Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
WOg4/~6~ 21 ~ 14 7 3 PCT~S93/12013
~YNl~SIS OF ENCODED POLYMERS
Field of the Invention
This invention relates to the fields of
biopolymer synthesis and drug design. More
particularly, the invention relates to methods for
synthesizing libraries of biologically active polymers
in association with an included polymer which is
~noo~ to facilitate deciphering.
Back~,~ulld of the Invention
Modern pharmaceutical technology has taken
two divergent paths in pursuit of new therapeutic
compounds. Rational drug design achieves results by
intensive analysis of the molec~ r structure of
b;n~ing sites, and designing compounds specifically to
complement a desired b; n~; ng site. For example, one
interested in preparing new antihypertensive compounds
might analyze the mol~clll A~ structure of the ~-
adrenergic receptor bi n~ i ng site using X-ray
crystallography and/or advanced NMR techniques, and
then synthesize compounds calculated to fit within the
binding site and complement the charge distribution.
The other approach is to prepare an enormous
library of compounds and select only those compounds
which exhibit a desired activity. This approach
differs from the traditional pharmaceutical cycle of
design/synthesize/test/synthesize variants by
conducting the scr~n; ng step in a massively parallel
fashion, scree~;ng an enormous number of different
compounds simultaneously. The challenge to this
W094/~6~ PCT~S93/~013
~$ approach is first to provide a group of compounds for
scr~n; ng that is sufficiently numerous and diverse to
insure that the activity sought is represented in the
group, and c~con~ to identify the active compounds at
low concentration within the group.
Rutter et al., US 5,010,175 disclosed a
method of making diverse mi~L~es of peptides by
adjusting the con~entration of each activated peptide
in PLU~Vr Lion to its reaction rate, in order to obtain
a substantially equimolar mixture of peptides. Rutter
also disclosed the ~L G~e~S of providing a mixture of
peptides (having at least 50 different peptides), and
selecting one or more peptides having a desired
property and separating them from the rest of the
peptides.
Zuckermann et al., PCT WO91/17823 disclosed
an alternative method for preparing diverse mixtures
of oligopeptides on solid-phase resins, and a robotic
device for performing the nececs~ry ma~;p~ tions. In
this method, a pool of resin particles is separated
into a number of groups (wherein each group is defined
as one or more separate reactions), and a different
amino acid coupled to the resin in each group. The
groups are then mixed together, separated into a
number of yLOU~ and again coupled with a different
amino acid for each group. This cycle is repeated
until the desired number of amino acids per
oligopeptide is obt~i n~ . One advantage of this
approach is that each coupling reaction occurs in
isolation from other reactants, which permits one to
drive each reaction to completion without carefully
adjusting the initial conc~tration of each reactant.
This method also facilitates the preparation of
oligopeptides wherein some positions within the
peptide chain are held constant, and where some posi~
tions are restricted to less than all amino acids.
For example, one may use this method to prepare a pep-
tide of the formula X1-~-X3-Glu-Ala-X4-Xs-~, where Xn
wo 941~6~ 21~ 1 4 7 3 PCT~S93/~0~
~ ~. , .
can be any amino acid. If desired, one could limit,
for example, X3 and X5 to l.yd~hobic residues.
Zuckermann also disclosed that this method may be
applied to the synthesis of oligonucleotides, which
may then be inserted into cloning and expression
vectors for biological expression.
Bartlett et al., PCT WO91/19735 disclosed a
variation of the Zuckermann et al. method i~ which a
diverse set of non-amino acid monomers is employed to
form mixtures of com~o~ C called "peptoids."
Peptoids sample a different region of physico-chemical
parameter space than traditional oligopeptides,
~Pp~nA; ng on the type of linkage between monomers, and
may be able to exhibit activities unavailable to
peptide libraries due to the diversity (or difference)
in side ch~; nc .
Houghten, US 4,631,211 disclosed a "tea-bag"
peptide synthesis method. The "tea bags" are mesh
bags cont~i n; ng resin beads for peptide synthesis.
Houghten's method enables one to add the same amino
acid to a number of different oligopeptides without
mixing the products: a number of "tea bags~' may be
reacted with an amino acid in a common pot, then
separated physically.
Cook, EP 383620 described synthesis of COP-
1, a random polymer of Ala, Glu, Lys, and Tyr, having
an average mol~c~ r weight of 23 kDa h~ving activity
in the treatment of multiple sclerosis. COP-l is made
in the prior art by chemical polymerization of the
amino acids. However, Cook described expression from
genes made by random polymerization of
oligonucleotides, and selection for those clones
expressing COP-1 with the highest activity.
-Lebl et al., EP 445915 described a machine
for performing multiple simultaneous peptide syntheses
using a planar support surface. The planar support
is, for example, paper or cotton.
W094/~ ~ PCT~S93/12013
~S~ 4~ -4-
Kauf~man et al., W086/05~03 disclosed
production of peptide libraries by expression from
synthetic genes which are partially or wholly
"s~o~ tic." S~o~h~tic genes are prepared by polym-
erizing a mixture of at least three oligonucleotides
(at leaæt heptamers) to form a double-stranded
sto~-hActic sequence, and ligating ~he stochastic
sequence into an expression vectorO
Lam et al., WO92/00091 disclosed libraries
of oligonucleotides, oli~o~e~Lides, and
peptide/nucleotide chimeras, and methods for screening
the librarieæ for active compollnA~. However, Lam did
not disclose conjugates having an active seguence and
a coA; ~ equence.
K.M. Derbyshire et al., Gene (1986) 46:145~
52 disclosed a method for "saturation mutagenesis" of
a segment of DNA, by synthesizing oligonucleotides
using contaminated pools of monomer. Each A, C, G,
and T reservoir con~ e~ 1/54 parts of each of the
other bases. The object was to prepare a DNA segment
mixture having one or two mutationC per sequence.
They did not observe equal frequencies of mutation,
presumably due to differences in coupling efficiency.
The authors suggested synthesizing seq~ seC using
four reservoirs cont~in;n~ pure bases, and one
reservoir cont~;ni~q a mixture of all four bases in
the co~cPntrations npcesc~ry to h~lAnce the coupling
efficiencies.
J.F. Rei~h~r-olson et al., Science (1988)
241:53-57 disclosed the generation of mutant ~
repressor proteins by replacing two co~o~c with random
nucleotides (NNG/C). The resulting mutant proteins
were assayed for activity to determine which amino
acid positions were critical, and which positions
should be conserved.
I.S. Dunn et al., Prot Enq (1988) 2:283-91
disclosed the use of random polynucleotides to gen-
erate mutant ~-lactamase ~-peptides, some of which
WO94/136~ ~1 S 14 7 3 PCT~S93/~013
~ ^ , .
-5-
exhibited properties superior to the native sequence
~-peptide.
A.R. Oli~h~nt & K. Struhl, Nuc Acids Res
(1988) 16:7673-83 disclosed the use of random poly-
nucleotides to investigate promoter function. A
section of random polynucleotide was inserted into the
-35 to -10 region of a gene conferring drug resistance
in E. coli, and the transformants screened for resis-
tance. Survivors were cloned and sequenced to provide
a functional con~pn~lc sequence.
F.W. Studier, Proc Natl Acad Sci USA (1989)
86:6917-21 disclosed a method for sequencing large
volumes of DNA by random priming of cosmid libraries.
A.R. Olirh~nt et al., Proc Natl Acad Sci USA
(1989) 86:9094-98 disclosed the generation of ~-
lactamase mutants having altered properties, by
cloning a random polynucleotide into the ~-lactamase
gene.
D.K. Dube et al., Biochem (1989) 28:5703-07
disclosed the generation of ~-lactamase mutants having
altered properties, by cloning a random polynucleotide
into the ~-lactamase gene.
R.A. Owens et al., Biochem Biophys Res Comm
(1991) 81:402-08 disclosed the selection of an HIV
protease inhibitor from a library of 240,000 tetrapep-
tides (in 22 mixtures). The mixtures were prepared by
the "mixed resin" t~h~ i ~ue.
These ~e~h~ i ques enable one to prepare
libraries of diverse compounds. However, the problem
of identifying the resulting compounds has ~eldom been
addressed. Oligopeptides are typically se~Pnc~ by
stepwise cleavage of each amino acid from tle parent
comro~ln~ (which is 11~ 1 ly immobilized on a resin),
with chromatographic analysis of the cleaved moiety.
Sensitive tec~n;ques are required to distinguish
between twenty or more amino acids. Analysis is
further complicated when uncommon amino acids are
W094/136~ PCT~S93/~0~
3 -6-
employed (using ~ulL~l~L tec~ni~ues)~ especially when
monomers are 1ink~ without using amide bonds.
SummarY of the Invention
The present invention provides a method of
synthesizing true mixtures of diverse oligopeptides
and/or peptide-like compolln~c along with an associated
enco~; ng polymer making it po sible to easily analyze
those compounds exhibiting a desired activity. The
invention involves synthesizing an ~nCOA i ng DNA strand
simultaneously with the peptide/peptoid. Each unique
peptide/peptoid sequence associated with its own
unique DNA strand to provide the conjugates of the
invention. These conjugates are screened to determine
which peptide/
peptoid compounds exhibit a desired activity, and the
active conjugates analyzed by DNA sequencing methods
to determine the att~hPA peptide/peptoid sequence by
deduc-tion, i.e., since each DNA sequence is
associated with a known peptide/peptoid, once the DN~
sequence is deter-
mined, the sequence of the peptide/peptoid can
be ~ c~.
Another aspect of the invention is a
conjugate comprising a peptide or peptoid coupled to
and/or directly associated with a coAi~g polymer (CP),
e.g. a nucleic acid (NA). The peptide/peptoid/CP
conjugate may be l;nke~ directly (i.e., covalently
bound either directly or through a small organic mol-
ecule), or by linkage to the same ~u~olL (e.g., bysynthesizing both peptide/peptoid and CP strand on the
same particle or bead of resin).
An important object of the invention is to
provide a chemical synthesis method which allows the
production of libraries of peptides and/or peptoids
along with a unique ~nco~ polymer such as a DNA
strand which makes it possible to readily determine
the sequence of the peptide or peptoid.
WOg4/~6~ 215 14 ~ 3 PCT~S93/~013
~ .
~ 7
An advantage of the present invention is
that the methodology makes it possible to readily
identify and sequence peptides and/or peptoids having
desirable biological activities.
A feature of the present invention is that
se~lPnc~c of peptoidæ or peptides which contain
nonconventional amino acids can still be readily
determined by seguencing associated polymers such as
DNA se~l~nce~ which are simult~n~o~lcly synthesized
with the peptoids and enco~e them.
These and other objects, advantages and
features of the present invention will become apparent
to those persons skilled in the art upon reading the
details of the structure, synthesis and use as more
fully set forth below, reference being made to the
accompanying figures forming a part hereof.
Brief Description of the Drawinqs
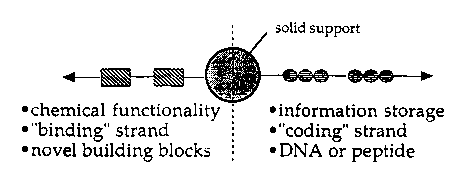
Figure 1 is a schematic diagram showing a
specific embodiment of a conjugate of the invention
which conjugate includes a "b; n~; ng~ strand or active
polymer attached to a solid-~u~u~L substrate which
substrate is also ho~ to an i~formation storage or
"co~;ng" strand;
Figure 2 is a schematic flow diagram
demonstrating how ~nco~ libraries can be synthesized
on beads as the solid _U~l L substrate;
Figure 3 is a schematic diagram showing
methods of the synthesis of both solid-phase and
solution-phase libraries;
Figure 4 is a schematic diagram showing
resin-bound libraries generated by the derivatization
of non-hydrolyzable resins;
Figure 5 is an HPLC chromatogram of binding
and co~;ng peptide strands simultaneously synthesized
via non-hydrolyzable resin l;nk~e;
W094/~6~ PCT~S93/~013
~ ~5~4~3 -8~
Figure 6 is an HPLC chromatogram of a coding
and binding strand adduct which was synthesized via a
hydrolyzable resin l;nk~;
Figure 7 iæ a plotted graph resulting from
ELISA competition of bi n~ i ng se~l~nc~s versus
bi n~ i ng/ enCoA; ng se~l~ec;
Figure 8 is a schematic diagram showing the
analysis of a solid-phase amptide; and
Figure 9 is a schematic flow diagram showing
the analysis of a solution-phase amptide.
Detailed Description of Preferred Embodiments
Before the present method of synthesis,
conjugates and methods of using such are described, it
is to be understood that this invention is not limited
to the part;c~ r methodologies, conjugates, or
methods of use described as such may, of course, vary.
It is also to be understood that the terminology used
herein is for the ~ul~o~e of describing particular
emho~;ments only and is not inten~ to be limiting
since the scope of the present invention will be
limited only by the apr~n~ claims.
It must be noted that as used herein and in
the apr~nA~A claims, the singular forms "a," "and,"
and "the" include plural referents unless the context
clearly dictates otherwise. Thus, for example,
reference to "a peptide" includes mixtures of
peptides, reference to "an amino acid" includes
mixtures thereof, and reference to "the reaction"
includes one or more reactions of the same type as
generally understood by those skilled in the art, and
so forth.
Unless defined all otherwise, all ~c-h~; cal
and scientific terms used herein have the same meaning
as commonly understood by one of ordinary skill in the
art to which this invention belongs. Although any
methods and materials similar or equivalent to those
described herein can be used in the practice or
WO94/136~ 21514 7 3 PCT~S93/~013
~ 9
testing of the present invention, the preferred
methods and materials are now described. All
publications mentioned herein are incorporated herein
by reference to describe and disclose specific
S information for which the reference was cited in
ro~ection with.
In general, the invention provides a rapid
method of synthesizing large numbers of conjugates
which conjugates are comprised of a peptide/peptoid
sequence, e.g., an amino acid sequence associated with
a unique ~nco~ i ng sequence, e.g., a DNA sequence. The
conjugates can be readily synthesized and thereafter
screened for biological activity, and when activity is
found, the particular peptide/peptoid sequence found
to be active can be readily identified by its
associated ~nco~;ng (DNA) strand. Each conjugate of
the invention is comprised of at least two aomponents
with one of the comron~nts being the peptide or
peptoid sequence which binds to a receptor of interest
and the other sequence being a polymer which encodes
the bi n~; n~ sequence. The invention may utilize
st~n~rd amino acids and DNA as encoding monomers to
produce a chemically diverse library of solution-phase
or solid-phase conjugates. In order to further
describe the invention in detail, the following
definitions are provided.
A. Definitions
The terms "nucleic acid" and "NA" refer to
oligomers constructed from DNA and/or RNA bases which
may be se~l~nc~A using st~n~-rd DNA sequencing tech-
niques. The NAs used herein may include uncommon
bases so long as such bases are distingll;~hAhle from
the other bases employed under the DNA sequencing
methods to be used and include peptide-nucleic acids
(PNAs) (disclosed by Nielsen, P.E., Egholm, M., Berg,
R.H. & Rll~h~rdt, O., Science (1991) 254, 1497-lSO0).
Such PNAs could serve as co~i ng ctrands and the
W094/~6~ PCT~S93/~013
~4~3 lo
detection would be by hybridi-zation. NAs will
usually be constructed from monomers linke~ by phos-
phodiester bonds, but other similar linkages may be
substituted if desired. For example,
phosphorothioates may be employed to reduce lability.
The term Upeptide'' as used herein refers to
the 20 commonly oc~ ing amino acids: Al~nin~ (A),
cysteine (C), aspartic acid (D), glutamic acid (E),
phenylAlAni~ (F), glycine (G), histi~in~ (H),
isoleucine (I), lysine (R), l~l~cine (L), methionine
(M), asparagine (N), proline (P), glutamine (Q),
arginine (R), serine (S), thr~Qnin~ (T), valine (V),
tryptophan (W), and tyrosine (Y).
The term "peptoid~ as used herein refers to
a non-peptide monomer of the general formula (R)~-X-
(L)m~ where R is a side chain group, n is at least 1,
L is a linkin~ group, m i8 at least 2, and X is a
small organic radical. It is preferred to select L
radicals that may be indivi~ y protected and
deprotected. Preferably n will be 1 or 2 and m will
be 2. Monomers wherein m is 3 or greater may be used
to form brAn~h~ active poly~ers. Presently preferred
monomers are N-substituted glycine derivatives of the
formula
~ x
wherein R is alkyl of 2-6 carbon atoms, haloalkyl of
1-6 carbon atoms wherein halo is F, Cl, Br, or I,
alkenyl of 2-6 carbon atoms, alkynyl of 2-6 carbon
atoms, cyclolkyl of 3-8 carbon atoms, alkoxyalkyl of
2-8 carbon atoms, aryl of 6-10 carbon atoms, arylalkyl
of 7-12 carbon atoms, arylalkyl of 7-12 carbon atoms
substituted with 1-3 r~icA~ epPnApntly selected
from halo and nitro and hY~LGXY~ am;~Q~1~Y1 Of 1-6
carbon atoms, ~ly~Loxyalkyl of 1-6 carbon atoms,
WOg4/~6~ 21 51 4 7 3 PCT~Ss3/~013
carboxy, carboxyalkyl of 2-6 carbon atoms,
carho~lk~y-alkyl of 3-10 carbon atoms, carbamyl,
carbamylalkyl of 2-6 carbon atoms, imidazolyl, imid-
azolylalkyl of 4-10 carbon atoms, pyridyl,
pyridylalkyl of 6-10 carbon atoms, piperidyl, pip-
eridylalkyl of 5-10 carbon atoms, indolyl, or
indolyalkyl of 9-15 carbon atoms. Thus, active
polymers composed of these monomers are equivalent to
polyglycine having side ChA; nc att~ at each
nitrogen. These and other monomers are described in
cop~n~;~g application USSN 07/715,823, incorporated
herein by reference, and PCT W091/19735.
The terms ~coA i ng~ and ~e~coAing~ indicated
that one or more coAi ng monomers corresponds directly
and uniquely to a given active monomer, e.g.,
conventional
nucleic acids ~n~o~ (in y~Ou~S of three) the 20 natural
amino
acids. The number of co~i ng monomers used for each
code ~p~nAc on the number of different coAing mono-
mers and the number of different active monomers.
Typically, the number of different active monomers
used will range from about 5 to about 30. A basis set
of 4 coding monomers can ~ncoA~ up to 1~ active
monomers taken in ~co~nc~ of 2 co~i ng monomers. By
increasing the co~i ng monomer basis set to five
distinct monomers, one can encode up to 25 different
peptide/peptoid monomers. A basis set of 4 coA i ng
monomers can ~nco~e up to 64 peptide/peptoid monomers
taken in "codons" of 3 co~;~g monomers. Note that one
can make the code degenerate or nonA~generate, and can
insert additional co~i ng information into the
sequence. For example, one may wish to begin each
codon with the same base (e.g., G), using that base
only in the first position, thus unambiguously identi-
fying the beginning of each codon. As a practical
matter, the group of monomers selected for use as
coding monomers will form polymers that are easier to
W094/~6~ PCT~S93/~0
12-
sequence than the active polymers, i.e., the co~in~
monomers may be more readily identified using present
day sequencing ~r~nology as compared to the monomer
of the active poly~ers. With current technology, the
order of preference for co~in~ monomers is nucleic
acids > peptides > peptoids. Nucleic acids have the
additional advantage that the roA i n~ sequence may be
amplified by cloning or PCR (polymerase chain
reaction) methods known in the art.
The term "active polymer" and/or "bin~in~
polymer" refers to a polymer having a desired
biological activity. Suitable biological activities
include bin~;n~ to natural receptors, pharmaceutical
effects, immunogenicity/antigenicity, and the like.
"Immunogenicity" refers to the ability to stimulate an
immune response (whole or partial serum-mediated
i -unity and/or cell-mediated immunity) in a bird or
mammal following administration. Antigenicity
reguires only that the active polymer bind to the
antigen-binAi n~ site of an antibody. Pharmaceutical
activities, for the ~hL~o~es of this invention, will
generally ~p~n~ on the ability of the active polymer
to bind a protein, carbGl~d~ate, lipid, nucleic acid,
or other com~ present in the subject. For
example, an active poly~er may bind to a cell surface
receptor and compete with the receptor's natural
ligand, with or ~ithout activation of the receptor.
Other useful pharmaceutical activities include
cleavage of endogenous molecules (e.g., protease
activity, nuclease activity, and the like), catalysis
of reactions either primarily or as a cofactor,
donation of functional yLo~ (e.g., acyl, ATP, alkyl,
and the like), pore formation, and the like. Active
polymers comprise a series of monomers which are
1inkP~ sequentially. The monomers will generally be
peptides, peptoids, or carbohydrates in the practice
of the instant invention.
W094/~6~ 215 1~ 7 3 PCT~S93/~013
~ .
- -13-
The term "mixture" as used herein refers to
a composition having a plurality of similar components
in a single vessel.
The term "couple" as uæed herein refers to
formation of a covalent bond.
The term "coupling moiety" refers to a
soluble or insoluble ~ u~ to which can be attached
one or more active monomers and the ~o~Le~pon~ing
~ncoA;~g monomers. Insoluble ~u~GLLs ("solid ~u~o~
lo means") may be any solid or semi-solid eurface which
is stable to the reaction conditions required for
synthesis of the active and ro~ing polymers, and is
suitable for covalently att~chin~ and immobilizing
both polymers, for example, most resins commonly
employed in DNA and peptide synthesis, such as MBHA,
Rink, and the like. The particular resin used will
~p~n~ upon the choice of coA i n~ and active polymers
and their associated synthetic chemistries. Soluble
coupling moieties are mol~c~ R having functional
~L Ou~A to which active and co~; ng monomers may be
attached. Each soluble coupling moiety must be able
to accommodate at least one coAing polymer and at
least one active polymer, although the active and
co~ing polymers need not be present in a 1:1 ratio.
The soluble coupling moiety may be as simple as an
amino acid having an functional group in its side
chain, or may be as complex as a function~ ed
(soluble) polymer.
The term "conjugate" as used here n refers
to the combination of any "active polymer" and its
associated "co~i ng~ polymer. The conjugate may be
formed using a "coupling moiety" or by b;n~in~ both
the "active polymer" and "~nco~ing polymer" to the
same ~lr~lJorL surface in close proximity with each
other so that the two polymers are "associated" with
each other. When both polymers are bound to the same
support surface, æuch as a small bead, the ~nro~;ng
polymer can be readily se~l~nce~ off of the bead and
W094/~6~ - PCT~S93/~013
i
~5 the othér "active polymers" remaining on the bead will
be identified once the enco~;ng sequence is known.
B. Related T.i hraxies and SYnthesis Mçthodologies for
producin~ same.
There are many limitations with the current
t~hnologies for probing the receptor-bi n~; ng
properties of peptide libraries. Filamentous -
bacteriophage libraries offer the largest source of
peptide diversity (~107-l08 different components) of
any current te~hnology to date (Scott, J. & Smith, G.,
Science, (1990), 249, 386-390; Devlin, J., PangAn;h~n,
L. & Devlin, P., Science, (1990), 249, 404-406;
Cwirla, S., Peters, E., Barret, R. & Dower, W., Proc.
Natl. Acad. Sci. U.S.A., (1990), 87, 6378-6382).
These libraries, however, are limited to the natural
set amino acids, suffer from biological biases (i.e.,
varying rates of growth, proteolysis, etc.) and also
suffer, in practice, from high levels of backy~o~.d
2 0 bi n~ i ng . The present invention is designed to
overcome these difficulties.
Multiple-peptide æynthesis te~hnology has
substantially increased the ability to generate
individual peptides (Geyæen, H., ~eloen, R. &
Bartel;~g, S., Proc. Natl. Acad. Sci. U.S.A., (1984),
81, 3998-4002; Houghten, R., Proc. Natl. Acad. Sci.
U.S.A., (1984), 5131-5135; !e~hn~rrenberg, G. &
Gerhardt, H., Tetrahedron, (1989), 45, 7759-7764;
Gausepohl, H., Kraft, ~., Rolll;~, C. & Frank, R. in
Peptides: Chemistry. Structure and BioloqY
(Proc~;n~s of the 11th American Peptide Symposium,
(1990), eds. Rivier J. & Marshall, G., (ESCOM,
Leiden), pp. 1003-1004; Frank, R. & Doring, R.,
Tetrahedron, (1988), 44, 6031-6040; Fodor, S., Read,
J., Pirrung, N., Stryer, L., Lu, A. & Solas, D.,
Science, (1991), 251, 767-773). The synthesis of ~104
individual peptides per cm2 of glass wafer represents
the diversity limit of this technology (Fodor, S.,
WOg4/~6~ 215 1~ 7 3 PCT~S93/~013
-15-
Read, J., Pirrung, M., Stryer, L., Lu, A. & Solas, D.,
Science, (1991), 251, 767-773). A mixed-resin
algorithm method (Furka, A., Sebestyén, M., Asgedom M.
& Dibo, G., Int. J. Petide Protein Res., (1991), 37,
487-493) has recently been used to generate solution-
phase libraries (Houghten, R., Pi ni 1 1~ ~ C., B1O~P1 1 e,
S., Appel, J., Dooley, C. & Cuervo, J., Nature,
(1991), 354, 84-86) and resin-bound peptide libraries
(Lam, K., Salmon, ~., Hersh, E., Hruby, V.,
Kazmiersky, W. & Knapp, R., Nature, (1991), 354, 82-
84) that contain ~106 and ~107 comrQn~nts,
respectively. The solution-phase libraries offer the
advantage of providing quantitative receptor-binding
information (Zuckermann, R., Kerr, J., Siani, M.,
Banville, S. & Santi, D.V., Proc. Natl. Acad. Sci.
U.S.A., 89, 4505-4509 (1992)). Furthermore~ these
libraries allow the affinity of the solution
conformation of a ligand to be determined, a quantity
that is essential for rational drug design. An
apparatus for the automated synthesis of equimolar
peptide mixtures is disclosed in Zuckermannr R.N.,
Kerr, J.M., Siani, M.A. & Banville, S.C., Int. J. Pep.
Pro. Res., (1992), 40, 498-507.
The publications cited and ~ e~ above
can be used in producing the active or bi n~; ng polymer
which is used in producing the conjugate of the
present invention. Accordingly, the disclosures of
all of these publications are in~o~o~ated herein by
reference in order to disclose peptide and peptoid
synthesis methodology. Al~holyh the methodology
~;~c~l~ced within these references is extremely
valuable with respect to the production of large
amounts of different types of bin~in~ polymers, the
mixtures of polymers pro~llc~ by this methodology are
often so large and complex that there are many
practical limitations with respect to their actual
analysis and use. The present invention can be
readily applied with such synthesis methodologies in
wos4/~6~ PCT~S93/~013
~$ ~ 16- i
order to provide an ef~icient, commercially practical
method of analyzing the proteins produced using such
methodology.
Both the mixed-resin and solution-phase
methods, however, do not allow incorporation of many
non-st~n~rd amino acids heC~l~c~ of the limitations of
peptide analysis. Resin-bound peptide libraries, in
particular, suffer from a relatively slow rate of
analysis (peptide se~"ci~ at 3 beads per day) and
are limited in complexity to ~107 beads/ml. In order
to generate a "complete" peptide library, there must
be multiple copies (>lO) of any given peptide
sequence. This becomes problematic at the sequenciny
stage because the same "hit" sequence may ~rr~r
multiple times. The alternative is to work with
libraries that are not complete at the risk of losing
se~l~nceC that bind.
When using the methodoloyy of the present
invention, the sequence of a biologically active
protein can be determined even without isolating the
protein of interest. This can be done by synthesizing
large numbers of different proteins on large numbers
of different ~U~OL ~ ~urfaces such as small beads. An
~nco~i n~ polymer is attached to beads to identify each
protein. A sample to be tested is then brought into
contact with the beads and the beads are observed with
respect to which proteins bind to a receptor site in
the sample. The bead having the receptor bound
thereon is analyzed by sequencing the coding polymer
which has also been synthesized on the bead. When the
encoding polymer has been se~nce~, the sequence of
the active polymer, which may be a peptide, can be
readily ~ ce~. Thus, the present invention makes it
possible to determine the activity and seguence an
active polymer, such as a biologically active peptide,
without ever isolating the peptide.
W094/~6~ 21 ~1 4 7 3 PCT~Ss3/~013
-17-
C. General Methodolooy
This invention describes a methodology for
the synthesis and scre~ni ng of large synthetic polymer
libraries that contain non-stAnA~rd amino acids and
even non-amide h~C~A polymers. The strategy utilizes
a modified mixed-resin peptide synthesis methodology
to simult~nesll~ly ~ynthesize two polymer ~e~nceC
one polymer strand (the 'lbinAi ng~l strand) is
synthesized for the in~Dn~A purpose of receptor
b;nAing, and the cecon~ strand (the ~oAing~ strand)
contains st~nA~rd amino acids or deoxyrihQnl~cleotides
that ~ncoA~ for the binAln~ strand (Figure 1). The
ability to decipher the bi nA i ng sequence by analysis
of the coA i n~ strand with s~nA~rd peptide or
oligonucleotide te~ni~ues allow the inclusion of a
wide variety of novel h~ ing blocks and
conformational constraints into a diverse ligand
library.
This invention also describes a methodology
to increase the size t~l8) and scr~ni n~ rate of a
ligand library. The method uses two polymers as
above, but specifically utilizes an
oligodeoxyrihomlcleotide for the ~coA ing" strand. The
use of DNA as the coA i ng strand allows for an
incr~eA sensitivity of detection (fmol vs pmol for
peptide analysis). This increased sensitivity allows
for a larger library size since the amount of polymer
ne~A for detection is reAnceA dramatically. The
rate of sequence det~r~in~tion of receptor binders is
increased since many samples can be analyzed in
parallel.
In order to couple a polymer's sequence
information with a peptide or oligonucleotide
sequence, there needs to be a method that
unambiguously correlates each polymer to each other.
Thus, when any particular non-st~nA~rd amino acid (or
other monomer) is added to a "bi n~ i ~g~ polymer chain,
the correcronAing information (amino acid or
W094/~6~ PCT~S93/~013
~S~4~ 3 -18-
nucleotide monomer) must also be added to the "coding"
strand. A "genetic code" is thus establi ~h~ (Table
1) where each b;n~;n~ monomer corre~ron~c to (a
multiple) of st~n~rd amino acids or nucleotides on
the co~; n~ strand. For example, the use of three
s~An~rd amino acids or nucleotide , in a 3:1 ratio
with a novel monomer, would allow for the unambiguous
representation of 27 novel ~onomers.
TABLE 1. Custom genetic code.
# of bases "codon" # of monomers
length coded for
2 3 8
2 4 16
2 5 32
3 2 9
3 3 27
4 2 16
4 3 64
The synthesis of coded libraries requires a
modified mixed-resin algorithm (Figure 2). The resin
beads are divided into equal portions, a unique
monomer is added to the "bi nA i~g" strand, followed by
the coupling of a ~ o~ g amino acid or
nucleotide to the '~co~in~ strand. The resin aliquots
are then combined to generate a mixture. A set of
compatible protecting yL OU~S iS thus required to
preferentially deprotect and extend each strand
independently
Two synthesis formats are possible for
amptide libraries, one that generates resin-bound
libraries and one that generates solution-phase
libraries (Figure 3). Resin-bound libraries can be
synthesized using non-hydrolyzable linkers that are
derivatized with the "bin~;ng" and l'co~;ng" monomers
strands. Solution-phase libraries can be synthesized
W094/~6~ PCT~S93/~013
2151~73
.. ,. ,, , ;, , . . 1 9
as a 1:1 polymer:peptide/DNA conjugate via a
hydrolyzable link~r attached to the resin.
Pe~tide as the "Codinq" strand
The use of base-labile Fmoc-protected
- monomers and acid-labile (N~-Ddz-protected ~mino acids
(Birr, C., Nassal, M., Pipkorn, R., Int. J. PeDtide
Protein Res., (1979), 13, 287-295), for example, allow
for selective deprotection and coupling to two
individual polymer strands. Resin-bound libraries can
be generated by the derivatization of non hydLolyzable
resins with a 1:1 ratio (or any desired ratio) of
Fmoc:Ddz monomers tFigure 4). This i~lLLGl.ce-c two
differently protected amino acids that an be ext~n~
;~p~n~ntly. Solution-phase libraries that contain
a 1:1 ratio of bin~;ng:co~;ng strands can be
synthesized by using a hydrolyzable Fmoc-Lysine(Moz)-
OH l;nk~ that allowed for chain growth at both the ~-
and ~-amino ~-~u~. Amino acids which do not contain
functional yLOu~s are preferred for the "coding"
strand in order to minimize unwanted binding
interactions.
The receptor-bi~ing ligand can be
identified by bead st~i~ing ~echn;ques (Lam, K.,
Salmon, S., Hersh, E., Hruby, V., Kazmiersky, W.
~ rr, R-, Nature, (1991), 354, 82-84) and the
sequence determined by N-terminal Edman degradation.
In order to ensure that only the '~oo~ing~ strand is
se~l~nc~, it is essential that the N-terminus of the
"b;n~ing" strand be acetylated or otherwise made non-
seq~ nc~hle.
DNA as the "Coding" Strand
The construction of libraries with DNA as
the co~ing strand is similar to those with peptides
but offers several advantages: the information
storage and replicative properties of DNA allow for
W094/~6~ - PCT~S93/~0
increased sensitivity of detection, a larger library
size and an increased rate of sequence determinationO
The synthesis of DNA as ~he co~; ng polymer
requires compatibility between the as~embly of Fmoc-
5 h~ ~ monomers and st~n~d DNA chemistry. Thesesynthesis strategies are likely to be compatible ((a)
Juby, C., Richardson, C. & Brous eau, R., ~et.
Letters, (1991), 32, 879-882. (b) Haralambidis, J.,
Duncan, L., Angus, B. & Tregear W.~ Nucleic Acid Res ,
(1990), 18, 493-499) (see Table 2)o Alternatively,
allyl-based protection strategies existæ for both
peptide (Lyttle, M.H.; ~l-A~on, D., Pe~tides: ChemistrY
and Biolo~y (Proc~inqs of the 12th American Peptide
Symposium): Smith, J. ~nd Rivier, J.E., Eds.; ESCOM,
T.~ , 1992, pp. 583-584) and
oligodeoxyr;honl~cleotide (Hayakawa, y., I~~k~h~yashi,
S., Kato, H. & Noyori, R., J. Am. Chem. Soc., (1990),
112, 1691-1696) synthesis. The assay of æolution-
phase libraries can be facilitated by using only
pyrimi~;n~c in the coAing strand, thereby avoiding the
potential problem of base pairing between individual
strands.
Strategies for the synthesis of co~i ng and
active polymers, as well as matc~i ng the active
polymer with a ~o~;ng ~equence to provide a genetic
tag are described in Brenner et al. Proc. Natl. Acad.
Sci. U.S.A., 89:5381-5383 (June 1992) which is
incorporated herein by reference.
wo g4/~6~ 2 1 ~ 1 4 7 3 PCT~S93/~0~
- 21 : . .~. .
TABL2 2
Compatibility of DNA vs. Peptide Synthesis Chemistry
Peptide r1lPmi.stry Oligonucleotide Chemistry
~ o ~ ~0
P~rmanent Protectin~ ~ v~
-OH t-butyl ether A,C benzoyl amide
-CO2H t-butyl ester G isob~Ly~l
-NH2 t-boc -P=O cyanoethyl, methyl
his, cys trityl
arg sulfonyl
remov~d by:
85% trifluoroacetic acid conc. NE~OH 55C S hours
2 hours @ room temp
T~m~orarY ~rot~ti~a ~ G~
9-fluorenylmethoxycaL~oll~l 4,4-dimethoxytrityl
rQmove~ by:
20% piperidine 3% trichloroacetic acid
CleavaqQ fro~ 801i~ ~UP~oXt:
85% trifluoroacetic acid conc. NE~OH 55C S hours
2 hours ~ room temp
SU~;TI ~ UTE:~ S3HEElr
W094/136~ PCT~S93/~OL3
22-
~ABLB 2 tCo~t)
Compatibility of DNA vs. Peptide Symthesis Chemistry
Std. conditions ~ ;ve - Problem addressed
t-Butyl carbamate/ allyl amino acid
ester deprotection from
detritylation
reagent
20% Piperidine 2% DBU substitution at
exocyclic amine
cyanoethyl allyl acylation of
phosphate phosphate
NH40H ethyl~neAi~mine/ racemization
EtOH
iodine t Butyll~ o Tyr, Met, Cys
peroxide oxidation
controlled pore poly~yl~ne resin transferring
glass resin
T, C & G T, C, G & A depurination
T, C T, C, G & A base pairing
SUBSTITUTE Sl~,EET
WOg4/~6~ 21514 7 3 PCT~S93/~013
.--
- -23-
Resin-bound libraries can be synthesized by
using non-hydrolyzable linkers to attach both the C-
terminus of the peptide and the 3'-end of the
oligonucleotide to the same bead. Solution-phase
libraries can be synthesized as a 1:1 peptide-
oligonucleotide conjugate, in which the C-terminus of
the peptide is attached to the 3'-end of the
oligonucleotide through a hydrolyzable Fmoc-Ser(O-Dmt)
linker which is attached to the resin.
The identification of binders in the resin-
bound peptide libraries can be detected by the bead
stA;n;ng methodology (Lam, K., Salmon, S., Hersh, E.,
Hruby, V., Kazmiersky, W. & Knapp, R., Nature, (1991),
354, 82-84). Although the peptides are bound to a
solid-phase, there does not have to be a 1:1 peptide-
oligonucleotide ratio since the DNA can be amplified
prior to the determination of its sequence. In fact,
less DNA is preferred so that there will be less
interference with the polymer's binding properties.
Once a bead is identified, the DNA sequence is
determined (Stahl, S., Hultman, T., Olsson, A., Mois
T., et al., Nucleic Acid Res., (1988), 16, 3025-3038)
after PCR amplification or by thermal-cycle sequencing
(Figure 8). This requires the inclusion of one or
more primer sites neighboring the coA; ng region of the
oligonucleotide. Similarly, the use of solution-phase
libraries requires isolation of each sequence from
each other. This can be accompl; ~h~ by restricting
the DNA after PCR amplification and inserting it into
M13 (or other suitable vector) for clonal isolation
and sequencing (Figure 9).
EXAMPLES
The following examples will provide those
skilled in the art with a complete disclosure of how
to make and use the invention and are not intended to
limit the scope of the invention. Efforts have been
made to insure accuracy with respect to numbers used
W094/~6~ PCT~S93/12013
24-
(e.g. amounts, temperature, etc.), but some
experimental error and deviation should be accounted
for. Unless indicated otherwise, parts or parts by
weight, mol~c~ ~ weight is weight average molecular
weight, temperature is in degrees centigrade and
presæure is at or near atmospheric.
Example 1
The in~p~n~pnt synthesis of two
unambiguously correlated se~lPnc~c has been
sllccessfully completed. The C~lhc~uent sequence
analysis of the "co~i n~l strand has also been
demonstrated. For ~ol.ve~ience, two peptide se~lPnc~s
were chosen. The "b; nA; ng" strand was synthesized
with N~-Fmoc-protected amino acids and the "co~i ng~
strand was synthesized with N~-Ddz-protected amino
acids. The simultaneous synthesis of the two peptide
strands was tested in two formats, 1) resin-bound
peptide library synthesis and 2) solution-phase
peptide libraries using a hydrolyzable Fmoc-Lys~Moz)
OH 1 ;nker (Wang, S.S.; Chen, S.T, Wang, K.T., and
Merrifield, R.B., Int. J. Peptide Protein Res.,
(1987), 30, 662-667). These synth~c~fi were performed
on single peptides (not libraries) as a demonstration
of research concept and in order to allow the full
characterization of the synthesis products.
WO94/136~ 215 1~ 7 3 PCT~S93/~013
- -25-
A. SYnthesis of a Resin-Bound Library Model
"B;n~;ng" Sequence: Ac-Arg-Leu-Val-Thr-His
(Fmoc peptide)
"Coding" Sequence: H~-Ala-Ser-Gly-Glu-Phe-Ala
(Ddz peptide)
Synthesis Scheme:
Step # Description
1 Derivatization of MBHA resin with l:1-Ddz-
Ala-OH: Fmoc-His(Trt)-OH
2 De~ection of "bin~;ng" strand with 20%
piperidine/DMF
3 Coupling of ~ConA ~b;n~;ng" amino acid,
Fmoc-Thr(But)-OH
4 Deprotection of "co~;ng strand" with 7.5%
TFA/ CH2Cl2
Coupling of c~cQn~ "coding" amino acid, Ddz-
Phe-OH
6 Repeat steps 2-5 with alternative Fmoc and
Ddz deprotection and corresponding amino acid
coupling
7 Final Deprotection of Fmoc followed by
acetylation
8 TFA deprotection of side-chain yroups
After the TFA deprotection, the model
library bead has two in~p~n~ntly synthesized
se~l~c~c and is ready for assay. Only the coding
strand has a free ~-amino group and can be
characterized by N-terminal Edman degradation. The
binding strand is acetylated and there-fore will not
interfere with the sequencing. The two peptides were
cleaved from the resin with HF thereby providing both
the "binding" and "ro~i ng~ se~l~nc~s as free peptides.
The amino acid composition, mass spectro-scopy and N-
terminal sequencing data are consistent with thecorrect products. (See Figures 5, 6 and 7.)
PCT~S93/~013
WO g4/~3623
3 ~
-26-
Mass Spectrometry:
theoretical observed
Ddz peptide 580.2 580.2
Fmoc peptide 666.9 666.4
Amino Acid Composition:
Fmoc Peptide Ddz Peptide
10 Theor. Observ. Theor. Observ.
(%) (%) (%) (%)
His 20 21 Ala 33 33
Thr 20 20 Glu 17 17
Val 20 19 Phe 17 17
15Leu 20 29 Gly 17 15
Arg 20 20 Ser 17 15
N-Terminal Edman Se~uencinq of Resin beads fcoding
peptide only):
Cycle # amino acid pmol
1 Ala 45
2 Ser 12
3 Gly 30
254 Glu 28
Phe 28
6 Ala 29
Example 2
Synthesis of a Solution-Phase Peptide Library Model
In this example, a 1:1 solution-phase adduct
between a "b;n~;n~'~ and a "coA;n~" strand was
synthesized and fully characterized. The "binding"
strand was assembled wi~h Fmoc-protected monomers, and
the "co~ " strand was assembled with Ddz-protected
monomers.
W094/~6~ 2 I 5 1 ~ 7 3 PCT~S93/~013
.
- --27--
~B; nA; ng~ sequence: ~c-Glu-Ser-Thr-Arg-Pro~nLeu-Lys-B-
(Fmoc peptide) Ala-NH2
~CoA; nq~ sequence: ~N-Gly-Ala-Phe-Gly-Ala-Phe-CONH
(Ddz peptide)
SYnthesis Scheme:
Step # Description
1 Derivatization of Rink Resin with Fmoc-B-Ala-
OH spacer
2 Fmoc deprotection with 20% piperidine/DMF
3 Coupling of Fmoc-Lys (4-
methOXYL~11ZY1OXYCa ~ LG11Y1 )-OH
4 Fmoc deprotection of '~b;n~tnq~ ~trand with
2 0~ piperidine/DMF
Coupling of first ''b;nAin~ monomer Fmoc-
nLeu-OH
6 Cleavage of ~coA; nq~ strand 4-
Methoxybenzyloxycarbonyl (Moz) group with
7.5% TFA/ ~ C12
7 Coupling of first ~C~A i nq~ amino acid Ddz-
2 0 Phe-OH
8 Repeat steps 4-7 With ~ re~;ron~;n~ amino
acids
9 Final Fmoc deprotection and acetylation
Cleavage and deprotection of resin sample
with TFA
Following TFA cleavage and deprotection, the
model solution-phase library contains a l:1 Fmoc/Ddz
conjugate peptide. One peptide sequence was
synthesized and not a mixture in order to fully
characterize the reaction product. The amino acid
composition and mass spectroscopy data are consistent
with the correct product. In addition, the "bi n~; n~
and "coA;~q" hybrid peptides were tested in a
competition T'T.T,~ format. The ELSTRPnL "binding"
sequence binds to an anti-gpl20 antibody with
submicromolar affinity. This value was not affected
by the presence of the "coding" peptide.
W094/1~ PCT~S931~013
-28-
Mass S~ectrosco~y:
Theoretical Observed
Fmoc/Ddz peptide conjugate: 1492.7 1492.6
S Amino Acid Com~osition:
Fmoc Peptide Ddz Peptide
Theor. Observ. Theor. Observ.
(%) (%) (~) (%)
Glu 7.1 7.0 Phe 4.2 14.7
Ser 7.1 7.1 Ala 14.2 14.4
Thr 7.1 702 Gly 14.2 13.8
Arg 7.1 7.2
Pro 7.1 7.9
Nleu 7.1 6.3
The instant invention is shown and described
herein in what is considered to be the most practical,
and preferred embodiments. It is reco~n;~ed~ however,
that departures may be made therefrom which are within
the scope of the invention, and that obvious
modifications will occur to one skilled in the art
upon reading this disclosure.