the COV1D-19 diagnostic apparatus, the one or more
metabolites
are kynurenine and arginine, and wherein the diagnosis of COV1D-19 positive is
indicated when
a ratio of arginine/kynurenine levels is statistically decreased from known
normal ratio levels of
arginine/kynurenine.
In another embodiment of the COV1D-19 diagnostic apparatus, the one or more
metabolite
is kynurenine, and wherein the diagnosis of COV1D-19 positive is indicated
when the levels of
kynurenine in the sample is statistically elevated from the known normal
levels of kynurenine.
In another embodiment of the COV1D-19 diagnostic apparatus, the one or more
metabolites
is arginine, and wherein the diagnosis of COV1D-19 positive is indicated when
the levels of
arginine in the sample is statistically decreased from the known normal levels
of arginine.
In another embodiment of the COV1D-19 diagnostic apparatus, only when the
subject is
COV1D-19 positive, the instruction further includes providing an output for
treating the subject
for COV1D-19
In another embodiment, the present invention provides for a kit for a COV1D-19
diagnostic
or quantitation assay, the kit comprising one or more internal standards
suitable for mass
spectrometry, packaging material, and instructions, wherein the one or more
internal standards
include the metabolites listed in Figs. 1B, 2A and 3B.
In one embodiment of the kit, the one or more internal standards include one
or a
combination of kynurenine, arginine, lysophospholipds and creatinine.
In another embodiment of the kit, the one or more internal standards is
labelled.
In another embodiment, the present invention relates to a method of treating
COV1D-19,
the method comprising administering to a subject in need an effective amount
of tryptophan,
arginine, sarcosine and/or LysoPCs or any combinations thereof
In one embodiment, the present invention is a method of treating COV1D-19 in a
patient,
the method comprising administering to the patient an agent that reduces the
levels of syndecan-1
degradation product in plasma or protects/restores vascular syndecan-1.
In one embodiment, the agent hat reduces the levels of syndecan-1 degradation
product in
6
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
plasma or protects/restores vascular syndecan-1 is sulodexide,
In another embodiment the agent hat reduces the levels of syndecan-1
degradation product
in plasma or protects/restores vascular syndecan-1 is heparin/heparan. In one
aspect the heparin
is a low molecular weight heparin.
In another embodiment, the agent that reduces the levels of syndecan-1
degradation product
in plasma or protects/restores vascular syndecan-1 is an inhibitor of a
syndecan-1 sheddase.
In another aspect, the agent that reduces the levels of syndecan-1 degradation
product in
plasma or that protects and/or restores vascular syndecan-1 is a protease
inhibitor, including a
soybean-based protease inhibitor. In aspects, the protease inhibitor is an
inhibitor of serine protease
activity.
In another embodiment, the agent that reduces the levels of syndecan-1
degradation product
in plasma or that protects and/or restores vascular syndecan-1 is an inhibitor
of metalloproteinase
(MMP) activity. In one aspect the MMP is 1VIMP2, 1VIMP7 or MMP9. In one
aspect, the inhibitor
of MMP activity is sphingosine-1- phosphate or a protease inhibitor, including
a soybean-based
protease inhibitor. In another aspect, the agent that reduces the levels of
syndecan-1 degradation
product in plasma or protects and/or restores vascular syndecan-1 is an
inhibitor of granzyme B or
an inhibitor of elastase 2. In one aspect, the inhibitor of granzyme B or the
inhibitor of elastase 2
is a protease inhibitor, including soybean-based protease inhibitors.
In another embodiment, the agent that reduces the levels of syndecan-1
degradation product
in plasma or protects/restores vascular syndecan-1 is a heparinase inhibitor.
In another embodiment, the patient is further treated with at least one
additional agent. In
one aspect, the at least one additional agent is an agent which blocks
platelet aggregation or an
anticoagulant or an agent which enhances thrombolysis or an agent which
prevents glycocalyx
degradation.
In another embodiment, the present invention is a method of diagnosing COVID-
19 in a
patient, the method comprising: (a) obtaining a test sample from the patient,
(b) performing one or
more assays configured to detect one or more biomarkers in the test sample,
(c) obtaining the levels
7
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
of the one or more biomarkers in the test sample, (c) comparing levels of the
one or more
biomarkers in the test sample with a normal control reference value of said
one or more disease
severity biomarkers, wherein an increase in the level of the one or more
biomarkers in the test
sample relative to the normal control reference value of said one or more
biomarkers is indicative
of COV1D-19 diagnosis, wherein the one or more biomarkers are syndecan-1,
hyaluronic acid
(HA), chondroitin sulfate ADAMTS13, heparin sulfate, Protein C, soluble P-
selectin (sP-selectin)
and von Willebrand factor (vWF).
In another embodiment, the present invention is a method of predicting disease
severity for
a COV1D-19 a patient, the method comprising: (a) obtaining a test sample from
the patient, (b)
performing one or more assays configured to detect one or more disease
severity biomarkers in
the test sample, (c) obtaining the levels of the one or more disease severity
biomarkers in the test
sample, (c) comparing levels of the one or more disease severity biomarkers in
the test sample
with a normal control reference value of said one or more disease severity
biomarkers, wherein an
increase in the level of the one or more disease severity biomarkers in the
test sample relative to
the normal control reference value of said one or more disease severity
biomarkers is indicative of
disease severity of the COV1D-19 patient, wherein the one or more biomarkers
are syndecan-1,
HA, chondroitin sulfate ADAMTS13, heparin sulfate, Protein C, sP-selectin and
vWF.
In another embodiment, this invention is a use of an agent that reduces the
levels of
syndecan-1 degradation product in plasma or protects/restores vascular
syndecan-1 for the
treatment of COV1D-19.
In another embodiment, this invention is a use of an agent that reduces the
levels of
syndecan-1 degradation product in plasma or protects/restores vascular
syndecan-1 in combination
with at least one additional agent which blocks platelet aggregation, or an
anticoagulant or which
enhances thrombolysis for the treatment of COVID-19 or which prevents
glycocalyx degradation.
In another embodiment, this invention is a use of an agent that reduces the
levels of
syndecan-1 degradation product in plasma or protects/restores vascular
syndecan-1 for the
preparation of a medicament for the treatment of COVID-19.
8
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
In one embodiment, the agent that reduces the levels of syndecan-1 degradation
product in
plasma or protects/restores vascular syndecan-1 is one or more of the agents
cited in previous
embodiments.
In one embodiment, the present invention is a method of predicting disease
severity,
including a mortality outcome for a COVID-19 patient with the method
comprising: (a) obtaining
a test sample from the patient, (b) performing one or more assays configured
to detect one or more
disease severity biomarkers in the test sample, (c) obtaining the levels of
the one or more disease
severity biomarkers in the test sample, (c) comparing levels of the one or
more disease severity
biomarkers in the test sample with a normal control (i.e. healthy) reference
value of said one or
more disease severity biomarkers, wherein an increase in the level of the one
or more disease
severity biomarkers in the test sample relative to the normal control
reference value of said disease
severity biomarker is indicative of disease severity, including mortality
outcome, of the COVID-
19 patient. The patient may be followed up to see if the one or more disease
severity markers return
to a normal level.
In one embodiment, step (c) comprises (i) measuring the expression levels of
the one or
more disease severity biomarkers in the test sample to form a set of raw
expression data, (ii)
normalizing the expression level for each of the one or more disease severity
biomarkers, to form
a set of normalized expression data, (iii) determining for the patient a risk
of disease severity,
including mortality, by comparing a divergence of the one or more disease
severity biomarkers in
the normalized expression data to reference expression data from the normal
controls.
In another embodiment, the disease severity biomarker is selected from the
analytes
included in Table 4, 7 and 8. In one aspect, the disease severity marker is
one or more analytes
having an AUC of 0.7 or greater.
In another embodiment, the disease severity biomarker is one or more of HSP70,
IL-1RA,
IL 10, MIG, CLM-1, IL12RB1, CD83, FAM3B, IGF1R and OPTC.
In another embodiment, the present invention is a COVID-19 diagnostic
apparatus, the
COVID-19 diagnostic apparatus including a computer readable storage medium and
a computer
9
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
program mechanism embedded therein, the computer program mechanism comprising
executable
instructions for performing a method of predicting disease severity, including
mortality outcome,
for a COVID-19 patient, said executable instructions comprising: (a) comparing
levels of one or
more disease severity biomarker in a test sample of the subject, with known
normal (i.e. healthy)
reference levels of the one or more disease severity biomarker, and (b)
providing a risk of mortality
positive signal when there is an increase in the level of the biomarker the
test sample relative to
the known normal reference.
In one embodiment of the apparatus, the disease severity biomarker is selected
from the
analytes included in Table 4, 7 and 8.
In another embodiment of the apparatus, the disease severity biomarker is one
or more of
HSP70, IL-1RA, IL 10, MIG, CLM-1, IL12RB1, CD83, FAM3B, IGF1R and OPTC.
In another embodiment, the invention is a method for determining the
likelihood that a
COVID-19 patient is at risk of disease severity, including mortality,
comprising: (a) measuring the
patient's concentration of one or more markers listed in Table 4, 7 and 8 in
absolute weight or
absolute moles per volume; (b) comparing the measured concentration to a
threshold level of
concentration in absolute weight or absolute moles per volume corresponding to
the measured one
or more markers; and, (c) determining from the comparison the likelihood that
the COVID-19
patient is at risk of mortality, wherein levels above the threshold level for
the one or more markers
concentration in absolute weight or absolute moles per volume indicate that
the patient is at risk
severe disease, including mortality.
In one embodiment, the present invention is a method of diagnosing COVID-19
infection
in a subject. The method, in one embodiment, includes (a) obtaining a test
sample from the subject
(b) comparing levels of a biomarker in the test sample with a known reference
value of said
biomarker, wherein an increase in the level of the biomarker in the test
sample relative to the
known reference value of said biomarker is indicative of positive COVID-19
diagnosis in the
subject. In one aspect, the biomarker is one or more of granzyme B, TNF, HSP70
and IL18. In
another aspect the biomarker is one or more of granzyme B, TNF, HSP70 IL18,
interferon-gamma-
inducible protein 10 (IP-10) and elastase 2. In one aspect the levels are
obtained using quantitative
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
measurements. In another aspect the known reference value of the biomarker is
a known normal
reference. In another aspect the known reference value of the biomarker is a
known abnormal
reference of COVID-19 negative (COVID-19-) subjects. In another aspect the
known reference
value of the biomarker is the level of said biomarker obtained from a normal
sample. In another
aspect, the known reference value of the biomarker is the level of the
biomarker from an abnormal
sample.
In one embodiment according to the previous embodiment, the method further
comprises
(c) treating the subject for COVID-19 infection when the level of the
biomarker in the test sample
is increased relative to the normal or abnormal control sample with an
inhibitor or antagonist of
the biomarker.
In one embodiment of the method of diagnosing COVID-19 of the present
invention, the
method further includes obtaining a sample from the subject during the
subject's recovery for
COVID-19 (i.e. during the subject's rehabilitation therapy), wherein decrease
in the levels of the
biomarker in the recovery sample relative to the levels obtained in the test
sample is indicative of
a normalization of the subject.
In another embodiment, the present invention is a method for the diagnosis of
COVID-19
in a subject comprising (a) comparing the proteomic profile of a test sample
of a biological fluid
of the subject with a control proteomic profile, wherein the control proteomic
profile is a normal
sample, an abnormal sample, a normal reference proteomic profile or an
abnormal reference
proteomic profile comprising at least one protein biomarker of the present
invention; and (b)
diagnosing said subject with COVID-19 if the proteomic profile of the test
sample shows a unique
expression of the at least one protein biomarker; wherein said at least one
protein biomarker is
granzyme B, TNF, Hsp 70, IL-18, IP-10 and elastase 2, and wherein said test
sample proteomic
profile and said control proteomic profile comprise information of the
expression of one or more
of granzyme B, TNF, Hsp 70, IL-18, IP-10 and elastase 2.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of
granzyme B. In one aspect, the inhibitor or antagonist of granzyme B is a
protease inhibitor. In
11
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
another aspect the protease inhibitor is a soybean-based protease inhibitor.
Protease inhibitors
include Kunitz-type protease inhibitor and Bowman-Birk type protease
inhibitors.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of TNF.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of
HSP70.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of IL-
18.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of IP-
10.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of
elastase 2.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of IL-
10.
In another embodiment, the present invention is a method of treating COVID-19
infection
in a subject, the method comprising administering to the subject an inhibitor
or antagonist of
elastase 2. In one aspect, the inhibitor or antagonist of elastase 2 is a
protease inhibitor. In another
aspect the protease inhibitor is a soybean-based protease inhibitor.
In another embodiment, the present invention is a use of the level of a
biomarker in the
diagnosis of COVID-19, wherein the biomarker is one or more of granzyme B,
TNF, HSP70, IL-
18, IP-10 and elastase 2. In one aspect the biomarker is one or more of
granzyme B, TNF, HSP70
12
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
and IL-18.
In another embodiment, the present invention is a use of a granzyme B
inhibitor or
antagonist in the treatment of COVID-19. In one aspect, the granzyme B
inhibitor or antagonist is
a protease inhibitor. In another aspect the protease inhibitor is soybean-
based.
In another embodiment, the present invention is a use of a TNF inhibitor or
antagonist in
the treatment of COVID-19.
In another embodiment, the present invention is a use of a HSP 70 inhibitor or
antagonist
in the treatment of COVID-19.
In another embodiment, the present invention is an IL-18 inhibitor or
antagonist in the
treatment of COVID-19.
In another embodiment, the present invention is an IP-10 inhibitor or
antagonist in the
treatment of COVID-19.
In another embodiment, the present invention is a use of an elastase 2
inhibitor or antagonist
in the treatment of COVID-19. In one aspect, the elastase-2 inhibitor or
antagonist is a protease
inhibitor. In another aspect the protease inhibitor is soybean-based.
In another embodiment, the present invention is a sue of a protease inhibitor
in the
treatment of COVID-19. In aspects, the protease inhibitor is a soybean-based
protease inhibitor.
In another embodiment, the present invention is a COVID-19 diagnostic
apparatus, the
COVID-19 diagnostic apparatus including a computer readable storage medium and
a computer
program mechanism embedded therein, the computer program mechanism comprising
executable
instructions for performing a method of diagnosing COVID-19 in a subject, said
executable
instructions comprising: (a) comparing levels of a biomarker in a test sample
of the subject, with
known reference levels of the biomarker, and (b) providing a COVID-19 positive
signal when
there is an increase in the level of the biomarker the test sample relative to
the control sample, the
biomarker being one or more of granzyme B, TNF, HSP70, IL-18, IP-10 and
elastase 2. In one
aspect of this embodiment the known reference levels of the biomarker is a
known abnormal level
13
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
of the biomarker in a COVID-19- subject. In another aspect of this embodiment
the known
reference levels of the biomarker is a level of the biomarker in a normal
subject.
In one embodiment of the COVID-19 diagnostic apparatus of the present
invention, the
biomarker is one or more of granzyme B, TNF, HSP70, IL18, IP10 and elastase 2.
In another embodiment of the COVID-19 diagnostic apparatus of the present
invention, the
instructions further include comparing the levels of the biomarker in the test
sample, with the levels
of the biomarker in a sample obtained from the subject during the subject's
treatment of COVID-
19, wherein a decrease in the level of biomarker during the treatment relative
to the levels of the
biomarker in the test ample is indicative of a normalization of the subject.
BRIEF DESCRIPTION OF THE DRAWINGS
The following figures illustrate various aspects and preferred and alternative
embodiments
of the invention.
Figs. 1A-1B: (A) Subjects plotted in two dimensions following dimensionality
reduction
of their respective metabolites by stochastic neighbor embedding. Green
circled dots represent
healthy control subjects, while orange dots represent age- and sex-matched
COVID-19+ ICU
patients (ICU day 1 plasma). The dimensionality reduction shows that based on
the plasma
metabolites the two cohorts are distinct and easily separable. The axes are
dimension-less. (B)
ROC analysis of healthy control subjects versus COVID-19+ patients, using an
Arginine/Kynurenine ratio, demonstrates an AUC of 1.00 (P = 0.0002). The
cutoff value is 15.6.
The diagonal broken blue line represents chance (AUC 0.50).
Figs. 2A-2B: (A) ROC analysis of COVID-19+ versus COVID-19- ICU patients,
using an
Arginine/Kynurenine ratio, demonstrates an AUC of 0.98 (P = 0.005). The
diagonal broken blue
line represents chance (AUC 0.50). (B) A time plot demonstrating the
Arginine/Kynurenine ratio
for both COVID-19+ (orange dots) and COVID-19- (circled blue dots) patients
over 10 ICU days.
The two cohorts are significantly different on ICU days 1 and 3 (*** P =
0.005). Healthy control
range values are represented by green shading.
Fig. 3A-3B (A) COVID-19+ ICU patients plotted in two dimensions following
14
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
dimensionality reduction of their respective metabolites by stochastic
neighbor embedding. Blue
(not circled) dots represent COVID-19+ ICU patients that survived their ICU
stay, while circled
orange dots represent COVID-19+ ICU patients that died (ICU day 1 plasma). The
dimensionality
reduction shows that based on the plasma metabolites the two cohorts are
distinct and easily
separable. The axes are dimension-less. (B) A time plot demonstrating the
Creatinine/Arginine
ratio for COVID-19+ ICU patients over 10 ICU days that either survived (blue
dots) or died
(circled orange dots). The two cohorts are significantly different on ICU days
1 and 3 (** P =
0.01). Healthy control range values are represented by green shading.
Figs. 4A to 4B. (4A) Subjects plotted in two dimensions following
dimensionality
reduction by stochastic neighbor embedding. Red dots represent COVID-19+
subjects (n=10, days
1-3) and circled green dots healthy control subjects (n=10). The
dimensionality reduction shows
that based on daily thrombotic factor and endothelial injury marker
concentrations, the two cohorts
are distinct and easily separable. The axes are dimension-less. (4B) ICU
sepsis patients plotted in
two dimensions following dimensionality reduction by stochastic neighbor
embedding. Red dots
represent COVID-19+ subjects (n=10, days 1-3) and circled green dots represent
COVID-19-
subj ects (n=10, days 1-3). The dimensionality reduction shows that based on
daily thrombotic
factor and endothelial injury marker concentrations, the two cohorts are
distinct and easily
separable. The axes are dimension-less.
Fig. 5: Time course for 3 endothelial injury markers between COVID-19+ and
COVID-
19¨ ICU patients. sP-selectin, hyaluronic acid and syndecan-1 remained
elevated until the final
plasma measurements on ICU day 7. Daily values are represented as means ( SEM;
*P<0.05).
Fig. 6 illustrates basal nitric oxide in human hPMVEC untreated (control "-
Hyaluronidase') and treated with hyaluronidase (+ Huyaluronidase).
Hyaluronidase treatment
decreased basal intracellular nitric oxide production by 98% to 64 87.5.
Fig. 7: The COVID-19 case patient and 20 healthy control subjects plotted in
two
dimensions following dimensionality reduction by stochastic neighbor
embedding. The Purple dot
represents the COVID-19 patient, while the circled yellow dots represent the
healthy controls. The
dimensionality reduction shows that based on 59 plasma analyte concentrations,
the COVID-19
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
patients is distinct and easily separable. The axes are dimension-less.
Figs. 8. Subjects plotted in two dimensions following dimensionality reduction
by
stochastic neighbor embedding. Purple dots represent COVID-19+ subjects,
yellow dots (circled)
healthy controls. The dimensionality reduction shows that based on daily
plasma analyte
concentrations, the two cohorts are distinct and easily separable. The axes
are dimension-less.
Figs. 9. Subjects plotted in two dimensions following dimensionality reduction
by
stochastic neighbor embedding. Purple dots represent COVID-19+ subjects,
yellow dots (circled)
COVID-19-. The dimensionality reduction shows that based on daily plasma
analyte
concentrations, the two cohorts are distinct and easily separable. The axes
are dimension-less.
Fig. 10A. tSNE plot demonstrating that the proteome between COVID-19+ patients
on
ICU day 1 that either survived or died are distinct and easily separable
(circled dots patients that
survived, non-circled dots patients that died).
Fig. 10B. tSNE plot demonstrating that the proteome between COVID-19+ patients
on ICU
day 3 that either survived or died are distinct and easily separable (circled
dots patients that
survived, non-circled dots patients that died).
Figs. 11A-11F. Time course for the top 6 inflammatory analytes between COVID-
19+ and
COVID-19¨ ICU patients. Daily values are represented as means ( SEM). *p<0.01.
11A: CLM-
1; 11B: lL12RB1; 11C: CD83; 11D: FAM3B; 11E: IGF1R 3.8, and 11F: OPTC.
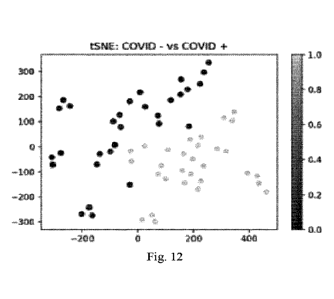
Fig. 12. Subjects plotted in two dimensions following dimensionality reduction
by
stochastic neighbor embedding. Purple (dark) dots represent coronavirus
disease 2019 positive
(COVID-19+) subjects, yellow (light) dots COVID-19¨. The dimensionality
reduction shows that
based on daily plasma analyte concentrations, the two cohorts are distinct and
easily separable.
The axes are dimension less.
Figs. 13A-F: Time course for the top six inflammatory analytes between COVID-
19+ and
COVID-19¨ ICU patients. Daily values are represented as mean ( sem). *p <
0.01. 13A: TNF;
13B: Granzyme B; 13C: HSP70; 13D: lt18: 13E: IP10; 13F: Elastase.
16
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Figs. 14A to 14J: Time course for inflammatory analytes between COVID-19+ and
COVID-19¨ ICU patients. 14A) IL10; 14B) MIG; 14C) M-CSF; 14D) IFNy; 14E) 11,8;
14F)
MMP8; 14G) IL2; 13H) IL15; 141) ILO1RA; 14J) MMPl. Daily values are
represented as means
( SEM). *p<0.01.
DESCRIPTION OF THE INVENTION
Abbreviations
Unless defined otherwise, all technical and scientific terms used herein have
the same
meaning as commonly understood by one of ordinary skill in the art to which
this invention
belongs. Also, unless indicated otherwise, except within the claims, the use
of "or" includes "and"
and vice versa. Non-limiting terms are not to be construed as limiting unless
expressly stated or
the context clearly indicates otherwise (for example "including", "having" and
"comprising"
typically indicate "including without limitation"). Singular forms including
in the claims such as
"a", "an" and "the" include the plural reference unless expressly stated
otherwise. "Consisting
essentially of' means any recited elements are necessarily included, elements
that would materially
affect the basic and novel characteristics of the listed elements are
excluded, and other elements
may optionally be included. "Consisting of' means that all elements other than
those listed are
excluded. Embodiments defined by each of these terms are within the scope of
this invention.
The contents of all documents (including patent documents and non-patent
literature) cited
in this application are incorporated herein by reference.
All numerical designations, e.g., levels, amounts and concentrations,
including ranges, are
approximations that typically may be varied (+) or (-) by increments of 0.1,
1.0, or 10.0, as
appropriate. All numerical designations may be understood as preceded by the
term "about".
"COVID-19¨ subjects" or "COVID-19 negative subjects" are subjects who are
septic with
Acute lung injury (ALT), but are confirmed SARS-CoV-2 negative.
"COVID-19+ subjects" (or patients) or "COVID-19 subjects" are subjects who are
septic
with Acute lung injury (ALT) and positive for SARS-CoV-2.
"Metabolome" refers to the collection of all metabolites in a biological cell,
tissue, organ
17
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
or organism, which are the end products of cellular processes. "Metabolome"
includes lipidome,
sugars, nucleotides and amino acids.
"Metabolomic profiling" refers to the characterization and/or measurement of
the small
molecule metabolites in biological specimen or sample, including cells,
tissue, organs, organisms,
or any derivative fraction thereof and fluids such as blood, blood plasma,
blood serum, capillary
blood, venous blood, saliva, synovial fluid, spinal fluids, urine,
bronchoalveolar lavage, tissue
extracts, tears, volatile organic compounds (VOCs), breath samples, sweat and
so forth. This
characterization may be targeted (limited to a defined number of specific
compounds) or
untargeted/nontargeted in nature (not limited to a defined or known number of
compounds).
The metabolite profile may include information such as the quantity and/or
type of small
molecules present in the sample. The ordinarily skilled artisan would know
that the information
which is necessary and/or sufficient will vary depending on the intended use
of the "metabolite
profile." For example, the "metabolite profile," can be determined using a
single technique for an
intended use but may require the use of several different techniques for
another intended use
depending on such factors as the disease state involved, the types of small
molecules present in a
particular targeted cellular compartment, the cellular compartment being
assayed per se., and so
forth.
The relevant information in a "metabolite profile" may also vary depending on
the intended
use of the compiled information, e.g., spectrum. For example, for some
intended uses, the amounts
of a particular metabolite or a particular class of metabolite may be
relevant, but for other uses the
distribution of types of metabolites may be relevant.
Metabolite profiles may be generated by several methods, e.g., HPLC, thin
layer
chromatography (TLC), electrochemical analysis, Mass Spectroscopy (MS),
refractive index
spectroscopy (RI), Ultra-Violet spectroscopy (UV), fluorescent analysis,
radiochemical analysis,
Near-InfraRed spectroscopy (Near-1R), Nuclear Magnetic Resonance spectroscopy
(NMR),
fluorescence spectroscopy, dual polarisation interferometry, computational
methods, liquid
chromatography (LC) Light Scattering analysis (LS), gas chromatography (GC),
or GC coupled
with MS, direct injection (DI) coupled with LC-MS/MS and/or other methods or
combination of
18
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
methods known in the art.
The term "subject" as used herein refers all members of the animal kingdom
including
mammals, preferably humans.
The term "patient" as used herein refers to a subject that has or is suspected
of having
COV1D-19.
The terms "test sample" or "sample" include biological specimen, including
cells, tissue,
organs, organisms, or any derivative fraction thereof and fluids such as
blood, blood plasma, blood
serum, capillary blood, venous blood, saliva, synovial fluid, spinal fluids,
urine, bronchoalveolar
lavage, tissue extracts, tears, volatile organic compounds (VOCs), breath
samples, sweat and so
forth.
"Plasma" is the clear, straw-colored liquid portion of blood that remains
after red blood
cells, white blood cells, platelets and other cellular components are removed.
The term "proteome" is used herein to describe a significant portion of
proteins in a
biological sample at a given time. The concept of proteome is fundamentally
different from the
genome. While the genome is virtually static, the proteome continually changes
in response to
internal and external events.
The term "proteomic profile" is used to refer to a representation of the
expression pattern
of a plurality of proteins in a biological sample, e.g., a biological fluid at
a given time. The
proteomic profile can, for example, be represented as a mass spectrum, but
other representations
based on any physicochemical or biochemical properties of the proteins are
also included. Thus,
the proteomic profile may, for example, be based on differences in the
electrophoretic properties
of proteins, as determined by two-dimensional gel electrophoresis, e.g. by 2-D
PAGE, and can be
represented, e.g. as a plurality of spots in a two-dimensional electrophoresis
gel. Proteins can be
measured with antibody tests (ie. Western blotting, Luminex bead-based assays,
Proximity
Extension Assay (PEA), planar multiplex assays, electrochemiluminescence,
proximal extension
assay with oligonucleotide-labeled antibodies, ELISA and RIA), flow cytometry
or mass spec
techniques. Enzymes can be measured with enzyme assays that measure either the
consumption
19
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
of a substrate or production of product over time. Differential expression
profiles may have
important diagnostic value, even in the absence of specifically identified
proteins. Single protein
spots can then be detected, for example, by immunoblotting, multiple spots or
proteins using
protein microarrays. The proteomic profile typically represents or contains
information that could
range from a few peaks to a complex profile representing 50, 1,000 or more
peaks. Thus, for
example, the proteomic profile may contain or represent at least 2, or at
least 5 or at least 10 or at
least 15, or at least 20, or at least 25, or at least 30, or at least 35, or
at least 40, or at least 45, or at
least 50 proteins, or over 1,000 proteins.
"Disease severity" is used in this document to characterize the impact that a
disease process
has on the utilization of resources, comorbidities, and mortality. Disease
severity in COVID-19
patients show sequential organ dysfunction, requirement of higher or greater
levels of support and
morbidity.
The term "pharmaceutically acceptable carrier", "pharmaceutically acceptable
excipient",
"physiologically acceptable carrier", or "physiologically acceptable
excipient" refers to a
pharmaceutically-acceptable material, composition, or vehicle, such as a
liquid or solid filler,
diluent, excipient, solvent, or encapsulating material. Each component must be
"pharmaceutically
acceptable" in the sense of being compatible with the other ingredients of a
pharmaceutical
formulation. It must also be suitable for use in contact with the tissue or
organ of humans and
animals without excessive toxicity, irritation, allergic response,
immunogenicity, or other
problems or complications, commensurate with a reasonable benefit/risk ratio.
See, Remington:
The Science and Practice of Pharmacy, 21st Edition; Lippincott Williams &
Wilkins: Philadelphia,
Pa., 2005; Handbook of Pharmaceutical Excipients, 5th Edition; Rowe et al.,
Eds., The
Pharmaceutical Press and the American Pharmaceutical Association: 2005; and
Handbook of
Pharmaceutical Additives, 3rd Edition; Ash and Ash Eds., Gower Publishing
Company: 2007;
Pharmaceutical Preformulation and Formulation, Gibson Ed., CRC Press LLC: Boca
Raton, Fla.,
2004).
The terms "active ingredient", "active compound", and "active substance" refer
to a
compound, which is administered, alone or in combination with one or more
pharmaceutically
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
acceptable excipients or carriers, to a subject for treating, preventing, or
ameliorating one or more
symptoms of COV1D-19 pathology.
The terms "agent", "drug", "therapeutic agent", and "chemotherapeutic agent"
refer to a
compound, or a pharmaceutical composition thereof, which is administered to a
subject for
treating, preventing, or ameliorating one or more symptoms of COV1D-19
pathology.
By "inhibitor" is meant any molecule that inhibits, suppresses or causes the
cessation of at
least one biological activity of a biomarker of the present invention, e.g. by
reducing, interfering
with, blocking, or otherwise preventing the interaction or binding of the
biomarker to its natural
target. Inhibitors include low molecular weight antagonists, antibodies,
proteins, peptides or
ligands that impair the biological action of the biomarker, antisense
oligonucleotides, including
anti-sense RNA molecules and anti-sense DNA molecules that are complimentary
to a nucleic acid
sequence from a gene or genes that encode the biomarker may be used in the
methods of the present
invention to block the translation of mRNA and inhibit protein synthesis, or
increasing mRNA
degradation, thus decreasing the level of biomarker protein, and thus
activity, in a cell. Small
inhibitory RNA (siRNA) is a form of gene silencing triggered by double-
stranded RNA (dsRNA).
In siRNA sequence-specific, post-transcriptional gene silencing in animals and
plants may be
initiated by double-stranded RNA (dsRNA) that is homologous in sequence to the
silenced gene.
A siRNA (small interfering RNA) is designed to target and thus to degrade a
desired mRNA (in
this case encoding mRNA of a suitable biomarker of the present invention) in
order not to express
the encoded protein.
Ribozymes may also function as inhibitors of protein expression for use in the
present
invention. Ribozymes are enzymatic RNA molecules capable of catalyzing the
specific cleavage
of RNA.
The compositions of the present invention include those suitable for oral,
parenteral
(including subcutaneous, intradermal, intramuscular, intravenous,
intraarticular, and
intramedullary), intraperitoneal, transmucosal, transdermal, rectal and
topical (including dermal,
buccal, sublingual and intraocular) administration. The compositions may
conveniently be
21
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
presented in unit dosage form and may be prepared by any of the methods well
known in the art
of pharmacy.
Formulations of the compounds disclosed herein suitable for oral
administration may be
presented as discrete units such as capsules, cachets or tablets each
containing a predetermined
.. amount of the active ingredient; as a powder or granules; as a solution or
a suspension in an
aqueous liquid or a non-aqueous liquid; or as an oil-in-water liquid emulsion
or a water-in-oil
liquid emulsion. The active ingredient may also be presented as a bolus,
electuary or paste.
Pharmaceutical preparations which can be used orally include tablets, push-fit
capsules
made of gelatin, as well as soft, sealed capsules made of gelatin and a
plasticizer, such as glycerol
or sorbitol.
The compounds may be formulated for parenteral administration by injection,
e.g., by
bolus injection or continuous infusion. Formulations for injection may be
presented in unit dosage
form, e.g., in ampoules or in multi-dose containers, with an added
preservative. The compositions
may take such forms as suspensions, solutions or emulsions in oily or aqueous
vehicles, and may
contain formulatory agents such as suspending, stabilizing and/or dispersing
agents. The
formulations may be presented in unit-dose or multi-dose containers, for
example sealed ampoules
and vials, and may be stored in powder form or in a freeze-dried (lyophilized)
condition requiring
only the addition of the sterile liquid carrier, for example, saline or
sterile pyrogen-free water,
immediately prior to use. Extemporaneous injection solutions and suspensions
may be prepared
from sterile powders, granules and tablets of the kind previously described.
In addition to the formulations described previously, the compounds may also
be
formulated as a depot preparation. Such long-acting formulations may be
administered by
implantation (for example subcutaneously or intramuscularly) or by
intramuscular injection. Thus,
for example, the compounds may be formulated with suitable polymeric or
hydrophobic materials
(for example as an emulsion in an acceptable oil) or ion exchange resins, or
as sparingly soluble
derivatives, for example, as a sparingly soluble salt.
For buccal or sublingual administration, the compositions may take the form of
tablets,
22
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
lozenges, pastilles, or gels formulated in conventional manner. Such
compositions may comprise
the active ingredient in a flavored basis such as sucrose and acacia.
The compounds may also be formulated in rectal compositions such as
suppositories or
retention enemas, e.g., containing conventional suppository bases such as
cocoa butter,
polyethylene glycol, or other glycerides.
Certain compounds disclosed herein may be administered topically, that is by
non-systemic
administration. This includes the application of a compound disclosed herein
externally to the
epidermis or the buccal cavity and the instillation of such a compound into
the ear, eye and nose,
such that the compound does not significantly enter the blood stream. In
contrast, systemic
administration refers to oral, intravenous, intraperitoneal and intramuscular
administration.
Formulations suitable for topical administration include liquid or semi-liquid
preparations
suitable for penetration through the skin to the site of inflammation such as
gels, liniments, lotions,
creams, ointments or pastes, and drops suitable for administration to the eye,
ear or nose.
For administration by inhalation, compounds may be delivered from an
insufflator,
nebulizer pressurized packs or other convenient means of delivering an aerosol
spray. Pressurized
packs may comprise a suitable propellant such as dichlorodifluoromethane,
trichlorofluoromethane, dichlorotetrafluoroethane, carbon dioxide or other
suitable gas. In the case
of a pressurized aerosol, the dosage unit may be determined by providing a
valve to deliver a
metered amount. Alternatively, for administration by inhalation or
insufflation, the compounds
according to the invention may take the form of a dry powder composition, for
example a powder
mix of the compound and a suitable powder base such as lactose or starch. The
powder
composition may be presented in unit dosage form, in for example, capsules,
cartridges, gelatin or
blister packs from which the powder may be administered with the aid of an
inhalator or
insufflator.
Preferred unit dosage formulations are those containing an effective dose, as
herein below
recited, or an appropriate fraction thereof, of the active ingredient.
The amount of active ingredient that may be combined with the carrier
materials to produce
23
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
a single dosage form will vary depending upon the host treated and the
particular mode of
administration.
The compounds can be administered in various modes, e.g., orally, topically,
or by
injection. The precise amount of compound administered to a patient will be
the responsibility of
the attendant physician. The specific dose level for any particular patient
will depend upon a
variety of factors including the activity of the specific compound employed,
the age, body weight,
general health, sex, diets, time of administration, route of administration,
rate of excretion, drug
combination, the precise disorder being treated, and the severity of the
disorder being treated. Also,
the route of administration may vary depending on the disorder and its
severity.
In the case wherein the patient's condition does not improve, upon the
doctor's discretion
the administration of the compounds may be administered chronically, that is,
for an extended
period of time, including throughout the duration of the patient's life in
order to ameliorate or
otherwise control or limit the symptoms of the patient's disorder.
In the case wherein the patient's status does improve, upon the doctor's
discretion the
administration of the compounds may be given continuously or temporarily
suspended for a certain
length of time (i.e., a "drug holiday").
Once improvement of the patient's conditions has occurred, a maintenance dose
is
administered if necessary. Subsequently, the dosage or the frequency of
administration, or both,
can be reduced, as a function of the symptoms, to a level at which the
improved disorder is retained.
Patients can, however, require intermittent treatment on a long-term basis
upon any recurrence of
symptoms.
Overview
The present invention relates to the diagnosis, assessing disease severity and
treatment of
COV1D-19 patients and follow-up the recovery of the patients.
Diagnosis
In one embodiment, a method of diagnosing COV1D-19 infection in a patient
includes
24
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
comparing the levels of a single biomarker or a cohort of biomarkers (i.e. one
or more biomarkers)
in a subject's sample using quantitative or non-quantitative measurements of
said biomarker or
cohort of biomarkers to the levels of said single biomarker or cohort of
biomarker in a known
normal reference range, or in a normal population. In the case of non-
quantitative measurements,
the levels of the one or more biomarkers can be normalized and compared by
reference to a known
reference value. A change, an increase or decrease, in the level of the single
biomarker or cohort
of biomarkers in the subject's sample relative to the normal reference range
being indicative of the
subject having COV1D-19 infection.
A method of diagnosing COV1D-19 infection in a patient includes: (a) obtaining
a test
sample from the patient, (b) performing one or more assays configured to
detect one or more
biomarkers in the test sample, (c) obtaining the levels of the one or more
biomarkers in the test
sample, (c) comparing levels of the one or more biomarkers in the test sample
with a normal control
(i.e. healthy) reference value of said one or more disease severity
biomarkers, wherein an increase
or decrease in the level of the one or more biomarkers in the test sample
relative to the normal
control reference value of said one or more biomarkers is indicative of
positive COV1D-19
diagnosis.
In one embodiment, the one or more biomarkers are those listed in Tables 2 and
3, and an
increase in the levels of the one or more biomarkers listed in Table 2 and 3
relative to a normal
control being indicative of positive COV1D-19 diagnosis.
In one embodiment of the present invention, the one or more biomarkers are
granzyme B,
TNF, HSP 70, IL 18, IL 10 and elastase 2, and an increase in the levels of any
one of granzyme B,
TNF, HSP 70, IL 18, IL 10 and elastase 2 relative to the normal control being
indicative of positive
COV1D-19 diagnosis.
In another embodiment, the one or more biomarkers are granzyme B, TNF, HSP 70
and IL
18.
In another embodiment, the one or more biomarkers are syndican-1, hyaluronic
acid (HA),
chondroitin sulfate ADAMTS13, heparin sulfate, Protein C, sP-selectin and von
Willebrand factor
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
(vWF).
In another embodiment, the method of diagnosing COVID-19 in a subject
comprises (a)
measuring an amount of one or more metabolites in a sample from the subject,
(b) determining a
parameter from the amount of each of the one or more metabolites, (c)
comparing the parameter
.. to one or more cutoff values, and based on the comparison, determining
whether the subject is
COVID-19 positive. For example, an increase in the levels of kynurenine in
COVID-19 positive
patients relative to healthy control subjects, or a decrease in lysoPCs in
COVID-19 positive
patients relative to healthy control subjects or a decrease in arginine levels
in COVID-19 patients
relative to healthy controls. The changes in kynurenine, lysoPCs and arginine,
taken alone or in
any combination thereof (i.e. levels of kynurenine alone, levels of lysoPCs
alone, levels of arginine
alone, levels of kynurenine and lysoPCs, levels of kynurenine and arginine,
levels of arginine and
lysoPCs) can be used to discriminate between COVDID-19 positive patients and
healthy control
subj ects.
In one embodiment, the one or more diagnostic biomarkers are those listed in
Table 13. In
.. another embodiment, the one or more biomarkers are those listed in Tables
14, 15 and 16. In
another embodiment, the one or more biomarkers are those listed in Table 14.
In another
embodiment, the one or more biomarkers are those listed in Table 15. In
another embodiment the
one or more biomarkers are those listed in Table 16. In another embodiment,
the one or more
biomarkers of the present disclosure are arginine, kynurenine, sarcosine,
lysophosphatidylcholines
and creatinine. In another embodiment the biomarker is kynurenine and/or
arginine. In another
embodiment the biomarker is one or more lysophosphatidylcholines, kynurenine
and/or arginine
In another embodiment, the biomarker is arginine. In another embodiment, the
biomarker is
kynurenine. In another embodiment, the biomarker is creatinine. In another
embodiment, the
biomarker is sarcosine. In another embodiment, the biomarker is creatinine. In
another
embodiment, the biomarker is one or more lysophosphatidylcholines.
In embodiments, the one or more metabolites is one or more of kynurenine,
arginine,
sarcosine, lysoPC18:1, lysoPC20:4, lysoPC14:0, lysoPC17:0, lysoPC18:2,
creatinine, creatine,
C3OH, PC40:6AA, C5, C6:1, C3:1 and methylmalonic acid, or the one or more
metabolites is one
26
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
more of kynurenine, arginine, sarcosine, lysoPC18:1, lysoPC20:4, lysoPC14:0,
lysoPC17:0, and
lysoPC18:2, or the one or more metabolites is one more of creatinine,
creatine, C3OH, PC40:6AA,
C5, C6:1, C3:1 and methylmalonic acid, or the one or more metabolites are
kynurenine and
arginine, and the parameter is an arginine/kynurenine ratio. The cutoff value
is equal or larger
than 11.6. The cutoff value is equal or larger than 15.7.
The methods and computer programs of the present invention may be used in
point-of-care
metabolomics testing with portable, table/counter-top or hand-held instruments
that generate
metabolite profiles.
The diagnostic methods may be used during the treatment of a COVID-19 patient.
Returns
to a normal level of the biomarkers may serve as an aid in following medical
interventions of
individuals affected by COVID-19.
When following the diagnosis, and a subject is COVID-19 positive, the method
further
comprising treating the subject for COVID-19 with any known method for
treating COVID-19 or
with a method of the present invention.
Assessing Disease Severity
In embodiments the present invention relates to biomarkers measured that serve
to assess
or predict the severity of the illness and mortality for COVID-19 patients,
particularly for patients
in ICUs, shortly after their admissions.
Biomarkers of the present invention also serve as predictors of disease
severity, including
mortality in COVID-19 patients. In embodiments, the one or more biomarkers are
those listed in
Table 7, Table 8 and Table 4. In other embodiments, the one or more biomarkers
is one or more
of HSP70, IL-1RA, IL10, MIG, CLM-1, IL12RB1, CD83, FAM3B, IGF1R and OPTC. In
embodiments, the one or more biomarkers are syndican-1, HA, chondroitin
sulfate ADAMTS13,
heparin sulfate, Protein C, sP-selectin and vWF.
In one embodiment, the present invention involves comparing the levels of a
single
biomarker or a cohort of biomarkers in a subject's sample, using quantitative
or non-quantitative
measurements of said biomarker or cohort of biomarkers to the levels of said
single biomarker or
27
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
cohort of biomarker in a known normal reference range, or in a normal
population. In the case of
non-quantitative measurements, the levels of the one or more biomarkers can be
normalized and
compared by reference to a known reference value. An increase in the level of
the one or more
biomarker in the subject's sample relative to the known reference values for
the one or more
biomarker in a normal healthy control being indicative of disease severity,
including mortality
outcome. Blood is taken from a COVID-19 patient and analytes are measured in a
sample taken
from the patient. The analytes are compared to a known reference value of
cutoff value established
in COVID-19 negative controls. An increase in the measurements, such as
absolute concentrations
(absolute weight, absolute moles per volume, etc.), of the analytes in the
sample of the patient is
indicative of COVID-19 disease severity or risk of death of the patient.
Since metabolites exist in a very broad range of concentrations and exhibit
chemical
diversity, there is no one instrument that can reliably measure all of the
metabolites in the non-
human or human metabolome in a single analysis. Instead, practitioners of
metabolomic profiling
generally use a suite of instruments, most often involving different
combinations of liquid
chromatography (LC) or gas chromatography (GC) coupled with MS, to obtain
broad metabolic
coverage [Circulation. 2012; 126: 1110-1120] Other instruments such as
electrochemical analysis,
RI, UV, near-lR, LS, GC and so forth may also be used.
Point-of-care testing (e.g., hand-held or table-top antibody testing, lateral
flow device, chip
or MS) could be developed to identify COVID-19 patients, and to prognosticate
outcome and/or
stratify to treatment.
A library of the measurements of the biomarkers of the present invention may
be
established for diagnosed COVID-19 cases. This library may be used as the
predetermined,
control set of biomarker measurements of COVID-19. Similarly, a predetermined
set of normal
biomarker measurements may be obtained from subjects known not to have COVID-
19. A
comparison may be made of the patient's biomarker's measurements, the
predetermined biomarker
measurements of COVID-19 and the predetermined biomarker measurements of
normal or control
samples to determine not only if the patient has COVID-19 but also the
prognosis.
The libraries of predetermined biomarker measurements may be provided in a
computer
28
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
product (memory sticks, as an app for hand-held devices such as pads and
cellular phones and so
forth), or they may be uploaded to the memory of a computer system, including
main frames, desk-
tops, lab tops, hand-held devices such as pads and cellular phones. Blood or
any other bodily fluid,
for example whole blood, blood plasma, blood serum, capillary blood, venous
blood, saliva,
synovial fluid, urine, spinal fluid, bronchoalveolar lavage, tears, volatile
organic compounds
(VOCs), breath samples sweat, extracts and so forth, may be taken from a
patient. Biomarker
measurements may be obtained from the patient's sample using any known
technology (for
example, high performance liquid chromatography, thin layer chromatography,
electrochemical
analysis, mass spectroscopy (MS), refractive index spectroscopy, ultra-violet
spectroscopy,
fluorescent analysis, radiochemical analysis, near-infrared spectroscopy,
light scattering analysis,
gas chromatography (GC), or GC coupled with MS, direct injection (DI) coupled
with LC-MS/MS
and so forth) or antibody tests (ie. Western blotting, Luminex bead-based
assays, Proximity
Extension Assay (PEA), planar multiplex assays, electrochemiluminescence,
proximal extension
assay with oligonucleotide-labeled antibodies, ELISA and RIA). The patient's
biomarker
measurements may then be uploaded to the computer system (main frames, desk-
tops, lab tops,
hand-held devices and so forth). An operator may then compare the patient's
biomarker
measurements with the predetermined set of biomarker measurements of COVID-19
and/or the
predetermined biomarker measurements of a control or normal to determine not
only if the patient
has COVID-19, but also the prognosis, or whether a treatment is efficient.
Treatment
The present invention relates also to the treatment of COVID-19 patients.
In embodiments, the methods of the present invention comprise administering to
a patient
in need an effective amount of an agent that reduces the levels of syndecan-1
degradation product
in plasma or protects and/or restores vascular syndecan-1. In embodiments, the
methods of the
present invention involve administering to the patient an effective amount of
an agent that inhibits
syndican-1 shedding. In embodiments, examples of the agent include sulodexide
(inhibitor of
platelet aggregation/reconstruction of glycocalyx) and inhibitors of
metalloproteinase (MMP)
activity such as sphingosine-1- phosphate or a protease inhibitor. In
embodiments, the agent that
29
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
reduces the levels of syndecan-1 degradation product or protects and/or
restores vascular
syndecan-1 is protease inhibitor, including soybean-based protease inhibitor.
Soybean-based
protease inhibitors inhibit the activity of M1VIPs, granzyme B and/or elastase-
2 that can also cleave
the glycocalyx. The compounds disclosed herein may also be combined or used in
combination
with other agents useful in the treatment of COVID-19.
It has now been found that the administration of pharmaceutical compositions
containing
therapeutically effective amounts of an agent that reduces the levels of
syndican-1 in plasma, or
protects/restores vascular syndecan-1, can be used to treat COVID-19 positive
(+) patients,
particularly C OVID-19 + patients suffering thrombosis.
Sulodexide is a glycosaminoglycan of natural origin extracted from mammalian
intestinal
mucosa having a sulfation degree and an anticoagulant activity lower than
those of the heparin.
The present invention includes the use of sulodexide and of the medicinal
compositions containing
it, in the treatment of COVID-19 + patients suffering thrombosis.
The pharmaceutical compositions having sulodexide can be administered by oral
route
preferred in carrying out the present invention are capsules, made by soft or
hard gelatine,
gastroresistant capsules, tablets, controlled release tablets, gastroresistant
tablets, granulates and
syrups.
The sulodexide dosage, depending on the body weight and the seriousness of the
illness, is
comprised between 500 L.S.U. (lipasaemic units) and 2000 L.S.U. a day.
The present invention also relates to the use of an inflammation inhibitory
amount of an
agent selected from the phospholipid sphingosine- 1 -phosphate (S1P)
derivatives of S1P, and
mimetics of the S113 or of the derivatives, and pharmaceutically acceptable
salts thereof and
derivatives thereof, in the treatment of COVID-19 infections. Derivatives of
S113 include, without
limitation, those disclosed in US Pat. No. 5260288: N,N-dimethylsphingosine- 1
-phosphate,
N,N,N,-trimethylsphingosine-l-phosphate, N-acylsphingosine-l-phosphate,
sphingosine-1,3 -
diphosphate, sphingosine-3-phosphate, sphingosine- 1 -thiophosphate, N,N-
dimethylsphingosine-
l-thiophosphate, N-acylsphingosine- 1 -thiophosphate and N,N,N-trimethyl
sphingosine-1-
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
thiophosphate.
In one embodiment, the present invention provides for a composition for
treating a
COVID-19 infection, the composition including one or more inhibitors of
granzyme B, TNF, HSP
70, interleukin-18 (IL-18 or IL 18), interleukin-10 (IL-10 or IL 10) or
elastase 2, and a
pharmaceutically acceptable carrier. In aspects of the present invention, the
composition may also
be used for treating, preventing or minimizing complications associated with
COVID-19.
This invention provides a method for treating COVID-19 infections in a subject
by
administering to the subject one or more inhibitors of one or more of the
biomarkers of the present
invention, in combination with a second agent. One or more inhibitors of
granzyme B, TNF, HSP
70, interleukin-18 (IL-18 or IL 18), interleukin-10 (IL-10 or IL 10) or
elastase 2. The inhibitors
may be formulated for oral administration, for administration by injection,
for topical
administration, inhalation.
An inhibitor of granzyme B is a substance that will inhibit or slow down the
cleavage of
extracellular proteins by granzyme B (United States Patent No. 9060960). For
example, a
compound or composition that prevents granzyme B from cleaving fibronectin,
elastin and/or
fibrillin is a granzyme B inhibitor. In many cases, inhibitors are referred to
as antagonists.
Examples of inhibitors of granzyme B described in international patent
application published
under WO 03/065987 and United States patent application published under US
2003/0148511;
Willoughby C A. et al. Bioorg. Med. Chem. Lett. 12:2197-2200 (2002); Hill GE.
et al. J. Thorac.
.. Cardiovasc. Surg. 110:1658-1662 (1995); Sun J. et al. J. Biol. Chem.
271:27802-27809 (1996);
Sun J. et al. J. Biol. Chem. 272:15434-15441 (1997); Bird et al. Mol. Cell.
Biol. 18, 6387-6398
(1998); Kam et al. Biochim. Biophys. Acta 1477:307:23 (2000); and Bio-x-IEPDP-
(0Ph)2 as
described in Mahrus S. and Craik C S. Chemistry &Biology 12:567-577 (2005).
Antisense
oligonucleotides directed against granzyme B have been designed and
manufactured by Biognostik
(Euromedex, Mundolshei, France) and are described in Hernandez-Pigeon, et al.,
J. Biol. Chem.
281: 13525-13532 (2006) and Bruno, et al., Blood, 96: 1914-1920 (2000).
Further examples of
granzyme B inhibitors are: Z-AAD-CMK (IUPAC name: 5-chloro-4-oxo-2-[2-[2-
(phenylmethoxycarbonylamino)propanoylamino]propanoylamino]pentanoic acid)
MF:
31
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Cl9H24C1N307 CID: 16760474; Ac-IEPD-CHO; Granzyme B Inhibitor IV or Caspase-8
inhibitor III (IUPAC: (4S)-4-[[(2S)-2-acetamido-4-methylpentanoyl]amino]-542-
[[(2S)-4-
hydroxy-1,4-dioxobutan-2-yl]carb am oyl]pyrroli di n-1-y1]-5 -oxop entanoi c
acid)
C22H34N409 OD: 16760476; and Ac-IETD-CHO; Caspase-8 Inhibitor 1 or Granzyme B
Inhibitor II (IUPAC: (4S)-4-[[(2S,3S)-2-acetamido-3-methylpentanoyl]amino]-5-
[[(2S,3 S)-3-
hydroxy-1-[[(2S)-4-hydroxy-1,4-dioxobutan-2-yl]amino]-1-oxobutan-2-yl]amino]-5-
oxopentanoic acid) WIF: C21H34N4010 OD: 16760475. Methods of identifying a
granzyme B
inhibitor are described in United States Patent No. 9060960.
Granzyme B inhibitors are described in the following documents: US 7,326,692,
US
2006/0019945, US 2007/0104699 Al, US 9,060,960 B2, US 10,537,652, US
10,246,487, US
9,458,193, US 9,458,192, US 9,458,138, US 9,969,772, US 9,969,770, US
9,849,112, US
2019/0038602, US 2020/0016125.
Granzyme B inhibitors include also protease inhibitors, including soybean-
based protease
inhibitors. Non-limiting examples of protease inhibitors include Kunitz-type
protease inhibitor,
Bowman-Birk type protease inhibitors.
TNF inhibitors are well known in the art, and include soluble cytokine
receptor that blocks
TNF-a activity, (ii) a monoclonal antibody that blocks TNF-a activity, or
(iii) a tetracycline or a
chemically modified tetracycline that blocks TNF-a activity. Examples of TNF
inhibitors include
Humira, AP301, OPRX-106.
Example of Hsp70 inhibitors include MAL3-101, MKT-077, VER-155008, Apoptozole,
Pifithrin- , NSC 630668-R/1, the Fatty acid synthase inhibitor, FASNALL, the
DAPK3 inhibitor
H538 and H572, an allosteric inhibitor selective for Hsp70i (Haystead, T.A.J.,
Methods Mol Biol.
2018; 1709: 75-8). Other inhibitors include: Elesclomol (STA-4783) is a small
molecule,
Minnelide, a water-soluble pro-drug of triptolide (active compound from a
Chinese herb), PAT-
5M6 is an anti-GRP78 monoclonal antibody. Spanidin is a small molecule that
binds HSP70.
Inhibitors of Hsp 70 are described in US 2006/0074063, US 8,754,094 B2, US
8,486,697, WO
2012/018862, US 9,878,987, US 10,221,171, US 2016/0368975, WO 2019/173394, US
2019/0282600.
32
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Inhibitors of IL-18 include IL-18 binding protein, an antibody against IL-18,
an antibody
against an IL-18 receptor subunits, an inhibitor of the IL-18 signaling
pathway, an antagonist of
IL-18 which competes with IL-18 and blocks the IL-18 receptor, an inhibitor of
caspase-1 (ICE),
an IL-18 isoform, an IL-18 mutein, an IL-18 fused protein, an IL-18 functional
derivative, an IL-
18 active fraction, and an IL-18 circularly permutated derivative thereof
inhibiting the biological
activity of IL-18. Examples include CERC-007 fully humanized anti-IL-18
monoclonal antibody,
GSK 1070806, a humanised IgGl/kappa, anti-interleukin 18 monoclonal antibody,
Tadekinig alfa
is a recombinant human interleukin-18 binding protein, VT-384 is a protein
that has been derived
from Yatapoxvirus. Example of an IL-18 inhibitor are found in US 8,431,130, US
2016/0215048,
US 2018/0127494, US 7,655,616, US 2004/0076628.
Inhibitors of elastase 2 (human neutrophil elastase) include BAY85-8501 small
molecule
inhibitor, CHF6333 small molecule inhibitor, Debio 9701 engineered protein
inhibitor, Elafin
protein inhibitor, MPH-966 is a small molecule inhibitor, P0L6014 is peptide
inhibitor. Inhibitors
of elastase 2 include also protease inhibitors, including soybean-based
protease inhibitors. Non-
limiting examples of protease inhibitors include Kunitz-type protease
inhibitor, Bowman-Birk
type protease inhibitors.
Inhibitors of IL10 include Rituximab is a chimeric mouse antihuman CD20
antibody.
In another embodiment, a method of treating COVID-19 in a patient comprises
administering to the patient an effective amount of tryptophan, arginine,
sarcosine and/or LysoPCs
or any combination thereof. In embodiments, a subject diagnosed with COVID-19
may be treated
with an effective amount of tryptophan, an effective amount of arginine, an
effective amount of
sarcosine and/or an effective amount of LysoPCs, or any possible combination
thereof
The one or more biomarker of the present invention may be used to
prognosticate: Patients
die from withdrawal of care after there has been no improvement in lung
function (2-3 weeks).
These biomarkers help determine who will have a bad outcome earlier and aid
end of life decision
making, or determine whom will do well and guide persistent management.
The one or more biomarker of the present invention may be used for disease
stratification:
33
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
biomarkers will aid, which is critically important for clinical trials (i.e.,
who should be enrolled ¨
if their likelihood of death is high regardless, enrolling them will skew the
data and therapies may
not appear to work ¨ but, they were going to die regardless of treatment).
In order to aid in the understanding and preparation of the within invention,
the following
illustrative, non-limiting, examples are provided.
EXAMPLES
General Methods for All Examples
Study participants and clinical data: The following studies were approved by
the Western
University, Human Research Ethics Board. We enrolled consecutive patients who
were admitted
to our level-3 academic ICU at London Health Sciences Centre (London, Ontario)
and were
suspected of having COVID-19 based on standard hospital screening procedures
(13). We
collected daily blood samples starting on admission and up to 3 days in COVID-
19- patients, or 7
days in COVID-19+ patients (1 additional blood draw on day 10). COVID-19
status was confirmed
as part of standard hospital testing by detection of two SARS-CoV-2 viral
genes using polymerase
chain reaction (14). Patient baseline characteristics were recorded on
admission and included age,
sex, comorbidities, medications, hematologic labs, creatinine, arterial
partial pressure to inspired
oxygen (P/F) ratio, and chest x-ray findings. We calculated Multiple Organ
Dysfunction Score
(MODS) (15) and Sequential Organ Failure Assessment (SOFA) Score (19) for both
COVID-19+
and COVID-19- patient groups to enable objective comparison of their illness
severity. We
categorized both patient groups as having confirmed or suspected sepsis
diagnosis using Sepsis
3.0 criteria (16). We also recorded clinical interventions received during the
observation period
including use of antibiotics, anti-viral agents, systemic corticosteroids,
vasoactive medications,
VTE prophylaxis, anti-platelet or anti-coagulation treatment, renal
replacement therapy, high flow
oxygen therapy, and mechanical ventilation (invasive and non-invasive). Final
participant groups
were constructed by age- and sex-matching COVID-19+ patients with COVID-19-
patients and
healthy controls that were previously banked in the Translational Research
Centre, London,
Ontario (https ://translationalresearchcentre. com/) (17, 18).
34
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
Blood draws: Standard operating procedures were used to ensure all samples
were treated rapidly
and equally. Blood was obtained from critically ill ICU patients via
indwelling catheters daily in
the morning and placed immediately on ice. If a venipuncture was required,
research blood draws
were coordinated with a clinically indicated blood draw. In keeping with
accepted research
phlebotomy protocols for adult patients, blood draws did not exceed maximal
volumes (19).
Once transferred to a negative pressure hood, blood was centrifuged and plasma
isolated,
aliquoted at 250 L, and frozen at -80C. All samples remained frozen until use
and freeze/thaw
cycles were minimized.
EXAMPLE 1
METHODS
Analyte Measurements: Levels of 57 inflammatory analytes were elucidated using
multiplexed biomarker immunoassay kits according to manufacturers'
instructions
(MilliporeSigma, 400 Summit Drive, Burlington, MA, 01803, USA) or enzyme-
linked
immunosorbent assay (ELISA). For the former, plasma inflammatory analytes were
measured
using a Bio-PlexTM 200 Suspension Array system (Bio-Rad Laboratories, 1000
Alfred Nobel
Drive, Hercules, CA, 94547, USA), which utilized Luminex xMAPTM fluorescent
bead-based
technology (Luminex Corp., 12212 Technology Blvd, Austin, TX, 78727, USA).
Bioanalyte
concentrations werecalculated from standard curves using 5-parameter logistic
regression in Bio-
Plex Manager 6.1 software. For the latter, plasma levels of TIMP1 (R&D Systems
Duo Set
#DY970-05, diluted 1:100 or 1:200), TIMP2 (R&D Systems Duo Set #DY971, diluted
1:100) and
TIMP3 (R&D Systems Duo Set #DY973, diluted 1:3 or 1:4) were measured with
ELISA.
Analyses: Medians (IQRs) and frequency (%) were used to report ICU patient
baseline
characteristics for continuous and categorical variables, respectively;
continuous variables were
compared using Mann-Whitney U tests (or Kruskal-Wallis tests, as appropriate),
and categorical
variables were compared using Fisher's exact chi-square, with P-values <0.05
considered
statistically significant. Given the number of analytes processed, we used 2
complimentary
methods, traditional population statistics (M.M.) and machine learning (M.D.).
Daily analyte
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
concentrations were also reported as medians (IQRs), and comparisons between
groups were
examined using Mann-Whitney U tests. Given the number of analytes analyzed and
the risk of
false positives, a P-value of <0.01 was used as our standard for statistical
significance. All analyses
were conducted using SPSS version 26 (IBM Corp., Armonk, NY, USA).
Receiver operating characteristic (ROC) curves were conducted to determine
sensitivity
and specificity of individual proteins for predicting outcome (alive or dead).
Area-under-the-curve
(AUC) was calculated as an aggregate measure of protein performance across all
possible
classification thresholds. All analyses were conducted using SPSS version 26
(IBM Corp.,
Armonk, NY, USA).
Machine Learning: COVID-19 analyte data of the 57 inflammatory analytes
elucidated
using multiplexed biomarker immunoassay were visualized with a nonlinear
dimensionality
reduction on the full data matrix using the t-distributed stochastic nearest
neighbor (t-SNE)
embedding algorithm (20). t-SNE assumes that the "optimal" representation of
the data lies on a
manifold with complex geometry, but low dimension, embedded in the full
dimensional space of
the raw data. For feature selection, we pooled analyte data across 1-3 ICU
days for each of the
COVID-19+ and COVID-19¨ cohorts and normalized observations within analyte. A
random
forest classifier was trained on the variables to predict COVID-19 status. A
random forest is a set
of decision trees and, consequently, we were able to interrogate this
collection of trees to identify
the features that have the highest predictive value (viz., those features that
frequently appear near
the top of the decision tree). We limited the decision trees to a maximum
depth of five levels and
constrained the forest to 50 trees to avoid overfitting the small dataset. We
further explored the
ability to perform automated classification of COVID-19+ versus COVID-19¨
patients from their
analyte spectra, conservatively employing only a single decision tree and
limiting the maximum
tree depth to three levels. We trained and tested the classifier using a five-
fold cross-validation
approach.
RESULTS
36
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
We measured 57 inflammatory analytes in plasma using either fluorescent bead-
based multiplex
technology or ELISAs. Table 2 shows that 20 inflammatory analytes were
significantly different
between COVID-19+ ICU patients and healthy controls (the remaining 37
nonsignificant analytes
are not shown). All significantly different analytes were elevated in COVID-
19+ ICU patients
relative to healthy controls except M1VIP2 that was decreased. COVID-19+ and
COVID-19¨
cohorts were then plotted in two dimensions following dimensionality reduction
by stochastic
neighbor embedding (Fig. 12). The dimensionality reduction shows that the
daily analyte
measurements (ICU days 1-3) between the two cohorts are distinct and easily
separable. To
determine which analytes were most informative for COVID-19 status
classification, we
performed feature selection with a random forest classifier. The top six
features were identified
for the binary outcome of COVID-19+ versus COVID-19¨ in the following order:
tumor necrosis
factor (TNF), granzyme B, heat shock protein 70 (HSP70), interleukin-18 (IL-
18), interferon-
gamma-inducible protein 10 (IP-10), and elastase 2 (Table 21). We then trained
and tested a simple
decision-tree classifier that yielded a classifier accuracy, or the ability of
the analytes to predict
COVID-19 status, of 98% (p < 0.001, five-fold cross-validation). Table 3 lists
17 inflammatory
analytes that were significantly different between COVID-19+ and COVID-19¨
patients on any
or all of ICU days 1-3 (the remaining 40 nonsignificant analytes for ICU days
1-3 are not shown).
All significant analytes were elevated in COVID-19+ ICU patients relative to
COVID-19¨ ICU
patients. While many analytes were significantly different between COVID-19+
and COVID-19-
patients over time, the top six analytes determined by feature classification
over ICU days 1-3 are
listed first, and were TNF, granzyme B, HSP70, and IL-18. IP-10 and elastase 2
were also
significantly different between COVID-19+ and COVID-19¨ patients but starting
on ICU day 2.
A time course for these six markers is shown in Fig. 13A to 13F over ICU days
1-3 for 5 COVID-
19¨ patients and over ICU days 1-7 for COVID-19+ patients. The mean values for
these six
analytes remained elevated in COVID-19+ patients across all seven ICU days.
The remainder of
the analytes measured are shown in Figs. 14A to 14J, with some analytes
increasing (e.g., 1V1_MP1,
Fig. 14J) and some decreasing (e.g., IFNy (Fig. 14D) and IL-1RA (Fig. 141))
over seven ICU days.
The feature matrix for day 1 COVID-19+ ICU patients was classified for
mortality using a Random
Forest classifier (thousand trees) and three-fold cross-validation. As HSP70
was the leading
37
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
analyte associated with COVID-19+ death, a ROC curve was then conducted to
determine the
sensitivity and specificity of HSP70 for predicting mortality. As shown in
Table 4, AUC for HSP70
was 1.00, indicating perfect sensitivity and specificity for our 10 COVID-19+
ICU patients. Using
Youden's Index, the HSP70 cutoff value for predicting mortality was >264,380
pg/mL. Of note,
with the addition of the 10 COVID-19¨ cases to the analysis, the AUC and the
cutoff for HSP70
remained the same.
The AUC for IL-1RA, IL10 and MIG was also 1. As such, HSP10, IL-1RA, IL10 and
MIG provide
perfect sensitivity and specificity for predicting death on ICU day 1 of
critically ill COVID-19
patients. These 4 analytes may be taken alone or in combination. Note that
other analytes included
in Table 4 predicted mortality with high accuracy, such as M-CSF (98%), IL6
(97%) and so forth.
As such, in embodiments, a combination of analytes included in Table 4 may be
used to determine
mortality.
In addition, as seen in Example 2 below, 6 analytes, CLM-1, IL12RB1, CD83,
FAM3B, IGF1R
and OPTC predicted mortality with high degree of certainty. In other
embodiments, any one of the
.. 10 analytes, HSP10, IL-1RA, IL10, MIG, CLM-1, IL12RB1, CD83, FAM3B, IGF1R
and OPTC,
alone or combination, can be used to predict mortality of COVID-19 patients.
In another
embodiment, the analytes in Table 4 can be used to predict mortality of COVID-
19 patients.
EXAMPLE 2
METHODS
Proximity Extension Assay (PEA): A total of 1,161 plasma proteins were
measured using an
immunoassay based on PEA technology (Olink) (21, 22). A 0.25 mL aliquot of
citrate plasma
obtained from each subject was transported frozen on dry ice to the Clinical
Research Laboratory
and Biobank (Hamilton, ON). The data generated were expressed as relative
quantification on the
1og2 scale of normalized protein expression (NPX) values. Individual samples
were screened based
on quality controls for immunoassay and detection, as well as degree of
hemolysis. NPX values
were rank-based normal transformed for further analyses. Following proteomic
quality control, all
participants were deemed suitable for analysis.
38
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Machine Learning: COV1D-19 analyte data of 1,161 plasma proteins measured
using an
immunoassay based on PEA technology, was visualized with a nonlinear
dimensionality reduction
on the full data matrix using the t-distributed stochastic nearest neighbor
embedding (t-SNE)
algorithm (23). t-SNE assumes that the 'optimal' representation of the data
lies on a manifold with
complex geometry, but low dimension, embedded in the full dimensional space of
the raw data
(20). For feature selection, the raw data for each subject were ingested and
normalized within each
feature, across subjects. More specifically, the data for each marker was
scaled to have unit norm.
A random forest classifier was trained on the variables to predict COVID
status. A random forest
is a set of decision trees and, consequently, we were able to interrogate this
collection of trees to
identify the features that have the highest predictive value (viz., those
features that frequently
appear near the top of the decision tree). The feature matrix for day one
COV1D-19 positive ICU
patients was classified for patient outcome using a three-fold cross
validation with a Random
Forest of hundred trees and max depth of 6 to reduce overfitting.
RESULTS
1,161 plasma proteins were measured using PEA immunoassays. Fig. 8 shows a
Tsne plot
illustrating that the COV1D-19+ ICU patient proteome (a circle was used to
better visualize the
COV1D-19+ patients) was distinct and easily separable from age- and sex-
matched healthy control
subjects. Feature classification identified the top 20 proteins underlying
these differences between
cohorts and are shown in Table 17 with their associated importance.
Classification accuracy was
100%. The biological functions of these leading 20 proteins are described in
Table 5.
Fig. 9 shows a tSNE plot illustrating that the COV1D-19+ ICU patient proteome
was distinct and
easily separable from age- and sex-matched COV1D-19- ICU patients. Feature
classification
identified the top 20 proteins underlying these differences between cohorts
and are shown in Table
18 with their associated importance. Classification accuracy was 100%. The
biological functions
of these proteins are described in Table 6.
39
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
We then determined the ability of the plasma proteome (all 1,161 plasma
proteins) to predict
mortality in COVID-19+ patients on either ICU days 1 or 3. Fig. 10A shows a
tSNE plot
demonstrating that the proteome between COVID-19+ patients on ICU day 1 that
either survived
or died are distinct and easily separable. The top 21 proteins underlying
these outcome differences
are shown in Table 19, and their biological functions are described in Table
7. Fig. 10B shows a
tSNE plot demonstrating that the proteome between COVID-19+ patients on ICU
day 3 that either
survived or died are distinct and easily separable. The top 21 proteins
underlying these outcome
differences are shown in Table 20, and their biological functions are
described in Table 8. The
classification accuracy to predict outcome with the entire 1,161 proteins in
COVID-19+ patients
on ICU days 1 and 3 was 92% and 83%, respectively.
To optimize outcome prediction in COVID-19+ patients, we then narrowed the
number of proteins
from 1,161 using ROC analyses. The top 6 proteins for predicting ICU
survival/death using only
a day 1 plasma sample are shown in Figs. 11A to 11F; also shown is their
associated time course
over ten ICU days. There were no deaths during the 10 ICU days for either
cohort; however, one
COVID-19+ ICU survivor was discharged on day 7 with no further plasma
measurements. With
all 6 proteins, the COVID-19+ patients that died had elevated levels relative
to those COVID-19+
patients that survived to ICU discharge. All 6 proteins provided 100%
classification accuracy with
the following cutoffs: CLM-1 7.8, 11,12RB1 3.3, CD83 3.3, FAM3B 4.7, IGF1R
3.8, and OPTC
3.6.
In this study, we measured 1,161 proteins in plasma obtained from ICU
patients, both COVID-
19+ and COVID-19-, as well as age- and sex-matched healthy controls. Given the
number of
analytes measured, we analyzed the data with state-of-the-art machine
learning. Our data indicate
the presence of a unique COVID-19 proteome with 6 proteins predicting ICU
mortality with 100%
accuracy.
CMRF-35-like molecule 1 (CLM-1), a type-1 transmembrane glycoprotein with an
extracellular
IgG domain, accurately predicted COVID-19 ICU outcome. CLM-1 is expressed
predominantly
in myeloid cells where it can impair IL-6 production in bone marrow derived
mast cells and
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
promotes phagocytosis of dead cells by binding phosphatidylserine, which
serves as a common
apoptotic cell surface recognition cue. The removal of apoptotic cells by CLM-
1 expressing
macrophages may prevent the generation of secondary necrosis and the release
of potentially toxic
or immunogenic components from necrotic cells, reducing the likelihood of an
inflammatory
reaction. IL12RB1, one of two subunits within the IL-12 receptor, is expressed
on natural killer
(NK) cells and T-cells cells. Essential for resistance to intracellular
pathogens, IL12RB mediates
the proinflammatory response to IL-12 that is released by antigen presenting
cells (24). Individual
variability in IL12RB1 function is introduced at the epigenetic, genomic
polymorphism, and
mRNA splicing levels, thereby inferring disease susceptibility and variable
outcomes (25). CD83,
a member of the immunoglobulin superfamily, is expressed on a variety of
activated immune cells
(26). Inferring selective immunosuppression when membrane bound on antigen
presenting cells,
soluble CD83 infers powerful immunosuppressive properties by inhibiting
proliferation and
function of T-cells. Viral infection leads to the degradation of dendritic
cell CD83, a mechanism
described as a viral immune escape mechanism (27). FAM3B, expressed at high
levels in the islets
of Langerhans of the endocrine pancreas, is a secreted cytokine that induces
apoptosis (28).
Increased CD38 is associated with pancreatic I cell dysfunction, hyperglycemia
and insulin
resistance, suggesting a role in the regulation of glucose and lipid
metabolism (29). IGF1R, a
transmembrane tyrosine kinase receptor that is activated insulin-like growth
factor 1, is expressed
on lymphocytes and macrophages and can mediate lung injury in response to
infectious pathogen
or chemical insult. In particular, phosphorylation of the IGF1R receptor
exaggerates inflammation
and its over-expression aggravates cytokine levels during influenza infection
(30). Conversely,
IGF1R deficiency attenuates acute inflammatory response in a lung injury mouse
model (31).
OPTC, also called opticin, is highly expressed in the eye nonpigmented ciliary
epithelium that
secretes it into the vitreous cavity where it associates with vitreous
collagen and adjacent basement
membranes (32). As a small leucine-rich protein, OPTC binds collagen fibrils
and regulates
extracellular matrix adhesiveness to suppress capillary morphogenesis and
inhibit endothelial
invasion (33). OPTC is also expressed in lymphocytes (34), but its role in
infection and
inflammation is unknown.
41
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
EXAMPLE 3
Quantitative assays are used to determine the absolute concentration of the
one or more markers
listed in Tables 4, 7 and 8, including each of HSP70, IL-1RA, IL 10, MIG, CLM-
1, IL12RB1,
CD83, FAM3B, IGF1R and OPTC in COVID-19 patients admitted to ICU and in
healthy controls
in absolute weight or absolute moles per volume. The absolute concentrations
in the healthy
controls are used to determine absolute concentration threshold that predicts
disease severity and
mortality. Levels of the one or more markers above the threshold level for the
one or more markers
concentration in absolute weight or absolute moles per volume indicate that
the patient is at risk
disease severity including at risk of death.
EXAMPLE 4
METHODS
Enzyme-Linked Immunosorbent Assay (ELISA): All plasma analytes were measured
with
immunoassays in duplicate as per the manufacturer's recommendation. Analytes
measured include
ADAMTS13 (Abcam #ab234559, diluted 1:200), Protein C (Assaypro #EP1311-7,
diluted 1:8),
von Willebrand factor (vWF; Thermo Fisher #EHVWF, diluted 1:8000), soluble
platelet selectin
(sP-selectin; Abcam # ab100631, diluted 1:50 or 1:20), heparan sulfate (TSZ
ELISA #H1J8718,
diluted 1:5), chondroitin sulfate (TSZ ELISA # H1J8720, diluted 1:2),
hyaluronic acid (R&D
Systems #DHYALO, diluted 1:20), and syndecan-1 (Abcam, #ab46506, diluted 1:2).
Isolation and culture of human pulmonary microvascular endothelial cells
(hPMVEC):
hPMVEC were isolated from resected human lung as previously described (35,
36). Briefly,
human peripheral lung tissue was finely minced, and digested in 0.3% type II
collagenase at 37 C.
The digested suspension was filtered, centrifuged, and washed in PBS.
Endothelial cells were then
isolated using magnetic Dynabeads coated with anti-human CD31 antibody.
Isolated cells were
resuspended in EGM-2 (Lonza # CC-3162) with 10% fetal bovine serum and placed
at 37 C in
5% CO2 until 50% confluent, then harvested and re-purified using anti-CD31-
coated magnetic
microbeads as above. PMVEC were propagated in EGM-2 + 10% FBS and 20 mM HEPES
on
fibronectin-coated flasks and passages 4-9 used.
42
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Hyaluronidase treatment of hPMVEC: PMVEC (2.5 x104 / well) were plated on
fibronectin-
coated 4-well plates in EGM-2 + 10% FBS and 20 mM HEPES. After 2 days, media
was changed
to HBSS (+100 mM HEPES, no bicarb) + 0.01% BSA and hPMVEC were treated for lh
with
hyaluronidase (0.5mg/mL; Sigma #H3506). Following treatment, hPMVEC were
loaded with a
nitric oxide-sensitive fluorochrome (2 [tM DAF-FM DA (Thermo Fisher # D23844)
for lh before
lysing with 0.5% SDS in PBS. After centrifugation for 10 min at 1x104RCF, the
fluorescence of
triplicate aliquots of supernatant were measured using a Victor3 multilabel
fluorescence
microplate reader (Wallac Oy; Perkin Elmer, Inc.) at 485/520 for DAF-FM. The
nitric oxide donor
DETA NONOate (20 [tM; Cayman #82120) was used as the positive control.
Population Statistics: Medians (IQRs) and frequency (%) were used to report
continuous and
categorical variables, respectively. Continuous variables were compared using
either the Mann-
Whitney U test or the Kruskal-Wallis test, as appropriate, and categorical
variables were compared
using Fisher's exact chi-square. P-values < 0.05 considered statistically
significant. All population
statistics were conducted using SPSS version 26 (IBM Corp., Armonk, NY, USA).
For data
comparison that were non-significant, G*Power version 3.1.9.4 was used to
determine the number
of patients per cohort required to potentially reach statistical significance
based on measured
values and 80% power (37).
Machine Learning: Nonlinear dimensionality reductions on the full datasets to
only two
dimensions was completed using the t-distributed stochastic nearest neighbor
(t-SNE) algorithm
(38). For classification, we pooled analyte data across days 1-3 for each of
the COVID-19+ and
COVID-19- cohorts and normalized observations within-analyte. A random forest
classifier was
trained on the variables to predict COVID status. In addition, another random
forest classifier was
trained on pooled analyte data for COVID-19+ patients for days 1-3 to predict
patient mortality.
A random forest is a set of decision trees that we can interrogate to identify
the features with the
highest predictive value. We limited the decision trees to a maximum depth of
6 levels and
constrained the forest to 10 trees in order to avoid overfitting the small
dataset. We trained and
tested the classifier using a 5-fold cross-validation approach.
43
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
RESULTS
We investigated 10 patients with a positive diagnosis of COVID-19 (median
years of age=61.0,
IQR 54.8, 67.0), 10 age- and sex-matched patients with a negative diagnosis of
COVID-19
(median years of age=58.0, IQR 52.5, 63.0), and 10 age- and sex-matched
healthy controls (median
years of age=57.5, IQR 52.8, 62.8; P=0.686). Baseline demographic
characteristics, comorbidities,
labs, and chest x-ray findings are reported in Table 1. COVID-19+ patients
relative to COVID-19-
patients were more likely to have bilateral pneumonia (P=0.001). Pathogens
were confirmed in
only 2 of the COVID-19- patients (P=0.001). All other reported baseline
measures were non-
significant between patients.
We measured 3 thrombosis factors and 5 endothelial cell injury markers in
plasma using ELISAs.
Table 9 shows that 3 markers (vWF, chondroitin sulfate, and syndecan-1) were
significantly
elevated in COVID-19+ ICU patients relative to healthy controls. Table 10
lists the plasma
measurements for 8 markers between COVID-19+ and COVID-19- patients on ICU
days 1-3.
Significant elevations were only observed in endothelial injury biomarkers,
including sP-selectin
(ICU day 3), heparan sulfate (ICU day 2), hyaluronic acid (ICU day 3) and
syndecan-1 (ICU days
1-3).
We then reduced the data to two-dimensions using t-SNE to visualize
differences between healthy
controls and COVID-19+ patients (ICU days 1-3; Fig. 4A), as well as COVID-19-
and COVID-
19+ patients (ICU days 1-3; Fig. 4B). In both cases, the COVID-19+ patients
were easily
distinguishable from either healthy controls or COVID-19- patients. We then
trained and tested a
random forest classifier that yielded a classifier accuracy, or the ability of
the markers to predict
COVID-19 status, of 85% (5-fold cross-validation). To determine which of the 8
markers were
most informative for COVID-19 status classification, we undertook feature
selection with the
random forest classifier. For ICU days 1-3, the top features in rank order
were identified for the
binary outcome of COVID-19+ versus COVID-19- as: syndecan-1 > hyaluronic acid
> chondroitin
sulfate > ADAMTS13 > heparan sulfate > Protein C> sP-selectin > vWF. However,
for ICU day
44
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
3 only, the top features in rank order were hyaluronic acid > sP-selectin >
syndecan-1 >>
ADAMTS13 > chondroitin sulfate = heparan sulfate > vWF > Protein C.
Given the significant elevation in plasma hyaluronic acid, sP-selectin and
syndecan-1 on ICU day
3, we continued daily plasma measurements until ICU day 7 (Fig. 5). For all
three endothelial
injury biomarkers, the plasma levels remained elevated suggesting ongoing
glycocalyx
degradation.
To determine a relationship between the thrombotic state and outcome, we
trained and tested a
random forest classifier to determine the ability of the 8 markers on ICU days
1-3 to predict
mortality in COVID-19+ patients. The thrombosis profile yielded a classifier
accuracy, or the
ability of the markers to predict mortality, of 86% (5-fold cross-validation).
Given the reliance of the classification accuracy on hyaluronic acid
degradation, and the reports
of injury to the pulmonary endothelium with COVID-19, we specifically removed
hyaluronic acid
from human hPMVEC with hyaluronidase treatment (Fig. 6). Hyaluronidase
treatment decreased
basal intracellular nitric oxide production by 98% to 64 87.5 RFUs, compared
to untreated
human PMVEC (P=0.008, n=5 separate experiments). The positive control DETA
NONOate (20
M) increased nitric oxide production by 16% compared to untreated controls
(data not shown).
Table 11 shows a comparison of healthy controls, COVID-19- and COVID-19+
patients on ICU
day 3. HA and syndecan-1 were significantly elevated in COVID-19+ patients on
ICU day 3.
In this study, we measured 3 thrombotic factors and 5 endothelial cell injury
markers in plasma
obtained from ICU patients, both COVID-19+ and COVID-19-, as well as age- and
sex-matched
healthy controls. Our data indicate increased vWF in COVID-19+ patients
relative to healthy
controls elevated sP-selectin, hyaluronic acid and syndecan-1.
Our COVID-19+ ICU patients were similar to those reported in earlier cohorts
from multiple
countries with respect to age, comorbidities and clinical presentation (39, 40-
42). In contrast to
COVID-19- ICU patients, our COVID-19+ ICU patients had a higher incidence of
bilateral
pneumonia (43). The COVID-19+ patients in our study appeared to have lower
illness severity
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
scores than the COVID-19- patients, yet mortality was high at 40%. In
contrast, all COVID-19-
ICU patients survived. Although these differences were not statistically
significant, the findings
suggest that acute respiratory distress syndrome in COVID-19+ patients has
worse outcomes,
perhaps due to the persistently high levels of plasma serine proteases (43).
Our data suggest that COVID-19 results in endothelial injury. Specifically, sP-
selectin, hyaluronic
acid, and syndecan-1 were all significantly elevated by ICU day 3 in plasma of
COVID-19+
patients relative to COVID-19- patients and remained persistently elevated in
plasma up to ICU
day 7.
Syndecan-1 is a proteoglycan containing both heparan- and chondroitin-sulfate
chains that
mediates cellular responses to signaling molecules as well as cell-cell and
cell-matrix interactions
(44). During inflammation, syndecan-1 functions to inhibit neutrophil adhesion
and migration.
Shedding of syndecan-1 from the cell surface is initiated by heparanase-
dependent removal of the
heparan-sulfate side chains (45), thereby instigating subsequent cleavage of
the core syndecan-1
protein by enzymes such as matrix metalloproteinases. Importantly, moderate
syndecan-1
shedding is thought to aid in resolving inflammation; however, excessive
shedding is likely
pathogenic as complete loss of syndecan-1 allows for increased leukocyte
adhesion and
recruitment across the endothelial monolayer, as well as enhanced platelet
aggregation and
adhesion. Different sheddases are able to cleave syndecans on the
extracellular side, releasing a
soluble syndecan consisting of the extracellular domain and the attached GAG
chains (syndecan-
degradation products).
Our study has identified a unique pro-thrombotic state in critically ill COVID-
19 patients that may
be amenable to therapeutic targeting.
Our study, taken in the context of the current literature, suggests that 'not
all coagulopathy is
created equal'. While some patients may develop an extreme pro-thrombotic
state secondary to
the development of either anticardiolipin antibodies (46) or activated
plasminogen (47), others
may be pro-thrombotic on the basis of alveolar-capillary membrane denudation
and exposure of
tissue factor (48). Anticoagulants are one treatment strategy; however, low
molecular weight
heparin did not confer an overall survival advantage in COVID-19 patients
(49). The beneficial
46
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
effects of specific therapeutic strategies may be diluted by patient and
disease heterogeneity,
suggesting that a personalized treatment approach is required.
Example 5
Preamble
COVID-19 presents clinical symptoms that share features of Kawasaki's Disease
(KD) and are
attributable in part to an acute vasculopathy. A `cytokine storm' has been
suggested to underlie
the syndrome, with tissue injury secondary to the host innate response (50).
The inflammatory and
endothelial injury mediators have not yet been described, but knowledge of
these analytes is
critically important for earlier syndrome recognition and for potential
interventions.
Results
A 15-year-old female presented to hospital to a tertiary care emergency
department with a history
of malaise, dry cough, strawberry tongue, rash and jaundice. COVID-19 was
confirmed by
detection of two SARS-CoV-2 viral genes using polymerase chain reaction. Her
complete blood
count, electrolytes, coagulation profile and blood gas were normal. C-reactive
protein and ferritin
were mildly elevated at 25.7 mg/L and 302 g/L, respectively. She had a mild
hepatitis with
alanine aminotransferase 142 U/L, aspartate aminotransferase 87 U/L, alkaline
phosphate 405 U/L,
total bilirubin 92.6 mon. She was admitted to hospital with a presumptive
diagnosis of atypical
KD and treated with intravenous immunoglobulin (IVIg) and Aspirin. Her
inpatient
electrocardiogram and echocardiogram were normal.
Blood was drawn for inflammation/endothelial injury profiling after the
patient's COVID-19 status
was confirmed, but IVIg had already been administered approximately 48 hours
earlier. Thus,
analyte measurements must be evaluated in the context of this immune modulator
(see below).
Nonetheless, 59 inflammation- and endothelium-related analytes were measured
using
multiplexed biomarker immunoassay kits or enzyme-linked immunosorbent assay
(ELISA). As
only one COVID-19 pediatric patient was admitted to our hospital, we compared
the measured
analyte values from this COVID-19 case patient to analyte reference ranges
that we obtained from
a cohort of 20 pediatric healthy control subjects [median 15 years of age (IQR
8)].
47
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
The analyte data from the COVID-19 patient and the 20 healthy control subjects
were first
visualized with a nonlinear dimensionality reduction on the full data matrix
using the t-distributed
stochastic nearest neighbour (t-SNE) embedding algorithm (Fig. 7). t-SNE
assumes that the
'optimal' representation of the data lies on a manifold with complex geometry,
but low dimension,
embedded in the full dimensional space of the raw data. Based on analyte
measurements, the
COVID-19 case patient is a clear outlier with respect to her inflammation and
endothelial injury
profile.
We then generated confidence intervals (CIs) for the expected value of each
analyte using the
plasma measurements from the 20 healthy pediatric controls. The plasma values
for each analyte
were not normally distributed, so we computed 99.9% (95%, Bonferonni corrected
for comparison
across 59 plasma analytes) CIs via the bias corrected and accelerated
bootstrap. Plasma analyte
values in the COVID-19 case patient that were outside the CIs for healthy
control subjects were
therefore considered significant (p<0.05, corrected; Table 12. We found
significant elevations in
21 inflammation and endothelial analyte markers, while 1 endothelial
glycocalyx degradation
marker (heparan sulfate) was significantly depressed (Table 12).
After 3 days of observation, and partial resolution of her symptoms, the COVID-
19 case patient
was discharged home on Aspirin (3 mg/kg/day) with a 2-week follow up
echocardiogram.
Matrix metalloproteinase 7 (MMP7) was the most elevated analyte in the COVID-
19 case patient
relative to healthy control subjects. Also called matrilysin, WM37 is
expressed in endothelial
cells, monocytes and macrophages and it is capable of degrading multiple
extracellular membrane
components (proteoglycans, laminin, fibronectin, casein and basement membrane
collagen type
IV). M1VIIP7 is significantly upregulated in KD and it is implicated in acute
vasculopathy (51).
Specifically, WM37 degrades endothelial junctions, which can promote vascular
leak/edema
and/or leukocyte migration into tissues (52). WM37 has been identified as a
syndecan-1 sheddase
in lung mucosa (62).
Interferon-y-inducible protein 10 (IP-10), an inflammatory cytokine secreted
primarily by
monocytes and endothelial cells in response to interferon-y (IFNy), was also
significantly elevated
in the COVID-19 case patient. IP-10 has multiple roles including lymphocyte
chemoattraction and
adhesion to endothelial cells. IP-10 is a promising target for the treatment
of infectious diseases as
48
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
it aids cellular targeting to threatened tissues where it modulates innate and
adaptive immune
responses. High serum IP-10 is found in KD, and it has been suggested as a KD
biomarker (53).
Resistin is highly expressed in macrophages, bone marrow and the non-fat
fraction of adipose
tissue, and it stimulates several pro-inflammatory pathways and cytokines.
Microvascular tone, as
well as endothelial cell barrier function and nitric oxide production, are all
altered by resistin.
Similar to our COVID-19 case patient, elevated resistin is found in plasma
from KD patients (54,
55).
Interleukin 3 (IL-3), released by activated T-cells, was elevated in our COVID-
19 case patient. IL-
3 promotes the production of inflammatory monocytes and neutrophils, thereby
contributing to the
.. cytokine storm that is implicated in sepsis from multiple etiologies. The
microvascular endothelial
cell response to inflammation and immunity is also regulated by IL-3 (56) and
vasculopathy is
suggested to be a primary feature of the novel multi-system inflammatory
syndrome.
Hyaluronic acid is a major constituent of the microvascular glycocalyx, an
extracellular matrix
that coats the luminal surface of the endothelium (57). Hyaluronic acid
degradation products are
significantly elevated in plasma from the COVID-19 case patient, suggesting
that the
microvascular endothelial cell luminal surface has been pathologically
altered. Disruption of the
endothelial glycocalyx is associated with vascular lesions in KD (58), as well
as decreased
endothelial nitric oxide production and increased platelet/endothelium
adhesion (57), Endothelial
cell injury was supported in the COVID-19 case patient by the parallel
elevation of soluble P-
selectin, an endothelial glycoprotein that mediates adhesive intercellular
interactions (59).
Our measurements showed minimal alterations in 37 inflammation and endothelial
analyte
markers: epidermal growth factor (EGF), granulocyte-colony stimulating factor
(G-CSF),
granulocyte-macrophage colony-stimulating factor (GM-C SF), IFNy, interleukin
la (IL-la), IL-
lb, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12(p40), IL-12(p70), IL-13,
IL-15, IL-17a,
IL17e/lL25, IL-17f, IL-22, macrophage colony-stimulating factor (M-CSF),
macrophage
inflammatory protein 1 a (MIP-1a), tumor necrosis factor a (TNFa), TNFI3,
vascular endothelial
growth factor A (VEGFA), regulated upon activation, normal T Cell expressed
and presumably
secreted (RANTES), MMP2, MMP3, MMP9, MIN/1P12, MMP13, neutrophil gelatinase-
associated
lipocalin (NGAL), Granzyme B, heat shock protein 70 (HSP-70), chondroitin
sulfate and
49
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
syndecan-1. As some of these measurements may have been depressed by IVIg
administration
(60), no significant conclusions can be made with regards to their pre-
treatment level. It is also
plausible that some inflammatory analytes were transiently increased with
inflammation onset,
with TNF and IL-6 as typical examples (61). TNF-a is a pro-inflammatory
cytokine released
primarily by monocytes and macrophages that enhances the adaptive immune
response. IL-6 is
produced by monocytes and macrophages, and induces T-cell activation, B cell
proliferation and
stimulates the acute phase reaction, all of which lead to augmentation of the
immune response.
In summary, pediatric COVID-19 patients can present with a novel multi-system
inflammatory
syndrome with some features similar to KD. The analyte measurements presented
in this study,
albeit post IVIg treatment, support a systemic inflammatory process that
resulted in significant
endothelial injury. These data should aid future hypothesis-generating
research, as some of the
identified analytes might be putative disease biomarkers and/or potential
therapeutic targets.
EXAMPLE 6
METHODS
DI-LC-MS/MS: A targeted quantitative metabolomics approach was used to analyze
the
samples using a combination of direct injection mass spectrometry with a
reverse-phase LC-
MS/MS custom assay. This custom assay, in combination with an ABSciex 4000
QTrap (Applied
Biosystems/MDS Sciex) mass spectrometer, can be used for the targeted
identification and
quantification of up to 150 different endogenous metabolites including amino
acids, acylcarnitines,
biogenic amines & derivatives, uremic toxins, glycerophospholipids,
sphingolipids and sugars (63,
64). The method combines the derivatization and extraction of analytes, and
the selective
spectrometric detection using multiple reaction monitoring (MRM) pairs.
Isotope-labeled internal
standards and other internal standards are used for metabolite quantification.
The custom assay
contains a 96 deep-well plate with a filter plate attached with sealing tape,
and reagents and
solvents used to prepare the plate assay. First 14 wells were used for one
blank, three zero samples,
seven standards and three quality control samples. For all metabolites except
organic acid, samples
were thawed on ice and subsequently vortexed and centrifuged at 13,000x g; 10
uL of each sample
was then loaded onto the center of the filter on the upper 96-well plate and
dried in a stream of
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
nitrogen. Subsequently, phenyl-isothiocyanate was added for derivatization.
After incubation, the
filter spots were dried again using an evaporator. Extraction of the
metabolites was then achieved
by adding 300 pL of extraction solvent. The extracts were obtained by
centrifugation into the lower
96-deep well plate, followed by a dilution step with MS running solvent.
For organic acid analysis, 150 pL of ice-cold methanol and 10 pL of isotope-
labeled
internal standard mixture was added to 50 pL of serum sample for overnight
protein precipitation.
Then it was centrifuged at 13000x g for 20 min. 50 pL of supernatant was
loaded into the center
of wells of a 96-deep well plate, followed by the addition of 3-
nitrophenylhydrazine (NPH)
reagent. After incubation for 2h, BHT stabilizer and water were added before
LC-MS injection.
Mass spectrometric analysis was performed on an ABSciex 4000 Qtrap tandem
mass
spectrometry instrument (Applied Biosystems/MDS Analytical Technologies,
Foster City, CA)
equipped with an Agilent 1260 series UHPLC system (Agilent Technologies, Palo
Alto, CA). The
samples were delivered to the mass spectrometer by a LC method followed by a
direct injection
(DI) method. Data analysis was done using Analyst 1.6.2.
lEINMR: Plasma and serum samples contain a significant concentration of large
molecular
weight proteins and lipoproteins which affects the identification of the small
molecular weight
metabolites by NMR spectroscopy. A deproteinization step, involving ultra-
filtration as previously
described (65), was therefore introduced in the protocol to remove plasma
proteins. Prior to
filtration, 3 KDa cut-off centrifugal filter units (Amicon Microcon YM-3),
were rinsed five times
each with 0.5 mL of H20 and centrifuged (10,000 rpm for 10 minutes) to remove
residual glycerol
bound to the filter membranes. Aliquots of each plasma sample were then
transferred into the
centrifuge filter devices and spun (10,000 rpm for 20 minutes) to remove
macromolecules
(primarily protein and lipoproteins) from the sample. The filtrates were
checked visually for any
evidence that the membrane was compromised and for these samples the
filtration process was
repeated with a different filter and the filtrate inspected again. The
subsequent filtrates were
collected and the volumes were recorded. If the total volume of the sample was
under 250 [IL an
appropriate amount from a 150 mM KH2PO4 buffer (pH 7) was added until the
total volume of
the sample was 173.5 pL. Any sample that had to have buffer added to bring the
solution volume
51
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
to 173.5 [IL, was annotated with the dilution factor and metabolite
concentrations were corrected
in the subsequent analysis. Subsequently, 46.5 [IL of a standard buffer
solution (54% D20:46%
1.75 mM KH2PO4 pH 7.0 v/v containing 5.84 mM DSS (2,2-dimethy1-2-silcepentane-
5-
sulphonate), 5.84 mM 2-chloropyrimidine-5 carboxylate, and 0.1% NaN3 in H20)
was added to
the sample. The plasma sample (250 [IL) was then transferred 3mm SampleJet NMR
tube for
subsequent spectral analysis. All 1H-NMR spectra were collected on a 700 MHz
Avance III
(Bruker) spectrometer equipped with a 5 mm HCN Z-gradient pulsed-field
gradient (PFG)
cryoprobe. lEINMR spectra were acquired at 25 C using the first transient of
the NOESY pre-
saturation pulse sequence (noesyldpr), chosen for its high degree of
quantitative accuracy (66).
All HD's (free induction decays) were zero-filled to 250 K data points. The
singlet produced by
the DSS methyl groups was used as an internal standard for chemical shift
referencing (set to 0
ppm) and for quantification all 1H-NMR spectra were processed and analyzed
using an in-house
version of the MAGMET automated analysis software package using a custom
metabolite library.
MAGMET allows for qualitative and quantitative analysis of an NMR spectrum by
automatically
fitting spectral signatures from an internal database to the spectrum. Each
spectrum was further
inspected by an NMR spectroscopist to minimize compound misidentification and
mis-
quantification. Typically, all of visible peaks were assigned. Most of the
visible peaks are
annotated with a compound name. It has been previously shown that this fitting
procedure provides
absolute concentration accuracy of 90% or better (67).
Population Statistics: Medians (IQRs) and frequency (%) were used to report
ICU patient
baseline characteristics for continuous and categorical variables,
respectively; continuous
variables were compared using Mann-Whitney U tests (or Kruskal-Wallis tests,
as appropriate),
and categorical variables were compared using Fisher's exact chi-square, with
P-values <0.05
considered statistically significant. Receiver operating characteristic (ROC)
curves were
conducted to determine sensitivity and specificity of individual metabolite
ratios for predicting a
binary outcome. Area-under-the-curve (AUC) was calculated as an aggregate
measure of
metabolite ratio performance across all possible classification thresholds.
All analyses were
conducted using SPSS version 26 (IBM Corp., Armonk, NY, USA).
52
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Machine Learning: COVID-19 analyte data were visualized with a nonlinear
dimensionality reduction on the full data matrix using the t-distributed
stochastic nearest neighbor
embedding (t-SNE) algorithm (68). t-SNE assumes that the 'optimal'
representation of the data
lies on a manifold with complex geometry, but low dimension, embedded in the
full dimensional
space of the raw data. For feature selection, the raw data for each subject
were ingested within
each feature, across subjects. A random forest classifier was trained on the
variables to predict
COVID-19 status or COVID-19 outcome. A random forest is a set of decision
trees and,
consequently, we were able to interrogate this collection of trees to identify
the features that have
the highest predictive value (viz., those features that frequently appear near
the top of the decision
tree). To reduce overfitting, COVID-19 status was determined using a six-fold
cross validation
with a random forest of ten trees, whereas, patient outcome was determined
using a three-fold
cross validation with a random forest of ten trees and max depth of 6 (69).
RESULTS
We investigated 10 COVID-19+ patients (median years of age=61.0, IQR= 54.8,
67.0), 10
age- and sex-matched COVID-19- patients (median years of age=58.0, IQR=52.5,
63.0), and 10
age- and sex-matched healthy controls (median years of age=57.5, IQR=52.8,
62.8; P=0.686).
Baseline demographic characteristics, comorbidities, laboratory values, and
chest x-ray findings
are reported in Table 1. The COVID-19- patients had significantly higher
unilateral pneumonia,
while COVD19+ patients were more likely to have bilateral pneumonia. Sepsis
was 'confirmed'
by infectious pathogen identification in only 20% of COVID-19- patients,
whereas sepsis was
'suspected' in the remaining 80%. A mortality rate of 40% was determined for
COVID-19+
patients.
We measured a total of 183 plasma metabolites using both DI-LC-MS/MS and 1I-1
NMR.
In the event of metabolite repeats measured with both techniques (21
metabolites), the 1I-1 NMR
metabolite repeat measurements were deleted from the combined metabolite
database yielding a
final number of 162 metabolites analyzed, which are listed in Table 13.
Fig. 1A shows a tSNE plot illustrating that the ICU day 1 COVID-19+ patient
metabolome
was distinct and easily separable from age- and sex-matched healthy control
subjects. In fact,
53
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
classification accuracy was 100% when comparing the 2 metabolomes. We then
identified the top
8 metabolites underlying these differences between cohorts and are shown in
Table 14 with their
associated % importance. In the COVID-19+ cohort, relative to the healthy
control subjects,
kynurenine increased 5.1-fold while arginine decreased 0.5-fold, sarcosine
decreased 0.6-fold and
lysophosphatidylcholines (LysoPCs) all decreased 0.3-fold on average. The
least number of
metabolites that were required to maintain a 100% classification accuracy
between cohorts was
then determined, with only arginine (cutoffs 52.8 [tM) and kynurenine (cutoff
> 3.1 [tM) required.
The excellent predictive ability of an arginine/kynurenine ratio for
discriminating a COVID-19
patient from a healthy control subject (cutoffs 15.7) is shown with ROC
analysis in Fig. 1B (AUC
1.00; P=0.0002).
A comparison of COVID-19+ and COVID-19- patient cohorts revealed distinct
metabolomes. Feature classification again identified kynurenine as one of the
leading metabolites
underlying the differences between COVID-19+ and COVID-19- cohorts (Table 15).
We then
determined that an arginine/kynurenine ratio again showed an excellent
discriminative ability to
determine COVID-19 status on ICU day 1 (5 cutoff < 11.6) via ROC analyses (AUC
0.98;
P=0.005; Fig. 2A). Fig. 2B shows an arginine/kynurenine ratio time plot for
the COVID-19+ and
COVID-19- patients over 10 ICU days. The cohorts' ratios were significantly
different on ICU
days 1 and 3 (P=0.005).
Fig. 3A shows a tSNE plot for COVID-19+ patients that either survived or died,
and
demonstrates that the outcomes were distinct and separable. To optimize
outcome prediction in
COVID-19+ patients, the number of metabolites were narrowed using feature
selection (Table 16).
Creatinine was the leading metabolite and it could predict death with 100%
accuracy on both ICU
days 1 (cutoff > 126 [tmo1/1) and 3 (cutoff > 174 [tmo1/1). To improve the
variation in patient
creatinine values, we then tested the ability of a creatinine/arginine ratio
to predict death; the
corresponding time plot is shown in Fig. 3B. Death could be predicted with
100% accuracy on
both ICU days 1 (cutoff > 3.4) and 3 (cutoff > 3.7) as the creatinine/arginine
ratios were
significantly different between COVID-19 patients that lived or died at both
time points (P=0.01).
There were no deaths during the 10 ICU days.
54
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
162 metabolites in plasma obtained from ICU patients were measured, both COV1D-
19+
and COV1D-19-, as well as age- and sex-matched healthy control subjects (see
Table 13). Given
the number of metabolites measured, the data was analyzed with machine
learning. The data
indicate the presence of a unique COV1D-19 plasma metabolome dominated by
changes in
kynurenine, arginine, sarcosine and LysoPCs. Moreover, either creatinine alone
or a
creatinine/arginine ratio predicted ICU mortality with 100% accuracy.
Previous work in these same patients have determined a unique inflammatory
profile
characterized by elevated TNF and serine proteases (70), and a thrombotic
profile associated with
endothelial activation and glycocalyx degradation (71). We have also
identified 6 novel protein
immune biomarkers that predict COV1D-19 associated death (72). Taken together
with the data
from this study, COV1D-19 represents a severe illness with a unique
pathophysiological signature,
as well as a high mortality rate. Indeed, in our cohort of COV1D-19 patients,
ICU death was 40%
with standardized ICU care.
The metabolites required for COV1D-19 diagnosis (arginine, kynurenine, and/or
arginine/kynurenine ratio) and outcome (either creatinine alone or
creatinine/arginine ratio) can be
easily measured using only mass spectrometry or immune assay, making their use
as COV1D-19
biomarkers affordable and easily available. Point-of-care analyses for these
metabolites could be
rapidly developed, such as a lateral flow immunochromatographic assay.
Moreover, the results
presented herein support the use of dietary supplementation of tryptophan,
arginine, sarcosine and
LysoPCs as adjunctive therapies for COV1D-19.
COV1D-19 status relied heavily on increased plasma kynurenine. The essential
amino acid
tryptophan is metabolized to elevate the energy producing cofactor
nicotinamide adenosine
dinucleotide, with kynurenine as the first stable intermediate to be formed
(73). Increased
degradation of tryptophan, with a consequential increase in kynurenine, occurs
during an immune
response and is driven by the release of interferon-gamma from activated T-
cells. COV1D-19
caused intense T-cell activation (74, 75) with an approximate 11-fold increase
in plasma
interferon-gamma in critically ill COV1D-19 patients (70).
While plasma kynurenine effectively discriminated COV1D-19+ patients from
healthy
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
control subjects, determination of COV1D-19 status in ICU patients required
further specificity
that was optimally provided by an arginine/kynurenine ratio. Arginine, an
amino acid precursor
for nitric oxide, was significantly depressed in COV1D-19+ patients. Arginine
depletion is likely
secondary to the intense requirement during COV1D-19 for nitric oxide
signaling and
antimicrobial activity (76), as well as consumption by the enzyme arginase 1
(ARG1) that
represents a macrophage immunoregulatory mechanism (77).
Sarcosine, an amino acid that helped discriminate COV1D-19+ patients from
healthy
control subjects, was also significantly depressed. While not superior to the
arginine/kynurenine
ratio for diagnosing COV1D-19 status, sarcosine sequestration may have a
critical role in COV1D-
19 pathology. Sarcosine enhances the activity of antigen presenting cells (78)
and activates
autophagy (79), or the body's removal of damaged cells and their
immunostimulatory debris. As
a protective catabolic process during COV1D-19, autophagy is critical to the
antiviral response by
direct elimination of virus, the presentation of viral antigens and the
inhibition of excessive
inflammation (80). Sarcosine levels decrease with age (79), and the elderly
are most susceptible
to COV1D-19 morbidity and mortality.
Depressed plasma LysoPCs also helped discriminate COV1D-19+ patients from
healthy
control subjects. The partial hydrolysis of phosphatidylcholines by
phospholipase A2 produces
LysoPCs, which are subsequently implicated in endothelial activation (81) and
phagocytosis of
cellular debris (82). Decreased plasma LysoPCs has been observed in sepsis
(83), where LysoPCs
may aid pathogen elimination, and therapeutic replacement has been suggested
to improve sepsis
outcome (84).
Acute renal dysfunction is strongly associated with high mortality in ICU
patients (85).
Plasma creatinine, a marker of renal dysfunction, was an excellent
discriminator for COVID-19
patients that either lived or died. In our COV1D-19+ cohort, 2 patients had
chronic kidney disease
and 2 patients required renal replacement therapy. The angiotensin-converting
enzyme 2 receptor
that is essential for SARS-CoV-2 uptake is highly expressed on tubule
epithelial cells (86). Acute
kidney injury is reported to occur in up to 37% of COVID-19 patients (87) and
is secondary acute
tubular injury from direct viral infection (88).
56
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
The data presented in this disclosure suggest that COVID-19 diagnosis
(arginine/kynurenine ratio) and outcome (creatinine alone or
creatinine/arginine ratio) can be
easily determined with point-of-care measurements of kynurenine, arginine and
creatinine, and
that this rapid and affordable biomarker approach may be complimentary to the
more expensive
and time-consuming diagnostic tools currently employed (e.g. polymerase chain
reaction and
antigen immunoassay). Moreover, dietary supplementation of tryptophan,
arginine, sarcosine and
LysoPCs can aid COVID-19 outcome as therapies or adjunctive therapies.
In summary, we report a unique metabolome in COVID-19+ ICU patients, with
identification of 3 metabolites that appear to be accurate
diagnostic/prognostic biomarkers for
future studies. Given the rapid spread of COVID-19 and the critical need for
rapid and affordable
diagnostics, our data may be invaluable for future testing. In addition, our
exploratory data may
be invaluable for guiding resource mobilization and/or goals of care
discussion, but only after
validation in larger COVID-19+ cohorts. Furthermore, patient stratification is
critically important
for future COVID-19 interventional trials.
Table 1 - Subject demographics and clinical data.
Healthy COVID19- COVID19+
Variable Controls Patients Patients
P-value
10 10 10
1.000
Age in years 57.5 (52.8, 62.8) 58.0 (52.5, 63.0) 61.0 (54.8,
67.0) 0.686
Sex 7F:3M 7F:3M 7F:3M
1.000
MODS 6.0 (3.8, 8.0) 4.0 (2.5,
7.3) 0.251
SOFA 7.5 (4.8, 11.0) 4.5 (2.8,
9.3) 0.160
Comorbidities
Hypertension 8 (80) 6 (60)
0.628
Diabetes 4 (40) 3 (30)
1.000
Chronic kidney disease 1(10) 2 (20)
1.000
Cancer 1(10) 2 (20)
1.000
57
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
COPD 1(10) 0(0) 1.000
Baseline Medications
Antiplatelet agents 6 (60) 2 (20) 0.170
Anticoagulants 1(10) 0 (0) 1.000
Baseline labs
WBC 15.3 (11.1,23.0) 8.5
(6.3, 16.1) 0.064
Neutrophils 12.2 (8.1, 15.2) 7.7
(5.7, 13.3) 0.197
Lymphocytes 1.6 (0.5, 2.3) 0.7 (0.6,
1.0) 0.141
Platelets 184 (159, 245) 206 (109,
294) 0.623
Hemoglobin 130 (104, 142) 122 (102,
136) 0.364
Creatinine 80 (54, 147) 107 (55,
288) 0.571
Chest X-ray findings
Bilateral pneumonia 1(10) 9 (90)
0.001*
Unilateral pneumonia 5 (50) 0 (0)
0.033*
Interstitial infiltrates 1(10) 1(10) 1.000
Normal 3 (30) 0 (0) 0.211
P:F ratio 172 (132, 304) 124 (69,
202) 0.153
Sepsis diagnosis
Suspected 8 (80) 0 (0)
0.001*
Confirmed 2(20) 10 (100)
0.001*
Interventions during study
Antibiotics 10 (100) 10 (100) 1.000
Anti-virals 0(0) 3 (30) 0.211
Steroids 3 (30) 2 (20) 1.000
Vasoactive medications 6 (60) 7 (70) 1.000
VTE prophylaxis 10 (100) 10 (100) 1.000
New antiplatelets 0(0) 1(10) 1.000
New anticoagulation 2 (20) 1(10) 1.000
58
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Renal replacement therapy 1(10) 2(20)
1.000
High-flow nasal cannul a 2 (20) 5 (50)
0.350
Non-invasive MV 8 (80) 6 (60)
0.628
Invasive MV 8 (80) 7 (70)
1.000
Patient Outcome
New VTE/ischemic stroke 2 (20) 1(10)
1.000
Survived 10 (100) 6(60)
0.087
Continuous data are presented as medians (IQRs), and categorical data are
presented as n (%).
MODS = Multiple Organ Dysfunction Score; SOFA= Sequential Organ Failure
Assessment Score;
COPD = Chronic Obstructive Pulmonary Disease; VTE = venous thromboembolism; MV
=
mechanical ventilation; VTE prophylaxis = number of patients receiving venous
thromboembolism prophylaxis with regular or low molecular heparin; new
antiplatelets = number
of patients who were started on aspirin or clopidogrel during ICU stay; new
anticoagulation =
number of patients who were started on therapeutic anticoagulation with
regular or low molecular
heparin, or novel oral anticoagulants.
59
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
Table 2 - Comparison of COVID19+ patients on ICU day 1 to healthy age- and sex-
matched control patients.
Ambito COVID-19+ Patients (n = 10)
Healthy Controls in =10) P
El astas e 2 40.2 (19.0, 69.9) 2.5 (1.7, 3.2)
<0.001
HSP70 208135 (14225a 318061) 26914 (24981.
30710) <0.001
IL-1 RA 123.84 (24.43. 1037.93) 4.30 (3.27, 4.77)
<0.001
IL-6 sal 3 (39.35, 306.70) 0.70 (0.30. 1.56)
<0.001
IL-8 a84 (5.67 18.64) 2.04 (1.48. 271)
<00O1
MCP-1 696.6 (439_9, 1093_2) 251.7 (209.0,
336.6) <0.001
MIG 10221 (6285, 41017) 1717 (1126, 2294)
<0.001
MMP8 2165 (1379, 4-173) 255 (128, 301)
<0.001
Resi sti n 39.15(30.26 118.81) 11.88 (9.23,
14.09) <0.001
TN F 194.4 (124.3, 251.8) 14.7 (10.3. 25.5)
<0.001
IL-10 44.26 (17.80, 170,55) 0 (0.4.95)
0.001
IL-18 141A- (84.6. 252.9) 34.63 (16.16,
44.92) 0.001
M-C SF 184.2 (127.6, 288.2) 21.7 (0, 38.0)
0.001
Granzyme B 9.61 (5,33. 23.12) 2.27 (1.65. 3.30)
0.002
Thrornbospondin-1 1294 (565, 2185) 188 (132460)
0.002
MI P-113 44.78 (35.88, 58.30) 31.09
(24.13,33.51) 0.003
MMP2 71040 (58159, 88142) 120458 (99649,
133271) 0.004
NGAL 117.5 (92.7, 506.7) 74.90 (62.92,
90.64) 0.004
IL-15 21.96 (12.78, 49.86) 6.69 (4.79, 9.33)
0.005
IFN-y 1 al 5 (7.82 144.80) 1.69 (0, 4.91)
0.006
Table 3 - Comparison of COVID19+ and COVID19- ICU patients (days 1-3).
COVID19- Patients
Analyte ICU Day COVID19+ Patients (n=10) (n=10)
P-value
TNF 1 194.4 (124.3, 251.8) 21.0 (6.4, 40.5)
<0.001*
2 141.2 (103.7, 216.9) 18.0 (9.0, 39.8) <0.001*
3 149.2 (94.8, 206.6) 16.4 (2.0, 44.6) 0.001*
Granzyme 1 9.61 (5.33, 23.12) 1.51 (1.11,2.98)
<0.001*
B
2 7.97 (5.72, 13.63) 1.26 (0.74, 1.94) <0.001*
3 8.31 (5.02, 12.98) 0.89 (0.63, 1.80) <0.001*
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
HSP70 1 208135 (142253, 318061) 106995 (32750, 0.002*
116705)
2 206109 (134528, 308362) 65791 (40991, 116411) 0.002*
3 236000 (130712, 362185) 62810 (31960, 100732) 0.001*
11,18 1 141.4 (84.6, 252.9) 33.8 (17.5, 64.3) 0.005*
2 140.8 (86.8, 205.0) 31.8 (10.8, 65.7) 0.001*
3 123.7 (110.0, 189.8) 48.2 (21.5, 69.8) <0.001*
1P10 1 3526 (1407, 19503) 165 (41, 371) 0.023
2 2496 (1081, 80080) 94 (54, 381) 0.004*
3 4289 (1108, 40564) 149(65, 526) <0.001*
Elastase 2 1 40.2 (19.0, 69.9) 21.6 (15.6, 35.0) 0.290
2 71.78 (33.92, 92.39) 14.19 (10.31, 23.04) 0.002*
3 75.00 (43.24, 121.40) 24.98 (10.65, 34.78) 0.002*
1L10 1 44.26 (17.80, 170.55) 14.56 (2.90, 28.17) 0.010*
2 46.95 (23.29, 156.33) 4.21 (0.14, 11.03) 0.001*
3 34.26 (23.16, 81.40) 2.23 (0, 9.42) <0.001*
MIG 1 10221 (6285, 41017) 2116 (1268, 3975) 0.001*
2 11180 (4948, 37212) 2684 (1089, 4606) 0.002*
3 12237 (5795, 33781) 1929 (998, 6329) 0.003*
M-CSF 1 184.2 (127.6, 288.2) 13.1 (0, 66.1) 0.002*
2 162.1 (91.2, 311.3) 26.2 (0, 102.0) 0.004*
3 149.2 (107.6, 284.1) 16.0 (0, 101.3) 0.003*
IFNg 1 18.15 (7.82, 144.80) 0 (0, 1.25) 0.001*
2 7.36 (5.56, 26.24) 0 (0, 2.04) 0.001*
3 7.95 (5.42, 48.60) 0.43 (0, 3.35) 0.005*
11,8 1 8.84 (5.67, 18.64) 2.51 (1.13, 4.45) 0.001*
2 6.51 (2.96, 10.06) 1.93 (1.43, 3.32) 0.005*
3 6.58 (4.68, 12.21) 1.67 (0.65, 4.42) 0.010*
61
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
MMP8 1 2165 (1379, 4173) 1666 (910, 4323)
0.364
2 4308 (2149, 7385) 1219 (521, 2753) 0.008*
3 5745 (2624, 10956) 1812 (460, 2503) 0.002*
IL2 1 0.33 (0, 1.87) 0 (0, 0)
0.005*
2 0(0, 0.57) 0(0, 0) 0.031
3 0.43 (0, 0.75) 0 (0, 0) 0.005*
IL15 1 21.96 (12.78, 49.86) 6.86 (2.69, 11.97)
0.008*
2 16.40 (9.18, 31.76) 8.06 (3.17, 13.17) 0.016
3 17.10 (10.37, 44.19) 5.49 (2.86, 12.69) 0.010*
IL-1RA 1 123.84 (24.43, 1037.93) 12.18 (5.69, 29.66)
0.019
2 36.55 (22.41, 622.70) 5.72 (3.26, 51.79) 0.019
3 55.44 (21.86, 321.47) 6.67 (2.27, 16.22) 0.003*
MMP1 1 1065 (414, 2330) 804 (474, 1349)
0.326
2 1424 (828, 3111) 670 (301, 1115) 0.034
3 1679 (800, 3382) 704 (490, 1211) 0.007*
MCP-1 1 696.6 (439.9, 1093.2) 356.7 (215.4, 481.7)
0.007*
2 548.0 (453.3, 938.6) 327.1 (193.2, 530.0) 0.023
3 767.6 (498.3, 1032.4) 237.6 (118.6, 748.4) 0.041
Only analytes with statistically significant data on one or more days are
shown. Data are presented
as medians (IQRs) with analyte concentration in pg/ml (*p<0.01). Analytes are
ordered by top 6
as found through machine learning analysis, then analytes with all 3 days
significant most to least
by day 3, then analytes significant on days 2 and 3 only most to least by day
3, then analytes
significant on days 1 and 3 only most to least by day 3, then analytes most
to least significant on
day 3 only, then analytes significant on day 1 only.
62
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Table 4 - Summary of receiver operating characteristic curve (ROC) analyses
for
predicting Death in COVID10. All plasma analytes were measured on ICU Day 1.
Variable AUC: 950..6 CI P-value
HSP70 1.00 1_00 - 1_00 0.002*
IL-1RA LOO 1_00 - 1_00 0_002*
IL10 1_00 LOU - LOO 0_002*
MIG 1.00 1.00 - 1.00 0.002*
M-CSF 0.98 0.94 - LOO 0.003*
IL6 0.97 0.89 - 1.00 0.005*
IFNg 0_95 0_86 - LOO 0_006*
IL8 0.95 0.86 - 1.00 0.006*
T1CFa 0.95 0.85 - 1.00 0.006*
MCP-1 0.94 0.83 - LOO 0.008*
P-selectin 0.94 0.82 - 1.00 0.008*
NBIP10 0_91 0_77 - 1_00 0.014
MHP-1i3 0.88 0.70 - 1.00 0.023
nastase 2 0.86 0.66 - 1.00 0.030
IL15 0.86 0.68 - LOO 0.030
NEAPS 0_86 0_67 - LOO 0.030
NGAL 0.86 0_66 - 1_00 0.030
Resistm 0.86 0.63 - 1_00 0.030
63
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
IL2 0.85 0.57 - LOU 0.033
IL3 0.82 0.54 - LOU 0.053
Chondroitin 0_81 0_62 - 1_00 0.059
I'vEMP 2 0_81 0.58- LOU 0.059
Lactofernii 0.80 0.48 - LOU 0.073
IL12(p40) 0.78 0.57 - 0.99 0.089
IL18 0.78 0.51 - 1.00 0.089
NLMIP7 0_78 0.51- 1_00 0.089
v-WF 0.78 0.49 - LOU 0.089
Heparan. 0.77 0.53 - 1.00 0.098
MIP-1a 0.77 0.47 - 1.00 0.098
N'L\1133 0.75 0.46 - 1.00 0.131
Eotaxm 0_73 0.46 - 1.00 0.156
Granrone B 0_73 0_49 - 0_98 0.156
Protem C 0.73 0.48 - 0_99 0.156
NEAP13 0.73 0.51 - 0.95 0.171
IL12(p70) 0.71 0.41 - 1.00 0.202
G-CSF 0.70 0.34 - 1.00 0.219
IL17F 0_70 0_46 - 0_94 0.219
IFNa2 0.70 0.47 - 0.92 0.238
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
IL5 0_69 0_46 - 0_92 0.757
Syn.dec an-1 0.69 036- 1.00 0.257
TN-Fp 0.69 0.36- LOU 0.757
IP10 0.68 028- LOU 0.277
NLMP9 0.67 0.33 - 1.00 0.299
Hyahironic acid 0.66 034 - 0.97 0.345
NEAP1 0.66 0.36 - 0.95 0.345
Thrombospondth-1 0.66 0.40 - 0.91 0.345
EGF 0.64 0.32 - 0.96 0.395
RANTES 0.64 035 - 0.93 0.395
IL7 0.63 030 - 0.95 0.450
NniiP 12 0.63 0.24- 1.00 0.450
IL 1 a 0.62 0.34 - 0.90 0.479
PDGF-AA 0.61 035 - 0.87 0.508
GM-CSF 0.60 0.25 - 0.95 0.539
'up 0.60 030 - 0.90 0.539
IL17A 0.60 0.29 - 0.91 0.539
IL4 0.59 0.28 - 0.91 0.571
PDGF-ARIBB 0_59 024 - 0_95 0.571
ADAMTS13 0.58 0.30 - 0.86 0.637
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
0.56 0.23 - 0.90 0.705
VEGFA 0_56 025 - 0_87 0.705
IL17E1125 0.53 0.24 - 0.82 0.850
IL13 0.52 0.18 - 0.85 0.925
Table 5 - ICU Day 1 analytes predict COVID19 patients versus healthy controls.
Unipro
Num Assay ID Function
TYMP P19971 Thymidine phosphorylase also called
platelet-
derived
endothelial cell growth factor is an
1. intracellular protein whose major source is
platelets. It may be involved in platelet
activation and its secreted metabolites may
potentiate thrombosis.
CXCL10 P02778 Induced by IFNy, produced by
endothelial
cells, monocytes, fibroblasts and
keratinocytes. Agonist for CXCR3 which is
2. expressed on some T, B, and NK cells.
Promotes Thl recruitment induces T cell
adherence to endothelial cells,
chemoattractant for monocytes, T cells, NKs.
ClQA P02745 Complement Clq subunit A is one of 3
3. subunits making up Clq, part of the classical
complement system.
AGR2 095994 Anterior gradient protein 2 homolog is
a
member of the protein disulfide isomerase
4. family normally located in the endoplasmic
reticulum of intestinal cells, as well as the
lung, stomach, colon and prostate; tissues
with mucus secreting or endocrine functions.
IL-18R1 Q13478 One of the heterodimers of the IL-18
receptor
5. complex, it is a type I transmembrane protein.
Expressed on NKs, T cells especially
66
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
activated Thl cells; it induces IFNy in concert
with IL-12 receptor.
CDON Q4KMGO Cell adhesion molecule-related, down-
regulated by oncogenes is a transmembrane
6. glycoprotein that acts as a cell adhesion
molecule and binds members of the hedgehog
family. It seems to normally be involved in
development and proliferating cells.
DDX58 095786 Retinoic acid-inducible gene 1, also
called
DEAD box protein 58, is a transmembrane
pattern recognition receptor that recognizes
7. viral replicative intermediates in the cytosol
during RNA virus infections. It is expressed
in endothelial cells and activates the type 1
interferon response.
CLEC6A Q6EIG7 C-type lectin domain family 6 member A,
also called dectin-2 is a transmembrane
pattern-recognition receptor highly expressed
8. on macrophages as well as monocytes,
Kupffer, Langerhans and some dendritic
cells. It binds surface polysaccharides of
pathogens and ultimately causes cytokine
production to direct a Th17 response.
CLM-6 Q08708 CMRF35-like molecule 6 also called
leukocyte mono-immunoglobulin-like
receptor 8 (LMIR8) and CD300c is a
9. transmembrane receptor expressed on almost
all leukocytes and plasmacytoid dendritic
cells it recognizes phosphatidylethanolamine
on apoptotic cells.
PXN P49023 Paxillin is a cytosolic scaffolding
protein
10. involved in focal adhesions and integrin-
mediated signal transduction.
LAG3 P18627 Lymphocyte activation gene 3 is a
11. transmembrane receptor expressed on several
types of T cells that regulates their function.
APLP1 P51693 Amyloid-like protein 1 is expressed
12 exclusively in the central nervous
system and
is a cytosolic protein thought to be involved
in cell-cell contacts in synapses.
13. LIF-R P42702 Leukemia inhibitory factor receptor is
a
transmembrane protein that along with gp130
67
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
forms receptor for LIE, a member of the IL-6
family. LIF-R is expressed in several organs
as well as monocytes and macrophages.
B4GALT1 P15291 Beta-1,4-galactosyltransferase 1 is a
trans-
golgi membrane protein that transfers a sugar
14 nucleotide to acceptors. When on the
plasma
membrane, it functions as a cell-adhesion
molecule involved in cell-cell and cell-matrix
interaction.
ASGR1 P07306 1 of two subunits of the
asialoglycoprotein
receptor, which is mainly expressed in liver,
that facilitates uptake of desialylated
15. glycoproteins It is a member of the C-
type
lectin family of receptors and can clear
hyposialylated vonWillebrand factor from
plasma.
CREVI1 Q9NZV1 Cysteine-rich motor neuron 1 is a
16 transmembrane receptor that regulates
growth
factor signaling in a number of organs during
organogenesis.
CD300E Q496F6 Also called CLM-2, it is a
transmembrane
receptor expressed on the surface of
17 monocytes and circulating myeloid
dendritic
cells. It appears to be involved in inducing
cytokine release, reactive oxygen species
production.
CDKN1A P38936 Cyclin-dependent kinase inhibitor 1
also
called p21 and CIP1 is a nuclear and
18. cytoplasmic protein which can arrest the cell
cycle due to DNA damage and is anti-
apoptotic.
CXCL11 014625 A ligand for CXCR3 which is expressed
on
19. some T, B, and NK cells. It promotes Thl
recruitment/chemotaxis.
IL6 P05231 A key pro-inflammatory cytokine it can
directly stimulate cells through its membrane-
bound receptor on hepatocytes, neutrophils,
20. monocytes, and some lymphocytes. In
concert with its soluble receptor, it can also
stimulate a wide-variety of cells that
expresses gp130.
68
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
CLM - CMRF35-like molecule
CXCR ¨ CXC receptor
IFNy ¨ interferon gamma
IL ¨ interleukin
NK ¨ natural killer cell
Thl ¨ type 1 T-helper cell
Table 6 - ICU Day 1 analytes that predict COVID19 status.
Unipro
Num Assay ID Function
DDX58 095786 Retinoic acid-inducible gene 1, also called
DEAD box
protein 58, is a transmembrane pattern recognition
1. receptor that recognizes viral replicative intermediates
in the cytosol during RNA virus infections. It is
expressed in endothelial cells and activates the type 1
interferon response.
RR1VI2B Q7LG56 Ribonucleoside-diphosphate reductase
subunit M2 B
2. also called p53R2, is a nuclear protein thought to be
involved in DNA repair after damage.
1RF9 Q00978 Interferon regulatory factor 9 is a nuclear
transcription
3. factor that is part of a transcription complex that
responds to type 1 interferon signaling.
NPM1 P06748 Nucleophosmin is a nucleolar protein found
in
proliferating cells, it has functions in mitotic spindle
4. assembly, ribosome synthesis, DNA repair,
embryogenesis and chromatin remodeling. It can bind
HEXEVI1 (see entry 10 below).
MCP-3 P80098 Monocyte chemotactic protein 3, also called
C-C motif
chemokine 7 (CCL7), is the ligand for CCR1, CCR2,
5.
CCR3 and CCR5. It attracts monocytes, T cells, NKs
immature DCs, basophils and eosinophils.
Gal-9 000182 Galectin-9 is a pattern recognition
receptor that binds 13-
galactosides. Its expressed on various immune cells,
6. especially T cells, and expression is increased in
response to various stimuli such as mitogen, TLR
activation, pro-inflammatory cytokines and viral
infection.
NADK 095544 nicotinamide adenine dinucleotide (NAD)
kinase is a
7. ubiquitously expressed cytosolic protein that
phosphorylates NAD.
8.
BRK1 Q8WUW1 Brickl also called Hspc300, is a cytoskeleton-
associated component of the Wave protein complex.
69
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
PFDN2 Q9UHV9 Prefoldin subunit 2 is a cytosolic protein, part of
the
9. hexameric prefoldin complex that captures unfolded
proteins and transfers them to a chaperonin.
HEXIIVI1 094992 Hexamethylene bis-acetamide-inducible protein 1
is a
nuclear transcriptional regulator. It can bind to
10. nucleophosmin (see entry 4 above) and hypoxia-
inducible factor a; may be recruited by NE-KB for
transcription of inflammation-responsive genes.
TCN2 P20062 Transcobalamin II is a serum protein that binds
vitamin
11. B12 for transport. It is synthesized in the intestinal
mucosa, liver, seminal vesicles, fibroblasts, bone
marrow, and macrophages.
12. BLM Q13867 Bleomycin hydrolase is a
cytosolic cysteine
hydrolase aminopeptidase which breaks down homocysteine.
13. KRT19 P08727 Keratin 19 also called
cytokeratin 19 is a cytoskeletal
protein expressed in laminated epithelium.
FUS P35637 Fused in sarcoma is a ubiquitously expressed
nuclear
14. RNA and DNA binding protein, though it also is found
in the cytoplasm.
RC0R1 Q9UKLO REST corepressor 1 is a nuclear protein that is
part of
15. protein complexes that modify chromatin to repress
gene expression.
PSME1 Q06323 Proteasome activator complex subunit 1, also
called
PA28a, is a subunit of PA(proteasome activator)28 that
16. binds to and activates the 20S proteasome. It is a
cytosolic protein expressed in most tissues and induced
by IFNy.
17. CXCL11 014625 A ligand for CXCR3 which is
expressed on some T, B,
and NK cells. It promotes Thl recruitment/chemotaxis.
CLSPN Q9HAW4 Claspin is a nuclear protein that associates with
DNA
18 replication stalled due to DNA damage. It's
expression
is tightly regulated during the cell cycle with high levels
in late S phase and G2.
S100A11 P31949 A cytosolic calcium-binding protein of the S100
family,
it is involved in growth arrest in contact inhibition. It is
19 expressed in a wide variety of cells and is
secreted by
an unconventional pathway. It is involved in cell-cell
contacts and can promote cell migration in response to
hypoxia-induced mitogenic factor.
CDON Q4KMGO Cell adhesion molecule-related, down-regulated by
20. oncogenes is a transmembrane glycoprotein that
acts as
a cell adhesion molecule and binds members of the
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
hedgehog family. It seems to normally be involved in
development and proliferating cells.
CXCR ¨ C-X-C motif receptor
DC ¨ dendritic cell
DEAD ¨ Asp-Glu-Ala-Asp
IFNy ¨ interferon gamma
NF-KB ¨ nuclear factor kappa B
NK ¨ natural killer cell
REST ¨ repressor for element-1 silencing transcription
Th# ¨ type # T-helper cell
TLR ¨ toll-like receptor
Table 7 - ICU Day 1 analytes that predict outcome.
Num Assay Unipro ID Function
CXCL9 Q07325 A chemokine produced by airway epithelial
cells in
response to infection, also induced by IFNy and in
1. endothelial cells by TNFa. An agonist for CXCR3 on T
cells and natural killer cells. Promotes NK, Thl, monocyte,
DC, neutrophil, and eosinophil recruitment.
ICOSLG 075 144 Inducible costimulator ligand is expressed by
B cells,
2. monocytes, DCs T cells and endothelial cells; TNFa is
required for induction. It activates the inducible
costimulator in the thymus and on activated T cells.
CLM- 1 Q8TDQ1 CMRF35-like molecule 1 is a receptor for
3. phosphatidylserine presented on the outer membrane
surface of apoptotic cells, that promotes macrophage and
inhibits DC efferocytosis.
IL 12RB 1 P42701 One of the two subunits that compose the IL-
12 receptor, its
signaling pathway activates STAT4. Expressed primarily
4. on activated T cells and NK cells, less so on dendritic cells
and some B-cells. This subunit is also shared with the IL-23
receptor.
CD83 Q01151 Expressed on B and T cells, monocytes, DCs,
microglia and
neutrophils, and has soluble and membrane-bound forms.
5. Membrane-bound CD83 is essential for CD4+ T cell
development and inhibiting autoimmunity, soluble CD83
induces regulatory mechanisms for tolerance.
CA12 043570 Carbonic anhydrase 12 is membrane-associated
6. glycoprotein that catalyzes the reversible hydration of
carbon dioxide. CA12 is up-regulated by hypoxia, at least in
71
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
tumor environments and CA activity is associated with
sleep apnea-related hypoxemia.
FLRT2 043155 Fibronectin leucine rich transmembrane protein 2
was first
7. discovered in a screen for extracellular matrix proteins and
participates in homotypic cell-cell adhesion and with
fibroblast growth factor receptor.
ROR1 Q01973 A transmembrane receptor tyrosine kinase that is
activated
by Wnt family ligands and is mainly thought to be involved
8. in organ/tissue genesis during development. Though recent
evidence suggests it may be involved in pro-inflammatory
p65 activation, at least in cancer.
11,32 P24001 Expressed by PBMCs, epithelial cells and NKs, it up-
9 regulates other pro-inflammatory cytokines and has
several
isoforms. Airway epithelial cell production is increased by
viral infections and oxididative stress.
NCS1 P62166 Neuronal calcium sensor 1 is a cytosolic protein
involved in
several cellular functions through binding partners and
intracellular Ca2+ regulation. It is highly expressed in
neurons, but is not neuron-specific.
S100A11 P31949 A cytosolic calcium-binding protein of the S100
family, it is
involved in growth arrest in contact inhibition. It is
11 expressed in a wide variety of cells and is
secreted by an
unconventional pathway. It is involved in cell-cell contacts
and can promote cell migration in response to hypoxia-
induced mitogenic factor.
ANGPTL7 043827 Angiopoietin-like protein 7 is an orphan ligand,
but appears
to be involved in hematopoietic stem cell regulation and
12. self-renewal. Its serum concentration is higher in
obese
subjects compared to non-obese controls and can be
lowered with exercise.
CLMP Q9H6B4 coxsackievirus and adenovirus receptor-like
membrane
13 protein is a transmembrane glycoprotein involved in
homophilic cell-cell adhesion and is expressed in a wide
variety of tissues.
IGF1R P08069 A tyrosine kinase receptor expressed on T and B
cells,
macrophages, NK cells and granulocytes where its ligands,
14. insulin-like growth factor 1 and 2, causes various effects
such as proliferation, cytokine production and
priming/activation.
15. TOP2B Q02880 DNA topoisomerase II beta is
expressed in a wide variety of
tissues and throughout the cell cycle. Mostly found in the
72
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
cell's nucleus, it is one of the enzymes that catalyzes
topological changes in DNA.
FAM3B P58499 Also called pancreatic derived factor
(PANDER), it is
16 highly expressed in pancreatic islets and
high serum levels
are associated with the progression of metabolic syndrome
and type 2 diabetes.
11,10.1 P22301 An important anti-inflammatory cytokine,
expressed in
17. virtually all immune cells except plasmacytoid DCs, to limit
immune responses and prevent host damage.
18. ILlO P22301 See "Th10.1" above.
THY 1 P04216 Thymocyte differentiation antigen 1 is a
glycoprotein
expressed on the outer surface of many cell types including
19. fibroblasts, T cells and activated endothelial cells and has a
soluble form. Its function is cell and tissue-dependent, but is
pro-fibrotic in pulmonary fibroblasts in pulmonary fibrosis.
PVRL4 Q96NY8 Poliovirus receptor-related protein 4 also
called nectin-4 is a
20. cell-cell adhesion molecule in aherens junctions,
overexpressed in several cancers.
OPTC Q9UBM4 Opticin is an extracellular matrix protein
associated with
21. collagen in the vitreous humor where it binds heparan and
chondroitin sulfate. It is an anti-angiogenic factor in retinas.
CXCR ¨ CXC receptor
DC ¨ dendritic cell
IFNy ¨ interferon gamma
IL ¨ interleukin
IFNy ¨ interferon gamma
NK ¨ natural killer cell
PBMC ¨ peripheral blood monocyte cell
STAT ¨ signal transducer and activator of transcription
Th# ¨ type # T helper cell
TNFa ¨ tumor necrosis factor alpha
Table 8 - ICU Day 3 analytes that predict outcome.
Num Assay Uniprot ID Function
One of the two subunits that form the IL-12 receptor.
1 IL12RB1 P42701 Expressed primarily on activated T cells and
NK cells, less
.
so on dendritic cells and some B-cells. This subunit is also
shared with the IL-23 receptor.
2 CLM-1 Q8TDQ1 CMRF35-like molecule 1 also called CD300f,
is a receptor
.
for phosphatidylserine presented on the outer membrane of
73
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
apoptotic cells that promotes macrophage and inhibits DC
efferocytosis.
A chemokine produced by airway epithelial cells in
response to infection, also induced by IFNy and in
3. CXCL9 Q07325 endothelial cells by TNF. An
agonist for CXCR3 on T
cells and natural killer cells. Promotes NK, Thl,
monocyte, DC, neutrophil, and eosinophil recruitment.
Family with sequence similarity 3, also called pancreatic
derived factor (PANDER), is a cytokine-like protein that is
4. FAM3B P58499 highly expressed in pancreatic
islets and high serum levels
are associated with the progression of metabolic syndrome
and type 2 diabetes.
Opticin is an extracellular matrix protein associated with
5. OPTC Q9UBM4 collagen in the vitreous humor
where it binds heparan and
chondroitin sulfate. It is an anti-angiogenic factor in
retinas.
Thymocyte differentiation antigen 1 is a glycoprotein
expressed on the outer surface of many cell types
6 THY 1 P04216 including fibroblasts, T cells and activated
endothelial
.
cells and has a soluble form. Its function is cell and tissue-
dependent, but is pro-fibrotic in pulmonary fibroblasts in
pulmonary fibrosis.
Inducible costimulator ligand is a transmembrane protein
expressed by B cells, monocytes, DCs T cells and
7. ICOSLG 075144 endothelial cells; TNFa is
required for induction. It
activates the inducible costimulator in the thymus and on
activated T cells.
A tyrosine kinase receptor expressed on T and B cells,
macrophages, NK cells and granulocytes where its ligands,
8. IGF1R P08069 insulin-like growth factor 1
and 2, causes various effects
such as proliferation, cytokine production and
priming/activation.
An important anti-inflammatory cytokine, expressed in
9. 1 P22301
virtually all immune cells except plasmacytoid DCs, it
IL10.
works to limit immune responses and prevent host
damage.
coxsackievirus and adenovirus receptor-like membrane
10. CLMP Q9H6B4 protein is a transmembrane
glycoprotein involved in
homophilic cell-cell adhesion and is expressed in a wide
variety of tissues.
11. IL10 P22301 See "Th10.1" above.
74
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
Expressed on B and T cells, monocytes, DCs, microglia
and neutrophils, and has soluble and membrane-bound
12. CD83 Q01151 forms. Membrane-bound CD83 is
essential for CD4+ T
cell development and inhibiting autoimmunity, soluble
CD83 induces regulatory mechanisms for tolerance.
A transmembrane receptor tyrosine kinase that is activated
by Wnt family ligands and is mainly thought to be
13. ROR1 Q01973 involved in organ/tissue genesis
during development.
Though recent evidence suggests it may be involved in
pro-inflammatory p65 activation, at least in cancer.
Poliovirus receptor-related protein 4 also called nectin-4 is
14. PVRL4 Q96NY8 a cell-cell adhesion molecule
in aherens junctions,
overexpressed in several cancers.
Expressed by PBMCs, epithelial cells and NKs, it up-
15 IL32 P24001 regulates other pro-inflammatory cytokines and has
several
.
isoforms. Airway epithelial cell production is increased by
viral infections and oxididative stress.
Carbonic anhydrase 12 is membrane-associated
glycoprotein that catalyzes the reversible hydration of
16. CA12 043570 carbon dioxide. CA12 is up-
regulated by hypoxia, at least
in tumor environments and CA activity is associated with
sleep apnea-related hypoxemia.
Neuronal calcium sensor 1 is a cytosolic protein involved
in several cellular functions through binding partners and
17. NCS1 P62166
intracellular Ca 2+ regulation. It is highly expressed in
neurons, but is not neuron-specific.
Fibronectin leucine rich transmembrane protein 2 was first
18 FLRT2 043155 discovered in a screen for extracellular matrix
proteins and
.
participates in homotypic cell-cell adhesion as well as
fibroblast growth factor receptor signaling.
A cytosolic calcium-binding protein of the S100 family, it
is involved in growth arrest in contact inhibition. It is
19 Si0All P31949 expressed in a wide variety of cells and is
secreted by an
.
unconventional pathway. It is involved in cell-cell contacts
and can promote cell migration in response to hypoxia-
induced mitogenic factor.
DNA topoisomerase II beta is expressed in a wide variety
20 TOP2B Q02880 of tissues and throughout the cell cycle. Mostly
found in
.
the cell's nucleus, it is one of the enzymes that catalyzes
topological changes in DNA.
21. ANGPTL7 043827 Angiopoietin-like protein 7 is an orphan ligand,
but
appears to be involved in hematopoietic stem cell
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
regulation and self-renewal. Its serum concentration is
higher in obese subjects compared to non-obese controls
and can be lowered with exercise.
CD ¨ cluster of differentiation
CXCL ¨ CXC ligand
CXCR ¨ CXC receptor
DC ¨ dendritic cell
IFNy ¨ interferon gamma
IL ¨ interleukin
IFNy ¨ interferon gamma
NK ¨ natural killer cell
PBMC ¨ peripheral blood monocyte cell
STAT ¨ signal transducer and activator of transcription
Th# ¨ type # T helper cell
TNFa ¨ tumor necrosis factor alpha
Table 9 - Comparison of COVID-19+ patients on ICU day 1 to healthy age- and
sex-
matched control patients.
Variable Healthy Controls COVID-19+ Patients
P-value
Thrombosis factors
ADAMTS13 (ng/ml) 788 (609, 1075) 633 (463, 794) 0.151a
Protein C ( g/m1) 12.1 (3.2, 15.5) 6.2 (0.9, 10.5)
0.131b
vWF (ng/ml) 1536 (927, 3320) 7018 (4916, 22821)
<0.001
Endothelial injury
sP-selectin (ng/ml) 20.7 (16.2, 43.4) 33.3 (16.2, 44.5)
0.623'
Heparan Sulfate (ng/ml) 2.4 (2.0, 4.5) 3.4 (2.7, 9.8)
0.450d
Chondroitin Sulfate 1.6 (1.0, 5.0) 7.0 (5.1, 11.0)
(pg/ml) 0.004
Hyaluronic acid (ng/ml) 54.6 (29.3, 73.4) 307.7 (39.8, 633.7)
0.226'
Syndecan-1 (ng/ml) 76.0 (26.3, 97.6) 181.9 (103.6, 313.3)
0.004
76
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Data are presented as medians (IQRs). vWF = von Willebrand factor. Based on
measured values,
the following number of patients per cohort would be required to potentially
reach statistical
significance (80% power): a40; h30; '76136; d121; '13.
Table 10 - Comparison of COVID-19- and COVID-19+ patients on ICU days 1-3.
ICU COVID-19- COVID-19+
Variable Day Patients Patients P-
value
Thrombosis factors
ADAMST13 (ng/ml) 1 713 (357, 1005) 633 (463,
794) 0.880a
2 626 (343, 885) 637 (581,
862) 0.597h
3 661 (397, 968) 616 (526,
733) 0.940'
Protein C ( g/m1) 1 10.2 (2.2, 11.9) 6.2 (0.9, 10.5)
0.257d
2 8.8 (1.3, 12.1) 8.1 (0.6, 9.8)
0.406'
3 11.2 (1.2, 14.3) 6.0 (0.9, 12.1)
0.326f
vWF (ng/ml) 1 7203 (3718, 14500)
7018 (4916, 22821) 0.199g
2
7319 (4067, 13450) 11833 (4422, 21872) 0.406'
10027 (5747, 7128 (4410,
21614)
3 15794)
0.6501
Endothelial injury
sP-selectin (ng/ml) 1 23.7 (16.0, 34.1) 33.3
(16.2, 44.5) 0.4061
2 30.1 (16.4, 42.0) 25.7
(17.9, 29.3) 0.496'
3 22.0 (16.5, 31.6) 47.0
(25.0, 57.8) 0.028
Heparan Sulfate (ng/ml) 1 2.8 (1.5, 4.2) 3.4 (2.7, 9,8)
0.1861
2 1.8 (1.2,4.1) 4.3 (2.4, 8.2)
0.049
3 1.9 (1.2, 2.6) 3.2 (1.9, 6.0)
0.070'
Chondroitin Sulfate 4.4 (3.4, 10.7) 7.0 (5.1, 11.0)
(pg/ml) 1
0.082n
77
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
2 5.2 (2.3, 13.7) 5.9 (4.9, 6.5)
0.762
3 5.2 (2.8, 7.0) 7.0 (5.1, 9.6)
0.226P
Hyaluronic acid (ng/m1) 1 71.7 (28.7, 153.2) 307.7
(39.8, 633.7) 0.315q
2 107.1 (39.4, 192.1)
340.5 (58.9, 745.5) 0.199'
3 78.5 (37.9, 168.1) 354.9
(67.6, 874.2) 0.010
Syndecan-1 (ng/m1) 1 46.9 (1.9, 89.0) 181.9
(103.6, 313.3) 0.010
2 53.6 (13.5, 101.4) 296.7
(142.2, 743.4) 0.005
3 54.0 (3.2, 98.8) 413.5
(139.7, 755.9) 0.004
Data are presented as medians (IQRs). vWF = von Willebrand factor. Based on
measured values,
the following number of patients per cohort would be required to potentially
reach statistical
significance (80% power): al111; h1972; '3568; d184; '1266; f73; g115;
h60;13236; J167; k110;132;
m22; P29195; '277; P42; '124; r20.
Table 11 - Comparison of Healthy Controls, COVID-19- and COVID-19+ patients on
ICU
day 3.
Healthy COVID-19- ICU COVID-19+ ICU P-
Post-Hoc
Controls Day 3 Day 3 value Analysis
sP-selectin 20.7 (16.2' 22.0 (16.5, 31.6) 47.0 (25.0,
57.8) 0.080 N/A
43.4)
HC vs
CoV+ =
Hyaluronic 54.6 (29.3, 354.9 (67.6,
0.004
78.5 (37.9, 168.1) 0.003
acid 73.4) 874.2) CoV- vs
CoV+ =
0.012
HC vs
CoV+ =
Syndecan- 76.0 (26.3, 413.5 (139.7
0.004
54.0 (3.2, 98.8) ' 0.002
1 97.6) 755.9) CoV- vs
CoV+ =
0.003
78
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Table 12
Plasma Analyte Units Healthy Controls: CI: 5%, Case
Patient
95%
1. MMP7 pg/m1 3356,
4424 51788
2. IP-10 pg/m1 86,242
1098
3. Resistin pg/m1 7.3,
11.1 41.8
4. IL-3 pg/m1 0.1,2.4
7.3
5. Hyaluronic acid (EC) ng/ml 17.6,
40.4 119.2
6. Thrombospondin-1 pg/m1 620,
1275 3286
7. Elastase 2 pg/m1 2.1, 4.4
11.3
8. PDGF-AB/BB pg/m1 769,
2537 6390
9. MIG pg/m1 1205,
2684 6531
10. MCP-1 pg/m1 190.3,
269.5 529.4
11. MMP1 pg/m1 384, 709
1286
12. Lactoferrin pg/m1 338.3,
521.3 845.8
13. IL-1RA pg/m1 8.1,
73.3 112.0
14. IL-18 pg/m1 30.3,
64.6 98.3
15. IFNa2 pg/m1 13.2,
120.7 179.6
16. P-selectin (EC) ng/ml 16.3,
22.4 30.4
17. MIP-113 pg/m1 22.8,
72.0 89.16
18. Eotaxin pg/m1 49.4,
81.6 97.4
19. MMP8 pg/m1 288.4,
643.6 762.7
20. PDGF-AA pg/m1 84.4,
652.9 766.8
21. MMP10 pg/m1 385.2,
751.4 876.1
22. Heparin sulfate (EC) ng/ml 22.7,
294.5 20.6
Table 13 - List of Metabolites
Creatinine Sarcosine LYSOC24:0 C6:1
Dimethylglycine
79
CA 03180489 2022-10-17
WO 2021/207858 PCT/CA2021/050526
Glycine Diacetylspermine LYSOC26: 1 C6 Ethanol
Alanine Creatine LYSOC26:0 C5OH Glycerol
Serine Betaine LYSOC28:1 C5:1DC Formate
Proline Choline LYSOC28:0 C5DC Hypoxanthine
Valine Trimethylamine 14:1SMOH C8 D-Mannose
N-oxide
Threonine Methylhistidine 16:1SM C5MDC L-Acetylcarnitine
Phenylethylamine Homocysteine 16:0SM C9 Oxoglutarate
Taurine Lactic acid 16:1SMOH C7DC Urea
Putrescine beta- 18:1SM C10:2 3-Hydroxybutyric
Hydroxybutyric acid
acid
trans- alpha- PC32:2AA C10:1 2-
Hydroxyproline Ketoglutaric acid hydroxyisovalerate
Leucine Citric acid 18:0SM C10 L-Alpha-
aminobutyric acid
Isoleucine Butyric acid 20:2SM C12:1 3-Methy1-2-
oxovaleric acid
Asparagine Propionic acid PC36:0AE C12 Malonate
Aspartic acid HPHPA PC36:6AA C14:2 Ketoleucine
Glutamine P- PC36:0AA C14:1 3-
Hydroxyhippuric Hydroxyisovaleric
acid acid
Glutamic acid Succinic acid 22:2SMOH C14 Isopropanol
Methionine Fumaric acid 22:1SMOH C12DC Acetone
Dopamine Pyruvic acid PC38:6AA C14:20H Methanol
Histidine Isobutyric acid PC38: OAA C14:10H Propylene glycol
Phenylalanine Hippuric acid PC40:6AE C16:2 Dimethyl sulfone
Methionine- Methylmalonic 24:1SMOH C16:1
sulfoxide acid
Arginine Homovanillic acid PC40:6AA C16
Acetyl-ornithine Indole-3-acetic PC40:2AA C16:20H
acid
Citrulline Uric acid PC40:1AA C16:10H
Serotonin Glucose CO C160H
Tyrosine LYSOC14:0 C2 C18:2
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Asymmetric LYSOC16:1 C3:1 C18:1
dimethylarginine
Total LYSOC16:0 C3 C18
dimethylarginine
Tryptophan LYSOC17:0 C4:1 C18:10H
Kynurenine LYSOC18:2 C4 2-
Hydroxybutyrate
Ornithine LYSOC18:1 C3OH Acetic acid
Lysine LYSOC18:0 C5:1 Acetoacetate
Spermidine LYSOC20:4 C5 L-Carnitine
Spermine LYSOC20:3 C4OH Dimethylamine
Table 14 - Feature classification demonstrating the top 8 plasma metabolites
that classify
COVID-19+ status versus healthy control subjects with their "A association.
Metabolite Importance ("/0)
1. Kynurenine 10
2. Arginine 10
3. Sarcosine 10
4. LYSOPC18:1 10
5. LYSOPC20:4 10
6. LYSOPC14:0 10
7. LYSOPC17:0 10
8. LYSOPC18:2 10
Table 15 - Feature classification demonstrating the top 8 plasma metabolites
that classify
COVID-19+ status versus COVID-19- patients with their % association.
Metabolite Importance ("/0)
1. Kynurenine 10
2. LysoPC17:0 10
3. LysoPC20:3 10
4. C5:1DC 10
5. C6:1 10
6. Glycine 8
7. Threonine 8
8. Hi stidine 6
81
SUBSTITUTE SHEET (RULE 26)
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Table 16 - Feature classification demonstrating the top 8 plasma metabolites
that classify
COVID-19+ ICU patient outcome as alive or dead with their `)/0 association.
Plasma
creatinine was 25 the leading outcome predictor metabolite.
Metabolite Importance
(%)
1. Creatinine 20
2. Creatine 10
3. C3OH 10
4. PC40:6AA 10
5. C5 10
6. C6:1 10
7. C3:1 10
8. Methylmalonic 10
acid
Table 17 - Feature classification demonstrating the top 20 inflammatory
analytes that
classify COVID-19 status in ICU patients' days 1-3 with their % association
(Fig. 8).
Num Assay Unipro Importance
Id
1. TYMP P19971 (0.036364)
2. CXCL10 P02778 (0.021818)
3. ClQA P02745 (0.020000)
4. AGR2 095994 (0.020000)
5. IL-18R1 Q13478 (0.020000)
6. CDON Q4KMGO (0.020000)
7. DDX58 095786 (0.020000)
8. CLEC6A Q6EIG7 (0.019939)
9. CLM-6 Q08708 (0.016364)
10. PXN P49023 (0.016364)
11. LAG3 P18627 (0.016364)
12. APLP1 P51693 (0.014848)
13. LIF-R P42702 (0.014848)
14. B4GALT1 P15291 (0.013333)
15. ASGR1 P07306 (0.012468)
16. CRIM1 Q9NZV1 (0.011818)
17. CD300E Q496F6 (0.011818)
18. CDKN1A P38936 (0.011515)
19. CXCL11 014625 (0.010000)
20. IL6 P05231 (0.010000)
82
SUBSTITUTE SHEET (RULE 26)
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Table 18 - Feature classification demonstrating the top 20 inflammatory
analytes that
classify COVID-19 status in ICU patients' days 1-3 with their A association
(Fig. 9).
Num Assay Uniprot ID Importance
1. DDX58 095786 (0.040045)
2. RRM2B Q7LG56 (0.022396)
3. IRF9 Q00978 (0.020178)
4. NPM1 P06748 (0.019699)
5. MCP-3 P80098 (0.019348)
6. Ga1-9 000182 (0.019016)
7. NADK 095544 (0.017407)
8. BRK1 Q8WUW1 (0.017145)
9. PFDN2 Q9UHV9 (0.017051)
10. HEXIM1 094992 (0.016900)
11. TCN2 P20062 (0.016566)
12. BLM hydrolase Q13867
(0.015594)
13. KRT19 P08727 (0.014615)
14. FUS P35637 (0.014505)
15. RCOR1 Q9UKLO (0.014103)
16. PSME1 Q06323 (0.013333)
17. CXCL11 014625 (0.012911)
18. CLSPN Q9HAW4 (0.012748)
19. S100A1 1 P31949 (0.012333)
20. CDON Q4KMGO (0.011295)
83
SUBSTITUTE SHEET (RULE 26)
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Table 19 - The top 21 proteins underlying the outcome differences shown in
Fig. 10A
Num Assay Unipro Id Importance
1. CXCL9 Q07325 (0.071033)
2. ICOSLG 075144 (0.068896)
3. CLM-1 Q8TDQ1 (0.052505)
4. IL12RB1 P42701 (0.052471)
5. CD83 Q01151 (0.049965)
6. CA12 043570 (0.049572)
7. FLRT2 043155 (0.049366)
8. ROR1 Q01973 (0.048488)
9. IL32 P24001 (0.048049)
10. NCS1 P62166 (0.047003)
11. S100A11 P31949 (0.045890)
12. ANGPTL7 043827 (0.044741)
13. CLMP Q9H6B4 (0.044442)
14. IGF1R P08069 (0.043656)
15. TOP2B Q02880 (0.043410)
16. FAM3B P58499 (0.042812)
17. IL10.1 P22301 (0.041501)
18. IL10 P22301 (0.041058)
19. THY 1 P04216 (0.040373)
20. PVRL4 Q96NY8 (0.038997)
21. OPTC Q9UBM4 (0.035772)
84
SUBSTITUTE SHEET (RULE 26)
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Table 20 - Top 21 proteins underlying the outcome differences shown in Fig.
10B.
Num Assay Unipro Id Importance
1. IL12RB1 P42701 (0.090633)
2. CLM-1 Q8TDQ1 (0.089183)
3. CXCL9 Q07325 (0.077083)
4. FAM3B P58499 (0.076017)
5. OPTC Q9UBM4 (0.071080)
6. THY 1 P04216 (0.070551)
7. ICOSLG 075144 (0.060209)
8. IGF1R P08069 (0.055931)
9. IL10.1 P22301 (0.054822)
10. CLMP Q9H6B4 (0.050275)
11. IL10 P22301 (0.049304)
12. CD83 Q01151 (0.046050)
13. ROR1 Q01973 (0.044522)
14. PVRL4 Q96NY8 (0.042428)
15. IL32 P24001 (0.036729)
16. CA12 043570 (0.028727)
17. NCS1 P62166 (0.024985)
18. FLRT2 043155 (0.015163)
19. S100A1 1 P31949 (0.006715)
20. TOP2B Q02880 (0.006509)
21. ANGPTL7 043827 (0.003084)
SUBSTITUTE SHEET (RULE 26)
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
Table 21 ¨ Feature classification demonstrating the top 15 inflammatory
analytes that
classify COVID-19 status in ICU patients' days 1-3 with their `)/0 association
(see Fig. 12).
Rank Analyte % Association
1 TNF 10.1
2 Granzyme B 7.8
3 HSP70 7.6
4 IL-18 6.4
IP-10 4.2
6 Elastase 2 3.9
7 MIG 3.2
8 IL-8 3.2
9 IL-17A 3.2
IFNa2 2.9
11 M-CSF 2.8
12 IL-2 2.7
13 IL-15 2.6
14 IL-10 2.4
IL-1I3 2.3
References
5 1. Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, et al: Covid-19 in
Critically Ill
Patients in the Seattle Region - Case Series. N Engl J Med 2020
2. Zhou F, Yu T, Du R, Fan G, et al: Clinical course and risk factors for
mortality of
adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study.
Lancet 2020;
395(10229):1054-1062
10 3. Wu C, Chen X, Cai Y, Xia J, et al: Risk Factors Associated With
Acute Respiratory
Distress Syndrome and Death in Patients With Coronavirus Disease 2019
Pneumonia in Wuhan,
China. JAMA Intern Med 2020
4. Grasselli G, Zangrillo A, Zanella A, Antonelli M, et al: Baseline
Characteristics
and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the
Lombardy
15 Region, Italy. JAMA 2020
5. Auld SC, Caridi-Scheible M, Blum JM, Robichaux C, et al: ICU and
Ventilator
Mortality Among Critically Ill Adults With Coronavirus Disease 2019. Crit Care
Med 2020.
86
SUBSTITUTE SHEET (RULE 26)
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
6. Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the
eye of
the cytokine storm. Microbiol Mol Biol Rev 2012;76:16-32.
7. Mehta P, McAuley DF, Brown M, et al. COVID-19: consider cytokine storm
syndromes
and immunosuppression. Lancet 2020;395:1033-4.
8. Zhou F, Yu T, Du R, etal. Clinical course and risk factors for mortality of
adult inpatients
with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet
2020;395:1054-62.
9. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with
2019 novel
coronavirus in Wuhan, China. Lancet 2020;395:497-506.
10. Feldmann M, Maini RN, Woody JN, et al. Trials of anti-tumour necrosis
factor therapy
for COVID-19 are urgently needed. Lancet 2020.
11. Ritchie AT, Singanayagam A. Immunosuppression for hyperinflammation in
COVID-
19: a double-edged sword? Lancet 2020;395:1111.
12. Mokra D, Mikolka P, Kosutova P, Mokry J. Corticosteroids in Acute Lung
Injury: The
Dilemma Continues. Int J Mol Sci 2019;20.
13. https ://www.cdc. gov/coronavi rus/2019-nC oV/hcp/clini cal-criteri a.
html :
14. https ://www.fda.gov/media/134922/download:
15. Priestap F, Kao R, Martin CM: External validation of a prognostic model
for
intensive care unit mortality: a retrospective study using the Ontario
Critical Care Information
System. Can J Anaesth 2020
16. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, et al: The Third
International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3).
JAIVIA 2016;
315(8): 801-810
17. Brisson AR, Matsui D, Rieder MJ, Fraser DD: Translational
research in pediatrics:
tissue sampling and biobanking. Pediatrics 2012; 129(1):153-162
18. Gillio-Meina C, Cepinskas G, Cecchini EL, Fraser DD: Translational
research in
pediatrics II: blood collection, processing, shipping, and storage. Pediatrics
2013; 131(4):754-766
19. Program NHRP: POLICY: Guidelines for Limits of Blood Drawn for Research
Purposes in the Clinical Center M95-9 (rev) 2009; June 5
20. van der Maaten L, Hinton G: Visualizing data using t-SNE. J Mach Learn
Res 2008;
9(11):2579-2605.
21. Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, et al: Homogenous
96-plex
PEA immunoassay exhibiting high sensitivity, specificity, and excellent
scalability. PLoS One
2014; 9(4):e95192
22. Lundberg M, Eriksson A, Tran B, Assarsson E, eta!: Homogeneous antibody-
based
proximity extension assays provide sensitive and specific detection of low-
abundant proteins in
human blood. Nucleic Acids Res 2011; 39(15):e102.
23. Fraser DD, Cepinskas G, Slessarev M, Martin C, et al: Inflammation
profiling of
critically ill COVID19 patients. Crit Care Explor 2020; 2:e0144.
87
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
24. Gillio-Meina C, Cepinskas G, Cecchini EL, Fraser DD. Translational
research in
pediatrics II: blood collection, processing, shipping, and storage. Pediatrics
2013;131:754-66.
25. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry
Depends
on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor.
Cell 2020.
26. Cullen SP, Brunet M, Martin SJ. Granzymes in cancer and immunity. Cell
Death Differ
2010;17:616-23.
27. Lauw FN, Simpson AJ, Hack CE, et al. Soluble granzymes are released during
human
endotoxemia and in patients with severe infection due to gram-negative
bacteria. J Infect Dis
2000;182:206-13.
28. Wensink AC, Hack CE, Bovenschen N. Granzymes regulate proinflammatory
cytokine
responses. J Immunol 2015;194:491-7.
29. Buzza MS, Zamurs L, Sun J, et al. Extracellular matrix remodeling by human
granzyme
B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem
2005;280:23549-58.
30. Omoto Y, Yamanaka K, Tokime K, et al. Granzyme B is a novel interleukin-18
converting enzyme. J Dermatol Sci 2010;59:129-35.
31. Akeda T, Yamanaka K, Tsuda K, Omoto Y, Gabazza EC, Mizutani H. CD8+ T cell
granzyme B activates keratinocyte endogenous IL-18. Arch Dermatol Res
2014;306:125-30.
32. Morel JC, Park CC, Woods JIM, Koch AE. A novel role for interleukin-18 in
adhesion
molecule induction through NF kappa B and phosphatidylinositol (PI) 3-kinase-
dependent signal
transduction pathways. J Biol Chem 2001;276:37069-75.
33. Pinsky MR, Vincent JL, Deviere J, Alegre M, Kahn RJ, Dupont E. Serum
cytokine
levels in human septic shock. Relation to multiple-system organ failure and
mortality. Chest
1993;103:565-75.
34. Abraham E, Anzueto A, Gutierrez G, et al. Double-blind randomised
controlled trial of
monoclonal antibody to human tumour necrosis factor in treatment of septic
shock. NORASEPT
II Study Group. Lancet 1998;351:929-33.
35. Wang L, Chung J, Gill SE, Mehta S: Quantification of adherens junction
disruption and
contiguous paracellular protein leak in human lung endothelial cells under
septic conditions.
Microcirculation 2019; 26(3):e12528
36. Wang L, Mehta S, Ahmed Y, Wallace S, et al: Differential Mechanisms of
Septic Human
Pulmonary Microvascular Endothelial Cell Barrier Dysfunction Depending on the
Presence of
Neutrophils. Front Immunol 2018; 9:1743
37. Faul F, Erdfelder E, Buchner A, Lang AG: Statistical power analyses
using G*Power 3.1:
tests for correlation and regression analyses. Behav Res Methods 2009;
41(4):1149-1160
38. van der Maaten L, Hinton G: Visualizing data using t-SNE. J Mach Learn
Res 2008;
9(11):2579-2605
88
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
39. Zhou F, Yu T, Du R, Fan G, et al: Clinical course and risk factors for
mortality of adult
inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet
2020;
395(10229): 1054-1062
40. Wu C, Chen X, Cai Y, Xia J, et al: Risk Factors Associated With Acute
Respiratory
Distress Syndrome and Death in Patients With Coronavirus Disease 2019
Pneumonia in Wuhan,
China. JAIVIA Intern Med 2020
41. Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, et al: Covid-19 in
Critically Ill Patients
in the Seattle Region - Case Series. N Engl J Med 2020
42. Grasselli G, Zangrillo A, Zanella A, Antonelli M, et al: Baseline
Characteristics and
Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the
Lombardy
Region, Italy. JAMA 2020
43. Fraser DD, Cepinskas G, Slessarev M, Martin C, et al: Inflammation
profiling of critically
ill COVID19 patients. Crit Care Explor 2020; In press.
44. Teng YH, Aquino RS, Park PW: Molecular functions of syndecan-1 in
disease. Matrix Biol
2012; 31(1):3-16
45. Fitzgerald ML, Wang Z, Park PW, Murphy G, et al: Shedding of syndecan-1
and -4
ectodomains is regulated by multiple signaling pathways and mediated by a TIMP-
3-sensitive
metalloproteinase. J Cell Biol 2000; 148(4):811-824
46. Zhang Y, Xiao M, Zhang S, Xia P, et al: Coagulopathy and
Antiphospholipid Antibodies
in Patients with Covid-19. N Engl J Med 2020; 382(17):e38
47. Ji HL, Zhao R, Matalon S, Matthay MA: Elevated Plasmin(ogen) as a
Common Risk Factor
for COVID-19 Susceptibility. Physiol Rev 2020; 100(3):1065-1075
48. Hofstra JJ, Haitsma JJ, Juffermans NP, Levi M, et al: The role of
bronchoalveolar
hemostasis in the pathogenesis of acute lung injury. Semin Thromb Hemost 2008;
34(5):475-484
49. Tang N, Bai H, Chen X, Gong J, et al: Anticoagulant treatment is
associated with decreased
mortality in severe coronavirus disease 2019 patients with coagulopathy. J
Thromb Haemost 2020;
18(5): 1094-1099
50. Mehta P, McAuley DF, Brown M, Sanchez E, et al: COVID-19: consider
cytokine storm
syndromes and immunosuppression. Lancet 2020; 395(10229):1033-1034
51. Reindel R, Baker SC, Kim KY, Rowley CA, Shulman ST, Orenstein JM, et
al. Integrins
a1pha4 and alphaM, collagen1A1, and matrix metalloproteinase 7 are upregulated
in acute
Kawasaki disease vasculopathy. Pediatr Res. 2013;73(3):332-6.
52. Swee M, Wilson CL, Wang Y, McGuire JK, Parks WC. Matrix
metalloproteinase-7
(matrilysin) controls neutrophil egress by generating chemokine gradients. J
Leukoc Biol.
2008;83(6):1404-12.
53. Ko TM, Kuo HC, Chang JS, Chen SP, Liu YM, Chen HW, et al. CXCL10/IP-10
is a
biomarker and mediator for Kawasaki disease. Circ Res. 2015;116(5):876-83.
54. Kemmotsu Y, Saji T, Kusunoki N, Tanaka N, Nishimura C, Ishiguro A, et
al. Serum
adipokine profiles in Kawasaki disease. Mod Rheumatol. 2012;22(1):66-72.
89
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
55. Nozue H, Imai H, Saitoh H, Aoki T, Ichikawa K, Kamoda T. Serum resistin
concentrations
in children with Kawasaki disease. Inflamm Res. 2010;59(11):915-20.
56. Korpelainen El, Gamble JR, Vadas MA, Lopez AF. IL-3 receptor
expression, regulation
and function in cells of the vasculature. Immunol Cell Biol. 1996;74(1):1-7.
57. Kolarova H, Ambruzova B, Svihalkova Sindlerova L, Klinke A, Kubala L.
Modulation of
endothelial glycocalyx structure under inflammatory conditions. Mediators
Inflamm.
2014;2014:694312.
58. Ohnishi Y, Yasudo H, Suzuki Y, Furuta T, Matsuguma C, Azuma Y, et al.
Circulating
endothelial glycocalyx components as a predictive marker of coronary artery
lesions in Kawasaki
disease. Int J Cardiol. 2019;292:236-40.
59. Schrijver IT, Kemperman H, Roest M, Kesecioglu J, de Lange DW. Soluble
P-selectin as
a Biomarker for Infection and Survival in Patients With a Systemic
Inflammatory Response
Syndrome on the Intensive Care Unit. Biomark Insights.
2017;12:1177271916684823.
60. Gupta M, Noel GJ, Schaefer M, Friedman D, Bussel J, Johann-Liang R.
Cytokine
modulation with immune gamma-globulin in peripheral blood of normal children
and its
implications in Kawasaki disease treatment. J Clin Immunol. 2001;21(3):193-9.
61. Andreasen AS, Krabbe KS, Krogh-Madsen R, Taudorf S, Pedersen BK, Moller
K. Human
endotoxemia as a model of systemic inflammation. Curr Med Chem.
2008;15(17):1697-705.
62. Chen, Peter et al. "MMP7 shedding of syndecan-1 facilitates re-
epithelialization by affecting
alpha(2)beta(1) integrin activation." PloS one vol. 4,8 e6565. 10 Aug. 2009,
doi:10.1371/j ournal.pone .0006565.
63. Foroutan A, Fitzsimmons C, Mandal R, Piri-Moghadam H, et al: The Bovine
Metabolome.
Metabolites 2020; 10(6)
64. Foroutan A, Guo AC, Vazquez-Fresno R, Lipfert M, et al: Chemical
Composition of
Commercial Cow's Milk. J Agric Food Chem 2019; 67(17):4897-4914
65. Psychogios N, Hau DD, Peng J, Guo AC, et al: The human serum
metabolome. PLoS One
2011; 6(2):e16957
66. Saude EJ, Slupsky CM, Sykes BD: Optimization of NMR analysis of
biological fluids for
quantitative accuracy. Metabolomics 2006; 2(3): 113-123
67. Ravanbakhsh S, Liu P, Bjorndahl TC, Mandal R, et al: Accurate, fully-
automated NMR
spectral profiling for metabolomics. PLoS One 2015; 10(5):e0124219
68. van der Maaten L, Hinton G: Visualizing data using t-SNE. J Mach Learn
Res 2008;
9(11):2579-2605
69. Tang C, Garreau D, von Luxburg U: When do random forests fail?
Proceedings of the 32nd
International Conference on Neural Information Processing Systems 2018;
December 2018 : 2987-
2997
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
70. Fraser DD, Cepinskas G, Slessarev M, Martin C, eta!: Inflammation
Profiling of Critically
Ill Coronavirus Disease 2019 Patients. Critical Care Explorations 2020;
2(6):e0144
71. Fraser DD, Patterson EK, Slessarev M, Gill SE, et al: Endothelial
Injury and Glycocalyx
Degradation in Critically Ill Coronavirus Disease 2019 Patients: Implications
for Microvascular
Platelet Aggregation. Critical Care Explorations 2020; In press.
72. Fraser DD, Cepinskas G, Patterson EK, Slessarev M, et al: Novel Outcome
Biomarkers
Identified with Targeted Proteomic Analyses of Plasma from Critically Ill
Coronavirus Disease
2019 Patients. Critical Care Explorations 2020; In press
73. Chen Y, Guillemin GJ: Kynurenine pathway metabolites in humans: disease
and healthy
States. Int J Tryptophan Res 2009; 2:1-19
74. De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, et al: Marked T
cell activation,
senescence, exhaustion and skewing towards TH17 in patients with COVID-19
pneumonia. Nat
Commun 2020; 11(1):3434
75. Chen Z, John Wherry E: T cell responses in patients with COVID-19. Nat
Rev Immunol
2020
76. Akerstrom S, Mousavi-Jazi M, Klingstrom J, Leij on M, et al: Nitric
oxide inhibits the
replication cycle of severe acute respiratory syndrome coronavirus. J Virol
2005; 79(3):1966-1969
77. Mondanelli G, Iacono A, Allegrucci M, Puccetti P, et al:
Immunoregulatory Interplay
Between Arginine and Tryptophan Metabolism in Health and Disease. Front
Immunol 2019;
10:1565
78 5. Dastmalchi F, Karachi A, Yang C, Azari H, et al: Sarcosine promotes
trafficking of
dendritic cells and improves efficacy of anti-tumor dendritic cell vaccines
via CXC chemokine
family signaling. J Immunother Cancer 2019; 7(1):321
79. Walters RO, Arias E, Diaz A, Burgos ES, eta!: Sarcosine Is Uniquely
Modulated by Aging
and Dietary Restriction in Rodents and Humans. Cell Rep 2018; 25(3):663-676
e666
80. Carmona-Gutierrez D, Bauer MA, Zimmermann A, Kainz K, et al: Digesting
the crisis:
autophagy and coronaviruses. Microb Cell 2020; 7(5):119-128
81. Li X, Fang P, Li Y, Kuo YM, et al: Mitochondria! Reactive Oxygen
Species Mediate
Lysophosphatidylcholine-Induced Endothelial Cell Activation. Arterioscler
Thromb Vasc Biol
2016; 36(6):1090-1100
82. Lauber K, Bohn E, Krober SM, Xiao YJ, et al: Apoptotic cells induce
migration of
phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell
2003; 113(6):717-730
83. Mecatti GC, Fernandes Messias MC, Sant'Anna Paiola RM, Figueiredo
Angolini CF, et al:
Lipidomic Profiling of Plasma and Erythrocytes From Septic Patients Reveals
Potential Biomarker
Candidates. Biomark Insights 2018; 13:1177271918765137
84. Cunningham TJ, Yao L, Lucena A: Product inhibition of secreted
phospholipase A2 may
explain lysophosphatidylcholines' unexpected therapeutic properties. J Inflamm
(Lond) 2008; 5:17
85. Clermont G, Acker CG, Angus DC, Sirio CA, et al: Renal failure in the
ICU: comparison
of the impact of acute renal failure and end-stage renal disease on ICU
outcomes. Kidney Int 2002;
62(3):986-996
86. Wysocki J, Lores E, Ye M, Soler MJ, et al: Kidney and Lung ACE2
Expression after an
ACE Inhibitor or an Ang II Receptor Blocker: Implications for COVID-19. J Am
Soc Nephrol
2020
91
CA 03180489 2022-10-17
WO 2021/207858
PCT/CA2021/050526
87. Hirsch JS, Ng JH, Ross DW, Sharma P, et al: Acute kidney injury in
patients hospitalized
with COVID-19. Kidney Int 2020; 98(1):209-218
88. Farkash EA, Wilson AM, Jentzen JIM: Ultrastructural Evidence for Direct
Renal Infection
with SARS-CoV-2. J Am Soc Nephrol 2020; 31(8):1683-1687
Through the embodiments that are illustrated and described, the currently
contemplated
best mode of making and using the invention is described. Without further
elaboration, it is
believed that one of ordinary skill in the art can, based on the description
presented herein, utilize
the present invention to the full extent. All publications cited herein are
incorporated by reference.
Although the description above contains many specificities, these should not
be construed
as limiting the scope of the invention, but as merely providing illustrations
of some of the presently
embodiments of this invention.
92