Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
W095/03~4 PCT~3/019~
21 67701
DNA VIRUS RIBOZYME8
The present invention is directed generally to
synthetic ribozymes and their mutants and derivatives
capable of catalysing cleavage of virus specified mRNA
and, more particulary, DNA virus specified mRNA. The
present invention further contemplates the use of
these ribozymes in the generation of transgenic cells
and, in particular, transgenic plant cells and plants
capable of inhibiting infection by the viruses.
Viruses are the causative agents of a large
number of serious and potentially serious diseases in
humans, animals and plant. Plant viruses in particular
have the potential to destroy or reduce crop yield and
to otherwise have a deleterious effect on agricultural
and horticultural industries to econ~;cally
significant levels.
Particularly important viruses in this regard are
the DNA viruses, including the Caulimo- and the
Geminiviruses and two unclassified viruses, the
subterranean Clover Stunt Virus (SCSV, Shu and helms,
Virology, 1986, 167, 38-49) and Coconut Foliar Decay
Virus (CFDV, Rohde et al, Virology, 1990, 176,
648-51). The geminiviruses are a group of small
viruses which infect plant cells. They contain a
single stand of circular DNA referred to as "virion-
sense" or "positive-sense" DNA of less than about 2900
base pairs. Upon infection of a host cell by a
geminivirus, the viral coat protein is removed and a
double stranded replicative form of DNA is synthesised
comprising the virion-sense strand and a
"complementary-sense" strand. Transcription occurs
from both the virion-sense strand and from the
WOg5/0~4 2 1 6 7 7 0 1 PCT~31019~
"complementary sense" strand, giving rise to (~) and
(-) sense RNA transcripts respectively.
Examples of geminiviruses include Maize Streak
Virus (MSV), Bean Golden Mosaic Virus (BGMV), African
Cassava Mosaïc Virus (ACMV), Chloris Striate Mosaic
Virus (CSMV), Tomato Yellow Leaf Curl Virus (TYLCV),
Tomato Golden Mosaic Virus (TGMV), Beet Curly Top
Virus (BCTV) and Tomato Leaf Curl Virus (TLCV) amongst
many others (see also the viruses mentioned in table l
below). These viruses cause economically important
c~s in cultivated plants including leaf curling
and other leaf distortion. TLCV in particular causes
severe crop damage in northern parts of Australia and
infected plants show symptoms similar to those caused
by TYLCV in the Middle East and Europe. These tomato
viruses are transmitted by the white-fly Bemisia
tabaci and constitute a major disease problem world-
wide. For convenience, and unless otherwise specified,
TLCV and TYLCV are used interchangeably and reference
herein to Israeli and Sardinian isolates of TLCV
should be taken as TYLCV isolates. ~n general, "TLCV"
refers to the Australian isolate.
Geminiviruses replicate in the nucleus of the
infected cell and are thought to employ a rolling-
circle mech~ni~ similar to one used by the single-
stranded DNA cont~in;nq coliphages (e.g. QX1~4) and
certain StaPhylococcus aureus and Bacillus subtilis
plasmids. The other known plant viruses, including
other plant DNA viruses, replicate via RNA
intermediates.
Geminivirus particles accumulate in the nuclei of
infected cells where DNA replication and- virus
assembly probably take place tDavies et al., J. Cell.
Sci (Suppl) (1987) 7 : 95-107). Their putative
replicative forms are doublc ~Llanded covalently
- WO95/03~ 2 1 6 7 7 0 1 PCT~3/019~
closed circular DNA Ca. 2,7 Kb in chromatine-like
structures and are likely to be the transcriptionally
active forms of the virus (Abouzid et al., Mol. Gen.
Genet. tl988) 212 : 252-258). Based on host range and
insect vectors, they may be conveniently divided into
three suLyLOu~ : the viruses which infect
dicotyledonous hosts, have bipartite genomes and are
transmitted by white-fly vectors Bemisia tabaci
tGenn.) tACMV, African Cassava Mosaic Virus (Stanley
and Gay, Nature (1983) 301 : 260) ; TGMV, Tomato
Golden Mosaic Virus (Hamilton et al, EMBO J. (1984) 3
: 2197) ; BGMV, Bean Golden Mosaic Virus (Horwarth ~et
al, PNAS (1985) 82 : 3512) ; those which have
monopartite genomes infect graminaceous hosts, and are
transmitted by leafhopper vectors (NSV, Maize Streak
Virus (Nigeria strain, Mullinaux et al., EM80 J.
(1984) 3 : 3063 ; Kenyan strain, Havel NAR (1984) 12 :
7359) ; DSV, Digitarian Streak Virus (Donson et al.,
Virology (1987) 161 : 160) ; WDV Wheat Dwarf Virus
(Mac Dowell et al., EMBO J. (1985) 4 : 2173)) and
those which possess monopartite genomes and have
members which can be transmitted by leafhoppers (BCTV,
Beet Curly Top Virus (Stanley et al., EMBO J. (1986) 5
: 1761) ; ToYDV, Toh~c~o Yellow Dwarf Virus (Maris et
al., Virology (1992) 187 : 633)) or by whiteflies
(TYLCV, Tomato Yellow Leaf Curl Virus (Israeli
isolate, Navet et al., Virology (19gl) 185 : 151 ;
Sardinian isolate, Kheyr-Pour et al. NAT (1991) 19 :
6763), TLCV, Tomato Leaf Curl Virus (Australian
isolate, Dry et al., J. Gen. Virol. (1993) 74 : 147)).
Due to economic importance of plant DNA viNses
and, in particular, geminiviruses, there is a need for
resistance strategy to be developed.
One approach is to artificially ~ ess gene
expression by antisense RNA which targets
wOg5/03~4 2 1 6 7 7 0 1 PCT~P93/019~
complementary RNA (Helene, C. & Tolume, J. J. Biochim.
Biophys. Acta 1049:99-125, 1990 : Vander Kro et al
Biotechniques 6:958-976, 1988). Although anti~ence RNA
has been s~lcce~fully used in suppressing viral
infection in bacteria and animal tissue (Helene, C &
Tolume, J. J. Supra), its use in plants has met with
only limited success (Van Del Elzen, P. J. M. et al
Plant Mol. Biol. 13:337-346, 1989 ; Powell, P A et al
Proc. Nad. Acad. Sci. USA 86:6949-6952, 1989).
TnAe~, transgenic plants expressing Cllc~~rher
Mosaic Virus (CMY) antisense RNA to different regions
were generally not resistant to CMV infection, except
one line, which paradoxically produced only very low
levels of antisense RNA (Rezaian et al., Plant Mol.
Biol. (1988) 11- : 463). There was no proof that
antisense was the cause of this resistance. In potato,
anti~ nc~ to PVX coat protein RNA sequence in
transgenic plants protected against PVX only at low
levels of inoculum (Hemenway et al., EMB0 J. (}988`; 7
: 1273).
Two major problems appear to be the quantity of
antisense n~eAe~ to cope with a replicating system
that can rapidly produce large amounts of viral
nucleic acid, and viral RNA is largely in the
cytoplasm whereas ant;~ nce transcripts are produced
in the nucleus.
With respect to geminiviruses, antisense
technology has been applied to tobacco plants
expressing an antisense DNA sequence of virally
enso~ ALl gene of TGMV transcriptionally fused to a
drug resistance gene (Day, A. G. et al Proc. Nad.
Acad. Sci. USA 88:6721-6725, 1991). Although
transgenic plant lines expressing the antisense
transcript showed r ~ce~ symptom development, it is
possible that the transgenic plants were infertile
W095/~4 PCT~31019~
21 67701
which would prevent large scale production of virus
resistant plants.
In work leading up to the present invention, the
inventors sought to overcome the disadvantages of the
antis~nce approach to generating plant DNA virus-
resistant plants by employing the strategy of
ribozymes.
Ribozymes are synthetic RNA molecules which
possess highly specific endoribonuclease activity. In
particular, they comprise a hybridising region which
is complementary in nucleotide sequence to at least
part of a target RNA and a catalytic region which is
adapted to cleave the target RNA. Ribozymes are well
described by Haseloff J. and Gerlach L. Nature
334:~86-59l, 1988 and in International Patent
Application No. W089/05852.
Howewer, despite the existence of ribozyme
technology, heretofore, it has not been used on plant
DNA viruses and in particular plant single stranded
DNA viruses like geminiviruses. In fact, the s~lcc~cc
of the use of ribozymes on DNA viruses was
unpredictable in view of their extremely complex
replication and transcription mechAn; ~t~ and the lack
of information presently available ronc~rning the
compartmentalization of these different operations and
the precise level of intervention of the different
target seqt~ences in the virus life cycle. For example,
it is postulated that the replicase C1 intervenes in
the "rolling circle" replication step but not in the
prec~ing step of the formation of the double-stranded
replicative form of the virus (Elmer et al, Nucl.
Acid. Res., 1988, 16, p 7043-7060).
In accordance with the present invention, the
inventors have s~tcceccfully developed ribozymes
capable of targeting plant DNA viruses thereby
WOg5/03~ PCT~P93tOl9~
21 67701
providing an economically viable and efficient means
of cG"LLolling such viruses.
Accordingly, one aspect of the present invention
contemplates a ribozyme comprising a hybridising
region and a catalytic region wherein the hybridising
region is capable of hybridising to at least part of a
target RNA transcribed from a DNA molecule associated
with a DNA virus, wherein said catalytic region is
capable of cleaving said target mRNA sequence thereby
substantially reducing replication, infection and/or
assembly of said DNA virus.
More particulary, the present invention is
directed to a ribozyme comprising a hybridising region
and a catalytic region wherein the hybridising region
is capable of hybridising to at least part of a target
mRNA sequence transcribed from a virion-sense DNA
strand or a complementary-sense DNA strand of a plant
single stranded DNA virus, wherein said catalytic
region is capable of cleaving said target mRNA
sequence thereby substantially reducing replication,
infection and/or assembly of said DNA virus.
For convenience, reference to a plant single
stranded DNA virus, includes reference to its double
stranded replicative form.
In a preferred embodiment, the plant single
stranded DNA virus is a geminivirus and the target
mRNA en~oAe~ a protein essential for replication. By
"essential" is meant a protein specified by the viral
genome (or a complementary form thereof) and required
or substantially required for a particular function
such as replication, infection and/or viral assembly.
A decrease in the level of the essential protein
results in a reduction in the level of that function.
The decrease in the level of the particular function.
Preferably, the ~evel of essential protein (or level
WO95/0~4 2 1 6 7 7 0 1 PCT~3/019~
of particular function) is 20 % less, more preferably
at least 30 % less, even more preferably at least 45 ~
less, still more preferably at least 55 % less and
still even more preferably 65-7 5% less and yet more
preferably more than 85-90 % less relative to the
level of essential protein (or level of particular
function) in the absence of the ribozyme.
Conveniently, the level of essential protein (or
level of particular function) can be tested- in vitro
within the limits of biological variation in vivo.
Such a test, for example, can be by analysis of mRNA
degradation products in vitro.
According to one preferred aspect of the present
invention, there is provided a ribozyme comprising a
hybridising region and a catalytic region wherein the
hybridising region is capable of hybridising to at
least part of a target mRNA sequence transcri~ed from
a virion-sense DNA molecule of a geminivirus or a DNA
molecule complementary to said virion-sense DNA
molecule, said mRNA enco~;~g a protein essential for
replication of said geminivirus in a plant cell
wherein said catalytic region is capable of cleaving
said target mRNA sequence thereby substantially
reducing replication of said geminivirus.
In an even more preferred emho~iment~ the target
mRNA sequence is present in low ab~ln~ce within the
cell. An example of such a mRNA molecule is the
transcript of the Cl gene or its equivalent in
geminiviruses, such as in TLCV or TYLCV and its
mutants, derivatives and variants. The single stranded
DNA of TLCV contains six open r~;ng frames (ORFs),
two on the virion-sense strand and four on the
complementary-sense strand of the replicative form DNA
produced in leaf curl infected plants. Analysis of the
viral mRNAs in infected plant cells reveal that t~e
W095/03~4 2 1 6 7 7 01 ~CT~3/019~
mRNA ~n50~; ng the C1 gene, which is essential f~r
replication, occurs at lower ab~ nc~ than the other
mRNA transcripts specified by the TLCV genome.
Equivalent forms of the C1 gene include the genes L1,
AL1 and ACl and reference herein to the "Cl gene" is
taken as including eguivalents thereof.
Other targets include the C2, C3 and C4
transcripts which play roles in transactivation of
virion-sense gene expression and spread of the virus,
expression of the disease phenotype, and development
of the disease respectively.
According to another preferred e~ho~;ment, the
present invention is directed to a ribozyme comprising
a hybridising region and a catalytic region wherein
the hybridising region is capable of hybridising to at
least part of a target mRNA sequence enco~; ng the Cl
gene of geminivirus and in particular TLCV and wherein
said catalytic region is capable of cleaving said
target mRNA sequence thereby substantialy reducing
replication of said geminivirus (e, g, TLCV).
In a preferred variant, the ribozymes of t-~e
present invention can be defined as nucleic acid
compounds having endoribonuclease activity and being
capable of inhibiting the replication, infection or
assembly of a DNA virus, said compound comprising at
least one unit, "R", having the general formula :
5' (X-Y-X') 3'
in which :
- X and X' each represent, independently, a
nucleotide sequence having at least 4 bases and being
complementary to at least a part of either the viral
genome, .or the (-)sense RNA transcript of a DNA
virus ;
WO95/03W~ PCT~3/019~
21 67701
- Y represents the catalytic region of a
ribozyme.
According to this variant, the target RNA is
preferably the (-)sense RNA transcript, i.e. the mRNA
originating from the transcription of the
complementary sense strand of the double-stranded
replicative form of the DNA virus genome, particularly
a geminivirus. The sequences X and X' are therefore
complementary to at least a part of the (-)sense RNA
transcript (which is equivalent to being complementary
to at least a part of the single stranded genomic
DNA).
The sequences X and X', hereinafter referred to
as "hybridising arms" are sufficiently long to allow
both stable hybridisation of the ribozyme to the
substrate RNA, and efficient cleavage of the
substrate. Normally, X and X' will have a length of at
least 4 nucleotides each, and preferably at least 6.
The upper limit of the lengths of the hybridising arms
is generally determined by the length of the targetted
substrate and can be as long as 800 or lOoO bases or
more. Preferred lengths are between 6 and 300 bases,
for example 10 to 150. The complementarity of the
hybridising arm to the substrate or to a part of the
! substrate, normally extends along the entire length of
the hybridisisng a~m.
The catalytic region of the ribozyme, "Y" is
derived from any type of suitable ribozyme, for
example "hammer~ " or "hairpin". These ribozymes àre
described in patent applications EP-A-321201 and EP-
A-360257 respectively. Particularly preferred are
"hammerhead" catalytic regions (see for example,
figure 7), and which will be described in detail
hereafter.
WOgS/03~ 2 1 6 7 7 0 1 PCT~31019~
The ribozymes may be either "monoribozymes"
possessing one single "R" unit of the formula [X-Y-
X'~, and having only one catalytic region.
Alternatively, the ribozymes may be
"polyribozymes" comprising a plurality of "R" units
and having the general formula : R1-R2-----Rn, wherein
n ~ l, and may have a value as high as 50 or l00. In
practice, a value for "n" of about 5 to lO is
frequent. The polyribozymes are thus constituted by a
head-to-tail series of ribozymes, the monoribozyme "^R"
or ~X-Y-X'] unit, being the unit moti~. In other
words, it is a series of catalytic regions connected
together by hybridising arms. Hereinafter, unless
otherwise indicated, the ayy~e~dte of the X and X'
motifs contributed by all the "R" units of a
polyri~ozyme, will be designated collectively as the
ncomplementary" or "hybridising" sequence. The
polyribozyme normally acts as a "uni-molecule" against
a single transcript, i.e. the cleavage sites of each
of the catalytic regions are located on the same
transcript.
The precise structure of the polyribozyme is in
fact primarily determined by the nature of the
substrate it is designed to cleave. The number of
catalytic regions, their position in the complementary
sequence and the distance between two consecutive
catalytic regions are dependent to a large degree on
the position of the "three-base target" sites in the
substrate.
The "hammerhead" ribozymes cleave the substrate
immediately downstream from a "target" site XXX,
preferably XUX, in which X represents one of the 4
bases A, C, ~, U and U represents uracil. One
particularly preferred target sequence is XUY in
which X is any ribonucleotide, U is uracil and Y
W095/0K~4 PCT~31019~
2 1 67701
represents adenine, cytosine or uracil. XCUY forms
part of a base pair flanking region and Y is not base
paired. Preferred triplets defining X~OUY include, but
are not limited to GUC, GUA, GW, WC and CUC. Other
target sites are possible , but less efficient, for
example CAC, UAC and AAC. Perriman et al (Gene, 1992,
113:157) have studied ext~n~e~ target sites for
hammerhead ribozymes. It has been found that GUC, W C,
CUC, GUA and GW targets show equivalent rates of
studies. For GUG, the normal ribozyme cannot induce
cleavage, but an alteration of the stem-loop in the
catalytic domain leads to the formation of a weakly
active ribozyme. There are some targets with
nucleotides other than U in the center position which
show significant, discernable cleavage.
In the case of the ribozymes of the "hairpin"
type, a preferred target sequence is AGUC.
These target sequences are important in the
construction and functioning of the polyribozymes, not
only because they indicate the positions of cleavage
of the substrate but also because they define the
position at which the catalytic region must be
inserted in the complementary sequence. In fact, each
catalytic region of the polyribozyme must be situated
at a site in the complementary sequence which
corresponds to a XUX site of the transcript. For
example, if one XUX site is situated at position 412
of the C~ gene and another at position 711, a
catalytic region is inserted at the position
corresponding to 412 in the complementary sequence,
and another at 711. In other words, the X and X'
hybridising sequences which flank the catalytic domain
in each 'IRu unit, are chosen so as to be complementary
to the bases surrounding the XUX site targeted in the
substrate.
WOg5/0~4 2 1 6 7 7 0 1 PCT~3/019~
The motif XUX is a motif which occurs very
frequently in the RNA sequences. For example, on
average there is a GUC motif every 64 bases in a
sequence having a random and equal distribution of
bases. This signifies that the substrate usually
contains a plurality of XUX cleavage sites. As
indicated above, the catalytic regions of the
polyribozymes are situated at positions of the
complementary sequence which correspond to the XUX
sites. However, it is not necesc~ry to include a
catalytic region for each XUX target sequence of the
substrate in order to obtain an efficient cleavage
according to the invention. The total number of
catalytic regions included in the polyribozyme is
normally equal to or smaller than the total number of
XUX sites present in the transcript target. The
polyribozyme variant of the invention may thus contain
a very variable number of catalytic regions. For
example, in the case of C~ of TLCV, the polyribozyme
may contain from about 2 to about 6 catalytic regions,
when the target sequence is GUC. In the case in which
it is decided to include a smaller number of catalytic
regions in the complementary sequence than the number
of XUX sites in the substrate, the choice of the sites
selected may be made by respecting the following
criteria :
a) the distance between the 2 XUX sites targeted
and, conc~quently, between 2 catalytic regions in the
polyribozyme must be long enough to enable the
hybridizing arms of the polyribozyme situated between
the corresponding catalytic regions to hybridize wi$h
the substrate in a stable manner and to prevent the
catalytic regions hybridizing with themselves. A
distance of at least 8 bases, and preferably at least
WOg5/03~ PCT~3/019~
2 1 6770 1
14 bases, for example about 20 bases is particularly
advantageous. Of course, this criterion must only be
taken into consideration when the substrate
contains XUX sites very close together. Otherwise, if
the XUX sites of the substrate are separated from each
other by more than 8 to 20 bases, this selection
criterion is not important.
b) the XUX sites targeted are preferably situated
in a part of the substrate which does not have
significant secondary structure. This faciIitates the
access of the polyribozyme to the substrate and
increases its efficacy. Three-dimensional folding
analyses may be carried out.
c~ the XUX sites targeted may form part of the
regions of homology conserved ~etween different
strains or variants or mutants of one and the same
virus, or between different related viruses. This
aspect of the invention is considered in detail later.
d) another selection criterion of the XUX sites
targeted is the absence of homology with endogenous
genes of the plant to be transformed. In fac~,
although they are rare, some viruses rose~c~ se~lPnoes
which find a homo~ogy in the genome of plants. It is
thus important to avoid XUX sites situated within such
a se~uence.
In the ribozymes of the invention, the
hybridising X' and X motifs withi~ any one "R" unit
are often complementary to adjacent, contiguous
partions of the substrate. In this case, the ribozyme
can be thought of as "symetric", and the catalytic
domain acts at a unique site determined by the
complementarity of its two flanking arms. A preferred
WOgs/0~4 2 1 6 7 7 0 1 PCT~31019~
example of this symetric type of ribozyme is the
"purely complementary" polyribozyme in which the 3'
hybridising arm of a given "R" unit and the S'
hybridising arm of the adjacent "R" unit are
complementary to sequences which are contiguous in the
(-~sense RNA transcript or in the DNA genome.
In other words, in this embodiment, the a~y-eyate
of the hybridising arms gives a sequence of
uninterrupted complementarity to the target sequence.
This type of polyribozyme may be considered as a
complementary sequence having, inserted at distinct
sites in that sequence, a plurality of ribozyme
catalytic regions. The complementary sequence may
hybridise with the entire length of the target
transcript. On the other hand, it may hybridise with
only a fragment of the targeted transcript. The
fragment in question must be long enough to allow the
inclusion of at least 2 catalytic regions in the
~G~L~l,o,~ing sequence of the polyribozyme. In
general, for this variant, the length of the
complementary sequence, not counting the catalytic
regions ~the sum of the hybridising arms), may vary
from about 40 to 2000 bases. Lengths of 400 to lO00 is
particularly preferred.
According to a variant of the invention, rather
than being "symetrical" as described above, the
ribozyme may be, or may possess at least one "R" unit
which is "asymetrical". This signifies that the X and
X' hybridising arms within a given "R" unit are
complemèntary to two se~encec which are non-
contiguous in the substrate. This in fact means that
the catalytic domain of the "asymetric" nR" unit can
act at two distinct XUX sites in the substrate, one
being the XUX site flanked by the sequence
complementary to its 5' arm, and the other being the
WO9S/03~4 PCT~3/019~
21 677QI
XUX site flanked by the sequence complementary to its
3' arm. This type of ribozyme may be specifically
constructed, or may be the result of a spontaneous
re-arrangement during synthesis. The locations of the
sequence to which, on the one hand, the 3' flanking
arm hybridises, and the sequence to which, on the
other hand, the 5' flan~ing arm hybridises, may be
separated by hundreds of bases or by as little as 10
or 20 bases. This type of structure is advantageous
since it allows a significant "economy" in c~talytic
regions : for example five ~asymetriCn nRII units can
achieve the same result as 10 symetric units.
According to another highly preferred embodiment
of the invention, in the polyribozyme having the
formula :
R1 ~R2~R3~~~~~Rn ~
the 3' hybridising arm (X') of a given "R" unit and
the 5' hybridising arm of the adjacent "R" unit are
complementary to non-contiguous portions of the genome
or of the (-~sense RNA transcript. This type of
polyribozyme will be referred to hereinafter as a
"short arm polyribozyme".
The structure of these polyribozymes is basically
a sequence complementary to the target sequence in
which ribozyme catalytic regions are embedded and
which has undergone deletions in the complementary
sequence. The distance between two catalytic regions
in the polyribozyme is shorter than the distance
between the two correspon~;~g XUX sites in the target
sequence. The distance between ribozymes can be
re~llce~ significantly with respect to the distance
between the coLlea~G..~;ng target sites in the
substrate, for example, if two adjacent targeted XUX
WOgS/03~ PCT~ ~3/019~
21 67701
16
sites in the substrate are separated by a distance of
1000 nucleotides, the distance between the catalytic
regions of the !'short arm ribozyme" may be as little
as 30 or 40 nucleotides. Reductions of over 90 % of
the length of the hybridising sequence may therefore
be made. The minimum length between two adjacent
catalytic regions is normally about 8 nucleotides and
preferably at least 20. The present inventors have
shown that such shortened hybridising se~ncPs do
not, surprisingly, pr~ve"t the polyribozyme from
efficiently cleaYing the substrate and can, in fact,
in many cases improve the efficiency of the
inactivation.
For example, the reduction in the length of the
complementary sequence avoids problems associated with
the formation of ceco~Ary structure leA~ing to ~he
inactivation of the ribozyme. Thus, for a given length
of ribozyme molecule, more catalytic regions may be
inserted targeting more cleavage sites in the
substrate.
Furthermore, the use of the short arm
polyribozyme means that efficient polyribozymes may be
constructed even if only parts of the sequence of the
substrate are known. It is sufficient to know the 10
or 15 bases surrounding an XUX target site.
Another important advantage is the possibility to
use, as the shortened hybridising arms, exclusively
those se~nces which are conserved in different
viruses, for example viruses of the same type. In this
way, different viruses may be inactivated by the same
polyribozyme even if the se~)ences between the
conserved regions are totally dissimilar.
Final~y the smaller molecule allows an easier and
more economical synthesis. The functional nature of
the "short arm" ribozymes of the invention prove that,
W095/0~ 2 1 6 7 7 0 1 PCT~3/019~
in a polyribozyme, the different catalytic regions can
act in~Pp~ndently of each other.
Any one polyribozyme of the invention may include
a mixture of "long arm" and "short arm" ribozymes, and
also of "symetric" and asymetric" ribozymes.
Alternatively, it may include only long arm ribozymes,
or only short arm, or only asymetric, etc... The
structure of the ribozyme can therefore be optimised,
and tested.
According to yet a further embodiment of the
invention, the hybridising arms of the polyribozyme
may be derived from at least two different types of
virus. Such polyribozymes are referred to hereinafter
as "chimeric polyribozymes". As an example of this
type of construction, mixtures of TLCV and TYLCV
derived sequences can be cited, for example : the
Sar~ini~n, Israeli or Tunisian strain. This type of
polyribozyme ensures the inactivation of both types of
virus, if present together, or allows the use of a
single construction in different geographical areas.
According to a further variant of the invention,
the polyribozymes may include, between two consecutive
"R" units, a non-complementary sequence (A) which has
a length of at least 4 bases, and which is non-
complementary to the (-)sense RNA transcript or DNA
genome.
The non-complementary sequence of the
polyribozyme may have a precise function, for example,
it may be constituted by a coding sequence which can
be used to select transformants or also a sequence
cont~;ning a ribozyme which acts on a substrate other
than the primary target of the DNA ~irus or which is
cis acting on a part of the polyribozyme. It typically
contains a polylinker for cloning, or an adaptor to
facilitate construction of chimeric polyribozyme. The
W095/03~ PCT~P93/019~
21 67701
non-complementary seguence usually has a length
comprised between 4 and 500 bases, for example 20 to
100 bases. When there is a plurality of non-
complementary se~lPn~es, they may together constitute
as much as about 90% of the length of the
polyribozyme, for example 50%. These non-complementary
sequences normally replace portions of the
complementary se~nceC X and X', but leave at least 4
complementary bases immediately next to the catalytic
domain.
According to a most preferred embodiment, the
ribozymes of the invention are constructed so that t;~e
hybridising arms are complementary to a region or to
regions which is (are) highly conserved between
variants or mutants of the same strain, between
strains of the same virus type or even between viruses
of different types.
Comparisons of ~llc~ amino-acid se~l~ncDs
~o~e~ by a different open-reading frames of
geminiviruses, that is V1, v2, Cl, C2, C3 and C4, has
been made (see for example references cited in table
1). For the Cl replicase polypeptide, the highest
homology occurs between TLCV and TYLCV-I with a
homology of 83 %. Between TLCV and TYLCV-S there is
79 % amino-acid identity over the Cl open-reading
frame.
If this comparison is made for the Israeli,
Sar~;~iAn and Tunisian isolates of TYLCV against TLCV,
similar homologies are found (see table 1).
WOg5/03~4 2 1 6 7 7 01 PCT~ ~31019~
~ABLE 1 : PAIRWISE COMP~T8Q~ OF v~u~v AMINO-ACID
8EQUENCES ~:rL~r~ BY Cl ORF OF ~MI ~ ~ VlKU~ :
Virus Amino-acid identity
(%)
1 : TYLCV-S/TYLCV-I 77
2 : TYLCV-S/TYLCV-T 75
3 : TYLCV-S/ACMVA 74 .
4 : TYLCV-S/ICMVA 76
5 : TYLCV-S/ABMVA 67
6 : TYLCV-S/TGMVA 67
7 : TYLCV-S/BGMVA 63
8 : TYLCV-S/SqLCV 53
9 : TYLCV-I/TYLCV-T 78
10 : TYLCV-I/ACMVA 73
11 : TYLCV-I/ICMVA 75
12 : TYLCV-T/ACMVA 72
13 : TYLCV-T/ICMVA 77
14 : AC~VA/ICMVA 74
15 : TLCV/TYLCV-I 83
16 : TLCV/TYLCV- Q 79
17 : TLCV/BGMVA 64
18 : TLCV/PYMVA 68
19 : TLCV/ABMVA 68
20 : TLCV/TGMVA 69
21 : TLCV/ACMVA 78
22 : TLCV/BCTV 61
23 : TLCV/TobY~V 25
24 : TLCV/MSV 25
25 : TLCV/WDV 24
Leqend for Table 1 :
For 1 to 14, amino-acid identity is given in
percent derived with the program BestFit of the
UWGCG-Sequence analysis package. The word size (K-
tuple) was 2, the gap weight was 2.0 and gaplength
weight was 0.1. The program FastA yielded the same
relationship.
For 15 to 25, amino-acid identity was calculated
using PALIGN program of the PC/GENE sequence analysis
package using the stru~ enetic comparison matrix
.(.open gap cost value, 10 ; unit gap cost value, 10).
ABMVA : Abutilon Nosaic Virus DNA A : ACMVA :
African Cassava Mosaïc Virus DNA A ; BGMVA : Bean
Golden Mosaïc Virus DNA A ; ICMV : Tn~ Cassa~a
W095l03~ 2 1 6 7 7 0 1 PCT~ ~3/019~
Mosaïc Virus DNA A ; SqlCV : Squash Leaf Curl Virus
DNA A ; PYMVA : Potato Yellow Mosaïc Virus DNA A ;
TGMVA : Tomato Golden Mosaïc Virus DNA A ; TYLCV-I :
Tomato Yellow Leaf Curl Virus Israeli isolate ;
TYLCV-S : Tomato Yellow Leaf Curl Virus Thai isolate ;
TLCV : Tomato Leaf Curl Virus ; BCTV : Beet Curly Top
Virus ; TobYDV : Tobacco Yellow ~warf Virus ; MSV :
Maize Streak Virus : WDV : Wheat Dwarf Virus.
The results su arized in the table have been
given by DrY et al (J. Gen. Virol. ~1993) 74:147) and
Kheyr-pour et al. (Nucl. Acid. Res. (1991) 19:6763).
The degree of sequence d~versity between the 3
isolates of TYLCV and TLCV is in pronounced constr~-t
to the usual very high sequence similarity between
different geminivirus isolates or strains, for
example : the DNA sequences of the Kenyan and Nigerian
isolates of ACMV show 96 ~ identity and Kenyan,
Nigerian and South African isolates of NSV vary
maximally by about 2 %. In spite of the pronounced
seguence diversity between the 3 isolates of TYLCV and
TLCV, the present inventors carried out a comparison
of the Cl RNA transcript and identified three very
highly conserved regions, two of which are shown in
figure 9. These regions are the following (the
following numerotation is the TLCV numerotation of the
complementary sense transcript) :
1. 365 to 435 a~Lo~imately
2. 634 to 737
3. 895 to 972 n
The nucleotide se~P~c~C of the Cl gene in three
TLCV isolates (from Australia, Israel and Sardinia,
respectively) are shown in Figure 1. The complete
nucleotide sequence of the Cl gene from TLCV is
disclosed in Mullineaux et al., Virology, 193 :
414-423, 1993 and Dry et al, J. Gen. Virol., 74 :
147-151, 1993. The Israeli isolate is disclosed by
Navot et al in Virology, 185 : 151-161, 1991 and the
WOg~/03~ 2 1 6 7 7 0 1 PCT/~1,3/019~
Sardinian isolate is disclosed by Rhyer-Pour et al,
Nucl. Acid. Res. 19 : 6763-6769, 1991.
Polyribozymes constructed with target sites in
these conserved regions may therefore be used to
ensure that all related strains or variants are
inactivated. As pointed out above, if the polyribozyme
is the deleted short arm version, whose hybridising
arms correspond exclusively to the conserved regions,
wide sequence diversity between the conserved regions
does not affect the efficient hybridisation of the
polyribozyme to the substrate.
According to this aspect of the invention, a
"conserved region" is considered as having a 100 %
homology over a distance of about 20 bases. Generally
speaking,.the homology may be around so % over 30
bases. Ribozymes Rl, R4, RS and R2 are designed so
that the target sites lie within these conserved
regions, as shown in the examples below.
According to this embodiment, therefore, the
present invention further relates to a ribozyme
comprising a hybridising region and a catalytic region
wherein the hybridising region is capable of
hybridising to at least one part of a target mRNA
sequence encoding the Cl gene of at least two variants
of particular geminivirus and in particular at least
two variants of TLCV, and wherein said catalytic
region is capable of cleaving said target mRNA
sequence from each of said variants thereby
susbtantially reducing replication of said geminivirus
(e.g. TLCV, TYLC and their mutants, derivatives and
variants).
The polyribozymes of the invention are usually
constituted -of RNA. Nonetheless, it is possible to
replace some parts of the polyribozyme by DNA, for
example the hybridizing arms or parts of these arms,
rB
WO95/03~ 2 1 6 7 7 0 1 PCT~P93/019~
or also a part of the catalytic region, in particular
the "loopl', provided that the catalytic activity is
maintained (see, for example, the substitution of the
RNA by DNA described in the international patent
application WO-9119789).
As far as the catalytic region is concerned, the
ribozymes of the present invention can be represented
as a molecule of formula (I) : .
3~ 5'
(X) n Y Y (X) n~
A C
A U
G lA G
C * G G A
X
y- yl I
5l 3' (I)
wherein, (X)n and (X)n' represent ribonucleotide
sequences substantially complementary to a mRNA
sequence of a DNA virus; the sum of n and n' may be
the same or different; an (*) ~e~ ents a base pair
between complementary ribonucleotides ; Y represents
any ribonucleotide and each Y may be the same or
different ; Y' and Y" represent ribonucleotides of
complementary sequence along at least part of their
length to allow base pairing between the
oligoribonucleotides, or Y' and Y" together form a
single RNA sequence wherein at least part of said
sequence comprises a stem formed by base pairing
between complementary nucleotides; and optionally, an
additional nucleotide selected from any one of A, G, C
or U is inserted after lA.
In another embodiment, the ribozyme is as set
forth in formula (II) :
.
W095/03~ 2 1 6 7 7 0 I PCT~3/019~
3' ~'
(X) nA Y (X) n'
A C
A U
G 1A G
C * G G A
Y * Y Y
Y * Y
(Y) m * (Y) m~
B (II)
wherein (X) n and (X) n ~ represent ribonucleotide
sequences substantially complementary to a mRNA of a
DNA virus and ~n particular a geminivirus such as
TLCV ; Y represents any ribonucleotide and each Y
residue may be the same or different ; an (*)
represents a base pair between complementary
ribonucleotides; n and n' are as previously defined, B
represents a bond, a base pair, a ribonucleotide, or
an oligoribonucleotide containing at least 2
ribonucleotides ; m and m' are l or more and may be
the same or different ; and optionally, an additional
nucleotide selected from any one of A, G, C or U is
inserted after ~A.
In a preferred embodiment, the ribozyme is as set
forth in formula (III) :
5'
(X) nA Y (X) n~
A C
A U
G A G
C * G G A
A * U U
G* C
G* C
A G
G U . (III)
wherein (X) n and (X) n I represent a ribonucleotide
sequence substantially complementary to a mRNA of a
DNA virus and in particular a geminivirus such a
,,
W095/03~4 2 1 6 7 7 01 ~CT~ ~3/019~
TLCV ; Y represents any ribonucleotide and each Y
residue may be the same or different an (*) represents
a base pair between complementary ribonucleotides ; n
and n' are as previously defined.
Preferably, the DNA virus is a single stranded
DNA virus. Preferably, the single stranded ~NA virus
is a geminivirus. Most preferably, the geminivirus is
TLCV and (X) n and (X) n~ represent a sequence capable of
hybridising to a region of Cl gene mRNA.
In a most preferred embodiment, (X) n and (X) n~
hybridise to the Cl gene mRNA transcript of TLCV or
TYLCV as set forth in Figure 1.
Particulary preferred ribozymes of the present
invention are specifie to the Cl gene of TLCV and its
variants and comprise the nucleotide sequences set
forth below :
3'- GUCCAGUCGUGUAAAAAG~AGGAGUGCCUGAGUAGUCGUAGGCUU
GUAAGUC-5' ;
3'- GAGG~A~AAAUA~,AAAAGCAGGAGUGCCUGAGUAGUCA~.AAA~C
UAGCUCA-5' ;
3'- GAG W GUAAACUACAAAGCAGGAGUGCCUGAGUAGUCUAACUUCU
AGUACUA-5'.
and
5'- GATTTAGCTCC~~ ATGAGTCCGTGAGGACGAAAATGTTCGGATG
GAAATGTGCTCT~AT&AGTCCGTGAGGACGAAACCTGGTTGGGGAT-3'
Although the catalytic regions illustrated above
and in Figure 7 have a conserved structure and
sequence, it has been observed that some nucleotides
~ay be deleted, inserted, substituted or modified
without prejudice to the activity of the ribozyme. The
invention comprises the use of these modified
catalytic regions in the polyribozyme provided that
their catalytic activity is conserved. This activity
can be verified by using the tests described below. ~
- WO95/03~4 PCT~3/019~
21 67701
For example, one or more nucleotides of the
catalytic region II illustrated in formula I, II and
III may be replaced by nucleotides containing bases
such as adenine, guanine, cytosine, methylcytosine,
uracil, thymine, xanthine, hypoxanthine, inosine or
other methylated bases. The "conserved" bases C-G
which together form the first base pair of the
catalytic loop, can be replaced by U-A (Koizumi et
al., FEBS Letts. 228, 2, 228-230, 1988).
The nucleotides of the catalytic region
illustrated in Figure 7 can also be modified
chemically. $he nucleotides are composed of a base, a
sugar and a monophosphate group. Each of these groups
can thus be modified. Such modifications are describ~d
in "Principles of Nucleic Acid Structure" (Ed. Wolfram
Sanger, Springer Verlag, New York, 1984). For example,
the bases may bear substituents such as halogeno,
hydroxy, amino, alkyl, azidol nitro, phenyl groups,
etc.. The sugar moiety of the nucleotide may also be
subjected to modifications such as the replacement of
the secondary hydroxyl groups by halogeno, amino or
azido groups or even to 2' methylation.
The phosphate group of the nucleotides may be
modified by the replacement of an oxygen by N, S or C,
giving rise to a phosphoramidate, phosphorothioate and
phosphonatel respectively. These latter may exhibit
useful pharmacokinetic properties.
The bases and/or the nucleotides of the catalytic
region may also bear substituents such as amino acids,
for example, tyrosine or histidine.
- According to a variant of the invention, the
-ribozyme may comprise as catalytic region one or more
structures such as those illustrated in Figure 17.
This structure, cal}ed "minizyme", is described in the
international patent application WO-A-9119789. It
WO95/0~4 2 1 6 7 7 0 1 PCT~ ~3/019~
26
represents a catalytic region of the "hammerhead"
type, the "loop" of which has been replaced by a "P"
group. P may be a covalent link between G and 1A, one
or more nucleotides ~RNA or DNA, or a mixture, or even
derivatives described above) or any atom or group of
atoms other than a nucleotide which does not- affect
the catalytic activity. When P represents a plurality
of nucleotides, it may contain internal base pairings.
The sequence and the number of nucleotides
constituting the group "P" is not critical and may
vary from l to 20 nucleotides for example, a~d
preferably ~rom 1 to 6. It is preferable to select a
sequence lacking internal base pairings of the
Watson-Crick type.
The catalytic activity of the ri~ozymes of the
invention may be verified in vitro by placing the
ribozyme, or a seguence which after transcription will
give rise to the ribozyme, in contact with the
substrate, followed by demonstration of the cleavage.
The experimental conditions for the in vitro cleavage
reaction are the following : a temperature comprised
between 4 and 60~C, and preferably between 20 and
5SC, a pH comprised between about 7.0 and 9.0, in
the pr~e~c-e of divalent metals, such as Mg2', at a
concen~ration of 1 to 100 mM ~preferably 1 to 20 mM).
The polyribozyme is usually present in an equimolar
ratio with the substrate, or in excess. The in vitro
cleavage reactions are advantageously carried out
according to the procedure described by Lamb and Hay
(J. Gen. Virol., l990, 71, 2257-2264). This article
also describes suitable conditions for in vitro
transcription for the production of ribozymes from
oligodeoxyribonucleotides inserted into plasmids.
The in vivo cleavage conditions are those
existing naturally in the c~ll.
. . .
W0951OE~n4 2 1 6 7 7 0 1 PCT~P93/019~
In accordance with the emboA;~?nts and aspects of
the invention as set forth above, the ribozyme is said
to "hybridise" to a target mRNA sequence or to have
"complementarity" hybridising arms. Such hybridisation
and complementarity is definied herein as an extent of
duplex formation sufficient to form a stable enough
structure so that cleavage with the target triplet can
occur. Depending on the target sequence, the ribozyme
and the secondary structure of the mRNA, preferably
less than about 5-10, more preferably less than about
3 to 5 and even more preferably less than 1 to 3 bases
would be mismatched between the target mRNA sequence
and one or both hybridising arms of the ribozyme. Over
the whole length of the hy~ridising arm, not more than
10 % of mismatches should occur. Accordingly, the
present invention extends to ribozymes described
herein and in particular Rzl, Rz2, Rz3, Rz4 and Rz5,
to the polyribozymes contA;n;ng them, and to any or
all functional mutants, deri~atives, parts, homologues
or analogues thereof. Examples of such mutants include
single or multiple nucleotide substitutions, deletions
and/or additions to the hybridising regions and/or
catalytic regions of the ribozymes provided such
modified rihozymes are still functional in cleaving
the target ~RNA sequence.
Put in alternative terms, the present invention
extends to a ri~ozyme having a hybridising domain and
a catalytic domain, wherein the hybridising domain
comprises a sequence of nucleotides substantially
complementary to a sequence of nucleotides in a mRNA
molecule encoding the Cl gene of TLCV or TYLCV or
which hy~ridises to said TLCV or TYLCV mRNA sequence
in vitro under high stringency conditions and wherein
said catalytic region is capa~le of cleaving said
target mRNA sequence. This aspect of the present
WOg5/03~ 2 1 6 7 7 0 1 PCT~3/019~
28 -
invention is conveniently determined by the extent of
cleavage of the target mRNA se~uence in vitro.
For the purposes of the present invention, the
stringency conditions set forth in Sambrook et al,
Molecular Cloning : A laboratory Manual, Cold Spring
Harbor Laboratories, Cold Spring Harbor, NY, USA,
pages 387-389 at paragraph 11 are considered high
stringent conditions. Alternatively, medium stringency
may be used involving 0.25-0.5 % w/v SDS at 45 C for
2-3 hours. The alternative conditions are applicable
depending on concentration, purity and source of
nucleic acid molecules.
The present invention extends to genetic
constructs comprising the ribozymes herein described.
These genetic constructs generally comprise a vector
sequence as a vehicle for transferring the ribozyme to
a host cell and a promoter sequence to direct
transcription of the ribozyme and a terminator. A
preferred promoter includes but is not limited to a
35S Cauliflower Mosaïc Virus tCaMV) or to a double 3~S
CaMV promoter. The CaMV promoter is described by Odell
et al, Nature 313 : 810-812, 1985. The ribozymes are
generally in the complementary form such that upon
transcription, the ribozyme is produced. The yenetic
constructs of the present invention are used to
generate transgenic plants capable of constitutively,
developmentally or inducibly (e.g. in response to
certain stimuli) expressing the ribozymes herein
described. Most preferred transgenic cells are
rendered sukstantially resistant to TLCV infection.
The ribozymes and genetic constructs of the
present invention can be conveniently tested in vivo
using virus-sensitive plants. A particularly useful
indicator plant is the tobacco plant. The analysis may
be qualitative such as by screening for severity of
WOgS/03~ 2 1 6 7 7 0 1 PCT~3tOl9~
symptoms or quantitative where samples are analysed,
for example, by measuring the level ribozyme
expression or level of target mRNA. A combination of
qualitative cr quantitative analyses may also be
employed.
All of the known means for introducing forei~n
DNA into plants may be used, for example
Aqrobacterium, electroporation, protoplast fusion,
bombardment with a particle gun, or penetration of DNA
into cells such as pollen, the microspore, the seed
and the immature embryo, viral vectors such as the
Geminiviruses or the satellite viruses. Aqrobacterium
tumefaciens and rhizoqenes constitute the preferred
means. In this case, the sequence ~oAing for the
polyribozyme is i.-L~Gd~ced into a suitable vector
together with all of the regulatory sequenc~s
necessary such as promoters, terminators, etc....as
well as any sequence nec~c~ry for selecting the
transformants.
In one approach, tobacco cells are transformed
using standard procedures employing Aqrobacterium with
the genetic construct to be tested. Generally,
Aqrobacterium tumefaciens mediated transformation of
tobacco is carried out essentially as described by
Horsch et al., Science 227 : 1229-1231, 1985, although
conveniently strain LBA 4404 is used and the plantlets
are selected on Kanamycin at a concentration of
250 ~l/ml. The transformed cells are regenerated into
small plants and samples remo~ed to screen for
expression. The plants are allowed to self-fertilise
and to seed and the transgenic seeds are then used to
generate new plants. The transgenic plants along with
suitable controls are then exposed to the virus and
the degree of symptom development analysed.
Alternatively or in addition to, a quantitative
,
WOg5/03~4 2 1 6 7 7 01 PCT~31019
3~
assessment is made by biochemical analysis. This
approach is particularly useful for screening for
ribozymes against geminiviruses and in particular
TLCV. Once suitable ribozyme genetic constructs are
identified using this procedure, these particular
construct can then be used to generate, for example,
in the case of TLCV and TYLCV, transgenic tomato
plants. Similar transformation and screening
procedures are employed.
The resistant nature o~ the transgenic plants can
be tested by carrying out a self-fertilisation, or a
cross with a non transformPd genotype, on a prima~y
descendant to obtain Tl. Subsequently, Tl plants are
inoculated with the DNA virus by Agroinoculation.
Degree o~ resistance (i.e. "complete resistance"
meaning total absence of symptoms, "tolerance" meaning
that the plant can be infected and shows symptoms but
recovers, "sensitive" meaning that the plant exhibits
symptoms and replicates the virus) is then noted.
The present invention is particularly directed to
transgenic tomato plants which exhibit at least some
resistance to TLCY and TYLCV infection. By resistance
to infection is meant a gualitative reduction in
symptoms ~generally due to inhibition of virus
replication, by more than approximately 30 %,
preferably more than approximately 50 % and even more
than approximately 70 %. ~ue to the variability of
biological systems, 100 % inhibition of TLCV
replication is difficult to achieve although the
effectiveness of the ribozyme can be ~h~n~ ,?,,~y
various molecular biological techniques.
According to this aspect of the present
invention~ there is provided a method of generating
transgenic tomato plants exhibiting reduced symptoms
following exposure to TLCV comp~red to non-transgenic
WO95/03~4 PCT~31019~
21 67701
plants, said method comprising transforming tomato
plant cells with a genetic construct encoding a
ribozyme as hereinbefore described capable of cleaving
C1 mRNA from TLCV, regenerating plants from said
transformed cells, self-fertilising said regenerated
plants and obt~i n; ng seeds there from and then growing
transgenic plants from said seeds.
The present invention extends to transgenic
plants exhibiting resistance to a plant DNA virus,
more particularly a geminivirus and even more
particularly TLCV. In one preferred embodiment, the
transgenic plants are tomato plants. The subject
invention also extends to reproductive material and
seeds from such transgenic plants (e.g. tomato
plants). The ribozymes may be expressed
constitutively, developmentally or inducibly.
The present invention is further described by
reference to the following non-limiting figures and
examples.
In the f~gures :
- Figure 1 is a representation of the nucleotide
sequences of the complementary-sense DNA strands from
three different isolates of TLCV : top row, TLCV
(Australia) ; middle row, TLCV (Israel) ; bottom row
TLCV (Sardinia~. The sequence is represented in a
5'->3' direction.
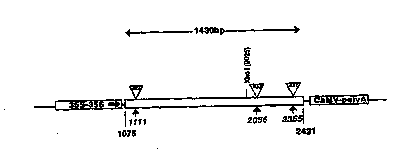
- Figure 2 is a schematic representation of
pRIB05BIN comprising a CaMV35S-1568bp TLCV fragment
(containing Rzl, Rz2 and Rz3)-CaMVpolyA expression
cassette cloned into BamHI cut Binl9. The insertion
sites of the 3 ribozymes Rzl, Rz2 and Rz3 are
indicated at positions 1111, 2056 and 2355. This
numbering is the vir on-sense numbering and
corresponds to positions 1654, 711 and 412,
WO95/03~4 2 1 6 7 7 01 PCT~3/019~
respectively, using the (-)sense numbering as
illustrated in figures 8 and 10.
- Figure 3 is a schematic representation of
pRIB05BINDel comprising a CaMV35S-1346bp TMCV fragment
(containing Rzl, Rz2 and Rz3)-CaMVpolyA expression
cassette c~oned into Bam~I cut Binl9.
- Figure 4 is a schematic representation 3f
pRIB07BIN comprising a CaMV35S-396bp TLCV fragment
(containing Rzl and Rz2)-CaMVpolyA expression cassette
cloned into BamHI cut Binl9.
- Figure 5 is a schematic representation of
pRIBO2BIN comprising a CaV35S-30bp TLCV fragment
(containing ~zl)-CaMVpolyA expression cassette cloned
into BamHI cut Binl9.
- Figure 6 is a schematic representation of the
location of primers used for RT-PCR analysis of
ribozyme construct expression in transgenic tobacco.
- Fig~re 7 is a schematic representation of a
ribozyme model (from Haselhoff and Gerlach, Nature,
1988). Substrate RNA can have any sequence (X)
flanking GUC at the cleavage site, as long as base
pairing forms with the ribozyme as shown. The arrow
indicates the cleavage site. Conserved bases in
ribozymes are boxed. In naturally occuring
h~merheads, the substrate and ribozyme portions are
connected by a loop, resulting in an intramolecular
cleavage.
- Figure 8 shows the nucleotide sequence of the
complement2ry-sense strand of TYLCV-S. Lower case
letters indicate the ribozyme cleavage site. The
sequence of TYLCV-S was published by Kher-Pour et al.
(Nucl. Acid Res. (1991) 19 : 6763)
- Figure 9 shows the regions conserved in the
Replicase gene, between TLCV and TYLCV-S : Figure 9(a)
shows the regions highly conserved around ribozymes
WOg5/0~4 2 1 6 7 7 0 1 PCT~ ~3/019~
Rl, R4 and R5 ; Figure 9(b) shows the regions highly
conserved ~round ribozyme R2. ":" means a mismatch, a
gap means ro nucleotide. Lower case letters indicate
the ribozyme cleavage site.
- Figure 10 illustrates the nucleotide sequence
of the complementary strand of TLCV. Dry et al. (J.
Gen. Virol. (1993) 74 : 147) Lower case letters
indicate the ribozyme cleavage site.
- Figure 11 shows the ~arget sequences for
polyribozymes D, F and E. Lower case letters indicate
the ribozyme cleavage site. Upper case letters
correspond to the complementary sequences of ribozyme
arms.
- Figure 12 is a schematic representation of
ribozyme constructs. A hatched line represents the
TYLCV-S complementary-sense strand, and a solid- line
repre~ents TLCV complementary sense strand. R
ribozyme.
- Figure 13 is a schematic physical map of binary
vector pGA2E.
- Figure 14 is a schematic physical map of
riboz~me plasmids, p~AZE-RzA, pGAZE-polyRzC, pGAZ~-
polyRzD, p~AZE-polyRzE, pGAZE-polyRzF.
- Figure 15(a) is a schematic representation of
pRIBO5BS : 1690bp TLCV Cl/C2/C3 complementary fragment
(containing 3 ribozymes) blunted into EcoRV cut
Bluesrript SK+ : ~igure 15(b) is a schematic
representation of pRIB05BIN : 2660bp Bgl II 35S-
C1/C2/C3 complementary (cont~;n;n~ 3 ribozymes)
CaMVpolyA cassette cloned into BamHI cut BIN 19; open
triangles show the position of 22bp ribozyme
insertions, E : EcoRI ; H : HindIII ; K : KpnI ; P :
PstI ; Sm : SmaI ; B : BamI : Sl : SalI : St : SstI :
X : XhoI ~ Xb : XbaI : C : ClaI.
WOg5/03~4 2 1 6 7 7 01 PCT~3/019
34
- Figure 16 is a schematic representation of the
ribozyme constructs, showing their inter-relationship
and the deletions carried out to obtain the "Short-
arm" ribozymes, polyE, polyF and polyD.
- Fig~re 17 illustrates the "Minizyme" structure
(WO-A-9119789) wherein the loop of the catalytic
region is replaced by an element "P". P may be at
least 1 nucleotide (ribonucleotides, deoxyribo-
nucleotides, derivatives or a mixture), provided that
the ribonucleotides of the "P" group are not bc~e
paired by "Watson -Crick" base pairings when the
sequenCeS ~X I ) n and (X) n~ and P are constituted
excl~sively of ribonucleotides. "P" may also be a bond
or any atom or group of atoms which do not affect the
catalytic activity of the ribozyme. X has the same
meaning as in formula I, II, III.
.
EX~MPI.ES
EXAMPI.E I - SELECTION OF TARGET mRNA OF TLCV:
To enable selection of a suitable target RNA, the
major mR~T~ transcripts were identified and mapped on
the circular genome of an Australian isolate of TLCV
~TLCV (Australia~, Mullineaux et al., Supra ; Dry et
al, Supra~ One of the viral mRNAs derived from the
complementary sense strand of the viral DNA occurred
at a lower abundance than the other TLCV transcripts.
This low ablln~c~ mRNA transcript was identified as
encoding the C1 gene, the product of which, -is
essential for replication of the virus. [Davies et al,
Trends in ~enetics, 5 : 77-81, 1989). The mRNA
trans¢ript also encodes three other gene products,
designated C2, C3 and C4, whose functions are not
W095/03~4 2 1 6 7 7 0 1 PCT~3/019~
conclusively known. Accordingly, the low abundance
mRNA encoding the Cl gene was selected as the target
mRNA. The ~ow abundance cf this transcript means a
higher ratio of ribozyme target sequence, thereby
potentially permitting a more substantial decrease in
Cl gene product.
EXAMP~E II - SELECTION OF RIBOZYMES BASED ON CONS~Kv~
REGIONS OF C1 GENE B~lw~ TLCV ISOLATES :
The nucleotide sequences of the complementary
sense strands of three TLCV isolates [TLVC
(Australia), TLCV (Israel) and TLVC (Sardinia)] were
co~r~red in order to identify conserved regions for
targeting by ribozymes. The sequence ~omrArison is
shown in ~igure 1. Three conserved regions were
identified for design of ribozymes. Two of these
conserved regions are shown in figure 1. In addition
to se~uen~e comparisons, a computer ~oy~am can be
used to pre~ict secondary structure of the target site
to ena~le greater efficacy of the ribozyme.
EXAMP~E II~ YNln~IS OF RIBOZYMES TARGETING
CONSE~VED SE~UENCES OF C1 GENE OF TLCV :
Riboz-~me 1 ~Rzl) :
3'- GUCCAGUCGUGUAAAAAGCAGGAGUGCCUGAGUAGUCGUAGGCUU
GUAAGUC - 5'
Ri~ozy~e 2 ~Rz21 :
3'- GAGG~AAAA~U~-AAAAGCAGGAGUGCCUGAGUAGUrA~AAAACU
AGCUCA - 5'
Ribozyme 3 ~R~3) :
3'- ~AGU~GUAAACUACAAAGCAGGAGUGCCUGAGUAGUCUAArUUCUA
GUACUA - 5'
W095/0~4 PCT~ ~3/019~
21 67701
EXAMPLE IV ~ llC CONSTRUCTS CARRYING Rzl, RzZ
and/or Rz3 ~ pRIBO5BIN :
A 1568 fragment of TLCV from nucleotide 1075-2643
covering the Cl, C2, C3 & C4 reading frames (ORPS) was
cloned into pALTER-l (Promega). Ribozyme sequences 1,
2 & 3 (Rzl, Rz2 and Rz3, respectively) were chemically
synthesised as described in Example III and introduced
into thei TLCV sequence using the Altered sites in
vitro mutagenesis system ~Promega). The integrity of
introduced ribozyme sequences was confirmed by
sequencing using the dideoxy chain termination method.
Rzl, Rz2 and Rz3 were shown to catalyse in vitro
cleavage of a complementary-sense viral RNA transcript
generated with 57 RNA polymerase at 50 C and 37- C.
The modified 1634 bp complementary-sense TLCV
fragment containing Rzl, Rz2 and Rz3 was cloned in the
anti~nse orientation (relative to C1 ORF
transcription) between a double 35S CaMV promoter and
a CaMV polyA termination sequence and the whole
expression cassette introduced into the BamHI site of
the m~ltip~ cloning site of the binary vector Binl9.
The resulting construct is pRIBOSBIN and is shown in
figure 2.
EXAMPLE V - ~N~ C CONSTRUCTS CARRYING Rzl, Rz2
and/or Rz3 : ~RIBO5BINDel :
This construct was derived from pRIBO5BIN
(Example IV~ by deletion of a 216 bp region from the
BstBI site ~nucleotide 2424~ to the 3'-end of the TLCV
fragment ~nucleotide 2643). The resulting construction
is pRIBO5BINDel and is shown in figure 3.
rB
W095tO3~ 2 1 6 7 7 0 1 PCT~P93/0194'7
EXAMP~E VI ~ N~ C CONSTRUCTS CAKKYl~G Rzl, Rz2
and/or Rz3 : pRIB07BIN :
This construct was derived from pRIB05BINDel
(Example V) by removal of a 9SO bp region from the
5'-end of the TLCV fragment (nucleotide 1075) to the
Xhol site ~nucleotide 202S). The resulting construct
is pRIB07BIN and is shown in figure 4.
EXAMPLE VII - &ENETIC CONSTRUCTS CARRYING Rzl, Rz2
and/or Rz3 pRIB02BIN :
A 52 bp fragment containing Rzl was amplified by
polymerase chain reaction (PCR) from pR~B05BIN
(Example ~V) and cloned in the antisense orientation
(relative to Cl ORF transcription) between a double
35S CaMV promoter and a CaMV polyA termination
sequ~nce and th~ whole expression cassette introduced
into the Bam~I site of the multiple cloning site of
the binary v~ctor Binl9. The resulting construct is
pRIB02BIN and is shown in figure 5.
EXAMPLE VI~I - EXPRESSION OF RIBOZYME ~N~ C
CONS~RUCTS IN TRANSGENIC TOBACCO CELL LINES :
The ribozyme constructs described in Examples 5
to 8 were tested for expression in transgenic tobacco
- plants using PCR analysis.
Leaf samples (150 mg) were collected from
transgenic to~acco lines prior to potting out in the
glasshouse and frozen in liquid N2. Samples were
grour.d to a fine powder and vortexed for 30s with
0.5 ml extraction buffer (contA;nin~ 0.1 M Tris-HCl
(p~ ~, 0.~ M ~aCl, 10 mM EDTA, 1 % w/v SDS and 1 %
v/v ~eta-mercaptoethanol) and 0.5 ml phenol. The
W095/0~4 2 1 6 7 7 0 1 PCT~ ~3/019~
38
extract was centrifuged at 12 000 g for 10 mn and the
supernatant fraction collected. The supernatant
fraction was extracted a further time with phenol a'nd
then with phenol/chloroform (1:1). The tot21 nucleic
acid fraction was precipitated at -2~ C with sodium
acetate/ethanol and centrifuged at 12 000 g for 10 mn.
The pellet was washed with 80 % v/v ethanol, dried and
resuspended in 50 ~1 sterile water.
First strand cDNA reactions were carried as
follcws. ~ucleic acid samples were annealed at 65 C
for 5 mn with 100 mg of CaMV Term primer (5'-
TTTAC~A~TACAAATACATACTAAGGGT in a total volume of
5 ~1. This primer is complementary to a region within
the CaMV termination sequence present in all ribozyme
construct ~see figure 6) transcripts produced from
these transcriptions.
To t~e annealed template was added S ~1 of
reverse trascriptase mix cont~;n;ng 50 mg Tris-HC1 (pH
8.3), 50 m~ ~Cl, 10 mM MgG12, O.S mM spermidine, 10 mM
DTT, unit ribonuclease inhibitor and 2.25 units AMV
reverse transcriptase. The mixture was incllhAted at
42- C for 60 mn followed by 94 C for 2 mn. Where
indicated~ control reactions were performed in wh-A_h
the ~MV reverse transcriptase was omitted from the
reaction mM Txis-HC1 (pH 9), 1.5 mM MgC12, 1.5 mM
MgCl~, 0.1 ~ v/v Triton X-loo, 200 ~M dNTPs, 250 ~M
CaMV Term primer (see figure 6), 0.5 units of Taq DNA
polymerase ~nd 2 ~1 of first strand cDNA reaction mix.
Two alternztive 5'-end primers were used, the
positions of which are shown in the attached figure :
C3 primer : 5'-ACTGATCTAATTACA~ AATGC
RlB primer : S'-GCAAGCTTCCTGAATGTTCGGAT
W095/03~4 2 1 6 7 7 0 1 PCT~3/olg~
39
PCR cycling conditicns were 94c C for 1 mn,
50-55' C for 1 mn, 72 C for 3 mn for 15-30 cycles.
PCR products were analysed on 1.2-1.8 % w/v agarose
gels.
Reverse transcriptase (RT)-PCR analysis of
RIBOSBIN an~ RIB05BINDel lines using the C3/CaMV Term
primer combination produces DNA products approximately
the predicted sizes i.e. 1690bp and 1450bp
respectively.
RT-PCR analysis o~ RIB05BIN, RIB05BINDel and
RIB02BIN lines- using the RlB/CaMV Term primer
combination produces DNA products approximating the
predicted sizes i.e. 504bp, 264bp and 235 bp,
respectivel~.
No PC~ products were observed in control
reactions : ~a) no reverse transcriptase (b) nucleic
acid samples from plants transformed with vector
sequenres alone i.e. not containing ribozyme
sequences. This demonstrates tha~ the PCR produ~s
observed wIth these primers were derived specifically
from RNA transcripts of the introduced ribozyme
constructs.
Predicted PCR product sizes using the C3/CaMV
Term primer combination are as follows :
1. RI~05BIN 1690 bp
2. RI~05BINDel 1450 bp
3. RIB07BIN
4. RIBO~BIN
Predicted PCR product sizes using the RIB/CaMV
Term primer comkination are as follows :
.
W095/0~4 2 1 6 7 7 0 1 PCT~3/019~
1. RIB05BIN 504 bp
2. RIB05BINDel 264 bp
3. RIB07BIN 264 bp
4. RIBOZBIN 235 bp
EXAM~LE IX - CONSTRUCTION OF RIBOZYME GENES :
Pairwise comparisons of deduced amino-acid
sequences encoded by C1 ORF of geminiviruses were
investigated ~table 1).
Alignment of nucleotide sequences of TYLCV genomes
isol~tes frsm ~ardinia (Kheyr-Pour et al., NAR (1991)
19 : 6763)~ Thai (Rochester et al., Virology (1990)
178 : 520~, Israeli (Navot et al., Virology (199;1)
185 : 1513, Australia (Dry et al., J. Gen. Vir. (1993)
74 : 147) snows that there are three conserved regions
in an internal 4ragment of the Cl transcript enco~;ng
the polypeptide essential to replication of the virus.
This BstBI ~ Eco OlO9I internal fragment (nt 346 to nt
982) of TYLCV-S genome was cloned into a Dra II/ Sma I
diges~ed pBluescript II SK~ phagemid (STRATAGENE).
This clone was used as template for the il,LLo~ction
of 2 ribozymes in the most highly conserved region.
Two and three dimensional folding analyses were also
made to retain the more theoretical accessible target
sites for clea~age with ribozyme.
The catalytic domain of hammerhead-type RNA
(figure 1), in this case satellite RNA of tobacco
ril~y~OL vtrus (STobRV), was used to design ribozymes
that form, with a completely unrelated substrate RNA,
a h~m~head structure in trans, so that the target
RNA is specifically cleaved at the predicted site
~Haseloff ~nd Gerlach (1989), Gene, 82 : 43-52).
According to the general rules described by these
authors, the only requirements for a su~strate RNA to
WOg5/0~4 PCT~W31019~
21 67701
be cleaved ~y hammerhead-type ribozymes is the
presence of the trinucleotide sequence for which
self-cleavage has been described (Perriman et al.,
Gene (1992) 113 : 157~. The two positions for the
hammerhead-type ribozymes retained on the most highly
cons~rved region of the C1 transcript of TYLCV-S are
shown in figures 2 and 3 :
* position R4, nucleotide 407 (target site GUC) ;
* position R5, nucleotide 430 (target site WC) ;
To introduce the 22 bases of the hammerhead
sequence of the satellite RNA of TobRV at these
positions, the strategy of rapid insertional
mutagenesis of DNA by polymerase chain reaction (PCR)
described by Kammann et al. (Nucl. Acid. Res. (1989)
17 : 54043 was used.
This method requires the following synthetic
oligonucleotide
Oligo ~4-R5 carrying the 2 hammerhead-type
ri~ozymes :
5' GATTTA&CTCCCTCTGATGAGTCCGTGAGGACGAA~ATGTTCGGATG
GAAATGTGC CTGATGAGTCCGTGAGGACGAAACCT>TGGGGAT 3'
The ~r.derlined sequences correspond to the
hammQrhead structure of the satellite of ToRSV. Bold
letters correspond to the target site.
This oligo R4-R5 was synthesized on an Applied
Biosystems Model 380B DNA synthesizer.
Oliqo T7 se~uencinq primer (Pharmacia) :
5' TTAATACGACTCACTAT 3'
Oliq~ T3 se~uerlcinq primer (Pharmacia) :
5' ATTAA-CCTCACTAAAG 3'
W095/03~ 2 1 6 7 7 0 1 PCT~P93/019~
42
In the first PCR ~PCR I) step, the sequence
between oligo R4-R5 and oligo T3 sequencing primer was
amplified. The amplified fragment was purified on a
molecular sieving column tPrimeErase Quick TM kit,
STRATAGENEj. In the second PCR (PCR II) step, this
ampl~fied frag~ent was used itself as a primer in
comb1nat~on with oligo T7 sequencing primer. The PCR
conditions were as follows :
PCR was carried out in a total reaction volume of
100 ~1 containing 50 mM KCl, 10 mM Tris-HCl
pH 8.3,1.5 m~ MgC12, 3GO ~M dNTP each, 2 U Taq
polymeras~ (Perkin Elmer Cetus), 40 pg template DNA,
100 pM primer each. PCR II was the same as PCR I,
exce~t tk.at 5 ~1 ~300 ng of amplified fragment
estinated fro~ agarose gel~ assay volume of PCR I was
directly used for priming. Denaturation : 2 min -94C ;
anne~lin~ 2 m-n 40C ; synthesis ; 3 min 72C with 30
cycles for each step.
After purification using agarose electrophoresis,
the PCR product was digested with BstBI, filled in
using Klenow polymerase and finally digested with
EcoQ:09I. h~ resulting 679 nt fragment was cloned
intc a Sma ~ /Dra II digested Bluescript SK+ vector.
The clon~ng vector containing the cloned sequences was
tran~for~ed in~o, and propagated in, the bacterial
hos~ XL1 Blue (STRATAGENE), using techniques known in
the art ~aniatis, Molecular cloning, A laboratory
guide, ~nd edition, 'g89, Cold Spring Harbor
Laboratory publishers).
The presen-e of the ~ hammerhead sequences of the
satellite ~A of TobRV was verified by sequence
anaiysis using T7 sequencing kit (Pharmacia). The
resu~t shows a perfect insertion of catalytic R4 and
R5 sites~
W095~0~U~ PCT~31019~
21 67701
EXAMPLE X CONSTRUCTION OF RIBOZYME PLASMIDS :
Plasmid pRIBO5BS shown in figure 15 was the base
point for the construction of the other plasmids. This
plasmid is a pBluescript SK~ vector carrying a 1690 bp
5' end - Ase I fragment which corresponds to the
C1/C4/C2/C3 complementary fragment of TLCV (figure 4)
bearing 3 hammerhead-type ribozymes at positions :
- Rl (nucleotide 412, target site CUU),
- R2 ~'n~cleotide 711, target site CUU) and,
-'R3 ~nucleotide 1654, target site GUA).
Plasmid pPolyrihozyme E is derived from plasmid
pRIBO5BS by deleting the 877bp l~filled-in using Klenow
Poly~erase Xho I - Bcl It' fragment corresponding to
the region between 2 ha~merhead-type sequences at
positions R2 and R3, and the 264 bp "5' end - Tth
lllI:; fragment. The Tth l~lI site was filled-in us'ng
klenow pol~merase. The "Tth III I - HindIII (site of
pBluescript SK+ vector") fragment was isolated after
separation by electrophore~is on agarose gel. This
fragment wa~ cloned into EcoRI filled-in using Klenow
Polymerase and ~ind III sites of pBluescript SK+
vector, deleted previously from Sac I-Xba I digested
using Mung Bean Nuclease fragment.
The polyriboz~ne E contains 434 bases and
includes 3 ribozymes : R1 (position 28), R2 (position
327) and R3 ~po~ition 3g9~ as shown in figures 5 and 6
(polyR2E-R2). The size of the arms flanking the
catalytic domains of the ribozymes is :
* 27 nLcleotides from 5' end to ribozymeRl ;
* 298 nucleotides between R1 and R2 ;
* 71 nuoleotides between R2 and R3 ;
.. . . .
* 35 nuoleotides from ribozyme R3 to 3'end.
The pGlyribozyme F plasmid is derived from
pPolyribozyme ~ plasmid. The 56bp "SmaI (site in
W095/03~ 2 1 6 7 7 0 1 PCT~3/019~
pBluescript SK + vector near 5' end) - Sac I" fragment
of the replicase gene of TLCV containing the ribozyme
Rl was replaced with the 98bp "BstBI filled-in using
klenow polymerase - Sac I" fragment of replicase gene
of T~LCV - 5ard~nian isolate bearing the two ribozymes
R4 and R as described previously. The exchanged
fragments possess a region which is very highly
conserved between different strains of TYLCV and TLCV.
The nucleotide sequence identity corresponds to 97 %
betw en the 365 nt to 435 nt region of TLCV and the
370 nt to 4~0 nt region of TY1CV-Sardinian isolate
(Figure 3~.
The chimeric polyribozyme F contains replicase
gene fragments which hav~ two different origins
TYLCV-Sardinian isolate and TLCV-australian isolate.
The ~ssem~ly of these fragments was made at the Sac I
site. It ic also possible ~o assemble the two
frasments using an artificial polylinker. The
inclusion of this non-complementary sequence has no
dele~erious effect on the catalytic activity of the
polyr~bozym.e.
The polyribozyme F contains 479 bases and
incl-~des 4 ribozymes : R4 (pGsition 59), R5 (position
83), R2 (positiQn 372) and R3 (position 444), as
shown in ~igures 5 and 6.
The size of the arms flanking the catalytic
domains of the ribozy~es is :
* 59 nucleotides from 5' end to ribozyme R4 ;
* 22 nuoieotides between ribozymes ~4 and R5 ;
* 288 nucleotides between ribozymes R5 and R2 ;
* 71 nuc~estides between ribozymes R2 and R3 ;
- ~ 3S nucleotides from ri~ozyme R3 to 3' end.
The ~hi~eriC pPolyribozyme D is derived from
pPolyriboz~me F. The 206bp i'SacI-SwaI" fragment
originatin~ from TLCV was deleted. The Sac I site was
WOg5/0~4 PCT~3/019~
21 67701
destroyed ~y ction of T4-DNA polymerase. The
polyri~ozyme D contains 270 bases and includes 4
ribozymes, R4 ~position 5g), R5 (position 83), R2
(position 16 ) and R3 (position 235) as shown in
figures 5 and 6~
The si2e of the arms flanking the catalytic
domains of the ribozymes is :
* 59 nucleotides from 5' end to ribozyme R4 ;
* 22 nucleotides between ribozymes R4 and R5 ;
79 nuclPotides between ribozymes R5 and R2 ;
* 71 nucleQ~.ides between ribozymes R2 and R3 ;
~ 35 nuc~eotides from ribozyme R3 to 3' end.
The se~uenc2s of poIyribozyme E, F and D were
verified uslng the T7 sequencing kit supplied by
Pharmacia.
The functioning of polyribozymes E, F and D was
tested in vitro in accordance with the protocol
described in '~Protocol and Application Book" from
Promega.
No expresslon signals are contained in the
polyribozymes. For this reason, they were placed under
the contrGl of double 35S promoter and poly(A) of 35S
RNA of CaMV.
To raalize these constr~ctions, the expression
cassette camp3sed of a douhle 3SS promoter,
monoribozyme Rl and poly (A) of 35S of CaMV was
isolated frGm pR~B02BIN plasmid described a~ove.
The p~IB02BIN plasmid was digested with Sac I.
The ac I ~;tes were made blunt-ended using T4-r~A
polymeras
The linearized pRI3~2BIN plasmid was then
digest~d with Sal I, the "Sac I filled-in Sal I"
fragm~nt wl~ich contains the expression cassette was
purified on agarose gel and cloned into pBluescript
WO9Sl0~4 PCT~3/019~
21 67701
46
SK+ vecto in 'IXba I filled-in using Klenow Polymerase
and Sal I" sit~s.
The Hlnd III - Bam HI fragment corresponding to
mono~iboz~tte R1 was then replaced with HindIII - BamHI
frag~ents ^ont2ining the polyribozymes E, F and D
isolated from pPolyribozymes E, F and D respective'y.
The Hind III - BamHI fragment is in inverse
orientation in the expression vector.
The expression cassette with polyribozymes E, F
or ~ were cloned into the binary vector pGAZE (figure
7) derived rcm pGA492 (An~ Plant Physiol. (1986) 81 :
86). The pGA 4g2 vector was double digested with Sac I
and Sca I, made blunt-ended using T4-DNA polymerase
and ligated after agarose gel purification.
The e7iminated fragment corresponds to nearly the
entire chloramphenicol acetyl transferase gene. The
Hind III sit~ was also converted into EcoRI site with
HindTII - E~oRI adaptors supplied by Stratagene. As
for pGA4g2~ pGAZE contains ~he cis-acting elements
requlred for plant transformation, including T-DNA
border, a selectable marker expressible in plants
(kanamycin res~stance), cloning sites, and a wide host
range rep'-icon. The kanamycin resistant marker is
cons_ructe~ from nopaline synthase control regions and
neom~cin p~losphotransferase coding region of Tn5.
The pGAZ~ vector was digested with EcoRI and
filled-in using Klenow-poiymerase. The "SacI filled-in
- Sal I" f,agments containing the expression cassettes
with polyriboz~es E, F or D were cloned into the
EcoRI filled~in site of pGAZ~
The ob~al~ed recombillant binary vectors, pGAZE-
polyribozyme E, pGAZE-polyribozyme F and pGAZE-
polyribozy~e D (figure 7) were introduced into the
desarmed strain LBA4404 of Aqrobacterium tumefaciens
(Hoekema et al., Nature f~983) 303 : 179) by direct
W095/03~ PCT~W3/019~
21 67701
47
transformaticn (protocol derived from Holsters et al.,
Mol. Gen. Genet~ (1978) 163 : 181)
For ri~ozyme A, which corresponds to monoribozyme
R1 includlng the catalytic domainand flanking arms 15
base~ long, ~nd polyribozyme C described above
(example V'f ex~ression cassettes were introduced in
pGAZE and/or p~-h492 as mentioned above.
Thus, pGAZE-ribozyme A, pGA492-ribozyme A and
pGAZE-polyribozyme C (Figure 8) were obtained. The
genes of in~erest of all the constructions described
above were transferred into tomato using Agrobacterium
based co-cultiv~tion system. Tomato transformation a~nd
regeneraticn was carried out as described by Fillatti
et al (Bictechnclogy (1987) 5 : 726).
EXAMPLE XI ~ MOLECU~AR ANALYSIS OF TRANSGENIC PLANTS
* RN~ analy~
Total ~NA is isolat2d from young leaves of
transgenic and non-transgenic plants grown in the
greenhouse according to Chandler et al. (Plant
Physiol. ~1983~ 74 : 47). RNA is separated on
agarose-formaldehyde gels and transferred to a
nitrocellulæse membrane. The blot is hybridiæed with
}abelled fr~Lgment or entir~ gene of interest. To chec~
whether equa ~mounts of RNA were applied to each
line, the nltrocellulose membrane is stained with
methylene blue.
* DNA analy~
; For Southern blot analysis, total DNA is isolated
from young leaves of transgenic and non-transgenic
plants grown in the greenhouse according to Dellapor~a
et al. (P~ant Mol. Biol. Re~. (19~3) 1 : 19), digested
with restriction enzymes according to the
WOs5/03~ PCT~ ~31019~
21 67701
~8
manufacturer's instructions, separated ~by
electropho~esis on agarose gel and transferred to
nitrocellulose ~y capillary action. The blot is probed
with labelled fragment or entire gene of interest.
EXAMPLE XIT - EVA~UATION OF RESISTANCE TO TYLCV FOR
TRANSFORMED T3MATO PLANTS WITH THE DIFFERENT
CONSTRUCTI~i~S BY AGRO~NOCNLATIONS
Since TYLCV or its cloned DNA are not transmitted
mech~nically/ the agroinoculation tPr-hnique (Grimsley
et al., Plar.t DNA Infections Agents, Hohn and Schell
(edsJ Springerl Wien, New York ~1987) pp 87-107) is
used lo assa4~ to infectivity of the cloned DNA.
The r~ Y~CV-Sardillian st:rain genome was inserted
into Sst ~ si_e of the b~nary plant transformation
vector pBin lg (Stanley et al., EM80 J. (1986)5 :
1761~ and a B~HI-fragment of about 600 bp extending
from polylinker of pEin 19 through the intergenic
region of TY~CV to the BamHI site at map position 152
of TV~rJ~V was deleted. Su~sequently a complete genome
unit of TY~ i was inserted intc the remaining Sst I
site yield~rig a 1,8 mer of the TYLCV-genome in pBin
19. This pl smid was introd~ced directly into
Aqrobacteri~m tumefaciens strain LBA4404.
Aqro~acterium tumefaciens cultures were grown at 28C
for about 4~ hours. The cells were pelleted and washed
twice with w~ter and re uspended in 1/10 th of the
initial volu~ne of sterile water~
Young transformed tomato plants, at the 3 to 4
leaf stage are inoculated into the petioles of the
three yo~r.ge~t leaves with the concentrated
Agroba~ter~- us~ng a 18-gauge needle.
WO 95/03404 2 1 6 7 7 0 1 PCT/EP93/01946
49
The pl ai..ts are placed in a closed growth chamber
with 16 h ~ day 1 i ~ht at 24 C under 70 % relative
humidi ty.
Those killed in the art will appreciate that the
invention described herein is susceptible to
variations and modifications other than those
specifically described. I t is to be understood that
the ~nvention inclucles all of the steps, features,
compositions and compounds referred to or indicated in
this specifi-ation, individually or collectively, and
any and all colhbinat:ions of any two or more of said
steps or featur2s.