Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
DNA mutagenesis by random fragmentation and reassembly.
BACKGROUND OF THE INVENTION
Field of the Invention
The present invention relates to a method for the production
of polynucleotides conferring a desired phenotype and/or encoding
a protein having an advantageous predetermined property which is
selectable. In an aspect, the method is used for generating and
selecting nucleic acid fragments encoding mutant proteins.
Description of the :Related Art
The complexity of an active sequence of a biological
macromolecule, e.g.. proteins, DNA etc., has been called its
information content. ("IC"; 5-9). The information content of a
protein has been defined as the resistance of the active protein
to amino acid sequence variation, calculated from the minimum
number of invariable amino acids (bits) required to describe a
family of related sequences with the same function (9, 10).
Proteins that are Sensitive to random mutagenesis have a high
information content. 'In 1974, when this definition was coined,
protein diversity existed only as taxonomic diversity.
Molecular biology developments such as molecular libraries
have allowed the :identification of a much larger number of
variable bases, and even to select functional sequences from
random libraries. Most residues can be varied, although
typically not all at the same time, depending on compensating
changes in the context. Thus a 100 amino acid protein can
contain only 2,000 different mutations, but 20100 possible
combinations of mutations.
Information density is the Information Content/unit length
of a sequence. Active sites of enzymes tend to have a high
information density. By contrast, flexible linkers in enzymes
have a low information density (8).
Current methods in widespread use for creating mutant
proteins in a library format are error-prone polymerase chain
reaction (11, 12, 1.9) and cassette mutagenesis (8, 20, 21, 22,
40, 41, 42), in which the specific region to be c-timized is
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
2
replaced with a synthetically mutagenized oligonucleotide. In
both cases, a 'mutant cloud' (4) is generated around certain
sites in the original sequence.
Error-prone PCR uses low-fidelity polymerization conditions
to introduce a low level of point mutations randomly over a long
sequence. Error prone PCR can be used to mutagenize a mixture of
fragments of unknown sequence. However, computer simulations
have suggested that point mutagenesis alone may often be .too
gradual to allow the block changes that are required for
continued sequence evolution. The published error-prone PCR
protocols do not allow amplification of DNA fragments greater
than 0.5 to 1.0 kb, limiting their practical application.
Further, repeated cycles of error-prone PCR lead to an
accumulation of neutral mutations, which, for example, may make
a protein immunogenic.
In oligonucleot.ide-directed mutagenesis, a short sequence is
replaced with a synthetically mutagenized oligonucleotide. This
approach does not generate combinations of distant mutations and
is thus not combinatorial. The limited library size relative to
the vast sequence length means that many rounds of selection are
unavoidable for protein optimization. Mutagenesis with synthetic
oligonucleotides requires sequencing of individual clones after
each selection round followed by grouping into families,
arbitrarily choosing a single family, and reducing it to a
consensus motif, which is resynthesized and reinserted into a
single gene followed by additional selection. This process
constitutes a statistical bottleneck, it is labor intensive and
not practical for many rounds of mutagenesis.
Error-prone PCR and oligonucleotide-directed mutagenesis are
thus useful for single cycles of sequence fine tuning but rapidly
become limiting when applied for multiple cycles.
Error-prone PCR can be used to mutagenize a mixture of
fragments of unknown sequence (11, 12). However, the published
error-prone PCR protocols (11, 12) suffer from a low processivity
of the polymerase. Therefore, the protocol is unable to result
in the random mutagenesis of an average-sized gene. This
inability limits the practical application of error-prone PCR.
Another serious limitation of error-prone PCR is that the
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
3
rate of down-mutations grows with the information content of the
sequence. At a certain information content, library size, and
mutagenesis rate, the balance of down-mutations to up-mutations
will statistically prevent the selection of further improvements
(statistical ceiling).
Finally, repeated cycles of error-prone PCR will also lead
to the accumulation of neutral mutations, which can affect, for
example, immunogenic:ity but not binding affinity.
Thus error-prone PCR was found to be too gradual to allow
the block changes that are required for continued sequence
evolution (1, 2).
In cassette mutagenesis, a sequence block of a single
template is typically replaced by a (partially) randomized
sequence. Therefore, the maximum information content that can be
obtained is statistically limited by the number of random
sequences (.i.e., library size). This constitutes a statistical
bottleneck, eliminating other sequence families' which are not
currently best, but which may have greater long term potential.
Further, mutagenesis with synthetic oligonucleotides
requires sequencing of individual clones after each selection
round (20). Therefore, this approach is tedious and is not
practical for many rounds of mutagenesis.
Error-prone PCR and cassette mutagenesis are thus best
suited and have been widely used for fine-tuning areas of
comparatively low information content. One apparent exception is
the selection of an RNA ligase ribozyme from a random library
using many rounds of amplification by error-prone PCR and
selection (13).
It is becoming increasingly clear that the tools for the
design of recombinant linear biological sequences such as
protein, RNA and DNA are not as powerful as the tools nature has
developed. Finding better and better mutants depends on
searching more and more sequences within larger and larger
libraries, and increasing numbers of cycles of mutagenic
amplification and selection are necessary. However as discussed
above, the existing mutagenesis methods that are in widespread
use have distinct limitations when used for repeated cycles.
Evolution of most organisms occurs by natural selection and
CA 02497384 1995-02-17
= WO 95/22625 PCT/US95/02126
4
sexual reproduction. Sexual reproduction ensures mixing and
combining of the genes of the offspring of the selected
individuals. During meiosis, homologous chromosomes from the
parents line up with one another and cross-over part way along
their length, thus swapping genetic material. Such swapping or
shuffling of the DNA allows organisms to evolve more rapidly (1,
2). In sexual recombination, because the inserted sequences were
of proven utility in a homologous environment, the inserted
sequences are likely to still have substantial information
content once they are inserted into the new sequence.
Marton et al.,(27) describes the use of PCR in vitro to
monitor recombination in a plasmid having directly repeated
sequences. Marton et al. discloses that recombination will occur
during PCR as a result of breaking or nicking of the DNA. This
will give rise to recombinant molecules. Meyerhans et al. (23)
also disclose the existence of DNA recombination during in vitro
PCR.
The term Applied Molecular Evolution ("AME") means the
application of an evolutionary design algorithm to a specific,
useful goal. While many different library formats for AME have
been reported for polynucleotides (3, 11-14), peptides and
proteins (phage (15-17), lacl (18) and polysomes, in none of
these formats has recombination by random cross-avers been used
to deliberately create a combinatorial library.
Theoretically there are 2,000 different single mutants
of a 100 amino acid protein. A protein of 100 amino acids has
20100 possible combinations of mutations, a number which is too
large to exhaustively explore by conventional methods. It would
be advantageous to develop a system which would allow the
generation and screening of all of these possible combination
mutations.
Winter and coworkers (43,44) have utilized an in vivo site
specific recombination system to combine light chain antibody
genes with heavy chain antibody genes for expression in a phage
system. However, their system relies on specific sites of
recombination and thus is limited. Hayashi et al. (48) report
simultaneous mutagenesis of antibody CDR regions in single chain
antibodies (scFv) by overlap extension and PCR.
CA 02497384 1995-02-17
79735-1D
Caren et al. (45) describe a method for generating a
large population of multiple mutants using random in vivo
recombination. However, their method requires the
recombination of two different libraries of plasmids, each
5 library having a different selectable marker. Thus the method
is limited to a finite number of recombinations equal to the
number of selectable markers existing, and produces a
concomitant linear increase in the number of marker genes
linked to the selected sequence(s).
Calogero et al. (46) and Galizzi et al. (47) report
that in vivo recombination between two homologous but truncated
insect-toxin genes on a plasmid can produce a hybrid gene.
Radman et al. (49) report in vivo recombination of
substantially mismatched DNA sequences in a host cell having
defective mismatch repair enzymes, resulting in hybrid molecule
formation.
CA 02497384 1995-02-17
79735-1D
6
It would be advantageous to develop a method for the
production of mutant. proteins which method allowed for the
development of large libraries of mutant nucleic acid sequences
which were easily searched. The invention described herein is
directed to the use of repeated cycles of point mutagenesis,
nucleic acid shuffling and selection which allow for the
directed molecular evolution in vitro of highly complex linear
sequences, such as proteins through random recombination.
CA 02497384 1995-02-17
79735-1D
7
Accordingly, it would be advantageous to develop a
method which allows for the production of large libraries of
mutant DNA, RNA or proteins and the selection of particular
mutants for a desired goal. The invention described herein is
directed to the use of repeated cycles of mutagenesis, in vivo
recombination and selection which allow for the directed
molecular evolution in vivo of highly complex linear sequences,
such as DNA, RNA or proteins through recombination.
Further advantages of the present invention will
become apparent from the following description of the invention
with reference to the attached drawings.
CA 02497384 2011-04-28
77471-78D
8
SUMMARY OF THE INVENTION
According to one aspect of the present invention, there is provided a
population of polynucleotides resulting from shuffling sequences from
substrate
polynucleotides encoding antibody chains, whereby the population of
polynucleotides
comprises recombinant polynucleotides encoding antibody chains having
different
combinations of three heavy chain CDRs and three light chain CDRs than are
encoded by the substrate polynucleotides.
In another aspect, the present invention further provides an antibody
comprising a heavy chain comprising three nonnaturally occurring
complementarity
determining regions (CDRs), and a light chain comprising three non-naturally
occurring CDR regions, wherein the antibody specifically binds to a
predetermined
antigen.
CA 02497384 1995-02-17
79735-1D
9
BRIEF DESCRIPTION OF THE DRAWINGS
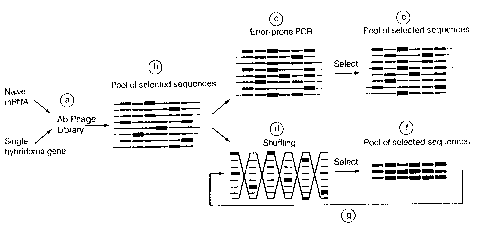
Figure 1 is a schematic diagram comparing mutagenic
shuffling over error-prone PCR; (a) the initial library; (b)
pool of selected sequences in first round of affinity
selection; (d) in vitro recombination of the selected sequences
('shuffling'); (f) pool of selected sequences in second round
of affinity selection after shuffling; (c) error-prone PCR; (e)
pool of selected sequences in second round of affinity
selection after error-prone PCR.
CA 02497384 1995-02-17
79735-1D
Figure 2 illustrates the reassembly of a 1.0 kb LacZ
alpha gene fragment from 10-50 bp random fragments. (a)
Photograph of a gel of PCR amplified DNA fragment having the
LacZ alpha gene. (b) Photograph of a gel of DNA fragments
5 after digestion with
CA 02497384 1995-02-17
WO 95/22625 rt- l i u bywuz i.zo
11
DNAseI. (c) Photograph of a gel of DNA fragments of 10-50 bp
purified from the digested LacZ alpha gene DNA fragment; (d)
Photograph of a gel. of the 10-50 bp DNA fragments after the
indicated number of cycles of DNA reassembly; (e) Photograph of
a gel of the recombination mixture after amplification by PCR
with primers.
Figure 3 is a schematic illustration of the LacZ alpha gene
stop codon mutants and their DNA sequences. The boxed regions
are heterologous areas, serving as markers. The stop codons are
located in smaller boxes or underlined. "+" indicates a wild-
type gene and "-" indicates a mutated area in the gene.
Figure 4 is a schematic illustration of the introduction or
spiking of a synthetic oligonucleotide into the reassembly
process of the LacZ alpha gene.
Figure 5 illustrates the regions of homology between a
murine IL1-B gene (]K) and a human IL1-B gene (H) with E. coli
codon usage. Regions of heterology are boxed. The
indicate crossovers obtained upon the shuffling of the two genes.
Figure 6 is a schematic diagram of the antibody CDR
shuffling model system using the scFv of anti-rabbit IgG antibody
(AlOB).
Figure 7 illustrates the observed frequency of occurrence of
certain combinations, of CDRs in the shuffled DNA of the scFv of
anti-rabbit IgG antibody (AlOB).
Figure 8 illustrates the improved avidity of the scFv anti-
rabbit antibody after DNA shuffling and each cycle of selection.
Figure 9 schematically portrays pBR322-Sfi-BL-LA-Sfi and in
vivo intraplasmidic recombination via direct repeats, as well as
the rate of generation of ampicillin-resistant colonies by
intraplasmidic recombination reconstituting a functional beta-
lactamase gene.
Figure 10 schematically portrays pBR322-Sfi-2Bla-Sfi and in
vivo intraplasmidic recombination via direct repeats, as well as
the rate of generation of ampicillin-resistant colonies by
intraplasmidic recombination reconstituting a functional beta-
lactamase gene.
Figure 11 illustrates the method for testing the efficiency
of multiple rounds of homologous recombination after the
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
12
introduction of polynucleotide fragments into cells for the
generation of recombinant proteins.
Figure 12 schematically portrays generation of a library of
vectors by shuffling cassettes at the following loci: promoter,
leader peptide, terminator, selectable drug resistance gene, and
origin of replication. The multiple parallel lines at each locus
represents the multiplicity of cassettes for that cassette.
Figure 13 schematically shows some examples of cassettes
suitable at various loci for constructing prokaryotic vector
libraries by shuffling.
DESCRIPTION OF THE PREFERRED EMBODIMENTS
The present invention relates to a method for nucleic acid
molecule reassembly after random fragmentation and its
application to mutagenesis of DNA sequences. Also described is
a method for the production of nucleic acid fragments encoding
mutant proteins having enhanced biological activity. In
particular, the present invention also relates to a method of
repeated cycles of mutagenesis, nucleic acid shuffling and
selection which allow for the creation of mutant proteins having
enhanced biological activity.
The present invention is directed to a method for
generating a very large library of DNA, RNA or protein mutants.
This method has particular advantages in the generation of
35 related DNA fragments from which the desired nucleic acid
fragment(s) may be selected. In particular the present invention
also relates to a method of repeated cycles of mutagenesis,
homologous recombination and selection which allow for the,
creation of mutant proteins having enhanced biological activity.
However, prior to discussing this invention in further
detail, the following terms will first be defined.
Definitions
As used herein, the following terms have the following
meanings:
The term "DNA reassembly" is used when recombination occurs
between identical sequences.
By contrast, the term "DNA shuffling" is used herein to
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
13
indicate recombination between substantially homologous but non-
identical sequences, in some embodiments DNA shuffling may
;involve crossover via nonhomologous recombination, such as via
cre/lox and/or flp/frt systems and the like.
The term "amplification" means that the number of copies of
a nucleic acid fragment is increased.
The term "identical" or "identity" means that two nucleic
acid sequences have the' same sequence or a complementary
sequence. Thus, "areas of identity" means that regions or areas
of a nucleic acid fragment or polynucleotide are identical or
complementary to another polynucleotide or nucleic acid fragment.
The term "corresponds to" is used'herein to mean that
a polynucleotide sequence is homologous (i.e., is identical, not
strictly evolutionarily related) to all or a portion of a
reference polynucleotide sequence, or that a polypeptide sequence
is identical to a reference polypeptide sequence. In
contradistinction, the term "complementary to" is used herein to
mean that the complementary sequence is homologous to all or a
portion of a reference polynucleotide sequence. For illustration,
the nucleotide sequence "TATAC" corresponds to a reference
sequence "TATAC" and is complementary to a reference sequence
"GTATA".
The following terms.are used to describe the sequence
relationships between two or more polynucleotides: "reference
sequence", "comparison window", "sequence identity", "percentage
of sequence identity", and "substantial identity". A "reference
sequence" is a defined sequence used as a basis for a sequence
comparison; a reference sequence may be a subset of a larger
sequence, for example, as a segment of a full-length cDNA or gene
sequence given in a sequence listing, such as a polynucleotide
sequence of Fig. 1 or Fig. 2(b), or may comprise a complete cDNA
or gene sequence. Generally, a reference sequence is at least 20
nucleotides in length, frequently at least 25 nucleotides in
length, and often at least 50 nucleotides in length. Since two
polynucleotides may each (1) comprise a sequence (i.e., a portion
of the complete polynucleotide sequence) that is similar between
the two polynucleotides, and (2) may further comprise a sequence
that is divergent. between the two polynucleotides, sequence
CA 02497384 1995-02-17
WO 95/22625 PCT1US95/02126
14
comparisons between two (or more) polynucleotides are typically
performed by comparing sequences of the two polynucleotides over
a "comparison window" to identify and compare local regions of
sequence similarity.
A "comparison window", as used herein, refers to a
conceptual segment of at least 20 contiguous nucleotide positions
wherein a polynucleotide sequence may be compared to a reference
sequence of at least 20 contiguous nucleotides and wherein the
portion of the polynucleotide sequence in the comparison window
I0 may comprise additions or deletions (i.e., gaps) of 20 percent or
less as compared to the reference sequence (which does not
comprise additions or deletions) for optimal alignment of the two
sequences. Optimal alignment of sequences for aligning a
comparison window may be conducted by the local homology
algorithm of Smith and Waterman (1981) Adv. ApDl. Math.'2: 482,
by the homology alignment algorithm of Needleman and Wunsch
(1970) J. Mol. Biol._ 48: 443, by the search for similarity method
of Pearson and Lipman (1988) Proc. Natl. Acad. Sci. (U.S.A.) 85:
2444, by computerized implementations of these algorithms (GAP,
BESTFIT, FASTA, and TFASTA in the Wisconsin Genetics Software
Package Release 7.0, Genetics Computer Group, 575 Science Dr.,
Madison, WI), or by inspection, and the best alignment (i.e.,
resulting in the highest percentage of homology over the
comparison window) generated by the various methods is selected.
The term "sequence identity" means that two
polynucleotide sequences are identical (i.e., on a nucleotide-by-
nucleotide basis) over the window of comparison. The term
"percentage of sequence identity" is calculated by comparing two
optimally aligned sequences over the window of comparison,
determining the number of positions at which the identical
nucleic acid base (e.g., A, T, C, G, U, or I) occurs in both
sequences to yield the number of matched positions, dividing the
number of matched positions by the total number of positions in
the window of comparison (i.e., the window size), and multiplying
the result by 100 to yield the percentage of sequence identity.
The terms "substantial identity" as used herein denotes a
characteristic of a polynucleotide sequence, wherein the
polynucleotide comprises a sequence that has at least 80 percent
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
sequence identity, preferably at least 85 percent identity and
often 90 to 95 percent sequence identity, more usually at least
99 percent sequence identity as compared to a reference sequence
over a comparison window of at least 20 nucleotide positions,
5 frequently over a window of at least 25-50 nucleotides, wherein
the percentage of sequence identity is calculated by comparing
the reference sequence to the polynucleotide sequence which may
include deletions or additions which total 20 percent or less of
the reference sequence over the window of comparison.
10 Conservative amino acid substitutions refer to the
interchangeability of residues having similar side chains. For
example, a group of amino acids having aliphatic side chains is
glycine, alanine, valine, leucine, and isoleucine; a group of
amino acids having aliphatic-hydroxyl side chains is serine and
15 threonine; a group of amino acids having amide-containing side
chains is asparagine and glutamine; a group of amino acids having
aromatic side chains; is phenylalanine, tyrosine, and tryptophan;
a group of amino acids having basic side chains is lysine,
arginine, and histidine; and a group of amino acids having
sulfur-containing side chains is cysteine and methionine.
Preferred conservative amino acids substitution groups are:
valine-leucine-isoleucine, phenylalanine-tyrosine, lysine-
arginine, alanine-valine, and asparagine-glutamine.
The term "homologous" or "homeologous" means that one
single-stranded nucleic acid sequence may hybridize to a
complementary single-stranded nucleic acid sequence. The degree
of hybridization may depend on a number of factors including the
amount of identity between the sequences and the hybridization
conditions such as temperature and salt concentration as
discussed later. Preferably the region of identity is greater
than about 5 bp, more preferably the region of identity is
greater than 10 bp.
The term "het:erologous" means that one single-stranded
nucleic acid sequence is unable to hybridize to another single-
stranded nucleic acid sequence or its complement. Thus areas of
heterology means that nucleic acid fragments or polynucleotides
have areas or regions in the sequence which are unable to
hybridize to another nucleic acid or polynucleotide. Such
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
16
regions or areas are, for example, areas of mutations.
The term "'cognate" as used herein refers to a gene
sequence that is evolutionarily and functionally related between
species. For example but not limitation, in the human genome,
the human CD4 gene is the cognate gene to the mouse CD4 gene,
since the sequences and structures of these two genes indicate
,that they are highly homologous and both genes encode a protein
which functions in signaling T cell activation through.MHC class
II-restricted antigen recognition.
The term "wild-type" means that the nucleic acid fragment
does not comprise any mutations. 'A "wild-type" protein means
that the protein will be active at a level of activity found in
nature and will comprise the amino acid sequence found in nature.
The term "related polynucleotides" means that regions or
areas of the polynucleotides are identical and regions or areas
of the polynucleotides are heterologous.
The term "chimeric polynucleotide" means that the
polynucleotide comprises regions which are wild-type and regions
which are mutated. It may also mean that the polynucleotide
200 comprises wild-type regions from one polynucleotide and wild-type
regions from another related polynucleotide.
The term "cleaving" means digesting the polynucleotide with
enzymes or breaking the polynucleotide.
The term "population" as used herein means a collection of
components such as polynucleotides, nucleic acid fragments or
proteins. A "mixed population" means a collection of components
which belong to the same family of nucleic acids or proteins
(i.e. are related) but which differ in their sequence (i.e. are
not identical) and hence in their biological activity.
The term "specific nucleic acid fragment" means a nucleic
acid fragment having certain end points and having a certain
nucleic acid sequence. Two nucleic acid fragments wherein one
nucleic acid fragment has the identical sequence as a portion of
the second nucleic acid fragment but different ends comprise two
different specific nucleic acid fragments.
The term "mutations" means changes in the sequence of a
wild-type nucleic acid sequence or changes in the sequence of a
peptide. Such mutations may be point mutations such as
CA 02497384 1995-02-17
WO 95/22625 PCTIUS9S/02126
17
transitions or transversions. The mutations may be deletions,
insertions or duplications.
In the polypeptide notation used herein, the lefthand
direction is the amino terminal direction and the righthand
direction is the carboxy-terminal direction, in accordance with
standard usage and convention. Similarly, unless specified
otherwise, the lefthand end of single-stranded polynucleotide
sequences is the 511 end; the lefthand direction of doub=le-
stranded polynucleot:ide sequences is referred to as the 5'
direction. The direction of 5' to 3' addition of nascent RNA
transcripts is referred to as the transcription direction;
sequence regions on the DNA strand having the same sequence as
the RNA and which are 5' to the 5' end of the RNA transcript are
referred to as "upstream sequences"; sequence regions on the DNA
strand having the same sequence as the RNA and which are 3' to
the 3' end of the coding RNA transcript are referred to as
"downstream sequences".
The term "naturally-occurring" as used herein as
applied to an object: refers to the fact that an object can be
found in nature. For example, a polypeptide or polynucleotide
sequence that is present in an organism (including viruses) that
can be isolated from a source in nature and which has not been
intentionally modified by man in the laboratory is naturally-
occurring. Generally, the term naturally-occurring refers to an
object as present in a non-pathological (undiseased) individual,
such as would be typical for the species.
The term "agent" is used herein to denote a chemical
compound, a mixture of chemical compounds, an array of spatially
localized compounds (e.g., a VLSIPS peptide array, polynucleotide
array, and/or combinatorial small molecule array), a biological
macromolecule, a bacteriophage peptide display library, a
bacteriophage antibody (e.g., scFv) display library, a polysome
peptide display library, or an extract made from biological
materials such as bacteria, plants, fungi, or animal
(particularly mammalian) cells or tissues. Agents are evaluated
for potential activity as antineoplastics, anti-inflammatories,
or apoptosis modulators by inclusion in screening assays
described hereinbelow. Agents are evaluated for potential
CA 02497384 1995-02-17
18
activity as specific protein interaction inhibitors (i.e., an
agent which selectively inhibits a binding interaction between
two predetermined polypeptides but which does not substantially
interfere with cell viability) by inclusion in screening assays
described hereinbelow.
As used herein, "substantially pure" means an object
species is the predominant species present (i.e., on a molar
basis it is more abundant than any other individual
macromolecular species in the composition), and preferably a
substantially purified fraction is a composition wherein the
object species comprises at least about 50 percent (on a molar
basis) of all macromolecular species present. Generally, a
substantially pure composition will comprise more than about 80
to 90 percent of all macromolecular species present in the
composition. Most preferably, the object species is purified to
essential homogeneity (contaminant species cannot be detected in
the composition by conventional detection methods) wherein the
composition consists essentially of a single macromolecular
species. Solvent species, small molecules (<500 Daltons), and
elemental ion species are not considered macromolecular species.
As used herein the term "physiological conditions"
refers to temperature, pH, ionic strength, viscosity, and like
biochemical parameters which are compatible with a viable
organism, and/or which typically exist intracellularly in a
viable cultured yeast cell or mammalian cell. For example, the
intracellular conditions in a yeast cell grown under typical
laboratory culture conditions are physiological conditions.
Suitable in vitro reaction conditions for in vitro transcription
cocktails are generally physiological conditions. In general, in
vitro physiological conditions comprise 50-200 mM NaCl or KC1, pH
6.5-8.5, 20-45 C and 0.001-10 mM divalent cation (e.g., Mg++,
Ca++) ; preferably about 150 mM NaCl or KC1, pH 7.2-7.6, 5 mM
divalent cation, and often include 0.01-1.0 percent nonspecific
protein (e.g., BSA). A non-ionic detergent (Tween~, NP-40, Triton
X-100) can often be present, usually at about 0.001 to 2%,
typically 0.05-0.2% (v/v). Particular aqueous. conditions may be
selected by the practitioner according to conventional methods.
For general guidance, the following buffered aqueous conditions
*trade-mark
CA 02497384 1995-02-17
19
may be applicable: 10-250 mM NaCl, 5-50 mM Tris HCl, pH 5-8, with
optional addition of divalent cation(s) and/or metal chelators
and/or nonionic detergents and/or membrane fractions and/or
antifoam agents and/or scintillants.
Specific hybridization is defined herein as the
formation of hybrids between a first polynucleotide and a second
polynucleotide (e.g., a polynucleotide having a distinct but
substantially identical sequence to the first polynucleotide),
wherein the first polynucleotide preferentially hybridizes to the
second polynucleotide under stringent hybridization conditions
wherein substantially unrelated polynucleotide sequences do not
form hybrids in the mixture.
As used herein, the term "single-chain antibody" refers
to a polypeptide comprising a VH domain and a VL domain in
polypeptide linkage, generally linked via a spacer peptide (e.g.,
[Gly-Gly-Gly-Gly-Ser],), and which may comprise additional amino
acid sequences at the amino- and/or carboxy- termini. For
example, a single-chain antibody may comprise a tether segment
for linking to the encoding polynucleotide. As an example, a
scFv is a single-chain antibody. Single-chain antibodies are
generally proteins consisting of one or more polypeptide segments
of at least lo'contiguous amino'ac'ids substantially encoded by
genes of the immunoglobulin superfamily (e.g., see The
Immunoglobulin Gene Superfamily, A.F. Williams and A.N. Barclay,
in Immunoalobulin Genes, T. Honjo, F.W. Alt, and T.H. Rabbitts,
eds., (1989) Academic Press: San Diego, CA, pp.361-387),
most frequently encoded by a
rodent, non-human primate, avian, porcine, bovine, ovine, goat,
or human heavy chain or light chain gene sequence. A functional
single-chain antibody generally contains a sufficient portion of
an immunoglobulin superfamily gene product so as to retain the
property of binding to a specific target molecule, typically a
receptor or antigen (epitope).
As used herein, the term "complementarity-determining
region" and "CDR" refer to the art-recognized term as exemplified
by the Kabat and Chothia CDR definitions also generally known as
hypervariable regions or hypervariable loops (Chothia and Lesk
(1987) J. Mol. Biol. 196: 901; Chothia et al. (1989) Nature 342:
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
877; E.A. Kabat et al., Sequences of Proteins of Immunological
Interest (National Institutes of Health, Bethesda, MD) (1987);
and Tramontano et al. (1990) J. Mol. Biol. 235: 175). Variable
region domains typically comprise the amino-terminal
5 approximately 105-1:15 amino acids of a naturally-occurring
immunoglobulin chain (e.g., amino acids 1-110), although variable
domains somewhat shorter or longer are also suitable for forming
single-chain antibodies.
An immunoglobulin light or heavy chain variable region
10 consists of, a "framework" region interrupted by three
hypervariable regions, also called CDR's. The extent of the
framework region and CDR's have been precisely defined (see
"Sequences of Proteins of Immunological Interest," E. Kabat et
al., 4th Ed., U.S. Department of Health and Human Services,
15 Bethesda, MD (1987)). The sequences of the framework regions of
different light or heavy chains are relatively conserved within
a species. As used herein, a "human framework region" is a
framework region that is substantially identical (about 85% or
more, usually 90-95% or more) to the framework region of a
20 naturally occurring human immunoglobulin. The framework region
of an antibody, that is the combined framework regions of the
constituent light and heavy chains, serves to position and align
the CDR's. The CDR's are primarily responsible for binding to an
epitope of an antigen.
35 As used herein, the term "variable segment" refers to
a portion of a nascent peptide which comprises a random,
pseudorandom, or defined kernal sequence. A variable segment can
comprise both variant and invariant residue positions, and the
degree-of residue variation at a variant residue position may be
limited; both options are selected at the discretion of the
practitioner. Typically, variable segments are about 5 to 20
amino acid residues in length (e.g., 8 to 10), although variable
segments may be longer and may comprise antibody portions or
receptor proteins, such as an antibody fragment, a nucleic acid
binding protein, a receptor protein, and the like.
As used herein, "random peptide sequence" refers to an
amino acid sequence composed of two or more amino acid monomers
and constructed by a stochastic or random process. A random
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
21
peptide can include framework or scaffolding motifs, which may
comprise invariant sequences.
As used herein "random peptide library" refers to a set
of polynucleotide sequences that encodes a set of random
peptides, and to the set of random peptides encoded by those
polynucleotide sequences, as well as the fusion proteins
containing those random peptides.
As used herein, the term "pseudorandom" refers to a set
of sequences that have limited variability, so that for example
the degree of residue variability at one position is different
than the degree of residue variability at another position, but
any pseudorandom position is allowed some degree of residue
variation, however circumscribed.
As used herein, the term "defined sequence framework"
refers to a set oi: defined sequences that are selected on a
nonrandom basis, generally on the basis of experimental data or
structural data; for example, a defined sequence framework may
comprise a set of amino acid sequences that are predicted to form
a fl-sheet structure or may comprise a leucine zipper heptad
repeat motif, a zinc-finger domain, among other variations. A
"defined sequence kernal" is a set of sequences which encompass
a limited scope of variability. Whereas (1) a completely random
10-mer sequence of the 20 conventional amino acids can be any of
(20)10 sequences, and (2) a pseudorandom 10-mer sequence of the
20 conventional amino acids can be any of (20) 10 sequences but
will exhibit a bias for certain residues at certain positions
and/or overall, (3) a defined sequence kernal is a subset of
sequences which is less that the maximum number of potential
sequences if each residue position was allowed to be any of the
allowable 20 conventional amino acids (and/or allowable
unconventional amino/imino acids). A defined sequence kernal
generally comprises variant and invariant residue positions
and/or comprises variant residue positions which can comprise a
residue selected from a defined subset of amino acid residues),
and the like, either segmentally or over the entire length of the
individual selected, library member sequence. Defined sequence
kernals can refer to either amino acid sequences or
polynucleotide sequences. For illustration and not limitation,
CA 02497384 1995-02-17
WO 95/22625
22
the sequences (NNK)10 and (NNM) 10, where N represents A, T, G, or
C; K represents G or T; and M represents A or.C, are defined
sequence kernals.
As used herein "epitope" refers to that portion of an
antigen or other macromolecule capable of forming a binding
interaction that interacts with the variable region binding
pocket of an antibody. Typically, such binding interaction is
manifested as an intermolecular contact with one or `more amino
acid residues of a CDR.
As used he-rein, "receptor" refers to a molecule that
has an affinity for a given ligand. Receptors can be naturally
occurring or synthetic molecules. Receptors can be employed in
an unaltered state or as aggregates with other species.
Receptors can be attached, covalently or noncovalently, to a
binding member, either directly or via a specific binding
substance. Examples of receptors include, but are not limited
to, antibodies, including monoclonal antibodies and antisera
reactive with specific antigenic determinants (such as on
viruses, cells, or other materials), cell membrane receptors,
complex carbohydrates and glycoproteins, enzymes, and hormone
receptors.
As used herein "ligand" refers to a molecule, such as
a random peptide or variable segment sequence, that is recognized
by a particular receptor. As one of skill in the art will
recognize, a molecule (or macromolecular complex) can be both a
receptor and a ligand. In general, the binding partner having a
smaller molecular weight is referred to as the ligand and the
binding partner having a greater molecular weight is referred to
as a receptor.
As used herein, "linker" or "spacer" refers to a
molecule or group of molecules that connects two molecules, such
as a DNA binding protein and a random peptide, and serves to
place the two molecules in a preferred configuration, e.g., so
that the random peptide can bind to a receptor with minimal
steric hindrance from the DNA binding protein.
As used herein, the term "operably linked" refers to a
linkage of polynucleotide elements in a functional relationship.
A nucleic acid is "operably linked" when it is placed into a
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
23
functional relationship with another nucleic acid sequence. For
instance, a promoter or enhancer is operably linked to a coding
sequence if it affects the transcription of the coding sequence.
Operably linked means that the DNA sequences being linked are
typically contiguous and, where necessary to join two protein
coding regions, contiguous and in reading frame.
Methodology
Nucleic acid shuffling is a method for in vitro or in vivo
homologous recombination of pools of nucleic acid fragments or
polynucleotides. Mixtures of related nucleic acid sequences or
polynucleotides are randomly fragmented, and reassembled to yield
a library or mixed _population of recombinant nucleic acid
molecules or polynucleotides.
In contrast to cassette mutagenesis, only shuffling and
error-prone PCR allow one to mutate a pool of sequences blindly
(without sequence information other than primers).
The advantage of the mutagenic shuffling of this invention
over error-prone PCR alone for repeated selection can best be
explained with an example from antibody engineering. In Figure
1 is shown a schematic diagram of DNA shuffling as described
herein. The initial library can consist of related sequences of
diverse origin (i.e. antibodies from naive mRNA) or can be
derived by any type of mutagenesis (including shuffling) of a
single antibody gene. A collection of selected complementarity
determining regions ("CDRs") is obtained after the first round of
affinity selection (Fi_g.l). In the diagram the thick CDRs confer
onto the antibody molecule increased affinity for the antigen.
Shuffling allows the free combinatorial association of all of the
CDRIs with all of the CDR2s with all of the CDR3s, etc. (Fig.1).
This method differs from PCR, in that it is an inverse chain
reaction. In PCR, the number of polymerase start sites and the
number of molecules grows exponentially. However, the sequence
of the polymerase start sites and the sequence of the molecules
remains essentially the same. In contrast, in nucleic acid
reassembly or shuffling of random fragments the number of start
sites and the number (but not size) of the random fragments
decreases over time. For fragments derived from whole plasmids
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
24
the theoretical endpoint is a single, large concatemeric
molecule.
Since cross-overs occur at regions of homology,
recombination will primarily occur between members of the same
sequence family. This discourages combinations of CDRs that are
grossly incompatible (eg. directed against different epitopes of
the same antigen). It is contemplated that multiple families of
sequences can be shuffled in the same reaction. Further,
shuffling conserves the relative order, such that, for example,
CDR1 will not be found in the position of CDR2.
Rare shufflants will contain a large number of the best (eg.
highest affinity) CDRs and these rare shufflants may be selected
based on their superior affinity (Fig. 1).
CDRs from a pool of 100 different selected antibody
sequences can be permutated in up to 1006 different ways. This
large number of permutations cannot be represented in a single
library of DNA sequences. Accordingly, it is contemplated that
multiple cycles of DNA shuffling and selection may be required
depending on the length of the sequence and the sequence
22 diversity desired.
Error-prone PCR, in contrast, keeps all the selected CDRs in
the same relative sequence (Fig. 1), generating a much smaller
mutant cloud.
The template polynucleotide which may be used in the methods
of this invention may be DNA or RNA. It may be of various
lengths depending on the size of the gene or DNA fragment to be
recombined or reassembled. Preferably the template
polynucleotide is from 50 bp to 50 kb. It is contemplated that
entire vectors containing the nucleic acid encoding the protein
of interest can be used in the methods of this invention, and in
fact have been successfully used.
The template polynucleotide may be obtained by amplification
using the PCR reaction (U.S. Patent No. 4,683,202 and 4,683,195)
or other amplification or cloning methods. However, the removal
of free primers from the PCR product before fragmentation
provides a more efficient result. Failure to adequately remove
the primers can lead to a low frequency of crossover clones.
The template polynucleotide often should be double-stranded.
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95102126
A double-stranded nucleic acid molecule is required to ensure
that regions of the resulting single-stranded nucleic acid
fragments are complementary to each other and thus can hybridize
to form a double-stranded molecule.
5 It is contemplated that single-stranded or double-stranded
nucleic acid fragments having regions of identity to the template
polynucleotide and regions of heterology to the template
polynucleotide may be added to the template polynucleotide at
this. step. It is also contemplated that two different but related
10 polynucleotide templates can be mixed at this step.
The double-stranded polynucleotide template and any added
double-or single-stranded fragments are randomly digested into
fragments of from about 5 bp to 5 kb or more. Preferably the
size of the random fragments is from about 10 bp to 1000 bp, more
15 preferably the size of the DNA fragments is from about 20 bp to
500 bp.
Alternatively, it is also contemplated that double-stranded
nucleic acid having multiple nicks may be used in the methods of
this invention. A nick is a break in one strand of the double-
20 stranded nucleic acid. The distance between such nicks is
preferably 5 bp to 5 kb, more preferably between 10 bp to 1000
bp.
The nucleic acid fragment, may be digested by a number of
different methods. The nucleic acid fragment may be digested
25 with a nuclease, such as DNAseI or RNAse. The nucleic acid may
be randomly sheared by the method of sonication or by passage
through a tube having a small orifice.
It is also contemplated that the nucleic acid may also be
partially digested with one or more restriction enzymes, such
that certain points of cross-over may be retained statistically.
The concentration of any one specific nucleic acid fragment
will not be greater than 1% by weight of the total nucleic acid,
more preferably the concentration of any one specific nucleic
acid sequence will not be greater than 0.1% by weight of the
total nucleic acid.
The number of different specific nucleic acid fragments in
the mixture will be at least about 100, preferably at least about
500, and more preferably at least about 1000.
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
26
At this step single-stranded or double-stranded nucleic acid
fragments, either synthetic or natural, may be added to the
random double-stranded nucleic acid fragments in order to
increase the heterogeneity of the mixture of nucleic acid
fragments.
It is also contemplated that populations of double-stranded
randomly broken nucleic acid fragments may be mixed or combined
at this step.
Where insertion of mutations into the template
polynucleotide is desired, single-stranded or double-stranded
nucleic acid fragments having a region of identity to the
template polynucleotide and a region of heterology to the
template polynucleotide may be added in a 20 fold excess by
weight as compared to the total nucleic acid, more preferably the
single-stranded nucleic acid fragments may be added in a 10 fold
excess by weight as compared to the total nucleic acid.
Where a mixture of different but related template
polynucleotides is desired, populations of nucleic acid fragments
from each of the templates may be combined at a ratio of less
than about 1:100, more preferably the ratio is less than about
1:40. For example, a backcross of the wild-type polynucleotide
with a population of mutated polynucleotide may be desired to
eliminate neutral mutations (e.g., mutations yielding an
insubstantial alteration in the phenotypic property being
selected for). In such an example, the ratio of randomly
digested wild-type polynucleotide fragments which may be added to
the randomly digested mutant polynucleotide fragments is
approximately 1:1 to about 100:1, and more preferably from 1:1 to
40:1.
The mixed population of random nucleic acid fragments are
denatured to form single-stranded nucleic acid fragments and then
reannealed. Only those single-stranded nucleic acid fragments
having regions of homology with other single-stranded nucleic
acid fragments will reanneal.
The random nucleic acid fragments may be denatured by
heating. One skilled in the art could determine the conditions
necessary to completely denature the double stranded nucleic
acid. Preferably the temperature is from 80 C to 100 C, more
CA 02497384 1995-02-17
WU YJ/1Lb15 YC'17U595/02126
27
preferably the temperature is from 90 C.to 96 C. Other methods
which may be used to denature the nucleic acid fragments include
pressure (36) and pH.
The nucleic acid fragments may be reannealed by cooling-
Preferably the temperature is from '20 C to 75 C, more
preferably the temperature is from 40 C to 65 C. If a high
frequency of crossovers is needed based on an average of only 4
consecutive bases of homology, recombination can be - forced by
using a low annealing temperature, although the process becomes
more difficult. The degree of renaturation which occurs will
depend on the degree of homology between the population of
single-stranded nucleic acid fragments.
Renaturation can be accelerated by the addition of
polyethylene glycol ("PEG") or salt. The salt concentration is
preferably from 0 mM to 200 mM, more preferably the salt
concentration is from 10 mM to 100 mM. The salt may be KC1 or
NaCl. The concentration. of PEG is preferably from 0% to 20%,
more preferably from 5% to 10%.
The annealed nucleic acid fragments. are next incubated in
the presence of a nucleic acid polymerase and dNTP's (i.e. dATP,
dCTP, dGTP and dTTP). The nucleic acid polymerase may be the
Klenow fragment, the Taq polymerase or any other DNA polymerase
known in the art.
The approach to be used for the assembly depends on the
minimum degree of homology that should still yield crossovers. If
the areas of identity are large, Taq polymerase can be used with
an annealing temperature of between 45-65 C. If the areas of
identity are small, Klenow polymerase can be used with an-
annealing temperature of between 20-30 C. One skilled in the art
could vary the temperature of annealing to increase the number of
cross-overs achieved.
The polymerase may be added to the random nucleic acid
fragments prior to annealing, simultaneously with annealing or
after annealing.
The cycle of denaturation, renaturation and incubation in
the presence of polymerase is referred to herein as shuffling or
reassembly of the nucleic acid. This cycle is repeated for a
desired number of times. Preferably the cycle is repeated from
CA 02497384 1995-02-17
WO 95/22625 YC:1/U595/u2126
28
2 to 50 times, more ;preferably the sequence is repeated from 10
to 40 times.
The resulting nucleic acid is a larger double-stranded
polynucleotide of from about 50 bp to about 100 kb, preferably
the larger polynucleotide is from 500 bp to 50 kb.
This larger polynucleotide fragment may contain a number of
copies of a nucleic acid fragment having the same size as the
template polynucleotide in tandem. This concatemeric fragment is
then digested into single copies of the template polynucleotide.
The result will be a population of nucleic acid fragments of
approximately the same size as the template polynucleotide. The
population will be a mixed population where single or double-
stranded nucleic acid fragments having an area of identity and an
area of heterology have been added to the template polynucleotide
prior to shuffling.
These fragment are then cloned into the appropriate vector
and the ligation mixture used to transform bacteria.
It is contemplated that the single nucleic acid fragments
may be obtained from the larger concatemeric nucleic acid
fragment - by amplification of the single nucleic acid fragments
prior to cloning by a variety of methods including PCR (U.S.
Patent No. 4,683,195 and 4,683,202) rather than by digestion of
the concatemer.
The vector used for cloning is not critical provided that it
will accept a DNA fragment of the desired size. If expression of
the DNA fragment is desired, the cloning vehicle should further
comprise transcription and translation signals next to the site
of insertion of the DNA fragment to allow expression of the DNA
fragment in the host: cell. Preferred vectors include the pUC
series and the pBR series of plasmids.
The resulting bacterial population will include a number of
recombinant DNA fragments having random mutations. This mixed
population may be tested to identify the desired recombinant
nucleic acid fragment. The method of selection will depend on
the DNA fragment desired.
For example, if a DNA fragment which encodes for a protein
with increased binding efficiency to a ligand is desired, the
proteins expressed by each of the DNA fragments in the population
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
29
or library may be tested for their ability to bind to the ligand
by methods known in the art (i.e. panning, affinity
chromatography). If a DNA fragment which encodes for a protein
with increased drug resistance is desired, the proteins expressed
by each of the DNA fragments in the population or library may be
tested for their ability to confer drug resistance to the host
organism. One skilled'in the art, given knowledge of the desired
protein, could readily test the population to identify DNA
fragments which confer the desired properties onto the protein.
It is contemplated that one skilled in the art could use a
phage display system in which fragments of the protein are
expressed as fusion proteins on the phage surface (Pharmacia,
Milwaukee WI). The recombinant DNA molecules are cloned into the
phage DNA at a site which results in the transcription of a
fusion protein a portion of which is encoded by the recombinant
DNA molecule. The phage containing the recombinant nucleic acid
molecule undergoes replication and transcription in the cell.
The leader sequence of the fusion protein directs the transport
of the fusion protein to the tip of the phage particle. Thus the
fusion protein which is partially encoded by the recombinant DNA
molecule is displayed on the phage particle for detection and
selection by the methods described above.
It is further contemplated that a number of cycles of
nucleic acid shuffling may be conducted with nucleic acid
fragments from a subbpopulation of the first population, which
subpopulation contains DNA encoding the desired recombinant
protein. In this manner, proteins with even higher binding
affinities or enzymatic activity could be achieved.
It is also contemplated that a number of cycles of nucleic
acid shuffling may be conducted with a mixture of wild-type
nucleic acid fragments and a subpopulation of nucleic acid from
the first or subsequent rounds of nucleic acid shuffling in order
to remove any silent mutations from the subpopulation.
Any source of nucleic acid, in purified form can be utilized
5 as the starting nucleic acid. Thus the process may employ DNA or
RNA including messenger RNA, which DNA or RNA may be single or
double stranded. In addition, a DNA-RNA hybrid which contains
one strand of each may be utilized. The nucleic acid sequence
CA 02497384 1995-02-17
W u 7J/LLOLJ Y1.1 / U JYJ/ V t 1,j0
may be of various lengths depending on the size of the nucleic
acid sequence to be mutated. Preferably the specific nucleic
acid sequence is from 50 to 50000 base pairs. It is contemplated
that entire vectors containing the nucleic acid encoding the
5 protein of interest may be used in the methods of this invention.
The nucleic acid may be obtained from any source, for
example, from plasmids such a pBR322, from cloned DkA or RNA or
from natural DNA or RNA from any source including bacteria,
10 yeast, viruses and higher organisms such as plants or animals.
DNA or RNA may be extracted from blood or tissue material. The
template polynucleotide may be obtained by amplification using
the polynucleotide chain reaction (PCR) (U.S. Patent no.
4,683,202 and 4,683,195). Alternatively, the polynucleotide may
15 be present in a vector present in a cell and sufficient nucleic
acid may be obtained by culturing the cell and extracting the
nucleic acid from the cell by methods known in the art.
Any specific nucleic acid sequence can be used to produce
the population of mutants by the present process. It is only
200 necessary that a small population of mutant sequences of the
specific nucleic acid sequence exist or be created prior to the
present process.
The initial small population of the specific nucleic acid
sequences having mutations may be created by a number of
25 different methods. Mutations may be created by error-prone PCR.
Error-prone PCR uses low-fidelity polymerization conditions to
introduce a low level of point mutations randomly over a long
sequence. Alternatively, mutations can be introduced into the.
template polynucleotide by oligonucleotide-directed mutagenesis.
30 In oligonucleotide-directed mutagenesis, a short sequence of the
polynucleotide is removed from the polynucleotide using
restriction enzyme digestion and is replaced with a synthetic
polynucleotide in which various bases have been altered from the
original sequence. The polynucleotide sequence can also be
altered by chemical mutagenesis. Chemical mutagens include, for
example, sodium bisulfite, nitrous acid, hydroxylamine, hydrazine
or formic acid. Other agents which are analogues of nucleotide
precursors include nitrosoguanidine, 5-bromouracil, 2-
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
31
aminopurine, or acridine. Generally, these agents are added to
the PCR reaction in place of the nucleotide precursor thereby
mutating the sequence. Intercalating agents such as proflavine,
acriflavine, quinacrine and the like can also be used. Random
mutagenesis of the polynucleotide sequence can also be achieved
by irradiation with X-rays or ultraviolet light. Generally,
plasmid DNA or DNA fragments so mutagenized are introduced into
E. coli and propagated as a pool or library of mutant plasmids.
Alternatively the small mixed population of specific nucleic
1 acids may be found in nature in that they may consist of
different alleles of the same gene or the same gene from
different related species (i.e., cognate genes). Alternatively,
they may be related DNA sequences found within one species, for
example, the immunoglobulin genes.
Once the mixed population of the specific nucleic acid
sequences is generated, the polynucleotides can be used directly
or inserted into an appropriate cloning vector, using techniques
well-known in the art.
The choice of vector depends on the size of the
polynucleotide sequence and the host cell to be employed in the
methods of this invention. The templates of this invention may
be plasmids, phages, cosmids, phagemids, viruses (e.g.,
retroviruses, parainfluenzavirus, herpesviruses, reoviruses,
paramyxoviruses, and the like), or selected portions thereof
(e.g., coat protein, spike glycoprotein, capsid protein). For
example, cosmids and phagemids are preferred where the specific
nucleic acid sequence to be mutated is larger because these
vectors are able to stably propagate large nucleic acid
fragments.
If the mixed population of the specific nucleic acid
sequence is cloned into a vector it can be clonally amplified by
inserting each vector into a host cell and allowing the host cell
to amplify the vector. This is referred to as clonal
amplification because while the absolute number of nucleic acid
sequences increases, the number of mutants does not increase.
Utility
The DNA shuffling method of this invention can be performed
blindly on a pool of unknown sequences. By adding to the
CA 02497384 1995-02-17
WO 95122625 YC'I/UJ95/02126
32
reassembly mixture oligonucleotides (with ends that are
homologous to the sequences being reassembled) any sequence
mixture can be incorporated at any specific position into another
sequence mixture. Thus, it is contemplated. that mixtures of
synthetic oligonucleotides, PCR fragments or even whole genes can
be mixed into another sequence library at defined positions. The
insertion of one sequence (mixture) is independent from the
insertion of a sequence in another part of the template. Thus,
the degree of recombination, the homology required, and the
diversity of the library can be independently and simultaneously
varied along the length of the reassembled DNA.
This approach of mixing two genes may be useful for the
humanization of antibodies from murine hybridomas. The approach
of mixing two genes or inserting mutant sequences into genes may
be useful for any therapeutically used protein, for example,
interleukin I, antibodies, tPA, growth hormone, etc. The approach
may also be useful in any nucleic acid for example, promoters or
introns or 3' untranslated region or 5' untranslated regions of
genes to increase expression or alter specificity of expression
of proteins. The approach may also be used to mutate ribozymes
or aptamers.
Shuffling requires the presence of homologous regions
separating regions of diversity. Scaffold-like protein
structures may be particularly suitable for shuffling. The
conserved scaffold determines the overall folding by self-
association, while displaying relatively unrestricted loops that
mediate the specific binding. Examples of such scaffolds are the
immunoglobulin beta-barrel, and the four-helix bundle (24). This
shuffling can be used to create scaffold-like proteins with
various combinations of mutated sequences for binding.
In Vitro Shuffling
The equivalents of some standard genetic matings may also be
performed by shuffling in vitro. For example, a 'molecular
backcross' can be performed by repeated mixing of the mutant's
nucleic acid with the: wild-type nucleic acid while selecting for
the mutations of interest. As in traditional breeding, this
approach can be used to combine phenotypes from different sources
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
33
into a background of choice. It is useful, for example, for the
removal of neutral mutations that affect unselected
characteristics (i.e. immunogenicity). Thus it can be useful to
determine which mutations in a protein are involved in the
enhanced biological activity and which are not, an advantage
which cannot be achieved by error-prone mutagenesis or cassette
mutagenesis methods.
Large, functional genes can be assembled correctly from a
mixture of small random fragments. This reaction may be of use
for the reassembly of genes from the highly fragmented DNA of
fossils (25). In addition random' nucleic acid fragments from
fossils may be combined with nucleic acid fragments from similar
genes from related species.
It is also contemplated that the method of this invention
can. be used for the In vitro amplification of a whole genome from
a single cell as is needed for a variety of research and,
diagnostic applications. DNA amplification by PCR is in practice
limited to a length of about 40 kb. Amplification of a whole
genome such as that of E. coli (5,000 kb) by PCR would require
about 250 primers yielding 125 forty kb fragments. This approach
is not practical due to the unavailability of sufficient sequence
data. On the other hand, random digestion of the genome with
DNAseI, followed by gel purification of small fragments will
provide a multitude of possible primers. Use of this mix of
random small fragments as primers in a PCR reaction alone or with
the whole genome as the template should result in an inverse
chain reaction with the theoretical endpoint of a single
concatemer containing many copies of the genome.
100 fold amplification in the copy number and an average
fragment size of greater than 50 kb may be obtained when only
random fragments are used (see Example 2). It is thought that
the larger concatemer is generated by overlap of many smaller
fragments. The quality of specific PCR products obtained using
synthetic primers will be indistinguishable from the product
obtained from unamplified DNA. It is expected that this approach
will be useful for the mapping of genomes.
The polynucleotide to be shuffled can be fragmented randomly
or non-randomly, at the discretion of the practitioner.
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
34
In Vivo Shuffling
In an embodiment often vivo shuffling, the mixed population
of the specific nucleic acid sequence is introduced into
bacterial or eukaryotic cells under conditions such that at least
two different nucleic acid sequences are present in each host
cell. The fragments can be introduced into the host cells by a
variety of different methods. The host cells can be transformed
with the fragments using methods known in the art, for example
treatment with calcium chloride. If the fragments are inserted
into a phage genome, the host cell can be transfected with the
recombinant phage cgenome having the specific nucleic acid
sequences. Alternatively, the nucleic acid sequences can be
introduced into the host cell using electroporation,
transfection, lipofec:tion, biolistics, conjugation, and the like.
In general, in this embodiment, the specific nucleic acids
sequences will be present in vectors which are capable of stably
replicating the sequence in the host cell. In addition, it is
contemplated that the vectors will encode a marker gene such that
host cells having the vector can be selected. This ensures that
20' the mutated specific; nucleic acid sequence can be recovered after
introduction into the host cell. However, it is contemplated
that the entire mixed population of the specific nucleic acid
sequences need not be present on a vector sequence. Rather only
a sufficient number of sequences need be cloned into vectors to
ensure that after introduction of the fragments into the host
cells each host cell, contains one vector having at least one
specific nucleic acid sequence present therein. It is also
contemplated that rather than having a subset of the population
of the specific nucleic acids sequences cloned into vectors, this
subset may be already stably integrated into the host cell.
It has been found that when two fragments which have
regions of identity are inserted into the host cells homologous
recombination occurs between the two fragments. Such
recombination between the two mutated specific nucleic acid
sequences will result in the production of double or triple
mutants in some situations.
It has also been found that the frequency of recombination
is increased if some of the mutated specific nucleic acid
CA 02497384 1995-02-17
It WU 95/22625 PCTIUS95/02126
sequences are present on linear nucleic acid molecules.
Therefore, in a preferred embodiment, some of the specific
nucleic acid sequences are present on linear nucleic acid
fragments.
5 After transformation, the host cell transformants are placed
under selection to identify those host cell transformants which
contain mutated specific nucleic acid sequences having the
qualities desired. For example, if increased resistance to a
particular drug is desired then the transformed host cells may be
0 subjected to increased concentrations of the particular drug and
those transformants producing mutated proteins able to confer
increased drug resistance will be selected. If the enhanced
ability of a particular protein to bind to a receptor is desired,
then expression of the protein can be induced from the
35 transformants and the resulting protein assayed in a ligand
binding assay by methods known in the art to identify that subset
of the mutated population which shows enhanced binding to the
ligand. Alternatively, the protein can be expressed in another
system to ensure proper processing.
20 Once a subset of the first recombined specific nucleic acid
sequences (daughter sequences) having the desired characteristics
are identified, they are then subject to a second round of
recombination.
In the second cycle of recombination, the recombined
25 specific nucleic acid sequences may be mixed with the original
mutated specific nucleic acid sequences (parent sequences) and
the cycle repeated as described above. In this way a set of
second recombined specific nucleic acids sequences can be
identified which have enhanced characteristics or encode for
30 proteins having enhanced properties. This cycle can be repeated
a number of times as desired.
It is also contemplated that in the second or subsequent
recombination cycle, a backcross can be performed. A molecular
backcross can be performed by mixing the desired specific nucleic
35 acid sequences with a large number of the wild-type sequence,
such that at least one wild-type nucleic acid sequence and a
mutated nucleic acid sequence are present in the same host cell
after transformation. Recombination with the wild-type specific
CA 02497384 1995-02-17
wv y~it~o~a ra.iiu~yaiuhiho
36
nucleic acid sequence will eliminate those neutral mutations that
may affect unselected characteristics such as immunogenicity but
not the selected characteristics.
In another embodiment of this invention, it is contemplated
that during the first round a subset of the specific nucleic acid
sequences can be fragmented prior to introduction into the host
cell. The size of the fragments must be large enough to contain
some regions of identity with the other sequences so as to
homologously recombine with the other sequences. The size of the
fragments will range from 0.03 kb to 100 kb more preferably from
0.2 kb to 10 kb. It is also contemplated that in subsequent
rounds, all of the specific nucleic acid sequences other than the
sequences selected from the previous round may be cleaved into
fragments prior to introduction into the host cells.
Fragmentation of the sequences can be accomplished by a
variety of method known in the art. The sequences can be
randomly fragmented or fragmented at specific sites in the
nucleic acid sequence. Random fragments can be obtained by
breaking the nucleic: acid or exposing it to harsh physical
treatment (e.g., shearing or irradiation) or harsh chemical
agents (e.g., by free radicals; metal ions; acid treatment to
depurinate and cleave). Random fragments can also be obtained,
in the case of DNA by the use of DNase or like nuclease. The
sequences can be cleaved at specific sites by the use of
restriction enzymes. The fragmented sequences can be single-
stranded or double-stranded. If the sequences were originally
single-stranded they can be denatured with heat, chemicals or
enzymes prior to insertion into the host cell. The reaction
conditions suitable for separating the strands of nucleic acid
are well known in the art.
The steps of this process can be repeated indefinitely,
being limited only by the number of possible mutants which can be
achieved. After a certain number of cycles, all possible mutants
will have been achieved and further cycles are redundant.
In an embodiment the same mutated template nucleic acid is
repeatedly recombined and the resulting recombinants selected for
the desired characteristic.
Therefore, the initial pool or population of mutated
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
37
template nucleic acid is cloned into a vector capable of
replicating in a bacteria such as E. coli. The particular vector
is not essential, so long as it is capable of autonomous
replication in E. coli. In a preferred embodiment, the vector is
designed to allow the expression and production of any protein
encoded by the mutated specific nucleic acid linked to the
vector. It is also preferred that the vector contain a gene
encoding for a selectable marker.
The population of vectors containing the pool of mutated
nucleic acid sequences is introduced into the E. coli host cells.
The vector nucleic: acid sequences may be introduced by
transformation, transfection or infection in the case of phage.
The concentration of vectors used to transform the bacteria is
such that a number of vectors is introduced into each cell. Once
present in the cell, the efficiency of homologous recombination
is such that homologous recombination occurs between the various
vectors. This results in the generation of mutants (daughters)
having a combination of mutations which differ from the original
parent mutated sequences.
The host cells are then clonally replicated and selected for
the marker gene present on the vector. Only those cells having
a plasmid will grow under the selection.
The host cells which contain a vector are then tested for
the presence of favorable mutations. Such testing may consist of
placing the cells under selective pressure, for example, if the
gene to be selected is an improved drug resistance gene. If the
vector allows expression of the protein encoded by the mutated
nucleic acid sequence, then such selection may include allowing
expression of the protein so encoded, isolation of the protein
and testing of the protein to determine whether, for example, it
binds with increased efficiency to the ligand of interest.
Once a particular daughter mutated nucleic acid sequence
has been identified which confers the desired characteristics,
the nucleic acid is isolated either already linked to the vector
or separated from the vector. This nucleic acid is then mixed
with the f irst or parent population of nucleic acids and the
cycle is repeated.
It has been shown that by this method nucleic acid sequences
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
38
having enhanced desired properties can be selected.
In an alternate embodiment, the first generation of mutants
are retained in the cells and the parental mutated sequences are
added again to the cells. Accordingly, the first cycle of
Embodiment I is conducted as described above. However, after the
daughter nucleic acid sequences are identified, the host cells
containing these sequences are retained.
The parent-mutated specific nucleic acid population, either
as fragments or cloned into the same vector is introduced into
the host cells already containing the daughter nucleic acids.
Recombination is allowed to occur' in the cells and the next
generation of recombinants, or granddaughters are selected by the
methods described above.
This cycle can be repeated a number of times until the
nucleic acid or peptide having the desired characteristics is
obtained. It is contemplated that in subsequent cycles, the
population of mutated sequences which are added to the preferred
mutants may come from the parental mutants or any subsequent
generation.
In an alternative embodiment, the invention provides a
method of conducting a "molecular" backcross of the obtained
recombinant specific nucleic acid in order to eliminate any
neutral mutations. Neutral mutations are those mutations which
do not confer onto the nucleic acid or peptide the desired
properties. Such mutations may however confer on the nucleic
acid or peptide undesirable characteristics. Accordingly, it is
desirable to eliminate such neutral mutations. The method of
this invention provide a means of doing so.
In this embodiment, after the mutant nucleic acid, having
the desired characteristics, is obtained by the methods of the
embodiments, the nucleic acid, the vector having the nucleic acid
or the host cell containing the vector and nucleic acid is
isolated.
The nucleic acid or vector is then introduced into the host
cell with a large excess of the wild-type nucleic acid. The
nucleic acid of the mutant and the nucleic acid of the wild-type
sequence are allowed, to recombine. The resulting recombinants
are placed under the same selection as the mutant nucleic acid.
CA 02497384 1995-02-17
WO 95122625 PCTIUS95/02126
39
Only those recombinants which retained the desired
characteristics will. be selected. Any silent mutations which do
not provide the desired characteristics will be lost through
recombination with the wild-type DNA. This cycle can be repeated
a number of times until all of the silent mutations are
eliminated.
Thus the methods of this invention can be used in a
molecular backcross to eliminate unnecessary or silent mutations.
Utility
The in vivo recombination method of this invention can be
performed blindly on a pool of unknown mutants or alleles of a
specific nucleic acid fragment or sequence. However, it is not
necessary to know the actual DNA or RNA sequence of the specific
155 nucleic acid fragment.
The approach of using recombination within a mixed
population of genes can be useful for the generation of any
useful proteins, for example, interleukin 1, antibodies, tPA,
growth hormone, etc. This approach may be used to generate
2200 proteins having altered specificity or activity. The approach
may also be useful for the generation of mutant nucleic acid
sequences, for example, promoter regions, introns, exons,
enhancer sequences, 3' untranslated regions or 5' untranslated
regions of genes. Thus this approach may be used to generate
25 genes having increased rates of expression. This approach may
also be useful in the study of repetitive DNA sequences.
Finally, this approach may be useful to mutate ribozymes or
aptamers.
Scaffold-like regions separating regions of diversity in
30 proteins may be particularly suitable for the methods of this
invention.) The conserved scaffold determines the overall
folding by self-association, while displaying relatively
unrestricted loops that mediate the specific binding. Examples
of such scaffolds are the immunoglobulin beta barrel, and the
35 four-helix bundle. The methods of this invention can be used to
create scaffold-like proteins with various combinations of
mutated sequences for binding.
The equivalents of some standard genetic matings may also be
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
performed by the methods of this invention. For example, a
"molecular" backcross can be performed by repeated mixing of the
mutant's nucleic acid with the wild-type nucleic acid while
selecting for the mutations of interest. As in traditional
5 breeding, this approach can be used to combine phenotypes from
different sources into a background of choice. It is useful, for
example, for the removal of neutral mutations that affect
unselected characteristics (i.e. immunogenicity). Thus it can be
useful to determine which mutations in a protein are involved in
10 the enhanced biological activity and which are not.
Peptide Display Methods
The present method can be used to shuffle, by in vitro
and/or in vivo recombination by any of the disclosed methods, and
3.5 in any combination, polynucleotide sequences selected by,peptide
display methods, wherein an associated polynucleotide encodes a
displayed peptide which is screened for a phenotype (e.g., for
affinity for a predetermined receptor (ligand).
An increasingly important aspect of biopharmaceutical
20, drug development and molecular biology is the identification of
peptide structures, including the primary amino acid sequences,
of peptides or peptidomimetics that interact with biological
macromolecules. One method of identifying peptides that possess
a desired structure or functional property, such as binding to a
25 predetermined biological macromolecule (e.g., a receptor),
involves the screening of a large library or peptides for
individual library members which possess the desired structure or
functional property conferred by the amino acid sequence of the.
peptide.
30 In addition to direct chemical synthesis methods for
generating peptide libraries, several recombinant DNA methods
also have been reported. One type involves the display of a
peptide sequence, antibody, or other protein on the surface of a
bacteriophage particle or cell. Generally, in these methods each
35 bacteriophage particle or cell serves as an individual library
member displaying a single species of displayed peptide in
addition to the natural bacteriophage or cell protein sequences.
Each bacteriophage or cell contains the nucleotide sequence
CA 02497384 1995-02-17
WO 95/22625 YCI'/US95/U2126
41
information encoding the particular displayed peptide sequence;
thus, the displayed peptide sequence can be ascertained by
pucleotide sequence determination of an isolated library member.
A well-known peptide display method involves the
presentation of a peptide sequence on the surface of a
filamentous bacteriophage, typically as a fusion with a
bacteriophage coat -protein. The bacteriophage library can be
incubated with an immobilized, predetermined macromolecule or
small molecule (e.g., a receptor) so that bacteriophage particles
which present a peptide sequence that binds to the immobilized
macromolecule can be differentially partitioned from those that
do not present peptide sequences that bind to the predetermined
macromolecule. The bacteriophage particles (i.e., library
members) which are bound to the immobilized macromolecule are
then recovered and replicated to amplify the selected
bacteriophage subpopulation for a subsequent round of affinity
enrichment and phage replication. After several rounds of
affinity enrichment. and phage replication, the bacteriophage
library members that are thus selected are isolated and the
nucleotide sequence encoding the displayed peptide sequence is
determined, thereby identifying the, sequence(s) of peptides that
bind to the predetermined macromolecule (e.g., receptor). Such
methods are further described in PCT patent publication Nos.
91/17271, 91/18980, and 91/19818 and 93/08278.
2 The latter PCT publication describes a recombinant DNA
method for the display of peptide ligands that involves the
production of a library of fusion proteins with each fusion
protein composed of a first polypeptide portion, typically
comprising a variable sequence, that is available for potential
binding to a predetermined macromolecule, and a second
polypeptide portion that binds to DNA, such as the DNA vector
encoding the individual fusion protein. When transformed host
cells are cultured under conditions that allow for expression of
the fusion protein,, the fusion protein binds to the DNA vector
encoding it. Upon lysis of the host cell, the fusion
protein/vector DNA complexes can be screened against a
predetermined macromolecule in much the same way as bacteriophage
particles are screened in the phage-based display system, with
CA 02497384 1995-02-17
wv ~a wio
42
the replication and sequencing of the DNA vectors in the selected
fusion protein/vector DNA complexes serving as the basis for
identification of the selected library peptide sequence(s).
Other systems for generating libraries of peptides and
like polymers have aspects of both the recombinant and in vitro
chemical synthesis methods. In these hybrid methods, cell-free
enzymatic machinery is employed to accomplish the in vitro
synthesis of the library members (i.e., peptides or
polynucleotides). In one type of method, RNA molecules with the
ability to bind a predetermined protein or a predetermined dye
molecule were selected by alternate rounds of selection and PCR
amplification (Tuerk and Gold (1990) Science 249: 505; Ellington
and Szostak (1990) Nature 346: 818). A similar technique was
used to identify DNA sequences which bind a predetermined human
transcription factor (Thiesen and Bach (1990) Nucleic Acids Res.
18: 3203; Beaudry and Joyce (1992) Science 257; 635; PCT patent
publication Nos. 92/05258 and 92/14843). In a similar fashion,
the technique of in vitro translation has been used to synthesize
proteins of interest and has been proposed as a method for
generating large libraries of peptides. These methods which rely
upon in vitro translation, generally comprising stabilized
polysome complexes, are described further in PCT patent
publication Nos. 88/08453, 90/05785, 90/07003, 91/02076,
91/05058, and 92/02536. Applicants have described methods in
which library members comprise a fusion protein having a first
polypeptide portion with DNA binding activity and a second
polypeptide portion having the library member unique peptide
sequence; such methods are suitable for use in cell-free in vitro
selection formats, among others.
The displayed peptide sequences can be of varying
lengths, typically from 3-5000 amino acids long or longer,
frequently from 5-:L00 amino acids long, and often from about 8-15
amino acids long. A library can comprise library members having
varying lengths of displayed peptide sequence, or may comprise
library members having a fixed length of displayed peptide
sequence. Portions or all of the displayed peptide sequence(s)
can be random, pseudorandom, defined set kernal, fixed, or the
like. The present display methods include methods for in vitro
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
43
and in vivo display of single-chain antibodies, such as nascent
scFv on polysomes or sscFv displayed on phage, which enable large-
scale screening of scFv libraries having broad diversity of
variable region sequences and binding specificities.
The present invention also provides random,
pseudorandom, and defined sequence framework peptide libraries
and methods for generating and screening those libraries to
identify useful compounds (e.g., peptides, including single-chain
antibodies) that bind to receptor molecules or epitopes of
interest or gene products that modify peptides or RNA in a
desired fashion. The random, pseudorandom, and defined sequence
framework peptides are produced from libraries of peptide library
members that comprise displayed peptides or displayed single-
chain antibodies attached to a polynucleotide template from which
the displayed peptide was synthesized. The mode of attachment
may vary according to the specific embodiment of the invention
selected, and can include encapsidation in a phage particle or
incorporation in a cell.
A method of affinity enrichment allows a very large
library of peptides and single-chain antibodies to be screened
and the polynucleotide sequence encoding the desired peptide(s)
or single-chain antibodies to be selected. The polynucleotide
can then be isolated and shuffled to recombine combinatorially
the amino acid sequence of the selected peptide(s) (or
predetermined portions thereof) or single-chain antibodies (or
just VH, VL, or CDR portions thereof). Using these methods, one
can identify a peptide or single-chain antibody as having a
desired binding affinity for a molecule and can exploit the
process of shuffling to converge rapidly to a desired high-
affinity peptide or scFv. The peptide or antibody can then be
synthesized in bulk by conventional means for any suitable use
(e.g., as a therapeutic or diagnostic agent).
A significant advantage of the present invention is
that no prior information regarding an expected ligand structure
is required to isolate peptide ligands or antibodies of interest.
The peptide identified can have biological activity, which is
meant to include at least specific binding affinity for a
selected receptor molecule and, in some instances, will further
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
44
include the ability to block the binding of other compounds, to
stimulate or inhibit metabolic pathways, to act as a signal or
messenger, to stimulate or inhibit cellular activity, and the
like.
The present invention also provides a method for
shuffling a pool of polynucleotide sequences selected by affinity
screening a library of polysomes displaying nascent peptides
(including single-chain antibodies) for library members which
bind to a predetermined receptor (e.g., a mammalian proteinaceous
receptor such as, for example, a peptidergic hormone receptor, a
cell surface receptor, an intracellular protein which binds to
other protein(s) to form intracellular protein complexes such as
heterodimers and the like) or epitope (e.g., an immobilized
protein, glycoprotein, oligosaccharide, and the like).
Polynucleotide sequences selected in a first selection
round (typically by affinity selection for binding to a receptor
(e.g., a ligand) by any of these methods are pooled and the
pool(s) is/are shuffled by iM vitro and/or in vivo recombination
to produce a shuffled pool comprising a population of recombined'
220 selected polynuclectide sequences. The recombined selected
polynucleotide sequences are subjected to at least one subsequent
selection round. The polynucleotide sequences selected in the
subsequent selection round(s) can be used directly, sequenced,
and/or subjected to one or more additional rounds of shuffling
and subsequent selection. Selected sequences can also be
backcrossed with :polynucleotide sequences encoding neutral
sequences (i.e., having insubstantial functional effect on
binding), such as for example by backcrossing with a wild-type or
naturally-occurring sequence substantially identical to a
selected sequence to produce native-like functional peptides,
which may be less immunogenic. Generally, during backcrossing
subsequent selection is applied to retain the property of binding
to the predetermined receptor (ligand).
Prior to or concomitant with the shuffling of selected
sequences, the sequences can be mutagenized. In one embodiment,
selected library members are cloned in a prokaryotic vector
(e.g., plasmid, phagemid, or bacteriophage) wherein a collection
of individual colonies (or plaques) representing discrete library
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
members are produced. Individual selected library members can
then be manipulated (e.g., by site-directed mutagenesis, cassette
mutagenesis, chemical mutagenesis, PCR mutagenesis, and the like)
to generate a collection of library members representing a kernal
5 of sequence diversity based on the sequence of the selected
library member. The sequence of an individual selected library
member or pool can be manipulated to incorporate random mutation,
pseudorandom mutation, defined kernal mutation (i.e., comprising
variant and invariant residue positions and/or comprising variant
10, residue positions which can comprise a residue selected from a
defined subset of amino acid residues), codon-based mutation, and
the like, either segmentally or over the entire length of the
individual selected library member sequence. The mutagenized
selected library members are then shuffled by in vitro and/or in
15, vivo recombinatorial shuffling as disclosed herein.
The invention also provides peptide libraries
comprising a plurality of individual library members of the
invention, wherein (1) each individual library member of said
plurality comprises a sequence produced by shuffling of a pool of
20, selected sequences, and (2) each individual library member
comprises a variable peptide segment sequence or single-chain
antibody segment sequence which is distinct from the variable
peptide segment sequences or single-chain antibody sequences of
other individual library members in said plurality (although some
25 library members may be present in more than one copy per library
due to uneven amplification, stochastic probability, or the
like).
The invention also provides a product-by-process,
wherein selected polynucleotide sequences having (or encoding a
30 peptide having) a predetermined binding specificity are formed by
the process of: (1) screening a displayed peptide or displayed
single-chain antibody library against a predetermined receptor
(e.g., ligand) or epitope (e.g., antigen macromolecule) and
identifying and/or enriching library members which bind to the
35 predetermined receptor or epitope to produce a pool of selected
library members, (2) shuffling by recombination the selected
library members (or amplified or cloned copies thereof) which
binds the predetermined epitope and has been thereby isolated
CA 02497384 1995-02-17
WO 95/22625 Yt;17US9aiU111b
46
and/or enriched from the library to generate a shuffled library,
and (3) screening the shuffled library against the predetermined
receptor (e.g., ligand) or epitope (e.g., antigen macromolecule)
and identifying and/or enriching shuffled library members which
bind to the predetermined receptor or epitope to produce a pool
of selected shuffled library members.
Antibody Display and Screening Methods
The present method can be used to shuffle, by in vitro
and/or in vivo recombination by any of the disclosed methods, and
in any combination, polynucleotide sequences selected by antibody
display methods, wherein an associated polynucleotide encodes a
displayed antibody which is screened for a phenotype (e.g., for
affinity for binding a predetermined antigen (ligand).
Various molecular genetic approaches have been devised
to capture the vast immunological repertoire represented by the
extremely large number of distinct variable regions which can be
present in immunoglobulin chains. The naturally-occurring
germline immunoglobulin heavy chain locus is composed of separate'
tandem arrays of variable (V) segment genes located upstream of
a tandem array of diversity (D) segment genes, which are
themselves located upstream of a tandem array of joining (J)
region genes, which are located upstream of the constant (CH)
region genes. During B lymphocyte development, V-D-J
rearrangement occurs wherein a heavy chain variable region gene
(VH) is formed by rearrangement to form a fused D-J segment
followed by rearrangement with a V segment to form a V-D-J joined
product gene which, if productively rearranged, encodes a
functional variable region (VH) of a heavy chain. Similarly,
light chain loci rearrange one of several V segments with one of
several J segments to form a gene encoding the variable region
(VL) of a light chain.
The vast. repertoire of variable regions possible in
immunoglobulins derives in part from the numerous combinatorial
possibilities of joining V and J segments (and, in the case of
heavy chain loci, D segments) during rearrangement in B cell
development. Additional sequence diversity in the heavy chain
variable regions arises from non-uniform rearrangements of the D
CA 02497384 1995-02-17
WO 95/22625 Yc 1/USy~/UZ11b
47
segments during V-D=-J joining and from N region addition.
,Further, antigen-selection of specific B cell clones selects for
higher affinity variants having nongermline mutations in one or
both of the heavy and light chain variable regions; a phenomenon
referred to as "affinity maturation" or "affinity sharpening".
Typically, these "affinity sharpening" mutations cluster in
specific areas of the variable region, most commonly in the
complementarity-determining regions (CDRs).
In order to overcome many of the limitations in
producing and identifying high-affinity immunoglobulins through
antigen-stimulated B cell development (i.e., immunization),
various prokaryotic expression systems have been developed that
can be manipulated to produce combinatorial antibody libraries
which may be screened for high-affinity antibodies to specific
antigens. Recent advances in the expression of antibodies in
Escherichia coli and bacteriophage systems (see, "Alternative
Peptide Display Methods", infra) have raised the possibility that
virtually any specificity can be obtained by either cloning
antibody genes from characterized hybridomas or by de novo
selection using antibody gene libraries (e.g., from Ig cDNA).
Combinatorial libraries of antibodies have been
generated in bacteriophage lambda expression systems which may be
screened as bacteriophage plaques or as colonies of lysogens
(Huse et al. (1989) Science 246: 1275; Caton and Koprowski (1990)
Proc. Natl. Acad. Sci. (U.S.A.) 87: 6450; Mullinax et al (1990)
Proc. Natl. Acad. Sci. (U.S.A.) 87: 8095; Persson et al. (1991)
Proc. Natl. Acad. Sci. (U.S.A.) U: 2432). Various embodiments
of bacteriophage antibody display libraries and lambda phage
expression libraries have been described (Kang et al. (1991)
Proc. Natl. Acad. Sc.. (U.S.A.) 88: 4363; Clackson et al. (1991)
Nature 352: 624; McCafferty et al. (1990) Nature 348: 552; Burton
et al. (1991) Proc. Natl. Acad. Sci. (U.S.A.) 85: 10134;
Hoogenboom et al. (1991) Nucleic Acids Res. 15: 4133; Chang et
al. (1991) J. Immunol. 147: 3610; Breitling et al. (1991) Gene
304: 147; Marks et al. (1991) J. Mol. Biol. 222: 581; Barbas et
al. (1992) Proc. Natl. Acad. Sci. (U.S.A.) 89: 4457; Hawkins and
Winter (1992) J. Immunol. 22: 867; Marks at al. (1992)
Biotechnology 10: 779; Marks et al. (1992) J. Biol. Chem. 267:
CA 02497384 1995-02-17
48
16007; Lowman et al (:1991) Biochemistry 30: 10832; Lerner et al.
(1992) Science 258: 1313).
Typically, a bacteriophage antibody display library is screened
with a receptor (e.g., polypeptide, carbohydrate, glycoprotein,
nucleic acid) that is. immobilized (e.g., by covalent linkage to
a chromatography resin to enrich for reactive phage by affinity
chromatography) and/or labeled (e.g., to screen plaque or colony
lifts).
One particularly advantageous approach has been the use
of so-called single--chain fragment variable (scFv) libraries
(Marks et al. (1992) Biotechnolocry 10: 779; Winter G and Milstein
'C (1991) Nature 349: 293; Clackson et al. (1991) op.cit.; Marks
et al. (1991) J. Mol. Biol. 222: 581; Chaudhary et al. (1990)
Proc. Natl. Acad. Sc:i. (USA) 87: 1066; Chiswell et al. (1992)
TIBTECH 10: 80; McCafferty et al. (1990) op.cit.; and Huston et
al. (1988) Proc. Natl. Acad. Sci. (USA) 85: 5879). Various
embodiments of scFv. libraries displayed on bacteriophage coat
proteins have been described.
Beginning in 1988, single-chain analogues of Fv
fragments and their fusion proteins have been reliably generated
by antibody engineering methods. The first step generally
involves obtaining the genes encoding VH and VL domains with
desired binding properties; these V genes may be isolated from a
specific hybridoma cell line, selected from a combinatorial
25, V-gene library, or made by V gene synthesis. The single-chain Fv
is formed by connecting the component V genes with an
oligonucleotide that encodes an appropriately designed linker
peptide, such as (Gly-Gly-Gly-Gly-Ser)3 or equivalent linker
peptide(s). The linker bridges the C-terminus of the first V
region and N-terminus of the second, ordered as either
VH-linker-VL or VL-Linker-VH. In principle, the scFv binding site
can faithfully replicate both the affinity and specificity of its
parent antibody combining site.
Thus, scFv fragments are comprised of VH and VL domains
linked into a single polypeptide chain by a flexible linker
peptide. After the scFv genes are assembled, they are cloned
into a phagemid and expressed at the tip of the M13 phage (or
similar filamentous; bacteriophage) as fusion proteins with the
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
49
bacteriophage pIII (gene 3) coat protein. Enriching for phage
expressing an antibody of interest is accomplished by panning the
recombinant phage displaying a population scFv for binding to a
predetermined epitope (e.g., target antigen, receptor).
The linked polynucleotide of a library member provides
the basis for replication of the library member after a screening
or selection procedure, and also provides the basis for the
determination, by nucleotide sequencing, of the identity of the
displayed peptide sequence or VH and VL amino acid sequence. The
displayed peptide(s) or single-chain antibody (e.g., scFv) and/or
,its VH and VL domains or their CDRs can be cloned and expressed
in a suitable expression system. Often polynucleotides encoding
the isolated VH and VL domains will be ligated to polynucleotides
encoding constant regions (CH and CL) to form polynucleotides
155 encoding complete antibodies (e.g., chimeric or fully-human),
antibody fragments, and the like. Often polynucleotides encoding
the isolated CDRs will be grafted into polynucleotides encoding
a suitable variable region framework (and optionally constant
regions) to form polynucleotides encoding complete antibodies
(e.g., humanized or fully-human), antibody fragments, and the
like. Antibodies can be used to isolate preparative quantities
of the antigen by immunoaffinity chromatography. Various other
uses of such antibodies are to diagnose and/or stage disease
(e.g., neoplasia), and for therapeutic application to treat
disease, such as for example: neoplasia, autoimmune disease,
AIDS, cardiovascular disease, infections, and the like.
Various methods have been reported for increasing the
combinatorial diversity of a scFv library to broaden the
repertoire of binding species (idiotype spectrum). The use of
PCR has permitted the variable regions to be rapidly cloned
either from a specific hybridoma source or as a gene library from
non-immunized cells, affording combinatorial diversity in the
assortment of VH and VL cassettes which can be combined.
Furthermore, the VH and VL cassettes can themselves be
diversified, such as by random, pseudorandom, or directed
mutagenesis. Typically, V. and VL cassettes are diversified in
or near the complementarity-determining regions (CDRs), often the
third CDR, CDR3. Enzymatic inverse PCR mutagenesis has been
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
shown to be a simple and reliable method for constructing
relatively large libraries of scFv site-directed mutants (Stemmer
et al. (1993) Biotech.nigues 14: 256), as has error-prone PCR and
chemical mutagenesis (Deng et al. (1994) J. Biol. Chem. 269:
5 9533). Riechmann et al. (1993) Biochemistry 2a: 8848 showed
semirational design of an antibody scFv fragment using site-
directed randomization by degenerate oligonucleotide PCR and
subsequent phage display of the resultant scFv mutants. Barbas
et al. (1992) on.cit. attempted to circumvent the problem of
10 limited repertoire sizes resulting from using biased variable
region sequences by randomizing the sequence in a synthetic CDR
region of a human tetanus toxoid-binding Fab.
CDR randomization has the potential to create
approximately 1 x 1020 CDRs for the heavy chain CDR3 alone, and
=15 a roughly similar number of variants of the heavy chain CDR1 and
CDR2, and light chain CDR3.-3 variants. Taken individually or
together, the combinatorics of CDR randomization of heavy and/or
light chains requires generating a prohibitive number of
bacteriophage clones to produce a clone library representing all
20 possible combinations, the vast majority of which will be non-
binding. Generation of such large numbers of primary
transformants is not feasible with current transformation
technology and bacteriophage display systems. For example,
Barbas et al. (1992) op. cit. only generated 5 x 107 transformants,
25 which represents only a tiny fraction of the potential diversity
of a library of thoroughly randomized CDRs.
Despite these substantial limitations, bacteriophage
display of scFv have already yielded a variety of useful
antibodies and antibody fusion proteins. A bispecific single
30 chain antibody has been shown to mediate efficient tumor cell
lysis (Gruber et al. (1994) J. Immunol. 152: 5368).
Intracellular expression of an anti-Rev scFv has been shown to
inhibit HIV-1 virus replication in vitro (Duan et al. (1994)
Proc. Natl. Acad. Sci. (USA) 91: 5075), and intracellular
35 expression of an anti-p2lras scFv has been shown to inhibit
meiotic maturation of Xenopus oocytes (Biocca et al. (1993)
Biochem. Biophys. R.es. Commun. 197: 422. Recombinant scFv which
can be used to diagnose HIV infection have also been reported,
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
51
demonstrating the diagnostic utility of scFv (Lilley et al.
(1994) J. Immunol. Meth. 171: 211). Fusion proteins wherein an*
fcFv is linked to a second polypeptide, such as a toxin or
fibrinolytic activator protein, have also been reported (Holvost
et al. (1992) Eur. J. 210: 945; Nicholls et al. (1993)
J. Biol. Chem. 268: 5302).
If it were possible to generate scFv libraries having
broader antibody diversity and overcoming many of the limitations
of conventional CDR mutagenesis and randomization methods which
can cover only a very tiny fraction of the potential sequence
combinations, the number and quality of scFv antibodies suitable
for therapeutic and diagnostic use could be vastly improved. To
address this, the jn vitro and in vivo shuffling methods of the
invention are used to recombine CDRs which have been obtained
(typically via PCR amplification or cloning) from nucleic acids
obtained from selected displayed antibodies. Such displayed
antibodies can be displayed on cells, on bacteriophage particles,
on polysomes, or any suitable antibody display system wherein the
antibody is associated with its encoding nucleic acid(s). In a
variation, the CDRs are initially obtained from mRNA (or cDNA)
from antibody-producing cells (e.g., plasma cells/splenocytes
from an immunized wild-type mouse, a human, or a transgenic mouse
capable of making a human antibody as in W092/03918, W093/12227,
and W094/25585), including hybridomas derived therefrom.
Polynucleotide sequences selected in a first selection
round (typically by affinity selection for displayed antibody
binding to an antigen (e.g., a ligand) by any of these methods
are pooled and the pool(s) is/are shuffled by vitro and/or
vivo recombination, especially shuffling of CDRs (typically
shuffling heavy chain CDRs with other heavy chain CDRs and light
chain CDRs with other light chain CDRs) to produce a shuffled
pool comprising a population of recombined selected
polynucleotide sequences. The recombined selected polynucleotide
sequences are expressed in a selection format as a displayed
antibody and subjected to at least one subsequent selection
round. The polynucleotide sequences selected in the subsequent
selection round(s) can be used directly, sequenced, and/or
subjected to one or more additional rounds of shuffling and
CA 02497384 1995-02-17
WO 95/22625 Yl 1 / U'9,/Ulllb
52
subsequent selection until an antibody of the desired binding
affinity is obtained. Selected sequences can also be backcrossed
with polynucleotide sequences encoding neutral antibody framework
sequences (i.e., having insubstantial functional effect on
antigen binding), such as for example by backcrossing with a
human variable region framework to produce human-like sequence
antibodies. Generally, during backcrossing subsequent selection
is applied to retain the property of binding to the predetermined
antigen. '
Alternatively, or in combination with the noted
variations, the valency of the target epitope may be varied to
control the average binding affinity of selected scFv library
members. The target epitope can be bound to a surface or
substrate at varying densities, such as by including a competitor
35 epitope, by dilution, or by other method known to those, in the
art. A high density (valency) of predetermined epitope can be
used to enrich for scFv library members which have relatively low
affinity, whereas a low density (valency) can preferentially
enrich for higher affinity scFv library members.
For generating diverse variable segments, a collection
of synthetic oligonucleotides encoding random, pseudorandom, or
a defined sequence kernal set of peptide sequences can be
inserted by ligation into a predetermined site (e.g., a CDR).
Similarly, the sequence diversity of one or more CDRs of the
single-chain antibody cassette(s) can be expanded by mutating the
CDR(s) with site-directed mutagenesis, CDR-replacement, and the
like. The resultant DNA molecules can be propagated in a host
for cloning and amplification prior to shuffling, or can be used
directly (i.e., may avoid loss of diversity which may occur upon
propagation in a host cell) and the selected library members
subsequently shuffled.
Displayed peptide/polynucleotide complexes (library
members) which encode a variable segment peptide sequence of
interest or a single-chain antibody of interest are selected from
the library by an affinity enrichment technique. This is
accomplished by means of a immobilized macromolecule or epitope
specific for the peptide sequence of interest, such as a
receptor, other macromolecule, or other epitope species.
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
53
Repeating the affinity selection procedure provides an enrichment
of library members encoding the desired sequences, which may then
be isolated for pooling and shuffling, for sequencing, and/or for
further propagation and affinity enrichment.
The library members without the desired specificity are
removed by washing. The degree and stringency of washing
required will be determined for each peptide sequence or single-
chain antibody of interest and the immobilized predetermined
macromolecule or epitope. A certain degree of control can be
exerted over the binding characteristics of the nascent
peptide/DNA complexes recovered by adjusting the conditions of
the binding incubation and the subsequent washing. The
temperature, pH, ionic strength, divalent cations concentration,
and the volume and duration of the washing will select for
nascent peptide/DNA complexes within particular ranges of
affinity for the immobilized macromolecule. Selection based on
slow dissociation rate, which is usually predictive of high
affinity, is often the most practical route. This may be done
either by continued incubation in the presence of a saturating
amount of free predetermined macromolecule, or by increasing the
volume, number, and length of the washes. In each case, the
rebinding of dissociated nascent peptide/DNA or peptide/RNA
complex is prevented, and with increasing time, nascent
peptide/DNA or peptide/RNA complexes of higher and higher
affinity are recovered.
Additional modifications of the binding and washing
procedures may be applied to find peptides with special
characteristics. The affinities of some peptides are dependent
on ionic strength or cation concentration. This is a useful
characteristic for peptides that will be used in affinity
purification of various proteins when gentle conditions for
removing the protein from the peptides are required.
One variation involves the use of multiple binding
targets (multiple epitope species, multiple receptor species),
such that a scFv library can be simultaneously screened for a
multiplicity of scFv which have different binding specificities.
Given that the size of a scFv library often limits the diversity
of potential scFv sequences, it is typically desirable to us scFv
CA 02497384 1995-02-17
WO 95/22625 rL 11U5!PWutito
54
libraries of as large a size as possible. The time and economic
considerations of generating a number of very large polysome
scFv-display libraries can become prohibitive. To avoid this
substantial problem,, multiple predetermined epitope species
(receptor species) can be concomitantly screened in a single
library, or sequential screening against a number of epitope
species can be used. In one variation, multiple target epitope
species, each encoded on a separate bead (or subset of beads),
can be mixed'and incubated with a polysome-display scFv library
under suitable binding conditions. The collection of beads,
comprising multiple epitope species, can then be used to isolate,
by affinity selection, scFv library members. Generally,
subsequent affinity screening rounds can include the same mixture
of beads, subsets thereof, or beads containing only one or two
35 individual epitope species. This approach affords efficient
screening, and is compatible with laboratory automation, batch
processing, and high, throughput screening methods.
A variety of techniques can be used in the present
invention to diversify a peptide library or single-chain antibody
library, or to diversify, prior to or concomitant with shuffling,
around variable segment peptides or VH1 VL, or CDRs found in early
rounds of panning to have sufficient binding activity to the
predetermined macromolecule or, epitope. In one approach, the
positive selected peptide/polynucleotide complexes (those
identified in an early round of affinity enrichment) are
sequenced to determine the identity of the active peptides.
Oligonucleotides are then synthesized based on these active
peptide sequences, employing a low level of all bases
incorporated at each step to produce slight variations of the
primary oligonucleotide sequences. This mixture of (slightly)
degenerate oligonucleotides is then cloned into the variable
segment sequences at the appropriate locations. This method
produces systematic, controlled variations of the starting
peptide sequences, which can then be shuffled. It requires,
however, that individual positive nascent peptide/polynucleotide
complexes be sequenced before mutagenesis, and thus is useful for
expanding the diversity of small numbers of recovered complexes
and selecting variants having higher binding affinity and/or
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
higher binding specificity. In a variation, mutagenic PCR
amplification of positive selected peptide/polynucleotide
complexes (especially of the variable region sequences, the
amplification products of which are shuffled ij vitro and/or in
5 vivo and one or more additional rounds of screening is done prior
to sequencing. The same general approach can be employed with
single-chain antibodies in order to expand the diversity and
enhance the binding affinity/specificity, typically . by
diversifying CDRs or adjacent framework regions prior to or
10 concomitant with shuffling. If desired, shuffling reactions can
be spiked with mutagenic oligonucleotides capable of in vitro
recombination with the selected library members can be included.
Thus, mixtures of synthetic oligonucleotides and PCR fragments
(synthesized by error-prone or high-fidelity methods) can be
15 added to the in vitro shuffling mix and be incorporated into
resulting shuffled library members (shufflants).
The present invention of shuffling enables the
generation of a vast library of CDR-variant single-chain
antibodies. One way to generate such antibodies is to insert
20 synthetic CDRs into the single-chain antibody and/or CDR
randomization prior to or concomitant with shuffling. The
sequences of the synthetic CDR cassettes are selected by
referring to known sequence data of human CDR and are selected in
the discretion of the practitioner according to the following
25 guidelines: synthetic CDRs will have at least 40 percent
positional sequence identity to known CDR sequences, and
preferably will have at least 50 to 70 percent positional
sequence identity to known CDR sequences. For example, a=
collection of synthetic CDR sequences can be generated by
30 synthesizing a collection of oligonucleotide sequences on the
basis of naturally-occurring human CDR sequences listed in Kabat
et al. (1991) op.cit.; the pool(s) of synthetic CDR sequences are
calculated to encode CDR peptide sequences having at least 40
percent sequence identity to at least one known naturally
35 occurring human CDR sequence. Alternatively, a collection of
naturally-occurring CDR sequences may be compared to generate
consensus sequences so that amino acids used at a residue
position frequently (i.e., in at least 5 percent of known CDR
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
56
sequences) are incorporated into the synthetic CDRs at the
corresponding position(s). Typically, several (e.g., 3 to about
50) known CDR sequences are compared and observed natural
sequence variations between the known CDRs are tabulated, and a
collection of oligonucleotides encoding CDR peptide sequences
encompassing all or most permutations of the observed natural
sequence variations is synthesized. For example but not for
limitation, if a collection of human VH CDR sequences have
carboxy-terminal amino acids which are either Tyr, Val, Phis, or
Asp, then the pool(s) of synthetic CDR oligonucleotide sequences
are designed to allow the carboxy-terminal CDR residue to be any
of these amino acids. In some embodiments, residues other than
those which naturally-occur at a residue position in the
collection of CDR sequences are incorporated: conservative amino
1 acid. substitutions are frequently incorporated and up to 5
residue positions may be varied to incorporate non-conservative
amino acid substitutions as compared to known naturally-occurring
CDR sequences. Such CDR sequences can be used in primary
library members (prior to first round screening) and/or can be
used to spike in vitro shuffling reactions of selected library
member sequences. Construction of such pools of defined and/or
degenerate sequences will be readily accomplished by those of
ordinary skill in the art.
The collection of synthetic CDR sequences comprises at
least one member that is not known to be a naturally-occurring
CDR sequence. It is within the discretion of the practitioner to
include or not include a portion of random or pseudorandom
sequence corresponding to N region addition in the heavy chain
CDR; the N region sequence ranges from 1 nucleotide to about 4
nucleotides occurring at V-D and D-J junctions. A collection of
synthetic heavy chain CDR sequences comprises at least about 100
unique CDR sequences, typically at least about 1,000 unique CDR
sequences, preferably at least about 10,000 unique CDR sequences,
frequently more than 50,000 unique CDR sequences; however,
usually not more than about 1 x 106 unique CDR sequences are
included in the collection, although occasionally 1 x 107 to 1 x
108 unique CDR sequences are present, especially if conservative
amino acid substitutions are permitted at positions where the
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
57
conservative amino acid substituent is not present or is rare
(i.e., less than 0.1 percent) in that position in naturally-
occurring human CDRs. In general, the number of unique CDR
sequences included in a library should not exceed the expected
number of primary t:ransformants in the library by more than a
factor of 10. Such single-chain antibodies generally bind
to a predetermined antigen (e.g., the immunogen) with an affinity
of about at least 1 x 107 M-1, preferably with anyaffinity of
about at least 5 x 107 M-1, more preferably with an affinity of
at least 1 x 108 M-'` to 1 x 109 M-1 or more, sometimes up to 1 x
101OM-1 or more.. Frequently, the predetermined antigen is a
human protein, such as for example a human cell surface antigen
(e.g., CD4, CD8, IL-2 receptor, EGF receptor, PDGF receptor),
other human biological macromolecule (e.g., thrombomodulin,
protein C, carbohydrate antigen, sialyl Lewis antigen, L-
selectin), or nonhuman disease associated macromolecule (e.g.,
bacterial LPS, virion capsid protein or envelope glycoprotein)
and the like.
High affinity single-chain antibodies of the desired
specificity can be engineered and expressed in a variety of
systems. For example, scFv have been produced in plants (Firek
et al. (1993) Plan, Mol. Biol. 23: 861) and can be readily made
in prokaryotic systems (Owens RJ and Young RJ (1994) J. Immunol.
Meth. 168: 149; Johnson S and Bird RE (1991) Methods Enzymol.
203: 88). Furthermore, the single-chain antibodies can be used
as a basis for constructing whole antibodies or various fragments
thereof (Kettleborough et al. (1994) Eur. J. Immunol. 24: 952).
The variable region encoding sequence may be isolated (e.g., by
PCR amplification or subcloning) and spliced to a sequence
encoding a desired human constant region to encode a human
sequence antibody more suitable for human therapeutic uses where
immunogenicity is preferably minimized. The polynucleotide(s)
having the resultant fully human encoding sequence(s) can be
expressed in a host cell (e.g., from an expression vector in a
mammalian cell) and purified for pharmaceutical formulation.
The DNA expression constructs will typically include an
expression control DNA sequence operably linked to the coding
sequences, including naturally-associated or heterologous
CA 02497384 1995-02-17
58
promoter regions. Preferably, the expression control sequences
will be eukaryotic promoter systems in vectors capable of
transforming or transfecting eukaryotic host cells. Once the
vector has been incorporated into the appropriate host, the host
is maintained under conditions suitable for high level expression
of the nucleotide sequences, and the collection and purification
of the mutant "engineered" antibodies.
As stated previously, the DNA sequences, will -be
expressed in hosts after the sequences have been operably linked
to an expression control sequence (i.e., positioned to ensure the
transcription and translation of the structural gene). These
expression vectors are typically replicable in the host organisms
either as episomes or as an integral part of the host chromosomal
DNA. Commonly, expression vectors will contain selection
markers, e.g., tetracycline or neomycin, to permit detection of
those cells transformed with the desired DNA sequences (see,
e.g., U.S. Patent 4,704,362).
In addition to eukaryotic microorganisms such as yeast,
mammalian tissue cell culture may also be used to produce the
polypeptides of the present invention (see, Winnacker, "From
Genes to Clones," VCH Publishers, N.Y., N.Y. (1987)).
Eukaryotic cells are, actually
preferred, because a number of suitable host cell lines capable
of secreting intact immunoglobulins have been developed in the
art, and include the CHO cell lines, various COS cell lines, HeLa
cells, myeloma cell. lines, etc, but preferably transformed B-
cells or hybridomas. Expression vectors for these cells can
include expression control sequences, such as an origin of
replication, a promoter, an enhancer (Queen et al. (1986)
Immunol. Rev. 89: 49), and, necessary processing information
sites, such as ribosome binding sites, RNA splice sites,
polyadenylation sites, and transcriptional terminator sequences.
Preferred expression control sequences are promoters derived from
immunoglobulin genes, cytomegalovirus, SV40, Adenovirus, Bovine
Papilloma Virus, and the like.
Eukaryotic DNA transcription can be increased by
inserting an enhancer sequence into the vector. Enhancers are
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
59
cis-acting sequences of between 10 to 300bp that increase
transcription by a promoter. Enhancers can effectively increase
transcription when either 5' or 3' to the transcription unit.
They are also effective if located within an intron or within the
coding sequence itself. Typically, viral enhancers are used,
including SV40 enhancers, cytomegalovirus enhancers, polyoma
enhancers, and adenovirus enhancers. Enhancer sequences from
mammalian systems are also commonly used, such as the mouse
immunoglobulin heavy chain enhancer.
Mammalian expression vector systems will also typically
include a selectable marker gene. Examples of suitable markers
include, the dihydrofolate reductase gene (DHFR), the thymidine
kinase gene (TK), or prokaryotic genes conferring drug
resistance. The first two marker genes prefer the use of mutant
cell lines that lack the ability to grow without the addition of
thymidine to the growth medium. Transformed cells can then be
identified by their ability to grow on non-supplemented media.
Examples of prokaryotic drug resistance genes useful as markers
include genes conferring resistance to G418, mycophenolic acid
and hygromycin.
The vectors containing the DNA segments of interest can
be transferred into the host cell by well-known methods,
depending on the type of cellular host. For example, calcium
chloride transfect:ion is commonly utilized for prokaryotic cells,
whereas calcium phosphate treatment. lipofection, or
electroporation may be used for other cellular hosts. Other
methods used to 'transform mammalian cells include the use of
Polybrene, protoplast fusion, liposomes, electroporation, and
microinjection (Age, generally, Sambrook et al., supra).
Once expressed, the antibodies, individual mutated
immunoglobulin chains, mutated antibody fragments, and other
immunoglobulin polypeptides of the invention can be purified
according to standard procedures of the art, including ammonium
sulfate precipitation, fraction column chromatography, gel
electrophoresis and the like (see, generally, Scopes, R., Protein
Purification, Springer-Verlag, N.Y. (1982)). Once purified,
partially or to homogeneity as desired, the polypeptides may then
be used therapeutically or in developing and performing assay
CA 02497384 1995-02-17
WO 95122625 PCTIUS95/02126
procedures, immunof].uorescent stainings, and the like (see,
generally, Immunological Methods, Vols. I and II, Eds. Lefkovits
and Pernis, Academic Press, New York, N.Y. (1979 and 1981)).
The antibodies generated by the method of the present
5 invention can be used for diagnosis and therapy. By way of
illustration and not limitation, they can be used to treat
cancer, autoimmune diseases, or viral infections. For treatment
of cancer, the antibodies will typically bind to an antigen
expressed preferentially on cancer cells, such as erbB-2, CEA,
10, CD33, and many other antigens and binding members well known to
,those skilled in the art.
Yeast Two-Hybrid Screening Assays
Shuffling can also be used to recombinatorially
15 diversify a pool of selected library members obtained by
screening a two-hybrid screening system to identify library
members which bind a predetermined polypeptide sequence. The
selected library members are pooled and shuffled by in vitro
and/or in vivo recombination. The shuffled pool can then be
20, screened in a yeast two hybrid system to select library members
which bind said predletermined polypeptide sequence (e.g., and SH2
domain) or which bind an alternate predetermined polypeptide
sequence (e.g., an SH2 domain from another protein species).
An approach to identifying polypeptide sequences which
25, bind to a predetermined polypeptide sequence has been to use a
so-called "two-hybrid" system wherein the predetermined
polypeptide sequence is present in a fusion protein (Chien et al.
(1991) Proc. Natl. Acad. Sci. (USA) 88: 9578). This approach
identifies protein-protein interactions in vivo through
30 reconstitution of a, transcriptional activator (Fields S and Song
0 (1989) Nature 340.: 245), the yeast Ga14 transcription protein.
Typically, the method is based on the properties of the yeast
Ga14 protein, which. consists of separable domains responsible for
DNA-binding and transcriptional activation. Polynucleotides
35 encoding two hybrid proteins, one consisting of the yeast Ga14
DNA-binding domain fused to a polypeptide sequence of a known
protein and the other consisting of the Ga14 activation domain
fused to a polypeptide sequence of a second protein, are
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
61
constructed and introduced into a yeast host cell.
Intermolecular binding between the two fusion proteins
reconstitutes the Gal4 DNA-binding domain with the Ga14
activation domain, which leads to the transcriptional activation
of a reporter gene (e.g., lacZ, HIS3) which is operably linked to
a Ga14 binding site. Typically, the two-hybrid method is used to
identify novel polypeptide sequences which interact with a known
protein (Silver SC and Hunt SW (1993) Mol. Biol. Rep. 17: 155;
Durfee et al. (1993) Genes Devel. 7; 555; Yang et al. (1992)
Science 257: 680; Luban et al. (1993) Cell 73: 1067; Hardy et al.
(1992) Genes Devel. 6; 801; Bartel et al. (1993) Biotechnictues
14: 920; and Vojtek et al. (1993) Cell 74: 205). However,
variations of the two-hybrid method have been used to identify
mutations of a known protein that affect its binding to a second
known protein (Li B and Fields S (1993) FASEB J. 7: 957;' Lalo et
al. (1993) Proc. Natl. Acad. Sci. (USA) 90: 5524; Jackson et al.
(1993) Mol. Cell. Biol. 13; 2899; and Madura et al. (1993) J.
Biol. Chem. 268: 12046). Two-hybrid systems have also been used
to identify interacting structural domains of two known proteins
(Bardwell et al. (1993) med. Microbiol. 8: 1177; Chakraborty et
al. (1992) J. Biol. Chem. 267: 17498; Staudinger et al. (1993) J.
Biol. Chem. 268: 4608; and Milne GT and Weaver DT (1993) Genes
Devel. 7; 1755) or domains responsible for oligomerization of a
single protein (Iwabuchi et al. (1993) Oncogene 8; 1693; Bogerd
et al. (1993) J. Virol. 67: 5030). Variations of two-hybrid
systems have been used to study the in vivo activity of a
proteolytic enzyme (Dasmahapatra et al. (1992) Proc. Natl. Acad.
Sci. (USA) 89: 4159). Alternatively, an E. coli/BCCP interactive
screening system (Germino et al. (1993) Proc. Natl. Acad. Sci.
(U.S.A.) 90: 933; Guarente L (1993) Proc. Natl. Acad. Sci.
(U.S.A.) 90: 1639) can be used to identify interacting protein
sequences (i.e., protein sequences which heterodimerize or form
higher order heteromultimers). Sequences selected by a two-
hybrid system can be pooled and shuffled and introduced into a
two-hybrid system for one or more subsequent rounds of screening
to identify polypeptide sequences which bind to the hybrid
containing the predetermined binding sequence. The sequences
thus identified can be compared to identify consensus sequence(s)
CA 02497384 1995-02-17
.62
and consensus sequence kernals.
As can be appreciated from the disclosure above, the present
invention has a wide variety of applications. Accordingly, the
following examples are offered by way of illustration, not by way
of limitation.
In the examples below, the following abbreviations have the
following meanings. If not defined below, then the abbreviations
have their art recognized meanings.
ml = milliliter
F1 = microliters
M = micromolar
nM = nanomolar
PBS = phosphate buffered saline
ng = nanograms
g = micrograms
IPTG = isopropylthio-13-D-galactoside
bp = basepairs
kb = kilobasepairs
dNTP = deoxynucleoside triphosphates
PCR = polymerase chain reaction
X-gal = 5-bromo-4-chloro-3-indolyl-/3-D-galactoside
DNAseI = deoxyribonuclease
PBS = phosphate buffered saline
CDR = complementarity determining regions
MIC = minimum inhibitory concentration
scFv = single-chain Fv fragment of an antibody
In general, standard techniques of recombination DNA
technology are described in various publications, e.g. Sambrook
et al., 1989, Molecular Cloning: A Laboratory Manual, Cold Spring
Harbor Laboratory; Ausubel et al., 1987, Current Protocols in
Molecular Biology, vols. 1 and 2 and supplements, and Berger and
Kimmel, Methods in Enzymology. Volume 152, Guide to Molecular
Cloning Techniques (1987), Academic Press, Inc., San Diego, CA.
Restriction enzymes and polynucleotide modifying
enzymes were used according to the manufacturers recommendations.
Oligonucleotides were synthesized on an Applied Biosystems Inc.
Model 394 DNA synthesizer using ABI chemicals. If desired, PCR
amplimers for amplifying a predetermined DNA sequence may be
selected at the discretion of the practitioner.
EXAMPLES
Example 1. LacZ alpha gene reassembly
CA 02497384 1995-02-17
63
1) Substrate preparation
The substrate for the reassembly reaction was the dsDNA
polymerase chain reaction ("PCR") product of the wild-type LacZ
alpha gene from pUC18. (Fig. 2) (28; Gene Bank No. X02514) The
primer sequences were 5'AAAGCGTCGATTTTTGTGAT3' (SEQ ID NO:1) and
5'ATGGGGTTCCGCGCACATTT3' (SEQ ID NO:2). The free primers were
removed from the PCR product by Wizard PCR prep (Promega, Madison
WI) according to the manufacturer's directions. The removal of
the free primers was found to be important.
2) DNAseI digestion
About 5 Ag of the DNA substrate was digested with 0.15 units
of DNAseI (Sigma, St. Louis MO) in 100 Al of [50 mM Tris-HC1 pH
7.4, 1 mM MgC12]1 for 10-20 minutes at room temperature. The
digested DNA was run on a 2% low melting point agarose gel.
Fragments of 10-70 basepairs (bp) were purified from the 2% low
melting point agarose gels by electrophoresis onto DE81 ion
exchange paper (Whatman, Hillsborough OR). The DNA fragments
were eluted from the paper with 1 M NaCl and ethanol
precipitated.
3) DNA Reassembly
The purified fragments were resuspended at a concentration
of 10 - 30 ng/ l in PCR Mix (0.2 mM.each dNTP, 2.2 mM MgC12, 50
mM KC1, 10 mM Tris-HC1 pH 9.0, 0.1% Triton X-100, 0.3 gl Taq DNA
polymerase, 50 gl total volume). No primers were added at this
point. A reassembly program of 94 C for 60 seconds, 30-45 cycles
of [94 C for 30 seconds, 50-55 C for 30 seconds, 72 C for 30
seconds] and 5 minutes at 72 C was used in an MJ Research
(Watertown MA) PTC-150 thermocycler. The PCR reassembly of small
fragments into larger sequences was followed by taking samples of
the reaction after 25, 30, 35 ,40 and 45 cycles of reassembly
(Fig. 2).
Whereas the reassembly of 100-200 bp fragments can yield a
single PCR product of the correct size, 10-50 base fragments
typically yield some product of the correct size, as well as
products of heterogeneous molecular weights. Most of this size
heterogeneity appears to be due to single-stranded sequences at
the ends of the products, since after restriction enzyme
digestion a single band of the correct size is obtained.
*trade-mark
CA 02497384 1995-02-17
= WO 95/22625 PCTIUS95/02126
64
4) PCR with primers
After dilution of the reassembly product into the PCR Mix
with 0.8 M of each of the above primers (SEQ ID Nos: 1 and 2)
and about 15 cycles of PCR, each cycle consisting of [94 C for 30
seconds, 50 C for 30 seconds and 72 C for 30 seconds], a single
product of the correct size was obtained (Fig. 2).
5) Cloning and analysis
The PCR product from step 4 above was digeste3 with the
terminal restriction enzymes BamHI and EcoO109 and gel purified
as described above in step 2. The reassembled fragments were
ligated into pUC18 digested with BamHI and EcoO109. E. coli were
transformed with the ligation mixture under standard conditions
as recommended by the manufacturer (Stratagene, San Diego CA) and
plated on agar plates having 100 g/ml ampicillin, 0.004% X-gal
and 2mM IPTG. The resulting colonies having the HinDIII-NheI
fragment which is diagnostic for the ++ recombinant were
identified because they appeared blue.
This Example illustrates that a 1.0 kb sequence carrying the
LacZ alpha gene can be digested into 10-70 bp fragments, and that
these gel purified 10-70 bp fragments can be reassembled to a
single product of the correct size, such that 84% (N=377) of the
resulting colonies are LacZ+ (versus 94% without shuffling; Fig.
2).
The DNA encoding the LacZ gene from the resulting LacZ-
colonies was sequenced with a sequencing kit (United States
Biochemical Co., Cleveland OH) according to the manufacturer's
instructions and the genes were found to have point mutations due
to the reassembly process (Table 1). 11/12 types of
substitutions were found, and no frameshifts.
CA 02497384 1995-02-17
............~., rL I/U,Y~/UZ1Z6
TABLE 1
Mutations introduced by mutaaenic shuffling
Transitions Frequency Transversions Frequency
G- A 6 A T 1
5 A- G 4 A- C 2
C- T 7 C- A 1
T C 3 C G 0
G - C 3
G - T 2
100 T - A 1
T - G 2
A total of 4,437 bases of shuffled lacZ DNA were sequenced.
15 The rate of point mutagenesis during DNA reassembly from 10-
bp pieces was determined from DNA sequencing to be 0.7 %
(N=4,473), which is similar to error-prone PCR. Without being
limited to any theory it is believed that the rate of point
20 mutagenesis may be lower if larger fragments are used for the
reassembly, or if a proofreading polymerase is added.
When plasmid DNA from 14 of these point-mutated LacZ-
colonies were combined and again reassembled/shuffled by the
method described above, 34% (N=291) of the resulting colonies
255 were LacZ+, and these colonies presumably arose by recombination
of the DNA from different colonies.
The expected rate of reversal of a single point mutation by
error-prone PCR, assuming a mutagenesis rate of 0.7% (10), would
be expected to be <1%.
30 Thus large DNA sequences can be reassembled from a random
mixture of small fragments by a reaction that is surprisingly
efficient and simple. One application of this technique is the
recombination or shuffling of related sequences based on
homology.
Example 2. LacZ gene and whole plasmid DNA shuffling
1) LacZ gene shuffling
Crossover between two markers separated by 75 bases was
measured using two LacZ gene constructs. Stop codons were
inserted in two separate areas of the LacZ alpha gene to serve as
negative markers. Each marker is a 25 bp non-homologous sequence
with four stop codons, of which two are in the LacZ gene reading
frame. The 25 bp non-homologous sequence is indicated in Figure
CA 02497384 1995-02-17
WO 95/22625 PCT7US95/02126
66
3 by a large box. The stop codons are either boxed or
underlined. A 1:1 mixture of the two 1.0 kb LacZ templates
containing the +- and -+ versions of the LacZ alpha gene (Fig. 3)
was digested with DNAseI and 100-200 bp fragments were purified
as described in Example 1. The shuffling program was conducted
under conditions similar to those described for reassembly in
Example 1 except 0.5 l of polymerase was added and the total
volume was 100 Al.
After cloning, the number of blue colonies obtained was 24%;
(N=386) which is close to the theoretical maximum number of blue
,colonies (i.e. 25%), indicating that recombination between the
two markers was complete. All of the 10 blue colonies contained
the expected HindIII-NheI restriction fragment.
2) Whole plasmid DNA shuffling
Whole 2.7 kb plasmids (pUC18-+ and pUC18+-) were also
tested. A 1:1 mixture of the two 2.9 kb plasmids containing the
+- and -+ versions of the LacZ alpha gene (Fig. 3) was digested
with DNAseI and 100-200 bp fragments were purified as described
in Example 1. The shuffling program was conducted under'
conditions similar to those described for reassembly in step (1)
above except the program was for 60 cycles [94 C for 30 seconds,
55 C for 30 seconds, 72 C for 30 seconds]. Gel analysis showed
that after the shuffling program most of the product was greater
than 20 kb. Thus, whole 2.7 kb plasmids (pUC18 -+ and pUC18 +-)
were efficiently reassembled from random 100-200 bp fragments
without added primers.
After digestion with a restriction enzyme having a unique
site on the plasmid (EcoO109), most of the product consisted of
a single band of the expected size. This band was gel purified,
religated and the DNA used to transform E. coll. The
transformants were plated on 0.004% X-gal plates as described in
Example 1. 11% (N= 328) of the resulting plasmids were blue and
thus ++ recombinants.
3) Spiked DNA Shuffling
Oligonucleotides that are mixed into the shuffling mixture
can be incorporated into the final product based on the homology
of the flanking sequences of the oligonucleotide to the template
DNA (Fig. 4). The LacZ- stop codon mutant (pUC18 -+) described
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
67
above was used as the DNAseI digested template. A 66 mer
oligonucleotide, including 18 bases of homology to the wild-type
LacZ gene at both ends was added into the reaction at a 4-fold
molar excess to correct stop codon mutations present in the
original gene. The shuffling reaction was conducted under
conditions similar to those in step 2 above. The resulting
product was digested, ligated and inserted into E. coli as
described above.
Table 2
% blue colonies
Control 0.0 (N>1000)
Top strand spike 8.0 (N=855)
Bottom strand spike 9.3 (N=620)
Top and bottom strand spike 2.1 (N=537)
ssDNA appeared to be more efficient than dsDNA, presumably
due to competitive hybridization. The degree of incorporation
can be varied over a wide range by adjusting the molar excess,
annealing temperature, or the length of homology.
Example 3. DNA reassembly in the complete absence of primers
Plasmid pUC18 was digested with restriction enzymes EcoRI,
EcoO109, XmnI and AlwNI, yielding fragments of approximately 370,
460, 770 and 1080 bp. These fragments were electrophoresed and
separately purified from a 2% low melting point agarose gel (the
370 and 460 basepair bands could not be separated), yielding a
large fragment, a medium fragment and a mixture of two small
fragments in 3 separate tubes.
Each fragment was digested with DNAseI as described in
Example 1, and fragments of 50-130 bp were purified from a 2% low
melting point agarose gel for each of the original fragments.
35 PCR mix (as described in Example 1 above) was added to the
purified digested fragments to a final concentration of 10 ng/ l
of fragments. No primers were added. -A reassembly reaction was
performed for 75 cycles [94 C- for 30 seconds, 60 C for 30
seconds] separately on each of the three digested DNA fragment
mixtures, and the products were analyzed by agarose gel
electrophoresis.
The results clearly showed that the 1080, 770 and the 370
CA 02497384 1995-02-17
WO 95/22625 PC1/U595IU212b
68
and 460 bp bands reformed efficiently from the purified
fragments, demonstrating that shuffling does not require the use
of any primers at all.
Example 4. IL-18 gene shuffling
This example illustrates that crossovers based on homologies
of less than 15 bases may be obtained. As an example, a human
and a murine IL-113 gene were shuffled.
A murine IL1-B gene (BBG49) and a human IL1-B gene with E.
coli codon usage (BBG2; R&D Systems, Inc., Minneapolis MN) were
used as templates in the shuffling reaction. The areas of
complete homology between the human and the murine IL-113
sequences are on average only 4.1 bases long (Fig. 5, regions of
heterology are boxed).
Preparation of dsDNA PCR products for each of the genes,
removal of primers, DNAseI digestion and purification of 10-50 bp
fragments was similar to that described above in Example 1. The
sequences of the primers used in the PCR reaction were
5'TTAGGCACCCCAGGCTTT3' (SEQ ID NO:3) and 5'ATGTGCTGCAAGGCGATT3'
20, (SEQ ID NO:4).
The first 15 cycles of the shuffling reaction were performed
with the Klenow fragment of DNA polymerase I, adding 1 unit of
fresh enzyme at each cycle. The DNA was added to the PCR mix of
Example 1 which mix lacked the polymerase. The manual program
25, was 94 C for 1 minute, and then 15 cycles of: [95 C for 1 minute,
10 seconds on dry ice/ethanol (until frozen), incubate about 20
seconds at 25 C , add 1U of Klenow fragment and incubate at 25 C
for 2 minutes]. In each cycle after the denaturation step, the
tube was rapidly cooled in dry ice/ethanol and reheated to the
30 annealing temperature. Then the heat-labile polymerase was
added. The enzyme needs to be added at every cycle. Using this
approach, a high level of crossovers was obtained, based on only
a few bases of uninterrupted homology (Fig. 5, positions of
cross-overs indicated by
35 After these 15 manual cycles, Taq polymerase was added and
an additional 22 cycles of the shuffling reaction [94 C for 30
seconds, 35 C for 30 seconds] without primers were performed.
The reaction was then diluted 20-fold. The following
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
69
primers were added to a final concentration of 0.8 M:
5'AACGCCGCATGCAAGCTTGGATCCTTATT3' (SEQ ID NO:5) and
5'AAAGCCCTCTAGATGATTACGAATTCATAT3' (SEQ ID NO:6) and a PCR
reaction was performed as described above in Example 1. The
second primer pair differed from the first pair only because a
change in restriction sites was deemed necessary.
After digestion of the PCR product with XbaI and SphI, the
fragments were ligated into XbaI-SphI-digested 'pUC18. The
sequences of the inserts from several colonies were determined by
a dideoxy DNA sequencing kit (United States Biochemical Co.,
Cleveland OH) according to the manufacturer's instructions.
A total of 17 crossovers were found by DNA sequencing of
nine colonies. Some of the crossovers were based on only 1-2
bases of uninterrupted homology.
It was found that to force efficient crossovers based on
short homologies, a very low effective annealing temperature is
required. With any heat-stable polymerase, the cooling time of
the PCR machine (94 C to 25 C at 1-2 degrees/second) causes the
effective annealing temperature to be higher than the set
annealing temperature. Thus, none of the protocols based on Taq
polymerase yielded crossovers, even when a ten-fold excess of one
of the IL1-2 genes was used. In contrast, a heat-labile
polymerase, such as the Klenow fragment of DNA polymerase I, can
be used to accurately obtain a low annealing temperature.
Example 5. DNA shuffling of the TEM-1 betalactamase gene
The utility of mutagenic DNA shuffling for directed
molecular evolution was tested in a betalactamase model system.
TEM-1 betalactamase is a very efficient enzyme, limited in its
reaction rate primarily by diffusion. This example determines
whether it is possible to change its reaction specificity and
obtain resistance to the drug cefotaxime that it normally does
not hydrolyze.
The minimum inhibitory concentration (MIC) of cefotaxime on
bacterial cells lacking a plasmid was determined by plating 10 Al
of a 10-2 dilution of an overnight bacterial culture (about 1000
cfu) of E. coli XL1-blue cells (Stratagene, San Diego CA) on
plates with varying levels of cefotaxime (Sigma, St. Louis MO),
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
followed by incubation for 24 hours at 37 C.
Growth on cefotaxime is sensitive to the density of cells,
and therefore similar numbers of cells needed to be plated on
each plate (obtained by plating on plain LB plates). Platings of
5 1000 cells were consistently performed.
1) Initial Plasmid Construction
A pUC18 derivative carrying the bacterial TEM-1
betalactamase gene was used (28). The TEM-1 betalactamase gene
confers resistance to bacteria against approximately 0.02 g/ml
10 of cefotaxime. Sfil restriction sites were added 5' of the
promoter and 3' of the end of the gene by PCR of the vector
sequence with two primers:
P r i m e r A ( S E Q I D N 0. 7).
5'TTCTATTGACGGCCTGTCAGGCCTCATATATACTTTAGATTGATTT3' and Primer B
15 ( S E Q I D N 0 8. )
5' TTGACGCACTGGCCATGGTGGCCAAAAATAAACAAATAGGGGTTCCGCGCACATTT3'
and by PCR of the betalactamase gene sequence with two other
primers:
Primer C (SEQ ID NO:9):
20 5' AACTGACCACGGCCTGACAG CCCGGTCTGACAGTTACCAATGCTT, and
Primer D (SEQ ID NO:10):
5'AACCTGTCCTGGC ACCATGGCCTAAATACATTCAAATATGTAT.
The two reaction products-were digested with Sf11, mixed,
ligated and used to transform bacteria.
25 The resulting plasmid was pUC182Sfi. This plasmid contains
an Sfil fragment carrying the TEM-1 gene and the P-3 promoter.
The minimum inhibitory concentration of cefotaxime for E.
colt XL1-blue (Stratagene, San Diego CA) carrying this plasmid
was 0.02 g/ml after 24 hours at 37 C.
30 The ability to improve the resistance of the betalactamase
gene to cefotaxime without shuffling was determined by stepwise
replating of a diluted pool of cells (approximately 107 cfu) on
2-fold increasing drug levels. Resistance up to 1.28 g/ml could
be obtained without shuffling. This represented a 64 fold
35 increase in resistance.
2) DNAseI digestion
The substrate for the first shuffling reaction was dsDNA of
0.9 kb obtained by PCR of pUC182Sfi with primers C and D, both of
CA 02497384 1995-02-17
71
which contain a Sfil site.
The free primers from the PCR product were removed by Wizard
PC#2 prep (Promega, Madison WI) at every cycle.
About 5 gg of the DNA substrate(s) was digested with 0.15
units of DNAseI (Sigma, St. Louis MO) in 100 Al of 50 mM Tris-HC1
pH 7.4, 1 mM MgC12, for 10 min at room temperature. Fragments of
100-300 bp were purified from 2% low melting point agarose gels
by electrophoresis onto DE81 ion exchange paper (Whatman,
Hillsborough OR), elution with 1 M NaCl and ethanol precipitation
by,the method described in Example 1.
3) Gene shuffling
The purified fragments were resuspended in PCR mix (0.2 mM
each dNTP, 2.2 mM MgC12, 50 mM KC1, 10 mM Tris-HC1 pH 9.0, 0.1%
Triton X-100), at a concentration of 10 - 30 ng/ l. No primers
were added at this point. A reassembly program of 94 C for 60
seconds, then 40 cycles of [94 C for 30 seconds, 50-55 C for 30
seconds, 72 C for 30 seconds] and then 72 C for 5 minutes was
used in an MJ Research (Watertown MA) PTC-150 thermocycler.
4) Amplification,of Reassembly Product with primers
After dilution of the reassembly product into the PCR mix
with 0.8 gM of each primer (C and D) and 20 PCR cycles [94 C for
seconds, 50 C for 30 seconds, 72 C for 30 seconds] a single
product 900 bp in size was obtained.
5) Cloning and analysis
25 After digestion of the 900 bp product with the terminal
restriction enzyme SfiI and agarose gel purification, the 900 bp
product was ligated into the vector pUC182Sfi at-the unique SfiI
site with T4 DNA ligase (BRL, Gaithersburg MD). The mixture was
electroporated into E. coli XL1-blue cells and plated on LB
30 plates with 0.32-0.64 g/ml of cefotaxime (Sigma, St. Louis MO).
The cells were grown for up to 24 hours at 37 C and the resulting
colonies were scraped off the plate as a pool and used as the PCR
template for the next round of shuffling.
6) Subsequent Reassembly Rounds
The transformants obtained after each of three rounds of
shuffling were plated on increasing levels of cefotaxime. The
colonies (>100, to maintain diversity) from the plate with the
highest level of cefotaxime were pooled and used as the template
*trade-mark
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
72
for the PCR reaction for the next round.
A mixture of the cefotaximer colonies obtained at 0.32-0.64
g/ml in Step (5) above were used as the template for the next
round of shuffling. 10 ul of cells in LB broth were used as the
template in a reassembly program of 10 minutes at 99 C, then 35
cycles of [94 C for 30 seconds, 52 C for 30 seconds, 72 C for 30
seconds) and then 5 minutes at 72 C as described above.
The reassembly products were digested and ligated into
pUC182Sfi as described in step (5) above. The mixture was
electroporated into E. coli XL1-blue cells and plated on LB
plates having 5-10 g/ml of cefotaxime.
Colonies obtained at 5-10 g/ml were used for a third round
similar to the first and second rounds except the cells were
plated on LB plates having 80-160 g/ml of cefotaxime. After the
third round, colonies were obtained at 80-160 g/ml, and after
replating on increasing concentrations of cefotaxime, colonies
could be obtained at up to 320 g/ml after 24 hours at 37 C
(MIC=320 g/ml).
Growth on cefotaxime is dependent on the cell density,
requiring that all the MICs be standardized (in our case to about
1,000 cells per plate). At higher cell densities, growth at up to
1280 g/ml was obtained. The 5 largest colonies grown at 1,280
g/ml were plated for single colonies twice, and the Sfil inserts
were analyzed by restriction mapping of the colony PCR products.
One mutant was obtained with a 16,000 fold increased
resistance to cefotaxime (MIC=0.02 g/ml to MIC=320 g/ml).
After selection, the plasmid of selected clones was
transferred back into wild-type E. coli XL1-blue cells
(Stratagene, San Diego CA) to ensure that none of the measured
drug resistance was due to chromosomal mutations.
Three cycles of shuffling and selection yielded a 1.6 x 104-
fold increase in the minimum inhibitory concentration of the
extended broad spectrum antibiotic cefotaxime for the TEM-1
betalactamase. In contrast, repeated plating without shuffling
resulted in only a 16-fold increase in resistance (error-prone
PCR or cassette mutagenesis).
7) Sequence analysis
All 5 of the largest colonies grown at 1,280 g/ml had a
CA 02497384 1995-02-17
WO 95/22625 rt iiu~y~,uh1hu
73
restriction map identical to the wild-type TEM-i enzyme. The
SfiI insert of the plasmid obtained from one of these colonies
was sequenced by dideoxy DNA sequencing (United States
Biochemical Co., Cleveland OH) according to the manufacturer's
instructions. All the base numbers correspond to the revised
pBR322 sequence (29), and the amino acid numbers correspond to
the ABL standard numbering scheme (30). The amino acids are
designated by their three letter codes and the nucleotides by
their one letter codes. The term G4205A means that nucleotide
4205 was changed from guanidine to adenine.
Nine single base substitutions were found. G4205A is
located between the -35 and -10 sites of the betalactamase P3
promoter (31) . The promoter up-mutant observed by Chen and
Clowes (31) is located outside of the Sfil fragment used here,
and thus could not have been detected. Four mutations were
silent (A3689G, G3713A, G3934A and T3959A), and four resulted in
an amino acid change (C3448T resulting in Gly238Ser, A3615G
resulting in Met182Thr, C3850T resulting in Glu104Lys, and G4107A
resulting in Alal8Val).
8) Molecular Backcross
Molecular backcrossing with an excess of the wild-type DNA
was then used in order to eliminate non-essential mutations.
Molecular backcrossing was conducted on a selected plasmid
from the third round of DNA shuffling by the method identical to
normal shuffling as described above, except that the DNAseI
digestion and shuffling reaction were performed in the presence
of a 40-fold excess of wild-type TEM-1 gene fragment. To make
the backcross more efficient, very small DNA fragments (30 to 100'
bp) were used in the shuffling reaction. The backcrossed mutants
were again selected on LB plates with 80-160 g/ml of cefotaxime
(Sigma, St. Louis MO).
This backcross shuffling was repeated with DNA from
colonies from the first backcross round in the presence of a 40-
fold excess of wild-type TEM-1 DNA. Small DNA fragments (30-100
bp) were used to increase the efficiency of the backcross. The
second round of backcrossed mutants were again selected on LB
plates with 80-160 gg/ml of cefotaxime.
The resulting transformants were plated on 160 g/ml of
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
74
cefotaxime, and a pool of colonies was replated on increasing
levels of cefotaxime up to 1,280 g/ml. The largest colony
obtained at 1,280 g/ml was replated for single colonies.
This backcrossed mutant was 32,000 fold more resistant than
wild-type. (MIC=640 g/ml) The mutant strain is 64-fold more
resistant to cefotaxime than previously reported clinical or
engineered TEM-1-derived strains. Thus, it appears that DNA
shuffling is a fast and powerful tool for at least several cycles
of directed molecular evolution.
The DNA sequence of the SfiI insert of the backcrossed
mutant was determined using a dideoxy DNA sequencing kit (United
States Biochemical Co., Cleveland OH) according to the
manufacturer's instructions (Table 3). The mutant had 9 single
base pair mutations. As expected, all four of the previously
15, identified silent mutations were lost, reverting to the sequence
of the wild-type gene. The promoter mutation (G4205A) as well as
three of the four amino acid mutations (Glu104Lys, Met182Thr, and
Gly238Ser) remained in the backcrossed clone, suggesting that
they are essential for high level cefotaxime resistance.
However, two new silent mutations (T3842C and A3767G), as well as
three new mutations resulting in amino acid changes were found
(C3441T resulting in Arg241His, C3886T resulting in Gly92Ser, and
G4035C resulting in Ala42Gly). While these two silent mutations
do not affect the protein primary sequence, they may influence
protein expression level (for example by mRNA structure) and
possibly even protein folding (by changing the codon usage and
therefore the pause site, which has been implicated in protein
folding).
CA 02497384 1995-02-17
WO 95/22625 YCl'/UJ9,/Ull2b
Table 3
Mutations in Betalactamase
Mutation Type Non-Backcrossed Backcrossed
5
amino acid Alal8Lys -
change Glu104Lys Glu104Lys
Met182Thr Met182Thr
Gly238Ser Gly238Ser
10, - Ala42Gly
Gly92Ser
silent T3959A -
G3934A -
G3713A -
A3689G -
T3842C
A3767G
20 promoter G4205A G4205A
Both the backcrossed and the non-backcrossed mutants have a
promoter mutation (which by itself or in combination results in
a 2-3 fold increase in expression level) as well as three common
25 amino acid changes (Glu104Lys, Met182Thr and Gly238Ser).
Glu104Lys and Gly238Ser are mutations that are present in several
cefotaxime resistant or other TEM-1 derivatives (Table 4).
9) Expression Level Comparison
The expression level of the betalactamase gene in the wild-
30 type plasmid, the non-backcrossed mutant and in the backcrossed
mutant was compared by SDS-polyacrylamide gel electrophoresis (4-
20%; Novex, San Diego CA) of periplasmic extracts prepared by
osmotic shock according to the method of Witholt, B. (32).
Purified TEM-1 betalactamase (Sigma, St. Louis MO) was used
35 as a molecular weight standard, and E. coli XL1-blue cells
lacking a plasmid were used as a negative control.
The mutant and the backcrossed mutant appeared to produce a
2-3 fold higher level of the betalactamase protein compared to
the wild-type gene. The promoter mutation appeared to result in
40 a 2-3 times increase in betalactamase.
Example 6. Construction of mutant combinations of the TEM-1
betalactamase gene
To determine the resistance of different combinations of
45 mutations and to compare the new mutants to published mutants,
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
76
several mutants were constructed into an identical plasmid
background. Two of the mutations, Glu104Lys and G1y238Ser, are
known as cefotaxime mutants. All mutant combinations constructed
had the promoter mutation, to allow comparison to selected
mutants. The results are shown in Table 4.
Specific combinations of mutations were introduced into the
wild-type pUC182Sfi by PCR, using two oligonucleotides per
mutation. '
The oligonucleotides to obtain the following mutations were:
Ala42Gly
(SEQ ID NO: 11) AGTTGGGTGGACGAGTGGGTTACATCGAACT and (SEQ ID NO:12)
AACCCACTCGTCCACCCAACTGATCTTCAGCAT;
Gln39Lys:
(SEQ ID NO:13) AGTAAAAGATGCTGAAGATAAGTTGGGTGCAC GAGTGGGTT and
(SEQ ID NO:14) ACTTATCTTCAGCATCTTTTACTT;
Gly92Ser:
(SEQ ID NO:15) AAGAGCAACTCAGTCGCCGCATACACTATTCT and (SEQ ID
NO:16) ATGGCGGCGACTGAGTTGCTCTTGCCCGGCGTCAAT;
Glu104Lys:
(SEQ ID NO: 17) TATTCTCAGAATGACTTGGTTAAGTACTCACCAGT CACAGAA and
(SEQ ID N0:18) TTAACCAAGTCATTCTGAGAAT;
Met182Thr:
(SEQ ID NO: 19) AACGACGAGCGTGACACCACGACGCCTGTAGCAATG and (SEQ ID
N0:20) TCGTGGTGTCACGCTCGTCGTT;
Gly2388er alone:
(SEQ ID NO:21) TTGCTGATAAATCTGGAGCCAGTGAGCGTGGGTCTC GCGGTA and
(SEQ ID NO:22) TGGCTCCAGATTTATCAGCAA;
Gly2388er and Arg241His (combined):
(SEQ ID NO:23) ATGCTCACTGGCTCCAGATTTATCAGCAAT and
(SEQ ID NO:24) TCTGGAGCCAGTGAGCATGGGTCTCGCGGTATCATT; G4205A:
(SEQ ID NO:25) AACCTGTCCTGGCCACCATG CCCTAAATACAATCAAA
TATGTATCCGCTTATGAGACAATAACCCTGATA.
These separate PCR fragments were gel purified away from the
synthetic oligonucleotides. 10 ng of each fragment were combined
and a reassembly reaction was performed at 94 C for 1 minute and
then 25 cycles; [94 C for 30 sec, 50 C for 30 seconds and 72 C
for 45 seconds]. PCR was performed on the reassembly product for
CA 02497384 1995-02-17
.. ~- r u~~ PCTIUS95/02126
77
25 cycles in the presence of the Sfil-containing outside primers
(primers C and D from Example 5). The DNA was digested with Sfil
and inserted into the wild-type pUC182Sfi vector. The following
mutant combinations were obtained (Table 4).
Table 4
Name Genotype MIC Source
of MIC
TEM-1 Wild-type 0.02
Glu104Lys 0.08 10
Gly238Ser 016 10
TEM-15 Glul04Lys/Gly238Ser* 10
TEM-3 Glul04Lys/Gly238Ser/Gln39Lys 10 37, 15
2-32
ST-4 Glu104Lys/Gly238Ser/Met182 10
Thr*
ST-1 Glu104Lys/Gly238Ser/Met182 320
Thr/Alal8Val/T3959A/G3713A/
G3934A/A3689G*
ST-2 Glul04Lys/Gly238Ser/Metl82Thr 640
/Ala42Gly/Gly92Ser/Arg24lHis/
T3842C/A3767G*
ST-3 Glu104Lys/Gly238Ser/Met182Thr 640
/Ala42Gly/Gly92Ser/Arg24lHis*
* All of these mutants additionally contain the G4205A promoter
mutation.
It was concluded that conserved mutations account for 9 of
15 doublings in the MIC.
Glu104Lys alone was shown to result only in a doubling of
the MIC to 0.08 g/ml, and Gly238Ser (in several contexts with
one additional amino acid change) resulted only in a MIC of 0.16
g/ml (26). The double mutant Glul04Lys/Gly238Ser has a MIC of
10 g/ml. This mutant corresponds to TEM-15.
These same Glu104Lys and Gly238Ser mutations-, in combination
with Gln39Lys (TEM-3) or Thr263Met (TEM-4) result in a high level
of resistance (2-32 gg/ml for TEM-3 and 8-32 Ag/ml for TEM-4 (34,
35).
A mutant containing the three amino acid changes that were
conserved after the backcross (Glu104Lys/Met182Thr/Gly238Ser)
also had a MIC of 10 Ag/ml. This meant that the mutations that
CA 02497384 1995-02-17
WV YJIShOL, a a i uo iv~ uv
78
each of the new selected mutants had in addition to the three
known mutations were responsible for a further 32 to 64-fold
increase in the resistance of the gene to cefotaxime.
The naturally occurring, clinical TEM-1-derived enzymes
(TEM-1-19) each contain a different combination of only 5-7
identical mutations (reviews). Since these mutations are in well
separated locations in the gene, a mutant with high cefotaxime
resistance cannot be obtained by cassette mutagenesis of a single
area. This may explain why the maximum MIC that was obtained by
the standard cassette mutagenesis approach is only 0.64 g/ml
(26). For example, both the Glu104Lys as well as the Gly238Ser
mutations were found separately in this study to have MICs below
0.16 g/ml. Use of DNA shuffling allowed combinatoriality and
thus the Glul04Lys/Gly238Ser combination was found, with a MIC of
10 g/ml.
An important limitation of this example is the use of a
single gene as a starting point. It is contemplated that better
combinations can be found if a large number of related, naturally
occurring genes are shuffled. The diversity that is present in
such a mixture is more meaningful than the random mutations that
are generated by mutagenic shuffling. For example, it is
contemplated that one could use a repertoire of related genes
from a single species, such as the pre-existing diversity of the
immune system, or related genes obtained from many different
species.
Example 7. Improvement of antibody A1OB by DNA shuffling of a
library of all six mutant CDRs.
The A10B scFv antibody, a mouse anti-rabbit IgG, was a gift
from Pharmacia (Milwaukee WI). The commercially available
Pharmacia phage display system was used, which uses the pCANTAB5
phage display vector.
The original A10B antibody reproducibly had only a low
avidity, since clones that only bound weakly to immobilized
antigen (rabbit IgG), (as measured by phage ELISA (Pharmacia
assay kit) or by phage titer) were obtained. The concentration
of rabbit IgG which yielded 50% inhibition of the A10B antibody
binding in a competition assay was 13 picomolar. The observed
CA 02497384 1995-02-17
79
low avidity may also be due to instability of the A10B clone.
The A10B scFv DNA was sequenced (United States Biochemical'
Co., Cleveland OH). according to the manufacturer's instructions.
The sequence was similar to existing antibodies, based on
comparison to Kabat (33).
1) Preparation of phage DNA
Phage DNA having the A10B wild-type antibody gene (10 ul)
was incubated at 99 C for 10 min, then at 72 C for 2. min. PCR
mix (50 mM KC1, 10 mM Tris-HC1 pH 9.0, 0.1% Triton X-100, 200 M
each dNTP, 1.9 mM MgCl), 0.6 pm of each primer and 0.5 Al Taq DNA
Polymerase (Promega, Madison WI) was added to the phage DNA. A
PCR program was run for 35 cycles of (30 seconds at 94 C, 30
seconds at 45 C, 45 seconds at 72 C). The primers used were:
5' ATGATTACGCCAAGCTTT 3' (SEQ ID NO:26) and
5' TTGTCGTCTTTCCAGACGTT 3' (SEQ ID NO:27).
The 850 bp PCR product was then electrophoresed and purified
from a 2% low melting point agarose gel.
2) Fragmentation
300 ng of the gel purified 850 bp band was digested with
0.18 units of DNAse I (Sigma, St. Louis MO) in 50 mM Tris-HC1 pH
7.5, 10 mM MgC1 for 20 minutes at room temperature. The digested
DNA was separated on a 2% low melting point agarose gel and bands
between 50 and 200 bp were purified from the gel.
3) Construction of Test Library
The purpose of this experiment was to test whether the
insertion of the CDRs would be efficient.
The following CDR sequences having internal restriction
enzyme sites were synthesized. "CDR H" means a CDR in the heavy
chain and "CDR L" means a CDR in the light chain of the antibody.
CDR Oligos with restriction sites:
CDR H1 (SEQ ID NO:34)
5' TTCTGGCTACATCTTCACAGAATTCATCTAGATTTGGGTGAGGCAGACGCCTGAA3'
CDR H2 (SEQ ID NO:35)
5' ACAGGGACTTGAGTGGATTGGAATCACAGTCAAGCTTATCCTTTATCTCAGGTCTCGAGTT
CCAAGTACTTAAAGGGCCACACTGAGTGTA 3'
CDR H3 (SEQ ID NO:36)
5' TGTCTATTTCTGTGCTAGATCTTGACTGCAGTCTTATACGAGGATCCATTGGGGCCAAGGG
*trade-mark
CA 02497384 1995-02-17
ACCAGGTCA 3'
CDR Ll (SEQ ID NO:37)
5'AGAGGGTCACCATGACCTGCGGACGTCTTTAAGCGATCGGGCTGATGGCCTGGTACCAACA
GAAGCCTGGAT 3'
5 CDR L2 (SEQ ID NO:38)
5' TCCCCCAGACTCCTGATTTATTAAGGGAGATCTAAACAGCTGTTGGTCCCTTTTCGCTTCAGT
3'
CDR L3 (SEQ ID NO:39) V~
5' ATGCTGCCACTTATTACTGCTTCTGCGCGCTTAAAGGATATCTTCATTTCGGAGGGGGGAC
10 CAAGCT 3'
The CDR oligos were added to the purified A10B antibody DNA
fragments of between 50 to 200 bp from step (2) above at a 10
fold molar excess. The PCR mix (50 mM KC1, 10 mM Tris-HC1 pH
9.0, 0.1% Triton* x-100, 1.9 mM MgCl, 200 Am each dNTP, 0.3 pl
15 Taq DNA polymerase (Promega, Madison WI), 50 gl total volume) was
added and the shuffling program run for 1 min at 94 C, 1 min at
72 C, and then 35 cycles: 30 seconds at 94 C, 30 seconds at 55 C,
30 seconds at 72 C.
1 Al of the shuffled mixture was added to 100 Al of a PCR
20 mix (50 mM KC1, 10 mM Tris-HC1 pH 9.0, 0.1% Triton X-100, 200 in
each dNTP, 1.9 mM MgCl, 0.6 MM each of the two outside primers
(SEQ ID NO:26 and 27, see below), 0.5 gl Taq DNA polymerase) and
the PCR program was run for 30 cycles of [30 seconds at 94 C, 30
seconds at 45 C, 45 seconds at 72 C]. The resulting mixture of
25 DNA fragments of 850 basepair size was phenol/chloroform
extracted and ethanol precipitated.
The outside primers were:
Outside Primer 1: SEQ ID N0:27
5' TTGTCGTCTTTCCAGACGTT 3'
30 Outside Primer 2: SEQ ID NO:26
5' ATGATTACGCCAAGCTTT 3'
The 850 bp PCR product was digested with the restriction
enzymes SfiI and NotI, purified from a low melting point agarose
gel, and ligated into the pCANTAB5 expression vector obtained
35 from Pharmacia, Milwaukee WI. The ligated vector was
electroporated according to the method set forth by Invitrogen
(San Diego CA) into TG1 cells (Pharmacia, Milwaukee WI) and
plated for single colonies.
*trade-mark
CA 02497384 1995-02-17
81
The DNA from the resulting colonies was added to 100 gl of
a PCR mix (50 mM KC1, 10 mM Tris-HC1 pH 9.0, 0.1% Triton-X-100,
200 pm each dNTP, 1.9 mM MgCl, 0.6 pM of Outside primer 1 (SEQ ID
No. 27; see below) six inside primers (SEQ ID NOS:40-45; see
below), and 0.5 pl Taq DNA polymerase) and a PCR program was run
for 35 cycles of [30 seconds at 94 C, 30 seconds at 45 C, 45
seconds at 72 C). The sizes of the PCR products were determined
by agarose gel electrophoresis, and were used to determine which
CDRs with restriction sites were inserted.
CDR Inside Primers:
H 1 (SEQ ID NO:40) 5' AGAATTCATCTAGATTTG 3',
H 2 (SEQ ID NO:41) 5' GCTTATCCTTTATCTCAGGTC 3',
H 3 (SEQ ID NO:42) 5' ACTGCAGTCTTATACGAGGAT 3'
L 1 (SEQ ID NO:43) 5' GACGTCTTTAAGCGATCG 3',
L 2 (SEQ ID NO:44) 5' TAAGGGAGATCTAAACAG 31,
L 3 (SEQ ID NO:45) 5' TCTGCGCGCTTAAAGGAT 3'
The six synthetic CDRs were inserted at the expected
locations in the wild-type A10B antibody DNA (Figure 7). These
studies showed that, while each of the six CDRs in a specific
2200 clone has a small chance of being a CDR with a restriction-site,
most of the clones carried at least one CDR with a restriction
site, and that any possible combination of CDRs with restriction
sites was generated.
4) Construction of Mutant Complementarity Determining Regions
("CDRs")
Based on our sequence data six oligonucleotides
corresponding to the six CDRs were made. The CDRs (Kabat
definition) were synthetically mutagenized at a ratio of 70
(existing base) :10:10:10, and were flanked on the 5' and 3' sides
by about 20 bases of flanking sequence, which provide the
homology for the incorporation of the CDRs when mixed into a
mixture of unmutagenized antibody gene fragments in a molar
excess. The resulting mutant sequences are given below.
Oligos for CDR Library
CDR H1 (SEQ ID NO:28)
5' TTCTGGCTACATCTTCACAACTTATGATATAGACTGGGTGAGGCAGACGCCTGAA 3'
CDR H2 (SEQ ID NO:29)
5'ACAGGGACTTGAGTGGATTGGATGGATTTTTCCTGGAGAGGGTGGTACTGAATACAATGAG
*trade-mark
CA 02497384 1995-02-17
82
AAGTTCAAGGGCAGGGCCACACTGAGTGTA 3'
CDR H3 (SEQ ID NO:30)
5' TGTCTATTTCTGTGCTAGAGGGGACTACTATAGGCGCTACTTTGACTTGTGGGGCCAAGGG
ACCACGGTCA 3'
CDR LI (SEQ ID NO:31)
5' AGAGGGTCACCATGACCTGCAGTGCCAGCTCAGGTATACGTTACATATATTGGTACCAACA
GAAGCCTGGAT 3'
CDR L2 (SEQ ID NO:32)
5' TCCCCCAGACTCCTGATTTATGACACATCCAACGTGGCTCCTGGAGTCCCTTTTCGCTTCAGT
3'
C D R L 3 ( S E Q I D N 0 3 3
'5'ATGCTGCCACTTATTACTTGCCAGGAGTGGAGTGGTTATCCGTACACGTTCGGAGGGGGGA
CCAAGCT 3' .
Bold and underlined sequences were the mutant sequences
synthesized using a mixture of nucleosides of 70:10:10:10 where
70% was the wild-type nucleoside.
A 10 fold molar excess of the CDR mutant oligos were added
to the purified A1OB antibody DNA fragments between 50 to 200 bp
in length from step (2) above. The PCR mix (50 mM KC1, 10 mM
Tris-HC1 pH 9.0, 0.1% Triton x-100, 1.9 mM MgCl, 200 m each
dNTP, 0.3 gl Taq DNA polymerase (Promega, Madison WI), 50 IL1
total volume) was added and the shuffling program run for 1 min
at 94 C, 1 min at 72 C, and then 35 cycles: [30 seconds at 94 C,
seconds at 55 C, 30 seconds at 72 C].
25 1 Al of the shuffled mixture was added to 100 Al of a PCR
mix (50 mM KC1, 10 mM Tris-HC1 pH 9.0, 0.1% Triton X-100, 200 m
each dNTP, 1.9 mM MgCl, 0.6 AM each of the two outside primers
(SEQ ID NO:26 and 27, see below), 0.5 Al Taq DNA polymerase) and
the PCR program was run for 30 cycles of [30 seconds at 94 C, 30
30 seconds at 45 C, 45 seconds at 72 C]. The resulting mixture of
DNA fragments of 850 basepair size was phenol/chloroform
extracted and ethanol precipitated.
The outside primers were:
Outside Primer 1: SEQ ID NO:27 5' TTGTCGTCTTTCCAGACGTT 3'
Outside Primer 2: SEQ ID NO:26 5' ATGATTACGCCAAGCTTT 3'
5) Cloning of the scFv antibody DNA into pCANTAB5
The 850 bp PCR product was digested with the restriction
enzymes SfiI and NotI, purified from a low melting point agarose
*trade-mark
CA 02497384 1995-02-17
83
gel, and ligated into the pCANTAB5 expression vector obtained
from Pharmacia, Milwaukee Wi. The ligated vector was
electroporated according to the method set forth by Invitrogen
(San Diego CA) into TG1 cells (Pharmacia, Milwaukee WI) and the
phage library was grown up using helper phage following the
guidelines recommended by the manufacturer.
The library that was generated in this fashion was screened
for the presence of improved antibodies, using six- cycles.of
selection.
6) Selection of high affinity clones
wells of a 96 well microtiter plate were coated with
Rabbit IgG (Jackson Immunoresearch, Bar Harbor ME) at 10 g /well
for 1 hour at 37 C, and then blocked with 2% non-fat dry milk in
PBS for 1 hour at 37 C.
15 100 Al of the phage library (1x1010 cfu) was blocked with 100
gl of 2% milk for 30 minutes at room temperature, and then added
to each of the 15 wells and incubated for 1 hour at 37 C.
Then the wells were washed three times with PBS containing
0.5% Tween-+20 at 37 C for 10 minutes per wash. Bound phage was
eluted with 100 gl elution buffer (Glycine-HC1, pH 2.2), followed
by immediate neutralization with 2M Tris pH 7.4 and transfection
for phage production. This selection cycle was repeated six
times.
After the sixth cycle, individual phage clones were picked
and the relative affinities were compared by phage ELISA, and the
specificity for the rabbit IgG was assayed with a kit from
Pharmacia (Milwaukee WI) according to the methods recommended by
the manufacturer.
The best clone has an approximately 100-fold improved
expression level compared with the wild-type A1OB when tested by
the Western assay. The concentration of the rabbit IgG which
yielded 50% inhibition in a competition assay with the best clone
was 1 picomolar. The best clone was reproducibly specific for
rabbit antigen. The number of copies of the antibody displayed
by the phage appears to be increased.
Example 8. In vivo recombination via direct repeats of
partial genes
A plasmid was constructed with two partial, inactive copies
*trade-mark
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
84
of the same gene (beta-lactamase) to demonstrate that
recombination between the common areas of these two direct
repeats leads to full-length, active recombinant genes.
A pUC18 derivative carrying the bacterial TEM-1
betalactamase gene was used (Yanish-Perron et al., 1985, Gene
33:103-119). The TEM-1 betalactamase gene ("Bla") confers
resistance to bacteria against approximately 0.02 g/ml of
cefotaxime. Sfil restriction sites were added 5' of the promoter
and 3' of the end of the betalactamase gene by PCR of the vector
sequence with two primers:
Primer A (SEQ ID NO: 46)
5' TTCTATTGACGGCCTGTCAGGCCTCATATATACTTTAGATTGATTT 3'
PRIMER B (SEQ ID NO: 47)
5' TTGACGCACTGGCCATGGTGGCCAAAAATAAACAAATAGGGGTTCCGCGCAC
ATTT 3'
and by PCR of the beta-lactamase gene sequence with two other
primers:
Primer C (SEQ ID NO: 48)
5' AACTGACCACGGCCTGACAGGCCGGTCTGACAGTTACCAATGCTT 3'
Primer D (SEQ ID NO: 49)
5' AACCTGTCCTGGCCACCATGGCCTAAATACATTCAAATATGTAT 3'
The two reaction products were digested with Sfil, mixed,
ligated and used to transform competent E. coli bacteria by the
procedure described below. The resulting plasmid was pUC182Sfi-
Bla-Sfi. This plasmid contains an Sfil fragment carrying the Bla
gene and the P-3 promoter.
The minimum inhibitory concentration of cefotaxime for E.
coli XL1-blue (Stratagene, San Diego CA) carrying pUC182Sfi-Bla-
Sfi was 0.02 g/ml after 24 hours at 37 C.
The tetracycline gene of pBR322 was cloned into pUC18Sfi-
Bla-Sfi using the homologous areas, resulting in pBR322TetSfi-
Bla-Sfi. The TEM-i gene was then deleted by restriction
digestion of the pBR322TetSfi-Bla-Sfi with SspI and FspI and
blunt-end ligation, resulting in pUC322TetSfi-Sfi.
Overlapping regions of the TEM-1 gene were amplified using
standard PCR techniques and the following primers:
Primer 2650 (SEQ ID NO: 50) 5' TTCTTAGACGTCAGGTGGCACTT 3'
Primer 2493 (SEQ ID NO: 51) 5' TTT TAA ATC AAT CTA AAG TAT 3'
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
P r i m e r 2 6 5 1 ( S E Q I D N O: 5 2) 5'
TGCTCATCCACGAGTGTGGAGAAGTGGTCCTGCAACTTTAT 3,'and
Primer 2652 (SEQ ID NO: 53)
ACCACTTCTCCACACTCGTGGATGAGCACTTTTAAAGTT
5 The two resulting DNA fragments were digested with Sfil and
BstXl and ligated into the Sfi site of pBR322TetSfi-Sfi. The
resulting plasmid was called pBR322Sfi-BL-LA-Sfi. A map of the
plasmid as well as a schematic of intraplasmidic recombination
and reconstitution of functional beta-lactamase is shown in
10 Figure 9.
The plasmid was electroporated'into either TG-1 or JC8679 E.
coli cells. E. coli JC8679 is RecBC sbcA (Oliner et al., 1993,
NAR 21:5192). The cells were plated on solid agar plates
containing tetracycline. Those colonies which grew, were then
15 plated on solid agar plates containing 100 g/ml ampicillin and
the number of viable colonies counted. The beta-lactamase gene
inserts in those transformants which exhibited ampicillin
resistance were amplified by standard PCR techniques using
Primer 2650 (SEQ ID NO: 50) 5' TTCTTAGACGTCAGGTGGCACTT 3' and
20 Primer 2493 (SEQ ID NO: 51) 5' TTTTAAATCAATCTAAAGTAT 3' and the
length of the insert measured. The presence of a 1 kb insert
indicates that the gene was successfully recombined, as shown in
Fig. 9 and Table 5.
TABLE 5
25 Cell Tet Colonies Amp colonies Colony PCR
TG-1 131 21 3/3 at 1 kb
JC8679 123 31 4/4 at 1 kb
vector 51 0
control
30,
About 17-25% of the tetracycline-resistant colonies were
also ampicillin-resistant and all of the Ampicillin resistant
colonies had correctly recombined, as determined by colony PCR.
Therefore, partial genes located on the same plasmid will
35 successfully recombine to create a functional gene.
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
86
Example 9. In vivo recombination via direct repeats of
full-length genes.
A plasmid with two full-length copies of different alleles
of the beta-lactamase gene was constructed. Homologous
recombination of the two genes resulted in a single recombinant
full-length copy of that gene.
The construction of pBR322TetSfi-Sfi and pBR322TetSfi-Bla-
Sfi was described above.
The two alleles of the beta-lactamase gene were constructed
as follows. Two PCR reactions were conducted with pUC18Sfi-Bla-
Sfi as the template. One reaction was conducted with the
following primers.
Primer 2650 (SEQ ID NO: 50) 5' TTCTTAGACGTCAGGTGGCACTT 3'
Primer 2649 (SEQ ID NO: 51)
5' ATGGTAGTCCACGAGTGTGGTAGTGACAGGCCGGTCTGACAGTTA
CCAATGCTT 3'
The second PCR reaction was conducted with the following primers:
Primer 2648 (SEQ ID NO: 54)
5' TGTCACTACCACACTCGTGGACTACCATGGCCTAAATACATTCAAA
TATGTAT 3'
Primer 2493 (SEQ ID NO: 51) 5' TTT TAA ATC AAT CTA AAG TAT 3'
This yielded two Bla genes, one with a 5' Sfil site and a 3'
BstX] site, the other with a 5' BstX] site and a 3' Sfil site.
After digestion of these two genes with BstXl and Sfil, and
ligation into the Sfil-digested plasmid pBR322TetSfi-Sfi, a
plasmid (pBR322-Sfi-2BLA-Sfi) with a tandem repeat of the Bla
gene was obtained. (See Figure 10).
The plasmid was electroporated into E. coli cells. The
cells were plated on solid agar plates containing 15 g/ml
tetracycline. Those colonies which grew, were then plated on
solid agar plates containing 100 gg/ml ampicillin and the number
of viable colonies counted. The Bla inserts in those
transformants which exhibited ampicillin resistance were
amplified by standard PCR techniques using the method and primers
described in Example 8. The presence of a 1 kb insert indicated
that the duplicate genes had recombined, as indicated in Table 6.
CA 02497384 1995-02-17
WO 95/22625 r~ i/U YzI,~cicv
87
TABLE 6
Cell Tet Colonies Amp Colonies Colony PCR
TG-1 28 54 7/7 at lkb
JC8679 149 117 3/3 at lkb
vector 51 0
control
Colony PCR confirmed that the tandem repeat was efficiently
recombined to form a single recombinant gene.
Example 10. Multiple cycles of direct repeat
recombination - Interplasmidic
In order to determine whether multiple cycles of
recombination could be used to produce resistant cells more
quickly, multiple cycles of the method described in Example 9
were performed.
The minus recombination control consisted of a single copy
of the betalactamase gene, whereas the plus recombination
experiment consisted of inserting two copies of betalactamase as
a direct repeat. The tetracycline marker was used to equalize
the number of colonies that were selected for cefotaxime
resistance in each round, to compensate for ligation
efficiencies.
In the first round, pBR322TetSfi-Bla-Sfi was digested with
EcrI and subject to PCR with a 1:1 mix (1 ml) of normal and
Cadwell PCR mix (Cadwell and Joyce (1992) PCR Methods and
Applications 2: 28-33) for error prone PCR. The PCR program was
70 C for 2 minutes initially and then 30 cycles of 94 C for 30
seconds, 52 C for 30 second and 72 C for 3 minutes and 6 seconds
per cycle, followed by 72 C for 10 minutes.
The primers used in the PCR reaction to create the one Bla
gene control plasmid were Primer 2650 (SEQ ID NO: 50) and Primer
2719 (SEQ ID NO: 55) 5' TTAAGGGATTTTGGTCATGAGATT 3'. This
resulted in a mixed population of amplified DNA fragments,
designated collectively as Fragment #59. These fragments had a
number of different mutations.
CA 02497384 1995-02-17
WO 95/22625 PCT/US95/02126
88
The primers used in two different PCR reactions to create
the two Bla gene plasmid were Primer 2650 (SEQ ID NO: 50) and
Primer 2649 (SEQ ID NO: 51) for the first gene and Primers 2648
(SEQ ID NO: 54) and Primer 2719 (SEQ ID NO: 55) for the second
gene. This resulted in a mixed population of each of the two
amplified DNA fragments: Fragment #89 (amplified with primers
2648 and 2719) and Fragment #90 (amplified with primers 2650 and
2649). In each case a number of different mutations had been
introduced the mixed population of each of the fragments.
After error prone PCR, the population of amplified DNA
fragment #59 was digested with Sfil, and then cloned into
pBR322TetSfi-Sfi to create a mixed population of the plasmid
pBR322Sfi-Bla-Sf il.
After error prone PCR, the population of amplified DNA
fragments #90 and #89 was digested with Sfil and BstXI at 50 C,
and ligated into pBR322TetSfi-Sfi to create a mixed population of
the plasmid pBR322TetSfi-2Bla-Sfil (Fig. 10).
The plasmids pBR322Sfi-Bla-Sfi' and pBR322Sfi-2Bla-Sfi' were
electroporated into E. coli JC8679 and placed on agar plates
having differing concentrations of cefotaxime to select for
resistant strains and on tetracycline plates to titre.
An equal number of colonies (based on the number of colonies
growing on tetracycline) were picked, grown in LB-tet and DNA
extracted from the colonies. This was one round of the
recombination. This DNA was digested with EcrI and used for a
second round of error-prone PCR as described above.
After five rounds the MIC (minimum inhibitory concentration)
for cefotaxime for the one fragment plasmid was 0.32 whereas the
MIC for the two fragment plasmid was 1.28. The results show that
after five cycles the resistance obtained with recombination was
four-fold higher in the presence of in vivo recombination.
Example 11. In vivo recombination via electroporation of
fragments
Competent E. coli cells containing pUC18Sfi-Bla-Sfi were
prepared as described. Plasmid pUC18Sfi-Bla-Sfi contains the
standard TEM-1 beta-lactamase gene as described, supra.
A TEM-1 derived cefotaxime resistance gene from pUCi8Sfi-
cef-Sfi, (clone ST2) (Stemmer WPC (1994) Nature 370: 389-91,
CA 02497384 1995-02-17
89
which confers on E. coli
carrying the plasmid an MIC of 640 ig/ml for cefotaxime, was
obtained. In one experiment the complete plasmid pUC18Sfi-cef-
Sfi DNA was electroporated into E. coli cells having the plasmid
pUC18Sfi-Bla-Sfi.
In another experiment the DNA fragment containing the
cefotaxime gene from pUC18Sfi-cef-Sfi was amplified by PCR using
the primers 2650 (SEQ ID NO: 50) and 2719 (SEQ ID NO:.55). The
resulting 1 kb PCR product was digested into DNA fragments of
<100 bp by DNase and these fragments were electroporated into the
competent E. coli cells which already contained pUC18Sfi-Bla-Sfi.
The transformed cells from both experiments were then
assayed for their resistance to cefotaxime by plating the
transformed cells onto agar plates having varying concentrations
of cefotaxime. The results are indicated in Table 7.
TABLE 7
Colonies/ Cefotaxime Concentration
0.16 0.32 1.28 5.0 10.0
no DNA control 14
ST-2 mutant, whole 4000 2000 800 400
ST-2 mutant, fragments 1000 120 22 7
Wildtype, whole 27
Wildtype, fragments 18
From the results it appears that the whole ST-2 Cef gene was
inserted into either the bacterial genome or the plasmid after
electroporation. Because most insertions are homologous, it is
expected that the gene was inserted into the plasmid, replacing
the wildtype gene. The fragments of the Cef gene from St-2 also
inserted efficiently into the wild-type gene in the plasmid. No
sharp increase in cefotaxime resistance was observed with the
introduction of the wildtype gene (whole or in fragments) and no
DNA. Therefore, the ST-2 fragments were shown to yield much
greater cefotaxime resistance than the wild-type fragments.
It was contemplated that repeated insertions of fragments,
CA 02497384 1995-02-17
WO 95/22625 rL I/USY /U11LO
prepared from increasing resistant gene pools would lead to
increasing resistance.
Accordingly, those colonies that produced increased
cefotaxime resistance with the St-2 gene fragments were isolated
5 and the plasmid DNA extracted. This DNA was amplified using PCR
by the method described above. The amplified DNA was digested
with DNase into fragments (<100 bp) and 2-4 g of the fragments
were electroporated into competent E. coli cells already
containing pUC322Sfi-Bla-Sfi as described above. The transformed
10 cells were plated on agar containing varying concentrations of
cefotaxime.
As a control, competent E. coli cells having the plasmid
pUC18Sfi-Kan-Sfi were also used. DNA fragments from the
digestion of the PCR product of pUC18Sfi-cef-Sfi were
15 electroporated into these cells. There is no homology between
the kanamycin gene and the beta-lactamase gene and thus
recombination should not occur.
This experiment was repeated for 2 rounds and the results
are shown in Table 8.
20 TABLE 8
Round Cef conc. KAN control Cef resistant
colonies
1 0.16-0.64 lawn lawn
replate 0.32 10 small 1000
2 10 10 400
25 Replate 100sm @ 2.5 50 @ 10
3 40 100 sm
1280 100 sm
Example 12 Determination of Recombination Formats
30 This experiment was designed to determine which format of
recombination generated the most recombinants per cycle.
In the first approach, the vector pUC18Sfi-Bla-Sfi was
amplified with PCR primers to generate a large and small
fragment. The large fragment had the plasmid and ends having
CA 02497384 1995-02-17
WO 95/22625 YC'1 /US9 Iu211b
91
portions of the Bla gene, and the small fragment coded for the
middle of the Bla gene. A third fragment having the complete Bla
gene was created using PCR by the method in Example 8. The
larger plasmid fragment and the fragment containing the complete
Bla gene were electroporated into E. coli JC8679 cells at the
same time by the method described above and the transformants
plated on differing concentrations of cefotaxime,.
In approach 2, the vector pUC18Sfi-Bla-Sfi was amplified to
produce the large plasmid fragment isolated as in approach 1
above. The two fragments each comprising a portion of the
complete Bla gene, such that the two fragments together spanned
the complete Bla gene werealso obtained by PCR. The large
plasmid fragment and the two Bla gene fragments were all
electroporated into competent E. coli JC8679 cells and the
transformants plated on varying concentrations of cefotaxime.
In the third approach, both the vector and the plasmid were
electroporated into E. coli JC8679 cells and the transformants
were plated on varying concentrations of cefotaxime.
In the fourth approach, the complete Bla gene was
electroporated into E. coli JC8679 cells already containing the
vector pUCSfi-Sfi and the transformants were plated on varying
concentrations of cefotaxime. As controls, the E. coli JC8679
cells were electroporated with either the complete Bla gene or
the vector alone.
The results are presented in Figure 11. The efficiency of
the insertion of two fragments into the vector is 100 X lower
than when one fragment having the complete Bla gene is used.
Approach 3 indicated that the efficiency of insertion does depend
on the presence of free DNA ends since no recombinants were
obtained with this approach. However, the results of approach 3
were also due to the low efficiency of electroporation of the
vector. When the expression vector is already in the competent
cells, the efficiency of the vector electroporation is not longer
a factor and efficient homologous recombination can be achieved
even with uncut vector.
Example 12. Kit for cassette shuffling to optimize vector
performance
In order to provide a vector capable of conferring an
CA 02497384 1995-02-17
WO 95/22625 PCTIUS95/02126
92
optimized phenotype (e.g., maximal expression of a vector-encoded
sequence, such as a cloned gene), a kit is provided comprising a
variety of cassettes which can be shuffled, and optimized
shufflants can be selected. Figure 12 shows schematically one
embodiment, with each loci having a plurality of cassettes. For
example, in a bacterial expression system, Figure 13 shows
example cassettes that are used at the respective loci. Each
cassette of a given locus (e.g., all promoters in this example)
are flanked by substantially identical sequences capable of
1 overlapping the flanking sequence(s) of cassettes of an adjacent
locus and preferably also capable of participating in homologous
recombination or non-homologous recombination (e.g., lox/cre or
flp/frt systems), so as to afford shuffling of cassettes within
a locus but substantially not between loci.
Cassettes are supplied in the kit as PCR fragments,
which each cassette type or individual cassette species packaged
in a separate tube. Vector libraries are created by combining
the contents of tubes to assemble whole plasmids or substantial
portions thereof by hybridization of the overlapping flanking
sequences of cassettes at each locus with cassettes at the
adjacent loci. The assembled vector is ligated to a
predetermined gene of interest to form a vector library wherein
each library member comprises the predetermined gene of interest
and a combination of cassettes determined by the association of
cassettes. The vectors are transferred into a suitable host cell
and the cells are cultured under conditions suitable for
expression, and the desired phenotype is selected.
While the present invention has been described with
reference to what are considered to be the preferred examples, it
is to be understood that the invention is not limited to the
disclosed examples. To the contrary, the invention is intended
to cover various modifications and equivalent arrangements
included within the spirit and scope of the appended claims.
35,
CA 02497384 1995-02-17
WO 95/22625 r l.l/ uJ7J va avv
93
REFERENCES
The following references are cited in this application at
the relevant portion of the application.
1. Holland, J. H. (1992) Sci. Am. July, 66-72.
2. Holland, J. H. (1992) "Adaptation in natural and artificial
systems". Second edition, MIT Press, Cambridge.
3. Joyce, G. F. (1992) Scientific American, December, 90-97.
4. Kauffman, S. A. (1993) "The origins of order". Oxford
University Press, New York.
5. Stormo, G. D. (1991) Methods Frnzymol. 208:458-468.
6. Schneider, T. D. et al., (1986) J. Mot. Biol. 188:415-431.
7. Reidhaar-Olson, J. F and Sauer, R.T. (1988) Science 241:53-
57.
8. Stemmer, W. P. C. et al., (1992) Biotechniques 14:156-265.
9. Yockey, H. P. (1977) J. Theor. Biol. 67:345-376.
10. Yockey, H. P. (1974) J. Theor. Biol. 46:369-380.
11. Leung, D. W. at al., (1989) Technique 1:11-15.
12. Caldwell, R. C. and Joyce, G. F. (1992) PCR Methods and
Applications 2:28-33.
13. Bartel, D. P., and Szostak, J. W. (1993) Science 261:1411-
1418.
14. Bock, L. C. et al., (1992) Nature 355:564-566.
15. Scott, J. K. and Smith, G. P. (1990) Science 249:386-390.
16. Cwirla, S. E. et al. (1990) Proc. Natl. Acad. Sci. USA
87:6378-6382.
17. McCafferty, J. at al. (1990) Nature 348:552-554.
18. Cull, M. G. et al., (1992) Proc. Natl. Acad. Sci. USA
89:1865-1869.
19. Gramm, H. et al., (1992) Proc. Natl. Acad. Sci. USA 89:3576-
3580.
50, 20. Arkin, A. and Youvan, D. C. (1992) Proc. Natl. Acad. Sci.
USA 89:7811-7815.
21. Oliphant, A. R. et al., (1986) Gene 44:177-183.
22. Hermes, J. D. at al., (1990) Proc. Natl. Acad. Sci. USA
87:696-700.
CA 02497384 1995-02-17
WO 95/22625 YC:T/US95/U212b
94
23. Meyerhans, A. et al., (1990) Nucleic Acids Res. 18:1687-
1691.
24,. Osterhout, J. J. et al., (1992) J. Am. Chem. Soc. 114:331-
337.
25. Cano, R. J. et al., (1993) Nature 363:536-538.
26. Palzkill and Botstein, (1992) J. Bacteriol. 174:5237-5243.
27. Marton et al., Nucleic Acids Res. 19:2423.
28. Yanish-Perron et al., [1985] Gene 33:103-119.
.29. Watson (1988) Gene 70:399-403.
30. Ambler et al. (1991) Biochem J. 276:269-272.
31. Chen and Clowes, (1984) Nucleic Acid Res. 12:3219-3234.
32. Witholt, B. ([1987] Anal. Biochem. 164(2):320-330
33. Kabat et al., (1991) "Sequences of Proteins of Immunological
Interest" U.S. Department of Health and Human Services, NIH
Publication 91-3242.
34. Philippon et al., (1989) Antimicrob Agents Chemother
33:1131-1136.
35. Jacoby and Medeiros (1991) Antimicrob. Agents Chemother.
35:167-1704.
36. Coelhosampaio (1993) Biochem. 32:10929-10935
37. Tuerk, C. et al., (1992) Proc. Natl. Acad. Sci. USA 89:6988-
6992.
38. United States Patent No. 4,683,195
10 39. United States Patent No. 4,683,202
40. Delagrave et al. (1993) Protein Engineering 6: 327-331
41. Delgrave et al. (1993) Bio/Technology 11: 1548-1552
42. Goldman, ER and Youvan DC (1992) Bio/Technology 10:1557-1561
43. Nissim et al. (1994) EMBO J. 13: 692-698
44. Winter et al. (1994) Ann. Rev. Immunol. 12: 433-55
45. Caren et al. (1994) Bio/Technology 12: 517-520
46. Calogero et al. (1992) FEMS Microbiology Lett. 97: 41-44
47. Galizzi et al. W091/01087
CA 02497384 1995-02-17
48. Hayashi et al. (1994) Biotechniques 17: 310-315
49. Radman et al. W090/07576