Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02378657 2002-O1-08
1
IDENTIFICATION AND OVEREXPRESSION OF A DNA SEQUENCE
CODING FOR 2-METHYL-6-PHYTYLHYDROQUINONE
METHYLTRANSFERASE IN PLANTS
The invention relates to a DNA encoding a polypeptide with
2-methyl-6-phytylhydroquinone methyltransferase activity. Also,
the invention relates to the use of DNA sequences encoding a
polypeptide with 2-methyl-6-phytylhydroquinone methyltransferase
activity for the generation of plants with an elevated tocopherol
and tocotrienol content, specifically to the use of the DNA
sequence SEQ ID No. 1 or SEQ ID No. 7 or DNA sequences
hybridizing herewith or DNA sequences which are homologous to the
full sequence or to subsequences, to a method for the generation
of plants with an elevated tocopherol and tocotrienol content,
and to the resulting plant itself.
The generation of plants with an elevated sugar, enzyme and amino
acid content has hitherto been an important objective in plant
molecular genetics. The development of plants with an elevated
vitamin content, such as, for example, an elevated tocopherol and
tocotrienol content, is, however, also of economic interest.
The naturally occurring eight compounds with vitamin E activity
are derivatives of 6-chromanol (Ullmann's Encyclopedia of
Industrial Chemistry, Vol. A 27 (1996), VCH Verlagsgesellschaft,
Chapter 4., 478-488, Vitamin E). The first group (la-d) is
derived from tocopherol, while the second group is composed of
tocotrienol derivatives (2a-d):
R1
HO
,/~5 I 3
R2 ~ 0 ~ ~ _. 4, ~ 8,
R3
1a, oc-tocopherol: R1 = R2 = R3 = CH3
1b, (3-tocopherol [148-03-8] : Rl = R3 = CH3, Rz = H
1c, y-tocopherol [54-28-4] : Rl = H, RZ = R3 = CH3
1d, 8-tocopherol [119-13-1]: Rl = RZ = H, R3 = CH3
R1
HO
R2 T 0 I v 3~ 7, 11,
R3
~817/~~~~1. CA 02378657 2002-O1-08
2
2a, a.-tocotrienol (1721-51-3): R1 = R2 = R3 = CH3
2b, (~-tocotrienol [490-23-3] : R1 = R3 = CH3, R2 = H
2c, .~y-tocotrienol [14101-61-2) : Rl = H, R2 = R3 = CH3
2d, 8-tocotrienol [25612-59-3 ) : Rl = R2 = H, R3 = CH3
oc-Tocopherol has great economic importance.
The development of crop plants with an elevated tocopherol and
tocotrienol content by means of conventional breeding is set a
limit.
The genetic engineering approach of isolating essential
biosynthesis genes which encode, for example, tocopherol
synthesis performance and introducing them into crop plants in a
directed fashion is a meaningful alternative. Knowledge of the
biosynthesis and its regulation, and identification of genes
which affect biosynthesis performance, are prerequisites for this
method.
Isoprenoids or terpenoids are composed of a variety of classes of
lipid-soluble molecules, and they are formed partially or
exclusively from C5-isoprene units. Pure prenyl lipids (for
example carotenoids) are composed of C skeletons based
exclusively on isoprene units, while mixed prenyl lipids (for
example chlorophylls, tocopherols and vitamin K) have an
isoprenoid side chain linked to an aromatic nucleus.
The biosynthesis of prenyl lipids starts with 3 x acetyl-CoA
units which are converted into the starting isoprene unit (C5),
namely isopentenyl pyrophosphate (IPP), via
f3-hydroxymethylglutaryl-CoA (HMG-CoA) and mevalonate. Recent in
vivo C13 feeding experiments have demonstrated that the IPP
formation pathway in various eubacteria, green algae and plant
chloroplasts is mevalonate-independent. In this pathway,
hydroxyethylthiamine, which is formed by decarboxylation of
pyruvate, and glycerolaldehyde-3-phosphate (3-GAP) are first
converted into 1-deoxy-D-xylulose-5-phosphate in a
"transketolase" reaction mediated by
1-deoxy-D-xylulose-5-phosphate synthase (Lange et al., 1998;
Schwender et al., 1997; Arigoni et al., 1997; Lichtenthaler et
al., 1997; Sprenger et al., 1997). In an intramolecular
rearrangement reaction, this 1-deoxy-D-xylulose-5-phosphate is
then converted into 2-C-methyl-D-erythritol-4-phosphate and then
into IPP (Arigoni et al., 1997; Zeidler et al., 1998).
Eiochemical data suggest that the mevalonate pathway operates in
the cytosol and leads to the formation of phytosterols. The
antibiotic mevinolin, a specific mevalonate formation inhibitor,
, 0$Z7/000d~.. CA 02378657 2002-O1-08
3
only leads to sterol biosynthesis inhibition in the cytoplasma,
while prenyl lipid formation in the plastids remains unaffected
(Bach and Lichtenthaler, 1993). In contrast, the
mevalonate-independent pathway is located in the plastids and
leads predominantly to the formation of carotenoids and plastid
prenyl lipids (Schwender et al., 1997; Arigoni et al:, 1997).
IPP is in equilibrium with its isomer, dimethylallyl
pyrophosphate (DMAPP). Condensation of IPP with DMAPP head to
l0 tail results in the monoterpene (Clp) geranyl pyrophosphate (GPP).
Addition of further IPP units results in the sesquiterpene (C15)
farnesyl pyrophosphate (FPP), and the diterpene (C2o)
geranylgeranyl pyrophosphate (GGPP). Bonding between two GGPP
molecules results in the formation of the C4p precursors of
carotenoids.
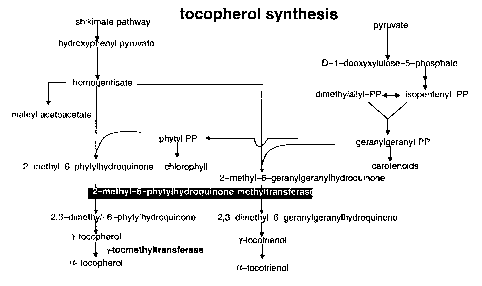
In the case of, mixed prenyl lipids, the isoprene side chain,
whose length varies, is linked to non-isoprene rings such as, for
example, a porphyrine ring in the case of chlorophylls a and b.
The chlorophyl.ls and phylloquinones contain a C2p phytyl chain, in
which only the first isoprene unit contains a double bond. GGPP
is converted by geranylgeranyl pyrophosphate oxidoreductase
(GGPPOR) to give phytyl pyrophosphate (PPP), the starting
material for the subsequent formation of tocopherols.
The ring structures of the mixed prenyl lipids which lead to the
formation of vitamins E and K are quinones whose starting
metabolites are derived from the shikimate pathway. The aromatic
amino acids phenylalanine or tyrosine are converted into
hydroxyphenyl pyruvate, which is dioxygenated to give
homogentisic acid. Starting from erythrose-4-phosphate and
phosphoenol pyruvate (PEP), the chorismate is formed by their
condensation to give 3-deoxy-D-arabinoheptulosonate-7-phosphate
(DAHP) via the intermediates of the shikimate pathway,
3'-dehydroquinate, 3'-dehydroshikimate, shikimate,
shikimate-3-phosphate and 5'-enolpyruvylshikimate-3-phosphate.
During this process, the erythrose-4-phosphate is formed in the
Calvin cycle and the PEP is provided during glycolysis. The
above-described homogentisic acid is subsequently bound to phytyl
pyrophosphate (PPP) or geranylgeranyl pyrophosphate to form the
precursors of a-tocopherol and a-tocotrienol, namely
2-methyl-6-phytylhydroquinone and 2-methyl-6-geranylgeranyl
hydroquinone, respectively. Methylation steps with
S-adenosylmethionine as methyl group donor lead first to
2,3-dimethyl-6-phytylquinol, subsequent cyclization leads to
y-tocopherol and further methylation to oc-tocopherol (Richter,
~81.7/Q~~dZ CA 02378657 2002-O1-08
4
Biochemie der Pflanzen [Plant biochemistry], Georg Thieme Verlag
Stuttgart, 1996;).
Examples which demonstrate that manipulation of an enzyme may
directionally affect metabolite flow can be found in the
literature. A direct effect on the quantities of carotenoids in
these transgenic tomato plants was measured in experiments on an
altered expression of phytoene synthase, which links two GGPP
molecules to give 15-cis-phytoene (Fray and Grierson, Plant Mol.
Biol. 22(4) (1993), 589-602; Fray et al., Plant J. 8 (1995),
693-701). As expected, transgenic tobacco plants which have
reduced quantities of phenylalanine-ammonium lyase show reduced
quantities of phenylpropanoid. The enzyme phenylalanine-ammonium
lyase catalyzes the degradation of phenylalanine and thus
withdraws it from phenylpropanoid biosynthesis (Bate et al.,
Proc. Natl. Acad. Sci USA 91 (16) (1994): 7608-7612; Howles et
al., Plant Physiol. 112 (1996), 1617-1624; ).
Little is known to date on increasing the metabolite flow for
elevating the tocopherol and tocotrienol contents in plants by
overexpression of individual biosynthesis genes. Only WO 97/27285
describes a modification of the tocopherol content by stronger
expression or down-regulation of the enzyme p-hydroxyphenyl
pyruvate dioxygenase (HPPD). WO 99/04622 describes a gene
sequence encoding a y-tocopherol methyltransferase from a
photosynthetically active organism. WO 99/23231 demonstrates that
the expression of a geranylgeranyl reductase in transgenic plants
results in an increased tocopherol biosynthesis. .
It is an object of the present invention to develop a transgenic
plant with an elevated tocopherol and tocotrienol content.
We have found that this object is achieved by overexpressing a
2-methyl-6-phytylhydroquinone methyltransferase gene in plants.
To this end, the activity of 2-methyl-6-phytylhydroquinone
methyltransferase (MPMT) was increased by overexpressing the
Synechocystis spec. PCC6803 MPMT gene in transgenic plants. This
may be achieved in principle by the expression of homologous or
heterologous MPMT genes.
Example 2 describes for the first time the cloning of an MPMT DNA
sequence (SEQ ID No. 1) from Synechocystis spec. PCC6803. To
ensure localization in the plastids, a transit signal sequence is
arranged upstream of the Synechocystis MPMT nucleotide sequence
(Fig. 3, Fig. 4). Another suitable expression cassette is a DNA
sequence encoding an MPMT gene which hybridizes with SEQ ID No. 1
0817~~00~1 CA 02378657 2002-O1-08
or which is homologous to the full sequence or to subsequences
and which is derived from other organisms or plants.
2,3-Dimethyl-6-phytylhydroquinone, of which larger quantities are
5 now available owing to the additional expression of the MPMT
gene, is reacted further toward tocopherols and tocotrienol
(Figure 1).
The transgenic plants are generated by transforming the plants
with a construct comprising the MPMT gene. Model plants employed
for the production of tocopherols and tocotrienols were
Arabidopsis thaliana, Brassica napes and Nicotiana tabacum.
Measurements on MPMT Synechocystis knock-out mutants showed a
drastic decrease regarding the tocopherol and tocotrienol
contents. This confirms the direct effect of plastid plant MPMT
on tocopherol and tocotrienol synthesis.
The invention relates to the use of a Synechocystis spec. PCC6803
DNA sequence SEQ ID No. 1 which encodes an MPMT or its functional
equivalents for the generation of a plant with an elevated
tocopherol and tocotrienol content. The nucleic acid sequence may
be, for example, a DNA or cDNA sequence. Encoding sequences which
are suitable for insertion into an expression cassette are, for
example, those which encode an MPMT and which allow the host to
overproduce tocopherols and tocotrienols.
The expression cassettes also comprise regulatory nucleic acid
sequences which govern the expression of the encoding sequence in
the host cell. In a preferred embodiment, an expression cassette
comprises a promoter upstream, i.e. on the 5'-end of the encoding
sequence, and a polyadenylation signal downstream, i.e. on the
3'-end, and, if appropriate, further regulatory elements which v
are linked operatively with the sequence in between which encodes
the MPMT gene. Operative linkage is to be understood as meaning
the sequential arrangement of promoter, encoding sequence,
terminator and, if appropriate, further regulatory elements in
such a way that each of the regulatory elements can fulfill its
function as intended when the encoding sequence is expressed. The
sequences preferred for operative linkage, but not restricted
thereto, are targeting sequences for guaranteeing subcellular
localization in the apoplast, in the vacuole, in plastids, in the
mitochondrion, in the endoplasmatic reticulum (ER), in the
nucleus, in elaioplasts or in other compartments, and translation
enhancers such as the tobacco mosaic virus 5'-leader sequence
(Gallie et al., Nucl. Acids Res. 15 (1987), 8693-8711).
~817/~~~OZ CA 02378657 2002-O1-08
6
As an example, the plant expression cassette can be incorporated
into a derivative of the transformation vector pain-19 with 35S
promoter (Bevan, M., Nucleic Acids Research 12 (1984):
8711-8721). Figure 4 shows a derivative of the transformation
vector pain-19 with the seed-specific legumin B4 promoter.
A suitable promoter of the expression cassette is, in principle,
any promoter which is capable of governing the expression of
foreign genes in plants. In particular, a plant promoter or a
promoter derived from a plant virus is preferably used.
Particularly preferred is the CaMV 35S promoter from cauliflower
mosaic virus (Franck et al., Cell 21 (1980), 285 - 294). As is
known, this promoter contains various recognition sequences for
transcriptional effectors which in their totality lead to
permanent and constitutive expression of the introduced gene
(Benfey et al., EMBO J. 8 (1989), 2195-2202).
The expression cassette may also comprise a chemically inducible
promoter which allows expression of the exogenous MPMT gene in
the plant to be governed at a particular point in time. Examples
of such promoters which can be used are, inter alia, the PRP1
promoter (Ward et al., Plant. Mol. Biol. 22 (1993), 361-366), a
salicylic-acid-inducible promoter (WO 95/19443), a
benzenesulfonamide-inducible promoter (EP-A 388186), a
tetracyclin-inducible promoter (Gatz et al., (1992) Plant J. 2,
397-404), an abscisic-acid-inducible promoter (EP-A 335528) or an
ethanol- or cyclohexanone-inducible promoter (WO 93/21334).
Furthermore, particularly preferred promoters are those which
ensure expression in tissues or parts of the plant in which, for
example, the biosynthesis of tocopherol or its precursors takes
place. Promoters which ensure leaf-specific expression should be
mentioned in particular. Promoters which should be mentioned are
the potato cytosolic FBPase or the potato ST-LSI promoter
(Stockhaus et al., EMBO J. 8 (1989), 2445 - 245).
A foreign protein was expressed stably in the seeds of transgenic
tobacco plants to an extent of 0.67 of the total soluble seed
protein with the aid of a seed-specific promoter (Fiedler and
Conrad, Bio/Technology 10 (1995), 1090-1094). The expression
cassette can therefore contain, for example, a seed-specific
promoter (preferably the phaseolin promoter (US 5504200), the USP
promoter (Baumlein, H. et al., Mol. Gen. Genet. (1991) 225 (3),
459 - 467) or the LEB4 promoter (Fiedler and Conrad, 1995)), the
LEB4 signal peptide, the gene to be expressed and an ER retention
signal.
CA 02378657 2002-O1-08
7
An expression cassette is generated by fusing a suitable promoter
with a suitable MPMT DNA sequence and, preferably, a DNA which is
inserted between promoter and MPMT DNA sequence and which encodes
a chloroplast-specific transit peptide, and with a
polyadenylation signal, using customary recombination and cloning
techniques as they are described, for example, by T. Maniatis,
E.F. Fritsch and J. Sambrook, Molecular Cloning: A Laboratory
Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
(1989) and by T.J. Silhavy, M.L. Berman and L.W. Enquist,
Experiments with Gene Fusions, Cold Spring Harbor Laboratory,
Cold Spring Harbor, NY (1984) and by Ausubel, F.M. et al.,
Current Protocols in Molecular Biology, Greene Publishing Assoc.
and Wiley-Interscience (1987).
Particularly preferred sequences are those which ensure targeting
into the plastids.
Other expression cassettes which can be used are those whose DNA
sequence encodes an MPMT fusion protein, part of the fusion
protein being a transit peptide which governs translocation of
the polypeptide. Chloroplast-specific transit peptides which are
cleaved off enzymatically from the MPMT residue after
translocation of the MPMT gene into the chloroplasts are
preferred. Particularly preferred is the transit peptide derived
from plastid Nicotiana tabacum transketolase or another transit
peptide (for example the transit peptide of the Rubisco small
subunit or of ferredoxin NADP oxidoreductase) or its functional
equivalent.
Especially preferred are DNA sequences of three cassettes of the
plastid transit peptide of tobacco plastid transketolase in three
reading frames as KpnI/BamHI fragments with an ATG codon in the
NcoI cleavage site:
pTP09
KpnI GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC
CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA
ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG
TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGGGA
TCC BamHI
pTFlO
KpnI GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC
CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA
ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG
~
~$1'~/~00~1 CA 02378657 2002-O1-08
TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGCTG
GATCC_BamHI
pTPl1
KpnI GGTACCATGGCGTCTTCTTCTTCTCTCACTCTCTCTCAAGCTATCCTCTCTCGTTCTGTC
CCTCGCCATGGCTCTGCCTCTTCTTCTCAACTTTCCCCTTCTTCTCTCACTTTTTCCGGCCTTAA
ATCCAATCCCAATATCACCACCTCCCGCCGCCGTACTCCTTCCTCCGCCGCCGCCGCCGCCGTCG
TAAGGTCACCGGCGATTCGTGCCTCAGCTGCAACCGAAACCATAGAGAAAACTGAGACTGCGGGG
ATCC BamHI
The inserted nucleotide sequence encoding an MPMT can be prepared
synthetically, obtained naturally or contain a mixture of
synthetic and natural DNA constituents, and may be composed of
various heterologous MPMT gene segments of a variety of
organisms. In general, synthetic nucleotide sequences are
produced which are equipped with codons which are preferred by
plants. These codons which are preferred by plants can be
determined from codons with the highest protein frequency
expressed in the majority of interesting plant species. When
preparing an expression cassette, a variety of DNA fragments may
be manipulated in order to obtain a nucleotide sequence which
expediently reads in the correct direction and which is equipped
with a correct reading frame. Adaptors or linkers may be added to
the fragments in order to link the DNA fragments to each other.
The promoter and terminator regions may expediently be provided,
in the direction of transcription, with a linker or polylinker
containing one ar more restriction sites for insertion of this
sequence. As a rule, the linker has 1 to 10, in most cases 1 to
8, preferably 2 to 6, restriction sites. In general, the linker
within the regulatory regions has a size less than 100 bp,
frequently less than 60 bp, but at least 5 bp. The promoter may
be native, or homologous, or else foreign, or heterologous, to
the host plant. The expression cassette comprises, in the 5'-3'
direction of transcription, the promoter, a DNA sequence encoding
an MPMT gene, and a region for transcriptional termination.
Various termination regions may be exchanged for each other as
desired.
Manipulations which provide suitable restriction cleavage sites
or which eliminate the excess DNA or restriction cleavage sites
may also be employed. In vitro mutagenesis, primer repair,
restriction or ligation may be used in cases where insertions,
deletions or substitutions such as, for example, transitions and
transversions are suitable. Complementary ends of the fragments
may be provided for ligation in the case of suitable
~
Q$1'J/000~1 CA 02378657 2002-O1-08
S
manipulations such as, for example, restriction, chewing back or
filling in overhangs for blunt ends.
Preferred polyadenylation signals are plant polyadenylation
signals, preferably those which correspond essentially to
Agrobacterium tumefaciens T-DNA-polyadenylation signals, in
particular those of gene 3 of the T-DNA (octopine synthase) of
the Ti-plasmid pTiACH5 (Gielen et al., EMBO J. 3 (1984), 835 et
seq.), or functional equivalents.
The fused expression cassette which encodes an MPMT gene is
preferably cloned into a vector, for example pBinl9, which is
suitable for transforming Agrobacterium tumefaciens. Agrobacteria
transformed with such a vector can then be used in a known manner
for transforming plants, in particular crop plants, such as, for
example, tobacco plants, for example by bathing wounded leaves or
leaf sections in an agrobacterial suspension and subsequently
growing them in suitable media. The transformation of plants by
agrobacteria is known, inter alia, from F.F. White, Vectors for
Gene Transfer in Higher Plants; in Transgenic Plants, Vol. 1,
Engineering and Utilization, edited by S.D. Kung and R. Wu,
Academic Press, 1993, pp. 15 - 38. Transgenic plants which
comprise, integrated into the expression cassette, a gene for
expressing an MPMT gene can be regenerated in a known manner from
the transformed cells of the wounded leaves or leaf sections.
To transform a host plant with a DNA encoding an MPMT, an
expression cassette is inserted into a recombinant vector whose
vector DNA comprises additional functional regulatory signals,
for example sequences for replication or integration. Suitable
vectors are described, inter alia, in "Methods in Plant Molecular
Biology and Biotechnology" (CRC Press), chapter 6/7, (1993), pp.
71 - 119.
Using the above-cited recombination and cloning techniques, the
expression cassettes can be cloned into suitable vectors which
allow their multiplication, for example in E. coli. Suitable
cloning vectors are, inter alia, pBR332, pUC series, Ml3mp series
and pACYC184. Especially suitable are binary vectors which are
capable of replication in E. coli and in agrobacteria.
The invention furthermore relates to the use of an expression
cassette comprising a DNA sequence SEQ ID No. 1 or a DNA sequence
hybridizing herewith for transforming plants, plant cells, plant
tissues or parts of plants. The preferred object of the use is an
elevated tocopherol and tocotrienol content of the plant.
0817/a0~01 CA 02378657 2002-O1-08
Depending on the choice of promoter, expression may take place
specifically in the leaves, in the seeds, the petals or in other
parts of the plant. Such transgenic plants, their propagation
material and the cells, tissues or parts of such plants are a
5 further subject of the present invention.
In addition, the expression cassette may also be employed for
transforming bacteria, cyanobacteria, yeasts, filamentous fungi
and algae for the purpose of increasing the tocopherol and
10 tocotrienol content.
The transfer of foreign genes into the genome of a plant is
termed transformation. It exploits the above-described methods of
transforming and regenerating plants from plant tissues or plant
cells for transient or stable transformation. Suitable methods
are protoplast transformation by polyethylene-glycol-induced DNA
uptake, the biolistic method using the gene gun - the so-called
particle bombardment method, electroporation, incubation of dry
embryos in DNA-containing solution, microinjection and
agrobacterium-mediated gene transfer. The abovementioned methods
are described, for example, in B. Jenes et al., Techniques for
Gene Transfer, in: Transgenic Plants, Vol. 1, Engineering and
Utilization, edited by S.D. Kung and R. Wu, Academic Press
(1993), 128 - 143, and in Potrykus, Annu. Rev. Plant Physiol.
Plant Molec. Biol. 42 (1991), 205 - 225. The construct to be
expressed is preferably cloned into a vector which is suitable
for the transformation of Agrobacterium tumefaciens, for example
pBinl9 (Bevan et al., Nucl. Acids Res. 12 (1984), 8711).
Agrobacteria transformed with an expression cassette can equally
be used in a known manner for transforming plants, in particular
crop plants such as cereals, corn, oats, soya, rice, cotton,
sugar beet, canola, sunflower, flax, hemp, potato, tobacco,
tomato, oilseed rape, alfalfa, lettuce and the various tree, nut
and grapevine species, for example by bathing wounded leaves or
leaf sections in an agrobacterial suspension and subsequently
growing them in suitable media.
Functionally equivalent sequences which encode an MPMT gene are
those sequences which still have the desired functions, despite a
differing nucleotide sequence. Functional equivalents thus
encompass naturally occurring variants of the sequences described
herein, and synthetic nucleotide sequences, for example those
obtained by chemical synthesis and adapted to suit the codon
usage of a plant.
0$17/~~~01 CA 02378657 2002-O1-08
11
Functional equivalents are also to be understood as meaning, in .
particular, natural or artificial mutations of an originally
isolated sequence encoding an MPMT which continue to show the
desired function. Mutations encompass substitutions, additions,
deletions, exchanges or insertions of one or more nucleotide
residues. Thus, the present invention also encompasses, for
example, those nucleotide sequences which are obtained by
modifying the MPMT nucleotide sequence. The purpose of such a
modification may be, for example, the further limitation of the
encoding sequence contained therein or else, for example, the
insertion of further restriction enzyme cleavage sites.
Example 8 describes a deletion clone of the MPMT gene, see SEQ ID
No. 7.
Functional equivalents are also those variants whose function is
attenuated or.increased compared with the starting gene, or gene
fragment.
Also suitable are artificial DNA sequences as long as they
mediate the desired characteristic, for example an elevated
tocopherol content in the plant, by overexpression of an MPMT
gene in crop plants, as described above. Such artificial DNA
sequences can be identified, for example, by backtranslation of
proteins with MPMT activity which have been constructed by means
of molecular modeling, or else by in vitro selection. Especially
suitable are encoding DNA sequences which have been obtained by
backtranslating a polypeptide sequence in accordance with the
host-plant-specific codon usage. An expert skilled in the art of
plant genetic engineering methods will readily be able to
identify the specific codon usage by computer evaluations of
other known genes of the plant to be transformed.
Further suitable equivalent nucleic acid sequences which should
be mentioned are sequences which encode fusion proteins, an MPMT
polypeptide or a functionally equivalent portion thereof being a
constituent of the fusion protein. The second part of the fusion
protein may be, for example, another enzymatically active
polypeptide, or an antigenic polypeptide sequence with the aid of
which detection of MPMT expression is possible (for example
myc-tag or his-tag). However, it is preferably a regulatory
protein sequence such as, for example, a transit peptide which
leads the MPMT protein to the plastids.
An elevated tocopherol and tocotrienol content is to be
understood as meaning for the purposes of the present invention
the artificially acquired ability to increase biosynthetic
0817/04001 CA 02378657 2002-O1-08
12
performance regarding these compounds by functional
overexpression of an MPMT gene SEQ ID No. 1 or SEQ ID No. 7 in
the.plant in comparison with the non-genetically modified plant
for at least one plant generation.
'
Both the tocopherol content and the tocotrienol content can be
increased. It is preferred to increase the tocopherol content.
However, under certain conditions, it is also possible
preferentially to increase the tocotrienol content.
The tocopherol biosynthesis site, for example, is inter alia the
leaf tissue, so that leaf-specific expression of the MPMT gene is
meaningful. However, it is obvious that tocopherol biosynthesis
need not be limited to the leaf tissue but may also take place in
a tissue-specific fashion in all of the other remaining parts of
the plant, for example in fatty seeds'.
The constitutive expression of the exogenous MPMT gene is also
advantageous. On the other hand, inducible expression may also be
desirable.
The expression efficacy of the transgenically expressed'MPMT gene
can be determined, for example, in vitro by shoot meristem
propagation. In addition, altered expression of the MPMT gene
with regard to type and level, and its effect on tocopherol
biosynthesis performance may be tested on test plants in
greenhouse experiments.
The invention furthermore relates to transgenic plants,
transformed with an expression cassette comprising the sequence
SEQ ID No. 1 or SEQ ID No. 7 or a DNA sequence hybridizing
herewith or a DNA sequence which is homologous to the full
sequence or to subsequences, and to transgenic cells, tissues,
parts and propagation material of such plants. Especially
preferred are transgenic crop plants such as, for example,
barley, wheat, rye, corn, oats, Soya, rice, cotton, sugar beet,
canola, sunflower, flax, hemp, potato, tobacco, tomato, oilseed
rape, alfalfa, tagetes, lettuce and the various tree, nut and
grapevine species.
Plants for the purposes of the invention are mono- and
dicotyledonous plants.
The invention furthermore relates to photosynthetically active
organisms transformed with an expression cassette containing the
sequence SEQ ID No. 1 or SEQ ID No. 7 or a DNA sequence
hybridizing herewith or a DNA sequence which is homologous to the
0817/00001, CA 02378657 2002-O1-08
13
full sequence or to subsequences. Examples of photosynthetically
active organisms'are, besides the plants, cyanobacteria, mosses
and algae.
Since this biosynthetic pathway is a metabolic pathway which is
exclusively located in the plastids, it offers optimal target
enzymes for the development of inhibitors. Since, according to
current knowledge, no enzyme which is identical or similar to
Synechocystis MPMT is present in human and animal organisms, it
can be assumed that inhibitors should have a very specific effect
on plants.
As already mentioned, MPMT is a potential target for herbicides.
To find efficient MPMT inhibitors, it is necessary to provide
suitable test systems with which inhibitor-enzyme binding studies
can be carried out. To this end, for example, the complete cDNA
sequence of the Synechocystis MPMT is cloned into an expression
vector (pQE, Qiagen) and overexpressed in E. coli.
The MPMT protein which is expressed with the aid of the
expression cassette according to the invention is particularly
suitable for finding MPMT-specific inhibitors.
To this end, MPMT can be employed, for example, in an enzyme test
in which the MPMT activity is determined in the presence and
absence of the active ingredient to be tested. A qualitative and
quantitative statement on the inhibitory behavior of the active
ingredient to be tested can be made by comparing the two activity
determinations.
The test system according to the invention allows a large number
of chemical compounds to be checked rapidly and simply for
herbicidal properties. The method allows reproducibly and
specifically to select, from a large number of substances, those
with high potency in order subsequently to carry out, with these
substances, further, in-depth tests with which the skilled worker
is familiar.
The invention furthermore relates to herbicides which can be
identified with the above-described test system.
Overexpression of the MPMT-encoding gene sequence SEQ ID No. 1 or
SEQ ID No. 7 in a plant allows an improved resistance to MPMT
inhibitors to be achieved. Transgenic plants generated thus are
also a subject of the invention.
~81'j/~~~~1 CA 02378657 2002-O1-08
1~
The MPMT protein prepared using the DNA sequence SEQ ID No. 1 or
SEQ ID No. 7 is also suitable for carrying out biotransformations
for providing substantial amounts of
2,3-dimethyl-6-phytylhydroquinone. To do this,
2-methyl-6-phytylhydroquinone is converted in the presence of the
enzyme MPMT and the co-substrate S-adenosyl-L-methionine to give
2,3-dimethyl-6-phytylhydroquinone. In principle, the
biotransformation can be carried out on entire cells which
express the enzyme MPMT, or on cell extracts of these cells, or
else on purified or ultrapure MPMT in the presence of
S-adenosyl-L-methionine.
The invention furthermore relates to:
- Methods for the transformation of a plant, which comprise
introducing, into a plant cell or protoplasts of plants,
expression cassettes containing a DNA sequence SEQ ID No. 1
or SEQ ID No. 7 or a DNA sequence hybridizing herewith or a
DNA sequence which is homologous to the full sequence or to
subsequences, and regenerating these to give entire plants.
- The use of the DNA sequence SEQ ID No. 1 or SEQ ID No. 7 or a
DNA sequence hybridizing herewith for the generation of
plants with an elevated tocopherol and tocotrienol content by
expressing, in plants, an MPMT DNA sequence.
The invention is illustrated by the examples which follow, but
not limited thereto.
Sequence analysis of recombinant DNA
Recombinant DNA molecules were sequenced using a laser
fluorescence DNA sequencer by Licor (sold by MWG Biotech,
Ebersbach) using the method of Sanger (Sanger et al., Proc. Natl.
Acad. Sci. USA 74 (1977), 5463 - 5467).
Example 1
Identification of a Synechocystis~spec. PCC6803
2-methyl-6-phytylhydroquinone methyltransferase
The Synechocystis spec. PCC6803 2-methyl-6-phytylhydroquinone
methyltransferase was cloned and identified as follows:
Using a sequence motif conserved in S-adenosyl-L-methionine
methyltransferases which is responsible for binding
S-adenosyl-L-methionine (SAM) (C. P. Joshi and V.L. Chiang. PMB.
~
p81'~/pp~p=, CA 02378657 2002-O1-08
37 (1998): 663-374), a Synechocystis spec. PCC6803 genomic DNA
database was screened (Kaneko et al., DNA Res. 34 (1996):
109-136). The hypothetical proteins identified during this
screening which contained the SAM binding motif were compared
5 with the primary sequences of the Synechocystis spec. PCC6803
'y-tocopherol methyltransferase (termed s1r0089) and the
Arabidopsis thaliana ~-tocopherol methyltransferase (David
Shintani and Dean DellaPenna. Sience. 282 (1998): 2098-2100).
10 It was possible to identify a hypothetical protein (termed
s110418 SEQ.-ID No. 2) whose amino acid sequence corresponded to
a low degree with the 'y-tocopherol methyl transferases from
Synechocystis spec. PCC6803 and Arabidopsis thaliana (36~ and 28~
identity, respectively).
Further studies of the primary sequence of the hypothetical
protein s110418 confirmed the existence of-a putative prokaryotic
signal sequence within the first 20 amino acids (PSIGNAL,
PC/GENET"' IntelliGenetics, Inc (1991). Such a sequence was also
identified.in Synechocystis spec. PCC6803 y-tocopherol
methyltransferase (s1r0089) (D. Shintani and D. DellaPenna.
Sience. 282 (1998): 2098-2100) and suggests the identical
localization of the two proteins..
The predicted molecular weight of the unprocessed protein is
34.9 kDa and is thus within a range which had also been
determined for the Synechocystis spec. PCC6803 Y-tocopherol
methyltransferase (David Shintani and Dean DellaPenna, Sience.
282 (1998): 2098-2100) and for the 'y-tocopherol methyltransferase
purified from bell peppers (d'Harlingue and Camara, Plastid
enzymes of terpenoid biosynthesis: Purification of y-Tocopherol
Methyltransferase from Capsicum Chromoplasts. Journal of
Biological Chemistry, Vol. 269 (1985), No.28, 15200-15203).
Taking into consideration the facts, we concluded that the
hypothetical protein s110418 might be a tocopherol
methyltransferase.
Example 2
Amplification and cloning of the Synechocystis spec. PCC 6803
2-methyl-6-phytylhydroquinone methyltransferase
The DNA encoding the ORF (open reading frame) s110418 was
amplified by means of polymerase chain reaction (PCR) from
Synechocystis spec. PCC6803 following the method of Crispin A.
Howitt (BioTechniques 21, July 1996:32-34) using a sense-specific
~
~8Z')/~~~~1 CA 02378657 2002-O1-08
16
primer (s1104185' SEQ ID No. 5) and an antisense-specific primer
(s1104183' SEQ ID No. 6).
The PCR conditions were as follows:
The PCR was carried out in a 50 ~,1 reaction batch which
contained:
5 ~1 of a Synechocystis spec. PCC6803 cell suspension
- 0.2 mM dATP, dTTP, dGTP, dCTP
- 1.5 mM Mg(OAc)2
- 5 ~.g bovine serum albumin
- 40 pmol s1104185'
- 40 pmol s1104183'
- 15 X11 3.3 X rTth DNA polymerase XL buffer (PE Applied
Biosystems)
- 5U rTth DNA polymerase XL (PE Applied Biosystems)
The PCR was carried out under the following cycle conditions:
Step 1: 5 minutes at 94°C (denaturing)
Step 2: 3 seconds at 94°C
Step 3: 2 minutes at 58°C (annealing)
Step 4: 2 minutes at 72°C (elongation)
40 repetitions of steps 2-4
Step 5: 10 minutes at 72°C (post-elongation)
Step 6: 4°C (waiting loop)
The amplicon was cloned into the PCR cloning vector pGEM-T
(Promega) using standard methods. The identity of the amplicon
generated was confirmed by sequencing using the M13F (-40)
primer.
Example 3
Generation of an s110418 knock-out mutant
A DNA construct to generate a deletion mutant of the ORF s110418
in Synechocystis spec. PCC6803 was generated using standard
cloning techniques.
The vector pGEM-T/s110418 was digested using the restriction
enzyme Ball. The presence of two Ball cleavage sites within the
s110418 sequence (position by 109 and by 202, respectively)
resulted in the loss of a 93-by internal fragment. The
aminoglycoside-3'-phosphotransferase of the transposon Tn.903 was
cloned into the Ball cleavage sites of the s110418 ORF. To this
0$17/~~Das. CA 02378657 2002-O1-08
17
end, the Tn903 was isolated as an EcoRI fragment from the vector
pUC4k (Vieira, J and Messing, J, Gene: 19 (1982), 259-268), the
overhangs of the restriction digest were made blunt-ended by
standard methods and ligated into the Ball-cut vector
pGEM-T/s110418. The ligation batch was used for the
transformation of E. coli X11 blue cells. Transformants were
selected by using kanamycin and ampicillin. A recombinant plasmid
(pGEM-T/s110418::tn903) was isolated and employed in the
transformation of Synechocystis spec. PCC6803 following the
method of Williams (Methods Enzymol. 167 (1987), 776-778).
Synechocystis spec. PCC6803 transformants were selected on
kanamycin-containing (kan) solid BG-11 medium (Castenholz,
Methods in Enzymology (1988), 68-93) at 28°C and 30 Eunol photons
x (m2x s)-1. After five selection cycles (passages of single
colonies onto fresh BG-11 kan medium), four independent knock-out
mutants were generated.
The complete loss of the s110418 endogene or the exchange for the
recombinant s110418::tn903 DNA was confirmed by PCR analyses.
Example 4
Comparison of the tocopherol production in Synechocystis spec.
PCC6803 wild-type cells and the generated knock-out mutants of
ORF s110418
The cells of the four independent Synechocystis spec. PCC68033
knock-out mutants of ORF s110418 which were cultured on the BG-11
kan agar medium and untransformed wild-type cells were used to
inoculate liquid cultures. These cultures were grown for
approximately 3 days at 28°C and 30 ~nol photons x (m2x s)-1
(30 ~,E). After determining the OD73o of the individual cultures,
the OD73p of all cultures was synchronized by suitable dilutions
with BG-11 (wild types) or BG-11 kan (mutants). These
cell-density-synchronized cultures were used to inoculate three
cultures per mutant and the wild-type controls. Thus, the
biochemical analyses were carried out using in each case three
independently grown cultures of a mutant and the corresponding
wild types. The cultures were grown to an optical density of
OD73o=0.3. The cell culture medium was removed by two
centrifugation steps at 14,000 rpm in an Eppendorf bench
centrifuge. The cells were subsequently disrupted by four
incubations for 15 minutes in an Eppendorf shaker at 30°C,
1000 rpm, in 100 methanol, and the supernatants obtained in each
~817/00~~~. CA 02378657 2002-O1-08
18
case were combined. Further incubation steps resulted in no
further release of tocopherols or tocotrienols.
To avoid oxidation, the resulting extracts were analyzed directly
after extraction with the aid of a Waters Allience 2690 HPLC
system. Tocopherols and tocotrienols were separated over a
' reverse phase column (ProntoSil 200-3-C30, Bischoff) using a
mobile phase of 100 methanol, and identified with reference to
standards (Merck). The detection system used. was the fluorescence
of the substances (excitation 295 nm, emission 320 nm), which was .
detected with the aid of a Jasco fluorescence detector FP 920.
In the Synechocystis spec. PCC6803 knock-out mutants of ORF
s110418, no tocopherols or tocotrienols were found. However,
tocopherols and tocotrienols were measured in the Synechocystis
spec. PCC6803 wild-type cells.
The loss of the ability to produce tocopherols and tocotrienols
within the knock-out mutants of ORF s110418 in comparison with
the Synechocystis spec. PCC6803 wild-type cells demonstrates. that
the gene s110418 encodes a 2-methyl-6-phytylhydroquinone
methyltransferase.
Example 5
Functional characterization of the Synechocystis spec. PCC6803
2-methyl-6-phytylhydroquinone methyltransferase by heterologous
expression in E. coli
The hypothetic Synechocystis spec. PCC6803 protein s110418 was
identified by functional expression in E. coli as 2-methyl-6-
phytylhydroquinone methyltransferase.
The gene s110418 which had been amplified from Synechocystis
spec. PCC6803 was subcloned into the expression vector pQE-30
(Qiagen) in the correct reading frame. The primers s1104185' and
s1104183' (SEQ ID No. 5 and 6, respectively) which had been used
for amplifying the ORF s110418 from Synechocystis spec. PCC6803,
were constructed in such a way that BamI-iI restriction cleavage
sites were added to the 5' end and the 3' end of the amplicon;
see SEQ ID No. 3. Using these flanking BamHI restriction cleavage
sites, the s110418 fragment was isolated from the recombinant
plasmid pGEM-T/s110418 and ligated into a BamHI-cut pQE-30 using
standard methods. The ligation batch was used for the
transformation of M15 E. coli cells, and kanamycin- and
ampicillin-resistant transformants were analyzed. The kanamycin
resistance is mediated by the pREP-4 plasmid, which is contained
. ~817/~~~01 CA 02378657 2002-O1-08
19
in the M15 cells. A recombinant plasmid (pQE-30/s110418) which
carried the s110418 fragment in the correct orientation was
isolated. Identity and orientation of the insert were confirmed
by sequencing.
The recombinant plasmid pQE-30/s110418 was used for the
transformation of M15 E. coli cells in order to generate
recombinant s110418 protein. Using a colony which originated from
the transformation, an overnight culture in Luria broth medium
supplemented with 200 ~,g/ml ampicillin (Amp) and 50 ~,g/ml
kanamycin (Kan) was inoculated. Starting from this culture, a
100 ml Luria broth culture (Amp/Kan) was inoculated the morning
thereafter. This culture was incubated at 28°C on a shaker
incubator until an OD6oo of 0.35-0.4 was reached. Then, production
of the recombinant protein was induced by adding 0.4 mM
isopropyl-f3-D-thiogalactopyranoside (IPTG). The culture was
shaken for a further 3 hours at 28°C and the cells were
subsequently pelleted by centrifugation at 8000 g.
The pellet was resuspended in 600 ~1 lysis buffer (approx.
1-1.5 ml/g pellet moist weight, 10 mM HEPES KOH pH 7.8, 5 mM
dithiothreitol (DTT), 0.24 M sorbitol). Then, PMSF (phenyl
methylsulfonate) was added to a final concentration of 0.15 mM
and the batch was placed on ice for 10 minutes. The cells were
disrupted by a 10-second ultrasonic pulse using an ultrasonic
processor. After addition of Triton X140 (final concentration
0.1~), the cell suspension was incubated on ice for 30 minutes.
The batch was subsequently centrifuged for 30 minutes at
25,000 x g and the supernatant was employed in the assay.
The 2-methyl-6-phytylhydroquinorie methyltransferase activity was
determined by detecting the radiolabeled reaction product
2,3-dimethyl-6-phytylhydroquinone.
To this end, 135 ~,l of the enzyme (approx. 300-600 ~.g) together
with 20 ~1 of substrate (2-methyl-6-phytylhydroquinone) and 15 ~,1
(0.46 mM SAM 14C) methyl group donor were incubated for 4 hours in
the dark at 25°C in the following reaction buffer: 200 ~,1 (125 mM)
tricine-NaOH pH 7.6, 100 ~tl (1.25 mM) sorbitol, 10 ~1 (50 mM)
MgCl2 and 20 ~tl (250 mM) ascorbate.
The reaction was quenched by adding 750 ~l of chloroform/methanol
(1:2) + 150 ~l of 0.9~ NaCl. The mixed batch was centrifuged
briefly, and the top phase was discarded. The bottom phase is
transferred into a new reaction vessel and evaporated under
nitrogen. The residues were taken up in 20 X11 of ether and
applied to a thin-layer plate in order to separate the substances
~
0$1,7~~~001 CA 02378657 2002-O1-08
by chromatography (solid phase: HPTLC plates: silica gel 60 F2s4
(Merck), liquid phase: toluene). The radiolabeled reaction
product is detected by using a phosphoimager.
5 These experiments confirm that the protein encoded by the
Synechocystis spec. PCC6803 gene s110418 (SEQ ID No.1) is a
2-methyl-6-phytylhydroquinone methyltransferase since it has the
enzymatic activity to convert 2-methyl-6-phytylhydroquinone into
2,3-dimethyl-6-phytylhydroquinone.
Figure 2 shows a sequence comparison at amino acid level between
the y-tocopherol methyltransferases from Synechocystis spec. PCC
Synechocystis spec. PCC6803 (s1r0089) and A.thaliana (aratmt}
with the 2-methyl-6-phytylhydroquinone methyltransferase
(s1104189) from Synechocystis spec. PCC6803. The correspondence
with the 'y-tocopherol methyltransferases from Synechocystis spec.
PCC6803 and Arabisopsis thaliana is 36 and 28~ identity,
respectively. .
Example 6
Substrate specificity of the 2-methyl-6-phytylhydroquinone
methyltransferase
Enzymatic studies as carried out in Example 5 confirm that the
enzyme MPMT - encoded by the Synechocystis spec. PCC6803 gene
s110418 (SEQ-ID No. 1) - converts 2-methyl-6-phytylhydroquinone
in 2,3-dimethyl-6-phytylhydroquinone.
In addition, the enzyme MPMT has a 2-methyl-6-geranyl-
geranylhydroquinone methyltransferase activity, while a
'y-tocopherol methyltransferase activity could not be detected.
This confirms that the enzyme 2-methyl-6-phytylhydroquinone
methyltransferase participates in tocotrienol biosynthesis since
it converts 2-methyl-6-geranylgeranylhydroquinone into
2,3-dimethyl-6-geranylgeranylhydroquinone. This clearly
demonstrates that the enzyme activity of the 2-methyl-6-
phytylhydroquinone methyltransferase differs from that of
Y-tocopherol methyltransferase.
Example 7
Generation of expression cassettes containing the MPMT gene
Transgenic plants were generated which express the Synechocystis
spec. PCC6803 2-methyl-6-phytylhy droquinone methyltransferase
firstly under the control of the constitutive CaMV (cauliflower
' 0817/0001 CA 02378657 2002-O1-08
21
mosaic virus) 35S promoter (Franck et al., cell 21 (1980),
285-294) and secondly under the control of the seed-specific
promoter of the Vicia faba legumin gene (Kafatos et al., Nuc.
Acid. Res., 14(6) (1986), 2707-2720). The basis of the plasmid
generated for the constitutive expression of the Synechocystis
spec. PCC6803 2-methyl-6-phytylhydroquinone methyltransferase was
pBinAR-TkTp-9 (Ralf Badur, PhD thesis, University of Gottingen,
1998). This vector is a derivative of pBinAR (Hofgen and
Willmitzer, Plant Sci. 66 (1990),-221-230) and contains the CaMV
(cauliflower mosaic virus) 35S promoter (Franck et al., 1980),
the termination signal of the octopine synthase gene (Gielen et
al., EMBO J. 3 (1984), 835-846) and the DNA sequence encoding the
transit peptide of the Nicotiana tabacum plastid transketolase
(Ralf Badur, PhD thesis, University of Gottingen, 1998). Cloning
the Synechocystis spec. PCC6803 2-methyl-6-phytylhydroquinone
methyltransferase into this vector taking into consideration the
correct reading frame generates a translational fusion of the
2-methyl-6-phytylhydroquinone methyltransferase with the plastid
transit peptide. The transgene is thus transported to the
plastids.
To generate this plasmid, the gene s110418 was isolated from
plasmid pGEM-T/s110418 using the flanking BamFiI restriction
cleavage sites. This fragment was ligated into a BamHI-cut
pBinAR-TkTp-9 using standard methods (see Figure 3). This plasmid
(pBinAR-TkTp-9/s110418) was used for the generation of transgenic
Arabidopsis thaliana, Brassica napus and Nicotiana tabacum
plants. Fragment A (529 bp) in Figure 3 contains the CaMV 35S
promoter (nucleotides 6909 to 7437 of cauliflower mosaic virus),
fragment B (245 bp) encodes the transit peptide of the Nicotiana
tabacum transketolase, fragment C (977 bp) encodes the
Synechocystis spec. PCC6803 ORF s110418, and fragment D (219 bp)
encodes the termination signal of the octopine synthase gene.
To generate a plasmid which allows the seed-specific expression
of the Synechocystis spec. PCC6803 2-methyl-6-phytylhydroquinone
methyltransferase in plants, the seed-specific promoter of the
legumin B4 gene (Kafatos et al., Nuc. Acid. Res., 14(6) (1986),
2707-2720), was used. The 2.7 kb fragment of the legumin B4 gene
promoter was isolated from plasmid pCR-Script/lePOCS using the
EcoRI cleavage site and the KpnI cleavage site which flank the
promoter at the 5' and at the 3' end, respectively. Plasmid
pBinAR-TkTp-9/s110418 was also treated with the restriction
enzymes EcoRI and KpnI. As a consequence, the CaMV 35S promoter
was excised from this plasmid. The promoter of the legumin gene
was subsequently cloned into this vector as EcoRI/KpnI fragment,
thus generating a plasmid which placed the expression of gene
~
~$1,'~/~~~~1 CA 02378657 2002-O1-08
22
s110418 under the control of this seed-specific promoter, see
Figure 4. This plasmid (pBinARleP-TkTp-9/s110418) was used for
the generation of transgenic Arabidopsis thaliana, Brassica napus
and Nicotiana tabacum plants.
Fragment A (2700 bp) in Figure 4 contains the promoter of the
Vicia faba legumin B4 gene, fragment B (245 bp) encodes the
transit peptide of the Nicotiana tabacum transketolase, fragment
C (977 bp) encodes the Synechocystis spec. PCC6803 ORF s110418,
l0 and fragment D (219 bp) encodes the termination signal of the
octopine synthase gene.
Example 8
Generation of expression cassettes containing a deletion clone of
the MPMT gene
Based on a computer analysis, a putative prokaryotic secretion
signal was identified in the primary sequence of ORF s110418. To
ensure that this has no adverse effect on the import of the
protein into the plastids upon expression in plants, a derivative
of the sequence of s110418 was generated in which the putative
secretion signal was deleted (SEQ ID No. 7). This deletion was
carried out using PCR technology. By virtue of the primers used
for this purpose (s110418DSp5', SEQ ID No. 9 and s110418DSp3',
SEQ ID No. 10), an EcoRV restriction cleavage site was added onto
the 5' end of the sequence and an SalI restriction cleavage site
onto the 3' end, thus allowing directed cloning into vector
pBinAR-TkTp-9. The resulting plasmid pBinAR-TkTp-9/s110418~SP is
described in Figure 5. Fragment A (529 bp) in Figure 5 contains
the CaMV 35S promoter (nucleotides 6909 to 7437 of the
cauliflower mosaic virus), fragment B (245 bp) encodes the
transit peptide cf the Nicotiana tabacum transketolase, fragment
C (930 bp) encodes the Synechocystis spec. PCC6803 ORF
s1104184SP, and fragment D (219 bp) encodes the termination
signal of the octopine synthase gene.
To generate a plasmid which allows the seed-specific expression
of the deletion clone of the Synechocystis spec. PCC6803
2-methyl-6-phytylhydroquinone methyltransferase in plants, the
seed-specific promoter of the legumin B4 gene which has already
been described was used again (Kafatos et al., Nuc. Acid. Res.,
14(6) (1986), 2707-2720). The 2.7 kb fragment of the legumin B4
gene promoter was isolated from plasmid PCR-Script/lePOCS using
the EcoR2 and the KpnI cleavage sites which flank the promoter at
the 5' end and at the 3' end, respectively. Plasmid
pBinAR-TkTp-9/s1104180SP was also treated with the restriction
~
~81,7/0~~~1 CA 02378657 2002-O1-08
23
enzymes EcoRI and KpnI. As a consequence, the CaMV 35S promoter.
was excised from this plasmid. The promoter of the legumin gene
was. subsequently cloned into this vector as an EcoRI/KpnI
fragment, thus generating a plasmid which placed the expression
of the deletion clone of the s110418 gene under the control of
this seed-specific promoter, see Figure 6. Fragment A (2700 bp)
in Figure 6 contains the promoter of the Vicia faba legumin B4
gene, fragment B (245 bp) encodes the transit peptide of
Nicotiana tabacum transketolase, fragment C (930 bp) encodes the
Synechocystis spec. PCC6803 ORF s110418~SP, and fragment D
(219 bp) encodes the termination signal of the octopine synthase
gene.
This plasmid (pBinARIeP-TkTp-9/s1104180SP) was used for the
generation of transgenic Arabidopsis thaliana, Brassica napus and
Nicotiana tabacum plants.
An increase in the tocopherol and tocotrienol contents was also
measured by expressing the DNA sequence SEQ-ID No. 7 in
transgenic plants.
Example 9
Generation of transgenic Arabidopsis thaliana plants
Wild-type Arabidopsis thaliana plants (Columbia) were transformed
with the Agrobacterium tumefaciens strain (EHA105) using a
modified vacuum infiltration method as the basis (Steve Clough
and Andrew Bent. Floral dip: a simplified method for
Agrobacterium-mediated transformation of Arabidopsis thaliana.
Plant J 16(6):735-43, 1998; Bechtold, N., Ellis, J. and Pelltier,
G., in: Planta Agrobacterium-mediated gene transfer by
infiltration of adult Arabidopsis thaliana plants. CRAcad Sci
Paris, 1993, 1144(2):204-212). The Agrobacterium tumefaciens
cells used had previously been transformed with plasmids
pBinAR-TkTp-9/s110418 or pBinARIeP-TkTp-9/s110418 (Figure 3 and
4) .
Seeds of the primary transformants were selected on the basis of
antibiotic resistance. Antibiotic-resistant seedlings were
planted into soil and used for biochemical analysis as fully
developed plants.
' 0817/~~~~1 CA 02378657 2002-O1-08
24
Example 10
Generation of transgenic Brassica napus plants
The generation of transgenic oilseed rape plants followed in
principle a procedure described by Bade, J.B. and Damm, B. (in
Gene Transfer to Plants, Potrykus, I. and Spangenberg, G., eds,
Springer Lab Manual, Springer Verlag, 1995, 30-38), which also
gives the composition of the media and buffers used.
The transformations were performed with Agrobacterium tumefaciens
strain EHA105. Plasmids pBinAR-TkTp-9/s110418 and
pBinARIeP-TkTp-9/s110418 were used for the transformation. Seeds
of Brassica napus var. Westar were surface-sterilized with 70~
I5 etrhanol (vlv), washed in water for 10 minutes at 55°C,
incubated
for 20 minutes in 1~ strength hypochlorite solution (25~ v/v
Teepol, 0.1~ v/v Tween 20) and washed six times with sterile
water for in each case 20 minutes. The seeds were dried on filter
paper for three days, and 10-15 seeds were germinated in a glass
flask containing 15 ml of germination medium. The roots and
apices were removed from several seedlings (approximate size
10 cm), and the hypocotyls which remained were cut into sections
of approximate length 6 mm. The approximately 600 explants thus
obtained were washed for 30 minutes with 50 ml of basal medium
and transferred into a 300 ml flask. After addition of 100 ml of
callus induction medium, the cultures were incubated for 24 hours
at 100 rpm.
An overnight culture of the agrobacterial strain was established
in Luria broth medium supplemented with kanamycin (20 mg/1) at
29°C, and 2 ml of this were incubated in 50 ml of Luria broth
medium without kanamycin for 4 hours at 29°C to an ODsoo of
0.4 - 0.5. After the culture had been pelleted for 25 minutes at
2000 rpm, the cell pellet was resuspended in 25 ml of basal
medium. The bacterial concentration in the solution was brought
to an OD6oo of 0.3 by adding more basal medium.
The callus induction medium was removed from the oilseed rape
explants using sterile pipettes, 50 ml of agrobacterial
suspension were added, the cultures were mixed carefully and
incubated for 20 minutes..The agrobacterial suspension was
removed, the oilseed rape explants were washed for 1 minute with
ml of callus induction medium, and 100 ml of callus induction
medium were subsequently added. Cocultivation was performed for
45 24 hours on an orbital shaker at 100 rpm. Cocultivation was
stopped by removing the callus induction medium, and the explants
were washed twice with 25 ml of wash medium for 1 minute each
~
081,7/0~~01 CA 02378657 2002-O1-08
time and twice for 60 minutes with 100 ml of wash medium each
time, at 100 rpm. The wash medium together with the explants was
transferred into 15 cm Petri dishes, and the medium was removed
using sterile pipettes.
5
For regeneration, batches of 20-30 explants were transferred into
90 mm Petri dishes containing 25 ml of shoot induction medium
supplemented with kanamycin. The Petri dishes were sealed with
two layers of Leukopor and incubated at 25°C and 2000 lux at
10 photoperiods of 16 hours light/8 hours dark. Every 12 days, the
developing calli were transferred to fresh Petri dishes
containing shoot induction medium. All further steps for
regenerating entire plants were carried out as described by Bade,
J.B. and Damm, B. (in Gene Transfer to Plants, Potrykus, I. and
15 Spangenberg, G., eds, Springer Lab Manual, Springer Verlag, 1995,
30-38)..
Example 11
20 Generation of transgenic Nicotiana tabacum plants
10 ml of YEB medium supplemented with antibiotic (5 g/1 beef
extract, 1 g/1 yeast extract, 5 g/1 peptone, 5 g/1 sucrose and
2 mM MgS04) were inoculated with a colony of Agrobacterium
25 tumefaciens and the culture was grown overnight at 28°C. The cells
were pelleted for 20 minutes at 4°C, 3500 rpm, using a bench
centrifuge and then resuspended under sterile conditions in fresh
YEB medium without antibiotics. The cell suspension was used for
the transformation.
The sterile-grown wild-type plants were obtained by vegetative
propagation. To this end, only the tip of the plant was cut off
and transferred to fresh 2M5 medium in a sterile preserving jar.
As regards the rest of the plant, the hairs on the upper side of
the leaves were removed and the central veins of the leaves were
removed. Using a razor blade, the leaves were cut into sections
of approximate size 1 cmz. The agrobacterial culture was
transferred into a small Petri dish (diameter 2 cm). The leaf
sections were briefly drawn through this solution and placed with
the underside of the leaves on 2M5 medium in Petri dishes
(diameter 9 cm) in such a way that they touched the medium. After
two days in the dark at 25°C, the explants were transferred to
plates with callus induction medium and warmed at 28°C in a
controlled-environment cabinet. The medium had to be changed
every 7-10 days. As soon as calli formed, the explants were
transferred into sterile preserving jars on shoot induction
medium supplemented with claforan (see above). Organogenesis
(~8Z'~/~~Q~1 CA 02378657 2002-O1-08
26
started after approximately one month and it was possible to cut
off the shoots formed. The shoots were grown on 2MS medium
supplemented with claforan and selection marker. As soon as a
substantial root ball had developed, it was possible to pot up
the plants in seed compost.
Example 12
Characterization of the transgenic plants
To confirm that expression of the Synechocystis spec. PCC6803
2-methyl-6-phytylhydroquinone methyltransferase increased vitamin
E biosynthesis in the transgenic plants, the tocopherol and
tocotrienol contents in leaves and seeds of the plants
transformed with the constructs pBinARIeP-TkTp-9/s110418 or
pBinAR-TkTp-9/s110418 (Arabidopsis thaliana, Brassica napus and
Nicotiana tabacum) were analyzed. To,this end, the transgenic
plants were grown in the greenhouse, and plants which express the
gene encoding the Synechocystis spec. PCC6803
2-methyl-6-phytylhydroquinone methyltransferase were analyzed at
Northern level. The tocopherol content and the tocotrienol
content in leaves and seeds of these plants were determined. In
all cases, the tocopherol or tocotrienol concentration in
transgenic plants which additionally express a DNA sequence SEQ
ID No. 1 or SEQ ID No. 7, was elevated in comparison with
untransformed plants.
35
45
CA 02378657 2002-O1-08
SEQUENCE LISTING
<110> SunGene GmbH & Co.KGaA
<120> Overexpression of a DNA sequence encoding a
2-methyl-6-phytylhydroquinone methyltransferase in
plants.
<130> MPMTSynechocystis
<140>
<141>
<160> 10
<170> PatentIn Vers. 2.0
<210> 1
<211> 957
<212> DNA
<213> Synechocystis PCC6803
<220>
<221> CDS
<222> (1)..(957)
<400> 1
atg ccc gag tat ttg ctt ctg ccc get ggc cta att tcc ctc tcc ctg 48
Met Pro Glu Tyr Leu Leu Leu Pro Ala Gly Leu Ile Ser Leu Ser Leu
1 5 10 15
gcg atc gcc get gga ctg tat ctc cta act gcc cgg ggc tat cag tca 96
Ala Ile Ala Ala Gly Leu Tyr Leu Leu Thr Ala Arg Gly Tyr Gln Ser
20 25 30
tcg gat tcc gtg gcc aac gcc tac gac caa ,tgg aca gag gac ggc att 144
Ser Asp Ser Val Ala Asn Ala Tyr Asp Gln Trp Thr Glu Asp Gly Ile
35 40 45
ttg gaa tat tac tgg ggc gac cat atc cac ctc ggc cat tat ggc gat 192
Leu Glu Tyr Tyr Trp Gly Asp His Ile His Leu Gly His Tyr Gl.y Asp
50 55 60
ccg cca gtg gcc aag gat ttc atc caa tcg aaa att gat ttt gtc cat 240
Pro Pro Val Ala Lys Asp Phe Ile Gln Ser Lys Ile Asp Phe Val His
65 70 75 80
gcc atg gcc cag tgg ggc gga tta gat aca ctt ccc ccc ggc aca acg 288
1
CA 02378657 2002-O1-08
Ala Met Ala Gln Trp Gly Gly Leu Asp Thr Leu Pro Pro Gly Thr Thr
85 90 95
gtattggatgtg ggttgcggc attggcggt agcagtcgc attctcgcc 336
ValLeuAspVal GlyCysGly IleGlyGly SerSerArg IleLeuAla
100 105 110
aaagattatggt tttaacgtt accggcatc accattagt ccccaacag 384
LysAspTyrGly PheAsnVal ThrGlyIle ThrIleSer ProGlnGln
115 120 125
gtgaaacgggcg acggaatta actcctccc gatgtgacg gccaagttt 432
ValLysArgAla ThrGluLeu ThrProPro AspValThr AlaLysPhe
130 135 140
gcggtggacgat getatggct ,ttgtctttt cctgacggt agtttcgac 480
AlaValAspAsp AlaMetAla LeuSerPhe ProAspGly SerPheAsp
145 150 155 160
gtagtttggtcg gtggaagca gggccccac atgcctgac aaagetgtg 528
ValValTrpSer ValGluAla GlyProHis MetProAsp LysAlaVal
165 170 175
tttgccaaggaa ttactgcgg gtcgtgaaa ccagggggc attctggtg 576
PheAlaLysGlu LeuLeuArg ValValLys ProG1yGly IleLeuVal
180 185 190
gtg gcg gat tgg aat caa cgg gac gat cgc caa gtg ccc ctc aac ttc 624
Val Ala Asp Trp Asn Gln Arg Asp Asp Arg Gln Val Pro Leu Asn Phe
195 200 205
tgg gaa aaa cca gtg atg cga ~caa ctg ttg gat caa tgg tcc cac. cct 672
Trp Glu Lys Pro Val Me_t Arg Gln Leu Leu Asp Gln Trp Ser His Pro.
210 215 220
gcc ttt gcc agc att gaa ggt ttt gcg gaa aat ttg gaa gcc acg ggt 720
Ala Phe Ala Ser Ile Glu Gly Phe Ala Glu Asn Leu Glu Ala Thr Gly
225 230 235 240
ttg gtg gag ggc cag gtg act act get gat tgg act gta ccg acc ctc 768
Leu Val Glu Gly Gln Val Thr Thr Ala Asp Trp Thr Val Pro Thr Leu
245 250 255
ccc get tgg ttg gat acc att tgg cag ggc att atc cgg ccc cag ggc 816
Pro Ala Trp Leu Asp Thr Ile Trp Gln Gly Ile Ile Arg Pro Gln Gly
260 265 270
tgg tta caa tac ggc att cgt ggg ttt atc aaa tcc gtg cgg gaa gta 864
2
CA 02378657 2002-O1-08
Trp Leu Gln Tyr Gly Ile Arg Gly Phe Ile Lys Ser Val. Arg Glu Val
275 280 285
ccg act att tta ttg atg cgc ctt gcc ttt ggg gta gga ctt tgt cgc 912
Pro Thr Ile Leu Leu Met Arg Leu Ala Phe Gly Val Gly Leu Cys Arg
290 295 300
ttc ggt atg ttc aaa gca gtg cga aaa aac gcc act caa get taa 957
Phe Gly Met Phe Lys Ala Val Arg Lys Asn Ala Thr Gln Ala
305 310 315
<210> 2
<211> 318
<212> PRT
<213> Synechocystis PCC6803
<400> 2
Met Pro Glu Tyr Leu Leu Leu Pro Ala Gly Leu Ile Ser Leu Ser Leu
1 5 10 15
Ala Ile Ala Ala Gly Leu Tyr Leu Leu Thr Ala Arg Gly Tyr Gln Ser
20 25 30
Ser Asp Ser Val Ala Asn Ala Tyr Asp Gln Trp Thr Glu Asp Gly Ile
35 40 45
Leu Glu Tyr Tyr Trp Gly Asp His Ile His Leu Gly His Tyr Gly Asp
50 55 60
Pro Pro Val Ala Lys Asp Phe Ile Gln Ser Lys Ile Asp Phe Val His
65 70 75 80
Ala Met Ala G1n Trp Gly Gly Leu Asp Thr Leu Pro Pro Gly Thr Thr
85 . 90 95
Val Leu Asp Val Gly Cys Gly Ile Gly Gly Ser Ser Arg Ile Leu Ala
100 105 110
Lys Asp Tyr Gly Phe Asn Val Thr Gly Ile Thr Ile Ser Pro Gln Gln
115 120 125
Val Lys Arg Ala Thr Glu Leu Thr Pro Pro Asp Val Thr Ala Lys Phe
130 135 140
Ala Val Asp Asp Ala Met Ala Leu Ser Phe Pro Asp Gly Ser Phe Asp
145 150 155 160
3
CA 02378657 2002-O1-08
Val Val Trp Ser Val Glu Ala Gly Pro His Met Pro Asp Lys Ala Val
165 170 175
Phe'Ala Lys Glu Leu Leu Arg Val Val Lys Pro Gly Gly Ile Leu Val
180 185 190
Val Ala Asp Trp Asn Gln Arg Asp Asp Arg Gln Val Pro Leu Asn Phe
195 200 205
Trp Glu Lys Pro Val Met Arg Gln Leu Leu Asp Gln Trp Ser His Pro
210 215 220
Ala Phe Ala Ser Ile Glu Gly Phe Ala Glu Asn Leu Glu Ala Thr Gly
225 230 235 240
Leu Val Glu Gly Gln Val Thr Thr Ala Asp Trp Thr Val Pro Thr Leu
245 250 255
Pro Ala Trp Leu Asp Thr Ile Trp Gln Gly Ile Ile Arg Pro Gln Gly
260 265 270
Trp Leu Gln Tyr Gly Ile Arg Gly Phe Ile Lys Ser Val Arg Glu Val
275 280 285
Pro Thr Ile Leu Leu Met Arg Leu Ala Phe Gly Val Gly Leu Cys Pxg
290 295 300
Phe Gly Met Phe Lys Ala Val Arg Lys Asn Ala Thr Gln Ala
~3 05 310 315
<210> 3
<211> 974
<212> DNA - . ..
<213> Synechocystis PCC6803
<220>
<221> CDS
<222> (7)..(963)
<400> 3
ggatcc atg ccc gag tat ttg ctt ctg ccc get ggc cta att tcc ctc 48
Met Pro Glu Tyr Leu Leu Leu Pro Ala Gly Leu Ile Ser Leu
1 5 10
tcc ctg gcg atc gcc get gga ctg tat ctc cta act gcc cgg ggc tat 96
Ser Leu Ala Ile Ala Ala Gly Leu Tyr Leu Leu Thr Ala Arg Gly Tyr
15 20 25 30
4
~
CA 02378657 2002-O1-08
cag tca tcg gat tcc gtg gcc aac gcc tac gac caa tgg aca gag gac 144
Gln Ser Ser Asp Ser Val Ala Asn Ala Tyr Asp Gln Trp Thr Glu Asp
35 40 45
ggc att ttg gaa tat tac tgg ggc gac cat atc cac ctc ggc cat tat 192
Gly Ile Leu Glu Tyr Tyr Trp Gly Asp His Ile His Leu Gly His Tyr
50 55 60
ggc gat ccg cca gtg gcc aag gat ttc atc caa tcg aaa att gat ttt 240
Gly Asp Pro Pro Val Ala Lys Asp Phe Ile Gln Ser Lys Ile Asp Phe
65 70 75
gtc cat gcc atg gcc cag tgg ggc gga tta gat aca ctt ccc ccc ggc 288
Val His Ala Met Ala Gln Trp Gly Gly Leu Asp Thr Leu Pro Pro Gly
80 85 90
aca acg gta ttg gat gtg ggt tgc ggc att ggc ggt agc agt cgc att 336
Thr Thr Val Leu Asp Va1 Gly Cys Gly Ile Gly Gly Ser Ser Arg Ile
95 100 105 110
ctc gcc aaa gat tat ggt ttt aac gtt acc ggc atc acc att agt ccc 384
Leu Ala Lys Asp: Tyr Gly Phe Asn Val Thr Gly Ile Thr Ile Ser Pro
115 120 125
caa cag gtg aaa cgg gcg acg gaa tta act cct ccc gat gtg acg gcc 432
Gln Gln Val Lys Arg Ala Thr Glu Leu Thr Pro Pro Asp Val Thr Ala
130 135 140
aag ttt gcg gtg gac gat get atg get ttg tct ttt cct gac ggt agt 480
Lys Phe Ala Val Asp Asp Ala Met Ala Leu Ser Phe Pro Asp Gly Ser
145 150 155
ttc gac gta gtt tgg tcg gtg gaa gca ggg ccc cac atg cct gac aaa 528
Phe Asp Val Val Trp Ser Val Glu Ala Gly Pro His Met Pro Asp Lys
160 165 170
get gtg ttt gcc aag gaa tta ctg cgg gtc gtg aaa cca ggg ggc att 576
Ala Val Phe Ala Lys Glu Leu Leu Arg Val Val Lys Pro Gly Gly Ile
175 180 185 190
ctg gtg gtg gcg gat tgg aat caa cgg gac gat cgc caa gtg ccc ctc 624
Leu Val Val Ala Asp Trp Asn Gln Arg Asp Asp Arg Gln Val Pro Leu
195 200 205
aac ttc tgg gaa aaa cca gtg atg cga caa ctg ttg gat caa tgg tcc 672
Asn Phe Trp Glu Lys Pro Val Met Arg Gln Leu Leu Asp Gln Trp Ser
210 215 220
CA 02378657 2002-O1-08
cac cct gcc ttt gcc agc att gaa ggt ttt gcg gaa aat ttg gaa gcc 720
His Pro Ala Phe Ala Ser Ile Glu Gly Phe Ala Glu Asn Leu Glu A1a
225 230 235
acg ggt ttg gtg gag ggc cag gtg act act get gat tgg act gta ccg 768
Thr Gly Leu Val Glu Gly Gln Val Thr Thr Ala Asp Trp Thr Val Pro
240 245 250
acc ctc ccc get tgg ttg gat acc att tgg cag ggc att atc,cgg ccc 816
Thr Leu Pro Ala Trp Leu Asp Thr Ile Trp Gln Gly Ile Ile Arg Pro
255 260 265 270
cag ggc tgg tta caa tac ggc att cgt ggg ttt atc aaa tcc gtg cgg 864
Gln Gly Trp Leu Gln Tyr Gly Ile Arg Gly Phe Ile Lys Ser Val Arg
275 280 285
gaa gta ccg act att tta ttg atg cgc ctt gcc ttt ggg gta gga ctt 912
Glu Val Pro Thr Ile Leu Leu Met Arg Leu Ala Phe Gly Val Gly Leu
290 295 300
tgt cgc ttc ggt atg ttc aaa gca gtg cga aaa aac gcc act caa get 960
Cys Arg Phe Gly Met Phe Lys Ala Val Arg Lys Asn Ala Thr Gln Ala
305 310 315
taa attgcggatc c 974
<210> 4
<211> 318
<212> PRT
<213> Synechocystis PCC6803
<400> 4
Met Pro Glu Tyr Leu Leu Leu Pro Ala Gly Leu Ile Ser Leu Ser Leu
1 5 10 15
Ala Ile Ala Ala Gly Leu Tyr Leu Leu Thr Ala Arg Gly Tyr Gln Ser
20 25 30
Ser Asp Ser Val Ala Asn Ala Tyr Asp Gln Trp Thr Glu Asp Gly Ile
35 40 45
Leu Glu Tyr Tyr Trp Gly Asp~His Ile His Leu Gly His Tyr Gly Asp
50 55 60
Pro Pro Val Ala Lys Asp Phe Ile Gln Ser Lys Ile Asp Phe Val His
6
CA 02378657 2002-O1-08
65 70 75 80
Ala Met Ala Gln Trp Gly Gly Leu Asp Thr Leu Pro Pro Gly Thr Thr
85 90 95
Val Leu Asp Val Gly Cys Gly Ile Gly Gly Ser Ser Arg Ile Leu Ala
100 105 110
Lys Asp Tyr Gly Phe Asn Val Thr Gly Ile Thr Ile Ser Pro Gln Gln
115 120 125
Val Lys Arg Ala Thr Glu Leu Thr Pro Pro Asp Val Thr Ala Lys Phe
130 135 140
Ala Val Asp Asp Ala Met Ala Leu Ser Phe Pro Asp Gly Ser Phe Asp
145 150 155 160
Val Val Trp Ser Val Glu Ala Gly Pro His Met Pro Asp Lys Ala Val
165 170 175
Phe Ala Lys Glu Leu Leu Arg Va1 Val Lys Pro Gly Gly Ile Leu Val
180 185 190
Val Ala Asp Trp Asn Gln Arg Asp Asp Arg Gln Val Pro Leu Asn Phe
195 200 205
Trp Glu Lys Pro Val Met Arg Gln Leu Leu Asp Gln Trp Ser His Pro
210 215 220
Ala Phe Ala Ser Ile Glu Gly Phe Ala Glu Asn Leu Glu Ala Thr Gly
225 230 235 240
Leu Val Glu Gly Gln Val Thr Thr Ala Asp Trp Thr Val Pro Thr Leu
245 250 255
Pro Ala Trp Leu Asp Thr Ile Trp Gln Gly Ile Ile Arg Pro Gln Gly
260 265 270
Trp Leu Gln Tyr Gly Ile Arg Gly Phe Ile Lys Ser Val Arg Glu Val
275 280 285
Pro Thr Ile Leu Leu Met Arg Leu Ala Phe Gly Val Gly Leu Cys Arg
290 295 300
Phe Gly Met Phe Lys Ala Val Arg Lys Asn Ala Thr Gln Ala
305 310 3i5
7
w CA 02378657 2002-O1-08
<210> 5
<211> 27
<212> DNA
<213> Synechocystis PCC6803
<220>
<221> primer_bind
<222> (1)..(27)
<400> 5
ggatccatgc ccgagtattt gcttctg 27
<210> 6
<211> 26
<212> DNA
<213> Synechocystis PCC6803
<220>
<221> primer_bind
<222> (1)..(26)
<400> 6
ggatccgcaa tttaagcttg agtggc 26
<210> 7
<211> 930
<212> DNA
<213> Synechocystis PCC6803
<220>
<221> CDS
<222> (i0)..(915)
<400> 7
gatatcacc atg gcc get gga ctg tat ctc cta act gcc cgg ggc tat cag 51
Met Ala Ala Gly Leu Tyr Leu Leu Thr Ala Arg Gly Tyr Gln
1 5 10
tca tcg gat tcc gtg gcc aac gcc tac gac caa tgg aca gag gac ggc 99
Ser Ser Asp Ser Val Ala Asn Ala Tyr Asp Gln Trp Thr Glu Asp Gly
15 20 25 30
att ttg gaa tat tac tgg ggc gac cat atc cac ctc ggc cat tat ggc 147
Ile Leu Glu Tyr Tyr Trp Gly Asp His Ile His Leu Gly His Tyr Gly
35 40 45
8
CA 02378657 2002-O1-08
r
gat ccg cca gtg gcc aag gat ttc atc caa tcg aaa att gat ttt gtc 195
Asp Pro Pro Val Ala Lys Asp Phe Ile Gln Ser Lys Ile Asp Phe Val
50 55 60
cat gcc atg gcc cag tgg ggc gga tta gat aca ctt ccc ccc ggc aca 243
His Ala Met Ala Gln Trp Gly Gly Leu Asp Thr Leu Pro Pro Gly Thr
65 70 75
acg gta ttg gat gtg ggt tgc ggc att ggc ggt agc agt cgc att ctc 291
Thr Val Leu Asp Val G1y Cys Gly Ile Gly Gly Ser Ser Arg Ile Leu
80 85 90
gcc aaa gat tat ggt ttt aac gtt acc ggc atc acc att agt ccc caa 339
Ala Lys Asp Tyr Gly Phe Asn Val Thr Gly Ile Thr Ile Ser Pro Gln
95 100 105 110
cag gtg aaa cgg gcg. acg gaa tta act cct ccc gat.gtg acg gcc aag 387
Gln Val Lys Arg Ala Thr Glu Leu Thr Pro Pro Asp Val Thr Ala Lys
115 120 125
ttt gcg gtg gac gat get atg get ttg tct ttt cct gac ggt agt ttc 435
Phe Ala Val Asp Asp Ala Met Ala Leu Ser Phe Pro Asp Gly Ser Phe
130 135 140
gac gta gtt tgg tcg gtg gaa gca ggg ccc cac atg cct gac aaa get 483
Asp Val Val Trp Ser Val Glu Ala Gly Pro His Met Pro Asp Lys Ala
145 150 155
gtg ttt gcc aag gaa tta ctg cgg gtc gtg aaa cca ggg ggc att ctg 531
Val Phe Ala Lys Glu Leu Leu Arg Val Val Lys Pro Gly Gly Ile Leu
160 165 170
gtg gtg gcg gat tgg aat caa cgg gac gat cgc caa gtg ccc ctc aac 579
Val Val Ala Asp Trp Asn Gln Arg Asp Asp Arg Gln Val Pro Leu Asn
175 180 185 190
ttc tgg gaa aaa cca gtg atg cga caa ctg ttg gat caa tgg tcc cac 627
Phe Trp Glu Lys Pro Val Met Arg Gln Leu Leu Asp Gln Trp Ser His
195 200 205
cct gcc ttt gcc agc att gaa ggt ttt gcg gaa aat ttg gaa gcc acg 675
Pro Ala Phe Ala Ser Ile Glu Gly Phe Ala Glu Asn Leu Glu Ala Thr
210 215 220
ggt ttg gtg gag ggc cag gtg act act get gat tgg act gta ccg acc 723
Gly Leu Val Glu Gly Gln Val Thr Thr Ala Asp Trp Thr Val Pro Thr
225 230 235
9
", CA 02378657 2002-O1-08
ctc ccc get tgg ttg gat acc att tgg cag ggc att atc cgg ccc cag 771
Leu Pro Ala Trp Leu Asp Thr Ile Trp Gln Gly Ile Ile Arg Pro Gln
240 245 250
ggc tgg tta caa tac ggc att cgt ggg ttt atc aaa tcc gtg cgg gaa 819
Gly Trp Leu Gln Tyr Gly Ile Arg Gly Phe Ile Lys Ser Val Arg Glu
255 260 265 270
gta ccg act att tta ttg atg cgc ctt gcc ttt ggg gta gga ctt tgt 867
Val Pro Thr Ile Leu Leu Met Arg Leu Ala Phe Gly Val Gly Leu Cys
275 280 285
cgc ttc ggt atg ttc aaa gca gtg cga aaa aac gcc act caa get taa 915
Arg Phe Gly Met Phe Lys Ala Val Arg Lys Asn Ala Thr Gln Ala.
290 295 300
attcttaagg tcgac 930
<210> 8
<211> 301
<212> PRT
<213> Synechocystis PCC6803
<400> 8
Met Ala Ala Gly Leu Tyr Leu Leu Thr Ala Arg Gly Tyr Gln Ser Ser
1 5 10 15
Asp Ser Val Ala Asn Ala Tyr Asp Gln Trp Thr Glu Asp Gly Ile Leu
20 25 30
Glu Tyr Tyr Trp Gly Asp His Ile His Leu Gly His Tyr Gly Asp Pro
35 40 45
Pro Val Ala Lys Asp Phe Ile Gln Ser Lys Ile Asp Phe Val His Ala
50 55 60
Met Ala Gln Trp Gly Gly Leu Asp Thr Leu Pro Pro Gly Thr Thr Val
65 70 75 80
Leu Asp Val Gly Cys Gly Ile Gly Gly Ser Ser Arg Ile Leu Ala Lys
85 90 , 95
Asp Tyr Gly Phe Asn Val Thr Gly Ile Thr Ile Ser Pro Gln Gln Va1
100 105 110
Lys Arg Ala Thr Glu Leu Thr Pro Pro Asp ~Val Thr Ala Lys Phe Ala
115 120 125
CA 02378657 2002-O1-08
Val Asp Asp Ala Met Ala Leu Ser Phe Pro Asp Gly Ser, Phe Asp Val
130 135 140
Val Trp Ser Val Glu Ala Gly Pro His Met Pro Asp Lys Ala Val Phe
145 150 155 160
Ala Lys Glu Leu Leu Arg Val Val Lys Pro Gly Gly Ile Leu Val Val
165 170 175
Ala Asp Trp Asn Gln Arg Asp Asp Arg Gln Val Pro Leu Asn Phe Trp
180 185 190
Glu Lys Pro Val Met Arg Gln Leu Leu Asp Gln Trp Ser His Pro Ala
195 200 205
Phe Ala Ser Ile Glu Gly Phe Ala Glu Asn Leu Glu Ala Thr Gly Leu
210 215 220
Val Glu Gly Gln Val Thr Thr Ala Asp Trp Thr Val Pro Thr Leu Pro
225 230 235 240
Ala Trp Leu Asp Thr Ile Trp Gln Gly Ile Ile Arg Pro Gln Gly Trp
245 250 255
Leu Gln Tyr Gly Ile Arg Gly Phe Ile Lys Ser Val Arg Glu Val Pro
260 265 270
Thr Ile Leu Leu Met Arg Leu Ala Phe Gly Val Gly Leu Cys Arg Phe
275 280 285
Gly Met Phe Lys Ala Val Arg Lys Asn Ala Thr Gln Ala
290 295 300
<210> 9
<211> 31
<212> DNA
<213> Svnechocystis PCC6803
<220>
<221> primer_bind
<222> (1)..(31)
<400> 9
gatatcacca tggccgctgg actgtatctc c 31
11
CA 02378657 2002-O1-08
<210> 10
<211> 31
<212> DNA
<213> Synechocystis PCC6803
<220>
<221> primer_bind
<222> (1)..(31)
<400> 10
gtcgacctta agaatttaag cttgagtggc g 31
12