Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
CONTENANT LES PAGES 1 A 71
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 71
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME:
NOTE POUR LE TOME / VOLUME NOTE:
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
1
TITLE OF THE INVENTION
METHODS FOR ASSESSING PATIENTS WITH ACUTE MYELOID LEUKEMIA
A "Sequence Listing" listing appendix is hereby incorporated by reference
herein.
BACKGROUND OF THE INVENTION
This invention relates to diagnostics, prognostics, and treatments for acute
myeloid leukemia (AML) based on the detection of molecular markers and/or gene
expression analysis.
Karyotyping is currently effective in providing prognostic value while it also
serves to identify biologically distinct subtypes of AML. In addition,
mutations in
genes such as FLT3, c-KIT, AML1, GATA1, CEBPA and N-RAS are implicated in
the pathogenesis of the disease. It is clear- that screening for FLT3 and
CEBPA
mutations can stratify groups that have different risks of relapse. Effective
risk
stratification can allow for the appropriate use of allogeneic stem cell
transplantation
or other adjuvant therapies. Two papers published recently describe gene
expression
profiling of newly diagnosed adult AML patients and its use in predicting
clinical
outcome. Bullinger et al. (2004); and Valk et al. (2004). These studies show
how
gene-expression profiling can further refine clinical outcome prediction.
Valk et al. (2004) evaluated 285 patients (bone marrow or peripheral blood) on
the Affymetrix U133A chip. The patient samples encompassed a wide range of
cytogenetic and molecular abnormalities. Only 16 clusters were identified
indicating AML may not be as heterogeneous as previously thought. Several of
the
clusters corresponded well with the cytogenetically and molecularly defined
sub-
types of AML thus supporting their use in the WHO classification system. These
clusters were also seen by Bullinger et al. (2004) and other previously
published
smaller studies. Schoch et al. (2002); Debemardi et al. (2003); and Kohlmann
et al.
(2003). These clusters, not surprisingly, correlated with prognosis since they
were
associated with well known prognostic karyotypes.
Bullinger et al. (2004) investigated expression profiles from 116 adult
patients
(65 peripheral blood and 54 bone marrow) using cDNA arrays. In addition to the
work done by Valk et al. (2004) they also developed a133 gene classifier for
predicting clinical outcome across all cytogenetic risk groups. Using a
training set
of 59 samples and a testing set of 57 samples they showed that the 133 genes
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
2
clustered patients into poorand good outcome.groups (p = 0.0061og rank; odds
ratio, 10, 95% CI, 2.6-29.3).
Notably, the genes identified in both these studies overlap, only in part, to
predictor genes previously identified in childhood leukemia. Yagi et al.
(2003).
Also, there is no overlap between the prognostic gene set identified by
Bullinger et
al. (2004) and the 3 genes recently identified that predict response to
tipifarnib. US
patent application serial no. 10/883,436.
The farnesyl transferase (FTase) enzyme mediates the covalent attachment of a
carbon farnesyl moiety to the C-terminal CAAX (C, cysteine; A, aliphatic
residue;
X, any amino acid) recognition motif. Reiss et al. (1990). This farnesylation
is
further processed by cleavage of the 3 terminal amino acids (AAX) and
methylation
of the C-terminal isoprenyl-cysteine. The inhibition of protein famesylation
abrogates the correct subcellular localization required for protein function.
Originally, the oncogenic Ras protein was thought to be the target for the
antiproliferative effects of FTIs in cancer biology. Reuter et al. (2000).
However, it
has since been shown that inhibition of Ras farnesylation does not account for
all of
actions of tipifarnib. For example, FTIs do not always require the presence of
mutant Ras protein to produce antitumor effects. Karp et al. (2001). Indeed,
while
early clinical studies were designed around populations with a high frequency
of ras
mutations, such as advanced colorectal and pancreatic cancer, no significant
difference in response rates were seen when compared to placebo. Van Cutsem et
al. (2004); and Rao et al. (2004).
Several other famesylated proteins have been implicated as candidate targets
that
may mediate the antitumorigenic effects of FTIs including the small GTPase
proteins Rho B, the centromere proteins CENP-E and CENP-F, the protein
tyrosine
phosphatase PTP-CAAX, and the nuclear membrane structural lamins A and B. The
inhibition of famesylation of these proteins may lead to the antiproliferative
effect of
FTIs and also indirectly modulate several important signaling molecules
including
TGFORII, MAPK/ERK, PI3K/AKT2, Fas (CD95), NF-KB, and VEGF. Adnane et
al. (2000); Morgan et al. (2001); Jiang et al. (2000); Na et al. (2004);
Takada et al.
(2004); and Zhang et al. (2002). Regulation of these signaling pathways leads
to the
modulation of cell growth, proliferation, and apoptosis. Thus, FTIs may have
complex inhibitory effects on several cellular events and pathways.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
3
There are currently no methods for determining status or predicting overall
survival of these patients.
BRIEF SUMMARY OF THE INVENTION
The invention is a method of using one or more gene signatures for predicting
prognosis in patients with acute myeloid leukemia (AML). These signatures can
be
used alone or in combination depending upon the type of drug treatment.
The present invention provides a method of assessing acute myeloid leukemia
(AML) status by obtaining a biological sample from an AML patient; and
measuring
Biomarkers associated with Marker genes corresponding to those selected from
Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9 where the expression
levels of
the Marker genes above or below pre-determined cut-off levels are indicative
of
AML status.
The present invention provides a method of staging acute myeloid leukemia
(AML) patients by obtaining a biological sample from an AML patient; and
measuring Biomarkers associated with Marker genes corresponding to those
selected
from Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9 where the
expression
levels of the Marker genes above or below pre-determined cut-off levels are
indicative of AML survival.
The present invention provides a method of determining acute myeloid leukemia
(AML) patient treatment protocol by obtaining a biological sample from an AML
patient; and measuring Biomarkers associated with Marker genes corresponding
to
those selected from Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9
where the
expression levels of the Marker genes above or below pre-determined cut-off
levels
are sufficiently indicative of response to therapy to enable a physician to
determine
the degree and type of therapy recommended to provide appropriate therapy.
The present invention provides a method of treating a acute myeloid leukemia
(AML) patient by obtaining a biological sample from an AML patient; and
measuring Biomarkers associated with Marker genes corresponding to those
selected
from Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9 where the
expression
levels of the Marker genes above or below pre-determined cut-off levels are
indicate
a response to therapy and; treating the patient with adjuvant therapy if they
have a
responder profile.
The present invention provides a method of determining whether a acute
myeloid leukemia (AML) patient is high or low risk of mortality by obtaining a
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
4
biological sample from an AML patient; and measuring Biomarkers associated
with
Marker genes corresponding to those selected from Table 3 where the expression
levels of the Marker genes above or below pre-determined cut-off levels are
sufficiently indicative of risk of mortality to enable a physician to
determine the
degree and type of therapy recommended.
The present invention provides a method of generating an acute myeloid
leukemia (AML) prognostic patient report by determining the results of any one
of
the above-described methods; and preparing a report displaying the results and
reports generated thereby.
The present invention provides a kit for conducting an assay to determine
acute
myeloid leukemia (AML) prognosis in a biological sample comprising: materials
for
detecting isolated nucleic acid sequences, their complements, or portions
thereof of a
combination of genes selected from the group consisting of Marker genes
corresponding to those selected from Table 3, Table 4, Table 5, Table 7, Table
8 or
Table 9.
The present invention provides articles for assessing acute myeloid leukemia
(AML) status comprising: materials for detecting isolated nucleic acid
sequences,
their complements, or portions thereof of a combination of genes selected from
the
group consisting of Marker genes corresponding to those selected from Table 3,
Table 4, Table 5, Table 7, Table 8 or Table 9.
The present invention provides a microarray or gene chip for performing the
above-described methods.
The present invention provides a diagnostic/prognostic portfolio comprising
isolated nucleic acid sequences, their complements, or portions thereof of a
combination of genes selected from the group consisting of Marker genes
corresponding to those selected from Table 3, Table 4, Table 5, Table 7, Table
8 or
Table 9.
BRIEF DESCRIPTON OF DRAWINGS
Figure 1. Unsupervised clustering of relapsed and refractory AML patients. The
dendogram shows the unsupervised k-means clustering of 58 relapsed or
refractory
AML patients, where each column represents a patient and each row represents a
gene. The expression ratio for each gene was calculated by dividing the
expression
level of that gene in a patient by the mean of all other patients. The color
bar
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
indicates the fold-change (log2). Red is upregulated, blue is down-regulated.
White
indicates no change. The presence of 6 main clusters is shown.
Figure 2. Real-time RT-PCR of 2 genes. AHR and AKAP13 were measured by
real-time RT-PCR. The HPRT or PBGD control genes were used to normalize gene
5 expression values. Error bars are standard deviations. The resulting values
were
plotted against the corresponding microarray data and linear regression
analysis was
performed.
Figure 3 depicts the predictive value of the AKAP 13 gene. Panel A shows a 2x2
table generated from a LOOCV performed using AKAP 13 expression as a
classifier
on the responders (R) and non-responders (NR). Panel B shows the AKAP 13
expression values for the same 58 patients. The P value indicates a
significant
difference in the gene expression between the mean values of each response
group.
Panel C shows the Kaplan-Meier curves generated from patients classified by
the
AKAP13 gene as being responders and non-responders.
Figure 4 provides identification of a minimal set of predictive markers. In
Panel
A, a LOOCV was performed using a sensitivity of 100%. Independent classifiers
were tested that contained from 1 to 19 genes. The resulting error rate is
plotted.
Panel B shows a 2x2 table generated from a LOOCV performed using the 3-gene
signature as a classifier on the responders (R) and non-responders (NR). Panel
C
shows the scores generated from the 3-gene classifier. The P value indicates a
significant difference in the gene expression between the response groups.
Panel D
is the Kaplan-Meier curves generated from patients classified by the 3-gene
signature as being responders and non-responders. Median survival times are
also
indicated.
Figure 5. A Kaplan-Meier analysis was performed on patients classified by the
3-gene signature as being predicted responders and non-responders. The
survival
curve of patients who were clinically defined as non-responders but classified
as
responders using the 3-gene signature is shown. Median survival times are also
indicated.
Figure 6 depicts over-expression of AKAP 13 in an AML cell line. Cell counts
were normalized to cultures with no drug (indicated at -12 log units) to give
a
percentage of control. Error bars indicate standard errors of the mean. Open
data
points indicate results from a second experiment exploring higher
concentrations of
drug.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
6
Figure 7 provides a model of FTI action in relapsed or refractory AML. A. In
responders the IL3RA and AKAP13 genes are lowly expressed allowing for
down-regulation of the ras, and RhoA, and lamin B pathways, respectively.
Up-regulation of RhoH leads to increased inhibition of cellular transformation
5 pathways. Together this allows for greater efficacy in FTI
antitumorigenicity. B.
The opposite expression profile is seen in non-responders allowing for the
expression of compensatory pathways.
Figure 8. The Zarnestra predictive gene signature has superior utility to an
independent prognostic gene signature. In panel A columns represent AML
samples
from relapsed or refractory patients and rows represent 167 probe sets that
correspond to 103 of the 133 prognostic genes identified by Bullinger et al.
ordered according to hierarchical clustering. Panel B shows Kaplan-Meier
survival
estimates of the cluster-defined groups of patients. In panel C the 3-gene
classifier
has been used to identify responders of tipifarnib in the good and poor
prognostic
groups defined by the Bullinger signature. Kaplan-Meier survival curves are
shown
for patients identified as being responders to tipifarnib in the good
(Zn+.clusterl)
and poor (Zn+.cluster2) prognostic groups. The median survival times for each
group are indicated.
Figure 9 is a flow chart depicting how the genes from Bullinger et al. (2004)
were matched to 167 probe sets (103 unique genes) on the Affymetrix U133A
chip.
Figure 10 shows the utility of the 167 probe set signature in relapsed or
refractory AML patients. In panel A columns represent AML samples from
relapsed
or refractory patients and rows represent 167 probe sets that correspond to
103 of the
133 prognostic genes identified by Bullinger et al. (2004), ordered according
to
hierarchical clustering. Panel B shows Kaplan-Meier survival estimates of the
cluster-defined groups of patients.
Figure 11 provides comparisons of prognostic and Zamestra predictive gene
signatures. Panel A shows the Kaplan-Meier survival curves for the good and
poor
prognostic clusters as defined by the subset of 103 Bullinger et al. (2004)
genes.
Panel B shows the Kaplan-Meier survival curves for the good and poor
prognostic
clusters as defined by the 3-gene signature that predicts response to
Zarnestra. Panel
C shows the Kaplan-Meier survival curves for the good and poor prognostic
clusters
from Panel A further stratified by the 3-gene Zamestra signature. Panel D
shows the
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
7
Kaplan-Meier survival curves for patients who are predicted to have a poor
prognosis and not respond to Zarnestra versus the remainder of patients.
Figure 1.2 Identification of a minimal set of predictive markers. a) A LOOCV
was performed selecting for genes with a sensitivity of 100%, specificity of
40% and
fold-change > 2. Independent classifiers were tested that contained from 1 to
8
genes ranked by the AUC . The resulting error rate is plotted. b) A 2x2 table
generated from a LOOCV performed using AKAP 13 as a classifier on the
responders (R) and non-responders (NR). c) The gene-expression values of
AKAP 13. The P value indicates a significant difference in the gene expression
between the response groups. d) The Kaplan-Meier curves generated from
patients
classified by AKAP 13 as being responders and non-responders. Median survival
times are also indicated.
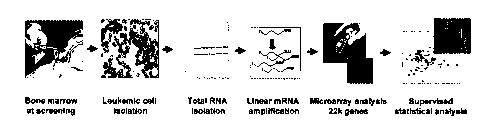
Figure 13 depicts an overview of gene expression analysis.
Figure 14 depicts AML samples maintain FTI-mediated global gene expression
changes following termination of tipifarnib treatment.
Figure 15 depicts predictive expression profiles and testing of predictive
classifiers in newly diagnosed AML.
Figure 16 depicts the 6-gene classifier stratifies newly diagnosed AML.
DETAILED DESCRIPTION OF THE INVENTION
A subset of genes previously described to have prognostic value in newly
diagnosed AML is shown here to have utility in relapsed and refractory AML
patients
treated with a molecularly targeted therapy (Zarnestra). Currently there is no
method
for predicting response to farnesyl transferase inhibitors (such as
Zarnestra). Also,
current methods for understanding the prognosis of patients with AML is
limited to
histological subtype and karyotyping, both of which are not ideal markers for
determining clinical outcome. The current signatures expand upon these
traditional
technologies by providing better stratification of prognostic high risk and
low risk
patients.
US Patent Application Serial No. 10/883,436 demonstrates that a 3-gene
classifier (including AHR, AKAP13 and MINA53) predicts relapsed, refractory
AML patient response to tipifarnib (Zarnestra , Rl 15777) with the lowest
error rate.
This was also seen when a leave-five-out cross validation was performed. When
more genes were added the error rate increased indicating that additional
genes
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
8
introduced noise to the classifier. For the 3-gene classifier the LOOCV
demonstrated a sensitivity of 86% and specificity of 70% with an overall
diagnostic
accuracy of 74%. Kaplan-Meier analysis again showed a significant difference
in
survival between the predicted responder group and the non-responder group.
Moreover, comparing the incorrectly classified non-responders to the correctly
classified non-responders, the misclassified non-responders showed a better
overall
survival.
Zarnestra is an orally available non-peptidomimetic competitive farnesyl
transferase inhibitor (FTI) that has been shown to inhibit the proliferation
of a
variety of human tumor cell lines both in vitro and in vivo. End et al.
(2001); and
Cox et al. (2002). A phase I clinical trial of tipifarnib demonstrated a 32%
response
rate in patients with refractory or relapsed acute myeloid leukemia. Karp et
al.
(2001). Activity has also been seen in early clinical trials for
myelodysplastic
syndrome (MDS) (Kurzrock et al. (2004)), multiple myeloma (MM) (Alsina et al.
(2003)) and chronic myeloid leukemia (CML). Cortes et al. (2003). Complete
remission was defined as less than 5% bone marrow blasts with a neutrophil
count
greater than 1000/ L, a platelet count less than 100,000/ L, and no
extramedullary
disease. While it is clear that FTIs function by inhibiting protein
farnesylation, it is
still not known what genes are implicated in the antitumor effects of
tipifarnib in
hematopoietic malignancies. Microarray technology allows for the measurement
of
the steady-state mRNA level of thousands of genes simultaneously, thereby
representing a powerful tool for identifying genes and gene pathways that
correlate
with FTI action. Global gene expression monitoring was therefore employed in a
phase 2 clinical study of tipifarnib in relapsed and refractory AML to
identify genes
that predict response to this FTI in hematologic malignancies.
The mere presence of nucleic acid sequences having the potential to express
proteins or peptides ("genes") within the genome is not determinative of
whether a
protein or peptide is expressed in a given cell. Whether or not a given gene
capable
of expressing proteins or peptides or transcribing RNA does so and to what
extent
such expression or transcription occurs, if at all, is determined by a variety
of
complex factors. Nevertheless, assaying gene expression can provide useful
information about the cellular response to a given stimulus such as the
introduction
of a drug or other therapeutic agent. Relative indications of the degree to
which
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
9
genes are active or inactive can be found in such gene expression profiles. In
some
instances, the presence of a molecular marker can, by itself or with the use
of gene
expression information, provide useful information about treatment efficacy
too.
The gene expression profiles and molecular markers of this invention are used
to
identify and treat AML patients.
Cancers, including hematological malignancies, typically arise from mutations
in
a variety of genes. The same type of cancer may arise as a result of, or
coincident
with, one or more mutations that differ from those of another patient having
the
same type of cancer. The fact that there are often multiple molecular bases
underlying the same cancers is consistent with the observation that some
therapies
that affect one patient do not necessarily equally affect another patient with
the same
type of cancer. Further, from a diagnostic point of view, the presence of
particular
mutations such as translocations, deletions, or SNPs can have powerful
implications.
In some instances, such molecular markers are themselves useful indicators for
diagnosis, prognosis, or treatment response determinations. This is
particularly true
where the molecular mutations can be associated with response to particular
treatments.
A Biomarker is any indicia of the level of expression of an indicated Marker
gene. The indicia can be direct or indirect and measure over- or under-
expression of
the gene given the physiologic parameters and in comparison to an internal
control,
normal tissue or another carcinoma. Biomarkers include, without limitation,
nucleic
acids (both over and under-expression and direct and indirect). Using nucleic
acids
as Biomarkers can include any method known in the art including, without
limitation, measuring DNA amplification, RNA, micro RNA, loss of
heterozygosity
(LOH), single nucleotide polymorphisms (SNPs, Brookes (1999)), microsatellite
DNA, DNA hypo- or hyper-methylation. Using proteins as Biomarkers can include
any method known in the art including, without limitation, measuring amount,
activity, modifications such as glycosylation, phosphorylation, ADP-
ribosylation,
ubiquitination, etc., imunohistochemistry (IHC). Other Biomarkers include
imaging, cell count and apoptosis markers.
The indicated genes provided herein are those associated with a particular
tumor
or tissue type. A Marker gene may be associated with numerous cancer types but
provided that the expression of the gene is sufficiently associated with one
tumor or
tissue type to be identified using the algorithm described herein to be
specific for a
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
lung cancer cell, the gene can be used in the claimed invention to determine
cancer
status and prognosis. Numerous genes associated with one or more cancers are
known in the art. The present invention provides preferred Marker genes and
even
more preferred Marker gene combinations. These are described herein in detail.
5 A Marker gene corresponds to the sequence designated by a SEQ ID NO when it
contains that sequence. A gene segment or fragment corresponds to the sequence
of
such gene when it contains a portion of the referenced sequence or its
complement
sufficient to distinguish it as being the sequence of the gene. A gene
expression
product corresponds to such sequence when its RNA, mRNA, or cDNA hybridizes
10 to the composition having such sequence (e.g. a probe) or, in the case of a
peptide or
protein, it is encoded by such mRNA. A segment or fragment of a gene
expression
product corresponds to the sequence of such gene or gene expression product
when
it contains a portion of the referenced gene expression product or its
complement
sufficient to distinguish it as being the sequence of the gene or gene
expression
product.
The inventive methods, compositions, articles, and kits of described and
claimed
in this specification include one or more Marker genes. "Marker" or "Marker
gene"
is used throughout this specification to refer to genes and gene expression
products
that correspond with any gene the over- or under-expression of which is
associated
with a tumor or tissue type. The preferred Marker genes are described in more
detail
in Table 8.
The present invention provides a method of assessing acute myeloid leukemia
(AML) status by obtaining a biological sample from an AML patient; and
measuring
Biomarkers associated with Marker genes corresponding to those selected from
Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9 where the expression
levels of
the Marker genes above or below pre-determined cut-off levels are indicative
of
AML status.
The present invention provides a method of staging acute myeloid leukemia
(AML) patients by obtaining a biological sample from an AML patient; and
measuring Biomarkers associated with Marker genes corresponding to those
selected
from Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9 where the
expression
levels of the Marker genes above or below pre-determined cut-off levels are
indicative of AML survival.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
11
The present invention provides a method of determining acute myeloid leukemia
(AML) patient treatment protocol by obtaining a biological sample from an AML
patient; and measuring Biomarkers associated with Marker genes corresponding
to
those selected from Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9
where the
expression levels of the Marker genes above or below pre-determined cut-off
levels
are sufficiently indicative of response to therapy to enable a physician to
determine
the degree and type of therapy recommended to provide appropriate therapy.
The present invention provides a method of treating a acute myeloid leukemia
(AML) patient by obtaining a biological sample from an AML patient; and
measuring Biomarkers associated with Marker genes corresponding to those
selected
from Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9 where the
expression
levels of the Marker genes above or below pre-determined cut-off levels are
indicate
a response to therapy and; treating the patient with adjuvant therapy if they
have a
responder profile.
The present invention provides a method of determining whether a acute
myeloid leukemia (AML) patient is high or low risk of mortality by obtaining a
biological sample from an AML patient; and measuring Biomarkers associated
with
Marker genes corresponding to those selected from Table 3 where the expression
levels of the Marker genes above or below pre-determined cut-off levels are
sufficiently indicative of risk of mortality to enable a physician to
determine the
degree and type of therapy recommended.
The method provided herein may further include, contain or utilize measuring
the expression level of at least one gene constitutively expressed in the
sample.
Preferably, the method provided herein results in a specificity of at least
about 40%.
Preferably, the method provided herein results in a sensitivity of at least at
least
about 80%. Preferably, the method provided herein results in a p-value of less
than
0.05.
The method provided herein may be performed by measuring gene expression on
a microarray or gene chip. The microarray can be a cDNA array or an
oligonucleotide array and may further contain one or more internal control
reagents.
The method provided herein may be performed by determining gene expression
by nucleic acid amplification conducted by polymerase chain reaction (PCR) of
RNA extracted from the sample. The PCR can be reverse transcription polymerase
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
12
chain reaction (RT-PCR) and can further contain one or more internal control
reagent.
The method provided herein may be performed by measuring or detecting a
protein encoded by the gene. Te protein can be detected by an antibody
specific to
the protein.
The method provided herein may be performed by measuring a characteristic of
the gene. Characteristics include, without limitation, DNA amplification,
methylation, mutation and allelic variation.
The present invention provides a method of generating an acute myeloid
leukemia (AML) prognostic patient report by determining the results of any one
of
the above-described methods; and preparing a report displaying the results and
reports generated thereby. The report may contain an assessment of patient
outcome
and/or probability of risk relative to the patient population and/or
likelihood or
response to chemotherapy.
The present invention provides a kit for conducting an assay to determine
acute
myeloid leukemia (AML) prognosis in a biological sample comprising: materials
for
detecting isolated nucleic acid sequences, their complements, or portions
thereof of a
combination of genes selected from the group consisting of Marker genes
corresponding to those selected from Table 3, Table 4, Table 5, Table 7, Table
8 or
Table 9. The kit can further contain reagents for conducting a microarray
analysis
and/or a medium through which said nucleic acid sequences, their complements,
or
portions thereof are assayed.
The present invention provides articles for assessing acute myeloid leukemia
(AML) status containing materials for detecting isolated nucleic acid
sequences,
their complements, or portions thereof of a combination of genes selected from
the
group consisting of Marker genes corresponding to those selected from Table 3,
Table 4, Table 5, Table 7, Table 8 or Table 9. The articles can further
contain
reagents for conducting a microarray analysis and/or a medium through which
said
nucleic acid sequences, their complements, or portions thereof are assayed.
The present invention provides a microarray or gene chip for performing the
above-described methods. The microarray may contain isolated nucleic acid
sequences, their complements, or portions thereof of a combination of genes
selected
from the group consisting of Marker genes corresponding to those selected from
Table 3, Table 4, Table 5, Table 7, Table 8 or Table 9. Preferably, the
microarray
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
13
provides a measurement or characterization at least 1.5-fold over- or
under-expression. Preferably, the microarray provides a measurement with a
statistically significant p-value over- or under-expression. More preferably,
the
p-value is less than 0.05. The microarray can be any known in the art
including,
without limitation, cDNA array or an oligonucleotide array and can further
contain
internal control reagents.
The present invention provides a diagnostic/prognostic portfolio comprising
isolated nucleic acid sequences, their complements, or portions thereof of a
combination of genes selected from the group consisting of Marker genes
corresponding to those selected from Table 3, Table 4, Table 5, Table 7, Table
8 or
Table 9. Preferably, the measurement or characterization is at least 1.5-fold
over- or
under-expression. Preferably, the measurement provides a statistically
significant
p-value over- or under-expression. More preferably, the p-value is less than
0.05.
Preferred methods for establishing gene expression profiles include
determining
the amount of RNA that is produced by a gene that can code for a protein or
peptide.
This is accomplished by reverse transcriptase PCR (RT-PCR), competitive RT-
PCR,
real time RT-PCR, differential display RT-PCR, Northern Blot analysis and
other
related tests. While it is possible to conduct these techniques using
individual PCR
reactions, it is best to amplify complementary DNA (cDNA) or complementary
RNA (cRNA) produced from mRNA and analyze it via microarray. A number of
different array configurations and methods for their production are known to
those
of skill in the art and are described in U.S. Patents such as: 5,445,934;
5,532,128;
5,556,752; 5,242,974; 5,384,261; 5,405,783; 5,412,087; 5,424,186; 5,429,807;
5,436,327; 5,472,672; 5,527,681; 5,529,756; 5,545,531; 5,554,501; 5,561,071;
5,571,639; 5,593,839; 5,599,695; 5,624,711; 5,658,734; and 5,700,637.
Microarray technology allows for the measurement of the steady-state mRNA
level of thousands of genes simultaneously thereby presenting a powerful tool
for
identifying effects such as the onset, arrest, or modulation of uncontrolled
cell
proliferation. Two microarray technologies are currently in wide use. The
first are
cDNA arrays and the second are oligonucleotide arrays. Although differences
exist
in the construction of these chips, essentially all downstream data analysis
and
output are the same. The product of these analyses are typically measurements
of
the intensity of the signal received from a labeled probe used to detect a
cDNA
sequence from the sample that hybridizes to a nucleic acid sequence at a known
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
14
location on the microarray. Typically, the intensity of the signal is
proportional to
the quantity of cDNA, and thus mRNA, expressed in the sample cells. A large
number of such techniques are available and useful. Preferred methods for
determining gene expression can be found in US Patents 6,271,002; 6,218,122;
6,218,114; and 6,004,755.
Analysis of the expression levels is conducted by comparing such signal
intensities. This is best done by generating a ratio matrix of the expression
intensities of genes in a test sample versus those in a control sample. For
instance,
the gene expression intensities from a diseased tissue can be compared with
the
expression intensities generated from benign or normal tissue of the same
type. A
ratio of these expression intensities indicates the fold-change in gene
expression
between the test and control samples.
Gene expression profiles can also be displayed in a number of ways. The most
common method is to arrange raw fluorescence intensities or ratio matrix into
a
graphical dendogram where columns indicate test samples and rows indicate
genes.
The data are arranged so genes that have similar expression profiles are
proximal to
each other. The expression ratio for each gene is visualized as a color. For
example,
a ratio less than one (down-regulation) may appear in the blue portion of the
spectrum while a ratio greater than one (indicating up-regulation) may appear
as a
color in the red portion of the spectrum. Commercially available computer
software
programs are available to display such data including "GENESPRING" from
Silicon
Genetics, Inc. and "DISCOVERY" and "INFER" software from Partek, Inc.
In the case of measuring protein levels to determine gene expression, any
method known in the art is suitable provided it results in adequate
specificity and
sensitivity. For example, protein levels can be measured by binding to an
antibody
or antibody fragment specific for the protein and measuring the amount of
antibody-bound protein. Antibodies can be labeled by radioactive, fluorescent
or
other detectable reagents to facilitate detection. Methods of detection
include,
without limitation, enzyme-linked immunosorbent assay (ELISA) and immunoblot
techniques.
Modulated Markers used in the methods of the invention are described in the
Examples. The genes that are differentially expressed are either up regulated
or
down regulated in patients with various lung cancer prognostics. Up regulation
and
down regulation are relative terms meaning that a detectable difference
(beyond the
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
contribution of noise in the system used to measure it) is found in the amount
of
expression of the genes relative to some baseline. In this case, the baseline
is
determined based on the algorithm. The genes of interest in the diseased cells
are
then either up- or down-regulated relative to the baseline level using the
same
5 measurement method.
Assays for the gene expression status of a cell also can determine
normal/abnormal tissue distribution for diagnostic purposes using techniques
such as
immunohistochemical analysis (IHC). Any method known in the art can be used,
for
example in the case of the LBC oncogene, the antibodies to LBC protein may be
used
10 in conjunction with both fresh-frozen and formalin-fixed, paraffin-embedded
tissue
blocks prepared for study by IHC. Each tissue block may consist of 50 mg of
residual
"pulverized" tumor.
Briefly, frozen-sections may be prepared by rehydrating 50 ng of frozen
pulverized tumor at room temperature in phosphate buffered saline (PBS) in
small
15 plastic capsules; pelleting the particles by centrifugation; resuspending
them in a
viscous embedding medium (OCT); inverting the capsule and pelleting again by
centrifugation; snap-freezing in -70 C isopentane; cutting the plastic capsule
and
removing the frozen cylinder of tissue; securing the tissue cylinder on a
cryostat
microtome chuck; and cutting 25-50 serial sections containing intact tumor
cells.
Permanent-sections may be prepared by a similar method involving rehydration
of
the 50 mg sample in a plastic microfuge tube; pelleting; resuspending in 10%
formalin for 4 hr fixation; washing/pelleting; resuspending in warm 2.5% agar;
pelleting; cooling in ice water to harden the agar; removing the tissue/agar
block from
the tube; infiltrating and embedding the block in paraffin; and cutting up to
50 serial
permanent sections.
For the IHC assay, the sections are overlaid with a blocking solution
containing:
3% bovine serum albumin (BSA) in PBS or other blocking reagents. The blocking
reagents include non-specific serum or dry milk. Blocking is allowed to
proceed for 1
hr at room temperature. Anti-LBC protein antibody is diluted with PBS buffer
containing 3% BSA, 0.1% TritonXTM-100 and t-octylphenoxypolyethoxyethanol, at
a
ratio of 1:100. The sample sections are generally overlaid with the antibody
solution
for 16 hr at 4 C. The duration and temperature conditions may be varied
according to
the antibody selected and the material tested. The optimal conditions are
determined
empirically. The antibody treated sections are then washed three times in PBS
for 15
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
16
min, each to remove unbound antibody and then overlaid with PBS containing 3%
BSA and a secondary antibody at a dilution of 1:2000. The secondary antibodies
may
be coupled to a chromogenic enzyme such as: horseradish peroxidase, alkaline
phosphatase, fluorescein isothiocyanate, or other suitable enzymes.
Alternatively, the
secondary antibody may be conjugated to biotin and used in conjunction with
chromophore-labeled avidin.
Another exemplary method for detecting the presence of a gene is via in situ
hybridization. Generally, in situ hybridization comprises the following major
steps:
(1) fixation of tissue or biological structure to be analyzed; (2)
prehybridization
treatment of the biological structure to increase accessibility of target DNA,
and to
reduce nonspecific binding; (3) hybridization of the mixture of nucleic acids
to the
nucleic acid in the biological structure or tissue; (4) post-hybridization
washes to
remove nucleic acid fragments not bound in the hybridization and (5) detection
of the
hybridized nucleic acid fragments. The reagent used in each of these steps and
the
conditions for use vary depending on the particular application.
In this case, a hybridization solution comprising at least one detectable
nucleic
acid probe capable of hybridizing to a gene (at its chromosomal locus) is
contacted
with the cell under hybridization conditions. Any hybridization is then
detected and
compared to a predetermined hybridization pattern from normal or control
cells.
Preferably, the probes are alpha-centromeric probes. Such probes can be made
commercially available from a number of sources (e.g., from Visys Inc.,
Downers
Grove, IL). In a preferred embodiment, the hybridization solution contains a
multiplicity of probes, specific for an area on the chromosome that
corresponds to the
translocation of the sequences that make up the chimera (e.g., 15q24-25).
Hybridization protocols suitable for use with the methods of the invention are
described, e.g., in Albertson (1984); Pinkel (1988); EP No. 430,402; and
Methods in
Molecular Biology, Vol. 33: In Situ Hybridization Protocols, Choo, ed., Humana
Press, Totowa, NJ (1994), etc. In one particularly preferred embodiment, the
hybridization protocol of Pinkel et al. (1998) or of Kallioniemi (1992) is
used.
Methods of optimizing hybridization conditions are well known (see, e.g.,
Tijssen
(1993) Laboratory Techniques in Biochemistry and Molecular Biology, Vol. 24:
Hybridization With Nucleic Acid Probes, Elsevier, NY).
In a preferred embodiment, background signal is reduced by the use of a
detergent
(e.g., C-TAB) or a blocking reagent (e.g., sperm DNA, cot-1 DNA, etc.) during
the
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
17
hybridization to reduce non-specific binding. Preferably, the hybridization is
performed in the presence of about 0.1 to about 0.5 mg/ml DNA (e.g., cot-1
DNA).
The probes may be prepared by any method known in the art, including
synthetically or grown in a biological host. Synthetic methods include but are
not
limited to oligonucleotide synthesis, riboprobes, and PCR.
The probe may be labeled with a detectable marker by any method known in the
art. Methods for labeling probes include random priming, end labeling, PCR and
nick
translation. Enzymatic labeling is conducted in the presence of nucleic acid
polymerase, three unlabeled nucleotides, and a fourth nucleotide which is
either
directly labeled, contains a linker arm for attaching a label, or is attached
to a hapten
or other molecule to which a labeled binding molecule may bind. Suitable
direct
labels include radioactive labels such as "P, 3H, and 35S and non-radioactive
labels
such as fluorescent markers, such as fluorescein, Texas Red, AMCA blue,
lucifer
yellow, rhodamine, and the like; cyanin dyes which are detectable with visible
light;
enzymes and the like. Labels may also be incorporated chemically into DNA
probes
by bisulfite-mediated transamination or directly during oligonucleotide
synthesis.
Fluorescent markers can readily be attached to nucleotides with activated
linker
arms incorporated into the probe. Probes may be indirectly labeled by the
methods
disclosed above, by incorporating a nucleotide covalently linked to a hapten
or other
molecule such as biotin or digoxygenin, and performing a sandwich
hybridization
with a labeled antibody directed to that hapten or other molecule, or in the
case of
biotin, with avidin conjugated to a detectable label. Antibodies and avidin
may be
conjugated with a fluorescent marker, or with an enzymatic marker such as
alkaline
phosphatase or horseradish peroxidase to render them detectable. Conjugated
avidin
and antibodies are commercially available from companies such as Vector
Laboratories (Burlingame, CA) and Boehringer Mannheim (Indianapolis, IN).
The enzyme can be detected through a colorimetric reaction by providing a
substrate for the enzyme. In the presence of various substrates, different
colors are
produced by the reaction, and these colors can be visualized to separately
detect
multiple probes. Any substrate known in the art may be used. Preferred
substrates
for alkaline phosphatase include 5-bromo-4-chloro-3-indolylphosphate (BCIP)
and
nitro blue tetrazolium (NBT). The preferred substrate for horseradish
peroxidase is
diaminobenzoate (DAB).
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
18
Fluorescently labeled probes suitable for use in the in situ hybridization
methods
of the invention are preferably in the range of 150-500 nucleotides long.
Probes may
be DNA or RNA, preferably DNA.
Hybridization of the detectable probes to the cells is conducted with a probe
concentration of 0.1-500 ng/ L, preferably 5-250 ng/ L. The hybridization
mixture
will preferably contain a denaturing agent such as formamide. In general,
hybridization is carried out at 25 C-45 C, more preferably at 32 C-40 C, and
most
preferably at 37 C-38 C. The time required for hybridization is about 0.25-96
hours,
more preferably 1-72 hours, and most preferably for 4-24 hours. Hybridization
time
will vary based on probe concentration and hybridization solution content
which may
contain accelerators such as hnRNP binding protein, trialkyl ammonium salts,
lactams, and the like. Slides are then washed with solutions containing a
denaturing
agent, such as formamide, and decreasing concentrations of sodium chloride or
in any
solution that removes unbound and mismatched probe.
The temperature and concentration of salt will vary depending on the
stringency
of hybridization desired. For example, high stringency washes may be carried
out at
42 C-68 C, while intermediate stringency may be in the range of 37 C-55 C, and
low
stringency may be in the range of 30 C-37 C. Salt concentration for a high
stringency wash may be 0.5-1 times SSC (0.15M NaC1, 0.015M Na citrate), while
medium stringency may be 1-4 times, and low stringency may be 2-6 times SSC.
The detection incubation steps, if required, should preferably be carried out
in a
moist chamber at 23 C-42 C, more preferably at 25 C-38 C and most preferably
at
37-38 C. Labeled reagents should preferably be diluted in a solution
containing a
blocking reagent, such as BSA, non-fat dry milk, or the like. Dilutions may
range
from 1:10-1:10,000, more preferably 1:50-1:5,000, and most preferably at 1:100-
1:1,000. The slides or other solid support should be washed between each
incubation
step to remove excess reagent.
Slides may then be mounted and analyzed by microscopy in the case of a visible
detectable marker, or by exposure to autoradiographic film in the case of a
radioactive
marker. In the case of a fluorescent marker, slides are preferably mounted in
a
solution that contains an antifade reagent, and analyzed using a fluorescence
microscope. Multiple nuclei may be examined for increased accuracy of
detection.
Additionally, assays for the expression product of the LBC oncogene can also
be
used to determine whether the LBC oncogene mutation has occurred. Most
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
19
preferably, such assays are immunoassays. Immunoassays, in their most simple
and
direct sense, are binding assays. Certain preferred immunoassays are the
various
types of enzyme linked immunosorbent assays (ELISAs) and radioimmunoassays
(RIA) known in the art. IHC detection using tissue sections is also
particularly useful
as are in situ hybridization and enzyme immunoassay.
In one exemplary ELISA, protein-specific antibodies are immobilized onto a
selected surface exhibiting protein affinity, such as a well in a polystyrene
microtiter
plate. Then, a test composition containing the desired antigen, such as a
clinical
sample, is added to the wells. After binding and washing to remove non-
specifically
bound immune complexes, the bound antigen may be detected. Detection is
generally
achieved by the addition of another antibody, specific for the desired
antigen, that is
linked to a detectable label. This type of ELISA is a simple "sandwich ELISA."
Detection may also be achieved by the addition of a second antibody specific
for the
desired antigen, followed by the addition of a third antibody that has binding
affinity
for the second antibody, with the third antibody being linked to a detectable
label.
Variations of ELISA techniques are well known. In one such variation, the
samples containing the desired antigen are immobilized onto the well surface
and then
contacted with the antibodies of the invention. After binding and appropriate
washing, the bound immune complexes are detected. Where the initial antigen
specific antibodies are linked to a detectable label, the immune complexes may
be
detected directly. Again, the immune complexes may be detected using a second
antibody that has binding affinity for the first antigen specific antibody,
with the
second antibody being linked to a detectable label.
In embodiments of the invention in which gene expression is detected for
determining AML prognosis or status, the use of gene expression portfolios is
most
preferred. A portfolio of genes is a set of genes grouped so that expression
information obtained about them provides the basis for making a clinically
relevant
judgment such as a diagnosis, prognosis, or treatment choice. In this case,
gene
expression portfolios can be fashioned to help make therapeutic decisions
regarding
AML patients.
Diseased, in this context, refers to an alteration of the state of a body that
interrupts or disturbs, or has the potential to disturb, proper performance of
bodily
functions as occurs with the uncontrolled proliferation of cells. Someone is
diagnosed with a disease when some aspect of that person's genotype or
phenotype
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
is consistent with the presence of the disease. However, the act of conducting
a
diagnosis or prognosis may include the determination of disease/status issues
such
as determining the likelihood of relapse, type of therapy and therapy
monitoring. In
therapy monitoring, clinical judgments are made regarding the effect of a
given
5 course of therapy by comparing the expression of genes over time to
determine
whether the gene expression profiles have changed or are changing to patterns
more
consistent with normal tissue.
Genes can be grouped so that information obtained about the set of genes in
the
group provides a sound basis for making a clinically relevant judgment such as
a
10 diagnosis, prognosis, or treatment choice. These sets of genes make up the
portfolios of the invention. As with most diagnostic markers, it is often
desirable to
use the fewest number of markers sufficient to make a correct medical
judgment.
This prevents a delay in treatment pending further analysis as well
unproductive use
of time and resources.
15 One method of establishing gene expression portfolios is through the use of
optimization algorithms such as the mean variance algorithm widely used in
establishing stock portfolios. This method is described in detail in US patent
publication number 20030194734. Essentially, the method calls for the
establishment of a set of inputs (stocks in financial applications, expression
as
20 measured by intensity here) that will optimize the return (e.g., signal
that is
generated) one receives for using it while minimizing the variability of the
return.
Many commercial software programs are available to conduct such operations.
"Wagner Associates Mean-Variance Optimization Application," referred to as
"Wagner Software" throughout this specification, is preferred. This software
uses
functions from the "Wagner Associates Mean-Variance Optimization Library" to
determine an efficient frontier and optimal portfolios in the Markowitz sense
is one
option. Use of this type of software requires that microarray data be
transformed so
that it can be treated as an input in the way stock return and risk
measurements are
used when the software is used for its intended financial analysis purposes.
The process of selecting a portfolio can also include the application of
heuristic
rules. Preferably, such rules are formulated based on biology and an
understanding
of the technology used to produce clinical results. More preferably, they are
applied
to output from the optimization method. For example, the mean variance method
of
portfolio selection can be applied to microarray data for a number of genes
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
21
differentially expressed in subjects with cancer. Output from the method would
be
an optimized set of genes that could include some genes that are expressed in
peripheral blood as well as in diseased tissue. If samples used in the testing
method
are obtained from peripheral blood and certain genes differentially expressed
in
instances of cancer could also be differentially expressed in peripheral
blood, then a
heuristic rule can be applied in which a portfolio is selected from the
efficient
frontier excluding those that are differentially expressed in peripheral
blood. Of
course, the rule can be applied prior to the formation of the efficient
frontier by, for
example, applying the rule during data pre-selection.
Other heuristic rules can be applied that are not necessarily related to the
biology
in question. For example, one can apply a rule that only a prescribed
percentage of
the portfolio can be represented by a particular gene or group of genes.
Commercially available software such as the Wagner Software readily
accommodates these types of heuristics. This can be useful, for example, when
factors other than accuracy and precision (e.g., anticipated licensing fees)
have an
impact on the desirability of including one or more genes.
The gene expression profiles of this invention can also be used in conjunction
with other non-genetic diagnostic methods useful in cancer diagnosis,
prognosis, or
treatment monitoring. For example, in some circumstances it is beneficial to
combine the diagnostic power of the gene expression based methods described
above with data from conventional markers such as serum protein markers (e.g.,
Cancer Antigen 27.29 ("CA 27.29")). A range of such markers exists including
such
analytes as CA 27.29. In one such method, blood is periodically taken from a
treated patient and then subjected to an enzyme immunoassay for one of the
serum
markers described above. When the concentration of the marker suggests the
return
of tumors or failure of therapy, a sample source amenable to gene expression
analysis is taken. Where a suspicious mass exists, a fine needle aspirate
(FNA) is
taken and gene expression profiles of cells taken from the mass are then
analyzed as
described above. Alternatively, tissue samples may be taken from areas
adjacent to
the tissue from which a tumor was previously removed. This approach can be
particularly useful when other testing produces ambiguous results.
Kits made according to the invention include formatted assays for determining
the gene expression profiles. These can include all or some of the materials
needed
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
22
to conduct the assays such as reagents and instructions and a medium through
which
Biomarkers are assayed.
Preferred methods for establishing gene expression profiles (including those
used
to arrive at the explication of the relevant biological pathways) include
determining
the amount of RNA that is produced by a gene that can code for a protein or
peptide
or transcribe RNA. This is best accomplished by reverse transcription PCR (RT-
PCR), competitive RT-PCR, real time RT-PCR, differential display RT-PCR,
Northern Blot analysis and other related tests. While it is possible to
conduct these
techniques using individual PCR reactions, it is often desirable to amplify
copy DNA
(cDNA) or copy RNA (cRNA) produced from mRNA and analyze it via microarray.
A number of different array configurations and production methods are known to
those of skill in the art and are described in US Patents such as: 5,445,934;
5,532,128;
5,556,752; 5,242,974; 5,384,261; 5,405,783; 5,412,087; 5,424,186; 5,429,807;
5,436,327; 5,472,672; 5,527,681; 5,529,756; 5,545,531; 5,554,501; 5,561,071;
5,571,639; 5,593,839; 5,599,695; 5,624,711; 5,658,734; and 5,700,637.
Microarray technology measures steady-state mRNA levels of thousands of genes
"
simultaneously thereby presenting a powerful tool for identifying AML patient
gene
expression profiles. Two microarray technologies are currently in wide use.
The first
are cDNA arrays and the second are oligonucleotide arrays. Although
differences
exist in the construction of these chips, essentially all downstream data
analysis and
output are the same. The products of these analyses are typically measurements
of the
intensity of the signal received from a labeled probe used to detect a cDNA
sequence
from the sample that hybridizes to a nucleic acid sequence at a known location
on the
microarray. Typically, the signal intensity is proportional to the cDNA
quantity, and
thus mRNA, expressed in the sample cells. A large number of such techniques
are
available and useful. Preferred methods can be found in US Patents 6,271,002;
6,218,122; 6,218,114; and 6,004,755.
Analysis of the expression levels is conducted by comparing such intensities.
This is best done by generating a ratio matrix of the expression intensities
of genes in
a test sample versus those in a control sample. For instance, the gene
expression
intensities from a tissue that has been treated with a drug can be compared
with the
expression intensities generated from the same tissue that has not been
treated with
the drug. A ratio of these expression intensities indicates the fold-change in
gene
expression between the test and control samples.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
23
Gene expression profiles can be displayed in a number of ways. A common
method is to arrange a ratio matrix into a graphical dendogram where columns
indicate test samples and rows indicate genes. The data are arranged so genes
that
have similar expression profiles are proximal to each other. The expression
ratio for
each gene is visualized as a color. For example, a ratio less than one
(indicating
down-regulation) may appear in the blue portion of the spectrum while a ratio
greater
than one (indicating up-regulation) may appear as a color in the red portion
of the
spectrum. Commercially available computer software programs are available to
display such data including "GENESPRINT" from Silicon Genetics, Inc. and
"DISCOVERY" and "INFER" software from Partek, Inc.
The differentially expressed genes are either up regulated or down regulated
in
diseased cells, as deduced by an assessment of gene expression as described
above.
Up regulation and down regulation are relative terms meaning that a detectable
difference (beyond the contribution of noise in the system used to measure it)
is found
in the amount of expression of the genes relative to some baseline. In this
case, the
baseline is the measured gene expression of a normal cell. The genes of
interest in the
diseased cells are then either up regulated or down regulated relative to the
baseline
level using the same measurement method. Preferably, levels of up and down
regulation are distinguished based on fold changes of the intensity
measurements of
hybridized microarray probes. A 1.5 fold difference is preferred for making
such
distinctions. That is, before a gene is said to be differentially expressed in
treated
versus untreated diseased cells, the treated cell is found to yield at least
1.5 times
more, or 1.5 times less intensity than the untreated cells. A 1.7 fold
difference is more
preferred and a 2 or more fold difference in gene expression measurement is
most
preferred.
One method of the invention involves comparing gene expression profiles for
various genes to determine whether a person is likely to respond to the use of
a
therapeutic agent. Having established the gene expression profiles that
distinguish
responder from non-responder, the gene expression profiles of each are fixed
in a
medium such as a computer readable medium as described below. A patient sample
is obtained that contains diseased cells (such as hematopoietic blast cells in
the case of
AML) is then obtained. Most preferably, the samples are of bone marrow and are
extracted from the patient's sternum or iliac crest according to routine
methods.
Preferably the bone marrow aspirate is processed to enrich for leukemic blast
cells
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
24
using routine methods. Sample RNA is then obtained and amplified from the
diseased patient cells and a gene expression profile is obtained, preferably
(in the case
of a large gene portfolio) via micro-array, for genes in the appropriate
portfolios. The
expression profiles of the samples are then compared to those previously
analyzed for
prognostic outcome. When a small number of genes are used in the portfolio
such as
when the three gene profile is used, a simple nucleic acid amplification and
detection
scheme is the most preferred method of ineasuring gene modulation. In such a
case,
PCR, NASBA, rolling circle, LCR, and other amplification schemes known to
skilled
artisans can be used with PCR being most preferred. Where the portfolios
include a
large number of genes or it is desirable to measure the expression of numerous
other
genes then it is preferred to assess the expression patterns based on
intensity
measurements of microarrays as described above.
In similar fashion, gene expression profile analysis can be conducted to
monitor
treatment response. In one aspect of this method, gene expression analysis as
described above is conducted on a patient treated with any suitable treatment
at
various periods throughout the course of treatment. If the gene expression
patterns
are consistent with a positive outcome the patient's therapy is continued. If
it is not,
the patient's therapy is altered as with additional therapeutics, changes to
the dosage,
or elimination of the current treatment. Such analysis permits intervention
and
therapy adjustment prior to detectable clinical indicia or in the face of
otherwise
ambiguous clinical indicia.
With respect to the molecular markers of the invention, a number of other
formats
and approaches are available for diagnostic use. Methylation of genomic
regions can
affect gene expression levels. For example, hypermethylation of gene promoter
regions can constitutively down-regulate gene expression whereas
hypomethylation
can lead to an increase in steady-state mRNA levels. As such, detection of
methylated regions associated with genes predictive of drug response,
prognosis or
status can be used as an alternative method for diagnosing gene expression
levels.
Such methods are known to those skilled in the art. Alternatively, single
nucleotide
polymorphisms (SNPs) that are present in promoter regions can also affect
transcriptional activity of a gene. Therefore, detection of these SNPs by
methods
known to those skilled in the art can also be used as a diagnostic for
detecting genes
that are differentially expressed in different prognostic outcomes.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
Articles of this invention are representations of the gene expression profiles
useful
for treating, diagnosing, prognosticating, staging, and otherwise assessing
diseases.
Preferably they are reduced to a medium that can be automatically read such as
computer readable media (magnetic, optical, and the like). The articles can
also
5 include instructions for assessing the gene expression profiles in such
media. For
example, the articles may comprise a CD ROM having computer instructions for
comparing gene expression profiles of the portfolios of genes described above.
The
articles may also have gene expression profiles digitally recorded therein so
that they
may be compared with gene expression data from patient samples. Alternatively,
the
10 profiles can be recorded in different representational format. Clustering
algorithms
such as those incorporated in "DISCOVERY" and "1NFER" software from Partek,
Inc. mentioned above can best assist in the visualization of such data.
Additional articles according to the invention are kits for conducting the
assays
described above. Each such kit would preferably include instructions in human
or
15 machine readable form as well as the reagents typical for the type of assay
described.
These can include, for example, nucleic acid arrays (e.g. cDNA or
oligonucleotide
arrays), as described above, configured to discern the gene expression
profiles of the
invention. They can also contain reagents used to conduct nucleic acid
amplification
and detection including, for example, reverse transcriptase, reverse
transcriptase
20 primer, a corresponding PCR primer set, a thermostable DNA polymerase, such
as
Taq polymerase, and a suitable detection reagent(s), such as, without
limitation, a
scorpion probe, a probe for a fluorescent probe assay, a molecular beacon
probe, a
single dye primer or a fluorescent dye specific to double-stranded DNA, such
as
ethidium bromide. Kits for detecting surface antigens contain staining
materials or
25 are antibody based including components such as buffer, anti-antigenic
antibody,
detection enzyme and substrate such as Horse Radish Peroxidase or biotin-
avidin
based reagents. Kit components for detecting blast cells generally include
reagents
for conducting flow cytometry, blast cell adhesion assays, and other common
blast
cell assays.
Conventional anti-cancer agents include, without limitation, tyrosine kinase
inhibitors, MEK kinase inhibitors, P 13K kinase inhibitors, MAP kinase
inhibitors,
apoptosis modulators and combinations thereof. Exemplary drugs that are most
preferred among these are the "GLEEVEC" tyrosine kinase inhibitor of Novartis,
U-0126 MAP kinase inhibitor, PD-098059 MAP kinase inhibitor, SB-203580 MAP
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
26
kinase inhibitor, and antisense, ribozyme, and DNAzyme, Bcl-XL, and anti-
apoptotics. Examples of other useful drugs include, without limitation, the
calanolides of US Patent 6,306,897; the.substituted bicyclics of US Patent
6,284,764; the indolines of US Patent 6,133,305; and the antisense
oligonucleotides
of US Patent 6,271,210; platinum coordination compounds for example cisplatin
or
carboplatin, taxane compounds for example paclitaxel or docetaxel,
camptothecin
compounds for example irinotecan or topotecan, anti-tumor vinca alkaloids for
example vinblastine, vincristine or vinorelbine, anti-tumor nucleoside
derivatives for
example 5-fluorouracil, gemcitabine or capecitabine, nitrogen mustard or
nitrosourea alkylating agents for example cyclophosphamide, chlorambucil,
carmustine or lomustine, anti-tumor anthracycline derivatives for example
daunorubicin, doxorubicin or idarubicin; HER2 antibodies for example
trastzumab;
and anti-tumor podophyllotoxin derivatives for example etoposide or
teniposide; and
antiestrogen agents including estrogen receptor antagonists or selective
estrogen
receptor modulators preferably tamoxifen, or alternatively toremifene,
droloxifene,
faslodex and raloxifene, or aromatase inhibitors such as exemestane,
anastrozole,
letrazole and vorozole.
Anti-cancer agents can also include therapeutics directed to gene therapy or
antisense therapy or RNA interference. These include, without limitation,
oligonucleotides with sequences complementary to an mRNA sequence can be
introduced into cells to block the translation of the mRNA, thus blocking the
function of the gene encoding the mRNA. The use of oligonucleotides to block
gene
expression is described, for example, in, Strachan and Read, Human Molecular
Genetics, 1996. These antisense molecules may be DNA, stable derivatives of
DNA
such as phosphorothioates or methylphosphonates, RNA, stable derivatives of
RNA
such as 2'-O-alkylRNA, or other antisense oligonucleotide mimetics. Antisense
molecules may be introduced into cells by microinjection, liposome
encapsulation or
by expression from vectors harboring the antisense sequence.
In gene therapy, the gene of interest can be ligated into viral vectors that
mediate
transfer of the therapeutic DNA by infection of recipient host cells. Suitable
viral
vectors include retrovirus, adenovirus, adeno-associated virus, herpes virus,
vaccinia
virus, polio virus and the like. Alternatively, therapeutic DNA can be
transferred
into cells for gene therapy by non-viral techniques including receptor-
mediated
targeted DNA transfer using ligand-DNA conjugates or adenovirus-ligand-DNA
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
27
conjugates, lipofection membrane fusion or direct microinjection. These
procedures
and variations thereof are suitable for ex vivo as well as in vivo gene
therapy.
Protocols for molecular methodology of gene therapy suitable for use with the
gene
is described in Gene Therapy Protocols, edited by Paul D. Robbins, Human
press,
Totowa NJ, 1996.
Compounds identified according to the methods disclosed herein may be used
alone at appropriate dosages defined by routine testing in order to obtain
optimal
inhibition or activity while minimizing any potential toxicity. In addition,
co-
administration or sequential administration of other agents may be desirable.
The invention is further illustrated by the following nonlimiting examples.
All
references cited herein are hereby incorporated herein by reference.
Example 1
Clinical Evaluation and Response Definitions
The current study was part of an open label, multicenter, non-comparative
phase
2 clinical study in which patients with relapsed or refractory AML (Harousseau
et al.
(2003)) were treated with tipifarnib at a starting oral dose of 600 mg bid for
the first
21 consecutive days of each 28-day cycle. Patients were enrolled into 2
cohorts,
those with relapsed AML and those with refractory AML. A total of 252 patients
(135 relapsed and 117 refractory) were treated. Eighty patients chose to
provide
bone marrow samples for RNA microarray analysis, for which a separate informed
consent was required. The overall response rate was relatively low in this
study.
Therefore, for the purposes of the gene expression profiling, response to
tipifarnib was defined as patients who had an objective response (complete
remission [CR], complete remission with incomplete platelet recovery [CRp] or
partial remission [PR]), a hematological response (decrease of >50% of
leukemic
blast cells in bone marrow) as determined by either central review or by the
clinical
site, or stable disease (no hematological response but no progression of the
disease)
as determined by both central review and the clinical site. Complete remission
with
incomplete platelet recovery was defined similarly, except for a platelet
count less
than 100,000/ L sufficient to ensure transfusion independence. Partial
remission
was defined as at least a 50% decrease in bone marrow blasts with partial
neutrophil
(>500/ L) and platelet count (>50,000/ L) recovery. Response had to be
confirmed
at least 4 weeks after first documentation.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
28
Sample Collection and Microarray Processing
Bone marrow samples were collected from patients before treatment with
tipifarnib, diluted with phosphate buffered saline (PBS) and centrifuged with
Ficoll-
diatrizoate (1.077 g/mL). White blood cells were washed twice with PBS,
resuspended in fetal bovine serum (FBS) with 10% dimethyl sulfoxide (DMSO) and
immediately stored at -80 C. Cells were thawed and total RNA was extracted
from
cell samples using the RNeasy Kit (Qiagen, Valencia, CA). RNA quality was
checked using the Agilent Bioanalyzer. Synthesis of cDNA and cRNA was
performed according to Affymetrix (Santa Clara, CA) protocols.
Microarray Processing
Two rounds of linear amplification were performed because the RNA yield for
several samples was too low to obtain enough labeled cRNA for chip
hybridization
using one round of amplification. For hybridization, 11 g of cRNA were
fragmented randomly by incubation at 94 C for 35 minutes in 40 mM Tris-
acetate,
pH 8.1, 100 mM potassium acetate, and 30 mM magnesium acetate. Fragmented
cRNA was hybridized to U133A arrays at 45 C for 16 hours in a rotisserie oven
set
at 60 rpm. Following hybridization, arrays were washed (with 6x SSPE and 0.5x
SSPE containing Triton X-100 [0.005%]), and stained with streptavidin-
phycoerythrin (SAPE; Molecular Probes, Eugene, OR). Quantification of bound
labeled probe was conducted using the Agilent G2500A GeneArray scanner
(Agilent
Technologies, Palo Alto, CA).
The total fluorescence intensity for each array was scaled to the uniform
value of
600. Chip performance was quantitated by calculating a signal to noise ratio
(raw
average signal/noise). Chips were removed from further analysis if their
signal-to-
noise ratio was less than 5. Genes were only included in further analysis if
they
were called "present" in at least 10% of the chips. Eleven thousand seven
hundred
twenty three Affymetrix probe sets remained following this cut-off. The
quality of
the gene expression data were further controlled by identifying outliers based
on
principal components analysis and by analyzing the normal distributions of the
gene
intensities (Partek Pro V5.1).
Statistical Analysis
To identify genes that predict response with high sensitivity, a percentile
analysis was employed. Genes that were up- or down-regulated in 100% of
responders compared to at least 40% of non-responders were identified. The chi-
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
29
squared test and Student's t-test were then used to test the significance of
the
correlations between patient response and patient co-variates, including ras
mutation
status, and gene expression. Unsupervised k-means and hierarchical clustering
were
performed in Omniviz. The predictive value of the selected genes was then
analyzed by leave-one-out and leave-five-out cross validation methods. Here,
one
(or five) sample(s) was (were) removed from the data set and the marker was
reselected from 11,723 genes. The predictive value of this gene was then
tested on
the left-out sample(s) using a linear discriminant analysis. Sensitivity was
calculated as the number of true positives detected by the test divided by the
sum of
true positives plus false negatives. Specificity was calculated as the number
of true
negatives detected by the test divided by the sum of true negatives and false
positives. Positive predictive value was calculated as the number of true
positives
divided by the number of true positives and false positives. Negative
predictive
value was calculated as the number of true negatives divided by the number of
true
negatives and false negatives. The positive likelihood ratio of a patient
responding
to treatment is sensitivity divided by 1 minus specificity. Receiver operator
curves
(ROC) were used to choose appropriate thresholds for each classifier,
requiring a
sensitivity of 100%. The ROC diagnostic calculates the sensitivity and
specificity
for each parameter.
Real-Time RT-PCR Validation
TaqMan real-time RT-PCR was employed to verify the microarray results of
the AHR and AKAP13 genes. For each 1 g sample of amplified RNA, cDNA was
produced using T7 oligo(dT) primer and Superscript II reverse transcriptase
according to the manufacturer's instructions (Invitrogen). Primers and MGB-
probes
for AKAP 13 gene and control gene PBGD were designed using Primer Express
(Applied Biosystems), while those for AHR gene and control gene HPRT were
available as Assays-on-Demand from ABI. Primer/probe sequences for AKAP 13
were as follows: AKAP13 forward, 5'ggtcagatgtttgccaaggaa3' (SEQ ID NO: 1);
AKAP 13 reverse, 5'tcttcagaaacacactcccatc-3' (SEQ ID NO: 2); AKAP 13 probe,
6FAM-tgaaacggaagaagcttgtA-3' (SEQ ID NO: 3).
All primers and probes were tested for optimal amplification efficiency above
90%. The relative standard curve was composed of 5 dilutions (10-fold each) of
HeLa cDNA (in most cases, ranging from 25 ng to 2.5 pg). RT=PCR amplification
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
mixtures (25 L) contained 100 ng template cDNA, 2x TaqMan universal PCR
master mix (12.5 L; Applied Biosystems), 500 nM forward and reverse primers,
and 250 nM probe. Reactions were run on an ABI PRISM 7900HT Sequence
Detector (Applied Biosystems). The cycling conditions were: 2 min of AmpErase
5 UNG activation at 50 C, 10 min of polymerase activation at 95 C and 50
cycles at
95 C for 15 sec and annealing temperature (59 C or 60 C) for 60 sec. In each
assay,
a standard curve and a no-template control along with template cDNA were
included
in triplicates for the gene of interest and control gene. The relative
quantity of each
gene was calculated based on the standard curve, and was normalized with the
10 quantity of the control gene. The median coefficient of variation (based on
calculated quantities) of triplicate samples was 8%. The correlation between
repeated runs using independently diluted templates from the stock was above
0.95.
Samples were only compared with microarray data if duplicate TaqMan
experiments showed reproducible results.
15 Cell Line Culture and AKAP 13 Over-expression Assay
The AKAP13 vectors, oncoLBC and protoLBC, and vector control (pSRalpha-
neo) were obtained from Dr. Deniz Toksoz. Zheng et al. (1995). The HL60 cell
line
was obtained from the American Tissue Culture Collection and grown in RPMI
1640 with 10% FBS. Cells were transiently transfected with each vector using
the
20 Effectene reagent (Qiagen) according to the manufacturer's instructions and
kept
under G418 (600 g/mL) for 7 days. Tipifamib was then added in various
concentrations (0, 1.5, 3.1, 6.3, 13, 25, 50, 100, 200, 1000, and 10,000 nM)
to
duplicate cultures (1.5x105 cells/mL). Cells were counted at Day 6. Cell
counts
were normalized to cultures with no drug to give a percent of viable control
cells.
25 Results
Expression Profiling of Relapsed and Refractory AML
FTIs were originally designed to specifically inhibit FTase activity, thereby
blocking the oncogenic ras pathways. Therefore, we initially analyzed DNA from
the bone marrow of 80 patients with relapsed or refractory AML for activating
ras
30 mutations and investigated the possible correlation between ras mutation
and the
response to tipifarnib. While 26% of the AML samples harbored N-ras mutations,
mutation status did not correlate with objective response or overall survival.
Harousseau et al. (2003). We therefore performed gene expression profiling to
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
31
identify novel signatures that could be used to predict response to the FTI
tipifamib.
Bone marrow samples were obtained for gene expression analysis from 80
patients
prior to treatment with tipifarnib. Table 1 shows the patient information.
Table 1. Patient Information
Patient ID AML CLASS SEX AGE Best Response* Survival Time Response
IA30060 REFRACTC)IZI \1ALL 7Q CR 355 A300i ] RELAPSED V'[ALL 70 PR 154 A30059
RCFRAC'T( K f EMALL r;? SI) 119 es
A30095 REFRACT(KY \I.ALE r;l ( R i) vc
130I77 RELAPSEL) I EMALE 03 SE) 7 vrs
\3(11'>_' RLL,AI'SI U \i:\I F. 67 tiL) ?60
vcs
A3O73~ RFI.IFSE:E) FFRtAE_F. ~1~ (R 270
vcs A;0246 Kf,:FEZ:AC[( iKY NfALI-, 7 4 II I 2 13
A~U353 ftLLAf'S11_) 1I7T'1ALL 3o III l~) vc,
\30-"5-; KEE_aPSEI-) 1+A111 E 44 IIl T7 ~c5 130360 UFEt,ACTUIZI" \1~AI F w0
llI 3~ vc~ A',(i3h4 KI~IAPSEG Ki.ti1 AL1=- ~4 HI 6 7
-A_')(371) KE-:I_:AP~HL) IE:%lr\LL (1 3 SE) \;03h0 KLFK:ACl(>KY 1, E.:%1-ALF
71 Ill 71
A30007 REFRACTORY FEMALE 54 NR 106 no
A30008 REFRACTORY MALE 52 NR 27 no
A30053 RELAPSED MALE 51 NR 48 no
A30057 REFRACTORY MALE 74 NR 102 no
A30060 REFRACTORY FEMALE 33 NR 175 no
A30096 REFRACTORY MALE 69 NR 182 no
A30179 REFRACTORY FEMALE 70 NR 148 no
A30182 REFRACTORY MALE 70 NR 92 no
A30190 RELAPSED FEMALE 54 NR 51 no
A30191 RELAPSED FEMALE 67 NR 78 no
A30245 RELAPSED MALE 63 NR 366 no
A30300 RELAPSED MALE 47 NR 414 no
A30302 RELAPSED MALE 62 NR 234 no
A30308 RELAPSED MALE 66 NR 71 no
A30311 RELAPSED FEMALE 61 NR 115 no
A30377 RELAPSED MALE 68 NR 364 no
A30047 RELAPSED FEMALE 63 NR 94 no
A30055 RELAPSED FEMALE 71 NR 56 no
A30063 RELAPSED MALE 46 NR 220 no
A30090 REFRACTORY FEMALE 85 NR 56 no
A30091 REFRACTORY FEMALE 67 NR 56 no
A30092 REFRACTORY FEMALE 54 NR 40 no
A301 11 REFRACTORY FEMALE 71 NR 38 no
A30112 RELAPSED FEMALE 61 NR 12 no
A30113 REFRACTORY MALE 75 NR 177 no
A30119 REFRACTORY MALE 19 NR 36 no
A30153 RELAPSED FEMALE 68 NR 105 no
A30176 REFRACTORY MALE 75 NR 54 no
A30178 RELAPSED FEMALE 70 NR 39 no
A30180 REFRACTORY MALE 62 NR 72 no
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
32
A30183 REFRACTORY MALE 63 NR 64 no
A30244 RELAPSED FEMALE 34 NR 35 no
A30247 REFRACTORY FEMALE 72 NR 35 no
A30248 RELAPSED MALE 46 NR 61 no
A30304 RELAPSED MALE 65 NR 44 no
A30306 RELAPSED FEMALE 28 NR 74 no
A30349 REFRACTORY MALE 58 NR 22 no
A30354 REFRACTORY FEMALE 31 NR 103 no
A30359 RELAPSED MALE 65 NR 8 no
A30363 RELAPSED MALE 64 NR 37 no
A30376 RELAPSED FEMALE 24 NR 383 no
A30378 RELAPSED FEMALE 76 NR 184 no
A30381 REFRACTORY FEMALE 70 NR 128 no
A30395 REFRACTORY MALE 61 NR 83 no
* Stable disease (SD) only included if confirmed by independent investigators
HI = hematological improvement
CR = complete response
PR = partial response
NR = no response
Fifty-eight of the 80 samples passed quality control measures including RNA
quality and chip performance. There were no significant differences in age,
sex,
AML class (relapsed or refractory), cytogenic risk factors, baseline blast
counts,
response, and overall survival between these 58 patients and the remainder of
the
clinical study population (N = 194; Table 2).
Table 2
co-variate 58 subset 194 remainder p-value
response 14 28 0.1237
male 28 119 0.1055
average age 60 56 0.1046
relapsed 31 104 0.8977
cytogenetic risk 34 5 0.1503
average blasts 55% 50% 0.1629
The gene expression data were integrated with the clinical information and
retrospective analyses were performed to identify genes that could separate
responders from non-responders with a high level of sensitivity. The data went
through several filtering steps before identification of differentially
expressed genes.
First, genes that were not expressed in at least 10% of the samples were
removed.
This reduced the number of genes from approximately 22,000 to 11,723 genes.
For
unsupervised analyses genes that showed little variation in expression across
the
dataset (coefficient of variance of <45% across all the samples) were also
excluded
and quantile normalization was applied to the remaining 5,728 genes. At this
stage
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
33
an unsupervised k-means clustering analysis was performed to identify any
differences between patients based on their global gene expression profiles.
Six
main clusters of patients were identified using this technique. No separation
between responders and non-responders was observed (Figure 1). Only a handful
of
genes may be associated with the anti-tumor effect of FTIs, for example, it is
possible that the differential expression of a single gene that is involved in
FTI
biology impacts clinical response and this would be masked by the noise
introduced
from the other 11,722 genes.
Example 2
Identification of Genes that Are Differentially Expressed Between Responders
and
Non-responders
We next performed supervised analysis using the gene expression data to
identify genes that were differentially expressed between all responders and
at least
40% of non-responders. These criteria were chosen to identify genes that could
predict response to tipifarnib with the highest level of sensitivity possible.
From
11,723 genes, a total of 19 genes were identified that could stratify
responders and
non-responders (Table 3 and Table 10 for more detail) and that gave
significant P
values in a t-test (P <0.05). The genes included those involved in signal
transduction, apoptosis, cell proliferation, oncogenesis, and potentially, FTI
biology
(ARHH, AKAP13, IL3RA).
Table 3 Top 19 Genes that Predict Response to tipifarnib and Results of
Analysis
SEQ ID
NO Symbol Specificity P value Functional description
151 AHR 0.52 0.00000255 Apoptosis, cell cycle, signal transduction
309 AKAP13 0.63 0.00006133 Small GTPase mediated signal transduction,
oncogenesis
488 MINA53 0.50 0.00006934 Cell proliferation
411 IDS 0.50 0.00023964 Glycosaminoglycan degradation
632 OPN3 0.40 0.00064297 G-protein coupled receptor protein signaling
280 GPR105 0.43 0.00087608 G-protein coupled receptor protein signaling
582 TENCI 0.43 0.0010309 Signal transduction
376 TNFSF13 0.40 0.00104219 Cell proliferation
134 SVIL 0.45 0.00145723 Cytoskeletal anchoring
272 IL3RA 0.40 0.00198392 Receptor signaling
209 C6orf56 0.40 0.00261553 -
697 FRAG1 0.45 0.00298989 Tumor suppressor
476 GOSRI 0.45 0.01201057 Intra-Golgi transport
204 KIAA1036 0.43 0.01262079 -
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
34
483 BTG3 0.47 0.01659402 Regulation of cell cycle
487 MAPK8IP3 0.40 0.01817428 Regulation of JNK cascade
419 LILRB3 0.41 0.02374898 Immune response
242 ARHH 0.40 0.02721922 Small GTPase mediated signal transduction
496 NPTX2 0.45 0.03346833 Heterophilic cell adhesion
Real Time RT-PCR Validation of Gene Markers
To verify the microarray gene expression data, TaqMan real time RT-PCR was
performed on cDNA that was used for generating the labeled target cRNA for
microarray hybridization. Two genes were selected to verify the gene
expression
data. The AHR and AKAP 13 genes were chosen because the use of these genes
resulted in the highest level of specificity for responders. The correlation
coefficient
was 0.74 for AHR and 0.94 for AKAP 13 indicating that the microarray gene
expression data could be validated by PCR (Figure 2).
Identification of the AKAP 13 Gene as a Marker of Resistance
AKAP13 was over-expressed in patients who were resistant to tipifarnib. The
predictive value of this gene was calculated for the 58 samples using a leave-
one-out
cross validation (LOOCV; Figure 3A). AKAP13 gene expression predicted non-
response with a negative predictive value (NPV) of 96%, and low expression
levels
mediated response with a positive predictive value (PPV) of 43% (x2 = 13.7; P
=
0.0022). The overall diagnostic accuracy was 69% and positive likelihood ratio
of
responding was 2.4. Therefore, stratification of this patient population based
on
AKAP13 gene expression increased the response rate from 24% (14/5 8) in the
entire
group to 43% (13/30) among those patients with low expression of the gene.
Expression values for the AKAP13 gene in each patient are shown in Figure 3B.
When survival was analyzed by Kaplan-Meier analysis, the median survival of
patients with low expression of this gene was 90 days longer than those
patients who
had high expression levels (P = 0.008; Figure 3C).
Identification of a Minimal Set of 3 Gene Markers
LOOCV was used to identify a candidate set of gene markers that could predict
response to tipifarnib with an improved accuracy compared to AKAP 13 alone.
Classifiers were built with an increasing number of genes based on t-test P
values,
and the error rate of these classifiers was calculated using LOOCV while
keeping
the sensitivity of predicting response at 100% (Figure 4A).
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
The 3-gene classifier could predict response with the lowest error rate
(Figure
4A). This was also observed when a leave-five-out cross validation was
performed.
When more genes were added the error rate increased, indicating that
additional
genes were introducing noise to the classifier. For the 3-gene classifier, the
LOOCV
5 demonstrated a NPV of 94% and a PPV of 48%, with an overall diagnostic
accuracy
of 74% and positive likelihood ratio of 2.9 (Figure 4B). The combined
expression
values for the 3 genes in each patient are shown in Figure 4C. Therefore, for
the
group of patients with this gene signature, the response rate to tipifamib was
48%
(12/25) compared to 24% (14/58) in this patient population.
10 Using the 3-gene signature (AHR, AKAP 13 and MINA53), Kaplan-Meier
analysis again showed a significant difference in survival between the
predicted
responder group and the non-responder group (Figure 4D). The 13 patients who
were incorrectly classified as responders had a better overall survival
compared to
the 31 patients correctly classified as non-responders (Figure 5).
Interestingly, the 2
15 patients that were misclassified as non-responders only demonstrated
hematological
improvements with relatively short overall survival times (71, 87 days).
Over-expression of AKAP13 Increases Resistance to tipifarnib in AML
The AKAP 13 gene was the most robust marker of resistance to tipifamib. We
therefore investigated its involvement in FTI biology by over-expressing the
20 oncoLBC and protoLBC variants of this gene in the HL60 cell line. Transient
transfectants were then tested for sensitivity to tipifarnib. Over-expression
of both
AKAP 13 variants in this AML cell line model led to an approximate 20-fold
increase in resistance to tipifarnib compared to control cells (Figure 3).
Both the
LBC oncogene and proto-oncogene increased the resistance to tipifarnib to the
same
25 extent, as seen by a parallel rightward shift of the kill curves by more
than one log-
unit compared to control.
Discussion
Two groups recently identified gene expression profiles of newly diagnosed
adult AML patients that are useful for predicting clinical outcome. Bullinger
et al.
30 (2004); and Valk et al. (2004). These profiles seem to be more powerful
than
currently used prognostic markers such as karyotyping. Moreover, expression
profiles have been found that predict response to anticancer compounds
including
standard chemotherapeutics (Chang et al. (2003); Okutsu et al. (2002); and
Cheok et
al. (2003)) and novel selective anticancer agents. Hofmann et al. (2002); and
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
36
McLean et al. (2004). Similarly, pharmacogenetic profiles have recently been
found
that correlate with patient response to the tyrosine kinase inhibitor
gefitinib. Paez et
al. (2004) and Lynch et al. (2004). In that study, a subgroup of non-small
cell lung
cancer patients had activating mutations within the target epidermal growth
factor
receptor that correlated with clinical response to the targeted therapy.
In a phase 2 study of relapsed and refractory AML patients, we have identified
gene expression profiles that predict response to tipifamib, a novel farnesyl
transferase inhibitor. This class of compounds is showing promise in the
treatment
of hematological malignancies (Karp et al. (2001); Kurzrock et al. (2004);
Alsina et
al. (2003); Cortes et al. (2003); and Thomas et al. (2001)) and solid tumors
such as
breast cancer (Johnston et al. (2003)) and recurrent glioma. Brunner et al.
(2003).
However, while clinical responses are being demonstrated, there is a growing
need
to tailor therapy by identifying patients who are most likely to respond to
the drug
and are, therefore, the best candidates for treatment. Furthermore, while ras
was
considered to be a primary target of this class of drugs, several clinical
studies have
shown that they are not necessarily effective in populations with a high
frequency of
ras mutations. Van Cutsem et al. (2004); and Rao et al. (2004).
Several gene markers were identified that have the potential to predict
response
to tipifarnib. A subset of these markers was both predictive of drug response
and
also thought to have the potential to be involved in FTI biology. One of the
top
candidates discovered from the microarray studies was the lymphoid blast
crisis
oncogene (oncoLBC or AKAP13). This gene functions as a guanine nucleotide
exchange factor for the Rho proteins (Zheng et al. (1995)) and as a protein
kinase A
anchoring protein. Carr et al. (1991). AKAP13 contains a region that is
homologous to an a-helical domain that is known to interact with lamin B.
Foisner
et al. (1991). This association could lead to lamin B activation via protein
kinase A,
consequently increasing mitotic activity. Both RhoB and lamin B are
farnesylated
and are candidate targets of FTIs. AKAP 13 is also a proto-oncogene, because
loss
of its 3-prime end causes cellular transformation. Sterpetti et al. (1999).
While it
was originally identified from a patient with chronic myeloid leukemia, its
expression has not been documented in AML.
The identification of several genes that are potentially involved in FTI
biology
(ARHH, AKAP13, IL3RA) support the idea that the interaction of multiple
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
37
pathways can affect how FTIs function in this population of AML patients
(Figure
7). These genes interact with several farnesylated proteins including ras,
rho, and
potentially lamin B. Rho proteins are potentially important antitumorigenic
targets
for FTIs. Sahai et al. (2002); and Lancet et al. (2003). RhoB, RhoA, and RhoC
have been found to be over-expressed in multiple cancer types. Sahai et al.
(2002).
In addition, RhoH (ARHH) is frequently re-arranged in tumors of myeloid
origin,
and this may lead to its over-expression. Pasqualucci et al. (2001). While
most of
these Rho proteins are geranygeranylated, they interact closely with each
other and
the farnesylated ras, RhoE, and RhoB small GTPases. Sahai et al. (2002); and
Li et
al. (2002). Furthermore, it has been shown that RhoH, RhoB, and RhoE can act
in
an antagonistic fashion to the transforming abilities of RhoA and RhoG. Li et
al.
(2002). The activity of RhoA, and possibly other related small GTPases, is
increased by the guanine nucleotide exchange factor lymphoid blast crisis
oncogene
(AKAP13). Sterpetti et al. (1999); and Toksoz et al. (1994). In addition,
AKAP13
may increase mitotic activity by activating lamin B via protein kinase A.
Foisner et
al. (1991). It is also well known that the IL3 receptor activates ras
pathways. Testa
et al. (2004). Therefore, as indicated in Figure 7, the increased activity of
IL3RA
and AKAP13, and the decrease in RhoH expression could lead to an increased
cellular profile of transformation. This might allow for the leukemic blast
cell to
overcome the anti-tumorigenic effects of FTIs through compensatory pathways.
In
contrast, when IL3RA and AKAP13 are under-expressed and there is an increase
in
RhoH activity, FTIs may be more effective in blocking these pathways.
Finally, we demonstrated that over-expression of AKAP 13 (both oncoLBC and
protoLBC variants) increased the IC50 of the HL60 AML cell line by
approximately
20-fold. This indicates that over-expression of AKAP 13 is a relevant marker
of
resistance and that it may also be a useful alternative drug target for
patients who are
resistant to tipifarnib.
Overall, our findings allow development of a gene expression based diagnostic
assay to identify patients likely respond to tipifarnib. This information
could be
used to better direct treatment to the appropriate patient population. Using
survival
as the gold standard, the gene signature predicts a level of response to
therapy that
cannot be predicted by using conventional clinical response criteria.
Alternatively,
this raises the question of whether the gene signature for predicting response
to FTI
treatment also has prognostic value irrespective of FTI therapy. We thus
evaluated a
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
38
prognostic signature previously identified in newly diagnosed AML patients who
were treated with conventional chemotherapy. Bullinger et al. (2004). While
this
signature showed utility in the current patient population our 3-gene
signature
further stratified these poor and good prognostic groups showing that it is a
predictor
of response to FTIs.
Example 3
Analysis of an AML prognostic gene signature
The 3-gene signature can predict prognosis irrespective of the type of drug
treatment. To determine this, we first evaluated a gene-expression signature
recently
identified in newly diagnosed AML patients who were treated with conventional
chemotherapy. Bullinger et al. (2004). This signature was defined using a cDNA
array and therefore we first matched these genes with the probes present on
the
Affymetrix gene chip. Of the 133 predictive genes identified by Bullinger et
al., 167
probe sets (corresponding to 103 unique genes) were matched to the Affymetrix
U133A chip. The 3 genes identified in our present analysis are not present in
the
Bullinger et al. 133 gene list. SEQ ID NOs: shown in Table 4. Two main groups
of
patients were defined by hierarchical clustering using these 167 probe sets
(Fig 8A).
Kaplan-Meier analysis showed a clear stratification of these clusters into
patients
with good and poor prognosis (Fig 8B, p = 0.000003). Our data therefore show
that
a subset of the 133-gene prognostic signature identified by Bullinger et al.
(2004)
can also be used in a relapsed and refractory cohort of patients.
Consequently, this
indicates that the prognostic gene profile is surprisingly robust across
different
microarray platforms, and different classes of AML.
Neither of the clusters defined by the prognostic gene signature had
significantly
more responders. However, when the tipifarnib 3-gene signature was applied to
the
good and poor prognostic groups, patients who responded to tipifarnib were
further
stratified from both prognostic groups (Fig 8C). Therefore, the 3-gene
signature has
independent utility to the prognostic signature and that it is specific for
FTI
treatment in this population of patients.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
39
Example 4
Clinical Evaluation and Response Definitions
The current study was an open label, multicenter, non-comparative Phase 2
study
investigating the efficacy and safety of farnesyl transferase inhibition with
tipifarnib
administered as a single agent, at a starting oral dose of 600 mg b.i.d. for
the first 21
days of each 28 day cycle in AML. Subjects were enrolled into two cohorts,
those
with relapsed AML and those with refractory AML. A total of 252 patients (135
relapsed and 117 refractory) were treated. For the purposes of the gene
expression
profiling response to tipifarnib was defined as patients who had an objective
response (CR, CRp, or PR) (as described above), or patients who demonstrated
confirmed stable disease, or a hematological response (decrease of >50% of
leukemic blast cells) as determined by either central review or by the
clinical site at
any time during follow up.
Sample Collection and Microarray Processing
All samples were obtained from patients who had consented to the described
processing and analyses. Bone marrow samples were collected from patients
before
treatment with tipifarnib, diluted with PBS and centrifuged with Ficoll-
diatrizoate
(1.077g/ml). White blood cells were washed twice with PBS, resuspended in FBS
with 10% DMSO and immediately stored at -80 C. Cells were thawed and total
RNA was extracted from cell samples using the RNeasy Kit (Qiagen, Valencia,
CA).
RNA quality was checked using the Agilent Bioanalyzer. Synthesis of cDNA and
eRNA were performed according to Affymetrix (Santa Clara, CA) protocols. Two
rounds of linear amplification were performed because the RNA yield for
several
samples was too low to obtain enough labeled cRNA for chip hybridization using
one round of amplification.
For hybridization, 11 g of cRNA were fragmented randomly by incubation at
94 C for 35 min in 40 mM Tris-acetate, pH 8.1, 100 mM potassium acetate, and
30
mM magnesium acetate. Fragmented cRNA was hybridized to U133A arrays at
45 C for 16 h in a rotisserie oven set at 60 rpm. Following hybridization,
arrays
were washed (with 6x SSPE and 0.5x SSPE containing Triton X-100 (0.005%)), and
stained with streptavidin-phycoerythrin (SAPE; Molecular Probes, Eugene, OR).
Quantification of bound labeled probe was conducted using the Agilent G2500A
GeneArray scanner (Agilent Technologies, Palo Alto, CA).
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
The total fluorescence intensity for each array was scaled to the uniform
value of
600. Chip performance was quantitated by calculating a signal to noise ratio
(raw
average signal/noise). Chips were removed from further analysis if their
signal-to-
noise ratio was less than 5. Genes were only included in further analysis if
they
5 were called "present" in at least 10% of the chips. Approximately 12,000
Affymetrix probe sets remained following this cut-off. Gene expression data
quality
was further controlled by identifying outliers based on principal components
analysis and by analyzing the normal distributions of the gene intensities
(Partek Pro
V5.1).
10 Statistical Analysis
Unsupervised hierarchical clustering and clustering was performed in Omniviz.
Kaplan-Meier analysis was performed using S-Plus.
Example 5
A prognostic signature identified in de novo AML has utility in relapsed and
15 refractory_patients
Two papers were recently published describing gene-expression profiling of
newly diagnosed adult AML patients and its use in predicting clinical outcome.
Bullinger et al. (2004); and Valk et al. (2004). We have profiled 58 patients
with
relapsed and refractory AML using the Affymetrix U133A gene chip. Of the 133
20 predictive genes identified by Bullinger et al. 167 probe sets
(corresponding to 103
unique genes) were identified on the U133A chip (Fig 9). Bullinger et al.
(2004).
The 167 probe sets are listed in the Sequence Listing Table and designated the
SEQ
ID NOs: shown in Table 4.
Two main groups of patients were defined by hierarchical clustering using
these
25 167 probe sets (Fig l OA). Kaplan-Meier analysis showed a clear
stratification of
these clusters into patients with good and poor prognosis (Fig. l OB, p =
0.0000219).
Our data therefore shows that a 103 gene subset of the 133-gene prognostic
signature identified by Bullinger et al. (2004) can also be used in a relapsed
and
refractory cohort of patients. Table 4. Consequently, this indicates that the
30 prognostic gene profile is surprisingly robust across different microarray
platforms,
different classes of AML, and for different treatment algorithms.
Table 4
SEQ ID NO clusterl mean J cluster2 mean ratio good rognostic group
44 2.101613636 0.980102944 2.144278466 u
51 1.574545244 0.899302214 1.750852182 u
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
41
52 2.459568465 1.161727897 2.117163987 up
55 1.752902097 0.968494897 1.809923937 up
56 2.395730325 0.72630844 3.298502667 up
68 1.319861385 1.36567127 0.96645614 down
76 1.269874887 1.430330507. 0.8878192 down
90 0.986377684 1.432950683 0.688354244 down
91 1.177005842 1.428309412 0.824055231 down
93 1.095239589 0.99028255 1.105986962 up
104 1.798024918 0.806182842 2.230294202 up
108 5.041411016 0.60238936 8.369024013 up
111 1.159807609 1.639962342 0.707216001 down
114 1.300310388 1.324114868 0.982022345 down
115 1.342190037 2.306688065 0.581868896 down
116 1.320735142 1.367135334 0.966060279 down
118 3.891659096 1.14773151 3.390739962 up
119 1.519690498 1.219697115 1.245957278 up
120 1.328167684 1.250925424 1.061748094 up
124 4.052024058 0.844109372 4.800354307 up
128 2.091653255 0.964077201 2.169591038 up
129 1.991638259 1.045332875 1.905267027 up
130 2.231280889 0.889145566 2.509466362 up
131 1.815199475 0.992602441 1.828727596 up
142 1.3198052 1.283492456 1.028292136 up
143 2.222738653 0.801693589 2.772553857 up
144 1.591280353 1.037860182 1.533231913 up
147 1.050088807 1.267434876 0.828515 down
148 1.07046517 1.621321185 0.660242511 down
150 1.768390061 0.83169323 2.126252803 up
153 1.253048826 1.17309932 1.068152376 up
157 1.24240959 1.057597528 1.174747063 up
158 1.039468561 1.565715718 0.663893546 down
159 0.897290104 2.462204436 0.364425509 down
160 2.042794407 1.039120916 1.965887103 up
161 2.935364557 1.109613717 2.645393178 up
167 0.771931017 1.585630652 0.486829021 down
174 1.206470925 1.279664724 0.942802363 down
200 1.099482264 1.491611294 0.737110445 down
207 4.901329448 0.871582063 5.623485909 up
208 2.346190152 0.930016604 2.522740069 up
211 0.774893647 2.117870914 0.365883323 down
212 8.626480573 0.518118436 16.64963061 up
220 2.002466652 2.360710711 0.84824737 down
221 1.031573096 1.410790966 0.731201943 down
222 1.236611762 1.579356606 0.782984512 down
224 4.252841813 1.038446727 4.095387566 up
227 2.330630017 0.740455045 3.147564506 u
229 1.271065192 1.275443314 0.996567372 down
230 4.079291876 0.770236845 5.296152606 up
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
42
231 1.613398862 1.053542946 1.531403032 up
234 2.462533188 0.694618451 3.54515948 up
235 1.736866874 1.312450625 1.323376926 up
236 0.973964566 1.442647881 0.675122862 down
237 2.146894817 1.117648799 1.920902898 up
238 1.121767102 1.496868758 0.749409122 down
241 1.09122088 1.120796083 0.973612325 down
243 1.563910635 1.102945516 1.417940063 u
244 1.473673649 1.474149259 0.999677367 down
245 1.148794849 1.399237842 0.821014709 down
246 1.350310245 1.327639719 1.017075812 u~
249 1.44906906 1.424150806 1.017496921 up
256 1.790625889 1.255091645 1.426689354 up
260 1.423005429 1.45961841 0.97491606 down
262 1.09603256 1.181485007 0.927673693 down
263 1.125580943 1.178287994 0.955268108 down
264 0.891946685 1.753005455 0.508809988 down
265 1.2942793 2.776461422 0.466161456 down
277 1.102290438 1.391074208 0.792402326 down
278 1.186998217 2.302906551 0.515434817 down
281 0.79821098 1.919103099 0.415929181 down
288 1.236023874 1.187331666 1.041009778 up
301 1.087313678 1.212772631 0.896551959 down
303 1.820970134 1.219911387 1.492706891 up
306 0.893432913 1.529980396 0.583950563 down
310 1.076214323 1.702252048 0.632229713 down
314 1.407137316 1.229815501 1.144185705 up
315 2.943883289 0.906317573 3.248180746 up
316 1.666394606 0.87032571 1.914679281 up
340 1.499937462 0.921725287 1.627315083 up
346 1.035229803 1.508152041 0.686422705 down
347 1.317050838 1.350558887 0.975189495 down
348 1.481608791 1.161107338 1.276030856 up
351 1.177285398 1.293627618 0.910065139 down
352 1.328716012 1.354505535 0.980960194 down
353 1.245636653 1.388354047 0.897203891 down
354 1.057630522 1.407147783 0.751612968 down
355 1.394737541 1.22633541 1.137321429 up
356 1.194410314 1.007530697 1.185482803 up
359 1.653993358 1.201194991 1.37695659 up
361 1.318561503 1.233384342 1.069059707 up
364 1.045812959 1.463795973 0.714452682 down
365 1.721871636 1.213128563 1.419364517 up
366 1.437978693 1.211203408 1.18723138 up
369 1.480402866 1.289306967 1.148215983 up
373 1.750186104 1.199777316 1.458759122 up
379 1.987020453 2.331667877 0.852188458 down
383 1.419274906 1.587359923 0.894110331 down
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
43
385 1.412275401 1.410313065 1.001391419 up
387 1.339345508 1.331753924 1.005700441 up
395 1.747595879 1.473302004 1.186176272 up
401 1.237925661 1.240617377 0.997830341 down
404 1.176676928 1.14697739 1.025893743 up
407 1.256304659 1.357899094 0.925182633 down
415 2.013680769 0.801461983 2.512509405 up
418 1.405 740421 1.158990553 1.212900673 up
423 1.346444485 1.136343796 1.184891835 up
426 9.592759013 1.560596128 6.146855576 up
430 1.768393033 1.23257422 1.434715252 up
437 1.40453464 1.860231401 0.755032218 down
438 1.673447728 3.930927905 0.425713157 down
445 1.283792446 1.501667999 0.85491097 down
446 1.214492279 1.091382628 1.11280155 up
448 1.561740275 1.228333636 1.271430033 up
450 1.734251715 1.844252606 0.940354759 down
452 1.408580649 1.582858413 0.889896807 down
453 1.1383915 1.741970797 0.653507798 down
454 1.079716972 1.450127515 0.744566917 down
457 1.783529874 1.314108612 1.357216487 up
460 1.136773942 1.003606776 1.132688588 up
462 1.220645614 1.113413039 1.096309789 up
466 1.183017496 1.211183566 0.976745003 down
467 1.113306085 1.0674171.58 1.042990621 up
469 1.211346488 1.1871952 1.020343148 up
484 0.998901987 1.654269584 0.60383265 down
485 1.133736575 1.914231031 0.592267368 down
489 1.412382921 1.384849519 1.019881873 up
493 3.988440713 0.965026485 4.132985753 up
495 1.187128423 1.462785203 0.81155348 down
498 3.691405839 0.765303503 4.823453473 up
499 2.216130138 0.848821568 2.610831558 up
507 2.199861346 0.773086467 2.845556663 up
520 1.67633492 1.194585675 1.403277265 up
524 7.743976211 0.841225224 9.205592025 up
529 2.591731743 1.08342825 2.392158173 up
530 1.273363236 1.097292728 1.160459013 up
535 1.095 8 79149 1.302490014 0.8413724 down
546 0.987046127 2.124625445 0.464574181 down
550 2.103990301 0.929475745 2.263631205 up
555 1.439273076 1.211738853 1.187774967 up
557 1.121983318 1.251788385 0.896304305 down
559 1.197910138 1.34693432 0.889360468 down
560 1.144102989 1.261720239 0.906780246 down
565 1.078544278 1.162115183 0.928087244 down
568 1.46401688 0.960152377 1.524775561 up
569 1.262437604 1.105379077 1.142085669 u
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
44
585 1.349621283 1.10729713 1.218842935 up
590 1.203037349 1.068047506 1.126389361 up
609 1.395352279 1.172354292 1.190213819 up
624 1.131210862 1.399636303 0.80821772 down
630 0.859929427 1.37477642 0.625504929 down
637 0.72293765 2.343345902 0.308506588 down
641 1.340847351 1.601484153 0.837252962 down
656 1.259818484 1.213892186 1.037833918 up
658 0.60640428 2.092078796 0.289857285 down
659 1.099549227 1.166865002 0.942310572 down
665 1.615399946 1.733324276 0.931966378 down
684 0.734401434 2.496195783 0.294208267 down
685 2.158835355 0.765517273 2.82010012 up
686 0.957971846 1.386779118 0.690789061 down
687 0.982131268 1.162818279 0.844612856 down
690 2.421692274 0.791473723 3.059725425 up
691 1.268020946 0.973474927 1.302571757 up
692 1.054213523 1.147045848 0.91906834 down
693 1.251020003 1.271391137 0.983977288 down
694 1.122443987 1.153661375 0.972940597 down
696 1.301034813 1.029185966 1.264139675 up
The prognostic signature is independent of a 3-gene signature that predicts
response
to tipifamib.
We identified a 3-gene signature (AHR, AKAP13, MINA53) that predicts
response to tipifarnib in relapsed and refractory AML patients. These genes
can
stratify patients into good and poor prognostic outcome groups (Fig 11 B, p =
0.002).
The question arises as to whether this gene signature is predicting response
to FTI
treatment or merely identifying patients who have a generally good prognosis.
When the 3-gene signature was applied to the good and poor prognostic groups,
responders were further stratified from the prognostic groups (Fig 11C, p =
0.000003). Following the application of both gene signatures there is clear
stratification of a group of patients that do not respond to tipifamib and
have a poor
prognosis irrespective of treatment type (Fig 11 D, p = 0.0000005). Therefore,
the 3-
gene signature seems to be independent of the prognostic signature that has
been
identified and it is specific for FTI treatment in this population of
patients. As a
result we suggest that the prognostic signature maybe used in conjunction with
drug-
specific signatures (such as the tipifarnib predictive profile) to better
manage patient
therapy.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
Example 6
Identification of genes that are differentially expressed
between responders and non-responders (not including stable disease patients)
Four patients were removed from the analysis since they were classified as
5 having stable disease and these patients cannot be clearly defined as either
responders or non-responders. Inclusion of stable disease patients may bias
the
analysis for selecting genes associated with prognosis irrespective of drug
treatment.
This resulted in comparing 10 responders with 44 non-responders. Selected
genes
were required to show a specificity of 40% and a minimum mean fold-change of
2Ø
10 These criteria were chosen to identify genes that could predict response to
tipifarnib
with the highest level of sensitivity possible. From 11,723 genes, a total of
8 genes
were identified that could stratify responders and non-responders (Table 5)
and that
gave significant P values in a t-test (P <0.05). The genes included those
involved in
signal transduction, apoptosis, cell proliferation, oncogenesis, and
potentially, FTI
15 biology. AKAP 13 is the most robust marker
We next aimed at identifying a minimal set of genes that would provide the
best
diagnostic accuracy from the 8 selected genes. Classifiers were built with an
increasing number of genes based on the AUC values from receiver operator
characteristic analysis, and the error rate of these classifiers was
calculated using
20 LOOCV while keeping the sensitivity of predicting response at 100% (Fig.
12a).
The AKAP 13 gene could predict response with the lowest error rate of less
than
40% (Fig. 12a). The error rate increased to more than 50% when more than 2
genes
were used in the classifier. For the AKAP 13 the LOOCV demonstrated a NPV of
93% and a PPV of 31%, with an overall diagnostic accuracy of 63% and positive
25 likelihood ratio of 2.0 (Fig. 12b). The expression value for AKAP 13 in
each patient
is shown in Fig. 12c. Therefore, for the group of patients with low expression
of
AKAP 13, the response rate to tipifarnib was 31 %(8/26) compared to 18%
(10/54)
in the current patient population. Using the AKAP13 gene, Kaplan-Meier
analysis
showed a significant difference in survival between the predicted responder
group
30 and the non-responder group (Fig. 12d).
Table 5. List of Top 8 Genes that Predict Response to Tipifarnib
SEQ ID Symbol AUC fold P value Functional Description
NO: change
309 AKAP13 0.830 0.491 0.00007 intracellular signaling, oncogenesis
151 AHR 0.807 0.446 0.00019 signal transduction, a o tosis
222 SCAP2 0.777 0.431 0.00007 signal transduction
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
46
496 NPTX2 0.738 0.115 0.02934 cell adhesion
451 BAT1 0.725 0.458 0.00097 cellular biosynthesis
272 IL3RA 0.705 0.375 0.00226 receptor signalling
411 IDS 0.645 0.395 0.00069 metabolism
280 P2RY14 0.627 0.369 0.00145 signal transduction
AUC = area under the curve from receiver operator characteristic analysis.
This is
an indication of the overall diagnostic accuracy.
Example 7
Gene Expression Profiling Predictive of Tipifarnib (ZARNESTRA , RI 15777)
Response in Patients With Newly Diagnosed Acute Myeloid Leukemia
Tipifarnib (ZARNESTRA , R115777), has demonstrated clinical response in
patients with hematological disease. While the inhibition of protein
farnesylation is
the primary mechanism of action (MOA), the level of farnesyl inhibition is not
a
reliable pharmacodynamic marker of response, nor is it clear what genetic
markers
can be employed to predict response. This prospectively designed study was
conducted to identify potential genetic markers and expression signatures that
may
be surrogate predictors of response for tipifarnib in patients with acute
myeloid
leukemia (AML). Bone marrow samples were collected and gene expression
profiles analyzed from a single arm phase 2 clinical study of tipifarnib in
poor-risk
patients with newly diagnosed AML. Lancet et al. (2004). In total, 79 samples
were
profiled before (n = 25), during (n = 30), and after (n = 24) tipifarnib
treatment.
Bone marrow samples were analyzed using the Affymetrix U133A GeneChip
array. Global gene expression signatures revealed that tipifarnib treatment
resulted
in gene expression changes that were maintained for up to 120 days following
treatment termination. Pretreatment vs post-treatment samples identified
approximately 500 genes that had significant changes (False Discovery Rate
[FDR]
<0.005) in gene expression following farnesyl transferase inhibition,
including
several genes associated with famesylation (eg K-ras, FNTA). Many of the
modulated genes were identified as those significantly involved in protein
biosynthesis, DNA replication, intracellular signaling, and cell cycle
pathways, thus,
reflective of inhibition of cellular proliferation. A subset of 27 genes
(including
genes associated with signal transduction and cell cycle) was also identified
as being
differentially modulated between responders and non-responders (P < 0.01).
Gene
expression signatures previously identified from a phase 2 clinical trial in
relapsed
and refractory AML were also tested in pretreatment samples to examine their
ability to predict response. Raponi et al. (2004). A combination of 6 genes
was
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
47
found to have significant predictive accuracy in this independent set of
samples (P =
0.05). The genes identified from these studies might be used as surrogate
biomarkers of tipifarnib activity.
Patient Samples
In a phase 2 study, patients with newly diagnosed AML received
ZARNESTRA , 600 mg bid21dQ4wks. Bone marrow samples were obtained
before, during, and after treatment with ZARNESTRA . Mononuclear cells
isolated by Ficoll-Hypaque density centrifugation and viably frozen.
Microarray Analysis
Message RNA was amplified from patient blast cells and hybridized to the
Affymetrix U133A chip, which can probe for approximately 22,000 genes (Fig.
13).
Chip data were pre-filtered to remove poor quality data and genes that were
not
expressed in at least 10% of the patient samples. In addition genes that did
not vary
across the dataset were removed (CV < 40%). Approximately, 8000 genes remained
for further analysis. A total of 79 chips passed quality control measures and
also
had associated clinical response data.
Statistical Analysis
Analysis of variance (ANOVA) and t-tests were used to investigate the effect
of
drug treatment and time and their interactions for each gene. Multiple
hypotheses
testing was controlled by applying the false discovery rate (FDR) algorithm.
All
statistical analyses were performed in S-Plus 6.1 (Insightful Corporation).
Principal
components analysis was performed in Partek Pro. Hierarchical clustering was
performed using a correlation metric and complete linkage (OmniViz ProTM,
OmniViz, Maynard, MA). Pathway analysis was performed using Gene Ontology
functional classifications. Table 6 shows the results of the analysis.
Table 6. Functional Gene Classes significantly modulated by tipifarnib
GO.ID GO.Class p.value
6886 Intracellular protein transport 5.75E-05
6951 Heatshock response 8.32E-05
6913 Nucleocytoplasmic transport 1.18E-04
6207 De novo pyrimidine base biosynthesis 1.58E-04
6809 Nitric oxide biosynthesis 1.58E-04
6376 mRNA splice site selection 2.05E-04
245 Spliceosome assembly 2.05E-04
6259 DNA metabolism 4.86E-04
6371 mRNA splicing 4.86E-04
6607 NLS-bearing substrate-nucleus import 5.64E-04
15031 Protein transport 6.05E-04
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
48
6338 Chromatin remodeling 9.87E-04
6397 mRNA processing 0.003201
7050 Cell cycle arrest 0.003685
8380 RNA splicing 0.003992
6512 Ubi uitin cycle 0.004639
6396 RNA processing 0.005096
398 Nuclear mRNA splicing, via spliceosome 0.007120
6916 Anti-a o tosis 0.007395
6118 Electron transport 0.010004
7049 Cell cycle 0.019631
6457 Protein folding 0.021817
7264 Small GTPase mediated signal 0.025281
transduction
8285 Negative regulation of cell proliferation 0.026482
6366 Transcription from Poll promoter 0.048613
Approximately 8000 genes were used for global unsupervised clustering.
Pretreatment 'samples clustered distally from during- and after-treatment
samples.
After-treatment samples included those that were from patients up to 120 days
following treatment termination. Figure 14 shows that AML samples maintain
FTI-mediated global gene expression changes following termination of
tipifarnib
treatment.
502 genes were found to be differentially expressed after treatment with
tipifarnib (p<0.005). These genes are listed in Tables 7A-7C and in more
detail in
Table 9.
Table 7A
SEQ ID NO psid SEQ ID NO psid SEQ ID NO psld 1SEQ ID NO pgld SEQ ID NO psld
4 200002 47 201104 97 202029 165 203133 215 204158
5 200008 48 201118 98 202077 166 203138 216 204168
6 200010 49 201134 99 202078 168 203140 217 204170
7 200013 50 201163 100 202090 169 203142 219 204294
8 200017 53 201227 101 202110 170 203211 220 204351
9 200018 54 201244 102 202114 171 203213 223 204411
200025 57 201273 103 202141 172 203255 225 204528
11 200026 58 201277 105 202163 173 203262 226 204640
12 200041 59 201300 106 202187 175 203316 228 204652
14 200056 60 201305 107 202197 176 203332 232 204766
200061 61 201317 109 202231 177 203362 233 204767
16 200073 62 201324 110 202233 178 203371 239 204905
17 200086 63 201352 112 202275 179 203385 240 204923
18 200087 64 201381 113 202279 180 203396 247 205033
19 200091 65 201393 117 202298 181 203437 248 205087
200634 66 201403 121 202324 182 203460 250 205133
21 200640 67 201429 122 202325 183 203484 251 205176
22 200643 69 201455 123 202349 184 203514 252 205213
24 200718 70 201472 125 202423 185 203528 253 205270
ZZOL6Z L95 bSL~2 505 LZOZ6Z 6~b 6L960Z ~8~ 6L980Z ~ZE
LL696Z 855 LZL~LZ 'VOS LOOZI.Z 9~t' Z9960Z Z8~ ZL980Z ZZ~
~~896Z 999 OOLE1- Z E05 98646Z 5~1 8Z960Z 1-8~ L9980Z 1-3~
t,8~91- Z t755 L89~6Z 305 9L61-6Z 17~17 589603 08~ 9S9803 03~
8b~92 ~S5 669E 2 605 8E6~2 ~~t, OZ960Z 8L~ ~V980Z 66~
LOZ96Z Z5S L99~6Z 009 Z~666Z Z~t' L0560Z LLC ~E980Z 8L~
1.699~Z 8bS Z09~6Z L6t, 6Z6~2 6Et7 Z6b60Z 9LC 6Z980Z LL~
~6756Z L'V5 V6b~6Z t76t, 85862 6Zb 4Lt,60Z tL~ 86580Z ~L~
66E56Z 5t,9 t.~~~6Z Z6t, LbL62 LZV 66760Z ZL~ 6ti580Z Z6~
08~52 t'ti5 L8Z~LZ G6t, 0~L1.6Z 5Zb 58~60Z LLE OZV80Z LL~
6L~9~Z ~tI5 ~SZ~6Z 0617 5b912 tZV LLfi60Z OL~ 06~80Z 80~
LZZ92 ZtlS 656~6Z 98t, L8b66Z ZZJF 69860Z 89E OLZ80Z LOC
6L694Z L b5 6Z6~6Z Z8t 517tl~ I.Z 6Zt, Z9~60Z L9C L6080Z 50E
Lb696Z 0tl5 bL08l.Z 68b OEv~6Z 0Zt' ~0~60Z C9C bL6LOZ b0E
9Eb92 6C9 Z90~6Z 08tl Z8602 LLt, 96Z60Z Z9~ St8LOZ ZO~
LZ~SIZ 8~5 690E6Z 6LV 69806Z 96t, Z8Z60Z 09C LLLLOZ 00~
LZ694Z L~5 Lti0~LZ BLt, Ob806Z KV 89Z60Z 85E ~LSLOZ 66Z
96096Z 9~5 8ZOE6Z LLt, 98LOLZ ELt, L5Z60Z L5~ 4L5LOZ 86Z
8EO9~Z bES b96Z2 5Lt, 98L06Z Z6b ZL~60Z 09C 899LOZ L6Z
OZ8tl6Z Z~5 LZ6Z2 tLt, 6b902 06V 8E1.60Z 6K 699LOZ 96Z
008tII.Z LE5 L06Z2 ELt' 6E902 60t, 68060Z 5'VE Ob9L0Z 56Z
699V2 8Z5 8L8Zt.Z ZLt, ~~901.Z 80t, 99060Z tltt'~ SE~LOZ t,6Z
L69b6Z LZ5 098Z2 LLt, 86902 90t, ~9060Z EtIE L8ZLOZ ~6Z
661717 6Z 9Z5 ~E8Z6Z OLt, ~95NZ 50t, ZZ060Z Zt'~ 99ZLOZ Z6Z
V6bb~Z 5Z5 L9LZ6Z 89b' ~5b06Z ~0t, 68680Z 6bE 59~LOZ WZ
LL~V6Z ~Z9 0~9UZ 99t, 8bbUZ Z0'V 9L680Z 6CC ~96LOZ 06Z
Z5~V2 ZZ5 E8SZ6Z t9t' b8Z0~Z 00t' 99680Z 8CC LZ4LOZ 68Z
88Zb~Z 6L5 8LSZ2 E9t, ~8Z06Z 66C 99680Z LEE ZLOLOZ L8Z
17931743 81.5 6L9321-917 5~3063 86~ 61.6803 5EE 8S6903 983
plsd ON a1 O3S P!sd ON aI 03s plsd ON ai 03s p!sd ION 01036Plsd ON al 03S
8L alqel
bL890Z 58Z 9ti6bOZ tlZ ~50~OZ ~96 600ZOZ 96 b601.OZ 9t,
89890Z tl8Z 5b6bOZ ELZ bb0~OZ ~9L L8660Z 96 6t060Z 5t,
06L90Z E8Z L90VOZ 0LZ L56ZOZ Z9L 8~660Z t,6 LZ060Z ~t,
~ZL90Z Z8Z 996~OZ 90Z bSBZOZ 95L IZ640Z Z6 66060Z Ztl
869903 6LZ 096~03 503 8178303 551- 068WZ 68 666003 617
SbZ90Z 9LZ 9~6~OZ ~0Z ~tBZOZ t,56 M60Z 88 W600Z 0t,
66Z90Z 5LZ ZZ6~OZ Z0Z vZBZOZ Z51. Z1860Z L8 68600Z 6C
LL6903 tLZ ~68~03 1-03 69LZOZ 6171- 508603 98 6L6003 8~
09L90Z ~LZ Z88EOZ 666 bSLZOZ 9t, L bSL60Z V8 b9600Z L~
99090Z ~LZ LZ8~OZ L6L SELZOZ 5tl 6 8~L60Z E8 6b600Z 9~
L969OZ 0LZ 69L~OZ 966 Z69ZOZ 6tL 96L60Z Z8 V~600Z 5~
6178903 69Z 817L~03 566 ~L9303 0176 51L60Z 68 936003 v~
L089OZ 89Z EtiL~OZ b6L 6b9ZOZ 6~L 6691OZ 08 Z0600Z ~~
1L950Z L9Z bL9~OZ ~66 Zt79ZOZ 8~L 9L960Z 6L 89800Z Z~
bb990Z 99Z L99~OZ Z6l ZZ9ZOZ L~l 599WZ 8L L5800Z L~
L9b90Z 69Z 999~OZ (-66 665ZOZ 9~L 5~9LOZ LL C9800Z 0~
~0tl50Z 65Z 6Z9EOZ 06L Z85ZOZ 5EL OZ960Z 5L 9ti800Z 6Z
L9~SOZ 85Z u9~OZ 686 bbSZOZ ~U L6540Z t7L V~800Z 8Z
6~~50Z L5Z 069~OZ 886 cO9ZOZ Z~L 88960Z CL 10800Z LZ
SE~SOZ 55Z 685EOZ L8~ ZtbZOZ LZL 895LOZ ZL 08LOOZ 9Z
~Z~90Z b5Z l~S~OZ 986 ZEbZOZ 9ZL ~BbLOZ lL ZLLOOZ 5Z
6t
OO19b0/SOOZSII/13d ObZ990/900Z OAd
0~-SO-LOOZ 99068930 FIO
tIEKZZ 699 9L58~Z 81.9
Z~b6ZZ 899 E9986Z L1.9
ESZI.ZZ L99 E1758LZ 969
~tiL 4ZZ 999 Z8782 5l9
ZV60ZZ V99 L9b84Z b19
t,980ZZ ~99 Ltlb86Z E~9
99LOZZ Z99 L6E82 Z19
6bLOZZ 199 56~82 1~69
bOibOZZ L99 EL~86Z Ol-9
6660ZZ 999 L9~81.Z 809
951OZZ t,59 O9~82 L09
0900ZZ E59 6~~82 909
Z500ZZ Z59 vE~8LZ 909
EZOOZZ L59 88Z8t.Z t709
LOOOZZ 099 ~8Z86Z ~09
90664Z 6t,9 08Z86Z Z09
66862 8t,9 9LZ8lZ 609
91861Z Lt,9 t~LZ82 009
59L66Z 9t,9 69Z86Z 669
65L62 9b9 95Z8bZ 869
9b561Z ~t,9 ~tiZ86Z L69
L0966Z ~V9 ~6Z86Z 969
90966Z Zt,9 886W 569
~-8o!q-03-U-X=I=Id 90L LtE66Z 0t,9 5L1.86Z t,65
8W 9~6L6W/'d~-~JSIWfIH-X=I=Ib' V0L E6Z62 6C9 L6682 ~65
W 65EOOX/L0O'dSH-X=IJ'd EOL 98Z66Z 8~9 961.82 Z65
~ 6S~OOX/LOOt/SH-X=I=Ib' Z0L ~9462 9C9 ~0686Z L69
bb969 WL 06664Z 5E9 85086Z 685
Z695S OOL 90662 17~9 6E086Z 885
60ZL~ S69 99062 ~~9 ~0082 L85
08ZZZZ 689 0~066Z 6E9 LL8L2 98S
~OZZZZ 889 9t,682 6Z9 998L2 1785
6996ZZ LL9 LE684Z 8Z9 098L6Z ~85
OZ92Z 9L9 9E6W LZ9 Et,8L2 L85
6696ZZ 5L9 EL882 9Z9 8E8L4Z 085
E692Z VL9 0~882 SZ9 ~EBL2 6L9
LL52Z ~L9 6L984Z CZ9 SZ8L2 8L5
8Z92Z ZL9 09982 ZZ9 Z68L2 LL9
6092Z 6L9 Ev98tZ LZ9 t,08L29L9
plSd :ON QI 03S p!Sd :ON dl 03S plSd :ON 41 b3S
OL alqel
ELLL<-Z 5LS bZZKZ L65 499Z2 65t, ~EZ06Z L6E 50680Z b~E
69LL~Z tLS Eb6b6Z 965 6b5Z2 8517 08606Z 96C 170680Z ~E~
VSLL6Z ELS 6Z6ti6Z S65 45bZ2 95t, L~106Z V6~ 00680Z ZEE
05LL6Z ZL5 bZ6t,6Z bl5 9ZtZ6Z S5t, L6002 ~6~ b6880Z LEE
6t,LL2 6L5 L60ti6Z ~65 46~Z2 6tt, E6006Z Z6~ 4E880Z OE E
~6bLI.Z 0L5 6S0t,6Z Zl-5 L8ZZ6Z Lbtl L0660Z 66~ 61880Z 6Z~
9E~LI.Z L99 ~00KZ 6l5 LLZZ6Z tltltl 89860Z 06C 80880Z 8ZE
6ZEL2 999 6b6EtZ 0L5 69ZZ6Z E~tl 8E860Z 688 V9L80Z LZC
6tZLLZ V95 t-66E6Z 609 8tZZ6Z Zt7t7 LZ860Z 888 9VL80Z 9ZE
ZZ6LI.Z ~95 L98~6Z 805 40ZZ2 L tl'V t~L60Z 988 SbL80Z 5Z~
906LLZ Z99 1-68uZ 909 6822 01717 L8960Z tl8E 56980Z tlZE
OS
OO19b0/SOOZSII/13d ObZ990/900Z OAd
0~-SO-LOOZ 99068930 FIO
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
51
620 218605 670 221505
Included in this list were many genes involved in FTI biology including AKT1,
CENPF, KRAS, RAF1, STATs and farnesyltransferase. Several gene functional
categories were found to be significantly enriched in this gene list (Table
8).
Table 8.
Genes differentially expressed between responders and non-responders after
treatment with ti ifarnib
SEQ Probe set ID Gene function (GO) RE pre v PD pre v PD post
ID NO: RE post
13 200044 RNA splicing U No difference
23 200661 Protein U No difference
85 201804 Protein folding Up No difference
123 202349 Protein folding Up No difference
137 202622 Nuclear Up No difference
149 202761 Cytoskeleton Up No difference
198 203845 transcription Down No difference
210 204067 Electron transport U No difference
218 204215 Membrane Down No difference
257 205339 Cell proliferation Down No difference
266 205644 RNA splicing Down No difference
268 205807 Bone Down No difference
mineralization
290 207163 Signal transduction Up No difference
329 208819 Signal transduction Up No difference
336 208927 RNA U No difference
362 209295 Signal transduction Down No difference
428 211762 Cell cycle Down No difference
470 212833 Transport Down Down (more in PD)
521 214298 Cell cycle U U(more in PC)
549 215764 Transport Up Up (more in PD)
551 215905 RNA splicing Up No difference
598 218256 Transport Down No difference
610 218373 A o tosis Up No difference
619 218603 Cell cycle Down No difference
660 220671 transcription No down
difference
698 44696 Signal transduction No Up
difference
699 49485 Transcription U No difference
We have previously identified 8 genes that were predictive of resistance to
tipifarnib in relapsed and refractory AML. The most predictive gene in that
dataset
was AKAP-13 (AUC = 0.83).
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
52
The predictive value of these genes was tested in the current set of newly
diagnosed AML samples (CTEP20). Samples from patients with complete response
or progressive disease were used. The results are shown in Figure 15.
A predictive classifier was built from the top 6 genes since these gave less
than a
50% error rate in the training set. This 6 gene classifier is shown in Table 9
and its
ability to stratify newly diagnosed AML is shown in Figure 16.
Table 9
SEQ ID NO: PSID Symbol NTI7AUC CTEP20AUC
309 208325 AKAP13 0.830 0.464
151 202820 AHR 0.807 0.750
222 204362 SCAP2 0.777 0.893
496 213479 NPTX2 0.738 0.766
451 212384 BAT1 0.725 0.288
272 206148 IL3RA 0.705 0.714
411 210666 IDS 0.645 0.446
280 206637 P2RY14 0.627 0.589
Conclusions
= 27% of the clinical bone marrow samples could be used indicating
optimization
of sample collection is required.
= Global gene expression signatures of AML cells indicated FTI-treatment
resulted in stable gene expression changes following treatment termination.
= Approximately 500 genes (p < 0.005) were affected by tipifarnib reflecting
the
multiple pathways that are targeted by FTIs. These included many pathways
previously associated with FTI biology.
= Differential expression of 27 genes was found between responders and non-
responders following treatment with tipifarnib (Table 8). These are candidate
PD
markers.
= A 6-gene classifier identified in relapsed and refractory AML was found to
have predictive value in newly diagnosed AML
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
53
Example 8
Antibodies (Pro hn etic)
An LBC oncogene-derived peptide is synthesized, coupled to keyhole limpet
hemocyanin, and used to immunize rabbits for production of polyclonal
antibodies.
The sera are tested for reactivity against the corresponding peptide with
ELISA, and
the positive batches are affinity-purified. The purified antibody specifically
detects
the peptide that has the epitope in tissue sections. This is verified by
complete
abolishment of the signal if the corresponding peptide is added simultaneously
with
the antibody. In addition to this polyclonal antibody, which works well in
IHC,
monoclonal antibodies able to detect the protein in its natural fold are
produced. To
produce monoclonal antibodies, a purified antigen, produced in mammalian cells
to
ensure natural fold and posttranslational modifications, is generated. The
antigen,
LBC onco protein-IgG constant part fusion protein, is expressed in mouse
myeloma
cells, and the protein is purified using the Fc part as bait. This purified
antigen is
recognized in Western blot by the C-terminal polyclonal antibody. The antigen
is
used to generate mouse monoclonal antibodies against LBC peptides by selecting
out of the positive clones those that produce antibodies that react against
LBC
peptide instead of the IgG constant part. Kits for the clinical identification
of LBC
oncogene can be readily fashioned employing these and similar antibodies. Such
kits would include antibodies directed to LBC peptide identification (and
hence,
LBC oncogene), appropriate indicator reagents (e.g., enzymes, labels, and the
like),
and (optionally) other reagents useful in the clinical application of such a
kit such as
dilution buffers, stabilizers, and other materials typically used in such
assays.
Example 9
Immunohistochemistry (Prophetic)
An affinity-purified polyclonal antibody against the C-terminal peptide of LBC
oncogene is used for the IHC detection and localization of LBC oncogene. Four
m
sections from formalin-fixed and paraffin embedded normal and tumor tissue is
mounted on 3-aminopropyl-triethoxy-silane (APES, Sigma, St. Louis, MO) coated
slides. The sections are deparaffinized and rehydrated in graded
concentrations of
ethanol and treated with methanolic peroxide (0.5% hydrogen peroxide in
absolute
methanol) for 30 minutes at room temperature to block the endogenous
peroxidase
activity. Antigen retrieval is done in a microwave oven twice for 5 minutes
(650W).
An Elite ABC Kit (Vectastain, Vector Laboratories, Burlingame, CA) is used for
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
54
immunoperoxidase staining. The LBC peptide antibody is used at an optimal
dilution of 1:2000. Both the biotinylated second antibody and the peroxidase-
labeled avidin-biotin complex are incubated on the sections for 30 minutes.
The
dilutions are made in PBS (pH 7.2), and all incubations are carried out in a
moist
chamber at room temperature. Between the different staining steps the slides
are
rinsed three times with PBS. The peroxidase staining is visualized with a 3-
amino-
9-ethylcarbazole (Sigma) solution (0.2 mg/ml in 0.05 M acetate buffer
containing
0.03% hydrogen peroxide, pH 5.0) at room temperature for 15 minutes. Finally,
the
sections are lightly counterstained with Mayer's haematoxylin and mounted with
aqueous mounting media (Aquamount, BDH). In control experiments the primary
antibodies are replaced with the IgG fraction of normal rabbit serum or the
primary
antibody was preabsorbed with the LBC peptide. These stainings indicate the
presence of the LBC oncogene in a subset of cells.
Although the foregoing invention has been described in some detail by way of
illustration and example for purposes of clarity of understanding, the
descriptions
and examples should not be construed as limiting the scope of the invention.
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
Table 10
Sequence Listing Description sEQ psid Description Name Accession
ID
NO
1 AKAP13 forward primer NM 007209
2 AKAP13 reverse primer NM 001494
3 AKAP13 probe NM 000975
4 200002 ribosomal protein L35 RPL35 NM 007209
5 200008 GDP dissociation inhibitor 2 NM 001494
6 200010 ribosomal protein L11 NM 000975
7 200013 ribosomal protein L24 RPL24 NM 000986
8 200017 ribosomal protein S27a RPS27A NM 002954
9 200018 ribosomal protein S13 RPS13 NM 001017
10 200025 ribosomal protein L27 RPL27 NM 000988
11 200026 ribosomal protein L34 RPL34 NM 000995
12 200041 HLA-B associated transcri t-1 D6S81 E NM 004640
13 200044 s licin factor, arginineserine-rich 9 SFRS9 NM 003769
14 200056 hi hl similar to integrin alpha-7 FLJ12486 AK022548
15 200061 Similar to ribosomal protein S24 BC000523
16 200073 hnRNP-C like protein M94630
17 200086 cytochrome c oxidase subunit IV AA854966
18 200087 FLJ21323 FLJ21323 AK024976
19 200091 ribosomal protein S25 AA888388
20 200634 profilin 1 PFN1 NM 005022
21 200640 tyrosine 3-monooxygenasetryptophan 5- YWHAZ NM_003406
monooxygenase activation protein, zeta polypeptide
22 200643 high density li o rotein binding protein HDLBP NM 005336
23 200661 protective protein for beta-galactosidase PPGB NM 000308
24 200718 transcri tion elongation factor B SIII , polypep 1-like NM 003197
25 200772 prothymosin, alpha (gene sequence 28) NM 002823
26 200780 guanine nucleotide binding protein (G protein), alpha GNAS1
NM_000516
stimulating activity ol e tide 1
27 200801 actin, beta ACTB NM 001101
28 200834 ribosomal protein S21 RPS21 NM 001024
29 200846 protein phosphatase 1, catalytic subunit, a isoform PPP1 CA NM
002708
30 200853 H2A histone family, member Z H2AFZ NM 002106
31 200857 nuclear receptor co-repressor 1 NCOR1 NM 006311
32 200858 ribosomal protein S8 RPS8 NM 001012
33 200902 15 kDa selenoprotein SEP15 NM 004261
34 200925 cytochrome c oxidase subunit Via polypeptide 1 COX6A1 NM 004373
35 200934 DEK oncogene (DNA binding) DEK NM 003472
36 200949 ribosomal protein S20 RPS20 NM 001023
37 200964 ubi uitin-activatin enzyme El UBE1 NM 003334
38 200971 stress-associated endoplasmic reticulum protein 1 SERP1 NM 014445
39 200981 neuroendocrine secretory protein 55 NESP55 NM 016592
40 200991 KIAA0064 gene product KIAA0064 NM 014748
41 200999 transmembrane protein (63kD), endoplasmic P63 NM_006825
reticulum Golgi intermediate compartment
42 201019 eukaryotic translation initiation factor 1A EIF1A NM 001412
43 201027 KIAA0741 gene product IF2 NM 015904
44 201042 trans lutaminase 2 AL031651
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
56
45 201049 ribosomal protein S18 RPS18 NM 022551
46 201094 ribosomal protein S29 RPS29 NM 001032
47 201104 DJ328E19.C1.1 NM 015383
48 201118 hos ho luconate deh dro enase PGD NM 002631
49 201134 c ochrome c oxidase subunit Vllc COX7C NM 001867
50 201163 insulin-like growth factor binding protein 7 IGFBP7 NM 001553
51 201195 L-t e amino acid trans orter 1 NM 003486
52 201212 protease, cysteine, 1 le umain NM 005606
53 201227 NADH deh dro enase 1 beta subcomplex, 8 NDUFB8 NM 005004
54 201244 v-raf-1 murine leukemia viral oncogene hom 1 RAF1 NM 002880
55 201249 solute carrier family 2 member 1 SLC2A1 NM 006516
56 201250 solute carrier family 2 member 1 SLC2A1 NM 006516
57 201273 signal recognition particle 9kD SRP9 NM 003133
58 201277 heterogeneous nuclear ribonucleoprotein AB HNRPAB NM 004499
59 201300 prion protein (p27-30) PRNP NM 000311
60 201305 acidic protein rich in leucines NM 006401
61 201317 roteasome (prosome, macro ain sub a type 2 PSMA2 NM 002787
62 201324 epithelial membrane protein 1 EMP1 NM 001423
63 201352 YME1 S. cerevisiae)-like 1 YME1 L1 NM 014263
64 201381 calcyclin binding protein NM 014412
65 201393 insulin-like growth factor 2 receptor IGF2R NM 000876
66 201403 microsomal glutathione S-transferase 3 MGST3 NM 004528
67 201429 ribosomal protein L37a RPL37A NM 000998
68 201445 calponin 3, acidic CNN3 NM 001839
69 201455 puromycin sensitive amino e tidase NM 006310
70 201472 von Hippel-Lindau binding protein 1 VBP1 NM 003372
71 201483 su ressor of Ty S. cerevisiae) 4 homolog I NM 003168
72 201568 low molecular mass ubiquinone-binding protein QP-C NM 014402
73 201588 thioredoxin-like, 32kD TXNL NM 004786
74 201597 c tochrome c oxidase subunit Vlla ol e 2 COX7A2 NM 001865
75 201620 membrane-bound transcription factor protease, site 1 MBTPS1 NM
003791
76 201621 neuroblastoma, suppression of tumorigenicity 1 NBL1 NM 005380
77 201635 fra ile X mental retardation, autosomal hom 1 U25165
78 201665 ribosomal protein S17 RPS17 NM 001021
79 201675 A kinase PRKA anchor protein 1 AKAP1 NM 003488
80 201699 proteasome 26S sub ATPase 6 PSMC6 NM 002806
81 201715 KIAA0670 KIAA0670 NM 014977
82 201716 sortin nexin 1 SNX1 NM 003099
83 201738 translation factor suil homolog GC20 NM 005875
84 201754 cytochrome c oxidase subunit Vlc COX6C NM 004374
85 201804 cytoskeleton-associated protein 1 CKAP1 NM 001281
86 201805 protein kinase, AMP-activated, y 1 non-catalytic sub PRKAG1 NM
002733
87 201812 6.2 kd pro LOC54543 NM 019059
88 201818 hypothetical protein FLJ12443 FLJ12443 NM 024830
89 201890 ribonucleotide reductase M2 polypeptide NM 001034
90 201910 RhoGEF (ARHGEF) and pleckstrin domain protein 1 FARP1 NM 005766
91 201911 RhoGEF ARHGEF and pleckstrin domain protein 1 FARP1 NM 005766
92 201921 guanine nucleotide binding protein 10 GNG10 NM 004125
93 201934 hypothetical protein PR02730 NM 025222
94 201938 deleted in oral cancer 1 DOC1 NM 004642
95 201987 thyroid hormone receptor-assoc protein, 240 kD sub NM 005121
96 202001 NADH deh dro enase 1 a subcom lex, 6 NDUFA6 NM 002490
97 202029 ribosomal protein L38 RPL38 NM_000999
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
57
98 202077 NADH deh dro enase 1, alphabeta subcomplex, 1 NDUFAB1 NM 005003
99 202078 COP9 subunit 3 COPS3 NM 003653
100 202090 ubi uinol-c tochrome c reductase (6.4kD) sub UQCR NM 006830
101 202110 cytochrome c oxidase subunit Vllb COX7B NM 001866
102 202114 sorting nexin 2 SNX2 NM 003100
103 202141 COP9 homolog NM 006710
104 202154 tubulin, beta, 4 TUBB4 NM 006086
105 202163 CCR4-NOT transcription complex, subunit 8 CNOT8 NM 004779
106 202187 rotein hos hatase 2 re ulato subunit B a isoform PPP2R5A NM 006243
107 202197 m otubularin related protein 3 MTMR3 NM 021090
108 202219 solute carrier family 6, member 8 SLC6A8 NM 005629
109 202231 dendritic cell protein GA17 NM 006360
110 202233 ubi uinol-c ochrome c reductase hinge protein UQCRH NM 006004
111 202242 transmembrane 4 su erfamily member 2 TM4SF2 NM 004615
112 202275 lucose-6- hos hate deh dro enase G6PD NM 000402
113 202279 chromosome 14 open reading frame 2 C14ORF2 NM 004894
114 202285 tumor-associated calcium signal transducer 2 NM 002353
115 202286 tumor-associated calcium signal transducer 2 NM 002353
116 202287 tumor-associated calcium signal transducer 2 NM 002353
117 202298 NADH deh dro enase 1 alpha subcomplex, 1 NDUFAI NM 004541
118 202310 proalpha 1 I chain of type I procollagen NM 000088
119 202311 proalpha 1 I chain of type I procoliagen NM 000088
120 202312 colla en, type I, alpha 1 COL1A1 NM 000088
121 202324 ol i resident protein GCP60 GCP60 NM 022735
122 202325 ATP synthase, H+ transporting, mitochondrial FO ATP5J NM_001685
complex, subunit F6
123 202349 d stonia 1, torsion DYT1 NM 000113
124 202411 interferon, alpha-inducible protein 27 IF127 NM 005532
125 202423 zinc finger protein 220 ZNF220 NM 006766
126 202432 protein phosphatase 3 catalytic subunit, beta isoform PPP3CB NM
021132
127 202442 adaptor-related protein complex 3, sigma 1 sub AP3S1 NM 001284
128 202458 protease, serine, 23 SPUVE NM 007173
129 202468 catenin a-like 1 CTNNALI NM 003798
130 202478 GS3955 protein GS3955 NM 021643
131 202481 short-chain deh dro enasereductase 1 SDR1 NM 004753
132 202503 KIAA0101 KIAA0101 NM 014736
133 202544 glia maturation factor, beta GMFB NM 004124
134 202565 supervillin, transcript variant 1 SVIL NM 003174
135 202582 RAN binding protein 9 RANBPM NM 005493
136 202591 sin le-stranded DNA-binding protein SSBP NM 003143
137 202622 spinocerebellar ataxia 2 SCA2 NM 002973
138 202642 transformationtranscription domain-assoc protein TRRAP NM 003496
139 202649 ribosomal protein S19 RPS19 NM 001022
140 202673 dolichyl-phosphate mannosyltransferase polypeptide DPM1 NM003859
1, catal tic subunit
141 202692 upstream binding transcription factor, RNA UBTF NM_014233
polymerase I
142 202712 creatine kinase, mitochondrial 1 CKMT1 NM 020990
143 202723 forkhead box O1A FOXOIA NM 002015
144 202724 forkhead box 01 A FOXO 1 A NM 002015
145 202735 emo binding protein EBP NM 006579
146 202754 KIAA0029 protein KIAA0029 NM 015361
147 202759 A kinase (PRKA) anchor protein 2 AKAP2 NM 007203
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
58
148 202760 A kinase (PRKA) anchor protein 2 AKAP2 NM 007203
149 202761 s na tic nuclei expressed gene 2 KIAA1011 NM 015180
150 202789 hos holi ase C, y 1 NM 002660
151 202820 aryl hydrocarbon receptor AHR NM 001621
152 202824 transcription elongation factor B SIII , polypeptide 1 TCEB1 NM
005648
153 202834 serine (or cysteine) proteinase inhibitor, clade A SERPINA8
NM000029
member 8
154 202841 7-60 protein 7-60 NM 007346
155 202848 G protein-coupled receptor kinase 6 NM 002082
156 202854 hypoxanthine hos horibos Itransferase 1 HPRT1 NM 000194
157 202860 KIAA0476 gene product KIAA0476 NM 014856.
158 202889 microtubule-associated protein 7 NM 003980
159 202890 microtubule-associated protein 7 NM 003980
160 202947 I co horin C, transcript variant 1 GYPC NM 002101
161 202949 four and a half LIM domains 2 FHL2 NM 001450
162 202957 hematopoietic cell-specific Lyn substrate 1 HCLS1 NM 005335
163 203044 KIAA0990 protein KIAA0990 NM 014918
164 203053 breast carcinoma amplified sequence 2 BCAS2 NM 005872
165 203133 protein translocation complex beta SEC61 B NM 006808
166 203138 histone acetyltransferase 1 HAT1 NM 003642
167 203139 death-associated protein kinase 1 DAPK1 NM 004938
168 203140 B-cell CLLI m homa 6 BCL6 NM 001706
169 203142 adaptor-related protein complex 3, 131 subunit AP3B1 NM 003664
170 203211 KIAA1073 protein KIAA1073 NM 016156
171 203213 cell division cycle 2, G1 to S and G2 to M NM 001786
172 203255 vitiligo-associated protein VIT-1 VIT1 NM 018693
173 203262 chromosome X uni ue 9928 expressed sequence DXS9928E NM 004699
174 203287 ladinin 1 LAD1 NM 005558
175 203316 small nuclear ribonucleoprotein ol e tide E SNRPE NM_003094
176 203332 inositol pol hos hate-5- hos hatase, 145kD INPP5D NM 005541
177 203362 MAD2 mitotic arrest deficient, yeast, homolo -like 1 MAD2L1 NM
002358
178 203371 NADH deh dro enase 1 beta subcomplex, 3 NDUFB3 NM 002491
179 203385 diac I I cerol kinase, alpha DGKA NM 001345
180 203396 proteasome (prosome, macro ain subunit, a type, 4 PSMA4 NM 002789
181 203437 putative receptor protein PMI NM 003876
182 203460 presenilin 1, transcript variant 1-463 PSEN1 NM 007318
183 203484 Sec6l gamma SEC61 G NM 014302
184 203514 mitogen-activated protein kinase kinase kinase 3 NM 002401
185 203528 sema domain, Ig domain, TM domain and short SEMA4D NM_006378
c o lasmic domain, 4D
186 203531 cullin 5 NM 003478
187 203581 RAB4, member RAS onco ene family NM 004578
188 203610 ring finger protein 15 NM 006355
189 203613 NADH deh dro enase 1 beta subcomplex, 6 NDUFB6 NM 002493
190 203621 NADH deh dro enase 1 beta subcomplex, 5 NDUFB5 NM 002492
191 203666 stromal cell-derived factor 1 SDF1 NM 000609
192 203667 tubulin-specific chaperone a TBCA NM 004607
193 203674 KIAA0054 gene product; Helicase KIAA0054 NM 014877
194 203743 thymine-DNA I cos lase TDG NM 003211
195 203748 RNA binding motif, single stranded interacting RBMS1 NM_016839
rotein 1, transcript variant MSSP-2
196 203761 Src-like-adapter SLA NM 006748
197 203827 hypothetical protein FLJ10055 FLJ10055 NM 017983
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
59
198 203845 p300CBP-associated factor NM 003884
199 203882 interferon-stimulated transcription factor 3, y ISGF3G NM 006084
200 203886 fibulin 2 FBLN2 NM 001998
201 203893 adrenal gland protein AD-004 LOC51578 NM 016283
202 203922 cytochrome b-245, beta polypeptide NM 000397
203 203936 matrix metalloproteinase 9 MMP9 NM 004994
204 203940 KIAA1036 protein KIAA1036 NM 014909
205 203960 HSPCO34 protein LOC51668 NM 016126
206 203965 ubi uitin s ecific protease 20 USP20 NM 006676
207 204014 dual specificity phosphatase 4 DUSP4 NM 001394
208 204015 dual specificity phosphatase 4 DUSP4 NM 001394
209 204049 KIAA0680 ene product KIAA0680 NM 014721
210 204067 sulfite oxidase NM 000456
211 204082 pre-B-cell leukemia transcription factor 3 PBX3 NM 006195
212 204141 tubulin, beta polypeptide TUBB NM 001069
213 204145 FSHD re ion ene 1 FRG1 NM 004477
214 204146 RAD51-interactin protein NM 006479
215 204158 T-cell, immune regulator 1 TCIRG1 NM 006019
216 204168 microsomal glutathione S-transferase 2 MGST2 NM 002413
217 204170 CDC28 protein kinase 2 CKS2 NM 001827
218 204215 hypothetical protein MGC4175 NM 024315
219 204294 aminomethyltransferase AMT NM 000481
220 204351 S100 calcium-binding protein P S100P NM 005980
221 204361 SKAP55 homologue SKAP-Hom NM 003930
222 204362 SKAP55 homologue SKAP-Hom NM 003930
223 204411 KIAA0449 protein KIAA0449 NM 017596
224 204416 a o{i o rotein C-I APOC1 NM 001645
225 204528 nucleosome assembly protein 1-like 1 NAP1L1 NM 004537
226 204640 s eckle-t e POZ protein SPOP NM 003563
227 204642 endothelial differentiation, sphingolipid G-protein- EDG1 NM_001400
cou led rece tor, 1
228 204652 nuclear res irato factor 1 NRF1 NM 005011
229 204694 al ha-feto rotein AFP NM 001134
230 204698 interferon stimulated gene ISG20 NM 002201
231 204729 syntaxin 1A STX1A NM 004603
232 204766 nudix -t pe motif 1 NUDT1 NM 002452
233 204767 flap structure-specific endonuclease 1 NM 004111
234 204777 T-cell differentiation protein, transcript variant a MAL NM 002371
235 204863 interleukin 6 signal transducer NM 002184
236 204864 interleukin 6 signal transducer IL6ST NM 002184
237 204881 UDP-glucose ceramide glucosyltransferase UGCG NM 003358
238 204885 mesothelin, transcript variant 1 MSLN NM 005823
239 204905 eukaryotic translation elongation factor 1 epsilon 1 EEF1 E1 NM
004280
240 204923 753P9 on chromosome Xq25-26.1 NM 018990
241 204950 KIAA0955 protein KIAA0955 NM 014959
242 204951 ras homolog gene family, member H ARHH NM 004310
243 204955 sushi-re eat-containin protein, X chrom SRPX NM 006307
244 204966 brain-specific an io enesis inhibitor 2 BA12 NM 001703
245 204989 integrin, beta 4 ITGB4 NM 000213
246 204990 integrin, beta 4 ITGB4 NM 000213
247 205033 defensin, alpha 1, myeloid-related sequence DEFA1 NM 004084
248 205087 DKFZP566KO23 protein NM 015485
249 205108 a oli o rotein B APOB NM 000384
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
250 205133 heat shock lOkD protein 1 HSPE1 NM 002157
251 205176 integrin beta 3 binding protein ITGB3BP NM 014288
252 205213 KIAA0050 gene product ACAP1 NM 014716
253 205270 I m hoc te cytosolic protein 2 LCP2 NM 005565
254 205323 metal-re ulato transcription factor 1 MTF1 NM 005955
255 205335 signal recognition particle 19kD SRP19 NM 003135
256 205336 parvalbumin PVALB NM 002854
257 205339 TALl (SCL) interru tin locus SIL NM 003035
258 205361 prefoldin 4 U41816
259 205403 interleukin 1 receptor, type II IL1 R2 NM 004633
260 205453 homeo box B2 HOXB2 NM 002145
261 205467 caspase 10 CASP10 NM 001230
262 205600 homeo box B5 HOXB5 NM 002147
263 205601 homeo box B5 HOXB5 NM 002147
264 205608 an io oietin 1 NM 001146
265 205609 an io oietin 1 ANGPT1 NM 001146
266 205644 small nuclear ribonucleoprotein polypeptide G SNRPG NM 003096
267 205671 MHC, class II DO HLA-DOB NM 002120
268 205807 tuftelin 1 TUFT1 NM 020127
269 205849 gamma-aminobutyric acid A receptor 3, trans var 1 GABRB3 NM 006294
270 205967 H4 histone family, member G H4FG NM 003542
271 206066 RAD51 S. cerevisiae) homolog C RAD51C NM 002876
272 206148 interleukin 3 receptor, alpha IL3RA NM 002183
273 206150 tumor necrosis factor receptor su erfamil , mem 7 TNFRSF7 NM 001242
274 206177 arginase, liver ARG1 NM 000045
275 206219 vav 1 oncogene VAV1 NM 005428
276 206245 NS1-bindin protein NS1-BP NM 006469
277 206289 homeo box A4 HOXA4 NM 002141
278 206298 hypothetical protein from s 23549 and 23762 LOC58504 NM 021226
279 206618 interleukin 18 receptor 1 IL18R1 NM 003855
280 206637 KIAA0001 KIAA0001 NM 014879
281 206674 fms-related tyrosine kinase 3 FLT3 NM 004119
282 206723 G protein-coupled receptor Edg-4 AF233092
283 206790 NADH deh dro enase 1 beta subcomplex, 1 NDUFB1 NM 004545
284 206868 KIAA0189 gene product KIAA0189 NM 014725
285 206874 Ste20-related serinethreonine kinase NM 014720
286 206958 UPF3 UPF3 NM 023011
287 207072 interleukin 18 receptor accessory protein IL18RAP NM 003853
288 207111 egf-like module containing, mucin-like, hormone EMR1 NM001974
receptor-like sequence 1
289 207127 heterogeneous nuclear ribonucleoprotein H3 HNRPH3 NM 021644
290 207163 v-akt murine thymoma viral oncogene hom 1 AKT1 NM 005163
291 207165 hyaluronan-mediated motility receptor trans var 2 HMMR NM 012485
292 207266 RNA binding motif, single stranded interacting RBMS1 NM_016837
protein 1, transcript variant MSSP-3
293 207287 h othetical protein FLJ14107 FLJ14107 NM 025026
294 207335 solute carrier family 1 member 7 SLC1A7 NM 006671
295 207540 spleen tyrosine kinase SYK NM 003177
296 207551 male-specific lethal-3 Droso hila -like 1 MSL3L1 NM 006800
297 207568 choliner ic receptor, nicotinic, a polypeptide 6 CHRNA6 NM 004198
298 207571 basement membrane-induced gene ICB-1 NM 004848
299 207573 ATP synthase, H+ transporting, mitochondrial Fl FO, ATP5JG
NM_006476
subunit
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
61
300 207777 nuclear body protein Sp140 SP140 NM 007237
301 207826 inhibitor of DNA binding 3, dominant negative helix- ID3 NM_002167
loo -helix protein
302 207845 anaphase-promoting complex 10 APC10 NM 014885
303 207935 keratin 13 KRT13 NM 002274
304 207974 S-phase kinase-associated protein 1A 19A SKP1A NM 006930
305 208091 h othetical protein DKFZ 564K0822 NM 030796
306 208130 thromboxane A synthase 1 TBXAS1 NM 030984
307 208270 arginyl amino e tidase RNPEP NM 020216
308 208310 follistatin-like 1 FSTLI NM 007085
309 208325 I m hoid blast crisis oncogene LBC NM 006738
310 208414 homeo box B3 HOXB3 NM 002146
311 208420 su ressor of Ty (S.cerevisiae) 6 homolog SUPT6H NM 003170
312 208549 roth mosin a14 LOC51685 NM 016171
313 208598 upstream re ulato element binding protein 1 UREB1 NM 005703
314 208621 villin 2 ezrin NM 003379
315 208622 villin 2 ezrin NM 003379
316 208623 cytovillin 2 VIL2 NM 003379
317 208629 hydroxyacyl-Coenzyme A dehydrogenase3-ketoacyl- U04627
Coenzyme A thiolaseenoyl-Coenzyme A hydratase
al ha subunit
318 208633 actin binding protein; macrophin AB029290
319 208643 Ku autoimmune antigen gene J04977
320 208656 cyclin I CYC1 AF135162
321 208667 putative tumor suppressor ST13 ST13 U17714
322 208672 s licin factor, arginineserine-rich 3 BC000914
323 208679 PNAS-139 AF279893
324 208695 ribosomal protein L39 RPL39 BC001019
325 208745 ATP synthase, H+ transporting, mitochondrial Fl FO, AA917672
subunit
326 208746 Fl FO-type ATP synthase subunit AF070655
327 208754 DKFZp 762G106 AL162068
328 208808 hi ility group protein 2 BC000903
329 208819 mel transforming oncogene - RAB8 homolog BC002977
330 208831 KIAA0162 D79984
331 208894 MHC class II HLA-DR-alpha M60334
332 208900 topoisomerase I W025108
333 208904 ribosomal protein S28 RPS28 BC000354
334 208905 cytochrome c BC005299
335 208919 Similar to hypothetical protein FLJ13052 BC001709
336 208927 s eckle-t e POZ protein BC001269
337 208956 deoxyuridine tri hos hatase DUT U62891
338 208966 interferon, gamma-inducible protein 16 AF208043
339 208975 importin beta subunit L38951
340 208977 tubulin, beta, 2 BC004188
341 208981 plateletendothelial cell adhesion molecule NM 000442
342 209022 hi hl similar to autonomously re licatin sequence ARS AK026678
343 209063 ol aden late binding protein-interacting protein 1 BF248165
344 209066 mitochondrial ubi uinone-bindin protein M26700
345 209089 RAB5A, member RAS oncogene family BC001267
346 209119 nuclear receptor subfamily 2, group F, mem 2 AV703465
347 209120 nuclear receptor subfamily 2, group F, mem 2 AV703465
348 209122 adi ose differentiation-related protein NM 001122
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
62
349 209138 immunoglobulin lambda locus FLM87790
350 209172 centromere protein F U30872
351 209191 Similar to tubulin, beta, 4 BC002654
352 209209 mito en inducible gene mig-2 AW469573
353 209210 mitogen inducible gene_ mig-2 Z24725
354 209227 Putative prostate cancer tumor suppressor NM 006765
355 209228 Putative prostate cancer tumor suppressor NM 006765
356 209239 nuclear factor of kappa light polypeptide gene p105 M55643
enhancer in B-cells 1
357 209257 chondroitin sulfate proteoglycan 6 bamacan NM 005445
358 209258 chondroitin sulfate roteo I can 6 bamacan NM 005445
359 209270 laminin S B3 chain LAMB3 NM 000228
360 209282 protein kinase D2 NM 016457
361 209289 nuclear factor IB NM 005596
362 209295 TRAIL receptor 2 AF016266
363 209303 NADH deh dro enase Fe-S protein 4 NM 002495
364 209309 zinc-al ha2- I co rotein D90427
365 209324 retinally abundant regulator of G-protein signalling 16 hRGS-r NM
002928
366 209325 retinally abundant regulator of G-protein si nalin hRGS-r NM 002928
367 209362 suppressor of RNA polymerase B, yeast homolog SRB7 NM 004264
368 209369 1,2-c clic-inositol- hos hate phosphodiesterase ANX3 NM 005139
369 209372 tubulin, beta polypeptide BC001352
370 209377 thyroid hormone receptor interactor 7 AF274949
371 209385 proline synthetase co-transcribed AL136616
372 209411 ADP-ribosylation factor binding protein GGA3 AF219139
373 209443 proteinase inhibitor, clade A member 5 NM 000624
374 209471 farnes I rotein transferase alpha-subunit NM 002027
375 209492 ATP synthase, H+ transporting, mitochondrial FO BC003679
complex, subunit e
376 209500 tumor necrosis factor su erfamil , member 13 TNSF13 NM 003808
377 209507 replication protein A3 NM 002947
378 209520 nuclear cap binding protein subunit 1 NM 002486
379 209560 delta-like homolog NM 003836
380 209585 multiple inositol ol hos hate phosphatase NM 004897
381 209628 hypothetical protein P15-2 NM 018698
382 209662 centrin, EF-hand protein, 3 NM 004365
383 209679 hypothetical protein from 643 NM 020467
384 209687 stromal cell-derived factor 1 U19495
385 209730 sema domain, Ig, short basic domain, secreted, 3F U38276
386 209734 hemato oietic protein I NM 005337
387 209810 pulmonary surfactant-associated protein B SP-B NM 000542
388 209827 interleukin 16 IL16 NM 004513
389 209838 thyroid receptor interacting protein 15 AF212227
390 209868 RNA binding motif, single stranded interacting prot 1 SCR2 NM
002897
391 209907 intersectin 2 long isoform ITSN2 AF182198
392 210093 ma o-nashi homolog, proliferation-associated NM 002370
393 210097 retinoic acid repressible protein AF130102
394 210137 dCMP deaminase BC001286
395 210139 peripheral myelin protein 22 GAS3 L03203
396 210180 s licin factor, arginineserine-rich 10 U87836
397 210233 interleukin 1 receptor accessory protein IL1 RAP AF167343
398 210235 LAR-interactin protein la U22815
399 210283 Sim to poly aden late binding protein-interacting I BC005295
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
63
400 210284 TAK1-binding protein 2 AF241230
401 210347 C2H2-t e zinc-finger protein AF080216
402 210448 ionotropic ATP receptor P2X5b U49396
403 210453 DKFZp566GO13 AL050277
404 210510 soluble neuropilin-1 AF145712
405 210563 FLICE-like inhibitory protein short form U97075
406 210598 AF130051
407 210615 neuropilin-1 soluble isoform 11 NRP1 AF280547
408 210633 acidic keratin-10 KRT10 M19156
409 210639 a o tosis-related protein APG5L AF293841
410 210649 BRG1-Associated Factor 250a BAF250a AF231056
411 210666 iduronate-2-sulfatase IDS NM 000202
412 210785 basement membrane-induced gene ICB-1 beta AB035482
413 210786 Friend leukemia virus integration 1 FLI-1 M93255
414 210840 IQ motif cont'g GTPase activating protein 1 D29640
415 210854 GABAnoradrenaline transporter U17986
416 210859 CLN3 protein CLN3 AF077973
417 210982 MHC class II HLA-DRA HLA-DRA M60333
418 211000 gp130 of the rheumatoid arthritis antigenic peptide- gp130-RAPS
AB015706
bearing soluble form
419 211133 leukocyte lg-like receptor subfamily B member 3 LILRB3 NM 00864
420 211430 anti-hepatitis A I G M87789
421 211445 FKSG17 FKSG17 AF315951
422 211487 ribosomal protein S17 BC004886
423 211535 fibroblast growth factor receptor FGFR M60485
424 211645 immuno lobulin kappa-chain VK-1 I K M85256
425 211730 ol merase (RNA) II (DNA directed) ol e tide L BC005903
426 211743 roteo I can 2, bone marrow BC005929
427 211747 U6 snRNA-associated Sm-like protein BC005938
428 211762 ka o herin alpha 2 BC005978
429 211858 guanine nucleotide-binding protein Gs a sub iso L2 AF088184
430 211915 beta-tubulin TUB4 U83110
431 211921 fetal thymus prothymosin alpha AF348514
432 211932 heterogeneous nuclear protein similar to rat helix BE867771
destabilizing protein
433 211938 hypothetical protein PR01843 BF247371
434 211976 FLJ21862fis AK026168
435 211985 matrix Gla protein A1653730
436 212007 UBX domain-containing 1 D87684
437 212012 KIAA0230 AF200348
438 212013 KIAA0230 AF200348
439 212027 S164 protein BE466128
440 212181 di hos hoinositol polyphosphate hos hoh drolase2 NUDT4 AF191654
441 212201 KIAA0692 protein AB014592
442 212248 FLJ20738 fis A1972475
443 212269 minichromosome maintenance deficient 3-assoc GANP AJ010089
444 212277 KIAA0647 protein AB014547
445 212283 est:t 49b04.x1 AF016903
446 212285 est:t 49b04.x1 AF016903
447 212287 KIAA0160 protein BF382924
448 212298 neuropilin 1 NM 003873
449 212311 KIAA0746 protein AB018289
450 212382 FLJ11918fis AK021980
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
64
451 212384 HLA-B associated transcri t-1 BG341380
452 212385 FLJ11918 fis AK021980
453 212386 FLJ11918fis AK021980
454 212387 FLJ11918 fis AK021980
455 212426 tyrosine 3-monooxygenasetryptophan 5- BF033313
monooxygenase activation protein, theta polypeptide
456 212451 KIAA0256 D87445
457 212531 lipocalin 2 LCN2 NM 005564
458 212549 DKFZp586N1323 A1149535
459 212561 KIAA1091 AA349595
460 212570 KIAA0830 AL573201
461 212571 KIAA1564 U00955
462 212573 KIAA0830 AL573201
463 212578 ribosomal protein S17 BF026595
464 212583 KIAA0560 gene product AB011132
465 212630 sec6 homolog AF055006
466 212664 tubulin, beta, 5 NM 006087
467 212740 hos hoinositide-3-kinase, re ulato sub 4 p150 BF740111
468 212757 FLJ22656fis BF111268
469 212771 DKFZp564AO26 AU150943
470 212833 TB1 ene M74089
471 212860 DKFZ 66702416 BG168720
472 212878 kinesin 2 AA284075
473 212907 hbc647 A1972416
474 212927 KIAA0594 AB011166
475 212964 DKFZ 434D1023 AB028943
476 213020 DKFZp566B213 GOSR1 NM 004871
477 213028 nuclear factor related to kappa B binding protein A1887378
478 213047 SET translocation AI278616
479 213061 KIAA0251 AA643304
480 213062 KIAA0251 AA643304
481 213074 DKFZ 564D156 BG545769
482 213129 I cine cleava e system protein H BE908931
483 213134 BTG family, member 3 BTG3 NM -006806
484 213147 homeo box A10 NM 018951
485 213150 homeo box A10 NM 018951
486 213159 KIAA0805 AB018348
487 213178 MAPK8IP3 NM 015133
488 213188 Weakly similar to T15138 hyp protein T28F2.4 NM 032778
489 213231 d stro hia myotonica-containing WD repeat motif L19267
490 213253 structural maint of chromosomes 2 yeast-like 1 SMC2L1 AU154486
491 213287 ene for acidic (type I cytokeratin 10 X14487
492 213311 KIAA1049 protein BF000251
493 213338 DKFZP586E1621 BF062629
494 213414 ribosomal protein S19 BE259729
495 213423 Putative prostate cancer tumor suppressor A1884858
496 213479 neuronal pentraxin li NPTX2 NM 002523
497 213502 immuno lobulin lambda-like polypeptide 3 X03529
498 213539 CD3D antigen, delta polypeptide (TiT3 com lex CD3D NM 000732
499 213553 a oli o rotein C-1 W79394
500 213567 23728 sequence BF431965
501 213599 O a-interactin protein 5 BE045993
502 213687 uncharacterized hypothalamus protein HSMNP1 BE968801
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
503 213700 hypothetical protein FLJ10803 AA554945
504 213727 h othetical protein FLJ1 1585 A1743654
505 213754 ol aden late binding protein-interacting protein 1 AW613203
506 213811 transcri tion factor 3 BG393795
507 213843 accessory proteins BAP31 BAP29 AW276522
508 213867 actin, AA809056
509 213911 H2A histone famil , member Z BF718636
510 213941 ribosomal protein S7 A1970731
511 214003 ribosomal protein S20 BF184532
512 214051 thymosin, beta BF677486
513 214097 ribosomal protein S21 AW024383
514 214124 DKFZ 434132027 AL043487
515 214129 KIAA0477 A1821791
516 214143 ribosomal protein L24 A1560573
517 214224 rotein NIMA-interacting, 4 parvulin BE674061
518 214264 est:tt46h03 A1656610
519 214288 proteasome (prosome, macro ain subunit, type, 1 W86293
520 214292 inte rin, beta 4 AA808063
521 214298 se tin 2 AL568374
522 214352 v-Ki-ras2 Kirsten rat sarcoma 2 viral oncogene hom BF673699
523 214377 est:Ul-H-B14-ao -e-12-0-UI.s1 BF508685
524 214433 selenium binding protein 1 SELENBP1 NM 003944
525 214494 cell matrix adhesion regulator CMAR NM 005200
526 214499 Bcl-2-associated transcription factor short form AF249273
527 214617 erforin 1 A1445650
528 214661 near HD on 4 16.3 with hom to hyp S. pombe gene R06783
529 214696 24659 sequence AF070569
530 214721 DKFZ 762L106 AL162074
531 214800 basic transcription factor 3 R83000
532 214820 transcri tional unit N143 AJ002572
533 214905 EUROIMAGE 46866 AL109674
534 215038 Huntin tin interacting protein AF049103
535 215073 nuclear receptor subfamily 2, group F, mem 2 AL554245
536 215096 esterase Dform I lutathione hydrolase AU145746
537 215121 immunoglobulin lambda locus AA680302
538 215127 RNA binding motif, single stranded interacting prot 1 AL517946
539 215136 DKFZ 56400482 AL050353
540 215147 23712 se uence AF007147
541 215171 NT2RP3000341 AK023063
542 215227 acid hos hatase 1, soluble BG035989
543 215379 immunoglobulin lambda joining 3 AV698647
544 215380 FLJ11717 fis AK021779
545 215399 amplified in osteosarcoma A1683900
546 215446 I s I oxidase LOX L16895
547 215493 3 end of the BTN2A1 gene encoding but ro hilin 2A AL121936
548 215691 HSPCO34 protein AV702994
549 215764 ada tor-related protein complex 2, alpha 2 subunit AA877641
550 215812 creatine trans orter SLC6A1O U41163
551 215905 U5 snRNP-specific 40 kDa protein AL157420
552 216207 immunoglobulin kappa variable 1-13 AW408194
553 216348 transcri tion factor AP-2 beta AL049693
554 216384 roth mosin alpha PTMA AF257099
555 216641 ladinin LAD U58994
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
66
556 216833 I co horin HeP2 U05255
557 216899 PAC RP5-1139P1 from 7 15- 21 AC003999
558 216977 U2 snRNP-specific A protein, alternative transcript 3 AJ130972
559 217013 PAC RP4-604G5 from 7 22- 31.1 AC004522
560 217014 PAC RP4-604G5 from 7 22- 31.1 AC004522
561 217022 1 A1-A2 lambda hybrid GAU heavy chain S55735
562 217106 utative dimethyladenosine transferase AF091078
563 217122 Matrix Metallo roteinase Female Rep tract MIFR1 2 MMP2122A AL031282
564 217249 BAC CTB-162B4 from 4 AC004544
565 217293 an io oietin 1 AF209975
566 217329 cytochrome c oxidase subunit Vllb pseudogene COX7BP1 AF042164
567 217336 RP5-858M22 on chromosome 20 AL118510
568 217410 FLJ11524 fis AK021586
569 217419 FLJ 11524 fis AK021586
570 217491 cytochrome c oxidase subunit Vllc pseudogene COX7CP1 AF042165
571 217749 coat rotein gamma-cop LOC51137 NM 016128
572 217750 hypothetical protein FLJ13855 NM 023079
573 217754 putative nucleolar RNA helicase NOH61 NM 019082
574 217769 h othetical protein HSPCO14 NM 015932
575 217773 NADH deh dro enase 1 alpha subcomplex, 4 NDUFA4 NM 002489
576 217801 ATP synthase, H+ transporting, mitochondrial Fl ATP5E NM006886
com lex, epsilon subunit
577 217812 hi h lucose-re ulated protein 8 HGRG8 NM 016258
578 217825 CGI-76 protein AF151039
579 217833 NS1-associated protein I NM 006372
580 217838 RNB6 RNB6 NM 016337
581 217843 HSPC126 protein HSPC126 NM 014166
582 217853 h othetical protein FLJ13732 similar to tensin TENC1 NM 022748
583 217860 NADH deh dro enase 1 alpha subcomplex, 10 NDUFAIO NM 004544
584 217866 h othetical protein FLJ12529 FLJ12529 NM 024811
585 217875 transmembrane, prostate androgen induced RNA TMEPAI NM 020182
586 217877 hypothetical protein SP192 SP192 NM 021639
587 218003 FK506-binding protein 3 FKBP3 NM 002013
588 218039 HQ0310 PR00310 1 LOC51203 NM 016359
589 218058 CpG binding protein CGBP NM 014593
590 218062 Cdc42 effector protein 4; binder of Rho GTPases 4 CEP4 NM 012121
591 218103 h othetical protein FLJ20062 FLJ20062 NM 017647
592 218116 hepatocellular carcinoma-associated anti en 59 LOC51759 NM 016520
593 218117 ring-box 1 RBX1 NM 014248
594 218175 hypothetical protein FLJ22471 FLJ22471 NM 025140
595 218188 translocase of inner mitochondrial membrane 13 TIMM13B NM012458
(yeast) homolog B
596 218213 chromosome 11 open reading frame 10 C11orf10 NM 014206
597 218241 ol i autoantigen, golgin subfamily a, 5 GOLGA5 NM 005113
598 218256 nucleo orin p54 NUP54 NM 017426
599 218259 KIAA1243 protein KIAA1 243 NM 014048
600 218274 h othetical protein FLJ10415 FLJ10415 NM 018089
601 218276 WW Domain-Containing Gene WW45 NM 021818
602 218280 H2A histone family, member 0 H2AFO NM 003516
603 218283 kiaa-iso protein LOC51188 NM 016305
604 218288 h othetical protein MDS025 MDS025 NM 021825
605 218334 hypothetical protein FLJ23445 FLJ23445 NM 025075
606 218339 HSPC158 protein HSPC158 NM 014180
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
67
607 218350 geminin LOC51053 NM 015895
608 218367 ubi uitin specific protease 21 USP21 NM 012475
609 218368 type I transmembrane protein Fn14 FN14 NM 016639
610 218373 hyp protein FLJ13258 similar to fused toes FLJ13258 NM 022476
611 218395 h othetical protein FLJ13433 FLJ13433 NM 022496
612 218397 h othetical protein FLJ10335 FLJ10335 NM 018062
613 218447 DC13 rotein DC13 NM 020188
614 218467 x 003 rotein MDS003 NM 020232
615 218482 DC6 rotein DC6 NM 020189
616 218543 h othetical protein FLJ22693 FLJ22693 NM 022750
617 218563 NADH deh dro enase 1 alpha subcomplex, 3 NDUFA3 NM 004542
618 218576 dual s ecificity phosphatase 12 DUSP12 NM 007240
619 218603 homolog of Drosophila headcase hHDC NM 016217
620 218605 hypothetical protein FLJ23182 FLJ23182 NM 022366
621 218643 postsyn a tic protein CRIPT CRIPT NM 014171
622 218660 dysferlin, limb girdle muscular dystrophy 2B DYSF NM 003494
623 218671 ATPase inhibitor precursor LOC51189 NM 016311
624 218801 UDP- lucose: I co rotein glucosyltransferase 2 FLJ10873 NM 020121
625 218830 ribosomal protein L26 homolog LOC51121 NM 016093
626 218873 h othetical protein FLJ20203 FLJ20203 NM 017710
627 218936 HSPC128 protein HSPC128 NM 014167
628 218937 hypothetical protein FLJ20417 FLJ20417 NM 017810
629 218946 HIRIP5 rotein HIRIP5 NM 015700
630 219008 h othetical protein FLJ21820 FLJ21820 NM 021925
631 219030 CGI-121 protein LOC51002 NM 016058
632 219032 o sin 3 ence halo sin OPN3 NM 014322
633 219056 h othetical protein FLJ11712 FLJ11712 NM 024570
634 219105 ori in reco nition complex, subunit 6-like ORC6L NM 014321
635 219110 GAR1 rotein GAR1 NM 018983
636 219163 h othetical protein FLJ20079 FLJ20079 NM 017656
637 219218 h othetical protein FLJ23058 FLJ23058 NM 024696
638 219286 h othetical protein FLJ12479 FLJ12479 NM 022768
639 219293 h othetical protein PTD004 NM 013341
640 219347 h othetical protein FLJ10956 FLJ10956 NM 018283
641 219452 putative di e tidase LOC64174 NM 022355
642 219506 hypothetical protein FLJ23221 FLJ23221 NM 024579
643 219507 hypothetical protein LOC51319 NM 016625
644 219546 hypothetical protein DKFZ 434P0116 NM 017593
645 219759 amino e tidase LOC64167 NM 022350
646 219765 hypothetical protein FLJ12586 FLJ12586 NM 024620
647 219816 hypothetical protein FLJ10482 FLJ10482 NM 018107
648 219819 HSPCO07 protein HSPCO07 NM 014018
649 219906 hypothetical protein FLJ10213 FLJ10213 NM 018029
650 220001 peptidyl arginine deiminase, type V PAD NM 012387
651 220023 a oli o rotein B48 receptor APOB48R NM 018690
652 220052 TERF1 TRF1 )-interacting nuclear factor 2 TINF2 NM 012461
653 220060 hypothetical protein FLJ20641 FLJ20641 NM 017915
654 220155 h othetical protein FLJ13441 FLJ13441 NM 023924
655 220199 hypothetical protein FLJ12806 FLJ12806 NM 022831
656 220386 chromosome 2 open reading frame 2 C2ORF2 NM 019063
657 220404 PR00611 protein PR00611 NM 014076
658 220416 hypothetical protein FLJ21472 FLJ21472 NM_024837
659 220558 pan-hematopoietic expression PHEMX NM 005705
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
68
660 220671 carbon catabolite repression 4-like CCRN4L NM 012118
661 220741 inor anic ro hos hatase SID6-306 NM 006903
662 220755 G8 protein G8 NM 016947
663 220864 CGI-39 rotein; cell death-re ulato protein GRIM19 LOC51079 NM
015965
664 220942 hypothetical protein, estradiol-induced E21G5 NM 014367
665 221009 PPAR amma an io oietin related protein PGAR NM 016109
666 221143 re lication protein A comp 34 kd subunit hom Rpa4 HSU24186 NM
013347
667 221253 hypothetical protein MGC3178 MGC3178 NM 030810
668 221432 hypothetical protein NPDO16 NPD016 NM 031212
669 221434 hypothetical protein DC50 DC50 NM 031210
670 221505 h othetical protein MGC5350 AW612574
671 221509 densit -re ulated protein SMAP-3 AB014731
672 221528 Similar to hypothetical protein FLJ11656 BC000143
673 221577 rostate differentiation factor AF003934
674 221593 Similar to ribosomal protein L31 BC001663
675 221599 Similar to PTD015 protein BC002752
676 221620 brain my025 AF061264
677 221669 ac I-Coenz me A deh dro enase family, member 8 BC001964
678 221776 bromodomain-containing 7 A1885109
679 221791 hypothetical protein BG167522
680 221853 FLJ23491 N39536
681 221860 DKFZ 4341-2128 AL044078
682 221932 FLB4739 AA133341
683 221935 FLJ13078 fis AK023140
684 221942 sec6l homolog A1719730
685 222108 BAC GS1-99H8 AC004010
686 222192 FLJ11610fis AK021672
687 222193 FLJ11610fis AK021672
688 222203 FLJ13563fis AK023625
689 222280 Weakl similar to ALUC BG491393
690 33304 HEM45 U88964
691 33768 59 protein L19267
692 35666 semaphorin III family homolog U38276
693 37004 ulmona surfactant-associated protein B SP-B J02761
694 37005 Unknown product D28124
695 37201 PK-120 D38535
696 37996 m otonic dystrophy kinase DM kinase L08835
697 41858 DKFZp564EO53 FRAG1 NM 014489
698 44696 IMAGE-1473925 AA915989
699 49485 W22625:71 E5 Homo sapiens cDNA W22625
700 55692 75H3 / not-directional W22924
701 59644 IMAGE-2354728 A1735391
702 AFFX-HSAC07/X00351 3 X00351
703 AFFX-HSAC07/X00351 M beta-actin X00351
704 AFFX-HUMISGF3A/M97935 MB transcription factor ISGF-3 M97935
705 AFFX-r2-Ec-bioB-3 E coli bioB gene biotin synthetase J04423
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
69
Reference list
5,242,974 5,561,071
5,384,261 5,571,639
5,405,783 5,593,839
5,412,087 5,599,695
5,424,186 5,624,711
5,429,807 5,658,734
5,436,327 5,700,637
5,445,934 6,004,755
5,472,672 6,133,305
5,527,681 6,218,114
5,529,756 6,218,122
5,532,128 6,271,002
5,545,531 6,271,210
5,554,501 6,284,764
5,556,752 6,306,897
Adnane et al. (2000) "Inhibition of farnesyltransferase increases TGFbeta type
II receptor
expression and enhances the responsiveness of human cancer cells to TGFbeta"
Oncogene 19:5525-
5533
Albertson (1984) EMBO J. 3:1227-1234
Alsina et al. (2003) "Famesyltransferase inhibitor FTI-R115777 is well
tolerated, induces
stabilization of disease, and inhibits farnesylation and oncogenic/tumor
survival pathways in patients
with advanced multiple myeloma" Eur. J. Haematol. 70:269
Brunner et al. (2003) "Farnesyltransferase inhibitors: an overview of the
results of preclinical and
clinical investigations" Cancer Res. 63:5656-5668
Bullinger et al. (2004) "Use of gene-expression profiling to
identify.prognostic subclasses in adult
acute myeloid leukemia" N. Engl. J. Med. 350:1605-1616
Carr et al. (1991) "Interaction of the regulatory subunit (RII) of cAMP-
dependent protein kinase
with RII-anchoring proteins occurs through an amphipathic helix binding motif'
J. Biol. Chem.
266:14188-14192
Chang et al. (2003) "Gene expression profiling for the prediction of
therapeutic response to
docetaxel in patients with breast cancer" Lancet 362:362-369
Cheok et al. (2003) "Treatment-specific changes in gene expression
discriminate in vivo drug
response in human leukemia cells" Nat. Genet. 34:85-90
Cortes et al. (2003) "Efficacy of the farnesyl transferase inhibitor R115777
in chronic myeloid
leukemia and other hematologic malignancies" Blood 101:1692-1697
Cox et al. (2002) "Farnesyltransferase inhibitors: promises and realities"
Curr. Opin. Pharmacol.
2:388-393
Debernardi et al. (2003) "Genome-wide analysis of acute myeloid leukemia with
normal karyotype
reveals a unique pattern of homeobox gene expression distinct from those with
translocation-mediated
fusion events" Genes Chrom. Cancer 37:149-158
End et al. (2001) "Characterization of the antitumor effects of the selective
farnesyl protein
transferase inhibitor RI 15777 in vivo and in vitro" Cancer Res. 61:131-137
EP 430,402
Esteva et al. (2003) "Phase III study of weekly docetaxel and trastuzumab for
patients with HER-2-
overexpressing metastatic breast cancer" J Clin Oncol. 20:1800-1808.
Foisner et al. (1991) "Protein kinase A- and protein kinase C-regulated
interaction of plectin with
lamin B and vimentin" Proc. Natl. Acad. Sci. USA 88:3812-3816
Gene Therapy Protocols, edited by Paul D. Robbins, Human press, Totowa NJ,
1996
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
Harousseau et al. (2003) "ZarnestraTM (R115777) in patients with relapsed or
refractory acute
myelogenous leukemia (AML): results of a multicenter phase 2 study" Blood
102:176a Abstract 614
Hofmann et al. (2002) "Relation between resistance of Philadelphia-chromosome-
positive acute
lymphoblastic leukaemia to the tyrosine kinase inhibitor ST1571 and gene-
expression profiles: a gene-
expression study" Lancet 359:481-486
Jensen et al. (2001) "Estrogen receptors and proliferation markers in primary
and recurrent breast
cancer" Proc Natl Acad Sci U S A. 98:15197-15202.
Jiang et al. (2000) "The phosphoinositide 3-OH kinase/AKT2 pathway as a
critical target for
farnesyltransferase inhibitor-induced apoptosis" Mol. Cell. Biol. 20:139-148
Johnston et al. (2003) "Phase II study of the efficacy and tolerability of two
dosing regimens of the
famesyl transferase inhibitor, R115777, in advanced breast cancer" J. Clin.
Oncol. 21:2492-2499
Kallioniemi (1992) Proc. Natl. Acad. Sci. USA 89:5321-5325
Karp et al. (2001) "Clinical and biologic activity of the farnesyltransferase
inhibitor Rl 15777 in
adults with refractory and relapsed acute leukemias: a phase I clinical-
laboratory correlative trial"
Blood 97:3361-3369
Kohlmann et al. (2003) " Molecular characterization of acute leukemias by use
of microarray
technology" Genes Chrom. Cancer Genes Chrom. Cancer 37:396-405
Kurzrock et al. (2004) "Phase II study of R115777, a farnesyl transferase
inhibitor, in
myelodysplastic syndrome" J. Clin. Oncol. 22:1287-1292
Lancet et al. (2003) "Farnesyltransferase inhibitors in hematologic
malignancies: new horizons in
therapy" Blood 102:3880-3889
Lancet et al. (2004) "Tipifarnib (ZARNESTR.ATM in Previously Untreated Poor-
Risk AML of the
Elderly: Updated Results of a Multicenter Phase 2 Trial" Blood 104:874a
Li et al. (2002) "The hematopoiesis-specific GTP-binding protein RhoH is
GTPase deficient and
modulates activities of other Rho GTPases by an inhibitory function" Mol.
Cell. Biol. 22:1158-1171
Lynch et al. (2004) "Activating mutations in the epidermal growth factor
receptor underlying
responsiveness of non-small-cell lung cancer to Gefitinib" N. Engl. J. Med.
350:2129-2139
McLean et al. (2004) "Pharmacogenomic analysis of cytogenetic response in
chronic myeloid
leukemia patients treated with imatinib" Clin. Cancer Res. 10:155-165
Methods in Molecular Biology, Vol. 33: In Situ Hybridization Protocols, Choo,
ed., Humana Press,
Totowa, NJ (1994)
Morgan et al. (2001) "Cell-cycle-dependent activation of mitogen-activated
protein kinase kinase
(MEK-1/2) in myeloid leukemia cell lines and induction of growth inhibition
and apoptosis by
inhibitors of RAS signaling" Blood 97:1823-1834
Na et al. (2004) "Inhibition of farnesyltransferase prevents collagen-induced
arthritis by down-
regulation of inflammatory gene expression through suppression of p21(ras)-
dependent NF-kappaB
activation" J Immunol. 173:1276-1283.
Okutsu et al. (2002) "Prediction of chemosensitivity for patients with acute
myeloid leukemia,
according to expression levels of 28 genes selected by genome-wide
complementary DNA microarray
analysis" Mol. Cancer Ther. 1:1035-1042
Paez et al. (2004) "EGFR mutations in lung cancer: correlation with clinical
response to gefitinib
therapy activating mutations in the epidermal growth factor receptor
underlying responsiveness of non-
small-cell lung cancer to Gefitinib" Science 304:1497-1500
Pasqualucci et al. (2001) "Hypermutation of multiple proto-oncogenes in B-cell
diffuse large-cell
lymphomas" Nature 412:341-346
Pinkel (1988) " Fluorescence in situ Hybridization with Human Chromosome-
Specific Libraries:
Detection of Trisomy 21 and Translocations of Chromosome 4" Proc Natl Acad Sci
USA 85:9138-
9142
Pinkel et al. (1998) "High resolution analysis of DNA copy number variation
using comparative
genomic hybridization to microarrays" Nature Genetics 20:207-211
CA 02589055 2007-05-30
WO 2006/066240 PCT/US2005/046100
71
Rao et al. (2004) "Phase III Double-Blind Placebo-Controlled Study of Farnesyl
Transferase
Inhibitor R115777 in Patients With Refractory Advanced Colorectal Cancer" J
Clin Oncol
22:3950-3957
Raponi, et al. (2004) "Identification of Molecular Predictors of Response to
ZARNESTRATM
(Tipifarnib, R115777) in Relapsed and Refractory Acute Myeloid Leukemia" Blood
104:861a
Reiss et al. (1990) "Inhibition of purified p2lras farnesyl:protein
transferase by Cys'AAX
tetrapeptides" Cell 62:81-88
Reuter et al. (2000) "Targeting the Ras signaling pathway: a rational,
mechanism-based treatment
for hematologic malignancies?" Blood 96:1655-1669
Sahai et al. (2002) "RHO-GTPases and cancer" Nat. Rev. Cancer 2:133-142
Schoch et al. (2002) "Acute myeloid leukemias with reciprocal rearrangements
can be distinguished
by specific gene expression profiles" Proc. Natl. Acad. Sci. USA 99:10008-
10013
Sterpetti et al. (1999) "Activation of the Lbc Rho exchange factor proto-
oncogene by truncation of
an extended C terminus that regulates transformation and targeting" Mol. Cell.
Biol. 19:1334-1345
Strachan and Read, Human Molecular Genetics, 1996
Takada et al. (2004) "Protein farnesyltransferase inhibitor (SCH 66336)
abolishes NF-kappaB
activation induced by various carcinogens and inflammatory stimuli leading to
suppression of NF-
kappaB-regulated gene expression and up-regulation of apoptosis" J Biol Chem.
279:26287-26299.
Testa et al. (2004) "Interleukin-3 receptor in acute leukemia" Leukemia 18:219-
226
Thomas et al. (2001) "R115777, a farnesyl transferase inhibitor (FTI), has
significant anti-leukemia
activity in patients with chronic myeloid leukemia (CML)" Blood 98
Tijssen (1993) Laboratory Techniques in Biochemistry and Molecular Biology,
Vol. 24:
Hybridization With Nucleic Acid Probes, Elsevier, NY
Toksoz et al. (1994) "Novel human oncogene lbc detected by transfection with
distinct homology
regions to signal transduction products" Oncogene 9:621-628
Valk et al. (2004) "Prognostically useful gene-expression profiles in acute
myeloid leukemia" N.
Engl. J. Med. 350:1617-1628
Van Cutsem et al. (2004) "Phase III trial of gemcitabine plus tipifamib
compared with gemcitabine
plus placebo in advanced pancreatic cancer" J. Clin. Oncol. 22:1430-1438
Yagi et al. (2003) "Identification of a gene expression signature associated
with pediatric AML
prognosis" Blood 102:1849-1856
Zhang et al. (2002) "Farnesyltransferase inhibitors reverse Ras-mediated
inhibition of Fas gene
expression" Cancer Res. 62:450-458
Zheng et al. (1995) "Direct involvement of the small GTP-binding protein Rho
in Ibc oncogene
function" J. Biol. Chem. 270:9031-9034
DEMANDE OU BREVET VOLUMINEUX
LA PRESENTE PARTIE DE CETTE DEMANDE OU CE BREVET COMPREND
PLUS D'UN TOME.
CECI EST LE TOME 1 DE 2
CONTENANT LES PAGES 1 A 71
NOTE : Pour les tomes additionels, veuillez contacter le Bureau canadien des
brevets
JUMBO APPLICATIONS/PATENTS
THIS SECTION OF THE APPLICATION/PATENT CONTAINS MORE THAN ONE
VOLUME
THIS IS VOLUME 1 OF 2
CONTAINING PAGES 1 TO 71
NOTE: For additional volumes, please contact the Canadian Patent Office
NOM DU FICHIER / FILE NAME:
NOTE POUR LE TOME / VOLUME NOTE: