Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02356492 2001-09-04
62451-860D
1
NEMATODE RESISTANT PLANTS AND SEEDS
CROSS-REFERENCE TO RELATED APPLICAT10N
This application is related to international
publication number WO 99/60141, and is a divisional of
Canadian patent application SN 2,323,312.
FIELD OF THE INVENTION
The present invention relates to isolated DNA sequences involved in nematode
resistance inplants.The invention also relates to~methods for improving the
genetic
traits for nematode resistance in plants b~~ utilizing such isolated DNA
sequences.
BACKGROUND
Plants are continually attacked by a diverse range of phytopathogenic
organisms. These organisms cause substantial losses to crops each year.
Traditional
approaches for control of plant diseases have been the use of chemical
treatment arid
the construction of interspecific hybrids between resistant crops and their
wild-type
relatives as sources of resistant germplasm. However, environmental and
economic
concerns make chemical pesticides undesirable, while the traditional
interspecific
breeding is inefEcient and often cannot eliminate the undesired traits of the
wild
species. Thus, the discovery of pest and pathogen-resistant genes provides a
new
approach to control plant disease.
Several genes responsible for disease resistance have been identified and
' isolated from plants. See Staskawicz et al. (1995) Science 268:661-667.
Recently,
2 o the sugar beet Hsl°'°'~ gene that confers resistance to the
beet cyst nematode was
cloned. See Cai et al: (1997) Science 275:832-834; and Moffat (1997) Science
275:77: Transformation of plants or plant tissues with the resistance genes
can
confer disease resistance to susceptible strains. See, for example, PCT
Publication
W093/19181; and Cai et al. (1997) Science 275:832-834.
2 5 Nematode infection is prevalent in many crops. For example, soybean cyst
nematode (Heterodera glycines) is a widespread pest that causes substantial
damage
CA 02356492 2001-09-04
62451-860 (D)
2
to soybeans every year. Such damage is the result of the
stunting of the soybean plant caused by the cyst nematode.
The stunted plants have smaller root systems, show symptoms
of mineral deficiencies in their leaves, and wilt easily.
The soybean cyst nematode is believed to be responsible for
yield losses in soybeans that are estimated to be in excess
of $500 million per year.
Nematicides such as Aldicarb and its breakdown
products are known to be highly toxic to mammals. As a
result, government restrictions have been imposed on the use
of these chemicals. Thus, there is a great need for the
isolation of genes that can provide an effective method of
controlling nematodes without causing health and
environmental problems.
SUMMARY OF THE INVENTION
This invention relates to DNA sequences isolated
from soybean and maize. The sequences alone, or in
combination with other sequences, confer nematode resistance
in a plant. The sequences are useful in methods for the
protection of plants from nematodes. Additionally, allelic
variants of the resistance gene from a susceptible plant are
included. In another aspect of the present invention,
expression cassettes and transformation vectors comprising
the isolated nucleotide sequences are disclosed. The
transformation vectors can be used to transform plants and
express the nematode resistance genes in the transformed
cells. Plants susceptible to nematode infection can be
targeted to confer nematode resistance. The transformed
cells as well as the regenerated transgenic plants
containing and expressing the isolated DNA and protein
sequences are also disclosed.
CA 02356492 2001-09-04
62451-860(D)
2a
In one aspect, the invention provides a plant
which has been transformed with a transformation vector
comprising a DNA sequence that encodes an amino acid
sequence selected from the group consisting of the sequences
set forth in SEQ ID NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, and
SEQ ID NO: 8.
Another aspect of the invention provides a plant
which has been transformed with a transformation vector
comprising a nucleotide sequence selected from the group
consisting of: (a) a nucleotide sequence having at least 70%
identity to the nucleotide sequence set forth in SEQ ID NO:
1, SEQ ID NO: 3, SEQ ID NO: 5, or SEQ ID NO: 7; and (b) a
nucleotide sequence having at least 70% sequence identity to
a nucleotide sequence encoding a plant protein, wherein said
sequence encoding said plant protein is contained in a
plasmid having ATCC accession number 209366, 209365, 209614,
209363, or 209364.
Another aspect of the invention provides a plant
which has been transformed with a transformation vector
comprising a nucleotide sequence selected from the group
consisting of: (a) the nucleotide sequence set forth in SEQ
ID NO: 1, SEQ ID NO: 3, SEQ ID NO: 5 or SEQ ID NO: 7; (b) a
nucleotide sequence encoding a plant protein, wherein said
sequence is contained in a plasmid having ATCC accession
number 209366, 209365, 209614, 209363, or 209364; and (c) a
nucleotide sequence having at least 85% identity to the
sequence of (a) or (b).
Another aspect of the invention provides a plant
transformed with a DNA sequence encoding a protein
comprising an amino acid sequence selected from the group
consisting of the amino acid sequences set forth in SEQ ID
NO: 2, SEQ ID NO: 4, SEQ ID NO: 6, and SEQ ID NO: 8, wherein
CA 02356492 2001-09-04
62451-860(D)
2b
said plant exhibits improved resistance to nematodes over
the native untransformed plant.
BRIEF DESCRIPTION OF THE DRAWINGS
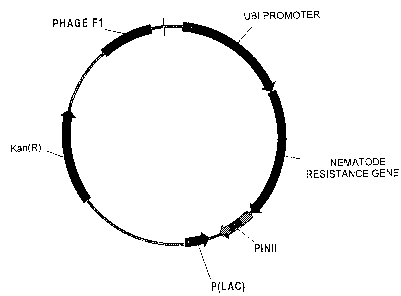
Figure 1 schematically illustrates the plasmid
vector comprising a nematode resistance DNA sequence of the
present invention operably linked to the ubiquitin promoter.
Constitutive expression of this sequence confers resistance
to nematodes in a transformed plant.
DETAILED DESCRIPTION OF THE INVENTION
Compositions and methods for the control of
nematodes in susceptible plants
CA 02356492 2001-09-04
are provided. The compositions comprise isolated proteins and DNA sequences
encoding such proteins involved in nematode resistance. Such isolated 'DNA
sequences can be transferred into plants to confer or improve nematode
resistance in
the transformed plants. Sequences of the invention have been isolated from
maize
and soybean. By "involved in nematode resistance" is intended that the
proteins or
sequences, either alone or in combination with other proteins or sequences,
confer
nematode resistance in a plant. In this manner, resistance to nematodes can be
enhanced or improved in the transformed plant as at least one of the sequences
required for nematode resistance is provided.
DNA sequences isolated from the genomes of maize and soybean are
disclosed. The nucleotide sequences and amino acid sequences from two maize
isolates are set forth in SEQ ID NOs: 1-2 and 3-4, and the corresponding
sequences
from two soybean isolates are set forth in SEQ ID NOs: 5-6 and 7-8. The
nucleotide
sequences in accordance with this invention are involved in nematode
resistance in
1 S plants and may confer, alone or in combination with other sequences,
nematode
resistance in plants. Also discussed are DNA sequences isolated from a
susceptible
genotype of soybean. The nucleotide and amino acid sequences for this isolate
are set
forth in SEQ ID NOs: 9-10. Nucleotide sequences of the invention also include
the
maize and soybean nematode resistance gene sequences as contained in plasmids
deposited with American Type Culture Collection (ATCC) and assigned Accession
Numbers 209366, 209365, 209614, 209363, and 209364.
Using the sequence information set forth in the SEQ ID NOs or the sequences
as contained in ATTC deposits assigned Accession Numbers 209366, 209365,
209614, 209363, and 209364, other plant DNA sequences comprising the
nucleotide
sequences disclosed above can be isolated based on sequence homology at either
the
amino acid or nucleotide sequence level. Any suitable molecular cloning method
can
be used including, but not limited to, PCR'amplification and DNA
hybridization. In
the same manner, synthetic nucleotide sequences can be designed based on the
amino
acid sequences of the invention. Methods to design and make such synthetic
sequences are available in the art.
In a hybridization method, the hybridization probes may be genomic DNA
fragments, cDNA fragments, RNA fragments, or other oligonucleotides, and may
be
CA 02356492 2001-09-04
62451-860D
labeled with a detectable group such as 3ZP, or any other detectable marker.
Probes
for hybridization can be made by labeling synthetic oligonucleotides based on
the
sequence of the soybean and/or maize sequence. Degenerate primers designed on
the
basis of conserved nucleotide or amino acid sequences in the maize and soybean
sequences can additionally be used. Preparation of probes for hybridization is
generally know in the art and is disclosed in Sambrook et al. ( 1989)
Molecular
Cloning: A Laboratory Manual (2d ed.. Cold Spring Harbor Laboratory Press.
Plainview, New Yorkj.~ The labeled probes can be
used to screen cDNA or genomic libraries made from nematode resistant plants.
Methods for construction of such cDNA and genomic libraries are generally
known in
the art and are disclosed in Sambrook et al. (1989).Molecular Cloninh: A
Laboratort:
Manual (2d ed.. Cold Spring Harbor Laboratory Press. Plainviev-. New York).
In a PCR method. the DNA or amino acid sequence encoded by the soybean
or maize sequences of the invention can be aligned with each other. Nucleotide
1 S primers can be designed based on any conserved short stretches of amino
acid
sequences or nucleotide sequences. Pairs of primers can be used in PCR
reactions for
amplification of DNA sequences from cDNA or genomic DNA extracted from plants
of interest. In addition. a single specific primer with a sequence
corresponding to one
of the nucleotide sequences disclosed herein can be paired with a primer
having a
sequence of the DNA vector in the cDNA or genomic libraries for PCR
amplification
of the sequences ~' or 3' to the nucleotide sequences disclosed herein.
Similarly.
nested primers may be used instead of a single specific primer for the
purposes of the
invention. Methods for designing PCR primers and PCR cloning are generally
known
in the art and are disclosed in Sambrook et al. (1989) Molecular Cloning: A
Laboratory Manual (2d ed.. Cold Spring Harbor Laboratory Press, Plainview, New
York).
The sequences of the invention comprise coding sequences from other plants
that may be isolated according to well-known techniques based on their
sequence
homology to the maize or soybean coding sequences set forth herein. In these
techniques, all or part of the known coding sequence is used as a probe that
selectively hybridizes to other possible nematode resistance coding sequences
present
in a population of cloned genomic DNA fragments or cDNA fragments (i. e.,
genomic
CA 02356492 2001-09-04
WO 99/60141 PCT/US98/27456
-5-
or cDNA libraries) from a chosen organism. To achieve specific hybridization
under
a variety of conditions, such probes include sequences that are unique and are
preferably at least about 10 nucleotides in length. and most preferably at
least about
20 nucleotides in length. Such probes may be used to amplify corresponding
coding
S sequences from a chosen organism by PCR. This technique may be used to
isolate
other possible nematode resistance coding sequences from a desired organism or
as a
diagnostic assay to determine the presence of the nematode resistance coding
sequence m an organism.
Such techniques include hybridization screening of plated DNA libraries
(either plaques or colonies; see, e.g., Innis et al.. eds. (1990) PCR
Protocols: A Guide
to Methods and Applications (Academic Press. New York)).
The isolated DNA sequences further comprise DNA sequences isolated from
other plants by hybridization with partial sequences obtained from maize and
soybean. Conditions that will permit other DNA sequences to hybridize to the
DNA
1 S sequences disclosed herein can be determined in accordance with techniques
generally known in the art. For example, hybridization of such sequences may
be
carried out under conditions of reduced stringency. medium stringency. or high
stringency conditions (e.g., conditions represented by a wash stringency of 3~-
40°ro
Formamide with Sx Denhardt's solution. 0.~% SDS. and lx SSPE at 37°C;
conditions
represented by a wash stringency of 40-45% Forniamide with Sx Denhardt's
solution.
0.5% SDS. and lx SSPE at 42°C; and conditions represented by a wash
stringenc~~ of
~0% Formamide with Sx Denhardt's solution, 0.~% SDS, and lx SSPE at
42°C.
respectively. See Sambrook et al. (1989) Molecular Cloning: A Laboratory
Manual
(2d ed., Cold Spring Harbor Laboratory Press, Plainview, New York). In
general.
sequences that confer nematode resistance and hybridize to the DNA sequences
disclosed herein will be at least 70-75% homologous, 80-85% homologous. and
even
90-95% homologous or more.
The following terms are used to describe the sequence relationships bet«~een
two or more nucleic acids or polynucleotides: (a) "reference sequence", (b)
"comparison window", (c) "sequence identity". (d) "percentage of sequence
identiy",
and (e) "substantial identity".
(a) As used herein, "reference sequence" is a defined sequence
CA 02356492 2001-09-04
WO 99/G0141 PCT/US98/27456
-6-
used as a basis for sequence comparison. A reference sequence may be a subset
of or
the entire specified sequence; for example, as a segment of a full-length cDNA
or
gene sequence, or the complete cDNA or gene sequence.
(b) As used herein, "comparison window" makes reference to a
contiguous and specified segment of a polynucleotide sequence, wherein the
polynucleotide sequence in the comparison window may comprise additions or
deletions (i.e., gaps) compared to the reference sequence (which does not
comprise
additions or deletions) for optimal alignment of the two sequences. Generally,
the
comparison window is at least 20 contiguous nucleotides in length, and
optionally can
be 30, 40, 50, 100, or more contiguous nucleotides in length. Those of skill
in the art
understand that to avoid a high similarity to a reference sequence due to
inclusion of
gaps in the polynucleotide sequence a gap penalty is typically introduced and
is
subtracted from the number of matches.
Methods of alignment of sequences for comparison are well-known in the art.
I S Optimal alignment of sequences for comparison may be conducted by the
local
homology algorithm of Smith and Waterman ( 1981 ) Adv. Appl. Math. 2:482; b~~
the
homology alignment algorithm of Needleman and Wunsch ( 1970) J. Mol. Biol.
48:443; by the search for similarity method of Pearson and Lipman ( 1988)
Proc. Natl.
Acad Sci. 85:2444; by computerized implementations of these algorithms.
including,
but not limited to: CLUSTAL in the PC/Gene program by Intelligenetics
(Mountain
View, California). GAP. BESTFIT. BLAST, FASTA, and TFASTA in the Wisconsin
Genetics Software Package, Genetics Computer Group (GCG) (57~ Science Drive,
Madison, Wisconsin); the CLUSTAL program is well described by Higgins and
Sharp (1988) Gene 73:237-244; Higgins and Sharp (1989); CABIOS S:l~l-153:'
Corpet et al. ( I 988) Nucleic Acids Res. 16:10881-90; Huang et al. ( 1992)
Computer
Applications in the Biosciences 8:155-65; and Person et al. ( 1994) Meth. of
Mol. Biol.
24:307-331; preferred computer alignment methods also include the BLASTP.
BLASTN, and BLASTX algorithms. See Altschul et al. (1990) J. Mol. Biol. 21
~:403-
410. Alignment is also often performed by visual inspection and manual
alignment.
(c) As used herein. "sequence identity" or "identity" in the context
of two nucleic acid or polypeptide sequences includes reference to the
residues in the
two sequences that are the same when aligned for maximum correspondence over a
CA 02356492 2001-09-04
v~0 99/60141 PCT/US98/27450
specified comparison window. When percentage of sequence identity is used in
reference to proteins, it is recognized that residue positions that are not
identical often
differ by conservative amino acid substitutions, where amino acid residues are
substituted for other amino acid residues with similar chemical properties
(e.g., charge
or hydrophobicity) and therefore do not substantially change the functional
properties
of the molecule. Where sequences differ in conservative substitutions, the
percentage
of sequence identity may be adjusted upward to correct for the conservative
nature of
the substitution. Sequences that differ by such conservative substitutions are
said to
have "sequence similarity" or "similarity." Means for making this adjustment
are
well-known to those of skill in the art. Typically this involves scoring a
conservative
substitution as a partial rather than a full mismatch. thereby increasing the
percentage
of sequence identity. Thus, for example, where an identical amino acid is
given a
score of 1 and a non-conservative substitution is given a score of zero, a
conservative
substitution is given a score between zero and I . The scoring of conservative
1 S substitutions is calculated, e.g., as implemented in the program PC/GENE
(Intelligenetics, Mountain View, California).
(d) As used herein, "percentage of sequence identity" means the
value determined by comparing two optimally aligned sequences over a
comparison
windov~~, wherein the portion of the polynucleotide sequence in the comparison
window may comprise additions or deletions (i. e., gaps) as compared to the
reference
sequence (which does not comprise additions or deletions) for optimal
alignment of
the two sequences. The percentage is calculated by determining the number of
positions at which the identical nucleic acid base or amino acid residue
occurs in both
sequences to yield the number of matched positions, dividing the number of
matched
positions by the total number of positions in the window of comparison and
multiplying the result by 100 to yield the percentage of sequence identity.
(e)(i) The term "substantial identity" of polynucleotide sequences
means that a polynucleotide comprises a sequence that has at least 70%
sequence
identity, preferably at least 80%, more preferably at least 90% and most
preferably at
least 95%, compared to a reference sequence using one of the alignment
programs
described using standard parameters. One of skill in the art will recognize
that these
values can be appropriately adjusted to determine corresponding identity of
proteins
CA 02356492 2001-09-04
WO 99/60141 PCT/US98/27456
_g_
encoded by two nucleotide sequences by taking into account codon degeneracy
amino acid similarity, reading frame positioning, and the like. Substantial
identity of
amino acid sequences for these purposes normally means sequence identity of at
least
60%, more preferably at least 70%, 80%, 90%, and most preferably at least 95%.
Another indication that nucleotide sequences are substantially identical is if
two molecules hybridize to each other under stringent conditions. Generally,
stringent temperature conditions are selected to be about 5°C to about
2°C lower than
the melting point (Tm) for the specific sequence at a defined ionic strength
and pH.
The denaturation or melting of DNA occurs over a narrow temperature range and
represents the disruption of the double helix into its complementary single
strands.
The process usually is characterized by the temperature of the midpoint of
transition,
T~,. which is sometimes described as the melting temperature. Formulas are
available
in the art for the determination of melting temperatures. Typically. stringent
wash
conditions are those in which the salt concentration is about 0.02 molar at pH
7 and
I S the temperature is at 50, 55, or 60°C. However, nucleic acids that
do not hybridize to
each other under stringent conditions are still substantially identical if the
polypeptides that they encode are substantially identical. This may occur, for
example, when a copy of a nucleic acid is created using the maximum codon
degeneracy permitted by the genetic code. One indication that two nucleic acid
sequences are substantially identical is that the poIypeptide that the first
nucleic acid
encodes is immunologically cross reactive with the polypeptide encoded by the
second nucleic acid.
(e)(ii) The terms "substantial identity" in the context of a peptide
indicates that a peptide comprises a sequence with at least 70% sequence
identity to a
reference sequence, preferably 80%, more preferably 85%, most preferably at
least
90°io or 95% sequence identity to the reference sequence over a
specified comparison
windov<-. Preferably, optimal alignment is conducted using the homology
alignment
algorithm of Needleman and Wunsch ( 1970) J. Mol. Biol. 48:443. An indication
that
two peptide sequences are substantially identical is that one peptide is
immunologically reactive with antibodies raised against the second peptide.
Thus, a
peptide is substantially identical to a second peptide, for example, where the
two
peptides differ only by a conservative substitution. Polypeptides that are
CA 02356492 2001-09-04
. ,'s
WO 99/60141 PCT/US98/27456
-9-
"substantially similar" share sequences as noted above except that residue
positions
that are not identical may differ by conservative amino acid changes.
The present invention also encompasses the proteins and peptides encoded by
the nucleotide sequences of this invention. It is recognized that the proteins
of the
invention may be oligomeric and will vary in molecular weight. component
peptides,
activity, and in other characteristics. The proteins of the invention can be
used to
protect plants against nematodes. Such methods are described in more detail
below.
Fragments and variants of the disclosed nucleotide sequences and proteins
encoded thereby are also encompassed by the present invention. By "fragment"
is
intended a portion of the nucleotide sequence or a portion of the amino acid
sequence
and hence protein encoded thereby. Fragments of a nucleotide sequence may
encode
protein fragments that retain the biological activity of the native protein
and hence
confer resistance to nematodes. Alternatively. fragments of a nucleotide
sequence
that are useful as hybridization probes generally do not encode fragment
proteins
retaining biological activity. Thus, fragments of a nucleotide sequence may
range
from at least about 20 nucleotides, about 50 nucleotides, about 100
nucleotides. and
up to the entire nucleotide sequence encoding the proteins of the invention.
By "variants" is intended substantially similar sequences. For nucleotide
sequences, conservative variants include those sequences that, because of the
degeneracy of the genetic code, encode the amino acid sequence of one of the
proteins
conferring resistance to nematodes. Generally. nucleotide sequence variants of
the
invention will have at least 70%, generally, 80%, preferably up to 90%
sequence
identity to its respective native nucleotide sequence.
By "variant" protein is intended a protein derived from the native protein by
deletion (so-called truncation) or addition of one or more amino acids to the
N-
terminal and/or C-terminal end of the native protein; deletion or addition of
one or
more amino acids at one or more sites~in the native protein; or substitution
of one or
more amino acids at one or more sites in the native protein. Such variants may
result
from. for example, genetic polymorphism or from human manipulation. Methods
for
such manipulations are generally known in the art.
Thus, the proteins of the invention may be altered in various ways including
amino acid substitutions, deletions, truncations, and insertions. Methods for
such
CA 02356492 2001-09-04
62451-860(S)
-10-
manipulations are generally known in the art. For example, amino acid sequence
variants of the proteins can be prepared by mutations in the DNA. Methods for
mutagenesis and nucleotide sequence alterations are well known in the art.
See_ for
example, Kunkel (1985) Proc. Natl. Acad. Sci. USA 82:488-49?; Kunkel et al.
(1987)
Methods in Enrymol. 154:367-382; U.S. Patent No. 4.873,192; Walker and
Gaastra,
eds. (1983) Techniques in Molecular Biology (MacMillan Publishing Company. New
York) and the references cited therein. Thus, the genes and nucleotide
sequences of
the invention include both the naturally occurring sequences as well as mutant
forms.
Likewise, the proteins of the invention encompass both naturally occurring
proteins as
_ w~el1 as variations, fragments, and modified forms thereof. Such variants
will;
continue to possess the desired activity of conferring resistance to
nematodes.
Obviously. the mutations.that will be made in the DNA encoding the variant
must not
place the sequence out of reading frame and preferably will not create
sequences
deleterious to expression of the gene.product. See, EP Patent Application
Publication
No.75,444.
The nematode resistance genes of the invention can be optimized for enhanced
expression in plants of interest. See, for example, EPA0359472; EPA0385962:
W091/16432; Perlak et al. (1991) Prnc. Natl. Acad. Sci. USA 88:3324-3328: and
Murray et al. (1989) Nucleic Acids Res. 17:477-498. In this manner, the genes
can be
synthesized utilizing.plant-preferred colons. See, for example, Murray et al.
(1989)
. Nucleic Acids Res. 17:477-498. In this manner, synthetic genes
can also be made based on the distribution
of colons a particular host uses for a particular amino acid. Thus, the
nucleotide
sequences can be optimized for expression in any plant. It is recognized that
ali or
any part of the gene sequence may be optimized or synthetic. That is,
synthetic or
partially optimized sequences may also be used.
The present invention also relates to a recombinant DNA transformation
construct comprising the isolated DNA sequences involved in nematode
resistance in
plants. The recombinant DNA transformation construct can be introduced into
plant
cells, prbtoplasts, calli, tissues, or whole plants to confer nematode-
resistance
properties in plants.
The sequences of the invention can be constructed in expression cassettes for
CA 02356492 2001-09-04
62451-860 (S)
-11-
expression in a plant. Such expression cassettes will comprise a
transcriptional
initiation region linked to the gene encoding the gene of interest. Such an
expression
cassette is provided with a plurality of restriction sites for insertion of
the gene of
interest behind the regulatory control of a designated promoter. The
expression
cassette may additionally contain selectable marker genes suitable for the
particular
host organism.
The transcriptional initiation region, the promoter, may be native or
analogous
or foreign or heterologous to the host. Additionally, the promoter may be the
natural
sequence or alternatively a synthetic sequence. By foreign is intended that
the
transcriptional initiation region is not found in the wild-type host into
which the
transcriptional initiation region is introduced. As used herein a chimeric
gene
comprises a coding sequence operably linked to a transcription initiation
region which
is heterologous to the coding sequence. While any promoter or promoter element
capable of driving expression of a coding sequence can be utilized. of
particular
interest for expression in plants are root promoters (Bevan et al. (1993) in
Gene
Conservation and Exploitation: Proceedings of the 20th Stadler Genetics
Symposium, ed. Gustafson et al. (Plenum Press, New York) pp. 109-129; Brears
et al.
( 1991 ) Plant J. 1:235-244; Lorenz et al. ( 1993) Plant J. 4:545-554; U.S.
Patent Nos.
5.459.252; 5,608,149; 5,599,670); pith promoter (U.S. Patent Nos. 5.466,785;
5.451.514; 5,391.725); or other tissue specific and constitutive promoters
(see, for
example, U.S. Patent Nos. 5.608.149; 5,608,144; 5,604.121; 5.569.597;
5,466.785;
5,399,680; 5,268,463; 5,608;142).
The ttanscriptional cassette will include in the 5'-to-3' direction of
transcription, transcriptional and translational initiation regions, a DNA
sequence of
interest, and transcriptional and translational termination regions functional
in plants.
The termination region may be native with the transcriptional initiation
region, may
be native with the DNA sequence of interest, or may be derived from another
source.
Convenient termination regions are available from the Ti-plasmid of A.
tumefaciens,
such as the octopine synthase and nopaline synthase termination regions. See
also,
Guerineau et al. ( 1991 ) Mol. Gen. Genet. 262:141-144; Proudfoot ( 1991 )
Cell 64:671-
674; Sanfacon et al. (1991) Genes Dev. 5:141-149; Mogen et al. (1990) Plant
Cell
2:1261-1272; Munroe et al. (1990) Gene 91:151-I58; Ballas et al. (1989)
Nucleic
CA 02356492 2001-09-04
WO 99/60141 PCT/US98/27456
-12-
Acids Res. 17:7891-7903; Joshi et al. (1987) Nucleic Acid Res. 15:9627-9639.
Methodologies for the construction of plant transformation constructs are
described in the art. The construct may include any necessary regulatory
elements
such as promoters, terminators (Guerineau et al. ( I 991 ) Mol. Gen. Genet.
226: I 41-144; Proudfoot ( 1991 ) Cell 64:671-674; Sanfacon et al. ( 1991 )
Genes Dev.
5:141-149; Molten et al. (1990) Plant Cell 2:1261-1272; Munroe et al. (1990)
Gene
91:151-158; Ballas et al. (1989) Nucleic Acids Res. 17:7891-7903; .Ioshi et
al. (1987)
Nucleic Acid Res. I 5:962?-9639); plant translational consensus sequences
(Joshi, C.P.
(1987) Nucleic Acids Research 15:6643-6653). enhancers, introns (Luehrsen and
Walbot (1991) Mol. Gen. Genet. 225:81-93) and the like, operably linked to the
nucleotide sequence. It may be beneficial to include 5' leader sequences in
the
transformation construct. Such leader sequences can act to enhance
translation. See.
for example, Elroy-Stein et al. ( 1989) PNAS USA 86:6126-6130; Allison et al.
( 1986):
Macejak and Sarnow (1991) Nature 353:90-94; Jobling and Gehrke (1987) Nature
325:622-625; Gallie et al. (1989) Molecular Biology of RNA, pp. 237-256;
Lommel et
al. (1991) Virology 81:382-385; and Della-Cioppa et al. (1987) Plant Physiol.
84:965-968.
Transcriptional and translational regulatory signals include but are not
limited
to promoters. transcriptional initiation start sites. operators, activators,
enhancers,
other regulatory elements, ribosomal binding sites. an initiation codon.
termination
signals, and the like. See, for example. U.S. Patent No. 5,039,523: U.S.
Patent No.
4,853.331; EPO 0480762A2; Sambrook et al. (1989) Molecular Cloning A
Laboratory Manual (2d .ed., Cold Spring Harbor Laboratory Press, Plainview,
New
York); Davis et al., eds. (1980) Advanced Bacterial Genetics (2d ed., Cold
Spring
Harbor Laboratory. Cold Spring Harbor, New York); and the references cited
therein.
For the expression of the proteins encoded by the isolated DNA sequences of
the present invention. a promoter capable of facilitating gene transcription
in plant
cells must be operable linked to the nematode resistance gene sequence. A
variety of
suitable promoters are generally known in the art. Both constitutive promoter
and
tissue-specific promoters can be used. A constitutive promoter is a promoter
that can
initiate RNA transcription in any tissue or cell in a plant, while tissue-
specific
promoters can do so only in specific tissues. Suitable promoters are known in
the art
CA 02356492 2001-09-04
WO 99/60141 PCT/US98t2745~b
-13-
and include 35S and I 9S promoter of CaMV. Agrobacterium NOS (nopaline
symthase) gene promoter, and the Agrobacterium mannopine synthase gene
promoter.
For tissue specific expression, the isolated DNA sequences of the invention
conferring
nematode resistance can be operably linked to tissue specific promoters.
In addition, a marker gene for identifying and selecting transformed cells.
tissues, or plants may be included in the transformation construct. By marker
gene is
intended to be either reporter genes or selectable marker genes.
Reporter genes are generally known in the an. The reporter gene used should
be exogenous and not expressed endogenously. Ideally the reporter gene will
exhibit
low- background activity and should not interfere with plant biochemical and
physiological activities. The products expressed by the.reporter gene should
be stable
and readily detectable. It is important that the reporter gene expression
should be able
to be assayed by a non-destructive., quantitative, sensitive, easy to perform
and
inexpensive method.
Examples of suitable reporter genes known in the art can be found in, for
example, Jefferson et al. (1991 ) in Plant Molecular- Biology Manual, ed.
Gelvin et al.
(Kiuwer Academic Publishers), pp. 1-33; (DeWet et al. (1987) Mol. Cell. Biol.
7:725-
737: Goff et al. (1990) EMBO J. 9:2517-2522: Kain et al. (1995) BioTechnigues
19:60-655; Chiu et al. (1996) Current Biology 6:32-330.
Selectable marker genes for selection of transformed cells or tissues can
include genes that confer antibiotic resistance or resistance to herbicides.
Examples
of suitable seiectable marker genes include, but are not limited to, genes
encoding
resistance to chloramphenicol (Herrera Estrella et al. (1983) EMBO,I. 2:987-
992:
methotrexate (Herrera Estrella et al. ( 1983) Nature 303:209-213; Meijer et
al. ( 1991 )
Plant Mol. Biol. 16:807-820); hygromycin Waldron et al. (1985) Plant Mol.
Biol.
x:103-108; Zhijian et al. (1995) Plant Science 108:219-227); streptomycin
(Jones et
al. ( 1987) Mol. Gen. Genet. 210:86-91; spectinomycin (Bretagne-Sagnard et al.
(1996) Transgenic Res. 5:131-137); bleomycin (Hille et al. {1990) Plant Mol.
Biol.
7:171-176); sulfonamide (Guerineau et al. (1990) Plant Mol. Biol. 15:127-136):
bromoxynil (Stalker et al .(1988) Science 242:419-423); glyphosate (Shaw et
al.
( 1986) Science 233:478-481 ); phosphinothricin (DeBlock et al. ( 1987) EMBO
J.
6:'_' ~ I 3-2518); kanomycin, and the like.
CA 02356492 2001-09-04
WO 99/60141 PCT/US98/27456
-14-
It is further recognized that the components of the transformation construct
may be modified to increase expression. For example, truncated sequences,
nucleotide substitutions or other modifications may be employed. See, for
example,
Perlak et al. (1991 ) Proc. Natl. Acad Sci. USA 88:3324-3328; Murray et al. (
1989)
Arcleic Acids Res. 17:477-498; and W091/16432.
In preparing the transformation construct, the various DNA fragments may be
manipulated, so as to provide for the DNA sequences in the proper orientation
and, as
appropriate in the proper reading frame. Toward this end, adapters or linkers
may be
employed to join the DNA fragments or other manipulations may be involved to
provide for convenient restriction sites, removal of superfluous DNA, removal
of
restriction sites. or the like. For this purpose, in vitro mutagenesis, primer
repair.
restriction. annealing, resection, ligation. PCR, or the like may be employed.
where
insertions, deletions. or substitutions, e.g., transitions and transversions,
may be
involved.
The present invention also relates to the introduction of the transformation
constructs into plant protoplasts, calli, tissues, or organ explants and the
regeneration
of transformed plants expressing the nematode resistance gene. The
compositions of
the present invention can be used to transform any plant. In this manner,
genetically
modified plants, plant cells, plant tissue, seed, and the like can be
obtained.
Transformation protocols may vary depending on the type of plant or plant
cell. i. e.,
monocot or dicot. targeted for transformation. Suitable methods of
transforming plant
cells include microinjection (Crossway et al. (1986) Biotechniques 7:320-334).
electroporation (Riggs et al. (1986) Proc. Natl. Acad. Sci. USA 83:5602-5606):
Agrobacterium-mediated transformation (Hinchee et al. (1988) Biotechnology
6:91 ~-921 ); direct gene transfer (Paszkowski et al. ( 1984) EMBO .I. 3:2717-
2722):
and ballistic panicle bombardment (see, for example, Sanford et al., U.S.
Patent
4.945,050; Tomes et al. (1995) in Plant Cell, Tissue and Organ Culture:
Fundamental Methods, ed. Gamborg and Phillips (Springer-Verlag, Berlin): and
McCabe et al. (1988) Biotechnology 6:923-926). Also see Weissinger et al.
(1988)
Ann. Rev. Genet. 22:421-477; Sanford et al. (1987) Particulate Science and
Technology x:27-37 (onion); Christou et al. (1988) Plant Physiol. 87:671-674
(soybean); McCabe et al. (1988) Biotechnolo~v 6:923-926 (soybean); Finer and
CA 02356492 2001-09-04
62451-860(5)
-15-
McMullen (1991) In Yitro Cell Deo. Biol. 27P:175-182 (soybean); Singh et al.
(1998)
Theor. Appl. Genet. 96:319-324 (soybean); Datta et al. (1990) Biotechnology
8:736-740 (rice); Klein et al. (1988) Proc. Natl. Acad Sci. USA 85:4305-4309
(maize); Klein et al. (1988) Biotechnology 6:559-563 (maize); Klein et al.
(1988)
Plant Physiol. 91:440-444 (maize); Fromm et al. ( 1990) Biotechnology 8:833-
839;
and 'Tomes et al. ( I 995) in Plant Cell, Tissue, and Organ Culture:
Fundamental
Methods, ed. Gamborg and Phillips (Springer-Veilag, Berlin) (maize); Hooydaas-
Van
Slogteren and Hooykaas (1984) Nature (London) 311:763-764; Bytebier et al.
(1987)
Proc. Natl. Acad Sci. USA 84:5345-5349 (Liliaceae); De Wet et al. (1985) in
The
Experimental Manipulation oJOvule Tissues, (G.H.P. Chapman et al., Longman. NY
eds. pp. I 97-209) ('pollen); Kaeppler et al. ( 1990) Plant Cell Reports 9.:41
~-418:
Kaeppler et al. ( 1992) Theor. Appl. Genet. 84:560-566 (whisker-mediated
transformation); DeHalluin et al. (1992) Plant Cell 4:1495-1505
(electroporation); Li
et al. (1993) Plant Cell Reports 1'':250-255, and Christou and Ford
(1995)Annals of
I 5 Botany 75:407-413 (rice); Osjoda et al. (1996) Nature Biotechnology 14:745-
750
(maize via Agrobacterium tumefaciens).
Plant tissues suitable for transformation include but are not limited to leaf
tissues, root tissues, shoots, meristems. and protoplasts. For soybean it is
often
preferred to utilize explants of cotyledons.
For example, the Agrobacterium tumefaciens strain A208 is known to be
highly virulent on soybean and to give rise to a higher rate of
transformation. See
Byme et al. (1987) Plant Cell Tissue and Organ Culture 8:3-I5. The
transformation
of soybean protoplasts by co-culturing them with Agrobacterium tumefaciens or
Agrobacterium rhizogenes has been known.
See Facciotti et al. (1985) Biotechnology (NeH~ y'ork) 3:241. Tissue explants
may be
inoculated with the bacterium for transformation. For example, U.S. Patent No.
5.569,834 issued to Hinchee et al. discloses a method for soybean
transformation and
regeneration by inoculating a cotyledon explant that is tom apart at the
cotyledonary
node.
Alternatively, plants can also be transformed successfully by the biolistic
technique. which involves using high velocity microprojectiles carrying
CA 02356492 2001-09-04
WO 99/60141 PCT/US98/27456
-16-
microparticles containing the transformation construct to propel the
microparticles
into a plant cell, protoplast, or tissue. The high velocity microprojectile
penetrates the
outer cell surface without destroying the cell and injects the microparticles
into the
cells. The transformation construct in the microparticles is thereafter
released and
incorporated into the cell genome. This technique is also known as particle
bombardment and is disclosed in U.S. Patent Nos. 4,945.050, 5,036,006, and
x.100,792, which are hereby incorporated by reference. The key advantage of
this
technique is that it works on virtually any plant tissue. An example of
successful
transformation of soybean using this particle bombardment technique is
demonstrated
in McCabe et al. (1988) Biotechnology 6:923-926.
In yet another method of transformation, protoplasts are transfected directly
with expression vector DNA that contains the nematode-resistance gene by
electroporation or DNA-protoplast co-precipitation in accordance with
procedures
generally known in the art. See Christou et al. (1987) Proc. Natl. Acad. Sci.
USA
84:3962-3966; Lin et al. (1987) Plant Physiol. 84:856-861.
Once the transformation construct containing the isolated DNA sequences of
this invention has been delivered, protoplasts, cells, or tissues expressing
the protein
encoded by the isolated nematode resistance gene are selected. Selection can
be
based on the selectable marker that is incorporated in the transformation
construct or
by culturing the protoplasts, cells, or tissues in media containing one of the
antibiotics
or herbicides. Alternatively, nematode-resistance may be directly selected by
inoculating nematodes into the transformed protoplasts, cells, or tissues.
Both
methods of selection are generally known in the art.
A further aspect of the present invention relates to the regeneration of
transgenic plants that express nematode resistance genes of the invention. The
cells
that have been transformed and selected for expression of the sequence of this
invention may be grown into plants in accordance with conventional ways. See.
for
example, McCormick et al. (1986) Plant Cell Reports 5:81-84. These plants may
then be grown. and either pollinated with the same transformed strain or
different
strains, the resulting hybrid having the desired genetic traits necessary for
nematode-
resistance.
For example. in soybean, transgenic soybean regeneration has been successful
CA 02356492 2001-09-04
62451-860(S)
-17-
from tissues such as nodal axillary buds transformed with elec~roporation-
mediated
gene transfer technique (Chowrira et al. (1996) Mol. Biotechnol. 5:85-96);
somatic
embryos transformed using microprojectile bombardment (Stewart et al. (1996)
Plafzt
Physiol. 112:121-129); and cotyledon explants that are tom apart at the
cotyledonary
node and are uansformed by Agrobacterium inoculation (LJ.S. Patent No.
5,569.834
issued to Hinchee et al.). Other methods for regenerating soybean plants are
disclosed in U.S. Patent No. 4,684,612 issued to Hemphill et al., and U.S.
Patent No.
4,992.375 issued to Wright.
The sequences of the invention are generally introduced into plants wherein
the plant in its native state does not contain the DNA sequences. However. it
is
recognized that in some plants the gene may occur but does not confer
resistance
because of aberrant expression, a mutation in the sequence, a nonfunctional
protein.
and the like. It will be beneficial to transform such plants with the
sequences of the
invention.
Using cells and tissues of the present invention that are resistant to
nematodes
helps to obviate the problem of nematode infection of the host cells and
tissues in the
culture. In addition, the cells and tissues according to the present invention
can also
be valuable in the elucidation of the mechanism underlying the plant
resistance to
pathogens. Such plants include maize, oats, wheat. rice, barley. sorghum,
alfalfa.
tobacco, cotton. sugar beet, sunflower, carrot, canola, tomato, potato,
oilseed rape.
cabbage, pepper. lettuce, brassicas. tobacco, and soybean.
It is recognized that resistance to nematodes may be multigenic and
quantitative in certain plants. Thus, the sequences disclosed herein may be
useful
alone or in combination with other sequences. Breeding programs have produced
mane genotypes that have varying numbers of the genes responsible for nematode
resistance.
Thus, the isolated DNA sequences of the invention are preferably used to
transform plants expressing one or more other nematode resistance genes. Such
plants may be naturally occurring, produced by breeding programs, or produced
b~~
transformation with other nematode resistance genes. The result of the
transformation
with the isolated nematode resistance gene of this invention improves the
plants
capacity for nematode resistance.
CA 02356492 2001-09-04
WO 99/60141 PCT/US98/27456
-18-
Cotransformation may be conducted to introduce the DNA sequences of this
invention into plants together with one or more other nematode resistance
genes. In
the transformation construct, the other known nematode resistance genes may be
contained on the same plasmid as the DNA sequence of this invention or may be
contained on a separate plasmid or DNA molecule. The methods for making
transformation constructs having the other known nematode resistance gene with
or
without a DNA sequence isolated in this invention are similar to the methods
described above and should be apparent to a person skilled in the art.
Several methods of cotransformation of plants have been developed.
Cotransformation is easily accomplished by DNA mediated processes, such as the
co-
precipitation method, biolistic method. and electroporation. Each of these
methods is
adequately suited for the introduction of the DNA sequences of this invention
and
other nematode resistance genes, on the same or separate plasmids, into the
plant
cells. Alternatively, Agrobacterium tumefaciens-mediated cotransformation
techniques can be employed. Examples of such techniques can be found in, for
example, Depicker et al. (1985) Mol. Gen. Genet. 201:477-484; McKnight et al.
(1987) Plant Mol. Biol. 8:439-445; De Block et al. (1991) Theor. Appl. Genet.
8?:257-263; de Framond et al. (1986) Mol. Gerz Genet. 202:125-131; and Komari
et
al. ( 1996) The Plant Journal 10: I 65-174. In an alternative method, multiple
transgenes may be brought together by breeding of separately transformed
parent
plants.
The following examples are offered by way of illustration and not by wav of
limitation.
CA 02356492 2001-09-04
-19-
EXAMPLES
Example 1: Incorporation of DNA Sequences Conferring
Nematode Resistance into Expression Vectors
Genomic DNA sequences spanning the full length coding regions of gene
fragments conferring nematode-resistance to maize and soybean were isolated
and
cloned. These sequences are set forth in SEQ ID NOs: 1 and 3 (maize) and 5 and
7
(soybean). Plasmids containing these sequences have been deposited with
American
Type Culture Collection (ATCC) on October 1 S, 1997, and on February 4, 1998
and
are assigned Accession Numbers 209366, 209365, 209614, 209363, and 209364.
Gene fragments are cloned into a plasmid vector, such as that shown in Figure
6, in the sense orientation so that they are under the transcriptional control
of a
constitutive promoter. The transformation construct is then available for
introduction
into soybean cells by bombardment methods as described in Example 2.
Example 2: Transformation of Soybean Cells and Regeneration
of Transgenic Plants Having Improved Nematode Resistance
Initiation and Maicitenance of EmbryoQenic Suspension Cultures
Embryogenic suspension cultures of soybean (Glycine max Merrill) are
initiated and maintained in a 10A40N medium supplemented with 5 mM asparagine
as described previously (Finer and Nagasowa (1988) Plant Cell Tissue Org.
Culi.
15:125-136). For subculture, two clumps of embryogenic tissue, 4 mm in
diameter,
are transferred to 35 ml of 10A40N medium in a 125-ml delong flask. High
quality
embryogenic material is selectively subcultured monthly at this low inoculum
density.
Preparation of DNA and Tungsten Pellets
Plasmid DNA from Example 1 is precipitated onto 1.1 ~tm (average diameter)
tungsten pellets using a CaCl2 precipitation procedure (Finer and McMullen
(1990)
Plant Cell Rep. 8:586-589). The pellet mixture containing the precipitated DNA
is
gently resuspended after precipitation, and 2 ~1 is removed for bombardment.
errneTr~rr~ cmr~r~r
CA 02356492 2001-09-04
.v
WO 99/60141 PCT/US98l27456
-20-
Preparation of Plant Tissue for Bombardment
Approximately 1 g of embryogenic suspension culture tissue (taken 3 weeks
after subculture) is transferred to a 3.5-cm-diameter petri dish. The tissue
is centered
in the dish, the excess liquid is removed with a pipette, and a sterile 500 pm
pore size
nylon screen (Tetko Inc., Elmsford, New York) is placed over the embryonic
tissue.
Open petri dishes are placed in a laminar-flow hood for 10 to 15 minutes to
evaporate
residual liquid medium from the tissue. The 3.5-cm-diameter petri dish is
placed in
the center of a 9-cm-diameter petri dish immediately before bombardment.
Bombardments are performed using a DuPont Biolistics Particle Delivery System
(model BPG). Each sample of embryogenic soybean tissue is bombarded once.
Selection for Transgenic Clones
Bombarded tissues are resuspended in the 1 OA40N maintenance medium.
One to two weeks after bombardment the clumps of embryogenic tissue are
resuspended in fresh 10A40N medium containing a selection agent, such as
kanomycin or hygromycin. The selection agent is filter-sterilized before
addition to
liquid media. The medium containing a selection agent is replaced with fresh
antibiotic-containing medium weekly for 3 additional weeks.
Six to eight weeks after the initial bombardment, brown clumps of tissue that
contain yellow-green lobes of embryogenic tissue are removed and separately
subcultured in 10A40N medium containing selection agent. After 3 to 4 months
of
maintenance in this medium, proliferating embryogenic tissues are maintained
by
standard subculture in 10A40N without added antibiotic. Embryogenic tissues
are
periodically removed from I OA40N medium containing selection agent and 10A40N
for embryo development and Southern hybridization analyses.
Embrvo Development and Germination
For embryo development. clumps of kanamycin-resistant embryogenic tissues
are placed at 23°C on the embryo development medium, which contains MS
salts
(Murashige and Skoog (1962) Physiol. Plant 15:474-497), B5 vitamins (Gamborg
et
al. (1968) Exp. Cell. Res. 50:151-158). 6% maltose, and 0.2% gelrite (pH 5.7).
One
CA 02356492 2001-09-04
WO 99/60141 PCT/US98I2745b
-21-
month after plating, the developing embryos are cultured as individual
embryos, 25
per 9-cm-diameter petri dish in fresh embryo development medium. After an
additional 4 weeks, the mature embryos are placed in dry petri dishes for 2 to
3 days.
After the desiccation treatment, the embryos are transferred to a medium
containing
MS salts, BS vitamins, 3% sucrose, and 0.2% Gelrite (pH 5.7). After root and
shoot
elongation, plantlets are transferred to pots containing a 1:1:1 mixture of
vermiculite,
topsoil. and peat, and maintained under high humidity. Plantlets are gradually
exposed to ambient humidity over a 2-week period and placed in the greenhouse.
where they are grown to maturity and monitored for expression of the nematode
resistance gene.
DNA Extraction and~Southern Hybridization Analysis
DNA is extracted from embryogenic tissue and leaves using the CTAB
procedure (Saghai-Maroof et al. (1984) Proc. Natl. Acad. Sci. USA 81:8014-
8018).
Digested DNAs are electrophoresed on a 0.8% agarose gel. The DNA in the gels
is
1 ~ treated with 0.2 N HCI, twice for 1 ~ minutes, followed with 0.5 M
NaOH/0.1 M 1.~
M NaCI, twice for 30 minutes, and finally 1 M NHaCzH302/0.1 M NaOH, for 40
minutes. The DNA is transferred (Vollrath et al. ( 1988) Proc. Natl. Acad.
Sci. USA
8:6027-6031 ) to nylon membranes (Zetaprobe-BioRad, Richmond, California)
overnight by capillary transfer using l M NH4C~H30z/0.1 M NaOH. The membranes
are baked at 80°C for 2 hours under vacuum and then prehybridized for 4
to 6 hours at
6~°C in SO mM Tris pH 8.0, Sx standard saline citrate (SSC). 2x
Denhardt's. 10 mM
Na~EDTA, 0.2% sodium dodecyl sulfate (SDS). and 62.~ ~tg/ml salmon sperm DNA.
All publications and patent applications mentioned in the specification are
indicative of the level of those skilled in the art to which this invention
pertains. All
publications and patent applications are herein incorporated by reference to
the same
extent as if each individual publication or patent application was
specifically and
individually indicated to be incorporated by reference.
Although the foregoing invention has been described in some detail by way of
illusuation and example for purposes of clarity of understanding. it will be
obvious
that certain changes and modifications may be practiced within the scope of
the
appended claims.
CA 02356492 2001-09-04
22
SEQUENCE LISTING
<110> Jessen, Holly J.
Meyer, Terry E.
<120> Genes and Methods for Control of Nematodes in Plants
<130> 5718-18-1, 035718/171690
<140> PCT/US/98/27456
<141> 1998-12-23
<160> 10
<170> PatentIn Ver. 2.0
< 210 > 1
<211> 1347
<212> DNA
<213> Zea mays
<220>
2 0 <221> CDS
<222> (146)..(991)
<400> 1
ccacgcgtcc gcggacgcgt gggtgcccgg gagcgccgcc gcggtcgtgt gccaggtcag 60
cgaggccagc ctgctcccgc gcctcgccgc gtgggacaag tccgagacgc tcgcggccaa 120
gatcatgtac gccatcgaga gccag atg cag ggc tgc gcc ttc acg ctc gga 172
Met Gln Gly Cys Ala Phe Thr Leu Gly
1 5
ctc ggc gag ccc aac ctc gcc ggc aag ccc gtg ctc gag tac gac cgc 220
3 0 Leu Gly Glu Pro Asn Leu Ala Gly Lys Pro Val Leu Glu Tyr Asp Arg
10 15 20 25
gtc gtg cgc ccg cac gag ctg cac gcg ctc aag ccc aag cca gcg ccg 268
Val Val Arg Pro His Glu Leu His Ala Leu Lys Pro Lys Pro Ala Pro
30 35 40
gag ccc aag tct ggg tac ctc aac agg gag aac gag acg ctg ttc acc 316
Glu Pro Lys Ser Gly Tyr Leu Asn Arg Glu Asn Glu Thr Leu Phe Thr
50 55
atg tac cag ata ctc gaa tcg tgg ctg cgc gcc gcg tcg caa ctc ctc 364
Met Tyr Gln Ile Leu Glu Ser Trp Leu Arg Ala Ala Ser Gln Leu Leu
60 65 70
gcc cgc ctc aac gaa cgg atc gaa gcc aag aac tgg gaa gcg gcg get 412
Ala Arg Leu Asn Glu Arg Ile Glu Ala Lys Asn Trp Glu Ala Ala Ala
75 80 85
gcc gac tgc tgg atc ctg gag cgc gtg tgg aag ctg ctc gcc gac gtc 460
Ala Asp Cys Trp Ile Leu Glu Arg Val Trp Lys Leu Leu Ala Asp Val
90 95 100 105
gag gac ctc cac ctg ctg atg gac ccg gac gac ttc ctg cgg ctc aag 508
Glu Asp Leu His Leu Leu Met Asp Pro Asp Asp Phe Leu Arg Leu Lys
110 115 120
ggc cag ctc get gta cga gcg get cca tgg tct gac gcg tcg ttc tgt 556
Gly Gln Leu Ala Val Arg Ala Ala Pro Trp Ser Asp Ala Ser Phe Cys
125 130 135
CA 02356492 2001-09-04
23
ttc cgg tcc agg gcg ctc ctg cac gtc get aac acc act agg gac ctc 604
Phe Arg Ser Arg Ala Leu Leu His Val Ala Asn Thr Thr Arg Asp Leu
140 145 150
aag aag cgt gtg ccc tgg gtg ctc ggt gtc gag gtg gac ccc aac ggc 652
Lys Lys Arg Val Pro Trp Val Leu Gly Val Glu Val Asp Pro Asn Gly
155 160 165
ggc ccg cgg gtg cag gag gca gcc atg atg ctg tac cac agc cgt agg 700
Gly Pro Arg Val Gln Glu Ala Ala Met Met Leu Tyr His Ser Arg Arg
170 175 180 185
cgc ggc gag ggc gag gag gcg ggc aag gtg gag ctg ctc cag gcc ttc 748
Arg Gly Glu Gly Glu Glu Ala Gly Lys Val Glu Leu Leu Gln Ala Phe
190 195 200
caa gca gtg gag gtg gcc gtg aga gga ttc ttc ttc gcg tac cgg cag 796
2 0 Gln Ala Val Glu Val Ala Val Arg Gly Phe Phe Phe Ala Tyr Arg Gln
205 , 210 _ 215
ctc gtg gcg gcg gtg atg ggc acg gcg gag gcg ttg ggc aac cgg gcg 844
Leu Val Ala Ala Val Met Gly Thr Ala Glu Ala Leu Gly Asn Arg Ala
220 225 230
ctg ttc gtg ccg gcg gag ggg atg gat cca ttg gcc cag atg ttc ctc 892
Leu Phe Val Pro Ala Glu Gly Met Asp Pro Leu Ala Gln Met Phe Leu
235 240 245
gag cca ccc tac tac ccc agc ctg gat gcc gcc aag acg ttc cta gcg 940
Glu Pro Pro Tyr Tyr Pro Ser Leu Asp Ala Ala Lys Thr Phe Leu Ala
250 255 260 265
gat tac tgg gtt cag cag atg gcg ggg gcc tct get ccg tca ata caa 988
Asp Tyr Trp Val Gln Gln Met Ala Gly Ala Ser Ala Pro Ser Ile Gln
270 275 280
agc tgaaacggcg aaatggcgcg gctggatagc gaccgaatcg cgcagttttg 1041
Ser
cagcctgaag atactatgta tgcatgcatc gtaatttcgc tgtggccttg tggtgataga 1101
gtgattcatt tctatagcga tcctgtacta gtgtagtaca tgtagcacta aattgtctta 1161
ttatcgttgt gcttgtgcac tgcgttgtgt tgtgttctac atagagattg attcagttag 1221
atgccatttg tcactctagg caagtgtttc aattgggcac cgtgtatata tagaactttt 1281
gtaaacactg gtagatggat tcatcaatta cagaatgttg atgttgacaa aaaaaaaaaa 1341
aaaaaa 1347
<210> 2
5 0 <211> 282
<212> PRT
<213> Zea mays
<400> 2
Met Gln Gly Cys Ala Phe Thr Leu Gly Leu Gly Glu Pro Asn Leu Ala
1 5 10 15
Gly Lys Pro Val Leu Glu Tyr Asp Arg Val Val Arg Pro His Glu Leu
20 25 30
60 His Ala Leu Lys Pro Lys Pro Ala Pro Glu Pro Lys Ser Gly Tyr Leu
35 40 45
CA 02356492 2001-09-04
24
Asn Arg Glu Asn Glu Thr Leu Phe Thr Met Tyr Gln Ile Leu Glu Ser
50 55 60
Trp Leu Arg Ala Ala Ser Gln Leu Leu Ala Arg Leu Asn Glu Arg Ile
65 70 75 80
Glu Ala Lys Asn Trp Glu Ala Ala Ala Ala Asp Cys Trp Ile Leu Glu
85 90 95
Arg Val Trp Lys Leu Leu Ala Asp Val Glu Asp Leu His Leu Leu Met
100 105 110
Asp Pro Asp Asp Phe Leu Arg Leu Lys Gly Gln Leu Ala Val Arg Ala
115 120 125
Ala Pro Trp Ser Asp Ala Ser Phe Cys Phe Arg Ser Arg Ala Leu Leu
130 135 140
His Val Ala Asn Thr Thr Arg Asp Leu Lys Lys Arg Val Pro Trp Val
145 150 155 160
Leu Gly Val Glu Val Asp Pro Asn Gly Gly Pro Arg Val Gln Glu Ala
165 ~ 170 175
Ala Met Met Leu Tyr His Ser Arg Arg Arg Gly Glu Gly Glu Glu Ala
180 185 190
3 0 Gly Lys Val Glu Leu Leu Gln Ala Phe Gln Ala Val Glu Val Ala Val
195 200 205
Arg Gly Phe Phe Phe Ala Tyr Arg Gln Leu Val Ala Ala Val Met Gly
210 215 220
Thr Ala Glu Ala Leu Gly Asn Arg Ala Leu Phe Val Pro Ala Glu Gly
225 230 235 240
Met Asp Pro Leu Ala Gln Met Phe Leu Glu Pro Pro Tyr Tyr Pro Ser
40 245 250 255
Leu Asp Ala Ala Lys Thr Phe Leu Ala Asp Tyr Trp Val Gln Gln Met
260 265 270
Ala Gly Ala Ser Ala Pro Ser Ile Gln Ser
275 280
<210> 3
<211> 1325
50 <212> DNA
<213> Zea mays
<220>
<221> CDS
<222> (126)..(980)
<400> 3
ccacgcgtcc gagcgccgcc gcggtcgtgt gccgggccag caaggccagc ctgctcccgc 60
gcctcgccgc gtgggagaag tctgaggcgc tcgcggccag gatcacgtac gccgtcgagg 120
gccag atg cag ggc tgc gcc tcc acg ctc ggc ctc ggc gag ccc aac ctc 170
Met Gln Gly Cys Ala Ser Thr Leu Gly Leu Gly Glu Pro Asn Leu
6 0 1 5 10 15
CA 02356492 2001-09-04
10
gccggcaagccc gtgctcgag tacgaccgc gtcgtgcgc ccgcacgag 218
AlaGlyLysPro ValLeuGlu TyrAspArg ValValArg ProHisGlu
20 25 30
ctgcacgcgctg aagcccgac cctgcgccg gagcccatg tccggctac 266
LeuHisAlaLeu LysProAsp ProAlaPro GluProMet SerGlyTyr
35 40 45
cgcaaccgggag ctcgagact ctgttcacc atgtaccag atactcgag 314
ArgAsnArgGlu LeuGluThr LeuPheThr MetTyrGln IleLeuGlu
50 55 60
tcctggctccgc gtcgcgtcg cagctgctc acccgcctc gacgagcgg 362
SerTrpLeuArg ValAlaSer GlnLeuLeu ThrArgLeu AspGluArg
65 70 75
atc gaa gac aag tgc tgg gag gcg gcg gcc ggc gac tgc tgg atc ctg 410
2 0 Ile Glu Asp Lys Cys Trp Glu Ala Ala Ala Gly Asp Cys Trp Ile Leu
80 ~ 85 90 95
gag cgc gtg tgg aag ctg ctc gcg gac gtc gag gac ctc cac ctg ctg 458
Glu Arg Val Trp Lys Leu Leu Ala Asp Val Glu Asp Leu His Leu Leu
100 105 110
atg gac ccg gac gag ttc cta cgg ctc aag agc cag ctc gcc gta cga 506
Met Asp Pro Asp Glu Phe Leu Arg Leu Lys Ser Gln Leu Ala Val Arg
115 120 125
gcg gcg ccg ggg tct gag tcc gcg tcc ttc tgt ttc cgg tcc acg gcg 554
Ala Ala Pro Gly Ser Glu Ser Ala Ser Phe Cys Phe Arg Ser Thr Ala
130 135 140
ctc ctg cac gtc get agc gcc act agg gac ctc aag aag cgt gtg ccc 602
Leu Leu His Val Ala Ser Ala Thr Arg Asp Leu Lys Lys Arg Val Pro
145 150 155
tgg gtg ctc ggt gtc gag gcg gac ccc agc ggc ggc cca cgg gtg cag 650
4 0 Trp Val Leu Gly Val Glu Ala Asp Pro Ser Gly Gly Pro Arg Val Gln
160 165 170 175
gag gcg gcc atg aag ctg tac cac agc cgt agg cgc ggt gag ggc gag 698
Glu Ala Ala Met Lys Leu Tyr His Ser Arg Arg Arg Gly Glu Gly Glu
180 185 190
gag gca ggc aag gtg gac ctg ctc cag gcc ttc cag gcg gtg gag gtg 746
Glu Ala Gly Lys Val Asp Leu Leu Gln Ala Phe Gln Ala Val Glu Val
195 200 205
gcc gtg aga gca ttc ttc ttc ggg tac cgg cag ctg gtg gcg gcg gtc 794
Ala Val Arg Ala Phe Phe Phe Gly Tyr Arg Gln Leu Val Ala Ala Val
210 215 220
atg ggc acg gcg gag gcg tcg ggc aac cgg gcg ctg ttc gtg ccg gcg 842
Met Gly Thr Ala Glu Ala Ser Gly Asn Arg Ala Leu Phe Val Pro Ala
225 230 235
gag gag atg gat ccg ctc gcc caa atg ttc ctg gag ccg cca tac tac 890
Glu Glu Met Asp Pro Leu Ala Gln Met Phe Leu Glu Pro Pro Tyr Tyr
240 245 250 255
CA 02356492 2001-09-04
26
cct agc ctg gac gcc gcc aag acg ttt cta gcg gat tac tgg gtt cag 938
Pro Ser Leu Asp Ala Ala Lys Thr Phe Leu Ala Asp Tyr Trp Val Gln
260 265 270
ctt cag cag atg gcg gag gcc tct get ccg tca aga caa agc 980
Leu Gln Gln Met Ala Glu Ala Ser Ala Pro Ser Arg Gln Ser
275 280 285
tgaaacggcg aaatggcacg gctgagccac cgaatcgcgc agttttgcag gactgaagat 1040
actatgcatg catttcgttg gggccttttg cccttgtggt gaatggtgat agagtgattc 1100
atttctatag cgatcatgta ctattgcagt acatgtcgca ctagaatact agattctctt 1160
actatcgttg tgcactgcgt tgtacgtgtt gtgttctacg tagatataga ttgattcagt 1220
tagatgtcat ttgtattgcc aagtaggtca attggatatg gaacttttgt aaataccgaa 1280
atactgttgt tgaaaaaaaa aaaaaaaaaa aaaaaaaaaa aaaaa 1325
<210> 4
<211> 285
2 0 <212> PRT
<213> Zea mat's - -
<400> 4
Met Gln Gly Cys Ala Ser Thr Leu Gly Leu Gly Glu Pro Asn Leu Ala
1 5 10 15
Gly Lys Pro Val Leu Glu Tyr Asp Arg Val Val Arg Pro His Glu Leu
25 30
His Ala Leu Lys Pro Asp Pro Ala Pro Glu Pro Met Ser Gly Tyr Arg
35 40 45
Asn Arg Glu Leu Glu Thr Leu Phe Thr Met Tyr Gln Ile Leu Glu Ser
55 60
Trp Leu Arg Val Ala Ser Gln Leu Leu Thr Arg Leu Asp Glu Arg Ile
65 70 75 80
Glu Asp Lys Cys Trp Glu Ala Ala Ala Gly Asp Cys Trp Ile Leu Glu
85 90 95
Arg Val Trp Lys Leu heu Ala Asp Val Glu Asp Leu His Leu Leu Met
100 105 110
Asp Pro Asp Glu Phe Leu Arg Leu Lys Ser Gln Leu Ala Val Arg Ala
115 120 125
Ala Pro Gly Ser Glu Ser Ala Ser Phe Cys Phe Arg Ser Thr Ala Leu
130 135 140
50 Leu His Val Ala Ser Ala Thr Arg Asp Leu Lys Lys Arg Val Pro Trp
145 150 155 160
Val Leu Gly Val Glu Ala Asp Pro Ser Gly Gly Pro Arg Val Gln Glu
165 170 175
Ala Ala Met Lys Leu Tyr His Ser Arg Arg Arg Gly Glu Gly Glu Glu
180 185 190
Ala Gly Lys Val Asp Leu Leu Gln Ala Phe Gln Ala Val Glu Val Ala
195 200 205
CA 02356492 2001-09-04
27
Val Arg Ala Phe Phe Phe Gly Tyr Arg Gln Leu Val Ala Ala Val Met
210 215 220
Gly Thr Ala Glu Ala Ser Gly Asn Arg Ala Leu Phe Val Pro Ala Glu
225 230 235 240
Glu Met Asp Pro Leu Ala Gln Met Phe Leu Glu Pro Pro Tyr Tyr Pro
245 250 255
Ser Leu Asp Ala Ala Lys Thr Phe Leu Ala Asp Tyr Trp Val Gln Leu
260 265 270
Gln Gln Met Ala Glu Ala Ser Ala Pro Ser Arg Gln Ser
275 280 285
<210> 5
<211> 1498
2 0 <212> DNA
<213> Glycine max
<220>
<221> CDS
<222> (69)..(1433)
<220>
<223> Immediate source: Clone- P12568
<400> 5
cgacaccaat ttctccatcc tctcattgaa aaacaaaatt aatcatctta cttatttatt 60
ctccgaaa atg gtt gat tta cat tgg aaa tca aag atg cca agt tcc gac 110
3 0 Met Val Asp Leu His Trp Lys Ser Lys Met Pro Ser Ser Asp
1 5 10
atg cct tcc aaa act cta aaa ctc tct ctc tcc gac aac aag tcc tta 158
Met Pro Ser Lys Thr Leu Lys Leu Ser Leu Ser Asp Asn Lys Ser Leu
15 20 25 30
ccc tct ttg caa cta ccc ttc cgc acc aca gat atc tct cac gcc gca 206
Pro Ser Leu Gln Leu Pro Phe Arg Thr Thr Asp Ile Ser His Ala Ala
35 40 45
cct tct gtt tgc gcc act tac gac tac tat ctc cgt ctt cct caa ctc 254
Pro Ser Val Cys Ala Thr Tyr Asp Tyr Tyr Leu Arg Leu Pro Gln Leu
55 60
aga aag ctt tgg aac tcc tca gat ttt cct aat tgg aac aac gaa cca 302
Arg Lys Leu Trp Asn Ser Ser Asp Phe Pro Asn Trp Asn Asn Glu Pro
65 70 75
atc tta aaa cct atc ttg caa get ctc gaa atc acc ttc cgc ttt ctc 350
50 Ile Leu Lys Pro Ile Leu Gln Ala Leu Glu Ile Thr Phe Arg Phe Leu
80 85 90
tcc att gtt ctc tcc gat cca aga cct tac tcc aac cac aga gaa tgg 398
Ser Ile Val Leu Ser Asp Pro Arg Pro Tyr Ser Asn His Arg Glu Trp
95 100 105 110
act cgc agg ata gag tct ctt atc aca cat caa att gaa atc att gcc 446
Thr Arg Arg Ile Glu Ser Leu Ile Thr His Gln Ile Glu Ile Ile Ala
115 120 125
CA 02356492 2001-09-04
28
atactttgtgaa gatgag gaacaaaat tccgacacacgt ggcact gca 494
IleLeuCysGlu AspGlu GluGlnAsn SerAspThrArg GlyThr Ala
130 135 140
ccaaccgetgat ctcagc aggaacaat agcagcgagagc agaagc tac 542
ProThrAlaAsp LeuSer ArgAsnAsn SerSerGluSer ArgSer Tyr
145 150 155
agcgaggcaagc ctgctt ccgcggctt gccacgtggtac aaatcc aag 590
SerGluAlaSer LeuLeu ProArgLeu AlaThrTrpTyr LysSer Lys
160 165 170
gacgtagcgcag aggatc cttctctca gttgaatgccaa atgagg agg 638
AspValAlaGln ArgIle LeuLeuSer ValGluCysGln MetArg Arg
175 180 185 190
tgt tcc tac acg ctg ggt ttg ggt gag ccg aac cta gcg ggc aaa ccg 686
2 0 Cys Ser Tyr Thr Leu Gly Leu Gly Glu Pro Asn Leu Ala Gly Lys Pro
195 . 200 . 205
agc ctg ctc tac gac ctc gtg tgt aag ccg aac gag atc cac gcg ctg 734
Ser Leu Leu Tyr Asp Leu Val Cys Lys Pro Asn Glu Ile His Ala Leu
210 215 220
aag acg acg ccg tac gat gag cgc gta gag aat cac gag aac cac gcg 782
Lys Thr Thr Pro Tyr Asp Glu Arg Val Glu Asn His Glu Asn His Ala
225 230 235
ttg cac gcg acg cac cag atc gcc gag tcg tgg atc cac gcg tcg cgg 830
Leu His Ala Thr His Gln Ile Ala Glu Ser Trp Ile His Ala Ser Arg
240 245 250
aag gtt cta gag agg atc gca gac gcg gtg ctc tcc aga acc ttc gag 878
Lys Val Leu Glu Arg Ile Ala Asp Ala Val Leu Ser Arg Thr Phe Glu
255 260 265 270
aag gcg get gag gac tgc tac gcc gtg gaa agg atc tgg aag ctt ctc 926
4 0 Lys Ala Ala Glu Asp Cys Tyr Ala Val Glu Arg Ile Trp Lys Leu Leu
275 280 285
gcg gag gtg gag gac ctc cac ctg atg atg gat ccg gac gat ttc ttg 974
Ala Glu Val Glu Asp Leu His Leu Met Met Asp Pro Asp Asp Phe Leu
290 295 300
aga ctg aag aat cag ctc tcg gtg aaa tcc tcc ggc ggc gaa acg get 1022
Arg Leu Lys Asn Gln Leu Ser Val Lys Ser Ser Gly Gly Glu Thr Ala
305 310 315
tcg ttc tgc ttc agg tcg aag gag ttg gtt gaa ctg acg aag atg tgc 1070
Ser Phe Cys Phe Arg Ser Lys Glu Leu Val Glu Leu Thr Lys Met Cys
320 325 330
aga gat ctg agg cac aag gtg ccg gag ata ttg gag gtg gag gtg gat 1118
Arg Asp Leu Arg His Lys Val Pro Glu Ile Leu Glu Val Glu Val Asp
335 340 345 350
ccg aag gga gga ccg agg att caa gag gcg gcg atg aag ctc tac gtt 1166
Pro Lys Gly Gly Pro Arg Ile Gln Glu Ala Ala Met Lys Leu Tyr Val
355 360 365
CA 02356492 2001-09-04
29
tcg aag agc gcg ttc gag aag gtt cac ttg ttg cag gcg atg cag gcg 1214
Ser Lys Ser Ala Phe Glu Lys Val His Leu Leu Gln Ala Met Gln Ala
370 375 380
att gag gcg gcg atg aag aga ttc ttc tac gcg tat aag cag gtg ttg 1262
Ile Glu Ala Ala Met Lys Arg Phe Phe Tyr Ala Tyr Lys Gln Val Leu
385 390 395
gcg gtg gtg atg gga agc tcc gag get aac ggt aac cga gtt ggg ttg 1310
Ala Val Val Met Gly Ser Ser Glu Ala Asn Gly Asn Arg Val Gly Leu
400 405 410
agt tgc gac tcg get gac tcg ttg act cag att ttc ctt gaa ccg acg 1358
Ser Cys Asp Ser Ala Asp Ser Leu Thr Gln Ile Phe Leu Glu Pro Thr
415 420 425 430
tat ttt cca agc ttg gat gcc gcc aag act ttt ctt gga tac ttg tgg 1406
2 0 Tyr Phe Pro Ser Leu Asp Ala Ala Lys Thr Phe Leu Gly Tyr Leu Trp
435 440 ~ 445
gat aat aac gat aat aac aaa tgg ata tgataaggga aaaaaaaaaa 1453
Asp Asn Asn Asp Asn Asn Lys Trp Ile
450 455
acggcacaaa aacgatggcc aaagtgagat tttcggtttg ggcac 1498
<210> 6
3 0 <211> 455
<212> PRT
<213> Glycine max
<400> 6
Met Val Asp Leu His Trp Lys Ser Lys Met Pro Ser Ser Asp Met Pro
1 5 10 15
Ser Lys Thr Leu Lys Leu Ser Leu Ser Asp Asn Lys Ser Leu Pro Ser
25 30
4 0 Leu Gln Leu Pro Phe Arg Thr Thr Asp Ile Ser His Ala Ala Pro Ser
35 40 45
Val Cys Ala Thr Tyr Asp Tyr Tyr Leu Arg Leu Pro Gln Leu Arg Lys
50 55 60
Leu Trp Asn Ser Ser Asp Phe Pro Asn Trp Asn Asn Glu Pro Ile Leu
65 70 75 80
Lys Pro Ile Leu Gln Ala Leu Glu Ile Thr Phe Arg Phe Leu Ser Ile
50 85 90 95
Val Leu Ser Asp Pro Arg Pro Tyr Ser Asn His Arg Glu Trp Thr Arg
100 105 110
Arg Ile Glu Ser Leu Ile Thr His Gln Ile Glu Ile Ile Ala Ile Leu
115 120 125
Cys Glu Asp Glu Glu Gln Asn Ser Asp Thr Arg Gly Thr Ala Pro Thr
130 135 140
Ala Asp Leu Ser Arg Asn Asn Ser Ser Glu Ser Arg Ser Tyr Ser Glu
145 150 155 160
CA 02356492 2001-09-04
x
,. _M
r
Ala Ser Leu Leu Pro Arg Leu Ala Thr Trp Tyr Lys Ser Lys Asp Val
165 170 175
Ala Gln Arg Ile Leu Leu Ser Val Glu Cys Gln Met Arg Arg Cys Ser
180 185 190
Tyr Thr Leu Gly Leu Gly Glu Pro Asn Leu Ala Gly Lys Pro Ser Leu
10 195 200 205
Leu Tyr Asp Leu Val Cys Lys Pro Asn Glu Ile His Ala Leu Lys Thr
210 215 220
Thr Pro Tyr Asp Glu Arg Val Glu Asn His Glu Asn His Ala Leu His
225 230 235 240
Ala Thr His Gln Ile Ala Glu Ser Trp Ile His Ala Ser Arg Lys Val
245 250 255
Leu Glu Arg I1e Ala Asp Ala Val Leu Ser Arg Thr Phe Glu Lys Ala
260 265 270
Ala Glu Asp Cys Tyr Ala Val Glu Arg Ile Trp Lys Leu Leu Ala Glu
275 . 280 285
Val Glu Asp Leu His Leu Met Met Asp Pro Asp Asp Phe Leu Arg Leu
290 295 300
3 0 Lys Asn Gln,Leu Ser Val Lys Ser Ser Gly Gly Glu Thr Ala Ser Phe
305 310 315 320
Cys Phe Arg Ser Lys Glu Leu Val Glu Leu Thr Lys Met Cys Arg Asp
325 330 335
Leu Arg His Lys Val Pro Glu Ile Leu Glu Val Glu Val Asp Pro Lys
340 345 350
Gly Gly Pro Arg Ile Gln Glu Ala Ala Met Lys Leu Tyr Val Ser Lys
4 0 355 360 365
Ser Ala Phe Glu Lys Val His Leu Leu Gln Ala Met Gln Ala Ile Glu
370 375 380
Ala Ala Met Lys Arg Phe Phe Tyr Ala Tyr Lys Gln Val Leu Ala Val
385 390 395 400
Val Met Gly Ser Ser Glu Ala Asn Gly Asn Arg Val Gly Leu Ser Cys
405 410 415
Asp Ser Ala Asp Ser Leu Thr Gln Ile Phe Leu Glu Pro Thr Tyr Phe
420 425 430
Pro Ser Leu Asp Ala Ala Lys Thr Phe Leu Gly Tyr Leu Trp Asp Asn
435 440 445
Asn Asp Asn Asn Lys Trp Ile
450 455
<210> 7
<211> 1418
<212> DNA
CA 02356492 2001-09-04
31
<213> Glycine max
<220>
<221> CDS
<222> (46)..(1398)
<400> 7
caccaaacaa aaaaatcaat cattttattt tatttttcta cgaaa atg gtt gat tta 57
Met Val Asp Leu
1
cat tgg aaa tca aag atg cct agt tcc aaa aca cca aaa ctc tct ctc 105
His Trp Lys Ser Lys Met Pro Ser Ser Lys Thr Pro Lys Leu Ser Leu
5 10 15 20
tcc gac aac aag tcc tta ccc tct ttg caa cta ccc ttc cgc acc aca 153
Ser Asp Asn Lys Ser Leu Pro Ser Leu Gln Leu Pro Phe Arg Thr Thr
25 30 35
2 0 gat atc tct ccc gcc get cct tcc gtt tgc gcc get tac gac tac tat 201
Asp Ile Ser Pro Ala Ala Pro Ser Val Cys Ala Ala Tyr Asp Tyr Tyr
40 45 50
ctc cgt ctt cct caa ctc aga aag ctt tgg aac tcc act gat ttt cct 249
Leu Arg Leu Pro Gln Leu Arg Lys Leu Trp Asn Ser Thr Asp Phe Pro
55 60 65
aat tgg aac aac gaa ccg att cta aaa cca att ttg caa get ctc gaa 297
Asn Trp Asn Asn Glu Pro Ile Leu Lys Pro Ile Leu Gln Ala Leu Glu
30 70 75 80
ata acg ttc cgc ttt ctt tcc att gtt ctc tcc gat ccc aga cct tac 345
Ile Thr Phe Arg Phe Leu Ser Ile Val Leu Ser Asp Pro Arg Pro Tyr
85 90 95 100
tcc aac cac aga gaa tgg act cgc cgg ata gag tct ctc atc atg cat 393
Ser Asn His Arg Glu Trp Thr Arg Arg Ile Glu Ser Leu Ile Met His
105 110 115
4 0 caa att gaa atc att gcc ata ctt tgt gaa gaa gag gaa caa aat tcc 441
Gln Ile Glu Ile Ile Ala Ile Leu Cys Glu Glu Glu Glu Gln Asn Ser
120 125 130
gac aca cgt ggc act gca cca acc get gat ctc agc agc agc aat agc 489
Asp Thr Arg Gly Thr Ala Pro Thr Ala Asp Leu Ser Ser Ser Asn Ser
135 140 145
agc gtg agc aga agc tac agc gag gcg agc ctg ctt cct cgg ctt gcc 537
Ser Val Ser Arg Ser Tyr Ser Glu Ala Ser Leu Leu Pro Arg Leu Ala
50 150 155 160
acg tgg tac aaa tcc agg gac gtg gcg cag agg atc ctt ctc tcc gtg 585
Thr Trp Tyr Lys Ser Arg Asp Val Ala Gln Arg Ile Leu Leu Ser Val
165 170 175 180
gaa tgc caa atg agg agg tgc tcc tac acg ctt ggt ttg ggc gag ccg 633
Glu Cys Gln Met Arg Arg Cys Ser Tyr Thr Leu Gly Leu Gly Glu Pro
185 190 195
60 aac cta gcg ggg aag ccg agc ctg ctc tac gac ctc gtg tgc aag ccg 681
Asn Leu Ala Gly Lys Pro Ser Leu Leu Tyr Asp Leu Val Cys Lys Pro
200 205 210
CA 02356492 2001-09-04
32
aatgagatc cacgcgctg aagacg acgccgtacgac gagcgcgtg gag 729
AsnGluIle HisAlaLeu LysThr ThrProTyrAsp GluArgVal Glu
215 220 225
aaccacgag aaccacgcg gtgcac gccacgcaccag atcgcggag tcg 777
AsnHisGlu AsnHisAla ValHis AlaThrHisGln IleAlaGlu Ser
230 235 240
tggattcac gcgtcgcgg aaggtt ctggagagaatc gcggacgcg gtg 825
TrpIleHis AlaSerArg LysVal LeuGluArgIle AlaAspAla Val
245 250 255 260
ctctccaga accttcctg aaagca gcagaggactgc tacgccgtg gag 873
LeuSerArg ThrPheLeu LysAla AlaGluAspCys TyrAlaVal Glu
265 270 275
agg atc tgg aag ctt ctc gcg gag gtg gag gac ctc cac ctg atg atg 921
2 0 Arg Ile Trp Lys Leu Leu Ala Glu Val Glu Asp Leu His Leu Met Met
280 285 . 290
gat ccg gac gat ttc ttg agg cta aag aat caa ctc tcg gtg aaa tcc 969
Asp Pro Asp Asp Phe Leu Arg Leu Lys Asn Gln Leu Ser Val Lys Ser
295 300 305
tcg agc ggc gaa acg gca tcg ttc tgc ttc aga tcg aat gag tta gtg 1017
Ser Ser Gly Glu Thr Ala Ser Phe Cys Phe Arg Ser Asn Glu Leu Val
310 315 320
gaa ctg acg aag atg tgc aga gat ctg agg cac aag gtg ccg gag ata 1065
Glu Leu Thr Lys Met Cys Arg Asp Leu Arg His Lys Val Pro Glu Ile
325 330 335 340
ttg gag gtg gag gtg gat ccg aag gga gga ccg agg att caa gag gcg 1113
Leu Glu Val Glu Val Asp Pro Lys Gly Gly Pro Arg Ile Gln Glu Ala
345 350 355
gcg atg aag ctc tac gtt tcg aag agc gag ttc gag aag gtt cac ttg 1161
4 0 Ala Met Lys Leu Tyr Val Ser Lys Ser Glu Phe Glu Lys Val His Leu
360 365 370
ttg cag gcg atg cag gcg att gag gcg gcg atg aag aga ttc ttc tac 1209
Leu Gln Ala Met Gln Ala Ile Glu Ala Ala Met Lys Arg Phe Phe Tyr
375 380 385
gcg tat aag cag gtg ttg gcg gtg gtg atg gga agt tca gag get aac 1257
Ala Tyr Lys Gln Val Leu Ala Val Val Met Gly Ser Ser Glu Ala Asn
390 395 400
ggt aac cga gtt ggg ttg agt tgc gac tcg get gac tcg ttg act cag 1305
Gly Asn Arg Val Gly Leu Ser Cys Asp Ser Ala Asp Ser Leu Thr Gln
405 410 415 420
att ttc ctt gaa ccg acg tat ttt cca agc ttg gat gcc gcc aag act 1353
Ile Phe Leu Glu.Pro Thr Tyr Phe Pro Ser Leu Asp Ala Ala Lys Thr
425 430 435
ttt ctt gga tac ctg tgg gat aat aac gat aat aac aaa tgg ata 1398
Phe Leu Gly Tyr Leu Trp Asp Asn Asn Asp Asn Asn Lys Trp Ile
440 445 450
CA 02356492 2001-09-04
33
tgaaaacgaa aaaaaaaaaa 1418
<210> 8
<211> 451
<212> PRT
<213> Glycine max
<400> 8
Met Val Asp Leu His Trp Lys Ser Lys Met Pro Ser Ser Lys Thr Pro
1 5 10 15
Lys Leu Ser Leu Ser Asp Asn Lys Ser Leu Pro Ser Leu Gln Leu Pro
25 30
Phe Arg Thr Thr Asp Ile Ser Pro Ala Ala Pro Ser Val Cys Ala Ala
35 40 45
Tyr Asp Tyr Tyr Leu Arg Leu Pro Gln Leu Arg Lys Leu Trp Asn Ser
2 0 50 55 60
Thr Asp PheProAsn TrpAsn AsnGluPro IleLeuLysPro IleLeu
65 70 . 75 80
Gln Ala LeuGluIle ThrPhe ArgPheLeu SerIleValLeu SerAsp
85 90 95
Pro Arg ProTyrSer AsnHis ArgGluTrp ThrArgArgIle GluSer
100 105 110
Leu Ile MetHisGln IleGlu IleIleAla IleLeuCysGlu GluGlu
115 120 125
Glu Gln AsnSerAsp ThrArg GlyThrAla ProThrAlaAsp LeuSer
130 135 140
Ser Ser AsnSerSer ValSer ArgSerTyr SerGluAlaSer LeuLeu
145 150 155 160
4 Pro Arg LeuAlaThr TrpTyr LysSerArg AspValAlaGln ArgIle
0
165 170 175
Leu Leu SerValGlu CysGln MetArgArg CysSerTyrThr LeuGly
180 185 190
Leu Gly GluProAsn LeuAla GlyLysPro SerLeuLeuTyr AspLeu
195 200 205
Val Cys LysProAsn GluIle HisAlaLeu LysThrThrPro TyrAsp
50 210 215 220
Glu Arg ValGluAsn HisGlu AsnHisAla ValHisAlaThr HisGln
225 230 235 240
Ile Ala GluSerTrp IleHis AlaSerArg LysValLeuGlu ArgIle
245 250 255
Ala Asp AlaValLeu SerArg ThrPheLeu LysAlaAlaGlu AspCys
260 265 270
60
Tyr Ala ValGluArg IleTrp LysLeuLeu AlaGluValGlu AspLeu
275 280 285
CA 02356492 2001-09-04
34
His Leu Met Met Asp Pro Asp Asp Phe Leu Arg Leu Lys Asn Gln Leu
290 295 300
Ser Val Lys Ser Ser Ser Gly Glu Thr Ala Ser Phe Cys Phe Arg Ser
305 310 315 320
Asn Glu Leu Val Glu Leu Thr Lys Met Cys Arg Asp Leu Arg His Lys
325 330 335
Val Pro Glu Ile Leu Glu Val Glu Val Asp Pro Lys Gly Gly Pro Arg
340 345 350
Ile Gln Glu Ala Ala Met Lys Leu Tyr Val Ser Lys Ser Glu Phe Glu
355 360 365
Lys Val His Leu Leu Gln Ala Met Gln Ala Ile Glu Ala Ala Met Lys
370 375 380
Arg Phe Phe Tyr Ala Tyr Lys Gln Val Leu Ala Val Val Mgt Gly Ser
385 390 395 400
Ser Glu Ala Asn Gly Asn Arg Val Gly Leu Ser Cys Asp Ser Ala Asp
405 410 ~ 415
Ser Leu Thr Gln Ile Phe Leu Glu Pro Thr Tyr Phe Pro Ser Leu Asp
420 425 430
3 0 Ala Ala Lys Thr Phe Leu Gly Tyr Leu Trp Asp Asn Asn Asp Asn Asn
435 440 445
Lys Trp Ile
450
<210> 9
<211> 1498
<212> DNA
<213> Glycine max
40 <220>
<221> CDS
<222> (69)..(1433)
<400> 9
cgacaccaat ttctccatcc tctcattgaa aaacaaaatt aatcatctta tttatttatt 60
ctccgaaa atg gtt gat tta cat tgg aaa tca aag atg cca agt tcc gac 110
Met Val Asp Leu His Trp Lys Ser Lys Met Pro Ser Ser Asp
1 5 10
atg cct tcc aaa act ctc aaa ctc tct ctc tcc gac aac aag tcc tta 158
50 Met Pro Ser Lys Thr Leu Lys Leu Ser Leu Ser Asp Asn Lys Ser Leu
15 20 25 30
ccc tct ttg caa cta ccc ttc cgc acc aca gat atc tct cac gcc gca 206
Pro Ser Leu Gln Leu Pro Phe Arg Thr Thr Asp Ile Ser His Ala Ala
35 40 45
cct tct gtt tgc gcc act tac gac tac tat ctc cgt ctt cct caa ctc 254
Pro Ser Val Cys Ala Thr Tyr Asp Tyr Tyr Leu Arg Leu Pro Gln Leu
50 55 60
CA 02356492 2001-09-04
10
aga aag ctt tgg aac tcc tca gat ttt cct aat tgg aac aac gaa cca 302
Arg Lys Leu Trp Asn Ser Ser Asp Phe Pro Asn Trp Asn Asn Glu Pro
65 70 75
atc tta aaa cct atc ttg caa get ctc gaa atc acc ttc cgc ttt ctc 350
Ile Leu Lys Pro Ile Leu Gln Ala Leu Glu Ile Thr Phe Arg Phe Leu
80 85 90
tcc att gtt ctc tcc gat cca aga cct tac tcc aac cac aga gaa tgg 398
Ser Ile Val Leu Ser Asp Pro Arg Pro Tyr Ser Asn His Arg Glu Trp
95 100 105 110
act cgc agg ata gag tct ctt atc aca cat caa att gaa atc att gcc 446
Thr Arg Arg Ile Glu Ser Leu Ile Thr His Gln Ile Glu Ile Ile Ala
115 120 125
ata ctt tgt gaa gat gag gaa caa aat tcc gac aca cgt ggc act gca 494
2 0 Ile Leu Cys Glu Asp Glu Glu Gln Asn Ser Asp Thr Arg Gly Thr Ala
130 . 135 140
cca acc get gat ctc agc agg aac aat agc agc gag agc aga agc tac 542
Pro Thr Ala Asp Leu Ser Arg Asn Asn Ser Ser Glu Ser Arg Ser Tyr
145 150 155
agc gag gca agc ctg ctt ccg cgg ctt gcc acg tgg tac aaa tcc aag 590
Ser Glu Ala Ser Leu Leu Pro Arg Leu Ala Thr Trp Tyr Lys Ser Lys
160 165 170
gac gta gcg cag agg atc ctt ctc tca gtt gaa tgc caa atg agg agg 638
Asp Val Ala Gln Arg Ile Leu Leu Ser Val Glu Cys Gln Met Arg Arg
175 180 185 190
tgt tcc tac acg ctg ggt ttg ggt gag ccg aac cta gcg ggc aaa ccg 686
Cys Ser Tyr Thr Leu Gly Leu Gly Glu Pro Asn Leu Ala Gly Lys Pro
195 200 205
agc ctg ctc tac gac ctc gtg tgc aag ccg aac gag atc cac gcg ctg 734
4 0 Ser Leu Leu Tyr Asp Leu Val Cys Lys Pro Asn Glu Ile His Ala Leu
210 215 220
aag acg acg ccg tac gat gag cgc gta gag aat cac gag aac cac gcg 782
Lys Thr Thr Pro Tyr Asp Glu Arg Val Glu Asn His Glu Asn His Ala
225 230 235
ttg cac gcg acg cac cag atc gcc gag tcg tgg atc cac gcg tcg cgg 830
Leu His Ala Thr His Gln Ile Ala Glu Ser Trp Ile His Ala Ser Arg
240 245 250
aag gtt cta gag agg atc gca gac gcg gtc ctc tcc aga acc ttc gag 878
Lys Val Leu Glu Arg Ile Ala Asp Ala Val Leu Ser Arg Thr Phe Glu
255 260 265 270
aag gcg get gag gac tgc tac gcc gtg gaa agg atc tgg aag ctt ctc 926
Lys Ala Ala Glu Asp Cys Tyr Ala Val Glu Arg Ile Trp Lys Leu Leu
275 280 285
gcg gag gtg gag gac ctc cac ctg atg atg gat ccg gac gat ttc ttg 974
Ala Glu Val Glu Asp Leu His Leu Met Met Asp Pro Asp Asp Phe Leu
290 295 300
CA 02356492 2001-09-04
36
aga ctg aag aat cag ctc tcg gtg aaa tcc tcc ggc ggc gaa acg get 1022
Arg Leu Lys Asn Gln Leu Ser Val Lys Ser Ser Gly Gly Glu Thr Ala
305 310 315
tcg ttc tgc ttc agg tcg aag gag ttg gtt gaa ctg acg aag atg tgc 1070
Ser Phe Cys Phe Arg Ser Lys Glu Leu Val Glu Leu Thr Lys Met Cys
320 325 330
aga gat ctg agg cac aag gtg ccg gag ata ttg gag gtg gag gtg gat 1118
Arg Asp Leu Arg His Lys Val Pro Glu Ile Leu Glu Val Glu Val Asp
335 340 345 350
ccg aag gga gga ccg agg att caa gag gcg gcg atg aag ctc tac gtt 1166
Pro Lys Gly Gly Pro Arg Ile Gln Glu Ala Ala Met Lys Leu Tyr Val
355 360 365
tcg aag agc gcg ttc gag aag gtt cac ttg ttg cag gcg atg cag gcg 1214
2 0 Ser Lys Ser Ala Phe Glu Lys Val His Leu Leu Gln Ala Met Gln Ala
370 375 3B0
att gag gcg gcg atg aag aga ttc ttc tac gcg tat aag cag gtg ttg 1262
Ile Glu Ala Ala Met Lys Arg Phe Phe Tyr Ala Tyr Lys Gln Val Leu
385 390 395
gcg gtg gtg atg gga agc tcc gag get aac ggt aac cga gtt ggg ttg 1310
Ala Val Val Met Gly Ser Ser Glu Ala Asn Gly Asn Arg Val Gly Leu
400 405 410
agt tgc gac tcg cgt gac tcg ttg act cag att ttc ctt gaa ccg acg 1358
Ser Cys Asp Ser Arg Asp Ser Leu Thr Gln Ile Phe Leu Glu Pro Thr
415 420 425 430
tat ttt cca agc ttg gat gcc gcc aag act ttt ctt gga tac ttg tgg 1406
Tyr Phe Pro Ser Leu Asp Ala Ala Lys Thr Phe Leu Gly Tyr Leu Trp
435 440 445
gat aat aac gat aat aac aaa tgg ata tgataaggga aaaaaaaaaa 1453
4 0 Asp Asn Asn Asp Asn Asn Lys Trp Ile
450 455
acggcacaaa aacgatggcc aaagtgagat tttcggtttg ggcac 1498
<210> 10
<211> 455
<212> PRT
<213> Glycine max
<400> 10
SO Met Val Asp Leu His Trp Lys Ser Lys Met Pro Ser Ser Asp Met Pro
1 5 10 15
Ser Lys Thr Leu Lys Leu Ser Leu Ser Asp Asn Lys Ser Leu Pro Ser
20 25 30
Leu Gln Leu Pro Phe Arg Thr Thr Asp Ile Ser His Ala Ala Pro Ser
40 45
Val Cys Ala Thr Tyr Asp Tyr Tyr Leu Arg Leu Pro Gln Leu Arg Lys
60 50 55 60
CA 02356492 2001-09-04
37
Leu Trp Asn Ser Ser Asp Phe Pro Asn Trp Asn Asn Glu Pro Ile Leu
65 70 75 80
Lys Pro Ile Leu Gln Ala Leu Glu Ile Thr Phe Arg Phe Leu Ser Ile
85 90 95
Val Leu Ser Asp Pro Arg Pro Tyr Ser Asn His Arg Glu Trp Thr Arg
100 105 110
Arg Ile Glu Ser Leu Ile Thr His Gln Ile Glu Ile Ile Ala Ile Leu
115 120 125
Cys Glu Asp Glu Glu Gln Asn Ser Asp Thr Arg Gly Thr Ala Pro Thr
130 135 140
Ala Asp Leu Ser Arg Asn Asn Ser Ser Glu Ser Arg Ser Tyr Ser Glu
145 150 155 160
Ala.Ser Leu Leu Pro Arg Leu Ala Thr Trp Tyr Lys Ser Lys Asp Val
165 170 175
Ala Gln Arg Ile Leu Leu Ser Val Glu Cys Gln Met Arg Arg Cys Ser
180 ~ 185 190
Tyr Thr Leu Gly Leu Gly Glu Pro Asn Leu Ala Gly Lys Pro Ser Leu
195 200 205
3 0 Leu Tyr Asp Leu Val Cys Lys Pro Asn Glu Ile His Ala Leu Lys Thr
210 215 220
Thr Pro Tyr Asp Glu Arg Val Glu Asn His Glu Asn His Ala Leu His
225 230 235 240
Ala Thr His Gln Ile Ala Glu Ser Trp Ile His Ala Ser Arg Lys Val
245 250 255
Leu Glu Arg Ile Ala Asp Ala Val Leu Ser Arg Thr Phe Glu Lys Ala
40 260 265 270
Ala Glu Asp Cys Tyr Ala Val Glu Arg Ile Trp Lys Leu Leu Ala Glu
275 280 285
Val Glu Asp Leu His Leu Met Met Asp Pro Asp Asp Phe Leu Arg Leu
290 295 300
Lys Asn Gln Leu Ser Val Lys Ser Ser Gly Gly Glu Thr Ala Ser Phe
305 310 315 320
Cys Phe Arg Ser Lys Glu Leu Val Glu Leu Thr Lys Met Cys Arg Asp
325 330 335
Leu Arg His Lys Val Pro Glu Ile Leu Glu Val Glu Val Asp Pro Lys
340 345 350
Gly Gly Pro Arg Ile Gln Glu Ala Ala Met Lys Leu Tyr Val Ser Lys
355 360 365
Ser Ala Phe Glu Lys Val His Leu Leu Gln Ala Met Gln Ala Ile Glu
370 375 380
CA 02356492 2001-09-04
3a
Ala Ala Met Lys Arg Phe Phe Tyr Ala Tyr Lys Gln Val Leu Ala Val
385 390 395 400
Val Met Gly Ser Ser Glu Ala Asn Gly Asn Arg Val Gly Leu Ser Cys
405 410 415
Asp Ser Arg Asp Ser Leu Thr Gln Ile Phe Leu Glu Pro Thr Tyr Phe
420 425 430
Pro Ser Leu Asp Ala Ala Lys Thr Phe Leu Gly Tyr Leu Trp Asp Asn
435 440 445
Asn Asp Asn Asn Lys Trp Ile
450 455