Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
_1_
Title: Porcine Adenovirus Vaccine
FIELD OF THE INVENTION
This invention relates to a novel porcine adenovirus, PadV-5, and its use
as a delivery vector or vaccine.
BACKGROUND OF THE INVENTION
Viruses in the family AdenoviYidae are widely distributed all over the
world. They have been isolated from humans, several other mammalian
species, birds, amphibians are also known to be present in fish. Adenoviruses
are generally very similar to each other with some variations, mostly
between avian and mammalian viruses, in genome size and genomic
organization.
The prototype strains of the first four porcine adenovirus (PAdV)
serotypes were isolated in the 1960s (Haig et aI. (1964); Clarke et al.
(1967);
Kasza (1966)). A new serotype was identified in Japan in 1990 and designated
as PAdV-5 (Hirahara et al. (1990)). The two reference strains, HNF-61 and
HNF-70 of PAdV-5 were isolated from the same swineherd. In 1995, again in
Japan, a different research group reported the isolation of another PAdV and
after serological comparisons with PAdV-1-4 but not with PAdV-5, it was also
described as PAdV-5 (Kadoi et al. (1995)). According to the limited data
provided by restriction endonuclease analysis of the genome of the 1995
PAdV-5 strain, it is likely that this virus is not identical to the PAdV-5
isolated.
in 1990. The viruses included in this application and referred to as PAdV-5
were the HNF-61 and HNF-70 strains identified in 1990.
Porcine adenoviruses (PAdV) generally do not cause disease in swine and
the proposed use of the PAdVs as viral vector vaccines (Tuboly et al. (1993)),
especially where mucosal immune response is required, led to the extensive
study of the genomes. So far 5 PAdV serotypes have been described (Haig et
al. (1964); Clarke et al. (1967); Kasza (1966); Hirahara et al. (1990)). The
restriction endonuclease physical maps of the genomes of PAdV-1-5 have
been established (Kleiboeker et al. (1993); Reddy et al. (1993); Tuboly et al.
(1995)) and the sequence analyses of the early regions E3, the site found most
suitable for foreign gene insertion, (PAdV-4, Kleiboeker (1994); PAdV-3
(Reddy et al. (1995); PAdV-1-2 (Reddy et al. (1997)), E1 (PAdV-4, (Kleiboeker
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-2-
1995)); PAdV-3, Reddy et al. (1998b)) and E4 (PAdV-3 (Reddy et al. (1997))
have also been carried out. Most recently, the sequence of the entire genome
of PAdV-3 has been published (Reddy et al. (1998a)).
Further, recently PAdV-3 was developed into helper-dependent (Reddy
et al. (1999a)) and -independent expression vectors (Reddy et al. (1999b);
Hammond et al. (2000)). The use of helper independent viral vectors as
vaccines is more practical. So far, two viral genes have been expressed by
such PAdV-3 vectors. The gD gene of the Aujeszky's disease virus was
inserted into the E3 region (Reddy et al. (1999b)), and the E2 gene of the
classical swine fever virus was inserted near the right hand terminus of the
viral genome (Hammond et al. (2000)). Both recombinant viruses expressed
the foreign gene to the some extent, proving that PAdVs could be used as
such vectors. Despite these results, the widespread occurrence of serotype 3
in the swine populations and the pre-existing PAdV-3 specific virus
neutralizing antibodies may restrict the use of this serotype as a vector
vaccine. Consequently, what is needed is an improved vector for
immunization of swine.
SUMMARY OF THE INVENTION
The virus which is the subject of the present invention belongs to
serotype 5 of porcine adenoviruses (PAdV-5), and was originally isolated in
Japan (Hirahara et al. (1990)). There is no further report on the presence of
PAdV-5 elsewhere around the world. The present inventors were the first to
sequence the genome of PAdV-5. The inventors further determined that at
Ieast 60% of the E3 region is not essential for virus replication, increasing
the
theoretical vector capacity of PAdV-5 to 2.9 kb, which is much larger than the
figure given for PAdV-3 (Reddy et al. (1999b)).
Accordingly, the present invention provides an isolated porcine
adenovirus serotype 5 (PAdV-5) having a nucleic acid sequence shown in
Figure 7 or SEQ.ID.N0.:1, or a homolog or analog thereof. In a preferred
embodiment, the nucleic acid sequence of the PAdV-5 comprises:
(a) a nucleic acid sequence as shown in Figure 7 (SEQ.ID.N0.:1),
wherein T can also be U;
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-3-
(b) a nucleic acid sequence that is complimentary to a nucleic
acid sequence of (a);
(c) a nucleic acid sequence that has substantial sequence
homology to a nucleic acid sequence of (a) or (b);
(d) a nucleic acid sequence that is an analog of a nucleic acid
sequence of (a), (b) or (c); or
(e) a nucleic acid sequence that hybridizes to a nucleic acid
sequence of (a), (b), (c) or (d) under stringent hybridization conditions.
The present inventors have identified the E3 region of PAdV-5 and
successfully inserted the TGEV S gene into the virus and successfully
generated TGEV specific antibodies in a recipient pig using the recombinant
PAdV-5 virus.
Accordingly, in another aspect, the present invention provides a
recombinant porcine adenovirus serotype 5, comprising a heterologous
nucleic acid sequence that is stably integrated into the recombinant porcine
adenovirus genome. Preferably the site of integration of the heterologous
nucleic acid sequence is in a non-essential region, such as the E3 region,
more
preferably between map units at about 75 and about 82 as shown in Figure 10
or in the E3 region as shown in Figure 13 (SEQ.ID.N0.:8) or 14
(SEQ.1D.N0.:9).
The present invention also includes modified forms of the isolated PAdV-
5 shown in Figure 7 (SEQ.1D.NO.:1) or modified forms of the analogs or
homologs as described above. Examples of modified forms include an
isolated PAdV-5 wherein the E3 region has been deleted.
According to one embodiment, the recombinant porcine adenovirus
serotype 5 (PAdV-5) includes a live porcine adenovirus having virion
structural proteins unchanged from those in a native porcine adenovirus
from which the recombinant porcine adenovirus is derived.
In one embodiment, the recombinant PAdV-5 of the invention comprises
a heterologous nucleic acid sequence that encodes an antigenic determinant
from an infectious agent. In another embodiment, the recombinant PAdV-5
further comprises a nucleic sequence encoding an immuno-potentiating
molecule where the molecule is preferably interleukin 3 (IL-3), porcine
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-4-
interleukin 4 (IL4), gamma interferon (yIFN), porcine granulocyte
macrophage colony stimulating factor (GM-CSF), or porcine granulocyte
colony stimulating factor (G-CSF).
In another aspect, the present invention provides a method of producing
a recombinant porcine adenovirus vector for use as a vaccine comprising
inserting into a non-essential region of a porcine adenovirus serotype 5
genome, at least one heterologous nucleic acid sequence preferably in
association with an effective promoter sequence. Preferably the heterologous
sequence encodes an antigenic polypeptide and/or an immuno-potentiating
molecule, preferably the heterologous nucleotide sequence encoding an
antigenic polypeptide encodes determinants of infectious agents and
preferably the nucleotide sequence encoding an immuno-potentiating
molecule is interleukin 3 (IL-3), porcine interleukin 4 (IL4), gamma
interferon
(yIFN), porcine granulocyte macrophage colony stimulating factor (GM-CSF),
or porcine granulocyte colony stimulating factor (G-CSF).
In yet another aspect, the present invention provides the use of the
recombinant PAdV-5 of the invention in the preparation of a vaccine for
generating and/or optimising antibodies or cell mediated immunity so as to
provide or enhance protection against infection by an infectious organism in
animals, where the vaccine includes recombinant porcine adenovirus
serotype 5 stably incorporating, at least one heterologous nucleotide
sequence, and suitable carriers and/or excipients. Preferably the at least one
heterologous nucleotide encodes an antigenic polypeptide, more preferably
antigenic determinants of infectious agents. Accordingly, the present
invention includes a vaccine for eliciting or enhancing an immune response to
an antigen comprising an effective amount of a recombinant porcine
adenovirus serotype 5 comprising a nucleic acid sequence encoding the
antigen, preferably in admixture with a suitable diluent or carrier.
In a further aspect the present invention provides a method of eliciting or
enhancing an immune response to an antigen comprising administering an
effective amount of a recombinant porcine adenovirus serotype 5 comprising
a nucleic acid sequence encoding the antigen to an animal in need thereof.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-5-
Other features and advantages of the present invention will become
apparent from the following detailed description. It should be understood,
however, that the detailed description and the specific examples while
indicating preferred embodiments of the invention are given by way of
illustration only, since various changes and modifications within the spirit
and
scope of the invention will become apparent to those skilled in the art from
this detailed description.
BRIEF DESCRIPTION OF THE DRAWINGS
The invention will now be described in relation to the drawings in which:
Figure 1A illustrates a PAdV-5 map of HindIII and MIuI restriction map of
the genome, m.u.: map unit,1 m.u.: 335 bp;
Figure 1B is an enlargement of sequenced region of the map of Figure
1A;
Figure 1C illustrates reading frames of the r strand of PAdV-5;
Figure 1D illustrates the portion of the DNA removed to generate the
clones;
Figure 2 illustrates the alignment of the predicted ORF2 amino acid
sequences of PAdV-5 HNF-70 (SEQ.ID.N0.:2) and some closely related animal
adenoviruses (SEQ.ID.NOs.:3-5);
Figure 3 illustrates the sequence alignment of the predicted ORF3
proteins of HNF-61 (SEQ.ID.N0.:6) and HNF-70 (SEQ.ID.N0.:7);
Figure 4 illustrates the unrooted phylogenetic tree of pVIII protein
homologues of selected animal adenovirus generated by the Clustal method;
Figure 5 illustrates the time course analysis of PAdV-5 HNF-70 nucleic
acid synthesis;
Figure 6 illustrates the restriction endonuclease analysis of the wild type
PAdV-5 HNF-70 strain (A) and its deletion mutant R-OHH (B) genomic DNA
in ethidium bromide stained 0.8% agarose gel.
Figure 7 (SEQ.ID.N0.:1) is the complete nucleotide sequence of porcine
adenovirus serotype 5 (PAV-5) strain HNF-70.
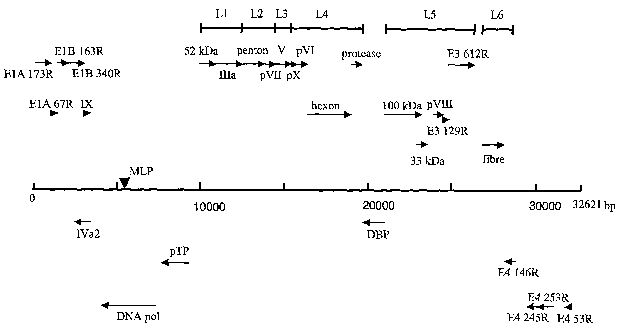
Figure 8 illustrates the genome organization and putative transcription
map of PadV-5. Arrows above and below the central line represent the
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-6-
locations of putative ORF's. The location of the major late promoter (MLP) is
indicated. Late regions (L1-L6) are indicated by lines.
Figure ~9' illustrates a phylogenetic analysis of the pVIII genes of selected
adenoviruses. The lengths of the branches indicate the phylogenetic distance
between the viruses. T'he scale bar represents 10 mutations per 100 sequence
positions. Virus names not defined elsewhere are: CELOV, CELO virus (fowl
adenovirus 1); EDSV, egg drop syndrome virus; FAdV, fowl adenovirus;
HEV, turkey haemorrhagic enteritis virus; MAdV, muriine adenovirus;
OAdV, ovine adenovirus.
Figure 10 illustrates the strategy for construction of the recombinant
transfer vectors.
Figure 11 is a Northern blot analysis of S gene expression showing total
RNA extracted from cells infected with recombinant virus Rl'AdV-2.2S and
ORPAdV-2.2Sc.
Figure 12 is a Western blot analysis the Rl'AdV-2.25 and ~IZPAdV-2.2Sc
recombinant virus infected cells were collected at 24 hours p..i.
Figure 13 illustrates the nucleotide sequence (SEQ.ID.N0.:8) of the E3
region of the same strain shown in Figure 7 (PAV-5, HNF-70).
Figure 14 illustrates the nucleotide sequence (SEQ.ID.N0.:9) of the E3
region of the HNF61 strain of porcine adenovirus serotype 5 (PAV-5).
DETAILED DESCRIPTION OF THE INVENTION
I. PAdV-5
As stated above, the present inventors have determined the complete
nucleotide sequence of PAdV-5 and constructed a putative genomic map.
Accordingly, the present invention provides an isolated porcine
adenovirus serotype 5 (PAdV-5) having a nucleic acid sequence shown in
Figure 7 or SEQ.ID.N0.:1, or a homolog or analog thereof.
The term "isolated" refers to a nucleic acid substantially free of cellular
material or culture medium when produced by recombinant DNA techniques
or chemical precursors or other chemicals when chemically synthesized.
The term "nucleic acid sequence" refers to a sequence of nucleotide or
nucleoside monomers consisting of naturally occurring bases, sugars and
intersugar (backbone) linkages. The term also includes modified or
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
_7-
substituted sequences comprising non-naturally occurring monomers or
portions thereof, which function similarly. The nucleic acid sequences of the
present invention may be ribonucleic (RNA) or deoxyribonucleic acids (DNA)
and may contain naturally occurring bases including adenine, guanine,
cytosine, thymidine and uracil. The sequences may also contain modified
bases such as xanthine, hypoxanthine, 2-aminoadenine, 6-methyl, 2-propyl,
and other alkyl adenines, 5-halo uracil, 5-halo cytosine, 6-aza uracil, 6-aza
cytosine and 6-aza thymine, pseudo uracil, 4-thiouracil, 8-halo adenine, 8-
amino adenine, 8-thiol adenine, 8-thio-alkyl adenines, 8-hydroxyl adenine and
other 8-substituted adenines, 8-halo guanines, 8-amino guanine, 8-thiol
guanine, 8-thioalkyl guanines, 8-hydroxyl guanine and other 8-substituted
guanines, other aza and deaza uracils, thymidines, cytosines, adenines, or
guanines, 5-trifluoromethyl uracil and 5-trifluoro cytosine.
In a preferred embodiment, the PAdV-5 nucleic acid sequence comprises:
(a) a nucleic acid sequence as shown in Figure 7 (SEC,~.ID.N0.:1),
wherein T can also be U;
(b) a nucleic acid sequence that is complimentary to a nucleic
acid sequence of (a);
(c) a nucleic acid sequence that has substantial sequence
homology to a nucleic acid sequence of (a) or (b);
(d) a nucleic acid sequence that is an analog of a nucleic acid
sequence of (a), (b) or (c); or
(e) a nucleic acid sequence that hybridizes to a nucleic acid
sequence of (a), (b), (c) or (d) under stringent hybridization conditions.
The term "sequence that has substantial sequence homology" means
those nucleic acid sequences which have slight or inconsequential sequence
variations from the sequences in (a) or (b), i.e., the sequences function in
substantially the same manner (e.g. useful as a vector or vaccine). The
variations may be attributable to local mutations or structural modifications.
Nucleic acid sequences having substantial homology include nucleic acid
sequences having at least 65%, more preferably at least 85%, and most
preferably 90-95% identity with the nucleic acid sequences as shown in Figure
7 (SEQ.ID.N0.:1).
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
_$_
The term "sequence that hybridizes" means a nucleic acid sequence that
can hybridize to a sequence of (a), (b), (c) or (d) under stringent
hybridization
conditions. Appropriate "stringent hybridization conditions" which promote
DNA hybridization are known to those skilled in the art, or may be found in
Current Protocols in Molecular Biology, John Wiley & Sons, N.Y. (1989), 6.3.1-
6.3.6. For example, the following may be employed: 6.0 x sodium
chloride/sodium citrate (SSC) at about 45°C, followed by a wash of 2.0
x SSC
at 50°C. The stringency may be selected based on the conditions used in
the
wash step. For example, the salt concentration in the wash step can be
selected from a high stringency of about 0.2 x SSC at 50°C. In
addition, the
temperature in the wash step can be at high stringency conditions, at about
65°C.
The term "a nucleic acid sequence which is an analog" means a nucleic acid
sequence which has been modified as compared to the sequence of (a), (b) or
(c) wherein the modification does not alter the utility of the sequence (i.e.
as a
vector or vaccine) as described herein. The modified sequence or analog may
have improved properties over the sequence shown in (a), (b) or (c). One
example of a modification to prepare an analog is to replace one of the
naturally occurring bases (i.e. adenine, guanine, cytosine or thymidine) of
the
sequence shown in Figure 7 with a modified base such as such as xanthine,
hypoxanthine, 2-aminoadenine, 6-methyl, 2-propyl and other alkyl adenines,
5-halo uracil, 5-halo cytosine, 6-aza uracil, 6-aza cytosine and 6-aza
thymine,
pseudo uracil, 4-thiouracil, 8-halo adenine, 8-aminoadenine, 8-thiol adenine,
8-
thiolalkyl adenines, 8-hydroxyl adenine and other 8-substituted adenines, 8-
halo guanines, 8 amino guanine, 8-thiol guanine, 8-thiolalkyl guanines, 8-
hydroxyl guanine and other 8-substituted guanines, other aza and deaza
uracils, thymidines, cytosines, adenines, or guanines, 5-trifluoromethyl
uracil
and 5-trifluoro cytosine.
Another example of a modification is to include modified phosphorous or
oxygen heteroatoms in the phosphate backbone, short chain alkyl or
cycloalkyl intersugar linkages or short chain heteroatomic or heterocyclic
intersugar linkages in the nucleic acid molecule shown in Figure 7. For
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-9-
example, the nucleic acid sequences may contain phosphorothioates,
phosphotriesters, methyl phosphonates, and phosphorodithioates.
A further example of an analog of a nucleic acid molecule of the invention
is a peptide nucleic acid (PNA) wherein the deoxyribose (or ribose) phosphate
backbone in the DNA (or RNA), is replaced with a polyamide backbone
which is similar to that found in peptides (P.E. Nielsen, et al Science 1991,
254,
1497). PNA analogs have been shown to be resistant to degradation by
enzymes and to have extended lives in vivo and in vitro. PNAs also bind
stronger to a complimentary DNA sequence due to the lack of charge
repulsion between the PNA strand and the DNA strand. Other nucleic acid
analogs may contain nucleotides containing polymer backbones, cyclic
backbones, or acyclic backbones. For example, the nucleotides may have
morpholino backbone structures (U.S. Pat. No. 5,034,506). The analogs may
also contain groups such as reporter groups, a group for improving the
pharmacokinetic or pharmacodynamic properties of nucleic acid sequence.
The present invention also includes modified forms of the isolated PAdV-
5 shown in Figure 7 (SEQ.ID.N0.:1) or modified forms of the analogs or
homologs as described above. Examples of modified forms include an
isolated PAdV-5 wherein the E3 region has been deleted. Such a modified
form can be used to insert a heterologous gene for the preparation of a
vaccine as described below. Accordingly, the present invention provides a
modified PAdV-5 wherein the E3 region, or a portion thereof, has been
deleted. In one embodiment, the E3 region that is deleted is as shown in
Figure 13 (SEQ.ID.N0.:8) or Figure 14 (SEQ.ID.N0.:9).
II. Recombinant PAdV-5
;. The isolated PAdV-5 of the invention is useful in preparing a recombinant
PAdV-5 vector for the insertion of heterologous nucleic acid sequences of
interest and the expression of the heterologous sequence in a host. In
particular, the inventors have identified the E3 region of PAdV-5 and
successfully inserted the TGEV S gene into the virus and successfully
generated TGEV specific antibodies in a recipient pig using the recombinant
PAdV-5 virus.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-10-
Accordingly, in another aspect, the present invention provides a
recombinant porcine adenovirus serotype 5, comprising a heterologous
nucleic acid sequence that is stably integrated into the recombinant porcine
adenovirus genome. Preferably the site of integration of the heterologous
nucleic acid sequence is in a non-essential region of the viral genome, most
preferably in the E3 region. When integrated in the E3 region, it is
preferably
between map units at about 75 and about 82 as shown in Figure 10 or in the
E3 region as shown in Figure 13 (SEQ.ID.N0.:8) or Figure 14 (SEQ.ID.NO.:9).
The terms "heterologous nucleic acid sequence" includes one or more
sequences that are not normally present in the PAdV-5 sequence in nature.
Preferably, the heterologous nucleic acid sequences encode the antigenic
determinants of infectious organisms against which the generation of
antibodies or cell-mediated immunity is desirable, such as antigenic
determinants of intestinal infections caused by gastrointestinal viruses; for
example rotavirus and parvovirus infections, or respiratory viruses, for
example influenza virus and porcine reproductive and respiratory syndrome
virus (PRRSV) or that of hog cholera virus (classical swine fever).
Heterologous nucleotide sequences which may be incorporated include,
but are not limited to, the antigenic determinants of the agents of: porcine
parvovirus; mycoplasma hyopneumonia; porcine influenza virus;
transmissible gastroenteritis virus (porcine coronavirus); porcine rotavirus;
hog cholera virus (classical swine fever); swine dysentery; African swine
fever
virus; pseudorabies virus (Aujeszky's disease virus), in particular the
glycoprotein D of the pseudorabies virus; porcine respiratory and
reproductive syndrome virus (PRRSV); and porcine circovirus (Postweaning
multisystemic wasting syndrome).
Heterologous nucleotide sequences more preferred for incorporation in
the vectors of the invention are those expressing antigenic determinants of
porcine parvovirus, porcine rotavirus, TGEV (porcine coronavirus) and
classical swine fever virus. Most preferred, are heterologous nucleotide
sequences expressing the antigenic determinants of TGEV.
The heterologous nucleic acid sequences incorporated may encode
immuno-potentiator molecules such as cytokines or growth promoters, for
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-11-
example porcine interleukin 4 (IL4), gamma interferon (yIFN), granulocyte
macrophage colony stimulating factor (GM-CSF), granulocyte colony
stimulating factor (G-CSF), FLT-3 ligand and interleukin 3 (IL-3).
It is to be understood that the heterologous nucleic acid sequence can
comprise both heterologous genes coding for antigenic determinants and
immuno-potentiator molecules.
Non-essential regions of the viral genome which may be suitable for the
purposes of replacement with or insertion of heterologous nucleic acid
sequences may for example be non-coding regions at the right terminal end
of the genome at map units between about 75 to about 82, preferably 75.7
and 81.7, of the genome-spanning parts of the HindIII F and D fragments.
The heterologous gene sequences may be associated with a promoter
and leader sequence in order that the nucleotide sequence may be expressed
in situ as efficiently as possible. Preferably the heterologous gene sequence
is
associated with the porcine adenoviral major late promoter and splice leader
sequence. The mammalian adenovirus major late promoter lies near 16-17
map units on the adenovirus genetic map and contains a classical TATA
sequence motif (Johnson, D.C., Ghosh-Chondhury, G., Smiley, J.R., Fallis, L.
and Graham, F.L. (1988), Abundant expression of herpes simplex virus
glycoprotein gB using an adenovirus vector. Virology 164,1-14).
Instead of the porcine adenoviral major late promoter, any other suitable
eukaryotic promoter may be used. For example, those of SV40 virus,
cytomegalovirus (CMV) or human adenovirus may be used.
The splice leader sequence of the porcine adenovirus serotype under
consideration is a tripartite sequence spliced to the 5' end of the mRNA of
all
late genes. The heterologous gene sequence may also be associated with a
poly-adenylation sequence.
One skilled in the art will appreciate that other components or sequences
may be included in the recombinant adenovirus and can be determined by
one of skill in the art. Examples of methods for constructing an adenovirus
vector encoding heterologous nucleic acid molecules are described in U.S.
Patent No. 4,920,209 and 6,086,890.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-12-
III. Uses
The invention includes all of the uses of the isolated PAdV-5, the modified
PAdV-5 and the recombinant PAdV-5 vectors of the inventions including the
use thereof as a vaccine.
Accordingly, in a further aspect of the invention there is provided a
recombinant PAdV-5 vaccine for generating and/or optimising antibodies or
cell-mediated immunity so as to provide or enhance protection against
infection with an infectious organism in animals, the vaccine comprising a
recombinant porcine adenovirus serotype 5 vector stably incorporating at
least one heterologous nucleic acid sequence formulated with suitable carriers
and excipients. Preferably the heterologous nucleic acid sequence encodes an
antigenic polypeptide and/or an immuno-potentiator molecule.
The recombinant vaccine may include a live recombinant porcine
adenovirus vector in which the virion structural proteins are unchanged from
that in the native porcine adenovirus from which the recombinant porcine
adenovirus is produced.
The vaccine may be directed against any infectious organism and/or
agent, for example, infectious organisms and/or agents causing respiratory
and/or intestinal infections. In order to direct the vaccine against a
specific
infectious organism, heterologous gene sequences encoding the antigenic
determinants of those infectious organisms may be incorporated into non
essential regions of the genome of the porcine adenovirus serotype 5
comprising the vector. If the vaccine is to be used to optimize protection
against disease, suitable heterologous nucleotide sequences may also be those
of immuno-potentiators such as cytokines or growth promoters.
In a further aspect of the invention, there is provided a method of
preparing a vaccine for generation and/or optimization of antibodies or cell-
mediated immunity so as to induce or enhance protection against an
infectious organism in an animal, which includes constructing a recombinant
porcine adenovirus serotype 5 vector stably incorporating at least one
heterologous nucleotide sequence, and placing said recombinant porcine
adenovirus vector in a form suitable for administration. Preferably the
nucleotide sequence encodes an antigenic polypeptide, although it may also
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-13-
be , an immuno-potentiator molecule. More preferably, the nucleotide
sequences may encode for and/or express, an antigenic polypeptide and an
immuno-potentiator molecule. The nucleotide sequence is conveniently
foreign to the host vector
Accordingly, the present invention includes a vaccine for eliciting or
enhancing an immune response to an antigen comprising an effective amount
of a recombinant porcine adenovirus serotype 5 comprising a nucleic acid
sequence encoding the antigen, preferably in admixture with a suitable
diluent or carrier. The present invention also provides a method of eliciting
or enhancing an immune response to an antigen comprising administering an
effective amount of a recombinant porcine adenovirus serotype 5 comprising
a nucleic acid sequence encoding the antigen to an animal in need thereof.
The term "enhancing or eliciting an immune response" is defined as
enhancing, improving or augmenting any response of the immune system,
for example, of either a humoral or cell-mediated nature. The enhancement
of an immune response can be assessed using assays known to those skilled
in the art including, but not limited to, antibody assays (for example ELISA
assays), antigen specific cytotoxicity assays and the production of cytokines
(for example ELISPOT assays).
Administration of an "effective amount" of the vaccine of the present
invention is defined as an amount effective, at dosages and for periods of
time necessary to achieve the desired result (e.g., to elicit or enhance an
immune response). The effective amount of a compound of the invention
may vary according to factors such as the disease state, age, sex, and weight
of the animal. Dosage regime may be adjusted to provide the optimum
therapeutic response. For example, several divided doses may be
administered daily or the dose may be proportionally reduced as indicated by
the exigencies of the therapeutic situation. The dose of the vaccine may also
be varied to provide optimum preventative dose response depending upon
the circumstances. For example an effective amount is an amount sufficient
to elicit an immune response, preferably at least 104 TCIDso per dose.
The term "animal" includes all members of the animal kingdom and is
preferably a pig.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-14-
The vaccine may be a multivalent vaccine and additionally contain
heterologous nucleic acid sequences encoding immunogens related to other
intracellular viral, parasitic and bacterial infectious diseases in a
prophylactically or therapeutically effective manner. Further, as will be
readily understood by those skilled in the art, any one recombinant
adenovirus can contain the expressible nucleic acid sequences of more than
one microbial antigen and/or more than one immuno-potentiator molecule.
The vaccines of the present invention may additionally contain suitable
diluents and/or carriers. Preferably, the vaccines contain one or more other
adjuvants, which can further enhance the immunogenicity of the vaccine in
vivo. These other one or more adjuvants may be selected from many known
adjuvants in the art including the lipid-A portion of the LPS from gram
negative bacteria (endotoxin), trehalose dimycolate of mycobacteria, the
phospholipid lysolecithin, dimethyldictadecyl ammonium bromide (DDA),
certain linear polyoxypropylene-polyoxyethylene (POP-POE) block polymers,
aluminum hydroxide, and liposomes. The vaccine may also contain
preservatives such as sodium azide, thimersol, beta propiolactone, and binary
ethyleneimine.
A vaccine of the invention is suitable for administration to subjects in a
biologically compatible form in vivo. The expression "biologically compatible
form suitable for administration in vivo" as used herein means a form of the
substance to be administered in which any toxic effects are outweighed by the
therapeutic effects.
The vaccines may be administered in a convenient manner such as by
injection (intradermal, subcutaneous, intravenous, intramuscular,
intraperitoneal, intranodal etc.), oral administration, inhalation,
transdermal
administration (such as topical cream or ointment, etc.), or suppository
applications.
Accordingly, the invention, provides (i) a vaccine vector such as a
recombinant porcine adenovirus serotype 5, containing DNA molecules of
the invention, placed under the control of elements required for expression
(if
necessary); (ii) a composition of matter containing a vaccine vector of the
invention, together with a diluent or carrier; particularly, (iii) a
pharmaceutical
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-15-
composition containing a therapeutically or prophylactically effective amount
of a vaccine vector; (iv) a method for inducing an immune response against
antigenic polypeptides in an animal, which involves administering to the
animal an immunogenically effective amount of a vaccine vector to elicit an
immune response, e.g., a protective or therapeutic immune response to the
antigenic polypeptide; particularly; (v) a method for preventing and/or
treating disease, which involves administering a prophylactic or therapeutic
amount of a vaccine vector containing DNA of the invention to an animal in
need; and (vi) a method for preventing and/or treating disease, which
involves administering a prophylactic or therapeutic amount of a
recombinant porcine adenovirus serotype 5 comprising at least one
heterologous nucleotide sequence, said heterologous nucleotide sequence
preferably encoding and/or expressing an antigenic determinant of an
infectious agent and/or an immuno-potentiating molecule.
The vaccine of the invention may of course be combined with vaccines
against other viruses or organisms such as parvovirus or Aujeszky's disease
at the time of its administration.
In vaccines of the present invention the recombinant adenovirus or
antigens may be in admixture with a suitable carrier, diluent, or excipient
such
as sterile water, physiological saline, glucose or the like. The vaccines can
also
be lyophilized. The vaccines can contain auxiliary substances such as wetting
or emulsifying agents, pH buffering agents, adjuvants, gelling or viscosity
enhancing additives, preservatives, flavoring agents, colors, and the like,
depending upon the route of administration and the preparation desired. The
vaccines can contain at least one adjuvant compound chosen from the
polymers of acrylic or methacrylic acid and the copolymers of malefic
anhydride and alkenyl derivative. Adjuvant compounds are the polymers of
acrylic or methacrylic acid which are cross-linked, especially with
polyalkenyl
ethers of sugars or polyalcohols. These compounds are known by the term
carbomer (Phameuropa Vol. 8, No. 2, June 1996). Persons skilled in the art can
also refer to U.S. Patent No. 2,909,462 (incorporated herein by reference)
which describes such acrylic polymers cross-linked with a polyhydroxylated
compound having at least 3 hydroxyl groups, preferably not more than 8, the
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-16-
hydrogen atoms of at least three hydroxyls being replaced by unsaturated
aliphatic radicals having at least 2 carbon atoms. The preferred radicals are
those containing from 2 to 4 carbon atoms, e.g. vinyls, allyls and other
ethylenically unsaturated groups. The unsaturated radicals may themselves
contain other substituents, such as methyl. The products sold under the name
Carbopol( (BF Goodrich, Ohio, USA) are particularly appropriate. They are
cross-linked with an allyl sucrose or with allyl pentaerythritol. Among then,
there may be mentioned Carbopol( 974P, 934P and 971P). Among the
copolymers of malefic anhydride and alkenyl derivative, the copolymers
EMA( (Monsanto) which are copolymers of malefic anhydride and ethylene,
linear or cross-linked, for example cross-linked with divinyl ether, are
preferred. Reference may be made to J. Fields et al., Nature, 186: 778-780, 4
June 1960, incorporated herein by reference. Adjuvants useful in any of the
vaccine compositions described herein are as follows.
Adjuvants for parenteral administration include aluminum compounds,
such as aluminum hydroxide, aluminum phosphate, and aluminum hydroxy
phosphate. The antigen can be precipitated with, or adsorbed onto, the
aluminum compound according to standard protocols. Other adjuvants, such
as RIBI (ImmunoChem, Hamilton, MT), can be used in parenteral
administration.
Adjuvants for mucosal administration include bacterial toxins (e.g., the
cholera toxin (CT), the E. coli heat-labile toxin (LT), the Clostridium
difficile toxin
A and the pertussis toxin (PT), or combinations, subunits, toxoids, or mutants
thereof). For example, a purified preparation of native cholera toxin subunit
B (CTB) can be of use. Fragments, homologs, derivatives, and fusions to any
of these toxins are also suitable, provided that they retain adjuvant
activity.
Preferably, a mutant having reduced toxicity is used. Suitable mutants have
been described (e.g., in WO 95/17211 (Arg-7-Lys CT mutant), WO 96/6627
(Arg-192-Gly LT mutant), and WO 95/34323 (Arg-9-Lys and Glu-129-Gly PT
mutant)). Additional LT mutants that can be used in the methods and
compositions of the invention include, for example Ser-63-Lys, Ala-69-Gly,
Glu-110-Asp, and Glu-112-Asp mutants. Other adjuvants (such as a bacterial
monophosphoryl lipid A (MPLA) of various sources (e.g., E. coli, Salmonella
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-17-
minnesota, Salmonella typhimurium, or Shigella flexneri). saponins, or
polylactide
glycolide (PLGA) microspheres), can also be used in mucosal administration.
Adjuvants useful for both mucosal and parenteral administrations include
polyphosphazene (for example, WO 95/2415), DC-chol (3 b-(N-(N',N'
dimethyl aminomethane)-carbamoyl) cholesterol (for example, U.S. Patent
No. 5,283,185 and WO 96/14831)) and QS-21 (for example, WO 88/9336).
The Th1 cell-mediated immune response is considered to play a pivotal
role in pig defense against mycobacterial infection and the development of
such immune responses is believed to involve (1) antigen presentation by
antigen-presenting cells (APC) including macrophages and dendritic cells to
antigen-specific T cells; (2) T cell activation and cytokine release; and (3)
enhanced bactericidal activities of macrophages by cytokines (immuno-
potentiator molecules) released from T cells (Monk et al. (1995)). Cytokines
including interleukin- 12 (IL-12), interferon (IFN) and granulocyte-
macrophage colony stimulating factor (GM-CSF) orchestrate in the
development of anti- mycobacterial Th1 immune responses. IL-12 is usually
released by APC upon interaction with infectious pathogens and is a crucial
Th1 differentiation and activation factor. Thus, antigen presentation and IL-
12
release by APC will result in the release of Th1 cytokine lFN from Th1
lymphocytes. IFN is a potent macrophage-activation cytokine capable of
enhancing bactericidal activities of macrophages. GM-CSF was originally
identified as a hematopoietic growth factor but recent evidence indicates that
it is a critical cytokine required for effective antigen presentation by
enhancing dendritic cell differentiation and APC activation via increasing
cell
surface expression of MHC II and B7 molecules (Peters et al. (1996)).
Recent studies have indicated that a prime/boost protocol, whereby
immunization with a adenovirus recombinant expressing a foreign gene
product is followed by a boost using a purified subunit preparation form of
that gene product, elicits an enhanced immune response relative to the
response elicited with either product alone. Accordingly, it is within the
scope
of the present invention to use a prime/boost protocol. A methodology of
prime/boost protocol is described in WO 98/58956, which is incorporated
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-18-
herein by reference. Alternatively, repeated vaccination with the same
recombinant adenovirus could be used as the boost.
The vaccines or compositions described herein can be prepared by per se
known methods for the preparation of pharmaceutically acceptable
compositions which can be administered to subjects, such that an effective
quantity of the active material is combined in a mixture with a
pharmaceutically acceptable vehicle. Suitable vehicles are described, for
example, in Remington's Pharmaceutical Sciences (Remington's
Pharmaceutical Sciences, Mack Publishing Company, Easton, Pa., USA 1985).
Ch1 this basis, the compositions include, albeit not exclusively, solutions of
the
active material in association with one or more pharmaceutically acceptable
vehicles or diluents, and contained in buffered solutions with a suitable pH
and iso-osmotic with the physiological fluids. In this regard, reference is
made to U.S. Patent No. 5,843,456, incorporated herein by reference, and
directed to rabies compositions and combination compositions and uses
thereof.
The utility of the compositions of the invention may be confirmed in
experimental model systems.
Animals are conveniently inoculated with vector vaccines according to
the invention at any age. For example, piglets may be vaccinated at 1 day
old, breeders may be vaccinated regularly up to point of giving birth and
thereafter.
Preferably animals are vaccinated while still not fully immunocompetent.
More conveniently, animals can be vaccinated for protection against re-
infection after a period of 4 weeks subsequent to initial vaccination.
In a preferred aspect of this embodiment of the invention, administration
is by oral delivery or infra-nasally.
The following non-limiting examples are illustrative of the present
invention:
EXAMPLES
MATERIALS AND METHODS FOR EXAMPLES 1-4
Viruses and viral DNA
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-19-
The origin of the PAdV-5 prototype strains HNF-61 and HNF-70 has
been described (Tuboly et al., 1995). Both strains were propagated in a
continuous swine testicle (ST) cell line (McClurkin et al., 1966) and the
intracellular viral DNA was extracted at the peak of infection by the method
of Hirt (1967).
DNA cloning and seguence analysis
The cloning of the MIuI and HindllI generated genomic DNA fragments
of HNF-61 and HNF-70 into the pGEM-7Zf(+) plasmid (Promega) has been
described (Tuboly et al., 1995). Nested set deletions of 300-400 basepairs
(bp)
of the HindIII F and D fragments were generated in both orientations by
using the ExoIII/S1 Deletion Kit (MBI, Fermentas, Lithuania) providing 200
300 by overlaps. The resulting clones were sequenced using the T7 and Sp6
specific primers at the Biological Research Institute, Szeged (Hungary), with
the PRISM ready reaction dye deoxy cycle protocol (Perkin Elmer) in an
automated DNA sequencer (Applied Biosystems Inc.).
Homology search of the GenBank database was performed by the Blast
program for the nucleotide and deduced amino acid (aa) sequences of the
predicted open reading frames (ORF). Sequence analysis was done with the
Seqaid II version 3.5 and the Clone Manager version 3.12 computer
programs. Sequence alignments were carried out with the Align program
version 1.02 and with the DNAstar MegAlign program. The distance matrix
phylogenetic analysis was carried out with the MegAlign program.
DNA and RNA time course
ST cells were grown to confluence in 3 cm Petri dishes and infected with
10 m.o.i. (multiplicity of infection) of the HNF-61 or HNF-70 strain of PAdV-5
and incubated for 2-24 hours at 37°C. Samples were collected every 2
hours.
For DNA dot blot analysis the supernatant was removed and 0.5 ml distilled
water was added to the cells and frozen to -70°C. Total cellular RNA
was
extracted every 2 hours post infection with an RNA extraction kit (RNeasy,
QIAGEN). RNA and DNA from mock-infected ST cells were similarly
collected. RNA and DNA samples were also collected from cells treated with
50 ~.g/ml cytosine arabinoside (AraC, Sigma) as described (Mittal et
a1.,1993).
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-20-
The start of DNA replication was detected by a dot blot method. The
frozen samples were thawed and mixed with an equal volume of 0.8 N
NaOH, incubated at 80°C for 20 minutes. Fifty ~,1 of the cell lysates
of each
time point were loaded into the wells of a dot blot filtration manifold and
blotted onto Nytran membranes (Schleicher and Schuell) as described
(Sambrook et al., 1989). The DNA was immobilized in a UV crosslinker
(Fischer) and probed with digoxigenin labeled (Boehringer Mannheim)
PAdV-5 HNF-70 genomic DNA according to the manufacturers' instructions.
The DNA was detected in a chemiluminescent reaction (Boehringer
Mannheim, Non Radioactive DNA Labeling and Detection Kit) and exposed
to Kodak X-Omat AR films.
The time course of RNA transcription was analyzed by Northern
blotting. Equal amounts of the total RNA extracted from mock and virus
infected cells of each time point were separated in 1.1% formaldehyde
agarose gels, transferred bidirectionally to Nytran membranes and
immobilized by W crosslinking. Prehybridization, hybridization in the
presence of 50% formamide and washing of the blots were carried out as
described (Sambrook et al., 1989). The E3 specific probe (fig. 1B) was the 1.9
kb EcoRI G fragment of the HNF-70 strain labeled with [a32P]dCTP (ICN) by
the random primer method (Random Primer Labeling Kit, Life Technologies).
Construction of E3 deleted PAdV 5
Plasmids containing the left (D) and right (C) terminal MluI fragments of
HNF-70 were used to construct full length genomic clones of the viral DNA
by homologuos recombination in E. coli. Briefly, the terminal fragments were
released from the plasmid by double digestions with BamHI-MIuI (MluI D
fragment) and HindIII-MluI (MIuI C fragment). These fragments were cloned
together into BamHI-HindIll digested pWEl5 cosmid vector (Stratagene,
modified by Ojkic et al., unpublished). The resulting clone was used in co-
transformation of competent E, coli BJ 5183 cells (Hanahan, 1983) together
with HNF-70 genomic DNA as described (Degryse,1996). The clones carrying
the entire PAdV-5 genome were selected and transformed into DHSa cells for
large scale DNA preparation with the Concert Nucleic Acid Purification
System (Life Technologies) according to the manufacturers' instructions. In
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-21-
order to introduce deletions into the E3 region the cloned MluI B fragment of
HNF-70 (Tuboly et al., 1995) was used. Two deletion clones were generated,
namely MluI B-x(992-2497) and MIuI B-0(1260-2497) by removing a 1505 by
PvuII-HpaI or a 1237 by HincII fragment from the original MIuI B fragment,
respectively (fig. 1D). The E3 region of the cloned full length HNF-70 DNA
was replaced by the deletion clones via homologous recombination in BJ 5183
E. coli resulting in full length genomic clones with deletions in the E3
region
(pR-Ol'H and pR-OHH). The cloned DNA was transfected into ST cells using
lipofectin reagent (Life Technologies) according to the instructions of the
manufacturer. The transfected cells were overlaid with 0.7% agarose in
DMEM supplemented with 10% fetal bovine serum. Plaque formation was
monitored daily and 6-7 days after transfection the plaques were transferred
to fresh ST cell monolayers. Viral DNA was extracted from the infected cells
by the Hirt method and analyzed with the appropriate restriction enzymes
(RE).
EXAMPLE 1
Seguence analysis
The putative E3 region of both strains of PAdV-5 was located between
75.7 and 81.7 map units (m.u.) of the genome (the full genome of PAV 5 is
shown in Figure 7), spanning parts of the HindIII F and D fragments. The
sequencing of these fragments revealed several ORFs on both the right (r)
and the left (1) strand. The 1 strand showed numerous ORFs but only five of
these had a capacity of more than 40 as and none of them exceeded 90 as (not
shown). Four complete ORFs of the r strand had a coding capacity of more
than 40 as (Figure 1C). The Blast analysis of these ORFs revealed sequence
homologies of different percentages with adenovirus sequences present in
Genbank. ORFs 1-4 of the right strand had theoretical coding capacities of
25.3,14.3, 68.4 and 5.5 kDa, respectively. The nucleotide sequence at the end
of ORF1 and ORF2 shared an 8-nucleotide overlap (ATGACTGA,
SEQ.ID.N0.:10), the stop codon of ORF1 embedded in the ORF2 coding gene.
ORF1 of 222 as between 73.7 and 75.7 m.u. (nt 1-666, Figure 1B) of the
genome showed high homology with several of the known human and
animal adenovirus pVIII protein sequences. The predicted ORF1 amino acid
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-22-
sequences of HNF-61 and HNF-70 were identical, and only one nucleotide
difference was detected at nt 546.
The deduced amino acid sequence of the ORF between 75.7 and 76.7 m.u.
(ORF2, 129 aa, nt 662-1048) was similar to the ORF5 of bovine adenovirus
type 1 (BAdV-1, 78.3%), the 13.7 kDa protein of PAdV-3 (63.7% similarity) and
also to the 13.3 kDa protein of canine adenovirus type 1 and 2 (61.2 and 58.9%
similarity, respectively). OItF2 also exhibited a strong similarity ranging
from
55.3% (human adenovirus, HAdV, type 7) to 61.6% (HAdV-40) to the 12.5
kDa E3 OItF of human adenoviruses. There was only a 2 as difference
between the predicted ORF2 sequences of the 2 PAdV-5 strains at as positions
31 and 35. The alignment of the nucleotide sequences of the HNF-61 and
HNF-70 strains showed 7 mismatches in this region. A comparison of the
predicted amino acid sequences of PAdV-5 HNF-70 ORF2 and the
homologous ORFs of the most closely related animal adenoviruses is shown
in Figure 2.
A large open reading frame (ORF3) was identified between 76.1 and 81.6
m.u. It was present in both strains of PAdV-5 (HNF-61: nt 798-2654, HNF-70:
nt 798-2633) with a coding capacity of 618 as and 612 aa, respectively. Figure
3 shows the alignment of the putative ORF3 as sequences of the strains HNF-
61 and HNF-70. These ORFs had 66.67% identity on the as level and 77.65%
homology at the nucleotide level. The first 300 as were almost completely
identical (97.3%) but from this point to approximately as 570 no significant
similarity could be detected. The last 40 as of the C-terminal exhibited again
a
high sequence identity (91.9%). ORF3 showed a weak similarity (39.3%) only
to the ORF10 of the E3 region of BAdV-1. A homology search of the
Genbank database for ORF3 did not reveal similarities with any other known
adenovirus sequences.
A short ORF of 50 as (ORF4) was present in the r strand of both strains,
starting at nt 1159. There was a 98.6% homology between the nucleotide
sequences of the ORF4 of the two strains of PAdV-5 and there was only a 2 as
difference between them. A Genbank search identified no related adenovirus
gene.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-23-
According to the sequence comparisons, the ORF starting at 81.7 m.u.
(ORF5, from nt 2709 of HNF-61 and nt 2690 of HNF-70) was the beginning of
the fiber protein coding region.
'The length of the putative E3 region of PAdV-5 starting at the end of
OIZF1 (pVIII homologue) and ending before the ATG signal of the fiber
coding ORF5 was 2039 by for HNF-61 and 2020 by for HNF-70. The full
nucletide sequence of the E3 region of each of the HNF-70 and HNF-61
strains are shown in figures 11 and 12 respectively.
One TATA box was identified (TATAA.A.A, SEQ.ID.N0.:11) at nt 351 of
both strains. No typical CCAAT was found upstream of the TATA box but a
CAGTT sequence was located at nt 322. CART sequences were identified
further upstream of the TATA box at nt 153 on both strains and also at nt 116
on HNF-61. A GC box was located starting at nt 322 (TTGGCGGGC,
SEQ.ID.N0.:12). Other GC rich sequences were identified before the TATA
box, GGCGG (SEQ.ID.N0.:13) at nts 57, 174, 324, 328 and GCCGG
(SEQ.ID.NO.:14) at nt 106 on both strains. Canonical (AATAAA,
SEQ.ID.N0.:15) poly-adenylation (poly-A) sequences were located on the
genome. Both strains had one poly-A signal at 10 (HNF-61) or 12 (HNF-70)
by downstream of the ORF3 stop codon at nts 2664 and 2648, respectively.
The HNF-61 E3 region contained another canonical poly-A signal at nt 1881,
which was not present in HNF-70. TTGTTT (SEQ.ID.N0.:16) signals were
identified at nt 1588 in both strains and at nt 2698 in HNF-61.
EXAMPLE 2
Phylogenetic analysis
The pVIII sequences of adenoviruses are generally considered to be
conserved to a certain extent at the amino acid level, and can be of interest
in
generating phylogenetic trees. The pVIII homologue of PAdV-5 (ORF1) was
used in such phylogenetic comparison with all the known animal and human
pVIII sequences. Figure 4 shows the result of the analysis with selected
representatives of animal adenoviruses. According to the alignment the
closest relative of PAdV-5 is BAdV-1 with 79% identity in this region. Canine
adenoviruses showed 58.9% (CAdV-1) and 61.2% (CAdV-2) similarity. PAdV
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-24-
1-3 serotypes clustered closely to PAdV-5, with approximately 64% similarity,
but PAdV-4 showed only 47.1% similarity.
EXAMPLE 3
DNA replication and transcription
To demonstrate that the putative E3 region, determined by sequence
analysis, between ORF1 (pVIII) and ORF5 (fiber) was indeed expressed at
early times of infection, DNA and RNA blotting assays were carried out.
Results for the HNF-70 strain are presented in Figure 5, the analysis of HNF-
61 gave similar results. The dot blot assay showed that DNA replication
started between 12-14 hours post infection (Figure 5A). No DNA replication
could be detected in AraC treated infected cells. The Northern blot analysis
indicated that RNA transcription within EcoRI G started between 4 and 6
hours post infection and the peak was reached at 12 hours from which point it
gradually declined (Figure 5B). At least five mRNA species could be detected.
The sizes of these transcripts were 3.2, 2.2, 1.6, 0.8 and 0.45 kb,
respectively.
The 0.8 kb transcript was the earliest mRNA detected. The 1.6 kb mRNA was
the most abundant and was transcribed for the longest period of time.
Transcription was not sensitive to AraC treatment.
EXAMPLE 4
Deletion of the E3 region
Two genomic clones with deletions in the E3 region (pR-Ol'H and pR
OHH) were generated by homologous recombination using the MIuI B-0(992-
2497) and MIuI B-0(1260-2497) clones, respectively (Fig.1D) and the DNA was
transfected into ST cells. The E3 portion deleted from pR-KPH started at nt
992
downstream from the ORF1 ATG and ended at nt 2497. This part of the E3
region contained the last 57 by of ORF2, the entire ORF4 and most of ORF3.
The deletion in pR-~HH affected a shorter segment of the E3 region leaving
ORF2 intact and removing the 3' 48 nts of ORF4 and most of ORF3 between
nts 1260 and 2497. After transfection into ST cells only the pR-OHH clone
resulted in infectious virus, pR-~I'H did not generate CPE in the cell culture
even after repeated attempts of transfections and several blind passages of
the transfected cells.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-25-
The pR-~HHH generated virus (R-OHH) was plaque purified three times
and the extracted viral DNA was analyzed with RE digestion. Figure 6 shows
the HpaI, EcoRI and HindllI RE patterns of wild type HNF-70 and R-~I-R-I
DNA. The original HpaI site was fused to the HincII site in R-~HH (Fig.lD)
and could be cleaved by HpaI so that the 5.9 kb HpaI C fragment of HNF-70
migrated at 4.7 kb in. R-~HHH. The size of the original 1.9 kb EcoRI fragment
(the same as the one used as E3 probe, Figure 2B) was 0.7 kb in R-~HH as
expected. Similarly, the size of the 4.2 kb Hindlll D fragment was decreased
to
3.0 kb by the 1.2 kb fragment deleted from R-~HH.
DISCUSSION OF EXAMPLES 1-4
The E3 region of adenoviruses is considered to be the most convenient
site for foreign gene insertion. Studies with HAdVs clearly demonstrated that
this region of the genome is not essential in virus replication, although it
may
play an important role in viral pathogenesis by helping the virus to evade
recognition by the immune system. Analysis of some animal adenovirus
genomes also showed that at least part of the E3 region could be deleted
without adverse effects on virus replication in vitro (Dragulev et al., 1991;
Mittal et a1.,1995; Evans et a1.,1998; Reddy et a1.,1999).
The sequenced region presented here comprised the entire pVIII gene
and the 5' portion of the fiber gene. By analogy with well characterized
adenoviruses it is suggested that the region of PAdV-5 between the pVIII and
fiber sequences is the E3 region. There are differences in size of the E3
region
of different mammalian adenoviruses. HAdVs in general have a longer E3
region than animal adenoviruses. The size of HAdV-5 E3 is for example 3 kb
(Cladaras and Wold, 1985), whereas it is only 0.8 kb in mouse adenovirus
type 1 (MAdV-1, Raviprakash et al., 1989), 1.1 kb in CAdV-1 (Dargulev et al.,
1991) and 1.8 kb in BAdV-1 (Evans et al., 1998). The size of PAdV-5 E3 was
determined and shown to be 2 kb, representing 6 % of the 33.5 kb genome
(Tuboly et al., 1995). It was also larger than the corresponding E3 of other
PAdVs, of approximately 1.2 kb in PAdV 1-3 (Reddy et a1.,1995 and 1996) and
1.8 kb in PAdV-4 (Kleiboeker,1994).
HAdVs have a complex E3 region and code for several overlapping
ORFs, the number of which vary among serotypes, whereas the E3 of animal
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-26-
adenoviruses is not only shorter but also more simple. The HAdV-5 E3 region
contains 10 ORFs (Cladaras et a1.,1985) and the short E3 of MAdV-1 has only
1 ORF (Raviprakash et al., 1989). All PAdV serotypes analyzed so far have 3
ORFs (Reddy et al., 1995 and 1996; Kleiboeker, 1994). There were also three
ORFs (ORF2-4) identified within the PAdV-5 E3 region. The pVIII gene was
followed by an ORF (ORF2) that was homologous to the 12.5 kDa HAdV E3
protein and its counterparts in PAdVs and several other mammalian
adenoviruses.
The long ORF3 of PAdV-5 was present in both strains. ORFs of such size
have not been reported for other animal adenovirus E3 regions and only
BAdV-1 ORF10 exhibited some homology with PAdV-5 ORF3 in the amino
terminal region. The comparison of ORF3 of HNF-61 and HNF-70 showed
almost perfect identity of the predicted amino acid sequences at the amino
terminal half and striking variability towards the C-terminal, although the
very ends of these proteins also appeared to be highly conserved between
the two strains. A large variation exists in HAdV E3a sequences among the
different subgenera and also among members of the same subgenus, but the
E3b region is usually more conserved (Bailey and Mautner, 1994). Porcine
adenoviruses however, show fewer variations in the E3 region. The E3
sequences of PAdV-1-3 serotypes are very similar to each other (Reddy et al.,
1996) and the analysis of several PAdV-4 isolates indicated that these viruses
were genetically stable (Kleiboeker et al., 1993). Despite the differences
detected in the ORF3 sequences of the PAdV-5 prototype strains, both viruses
proved to be genetically stable when subjected to several tissue culture
passages (results not shown). The viruses were isolated at the same time and
from the same herd and it was speculated earlier that the differences i11 the
RE
pattern were due to rapid genetic changes of the virus rather than a
concurrent infection with two different viruses of the same serotype (Tuboly
et a1.,1995).
ORF4 appeared to be unique to the PAdV-5 genome, but the putative
protein product has an unknown function.
The phylogenetic analysis of the established pVIII protein sequences
showed that PAdV-5 is more closely related to BAdV-1 than to any of the
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-
known PAdVs. The ORF2 similarity searches of the Genbank also showed a
closer relationship between PAdV-5 and BAdV-1 than PAdV-5 and other
PAdV-s. The pVIfI and ORF2 as comparisons indicated that PAdV-5 was
almost as similar to CAdVs as to PAdV-1-3. These data suggested that PAdV-
5 may not be a direct descendant of one of the well established porcine
adenovirus serotypes. It is more likely that either a strain of BAdV-1 or a
hypothetical common ancestor of PAdV-5, BAdV-1, PAdV-1-3 and CAdV-1-2
has recently been introduced to the swine species.
The early expression of the PAdV-5 E3 genes, as determined by Northern
blot analysis, further supported the identity of this region as an E3 region.
Although detailed transcription analysis was not done, the number of AraC
resistant transcripts showed that PAdV-5 has a more complex transcriptional
pattern than other porcine adenoviruses. The sequence data confirmed this
assumption. The promoter region of PAdV-5 E3 was localized between nts 95
and 364, with no typical CCAAT sequences but with several GC rich regions
preceding the TATA box. The poly-A signal for the E3 region was identified
downstream of the ORF3 stop codon but the presence of the alternative
TTGTTT signal in the middle part of the E3 and an additional canonical poly-A
signal at nt 1881 in HNF-61 indicated the potential for a more complex
transcriptional map.
The limited vector capacity of adenoviruses is an important issue in
vector development. Studies with helper independent HAdVs show that
approximately 105% of the original genome size can be accommodated inside
the virion (Bett et al., 1993). Viruses with larger packaged DNA are more
subject to genetic rearrangements and eventually loss of the inserted foreign
gene. The size of the PAdV-5 genome is approximately 33.5 kb (Tuboly et al.,
1995) which in theory means that a maximum of 1.7 kb foreign gene can be
inserted into a helper independent PAdV-5 vector without deleting any part
of the genome. It is possible to increase the size of the foreign DNA with the
size of a sequence that could be deleted from the PAdV-5 genome without
compromising its ability to replicate. In order to increase the vector
capacity
two genomic clones (pR-Ol'H and pR-~I~-i) with different sizes of E3
deletions were generated and the DNA transfected into ST cells. The
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-28-
transfection with the clone where a 1505 by part was deleted (pR-~l'H) did
not result in infectious virus particles after several repeated attempts. The
deletion removed part of the 12.5 kDa homologue (ORF2) of the E3 region.
This ORF is not essential for replication in cell culture in HAdVs (Hawkins
and
Wold, 1992) and BAdV-1 (Evans et al., 1998). The deletion of part of the 13.3
kDa protein coding gene and its fusion with the downstream ORF3 sequences
in the attenuated CLL strain of CAdV-1 (Dragulev et al., 1991) does not seem
to affect the virus replication in vitro. On the other hand the insertional
inactivation of the PAdV-3 ORF1 indicated that this gene may be essential for
replication of porcine adenoviruses (Reddy et al., 1999). These ORFs were
identical in the otherwise variable E3 regions of both prototype strains
indicating that at least in vivo ORF2 might have an important role during the
infection cycle. The smaller E3 deletion of 1237 by did not influence the
virus
replication. The viruses generated by the pR-OHH transfection replicated to
as high titers as the wild type virus. The size of the deletion was double
that
described for PAdV-3 (Reddy et al, 1999) and the deleted 1.2 kb fragment
increased the theoretical vector capacity (105%) of PAdV-5 to 2.9 kb.
EXAMPLE 5: THE COMPLETE NUCLEOTIDE SEQUENCE OF PAdV-5
The source and propagation of the PAdV-5 HNF-70 strain, extraction
and cloning of the viral DNA fragments have been described (Tuboly et al.,
1995). Nested set deletions were generated by exonuclease III and S1 nuclease
digestions (Henikoff,1984), and the clones were sequenced in both directions
with the SP6 and T7 promoter specific primers and primers based on the
obtained sequence data by the dideoxy nucleotide chain termination
technique. The contiguous sequence was assembled from overlapping
sequences using the Lasergene software package.
Homology search of the GenBank database for the deduced amino
acid sequence of each open reading frame (ORF) was done with the aid of the
BLAST program (Altschul et al., 1990). The promoters and splice sites were
predicted at the Berkeley Drosophila Genome Project website (Ohler & Reese,
1998; Reese et al., 1997). Polyadenylation (poly(A)) site prediction was done
with the POLYAH program (Salamov & Solovyev, 1997). Protein sequence
alignments were generated with CLUSTAL W (Thompson et al., 1994). The
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-29-
distance matrix analysis was carried out with the PHYLIl' (v.3.5) program
package (Felsenstein,1989).
Genome organization. The genome of PAdV-5 was 32621 by in length
(Figure 7), the G + C content was 50.5% and the genome structure and
arrangement (Figure 8) were similar to those of published mastadenoviruses.
Early regions. Early genes are required for the expression of other
viral genes, replication of viral DNA, transformation of cultured cells, and
influencing the immune response of the infected host (Gooding & Wold,
1990). Early regions E1 to E4 were identified in PAdV-5.
The E1A proteins were located between nt 418 and 1084. E1A 1738
(19.88 kDa) and E1A 67R (7.76 kDa) encoded by two overlapping ORFs were
similar in length to the canine adenovirus serotype 1 (CAdV-1) E1A proteins,
although in CAdV-1 these ORFs are not overlapping (Morrison et al., 1997).
The retinoblastoma susceptibility protein (pRb) binding motif LXCXE (Defeo-
Jones et al., 1991) with a slight difference (9'LDYPE), and the zinc finger
motif
(107CXZCXI3CXaC; Culp et al., 1988) were present in the E1A 1738 protein as in
PAdV-3 (Reddy et al., 1998a), or human adenovirus (HAdV) E1A proteins
(Culp et al.,1988). The E1B region (between nt 1180 and nt 2787) encoded two
proteins in two non-overlapping ORFs, the 1638 (19 kDa, small T antigen)
and the 3408 (38 kDa, large T antigen). The E1B 1638 protein showed the
highest homology to the BAdV-2 E1B ORF2159R protein (47% identity). The
E1B 3408 protein had 37% amino acid identity with the PAdV-3 E1B 4748
protein.
The mRNAs for DNA-binding protein (DBP) are transcribed from the
E2A region (located on the 1 strand between nt 21166 and 19841). The putative
DBP of PAdV-5 was 441 amino acids (49.8 kDa), and the N-terminal domain
contained two nuclear localization signals (4°PKPKK (SEQ.ID.N0.:17) and
4'RRRIC (SEQ.ID.N0.:18)) which were similar to those predicted for BAdV-3
(29PRKK (SEQ.ID.N0.:19) and 35RKRR (SEQ.ID.N0.:20)) and PAdV-3 (4'RRKR
(SEQ.ID.N0.:21), "RRK (SEQ.ID.N0.:22)) (Reddy et al., 1998a, b). Two
conserved zinc binding motifs were present: one at 199CXHX5ZCX15C and the
other at 311CXCX51CX15C exactly as in the corresponding proteins of HAdV-2
(Tucker et al.,1994) and PAdV-3 (Reddy et al.,1998a).
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-30-
According to amino acid sequence alignments with known terminal
protein precursors (pTP), ORF9~o_~542 encoded the main body of the pTP in the
E2B region with a predicted splice acceptor site at nt 9348. All known pTPs
have the sequence motif YSRLRYT (SEQ.ID.N0.:23) involved in protein
primed DNA replication initiation (Hsieh et al., 1990). In PAdV-5 the
$6YSRLKYT (SEQ.ID.N0.:24) motif was identified at the same location. The
nuclear localization signal (NLS) RLPI(R)4PRI of the pTP of PAdV-5 was
similar to that of PAdV-3 (RLPL(R)4PRP) (Reddy et al., 1998a). The serine
residue, involved in the initiation of DNA replication, and the flanking
residues (NSGD) were also well conserved (Smart & Stillman, 1982) at
512NSGD in the PAdV-5 pTP.
ORF~7lo-4309 with a predicted splice acceptor site at nt 7690 comprised
the main body of the predicted DNA polymerase (pol) gene of PAdV-5. The
conserved region I (YGDTDS (SEQ.ID.N0.:25)) and two possible zinc finger
motifs CEYC(X)~HTC(X)loHH and CETRCDKC(X)z3CSVC of PAdV-5 pol
were also present in PAdV-3 (Reddy et al.,1998a).
Based on the available sequence data, PAdV-5 has the largest E3 region
so far reported among PAdVs. Moreover, E3 ORF4 was unique to PAdV-5.
The E4 region of PAdV-5 was also larger, about 50%, than in most human
adenoviruses and in PAdV-3. In addition, 8 of the 11 ORFs were unique to
PAdV-5. The detailed analyses of PAdV-5 E3 and E4 regions have been
described (Tuboly & Nagy, 2000; Tuboly et al., 2000).
Intermediate regions. In HAdVs, two genes coding for the IX and
IVa2 proteins are classified as intermediate genes (Shenk, 1996). The minor
capsid component (IX) is needed for packaging the viral DNA (Ghosh
Choudhury et al.,198~) and is involved in activating the major late promoter
(MLP) (Lutz et al., 1997). The putative IX gene of PAdV-5 (126 aa, 13.7 kDa)
showed 42% identity to the BAdV-21188 ORF-4 protein. The IVa2 protein of
PAdV-5 (372 aa, 42.5 kDa) had 66% amino acid identity with the IVa2 protein
of HAdV-2. The entire potential nucleoside triphosphate binding site
GPTGCGKS (SEQ.ID.N0.:26) (Gorbalenya & Koonin, 1989) was present in
PAdV-5 IVa2.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-31-
Late regions. The late regions of the genome were characterized by
their predicted common poly(A) sites, and their products are mainly
structural proteins (Shenk,1996). Transcription starts from the MLP, and the
primary transcript is processed into several late mRNAs. For PAdV-5 six late
regions (L1-L6) were predicted (Fig.1). The putative MLP of PAdV-5 (nt 5077-
5273) was deduced by promoter prediction and sequence similarity with
known adenovirus MLP sequences. The canonical TATA box of the predicted
MLP was located at nt 5122-5128. An inverted CART box (nt 5084-5088), an
upstream promoter element (Sawadogo ~ Roeder, 1985) (nt 5104-5109),
initiator element (Lu et al., 1997) (nt 5150-5156), and two downstream
activating elements (Leong et al., 1990) DE1 (nt 5225-5235) and DE2 (nt 5240-
5255) were identified within this region.
The common poly(A) tail addition site of the L1 region was predicted
at nt 12173. The putative L1 52 kDa protein (354 aa, 40.4 kDa) was most
similar to the 55 kDa protein of HAdV-17 (62% identity) and pllIa (573 aa,
64.5 kDa) showed the highest identities to the pIIIa of HAdV-40
(61°l°).
'The putative L2 region had a common poly(A) tail addition site at nt
14172. The III (penton base; 471 aa, 52.7 kDa) and pVII (147 aa, 18.9 kDa)
proteins were predicted in this region. The RGD motif of protein III, which
interacts with surface integrins a~(33 and oc~[35 (Wickham et al., 1993) was
missing from the predicted penton protein of PAdV-5. However, the entire
LDV motif, which interacts with integrin oc4(31 (Komoriya et al., 1991), was
present at 264LDV. The fibre-interacting domain is highly conserved in the
penton base proteins of adenoviruses (Caillet-Boudin, 1989). In PAdV-5 the
ZsoSRLNIVLLGIRKR (SEQ.ID.N0.:27) motif was identical to the PAdV-3 fibre
interacting domain (Reddy et al., 1998a). Protein III of PAdV-5 was most
similar to that of PAdV-3 (77% identity), and pVII exhibited 54% similarity to
pVII of BAdV-2. One putative protease cleavage site was found in pVII at
z°MYGGA (SEQ.ID.N0.:28), exactly at the same position as in PAdV-3
(Reddy
et al.,1998a).
The L3 region had a predicted common poly(A) site at nt 15356.
Protein V of PAdV-5 (374 aa, 42.4 kDa) was most closely related to the
corresponding protein of BAdV-2 (57% identity).
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-32-
The common poly(A) tail addition site of the L4 region was located at
nt 19790. The predicted pX protein (70 aa, 7.8 kDa) had 79% amino acid
identity to pX of BAdV-2. There was only one protease cleavage site at
38MSGGF (Weber & Anderson, 1988). The pVI protein of PAdV-5 (233 aa,
25.2 kDa) showed the highest similarity to pVI of HAdV-40 (55% identity)
and contained two sequence motifs (3°MNGGAFNW (SEQ.ID.N0.:29) and
al9IVGLGVRS (SEQ.ID.N0.:30)) which corresponded to the consensus
protease cleavage site sequences (Russell & Kemp, 1995). In HAdV-2 the
protease requires a peptide (GVQSLI~;RRRCF (SEQ.ID.N0.:31)), which derives
from the C-terminus of pVI as a cofactor for its activity (Mangel et al.,
1993;
Webster et al., 1993). In pVI of PAdV-5 this peptide sequence was well
conserved at 2'~LGVRSVKItRRCF (SEQ.ID.N0.:32).
The predicted L5 region was characterized by a poly(A) tail addition
site at nt 26455. The 100 kDa protein (722 aa) and the 33 kDa protein (219 aa)
showed the highest similarity to the corresponding BAdV-3 proteins (59%
and 27% identity, respectively). The pVIII gene (222 aa, 24.1 kDa) has been
previously described (Tuboly & Nagy, 2000). After alignment to the
corresponding BAdV-3 protein (Reddy et al., 1998b), two putative protease
cleavage sites were found in the pVIII protein at 1°BLAGGGRTT
(SEQ.ID.N0.:33) and at 148LAGGSRSS (SEQ.ID.N0.:34). Figure 9 shows the
phylogenetic analysis of the putative pVIII protein compared to
representative adenoviruses. PAdV-1-3 had an inferred common ancestor,
whereas PAdV-4 and PAdV-5 were in two additional separate lineages.
Similar relationships were noted for the hexon proteins. It seemed that
PAdV-5 was phylogenetically closer to certain bovine adenoviruses,
specifically to BAdV-1 (based on pVIII) and BAdV-2 (based on hexon,
sequences provided by D. Ojkic, Guelph, Canada; no BAdV-1 sequences were
available) than to other described porcine adenoviruses. All these findings
underline the recent classification of PAdVs (Benko et al.,1999).
The poly(A) site for the L6 region was located at nt 28688. The N-
terminal region of the PAdV-5 fibre protein (500 aa, 53 kDa) encoded here,
contained a nuclear localisation signal (2KRAKR (SEQ.ID.N0.:35)) motif
(Hong & Engler, 1991) and a penton base interacting 11FDPVYPYG
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-33-
(SEQ.ID.N0.:36) sequence (Caillet-Boudin, 1989). Nineteen so-called
pseudorepeats (Green et al., 1983) were observed in the shaft region of the
PAdV-5 fibre protein. The sequence of the last complete motif before the
head (KLGXGLXFD/N) (Chroboczek et al., 1995) was well conserved at
3lsKLGAGLIFD (SE(~.ID.N0.:37). Interestingly, the TLWT motif, which in
most cases indicates the N-terminus of the fibre head (Chroboczek et al.,
1995) was not found in the PAdV-5 fibre.
Virus-associated RNA (VA RNA). Some adenoviruses encode low
molecular weight RNAs (VA RNAs) transcribed by RNA polymerase-III,
required for the efficient translation of viral mRNAs late after infection
(Larsson et al., 1986). In the mammalian adenoviruses studied by Ma and
Mathews (1996), there are either one or two genes for VA RNA, known as
VA RNAI and VA RNAa, located between the pTP and the 52 kDA ORFs.
Based on sequence analysis and RNA secondary structure prediction only, the
presence of VA RNAs in this region could not be predicted for PAdV-5 as was
also observed for non-primate mastadenoviruses.
The analysis of the PAdV-5 genome summarized herein indicate that
the size and the genome organization of this adenovirus are similar to that of
mastadenoviruses. However, unique characteristics of PAdV-5 were also
identified. Most importantly the RGD motif of the penton base protein and
the- TLWT motif of the fibre protein were not present. Only one protease
cleavage site was found in pX. Phylogenetic analysis of pVIII and hexon
proteins showed that PAdV-5 was well separated from the other PAdVs but
was more closely related to BAdV-1 and BAdV-2.
MATERIALS AND METHODS FOR EXAMPLE 6 TO 8
Cells, viruses, viral DNA and cDNA
The HNF-70 strain of PAdV-5 and the cell culture adapted Purdue115
strain of TGEV were propagated in continuous swine testicle (ST) cells
(McClurkin et al., 1966) as described (Tuboly et al., 1995). Virus titrations,
plaque purifications and virus neutralization assays were also performed in
ST cells as described (Tuboly et a1.,1993).
Adenoviral DNA was extracted from HNF-70 infected ST cells by the
method of Hirt (1967) at the peak of the cytopathic effects (CPE). The TGEV S
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-34-
gene cDNA synthesis and cloning have been described elsewhere (Tuboly et
a1.,1995).
Transfer' vector construction
Full length genomic PAdV-5 clones of the HNF-70 strain were
constructed by homologous recombination in E. coli BJ 5183 (Hanahan, 1983)
cells as described (Degryse, 1996). The strategy for the construction of the
recombinant transfer vectors is summarized in Figure 10. Plasmid Rpac+ was
generated by replacing the 1.9 kb SaII-HpaI fragment (spanning part of the
pVIII protein and the majority of the E3 coding region) with a unique PacI
restriction enzyme (RE) site. The MIuI B fragment of PAdV-5 (Tuboly et al.,
1995) was used for the insertion of the S gene. Five different S gene-
containing MluI B fragments were generated: one construct contained the
entire E3 region, and in four constructs a 1.2 kb piece was deleted in the E3
region between the HincII and the HpaI sites. i) MIuIB-2.2S: the 2.2 kb 5'-end
of the S gene was inserted into the HpaI site of the E3 region in left to
right (1-
r) orientation; ii) OlVIIuIB-2.2Sc: the 1.2 kb HincII-HpaI fragment of the E3
region was replaced by the 2.2 kb 5' S fragment, also in 1-r orientation; iii)
dlVIIuIB-2.2Sr: the same deletion in the E3 as in ii) construct but the 2.2 S
gene
was inserted in the reverse, r-1 orientation; iv) OIVIIuIB-2x2.2S: the 2.2 kb
S
gene was inserted in 1-r and subsequently in r-1 orientation; v) ~lVIluIB-
4.4S:
the entire 4.4 kbp S gene was inserted into the E3 region in 1-r orientation.
The transfer vectors were generated in bacteria by homologous
recombinations of the modified MluIB fragments carrying the S gene and the
Rpac+ genomic clone linearized with PacI. The recombinant clones were
analyzed and selected by standard miniprep and RE digestion methods
(Sambrook et al., 1989). Large scale DNA preparation of the clones selected
for ST cell transformation was done with the Concert Nucleic Acid
Purification System (Life Technologies), according to the instructions of the
manufacturer.
DNA tYansfection and selection of recombinant viruses
Lipofectin (Life Technologies) mediated ST cell transfections were done as
described earlier (Tuboly and Nagy, 2000) following the manufacturers'
instructions. The transfected cells were covered with 0.7 % agarose in DMEM
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-35-
supplemented with 10% fetal bovine serum. Plaque formation was monitored
daily and 10 individual plaques from each transfection were transferred to
Eppendorf centrifuge tubes with 1 ml of DMEM on day 7 post transfection.
The tubes were frozen to -70°C and thawed on ice. The contents
were used
for the inoculation of duplicate wells of ST cell monolayers in 6-well tissue
culture plates.
The cell culture supernatant from each well was collected at the peak of
CPE, about 6-7 days after inoculation and stored at -70°C until the
next round
of plaque purification. Cells were harvested for DNA RE analysis and for
Western blotting. Only those viruses -one from each lineage- were included in
further rounds of plaque purification that contained the entire expected S
gene insert. Viruses, selected after three rounds of such plaque purification,
were designated as RPAdV-2.25, ORPAdV-2.2Sc, RPAdV-2.2Sr, ~Rl'AdV-
2x2.2Sc and ~Rl'AdV-4.4S, and were used for large scale virus propagation.
Western blot analysis of recombinant S proteins
Wild type and recombinant adenovirus infected, together with uninfected
ST cells were harvested at the peak of CPE formation, the proteins were
separated in 10% SDS-polyacrylamide gels as described (Laemmli 1970) and
transferred to nitrocellulose membranes (Sambrook et al., 1989). TGEV
specific pig polyclonal antibodies (Tuboly et al., 1994) were used in 1:500
dilution to detect the proteins. The reaction was developed by the Boehringer
Mannheim chemiluminescent detection kit according to the instructions of the
manufacturer.
S gene mRNA time course
ST cells grown in 6 well dishes were infected at a multiplicity of infection
of 10 (M.O.L) with the wild type and the recombinant viruses. RNA was
extracted with the total RNA extraction kit (RNeasy, QIAGEN) every 4 hours
between 2-24 hours post infection (p.i.) and frozen to -70°C until use.
RNA
from mock-infected ST cells were also collected. Equal amounts of the total
RNA extracted from each time point were separated on 1.1% formaldehyde-
agarose gels, transferred to Nytran membranes (Sambrook et al., 1989) and
immobilized by W crosslinking (W Crosslinker, Fisher Scientific).
Prehybridization, hybridization in the presence of 50% formamide and
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-36-
washing of the blots were carried out 'as described by Sambrook et al. (1959).
3'
The cloned 2.2 kb TGEV S gene was released from the plasmid, labelled with
[s2P]dCTP (ICN Pharmaceuticals) by the random primer method (Random
Primer Labeling Kit, Life Technologies) and used as a probe.
Animal experiments
Fifteen Yorkshire piglets from a TGEV- and PAdV-5-seronegative herd
were weaned 21 days after birth and divided into 5 groups and housed
separately. One group received uninfected ST cell supernatant, one group
was immunized with wild type PAdV-5 and 3 groups were immunized with
the selected recombinant viruses (RPAdV-2.25, ORPAdV-2.2Sc and ORPAdV-
2.2Sr). Each pig received a single oral dose of 1 ml, with a virus titer of
5x106
p.f.u./ml (repeated vaccination with the same recombinant adenovirus could
be used as the boost). Blood samples were collected weekly and the clinical
signs were monitored daily. The pigs were euthanized after 3 weeks and
subjected to post-mortem examination. Contents from the small intestine
and parts of the lung were collected, processed as described (Tuboly et al.,
1993) and tested for the presence of virus and sIgA antibodies. For antibody
detection, the serum samples and the filtered intestinal and lung contents
were heat inactivated at 56°C for 1 hour. Samples were tested in a TGEV
specific IgG or IgA ELISA as described earlier (Tuboly et al., 1993) and in a
TGEV-specific virus neutralization (VN) microtiter assay (Tuboly et al.,
2000).
The serum samples were also tested for the presence of PAdV-5 specific
antibodies by a VN assay (Tuboly et a1.,1993).
Rectal swabs were collected daily to monitor virus shedding. The swabs
were processed as described (Tuboly et al., 1995) and the viral titers were
determined in 96 well plates with ST cells. The viruses isolated at day 5 p.i.
were pooled in each group, and the virus was propagated in ST cells for DNA
extraction and RE analysis of the viral DNA.
EXAMPLE 6
Transfer vectors
Five full-length genomic PAdV-5 clones were generated by
recombination in E. coli strain BJ5153 cells, each one carrying either the
full or
a partial S gene. The structure of these vectors, derived from the MIuIB and
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-37-
OlVIluIB clones is shown in Figure 10A. The detailed RE analysis of the
transfer vectors indicated that the orientation, location and size of the
inserts
were as expected.
Construction of recombinant PAdV
Recombinant PAdVs were plaque purified 3 times and the presence and
orientation of the S gene were confirmed by RE analysis of the viral genome
(data not shown) after each round of plaque purification. The expression of
the recombinant proteins was monitored by Western blots. Table 1
summarizes the stability data of each recombinant virus in tissue culture.
Following transfection with RPAdV-2.2S (no deletion in the E3 region)
70% of the plaques carried the 2.2 kb S gene in the correct orientation and
expressed the S gene. Stable virus clones could be selected after the first
plaque purification, with neither the insert nor parts of the E3 region being
lost. Those recombinant viruses that carried a single copy of the same 2.2 kb
insert in either orientation (~RPAdV-2.2Sc and ~RPAdV-2.2Sr) produced
plaques immediately after the transfection and were all positive in the RE
digestion. Western blots showed that the S protein was expressed only in
those viruses in which the S gene was inserted in left to right orientation
(dRPAdV 2.2Sc). These viruses remained stable during further plaque
purification.
Transfection with the vector containing two duplicate 2.2 kb S genes
inserted in opposite orientations (~Rl'AdV-2x2.2S), carrying altogether a 4.4
kb foreign DNA insert, yielded eight positive plaques in the first round of
purification. All of which expressed the S protein and the insert and S gene
expression remained stable during subsequent purification.
Only 20% of the viruses in which the complete S gene was inserted into
the PAdV E3 region (~RPAdV-4.4S) retained the S gene after the transfection,
and the ratio remained low throughout further plaque purifications. In
contrast with the rest of the recombinant viruses, the RE digests of the DNA
always indicated that only part of the virus population from a single plaque
carried the entire S gene, smaller DNA bands also appeared even after the
third plaque purification (not shown). Similarly, many bands were observed
in the Western blots of cells infected by these virus clones (not shown).
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-38-
EXAMPLE 7
Expression of the S gene and S protein
S gene expression was monitored by Northern blot analysis of total RNA
extracted at 2 h p.i. and every 4 h thereafter from recombinant virus infected
cells and blots were probed with radioactively labeled 2.2 kb S gene DNA.
RPAdV-2.2S and dRPAdV-2.2Sc expressed TGEV S gene specific mRNA at
approximately the same level. The S gene mRNA synthesis in RPAdV-2.25
infected cells was undetected during early times of virus replication and
could
be detected first only at 18 hours p.i. whereas S gene specific mRNA appeared
somewhat earlier in ORPAdV-2.2Sc infected cells, at 14 hours p.i. (Figure 11).
No S gene specific mRNA detected in ORPAdV-2.2Sr infected cells (data
not shown). The temporal pattern of transcription in ~RPAdV-2x2.2S infected
cells was similar to that for ORPAdV-2.2Sc. However Northern blot analysis
of replicates of ORPAdV-4.4S infected ST cell cultures indicated different
sizes
of transcripts and the ratios of the transcripts were not consistent (data not
shown).
For Western blot analysis, cells infected with the different recombinant
viruses were collected at 24 hours p.i. RPAdV-2.2S and ~RPAdV-2.2Sc
expressed the S protein of the expected size,110 kDa (Fig.3; lanes 2 ~ 3) and
a
similar result was obtained with the ORl'AdV-2x2.2S recombinant virus
(Figure 12; lane 4). No S protein was detected in cells infected with dRPAdV-
2.2Sr virus. S protein specific bands with a wide range of sizes (30 - 220
kDa)
were seen on the blots of samples collected from ORPAdV-4.4S infected cells
(Figure 12, lane 5).
EXAMPLE 8
Animal experiments
Pigs orally inoculated with the recombinant viruses, namely RPAdV-2.2S,
ORPAdV-2.2Sc and ORPAdV-2.2Sr, remained healthy throughout the
experiment and no signs of diarrhea or respiratory distress were observed.
The titre of virus in the rectal swabs collected daily was determined and the
results are summarized in Table 2. Virus was detected from day 1 to 7 but
was not detected in any of the samples by day 8 p.i. Virus was not recovered
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-39-
from the lungs or the small intestine of the euthanised pigs at 3 weeks p.i.
(not shown). A sample was considered negative after 3 blind passages in
tissue culture.
y All three types of recombinant viruses isolated at day 5 p.i. were tested
by RE analysis of the extracted DNA with several of the characteristic REs.
The DNA fragment patterns of viruses recovered from inoculated pigs were
indistinguishable from those observed for the inocula before the animal
"passages" (data not shown) indicating that the recombinant viruses were
stable.
ELISA and VN assays were conducted to detect TGEV- and PAdV-5
specific antibodies in the infected pigs. The results are shown in Table 3.
VN test of the sera collected at the end of the experiment showed
relatively high TGEV neutralizing titres (up to 1: 64) in groups injected with
RPAdV-2.25 and ORl'AdV-2.2Sc. No TGEV specific VN antibodies were
detected in the samples from pigs injected with ~RI'AdV-2.2Sr or wild type
PAdV-5, or from the mock-infected group. Similar results were obtained in
the TGEV specific ELISA to detect serum IgG. PAdV-5 specific VN antibodies
were present in the sera of all animals immunized with recombinant or wild
type PAdV-5 but there was no evidence of such antibodies in the mock-
infected group. TGEV specific antibodies of class A were detected in both the
lungs and the intestinal contents of the pigs immunized with RPAdV-2.25 and
ORPAdV-2.2Sc. The intestinal sIgA was present in all of the animals by ELISA.
However the sIgA titres measured in the lungs were lower in all animals and
pig #2 of the group injected with RPAdV-2.2S was negative.
DTSCUSSION OF EXAMPLES 6 TO 8
'Recombinant human adenoviruses have been shown to be efficient
vector systems for the delivery of porcine coronavirus antigens like those of
the TGEV or porcine respiratory coronavirus S protein (Torres et al., 1996;
Calleabut et a1.,1996). Their widespread use of human adenovirus in domestic
animals may be limited, mainly because of safety concerns. Animal
adenoviruses, however, are mostly species-specific, presenting almost
negligible risk for humans or other animal species and replicate more
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-40-
efficiently. that human adenovirus in the native porcine host, thereby
providing a safer and more efficient delivery system in animals.
PAdV-3 carrying the gD gene of the Aujeszky's disease virus (Reddy et
al., 1999b) and the E2 gene of the classical swine fever virus (Hammond et
al.,
2000) has already been developed as a recombinant virus vector. However
the wide prevalence of PAdV-3 may be a limiting factor in their use as
recombinant vaccines because of widespread preexisting PAdV-3 neutralizing
antibodies.
In contrast, PAdV-5, to our knowledge is not present in pig populations,
and has been reported only once from Japan (Hirahara et al., 1990). The
development of PAdV-5 into a recombinant TGEV vaccine is described in
herein. Five helper independent recombinant porcine adenoviruses have
been constructed and tested for their stability and their ability to express
the
entire or the 5' 2.2 kb half of the TGEV S gene. Three of the recombinant
viruses carrying the 2.2 kb S gene were selected and their ability to induce
TGEV neutralizing antibodies was tested by oral immunization of pigs. Two
of these viruses carrying the foreign gene in left to right orientation
expressed the protein in vitro and induced humoral immune response in pigs,
not,only systemic but also of a local nature.
The same approach was used for the construction of all the recombinant
adenoviruses. One of the purposes was to test whether it is necessary to
include foreign promoter sequences upstream of the insert or if the PAdV
promoters are sufficient to express the gene. In one construct (RPAdV-2.2S)
no E3 sequences were removed and the 2.2 kb S gene fragment was inserted
in a left to right orientation near the 3' end of the E3 region, more than 1.8
kb
downstream of the putative E3 promoter (see Example 1). S gene specific
transcripts in Northern blots were detected from 18 hours p.i., reaching the
peak between 18 and 24 hours p.i. The S protein in Western blots was
detected indicating that the native adenovirus promoters were sufficient for
foreign gene expression (Torres et al., 1996). Although, together with the 2.2
kb foreign gene, the genome size of RPAdV-2.2S was 106.6% of the original
wild type genome, neither the insert nor parts of the E3 region were lost
during the plaque purifications or the several replication cycles in the pig
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-41-
intestine. The stable insertion of such a large foreign DNA is in accordance
with the findings of Hammond et al. (2000) who increased the genome size of
PAdV-3 to 106.8% of the original, despite earlier findings of a maximum of
105% for human adenoviruses (Bett et a1.,1993).
As a result of the 1.2 kb deletion of the E3 region, the other recombinant
viruses carried the S gene closer to the E3 promoter than in RPAdV-4.4S (only
594 by downstream of the putative E3 promoter. Those recombinant viruses
that had the insert in left to right orientation (ORPAdV-2.2Sc, ORl'AdV-
2x2.2S,
~RPAdV-4.4S) started to express the gene at the end of early times, between
14 and 18 hours p.i., as detected by Northern blot analysis, whereas the virus
with the S gene in reverse orientation (~RPAdV-2.2Sr) showed no signs of S
gene expression in vitro (Northern and Western blot analyses) or in vivo as
judged by the lack of TGEV specific antibodies in the immunized pigs.
Those ~RPAdVs that carried a single copy of the 2.2 kb S gene appeared
to be stable right after the transfection and all of the plaques tested had
the
inserted gene of the expected size at the expected position. ORPAdV-2x2.25,
with two sets of the 3' truncated S gene, produced 7 out of 10 plaques that
carried both inserts right after the transfection and became stable during
further rounds of plaque purifications. The virus did not lose any of the
inserts or the PAdV sequences as detected by RE analysis (not shown). The
size of the genome of this recombinant virus was 109.6% of the original
genome size, exceeding the expected maximum of 106.8 % (Hammond et al.,
2000) described for PAdV-3. The ~RPAdV-4.4S virus with the full-length S
gene did not yield a stable lineage, despite several rounds of plaque
purification of the positive viruses. The expected genome size of this virus
was also 109.6% of the wild type genome but unlike the dRPAdV-2x2.2S,
parts or all of the insert or the PAdV genome were constantly being lost
during virus replication. This phenomenon raised questions about current
theories of adenovirus genome stability. The main limiting factor of the
stability is believed to be the packaging capacity determined by the size and
icosahedral structure of the virion. According to experiments reported
herein, the size of the insert is not the only important factor influencing
the
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-42-
stability of the genome. The sequence, structure or the orientation of the
insert may also play an important role.
The recombinant viruses were analyzed by Western blotting to
determine the size of the recombinant proteins. All of the viruses with the
gene in a left to right orientation expressed a protein of the predicted size.
The estimated size of the S protein in RT'AdV-2.2S, ~Rl'AdV-2.2Sc and
~RI'AdV-2x2.25 was 110 kD. The ~RI'AdV-4.4S virus preparation also
expressed the expected 200 kD protein but smaller S protein fragments were
also detected.
Direct measurement of the amount of recombinant proteins was not
carried out but from comparisons to known amounts of baculovirus and
transgenic plant expressed S proteins (Tuboly et al., 1994; Tuboly et al.,
2000)
it was estimated that approximately 5-10 ~.g protein/106 cells can be obtained
at 24 hours p.i. This figure is in accordance with that of Torres et al.
(1995) for
the expression of TGEV S gene and gene fragments in a human adenovirus
vector without the use of additional external promoters.
~w Three recombinant viruses were tested for their ability to induce TGEV
specific immune responses in pigs. Those that carried the 2.2S gene in a left
to
right orientation induced a TGEV-specific response. It was concluded that a
single oral dose of the recombinant virus was sufficient to induce both a
systemic and a local humoral immune response. The efficiency of live vaccines
is dependant on their ability to induce the required immune response in one
dose, as the second injection may be less effective due to the immune
response induced by the first injection. The antibodies induced by the
recombinant viruses were capable of neutralizing both the PAdV-5 and the
TGEV. The presence of TGEV specific IgA antibodies in the small intestine
indicated that a local immune response was also induced. This is very
important for TGEV arid other viruses with replication localized to surfaces.
A local immune response is particularly important against TGEV where the
survival of the piglets depends on the sIgA and sIgA secreting plasma cells in
the colostrum of the sow.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-43-
Although challenge experiments were not done, it was concluded that
recombinant PAdV-5 carrying the 2.~ kb S gene fragment could be a useful
tool in the protection of swine herds against TGEV.
While the present invention has been described with reference to what
are presently considered to be the preferred examples, it is to be understood
that the invention is not limited to the disclosed examples. To the contrary,
the invention is intended to cover various modifications and equivalent
arrangements included within the spirit and scope of the appended claims.
All publications, patents and patent applications are herein incorporated
by reference in their entirety to the same extent as if each individual
publication, patent or patent application was specifically and individually
indicated to be incorporated by reference in its entirety.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-44-
Table 1. Analysis of the recombinant viruses.
irus Rounds
of
la
ue
urifications
1 2
3
Ea b b
RPAdv-2.2s 7/10' 7/10 10/10 10/10 10/10 10/10
dRPAdV-2.2Sc 10/10 10/10 10/10 10/10 10/10 10/10
~RPAdV-2.2Sr 10/10 0/10 10/10 0/10 10/10 0/10
~RPAdV-2x2.258/10 8/10 10/10 10/10 10/10 10/10
~RPAdV-4.45 /10 /10 /10 /10 /10 /10
aDNA profile based on Restriction endonuclease digestion,
bExpression of the S protein analyzed by Western blot,
'No. of S gene (protein) positive plaques/No. of plaques tested.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-45-
Table 2. Shedding of wild type and recombinant PAdV-5 viruses.
Virus Pig# Virus
titers
Rectal Int.Lung
swabs
(days
p.i.)
1 2 4 5 6 7 8 9 1 3
3 weeks
0 p.i.
1 1 3 1 1 1 0 0 0 0 0 0
2
RPAdV-2.25 2 1 3 2 2 1 0 0 0 0 0 0
2
3 2 3 2 1 0 0 0 0 0 0 0
3
4 1 3 1 0 0 0 0 0 0 0 0
2
dRPAdV-
2.2.Sc S 2 2 1 1 1 0 0 0 0 0 0
1
6 2 2 1 1 1 0 0 0 0 0 0
1
7 1 3 1 1 1 0 0 0 0 0 0
2
ORPAdV-
2.2Sr 8 2 2 1 1 1 1 0 0 0 0 0
1
9 1 3 1 1 1 0 0 0 0 0 0
2
10 2 2 2 2 1 0 0 0 0 0 0
3
PAdV-5 11 3 4 2 2 2 1 0 0 0 0 0
3
12 2 1 2 1 1 0 0 0 0 0 0
2
Titers as 1og10 dilutions in 0.1 ml
p.i.= post infection
30
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
46 -
Table 3. TGEV and
PAdV-5 specific antibody
titers of pigs immunized
with
recombinant PadV and
measured by virus
neutralization and
ELISA.
Virus Pig# VN TGEV specific ELIS A
TGEV PAdV-5 Serum IgG Intestinal
IgA Lung
IgA 1 5 6 7 2 1
RPAdV-2.2S 2 6 7 7 2 0
3 5 6 8 3 2
4 4 7 7 2 1
~RPAdV-2.2Sc 5 5 8 6 2 1
6 5 6 7 2 1
~ 0 6 0 0 0
ORPAdV-2.2Sr 8 0 4 0 0 0
9 0 6 0 0 0
10 0 8 0 0 0
PAdV-5 11 0 7 0 0 0
12 0 7 0 0 0
Titers are expressed as log2 dilutions of the samples.
Dilutions for VN started with 1:2, and for ELISA with 1:10.
Mock infected pigs were negative in all tests.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-47-
FULL CITATIONS FOR REFERENCES REFERRED TO IN THE
SPECIFICATION
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990).
Basic
local alignment search tool. Journal of Molecular Biology 215, 403-410.
Bailey A. and Mautner V., 1994 Phylogenetic relationships among adenovirus
serotypes. Virology 205, 438-452.
Bett AJ, Prevec L, Graham FL. Packaging capacity and stability of human
adenovirus type 5 vectors. J Virol 1993; X7(10):5911-21.
Benko, M., Harrach, B. & Russell, W.C. (1999). Family Adenoviridae. In Virus
Taxonomy. Seventh Report of the International Committee on Taxonomy of
Viruses,
pp. 227-238. Edited by M.H.V. van Regenmortel, C.M. Fauquet, D.H.L.
Bishop, E.B. Carstens, M.K. Estes, S.M. Lemon, J. Maniloff, M.A. Mayo, D.J.
McGeoch, C.R. Pringle, R.B. Wickner. New York, San Diego, Academic Press.
Caillet-Boudin, M. L. (1989). Complementary peptide sequences in partner
proteins of the adenovirus capsid. Journal of Molecular Biology 208,195-198.
Chroboczek, J., Ruigrok, R. W. H. & Cusack, S. (1995). Adenovirus fiber.
Current Topics in Microbiology and Immunology 199,163-200.
Cladaras C. and Wold W.S., 1985 DNA sequence of the early E3 transcription
unit of adenovirus 5. Virology 140, 28-43.
Clarke M.C., Sharpe H.B. and Derbyshire J.B., 1967 Some characteristics of
three porcine adenoviruses. Arch. Gesamte Virusforsch. 21, 91-97.
Correa I, Gebauer F, Bullido MJ et al. Localization of antigenic sites of the
E2
glycoprotein of transmissible gastroenteritis coronavirus. J Gen Virol 1990;
71 (2):271-9.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-48-
Culp, J. S., Webster, L. C., Friedman, D. J., Smith, C. L., Huang, W. J., Wu,
F.
Y., Rosenberg, M. & Ricciardi, R. P. (1988). The 289-amino acid E1A protein of
adenovirus binds zinc .in a region that is important for traps-activation.
Proceedings of the National Academy of Sciences, USA 85, 6450-6454.
Defeo-Jones, D., Huang, P. S., Jones, R. E., Haskell, K. M., Vuocolo, G. A.,
Hanobik, M. G., Huber, H. E. & Oliff, A. (1991). Cloning of cDNAs for cellular
proteins that bind to the retinoblastoma gene product. Nature 352, 251-254.
Degryse E. In vivo intermolecular recombination in Escherichia coli:
application
to plasmid constructions. Gene 1996;170(1):45-50.
Delmas B, Rasschaert D, Godet M, Gelfi J, Laude H. Four major antigenic sites
of the coronavirus transmissible gastroenteritis virus are located on the
amino-terminal half of spike glycoprotein S. J Gen Virol 1990; 71(6):313-23.
Derbyshire J.B., Clarke M.C. and Collins A.P., 1975 Serological and
pathogenicity studies with some unclassified porcine adenoviruses. J. Comp.
Pathol. 85, 437-443.
Dragulev B.P., Sira S., Abouhaidar M.G. and Campbell J.B., 1991 Sequence
analysis of putative E3 and fiber genomic regions of two strains of canine
adenovirus type 1. Virology 183, 298-305.
Evarts P.S., Benko M., Harrach B. and Letchworth G.J., 1998 Sequence,
transcriptional analysis, and deletion of the bovine adenovirus type 1 E3
region. Virology 244, 173-185.
Felsenstein, J. (1989). PHYLIP-Phylogeny inference package (version 3.2).
Cladistics 5,164-166.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-49-
Garwes DJ, ,Lucas MH, Higgins DA, Pike BV, Cartwright SF. Antigenicity of
structural components from transmissible gastroenteritis virus. Vet Microbiol
1978; 3:179-90.
Ghosh-Choudhury, G., Haj-Ahmad, Y. & Graham, F. L. (1987). Protein I7C, a
minor component of the human adenovirus capsid, is essential for the
packaging of full length genomes. EMBO Journal 6,1733-1739.
Godet M, Rasschaert D, Laude H. Processing and antigenicity of entire and
anchor-free spike glycoprotein S of coronavirus TGEV expressed by
recombinant baculovirus. Virology 1991;185(2):732-40.
Gomez N, Carrillo C, Saunas J, Parra F, Borca MV, Escribano JM. Expression
of immunogenic glycoprotein S polypeptides from transmissible
gastroenteritis coronavirus in transgenic plants. Virology 1998; 249(2):352-8.
Gooding, L. R. & Wold, W. S. M. (1990). Molecular mechanisms by which
adenoviruses counteract antiviral immune defenses. Critical Reviews in
Immunology 10, 53-71.
Gorbalenya, A. E. ~ Koonin, E. V. (1989). Viral proteins containing the purine
NTP-binding sequence pattern. Nucleic Acids Research 17, 8413-8440.
Haig DA, Clarke MC, Pereira MS. Isolation of an adenovirus from a pig. J
Comp Pathol 1964; 74, 81-85.
Hammond JF, McCoy RJ, Jansen ES, Morrissy CJ, Hodgson ALM Johnson
MA. Vaccine 2000;18: 1040-1050.
Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol
Biol 1983;166(4):557-80.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-50-
Hawkins L.K. and Wold W.S., 1992 A 12,500 MW protein is coded by region
E3 of adenovirus. Virology 188, 486-494.
Henikoff, S. (1984). Unidirectional digestion with exonuclease III creates
targeted break points for DNA sequencing. Gene 28, 351-359.
Hirt B. Selective extraction of polyoma DNA from infected mouse cell
cultures. jMol Biol 1967; 26(2):365-9.
Hong, J. S. ~ Engler, J. A. (1991). The amino terminus of the adenovirus fiber
protein encodes the nuclear localization signal. Virology 185, 758-767.
Hsieh, J. C., Yoo, S. K. & Ito, J. (1990). An essential arginine residue for
initiation of protein-primed DNA replication. Proceedings of the National
Academy of Sciences, USA 87, 8665-8669.
Hu S, Bruszewski J, Smalling R, Browne JK. Studies of TGEV spike protein
gp195 expressed in. E. coli and by a TGE-vaccinia virus recombinant. Adv Exp
Med Biol 1985;185:63-82.
Jimenez G, Correa I, Melgosa MP, Bullido Mj, Enjuanes L. Critical epitopes in
transmissible gastroenteritis virus neutralization. J Virol 1986; 60(1):131-9.
Kadoi K., moue Y., Ikeda T., Kamata H., Yukawa M., Iwabuchi M. and Inaba
Y., 1995 Isolation of porcine adenovirus as a candidate of 5th serotype. J.
Basic. Microbiol. 35,195-204.
Kasza L .Isolation of an adenovirus from the brain of a pig. Am J Vet Res
1966;
27(118):751-8.
Kleiboeker SB. Identification and sequence analysis of the E1 genomic region
of a porcine adenovirus. Virus Res 1995 36(2-3):259-68
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-51-
Kleiboeker S.B., 1994 Sequence analysis of putative E3, pVIII, and fiber
genomic regions of a porcine adenovirus. Virus Res. 31,17-25.
Kleiboeker S.B., Seal B.S. and Mengeling W.L., 1993 Genomic cloning and
restriction site mapping of a porcine adenovirus isolate: demonstration of
genomic stability in porcine adenovirus. Arch. Virol. 133, 357-368.
Komoriya, A., Green, L. j., Mervic, M., Yamada, S. S., Yamada, K. M. &
Humphries, M. J. (1991). The minimal essential sequence for a major cell type-
specific adhesion site (CS1) within the alternatively spliced type III
connecting
segment domain of fibronectin is leucine-aspartic acid-valine. Journal of
Biological Chemistry 266,15075-15079.
Laemmli UK. Cleavage of structural proteins during the assembly of the head
of bacteriophage T4. NatuYe 1970; 227(259):680-5.
Larsson, S., Bellett, A. & Akusjarvi, G. (1986). VA RNAs from avian and
human adenoviruses: dramatic differences in length, sequence, and gene
location. Journal of Virology 58, 600-609.
Leong, K., Lee, W. & Berk, A. J. (1990). High level transcription from the
adenovirus major late promoter requires downstream binding sites for late-
phase-specific factors. Journal of Virology 64, 51-60.
Lu, H., Reach, M. D., Minaya, E. & Young, C. S. H. (1997). The initiator
element of the adenovirus major late promoter has an important role in
transcription initiation in vivo. Journal of Virology 71,102-109.
Lutz, P., Rosa-Calatrava, M. & Kedinger, C. (1997). The product of the
adenovirus intermediate gene IX is a transcriptional activator. Journal of
Virology 71, 5102-5109.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-52-
Ma, Y. & Mathews, M. B. (1996). Structure, function, and evolution of
adenovirus-associated RNA: a phylogenetic approach. journal of Virology 70,
5083-5099.
Mangel, W. F., McGrath, W. J., Toledo, D. L. & Anderson, C. W. (1993). Viral
DNA, and viral peptide can act as cofactors of adenovirus virion proteinase
activity. Nature 361, 274-275.
McClurkin AW, Norman JO. Studies on transmissible gastroenteritis of swine.
II. Selected characteristics of a cytopathogenic virus common to five isolates
from transmissible gastroenteritis. Can J Comp Med Vet Sci 1966; 30(7):190-8.
Mittal S.K., McDermott M.R., Johnson D.C., Prevec L. and Graham F.L., 1993
Monitoring foreign gene expression by a human adenovirus-based vector
using the firefly luciferase gene as a reporter. Virus Res. 28, 67-90.
Mittal S.K., Prevec L., Graham F.L. and Babiuk L.A., 1995 Development of a
bovine adenovirus type 3-based expression vector. J. Gen. Virol. 7f, 93-102:
Morrison, M. D., Onions, D. E. & Nicolson, L. (1997). Complete DNA sequence
of canine adenovirus type 1. Journal of General Virology 78, 873-878.
Ohler, U. & Reese, M. G. (1988). Detection of eukaryotic promoter regions
using polygrams. In Molekulare BioinfoYmatik, pp. 89-100. Edited by R.
Hofestadt. Aachen, Shaker.
Raviprakash K.S., Grunhaus A., e1 Kholy M.A. and Horwitz M.S., 1989 The
mouse adenovirus type 1 contains an unusual E3 region. J. Virol. 63, 5455-
5458.
Reddy PS, Idamakanti N,Babiuk L, Mehtali M, Tikoo SK. Porcine adenovirus
as a helper-dependent expression vector. J Gen Virol 1999a; 80: 2909-291f .
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-53-
Reddy PS, Idamakanti N, Hyun BH, Tikoo SK, Babiuk LA. Development of
porcine adenovirus-3 as an expression vector. J Gen Virol 1999b; 80 (3):563-
70.
Reddy PS, Nagy E, Derbyshire JB. Sequence analysis of putative pVIIf, E3 and
fibre regions of porcine adenovirus type 3. Virus Res 1995; 36(1):97-106
Reddy P.S., Idamakanti N., Derbyshire J.B.and Nagy E., 1996 Porcine
adenoviruses types 1, 2 and 3 have short and simple early E-3regions. Virus
Res. 43, 99-109.
Reddy P.S., Idamakanti N., Derbyshire J.B. and Nagy E., 1997
Characterization of the early region 4 of porcine adenovirus type 3. Virus
Genes 15, 87-90.
Reddy P.S., Idamakanti N., Song J.Y., Lee J.B., Hyun B.H., Park J.H., Cha
S.H.,
Bae Y.T., Tikoo S.K. and Babiuk L.A., 1998a Nucleotide sequence and
transcription map of porcine adenovirus type 3. Virology 251, 414-426.
Reddy P.S., Idamakanti N., Song J.Y., Lee J.B., Hyun B.H., Park J.H., Cha
S.H.,
Tikoo S.K. and Babiuk L.A.,1998b Sequence and transcription map analysis of
early region-1 of porcine adenovirus type-3. Virus Res. 58, 97-106.
Reddy, P. S., Idamakanti, N., Zakhartchouk, A. N., Baxi, M. K., Lee, J. B.,
Pyne,
C., Babiuk, L. A. ~ Tikoo, S. K. (1998b). Nucleotide sequence, genome
organization, and transcription map of bovine adenovirus type 3. Journal of
ViYOlogy 72,1394-1402.
Reddy P.S., Nagy E. and Derbyshire J.B., 1993 Restriction endonuclease
analysis and molecular cloning of porcine adenovirus type 3. Intervirology 36,
161-168.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-54-
Reddy P.S., Tuboly T., Nagy E. and Derbyshire J.B., 1995 Molecular cloning
and physical mapping of porcine adenovirus types 1 and 2. Arch. Virol. 140,
195-200.
Russell, W. C. & Kemp, G. D. (1995). Role of adenovirus structural
components in the regulation of adenovirus infection. Current Topics in
Microbiology and Immunology 199, 81-98.
Salarnov, A. A. & Solovyev, V. V. (1997). Recognition of 3'-end processing
sites of human mRNA precursors. Comput Appl Biosci 13, 23-28.
Sambrook J., Fritsch E.F. and Maniatis T. 1989 Molecular cloning: a laboratory
manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor,
New York.
Sawadogo, M. & Roeder, R. G. (1985). Interaction of a gene-specific
transcription factor with the adenovirus major late promoter upstream of fhe
TATA box. Cell 43,165-175.
Shenk, T. (1996). Adenoviridae: the viruses and their replication. In Fields
Virology, 3rd edn, pp. 2111-2148. Edited by B. N. Fields, D. M. Knipe, P.M.
Howley et aI. Raven Press, N.Y.
Smart, J. E. & Stillman, B. W. (1982). Adenovirus terminal protein precursor.
Partial amino acid sequence and the site of covalent linkage to virus DNA.
Journal of Biological Chemistry 257,13499-13506.
Smerdou C, Anton IM, Plana J, Curtiss R 3rd, Enjuanes L. A continuous
epitope from transmissible gastroenteritis virus S protein fused to E. coli
heat-
labile toxin B subunit expressed by attenuated Salmonella induces serum and
secretory immunity. Virus Res 1996; 41(1):1-9.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-55-
Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994). CLUSTAL W:
improving the sensitivity of progressive multiple sequence alignment
through sequence weighting, position-specific gap penalties and weight
matrix choice. Nucleic Acids Research 22, 4673-4680.
Torres JM, Alonso C, Ortega A, Mittal S, Graham F, Enjuanes L. Tropism of
human adenovirus type 5-based vectors in swine and their ability to protect
against transmissible gastroenteritis coronavirus. J Virol 1996; 70(6):3770-
80.
Tuboly T, Nagy E, Dennis JR, Derbyshire JB. Immunogenicity of the S protein
of transmissible gastroenteritis virus expressed in baculovirus. Arch Virol
1994;137(1-2):55-67.
Tuboly T, Nagy E, Derbyshire JB. Potential viral vectors for the stimulation
of
mucosal antibody responses against enteric viral antigens in pigs. Res Vet Sci
1993; 54(3):345-50.
Tuboly T, Nagy E, Derbyshire JB. Passive protection of piglets by
recombinant baculovirus induced transmissible gastroenteritis virus specific
antibodies. Can J Vet Res 1995; 59(1):70-2.
Tuboly T, Yu W, Bailey A, Degrandis S, Du S, Erickson L, Nagy E.
Immunogenicity of porcine transmissible gastroenteritis virus spike protein
expressed in plants. Vaccine 2000;18:2023-2028.
Tuboly T, Reddy PS, Nagy E, Derbyshire JB. Restriction endonuclease
analysis and physical mapping of the genome ofporcine adenovirus type 5.
ViYUS Res 1995; 37(1):49-54.
Tuboly, T. & Nagy, E. (2000). Sequence analysis and deletion map of porcine
adenovirus serotype 5 E3 region. Virus Research 68,109-117.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-56-
Tuboly, T., Nagy, M. & Nagy, E. (2000). Characterization of early region 4 of
porcine adenovirus serotype 5. Virus Genes 20, 217-219.
Tucker, P. A., Tsernoglou, D., Tucker, A. D., Coenjaerts, E. J., Leenders, H.
&
Van der Vliet, P. C. (1994). Crystal structure of the adenovirus DNA binding
protein reveals a hook-on model for cooperative DNA binding. EMBO Journal
13,2994-3002.
Weber, J.M. & Anderson, C.W. (1988). Identification of the gene coding for
the precursor of adenovirus core protein X. Journal of ViYOlogy 62,1741-1745.
Webster, A., Hay, R. T. & Kemp, G. (1993). The adenovirus protease is
activated by a virus-encoded disulphide-linked peptide. Cell 72, 97-104.
Wickham, T. J., Mathias, P., Cheresh D. A. & Nemerow, G. R. (1993). Integrins
a~(33 and oc~(35 promote adenovirus internalization but not virus attachment.
Cell 73, 309-319.
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-57-
DETAILED LEGENDS FOR VARIOUS FIGURES
Figure 1. PAdV-5 maps. (A) HindIII and MluI restriction map of the genome,
m.u.: map unit, 1 m.u.: 335 bp. (B) The sequenced region enlarged. Bar
indicates the EcoRI DNA fragment (between nts 616 and 2569) used as the E3
probe in Northern blotting. (C) RF: reading frames of the r strand. The open
reading frames are shown by the boxes. Numbers above the boxes are the
start and stop positions of each ORF. The numbers inside the boxes indicate
the putative number of amino acids encoded by each ORF of HNF-70. (D) The
dotted boxes represent the part and size (indicated below the box) of the
DNA removed to generate pR-KPH and pR-OHH genomic clones.
Figure 2. Alignment of the predicted ORF2 amino acid sequences of PAdV-5
HNF-70 and some closely related animal adenoviruses. The adenovirus
serotypes are indicated on the left. The sequence identities are indicated by
dots.
Figure 3. Sequence alignment of the predicted ORF3 proteins of HNF-61 and
HNF-70. The number of amino acids is indicated on the right, identities are
shown by dots.
Figure 4. Unrooted phylogenetic tree of pVIII protein homologues of selected
animal adenoviruses generated by the Clustal method. The length of
branches represents the distance between sequence pairs. Units at the bottom
indicate the number of substitution events.
Figure 5. Time course analysis of PAdV-5 HNF-70 nucleic acid synthesis.
Numbers in the middle indicate hours p.i. (A) DNA dot blot, in the absence
(AraC-) and presence (AraC+) of AraC, probed with digoxigenin labeled
genomic DNA. (B) Northern blot of E3 region transcripts, probed with the
EcoRI G fragment (Figure 1). Lines and numbers on the right show the
position and size (kb) of the RNA molecular weight marker (M).
CA 02413265 2002-12-23
WO 01/83737 PCT/CA01/00598
-58-
Figure 6. Restriction endonuclease analysis of the wild type PAdV-5 HNF-70
strain (A) and its deletion mutant R-OHH (B) genomic DNA in ethidium
bromide stained 0.8% agarose gel. Lanes 1: HpaI, lanes 2: EcoRI, lanes 3:
HindIII. Arrowheads indicate the corresponding fragments for each digest. M:
1 kb DNA ladder.