Note: Descriptions are shown in the official language in which they were submitted.
WO 2022/040589
PCT/US2021/046997
IN THE UNITED STATES PATENT AND TRADEMARK OFFICE
Title: SYSTEMS, DEVICES AND METHODS FOR DETERMINING MOST
PROBABLE NUMBER IN BIOLOGICAL SAMPLE ANALYSIS
RELATED APPLICATIONS
100011 The current disclosure claims benefit of and priority to US provisional
application no.
63/068,884, filed August 21, 2020, and is also related to U.S. patent nos.
8961878, and 9999855,
PCT publication nos. W02014/144340, W02014/144782, W02014/144810,
W02014/145765,
W02014/165317, W02016/210348, W02017/004595, W02018/026605, W02019/117877,
W02019/103741 and PCT application no. PCT/US2021/041616. Each of the foregoing
disclosures is incorporated by reference herein in its entirety.
BACKGROUND
100021 Most-probable-number, or MPN, is an approach in microbiology used to
determine a
number of colony-forming-units (CFUs) of bacteria being present in a sample
(i.e., one (1) CFU
corresponds to one viable bacteria cell). MPN methods involve, first,
preparing a sample (such as
a rinsate of poultry parts, a dilution of a ground sample, a dilution of a
boot-swab, a dilution of
growth area litter, rinse of an environmental swab, etc.), then serially
diluting and enriching the
sample to increase the pathogen count in each dilution, often in duplicate,
triplicate or at higher
replicates. In classic MPN analysis, a second set of dilutions and enrichments
is performed. The
1
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
presence or absence result from the series of dilutions after the enrichment
period is used to
calculate the most probable number of organisms based on statistics.
SUMMARY
100031 Embodiments of the present disclosure present and use automated methods
via an assay
system, e.g., such Ancera LLC's PIPER system ("PIPER), flow cytometry (cell
counts), PCR
(cycle times), and/or immunoassay (intensity) in combination with a serial
dilution to generate a
series of results where the concentration of cells (CFU level) in the original
sample can be
extrapolated from the dilution factor of at least two (last) positive dilution
in the dilution series.
In some embodiments, duplicate (or triplicate, and more) dilution sets can be
performed.
100041 The PIPER system is an assay system which uses a multi-lane (e.g., 12-
lane) microfluidic
cartridge and a processing instrument that is used to process a sample(s)
loaded into a cartridge
(see W02018026605, herein incorporated by reference). The system uses
magnetophoresis to
manipulate particles or cells with a ferrofluid in microfluidic channels. The
cartridge is arranged
within the Piper instrument in close proximity to a printed circuit board
(PCB) that generates a
magnetic field(s). The ferrofluid is a colloidal suspension of polymer or
surfactant stabilized
superparamagnetic nanoparticles, which, under the influence of a magnetic
field generated by the
PCB, as well as a pumping system built into the cartridge, particles or cells
and be concentrated or
otherwise focused and flowed along a surface of capture zone coated with
binders specific for a
particular target particle or cell. The PCB power and frequency, as well as
the flow rate of the
system, can be optimized for specific sized particles or cells for optimal
assay performance The
capture zone(s) can be designed with between one and a plurality of capture
areas (e.g., eight
unique capture areas, or more) depending on the assay format. After capture,
the particles or cells
can be labeled for fluorescence detection, via, for example, fluorescent DNA
intercalating dyes,
labeled target specific binders such as labeled antibodies, enzyme substrates
specific for
intracellular enzymes, and labeled nucleic acid probes that enable
fluorescence in-situ
hybridization detection of target specific DNA, mRNA or rRNA sequences. The
processing
instrument (PIPER instrument) can include an optical system that enables
detection and counting
of fluorophore labeled particles or cells attached to the capture zone. The
optics can include a
2
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
camera and microscope objective along with, for example, a specific LED and/or
a filter set that
enables excitation and detection of emission of the label fluorophores. The
PIPER system can be
configured with custom imaging algorithms to facilitate accurate imaging and
counting of labeled
cells In addition, each lane of the Piper microfluidic cartridge can at least
one of
- contain a sample well and a dye chamber (used to add the specific label
to the captured
cells); and
- including one or more target specific capture zones.
[0005] In some cases, assays can be designed that do not require particle or
cell capture. In such
assays, pre-labeled particles or cells are simply pushed to the surface and
imaged. The PIPER
system is configured such that samples recirculate back to a sample chamber
during a pumping
cycle. Specifically, the cartridge includes a peristaltic pumping system that
is actuated by the
PIPER instrument and controls valves to accomplish such recirculation, as well
as enable the
addition of labeling reagents.
[0006] The PCB of PIPER is mated to a stage that controls temperature of the
microfluidic
cartridge during the assay.
[0007] In some embodiments, assay precision can be further improved by using a
calibrated count
assay at several dilutions to calculate the original quantity.
[0008] Calibrated assay counts, according to some embodiments, can be obtained
when an assay
instrument is pre-loaded (or can be later converted to CFUs from PIPER, in the
cloud, a secondary
worksheet, and the like) with standard curves generated from the enrichment of
samples having
known measured CFUs of the assay target resulting in a standard curve that can
be used to convert
assay counts (in our case PIPER counts) into actual CFU levels in the tested
sample. This,
according to some embodiments, defines the range of CFU levels over which
accurate quantitation
can be achieved (dynamic range). The dynamic range of calibrated counts may
vary based on the
enrichment time. When such a standard curve is loaded into an instrument (or
converted to CFUs,
e.g., in the cloud, secondary worksheet, and the like), the instrument can
display a calibrated count
which is proportional to a CFU number in the sample being tested. In this
application, we can
apply the standard curve to the instrument counts of the last positive
dilution to further increase
3
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
the accuracy of the measurement beyond the number provided solely by the
original dilution factor
of the sample.
100091 For embodiments of the present disclosure, sample types include (and
for example), poultry
rinsates, feces, boot swabs, boot socks, feed, grain, carcass swabs, dairy
products, water and juice
samples, prepared meats, produce, food production samples, and the like.
100101 Accordingly, in some embodiments, a method for quantifying a number of
pathogens or
microbes present in a food sample in a microfluidic based assay is provided
and includes providing
a food sample (optional), establishing a first dilution of the food sample by
adding a first volume
of media serially diluting the first dilution at least one additional time or
a plurality of times, and
enriching the food sample after each dilution. Subsequent dilutions are
increased by a dilution
factor, and such dilutions range from between 1 part sample with 1 part media,
to 1 part sample
with 1000 parts media.
100111 In some embodiments, dilutions range from between:
- 1 part sample with 1 part media, or 1 part sample with greater than 1
part media,
- 1 part sample and up to 5 parts media,
- 1 part sample and up to 6 parts media,
- 1 part sample and up to 7 parts media,
- 1 part sample and up to 8 parts media,
- 1 part sample and up to 9 parts media,
- 1 part sample and up to 10 parts media,
- 1 part sample and up to 20 parts media,
- 1 part sample and up to 25 parts media,
- 1 part sample and up to 50 parts media,
- 1 part sample and up to 75 parts media,
- 1 part sample and up to 100 parts media,
- 1 part sample and up to 150 parts media,
4
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
- 1 part sample and up to 200 parts media,
- 1 part sample and up to 300 parts media,
- 1 part sample and up to 400 parts media,
- 1 part sample and up to 500 parts media,
- 1 part sample and up to 600 parts media,
- 1 part sample and up to 700 parts media,
- 1 part sample and up to 800 parts media,
- 1 part sample and up to 900 parts media,
- 1 part sample and up to 1000 parts media,
- 1 part sample and up to 5000 parts media,
- 1 part sample and up to 10000 parts media, and
- 1 part sample and greater than 10000 parts media.
and ranges between any of the foregoing.
100121 In the method, according to the above-noted embodiments, the plurality
of dilutions can be
between: 2-3 times, 2-4 times, 2-5 times, 2-6 times, 2-7 times, 2-8 times, 2-9
times, 2-10 times, 2-
12 times, 2-13 times, 2-14 times, 2-15 times, 2-16 times, 2-17 times, 2-18
times, 2-19 times, and
2-20 times (and ranges therebetween of any of the foregoing).
100131 Some embodiments (e.g., those above) may include one and/or another
(and in some
embodiments, a plurality, a majority, substantially all of, or in some
embodiments, all) of the
following features, functionality, steps, structure, or clarifications,
yielding yet further
embodiments:
- after each enrichment, the method further comprises incubating each
dilution at a
predetermined temperature for a predetermined amount of time;
- after incubation, testing each sample for presence of at least one food
pathogen,
producing a result;
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
- the at least one food pathogen or microbes (such terms being used
interchangeably
herein) comprises at least one of Salmonella, Campylobacter, E. coli,
Listeria, and
Clostridium perfringens
- a result is proportional to the dilution of a last positive test (i.e., a
pathogen count is
greater than a predetermined amount);
- testing is performed on an assay system;
- testing is performed on a ferrofluidic based assay system;
- testing is performed on a cartridge based assay system;
- testing is performed on a ferrofluidic, cartridge based assay system;
- after enrichment (and in some embodiments, incubation) of each individual
dilution,
the method further comprises at least one of (and in some embodiments, a
plurality of,
a majority of, substantially all of, or in some embodiments, all of):
o processing each sample with an assay system (e.g., according to at least
one of
the assay processing embodiments disclosed in the incorporated by reference
patents, PCT publications, and PCT application);
o processing each sample with an assay system using a single "lane" of a
cartridge, of a cartridge based assay system (e.g., according to at least one
of
the assay processing embodiments disclosed in the incorporated by reference
patents, PCT publications, and PCT application), or in some embodiments, a
plurality of lanes;
o combining the processed samples of each individual dilution to produce a
combined sample; and
o processing the combined sample with an assay system (e.g., according to
at
least one of the assay processing embodiments disclosed in the incorporated by
reference patents, PCT publications, and PCT application);
6
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
- a combined sample is tested via a cartridge based assay system having at
least one lane
with a plurality of unique capture zones configured to capture labeled cells
from each
dilution;
- using a result from at least a last two positive dilutions to determine
an MPN value;
- using a result from at least a last positive dilution to determine an MPN
value;
- a calibrated assay system for use with a method (according to some
embodiments)
includes standard test information or a curve(s) generated from an enrichment
of food
samples having known measured CFUs of one or more types of pathogens;
- determining a cell concentration by analysis of presence-absence data
(i.e., is the
pathogen present or not),
- the factor comprises between 1:2 to 1:1000 the amount of sample of a
previous dilution.
a minimum detected amount is at least one cell or CFU;
- applying calibrated assay system counts to last positive dilution sample,
processing and
averaging the results from duplicate sample sets;
- the plurality of dilutions comprise at least three dilutions A, B and C,
with dilution
factors of 1.000x, 0.010x and 0.001x, respectively;
- testing for the presence of at least one food pathogen, and producing a
result thereof;
- a dilution is positive if the cell count is greater than the assay
background;
- a dilution is negative if the cell count is less than the assay
background;
- upon dilutions A, B, and C being positive, assay input is greater than 1
cell x lowest
dilution factor;
- upon the lowest dilution factor being 1:100 (i.e., 0.01x), assay input is
> 100 cell;
- upon the lowest dilution factor being 1:1000 (i.e., 0.001x), assay input
is > 1000 cell;
- upon the dilutions A and B being positive, and dilution C being negative,
assay input
is between 1 cell x lowest dilution factor that is positive, and 1 cell x next
lowest
dilution factor;
7
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
- upon the lowest positive dilution factor being 1:100 (i.e., 0.01x) and a
next lowest
dilution factor is 1:1000 (i.e., 0.001x), assay input is between 100 and 1000
cells;
- upon the lowest positive dilution factor is 1:100 (i.e., 0.01x) and a
next lowest dilution
factor is 1:10,000 (i.e., 0.0001x), assay input must be between 100 and 10,000
cells;
- upon dilution A being positive and dilutions B and C being negative,
assay input is
between 1 cell x lowest dilution factor that is positive and 1 cell x next
lowest dilution
factor; and
- upon:
o the lowest positive dilution factor being the undiluted sample and a next
lowest
dilution factor is 1.100 (i.e., 0.01x), the assay input is between 1 and 100
cells,
or
o no dilutions being positive, assay input is less than minimum detected
amount
of 1 cell.
100141 These and other embodiments, objects, advantages, features,
functionality, steps, structure,
and clarifications of the disclosure will be even more evident with reference
to the following details
noted below, and accompanying figures, a brief description of which is
immediately set out below.
100151 Fig. 1 illustrates a method for processing food samples and performing
dilutions for
determining MPN according to some embodiments of the disclosure.
100161 Fig. 2 illustrates dilutions according to four (4) different groups (A1-
3, B1-3, C1-3, and
D1-3), according to some embodiments of the disclosure.
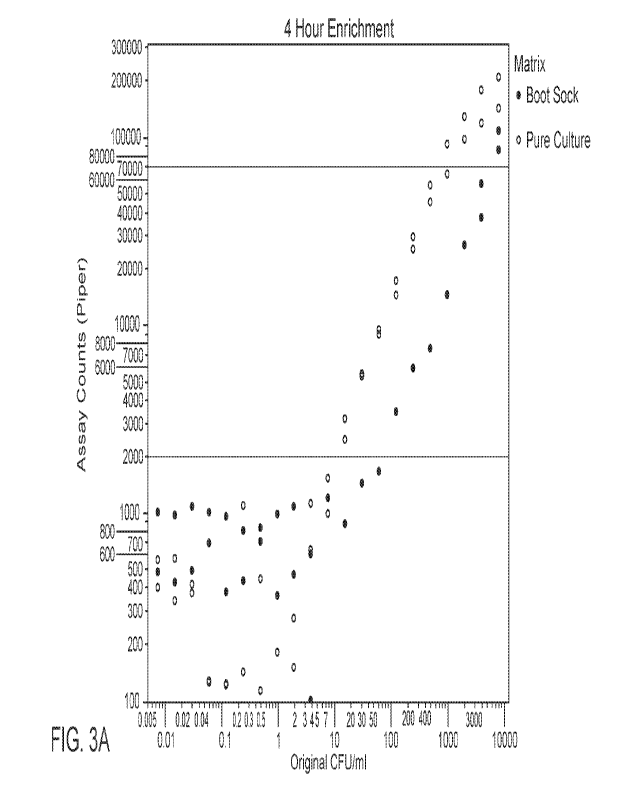
100171 Fig. 3A is a calibration curve obtained for a sample at a first
incubation time point,
according to some embodiments.
100181 Fig. 3B is a calibration curve obtained for a sample at a second
incubation time point,
according to some embodiments.
8
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
DETAILED DESCRIPTION
100191 Accordingly, in some embodiments, the accuracy of the MPN when using
assay
instrumentation (such as PIPER, for example) can be increased by converting
counts from the
assay instrument to calibrated counts (such a system can be considered a
"calibrated assay system"
or "calibrated count assay system"). This can be accomplished, according to
some embodiments,
by pre-loading the assay system (e.g., PIPER) with information on
dilutions/MPN with respect to
dilutions that have been enriched for a predetermined amount of time; this is
shown in the example
results shown in Figs. 3A and 3B. Specifically, serial dilutions (21, 2-fold
dilutions) of Salmonella
Typhimurium (s. typhimurium) were made in both Romer Rapid Check Salmonella
growth media
and a boot sock sample that had been prepared in 250 mL of the same media.
Final concentrations
of cells in each sample were as indicated (in the figures). After dilutions
were prepared, they were
incubated at 42 C for 4 hours (Figure A) and 6 hours (Figure B). After
incubation, samples, in
duplicate, were removed, mixed with ferrofluid (EMG 700), and added to sample
wells of a
microfluidic cartridge. The cartridges were processed on a PIPER instrument
and the captured
cells were FISH (fluorescence, in-situ hybridization) labeled using labeling
conditions as described
in co-pending PCT application no. PCT/US2021/041616. The graphs of Figs. 3A
and 3B show the
calibration curves obtained for both samples, pure culture (cells diluted in
Rapid Check Media)
and in Boot Socks (cells diluted in a prepared boot sock sample), at 2
incubation (enrichment) time
points. Average PIPER counts for each dilution of s. typhinmrium are shown and
the linear range
of the assay is shown.
100201 Accordingly, in some embodiments, a standard curve of this type can be
used convert the
assay system (e.g., PIPER) counts to the actual number of CFUs of salmonella
that are present in
the last positive dilution of the multi-lane MPN assay. Using this number,
plus the actual dilution
value for the sample, allows for an accurate determination of the number of
CFUs present in a
sample. One of skill in the art will appreciate that in the noted example
above, the 6 hour graph
in Fig. 3B shows increased sensitivity than that of the 4 hour graph in Fig.
3A, indicating that each
sample incubation/enrichment time corresponds to a standard curve prepared
with the same
incubation time. In some embodiments, a similar type of calibration curve can
be prepared for
assays using combined barcoded enriched sample dilutions that are processed
with a multi-zone
capture assay microfluidic cartridge (as described in PCT/US2021/041616).
9
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
[0021] Accordingly, when this approach is used for each sample, the value from
at least one last
positive dilution (preferably two or more) and the calibrated assay system
(e.g., PIPER) count can
both be used to provide a more accurate CFU per sample value than would be
determined from
just the last positive dilution alone Additional increased accuracy can be
obtained by also
examining the calibrated count at a next most concentrated sample (if that
count in within the
calibration range).
[0022] To this end, and according to some embodiments, a lower number of
dilutions can be
performed when compared to a typical MPN method. For example, a 3-tube MPN,
corresponding
to 3x5 tubes (5 replicates per dilution), results in 15 dilutions/tests in
total. However, according to
some embodiments of the present disclosure, only 2-5 dilutions, as opposed to
15 dilutions, need
be performed to obtain the same MPN result.
[0023] Typically, lag times and growth times are the largest variables in
allowing single
enrichment methods from being quantitative. However, irrespective of growth
rate, lag time, cell
surface protein, immune-target, or nucleic acid count in cells, methods
according to some
embodiments of the present disclosure work accurately. Moreover, the number of
dilutions and
the dilution ratios can be changed to add additional granularity, and/or to
provide a broader
dynamic range. For example, when analyzing 1-10 dilutions, best accuracy is
within 10-fold; when
analyzing 1-2 dilutions, the best accuracy is within 2 fold, however, to cover
the same range as 1-
dilutions, more 1-2 dilutions should be performed
[0024] Accordingly, the approach used according to some embodiments, allows
for combining a
presence-absence assay (i.e., is the pathogen present) with a quantification
assay (i.e., how much
of the pathogen is present, e.g., how many CFUs are present in a sample),
which is not possible
even for standard MPN. Moreover, MPN and calibration-based calculations,
according to
embodiments of the present disclosure, can be automated via the control system
of, for example,
an assay system (e.g., PIPER). Additionally, in some cases, such methods can
be accomplished
via a single-lane/channel assay system via, for example, embodiments in the
patents, PCT
publication, and PCT application incorporated by reference in the present
disclosure. The
method(s), according to some embodiments described herein, can enable the
generation of
quantitative results with only one set of enrichments. For example, with
PIPER, the method(s)
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
allow for a rapid sensitive determination of presence/absence that can be
coupled with use of a
calibration curve. Results can be available to the customer within a short
period of time (e.g.,
within 5 to 20 hours from receipt of samples), depending on the length of
enrichment.
100251 As an example, and according to some embodiments, glucose pyruvate can
be used for
resuscitation for non-enrichment quantification, and/or robustness for
enrichment based an MPN
system for samples that have reached a plateau in enrichment.
100261 Sample Outcome Data via an assay system (e.g., using PIPER and
according to some
embodiments).
= If assay system counts are > 400*, sample should be considered positive;
= If assay system counts are < 400*, algorithm should be considered
negative,
* Minimum threshold to call a sample positive may vary dependent on the
instrument and imaging algorithm
= Positive sample = 1
= Negative sample = 0
Dilution Outcome #1 Outcome #2 Outcome #3 Outcome #4
A(lx) 1 1 1 0
B (0.01x) 1 1 0 0
C (0.001x) 1 0 0 0
Estimated
> 1000 cells 100-1000 cells 1-100 cells <1 cell
assay input
100271 Sample Outcome Calculations (from PIPER and according to some
embodiments)
= Note: Minimum detected amount is ¨1 cell
11
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
= If all 3 dilutions (A, B, C) are positive, assay input should be greater
than (1 cell x lowest
dilution factor, i.e., volume of stock solution taken divided by the final or
total volume of
the diluted solution (stock solution + diluent);
a. If lowest dilution factor is 1:100 (i.e., 0.01x), assay input should be
> 100 cells (i.e.,
1 x 100);
b. If lowest dilution factor is 1:1000 (i.e., 0.001x), assay input should
be > 1000 cells
(i.e., 1 x 1000);
= If dilutions A and B are positive (i.e., assay system count greater than
400 see above) but
dilution 3 is negative, assay input should be between (1 cell x lowest
dilution factor that is
positive) and (1 cell x next lowest dilution factor):
a. If lowest positive dilution factor is 1:100 (i.e., 0.01x) and the next
lowest dilution
factor is 1:1000 (i.e., 0.001x), assay input should be between 100 (i.e., 1 x
100) and
1000 (i.e., 1 x 1000) cells;
b. If lowest positive dilution factor is 1:100 (i.e., 0.01x) and next
lowest dilution factor
is 1:10,000 (i.e., 0.0001x), assay input should be between 100 (i.e., 1 x 100)
and
10,000 (i.e., 1 x 10,000) cells;
= If dilution A is positive and dilutions B & C are negative, assay input
should be between
(1 cell x lowest dilution factor that is positive) and (1 cell x next lowest
dilution factor);
a. If lowest positive dilution factor is the undiluted sample & next lowest
dilution
factor is 1:100 (i.e., 0.01x), assay input should be between 1 (i.e., 1 x 1)
and 100
(i.e., lx 100) cells;
= If no dilutions are positive, assay input should be less than minimum
detected amount of 1
cell.
Example
100281 As shown in the process outlined in Fig. 1, aseptically poultry parts
were removed from
packaging and placed into a 55 oz Whirl-Pak Homogenizer Blender Filter Bag
(Fisher
Scientific). A pre-warmed media was added to the bag containing the poultry
parts at a ratio of 1
12
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
mL of enrichment media to 5 g of poultry (e.g. 100 mL of media per 500 g of
chicken parts). The
bag was then closed and massaged gently for 1 minute to ensure that all
surfaces of the sample
were adequately rinsed. Then:
- 30 mL of the sample rinse were aseptically transferred into 3, 7 oz,
sterile Whirl-Pak
bags containing 30 mL of prewarmed media and then labeled Al, Bl and Cl.
- Six (6) other sterile Whirl-Pak bags were then filled with 54 mL of media
and labeled
A2, A3, B2, B3 and C2, C3.
- Three (3) other sterile Whirl-Pak bags were filled with 60 mL of media
and labeled D1-
3 (media only for control).
100291 The bags were then separated into groups A, B, C and D and inoculated
according to Fig.
2, and TABLE 1 (below). Specifically, bag Al was not inoculated, bag B1 was
inoculated with 3
CFU, and bags C and D were inoculated with 300 CFU. The first bag of each set
(bags #1) were
massaged, and then 6 mL of the contents of each (Al, Bl, Cl, D1) was
transferred into the
corresponding second bag of each set (A2, B2, C2, D2, respectively). The
second bag of each set
(bags #2) was then massaged and then 6 mL of the contents of each (A2, B2, C2,
D2) was
transferred into the corresponding third bag of each set (A3, B3, C3, D3,
respectively).
TABLE 1
Bag Group A Bag
Group D
Bag Group B Bag Group C
(rinsate contamination .
(media only
Series: control) (rmsate presence/absence) (rinsate
MPN)
control)
Bag/Dilution
0 CFU 3 1. CFU
300 100 CFU 300 100 CFU
#1
Bag/Dilution 30 10 CFU 30
10 CFU
0 CPU expected 0 MT expected
#2 (10x) expected
expected
Bag/Dilution
#3 0 CFU expected 0 CFU expected 3 - 1 CFU 3 -
1 CFU
expected
expected
(100x)
13
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
[0030] Bag Group A ¨ control for natural contamination of the rinsate with
Salmonella initial
sample containing 50% poultry rinse.
100311 Bag Group B ¨ simulates MPN with very low levels of contamination.
[0032] Bag Group C ¨ simulation of MPN with high levels of contamination.
[0033] Bag Group D ¨ no-sample control (optional); comparison of Salmonella
cell counts post
enrichment in groups A, B, C with this series will show if presence of the
poultry rinse affects
Salmonella growth.
[0034] Accordingly, Bag #1 of each group comprises an initial dilution, Bag #2
of each group
comprises 10x dilution, and Bag #3 of each group comprises 100x dilution.
[0035] The samples were then incubated at 42 C for 11 hours. Thereafter, an
aliquot for testing is
removed from the third bag of each set.
[0036] TABLE 2 illustrates the results of the assay system (PIPER):
TABLE 2
Bag Group A
Bag Group B
(rinsate Bag Group C Bag Group D
(rinsate
Series: contamination
presence/absence) (rinsate MPN)
(media only control)
control)
Bag/Dilution 88,865
2,942 344,669 31,920
636 + 43 CFU 7,892 782 CFU
#1 CFU
CFU
Bag/Dilution 514 51 CFI' 885 714
(7(J57,137 2,129 143,238 36,985
#2 (10x) CFU
CFU
Bag/Dilution
#3 601 + 222 CFU 505 147 CFU 38,807
1,643 109,193 7,623
CFU
CFU
(100x)
100371 From such result counts, counts less than 2,000 were considered
negative, and counts
greater than 2,000 were considered positive, thus, as shown in TABLE 3:
14
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
TABLE 3
Bag Group A
Bag Group B
(rinsate Bag Group C Bag Group D
(rinsatc
Series: contamination
presence/absence) (rinsate MPN)
(media only control)
control)
Bag/Dilution
negative positive positive#1
positive
Bag/Dilution
negative negative positive
positive
Bag/Dilution
#3 negative negative positive
positive
(100x)
[0038] With the calculated input in the original samples being: Bag Group A:
0; Bag Group B: 1-
10, and Bag Groups C and D. greater than 100.
[0039] While various inventive embodiments have been described and illustrated
herein, those of
ordinary skill in the art will readily envision a variety of other means
and/or structures for
performing the function, and/or obtaining the results and/or one or more of
the advantages
described herein, and each of such variations and/or modifications is deemed
to be within the scope
of the inventive embodiments described herein. More generally, those skilled
in the art will readily
appreciate that all parameters, dimensions, materials, steps, time periods,
temperatures (e.g.,
incubation times and temperatures), and configurations described herein are
meant to be merely
an example and that the actual parameters, dimensions, materials, steps, time
periods, temperatures
(e.g., incubation times and temperatures) and configurations will depend upon
the specific
application or applications for which the inventive teachings is/are used.
Those skilled in the art
will recognize, or be able to ascertain using no more than routine
experimentation, many
equivalents to the specific inventive embodiments described herein. It is
therefore to be understood
that the foregoing embodiments are presented by way of example only and that,
within the scope
of claims supported by the subject disclosure and equivalents thereto, and
inventive embodiments
may be practiced otherwise than as specifically described and claimed.
Inventive embodiments of
the present disclosure are directed to each individual feature, device,
system, article, material, kit,
step, function/functionality, and method described herein. In addition, any
combination of two or
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
more such features, devices, systems, articles, materials, kits, steps,
functions/functionality, and
methods, if such features, systems, articles, materials, kits, steps,
functions/functionality, and
methods are not mutually inconsistent, is included within the inventive scope
of the present
disclosure, and considered embodiments
100401 Embodiments disclosed herein may also be combined with one or more
features, as well
as complete systems, devices, and/or methods, to yield yet other embodiments
and inventions.
Moreover, some embodiments, may be distinguishable from the prior art by
specifically lacking
one and/or another feature disclosed in the particular prior art reference(s),
i.e., claims to some
embodiments may be distinguishable from the prior art by including one or more
negative
limitations.
100411 Also, as noted, various inventive concepts may be embodied as one or
more methods, of
which one or more examples have been provided. The acts performed as part of
the method(s) may
be ordered in any suitable way. Accordingly, embodiments may be constructed in
which acts are
performed in an order different than illustrated, which may include performing
some acts
simultaneously, even though shown as sequential acts in illustrative
embodiments.
100421 Any and all references to publications or other documents, including
but not limited to,
patents, patent applications, articles, webpages, books, etc., presented
anywhere in the present
application, are herein incorporated by reference in their entirety. Moreover,
all definitions, as
defined and used herein, should be understood to control over dictionary
definitions, definitions in
documents incorporated by reference, and/or ordinary meanings of the defined
terms
100431 The indefinite articles "a" and "an," as used herein in the
specification and in the claims,
unless clearly indicated to the contrary, should be understood to mean "at
least one."
100441 The terms "can" and "may" are used interchangeably in the present
disclosure, and indicate
that the referred to element, component, structure, function, functionality,
objective, advantage,
operation, step, process, timing, amount, apparatus, system, device, result,
or clarification, has the
ability to be used, included, or produced, or otherwise stand for the
proposition indicated in the
statement for which the term is used (or referred to).
100451 The phrase "and/or," as used herein in the specification and in the
claims, should be
understood to mean "either or both" of the elements so conjoined, i.e.,
elements that are
16
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
conjunctively present in some cases and disjunctively present in other cases.
Multiple elements
listed with "and/or" should be construed in the same fashion, i.e., "one or
more" of the elements
so conjoined. Other elements may optionally be present other than the elements
specifically
identified by the "and/or" clause, whether related or unrelated to those
elements specifically
identified. Thus, as a non-limiting example, a reference to "A and/or B", when
used in conjunction
with open-ended language such as "comprising" can refer, in one embodiment, to
A only
(optionally including elements other than B); in another embodiment, to B only
(optionally
including elements other than A); in yet another embodiment, to both A and B
(optionally
including other elements); etc.
100461 As used herein in the specification and in the claims, "or" should be
understood to have the
same meaning as "and/or" as defined above. For example, when separating items
in a list, "or"
or "and/or" shall be interpreted as being inclusive, i.e., the inclusion of at
least one, but also
including more than one, of a number or list of elements, and, optionally,
additional unlisted items.
Only terms clearly indicated to the contrary, such as only one of or "exactly
one of, or, when
used in the claims, "consisting of," will refer to the inclusion of exactly
one element of a number
or list of elements. In general, the term or as used herein shall only be
interpreted as indicating
exclusive alternatives (i.e. one or the other but not both") when preceded by
terms of exclusivity,
such as "either," one of," "only one of," or "exactly one of' "Consisting
essentially of" when used
in the claims, shall have its ordinary meaning as used in the field of patent
law.
100471 As used herein in the specification and in the claims, the phrase "at
least one," in reference
to a list of one or more elements, should be understood to mean at least one
element selected from
any one or more of the elements in the list of elements, but not necessarily
including at least one
of each and every element specifically listed within the list of elements and
not excluding any
combinations of elements in the list of elements. This definition also allows
that elements may
optionally be present other than the elements specifically identified within
the list of elements to
which the phrase "at least one" refers, whether related or unrelated to those
elements specifically
identified. Thus, as a non-limiting example, "at least one of A and B" (or,
equivalently, "at least
one of A or B," or, equivalently "at least one of A and/or B") can refer, in
one embodiment, to at
least one, optionally including more than one, A, with no B present (and
optionally including
elements other than B); in another embodiment, to at least one, optionally
including more than
17
CA 03188680 2023- 2-7
WO 2022/040589
PCT/US2021/046997
one, B, with no A present (and optionally including elements other than A); in
yet another
embodiment, to at least one, optionally including more than one, A, and at
least one, optionally
including more than one, B (and optionally including other elements); etc.
100481 In the claims, as well as in the specification above, all transitional
phrases such as
"comprising," "including," "carrying," "having," "containing," "involving,"
"holding," "composed
of," and the like are to be understood to be open-ended, i.e., to mean
including but not limited to.
Only the transitional phrases "consisting of and "consisting essentially of'
shall be closed or semi-
closed transitional phrases, respectively, as set forth in the United States
Patent Office Manual of
Patent Examining Procedures, Section 2111.03.
18
CA 03188680 2023- 2-7