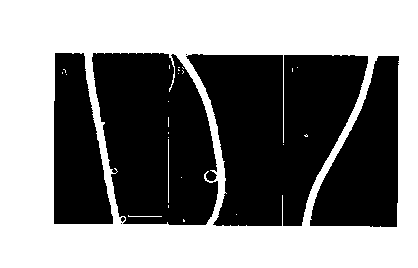Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02301257 2000-03-24
SPECIFICATION
The IRE gene regulating the root-hair growth in Arabidopsis
BACKGROUND OF THE INVENTION
Field of the Invention
The present invention relates to a gene coding for a
protein that is involved in regulation of tip growth in plants,
and also to the promoter of the gene.
Prior Art
Unlike other cells, it is known that root hairs of plants
elongate in a particular manner called tip growth. Pollen tubes
of higher plants also perform tip growth. Tip growth is also
observed in those eukaryotic cells that hold cell walls in ferns,
mosses, algae, fungi and so forth. A number of mutants that
show abnormalities in the number or shape of root hairs have
been isolated from Arabidopsis. hy5, phyB, rhd2 and tipl are
known as mutants with abnormal root-hair lengths (Genes &
Development, Vol. 11, p. 2983-2995 (1997); Plant Cell, Vol. 5, p.
147-157 (1993); Plant Cell, Vol. 2, p. 235-243 (1990); Plant
Physiology, Vol. 103, p. 979-985 (1993)). These mutants except
tipl show various phenotypes in addition to root-hair elongation,
suggesting that roles of these genes are not restricted just to
root hairs or to tip growth. It is therefore unclear how these
genes are involved in root-hair elongation. Since the tipl
mutant shows abnormalities in elongation both of root hairs and
1
CA 02301257 2000-03-24
pollen tubes, tipl is considered as a tip growth-impaired mutant.
The molecular function of this gene product has not yet been
revealed. As described above, those genes that specifically
function in regulation of the root-hair elongation or tip growth
have remained to be seen.
SUMMARY OF THE INVENTION
The present invention aims to isolate a novel gene
regulating tip growth such as root-hair elongation and to use
the gene for providing plants that show altered tip growth rates.
The present inventors have made great efforts to solve the
above mentioned problems and isolated a short root-hair mutant
of Arabidopsis. They have cloned the gene responsible for the
phenotype. Furthermore, the cloned gene has been introduced
into plants and the effect of the gene has been confirmed. Thus,
the present invention has been completed.
The first aspect of the present invention relates to a
gene coding for a protein (a) or (b) mentioned below:
(a) a protein represented by the amino acid sequence as set
forth in SEQ ID NO.: 2; or
(b) a protein represented by an amino acid sequence having one
or more amino acids deleted from, substituted in, modified in or
added to the amino acid sequence as set forth in SEQ ID NO.: 2,
and having a tip growth regulating activity.
The second aspect of the present invention relates to a
DNA (a) or (b) as mentioned below:
(a) a DNA represented by the nucleotide sequence as set forth in
SEQ ID NO.: 3; or
2
CA 02301257 2000-03-24
(b) a DNA represented by a nucleotide sequence having one or
more nucleotides deleted from, substituted in or added to the
nucleotide sequence as set forth in SEQ ID NO.: 3, and having a
function as a promoter.
This specification includes part or all of the contents as
disclosed in the specification and/or drawings of Japanese
Patent Application No.82402/1999, which is a priority document
of the present application.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 shows root hairs of Arabidopsis: wild type (A),
the ire mutant (B), and an ire mutant in which the wild-type IRE
gene is introduced (C).
Figure 2 shows elongation properties of root hairs of wild
type (A) and the ire mutant (B).
Figure 3 shows a physical map of the genome around the IRE
locus.
Figure 4 shows the exon regions of the IRE gene.
Figure 5 shows expression patterns of the GUS gene under
the control of the IRE promoter.
Figure 6 shows the nucleotide sequence of the IRE promoter.
DESCRIPTION OF THE INVENTION
The present invention is hereinafter described in detail.
(1) First Invention (Structural Gene)
The first invention relates to a gene coding for a protein
(a) or (b) as mentioned below and encompasses the protein
encoded by the gene, mutant genes, organisms carrying these
3
CA 02301257 2000-03-24
genes:
(a) a protein represented by the amino acid sequence as set
forth in SEQ ID NO.: 2; or
(b) a protein represented by an amino acid sequence having one
or more amino acids deleted from, substituted for, modified in
or added to the amino acid sequence as set forth in SEQ ID NO.:
2, and having a tip growth regulating activity.
The term "one or more" used herein means the number of
amino acids which can be deleted or otherwise altered according
to the technology conventionally used at the time of filing the
present application such as the site-directed mutagnenesis.
Chemical modification of amino acid residues in a protein
is commonly carried out in the art: see, for example, Hirs,
C.H.W. and Timasheff, S.N., eds, (1977) Methods in Enzymology,
Vol. 47, p. 407-498, Academic Press, New York.
The gene coding for the amino acid sequence as set forth
in SET ID No.: 2 modifies the elongation of root hairs. This
is shown by the fact that the mutant impaired this gene displays
shorter root hairs. This effect is restricted in root hairs
because the mutant shows the normal elongation of root hair-
bearing cells. Thus we have named this gene IRE (Incomplete
Root-hair Elongation). No gene products that have a tip growth
specific function have not been found.
The IRE gene can be cloned according to the method of
Japanese Patent Application Laying Open No. 10-4978 as described
below.
First, genomic DNA of Arabidopsis is fragmented with an
appropriate restriction enzyme and the DNA fragments are ligated
4
CA 02301257 2000-03-24
into an appropriate vector for a genomic DNA library. This
library is amplified by transforming into a host microorganism.
The isolation of DNA may be carried out in a conventional method
using cesium chloride and ethidum bromide. The restriction
enzyme is not particularly limited and may include, for example,
Sau3AI. The vector is also not particularly limited and may
include, for example, a DASH II. The host microorganism may be
determined depending upon the vector used. For example, 1 Dash
II requires Escherichia coli XL-1 Blue MRA (P2).
Then, the above mentioned genomic DNA library is screened
for the genomic region of the IRE locus. A DNA fragment that is
adjacent to the T-DNA insertion in the ire mutant is used for
the probe. Since the ire gene is disrupted by the insertion in
the genome of the ire mutant, the adjacent region includes a
part of the IRE gene.
Positive clones selected by the above mentioned probe are
purified and the insert genomic DNA fragments are separated. A
genomic fragment is used for a probe to screen a cDNA library.
The cDNA library may be prepared according to a conventional
procedure. That is, the total RNA is isolated from plant
tissues and polyA+ RNA is purified using oligo(dT) resins. cDNA
is synthesized from the template polyA+ using a reverse
transcriptase. The synthesized cDNA is ligated into an
appropriate vector for a cDNA library. The library is amplified
by transforming into a host microorganism. Positive clones in
the cDNA library contain the IRE gene.
Capabilities of the hybridization between one DNA sequence
and others under stringent conditions may be determined, for
CA 02301257 2000-03-24
example, by the following method. First one DNA fragment is
blotted and immobilized onto a nylon membrane. Then, this
membrane is prehybridized by the solution containing 6X SSC,
O.O1M EDTA, 5X Denhardt's solution, 0.5% SDS, 100 ~g/ml
denatured salmon DNA at 65~C for one hour or more. A hybridized
solution is prepared by adding labeled DNA fragments or labeled
RNA transcripts of a DNA template in the prehybidization
solution. The prehybridized nylon membrane is hybridized in the
hybridization solution at 65 ~C for 3 to 16 hours. Thereafter,
it is washed in a solution containing 2X SSC and 0.1% SDS at 65
'C for 30 minutes. Then, it is further washed in 2X SSC, 0.1%
SDS solution at room temperature for 15 minutes. Further, it
washed in 0.1X SSC, 0.1% SDS solution at 65 ~C for 30 minutes.
Then, the signal on the membrane is detected by a suitable
method depending the labeling reagent.
In order to express genes of the first invention (the IRE
gene and modified genes) in a host, the genes may be inserted
into an appropriate expression vector and the recombinant vector
may be introduced into the host. Hosts are not particularly
limited. Plants are preferable but bacteria, yeast and animals
may be used. Species of plants used as the host are not
particularly limited but those species of soybean, rape, and
cotton may be preferable. Any expression vector may be used as
far as it contains both a promoter for the expression and a
marker gene that enable the handling the host. For example,
plant tissues preferably require vectors that carry the
cauliflower mosaic virus 35S promoter because of the wide
application of species. pBI121 vector (Clontech) is an example.
6
CA 02301257 2000-03-24
Methods to introduce a vector into the host are not particularly
limited and may be done depending upon the host organism. An
Agrobacterium-mediated method is preferable for introducing into
plant tissues although those methods using electroporation,
particle gun may also be applicable.
Since the gene of the first invention has a function to
modify tip growth such as elongation or root hairs, the
following utilities may be considered.
I) Applications using the tip-growth modifying activity in root
hairs.
Introducing the gene of the first invention into a plant
and expressing it therein can let the plant have root hairs with
different lengths which can provide drought resistance of the
plant. Bacteria in Rhizobium are parasites of leguminous plants
and they enable the plants nitrogen fixation. Those bacteria
are shown to infect only elongating root hairs. Thus plants
that have altered root-hair growth by using the gene can be
expected to improve the infectious efficiency of the bacteria.
II) Applications using the tip-growth modifying activity in
other than root hairs.
In higher plants, pollen tubes emerge from pollen grains
(male gametophytes) and elongate to female gametophytes while
the fertilization. It is expected that the manipulation of the
gene expression in pollen tubes may alter the fertilization
timing and efficiency. Namely, plants (male gametophytes) with
either promoting or retarding fertilization ability may be
created. Cotton fibers, which are the raw material of the
common fiber, cotton yarn, elongate with a kind of tip growth in
7
CA 02301257 2000-03-24
cotton fruits. Thus it is also expected to improve the quality
of cotton fiber.
(2) Second Invention (Promoter)
The second invention relates to a DNA (a) or (b) as
mentioned below and encompasses promoters, expression vectors,
organisms carrying these DNA sequences:
(a) a DNA represented by the base sequence as set forth in SEQ
ID NO.: 3; or
(b) a DNA represented by a base sequence having one or more
bases deleted from, substituted for or added to the base
sequence as set forth in SEQ ID NO.: 3, and having a function as
a promoter.
The term "one or more" used herein means the number of
nucleotide sequences which can be deleted or otherwise altered
according to the technology conventionally used at the time of
filing the present application such as the site-directed
mutagnenesis.
Since the DNA of the second invention exists in the
upstream region of the gene of the first invention, it may be
cloned according to the method of Japanese Patent Application
Laying Open No. 10-4978 as in the case of the gene of the first
invention.
The DNA of second invention function as a promoter in such
tip-growing tissues as root hairs and pollen grains (pollen
tubes). Recombinant genes that have any gene under this DNA
sequence can be expressed specifically in root hairs and/or
pollen grains. Thus useful biological functions depending upon
8
CA 02301257 2000-03-24
root hairs and/or pollen grains can be introduces.
EXAMPLES
The present inventions are illustrated in more detailed by
way of examples which in no way limit the scope of the present
invention.
Example 1: Production of transgenic plants for the isolation of
IRE
Transgenic plants for the isolation of IRE were created
according to the method of Example 1 of Japanese Patent
Application Laying Open No. 10-4978.
Seeds of Arabidopsis (variety: Wassilewskija [WS]) were
sterilized and sown on an agar medium. These seeds were
incubated at 4 'C in darkness for two to four days. This cold
treatment prompted breakage of seed dormancy and made the
germination uniform and enhanced the flowering timing.
Thereafter, seeds were germinated and grown under continuous
light at 22 ~C. The seed sterilization was carried out by using
a sterilization solution containing 10 ~ of Hitar (Kao) and
0.02 Triton-X 100. Seeds were mixed and vortexed in the
sterilization solution and allowed to stand at room temperature
for three to five minutes, and washed five times with sterilized
water. The agar medium contained 1/2 x Arabidopsis nutrient
salt solution and 1.5 ~ agar (Nakarai, special grade). lx
Arabidopsis nutrient salts solution was prepared by following:
985 ml distilled water or deionized water, 5 ml of 1M KN03, 2 ml
of 1M MgS04, 2 ml of 1M Ca(N03)2, 2.5 ml of 20mM Fe-EDTA, 1 ml of
trace element solution, 2.5 ml of K-P04 buffer (pH 5.5)]
9
CA 02301257 2000-03-24
(Hideaki Shiroishi et al., (1991), Gendai-Kagaku vo120, Plant
Biotechnology II, p. 38). The agar medium was autoclaved and
poured into petri dishes. This concentration of agar prevented
Arabidopsis roots from penetrating in the agar medium. It made
he observation of the root morphology easier. The light source
was built with two commonly used 40 W fluorescent lamps and one
Homolux fluorescent lamp (National). Plants were grown under
this light source at an about 30cm distance. The intensity of
the light was about 3000 lux.
Forty individuals of Arabidopsis at three weeks after the
seed sowing were inoculated with Agrobacterium. The inoculation
was carried out according to the modified in planta method
(Plant Journal, Vol. 5, p. 551-558 (1994)). In this method,
floral stems of plants were wounded in order to promote the
infection. Agrobacterium tumefaciens strain C58C1rif (Nucleic
Acids Research, Vol. 13, p. 6981-6998 (1985)) was obtained from
Velten, J. et al. and used. This strain carries pGV3850 HPT as
the intermediate Ti plasmid and between the right- and left-
borders, there is a hygromycin phosphotransferase gene driven by
the cauliflower mosaic virus 35S promoter on this vector that
acts as the selection marker for plants.
After the inoculation of Agrobacterium, those plants were
transplanted on soil of the 1:1 mixture of vermiculite and
perlite. After 1.5 to 2 months of transplantation, seeds (T1
seeds) were harvested. Those seeds were sterilized in a similar
manner to the above mentioned, and sown on a hygromycin
containing medium (1X Ganborg B5 mixed salts for culture medium,
1~ sucrose, 0.8~ agar, 10 mg/1 hygromycin B). Individuals
CA 02301257 2000-03-24
showing hygromycin resistance were selected and transplanted on
soil. The soil was mentioned above. After 1.5 to 2 months from
the transplantation, self-pollinated seeds (T2 seeds) were
harvested.
Example 2: Screening for root-morphology mutants
The screening was carried out according to Example 2 of
Japanese Patent Application Laying Open No. 10-4978. The T2
seeds obtained in Example 1 were sterilized, sown on an agar
medium and grown. The sterilization and the agar medium were as
above mentioned in Example 1. To facilitate morphological
observation of roots, the agar medium was placed in a
transparent plastic Petri dish (Eiken Kizai Kabushiki Kaisha, No.
2 square Petri dish, 14 cm x 10 cm).
The morphology of roots was observed through transmitted
light by means of a stereoscopic microscope OLYMPUS SZH-IDDL.
Morphologies of roots in about 300 lines of transgenic
Arabidopsis were examined and ire was isolated as an abnormal
root-hair length mutant. The length of root hairs was smaller
in the ire mutant (Fig. 1B) than in wild type (Fig. 1A).
However, the density of root hairs and the distance between root
hairs were identical, indicating that this mutation specifically
reduces the length of root hairs. A detailed analysis revealed
that the elongation of root hairs ceased earlier in the mutant
(Fig. 2B) than in wild type (Fig. 2A). This cessation in the
mutant resulted in 60 ~ of root-hair length of wild type.
Example 3: Genetic studies of the ire mutant
11
CA 02301257 2000-03-24
According to the method in Japanese Patent Application
Laying Open No. 10-4978, the ire mutant and wild type were
cross-pollinated. The phenotype (length of root hairs) of wild
type, ire and their F1 progeny was examined (Table 1).
Length of root hairs
t i ne (11m + SE)
Wild type 396 ~ 17
Ire 232 ~ 25
F1 progeny and wild type 452 ~ 32
As shown in Table 1, the Fl progeny showed root hairs of a
normal length like wild type, indicating that the ire mutation
is recessive. Among 711 individuals of selfed progeny of the F1
plants (F2 progeny), 165 individuals (23~) showed the short
root-hair phenotype, suggesting that this phenotype is caused by
a single recessive locus.
Example 4: Preparatory studies for the cloning of the IRE gene
The ire mutant was isolated as a T-DNA insertion line
carrying the selection marker. In order to examine if the
inserted T-DNA disrupted the IRE gene or not, selfed F3 seeds of
213 mutants in the F2 progeny were subjected to the linkage
analysis between the phenotype and the selection marker gene
(hygromycin resistance gene). All F3 progeny of every mutant
showed the hygromycin resistance on the selection medium (1x
12
CA 02301257 2000-03-24
Gamborg B5 mixed salts for medium, 1~ sucrose, 0.8~ agar, 10
mg/1 hygromycin B). Namely, each mutant in the F2 progeny
carried the T-DNA homozygously. Thus it was suggested that the
inserted T-DNA is closely linked with the IRE locus and probably
disrupts the gene. Therefore, genomic regions flanking the T-
DNA insertion should include the IRE gene.
Example 5: Cloning of the IRE gene
A genomic DNA fragment adjacent to the inserted T-DNA was
amplified and cloned by the TAIL-PCR method using the genome DNA of
the ire mutant as a template (Plant Journal, Vol.8, p.457-463
(1995)). The sequences used in the PCR are below: a first specific
primer 5'-CACATCATCTCATTGATGCTTGGT-3' (24mer); a second specific
primer 5'-CATAGATGCACTCGAAATCAGCC-3' (23mer); a third specific
primer 5'-GTGTTATTAAGTTGTCTAAGCGTC-3' (24mer); and arbitrary
primers 5'-(A/T)GTG(A/T/G/C)AG(A/T)A(A/T/G/C)CA(A/T/G/C)AGA-3'
(l6mer). A 0.7 kb PCR fragment was amplified using this set of
primers. The template genomic DNA was extracted according to
Example 4 of Japanese Patent Application Laying Open No. 10-4978.
An Arabidopsis genomic library was screened using the amplified 0.7
kb PCR fragment as a probe. Those genomic clones were isolated
that covered an about 10 kb region in which a sequence hybridized
by the PCR fragment was contained. This genomic region was divided
into several fragments and subcloned into pBluescript II KS+
(Stratagene) to determine the nucleotide sequence. The genomic
region underlined in Fig. 3 was used as a probe to screen a cDNA
library. A cDNA clone was isolated in this screening. The
screening of the genomic and cDNA libraries as well as the method
13
CA 02301257 2000-03-24
of determination of nucleotide sequences was as described in
Example 7 of Japanese Patent Application Laying Open No. 10-4978.
The isolated cDNA was only about 1 kb in length. In order to
isolate a full length cDNA, a PCR method was tried. cDNA that had
been synthesized for the cDNA library was used as a template and
several primer sets designed based on the genome sequence were used
for the PCR. Two PCR fragments were amplified when using the
following sets of primers and the PCR fragments and the isolated
cDNA were joined into a full length cDNA. One of the PCR fragments,
which is composed of 2.2 kb at the 5' region of the full length
cDNA, was amplified using following primers: f2384 f2384:
CAACCGCTTCTCTGTAATC and r4950: AGCCTTCCTATCCTGAATG. The other
fragment, which is composed of 1.2 kb at the 3' region of the full
length cDNA was amplified using following primers: f4897:
TCATGATTGAGCAGTTGGA and r6898: CCGAGCAAGTGTGTCC.
The nucleotide sequence of the full length cDNA was
determined. The full length except for polyA sequence consists
of 3842 by (SEQ ID NO.: 1) and the possible largest ORF (Open
Reading Frame) consists of 1168 amino acid residues (SEQ ID NO.:
2). A stop codon locates in front of the start codon in frame
of this ORF. The correspondence between the cDNA and the genome
of IRE gene is shown in Fig. 4. The IRE gene consists of 17
exons and the T-DNA was inserted into the promoter region in the
ire mutant. The wild-type genomic region as shown in Fig. 4 was
introduced into the ire mutant. A vector was reconstructed from
pBI121 (Clontech) by removing the b-Glucuronidase (hereinafter
referred to GUS) gene and replaced with the genomic region. pBI
121 carries a gene for kanamycin resistance and transformed
14
CA 02301257 2000-03-24
plants show the resistance against kanamycin. The construction
of vector and gene transfer into plants were carried out
according to the method of Example 8 of Japanese Patent
Application Laying Open No. 10-4978; however, the media for
selecting bacteria and plants were supplemented with 50 mg/1
kanamycin sulfate (Meiji Seika). As shown in Fig. 1C, the
transformed plant showed longer root hairs than the
untransformed plant (the ire mutant), indicating that this gene
has an ability to increase the length of root hairs.
Example 6: Tissue-specific activity of the promoter region of
the IRE gene
The DNA fragment as set forth in Fig. 6 (SEQ ID NO.: 3)
was obtained by digesting with restriction enzymes XhoI and
BamHI. This fragment was ligated into SalI-BamHI sites of
pBI101-2 vector (Clontech). This recombinant vector was used
for the transformation into wild-type plants (using the method
of Example 5 above). Restriction enzymes used were purchased
from Takara Shuzo. The GUS gene was to be expressed under the
control of the IRE gene promoter. The sequence as shown in Fig.
6 corresponds to the region spanning between XhoI and BamHI
sites in Fig. 4. The sequence of white letters in Fig. 6
corresponds to the first exon of the IRE gene where the start
codon ATG is enclosed with the rectangle. The sequences with
dotted underlines show exons of another gene adjacent to the IRE
gene. To observe the activity of the expressed GUS, the
transformants were treated with an X-Gluc solution (5.7 mM X-
Gluc (5-bromo-4-chloro-3-indolyl-~-glucuronide), 1.5 mM
CA 02301257 2000-03-24
K3Fe (CN) 6, 1 . 5 mM K4Fe (CN) 6, 0. 9~ Triton X-100) . Transformant
plants were soaked in the X-Gluc solution, subjected to a vacuum
treatment, and then incubated at 37 ~C overnight to develop the
color. Reagents used were purchased from Wako Pure Chemical.
The GUS activities were substantially shown in only the root and
pollen grains. As shown by arrows in Fig. 5B, the GUS activity
was found in pollen grains in the anther. Pollen grains are
germinated on a stigma and elongate pollen tubes. The GUS
activity was also found in elongating pollen tubes (Fig. 5C,
arrow). The foregoing indicates that the promoter of IRE gene
as set forth in SEQ ID NO.: 3 has activities in such tip-growing
cells as root hairs and pollen tubes.
The present inventions) provides) a structural gene
coding a protein that is involved in regulation of tip growth in
plants, and its promoter. Various plants with different
morphologies such as those showing prolonged root hairs, can be
created by utilizing the structural gene of the present
invention. Further, any gene may be expressed specifically in
tissues that are tip-growing by utilizing the promoter of the
present invention.
All publications, patents and patent applications cited
herein are incorporated herein by reference in their entirety.
16
CA 02301257 2000-06-13
SEQUENCE LISTING
(1) GENERAL INFORMATION:
(i) APPLICANT: BIOMOLECULAR ENGINEERING RESEARCH INSTITUTE
(ii) TITLE OF INVENTION: THE IRE GENE REGULATING THE ROOT-JAIR GROWTH IN
ARABIDOPSIS
(iii) NUMBER OF SEQUENCES: 4
(iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: SMART & BIGGAR
(B) STREET: P.O. BOX 2999, STATION D
(C) CITY: OTTAWA
(D) STATE: ONT
(E) COUNTRY: CANADA
(F) ZIP: K1P 5Y6
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Floppy disk
(B) COMPUTER: IBM PC compatible
(C) OPERATING SYSTEM: PC-DOS/MS-DOS
(D) SOFTWARE: ASCII (text)
(vi) CURRENT APPLICATION DATA:
2 O (A) APPLICATION NUMBER: CA 2,301,257
(B) FILING DATE: 24-MAR-2000
(C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER: JP 82402/1999
(B) FILING DATE: 25-MAR-1999
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: SMART & BIGGAR
(B) REGISTRATION NUMBER:
(C) REFERENCE/DOCKET NUMBER: 72813-119
3O (ix) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: (613)-232-2486
(B) TELEFAX: (613)-232-8440
17
CA 02301257 2000-06-13
(2) INFORMATION ID N0:1:
FOR
SEQ
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 842base rs
3 pai
(B) TYPE: leicacid
nuc
(C) STRANDEDNESS: double
(D) TOPOLOGY:linear
(ii)MOLECULE TYPE:cDNA
(vi)ORIGINAL SOURCE:
(A) ORGANISM:Arabidopsisthaliana
lO (ix)FEATURE:
(A) NAME/KEY:CDS
(B) LOCATION:124..3627
(xi)SEQUENCE DESCRIPTI ON: ID
SEQ N0:1:
CAACCGCT TC TCTGTAATCG TTTTG AAAACAAACA 60
TTATG ATAAAATCAC AACAAACAAA
AATTAAAAGA TTCAA ACAAATTTTT CTATTAATAA 120
GTGTAAATAT ATAATTCTCT
GAAAA
CAA ATG TCT ACG ACG CCGTCG GAGAATGAT CGGGATCCA CAA 168
GAA CCT
Met Ser Thr Thr ProSer GluAsnAsp ArgAspPro Gln
Glu Pro
1 5 10 15
2 CCG ACC ACC ATA TCT CCGACT ACCAATGCG AAGCTTCTG AAG 216
O ACT TCC
Pro Thr Thr Ile Ser ProThr ThrAsnAla LysLeuLeu Lys
Thr Ser
20 25 30
AAG ATC CCT GCG ATT TTCCGC TCGGATAAG GAAGGAGAA GAT 264
CCT CAT
Lys Ile Pro Ala Ile PheArg SerAspLys GluGlyGlu Asp
Pro His
35 40 45
GAA CAA GCG AAG ACG GAGGTT ACGGAATTA GCTGGGGAA GGT 312
GAC ACT
Glu Gln Ala Lys Thr GluVal ThrGluLeu AlaGlyGlu Gly
Asp Thr
30 50 55 60
CCG ATG AGT CAC GAT CCGGAA CTTGCTCCT TCTTCCTTA GGT 360
TCG ATT
Pro Met Ser His Asp ProGlu LeuAlaPro SerSerLeu Gly
Ser Ile
65 70 75
TTG AAT CAT ATC AGA AAATCG CCTGCGCCG TCTCCTTTG AGA 408
ACG TCT
Leu Asn His Ile Arg LysSer ProAlaPro SerProLeu Arg
Thr Ser
80 85 90 95
40 TTT TCA TCT GCC ACG TTGATA CCGGGTCAA GATGATAAA GAC 456
CCT TCT
Phe Ser Ser Ala Thr LeuIle ProGlyGln AspAspLys Asp
Pro Ser
100 105 110
GTC GCC AAG GAG AAA AGGGTT GTTGTTGAT GCTCGTGCT GAT 504
CCG GGT
Val Ala Lys Glu Lys ArgVal ValValAsp AlaArgAla Asp
Pro Gly
115 120 125
18
CA 02301257 2000-06-13
GCT CGT GCC AGA TGG CCG ATT CCT CCA CAT CAA CCC GAT CAA GGG AAA 552
Ala Arg Ala Arg Trp Pro Ile Pro Pro His Gln Pro Asp Gln Gly Lys
130 135 140
AAA GTT CAG TGG AGT CAA TCA AAA TCT CAA CGA GTG CCT GCA AAT TCA 600
Lys Val Gln Trp Ser Gln Ser Lys Ser Gln Arg Val Pro Ala Asn Ser
145 150 155
AAT CCA GGG GTA GAG AGT ACT CAT GTA GGT CTT GCC AAG GAA ACA CAA 648
Asn Pro Gly Val Glu Ser Thr His Val Gly Leu Ala Lys Glu Thr Gln
160 165 170 175
TCT CCA CGT TTT CAG GCG ATA TTG CGT GTC ACA AGT GGA AGG AAG AAG 696
Ser Pro Arg Phe Gln Ala Ile Leu Arg Val Thr Ser Gly Arg Lys Lys
180 185 190
AAA GCT CAT GAC ATT AAG AGC TTC TCT CAT GAA CTT AAT TCT AAA GGT 744
Lys Ala His Asp Ile Lys Ser Phe Ser His Glu Leu Asn Ser Lys Gly
195 200 205
GTG CGA CCT TTT CCC GTG TGG AGA TCT CGC GCA GTT GGT CAC ATG GAG 792
Val Arg Pro Phe Pro Val Trp Arg Ser Arg Ala Val Gly His Met Glu
210 215 220
GAG ATT ATG GCG GCG ATC AGA ACG AAG TTT GAT AAG CAA AAG GAA GAT 840
Glu Ile Met Ala Ala Ile Arg Thr Lys Phe Asp Lys Gln Lys Glu Asp
225 230 235
GTT GAT GCT GAT TTA GGA GTT TTT GCT GGC TAT TTA GTG ACA ACT CTA 888
3~ Val Asp Ala Asp Leu Gly Val Phe Ala Gly Tyr Leu Val Thr Thr Leu
240 245 250 255
GAG AGC ACA CCA GAA TCT AAC AAA GAA TTA AGA GTG GGT CTA GAG GAT 936
Glu Ser Thr Pro Glu Ser Asn Lys Glu Leu Arg Val Gly Leu Glu Asp
260 265 270
TTG TTA GTT GAG GCT CGG CAA TGT GCA ACC ATG CCA GCT AGT GAG TTT 984
Leu Leu Val Glu Ala Arg Gln Cys Ala Thr Met Pro Ala Ser Glu Phe
275 280 285
TGG TTG AAA TGT GAA GGT ATT GTT CAG AAG CTT GAT GAT AAA CGT CAG 1032
Trp Leu Lys Cys Glu Gly Ile Val Gln Lys Leu Asp Asp Lys Arg Gln
290 295 300
GAG CTA CCC ATG GGA GGA CTG AAA CAG GCT CAT AAT CGT CTT CTT TTC 1080
Glu Leu Pro Met Gly Gly Leu Lys Gln Ala His Asn Arg Leu Leu Phe
305 310 315
ATT CTT ACT CGT TGC AAT AGG CTT GTG CAA TTT CGT AAA GAG AGT GGT 1128
Ile Leu Thr Arg Cys Asn Arg Leu Val Gln Phe Arg Lys Glu Ser Gly
320 325 330 335
TAT GTT GAA GAA CAC ATT CTG GGA ATG CAC CAG CTA AGT GAT CTT GGA 1176
Tyr Val Glu Glu His Ile Leu Gly Met His Gln Leu Ser Asp Leu Gly
340 345 350
GTT TAT CCT GAA CAG ATG GTG GAA ATC TCG CGA CAA CAG GAC CTT CTT 1224
Val Tyr Pro Glu Gln Met Val Glu Ile Ser Arg Gln Gln Asp Leu Leu
355 360 365
CGA GAG AAG GAA ATC CAA AAG ATA AAT GAA AAG CAA AAT CTA GCT GGT 1272
Arg Glu Lys Glu Ile Gln Lys Ile Asn Glu Lys Gln Asn Leu Ala Gly
370 375 380
19
CA 02301257 2000-06-13
AAA CAA GAT GAT CAG AAC TCG AAC AGC GGA GCT GAT GGA GTG GAA GTA 1320
Lys Gln Asp Asp Gln Asn Ser Asn Ser Gly Ala Asp Gly Val Glu Val
385 390 395
AAT ACT GCT AGA AGT ACT GAT TCA ACT TCG AGC AAT TTT CGG ATG TCA 1368
Asn Thr Ala Arg Ser Thr Asp Ser Thr Ser Ser Asn Phe Arg Met Ser
400 405 410 415
TCT TGG AAG AAG CTT CCA TCT GCT GCC GAG AAA AAC CGT AGT CTT AAT 1416
Ser Trp Lys Lys Leu Pro Ser Ala Ala Glu Lys Asn Arg Ser Leu Asn
420 425 430
AAC ACT CCC AAG GCT AAG GGG GAG AGC AAA ATC CAA CCA AAA GTT TAT 1464
Asn Thr Pro Lys Ala Lys Gly Glu Ser Lys Ile Gln Pro Lys Val Tyr
435 440 445
GGT GAT GAA AAC GCT GAA AAC TTG CAT AGC CCG TCA GGC CAG CCT GCA 1512
Gly Asp Glu Asn Ala Glu Asn Leu His Ser Pro Ser Gly Gln Pro Ala
450 455 460
TCT GCA GAC AGA AGT GCT TTG TGG GGT TTC TGG GCG GAC CAT CAA TGT 1560
Ser Ala Asp Arg Ser Ala Leu Trp Gly Phe Trp Ala Asp His Gln Cys
465 470 475
GTG ACA TAC GAT AAT TCT ATG ATT TGT CGT ATC TGT GAA GTT GAA ATA 1608
Val Thr Tyr Asp Asn Ser Met Ile Cys Arg Ile Cys Glu Val Glu Ile
480 485 490 495
CCA GTT GTA CAT GTA GAA GAG CAC TCT CGA ATA TGC ACA ATC GCT GAT 1656
Pro Val Val His Val Glu Glu His Ser Arg Ile Cys Thr Ile Ala Asp
500 505 510
AGA TGT GAC TTG AAG GGA ATA AAT GTA AAC TTA AGA CTT GAA AGA GTA 1704
Arg Cys Asp Leu Lys Gly Ile Asn Val Asn Leu Arg Leu Glu Arg Val
515 520 525
GCT GAA AGT CTT GAG AAA ATT CTG GAG TCA TGG ACG CCA AAG AGC AGC 1752
Ala Glu Ser Leu Glu Lys Ile Leu Glu Ser Trp Thr Pro Lys Ser Ser
530 535 540
GTA ACT CCA AGA GCA GTT GCT GAT AGC GCA AGA TTA TCA AAT TCC AGT 1800
Val Thr Pro Arg Ala Val Ala Asp Ser Ala Arg Leu Ser Asn Ser Ser
545 550 555
AGA CAA GAA GAT CTG GAT GAA ATC TCT CAG AGA TGT TCA GAT GAC ATG 1848
Arg Gln Glu Asp Leu Asp Glu Ile Ser Gln Arg Cys Ser Asp Asp Met
560 565 570 575
CTT GAT TGC GTT CCT CGT TCA CAG AAT ACA TTT TCT TTG GAT GAA CTG 1896
Leu Asp Cys Val Pro Arg Ser Gln Asn Thr Phe Ser Leu Asp Glu Leu
580 585 590
AAT ATC TTG AAT GAA ATG TCT ATG ACC AAT GGA ACA AAG GAC TCG TCA 1944
Asn Ile Leu Asn Glu Met Ser Met Thr Asn Gly Thr Lys Asp Ser Ser
595 600 605
GCA GGA AGC TTG ACA CCG CCA TCA CCA GCA ACA CCA AGG AAT AGC CAA 1992
Ala Gly Ser Leu Thr Pro Pro Ser Pro Ala Thr Pro Arg Asn Ser Gln
610 615 620
GTA GAT TTG CTA CTA AGT GGT CGA AAA ACA ATA TCA GAG CTT GAG AAT 2040
Val Asp Leu Leu Leu Ser Gly Arg Lys Thr Ile Ser Glu Leu Glu Asn
625 630 635
CA 02301257 2000-06-13
TAT CAG CAG ATA AAC AAG TTG CTT GAT ATT GCT CGT TCG GTG GCA AAT 2088
Tyr Gln Gln Ile Asn Lys Leu Leu Asp Ile Ala Arg Ser Val Ala Asn
640 645 650 655
GTG AAT GTA TGT GGA TAC AGT TCA CTG GAC TTC ATG ATT GAG CAG TTG 2136
Val Asn Val Cys Gly Tyr Ser Ser Leu Asp Phe Met Ile Glu Gln Leu
660 665 670
GAT GAG CTC AAG TAC GTC ATT CAG GAT AGG AAG GCT GAT GCC CTT GTG 2184
Asp Glu Leu Lys Tyr Val Ile Gln Asp Arg Lys Ala Asp Ala Leu Val
675 680 685
GTA GAA ACG TTT GGA AGA CGA ATC GAG AAG CTA CTG CAG GAG AAG TAC 2232
Val Glu Thr Phe Gly Arg Arg Ile Glu Lys Leu Leu Gln Glu Lys Tyr
690 695 700
ATT GAA CTC TGT GGA CTG ATA GAT GAT GAA AAA GTA GAC TCA TCC AAT 2280
Ile Glu Leu Cys Gly Leu Ile Asp Asp Glu Lys Val Asp Ser Ser Asn
705 710 715
GCC ATG CCG GAT GAA GAA AGC TCA GCA GAT GAG GAT ACA GTA CGA AGC 2328
Ala Met Pro Asp Glu Glu Ser Ser Ala Asp Glu Asp Thr Val Arg Ser
720 725 730 735
TTA CGG GCA AGC CCA CTT AAT CCA CGT GCT AAA GAT CGA ACA TCA ATA 2376
Leu Arg Ala Ser Pro Leu Asn Pro Arg Ala Lys Asp Arg Thr Ser Ile
740 745 750
GAA GAT TTT GAA ATT ATA AAA CCA ATT AGC CGT GGT GCA TTT GGA AGA 2424
Glu Asp Phe Glu Ile Ile Lys Pro Ile Ser Arg Gly Ala Phe Gly Arg
755 760 765
GTT TTT CTT GCA AAA AAG AGA GCT ACC GGT GAT TTG TTC GCC ATA AAG 2472
Val Phe Leu Ala Lys Lys Arg Ala Thr Gly Asp Leu Phe Ala Ile Lys
770 775 780
GTT TTA AAG AAG GCT GAT ATG ATC CGT AAG AAT GCT GTT GAA AGT ATT 2520
Val Leu Lys Lys Ala Asp Met Ile Arg Lys Asn Ala Val Glu Ser Ile
785 790 795
TTA GCT GAG CGT AAC ATC CTT ATA TCA GTT CGT AAT CCA TTC GTG GTT 2568
Leu Ala Glu Arg Asn Ile Leu Ile Ser Val Arg Asn Pro Phe Val Val
800 805 810 815
CGT TTT TTC TAT TCT TTC ACA TGC CGG GAA AAT CTC TAT CTG GTC ATG 2616
Arg Phe Phe Tyr Ser Phe Thr Cys Arg Glu Asn Leu Tyr Leu Val Met
820 825 830
GAG TAC TTG AAT GGT GGA GAT CTC TTT TCC TTG TTG AGA AAT CTT GGT 2664
Glu Tyr Leu Asn Gly Gly Asp Leu Phe Ser Leu Leu Arg Asn Leu Gly
835 840 845
TGC TTG GAC GAA GAC ATG GCC CGC ATT TAT ATT GCT GAA GTG GTG CTT 2712
Cys Leu Asp Glu Asp Met Ala Arg Ile Tyr Ile Ala Glu Val Val Leu
850 855 860
GCT CTG GAG TAT CTG CAT TCT GTA AAT ATC ATT CAC AGA GAC TTA AAG 2760
Ala Leu Glu Tyr Leu His Ser Val Asn Ile Ile His Arg Asp Leu Lys
865 870 875
CCA GAC AAT TTG TTG ATC AAT CAG GAT GGT CAC ATC AAG TTG ACA GAT 2808
Pro Asp Asn Leu Leu Ile Asn Gln Asp Gly His Ile Lys Leu Thr Asp
880 885 890 895
21
CA 02301257 2000-06-13
TTC GGG CTT TCC AAG GTT GGT CTT ATC AAT AGC ACA GAT GAC TTA TCA 2856
Phe Gly Leu Ser Lys Val Gly Leu Ile Asn Ser Thr Asp Asp Leu Ser
900 905 910
GGT GAA TCA TCA TTG GGA AAC AGT GGA TTT TTC GCA GAA GAT GGA TCA 2904
Gly Glu Ser Ser Leu Gly Asn Ser Gly Phe Phe Ala Glu Asp Gly Ser
915 920 925
AAA GCT CAA CAT TCA CAA GGC AAA GAT AGT CGT AAG AAA CAT GCA GTT 2952
Lys Ala Gln His Ser Gln Gly Lys Asp Ser Arg Lys Lys His Ala Val
930 935 940
GTT GGA ACC CCT GAT TAT CTA GCA CCT GAA ATA CTT CTT GGA ATG GGT 3000
Val Gly Thr Pro Asp Tyr Leu Ala Pro Glu Ile Leu Leu Gly Met Gly
945 950 955
CAT GGT AAA ACC GCT GAT TGG TGG TCA GTA GGT GTT ATT CTC TTT GAG 3048
His Gly Lys Thr Ala Asp Trp Trp Ser Val Gly Val Ile Leu Phe Glu
960 965 970 975
GTT CTC GTT GGT ATT CCT CCT TTC AAT GCA GAA ACC CCA CAG CAA ATT 3096
Val Leu Val Gly Ile Pro Pro Phe Asn Ala Glu Thr Pro Gln Gln Ile
980 985 990
TTT GAA AAT ATA ATC AAC AGA GAT ATA CCA TGG CCA AAT GTG CCA GAG 3144
Phe Glu Asn Ile Ile Asn Arg Asp Ile Pro Trp Pro Asn Val Pro Glu
995 1000 1005
GAG ATA TCT TAT GAA GCA CAT GAT CTG ATC AAC AAG CTG CTA ACT GAA 3192
Glu Ile Ser Tyr Glu Ala His Asp Leu Ile Asn Lys Leu Leu Thr Glu
1010 1015 1020
AAT CCT GTC CAA AGA CTA GGG GCT ACG GGA GCT GGA GAG GTG AAA CAA 3240
Asn Pro Val Gln Arg Leu Gly Ala Thr Gly Ala Gly Glu Val Lys Gln
1025 1030 1035
CAT CAT TTT TTC AAA GAT ATT AAC TGG GAC ACA CTT GCT CGG CAA AAG 3288
His His Phe Phe Lys Asp Ile Asn Trp Asp Thr Leu Ala Arg Gln Lys
1040 1045 1050 1055
GCT ATG TTT GTA CCA TCA GCT GAA CCA CAA GAC ACT AGT TAT TTC ATG 3336
Ala Met Phe Val Pro Ser Ala Glu Pro Gln Asp Thr Ser Tyr Phe Met
1060 1065 1070
AGC CGA TAT ATA TGG AAC CCG GAA GAC GAA AAT GTT CAT GGA GGC AGC 3389
Ser Arg Tyr Ile Trp Asn Pro Glu Asp Glu Asn Val His Gly Gly Ser
1075 1080 1085
GAT TTT GAT GAC CTT ACA GAC ACA TGC AGT AGC AGC TCC TTT AAT ACA 3432
Asp Phe Asp Asp Leu Thr Asp Thr Cys Ser Ser Ser Ser Phe Asn Thr
1090 1095 1100
CAG GAA GAA GAT GGT GAT GAG TGT GGT AGC TTA GCA GAA TTT GGA AAT 3480
Gln Glu Glu Asp Gly Asp Glu Cys Gly Ser Leu Ala Glu Phe Gly Asn
1105 1110 1115
GGA CCA AAT CTT GCT GTG AAG TAT TCC TTC AGC AAT TTT TCG TTC AAG 3528
Gly Pro Asn Leu Ala Val Lys Tyr Ser Phe Ser Asn Phe Ser Phe Lys
1120 1125 1130 1135
AAC CTC TCA CAA CTG GCT TCG ATC AAC TAC GAT CTT GTC CTA AAG AAC 3576
Asn Leu Ser Gln Leu Ala Ser Ile Asn Tyr Asp Leu Val Leu Lys Asn
1140 1145 1150
22
CA 02301257 2000-06-13
GCA AAG GAA TCA GTA GAA GCT TCG AAC CAG TCA GCC CCT CGA CCC GAA 3624
Ala Lys Glu Ser Val Glu Ala Ser Asn Gln Ser Ala Pro Arg Pro Glu
1155 1160 1165
ACA TGATGACTAC TAATATGTAT GGATCATTTA TGTAACTTCA AGAGCTGATT 3677
Thr
GGTTCCTGTT CAAGCCATCA AAGAAAGTAA TAATAACTTG GAAGCATAGC ATAGCTAGGA 3737
AGCAAAAGAG CTACAGAGAT TTGAGTATGA TATTTTATGT AATTACATGA TACACTTACT 3797
lO GAAATTACAA GATTTTTTCA AATAAATCTA AAAAAATCTT GAATT 3842
(2) INFORMATION FOR SEQ ID N0:2:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1168 amino acids
(B) TYPE: amino acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: unknown
(ii) MOLECULE TYPE: peptide
2 O (vi) ORIGINAL SOURCE:
(A) ORGANISM: Arabidopsis thaliana
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:2:
Met Ser Thr Thr Glu Pro Ser Pro Glu Asn Asp Arg Asp Pro Gln Pro
1 5 10 15
Thr Thr Ile Ser Thr Pro Thr Ser Thr Asn Ala Lys Leu Leu Lys Lys
20 25 30
Ile Pro Ala Ile Pro Phe Arg His Ser Asp Lys Glu Gly Glu Asp Glu
30 35 40 45
Gln Ala Lys Thr Asp Glu Val Thr Thr Glu Leu Ala Gly Glu Gly Pro
50 55 60
Met Ser His Asp Ser Pro Glu Ile Leu Ala Pro Ser Ser Leu Gly Leu
65 70 75 80
Asn His Ile Arg Thr Lys Ser Ser Pro Ala Pro Ser Pro Leu Arg Phe
85 90 95
Ser Ser Ala Thr Pro Leu Ile Ser Pro Gly Gln Asp Asp Lys Asp Val
100 105 110
Ala Lys Glu Lys Pro Arg Val Gly Val Val Asp Ala Arg Ala Asp Ala
115 120 125
Arg Ala Arg Trp Pro Ile Pro Pro His Gln Pro Asp Gln Gly Lys Lys
130 135 140
23
CA 02301257 2000-06-13
Val Gln Trp Ser Gln Ser Lys Ser Gln Arg Val Pro Ala Asn Ser Asn
145 150 155 160
Pro Gly Val Glu Ser Thr His Val Gly Leu Ala Lys Glu Thr Gln Ser
165 170 175
Pro Arg Phe Gln Ala Ile Leu Arg Val Thr Ser Gly Arg Lys Lys Lys
180 185 190
Ala His Asp Ile Lys Ser Phe Ser His Glu Leu Asn Ser Lys Gly Val
195 200 205
Arg Pro Phe Pro Val Trp Arg Ser Arg Ala Val Gly His Met Glu Glu
210 215 220
Ile Met Ala Ala Ile Arg Thr Lys Phe Asp Lys Gln Lys Glu Asp Val
225 230 235 290
Asp Ala Asp Leu Gly Val Phe Ala Gly Tyr Leu Val Thr Thr Leu Glu
245 250 255
Ser Thr Pro Glu Ser Asn Lys Glu Leu Arg Val Gly Leu Glu Asp Leu
260 265 270
Leu Val Glu Ala Arg Gln Cys Ala Thr Met Pro Ala Ser Glu Phe Trp
275 280 285
Leu Lys Cys Glu Gly Ile Val Gln Lys Leu Asp Asp Lys Arg Gln Glu
290 295 300
Leu Pro Met Gly Gly Leu Lys Gln Ala His Asn Arg Leu Leu Phe Ile
305 310 315 320
Leu Thr Arg Cys Asn Arg Leu Val Gln Phe Arg Lys Glu Ser Gly Tyr
325 330 335
Val Glu Glu His Ile Leu Gly Met His Gln Leu Ser Asp Leu Gly Val
340 395 350
Tyr Pro Glu Gln Met Val Glu Ile Ser Arg Gln Gln Asp Leu Leu Arg
355 360 365
Glu Lys Glu Ile Gln Lys Ile Asn Glu Lys Gln Asn Leu Ala Gly Lys
370 375 380
Gln Asp Asp Gln Asn Ser Asn Ser Gly Ala Asp Gly Val Glu Val Asn
385 390 395 400
Thr Ala Arg Ser Thr Asp Ser Thr Ser Ser Asn Phe Arg Met Ser Ser
405 410 415
Trp Lys Lys Leu Pro Ser Ala Ala Glu Lys Asn Arg Ser Leu Asn Asn
420 425 430
Thr Pro Lys Ala Lys Gly Glu Ser Lys Ile Gln Pro Lys Val Tyr Gly
435 440 445
Asp Glu Asn Ala Glu Asn Leu His Ser Pro Ser Gly Gln Pro Ala Ser
450 455 460
Ala Asp Arg Ser Ala Leu Trp Gly Phe Trp Ala Asp His Gln Cys Val
465 470 975 480
24
CA 02301257 2000-06-13
Thr Tyr Asp Asn Ser Met Ile Cys Arg Ile Cys Glu Val Glu Ile Pro
485 490 495
Val Val His Val Glu Glu His Ser Arg Ile Cys Thr Ile Ala Asp Arg
500 505 510
Cys Asp Leu Lys Gly Ile Asn Val Asn Leu Arg Leu Glu Arg Val Ala
515 520 525
Glu Ser Leu Glu Lys Ile Leu Glu Ser Trp Thr Pro Lys Ser Ser Val
530 535 540
Thr Pro Arg Ala Val Ala Asp Ser Ala Arg Leu Ser Asn Ser Ser Arg
545 550 555 560
Gln Glu Asp Leu Asp Glu Ile Ser Gln Arg Cys Ser Asp Asp Met Leu
565 570 575
Asp Cys Val Pro Arg Ser Gln Asn Thr Phe Ser Leu Asp Glu Leu Asn
580 585 590
Ile Leu Asn Glu Met Ser Met Thr Asn Gly Thr Lys Asp Ser Ser Ala
595 600 605
Gly Ser Leu Thr Pro Pro Ser Pro Ala Thr Pro Arg Asn Ser Gln Val
610 615 620
Asp Leu Leu Leu Ser Gly Arg Lys Thr Ile Ser Glu Leu Glu Asn Tyr
625 630 635 640
Gln Gln Ile Asn Lys Leu Leu Asp Ile Ala Arg Ser Val Ala Asn Val
645 650 655
Asn Val Cys Gly Tyr Ser Ser Leu Asp Phe Met Ile Glu Gln Leu Asp
660 665 670
Glu Leu Lys Tyr Val Ile Gln Asp Arg Lys Ala Asp Ala Leu Val Val
675 680 685
4 0 Glu Thr Phe Gly Arg Arg Ile Glu Lys Leu Leu Gln Glu Lys Tyr Ile
690 695 700
Glu Leu Cys Gly Leu Ile Asp Asp Glu Lys Val Asp Ser Ser Asn Ala
705 710 715 720
Met Pro Asp Glu Glu Ser Ser Ala Asp Glu Asp Thr Val Arg Ser Leu
725 730 735
Arg Ala Ser Pro Leu Asn Pro Arg Ala Lys Asp Arg Thr Ser Ile Glu
50 740 795 750
Asp Phe Glu Ile Ile Lys Pro Ile Ser Arg Gly Ala Phe Gly Arg Val
755 760 765
Phe Leu Ala Lys Lys Arg Ala Thr Gly Asp Leu Phe Ala Ile Lys Val
770 775 780
Leu Lys Lys Ala Asp Met Ile Arg Lys Asn Ala Val Glu Ser Ile Leu
785 790 795 800
Ala Glu Arg Asn Ile Leu Ile Ser Val Arg Asn Pro Phe Val Val Arg
805 810 815
CA 02301257 2000-06-13
Phe Phe Tyr Ser Phe Thr Cys Arg Glu Asn Leu Tyr Leu Val Met Glu
820 825 830
Tyr Leu Asn Gly Gly Asp Leu Phe Ser Leu Leu Arg Asn Leu Gly Cys
835 840 845
Leu Asp Glu Asp Met Ala Arg Ile Tyr Ile Ala Glu Val Val Leu Ala
850 855 860
Leu Glu Tyr Leu His Ser Val Asn Ile Ile His Arg Asp Leu Lys Pro
865 870 875 880
Asp Asn Leu Leu Ile Asn Gln Asp Gly His Ile Lys Leu Thr Asp Phe
885 890 895
Gly Leu Ser Lys Val Gly Leu Ile Asn Ser Thr Asp Asp Leu Ser Gly
900 905 910
Glu Ser Ser Leu Gly Asn Ser Gly Phe Phe Ala Glu Asp Gly Ser Lys
915 920 925
Ala Gln His Ser Gln Gly Lys Asp Ser Arg Lys Lys His Ala Val Val
930 935 940
Gly Thr Pro Asp Tyr Leu Ala Pro Glu Ile Leu Leu Gly Met Gly His
945 950 955 960
Gly Lys Thr Ala Asp Trp Trp Ser Val Gly Val Ile Leu Phe Glu Val
965 970 975
Leu Val Gly Ile Pro Pro Phe Asn Ala Glu Thr Pro Gln Gln Ile Phe
980 985 990
Glu Asn Ile Ile Asn Arg Asp Ile Pro Trp Pro Asn Val Pro Glu Glu
995 1000 1005
Ile Ser Tyr Glu Ala His Asp Leu Ile Asn Lys Leu Leu Thr Glu Asn
1010 1015 1020
4 0 Pro Val Gln Arg Leu Gly Ala Thr Gly Ala Gly Glu Val Lys Gln His
1025 1030 1035 1040
His Phe Phe Lys Asp Ile Asn Trp Asp Thr Leu Ala Arg Gln Lys Ala
1045 1050 1055
Met Phe Val Pro Ser Ala Glu Pro Gln Asp Thr Ser Tyr Phe Met Ser
1060 1065 1070
Arg Tyr Ile Trp Asn Pro Glu Asp Glu Asn Val His Gly Gly Ser Asp
50 1075 1080 1085
Phe Asp Asp Leu Thr Asp Thr Cys Ser Ser Ser Ser Phe Asn Thr Gln
1090 1095 1100
Glu Glu Asp Gly Asp Glu Cys Gly Ser Leu Ala Glu Phe Gly Asn Gly
105 1110 1115 1120
Pro Asn Leu Ala Val Lys Tyr Ser Phe Ser Asn Phe Ser Phe Lys Asn
1125 1130 1135
Leu Ser Gln Leu Ala Ser Ile Asn Tyr Asp Leu Val Leu Lys Asn Ala
1140 1145 1150
26
CA 02301257 2000-06-13
Lys Glu Ser Val Glu Ala Ser Asn Gln Ser Ala Pro Arg Pro Glu Thr
1155 1160 1165
(2) INFORMATION FOR SEQ ID N0:3:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1270 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: single
(D) TOPOLOGY: linear
(ii) MOLECULE TYPE: genomic DNA
(vi) ORIGINAL SOURCE:
(A) ORGANISM: Arabidopsis thaliana
(xi) SEQUENCE DESCRIPTION: SEQ ID N0:3:
CTCGAGTGGC TCTTCCAAAC CAGCCTTCCC TGGTTTCATT GGTTGATACG CTGTCGCCAT 60
ATTTGCAGCT TCTTCTGAAA ACAATCACAA ACACACCAAC CAATCCATCA CATTAACAAA 120
TGATCCGATA AACTCATGTA AACATGAAAA ATATTAAAAC TTGGATCAAG TGAAGAGAAC 180
CTGAGAGACG AACGAAGCTC GACGGCGGAT TGCTTTTTGC TGCAAGGAAG GAAGGAGTCG 240
AATGATTTAA CCTAAGAGAC GGTTGATTTA ATATGAAAAC CCTAATTTAT ATGGACTTGT 300
2 O TGGGCCCAAA CAATATTCAG CCCAATAACT ACAAAAATAT TTGCTCTGTA TTTTTCTTTT 360
GTGTTTTGAC CCAATTACTT GGAAAACAAT TCATTTTATT AAAAAATACA AACCATGTTT 420
GATTTAATTA CATAAATTTT TGTAATGAAT AAACGTGTTA TTATAAAAAA CAAATTAATA 480
TTTTTTAGAA GTTTTAACCA AAAATATCTT GGTACCGTGT TTTTTAGTCA ACCCAGAGAA 540
AACATCACGA ACGGCTATTT TTTAAAATTA AACAAAAAAA AATTAAGTTC TTTGTTATTT 600
CTAGAAAATG GGGAAGACAT GCATTTGTTG GAAGCACGTA GTTTCGATAT GGACACTCTC 660
CCACATGATT ATTCTTTAAG AACTATACAA ATCATAAAAT ATCACCATTC ATTAATAACT 720
TAAGGTGTTG TAAGAATAAT AAAGTCAGAA ATTATAACTT TAGCACTATG GATATAAGAA 780
TACAACGAAT AATCACAAAT CACAACCATA ATAAAATCTC TGTCTGACTT TTGAATTTGG 840
AACTAATTTA GAATTATTTT TTGTTGACAG CAAATCTTTA TAATTTAAAG ATATTAAGTT 900
3O AACATCAAAT AGAAGTATCA TACTATAACT ATAGTCATAT TGAATTAAAG ATAACAAATT 960
ACCATAAAAT TTAGATGTAA ATAGTCAAAT TTATTTATCT GCAAGAAGAA CAAAAAAAAT 1020
GGCGAATGAG CCAATTTTTG CATCAAGTAG TAAAAACGGA GTCCCCCACT TCCCGCGAAA 1080
ACCTCAGTCT CTCTCTCTCT CTCTCGCAAC CGCTTCTCTG TAATCGTTAT GTTTTGATAA 1140
27
CA 02301257 2000-06-13
AATCACAAAA CAAACAAACA AACAAAAATT AAAAGAGTGT AAATATGAAA ATTCAAATAA 1200
TTCTCTACAA ATTTTTCTAT TAATAACAAA TGTCTACGAC GGAACCGTCG CCTGAGAATG 1260
ATCGGGATCC 1270
(2) INFORMATION FOR SEQID
N0:4:
(i) SEQUENCE ARACTERISTIC S:
CH
(A) : pairs
LENGTH 3504
base
(B) nucleic acid
TYPE:
(C) single
STRANDEDNESS:
(D) linear
TOPOLOGY:
(ii)MOLECULE cDNA
TYPE:
(vi)ORIGINAL (A) Arabidopsis thaliana
SOURCE: ORGANISM:
(xi)SEQUENCE EQ ID
DESCRIPTION: N0:4:
S
ATG TCT ACG GAA CCGTCGCCT GAGAATGAT CGGGATCCA CAACCG 48
ACG
Met Ser Thr Glu ProSerPro GluAsnAsp ArgAspPro GlnPro
Thr
1 5 10 15
ACC ACC ATA ACT CCGACTTCC ACCAATGCG AAGCTTCTG AAGAAG 96
TCT
2 Thr Thr Ile Thr ProThrSer ThrAsnAla LysLeuLeu LysLys
0 Ser
20 25 30
ATC CCT GCG CCT TTCCGCCAT TCGGATAAG GAAGGAGAA GATGAA 144
ATT
Ile Pro Ala Pro PheArgHis SerAspLys GluGlyGlu AspGlu
Ile
35 40 45
CAA GCG AAG GAC GAGGTTACT ACGGAATTA GCTGGGGAA GGTCCG 192
ACG
Gln Ala Lys Asp GluValThr ThrGluLeu AlaGlyGlu GlyPro
Thr
50 55 60
30
ATG AGT CAC TCG CCGGAAATT CTTGCTCCT TCTTCCTTA GGTTTG 240
GAT
Met Ser His Ser ProGluIle LeuAlaPro SerSerLeu GlyLeu
Asp
65 70 75 80
AAT CAT ATC ACG AAATCGTCT CCTGCGCCG TCTCCTTTG AGATTT 288
AGA
Asn His Ile Thr LysSerSer ProAlaPro SerProLeu ArgPhe
Arg
85 90 95
TCA TCT GCC CCT TTGATATCT CCGGGTCAA GATGATAAA GACGTC 336
ACG
4 Ser Ser Ala Pro LeuIleSer ProGlyGln AspAspLys AspVal
0 Thr
100 105 110
GCC AAG GAG CCG AGGGTTGGT GTTGTTGAT GCTCGTGCT GATGCT 384
AAA
Ala Lys Glu Pro ArgValGly ValValAsp AlaArgAla AspAla
Lys
115 120 125
CGT GCC AGA CCG ATTCCTCCA CATCAACCC GATCAAGGG AAAAAA 432
TGG
Arg Ala Arg Pro IleProPro HisGlnPro AspGlnGly LysLys
Trp
130 135 140
50
28
CA 02301257 2000-06-13
GTT CAG TGG AGT CAA TCA AAA TCT CAA CGA GTG CCT GCA AAT TCA AAT 480
Val Gln Trp Ser Gln Ser Lys Ser Gln Arg Val Pro Ala Asn Ser Asn
145 150 155 160
CCA GGG GTA GAG AGT ACT CAT GTA GGT CTT GCC AAG GAA ACA CAA TCT 528
Pro Gly Val Glu Ser Thr His Val Gly Leu Ala Lys Glu Thr Gln Ser
165 170 175
CCA CGT TTT CAG GCG ATA TTG CGT GTC ACA AGT GGA AGG AAG AAG AAA 576
Pro Arg Phe Gln Ala Ile Leu Arg Val Thr Ser Gly Arg Lys Lys Lys
180 185 190
GCT CAT GAC ATT AAG AGC TTC TCT CAT GAA CTT AAT TCT AAA GGT GTG 624
Ala His Asp Ile Lys Ser Phe Ser His Glu Leu Asn Ser Lys Gly Val
195 200 205
CGA CCT TTT CCC GTG TGG AGA TCT CGC GCA GTT GGT CAC ATG GAG GAG 672
Arg Pro Phe Pro Val Trp Arg Ser Arg Ala Val Gly His Met Glu Glu
210 215 220
ATT ATG GCG GCG ATC AGA ACG AAG TTT GAT AAG CAA AAG GAA GAT GTT 720
Ile Met Ala Ala Ile Arg Thr Lys Phe Asp Lys Gln Lys Glu Asp Val
225 230 235 240
GAT GCT GAT TTA GGA GTT TTT GCT GGC TAT TTA GTG ACA ACT CTA GAG 768
Asp Ala Asp Leu Gly Val Phe Ala Gly Tyr Leu Val Thr Thr Leu Glu
245 250 255
AGC ACA CCA GAA TCT AAC AAA GAA TTA AGA GTG GGT CTA GAG GAT TTG 816
Ser Thr Pro Glu Ser Asn Lys Glu Leu Arg Val Gly Leu Glu Asp Leu
260 265 270
TTA GTT GAG GCT CGG CAA TGT GCA ACC ATG CCA GCT AGT GAG TTT TGG 864
Leu Val Glu Ala Arg Gln Cys Ala Thr Met Pro Ala Ser Glu Phe Trp
275 280 285
TTG AAA TGT GAA GGT ATT GTT CAG AAG CTT GAT GAT AAA CGT CAG GAG 912
Leu Lys Cys Glu Gly Ile Val Gln Lys Leu Asp Asp Lys Arg Gln Glu
290 295 300
CTA CCC ATG GGA GGA CTG AAA CAG GCT CAT AAT CGT CTT CTT TTC ATT 960
Leu Pro Met Gly Gly Leu Lys Gln Ala His Asn Arg Leu Leu Phe Ile
305 310 315 320
CTT ACT CGT TGC AAT AGG CTT GTG CAA TTT CGT AAA GAG AGT GGT TAT 1008
Leu Thr Arg Cys Asn Arg Leu Val Gln Phe Arg Lys Glu Ser Gly Tyr
325 330 335
GTT GAA GAA CAC ATT CTG GGA ATG CAC CAG CTA AGT GAT CTT GGA GTT 1056
Val Glu Glu His Ile Leu Gly Met His Gln Leu Ser Asp Leu Gly Val
340 345 350
TAT CCT GAA CAG ATG GTG GAA ATC TCG CGA CAA CAG GAC CTT CTT CGA 1104
Tyr Pro Glu Gln Met Val Glu Ile Ser Arg Gln Gln Asp Leu Leu Arg
355 360 365
GAG AAG GAA ATC CAA AAG ATA AAT GAA AAG CAA AAT CTA GCT GGT ATA 1152
Glu Lys Glu Ile Gln Lys Ile Asn Glu Lys Gln Asn Leu Ala Gly Ile
370 375 380
CAA GAT GAT CAG AAC TCG AAC AGC GGA GCT GAT GGA GTG GAA GTA AAT 1200
Gln Asp Asp Gln Asn Ser Asn Ser Gly Ala Asp Gly Val Glu Val Asn
385 390 395 400
29
CA 02301257 2000-06-13
ACT GCT AGA AGT ACT GAT TCA ACT TCG AGC AAT TTT CGG ATG TCA TCT 1248
Thr Ala Arg Ser Thr Asp Ser Thr Ser Ser Asn Phe Arg Met Ser Ser
405 410 415
TGG AAG AAG CTT CCA TCT GCT GCC GAG AAA AAC CGT AGT CTT AAT AAC 1296
Trp Lys Lys Leu Pro Ser Ala Ala Glu Lys Asn Arg Ser Leu Asn Asn
420 425 430
ACT CCC AAG GCT AAG GGG GAG AGC AAA ATC CAA CCA AAA GTT TAT GGT 1344
Thr Pro Lys Ala Lys Gly Glu Ser Lys Ile Gln Pro Lys Val Tyr Gly
435 440 445
GAT GAA AAC GCT GAA AAC TTG CAT AGC CCG TCA GGC CAG CCT GCA TCT 1392
Asp Glu Asn Ala Glu Asn Leu His Ser Pro Ser Gly Gln Pro Ala Ser
450 455 460
GCA GAC AGA AGT GCT TTG TGG GGT TTC TGG GCG GAC CAT CAA TGT GTG 1440
Ala Asp Arg Ser Ala Leu Trp Gly Phe Trp Ala Asp His Gln Cys Val
465 470 475 480
ACA TAC GAT AAT TCT ATG ATT TGT CGT ATC TGT GAA GTT GAA ATA CCA 1488
Thr Tyr Asp Asn Ser Met Ile Cys Arg Ile Cys Glu Val Glu Ile Pro
485 490 495
GTT GTA CAT GTA GAA GAG CAC TCT CGA ATA TGC ACA ATC GCT GAT AGA 1536
Val Val His Val Glu Glu His Ser Arg Ile Cys Thr Ile Ala Asp Arg
500 505 510
TGT GAC TTG AAG GGA ATA AAT GTA AAC TTA AGA CTT GAA AGA GTA GCT 1584
Cys Asp Leu Lys Gly Ile Asn Val Asn Leu Arg Leu Glu Arg Val Ala
515 520 525
GAA AGT CTT GAG AAA ATT CTG GAG TCA TGG ACG CCA AAG AGC AGC GTA 1632
Glu Ser Leu Glu Lys Ile Leu Glu Ser Trp Thr Pro Lys Ser Ser Val
530 535 540
ACT CCA AGA GCA GTT GCT GAT AGC GCA AGA TTA TCA AAT TCC AGT AGA 1680
Thr Pro Arg Ala Val Ala Asp Ser Ala Arg Leu Ser Asn Ser Ser Arg
545 550 555 560
CAA GAA GAT CTG GAT GAA ATC TCT CAG AGA TGT TCA GAT GAC ATG CTT 1728
Gln Glu Asp Leu Asp Glu Ile Ser Gln Arg Cys Ser Asp Asp Met Leu
565 570 575
GAT TGC GTT CCT CGT TCA CAG AAT ACA TTT TCT TTG GAT GAA CTG AAT 1776
Asp Cys Val Pro Arg Ser Gln Asn Thr Phe Ser Leu Asp Glu Leu Asn
580 585 590
ATC TTG AAT GAA ATG TCT ATG ACC AAT GGA ACA AAG GAC TCG TCA GCA 1824
Ile Leu Asn Glu Met Ser Met Thr Asn Gly Thr Lys Asp Ser Ser Ala
595 600 605
GGA AGC TTG ACA CCG CCA TCA CCA GCA ACA CCA AGG AAT AGC CAA GTA 1872
Gly Ser Leu Thr Pro Pro Ser Pro Ala Thr Pro Arg Asn Ser Gln Val
610 615 620
GAT TTG CTA CTA AGT GGT CGA AAA ACA ATA TCA GAG CTT GAG AAT TAT 1920
Asp Leu Leu Leu Ser Gly Arg Lys Thr Ile Ser Glu Leu Glu Asn Tyr
625 630 635 640
CAG CAG ATA AAC AAG TTG CTT GAT ATT GCT CGT TCG GTG GCA AAT GTG 1968
Gln Gln Ile Asn Lys Leu Leu Asp Ile Ala Arg Ser Val Ala Asn Val
645 650 655
CA 02301257 2000-06-13
AAT GTA TGT GGA TAC AGT TCA CTG GAC TTC ATG ATT GAG CAG TTG GAT 2016
Asn Val Cys Gly Tyr Ser Ser Leu Asp Phe Met Ile Glu Gln Leu Asp
660 665 670
GAG CTC AAG TAC GTC ATT CAG GAT AGG AAG GCT GAT GCC CTT GTG GTA 2064
Glu Leu Lys Tyr Val Ile Gln Asp Arg Lys Ala Asp Ala Leu Val Val
675 680 685
GAA ACG TTT GGA AGA CGA ATC GAG AAG CTA CTG CAG GAG AAG TAC ATT 2112
Glu Thr Phe Gly Arg Arg Ile Glu Lys Leu Leu Gln Glu Lys Tyr Ile
690 695 700
GAA CTC TGT GGA CTG ATA GAT GAT GAA AAA GTA GAC TCA TCC AAT GCC 2160
Glu Leu Cys Gly Leu Ile Asp Asp Glu Lys Val Asp Ser Ser Asn Ala
705 710 715 720
ATG CCG GAT GAA GAA AGC TCA GCA GAT GAG GAT ACA GTA CGA AGC TTA 2208
Met Pro Asp Glu Glu Ser Ser Ala Asp Glu Asp Thr Val Arg Ser Leu
725 730 735
CGG GCA AGC CCA CTT AAT CCA CGT GCT AAA GAT CGA ACA TCA ATA GAA 2256
Arg Ala Ser Pro Leu Asn Pro Arg Ala Lys Asp Arg Thr Ser Ile Glu
740 745 750
GAT TTT GAA ATT ATA AAA CCA ATT AGC CGT GGT GCA TTT GGA AGA GTT 2304
Asp Phe Glu Ile Ile Lys Pro Ile Ser Arg Gly Ala Phe Gly Arg Val
755 760 765
TTT CTT GCA AAA AAG AGA GCT ACC GGT GAT TTG TTC GCC ATA AAG GTT 2352
Phe Leu Ala Lys Lys Arg Ala Thr Gly Asp Leu Phe Ala Ile Lys Val
770 775 780
TTA AAG AAG GCT GAT ATG ATC CGT AAG AAT GCT GTT GAA AGT ATT TTA 2400
Leu Lys Lys Ala Asp Met Ile Arg Lys Asn Ala Val Glu Ser Ile Leu
785 790 795 800
GCT GAG CGT AAC ATC CTT ATA TCA GTT CGT AAT CCA TTC GTG GTT CGT 2448
Ala Glu Arg Asn Ile Leu Ile Ser Val Arg Asn Pro Phe Val Val Arg
805 810 815
TTT TTC TAT TCT TTC ACA TGC CGG GAA AAT CTC TAT CTG GTC ATG GAG 2496
Phe Phe Tyr Ser Phe Thr Cys Arg Glu Asn Leu Tyr Leu Val Met Glu
820 825 830
TAC TTG AAT GGT GGA GAT CTC TTT TCC TTG TTG AGA AAT CTT GGT TGC 2544
Tyr Leu Asn Gly Gly Asp Leu Phe Ser Leu Leu Arg Asn Leu Gly Cys
835 890 845
TTG GAC GAA GAC ATG GCC CGC ATT TAT ATT GCT GAA GTG GTG CTT GCT 2592
Leu Asp Glu Asp Met Ala Arg Ile Tyr Ile Ala Glu Val Val Leu Ala
850 855 860
CTG GAG TAT CTG CAT TCT GTA AAT ATC ATT CAC AGA GAC TTA AAG CCA 2690
Leu Glu Tyr Leu His Ser Val Asn Ile Ile His Arg Asp Leu Lys Pro
865 870 875 880
GAC AAT TTG TTG ATC AAT CAG GAT GGT CAC ATC AAG TTG ACA GAT TTC 2688
Asp Asn Leu Leu Ile Asn Gln Asp Gly His Ile Lys Leu Thr Asp Phe
885 890 895
GGG CTT TCC AAG GTT GGT CTT ATC AAT AGC ACA GAT GAC TTA TCA GGT 2736
Gly Leu Ser Lys Val Gly Leu Ile Asn Ser Thr Asp Asp Leu Ser Gly
900 905 910
31
CA 02301257 2000-06-13
GAA TCA TCA TTG GGA AAC AGT GGA TTT TTC GCA GAA GAT GGA TCA AAA 2784
Glu Ser Ser Leu Gly Asn Ser Gly Phe Phe Ala Glu Asp Gly Ser Lys
915 920 925
GCT CAA CAT TCA CAA GGC AAA GAT AGT CGT AAG AAA CAT GCA GTT GTT 2832
Ala Gln His Ser Gln Gly Lys Asp Ser Arg Lys Lys His Ala Val Val
930 935 940
GGA ACC CCT GAT TAT CTA GCA CCT GAA ATA CTT CTT GGA ATG GGT CAT 2880
Gly Thr Pro Asp Tyr Leu Ala Pro Glu Ile Leu Leu Gly Met Gly His
945 950 955 960
GGT AAA ACC GCT GAT TGG TGG TCA GTA GGT GTT ATT CTC TTT GAG GTT 2928
Gly Lys Thr Ala Asp Trp Trp Ser Val Gly Val Ile Leu Phe Glu Val
965 970 975
CTC GTT GGT ATT CCT CCT TTC AAT GCA GAA ACC CCA CAG CAA ATT TTT 2976
Leu Val Gly Ile Pro Pro Phe Asn Ala Glu Thr Pro Gln Gln Ile Phe
980 985 990
GAA AAT ATA ATC AAC AGA GAT ATA CCA TGG CCA AAT GTG CCA GAG GAG 3024
Glu Asn Ile Ile Asn Arg Asp Ile Pro Trp Pro Asn Val Pro Glu Glu
995 1000 1005
ATA TCT TAT GAA GCA CAT GAT CTG ATC AAC AAG CTG CTA ACT GAA AAT 3072
Ile Ser Tyr Glu Ala His Asp Leu Ile Asn Lys Leu Leu Thr Glu Asn
1010 1015 1020
CCT GTC CAA AGA CTA GGG GCT ACG GGA GCT GGA GAG GTG AAA CAA CAT 3120
Pro Val Gln Arg Leu Gly Ala Thr Gly Ala Gly Glu Val Lys Gln His
1025 1030 1035 1040
CAT TTT TTC AAA GAT ATT AAC TGG GAC ACA CTT GCT CGG CAA AAG GCT 3168
His Phe Phe Lys Asp Ile Asn Trp Asp Thr Leu Ala Arg Gln Lys Ala
1045 1050 1055
ATG TTT GTA CCA TCG GCT GAA CCA CAA GAC ACT AGT TAT TTC ATG AGC 3216
Met Phe Val Pro Ser Ala Glu Pro Gln Asp Thr Ser Tyr Phe Met Ser
1060 1065 1070
CGA TAT ATA TGG AAC CCG GAA GAC GAA AAT GTT CAT GGA GGC AGC GAT 3264
Arg Tyr Ile Trp Asn Pro Glu Asp Glu Asn Val His Gly Gly Ser Asp
1075 1080 1085
TTT GAT GAC CTT ACA GAC ACA TGC AGT AGC AGC TCC TTT AAT ACA CAG 3312
Phe Asp Asp Leu Thr Asp Thr Cys Ser Ser Ser Ser Phe Asn Thr Gln
1090 1095 1100
GAA GAA GAT GGT GAT GAG TGT GGT AGC TTA GCA GAA TTT GGA AAT GGA 3360
Glu Glu Asp Gly Asp Glu Cys Gly Ser Leu Ala Glu Phe Gly Asn Gly
1105 1110 1115 1120
CCA AAT CTT GCT GTG AAG TAT TCC TTC AGC AAT TTT TCG TTC AAG AAC 3408
Pro Asn Leu Ala Val Lys Tyr Ser Phe Ser Asn Phe Ser Phe Lys Asn
1125 1130 1135
CTC TCA CAA CTG GCT TCG ATC AAC TAC GAT CTT GTC CTA AAG AAC GCA 3456
Leu Ser Gln Leu Ala Ser Ile Asn Tyr Asp Leu Val Leu Lys Asn Ala
1140 1145 1150
AAG GAA TCA GTA GAA GCT TCG AAC CAG TCA GCC CCT CGA CCC GAA ACA 3504
Lys Glu Ser Val Glu Ala Ser Asn Gln Ser Ala Pro Arg Pro Glu Thr
1155 1160 1165
32