Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Differential Diagnosis of Colorectal Cancer and other Diseases of the Colon
The present invention provides biomolecules and the use of these biomolecules
for the differential
diagnosis of colorectal cancer or a non-malignant disease of the large
intestine. In specific
embodiments, the biomolecules are characterised by mass profiles generated by
contacting a test
and/or biological sample with an anion exchange surface under specific binding
conditions and
detecting said biomolecules using gas phase ion spectrometry. The biomolecules
used according to the
invention are preferably proteins or polypeptides. Furthermore, . preferred
test and/or biological
samples are blood serum samples and are of human origin.
BACKGROUND TO THE INVENTION
Colorectal cancer is the fourth most common cancer in the world to date, and
accounts for
approximately 20b,000 deaths per year in Europe and the US alone. Although
colorectal cancer
generally affects both men and women equally (currently at 9.4% and 10.1% of
incident cancer,
respectively), its distribution as a leading cause of death in men and women
is disproportionate.
Whereas colorectal cancer is the fourth leading cancer-related cause of death
in men (following lung,
stomach and prostate cancer), in women it takes second place to breast cancer.
Furthermore, colorectal
cancer is more prevalent in developed countries exhibiting more westernised
lifestyle practices.
Familial and hereditary factors have been observed to play primary roles in
the cause of colorectal
cancers. In addition, a number of other factors have been shown to be
associated with an~increased risk
of developing colorectal cancer namely the presence of adenomatous polyps,
history/presence of
inflammatory bowel disease, diets rich in animal fats and significantly
decreased consumption of raw
or fresh vegetables (especially leafy green vegetables, . cruciferous
vegetables, as well as album
vegetables such as garlic, onions, chives).
Significant differences exist regarding the survival of patients affected by
colorectal cancer according
to the stages at which the disease is diagnosed. Most patients exhibit
symptoms such as rectal
bleeding, pain, abdominal distension or weight loss only after the disease is
in its advanced stages,
leaving little therapeutic options available. Clearly, early detection of
primary, metastatic, and
recurrent disease can significantly impact the prognosis of individuals
suffering from colorectal
cancer. Diagnosis at an early stage, prior to lymph-node spread, can
significantly improve the rate of
survival as compared to a diagnosis established at a later stage of the
disease, since the therapies used
to treat colorectal cancer are stage-dependent.
In date, fecal occult blood test (FOBT), flexible sigmoidoscopy, double
'contrast barium enema, and
colonoscopy are the primary tools utilised to detect colorectal cancer at its
early stages. Among these
1
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
only FOBT, which is based on the high probability that blood found .within a
patients' fecal (heme-
positive) sample arises from tumours found within the large intestine, is non-
invasive, simple and
relatively inexpensive. Unfortunately, this method of early detection has
several drawbacks.
Firstly, a positive FOBT result leads to further examination, mainly
colonoscopy - an extremely
discomforting, invasive diagnostic method which is expensive and carries a
serious complication rate
of one per 5,000 examinations. Colonoscopy, as a follow-up diagnostic method,
might prove to be
effective in confirming colorectal cancer within a patient provided that the
FOBT results indeed reflect
the presence of the disease. Unfortunately this is more often not the case,
since only 12% of the
patients with a heme-positive fecal sample are diagnosed with cancer or large
polyps at the time of
colonoscopy. Furthermore, physicians frequently fail to properly instruct
their patients on how fecal
samples should be collected. Normally, patients are told to adhere to specific
dietary guidelines and to
avoid taking medication known to induce gastrointestinal bleeding. Should the
patient not be
instructed properly, nor adhere to the strict protocol, the chance of
obtaining a false-positive FOBT
result is greatly increased. The false positive-FOBT result will subsequently
send the patient for a
confirmatory diagnosis, which is neither necessary, inexpensive, or pleasant.
Secondly, a
false-negative result holds even greater consequence since a patient
possessing colorectal cancer, in
this case, would not be diagnosed as having the disease and would be sent home
without proper
therapy.
Currently, many groups are utilising proteomic technologies to comparatively
analyse the differences
in protein levels in colorectal cancers vs. normal large intestinal tissue in
the hopes of developing
diagnostic markers that could assist the practicing clinician in the
management of colorectal cancer.
Currently, the standard method of proteome analysis , has been two,
dimensional (2D) gel
electrophoresis, which has been an invaluable tool~for the separation and
identification of proteins.
This method is also effective in identifying aberrantly expressed proteins in
a variety of tissue
samples: Unfortunately, the analysis of data generated by 2D-gel
electrophoresis is labour-intensive
and requires large quantities of material for protein analysis, thereby
rendering it impractical for
routine clinical use.
Through the introduction of SELDI (surface enhanced laser desorption
ionization), a modification of
~MALDI-TOF (matrix-assisted laser desorption ionization/time of flight) which
is a mass spectrometry
technique that allows for the simultaneous analysis of multiple proteins in
one sample, this tool has
been achieved. Small amounts of proteins can be directly bound to a biochip,
carrying spots with
different types of chromatographic material, including those with hydrophobic,
hydrophilic, cation-
exchanging and anion-exchanging characteristics. This approach has been proven
to be very useful to
identify proteins and protein patterns (profiles) in various biological
fluids, including serum, urine or
2
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
pancreatic juice.
To date, specific biomarkers for the detection of breast and prostate cancers
(patents W00223200,
W003058198 and W00125791 from Ciphergen, respectively) have been identified
using the above
mentioned SELDI technology. Unfortunately, due to the nature of sample
testing, the biomarkers
identified can only be used to diagnose a patient as having a specific cancer
(either breast or prostate)
versus not having the disease at all..For example, whereas the test samples
analysed in W003058198
(Ciphergen) and W00223200 (Ciphergen) were taken from patients with late-stage
breast cancer
(stages III and IV), the control samples were taken from patients with
undetectable breast cancer. The
biomarkers identified are neither grade-specific nor can they detect the
disease at its earliest stages
(stage I and II), and thereby would not allow for effective patient-specific
treatment of the disease.
Moreover, biomarkers that can differentiate between the presence of a
colorectal cancer, a non-
malignant disease of the large intestine, or an acute and chronic inflammation
of the epithelium have
not yet been identified.
Accordingly, there is a critical need to develop a simple, non-invasive,
reliable and inexpensive
method for the effective detection of colorectal cancer at its early stages.
Preferably, such a diagnostic
method should be able to detect early-stage colorectal cancer, as well as
distinguish between the later
stages or grades of the disease. With such valuable information, medical
practitioners would be able to
tailor patient therapies for optimum treatment of the disease.
The present invention addresses this difficulty with the development of a non-
invasive diagnostic tool
for the differential diagnosis of colorectal cancer and non-malignant diseases
of the large intestine.
SUMMARY OF THE INVENTION
The present invention relates to methods for the differential diagnosis of
colorectal cancer or non-
malignant disease of the large intestine by detecting one or more
differentially expressed biomolecules
within a test sample of a given subject, comparing results with samples from
healthy subjects, subjects
having a precancerous, lesion of the large intestine, subjects having a
colorectal cancer, subjects having
a metastasised colorectal cancer, or subjects having a non-malignant disease
of the large intestine,
wherein the comparison allows for the differential diagnosis of a subject as
healthy, having a
precancerous lesion of the large intestine, having a colorectal cancer, having
a metastasised colorectal
cancer or a non=malignant disease of the large intestine.
The present invention provides a method for the differential diagnosis of a
colorectal cancer and/or a
non-malignant disease of the large intestine, in vitro, comprising obtaining a
test sample from a
subject, contacting test sample with a biologically active surface under
specific binding conditions,
3
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
allowing for biomolecules present within the test sample to bind to the
biologically active surface,
detecting one or more bound biomolecules using mass spectrometry thereby
generating a mass profile
of said test sample, transforming data into a computer-readable form, and
comparing said mass profile
against a database containing mass profiles specific for healthy subjects,
subjects having a
precancerous lesion of the large intestine, subjects having colorectal cancer,
subjects having
metastasised colorectal cancers, or , subjects having a non-malignant disease
of the large intestine,
wherein the - comparison allows for the differential diagnosis of a subj ect
as healthy, having a
precancerous lesion of the large intestine, having a colorectal cancer, having
a metastasised colorectal
cancer or a non-malignant disease of the large intestine.
In one embodiment the invention provides a database comprising of mass
profiles of biological
samples from healthy subjects, subjects having a precancerous lesion of the
large intestine, subjects
having a colorectal cancer, subjects having a metastasised colorectal cancer,
or subjects having a non-
malignant disease of the large intestine.
Within the same embodiment the database is generated by obtaining biological
samples from healthy
subjects, subjects having a precancerous lesion of the large intestine,
subjects having a colorectal
cancer, subjects having a metastasised colorectal cancer, and subjects having
a non-malignant disease
of the large intestine, contacting said biological samples with a biologically
active surface under
specific binding conditions, allowing the biomolecules within the biological
sample to bind to said
biologically active surface, detecting one or more bound biomolecules using
mass spectrometry
thereby generating a mass profile of said biological samples, transforming
data into a
computer-readable form, and applying a mathematical algorithm to classify the
mass profiles as
specific for healthy subjects, subjects having a precancerous lesion of the
large intestine, subjects
having colorectal cancer, subjects having metastasised colorectal cancer, and
subjects having a.non-
malignant disease of the large intestine.
In specific embodiments, the present invention provides biomolecules having a
molecular mass
selected from the group consisting of 2020 Da ~ 10 Da, 2049 Da ~ 10 Da, 2270
Da ~ 11 Da, 2508 Da
~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15 Da, 3227 Da ~ 17 Da, 3326 Da ~ 17 Da,
3456 Da ~ 17 Da,
3946 Da .~ 20 Da, 4103 Da ~ 21 Da, 4242 Da ~ ~21 Da, 4295 Da ~ 21 Da, 4359 Da
~ 22 Da, 4476 Da ~
22 Da, 4546 Da ~ 23 Da, 4607 Da ~ 23 Da, 4719 Da ~ 24 Da, 4830 Da ~ 24 Da,
4865 Da ~ 24 Da,
4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226 Da ~ 26 Da, 5493 Da ~ 27 Da, 5648 Da ~
28 Da, 5772 Da ~
29 Da, 5854 Da ~ 29 Da, 6446 Da~~ 32 Da, 6644 Da ~ 33 Da, 6852 Da ~ 34 Da,
6897 Da ~ 34 Da,
6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38 Da, 8076 Da ~ 40 Da, 8215 Da ~
41 Da, 8474 Da ~
42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44 Da, 8780 Da ~ 44 Da, 8922 Da ~ 45 Da,
9078 Da ~ 45 Da.,
9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359 Da ~ 47 Da, 9425 Da ~ 47 Da, 9581 Da ~
48 Da, 9641 Da ~
4
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50 Da, 10215 Da ~ 51 Da, 1'0369 Da ~ 52 Da,
10440 Da ~ 52 Da,
10594 Da ~ 53 Da, 11216 Da ~ 56 Da, 11464 Da ~ 57 Da, 11547 Da ~ 58 Da, 11693
Da ~ 58 Da,
11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619 Da ~ 63 Da, 12828 Da ~ 64 Da, 13290
Da ~ 66 Da,
13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005
Da ~ 75 Da,
S 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879 Da ~ 79 Da, 15957 Da ~ 80 Da,
16104 Da ~ 81 Da,
16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263 Da ~ 86 Da, 17397 Da t 87 Da, 17617
Da ~ 88 Da,
17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338
Da ~ 112 Da,
22466 Da ~ 112 Da, 22676 Da ~ 113 Da, 22951 Da ~ 115 Da, 24079 Da ~ 120 Da,
28055 Da ~ 140
Da, and 28259 Da ~ 141 Da. The biomolecules having said molecular masses are
detected by
contacting a test and/or biological sample with a biologically active surface
comprising an adsorbent
under specific binding conditions and further analysed by gas phase ion
spectrometry. Preferably the
adsorbent used is comprised of positively charged quaternary ammonium groups
(anion exchange
surface).
In. specific embodiments, the invention provides specific binding conditions
for the detection of
biomolecules within a sample. In preferred embodiments, a sample is diluted
1:5 in a denaturation
buffer consisting of 7 M urea, 2 M thiourea, 4% CHAPS, 1% DTT', and 2%
Ampholine, and then
diluted again 1:10 in binding buffer consisting of 0.1 M Tris-HCI, 0.02%
Triton X-100 at a pH 8.5 at 0
to 4°C. The treated sample is then contacted with a biologically active
surface comprising of positively
charged (cationic) quaternary ammonium groups (anion exchanging), incubated
for 120 minutes at 20
to 24°C, and the bound biomolecules are detected using gas phase ion
spectrometry.
In an alternative embodiment, the invention provides a method for the
differential diagnosis of a
colorectal cancer and/or a non-malignant disease of the large intestine
comprising detecting of one or
more differentially expressed biomolecules within a sample. This method
comprises obtaining a test
sample from a subject, contacting said sample with a binding molecule specific
for a differentially
expressed polypeptide, detecting an interaction between the binding molecule
and its specific
polypeptide, wherein the detection of an interaction indicates the presence or
absence of said
polypeptide, thereby allowing for the differential diagnosis of a subject as
healthy, having a
precancerous lesion of the large intestine, having a colorectal cancer, having
a metastasised colorectal
cancer andlor a non-malignant disease of the large intestine. Preferably,
binding molecules are
antibodies specific for said polypeptides.
The biomolecules related to the invention, having a molecular mass selected
from the group consisting
of 2020 Da ~ 10 Da, 2049 Da ~ 10 Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732
Da, ~ 14 Da, 3026
Da ~ 15 Da, 3227 Da ~ 17 Da, 3326 Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20
Da, 4103 Da ~ 21
Da, 4242 Da t 21 Da, 4295 Da ~ 21 Da, 459 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da
~ 23 Da, 4607
5
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Da ~ 23 Da, 4719 Da ~ 24 Da, 4830 Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25
Da, 5112 Da ~ 26
Da, 5226 Da ~ 26 Da, 5493 Da ~ 27 Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854
Da t 29 Da, 6446
Da ~ 32 Da, 6'644 Da ~ 33 Da, 6852 Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35
Da, 7575 Da ~ 38
Da, 7657 Da ~ 38 Da, 8076 Da ~ 40 Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da; 8574
Da ~ 43 Da, 8702
Da ~ 44 Da, 8780 Da ~ 44 Da, 8922 Da t 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46
Da, 9201 Da ~ 46
Da, 9359 Da ~ 47 Da, 9425 Da ~ 47 Da, 9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718
Da ~ 49 Da, 9930
Da ~ 50 Da, 10215 Da ~ 51 Da, 10369 Da ~ 52 Da, 10440 Da ~ 52, Da, 10594 Da ~
53 Da, 11216 Da
~ 56 Da, 11464 Da ~ 57 Da, 11547 Da ~ 58 Da, ~ 11693 Da ~ 58 Da, 11905 Da ~ 60
Da, 12470 Da ~ 62
Da, 12619 Da ~ 63 Da, 12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da,
13784 Da ~ 69 Da,
13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350
Da ~ 77 Da,
15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953
Da ~ 85 Da,
17263 Da ~ 86 Da, 17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890
Da ~ 89 Da,
18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da,
22676 Da ~ 113 Da,
22951 Da ~ 115 Da, 24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da
, and may
include, but are not limited to, molecules comprising,nucleotides, amino
acids, sugars, fatty acids,
steroids, nucleic acids, polynucleotides (DNA or RNA), polypeptides, proteins,
antibodies,
carbohydrates, lipids, and combinations thereof (e.g., glycoproteins,
ribonucleoproteins, lipoproteins).
Preferably said biomolecules are proteins, polypeptides, or fragments thereof.
' In yet another embodiment, the invention provides a method for the
identification of biomolecules
within a sample, provided that the biomolecules are proteins, polypeptides or
fragments thereof,
comprising: chromatography and fractionation, analysis of fractions for the
presence of said
differentially expressed proteins and/or fragments thereof, using a
biologically active surface, further
analysis using mass spectrometry to obtain amino acid sequences encoding said
proteins andlor
fragments thereof, and searching amino acid sequence databases of known
proteins to identify said
differentially expressed proteins by amino acid sequence comparison.
Preferably the method of
chromatography is high performance liquid ichromatography (HPLC) or fast
protein liquid
chromatography (FPLC). Furthermore, the mass spectrometry used is selected
from the group of
matrix-assisted laser desorption ionization/time of flight (MALDI-TOF),
surface enhanced laser
desorption ionisation/time of flight (SELDI-TOF), liquid chromatography, MS-
MS, or ESI-MS.
Furthermore, the invention provides kits for the differential diagnosis of a
colorectal cancer and/or a
non-malignant disease of the colon.
The test or biological samples used according to the invention may be of
blood, blood serum, plasma,
nipple aspirate, urine, semen, seminal fluid, seminal plasma, prostatic fluid,
excreta, tears, saliva,
sweat, biopsy, ascites, cerebrospinal fluid, milk, lymph, or tissue extract
origin. Preferably, the test
6
CA 02525743 2005-11-14
WO 2004/102190 ' PCT/EP2004/005294
and/or biological samples are blood serum samples, _and are isolated from
subjects of mammalian
origin, preferably of human origin.
A colorectal cancer of the invention is a cancer of the large intestine, and
may include cancers of the
colon, rectum etc. Furthermore, a colorectal cancer, as intended by the
invention, may be of various
stages and/or grades.
DESCRIPTION OF FIGURES
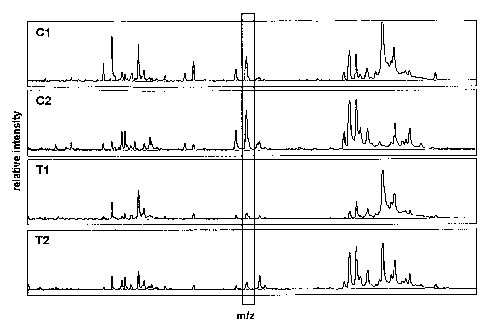
Figure 1. Comparison of protein mass spectra processed on the anion exchange
surface of a SAX2
ProteinChip array comprised of cationic quaternary ammonium groups. Protein
mass spectra obtained
from sera of endoscopy control patients (Cl and C2), suffering from non-
malignant diseases of the
large intestine (e.g., acute or chronic inflammation, adenoma) and of patients
with colon cancer (Tl
and T2) are shown. Scattered boxes indicate differentially expressed proteins
with high diagnostic
significance. A representative differentially expressed protein (m/z= 6645 Da)
is highlighted
possessing high importance within the generated classifiers (ensemble of
decision trees) according to
overall improvement, see Tables 1-4. The X-axis shows the mass/charge (m/z)
ratio, which is
equivalent to the apparent molecular mass of the corresponding biomolecule.
The Y-axis shows the
normalized relative signal intensity of the peak in the examined serum
samples.
Figure 2A - F. Scatter plots of clusters (peaks, variables), belonging to
differentially expressed
proteins included in the four classifiers. The X-axis shows the mass/charge
(m/z) ratio, which is
equivalent to the apparent molecular mass of the corresponding biomolecule.
The Y-axis shows the
logarithmic normalized relative signal intensity of the peaks in the examined
serum samples. First,
intensities were shifted to yield entirely positive values. Then, for each
mass, intensities .were
normalized by dividing the intensity values by the average intensity of that
mass. Finally, the natural
logarithm was taken. o T (Tumour): Colon cancer patients' serum samples. o N
(Normal): Endoscopy
control patients' serum samples.
Figure 3A - F. Additionally scaled scatter plots of clusters (peaks,
variables), belonging to
differentially expressed proteins included in the four classifiers. The X-axis
shows the mass/charge
(m/z) ratio, which is equivalent to the apparent molecular mass of the
corresponding biomolecule. As
in Figure 2, the Y-axis shows the logarithmic normalized relative signal
intensity of the peaks in the
examined serum samples. However, intensities were additionally (shifted and)
scaled so that the
intensities of each mass cover the entire range of the Y-axis. Thereby, the
minimum and maximum
intensities of all masses are aligned on the lower and upper edge of the plot,
respectively. This allows
to better visualize the extend of class_overlap.. o T (Tumour): Colon cancer
patients' serum samples.
o N (Normal): Endoscopy control patients', serum samples.
7
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Figure 4. Complexity of proof of principle classifier. The histogram
visualizes the distribution of the
number of decision tree variables (peaks, clusters) for the obtained proof of
principle classifier for
gastric cancer. 6 variables per decision tree are typical.
Figure 5. Variable importance of the._proof of principle classifier. The
histograms visualize how often
a variable (mass) is employed in the proof of principle classifier. The
frequency of variable selection
is presented in histogram form for each hierarchical level (a-j) and for all
hierarchical levels taken
together (k).
Figure 6. Complexity of lst final classifier. The histogram visualizes the
distribution of the number of
decision tree variables (peaks, clusters) for the obtained 1St final
classifier in the range of 1 to 10
decision tree variables. 9 variables per decision tree are typical.
Figure 7. Variable importance of 1St final classifier. The histogram
visualizes how often a variable
(mass) is employed in the final classifier. The frequency of variable
selection is presented in histogram
form for each of the first 10 hierarchical levels (a-j) and for the first ten
hierarchical levels taken
together (k).
Figure 8. Complexity of 2nd final classifier. The histogram visualizes the
distribution of the number of
decision tree variables (peaks, clusters) for the obtained 2nd final
classifier in the range of 1 to 10
decision tree variables. As many as 10 variables per decision tree are
typical.
Figure 9. Variable importance of 2nd final classifier. The histogram
visualizes how often a variable
(mass) is employed in the 2nd final classifier. The frequency of variable
selection is presented in,
histogram form for each of the first 10 hierarchical levels.(a-j) and for the
first ten hierarchical levels
taken together (k).
Figure 10. Complexity of 3'd final classifier. The histogram visualizes the
distribution of the number
of decision tree variables (peaks, clusters) for the obtained 3rd final
classifier in the range of 1 to 10
decision tree variables. As many as 10 variables per decision tree are
typical.
Figure 11. Variable importance of 3~d final classifier. The histogram
visualizes how often a variable
(mass) is employed in the 3'~ final classifier. The frequency of variable
selection is presented in
histogram form for each of the first 10 hierarchical levels (a-j) and for the
first ten hierarchical levels
taken together (k).
8
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
DESCRIPTION OF THE INVENTION
It is to be understood that the present invention is not limited to the
particular materials and methods
described or equipment, as these may vary. It is also to be understood that
the terminology used herein
is for the purpose of describing particular embodiments only, and is not
intended to limit the scope of
the present invention, which will be limited only by the appended claims.
It should be noted that as used herein and in the appended claims, the
singular forms "a," "an," and
"the" include plural reference unless the context clearly dictates otherwise.
Thus, for example, a
reference to "an antibody" is a reference to one or more antibodies and
derivatives thereof known to
those skilled in the art, and so forth.
Unless defined otherwise, all technical and scientific terms used herein have
the same meanings as
commonly understood by one of ordinary skill in the art. Although any
materials and methods, or
equipment comparable to those specifically described herein can be used to
practice or test the present
invention, the preferred equipment, materials and methods are described below.
All publications
mentioned herein are cited for the purpose of describing and disclosing
protocols, reagents, and
current state of the art technologies that might be used in connection with
the invention. Nothing
herein is to be construed as an admission that the invention is not entitled
to precede such disclosure
by virtue of prior invention.
Definitions
The term "biomolecule" refers to a molecule produced by a cell or living
organism. Such molecules
include, but are not limited to, molecules comprising nucleotides, amino
acids, sugars, fatty acids,
steroids, nucleic acids, polynucleotides, polypeptides, proteins,
carbohydrates, lipids, and
combinations thereof (e.g., glycoproteins, ribonucleoproteins, lipoproteins).
Furthermore, ~ the terms
"nucleotide" or polynucleotide" refer to a nucleotide, oligonucleotide,
polynucleotide, or any fragment
thereof. These phrases also refer to DNA or RNA of genomic or synthetic origin
which may be single
stranded or double-stranded and may represent the sense, or the antisense
strand, to peptide
polynucleotide sequences (i.e. peptide nucleic acids; PNAs), or to any DNA-
like or RNA-like
material.
The term "fragment" refers to a portion of a polypeptide (parent) sequence
that comprises at least 10
consecutive amino acid residues and retains a biological activity and/or some
functional characteristics
of the paxent polypeptide e.g. antigenicity or structural domain
characteristics.
The terms "biological sample" and "test sample" refer to all biological fluids
and excretions isolated
from any given subject. In the context of the invention such samples include,
but are not limited to,
9
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
blood, blood serum, plasma, nipple aspirate, urine, semen, seminalvfluid,
seminal plasma, prostatic
fluid, excreta, tears, saliva, sweat, biopsy, ascites, cerebrospinal fluid,
milk,. lymph, or tissue extract
samples.
The term "specific binding" refers to the binding reaction between a
biomolecule and a specific
"binding molecule". Related to the invention are binding molecules that
include, but are not limited to,
proteins, peptides, nucleotides, nucleic acids, hormones, amino acids, sugars,
fatty acids, steroids,
polynucleotides, carbohydrates, lipids, or a combination thereof (e.g.
glycoproteins,
ribonucleoproteins, lipoproteins). Furthermore, a binding reaction is
considered to be specific when
the interaction between said molecules is substantial. In the context of the
invention, a binding
reaction is considered substantial when the reaction that takes place between
said molecules is at least
two times the background. Moreover, the term "specific binding conditions"
refers to reaction
conditions that permit the binding of said molecules such as pH, salt,
detergent and other conditions
known to those skilled in the art.
The term "interaction" relates to the direct or indirect binding or alteration
of biological activity of a
biomolecule.
The term "differential diagnosis" refers to a diagnostic decision between a
healthy and different
~ disease states, including various stages of a specific disease. A subject is
diagnosed as healthy or to be
suffering from a specific disease, or a specific stage of a disease based on a
set of hypotheses that
allow for the distinction between healthy and one or more stages of the
disease. The choice between
healthy and one or more stages of disease depends on a significant difference
between each
hypothesis. Under the same principle, a "differential diagnosis" may also
refer to a diagnostic decision
between one disease type as compared to another (e.g. colon cancer vs.
diverticulosis).
y
The term "colorectal cancer" refers to a cancer state associated with the
large intestine of any given
subject, wherein the cancer state is defined according to its stage andlor
grade. The various stages of a
cancer may be identified using staging systems known to those skilled in the
art [e.g. Union
Internationale Contre Cancer (ITICC) system or American Joint Committee on
Cancer (AJC)]. In the
context of the invention colorectal cancers include but are not limited to
colon and rectal cancers.
The term "non-malignant disease of the large intestine" refers to alterations
in the physiological,
functional andlor anatomical state of the large intestine, wherein the
alterations deviate from normal.
In addition, this term encompasses alterations in the physiological,
functional and/or anatomical state
of the large intestine that cannot be staged or graded according to cancer
staging systems known to
those skilled in the art [e.g. Union Internationale Contre Cancer (UICC)
system or American Joint
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Committee on Cancer (AJC)]. Such non-malignant disease include but are not
limited to the acute and
chronic inflammation of the large intestinal epithelium, diverticular disease
including diverticulosis
and diverticulitis, colitis, ulcerative colitis, pancolitis, Crohn's disease
(ileitis), proctitis, intestinal
polyps including hyperplastic polyps, hamartomatous polyps (i.e. Juvenile
polyps, Peutz-Jeghers
polyps), inflammatory polyps, and lymphoid polyps, adenomatous polyps.
The term "healthy individual" refers to a subject possessing good health. Such
a subject demonstrates
an absence of any disease within the large intestine, preferably a colorectal
cancer or a non-malignant
disease of the large intestine.
The term "precancerous lesion of the large intestine" refers to a biological
change within a cell and/or
tissue of the large intestine such that said cell and/or tissue becomes
susceptible to the development of
a cancer. More specifically, a precancerous lesion of the large intestine is a
preliminary stage of a
colorectal cancer (i.e. dysplasia). Causes of a precancerous lesion of the
larger intestine may include,
but are not limited to, genetic predisposition and exposure to cancer-causing
agents (carcinogens);
such cancer causing agents include agents ~ that cause genetic damage and
induce neoplastic
transformation of a cell. Furthermore, the phrase "neoplastic transformation
of a cell" refers an
alteration in normal cell physiology and includes, but is not limited to, self
sufficiency in growth
signals, insensitivity to growth-inhibitory (anti-growth) signals, evasion of
programmed cell death
(apoptosis), limitless replicative potential, sustained angiogenesis, and
tissue invasion and metastasis.
The term "dysplasia" refers to morphological alterations within a tissue,
which are characterised by a
loss in the uniformity of individual cells, as well as a loss in their
architectural orientation.
Furthermore, dysplastic cells also exhibit a variation in size and shape.
The phrase "differentially present" refers to differences in the quantity of a
biomolecule (of a
particular apparent molecular mass) present in a sample from a subject as
compared to a comparable
sample. For example, a biomolecule is present at an elevated level, a
decreased level or absent in
samples of subjects having colorectal cancer compared to samples of subjects
who do not have a
cancer of the large intestine. Therefore in the context of the invention, the
term "differentially present
biomolecule" refers to the quantity biomolecule (of a particular apparent
molecular mass) present
within a sample taken from a subject having a disease or cancer of the large
intestine as compared to a
comparable sample taken from a healthy subject. Within the context of the
invention, a biomolecule is
differentially present between two samples if the quantity of said biomolecule
in one sample is
statistically significantly different from the quantity of said biomolecule in
another sample.
The term "diagnostic assay" can be used interchangeably with "diagnostic
method" and refers to the
11
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
detection of the presence or nature. of a pathologic condition. Diagnostic
assays differ in. their
sensitivity and specificity. Within the context of the invention the
sensitivity of a diagnostic assay is
defined as the percentage of diseased subjects who test positive for a
colorectal cancer or a non-
malignant disease of the large intestine and are considered "true positives".
Subjects having a
colorectal cancer or a non-malignant disease of the large intestine but not
detected by the diagnostic
assay are considered "false negatives". Subjects who are not diseased and who
test negative in the
diagnostic assay are considered "true negatives". Furthermore, the term
specificity of a diagnostic
assay, as used herein, is defined as 1 minus the false positive rate, where
the "false positive rate" is
defined as the proportion of those subjects devoid of' a colorectal cancer or
a non-malignant disease of
the large intestine but who test positive in said assay.
The term "adsorbent" refers to any material that is capable of accumulating
(binding) a biomolecule.
The adsorbent typically coats a biologically active surface and is composed of
a single material or a
plurality of different materials that are capable of binding a biomolecule.
Such materials include, but
are not limited to, anion exchange materials, cation exchange materials, metal
chelators,
polynucleotides, oligonucleotides, peptides, antibodies, metal chelators etc.
The term "biologically active surface" refers to any two- or three-dimensional
extension of a material
that biomolecules can bind to, or interact with, due to the specific
biochemical properties of this
material and those of the biomolecules. Such biochemical properties include,
but are not limited to,
ionic character (charge), hydrophobicity, or hydrophilicity.
The term "binding molecule" refers to a molecule that displays an affinity for
another molecule. With
in the context of the invention such molecules may include, but are not
limited to nucleotides, amino
acids, sugars, fatty acids, steroids, nucleic acids, polypeptides,
carbohydrates, lipids, and combinations
thereof (e.g. glycoproteins, ribonucleoproteins, lipoproteins). Preferably,
such binding molecules are
antibodies.
The term "solution" refers to a homogeneous mixture of two or more substances.
Solutions may
include, but are not limited to buffers, substrate solutions, elution
solutions, wash solutions, detection
solutions, standardisation solutions, chemical solutions, solvents, etc.
Furthermore, other solutions
known to those skilled in the art are also included herein.
The term "mass profile" refers to a mass spectrum as a characteristic property
of a given sample or a
group of samples, especially when compared to the mass profile ,of a second
sample or group of
samples in any way different from the first sample or group of sample. In the
context of the invention,
the mass profile is obtained by treating the biological sample as follows. The
sample is diluted it 1:5 in
12
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
a denaturation buffer consisting of 7 M urea, 2 M thiourea, 4% CHAPS, 1% DTT,
and 2% ampholine
anal subsequently diluted 1:10 in binding buffer consisting of 0.1 M Tris-HCl,
0.02% Triton X-100 at
pH 8.5. Thus pre-treated sample is applied to a biologically active surface
comprising positively
charged quaternary ammonium groups (anion exchange surface) and incubated for
120 minutes: The
biomolecules bound to the surface are analysed by gas phase ion spectrometry
as described in another
section. All but the dilution steps are performed at 20 to 24°C.
Dilution steps are performed at 0 to
4°C.
The phrase "apparent molecular mass" refers to the molecular mass value in
Dalton (Da) of a
~ biomolecule as it may appear in a given method of investigation, e.g. size
exclusion chromatography,
gel electrophoresis, or mass spectrometry.
The term "chromatography" refers to any method of separating biomolecules
within a given sample
such that the original native state of a given biomolecule is retained.
Separation of a biomolecule from
other biomolecules within a given sample for the purpose of enrichment,
purification and/or analysis,
may be achieved by methods including, but not limited to, size exclusion
chromatography, ion
exchange chromatography, hydrophobic and hydrophilic interaction
chromatography, metal affinity
chromatography, wherein "metal" refers to metal ions (e.g. nickel, copper,
gallium, or zinc) of all
chemically possible valences, or ligand affinity chromatography wherein
"ligand" refers to binding
molecules, preferably proteins, antibodies, or DNA. Generally, chromatography
uses biologically
active surfaces as adsorbents to selectively accumulate certain biomolecules.
The term "mass spectrometry" refers to a method comprising employing an
ionization source to
generate gas phase ions from a biological entity of a sample presented on a
biologically active surface
and detecting the gas phase ions with a mass spectrometer.
The phrase "laser desorption mass spectrometry" refers to a method comprising
the use of a laser as an
ionization source to generate gas phase ions from a biomolecule presented on a
biologically active
surface and detecting the gas phase ions with a mass spectrometer.
The term "mass spectrometer" refers to a gas phase ion spectrometer that
includes an inlet system, an
ionisation source, an ion optic assembly, a mass analyser, and a detector:
Within the context of the invention, the terms "detect", "detection" or
"detecting" refer to the
identification of the presence, absence, or quantity of a biomolecule.
The term "energy absorbing molecule" or "EAM" refers to a molecule that.
absorbs energy from an
13
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
energy source in a mass spectrometer thereby enabling desorption of a
biomolecule from a
biologically active surface. Cinnamic acid derivatives, siriapinic acid and
dihydroxybenzoic acid are
frequently used as energy-absorbing molecules in laser desorption of
biomolecules. See U.S. Pat. No.
5,719,060 (Hutchens & Yip) for a further description of energy absorbing
molecules.
The term "training set" refers to a subset of the respective entire available
data set. This subset is
typically randomly selected, and is solely used for the purpose of classifier
construction.
The term "test set" refers to a subset of the entire available data set
consisting of those entries not
included in the training set. Test data is applied to evaluate classifier
performance.
The term "decision tree" refers to a flow-chart-like tree structure employed
for classification. Decision
trees consist of repeated splits of a data set into subsets. Each split
consists of a simple rule applied to
one variable, e.g., "if value of 'variable 1' larger than 'threshold 1' then
go left else go right".
Accordingly, the given feature space is partitioned into a set of rectangles
with each rectangle assigned
to one class.
The terms "ensemble", "tree ensemble" or "ensemble classifier" can be used
interchangeably and refer
to a classifier that consists of many simpler elementary classifiers, e.g., an
ensemble of decision trees
is a classifier consisting of decision trees. The result of tie ensemble
classifier is obtained by
combining all the results of its constituent classifiers, e.g., by majority
voting that weights all
constituent classifiers equally. Majority voting is especially reasonable in
the case of bagging, where
constituent classifiers are then naturally weighted by the frequency with
which they are generated.
The term "competitor" refers to a variable (in our ~~case: mass) that can~~be
used as an alternative
splitting rule in a decision tree. In each step of decision tree construction,
only the variable yielding
best data splitting is selected. Competitors are non-selected variables with
similar but lower
performance than the selected variable. They point into the direction of
alternative decision trees.
The term "surrogate" refers to a splitting rule that closely mimics the action
of the primary split. A
surrogate is a variable that can substitute a selected decision tree variable,
e.g. in the case of missing
values. Not only must a good surrogate split the parent node into descendant
nodes similar in size and
composition to the primary descendant nodes. In addition, the surrogate must
also match the primary
split on the specific cases that go to the left child and right child nodes.
The terms "peak" and "signal" may be used interchangeably and refer to any
signal which is generated
by a biomolecule when under investigation using a specific method, for example
chromatography,
14
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
mass spectrometry, or any type of spectroscopy like Ultraviolet/Visible Light
(LJV/Vis) spectroscopy,
Fourier Transformed Iufrared (FTIR) spectroscopy, Electron Paramagnetic
Resonance (EPR)
spectroscopy, or Nuclear Mass Resonance (NMR) spectroscopy.
Within the context of the invention, the terms "peak" and "signal" refer to
the signal generated by a
biomolecule of a certain molecular mass hitting the detector of a mass
spectrometer, thus generating a.
signal intensity which correlates with the amount or concentration of said
biomolecule of a given
sample. A "peak" and "signal" is defined by two values: an apparent molecular
mass value and an
intensity value generated as described. The mass value is an elemental
characteristic of a biological
entity, whereas the intensity value accords to a certain amount or
concentration of a biological entity .
with the corresponding appaxent molecular mass value, and thus "peak" and
"signal" always refer to
the properties of this biological entity.
The term "cluster" refers to a signal or peak present in a certain set of mass
spectra or mass profiles
obtained from different samples belonging to two or more different groups
(e.g. cancer and non
cancer). Within the set, signals belonging to~ cluster can differ in their
intensities, but not in the
apparent molecular masses.
The term "variable" refers to a cluster which is subjected to a statistical
analysis aiming towards a
classification of samples into two or more different sample groups (e.g.
cancer and non cancer) by
using decision trees, wherein the sample feature relevant for classification
is the intensity value of the
variables in the analysed samples.
Detailed Description.of the invention
a) Diagnostics
The present invention relates to methods for the differential diagnosis of
colorectal cancers or a non-
malignant disease of the large intestine by detecting one or more
differentially expressed biomolecules
within a test sample of a given subject, comparing results with samples from
healthy subjects, subjects
having a precancerous lesion of the large intestine, subjects having a
colorectal cancer, subjects having
a metastasised colorectal cancer, or subjects having a non-malignant disease
of the large intestine,
wherein the comparison allows for the differential diagnosis of a subject as
healthy, having a
precancerous lesion of the large intestine, having a colorectal cancer, having
a metastasised colorectal
cancer or a non-malignant disease of the large intestine.
In one aspect of the invention, a method for the differential diagnosis of a
colorectal cancer or a non-
malignant disease of the large intestine comprises obtaining a test sample
from a. given subject,
contacting said sample with an adsorbent present on a biologically active
surface under specific
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
binding conditions, allowing the biomolecules within the test sample to bind
.to said adsorbent,
detecting one or more bound biomolecules using a detection method, wherein the
detection method
generates a mass profile of said sample, transforming mass profile data into a
computer-readable form
comparing the mass profile of said sample with a database containing mass
profiles from comparable
samples specific for healthy subjects, subjects having a precancerous lesion
of the large intestine,
subjects having a colorectal cancer, subjects having a metastasised colorectal
cancer, ..or subjects
having a non-malignant disease of the large intestine. A comparison of mass
profiles allows for the
medical practitioner to determine if a subject is healthy, has a precancerous
lesion of the large
intestine, a colorectal cancer, a metastasised colorectal cancer or a non-
malignant disease of the large
intestine based on the presence, absence or quantity of specific biomolecules.
In more than one embodiment, a single biomolecule or a combination of more
than one biomolecule
selected from the group having an apparent molecular mass of 2020 Da ~ 10 Da,
2049 Da ~ 10 Da,
2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15 Da, 3227 Da ~
17 Da, 3326 Da ~
17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20 Da, 4103 Da ~ 21 Da, 4242 Da ~ 21 Da,
4295 Da ~ 21 Da,
4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da W 23 Da, 4607 Da ~ 23 Da, 4719 Da ~
24 Da, 4830 Da ~
24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226 Da ~ 26 Da,
5493 Da ~ 27 Da,
5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32 Da, 6644 Da ~
33 Da, 6852 Da ~
34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38 Da,
8076 Da ~ 40 Da,
8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574 Da ~ 43 Da~, 8702 Da t 44 Da, 8780 Da ~
44 Da, 8922 Da ~
45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359 Da ~ 47 Da,
9425 Da ~ 47 Da,
9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50 Da, 10215 Da ~
51 Da, 10369
Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da, 11464 Da ~
57 Da, 11547,Da
~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619 Da ~ 63
Da, 12828 Da ~ 64
Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983 Da ~ 70 Da,
14798 Da ~ 74 Da,
15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879 Da ~ 79 Da, 15957
Da ~ 80 Da,
16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953 Da t 85 Da, 17263 Da ~ 86 Da, 17397
Da t 87 Da,
17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115 Da ~ 91 Da, 18390
Da ~ 92 Da,
22338 Da ~ 112 Da, 22466 Da ~ 112 Da, 22676 Da ~ 113 Da, 22951 Da ~ 115 Da,
24079 Da ~ 120
Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da may be detected within a given
sample. Detection of a
single or a combination of more than one biomolecule of the invention is based
on specific sample
pre-treatment conditions, the pH of binding conditions, and the type of
biologically active surface used
for the detection of biomolecules. For example, prior to the detection of the
biomolecules described
herein, a given sample is pre-treated by diluting 1:5 in a denaturation buffer
consisting of 7 M urea, 2
M thiourea, 4% CHAPS, 1% DTT, and 2% ampholine. The denatured sample is then
diluted 1:10 in a
specific binding buffer (0.1 M Tris-HCI, 0.02% Triton X-100, pH 8.5), applied
to a biologically active
surface comprising of positively-charged quaternary ammonium groups (cationic)
and incubated using
16
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
specific buffer conditions (0.1 M Tris-HCl, 0.02% Triton X-100, pH~8.5) to
allow for binding of said
biomolecules to the above-mentioned biologically active surface.
According to the invention, a biomolecule with the molecular mass of 2020 Da ~
10 Da, 2049 Da ~ 10
Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15 Da, 3227
Da ~ 17 Da, 3326
Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da t 20 Da, 4103 Da ~ 21 Da, 4242 Da ~ 21
Da, 4295 Da ~ 21
Da, 4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da ~ 23 Da, 4607 Da ~ 23 Da, 4719
Da ~ 24 Da, 4830
Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226 Da ~ 26
Da, 5493 Da ~ 27
Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29 I7a, 6446 Da ~ 32 Da, 6644
Da ~ 33 Da, 6852
Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38
Da, 8076 Da ~ 40
Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44 Da, 8780
Da ~ 44 Da, 8922
Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359 Da ~ 47
Da, 9425 Da ~ 47
Da, 9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50 Da, 10215
Da ~ 51 Da,
10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da, 11464
Da ~ 57 Da,
11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619
Da ~ 63 Da,
12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983
Da ~ 70 Da,
14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879
Da ~ 79 Da,
15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263
Da ~ 86 Da,
17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890 Da ~ 89.Da, 18115
Da ~ 91 Da,
18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da, X2676 Da ~ 113 Da,
22951 Da ~ 115 Da,
24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da is detected by
diluting the biological
sample 1:5 in a denaturation buffer consisting of 7 M urea, 2 M thiourea, 4%
CHAPS, 1% DTT, and
2% Ampholine, and then 1:10 in binding buffer consisting of 0.1 M Tris-HCI,
0.02% Triton X-100 at
pH 8.5 at 0 to 4°C, applying thus treated sample to a biologically
active surface comprising positively
charged (cationic)quaternary ammonium groups (anion exchanging), incubating
for 120 minutes at 20
to 24°C, and subjecting the bound biomolecules to gas phase ion
spectrometry as described in another
section.
A biomolecule of the invention may include any molecule that is produced by a
cell or living
organism, and may have any biochemical property (e,g. phosphorylated proteins,
positively charged
molecules, negatively charged molecules, hydrophobicity, hydrophilicity), but
preferably biochemical
properties that allow binding of the biomolecule to a biologically active
surface comprising positively
charged quaternary ammonium groups after denaturation in 7 M urea, 2 M
thiourea, 4% CHAPS, 1%
DTT, and 2% Ampholine and dilution in 0.1 M Tris-HCI, 0.02% Triton X-100 at pH
8.5 at 0 to 4°C
followed by incubation on said biologically active surface for 120 minutes at
20 to 24°C. Such
molecules include, but are not limited to, molecules comprising nucleotides,
amino acids, sugars, fatty
acids, steroids, nucleic acids, polynucleotides (DNA or RNA), polypeptides,
proteins, antibodies,
17
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
carbohydrates, lipids, and combinations thereof (e.g., glycoproteins,
ribonucleoproteins, lipoproteins).
Preferably a biomolecule may be a nucleotide, polynucleotide, peptide, protein
or fragments thereof.
Even more preferred are peptide or protein biomolecules or fragments thereof.
The methods for detecting these biomolecules have many applications. For
example, a single
biomolecule or. a combination of more than one biomolecule selected from the
group having an
apparent molecular mass of 2020 Da ~ 10 Da, 2049 Da ~ 10 Da, 2270 Da ~ 11 Da,
2508 Da ~ 13 Da,
2732 Da ~ 14 Da, 3026 Da ~ 15 Da, 3227 Da ~ 17 Da, 3326 Da ~ 17 Da, 3456 Da ~
17 Da; 3946 Da ~
20 Da, 4103 Da ~ 21 Da, 4242 Da ~ 21 Da, 4295 Da ~ 21 Da, 4359 Da ~ 22 Da,
4476 Da ~ 22 Da,
4546 Da ~ 23 Da, 4607 Da ~ 23 Da, 4719 Da ~ 24 Da, 4830 Da ~ 24 Da, 4865 Da ~
24 Da, 4963 Da ~
25 Da, 5112 Da ~ 26 Da, 5226 Da ~ 26 Da, 5493 Da ~ 27 Da, 5648 Da ~ 28 Da,
5772 Da ~ 29 Da,
5854 Da ~ 29 Da, 6446 Da ~ 32 Da, 6644 Da ~ 33 Da, 6852 Da ~ 34 Da, 6897 Da ~
34 Da, 6999 Da ~
35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38 Da, 8076 Da ~ 40 Da, 8215 Da ~ 41 Da,
8474 Da ~ 42 Da,
8574 Da ~ 43 Da, 8702 Da ~ 44 Da, 8780 Da ~ 44 Da, 8922 Da ~ 45 Da, 9078 Da ~
45 Da, 9143 Da ~
46 Da, 9201 Da ~ 46 Da, 9359 Da ~ 47 Da, 9425 Da ~ 47 Da, 9581 Da ~ 48 Da,
9641 Da ~ 48 Da,
9718 Da ~ 49 Da, 9930 Da ~ 50 Da, 10215 Da ~ 51 Da, 10369 Da ~ 52 Da, 10440 Da
~ 52 Da, 10594
Da ~ 53 Da, 11216 Da ~~ 56 Da, 11464 Da ~ 57 Da, 11547 Da ~ 58 Da, 11693 Da ~
58 Da, 11905 Da
~ 60 Da, 12470 Da ~ 62 Da, 12619 Da ~ 63'Da, 12828 Da ~ 64 Da, 13290 Da ~ 66
Da, 13632 Da ~ 68
Da, 13784 Da ~ 69 Da, 13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005 Da ~ 75 Da,
15140 Da t 76 Da,
15350 Da ~ 77 Da, 15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164
Da ~ 81 Da,
16953 Da ~ 85 Da, 17263 Da ~ 86 Da, 17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766
Da ~ 89 Da,
17890 Da ~ 89 Da, 18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466
Da ~ 112 Da,
22676 Da ~ 113 Da, 22951 Da ~ 115 Da, 24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or
28259 Da ~ 141
Da can be measured to differentiate between healthy subjects, subjects having
a precancerous lesion of
the large intestine, subjects having colorectal cancer, subjects having a
metastasized colorectal cancer
or subjects with a non-malignant disease of the large intestine, and thus are
useful as an aid in the
diagnosis of a colorectal cancer and/or a non-malignant disease of the large
intestine within a subject.
Alternatively, said biomolecules may be used to diagnose a subject as healthy.
For example, a biomolecule having the apparent molecular mass of about e.g.
4242 Da is present only
in biological samples from patients having a metastasised colorectal cancer.
Mass profiling of two test
samples from different subjects, X and Y, reveals the presence of a
biomolecule with the apparent
molecular mass of about 4242 Da in a sample from test subject X, and the
absence of said biomolecule
in test sample from subject Y. The medical practitioner is able to diagnose
subject X as having a
metastasised colorectal cancer and subject Y as not having a metastasised
colorectal cancer. In yet
another example, three biomolecules having the apparent molecular mass of
about 5772 Da, 2020 Da
and 22951 Da are present in varying quantities in samples specific for
precancerous lesions and
18
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
"early" colorectal cancers. The biomolecule having the apparent molecular mass
of 5772 Da is more
present in samples specific for precancerous lesions of the large intestine
than for "early" colorectal
cancers. A biomolecule having an apparent molecular mass of 2020 Da is
detected in samples from
subjects having "early" colorectal cancers but not in those having a
precancerous lesion, whereas the
biomolecule having the molecular mass of 22951 Da is present in about the same
quantity in both
sample types. Such biomolecules are not present in samples from healthy
subjects, only those of
apparent molecular mass of 8780 Da and 16104 Da. Analysis of a test sample
reveals the presence of
biomolecules having the molecular mass of 22951 Da, 5772 Da and 2020 Da.
Comparison of the
quantity of the biomolecules within said sample reveals that the biomolecule
with an apparent
molecular mass of 5772 Da is present at lower levels than those found in
samples from subjects having
a precancerous lesion. The medical practitioner is able to diagnose the test
subject as having an "early"
colorectal cancer. These examples are solely used for the purpose of
clarification and are not intended
to limit the scope of this invention. .
In another aspect of the invention, an immunoassay can be used to determine
the presence or absence
of a biomolecule within a test sample of a subject. First, the presence or
absence of a biomolecule
within a sample can be detected using the various immunoassay methods known to
those skilled in the
art (i.e. ELISA, western blots). If.a biomolecule is present in the test
sample, it will form an antibody-
marker complex with an antibody that specifically binds a biomolecule under
suitable incubation
conditions. The amount of an antibody-biomolecule comple~c can be determined
by comparing to a
standard.
Thus, the invention provides a method for the differential~diagnosis of a
colorectal cancer and/or a
non-malignant disease of the large intestine comprising detecting of one or
more differentially
expressed biomolecules within a sample. This method comprises obtaining a test
sample from a
subject, contacting said sample with a binding molecule specific for a
differentially expressed
polypeptide, detecting an interaction between the binding molecule and its
specific polypeptide,
wherein the detection of an interaction indicates the presence or absence of
said polypeptide, thereby
allowing for the difFerential diagnosis of a subject as healthy, having a
precancerous lesion of the large
intestine, having a colorectal cancer, having a metastasised colorectal cancer
and/or a non-malignant
disease of the large intestine. Binding molecules include, but are not limited
to, proteins, peptides,
nucleotides, nucleic acids, hormones, amino acids, sugars, fatty acids,
steroids, polynucleotides,
carbohydrates, lipids, or a combination thereof (e.g.' glycoproteins,
ribonucleoproteins, lipoproteins),
compounds or synthetic molecules. Preferably, binding molecules are antibodies
specific for
biomolecules selected from the group of having an apparent molecular mass of
2020 Da ~ 10 Da, 2049
Da ~ 10 Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15
Da, 3227 Da ~ 17
Da, 3326 Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20 Da, 4103 Da ~ 21 Da, 4242
Da ~ 21 Da, 4295
19
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Da ~ 21 Da, 4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da ~ 23~ Da, 4607 Da ~ 23
Da, 4719 Da ~ 24
Da, 4830 Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226
Da ~ 26 Da, 5493
Da ~ 27 Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32
Da, 6644 Da ~ 33
Da, 6852 Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657
Da ~ 38 Da, 8076
Da ~ 40 Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44
Da, 8780 Da ~ 44
Da, 8922 Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359
Da ~ 47 Da, 9425
Da ~ 47 Da, 9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50
Da, 10215 Da ~ 51
Da, 10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da,
11464 Da ~ 57 Da,
11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619
Da ~ 63 Da,
12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983
Da ~ 70 Da,
14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879
Da ~ 79 Da,
15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263
Da ~ 86 Da,
17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115
Da ~ 91 Da,
18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da, 22676 Da ~ 113 Da,
22951 Da ~ 115 Da,
24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da
~In another aspect of the invention, a method for detecting the differential
presence of one or more
biomolecules selected from the group having an apparent molecular mass of 2020
Da ~ 10 Da, 2049
Da ~ 10 Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15
Da, 3227 Da ~ 17
Da, 3326 Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20 Da, 403 Da ~ 21 Da, 4242 Da
~ 21 Da, 4295
Da ~ 21 Da, 4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da ~ 23 Da, 4607 Da ~ 23
Da, 4719 Da ~ 24
Da, 4830 Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226
Da ~ 26 Da, 5493
Da ~ 27 Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32
Da, 6644 Da ~ 33
Da, 6852 Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657
Da ~ 38 Da, 8076
Da ~ 40 Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44
Da, 8780 Da ~ 44
Da, 8922 Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359
Da ~ 47 Da, 9425
Da ~ 47 Da, 9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50
Da, 10215 Da ~ 51
Da, 10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da,
11464 Da ~ 57 Da,
11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619
Da ~ 63 Da,
12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983
Da ~ 70 Da,
14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879
Da ~ 79 Da,
15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263
Da ~ 86 Da,
17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115
Da ~ 91 ~Da,
18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da, 22676 Da ~ 113 Da,
22951 Da ~ 115 Da,
24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da in a test sample of
a subject involves
contacting the test sample with a compound or agent capable of detecting said
biomolecule such that
the presence of said biomolecule is directly and/or indirectly labelled. For
example a fluorescently
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
labelled secondary antibody can be used to detect a primary antibody bound to
its specific
biomolecule. Furthermore, such detection methods can be used to detect a
variety of biomolecules
within a test sample both in vitro as well as in vivo.
For example, in vivo, antibodies or fragments thereof may be utilised for the
detection of a
biomolecule in a biological sample comprising: applying a labelled antibody
directed against a given
biomolecule of the invention to said sample under conditions that favour an
interaction between the
labelled antibody and its corresponding protein. Depending on the nature of
the biological sample, it is
possible to determine not only the presence of a biomolecule, but also its
cellular distribution. For
example, in a blood serum sample, only the serum levels of a given biomolecule
can be detected,
whereas its level of expression and cellular localisation can be detected in
histological samples. It will
be obvious to those skilled in the art, that a wide variety of methods can be
modified in order to
achieve such detection.
For example, an antibody coupled to an enzyme is detected using a chromogenic
substrate that is
recognised and cleaved by the enzyme to produce a chemical moiety, which is
readily detected using
spectrometric, fluorimetric or visual means. Enzymes used to for labelling
include, but are not limited
to, malate dehydrogenase, staphylococcal nuclease, delta-5-steroid isomerase,
yeast alcohol
dehydrogenase, alpha-glycerophosphate, dehydrogenase, triose phosphate
isomerase, horseradish
peroxidase, alkaline phosphatase, asparaginase, glucose oxidase, beta-
galactosidase, ribonuclease,
crease, catalase, glucose-6-phosphate dehydrogenase, glucoamylase and
acetylcholinesterase.
Detection may also be accomplished by visual comparison of the extent of the
enzymatic reaction of a
substrate with that of similarly prepared standards. Alternatively,
radiolabelled antibodies can.be
detected using a gamma or a scintillation counter, or they ,can be detected
.using autoradiography. In
another example, fluorescently labelled antibodies are detected based on the
level at which the
attached compound fluoresces following exposure to a given wavelength:
Fluorescent compounds
typically used in antibody labelling include, but are not limited to,
fluorescein isothiocynate,
rhodamine, phycoerthyrin, phycocyanin, allophycocyani, o-phthaldehyde and
fluorescamine. In yet
another example, antibodies coupled to a chemi- or bioluminescent compound can
be detected by
determining the presence of luminescence. Such compounds include, but are not
limited to, luminal,
isoluminal, heromatic acridinium ester, imidazole, acridinium salt, oxalate
ester, luciferin, luciferase
and aequorin.
Furthermore, in vivo techniques for the detection of a biomolecule of the
invention include introducing
into a subject a labelled antibody directed against a given polypeptide or
fragment thereof.
21
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
In more than one embodiment of the invention, the test sample used for the
differential diagnosis of a
colorectal cancer and/or a non-malignant disease of the large intestine of a
subject may be of blood,
blood serum, plasma, nipple aspirate, urine, semen, seminal fluid, seminal.
plasma, prostatic fluid,
excreta, tears, saliva, sweat,. biopsy, ascites, cerebrospinal fluid, milk,
lymph, or tissue extract origin.
Preferably, test samples are of blood, blood serum, plasma, urine, excreta,
prostatic fluid, biopsy,
ascites, lymph or tissue extract origin. More preferred are blood, blood
serum, plasma, urine, excreta,
biopsy, lymph or tissue extract samples. Even more preferred are blood serum,
urine, excreta or biopsy
samples. Overall preferred are blood serum samples.
Furthermore, test samples used .for the methods of the invention are isolated
from subjects of
mammalian origin, preferably of primate origin. Even more preferred are
subjects of human origin.
In addition, the methods of the invention for the differential diagnosis of
healthy subjects, subjects
having a precancerous lesion of the large intestine, subjects having a
colorectal cancer, subjects having
a metastasized colorectal cancer or subjects having a non-malignant disease of
the large intestine
described herein may be combined with other diagnostic methods to improve the
outcome of the
differential diagnosis. Other diagnostic methods are known to those skilled in
the art.
b) Database
In another aspect of the invention, a database comprising of mass profiles
specific for healthy subjects,
subjects having a precancerous lesion of the large intestine, subjects having
a colorectal cancer,
subjects having a metastasised colorectal cancer, or subjects having a non-
malignant disease of the
large intestine is generated by contacting biological samples isolated from
above-mentioned subjects
with an adsorbent on a biologically active surface under specific binding
conditions, allowing the
biomolecules with said sample to bind .said adsorbent, detecting one or more
bound ~biomolecules
using a detection method wherein the detection method generates a mass profile
of said sample,
transforming the mass pxofile data into a computer-readable .form and applying
a mathematical
algorithm to classify the mass profile as specific for healthy subjects,
subjects having a precancerous
lesion of the large intestine, subjects having a colorectal cancer, subjects
having a metastasised
colorectal cancer, ox subjects having a non-malignant disease of the large
intestine.
According to the invention, the classification of said mass profiles is
performed using the "CART"
decision tree approach (classification and regression trees; Breiman et al.,
194) and is known to those
skilled in the art. Furthermore, bagging of classifiers is applied to overcome
typical instabilities of
forward variable selection procedures, thereby increasing overall classifier
performance (Breiman,
1994).
22
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
In more than one embodiment, one or more biomolecules selected from the group
having an apparent
molecular mass of 2020 Da ~ 10 Da, 2049 Da ~ 10 Da, 2270 Da ~ 11 Da, 2508 Da ~
13 Da, 2732 Da
~ 14 Da, 3026 Da ~ 15 Da, 3227 Da ~ 17 Da, 3326 Da ~ 17 Da, 3456 Da ~ 17 Da,
3946 Da ~ 20 Da,
4103 Da ~ 21 Da, 4242 Da~~ 21 Da, 4295 Da ~ 21.Da, 4359 Da ~ 22 Da, 4476 Da ~
22 Da, 4546 Da ~
23 Da, 4607 Da ~ 23 Da, 4719 Da ~ 24 Da, 4830 Da ~ 24 Da, 4865 Da ~ 24 Da,
4963 Da ~ 25 Da,
5112 Da ~ 26 Da, 5226 Da ~ 26 Da, 5493 Da ~ 27 Da, 5648 Da ~ 28 Da, 5772 Da ~
29 Da, 5854 Da ~
29 Da, 6446 Da ~ 32 Da, 6644 Da ~ 33 Da, 6852 Da ~ 34 Da, 6897 Da ~ 34 Da,
6999 Da ~ 35 Da,
7575 Da ~ 38 Da, 7657 Da ~ 38 Da, 8076 Da ~ 40 Da, 8215 Da ~ 41 Da, 8474 Da ~
42 Da, 8574 Da ~
43 Da, 8702' Da ~ 44 Da, 8780 Da ~ 44 Da, 8922 Da ~ 45 Da, 9078 Da ~ 45 Da,
9143 Da ~ 46 Da, .
9201 Da ~ 46 Da, 9359 Da ~ 47 Da, 9425 Da ~ 47 Da, 9581 Da ~ 48 Da, 9641 Da ~
48 Da, 9718 Da ~
49 Da, 9930 Da ~ 50 Da, 10215 Da ~ 51 Da, 10369 Da ~ 52 Da, 10440 Da ~ 52 Da,
10594 Da ~ 53
Da, 11216 Da ~ 56 Da, 11464 Da ~ 57 Da, 11547 Da ~ 58 Da, 11693 Da ~ 58 Da,
11905 Da ~ 60 Da,
12470 Da ~ 62 Da, 12619 Da ~ 63 Da, 12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632
Da ~ 68 Da,
13784 Da ~ 69 Da, 13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140
Da ~ 76 Da,
15350 Da ~ 77 Da, 15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164
Da ~ 81 Da,
16953 Da ~ 85 Da, 17263 Da ~ 86 Da, 17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766
Da ~ 89 Da,
17890 Da ~ $9 Da, 18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466
Da ~ 112 Da,
22676 Da ~ 113 Da, 22951 Da ~ 115 Da, 24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or
28259 Da ~ 141
Da may be detected within a given biological sample. Detection of said
biomolecules of the invention
is based on specific sample pre-treatment conditions, the pI3 of binding
conditions, and the type of
biologically active surface used for the detection of biomolecules.
Within the context of the invention, biomolecules within a given sample are
bound to an adsorbent on
a biologically active surface under specific binding conditions, for example,
the biomolecules within a -
given sample are applied to a biologically active suiface comprising
positively-charged quaternary
ammonium groups (cationic) and incubated with 0.1 M Tris=HCI, 0.02% Triton X-
100 at a pH of 8.5
to allow for specific binding. Biomolecules that bind to said biologically
active surface under these
conditions are negatively charged molecules. It should be noted that although
the biomolecules of the
invention are bound to a cationic adsorbent comprising of positively-charged
quaternary ammonium
groups, the biomolecules are capable of binding other types of adsorbents, as
described in another
section using binding conditions known to those skilled in the art.
Accordingly, some embodiments of
the invention are not limited to the use of cationic adsorbents
According to the invention, a biomolecule with the molecular mass of 2020 Da ~
10 Da, 2049 Da ~ 10
Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ l4.Da, 3026 Da ~ 15 Da, 3227
Da ~ 17 Da, 3326
Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20 Da, 4103 Da ~ 21 Da, 4242 Da ~ 21
Da, 4295 Da ~ 21
Da, 4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da ~ 23 Da, 4607 Da t 23 Da, 4719
Da ~ 24 Da, 4830
23
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112 Da ~ 26~ Da, 5226 Da ~ 26
Da, 5493 Da ~ 27
Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32 Da, 6644
Da ~ 33 Da, 6852
Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38
Da, 8076 Da ~ 40
Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574. Da ~ 43 Da, 8702 Da ~ 44 Da, 8780
Da ~ 44 Da, 8922
Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201 Da t 46 Da, 9359 Da ~ 47
Da, 9425 Da ~ 47
Da, 9581 Da t 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50 Da, 10215
Da ~ 51 Da,
10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da, 11464
Da ~ 57 Da,
11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619
Da ~ 63 Da,
12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983
Da ~ 70 Da,
14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879
Da ~ 79 Da,
15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263
1Da ~ 86 Da,
17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115
Da ~ 91 Da,
18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da, 22676 Da ~ 113 Da,
22951 Da ~ 115 Da,
24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da is detected by
diluting the biological
sample 1:5 in a denaturation buffer consisting of 7 M urea, 2 M thiourea, 4%
CHAPS, 1% DTT, and
2% Ampholine, and then 1:10 in binding buffer consisting of 0.1 M Tris-HCI,
0.02% Triton X-100 at
pH 8.5 at 0 to 4°C, applying thus treated sample to a biologically
active surface comprising positively
charged (cationic) quaternary ammonium groups (anion exchanging), incubating
for 120 minutes at 20
to 24°C, and subjecting the bound biomolecules to gas phase ion
spectrometry as described in another
section.
In one embodiment of the invention, biological samples used to generate a
database of mass profiles
for healthy subjects, subjects having a precancerous lesion of the large
intestine, subjects having a
colorectal cancer, subjects having a metastasised colorectal cancer or
subjects having a non-malignant
disease of the large intestine, may be of blood, blood serum, plasma, nipple
aspirate, urine, semen,
seminal fluid, seminal plasma, prostatic fluid, excreta, tears, saliva, sweat,
biopsy, ascites,
cerebrospinal fluid, milk, lymph, or tissue extract origin. Preferably,
biological samples are of blood,
blood serum, plasma, urine, excreta, prostatic fluid, biopsy, ascites, lymph
or tissue extract origin.
More preferred are blood, blood serum, plasma, urine, excreta, biopsy, lymph
or tissue extract
samples. Even more preferred are blood serum, urine, excreta or biopsy
samples. Overall preferred are
blood serum samples.
Furthermore, the biological samples related to the invention are isolated from
subjects considered to
be healthy, having a precancerous lesion of the large intestine, having a
colorectal cancer, having a
metastasised colorectal cancer or having a non-malignant disease of the large
intestine. Said subjects
are of mammalian. origin, preferably of primate origin. Even more preferred
are subjects of human
origin.
24
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
A subject of the invention that is said to have a precancerous lesion of the
large intestine, displays
preliminary stages of a cancer (i.e, dysplasia), wherein a cell and/or tissue
has become susceptible to
the development of a cancer as a result of either a genetic predisposition,
exposure to a cancer-causing
agent (carcinogen) or both.
A genetic pre-disposition may include a predisposition for an autosomal
dominant inherited cancer
syndrome which is generally indicated by a strong family history of uncommon
cancer andlor an
association with a specific marker phenotype (e.g. familial adenomatous polyps
of the colon), a
familial cancer wherein an evident clustering of cancer is observed but the
role of inherited
predisposition may not be clear (e.g. breast cancer, ovarian cancer, or colon
cancer), or an autosomal
recessive syndrome characterised by chromosomal or DNA instability. Whereas,
cancer-causing
agents include agents that cause genetic damage and induce neoplastic
transformation of a cell. Such
agents fall into three categories: 1) chemical carcinogens such as alkylating
agents, polycyclic
aromatic hydrocarbons, aromatic amines, azo dyes, nitrosamines and amides,
asbestos, vinyl chloride,
chromium, nickel, arsenic, and naturally occurring carcinogens (e.g. aflotoxin
B1); 2) radiation such as
ultraviolet (iJV) and ionisation radiation including electromagnetic (e.g. x-
rays, y-rays) and particulate
radiation (e.g. a and [3 particles, protons, neutrons); 3) viral and microbial
carcinogens such as human
Papillomavirus (HI'V), Epstein-Barr virus (EBV), hepatitis B virus (HBV),
human T-cell leukaemia
virus type 1 (HTLV-1), or Helicobacter pylori.
Alternatively, a subject within the invention that is said to have a
colorectal cancer possesses a cancer
that arises from the large intestine (interchangebly referred ~to as
colorectal cancers within the
invention). Such cancers may include, but are not limited to, colon and rectal
cancers.
Within the context of the invention, cancers of large intestine
(interchangebly referred to as colorectal
cancers within the invention) may also be of various stages, wherein the
staging is based on the size of
the primary lesion; its extent of spread to regional lymph nodes, and the
presence or absence of
blood-borne metastases (metastatic colorectal cancers. The various stages of a
cancer may be
identified using staging systems known to those skilled in the art [e.g. Union
Internationale Contre
Cancer (UICC) system or American Joint Committee on Cancer (AJC)]. Also
included are different
grades of said cancers, wherein the grade of a cancer is based on the degree
of differentiation of the
epithelial cells within the lining of the large intestine and the number of
mitoses as a correlation to a
neoplasm's aggression.
Healthy individuals, as related to certain embodiments of the invention, are
those that possess good
health, and demonstrate an absence of a .colorectal cancer or a non-malignant
disease of the large
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
intestine.
c) Biomolecules
The differential expression of biomolecules in samples from healthy subjects,
subjects having a
precancerous lesion of the large intestine, subjects having a colorectal
cancer, subjects having
metastasised colorectal cancer, and subjects having a non malignant disease of
the large intestine,
allows for the differential diagnosis of a non-malignant disease or a cancer
of the large intestine wihin
a subject.
Biomolecules are said to be specific for a particular clinical state (e.g.
healthy, precancerous lesion of
the large intestine, colorectal cancer, metastasised colorectal cancer, a non-
malignant disease of the
large intestine) when they are present at different levels within samples
taken from subjects in one
clinical state as compared to samples taken from subjects from other clinical
states (e.g. in subjects
with a precancerous lesion of the large intestine vs. in subjects with a
metastasised colorectal cancer).
Biomolecules may be present at elevated levels, at decreased levels, or
altogether absent within a
sample taken from a subject in a particular clinical state (e.g. healthy,
precancerous lesion of the large
intestine, colorectal cancer, metastasised colorectal cancer, a non-malignant
disease of the large
intestine). For example, biomolecules A and B are found at elevated levels in
samples isolated from
healthy subjects as compared to samples isolated from subjects having a
precancerous lesion of the
large intestine, a colorectal cancer, a metastatic colorectal cancer or a non-
malignant disease of the
large intestine. Whereas, biomolecules X, Y, Z are found at elevated levels
and/or more frequently in
samples isolated from subjects having a precancerous lesion of the large
intestine as opposed to
subjects in good health, having a colorectal cancer, a metastasised colorectal
cancer or a non-
malignant disease of the large intestine. Biomolecules A and B are said to be
specific for healthy
subjects, whereas biomolecules X, Y, Z are specific for subjects having a
precancerous lesion of the
large intestine.
Accordingly, the differential presence of one or more biomolecules found in a
test sample compared to
samples from healthy subjects, subjects with a precancerous lesion of the
large intestine, a colorectal
cancer, a metastasized colorectal cancer, or a non-malignant disease of the
large intestine, or the mere
detection of one or more biomolecules in the test sample provides useful
information regarding
probability of whether a subject being tested has a precancerous lesion of the
large intestine, a
colorectal cancer, a metastasized colorectal cancer or a non-malignant disease
of the large intestine.
The probability that a subject being tested has a precancerous lesion of the
large-intestine, a colorectal
cancer, a metastasized colorectal cancer or a non-malignant disease of the
large intestine depends on
whether the quantity of one or more biomolecules in a test sample taken from
said subject is
statistically significantly different from the quantity of one or more
biomolecules in a biological
26
CA 02525743 2005-11-14
WO 2004/102190 ~ PCT/EP2004/005294
sample taken from healthy subjects, subjects having a precancerous lesion of
the large intestine, a
colorectal cancer, a metastasised colorectal cancer, or a non-malignant
disease of the large intestine.
A biomolecule of the invention may be any molecule that is produced by a cell
or living organism, and
may have any biochemical property (e.g. phosphorylated proteins, positively
charged molecules,
negatively charged molecules, hydrophobicity, hydrophilicity), but preferably
biochemical properties
that allow binding of the biomolecule to a biologically active surface
comprising positively charged
quaternary ammonium groups after denaturation in 7 M urea, 2 M thiourea, 4%
CHAPS, 1% DTT, and
2% Ampholine and dilution in 0.1 M Tris-HCI, 0.02% Triton X-100 at pH 8.5 at 0
to 4°C followed by
incubation on said biologically active surface for 120 minutes at 20 to
24°C. Such molecules include,
but are not limited to, molecules comprising nucleotides, amino acids, sugars,
fatty acids, steroids,
nucleic acids, polynucleotides (DNA or RNA), polypeptides, proteins,
antibodies, carbohydrates,
lipids, and combinations thereof (e.g., glycoproteins, ribonucleoproteins,
lipoproteins). Preferably a
biomolecule may be a nucleotide, polynucleotide, peptide, protein or fragments
thereof. Even more
preferred are peptide or protein biomolecules.
The biomolecules of the invention can be detected based on specific sample pre-
treatment conditions,
the pH of binding conditions, the type of biologically active surface used for
the detection of
biomolecules within a given sample and their molecular mass. For example,
prior to the detection of
the biomolecules described herein, a given sample is pre-treated by diluting
1:5 in a denaturation
buffer consisting of 7 M urea, 2 M thiourea, 4% CHAPS, 1% DTT, and 2%
ampholine. The denatured
sample is then diluted 1:10 in 0.1 M Tris-HCl, 0.02% Triton X-100, pH 8.5,
applied,to a biologically
active surface comprising positively-charged quaternary ammonium groups
(cationic) and incubated
using specific buffer conditions (0.1 M Tris-HCI, 0.02% Triton X-100, pH 8.5)
to allow for binding of
said biomolecules to the above-mentioned biologically active surface. It
should be noted that although
the biomolecules of the invention are detected using a cationic adsorbent
positively charged
quaternary ammonium groups, as well as specific pre-treatment and binding
conditions, the
biomolecules are capable of binding other types of adsorbents, as described
below, using alternative
pre-treatment and binding conditions known to those skilled in the art.
Accordingly, some
embodiments of the invention are not limited to the use of cationic
adsorbents.
The biomolecules of the invention include biomolecules having a molecular mass
selected from the
group consisting of 2020 Da ~ 10 Da, 2049 Da ~ 10 Da, 2270 Da x'11 Da, 2508 Da
~ 13 Da, 2732 Da
~ 14 Da, 3026 Da ~ 15 Da, 3227 Da ~ 17 Da, 3326 Da ~ 17 Da, 3456 Da ~ 17 Da,
3946 Da ~ 20 Da,
4103 Da ~ 21 Da, 4242 Da ~ 21 Da, 4295 Da ~ 21 Da, 4359 Da ~ 22 Da, 4476 Da ~
22 Da, 4546 Da t
23 Da, 4607 Da t 23 Da, 4719 Da ~ 24 Da, 4830 Da ~ 24 Da, 4865 Da ~ 24 Da,
4963 Da ~ 25 Da,
5112 Da ~ 26 Da, 5226 Da ~ 26 Da, 5493 Da ~ 27 Da, 5648 Da ~ 28 Da, 5772 Da ~
29 Da, 5854 Da ~
27
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
29 Da, 6446 Da ~ 32 Da, 6644 Da ~ 33 Da, 6852 Da ~ 34 Da, 6897 Da ~ 34 Da,
6999 Da ~ 35 Da,
7575 Da ~ 38 Da, 7657 Da ~ 38 Da, 8076 Da t 40 Da, 8215 Da ~ 41 Da, 8474 Da ~
42 Da, 8574 Da ~
43 Da, 8702 Da ~ 44 Da, 8780 Da ~ 44 Da, 8922 Da ~ 45 Da, 9078 Da ~ 45 Da,
9143 Da ~ 46 Da,
9201 Da ~ 46 Da, 9359 Da ~ 47 Da, 9425 Da ~ 47 Da, 9581 Da ~ 48 Da, 9641 Da ~
48 Da, 9718 Da ~
49 Da, 9930 Da ~ 50 Da, 10215 Da ~ 51 Da, 10369 Da ~ 52 Da, 10440 Da ~ 52 Da,
10594 Da ~ 53
Da, 11216 Da ~ 56 Da, 11464 Da ~ 57 Da, 11547 Da ~ 58 Da, 1.1693 Da ~ 58 Da,
11905 Da ~ 60 Da,
12470 Da ~ 62 Da, 12619 Da ~ 63 Da, 1.2828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632
Da ~ 68 Da,
13784 Da ~ 69 Da, 13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140
Da ~ 76 Da,
15350 Da ~ 77 Da, 15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164
Da ~ 81 Da,
16953 Da ~ 85 Da, 17263 Da ~ 86 Da, 17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766
Da ~ 89 Da,
17890 Da ~ 89 Da, 18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466
Da ~ 112 Da,
22676 Da ~ 113 Da, 22951 Da ~ 115 Da, 24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or
28259 Da ~ 141
Da.
1 S According to the invention, a biomolecule with the molecular mass of 2020
Da ~ 10 Da, 2049 Da ~ 10
Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15 Da, 3227
Da ~ 17 Da, 3326
Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20 Da, 4103 Da ~ 21 Da, 4242 Da ~ 21
Da, 4295 Da ~ 21
Da, 4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da ~ 23 Da, 4607 Da ~ 23 Da, 4719
Da ~ 24 Da, 4830
Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112 Da t 26 Da, 5226 Da ~ 26
Da, 5493 Da ~ 27
Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32 Da, 6644
Da ~ 33 Da, 6852
Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38
Da, 8076 Da ~ 40
Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44 Da, 8780
Da ~ 44 Da, 8922
Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359 Da ~ 47
Da, 9425 Da ~ 47
Da, 9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50 Da, 10215
Da ~ 51 Da,
10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da, 11464
Da ~ 57 Da,
11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619
Da ~ 63 Da,
12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da; 13784 Da ~ 69 Da, 13983
Da ~ 70 Da,
14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879
Da ~ 79 Da,
15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263
Da ~ 86 Da,
17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115
Da ~ 91 Da,
18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da, 22676 Da ~ 113 Da,
22951 Da ~ 115 Da,
24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da is detected by
diluting the biological
sample 1:5 in a denaturation buffer consisting of 7 M urea, 2 M thiourea, 4%
CHAPS, 1% DTT, and
2% Ampholine, and then 1:10 in binding buffer consisting of 0.1 M Tri's-HCI,
0.02% Triton X-100 at
pH 8.5 at 0 to 4°C, applying thus treated sample to a biologically
active surface comprising positively
charged (cationic) quaternary ammonium groups (anion exchanging), incubating
for 120 minutes at 20
to 24°C, and subjecting the bound biomolecules to gas phase ion
spectrometry as described in another
28
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
section.
Although said biomolecules were first identified in blood serum samples, their
detection is not Limited
to said sample type. The biomolecules may also be detected in other samples
types, such as blood,
blood serum, plasma, nipple aspirate, urine, semen, seminal fluid, seminal
plasma, prostatic fluid,
excreta, tears, saliva, sweat, biopsy, ascites, cerebrospinal fluid,. milk,
lymph, or tissue extract.
Preferably, samples are of blood, blood serum, plasma, urine, excreta,
prostatic fluid, Biopsy, ascites,
lymph or tissue extract origin. More preferred are blood, blood serum, plasma,
urine, excreta, biopsy,
lymph or tissue extract samples. Even more preferred are blood serum, urine,
excreta or biopsy
samples. Overall preferred are blood serum samples.
Since the biomolecules can be sufficiently characterized by their mass and
biochemical characteristics
such as the type of biologically active surface they bind to or the pH of
binding conditions, it is not
necessary to identify the biomolecules in order to be able to identify them in
a sample. It should be
noted that molecular mass and binding properties are characteristic properties
of these biomolecules
and not limitations on the means of detection or isolation. Furthermore, using
the methods described
herein, or other methods known in the art, the absolute identity of the
markers can be determined. This
is important when one wishes to develop and/or screen for specific binding
molecules, or to develop a
an assay for the detection of said biomolecules using specific binding
molecules.
d) Biologically Active Surfaces
In one embodiment of the invention, biologically active surfaces include, but
are not restricted to,
surfaces that contain adsorbents such as quaternary ammonium 'groups (anion
exchange surfaces),
carboxylate groups (cation exchange surfaces), alkyl or aryl chains
(hydrophobic interaction, reverse
phase chemistry), groups such as ~nitriloacetic acid that immobilize metal
ions such as nickel, gallium,
copper, or zinc (metal affinity interaction), or biomolecules such as
proteins, preferably antibodies, or
nucleic acids, preferably protein binding sequences, covalently bound to the
surface via carbonyl
diimidazole moieties or epoxy groups (specific affinity interaction).
Preferred are adsorbents
comprising anion exchange surfaces.
These surfaces may be located on matrices like polysaccharides such as
sepharose, e.g. anion
exchange surfaces or hydrophobic interaction surfaces, or solid metals, e.g.
antibodies coupled to
magnetic beads. Surfaces may also include gold-plated surfaces such as those
used for Biacore Sensor
Chip technology. Other surfaces known to those skilled in the art are also
included within the scope of
the invention.
Biologically active surfaces are able to adsorb biomolecules like amino acids,
sugars, fatty acids,
29
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
steroids, nucleic acids, polynucleotides, polypeptides, carbohydrates, lipids,
and combinations thereof
(e.g., glycoproteins, ribonucleoproteins, lipoproteins).
In another embodiment, devices that use biologically active surfaces to
selectively adsorb
biomolecules may be chromatography columns for Fast Protein Liquid
Chromatography (FPLC) and
High Pressure Liquid Chromatography (HI'LC), where the matrix, e.g. a
polysaccharide, carrying the
biologically active surface, is filled into vessels (usually referred to as
"columns") made of glass, steel,
or synthetic materials like polyetheretherketone (PEEK).
Iu yet another embodiment, devices that use biologically active surfaces to
selectively adsorb
biomolecules may be metal strips carrying thin layers of the biologically
active surface on one or more
spots of the strip surface to be used as probes for gas phase ion spectrometry
analysis, for example the
SAX2 ProteinChip array (Ciphergen Biosystems, Inc.) for SELDI analysis.
e) Mass Profiling
In one embodiment, the mass profile of a sample may be generated using an
array-based assay in
which the biomolecules of a given sample are bound by biochemical or affinity
interactions to an
adsorbent present on a biologically active surface located on a solid platform
("array" or "probe"):
After the biomolecules have bound to the adsorbent, they are detected using
gas phase ion
spectrometry. Biomolecules or other substances bound to the adsorbents on the
probes can be analyzed
using a gas phase ion spectrometer. This includes, e.g., mass spectrometers,
ion mobility
spectrometers, or total ion current measuring devices. The quantity and
characteristics of the
biomolecule can be determined using gas phase ion spectrometry. Other
substances in addition to the
biomolecule of interest can also be detected by gas phase ion spectrometry.
In one embodiment, a mass spectrometer can be used to detect biomolecules on
the probe. In a typical
mass spectrometer, a probe with a biomolecule is introduced into an inlet
system of the mass
spectrometer. The biomolecule is then ionized by an ionization source, such as
a laser, fast atom
bombardment, or plasma. The generated ions are collected by an ion optic
assembly, and then a mass
analyzer disperses and analyzes the passing ions. Within the scope of this
invention, the ionisation
course that ionises the biomolecule is a laser.
The ions exiting the mass analyzer are detected by a ion detector. The ion
detector then translates
information of the detected ions into mass-to-charge ratios. Detection of the
presence of a biomolecule
or other substances will typically involve detection of signal intensity.
This, in turn, can reflect the
quantity and character of a biomolecule bound to the piobe.
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
In another embodiment, the mass profile of a sample may be generated using a
liquid-chromatography
(LC)-based assay in which the biomoleoules of a given sample are bound by
biochemical or affinity
interactions to an adsorbent located in a vessel made of glass, steel, or
synthetic material; known to
those skilled in the art as a chromatography column. The biomolecules are
eluted from the biologically
active surface by washing the vessel with appropriate solutions known to those
skilled in the art. Such
solutions include but are not limited. to, buffers, . e.g. Tris
(hydroxymethyl) aminomethane
hydrochloride' (TRIS-HCl), buffers containing salt, e.g. sodium chloride
(NaCl), or organic solvents,
e.g. acetonitrile. Biomolecule mass profiles are generated by application of
the eluting biomolecules of
the sample by direct connection via an electrospray device to a mass
spectrometer (LC/ESI-MS).
Conditions that promote binding of biomolecules to an adsorbent are known to
those skilled in the art
(reference) and ordinarily include parameters such as pH, the concentration of
salt, organic solvent, or
other competitors for binding of the biomolecule to the adsorbent. Within the
scope of the invention,
incubation temperatures are of at least 0 to 100°C, preferably of at
least 4 to 60°C, and most preferably
of at least 15 to 30°C. Varying additional parameters, such as
incubation time, the concentration of
detergent, e.g., 3-[(3-Cholamidopropyl) dimethylammonio~-2-hydroxy-1-
propanesulfonate (CHAPS),
or reducing agents, e,g. dithiothreitol (DTT), are also known to those skilled
in the art. Various
degrees of binding can be accomplished by combining the above stated
conditions as needed, and will
be readily apparent to those skilled in the art.
f) Methods for detecting biomolecules within a sample
In yet another aspect, the invention relates to methods for detecting
differentially present biomolecules
in a test sample and/or biological sample. Within the context of the
invention, any suitable method can
be used to detect one or more of the biomolecules described herein. For
example, gas phase ion
spectrometry can be used. This technique includes, e,g., laser
desorption/ionization mass spectrometry.
Preferably, the test and/or biological sample is prepared prior to gas phase
ion spectrometry, e.g.,
pre-fractionation, two-dimensional gel chromatography, high performance liquid
chromatography, etc,
to assist detection of said biomolecules. Detection of said biomolecules can
also be achieved using
methods other than gas phase ion spectrometry. For example, immunoassays can
be used to detect the
biomolecules within a sample.
In one embodiment, the test and/or biological sample is prepared prior to
contacting a biologically
active surface and is in aqueous form. Examples said samples include, but are
not limited to, blood,
blood serum, plasma, nipple aspirate, urine, semen, seminal fluid, seminal
plasma, prostatic fluid,
tears, saliva, sweat, ascites, cerebrospinal fluid, mills, lymph, or tissue
extract samples. Furthermore,
solid test and/or biological samples, such as excreta or biopsy samples can be
solubilised in or
admixed with an eluent using methods known to those skilled in the art such
that said samples may be
31
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
easily applied to a biologically active surface. Test and/or biological
samples in the aqueous form can
be further prepared using specific solutions for denaturation (pre-treatment)
like sodium dodecyl
sulfate,.mercaptoethanol, urea, etc. For example, a test and/or biological
sample of the invention can
be denatured prior to contacting a biologically active surface comprising of
quaternary ammonium
groups by diluting said sample 1:5 with a buffer consisting of 7 M urea, 2 M
thiourea, 4% CHAPS,
1% DTT and 2% ampholine.
The sample is contacted with a biologically active surface using any
techniques including bathing,
soaking, dipping, spraying, washing over, or pipetting, etc. Generally, a
volume of sample containing
from a few atomoles to 100 picomoles of a biomolecule in about 1 to 500 ~1 is
sufficient for detecting
binding of the biomolecule to the adsorbent.
The pH value of the solvent in which the sample contacts the biologically
active surface is a function
of the specific sample and the selected biologically active surface.
Typically, a sample is contacted
with a biologically active surface under pH values between 0 and 14,
preferably between about 4 and
10, more preferably between 4.5 and 9.0, and most preferably, at pH 8.5. The
pH value depends on the
type of adsorbent present on a biologically active surface and can be adjusted
accordingly.
The sample can contact the adsorbent present on a biologically active for a
period of time sufficient to
allow the marker to bind to the adsorbent. Typically, the sample and the
biologically active surface are
contacted for a period of between about 1 second and about 12 hours,
preferably, between about 30
seconds and about 3 hours, and most preferably for 120 minutes.
The temperature at which the sample contacts the biologically active surface
(incubation temperature)
is a function of the specific sample and the selected biologically active
surface. Typically, the washing
solution can be at a temperature of between 0 and 100°C, preferably
between 4 and 37°C, and most
preferably between 20 and 24°C.
For example, a biologically active surface comprising of quaternary ammonium
groups (anion
exchange surface) will bind the biomolecules described herein~when the pH
value is between 6.5 and
9Ø Optimal binding of the biomolecules of the present invention occurs at a
pH of 8.5. Furthermore, a
sample is contacted with said biologically active surface for 120 min, at a
temperature of 20 - 24 °C.
Following contacting a sample or sample solution with a biological surface, it
is preferred to remove
any unbound biomolecules so that only the bound biomolecules remain on the
biologically active
surface. Washing unbound biomolecules are removed by methods known to those
skilled in. the art
such as bathing, soaking, dipping, rinsing, spraying, or washing the
biologically active surface with an
32
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
eluent or a washing solution. A microfluidics process is preferably used when
a washing solution such
as an eluent is introduced to small spots of adsorbents on the biologically
active surface. Typically, the
washing solution can be at a temperature of between 0 and 100°C,
preferably between 4 and 37°C, and
most preferably between 20 and 24°C.
Washing solution or eluents used to wash the unbound biomolecules from a
biologically active surface
include, but are not limited to, organic solutions, aqueous solutions such as
buffers wherein a buffer
may contain detergents, salts, or reducing agents in appropriate
concentrations as those known to those
skilled in the art.
Aqueous solutions are preferred for washing biologically active surfaces.
Exemplary aqueous
solutions include, but not limited to, HEPES buffer, Tris buffer, phosphate
buffered saline (PBS), and
modifications thereof. The selection of a particular washing solution or an
eluent is dependent on other
experimental conditions (e. g., types of adsorbents used or biomolecules to be
detected), and can be
determined by those of skill in the art. For example, if a biologically active
surface comprising a
quaternary ammonium group as adsorbent (anion exchange surface) is used, then
an aqueous solution,
such as a Tris buffer, may be preferred. In. another example, if a
biologically active surface comprising
a carboxylate group as adsorbent (cation exchange surface) is used, then an
aqueous solution, such as
an acetate buffer, may be preferred.
Optionally, an energy absorbing molecule (EAM), e.g. in solution, can be
applied to biomolecules or
other substances bound on the biologically active surface by spraying,
pipetting or dipping. Applying
an EAM can be done after unbound materials are washed off of the biologically
active surface.
Exemplary energy absorbing molecules include, but are not limited to, cinnamic
acid derivatives,
sinapinic acid and dihydroxybenzoic acid.
t
Once the biologically active surface is free of any unbound biomolecules,
adsorbent-bound
biomolecules are detected using gas phase ion spectrometry. The quantity and
characteristics of a
biomolecule can be determined using said method. Furthermore, said
biomolecules can be analyzed
using a gas phase ion spectrometer such as mass spectrometers, ion mobility
spectrometers, or total
ion current measuring devices. Other gas phase ion spectrometers known to
those skilled in the art are
also included.
In one embodiment, mass spectarometry can be used to detect biomolecules of a
given sample present
on a biologically active surface. Such methods include, but are not limited
to, matrix-assisted laser
desorption ionization/time-of flight (MALDI-TOF), surface-enhanced laser
desorption
ionization/time-of flight (SELDI-TOF), liquid chromatography coupled with MS,
MS-MS, or
33
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
ESI-MS. Typically, biomolecules are analysed by introducing a biologically
active surface containing
said biomolecules, ionizing said biomolecules to generate ions that are
collected and analysed.
In a preferred embodiment, the biomolecules present in a sample are detected
using gas phase ion
spectrometry, and more preferably, using mass spectrometry. In one embodiment,
matrix-assisted laser
desorption/ionization ("MALDI") mass spectrometry can be used. In MALDI, the
sample is typically
quasi-purified to obtain a fraction that.essentially consists of a marker
using separation methods such
as two-dimensional gel electrophoresis or high performance liquid
chromatography (HPLC).
In another embodiment, surface-enhanced laser desorption/ionization mass
spectrometry ("SELDI")
can be used. SELDI uses a substrate comprising adsorbents to capture
biomolecules, which can then
be directly desorbed and ionized from the substrate surface during mass
spectrometry. Since the
substrate surface in SELDI captures biomolecules, a sample need not be quasi-
purified as in MALDI.
However, depending on the complexity of a sample and the type of adsorbents
used, it may be
desirable to prepare a sample to reduce its complexity prior to SELDI
analysis.
For example, biomolecules bound to a biologically active surface can be
introduced into an inlet
system of the mass spectrometer. The biomolecules are then ionized by an
ionization source such as a
laser, fast atom bombardment, or plasma. The generated ions are then collected
by an ion optic
assembly, and then a mass analyzer disperses the passing ions. The ions
exiting the mass analyzer are
detected by a detector and translated into mass-to-charge ratios. Detection of
the presence of a
biomolecule typically involves detection of its specific signal intensity, and
reflects the quantity and
character of said biomolecule.
In a preferred embodiment, a laser desorption time-of flight mass
spectrometeris used with the probe
of the present invention. In laser desorption mass spectrometry, biomolecules
bound to a biologically
active surface are introduced into an inlet system. Biomolecules are desorbed
and ionized into the gas
phase by a laser. The ions generated are then collected by an ion optic
assembly. These ions are
accelerated through a short high voltage field and let drift into a high
vacuum chamber of a time-of
flight mass analyzer. At the far end of the high vacuum chamber, the
accelerated ions strike a sensitive
detector surface at a different time. Since the time-of flight is a function
of the mass of the ions, the
elapsed time between ionization and impact can be used to identify the
presence or absence of
molecules of a specific mass.
The detection of biomolecules described herein can be enhanced using certain
selectivity conditions
(e. g., types of adsorbents used or washing solutions). In a preferred
embodiment, the same or
substantially the same selectivity conditions that were used to discover the
biomolecules can be used
34
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
in the methods for detecting a biomolecule in a sample.
Combinations of the laser desorption time-of flight mass spectrometer with
other components
described herein, in the assembly of mass spectrometer that employs various
means of desorption,
acceleration, detection, measurement of time, etc., are known to those skilled
in the art.
Data generated by desorption and detection of markers can be analyzed with the
use. of a
programmable digital computer. The computer program generally contains a
readable medium that
stores codes. Certain codes can be devoted to memory that include the location
of each feature on a
biologically active surface, the identity of the adsorbent at that feature and
the elution conditions used
to wash the adsorbent. Using this information, the program can then identify
the set of features on the
biologically active surface defining certain selectivity characteristics (e.
g. types of adsorbent and
eluents used). The computer also contains codes that receive as data (input)
on the strength of the
signal at various molecular masses received from a particular addressable
location on the biologically
active surface. This data can indicate the number of biomolecules detected, as
well as the strength of
the signal and the determined molecular mass for each biomolecule detected.
Data analysis can include the steps of determining signal strength (e. g.,
height of peaks) of a
biomolecule detected and removing "outliers" (data deviating from a
predetermined statistical
distribution). For example, the observed peaks can be normalized, a process
whereby the height of
each peak relative to some reference is calculated. For example, a reference
can be background noise
generated by instrument and chemicals (e. g., energy absorbing molecule),
which is set as zero in the
scale. Then the signal strength detected for each biomolecule can be displayed
in the form of relative
intensities in the scale desired (e. g., 100). Alternatively, a standard may
be admitted with the sample
so that a peak from the standard can be used as a reference to calculate
relative intensities of the
signals observed for each biomolecule or other biomolecules detected.
The computer can transform the resulting data into various formats for
displaying. In. one format,
referred to as "spectrum view", a standard spectral view can be displayed,
wherein the view depicts
the quantity of a biomolecule reaching the detector at each particular
molecular mass. In another
format, referred to as "scatter plot" only the peak height and mass
information are retained from the
spectrum view, yielding a cleaner image and enabling biomolecules with nearly
identical molecular
mass to be more visible.
Using any of the above display formats, it can be readily determined from the
signal display whether a
biomolecule having a particular molecular mass is detected from a sample.
Preferred biomolecules of
the invention are biomolecules with an apparent molecular mass of about 2020
Da ~ 10 Da, 2049 Da ~
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14 Da, 3026: Da ~ 15 Da, 3227
Da ~ 17 Da,
3326 Da ~ 17 Da,. 3456 Da ~ 17 Da, 3946 Da x_20 Da, 4103 Da ~ 21 Da, 4242 Da ~
21 Da, 4295 Da ~
21 Da, 4359 Da ~ 22 Da, 4476 Da ~ 22 Da; 4546 Da ~ 23 Da, 4607 Da ~ 23 Da,
4719 Da ~ 24 Da,
4830 Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226 Da ~
26 Da, 5493 Da ~
5 27 Da, 5648 Da ~ 28 Da; 5772 Da ~ 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32 Da,
6644 Da ~ 33 Da,
6852 Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~
38 Da, 8076 Da ~
40 Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44 Da,
8780 Da ~ 44 Da,
8922 Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359 Da ~
47 Da, 9425 Da ~
47 Da, 9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50 Da,
10215 Da ~ 51 Da,
10 10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da,
11464 Da ~ 57 Da,
11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619
Da ~ 63 Da,
12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784 Da w 69 Da, 13983
Da ~ 70 Da,
14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879
Da ~ 79 Da,
15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263
Da ~ 86 Da,
17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115
Da ~ 91 Da,
18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 .Da ~ 112 Da, 22676 Da ~ 113 Da,
22951 Da ~ 115 Da,
24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da. Moreover, from the
strength of
signal, the amount of a biomolecule bound on the biologically active surface
can be determined.
g) Identification of proteins
In. case the biomolecules of the invention are proteins, the present invention
comprises a method for
the identification of these proteins, especially by obtaining their amino acid
sequence. This method
comprises the purification of said proteins from the complex biological sample
(blood, blood serum,
plasma, nipple aspirate, urine, semen, seminal fluid, seminal plasma,
prostatic fluid, tears, saliva,
sweat, ascites, cerebrospinal fluid, mills, lymph, or tissue.extraot samples)
by fractionating said sample
using techniques known by the one of ordinary skill in the art, most
preferably protein
chromatography (FPLC, HPLC).
The biomolecules of the invention include those proteins with a molecular mass
selected from 2020
Da ~ 10 Da, 2049 Da ~ 10 Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14
Da, 3026 Da ~ 15
Da, 3227 Da ~ 17 Da, 3326 Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20 Da, 4103
Da ~ 21 Da, 4242
Da ~ 21 Da, 4295 Da ~ 21 Da; 4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da ~ 23
Da, 4607 Da ~ 23
Da, 4719 Da ~ 24 Da, 4830 Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112
Da ~ 26 Da, 5226
Da ~ 26 Da, 5493 Da ~ 27 Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29
Da, 6446 Da ~ 32
Da, 6644 Da ~ 33 Da, 6852 Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575
Da ~ 38 Da, 7657
Da ~ 38.Da, 8076 Da ~ 40 Da, 8215 Da ~ 41 Da, 8474 Da ~ 42 Da, 8574 Da ~ 43
Da, 8702 Da ~ 44
Da, 8780 Da ~ 44 Da, 8922 Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201
Da ~ 46 Da, 9359
36
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Da ~ 47 Da, 9425 Da ~ 47 Da, 9581 Da ~ 48 Da, 9641 Da ~ 48v Da, 9718 Da ~ 49
Da, 9930 Da ~ 50
Da, 10215 Da ~ 51 Da, 10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da,
11216 Da ~ 56 Da,
11464 Da ~ 57 Da, 11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470
Da t 62 Da,
12619 Da ~ 63 Da, 12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784
Da ~ 69 Da,
13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350
Da ~ 77 Da,
15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953
Da ~ 85 Da,
17263 Da ~ 86 Da, 17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890
Da ~ 89 Da,
18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da,
22676 Da ~ 113 Da,
22951 Da ~ 115 Da, 24079 Da ~ 120 Da, 28055 Da ~ 140 Da, and 28259 Da ~ 141
Da.
Furthermore, the method comprises the analysis of the fractions for the
presence and purity of said
proteins by the method which was used to identify them as differentially
expressed biomolecules, for
example two-dimensional gel electrophoresis or SELDI mass spectrometry, but
most preferably
SELDI mass spectrometry. The method also comprises an analysis of the purified
proteins 'aiming
towards the revealing of their amino acid sequence. This analysis may be
performed using techniques
in mass spectroscopy known to those skilled in~the art.
In one embodiment, this analysis may be performed using peptide mass
fingerprinting, revealing
information about the specific peptide mass profile after proteolytic
digestion of the investigated
protein.
In another embodiment, this analysis may be preferably performed using post-
source-decay (PSD), or
MSMS, but most preferably MSMS, revealing mass information about all possible
fragments of the
investigated protein or proteolytic peptides thereof leading to the amino acid
sequence of the
~ investigated protein of proteolytic peptide thereof.
The information revealed by the aforementioned techniques can be used to feed
world-wide-web
search engines, such as MS Fit (Protein Prospector,
http://prospector.ucsf.edu) for information
obtained.from peptide mass fingerprinting, or MS Tag (Protein Prospector,
http://prospector.ucsf.edu)
for information obtained from PSD, or mascot (www.matrixscience.com) for
information obtained
from MSMS and peptide mass fingerprinting, for the alignment of the obtained
results with data
available in public protein sequence databases, such as SwissProt
(http://us.expasy.org/sprot~, NCBI
(http://www.ncbi.nhn.nih.govBLAST~, EMBL (http://srs.embl-
heidelberg.de:8000/srs5n which leads
to a confident information about the identity of said proteins.
37
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
This information may comprise, if available, the complete amino acid sequence,
the calculated
molecular mass, the structure, the enzymatic activity, the physiological
function, and gene expression
of the investigated proteins.
h Kits
In yet another aspect, the invention provides kits using the methods of the
invention as described in the
section Diagnostics for the differential diagnosis of colorectal cancer or a
non-malignant disease of the
large intestine, wherein the kits are used to detect the biomolecules of the
present invention.
The methods used to detect the biomolecules of the invention can also be used
to determine whether a
subject is at risk of developing colorectal cancer or a non-malignant disease
of the large intestine, or
has developed a colorectal cancer or a non-malignant disease of the large
intestine. Such methods may
also be employed in the form of a diagnostic kit comprising an antibody
specific to a biomolecule of
the invention or a biologically active surface described herein, which may be
conveniently used, for
example, in clinical settings to diagnose patients exhibiting symptoms or a
family history of a
non-steroid dependent cancer. Such diagnostic kits also include solutions and
materials necessary for
the detection of a biomolecule of the invention, and instructions to use the
kit based on the
above-mentioned methods.
The biomolecules of the invention include those proteins wit~l a molecular
mass selected from 2020
Da ~ 10 Da, 2049 Da ~ 10 Da, 2270 Da ~ 11 Da, 2508 Da ~ 13 Da, 2732 Da ~ 14
Da, 3026 Da ~ 15
Da, 3227 Da ~ 17 Da, 3326 Da ~ 17 Da, 3456 Da ~ 17 Da, 3946 Da ~ 20 Da, 4103
Da ~ 21 Da, 4242
Da ~ 21 Da, 4295 Da ~ 21 Da, 4359 Da ~ 22 Da, 4476 Da ~ 22 Da, 4546 Da ~ 23
Da, 4607 Da ~ 23
Da, 4719 Da ~ 24 Da, 4830 Da ~ 24 Da, 4865 Da ~ 24 Da, 4963 Da ~ 25 Da, 5112
Da ~ 26 Da, 5226
Da ~ 26 Da, 5493 Da ~ 27 Da, 5648 Da ~ 28 Da, 5772 Da ~ 29 Da, 5854 Da ~ 29
Da, 6446 Da ~ 32
Da, 6644 Da ~ 33 Da, 6852 Da ~ 34 Da, 6897 Da ~ 34 Da, 6999 Da ~ 35 Da, 7575
Da ~ 38 Da, 7657
Da ~ 38 Da, 8076 Da ~ 40 Da, 8215 Da ~ 41 Da,~ 8474 Da ~ 42 Da, 8574 Da ~ 43
Da, 8702 Da ~ 44
Da, 8780 Da ~ 44 Da, 8922 Da ~ 45 Da, 9078 Da ~ 45 Da, 9143 Da ~ 46 Da, 9201
Da ~ 46 Da, 9359
Da ~ 47 Da, 9425 Da ~ 47 Da, 9581 Da ~ 48 Da, 9641 Da ~ 48 Da, 9718 Da ~ 49
Da, 9930 Da ~ 50
Da, 10215 Da ~ 51 Da, 10369 Da ~ 52 Da, 10440 Da ~ 52 Da, 10594 Da ~ 53 Da,
11216 Da ~ 56 Da,
11464 Da ~ 57 Da, 11547 Da ~ 58 Da, 11693 Da ~ 58 Da, 11905 Da ~ 60 Da, 12470
Da ~ 62 Da,
12619 Da ~ 63 Da, 12828 Da ~ 64 Da, 13290 Da ~ 66 Da, 13632 Da ~ 68 Da, 13784
Da ~ 69 Da,
13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005 Da ~ 75 Da, 15140 Da ~ 76 Da, 15350
Da ~ 77 Da,
15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104 Da ~ 81 Da, 16164 Da ~ 81 Da, 16953
Da ~ 85 Da,
17263 Da ~ 86 Da, 17397 Da ~ 87 Da, 17617 Da ~ 88 Da, 17766 Da ~ 89 Da, 17890
Da ~ 89 Da,
18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338 Da ~ 112 Da, 22466 Da ~ 112 Da,
22676' Da ~ 113 Da,
22951 Da ~ 115 Da, 24079 Da ~ 120 Da, 28055 Da ~ 140 Da, or 28259 Da ~ 141 Da.
38
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
For example, the kits can be used to detect one or more of differentially
present biomolecules as
described above in a test sample of subject. The kits of the invention have
many applications. For
example, the kits can.be used to differentiate if a subject is healthy, having
a precancerous lesion of
the large intestine, a colorectal cancer, a metastasized colorectal cancer or
a non-malignant disease of
the large intestine. Thus aiding the diagnosis of colorectal cancer or a non-
malignant disease of the
large intestine. In another example, the kits can be used to identify
compounds that modulate
expression of said biomolecules.
In one embodiment, a kit comprises an adsorbent on a biologically active
surface, wherein the
adsorbent is suitable for binding one or more biomolecules of the invention, a
denaturation solution for
the pre-treatment of a sample, a binding solution, a washing solution or
instructions for making a
denaturation solution, binding solution, or washing solution, wherein the
combination allows for the
detection of a biomolecule using gas phase ion spectrometry. Such kits can be
prepared from the
materials described in other previously detailed sections (e. g., denaturation
buffer, binding buffer,
adsorbents, washing solutions, etc.).
In some embodiments, the kit may comprise a first substrate comprising an
adsorbent thereon (e. g., a
particle functionalized with an adsorbent) and a second substrate onto which
the first substrate can be
positioned to form a probe, which is removably insertable into a gas phase ion
spectrometer. In other
embodiments, the kit may comprise a single substrate, which is in the form of
a removably insertable
probe with adsorbents on the substrate.
In another embodiment, a kit comprises a binding molecule that specifically
binds to a biomolecule
related to the invention, a detection reagent, appropriate solutions and
instructions on how to use the
kit. Such kits can be prepared from the materials described above, and other
materials known to those
skilled in the art. A binding. molecule used within such a kit may include,
but is not limited to,
proteins, peptides, nucleotides, nucleic acids, hormones, amino acids, sugars,
fatty acids, steroids,
polynucleotides, carbohydrates, lipids, or a combination thereof (e.g.
glycoproteins,
ribonucleoproteins, lipoproteins), compounds or synthetic molecules.
Preferably, a binding molecule
used in said kit is an antibody.
In either embodiment, the kit may optionally fuxther comprise a standard or
control information so that
the test sample can be compared with the control information standard to
determine if the test amount
of a marker detected in a sample is a diagnostic amount consistent with a
diagnosis of colorectal
cancer.
39
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
The present invention also relates to use 2020 Da ~ 10 Da, 2049 Da ~ 10 Da,
2270 Da ~ 11 Da, 2508
Da ~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15 Da, 3227 Da ~ 17 Da, 3326 Da ~ 17
Da, 3456 Da ~ 17
Da, 3946 Da ~ 20 Da, 4103 Da ~ 21 Da, 4242 Da ~ 21 Da, 4295 Da ~ 21 Da, 4359
Da ~ 22 Da, 4476
Da ~ 22 Da, 4546 Da ~ 23 Da, 4607 Da ~ 23 Da, 4719 Da ~ 24 Da, 4830 Da ~ 24
Da, 4865 Da ~ 24
Da, 4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226 Da ~ 26 Da, 5493 Da ~ 27 Da, 5648
Da ~ 28 Da, 5772
Da ~ 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32 Da, 6644 Da ~ 33 Da, 6852 Da ~ 34
Da, 6897 Da ~ 34
Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38 Da, 8076 Da ~ 40 Da, 8215
Da ~ 41 Da, 8474
Da ~ 42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44 Da, 8780 Da ~ 44 Da, 8922 Da~~ 45
Da, 9078 Da ~ 45
Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359 Da ~ 47 Da, 9425 Da ~ 47 Da, 9581
Da ~ 48 Da, 9641
Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ SO Da, 10215 Da ~ 51 Da, 10369 Da ~ 52
Da, 10440 Da ~
52 Da; 10594 Da ~ 53 Da, 11216 Da ~ 56 Da, 11464 Da ~ 57 Da, 11547 Da ~ 58 Da,
11693 Da ~ 58
Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619 Da ~ 63 Da, 12828 Da ~ 64 Da,
13290 Da ~ 66 Da,
13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983 Da ~ 70 Da, 14798 Da ~ 74 Da, 15005
Da ~ 75 Da,
15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104
Da ~ 81 Da,
16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263 Da ~ 86 Da, 17397 Da ~ 87 Da, 17617
Da ~ 88 Da,
17766 Da.~ 89 Da, 17890 Da ~ 89 Da, 18115~Da ~ 91 Da, 18390 Da ~ 92 Da, 22338
Da ~ 112 Da,
22466 Da ~ 112 Da, 22676 Da ~ 113 Da, 22951 Da ~ 115 Da, 24079 Da ~ 120 Da,
28055 Da ~ 140
Da, or 28259 Da ~ 141 Da for manufacture of an agent for diagnosis,
prophylactic and/or therapeutic
treatment of non-steroid dependent cancer, preferably colorectal cancer.
_
The invention also relates to a method for aiding non-steroid dependent cancer
diagnosis especially
colorectal cancer, the method comprising (a) detecting at least one protein
marker in a sample,
wherein the protein marker is selected from 2020 Da ~ 10 Da, 2049 Da ~ 10 Da,
2270 Da ~ 11 Da,
2508 Da ~ 13 Da, 2732 Da ~ 14 Da, 3026 Da ~ 15 Da, 3227 Da ~ 17 Da, 3326 Da ~
17 Da, 3456 Da ~
17 Da, 3946 Da ~ 20 Da, 4103 Da ~ 21 Da, 4242 Da ~ 21 Da, 4295 Da ~ 21 Da,
4359 Da ~ 22 Da,
' 4476 Da ~ 22 Da, 4546 Da ~ 23 Da, 4607 Da ~ 23 Da, 4719 Da ~ 24 Da, 4830 Da
~ 24 Da, 4865 Da ~
24 Da, 4963 Da ~ 25 Da, 5112 Da ~ 26 Da, 5226 Da ~ 26 Da, 5493 Da ~ 27 Da,
5648 Da ~ 28 Da,
5772 Da t 29 Da, 5854 Da ~ 29 Da, 6446 Da ~ 32 Da, 6644 Da ~ 33 Da, 6852 Da ~
34 Da, 6897 Da ~
34 Da, 6999 Da ~ 35 Da, 7575 Da ~ 38 Da, 7657 Da ~ 38 Da, 8076 Da ~ 40 Da,
8215 Da ~ 41 Da,
8474 Da ~ 42 Da, 8574 Da ~ 43 Da, 8702 Da ~ 44 Da, 8780 Da ~ 44 Da, 8922 Da ~
45 Da, 9078 Da ~
45 Da, 9143 Da ~ 46 Da, 9201 Da ~ 46 Da, 9359 Da ~ 47 Da, 9425 Da ~ 47 Da,
9581 Da ~ 48 Da,
9641 Da ~ 48 Da, 9718 Da ~ 49 Da, 9930 Da ~ 50 Da, 10215 Da ~ S 1 Da, 10369 Da
~ 52 Da, 10440
Da ~ 52 Da, 10594 Da ~ 53 Da, 11216 Da ~ 56 Da, 11464 Da ~ 57 Da, 11547 Da ~
S8 Da, 11693 Da
~ 58 Da, 11905 Da ~ 60 Da, 12470 Da ~ 62 Da, 12619 Da ~ 63 Da, 12828 Da ~ 64
Da, 13290 Da ~ 66 .
Da, 13632 Da ~ 68 Da, 13784 Da ~ 69 Da, 13983 Da ~ 70 Da, 14798 Da ~ 74 Da,
15005 Da ~ 75 Da,
15140 Da ~ 76 Da, 15350 Da ~ 77 Da, 15879 Da ~ 79 Da, 15957 Da ~ 80 Da, 16104
Da ~ 81 Da,
16164 Da ~ 81 Da, 16953 Da ~ 85 Da, 17263 Da ~ 86 Da, 17397 Da ~ 87 Da, ~
17617 Da ~ 88 Da,
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
17766 Da ~ 89 Da, 17890 Da ~ 89 Da, 18115 Da ~ 91 Da, 18390 Da ~ 92 Da, 22338
Da ~ 112 Da,
22466 Da ~ 112 Da, 22676 Da ~ 113 Da, 22951 Da ~ 115 Da, 24079 Da ~ 120 Da,
28055 Da ~ 140
Da, or 28259 Da ~ 141 Da and (b) correlating the detection of the or protein
marker with a probable
diagnosis of non-steroid cancer especially colorectal cancer..
Each recorded measurement reading is accompanied by a margin of deviation. The
latter
statistical imprecision is well-known to those skilled in the art. In the
scope of the present
invention, the margin of deviation is exclusively device-specific. That means
it is caused by
the type of analytical device used which is preferably a mass spectrometer.
The accuracy of
the recorded measurement reading is specified by a fixed percentage. In the
meaning of the
present 'invention, each disclosed molecular mass represents the averaged
value of that range
which deviates from the averaged value about ~ 0.5 %.
Furthermore, slight differences appear in the molecular mass value itself
which concerns the
. same protein in parallel patent applications disclosing the matter of cancer
biomaxkers. There
are three reasons to be considered. First, each molecular mass results from
the analysis of
samples belonging to another type of cancer. The origin of sample, the
cellular status, the
environmental conditions of the gathered tissue etc. exert an influence on the
measurements.
Secondly, the given molecular mass of the biomarkers represents the averaged
value which is
calculated from the data of numerous samples of each cancer species. Thirdly,
measuring
errors might be also imaginable, for example due to the sample preparation.
Above statements are further illustrated by examples which should not be
construed as
limiting with regard to the type of disease, the number of given molecular
masses or in any
other way. The following molecular masses of biomolecules are regarded as
equivalent:
(i) 2020 10 (epithelial cancer) and 2020 10
(colorectal cancer)
(ii) 2050 10 (epithelial cancer) and 2049 10
(colorectal cancer)
(iii) 3946 20 (epithelial cancer) and 3946 20
(colorectal cancer)
(iv) 4104 21 (epithelial cancer) and 4103 21
(colorectal cancer)
(v) 4298 21 (epithelial cancer) and 4295 21
(colorectal cancer)
(vi) 4360 ~ 22 (epithelial cancer) and 4359 ~ 22 (colorectal cancer)
(vii) 4477 ~ 22 (epithelial cancer) and 4476 ~ 22 (colorectal cancer)
(viii) 4867 ~ 24 (epithelial cancer) and 4865 ~ 24 (colorectal cancer)
(ix) 4958 ~ 25 (epithelial cancer) and 4963 ~ 25 (colorectal cancer)
41
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
(x) 5491 27 (epithelial cancer) and 5493 27
(colorectal cancer)
(xi) 5650 28 (epithelial cancer) and 5648 28
(colorectal cancer)
(xii) 6449 32 (epithelial cancer) and 6446 32
(colorectal cancer.)
(xiii) 6876 34 (epithelial cancer) and 6852 34
(colorectal cancer)
(xiv) 7001 35 (epithelial cancer) and 6999 35
(colorectal cancer)
(xv) 8232 41 (epithelial cancer) and 8215 41
(colorectal cancer)
(xvi) 8711 44 (epithelial cancer) and 8702 44
(colorectal cancer)
(xvii) 12471 62 (epithelial cancer) and 12470
62 (colorectal cancer)
(xviii) 12669 63 (epithelial cancer) and 12619
63 (colorectal cancer)
(xix) 13989 70 (epithelial cancer) and 13983 70
(colorectal cancer)
(xx) 15959 80 (epithelial cancer) and 15957 80
(colorectal cancer)
(xxi) 16164 81 (epithelial cancer) and 16164 81
(colorectal cancer)
(xxii) 17279 86 (epithelial cancer) and 17263 86
(colorectal cancer)
(xxiii) 17406 87 (epithelial cancer) and 17397 87
(colorectal cancer)
(xxiv) 17630 88 (epithelial cancer) and 17617 88
(colorectal cancer)
(xxv) 18133 91 (epithelial cancer) and 18115 91
(colorectal cancer)
In all examples, each recorded measurement reading is overlapping with any
others within its
margin of deviation.
A further calculation of averaged values which incorporates the matching
molecular masses
of each type of cancer is known to those skilled in the art. By applying
formulas which the
method of error calculation by means of weights (weighted average) is based
upon, the
following generalized results are obtained for the aforementioned examples:
(i) 2020 ~ 10
(ii) 2050 ~ 10
(iii) 3946 ~ 20
(iv) 4104 ~ 21
(v) 4297 ~ 21
(vi) 4360 ~ 22
(vii) 4477 ~ 22
(viii) 4866 ~ 24
42
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
(ix) 4961
25
(x) 5492
27
(xi) 5679
28
(xii) 6448
32
(xiii) 6864
34
(xiv) 7000
35
(xv) 8224
41
(xvi) 8707
44
(xvii) 12471 ~ 62
(xviii) 12644
63
(xix) 13986
70
(xx) 15958
80
(xxi) 16164
81
(xxii) 17271
86
(xxiii) 14402
87
(xxiv) 17624 ~ 88
y (xxv) 18124 ~ 91
The present invention is fixrther illustrated by the following examples, which
should not be construed
as limiting in auy way. The contents of all cited references (including
literature references, issued
patents, published patent applications), as cited throughout this application,
are hereby expressly
incorporated by reference. The practice of the present invention will employ,
unless otherwise
indicated, conventional techniques of cell biology, cell culture, molecular
biology, transgenic biology,
microbiology, recombinant DNA, and immunology, which are known to those
skilled in the art. Such
techniques are explained fully in the literature.
Example 1. Sample collection for colon cancer evaluation.
Serum samples were obtained from a total of 151 individuals, which included
two different groups of
subjects. In the first group (group I), sera were drawn from 57 colon cancer
patients, undergoing
diagnosis and treatment of colon cancer at the Departments of Gastroenterology
and Surgery of the
Universities of Magdeburg, Erlangen, and Cottbus (all Germany). Serum samples
were collected from
the patients directly before surgery. At this time, a primary diagnosis was
made based on endoscopy,
ultrasonic testing, and/or other means for the detection of colorectal cancer.
In all cases the diagnosis
was confirmed by histological evaluation after surgery. Follow-up data for all
colon cancer patients
are currently collected and will be available for later studies.
43
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
The non-cancer control group (group lI) consisted of 94 subjects with non-
malignant disease
symptoms of the large intestine (adenoma, inflammation, diverticulosis), which
were recruited from
the University Hospitals in Magdeburg, Cottbus, and Erlangen. Serum from each
subject was taken
following colorectal endoscopy, wherein the absence of colorectal cancer was
oonfirmed.
Furthermore, all subjects denied a personal history of cancer and were
otherwise healthy. Follow-up
data for all non-cancer controls are currently collected and will be available
for later studies. In
addition, 77 serum samples from healthy blood donors was also collected for
test-set analysis. Blood
donors are considered to be healthy individuals not suffering from severe
diseases.
Example 2. ProteinChip Array analysis.
ProteinChip Arrays of the SAX2-type (strong anion exchanger) were arranged
into a bioprocessor
(Ciphergen Biosystems, Inc.), a device that contains up to 12 ProteinChips and
facilitates processing
of the ProteinChips. The ProteinChips were pre-incubated in the bioprocessor
with 200 pl binding
buffer (0.1 M Tris-HCI, 0.02% Triton X-100, pH 8.5). 10 wl of serum sample was
diluted 1:5 in a
buffer (7 M urea, 2 M thiourea, 4% CHAPS, I% DTT, 2% ampholine) and again
diluted 1:10 in the
binding buffer. Then, 300 ~,l of this mixture (equivalent to 6 p,l original
serum sample) were directly
applied onto the spots of the SAX2 ProteinChips. In between dilution steps and
prior to the application
to the spots, the sample was kept on ice (at 0°C). After incubation for
120 minutes at 20 to 24 °C the
chips were incubated with 200 ~1 binding buffer, before 2 x 0.5 ~,l EAM
solution (20 mg/ml sinapinic
acid in 50% acetonitrile and 0.5% trifluoroacetic acid) was applied to the
spots. After air-drying for 10
min, the ProteinChips were placed in the ProteinChip Reader (ProteinChip
Biology System II,
Ciphergen Biosystems, Inc.) and time-of flight spectra were generated by laser
shots collected in the
positive mode at laser intensity 215, with the detector sensitivity of 8.
Sixty laser shots per average
spectra were performed.
Calibration of mass accuracy was performed by using the following mixture of
mass standard calibrant
proteins: Dynorphin A (porcine, 209 - 225, 2147.50 Da), Beta-endorphin (human,
61 - 91, 3465.00
Da), Insulin (bovine, 5733.58 Da), and Cytochrome c (bovine, 12230.90 Da) at a
concentration of 1.21
pmol/~ul, and Myoglobin (equine cardiac, 16951.50 Da) at a concentration of
5.16 pmol/p,l. 0.5,1 of
this mixture was applied to a single spot of a H4 ProteinChip array. After air-
drying of the drop, 2 x 1
p,l matrix solution (a saturated solution of sinapinic acid in 50%
acetonitrile 0.5% trifluoracetic acid)
was applied to the spot. The drop was allowed to air-dry for 10 min after each
application of matrix
solution.
The ProteinChip was placed in the ProteinChip Reader (Biology System II,
Ciphergen Biosystems,
Inc.) and time-of flight spectra were generated by laser shots collected in
the positive mode at laser
44
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
intensity 210, with the detector sensitivity of 8. Sixty laser shots per
average spectra were performed.
Subsequently, Time-Of Flight values were correlated to the molecular masses of
the standard proteins,
and calibration was performed according to the instrument manual.
Example 3. Peak detection and data analysis.
The analysis of the data was performed by automatic peak detection and
alignment using the operating
software of the ProteinCliip. Biology System II, the ProteinChip Software
Version 3.1 (Ciphergen
Biosystems, Inc.). Figure 1 shows a comparison of protein mass spectra
detected using the above
mentioned SAX2 ProteinChip arrays for samples isolated from patients suffering
from non-malignant
diseases of the large intestine (e.g., acute or chronic inflammation, adenoma)
(Cl and C2) and of
patients with colon cancer (Tl and T2).
The complete set of patients was randomly divided into a training set and a
test set. The train set
comprised of 54 randomly selected patients with colon cancer and 75 randomly
selected patients
without colon cancer. The test set comprised of 14 randomly selected patients
with colon cancer and
19 randomly selected patients without colon cancer. Additionally, a test set
comprising of 77 sera
obtained from healthy blood donors was compiled. This was done in order to
test the classification
algorithm generated on the basis of the spectra of the subgroup of healthy
individuals (see below).
The m/z values of all mass spectra selected for the analysis ranged between
2000 Da and 30000 Da,
wherein smaller masses were not used since artefacts with the '.'Energy
Absorbing Molecule, EAM"
("Matrix") could not be excluded, and higher masses were not detected under
the chosen experimental
conditions. The spectra within the train set were normalised according to the
intensity of the total ion
current, followed by baseline subtraction, and automatic peak detection as
previously described by
Adam et al. (2002) Cancer Research 62: 3609-3614, using the "Bioinarker
Wizard" tool of the
ProteinChip Software Version 3.1 (Ciphergen Biosystem, Inc.). The following
settings were chosen
for peak detection by "Biomarker Wizard": a) auto-detect peaks to cluster, b)
first pass: 5 signal/noise,
c) minimum peak threshold: 5% of all spectra, d) deletion of user-detected
peaks below threshold, e)
cluster mass window: +/- 0.3% of mass. Using these settings, 90 signal
clusters were identified.
The normalization coefficient generated by normalizing the spectra of the
train sets and the cluster
information of the train sets generated by the "Biomarker Wizaxd" tool of the
software were saved and
used to externally normalize the spectra of the corresponding test sets and to
cluster the signals of the
corresponding test sets according to the normalization and peak identification
of the train sets.
The cluster information fox each train and test set (containing sample lD and
sample group, cluster
mass values and cluster signal intensities for each spectrum within the sets)
was transformed into an
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
interchangeable data format (a .csv table) using the "Sample group statistics"
function of the
"Biomarker Wizard" tool of the ProteinChip Software Version 3.1. In this
format, the data can be
analysed by a specific software for the generation of regression and
classification trees (see examples
to 7).
5
Example 4. Construction of classifiers.
Four classifiers with binary target variable (cancer versus nor-cancer) were
constructed: First, as a
proof. of principle, a classifier was constructed only on the basis of the
training set described above.
Second, a final classifier was constructed on the basis of all available mass
peaks and all colon cancer
samples, fusing the corresponding training and test data sets. Third, a
2°d final colon classifier was
constructed analogously to the first final colon cancer classifier but
excluding the most informative
and dominating mass of the first final colon classifier. Fourth, a 3'~ final
colon classifier was
constructed analogously to the first final colon cancer classifier but
excluding the most informative
and dominating masses of the first and 2"d final colon classifier.
Forward variable selection was applied in order to determine highly
informative sets . of variables
("patterns") for classification. The results of the present invention were
generated using the "CART"
decision tree approach (classification and regression trees; Breiman et al.,
1984). Moreover, bagging
of classifiers was applied to overcome typical instabilities of forward
variable selection procedures,
thereby increasing overall classifier performance (Breiman, 1994).
More precisely, for the training set 50 bootstrap samples were generated
(sampling with replacement,
maximal 3 sample redraws). For each bootstrap sample an exploratory decision
tree was generated.
Nodes were split using the Gini rule until all final nodes were either pure,
i.e., contained only samples
of one class, or until one of the following stopping rules was met: no nodes
comprising less than 4
cases were split and no splits were considered resulting in a node comprising
only one sample. The
such obtained 50 single classifiers, one for each bootstrap sample, were
combined to constitute an
ensemble of classifiers predicting class membership by plurality vote.
The procedure of classifier construction was conducted four times to obtain
one proof of principle
classifier and three final classifiers for colon cancer detection.
Example 5. Classifier structure.
The proof of principle classifier employed 71 masses (variables) out of 90
determined signal clusters.
Single decision trees consisted of 4 to 9 variables (5 to 10 end nodes), 6
variables being typical, see
histogram of Figure 4. Variable importance was roughly deduced by overall
improvement, i.e., for
each mass we summed the improvement values achieved in the generation of all
50 decision trees of
46
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
the decision tree ensemble. The masses used by the proof of principle
classifier are listed in Table 1
(starting with most important masses having high improvement). An overview of
the distribution of
masses is given in Figure 5. .
The 1St final classifier for colon cancer employed 75 masses out of 90
determined signal clusters.
Single decision trees consisted of more.variables than in the proof of
principle classifier: 9 variables
were typical, see histogram of Figure 6. Variable importance was roughly
deduced by overall
improvement. The masses used by the 1St final classifier are listed in Table 2
(starting with most
important masses, i.e. masses with highest improvement values). An overview of
the distribution of
masses of the lst final classifier is given in Figure 7.
The 2nd final classifier for colon cancer employed 77 masses out of 90
determined signal clusters.
Single decision trees consisted of even more variables than in 1St final
classifier: 10 variables were
typical, see histogram of Figure 8. Variable importance was roughly deduced by
overall improvement.
The masses used by the 2nd final classifier are listed in Table 3 (starting
with most important masses,
i.e, masses with highest improvement values): An overview of the distribution
of masses of the 2nd
final classifier is given in Figure 9.
The 3rd final classifier for colon cancer employed 80 masses out of 90
determined signal clusters.
Single decision trees consisted of even more variables than in lst final
classifier: 10 variables were
30
typical, see histogram of Figure 10. Variable importance was roughly deduced
by overall
improvement. The masses used by the 3'~ final classifier are listed in Table 4
(starting with most
important masses, i.e. masses with highest improvement values). An overview of
the distribution of
masses of the 3'd final classifier is given in Figure 11.
With the exception of mass 10722 Da, the classifiers include all of the
differentially expressed
biomolecules found in this study.
Example 6. Classification performance.
Classification performance is determined for the proof of principle classifier
on the colon cancer
versus endoscopy control test data set as well as on a separate test set
consisting of presumably healthy
blood donors. The classifier achieved 93% sensitivity and 84% specificity on
the cancer versus
endoscopy controls test data set and 94% specificity on 77 samples of blood
donors.
For the three final classifiers, we determined their specificity on 77 samples
of blood donors. We
obtained 92% specificity for the 1St final classifier, 100% specificity for
the 2nd final classifier, and
92% specificity for the 3'd final classifier.
47
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Table 1: Ranking of masses of proof of principle classifier by overall
improvement.
mass improvement mass Improvement mass improvement
5493 11.397 6447 0.193 11465 0.048
4964 0.915 15879 0.193 8703 0.046
6645 0.724 4719 0.188 13290 0.045
.
12619 0.589 3228 0.176 4607 0.041
8781 0.511 17263 0.17 3457 0.04
3947 0.483 15005 0.159 8215 0.039
7576 0.464 17617 0.157 3027 0.038
10595 0.446 2509 0.155 9360 0.038
22952 0.442 9078 0.153 5113 0.031
6852 0.415 -4104 0.132 4295 0.03
.3327 0.409 13633 0.127 17890 0.028
22467 0.405 7000 0.122 11694 0.027
24080 0.398 2733 0.105 11905 0.026
~
2021 0.359 9202 0.095 4546 0.025
.
12829 0.346 16105 0.086 16164 0.025
8575 0.342 18116 0.082 9642 0.014
2270 0.323 9718 0.08 22339 0.013
9143. 0.267 4242 0.069 15957 0.012
4866 0.229 6898 ~ 0.067 4830 0.011
4359 0.225 4476 0.066 5854 0.011
2049 0.223 8922 0.066 5773 0.009
8077 0.214 7658 0.062
13784 0.202 8474 0.058
22677 0.202 12470 0.058
_
17397 0.198 5648 0.052
48
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Table 2: Ranking of masses of 1St final classifier by overall improvement.
mass improvement mass improvement mass improvement
5493 12.849 17890 0.157 3947 0:056
6645 1.216 10595 . 0.156 2733 0.051
4964 0.907 7658 0.148 9581 0.046
8781 0.559 11216 0.147 28259 0.045
.
12829 0.494 2509 0.141 4607 0.044
15879 0.392 3228 0.141 4546 ~ 0.042
2021 0.363 16105 0.128 9930 0.039
22952 0.353 22467 0.112 17617 0.039
2270 0.323 9360 0.111 3457 0.038
28055 0,305 4476 0.099 22677 0.036
18116 0.3 4830 0.093 13633 0.033
8077 0.298 9143 0.088 11694 0.032
6852 0.268 10369 0.088 11905 0.031
2049 0.252 17767 0.085 8703 0.028
4359 0.239 4242 0.083 11465 0.024
8575 0.233 6447 0.078 13983 0.024
~
24080 0.232 22339 0.078 9078 0.022
12619 0.197 15005 ~ 0.075 14798 0.022
7576 0.179 4719 0.073 16953 0.021
12470 0.168 7000 0.064 13290 0.021
4104 0.166 5113 0.062 11547 0.02
15957 0.165 9202 0.062 5648 0.011
17263 0.165 4866 0.058 _ 5226 0.01
5854 0.161 16164 0.058 6898 0.01
3327 0.161 3027 0.057 5773 0.009
15
49
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Table 3: Ranking of masses of 2"d final classifier by overall improvement.
mass improvement mass improvement mass improvement
3947 5.672 9360 ~ 0.187 8575 0.068
12829 2.203 3027 0.179 10369 0.066
6645 1.472 4866 0.169 17767 0.063
4964 1.441 12470 0.163 15350 0.056
8077 1.158 9078 0.148 11216 0.046
28055 1.072 2509 0.147 1.7890 0.044
15957 0.912 6898 0.142 8703 0.039
6852 0.811 10595 0.139 4295 0.036
12619 0.539 7576 0.135 15005 0.036
24080 0.393 8781 0.116 22677 0.036
3327 0.385 22339 0.115 9581 0.031
28259 0.34 5854 0.114 9426 0.03
_
2021 0.337 2270 0.11 13290 0.027
16105 0.316 ~ 6447 0.106 15879 0.026
~
11694 0.315 22952 0.104 17397 0.023
4104 0.299 4242 0.092 5648 0.022
2049 0.293 10215 ~ 0.092 17617 0.022
4719 0.27 5113 0.09 8474 0.019
16164 0.25 9202 0.089 10440 0.016
3457 0.241 9143 0.086 4359 0.009
4546 0.238 13983 0.082 5226 0.008
17263 0.232 4830 0.081 - 7000 0.006
16953 0.228 4476 0.08 7658 0.006
2733 0.225 11465 0.072
22467 0.218 18116 0.071
5773 0.193 15140 0.07
3228 0.19 4607 0.068
15
CA 02525743 2005-11-14
WO 2004/102190 PCT/EP2004/005294
Table 4: Ranking of masses of 3rd final classifier by overall improvement..
mass improvement mass improvement mass improvement
4964 - 3.431 10595 . 0.187 15140 0.047
12829 2.166 7658 0.183 7000 0.046
6645 1.999 9078 0.183 22467 0.044
28055 1.288 8781 0.171 10369 0.042
.282591.152 5773 0.144 18390 0.042
6852 1.089 2270 0.134 13290 0.041
3327 0.781 5113 0.133 6898 0.038
16105 0.737 7576. 0.132 17767. 0.038
16953 0.736 9143 . 0.131 8703 0.036
15957 0.714 6447 0.128 13633 0.036
12619 0.705 2733 0.111 15005 0.036
8077 0.666 18116 0.109 15350 0.032
~
4830 0.615 4607 0.104 13784 0.031
4546 0.485 11694 0.104 17617 0.029
2021 0.403 . 15879 0.1 14798 0.027
4242 0.329 9202 0.099 17397 0.026
4719 0.304 10215 : 0.092 5226 0.026
12470 0.292 4476 0.089 9426 0.026
9360 0.283 9581 0.089 5648 0.022
3457 0.279 11905 0.086 8474 0.019
22952 0.275 4359 0.079 8575 0.019
2509 0.261 4295 ~ 0.075 10440 0.016
4104 0.245 4866 0.068 17263 0.009
2049 0.23 9718 0.068 11216 0.008
24080 0.219 11465 0.062
16164 0.201 13983 0.062
3228 0.198 22339 0.056
5854 0.192 3027 0.047
15
51