Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
GENETIC METHOD FOR THE EXPRESSION OF POLYPROTEWS IN PLANTS
The present invention relates to a method for increasing protein expression
levels. in
particular by the coexpression of two or more proteins in plants within a
single transcription
s unit, to the coexpression and secretion of two or more proteins in plants,
to linker sequences
for use in the method of the invention. to DNA constructs for use in the
invention and to plants
transformed with the constructs of the invention.
For many applications based on genetic modification of plants by transgenesis,
it is
desirable to express co-ordinately two or more transgenes. For instance,
coexpression in
1 o plants of transgenes encoding antimicrobial proteins with different
biochemical targets can
result in enhanced disease resistance levels, resistance against a broader
range of pathogens, or
resistance that is more difficult to overcome by mutational adaptation of
pathogens. Other
examples include those aimed at producing a particular metabolite in
transgenic plants by
coexpression of multiple transgenes that are involved in a biosynthetic
pathway. There are
15 different ways to obtain transgenic plants expressing multiple transgenes.
One frequently
chosen option is to introduce each transgene individually via separate
transformation events
and to cross the different single-transgene expressing lines. The drawback of
this method is
that the different transgenes in the resulting progeny will be inserted at
different loci, which
complicates the subsequent breeding process. Moreover, this method is not
applicable to
'o crops that are propagated vegetatively, such as for instance potato, many
ornamentals and fruit
tree species.
A second possibility is to introduce the different transgenes as linked
expression
cassettes, each with their own promoters and terminators, within a single
transformation
vector. Such a set of transgenes will in this case segregate as a single
genetic locus. It has
25 been obsec-ved, however, that the presence of multiple copies of the same
promoter within a
transgenic plant often results in transcriptional silencing of the transgenes
(Matzke, M.A. and
Matzke, A.J.M., 1998, Cellular and Molecular Life Sciences ~4. 94-103). In an
attempt to
introduce a vector containin; four linked transgenes each driven by a CaMV35S
promoter,
Van den Elzen P.J. et al. (Phil. Traps. R. Soc. Lon. B., 1993. 3~t2: 271-278)
obsen~ed that none
30 of the analysed transeenic lines expressed all four transgenes at a
reasonably high level. To
avoid this problem one could use different promoters for each of the
expression cassettes used
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-7-
in the construct. However, there is currently only a very limited choice of
promoter sets that
have comparable characteristics in terms of expression levels. cell-type and
developmental
specificity and response to environmental factors.
A third option would be to produce multiple proteins from one transcription
unit by
separating the distinct coding regions by so-called internal ribosomal entry
sites, which allow
ribosomes to reiterate translation at internal positions within a m1Z\'A
species. Although
imernal ribosomal entry sites are well documented in animal systems (Kaminski
A. et al.,
1994, Genet. Eng. 16, 11 ~-1 ~~) it is not known at present whether such sites
are also
functional in nuclear-encoded genes from plants. Polycistronic genes can be
expressed when
inserted in plant chloroplastic genomes (Daniell H. et al., 1998, Nature
Biotechnology 16,
345-348) but the gene products in this case are confined to the chloroplast,
which is not alwavs
the preferred site of deposition of foreign proteins.
A fourth strategy, finally, is based on the production of multiple proteins by
proteolytic
cleavage of a single polyprotein precursor encoded by a single transcription
unit. Potyviruses,
t 5 for instance, translate their genomic RNA into a single polyprotein
precursor that encompasses
proteolytic domains able to cleave the polyprotein precursor in cis
(Dougherty, W.G. and
Carrington, J.C., 1988, Annu. Rev. Phytopathol. 26, 123-143). Beck von Bodman,
S. et al.,
(1995, Bio/Technology 13, 587-591) have already exploited the potvviral system
to co-express
two enzymes involved in the biosynthesis of mannopine. The two biosynthetic
enzymes were
fused within one open reading frame together with a protease derived from a
potyviral
polyprotein precursor, and the adjoining regions were separated by 8 amino
acids long spacers
representing specific cleavage sites for the protease. Plants transformed with
this construct
synthesized mannopine, suggesting that the two enzymes had somehow been
produced in a
form that was at least partially functional, although direct evidence for the
presumed cleavage
35 events in planta was not presented. A disadvantage of this system is that a
viral protein needs
to be co-expressed with proteins of interest, which is not always desirable.
?More recently,
Urwin P.E. et al. (1998, Planta 204, 472-479) have shown that it is possible
to co-express two
different proteinase inhibitors joined by a protease-sensitive propeptide
derived from a plant
metallothionein-like protein. A polyprotein precursor consisting of a cysteine
protease
3o inhibitor (oryzacystatin from vice), a propeptide from pea metallothionein-
like protein and a
serine protease inhibitor (cowpea trypsin inhibitor), was found to be cleaved
in transgenic
SU8ST1TUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- 3 -
,4rabidopsis thaliana plants. The cleavage, however, was only partial, as
uncleaved
polyprotein precursor could also be detected in the transgenic plants. As the
polyprotein
precursor did not contain a leader peptide, the translation products are
predicted to be
deposited in the cytosol. The metallothionein from which the propeptide was
derived also does
not contain a leader peptide (Evans IM 1990, FEBS Lett. 262, 29-32) and hence
its processing
must occur in the cytosol.
For some applications, cytosolic processing and deposition is a drawback. Many
proteins, especially glycosvlated proteins or proteins with multiple disulfide
bridges, must be
synthesized in the secretory pathway (encompassing the endoplasmic reticulum
and Golgi
o apparatus) in order to be folded in a functional form (Bednarek and Raikhel
1992, Plant Mol.
Biol. 20, 133-1~0). In addition, for some applications such as fox instance
the expression of
antimicrobial proteins, the extracellular space is the preferred deposition
site, as most
microorganisms occur at least during the early stages of infection in the
extracelluiar space.
Proteins destined to the extracellular space are also synthesised via the
secretory pathway but
t 5 lack additional targeting information other than the leader peptide
(Bednarek and Raikhel
1992, Plant Mol. Biol. 20. 133-150). Other examples of the application of this
strategy are
described in WO 9/24486 and W095/17514.
The applicants have unexpectedly found that expression levels of plant
defensins in
plants transformed with a polyprotein precursor construct were much higher
compared to those
2o in plants transformed with single plant defensin constructs.
The invention therefore provides a method of improving expression levels of a
protein
in a transgenic plant comprising inserting into the genome of said plant a DNA
sequence
comprising a promoter region operably linked to two or more protein encoding
regions and a
3'-terminator region wherein said protein encoding regions are separated from
each other by a
25 DNA sequence coding for a linker propeptide said propeptide providing a
cleavage site
whereby the expressed polyprotein is post-translationally processed into the
component
protein molecules.
The processing system described here can be used not only to co-express two or
more
different proteins, but also to obtain higher expression levels of a protein,
particularly of small
3o proteins. The reason for the observed stimulatory effect on translational
efficiency is currently
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
unclear. It might be due to an effect of mRNA length or length of primary
translation product
on translational efficiency.
Preferably, a signal sequence is operatively interconnected with the protein
coding
regions.
As used herein the expression "signal sequence" is used to define a sequence
encoding
a leader peptide that allows a nascent polypeptide to enter the endoplasmic
reticulum and is
removed after this translocation.
The signal sequence may be derived from any suitable source and may for
example be
naturally associated with the promoter to which it is operably linked. We have
found the use
of signal sequences from the class of plant proteins known as defensins
(Broekaert et al, 1995
Plant Physiol 108. 1353-1358; Broekaert et al, 1997, Crit, Rev, Plant Sci. 16,
297-323) to be
particularly suitable for use in the method of the invention.
Thus, in a further preferred embodiment, there is provided a method of
improving
expression levels of a protein in a transgenic plant comprising inserting into
the genome of
said plant a DNA sequence comprising a promoter region operably linked to a
signal sequence
said signal sequence being operably linked to two or more protein encoding
regions and a 3'-
terminator region wherein said protein encoding regions are separated from
each other by a
DNA sequence coding for a linker propeptide said propeptide providing a
cleavage site
whereby the expressed polyprotein is post-translationally processed into the
component
2o protein molecules.
This method of the invention is particularly suitable for the expression of
proteins
which are 100 amino acids or less in length
The present invention provides a convenient and highly efficient method of co-
expressing two or more proteins in a plant as a single transcription unit
where the two proteins
35 are joined by a cleavable linker, the construct being designed such that
cleavage occurs in the
secretory pathway of the plant thereby releasing the proteins extracellularly.
According to a further aspect of the present invention, there is provided a
method for
the expression of multiple proteins in a transgenic plant comprising inserting
into the genome
of said plant a DNA sequence comprising a promoter region operably linked to a
signal
3o sequence said signal sequence being operably linked to two or more protein
encoding regions
and a 3'-terminator region wherein said protein encoding regions are separated
from each
SUBSTITUTE SHEET (RULE 26j
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_j_
other by a DNA sequence coding for a linker propeptide said propeptide
providing a cleavage
site whereby the expressed polvprotein is post-translationally processed into
the component
protein molecules.
The tvo or more protein encoding regions according to all aspects of the
invention
preferably do not encode identical proteins i.e. the method of the invention
allows the
production of different proteins in a single transcription unit. The DNA
sequence to be
e,cpressed according to the method of the invention is one which does not
occur naturally in
the plant used for the production of the multiple proteins i.e. one or more of
the components of
the DNA sequence will be heterologous to the plant host:
to The method for the expression of multiple proteins described herein does
not cover the
use of a linker propeptide as expressed by the Ib-AMP gene and as described in
SEQ ID Nos
14,1 S, 16, 17 or 18 of Published International Patent Application No. WO
95/24:186 separating
three protein encoding regions each of which encodes Rs-AFP2; nor the
insertion thereof into
a plant genome. Suitably, the method of the invention does not use a linker
propetide of the
I5 native Ib-AMP gene as shown in SEQ ID Nos 14, 15, 16, 17 or 18 of WO
95/24486.
In a further aspect, the present invention there is provided a method for the
expression
of multiple proteins in a transgenic plant comprising inserting into the
genome of said plant a
DNA sequence comprising a promoter region operably linked to a signal sequence
said signal
sequence being operably linked to two or more protein encoding regions and a
3'-terminator
2o region wherein said protein encoding regions are separated from each other
by a DNA
sequence coding for a linker propeptide said propeptide providing a cleavage
site whereby the
expressed polyprotein is post-translationally processed into the component
protein molecules
with the proviso that when the linker propeptide is derived from the Ib-AMP
gene as described
in SEQ ID Nos 14,15, 16, 17 or 18 of Published International Patent
Application No. WO
25 95/24486 it does not separate three protein encoding regions each of which
encodes Rs-AFP2.
The sequence of Rs-AFP2 is fully described in Published International patent
Application no. WO 93/05 l j3 published 18 March 1993.
The promoter sequence may for example be that naturally associated with the
signal
sequence, and/or it may be that naturally associated with the protein encoding
sequence to
3o which it is linked, or it may be any other promoter sequence conferring
transcription in plants.
It may be a constitutive promoter or it may be an inducible promoter.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-6-
The linker propeptide for use in all aspects and embodiments of the invention
described herein is preferably a linker propeptide which is cleaved on passage
of said DNA
encoding the polyprotein precursor through the secretory pathway of the plant
cells in which
the polyprotein -encoding DNA is expressed. The linker propeptide is
preferably designed or
chosen such that cleavage of the propeptide occurs by proteases which are
naturally present in
the secretory pathway of the plant cell in which the DNA encoding the
polyprotein is
expressed. Particular promoters of the cauliflower mosaic virus such as the
Penh 255
promoter of the 35S RNA, examples of such proteases include subtilisin-like
proteases, .
In a preferred embodiment the invention therefore provides a method for the
expression of multiple proteins in a transgenic plant comprising inserting
into the genome of
said plant a DNA sequence comprising a promoter region operably linked to a
signal
sequence, said signal sequence being operably linked to two or more protein
encoding regions
and a 3'-terminator region wherein said protein encoding regions are separated
from each
other by a DNA sequence coding for a linker propeptide said propeptide
providing a cleavage
15 site whereby the expressed polyprotein is post-translationally processed
into the component
protein molecules said linker propeptide being cleaved on passage of said DNA
encoding the
polyprotein precursor through the secretory pathway of the plant cells in
which the polyprotein
-encoding DNA is expressed.
The method for the expression of multiple proteins described herein does not
cover the
2o use of a linker propeptide derived from the Ib-AMP gene as described in SEQ
ID Nos 14, 1 S,
16, 17 or 18 of Published International Patent Application No. WO 95/24486
separating three
protein encoding regions each of which encodes Rs-AFP2 and the insertion
thereof into a plant
genome.
In some embodiments of the invention, the linker propeptide is not derived
from a
2s virus.
In a particularly preferred embodiment the invention provides a method for the
expression of multiple proteins in a transgenic plant comprising inserting
into the genome of
said plant a DNA sequence comprising a promoter region operably linked to a
signal sequence
said signal sequence being operably linked to two or more protein encoding
regions and a 3'-
30 terminator region wherein said protein encoding regions are separated from
each other by a
DNA sequence coding for a linker propeptide said propeptide providing a
cleavage site
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_7_
whereby the expressed polyprotein is post-translationally processed into the
component
protein molecules. said linker propeptide being cleaved on passage of said DNA
encoding the
polyprotein precursor through the secretory pathway of the plant cells in
which the polyprotein
-encoding DNA is expressed wherein cleavage of the propeptide occurs by
proteases which are
naturally present in the secretow pathway of said plant cell.
The linker propeptide may be a peptide which naturally contains processing
sites for
proteases occuring in the secretory pathway of plants such as the internal
propeptides derived
from the Ib-AMP gene which are described further herein, or may be a peptide
to which such a
protease processing site has been engineered at either or both ends thereof to
facilitate
t o cleavage of the sequence. Where a propeptide possesses one such protease
processing site a
further protease processing site may be added. If necessary or desired,
repeats of the
processing site. for example up to 6 repeats may be included.
For example, as described fully herein, a further protease processing site has
been
added to the 3' end of the DNA sequence coding for the C-terminal propeptides
from Dahlia
15 and Amaranthus which naturally possess a protease processing site at their
N-terminal end for
an unknown secretory pathway protease and these peptides are particularly
suitable for use
according to the method of the invention. Certain Dahlia sequences including C-
terminal
propeptide sequences are described and claimed in copending British Patent
Application No.
9818003.7.
20 Yet another strategy is based upon the use of virus e.g. picornovirus
sequences such
as 20 amino acid sequences called the 2A sequence of the foot-and-mouth
disease virus
(FMDV) RNA, which results in the cleavage of polyproteins (Ryan and Drew 1994,
EMPO
J., I3, 928-933). In this instance however, in order to avoid the retention of
unwanted amino
acids on the protein product, combined with a sequence which produces N-
terminal
35 sequence, for example a plant derived sequence or a fragment thereof to
form a chimeric
propeptide.
In the present invention. we have developed novel strategies for making
artificial
polyprotein precursors which are cleaved in the secretory pathway. The first
one was based on
the use of a propeptide derived from the IbAMP gene. IbAMP is a gene from the
plant
3o Impatiens balsamina which encodes a peculiar polyprotein precursor
featuring a leader peptide
and six consecutive antimicrobial peptides, each flanked by propeptides
ranging from 16 to 28
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-s-
amino acids in length (Tailor R.H. et al., 1997, J. Biol. Chem. 272, 24480-
24487). It is not
known how and where processing of the IbAMP precursor occurs in its plant of
origin. One of
the internal propeptides from IbAVIP was used to separate two distinct plant
defensin coding
regions, one originating from radish seed (RsAFP2, Tetras F.R.G. et al., 1992,
J. Biol. Chem.
267, 15301-15309; Tetras et al I99~ Plant Cell, 7, 573-X88) and one from
dahlia seed
(DmAMPI, Osborn R.W. etal., 1995, FEBS Lett. 368, 2~7-262).
An other strategy was based on the use of C-terminal propeptides from either
the
DmAMPI precursor or the AcAMP3 precursor (De Bolle M.F.C. et al., 1993, Plant
Mol. Biol.
22, 1187-1190) or fragments of these. These C-terminal propeptides were chosen
based on
io our previous observation that they apparently can be cleaved in transgenic
tobacco plants
without influencing extracellular deposition of the mature proteins to which
they are connected
in the precursor (R.W. Osborn and S. Attenborough, personal communication; De
Bolle
M.F.C. et al., 1996, Plant Mol. Biol. 31, 993-1008) implicating that such
cleavage is
performed by a protease present in the secretory pathway excluding the
vacuole. To convert
these C-terminal propeptides to internal propeptides, a subtilisin-like
protease processing site
was engineered at the C-terminal part of the propeptides.
Subtilisin-like proteases are enzymes that specifically cleave at recognition
sites of
which the last two residues are basic (Bart, P.J., 1991, Cell 66, 1-3; Park
C.M. et al., 1994.
Mol. Microbiol. I 1, 155-164). Although subtilisin-like. proteases are best
documented in funei
(e.g. Kex2-like proteases) and higher animals (e.g. furins), recent evidence
suggests that such
enzymes are also present in plants (Kinal H. et al., 1995, Plant Cell 7, 677-
688; Tornero P. et
al., 1997, J. Biol. Chem. 272, 14412-14419), including Arabidopsis (Ribeiro A.
et al., 1995,
Plant Cell 7, 785-794).
We have found that polyprotein precursors consisting of a leader peptide
followed by
two different plant defensins separated from each other by any of the above
described internal
propeptides can be processed in transgenic plants to release both plant
defensins
simultaneously. The cleavage does occur such that at least the major part of
the plant
defensins are deposited in the extracellular space. Hence processing of the
precursor occurred
either in the secretory pathway or in the extracellular space. The different
propeptides shown
3o to be cleaved in the transgenic plants do not reveal primary sequence
homology. However, the
sequences all appear to be rich in the small amino acids A, V, S and T and all
contain
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-9-
dipeptidic sequences consisting of either two acidic residues, two basic
residues or one acidic
and one basic residue. Although propeptide cleavage in the examples shown in
this invention
did apparently not occur within vacuoles, internal propeptides from vaeuolar
proteins (e.g. 2S
albumins) might also be used if vacuolar deposition of the proteins would be
desirable. In the
co-expression experiments described here two different plant defensins were
used but it is
predicted that similar results will be obtained when other types of proteins
would be used or
when more than two mature protein domains would be used in the polyprotein
precursor
structure.
Where it is desired to target the polyprotein to a particular cellular
organelle along the
secreton~ pathway a suitable targeting sequence may be added to one or more of
the multiple
protein encoding regions. For example, an endoplasmic reticulum targeting
sequence such as
that encoding KDEL (SEQ ID NO 6~) may be added to the 3' end of one or more of
the
mature protein encoding regions, or a vacuolar targeting sequence (Chispeels
and Raikhel
1992, Cell 68, 613-616) can be added to the 3' or ~' end of one or more of the
protein
15 encoding regions. An example of the latter is the barley lectin carboxy-
terminal propeptide
which has been shown to destine heterologous proteins that are otherwise
secreted to the
vacuoles (Bednarek and Raikhel 1991, Plant Cell 3, 1195-1206; De Bolle et al,
1996 Plant
MoI. Biol. 31, 993-1008).
At least 40% of the sequence of the linker propeptide for use in accordance
with all
2o aspects and methods of the invention as described herein preferably
consists of stretches of
either two to five consecutive hydrophobic residues selected from alanine,
valine, isoleucine,
methionine, leucine, phenylalanine, tryptophan and tyrosine or stretches of
two to five
hydrophilic residues selected from aspartic acid, glutamic acid, lysine,
arginine, histidine,
serine, threonine, glutamine and asparagine.
25 The said hydrophobic residues are preferably alanine, valine, leucine,
methionine
and/or isoleucine and the said hydrophilic residues are preferably aspartic
acid, glutamic acid,
lysine and/or arginine.
It is further preferred that the linker propeptide has within 7 residues of
its N- or C-
terminal cleavage site a sequence with two to five consecutive acidic
residues, two to five
3o basic residues or two to five consecutive intermixed acidic and basic
residues.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- 10-
It is especially preferred that at least 40% of the sequence of the linker
propeptide for
use in accordance with all aspects of the invention as described herein
preferably consists of
stretches of either two to five consecutive hydrophobic residues selected from
alanine, valine,
isoleucine, methionine. leucine, phenylalanine, tryptophan and tyrosine or
stretches of two to
five hydrophilic residues selected from aspartic acid, glutamic acid, lysine,
arginine, histidine,
serine, threonine, glutamine and asparagine and has within 7 residues of its N-
or C- terminal
cleavage site a sequence with two to five consecutive acidic residues, two to
five basic
residues or two to f ve consecutive intermixed acidic and basic residues.
The use of linker propeptides rich in the small amino acids A, V, S and T and
t 0 containing dipeptidic sequences consisting of either two acidic residues,
two basic residues or
one acidic and one basic residue which on translation provides a cleavage site
whereby the
expressed polyprotein is post-translationally processed into the component
protein molecules
is also preferred.
As used herein the term 'rich' is used to denote that the residues A,V, S and
T are
i 5 present more frequently than would be expected based on a random
distribution of amino
acids.
It is further preferred that the linker propeptides have a dipeptidic sequence
within
seven amino acids from the N- and/or C- terminal ends thereof, the said
dipeptidic sequences
consisting of either two acidic residues, two basic residues or an acidic and
a basic residue
2o wherein said dipeptidie sequences may be the same or different at each
terminus.
In a further preferred embodiment said dipeptidic sequences are selected from
the
following EE, ED and/or KK.
It is particularly desirable that the linker propeptide should hold the two
(or more)
protein domains sufficiently far apart so that they can fold appropriately and
independently.
25 For this purpose, the linker polypeptide is suitably at least 10 and
preferably at least 1 ~ amino
acids long. It is further advantageous that the linker propeptide should not
interact with any
secondary structural element in the two proteins which it links and should
therefore itself have
no particular secondary structure or form a solitary secondary structure
element such as an
alpha helix.
3o In this and all other aspects and embodiments of the invention described
herein the
linker propeptide sequence providing the cleavage site preferably comprises a
linker sequence
SUBSTITUTE SHEET (RULE 26~
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
which is isolatable from a natural source such as a plant or virus, or variant
thereof or a
frament of either of these. In particular the linker propeptide is isolatable
from a plant protein.
or a fragment, or variant or derivative thereof which can provide suitable
cleavage sites.
Particular examples include a cleavable linker derived from the C-terminal
propeptide region
of a Dahlia gene such as those described and claimed in copending British
Patent Application
No. 9818003.7.
Where a viral sequence is used, it is preferably an element of a chimeric
propeptide
sequence.
The expression "variant" refers to sequences of amino acids which differ from
the base
1o sequence from which they are derived in that one or more amino acids within
the sequence are
substituted for other amino acids. Amino acid substitutions may be regarded as
"conservative" where an amino acid is replaced with a different amino acid
with broadly
similar properties. Non-conservative substitutions are where amino acids are
replaced with
amino acids of a different type. Broadly speaking, fewer non-conservative
substitutions will
~5 be possible without altering the biological activity of the polypeptide.
Suitably variants have
at least 85% similarity and preferably at least 90% similarity to the base
sequence
In the context of the present invention, two amino acid sequences with at
least
85% similarity to each other have at least 85% similar (identical or
conservatively
replaced) amino acid residues in a like position when aligned optimally
allowing for up to
30 3 gaps, with the proviso that in respect of the gaps a total of not more
than 1 ~ amino acid
residues is affected. Likewise, two amino acid sequences with at least 90%
similarity to
each other have at least 90% identical or conservatively replaced amino acid
residues in a
like position when aligned optimally allowing for up to 3 gaps with the
proviso that in
respect of the gaps a total of not more than 1 ~ amino acid residues is
affected.
?5 For the purpose of the present invention, a conservative amino acid is
defined as
one which does not alter the activity/function of the protein when compared
with the
unmodified protein. In particular. conservative replacements may be made
between amino
acids within the following groups:
(i) Alanine, Serine, Glycine and Threonine
30 (ii) Glutamic acid and Aspartic acid
(iii) Arginine and Lysine
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- l2-
(iv) Isoleucine. Leucine, Valine and Methionine
(v) Phenylalanine, Tyrosine and Tryptophan
Sequence similariy may be calculated using sequence alignment algorithms
known in the art such as, for e,cample, the Clustal Method described by Myers
and Miller
(Comput. Appl. Biosci .4 11-17 (1988).) and Wilbur and Lipman (Proc. Natl.
Acad. Sci. USA
80, 726-30 ( 1983) ) and the Watterman and Eggert method (The Journal of
Molecular Biology
(1987) 197, 723-728). The MegAlign Lipman Pearson one pair method (using
default
parameters) which may be obtained from DNAstar Inc, 1228 Selfpark Street,
Madison,
Wisconsin, 53715, USA as part of the Lasergene system may also be used.
In particular the linker prapetide is a sequence isolatable from a plant
protein and more
preferably from the precursor of a plant antimicrobial protein such as a
defensin. or a hevein-
type antimicrobial peptide (Broekaert et al 1997, Crit. Rev. Plant Sci. 16,
297-323). The linker
propeptide is most preferably derivable from a defensin and/or a hevein type
antimicrobial
peptide, especially from the C-terminal propeptides from Dm-AMP l and Ac-AMP2
the
t5 sequences of which are as described in Figure 2 herein (SEQ ID NO S and SEQ
ID NO 8)
The use of a linker propeptide derived from an antimicrobial peptide derived
from the
genus Impatiens is also preferred. The Ib-AMP gene comprises five propeptide
regions all of
which are suitable for use in the. present invention and which are described
fully in Published
International Patent Application WO 95/24486 at pages 29 and 40 to 42, the
contents of
2o which are incorporated herein by reference. All or part of the C-terminal
propeptides derived
from the Dm-AMP and Ac-AMP gene may be used.
In a particularly preferred embodiment, the linker propeptide sequence used
comprises
a naturally occurring linker propeptide sequence which is modified so that
amino acids from
said sequence remaining attached to protein product after cleavage thereof is
reduced,
25 preferably so that none remain. Suitable modifications may be determined
using routine
methods as described hereinafter. In its simplest form, protein products of
the invention are
isolated and analyzed to see whether they include any residual amino acids
derived from the
propeptide linker. The linker sequence may then be modified to eliminate some
or ali of these
residues, provided the function of post-translational cleavage remains.
SUBSTITUTE SHEET (RULE 25)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-13-
The term "fragment" refers to sequences from which amino acids have been
deleted,
preferably from an end region thereof. Thus these include the modified forms
of the natural
sequences mentioned above.
A linker propeptide of the invention may comprise one or more such fragments
from
different sources provided it functions as a post-translational cleavage site.
Examples of
linker propeptide sequences are SEQ ID NOs 3, 4, 6, 7, 21, 22, 23, 24, 25, 26,
27. 28 and 29
as shown herein and variants therefore which act as a propeptide. Particular
examples of
these are SEQ ID NOs 3, 4, 6, 7, '' 1. 22, 23, 24, 25, 26, 27, 28 and. '.?9
themselves.
In particular, the propeptide sequences comprise SEQ ID NOs 3, 4, 6 or 7.
o According to a preferred embodiment the present invention further provides a
method
for the expression of multiple proteins in a transgenic plant comprising
inserting into the
genome of said plant a DNA sequence comprising a promoter region operably
linked to a
signal sequence said signal sequence being operably linked to tw-o or more
protein encoding
regions and a 3'-terminator region wherein said protein encoding regions are
separated From
~ 5 each other by a DNA sequence coding for a linker propeptide wherein the
linker propeptide is
derivable from a defensin and/or a hevein type antimicrobial peptide said
propeptide providing
a cleavage site whereby the expressed polyprotein is post-translationally
processed into the
component protein molecules.
The use of the C-terminal propeptides from Dm-AMP 1 and Ac-AMP2 as described
in
2o Figure 2 herein as cleavable linkers i.e. to provide a cleavable linkage
site, are particularly
preferred. Depending on the choice of propeptide it may be necessary to
engineer an
additional specific protease recognition site at either or both ends to
facilitate cleavage of the
sequence. Suitable specific protease recognition sites include for example,
recognition sites
for subtilisin -like proteases recognising either a dipeptidic sequence
consisting of two basic
z5 residues; tetrapeptidic sequences consisting of a hydrophobic residue, any
residue, a basic
residue and a basic residue or a tetrapeptidic sequence consisting of a basic
residue, any
residue, a basic residue and a basic residue. Subtilisin-like protease
recognition sites are
particularly preferred for use in the method of the invention.
According to a yet further preferred embodiment the present invention further
provides
3o a method for the expression of multiple proteins in a transgenic plant
comprising inserting into
the genome of said plant a DNA sequence comprising a promoter region operably
linked to a
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- 14-
signal sequence said signal sequence being operably linked to two or more
protein encoding
regions and a 3'-terminator region wherein said protein encoding regions are
separated from
each other by a DNA sequence coding for a linker propeptide said propeptide
providing a
cleavage site whereby the expressed polyprotein is post-translationally
processed into the
component protein molecules and wherein an additional specific protease
recognition site has
been engineered at either or both ends of said linker propeptide to facilitate
cleavage of the
sequence.
According to a vet further preferred embodiment the present invention further
provides
a method for the expression of multiple proteins in a transgenic plant
comprising inserting into
o the genome of said plant a DNA sequence comprising a promoter region
operably linked to a
signal sequence said signal sequence being operably linked to two or more
protein encoding
regions and a 3'-terminator region wherein said protein encoding regions are
separated from
each other by a DNA sequence coding for a linker propeptide wherein the linker
propeptide is
derivable from a defensin and/or a hevein type antimicrobial peptide said
propeptide providing
15 a cleavage site whereby the expressed polyprotein is post-translationally
processed into the
component protein molecules and wherein an additional specific protease
recognition site has
been engineered at either or both ends of said linker propeptide to facilitate
cleavage of the
sequence.
The invention further provides the use of propeptides isolatable from plant
derived
?o proteins as cleavable linkers in polyprotein precursors synthesised via the
secretory pathway in
transgenic plants. The propeptides are preferably isolatable from the
precursor of a plant
defensin or a hevein-type antimicrobial peptide (Broekaert et al 1997, Crit.
Rev. Plant Sci. 16,
297-323). The propeptides may also preferably be isolatable from an
antimicrobial peptide
derived from the genus Impatiens.
25 In a further aspect the invention provides the use of a propeptide wherein
at least 40%
of the sequence of the propeptide consists of stretches of either two to five
consecutive
hydrophobic residues selected from alanine, valine, isoleucine, methionine.
leucine,
phenylalanine, tryptophan and tyrosine or stretches of two to five hydrophilic
residues selected
from aspartic acid, glutarnic acid, lysine, arginine, histidine, serine,
threonine, glutamine and
3o asparagine as a cleavable linker in polyprotein precursors synthesised via
the secretory
pathway in transgenic plants.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-15-
It is further preferred that the linker propeptide has within 7 residues of
its N- or C-
terminal cleavage site a sequence with two to five consecutive acidic
residues. two to five
basic residues or two to five consecutive intermixed acidic and basic
residues.
It is especially preferred that at least 40% of the sequence of the linker
propeptide
consists of stretches of either two to five consecutive hydrophobic residues
selected from
alanine, valine, isoleucine, methionine, leucine, phenylalanine, tryptophan
and tyrosine or
stretches of two to five hydrophilic residues selected from aspartic acid,
glutamic acid, lysine.
arginine, histidine, serine, threonine, glutamine and asparagine and has
within 7 residues of its
N- or C- terminal cleavage site a sequence with two to five consecutive acidic
residues. two to
to five basic residues or two to five consecutive intermixed acidic and basic
residues.
In a further aspect the invention provides the use of a peptide sequence rich
in the
small amino acids A, V, S and T and containing dipeptidic sequences consisting
of either two
acidic residues, two basic residues or one acidic and one basic residue as a
cleavable linker
sequence wherein said sequence is isolatable from a plant defensin or a hevein-
type
15 antimicrobial protein.
The methods of the invention may be used to achieve efficient expression and
secretion
of any desired proteins and is particularly suitable for the expression of
proteins which must
naturally be synthesised in the secretory pathway in order to be folded in a
functional form
such as, for example, glycosylated proteins and those with disulphide bridges.
Additionally. it
2o is extremely advantageous for proteins involved in the defence of a plant
to attack by a
pathogen to be secreted efficiently to the extracellular space since this is
usually the initial site
of pathogen attack and the present methods of the invention provide an
effective means of
delivering multiple proteins extracellularly.
The method of the invention is also particularly suitable for producing small
peptides
25 which may then be used for immunisation purposes i.e. the transgenic plant
or a seed derived
therefrom may be used directly as a foodstuff thereby passively immunising the
recipient.
Examples of proteins which may be expressed according to the methods of the
present
invention include, for example, antifungal proteins described in Published
International Patent
Application Nos W092/15691, W092/21699, W093/05153, W093/04586, W094/I 1511,
3o W095/04754, W095/18229, W095/24486, W097/21814 and W097/21815 including Rs-
AFP 1, Rs-AFP2, Dm-AMP 1, Dm-AMP2, Hs-AFP 1, Ah-AMP I , Ct-AMP 1, Ct-AMP2, B n-
SU8ST1TUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- 16-
AFP 1, Bn-AFP2, Br-AFP 1, Br-AFP2, Sa-AFP I , Sa-AFP?, Cb-AiVIP 1, Cb-AMP?. Ca-
AMP 1,
Bm-AMP I , Ace-AMP I , Ac-AMP I , Ac-AMP2, Mj-A MP I , Mj-Al~iP2, Ib-AMP I .
Ib-AMP2,
Ib-AMP3, Ib-AMP=l, PR-I type proteins such as chitinases, glucanases such as
betal.3 and
betal,6 glucanases. chitin-binding lectins, zeamatins, osmotins, thionins and
ribosome-
inactivating proteins and peptides derived therefrom or antifungal proteins
showing 85%
sequence identity, preferably greater than 90% sequence identity; more
preferably greater than
95% sequence identity with any of said proteins where sequence identity is as
defined above.
The cleavable linkers are used to join two or more proteins of interest and
provide
cleavage sites whereby the polyprotein is post-translationally processed into
the component
protein molecules.
In a further aspect the invention provides a DNA construct comprising a DNA
sequence comprising a promoter region operably linked to a plant derived
signal sequence said
signal sequence being operably linked to two or more protein encoding regions
and a 3'-
terminator region wherein said protein encoding regions are separated from
each other by a
DNA sequence coding for a linker propeptide said prapeptide providing a post-
translational
cleavage site.
Suitably the protein encoding region encode different proteins. Preferred
examples of
propeptide linker sequences are as detailed above.
In a preferred embodiment of this aspect the invention provides a DNA
construct
wherein said DNA sequence encoding said linker propeptide encodes an internal
propeptide
from the Ib-AMP gene. In a further preferred embodiment of this aspect the
invention
provides a DNA construct wherein said DNA sequence encoding said linker
propeptide
encodes the C-terminal propeptide from the Dm-AMP or from the Ac-AMP gene.
In a particularly preferred embodiment the invention provides a DNA construct
as
described above wherein when the DNA sequence encoding the linker propeptide
is derived
from the Dm-AMP gene or from the Ac-AMP gene it additionally comprises one or
more
protease recognition sites at either or both ends thereof.
In a further aspect the invention provides a DNA construct comprising a DiV'A
sequence comprising a promoter region operably linked to two or more protein
encoding
3o regions and a 3' terminator-region wherein said protein encoding regions
are separated from
each other by a DNA sequence coding for a linker propeptide encoding the C-
terminal
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- 17-
propeptide from the Dm-AMP gene or the from the Ac-AMP gene said propeptide
providing a
post-translational cleavage site.
In a particularly preferred embodiment the invention provides a DNA construct
as
described above wherein the DNA sequence encoding the linker propeptide from
Dm-AMP or
Ac-AMP additionally comprises one or more protease recognition sites at either
or both ends
thereof.
In a yet further aspect the invention. provides a transgenic plant transformed
with a
DNA construct according to any of the above aspects of the invention.
In a further aspect the invention provides a transgenic plant transformed with
a DVA
o sequence comprising a promoter region operably linked to a signal sequence
said signal
sequence being operably linked to two or more protein encoding regions and a
3'-terminator
region wherein said protein encoding regions are separated from each other by
a DNA
sequence coding for a linker propeptide which on translation provides a
cleavage site.
In a preferred embodiment of this aspect at least 40% of the sequence of the
said linker
15 propeptide consists of stretches of either two to five consecutive
hydrophobic residues selected
from alanine, valine, isoleucine, methionine, leucine, phenylalanine,
tryptophan and tyrosine
or stretches of two to five hydrophilic residues selected from aspartic acid,
glutamic acid,
lysine, arginine; histidine, serine, threonine, glutamine and asparagine.
The said hydrophobic residues are preferably alanine, valine, leucine,
methionine
2o and/or isoleucine and the said hydrophilic residues are preferably aspartic
acid, glutamic acid,
lysine and/or arginine.
It is further preferred that the linker propeptide has within 7 residues of
its N- or C-
terminal cleavage site a sequence with two to five consecutive acidic
residues, two to five
basic residues or two to five consecutive intermixed acidic and basic
residues.
?5 It is especially preferred that at least 40% of the sequence of the linker
propeptide
consists of stretches of either two to five consecutive hydrophobic residues
selected from
alanine, valine, isoleucine, methionine, leucine, phenylalanine, tr~~ptophan
and tyrosine or
stretches of two to five hydrophilic residues selected from aspartic acid,
glutamic acid, lysine.
arginine, histidine, serine, threonine, glutamine and asparagine and has
within 7 residues of its
3o N- or C- terminal cleavage site a sequence with two to five consecutive
acidic residues, two to
five basic residues or two to five consecutive intermixed acidic and basic
residues.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- ls-
In a further preferred embodiment of this aspect of the invention the DNA
sequence
providing the cleavage site encodes a peptide sequence rich in the small amino
acids A, V, S
and T and containing dipeptidic sequences consisting of either two acidic
residues, two basic
residues or one acidic and one basic residue.
In a particularly preferred embodiment of this aspect of the invention the DNA
sequence providing the cleavage site encodes a propeptide derived from the Ib-
AMP gene such
as for example that described in Figure 2. In a further particularly preferred
embodiment of
this aspect of the invention the DNA sequence providing the cleavage site
encodes the C-
terminal propeptides from Dm-AMP1 and Ac-AMP2 as described in Figure 2 which
may
0 optionally be engineered to include a further DNA sequence encoding a
subtilisin-like protease
recognition site.
In a further aspect the invention provides a vector comprising a DNA construct
as
described above.
Certain linker sequences described herein are novel and theses and the coding
sequence
~ 5 for these form a further aspect of the invention. In particular therefore,
there is provided a
nucleic acid which encodes a linker peptide of SEQ ID NO 4, 6, 7, 29, 21, 22.
23, 24, 25, 26,
27, 28 or the linker peptide shown in Figure 34 as well as variants thereof.
Particular variants
will be those which have SEQ ID NO 77 linked at the C-terminal end.
As will be readily apparent to a man skilled in the art the sequence of the
individual
2o components of the DNA sequence i.e. the signal sequence, promoter sequence,
linker
sequence, protein sequence(s), terminator sequence for use in the methods
according to the
invention may be predicted from its known amino acid sequence and DNA encoding
the
protein may be manufactured using a standard nucleic acid synthesiser.
Alternatively, DNA
encoding the components of the invention may be produced by appropriate
isolation from
2~ natural sources.
The invention is further illustrated with reference to the following non-
limiting
examples and figures in which
Figure l: shows nucleotide sequence (SEQ ID NO 1) and corresponding amino acid
sequence (SEQ ID NO 2) of coding region of the DmAMPI gene. The amino acids
3o corresponding to mature DmAMPi are underlined. The nucleotides
corresponding to the
intron are double underlined.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- 19-
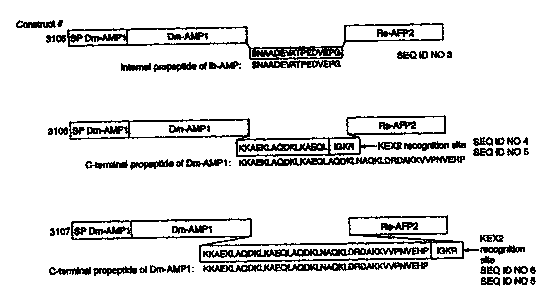
Figure 2: shows schematic representation of the coding regions from the vector
constructs
(SEQ ID NOS 3-8). Amino acids sequences below the internal propeptides
represent the
propeptide sequences from which the linker propeptides were derived.
Figure 3: shows schematic representation of plant transformation vector
pFAJ3105
Figure 4: shows schematic representation of plant transformation vector
pFAJ3106
Figure 5: shows schematic representation of plant transformation vector
pFAJ3107
Figure 6: shows schematic representation of plant transformation vector
pFAJ3108
Figure 7: shows schematic representation of plant transformation vector
pFAJ3109
Figure 8: shows nucleotide sequence (SEQ ID NO 9) and corresponding amino acid
sequence {SEQ ID NO 10) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pFAJ3105. The amino acids corresponding to
mature
DmAMPI and mature RsAFP2 are underlined and double-underlined, respectively.
Figure 9: shows nucleotide sequence (SEQ ID NO 11 ) and corresponding amino
acid
sequence (SEQ ID NO 12) of the open reading frame of the region comprised
between the
~ 5 NcoI and SacI sites of plasmid pFAJ3106. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP2 are underlined and double-underlined, respectively.
Figure I0: shows nucleotide sequence (SEQ ID NO 13) and corresponding amino
acid
sequence (SEQ ID NO 14) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pF AJ3107. The amino acids corresponding to
mature
2o DmAVIP 1 and mature RsAFP2 are underlined and double-underlined,
respectively.
Figure 11: shows nucleotide sequence (SEQ ID NO 15) and corresponding amino
acid
sequence (SEQ ID NO 16) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pF AJ3108. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP2 are underlined and double-underlined, respectively.
25 Figure 12: shows nucleotide sequence (SEQ ID NO 19) and corresponding amino
acid
sequence (SEQ ID NO 20) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pFAJ3109. The amino acids corresponding to
mature
DmAMP i are underlined.
Figure 13: shows the Dm-AMP1 e:cpression levels (as % of total soluble
protein) of a series
30 of transgenic individual plants transformed with construct pFAJ3105 and a
series of transgenic
individuals transformed with construct pFAJ3109.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-20-
Figure I=l: shows RP-HPLC analysis on a C8-silica column of crude extracts
from leaves
transformed with construct pFAJ3105 (A) or pFAJ3106 (B). Extracts were
prepared as
described in Materials and Methods. The column was eluted with a gradient of
acetonitrile in
0.1 % TFA (0-3~ min. I~ % - ~0 % acetonitrile in 0.1 % TFA). The eluate was
monitored on-
line for measurement of the absorbance at 214 nm (top trace), fractionated,
and subjected to
Elisa assays for DmAMP 1 (lower bar graph, black bars) and RsAFP2 (lower bar
Graph, white
bars). The elution position of authentic DmAMP l and RsAFP2 are indicated with
arrows on
the A," chromatograms.
Figure 1 ~: shows the results of reverse phase chromatography (RPC) of the
extracellular fluid
fraction of Arabidopsis plants transformed with construct 3105 (line 14). RPC
was performed
on a C8-silica column (Microsorb-MV, 4.6 x 250 mm, Rainin) equilibrated with
0.1
trifluoroacetic acid (TFA). After loading the column was eluted at a flow rate
of 1 mUmin for
20 min with 0.1 % TFA, whereafter a 35 min linear gradient was applied from 1~
to 50
acetonitrile in 0.1 % TFA. Absorbance (full line) was measured on-line at 280
nm and
acetonitrile concentration (dashed line) was measured on-line with a
conductivity monitor.
Fractions were collected and assessed for DmAMPI-CRP and RsAFP2-CRP using
ELISA
assays. Peak numbers in bold indicate presence of DmAMPl-CRP, peak numbers in
italic
indicate presence of RsAFP2-CRP.
Figure 16: shows the results of RPC of an extract of Arabidopsis plants
transformed with
2o construct 3105 (line 14). Samples were two different fractions from IEC
showing presence of
either DmAMPI-CRPs or RsAFP2-CRPs, namely those fractions eluting between O.I7
- 0.33
M NaCI (A), and 0.33 - 0.49 M NaCI (B). RPC was performed as in the legend to
Figure 14.
Absorbance (full line) was measured on-line at 280 nm and acetonitrile
concentration (dashed
line) was measured on-line with a conductivity monitor. Fractions were
collected and assessed
35 for DmAMPI-CRP or RsAFP2-CRP using ELISA assays. Peak numbers in bold
indicate
presence of DmAIvIP 1-CRP, peak numbers in italic indicate presence of RsAFP2-
CRP.
Figure 17: shows the amino acid sequence of the polyprotein precursors encoded
by constructs
pFAJ3105, pFAJ3106 and pFAJ3108. Dashes indicate omission from the full
sequence for
sake of brevity. The sequence in italic is the DmAMP 1 leader peptide, the
underlined
3o sequence is mature DmAMPI, the bold sequence is the linker peptide, the
double underlined
SU8ST1TUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-21 -
sequence is mature RsAFP2. Arrows indicate processing sites according to the N-
terminal
sequence and mass spectrometn~ analyses of purified DmAMP-CRPs and. RsAFP2-
CRPs.
Figure I 8: shows the RPC of the extracellular fluid fraction of Arabidopsis
plants
transformed with construct pFAJ3106 (line 9). RPC was performed and fractions
analysed
a as described in the legend to figure 15. Peak numbers in bold indicate
presence of
DmAMPI-CRP, peak numbers in italic indicate presence of RsAFP2-CRP.
Figure 19: shows the RPC results of an extract of Arabidopsis plants
transformed with
construct 3108 (line 9). The sample was a fraction from IEC showing presence
of either
DmAMPI-CRPs or RsAFP2-CRPs, namely those fractions eluting between 0.17 - 0.33
M
t0 NaCI and showing the presence of DmAMPI-CRPs. RPC was performed and
fractions
analysed as in the legend to Figure 15. Peak numbers in bold indicate presence
of DmAMPI-
CRP.
Figure 20: is a schematic representation of the coding region of constructs
pFAJ3105,
pFAJ3343, pFAJ3344, pFAJ334~, pFAJ3346, and pFAJ3369. Full arrowheads indicate
t5 experimentally determined cleavage sites. Open arrowheads indicate presumed
cleavage
sites. Abbreviations: SP DmAMP 1: signal peptide region of DmAMP 1 (see figure
1 );
DmAMP 1: mature protein region of DmAMP 1 (see figure 1 ); RsAFP2: mature
protein
region of RsAFP2 (Terms et al. 1995, Plant Cell, 7, 573-588). Linker peptide
sequences are
shown in full (SEQ ID NOS 3, 29. 21-24 respectively).
2o Figure 21: is a schematic representation of the coding region of constructs
pFAJ3367 with
linker peptide of SEQ ID NO 24. Abbreviations: SP DmAMPI : signal peptide
region of
DmAMP 1 (see figure 1 ); DmAMP 1: mature protein region of DmAMP 1 (see figure
1 );
RsAFP2: mature protein region of RsAFP2 (Terras et al. 1995, Plant Cell, 7,
573-588);
HsAFPl: mature protein region of HsAFPI (Osborn et al. 1995, FEBS Lett. 368,
257-262);
25 AceAMP 1; mature protein region of AceAMP 1 (Cammue et al. 1995. Plant
Physiol. 109,
445-455).
Figure 22: is a schematic representation of the coding region of constructs
pFAJ3106-2,
pFAJ3107-2, and pFAJ3108-2. Abbreviations: SP DmAMP 1: signal peptide region
of
DmAMP 1 (see figure 1 ); DmAMP 1: mature protein region of DmAMP 1 (see figure
1 );
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-22-
RsAFP2: mature protein region of RsAFP2 (Terras et al. 1995, Plant Cell, 7,
573-588); RS
Kex2p: recognition sequence (IGKR) of the Kex2 protease (Jiang and Rogers,
1999, Plant J.,
18, 23-32); AcAMP i : mature protein region of AcAMP 1 (De Bolle et al. Plant
Mol Biol, 31.
997-1008). The linker propeptide sequences are shown in full as SEQ ID NOS 25,
26 and 27
s respectively.
Figure 23: is a schematic representation of the coding region of constructs
pFAJ3368 and
pFAJ3370. Open arrowheads indicate presumed cleavage sites. Abbreviations: SP
DmAMP 1: signal peptide region of DmAMP 1 (see figure 1 ); DmAMP 1: mature
protein
region of DmAMP 1 (see figure 1 ); RsAFP2: mature protein region of RsAFP2
(Terras et al.
1995, Plant Cell, 7. X73-X88); 2A sequence: cleavage recognition site of the
Foot and Mouth
Disease Virus polyprotein. The linker propeptide sequence is shown in full as
SEQ ID NO
28.
Figure 24: shows nucleotide sequence (SEQ ID NO 30) and corresponding amino
acid
sequence (SEQ ID NO 31 ) of the open reading frame of the region comprised
between the
is rVcoI and SacI sites of plasmid pFAJ3343. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP2 are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 29).
Figure 25: shows the nucleotide sequence (SEQ ID NO 32) and corresponding
amino acid
sequence (SEQ ID NO 33) of the open reading frame of the region comprised
between the
~VcoI and SacI sites of plasmid pFAJ3344. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP2 are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 21 ).
Figure 26: shows the nucleotide sequence (SEQ ID NO 34) and corresponding
amino acid
sequence (SEQ ID NO 35) of the open reading frame of the region comprised
between the
.VcoI and Saci sites of plasmid pFAJ3345. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP2 are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 22).
Figure 27: shows the nucleotide sequence (SEQ ID NO 36) and corresponding
amino acid
sequence (SEQ ID NO 38) of the open reading frame of the region comprised
between the
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_23-
rVcol and SacI sites of plasmid pFAJ3346. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP? are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 23).
Figure 28: shows the nucleotide sequence (SEQ ID NO 38) and corresponding
amino acid
sequence (SEQ ID NO 39) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pFAJ3369. The amino acids corresponding to
mature
DmAMPI and mature RsAFP? are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 24) .
Figure 29: shows the nucleotide sequence and corresponding amino acid sequence
of the
open reading frame of the region comprised between the NcoI and Sacl sites of
plasmid
pFAJ3367. The amino acids corresponding to mature DmAMP, mature RsAFP2, mature
HsAFP 1 and mature AceAMP 1 are underlined, double-underlined, dashed-
underlined and
dotted-underlined, respectively. The amino acids corresponding to the internal
linker
peptides are in bold (SEQ ID NO 24).
is Figure 30: shows the nucleotide sequence (SEQ ID NO 42) and corresponding
amino acid
sequence (SEQ ID NO 43) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pFAJ3106-2. The amino acids corresponding to
mature
DmAMPI and mature RsAFP2 are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 4).
2o Figure 31: shows the nucleotide sequence (SEQ ID NO 44) and corresponding
amino acid
sequence (SEQ ID NO 45) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pFAJ3107-2. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP2 are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 6).
z5 Figure 32: shows the nucleotide sequence (SEQ ID NO 46) and corresponding
amino acid
sequence (SEQ ID NO 47) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pFAJ3108-2. The amino acids corresponding to
mature
DmAMP 1 and mature RsAFP2 are underlined and double-underlined, respectively.
The
amino acids corresponding to the internal linker peptide are in bold (SEQ ID
NO 7).
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_2:I_
Figure 33: shows the nucleotide sequence (SEQ ID NO 48} and corresponding
amino acid
sequence (SEQ ID NO 49) of the open reading frame of the region comprised
becveen the
NcoI and SacI sites of plasmid pFAJ3370. The amino acids corresponding to
mature
DmAMPI and mature RsAFP2 are underlined and double-underlined, respectivei~r.
The
linker sequence is indicated in bold type (SEQ ID NO 28} with the amino acids
corresponding to the 2A sequence indicated in bold italic.
Figure 34: shows the nucleotide sequence (SEQ ID 48) and corresponding amino
acid
sequence (SEQ ID NO 49) of the open reading frame of the region comprised
between the
NcoI and SacI sites of plasmid pFAJ3368. The amino acids corresponding to
mature
DmAMP l and mature RsAFP2 are underlined and double-underlined, respectively.
The
linker sequence is indicated in bold type with amino acids corresponding to
the 2A sequence
are indicated in bold italic.
The following Examples illustrate the invention.
1s
Example 1
Cloning of DmAMPl cDNA and DmAMPI gene
Cloning procedures and polymerase chain reaction (PCR) procedures were
performed
following standard protocols (Sambrook et al., 1989, Molecular Cloning: a
laboratory manual,
20 2"° edition, Cold Spring Harbor Laboratory Press, Cold Spring
Harbor, NY). A cDNA library
was constructed from near-dry seeds collected from flowers of Dahlia merckii.
Total RNA
was purified from the seeds using the method of Jepson I. et al. ( I991, Plant
Mol. Biol.
Reporter 9, 131-138). 0.6 mg of total RNA was obtained from 2 g of D. merckii
seed.
PolyATract magnetic beads (Promega) were used to isolate approximately 2 ug
poly-A+ RNA
25 from 0.2 mg of total RNA.
The poly-A+ RNA was used to construct a cDNA library using a ZAP-cDNA
synthesis
kit (Stratagene). Following first and second strand synthesis, cDNAs were
ligated with vector
DNA. After phage assembly using Gigapack Gold (Stratagene) packaging extracts,
approximately 1 x 105 plaque forming units (pfu) were obtained.
3o Using oligonucleotides AFP-S (5'-TG(T,C)GANAANGCN(A,T)(G,C)NAA(A,G)ACNTGG)
SUBSTITUTE SHEET (RUt.E 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_7j_
(SEQ ID NO 13) based on the N-terminal sequence CEKASKTW (SEQ ID NO 14) of
DmAMPI, Osborn R.W. er al., 1995, FEBS Lett. 368, 257-262) and AFP-3EX (~'-
CA(A,G)TT(A,G)AANTANCANAAA(A,G) CACAT) (SEQ ID ~i0 52) based on the C-
terminal sequence MCFCYFNC (SEQ ID NO 53) of DmAMPI) and genomic DNA isolated
from D. merckii leaves. a 144 by PCR product was produced and isolated from an
agarose gel.
The PCR product was cloned into pBluescript. The insert of 10 transformants
were
sequenced. The sequences represented 3 closely homologous DmAMPI-like genes
one of
which, PCR clone 4, encoded the observed mature DmAMP 1. The 144 by PCR
product
mixture labelled with''--P CTP was used to probe Hybond N (Amersham) filter
lifts made
o from plates containing a total of 6 x 10' pfu of the cDNA library. Thirty
potentially positive
signals were observed. 22 plaques were picked and taken through two further
rounds of
screening. After in vivo excision 13 clones were characterised by DNA
sequencing.
Four classes of DmAMP related peptides were encoded by the 13 cDNA clones.
Three
versions of the DmAMP mature protein region were represented in the four
classes. One of
the classes (Dm2.5 type) contained a mature protein region which may
correspond to
DmAMP2 (Osborn R.W. et al., 1995, FEBS Lett. 368, 257-262). None of the cDNAs
encoded
a mature protein region equivalent to the observed mature DmAMP 1 peptide
sequence.
Using the sequence of PCR clone 4 (above) and information from the N- and C-
terminal ends of the peptides deduced from cDNA sequences, two pairs of
oligonucleotides
?o were designed for amplification of a gene encoding DmAMPI. Genomic DNA from
D.
merckii was used in a PCR reaction with oIigonucleotides MATAFP-SP (5'-
ATGGC(C,G)AAN(A,C)(A,G)NTC (A,G)GTTGCNTT) (SEQ ID NO 66) and MATAFP-S
(5'- AAACACATGTGTTTCCCATT) (SEQ ID NO 54), the PCR product was cloned into
pBluescript and clones were sequenced. A clone containing the ~' half of a
DmAMP 1 gene
'S was identified. Genomic DNA from D. merckii was used in a PCR reaction with
MATAFP-3
(~'- AGCGTGTCATGTGCGTAAT) (SEQ ID NO 5~ ) and DM2WIAT-3 (5'- TAAAGA
AACCGACCCTTTCACGG) (SEQ ID NO 56), the PCR product was cloned into pBluescript
and clones were sequenced. A clone containing the 3' half of a DmAMP 1 gene
was identified.
The 5' and 3' sections of the mature gene were combined to assemble the
sequence of the
3o coding region of the DmAMP 1 gene (Figure 1 ).
SU8ST1TUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-26-
The DmAMP 1 gene encodes a precursor with a 28 amino acids leader peptide. a
50
amino acids mature protein and a 40 amino acids C-terminal propeptide. The
open reading
frame is interrupted by a 92 by intron located within the leader peptide
region.
To eliminate the intron from the DmAMP 1 gene sequence and to allow cloning of
the
DmAMP 1 encoding region, either with or without the C-terminal propeptide
region, into an
expression cassette vector, two PCR reactions were carried out with
respectively the primer
sets DMVEC-3 (~'- ATGCATCCATGGTGAATCGGTCGGTTGCGTTCTCCGCGTTCGTT
CTGATCCTTTTCGTGCTCGCCATCTCAGATATCGCATCCGTTAGTGGAGAACTATG
CGAGAAA) (SEQ ID NO 57) and DMVEC-2 (5'-
o AAACCGACCGAGCTCACGGATGTTCAACGTTTGGA AC) (SEQ ID NO 58), and
DMVEC-3 and DMVEC-1 (5'- AGC.AAGCTTTTCGGGAGCTCAACAATTGA
AGTAA)(SEQ ID NO ~9). DMVEC-3 primes at the top strand of the DmAMPI gene,
corresponds to the leader peptide region without the intron and introduces an
NcoI site at the
translation start. DMVEC-2 primes at the bottom strand of the DmAMPI gene at
the 3'-end
15 of the C-terminal propeptide region and introduces a SacI site behind the
translation stop
codon. DMVEC-4 primes at the bottom strand of the DMAMPI gene at the 3' end of
the
mature protein region, fuses a stop codon behind this region and introduces a
SacI site behind
the stop codon.
Both PCR products were cut with NcoI and SacI which cleaved the PCR products
in
20 two fragments due to an internal NcoI site in the mature protein region.
The resulting NcoI-
SacI and NcoI-NcoI fragments were cloned sequentially in plasmid pMJB 1. pMJB
1 is an
expression cassette vector containing in sequence a HindIII site, the enhanced
cauliflower
mosaic 35S RNA (CaMV35S) promoter (Kay R. et al., 1987, Science 236, 1299-
1302), aXhoI
site, the 5' untranslated leader sequence of tobacco mosaic virus (T~IV)
(Gallie D.R. and
25 WaIbot V., 1992, Nucl. Ac. Res. 20, 4631-4638) a polylinker including NcoI,
SmaI, KpnI and
SacI sites, the 3' untranslated terminator region of the Agrobacterium
tumefaciens nopaline
synthase gene {Bevan M.W. et al., 1983, Nature 304, 184-187) and an EcoRI
site. The
resulting plasmids were termed pDMAMPE (leader peptide region. mature protein
region and
C-terminal propeptide region) and pDMAMPD (leader peptide region and mature
protein
3o region), respectively. The coding regions were verified by DNA sequencing.
SU8ST1TUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-27_
Examine 2
Construction of plant transformation vectors
To explore the possibility of expressing polyprotein precursor genes in
plants, four different
plant transformation vectors were made with the aim to co-express two
different cysteine-rich
plant defensins with antifungal properties, namely RsAFP2 and DmAMP 1. The
polyprotein
precursor regions of these constructs all featured a leader peptide region
derived from the
DmAMPI cDNA, the mature protein domain of DmAMPI, an internal propeptide
region, and
the mature protein domain of RsAFP2. The four constructs differed only in the
internal
propeptides (Figure 2):
~ construct 3 I OS has one of the IbAMP internal propeptides as a propeptide
separating
DmAMP 1 and RsAFP2.
~ construct 3 I 06 has a propeptide consisting of a pan of the DmAMPl
propeptide and a
putative subtilisin-like protease processing site (IGKR) (SEQ ID NO 67) at its
C-terminus.
~ construct 3107 is identical to construct 3106 except that the entire DmAMPI
propeptide
was taken.
~ construct 3 I 08 has a propeptide consisting of the AcAMP2 propeptide and a
putative
subtilisin-like protease processing site (IGKR) at its C-terminus.
The rationale behind constructs 3106, 3107 and 3108 is based on our
observations that the C-
terminal propeptides of AcAMP2 and DmAMP I are cleaved off at their N-terminus
when
expressed as AcAMP2- and DmAMPI-preproproteins in tobacco, respectively, while
this
processing event does not detract the mature proteins from being sorted to the
apoplast (De
Bolle et al., 1996, Plant Mol. Biol. 31, 993-1008; R.W. Osborn and S.
Attenborough, personal
communication). This infers that the processing enzymes are either in the
secretory pathway
or in the apoplast. On the other hand, C-terminal cleavage of the internal
propeptide in these
constructs should be executed by a subtilisin-like protease, a member of which
in yeast (Kex2)
is known to occur in the Golgi apparatus (Wilcox C.A. and Fuller R.S., 1991,
J. Cell. Biol.
I 15, 297), while a member in tomato occurs in the apoplast (Tornero P. et
al., 1997, J. Biol.
Chem. 272, 14412-14419). Proteins deposited in the apoplast, the preferred
deposition site for
antimicrobial proteins engineered in transgenic plants (Jongedijk E. et al.,
1995, Euphytica 85.
173-180; De Bolle et al., 1996, Plant Mol. Biol. 31, 993-1008) are normally
synthesized via
the secretory pathway, encompassing the Golgi apparatus.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_~g_
A construct was also made for expression of only DmAMP 1 (construct 3109,
figure 7).
Schematic representations of the plant transformation vectors prepared,
pF.~J3105,
pFAJ3106, pFAJ3107, pFAJ3108 and pFAJ3 I09, are shown in Figures 3 to 7,
respectively.
T'he nucleotide sequences comprised between the XhoI and SacI sites of these
plasmids, which
encompass the regions encoding antimicrobial proteins, are presented in
Figures 8 to 13. The
regions comprised between the ~YhoI and SacI sites of plasmid pFAJ3105 (shown
in Figure 8)
was constructed following the two-step recombinant PCR protocol of Pont-Kindom
G.A.D.
(1994, Biotechniques I6, 1010-1011). Primers OWBI75
(5'AGGAAGTTCATTTCATTTGG) and (SEQ ID NO 68), OWB278 (S'-
to GCCTTTGGCACAACTTCTGT
CCTGGCTCCACGTCCTCTGGGGTAGCCACCTCGTCAGCAGCGTTGGAACAATTGA
AGTAACAGAAACAC) (SEQ ID NO 60) were used in a first PCR reaction with plasmid
pDMAMPE (see above) as a template. The second PCR reaction was done using as a
template
plasmid pFRG4 (Terras F.R.G. et al., 1995, Plant Cell 7, 573-588) and as
primers a mixture of
15 the PCR product of the first PCR reaction, primer OWB 175 and primer OWB
172
(5'TTAGAGCTCCTATTAACAAGGAAAGTAGC (SEQ ID NO 61), SacI site underlined).
The resulting PCR product was digested with XhoI and SacI and cloned into the
expression
cassette vector pMJB 1 (see above). The expression cassette in the resulting
plasmid, called
pFAJ3099, was digested with HindIII (flanking the 5' end of the CaMV35S
promoter) and
3o EcoRI (flanking the 3' end of the nopaline synthase terminator) and cloned
in the
corresponding sites of the plant transformation vector pGPTVbar (Becker D. et
al., 1992, Plant
Mot. Biol. 20, 1195-1197) to yield plasmid pFAJ3105.
Plasmids pFAJ3106, pFAJ3107 and pFAJ3108 were constructed analogously except
that primer OWB278 in the first PCR reaction was replaced by the following
primers,
35 respectively: OWB279 (5'-
GCCTTTGGCACAACTTCTGCCTCTTTCCGATGAGTTGTTCGGCTTT AAGTTTGTC);
(SEQ ID NO 62), OWB303 (5'-GCCTTTGGCACAACTTCTGCCTCTTTCCG
ATCGGATGTTCAACGTTTGGAACC) (SEQ ID NO 63) ; OWB304 (5'-
GCCTTTGGCACAACTTCTGCCT
~o CTTTCCGATAGTTTTGGTGGCAGCAACATCAGCTTGGTGATCCACAGTAGTACTGG
CACAATTGAAGTAACAGAAACAC) (SEQ ID NO 64).
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-29-
Plasmid pFAJ3109 was constructed by cloning the HindIII-EcoRI fragment of
plasmid
pDMAMPD (see above) into the corresponding sites of plant transformation
vector pGPTVbar
(see above).
Example 3
Plant transformation
Arabidopsis thaliana ecotype Columbia-O was transformed using recombinant
Agrobacteriurn tumefaciens by the inflorescence infiltration method of
Bechtoid N. et al.
(1993, C.R. Acad. Sci. 316, 1194-1199). Transformants were selected on a
sand/perlite
o mixture subirrigated with water containing the herbicide Basta (Agrevo) at a
final
concentration of 5 mg/1 for the active ingredient phosphinothricin.
Example 4
Assays for target proteins including Elisa assays and protein assays
Antisera were raised in rabbits injected with either RsAFP2 (purified as
described in
Terras F.R.G. et al., 1992, J. Biol. Chem. 267, 15301-15309) or DmAMPI
(purified as in
Osborn R. W. et al., 1995, FEBS Lett. 368, 257-262). ELISA assays were set up
as
competitive type assays essentially as described by Penninckx LA.M.A. et al.
(1996, Plant
Cell 8, 2309-2323). Coating of the ELISA microtiter plates was done with ~0
ng/ml RsAFP2
30 or DmAMP 1 in coating buffer. Primary antisera were used as I 000- and 2000-
fold diluted
solutions (DmAMPI and RsAFP2, respectively) in 3 % (w/v) gelatin in PBS
containing
0.05 % (v/v) Tween 20.
Total protein content was determined according to Bradford (1976, Anal.
Biochem. 72,
248-254) using bovine serum albumin as a standard.
Arabidopsis leaves were homogenized under liquid nitrogen and extracted with a
buffer consisting of 10 mM NaHZPO" 15 mM Na,HP04, 100 mM KCI, 1.5 M NaCI. The
homogenate was heated for 10 min at 85°C and cooled down on ice. The
heat-treated extract
was centrifuged for I 5 min at 15 000 x g and was injected on a reserved phase
high pressure
liquid chromatography column (RP-HPLC) consisting of C8 silica (0,46 cm x 25
cm; Rainin)
3o equilibrated with 0.1 % (v/v) trifluoroacetic acid (TFA). The column was
eluted at 1 ml/min
in a linear gradient in 3~ min from 1 ~ % to 50 % (v/v) acetonitrile in 0.1 %
(v/v) TFA. The
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
- j~ -
eluate was monitored for absorbance at 214 nm, collected as 1 ml fractions,
evaporated and
finally redissolved in water. The fractions were tested by ELISA assays.
Example ~
Preparation of intracellular extract
Intercellular fluid was collected from Arabidopsis leaves by immersing the
leaves in a
beaker containing extraction buffer ( 10 mM~ NaH,P04, 1 S mM Na,HPOa, 100 mM
KCI, 1.~ M
NaCI). The beaker with the lea~~es was placed in a vacuum chamber and
subjected to six
consecutive rounds of vacuum for 2 min followed by abrupt release of vacuum.
The infiltrated
leaves were gently placed in a centrifuge tube on a grid separated from the
tube bottom. The
intercellular fluid was collected from the bottom after centrifugation of the
tubes for 15 min at
1800 x g. The leaves were resubjected to a second round of vacuum infiltration
and
centrifugation and the resulting (extracellular) fluid was combined with that
obtained after the
first vacuum infiltration. After this step the leaves were extracted in a
Phastprep
. (BIO101/Savant) reciprocal shaker and the extract clarified by
centrifugation (10 min at
10,000 x g) and the resulting supernatant considered as the intracellular
extract.
Expression levels of DmAMPl and RsAFP2 were analysed in leaves taken from a
series of T1 transgenic Arabidopsis plants resulting from transformation with
the constructs
described above. The results of the expression analyses based on Elisa assays
as described
above are presented in Table 1.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-31 -
Table 1: Expression levels of Dm-AMP1 and Rs-AFP2 in transgenic Arabidopsis
Iines
constructlineExpressionexpression Expression e~cpression
level of level constructline level of level
Dm- of Rs- Dm- of Rs-
~MP 1 (%) .-~FP2 aMP 1 (%) .aFP2
(%) (r)
3105 1 0,77 0,29 3107 1 0,04 0,04
2 1,13 0,22 2 0,75 0,42
3 0,48 0,20 3 0,14 0.13
4 0,005 <0,001 4 0,01 0,01
5 0,36 0,05 ~ 0,27 0,29
6 0,99 0,25 3108 1 0,47 0,10
7 0,60 0,09 2 3,00 0,53
8 0,13 <0,001 3 0,91 0,24
9 0,25 0,08 4 2,04 0,22
10 4,1~ 0,85 ~ 0,17 0,04
lI 1,35 0,35 6 0,55 0,05
12 0,24 0,07 7 0,16 0,11
13 4,43 0,91 8 0,05 0,02
14 1,18 0,24 9 0,45 0,02
15 0,68 0,17 3109 1 0,19 nd
16 0,49 0,07 2 0,05 nd
3106 1 0,10 0,001 3 0,02 nd
2 1,82 0,008 4 0,20 nd
3 0,68 0,20 5 0,10 nd
4 1,15 0,38 6 0,06 nd
5 0,20 0,10 7 0,07 nd
6 0,10 0,05 8 0,003 nd
7 0,40 0,17 9 0,18 nd
8 2,64 0,50
9 0,40 0,1 S
10 0,21 0,07
11 0,06 0,03
12 0,24 0,09
In the above Table "nd" indicates not done.
SUBSTITUTE SHEET (RULE 26j
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
Most of the tested lines transformed with the polyprotein constructs 3105,
3106. 3 I07 and
3108 clearly expressed both DmAMPI-CRPs (DmAMPI-crossreactive proteins) and
RsAFP2-
CRPs (Rs-AFP2-crossreactive proteins). There was generally a good correlation
between
DmAMPI-CRP and RsAFP2-CRP levels. However, the RsAFP2-CRP levels were
generally
2 to 5-fold Lower than the DmAMP I -CRP levels. The Elisa assays for measuring
the
RsAFP2-CRPs in the extracts are, however, less reliable than those for the Dm-
AMP/-CRPs.
In Rs-AFP? Elisa assays, dilutions of extracts of transgenic plants yielded
dose-response
curves that deviated from those obtained for dilutions of standard solutions
containing
authentic Rs-AFP2, indicating that the majority of the Rs-AFP2 -CRPs in the
extracts were
to imunologically not identical to RsAFP2 itself: Deviations from RsAFP2
standard dose-
response curves were much more pronounced for extracts from plants transformed
with
constructs 3106, 3107, and 3108 than for those of plants transformed with 31
O5.
None of the extracts showed deviations from Dm-AMP 1 standards in dose
response
curves in Dm-AMP/ Elisa assays. The DmAMP-CRP levels in the lines transformed
with
2 5 the poiyprotein constructs 31 O5, 3106, 3107 or 3108 were generally much
higher compared to
those in the line transformed with the single protein construct 3I09. This is
also illustrated in
Figure I 3 where DmAMP 1-CRP expression levels are compared for plants
transformed with
the polyprotein construct 3105 and plants transformed with the single protein
construct 3109.
Expression levels as high as 4% of total protein (e.g. DmAMPI-CRP Level in
lines 3105-l~
2o and 3105 -18. see table I) have so far never been reported in the
literature for a peptide
expressed in transgenic plants. Hence, the use of polyprotein constructs
appears to result in
markedly enhanced expression, which is an unexpected finding.
Example 6
25 Separation of proteins processed from~olyprotein precursors
A transgenic line was selected among each of the populations transformed with
either
construct 3105 (line 1) or 3106 (line 2) and the selected lines were further
bred to obtain plants
homozygous for the transgenes. In order to analyse whether DmA~iP 1 and RsAFP2
were
correctly processed in these lines, extracts from the plants were prepared as
described in
3o Example 1 and separated by RP-HPLC on a C8-silica column. Fractions were
collected and
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
assessed for presence of compounds cross-reacting with antibodies raised
against either
DmAMP I or RsAFP2 using Elisa assays as described in Example 4.
As shown in figure 15. DmAlVIP 1- CRPs eluted at a position identical or very
close to that of
authentic DmAMP 1 in the line transformed with construct 31 OS as well as in
that transformed
with construct 3106. Likewise, RsAFP2-CRPs were detected in both the construct
3105 and
3106 lines at an elution position identical or very close to that of authentic
RsAFP2. None of
the fractions reacted with both the anti-DmAMPl and anti-RsAFP2 antibodies,
indicating that
an uncleaved fusion protein was not present in the extracts. No cross-reacting
compounds
were observed in a non-transformed line.
to Thus it appears that the primary translation products of the transcription
units of
construct 3105 (IbAMP internal propeptide as linker peptide) and construct 3 I
06 {partial
DmAMP 1 C-terminal propeptide with subtilisin-like protease site as a linker
peptide) are
somehow processed to yield separate DmAMPI-CRPs and RsAFP2-CRPs that appear to
be
identical or very closely related to DmAMPI and RsAFP2, respectively, based on
their
15 chromatographic behavior.
Example 7
Analysis of the subcellular location of coexeressed plant defensins
In order to determine whether the coexpressed plant defensins are either
secreted
3o extracellularly or deposited intracellularly, extracellular fluid and
intracellular extract fractions
were obtained from leaves of homozygous transgenic Arabidopsis lines
transformed with
either constructs 3105 (line 2), 3106 (line 2) or 3108 (line 12). The
cytosoIic enzyme glucose-
6-phosphate dehydrogenase was used as a marker to detect contamination of the
extracellular
fluid fraction with intracellular components. As shown in Table 2, glucose-6-
phosphate
25 dehydrogenase was partitioned in a ratio of about 80/20 between
intracellular extract fractions
and extracellular fluid fractions. In contrast, the majority of DmAMPI-CRP
and. RsAFP2-
CRP content in all transgenic plants tested was found in the extracellular
fluid fractions.
These results indicate that both plant defensins released from the polyprotein
precursors are
deposited primarily in the apoplast. Hence, all processing steps that result
in cleavage of the
3o polyprotein structure must occur either in the apoplast or along the
secretory pathway i.e. in
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_ 34 -
the endoplasmic reticulum. the Golgi apparatus or in vesicles trafficking
between Golgi and
apoplast.
s Table 2: Relative abundance of glucose-6-phosphate dehydrogenase activity
(GPD),
DmAMP 1 and RsAFP2 in the extracellular fluid (EF) and intracellular extract
(IE) fractions
obtained from transgenic .Arabidopsis plants:
Construct Relative abundance' (%) of
UYD DmAMP l RsAFP2
EF IE EF IE EF IE
pFAJ3105 17 83 93 7 92 g
pFAJ3106 17 83 94 6 60 40
pFAJ3108 20 80 98 2 75 25
'Relative abundance is expressed as % of the sum of the contents in the EF and
IE fractions.
1o Example 8
Purification of proteins processed from nolvprotein precursor construct 3105
Transgenic line 14 from the population transformed with construct 3105 was
further bred to
obtain plants homozygous for the transgene. The DmAMPI-CRPs and RsAFP2-CRPs
were
purified by reversed phase chromatography from extracellular fluid prepared
from leaves of
IS this line. To this end, leaves were vacuum infiltrated with a buffer
containing 50 mM MES
(pH6) and a mixture of protease inhibitors ( I mM
phenylmethylsulfonylfluoride, 1 mM N-
ethylmaleimide, ~mM EDTA and 0.02 mM pepstatin A), and the extracellular fluid
collected
by centrifugation. Using this procedure homogenization and hence exposing
DmAMPI-
CRPs and RsAFP2-CRPs to compartimentalized proteases was avoided. The
collected
?o extracellular fluid was analyzed by RP-HPLC on a C8-silica column
(Microsorb-MV, 4.6 x
250 mm, Rainin) and the fractions tested for presence of DmAMPI-CRPs and
RsAFP2-
CRPs by Elisa using antibodies raised against DmAMP l and RsAFP2,
respectively. The
result of this analysis for the Arabidopsis transgenic line 14 transformed
with construct 3105
is shown in figure 1~. DmAMPI-CRPs eluted in two peaks, the latter of which
eluted at a
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_3J-
position very close to that of authentic DmAMP I . RsAFP2-CRPs were found in a
single
peak that was well separated from the DmAMP 1-CRP peaks and eluted at a
position very
close to that of authentic RsAFP?. None of the fractions reacted with both the
anti-
DmAMPI and anti-RsAFP2 antibodies, indicating that an uncleaved fusion protein
was
absent from the extracellular fluid. Based on comparison of the peak areas of
the DmAMPI-
CRPs and RsAFP2-CRPs with those of a series of standards consisting of
authentic Dm-
AMP l and RsAFP2, respectively, it was judged that the extract for the line
transformed with
construct 3105 contained about equal amounts of DmAMPI-CRPs and RsAFP2-CRPs.
This
indicates that cleavage of the polyprotein precursor in this line results in
about equimolar
1o amounts of DmAMPI-CRPs and RsAFP2-CRPs. Very similar chromatograms were
obtained upon analysis of extracellular fluid prepared from transgenic line 2
(results not
shown), indicating that the chromatographic pattern of DmAMPI-CRPs and RsAFP2-
CRPs
is independent from the transgenic line tested.
To test whether the purification procedure based on extracellular fluid
preparation
~ 5 reflects the true composition in DmAMP-CRPs and RsAFP2-CRPs of the
transgenic
Arabidopsis leaves, an alternative purification procedure was developed
starting from a crude
leaf extract. To this end, leaves were homogenized under liquid nitrogen and
extracted with
50 mM MES (pH6) containing a mixture of protease inhibitors ( I mM
phenylmethylsulfonylfluoride, 1mM N-ethylmaleimide, ~mM EDTA and 0.02 mM
pepstatin
2o A). The homogenate was cleared by centrifugation (10 min at 10000 x g). The
supernatant
was then fractionated by ion exchange chromatography (IEC) and subsequently by
reversed
phase chromatography (RPC). After each separation, fractions were collected
and assessed
for DmAMP-CRPs and RsAFP2-CRPs using two different Elisa assays with
antibodies
raised against DmAMPI and RsAFP2, respectively. IEC was performed by passing
the
25 extract over a cation exchange column (Mono S, 5 x SO mm, Pharmacia) at pH
6. When the
column was eluted with a linear gradient of 0 to 0.5 M NaCI in 50 mM N-
morpholino ethane
sulfonic acid (MES) at pH 6, DmAMPI-CRPs were detected in fractions eluting
between
0.17 and 0.33 M NaCI, while RsAFP2-CRPs eluted between 0.24 and 0.49 M NaCI.
Fractions containing either DmAMPI-CRPs or RsAFP2-CRPs were pooled into two
3o fractions (0.17 to 0.33 M NaCI; and 0.33 to 0.49 M NaCI) which were each
subjected to RPC
on a C8-silica column (Microsorb-MV, 4.6 x 250 mm, Rainin) eluted with a
linear gradient
SUBSTITUTE SHEET (RUSE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-36-
of acetonitrile (Figure 16). DmAMPI-CRPs eluted in two peaks. the latter of
which eluted
at a position very close to that of authentic DmAMP 1. RsAFP2-CRPs were found
in a single
peak that was well separated from the DmAMP-CRP peaks and eluted at a position
very
close to that of authentic RsAFP2. Again, none of the fractions reacted with
both the anti-
DmAMP I and anti-RsAFP? antibodies, indicating that an uncleaved fusion
protein was not
present in the extracts.
The different DmA>ViPI-CRPs and RsAFP2-CRPs purified from extracellular fluid
were subjected to N-terminal amino acid sequence analysis (procedures as
described in
Cammue et al., 1992, J. Biol. Chem., 2228-2233) as well as to NL~LDI-TOF
(matrix-assisted
t o laser desorption ionization-time of flight) mass spectrometry (Mann and
Talbo, 1996, Curr.
Opinion Biotechnol. 7, 11-19). The C-terminal amino acid was determined based
on the best
approximation of the predicted theoretical mass by the experimentally
determined mass
(Table 3). Both the minor DmAMPl-CRPs, p3105EF1, and the major DmAMPI-CRP,
p3105EF2 (protein codes as in figure 15 and Table 3), had exactly the same N-
terminal
15 sequence as mature DmA.MP 1. p3 I OSEF 1 and p31 OSEF2 had masses that were
consistent
with the presence of a single additional serine residue at their C-terminal
end compared to
authentic DmAMPI. However, while the mass ofp3105EF2 corresponded exactly
{within
experimental error) to that calculated for a DmAMP 1 derivative with a C-
terminal serine
(hereafter called DmAMPI+S), that of p3I05EF1 was in excess b~~ about 8 dalton
relative to
2o the calculated mass for DmAMPl+S, Hence, this protein might be a DmAMPI+S
derivative
with reduced disulfide bridges. The RsAFP2-CRP fraction p3103EF3 represents,
based on
N-terminal sequence and mass data, an RsAFP2 derivative with the additional
pentapeptide
sequence DVEPG at its N-terminus. This protein is further referred to as
DVEPG+RsAFP2.
The different DmAMPI-CRPs and RsAFP2-CRPs purified from total leaf extract
were
25 analyzed in the same way. The analyses indicated that the same molecular
species were
present in the total leaf extract, i.e. DmAMPI+S, a putatively reduced form of
DmAMPI+S,
and DVEPG+RSAFP2 (Table 3 see Example 10 below).
The purified fractions containing the major processing products, DmAMPI+S and
DVEPG+RsAFP2 respectively, were subjected to an antimicrobial activity test
using the
30 fungus Fusarium culmorum according to the procedure outlined by Cammue et
al. (1992, J.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_ 37 -
Biol. Chem. 267, 2228-223.i). The specific antimicrobial activity, expressed
as protein
concentration required for ~0 % growth inhibition of the test organism, of
purified
DmAMPI+S was identical to that of authentic DmAMPI. The specific antimicrobial
activity of purified DVPEG-RsAFP2 was about 2-fold lower relative to that of
authentic
RsAFP2. The slight drop in specific antimicrobial activity of DVPEG+RsAFP2 is
most
likely due to the presence of s additional N-terminal amino acids.
Nevertheless, our data
prove that processing of the polyprotein precursors in transgenic plants can
result in the
release of bioactive proteins.
Analysis of the AFPs produced in transgenic plants transformed with construct
310
reveals that the precursor is apparently processed by three cleavage steps
(Figure 17):
(i) the precursor is cleaved at the C-terminal end of the leader peptide in
the same way as for
the authentic DmAMP I precursor; (ii) the precursor is cleaved at the C-
terminal end of the
first amino acid of the linker peptide, thus releasing DmAMPI+S; (iii) the
precursor is
i5 further processed at the N-terminal end of the fifth last residue of the
linker peptide, thus
releasing DVEPG+RsAFP2. It is not known which proteases effect the observed
cleavages,
nor how many different proteases are involved. Cleavages in the linker
peptides might
involve only endoproteinases or result from the coordinated action of
endoproteinases and
exopeptidases that further trim the cleavage products at their ends.
Processing at the C-
2o terminal side of the linker peptide occurs between the two acidic residues
E and D. The
acidic doublet might be a target sequence for a specifc endoproteinase. An
aspartic
endoproteinase that is able to cleave between two consecutive acidic residues
has previously
been purified from Arabidopsis seeds (D'Hondt et al. 1993, J. Biol. Chem. 268,
20884-
20891 ). It is worthwhile to mention that the sequence ED occurs at the very C-
terminal end
25 in five out of six internal propeptides of the IbAMP 1 polyprotein
precursor (Tailor et al.
1997, J. Biol. Chem. 272, 24480-24487). In one of the six internal IbAMP
propeptides, more
precisely the one that was used in construct 3105, the ED sequence does not
occur at the C-
terminal end of the propeptides but is separated by 4 amino acids from this
end. Processing
of this propeptide in Impatiens balsamina might involve cleavage of the ED
sequence
3o followed by partial N-terminal trimming of the resulting protein by an
aminopeptidease.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_~$_
It would be expected that an internal propeptide resembling the IbAMP 1
propeptide
used in construct 310 but in which the ED dipeptidic sequence is moved to the
C-terminal
end of the propeptide, would result in a cleavage product with only one or no
extra N-
terminal amino acids in the protein located C-terminally from the internal
propeptide.
Alternatively, another IbAMP 1 propeptide which already has an ED sequence at
its C-
terminal end (Tailor et al.. 1997, J. Biol. Chem. 272, 24480-24487) or a
related sequence
might give a similar improvement of processing accuracy.
Example 9
to Purification of proteins processed from nolvproteinprecursor construct
pFAJ3106
Transgenic line 9 from the population of Arabidopsis plants transformed with
construct
pFAJ3106 was further bred to obtain plants homozygous for the transgene. The
DmAMPI-
CRPs and RsAFP2-CRPs were purified by reversed phase chromatography from leaf
extracellular fluid prepared in the same way as described above in Example 8
for the line
15 transformed with construct pFAJ3 I O5. The chromatogram of this separation
is shown in
Figure 18. DmAMP I-CRPs eluted in two peaks, called p3106EF 1 and p3106EF2.
Both
fractions had the same N-terminal sequence as DmAMPI (Table 3 see Example 10
below).
The mass of p3106EF2 corresponded to that predicted for a DmAMPI derivative
with an
additional lysine. We therefore conclude that it represents the cleavage
product of the
2o precursor cleaved at the signal peptide cleavage site and C-terminally
behind the first residue
(lysine) of the linker peptide; This protein is further referred tows
DmAMPI+K.
The RsAFP2-CRP fraction was found by N-terminal amino acid sequencing to start
by the sequence LIGKRQK. Hence, this protein, called QLIGKR+ RsAFP2, is
derived from
cleavage of the precursor N-terminally from the sixth last residue (glutamine)
of the linker
35 peptide. The proposed cleavage steps involved in processing of the
precursor of construct
pFAJ3106 are shown in Figure 17.
Example 10
Purification of proteins processed from ~olvnrotein precursor construct
pFAJ3108
Transgenic line 9 from the population of Arabidopsis plants transformed with
construct
30 pFAJ3 I 08 was further bred to obtain plants homozygous for the transgene.
The DmAMP 1-
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCTlGB99/02716
-39-
CRPs and RsAFP2-CRPs were purified from a total crude leaf extract of this
line. :ollowing
a procedure based on IEC and RPC as described above in Example 8 for the line
transformed
with construct 310. The chromatograms of the IEC and RPC separations are shown
in
Figure 19. The IEC separation yielded two peaks containing DmAMPI-CRPs.
However. no
RsAFP2-CRPs could be detected in any of the eluate fractions. As RsAFP2-CRPs
were
clearly present in crude extracts and EF fractions of plants transformed with
construct
pFAJ3108 (see tables l and 2) the RsAFP2-CRPs must have been lost during the
separation.
The most likely explanation is that the RsAFP2-CRPs were not eluted from the
IEC column
to with 0.~ M NaCI, the highest concentration used in the elution gradient.
Fractions
containing DmAMPI-CRPs were separated by RPC, yielding two DmAMPI-CRP peak.
Analysis of this fraction by N-terminal sequencing and MALDI-TOF mass
determination
(Table 3) revealed that it represents a DmAMPI derivative with an additional
alanine at its
C-terminus (DmAMP 1+A), This protein results from cleavage of the precursor at
the signal
15 peptide cleavage site and C-terminally from the first residue (aIanine) of
the linker peptide
(Figure 17).
Table 3: Mass determined by MALDI-TOF-MS or EI-'HIS and N-terminal sequence
determined by automated Edman degradation of DmAMPI-CRP and RsAFP2-
CRP fractions purified as described in Figures 15, 16, 18 and 19. Also shown
are
20 the predicted C-terminal sequence that gives best correspondence between
experimental mass and theoretical mass.
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 - 40 - PCT/GB99/02716
V 'C N
m ~mn vmn c~ ~ m
~ v~ oo N
M
c~ N N ~ N N M M
~ ~
H V N ,~ "~ v1 ~ N V1 00
et N v1 00
O p N O O ~ G~
r1 ~
~O ~O N ~O N v1
~D N ~O v1
v1 v'1 ~O ~n vWD
V1 UWC
~~ ~'~'Uv~v~~ a~a~~ a
N
_ U U ~ U
~
~ ~a~ ww~~w~ ww~
, .
U U V U U U U U U
U
p v~ vWG r~ v~ v~ v~
~n ~G ~
~a~~
~ U U w U U U U ~ U
w
c'
~
w w p w w w
w
p ~
w
.J
b
~
~
M D A ~ !~
os
p D G7 O
Qozzzz zzz z
w
b
.
~ N M O ~' O M
~
~~o ~ ~~ z z
b a i u w
' ; w w w
i
~
c ,
c r
, c
V
O O O O O O O O O
O
U r.~D t3. O. G. O. O.
~" LL O O.
CY O
c,,.~""., .
~' ~O .
N
G
_
O O O
..w --.
M h ~ y.~r
w a b
c
z,
o, a. r.~ o
z
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_ :11 _
Example 11
Modifications to construct pFAJ3105
From the analysis of Arabidopsis plants transformed with construct pFAJ3105 it
is
clear that the polyprotein precursor is indeed cleaved (see Table 3, Figure
17). However,
cleavage occurs such that one amino acid from the linker peptide remains
attached to the
mature protein located N-terminally from the linker peptide, and that fiv a
amino acids remain
attached to the mature protein located C-terminally from the linker peptide
(see Figure 17).
In order to reduce the number of linker peptide-derived amino acids attached
to the mature
1o proteins, which could possibly interfere with the functional properties of
these mature
proteins, a number of constructs have been designed in order to obtain
cleavage occurnng
closer to (or even preferentially at) the borders of the mature proteins.
In construct pFAJ3343, the codon for the N-terminal residue of the linker
peptide
occurring in pFAJ3105 has been deleted. It is expected that cleavage of mature
DmAMPl
will occur without addition of any amino acid from the linker peptide (Figure
20). In
constructs pFAJ3344, pFAJ3345 and pFAJ3346, the codons at the carboxyl-
terminal end of
the linker peptide in pFAJ310~ have been modified such that the last two, four
and five
residues have been deleted, respectively. It is expected that the number of
residues
remaining attached to the N-terminal end of RsAFP2 after cleavage will be
respectively
2o three, one and zero in constructs pFAJ3344, pFAJ3345 and pFAJ3346 (Figure
20). Other
constructs can be made in which the number of residues at either the N- or C-
terminal end of
the linker peptide region in construct pFAJ3105 is reduced.
In construct pFAJ3105 the linker peptide is derived from the fourth internal
propeptide of the IbAMP precursor (Tailor R.H. et al., 1997, J. Biol. Chem.
272, 24480-
24487). In construct pFAJ3369, this linker peptide has been replaced by the
first internal
propeptide of the IbAMP precursor (Tailor R.H. et al., 1997, ibid.). In the
latter linker
peptide the doublet of acidic residues occurs at the C-terminus. It is
expected that the
cleavage will occur such that only one residue will remain attached to the N-
terminus of
RsAFP? (Figure 20).
Example 12
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-42-
Construction of a construct for expression of a polvprotein with four mature
protein
domains
The polyprotein region in construct pFAJ3367 consists of the signal peptide
region of
DmAMP 1 cDNA followed by the coding regions of four different antimicrobial
peptides,
each separated by the first internal propeptide region of the IbAMP precursor.
The coding
region for the four different antimicrobial proteins are, in order (see Figure
21 ):
1. The plant defensin DmAMPl (Osborn R.W. et al., 1995, FEBS Lett. 368, 257-
262)
2. The plant defensin RsAFP2 (Terras F.R.G. et al., 1995, Plant Cell 7, 573-
588)
3. The plant defensin HsAFPI (Osborn R.W. et al., 1995, FEBS Lett. 368, 257-
262)
t0 4. The lipid transfer protein-like protein AceAMPI (Cammue B.P.A. et al.,
1995, Plant
Physiol. 109, 445-455)
This construct will give rise to four different mature antimicrobial proteins
(DmAMPI,
RsAFP2, HsAFP 1 and AceAMP 1 ), each of which secreted to the extracellular
space.
Other constructs can be made other mature peptide regions and with any other
linker peptide
regions described above.
Example 13
Modifications to constructs nFAJ3106, nFAJ 3107 and pFAJ 3108
The polyprotein encoded by constructs pFAJ3106, pFAJ3107 and pFAJ3108 contain
linker
2o peptides with the Kex2 recognition site IGKR at their C-terminal ends.
Jiang L. and Rogers
J.C. (1999, Plant J. 18, 23-32) have shown that polyproteins containing a IGKR
site are not
or poorly cleaved in transgenic tobacco plants. improved cleavage was observed
in
polyproteins in which the IGKR sequence was replaced by the IGKRIGKRIGKR (SEQ
ID
NO 77) sequence.
?5 Constructs pFAJ3106-2, pFAJ3107-2 and pFAJ3108-2 are identical to
constructs pFAJ3106,
pFAJ3107 and pFAJ3108 except for the replacement of the IGKR coding region by
a region
coding for IGKRIGKRIGKR (Figure 22). Polyproteins encoded by these constructs
will be
efficiently cleaved both at the N-terminal end and the C-terminal end of the
linker peptide.
Other constructs can be made in which the number of residues at either the N-
or C-terminal
;o end of the linker peptide region in constructs pFAJ3106, pFAJ3107 or
pFAJ3108 is reduced.
SUBSTITUTE SHEET (RULE 26j
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
_ 43 _
Cxamnle 14
Polyprotein constructs based on hybrid linker peptides containing the 2A
sequence
The foot-and-mouth disease virus (FMDV) RNA is translated as a polyprotein
whose
cleavage depends on a 20 amino acids sequence called the 2A sequence (Ryan and
Drew
...1994, EMBO J. 13, 928-933). Cleavage of the polyproteins joined by the 2A
sequence
occurs between the 19'" amino acid (G) and the 20'" amino acid (P) of the 2A
sequence via a
process which is apparently independent of processing enzymes and which might
be due to
improper formation of the peptide bond between G and P (Halpin et al., 1999,
Plant J. 17,
453-459). Halpin C. et al. '1999 (Plant J. 17, 453-459) have shown that
polyproteins
to containing the FMDV 2A sequence as a linker peptide are efficiently cleaved
when
expressed in plants. One major drawback of the use of the FMDV 2A sequence as
a linker
peptide, however, is that cleavage does not occur at the N-terminus of the
linker peptide.
Hence, a relatively long stretch of 19 amino acids corresponding to the first
19 residues of
the FMDV 2A sequence remains attached to the C-terminus of the mature protein.
This
15 additional stretch of 19 residues may interfere with the functional
properties of the protein to
which it is attached.
In order to address this problem of incomplete removal of the linker peptide
after
cleavage, hybrid linker peptides consisting at their N-terminal part of a
linker peptide
described in constructs pFAJ3105, pFAJ3106, pFAJ3107 or pFAJ3108 (or a part of
such
20 peptide) and at their C-terminal part of the FMDV 2A sequence (or a part of
such peptide)
are proposed. Examples of constructs based on this principle are constructs
pFAJ3370 and
pFAJ3368 (Figure 23). Construct pFAJ3370 has a polyprotein region identical to
that of
construct pFAJ31 OS except that the linker peptide is a 29 amino acids peptide
consisting of
the first 9 amino acids of the fourth internal propeptide of the IbAMP
precursor (Tailor R.H.
35 et al., 1997, J. Biol. Chem. 272, 24480-24487) followed by the 20 amino
acids of the entire
FMDV 2A sequence. Cleavage of this linker peptide should release a mature
DmAMPl with
an additional serine at its C-terminus and a mature RsAFP2 with an additional
proline at its
N-terminus.
Construct pFAJ3368 is identical to construct pFAJ3370 except that the C-
terminal
3o mature protein domain (in this case encoding RsAFP2) is replaced by a
domain encoding this
SUBSTITUTE SHEET (RULE 26)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
-~.4-
mature protein domain preceded by a signal peptide domain (in this case
encoding RsAFP?
with its own signal peptide). If cleavage between G and P of the FMDV ?A
sequence occurs
prior to full translocation of the polyprotein into the endoplasmic reticulum
then it is
expected that construct pFAJ3368 will provide better targetting of both mature
proteins to
s the extracellular space in comparison to construct pFAJ3370. In this case,
the secreted
mature proteins will consist of DmAMPl with an additional serine at its C-
terminus and
RsAFP2 with no added amino acids. If cleavage between G and P of the FMDV 2A
sequence occurs after translocation of the polyprotein into the eridoplasmic
reticulum, then it
is expected that the signal peptide attached to RsAFP2 will not be efficiently
removed and in
to this case construct pFAJ3370 will be preferred over pFAJ3368.
SUBSTITUTE SHEET (RULE 2B)
CA 02335379 2001-04-04
1
SEQUENCE LISTING
<110> ZENECA Limited
Broekaert, Willem F
Francois, Isabelle EJ1?
Evans, Ian J
De Bolle, Miguel FC
Ray, John A
<120> Genetic Method
<130> CBB 1217
<140> CA 2335379
<141> 1999-08-17
<150> PCT/GB99/02716
<151> 1999-08-17
<160> 81
<170> PatentIn Ver. 2.1
<210> 1
<211> 446
<212> DNA
<213> Dahlia merckii
<220>
<221> CDS
<222> (1) . . (64)
<220>
<221> CDS
<222> (157) . . (446)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
2
<400> 1
atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt ttc 48
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
gtg ctc gcc atc tca g gttatcaaat ctttagttca tttattgaat atgatagtat 104
Val Leu Ala Ile Ser
ttatattctt ttatggtttt atgtgttctg acaagttgca aatattgagt ag at atc 161
Asp Ile
gca tcc gtt agt gga gaa cta tgc gag aaa get agc aag aca tgg tcg 209
Ala Ser Val Ser Gly Glu Leu Cys Glu Lys Ala Ser Lys Thr Trp Ser
30 35
gga aac tgt ggc aat acg gga cat tgt gac aac caa tgt aaa tca tgg 257
Gly Asn Cys Gly Asn Thr Gly His Cys Asp Asn Gln Cys Lys Ser Trp
40 45 50 55
gag ggt gcg gcc cat gga gcg tgt cat gtg cgt aac ggg aaa cac atg 305
Glu Gly Ala Ala His Gly Ala Cys His Val Arg Asn Gly Lys His Met
60 65 70
tgt ttc tgt tac ttc aat tgt aaa aaa gcc gaa aag ctt get caa gac 353
Cys Phe Cys Tyr Phe Asn Cys Lys Lys Ala Glu Lys Leu Ala Gln Asp
75 80 85
aaa ctt aaa gcc gaa caa ctc get caa gac aaa ctt aat gcc caa aag 401
Lys Leu Lys Ala Glu Gln Leu Ala Gln Asp Lys Leu Asn Ala Gln Lys
90 95 100
ctt gac cgt gat gcc aag aaa gtg gtt cca aac gtt gaa cat ccg 446
Leu Asp Arg Asp Ala Lys Lys Val Val Pro Asn Val Glu His Pro
105 110 115
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
3
<210> 2
<211> 118
<212> PRT
<213> Dahlia merckii
<400> 2
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Lys Lys
65 70 75 80
Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu Ala Gln
85 90 95
Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala Lys Lys Val Val
100 105 110
Pro Asn Val Glu His Pro
115
<210> 3
<211> 16
<212> PRT
<213> Artificial Sequence
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
4
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 3
Ser Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu Pro Gly
1 5 10 15
<210> 4
<211> 20
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 4
Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu
1 5 10 15
Ile Gly Lys Arg
<210> 5
<211> 40
<212> PRT
<213> Dahlia merckii
<400> 5
Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu
1 5 10 15
Ala Gln Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala Lys Lys
20 25 30
CA 02335379 2001-O1-17
WO 00/11175 PC'T/G899/02716
Val Val Pro Asn Val Glu His Pro
35 40
<210> 6
<211> 44
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 6
Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu
1 5 10 15
Ala Gln Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala Lys Lys
20 25 30
Val Val Pro Asn Val Glu His Pro Ile Gly Lys Arg
35 40
<210> 7
<211> 20
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 7
Ala Ser Thr Thr Val Asp His Gln Ala Asp Val Rla Ala Thr Lys Thr
1 5 10 15
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
6
Ile Gly Lys Arg
<210> 8
<211> 31
<212> PRT
<213> Amaranthus caudatus
<400> 8
Ala Ser Thr Thr Val Asp His Gln Ala Asp Val Ala Ala Thr Lys Thr
1 5 10 15
Ala Lys Asn Pro Thr Asp Ala Lys Leu Ala Gly Ala Gly Ser Pro
20 25 30
<210> 9
<211> 522
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (76)..(513)
<400> 9
ctcgagtatt tttacaacaa ttaccaacaa caacaaacaa caaacaacat tacaattact 60
atttacaatt acacc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt 111
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val
1 5 10
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
7
ctg atc ctt ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga 159
Leu Ile Leu Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly
15 20 25
gaa cta tgc gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac 207
Glu Leu Cys Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn
30 35 40
acg gga cat tgt gac aac caa tgt aaa tca tgg gag ggt gcg gcc cat 255
Thr Gly His Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His
45 50 55 60
gga gcg tgt cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc 303
Gly Ala Cys His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe
65 70 75
aat tgt tcc aac get get gac gag gtg get acc cca gag gac gtg gag 351
Asn Cys Ser Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu
80 85 90
cca gga cag aag ttg tgc caa agg cca agt ggg aca tgg tca gga gtc 399
Pro Gly Gln Lys Leu Cys Gln Arg Pro Ser Gly Thr Trp Ser Gly Val
95 100 105
tgt gga aac aat aac gca tgc aag aat cag tgc att aga ctt gag aaa 447
Cys Gly Asn Asn Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys
110 115 120
gca cga cat gga tct tgc aac tat gtc ttc cca get cac aag tgt atc 495
Ala Arg His Gly Ser Cys Asn Tyr Val Phe Pro Ala His Lys Cys Ile
125 130 135 140
tgc tac ttt cct tgt taa taggagctc 522
Cys Tyr Phe Pro Cys
145
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/0271G
8
<210> 10
<211> 145
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 10
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 ~ 10 15
Val L~u Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser Asn
65 70 75 80
Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu Pro Gly Gln Lys
85 90 95
Leu Cys Gln Arg Pro Ser Gly Thr Trp Ser Gly Val Cys Gly Asn Asn
100 105 110
Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly
115 120 125
Ser Cys Asn Tyr Val Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro
130 135 140
Cys
145
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
9
<210> 11
<211> 534
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (76)..(525)
<400> 11
ctcgagtatt tttacaacaa ttaccaacaa caacaaacaa caaacaacat tacaattact 60
atttacaatt acacc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt 111
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val
1 5 10
ctg atc ctt ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga 159
Leu Ile Leu Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly
15 20 25
gaa cta tgc gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac 207
Glu Leu Cys Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn
30 35 40
acg gga cat tgt gac aac caa tgt aaa tca tgg gag ggt gcg gcc cat 255
Thr Gly His Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His
45 50 55 60
gga gcg tgt cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc 303
Gly Ala Cys His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe
65 70 75
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
aat tgt aaa aaa gcc gaa aag ctt get caa gac aaa ctt aaa gcc gaa 351
Asn Cys Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu
80 85 90
caa ctc atc gga aag agg cag aag ttg tgc caa agg cca agt ggg aca 399
Gln Leu Ile Gly Lys Arg Gln Lys Leu Cys Gln Arg Pro Ser Gly Thr
95 100 105
tgg tca gga gtc tgt gga aac aat aac gca tgc aag aat cag tgc att 447
Trp Ser Gly Val Cys Gly Asn Asn Asn~Ala Cys Lys Asn Gln Cys Ile
110 115 120
aga ctt gag aaa gca cga cat gga tct tgc aac tat gtc ttc cca get 495
Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr Val Phe Pro Ala
125 130 135 140
cac aag tgt atc tgc tac ttt cct tgt taa taggagctc 534
His Lys Cys Ile Cys Tyr Phe Pro Cys
145
<210> I2
<211> 149
<2I2> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 12
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
CA 02335379 2001-O1-17
WO 00/11175 PGT/GB99/02716
11
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Lys Lys
65 70 ~ 75 80
Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu Ile Gly
85 90 95
Lys Arg Gln Lys Leu Cys Gln Arg Pro Ser Gly Thr Trp Ser Gly Val
100 105 110
Cys Gly Asn Asn Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys
115 120 125
Ala Arg His Gly Ser Cys Asn Tyr Val Phe Pro Ala His Lys Cys Ile
130 135 140
Cys Tyr Phe Pro Cys
145
<210> 13
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Oligonucleotide
<220>
<221> misc feature
<222> (6, 9, 12, 15, 21)
<223> n is any residue
<400> 13
tgyganaang cnwsnaarac ntgg 24
CA 02335379 2001-O1-17
WO 00/I 1175 PC'T/GB99/02716
12
<210> 14
<211> 8
<212> PRT
<213> Dahlia merckii
<400> 14
Cys Glu Lys Ala Ser Lys Thr Trp
1 5
<210> 15
<211> 606
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (76)..(597)
<400> 15
ctcgagtatt tttacaacaa ttaccaacaa caacaaacaa caaacaacat tacaattact 60
atttacaatt acacc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt 111
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val
1 5 10
ctg atc ctt ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga 159
Leu Ile Leu Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly
15 20 25
gaa cta tgc gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac 207
Glu Leu Cys Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn
30 35 40
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
13
acg gga cat tgt gac aac caa tgt aaa tca tgg gag ggt gcg gcc cat 255
Thr Gly His Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His
45 50 55 60
gga gcg tgt cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc 303
Gly Ala Cys His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe
65 70 75
aat tgt aaa aaa gcc gaa aag ctt get caa gac aaa ctt aaa gcc gaa 351
Asn Cys Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu
80 85 90
caa ctc get caa gac aaa ctt aat gcc caa aag ctt gac cgt gat gcc 399
Gln Leu Ala Gln Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala
95 100 105
aag aaa gtg gtt cca aac gtt gaa cat ccg atc gga aag agg cag aag 447
Lys Lys Val VaI Pro Asn Val Glu His Pro Ile Gly Lys Arg Gln Lys
110 lI5 120
ttg tgc caa agg cca agt ggg aca tgg tca gga gtc tgt gga aac aat 495
Leu Cys Gln Arg Pro Ser Gly Thr Trp Ser Gly Val Cys Gly Asn Asn
125 130 135 140
aac gca tgc aag aat cag tgc att aga ctt gag aaa gca cga cat gga 543
Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly
145 150 155
tct tgc aac tat gtc ttc cca get cac aag tgt atc tgc tac ttt cct 591
Ser Cys Asn Tyr Val Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro
160 165 170
tgt taa taggagctc 606
Cys
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
14
<210> 16
<211> 173
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 16
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Lys Lys
65 70 75 80
Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu Ala Gln
85 90 95
Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala Lys Lys Val Val
100 105 110
Pro Asn Val Glu His Pro Ile Gly Lys Arg Gln Lys Leu Cys Gln Arg
115 120 125
Pro Ser Gly Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys
130 135 140
Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr
145 150 155 160
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/0271b
Val Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
165 170
<210> 17
<211> 534
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (76)..(525)
<400> 17
ctcgagtatt tttacaacaa ttaccaacaa caacaaacaa caaacaacat tacaattact 60
atttacaatt acacc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt 111
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val
1 5 10
ctg atc ctt ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga 159
Leu Ile Leu Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Sir Gly
15 20 25
gaa cta tgc gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac 207
Glu Leu Cys Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn
30 35 40
acg gga cat tgt gac aac caa tgt aaa tca tgg gag ggt gcg gcc cat 255
Thr Gly His Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His
45 50 55 60
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
16
gga gcg tgt cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc 303
Gly Ala Cys His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe
65 70 75
aat tgt gcc agt act act gtg gat cac caa get gat gtt get gcc acc 351
Asn Cys Ala Ser Thr Thr Val Asp His Gln Ala Asp Val Ala Ala Thr
80 85 90
aaa act atc gga aag agg cag aag ttg tgc caa agg cca agt ggg aca 399
Lys Thr Ile Gly Lys Arg Gln Lys Leu Cys Gln Arg Pro Ser Gly Thr
95 100 105
tgg tca gga gtc tgt gga aac aat aac gca tgc aag aat cag tgc att 447
Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys Asn Gln Cys Ile
110 115 120
aga ctt gag aaa gca cga cat gga tct tgc aac tat gtc ttc cca get 495
Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr Val Phe Pro Ala
125 130 135 140
cac aag tgt atc tgc tac ttt cct tgt taa taggagctc 534
His Lys Cys Ile Cys Tyr Phe Pro Cys
145
<210> 18
<211> 149
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 18
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
17
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ala Ser
65 70 75 80
Thr Thr Val Asp His Gln Ala Asp Val Ala Ala Thr Lys Thr Ile Gly
85 90 95
Lys Arg Gln Lys Leu Cys Gln Arg Pro Ser Gly Thr Trp Ser Gly Val
100 105 110
Cys Gly Asn Asn Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys
115 120 125
Ala Arg His Gly Ser Cys Asn Tyr Val Phe Pro Ala His Lys Cys Ile
130 135 140
Cys Tyr Phe Pro Cys
145
<210> 19
<211> 316
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (76)..(312)
CA 02335379 2001-O1-17
WO 00/1115 PCT/GB99/02716
18
<400> 19
ctcgagtatt tttacaacaa ttaccaacaa caacaaacaa caaacaacat tacaattact 60
atttacaatt acacc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt 111
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val
1 5 10
ctg atc ctt ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga 159
Leu Ile Leu Phe Val Leu Ala Ile Ser,Asp Ile Ala Ser Val Ser Gly
15 20 25
gaa cta tgc gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac 207
Glu Leu Cys Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn
30 35 40
acg gga cat tgt gac aac caa tgt aaa tca tgg gag ggt gcg gcc cat 255
Thr Gly His Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His
45 50 55 60
gga gcg tgt cat gtg cgt aat ggg aaa cac atg tgt ttc tgt tac ttc 303
Gly Ala Cys His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe
65 70 75
aat tgt tga gctc 316
Asn Cys
<210> 20
<211> 78
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 20
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
19
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys
65 70 75
<210> 21
<211> 14
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
peptide
<400> 21
Ser Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu
1 5 10
<210> 22
<211> 12
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
peptide
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
<400> 22
Ser Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Asp
1 5 10
<210> 23
<211> 11
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
peptide
<400> 23
Ser Asn Ala Ala Asp Glu Val Ala Thr Pro Glu
1 5 10
<210> 24
<211> 28
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
peptide
<400> 24
Ala Asn Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu
1 5 10 15
Leu Ala Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp
20 25
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
21
<210> 25
<211> 28
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 25
Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu
1 5 10 15
Ile Gly Lys Arg Ile Gly Lys Arg Ile Gly Lys Arg
20 25
<210> 26
<211> 52
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 26
Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu
1 5 10 15
Ala Gln Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala Lys Lys
20 25 30
Val Val Pro Asn Val Glu His Pro Ile Gly Lys Arg Ile Gly Lys Arg
35 40 45
Ile Gly Lys Arg
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
22
<210> 27
<211> 28
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 27
Ala Ser Thr Thr Val Asp His Gln Ala Asp Val Ala Ala Thr Lys Thr
1 5 10 15
Ile Gly Lys Arg Ile Gly Lys Arg Ile Gly Lys Arg
20 25
<210> 28
<211> 29
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker
propeptide
<400> 28
Ser Asn Ala Ala Asp Glu Val Ala Thr Gln Leu Leu Asn Phe Asp Leu
1 5 10 15
Leu Lys Leu Ala Gly Asp Val Glu Ser Asn Pro Gly Pro
20 25
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
23
<210> 29
<211> 15
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Linker peptide
<400> 29
Asn Ala Ala Asp Glu Val AIa Thr Pro Glu Asp Val Glu Pro Gly
1 5 10 15
<210> 30
<211> 446
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3}..(437)
<400> 30
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag aeg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
CA 02335379 2001-O1-17
WO 00/11175 PGT/GB99102716
24
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt aac 239.
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Asn
65 70 75
gcg gcc gac gag gtg get acc cca gag gac gtg gaa cct ggt cag aag 287
Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu Pro Gly Gln Lys
80 85 90 95
ttg tgc caa agg cca agt cgt aca tgg tca gga gtc tgt gga aac aat 335
Leu Cys Gln Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn
100 105 i10
aac gca tgc aag aat cag tgc att aga ctt gag aaa gca cga cat gga 383
Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly
115 120 125
tct tgc aac tat cgt ttc cca get cac aag tgt atc tgc tac ttt cct 431
Ser Cys Asn Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro
130 135 140
tgt taa taggagctc 446
Cys
<210> 31
<211> 144
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 31
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
CA 02335379 2001-O1-17
WO 00/11175 PCT/G899/02716
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Asn Ala
65 70 75 80
Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu Pro Gly Gln Lys Leu
85 90 95
Cys Gln Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn
100 105 110
Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser
115 120 125
Cys Asn Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
130 135 140
<210> 32
<211> 443
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(434)
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
26
<400> 32
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt tcc 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser
65 70 75
aac gcg gcc gac gag gtg get acc cca gag gac gtg gaa cag aag ttg 287
Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu Gln Lys Leu
80 85 90 95
tgc caa agg cca agt cgt aca tgg tca gga gtc tgt gga aac aat aac 335
Cys Gln Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn
100 105 110
gca tgc aag aat cag tgc att aga ctt gag aaa gca cga cat gga tct 383
Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser
115 120 125
tgc aac tat cgt ttc cca get cac aag tgt atc tgc tac ttt cct tgt 431
Cys Asn Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
130 135 140
taa taggagctc 443
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
27
<210> 33
<211> 143
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 33
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser Asn
65 70 75 80
Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val Glu Gln Lys Leu Cys
85 90 95
Gln Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala
100 105 110
Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys
115 120 125
Asn Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
130 135 140
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
28
<210> 34
<211> 437
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(428)
<400> 34
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt tcc 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser
65 70 75
aac gcg gcc gac gag gtg get acc cca gag gac cag aag ttg tgc caa 287
Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Gln Lys Leu Cys Gln
gp 85 90 95
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
29
agg cca agt cgt aca tgg tca gga gtc tgt gga aac aat aac gca tgc 335
Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys
100 105 110
aag aat cag tgc att aga ctt gag aaa gca cga cat gga tct tgc aac 383
Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn
115 120 , 125
tat cgt ttc cca get cac aag tgt atc tgc tac ttt cct tgt taa 428
Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
130 135 140
taggagctc 437
<210> 35
<211> 141
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 35
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser Asn
65 70 75 80
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Gln Lys Leu Cys Gln Arg
85 90 95
Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys
100 105 ~ 110
Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr
115 120 125
Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
130 135 140
<210> 36
<211> 434
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(425)
<400> 36
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
I 5 10 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
40 45
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
31
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt tcc 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser
65 70 75
aac gcg gcc gac gag gtg get acc cca, gag cag aag ttg tgc caa agg 287
Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Gln Lys Leu Cys Gln Arg
80 85 90 95
cca agt cgt aca tgg tca gga gtc tgt gga aac aat aac gca tgc aag 335
Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys
100 105 110
aat cag tgc att aga ctt gag aaa gca cga cat gga tct tgc aac tat 383
Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr
115 120 125
cgt ttc cca get cac aag tgt atc tgc tac ttt cct tgt taa taggagctc 434
Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
130 135 140
<210> 37
<211> 140
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 37
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
32
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser Asn
b5 70 75 80
Ala Ala Asp Glu Val Ala Thr Pro Glu Gln Lys Leu Cys Gln Arg Pro
85 90 95
Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys Asn
100 105 110
Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr Arg
115 120 125
Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
130 135 140
<210> 38
<211> 485
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
seauence
<220>
<221> CDS
<222> (3)..(476)
<400> 38
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
33
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt get 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ala
65 70 75
aac get gag gaa get get get get att cct gaa get tct gaa gaa ctt 287
Asn Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu Leu
80 85 90 95
get caa gaa gaa get cct gtg tac agt gaa gat cag aag ttg tgc caa 335
Ala Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp Gln Lys Leu Cys Gln
100 105 110
agg cca agt cgt aca tgg tca gga gtc tgt gga aac aat aac gca tgc 383
Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys
115 120 125
aag aat cag tgc att aga ctt gag aaa gca cga cat gga tct tgc aac 431
Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn
130 135 140
tat cgt ttc cca get cac aag tgt atc tgc tac ttt cct tgt taa 476
Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro. Cys
145 150 155
taggagctc 485
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
34
<210> 39
<211> 157
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 39
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ala Asn
65 ?0 75 80
Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu Leu Ala
85 90 95
Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp Gln Lys Leu Cys Gln Arg
100 105 110
Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys
115 120 125
Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr
130 135 140
Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
145 150 155
CA 02335379 2001-O1-17
WO 00/11175 PCf/GB99/02716
<210> 40
<211> 1093
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(1085)
<400> 40
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 20 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aac tgc get 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ala
65 70 75
aac get gag gaa get get get get att cct gaa get tct gaa gaa ctt 287
Asn Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu Leu
g0 85 90 95
CA 02335379 2001-O1-17
WO 00/11175 PCT1GB99/02716
36
get caa gaa gaa get cct gtg tac agt gaa gat cag aag ttg tgc caa 335
Ala Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp Gln Lys Leu Cys Gln
100 105 110
agg cca agt cgt aca tgg tca gga gtc tgt gga aac aat aac gca tgc 383
Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys
115 120 125
aag aat cag tgc att aga ctt gag aaa gca cga cat gga tct tgc aac 431
Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn
130 135 140
tat cgt ttc cca get cac aag tgt atc tgc tac ttc cct tgt gcg aat 479
Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys Ala Asn
145 150 155
get gaa gaa get get get get att cct gaa get tct gaa gaa ctt get 527
Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu Leu Ala
160 165 170 175
caa gaa gaa gca ccg gtt tac tct gaa gat gac gga gtg aag ctc tgc 575
Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp Asp Gly Val Lys Leu Cys
180 185 190
gac gtg cca tcc gga acc tgg tcc gga cac tgc ggt tcc tcc agc aag 623
Asp Val Pro Ser Gly Thr Trp Ser Gly His Cys Gly Ser Ser Ser Lys
195 200 205
tgc agc caa caa tgc aag gac agg gag cac ttc get tac gga gga get 671
Cys Ser Gln Gln Cys Lys Asp Arg Glu His Phe Ala Tyr Gly Gly Ala
210 215 220
tgc cac tac caa ttc cca tcc gtg aag tgc ttc tgc aag agg caa tgc 719
Cys His Tyr Gln Phe Pro Ser Val Lys Cys Phe Cys Lys Arg Gln Cys
225 230 235
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
37
get aac get gag gaa get get get get att cct gaa get tct gaa gaa 767
Ala Asn Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu
240 245 250 255
ctt get caa gaa gaa get cct gtg tac agt gaa gat cag aac ata tgc 815
Leu Ala Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp Gln Asn Ile Cys
260 265 270
cca agg gtt aat cga att gtg aca ccc.tgt gtg gcc tac gga ctc gga 863
Pro Arg Val Asn Arg Ile Val Thr Pro Cys Val Ala Tyr Gly Leu Gly
275 280 285
agg gca cca atc gcc cca tgc tgc aga gcc ctg aac gat cta cgg ttt 911
Arg Ala Pro Ile Ala Pro Cys Cys Arg Ala Leu Asn Asp Leu Arg Phe
290 295 300
gtg aat act aga aac cta cga cgt get gca tgc cgc tgc ctc gta ggg 959
Val Asn Thr Arg Asn Leu Arg Arg Aia Ala Cys Arg Cys Leu Val Gly
305 310 315
gta gtg aac cgg aac ccc ggt ctg aga cga aac cct aga ttt cag aac 1007
Val Val Asn Arg Asn Pro Gly Leu Arg Arg Asn Pro Arg Phe Gln Asn
320 325 330 335
att cct cgt gat tgt cgc aac acc ttt gtt cgt ccc ttc tgg tgg cgt 1055
Ile Pro Arg Asp Cys Arg Asn Thr Phe Val Arg Pro Phe Trp Trp Arg
340 345 350
cca aga att caa tgc ggc agg att aac taa tagagctc 1093
Pro Arg Ile Gln Cys Gly Arg Ile Asn
355 360
<210> 41
<211> 360
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
38
<400> 41
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Iie Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile~Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Sex Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ala Asn
65 70 75 g0
Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu Leu Ala
85 90 95
Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp Gln Lys Leu Cys Gln Arg
100 105 110
Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys
115 120 125
Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr
130 135 140
Arg Phe Pro Ala His Lys Cys ile Cys Tyr Phe Pro Cys Ala Asn Ala
145 150 155 160
Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu Leu Ala Gln
165 170 175
Glu Glu Ala Pro Val Tyr Ser Glu Asp Asp Gly Val Lys Leu Cys Asp
180 185 190
CA 02335379 2001-O1-17
WO 00/11175 PCf/GB99/02716
39
Val Pro Ser Gly Thr Trp Ser Gly His Cys Gly Ser Ser Ser Lys Cys
195 200 205
Ser Gln Gln Cys Lys Asp Arg Glu His Phe Ala Tyr Gly Gly Ala Cys
210 215 220
His Tyr Gln Phe Pro Ser Val Lys Cys Phe Cys Lys Arg Gln Cys Ala
225 230 235 240
Asn Ala Glu Glu Ala Ala Ala Ala Ile Pro Glu Ala Ser Glu Glu Leu
245 250 255
Ala Gln Glu Glu Ala Pro Val Tyr Ser Glu Asp Gln Asn Ile Cys Pro
260 265 270
Arg Val Asn Arg Ile Val Thr Pro Cys Val Ala Tyr Gly Leu Gly Arg
275 280 285
Ala Pro Ile Ala Pro Cys Cys Arg Ala Leu Asn Asp Leu Arg Phe Val
290 295 300
Asn Thr Arg Asn Leu Arg Arg Ala Ala Cys Arg Cys Leu Val Gly Vai
305 310 315 320
Val Asn Arg Asn Pro Gly Leu Arg Arg Asn Pro Arg Phe Gln Asn Ile
325 330 335
Pro Arg Asp Cys Arg Asn Thr Phe Val Arg Pro Phe Trp Trp Arg Pro
340 345 350
Arg Ile Gln Cys Gly Arg Ile Asn
355 360
cca aga att caa tgc ggc
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
<210> 42
<211> 485
<2I2> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(476)
<400> 42
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt aaa 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Lys
65 70 75
aaa gcc gaa aag ctt get caa gac aaa ctt aaa gcc gaa caa ctc atc 287
Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu Ile
80 85 90 95
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
41
gga aag agg atc gga aag agg atc gga aag agg cag aag ttg tgc caa 335
Gly Lys Arg Ile Gly Lys Arg Ile Gly Lys Arg Gln Lys Leu Cys Gln
100 105 110
agg cca agt cgt aca tgg tca gga gtc tcjt gga aac aat aac gca tgc 383
Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys
115 120 125
aag aat cag tgc att aga ctt gag aaa gca cga cat gga tct tgc aac 431
Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn
130 135 140
tat cgt ttc cca get cac aag tgt atc tgc tac ttt cct tgt taa 476
Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
145 150 155
taggagctc 485
<210> 43
<211> 157
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 43
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
42
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Lys Lys
65 70 75 80
Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu Ile Gly
85 90 95
Lys Arg Ile Gly Lys Arg Ile Gly Lys Arg Gln Lys Leu Cys Gln Arg
100 105 110
Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys
115 120 125
Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr
130 135 140
Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
145 150 155
<210> 44
<211> 557
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(548)
<400> 44
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
43
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgtjggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt aaa 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Lys
65 70 75
aaa gcc gaa aag ctt get caa gac aaa ctt aaa gcc gaa caa ctc get 287
Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu Ala
80 85 90 95
caa gac aaa ctt aat gcc caa aag ctt gac cgt gat gcc aag aaa gtg 335
Gln Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala Lys Lys Val
100 105 110
gtt cca aac gtt gaa cat ccg atc gga aag agg atc gga aag agg atc 383
Val Pro Asn VaI Glu His Pro Iie Gly Lys Arg Ile Gly Lys Arg Ile
115 120 125
gga aag agg cag aag ttg tgc caa agg cca agt cgt aca tgg tca gga 431
Gly Lys Arg Gln Lys Leu Cys Gln Arg Pro Ser Arg Thr Trp Ser Gly
130 135 140
gtc tgt gga aac aat aac gca tgc aag aat cag tgc att aga ctt gag 479
Val Cys Gly Asn Asn Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu
145 150 155
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
44
aaa gca cga cat gga tct tgc aac tat cgt ttc cca get cac aag tgt 527
Lys Ala Arg His Gly Ser Cys Asn Tyr Arg Phe Pro Ala His Lys Cys
160 165 170 175
atc tgc tac ttt ~cct tgt taa taggagctc 557
Ile Cys Tyr Phe Pro Cys
180
<210> 45
<211> 181
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 45
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Lys Lys
65 70 75 80
Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala Glu Gln Leu AIa Gln
85 90 95
Asp Lys Leu Asn Ala Gln Lys Leu Asp Arg Asp Ala Lys Lys Val Val
100 105 110
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
Pro Asn Val Glu His Pro Ile Gly Lys Arg Ile Gly Lys Arg Ile Gly
115 120 125
Lys Arg Gln Lys Leu Cys Gln Arg Pro Ser Arg Thr Trp Ser Gly Val
130 135 ~ 140
Cys Gly Asn Asn Asn Ala Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys
145 150 155 160
Ala Arg His Gly Ser Cys Asn Tyr Arg Phe Pro Ala His Lys Cys Ile
165 170 175
Cys Tyr Phe Pro Cys
180
<210> 46
<211> 485
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(476)
<400> 46
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
46
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys AIa Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt gcc 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ala
65 70 75
agt act act gtg gat cac caa get gat gtt get gcc acc aaa act atc 287
Ser Thr Thr Val Asp His Gln Ala Asp Val Ala Ala Thr Lys Thr Ile
80 85 90 95
gga aag agg atc gga aag agg atc gga aag agg cag aag ttg tgc caa 335
Gly Lys Arg Ile Gly Lys Arg Ile Gly Lys Arg Gln Lys Leu Cys Gln
100 105 110
agg cca agt cgt aca tgg tca gga gtc tgt gga aac aat aac gca tgc 383
Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys
115 120 125
aag aat cag tgc att aga ctt gag aaa gca cga cat gga tct tgc aac 431
Lys Asn Gln Cys Ile Arg Leu GIu Lys Ala Arg His Gly Ser Cys Asn
130 135 140
tat ctg ttc cca get cac aag tgt atc tgc tac ttt cct tgt taa 476
Tyr Leu Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
145 150 155
taggagctc 485
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
47
<210> 47
<211> 157
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ala Ser
65 70 75 80
Thr Thr Val Asp His Gln Ala Asp Val Ala Ala Thr Lys Thr Ile Gly
85 90 95
Lys Arg Ile Gly Lys Arg Ile Gly Lys Arg Gln Lys Leu Cys Gln Arg
100 105 110
Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys
115 120 125
Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr
130 135 140
Leu Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
145 150 155
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
48
<210> 48
<211> 488
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(479)
<400> 48
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt tcc 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser
65 70 75
aac gcg gcc gac gag gtg get acc cag ctg ttg aat ttt gac ctt ctt 287
Asn Ala Ala Asp Glu Val Ala Thr Gln Leu Leu Asn Phe Asp Leu Leu
80 85 90 95
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
49
aag ctt gcg gga gac gtc gag tcc aac cct ggg ccc cag aag ttg tgc 335
Lys Leu Ala Gly Asp Val Glu Ser Asn Pro Gly Pro Gln Lys Leu Cys
100 105 110
caa agg cca agt cgt aca tgg tca gga gtc tgt gga aac aat aac gca 383
Gln Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala
115 I20 125
tgc aag aat cag tgc att aga ctt gag aaa gca cga cat gga tct tgc 431
Cys Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys
130 135 140
aac tat cgt ttc cca get cac aag tgt atc tgc tac ttt cct tgt taa 479
Asn Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
145 150 155
taggagctc 488
<210> 49
<211> 158
<212> PRT
<213> Artificial Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 49
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Sex Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser Asn
65 70 75 80
Ala Ala Asp Glu Val Ala Thr Gln Leu Leu Asn Phe Asp Leu Leu Lys
85 90 95
Leu Ala Gly Asp Val Glu Ser Asn Pro Gly Pro Gln Lys Leu Cys Gln
100 105 110
Arg Pro Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys
115 120 125
Lys Asn Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn
130 135 140
Tyr Arg Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
145 150 155
<210> 50
<211> 575
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<220>
<221> CDS
<222> (3)..(566)
<400> 50
cc atg gtg aat cgg tcg gtt gcg ttc tcc gcg ttc gtt ctg atc ctt 47
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu
1 5 10 15
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
51
ttc gtg ctc gcc atc tca gat atc gca tcc gtt agt gga gaa cta tgc 95
Phe Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys
20 25 30
gag aaa get agc aag acg tgg tcg ggc aac tgt ggc aac acg gga cat 143
Glu Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His
35 40 45
tgt gac aac caa tgt aaa tca tgg gag ggt gcg get cac gga gcg tgt 191
Cys Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys
50 55 60
cat gtg cgt aac ggg aaa cac atg tgt ttc tgt tac ttc aat tgt tcc 239
His Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser
65 70 75
aac gcg gcc gac gag gtg get acc cag ctg ttg aat ttt gac ctt ctt 287
Asn Ala Ala Asp Glu Val Ala Thr Gln Leu Leu Asn Phe Asp Leu Leu
80 85 90 95
aag ctt gcg gga gac gtc gag tcc aac cct ggg ccc atg get aag ttt 335
Lys Leu Ala Gly Asp Val Glu Ser Asn Pro Gly Pro Met Ala Lys Phe
100 105 110
gcg tcc atc atc gca ctt ctt ttt get get ctt gtt ctt ttt get get 383
Ala Ser Ile Ile Ala Leu Leu Phe Ala Ala Leu Val Leu Phe Ala Ala
115 120 125
ttc gaa gca cca aca atg gtg gaa gca cag aag ttg tgc caa agg cca 431
Phe Glu Ala Pro Thr Met Val Glu Ala Gln Lys Leu Cys Gln Arg Pro
130 135 140
agt cgt aca tgg tca gga gtc tgt gga aac aat aac gca tgc aag aat 479
Ser Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys Asn
145 150 155
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
52
cag tgc att aga ctt gag aaa gca cga cat gga tct tgc aac tat cgt 527
Gln Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr Arg
160 165 170 175
ttc cca get cac aag tgt atc tgc tac ttt cct tgt taa taggagctc 575
Phe Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
180 185
<210> 51
<211> 187
<212> PRT
<213> Artificial' Sequence
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 51
Met Val Asn Arg Ser Val Ala Phe Ser Ala Phe Val Leu Ile Leu Phe
1 5 10 15
Val Leu Ala Ile Ser Asp Ile Ala Ser Val Ser Gly Glu Leu Cys Glu
20 25 30
Lys Ala Ser Lys Thr Trp Ser Gly Asn Cys Gly Asn Thr Gly His Cys
35 40 45
Asp Asn Gln Cys Lys Ser Trp Glu Gly Ala Ala His Gly Ala Cys His
50 55 60
Val Arg Asn Gly Lys His Met Cys Phe Cys Tyr Phe Asn Cys Ser Asn
65 70 75 80
Ala Ala Asp Glu Val Ala Thr Gln Leu Leu Asn Phe Asp Leu Leu Lys
85 90 95
Leu Ala Gly Asp Val Glu Ser Asn Pro Gly Pro Met Ala Lys Phe Ala
100 105 110
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
53
Ser Ile Ile Ala Leu Leu Phe Ala Ala Leu Val Leu Phe Ala Ala Phe
115 120 125
Glu Ala Pro Thr Met Val Glu Ala Gln Lys Leu Cys Gln Arg Pro Ser
130 135 140
Arg Thr Trp Ser Gly Val Cys Gly Asn Asn Asn Ala Cys Lys Asn Gln
145 150 155 160
Cys Ile Arg Leu Glu Lys Ala Arg His Gly Ser Cys Asn Tyr Arg Phe
165 170 175
Pro Ala His Lys Cys Ile Cys Tyr Phe Pro Cys
180 185
<210> 52
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<221> misc feature
<222> (9, 12, 15y
<223> n is any residue
<220>
<223> Description of Artificial Sequence:
Oligonucleotide
<400> 52
carttraant ancanaaarc acat 24
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
54
<210> 53
<211> 8
<212> PRT
<213> Dahlia merckii
<400> 53
Met Cys Phe Cys Tyr Phe Asn Cys
1 5
<210> 54
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
Oligonucleotide
<400> 54
aaacacatgt gtttcccatt 20
<210> 55
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
Oligonucleotide
<400> 55
agcgtgtcat gtgcgtaat 19
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
<210> 56
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:
Oligonucleotide
<400> 56
taaagaaacc gaccctttca cgg 23
<210> 57
<211> 107
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 57
atgcatccat ggtgaatcgg tcggttgcgt tctccgcgtt cgttctgatc cttttcgtgc 60
tcgccatctc agatatcgca tccgttagtg gagaactatg cgagaaa 107
<210> 58
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 58
aaaccgaccg agctcacgga tgttcaacgt ttggaac 37
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
56
<210> 59
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 59
agcaagcttt tcgggagctc aacaattgaa gtaa 34
<210> 60
<211> 89
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 60
gcctttggca caacttctgt cctggctcca cgtcctctgg ggtagccacc tcgtcagcag 60
cgttggaaca attgaagtaa cagaaacac 89
<210> 61
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 61
ttagagctcc tattaacaag gaaagtagc 29
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
57
<210> 62
<211> 55
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 62
gcctttggca caacttctgc ctctttccga tgagttgttc ggctttaagt ttgtc 55
<210> 63
<211> 53
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 63
gcctttggca caacttctgc ctctttccga tcggatgttc aacgtttgga acc 53
<210> 64
<211> 101
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 64
gcctttggca caacttctgc ctctttccga tagttttggt ggcagcaaca tcagcttggt 60
gatccacagt agtactggca caattgaagt aacagaaaca c 101
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
58
<210> 65
<211> 4
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 65
Lys Asp Glu Leu
1
<210> 66
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<221> misc feature
<222> (9, 12, 21)
<223> n is any residue
<220>
<223> Description of Artificial Sequence:
Oligonucleotide
<400> 66
atggcsaanm rntcrgttgc ntt 23
<210> 67
<211> 4
<212> PRT
<213> Artificial Sequence
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
59
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 67
Ile Gly Lys Arg
1
<210> 68
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Primer
<400> 68
aggaagttca tttcatttgg 20
<210> 69
<211> 7
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Determined
N-terminal sequence
<400> 69
Glu Leu Cys Glu Lys Ala Ser
1 5
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
<210> 70
<211> 7
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Determined
N-terminal sequence
<400> 70
Asp Val Glu Pro Gly Gln Lys
1 5
<210> 71
<211> 7
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Determined
N-terminal sequence
<400> 71
Leu Ile Gly Lys Arg Gln Lys
1 5
<210> 72
<211> 6
<212> PRT
<2I3> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Predicted
C-terminal sequence
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
61
<400> 72
Cys Tyr Phe Asn Cys Ser
1 5
<210> 73
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Predicted
C-terminal sequence
<400> 73
Ile Cys Tyr Phe Pro Cys
1 5
<210> 74
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Predicted
C-terminal sequence
<400> 74
Cys Tyr Phe Asn Pro Ser
1 5
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
62
<210> 75
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Predicted
C-terminal sequence
<400> 75
Cys Tyr Phe Asn Cys Lys
1 5
<210> 76
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Predicted
C-terminal sequence
<400> 76
Cys Tyr Phe Asn Cys Ala
1 5
<210> 77
<211> 12
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
63
<400> 77
Ile Gly Lys Arg Ile Gly Lys Arg Ile Gly Lys Arg
1 5 10
<210> 78
<211> 6
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 78
Val Ser Gly Glu Leu Cys
1 5
<210> 79
<211> 22
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 79
Phe Asn Cys Ser Asn Ala Ala Asp Glu Val Ala Thr Pro Glu Asp Val
1 5 10 15
Glu Pro Gly Gln Lys Leu
CA 02335379 2001-O1-17
WO 00/11175 PCT/GB99/02716
64
<210> 80
<211> 26
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 80
Phe Asn Cys Lys Lys Ala Glu Lys Leu Ala Gln Asp Lys Leu Lys Ala
1 5 10 15
Glu Gln Leu Ile Gly Lys Arg Gln Lys Leu
20 25
<210> 81
<211> 26
<212> PRT
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence: Synthetic
sequence
<400> 81
Phe Asn Cys Ala Ser Thr Thr Val Asp His Gln Ala Asp Val Ala Ala
1 5 10 15
Thr Lys Thr Ile Gly Lys Arg Gln Lys Leu
20 25