Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
Processes for increasing the yield in plants
Description
The present invention relates to processes for increasing the yield in plants,
recombinant nucleic acid molecules used for these processes, their uses as
well as
to plants with an increased yield.
1n the field of agriculture and forestry constantly efforts are being made to
produce
plants with an increased yield, in particular in order to guarantee the supply
of the
constantly increasing word population with food and to guarantee the supply of
reproducible raw materials. Conventionally, it is tried to obtain plants with
an
increased yield by breeding, which is, however time-consuming and labor-
intensive.
Furthermore, appropriate breeding programs have to be performed for each
relevant
plant species.
Progress has partly been made by the genetic manipulation of plants, that is
by
introducing into and expressing recombinant nucleic acid molecules in plants.
Such
approaches have the advantage of usually not being limited to one plant
species but
being transferable to other plant species. In EP-A 0 511 979, e.g., it was
described
that the expression of a prokaryotic asparagine synthetase in plant cells
inter alia
leads to an increased biomass production.
In WO 96121737, e.g., the production of plants with an increased yield by the
expression of deregulated or unregulated fructose-1,6-bisphosphatase due to
the
increase of the photosynthesis rate is described.
Nevertheless, there still is a need of generally applicable processes for
improving the
yield in plants interesting for agriculture or forestry.
Therefore, the problem underlying the present invention is to provide further
processes for increasing the yield in plants.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
2
This problem is solved according to the invention by providing the embodiments
characterized in the claims.
Therefore, the present invention relates to a process for increasing the yield
in plants,
characterized in that recombinant DNA molecules stably integrated into the
genome
of plants are expressed, comprising
(a) a region allowing the transcription specifically in the companion cells;
and
operatively linked thereto
(b) a nucleotide sequence encoding a polypeptide selected from the group
consisting of:
(i) proteins with an enzymatic activity that cleaves sucrose;
(ii) sucrose transporters;
(iii) proteins the activity of which leads to the stimulation of the proton
gradient located at the plasma membrane of plant cells; and
(iv) citrate synthases (E.C. 4.1.3.7).
It was surprisingly found that the expression of the above-described proteins
specifically in the phloem of plants leads to a dramatic increase in yield.
The term "increase in yield" preferably relates to an increase of the biomass
production, in particular when determined as the fresh weight of the plant.
Such an increase in yield preferably refers to the so-called "sink" organs of
the plant,
which are the organs that take up the photoassimilates produced during
photosynthesis. Particularly preferred are parts of plants which can be
harvested,
such as seeds, fruits, storage roots, roots, tubers, flowers, buds, shoots,
stems or
wood. The increase in yield according to the invention is at least 3 % with
regard to
the biomass in comparison to non-transformed plants of the same genotype when
cultivated under the same conditions, preferably at least 10 % and
particularly
preferred at least 20 %.
The above-described proteins have in common that when they are expressed in
the
phloem their biological activity leads to an increased loading of the phloem
with
photoassimilates.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
3
In the context of the present invention photoassimilates are understood to be
sugars
and/or amino acids.
According to the invention the nucleotide sequence mentioned in (b) can
usually
encode a plant protein or a bacterial protein or a protein originating from
fungi or
animal organisms.
In a preferred embodiment the nucleotide sequence encodes a sucrose synthase
(E.C. 2.4.1.13), preferably a plant sucrose synthase, in particular from
Solanum
tuberosum, and particularly preferred the type expressed in the tubers of S.
tuberosum. Such sequences are, for example, described in Salanoubat and
Belliard
(Gene 60 (1987), 47-56) and are available in the EMBL gene bank under
accession
number X67125.
In a further preferred embodiment the nucleotide sequence encodes a sucrose
phosphorylase (E.C. 2.4.1.7).
Sequences encoding sucrose phosphorylase are, for example, known from
WO 96/24679.
In another preferred embodiment the nucleotide sequence encodes an invertase
(E.C. 3.2.1.26), preferably an invertase from a microorganism, in particular
from a
fungus of the genus Saccharomyces, preferably from S. cerevisiae. Particularly
preferred are sequences encoding a cytosolic invertase (Sonnewald et al.,
Plant J. 1
(1991 ), 95-106).
According to the invention a sucrose transporter is understood to be a
transporter
transporting sucrose in plant systems across a membrane. Such a transporter
preferably is of plant origin (for example EMBL gene bank accession number
G21319). Particularly preferred the sequence described in (b) encodes a
sucrose
transporter from spinach (Spinacia oleracea), in particular with the sequence
of the
clone SoSUT1, as, e.g., described in Riesmeier et al. (EMBO J. 11 (1992), 4705-
4713).
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
4
In a further preferred embodiment the protein that stimulates the proton
gradient
located at the plasma membrane is a proton ATPase.
In this case, the sequence described in (b) preferably encodes a protein from
a
microorganism, in particular a fungus of the genus Saccharomyces, preferably
from
S. cerevisiae.
In a particularly preferred embodiment the sequence encodes the proton ATPase
PMA1 from S. cerevisiae (Serrano et al., Nature 319 (1986), 689-693; EMBL gene
bank) or a version of this proton ATpase from S. cerevisiae which is truncated
at the
3' end, in particular the ATPase ~PAM1 as described in Example 3 of the
present
invention.
Alternatively, the nucleotide sequence can also encode a proton ATPase from
plants,
preferably a proton ATPase from Solanum tuberosum.
Particularly preferred are sequences encoding the proton ATPase PHA2 from
potato
(Harms et al, Plant Mol. Biol. 26 (1994), 979-988; EMBL gene bank X76535) or a
version of this proton ATPase from potato which is truncated at the 3' end, in
particular the ATPase OPHAZ as described in Example 4 of the present
invention.
According to the invention the citrate synthase can be any citrate synthase,
for
example those from bacteria, fungi, animals or plants. DNA sequences encoding
citrate synthase are known, for example, from the following organisms:
Bacillus
subtilis (U05256 and U05257), E. coli (V01501), R. prowazekii (M17149), P.
aeruginosa (M29728), A. anitratum (M33037) (see Schendel et al., Appl.
Environ.
Microbiol. 58 (1992), 335-345 and references cited therein), Haloferax
volcanii
(James et al., Biochem. Soc. Trans. 20 (1992), 12), Arabidopsis thaliana
(217455)
(Unger et al., Plant Mol. Biol. 13 (1989), 411-418), B. coagulans (M74818), C.
burnetti (M36338) (Heinzen et al., Gene 109 (1990), 63-69), M. smegmatis
(X60513),
T. acidophilum (X55282), T. thermophila (D90117), pig (M21197) (Bloxham et
al.,
Proc. Natl. Acad. Sci. USA 78 (1981), 5381-5385), N. crassa (M84187) (Ferea et
al.,
Mol. Gen. Genet. 242 (1994), 105-110), S. cerevisiae (211113, 223259, M14686,
M54982, X00782) (Suissa et al., EMBO J. 3 (1984), 1773-1781 ) and potato (EP
95
91 3066.7).
The numbers in brackets are the corresponding accession numbers in the GenEMBL
data base.
*rB
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
The nucleotide sequences according to the invention can generally encode any
appropriate proteins from any organism, in particular from plants, fungi,
bacteria or
animals. The sequences preferably encode proteins from plants or fungi.
Preferably,
the plants are higher plants, in particular starch or oil storing useful
plants, for
example potato or cereals such as rice, maize, wheat, barley, rye, triticale,
oat, millet,
etc., as well as spinach, tobacco, sugar beet, soya, cotton etc.
The fungi preferably are of the genus Saccharomyces, Schizosaccharomyces,
Aspergillus or Neurospora, in particular Saccharomyces cerevisiae,
Schizosaccharomyces pombe, Aspergillus flavus, Aspergillus niger or Neurospora
crassa.
In a preferred embodiment of the process according to the invention the region
mentioned in (a), which guarantees a companion cell specific transcription, is
the
promoter of the rolC gene from Agrobacterium rhizogenes.
This promoter is, for example, described in Schmulling et al. (Plant Cell
(1989), 665-
671 ) and Kuhn (Characterization and localization of the sucrose carrier SUT1
in
Solanaceae, Doctoral Thesis (1991 }, Freie Universitat Berlin, biology
department).
Preferably, the region of the promoter is used that has the nucleotide
sequence
described in Seq ID No. 1.
Apart from the rolC promoter mentioned above the person skilled in the art can
without further ado use other promoters for a companion cell specific
expression.
Further companion cell specific promoters are described in the literature,
such as the
promoter of the sucrose transporter from Arabidopsis thaliana (Truernit and
Sauer,
Planta 196 (1995), 564-570.
Furthermore, for different RNAs and proteins their specific occurrence in the
companion cells has been described in the literature (see, for example, Foley
et al.,
Plant Mol. Biol. 30 (1996), 687-695; DeWitt, Plant J. 1 (1991 ), 121-128;
Stadler et al.,
Plant Cell 7 (1995), 1545-1554). Starting from a known protein it is possible
for the
person skilled in the art without further ado to isolate the cDNA by means of
antibodies or by using oligonucleotides derived from the amino acid sequence
(cf.,
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
6
e.g., Sambrook et al, Molecular Cloning, A Laboratory Manual, Cold Spring
Harbor
Laboratory Press (1989), Cold Spring Harbor, NY.). Starting from the cDNAs
obtained this way it is furthermore possible to screen established genomic
libraries
from the corresponding organism and to identify genomic fragments. By
comparing
the nucleotide sequence of the cDNA and of the genomic clone the position of
the
promoter can be roughly determined. The specificity of the promoter can be
verified
in a transgenic situation by using chimeric genes consisting of the promoter
and
indicator genes, such as the ~i-glucuronidase (cf., e.g., Kertbundit et al.,
Proc. Natl.
Acad. Sci. USA 88 (1991 ), 5212-5216).
The process according to the invention can in principle be applied to any
plant.
Therefore, monocotyledonous as well as dicotyledonous plant species are
particularly suitable. The process is preferably used with plants that are
interesting
for agriculture, horticulture and/or forestry.
Examples thereof are vegetable plants such as, for example, cucumber, melon,
pumpkin, egg plant, zucchini, tomato, spinach, cabbage species, peas, beans,
etc.,
as well as fruits such as, for example, pears, apples, etc.
Furthermore, oil storing plants are suitable such as, for example, rape,
sunflower,
soya. In a particularly preferred embodiment starch storing plants are
suitable, in
particular such as cereals (rice, maize, wheat, rye, oats, triticale, millet,
barley),
potato, cassava, sweet potato, etc.
The process can also be applied for sucrose storing plants such as, for
example,
sugar beet and sugar cane, but also for other useful plants such as, for
example,
cotton, tobacco, types of wood, wine, hops etc.
The invention further relates to recombinant nucleic acid molecules,
containing
(a) a region allowing the transcription specifically in the companion cells of
plants;
and operatively linked thereto
(b) a nucleotide sequence encoding a polypeptide, selected from the. group
consisting of
(i} sucrose synthases;
(ii) sucrose phosphorylases;
(iii) sucrose transporters;
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
7
(iv) proteins the activity of which leads to the stimulation of the proton
gradient located at the plasma membrane of plant cells; and
(v) citrate synthases.
With regard to the preferred embodiments of such molecules, the same applies
to the
region mentioned in (a) and the nucleotide sequence mentioned in (b) what was
already mentioned above in connection with the process of the invention.
The invention also relates to vectors containing nucleic acid molecules of the
invention, in particular those which are suitable for the transformation of
plant cells as
well as for the integration of foreign DNA into the plant genome.
The present invention further relates to plant cells transformed with a
nucleic acid
molecule of the invention and containing it stably integrated into the genome.
These
cells differ from naturally occurring plant cells for example in that a
nucleic acid
molecule of the invention is integrated into the genome of the cell at a
location where
it does not naturally occur.
The invention further relates to transgenic plants containing plant cells of
the
invention and, due to the expression of the recombinant nucleic acid molecule
integrated into the genome in the companion cells of the plants, showing an
increased yield in comparison with corresponding non-transformed plants that
were
cultivated under the same conditions.
The present invention further relates to propagation material of plants of the
invention
containing the above-described plant cells of the invention. The term
"propagation
material" in particular comprises seeds, fruits, tubers, rhizomes, cuttings,
calli, cell
cultures, etc.
Finally, the present invention relates to the use of recombinant nucleic acid
molecules containing a region allowing the transcription specifically in the
companion
cells of plants and, operatively linked thereto, a nucleotide sequence
encoding a
polypeptide selected from the group consisting of:
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
s
(i) proteins with sucrose cleaving enzymatic activity;
(ii) sucrose transporters;
{iii) proteins the activity of which leads to the stimulation of the proton
gradient
located at the plasma membrane; and
(iv) citrate synthases
for the expression in transgenic plants for increasing the yield.
The encoded proteins preferably are the proteins further described above.
Processes for the transformation of monocotyledonous and dicotyledonous plants
are known to the person skilled in the art.
For the introduction of DNA into a plant host cell a variety of techniques is
available.
These techniques comprise the transformation of plant cells with T-DNA using
Agrobacterium tumefaciens or Agrobacterium rhizogenes as transformation means,
the fusion of protoplasts, the injection, the electroporation of DNA, the
introduction of
DNA by means of the biolistic method as well as further possibilities.
For the injection and electroporation of DNA in plant cells the plasmids do
not have to
fulfill specific requirements. Simple plasmids such as pUC derivatives can be
used.
The use of agrobacteria for the transformation of plant cells has extensively
been
examined and sufficiently disclosed in the specification of EP-A 120 516, in
Hoekema
(In: The Binary Plant Vector System Offsetdrukkerij Kanters B.V., Alblasserdam
(1985), Chapter V), Fraley et al. (Crit. Rev. Plant. Sci. 4, 1-46) and An et
al. (EMBO
J. 4 (1985), 277-287).
For the transfer of the DNA to the plant cell plant explants can be co-
cultivated with
Agrobacterium tumefaciens or Agrobacterium rhizogenes. From the infected plant
material (for example leaf explants, segments of stems, roots but also
protoplasts or
suspension cultivated plant cells) whole plants can be regenerated in a
suitable
medium which may contain antibiotics or biozides for the selection of
transformed
cells. The plants obtained that way can then be examined for the presence of
the
introduced DNA. Other possibilities for the introduction of foreign DNA using
the
biolistic method or by protoplast transformation are known (cf., e.g.,
Willmitzer, t-.,
1993 Transgenic plants. ln: Biotechnology, A Multi-Volume Comprehensive
Treatise
(H.J. Rehm, G. Reed, A. Piihler, P. Stadler, eds.), Vol. 2, 627-659, VCH
Weinheim-
New York-Basel-Cambridge).
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
9
The transformation of dicotyledonous plants via Ti-plasmid-vector systems with
the
help of Agrobacterium tumefaciens is well-established. Recent studies have
indicated
that also monocotyledonous plants can be transformed by means of vectors based
on Agrobacterium (Chan et ai., Plant Mol. Biol. 22 (1993), 491-506; Hiei et
al., Plant
J. 6 (1994), 271-282; Deng et al., Science in China 33 (1990), 28-34; Wilmink
et al.,
Plant Cell Reports 11 (1992), 76-80; May et al., Bio/Technology 13 (1995), 486-
492;
Conner and Domisse; Int. J. Plant Sci. 153 (1992), 550-555; Ritchie et al.,
Transgenic Res. 2 (1993), 252-265).
Alternative systems for the transformation of monocotyledonous plants are the
transformation by means of the biolistic method (Wan and Lemaux, Plant
Physiol.
104 (1994), 37-48; Vasil et al., BiolTechnology 11 (1993), 1553-1558; Ritala
et al.,
Plant Mol. Biol. 24 (1994), 317-325; Spencer et al., Theor. Appl. Genet. 79
(1990},
625-631 ), the protoplast transformation, the electroporation of partially
permeabilized
cells, as well as the introduction of DNA by means of glass fibers.
In particular the transformation of maize is described in the literature
several times
(cf., e.g., W095106128, EP 0 513 849; EP 0 465 875; Fromm et al.,
Biotechnology 8
(1990), 833-844; Gordon-Kamm et al., Plant Cell 2 (1990), 603-618; Koziel et
al.,
Biotechnology 11 (1993), 194-200). In EP 292 435 and in Shillito et al.
(Bio/Technology 7 (1989), 581 ) a process is described with the help of which
and
starting from a mucus-free, soft (friable) maize callus fertile plants can be
obtained.
Prioli and Sandahl (BioITechnology 7 (1989), 589) describe the regenerating
and
obtaining of fertile plants from maize protoplasts of the Cateto maize inbred
line Cat
100-1.
The successful transformation of other cereal species has also been described,
for
example for barley (Wan and Lemaux, see above; Ritala et al., see above) and
for
wheat (Nehra et al., Plant J. 5 (1994), 285-297).
Once the introduced DNA has been integrated into the genome of the plant cell,
it
usually is stable there and is also contained in the progenies of the
originally
transformed cell. It usually contains a selection marker which makes the
transformed
plant cells resistant to a biozide or an antibiotic such as kanamycin, G 418,
bieomycin, hygromycin or phosphinotricin and others. Therefore, the
individually
chosen marker should allow the selection of transformed cells from cells
lacking the
introduced DNA.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
The transformed cells grow within the plant in the usual way (see also
McCormick et
al., Plant Cell Reports 5 (1986), 81-84). The resulting plants can be cultured
normally. Seeds can be obtained from the plants.
Two or more generations should be cultivated to make sure that the phenotypic
feature is maintained stably and is transmitted. Seeds should be harvested to
make
sure that the corresponding phenotype or other properties are maintained.
Figure 1 schematically shows the construction of the plasmid pBinRoIC-SS.
Figure 2a shows the analysis of the sucrose synthase {SS) activity in leaves
of
transgenic potato plants which had been transformed with the RoiC-SS
construct. The enzyme activity was determined according to Zrenner et
al. (Plant J. 7 (1995), 97-107). The activity is indicated in wmol hexose
equivalentsl(min x g fresh weight).
The columns represent the average values of three samples per
genotype. The standard deviation is also indicated.
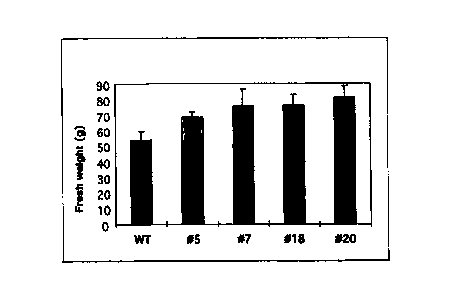
Figure 2b shows the analysis of the tuber yield of transgenic potato plants
which
had been transformed with the RoIC-SS construct. The columns
represent average values of ten to fifteen plants per genotype. The
standard deviation is also indicated. The tuber yield is indicated in g per
fresh weight.
Figure 2c shows the analysis of the tuber starch of transgenic potato plants
that
had been transformed with the RoIC-SS construct. For this purpose
tubers harvested from ten to fifteen plants per genotype were collected
and the starch content of the tubers was determined according to Von
Scheele et al. (Landw. Vers. Sta. 127 (1937), 67-96).
Figure 3 schematically shows the construction of the plasmid pBinRoIC-Suc2.
Figure 4 schematically shows the construction of the plasmid pBinRoIC-OPMA1 .
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
11
Figure 5 schematically shows the cloning strategy of oPMA1 .
Step from A to B:
The H'-ATPase OPMA1, which was truncated at the 3' end, was
amplified via PCR with the PMA1 cDNA as the matrix and
complementary internal primers (A). The flanking cleavage sites of the
PCR product (B) were introduced via the correspondingly synthesized
primers.
Step from B to C:
PstI/Notl digestion and cloning of the PCR fragment into the E. coli
vector SK- via PstI/Notl cleavage sites (C).
Step from C to D:
BcII/Spel digestion of the plasmid SK-OPMA1 and cloning of the
fragment into the compatible BamHIIXbaI cleavage sites of pBinRoIC
{D)
Figure 6 shows the results of the polymerase chain reaction with specific
oligonucleotides indicating the stable integration of oPMA1 in the
genome of transgenic plants which had been obtained by
transformation with the rolC-OPMA2 construct. Size of the PCR product
= 730 bp; WT = wildtype; M = marker.
Figure 7 schematically shows the construction of the plasmid pBinRoIC-~PHA2.
Figure 8 schematically shows the cloning strategy of OPHA2 .
Step from A to B:
The H''-ATPase ~PHA2, which was truncated at the 3' end, was
amplified via PCR with the PHA2 cDNA as the matrix and
complementary internal primers (A). The flanking cleavage sites of the
PCR product (B) were introduced via the correspondingly synthesized
primers.
Step from B to C:
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
12
PstI/EcoRl digestion and cloning of the PCR fragment into the E. coli
vector SK- via PstI/EcoRl cleavage sites (C).
Step from C to D:
BgIIIISpeI digestion of the plasmid SK-~PHA2 and cloning of the
fragment into the compatible BamHI/Xbal cleavage sites of pBinRoIC
(D)
Figure 9 shows the results of the polyermase chain reaction with specific
oligonucleotides indicating the stable integration of OPHA2 in the
genome of transgenic plants which had been obtained by
transformation with the roIC-dPHA2 construct. Size of the PCR product
= 758 bp; WT = wildtype; M = marker.
Figure 10 schematically shows the construction of the plasmid pBinRoIC-SoSUT1.
Figure 11 schematically shows the construction of the plasmid pBinRoIC-CiSy.
Figure 12 shows the results of the determination of the sucrose content in
parenchymatic samples of tubers of engrafted potato plants enriched
with vascular tissue. The genotypes used for engrafting are the lines
RoIC-Suc2-#25 (cytosolic invertase) and wildtype Solanum tuberosum
var. Desiree. The sucrose content was determined according to Stitt et
al. (Methods Enzymol. 174 (1989), 518-522). The columns represent
the average values of 12 samples per engrafted type. The standard
deviation is indicated. The sucrose content is indicated as ~mol hexose
equivalents/g fresh weight.
Figure 13 shows the analysis of phloem exudates of OPMA1 leaves which were
kept under light for six hours in a 14C02 atmosphere. The sucrose
content was determined according to Stitt et al. (loc. city. The columns
represent the average values of four to five samples per genotype. The
standard deviation is indicated.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
13
Figure 14 shows the tuber yield (in gram fresh weight) of oPMA1 plants. The
columns represent the average values of six plants per genotype. The
standard deviation is indicated. The tuber yield is indicated in g fresh
weight.
Figure 15 shows the tuber yield (in gram fresh weight) of OPHA2 plants. The
columns represent average values of four to five plants per genotype.
The standard deviation is indicated. The tuber yield is indicated in g
fresh weight.
The following examples illustrate the invention.
Example 1
Production of the plasmid pBinRoIC-SS and production of transgenic potato
plants
The plasmid pBinRoIC-SS contains the three fragments A, B and C in the binary
vector pBin19 (Bevan, Nucl. Acids Res. 12 (1984), 8711 ) (cf. Fig.1 ).
The fragment A comprises the rolC promoter from Agrobacterium rhizogenes. The
rolC promoter contains as an EcoRl/Asp718 DNA fragment of 1138 by (Lerchl et
al.,
Plant Cell 7 (1995), 259-270) the DNA region (position: 11306 to position
12432) of
the TL-DNA of the Ri-agropin-type plasmid from A. rhizogenes (Slightom et al.,
J.
Biol. Chem. 261 (1986), 108-121 ). The fragment A is inserted into the EcoRl
and
Asp718 cleavage sites of the polylinker of pBin19.
The fragment B contains the coding region (position: 76 to position 2493) of
the
cDNA of the sucrose synthase (SS) from Solanum tuberosum (Salanoubat and
Belliard, Gene 60 {1987), 47-56). The fragment B was obtained as BamHl
fragment
of 2427 by from the vector pBluescript SK-, in which it is inserted into the
BamHl
cleavage site of the polylinker. The fragment B was inserted in sense
orientation in
the vector pBin19 into the BamHl cleavage site, that is downstream of the rolC
promoter in an orientation allowing the transcription of a translatable RNA.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
14
The fragment C contains the polyadenylation signal of the Gene 3 of the T-DNA
of
the Ti plasmid pTi ACH 5 (Gielen et al., EMBO J. 3 (1984), 835-846), in
particular the
nucleotides 11749-11939, which was isolated as a PvuIIIHindlll fragment from
the
plasmid pAGV 40 (Herrera-Estrella et al., Nature 303 (1983), 209-213) and
which
upon addition of Sphl linkers was cloned into the Pvull cleavage site between
the
Sphl and the Hindlll cleavage site of the polylinker of pBin19.
The resulting plasmid pBinRoIC-SS was introduced into potato plant cells via
the
gene transfer mediated by Agrobacterium tumefaciens. For this purpose ten
small
leaves of a potato sterile culture (Solanum tuberosum L. cv. Desiree) wounded
with
the scalpel were put into 10 ml MS medium (Murashige and Skoog, Physiol.
Plant. 15
(1962)), 473 with 2% of sucrose containing 50 ~,1 of a Agrobacterium
tumefaciens
overnight culture grown under selection. After 3 to 5 minutes of gentle
shaking a
further incubation followed for two days in the dark. Then the leaves were put
on MS
medium with 1.6 % glucose, 5 mgll naphtyl acetic acid, 0.2 mgll
benzylaminopurin,
250 mgll claforan, 50 mg/l kanamycin and 0.8 % bacto-agar for callus
induction. After
an incubation of one week at 25 °C and 3000 lux the leaves were put on
MS medium
with 1.6 % glucose, 1.4 mg/l zeatin ribose, 20 p.gll naphtyl acetic acid, 20
p.gll
giberellic acid, 250 mg/l claforan, 50 mgll kanamycin and 0.8 % bacto-agar.
The analysis of the leaves of a number of plants transformed with this vector
system
unambiguously indicated the presence of an increased sucrose synthase
activity.
This is a result of the expression of the sucrose synthase gene from potato
contained
in pBinRoIC-SS (cf. Figure 2a).
The analysis of the tuber yield (tuber fresh weight in gram) of plants
transformed with
this vector system and showing an increased sucrose synthase activity
unambiguously showed an increased tuber yield. This is also a result of the
expression of the sucrose synthase gene from potato contained in pBinRoIC-SS
(cf.
Figure 2b).
The starch content of potato tubers is linearly dependent on the density of
the tubers
(von Sch~ele et al., Landw. Vers. Sta. 127 (1937), 67-96). The analysis of the
density
of transgenic tubers of plants which had been transformed with the vector
system
pBinRoIC-SS having an increased sucrose synthase activity surprisingly showed
an
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
increased starch content. This is a result of the expression of the sucrose
synthase
gene from potato contained in pBinRoIC-SS (cf. Figure 2c).
Example 2
Production of the ptasmid pBinRolC-Suc2 and production of transgenic potato
plants
The plasmid pBinRoIC-Suc2 contains the three fragments A, B and C in the
binary
vector pBin19 (Bevan, loc. cit.) and is illustrated in Figure 3.
The fragments A and C correspond to the fragments A and C as described in
Example 1.
The fragment B contains the coding region (position: 845 to position: 2384) of
the
gene of the cytosolic invertase from yeast (Saccharomyces cerevisiae). The
fragment
B was obtained as a BamHl fragment with a length of 1548 by from the vector
pBluescript SK' in which it is inserted in the BamHl cleavage site of the
polylinker.
The fragment B is inserted in sense orientation into pBin19 in the BamHl
cleavage
site.
The plasmid pBinRolC-Suc2 was introduced into potato plant cells via gene
transfer
mediated by Agrobacterium. From transformed cells whole plants were
regenerated.
Such plants show in comparison to non-transformed plants an increased yield
(increased biomass).
Example 3
Production of the plasmid pBinRoIC-OPMA1 and production of transgenic
potato plants
The plasmid pBinRoIC-OPMA1 contains the three fragments A, B and C in the
binary
vector pBin19 (Bevan, ioc. cit.) and is schematically illustrated in Figure 4.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
16
The fragments A and C correspond to the fragments A and C as described in
Example 1.
The fragment B contains the coding region (position: 937 to position: 3666) of
the
gene of the proton ATPase PMA1 from the yeast Saccharomyces cerevisiae
(Serrano et al., Nature 319 (1986), 689-693). The fragment B was obtained by
means of polymerase chain reaction (PCR). For this purpopse the 3' end of the
coding region of the gene PMA1 was truncated on purpose by 27 by and at the
same
time a necessary new stop codon was introduced. The DNA fragment modified this
way was called oPMA1. The fragment B was inserted, as a BcII/Spel fragment
with a
length of 2739 bp, in sense orientation into the BamHl (compatible insertion
site for
Bcll restriction sites) and Xbal (compatible insertion site for Spel
restriction sites)
cleavage sites of the vector pBin19.
The fragment B was obtained as BcII/Spel fragment from the vector pBluescript
SK-
in which it is inserted via the cleavage sites Notl and Pstl of the polylinker
(cf. Fig. 5).
The plasmid pBinRoIC-OPMA1 was introduced into potato plant cells via the gene
transfer mediated by Agrobacterium. Whole plants were regenerated from
transformed cells.
The stable integration of OPMA1 in the genome of transgenic plants which had
been
obtained by using the vector system pBinRolC-OPMA1 was detected by means of
polymerase chain reaction (PCR) (cf. Fig. 6).
The transformed plants show an increased yield (increased biomass) in
comparison
to non-transformed plants (see Figures 13 and 14).
Example 4
Production of the plasmid pBinRoIC-~PHA2 and production of transgenic
potato plants
The plasmid pBinRoIC-OPHA2 contains the three fragments A, B and C in the
bihary
vector pBin19 (Bevan, loc. cit.) and is schematically illustrated in Figure 7.
The fragments A and C correspond to the fragments A and C as described in
Example 1.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
17
The fragment B contains the coding region (position: 64 to position: 2672) of
the
cDNA of the proton-ATPase PHA2 (Harms et al., Plant Mol. Biol. 26 (1994), 979-
988). The fragment B was obtained by means of polymerase chain reaction (PCR).
For this purpose the 3' end of the coding region of the gene PHA2 was on
purpose
truncated by 249 bp, and at the same time two new stop codons were introduced.
The DNA fragment modified that way was called oPHA2. The fragment B was
inserted in sense orientation as a BgIII/Spel fragment with a length of 2631
by into
the BamHl (compatible insertion site for Bglll restriction sites) and Xbal
(compatible
insertion site for Spel restriction sites) cleavage sites of the vector
pBin19.
The fragment B was obtained as BgIII/Spel fragment from the vector pBluescript
SK',
in which it is inserted into the EcoRl and Pstl cleavage sites of the
polylinker
sequence (cf. Fig. 8: cloning strategy ~PHA2).
The plasmid pBinRoIC-~PHA2 was introduced into potato plant cells via the gene
transfer mediated by Agrobacterium. Whole plants were regenerated from
transformed cells.
The stable integration of OPHA2 in the genome of transgenic plants which had
been
obtained using the vector system pBinRolC-OPHA2 was detected by means of
polymerase chain reaction (PCR) (cf. Fig. 9).
The transformed plants show an increased yield (increased biomass) in
comparison
to non-transformed plants (see Figure 15).
Example 5
Production of the ptasmid pBinRotC-SoSUT1 and production of transgenic
potato plants
The plasmid pBinRoIC-SoSUTI contains the three fragments A, B and C in the
binary vector pBin19 (Bevan, loc. cit.) and is schematically illustrated in
Figure 10.
The fragments A and C correspond to the fragments A and C as described in
Example 1.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
18
The fragment B contains the cDNA (position: 1 to position: 1969) encoding a
sucrose
transporter from spinach (Spinacia oleracea} (Riesmeier et al., EMBO J. 11
(1992),
4705-4713; accession number X67125 and S51273). The fragment B was obtained
as a Notl fragment from the vector pBluescript SK-, in which it is inserted
via a Notl
linker sequence. For the cloning into the Smal cleavage site of the vector
pBin19 the
sticky ends of the fragment resulting from the Notl digestion were converted
to blunt
ends and inserted in sense orientation into the pBin19. The resulting plasmid
was
called pBinRoIC-SoSUT1.
It was introduced into potato plant cells via the gene transfer mediated by
Agrobacterium. Whole plants were regenerated from transformed cells.
Plants transformed that way show an increased yield (increased biomass) in
comparison to non-transformed plants.
Example 6
Production of the plasmid p8inRoIC-CiSy and production of transgenic potato
plants
The plasmid pBinRoIC-CiSy contains the three fragments A, B and C in the
binary
vector pBin19 {Bevan, Nucl. Acids Res. 12 {1984), 8711 ) modified according to
Becker (Nucl. Acids Res. 18 {1990), 203) (cf. Fig. 11 ).
The fragment A comprises the rolC promoter from Agrobacterium rhizogenes. The
rolC promoter contains as an EcoRI/Asp718 DNA fragment with a length of 1143
by
(Lerchl et al., The Plant Cell 7 (1995), 259-270) the DNA region (position:
11306 to
position 12432) of the TL-DNA of the Ri-agropin type plasmid from A.
rhizogenes
(Slightom et al., J. Biol. Chem. 261 (1986), 108-121 ). The fragment A is
inserted in
the EcoRl and Asp718 cleavage sites of the polylinker of pBin19.
The fragment B contains the coding region of the cDNA of the citrate synthase
(CiSy)
from the fission yeast Saccharomyces cerevisiae. The fragment B was obtained
as a
BamHl fragment with a length of 1400 by from the vector pBluescript SK-, in
which it
is inserted in the BamHl cleavage site of the polylinker (Landschutze, Studies
on the
influence of the acetyl-CoA synthesis and use in transgenic plants, Doctoral
Thesis,
Freie Universit~t Berlin, {1985) D83/FB15 No. 028).
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
19
The fragment C contains the polyadenylation signal of the gene 3 of the T-DNA
of the
Ti plasmid pTiACH 5 (Gielen et al., EMBO J. 3 (1984), 835-846), nucleotides
11749-
11939, which had been isolated as a PvulUHindlll fragment from the plasmid
pAGV
40 (Herrera-Estrella et al., Nature 303 (1983), 209-213) and which after
addition of
Sphl linkers to the Pvull cleavage site had been cloned between the Sphl and
Hindlll
cleavage site of the polylinker of pBin19.
The plasmid pBinRoIC-CiSy has a length of about 13 kb.
The plasmid pBinRoIC-CiSy was inserted into potato plants via the gene
transfer
mediated by Agrobacterium tumefaciens. Whole plants were regenerated from
transformed cells.
The analysis of a number of plants transformed with this vector system
unambiguously showed an increased biomass, which is a result of the expression
of
the CiSy cDNA from yeast contained in pBinRoIC-CiSy.
Example 7
Grafting experiment
For grafting the shoot of a receiver plant is replaced with the shoot of a
donor plant.
In this experiment the shoot of a transgenic plant (RoIC-Suc2 #25) is grafted
onto the
stock of a wildtype plant (Solarium tuberosum, var. Desiree). In a control
experiment
a wildtype shoot is grafted onto a wildtype stock in order to rule out
culturing
differences in the experiments (autografting). The aim of the experiment is to
examine the exclusive impact of the photosynthetic activity and
photoassimilate
distribution of a transgenic shoot on organs (in this case tubers) of a
wildtype stock.
Potato plants were transferred from a tissue culture to soil and placed into a
greenhouse. After approx. five weeks (the plants have not yet induced tuber
production at this stage) the plants are grafted. For this purpose the shoot
of the
receiver plant which is not needed is cut off, and a wedge is cut into the
stem of the
receiver plant. The donor shoot to be grafted is cut at the stem end in the
appropriate
way and is inserted into the wedge of the receiver plant. The grafting site is
fixed with
an adhesive tape.
CA 02299388 2000-02-04
WO 99/07863 PGT/EP98/04878
Then the grafted potato plants are kept under increased air humidity and in
shadow
for approx. one week. Within seven to ten days they are step by step adapted
to
normal greenhouse conditions. At this stage the plants are seven weeks old.
All leaves of the receiver plant are now removed, and the stem is covered from
light
with an aluminum sheet in order to guarantee that exclusively the
photosynthetic
activity and the photoassimilate distribution of the donor shoot nourishes the
stock of
the grafted plant.
The plants are kept in the greenhouse until the potato tubers are harvested
approx.
two months after the grafting and approx. three months after the planting into
soil.
The results of such a grafting experiment are illustrated in Figure 12.
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
21
Sequence Listing
(1) GENERAL INFORMATION:
(i) APPLICANT:
(A) NAME: Max-Planck-Gesellschaft zur Foerderung der
Wissenschaften e.V
(B) STREET: none
(C) CITY: Berlin
(E) COUNTRY: Germany
(F) POSTAL CODE: none
(ii) TITLE OF INVENTION: Process for increasing the yield in plants
(iii) NUMBER OF SEQUENCES: 1
(iv) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(H) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D} SOFTWARE: PatentIn Release #1.0, Version #1.30 (EPA)
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1138 base pairs
(B) TYPE: nucleotide
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: Genomic DNA
(iii) HYPOTHETICAL: NO
(iv) ANTISENSE: NO
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Agrobacterium rhizogenes
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 1:
AATTCGATACGAAAAAGGCAAGTGCCAGGG CCATTTAAAA TACGGCGTCGGAAACTGGCG 60
CCAATCAGACACAGTCTCTGGTCGGGAAAG CCAGAGGTAG TTTGGCAACAATCACATCAA 120
GATCGATGCGCAAGACACGGGAGGCCTTAA AATCTGGATC AAGCGAAAATACTGCATGCG 180
TGATCGTTCATGGGTTCATAGTACTGGGTT TGCTTTTTCT TGTCGTGTTGTTTGGCCTTA 240
GCGAAAGGATGTCAAAAAAGGATGCCCATA ATTGGGAGGA GTGGGGTAAAGCTTAAAGTT 300
GGCCCGCTATTGGATTTCGCGAAAGCGGCA TTGGCAAACG TGAAGATTGCTGCATTCAAG 360
ATACTTTTTCTATTTTCTGGTTAAGATGTA AAGTATTGCC ACAATCATATTAATTACTAA 420
CATTGTATATGTAATATAGTGCGGAAATTA TCTATGCCAA AATGATGTATTAATAATAGC 480
CA 02299388 2000-02-04
WO 99/07863 PCT/EP98/04878
22
AATAATAATATGTGTTAATCTTTTTCAATCGGGAATACGTTTAAGCGATT ATCGTGTTGA540
ATAAATTATTCCAAAAGGAAATACATGGTTTTGGAGAACCTGCTATAGAT ATATGCCAAA600
TTTACACTAGTTTAGTGGGTGCAAAACTATTATCTCTGTTTCTGAGTTTA ATAAAAAATA660
AATAAGCAGGGCGAATAGCAGTTAGCCTAAGAAGGAATGGTGGCCATGTA CGTGCTTTTA720
AGAGACCCTATAATAAATTGCCAGCTGTGTTGCTTTGGTGCCGACAGGCC TAACGTGGGG780
TTTAGCTTGACAAAGTAGCGCCTTTCCGCAGCATAAATAAAGGTAGGCGG GTGCGTCCCA840
TTATTAAAGGAAAAAGCAAAAGCTGAGATTCCATAGACCACAAACCACCA TTATTGGAGG900
ACAGAACCTATTCCCTCACGTGGGTCGCTAGCTTTAAACCTAATAAGTAA AAACAATTAA960
AAGCAGGCAGGTGTCCCTTCTATATTCGCACAACGAGGCGACGTGGAGCA TCGACAGCCG1020
CATCCATTAATTAATAAATTTGTGGACCTATACCTAACTCAAATATTTTT ATTATTTGCT1080
CCAATACGCTAAGAGCTCTGGATTATAAATAGTTTGGATGCTTCGAGTTA TGGGGTAC1138