Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02231330 1998-03-06
9~ PC~T/US96/14567
U~S 19 SEP 19~7
TITLE OF THE INVENTION
A HIGH THROUGHPUT ASSAY USING FUSION PROTEINS
BACK;GROUND OF THE INVENTION
Src homology 2 (SH2) domains are a family of homologous
protein domains that share the common property of recogni~in,~r
phosphorylated tyrosine residues in specific peptide contexts. They
have routinely been expressed in E. coli as fusion proteins with
glutathione-S-transferase (GST). This usually provides high level
expression and straightforward affinity purification on glutathione-
Sepharose. Ligand binding is then assayed by incubating the GST/SH2
with a radiolabeled phosphopeptide, precipitating the complex with
glutathione-Sepharose, washing the beads, and then counting the beads
to determine bound radioactivity [Isakov et al., J. Exp. Med., 181, 375-
380 (1995); Piccione et al., Biochemistry, 32, 3197-3202 (1993); Huyer
et al., Biochemistry, 34, 1040-1049 (1995)]. There are several
disadvantages to this procedure, particularly when applied to high-
throughput screening for agonists, antagonists, or inhibitors as new
leads for drug development. First, the radiolabeling of the peptide is
carried out either enzymatically with a kinase and [32P]ATP or
chemically with [125I]Bolton-Hunter reagent. In both cases, the
-- isotop~es are short-lived and thus require frequent preparation of
material. In the case of enzymatic phosphorylation, the appropriate
kinase must also be available in sufficient quantity to generate enough
material for screening purposes. Second, the protocol requires
separiation of bound complex from free phosphopeptide by washing of
the glutathione-Sepharose beads This is a nonequilibrium procedure
that riisks dissociation of the bound ligand, particularly when off-rates
are fast. Thus, there is the possibility of misleading results. Finally,
due to the number of manipulations and centrifugations involved, the
protocol is very tedious to conduct manually and is not readily adaptable
to robotic automation to increase throughput.
Two additional methods for measuring the interaction of
proteins and ligands that have been applied to SH2 domains are
biospecific interaction analysis using surface plasmon resonance and
AIUEI'~DED ~~ET
CA 02231330 1998-03-06
s ~ nr~1l)S 9 6 ~1 4 5 6 7
~US 19 SEP 1997
isothermal titration calorimetry (Felder et al., Mol. Cell. Biol., 13,
1449-1455 (1993); Panayotou et al., Mol. Cell. Biol., 13, 3567-3576
(1993); Payne et al., Proc. Natl. Acad. Sci. U.S.A., 90, 4902-4906
(1993); Morelock et al., J. Med. Chem. 38, 1309-18 (1995); Ladbury et
5 al., Proc. Natl. Acad. Sci. U.S.A., 92, 3199-3203 (1995); Lemmon et
al., Biochemist~y, 33, 5070-5076 (1994)). These techniques do not
require a particular fusion partner for the SH2 domain, but do require
sophisticated instrumentation that is not amenable to high throughput
screening.
--' 10
SUMMARY OF THE INVENTION
The instant invention covers a method of screening for
compounds capable of binding to a fusion protein which comprises
combiining a test compound, a tagged ligand, a fusion protein (target
15 protein, peptide linker and FK506-binding protein), and a radiolabeled
ligand~ in a coated microscintillation plate, and then measuring the
scinti]lation counts attributable to the binding of the tagged ligand to the
fusion protein in the presence of the test compound relative to a contro
assay in the absence of the test compound, so as to determine the effect
20 the test compound has on the binding of the tagged ligand. Also within
-- the scope of this invention are the processes for preparing and
expressing the recombinant DNA encoding a fusion protein. This
invention further relates to the recombinant DNA expression vector
capable of expressing the fusion protein. This invention further relates
25 to a process for purifying the recombinant fusion protein. This
invention provides an immediate means of making use of
micrc,scintillation plate technology for the functional assay of ligand
binding to a single or multiple signal transduction domain(s), for
example a phosphopeptide binding to an SH2 domain. The present
30 invention does not require specialized radiochemical synthesis and is
readi]y adaptable to robotic automation for high capacity screening for
agonists, antagonists, and/or inhibitors.
~MENDED StlEET
CA 02231330 1998-03-06 ~ r ~ ,
19.j~ IP~S 1 9 SEP 1997
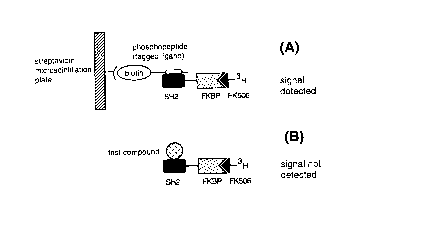
BRIEl~ DESCRIPTION OF THE FIGURES
Figure 1.
A.) Binding of the streptavidin microscintillation plate, biotinylated
ligand and the fusion protein (SH2:FKBP), which emits a detectable
signal; and
B.) Binding of the test compound and the fusion protein (SH2:FKBP),
which results in no signal detection .
DETAILED DESCRIPTION OF THE INVENTION
-~ 10 The present invention relates to a method of screening for~~ compounds which preferentially bind to a target protein.
An embodiment of this invention is a method of screening
for compounds capable of binding to a fusion protein which comprises
the steps of:
a) mixing a test compound, a tagged ligand, the fusion protein,
and a radiolabeled ligand;
b) ~lflin~ the mixture to a coated microscintillation plate;
c) incubating the mixture for between about 1 hour and about 24
hours;
d) measuring the plate-bound counts attributable to the binding
of the tagged ligand to the fusion protein in the presence of
the test compound using scintill~tion counting; and
e) determining the binding of the tagged-ligand to the fusion
protein in the presence of the test compound relative to a
control assay run in the absence of the test compound.
A second embodiment of this invention is a process for
preparing a recombinant DNA expression vector encoding for a fusion
protein comprising the steps of:
a) removing the stop codon on DNA encoding for an FK506-
binding protein;
b) synthesizing a modified DNA fragment on the DNA encoding
for the FK506-binding protein which encodes for a peptide
linker;
,r!~,3 CL~LI-~
CA 02231330 1998-03-06
~S 19 SÉP 1~97
c) digesting an expression vector at cloning sites;
d) cloning the modified DNA fragment encoding for the FK506-
binding protein with a peptide linker into the digested
expression vector to generate a recombinant DNA
expression vector encoding for FK506-binding protein with
a peptide linker; and
e) cloning DNA encoding for a target protein into a recombinant
DNA expression vector encoding for FK506-binding
protein with a peptide linker to produce the recombinant
--- 10 DNA expression vector encoding for the fusion protein.
A third embodiment of this invention is a process for
expressing recombinant DNA encoding for a fusion protein in an
expression vector comprising the steps of:
a) transforming a host cell with the fusion protein expression
vector;
b) inducing expression of the fusion protein in the host cell;
c) recovering the fusion protein from the host cell; and
d) purifying the fusion protein.
A fourth embodiment of this invention is a process for
purifying an isolated FBP-SH2 fusion protein, comprising the steps of:
a) preparing an affinity matrix consisting of biotinylated
phosphopeptide coupled to avidin or streptavidin
immobilized on a solid support;
b) preparing a freeze/thaw extract from cells expressing the
fusion protein;
c) loading the extract onto the affinity matrix and washing off
unbound protein; and
d) eluting the desired fusion protein with phenyl phosphate.
The terrn "fusion protein" refers to a "target protein" fused
to an "FK506-binding protein" (FKBP), the two proteins being
separated by a "peptide linker".
CA 02231330 1998-03-06
9~ P C T/U S 9 6 / 1 4 5 6 7
~P~JS 19 SEP 19g7
A "peptide linker" may consist of a sequence cont~ininp
from about 1 to about 20 arnino acids, which may or may not include
the sequence for a protease cleavage site. An example of a peptide
linker which is a protease cleavage site is represented by the amino acid
5 sequence GLVPRGS (SEQ.ID.NO. 7).
The terrn "target protein" refers to any protein that has a
defineld ligand. Included within this definition of target protein are
single and multiple signal transduction domains, such as, but not lirnited
to, Src homology 1 (SH1), Src homology 2 (SH2), Src homology 3
- 10 (SH3). and pleckstrin homology (PH) domains [Hanks & Hunter,
FASEirs J., 9, 576-596 (1995); Bolen, Curr. Opin. lmmunol., 7, 306-
311- (1995); Kuriyan & Cowburn, Curr. Opin. Struct. Biol., 3, 828-837
(1993)l; Cohen et al., Cell, 80, 237-248 (1995)]. The term "SH1
domain" refers to a farnily of homologous protein domains that bind
15 ATP and catalyze tyrosine phosphorylation of peptide and protein
substrates. The term "SH2 domain" refers to a farnily of homologous
protein domains that share the cornrnon property of recognizing
phosp]horylated tyrosine residues in specific peptide contexts. The term
"SH3 domain" refers to a family of homologous protein domains that
20 share the cornrnon property of recognizing polyproline type II helices.
The term "PH domain" refers to a family of homologous protein
domains that mediate both protein-protein and protein-lipid interactions.
Exam~ples of SH2 domains which may be utilized in the method of the
invention include, but are not limited to, the single and tandem SH2
25 domains present in the tyrosine kinases ZAP, SYK and LCK. The DNA
seque]nces were obtained from GenBank, National Center for
Biotechnology Inforrnation, National Library of Medicine, 8600
Rockville Pike, Bethesda, MD 20894. The Accession Numbers for the
sequences are: human ZAP (L05148); human SYK (L28824) and human
30 LCK (X13529). The sequences for ZAP, SYK and LCK are disclosed
in the sequence listing as follows: the isolated DNA encoding for a
fusion protein containing ZAP is (SEQ.ID.NO.1); the isolated DNA
encod~ing for a fusion protein containing SYK is (SEQ.ID.NO.2); the
isolated DNA encoding for a fusion protein cont~ining LCK is
~ ~ .r
CA 02231330 1998-03-06 PC
- 6 -
(SEQ.ID.NO.3); the sequence for the FKBP-ZAP:SH2 fusion protein is
(SEQ.ID.NO.4); the sequence for the FKBP-SYK:SH2 fusion protein is
(SEQ.ID.NO.5); and the sequence for the FKBP-LCK:SH2 fusion
protein is (SEQ.ID.NO.6).
The terrn "tagged ligand" refers to a biotinylated or epitope
tagged ligand for the target protein.
The term "radiolabeled ligand" refers to a [3H]-, [125I]-,
[14C]-, [35S]-, [32p], or [33P]-labeled ligand which binds to the FKBP.
An example of a radiolabeled ligand useful in the instant invention is
- 10 [3H]-dihydroFK506.
The terrn "coated microscintillation plates" refers to
strept;avidin-coated microscintillation plates when the tagged ligand is
biotinylated, and to anti-epitope antibody bound to anti-antibody-coated
or protein A-coated microscintillation plates when the tagged ligand is
epitope-tagged. Examples of coated microscintillation plates useful in
the instant invention are streptavidin-coated, sheep anti-rabbit-coated,
and goat anti-mouse-coated FlashPlate Plus (DuPont-NEN). Additional
coatings, including but not limited to protein A, may be applied to
uncoated FlashPlates by methods known to those skilled in the art.
The term "control assay" refers to the assay when
performed in the presence of the tagged ligand, the fusion protein, the
radio]Labeled ligand and the coated microscintill~tion plates, but in the
absence of the test compound.
The term FK506-binding proteins may include, but are not
25 limited to, the below listed FKBPs and FKBP homologues, which
include a citation to the references which disclose them. This list is not
intenlded to limit the scope of the invention.
l~n-m~li?n
FKB~P-12 Galat et al., Eur. J. Biochem., 216:689-
707 (1993)-
FKBP-12.6 Wiederrecht, G. and F. Etzkorn
Perspectives in Drug Discovery and
Design, 2:57-84 (1994).
AMEND~D SHE~
CA 02231330 1998-03-06
S 96/14567
S' 1 ~ S~~ 199~/
FKBP-13 Galat et al., supra; Wiederrecht and
Etzkorn, supra.
FKBP-25 Galat et al., supra; Wiederrecht and
Etzkorn, supra.
FKBP-38 Wiederrecht and Etzkorn, supra.
FKBP-51 B~-lghm~n et al., Mol. Cell. Biol., 8,
4395-4402(1995) .
FKBP-52 Galat et al., supra.
Bacte ria
Legionella pneumophilia Galat et al., supra.
Legionella micadei Galat et al., supra
Chlamydia trachomatis Galat et al., supra.
E. coli fkpa Horne, S.M. and K.D. Young, Arch.
Microbiol., 163:357-365 (1995).
E. coli slyD Roof et al., J. Biol. Chem. 269:2902-
2910 (1994).
E. coli orfl49 Trandinh et al., FASEB J. 6:3410-3420
(1992).
Neisseria meningitidis Hacker, J. and G. Fischer, Mol. Micro.,
10:445-456 (1993).
Streptomyces chrysomallus Hacker and Fischer, supra.
Fun~al
yeast FKBP-12 Cardenas et al., Perspectives in Drug
Discovery and Design, 2:103-126
(1994).
yeast FKBP-13 Cardenas et al., supra.
yeast NPRl(FPR3) Cardenas et al., supra.
Neurospora Galat et al., supra.
A variety of host cells may be used in this invention,
which include, but are not limited to, bacteria, yeast, bluegreen
algae, plant cells, insect cells and ~nim~l cells.
r ~ t~
CA 02231330 1998-03-06
~95'~ ' ~'J f~ f 7 7 r ~ 7
IPEA/US 1.~ ~FP 1997
E7cpression vectors are defined herein as DNA sequences
that are required for the transcription of cloned copies of genes and
the translation of their mRNAs in an appropriate host. Such vectors
can be used to express genes in a variety of host cells, such as,
bacteria, yeast, bluegreen algae, plant cells, insect cells and ~nimAl
cells.
Specifically designed vectors allow the shuttling of DNA
between hosts such as bacteria-yeast or bacteria-~nim~l cells. An
appropriately constructed expression vector may contain: an origin of
' 10 replication for autonomous replication in host cells, selectable markers,
a limited number of useful restriction enzyme sites, a potential for high
copy number, and active promoters. A promoter is defined as a DNA
sequence that directs RNA polymerase to bind to DNA and initiate RNA
synthesis. A strong promoter is one which causes mRNAs to be
initiated at high frequency Expression vectors may include, but are not
limited to, cloning vectors, modified cloning vectors, specifically
designed plasmids or viruses. Commercially available vectors suitable
for F~BP fusion protein expression include, but are not limited to
pBR322 (Promega), pGEX (Amersham), pl7 (USB), pET (Novagen),
pIBI (IBI), pProEX-1 (Gibco/BRL), pBluescript iI (Stratagene),
pTZl~R and pTZ19R (USB), pSE420 (Invitrogen), pVL1392
(Invitrogen), pBlueBac (Invitrogen), pBAcPAK (Clontech), pHIL
(Invitrogen), pYES2 (Invitrogen), pCDNA (Invitrogen), pREP
(Invitrogen) or the like.
The expression vector may be introduced into host cells via
any one of a number of techinques including but not limited to
transformation, transfection, infection, protoplast fusion, and
electroporation.
E. coli cont~inin~ an expression plasmid with the target
gene fused to FKBP are grown and appropriately induced. The cells
are then pelleted and resuspended in a suitable buffer. Although FKBP-
12 lacks sequences that specifically direct it to the periplasm, FKBP
fusions are primarily located there and can be released by a standard
freeze/thaw treatment of the cell pellet. Following centrifugation, the
CA 02231330 1998-03-06
95'~ fj 7
J
resulting supernatant contains >80% pure FKBP fusion, which if
desired can be purified further by conventional methods. Alterna~ively,
the assay is not dependent on pure protein and the initial periplasmic
preparation may be used directly. A thrombin site located between
S FKBF' and the target protein can be used as a means to cleave FKBP
from the fusion; such cleaved material may be a suitable negative
control for subsequent assays.
A fusion protein which contains a single or multiple SH2
domain(s) may be purified by preparing an affinity matrix consisting of
10 biotinylated phosphopeptide coupled to avidin or streptavidin
immobilized on a solid support. A freeze/thaw extract is prepared from
the cells which express the fusion protein and is loaded onto the affinity
matrix. The desired fusion protein is then specifically eluted with
phenyl phosphate.
To assay the formation of a complex between a target
protein and its ligand, the tagged ligand is mixed with the FKBP fusion
protein in a suitable buffer in the presence of the radiolabeled ligand.
After a suitable incubation period to allow complex formation to occur,
the mixture is transferred to a coated microscintillation plate to capture
20 the tagged ligand and any bound fusion protein. The plate is sealed,
incubated for a sufficient period to allow the capture to go to
completion, then counted in a multiwell scintillation counter. Screening
for agonists/antagonists/inhibitors is carried out by performing the
initial incubation prior to the capture step in the microscintillation plate
25 in the presence of a test compound(s) to determine whether they have an
effect upon the binding of the tagged ligand to the fusion protein. This
principle is illustrated in Figure 1.
The present invention can be understood further by the
following examples, which do not constitute a limitation of the
30 invention.
CA 02231330 1998-03-06
195~ ~ J
~ 9 9 7
- 10-
EXAMPLE 1
Process for Preparing the FKBP fusion clonin~ vector
General techniques for modifying and expressing genes in
5 various host cells can be found in Ausubel, F.M., Brent, R., Kingston,
R.E., Moore, D.D., Seidman, J.G., Smith, J.A. and Struhl, K. Current
Protocols in Molecular Biology (John Wiley & Sons, New York, New
York, 1989). Sequences for a 3'- altered FKBP fragment that contained
a glycine codon (GGT) in place of the stop (TGA) codon followed by a
-~- 10 sequence encoding a thrombin site (Leu-Val-Pro-Arg) and BamHI
restricltion site (GAAl-rC) were amplified using the polymerase chain
reaetion (PCR). The PCR reaction contained the following primers:5'-
GATCGCCATGGGAGTGCAGGTGGAAACCATCTCCCCA-3'
(SEQ..[D.NO.8); and 5'-TACGAATTCTGGCGTGGATCCACGC
15 GGAACCAGACCTTCCAGTTTTAG-3' (SEQ.ID.NO.9); and a plasmid
containing human FKBP-12 as the template. The resulting 367 base
pair amplification product was ligated into the vector pCRII
(Invitrogen) and the ligation mixture transformed into competent
Esche~richia coli cells. Clones cont~inin~ an insert were identified using
20 PCR with fl~nking vector primers. Dideoxy DNA sequencing
confirlmed the nucleotide sequence of one positive isolate. The altered
338 base pair FKBP fragment was excised from the pCRII plasmid
using NcoI and BamHI and ligated into NcoI andBamHI digested pET9d
(Novagen) plasmid. Competent E. coli were transformed with the
25 ligation mixture, and colonies containing the insert were identified using
PCR with primers encoding for fl~nking vector sequences. The FKBP
fusion cloning vector is called pET9dFKBPt.
EXAMPLE 2
Process for Preparin~ the FK-ZAP fusion expression vector
A DNA fragment encoding for the tandem SH2 domains of
ZAP-70 was prepared by PCR to contain a BamHI site at the 5'-end
such l:hat the reading frame was conserved with that of FKBP in the
35 fusion vector. At the 3'-end, the fragment also incorporated a stop
A~ENDE~ SHEEt
CA 02231330 1998-03-06
9 6 l 1 4 5 6 7
SI~ P 1997
codon followed by a BamHI site. The PCR reaction contained Molt-4
cDNA (Clontech) and the following primers:
S '-ATTAGGATCCATGCCAGATCCTGCAGCTCACCTGCCCT-3'
(SEQ.ID.NO.10) and 5'-ATATGGATCCTTACCAGAGGCGTTGCT-3'
(SEQ.ID.NO.11). The fragment was cloned into a suitable vector,
sequenced, digested with BamHI, and the insert cont~ininp the SH2
domains ligated to BamHI treated pET9dFKBPt, and transformed into
E. coli. Clones containing inserts in the correct orientation were
identi~led by PCR or restriction enzyme analysis. Plasmid DNA was
~~~10 prepared and used to transform BL21(DE3) cells.
..
EXAMPLE 3
Process for Preparin~ the FK-SYK fusion expression vector
The expression vector for the tandem SH2 domains of SYK
fused to FKBP was prepared as in Exarnple 2 except that the PCR
reacti~n contained Raji cell cDNA (Clontech) and the following
primers: 5'-CAATAGGATCCATGGCCAGCAGCGGCATGGCTGA-3'
(SEQ,ID.NO.12) and 5'-GACCTAGGATCCCTAATTAACATTTCC
CTGTGTGCCGAT-3' (SEQ.ID.NO.13).
EXAMPLE 4
Process for Preparin~ the FK-LCK fusion expression vector
The expression vector for the SH2 domain of LCK fused to
FKBP was prepared as in Example 2 except that the PCR reaction
contained Molt-4 cDNA (Clontech) and the following primers:
5'-ATATGGATCCATGGCGAACAGCCTGGAGCCCGAACCCT-3'
(SEQ.ID.NO.14) and
5'-ATTAGGATCCTTAGGTCTGGCAGGGGCGGCTCAACCGTG
TGCA-3' (SEQ.ID.NO.15).
CA 02231330 1998-03-06
PCT/US 9h/14567
lPEAlUS 19 SEP 1997
EXAMPLE 5
FK-ZAP
S Step A: Process for Expression of FK-ZAP
E. coli BL21(DE3) cells cont~ining the pET9dFKBPt/ ZAP
SH2 plasmid were grown in Luria-Bertani (LB) media containing 50
microgram/ml k~n~mycin at about 37 degrees C until the optical density
measured at 600 nm was about 0.5-1Ø Expression of the FK-ZAP
-- 10 fusion protein was induced with 0.1 mM isopropyl beta-
- thiogalactopyranoside and the cells were grown for another 3-5 hr at
about 30 degrees C. They were pelleted at 4400 x g for about 10 min at
about 4 degrees C and resuspended in 2% of the original culture volume
with 100 mM tris pH 8.0 containing 1 microgram/ml each aprotinin,
15 pepstatin, leupeptin, and bestatin. The resuspended pellet was frozen at
about -20 degrees C until further purification.
Step B: Process for Purification of FK-ZAP
The affinity matrix for purification of FK-ZAP was
20 prepared by combining agarose-immobilized avidin with excess
biotinylated phosphopeptide derived from the ~1 ITAM sequence of the
human T-cell receptor, biotinyl-GSNQLpYNELNLGRREEpYDVLDK,
(SEQ.ID.NO.16) and washing out unbound peptide. Frozen cells
cont~ining FK-ZAP were thawed in warm water, refrozen on dry ice
25 for about 25 min., then thawed again. After the addition of 0.1% octyl
glucoside, 1 mM dithiothreitol (DTT) and 500 mM NaCl, the extract
was centrifuged at 35,000 x g for approximately 30 minutes. The
supernatant was loaded onto the phosphopeptide affinity column, at
about 4~ and washed with phosphate buffered saline containing 1 mM
30 DTT and 0.1% octyl glucoside. FK-ZAP was eluted with 200 mM
phenyl phosphate in the same buffer at about 37~. The protein pool was
concentrated and the phenyl phosphate removed on a desalting column.
The purified FK-ZAP was stored at about -30~ in 10 mM HEPES/150
mM NaCl/1 mM DTT/0.1 mM EDTA/10% glycerol.
CA 02231330 1998-03-06
195~ 7 ~ _~
- ' ~ ?~
EXAMPLE 6
FK-SYK
E. coli BL21(DE3) cells containing the pET9dFKBPt/
5 SYKSH2 plasmid were grown, induced, and harvested as described in
Example 5. FK-SYK was purified using the same affinity matrix and
methodology described in Example 5.
EXAMPLE 7
~'. 10
- FK-LCK
E. coli BL21(DE3) cells containing the pET9dFKBPt/
LCKSH2 plasmid were grown, induced, and harvested as described in
Example 5. The affinity matrix for puri~lcation of FK-LCK was
prepared by combining agarose-immobilized avidin with excess
biotinyl- EPQpYEEIPIYL (SEQ.ID.NO.17) and washing out unbound
peptidLe. The rem~ining methodology for puri~lcation was the same as
Example 5.
EXAMPLE 8
Assay of phosphopeptide binding to FK-ZAP
Assays were conducted at ambient temperature in a buffer
consisting of 25 mM HEPES, 10 mM Dl-r, 0.01% TWEEN-20, pH 7Ø
300 Ill of a mixture of buffer and varying amounts of biotinyl-
phosphopeptide were combined with 25 ,ul of FK-ZAP protein and 50
,ul of [3H]-dihydroFK506 (DuPont NEN) in microfuge tubes. A 150 111
portion of each assay was then transferred to the well of a streptavidin-
coated FlashPlate Plus (DuPont-NEN) and an additional 50 111 of buffer
was added. Final concentrations of the assay components were:
0-50 nM biotinyl-GSNQLpYNELNLGRREEpYDVLDK
(SEQ.ID.NO. 16)
100 nM FK-ZAP fusion protein
25 nM [3H]-dihydroFK506
. . ~,r - '
CA 02231330 1998-03-06
' ~ ,7
195'~
~ t ~ 9q7~
- 14 -
The plate was sealed and incubated 20 hours. Plate-bound radioactivity
was measured at various timepoints in a Packard Topcount microplate
scintillation counter.
5EXAMPLE 9
Method of Screening for Antagonists of FK-ZAP
Assays are conducted at ambient temperature in a buffer
consisting of 25 mM HEPES, 10 mM DTT, 0.01% TWEEN-20, pH 7Ø
-~- 10 10 ~11 of a DMSO solution of test compound(s) and 120 111 of biotinyl-
phosphopeptide stock solution are dispensed into the wells of a standard
96-well plate. Next, 20 ~11 of a mixture of FK-ZAP protein and
[3H]-dihydroFK506 (DuPont NEN) are added to each test well. The
assays are then transferred to the wells of a streptavidin-coated
FlashPlate (DuPont NEN). Final concentrations of the assay
components are:
25 nM biotinyl-GSNQLpYNELNLGRREEpYDVLDK
(SEQ.ID.NO. 16)
25 nM FK-ZAP fusion protein
10 nM [3H]-dihydroFK506
5% DMSO
The plate is sealed and incubated between 1 and 8 hours. Bead-bound
radioactivity is then measured in a Packard Topcount microplate
scintill~ion counter.
EXAMPLE 10
Method of Screenin~ for Anta~onists of FK-SYK
The assays are conducted as set forth in Example 9, except
that FK-SYK replaces FK-ZAP.
~ O S~
CA 02231330 1998-03-06
19'i~ 9 ~ / 1 4 5 6 7
1 9 SirP 1997
EXAMPLE 1 1
Method of Screening for Antagonists of FK-LCK
The assays are conducted as set forth in Example 9, except
S that FK-LCK replaces FK-ZAP and the tagged ligand is 25 nM
biotinyl-EPQpYEEIPIYL (SEQ.ID.NO.17).
CA 0223l330 l998-03-06
l~524 PCT/US 9 6 / 14 5 6 7
-16- IPE~/US 19 SEP 1997
SEQUENCE LISTING
(1) GENERAL INFORMATION
(i) APPLICANT: MERCK & CO., INC.
(ii) TITLE OF THE INVENTION: A HIGH THROUGHPUT ASSAY USING
FUSION PROTEINS
(iii) NUMBER OF SEQUENCES: 17
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Merck & Co., Inc.
(B) STREET: P.O. Box 2000, 126 E. Lincoln Ave.
(C) CITY: Rahway
(D) STATE: NJ
(E) COUNTRY: USA
(F) ~IP: 07065-0900
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Diskette
(B) COMPUTER: IBM Compatible
(C) OPERATING SYSTEM: DOS
(D) SOFTWARE: FastSEQ for Windows Version 2.0
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER: PCT/US96/14567
(B) FILING DATE: 11-SEP-1996
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA:
(A) US APPLICATION NUMBER: 60/003.819
FILING DATE: 15 SEPTEMBER 1995
(B) GB APPLICATION NUMBER: 9605210.5
FILING DATE: 12 MARCH 1996
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: Camara, Valerie J
(B) REGISTRATION NUMBER: 35,090
(C) REFERENCE/DOCKET NUMBER: 19524
(ix) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: 908-594-3902
(B) TELEFAX: 908-594-4720
(C) TELEX:
;, .,,, ~,,
CA 0223l330 l998-03-06 ~T~VS 9 k / I 4 5 6 7
19.~'4
~ v v,
- 17-
(2) INFORMATION FOR SEQ ID NO:1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1137 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:1:
ATGGGAGTGC AGGTGGAAAC CATCTCCCCA GGAGATGGAC GCACCTTCCC CAAGCGCGGC 60
CAGACC'TGCG TGGTGCACTA CACCGGGATG CTTGAAGATG GAAAGAAATT TGATTCCTCC 120
CGGGAC'AGAA ACAAGCCCTT TAAGTTTATG CTAGGCAAGC AGGAGGTGAT CCGAGGCTGG 180
GAAGA~GGGG TTGCCCAGAT GAGTGTGGGT CAGAGAGCCA AACTGACTAT ATCTCCAGAT 240
TATGCCTATG GTGCCACTGG GCACCCAGGC ATCATCCCAC CACATGCCAC TCTCGTCTTC 300
GATGTC,GAGC TTCTAAAACT GGAAGGTCTG GTTCCGCGTG GATCCATGCC AGATCCTGCA 360
GCTCACCTGC CCTTCTTCTA CGGCAGCATC TCGCGTGCCG AGGCCGAGGA GCACCTGAAG 420
CTGGCGGGCA TGGCGGACGG GCTCTTCCTG CTGCGCCAGT GCCTGCGCTC GCTGGGCGGC 480
TATGTGCTGT CGCTCGTGCA CGATGTGCGC TTCCACCACT TTCCCATCGA GCGCCAGCTC 540
AACGG('ACCT ACGCCATTGC CGGCGGCAAA GCGCACTGTG GACCGGCAGA GCTCTGCGAG 600
TTCTA('TCGC GCGACCCCGA CGGGCTGCCC TGCAACCTGC GCAAGCCGTG CAACCGGCCG 660
TCGGG('CTCG AGCCGCAGCC GGGGGTCTTC GACTGCCTGC GAGACGCCAT GGTGCGTGAC 720
TACGT(,CGCC AGACGTGGAA GCTGGAGGGC GAGGCCCTGG AGCAGGCCAT CATCAGCCAG 780
GCCCC(,CAGG TGGAGAAGCT CATTGCTACG ACGGCCCACG AGCGGATGCC CTGGTACCAC 840
AGCAGCCTGA CGCGTGAGGA GGCCGAGCGT AAACTTTACT CTGGGGCGCA GACCGACGGC 900
AAGTTCCTGC TGAGGCCGCG GAAGGAGCAG GGCACATACG CCCTGTCCCT CATCTATGGG 960
AAGACGGTGT ACCACTACCT CATCAGCCAA GACAAGGCGG GCAAGTACTG CATTCCCGAG 1020
GGCACCAAGT TTGACACGCT CTGGCAGCTG GTGGAGTATC TGAAGCTGAA GGCGGACGGG 1080
CTCATCTACT GCCTGAAGGA GGCCTGCCCC AACAGCAGTG CCAGCAACGC CTCTTAA 1137
(2) INFORMATION FOR SEQ ID NO:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1155 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
CA 0223l330 l998-03-06
PCT/US 9 6 / 14 5 6 7
-18- ~ EP I~
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:2:
ATGG(,AGTGC AGGTGGAAAC CATCTCCCCA GGAGATGGAC GCACCTTCCC CAAGCGCGGC 60
CAGACCTGCG TGGTGCACTA CACCGGGATG CTTGAAGATG GAAAGAAATT TGATTCCTCC 120
CGGGACAGAA ACAAGCCCTT TAAGTTTATG CTAGGCAAGC AGGAGGTGAT CCGAGGCTGG 180
GAAGAAGGGG TTGCCCAGAT GAGTGTGGGT CAGAGAGCCA AACTGACTAT ATCTCCAGAT 240
TATGCCTATG GTGCCACTGG GCACCCAGGC ATCATCCCAC CACATGCCAC TCTCGTCTTC 300
GATGTGGAGC TTCTAAAACT GGAAGGTCTG GTTCCGCGTG GATCCATGGC CAGCAGCGGC 360
ATGGCTGACA GCGCCAACCA CCTGCCCTTC TTTTTCGGCA ACATCACCCG GGAGGAGGCA 420
GAAGATTACC TGGTCCAGGG GGGCATGAGT GATGGGCTTT ATTTGCTGCG CCAGAGCCGC 480
AACTACCTGG GTGGCTTCGC CCTGTCCGTG GCCCACGGGA GGAAGGCACA CCACTACACC 540
ATCGAGCGGG AGCTGAATGG CACCTACGCC ATCGCCGGTG GCAGGACCCA TGCCAGCCCC 600
GCCGACCTCT GCCACTACCA CTCCCAGGAG TCTGATGGCC TGGTCTGCCT CCTCAAGAAG 660
CCCTTCAACC GGCCCCAAGG GGTGCAGCCC AAGACTGGGC CCTTTGAGGA TTTGAAGGAA 720
AACCTCATCA GGGAATATGT GAAGCAGACA TGGAACCTGC AGGGTCAGGC TCTGGAGCAG 780
GCCATCATCA GTCAGAAGCC TCAGCTGGAG AAGCTGATCG CTACCACAGC CCATGAAAAA 840
ATGC('TTGGT TCCATGGAAA AATCTCTCGG GAAGAATCTG AGCAAATTGT CCTGATAGGA 900
TCAAAGACAA ATGGAAAGTT CCTGATCCGA GCCAGAGACA ACAACGGCTC CTACGCCCTG 960
TGCCTGCTGC ACGAAGGGAA GGTGCTGCAC TATCGCATCG ACAAAGACAA GACAGGGAAG 1020
CTCT('CATCC CCGAGGGAAA GAAGTTCGAC ACGCTCTGGC AGCTAGTCGA GCATTATTCT 1080
TATAAAGCAG ATGGTTTGTT AAGAGTTCTT ACTGTCCCAT GTCAAAAAAT CGGCACACAG 1140
GGAAATGTTA ATTAG 1155
(2) INFORMATION FOR SEQ ID NO:3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 675 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:3:
ATGGGAGTGC AGGTGGAAAC CATCTCCCCA GGAGATGGAC GCACCTTCCC CAAGCGCGGC 60
CAGACCTGCG TGGTGCACTA CACCGGGATG CTTGAAGATG GAAAGAAATT TGATTCCTCC 120
CGGGACAGAA ACAAGCCCTT TAAGTTTATG CTAGGCAAGC AGGAGGTGAT CCGAGGCTGG 180
GAAGAAGGGG TTGCCCAGAT GAGTGTGGGT CAGAGAGCCA AACTGACTAT ATCTCCAGAT 240
TATGCCTATG GTGCCACTGG GCACCCAGGC ATCATCCCAC CACATGCCAC TCTCGTCTTC 300
GATGTGGAGC TTCTAAAACT GGAAGGTCTG GTTCCGCGTG GATCCATGGC GAACAGCCTG 360
GAGCCCGAAC CCTGGTTCTT CAAGAACCTG AGCCGCAAGG ACGCGGAGCG GCAGCTCCTG 420
GCGCCCGGGA ACACTCACGG CTCCTTCCTC ATCCGGGAGA GCGAGAGCAC CGCGGGATCG 480
TTTTCACTGT CGGTCCGGGA CTTCGACCAG AACCAGGGAG AGGTGGTGAA ACATTACAAG 540
ATCCGTAATC TGGACAACGG TGGCTTCTAC ATCTCCCCTC GAATCACTTT TCCCGGCCTG 600
CATGAACTGG TCCGCCATTA CACCAATGCT TCAGATGGGC TGTGCACACG GTTGAGCCGC 660
CCCTGCCAGA CCTAA 675
AMENDED SilEET
CA 02231330 l998-03-06
19.524 ' ~ S~ 7
-19- ~PE~JUS 1 9 SEP 1~97
(2) INFOR~ATION FOR SEQ ID NO:4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 378 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:4:
Met Gly Val Gln Val Glu Thr Ile Ser Pro Gly Asp Gly Arg Thr Phe
1 5 10 15
Pro Lys Arg Gly Gln Thr Cys Val Val His Tyr Thr Gly Met Leu Glu
Asp Gly Lys Lys Phe Asp Ser Ser Arg Asp Arg Asn Lys Pro Phe Lys
Phe Met Leu Gly Lys Gln Glu Val Ile Arg Gly Trp Glu Glu Gly Val
Ala C;ln Met Ser Val Gly Gln Arg Ala Lys Leu Thr Ile Ser Pro Asp
Tyr Ala Tyr Gly Ala Thr Gly His Pro Gly Ile Ile Pro Pro His Ala
Thr I.eu Val Phe Asp Val Glu Leu Leu Lys Leu Glu Gly Leu Val Pro
100 105 110
Arg C,ly Ser Met Pro Asp Pro Ala Ala His Leu Pro Phe Phe Tyr Gly
115 120 125
Ser Ile Ser Arg Ala Glu Ala Glu Glu His Leu Lys Leu Ala Gly Met
130 135 140
Ala Asp Gly Leu Phe Leu Leu Arg Gln Cys Leu Arg Ser Leu Gly Gly
145 150 155 160
Tyr Val Leu Ser Leu Val His Asp Val Arg Phe His His Phe Pro Ile
165 170 175
Glu Arg Gln Leu Asn Gly Thr Tyr Ala Ile Ala Gly Gly Lys Ala His
180 185 190
Cys Gly Pro Ala Glu Leu Cys Glu Phe Tyr Ser Arg Asp Pro Asp Gly
195 200 205
Leu Pro Cys Asn Leu Arg Lys Pro Cys Asn Arg Pro Ser Gly Leu Glu
210 215 220
Pro Gln Pro Gly Val Phe Asp Cys Leu Arg Asp Ala Met Val Arg Asp
225 230 235 240
Tyr Val Arg Gln Thr Trp Lys Leu Glu Gly Glu Ala Leu Glu Gln Ala
245 250 255
Ile Ile Ser Gln Ala Pro Gln Val Glu Lys Leu Ile Ala Thr Thr Ala
260 265 270
His Glu Arg Met Pro Trp Tyr His Ser Ser Leu Thr Arg Glu Glu Ala
275 280 285
Glu Arg Lys Leu Tyr Ser Gly Ala Gln Thr Asp Gly Lys Phe Leu Leu
290 295 300
Arg Pro Arg Lys Glu Gln Gly Thr Tyr Ala Leu Ser Leu Ile Tyr Gly
305 310 315 320
Lys Thr Val Tyr His Tyr Leu Ile Ser Gln Asp Lys Ala Gly Lys Tyr
325 330 335
~ Li
CA 02231330 1998-03-06 ~ , ,
'' t ~~
19~24
-20- IPEA~lJS 1 9 SEP 1997
Cys Ile Pro Glu Gly Thr Lys Phe Asp Thr Leu Trp Gln Leu Val Glu
340 345 350
Tyr Leu Lys Leu Lys Ala Asp Gly Leu Ile Tyr Cys Leu Lys Glu Ala
355 360 365
Cys Pro Asn Ser Ser Ala Ser Asn Ala Ser
370 375
(2) INFORMATION FOR SEQ ID NO:5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 384 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:5:
Met Gly Val Gln Val Glu Thr Ile Ser Pro Gly Asp Gly Arg Thr Phe
1 5 10 15
Pro Lys Arg Gly Gln Thr Cys Val Val His Tyr Thr Gly Met Leu Glu
Asp Gly Lys Lys Phe Asp Ser Ser Arg Asp Arg Asn Lys Pro Phe Lys
Phe Met Leu Gly Lys Gln Glu Val Ile Arg Gly Trp Glu Glu Gly Val
Ala C;ln Met Ser Val Gly Gln Arg Ala Lys Leu Thr Ile Ser Pro Asp
Tyr Ala Tyr Gly Ala Thr Gly His Pro Gly Ile Ile Pro Pro His Ala
Thr Leu Val Phe Asp Val Glu Leu Leu Lys Leu Glu Gly Leu Val Pro
100 105 110
Arg Gly Ser Met Ala Ser Ser Gly Met Ala Asp Ser Ala Asn His Leu
115 120 125
Pro l?he Phe Phe Gly Asn Ile Thr Arg Glu Glu Ala Glu Asp Tyr Leu
130 135 140
Val Gln Gly Gly Met Ser Asp Gly Leu Tyr Leu Leu Arg Gln Ser Arg
145 150 155 160
Asn Tyr Leu Gly Gly Phe Ala Leu Ser Val Ala His Gly Arg Lys Ala
165 170 175
His His Tyr Thr Ile Glu Arg Glu Leu Asn Gly Thr Tyr Ala Ile Ala
180 185 190
Gly Gly Arg Thr His Ala Ser Pro Ala Asp Leu Cys His Tyr His Ser
195 200 205
Gln Glu Ser Asp Gly Leu Val Cys Leu Leu Lys Lys Pro Phe Asn Arg
210 215 220
Pro Gln Gly Val Gln Pro Lys Thr Gly Pro Phe Glu Asp Leu Lys Glu
225 230 235 240
Asn Leu Ile Arg Glu Tyr Val Lys Gln Thr Trp Asn Leu Gln Gly Gln
245 250 255
Ala Leu Glu Gln Ala Ile Ile Ser Gln Lys Pro Gln Leu Glu Lys Leu
260 265 270
Ile Ala Thr Thr Ala His Glu Lys Met Pro Trp Phe His Gly Lys Ile
~ , , ~ ; . . ~ ;
CA 02231330 l998-03-06
l9.524 ,
~r~ h 7
- 21 -
275 280 285
Ser Arg Glu Glu Ser Glu Gln Ile Val Leu Ile Gly Ser Lys Thr Asn
2~0 295 300
Gly L~s Phe Leu Ile Arg Ala Arg Asp Asn Asn Gly Ser Tyr Ala Leu
305 310 315 320
Cys Leu Leu His Glu Gly Lys Val Leu His Tyr Arg Ile Asp Lys Asp
325 330 335
Lys Thr Gly Lys Leu Ser Ile Pro Glu Gly Lys Lys Phe Asp Thr Leu
340 345 350
Trp Gln Leu Val Glu His Tyr Ser Tyr Lys Ala Asp Gly Leu Leu Arg
355 360 365
Val Leu Thr Val Pro Cys Gln Lys Ile Gly Thr Gln Gly Asn Val Asn
370 375 380
(2) INFORMATION FOR SEQ ID NO:6:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 224 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:6:
Met Gly Val Gln Val Glu Thr Ile Ser Pro Gly Asp Gly Arg Thr Phe
1 5 10 15
Pro L,ys Arg Gly Gln Thr Cys Val Val His Tyr Thr Gly Met Leu Glu
Asp Gly Lys Lys Phe Asp Ser Ser Arg Asp Arg Asn Lys Pro Phe Lys
Phe Met Leu Gly Lys Gln Glu Val Ile Arg Gly Trp Glu Glu Gly Val
Ala C:ln Met Ser Val Gly Gln Arg Ala Lys Leu Thr Ile Ser Pro Asp
Tyr Ala Tyr Gly Ala Thr Gly His Pro Gly Ile Ile Pro Pro His Ala
Thr Leu Val Phe Asp Val Glu Leu Leu Lys Leu Glu Gly Leu Val Pro
100 105 110
Arg (,ly Ser Met Ala Asn Ser Leu Glu Pro Glu Pro Trp Phe Phe Lys
115 120 125
Asn Leu Ser Arg Lys Asp Ala Glu Arg Gln Leu Leu Ala Pro Gly Asn
:L30 135 140
Thr His Gly Ser Phe Leu Ile Arg Glu Ser Glu Ser Thr Ala Gly Ser
145 150 155 160
Phe ',er Leu Ser Val Arg Asp Phe Asp Gln Asn Gln Gly Glu Val Val
165 170 175
Lys His Tyr Lys Ile Arg Asn Leu Asp Asn Gly Gly Phe Tyr Ile Ser
180 185 190
Pro Arg Ile Thr Phe Pro Gly Leu His Glu Leu Val Arg His Tyr Thr
195 200 205
Asn Ala Ser Asp Gly Leu Cys Thr Arg Leu Ser Arg Pro Cys Gln Thr
210 215 220
CA 02231330 1998-03-06
19.~24 ~ 5 S 7
-22- ~ t5~ i
(2) INFORMATION FOR SEQ ID NO:7:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 7 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:7:
Gly Leu Val Pro Arg Gly Ser
1 5
(2) INFORMATION FOR SEQ ID NO:8:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 37 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
~D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:8:
GATCGCCATG GGAGTGCAGG TGGAAACCAT CTCCCCA 37
(2) INFORMATION FOR SEQ ID NO:9:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 48 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:9:
TACGAATTCT GGCGTGGATC CACGCGGAAC CAGACCTTCC AGTTTTAG 48
(2) INFORMATION FOR SEQ ID NO:10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 38 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
AMEN-DED SltEET
CA 0223l330 l998-03-06
l9524 PCT/US 9 h / ' 4 5 6 7
199J
- 23 -
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:10:
ATTAGGATCC ATGCCAGATC CTGCAGCTCA CCTGCCCT 38
(2) INFORMATION FOR SEQ ID NO:11:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:11:
ATATGGATCC TTACCAGAGG CGTTGCT 27
. ,. ~
- (2) INFORMATION FOR SEQ ID NO:12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 34 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:12:
CAATAGGATC CATGGCCAGC AGCGGCATGG CTGA 34
(2) INFORMATION FOR SEQ ID NO:13:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:13:
GACCTAGGAT CCCTAATTAA CATTTCCCTG TGTGCCGAT 39
(2) INFORMATION FOR SEQ ID NO:14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 38 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
CA 0223l330 1998-03-06
PCT/US ~ ~ / '4 567
- 24 ~ A/Us 1 9 SEP 199~
(ii) MOLECULE TYPE: Genomic DNA
(xi3 SEQUENCE DESCRIPTION: SEQ ID NO:14:
ATATGGATCC ATGGCGAACA GCCTGGAGCC CGAACCCT 38
(2) INFORMATION FOR SEQ ID NO:15:
~i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 43 base pairs
~B) TYPE: nucleic acid
(C) STRANDEDNESS: single
~D3 TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:15:
ATTAGGATCC TTAGGTCTGG CAGGGGCGGC TCAACCGTGT GCA 43
(2) INFORMATION FOR SEQ ID NO:16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: protein
(ix) FEATURE:
(A) NAME/KEY: Other
(B) LOCATION: 6...6
(D) OTHER INFORMATION: Xaa = Phosphorylated Tyrosine
(A) NAME/KEY: Other
(B) LOCATION: 17...17
(D) OTHER INFORMATION: Xaa = Phosphorylated Tyrosine
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:16:
Gly Ser Asn Gln Leu Xaa Asn Glu Leu Asn Leu Gly Arg Arg Glu Glu
1 5 10 15
Xaa Asp Val Leu Asp Lys
(2) INFORMATION FOR SEQ ID NO:17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 11 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
CA 02231330 1998-03-06
l9524
-- 25 ~ v ~
(ii) MOLECULE ~-YPE: protein
(ix) FEATURE:
(A) NAME/KEY: Other
(B) LOCATION: 4...4
(D) OTHER INFORMATION: Xaa = Phosphorylated Tyrosine
(xi) SEQUENCE DESCRIPTION: SEQ ID NO:17:
Glu Pro Gln Xaa Glu Glu Ile Pro Ile Tyr Leu
l 5 l0
AMENDED SHEET