Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
GENETIC TRAIT BREEDING METHOD
FIELD OF THE INVENTION
This invention is in the field of plant molecular biology. In particular, this
invention relates to a method for breeding plants for improved agricultural
traits.
BACKGROUND OF THE INVENTION
To date, almost all improvements in agricultural crops have been achieved
using traditional plant breeding techniques. These techniques involve crossing
parental plants with different genetic backgrounds to generate progeny with
1o genetic diversity. The progeny are then selected to obtain those plants
that
express the desired traits. Desired traits are then fixed while deleterious
traits are
eliminated via multiple backcrossings or selfings to eventually yield progeny
with
the desired characteristics. Hybrid corn, low erucic acid oilseed rape, high
oil
corn, and hard white winter wheat are examples of significant agricultural
15 advances achieved with traditional breeding.
However, the amount of genetic diversity in the germplasm of a particular
crop limits what can be accomplished by breeding. Although traditional
breeding
has proven to be very powerful, as advances in crop yields over the last
century
demonstrate, recent data suggest that the rate of yield improvement is
tapering
20 off for major food crops (Lee (1998) Proc. Natl. Acad. Sci. USA 95: 2001-
2004).
The introduction of molecular mapping markers into breeding programs may
accelerate the process of crop improvement in the near term, but ultimately
the
lack of new sources of genetic diversity will become limiting. In particular,
traditional breeding has proved rather ineffective for improving many
polygenic
25 traits such as increased disease resistance.
In recent years, biotechnology approaches involving the expression of
single transgene in crops have resulted in the successful commercial
introduction
of new plant traits, including herbicide resistance (glyphosate or Roundup
resistance), insect resistance (expression of Bacillus thuringiensis toxins)
and
30 virus resistance (over expression of viral coat proteins). However, the
list of
single gene traits of significant value is relatively small. The greatest
potential of
biotechnology lies in engineering complex polygenic traits to for
environmental
stresses, disease, plant development and architecture, yield and quality
traits.
Presently, engineering such polygenic traits has proven extremely challenging.
35 The present invention provides a novel method for rapidly identifying
genes that are useful for modifying complex plant traits.
-1-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
SUMMARY OF THE INVENTION
In one aspect, the invention provides a method for systematically
screening for traits associated with the altered expression of a gene of
interest in
plants. The method comprises providing a first pool of donor vectors, wherein
each donor vector comprises a transactivator and a second pool of receptor
1o vectors, wherein each receptor vector comprises a transactivator binding
site
operably linked to a different gene of interest. Then a first plant is
transformed
with a member of the donor vector pool and a second plant transformed with a
member of the receptor vector pool to generate first transformed plants
comprising the donor vector and a second transformed plant comprising the
15 receptor vector. First and second transformed plants are crossed to
generate a
hybrid plant. Both first and second transformed plants have a wild type
phenotype because the expression levels of the gene of interest is not altered
from that in a nontransformed plant. However, the phenotype of the hybrid
plant
comprising both transactivator and the gene of interest may be different from
wild
2o type because the expression levels of the gene of interest is altered
compared
with that of a nontransformed plant. The phenotype of these plants is
investigated to identify a hybrid plant with an improved trait.
The transactivator may be operably linked to (1 ) a constitutive promoter,
(2) an inducible promoter, (3) a tissue active or specific promoter or (4) a
25 developmental-stage active or specific promoter. When the transactivator is
linked to a constitutive promoter, changes in expression of a gene will be
observed in all tissues and at all times and a broad overview of the effects
of the
expression of the gene on a plant will be observed. When the transactivator is
linked to a tissue specific promoter or an inducible promoter or developmental-
30 stage promoter, the expression of the gene may be turned on or off in a
particular
tissue such as seed, roots, flowers, leaves, shoots, fruits or stems, during a
particular period in development, such as early, middle or late stages in
development, or under particular conditions, such as specific environmental or
disease stresses. A plant may be transformed with more than one receptor
35 vector or with more than one donor vector.
-2-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
The gene of interest may be any gene, but is preferably a regulatory gene
such as a transcription factor, a phosphatase or a protein kinase. In one
preferred embodiment, the genes of interest are all the transcription factors
identified in a plant, such as those identified in Arabidopsis thaliana. These
genes
collectively control all gene expression in plants and thus control all plant
traits.
The gene may be in a sense orientation for overexpression analysis or in an
antisense orientation for underexpression analysis. Additionally, the gene may
be
a full length coding sequence for a gene or a fragment of a gene, in
particular a
fragment with biological activity.
io The donor and receptor vectors may also include first and second
selectable markers, respectively, to assist in selecting transformed hybrid
plants.
In a second aspect, the invention provides a method for breeding plants
for a desired or improved trait. The method involves crossing a member of a
first
pool of plants, each plant in this pool having been transformed with an donor
15 vector comprising a transactivator, with a member of a second pool of
plants,
each plant in this pool having been transformed with a receptor vector
comprising
a transactivator binding site operably linked to a different gene of interest
to
generate a collection of hybrid plants. Traditional plant breeding techniques
are
used to obtain transgenic plants having both donor and receptor vectors and
2o exhibiting a desired or improved trait. Additionally, the invention
provides a
transgenic plant generated by the method described above.
In yet another aspect, the invention is a plant breeding kit. The plant
breeding kit comprises (a) a pool of activator vectors, wherein each donor
vector
comprises a transactivator; and (b) a pool of receptor vectors, wherein each
25 receptor vector comprises a transactivator binding site operably linked to
a
different gene of interest. The vectors may also include first and second
selectable markers to assist in selecting transformed hybrid plants. The
transactivator may be operably linked to (1 ) a constitutive promoter, (2) an
inducible promoter, (3) a tissue active or specific promoter or (4) a
developmental-
3o stage active or specific promoter.
Individual donor vectors and receptor vectors are transformed into first and
second plants, respectively, and traditional plant breeding techniques are
employed to generate hybrid plants comprising first and second vectors with
agriculturally valuable traits.
35 In a further aspect, the invention is a method for modifying the patterns
of
gene expression in a plant. The method first entails providing a first pool of
donor
-3-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
vectors, wherein each activator vector member comprises a transactivator, and
a
second pool of receptor vectors, wherein each receptor vector member comprises
a transactivator binding site operably linked to a regulatory gene. Activator
and
receptor vector members are transformed into first and second plants.
Transformed first and second plants are crossed to generate a hybrid plant
with
modified patterns of gene expression.
DETAILED DESCRIPTION OF THE INVENTION
1o DEFINITIONS
A "transgenic or transformed plant" refers to a plant which contains a
recombinant polynucleotide introduced by transformation. Transformation means
introducing a nucleotide sequence in a plant in any manner to cause stable or
transient expression of the sequence. This may be achieved by transfection
with
viral vectors, transformation with plasmids, such as Agrobacterium-based
vectors,
or introduction of naked DNA by electroporation, lipofection, or particle gun
acceleration. A transformed plant may refer to a whole plant as well as to
seed,
plant tissue, plant cells or any other plant material, and to the plant's
progeny.
A "vector" is a nucleic acid construct, generated recombinantly or
2o synthetically, comprising nucleic acid elements that can cause expression
of a
gene. A "donor vector" is a construct for expression of a polynucleotide
sequence
for a transactivator gene. The transactivator gene is operably linked to a
promoter. The promoter region may include tissue active-or-specific promoters,
developmental stage active-or-specific promoters, inducible promoters or
constitutive promoters.
A "receptor vector" is a construct for expression of a gene of interest such
as regulatory gene. Typically, the receptor vector includes the sequence for a
transactivator binding site. The construct sequence may also include
promoters,
operators, enhancer regions, silencer regions, polyadenylation sites,
translation
initiation sites and the like.
A "gene of interest" is a polynucleotide sequence for a regulatory gene
such as a transcription factor, a protein kinase or a phosphatase. These
sequences may be in a sense or antisense orientation, or partial or complete
gene sequences.
A nucleotide sequence is "operably linked" when it is placed into a
functional relationship with another nucleotide sequence. For example, a
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
promoter or enhancer is operably linked to a gene coding sequence if the
presence of the promoter or enhancer increases the level of expression of the
gene coding sequence.
A "pool" entails a group of at least two members, preferably at least 10
members, more preferably at least 100 members. For example, a pool can be all
the identified genes of a certain type in a plant such as all identified
transcription
factors derived from a plant, as exemplified by up to 1,700 transcription
factors
identified in Arabidopsis thaliana.
The phrase "altered or modified expression" in reference to polynucleotide
or polypeptide expression refers to an expression pattern in a transgenic
plant
that is different from the expression pattern in the wild type plant or a
reference
plant; for example, by expression in a cell type other than a cell type in
which the
sequence is expressed in the wild type plant, or by expression at a time other
than at the time the sequence is expressed in the wild type plant, or by a
t5 response to different inducible agents, such as hormones or environmental
signals, or at different expression levels (either higher or lower) compared
with
those found in a wild type plant. The term also refers to lowering the levels
of
expression to below the detection level or completely abolishing expression.
The
resulting expression pattern may be transient or stable, constitutive or
inducible.
"Trait" refers to the physiological, morphological or physical
characteristics of a plant or particular plant material. These characteristics
may
be visible to the human eye, such as germination rates or seed size, or be
measurable by laboratory techniques, such as the protein, starch or oil
content of
seed by biochemical assays or changes in the expression level of genes by
employing Northerns, RT PCR or microarray gene expression assays.
Trait modifications or improvements of particular interest include those to
seed, fruit, root, flower, leaf, stem, shoot, seedling or the like, including:
enhanced
tolerance to environmental conditions including freezing, chilling, heat,
drought,
water saturation, radiation and ozone; enhanced resistance to microbial,
fungal or
3o viral diseases; resistance to nematodes, decreased herbicide sensitivity,
enhanced tolerance of heavy metals (or enhanced ability to take up heavy
metals), enhanced growth under poor photoconditions (e.g., low light and/or
short
day length), or changes in expression levels of genes of interest. Other
traits that
may modified relate to the production of plant metabolites, such as variations
in
the production of taxol, tocopherol, tocotrienol, sterols, phytosterols,
vitamins, wax
monomers, anti-oxidants, amino acids, lignins, cellulose, tannins,
prenyllipids
_b_
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
(such as chlorophylls and carotenoids), glucosinolates, and terpenoids,
enhanced
or compositionally altered protein or oil production (especially in seeds), or
modifiecJ sugar (insoluble or soluble) and/or starch composition. Physical
plant
characteristics that may be modified include cell development, fruit and seed
size
and number, yields of plant parts such as stems, leaves and roots, the
stability of
the seeds during storage, characteristics of the seed pod (e.g.,
susceptibility to
shattering), root hair length and quantity, internode distances, or the
quality of
seed coat. Plant growth characteristics that may be modified include growth
rate,
germination rate of seeds, vigor of plants and seedlings, leaf and flower
io senescence, male sterility, apomixis, flowering time, flower abscission,
rate of
nitrogen uptake, biomass or transpiration characteristics, as well as plant
architecture characteristics such as apical dominance, branching patterns,
number of organs, organ identity, organ shape or size.
"Hybrid plant" refers to a plant generated by crossing two plants of
15 interest, propagating by seed or tissue and then growing the plants. When
plants
are crossed sexually, the step of pollination may include cross pollination or
self
pollination or back crossing with an untransformed plant or another
transformed
plant. Hybrid plants include first generation and later generation plants.
2o The present invention provides a method to manipulate and improve a
plant trait. The method combines the power of genomics with plant breeding
techniques. In the method, the expression levels of known genes of interest in
a
plant can be altered constitutively, or altered selectively to monitor tissue
specific
expression, inducible expression, developmental-stage specific expression or
the
25 like in a high-throughput manner. Phenotypic changes resulting from
expressing
specific plant genes at different levels, at different times, under different
types of
stress, in different plant tissues or the like is then screened. Finally,
plants with
improved traits are selected.
The method entails transforming a first plant with a member of a first pool
30 of donor vectors. Each donor vector includes a transactivator that is
placed
under the control of a different promoter so that the expression of the
transactivator can be controlled under different conditions. Further, the
method
entails transforming a second plant with a member of a second pool of receptor
vectors. The receptor construct comprises a transactivator binding site for
35 binding a transactivator. The transactivator binding site is operably
linked to a
gene of interest which permits overexpression (for example, by using sense
-6-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
constructs) or underexpression (for example, by using antisense constructs) of
the gene when transactivator expression is turned on. Of particular interest
are
genes that affect polygenic traits, such as regulatory genes.
Then specific crosses are made in a combinatorial manner between
individual members from the two pools of plants: a first pool engineered to
contain
specific regulatory sequences (such as promoters) and a second pool engineered
to contain genes of interest (such as regulatory genes). The gene of interest
is
expressed only under control of each different promoter in the progeny plant,
providing the same effect as if each plant had been transformed initially with
the
1o specific gene-promoter combination. In this manner large numbers of
specific
gene-promoter combinations can therefore be made and the effect on
transcription expression and trait improvement investigated with minimal time
and
expense.
This method is an improvement of the method described in Liu et al. US
15 Patent No. 5,968,793 and Guyer et al. (1998) Genetics 149: 633-639, that
describe methods wherein both donor and receptor vectors are transformed into
the same plant to regulate gene expression and to observe trait improvements
in
a plant. Our method provides a greater degree of flexibility and speed for
observing the effects of selective gene expression on the traits of a plant.
2o Trait improvements for any plant may be investigated by this method. The
plant may be a crop plant such as soybean, wheat, corn, potato, cotton, rice,
oilseed rape (including canola), sunflower, alfalfa, sugarcane and turf; or a
fruit or
vegetable plant, such as apple, banana, blackberry, blueberry, strawberry, and
raspberry, cantaloupe, carrot, cauliflower, coffee, cucumber, eggplant, grape,
25 honeydew, lettuce, mango, melon, onion, papaya, peas, peppers, pineapple,
spinach, squash, sweet corn, tobacco, tomato, watermelon, rosaceous fruits
(such as peach, cherry and plum) and vegetable brassicas (such as broccoli,
cabbage, cauliflower, brussel sprouts and kohlrabi). Other crops, fruits and
vegetables whose trait may be improved include barley, sorghum, currant,
3o avocado, citrus fruits such as oranges, lemons, grapefruit and tangerines,
artichoke, currant, cherries, nuts such as the walnut and peanut, pear,
endive,
leek, roots, such as arrowroot, beet, cassava, turnip, radish, yam, sweet
potato
and beans.
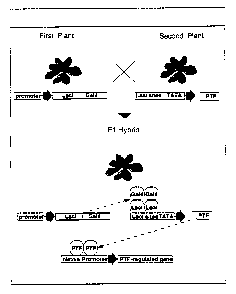
One embodiment of the present invention is illustrated in Figure 1. An
35 activator plant is grown from seed derived from plants that have been
transformed
with a construct containing one of ten different promoters linked to a
_7_
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
transactivator gene (in this Figure a fusion of the Lac I binding domain and
the
Gal4 transactivation domain). A target plant is transformed with a construct
containing the Lacl binding sites linked to one of over 1,700 transcription
factor
genes that have been identified from Arabidopsis thaliana. The target plant
does
not express the plant transcription factor unless the transactivator is
present.
Only after activator and target plants are crossed is the plant transcription
factor
over-or-underexpressed by having the transactivator bind to its binding site
on the
plant transcription factor gene construct. Thus, the traits associated with
selective
or constitutive over-or-underexpression of a plant transcription factor in
plant
1o tissue, depending on the promoter, can be easily controlled and screened.
The Donor Construct
The donor construct comprises a recombinant polypeptide sequence
which encodes a DNA binding domain fused to a transcription activation domain.
i5 This recombinant polynucleotide is the transactivator. A DNA binding domain
is a
sequence that binds to DNA with some degree of specificity. A common feature
of some activation domains is that they are designed to form amphiphilic alpha
helices with excess positive or negative charge (Giniger and Ptashne (1987)
Nature 330:670-672, Gill and Ptashne (1987) Cell 51:121-126, Estruch et al
20 (1994) Nucl. Acids Res. 22:3983-3989). Examples include the transcription
activation region of VP16 or GAL4 (Moore et al. (1998) Proc. Natl. Acad. Sci.
USA
95: 376-381; and Aoyama et al. (1995) Plant Ce117:1773-1785), peptides derived
from bacterial sequences (Ma and Ptashne (1987) Cell 51; 113-119) and
synthetic peptides (Giniger and Ptashne, supra). Exemplary transactivators are
25 those described in Brent and Ptashne, US Patent No. 4,833,080, herein
incorporated by reference or in Hasselhoff and Hodge, W097/30164.
Various promoter sequences are available which may be used to control
expression of the transactivator. Such promoters may be utilized to initiate
transcription of a nucleic acid sequence of interest operably linked to the
promoter
3o region.
For constitutive expression in plants viral promoters may be utilized in
plant expression vectors. These include the 35S RNA and 19S RNA promoters of
CaMV (Brissoh, et al., Nature, 310:511, 1984; Odell, et al., Nature, 313:810,
1985); the promoter from Figwort Mosaic Virus (FMV) (Gowda, et al., J. Cell
35 Biochem., 13D: 301, 1989) and the coat protein promoter of TMV (Takamatsu,
et
al., EMBO J. 6:307, 1987). Additional promoters include the nopaline synthase
_g_
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
promoter (An et al., (1988) Plant Physiol. 88:547); and the octopine synthase
promoter (Fromm et al., (1989) Plant Cell 1: 977).
The donor construct may include one or more inducible promoters.
Examples of inducible promoters useful in plants include those induced by
s chemical means, such as the yeast metallothionein promoter which is
activated by
copper ions (Mett, et al., (1993) Proc. Natl. Acad. Sci., U.S.A., 90:4567);
In2-1
and In2-2 regulator sequences which are activated by substituted
benzenesulfonamides, e.g., herbicide safeners (Hershey, et al., (1991 ) Plant
Mol.
Biol., 17:679); and the GRE regulatory sequences which are induced by
to glucocorticoids (Schena, et al. (1991), Proc. Natl. Acad. Sci., U.S.A.,
88:10421).
Plant promoters also include the light-inducible promoter from the small
subunit of
ribulose bis-phosphate carboxylase (ssRUBISCO) (Coruzzi, et al., (1984) EM80
J., 3:1671; Broglie, et al., (1984) Science, 224:838), promoters regulated by
heat
(Callis et al., (1988) Plant Physiol. 88:965; Ainley, et al. (1993) Plant Mol.
Biol. 22:
15 13-23; hormones, such as abscisic acid (Marcotte et al., (1989) Plant Cell
1: 969);
wounding (e.g., wunl, Siebertz et al., (1989) Plant Cell 1: 961; and chemicals
such
as methyl jasminate or salicylic acid (Gatz et al., (1997) Ann. Rev. Plant
Physiol.
Plant MoL Biol. 48: 89-108).
Tissue specific promoters may also be utilized for expression of genes in
2o plants. Tissue specific promoters useful in transgenic plants include the
cdc2a
promoter and cyc07 promoter (Ito, et al.,(1994) Plant MoL Biol., 24:863;
Martinez,
et al. (1992) Proc. Natl. Acad. Sci. USA, 89:7360; Medford, et al., (1991)
Plant
Cell, 3:359; Terada, et al. (1993) PIantJournal, 3:241; Wissenbach, et al.,
(1993)
Plant Journal, 4:411 ). Additional tissue specific promoters that are utilized
in
25 plants include the histone promoter (Atanassova, et al., (1992) Planf
Journal,
2:291 ); the cinnamyl alcohol dehydrogenase (CAD) promoter (Feuillet, et al.,
(1995) Plant Mol. Biol., 27:651 ); the mustard CHS1 promoter (Kaiser, et al.,
(1995) Plant Mol. Biol., 28:231 ); the bean grp 1.8 promoter (Keller, et al.,
(1994)
Plant Mol. Biol., 26:747); the PAL1 promoter (Ohl, et al. (1990) Plant Cell,
2:837);
30 and the chalcone synthase A promoter (Plant MoL Biol., (1990) 15:95-109).
In
addition, the timing of the expression can be controlled by using promoters
such as
those acting at senescence (Gan and Amasino (1995) Science 270: 1986-1988); or
late seed development (Odell et al. (1994) Plant Physiol. 106:447-458).
Other promoters include root-specific promoters such as root-specific
35 promoters disclosed in US Patent Nos. 5,618,988, 5,837,848 and 5,905,186 or
the prxEa promoter in Wanapu and Shinmyo (1996) Ann. N. Y. Acad. Sci.
_g_
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
782:107-113 or Miao et al. (1991 ) Plant Cell 3:11-22 or Hirel et al. (1992)
Plant
Mol. Biol. 20:207-218), seed-specific promoters such as the napin, phaseolin
or
DC3 promoter described in US Pat. No. 5,773,697, the oleate 12-hydroxylase:
desaturase promoter from Lesquerella (Brown et al (1998) Plant J. 13:201-210),
the oleosin promoter or Arabidopsis (Plant et al (1994 ) Plant Mol. Biol.
25:193-
205), a zein promoter of maize (Russet et al (1997) Transgenic Res. 6:157-
168),
the glutelin promoters of rice (Washida et al. Plant Mol Biol. (1999) 40:1-12)
and
maize (Thomas et al (1990) Plant Cell 2:1171-1180), fruit-specific promoters
such
as those active during fruit ripening (such as the dru 1 promoter (US Pat. No.
5,783,393), or the 2A11 promoter (US Pat. No. 4,943,674) and the tomato
polygalacturonase promoter (Bird et al. (1988) Planf Mol. Biol. 11:651 ),
pollen-
active promoters such as PTA29, PTA26 and PTA13 (US Pat. No. 5,792,929),
promoters active in vascular tissue (Ringli and Keller (1998) Plant Mol. Biol.
37:977-988), flower-specific (Kaiser et al, (1995) Plant Mol. Biol. 28:231-
243),
i5 pollen (Baerson et al. (1994) Plant Mol. Biol. 26:1947-1959), carpets (Ohl
et al.
(1990) Plant Cel12:837-848), pollen and ovules (Baerson et al. (1993) Plant
Mol.
Biol. 22:255-267).
Preferred inducible or tissue-specific promoters include Rd 29a
(Yamaguchi-Shinozaki and Shinozaki (1993) Plant Physiol. Mar;101:1119-20), ,
2o LTP1 (Thoma et al. (1994) Plant Physiol. 105(1 ):35-45), STM (Long et al.
(1996)
Nature. 1996 379:66-9), rbcS ( Krebbers (1988) Plant Mol Biol. 11: 745-759),
sucrose synthase (Martin et al. (1993) Plant J. 4:367-77), EIR1 (Luschnig et
al.
(1998) Genes Dev. 12:2175-87), IL (Bernhard, and Matile, GenBank Accession
Number M83534), PR1 (Lebel et al. (1998) Plant J. 16:223-33), AGL1 (Ma et al.
25 (1991 ) Genes 5:484-95), AP1 (Mandel et al. (1992) 360:273-7), E4 (Cordes
et al.
(1989) Plant Cell. 1 (10):1025-34), or GL2 (Rerie et al. (1994) Genes Dev.
8(12):1388-99).
The donor construct may also include additional sequences such as
selectable markers linked to a constitutive promoter for selecting plants
containing
3o the donor construct in field trials or tissue culture. These may include
the
acetoacetate synthase gene for chlorosulfuron resistance or the gene that
confers
resistance to cyanamide.
Plant transformation constructs may also include RNA processing signals,
for example, introns, which may be positioned upstream or downstream of the
35 open reading frame sequence (ORF). In addition, the expression constructs
may
also include additional regulatory sequences from the 3'-untranslated region
of
-10-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
plant genes, e.g., a 3' terminator region to increase mRNA stability of the
mRNA,
such as the PI-II terminator region of potato or the octopine or nopaline
synthase
3' terminator regions.
The Receptor Construct
The receptor construct comprises one or more DNA binding sites for one
of the transactivators described above, such as those disclosed in US Patent
No.
4833,080, operably linked to a gene of interest, such as a transcription
factor,
phosphatase or kinase. The transcription factors contained in the construct
may
io be derived from one or more of the transcription factor families described
below.
The plant transcription factors may belong to one of the following
transcription factor families: the AP2 (APETALA2) domain transcription factor
family (Riechmann and Meyerowitz (1998) J. Biol. Chem. 379:633-646); the MYB
transcription factor family (Martin and Paz-Ares, (1997) Trends Genet. 13:67-
73);
the MADS domain transcription factor family (Riechmann and Meyerowitz (1997)
J. Biol. Chem. 378:1079-1101 ); the WRKY protein family (Ishiguro and Nakamura
(1994) Mol. Gen. Genet. 244:563-571 ); the ankyrin-repeat protein family
(Zhang
et al. (1992) Planf Cell 4:1575-1588); the zinc finger protein (Z) family
(Klug and
Schwabe (1995) FASEB J. 9: 597-604); the homeobox (HB) protein family
(Duboule (1994) Guidebook to the Homeobox Genes, Oxford University Press);
the CART-element binding proteins (Forsburg and Guarente (1989) Genes Dev.
3:1166-1178); the squamosa promoter binding proteins (SPB) (Klein et al.
(1996)
Mol. Gen. Genet. 1996 250:7-16); the NAM protein family (Sower et al. (1996)
Cell
85:159-170); the IAA/AUX proteins (Rouse et al. (1998) Science 279:1371-1373);
the HLH/MYC protein family (Littlewood et al. (1994) Prot. Profile 1:639-709);
the
DNA-binding protein (DBP) family (Tucker et al. (1994) EMBO J. 13:2994-3002);
the bZIP family of transcription factors (Foster et al. (1994) FASEB J. 8:192-
200);
the Box P-binding protein (the BPF-1 ) family (da Costa a Silva et al. (1993)
Plant
J. 4:125-135); the high mobility group (HMG) family (Bustin and Reeves (1996)
3o Prog. Nucl. Acids Res. MoL Biol. 54:35-100); the scarecrow (SCR) family (Di
Laurenzio et al. (1996) Cell 86:423-4.33); the GF14 family (Wu et al. (1997)
Plant
Physiol. 114:1421-1431 ); the polycomb (PCOMB) family (Kennison (1995) Annu.
Rev. Genet. 29:289-303); the teosinte branched (TEO) family (Luo et al. (1996)
Nature 383:794-799; the AB13 family (Giraudat et al. (1992) Plant Cell 4:1251-
1261 ); the triple helix (TH) family (Dehesh et al. (1990) Science 250:1397-
1399);
the EIL family (Chao et al. (1997) Cell 89:1133-44); the AT-HOOK family
(Reeves
-11-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
and Nissen (1990) Journal of Biological Chemistry 265:8573-8582); the S1 FA
family (Zhou et al. (1995) Nucleic Acids Res. 23:1165-1169); the bZIPT2 family
(Lu and Ferl (1995) Plant Physiol. 109:723); the YABBY family (Bowman et al.
(1999) Development 126:2387-96); the PAZ family (Bohmert et al. (1998) EM80
J. 17:170-80); a family of miscellaneous (MISC) transcription factors
including the
DPBF family (Kim et al. (1997) Plant J. 11:1237-1251 ) and the SPF1 family
(Ishiguro and Nakamura (1994) Mol. Gen. Genet. 244:563-571 ); the golden (GLD)
family (Hall et al. (1998) Plant Cell 10:925-936).
Other transcription factors may be identified by screening polynucleotide
or polypeptide sequence databases, such as GenBank, using using sequence
alignment methods and homology calculations, such as those described in
Altschul et al. (1994) Nature Genetics 6: 119-129. For example, the NCBI Basic
Local Alignment Search Tool (BLAST) (Altschul et al. (1990) Basic local
alignment
search tool. J. Mol. Biol. 215:403-410) is available from several sources,
including
the National Center for Biotechnology Information (NCBI, Bethesda, MD, for use
in connection with the sequence analysis programs blastp, blastn, blastx,
tblastp,
tblastn and tblastx. Alternatively, a program that identifies particular
sequence
motifs may be employed along with specific characteristic consensus sequences,
such as FIND PATTERN (GCG, Madison, WI).
2o Exemplary transcription factors which can be employed in the invention
include those disclosed in Zhang et al. US Serial Application No. 09/394,519,
filed September 13, 1999, entitled "Plant Gene Sequences I", Keddie et al. US
Serial Application No. , filed February 17, 2000, entitled "Plant Gene
Sequences II", Keddie et al. US Serial Application No. , filed March
22, 2000, entitled "Polynucleotides for Seed Trait Alteration", Cai-Zhong et
al.
US Serial Application No. , filed March 22, 2000, entitled
"Polynucleotides for Root Trait Alteration", Heard et al. US Serial
Application No.
filed March 22, 2000, entitled "Disease-Induced Polynucleotides",
Samaha et al. US Serial Application No. , filed March 22, 2000,
3o entitled "Stress-Induced Polynucleotides", and Riechmann et al. US Serial
Application No. , filed March 22, 2000, entitled "Polynucleotides for
Flower Trait Alteration".
The transcription factors encompass the naturally occuring sequences as
well as non-naturally occurring sequences which are derivatives of the
transcription factors described above. For example, the amino acid sequence
encoding the binding protein may be a naturally occurring sequence such as the
-12-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00i09448
ones shown above or a non-naturally occuring sequence using domains of
transcription factors described above fused in frame, but not necessarily
adjacent,
with functional domains derived from other sequences or sources. Additionally,
the invention includes polypeptides derived from shuffling regions of
transcription
factors described above by methods described in Minshull and Stemmer, US
Patent No. 5,837,458, entitled "Methods and Compositions for Cellular and
Metabolic Engineering" and Stemmer and Crameri, US Patent No. 5,811,238,
entitled "Methods for Generating Polynucleotides having Desired
Characteristics
by Iterative Selection and Recombination".
1o The particular arrangement of the transcription factor sequence in the
transformation vector will be selected according to the type of expression of
the
sequence that is desired. Where enhanced transcription factor activity is
desired
in the plant, a transcription factor coding sequence may be operably linked to
a
constitutive high-level promoter such as the CaMV 35S promoter. Generally,
this
15 will require the full length sequence encoding the transcription factor. In
contrast,
a reduction of transcription factor activity in the transgenic plant may be
obtained
by introducing into plants antisense constructs based on the transcription
factor
cDNA. For antisense suppression, the transcription factor cDNA is arranged in
reverse orientation relative to the promoter sequence in the transformation
vector.
2o The introduced sequence need not be the full length transcription factor
cDNA or
gene, and need not be exactly homologous to the transcription factor cDNA or
gene found in the plant type to be transformed. Generally, however, where the
introduced sequence is of shorter length, a higher degree of homology to the
native transcription factor sequence will be needed for effective antisense
25 suppression.
Constructs in which RNA encoded by the transcription factor cDNA (or variants
thereof) is overexpressed may also be used to obtain co-suppression of the
endogenous
transcription factor gene in the manner described in U.S. Patent No. 5,231,020
to
Jorgensen. Such co-suppression (also termed sense suppression) does not
require that
30 the entire transcription factor cDNA be introduced into the plant cells,
nor does it require
that the introduced sequence be exactly identical to the endogenous
transcription factor
gene. However, as with antisense suppression, the suppressive efficiency will
be
enhanced as (1 ) the introduced sequence is lengthened and (2) the sequence
similarity
between the introduced sequence and the endogenous transcription factor gene
is
~5 increased. Alternatively, supression on a transcription gene activity may
be obtained by
double stranded RNA-mediated interference (Voinnet, et al.. (1998) Cell 95,
177-187,
Waterhouse, et al. (1998) Proc. Natl. Acad. Sci. USA 95, 13959-13964).
-13-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
The receptor vector may also include additional sequences such as
selectable markers linked to a constitutive promoter for selecting plants
containing
the activatorconstruct in field trials or tissue culture. These may include
the
acetoacetate synthase gene for chlorosuifuron resistance or the gene that
confers
resistance to cyanamide.
Any of the identified sequences may be incorporated into a cassette or
vector for expression in plants. A number of expression vectors suitable for
stable
1o transformation of plant cells or for the establishment of transgenic plants
have
been described including those described in Weissbach and Weissbach, (1989)
Methods for Plant Molecular Biology, Academic Press, and Gelvin et al., (1990)
Plant Molecular Biology Manual, Kluwer Academic Publishers. Specific examples
include those derived from a Ti plasmid of Agrobacterium tumefaciens, as well
as
Is those disclosed by Herrera-Estrella, L., et al., (1983) Nature 303: 209,
Bevan, M.,
Nucl. Acids Res. (1984) 12: 8711-8721, Klee, H. J., (1985) BiolTechnology3:
637-642, for dicotyledonous plants.
Alternatively, non-Ti vectors can be used to transfer the DNA into plants
and cells by using free DNA delivery techniques. Such methods may involve, for
2o example, the use of liposomes, electroporation, microprojectile
bombardment,
silicon carbide wiskers, and viruses. By using these methods transgenic plants
such as wheat, rice (Christou, P., (1991 ) Biotechnology 9: 957-962) and corn
(cordon-Kamm, W., (1990) Plant Cell2: 603-618) can be produced. An
immature embryo can also be a good target tissue for direct DNA delivery
25 techniques by using the particle gun (Weeks, T. et al., (1993) Plant
Physiol. 102:
1077-1084; Vasil, V., (1993) BiolTechnology 10: 667-674; Wan, Y. and Lemeaux,
P., (1994) Plant Physiol. 104: 37-48, and for Agrobacterium-mediated DNA
transfer (Ishida et al., (1996) Nature Biotech. 14: 745-750).
Typically, plant transformation vectors include one or more cloned plant
3o coding sequences (genomic or cDNA) under the transcriptional control of 5'
and
3' regulatory sequences and a dominant selectable marker. Such plant
transformation vectors typically also contain a promoter (e.g., a regulatory
region
controlling inducible or constitutive, environmentally-or developmentally-
regulated,
or cell- or tissue-specific expression), a transcription initiation start
site, an RNA
35 processing signal (such as intron splice sites), a transcription
termination site,
and/or a polyadenylation signal.
-14-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
Transformation
Standard techniques may be used to transform plants with the above
described vectors to overexpress or underexpress the genes of interest in
plants
in order to understand a gene's effect on a plant's phenotype. Additionally,
combinations of transactivator or donor rvectors may be used to transform
plants
to understand the contribution of multiple genes of interest to a trait.
Exemplary plants to be transformed may be any higher plant, including
monocotyledonous and dicotyledenous plants. Suitable protocols are available
for
io Leguminosae (alfalfa, soybean, clover, etc.), Umbelliferae (Carrot, celery,
parsnip), Cruciferae (cabbage, radish, rapeseed, broccoli, etc.),
Curcurbitaceae
(melons and cucumber), Gramineae (wheat, corn, rice, barley, millet, etc.),
Solanaceae (potato, tomato, tobacco, peppers, etc.), and various other crops
See protocols described in Ammirato et al. (1984) Handbook of Plant CeII
Culture
-Crop Species. Macmillan Publ. Co. Shimamoto et al. (1989) Nature 338:274-
276; Fromm et al. (1990) BiolTechnology 8:833-839; and Vasil et al. (1990)
BiolTechnology 8:429-434.
Transformation and regeneration of both monocotyledonous and
dicotyledonous plant cells is now routine, and the selection of the most
2o appropriate transformation technique will be determined by the
practitioner. The
choice of method will vary with the type of plant to be transformed; those
skilled in
the art will recognize the suitability of particular methods for given plant
types.
Suitable methods may include, but are not limited to: electroporation of plant
protoplasts; liposome-mediated transformation; polyethylene glycol (PEG)
mediated transformation; transformation using viruses; micro-injection of
plant
cells; micro-projectile bombardment of plant cells; vacuum infiltration; and
Agrobacterium tumeficiens (AT) mediated transformation.
Successful examples of the modification of plant characteristics by
transformation with cloned cDNA sequences which serve to illustrate the
current
3o knowledge in this field of technology, and which are herein incorporated by
reference, include: U.S. Patent Nos. 5,571,706; 5,677,175; 5,510,471;
5,750,386;
5,597,945; 5,589,615; 5,750,871; 5,268,526; 5,780,708; 5,538,880; 5,773,269;
5,736,369 and 5,610,042. Successful transformation of woody species, such as
poplar and aspen transformation using Agrobacterium tumefaciens, can be
performed as described by De Block, (1990) Plant Physiol. 93:1110-1116. Other
woody species that may be transformed include pine. Of particular interest is
the
-15-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
transformation of tomatoes, for example as illustrated in Filatti et al. US
Patent
No. 5,565,347.
Following transformation and regeneration of plants with the
transformation vector, transformed plants may be selected using a dominant
selectable marker incorporated into the transformation vector. Typically, such
a
marker will confer antibiotic or herbicide resistance on the seedlings of
transformed plants, and selection of transformants can be accomplished by
exposing the seedlings to appropriate concentrations of the antibiotic or
herbicide.
1o Crossing
Plants transformed with any of the donor vector may be crossed with
plants transformed with any of the receptor vectors as described in Figure 1.
Plants transformed with a receptor vector for a specific transcription factor
(in a
sense or antisense configuration) can be crossed with a variety of plants
transformed with donor vectors comprising a variety of different promoters,
including constitutive, inducible or tissue-specific vectors to investigate
the effects
of trancription factor expression throughout the plant, under specific
conditions
such as environmental stresses, disease, or the like, in specific tissues,
such as
seeds, roots, flowers, stems, leaves, fruits or the like. Seeds are collected
and
2o hybrid plants are grown to maturity. These plants can then be screened to
identify
plants with valuable traits.
These plants may be grown in the presence of first and second herbicide
resistant selectable markers. Seed obtained from herbicide resistant
regenerated
plants may be crossed further to generate later generation hybrid plants.
Additional details in crop breeding techniques, in particular those for
tomatoes' are
described in US Patent No. 5,817,913, herein incorporated by reference.
Addtionally, the present invention is a plant breeding kit. In this manner,
seed or plants are provided, wherein a first pool of seed or plants is
provided that
are transformed with the donor vector and a second pool of seed or plants is
3o provided that are transformed with the receptor vector. Then plant breeders
can
cross plants from first and second pools as described above to breed plants
with
improved traits.
The following examples are provided to better elucidate the practice of the
present invention and should not be interpreted in any way to limit the scope
of
the present invention. Those skilled in the art will recognize that various
-16-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
modifications can be made to the methods and genes described herein without
departing from the spirit and scope of the present invention.
EXAMPLES
The invention is illustrated by Examples wherein six different donor vectors
having a constitutive, an inducible promoter or a tissue specific promoter
operably
linked to either of two different transactivators are prepared. Additionally
the
invention is illustrated by the preparation of eight different receptor
vectors
comprising a transactivator binding site operably linked to a transcription
factor.
1o Donor and receptor vectors are then transformed into different tomato
plants.
Transformed tomato plants having different donor vectors are then crossed with
transformed tomato plants having different receptor vector. Improved traits
can
be screened for in the progeny.
Example 1. Cloning of Promoters in Donor Vector
The plasmid vector pMEN020 was the vector used for donor and receptor
vector construction. The pMEN020 plasmid construct is a binary cloning vector
that contains both E. toll and Agrobacterium fumefaciens origins of DNA
replication but no vir genes encoding proteins essential for the transfer and
2o integration of the target gene inserted in the T-DNA region. PMEN020
requires
the trfA gene product to replicate in Agrobacterium. The strain of
Agrobacterium
containing this trfA gene is called the ABI strain and is described in U.S.
Patent
Nos. 5,773,701 and 5,773,696. This cloning vector serves as an E. coli-
Agrobacterium fumefaciens shuttle vector. All of the cloning steps are carried
out
in E. toll before the vector is introduced into ABI strain of Agrobacterium
tumefaciens.
Two different sets of constructs, which are based on bacterial DNA binding
proteins LexA and Lacl, respectively, were prepared. For the LexA system, the
construct was prepared in two steps. First, an intermediate construct was
generated
3o containing the LexA protein and the gal4 activation domain. In the
intermediate
construct, four fragments were generated separately and fused by overlap
extension
PCR. The first fragment was the 35S minimal promoter and omega translation
enhancer;
The fragment was amplified from construct SLJ4D4 (Jones et al 1992 Transgenic
Research 1:285) using primers 011731 (SEQ ID No. 1 ):
GCCCAAGCTTTGAGCTCCGCGGCCGCAAGACCCTTCCTCTATATAAGGAAGTTCA
and 011733 (SEQ ID No. 2): ACGCTTCCATGGTAATTGTAAATGTAATTGTAATGTTGT .
-17-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
The second fragment was the IexA gene of E coli; it was amplified from ptasmid
pLexA (Clontech, Palo Alto, CA) using primers 011732 (SEQ ID No. 3):
TTACAATTACCATGGAAGCGTTAACGGCCAGGCAACAAGA
and 0117717 (SEQ ID No. 4):
TATTCCCACTTTGATTAAAATTGGGGAATTCCAGCCAGTCGCCGT.
The third fragment was the gat4 transcription activation domain; it was
amplified
from pGAD424 (aa 768-881, Clontech) by primers 011715 (SEQ ID No. 5):
GGCTGGAATTCCCCAATTTTAATCAAAGTGGGAA and 011718 (SEQ ID No. 6):
AAGCTCTAGCTACTCTTTTTTTGGGTTTGGTGGGGT. The fourth fragment was the
to E9 transcription terminator (Fluhr et al 1986 EMBO J. 5:2063); it was
amplified from
pMON10098 using primers 011716 (SEQ ID No. 7):
AAGAGTAGCTAGAGCTTTCGTTCGTATCA and 011719 (SEQ ID No. 8):
TGCTCTAGATTGATGCATGTTGTCAATCA. The four fragments were fused together
by PCR using primers 011731 and 011719. Note that restriction enzyme digest
sites
15 Hindlll and Xbal were added to the ends of 011731 and 011719, respectively.
Inserts from the intermediate construct were cloned into the Hindlll and
BamHl sites of pMEN020.
The lacl system was constructed in a similar fashion as the IexA system,
in two steps. The translation initiation of the lacl gene was changed to ATG
from
2o GTG and a mutation at position 17 (Y to H, Lehming et al 1987 EMBO J
6:3145)
was introduced. The lacl gene was cloned from E coli genomic DNA by PCR
amplification using primers 016400 (SEQ ID No. 9):
CATGCCATGGAACCAGTAACGTTATACGATGTCGCAGAGTATGCCGGTGTC
TCTCATCAGACCGTTTCCCGCG) and 016401 (SEO ID No. 10):
25 GGGGAATTCAAGGGTGGTTTTTCTTTTCACCAGTGA. Note that Ncot and
EcoRl sites were introduced with 016400 and 016401, respectively. The lacl
coding region, which is defined by the Ncol and EcoRl sites, was used to
replace
IexA coding region in the previous intermediate construct. Insets from the two
intermediate constructs were cloned into the Hindtlt and BamHl sites of
pMEN020
3o in a three way ligation.
A multiple cloning site was added upstream of the Lacl (LexA)/gal4 fusion
protein by 011731 to facilitate the cloning of promoter fragments. In order to
test
the functionality of the system, full 35S promoters may be cloned upstream of
the
Lacl (LexA)/gal4 fusion protein by using restriction enzymes Hind III and
Notl.
35 Inducible promoters from plant genomic DNAs can be isolated by PCR
-18-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
amplification using primers flanking the promoter region and containing
suitable
restriction sites for cloning into the activation vector. For example, the
rd29a
gene was characterized by Shinozaki's group in Japan (Yamaguchi-Shinozaki and
Shinozaki (1993) Plant Physiol. 101: 1119-1120). The rd29a gene expression is
induced by desiccation, salt stress and exogenous ABA treatment (Yamaguchi-
Shinozaki and Shinozaki (1993) Plant Physiol. 101: 1119-1120 (1993); Ishitani
et
al. (1998) Plant CeII 10: 1151-1161 ).
A genomic clone carrying the rd29a promoter was identified by using
rd29a as a search word at the www site of NCBI. The sequence for the rd29a
promoter is located in the region between nucleotide positions 3892 to 5390
(Accession No. D13044). The following two primers were designed to amplify
this
promoter region from Arabidopsis genomic DNA: rd29a-primer1 (SEQ ID No. 11 ):
GCCCAAGC7TGGTTGCTATGGTAGGGACTAT; and rd29a-primer2 (SEQ ID No.
12): ATAAGAATGCGGCCGCGAGAGATAAAGGGACACGTATGAAGC. The
rd29a-primer1 includes a Hind III (AAGCTT) restriction site near the 5'-end of
the
primer and rd29a-primer2 has a Notl (GCGGCCGC) restriction site near 5'-end of
the primer.
Total genomic DNA was isolated from Arabidopsis fhaliana (ecotype
Colombia) by using the CTAB method (Ausubel et al. (1992) Current Protocols in
Molecular Biology (Greene 8~ Wiley, New York)). Ten nanograms of the genomic
DNA was used as a template in a PCR reaction under conditions suggested by
the manufacturer (Boehringer Mannheim). The reaction conditions that were
used in this PCR experiment are as follows: Segment 1: 94°C, 2 minutes;
Segment 2: 94°C, 30 seconds; 60°C, 1 minute; 72°C, 3
minutes, for a total of 35
cycles; Segment 3: 72°C for 10 minutes. A PCR product of 1525 base pair
is
expected. The PCR products were subject to electrophoresis in a 0.8% agarose
gel and visualized by ethidium bromide staining. The DNA fragments containing
the inducible promoter were excised and purified using a Qiaquick gel
extraction
kit (Qiagen, CA). The purified PCR product can then be digested with Hindlll
and
3o Notl, and cloned into the Hindlll and Notl sites of LexA and Lacl based
donor
vectors.
Similarly, tissue specific promoters from plant genomic DNAs can also be
isolated by PCR amplification. For example, the non-specific lipid transfer
protein
(or LTP1 ) promoter is specific for the epidermis layer of plants (Thoma et
al, Planf
Physiol. 105(1 ) 35-45 (1994)). The sequence for the LTP1 promoter is located
in
the region between nucleotide positions 1 to 1130 (Accession No. M80567). The
-19-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
following two primers were designed to amplify this promoter region from
Arabidopsis genomic DNA: LTP1-primer1 (SEQ ID No. 13):
GCCCAAGCTTGATTAACTTGCATTACAGTTGGGAAGT; and LTP1-primer2
(SEQ ID No. 14):
ATAAGAATGCGGCCGCGGTACGTATATGTTATGTGGTGTGAATG. The
LTP1-primer1 includes a Hind III (AAGCTT) restriction site near the 5'-end of
the
primer, and LTP1-primer2 has a Notl (GCGGCCGC) restriction site near 5'-end of
the primer. The promoter fragment can then be cloned into the activation
vector
as described above
Example 2 Construction of Receptor Vector
The receptor vector includes a corresponding binding region for the
transactivator factor prepared above, in this case a LexA or Lacl binding
site, and
a gene of interest, such as a transcription factor.
Two versions of the cloning vector will be built, one each for Lexa and
Lacl, respectively. For the Lexa version, eight copies of the Lexa binding
site and
the 35S minimal promoter will be cloned upstream of the E9 terminator. To this
end, two fragments will be generated and then fused together by overlap PCR.
The first fragment will be generated by primers 016417 (SEQ ID No. 15):
2o GGCCCAAGCTTACATATCCATATCTAATCTTACCT
and 011723, using the IexA_OP construct (described in the previous section) as
template. The second will be generated by primers 011721 and 016416 (SEQ ID
No. 16): CTAGAGGATCCGGTACGAGGCCTGTCTA, using pMEN20 as
template. The two PCR products can be assembled together by PCR using
primers 016417 and 016416. Note that Hindlll recognition site is designed in
the
5' of 016417, and the pMEN20 multiple cloning site will be included in the
final
product. This product will be ligated into the Hindlll and BamHl sites of a
modified
pMEN20, which should have a different selectable marker (e.g. glyphosate
resistance marker). The lacl version will be identical to the above, except
that two
3o copies of the lacl binding site will replace the lexa binding site. The
lacl binding
sites will be included in 016415 (SEQ ID No. 17):
GGCCCAAGCTTAATTGTGAGCGCTCACAATTCATGAATTGTGAGCGCTCACA
ATT PCR product by primers 016415 and 016416, using pMEN20 as template,
will include two copies of the lacl binding sites, the 35S minimal promoter,
and the
multiple cloning site of pMEN20.
-20-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
The CBF1 coding region can be amplified by PCR using primers 011700
(SEQ ID No. 18): ACGCGTCGACGACTGAGAACTCTAGTAACTACGTA and
011702 (SEQ ID No. 19):
ATAAGAATGCGGCCGCCGACTATCGAATATTAGTAACTCCA.
The resulting PCR product, approximately 780 bp, can be digested with
Sall and Notl and cloned into the Sall and Notl sites of the receptor vector.
Additionally, a seed specific transcription factor ATML1 (Lu et al. (1996)
during embryonic pattern formation and defines a new class of homeobox genes
Plant Cell 8(12):2155-68, GenBank Accession No. 037589) can be amplified by
1o PCR using primers 05184 (SEQ ID No. 20):
CGGGGTACCCTTCTCCACAAGTAAGGGAACCAGA and 05185 (SEQ ID No.
21):ATAAGAATGCGGCCGCCCTCCCCTTTCACTCTTACCTTCCGAA.
The resulting PCR product, approximately 2,400 bp, can be digested with Kpnl
and Notl and cloned into the Kpnl and Notl sites of the receptor vector.
Furthermore, a root specific transcription factor PRL2 (Newman, et al. (1994)
Plant Physiol. 106, 1241-1255) was obtained from a full length expressed
sequence tag (GenBank Accession No. R86816). The cDNA can be isolated
from the library vector by using the Sall and Notl enzymes, and then it can be
cloned into the Sall and Notl sites of the receptor sites. In a fourth
example, a
2o ATHB-12 (Lee et al. (1998) Plant Mol BioL 37:377-84) was subcloned into the
receptor vector by using Sall and Notl as previously described.
Example 3: Transformation of Tomatoes
Tomato transformations are performed using the following procedure.
Tomato seeds are sterilized in 50 % bleach solution for 20-30 minutes and
rinsed
at least 3 times with sterile water. The seed are placed in magenta jars that
contain the following media: 1X MS Salts, 1X Gamborg's Vitamins, 2% sucrose,
and 0.8% phytagar. The seeds are germinated for 7-10 days at 25°C with
16
hours of light. Agrobacterium from glycerol stock is inoculated into 5 ml of
LB or
3o YEP with the appropriate antibiotics, and the culture grown at 28°C
overnight with
shaking at 250 rpm. When the optimal OD600 is reached (between 1.0 and 1.5),
the bacteria are spun down and resuspended in liquid germination media to a
concentration of 0.2 OD600. The cotyledons from 7-10 days old seedlings are
cut into 0.5cm pieces, and transferred to a co-cultivation plate with (D1-)
media
containing the following: 1X MS salts, 1X Gamborg's Vitamins, 3% glucose. 1
mg/L zeatin, 0.8% (w/v) phytagar, pH to 5.8, and 375 uM Acetosyringone. 10-15
-21-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
mi of diluted Agro solution are poured over the explants and incubated for 40
min.
The expiants are turned upside down on (D1-) cocultivation media for 48 hours
at
25°C with 16 hours of light. About 20-25 explants are transferred onto
D1 plates
that contain the following media: 1X MS salts, 1X Gamborg's Vitamins, 3%
glucose, 1 mglL zeatin, 0.8% (w/v) phytagar, pH 5.8, 300 u/ml Timentin, and
selected for the construct. After 10-15 days, whole explants are transferred
to D2
plates containing the following: 1X MS salts, 1X Gamborg's Vitamins, 3%
glucose, 0.1 mg/L zeatin, 0.8% (w/v) phytagar, pH 5.8, 300 u/ml Timentin, and
appropriate selection for the construct. And from then on, every 2-3 weeks the
1o explants are transferred to fresh D2 plates. Shoots form in a few weeks.
When
the shoots are 1-1.5 cm tall, the shoots are cut off from the calli and
transferred to
a medium containing the following: 1X MS salts, 1X Gamborg's Vitamins, 3%
glucose, 0.8% (w/v) phytagar, pH 5.8, 300 u/ml Timentin, and appropriate
selection for the construct. Rooted shoots are transferred to soil in 2-inch
pots
inside Magenta jars. Once the plants have established in the soil, they can be
transplanted to bigger pots and tested for the presence of the transgene.
Tomato Breeding for Trait Improvement
Using the above protocol, a first pool of tomatoes can be transformed with
2o vectors comprising a constitutive promoter (35S promoter) or an inducible
promoter (rd29a or Itp1 ) in combination with either IexA/Gal4 or lacl/Gal4
transactivators. A second pool of tomatoes can be transformed with vectors
comprising IexA or lacl binding sites in combination with either CBF, PRL2,
ATML1 or ATHB-1.
First and second pools of tomato plants are components of the plant breeding
kit.
The following definitions are employed: T0: the initial transgenic plant
produced from tissue culture, S1-S4 or F1-F4: The S numbers are for self
pollinations, while the F number is a cross between two lines. For example, a
TO
3o plant can be both self pollinated (S1 seeds produced) and outcrossed to an
donor
line initially (F1 seeds produced). F1 S2 would be the selfed progenies of a
F1
parent.
The transformed TO plants will be grown to maturity in greenhouses (the
plants can be scored with cyanamide herbicides if escapes are a problem). Some
of the flowers will be crossed to TO plants containing the donor vector. (The
F1
progeny of this cross will be segregating for both genes). Several flowers
will be
-22-
CA 02367408 2001-10-04
WO 00/60089 PCT/US00/09448
crossed to obtain at least 100 seeds. The self pollinated seeds (S1) will be
saved for later crosses to different activator promoters.
The F1 progeny will be screened by spraying chlorsulfuron and cyanamide
to find the progeny containing both the donor (chlorsulfuron resistant) and
receptor (cyanamide resistant) vectors (heterozygous for both). Both
chlorsulfuron resistance and cyanamide resistance are dominant in their
effects,
so phenotypes resistant to both herbicides and thus containing both genes
should
be observable in the F1 progeny. The presence of two distinguishable
herbicidal
markers greatly facilitates determining the genotype.
1o The F1 progeny may be screened by RT-PCR . F1 S2 plants are
segregating for both the activator and receptor genes. F1 S2 plants are grown
and
sprayed with chlorsulfuron and cyanamide to find plants containing both
transgenes (3/4 x'/4 = 9/16) and these plants are allowed to self-pollinate.
1/9'" of
these (1/16'" of original planting prior to spraying) F1 S2S3 seeds from these
F1 S2
plants will be homozygous for both transgenes and produce homozygous progeny
(100% resistant to both chlorsulfuron and cyanamide). Single plant analysis of
these F1 S2S3 plants can be performed. These are homozygous for both the
donor and receptor vectors and can be more extensively studied in this
generation. Alternatively, the seeds from the F1 S2S2 plants can be bulked up
for
2o detailed studies.
Plants are screened for desired plant phenotypes for each gene. Initially
studies may be preformed using two component systems where transcription
factor expression is activated with the CaMV 35S activator gene. Transcription
factors which appear to play a role in interesting phenotypes can then be
further
investigated by crossing specific receptor vectors with tissue-specific or
inducible
activator lines, such as fruit specific activator lines, or environmental or
disease-
inducible lines or developmental-stage specific lines.
Particular traits of interest in tomato include increased levels of
carotenoids (particularly lycopene), increased levels of soluble solids, and
3o enhanced disease resistance.
All references (publications and patents) are incorporated herein by
reference in their entirety for all purposes.
Although the invention has been described with reference to the
embodiments and examples above, it should be understood that various
modifications can be made without departing from the spirit of the invention.
Accordingly, the invention is limited only by the following claims.
-23-
CA 02367408 2001-10-04
WO 00/60089 1 PCT/US00/09448
SEQUENCE LISTING
< 110>Fromm, Michael
Zhang, James
<120> Genetic Trait Breeding Method
<130> MBI-0013
<140>
<141>
<150> 60/128,153
<151> 1999-04-07
<160> 21
<170> PatentIn Ver. 2.1
<210> 1
<211> 54
<212> DNA
<213> Artificial Sequence
<220>
<223> 011731
<400> 1
gcccaagctt gagctccgcg gccgcaagac ccttcctcta tataaggaag ttca 54
<210> 2
<211> 39
<212> DNA
<213> Artificial Sequence
<220>
<223> 011733
<400> 2
acgcttccat ggtaattgta aatgtaattg taatgttgt 39
<210> 3
<211> 40
<212> DNA
<213> Artificial Sequence
CA 02367408 2001-10-04
WO 00/60089 2 PCT/US00/09448
<220>
<223> 011732
<400> 3
ttacaattac catggaagcg ttaacggcca ggcaacaaga 40
<210> 4
<211> 45
<212> DNA
<213> Artificial Sequence
<220>
<223> 011717
<400> 4
tattcccact ttgattaaaa ttggggaatt ccagccagtc gccgt 45
<210> 5
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> 011715
<400> 5
ggctggaatt ccccaatttt aatcaaagtg ggaa 34
<210> 6
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> 011718
<400> 6
aagctctagc tactcttttt ttgggtttgg tggggt 36
<210> 7
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
CA 02367408 2001-10-04
WO 00/60089 3 PCT/US00/09448
<223> 011716
<400> 7
aagagtagct agagctttcg ttcgtatca 29
<210> 8
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> 011719
<400> 8
tgctctagat tgatgcatgt tgtcaatca 29
<210> 9
<211> 73
<212> DNA
<213> Artificial Sequence
<220>
<223> 016400
<400> 9
catgccatgg aaccagtaac gttatacgat gtcgcagagt atgccggtgt ctctcatcag 60
accgtttccc gcg 73
<210> 10
<211> 36
<212> DNA
<213> Artificial Sequence
<220>
<223> 016401
<400> 10
ggggaattca agggtggttt ttcttttcac cagtga 36
<210> 11
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
CA 02367408 2001-10-04
WO 00/60089 4 PCT/US00/09448
<223> rd29a-primerl
<400> 11
gcccaagctt ggttgctatg gtagggacta t 31
<210> 12
<211> 42
<212> DNA
<213> Artificial Sequence
<220>
<223> rd29a-primer2
<400> 12
ataagaatgc ggccgcgaga gataaaggga cacgtatgaa gc 42
<210> 13
<211> 37
<212> DNA
<213> Artificial Sequence
<220>
<223> LTP1-primerl
<400> 13
gcccaagctt gattaacttg cattacagtt gggaagt 37
<210> 14
<211> 44
<212> DNA
<213> Artificial Sequence
<220>
<223> LTP1-primer2
<400> 14
ataagaatgc ggccgcggta cgtatatgtt atgtggtgtg aatg 44
<210> 15
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> 016417
CA 02367408 2001-10-04
WO 00/60089 5 PCT/US00/09448
<400> 15
ggcccaagct tacatatcca tatctaatct tacct 35
<210> 16
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> 016416
<400> 16
ctagaggatc cggtacgagg cctgtcta 28
<210> 17
<211> 55
<212> DNA
<213> Artificial Sequence
<220>
w <223> 016415
<400> 17
ggcccaagct taattgtgag cgctcacaat tcatgaattg tgagcgctca caatt 55
<210> 18
<211> 35
<212> DNA
<213> Artificial Sequence
<220>
<223> 011700
<400> 18
acgcgtcgac gactgagaac tctagtaact acgta 35
<210> 19
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> 011702
<400> 19
CA 02367408 2001-10-04
WO 00/60089 6 PCT/US00/09448
ataagaatgc ggccgccgac tatcgaatat tagtaactcc a 41
<210> 20
<211> 34
<212> DNA
<213> Artificial Sequence
<220>
<223> 05184
<400> 20
cggggtaccc ttctccacaa gtaagggaac caga 34
<210> 21
<211> 43
<212> DNA
<213> Artificial Sequence
<220>
<223> 05185
<400> 21
ataagaatgc ggccgccctc ccctttcact cttaccttcc gaa 43