Note: Descriptions are shown in the official language in which they were submitted.
WO 2022/140254
PCT/US2021/064342
COMPOSITIONS AND METHODS FOR CAPTURING AND AMPLIFYING
TARGET POLYNUCLEOTIDES USING MODIFIED CAPTURE PRIMERS
CROSS-REFERENCE TO RELATED APPLICATIONS
[0001] This application claims the benefit of U.S. Provisional Patent
Application No.
63/128,675, filed December 21, 2020 and entitled "Compositions and Methods for
Capturing
and Amplifying Target Polynucleotides Using Modified Capture Primers," the
entire contents
of which are incorporated by reference herein.
SEQUENCE LISTING
[0002] The instant application contains a Sequence Listing which has been
submitted
electronically in ASCII format and is hereby incorporated by reference in its
entirety. Said
ASCII copy is named IP-2054-PCT_SL.txt, is 4,174 bytes in size.
FIELD
[0003] This application relates to cluster amplification.
BACKGROUND
[0004] Cluster amplification is an approach to amplifying polynucleotides, for
example for
use in genetic sequencing. Target polynucleotides are captured by primers
(e.g., P5 and P7
capture primers) coupled to a substrate surface in a flowcell, and form
"seeds" at random
locations on the surface. Cycles of amplification are performed to form
clusters on the
surface around each seed. The clusters include copies and complementary copies
(which
together may be referred as -amplicons") of the seed polynucleotides. For
example, FIG. 1
schematically illustrates amplification of a polynucleotide on a substrate.
Capture and
amplification of a single seed 111 on substrate region 101 may result in
monoclonal cluster
121 which may substantially fill the substrate region with amplicons of seed
111, and which
readily may be used for sequencing-by-synthesis, or SBS (the dashed and dotted
circles being
intended to represent expansion of the cluster overtime). In some
circumstances, the
substrate is patterned so as to define regions that bound different clusters,
such as wells that
may be filled with respective clusters.
1
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
SUMMARY
[0005] Examples provided herein are related to capturing and amplifying target
polynucleotides using modified capture primers. Compositions and methods for
performing
such capture and amplification are disclosed.
100061 In some examples, provided herein is a composition for capturing target
polynucleotides at a surface of a substrate. The composition may include a
plurality of
capture primers coupled to the surface of the substrate. Each capture primer
may include
modified nucleic acids. The composition also may include a plurality of
orthogonal capture
primers coupled to the surface of the substrate. Each orthogonal capture
primer may include
modified nucleic acids.
[0007] In some examples, the modified nucleic acids of the capture primers
include locked
nucleic acid (LNA), peptide nucleic acid (PNA), or super T. In some examples,
the capture
primers further include deoxyribonucleic acid (DNA). In some examples, the
modified
nucleic acids and the DNA are distributed between a 5' end and a 3' end of the
capture
primers. In some examples, the modified nucleic acids are disposed at a 5 end
of the capture
primers and the DNA is disposed at a 3' end of the capture primers.
[0008] In some examples, the modified nucleic acids of the orthogonal capture
primers
include locked nucleic acid (LNA), peptide nucleic acid (PNA), or super T. In
some
examples, the orthogonal capture primers further include deoxyribonucleic acid
(DNA). In
some examples, the modified nucleic acids and the DNA are distributed between
a 5' end and
a 3' end of the capture primers. In some examples_ the modified nucleic acids
are disposed at
a 5' end of the orthogonal capture primers and the DNA is disposed at a 3' end
of the
orthogonal capture primers.
[0009] In some examples, the composition further includes first target
polynucleotides, each
including a first adapter that is complementary to the capture primers, and a
second adapter
that is complementary to the orthogonal capture primers. The composition
further may
include second target polynucleotides that are complementary to respective
ones of the first
target polynucleotides.
[0010] In some examples, the first adapters of some of the first target
polynucleotides are
hybridized to respective ones of the capture primers to form first duplexes,
and the second
2
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
adapters of some of the first target polynucleotides are hybridized to
respective ones of the
orthogonal capture primers to form second duplexes. In some examples, the
first and second
duplexes each have a melting temperature (Tm) that is greater than a Tm of
third duplexes
that would be formed by hybridization of the second target polynucleotides to
the respective
ones of the first target polynucleotides to which the second target
polynucleotides are
complementary. In some examples, the first and second duplexes each have a Tm
that is
between about 80 C and about 110 C. In some examples, the first and second
duplexes each
have a Tm that is between about 85 C and about 105 C. In some examples, the
first and
second duplexes each have a Tm that is between about 90 C and about 100 C.
[0011] In some examples, substantially none of the second target
polynucleotides are
hybridized in solution to any of the first target polynucleotides.
[0012] In some examples, the composition further includes about 1% to about
100%
formami de (%v/v), or about 5% to about 80% formamide (%v/v).
[0013] In some examples, the composition further includes further includes
about 100 to
about 800 mM Na+, or about 200 to about 800 mM Na+.
[0014] In some examples, the capture primers are modified P5 capture primers,
and the
orthogonal capture primers are modified P7 capture primers.
[0015] In some examples, each of the capture primers includes between about
five and about
twenty of the modified nucleic acids, and each of the orthogonal capture
primers includes
between about five and about twenty of the modified nucleic acids.
[0016] In some examples, each of the capture primers includes at least about
nine of the
modified nucleic acids, and each of the orthogonal capture primers includes at
least about
nine of the modified nucleic acids; or each of the capture primers includes at
least about
twelve of the modified nucleic acids, and each of the orthogonal capture
primers includes at
least about twelve of the modified nucleic acids; or each of the capture
primers includes at
least about fifteen of the modified nucleic acids, and each of the orthogonal
capture primers
includes at least about fifteen of the modified nucleic acids.
[0017] In some examples, provided herein is a method for capturing and
amplifying target
polynucleotides at a surface of a substrate. The method may include contacting
a
3
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
composition with a fluid. The composition may include a plurality of capture
primers
coupled to the surface of the substrate, each capture primer including
modified nucleic acids;
and a plurality of orthogonal capture primers coupled to the surface of the
substrate, each
orthogonal capture primer including modified nucleic acids. The fluid may
include first
target polynucleotides, each including a first adapter that is complementary
to the capture
primers and a second adapter that is complementary to the orthogonal capture
primers; and
second target polynucleotides that are complementary to respective ones of the
first target
polynucleotides. The method may include, while inhibiting hybridization of the
second target
polynucleotides to the first target polynucleotides in the fluid: hybridizing
the first adapters of
some of the first target polynucleotides to respective ones of the capture
primers to form first
duplexes; hybridizing the second adapters of some of the first target
polynucleotides to
respective ones of the orthogonal capture primers to form second duplexes; and
then
amplifying the first target polynucleotides, the amplifying including
generating respective
amplicons of the first target polynucleotides.
[0018] In some examples, the modified nucleic acids of the capture primers
include locked
nucleic acid (LNA), peptide nucleic acid (PNA), or super T. In some examples,
the capture
primers further include deoxyribonucleic acid (DNA). In some examples, the
modified
nucleic acids and the DNA are distributed between a 5' end and a 3 end of the
capture
primers. In some examples, the modified nucleic acids are disposed at a 5' end
of the capture
primers and the DNA is disposed at a 3' end of the capture primers.
[0019] In some examples, the modified nucleic acids of the orthogonal capture
primers
include locked nucleic acid (LNA), peptide nucleic acid (PNA), or super T. In
some
examples, the orthogonal capture primers further include deoxyribonucleic acid
(DNA). In
some examples, the modified nucleic acids and the DNA are distributed between
a 5' end and
a 3' end of the capture primers. In some examples, the modified nucleic acids
are disposed at
a 5' end of the orthogonal capture primers and the DNA is disposed at a 3' end
of the
orthogonal capture primers.
[0020] In some examples, the first and second duplexes each have a melting
temperature
(Tm) that is greater than a Tm of third duplexes that would be formed by
hybridization of the
second target polynucleotides to the respective ones of the first target
polynucleotides to
which the second target polynucleotides are complementary. In some examples,
the first and
second duplexes each have a Tm that is between about 80 C and about 110 C. In
some
4
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
examples, the first and second duplexes each have a Tm that is between about
85 C and
about 105 C. In some examples, the first and second duplexes each have a Tm
that is
between about 90 C and about 100 C.
[0021] In some examples, substantially none of the second target
polynucleotides are
hybridized in solution to any of the first target polynucleotides.
[0022] In some examples, the hybridizing is performed in about 1% to about
100%
formamide (%v/v), or in about 5% to about 80% formamide (%v/v).
[0023] In some examples, the hybridizing is performed in about 100 to about
800 mM Na+,
or in about 200 to about 800 mM Na+.
[0024] In some examples, the capture primers are modified P5 capture primers,
and the
orthogonal capture primers are modified P7 capture primers.
100251 In some examples, each of the capture primers includes between about
five and about
twenty of the modified nucleic acids, and each of the orthogonal capture
primers includes
between about five and about twenty of the modified nucleic acids. In some
examples, each
of the capture primers includes at least about nine of the modified nucleic
acids, and each of
the orthogonal capture primers includes at least about nine of the modified
nucleic acids; or
each of the capture primers includes at least about twelve of the modified
nucleic acids, and
each of the orthogonal capture primers includes at least about twelve of the
modified nucleic
acids; or each of the capture primers includes at least about fifteen of the
modified nucleic
acids, and each of the orthogonal capture primers includes at least about
fifteen of the
modified nucleic acids.
100261 It is to be understood that any respective features/examples of each of
the aspects of
the disclosure as described herein may be implemented together in any
appropriate
combination, and that any features/examples from any one or more of these
aspects may be
implemented together with any of the features of the other aspect(s) as
described herein in
any appropriate combination to achieve the benefits as described herein.
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
BRIEF DESCRIPTION OF DRAWINGS
[0027] FIG. 1 schematically illustrates amplification of a polynucleotide on a
substrate.
[0028] FIGS. 2A-2C schematically illustrate compositions and operations in a
process flow
for capture of polynucleotides on a substrate using previously known capture
primers.
[0029] FIGS. 3A-3H schematically illustrate example compositions and
operations in an
example process flow for capture and amplification of polynucleotides on a
substrate using
the present capture primers.
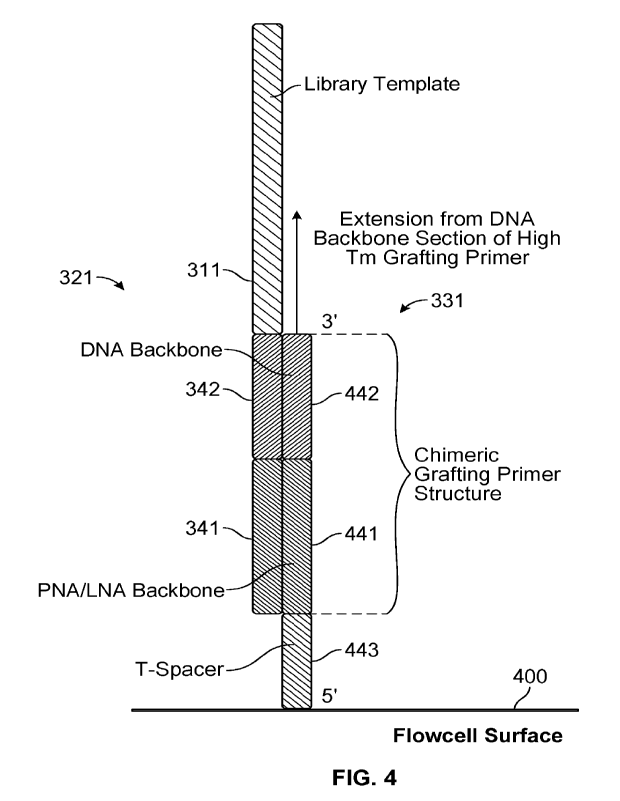
[0030] FIG. 4 schematically illustrates an example duplex between a
polynucleotide and one
of the present capture primers, according to some examples.
[0031] FIGS. 5A-5C are plots illustrating example effects of conditions on the
capture of
polynucleotides by the present capture primers.
[0032] FIG. 6 illustrates an example flow of operations in a method for
capturing and
amplifying a polynucleotide using the present capture primers.
DETAILED DESCRIPTION
[0033] Examples provided herein are related to capturing and amplifying
polynucleotides
using modified capture primers. Compositions and methods for performing such
capture and
amplification are disclosed.
[0034] The modified capture primers provided herein may hybridize more
strongly with
target polynucleotides than do previously known capture primers, and as such
may enhance
the efficiency with which target polynucleotides are captured on a substrate.
Illustratively,
because of the stronger binding, lower concentrations of target
polynucleotides may be used
to prepare clusters on a substrate, as compared to previously known capture
primers. For
example, in a manner such as described further below with reference to FIGS.
2A-2C,
polynucleotide capture and amplification using previously known primers may be
performed
under conditions in which surface hybridization and solution-based annealing
operate as
competing processes that render some of the polynucleotides unavailable for
capture or
amplification. In comparison, in a manner such as described with reference to
FIGS. 3A-3H,
4, 5A-5C, and 6, the present capture primers allow for polynucleotide capture
and
6
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
amplification to be performed under conditions that reduce or inhibit solution-
based
annealing as a competing process, and as such may substantially increase the
availability of
polynucleotides for capture and amplification, may permit use of higher
concentrations of
target polynucleotides to prepare clusters on a substrate, as compared to
previously known
capture primers, and may reduce the overall seeding time.
[0035] First, some terms used herein will be briefly explained. Then, some
example
compositions and example methods for capturing and amplifying polynucleotides
using the
present capture primers will be described.
Terms
100361 Unless defined otherwise, all technical and scientific terms used
herein have the same
meaning as is commonly understood by one of ordinary skill in the art. The use
of the term
"including" as well as other forms, such as "include," "includes," and
"included," is not
limiting. The use of the term -having" as well as other forms, such as -have,"
-has," and
"had," is not limiting. As used in this specification, whether in a
transitional phrase or in the
body of the claim, the terms -comprise(s)- and -comprising- are to be
interpreted as having
an open-ended meaning. That is, the above terms are to be interpreted
synonymously with
the phrases "having at least" or "including at least." For example, when used
in the context
of a process, the term "comprising" means that the process includes at least
the recited steps,
but may include additional steps. When used in the context of a compound,
composition, or
device, the term "comprising" means that the compound, composition, or device
includes at
least the recited features or components, but may also include additional
features or
components.
[0037] The terms "substantially," "approximately," and "about" used throughout
this
specification are used to describe and account for small fluctuations, such as
due to variations
in processing. For example, they may refer to less than or equal to +10%, such
as less than or
equal to +5%, such as less than or equal to +2%, such as less than or equal to
+1%, such as
less than or equal to +0.5%, such as less than or equal to +0.2%, such as less
than or equal to
0.1%, such as less than or equal to +0.05%.
[0038] As used herein, "hybridize" is intended to mean noncovalently
associating a first
polynucleotide to a second polynucleotide along the lengths of those
polynucleotides to form
a double-stranded "duplex." For instance, two DNA polynucleotide strands may
associate
7
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
through complementary base pairing. The strength of the association between
the first and
second polynucleotides increases with the complementarily between the
sequences of
nucleotides within those polynucleotides. The strength of hybridization
between
polynucleotides may be characterized by a temperature of melting (Tm) at which
50% of the
duplexes have polynucleotide strands that disassociate from one another.
Polynucleotides
that are "partially" hybridized to one another means that they have sequences
that are
complementary to one another, but such sequences are hybridized with one
another along
only a portion of their lengths to form a partial duplex. Polynucleotides with
an "inability" to
hybridize include those which are physically separated from one another such
that an
insufficient number of their bases may contact one another in a manner so as
to hybridize
with one another.
100391 As used herein, the term "nucleotide" is intended to mean a molecule
that includes a
sugar moiety, a backbone component that includes at least one phosphate group,
and in some
examples also includes a nucleobase. A nucleotide that lacks a nucleobase may
be referred to
as -abasic." Nucleotides include deoxyribonucleotides, modified
deoxyribonucleotides,
ribonucleotides, modified ribonucleotides, peptide nucleotides, modified
peptide nucleotides,
modified phosphate sugar backbone nucleotides, and mixtures thereof Examples
of
nucleotides include adenosine monophosphate (AMP), adenosine diphosphate
(ADP),
adenosine triphosphate (ATP), thymidine monophosphate (TMP), thymidine
diphosphate
(TDP), thymidine triphosphate (TTP), cytidine monophosphate (CMP), cytidine
diphosphate
(CDP), cytidine triphosphate (CTP), guanosine monophosphate (GMP), guanosine
diphosphate (GDP), guanosine triphosphate (GTP), uridine monophosphate (UMP),
uridine
diphosphate (UDP), uridine triphosphate (UTP), deoxyadenosine monophosphate
(dAMP),
deoxyadenosine diphosphate (dADP), deoxyadenosine triphosphate (dATP),
deoxythymidine
monophosphate (dTMP), deoxythymi dine diphosphate (dTDP), deoxythymi dine
triphosphate
(dTTP), deoxycytidine diphosphate (dCDP), deoxycytidine triphosphate (dCTP),
deoxyguanosine monophosphate (dGMP), deoxyguanosine diphosphate (dGDP),
deoxyguanosine triphosphate (dGTP), deoxyuridine monophosphate (dUMP),
deoxyuridine
diphosphate (dUDP), and deoxyuridine triphosphate (dUTP).
[0040] As used herein, the term "nucleotide" also is intended to encompass any
"nucleotide
analogue" which is intended to refer to a type of nucleotide that includes a
modified
nucleobase, sugar moiety, and/or backbone component (such as a phosphate or
amide)
8
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
compared to naturally occurring nucleotides. Nucleotide analogues also may be
referred to as
"modified nucleic acids." Example modified nucleobases include inosine,
xathanine,
hypoxathanine, isocytosine, isoguanine, 2-aminopurine, 5-methylcytosine, 5-
hydroxymethyl
cytosine, 2-aminoadenine, 6-methyl adenine, 6-methyl guanine, 2-propyl
guanine, 2-propyl
adenine, 2-thiouracil, 2-thiothymine, 2-thiocytosine, 15-halouracil, 15-
halocytosine, 5-
propynyl uracil, 5-propynyl cytosine, 6-azo uracil, 6-azo cytosine, 6-azo
thymine, 5-uracil, 4-
thiouracil, 8-halo adenine or guanine, 8-amino adenine or guanine, 8-thiol
adenine or
guanine, 8-thioalkyl adenine or guanine, 8-hydroxyl adenine or guanine, 5-halo
substituted
uracil or cytosine, 7-methylguanine, 7-methyladenine, 8-azaguanine, 8-
azaadenine, 7-
deazaguanine, 7-deazaadenine, 3-deazaguanine, 3-deazaadenine or the like. As
is known in
the art, certain nucleotide analogues cannot become incorporated into a
polynucleotide, for
example, nucleotide analogues such as adenosine 5'-phosphosulfate. The
backbone
components of nucleotides may include any suitable number of phosphates, e.g.,
three, four,
five, six, or more than six phosphates. Nucleotide analogues also include
locked nucleic
acids (LNA), peptide nucleic acids (PNA), and 5-hydroxylbutyn1-2'-deoxyuridine
("super
T"). LNA includes an RNA-like backbone in which ribose moieties include an
additional
bridge connecting the 2' oxygen and 4' carbon. PNA includes a backbone that
includes
repeating N-(2-aminoethyl)-glycine units linked by peptide bonds, and
nucleobases that are
coupled to the backbone via a methylene bridge and carbonyl group. Super T
includes a 5-
hydroxybutyn1-2'-deoxyuridine nucleobase. Example structures for LNA, PNA, and
super T
are shown below:
Base
T
0
.$
c$
=."
s;)
ease:
( 0
0."c)H 3'
0,ci' 0
looker.:(3C acki nt,E08k; StipEF
f.LNA) (PNA)
9
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
[0041] As used herein, the term "polynucleotide" refers to a molecule that
includes a
sequence of nucleotides that are bonded to one another. A polynucleotide is
one nonlimiting
example of a polymer. Examples of polynucleotides include deoxyribonucleic
acid (DNA),
ribonucleic acid (RNA), and analogues thereof such as locked nucleic acids
(LNA) and
peptide nucleic acids (PNA). A polynucleotide may be a single stranded
sequence of
nucleotides, such as RNA or single stranded DNA, a double stranded sequence of
nucleotides, such as double stranded DNA, or may include a mixture of a single
stranded and
double stranded sequences of nucleotides. Double stranded DNA (dsDNA) includes
genomic
DNA, and PCR and amplification products. Single stranded DNA (ssDNA) can be
converted
to dsDNA and vice-versa. Polynucleotides may include non-naturally occurring
DNA, such
as enantiomeric DNA, LNA, or PNA. The precise sequence of nucleotides in a
polynucleotide may be known or unknown. The following are examples of
polynucleotides: a
gene or gene fragment (for example, a probe, primer, expressed sequence tag
(EST) or serial
analysis of gene expression (SAGE) tag), genomic DNA, genomic DNA fragment,
exon,
intron, messenger RNA (mRNA), transfer RNA, ribosomal RNA, ribozyme, cDNA,
recombinant polynucleotide, synthetic polynucleotide, branched polynucleotide,
plasmid,
vector, isolated DNA of any sequence, isolated RNA of any sequence, nucleic
acid probe,
primer or amplified copy of any of the foregoing. A polynucleotide may have a
"chimera"
structure that includes adjoined sections of different types of
polynucleotides, such as
adjoined sections of DNA and PNA, of DNA and RNA, of PNA and RNA, or the like.
[0042] As used herein, a "polymerase" is intended to mean an enzyme having an
active site
that assembles polynucleotides by polymerizing nucleotides into
polynucleotides. A
polymerase can bind a primed single stranded target polynucleotide, and can
sequentially add
nucleotides to the growing primer to form a -complementary copy"
polynucleotide having a
sequence that is complementary to that of the target polynucleotide. Another
polymerase, or
the same polymerase, then can form a copy of the target nucleotide by forming
a
complementary copy of that complementary copy polynucleotide. Any of such
copies may
be referred to herein as -amplicons." DNA polymerases may bind to the target
polynucleotide and then move down the target polynucleotide sequentially
adding nucleotides
to the free hydroxyl group at the 3' end of a growing polynucleotide strand
(growing
ampli con). DNA polymerases may synthesize complementary DNA molecules from
DNA
templates and RNA polymerases may synthesize RNA molecules from DNA templates
(transcription). Polymerases may use a short RNA or DNA strand (primer), to
begin strand
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
growth. Some polymerases may displace the strand upstream of the site where
they are
adding bases to a chain. Such polymerases may be said to be strand displacing,
meaning they
have an activity that removes a complementary strand from a template strand
being read by
the polymerase. Exemplary polymerases having strand displacing activity
include, without
limitation, the large fragment of Bst (Bacillus stearothermophilus)
polymerase, exo-Klenow
polymerase or sequencing grade T7 exo-polymerase. Some polymerases degrade the
strand in
front of them, effectively replacing it with the growing chain behind (5'
exonuclease activity).
Some polymerases have an activity that degrades the strand behind them (3'
exonuclease
activity). Some useful polymerases have been modified, either by mutation or
otherwise, to
reduce or eliminate 3' and/or 5' exonuclease activity.
[0043] As used herein, the term -primer" is defined as a polynucleotide to
which nucleotides
may be added via a free 3' OH group. A primer may include a 3' block
preventing
polymerization until the block is removed. A primer may include a modification
at the 5'
terminus to allow a coupling reaction or to couple the primer to another
moiety. A primer
may include one or more moieties, such as 8-oxo-G, which may be cleaved under
suitable
conditions, such as UV light, chemistry, enzyme, or the like. The primer
length may be any
suitable number of bases long and may include any suitable combination of
natural and non-
natural nucleotides. A target polynucleotide may include an -adapter" that
hybridizes to (has
a sequence that is complementary to) a primer, and may be amplified so as to
generate a
complementary copy polynucleotide by adding nucleotides to the free 3' OH
group of the
primer. A "capture primer- is intended to mean a primer that is coupled to the
substrate and
may hybridize to a first adapter of the target polynucleotide, while an
"orthogonal capture
primer" is intended to mean a primer that is coupled to the substrate and may
hybridize to a
second adapter of that target polynucleotide. The first adapter may have a
sequence that is
complementary to that of the capture primer, and the second adapter may have a
sequence
that is complementary to that of the orthogonal capture primer. A capture
primer and an
orthogonal capture primer may have different and independent sequences than
one another.
Additionally, a capture primer and an orthogonal capture primer may differ
from one another
in at least one other property. For example, the capture primer and the
orthogonal capture
primer may have different lengths than one another; either the capture primer
or the
orthogonal capture primer may include a non-nucleic acid moiety (such as a
blocking group
or excision moiety) that the other of the capture primer or the orthogonal
capture primer
lacks; or any suitable combination of such properties. A -modified capture
primer" is a
11
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
capture primer, or orthogonal capture primer, that includes a plurality of
nucleic acid
analogues such as, but not limited to, LNA, PNA, or super T. A modified
capture primer
additionally may include a plurality of naturally occurring nucleic acids such
as, but not
limited to, DNA.
[0044] As used herein, the term "substrate" refers to a material used as a
support for
compositions described herein. Example substrate materials may include glass,
silica, plastic,
quartz, metal, metal oxide, organo-silicate (e.g., polyhedral organic
silsesquioxanes (POSS)),
polyacrylates, tantalum oxide, complementary metal oxide semiconductor (CMOS),
or
combinations thereof An example of POSS can be that described in Kehagias
etal.,
Microelectronic Engineering 86 (2009), pp. 776-778, which is incorporated by
reference in
its entirety. In some examples, substrates used in the present application
include silica-based
substrates, such as glass, fused silica, or other silica-containing material.
In some examples,
substrates may include silicon, silicon nitride, or silicone hydride. In some
examples,
substrates used in the present application include plastic materials or
components such as
polyethylene, polystyrene, poly(vinyl chloride), polypropylene, nylons,
polyesters,
polycarbonates, and poly(methyl methacrylate). Example plastics materials
include
poly(methyl methacrylate), polystyrene, and cyclic olefin polymer substrates.
In some
examples, the substrate is or includes a silica-based material or plastic
material or a
combination thereof In particular examples, the substrate has at least one
surface comprising
glass or a silicon-based polymer. In some examples, the substrates may include
a metal. In
some such examples, the metal is gold. In some examples, the substrate has at
least one
surface comprising a metal oxide. In one example, the surface comprises a
tantalum oxide or
tin oxide. Acrylamides, enones, or acrylates may also be utilized as a
substrate material or
component. Other substrate materials may include, but are not limited to
gallium arsenide,
indium phosphide, aluminum, ceramics, polyimide, quartz, resins, polymers and
copolymers.
In some examples, the substrate and/or the substrate surface may be, or
include, quartz. In
some other examples, the substrate and/or the substrate surface may be, or
include,
semiconductor, such as GaAs or ITO. The foregoing lists are intended to be
illustrative of,
but not limiting to the present application. Substrates may comprise a single
material or a
plurality of different materials. Substrates may be composites or laminates.
In some
examples, the substrate comprises an organo-silicate material. Substrates may
be flat, round,
spherical, rod-shaped, or any other suitable shape. Substrates may be rigid or
flexible. In
some examples, a substrate is a bead or a flow cell.
12
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
[0045] In some examples, a substrate includes a patterned surface. A
"patterned surface"
refers to an arrangement of different regions in or on an exposed layer of a
substrate. For
example, one or more of the regions may be features where one or more capture
primers are
present. The features can be separated by interstitial regions where capture
primers are not
present. In some examples, the pattern may be an x-y format of features that
are in rows and
columns. In some examples, the pattern may be a repeating arrangement of
features and/or
interstitial regions. In some examples, the pattern may be a random
arrangement of features
and/or interstitial regions. In some examples, substrate includes an array of
wells
(depressions) in a surface. The wells may be provided by substantially
vertical sidewalls.
Wells may be fabricated as is generally known in the art using a variety of
techniques,
including, but not limited to, photolithography, stamping techniques, molding
techniques and
microetching techniques. As will be appreciated by those in the art, the
technique used will
depend on the composition and shape of the array substrate.
[0046] The features in a patterned surface of a substrate may include wells in
an array of
wells (e.g., microwells or nanovvells) on glass, silicon, plastic or other
suitable material(s)
with patterned, covalently-linked gel such as poly(N-(5-azidoacetamidylpentyl)
acrylamide-
co-acrylamide) (PAZAM). The process creates gel pads used for sequencing that
may be
stable over sequencing runs with a large number of cycles. The covalent
linking of the
polymer to the wells may be helpful for maintaining the gel in the structured
features
throughout the lifetime of the structured substrate during a variety of uses.
However in many
examples, the gel need not be covalently linked to the wells. For example, in
some conditions
silane free acrylamide (SFA) which is not covalently attached to any part of
the structured
substrate, may be used as the gel material.
[0047] In particular examples, a structured substrate may be made by
patterning a suitable
material with wells (e.g. microwells or nanowells), coating the patterned
material with a gel
material (e.g., PAZAM, SFA or chemically modified variants thereof, such as
the azidolyzed
version of SFA (azido-SFA)) and polishing the surface of the gel coated
material, for
example via chemical or mechanical polishing, thereby retaining gel in the
wells but
removing or inactivating substantially all of the gel from the interstitial
regions on the surface
of the structured substrate between the wells. Primers may be attached to gel
material. A
solution including a plurality of target polynucleotides (e.g., a fragmented
human genome or
portion thereof) may then be contacted with the polished substrate such that
individual target
13
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
polynucleotides will seed individual wells via interactions with primers
attached to the gel
material; however, the target polynucleotides will not occupy the interstitial
regions due to
absence or inactivity of the gel material. Amplification of the target
polynucleotides may be
confined to the wells because absence or inactivity of gel in the interstitial
regions may
inhibit outward migration of the growing cluster. The process is conveniently
manufacturable, being scalable and utilizing conventional micro- or nano-
fabrication
methods.
[0048] A patterned substrate may include, for example, wells etched into a
slide or chip. The
pattern of the etchings and geometry of the wells may take on a variety of
different shapes
and sizes, and such features may be physically or functionally separable from
each other.
Particularly useful substrates having such structural features include
patterned substrates that
may select the size of solid particles such as microspheres. An exemplary
patterned substrate
having these characteristics is the etched substrate used in connection with
BEAD ARRAY
technology (Illumina, Inc., San Diego, Calif.).
[0049] In some examples, a substrate described herein forms at least part of a
flow cell or is
located in or coupled to a flow cell. Flow cells may include a flow chamber
that is divided
into a plurality of lanes or a plurality of sectors. Example flow cells and
substrates for
manufacture of flow cells that may be used in methods and compositions set
forth herein
include, but are not limited to, those commercially available from Illumina,
Inc. (San Diego,
Calif.).
[0050] As used herein, the term -directly" and the like, when used in
reference to a layer
covering the surface of a substrate is intended to mean that the layer covers
the substrate's
surface without a significant intermediate layer, such as, e.g., an adhesive
layer or a polymer
layer. Layers directly covering a surface may be attached to this surface
through any
chemical or physical interaction, including covalent bonds or non-covalent
adhesion.
100511 As used herein, the term "immobilized" when used in reference to a
polynucleotide is
intended to mean direct or indirect attachment to a substrate via covalent or
non-covalent
bond(s). In certain examples, covalent attachment may be used, or any other
suitable
attachment in which the polynucleotides remain stationary or attached to a
substrate under
conditions in which it is intended to use the substrate, for example, in
polynucleotide
amplification or sequencing. Polynucleotides to be used as capture primers or
as target
14
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
polynucleotides may be immobilized such that a 3'-end is available for
enzymatic extension
and at least a portion of the sequence is capable of hybridizing to a
complementary sequence.
Immobilization may occur via hybridization to a surface attached
oligonucleotide, in which
case the immobilized oligonucleotide or polynucleotide may be in the 3'-5'
orientation.
Alternatively, immobilization may occur by means other than base-pairing
hybridization,
such as covalent attachment. Illustratively, a chemical functionality may be
incorporated
onto the 5' end of one of the present capture primers via a chemical linker.
The chemical
functionality on the capture primer is capable of attachment to the substrate
surface via any
suitable combination of one or more non-covalent interaction(s) (e.g.,
electrostatic, metal-
ligand binding, hybridization, or the like) or covalent interaction(s) (e.g.,
copper click
chemistry reactions, copper free click chemistry reactions, or the like).
100521 As used herein, the term "array" refers to a population of substrate
regions that may
be differentiated from each other according to relative location. Different
molecules (such as
polynucleotides) that are at different regions of an array may be
differentiated from each
other according to the locations of the regions in the array. An individual
region of an array
may include one or more molecules of a particular type. For example, a
substrate region may
include a single target polynucleotide having a particular sequence, or a
substrate region may
include several polynucleotides having the same sequence (or complementary
sequences
thereof). The regions of an array respectively may include different features
than one another
on the same substrate. Exemplary features include without limitation, wells in
a substrate,
beads (or other particles) in or on a substrate, projections from a substrate,
ridges on a
substrate or channels in a substrate. The regions of an array respectively may
include
different regions on different substrates than each other. Different molecules
attached to
separate substrates may be identified according to the locations of the
substrates on a surface
to which the substrates are associated or according to the locations of the
substrates in a
liquid or gel. Exemplary arrays in which separate substrates are located on a
surface include,
without limitation, those having beads in wells.
100531 As used herein, the term -plurality" is intended to mean a population
of two or more
different members. Pluralities may range in size from small, medium, large, to
very large.
The size of small plurality may range, for example, from a few members to tens
of members.
Medium sized pluralities may range, for example, from tens of members to about
100
members or hundreds of members. Large pluralities may range, for example, from
about
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
hundreds of members to about 1000 members, to thousands of members and up to
tens of
thousands of members. Very large pluralities may range, for example, from tens
of thousands
of members to about hundreds of thousands, a million, millions, tens of
millions and up to or
greater than hundreds of millions of members. Therefore, a plurality may range
in size from
two to well over one hundred million members as well as all sizes, as measured
by the
number of members, in between and greater than the above exemplary ranges.
Exemplary
polynucleotide pluralities include, for example, populations of about 1x105 or
more, 5 x105 or
more, or 1x106 or more different polynucleotides. Accordingly, the definition
of the term is
intended to include all integer values greater than two. An upper limit of a
plurality may be
set, for example, by the theoretical diversity of polynucleotide sequences in
a sample.
[0054] As used herein, the term -double-stranded," when used in reference to a
polynucleotide, is intended to mean that all or substantially all of the
nucleotides in the
polynucleotide are hydrogen bonded to respective nucleotides in a
complementary
polynucleotide. A "partially" double stranded polynucleotide may have at least
about 10%, at
least about 25%, at least about 50%, at least about 60%, at least about 70%,
at least about
80%, at least about 90% or at least about 95% of its nucleotides, but fewer
than all of its
nucleotides, hydrogen bonded to nucleotides in a complementary polynucleotide.
[0055] As used herein, the term "single-stranded," when used in reference to a
polynucleotide, means that essentially none of the nucleotides in the
polynucleotide are
hydrogen bonded to a respective nucleotide in a complementary polynucleotide.
A
polynucleotide that has an "inability" to hybridize to another polynucleotide
may be single-
stranded.
[0056] As used herein, the term "target polynucleotide" is intended to mean a
polynucleotide
that is the object of an analysis or action, and may also be referred to as a
"library
polynucleotide," "template polynucleotide," or "library template." The
analysis or action
includes subjecting the polynucleotide to capture, amplification, sequencing
and/or other
procedure. A target polynucleotide may include nucleotide sequences additional
to a target
sequence to be analyzed. For example, a target polynucleotide may include one
or more
adapters, including an adapter that functions as a primer binding site, that
flank(s) a target
polynucleotide sequence that is to be analyzed. A target polynucleotide
hybridized to a
capture primer may include nucleotides that extend beyond the 5' or 3' end of
the capture
oligonucleotide in such a way that not all of the target polynucleotide is
amenable to
16
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
extension. In particular examples, target polynucleotides may have different
sequences than
one another but may have first and second adapters that are the same as one
another. The two
adapters that may flank a particular target polynucleotide sequence may have
the same
sequence as one another, or complementary sequences to one another, or the two
adapters
may have different sequences. Thus, species in a plurality of target
polynucleotides may
include regions of known sequence that flank regions of unknown sequence that
are to be
evaluated by, for example, sequencing (e.g., SBS). In some examples, target
polynucleotides
carry an adapter at a single end, and such adapter may be located at either
the 3' end or the 5'
end the target polynucleotide. Target polynucleotides may be used without any
adapter, in
which case a primer binding sequence may come directly from a sequence found
in the target
polynucleotide.
100571 The terms "polynucleotide" and "oligonucleotide" are used
interchangeably herein.
The different terms are not intended to denote any particular difference in
size, sequence, or
other property unless specifically indicated otherwise. For clarity of
description, the terms
may be used to distinguish one species of polynucleotide from another when
describing a
particular method or composition that includes several polynucleotide species.
[0058] As used herein, the term "amplicon," when used in reference to a
polynucleotide, is
intended to means a product of copying the polynucleotide, wherein the product
has a
nucleotide sequence that is substantially the same as, or is substantially
complementary to, at
least a portion of the nucleotide sequence of the polynucleotide. -
Amplification- and
"amplifying" refer to the process of making an amplicon of a polynucleotide. A
first
amplicon of a target polynucleotide may be a complementary copy. Additional
amplicons are
copies that are created, after generation of the first amplicon, from the
target polynucleotide
or from the first amplicon. A subsequent amplicon may have a sequence that is
substantially
complementary to the target polynucleotide or is substantially identical to
the target
polynucleotide. It will be understood that a small number of mutations (e.g.,
due to
amplification artifacts) of a polynucleotide may occur when generating an
amplicon of that
polynucleotide.
[0059] A substrate region that includes substantially only amplicons of a
given
polynucleotide may be referred to as -monoclonal," while a substrate region
that includes
amplicons of polynucleotides having different sequences than one another may
be referred to
as -polyclonal.- A polyclonal region of a substrate may include different
subregions therein
17
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
that respectively are monoclonal. Each such monoclonal region, whether within
a larger
polyclonal region or on its own, may correspond to a -cluster" generated from
a "seed." The
"seed" may refer to a single target polynucleotide, while the "cluster" may
refer to a
collection of amplicons of that target polynucleotide.
Compositions and methods for capturing and amplifying polynucleotides
[0060] Examples provided herein relate to compositions and methods capturing
and
amplifying target polynucleotides using modified capture primers that may be
used under
conditions that reduce or inhibit solution-based annealing while permitting
the modified
capture primers to capture target polynucleotides from solution. In
comparison, previously
known capture primers may be used under conditions in which solution-based
annealing
competes with capture of target polynucleotides from solution, and such
competition reduces
availability of the target polynucleotides for capture.
[0061] For example, FIGS. 2A-2C schematically illustrate compositions and
operations in a
process flow for capture of polynucleotides on a substrate using previously
known capture
primers. Composition 2000 illustrated in FIG. 2A includes substrate 200 having
capture
primers 231 and orthogonal capture primers 232 coupled thereto, as well as a
fluid (solution)
containing target polynucleotides, e.g., polynucleotides that it is intended
to capture, amplify,
and sequence using sequencing-by-synthesis (SBS). The target polynucleotides
may include
or may be provided in the form of duplexes (illustratively, duplexes D1, D2,
and D3) between
target polynucleotides having sequences that are complementary to one another.
For
example, duplex D1 may include a first target polynucleotide including
sequence 211, first
adapter 221, and second adapter 222, and a second target nucleotide that is
complementary to
the first target polynucleotide, e.g., including complementary sequence 211',
first
complementary adapter 221', and second complementary adapter 222'. Similarly,
duplex D2
may include a first target polynucleotide including sequence 212, first
adapter 221, and
second adapter 222, and a second target nucleotide that is complementary to
the first target
polynucleotide, e.g., including complementary sequence 212', first
complementary adapter
221', and second complementary adapter 222'. Similarly, duplex D3 may include
a first
target polynucleotide including sequence 213, first adapter 221, and second
adapter 222, and
a second target nucleotide that is complementary to the first target
polynucleotide, e.g.,
including complementary sequence 212', first complementary adapter 221', and
second
complementary adapter 222'. Sequences 211, 212, and 213 may be different than
one
18
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
another, and it may be desired to determine those sequences using SBS. First
adapters 221
may be complementary to capture primers 231 so as to be able to hybridize
thereto under
suitable conditions, and second adapters 222 may be complementary to
orthogonal capture
primers 232 so as to be able to hybridize thereto under suitable conditions.
Although not
specifically illustrated, each capture primer 231 or each orthogonal capture
primer 232 may
include a cleavable moiety such as 8-oxo-G.
[0062] Capture primers 231 may, for example, be P5 capture primers, and
orthogonal capture
primers 232 may, for example, be P7 capture primers. P5 capture primers, which
are
commercially available from Illumina, Inc. (San Diego, CA) have the sequence
5'-
AATGATACGGCGACCACCGA-3' (SEQ ID NO: 1). P7 capture primers, which also are
commercially available from Illumina, Inc., have the sequence 5'-
CAAGCAGAAGACGGCATACGA-3' (SEQ ID NO: 2). First adapters 221 may be, for
example, complementary P5 adapters (cP5) and second adapters 222 may be, for
example,
complementary P7 adapters (cP7). Complementary P5 adapters, which are
commercially
available from Illumina, Inc. (San Diego, CA), have the sequence 5'-
TCGGTGGTCGCCGTATCATT-3' (SEQ ID NO: 3). Complementary P7 adapters, which
are commercially available from Illumina, Inc. (San Diego, CA), may have the
sequence 5'-
TCGTATGCCGTCTTCTGCTTG-3' (SEQ ID NO: 4).
[0063] Before attempting to capture the target polynucleotides on substrate
200 for later
amplification and sequencing, duplexes D1, D2, and D3 are melted in a manner
such as
illustrated in FIG. 2B so as to obtain single-stranded target polynucleotides
having first
adapters 221 that are available to hybridize to capture primers 231, and
second adapters 222
that are available to hybridize to orthogonal capture primers 232. Such
melting may be
performed, for example, by changing the temperature and/or composition of the
solution in
which duplexes D1, D2, and D3 are disposed. For example, duplexes D1, D2, and
D3 may
be exposed to a sufficient amount of formamide in solution, e.g., about 1% to
100%
formamide (%v/v), or about 5% to about 80% formamide (%v/v), or about 10% to
about 50%
formamide (%v/v), or about 1% to about 20% formamide (%v/v), to cause the
duplexes to
dissociate at the current solution temperature. Additionally, or
alternatively, the temperature
of the solution may be increased above the melting temperature (Tm) of the
duplexes at the
current solution composition. It will be appreciated that the particular Tm of
a duplex may
depend on the composition of the solution (e.g., salt (Na+) concentration and
formamide
19
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
concentration, if any), as well as the length and sequences of the
polynucleotides in the
duplex.
[0064] After the duplexes D1, D2, and D3 are melted such as described with
reference to
FIG. 2B, first adapters 221 may be available to hybridize to capture primers
231, and second
adapters 222 may be available to hybridize to orthogonal capture primers 232.
However, the
solution and temperature conditions that caused such melting also may inhibit
first adapters
221 from hybridizing to capture primers 231, and may inhibit second adapters
222 from
hybridizing to orthogonal capture primers 232. So as to promote such
hybridization and thus
promote capture of the target polynucleotides on substrate 200, the
temperature and/or
composition of the solution in which the target polynucleotides are disposed
again may be
changed. For example, a sufficient amount of formamide may be removed from the
solution
at the current solution temperature. Additionally, or alternatively, the
temperature of the
solution may be decreased below the melting temperature (Tm) of duplexes
between (a) first
adapters 221 and capture primers 231 and (b) second adapters 222 and
orthogonal capture
primers 232, at the current solution composition.
[0065] In the nonlimiting example illustrated in FIG. 2C, this additional
change in
conditions, e.g., solution temperature and/or composition, facilitates
hybridization between
the second adapter 222 coupled to sequence 211 and one of capture primers 232;
and
hybridization between first adapter 221 coupled to sequence 213 and one of
capture primers
231. However, this additional change in conditions also facilitates annealing
between target
polynucleotides in solution. For example, the first and second target
polynucleotides
respectively including sequences that are complementary to one another may
reanneal with
each other along their lengths, illustratively, such as sequences 212 and 212
that reanneal
along their lengths to re-form duplex D2. Additionally, first and second
target
polynucleotides including sequences that are not complementary to one another
may partially
anneal with each other, e.g., at adapters 221 and 221' and at adapters 222 and
222',
illustratively, such as non-complementary sequences 214 and 211' that anneal
substantially
only at their adapters to form duplex D4 in a manner such as illustrated in
FIG. 2C. Target
polynucleotides that anneal in solution to form duplexes, illustratively, the
target
polynucleotides including sequences 212 and 214 in the nonlimiting example
shown in FIG.
2C, are unavailable to hybridize with capture primers 231 or orthogonal
capture primers 232
and as such may not be captured, let alone amplified or sequenced.
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
[0066] In comparison, the present modified capture primers may be hybridized
with the
adapters of target polynucleotides under conditions at which annealing in
solution
substantially may not occur, e.g., at a salt concentration, formamide content,
and/or
temperature at which annealing in solution substantially may not occur. As
such, target
polynucleotides substantially may not anneal in such solution to form duplexes
and therefore
may remain available to hybridize with the present capture primers, following
which they
may be amplified and sequenced or otherwise used as desired. Additionally, the
present
modified capture primers may be hybridized with the adapters of target
polynucleotides under
conditions that inhibit any non-specific or non-productive hybridization of
target
polynucleotides to capture primers or to adapters in solution, and as such it
may be expected
that substantially all hybridization events will be productive and will lead
to duplexes that
may be sequenced.
[0067] FIGS. 3A-3H schematically illustrate example compositions and
operations in an
example process flow for capture and amplification of polynucleotides on a
substrate using
the present capture primers. Composition 3000 illustrated in FIG. 3A includes
substrate 300
having capture primers 331 and orthogonal capture primers 332 coupled thereto,
as well as a
fluid (solution) containing target polynucleotides, e.g., polynucleotides that
it is intended to
capture, amplify, and sequence using sequencing-by-synthesis (SBS). The target
polynucleotides may include or may be provided in the form of duplexes
(illustratively,
duplexes D5, D6, and D7) between target polynucleotides having sequences that
are
complementary to one another. For example, in a manner similar to that
described with
reference to FIG. 2A, duplex D5 may include a first target polynucleotide
including sequence
311, first adapter 321, and second adapter 322, and a second target nucleotide
that is
complementary to the first target polynucleotide, e.g., including
complementary sequence
311', first complementary adapter 321', and second complementary adapter 322'.
Similarly,
duplex D6 may include a first target polynucleotide including sequence 312,
first adapter
321, and second adapter 322, and a second target nucleotide that is
complementary to the first
target polynucleotide, e.g., including complementary sequence 312', first
complementary
adapter 321', and second complementary adapter 322'. Similarly, duplex D7 may
include a
first target polynucleotide including sequence 313, first adapter 321, and
second adapter 322,
and a second target nucleotide that is complementary to the first target
polynucleotide, e.g.,
including complementary sequence 312', first complementary adapter 321', and
second
complementary adapter 322'. Sequences 311, 312, and 313 may be different than
one
21
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
another, and it may be desired to determine those sequences using SBS. First
adapters 321
may be complementary to capture primers 331 so as to be able to hybridize
under suitable
conditions, and second adapters 322 may be complementary to modified
orthogonal capture
primers 332 so as to be able to hybridize thereto under suitable conditions.
[0068] Each of capture primers 331 may include a plurality of nucleic acids
that increase the
Tm of duplexes between capture primers 331 and first adapters 321 as compared
to the Tm of
duplexes between first adapters 321 and complementary first adapters 321'. For
example, the
modified nucleic acids of capture primers 331 may include locked nucleic acid
(LNA),
peptide nucleic acid (PNA), or super T, each of which may be expected to
increase the Tm of
duplexes between capture primers 331 and first adapters 321 as compared to the
Tm of
duplexes between first adapters 321 and complementary first adapters 321'.
Capture primers
331 further may include DNA. The modified nucleic acids and the DNA may be
distributed
between a 5' end and a 3' end of the capture primers. For example, the
sequences of capture
primers 331 may include one or more DNA molecules, followed by one or more
modified
nucleic acids, followed by one or more DNA molecules, followed by one or more
modified
nucleic acids, and so on. Alternatively, the modified nucleic acids may be
disposed at a 5'
end of the capture primers and the DNA is disposed at a 3' end of the capture
primers, e.g., in
a manner such as described below with reference to FIG. 4. Although not
specifically
illustrated, each capture primer 331 also may include a cleavable moiety such
as 8-oxo-G.
[0069] Similarly, each of orthogonal capture primers 332 may include a
plurality of modified
nucleic acids that increase the Tm of duplexes between orthogonal capture
primers 332 and
second adapters 322 as compared to the Tm of duplexes between second adapters
322 and
complementary second adapters 322'. For example, the modified nucleic acids of
orthogonal
capture primers 332 may include locked nucleic acid (LNA), peptide nucleic
acid (PNA), or
super T, each of which may be expected to increase the Tm of duplexes between
orthogonal
capture primers 332 and second adapters 322 as compared to the Tm of duplexes
between
second adapters 322 and complementary second adapters 322'. Orthogonal capture
primers
332 further may include deoxyribonucleic acid (DNA). The modified nucleic
acids and the
DNA may be distributed between a 5' end and a 3' end of the orthogonal capture
primers. For
example, the sequences of orthogonal capture primers 332 may include one or
more DNA
molecules, followed by one or more modified nucleic acids, followed by one or
more DNA
molecules, followed by one or more modified nucleic acids, and so on.
Alternatively, the
22
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
modified nucleic acids may be disposed at a 5' end of the capture primers and
the DNA is
disposed at a 3' end of the capture primers, e.g., in a manner such as
described below with
reference to FIG. 4. Although not specifically illustrated, each orthogonal
capture primer 332
also may include a cleavable moiety such as 8-oxo-G.
[0070] Note that the modified nucleic acids of capture primers 331 may be, but
need not
necessarily be, the same type of modified nucleic acids of orthogonal capture
primers 332.
Illustratively, capture primers 331 and orthogonal capture primers 332 may
include LNA;
capture primers 331 and orthogonal capture primers 332 may include PNA; or
capture
primers 331 and orthogonal capture primers 332 may include super T.
Alternatively, capture
primers 331 may include LNA while orthogonal capture primers 332 may include
PNA or
super T; capture primers 331 may include PNA while orthogonal capture primers
332 may
include LNA or super T; or capture primers 331 may include super T while
orthogonal
capture primers 332 may include LNA or PNA. In still other examples,
orthogonal capture
primers 332 may include LNA while capture primers 331 may include PNA or super
T;
orthogonal capture primers 332 may include PNA while capture primers 331 may
include
LNA or super T; or orthogonal capture primers 332 may include super T while
capture
primers 331 may include LNA or PNA.
[0071] LNA closely mimics DNA with the only change being a linker that
connects the 2'
and 4' carbons. PNAs are structured similarly to a polypeptide, with a
carboxylic acid group
and an amino group. Super T includes a modified thymidine base with a butyne
group. In
the case of LNA and super T base modifications, increased rigidity of the
molecular structure
and stronger base stacking may increase the binding energy of the duplex
interaction (321-
331 or 322-332) as compared to DNA/DNA duplexes (321-321' or 322-322'). On the
other
hand, the melting point of duplexes containing un-charged PNA (321-331 or 322-
332) may
be increased compared DNA/DNA duplexes (321-321' or 322-322') due to reduced
electrostatic repulsion. When capture primer sequences are modified to include
such
modified nucleic acids, the melting point between the surface/solution duplex
(321-331 or
322-332) can be made significantly higher than the melting point of the
solution/solution
DNA duplex (321-321' or 322-322'). This allows for the target polynucleotides
311, 312,
313 to be seeded at a set of conditions (e.g., salt concentration, formamide
concentration, and
temperature) that does not allow for reannealing of adapters in solution. As
such, the target
23
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
polynucleotides 311, 312, 313 may be seeded with substantially no loss from
adapter
reannealing such as described with reference to FIG. 2C.
[0072] Before attempting to capture the target polynucleotides on substrate
300 for later
amplification and sequencing, duplexes D5, D6, and D7 are melted in a manner
such as
illustrated in FIG. 3B so as to obtain single-stranded target polynucleotides
having first
adapters 321 that are available to hybridize to capture primers 331, and
second adapters 322
that are available to hybridize to orthogonal capture primers 332. In a manner
similar to that
described with reference to FIG. 2B, such melting may be performed, for
example, by
changing the temperature and/or composition of the solution in which duplexes
D5, D6, and
D7 are disposed. For example, duplexes D5, D6, and D7 may be exposed to a
sufficient
amount of formamide in solution, e.g., about 1% to 100% formamide (%v/v) ), or
about 5%
to about 80% formamide (%v/v), or about 10% to about 50% formamide (%v/v), or
about 1%
to about 20% formamide (%v/v), to cause the duplexes to dissociate at the
current solution
temperature. Additionally, or alternatively, the temperature of the solution
may be increased
above the melting temperature (Tm) of the duplexes at the current solution
composition. It
will be appreciated that the particular Tm of a duplex may depend on the
composition of the
solution (e.g., salt (Na+) concentration and formamide concentration, if any),
as well as the
length and sequences of the polynucleotides in the duplex.
[0073] After the duplexes D5, D6, and D7 are melted such as described with
reference to
FIG. 3B, first adapters 321 may hybridize to capture primers 331, and second
adapters 322
may hybridize to orthogonal capture primers 332 at the same conditions that
were used to
melt duplexes D5, D6, and D7, in a manner such as illustrated in FIG. 3C. That
is, the
solution and temperature conditions that caused such melting may not inhibit
first adapters
321 from hybridizing to capture primers 331, and may not inhibit second
adapters 322 from
hybridizing to orthogonal capture primers 332. Instead, such conditions may
inhibit any
annealing between adapters 321 and complementary adapters 321' and any
reannealing
between adapters 322 and complementary adapters 322' and thus may inhibit
formation of
any duplexes such as described with reference to FIG. 2C. As such, each of the
target
polynucleotides may remain available for capture, and may be captured on
substrate, without
necessarily changing the conditions that were used to melt duplexes D5, D6,
and D7, e.g.,
without necessarily reducing the concentration of formamide and without
necessarily
decreasing the solution temperature.
24
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
[0074] In the nonlimiting example illustrated in FIG. 3C, at conditions that
inhibit annealing
between target polynucleotides in solution, the first adapters 321 of some of
the target
polynucleotides are hybridized to respective ones of the capture primers 331
to form first
duplexes, and the second adapters 322 of some of the target polynucleotides
are hybridized to
respective ones of the orthogonal capture primers 332 to form second duplexes.
The first and
second duplexes each may have a melting temperature (Tm) that is greater than
a Tm of third
duplexes that would be formed by hybridization of the second target
polynucleotides to the
respective ones of the first target polynucleotides to which the second target
polynucleotides
are complementary. Illustratively, first adapter 321 coupled to sequence 312
is hybridized to
one of capture primers 331 to form duplex D8; second adapter 322 coupled to
sequence 311
is hybridized to one of orthogonal capture primers 332 to form duplex D9; and
second
adapter 322 coupled to sequence 313 is hybridized to one of orthogonal capture
primers 332
to form duplex D10.
[0075] The Tms of duplexes between first adapters 321 and capture primers 331
may be
greater than the Tms of duplexes between first adapters 321 and complementary
first adapters
321', e.g., may exceed the Tms of duplexes between first adapters 321 and
complementary
first adapters 321' by at least about 5 C, by at least about 10 C, by at least
about 15 C, by at
least about 20 C, or by at least about 25 C. Similarly, the Tms of duplexes
between second
adapters 322 and orthogonal capture primers 332 may be greater than the Tms of
duplexes
between second adapters 322 and complementary second adapters 322', e.g., may
exceed the
Tms of duplexes between first adapters 321 and complementary first adapters
321' by at least
about 5 C, by at least about 10 C, by at least about 15 C, by at least about
20 C, or by at
least about 25 C. As such, the target polynucleotides suitably may be captured
on substrate
300 at a temperature that is below the Tms of duplexes between first adapters
321 and capture
primers 331 and below the Tms of duplexes between second adapters 322 and
orthogonal
capture primers 332, and that is above the Tms of duplexes between first
adapters 321 and
complementary first adapters 321' and that is above the Tms of duplexes
between second
adapters 322 and complementary second adapters 322'.
[0001] Illustratively, the Tms of duplexes D8, D9, and D10 illustrated in FIG.
3C may be
greater than the Tms of any of duplexes D1, D2, D3, and D4 described with
reference to
FIGS. 2A and 2C. For example, duplexes D8, D9, and D10 each may have a melting
temperature (Tm) that is between about 80 C and about 110 C, e.g., between
about 85 C and
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
about 105 C, or between about 90 C and about 100 C. In comparison, the Tms of
duplexes
D1, D2, D3, and D4 may be below about 80 C, e.g., below about 75 C, below
about 70 C,
below about 65 C, or below about 60 C. As a result of the significantly
different Tms of
duplexes between adapters and the surface primers as compared to the Tms of
duplexes
between adapters in solution, substantially none of the second target
polynucleotides are
hybridized in solution to any of the first target polynucleotides. As such,
duplexes D8, D9,
and D10 readily may form at certain conditions under which duplexes D1, D2,
D3, and D4
may not form. The duplexes may form on the surface of substrate 300 at
different locations,
e.g., in accordance with the Poisson distribution. Although not specifically
illustrated, it will
be appreciated that substrate 300 may be patterned, e.g., so as to define
different regions
within which a duplex respectively may form, and within which clusters
subsequently may be
formed using amplification. Additionally, although the examples described with
reference to
FIGS. 3A-3H are illustrated so as to suggest the use of flat substrates, it
should be apparent
that more complex substrates may be used, e.g., that include wells that
respectively are
seeded by target polynucleotides and within which substantially monoclonal
clusters may be
formed.
[0076] As illustrated in FIG. 3D, after the initial hybridizations described
with reference to
FIG. 3C, each of the target polynucleotides 311, 312, 313 may be amplified so
as to form
respective amplicons 311', 312', and 313', respectively. Following such
amplification, the
target polynucleotides 311, 312, 313 may be dehybridized in a manner such as
illustrated in
FIG. 3E, while amplicons 311', 312', and 313' remain covalently bound to
substrate 300.
Note that such dehybridization need not necessarily be performed. For example,
instead of
dehybridizing target polynucleotides 311, 312, 313, such polynucleotides may
remain
hybridized to the substrate and may be further amplified using a strand
invasion process such
as known in the art and may be referred to as ExAmp.
[0077] As illustrated in FIG. 3F, after the initial amplifications described
with reference to
FIGS. 3D-3E, the resulting amplicons may bend so as potentially to hybridize
to other
capture primers or orthogonal capture primers on substrate 300. For example,
complementary first adapter 321' of amplicon 311' may hybridize to one of
capture primers
331; complementary second adapter 322' of amplicon 312' may hybridize to one
of
orthogonal capture primers 332; and complementary first adapter 321' of
amplicon 313' may
hybridize to one of capture primers 331. The duplexes between the amplicons'
adapters and
26
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
respective capture primers or orthogonal capture primers may have similar or
the same Tms
as duplexes DR, D9, and D10, and thus may remain hybridized so as to promote
further
amplification of the target polynucleotides.
[0078] FIG. 3G illustrates the composition of FIG. 3F following another
amplification
operation. It may be seen that the composition includes an additional amplicon
311 of
amplicon 311'; an additional amplicon 312 of amplicon 312'; and an additional
amplicon 313
of amplicon 313'. Such additional amplicons may be hybridized to the amplicons
from
which they were generated. The solution may be increased so as to dehybridize
the
amplicon's adapters from the capture primers or orthogonal capture primers in
a manner such
as illustrated in FIG. 3H. The amplification operation may be repeated any
suitable number
of times so as to generate further amplicons of amplicons 311, 311', 312,
312', 313, and 313'.
Amplification operations may be formed any suitable number of times so as to
substantially
fill respective substrate regions (not specifically illustrated) with
substantially monoclonal
clusters, e.g., with amplicons of target polynucleotide 311, 312, or 313,
respectively. For
example, amplicons within each of the substrate regions each may include at
least about 60%
amplicons of one selected target polynucleotide, or at least about 70%
amplicons of one
selected target polynucleotide, or at least about 80% amplicons of one
selected target
polynucleotide, or at least about 90% amplicons of one selected target
polynucleotide, or at
least about 95% amplicons of one selected target polynucleotide, or at least
about 98%
amplicons of one selected target polynucleotide, or at least about 99%
amplicons of one
selected target polynucleotide, or about 100% amplicons of one selected target
polynucleotide.
[0079] As noted above, in some examples, certain capture primers and
orthogonal capture
primers may include non-nucleotide moieties. Such non-nucleotide moieties may
include,
but are not limited to, excision moieties via which a portion of the capture
primers selectively
may be removed. The excision moieties may be located at any suitable position
along the
length of any suitable primer(s) and may be, but need not necessarily be, the
same type of
excision moiety as one another. Following a desired number of additional
amplification
operations such as described with reference to FIGS. 3E-3H, portions of
capture primers 331
or orthogonal capture primers 332 may be removed by reacting suitable
enzyme(s) or
reagent(s) with the excision moieties.
27
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
[0080] The present capture primers and orthogonal capture primers may include
any suitable
number, type, and arrangement of modified nucleic acids that sufficiently
increases the Tms
of duplexes between those capture primers or orthogonal capture primers, and
adapters of the
target polynucleotides. FIG. 4 schematically illustrates an example duplex
between a
polynucleotide and one of the present capture primers, according to some
examples. In the
nonlimiting example illustrated in FIG. 4, modified nucleic acids 441 (such as
PNA or LNA)
are disposed at a 5' end of capture primer 331 and DNA 442 is disposed at a 3
end of the
capture primer. An optional T-spacer 443 is disposed between DNA 442 and the
surface of
substrate 400 (e.g., flowcell). Together, modified nucleic acids 441 and DNA
442 may be
considered to provide a "chimeric" grafting primer structure. The target
polynucleotide may
include library template 311 coupled to adapter 321 which includes a first
subsequence 341
that is complementary to the sequence of modified nucleic acids 441, and a
second
subsequence 342 that is complementary to the sequence of DNA 442.
Alternatively, the
modified nucleic acids (such as PNA, LNA, or super T) and the DNA are
distributed between
a 5' end and a 3' end of the capture primers in a manner such as described
further above.
Orthogonal capture primers 332 may be configured similarly as capture primer
331 illustrated
in FIG. 4, e.g., may include a chimeric grafting primer structure, but with
different sequences.
Alternatively, the modified nucleic acids (such as PNA, LNA, or super T) and
the DNA may
be distributed between a 5' end and a 3' end of the orthogonal capture primers
in a manner
such as described further above.
[0081] In some examples, each of the capture primers 331 may include between
about five
and about twenty of the modified nucleic acids, and each of the orthogonal
capture primers
332 may include between about five and about twenty of the modified nucleic
acids.
However, it will be appreciated that the number of modified nucleic acids
suitably may be
adjusted to obtain an appropriate Tm for use in conditions that inhibit duplex
formation in
solution while allowing duplex formation at the substrate surface, and that
the capture
primers and orthogonal capture primers need not include the same number, type,
or
distribution of modified nucleic acids as each other. Illustratively, each of
the capture
primers may include at least about nine of the modified nucleic acids, and
each of the
orthogonal capture primers may include at least about nine of the modified
nucleic acids; or
each of the capture primers may include at least about twelve of the modified
nucleic acids,
and each of the orthogonal capture primers may include at least about twelve
of the modified
nucleic acids; or each of the capture primers may include at least about
fifteen of the
28
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
modified nucleic acids, and each of the orthogonal capture primers may include
at least about
fifteen of the modified nucleic acids.
[0082] Note that although capture primers 331 and orthogonal capture primers
332 include
modified nucleic acids, they otherwise may have similar or the same sequences
as capture
primers 231 and 232. That is, capture primers 331 may be modified P5 capture
primers, and
wherein the orthogonal capture primers are modified P7 capture primers. For
example,
capture primers 331 may be or include P5 primers having the sequence provided
elsewhere
herein, in which at least some of the P5 DNA bases are replaced with their
PNA, LNA, or
super T analogues. Additionally, or alternatively, orthogonal capture primers
332 may be or
include P7 primers having the sequence provided elsewhere herein, in which at
least some of
the P7 DNA bases are replaced with their PNA, LNA, or super T analogues.
Nonlimiting
examples of chimeric P5 and P7 sequences including varying lengths of LNA, the
calculated
Tm for a duplex between the chimeric P5 or P7 sequence and their respective
adapter cP5 or
cP7 (750 mM Na+) are provided in Table 1 below in which bolded text indicates
LNA, and
unbolded text indicates DNA. The commercially available P5 and P7 sequences,
and the
calculated Tm for a duplex between the P5 or P7 sequence and their respective
adapter cP5 or
cP7, are provided for reference.
Table 1. High Tm DNA/LNA chimeric P5/P7
Type Sequence Tm
( C)
Standard P5 TTTTTT AATGA TACGG CGACC ACCGA GAUCT ACAC
78.1
(SEQ ID
NO: 5)
LNA-mod. TTTTTT AATGA TACGG CGACC ACCGA GAUCT ACAC
99.1
P5 (SEQ ID
NO: 6)
LNA-mod. TTTTTT AATGA TACGG CGACC ACCGA GAUCT ACAC
94.1
P5 (SEQ ID
NO: 7)
29
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
LNA-mod. TTTTTT AATGA TACGG CGACC ACCGA GAUCT ACAC
89.1
P5 (SEQ ID
NO: 8)
Standard P7 TTTTTT CAAGC AGAAG ACGGC ATAC(8-oxoG) AGAT
72.1
(SEQ ID
NO: 9)
LNA-mod. TTTTTT CAAGC AGAAG ACGGC ATAC(8-oxoG) AGAT
93.2
P7 (SEQ ID
NO: 10)
LNA-mod. TTTTTT CAAGC AGAAG ACGGC ATAC(8-oxoG) AGAT
89.1
P7 (SEQ ID
NO: 11)
LNA-mod. TTTTTT CAAGC AGAAG ACGGC ATAC(8-oxoG) AGAT
84.7
P7 (SEQ ID
NO: 12)
[0083] From the examples provided in Table 1, it may be understood that for
the P5
sequence, replacing fifteen DNA bases with LNA may increase the Tm of a duplex
between
that sequence and the cP5 adapter by about 21 C; replacing twelve DNA bases
with LNA
may increase the Tm of a duplex between that sequence and the cP5 adapter by
about 16 C;
and replacing nine DNA bases with LNA may increase the Tm of a duplex between
that
sequence and the cP5 adapter by about 11 C. Additionally, from the examples
provided in
Table 1, it may be understood that for the P7 sequence, replacing fifteen DNA
bases with
LNA may increase the Tm of a duplex between that sequence and the cP7 adapter
by about
21 C; replacing twelve DNA bases with LNA may increase the Tm of a duplex
between that
sequence and the cP7 adapter by about 17 C; and replacing nine DNA bases with
LNA may
increase the Tm of a duplex between that sequence and the cP7 adapter by about
12 C. It
will be appreciated that the number of modified nucleic acids in any sequence
in order to
provide any suitable Tm for use in a capture primer or orthogonal capture
primer such as
provided herein.
100841 One consideration for implementing the present modified capture primers
is that such
primers be compatible with suitable enzymes, such as recombinases, single-
stranded binding
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
proteins, and polymerases, as may be used for cluster generation and
sequencing. Chimera
structures including PNA or LNA such as described with reference to FIG. 4 are
expected to
be compatible with such enzymes in a manner similar to that described in the
following
references, the entire contents of which are incorporated by reference herein:
Levin et al.,
"Position-dependent effects of locked nucleic acid (LNA) on DNA sequencing and
PCR
primers," Nucleic Acids Research 34(20): e142, 11 pages (2006); and Misra et
al.,
"Polyamide nucleic acid-DNA chimera lacking the phosphate backbone are novel
primers for
polymerase reaction catalyzed by DNA polymerases," Biochemistry 37: 1917-1925
(1998).
Further information about LNA may be found in Braasch et al., "Locked nucleic
acid (LNA):
fine-tuning the recognition of DNA and RNA," Chemistry & Biology 8: 1-7
(2001), the
entire contents of which are incorporated by reference herein. Providing DNA
at the 3' end
of a chimera structure such as described with reference to FIG. 4 may help to
maintain
expected enzyme activity by locating the unmodified DNA bases at positions
with which the
enzymes are likely to interact, while providing the modified nucleic acids at
the 5 end of the
chimera structure may enhance the strength of binding to an adapter.
[0085] Additionally, as noted elsewhere herein and as will be appreciated by
those skilled in
the art, the Tm of any given duplex may vary based on the composition of the
solution in
which that duplex is disposed. For example, adding fonnamide to the solution
may decrease
the Tm of a given duplex, while adding salt to the solution may increase the
Tm of a given
duplex. As such, the example Tms provided herein (and differences between Tms)
are purely
illustrative and may vary based on the particular composition of the solution
in which the
duplexes are disposed. FIGS. 5A-5C are plots illustrating example effects of
conditions on
the capture of polynucleotides by the present capture primers. In the
nonlimiting example
shown in FIG. 5A, the percent of double-stranded DNA (dsDNA) is shown as a
function of
temperature for duplexes formed by hybridization between cP7 and the P7
standard library
capture primers (curve 501) and for duplexes formed by hybridization between
cP7 and
LNA-modified P7 capture primers (curve 502), in a solution including 0% v/v
formamide and
750 mM Na+. The LNA-modified P7 capture primers had the sequence CAACC AGAAG
ACGGC ATAC(8-oxoG) AGAT (SEQ ID NO: 13) in which bolded text indicates LNA,
and
were modeled to have a 20 C Tm when duplexed with cP7.
[0086] In FIG. 5A, it may be seen that a temperature of about 77 C corresponds
to the Tm of
duplexes formed by hybridization between cP7 and the P7 standard library
capture primers
31
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
(curve 501), while a temperature of about 91 C corresponds to the Tm of
duplexes formed by
hybridization between cP7 and LNA-modified P7 capture primers (curve 502). At
temperatures above about 85 C, curve 501 shows that the percent dsDNA becomes
insignificant (e.g., less than 1%) for duplexes formed by hybridization
between cP7 and the
P7 standard library capture primers. Additionally, at temperatures below about
90 C, curve
501 shows that the percent dsDNA is approximately 100% for duplexes formed by
hybridization between cP7 and LNA-modified P7 capture primers. Accordingly,
from FIG.
5A it may be understood that at a temperature of about 85 C-90 C,
hybridization between
cP7 and the P7 standard library capture primers may be substantially
completely inhibited,
while hybridization between cP7 and LNA-modified P7 capture primers forms
highly stable
duplexes. As such, within this temperature range, template polynucleotides may
be captured
from solution with high efficiency and with substantially no competition from
solution-based
annealing processes such as described with reference to FIG. 2C. It will be
appreciated that
the particular temperature range suitable for performing such operations may
vary depending
on the particular composition of the solution. It may be expected that any
other capture
primer and adapter sequences will exhibit a similar dependence of duplex
formation upon
temperature.
[0087] For example, in the nonlimiting example shown in FIG. 5B, the Tms for
duplexes
formed by hybridization between cP7 and the P7 standard library capture
primers (curve 503)
and for duplexes formed by hybridization between cP7 and LNA-modified P7
capture
primers (curve 504) are shown as a function of formamide concentration (%v/v),
for a salt
concentration of 750 rnlVINa+. Similarly as in FIG. 5A, it may be seen that
for 0% v/v
formamide, a temperature of about 77 C corresponds to the Tm of duplexes
formed by
hybridization between cP7 and the P7 standard library capture primers (curve
503), while a
temperature of about 91 C corresponds to the Tm of duplexes formed by
hybridization
between cP7 and LNA-modified P7 capture primers (curve 504). As the
concentration of
formamide increases, the Tms decrease both for duplexes formed by
hybridization between
cP7 and the P7 standard library capture primers and for duplexes formed by
hybridization
between cP7 and LNA-modified P7 capture primers. For example, at a
concentration of
about 5% v/v formamide, the Tm of duplexes formed by hybridization between cP7
and the
P7 standard library capture primers decreases to about 69 C, and the Tm of
duplexes formed
by hybridization between cP7 and LNA-modified P7 capture primers decreases to
about
88 C. At a concentration of about 10% v/v formamide, the Tm of duplexes formed
by
32
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
hybridization between cP7 and the P7 standard library capture primers
decreases to about
66 C, and the Tm of duplexes formed by hybridization between cP7 and LNA-
modified P7
capture primers decreases to about 85 C. At a concentration of about 15% v/v
formamide,
the Tm of duplexes formed by hybridization between cP7 and the P7 standard
library capture
primers decreases to about 63 C, and the Tm of duplexes formed by
hybridization between
cP7 and LNA-modified P7 capture primers decreases to about 82 C. At a
concentration of
about 20% v/v formamide, the Tm of duplexes formed by hybridization between
cP7 and the
P7 standard library capture primers decreases to about 58 C, and the Tm of
duplexes formed
by hybridization between cP7 and LNA-modified P7 capture primers decreases to
about
79 C.
[0088] Accordingly, from FIG. 5B it may be understood that at any given
concentration of
formamide (e.g., at a concentration of about 1% to about 20% formamide (%v/v),
or about
5% to about 20% formamide (%v/v)), there is a range of temperatures at which
hybridization
between cP7 and the P7 standard library capture primers may be substantially
completely
inhibited, while hybridization between cP7 and LNA-modified P7 capture primers
forms
highly stable duplexes. As such, within this temperature range, template
polynucleotides
may be captured from solution with high efficiency and with substantially no
competition
from solution-based annealing processes such as described with reference to
FIG. 2C. It may
be expected that any other capture primer and adapter sequences will exhibit a
similar
dependence of duplex formation upon formamide concentration.
[0089] As another example, in the nonlimiting example shown in FIG. 5C, the
Tms for
duplexes formed by hybridization between cP7 and the P7 standard library
capture primers
(curve 505) and for duplexes formed by hybridization between cP7 and LNA-
modified P7
capture primers (curve 506) are shown as a function of salt concentration (mM
Na+). It may
be seen that for 750 mM Na+, a temperature of about 72 C corresponds to the Tm
of
duplexes formed by hybridization between cP7 and the P7 standard library
capture primers
(curve 505), while a temperature of about 89 C corresponds to the Tm of
duplexes formed by
hybridization between cP7 and LNA-modified P7 capture primers (curve 506). As
the
concentration of salt decreases, the Tms decrease both for duplexes formed by
hybridization
between cP7 and the P7 standard library capture primers and for duplexes
formed by
hybridization between cP7 and LNA-modified P7 capture primers. For example, at
a
concentration of about 500 mM Na+, the Tm of duplexes formed by hybridization
between
33
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
cP7 and the P7 standard library capture primers decreases to about 70 C, and
the Tm of
duplexes formed by hybridization between cP7 and LNA-modified P7 capture
primers
decreases to about 87 C. At a concentration of about 100 mM Na+, the Tm of
duplexes
formed by hybridization between cP7 and the P7 standard library capture
primers decreases
to about 55 C, and the Tm of duplexes formed by hybridization between cP7 and
LNA-
modified P7 capture primers decreases to about 77 C. At a concentration of
about 0 mIVI
Na+, the Tm of duplexes formed by hybridization between cP7 and the P7
standard library
capture primers decreases to about 42 C, and the Tm of duplexes formed by
hybridization
between cP7 and LNA-modified P7 capture primers decreases to about 58 C.
[0090] Accordingly, from FIG. 5C it may be understood that at any given
concentration of
salt, (e.g., at a concentration of about 100 to about 800 mM Na+, or a
concentration of 200 to
about 800 mM Na+), there is a range of temperatures at which hybridization
between cP7 and
the P7 standard library capture primers may be substantially completely
inhibited, while
hybridization between cP7 and LNA-modified P7 capture primers forms highly
stable
duplexes. As such, within this temperature range, template polynucleotides may
be captured
from solution with high efficiency and with substantially no competition from
solution-based
annealing processes such as described with reference to FIG. 2C. It may be
expected that
any other capture primer and adapter sequences will exhibit a similar
dependence of duplex
formation upon salt concentration.
[0091] It will be appreciated that example compositions such as described
herein may be
used in any suitable method for capturing and amplifying a polynucleotide. For
example,
FIG. 6 illustrates an example flow of operations in a method 600 for capturing
and
amplifying a polynucleotide using the present modified primers. Although
method 600 may
be implemented using composition 3000 described with reference to FIGS. 3A-3H,
method
600 may be implemented using any other suitable composition.
[0092] Referring now to FIG. 6, method 600 may include providing a composition
including
(a) a plurality of capture primers coupled to the surface of the substrate,
each capture primer
including modified nucleic acids; and (b) a plurality of orthogonal capture
primers coupled to
the surface of the substrate, each orthogonal capture primer including
modified nucleic acids
(operation 610). The composition may be similar to composition 3000 described
with
reference to FIG. 3A, e.g., may include capture primers 331 and orthogonal
capture primers
332 coupled to substrate 300.
34
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
[0093] Method 600 illustrated in FIG. 6 further may include providing a fluid
including (a)
first target polynucleotides, each including a first adapter that is
complementary to the
capture primers and a second adapter that is complementary to the orthogonal
capture
primers; and (b) second target polynucleotides that are complementary to
respective ones of
the first target polynucleotides. The fluid may be configured similarly as
described with
reference to FIG. 3A, e.g., may include target polynucleotides 311, 312, 313,
each of which is
coupled to a first adapter 321 and a second adapter 322, and may include
complementary
target polynucleotides 311', 312' 313', each of which is coupled to a
complementary first
adapter 321' and a complementary second adapter 322'.
[0094] Method 600 illustrated in FIG. 6 further may include contacting the
composition with
the fluid (operation 630). For example, the composition provided in operation
610 may be
contacted with the fluid provided in operation 620. Illustratively, the
composition may be
provided in a flowcell, and the fluid flowed into contact with the composition
within the
flowcell.
[0095] Method 600 illustrated FIG. 6 further may include, while inhibiting
hybridization of
the second target polynucleotides to the first target polynucleotides in the
fluid, (a)
hybridizing the first adapters of some of the first target polynucleotides to
respective ones of
the capture primers to form first duplexes and (b) hybridizing the second
adapters of some of
the first target polynucleotides to respective ones of the orthogonal capture
primers to form
second duplexes (operation 640). For example, in a manner such as described
with reference
to FIGS. 3B-3C, the hybridizations of the first adapters to capture primers
and the of the
second adapters to orthogonal capture primers may be performed under
conditions that inhibit
the first and second target polynucleotides from annealing to each other.
Example conditions
are provided elsewhere herein. Note that the same set of conditions may be
used to cause
dissociation of the first and second polynucleotides from one another and to
promote
hybridization of the adapters of the first polynucleotides to the present
capture primers. In
comparison, using previously known capture primers may involve two sets of
conditions, the
first condition to cause dissociation of the first and second polynucleotides
from one another,
followed by the second condition to promote hybridization of the adapters of
the first
polynucleotides to the previously known capture primers.
[0096] Method 600 illustrated in FIG. 6 further may include amplifying the
first target
polynucleotides, the amplifying comprising generating respective amplicons of
the first target
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
polynucleotides (operation 650). Non-limiting examples of the manner in which
such
amplicons may be generated are provided with reference to FIGS. 3D-3H.
[0097] Accordingly, whereas previously known capture primers may used under
conditions
at which non-productive rehybridization complexes may form in solution, the
present
modified capture primers (including orthogonal capture primers) overcome this
problem by
using sequences that include modified nucleic acids that impart stronger
binding (and higher
Tm) for surface/solution duplexes as compared to solution/solution duplexes.
The difference
in Tm between surface/solution and solution/solution duplexes may be achieved
by using
modified capture primer sequences that may, for example, include LNA, PNA,
super T, or
any combination of these. By incorporating base modifications to the capture
primers, a
temperature regime exists in which the adapters of target polynucleotides are
able to
hybridize to the surface but not able to hybridize to other adapters in
solution.
[0098] As such, the present modified capture primers provide one or more of
the following
benefits:
100991 A) Library capture is more efficient. In appropriate cases, lower input
concentration
of target polynucleotides may be used;
[0100] B) Surface capture post "on flowcell" denaturation may be carried out
at higher
temperatures, which may reduce or eliminate the need to integrate active
cooling in order to
reach temperatures favoring hybridization in a timely manner; and/or
[0101] C) Flowcells which require a large about of library may utilize
multiple fluidic pushes
for seeding, which greatly increases turnaround time (TAT), e.g., time from
the beginning of
sample prep to obtaining sequencing data. With the higher seeding efficiency
provided by
the present capture primers, it is possible to use higher concentrations of
target
polynucleotides with fewer fluidic pushes. This may save time during seeding.
[0102] It will be appreciated that the present compositions and methods are
not limited to use
with the particular operations described above. For example, although FIGS. 3A-
3H may be
considered to described operations consistent with -bridge amplification- or -
surface-bound
polymerase chain reaction," it will be appreciated that the present
compositions and methods
readily may be adapted for use with other amplification modalities. One such
amplification
modality is "exclusion amplification," or ExAmp. Exclusion amplification
methods may
36
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
allow for the amplification of a single target polynucleotide per substrate
region and the
production of a substantially monoclonal population of amplicons in a
substrate region. For
example, the rate of amplification of the first captured target polynucleotide
within a
substrate region may be rapid relative to much slower rates of transport and
capture of target
polynucleotides at the substrate region. As such, the first target
polynucleotide captured in a
substrate region may be amplified rapidly and fill the entire substrate
region, thus inhibiting
the capture of additional target polynucleotide in the same substrate region.
Alternatively, if a
second target polynucleotide attaches to same substrate region after the first
polynucleotide,
the relatively rapid amplification of the first polynucleotide may fill enough
of the substrate
region to result in a signal that is sufficiently strong to perform sequencing
by synthesis. The
use of exclusion amplification may also result in super-Poisson distributions
of monoclonal
substrate regions; that is, the fraction of substrate regions in an array that
are substantially
monoclonal may exceed the fraction predicted by the Poisson distribution.
[0103] Increasing super-Poisson distributions of useful clusters is useful
because more
substantially monoclonal substrate regions may result in higher quality
signal, and thus
improved SBS; however, the seeding of target polynucleotides into substrate
regions may
follow a spatial Poisson distribution, where the trade-off for increasing the
number of
occupied substrate regions is increasing the number of polyclonal substrate
regions. One
method of obtaining higher super-Poisson distributions is to have seeding
occur quickly,
followed by a delay among the seeded target polynucleotide. The delay, termed
"kinetic
delay- because it is thought to arise through the biochemical reaction
kinetics, gives one
seeded target polynucleotide an earlier start over the other seeded targets.
Exclusion
amplification works by using recombinase to facilitate the invasion of primers
(e.g., primers
attached to a substrate region) into double-stranded DNA (e.g., a target
polynucleotide) when
the recombinase mediates a sequence match. The present compositions and
methods may be
adapted for use with recombinase to facilitate the invasion of the present
capture primers and
orthogonal capture primers into the target polynucleotides when the
recombinase mediates a
sequence match. Indeed, the present compositions and methods may be adapted
for use with
any surface-based polynucleotide amplification methods such as thermal PCR,
chemically
denatured PCR, and enzymatically mediated methods (which may also be referred
to as
recombinase polymerase amplification (RPA) or ExAmp).
37
CA 03183729 2022- 12- 21
WO 2022/140254
PCT/US2021/064342
Additional comments
[0104] While various illustrative examples are described above, it will be
apparent to one
skilled in the art that various changes and modifications may be made therein
without
departing from the invention. The appended claims are intended to cover all
such changes
and modifications that fall within the true spirit and scope of the invention.
[0105] It is to be understood that any respective features/examples of each of
the aspects of
the disclosure as described herein may be implemented together in any
appropriate
combination, and that any features/examples from any one or more of these
aspects may be
implemented together with any of the features of the other aspect(s) as
described herein in
any appropriate combination to achieve the benefits as described herein.
38
CA 03183729 2022- 12- 21