Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
~175587
WO 95112608 PCT/US9~1123~7
SYNTTTF-~T7r~G AND SCl~FFNT~G MOr FCUL~R DIVFRSITY
RELATED APPLICATIONS
This application is a rnnfin~ nn-in-part of U.S. Application Serial Nos.
08/146,886 and 08/149,675, both filed on November 2, 1993, each of whieh is
ill~,ulpl ' herein by reference for all purposes.
COPYRIGHT NOTICE
A portion of the disclosure of fhis patent document contains material which
is subject to copyright protection. The copyright owner has no objection to the facsimile
lc~JluJucLiull by anyone of the patent document or the patent disclosure as it appears in the
Patent and Trademark Office patent file or records, but otherwise reserves all copyright
rights whatsoever.
FIELD OF THE INVENTION
The present invention relates generally to methods and devices for
~yllLll~ g very large collections of diverse molecules and for identifying and isolating
,uu .-l~ with useful and desired activities from such enllPrtinnc The invention also
relates to the illl,Ull~Ul~iUI. of i~f ~ ;.." tags in such collçctions to facilitate
i~lPntifir~tinn of .-. .l~u~ with desired properties.
BACKGROUND OF THE INVENTION
Ligands for II~f~lu~l~ol_ ulfu receptors can be identified by screening diverse
collections of peptides produced through either molçcular biological or synthetic chemical
Rf~-~o,.,~ peptide libraries have been generated by inserting degenerate
30 nli~ ' " ' into genes encoding capsid proteins of fil~nnP~n~lc 1.- ~ .g~ and the
DNA-binding protein Lac I. See Cwirla et al., 1990, Prûc. Natl. ~a~. Sci. USA 87:
6378-6382; Scott & Smith, 1990, Science ~: 386-390; Devlin et al., 1990, Sciencç 249:
4.14-4û6; Cull et a~., 1992, PrQc. Natl. Acad. ~Çi. USA 89:1865-1869; and PCT
publication Nos. WO 91/17271, WO 9111981B, WO 93/08278, each of which is
35 illCUl~JI ' ' herein by reference. These random libraries may contain more than 109
different peptides, each fused to a larger protein sef~uence that is physically linked to the
WO 95/12608 2 ~ 7 5 5 8 7 PCTIIJS9J1123.17
genetic material encoding it. Such libraries are efficiently screened for inîeraction with a
receptor by several rounds of affinity purification, the selected exposition or display
vectors being amplified in E. coli and the DNA of individual clones sequenced to reveal
the identity of the peptide responsible for receptor binding. See also PCT publication Nos.
WO 91/05058 and WO 92/02536.
Chemical approaches to generating peptide or other molecular libraries are
not limited to syntheses using just the 20 genetically coded amino acids. By expanding the
building block set to include unnatural amino acids and other molecular building blocks,
the accessible sequence and structural diversity is tlr~ y increased. In several of the
strategies described for creating synthetic molecular libraries, the reaction products are
spatially segregated and the identity of individual library members is ~ ly
defined by the nature of the synthesis See Geysen et al., 1984, Proc. ~. ~. Sci.USA 81: 3998 4002; Geysen et al, 1986, in Synthetic Peptides as Antieens; Ciba
Foundation Symposium 119, eds. Porter, R. & Wheelan, J. (Wiley, New York) pp.
131-146; Fodor et al., 1991, Science 251: 767-773; U.S. Patent No. 5,143,854; and PCT
patent publication Nos WO 84/03564; 86/00991; 86/06487; 90/15070; dnd 92/10092,
each of which is i~. u~uld~d herein by reference.
Libraries of more than 30 million soluble peptides have been prepared by
the "tea-bag" method of multiple peptide synthesis. See Houghten, 1985, ~. ~1.
Acad. Sci. II~ 82: 5131-5135; and U.S. Patent No. 4,631,211, each of which is
;.., u.~ ' herein by reference. Each library is ,yll~ .l and screened as degenerate
peptide mixtures in which individual amino acids within the sequence are explicitly
defined. An iterative process of screening (e.g., in a ~ i.", binding assay) andresynthesis is used to fractionate these mixtures and define the most active peptides within
the library. See Houghten et al., 1991, Nature 354: 84-86; Pinilla et al., 1992, Pe~tide
~h _: 351-358; Blake, J. & Litzi-Davis, 1992, B;u~ullju~dt~ m. 3: 510-513; and
PCT patent publication No. WO 92/09300, each of which is ;-.w-~u-~. l herein by
reference.
Using the split-synthesis protocol of Furka et al., 1988, ~. 14th ~.
~n~. Biochem., Prague, Czech. 5: 47 (see also Furkd et al., 1991, In~ 1. ~ Prot~in
~- ~Z: 487~93; and Sebestyen et al., 1993, Bioor~. Med. Chem. Lett. ~: 413-418),Lam and coworkers have prepared libraries containing-- 106 peptides attached to 100-200
~m diameter resin beads. See Lam et al., 1991, Nature ~: 82-84; Lam et al., 1993,
~ WO 95/12608 2 ~ 7 5 5 8 7 PCT/US9~/123.17
Bioor~ . Chem. I~. 3: 419-424; and PCT paterlt publication No. WO 92/00091,
each oF which is il~,o-,uuldL~d herein by reference. The bead library is screened by
incubation with a labeled receptor: beads binding to the receptor are identified by visual
inspection and are selected with the aid of a mi~ ~ullldlli~uJl_~vl . Each bead contdins 50-200
5 pmol of a single peptide sequence which may be d~t~rminp~l directly either by Edman
~1. ~,..,.I~;r.,. or mass :,Ue~L~ul,l~ L-~ analysis. In principlel one could create libraries of
greater diversity using this approach by reducing the fiim~-ncinnc of the bedds. The
sensitivity of peptide sequencing techniques is limited to ~ I pmole, however, placing a
cledr limitation on the scope of direct peptide ~PqllPnrin~ analysis. Moreover, neither
10 analytical method provides for ~LIdi~llLrulwdld and ~ sequence analysis when
the library building block set is expanded to include D- or other non-natural amino acids
or other chemical building blocks.
High throughput screening ûf collections of chemically synthesized
molecules and of natural products (such as microbial fPrmPnt~ir,n broths) has traditionally
15 played a central role in the search for lead ~ for the dcv~lul of new
I,~...i, nlr.~ agents. The ~ dbl~ surge of interest in cul~lb 1 chemistry and
the associated ~Prhnr,ln~iPc for generating and evaludting molecular diversity represent
significant milestones in the evolution of this paradigm of drug discovery. See Pavia et
1-, 1993, ~iQ~e. ~. ~h~m. I~t. ~: 387-396, illco.pu-dL~d herein by reference. To20 date, peptide chemistry has been the principle vehicle for exploring the utility of
.,,,.1.;,~ u... ;~1 methods in ligand idPntifir~tinn See Jung & Beck-Sickinger, 1992, ~Q.
m. Inl. Ed. En~l. 31: 367-383, I ' herein by reference. This may be
ascribed to the availability of a large and structurally diverse range of amino acid
mnnnm~rc a relatively generic, high-yielding solid phase coupling chemistry and the
25 synergy with biological approaches for generating .e~ peptide libraries.
Moreover, the potent and specific biological activities of many low molecular weight
peptides make these molecules attractive starting points for therapeubc drug discovery.
See ~irc-hm~nn, 1991, ~ m. InI. Ed. ~gl. 30: 1278-1301, and Wiley & Rich,
. 1993, ~. Res. Rev. 13: 327-384, each of which is ;llCOl~JuldL~ herein by reference.
30 U~rdvu~dbl~ pll~ul.ld~od~rnamic properties such as poor oral bioavailability and rapid
clearance ~ yivû have limited the more widespread dc~luull~..,L of peptidic ~ u,....l~ as
drugs however. This realization has recently inspired workers to extend the concepts of
Cùlllb I organic synthesis beyond peptide chemistry to create libraries of known
WO9~/12608 2 1 7 5 5 8 7 PCTIS91/123~7 ~
r~ c~ like h~n7n~1io7rrinrC (see Bunin & Ellman, 1992, I Amer. Chem. Soc.
114: 10997-10998, illcul,uol~ herein by reference) as well as polymerie molecules sueh
as oligomeric N-substituted glyeines ("peptoids") and uli~;u~-du~a~ See Simon et al.,
1992, proc. Natl. Acad. Sci. USA 89: 9367-9371; 711rkl~rm:~nn et al., 1992, l. Amer.
Chem. Soc. 114: 10646-10647; and Cho et al., 1993, Science 261:1303-1305, each of
which is i~cvluùldLed herein by reference.
Despite the great value that large libraries of molecules can have for
identifying useful c~ ,u~ or improving the properties of a lead eomrollnrl~ the
difficulties of sereening sueh libraries, Lldl~iuul~ly large libraries, has limited the impaet
aecess to sueh libraries should have made in reducing the costs of, e.g., drug discovery
and dcv~ lu,u"~ . Cv~l~uc~llLly~ the d~iclv,u"l~ of methods for generating and screening
libraries of molecules in which each member of the library is tagged with a unique
identifier tag to facilitate i~lrntifinA~inn of, .,..,~ (see PCT patent publieation No. WO
93/06121, ill~Ull~Vl~t~ herein by referenee; see also U.S. patent application Serial Nos.
946,239, filed September 16, 1992, and 762,5æ, filed September 18, 1991, 5~) metwith great ~-n~h-~ci lcm In the method, produets of a chemieal synthesis procedure,
typically a ~.~.,.I,;,,-I..,iAl synthesis on resin beads, are explieitly specificd by attachment of
an identifier tag to the beads coincident with each coupling or othe} product generating
reaction step in the synthesis. Each tag specifies what happened in a reaction step of
interest, e.g., which amino aeid monomer was eoupled in a partieular step of a peptide
synthesis proeedure. The strueture or idenvity of a eompound, e.g., the sequenee of a
peptide, on any bead can be dedueed by reading the set of tags on that bead. Ideally, sueh
tags have a high i,-r~ eontent, are amenable to very high sensitivity detection and
deeoding, and are stable to reagents used in the synthesis. The concept of an
olignnl~nll~ot~ encoded ehemieal synthesis was also proposed by Brenner and Lerner,
1992, ~2Ç ~. Aead. Sci. USA 89: 5181-5183, i~l~u~ùldL~d herein by reference.
The encoding method has been employed to show that, starting with an
orthogonally differentiated diamine linker, parallel ~ A~ ;Al synthesis ean be used to
generate a library of soluble ehimeric peptides comprising a "binding" strand and a
"coding" strand. See Kerr et al., 1993, l- Amer. Chem. Soe. 115: 2529-2531,
ullJvl~L~d herein by reference. The coupling of either natural or unnatural amino acid
monomers to the binding strand was recorded by building an amino acid code comprised of
four L-amino acids on the "coding" strand. Compounds were selected from equimolar
WO95/12608 2 1 75587 PCT/US9J1123~7
peptide mixtures by affinity ~u~ ;.." on a receptor and were resolved by HPLC. The
sequence of the coding strand of individual purified molecules was then A~t~rmin~A by
Edman .1.~;"..1-l;..., to reveal the structure of the binding strand. An analogous peptidic
coding scheme was also recently reported by Nikolaiev et al., 1993, Peptide B~ç~h _:
161-170.
Cnnetr~inte on the sensitivity and throughput of the Edman procedure will
ultimately restrict the scope of tbis aspect of the encoding method to analyzing libraries of
limited diversity. The use of ~"~ ' '- tags offers greater promise, but imp}ovedmethods for ~J ' ~ llig~.. l~.ll;.l. -tagged molecular libraries are needed.10 Moreover, there remains a need for altemate m~thnAl-l~gy for ~y~ ;Liilg and screening
very large tagged molecular libraries.
Where it is desirable to synthesize diverse collections of molecules on a
plurality of solid supports such as beads, additional problems can arise. Examples of the
use of beads with diverse molecular products ~y~ ;L~J thereon are disclosed in, for
15 example, the following ~1.~.1;~-li~ , ' herein by reference for all purposes:U.S. Application Serial No. 07/876,792, filed on April 29, 1992; U.S. Application Serial
No. 07/762,522, filed on September 18, 1991; and U.S. Application Serial No.
07/946,239, filed on September 16, 1992.
While meeting with substantial success, the techniques described above have
also met with certain lin it~ti~-nc For example, when the synthesis of diverse products
takes place on beads, many manual . of such beads become necessary. For
example, in U.S. Application Serial No. 07/876,792, filed on April 29, 1992, ;l~by reference herein for all pum. oses, one must suspend a collection of beads in a carrier,
divide the beads, perform monomer addition reactions on the divided sets of beads,
sometimes redivide and selectively recombine the beads thus ~ l~iL~l, mix the
.l ' beads, and repeat the process. When large numbers of monomers are
involved and when the reactions involve many monomer addition steps, manual techniques
become extremely tedious. In addition, the "accounting" for the many products that have
been ~ ' ' becomes a daunting task.
From the above, it is seen that improved methods and devices for
~J ' ' and screening very large tagged molecular libraries are desired. The present
invention meets these and other needs.
wo 95/12608 ~ ~ 7 5~ 8 7 PCT/US9~1123.17
SUMMARY OF THE INVE~TION
The present invention provides improved methods for generating and
screening molecular libraries in which the individual molecules in the library are tagged
with unique, easily decoded identifler tags. Also provided is an apparatus and method for
5 rapidly and efficiently synthesi~ing diverse molecular products.
In one . ."l.o.~ the present invention provides methods and reagents for
tagging the products of C.,..~ chemical processes to construct encoded syntheticchemical libraries. In an important . .~.l.o~ the invention provides a method for
p~.rul",~ peptide and ,~l jg~"",~ synthesis on Illi~,~U:~,UIJic beads through an10 alternating and compatible synthetic procedure. The large ~ ..".. lff)~;~encoded
synthetic peptide library produced by this ~ulllb;~ Lul;dl synthesis is composecl of many
beads, each of which contains many copies of a single peptide (with a defined sequence)
and a single-stranded DNA tag whose se~quence artificially and ~ ly codes for
the structure of the associated peptide. The library can be efficiently illt~ llu~ ~i for
15 interaction with lluu~ ,lLly-labeled biological receptors by flow cytometry, and
individual beads selected by exploiting the ability of FACS il.aLIul--~.,Ld~ion to sort single
beads. The DNA tag on a sorted bead is amplified by the PCR and sequenced to
determine the structure of the encoded peptide ligand. The library can be used, for
example, to find high affinity (~l~lu~l~ol~u) ligands for a receptor such as an anti-peptide
LO m~nru-lr)n~l antibody.
A synthetic molecular library of the invention can be produced by
ayllLi.~;L;l.g on each of a plurality of solid supports a compound, the compound being
different for different solid supports. The compound is ayllll~ ;L~i in a process
cûmprising the steps of: (a) dlJ~)Uli' ' ,, the supports in a stochastic manner among a
25 plurality of reaction vessels; (b) exposing the supports in each reaction vessel to a first
chemical building block; (c) pooling the supports; (d) apportioning the supports in a
stochastic manner among the plurality of reaction vessels; (e) exposing the supports in each
reaction vessel to a chemical building block; and (fl repeating steps (a) through (e) from at
least one to twenty times. Typically, substantially equal numbers of solid supports will be
30 ~IJUlLiull~d to each reaction vessel. In one ~ bodilll~l,L of the method, the chemical
building blocks are chosen from the set of amino acids, and the resulting compound is a
peptide oligomer.
WO 95/12608 2 1 7 ~ 5 ~ 7 PCT/US9.11123-17
More particular~y. the invention relates to certain improvements in the
coupling l l- ";~ c associated with such methods. One such illl~)lU..,~ relates to the
chemistry used to remove the Fmoc protecting group from the alpha-amino group of a
bead, linker, or growing peptide chain in such syntheses. Preferably, such removal is
effected by treatment with 5 to 15%, preferably 105~o, piperidine for 5 to 60 minutes,
preferably 5 to 10 minutes, although other conditions may be employed, e.g.. 15 to 30%
piperidine for 5 to 30 minutes. Other improvements relate to the activation chemistry of
the peptide coupling reactions, in that when cenain automated i~ ulll~..lLdtiOn is used to
perform the synthesis of an r~ r~nl~riPrJti~lP tagged peptide library, the invention provides
10 for a simple mixture of HOBVHBTU to reduce reagent supply bottles.
In another aspect, the present invention relates to methods of synthesizing a
tagged molecular library. wherein each molecu~e in the library is covalently attached to a
solid suppOn and is tagged with one or more different chemically inert hydrocarbon tags,
wherein said tags comprise a variable l~ydlu~l/ull region and a molecular hook.
15 Preferably, such tags comprise a cleavable linker attaching said tag to said solid suppon, a
molecular hook, and a variable length lI,~I.hU~UIJUII chain linking said molecular hook to
said cleavable linker. More preferred are those PmhrriimPn~c wherein said tag comprises
the formula:
20 wherein n is from I to 10 or more, X is a cleavable linker and R is a molecular hook.
Such molecular hooks are preferably selected from the group consisting of biotin, a
of a hi~h association peptide pair and a protected activatable group, such as a
phu~u.l~LivdLdble group. Preferred cleavable linkers are photocleavable linkers
Also provided is a method of detecting the presence of such chemically inen
25 llydlU~ .lUII tags. The method comprises cleaving the one or more different tags from the
solid support, followed by immohili7in~ the tags to a second solid suppon. The
immo~hili7Po tags are then treated with an olipf n~ PoririP sequence whereby theoli~ ' ' sequence selectively binds to the immr~hili7P~ tag. The r~ nllrlpoti~ipsequence is then amplified, and its presence is detected, wherein the presence or absence
30 of the ~"~ ' ' sequence is indicative of the presence or absence of the tag.
21 755~7
WO 95/12608 PCT/US9~/123-17
Another ~ .o.~ of the present invention provides a method of
d~....;,l;.~g the sequence of synthesis steps for a synthesized molecule attached to a solid
support in tagged molecular library, tagged with chemically inert llydlu~ulJull tags. The
method comprises individually detecting the presence of one or more different tags on said
5 solid support, the presence or absence of said individual tags being indicative of the
occurrence of a particular synthesis step in the synthesis of said molecule.
In another aspect, the invention relates to methods and il.~L ul..~ ,.~Liù.l forencoded synthetic chemical libraries on beads too smaU to be separated on
cull~ "iunal flow cytometry ill:~LIu~ Such small beads aUow the resulting library
size to increase from the more typical range of 109 to 10'3 for bead based libraries up to a
size of 1013 members for bead-free libraries. The invention also relates to methods for
screening such libraries.
The invention also relates to methods for screening encoded synthetic
libraries to identify useful ~~ ~ In one important aspect, the invention provides
15 important advances in the field of natural product screening relating to methods for
generating, tagging, and screening natural product libraries to, l".,,.. t. . I'~ and identify
.u l~ with useful activity.
In another aspect, the invention relates to an improved process for rapidly
and efficiently identifying a pool of cu"~l,u~ from a molecular library of the invention.
20 In this method, the oli~u~u~ lcA Lide tags from a pool of tagged ~ that exhibit a
desired property (e.g., binding to a receptor) are ..)....~...,.. ;.. A and cloned to facilitate
sequencing of a plurality of tags in a single sequencing reaction. If the tagged cu~
are peptides, and an encoding scheme based on the genetic code is employed, then one can
subclone individual tags from the u u..~,..t~ "~ . into other selection and expression systems,
25 such as the plasmid and phage-based systems described in the ~ ;lUUII,I section above,
for further analysis of the peptide.
In another . 1~o-~ the present invention provides an apparatus and
method for rapidly and efficiently ~ "iLill~ diverse molecular products. According to
specific aspects of the invention, diverse polymers are synthesized on substrates such as
30 glass beads. Optionally, the beads may be ~ ln~ ly "tagged" during the synthesis
reactions with a molecular tag. Merely by way of example, the syl.~ll~iL~ molecules on
the beads may comprise peptides, while the molecular tags may comprise olig~ i
Of course, other molecular products may also be synthesized using the techniques
~ wo 95/12608 2 ~ 7 5 5 ~ 7 PCTNS9 1/12317
described herein whenever a molecule has a basic "building block" common to other
related molecules. Examples include l.~ ibl~ r~ , and beta turn
mimetics.
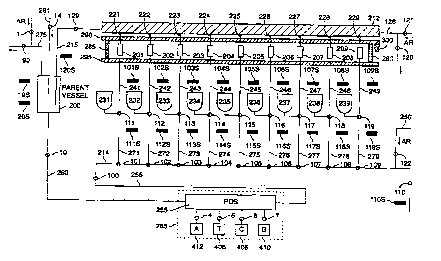
According to one r.llllO/1;ll~rlll of the invention, a parent vessel is used to
5 mix bead ~"~ The mixed beads are distributed through a common manifold to a
plurality of separate reaction vessels. In the reaction vessels, the beads are exposed to
different, selected mnnnmrrc, which react on the beads to be coupled thereto, preferably
covalently The beads may, optionally, be exposed to chemical "tags" which also couple,
covalently or otherwise, to the beads. The beads are then ~ d through the
10 manifold back to the parent vessel and mixed. The mixed bead suspension is then again
divided among the plurality of reaction vessels, and the process of monomer addition, bead
mixing, and ~ L~ tioll continues. The process results in the formation of a collection
of beads or other substrates with a diverse set of molecules formed on the surfaces thereof.
According to one aspect of the invention, the invention includes an apparatus
15 and a method for ~yll~h~siLil.g diverse molecules on substrates. The substrates are
distributed to selected reaction vessels from a parent vessel. Reagents are then introduced
into the reaction vessels to synthesize a portion of the molecules. The substrates are then
moved to the parent vessel for mixing. The substrates are then lrA,~l,il...lr~l to the
reaction vessels for further synthesis. The cycles continue until a desired set of molecules
are ~yllLll~iL~ During synthesis, the entire synthesizer is sealed from the external
dLIllU;llJIl~lt:.
In general, the invention provides apparatus and improved methods for
generating and screening molecular libraries in which the individual molecules in the
library are tagged with unique, easily decoded identifier tags.
A further l---d~ ~L~ of the nature and advantages of the invention may
be had with reference to the description and drawings below.
DESCRIPI`ION OF THE DRAWINGS
Fig. I shows a schematic diagram of the synthesizer of the present
invention;
Fig. 2 shows a schematic diagram of a reagent reservoir;
Fig. 3 shows a 3-port valve used in the synthesizer;
Fig. 4 shows a 2-port valve used in the ~yllLll.~
~VO 95112608 2 1 7 5 5 8 7 PCI`IUS9~11123 17
~
Fig. 6A shows an alternative ~ al.~ lL of a reaction vessel bank having a
rotatable carousel holding a plurality of groups of reagent reservoirs;
Fig. 7 shows the among a lower manifold valve, an
injcction valve, a reagent reservoir, and a reaction vessel;
Fig. 8 shows a ~ iC agitator;
Fig. 9 shows the upper portion of a reaction vessel bank;
Figs. lOA and IOB show a lower reaction vessel bracket;
Fig. llA-1 lD show an optical alignment block for use with optical sensors
to detect the presence of a liquid within a substantially translucent tube according to one
aspect of the present invention;
Fig. 12 shows a reaction vessel according to one aspect of the present
invention;
Fig. 12A shows a ~ r controlling jacket around the reaction vessel
of Fig. 12;
Fig. 12B shows a cross-sectional view of the jacket and vessel of Fig. 12A;
Fig. 12C shows an alternative; ' ~ " of reaction vessel having a
lLulc controlling jacket;
Fig. 12D shows a cross-sectional view of the jacket and vessel of Fig. 12C;
Fig. 13 shows a parent vessel;
Fig. 14 is a simplified diagram of the electronic hardware for controlling the
~y~ ,.;~"
hg. 15 shows a simplified diagram of the controller circuit;
Flg. 16 shows the steps taken by the control computer to drain the reaction
vessels of all liquids;
Fig. 17 shows the steps taken by the control computer to clear the bottom
manifold of material;
Fig. 18 shows the steps taken by the control computer to agitate the contents
of the parent vessels;
Figs. I9A through 19D show the steps taken by the control computer to
reallocate the bead suspension from the parent vessel to the reaction vessels;
-
~UBSTlTUrE SHEET (RULE 261
~ wo gS/12608 2 i 7 5 5 ~ 7 PCT/US9.1/123~7
Fig. 20 shows th~ steps taken by the control computer to fill the reaction
vessels with reagents from the ~ cd delivery system;
Figs. 21A-21D srhpmqrir~lly illustrate the steps taken by the control
computer to fill the reaction vessels with reagents from the ,ul~ cd delivery system;
Fig. Z2 shows the steps taken by the control computer to introduce amino
acid monomers into the reaction vessels;
Figs. 23A-23C 5rhPmq~irqlly illustrate the steps taken by the control
computer to introduce amino acid monomers into the reaction vessels;
Fig. 24 shows the steps taken by the contro', computer to transfer the bead
suspension from the reaction vessels to the parent vessel for mixing;
Fig. 25 ~rhPi~ir~lly illustrates the data flow among the major modules of
the control software;
Fig. 26 srhpm~rirqlly illustrates the command interpreter structure;
Fig. 27 crhPrrq~ir~lly illustrates the scheme used to access valve data; and
Fig. 28 crhPm~tir~lly illustrates the scheme used to access sensor data.
Fig. 29 shows a device for ~yilLll. ~;~;llg cu.,.l, I chemica', libraries on
IlI;~lUi~.Ui~;C beads. The device is composed of a vacuum manifold or magnetic plate
attached to a solid substrate having a synt,~,esis surface having an arr~,y of reaction sites at
which cù...l~uu~ can be ~yl-L~ cd. The partition block is composed of an array of
20 reaction wells Cullc~uul..'i;.lg to said reaction sites and is used to partition library members
after each mixing step. The device can a-,so be used to aid the synthesis of tagged
chemica-, libraries.
Fig. 30 illustrates the use of 13C NMR to monitor the stability of
ll,;~ .,li.l;"~ . . Panel C shows the 13C NMR spectrum of support bound i .
25 which has been doubly labeled with a 13C atom at the position 2 of the ring and at the
position a'ipha to the carbonyl of the ,inker (labeled positions are indicated with a "*").
Panel B shows the 13C NMR spectrum of support-bound doubly labeled rhi~7r,li~1inn~nP after
treatment with 95% TFA for 1 hour. Panel A shows 13C NMR spectrum of support bound
doubly labeled II.;r ~ ..J ~ after 40 cycles of DNA synthesis.
Fig. 31 further i,lustrates the use of 13c NMR to monitor the stability of
thi~7~ nl-nPc Panel C shows the 13C NMR spectrum of support-bound thi~7r.1i~' -
which has been doubly labeled with a 13C atom at the position 2 of the ring and at the
position aipha to the carbonyl of the linker (labeled positions are indicated with a "*").
2 1 7 5 5 8 7
WO 95/12608 ` PCT/US9.1/123~7
12
Panel B shows the 13C NMR spectrum of support-bound doubly labeled thi:;7~1ifiin~nP after
90 minute photolysis in PBS buffer. Panel A shows 13C N~IR spectrum of support bound
doubly labeled ~ l;,-.",f after 3 hours of photolysis in PBS buffer.
Fig. 32 shows an HPLC trace for the reaction mixture produced by
S subjecting a support-bound ~hi~7f~ innnf to 40 cycles of DNA synthesis and 3 hour
photolysis in PBS buffer.
Fig. 33 illustrates a graphic user interface ("GUI") as ;,,,~ ,t -~ on the
control computer. As shown, the GUI includes a rect~ngular window 1301 with a
workspace 1303. At the top of the window is a menu bar 1305 with user command
choices 1306-1313. Each of these command choices include additional submenus forcontrolling the operations of the synthesizer. A user can program the synthesizer by
selecting the ~ " command choice with the mouse.
Fig. 34 depicts the GUI, showing the submenu options in the Macro menu.
Fig. 35 depicts the dialog box for running a macro.
Eig. 36 depicts the GUI, showing the submenu options in the Groups menu.
Fig. 37 depicts the GUI, showing the submenu options in the Variables
menu.
Fig. 38 depicts the GUI, showing the submenu options in the Diagnostics
menu.
Fig. 39 depicts the valve diagnostic screen.
Fig. 40 depicts the sensor diagnostic screen.
Fig. 41 depicts the GUI, showing the submenu options in the File menu.
Figs. 42-44 show the dialog boxes involved in setting up a synthesis,
allowing the user to select the reaction vessels to be used in the synthesis (Fig. 42), select
the start, loop and end macros (Fig. 43) and enter the amino acid symbol and
olig~,,, " l~,l;,l~ code for each reaction vessel (Fig. 44).
Fig. 45 depicts the GUI, showing the submenu options in the Synthesis
menu.
Fig. 46 depicts the status display during a synthesis or macro execution. ,-Fig. 47 depicts the User Abort dialog box.
Fig. 48 depicts the GUI, showing the submenu options in the Edit menu.
Fig. 49 5~hPm~ti~ y illustrates the data flow among the major modules of
the control software in a Windowsn' environment.
WO95112608 587 PCTIIlS91/123.17
DESCRIPTION OF THE SPECIFIC EMBODIMENTS
The present invention relates generally to improved methods for generating
and screening tagged chemical libraries. The present invention also relates to a device
useful in synthesizing collections of diverse molecules, such as the described tagged
5 chemical libraries.
To appreciate the value of the improved methods, one must understand not
only the basic mr-thnrlr~logy for making and using tagged libraries but also how the various
steps of synthesis and screening interact and how the selection of reagents impacts the
results achieved. Tagged chemical libraries are often ~y~ iLcl on a solid support, and
10 the choice of support and linker is critical to success. A linker can be used to attach the
support to the tag, to attach the support to a library molecule, or, in an . .,.l-o.l;, where
there is no solid support, to attach the tag to a library molecule. The choices relating to
chemical building blocks, tags, and synthesis methods can be ef~ually critical and are also
impacted by the nature of the solid supports and linkers available. The assays and
15 ~ dLi~l~s for which the tagged libraries are intended also impact these choices, as well
as the il~ LdLiul~ and reagents available.
Although the apparatus and methods of the present invention are illustrated
primarily with regard to the synthesis of oli~ c and peptides, the invention is not
so limited. The invention will find application in the synthesis of materials such as
20 pol~ f ~ lCI~ r~ UI~LIIcLl~ f i, ~ , and beta
turn mimetics, and other materials. Cyclic materials may be formed as disclosed in U.S.
Patent No. 5,242,974 (Holmes), i.,.~ dL~d herein by reference.
The use and synthesis of diverse materials such as .~ c and
peptides is disclosed in further detail in the following copending applications, which are
25 i~c~ )oldl1d herein by reference for all purposes: U.S. Application Seria~ No.
07/876,792, filed on April 29, 1992; U.S. Application Serial No. 07/762,522, filed on
September 18, 1991; and U.S. Application Serial No. 07/946,239, filed on September 16,
1992.
The description of the invention is provided as indicated by the following
30 outline:
OUTLINE
I. Overview of a Synthesis of a Tagged Chemical Library
-
2 1 755~7
WO 9~;/12608 PCTI~S9~/123~7
14
Il. The Solid Support
A. Types
B. Linkers
C. Molecular Supports
S III. The Chemical Building Blocks
A. Oligomers and Monomers
B. Other Building Blocks
IV. The Tag
V. Synthesis Methods
A. Ol j~""~ Tagged Peptide Libraries
B. Improved Method for SyllLll.,;Lillg Oligonucleotide-
Tagged Peptide Libraries
C. Small Molecule Synthesis
D. Methods for Generating Soluble Libraries
15 V~I. Assay Methods
A. Screening Assays for Bead-based Libraries
B. Screening Soluble Molecules
C. Screening Natural Product Libraries
VII. I,l~Llu,,,~llLG~ion and Reagents
20 VlII.Apparatus for Parallel Coupling Synthesis Redctions
Examples
End of Outline
In addition to the outline abûve, the following glossary is provided to
25 facilitate the description of the invention, and a number of abbreviations and terms are
defined to have the general meanings indicdted as used herein to describe the invention.
Abbrevi~ n~ HBTU, O-QJ~,IlLULlilLOI-I-yl)-l,1 ,3,3-tetramethyluronium
dnuu~ u~ HOBt, l-hy~ u~.y~ HATU, [0-(7-dL~b~.LUll;dLul-l-yl)-
1,1,3,3 t~.ldUII~lllylUI~ n"~ r' ', TFA, L inuul~ua~Lic acid; TCA,
30 trichloroacetic acid; DIEA, dii~u,ulu~ylc;Lllyldlllill~, DMF, dimethylformamide; Fmoc,
9-fluorenylmethyloxycarbonyl; DMT, dimethoxytrityl; Trt, trityl; Bz, benzoyl; Pmc,
2,2,5,7,8-pentamethylchroman-6-sulfonyl; 'Boc, rer~-butyluAy, dubu--yl; PBS,
I ' ,' buffered saline; BSA, bovine serum albumin; mAb, mnno~ l antibody.
~ wo 9~/12608 2 ~ 7 5 5 8 7 Pcrluss~ll23~7
C~ f. ~ rl~ or Allhctf~nti~lly ~v~ llrll~ . These terms refer to the
ability of one compound to bind to another, e.g., as a ligand binds to its c,~,l,l,l.. ~I ~y
receptor. Typically, these terms are used in connection with a description of base pairing
between nllrlPntiflPc of nucleic acids, such as, for inshnce, between the two strands of a
5 double stranded DNA molecule or between an nli~nnllrlPoti~p primer and a primer binding
site on a single stranded nucleic acid to be sequenced or amplified. "C~ ,l. .llrlll- y"
: ' are, generally, A and T (or A and U), and C and G, but there are a wide
variety of synthetic or modified "". lr.JIif'f; with binding properties Icnown to those of skill
in the art. "Subshntial ccmnrlf.~ ;ly" exists when an RNA or DNA strand will
10 hybridize under selective hybridization conditions to a ~ ,lf.~r~ ,y nucleic acid.
Typically, llyblidi~Liu~ will occur when there is at least about 55% 1 ~",,1,1 ...... ~1 .l ily over
a stretch of at least 14 to 25 I,,,,I..ll;rlf c but more selective hybridization will occur as
CUII.I.I .1- I~ y increases to 655~o, 75%, 9û%, and 100%. See Kanehisa, 1984, ~Çl.
~ Res. 12:203, illCul,~)l ' ' herein by reference. Highly selective l~yblidi~Li
15 conditions are Icnown as "stringent hybridization conditions", defined below.Epitope: This term is used to describe a portion of an antigen molecule
delineated by the area of interaction with the subclass of receptors Icnown as antibodies.
IdPntifiPr t~ In the most general sense, this term is used to denote a
physical attribute that provides a means whereby one can identify a chemical reaction, such
20 as a monomer addition reaction an individual solid support has ~ PCI in the synthesis
of an oligomer on that solid support. The identifier hg serves to record a step in a series
of reactions used in the synthesis of a chemical library. The identifier hg may have any
rPcog~i7~hlP feature, including for example: a microscopically or otherwise ~ 1;. g,.;~ ,lf
shape, size, mass, color, optical density, etc.; a differential dbSVl~ G or emission of
25 light; chemically reactivity; magnetic or electronic properties; or any other distinctive
mark capable of encoding the required i,~rvl,lld~iull, and llf ;1~ at the level of one
~or a few) molecules. A preferred example of such an identifier tag is an f~l,g~"..fl. Jl;flf,
because the nucleotide sequence ûf an l.li~ lf,JL;~l~P is a robust form of encoded
;,lr..,... l;.~" An "identifier tag" can be coupled directly to the oligomer ~ .iL~d,
3û whether or not a solid support is used in the synthesis. In this latter ...,1,.,.1;,.,...1 the
identifier tag can ccmrPrtl~lly be viewed as also serving as the "support" for oligomer
synthesis.
WO 95112608 2 ~ 7 5 5 8 7 PCT/US9~1123.17
L~: This term is used to denote a molecule that is recogni~ed by,
typically by binding to, a particular receptor. The agent bound by or reacting with a
receptor is called a "ligand", a term which is ~iPt`initionqlly meaningful only in terms of its
~UUIIt~ UI receptor. The term "ligand" does not imply any particular molecular size or
5 other structural or ~.~."~ 1 feature other than that the substance in question is capable
of binding or otherwise interacting with the receptor. Also, a ~ligand" may serve either as
the natural ligand to which the receptor binds, or as a functional analogue that may act as
an agonist or antagonist. Ligands that can be investigated by this inYentiOn include, but
are not restricted to, agonists and antagonists for cell membrane receptors, toxins and
10 Yenoms, viral epitopes, hormones, sugars, cofactors, peptides, enzyme substrates,
cofactors, drugs (e.g., opiates, steroids, etc.), and proteins.
Monomer: This term is used to denote any member of a set of molecules
that can be joined together to form another molecule or set of molecules, such as a set of
oligomers or polymers. Sets of monomers useful in the present invention include, but are
15 not restricted to, for the example of peptide synthesis, the set of L-amino acids, D-amino
acids, or synthetic amino acids. As used herein, "monomer" refers to any member of a
basis set for synthesis of an oligomer. For example, dimers of L-amino acids form a basis
set of 400 "ll~olloll~ " for synthesis of polypeptides. Different basis sets of monomers
may be used at successive steps in the synthesis of a polymer. Those of skill in the art
20 will recognize that a "monomer" is simply one type of "chemical building block" and that
any type of chemical building block can be employed in the present method, regardless of
whether one is synthesizing an oligomer or a small organic molecule or some other
molecule.
Oli.~omer or Polymer: These terms are used to denote molecules that are
25 forrned by a process involving the chemical or enzymatic addition of monomer subunits.
Such oligomers include, for example, both linear, cyclic, and branched polymers of
nucleic acids, poly~ hqri~lr~ pt.~ and peptides having either alpha-, beta-, or
omega-amino acids, hct~luuolyll,~l~, polyul~LI,~ , polyesters, polyu~ubul,~t~, polyureas,
pol~lll";J~, polyethylrn~imin~ polyarylene sulfides, polysiloxanes, polyimides,
30 polyacetates, or other polymers, as will be readily apparent to one skilled in the art upon
review of this disclosure.
Pe~tide: This term is used to denote an oligomer in which the monomers
are alpha amino acids joined together through amide bonds. A "peptide" can also be
Wo 9S112608 2 ~ 7 5 5 8 7 PCTlUS9~ill23~7
17
referred to as a "polypeptide." In the context of this invention, one should appreciate that
the amino acids may be the L-optical isomer or the D-optical isomer. Peptides are more
than two amino acid monomers long, but more often are more than 5 to 10 amino acid
monomers long and can be even longer than 20 amino acids, although peptides longer than
5 20 amino acids are more likely to be called "polypeptides." Standard single letter
.~ abbreviations for amino acids are used (e.g., P for proline). These abbreviations are
included in Stryer, Bio~ liaLly. Third Ed. (1988), which is i,.~o.~o.~L~ herein by
reference.
Oli~ vl;d~s This term is used to denote a single-stranded DNA or
10 RNA molecule, typicaLIy prepared by synthetic means. Oliennl~rlpoti~ipc employed in the
present invention will usually be 50 to 150 nllrlPotir;Ps in length, preferably from 80 to
120 nl~rlPnîi~iPc although r,li~nnllrlP~ti iPc of different length may be .IL.~,u~ L~ in some
For instance, an olig.~ PI~I;fiP tag can be built nucleotide-by-nucleotide
in coordination with the monomer-by-monomer addition steps used to synthesize the
15 oLigomer. In addition, very short, i.e., 2 to 10 rlllrlPnti~i~c~ nlig~.,..l. l.'.~l;~iPC may be used
to extend an existing .~i;~...,..,l~.,li~P tag to identify a monomer coupling step. Suitable
oligrnllrlt oiiriPc may be prepared by the IJI .~L.llUI"".; iitP method described by Beaucage
and Carruthers, 1981,1~51. Lett. _: 1859-1862, or by the triester method, according to
Matteucci et al., 1981, I. Am. ~m. Soc. 103:3185, both il~u~L~u~LPd herein by
20 reference, or by other methods such as by using commercial automated r,l;~" ~ P
Operably linked: This terms refers to a functional l~.L.lic.,l~l,iLJ between onesegment of a nucleic acid and another. For instance, a promoter (or enhancer) is"operably linked" to a coding seouence if the promoter causes or otherwise positively
25 influences the l..,..~ .L;~ of the coding sequence. Generally, operably linked means that
the nucleic acid segments or sequences being linked are contiguous and, where necessary
to join two protein coding regions, contiguous and in reading frame.
p~r~llPI Cou~lin~: This phrase refers to the ~i""lll~ coupling of two
building block ~ u.~ to separate distinct points on a substrate. Such a substrate may
30 be a solid support having distinct groups to which these building blocks are attached, or
may constitute another chemical compound possessing two distinct groups where each
building block ;~ ..lly attaches. Sim~lt~nl OllC coupling refers to the coupling of two
c~ o~ c to such distinct points prior to the addition of a new building block compound
WO 95/12608 2 T 7 ~ 5 8 7 PcTlu59~l1123~J7 ~
18
to the first coupled building block. Thus, the term "cimlllt~n~n~ as used in this context
is not strict in the sense that the two building blocks are coupled with precise ~ rlll e.
Rece~tor: This term refers to a molecule that has a specific affinity for a
given ligand. Receptors may be naturally occurring or synthetic molecules. Receptors can
5 be employed in their unaltered natural or isolated state or as aggregates with other species.
Receptors may be attached, covalently or noncovalently, to other sllhctAnrpc Examples of ,.
receptors that can be employed in the method of the present invention include, but are not
restricted to, antibodies, cell membrane receptors, mnnnrlnnAl antibodies, antisera reactive
with specific antigenic .Irlrl,...,.- ,l~ (such as on viruses, cells, or other materials),
10 ~oly.,... 1~ C, nucleic acids, lectins, poly~ C, cells, cellular ~ U~ S~ and
organelles. Receptors are also known as "anti-ligands." A "ligand-receptor pair" is
formed when two molecules, typically Ill,~ ""nl~. "lPc, have combined through molecular
I~U~ iUl. to form a complex. Other examples of receptors include, but are not restricted
to specific transport proteins or enzymes essential to survival of l";..luùl~;Auli",.s for which
15 antibiotics are needed; the binding site of any enzyme; the ligand-binding site on an
antibody molecule; a nucleic acid; a catalytic polypeptides as described in Lemer et al.,
1991, Science 252: 659, incorporated herein by reference; and hormone receptors such as
the receptors for insulin and growth hormone.
Substr-AtP or Solid Support: These terms denote a material having a rigid or
20 semi-rigid surface. Such materials will preferably take the form of small beads, pellets,
dislcs, or other convenient forms, although other forms may be used. In some
clllbùdilllcllL~, at least one surface of the substrate can be sllhctAnti~ y flat. A roughly
spherical shape is preferred.
Strineent llylJli~ iul~ conditions: This phrase refers to highly selective
25 hybridization conditions in which nucleic acids remain stably bound in association with
other nucleic acids (or other segments of the same nucleic acid) only if the associated
sequences are perfectly or highly (i.e., greater than 80%) ~u..,l~l ,.,. .llAly. Such conditions
typically include salt rr~nrPntrltinnC of less than about I M, such as less than 50û mM, and
will often include salt ..~ ;n.~c of less than 200 mM. The hybridization l~,lll~ldLulc ,-
30 for oligomers will typically be greater than 22 C, such as greater than about 30'C, andwill often be in excess of about 37 C. Longer fragments may require higher hybridization
t~ Lulcs for specific hyhririi7AAtinn As other factors may dramatically affect the
stringency of hybridization (such factors include base ~I.Ull}lO~i~iOl~, length of the
~ wo 95/12608 1 7 5 5 8 7 PCT/US9~/12347
.y stlands, presence of organic solvents, and extent of base mi~m~rhin~), Lhe
~u".l,~ ;.." of factors is more important than the absolute measure of any one factor
alone.
Synthetic: A compound is "synthetic" when produced by in vitro chemical
5 or enzymatic synthesis. The synthetic libraries of the present invention may be contrasted
with those in viral or plasmid vectors, for instance, which may be ~ ' in bacterial,
yeast, or other living hosts.
I. Overview of the Synthesis of a Tagged Chemical Library
The present invention relates generally to methods for ~ -Lh~;,iLil-~ and
screening tagged chemical libraries. In essence, each "book" of a chemical library of the
invention consists of a chemical or molecule of interest, a tag identifying the chemical or
molecule of interest or some important aspect thereof, and a linkage between the chemical
15 or molecule of interest and the tag. In one important ~mho~iiml nt~ the chemical or
molecule of interest is an oligomer such as a peptide, the tag is an oligomer such as a
nucleic acid, and the linkage is a solid support or particles, from which oligomers and tags
may optionally be cleaved, e.g., to facilitate detection or to provide a soluble library.
Such libraries can be screened to isolatc individual oligomers that bind to a receptor or
20 possess some other desired property. A general method for producing a tagged chemical
library is illustrated by the production of a large, highly diverse collection of oligomers, in
which each different library member is an oligomer with a unique monomer sequence
relative to other library members (although the library will typically comprise duplicate
"books"). Such a library or collection may contain, for example, all ~ ;.."c of X
25 different monomers in a set of monomers assembled into length n oligomers yielding, X'
different o"~l,u,. ~1~ The collection may also contain oligomers having different
monomer units at, for example, only one or a small number of positions, while having an
identical sequence at all other positions.
A general method for syllLl~ such collections of oligomers typically
30 involves a random ,.~",1.;",.1...; ~l ("stochastic") approach and the chemical andtor
enzymatic assembly of monomer units. One process comprises the steps of: (a)
dl"JulLi~..i.~g a plurality of solid supports among a plurality of reaction vessels; (b)
coupling to the supports in each reaction vessel a first monomer and a first tag using
2i 75587
Wo gS/12608 PCTIIIS9~/12
different Flrst monomer and tag ,o...l,i"~iu-.~ in each different reaction vessel; (c) pooling
the supports; (d) ~IUUUlLiUllillg the supports among a plurality of reaction vessels; (e)
coupling to the first monomer a second monomer and coupling to either the solid support
or to the first tag a second tag using different second monomer and second tag
5 ~ c in each different reaction vessel; and optionally repeaùng the coupling and
ulLullillg steps with different tags and different monomers one to twenty or more times.
Typically, sl~hcrSm~ y equal numbers of solid supports will be ~yyulLiull~d to each
reaction vessel. Those of skill in the art recogmze that the same chemical building block
can be employed in different coupling steps and that the same chemical building block can
10 be employed in more than one coupling reaction (reaction vessel) of a single coupling step.
To visualize the method more readily, one might first consider the stochastic
synthesis of an untagged library of all oligomers three residues in length, assembled from a
monomer set of three different monomers: A, B, and C. Three aliquots of beads are
.I,Up~ iUII~ among three reaction vessels, and monomer A is coupled to the beads in the
15 first reaction vessel, B is coupled in the second, and C in the third. The beads from all
the reaction vessels are then pooled. The pool contains ~ lu,.illl..t~ly equal numbers of
three different types of beads, with each type . ~ by the monomer coupled to thebead. The pool is mixed and l~ I to the separate monomer reaction vessels, each
containing A, B, or C as the ne~t monomer to be coupled~
2û Following this coupling reaction, each reaction vessel now has beads withall three different monomers in position one and the monomer contained in each particular
second reaction vessel in position 2. All beads are pooled again, producing a mixture of
beads each bearing one of the nine possible dimers. The pool is again distributed among
the three reaction vessels and coupled to the three different monomers, producing the
complete set of all trimers of the three monomers (33 = 27). As can be readily
-luy~ L~I, the use of a ~u~rc;~-lLly large number of synthesis beads helps to ensure that
the æt completely represents the various comhin~irmc of monomers employed in this
random, stochastic, ~ ";~l synthesis scheme.
Mo.l;r. ~ of this completely random approach are also possible. For
3û example, the monomer æt may be expanded or contracted from step to step; or the
monomer set could be changed completely for the next step (e.g., amino acids in one step,
~' ~ ' in another step, ,~bollydl_'~,, in another step), if the coupling chemistry were
available. A monomer unit for peptide synthesis, for example, may include single amino
~ wo g~/1260g 2 ~ 7 5 5 8 7 PCTIUS9~1123~7
21
acids or larger peptide units, or both. One variation is to form several pools of various
sequences on solid supports to be distributed among different monomer sets at certain steps
of the synthesis. By this approach, one can also build oligomers of different lengths with
either related or unrelated sequences, and one can fix certain monomer residues at some
5 positions whi~e varying the other residues, to construct oligomer r.d..l~ ~.Jll~ wherein
certain residues or regions are altered to provide diversity.
The synthesis of a tagged chemical library often involves such . '
synthesis steps. Because the identifier tag can be easily decoded to report the identity of
each oligomer, however, tagged chemical libraries can be cienifi~ntly larger and more
10 complex than untagged libraries. In fact, the present methods for syllLI~ g encoded
synthetic libraries of ~u,.~l,uu~ makes possible the screening of large collections of
non~ u. ~1,1~ ~ul~lpuullJs produced by multi-step synthesis.
In particular, the use of oliennllrlPo~ p tags and olignnllrlpori~1p encryption
provides a powerful nn.~h~nicnn for recording the structural identity of every member of
15 vast library of tethered co"~ull-~J~, especially peptides, generated through a
synthesis. The methods are broadly applicable to encoding the ~u~ l assembly of
other non-peptidic structures, providing the parallel synthetic schemes remain orthogonal
and comr~ihlP The net outcome of a culllb;llalulidl synthesis is ~ ly def~ned
only for a sequence of reactions that each proceed in very high yield to afford single
2û products. This situation is ~ u~Jlll~t~J with standard peptide and DNA synthesis
, and the resulting product structures are explicitly specified by the order of the
building blocks andtor coupling reactions used in the synthesis.
However most synthetic organic reactions are more idiu~ LLi-, giving
variable yields and frequently multiple products (such as regio- and st~l~
25 structures). Using such chemistry to synthesize ~",~);",.1",;~l libraries on solid supports
yields a mixture of products on each bead in the library. In the most general case, the
encryption of a synthesis may not uniquely specify the chemical structure of an associated
entity. Rather, the encryption process may more accurately be viewed to encode the exact
synthetic protocol (e.g., reagents, reaction conditions, etc.) by which a member of the
30 library was constructed. The library is screened to identify "active recipes" that then can
be ~c~luJuucd on a plc~ Livc scale and r.~. I;n..,-t .1 (if necessary~ to isolate the bioactive
~iclll~iln~ (s)~ The encoded library ~Prhn~ giP~ have c~nci~lpr~hle potential to expand the
Scope of ~ul~ Luli~l chemistry and its applications to drug discovery and the
WO 95112608 2 i 7 5 5 8 7 PCTIUS911123.~7 ~
dc~lv~ and isolation of a wide variety of useful ~ Jr~ With this overview of
the synthesis of tagged molecular libraries, one can better appreciate important aspects of
the invention, such as the use and choice of solid supports in library synthesis.
5 II. The Solid Support
A. Types
Typically, the tagged chemical libraries of the invention are composed of a
collection of "solid supports", such as beads or particles. Such solid supports may be of
10 any shape, although they will preferably be roughly spherical. The supports need not
necessarily be l~ r.". ,.- ~ in size, shape, or romr~Cit;rm although the supports usually
and preferably will be uniform. In some e...l~oJi~ , supports that are very uniform in
size may be particularly preferred. In another ..~ , however, two or more
distinctly different p,.l~ul-';",.~ of solid supports may be used for certain purposes, i.e., the
15 solid supports may be composed of a single particle, or two or more linked particles.
Solid supports may consist of many materials, limited primarily by capacity
for derivatization to attach any of a number of chemically reactive groups and
comr~tihility with the chemistry of oligomer or other molecular synthesis and tag
:Itt~hmr-n~ Suitable support materials include glass, latex, heavily cross-linked
20 polystyrene or similar polymers, gold or other colloidal metal particles, and other materials
known to those skilled in the art. Except as otherwise noted, the chemically reactive
groups with which such solid supports may be derivatized are those commonly used for
solid state synthesis of the respective molecule or oligomer and thus will be well known to
those skilled in the art. The term "solid support" as used herein embraces a particle with
25 d,u~luulid~e sites for oligomer synthesis and, in some ~."I,orli",. ~ , tag attachment and~or
synthesis. There are various solid supports useful in ~lC~dUdLiUII of the synthetic oligomer
libraries of the present invention. Solid supports are commonly used for solid phase
synthesis of, for example, peptides and nucleic acids and other oligomers as;
above, and thus are well known to those skilled in the art. The solid supports of the '
30 present invention do not include living cells, viruses, or cloning vectors such as phage
vectors or plasmids. Mon~bP~icTY (commercially available from Pharmacia Fine
Chemicals AB, Uppsala Sweden) or their equivalent, are udlLicul~ly useful as solid
supports for the various aspects of the present invention. MonobeadsTY provide good size
~ wo 95/1261~8 2 1 / 5 5 8 7 PCT/US9~/123.f7
I-f ~ ~,. Iy and a small size, 10~m. Further, these MonobP~srA do not clump in eitber
organic or inorganic solvents, and provide a suitable support for both olieon~clf-otiflP and
peptide synthesis. Finally, M.. ~ provide very high loading of primary amines (100
nmolelmg).
S One important aspect of the particular solid support chosen for practicing the
invention is the size of the support. With enough solid supports and efficient coupling,
one can generate complete sets of cert,~in oligomers, if desired. In general, the solid
support size is in the range of 1 nm to 100 ~m, but a more massive solid support of up to
1 mm in size may sometimes be used. The ~ IUIJl;..'~, size of the solid support depends
10 on (1) the number of oligomer synthesis sites and identifler tag attachment sites desired;
~'2) the number of different ~ u l~ to be ~yl~ J (and the number of solid supports
bearing each oligomer that are needed for screening); and (3) the effect of the size of the
solid supports on the specific screening strategies [e.g., nuulc.. ~.~c~-activated cell sorters
~'FACS)] to be used.
As a specific example, solid supports of 1 ~m in diameter may be used in
the method. If each reaction vessel contains ~ 'y 0.2 mL of solid supports, and
the oligomers are ~yllLII~;~ from a set of 50 monomers (50 parallel reactions), then a
total of 10 mL of solid supports, or .,~ / 10~3 solid supports, would be re~uired.
If one wishes to make hexamers with these 50 rf~nnnmpr~ then there are over 1.5 x 10
20 possible se~uences, and each specific se~uence would be ~ t~ on about ld3 solid
supports. An estimated capacity of each bead, based on the capacity of commonly used
peptide syllth~.~iJ.g resins, is about 0.1 pg of peptide per bead. By this estimation, then,
each solid support would have about 100 amol or 10~ oligomer chains.
To improve washing . r,~ r - C, one could employ nonporous beads or5 other solid supports less porous than typical peptide synthesis; however, for certain
of the invention, quite porous beads or resins work well and are often
preferable. Nonporous supports will have a lower density of growing chains, but even
with a decrease in ~apacity of several orders of magnitude, sufficient oligomer densities
'~ can be produced for efficient screening. With the less porous supports, a greater
30 proportion of the oligomers will be accessible for binding to the receptor during the
screening process. Also, the less porous supports will reduce the carryover of tags from
one reaction to the next, thus improving the accuracy of reading the dominant (correct)
tags.
WO 95112608 2 ~ 7 5 5 ~ 7 PCT/US9-11123-17
24
As noted above, another . ,l-o~ involves the use of two solid supports,
such as beads, that are physically linked together, one with synthesis sites (or linkers) for
the molecule or oligomer and one with attachment sites (or linkers) for the identifier
tag(s). This ..",~ allows the sc~;,C~"lLiul~ of molecules or oligomers and identifier
tags into discrete "zones" and permits the use of widely different chemically reactive
~roups and nhpmic~rips for :~t~rhmrnt The solid supports can be derivatized sepaMtely
and then linked under conditions where all or nearly all of the synthesis solid supports will
have a tag-attachment solid support in tow. The solid supports can be of different sizes, as
for example a large synthesis bead with several (or many) smaller tag-attachment beads
linked. In one ~ o~ the first solid support will have at least one attached amino
acid and the second solid support will have at least one attached nucleotide.
The mode of linking the two beads is constMined by the chemistry of
oligomer synthesis. The most obvious means of linking the beads is with a
h~.t~ 1ubirullctional cross-linking agent (for examples of such agents, see Pierce
ImmunoTechnolo y Catalo~ and Handbook pp. E10-E18 (1991)) inteMcting with the
dominant chemically reactive groups on each species of solid support. Such cross-linking
agents can serve a variety of purposes, as indicated by the following section.
B. Linkers
When bound to a solid support, the oligomer and its associated tag arc
usually attached to the support by means of one or more molecular linkers. The linker
molecule, prior to ~t~rhm~n~. has an a~ lUp functional group at each end, one group
a~ Upl for attachment to the support and the other group àplJ1UIJ1id1C for attachment to
the oligomer or tag. In some ~ o~l;,.,. .11~, cleavable linkers will be used to facilitate an
assay or detection step.
Given the wide availability of diverse linking reagents, one can link the
identifier tags either to the oligomer or other libMry compound of ;nterest or to the solid
support or to a pre-existing tag. For instance, the identifier tag may be attached to a
monomer il1- u1~Ju1~cd into an oligomer or to a building block illCUI~l ' ' into a ,'
non-oligomeric compound. For peptidic oligomers, the side chain of a cysteine residue
provides a convenient site for tag ~t~f~hmrnt In other instances, the tag could even be
attached so as to cap a small number of the oligomer chains, providing the decreased
amount of net synthesis of the desired oligomer could be readily tolerated. One can attach
~ WO 95/12608 2 ~ 7 5 5 8 7 PCT/US9~1123J7
the tag directly to the linker that binds the oligomer (or other compound of interest) to the
solid support. In this ~ bodi~ , the linker has, pnor to ~tt~rhmPnt a third functional
group dyyLUylidt- for the attachment of the identifier tag.
One can of course illCuluOIcl~ a wide variety of linkers, depending upon the
5 application and effect desired. For instance, one can select linkers that impart
llydluyllobi--iLy~ hydrophilicity, or steric bulk to achieve desired effects on properties such
as coupling or binding efficiency. In one aspect of the invention, branched linkers, i.e.,
linkers with bulky side chains such as the linker Fmoc-Thr(tBu), are used to provide
rigidity to or to control spacing of the molecules on a solid support in a library or between
10 a molecule and tag in the library.
As noted abûve, cleavable linkers can be employed to useful effect.
Preferred photocleavable linkers of the invention include 6-nitrov~ lyluAy~l,ullyl
(NVOC) and other NVOC related linker .~ u~ (see PCT patent publication Nos. WO
90/15070 and WO 92/10092; see also U.S. patent application Serial No. 9?1,181, filed 2
15 Nov. 1992, ill~ ulyOI.ll~d herein by reference). In another ~ budil..e~L, the linkers are
nucleic acids with one or more restriction sites, so that one portion of a library member
(either the tag, the oligomer or other compound of interest or both, or the solid support)
can be selectively cleaved from another by the dyylUyli,lL~ restriction enzyme. This novel
nucleic acid linker illustrates the wide variety of linkers that may be employed to useful
20 effect for purposes of the present invention.
C. Molecular Supports
As noted above, the invention can also be carried out in a mode in which
there is no solid support, and the tag is attached directly (typically through a linker) to the
25 oligomer or other molecule being ~yllL~ ~. Alternatively, the oligomer or otber
molecule and its associated tag can be ~yllL~ on a solid support and then cleaved or
otherwise removed from the solid support prior to screening or other use. Such methods
are described more fully below. Regardless of whether a solid support is present, the size
~ and . u.. ~ .,. of the library will be determined by the number of coupling and mixing
30 steps and the monomers or other building blocks used during the synthesis.
WO 9S112608 2 1 7 5 5 8 7 pCT~lJss~llu~7 ~
26
m. The Chemical suilding Blocks
A. Oligomers and Monomers
The wide applicability of the present inventions is perhaps most readily
S grasped by ~ ;"~ the synthesis and screening of large libraries of diverse oligomers
and polymers. Oligomers are polymeric c.""l,u,~ composed of monomers; for
biological polymers, the sequence of the monomers in an oligomer often specifiesimportant biological properties. Preferred oligomers of interest include peptides,
olig.."l, ~ , oligo N-substituted glycines, and poly~,~ul,d~dL~. As noted above, for
10 purposes of the present invention a monomer is any member of a set of molecules that can
be joined together to form an oligomer or polymer, i.e., amino acids, carbamates,
sulfones, sulfoxides"l ~ ,dlbOllydldlc~ ureas, ~ , lipids, esters,
- c~ of the same, and the like. Thus, the monomers may be of any type that can
be ~ lUl~lidt~,ly activated for chemical coupling or accepted for enzymatic coupling.
This method of assembling oligomers from many types of monomers
requires using the d~ u~ L~ couFling chemistry for a given set of monomer units or
building blocks. Any set of building bloctcs that can be attached to one another in a
step-by-step fashion can serve as the monomer set. The attachment may be mediated by
chemical, enzymatic, or other mcans, or by a cnmhin~tinn of any of these means. The
20 resulting oligomers can be linear, cyclic, branched, or assume various other cullrullllaLiulls
as will be apparent to those skilled in the art.
B. Other Building Blocks
The invention is described herein primarily with regard to the lu-c~ -Liu-~ of
25 molecules containing sequences of amino acids, but the invention can readily be applied to
the ~ J~dLiu.~ of other oligomers and to any set of cnmrolln~c that can be ~YII~ ;L~ in
a component-by-component fashion, as can be ~ idt~ by those skilled in the art. For
instance, rnmro~n~lc such as t.. ~ 5, hydantoins, and peptidy'~' .' can
be prepared using the present methods (see U.S. patent application Serial No. 08/119,700, ,'
filed 9 Sept. 1993, which is a n nn~in~l~tinn-in-part of Serial No. 081,577, filed 21 June
1993, now ~h~n~lnnPri, which is a cnntinll~tinn-in-part of U.S. Patent No. 5,339,115, each
of which is i~l~,ul~u~dl~d herein by reference.
WO 9S/12608 L~ 1 7 5 5 8 7 PCTIUS9~/123 17
In one embodiment, the present method can be used to create libraries of
branched polymers. While in many instances libraries of lincar polymers, such aspeptides, are quite useful, with more than 3-4 residues, the shape of these linear molecules
becomes long and narrow. Most drugs do not have such an extended shape, perhaps due
5 in part to the high degree of flexibility of the molecules. Branched backbone polymers can
result in molecular shapes similar to known drugs. Thus, in one ~ bodilll~,.lL, the present
invention relates to the incorrJoration of monomers with at least three functional groups to
which other monomers ca~n be attached.
If one uses such monome}s exclusively, however, then the fully branched
10 synthesis will always result in a high ratio (relative to the other monomers used in the
synthesis) of the last monomer coupled. One could of course illuul~uldlr mixtures of
different branching monomers to alter this ratio, but then one might have more difficulty in
identifying the structure of a compound of interest, i.e., the more complex the mixture of
branched mt-nrnn~rC, the less i.lrull~ iull the tag may provide about the particular
15 compound syllLllQiL~I. In an improved method of the invention, one il~,Ul~lUI...I~ a
mixture of two monomers -- one capable of branching and one not -- at each monomer
coupling step, producing a library COIll~ g a great diversity of shapes with highly
informative tags. In this case, the tag would specify the monomers present at each
coupling step but not whether the monomer was capable of branching. However, a simple
20 resynthesis using only those monomers contained in the selected set of ~ u~ ,u .1~ from
the first library would readily identify the structure of those . ~.."I,u~
IV. The Tag
The identifier tag has a ~rl,u~ iLdblc feature that is, for example,
u~U~ ly or otherwise distinguishable in shape, size, mass, charge, or color. This
Ir~ug.~ r feature may arise from the optical, chemical, electronic, or magnetic
properties of the tag, or from some combination of such properties. In essence, the tag
serves to label a molecule and to encode illrull~ldLiùl~ at the level of one (or a
few~ molecules or solid supports. By using identifier tags to track the synthQis pathway
that each member of a chemical library has taken, one can deduce the structure of any
chemical in the library (i.e., the sequence of monomers of any oligomer) by reading the
identifier tag.
2' 75587
WO 9S/12608 PCT/1)59-11123-17
28
One can construct Illi-,lU~,U,UiCdlly ir::~n~ifi:~hl~ lags as small beads o~
.~u~ ly different sizes, shapes, or colors, or labeled with bar codes. The tags can be
"machine readable" l~.,..;nr~ 1 or radioactive labels. The identifier tag can also be an
encodable molecular structure. The i~rO~ Liull may be encoded in the size (the length of
S a polymer) or the c~rnCitinn of the molecule. Perhaps the best example of this latter
type of tag is a nucleic acid sequence, i.e., RNA or DNA assembled from natural or
modified bases.
To illustrate the role played by the tag in the,synthesis and screening of a
chemical library, consider for example, the use of ~ lu~u~i~lly l~cu~l~i~blc,
0 -~i 'r tags that are attached to each bead in an oligomer synthesis. The tag "A1"
means that a bead participated in the A-monomer reaction at step 1, "C2" means that a
bead ~?dlt; l ' in the C-monomer reaction at step 2, and "B3" means B-monomer was
added in step 3, and so on. At the end of a 3-step synthesis, one bead would have three
tags attached, e.g., A1, C2, and B3, indicating that the sequence of the peptides on the
15 bead is ACB. This scheme requires a number of distinct identifier tags equal to at most
the product of the number of different monomers and the number of synthesis steps (nine
in this example). The number of identifier tags is reduced if the symbols are attached to
one another in the order of the steps: A, A-C, A-C-B, in which case only as manyidentifier tags are needed as monomers, and the identifier tag is assembled in a way that
20 preserves the record of what monomer was added, and in which addition step.
In another example, the tag is comprised of a variety of light-add~ dl,lc
molecules, such as fluorescent or ,?I~,*,I.ul.~"~ cornrolln~c~ the spectral properties of
which can be changed (e.g., ~ b~ g) and therefore used to store ,l.l-l., .,..l;...,.
which are used to mark each bead or other solid support in the library. In one such mode,
25 a bead i,.. .,l~ t ~ a variety of lluolu~llu~a, each of which can be selectively
~h .~ 1, and so rendered incapable of nuulu~i~ ..l, e or of diminished fluorescence.
During each coupling or chemical reaction step, the bead is irradiated (or not) to
1 (or not) one or more particular types of lluulu~Jhol~, thus recording the
monomer identity in the oligomer synthesized. See Science 255: 1213 (6 Mar. 1992), A-
~UI~.~i herein by reference.
The identifier tags therefore identify each monomer coupling or other
reaction step that an individual library member or solid support has ~A~J~Ih,ll~,V,i and record
the step in the synthesis series in which each monomer was added or other chemical
2 ~ ~5587
WO 9~112608 PCT/US9-ill23-17
29
reaction performed. The tags may be attached i.~ before, during, or after the
monomer addition or other reaction, as convenient and compatible with the type of
identifier tag, modes of lltt:lrhmr~nt, and chemistry of oligomer or other molecular
- synthesis. As noted above, the identifier tag can be associated with the oligomer through a
S variety of ,~ n.i~ , either directly, through a linking molecule, or through a solid
,~ support upon which the oligomer is synthesized. In the ~atter mode, one could also attach
the tag to another solid support that, in turn, is bound to the solid support upon which the
oligomer is ~y~ d. The identifier tag is added when the solid supports that haveundergone a specific monomer addition or other chemical reaction step are physically
10 together and so can be tdgged as a group, i.e., prior to the next pooling step.
In some cases, of course, when only a small number of monomer units of
an oligomer are varied, one may need to identify only those monomers which vary among
the oligomers, as when one wants to vary only a few amino acids in a peptide. For
instance, one might want to change only 3 to 6 amino acids in peptides 6 to 12 amino
15 acids long, or one might want to change as few as 5 amino acids in polypeptides up to 50
amino acids long. One may uniquely identify the sequence of each peptide by providing
for each solid support an identifier tag specifying only the amino acids varied in each
sequence, as will be readily ~ ,id~-i by those skilled in the art. In such cases, all solid
supports may remain in the same reaction vessel for the addition of common monomer
20 units and d~lJuilh)1~e~ among different reaction vessels for the addition of rii~
monomer units.
Synthetic olig~d~u7.y,i~ f~11irlr-s are especlally preferred
i~ru1111d~iull-bearing identifier tags. Oli~on~ ti~ c are a natural, high density
i r .. -~-~1i. ." storage medium. The identity of monomer type and the step of addition or
25 any other i..ru~ LiU11 relevant to a chemical synthesis procedure is easily encoded in a
short r,li~ rif.J~ sequence. Oljgrm--rl~-oti ir-c, in turn, are reddily amenable for
attachment to a wide variety of solid supports, oligomers, linkers, and other molecules.
For example, an nli~nnllrlPrltjde can readily be attdched to a peptide synthesis bead.
One ~ advantage inherent in using an rlligr~m~lrlPI~ti~iP-based coding
3û scheme is the ability to achieve tremendous levels of target ~mrlifir~tir~m through the
pOI,ylll~.~d;~l; chain reaction (PCR, see PCR Prûtûcûls; A Guide tû MPthori~
~7~1ic~tion~ (Innis, M., Gelfand, D., Sninsky, J. and White, T., Academic Press, San
Diego 1990); see also U.S. Patent Nos. 4,683,202 and 4,965,18g, each of which is
Wo 9~112608 ~ ~ 7 5 5 ~ 7 PCT/USg~/123
~ uluu ' herein by reference) and other nucleic acid replication and ~ iri. -~;.."
techniques. Although the most commonly used irL vitro DNA :~mrlifir~tirln method is
PC~, suitab~e alternate ~mrlifir:ltion methods include, for example, nucleic acid
sequence-based amplification (Compton, 1991, Nature 350:91-92, il~Cu,L,ulAt_i herein by
5 reference) and amplified antisense RNA (Van Gelder P~t al., 1988, Proc. Nat. Acad. Sci.
~l~ 85:7652-7656, il.~Ul~Ju-dL~d herein by reference), and the self-sustained sequence
replication system (3SR, see Guatelli et al., 1990, Proc. Nat~. Acad. Sci. USA 87:
1874-1878, i,.~ul~u-~d herein by reference). Only tiny quantities (with highly selective
and efficient methods, even a single copy is sufficient) of DNA template is required for
10 PCR, enabling one to use solid supports of microscopic dimensions and obtain larger
libraries.
The use of nucleic acid tags facilitates the construction and screening of
synthetic libraries that far exceed the diversity accessible through other tethered library
techniques. Moreover, these libraries employ manageable quantities of bead material and
15 can therefore be assayed for receptor binding using practical volumes of biological
reagents. One improved method of the invention relates to a limiting step in theprocessing of ESL libraries with oli~.-"~ ,Li~p tags ~ the ~ , strand
separation, and sequencing of tags from individual beads. The method increases
sequencing efficiency by at least an order of magnitude, and relates to the i...,u,,uu,~Liu-- of
20 a tag ~ ",~, ;,,.~;nn (, ' " ) step, in which a number of different tags typically
amplified from a selected set of library members are ligated together prior to either cloning
or sequencing of the oligonucleotide tags.
In one ~ ,ll of the method, the amplified tags are ' and
then cloned as linear arrays of 10 to 20 (or even more) tags in a cull~ Liu.l.,l sequencing
25 vector. Preferably, ~ ulu~)~;dl~: restriction sites are installed adjacent to the "coding
regions" (sequences with information content) of the ..lic""",~ idP tags; after amplifying
the tags on a group of beads. the restriction sites are cut, and the fragments ligated to form
. The concatamers are then cloned into an dlJplupli.lt4 sequencing vector.
Each template can then be used for bidirectional sequencing of a total of, for example, 500
to 800 bases, allowing tlle identification of more than at least 10 tags per template. This
approach will also provide the option of avoiding the isolation of individual beads with
FACS. Beads or tagged compûunds can be sorted into pools, the pool of tags amplified,
c- i, and cloned for C~q~ n~in~. In addition, because the l~uil~ t to
WO 95/12(~08 ~ 1 7 5 ~ 8 7 P -r/US9J/123-17
manipulate individual beads is relieved, one can use beads smaller than l ~Lm (typically,
this size is too small for conventional FACS analysis) for library construction and
screening. The selection can be conveniently Arcf mrli~hPA by affinity ,~ irlcALiull
methods (panning, magnetic beads, etc.) and the enriched pools of beads then amplified
5 and cloned as above.
Oligrln~lrlpotifip identifier tags can be assembled base-by-base before,
during, or after the ,.~, l~ ~l,.,.,.l;,.~ monomer coupling (for oligomer synthesis) or other
chemical reaction step. In one case of base-by-base synthesis of an f~ f nllrlrl~tiflP tag,
the tag for each step is a single nucleotide, or at most a very few ~ lPU~ (i.e., a block
10 of 2 to 5 nllr1P/~tif~Ps) In the block-by-block approach, encoded sets of nucleotides
("codons") of 2 to 5 to 10 or more bases are added as protected, activated blocks. Each
block carries the monomer-type or other inff.~rmqtif n, and the order of addition of one tag ~-
block to the next represents the order of the monomer addition or ûther reactions.
AI~ lALiY~;ly, the block may encode the oligomer synthesis or other reaction step number
15 as well as the monomer-type or other building block ;,.r." ",_ti"" This strategy preserves
the order of the steps in the linear Al IAII~ of the r~liennllrlPotifip chain grown in
para]lel with the oligomer. To preserve the chemical compatibility of the parallel synthetic
steps (oligonucleotides and peptides, for example), one can modify the standard synthesis
rhPmictriP~, an important aspect of the present invention discussed in further detail below.
20 One can also attach protected (or u.. ~lu~e.,~td) f~l;g~ . .,Lides containing
A",~,liri. -li.", primer sites, monomer-specific i.,r."", ~ ", and order-of-reaction
information, from 50 to 150 bases (lll.. lf,Jl;fi. ~) in length, at each step. At the end of a
senes of n oligomer synthesis (monomer coupling) or other chemical synthesis steps, there
would be n differently encoded sets of oli~.. ,l, If.,lifif identifier tags associated with each
~5 oligomer sequence or other chemical in the library. After identifying the oligomers with
ligand activity, the associated oligul,uclcuLides could be amplified by PCR and sequenced
to decode the identity of the oligomer or other compound.
As discussed more fully below, the choice of bases used in an
oligu~ -lifl,P identifier tag is dictated by the chemistry of oligomer synthesis or other
30 chemica] reaction conditions to which the tag will be exposed. For example, the use of
strong acid can depurinate nucleic acids. Therefore, when . ~ l l if-~ requiring the use of
strong acid are employed, the use of an f~lie~mllr~pl~tide composed of only the pyrimidines
C and T and a binary code can prove of value. In similar fashion, the lability of purine
WO 95/lZ608 2 7 7 5 5 ~ 7 PcrluS9=i/lU=i7 ~
nucleotides to strong acid may be overcome through the use of the purine nucleoside
analogs, such as 7-dea~a-2'-dcuA~a~ osil-~ and 7-deaza-2'-deu~y~,ùaulo~ e (see Barr _
1., 1986, BioTechniques _:428-432, and Scheit, Nucleotide Analogs: Svnthesis andBiolo~ical Function pp. 64-65 aohn Wiley and Sons, New York), both of which are
5 i,~cul~oldtcd herein by reference). Use of these or other anaLogs would permit the use of a
quaternary or other, as opposed to a binary, encoding scheme. Thus, in a preferred ,.
t,~ c~l;",r"l, the identifier tag will be an oligrmllrit~nt~ about 50 to 150 nllrltr)tiri~ in
length and composed of pyrimidines or ~J~lillli~ill~i and punne analogs or any type of
nucleoside that will not degrade under the coupling conditions used to assemble the
lû oligomer library. The oligr~n~rlPnti~ir identifier tag may contain a 5' and a 3'
:~mriifir~ti~n site, and optionally a DNA sequencing primer site, which may be specific for
each step of the oligomer synthesis.
Encoding a, L I synthetic procedure with nli~on~lrll oti(i~ provides
a mechanism for addressing the major limitations of ambiguity and sensitivity encountered
15 in the direct structural analysis of minute quantities of ligands isolated from large libraries.
The high capacity of DNA for information storage can be exploited to archive the precise
details of a library's coll~LIu~Liul-. In Example I below, a "codon" structure of 2
contiguous nucleotides comprising three bases (c7dA, dC, T), capable of encoding a
synthesis ill~UllJUl.lLil~g up to 32 = 9 amino acid building blocks was used (only seven
20 building blocks were used in the synthesis of this library). If c7dG was also included in
the coding template, then a ~ l. ., ;,d synthesis employing 1000 different monomers
could be ~rcrlmmnfi~tf (i by using a "codon" size of iuSt 5 ~lu-,leuLidcs (45 = 1û24).
Information may be encoded in the length rather than, or in addition to, the
sequence of the olig~n~rl~nti~i~, or for that matter any other polymeric or oligomeric, tag.
25 If only length is utili~ed to represent each specific monomer addition to the oligomer, then
the identity of the oligomer can be decoded by, for example, amplifying an oligu,luclcu~ide
tag, as described above, and identifying the tags through any of a variety of size-separation
techniques, including polyacrylamide gel or capillary gel elc~lu~ u.csis. Each different
monomer added at a given step in an oligomer synthesis or each different chemical ~'
30 reaction step is Ir~Jlc~llt~i by an olig-""l~ IrlJL;~ir tag of unique length. The
uli~ullu~l~olide tag contains àlll~Jlirl~à~iull sites, such as PCR priming sequences, the
sequences of which are designed to be (.IIAI~I t~ of the given step-number in the
oligomer or other chemical synthesis. Drlrl ",;"-~;",, of the oligomer composition a~ any
WO 95112608 2 1 7 5 5 8 7 PCT/US91/12317
given position in the secLuence then involves amplifying the tag using the PCR priming
sequence rhqrq~fPricti~ for that step in the synthesis and size-separating the amplification
products utilizing techniques well known in the art, such as gel or capillary electrophoresis
(using the tagging f)li~ rl~--Litil~ as standards) This ~",l~ is particularly useful
5 when one desires to make a library of ~..,..l.~"",,i~ related to a lead sequence. One need
only tag during steps in which a site being analoged is ~y~ ,d.
In addition to length, oligomer sequence informqti~n can also be encoded in
the sequence of bases comprising the oliE-)n~rlP~-titlP tag. This type of encryption is of
value not only in the ~ hot~ l in which one attaches a different oligt~n~ lpoti~ip tag at
10 each coupling step but also in the ~ 1;",. ~1 in which one extends a pre-existing
;~,..",.~l. ..l,,i~ tag at each coupling step. For example, one may use oligrmll( l~oti~ip~ of
up to about 100 bases (or somewhat longer), each having seven (or more) regions, as
described below.
Region I is a 3'-PCR primer site (20 to 25 bases). This site is used in
15 ~-,-,ju,--,Liu~, with another PCR site (at the 5'-end of the oliEnnll~ lP~ti~iP) to prime
q~mrlifirqtion by PCR. Other ~",~ l,.,,. methods may also be used.
Region 2 is a "step-specific" DNA sequencing primer site (15-20 bases).
This site is specific for the particular numbered step in the synthesis series. All the
.,ii~....,l.-,~;~i. ~ added to all the beads at a particular step will have this sequence in
20 common. Each numbered step will have a highly specific primer site representing that
step.
Region 3 is a spacer (20-30 bases). A spacer segment of variable length,
but preferably 20 to 30 bases long~ places the coding site ~u[rlci~ Lly distant from the
sequencing primer site to give a good "read" through the monomer encoding or
25 i~iPntifi~qtir)n region.
Region 4 is a monomer i~ " region (8 bases). In this illustrative
, each base in the 8-bit string represents one bit of binary code, where, for
exarnple, T = 0 and C = 1. Each set of step-specific identifier tags consists of 8 bases
~ with a 1 (C) or a O CI') at each of the 8 positions. These may be thought of as switches
30 set to "on" or "off" at the different positions. Each monomer type is encoded by a
mixture of 1 to 8 of these "switches."
Region S is a step number . . ,. ,1; ", - ~ " region (4 bases plus 2 bases on
either side for region distinction). Four bits in this short stretch encode the step number.
WO 9~i/12608 2 1 7 5 5 8 7 Pf'T/US9-1/1~3-i7
34
This is redundant to the sef~uencing primer but can be used to csnfirm that the proper
pnmers were used and that the right step is decoded.
Region 6 is a repeat of the monome} i if ntifir~irln region (8 bases). This
region has the same information as region 4, and is used to confirm monomer identity.
S Installing this second monomer encoding region also increases the probability that a good
se ~uencing "read" will be obtained.
Region 7 is a 5'-PCR primer site (20 to 25 bases). This site serves as a site
for annealing the second PCR primer for ~ ,lir-- -~i..., of the se~uence. The length of
nli~fmllf lf ntides witll all seven of these features, some of which are optional, will
commonly be between 75 and 125 bases.
An 8 bit format can encode 256 different monomer types. The number of
steps that can be encoded is determined by the number of step-specific sets (8 per set) of
.li, .. ,.ll,lf~ , on hand. With 10 sets (80 nli~.. l~ ~li.l. ;~ one can encode up to 256
different monomers assembled into oligomers up to 10 units long (thus providing encoding
capability for up to 2561 = 1.2 x 1024 oligomer se~uences). The coded identif~er tags
may be used so that each monomer is assigned a speciftc binary number (e.g., Ala =
00000001, Gly = 000001~0, etc.). The ~ u~ iflf'C are combined to give
the correct binary code.
To facilitate l-l;~ lrJ~Iiflf- tag i~ iri, -li..." one has a variety of options.20 For instance, one could read the tag directly from the bead by sef~uencing or hybridi~ation.
One can also amplify II;g.. ,. lf.J~;tif~ tags to facilitate tag iflf-nfifir~rinn The
oligonllr~ identifier tags carried by a single solid support or oligomer can be
amplified in vivo, by cloning, or in vitrs, e.g., by PCR. If the limit of detection is on the
order of 100 molecules, then at least 100 or more copies of each oligsn~rlfr,~ifif~ tag on a
25 bead would be ref~uired. Copies of the tag are produced, either as single stranded
Uli~ lf-~-iiflPC double-stranded nucleic acids, or mixtures of single and double-stranded
nucleic acids, by any of a variety of methods, several of which are described below, and
the amplified material is se~uenced. In the ~ ..l of the invention in which a
separate and distinct olig.-.~ tag is added at each monomer addition step (as ~'
30 opposed to extending an existing tag at each step), one can amplify all tags at once and
then divide the amplified material into as many separate se~uencing reactions as there were
oligomer synthesis steps (employing a different se~uencing primer for each type of tag).
In this embodiment, one could also design the tags so that each tag could be amplified
WO 95/12~108 2 1 7 5 5 8 7 PCT/US9~/123-17
separatf ly from the other tags by .~ U~ . choice of primer sequences. The sequencing
reactions are performed and run on a standard sequencing gel, and the oligomer sequence
is deduced from the code revealed in the resulting sequence informatiûn
An altemative strategy is to use common PCR primers and common
5 sequencing primers (the sequencing primer may even overlap completely or partially with
. a PCR primer site) and identify the step by hybridization to oli~""~ i iP probes that are
comr~l~nnPntqry to each step-specific se~uence in the nli~nnllrlfntides from the bead. A
single set of sequencing reactions is performed on all of the amplified oli~nnllrlfotides
from a single bead, and the reaction products are run in a single set of lanes on a gel. The
10 reaction products are then transferred to a suitable hybridization membrane and hybridized
to a single step-specific probe (see Maniatis et al., Cold Spring Harbor Laboratory, Cold
Spring Harbor, NY (1982), which is i~-,ull~ui4l~d herein by reference). After detection of
the resulting signal, the probe is washed from the membrane and another step-specific
probe is hybridized. One could also use the procedure described in EPO publication No.
237,362 and PCT publication No. 89/11548, each of which is incorporated herein by
reference.
Parallel hybridization provides an altemative to sequential hyhri ii~inn
The sequencing reactions are divided into a number of aliquots ef;ual to the number of
peptide synthesis steps and run in a separate set of lanes for each on the sequencing gel.
After transfer of the reaction products to a suitable membrane, the membrane is cut to
separate the sets of lanes. Each lane set is then hybridized to one of a plurality of
step-specific ulig.,,,. lf~,Li l probes (see "Uniplex DNA ~ ;" and "Multiplex DNA
cf~ Pnring"" in Plex 1~ .; Kits Product Cqtqin~. Bedford, MA, 1990, i~ull~ul~d
herein by reference).
As noted above, a single synthesis solid support (or an attached bead beanng
a tag, or in solution in a "well") may only comprise a few hundred copies of each
f~1l;df tag These tags may be amplified, e.g., by PCR or other means well
known to those skilled in the art, to provide sufficient DNA to be sequenced accurately.
. The ability to decode the oligomers depends on the number of available olignnllrlf Oti~if
identifier tags. the level of :~mr~ifir~tinn that can be achieve~i from the available tags, and
the accuracy of sequencing that amplified DNA.
If PCR :~mrlifir~tinr of an oli~ lf~ if identifier tag is employed, one
may encounter "PCR product ~ ",;".l;~.,. " caused by the product of one PCR reaction
wo 9~ 608 ~ ~ 7 5 5 8 7 PCT/US9~ 3~7
...,.~,.,;..AI;~ a sU~sef~uent PCR reaction mixture designed to amplify other tags having
the same PCR primer binding sites. One may prevent this problem by ill~l.JJU~illg lability
into the product secluences and treating subsequent reactions so as to destroy potential
carried over from previous reactions. A specific example of this strategy,
5 for which commercial kits are sold by PECI and Life T~ c-gi~, is to introduce dUMP
into the product. Treating each new PCR reaction with uracil-N-glycosidase degrades any r
dU-containing DNA present, preventing :~mrlifir~fifm of the ~ lrl~ The template
DNA, which contains no dU (only dT) is not affected. Of course, the ~;ly~ .,siJd~e is
removed or inactivated before ~ ,. is begun.
Some of the tags described above for peptide synthesis have the unusual
of containing only L~Y ' This means that the uracil glycosidase
strategy (Perkin Elmer Cetus Il~LIU--I~I.L~ (PECI) Catalog, Alameda (l99l), incorporated
herein by reference) will work on only half of the strands produced-- those containing T's
(or U's). One cannot introduce dUMP into the ~1. j,l..,....~- y, purine-only strand;
15 however, the purine strand is highly vulnerable to acid fiPr~lrin~if n and alkaline-mediated
scission of the backbone. The ~llli. -1;l-ll of these treatments can greatly reduce
problems with product ~1Illl_ ll;ll_l;lll~ Another approach to preventing carryover
f-,,-t~ ;l, l;flll involves incorporation of a restriction site (~I could be used for
polypyrimidine tags) into the f~ rntide tag and digestion with the u u~ JIldil-grestriction enzyme prior to ~mriifit~tif n of a reaction suspected of being l ~ lf~
with the tag. This method only works if the tag to be amplified will not be cleaved by the
en~yme, as would generally be the case for a single stranded f,~ ,lif~f- tag.
For sequencing amplified DNA, one usually desires to generate sin~le
stranded templates. This generation may be ~ 1...1 by any of several means. One
such means is ~y"l,l,e~lic PCR, where an excess of one of the primers is used to amplify
one strand to a level 10 to 100-fold higher than the other (see, for example, U.S. Patent
No. 5,066,5~4, incorporated herein by reference). Another means of providing a single
stranded template is to by biotinylate one of the primers and purify or remove the resulting
strand by adsorption to immobilized streptavidin (Pierce Tmmllnot~prhnolo~y Cataloa and ~.
~n~ks2~ 1991, ill.oll,~,ldt.d herein by reference). Yet another means involves
generation of RNA transcripts (I~l~ illg only one of the strands) from an RNA
UOlylll.~ promoter and sequencing the transcripts with reverse Ll~l~ JL~c (Sommer et
1-, Chapter 25, In PCR Protocols A Gllif~P to Methods and Applications Supra.
WO 95/12608 2, 7 :~ 5 ~ 7 PCT/IIS9,-ill23.17
ill~ UlUUl_t~i herein by reference). If the tags dre cûmpûsed ûf only pyrimidinen-lrlf~tifiPc then all purine strands can be eliminated by acid/base treatment7 leaving the
pyrimidine strand for Cf~/plf~nrin~,
O The use of separate sequencing primers for each step-specific
olignnl.rl~otifi~ requires a separate, conventional sequencing reaction for each step-speci~lc
primer. Using primers that are differentially labe~ed would allow the identifier tags frûm a
single solid support to be sequenced in a single reaction and run in a single lane set (2
lanes if only polypyrimidines are used; 4 lanes if 4 different bases are used) on a gel.
There are now l U~ ,idlly available primers labeled with ~ ,Al)le fluorophores
that are suitable for this purpose (ABI Catalog, ill~.Ul,~l ' I herein by reference). Sets ûf
rhf-mil"l";"~ labels now distributed commercially may also be used (Bronstein et al.,
BioT~rhnicues 8: 310-314 (1990), illCUI~ herein by reference).
The amplified product can be easily sequenced or otherwise identified to
decode the identity of the peptide or other molecule on the bead or otherwise attached to
the oligr,n~lclf Q~ifip tag. For this purpose, one can use any of a variety of sequencing
methods, including sequencing by sequence-specific probe lIY~ DNA secuencing
enzymes which may be employed in the present invention include Taq DNA polymerase,
E. CQLL DNA pUIylll~ld~ I (or the Klenow fragment), T7 IJUIyll.~.d~C, Sequenase~ and
Sequenase Ir (Modified T7 DNA ~uly~ Ld~. ~), ~ DNA polymerase, and reverse
11dll~ )Ld5t~ (from AMV, MMLV, RSV, etc., see USB ~n7ymes foF DNA Se~ lpnrin~
U.S. l~irrhfmir-Al Corp, 1991, Cleveland OH, illCUI~ herein by reference). The
sequence of an olig~..,l, l~ ,~;fir tag may also be identified by a high f~delity DNA
hybridization technique. To this end, very large scale immobilized polymer synthesis with
oligrln~lrlf oti~if c may be useful (see PCT patent publication Nos. 92/10587 and 92/10588,
each of which is i.lcul},uld~cd herein by reference).
The choice of tag, whether the tag is an oli~nnllrlro~i~if or some other
molecular structure, depends upon the nature of the molecules of which the library is
composed and the method by which those molecules are to be ~yllLllcsi~cd, as discussed in
'A the following section.
Where synthesis rhf-nnic~rif~ involve the use of reagents and reaction
conditions incompatible with the above described ~ .,.",~ l;fif tags, it may be desirable
to utilize an alternate tagging method. Thus, the methods for synthesizing a tagged
molecular library of the present invention also envision utilizing chemically inert
wo gSrl2608 2 1 7 5 ~ 8 7 PCTrusgl/l23~7
38
hydrocarbon tagging molecules which are discretely resolvable by a variety of methods,
such as ~ methods.
The use of such inert l..y~ilu~l,on tags in molecular libraries has been
described. See, Michael H. J. Ohlmeyer, et al., Proc. Nat'l. Acad. Sci. 90:10922-26 -_
(December 1993), and published PCT Application No. WO 94/08051, both references
.r~,l herein by reference for all purposes.
The tags described utilize a binary coding scheme like that described herein.
Specificaily, a binary code is assigned to each chemicai building block, i.e., an amino
acid, to be added in the synthesis. The length of the code may be dependent upon the total
number of building blocks to be added. For example, where only seven totai building
blocks are to be added, a three bit binary code may be used. This ailows for seven
separate specific codes, 001 through 111, each one being assigned to one of the seven
building blocks, e.g., Iysine=001. Where the number of building blocks is greater, a
larger code can be utilized, e.g., an eight bit binary code, as described herein.
A number of tags are prepared, each having a different resolution or
separation pattem from the others using ~.1..~ ,. ,' - methods. If a particuiar tag is
present, it will represent a "1" in each position of the final tag code for a given
~ ' ' molecule. Thus, where a molecule has four building blocks and is coded in a
three bit coding system, there are 12 potentiai code digits, each building block having
three bits. For each step in the synthesis, the solid support upon which the molecule is to
be ~y~lLh~ ~;~i is tagged so as to indicate not oniy the building block added, but aiso the
step at which it was added. For example, the presence of tag no. 1 or "Tl" indicates that
there is a " I " in the ftrst position of the binary tag code for the building block added at the
first step. Similarly, the presence of T7 indicates that there is a " 1 " in the seventh
position of the overall code, which, in a three digit code, would also correspond to the
first position of the third added buiiding block. Thus, the building block assigned the code
111, if added at position one, would be encoded by the presence of tags Tl, T2 and T3.
Altematively, if added in step 2, the same building block would be encoded by the
presence of tags T4, TS and T6.
WO 9~/12608 392 1 7 5 5 8 7 PCT/U59S/123 i7
The specihc l~y iluwb~ll tags described by Ohlmeyer have the following
structure:
Linkcr Electrophoric Tar
HOO~ \~/ Ar
t NOC
Cl Cl Cl H Cl H
Ar = ~cl ~ci ~F ~
Cl Cl C H Cl H
where n is from I to 10. The varying length of the llyuiluwl,u., chain and the varied
S h~lnEJPn~tPA group allow physical separation or resolution of the tags by
met}tods, specificaily, electron capture gas .,;..~ ,y~ y.
Detection of these lly~ u~bull tags is, however, limited by t'ne sensitivity
of the detection method. Thus, to ensure detection of the tags, larger amounts of tag must
be used. This requires larger sized beads, thus reducing the size of the overall library
10 which can be ~yllLII~,~,;~, and increased reaction times to ensure maximum coupling of the
tags to the solid support. Finally, where larger molecules are screened, more IIY~iIU~LtbUII
hgs are added to the solid support. The i~.u~yvl<lLiun of large amounts of l~yui~uwlJul.s
on a solid support, e.g., for large synthetic ~n~ ull l l~ having numerous tags, will likely
have adverse effects on continued synthesis and/or screening of the compound on t~ e solid
15 support due to steric, lly~ilu~h~;c or ionic
nte problems of detectability, tagging time, and screening or synthesis
' associated with these lly~ilu~bull tags may be ~ . ' ' utilizing the
methods of the present invention. Specifically, the present invention, in one I --~i;,- ~,
provides a method of tagging using l'I,y~'ilU~LllJUII tags wherein such tags have a "molecular
2 t 75587
Pcr/uss~/l23~7
wo 95112608
hook," instead of a detectable electrophore as described in Ohlmeyer, et al.. This
"molecular hook" is defined herein as a functional group on the tag which allows for the
attachment of an dlllt lir,~d,le, detectable group, thus permitting detection of smaller
amounts of tag on the solid support. The hook will generally comprise a stable functional
5 group or molecule which will either form a covalent linkage with the amplifiable,
detectable group, or will have a high affinity for a portion of that group. Examples of ,
such hooks include, e.g., biotin, to which a streptavidin linked amplifiable, detectable
group may be bound or one i-"~ "~ of a high associahon peptide. Such high
association peptides generally comprise a ~u",~ .y pair of peptides each ha~/ing a
10 high affinity for the other. Thus, one, ",11~ .". .1 denotes one peptide of such a pair.
High association peptides are generally described in U.S. Application Serial No.08/321,933, filed October 12, 1994, which is a nnntinll~rinn-in-part of U.S. Application
No. 08/067,387, filed May 24, 1993, each of which is i.l~u.~o.~l~d herein by reference
for all purposes. Alternatively, the hook may comprise a protected activatable group,
which may be activated to covalently attach the ~ Jlirl(lblc, detectdble group to the tag.
Phu~vlJlvLe~ l activated groups are particularly useful in this ~rpli~inn Activated
groups are generally well known in the art, and include such groups as carboxyl,hydroxyl, amino, thiol and the like. These groups may be protected using photolabile
protecting groups such as those described in published PCT Application No. WO
93/22680, i~ul~u~lL~d herein by reference for all purposes. The resulting group is
photoactivatable .
In addition to the above, the molecular hooks may comprise .~ lir ~
groups. For example, the molecular hook may comprise two or more different functional
groups capable of being coupled to two separate entities. This may be the case where for
example, it is desirable to recouple the tag to another solid support for detection purposes,
e.g., a reaction well in a microtiter plate. The first functional group may be used to
selectively bind a ~nmrl~mf~nr~ry group on the solid support. Once coupled, the second
functional group may be used for selective coupling of the amplifiable detectdble group.
Such hooks may generally comprise ~vlllb;~ Livl~ of the functional groups described
herein, or other groupS which are capable of being selectively bound to another such
group. As an example, such a hook might comprise both biotin and digoxin, orthogonally
linked to the lly-l-v-~ul,v,, tag. Once separated, these tags may be contacted with a solid
support, e.g., a microtiter well, which is coated with a group capable of binding to one of
Wo 9~/12608 2 1 7 5 5 8 7 PCT~S9~1l23~7
41
the functional groups on the tag, e.g., an anti-digoxin antibody, and allowed to bind
thereto. After repeated washing steps, the solid support is contacted with the
oli~ ul;~i~ coupled to a group capable of binding to the second functional group, e.g.,
streptavidin linked olig-",~ as described previously. The bound oligl~ o~ is
5 then detected as previously described. The synthesis of the llydlu~ull tags of the
present invention may be carried out by methods well known in the art. See, e.g., March,
Advanced Organic Chemistry (John Wiley & Sons, 3rd Ed., 1985), Larock,
Cu~ lcll~.laive Organic Tl~l,ru~ Lions (VCH Publishers, 1989).
Because the inert l~ydlu~lJul~ tags of the present invention provide for more
lû sensitive detection of the l~ydlu~lJull t;igs, the amount of ~ particular tag on a solid
support may be reduced without affecting its detectability. Further, by reducing the
amount of tag on the solid support, time required for the cûupling of the tag to the support
may be reduced.
Tags useful in the present invention will generally comprise a variable
15 llydlu~dlbull region and a molecular hook. More preferably, such tags will comprise a
cleavable linker attaching the tag to the solid support, a molecular hook as described, and
a varied length lly~lu- ~u~oll chain connecting the molecular hook to the linker. Different
tags will have a different length l~ydlu~bull chain, or a different molecular hook, so as to
allow for their physical sepa;ation and detection.
21 75587
WO 95/12608 PCTI~S9`1/123 17
42
Tags having the following general structure are preferred:
!~ / \~/ ~ R
where n is from 1 to 10, or more, X is a cleavable linker and R is a molecular hook.
Preferred molecular hooks comprise, e.g., biotin, a high association peptide, and
S activatable groups, such as a yllU~Uà~,LiVdLdble group, or a combination thereof.
Examples of cleavable linkers which are useful in the present invention
include, for example, the yllvLucl~,avablC linkers describcd in U.S. Application No.
08/265,090, filed June 23, 1994, and ;IICul~ ~ herein by reference for all purposes.
Examples of such ~lluLuCI~,ava~lC linkers include those having the following structures:
0_'~0~ ~ ~~
~,,NOz ~$~No2
~N~ O~N~ ~--N~
H H
~0~ ~_
~o, ,¢,NOZ o
~ N~
1 O H
21 75587
WO 95/12608 PCT/US9~/lU-17
43
Follow;ng synthesis and tagging, the tags are removed from the solid
support, e.g., by photo~ysis of the linker. The tags are then separated from each other
using a method which preserves the separation pattem of the tags, e.g., HPLC with
fraction collection, or other ~ .;r methods, such as gel or capillary
5 ~ u~ ;s. Because the tags are generally present in amounts lln~l~pt~prt:~hlp by normal
means, e.g., absorbance, etc., they must be separated in a manner which allows
subsequent detection and correlation to their separation pattern. As an example, the tags
cleaved from a solid support, are separated on an HPLC column and collected in a fraction
collector. When the tags are eventually detected, as described in further detail below, the
10 fractions which indicate the presence of a tag are correlated against the known elution
profile, or separation pattem, for all of the tags used in the tagging/synthesis.
The separated tags are then immnhili7PA according to their separation
pattem. Such immobili7ation may take the form of spotting individual fractions, blotting
for gel based CPp~r~tion~ or immohili7:~tirn within reaction wells, i.e. on a microtiter
15 plate.
Once ;mm~-bili7PA the tags may be "hooked" to an a.l.~Jlirlablc, detectable
group. Amplifiable/detectable groups generally include a compound or structure which is
capable of being amplified, or produces a signal which is capable of being amplified.
~~omro~ln~ic capable of PYrnnPnh~l ~u ~ i,... are preferred. A ~dlLil ulculy useful
amplifiable, detectable group is an ~li~.,.. -lrlJI;rlf sequence. Hooking of the amplifiable,
detectable group may take a variety of fomms. For example, where the hook comprises a
biotin group, the ..li~...".. I~ ;AP may be coupled to streptavidin which will tightly bind the
biotin. Alternatively, the ~Ll~Ldvidin may be added in an ;"t . ".~ tr step followed by
addition of b;.)~ ' ' 1 nl;~ J~ P In altemative ....~.o~ , the tag may comprise
25 a ..,,1ll 1 .1l .1 of a high association peptide. In this case, the ol;L~ Lide is linked to
the other comrlPmPnt to that peptide so that the ~li~....l.~ I~.~l;~IP may tightly bind to the
tag. In yet another Clllb~dill.._.lL, where the tag comprises an activated group protected by
a photolabile protecting group such as thos~ described in published PCT Application No.
wo 93/æ680, previously il~uul~uldl~d by reference, this group may be activated, i.e., by
30 photolysis of the photolabile group, allowing the oli~..,... I~.J~;~P to then be coupled to the
tag by methods well known in the art. Where this is the case, it may be desireable to
select a photolabile protecting group having different photolysis, ~ lr~ from a the
WO 95112608 ~ ~ 7 5 5 8 7 PCTII~S9~/1231
photocleavable linker, if one is being used. This will allow selective cleavage of the tag
from the solid support without activation of the molecular hook.
Once hooked to the tag, the uli~;v~uclcvLi l~ sequence may be amplified
using the PCR techniques descnbed herein. Where the imml~hil;7:~fi(1n is in a blotting
S format, the ~mrlifi- :~tinn must be carried out so as to preserve the local concentration and
avoid diffusion of the amplified o~ r, thus allowing its detection and correlation
to the separation pattern. This may be ~comrlic~ A by performing the ~mrlifil ~ti~n
reaction in, for example, a gel overlay of the blot.
Detection of the hooked .,I;r,..-- ~ l..,l;A,~, and thus the tag, may be carried10 out by ill-,Ul~J(Jld~ g a label into the amplified olignnll~lrnfiA~ or by probing for the
amplified ~ ide sequence where such sequence is known. Detection of a tag is
correlated to the known separation pattern for the tags. A tag so identified is then
i-lcul~u~ d in the overall tag code for the syl~Lll~.,i~d molecule. For example purposes
only, if following HPLC separation, hooking, ~mplifir:~finn and probing, fraction #17
15 indicates the presence of a tag, this is correlated to the tag which is known to separate into
fraction #17. If, for example, this is tag # 5, then a "1" may be assigned to position # 5
of the overall binary code for the molecule ~yllLl,~ d on the particular support. The
coding schemes described herein are for example purposes, and those of skill in the art
will recognize that a variety of coding schemes may be applied to the methods of the
~0 present invention.
Those of skill in the art will also recogni2e that there is no ~Luil~,l,c..L that
the code reporting the sequence of a ~ h~5i~ molecule be contained in a single
polymeric sequence of individual tags. Instead, the code may be embodied by the
presence or absence of individual different tags on the solid support. While these tags may
~5 potentially be coupled sequentially to each other, those of skill will recogni2e the benefit of
having each different tag individually attached to the solid support. Specifically, only a
single coupling chemistry would be required for any and all tagging steps. Further,
complex protocols of protecting\~luL~Li--g reactions for tags having different reactive
groups may be avoided.
WO 95/12608 2 1 7 5 5 ~ 7 PCT,US9~/,23~7
V. Synthesis Methods
The method of the present invention can be applied to any set of synthetic
chemical reactions performed in a sequence to generate diverse cu~ uu-~do. While the
5 invention is typical~y illustrated using chemical building blûcks, more typically monomer
building blocks, the general nature of the invention should be d~ The majority of
synthetic chemical reactions proceed quite differently than the typical monomer coupling
reaction; the typical ûrganic chemical reaction gives variable yields and leads to multiple
products, such as regio- and ~Irlr~ structures. The present invention can be used
10 to identify useful products of such series of chemical reactions, because one can practice
the methods so that the tag encodes the protocol for sy~ ,Oi~i,.g the compound instead of
explicitly specifying the structure of the reaction product.
To simpliFy discussion, however, the.invention is most readily viewed as a
series of monomer coupling steps. Because the various coupling reactions of the present
15 method can be carried out in separate reaction vessels at separate times, even building
blocks, such as monomers, with very different coupling . ~ ;rC can be used to
assemble the ~ u.. A~ of interest in a library. While the invention can be practiced by
exposing solid supports to a building block and an identifier tag at the same time, or
sequentially (either building block and then tag or tag and then building block), the
20 sequential approach allows one additional flexibility with respect to coupling ~
In any event, the preferred ~ lrlll for conducting coupling reactions is one in which
the diverse coupling reactions are carried out in paraLiel.
After each parallel series of coupling steps is performed, the solid supports
on which the oli~omers or other ~,u~ uu~Jo of the library are or~ . i are pooled and
25 mixed prior to re-allocation to the individual vessels for the next coupling step. This
shuMing process produces a large library of c~ v~ with each distinct member of the
library on a distinct solid support. If each synthesis step has high coupling efftciency, then
sl~hcf~ti~lly ail the cu~ ,vull~is on a single solid support have the same structure or, if the
c are oligomers, monomer sequence. That structure or sequence is determined
30 by the synthesis pathway (type and sequence of monomer or other building block coupling
reactions) for any given solid support at the end of the synthesis. The maximum iength of
oligomers is typicaily less than about 20, usualiy from 3 to 15 monomers in length, but in
some cases a length of 8 to 12 monomers (residues) is preferred.
WO 95/12608 2 ~ 7 5 5 ~ j~ PCT/[)S9.1/123~7 ~
46
Given the diverse numbers of tags and building blocl~s suitable for use with
the present invention, there are a number of chemical methods by which one can prepare
chemicaL libraries of the invention. However, one must ensure that each coupling step,
whether of tag or o~igomer, does not produce unacceptable levels of unwanted reactions or
5 destroy tags or oligomers already present on the support. In one embodiment, one ensures
that only desired reactions occur by using solid supports with chemically reactive groups ,.
for tag and oligomer attachment that are protected using two different or "orthogonal"
types of protecting groups. The solid supports are exposed to a first d~lut~lio,~ agent or
activator, removing the first type of protecting group from, for example, the chemically
reactive groups that serve as oligomer synthesis sites. After reaction with a first
monomer, and after any optional blocking steps, the solid supports are then exposed to a
second activator that removes the second type of protecting group, exposing, for example,
the chemically reactive groups that serve as identifier tag attachment sites. The tag is then
coupled, and these steps are repeated, typically from one to about 20 times.
A. Oli~,vllucl~Lide Tagged Peptide Libraries
In one important ~ \o~ the present invention relates to the synthesis
of Itlrge libraries of diverse peptides. While many other c~ u~ and oligomers can be
made by the method (see Gait, Olil~onl~clf otid~ Synthesis: A Practical Approach. IRL
Press, Oxford (1984); Friesen and Danishefsky, 19~9, I- Ame}. Chem. Soc. 111:6656
and Paulsen, 1986, ~Ç~. Chem. Int. Ed. En~l. 25:212, all of which are il-~u~lJuldL~d
herein by reference), techniques for solid state synthesis of peptides are particularly
important and well Icnown (see Merrifield, 1963, I ~m Chem. Soc. 85:2149-2154,
u~dL~d herein by reference), and peptide libraries are highly useful for a variety of
purposes. Ln the Merrifield method, an amino acid is covalently bonded to a support made
of an insoluble polymer. Another amino acid with an alpha-amino protecting group is
reacted with the covalently bonded amino acid to form a dipeptide. The protective group
is removed, and a third amino acid with an alpha protectiYe group is added to the
dipeptide. This process is continued until a peptide of a desired length and sequence is ~'
obtained. Protective groups known to those skilled in the art may be used to prevent
spurious coupling (see The Peptides, Vols. I & 3 (eds. Gross, E., and J. Meinhofer,
Academic Press, Orlando (1979 & 1981), which is il~,UI~ ,' herein by reference) or to
allow one to control coupling. Phûtolabile, base-labile, and acid-labile protecting groups,
~ wo gs/12608 ~ 1 7 5 5 ~ 7 PCT/U59.11123.17
and ~.. ,l,;",,l,.",~ of the same can all be employed for various purposes of the present
invention.
Additionally, both L and D forms of arnino acids may be employed in
peptide synthesis methods described herein. ~mploying D-amino acids may be useful in
5 the synthesis of `'retro-inverso peptides," as described in U.S. Application Senal
No.08/309,451, filed September 21, 1994, ill~,UI~ ' ~ herein by reference for all
purposes. Such retro-inverso peptides will comprise the same amino acid sequence, but
having reversed ~ILlt~O~,ll.llli~Lly from a peptide which is ~y~ ~ using L-amino acids.
When the present invention is used to make and screen peptide libraries, the
10 tag of choice is a nucleic acid. There are a variety of compatible ~ c for peptide
synthesis and round by round attachment of ~' g ' '- identifier tags. However, to
maintain the integrity of an o~ p tag during peptide synthesis, one may need to
use different cù.~ la~io.ls of protecting groups and/or synthetic nl~rlPI-ti-iPc to avoid
"~ of the tag or the oligomer ~y~Lh. ,;~. In general, uolyAuy~ li-le
15 nlip""~ P tags are relatively stable under typical peptide synthesis conditions, as
opposed to olieonl~lP{ltidP tags that contain natural purine nllrl~oti~p~ but a
pOIy~uy~illli~i,lC nucleotide tag may be somewhat refractory to ~mrlifi~tinn by PCR. One
may need to illcUl,u, purine bases, or analogs such as 7-deaza-deoxy~ Pm~cinP, and
7-deaza-d~y~uallu~;..c, tested for ability to withstand peptide coupling (and d~:~lu~c~iull)
20 conditions, into the tag to achieve a desired efficiency of .' ~ For purposes of
the present invention, the tag optionally may contain from 10 to 90%, more preferably 35
to 50%, and most preferably 33 to 35%, purine or purine analog nllrlPoti~iPs Theoligonll~lpoti~lp tags may optionally ill~UllJI ' a biotin ûr other reporter group to facilitate
U~.~;1;. .1,l~l~ hybridizdtion, ~ nrlifi~ti-~n, or detection (see p;Prce Imm~nnTechnoloey
25 ('s~t~lo~ and T~n~honl~ 1991, iUl~ Ul,UOldt~ herein by reference).
Thus, in selecting the ~ used to create an .~ ~lrvl;~p-tagged
peptide library of the invention, one must (1) select a solid support with ~plu~JIiaLt:
functional groups; (2) select the amino acid coupling chemistry; (3) select the
nlit ll...,. l~U~ ,; tag coupling chemistry; (4) select the protechng groups for the various
30 tags, monomers, and oligomers; and (5) select the d.~lul~Lioll and, in some l-mho~limPn
cleavage chemistry (for either the tag or peptide). Those of skill in the art recognize that
not all of the above selections need be made in every case, as some applications may not
present the same issues as others. For instance, one or more protecting groups may not be
WO 95/12608 2 ~ ~ 3 ~ PCT/US91/123.17
48
required for all applications. In the generaL case, however, each of these selections is
important.
To consider factors relevant to the selection of coupling chemistries and
protecting groups for the synthesis of nli~nnllrlf~otiA~ tagged peptide libraries, consider a
S synthesis in which commercially available Fmoc protected amino acids are coupled using
standard Merrifield chemistry, and the . li~.""~ ,L;~I~ tags are coupled using standard
IIAI~ chemistry. The process can be viewed as having the following steps: (1)
removal of the amino-terminal Fmoc protecting group from the linker or peptide attached
to the bead; (2) coupling an Fmoc protected (the side chains may be protected as well)
amino acid to the free amino group produced in step (1); (3) optional capping of unreacted
free amino groups; (4) removal of the D~T protecting group from the hydroxyl group on
the bead or tag to which the nucleotide tag is to be attached; (S) coupling a nucleotide
,l"-,A,~idi~f~ with a 5'-DMT protecting group as well as protecting groups on the
phosphate and exocyclic amines of the bases; (6) optional capping of any unreacted free
hydroxyl groups; (7) oxidation of the ~l~u~ oluu~ of the nli~on~ o~id~ tag; and (8)
deprotection of the peptide and nlig,-"", 1 .J~ tag. Each of these steps is discussed
below.
(I) P~emoval of the amino-terminal Fmoc protecting group from the linker or
peptide attached to the bead is necessary prior to the attachment of the next amino acid
monomer. Typically, treatment with 30% piperidine in DMF for about one hour is used
to achieve this d.~luL~iull (see also step 8), but one aspect of the present invention relates
to the use of reduced ~ IC of piperidine or reduced d.,~-ut~iu~ times for the
synthesis of olignnl~nl~n~ tagged peptide libraries. Piperidine may cause deprotection of
methyl triester protected ~ " ~ Lid~ tags, and O-methyl phosphate protecting groups
have greater base stability tban the standard beta-cyanoethyl group, known to besusceptible to piperidine cleavage. Preferred Fmoc deprotection conditions of the
invention are 5 to 15%, preferably 10%, piperidine for 5 to 60 minutes, preferably 10 to
20 minutes, and 15 to 30% piperidine for 15 to 30 minutes. Another treatment known to
effect Fmoc removal is treatment with DBU (1,8-diazabicyclo[5.4.0]undec-7-ene), e.g., ~'
5% DBU for 5 min. However, a report by PaLom et al., Tetr. 1~- 34: 2195-2198,
uldlcd herein by reference, suggests that such treatment can result in methylation at
N-3 of thymidine. While DBU-mediated Fmoc removal can be effective in some
A~ C, the potential for base mnt'ifi~A~in~ should be recogni~ed.
_
~ Wo 9~12608 2 1 7 5 5 ~ 7 PCT/US9~ 7
49
(2) Coupling an Fmoc protected amino acid to the free amino group on the
bead or peptide can be achieved using standard BOP coupling chemistry (see ~
Peptides, supra~. Typically, a mixture of an Fmoc protected amino acid (l l0 mM),
- HBTU (l00 mM), HOBt (l00 mM), and DIEA (300 mM) in a solution composed of l:l
DMF/DCM is employed to effect amino acid coupling. Other activation chemistnes may
¢ also be applied in this instance, for example ~ " of HBTU/HOBt with HATU. In
one rmhl~rlimrnl of the invention, however, the reaction mixture is composed of 55 mM
Fmoc-protected amino acid, 50 mM HBTU, and 150 mM DIEA in a solution composed of3:1 DMF/DCM; this ~",1,o,1;",~"1 is preferred for use with ill~LIu..lcllL~ where reagent
delivery bottles may be limited. The side chains may be protected as well; Fmoc/'Bu
protection is preferred for most purposes, due to the commercial availability of building
blocks. Other useful amino acid building blocks with side chain protection included
Arg(Pmc), Gln(Trt), His(Trt), Asn(Trt), Asp(O'Bu), Glu(O'Bu), and Lys('Boc) and amino
acids with side chain protecting provided by photolabile protecting groups.
(3) Optional capping of unreacted free amino groups can be achieved by
treatment with acetic anhydride and l-methyl imida~ole or by other methods known in the
art.
(4) Removal of the DMT protecting group from the hydroxyl group on the
bead or tag to which the nucleotide tag is to be attached can be achieved by treatment w;th
trichloroacetic acid (TCA), i.e., l % TCA in CH2Cl2. If one uses acid-labile protecting
groups on phosphates and exocyclic amines of the '~: ' ~ (i.e., deoxycytidine,
7-deaza-deoxyadenosine, and 7-deaza-d~y~ o~l~ ), then those groups should be
sufficiently robust to resist the TCA (typically 1-3 %) used in 5'-0-eJcLIiLyldlio.,.
(5) Coupling a nucleotide ~ "",--,-~ with a 5'-DMT protecting group
can be achieved using standard ~ ",~ 1;1r chemistry, although one must take into,~,,ci,l. .A1;~... the need for protecting groups on the phosphate oxygen as well as on the
exocyclic amines of the bases of the Oli~;ol~uclcuLide_ tags. For photolabile protecting
groups for nucleic acids, see PCT patent publication WO 92/10092 and Baldwin et al.,
', 1990, :~. L~. 46: 6879-6884, each of which is ;-lw-~ul~l~d herein by reference. As
30 noted above, suitable phosphate protecting groups include the O-methyl and
beta-cyanoethyl groups, but O-allyl and/or N-allylù,-y~ 'uùllyl groups (i.e., byillCuluuldLil~g 3-(allyl N,N'-diisopropyl) ~ Ps) can also be used to protect
phosphate oxygens and the exocyclic amines of the nucleoside bases, respectively (see
Wo 9S/12608 2 1 7 5 5 ~ 7 PCT/liS9~1123~7 ~
Hayakawa et al., 1990, l. Amer. Chem. Soc. 112: 1691-1696, iL-~olL-ul LLt;d herein by
reference). Allylic protecting groups can be removed using THF containing tris
,yl;~ (,,.r) ~iir~ m-chloroform complex, LLi~ yl~ r~ and
n-butylamine/formic acid, followed by a THF wash, an aqueous sodium
S N,N-diethylrlithi~ wash, and a water wash. r~ it coupling is
mediated with agents such as lH-tetrazole; 4-~ luyll~.lyl tetrazole; pyridinium .
hydrochloride/imida201e. The latter L~ h ~A~ ;rl;l~ activator leads to se~ectiveS'-O-phu,~ yl,Lliull at the expense of low levels of spurious reaction at nitrogen on the
peptide or oli,,u--uclcvLid~ (see Gryaznov and Letsinger, 1992, ~Ii Acids Research 20:
1879-1882, ill- ul~Jul LLt;d herein by reference).
(6) Optional capping of any unreactecL free hydroxyl groups can be achieved
by treatment with acetic anhydride and l-methyl tetrazole or by treatment with acetic
anhydride/lutidine/DMAP.
(7) Oxidation of the ~ luu~ of the ~,l;g~ Lidc tag can be achieved
15 by treatment with iodine and pyridine or by treatment with 12, collidine, MeCN in H20.
Alternatively, by employing the mild oxidant 'BuOOH for oxidation at the phosphorous,
one can minimize oxidation of the amino acids m~-thirnin~, tryptophan, and histidine (see
Hayakawa et al., 1990, Tetr. Lett. 27:4191-4194, il~,ul~ol_Sd herein by reference).
(8) D~L,LuL~Liull of the peptide and ~lig.. - i~ lr tag can be effected by
20 se~tuential treatment with 1% TCA in dichlul, ' , then with thiophenol/NEt3/d;oxane
(1:2:2), then with cLl~yl~ ,..; /EtOH (1:1) at 55 degrees C, to remove the protecting
groups from the tag, and then Llill~lulu~c~Lic acid (95:5 TFA/water, with cationscavengers) is used to remove acid-labile amino acid protecting groups. The lability of
purine nllrlr~ C to strong acid (e.g., TFA) can be avoided by use of ~ho~,lluldll,idites
Z5 of the purine nucleoside analogs 7-deaza-2'-~v,.y~l.u~l.~ and 7-deaza-2'-deoxyguanosine
(see Barr et al., 1986, BioT~hni~llpc 4:428-432, and Scheit, Nucleotide Analogs:Synthesis and Biolo~ical Function pp. 64-65 (John W1ley and Sons, New York), both
illUUl~J~ ' ' herein by reference).
The next section illustrates one preferred ~:IllI,odilll~ llL for synthesizing
~ tagged peptide libraries.
wo ss/~2608 PCT~vS9~ 7
B. Improved Method for Syl~Ll.~u~g I;v ~ Tagged Peptide Libraries
r~ a practical bead-based ~,lie,.",~ .,Lide-encoded peptide library
mPthndnlngy demands that several key technical criteria be met. These include (i) the
d~lu~ ; of mutually compatible h 1~ for parallel assembly of peptides and
5 rl ' ' " fii) the selectiûn of bead material with d,U~JlUlJlid~: physical
..1,~ (iii) the facile isolation of small beads bearing ligands that bind a t~irget
receptor; and (iv) successful reading of the tags from a single bead, i.e., by PCR
a~rlifi--~tinn and sequencing of template tag DNA from single beads. The presentinvention provides an improved method for ~ll~ lg such libraries, as illustrated in this
section and Example 1, which show how to use single stranded nl;e~ l. uL;.~ tags to
encode a cu~ peptide synthesis on 10 ~m diameter polystyrene beads.
In this improved method, peptides and n~ lPnti~lPc are assembled in parallel.
alternating syntheses so that each bead bears many copies of both a single peptide sequence
and a unique ~ identifier tag. The ~ lP~ share common 5 - and
3'-PCR priming sites; the beads can therefore serve as templates for the PCR. Toillustrate the method, an encoded synthetic library of some 8.2 x 105 hepta-peptides was
generated and screened for binding to an anti-dynorphin B mnnnf~lnn~l antibody D32.39
(Cull et al., 1992, Proc. ~. Acad. ~. ~ ~:1865-1869"ll~ul~uuld~d herein by
reference), using a lluul~.,.,~ activated cell sorting (FACS) instrument to select
individual beads that strongly bind the antibody. After PCR ~mrlih~ inn of the
o1~ tags on sorted beads, the DNA was sequenced to determine the identity of
the peptide ligands, as is described more fully below.
One important aspect of this method, which is described in additional detail
in Example 1, below, is the solid support selected for synthesis of the peptide and tag.
Solid supports, i.e., 10 ~m diadmeter beads, fashioned from a macroporous
styrene~liviu,~ll,. .~l c copolymer and derivatized with a dOd~lddlllille linker are
preferred. The amino group loading of these beads was estimated to be ~ 100 ~lmol/g by
exhaustive acylation with Fmoc-glycine, followed by piperidine cleavage of the ~moc
group and ~ r- ' 'f' ~ '- " of the released piperidine-dibenzofulvene adduct
(e3C2 = 7,8001 mol~l cm~'). With 5 x 109 beads/g, this cu~ ,uul~d~ to a maximum peptide
load`ing of--20 L"~ dd. Acylation of the beads with a mixture of an d~Juluulid~ly
protected amino acid and an omega hydroxy acid provide orthogonally di~r~l~"~;d~d amino
and hydroxyl groups from which the peptide and nucleotide chains respectlvely can be
WO 9~i/12608 2 1 7 ~ 5 8 7 PCT/llS9~/123~7
52
extended. The average ~Ir~ i(""- Iy oFpeptide to n~ f ~ iP per bead is controlled
by varying the ratio of amino and hydroxy acids coupled to the initial bead mass (vide
infra). Test peptide syntheses (5-mers to 12-mers) on these beads equipped with a
Llinuulu~l,Li~ acid-cleavable Knorr linker (F .. i.~ ~1-, 1989, ~. Lett. 30: ;
4645-4648, i--cu,,ul ~ herein by reference) using standard Fmoc chemistry were found
to proceed with high fidelity that was i~d~Li~l~ui~ll~lc from syntheses performed on
conventionai peptide synthesis resin, as ~iPtPnninP~i by HPLC analysis of the crude cleaved
peptide carboxamides.
Parailel synthesis strategies require the use of a set of protecting groups on
the amino acids and nucleotide building blocks that are mutually orthûgonal, and that each
of the polymer chains be stable to the reagents used in the synthesis and d~JIut~Liul) of
the second chain. Although, in principle, a variety of protection/deprotection schemes can
be used (as discussed above), Fmûc/'Bu prûtection on the peptide building blocks is
preferred, because of the extensive commercial availability of natural and unnatural amino
acids protected in this manner. However, the 'Bu-based peptide side chain prûtecting
groups require treatment with strong acid (typically Llinuulua~.Lic acid) for removal,
conditions that can lead to rapid ,I.I,UI;ll,.Liull of ~ P..I;~iPS containing either
2'-.lcu~ (dA) or 2'-dw~.y~;L.~o~illc (dG) (see Capon, 1969, Chem. Rev. 69:
407-498, i--l ull~u~At~d herein by reference). This problem has been ~ u~ lLed by using
20 7-deaza-2'-d~o,.~ llo~;llf (c7 dA) in place of dA in the template r~ r,n-lrlPntiflP tag. The
glycûsidic bonds of fiP~Ar-~rin~P "", ~,~n~..lf~ are resistant to acid-catalyzed hydrolysis (see
Scheit, 1980, Nucleotide ~P~ and Biolo~ical E~i~ (John Wiley and
Sons, New York) pp. 64-65, i~,ul~J~ ' herein by reference), and oli~f~nllrlPl)tiflP~
UI~oldLi~g these monomers are faithfully copied by ~I,r""--~t~l,lr pOlylll~ t~ used in
the PCR (see McConlogue ct al., 1988, Nucl. ~i51 Re,s. 16: 9869, and Barr et ai., 1986,
BioTechniques 4: 428-432, each of which is i~u~lJu ' herein by reference).
Acid-resistant guanosine analogs can also be in~ , ' ' into the template DNA.
5'-O--lilll.,Lllu~-yLliLyl 2'-df ~y~lu~l osidf 3'-(O-methyl-N,N-diisopropyl)
Al~ were used in all parallel syntheses. The reagent
(12/collidinelH2O/aretr,nitri~P) used to convert the nucleotide phosphite intermediates tû
llU~IJllU~ t.l~ in the DNA synthesis protocol was not found to adversely affect either the
readily oxidized residues Trp and Met or any of the other protected amino acids.Complete removal of the 5'-O-DMT group from the growing r~iL~ JI;fiP chain was
~ Wo 95/12608 2 7 7 5 5 8 7 PCr~S9~1123~7
achieved in ~ 40 sec using 1% trichloroacetic acid (TCA) in dichloromethane, while a~l
of the acid-labile side chain protccting groups used conventionally in Fmoc/'Bu chemistry,
excepting thè 'Bu ether derivative of tyrosine, were inert to treatment with 1% TCA for I
hour. Fmoc-Tyr(O-B~) proved a suitable Ir~ in the synthesis of
5 tyrosine-containing peptides, the O-benzoyl ester being robust towards both TCA and the
piperidine used for removal of the alpha-N-Fmoc protecting group in peptide synthesis.
Quantitative d~vlu~Liu-~ of the alpha-amino residues required 5-10 minute treatment with
piperidine/DMF (10% v/v) and also resulted in partial d~ LI~ .Lion of the prûtected
polyl-llclcuLid~ phrlcrh~tri~-forc (tl,2 ~ 45 min). Control rlllrl ;" ..1~ indicated that any
10 aberrant ~11u~1~i~yldLiull of the resulting p~ `t spccies during subsequentnucleotide chain elongation was reversed by the final r~ Lide deprotection steps(see Lehmann et al., 1989, Nucl. ~i~ Bs- 17: 23n-2390, i.l. vl~u.a~e~ herein byreference). At the completion of the parallel synthesis, the DNA was fully d~-u~ L~d by
treatment with 11~;"~1,. nvldLl; (phosphate O-d~ l.illyldLiull) then ethanolic ethy1-~11rl1;A"",~P
15 (d~v~ uyldLion of protected cytidine and 7-deaza-adenine residues). These mild,
anhydrous aminolysis conditions did not adversely affect protected peptide sequences (see
Juby et al., 1991, ~ L~- 3: 879-882, ;II~,UII~u herein by reference), which weredeblocked using TFA under standard conditions.
The carboxy-terminal region of opioid peptide dynorphin B
20 (YGGFLRRQFKVVT) (SEQ ID NO: 1) has been previously shown to represent the epitope
of anti-dynorphin B mAb D32.39 (see Cull et al., ~): the soluble hepta-peptide
RQFKWT (SEQ ID NO:2) binds D32.39 with high affinity (Kd ~ I nM). A parallel
synthesis of this peptide and a 69 base oligodeu;~yllu~le~Lide was performed on
orthogonally di~f~ llLidL~ beads bearing an acid-cleavable Fmoc-protected carboxamide
'~ (Knorr) linker. After addition of the first 20 n~ Pmti-lPc the beads were treated with
piperidine/DMF and the first peptide residue (Fmoc-Thr('Bu)-OH) coupled to the free
amines. The beads were then subjected to two cycles of r1~v~ chemistry and
coupling of the next amino acid (Fmoc-Val-OH). This process was repeated until the
1~ .Lirlo sequence and nucleotide coding region had been fully elaborated, and then the
3û DNA was extended by a further 35 ., ~1. v~iri ~ to provide a spacer region and 5'-pnming
site for the PCR. The beads were finally exposed to full ~ --,1-- lr--li~P and then peptide
d~.-ut~Lio-~ conditions, and the TFA ~ 1 containing the cleaved peptide was
analyzed by reverse-phase HPLC. The HPLC results showed that the crude peptide from
WO 95112608 2 1 7 5 5 8 7 PCTIU59~1123 17 0
54
the parallel synthesis consists of a single major component (co-eluting with authentic
RQFKVVT, (SEQ ID NO:2)) and that this crude product is not significantly different from
that generated in a control peptide synthesis in which no ~ "".-lf"~ P chemistryoccurred.
The stability to parallel synthesis chPmistripc of template DNA containing T,
dC and c7 dA was compared with an analogous target containing the standard punnenucleotide dA. Using the single bead cloning capability of a FACStar Plus cytometer,
individual deprotected beads from the two syntheses were sorted into microfuge tubes and
the tethered illig,.",l. 1~ F template amplified through 45 cycles of the PCR. A "clean"
~mrlifjr~tinn product of the expected size and nucleotide sequence was obtained only from
template containing the ~ F Thus the integrity of this olignn~lPo~ P was
maintained through the course of a parailel peptide synthesis, ~l~".. l,,.li.,~ that template
from a single bead can be readily ampli6ed and sequenced.
An encoded library designed to contain 823,543 (77) different hepta-peptides
15 attached to 10 ~m beads was constructed by a ~..,..l.;.. ~ .,iAl synthesis using seven amino
acid building blocks Arg, Gln, Phe, Lys, Val, D-Val and Thr.
Alpha-N-Fmoc-Thr(tert-butyl)-o,~yl-~ .l,uLIi~ulc (protected threonine) and succinimidyl
4-O-DMT-u~ybu~yl~lL~; residues were first coupled to all the beads to provide the
orthogonally differentiated amino and hydroxyl groups for this synthesis. On average,
20 each bead bore 2û molecules of a single peptide sequence per molecule of DNA tag.
Every amino acid addition was encoded by building a . ~ ;cl;- fiin~lriP~tiriP unit, and
after the seventh cycle of peptide coupling the beads were pooled, and the DNA synthesis
completed. Starting with a total bead mass of 35 mg (1.75 x 10~ beads) ensured that each
peptide sequence appeared on ~ 200 different beads in the library. Peptide
25 micro-sequencing analysis of an aliquot of the library confirmed that the seven amino acids
were ctnr~h~ctic~lly distributed among every position of the degenerate hepta-peptide
mixture (note that L-valine and D-valine are not .I;~ in the Edman degradation
procedure).
The binding of mAb D32.39 to control beads and to the bead library was
30 analyzed by flow cytometry. Beads carrying the positive control sequence RQFKVVT
(SEQ ID NO:2) and a 69-mer nli~r~nllrlPotiriP tag were strongly stained by the antibody
whereas blank beads were unstained. By contrast, only a small fraction of the encoded
library bound D32.39. Analysis of 105 events indicateci ~ 2~o of the library st~ined above
~ wo 9~112608 2 1 7 ~ 5 ~ 7 PCT~S9VI23~7
background levels. .~ienifi~An~ly, this binding to D32.39 was specific for the combining
site as it could be completely blocked by ,~ the mAb with soluble RQ~KVVT
peptide (SEQ ID NO:2). Individual beads from the library having lluu.e~:e.~e intens;ties
comparable with the positive control beads were sorted into microfuge tubes for tag
5 ~lmrlifirA~ion by PCR (beads with nUUlG~ ,G in the top 0.17% of the population were
collected). The amplification reactions contained dUTP and uracil DNA ~ ly~,usid~sG to
prevent carryover ~ ;.." with soluble product from previous amplifications (see
Longo ç~aL . l990, Gene ~;125-128, i~l~;u~ ' herein by reference). Nucleotide
sequences were obtained from 12 sorted beads and the deduced peptide sequences are
lO given in Table 1. R~l.,e~ ,v~ peptide sequences obtained from single beads having
IrlUUi~ ,G which was not c,~";l;. - :ly above background are also tabulated for
~OI~ U~I.
:I~I~l
High Fluul~ ,G Intensity Beads
SeQuence ~I
(SEQ ID NO:3) TFRQFKVT 0.29
(SEQ ID NO:4) TTRRFRVT 4.3
(SEQ ID NO:5) TVRQFKTT 8.8
(SEQ ID NO:6) QvRQFKTT 16
(SEQ ID NO:7) RQFRTVQT 76
(SEQ ID NO: 8) KQFKVTKT 340
(SEQ ID NO:9) QQFKVVQT 370
(SEQ ID NO:10) KQFKVTQT 410
(SEQ ID NO: I l) TQFKVTKT 560
(SEQ ID No:i2) TFRvFRVT 1400
(SEQ ID NO:13) FRRQFRVT not teste~d
(SEQ ID NO:14) RQFKQVQT not tested
WO 95/12608 2 ~ 7 5 5 8 7 PCTIUS9`1/123.17
56
Low Fluv~ Intensity Beads
Sequence Kd. mM
(SEQ ID NO:15) QTvTvKKT ~1
(SEQ ID NO:16) QQVQRQTT >0 4
S (SEQ ID NO:17) KTQvVQFT not tested
(SEQ ID NO:18) QvTQvRVT not tested
(SEQ ID NO:19) FVVTVRVT not tested
The data in Table I is consistent with earlier studies ~ that the
preferred 1~0~ UII sequence of D32.39 is localized to the six amino acid fragment
RQFKW of dynorphin B (see Cull et al., supra). The positively charged residues
arginine and lysine are strongly preferred in the first and fourth positions of the epitope
and phenylalanine appears e.~ ly as the third residue of this motif. ~t the second
positiûn glutamine is the favored residue in tbis library, while the aliphahc b-branched
15 amino acids valine (L; only) and threonine are clearly preferred as the flhh and
six residues. D-Valine appears to be best tolerated at positions outside of the consensus
motif. The range of affinities ûf peptides that were selected ( K~ - 0.3 - 1400 nM) v,as
not unexpected given the design of the binding assay: bivalent primary antibody with
labeled second antibody detection. r' . l~ n of the binding valency (for example~ by
~0 using directly labeled monovalent receptor) and the stringency of wash conditions will
improve the capacity tû isolate only the highest affinity ligands.
C. Small Molecule Synthesis
Although primarily described in terms of the synthesis of peptide or other
25 large polymer libraries, as noted previously, the various aspects of the present invention
are equally applicable to other chemical syntheses. Specifically, the apparatus and
methods of the present invention may be used to carry out a variety of chemical synthesis
steps in many synthetic protocols, including, for example, the synthesis of small
molecules. The apparatus may be employed in a chemical synthesis scheme to selectively
30 add reagents in, r _ with the synthesis protocol for the particular molecule.Further, different synthesis protocols may be carried out in parallel, as with
the p~ptide synthesis described above, where different reagents are added at different steps
to yield a diverse library of small molecules on solid supports. These diverse libraries
wo ss~l2608 2 1 7 5 5 8 7 PCTi TSg~/123~7
may be screened for desired properties by the methods described herein. Additionally,
where such a diverse library of small molecules is desired, the library may be tagged so as
to encode the synthesis steps which were involved in the synthesis of each discreie member
of the library of molecules.
Examples of the synthesis of such smal~ molecules on solid supports include,
for example, fhi~7r~ iinnnP~, mPi~hi~7r~Ti~iin(~nP~, and derivatives thereof, as described in
Example 2, included herein, and U.S. Patent Application No. 08/265,090, filed June 23,
1994, and i-lcul~u~ d herein by reference for all purposes.
D. Methods for Generating Soluble Libraries
For some ~l,}li~ , one may desire a "bead-free" or "soluble" library of
molecules. Soluble molecules, both tagged and untagged, can be useful for a variety of
purposes, including assaying the activity of a compound (see Section VI.B, below) and
amplifying a tag. There are a variety of ways one to generate soluble molecular libraries,
both tagged and untagged, and to solubilize ~u.~ u ~ both tagged and untagged,
syllLl-.,;~i on a solid support. Typically, cleavable linkers are employed in such
methods.
For instance, and as noted above in Section II.B, cleavable linkers can be
used to cleave tagged or untagged molecules from a bead or other solid support, thus
~r~ hili7in~ the molecule of interest. To produce a soluble tagged molecule, the cleavable
linker will be attached to the bead or other solid support and have at least two functional
groups: one for ~yllLll~ the molecule of interest and the other for synthesizing the
tag. Thus, the molecule and tag are ~y~lLh.~i~i attached to a common linker, which, in
turn, is bound to the solid support. Once the molecule and tag are ~ylllll.s;~:~, the linker
is cleaved to provide a soluble tagged molecule.
A single, planar solid support can be used to synthesize the library, and the
members can be cleaved from the support prior to screerling using very large scale
immr~hili7P~l polymer synthesis (VLSIPSn') technology. See U.S. Patent No. 5,143,854
<~ and PCT patent publication No. 92/10092, each of which is ill-,ull~ùldL~d herein by
30 reference. In one ~ o,lil" ,1 an array of ~ lig".. 1~ is synthesized on the
VLSIPS~ chip, and each ~I;G~ rIr ~ iP is linked to the chip by a cleavable linker, such
as a disulfide (see U.S. patent application Serial No. 874,849, filed April 24, 1992,
illl,ul~ulal~d herein by reference). The ri;~ lr.JliriP tag has a free functional group,
wo 95/1~608 2 ~ 7 5 5 ~ 7 Pc rlUS9~1123~7
such as an amine, for attachment of the molecule to be tagged, which is typically an
oligomer and preferably a peptide. The tag may optionally contain only pyrimidine or
pyrimidine and purine analog bases. The tag also contains binding sites for amplification,
i.e., PCR primer sites optionally a sequencing primer site, and a short section uniquely
5 coding the monomer se~uence of the oligomer to be tagged. Then, the oligomer is
synthesized, i.e., from a free terminal amine groups on the tag or a linker linked to the
tag, so that each oligomer is linked to a tag. The collection of tagged oligomers can be
released from the chip by cleaving the linker, creating a soluble tagged oligomer library.
Other advantages can be realized by generating soluble libraries of
10 molecules. In any bead-based library, the size (mass) of the bead will impose practical
limits on the size of the library that can be assembled. For instance, several grams of
beads may be required to assemble a library containing 109 different tagged molecules.
The present invention provides an improved method for ,~ g tagged molecular
libraries that enables one to obtain much larger libraries much more practically. This
15 improved method provides a means whereby the c~mro~n~ic are released from the solid
support prior to the mixing steps but are reattached to the solid support pnor to each
coupling step.
In this method, the tagged molecule is imn~hi1i7P~i on a solid support in a
reversible manner, allowing one to release the tagged molecule from the support during
20 each of the mixing steps of the method. In one ~ I,o~ ; this reversible binding is
provided by an 1~1tr~filtr~itm membrane (for suitable ". ,.1" .". ~, see, e.g., the
"Commercial C--mr~tihility Chart" in the Millipore catalogue, which shows ",~ ..,1."~"~ ~
inert to a variety of solvents and chemicals used in synthesis methods). A membrane with
a molecular weight cut-off of about 2,000 to 10,000 daltons (such as the Amicon YM5
25 membrane) would be suitable for most libraries. During the coupling steps, the molecules
of the library would be retained by the membrane, while the coupling and other reagents
would be drawn through the membrane by vacuum suction. The vacuum would be
released to allow the molecules to be mixed during the mixing steps.
In another ~ .,\ho.i;-"~,.l of the method, a reversible covalent linkage is used30 to attach the tagged molecules to the support during the coupling steps. Examples of
suitable reversible chemical linkages include (I) a sulfoester linkage provided by, e.g., a
thiolated tagged-molecule and a N-hydroxy-~ l-idyl support, which linkage can becontrolled by the NH20H ~v,~ t;~ ; and (2) a disulfide linkage provided by, e.g., a
wo 95112608 2 1 7 5 5 8 7 PCTr~vS9J1123~7
thiolated tagged-molecule and a 2-pyridyl disulfide support (e.g 7 thiolsepharose from
Sigma), which linkage can be controlled by the DIT (diLlliuLlllriL~ .
VI. Assay Methods
s
s The utility of large: ' I libraries for ligand discovery depends
critically on the availability of robust and affinity-sensitive bio- h~mir~l assay
m~th~ r,gi~ The present invention provides a number of novel assays for use withencoded synthetic molecular libraries, which in tum have a wide variety ~l~uliu~lLiull~. By
way of example, such libraries can be used in assays to identify ligands that bind
receptors, such as peptides and nucleic acids that bind to proteins, drugs that bind
therapeutic t~rget receptors, and epitopes (both natural and synthetic) recognized by
antibodies, as well as to identify a variety of v~ )u~ with ~,1, .""~ ~,11;. ,.1, ~vnr~ lr~l
and medical diagnostic ~I,,uli~ Given these diverse ~ , there are a wide
variety of assay methods relevant to the present invention. Two important types of assays,
albeit with some overlap, include bead-based assays and assays of soluble molecules.
In general, however, such assays typically involve the following steps.
The libraries are screened by assays in each different molecule in the library is assayed for
ability to bind to a receptor of interest. The receptor is contacted with the library of
synthetic molecules, forming a bound member between the receptor and any molecule in
the library able to bind the receptor under the assay conditions. The bound molecule is
then identified by rAdlllill~LiUII of the tag associated with that molecule. In one
rll,l,,.l;".. .1, the receptûr tû which the library is exposed under binding conditions is a
mixture of receptors, each of which is associated with an identifier tag specifying the
25 receptor type, and ~ lly two tags are examined after Lhe binding assay.
A. Screening Assays for Bead-based Libraries
When specific beads are isolated in a receptor screening, the beads can be
segregated individually by a number of means, including infinite dilution,
30 Illi, l.".,~.~;l",l~tiorl or preferably, flow cytometry. Libraries of tethered ligands are most
effectively evaluated in binding assays with soluble labeled receptors. By adopting
cell-sized solid supports or beads, one can use flow cytometry for high sensitivity receptor
binding analysis and facile bead ~Il- l;ll.llrl;llll
WO 95~12608 2 1 7 5 5 8 7 PCT/U59~1123~7 ~
Flow cytometry, commonly referred to as 111,..,, ~ . f- activated cell sortingor FACS should be viewed as equivalent to "fluorescence activated molecular sorting" or
"fluorescence activated bead sorting" for purposes of the present invention. One of
ordinary skill in FACS methods for cloning mAm~ n oells expressing cell surface
S antigens or receptors can readily practice the assay methods of the present invention. In
general, these assays involve the binding of a receptor labeled with a fluorescent tag to a
mixture of beads displaying the diverse molecules of a molecular library. After washing
away unbound or non-specifically bound receptors, one then employs a FACS instrument
to sort the beads and to identify and isolate physically individual beads showing high
10 IIUU~ C~ See Methods in Cell Biolo~v~ Vol. 33 (Dar~ynkiewic~, Z. and Crissman,
H.A., eds., Academic Press); and Dangl and TT~..,- ..1.~.c, 1982, J. Immunol. Meth.
~:1-14, both il~ulL ' herein by reference. Once the desired beads have been
isolated, one identifies the tag to ascertain the identity (or molecular structure,
~ u,.,~ , or conditions of synthesis) of the molecule of interest on the bead.
Standard FACS i~ permits bead (cell) lluul~l en.e analysis
rates of ~ 104 events/sec. and, when operated in single bead cloning mode, sort rates that
are 5-10 fold slower. In assaying very large libraries (e.g., > > 10~beads) some form of
affinity-selective pre-screen can be used prior to individual bead isolation with the
cell-sorter. For example, receptor-coated sub-micron sized ~U~ A~A~ c~ particles are
frequently used to affinity purify specific cells from large, mixed ~U,U.11A1;11,.C by magnetic
activated sorting (see Miltenyi et al., l99û, Cvtometrv 11: 231-238, ill~,UlL~Uld~d herein by
reference). To have a high probability of detecting very rare binding events, each
different compound in the library should be present on many beads in the librar~. A
practical upper limit for the size of an encoded librdry constructed from 10 llm particles,
assuming a hundred-fold l~lullddll~,y, is probably 10~- 109cu.. l,uu ' ayll~ l on ~
101 - 101l beads. Even larger libraries can be preparefi using smaller beads, but
conventional cytometers are unlikely to detect or manipulate particles much less than ~ 1
IAm in diameter- Of course, as noted elsewhere herein, the present invention provides a
variety of Ar~ innc for such small beads in the synthesis and screening of libraries of
30 molecules. For instance, by using the ~1;c~ f-J~ f tag c~ A~ I- method of the
invention, one need not use FACS 1"r~ ^cy to sort the molecules in the library.
~nnP~hP1Pcc, one should not u,.~i. .r~ ^ the power of FACS
Llulll~lltflLion for purposes of the present invention. In one assay method of the
2t 7~5~7
Wo 95/12608 PCTIUS9~ 7
61
invention, the tagged molecular library is ~yllLl.~iL~ on fluorescent beads. The beads are
smaller than cells and composed of a fluorescent material. The library is incubated with a
suspension of cells expressing a high level of a cell surface receptor of interest, such as a
G-protein-linked receptor. Of course, one can also perform a variety of controls, such as
5 conducting all steps with cells that do not express a high level of the cell surface protein,
and use those controls to identify false positives.
In any event, cells expressing the receptor can bind to any library members
presenting a ligand for the receptor. Flu~ lly labeled cells can be readily
1 and separated from fluorescent unbound library beads and from unlabeled
10 cells with a FACS instrument based on light-scattering or another fluorescent signal, e.g.,
from a cell nucleus. After sorting, the tags from the beads attached to the cells are
examined to identify the ligands specific for the receptor. Depending on the application,
one would sort for cells expressing the highest level of the desired receptor, e.g., by
selecting only the brightest cells, and would adjust the binding conditions to maximize
15 specific binding events. To ,~ between ligands specific for the receptor of
interest and those specific for other cell surface receptors, one could examine tags
associated with beads binding to cells expressing high levels of the receptor of interest and
cells that do not.
The methods of the present invention also enable one to use FACS
20 m~LI, ' " to sort tagged molecular libraries ~ ;L~ on beads much smaller thanthe smallest beads current FACS , are capable of sorting. In this method,
encoded synthetic libraries are screened for effector activity on signal trAn~ r~ir,n
pathways. The synthetic library is c~ Llu~ with several !l.n~ .,,c- (a) the beads
are I l~m or smaller and nePd not be sortable in the FACS, allowing rather small beads to
25 be used in some instances; (b) the tags are rl;~"", lr.~ Pc resistant to the in~r~r~ r
(most IJcuLi~.ul~ly nuclease resistant), pl.o~L,l,uloLllioates are preferred for this
purpose; and (c) the peptides (or other diverse chemical entities) are attached to the bead
support via a linker that cleaves in the 'I~ r l,.lVilU..lll_.lL. Such linkers include
`- linkers that can be cleaved upon the application of an external factor, such as light, that
30 does not harm the cells and linkers labile to the in~r~rPl~ r Gllvi~ -"~llt, such as a
.l,n~ bond or a disulfide bond, but in any case, the cleavable linker must be
stable to the parallel synthesis process.
21 75587
WO 95~12608 PCT/IIS9~/1231
62
The library beads are introduced into the reporter cells preferably by a
rnr rh~nir~l process such as, for example, biolistic projection. In some cases, a
hir~rhrmin~lly-mediated process leading to irt nr ~1i7~iinn can be employed, but this route
usually results in il,cu.,uu.~lliul~ into an ~ cellular ,UIII~J~U~ (i.e., Iysozomal
5 lnr~li7~tinn). Once the beads are in the cells, the peptides or other cull,puullds of interest
are released. Given that 10 llm beads have a ~ i capacity of 10'peptide (or
other) synthesis sites, then if the capacity scales with volume, a 1 ~m bead of th~ same
material would contain 107 molecules of the ~yllLll~ l peptide. If all of the synthesized
peptide is released in a single (spherical) cell of ~ 10 ~m diameter ( a volume of ~ 0.5
10 pL), then a c~ l,u;.", of free peptide of--30 ~lM would result. This ~
would be controllable by the synthesis density on the beads, and a lower loading density
could provide for a more stringent screening format (i.e., a screen for more active
~U!'~l'O~ ). The recipient cells are engineered to generate a fluorescent signal upon
activation or inactivation of the pathway of interest. The individual cells producing the
15 desired effect are selected by FACS ill ,LI~ and the tags, which are still attached
to the beads and contained within the cells, are amplified and sequenced to identify the
active synthetic ~u..,l~r,~
In another r ,l~bud;~ , large beads are employed and used to screen a
population of cells that express a receptor (i.e., the enzyme beta-g~ rtn~ ) capable of
~0 generating a fluorescent or other detectable signal, i.e., by cleavage of a substrate to
produee a detectable eompound. The beads are then mixed with a population of the cells,
which are allowed to attach to the beads. If the receptor on the cell surface is stimulated
by the compound on the bead, then the detectable compound is produced, providing a basis
for sorting activated cells attached to the beads from u-l~ Liv~Lcd cells. One could employ
25 ~ JlU~ , reagents (i.e., free labeled or unlabeled receptor) to maximize selection of high
af~lnity ligands.
There are of course a variety of alternatives to flow cytometry for purposes
of screening and selecting for library molecules of interest. In one F..ll.o.i;l~.. ,ll, an
encoded synthetic library is sereened for ~ulLil~ -ubial activity to find ~....1l,.~,..,.1~ that
30 retard the growth or kill bacteria or any other Illi~rlUOl~Ulhllll that can be plated in two
riimFn~ n~, such as virus-infected cells, many eulcaryotic cells including cancer cells, and
some protozoa. Large libraries of related or unrelated chemical structures can be screened
2~ 75587
WO 9~/12608 PCTIUS9~/123.17
63
against cells in agar culture by controlled release of the peptide or compound from the
bead on which it was ~yllLi~
The steps of the method follow: (1) plate the cells of interest on agar
plates; (2) overlay the cells with another layer of agar in which the beads bearing the
5 ~y~ ~ peptide/drug are suspended at a dilution that provides for even dispersion so
that individual beads can be picked, e.g., with a capillary tube, from the solid agar; (3)
initiate release of the peptide/drug from the beads; (4) culture the plate to allow diffusion
of the peptide/drug from the bead immr-bili7PA in agar into the ~UllUul~ lg agar and into
the agar below containing the indicator cells; (5) read the extent to which the diffused test
10 r~ u~ from individual beads have affected the growth/.,lul~ul~olo~ y/~ uLy~ of the
indicator cells; (6) choose zones where the indicator cells exhibit the desired response
(e.g., death of a bacterial lawn) and using a capillary tube or similar, pick out the zone of
agar that contains the original bead from which the test drug had diffused; (7) read the tag,
e.g., by PCR amplification of the encoded material on the individual bead, to determine
15 the structure of the peptide/drug that elicited the desired response; and (8) optionally
chemically synthesize the ~I,U,~IU~) ' drug/peptide and verify desired effect.There are a variety of ways to release the test compound from beads. For
instance, one could partially cleave the peptides/drugs from beads using TFA and allowing
cleaved peptide to dry down onto the bead surface in such a form that subsequent20 Ir~ ", in water (agar) will allow release of the peptide/drug and Inr~1i7~tinn of the
released compound to the zone of agar around a particular bead. One could link the
drug/peptide to the bead using chemistry that is sensitive to a particular change in bead
environment that can be initiated upon plating onto the agar and indicator cells or after
plating and agar 5111itijfir:~ti(m, e.g., a l,l..,t~ e linkage, a thiol sensitive linkage, a
25 periodate sensitive linkage, etc. These chemical agents could themselves be diffused in
through another thin agar overlay, if necessary. Such release chemistry must be
compatible with the integrity of the test substance, integrity of the encryption on the bead,
and he Ith of the underlying indicator cells. The particular release chemistry used will
. also of course depend on the type of chemistry used for synthesizing the library and the
30 nature of the indicator cells. The method is especially preferred for screening libraries of
beta-lactam antibiotics for i~Pntifir~tinll of new antibiotics that might kill newly evolved
strains of bacteria resistant to existing beta-lactams and for screening peptide libraries of
analogues of known anti-bacterial peptides such as the magainins.
:
~ 1 755&7
Wo 95/12608 PCT/USg~/12317
v4
Other methods can also be used to screen bead-based molecular libraries.
Affinity adsorption techniques can be employed in l..Ulljl.ll~,LiUII with the libraries of the
invention. For example, the mixture of beads can be exposed to a surface on which a
receptor has been immobilized (see PCT patent publication No. 91/û7û~7, iliCUIL)vlcl~t~d
5 herein by reference). After washing the substrate to remove unbound beads, one can then
elute beads bound to the surface using conditiûns that reduce the avidity of theul;ZSvll1~.~l~Lul interaction (low pH, acid treatment, or base treatment, for example)
The process of afhnity adsorption can be repeated with the eluted beads, if desirable.
These methods, and related variants, such as the use of magnetic selecfion, describe~i
10 above, can be practiced in diverse ways; for instance the solid support can be a resin
packed into a ~ jC column.
In another mefhod of the invention, libraries of "tethered" compounds are
used as a source of structural diversity in a form suitable for affinity pllrifc:lfinn of
families of related molecules, such as families of receptors of,ul1~ ulogic interest. In
15 general, this method relates to the use of a tagged and tethered molecular library to screen
a second library of untagged molecules. The tagged, tethered library molecule serves as
an affinity pllrih~finn reagent to screen complex mixtures of soluble proteins,
ol;g,~ lP~,~I,vl,yd1~Lt~,antibodie5,etC. Subsequenttoaffinity,u ~ 'r~
molecules that bind to the . ..I..I.il. ,1..,;,,l library members are identified by elution and
20 .~,uu~u, ' separation and i,i.or,fih,~fi~m methods. The: ' I library is then divided
into smaller fractions of ~ ~ liy ,J..LI ~ u"-~ to determine, through
repeated cycles as necessary, reductively and precisely which compound(s) mediate the
binding process.
In similar fashion, ~ chemical libraries can be used to identify
25 and clone novel receptors. Many receptors are members of families of proteins that share
sequence homology (usually reflecting divergent evolution from an ancestral parent) but
exhibit differences in their specif.~i1y/drr....~y for structurally related sets of
ligands/cognate receptors. Each member of a receptor family (R~,) may represent a
separate target for specific ,UI~ UlOy,;C action and hence for drug discovery and
30 dcvclu~,--.~.-~ by virtue of their different properties, i.e., locations in the body, ~recir~ifirs
affinities for ligands, etc. If one identifies a receptor (R,) whose binding properties are of
sufficient interest so that the i~nfifi~fitln of other receptors in the same family would be
beneficial, then one can employ the following method to identify receptors related to R, in
wo g~/12608 PCT/USg~1123 17
their binding site properties. One first identifies a ligand that binds to R, and then creates
a tagged . . .".1,i1, I' ll iAI compound library of molecules closely related structurally to the
ligand.
Next, one prepares polysome ~ Ud~irVlls from cells believed likely to
5 express additional members of the receptor family. Such polysomes comprise ribosomes
attached to mRNA with pendant receptor in various stages of protein synthesis from
nascent peptide to almost fully elaborated protein. The receptor protein nearing completion
of synthesis will express the specific receptor property of binding to one or more members
of the c, ~ 1 library. Using the ~U~ library tethered to solid support,0 affinity Fllrifi~Atir n of polysomes bearing receptors with affinity for any member of the
iA1 library is performed. Such affinity ~u~ i ri~ may involve column
. ' . methods, batchwise separation of imn~-hili7r-~ ~;u~ -.r.~ from the
liquid phase, or aqueous two phase separation methods to achieve separation of the solid
phase bearing attached receptor and relevant mRNA encoding the receptor from
15 non-adherent polysomes.
Next, one performs cDNA synthesis from the mRNAs that encode the
cognate receptor population using standard technology (reverse LIAI'`' ~ ., etc.) and
clones the cDNA population into a vector suitable for rapid sequence analysis. Dependent
on prior knowledge of the receptor sequences that are li~ely and the degree of sequence
20 Cul,~.,vALiu-, that can be ~Antil-ir:~f~A, one may attempt to use PCR or another AnnrlifirAtirln
to amplify the cDNAs enriched by this method. By sequence analysis of a suitable number
of cDNA clones, one can identify cDNAs (whether full length or not) that show sufficient
sequence homology with the sequence of the already known receptor Rl to represent
putative additional members of the same receptor family (R~). One prepares ûptionally full
25 length cDNA clones of these novel cDNAs (or relevant portions thereof, such as the
portion encoding the e~tr~r~ Ar domain of relevance to ligand binding) by standard
cloning methods and expresses these cDNAs by standard methods (i.e., in eukaryotic
expression systems as soluble or membrane bound proteins as Al,lv,u~u ). Using
~, st;andard formats for testing receptor ligand int~rA~tir~n, one tests for binding of populations
30 of mixed~u~ -u~ fromthe IUlll~ I;Al libraryorindividual~ ,v~ Inthisway,
one can identify precisely which compound(s) from the library bind to the newly identifled
receptor.
21 755~7
WO 95/12608 PCT/IIS9 11123~7
66
Individual beads can be physically separated, for example, by limlted
dilution or by methods similar to those in which cells are incubated with a receptor
coupled to small ~ L~ beads and then cells expressing a ligand for the
receptor are extracted using a high power magnet (see Miltenyi et al., 1990, Cvtometrv
11: 231-238 ;II~.UI~JI ' ' herein by reference). As noted above, m~nPtil~lly selected
cells can be further analyzed and sorted using FACS. ~ P~ may also serve to
label a receptor, allowin~ one to identify and isolate beads by selecting beads that are
l~lio~ Li~ ly labeled.
B. Screenirlg Soluble Molecules
One can also employ tagged molecular libraries to useful effect in novel
assays of the invention in which a ligand is solubilized in either tagged or untagged form
prior to binding to a receptor of interest. For screening very large libraries of soluble
(bead-free) tagged molecules, one preferably employs affinity ~ n~ y under
conditions of weak affinity. For example, a 3û mg library of lûl8 molecules can be
screened with a simple 10 mL affinity clll -ll a..~ .l y column containing a few hundred
~g of a receptor of interest. O ~ IP5 are preferred tags for such libraries, being
readily PCR amplified and cloned into the commercially available TA cloning vector
(Invitrogen, Inc.), a convenient form for storing tag inf~rm~rion prior to analysis by DNA
sequencing. In addition,, l ~ ' '- tags can be, 1, as described above,
allowing one to collect pools of soluble tagged molecules, clone the .-~ 1 tags from
the selected pools, and then sequence the tags to identify the desired comr~ln~
Soluble tagged molecules can also be screened using an immllhili7P~i
receptor. After contacting the tagged molecules with the ' 1i7P~I receptor and
2'i washing away non-specifically bound molecules, bound, tagged molecules are released
from the receptor by amy of a wide variety of methods. The tags are optionally amplified
and then examined and decoded to identify the structure of the molecules that bind s
specifically to the receptor. A tagged oligomer in solution can be assayed using a
receptor ' '~ ' by attachment to a bead, for example, by a ~omrPti~inn assay with a
30 Il,--..r~ ly labeled ligand. One may recover the beads bearing imm~-bili7P~I receptors
and sort the beads using FACS to identify positives (diminished fluorescence caused by the
library molecule competing with the labeled ligand). The associated identifier tag is then
be amplified and decoded.
~ WO 95/12608 2 1 7 5 5 ~ 7 PCTIUS9.11123~7
The soluble molceules of the library ean be ~yllLi~ ~ on beads and then
cleaved prior to assay. In one c~lbvdilllc~L, the llliClU~ u~,;c beads of a molceular library
are placed in very small individual culll~uLlll~ or wells that have bcen "nanofabricated"
in a silicon or other suitable surface. Beads are loadcd in the wells by dispersing them in
5 a volume of loading buffer suffieient to produee an average of one bead per well. In one
~llll,odill.~,L, the solution of bcads is plaeed in a reservoir above the wells, and the bcads
are allowed to settle into the wells. Clcavage of the oligomers from the bcads may be
""~ A using chemical or thermal systems, but a l - 1 v~lc system is preferred.
The moleeules of interest ean be clcaved from the beads to produce either untagged
10 moleeules in solution (the tag remaining attaehed to the bead) or tagged moleeules in
solution. In either event, the moleeules of interest are eleaved from the beads but remain
eontained within the ~:ollll,~UL,,,~I~L along with the bcad and the identifier t~Lg(S).
In one _..lbo~i".~l.L, a surfaee or a portion of the surfaee of the well is
eoated with a rceeptor. Binding buffer and a n.,.,,. ,~ y labeled known ligand for the
15 receptor is added to the well to provide a solutiûn phase . . on assay for ligands
speeific for the receptor. The binding of the nuvlc~. . .lLly labeled ligand to the rcceptor
can in one embodiment be estimatcd by eonfoeal imaging of the monolayer of immobilized
reeeptor. Wells with deereased llu~lc ,~ ~.lcc on the reeeptor surfaee indieate that the
released ligand eompetes with the labelcd ligand. The beads or the tags in wells showing
20 eompetition are examined to reveal the identity of the Culll~LiLivc ligand.
Reeovery of identifier-tagged beads from positive wells may optionally be
effcctuated by a ll,i~,u,., ~ .. to pluek individual beads out of wells. Another mode
involves the use of beads that have illCUllJI ' ' a fluoreseent moleeule, either during bcad
~l~lur~cLulc or through labeling. A laser of the a~lllul vv~1. ..6LII is used to bleach
the resident beads in only the positive wells. All the bcads are then removed cn masse and
sorted by FACS to identify the bleaehed positives. The assoeiated tags may then be
amplified and dccodcd to identify the molceules that bind speeifieally to the reeeptor.
In another c.,Lodi,.,c,lL of the invention, one employs relatively large tagged
~- bcads, from whieh the moleeules of interest are eleaved in a series of reaetions. In this
method, the beads are 50 to 500 ~um in diameter, with eapaeities cquiva~ent to 100 to 50û
pmol of peptide per bead; preferably, one uses 100 ~Lm beads with a eapaeity of about 200
pmol, if c u~Llu~Lill6 a peptide library. The typical size of such a library is from about
106 to 10~, preferably 107 different molecules. The library is dividcd into about 100 poo~s,
21 755~7
wo 95112608 r~ Tluss~ll23~7
68
each containing about 100,000 beads. A certain p~r~ ntrgf-, about 25 7'o, of tlle molecule
of interest is cleaved from the pool, producing, in the case of a peptide library, for
example, each peptide at 50 nM in a volume of 1 mL.
The cleaved pool is then tested in a ~ i..,. or functional assay. One
5 identifies the pools with the highest activity, and then retrieves the remainder of the
original pool and aliquots the remainder into 100 pools of 1000 beads per pool. The
process is repeated until one has a single bead, from which one reads the tag and identifies
the compound of interest. This method avoids the resynthesis and frame limitations of the
Houghten method and is adv~l~..~ in that the pools are random, rather than related,
10 ~u~ ,u ,~l~ The chances of a mixture being active because of the cumulative potency of
many low affinity related molecules is reduced.
C. Screening Natural Product Libraries
With the automated high flux assays that are now available, the present
15 limitations in natural product screening are first, the ability to obtain and handle (dispense,
dissolve, label, etc.) the samples; and second, the substantial effort required to characterize
the active ~ of positive samples. The present invention provides methods for
generating and screening natural product libraries that can provide a huge number of
samples in readily screened form and to identify active ~..,,,1,.,,...,1~ in the samples. The
20 basis of the method is the ...."h;"~l;.", Of hiflfh,-mir~l and chemical diversity with
metabolic diversity from "natural products", i.e., from nature. The simplest example
involves feeding collections of peptides to cultures of ~ u~r,o~ IlC Each microbial
strain might create many modifled peptides (a metabolite library). Because each culture
would (potentially) contain a very complex mixture of I~ Lbol;t~s~ an efficient method of
~5 screening is required.
Several approaches are available and might be orthogonally classified as
factored or tagged, and soluble or tethered. For the sake of illl~ctr~tif)n, cons;der as the
"feedstock" a library of soluble peptides. An aliquot of the library is incubated with each
of the many strains typical of a IlliWUUl~ lll r~ td~iUII screening program, and the ~
30 media screened in typical fashion. Positive cultures are then incubated with subsets of the
libraries and rescreened. This process of factoring continues until the input peptides
generating the most active mrt:~hfllitf~c are identi~ied. The . ~ ", of the active
metabolites then proceeds aided by the knowledge of the likely precursor molecules.
~ wo 95112608 2 1 7 5 5 8 7 p~ S9~1123~7
69
Thus, the first screening identifies the active organism(s), subsequent steps identify the
active precursors, and finally, the active ~ dboli~ are identified by standard analytical
means.
In all its formats, however, factoring is a tedious process. Libraries
5 produced by split synthesis and cleaved free of the resin produce soluble uu~ Jùul~ds
amenable to cellular uptake and Illc~dbolialll by intact organisms. However, theff~nfPntrAfinnQ of the individual fnmrounfic is quite low (inversely related to the diversity
of the collection), leading to inefficient enzymatic turnover and very low cnnfrntr~innc of
the resulting ~ The ~ of the c.~ may be increased by
10 producing subsets of the libraries and fermenting each subset separately with each
microbial isolate. Sub-libraries are ~ull~Lluu~cd by fixing one or more of the positions and
" -ficl".;,;.,e the remaining positions. For example, there are 500 pentapeptide sublibraries
containing all pf-rml,t~tinnc of 2 fixed positions utilizing 50 building blocks. Each of these
...1,1;1" . ;, ~ contains 125,fLiOO ~ ùll 1c The use of tagged libraries offers a major
15 advantage in ease and sensitivity, but requires rnffiififAtinnc in the method of exposing the
compound collections to the metabolic activities. The .~"..1. . ~-II;AI feedstock need not be
only peptides but could consist of any type of c~ ;AI chemical collections.
Oligomer and other molecular libraries can be cullaLIu~Lc~i in a ~ulllbilldiulidl
process and each step encoded with identifying tags. This may be dfne via a direct
20 linkage and parallel synthesis of the oligomer to the tag. If f~ ifif-C are used as
the tags, then the complexes will be relatively large but small enough to insert actively into
the cells via liposome fusion, elc. L u~uu,dLiu.., solvent ~ I,;1;,AI;.,.., etc. Once inside,
the complexes would be subject to the metabolic machinery of the cells. One would avoid
the vulnerability of the olign~-lrlf-~ifif- tags to ri~lAllAI;- I~ by the use of modified
25 nllrl~rJtifl~s and nucleotide linkages. Upon recovery of the active " 1-l~ from the
culture of from Iysed cells, the samples are screened and the tags decoded to reveal the
precursor compound. Scaled-up fi-rmPr~ inn of the active organism with the active
precursors should produce sufficient quantities of the active m.ot~hr,litrc to ~~- Libraries of . u",l,uu,~ made by an encoded ~O~ IAI~ process on beads can be
30 exposed to Iysates of bacteria, fungi, plant cells, etc. With this format, the need to insert
the tagged complexes into intact cells is avoided, and only a relatively few of the many
molecules on the bead need be processed to be detected (e.g., in a nl,ulc~.c.l~c-activated
binding assay).
WO 9~112608 2 ~ 7 5 ~ ~ 7 PCT/US9.11123-17
Another useful method of the invention involves the utilization of the
products of one mlerobial culture as feed for another culture. To illustrate, consider a
collection of 100 different microbial isolates from large scale cultures (~ I liter). The
~U~ .L~u,L of each culture is recovered by filtration and divided into one hundred 10 mL
aliquots. Eaeh aliquot is inoculated with one of the 100 strairis and incubated. 10,000
samples (1 :-I,nl;lr~ of "~rlAI..,Iilr~) are thereby generated from the 100 microbial isolates.
This method of .~ ,L~Ib~ can be extended to sequential ~ LabOli,lll by
greatly different species: subjecting the product of microbial fPrmPnt~tinn to ineubation
with exotic plant Iysates or incubating extracted fractions of plant tissues with fungal
10 cultures, for example. These methods can be used in ~.,,,,1,;1,-l;..; any product of a
chemical diversity generating method can be subjected to these sequential l~l~ Llboli~."
product exposure steps.
In another aspect of the invention, natural product diversity is screened by
creatirlg a mixture of c, ' 'Iy-tagged liposomes, each liposome preferably
15 rl~ g only one member or a simple mixture of a natural product compound
library. The invention allows for the ~ P~ assay of lOOO's-lO,OOO's of chemical
UO~ JUUlldS or natural product extracts and assay of lOO's of ~ ly separated
fractions derived from natural product extracts that are signal-positive. In this rnnnPrtinn
~cimlllt~nPo~ means assayed together in the same tube with the cells of the readout
20 system.
The mixture of, I 'Iy tagged liposomes is prepared as follows.
For each individual natural product extract or cbemical from an inventory, one prepares
separate liposomes r~ lg the test substance in aqueous phase. A unique liposome
tag is il~co,l~o,_Lcd into the liposome IJlc~u~Liull at the time of Pnr~lrslllAtinn The
25 liposomes can be Iyophiliæd for long-term storage at lu.. t~ clALulC~ a significant
advantage to the collection and storage of natural product samples near the site of
colleetion, as well as for the long term storage of the natural product extract in a form
suitable for subse~uent ~ulll~ Lulidl ~ .l,r,;",r..lAI;nn The lipids in the liposome
preparations are preferably identical for all samples and chosen in terms of types and
30 rnmrn~itinn to produce unilamellar liposomes of the desired siæ and integrity. Agents
such as trehalose can be included at the time of liposome folmation to allow Iynrhili7Atinn
and subsequent rcronctitlltiorl of intact liposomes by addition of water. At the time of
generatiOn/lr~ of tagged liposomes Pnr~r~lllAtin~ the extract/chemical, one can
2~ 75587
WO 95/12608 PCT/US9~/122~ 17
71
also use a high pressure technique that allows for the r^nrA-rs~ ^n of greater volume of
aqueous phase than the calculated volume enclosed by the liposome. A 3 - 5 fold irlcrease
in volume-equivalent can be rn^AArclllAte~i by this pressure method, allowing greater
volume of test material to be tested, hence greater signal in the cell-based readout.
Existing liposome technology allows for creation of liposomes that
incorporate a high percentage (>80%) of the aqueous phase (relevant to the efficiency of
use of each test substance). U-lu-~oll,u-dlcd aqueous phase can be removed by diverse
"wash" methods. In addition, one can create liposomes that do not leak or exchange
eni^Arc~ aqueous phase (relevant to the specificity of tagging and absence of mixing
enclosed aqueous phases), as well as liposomes that do not exchange ~ inserted
into their lipid monolayer (glycolipid/protein antigens inserted as tags cannot be
exchanged).
A wide variety of tags can be employed with the method. For example, the
tags can be: (a) different lluulu~I-ol~, with excitation and emission properties that allow
the nuulu~ ulc~ of each to be measured in the presence of each of the others, or",~ thereof -- the nuulu,.l-ul~ can be selected to partition in the ~ J1
aqueous phase or in the membrane phase of the IC~J'I`l;ll'~' i liposomes, facing outwards;
(b) different metal cations of rare earth elements that can be distinguished individually by
atomic absorption ~e~lUIll~LIy -- the rare metal atoms would be designed to partition as
salts in the ~ ^i aqueous phase of the lc~ Iiposomes; ( c) different
antigens, that can be di~Lill~ui~ i by their specific reactions with d~ lUI rAi n îî I~^,nAI
antibodies and primary/secondary florescent detecting antibodies~nu<,~ ul- ^, as necessary
-- the antigens, bome on proteins, ~Iy~uL~Iut~ ;l., and/or glycolipids, can be selected to
partition in the membrane phase of the ~ ' liposomes, facing outwards; and (d)
~o-l-l,;,-a~ s of antigens, IIUUIU~ OICS~ and/or metal ions can greatly increase the number
of possible signatures for CimlllrAA~uc screening, and an additional level of tagging of
different liposomes (increased numbers) can result from use of different levels of
lI.,ulu~Jl.ulcs/metal ions/antigens, such that the different "quanta" of each comronent in the
signature mixture could be identified.
3û One can also employ a general fluorescent tag that shared by all liposomes
that enables rapid selection of cel~s fused with a liposome from those that did not fuse with
a lirosome. This tag is distinct from any tags used in ~ labelling of the
individual liposome ~ ,audLiul,~ and is mixed in with the liposome-generating lipids, the
WO 95112608 2 ~ 7 5 5 8 7 PCTlU59iill23 i7
72
signature tags, and the aqueous sample of drug/natural product at the time of liposome
generation. One can also employ a fluorescent tag that is self-quenched at high density
(i.e., in the liposome membrane) but that will exhibit lluu~ r 1l~ r in the outer cell
membrane of a cell after a liposome fusion event and lateral diffusion of the fLuorophore in
5the cell membrane. Depending on the mode of liposome fusion with cells, one can also
incorporate a fusogenic protein of viral origin, or a glycolipid, for example, that will F
mediate ti~ht adhesion of liposome to cell (dependent on a lectin like adhesion process
mcdiated by a suitable rceeptor ;- .,,1,l.,...1 as necessary into the cell line used for read
out). Such an element would not affect liposome-liposome irteraction (an event to be
10 avoidcd) but can enhance the efficiency of liposome-cell fusion.
The method can employ a cell read-out system that utilizes a cell line that
contains a reporter gene (e.g., luciferase, beta-g~ t~ s~) dU..~I~L1~III of a promoter
that is activated in response to addition of an exogenous ho}mone or ligand, such as a
steroid, cytokine, ~ ;." antibody, antigen, etc. to the cells. Binding of the
15 activating ligand to either a cell surface reccptor or an intr~rt ll~ r receptor activates a
signal cascade that leads ultimately to activation of the responsive promoter and
Ll~ul~ iL)L;ull of the signal gene. Exprcssion of the signal protein leads to generation of a
signal from the individual activated cell that can be deteeted LI~ LiL~ Li~Cly. In a search for
Ctlrnro~ln~iC that act on any part of the intr~tll~ r signal ~ ;.". cascade as
20 r~nr~goniC~ the entire population of cells ean be pretreated by addition of the exogenous
signal agonist (cytokine, hormone, ete.), and one measures a deerease in signal output on
an individual cell basis after the liposome fusion event. In a seareh for ct~mrt~llntl~ that act
on any part of the intraeellular signal l,~.lc.~i.,. ~;I~n easeade as agonists, no exogenous signal
nced be addcd to the cells, and one can measure the -l~u~-,.---- ~ of a signal on an
25 individual cell basis after the liposome fusion event.
The mixture of tagged liposomes is mixed with a very large number excess
of read-out cells. Cell number excess is eritic~al such that after liposome-cell fusion only
the following products will result: (i) cells that did not fuse with a liposome; and (ii) cells
that fused with one liposome (acceptor cells). Efficient mixing is essential at this step and
30 can be performcd using a ~..1.l;"l..7u~ly stirred or linear-flow cell suspension to which the
liposome mixture is added at a slow rate. Fusion is initiated by standard methods such as
addition ûf PEG or applieation of a high voltage. Fusion may be enhaneed if necessary by
inclusion of a fusogen or a ligand-receptor recognition pair into the eell-liposome
WO 95/12608 5 5 8 7 PCT/tJS9 1/123J7
",. .,.~ The ~usion step ef~ectively adds the aqueous phase ~ulllL/alL~ of a single
liposome to an acceptor cell. Hence, the aqueous natural product extract, test compound
from a chemical inventory, or fraction from .1.~ ".~ separation of a natural
product extract is now able to act at any point in the intr~t~Pillll~r signal tr~nc~ tif~n
5 pathway. The fusion step also adds the specific tags that provide the signature of the
particular test compound sample to the individual acceptor cell. If those tags were in the
lipid membrane of the liposome, then the tags are distributed in the outer cell membrane of
the acceptor cell. Antigens at this location are accessible to panels of specific mcmf~ n~
antibodies. Rare earth metal ions that were in the aqueous phase of the particular
10 liposome are in the acceptor cell cytoplasm. The fusion step also adds the shared liposome
tag that identifies cells that acted as acceptors from those, the excess, that did not undergo
a liposome fusion event. The tag can be a nuulu~llulc; that moves from the liposome
membrane to the acceptor cell membrane.
The mixture of cells, cells fused to individual liposomes, and any unfused
15 liposomes is next incubated with the exogenous ligand (e.g., in the case of testing for an
antagonist) or incubated without any addition (e.g., in the case of testing for an agonist).
The time of this incubation is dPtPrmi ~A using control c,~ u ,~ at defined
and incubation times.
Preferably, one uses FACS to select ~ u~; u~ (cells) of interest. For
20 instance, one can first use forward or side light scatter to sort cells (whether acceptors or
not) from any unfused liposomes. Large cells can be readily separated from smallliposomes. Next, one can sort cells that were liposome acceptors from those, the excess,
that were not liposome acceptors. Cells that were acceptors bear the shared
liposome-derived fluorescent label, whereas the non-acceptor cells are non-fluorescent with
25 this label. This step is of course optional but, if performed as a presort, allows separation
of the (typically) majority of cells that are irrelevant to subsequent analysis from the
minority that were acceptors. For i~iPntifi~ti~m of an antagonist, one can sort on the basis
of light emission from the reporter protein (e.g., beta-g~l~rt~ciA~cP or luciferase),
- separating the majority of lluul.,.,~ ~.lc~-positive cells (rendered such by addition of the
30 exogenous ligand earlier), from the minority of nuUI~ i~.., c-negative cells or low
lluUIc~ cells. The latter two cell categories result from presumed antagonist effects
of cu~ u~,,-A~ that were ~ 1 nr~l in the particular liposomes that fused with these
individual cells. For i~iPntifi~tit-n of an agonist, one can sort on the basis of light
wo g~/12608 2 1 7 5 5 8 7 PCT~s9~
emission from the reporter protein, separating the majority of lluu~ e-negative cells
from the minority of lluul~ cll~r-positive cells. The latter cells have resulted from a
presumed agonist effect of liposome-derived compounds.
In some ~ one can sort all cells of interest according to tlle
5criteria above as a population and collect occasional cells as cloned individuals using
standard FACS methods. These individual cells can be analyzed as single cells for the r
particular tags that they bear, allowing precise i~ l of the particular liposome that
mediated the desired effect. In other .~ , one can analyze the tag ~iicfrihufif~n in
the entire sorted event-positive population and dependent on the design of the experiment
10and particular tags that had been ill~Ul~JUl~i in samples from different
times/locationslinventories, be able in a first pass to determine the diversity of tag types in
the total event-positive population.
Collected single cells or pop~ til nc of cells can be analyzed by methods
d~lU~JlidLt: to the particular tag ~ c used. Fl~ tags can be analyzed by
15FACS and/or traditional a~e LIu~,l,u~u.l.~,Lly. Antigen tags can be analyzed by addition of
d~ JlU~Jl' ' Iy labeled mt)nll~ n:~l antibodies and ELISA, FACS, l.l-lio;~u~u~ic, or
!1lll.l..` ~. f"' C' assisted assays. Metal ion tags can be analyzed last by atomic absorption
alJ~Llulllc:~ly. After tag decoding, the tests can be repeated either with mixtures of only
those liposomes that yielded a positive event on first pass or with pure liposomes of each
20 member of interest added to separate cell samples.
These and other methods of the invention can be automated to facilitate
practice of the invention, as discussed in the following section.
VII. IllaLl Ulll~"lLdli
The coupling steps for some of the monomer sets (amino acids, for example)
can in some f lllbO.lilll.llLa require a relatively lengthy incubation time, and for this and
other reasons a system for ~,clrulllling many monomer additions in parallel is desirable.
The present invention relates to automated ill;,Llulll~..lhLiull for use in generating and
30 screening encoded synthetic molecular libraries. One preferred instrument, able to
perform 50 to 100 or more parallel reactions cim~ rPmlcly, is described in U.S. patent
application Serial No. 08/149,675, filed November 2, 1993, illcull-u~d~d herein by
reference. Such an instrument is capable of fiiclrjhlllin~ the reaction mixture or slurry of
WO 95/12608 ;~ ¦ 7 5 5 8 7 PCT/US9-11123~7
synthesis solid supports, under ,ulu~ "~ f control, to the various channels for pooling,
mixing, and l~diaLl;buLiull~
In general, however, the il.,Llu~ lL~lLiul~ for generating synthetic libraries of
tagged molecules rcquires plumbing typical of peptide ayllLh~ a, together with a large
S number of reservoirs for the diversity of monomers and the number of tags employed and
the number of si~ coupling reactions desired. The tag dispensing capability
translates simple ill~LIu-,Liulls into the proper mixture of tags and dispenses that mixture.
Monomer building blocks are dispensed, as desired, as specif~ed mixtures. Reaction
agitation, L~ dLulc:, and time controls are provided. An dl~,UlUI 1~ designed
lû instrument also serves as a multi-channel peptide a~lLi.~ ,i~. capable of producing 1 to 50
mgs (crude) of up to 100 specific peptides for assay purposes. Sce also PCT patent
publication 91/17823, illCul~ ' ~ herein by reference.
Typical ill~LIulll~,lLdLiull comprises (1) means for storing, mixing, and
delivering synthesis reagents, such as peptide and ..1;~ synthesis reagents; (2) a
sealcd chamber into which the various reagents are delivered and inside of which the
various reactions can proceed under an inert ~I~lllu~ , (3) a matrix of sealed reaction
vessels; (4) means for directing the flow of reagents to the d~JIJlU,UlidL~ reaction vessels: (5)
means for combining and ,u~uLiLiullillg small (û. 1-100 ~Im) beads; and (o) means for
washing the beads in each reaction vessel at the conclusion of each chemical reaction. The
matrix of reaction vessels can have any one of several designs. For example, the vessels
can be arranged in a circle so that the vessels can be made to rotate about a central axis
(i.e., a centrifuge). Alt~ Li~ ly the vessels can be arranged in a 12x8 matrix (96-well
microtiter plate fommat). Any iu ~ amenable to accessibility by robotic delivery,
aspiration, and transfer functions is useful for some arr1i~ ~til~n~
The system used for combining and ,~;~ .u~ particles can have one of
several designs. For instance, the beads can be suspended in a solvent of d,UI~lU~lidL~
surface tension and density such that a robotic pipetting instrument can be used to transfer
the beads to a combining vessel After mixing, the beads can be l~diaLIibut~ to the
-- rcaction vessels by the same robotic pipettor. Altematively, the beads can be combined by
using a special valved reaction chamber. The valve is opened to allow solvent flow to
transfer the beads to a combining vessel. After mixing, the beads are I~LiLiull~ by
reversing the flow to each reaction vessel.
76
In another embodiment, the beads are combined using closely spaced
reaction vessels with open top ends. Flooding the vessels allows the beads to mix. If the
beads are magnetic, then the beads are re-partitioned by pulling the beads back down to
the bottom of the vessels by application of a magnetic field. Non-magnetic beads are
re-partitioned by vacuum suction through the bottom of the reaction vessels. In yet
another embodiment, the beads are partitioned by distributing them on a flat surface and
then restricting them to certain sectors by covering them with a "cookie-cutter" shaped
device, described more fully below.
The system for washing the beads can also have one of several designs.
The beads can be washed by a combination of liquid delivery and aspiration tubing. Each
reaction vessel has its own set of tubing, or a single set can be used for all reaction
vessels. In the latter case, the liquid delivery and aspiration lines can be mounted on a
robotic arm to address each vessel individually. The beads in each vessel can be made to
form a single pellet by either centrifugation or the use of magnetic beads and application of
a magnetic field. One can also employ a reaction vessel with a bottom wall composed of a
chemically inert membrane so that reagents can be removed from the vessels by
application of a vacuum. Reagents can be also be removed from each vessel by using vessels
that can accommodate continuous flow through of reagents and washing solutions, i.e., a
vessel with luer fitting and membranes on each end.
Any automated combinatorial instrument that relies on an individual reaction
chambers, each connected to reagent delivery systems and to a "mother pot" to which the
beads are pumped for pooling and from which the beads are reallocated among the reaction
chambers for successive rounds of monomer addition faces a very important practical
limitation. There is a wealth of monomer or other building block units, and the difficulty
of partitioning beads and reagents among the potentially large number of reactions may
limit such instruments to fewer than 100 separate parallel reactions.
The present invention provides an instrument that avoids the need to pump
beads between chambers to mix and reallocate, simplifies reagent delivery, and allows the
simple and accurate partitioning of very small numbers of tiny beads. The basic design
consists of a plate with an array of reaction "sites" located on the surface; the surface may
be planar or may consist of an array of shallow wells that form reaction sites. In one
embodiment, there are 256 sites in a 16 x 16 array. Each reaction site is a spot, or well,
on the surface to which a group of synthesis beads is attracted. The attractive force may be
~ WO 95/1260~ 2 ~ 7 5 5 8 7 PCT/US9~/123~7
m~nrlicm, vacuum filtration, gravity with passive mPrh~ni~l sorting, or various other
simple means. The beads are initially applied as a dilute slurry in a shallow reservoir
evenly covering the array of reaction sites. Upon application of the attractive force, beads
are ,.... ~"~ A at each site.
After positioning all the beads on the reaction sites, the sites are then
separated by mPrh~nir~l partitions to create (temporarily) the individual reaction chambers
as shown in Figure 29. A variation provides partitions p~ lLly affixed to the surface
to form shallow wells. The reaction ~. \1.."....1~ are delivered to each chamber, the beads
released into Cllcr~Pncirm, and the reaction initiated. When desired, the beads can be
reattached to the surface and the reagents removed. After all steps for a coupling cycle
are completed, the chamber partitions are removed, and the beads are released into the
common reservoir above the array of sites.
Mixing of the beads is caused by induced convection of the reservoir fluid,
and the beads are then reattracted to the surface sites for the next round of coupling.
Subsecluent steps, including the wash steps, are ~ t~ i in a similar fashion.
Addition and removal of reagents is done with a c~.",l, ~ " of plumbing and automated
pipetting. Addition of reaction specific reagents (mrml-mPr~, for example) may be done
with robotic ""~ Addition of common reagents and the removal of all reagents
can be done with a fixed plumbing system not recluiring valving at each reaction chamber.
Some common steps such as washes can be done on the beads gl ma~, before installing
or after removing the chamber partitions.
The use of large numbers of monomers or other building bloclcs places an
additional burden on the encoding process. In one encoding scheme for nli~ lPolirlp
tags, a basis set of 1000 monomers might require a 5 base secluence to tag each reaction
step; a set of more than 1024 monomers could require 6 bases to encode. To reduce the
plumbing complexity of the synthesis instrument (i.e., to reduce the number of specific
reaction additions), a special encoding strategy is provided by the present invention. To
illustrate the method, consider an array of 16 x 16 reaction sites, an ,.,,~ ,c,. ..1~ .1l that
`- allows 256 different reactions to be carried out cim~ r,P~ucly To encode each reaction
individually with multiple base coupling is a difficult ul~d~k,l~ g~
The array consists of 16 rows and 16 columns, each site in the array having
a unique ~r,~",.l.l,;r:~l address. Each row of sites can be tagged as a group, and all 16
rows can be uniquely encoded with 2 base codons ("subcodons"). A striped template or
wo g~JI2608 2 1 7 5 5 8 7 PCTIUS9~/123~7 ~1
78
channel block can serve to form the 16 reaction chambers for these 2-base additions (note
that the bases are coupled as monomenc ~ if ~C not as dimers). See PCT
patent publication No. WO 93/0966g, iul~u~ cd herein by reference. This form of
addressing of the reaction sites is analogous to others; for example, an optical method can
5 be used to label the beads, as in the striped masking process described in U.S. Patent No.
5,143,854, il-~ul~JuldL~d herein by reference. If a template or channel block is employed,
then the template is lowered onto the synthesis surface just as is the grid template that
isolates the individual reactions during synthesis of the library molecule.
The beads are not released during the fagging reaction, however, as their
10 spatial ~,I~ALio,l must be maintained through the next step. When the rows have been
tagged with subcodons, the template is lifted, rotated 90, and lowered to form stripes
covering the columns. The 16 columns are then labeled with 2-base subcodons, resulting in
the unique tagging of each of the 256 reaction addresses with a 4-base "~u~ l"odu,l". By
identifying the reaction addresses, the au~ u~:lull~ also specify the monomer that waS
15 added in each reaction.
VllI.Apparatus for Parallel Coupling Synthesis Reactions
In general, the device of the present invention provides for the synthesis of
20 diverse materials on a solid substrate. By way of example, the present invention utilizes
beads such as those described herein. Examples of a fhe synthesis of diverse peptides are
discussed below to provide a framework for the discussion of the ~yl.tll~i~l.
The invention will utilize a plurality of substrates, referred to as "S," on
which synthesis reactions take place. The substrates are optionally provided with a linker
25 molecule "L" on which coupling reactions take place. The substrates are divided and
reacted with diverse monomers, such as "A" and "B" to form collections of, for example,
the following substrates:
S-A and S-B
Thereafter, the substrates are l`~U'I'I';l~''''l, mixed, and divided again. After such mixing
30 and dividing steps, two or more collections of substrates are formed, each containing both
S-A and S-B.
~ WO95/12608 2~ 75~ PCT/US9.1/123.17
79
Additional coupling reactions then take place. For example, the pooled
products above may be reacted with monomers C and D to form the following collections
of products:
1. S-A-C S-A-D
2. S-B-C S-B-D
t From even the above simple example it becomes apparent that such synthesis techniques
rapidly create large collections of diverse products. By carefully planning the synthesis of
such diverse collections of molecules and/or by providing for the parallel synthesis of tags
on such substrates, the substrates will fnd use in a variety of ::rplin ~tilm~ as described
above. In a particular ~ - the present invention provides devices and methods
that may eff1ciently generate substrates for these and other uses.
A. General
Fig. 1 illustrates a device used to synthesize diverse collections of
molecules. The device includes a parent mixing vessel 200 coupled to a plurality of
reaction vessels 201-209 by a top common manifold 212 and tubes 215, and 221-229. Top
common manifold 212 couples to tubes 221-æ9 and 215. Reaction vessels 201-209 also
selectively couple to monomer addition reagent supply reservoirs 231-239 via valves 111-
119 and tubes 241-299. A l~ delivery system (PDS) 265 is coupled to both
parent vessel 200 and reaction vessels 201-209 via tubes 260 and 256 respectively.
A synthesis reaction begins when a bead suspension is transferred from
parent vessel 200 to reaction tubes 201-209. A valve 129 opens (all valves are closed as
default), and the bead suspension enters top common manifold 212 from parent vessel 200
through tube 215. The bead suspension is thereafter distributed among reaction vessels
201-209 through tubes æl-229. Selected reagents from monomer reservoirs 231-239 then
enter respective reaction vessels 201-209 through respective tubes 241-249. Coupling
reactions then take place inside reaction vessels 201-209 on beads contained therein.
Fig. 1 shows ylw~ i~l delivery system 265 coupled to parent vessel 200
- via a tube 260. P~W~Uli~l delivery system 265 delivers pl~;a~ d reagents to parent
vessel 200 from delivery system 265 via tube 260 when valve 10 and vent valve 90 open.
Plw,u~ d delivery system 265 is also coupled to reaction vessels 201-209
through a tube 256, an isolation valve 100, lower manifold valves 101-109, lower tubes
271-279, injection valves 111-119, and tubes 241-249. To deliver a reagent from PDS
wo 95/12608 2 ~ 7 5 ~ ~ 7 PCT/US9~/123~7
265 to selected reaction vessels 201-209, the pressurized reagent enters tube 256 and a
lower manifold 214 through open valve 100. Thereafter, the ~ UI;L~ reagent is forced
up selected tubes 271-279 through selected open valves 101-109~ The ~JII...~UIi~l reagent
is then forced into selected reaction vessels 201-209 through selected tubes 241-249.
To deliver a reagent from, for example, a given monomer reservoir '231 to a
respective reaction vessel 201, a quantity of ~ D~u~ d activating solution from PDS 265
is forced into tube 256, into lower manifold 214, and up tube 271. At an d,~)~JlU~
moment, a quantity of monomer reagent from monomer reservoir 231 is injected into the
stream of activating solution travelling up tube 271. Following the reagent injection, the
stream of solution including the monomer reagent injected from reagent reservoir 231
enters reaction vessel 201 through a tube 241 to participate in the coupling reactions.
To optionally tag the beads inside selected reaction vessels 201-209 w;th a
monomer from monomer reservoirs 406-412, a ~ UIi~.i tag monomer reagent from
monomer reservoirs 406-412 enters a common manifold 255 of PDS 265 through an open
valve 4-7. Thereafter, the pressurized tag monomer and an dl~lJlUIJlidL~ activation reagent
enter lower manifold 214 through open valve 100 and tube 256. The pl~auliL~ monomer
tagging reagent and its d~lJlUIJlidlC activation reagent travel up selected tubes 271-279 and
241-249 through selected open valves 101-109 into selected reaction vessels 20~-209 where
the synthesis of tags on beads takes place.
After desired monomer and/or tag addition reactions, the bead suspension in
reaction vessels 201-209 is transferred back to parent vessel 200 for pooling and mixing.
To move the bead suspension from reaction vessels 201-209 to parent vessel 200, the bead
suspension is ~ ,auli~ d with argon from tube 250 via open valve 122 and 101-109.
Valves 100 and 110 are closed, thereby pushing the ~I-,a~Uli~i bead suspension into tubes
221-229, top common manifold 212, tube 215 through open valve 129, and finally parent
vessel 200. In parent vessel 200, the bead suspension is mixed in ~ JdUd~iUII for re-
allocation among reaction vessels 201-209 to further synthesize the desired set of
molecules.
In an alternative ~ ,of~ a plurality of three-way valves can be
provided between each of the reaction vessels 201-209 and tbe parent vessel 200. Such
valves will preferably be held in the top common mamifold 212. In this way, certain
vessels can be isolated from the parent vessel 200. This may be particularly advantageous
when l~di~LIibu~ g the bead suspension for further synthesis. For example, if the beads
WO g5/12608 2 1 7 5 5 8 7 PCT/US9~/123~7
81
were initially allocated to all the reaction vessels 201-209 for synthesis and then returned
to the parent vessel 200, during re-allocation only certain of the three-way valves could be
opened so that, for instance, only reaction vessels 201, 205 and 209 received the bead
suspension for further synthesis.
Fig. 1 also shows n~ ",f ~ agitators 280 and 285 coupled to a top
reaction vessel bracket 290 and a bottom reaction vessel bracket 295. Top reaction vessel
bracket 290 is held stationary while bottom reaction vessel bracket 295 is permitted to
follow the motion of n---,f~ agitators 280 and 285. Each n--,...-.... ,.I~ic agitator
cooperates with a vortexing motor 300 to exert an agitation force on bottom vessel bracket
295 and the bottom end of reaction vessels 201-209. Since the tubes between the brackets
are flexible, this agitation force causes the contents of each individual reaction vessel 201-
209 to vortex inside the reaction vessel thereby enhancing synthesis reactions.
Top common manifold 212 connects to tube 215 at one end to provide a
conduit for L.dll~rt,~ g material between parent vessel 200 and top common manifold 212.
At the other end, top common manifold 212 connects to a tube 126. Tube 216 provides
~lc~ cd argon to top common manifold 212 through a valve 121. Tube 216 also
allows top common manifold 212 to vent its contents through a valve 120.
Two capacitive sensors 90S and 99S are located near the exterior surface of
parent vessel 200 to detect the level of liquid in parent vessel 200. If a fluid exists within
the detection envelope of a capacitive sensor, that capacitive sensor is turned on.
Conversely, the capacitive sensor is off if no fluid exists within the detection envelope.
Sensors lOlS-119S are optical sensors for detecting the presence of a fluid
within a cllhct~nti~lly translucent tube. These optical sensors are on when a column of
fluid is present in the tube. The optical sensors are off when no fluid is detected.
Likewise, an acoustic sensor 120S detects the presence of a fluid in its
detection envelope. Fluid, including bead ~crPncirm flowing through a tube which has
been placed in the acoustic sensor's detection envelope turns acoustic sensor 120S on.
Conversely, acoustic sensor 120S is off when no fluid is present in the tube which has
~- been placed in the detection envelope of the sensors. Acoustic sensor is used for sensor
120S because optical sensors cannot reliably f~ictin~,.ich, under certain conditions, between
an empty translucent teflon tube and a translucent teflon tube containing a bead SllcrPncif~n
Further, although an acoustic sensor is chosen for sensor 120S, any sensor which can
distinguish the difference between an empty tube and a tube filled with either a fluid or a
wo 95/12608 ~ 1 7 ~ 5 8 7 PCTlUS9~/lZ3
82
bead suspension may be used. Because acoustic sensors are fairly costly relative to other
types of sensor such as optical sensors, there is only one acoustic sensor 120S per
synthesiær.
The reaction vessel bank is designed such that there is only one or fewer
5 valve between the parent vessel and the }eaction vessels. In fact, valve 129 is optional.
This design is advd,l~5cvu~ because it reduces the possibility of the mechanical opening
and closing action of valves damaging the fragile beads and the synthesiæd polymers.
A~so, if the size of the beads are large enough, they may become lodged in the valve and
clog the system. FUI~1-. .-11~111, some polymers are ~III~ld~UI~ sensitive and may be
10 adversely affected by the heat generated by the valves during operation. Accordingly, it is
desirable to reduce the number of valves through which the bead suspension must traverse.
If valve 129 is not included, ~ techniques can be used to prevent fluids from
flowing between the parent vessel and the reaction vessel banks.
As discussed earlier, the valves used in this r,.~l-9(1;.~ are closed in their
15 default state. Absent a specific command to open, the valves always remain in this default
closed state.
B. MPrh~nir~l t'nnnr~mPnt~
MPrh~rir~lly speaking, the automated synthesizer may be roughly divided
20 into three subsystems: the reaction vessel bank, the parent vessel, and the pressuriæd
delivery system. As mentioned above, coupling synthesis takes place at the reaction vessel
bank inside the reaction vessels. The parent vessel holds, pools and mixes the beads from
all reaction vessels. The delivery system ensures that the proper solvent and/or reagent
solution in an d~ Ul reagentlsolvent ~ . is delivered to either the parent
25 vessel or the reaction vessel bank at an d~ Jl;d~t~ step in the synthesis process.
Furthermore, the entire system is sealed during operation. plr~ " where
necessary, is done with an inert gas such as argon. Argon is the preferred ~ UIi .;llg
agent because of its availa~ility and low chemical reactivity.
For ease oF discussion, the automated synthesiær will now be described
30 with reference to a specific example. The specific example used throughout this disclosure
involves the synthesis on beads of a set of polypeptides. The beads are tagged for
i~l~ .lli~i. .l;.~,~ following each amino acid coupling reaction with four nucleotide monomers:
A, T, C, and G.
2 1 75587
WO 9S/12608 PCTIIJS9 ~/123~7
83
It must be recognized, however, that the automated synthesizer is neither
limited to the synthesis of the particular polymer described in the above specific example
nor to tagged polymer synthesis. Although reference will be made throughout thisdisclosure to the synthesis of polypeptides and the tagging of beads with the above
S nllnl~-o~ c utilizing a reaction vessel bank having nine reaction vessels, there is no
P inherent upper or lower limit in the number of reaction vessels which may be included in
each reaction vessel bank In fact, modular ~ aLlu~_liull of the device permits easy
addition of additional reaction vessel banks. Further, many other molecules and tags may
be ayllLl~ on the beads, or the tags may be eliminated entirely.
1. The P1c,au1i~ Delivery System
The detailed description of a ~ au~ ;l delivery system 265, which has
been specifically tailored to synthesize the polypeptides according to the specific example,
has been divided into three parts: reservoirs for use syllLl1.,,iL;11~ polypeptides, reservoirs
for use in tagging beads, and the delivery vaives.
a. Reservoirs for use in syllLl-~ g polypeptides
In addition to the nine amino acid monomers used to synthesize the peptides
of the specific example, several other additional "common" reagents will be employed in
the synthesis. In a typical peptide synthesis, for example, the following reagents may be
employed:
TABL~ 2
D~1u~t~Liu,l lû% Piperidine in DMF
Activation O.~M HBTU and 0.6M DIEA
in DMF/DCM mixture having a 3:1 ratio
Capping acetic anhydride in THF
~- n-methyl imidazole in THF
Washing DMF
THF
2 1 7 ~ 5 8 7
wo 9s/1260~ Pcrluss~ 7 ,~
84
b. Reservoirs for use in tagging beads
In the specific example, the beads are optionally tagged. In a peptide
synthesis reaction, the beads are in one . ' ' tagged with a nucleic acid comprising
n~ oti~ from tlle group A, T, C, and G. In addition to the four nucleotide reagents,
5 the following solvents and reagents are used during the synthesis of tags.
TABLE~ 3
Funcbon ~ ~ Chemical
n~ ut~Liu-- trichloroacetic acid in DCM
Oxidation I2, collidine, HlO, and MeCN
Activation 0.5M tetrazole in MeCN
Capping acetic anhydride in THF
N-methyl imidazole in THF
Washing MeCN
These nucleotide reagents are contained in reservoirs having suffic;ent
volume and quantity to ~rcn~rlich the synthesis of tags utilizing the automated
synthesizer.
Flg. 2 shows a lc~.. ,.~.l~live reseNoir 400 for containing, for example, the
MeCN solution in Table 3. There are two tubes associated with each reservoir listed in
Tables 2 and 3. As shown in Fig. 2, a tube 450 contains ~ ul;~d argon for
~Ui~:~U1;Lil1g the reservoir to force the contents of the reservoir up a second tube 452. In
some ~ d; ~ , the reagent reservoirs are always pressurized. In other embodiments,
the argon tube is controlled by a local on/off valve to pressurize a reagent reservoir only
when the contents of that reservûir are needed.
c. Delivery Valves
Fig. 3 shows a IC~ ' "YC 3-port solenoid valve in greater detail. The -~
valve may be, for example, a Model 2-110-900 by General Valve Colp. of Fairfield, New
Jersey. Further, the 3-port valve of Fig. 3 represents, for example, valve 4 which delivers
a reagent from reservoir 412. Three-port valves are used in the ~ u~ ,d deliverysystem 265 of Fig. 1. The 3-port solenûid valve employed in the present embodiment
2 1 755~7
WO 95/126~)8 PCT/US9~/123~7
includes a first port 454 and a second port 456. The 3-port solenoid valve also has a
channel 458 through its body that . u~ ; .UrC with first port 454 and second port 456
and always permits a fluid to flow freely between the first and second port. To fo}m a
common manifold 462, second port 456 is coupled with, for example, first port 454 of
5 another 3-port valve. The other 3-pc~rt valve may be, for example, valve 5 of Fig. 1. A
solenoid inside the valve, responsive to a control signal through wires 190, selectively
permits a third port 460 to c..., . l l~ - with channel 458. Third port 460 of valve 4 is
coupled to reservoir 412 of Fig. 1.
To control the injection of a reagent from reservoir 412 into manifold 462, a
line 464 carrying the~ u~ reagent is connected to third port 460 of valve 4. At an
d~ ulJliaL~ moment, the solenoid opens and permits third port 460 to ~nnnmllni~^ with
channel 458, thereby causing the ~ UI;LCd reagent from third port 460 to be injected
into channel 458 of valve 4 and into common manifold 462.
Fig. 4 shows a ~ lLaLiv~ on/off 2-port solenoid valve 10. This valve
may be, for example, a Model 2-17-900 by General Valve Corp. of Fairfield, New ~ersey,
for selectively permitting a fluid or a gas to flow in a channel between its two ports. As
shown in Fig. 4, valve 10 includes two ports 468 and 470. Valve 10 also has a channel
through its body that cnmn~ ni~ between a first port 468 and a second port 470 to
permit a fluid or a gas to flow between the two ports. A solenoid inside valve 10,
responsive to a control signal through wires 472, selectively permits first port 468 to
~u, . ,;l ~1~ with second port 470. When one port of valve 10 is connected to a tube
carrying a ~ UliLVd gas or fluid, valve 10 can be used to permit or inhibit flow from
that port to the other port of valve 10.
Fig. 5 shows in grc~ater detail a ~JIC~:~UIi~ delivery system 265 according
to one aspect of the present invention. Fig. 5 shows 24 valves 0-23 through which a
common manifold is fûrmed. The 2-port and 3-port valves are daisy-chained by coupling
their first and second ports together so as to form a common manifold through which
reagent flows. Three-port valves 1-7, 9-12, and 14-22 may be, for example, substantially
~^ similar to valve 4 of Fig. 3. Two-port valves 0, 8, 13, 23 may be, for example,
3û s~lhct:lnli~lly similar to valve 10 of Fig. 4. The common manifold includes the through
channels of the 3-port valves and of the on/off valves, as well as the coupling tubes
between adjacent valves. As previously mentioned, the third port of a 3-port valve is
controlled by a solenoid in the valve. The third port of each 3-port valve is coupled to a
2775
wo 95/12608 5 8 7 PC r/uss~ll23~7
86
tube from a reagent or solvent reservoir to transfer a reagent or solvent to and from PDS
265. Alternatively, the third port of a 3-port valve may serve as an exit port for
delivering reagents to, for example, valve 100 of a reaction vessel bank. Two-port valves
are used primarily as isolation valves or argon supply valves.
As shown in Fig. 5, the 24 valves are phystcally arranged in three separate
banks to save space. The delivery system of Fig. S also includes a tube 480 for ~r
connecting the left bank of valves with the center bank. A tube 482 cormects the right
bank with the center bank. Table 4 lists the valves used in the delivery system, specifying
the types of valves used and the reagent reservoir controlled by each valve in a typical
2t 75587
WO 95112608 PCT/US9 1/123~17
87
TABLE 4
Reservoir
Va~ve ~ Reservolr
- 5 0 On/Off -) Argon
FWO 60 400 MeCN
r 2 FWO 60 402 1% trichloroacetic
3 FWO 60 404 Tetrazole
4 FWO 30 406 C
10 5 FWO 30 408 T
6 FWO 30 410 G
7 FWO 30 412 A
8 On/Off -- --
9 FWO 30 414 Waste
1510 FWO 30 416 Bottom of Parent
Vessel
11 FWO 30 100 RV Banks
12 FWO 30 420 Waste
13 On/Off -- --
14 FWO 60 422 Top Parent
2015 FWO 60 424 ~ collidine, H~O,
16 FWO 60 426 Acetic anhydride in
THF
17 FWO 60 428 in THF
18 FWO 60 430 Piperidine
19 FWO 60 432 HBTU
2520 FWO 60 434 DIE~A
21 FWO 60 436 MeCN
22 FWO 60 43~ DMF
23 On/Off -- Argon
-, For example, valve 22 is shown to be a FWO60 valve or a fast wash out
(FWO) 3-port valve having a 60/1000-inch through channel. FulLllc;llllult, valve 22
controls a reagent from 1"~ reservûir 438 which, as indicated by Table 4, contains
DMF. As a further example, valve 23 is an on/off valve controlling the flow of
~f 75587
wo 95/12608 PCT~159VI23~7
88
Uli~ argon from an argon supply source (not shown~ to the common manifold of
PDS 265.
Fig. S shows a tube 484 connected to valve 0 for ~ Uli~illg the common
manifold of PDS 265 from one end. Anothe} tuoe 486 is connected to valve 23 and
5 pressurizes the common manifold of PDS 265 with argon from the other end.
As an ili1lc~r~ion, the operation of ~ Uli~.~ delivery system 265 during a
peptide synthesis deprotection cycle is described below. For d~.ut~Liul, of polypeptides,
a solution of 10% piperidine in D~F is delivered to the reaction vessels in the reaction
vessel (RV) bank. Table 4 indicates that valve 18 permits the flow of piperidine from
reservoir 430. Cu~s~L~,lLly1 valve 18 needs to open to permit piperidine from
~Ir~aaul;~l reservoir 430 to flow into the common manifold of PDS 265. To force the
solution to enter RV bank valve 11, isolation valve 8 is closed and isolation valve 13 opens
to force the ~ uli~cd piperidine to enter open RV bank valve 11.
As shown in Fig. 5 and Table 4, RV bank valve 11 and parent vessel valve
15 10 are centraily located in the chain of valves. This ~ f ~1 ad~UlL~eU~I~Iy
minimizes the length of the manifold section between these valves and a given reagent
valve. Cullac LU~..Lly, a smaller volume of reagent is recLuired to fill up this manifold
section. Isolation valves 8 and 13 can be closed to prevent the reagent from one end of
the manifold from uvc~ oULil,g RV bank valve 11 or parent vessel valve 10 and from
20 Il,,,,rr~ ly entering another portion of the common manifold.
Table 4 also shows valves 4-7 and 9-12 to be 3-port valves having a through
channel dimension of 3011000 inch. In contra3t, the remaining valves in the manifold have
a through channel dimension of 60/1000 inch. The reduced channel cross section further
reduces the volume in the respective portion of the manifold. Cu-ls~ucllLly, less reagent
25 is needed to fill up the manifold.
For example, nucleotides A, T, C, and G are relatively costly. It is
therefore desirable to keep the volume of reagent used to the necessary minimum.Nucleotide valves 4-7 are located proximate to RV bank exit valve 11 to keep the distance
between a nucleotide valve and RV bank exit valve 11 short and the required volume of
30 reagent low. The cross section of the manifold along the path from any nucleotide valve
to RV bank exit vaive 11 is aiso kept small to further reduce the volume of nucleoîide
reagent present in the manifold. In fact, tube 480 of Fig. S as well as the portion of the
21 75587
WO 95112608 PCT/US9-J/123~7
89
manifold between the nucleotide vaives and isolation valve 8 have a reduced cross section
of 30/1000 inch.
C. Reaction Vessel Banks
Fig. 6 shows a simplified reaction vessel bank 500 according to one aspect
of the present invention. For ease of iilllctr~til-n, tubes through which solution flows have
been partiaily deleted. Reaction vessel bank 500 includes a top bracket SOZ, two side
brackets 504 and 506, and two bases 508 and 510. A top reaction vessel bracket 290
attaches to side brackets 504 and 506. A vortexing motor 300 attaches to top reaction
vessel bracket 290 for supplying an agitation force to a pluraiity of reaction vessels 201-
209 via a drive belt 521 . The bottom of reaction vessels 201-209 are attached to a bottom
reaction vessel bracket 295. Brackets 502, 504, 506, 290, and bases 508 and 510 may be
constructed from any suitable material. For ease of machining, strength, and light weight,
aluminum is used to construct the above-mentioned brackets in the present ~mhQ~imPnt
lS Bottom reaction vessel bracket 295 iS attached to top reaction vessel bracket
290 by two n~ shafts 280 inside shaft housings 520. N~ l iC shahs 280
are rotatably mounted through aperhtres (not shown) in top reaction vessel bracket 291).
. ;. shafts 280 are u~.~ ly coupled to a vortexing motor 300 through belt
521. As will be discussed later, -- - shafts 280 translate the rotational force
supplied by vortexing motor 300 to an agitation force for urging bottom reaction vessel
bracket 295 to move in a circuiar pattern. This circular motion exerts a vortexing effect
upon the contents of reaction vessels 201-209. Because every reaction vessel 201-209 is
attached at its respective lower end to bottom reaction vessel bracket 295, ail reaction
vessels are agitated uniformly and ! ' ''
Fig. 6 aiso shows a bottom bracket 522. Bottom bracket 522 is attached to
side brackets 504 and 506 and may also be Cu~ ul,t~ from any suitable material,
including aiuminum. A pluraiity of amino acid reservoirs 231-239 are mounted beneath
bottom bracket 5æ The amino acid reagents in amino acid reservoirs 231-239 are used
~~ as building blocks for ~ Lil~ul~, the set of puly~l~Lidc~s of the specific example. The
present . ~ ' using nine different amino acid monomers per bank for
g the set of A~oly}~.d.,s.
Fig. 6 also shows an isolation valve bracket 526 attached to side brackets
504 and 506. Isolation valve bracket 526 includes a channel 530 for mounting a plurality
wo 9~12608 ~ ~ 7 ~ 5 ~ ~ PCT/U59~11
of lower manifold valves 101-109. As shown in Fig. 6, each lower manifold valve 101-
109 is secured within channel 530 in the present ~ ùd~ .lL. However, lower manifold
valves 101-109 may be secured to isolation bracket 526 using commercially available
mounting hardware or othe} mounting methods. Lower manifold valves 101-109 are 3-
5 port solenoid valves and are the same as the 3-port valve discussed earlier in connection
with Fig. 3. As used in reaction vessel bank 500, the through channels of lower manifold
valves 101-109 are coupled together to form a common lower manifold 214 through which
solution from the pressurized delivery system 265 flows.
Three 2-port valves 100, 110, and 122 are also shown in Fig. 6. Nine
valves 101-109 control the flow of solution from lower manifold 214 to the nine reaction
vessels 201-209. Isolation valve 110 at a first end of the common lower manifold 214
opens to a waste line (not shown in Fig. 6). Isolation valve 100 at a second end of
common lower manifold 214 selectively inhibits or permits the flow of solution from PDS
265 to the rest of the common lower manifold 214. An optional isolation valve 122
supplies local argon pressure to assist in the delivery of solution to and from various
portions of reaction vessel bank 500.
In another el--bo~ -l-.-L, the eleven isolation valves 100-110 and 122 are
provided for in an 1 l-valve block such as model P/N601374, by ABI of Foster City,
California. The block comes ,~ s~ "~1~1 1 and thus simplifies ~UII~Llu.Liol~. The eleven
valves of the 11-valve block function substantially as discussed above.
An injection valve bracket 532 made of a suitable material such as aluminum
is attached to side brackets 504 and 506. A plurality of injection valves 111-119 are
mounted through apertures in injection valve bracket 532. Fig. 6 shows a total of 9
injection valves 111-119 to control the injection of amino acids from nine amino acid
reservoirs 231-239 Injection valves 111-119 are 3-port solenoid valves and are the same
as the 3-port valve discussed earlier in connection with Fig. 3. The first port of each
injection valve couples to a reaction vessel 201-209 while the second port of each injection
valve is coupled to the third port of a lower manifold valve 101-109. The coupling is
a..u,..l,li,licd with ~ ,UI 'y sized tubes, such as the 1/16-inch teflon tubes employed
30 in the present ~ I,udi-,-..-L. As is apparent from the foregoing, the through channel of
each injection valve 111-119 permits a solution to flow freely between a reaction vessel
201-209 and the third port of a lower manifold valve 101-109. The third port of each
injection valve 111-119 is connected to an amino acid reservoir 231-239 to selectively
~ WO 95/12608 2 ~ 7 5 5 8 7 PCT/US9.J1123J7
91
inhibit or permit an amino acid to be injected into a stream of solution flowing between a
lower manifold valve 101-109 and a reaction vessel 201-209.
Fig. 6 also shows a top common manifold 212. Top common manifold 212
includes nine manifold ports 542 for connecting top common manifold 212 to the nine
reaction vessels 201-209. In the present ~ ~l;",. , 1/8-inch flexible teflon tubes are
r used to couple manifold ports 542 to the top end of reaction vessels 201-209 Top
common manifold 212 also includes a first end port 544 for connecting with a parent
vessel (not shown in Fig. 6) where the beads from individual reaction vessels 201-209 are
pooled and mixed together. A second end port 546 connects top common manifold 212
with a 3-port pressure/vent valve (not shown in Fig. 6). The pressure/vent valve and
second end port 546 provide another route through which lJlC~ UliLI~ argon, solutions,
reagents, etc., may be supplied to top common manifold 212. Altematively, the
pressure/vent valve and second end port 546 provide an additional route through which
aul;L~ argon, solutions, etc., may be vented from top common manifold 212 to thedylJIO~JIiALc reservoir.
An altemative Al IA I~ for the reservoirs 231-239 is shown in Fig. 6A.
Instead of employing a single group of reservoirs, e.g., reservoirs 231-239, a plurality of
groups of reservoirs can be provided, e.g., 231a-239a, 231b-239b, etc. The reservoirs are
held within a rotatable carousel 1000. The reservoirs are open at a top surface 1002 of the
carousel 1000 so that the reagents held in the reservoirs can be accessed from the top
surface 1002. The carousel 1000 is held within a pressure vessel (not shown) so that each
of the reservoirs are subjected to the same pressure within the pressure vessel. To transfer
the reagents in the reservoirs to the reaction vessels 201-209, a plurality of tubes in
ll with the tubes 241-249 are disposed within the pressure vessel. The tubes
within the pressure vessel are placed into the reservoirs of a selected group of reservoirs,
e.g., reservoirs 231a-239a. The tubes can be placed into the reservoirs by translating the
tubes toward the reservoirs or by translatung the carousel 1000 toward the tubes. The
carousel 1000 is rotated to align the tubes with the selected group of reservoirs. The
pressure within the pressure vessel is such that a pressure gradient exists between the
reservoirs and the tubes 241-249. When the tubes are placed into the reservoirs, and when
selected valves 111-119 are opened, the pressure gradient drives the reagents in the
reservoirs into the tubes 241-249 for delivery to the reaction vessels 201-209 as previously
described. The carousel 1000 thus provides flexibility to the ~y,.Ll~ L~. by allowing the
WOg5/12608 2 ~ 7 5 ~ 8 7 PCT~S9UI23~7 ~
92
reagents to be selectively chosen from a variety of different reagents, e.g., allowing a
different set of building blocks to be used at each synthesis step.
Fig. 7 shows in greater detail the i-~k~ ,r~lion~ among an amino acid
reservoir 231-239, an in~ection valve 111-119, a lower manifold valve 101-109, and a
reaction vessel 201-209. A lower manifold valve, for example, valve 101, which is a
three-port valve, is connected to common lower manifold 214 so as to permit a solution to r
flow freely between the Flrst port and the second port of lower manifold valve 101. The
third port of lower manifold valve 101 is connected via tube 271 to either the first or
second port of the 3-port injection valve 111. The other port of either the first or second
port is coMected to one end of reaction vessel 201 via tube 241. The other end of
reaction vessel 201 is connected to a manifold port 542 of top common manifold 12 (not
shown in Fig. 7) via a tube 2Z1. Tubes 271, 241, and æl are made from a chemically
resistant material such as teflon. In fact, the present ~ u~ employs translucentPTFE and FEP teflon tubes of various cross-sectional dimensions throughout because of
the low reactivity and optical .1,."-~ of the translucent teflon material.
Amino acid reservoir 231 is LJIC~ UI;~ with an inert gas such as a}gon via
tube 562. The UlC:~Uli~d amino acid solution in amino acid reservoir 231 enters the third
port of 3-port injection valve 111 through tube 560. Upon receipt of an d~ylu,ulialc
command, injection valve 111 opens to permit the l~ amino acid solution to enterthe through channel of injection valve 111.
l~ig. 7 also shows two optical sensors lllS and 101S. Optical sensors lllS
and 101S detect the presence or absence of a liquid within substantially translucent teflon
tubes 271 and 241. As shown in Fig. 7, optical sensor lllS is positioned below injection
valve 111 and optical sensor 101S is positioned below reaction vessel 201. Data from
optical sensors lllS and 101S are sent to a control computer (not shown in Fig. 7) for use
in controlling various phases of the synthesis reaction.
Fig. 8 shows in greater detdil the ~ C,--, IIlif agitator 280 of Fig. 1.
N""- o ,~ l "~ agitator 280 includes two cylindrical shafts 564 and 568 coupled to a
cylindrical knuckle 566. Shaft 564 aligns lt)n~itll~in~lly with the radial axis of cylindrical ~
knuckle 566 and is coupled at one of the two planar surfaces of cylindrical knuckle 566.
Shaft 568 is coupled to the other planar surface of cylindrical knuckle 566 and is offset
from the radial axis of cylindrical knuckle 566. In one emT-o~iimrnt, shafts 564 and 568,
and cylindrical knuckle 566 are machined from a single piece of metal stock.
WO 95/12608 t 7 5 5 ~ 7 PCTJUS9.1/123~7
When cylindrical shart 564 is torqued to rotate within a fixed rotary support
such as a roller bearing, cylindrical shaft 568, which is offset f}om the axis of rotation of
cylindrical shaft 564, moves in a circular path around the axis of rotation of cylindrical
shaft 564. More than one n~ u~ agitator 280 may be operatively coupled, for
S example, by a belt-and-pulley AIIAI~C" 1"-1ll to allow a plurality of ll~ ic
T agitators 280 to move in unison. In the present Pmho~imPn~, shafts 568 of two
n..,., .~ agitators 280 are connected to a single bracket to move the bracket in a
circular path when shafts 564 are rûtated. r..~ u-c, shafts 568 and vortexing motor
300 are des~igned to move the bottom of each reaction vessel in a circu~ar path at
0 ~),UIU~.illlakly 1500 lCVUIl,l~iUll~ per minute To prevent damage to beads, the circular path
of the present ~ bodil~ is preferably limited to a radius of ~lU~ .y 3.5 mm.
Fig. 9 shows in greater detail the upper portion of the reaction vessel bank
500 including reaction vessels 201-209 and vortexing motor 300 of Fig. 6. As discussed
in connection with Fig. 6, reaction vessel bank 500 includes a plurality of reaction vessels
201-209 connected to top bracket 290. Top bracket 290 has a plurality of apertures 630 at
which reaction vessels 201-209 connect. A teflon tube from above the aperture (not
shown) connects to the upper end of each reaction vessel 201-209 at aperture 630 in a
manner that permits a fluid to flow freely between the teflon tube and reaction tube 201-
209.
The lower ends of reaction vessels 201-209 are connected to lower reaction
vessel bracket 295. Figs. lOA and lOB show in greater detail a bottom view of the lower
reaction vessel bracket 295 of Fig. 6. Fig. lOA is a close-up bottom view of a portion of
lower reaction vessel bracket 295.
Fig. lOB shows a plurality of channels 65û in lower reaction vessel bracket
295. A flexible and ~llhst~n~iAlly translucent teflon tube (omitted from Fig. lOB for ease
of illllct~tir-n) extends from the ~ower end of each reaction vessel 201-209 (also omitted
from Fig. lOB for ease of illustration) and fits in a channel 650. A groove 652 for
mounting an optical sensor is associated with each channel 650. Groove 652 is clearly
illustrated in Fig. lOA.
Fig. lOB shows a plurality of mounting holes 654 for securely fastening
optical sensors to lower reaction vessel bracket 295. Also shown in Fig. lOB is a plurality
of optional holes 656 for reducing weight. As discussed earlier, lower reaction vessel
bracket 295 follows the motion of the nc,.,,-, r.llli< agitators in a circular path to vortex
~ 1 75587 7
wo 95/12608 PCT/U59~/123
94
the contents of the reaction vessels. Optional holes 656 may be machined through lower
reaction vessel bMcket 295 to reduce the mass of the bracket, thereby reducing the amount
of power needed to move the bracket.
A through hole 658 near each end of lower reaction vessel bracket 295
connects a ~ agitator 280 to lower reaction vessel bracket 295. The lower
ends of reaction vessels 201-209, which are extended by flexible teflon tubes 241-249 to ht
through channels 650 in lower reaction vessel bracket 295, follow the circular movement
of lower reaction vessel bracket 295. As lower reaction vessel bracket 295 moves in a
circular path, the contents of all reaction vessels 201-209 in a reaction vessel bank are
vortexed in a parallel manner.
Fig. 9 also shows optional 1~-. .. ~ . ..I, ;r agitator housings 520 for fittingover ~ - ir agitators 280. Optional n. .-~ agitator housings 520 enclose
the - shafts within a hollow cylindrical housing to prevent possible injury to
human users and damage to equipment when the - shafts are in motion.
The present ~--,1.~1;.-. ,1 uses a stepper motor (Model PX245-OlAA by
Oriental Motor U.S.A. Corp. of Torrance, California) along with a stepping motorcontroller (Model RD122 by Semix Corp., of Fremont, California) for supplying the
rotational force to the G 7 ~ ~ lt.;~ agitators. As shown in Fig. 9, three pulleys 6707
672, and 674 cooperate with vortexing motor 300 and two drive belts 521 and 676 to
20 rotate the two 1~- r- ~;- agitators 280 in unison inside optional n-.~ , agitator
housings 520. Although the present ~ ,1,~1;- .: utiliæs a stepper motor and a stepping
motor controller, the rotational force may be supplied by any other suitable type of
motors, including other electrical or pneumatic motors. rultl~ u-c, the forc~ supplied
by vortexing motor 300 may be i ' to n. . ~ agitators 280 by any suitable
I l ,."- ;~ n means including chains and sprockets, pulleys and belts, gears, etc.
Fig. llA shows a lC~ , optical sensor 680 for use in detecting the
presence or absence of a fluid within a t~_L 'Iy translucent teflon tube. Optical sensor
680 is the same as optical sensors 101S-1 l9S of the present ~mho~iim~-n~ Optical sensor
680 (Model EE-SX671 by Omron, Inc. of Srh~ mh~r~, Illinois) includes two forked ends
3û 682 and 684 for housing a light transmitter and a collector, I. ~IJaL;~y. Optical sensor
680 also has a body 686 for housing the ~ U~ t~ electronic circuitry to transmit sensor
data to a control computer and for attacbing optical sensor 680 to a bracket.
WO 95/12608 2 ~ 7 5 5 8 7 PCTIUS9~/123~7
95
Fig. 1 lB shows in greater detail forked ends 682 and 684 of optical sensor
680. Located at the interior surface of forked end 682 is a cl~hst ~n~ y rectangular
transmitter 685 for ~ light to a ~llhct~rti~lly rectangular collector (not shown in
Fig. 11B) in the direction of arrows 688. The collector is located at the interior surface of
5 forked end 684 and is likewise of a cllh-~n~i~lly ~ ul~u shape. To detect the presence
of a fluid within a substantially translucent teflon tube, the teflon tube is fitted through the
gap between the two forked ends 682 and 684. When a fluid is present within the
s~hct~n~i~lly translucent teflon tube, the collector is triggered signifying detection of a fluid
within the teflon tube.
In practice, it was discovered that the ~lb~t~nfi~lly translucent teflon
material may, when empty, cause optical sensor 680 to fail to trigger. To ~Iv~Lag~()u~ly
use common optical sensors to sense the presence of a liquid inside a $l~h$t~n~ y
translucent tube, a novel optical alignment block is used. Fig. 1 lC shows in greater detail
an optical alignment block 690 Optical alignment block 690 is made of an opaque
material which s~lhc~nti~lly blocks any light emitted by transmitter 685. Optical alignment
block 690 includes two retaining walls 692 and 694 at a first surface 696 for frictionally
engaging block 690 with one of the forked ends of optical sensor 680, and to securely hold
optical alignment block 690 between the forked ends. In one cllll,~di~ .li, retaining walls
692 and 694 are designed to engage the collector forked end 684 of Fig. 1 lB.
Optical alignment block 690 also includes a channel 698 built into a second
block surface 700. Second block surface 700 is the surface opposite the above-mentioned
first surface 696. The axis of channel 698 is orthogonal to retaining wall 692 and 694.
Channel 698 is sized to grip the teflon tube snugly. As a result, the teflon tube is secured
within channel 6g8 and is aligned at a right angle witb respect t~ the above-mentioned
transmitter strip 685 when optical alignment block 700 is fitted into the gap between
forked ends 682 and 684.
Fig. I ID shows an aperture 702 located along the center line of channel
698. Aperture 702 permits a small amount of light to travel through optical alignment
-- block 690 along its bore between first surface 696 and second surface 700. When a
cllhct:ln~ lly translucent teflon tube such as tubes 241-249 or 271-279 is fitted snugly
within channel 698, the axis of aperture 702 runs through the center of the teflon tube.
The shape and size of aperture 702 is a function of the optical properties of the tubes.
WO 95/12608 2 ~ 7 5 5 8 7 PCT/US9~1123~7
96
When optical alignment block 690 is fitted between forked ends 682 and 684
in the manner shown in Fig. llA, light from transmitter 685 in forked end 682 travels
through a cllhs~n~ y translucent tube 704. ~ost of the light is blocked by optical
alignment block 690 after passing through tube 704. Some of the light passing through
tube 704 reaches aperture 702 (hidden from view in Fig. llA) and travels along the bore
of aperture 702 to reach the collector within forked end 684.
Since the axis of aperture 702 runs through the center of substanhally
translucent tube 704, light passing through the center of the tube reaches a portion of the
collector in forked end 684. Since light going through empty tube 704 is diffracted, an
in~-~ffirirnt amount of light reaches the collector to trigger the sensor. When a fluid is
present within translucent teflon tube 704, light passing through the filled tube is focused
by the fluid within. The focused light enters aperture 702 from the direction of forked end
682 to trigger the collector in forked end 684. When a fluid is absent, the focusing effect
is less pronounced. C~ , less light enters aperture 702. In fact, when there is no
fluid in teflon tube 704, there is in~l~ffirirnt light passing through aperture 702 to trigger
the collector in forked end 684.
As discussed earlier, optical alignment block 690 iS sized to snugly grip
teflon tube 704. When optical alignment block 690 is fitted between forked ends 682 and
684, the teflon tube is securely gripped, as shown in Fig. llA, by optical sensor 680 and
block 690. By securing optical sensor 680 to a bracket, teflon tube 704 is thereby secured
to the bracket. In this manner, the teflon tubes 241-249 extending from the bottom of
reaction vessels 201-209 of this r--' ' ' are secured to the bottom reaction vessel
bracket 295.
As shown in Fig. 12, a reaction vessel such as reaction vessel 201 of the
present ~",1.,"1",.~"l consists of a segment of FEP teflon tube 710. Tube 710 has an
outside diameter of 1/4 inch and an inside diameter of 0.19 inch. As described in greater
detail hereinafter, the diameter of tube 710 can alL~ ivr,l~l be made much larger for
~IJplb ~l~iUl~s where two or more reagents are ' 'y mixed in the reaction vessel201. Tube 710 is sealingly coupled with a flexible teflon tube 271. Flexible teflon tube "
271 is secured to lower reaction vessel bracket 295 of the reaction vessel bank. As the
lower reaction vessel bracket 295 moves in a circular motion, the bottom of theflexible
teflon tube 271 fo~lows the circular motion described by the bottom reaction vessel bracket
295 to vortex the contents within tube 710.
~ WO 95/12608 2 1 7 5 5 ~ 7 PCT/I~S9-J/123J7
97
In this f .1lbo~ -1L, tube 271 has an outside diameter of 1/8 inch and an
inside diameter of 1/16 inch. Fig. 12 shows a tube connector comprising a first coupler
714, a second i11t~.~,O1111~U1 716, and a third coupler 718 for sealingly connecting tubes of
different cross-sectional ~limPncinnc together. The aforemPn~ionpJ1 tube connector is
5 available from Norton, Inc. of Akron, Ohio. There is a frit or filter 1102 (hidden from
view in Fig. 12) located at the bottom end of tube 710 for preventing substrates within
tube 710 from entering flexible tube 712. The frit may be, for example, 2 micron titanium
frit.
At the other end of tube 710, a fourth coupler 720, a fifth i~ ollll~Lul
721, and a sixth coupler 722 sealingly connect tube 710 to a tube 221. ~he couplers 720
and 722 as well as il~t_l-,UII..~Lul 721 are necessary because tube 221 of the present
has different cross-sectional ~limPnci~nc from tube 710. Tube 221 connects to
a manifold port 542 of top manifold 212 (not shown in Fig. 6).
A flexible O-ring 724 is fitted within a hole 726 in bracket 290 (shown in
Fig. 12 in a cutaway view). O-ring 724 flexibly grips coupler 722, thereby flexib]y
securing reaction vessel 201 to bracket 290. When the bottom end of reaction vessel 201
is agitated, O-ring 724 serves as a pivot point and holds the top end of reaction vessel 201
relatively immobile to enhance the vortex effects.
As shown in Figs. 12A and 12B, the reaction vessel 201 in one particular
20 f .IlI~o~i;ll.. 1 can optionally be provided with a t~,lllAu~.aLul~ control jacket 1100 for
controlling the ;.Ill~ LLul~ of the reaction vessels 201-209. The L~ Lul~ control
jacket 1100 in this P.,,~ ,l includes a fitting 1104 for connecting the reaction vessel
201 to the lines 221 and 241 (not shown). Inlet/outletports 1106, 1108 are provided for
supplying a thermally conductive fluid to the reaction vessel 201. The thermallyconductive fluid can be used to either heat or cool the reaction vessel 201. In this
~lllbodilll~llL~ the reaction vessel 201 is formed from a teflon tube lll0, preferably having
an outer diameter of 1/4 inch and a wall thickness of 1/32 inch. Spaced-apart from the
tube lll0 is an outer tube 1112, preferably ,ul-~Llu~d of teflon and having an outer
'~ diameter of 3/4 inch and a wall thickness of 1/32 inch. The outer tube 1 1 12 is positioned
over the fitting 1104 to form an annular space 1114 for receiving the thermally conductive
fluid. O-rings 1116, 1118 are placed between the outer tube 1112 and the fitting 1104 to
form a fluid tight se~l. In this way, the thermally conductive fluid can be introduced into
the annular space 1114 through either of the ports 1106, 1108 for heating or cooling the
WO 9S/1260X 2 t 7 5 5 8 7 PCT~S9~/123~7 ~
98
reaction vessel 201. The ports 1106, 1108 also allow the thermally conductive fluid to be
;""~ ycirculatedthroughtheannularspace 1114.
An altemative ~".I~Ii",. .1l of a reaction vessel 1120 haYing an integrally
formed ~.~ ul~ control jacket 1122 is shown in Figs. 12C and 12D. The reaction
vessel 1120 functions cl~hcf~nti~lly identical to the vessel 201 except that the reaction
vessel 1120 is preferably constructed of glass. The t~lll,U~,Ld~Ult: control jacket 1122 is also L
preferably COual~u-~ of glass Such a [-~mfi~ll~til~n allows the reaction vessel 1120 and
the t~ d~ul~ control jacket 1122 to be formed as an integral unit. The reaction vessel
1120 includes a frit 1124, and ~ l;""~ 1126 and 1128 for connection to lines 221 and
241 (not shown). Inletloutlet ports 1130, 1132 are provided for circulating thermally
conductive fluid through an annular space 1134 between the jacket 1122 and the reaction
vessel 1120. Hose barbs 1136, 1138 are ~,UII~ ' 'y provided for connection of the
inletloutlet ports 1130, 1132 to a suitable fluid source. In this way, thermally conductive
fluid can be circulated through the annular space 1134 to heat or cool the reaction vessel
1 120.
D. Parent Vessel
Fig. 13 shows in greater detail the pænt vessel of the present ~ o~
Parent vessel 200 has, for example, a volume of d~-~lu~dl~ t ly 30 mL. A tube 260 is
sealingly connected to the bottom end of parent vessel 200 for ~ r~ " i,~ a reagent or
argon to and from delivery system PDS 265 (not shown in Fig. 13). A frit or filter 746 is
fitted near the bottom of parent vessel 200 to prevent substrates from entering tube 260.
There is a removable cap 743 fitted on the top end of parent vessel 200 for adding and
removing material. Tube 215 fitted through cap 743 transfers material between parent
vessel 200 and top common manifold 212 in a reaction vessel bank (not shown in Fig. 13).
Tube 215 extends through cap 743. A pressuri~ed argon/vent line 275 also extendsthrough cap 743. An optional rinse line 281 connects to a solvent source for delivering
aaul~ solvent to the interior walls of parent vessel 5 to rinse the interior walls.
Fig. 13 shows two capacitive sensors 90S and 99S (Model 18-08 by
Electromatic Control Corp. of Hoffman Estates, Illinois) mounted near the exterior of
parent vessel 200. Each capacitive sensors 90S or 95S detects the presence of a liquid in
its vicinity and transmits sensor data to a control computer (not shown) via w;res 756 and
758 respectively. Capacitive sensor 99S is used for detecting the level of reagents added
wo 9~/12608 ~ t 7558 7 PCT/US9~/123~7
99
to the parent. Capacitive sensor 90S is for detecting the level of bead suspension for
LlibuLioll. Both sensor levels can be adjusted according to amount of beads and the
number of reaction vessels used. The capacitive sensor data is utilized by the software to
control various cycles of the synthesis process.
E. Control System
1. Control Computer
The automated ~yl~ utilizes a control computer to acquire data from
the sensors, and to control the valves and the vortexing motor during the various cycles of
the synthesis process. When used in ~;Ul.jU~ iO.l with the automated synthesizer, any
computer including those popularly known as III;~,IU~'''''Il''l~: ':., 1ll;ll;' '''''1''ll(-'~
wnrkctAtinnc, ~--A;.,r,A"~, and the like, may be used to process the sensor data and to
issue commands to control the valves and the vortexing motor.
Fu~ -l-ul~, sensor data from the sensors in the ~yll~hc~ may be
acquired by any number of commercially available data acquisition devices using common
data acquisition methods. Likewise, the valves and the vortexing motor may be
controlled, responsive to an d~ , ' computer command, by commercially available
input/output controllers.
In one ,-I-o~ , an IBM-compatible llli.,lU-,U~ JUt~l (also known as a
personal computer or PC) is used as the control computer (Model Gateway 2000 4DX2-
50V, by Gateway 2000 Inc. of No. Sioux City, South Dakota). Within the PC, there are a
plurality of expansion slots permitting the addition of various expansion boards. These
boards tap into the bus resources of the PC and permit the PC to ...,~.",~ Al~ with the
circuitry on the card to perform an electronic function, Certain expansion boards alsû
permit the PC to ~ with external devices and circuitry. For example, a board
popularly known as a modem board plugs into an expansion slot on a PC and permits the
PC to c~ ", ~ with another computer having a modem. The use of expansion boards
with a personal computer is a matter of common ~ knowledge.
'; In one ~--lbodi~ , the automated synthesizer .u~ A~ with the PC
via a m-lltirh~nn,-l digital I/O board (Model PCDIO120-P by Industrial Computer Source
of San Diego, Califomia). The crerifi~ tinn of the PCDIO120-P board is described in
detail in Product Manual No. 00431-050-20A which is also available from Industrial
Computer Source.
wo 95/12608 2 1 ~7 ~ ~ ~3 7 PCT/US9.U123-17
100
Each PCDIO120-P provides 120 channels of buffered inputs/outputs (I/O) in
five 24-channel groups. Each 24-channel group is controlled by a ~lu~;ld~ lblc
peripheral interface (PPI) 825~A chip. The channels are selectable in set of 8 For either
input or output.
S Fig. 14 shows a simplified diagram of a portion of the control hardware. Acomputer 760, such as the PC, having a plurality of expansion slots, connects to a display
monitor 762 and a keyboard 764. A PCDP120-P I/O board 766 is inserted into one of the
expansion slots to permit tbe PC to cnmm~r:^~t~ with five controller circuits 768. Each
controller circuit 768 rnmTn~ ir~ C with I/O board 766 via a conductor channel 770~ In
the present . ."1,.~.1;".. "1, conductor channel 770 includes a S0-conductor ribbon having the
capability to service 24 I/O channels.
2. Controller Circuits
The output signals from I/O board 766, typically 15 mA of source current
1~ and 24 mA of sink current, are inadequate to operate solenoid valves. Consequently,
controller circuits 768 convert the output signals coming from I/O board 766 into power
signals having adequate power to actually operate the solenoid valves. Controller circuit
768 has the capability to receive 24 output signals from T/O board 766 and in turn outputs
24 power signals 771 to control various devices of the synthesizer. The 24 power lines
771 of each controller circuit 768 are shown in Fig. 14.
Each controller circuit 768 also provides a central physical location into
which sensor data from up to 24 sensor lines, one for each sensor, may be gathered. Data
from up to 24 different sensors may be received by a sensor port 772 on each controller
circuit 768.
Thus, each controller circuit 768 can service up to 48 TIO channels, 24
inputs and 24 outputs, of I/O board 766. Fig. 15 shows a Ic~ ,e~ ivc diagram of a
controller circuit 768 according to one aspect of the present invention. A 50-pin header
781 connects the controller circuit 768 to I/O board 766 (not shown in Fig. 15). Header
781 is an input header and is connected via a bus 780 to a sensors port 772. Sensors port
772 comprises headers or connectors for connecting controller circuit 768 with up to 24
input devices such as sensors. T,ach conductor on bus 780 carries a signal from one sensor
to input header 781. Fig. 15 also shows an optional LED bus 784 for carrying LED
WO95/12608 2 ~ 75~ Q;7 PCT/US9~1123.17
101
indicator signals from sensors port 772 to an optional bank of light emitting diodes (LE~s)
186. Each conductor in optional LED bus 784 carries a signal to one LED in bank 786.
Fig. 15 also shows another 50-pin header 788 for connecting controller
circuit 768 to I/O board 766 (not shown in Fig. 15). Header 788 is an output header for
receiving output signals from I/O board 766. A bus 790 carries up to eight output signals
from header 788 to an octal inverter 74LS240 chip 792. Octal inverter 74LS240 chips 792
are ln~lur~Lulcd by Texas Instruments, Inc. of Dallas, Texas. A bus 794 carries the
inverted buffered output signals from chip 792 to an octal latch driver 796. Latch dnver
chips 796 are Model MIC59P50, ., ~ " r~ I lc~ by Micrel, Inc. of San Jose, California.
Output signals from each latch driver chip 796 are connected to an output port 798 via a
bus 800. An optional LED bus 802 carries LED indicator signals from chip 796 to an
optional bank of light emitting diodes (LEDs~ 804. Each conductor in optional LED bus
802 carries a signal to one LED in bank 804.
3. The Valves
The solenoid valves such as, for example, valves 4- 7, 10, 14, 90-91, 100-
121, and 129, used in the present r,,.,l-o~ are normally closed unless cf~mm:m~1~ to
open. C~ c~ Lly, the default state for all valves in the ~,IIL~ is off. Safety is
ensured because no material is permitted to flow when the synthesizer is in its default
state. When open, valves use power and heat up. Besides the obvious drain on the system
power, hot valves may adversely affect the chemicals passing through their ports. Because
it normally takes a greater amount of power to open a solenoid valve than to keep an
already opened valve open, a strike relay such as, for example, a model DlD20 byCrydom, Inc. of Long Beach, California, is used to operate the valves. A strike relay
such as the DlD20 supplies +12 volts to a valve for a specified period of time, typically
100 millicf~ronrlc to open the solenoid valve from the off state. The period of time during
which the strike relay supplies + 12 volts can be specified through software control, and
the strike is supplied via an I/O channel. Thereafter, the strike relay supplies a reduced
voltage, typically half the rated voltage or ~yylu~d~ L~I~ 6 volts in the present
embodiment, to keep the solenoid valve open. C.~ ,lly, less energy is required to
operate the valves and less heat is produced.
The present invention provides for four separate power supplies. A first
power supply outputs +5 volts to power the l~L chips such as those found on controller
wo 95/12608 2 ~ 7 5 5 8 7 PCrlUSg~1123~7 ~
102
circuits 76~. A second power supply outputs +32 volts for use by the stepper motor. A
third power supply provides + 12 volts to activate the solenoid valves. An optional fourth
power supply also provides +12 volts for use by the sensors. A separate fourth power
supply for the valves ensures that any noise ~enerated by the valves as they open and close
5 does not interfere with sensor operation.
In its default state, all valves are closed. As an additional safety measure,
the synthesizer further provides for a solid state watchdog relay to shut off all valves in the
event the control computer malfunctions. A solid state watchdog relay such as a Model
SM-WDT5 by Brentek Tntprn~ti~n~l (available from Industrial Computer Source of San
10 Diego, California) is interposed between the control computer and the power supply to the
valves. A softwarc ~ ' pulse is transmitted from the control computer to the
watchdog relay on one of the I/O channels. When the pulse is absent, e.g., upon CPU
failure, latch-up or power failure, the watchdog relay shuts down the power supply for the
valves, thereby closing all valves.
F~ Control Software
The control software will now be discussed in detail with reference to the
flow charts of Figs. 16-24. These flOw charts illustrate the commands issued by tlle
control computers or completing relevant phases of the synthesis process. To simplify the
20 discussion below, it is assumed that at all relevant times, the valves of the pressunzed
delivery system receive the ~ 1 commands from the control computer to deliver the
desired reagent to the reaction vessel bank valve
Fig 16 is a flow chart illustrating the ~.,,,,I,;,..U;..Il of commands issued bythe control computer for draining reaction vessels 201-209 of their contents. Before
draining, reaction vessels 201-209 contain a liquid or a bezd C~r~ n~ n An argon supply
valve 121, connected to top common manifold 212, receives an open command and opens
to pressurize top common manifold 212 with argon. At the same time, selected 3-port
valves 101-109 open to permit a liquid from reaction vessels 201-209 to enter lower
manifold 214. Only selected valves 201-209 open because not all reaction vessels 201-209
are used during some coupling reactions. If a reaction vessel sits empty throughout a
synthesis session, there is no ne~d to drain its contents. Waste valve 110 in bottom
manifold 214 opens to permit fluid exit. As a f~ u~ of the pressure different~al,
argon pressure pushes fluid from reaction vessels 201-209, through lower manifold 214,
~ WO 95/12608 2 1 7 5 5 8 7 PCTIU59~1123~7
103
and out waste valve 110. When sensor llOS turns off, signifying that no liquid is left to
drain, the valves stay open an additional 2-5 seconds to ensure all liquids are drained from
the reaction vessel bank.
Fig. 17 depicts the combination of commands issued by the control
computer to clear lower manifold 214. At the start step, there is material in lower
manifold 214. Thereafter, isolation Yalve 100 opens to permit ~ UliL~i argon from
PDS 265 to enter lower manifold 214. .S ly~ waste valve 110 opens to vent
material from lower manifold 214. Material exits from lower manifold 214 until no
material is left. The valves return to their default state when optical sensor 1 IOS detects
no material in the waste tube.
Fig. 18 shows the set of commands issued by the control computer to mix
the contents of parent vessel 200. The system introduces argon into parent vessel 200
from below to mix the contents. The argon bubbles agitate the contents of parent vessel
200 as they rise from the bottom of the parent vessel to the surface of the contents inside
~5 the parent vessel. Again, all valves which have not been expressly . ~ to stay
open are in their default state. Valve 100 orens to introduce pl~"u~;~l argon from PDS
265 to the bottom of the parent vessel to agitate the contents within. Valve 90 also opens
to vent argon from the parent vessel. Valves 100 and 90 return to their default state after
~ Iu~ ly 15 to 30 seconds.
Figs. l9A and l9B depict the sequence of commands issued by the control
computer to allocate bead suspension from the parent vessel to the various reaction vessels.
The sequence of commands i",l,l. .~,. ..lc a volumetric technique for filling the various
reaction vessels with the bead C..crl-n~irm Using this approach, the reactions vessels are
filled with little ~ e on flow rate. Thus, as will be seen, the bead suspension is
distributed evenly among the reaction vessels regardless of the distance between that
reaction vessel and the manifold port through which the bead suspension enters. At the
start of the allocation cycle, the reaction vessel bank contains only argon, and the parent
vessel contains a bead suspension at step 852.
- At step 854, the reaction vessel bank is partially filled with DMF to displace
the argon that exists in the reaction vessel bank prior to the reallocation phase. Isolation
valve 100 opens to permit JJI. ~Uli'~ DMF to enter the lower manifold from the
u~;~cd delivery system. Valves 101-109 open to permit DMF to rise toward the
reaction vessels. Valve 120 opens to vent argon from the top common manifold. Valves
2 1 75~87
WO 95112608 PCT/US9 1/123~7
104
100, 101-109, and 120 return, preferably in series, to their default state afler a
ulu~all~ll.cd time period, which may be about 3 seconds each~ Reaction vessels 201-
209 are then mixed to dislodge bubbles. Valves 100-109 open again for about 3 seconds.
At the expiration of this ,ulc~lugldl~ lcd time period, the level of DMF in each tube 241-
249 leading to the reaction vessels will be near the top common manifold 212.
The reaction vessel bank and parent vessel 200 are then filled with DMF at
step 858. Valve 100 opens to permit pressurized DMF to enter the reaction vessel bank
from PDS 265. Valves 101-109 open to continue filling the reaction vessel bank with
DMF. Valve 129 opens to permit the DMF which overflows the reaction vessel bank to
enter parent vessel 200. Valve 90 also opens to vent the displaced argon from parent
vessel 200. The filling continues until the level of fluid in parent vessel 200 rises to the
level of upper capacitive sensor 99S when sensor 99S detects DMF within its detection
envelope. The reaction vessel bank is then completely filled. Parent vessel 200 fills up to
a~J~nu~ -,.t~,ly the level of second capacitive sensor 99S. Valves 90, 100, 101-109 then
retum to their closed state at step 860.
Next, the top common manifold is cleared of DMF at step 862. Valve 121
opens to pressurize the top common manifold with pressurized argon. Valve 129 opens to
permit DMF to enter the parent vessel from the top common manifold. Valve 90 opens to
vent the displaced argon from the parent vessel. DMF from the top common manifold is
thereby transferred to the parent vessel. The parent vessel is designed to have a sufficient
volume to accept the additional DMF without ù._lnu..il.~,. After a pl=lJIu~;lallllllcd time
period of, for example, about five seconds, valves 90, 121, and 129 retum to their default
state at step 864. The ~.c~,u~;.a...,,led time period is variable but must equal or exceed
the time it takes to clear the top common manifold of DMF.
Had the reaction vessels not been prefilled with DMF prior to the
udLl~liù~ of the bead Cll~rPnCil~n, i.e., had the reaction vessels been empty, an uneven
~lictrihll~ion of bead suspension would occur. If the reaction vessels were empty, the
reaction vessel which is the closest to the manifold port through which the bead suspension
enters from the parent vessel would fill up first. There may be no bead suspension left for ~'
some reaction vessels if a few were allowed to fill up ~ cly.
The present invention employs a novel method for controlling the volume at
which a reaction vessel accepts the bead CllcrPncil~n First, at step 866, a small column of
argon is introduced to the top of each tube which connects the reaction vessels to the top
~, wo 95/12608 ~ 1 7 5 5 8 7 PCT/US9-11123~7
105
common manifold. To create this argon bubble, valve 121 opens to pemmit yl~ u~
argon to enter the top common manifold. Selected vaLves 101-109 open in series to permit
some DMF to drain from the reaction vessels to the lower manifold. Valve 110 opens to
vent the displaced DMF from the bottom manifold. After about 0.3 seconds, a small
5 column of argon appears at the top of the tube which connects the reaction vessel to the
top common manifold Valves 101-110, and 121 then retum to their default state at step
868.
At step 869, the bead suspension in the parent vessel is mixed in preparation
for ~;~l.;l.ul,.." among the reaction vessels. The commands associated with this step are
10 similar to those discussed in connection ~vith Fig. 18. Valves lO and 90 then return to
their default state at step 870. This step ensures that a I~UIIIO~-IIVJU~ bead suspension is
evenly distributed among the selected reaction vessels.
A portion of the bead suspension is then introduced to the top common
manifold at step 871. This step is timed according to a IJlti~JlU~ldUIIIII~ time period so that
15 the bead suspension that enters the top common manifold displaces most of the argon
existing within the top common manifold without flowing past the manifold port into which
the last reaction vessel tube, e.g., the reaction tube associated with sensor lO9S, connects.
A ~ ,Idllllll~,d period of about 0!5 seconds has been found to be ~dLi:~rd~ LUly. To
introduce this portion of the bead suspension to the top common manifold, valve 91 opens
20 to pressurize the parent vessel with argon. Valve 129 opens to permit the bead suspension
to flow into the top common manifold. Valve 120 opens to vent the argon existing in the
top common manifold. After the expiration of the previously discussed l,.~k".,~;.dl.ll,l~d
time period, valves 91 and 120 retum to their default state at step 872. Most illl~,olLd,llly,
valve 129 continues to stay open to prevent beads and polymers from being damaged due
to the closing action of the valve. In the present . .. ,~-~1;". .. : 2-port valve 129 continues
to receive the command signal from the control computer to stay open in the manner
discussed earlier. However, valve 129 may be a latch valve which toggles between the
open and shut states upon receipt of a command pulse from the control computer. If valve
129 is a latch valve and is already open, no action needs to be taken by the control
30 computer to keep latch valve 129 open.
Fig. l9B is a ~,."1,...,-l,"" of Fig. l9A. After some argon which existed in
top common manifold 212 has been displaced, tbe rest of the bead suspension is
transferred to top common manifold 212 at step 874. Valve 91 opens to pressun~e parent
21 75587
Wo 95/12608 Pc rlUSs~1123~7
106
vessel 200 with argon. Valve 129 which has been kept open permits the pressurized bead
suspension to enter top common manifold 212. Selected valves 101-109 open to permit the
DMF within reaction vessels 200 to exit into lower manifold 214. Valve 110 opens to
drain DMF from lower manifold 214. Partly due to the resistance of the bead 5~pPn~inn
DMF recedes down selected reaction tubes 201-209 relatively slowly. The DMF displaced
from selected tubes 221-æ9 connecting selected reaction vessels 201-209 with top common
manifold 212 is replaced with the bead sllcr--noinn The column of suspension-bubble-
DMF advances slowly downward towards lower manifold 214.
The column of bubble introduced earlier also ad~ ~euu~ly serves as a
volume marker, i.e., provides a way for the control computer to determine when each
reaction vessel has received a sufficient amount of bead s~srPn~inn The column of bubble
stays trapped between the column of DMF and the column of suspension because of the
surface properties of the teflon and the high contact angle between the DMF and teflon.
As discussed earlier, the optical sensors used in the present ~ bo~ can detect the
presence or absence of a liquid column inside a cllhct~n~i~lly translucent teflon tube. As
DMF is replaced by the downwardly advancing mass of bead C~lcrPncinn, the bubbles
move down into the reaction tubes and into the tubes which connect the reachon vessels to
the lower manifold. At the moment the argon bubble is detected by an optical detector
IOIS-109S on its downward movement, that detector is ly tumed off. The
sensor data, as discussed earlier, is ~ to the control computer which promptly
issues a command signal to shut off a respective valve 101-109. After all valves 101-109
have been turned off, the bead suspension transfer is restarted and continues at step 878
until acoustic sensor 120S detects no fluid in the tube connecting the parent vessel with the
top common manifold. At step 882, all valve3 except valve 129 return to their default
state. As discussed, valve 129 stays open to avoid damage to the beads and valve 129.
The synthesizer at this point has suspension in its reaction vessels, and
possibly some bead suspension residue in the parent vessel. To make sure all the beads
are transferred to the reaction vessels, a rinsing process comprising at least one rinse cycle
is employed. '
The rinsing cycle is initiated by showering the interior walls of the parent
vessel at step 890 with DMF sprays to loosen any bead suspension residue which may
have cling to the wall. Valve 14 opens to permit sprays of DMF to wash down the walls.
Valve 90 opens to vent the replaced argon from the parent vessel. The interior wall
~ WO 95/12608 2 i 7 5 5 8 7 PCT/US9~/123`17
107
continues to receive sprays of DMF until the level of DMF in the parent vessel nses to the
level of lower capacitive sensor 90S and turns it on. This sensor data is received by the
control computer which promptly issues a command at step 892 to return all valves except
valve 129 to their default state. The contents of the parent are then stirred by argon
5 bubbles in the manner previously discussed.
Altematively, the parent vessel may be rinscd by refilling it with DMF and
then mixing it to loosen any bead suspension residue which may have clung to the interior
walls or frit of the parent vessel. First, at step 884 the parent vessel is refilled with fresh
DMF. The refilling is - ~ ' ~' by opening valve 10 to permit DMF to enter the
10 parent vessel from the ~ Ul;~d delivery system. Valve 90 also opens to permitdisplaced argon to exit the parent vessel. When the level of DMF in the parent vessel
rises to the level of the top capacitive sensor 99S, top capacihve sensor 99S is turned on.
This sensor data is received by the control computer which promptly issues a command at
step 886 to return all valves except valve 129 to their default state. The parent vessel is
15 then mixed at step 888 by i~duc~ argon bubbles to the parent vessel in the manner
previously discussed.
The mixture of DMF and bead suspension is then transferred to the reaction
vessels at step 894 by opening valve 91 to pressurize the parent vessel with argon and to
transfer the mixture to the top common manifold through valve 129 which has remained
opened through out the rinsing process. Selected valves 101-109 open to permit fluid to
flow from the reaction vessels to the lower manifold. Valve 110 opens to drain DMF
from the bottom manifold. The frits at the bottom of the reaction vessels strain all beads
inside the reaction vessels. Eventually, the parent vessel is drained. Sensor 120 turns off
when no fluid is present in the tube connecting the parent vessel with the top common
manifold. This sensor data is ' to the control computer to signify that no
fluid is left in the parent vessel to transfer. The control computer continues to open valve
91 for another 5-10 seconds to pressurize the top common manifold and to move any
remaining mixture into the reaction vessels. After 5-10 seconds, all valves except valve
129 are returned to the default state. One rinse cycle is completed.
As discussed earlier, a plurality of rinse cycles may be employed to ensure
that ~ ' "y all bead suspension from the parent vessel is transferred to the reaction
vessels. Two to three rinse cycles have been found to be satisfactory.
21 75587
wo 95112608 PC rlUSs~1123~7
108
When all rinse cycles are completed, all valves including valve 129 are
returned to the default state at step 896. Note that valve 129 remains open throughout the
bead suspension reallocation process, including the rinsing process, to minimize any
damage to beads and polymers. The reaction vessel bank is then drained of all fluids in
S the manner earlier discussed at step 898.
From the steps in Figs. 19A-19B, it can be seen that the use of the argon
bubble allows the bead suspension to be distributed evenly among the reaction vessels
regardless of the flow rate of each ffow path between the parent vessel and reaction vessel.
As long as the argon bubble remains stable between the bead suspension and the DMF
reagent, a viable marker is provided, enabling the sensors to determine the status of the
reaction vessels. The ability to maintain a stable argon bubble, as previously mentioned,
is due to the favorable physical properties of DMF, i.e., high contact angle of DMF with
teflon.
However, it may be desirable to use fluids other than DMF when delivering
the beads to the reaction vessels. This may create problems, p~uL;.,ul~uly if the substitute
fluid does not have physical properties which are conducive to creating a stable argon
bubble. For instance, ~rPt~nitril~ (MeCN), which is used as a solvent for DNA synthesis,
is unable to produce a stable argon bubble. Thus, a different approach for disLI;l,uL;llg the
beads to the reaction vessels is required.
Figs. l9C-19D illustrate an altemative set of commands issued by the
control computer for mixing the contents of the parent vessel when a stable argon bubble
cannot be produced.
This technique is particularly useful when only a small number, e.g., up to four to five, of
reaction vessels are employed.
Steps 854a-860a are similar to steps 854-860 in Fig. 19 except that only a
smaller number of valves 101-109 are opened, i.e., only those ~u~ d;l,g to the
selected reaction vessels. The selected reaction vessels are partially filled with a solvent,
such as MeCN in order to displace the argon that exists in the reaction vessel bank pnor to
the reallocation phase. After which, the selected reaction vessels and the parent vessel are
filled with MeCN. The flow of MeCN is stopped when the amount of MeCN in parent
vessel 200 rises to the level of second capacitive sensor 99S, indicating that filling is
completed. Upon comrl~tinn~ the selected valves then return to their closed state.
~ WO 95/12608 2 1 7 5 5 8 7 PCT/US9.11123.17
109
Next, at step 872a, the bead suspension located in the parent vessel is mixed
in preparation for di~LLi~uLiul~ to the selected reaction vessels. At step 874a, the bead
suspension is then introduced to the top common manifold and distributed to the selected
reaction vessels by opening valves 91, 129, and selected ones of 101-109. These valves
5 remain open until the sensor 120S detects the absence of fluid in the tube connecting the
parent vessel with the top common manifold. Since no argon bubble is produced, sensors
lOlS-109S are not employed. Because only a small number of reaction vessels receive the
beads, the time required for the beads to reach each of the reaction vessels is
~,U~JIU~illldt~ ly equal, thereby ensuring a generally equal distribution for each of the
10 reaction vessels.
At step 890a, a rinsing process, such at those already discussed in Fig. l9A-
l9B, is employed to make sure all the beads are transferred to the reaction vessels. Upon
completion of the rinsing process, all valves including valve 129 are returned to the default
state at step 896a. The reaction vessel bank is then drained of all fluids at step 898a.
Fig. 20 is a flow chart showing the sequence of commands issued by the
control computer to fill the reaction vessel with the desired reagent from the de~ivery
system. The steps discussed in connection with Fig. 20 are also ~rh~rn ~ lly illustrated
in Figs. 21A-21D. This process assumes that the lower manifold is filled only with inert
argon at step 900. Fig. 21A graphicaLly shows a relevant portion of the reaction vessel
bank having an empty manifold. The lower manifold is first filled with a reagent at step
902. Valve 100 opens to permit the reagent to enter the lower manifold, and valve 110
opens to vent the argon displaced from the lower manifold. When the reagent is detected
by sensor llOS, all valves return to default at step 903.
In some instances, the sensors lOlS-109S may be tripped i-lauv~ .lLly or
prematurely while filling the tubes connecting the reaction vessels with the lower manifold.
For example, a sensor may be actuated by a stray droplet of reagent before the reagent
actually reaches it. This can cause an ,, ~,.11~;. .,l amount of reagent to be present in the
tubes for delivery into the reaction vessels.
To reduce or eliminate problems associated with premature sensor actuation,
the control computer, at step 904, can optionally be ~lu~ d to open valves 101-109
for a set amount of time in order to prefill the tubes. Generally, the time is set so as to
prefill the tubes to about 75%, before being detected by the sensors lOlS-109S. As
described, this step does not rely on the use of sensors lOlS-109S. Thus, preflling
21 75587
wo 95/12608 P~TIUS9~/123~7
110
ensures that there is at least a sufficient amount of reagent present in the tubes to inject
into the reaction vessels to ade~uately perform mixing, even if the sensors are ~ dLul~ly
actuated At step 905, the valves retum to default after the predefined time has expired in
preparation for filling the tubes.
At step 906, the tubes connecting the reaction vessel with the lower
manifold are filled to the sensors with reagent. The filling process may be carried out in
parallel to save time or in series. Valve 100 opens to pemmit ~lc~ul;~l argon from the
~ICaaUli~,_i delivery system to enter the lower manifold. Selected valves 101-109 open,
serially or in parallel, to permit the reagent to enter the tube connecting the reaction vessel
with lower manifold. When the level of reagent in a tube reaches the upper light sensors
IOlS-109S, the light sensors tum on, and the control computer promptly tums off an
associated valve 101-109. When all sensors lOlS-109S are on, all valves are returned to
the closed state at step 908. Fig. 21B srhrm ~ir~lly illustrates the result after this filling
step is completed.
The bottom manifold is then cleared in the manner earlier discussed at step
910. Fig. 21C shows a relevant portion of the reaction vessel bank having a cleared
manifold and a column of reagent inside the portion of the tube between a top optical
sensor, e.g., lOlS7 and a valve, e.g., 101. After the bottom manifold is cleared, all
valves are again retumed to default at step 912. The reagent in the tubes are then pushed
up into the reaction vessels through the frits.
To push the reagent into the reaction vessels, Argon valve 122 opens to
permit argon from the L,l~,u.i~ delivery system to pressurize the lower manifold. Vent
valve 120 also opens.
At step 913, the vortex motor is activated to commence agitating the
reaction vessel bank for a ~ i amount of time. The time period is sufficient
long to permit the contents within the reaction vessels to mix completely. Generally, a
time of about 4 has been found to be adequate, but may vary depending on the type of
synthesis.
During the above mentioned mixing period, at step 914, valves 101-109
open for a ~ uy,ldllllll~ d period of time to flow a small amount of reagent into the
reaction vessels. UsuaUy, the reaction vessels are filled and drained repetitively dunng a
synthesis process. Each time the reaction vessels are drained, the beads therein become
dry and clump together to form a "bead c~ke". Step 914 fluidizes the contents of the
~ WO 95/12608 2 1 7 5 5 ~ 7 PCTII~S9J1123.17
111 ~
reaction vessel and dissolves the bead cake. Since vortexing is most effective when the
beads are at or near the bottom of the reaction vessel, only a small amount of fluid should
be injected. Otherwise, the bead cake may float to the top of the reaction vessel, requinng
more time to dissolve it. At step 915, the valves retum to default after the predefined time
has expired.
At step 916, the remaining portion of the reagent is pushed up into the
reaction vessel by opening valves 101-109. Each vessel is filled until its associated sensor
101S-109S detects the absence of a fluid and the control computer tums off that valve.
When all sensors 101S-109S are off, all reaction vessels are filled. To improve mixing,
the vessel bank is agitated while being filled. As noted before, the reaction vessels may be
u~ la~ d so as to fill the vessels in parallel or in series by ûpening all the valves 101-
109 at once or sequentially.
Altematively, the reaction vessels may be filled without relying on sensors
101S-109S. For example, valves 101-109 may be opened for a pl~ ;ldlllll.Cd time
period which is sufficient to fill the vessels to the desired level. A time period of 0.5
seconds to l second has been found to be ~ILi~ri~Luly. However, this time period may
vary according to the number of vessels being used, i.e., the greater the number, the
longer the time required. Fig. 21D shows a diagram of the reaction vessel after being
filled~
Note that the volume of reagent to be pushed up into the reaction vessels
also can be easily changed by varying the length or diameter of tubing between valves 101-
109 and sensors 101S-109S. This change can be easily ~ ~c. ,~ "s,i by ~ the
tube which connects a reaction vessel to an injection valve, e.g., valve 111, with a tube
having a different length or cross-sectional dimension.
It may also be advantageous in some instances to increase the diameter of
reaction vessels themselves. For instance, it may be desirable to ciTr~llt~ oucly mix the
beads with two or more reagents. Such mixing can occur by following the steps descnbed
in Figs. 21A-21D to introduce the first reagent to the reaction vessels. A second reagent
is then introduced to the reaction vessel by repeating the steps described in Figs. 21B-21D.
In so doing, however, an argon bubble will be disposed between the bead suspension and
the second reagent due to the argon left in the tubes 241-249 and 271-279 beforeintroduction of the second reagent. To remove the argon bubble from the reaction vessel,
the inner diameter of the reaction vessel can be made larger to reduce the height of the
wo 95/1260~ 2 1 7 5 5 8 7 PCT~US9~1123~7 ~
112
bead suspension within the reaction vessel, thereby making escape of the argon bubble
through the bead suspension easier. After removal of the argon bubble, the bead
suspension and the second reagent are mixed together by vortexing as previously
described.
Fig. 22 is a flow chart showing the sequence of commands issued by the
control computer to inject activated amino acid reagents into the reaction vessels. Figs.
23A-23C ~1,. ..~ ~ll. ^lly illustrate the relevant steps of the amino acid injection process.
Again, it is assumed that the lower manifold is cleared, i~e., contains only argon, at the
start. Fig. 23A shows a diagram of the reaction vessel, the lower manifold, the valves,
10 the sensors, and associated tubes at the start.
An alternatiYe ~ n~--- for the reservoirs 231-239 is shown in Fig. 6A.
For . .,.1~.~.1",.. .,1~ E. a rotatable carousel for holding a plurality
of groups of reservoirs, the carousel is rotated to align the tubes with the selected group of
reservoirs at step 917. Once the ~ reservoirs are selected, the manifold is filled
at step 91~ with amino acid activating reagents such as 0.2M HBTU and 0.6M DIEA in a
so1ution of 3:1 DMF to DCM. The manifold is filled in the manner discussed in
connection witn Fig. 20, i.e., open valves 100 and 110 until sensor 110S is turned on.
Thereafter, aU valves close at step 920.
The activating reagents then enter the tubes connecting the reaction vessels
with the lower manifold. Valves 100 and 120 open to let ~ UI;L~i activating reagent
enter the lower manifold at step gæ. Selected ones of valves 101-109 open until the lower
optical sensors lllS-119S senses fluid presence. All valves again close at step 924. Fig.
23B graphically shows the presence of fluid in relevant portions of the reaction vessel bank
after this initial filling step.
: To ~rCrl~rlich the injection, valves 100 and 120 again open to let
1"' ' activating reagents enter the lower manifold at step 926. Selected ones ofvalves 101-109 open in parallel to permit a column of l ~ activating reagent to
advance up the ~u, .... ,1i.. ~.1 tube. c ' '~, associated ones of valves 111-119
open to inject amino acid into the upwardly advancing column of ~ LII;L~ activating
30 reagent and to mix with the activating reagent. Fig. 23C graphically shows this injection
step.
When each ænsor 101S-109S detects a fluid presence, the control computer
turns off a valve 101-109 which is associated with that sensor. When all valves 10 1 -1 09
~ WO 95/12608 2 ~ 7 5 5 8 7 PCT/US9.1/123~7
113
close at step 928, all valves of the reaction vessel bank return to the default mode. The
mixture of amino acid and activating reagent is preferably permitted to stay in the
afor~m-~n~inn~A tube for about two minutes to ensure proper activation at step 930.
Thereafter, the bottom manifold is cleared at step 932 in the manner
5 discussed earlier in connection with Fig. 17. The column of mixture between valves 101-
109 and upper sensors lOlS-109S are then pushed up into the reaction tubes in the manner
discussed in connection with Fig. 20 at steps 913-916.
Fig. 24 shows the sequence of commands issued by the control computer for
r~ the bead suspension within the reaction vessels back to the parent vessel. At10 the start, it is assumed that the reaction vessels have been drained, and the lower manifold
is filled with argon. The reaction vessels are first filled with a solvent at step 936 in the
manner discussed in connection with Fig. 20. Thereafter, all valves return to default.
Next, the mixture of beads and reagent is vortexed to create a suspension at
step 938. AlL~ tiv~ly, the mixture of beads can be vortexed while the reaction vessels
15 are being filled. Such a process improves the speed of suspension and helps prevent the
beads from clumping together at the bottom of the vessel. The contents of the reaction
tubes are transferred to the parent vessel at step 940. Valve 122 opens to pressurize the
lower manifold with argon. Valve 129 opens to permit the bead suspension to move from
the reaction vessel bank to the parent vessel. Selected ones of valves 101-109 open,
20 preferably in series, to permit argon to blow the contents of each reaction vessel up toward
the top common manifold and into the parent vessel. The contents of the reaction vessels
are thus transferred to the parent vessel serially. Although the above transfer may also be
performed in parallel by opening valves 101-lO9 ~ ,. Ju~ly~ serial transfer permits
argon pressure within the reaction vessel bank to remain high and is therefore preferable.
25 FUILI~ U~, each of valves lOl-lO9 preferably remains open for about four seconds to
ensure that s~bst~nti~lly all of the contents of a given reaction vessel are transferred to the
parent vessel. During this process, valves 90, 122, and 129 remain open.
After the contents of all reaction vessels are transferred to the parent vessel,~ the reaction vessels may be rinsed and another transfer process may occur. To rinse the
30 reaction vessels, the above steps are repeated, starting with the refilling of the reaction
vessels with DMF at step 936. As shown in steps 937 and 941, valve 129 is kept open
during the line cycles to prevent damage to the beads and valve 129. The parent vessel
preferably has volume for at least three transfers. At the end of the l~ul.lbi~ ion, the
wo gS/1260~ ~ 1 7 ~ ~ 8 7 Pcrluss~ll23~7 0
114
parent vessel is preferably drained by opening valves 110 and 91 until the level of f~uid in
the parent vessel reaches below lower capacitive sensor 90S and tums that sensor off.
Three cycles of rinse have been found to be satisfactory.
Thereafter, all valves including valve 129 return at step 942 to the default
S state. The contents of the parent vessel are agitated at step 944 to mix the beads from the
various vessels in the manner discussed in connection with Fig. 20. If the beads are to be
removed from the parent vessel, the parent vessel is preferably drained at step 946 by
opening valves 10 and 91 until the level of fluid in the parent vessel reaches below lower
capacitive sensor 90S and tums that sensor off. All valves are ~ub,,~u~llLly returned at
step 948 to their default state. The mixture containing beads may then be removed from
the parent vessel for use.
Alternatively, the beads may be reaUocated to the reaction vessels in the
manner discussed in connection with Figs. 21A-21B. FoUowing the rP~IIn~rinn all valves
retum to the default off state.
G. Overall Diagram of Software
Fig. 25 is a flow chart of the source code which is included herein as
Appendix I. Module 950 represents the user. A command interpreter 952 accepts the
textual commands from tbe user. Altematively, the user may enter commands to run the
synthesizer using a menu system 953. The commands received by menu system 953 are
either converted to a format usable by command interpreter 952, or call a support routine
in support routines module 962 directly. Command interpreter 952 also parses thecommands entered, textually or otherwise, by the user. Thereafter, the parsed commands
call and execute support routines in support routines module 962. ru-LI~ lulc, the parsed
commands are formatted by a display formatter 954 and displayed on a display 762.
Module 958 contains a plurality of macro files. A macro file defines, for
example, the sequence of steps that must actually take place to run a synthesis or build a
library. At its most basic level, a macro file contains, for example, macros which in turn
contains sets of discrete cûmmands for controlling valves and reading sensor information.
Macros may utilize other basic macros to perfomm higher level functions such as draining
reaction vessels 201-209.
The macros received by command interpreter 952 from macro files 958 are
passed into a synthesizer library controller 960. Synthesizer library controller 960 calls
~, WO 9S/12608 2 ~ 7 5 ~ 8 7 PCT~59~112317
115
the macros for actuai synthes~s. For example, one macro may specify the initial global
variables that must be set before synthesis begins.
Fig. 25 shows a support routines module 962 for running a variety of
support ~ubluuLill~s. One such subroutine is autofill, which is a subroutine for5 ~l~fom~tir~liy filling the reaction vessels until all sensors are on. Support routines 962
accept inputs from either command interpreter 952 or menu system 953. Option list 964
contain pointers to functions, etc. Fig. 26 shows the inputs and outputs of option list 964
and its,,1~ with command interpreter 952 and support routines module 962.
There are also; .;I; .I;,~ti"" files 966 for holding global variables and global10 settings. Tr~ li7~tilm hles 966 hold, for example, a value ~ ; e the amount of time
during which a strike voltage is supplied to a valve to open a closed valve, etc. A lookup
table file 968 cooperates with ~yllLII~;~, command library controller 960 to, for example,
permit a monomer to enter ~ u~lidLe selected reaction vessels. Lookup table file 968
may contain, for example, a listing of each reaction vessel, its ~.."~ .on.l;l,~ tag
monomer, and the list of monomers necessary for sy^~hPci7in~ the desired polymer.
Fig. 25 also shows a log file 970. Log file 970 accepts inputs from
command interpreter 952 and ~l-tll~. . library controller 960. Log file 970 contdins
npPrPtirn~l data for diagnostic purposes. An entry in log file 970 contains, for example,
;"r""" ~;.", relating to the macros cailed.
An associate file 972 contains a listing of each reaction vessel and its
associated valves and sensors. Associate file 972 cooperates with both synthesizer library
controller 960 and support routines 962 to simplify the task of addressing each reaction
vessel and its associated valves and sensors.
The digital commands outputted by support routines 962 enter a parallel
driver 974. Parallel driver 974 may be, for example, PCDIO120-P l/O board 766.
Parallel driver 974 outputs valve control signals 976 via its IlO channels to drive the
solenoid valves. The vaive control signals, as discussed, are further processed by
controller circuit 768. FulLil~ lui~, parallel driver 974 outputs stepping motor controller
- signals 978 to control the vortexing stepper motor. Sensor inputs 980 from the optical
sensors, the ultrasonic sensor, and the capacitive sensors of the ~yllLIl~ . are also
received by parallel driver 974 for processing by support routines 962 via sensor checicing
~UIJlUULil.~s 982.
wo 95/12608 2 1 7 5 5 8 7 Pcr~S9J/123~7 ~
116
Fig. 27 illustrates the data structures necessary to control valves. A valve
index VINDEX array 986 accepts a valve number 984 as input and provides a pointer to a
VALVES array 988. ~ach element of VALVES array 988 contains a pointer to a data
byte 990. Each data byte 990 contains 8 bits of valve data i"r ,""~ The bit
containing valve data i--rulll~Liu-- for a given valve is accessible by the address of its data
byte 990 and a shift value It~ g the relative location of that bit within dat~ byte
990. Each bit of data byte 990 may be m~nir~ t~fi to dp~Jlul~lidL~ly turn on or off a
valve. The valve data i"r,~ ;Oll in each bit corltrols a (;~II~,.JUlldil~g valve via output
port 992.
Fig. 28 iilustrates the data structures necessary for receiving sensor
i"r~,l...d~ from input port 994. The ;,.r,,"~ " representing the binary state of each
sensor is stored in one bit in a data byte 996. To access the information in data byte 996,
a sensor number is used to access SENSOR array 998. Each element of SE~SOR array998 contains a pointer to an d~ JlidL~ data byte. Each data byte 996 contains 8 bits of
15 sensor data information. The bit containing valve data ;r~-"~ ,. for a given valve is
accessible by the address of its data byte 996 and a shift value representing the relative
location of that bit within data byte 996.
H. Windows Interface
The control software may illl,Ol~ a Windows-type interface or
workspace. Generally, the Windows interface is a l~ ~ ' , graphical user interface
(GUI) providing one or more windows for display on the screen. Additional windowobjects may be displayed in various si~es and formats (e.g., tiled or cascaded), as desired.
At the top of the window is a menu bar with a plurality of user-command choices, each of
which may invoke additional submenus and software tools for use with application objects.
The window also includes an area for displaying and n ~nirlll~in~ screen objects. This
area is a workspace or viewport for the user to interact with data objects which reside in
the memory of the control computer system.
The windows interface includes a screen cursor or pointer for selecting and
otherwise invoking screen objects of interest. In response to user movement signals from
a pointing device such as a mouse, the cursor floats (i.e., freely moves) across the screen
to a desired screen location. During or after cursor movement, the user may generate
user-event signals (e.g., mouse button "clicks" and "drags") for selecting and manipulating
~ WO 95/12608 2 i 7 5 5 8 7 PCT/US9~1123.17
117
objects, as is known in the art. For example, the window may be closed, resized, or
scrolled by "clicking on" (selecting) screen ' ""'l'') " Keystroke equivalents, including
keyboard a 1 or "hot keys", are provided for pcl~ull~ g these and other user
operations through the keyboard. Thus, a Windows interface provides a more intuitive
5 approach to interacting with the control computer.
Underlying the Windows interface is a message or event-driven architecture.
This model is perhaps best described by contrasting its operation with that of a modal or
sequential al~ ll;t~ult; that has been traditionally employed, as ~ ~",l liri~l by the
command interpreter in Fig. 25. In this manner, the reader may appreciate the added
10 flexibility as well as complexity of an event-driven system.
A modal program comprises a series of discrete operating blocks or modes
having a well-defined beginning, middle, and end. Thus, the program follows a fairly
rigid sequence of operation with each step necessarily being completed before the program
proceeds to the next step.
While a modal program is relatively easy to design and implement, it is
generally not easy to use. The design certainly ensures that all required information is
entered, but only at the expense of forcing users to operate in a manner dictated by the
program. Specifically, since the program is built around a pre-arranged set of modes, a
user cannot get from one mode to another without first completing a previously-required
20 mode. Any deviation from this sequence by the user is simply not permitted. This
inflexibility of the modal programs may be inefficient for handling real-world tasks.
On the other hand, an event-driven ~ u,~ eschews a pre-selected
sequence, opting instead for an "event loop. n The event loop is a centralized mechanism
for processing messages about user and system events. It includes an event queue and
25 "~ for retrieving and ', ~ messages to various window classes.
Messages are how the operating system manages and ~ llul~ multiple
- C and hardware events, such as clicks of a mouse or presses of a keyboard,
which in MS-Windows are converted to messages by Windows event handlers. From a
^- UII ~ , a message is simply a data structure containing information
30 about a particular event. The message structure may include a message identifier which
serves as a symbolic constant for a particular event. For example, messages from a
window object might include i r.~ ;.... about creating, closing, moving, and re-sizing
the window. Additional event data are available as message r:~r~m-trr~ the exact
WO 95/12608 2 1 7 5 5 8 7 PCTIUS9~/123~7 ~
118
illL~I,ul~;L~Liull of a given parameter varies with each event type ~,UI~ d Input
messages are coLected in a system-wide queue and then directed to the proper window.
These messages, along with timer and screen paint (screen refresh) messages, are passed
to the target application(s) of interest.
A m,~nh~nicm is provided for retrieving messages from the system queue and
rii~r~t~hin~ them to the appropriate application which, in turn, may proceed to process any
message that arrives. Each window belongs to a particular window type which defines
certain ch~r~-~t~ri5fir~ common to all windows of that type. Associated with each type is a
Windows function which processes all messages sent to windows of its type. An
lû application queue is provided where Windows may place messages that belong to a specific
~rrli~"ti"n When the application is ready to receive input, it simply reads the awaiting
messages. If none are found or if there exists a message for other ~rFlir~-innc Witl~ higher
priority, Windows passes control to the other -l,r,li. ~li...,~
The general mechanism for retrieving and ~ messages in an event-
15 ~ased system, such as Microsoft~D WindowsW, is known in the art; see, e.g., Petzold, C.,
Plu,slu.,."..ng Wi~dûws, Second Edition, Microsoft Press, 1990 and Custer, H., Inside
Windows N7, Microsoft Press, 1993. Additional inf~lrm~tirm can be found in Microsoft's
Window Software D~velo~ lL, available from Microsoft Corp. of Redmond, WA. The
disclosures of each of the foregoing are hereby illCullJuldL~I by reference for all purposes.
Fig. 33 illustrates a GUI as ;, ~ on the control computer. The CUI
includes a rectangular window 1301 with a workspace 1303. At the top of the window is
a menu bar 1305 with user command choices 1306-1313. Each of these command choices
include additional submenus containing commands for controlling the operations of the
synthesizer. These command choices offer a user the flexibility of either pc~lrollllil~g a
synthesis ~llt~m~fir~lly or manually by invoking the ,~ lu, commands with the
mouse.
The GUI is designed with the intention of being a user friendly
~IIV;~UI~ , thus ",;,.;",;,;"~ efforts required for ~IU~ g the synthesizer. For
example, a dialog box object or a set of dialog box objects is associated with each
command. When a command is invoked, the ~,U~UIU~IidL~ dialog objects are displayed in
the workspace and interactively prompt the user to enter the necessary il~rû~ldliull. A
Help command 1313 provides ill~U~ Liull to assist a user through the process. Using the
2 1 75587
WO 9~/12608 PCT/US9-11123~7
119
dialog objects, the user may program the synthesizer without using traditional text
r,omm:~n~lc
Fig. 34 iliustrates a submenu 1320 that is associated with the Macros option.
A user may invoke either a Learn or Run command by clicking the mouse on submenuitem 1321 or 1322, respectively. In the Windows ~llvilUll~ , a macro is an object that
contains a set of discrete comm~nl1c such as those discussed in connection with Figs. 16-
24, for controlling the synthesizer. Using the Learn command 1321, the user can define a
macro to perform specific functions. The submenu may also identify the keystrokes
assigned to invoke the available rl.mrn-.n~C For example, "Alt+L" is used to execute the
Learn command.
Once the macro is "learned," the Run command 1322 may be selected to
execute the learned macro or any other macros which have been previously defined. Fig.
35 exemplifies a dialog box object that is displayed when the Run command is invoked.
The dialog box object includes an area 1325 (combination box) which lists the available
macros. To select a macro, the user enters the name of a macro in a Select Macro space
1324. Alternatively, the user may scroll, by clicking and dragging the mouse, until the
desired macro flle in space 1325 is selecoed. The user then enters the number of times the
macro is to be repeated in space 1326. Finally, to run the macro, the user clicks the
mouse on a RunMacro button 1327 or a RunSMacro button 1328. The RunSMacro
command instructs the system to perform the macro functions serially, i.e., one reaction
vessel at a time. The Cancel choice 1329, when selected, exits the dialog box object 1326.
A Help choice 1330 provides i~lru~ iul~ regarding the different choices in the dialog box
object.
Fig. 36 illustrates a submenu 1335 which is displayed when the Groups
option 1310 on the menu bar is chosen. Submenu 1330 includes a Define command 1331
which is employed to define a set of valves associated with a group of specific reaction
vessels. Once defined, the valve group is stored as a group object in memory. The
computer's memory may contain many group objects, each defining a unique valve group.
Submenu 1330 also includes an Open/Pulse command 1332 to fill and drain
the selected reaction vessels for a predefined time period. When invoked, a dialog object
box associated with the Open/Pulse command is displayed. The dialog object box,
somewhat akin to the one illustrated in Fig. 35, contains a ~,UIIliJ;l~Lioll box that lists the
SUBSTITUl~ SHEET (RUi~ 26)
Wo 95/12608 2 t 7 5 5 8 7 PCT~lSg~1l23~7 ~
120
available group objects from which to choose. The user selects the desired group and
inputs the desired time period to pulse the selected valves in an assigned space proYided by
the dialog object. To fill the reaction vessels, the user clicks an Open button to indicate
that the valves are to be opened. Next, the user initiates the filling process by clicking
either a Pulse or SPulse (to fill the reaction vessels serially) button. To drain the reaction
vessels, the user clicks a Close button and either the Pulse or SPulse button.
The user, using a Fill/Drain command 1333, can fill or drain the vessels
~llSnrr~sir~lly using sensors. The Fill/Drain command, when selected, displays a dialog
box object similar to that of the Open/Pulse command. The user selects the group of
valves and clicks either an AutoFill or SAutoFill button to perform a fill function or an
AutoDrain or SAutoDrain button to empty the reaction vessels. In some rmhotlin~.on~c, a
Time Delay option may be provided to delay the valves from being closed or opened after
triggering the sensors. This function is p~uLi~uld ly useful in situations where the reaction
vessels are not quite at their desired levels when the sensors æ activated. By setting the
delay option with a specific delay period, the reaction vessels can be filled d~lU,UlidL~Iy.
As can be seen, the commands associated with the Group menu afford the user flexibility
in choosing ~n"-l,i"~ , of reaction vessels to employ during synthesis.
Fig. 37 illustrates a submenu 1340 containing the options available for the
Variables menu selection. A create command 1341 defines a variable which may be
1l"~ 1 in the macros. Variables are typically employed, for example, in situations
where the value, such as time, may vary from one synthesis to another. Instead of
creating a macro for each time value, a variable is simply defined to correspond to time.
The variables are set to the desired value before each synthesis cycle using the set
command 1342.
Fig. 38 illustrates a submenu 1345 that is associated with the Diagnostics
option. Submenu 1345 includes Valves 1346, Sensors 1347, and Mix 1348 commands to
gives a user intimate control of the synthesizer and access to inform~Sinn regarding Ihe
synthesizer for diagnostic purposes. The Mix command 1348, when selected, activates the
vortex motor to mix the reaction vessels for a period of time as specified by the user.
Referring to Fig. 39, a Valve Diagnostic dialog box object 1390 is displayed
when the Valves command is selected. The user, via dialog box object 1390, may contro]
the operations of any valve in the synthesi~er by selecting the entry with which th~ desired
valve cull~uul.d~. For example, Valve 100 in Bank I is opened by clicking the cursor on
WO 9S/12608 2 1 7 ~ 5 8 7 PCTIUS9~1123-11
121
box 1391. For ~u.,~ , all the valves may be closed by selecting the Close All
command 1397. A Cancel command 1398 closes the dialog box object 1390.
Fig. 40 illustrates a Sensor Diagnostics dialog box object 1391
CUI~ pOlldil~g to the sensor command. As shown, Dialog box object 1391 contains a box
1396 associated with each sensor. Boxes 1396 inform the user of the status of the
1ull~ uildillg sensors. For example, if a box has an "X" in it, this would indiQte that the
sensor with which it is associated is on. Conversely, an empty box would indiQte that the
sensor is off.
Fig. 41 illustrates a submenu 1350 that is displayed when the File option is
selected. A New command 1351 creates a new synthesis setup file for Qrrying out a
synthesis and a Modify command 1352 allows a user to select a preexisting synthesis file
for editing. Once the synthesis setup file is completed, the user invokes either the Save
1354 or Save As command 1355 for saYing the file in memory. A Print command 1357prints the selecoed synthesis file. A Print Setup command 1358 configures the printer to
desired mode, such as printing the file in landsQpe mode. An Exit command 1359 is
invoked to leave the File option.
In some instances, such as prior to each synthesis, or when a new valve
group is selected, the user may wish to configure the system by invoking the Load CFG
files command 1356. The system loads the d~ U~ t~, files to inform which are thed~lU~Jl valves to use. In effect, CFG files map or "associate~ the Yalves with each
selected reaction vessel.
Figs. 42-44 illustrate the dialog objects used in creating and modifying a
synthesis setup file. Referring to Fig. 42, a Set Associate dialog box object 1360 allows
the user to select the desired reaction vessels by checking the dl)~lUIJl ' boxes contained
in space 1361. For Uu~ , a Check All button 1362 and Uncheck All button 1363
are provided to easily select or de-select all the reaction vessels. Check Bank buttons
1364a-1364d allow the user to select all the reaction vessels belonging to a specific bank.
A Cancel button 1368, when selected, aborts the synthesis setup process. A Help button
1369, as previously explained, provides information to assist the user through the process.
To continue the synthesis setup process, the user clicks the OK button 1365 which closes
the Set Associate dialog box object and displays a Synthesis Setup dialog box object.
Fig. 43 illustrates the Synthesis Setup dialog box object 1370 with which a
user defines a Start Macro 1371, a Loop Macro 1372, and an End Macro 1373 for the
SUBSTITUTE SHEET (RULE 26)
wo 95112608 2 ~ 7 5 5 ~ ~ Pf~/Us9~,l23~7 ~
122
synthesis. The Start Macro and End Macro, for example, include commands for washing
the reaction vessels before and after each synthesis process. The Loop Macro contains
commands to perform a synthesis. These commands include injecting the selected reaction
vessels witb amino acid building blocks and oligon~f leotiflrc~ pooling the contents of the
reaction vessels in the parent vessel and mixing them to form a bead Sll~r~n~ n, and
fi~ the bead suspension to the selected reaction vessels. The injecting, pooling,
and ~ ;b~ steps constituoe one synthesis cycle. The synthesis cycle is repeated
according to the value enoered in the Number Synthesis Steps parameter 1374. Once the
Macros are defined, the user saves the conoents of the Synthesis Setup dialog box object
and continues the setup process by clicking an OK button 1375. Again, the user may abon
the process by clicking a Cancel button 1376.
Fig. 44 illustraoes the Amino ~ Oligo Setup dialog box object 1380. As
shown, the box contains entries 1381 associaoed with each reaction vessel. The user enoers
the desired amino acid symbol and f~ fln~ f~otifif~ code in the entries. For example, if the
entry for reaction vessel 1 (RV01) contains "V ATGCCGA", this causes the synthesizer to
inject amino acid V and I;L, '~ . ' ATGCCGA into reaction vessel 1. After the
a~ r codes are entered, the user clicks the OK button 1382 to compleoe the setupprocess. A Cancel button 1383 is provided to abon the process. As can been seen, a
synthesis library can easily be creaoed using the synthesis setup procedure.
Once the synthesis setup is compleoed, the user may initiaoe the synthesis
process. Referring to Fig. 45, the user first selects the Synthesis option 1308 and then the
Go command 1391 to begin the process. As an aloernative, a user may build a single
macro to perform functions similar to those saved in the synthesis files.
Referring to Flg. 46, the GUI displays a status screen 1400 during the
synthesis or macro execution. As shown, the status screen is divided into two separate
areas 1401 and 1402. The first area 1401 displays information relating to the synthesis.
For example, the names of the Stan Macro, Loop Macro, and End Macro are lisoed.
Additionally, a Loop Number line 1408 informs the user as to the number of loopsremaining in the synthesis.
The second ar a 1402 displays the filename of the Macro currently being
execuoed. As previously described, a macro may be nesoed with other macros to perform
higher level functions. The starus screen may be designed to list up to 10 nested levels
1409a-1409j of macros being called. A Command line 1410, a Timing line 1411, Message
SUB~TITUTE SHEET ~RUL~ 26)
~ WO 95/12608 2 ~ 7 5 5 8 7 PCTIUS9 11123~7
123
line 1412, and RV-s line 1413 may be provided to display additional status or c~nfl~l~r?ri--r~
information, For example, the Command line may show which mæro command is being
executed; the Message line, when applicable, displays a text message as ~lc~lu~ .lcd by
the user. The Timing line displays the amount of time that remains in which the valves are
5 opened or closed. The RV's line informs the user which reaction vessels are used. A
Time to Complete bar 1414 informs the user as to the percentage of time remaining before
the synthesis is completed.
At anytime during synthesis or macro execution, the user may switch into
the "User Abort" mode by pressing a preassigned function key, i.e. F12. The User Abort
10 mode allows the user to pause and execute macros that are not defined as part of the
synthesis without having to abort the synthesis process. When invoked, the sysoem
temporarily pauses execution and displays the User Abort dialog box object 1420, as
illustrated in Fig. 47. At this point, the user may insert any macro for execution. This
can be done by typing the name of the desired macro in a Select Macro box 1421 and
15 clicking the mouse on a Run Macro button 1422.
The User Abort dialog box object also provides an Ignore button 1424 which
enables the user to continue the synthesis process as if the system had never entered into
the User Abort mode. A skip option 1423 instructs the system to first skip over the
current command in the macro before continuing the synthesis. A Retry button 1425
20 causes the system to execute the current macro from the beginning. An Abort button 1426
allows the user to cancel the synthesis.
Fig. 48 illustrates a submenu 1430 associated with the Edit command. The
Edit command permits the user to easily access and modify the various objects by selecting
the dp~lu~JI items listed in the submenu. An Associate command 1431, when selected,
25 displays the dialog box object which lists the Set Associate objects stored in memory. By
selecting the desired object, the Set Associate dialog box object is displayed. At this point,
the user may edit the contents of the dialog box object with the mouse. When finished, the
user clicks the OK button and the ".~l;ri. ,.li.",~ are saved. Likewise, the user may edit the
Group, Macro, Variable, and Code objects by selecting the desired object. The selected
30 objects are then placed in an editor, allowing the user to modify the text.
As previously mentioned, the sensors may be inadvertently triggered by the
presence of a drop of liquid. This may be a problem, particularly when the solution used
SUBSTITUTE SH~ET ~U~E 2~3
WO 95/12608 2 1 7 ~ 5 ~ 7 PCT/IIS9.1112317 ~
124
dunng the synthesis cycle contains a high rnn~Pntr:~tinn of bubbles. To avoid or mitigate
t .,1~ 1 triggering of the sensors, their sensitivity may be ~ P~I
The sensors are IJ.u~l~u~ d using a Machine Config command 1456. This
command displays a Config dialog object box containing seYeral entries, Fill%, Drain%,
5 TimeOut%, amd PtsAve. The Fill% specifies the percentage that the sensor remains on to
determine when the reaction vessels are full. For example, if 80% is entered, the sensor
must be on 80% of the time that it is read. The Drain% specifies the percentage that the
sensor remains off to determine when the reaction vessels are empty. As an example, if
20% is entered, the sensor must be off 80% of the time that it is read. Timeout specifies
10 the time period to delay before the sensors are switched on or off. PtsAve specifies the
number of points or readings to use to determine the percentage of time that the sensors
are on or off. The user may also specify the sensor or group of sensors from which to
read. Thereafter, the user cliclcs an OK button which ~lltnm~ti(~ y configures the system
accûrding to the p~r~mP~Prs.
Fig. 49 is a flow chart illustrating the event-driven ~Ul,~ UIt of the
control sûftware as described in Figs. 33-48. As shown, an l/O Object module 1450,
which is the heart of the ~yllLl.~ s GUI, facilitates c~ ..,. among the various
object modules comprising the GUI. When the control software is initiated, a mûdule
1460 loads the u.Jl~ ;ul~lLiull files into the I/O Object module. These files are used to
~0 configure a Variable Objects module 1456, Lookup Table Objects module 1457, Macro
Objects module 1458, Grûup Objects module 1459, Valve Object Array module 1461, and
Sensor Object Array module 1462.
The Lookup Table Objects module contains a file used for mapping the
valves and sensors to their ~ ~ull~ ; reaction vessels so as to eliminate the need to
25 manually address each individual valve or sensor relating to a specif~c reaction vessel.
The Macro Objects module stores the list of defined macro; the Variable Objects module
stores the defined variables while the defined group objects are stored in the Group Objects
modules.
The Valve Object Array module stores an array identifying each individua~ ~
30 valve in the system. To close or open a valve, the I/O Object module scans the Valve
Object Array module until if finds a match. When a match is found, the Valve Object
Array module outputs a signal tû control that specific valve. Tû cûntrol a grûup ûf valves,
-
~ WO95/L2608 21 7~587 PCTIUS9.11123.17
125
the I/O module cooperates with the Group Objects module to sc;m the Valve Object Array,
causing it to send the d~lU~ L~ control signals to the selected valves.
The Sensor Object Array module stores an array identifying each individual
sensor in the system. The 1/0 module may read a sensor by scanning the Sensor Object
Array module until a match is found, causing it to read the specified sensor. To read a
t group of sensors, the I/O and Group Objects modules scan the Sensor Object Array
module to determine the d~JlU~lidt~ sensors to read.
The GUI also includes a Dialog Box Objects module 1453 and a Synthesis
Object module 1454. The Dialog Box Objects module contains the dialog box objects
which are displayed when certain commands are invoked. The Synthesis Object module
stores the current synthesis setup file. To perform a different syntlnesis, the GUI reads the
desired synthesis setup file from memory into the Synthesis Object module. Conversely,
the contents in the Synthesis Object module are written to memory to save a synthesis
setup file.
A Main Window Object (MWO) module 1452 ro~nml~ni~ with a View
Object module 1455 which displays the GUI's main window (Fig. 33) on the screen. A
Message, in response to a user command, is sent tû the MWO. This message is parsed to
determine the ~ Jl command to execute. For example, if a Macros command is
received, the MWO instructs the I/O object to retrieve the macrûs dialog box object from
the Dialog Box Objects module and the lists of macrûs from the Macrûs Objects module.
The View Object module receives this ;"r," ", ~ "- frûm the I/O module and displays it on
the G~JI.
In one ~ . the I/O module contains a Watchdog routine to shut
down the ~y~ if the computer should m~lfiln~ tinn For example, the 1/0 object
25 module may be ~.u~ n~ to send a pulse every 2 seconds to the ~yllLllC~ If for any
reason the ~yll~ siL~,l fails to receive these pulses, it will assume that the control computer
has msllfilnrtionf-d and will power down.
~XAMPT F~
3û The following examples are provided as further illustration of the present
invention and not as a limitation.
wo 95112608 2 1 7 ~ 5 8 7 PCT/US9~123~7 ~
126
EXAMPLE 1: LIBRARY PREPARATION AND SCREENING
This Example illustrates how the products of a ulllbill~Lulidl peptide
synthesis on resin beads can be explicitly specified by attaching an ~ lrlrul;~lP
identifler tag to the beads coincident with each amino acid coupling step in the synthesis.
5 Each tag conveys which amino acid monomer was coupled in a particular step of the
synthesis, and the overall sequence of a peptide on any bOEd can be deduced by reading the
tag(s) on that bead. The collection of beads can be screened for binding to a
lluu~ lLly-labeled anti-peptide antibody using a lluulc~ activated cell sorting
(FACS) instrument. Those bOEds to which an antibody binds tightly can be isolated by
10 FACS, and the ,.li~,..",. lr.,li~lr- identifiers that j~re attached to individual sorted bOEds can
be amplifled by the PCR. The sequences of the amplified DNAs are determined to reveal
the identity of the peptide sequences which bind to the antibody with high affinity. By
combining high capacity, l~lir,~"".~ code-based i- r."" ~ storage, amrlihr~til-n
mrthrl~lrlngy, and lluuic,--.l- e-based sorting, the present method provides a means for
15 specifying the identity of OEch member of a vast library of molecules ~y~ from
both natural and unnatural chemical building blocks and for quickly and efficiently
isolating individua~ bOEds that bOEr high affinity ligands for biological receptors.
In this Example, single stranded ~ ,Lides are used to encode a
.ulllbilldLu-i~l peptide synthesis using both L- and D-amino acid building blocks and 10 ~m
20 diameter ~oly~y~ beads. The ~ "~ .Jl;~rl~ tags have a high information content, are
amenable to very high sensitivity detection and decoding, and, with the present method,
are stable to reagents used in peptide synthesis. Peptides and n~ ti~i~-s are assembled in
parallel, alternating syntheses so that each bead bOErs many copies of both a single peptide
sequence and a unique oligon~lrlPoti~ identifier tag. The olieom-rl~tides share common
25 5'- and 3'-PCR priming sites, and thus the bOEds can serve as templates for the PCR. The
encoded synthetic library contains about 8.2 x 105 hepta-peptides and is screened for
binding to an anti-dynorphin B ,.,~ 1 antibody D32.39 (see Barrett & Goldstein,
1985, Neurûpeptides : 113-120, illl,Ul,UI ' ' herein by reference), using a lluu-c:s-c,.
activated cell sorting (FACS) instrument to select individual beads that strongly bind the
30 antibody. After PCR ~mrlifir~tir~n of the ,~ ullllrl~lLide tags on sorted bOEds, the DNA
is sequenced to determine the identity of the peptide ligands.
~ WO 95/12608 2 1 7 5 5 8 7 PCT/US91/123.17
127
A. Reagents and General Methods
The Illu~lodi~zc~ 10 ,um diameter bead materiai used in this work was a
custom-sy-lLl,~,;~d "~a~lul~uluus styrene-divil.y~ lle copolymer filnrlirn~li7P~i with a
1,12-.l;~ ln~lf~ - linker purchased from Pharmacia. The beads are Pharmacia
S MflnnhP~fl~u that have not been derivati_ed with Pharmacia's Gene Assembler Support
t linker. See Ugelstad and Mork, 1980, Adv. ~Q~ 101-140,
i". u",, ~ herein by reference.
All protected amino acids were obtained from Bachem Bioscience Inc. PCR
and sequencing primers were ~yllLll..,;~J with an Applied Biosystems model 394
10 ~ ..";.1P ~yll~ .. Authentic samples of certain peptides were ~yllLll~ .d with
an Applied Biosystems model 431A peptide ~llLi.~,;~l using Fmoc-protected amino acids,
HBTU/HOBt in situ activation chemistry, and d~lut~Liull with 40:1:1
TFA/water/r~ l.;r,l. These peptides were purified by HPLC (>95% purity) on a
Rainin Cl8 reverse phase column using 'WdiCI/i" ~ P/0, 1 % TFA as eluant, and
15 structures were verified by mass ~ UIIl~Lly.
B. Parallel Synthesis of a 69-base Ol;~ ;flP and the Opioid Peptide
Dynorphin B
The C-terminal seven amino acid fragment of the opioid peptide dynorphin
20 B H-Arg-Gln-Phe-Lys-Val-Val-Thr-NHz (RQFKVVT) (SEQ ID NO:2) was ~y-l~l-e~i~l in
parallel with a 69-mer oliLudw~yl.J~l~;uLidc (ST08) on 10 ~m diameter beads. Thesequence of ST08 was 5'-ATC ('AA TCT CTC CAC (ATC TCT ATA CTA TCA) TCA
CC iTA TC CT AT TT TT AC] CTC ACT CAC TTC CAT TCC AC-3' (SEQ ID
NO:20) . Underlined portions of this sequence correspond to PCR-priming sites whi~e the
25 region in L~u~ es is l~u..lulo~;uu~ to the primer used fûr sequencing this template. The
14-base sequence enclosed in brackets represents the coding region of the template.
The beads were first treated with a mixture of succinimidyl
4-O-DMT-u~ybu~yldL~ (Molecular Probes) and the l-u~yb~ P ester of either
N-Fmoc-2,4-dimethoxy-4'-(~bu~y.l.~Li.ylo~:y)-b~l.Ll.yd,yla...;..~ (i.e. the acid-cleavable
30 Knorr ~ubu~d~;d~ linker) or N-Fmoc-Thr('Bu)-OH (for non-cleavable ~ s). The
ratio of Fmoc-protected amino groups to DMT-protected hydroxyl residues on the beads
was determined ~L..,~ y to be ~ .u,~..n~t~.ly 20:1. The beads were
subjected to 20 cycles of Dliel.l. lflr~liflP synthesis on an automated ~yllLI~ using
Wo 95/12608 2 ~ 7 5 5 8 7 Pcr/uss~ll23~ ~
128
3'-O-methyl-N,N-diisopropyl P~ A~ PC of the following nl~rlPo~i~Pc
N6-Bz-5'-O-DMT-('7-dea~a)-2'-deoxyadenosine (Berry and Associates, ~nn Arbor,
Michigan), N~-Bz-S'-O-DMT-2'-dcv,.y~ -e, and 5'-O-DMT-thymidine (Glen Researchl-The beads were then removed from the instrument and treated for 5 min.
5 with 10% piperidine in DMF to remove the Fmoc protecting group. After coupling the
first amino acid residue (N-Fmoc-Thr('Bu)-OH), the beads were treated with a DME~
solution of acetic anhydride and 1-methylimidazole to cap any unreacted amines. All
peptide coupling rcactions were run for 20 min. and contained 0.11 M Fmoc-amino acid,
0.1 M HBTU, 0.1 M HOBt, and 0.3 M DIEA in DMF. The beads were then subjected to
10 two cycles of nucleotide addition on the synthesizer (detritylation with TCA;tetrazole-catalyzed ~ u~l-;L~l.lLiu-~; capping with acetic anhydride; oxidation with iodine in
acetonitrile/water). Sequential steps of amino acid coupling and din~ e addition were
repeated until synthesis of the peptide sequence RQ~KWT (S~Q ID NO:2) and
~UIlsLlu- ~io.. of the oli~;~,-"~ P coding region had been completed. After p~lru~ g an
15 additional 35 cycles of oli~j~",~ lPul;-iF synthesis, the beads were treated sequentially with
piperidine~DMF (1:9 for 8 min), thiophenol/L.;~ ullillc/dioxane (1:2:2 for 4 hr),
ethyl. f-l; "..,.,r/ethanol (1:1 for 5 hr at 55 C), and TFA/water (20:1 for 1 hr) to
deprotect fully both the peptide and ~ F~ f chains. In ~ F ` using the
acid-cleavable linker, the ~ from the TFA d-~.,lu._Liu.~ reaction was conrrntr~tP~i
20 ~n vacuo, and the isolated crude peptide was then analyzed by HPLC.
C. Construction of an Encoded Library
The parallel synthesis chemistry outlined above was used in the construction
of the library. The sites of peptide synthesis were ~ir~ from DNA synthesis sites
25 in this P~rPrimrnt by coupling to all the beads a mixture of N-Fmoc-Thr~Bu)-OBt and
suc~i-,illlidyl 4-O-DMT-u~.ybuLyl~Lc; as described above. Sequences of oli~;ullucleuLide tags
in the library deviated from ST08 only within the coding region. The 3'-conserved region
of the ,~lig~ FvLidc ST08 was first ayllLII.,;~ on a total bead mass of 35 mg (-- 1.75
x 10~ beads). The E~moc protecting group was removed and the bead mass was divided
30 into seven equal parts. To each aliquot was coupled one of seven different
alpha-N-Fmoc-protected amino acids (side chain protecting groups are shown in
p~ut llLIl~ sia): Arg(Nr-Pmc), GlnF~Trt), Phe, Lys('Boc), Val, D-Val and Thr(tBu). Each
part was then subjcctcd to two rounds of automated oli~u,.~ P synthesis. The
~ WO95112608 2 1 75587 PCT/US9~/123-17
129
respcctive sef~uences of the appended (~ r J~ that specified uniquely each different
amino acid residue were TA, TC, CT, AT, TT, CA and AC The beads were then
pooled, mixed thoroughly, and the entire bead mass subjected to Fmoc d~luLc~Liu-l.
This cycle of bead partitioning, peptide coupling, nli~nnllrlf~)tiflf dimer
5 synthesis, bead l~r ,.~ i.... and Fmoc removal was repeated for a total of seven times.
The final Fmoc protecting group was not removed. Rather, the pooled bead mass was
subjected to 35 cycles of oli~nn-lrlfl~tifif synthesis. The library was then fully dc~ut~lcd
as describcd above.
D. Library Staining and FACS Analysis
A portion of a library (typically 0.5-2 mg of beads) was suspended in
blocking buffer ~PBS, 1% BSA, 0.05% Tween-20) and incubated at room t~llpcldlulr for
1 hr. The beads were pelleted by - .I ir~ ;-.,. and ~ lrll in a solution of mAbD32.39 (10 mg/mL in blocking buffer). The suspension was incubated on ice for 30 min.,
15 pelleted by ,f ,t, i rl,~f l i..", and washed with blocking buffer. The beads were then
suspended in a solution of pl~y~ u~ly~ -conjugated goat anti-mouse antibody (Molecular
Probes) for 20 min. on ice. The beads were washed in blocking buffer and diluted in PBS
for delivery into the nuulc~ activated cell sorting (FACS) instrument (Bccton
Dickinson FACStar Plus). Beads which had bound the mAb D32.39 were identified by20 their acquired nUUlL~ CllCe. Individual beads from both the most brightly stained û. 17%
of the library and from the region having the lowest lluu.c~ ~, ce (ca. 98%) were sorted
into PCR microfuge vials. Specific binding of D32.39 to the beads was blocked byplr;l~ ll of the mAb with the soluble peptide Ac-RQFKWT-OH (SEQ ID NO:2) at a
final - ..,. f ~Ill,U;l~ll of 10 ~M.
E. PCR of Bead-Bound Template
PCR ~, l.lir; ~ c were performed in the ... r Il~r~ supplied buffer
system (50 mM KCl, 10 mM Tris-HCI, pH 9.0, 0.1% Triton X-100, 2 mM MgCI2) with
0.2 mM dATP, dCTP, and dGTP, 0.8 mM dUTP, 2 mM each primer, 3 units TaQ
30 polymerase (Promega), and 1 unit of uracil DNA glycosylase (Gibco BRL) (total volume
70 L). The primer scquences, 5'-ATC CAA TCT CTC CAC-3' (SP13) (SEQ ID NO:21)
and 5'-f~biotin)-GTG GAA TGG AAG TGA-3' (SP14) (SEQ ID NO:22) were respectively
hnmnlf~go~ls and ~""1~1 ."~ ly to the template ST08. PCR reactions consisted of 45
WO 95112608 2 ~ 7 5 5 8 7 PCT/IJS9~ -5.17 ~
130
cycles of d~,,dLuld~iull at 95C for 30 sec., primer annealing at 50~C for I min., and
extension at 72C for I min. Reactions were analyzed by clc~luyllulc,;s in 20%
acrylamide or 2% low melting point agarose gels.
F. Sequencing of PCR Product
BiuL;Ily- ' PCR product from individual reactions was isolated with
streptavidin-coated magnetic beads (Dynal, Inc.). After alkaline elutiûn of the
non-biotinylated strdnd and washing, each bead sample was treated with sequencing
cocktail. Dideoxy sequencing was performed using the primer 5'-ATC TCT ATA CTA
TCA-3' (SP15) (SEQ ID NO:23) and Bst L)olylllrldac (Bio-Rad) according to the
r~ f~ tllrpr'5 ill~LIul,~iul-~, with the exception that a 1:100 ratio of deoxy- to
d;dco~y~ lcuLide Lli~llu~lJlldL~a (Pharmacia) was emplûyed.
G. Dr~ inl~ ûf Peptide Binding Affinities
The binding affinities ûf variûus peptides fûr the mcnn~lnn~l antibûdy
D32.39 were measured in a I ,-"~ inl~ binding PYrPrimpnt A tracer peptide
(I.RRASLGGGRRQFKWT (SEQ ID NO:24); 50 pM) containing the known epitope for
D32.39 fused to a consensus substrate sequence for cAMP-dependent protein kinase was
radiolabeled to high specific activity with [g -33P]ATP (see Li et al., 1989, Prûc. Natl.
Acad. Sci. USA 86: 558-562, iul~_UIAuo ' herein by reference) and mixed with various
cnnrPntr~tinnc of the peptide of interest (10 ~M-I pM). The peptide mixtures were added
to pOlyaLylcllc wells coated with D32.39 ( 0.1 Gmg/mL). Samples were incubated 2 hr. at
4C, the wells washed with PBS, and the radioactivity associated with each well was
counted and used to generate a CulllpCLi~ivc binding eurve. Under the conditions of the
assay the IC50 should be close to the 11;~ l const~nt (K~ for the peptide.
EXAMPLE 2: SYNTHESIS AND STABILITY STUDIES OF THIAZOLIDINONES
The following examples relate to the synthesis of thi~7nlitlinnnPc using the
methods ûf the present invention. This synthesis is described in greater detail in U.S.
Patent Application Nû.081265,090, filed June 23, 1994, and il,cull,uldLcd herein by
reference for all purposes.
~ Wo 95/12608 2 1 7 5 5 8 7 PCr/uss~ll23~7
131
A. P~ dldLiUII of Double-Labeled Thi~7r~ innnr-
H2N-S-~entaGel (500 mg), a cu~ "_;dlly available polystyrene based resin
(Rapp Polymere, Tubingen, Germany, lg, 0.30 mmol/g loading), was elaborated withFmoc-Gly-OH labeled at the r -carbon (2-l3C, 99% from Cambridge Isotope Laboratones,
5 Inc., Andover, MA). The resin was capped with Ac20, d~luL~d with piperidine, and
the Fmoc-rh~t~ nkpr coupled as its OBt-activated ester. The resin was again capped,
J~1uL~d, and reacted with unlabeled Fmoc-Glycine-OH as its anhydride. An additional
round of capping amd J~lu~:lio-l generated the free amine rc-sin. Reaction with 0.75 M
PhCHO labcled at the carbonyl (carbonyl-l3C, 99% from Cambridge Isotope Laboratones,
10 Inc., Andover, MA) and 2.0 M I~ dlJ~Ud. ~ liC acid in ACN containing 3A molecular
sieves for 2 hours at 70C generated the double labeled thi~7nlirlinrmr- resin. The resin
was washed extensively (3xs ml CH2Cl2 3X5 ml DMF, 3X5 ml CH2CI2, 3X5 ml MeOH,
3X5 ml CH2CI2, 3X5 ml Et2O) and dried under vacuum.
15 B. TFA Stability Studies
A portion (20mg) of the resin was treated with 95% TFA/5% H20 For I
hour followed by washing with CH2CI2, MeOH and Et2O. Gel-~3C NMR analysis of theresin indicatcd no loss of thi~l7rl1irlinrmP, as evidenced by relative integration of the two
labeled carbons. Sce Panel B, Fig. 30. Any destruction of either the photolinker or
20 thi37l~ 1inr~ne would be expected to result in the integration of the benzylic carbon to
decrease. This ~ 1 d--- ' that both the ll- -,.,li~l;....nr and the rhrltrllinkr-r
were stable to TFA treatment.
C. DNA Synthesis Stability Studies
A portion (20 mg) of the resin was loaded into the standard DNA synthesis
cartridge and subjected to 40 cycles of DNA synthesis with A, C and T n~mlr~ irlP~
employed as their rl~c,~ followed by iodine oxidation after every cycle.
"Mock" dimethoxytrityl (DMT) removal was ~(.,,,.l.l;~h-`il by treating the resin 2%
- TFA/CH2CI2 at tbe start of every cycle. The resin was removed from the cartridge,
washed with DMF, and analyzed by gel-l3C NMR *~L u~,u~,y. See Panel A, Fig. 30.
The spectrum obtained revealed little or no destruction of either the photolinker or
II,;,.,.,li,l;"~",e molecules. A portion (2 mg) of the resin was also photolyzed for 3 hours in
pH 7.4 PBS buffer and the liberated thi~7nlirlinr~nP analyzed by HPLC. See Panel A, Fig.
WO 95/12608 ~ 7 7 5 5 8 7 PCTIUS9~1113~7
132
31 and Fig. 32. The data revea'ed that the thi~7~ inrm~ was released in high punty and
that both the photolinker and thi~7,~ 1inone were not significantly altered upon treatment
with standard DNA synthesis reagents.
5 EXAMPLE 3: COME,INATORIAL SYNTHESIS
A ~.,,.,1,;.. ~,.;,-1 synthesis of YGGFL was performed using the synthesis
device. The synthesis was done in reaction vessels 1-6; 7-9 were ~ 1 The six
amino acids added to the beads were L, E, G, Y, A, and F. YGGFL was specified along
with other peptides. Beads were added to the parent vessel (29.5 mg) and suspended in
10 DMF~ Each synthesis cyc~e included steps of rerli~t7ih~ ,n, peptide coupling, capping,
amine d~lu~r~LiO-I, collection of dr~lut~Liull for FMOC, rinsing with DMF, and
c~ . in the parent.
Following synthesis, the labeled Hert7 antibody was introduced to the mixed
beads. The Hertz antibody binds mostly with YGGFL. FACS ana'lysis identified the15 presence of YGGFL, provJtg that this specified .-,~ 7- was synthesized by thesynthesizer. The ,-Yrr7-im!-nt shows that a diverse collection ot- peptides, including the
YGGFL chain, can be specified and 7yllL~I~,,~..I via the synthesizer.
Although the present invention has been described in some detail by way of
illustration and example for purposes of clarity and ~ , it will be apparent that
certain changes and ."~ may be practiced within the scope of the appended
claims.
EXAMPLE 4: DETERMINATION OF BEAD DISTRIBUTION USING THE
APPARATUS OF THE INVENTlON
To determine if mixed beads from the parent vessel were being evenly
distributed to the reaction vessels, some beads were biuLulyldlrd. The biotinylated beads
were manua',ly deposited in one reaction vessel. Non-'u;oLi"~' J beads were manually
deposited in the other 8 reaction vessels. The synthesi_er transfers all beads from 9
reaction vessels to the parent vessel. A sample was taken from the parent vessel, and
fluoresced streptavidin was allowed to bind with biotin on the l,;o~ hl~d beads.Florescence Activated Cell Sorter (FACS) analysis shows that ~yluHIl~ ly 9. l % of the
beads in the parent vessel were b;uLilly ' The beads were then rea',located to 9 reaction
vessels, and the percentage of biotinylated beads to tota'~ beads in each vessel was
wo gs/l2co8 2 1 7 5 5 ~ 7 PCT/USg~123~7
133
deternlined by a FACS analyzer. Table ~ shows that the mixed beads in each reaction
vessel have d~u~luxi~ .,ly the same ratio of biotinylated beads to total beads as the parent
vessel.
TABLE 5
% Bright Beads
Parent 9.1
9.7
2 9.7
3 9.4
4 8.7
9.4
6 8.9
7 9.1
8 8.9
9 9.3
Average 9.2
Standard dev. 0.32
It is to be understood that the above description is intended to be illustrativeand not restrictive. Many f ,~l~o~;",~ will be apparent to those of skill in the art upon
reviewing the above ~iPcrrir~irm The scope of the invention should, therefore, be
deterrnined not with reference to the above ~if~ rirtion~ but should instead be ~f ~f rnninf~l
with reference to the appended claims, along with the full scope of equivalents to which
such claims are entitled.
WO95112608 2 i 7 55 ~ 7 PCT~S9~/123~7 ~
134
SEQUENCE LISTING
(1) GENERRL INFORMRTION:
(i) APPLIQNT: AFFYMRX ,~, ,rrrr'fi N.V.
(ii) TITLE OF INVENTION: Synthesizing ~nd ScreenLng Molecular Diver5ity
( iii ) NUMBER OF SEQUENCES: 24
( iv ) ~:-JKeu~ ADDRESS:
A PnnDT`CqET': Townsend and Townsend Ehourie and Crew
B STREET: One Market Plaza, Steuart Tower, Suite 2000
C CITY: San Francisco
D STATE: Cali~ornia
E COUNTRY: USA
F ZIP: 94105
( v ) COMPUTER RERDABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC _ ~hl..
(C) OPERRTING SYSTEM: PC--DOS/MS--DOS
(D) SOFTWRRE: PatentIn Relea~e ~1.0, Version #1.25
~vi) CURRENT APPLICRTION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
(C) CLASSIFIQTION:
(vii) PRIOR APPLIQTION DATA:
(A) APPLICATION NUMBER: Us 08/146,886
(B) FILING DATE: 02--NOV-1993
(vii) PRIOR APPLIQTION DATA:
(A) APPLICATION NUMBER: US 08/149,675
(B) FILING DATE: 02--NOV-1993
(viii) ATTORNEY/AGENT INFOPMRTION:
(A) NAME: Norviel, Vernon R.
(B) REGISTRRTION NUMBER: 32,4B3
(C) REFERENCE/DOCICET NUMBER: 16528J--000740PC
(ix) TP'Tr'r`tMMllNrCATION INFORMRTION:
(A) TELEPHONE: 415--326--2400
(B) TELEFAX: 415-326--2422
(2) INFORMATION FOR SEQ ID NO:1:
(i) SEQUENCE -DPDarT~DT~TIcs:
(A) LENGTE~: 13 amino acids
( B ) TYPE: amino acid
(C) STDP~ FI'~'~qS; aingle
(D) TOPOLOGY: linear
(iL) MOLECULE TYPE: peptide
~ WO 95112608 2 t 7 ~ 5 8 7 PCTIUS9~ 3-17
135
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:l:
Tyr Gly Gly Phe Leu Arg Arg Gln Phe Lys Val Val Thr
2 ~ INFOR~5ATION FOR SEQ ID l O: 2:
(i) SEQUENCE rWl~RArT~RT.~TICS
A LENGTH: 7 a_ino acid~
B TYPE: arnino acid
, C STRr'~ n`----C ~ingle
D, TOPOLOGY: linear
( ii ) MOLECULE TYrE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:2:
Arg Gln Phe Lys Val V~l Thr
(2) l~rl mTr-N FOR SEQ ID NO:3:
(i) SEQUENCE rl~o~rT~RTqTICS
(A) LENGT~I: 8 unino acid~
(B) TYPE: amino ~Cid
(C) STR~Nnl;!nM~C ~ingle
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE l/~.~r~luN: SEQ ID NO:3:
Thr Phe A~g Gln Phe Lys Val Thr
(2) INFORMATION FOR SEQ ID NO:4:
( i ) SEQUENCE r~l~o~ lU n
(A) LENGTH: B amino ~cids
( B ) TYPE: ar~ino acld
(C) sTR~Nn~nM~.~c ~ingle
ID~ TOPOLOGY: linear
(ii) MOLECULE TYPE: peptLde
( xi ) SEQUENCE ~ lUli: SEQ ID NO: 4:
Thr Thr Arg Arg Phe Arg Val Thr
2 ~ 755~7
WO 95112608 PCTIUS9~/123~7
136
(2) INFOR~ATION FOR SEQ ID NO:5:
(i~ SEQUENCE ri~R~r~TpRTcTIcs:
(A) LENGTH: 8 amino acid~
(B) TYPE: a:nino acid
(C) s~rP~M~lPnNPCC: single
ID) TOPOLOGY: line~r
( ii ) ~OLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:5:
Thr V~l Arg Gln Phe Lys Thr Thr
(2) INFORMATION FOR SEQ ID NO:6:
(i) SEQUENCE rN~R~rTpRT~TIcs
(A) LENGTH: 8 amino ~Cid5
( B ) TYPE: ~mino acid
(C) .cTR;~npnNpcc: single
(D) TOPOLOGY: linear
(ii) ~IOLECULE TYPE: peptide
(xi) SEQUENOE DESCRIPTION: SEQ ID NO:6:
Gln Val Arg Gln Phe Ly~ Thr Thr
(2) INFOR~LaTION FOR SEQ ID NO:7:
(i) SEQUENCE CE~ARACTERISTICS:
(A) LENGTH: 8 ~nino ~cids
(B) TYPE: amino ~cid
(C) S~R~MI~pnNpcc- single
(D) TOPOLOGY: line21r
(ii) ~OLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:7:
Arg Gln Phe Arg Thr Val Gln Thr
21 75587
WO 95/12608 Pi'T/lJS9.1/123~7
137
i2) INFORMATION FOR SEQ ID NO:8:
(i) SEQUENCE ~ ARArT~T.~iTIcs
(A~ LENGTE~: 8 amino acids
(B) TYPE: amino acid
(C) 5'rRANn~nNl;'~.C: 5inyle
(D1 TOPOLOGY: linear
( ii ) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:8:
Lys Gln Phe Lys Val Thr Lys Thr
(2) INFORMATION FOR SEQ ID NO:9:
( i ) SEQUENCE CEIARACTERISTICS:
(A) LENGTH: 8 amino acidu
(B) TYPE: amino acid
(C) 5~12P~mFnM~CC single
(D) TOPOLOGY: lLnear
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:9:
Gln Gln Phe Lys Val Val Gln Thr
(2) INFORMATION FOR SEQ ID NO:10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGT~: 8 amino acids
( B ) TYPE: amino acid
(C) S~P~ nMI;qc: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:10:
Lys Gln Phe Lys Val Thr Gln Thr
-
2~755
WO95/12608 ~7 PCTIUS9~/123.17
138
(2) INFORMATION FOR SEQ ID NO:ll:
(i~ SEQUENCE CE~ARACTERISTIC5:
. A) LENGTH: B amino acids
B) TYPE: amino acid
C) STRANnEnMTi:cc: Dingle
D ) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID No:ll:
Thr Gln Phe Lys Val Thr Lyn Thr
(2) INFORMATION FOR SEQ ID NO:12:
( i ) SEQUENOE CBARACTERI ST I CS:
(A) LENGTB: 8 amino acids
(3) TYPE: amino acid
(C~ sT~ANn~nN~c~: single
(D) TOPOLOGY: linear
(il1 MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:12:
Thr Phe Arg Val Phe Arg Val Thr
(2) INFORMATION FOR SEQ ID No:13:
( i ) SEQUENCE rTTARAoTERTcTIcs
A) LENGTB: 8 amino acids
, B) TYPE: amino acid
, C) STRANDEDNESS: Gingle
D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:13:
Phe Arg Arg Gln Phe Arg Val Thr
2 1 75~87
WO 9~/12608 PCT/US9 1/123~7
139
(2) INFORMATION FOR SEQ ID NO:14:
(i) SEQUENCE rTlARPr~RTRTICS:
(A) LENGTH: 8 amino acl~
(B) TYPE: amino acid
(C) STRpMn~nMF~-R: ~ingle
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
r
(xi) SEQUENCE DESCRIPT~ON: SEQ ID NO:14:
Arg Gln Phe Lys Gln Val Gln Thr
(2) INFORI~ATION FOR SEQ ID NO:15:
(i) SEQUENCE ~ pRAr~ERT~TIcs:
(A) LENGTH: 8 amino acids
(B) TYPE: amino acid
(C) STRrMnT~nN~Rs: single
(D) TOPOLOGY: linear
(ii1 MOLECULE TYPE: peptide
(xi) SEQUENCE LI~CRl~.lu~: SEQ ID NO:15:
Gln Thr Val Thr Val Lys Ly~ Thr
( 2 ~ INFORMATION FOR SEQ ID NO: 16:
~i~ SEQUENCE ~'FlPRA-''rli:RTS~ICS:
~A~ LENGTH: 8 amino acid~
( B ~ TYPE: amino acid
(C~ s~RrMnRnNrqR: ginyle
(D~ TOPOLOGY: linear
(ii) MOLECULE TYPE: peptide
~xi) SEQUENCE DESCRIPTION: SEQ ID NO:16:
Gln Gln Val Gln Arg Gln Thr Thr
WO 9~/12608 ~ I 7 5 5 8 7 pCT~S9~1123~7 ~
l~o
(2) INFORXATION FOR SEQ ID NO:17:
(i) SEQUENCE oll~RprTFR~cTIcs
A LENGTH: 8 nmino ~cid~
B TYPE: amino ~cid
C I RTPP`~lFnNF~q: single
~ D TOPOLOGY: linear
(ii) MOLECIJLE TYPE: peptide
(xi) SEQUENCE L~:nlr~14N: SEQ ID NO:17:
Ly~ Thr Gln Val Val Gln Phe Thr
~2) INFOR~TION FOR SEQ ID NO:18:
(i) SEQUENCE r'l"P'rTFR~.CTICS-
. A) LENGTH: B ~ino acids
B) TYPE: ~mino ~cid
.C) STRr --lFn~TE~C: ~ingle
, D ) TOPOLOGY: linear
(ii) MOLECIJLE TYPE: peptide
(xi) SEQUENCE ~5~.nl~,l4N: SEQ ID NO:18:
Gln Val Thr Gln Val Ary Val Thr
(2) INFORMPTION FOR SEQ ID NO:19:
( i) SEQUENCE r~PPrTFRTCTICS
(A) LENGTH: 8 amino ~cid~
(B) TYPE: amino acid
(C) STRP~)FnrlFCC: ~ingle
(D) TOPOLQGY: linear
(ii) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:lg:
Phe Vnl V~l Thr Val Arg Val Thr
2175
WO 95112608 5 8 7 PCT/US9~/123~7
141
(2) INFOR~5ATION FOR SEQ ID No:20:
(i) SEQUENCE rTT~rT~RTgTICS
(A) LENGTH: 69 base pairs
(B) TYPE: nuclelc acid
(C) sTI:r~Mr~pnNrqs: L;ingle
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (olignn11~1~nfi,i )
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:20:
ATCCAATCTC TCQCATCTC TATACTATQ TCACCTATCC TATTTTTACC TCACTCACTT 60
CCATTCCAC 69
(2) INFORMATION FOR SEQ ID NO:21:
( i ) SEQUENCE CEaRACTERISTICS:
(A) LENGTH: lS baue pairs
(B) TYPE: nucleic acid
(C) STR~NI~nNT~c5: single
(D) TOPOLOGY: linear
( ii) MOLECULE TYPE: DNA (ol i ~nnllrl ~cf i ~ic.)
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:21:
ATCCAATCTC TCCAC lS
(2) INFORMATION FOR SEQ ID NO:22:
( i ) SEQUENCE r~l~T~rTT'RT.qTICS:
(A) LENGTP.: 15 ba~e paini
(B) TYPE: nucleLc acid
(C) STR~nFl~T'eS: f~ingle
( D ) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (~ qnn ~ ~ntide)
(xi) SEQUENCE DESCRIPTION: SEQ ID No:22:
GTGGAATGGA AGTGA l S
(2) INFORMATION FOR SEQ ID NO:23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: lS baie pairi
(B) TYPE: nucleic acid
(C) STT~ ~MnT~n~lT~cc: iingle
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: DNA (primer)
(xi) SEQUENCE DESCRIPTION: SEQ ID No:23:
ATCTCTATAC TATQ lS
WO 95/12608 2 ~ 7 5 5 8 7 PCT~ ~S9~1123~7 ~
142
(2) INFoRMaTIoN FOR SEQ ID NO:24:
(i~ SEQUENCE CHARACTERISTICS:
(A) LENGTd: 17 amino ~cLds
(B) TYPE: /ullino ~cld
(C) STRaNDEDNESS: single
(D) TOPOLOG~: linear
( ii ) MOLECULE TYPE: peptide
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:24:
Leu Ary Arg Ala Ser Leu Gly Gly Gly Arg Arg Gln Phe Lys Val Val
5 10 15
Thr