Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
1338410
The present invention relates to a process for the isolation
and characterization of a gene enzyme system for the
inactivation of the herbicide phenmedipham. The enzyme is a
carbamate hydrolase of Arthrobacter oxidans, which is
responsible for the cleavage of the carbamate bond between
the benzene rings of phenmedipham which is the common name
for the herbicide methyl 3-m-tolylcarbamoyloxyphenyl-
carbamate.
The invention will be more readily understood by reference to
the accompanying drawings, in which:
Abbreviations
DEAE Diethylaminoethyl
FPLC Fast ~rotein/~eptide/~olynucleotide liquid
Chromatography
SDS Sodium lauryl sulphate
DTT Dithiothreitol
1 x SSC 0.15 M NaCl 0.015 M trisodium citrate pH 7.0
20 1 x Denhardt 0.02% (w/v) Bovine serum albumin
(Sigma, Fraction V)
0.02% (w/v) Ficoll 400
0.02% polyvinylpyrrolidone
MCS Multiple cloning site
Abbreviation for restriction endonucleases
Bm = BamHI, Bs = BstEII, Cl = ClaI, EV = EcoRV,
HII = HindII, Kp = KpnI, Nc = NcoI, Nd = NdeI, Nh = NheI,
Ps = PstI, PvI = PvuI, PvII = PvuII, Sc = SacI, Sp = SphI,
St = StuI, Xb = XbaI.
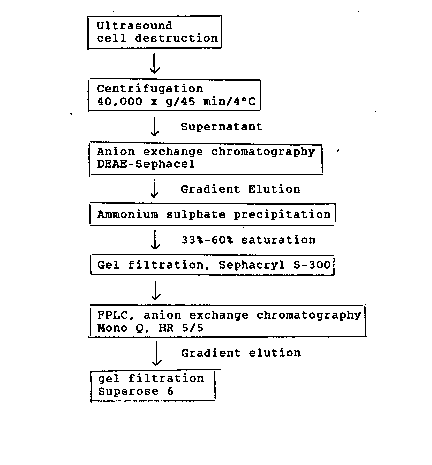
Figure 1 shows the process for isolating and subsequent
purification of phenmedipham cleaving carbamate hydrolase
from Arthrobacter oxidans (Example 2)
* Trade mark - 1 -
D
1338440
-
Figure 2 shows the electrophoretic separation of the crude
extract and the isolation and pooled protein fractions after
the individual purification steps on an SDS-polyacrylamide
gel, in which standard proteins (S) run alongside as markers
for the molecular weight.
A: Crude extract from the centrifuge supernatant of
the ultrasound cell destruction of Arthrobacter
oxidans.
B: Pooled protein fraction after gradient elution on a
DEAE Sephacel* column.
C: Ammonium sulphate precipitation of the fraction
from B.
D: Protein fraction after gel filtration of the
ammonium sulphate precipitation and subsequent
separation on Sephacryl* S-300 column.
E: Protein fraction after separation of the pooled
fraction from D by FPLC (anion exchange
chromatography).
F: Protein fraction after separation of the pooled
fraction E on a Superose* 6 column (Example 2).
Figure 3 shows a diagram from which the pH-optimum (pH 6.8)
of the carbamate hydrolase can be obtained. The pH optimum
can be established in such a way that the enzyme activity in
percentage against the pH value can be applied (Example 2).
Figure 4 shows the restriction map of the plasmid pHP52 from
the Arthrobacter oxidans (species P52).
- la -
* Trade mark
~,~
1338440
Figure 5 shows a scheme of cloning of the carbamate hydrolase
gene. To improve clarity the regions of the gene area with
hybridized oligonucleotides including the 5' and 3' flanking
regions are enlarged above the ring-forming plasmid map.
The recombinant clones pp52Pst and pp52Pst inv. are
represented in a linear manner. The transcription direction
of the lac Z gene is represented by an arrow (~
Figure 6 shows the restriction map of the cloned 3.3 kbPstI
restriction fragment which arises from the exact position of
the carbamate hydrolase gene (Start: GTG = Start codon. Stop:
TGA = Stop codon).
The sequenced areas are characterized by arrows under the
restriction map. From the length of the arrow can be read
the length of the individual sequenced areas.
The restriction fragments which contain the homology areas of
the obligonucleotides described in Example 4 are empathized
in the map (D .
The M13 clones are described as follows:
Clone Inserted Fraqment M13 Vector
A NdeI/SacI (pUC19)-500 mp 18
B NdeI/SacI 1289 bp mp 18
C PvuI/PstI ~700 bp mp 19
D PvuI/PvuI 564 bp mp 18
E BamHI/BamHI (pUC 19) mp 18
~1060 bp
F BamHI/BamHI 958 bp mp 18
G PvuI/PvuI 564 bp mp 18
H PvuII/SacI 476 bp mp 18
I ClaI/BamHI 561 bp mp 18
K ClaI/BamHI 397 bp mp 18
- lb -
~`
13384~0
-
Clone Inserted Fraqment M13 Vector
L PvuII/SacI476 bp mp 19
M KpnI/HindII 445 bp mp 19
N BamHI/BamHI 958 bp mp 18
O BamHI/BamHI 1100 bp mp 18
P KpnI/HindII 445 bp mp 18
Figure 7 shows the nucleotide sequence of the carbamate
hydrolase gene including the 5' and 3' flanking area.
Especially characterized are:
1. The GTG start codon
2. The ribosomal binding site
(Shine-/Dalgarno Box: S/D)
3. The homology area of the oligonucleotide
described in Example 4 (oligo I and
II/oligo III).
The coding nucleotide sequence was formally translated in an
amino acid sequence. The reading frame is determined clearly
by the protein level of the established amino acid partial
sequences.
In practice it is often necessary to use several herbicides
or herbicide mixtures for combating various weeds. These
problems can be avoided by biotechnical change to the plant
in which a resistance to a non-selective herbicide is
introduced.
The provision of herbicide-tolerant plants is now coming more
to the foreground in the plant protection area.
In order to produce herbicide-tolerant plants, it is
necessary first to have a process for isolation and
-- lc --
1338~0
_
subsequent purification of an enzyme which is able to
inactivate a herbicide, e.g. by metabolism, and then to have
a process for characterizing the gene enzyme system that
containing the DNA sequence that codes for the active enzyme.
Such a process for the isolation and subsequent
characterization of gene enzyme system which can inactivate
phenmedipham was not previously known.
It has now been found that a carbamate hydrolase can be
isolated from some microorganisms, such as Arthrobacter
oxidans, which responsible for the hydrolysis of the
- ld -
."
13384~0
_ - 2 -
carbamate bond between the two benzene rings of
phenmedipham. The hydrolysi~ of this bond leads to
herbicidally inactive compounds ~uch as methyl ~~
3-hydroxyphenylcarbamate and meta-toluidine, according to
the following reaction.
CH3 CH3
1~ ~ ~ ~ 2
~ m-~olu~dinc
- HN-COOCH3 HN-COOCH3
Phenmedipham phenyl-carbamat~
For isolation and sub6equent purification of a gene enzyme
system which can hydrolyse phenmedipham according to the
above described reaction, microorganisms of Arthrobacter
oxidans are cultivated in a nutrient medium. The carbamate
hydrolase re6ponsible for the cleavage of phenmedipham i8
isolated by ultrasound cell destruction, centrifugation
and purification by anion exchange chromotography,
gradient elution, ammonium sulphate precipitation and FPLC
separation until electrophoretic homogeneity is achieved.
From the purified carbamate hydrolase, two peptides are
isolated after BrCN cleavage whose sequence can be
estimated by Edman degradation. According to the sequence
information of these peptides, oligonucleotides can be
synthesised which can be used as hybridizationin probes
for the detection of the carbamate hyrolase gene.
In all the mentioned isolates of the soil bacteria
Arthrobacter oxidans, plasmids could be detected after
lysis of the cells and extraction of the nucleic acids.
For the species Arthrobacter oxidans P52, it can be shown
1338440
-- 3
that the carbamate hydrolase is coded by one plasmid.
By loss of the plasmid pHP52 the properties of this
6pecies to be able to hydrolytically cleave phenmedipham
is lost. A carbamate hydroysis cannot be biochemically
efitablished in the plasmid-free derivative of the specie6
P52.
The plasmid pHP52 can be preparatively isolated and mapped
with restriction endonucleases. The electrophoretically
separated restriction fragments can be transferred on
membrane filters and hybridised with the oligonucleotides.
From the data from the blot-hybridisations, it appears
that the carbamate hydrolase gene is localised on a PstI
restriction fragment of a size 3.3 kb.
This fragment can be preparatively isolated from the
plasmid pHP52 and inserted in the PstI position of the
vector pUC19C (Yanish-Perron, C., Vieira, J. & Mes6ing
(1985) Gene 33, 103 ff). After transformation of E coli
DH5a with the ligation preparation, two types of
recombinant E. coli clone are obtained (pp52Pst and
pp52Pst inv.) which contain the carbamate hydrolase gene
in different orientations to the lac-promoter of the
vector (see figure 5).
The carbamate hydrolase can be functionally expres6ed in
the presence of the inducer isopropyl-A-D-thiogalacto-
pyranoside from cultures of the clone of the type E. coli
DH5a (pp52Pst). In protein extracts of clones of the
type E. coli DH5a (pp52 inv.) which contain the
carbamate hydrolase gene in inverse orientation to the
lac-promoter, no expression of the carbamate hydrolyse
gene can be detected. It follows that the Arthrobacter
promoter could not be recognised by the E. coli
1338~0
~ - 4 _
RNA-polymera6e.
The nucleotide sequence of the carbamate hydrolase gene
can be determined according to the method of Sanger et al.
(Sanger, F., Nicklen, S. ~ Coul~on, A (1977), Proc. Natl.
Acad. Sci. USA 74, 5463-5468).
For thi6 15 cub-clones arising from the cloning of 3.3 kb
long P6tI rectriction fragments can be constructed in the
cingle 6train DNA bacteriophage~ M13 mp 18 and mp 19
(Messing, J. (1983) Methods in Enzymol. 101, 20-78). In
Figure 6 an exact restriction map of the coded area ic
represented from which the 6equencing 6trategy can be
seen.
The establi6hed nucleotide sequence with the protein
sequence thus derived i6 illu6trated in Figure 7. The
amino acid 6equences of both e~tabli6hed BrCN-cplitting
peptide6 (see Example 4) can be identified in the same
reading frame a6 the DNA level. Thi6 reading frame end6
with a TGA tran61ation ctop codon (cee Figure 7,
nucleotide po6itions 1789-1791) and begins very probably
with a GTG-6tart codon (figure 7 - nucleotide pocition6
340-342). Altogether a reading frame of 1479 ba6e pairs
result~. Upstream of the putative GTG-6tart codon, a
region with ~ignificant homology to the consensu6 sequence
for E. coli ribosome binding 6ites (~Shine-Dalgarno Box")
can be established (see Figure 7, nucleotide positions
298-30Z).
On the 24 March, 1987 ~he following micro-organisms were
depo6ited at the German Collection of Microorganism6 (DSM)
in Gottingen, Germany.
- 1338440
-- 5
Arthrobacter oxidans P 16/4/B (DSM 4038)
Arthrobacter oxidan6 P 67 (DSM 4039)
Arthrobacter oxidans P 75 (DSM 4040)
Arthrobacter oxidans P ll/l/-b (DSM 4041)
and on the 27 q~rch 1987~ th~ fallo~ g
micro-organ~sms were deposited,
Arthrobacter oxidan~ P 15/4/A (DSM 4045)
Arthrobacter oxidans P 21/2 (DSM 4046)
Arthrobacter oxidans P 52, containing the plasmid pHP52
(DSM 4044).
b 1338440
ExamPle 1
Isolation of microorgani~m~ that posses6 the ability to
inactivate the berbicide phenmedipham.
To identify microorganisms which possessed the ability to
inactivate the herbicide phenmedipham by metaboli~n,
variou~ microorganisms were screened. As ~ource for the
microorgani~ms, soil ~ample~ from various locations (field
test sites which had been treated several times with
phenmedipham) and also from settling sediment, were used.
Selection criteria for the identification of
microorganisms which can carry out a carbamate cleavage,
were as follows.
a) Growth in a nutrient medium with phenmedipham as a
single carbon or nitrogen source.
b) Breaking down of phenmedipham to highly water soluble
compounds according to the following reaction.
CH3 C,H3
OIC--NH ~ OH H2N~ C02
~ ~ ~ m-Tolu~di~
HN-COOCH3 HN-COOCH3
PhenmediphamMethyl 3-hydroxy-
phenyl-carbamat~
From the large number of microorganisms obtained from the
60il ~amples, which were capable of cleavage of
phenmedipham, seven representatives were chosen which
clearly showed a breakdown. These soil bacteria which are
all representatives of the Arthrobacter species and within
thi~ species, the sub-specie~ of oxidans, were cultivated
1338~40
in culture broths containing a synthetic medium
(M9-medium) having the following composition.
1.0 g/l NH4Cl
0.25 g/l MgS04.7H20
3.0 g/l KH2P04
7.0 g/l Na2HP04.2H20
2.0 g/l Glucose
and 0.5 g/l NaCl
The M9-medium, in addition, contained 1 mg/l thiamine
(vitamin Bl) as well as trace elements which were added in
the form of a stock solution (1 ml/l M9-medium). The trace
element stock solution contained:
0.5 Boric acid
0.04 g/l CuS04.5H20
0.2 g/l FeC13.6H20
0.4 g/l MnS04.7H20
0.4 g/l ZnC12
g/ ( 4)6 7 24 4H2
For shaking cultures in liquid mediums, the synthetic
medium was supplemented by 0.1% casamino acids (Difco ).
The soil bacteria were incubated in this M9 medium at 28C
with good aeration until the end of the logarithmic growth
phase. For enzyme purification, a total of 10 litres of
medium were inoculated with a stationary pre-culture
(1:100).
By HPLC analysis of the culture broth it was shown that in
the cultures of Arthrobacter oxidans, the desired cleavage
of phenmedipham to the herbicidally inactive products was
achieved.
13~8~40
Example 2
Isolation and purification of the carbamate hydrolase from
Arthrobacter oxidans.
The isolation and 6ubsequent purification of the carbamate
hydrolase to electrophoretic homogeneity was carried out
over a 6ix 6tage purification proce66. From 6 litres of an
end logarithmic culture of Arthrobacter oxidan6 (pHP52)
(DSM No 4044), 0.5 - 1 mg carbamate hydrolase wa6
reproducibly i601ated. For i601ation of the carbamate
hydrolase, the cell~ were harve6ted by centrifugation
(7000 x g) and resu6pended in about 40 ml of decomposition
buffer (10 mM sodium pho6phate pH 6.8/lmM DTT). The cell
6u6pen6ion wa6 di6rupted by ultrasound and homogeni6ed at
the same time. The homogenate was then centrifuged for 45
minute6 at 40000 x q, at 4C. The 6ediment wa6 removed and
the ~upernatant, equilibrated with lOOmM Tri6-HCl pH
7.2/100 mM NaCl/l mM DTT, wa6 applied to a DEAE Sephacel
column (column diameter 2.6 mm, height of the gel bed 20.5
cm, column volume about 100 ml). Before application to the
column, the cell extract wa6 diluted at a ratio of about
1:10 with 6tarting buffer (100 mM Tri~/100 mM NaCl/lmM
DTT). The column was then wa~hed with starting buffer in
order to remove the unbound material. The carbamate
hydrolase was then eluted with a linear gradient 100 mM
NaCl -~ 500 mM NaCl (5 x column volume). The enzymatically
active fraction6 were pooled and treated with dry ammonium
sulphate (NH4)2S04 to an end concentration of 33% of
the ~aturated 601ution. The re6ulting protein precipitate
was 6edimented by centrifugation (20000 x g/30 min6) and
discarded. The 6upernatant wa6 treated with 601id ammonium
6ulphate to an end concentration of 60% of the 6aturated
601ution and 6tirred for about 12 hour6 at 0C. The
~edimented protein wa6 collected by centrifugation
i 13384~0
-
(20000 x g /30 mins) and dissolved in about 1 ml starting
buffer, treated with 10% (w/v) saccharose and loaded to a
Sephacryl* S-300 column. The gel filtration was carried out
at a flow rate of 2.5 cm/h (elution buffer -: start buffer).
The column had a diameter of 2.6 cm, a height of 95 cm and a
volume of 475 ml. The enzymatically active fraction was
then worked up on an FPLC column (mono Q HR 5/5; anion
exchange). Gradient elution 100 mM NaCl -~ 300 mM NaCl;
flow rate: 0.5 ml/mm; application volume 2 ml).
The unbound protein was separated by isocratic elution with
19 ml starting buffer.
(Gradient elution 100 mM NaCl -~ 300 mM NaCl in 20 ml with
100 mM Tris/HCl pH 7.2/1 mM DTT~.
The enzymatically active fractions were concentrated by
ultra-filtration after electrophoretic analysis of the
purity (SDS-polyacrylamide-gel electrophoresis by the method
of Lammli), using an Amicon(R), Centrikon* 10 concentrator,
and put an FPLC gel-filtration column (Superose* 6-HR 10/30,
Pharmacia) (flow rate 0.2 ml/min; application volume 100 ul;
eluent: 100 mM Tris/HCl pH 7.2/10 mM NaCl).
*trademark
g
X
1338440
The active protein fractions which result from this step are
electrophoretically homogenous.
The isolated enzyme is active in buffered solutions (ie
buffers conventionally used in biochemical systems, such as
phosphate buffers, Tris buffers etc; pH 6.8). Co-factors or
metal ions are not necessary for the reaction. A
sensitivity against SH reagents is also not seen. The
optimum pH of the enzyme is 6.8.
- 9a -
~ 1338440
The molecular weight of the carbamate hydrolase is in the
range of 50-60, preferably 53-57 kd, both under
denaturing/dissociating conditions (SDS gel
electrophoresis) as well as under native conditions
(gel-filtration) From this it follows that the carbamate
hydrolase is a monomeric protein. The isoelectric point of
the carbamate hydrolase is at pI = 6.2.
Example 3
Process for detecting the carbamate hydrolase.
For a quick and sure determination of enzyme activity
during the purification of the protein crude extracts an
in vitro enzyme test was developed. The test is based on
the ability of the enzyme to change the highly water
insoluble phenmedipham into a soluble hydrolysis product.
For this, solid phenmedipham was suspended in water and
micronised by ultrasound. This micro-suspension was then
poured, with stirring at 50C, into an agarose solution
and this mixture put into a petri dish before it
solidified, where it formeds into a turbid gel matrix. The
enzyme solution was then put into wells which had been
punched in the solid matrix. After incubation of the test
plates for 2-4 hours at 30C, the enzyme activity was
demonstrated by observing clear zones in the matrix which
had been made opaque by the phenmedipham.
I~
1338~0
ExamPle 4
Identification of the amino acid 6equence of two BrCN
cleaving peptide6 and 6ynthe6is of oligonucleotide6 for
6pecific evidence of the carbamate hydrolase gene by
hybridization.
Resulting from the purified carbamate hydrolase, two
peptides were i601ated after BrCN cleavage, who6e partial
~equence wa6 e6tabli6hed by Edman degradation.
BrCN Peptide I:
H2N - Ser - ~sp - Glu - Phe - ~ Asn - Leu - Asp -
- Arg - Trp - T~r - Gly - Lys - Pro - Phe - V~l - Asp ~Val~ -
- 61y ~His) - Leu - Asp - 61u - Val - Al~ - Val - COOH
BrCN Peptide II:
N2H - 61u - His - Thr - Lys - Phe~Val) - Asn~61y) - 61u - Ar~Cys) -
- Pro - Leu - ~la - Phe - Tyr - Pro -Val - Phe - ~sn - 61u - COOH
Sing the amino acid 6equence information of the6e
peptide6, oligonucleotide6 were 6ynthesi6ed which could be
used a6 hybridization probes for the detection of the
carbamate hydrolase gene:
Oliqonucelotide I (17 mer "mixed probe") contains as the
single 6trand DNA fragment, the 6equence information of
the BrCN peptide I amino acid po6ition 10-15
(complementary 6trand).
5'- AA ¦66G ¦TTT 16CC ¦66T ¦CCA--3'
¦ T ¦ ¦ T ¦ T
la, 1338~40
Oligonucleotide II (42 mer) contains as a 6ingle 6trand
DNA fragment, the 6equence information of the BrCN peptide
I amino acid position 8-21 (complementary 6trand). The
codon 6election wa6 carried out under the as6umption of a
guanine(G) and cytosine(C) rich DNA sequence (thi6 takes
into consideration guanine(G) and cytosine(C) nucleotides
before adenine(A) and thiamine(T) nucleotides on the third
po6ition of the triplets).
5'- CTG I 6TC ¦ CAt ¦ GCC ¦ 6TC ¦ CAC ¦ 6AA
10 61n ¦ Asp I Leu 1 61y 1 ~sp I V~l Phe
C6G I CTT ¦ 6CC ¦ 66T ¦ CCA ¦ GC6
Pro ¦ Lys ¦ 61y I Thr I Trp I Arg
6TC ¦- 3
~p I
Oligonucleotide III contains as the 6ingle 6trand DNA
fragment sequence, information of the BrCN peptide II
(complementary 6trand).
61u ¦ Asn ¦ Phê ¦ V~l ¦ Pro ¦ Tyr ¦ Phê ¦ CGC ¦
By using these oligonucleotides it was possible to
localise the carbamate hydrolase gene within the plasmid
pHP25 by hybridization. For this, the plasmid DNA wa6
cleaved with restriction endonucleases and the resulting
fragments were separated by agarose gel electrophoresis
and then transferred according to the method of
E.M. Southern (J. Mol. Biol. 98, 503-517 (1975)) in single
strand form on membrane filters (Gene Screen PlusTM
hybridising membrane, Du Pont de Nemours/NEN Research
Products).
- ~ 1338440
The oligonucleotides were end marked by use of
T4-polynucleotide kinase (Boehringer Mannheim) and
~-32P]-adenosine-5'-triphosphate (>5000 Ci~mmol,
Du Pont de Nemours/NEN Research Products) u6ing the method
of R.B. Wallace and C.G. Miyada, Methods in Enzymology
152, 432-442 (1987) and treated without further
purificiation for the hybridisation.
The hybridization was carried out using standard proce6ses
(P.J. Mason & J.G. Williams in "Nucleic Acid
Hybridi6ation" p. 113-160 (1985) B.D. Hames & S.J. Higgins
Hrsg. IRL Press Oxford, Washington D.C.). Under the
conditions 6 x SSC, 10 x Denhardt, 0.5% w/v SDS and 100
u/ml t RNA (Backerhefe, Boehringer Mannheim), a6 well as
10 ng/ml marked oligonucleotides I/II/III at 41C
(= 6 hour6), a 6pecific hybridi6ation can be achieved. The
detection of the hybrids wa6 carried out by
autoradiography (T Maniati~, E F Fritsch & J Sambrook,
"Molecular Cloning", Cold Spring Harbor Laboratory (lg82)).
L
~338440
Example 5
Isolation and characterisation of the plasmid pHP52 from
Arthrobacter oxidans P52.
For isolation of plasmid pH52 from Arthrobacter, the
alkali extraction method of Birnboim and Doly
(Birnboim H.C. ~ Doly J. (1979) Nucl Acid Res.,
7, 1513-1523) was u6ed, with a modification by Brandsch
and Decker (Brandsch, R. & Decker, K. (1984) Arch.
Microbiol. 138, 15-17) i8 used. ~or plasmid preparation,
the bacteria were cultivated in 6 litres LB-medium
comprising:
Bacto-trypton (DifcoR) 10 g/l
Bacto-Yeast-Extract (DifcoR) 5 g/l
NaCl 10 g/l
to a cell density of OD550 = 1.4 and harvested by
centrifuging.
The cells were resuspended in a total of 210 ml 601ution I
(50 mM glucose: 10 mM EDTA: 25 mM Tris/HCl, pH 8.0:
1 mg/ml lysosyme) and incubated for 1 hour at room
temperature. Ly6i6 was carried out by addition of 360 ml
solution II (0.2 M NaOH: 1% SDS). After gentle but
thorough mixing and subseguent incubation for 5 minutes at
room temperature, followed by cooling on ice for 5
minutes, the mixture was neutralised by addition of 180 ml
601ution III (2 M Tris/HCl, pH 7.0/0.5 M KCl~ After
incubation for 1 hour on ice, the undi6601ved precipitate
was separated by centrifuging. The pla6mid DNA was
precipitated from the clear supernatant by addition of 0.6
volumes isopropanol and, after an incubation of 15 minutes
at room temperature, pelleted by centrifugation (15,000 x
g/30 minutes). The plasmid-containing precipitate was
dried in vacuo and dissolved in 24 ml 10 x TE buffer
- 1~38440
(100 mM Tris/HCl, pH 8.0; 10 mM EDTA). This plasmid
containing solution was then purified by isopycnic caesium
chloride density gradient centrifuging in the presence of ~
homidium bromide (Maniatis T., Fritsch E.F. & Sambrook J.
in "Molecular Cloning" (1982), Cold Spring Harbor N Y).
Purified plasmid DNA was mapped by restriction analysis
which cut the plasmid once or gave several fragments.
These fragments were re601ved by agarose gel
electrophoresis (0.8% w/v). Molecular weight standards
used in mapping plasmid DNA were Hind III or Hind III and
EcoRI digested bacteriophage DNA.
The restriction analysis data were consistent with a
circular map of pHP52 (Figure 4). The size of the plasmid
is the sum of individual restriction fragments.
All the processes were carried out in this Example
according to standard methods (cf. Maniatis T.,
Frit~ch E.F. & Sambrook J. in "Molecular Cloning" Cold
Spring Harbor, N Y (1982)).
1~38440
Example 6
Identification of the coding region of the carbamate ~
hydrolase gene by oligonucleotide hybridisation.
By hybridisation of the restriction fragments of the
plasmid pHP52 separated by gel electrophoresis and
transferred on membrane filters with the 32p marked
oligonucleotide described in Example 5, the position of
the coding region of the carbamate hydrolase gene can be
definitely correlated on the restriction map of the
plasmid pHP52. In Figure 5, the hybridizing area is
enlarged. All three oligonucleotides hybridize with the
central part of a PstI restriction fragment of size 3.3
kb. In Figure 6, a detailed restriction map of the
fragment is shown from which the exact positions of the
hybridising areas can be seen.
Example 7
Cloning of the carbamate hydrolase gene in E. coli and
demonstration of the genes' expression under lac promoter
control.
For cloning the carbamate hydrolase gene in E. coli the
vector pUCl9 (Yanish-Perron, C. Vicira, J. & Messing, J.
(1985) Gene 33, 103ff) was used. The pUCl9 DNA was
linearised by cleavage with restriction nuclease PstI and
treated with alkaline phosphatase. The DNA of the 3.3 kb
long PstI restriction fragment of the plasmid pHP52 was
isolated (after digesting the wild-type plasmid DNA with
PstI) by preparative agarose gel electrophoresis. The
linearised and dephosphorylated vector DNA and the 3.3 kb
long PstI fragment was then ligated with T4 DNA ligase. E.
coli DH5a was transformed with the ligation mixture,
- 1~ 13384~0
Two types of clones were obtained which contained the
fragment in different orientations to the transcription
direction of the lac Z' gene of the vector pUC 19. These ~~
are the clones pp52 Pst and ppS2 Pst inv. The restriction
map of both clones is shown in Figure 5. The clones of
type E. coli ppS2 Pst express carbamate hydrolase, after
addition of the inductor isopropyl-~-D-thiogalacto-
pyranosid to the culture medium. Without inducer addition
(repressed state of the lac promotor) to logarithmic
cultures of the clone ppS2 Pst as well as by repre6sed and
induced logarithmic cultures of the clone ppS2 Pst inv in
enzyme extracts, no enzyme activity was seen using the
assays described in Example 3.
This means that the carbamate hydrolase gene in clones of
type ppS2 Pst lies in the same transcription direction
(5'-3' orientation) as the lac Z' gene of the vector. The
Arthrobacter promoter is not or only slightly expressed in
E. coli.
~g- 1338440
Example 8
Nucleotide sequence of the carbamate hydrolase gene from
the Arthrobacter oxidan6 (specie6 P52) and the deduced
protein sequence.
The nucleotide 6equence of the carbamate hydrolase gene
was establi6hed by the method of Sanger (Sanger F.,
Nicklen S. & Coul60n A. (1977) Proc. Natl. Acad. Sci. USA,
74, 5463-5468).
For thi6, 15 6ub-clone6 in the 6ingle 6tranded DNA
bacteriophage M13 mpl8 and M13 mpl9 from the pp52 PST DNA
were constructed (Me66ing, J. (1983) Method6 in Enzymol.
101, 20-78). After tran6fection of E. coli DH5aF', the
6equence of the single stranded recombinant DNA was
establi6hed.
In Figure 6, the 6equencing 6trategy of the carbamate
hydrolase gene i6 6hown. Altogether the 6equence of 1864
base pairs was is establi6hed.
In Figure 7, the established nucelotide sequence i6 shown
with the deduced amino acid sequence of the carbamate
hydrola6e. The reading frame is clearly defined as
described by the amino acid sequence6 of two BrCN cleavage
peptides as described in Example 4. The reading frame
finishes with a TGA stop codon (nucleotide position
1789-1791 in Figure 7). As a translation start codon, a
GTG (nucleotide position 340-342) is suitable. This gives
the longest open reading frame of 1479 bp (= 493 amino
acids). All open reading frames which begin with the usual
ATG start codons give no protein of suitable size
(compared to the molecular weight determination of the
protein).
384 4 0
The hypothesi6 that translation start6 from GTG (position
340-342) i6 further supported by the exi6tence of a
definite homologou6 region to the con6en6us 6equence for ~
ribosomal E. coli binding 6ites 7 bp up6tream of the
putative GTG 6tart codon.
All cloning 6tep6 were carried out by 6tandard processe6
(cf Maniati6, T., Frit6ch, E.F. & Sambrook, J. (1982) in
"Molecular Cloning", Cold Spring Harbor, N Y). The
6equencing reaction6 were carried out u6ing Sequena6e
DNA Sequencing Kits (United States Biochemical
Corporation) according to information by the producer. The
separation of the marked reaction products was carried in
6% w/v polyacrylamide/urea gel (Maxam, A.M. ~ Gilbert, W.
(1980) Methods Enzymol. 65, 497-559).