Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02429860 2003-05-26
ENDOGENOUS AND NON-ENDOGENOUS VERSIONS OF
HUMAN G PROTEIN-COUPLED RECEPTORS
CROSS-REFERENCE TO RELATED APPLICATIONS
This application is a continuation-in-part of U.S. Serial Number 09/170,496,
filed
with the United States Patent and Trademark Office on October 13, 1998 and its
corresponding PCT application number PCT/LJS99/23938, published as WO 00/22129
on April 20, 2000. This document claims the benefit of priority from the
following
provisional applications, all filed via U.S.~ Express Mail with the United
States Patent and
Trademark Office on the indicated dates: U.S. Provisional Number 60/253,404,
filed
November 27, 2000; U.S. Provisional Number 60/255,366, filed December 12,
2000;
U.S. Provisional Number 60/270,286 filed February 20, 2001; U.S. Provisional
Number
60/282,356, filed April 6, 2001, which claims priority from U.S. Provisional
Number
60/270,266, filed February 20, 2001; U.S. Provisional Number 60/282,032, filed
April 6,
2001; U.S. Provisional Number 60/282,358, filed April 6, 2001; U. S.
Provisional
Number 60/282,365, filed April 6, 2001; U.S. Provisional Number 60/290,917,
filed
May 14, 2001; U.S. Provisional Number 60/309,208, filed July 31, 2001; the
disclosures
of which are incorporated in their entirety by reference.
FIELD OF THE INVENTION
The present invention relates to transmembrane receptors, in some embodiments
to
G protein-coupled receptors and, in some preferred embodiments, to endogenous
GPCRs
that are altered to establish or enhance constitutive activity of the
receptor. In some
embodiments, the constitutively activated GPCRs will be used for the direct
identification
of candidate compounds as receptor agonists or inverse agonists having
applicability as
therapeutic agents.
1
CA 02429860 2003-05-26
BACKGROUND OF THE INVENTION
PATENT
Although a number of receptor classes exist in humans, by far the most
abundant
and therapeutically relevant is represented by the G protein-coupled receptor
(GPCR) class.
It is estimated that there are some 30,000-40,000 genes within the human
genome, and of
these, approximately 2% are estimated to code for GPCRs. Receptors, including
GPCRs,
for which the endogenous ligand has been identified, are referred to as
"known" receptors,
while receptors for which the endogenous ligand has not been identified are
referred to as
"orphan" receptors.
GPCRs represent an important area for the development of pharmaceutical
products: from approximately 20 of the 100 known GPCRs, approximately 60% of
all
prescription pharmaceuticals have been developed. For example, in 1999, of the
top 100
brand name prescription drugs, the following drugs interact with GPCRs
(diseases and/or
disorders treated are indicated in parentheses):
Claritin~ (allergies)Prozac~ (depression)Vasotec~ (hypertension)
Paxil~ (depression) Zoloftfl (depression)Zyprexa ~ (psychotic
disorder)
Cozaar~ (hypertension)Imitrex~ (migraine)Zantac~ (reflux)
Propulsid~ (reflux Risperdal~ (schizophrenia)Serevent~ (astlnna)
disease)
Pepcid~ (reflux) Gaster~ (ulcers) Atrovenifl (bronchospasm)
Effexor~ (depression)Depakote~ (epilepsy)Cardura~ (prostatic
hypertrophy)
Allegra~ (allergies) Lupron~ (prostate Zoladex~ (prostate
cancer) cancer)
Diprivan~ (anesthesia)BuSpar~ (anxiety) Ventolin~ (bronchospasm)
Hytrin~ (hypertension)Wellbutrin~ (depression)Zyrtec~ (rhinitis)
Plavix~ (MI/stroke) Toprol-XL~ (hypertension)Tenormin~ (angina)
Xalatan~ (glaucoma)Singulair~ (asthma)Diovan~ (hypertension)
Harnal~ (prostatic hyperplasia)
(Med Ad News 1999 Data).
GPCRs share a common structural motif, having seven sequences of between 22 to
24 hydrophobic amino acids that form seven alpha helices, each of which spans
the
membrane (each span is identified by number, i.e., transmembrane-1 (TM-1),
transmebrane-2 (TM-2), etc.). The transmembrane helices are joined by strands
of amino
acids between transmembrane-2 and transmembrane-3, transmembrane-4 and
transmembrane-5, and transmembrane-6 and transmembrane-7 on the exterior, or
2
CA 02429860 2003-05-26
AREN-0309 PATENT
"extracellular" side, of the cell membrane (these are referred to as
"extracellular"
regions 1, 2 and 3 (EC-1, EC-2 and EC-3), respectively). The transmembrane
helices are
also joined by strands of amino acids between transmembrane-1 and
transmembrane-2,
transmembrane-3 and transmembrane-4, and transmembrane-5 and transmembrane-6
on the
interior, or "intracellular" side, of the cell membrane (these are referred to
as "intracellular"
regions 1, 2 and 3 (IC-1, IC-2 and IC-3), respectively). The "carboxy" ("C")
terminus of
the receptor lies in the intracellular space withili the cell, and the "amino"
("N") terminus of
the receptor lies in the extracellular space outside of the cell.
Generally, when an endogenous ligand binds with the receptor (often referred
to as
"activation" of the receptor), there is a change in the conformation of the
intracellular region
that allows for coupling between the intracellular region and an intracellular
"G-protein." It
has been reported that GPCRs are "promiscuous" with respect to G proteins, i.
e., that a
GPCR can interact with more than one G protein. See, I~enal~in, T., 43 Life
Sciences 1095
(1988). Although other G proteins exist, currently, Ga, GS, G;, GZ and Go are
G proteins that
have been identified. Ligand-activated GPCR coupling with the G-protein
initiates a
signaling cascade process (referred to as "signal transduction"). Under normal
conditions,
signal transduction ultimately results in cellular activation or cellular
inlubition. Although
not wishing to be bound to theory, it is thought that the IC-3 loop as well as
the carboxy
terminus of the receptor interact with the G protein.
Under physiological conditions, GPCRs exist in the cell membrane in
equilibrium
between two different conformations: an "inactive" state and an "active"
state. A receptor
in an inactive state is unable to link to the intracellular signaling
transduction pathway to
initiate signal transduction leading to a biological response. Changing the
receptor
conformation to the active state allows linkage to the transduction pathway
(via the G-
protein) and produces a biological response.
A receptor may be stabilized in an active state by a ligand or a compound such
as a
drug. Recent discoveries, including but not exclusively limited to
modifications to the
amino acid sequence of the receptor, provide means other than ligands or drugs
to promote
and stabilize the receptor in the active state conformation. These means
effectively stabilize
the receptor in an active state by simulating the effect of a ligand binding
to the receptor.
Stabilization by such ligand-independent means is termed "constitutive
receptor activation."
3
CA 02429860 2003-05-26
AREN-0309 PATENT
SITM1VIARY OF THE INVENTION
Disclosed herein are endogenous and non-endogenous versions of human GPCRs
and uses thereof.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.ID.N0.:2, non-endogenous,
constitutively activated versions of the same, and host cells comprising the
same..
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.N0.:1 and host cells comprising the same.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.ID.N0.:4, non-endogenous,
constitutively activated versions of the same, and host cells comprising the
same..
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.N0.:3, non-endogenous, constitutively activated
versions of the same, and host cells comprising the same.
Some embodiments of the present invention relate to G protein-coupled receptor
encoded by an amino acid sequence of SEQ.ID.N0.:6, non-endogenous,
constitutively
activated versions of the same, and host cells comprising the same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.NO.:S and host cells comprising the same.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.ID.NO.:B, non-endogenous,
constitutively activated versions of the same, and host cells comprising the
same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.N0.:7, non-endogenous, constitutively activated
versions of the same, and host cells comprising the same.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.ID.NO.:10, non-endogenous,
constitutively activated versions of the same, and host cells comprising the
same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.N0.:9 and host cells comprising the same.
4
CA 02429860 2003-05-26
AREN-0309 PATENT
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.ID.N0.:12, non-endogenous,
constitutively activated versions of the same, and host cells comprising the
same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.NO.:11, and host cells comprising the same.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.ID.N0.:14, constitutively
activated
versions of the same, and host cells comprising the same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.NO.:13 and host cells comprising the same.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.ID.N0.:16, constitutively
activated
versions of the same, and host cells comprising the same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.NO.:15 and host cells comprising the same.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.1D.N0.:18, constitutively
activated
versions of the same, and host cells comprising the same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.NO.:17 and host cells comprising the same.
Some embodiments of the present invention relate to a G protein-coupled
receptor encoded by an amino acid sequence of SEQ.1D.NO.:20, constitutively
activated
versions of the same, and host cells comprising the same.
Some embodiments of the present invention relate to a plasmid comprising a
vector and the cDNA of SEQ.ID.N0.:19 and host cells comprising the same.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure 1 is a graphic representation of activation of RUP32, Ga(del)/G, RUP32
co-transfected with Gg(del)/G;, and CMV (control; expression vector) in a
second
messenger assay measuring the accumulation of inositol phosphate (IP3)
utilizing 293 cells.
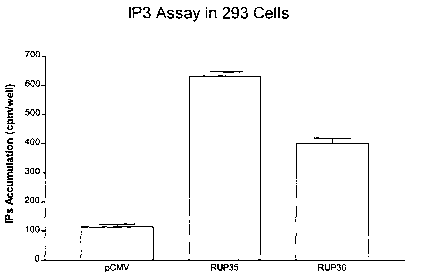
Figure 2 provides an illustration of second messenger IP3 production from
endogenous version RUP35 and RUP36 as compared with the control ("CMV").
5
CA 02429860 2003-05-26
AREN-0309 PATENT
DETA~ED DESCRIPTION
The scientific literature that has evolved around receptors has adopted a
number of
terms to refer to ligands having various effects on receptors. For clarity and
consistency,
the following definitions will be used throughout this patent document. To the
extent that
these definitions conflict with other definitions for these terms, the
following definitions
shall control:
AGONISTS shall mean materials (e.g., ligands, candidate compounds) that
activate
the intracellular response when they bind to the receptor, or enhance GTP
binding to
membranes. In some embodiments, AGONISTS are those materials not previously
known
to activate the intracellular response when they bind to the receptor or to
enhance GTP
binding to membranes.
AMINO ACID ABBREVIATIONS used herein are set out in Table A:
TABLE A
ALANINE ALA A
ARGININE ARG R
ASPARAGINE ASN N
ASPARTIC ACID ASP D
CYSTEINE CYS C
GLUTAMIC ACID GLU E
GLUTAM1NE GLN Q
GLYCINE GLY G
HISTIDINE HIS H
ISOLEUCINE ILE I
LEUC1NE LEU L
LYSINE LYS K
6
CA 02429860 2003-05-26
AREN-0309 PATENT
METHIONINE MET M
PHENYLALANINE PHE F
PROLINE PRO P
SERINE SER S
THREONINE THR T
TRYPTOPHAN TRP W
TYROSINE TYR Y
VALINE VAL V
ANTAGONIST shall mean materials (e.g., ligazids, candidate compounds) that
competitively bind to the receptor at the same site as the agonists but which
do not activate
the intracellular response initiated by the active form of the receptor, and
can thereby inhibit
the intracellular responses by agonists. ANTAGOI~IISTS do not diminish the
baseline
intracellular response in the absence of an agonist. In some embodiments,
ANTAGOI~IISTS are those materials not previously known to activate the
intracellular
response when they bind to the receptor or to enhance GTP binding to
membranes.
CANDIDATE COMPOUND shall mean a molecule (for example, and not
limitation, a chemical compound) that is amenable to a screening technique.
Preferably, the
phrase "candidate compound" does not include compounds which were publicly
known to
be compounds selected from the group consisting of inverse agonist, agonist or
antagonist
to a receptor, as previously determined by an indirect identification process
("indirectly
identified compound"); more preferably, not including an indirectly identified
compound
which has previously been determined to have therapeutic efficacy in at least
one mammal;
and, most preferably, not including an indirectly identified compound which
has previously
been determined to have therapeutic utility in humans.
COMPOSITION means a material comprising at least one component; a
"pharmaceutical composition" is an example of a composition.
COMPOUND EFFICACY shall mean a measurement of the ability of a
compound to inhibit or stimulate receptor functionality; i.e. the ability to
activate/inhibit a
7
CA 02429860 2003-05-26
AREN-0309 PATENT
signal transduction pathway, as opposed to receptor binding affinity.
Exemplary means
of detecting compound efficacy are disclosed in the Example section of this
patent
document.
CODON shall mean a grouping of three nucleotides (or equivalents to
nucleotides)
which generally comprise a nucleoside (adenosine (A), guanosine (G), cytidine
(C), uridine
(U) and thymidine (T)) coupled.to a phosphate group and which, when
translated, encodes
an amino acid.
CONSTITUTIVELY ACTIVATED RECEPTOR shall mean a receptor
subjected to constitutive receptor activation. A constitutively activated
receptor can be
endogenous or non-endogenous.
CONSTITUTIVE RECEPTOR ACTIVATION shall mean stabilization of a
receptor in the active state by means other than binding of the receptor with
its ligand or a
chemical equivalent thereof.
CONTACT or CONTACTING shall mean bringing at least two moieties together,
whether in an in vitro system or an in vivo system.
DIRECTLY IDENTIFYING or DIRECTLY IDENTIFIED, in relationship to
the phrase "candidate compound", shall mean the screening of a candidate
compound
against a constitutively activated receptor, preferably a constitutively
activated orphan
receptor, and most preferably against a coiistitutively activated G protein-
coupled cell
surface orphan receptor, and assessing the compound efficacy of such compound.
This
phrase is, under no circumstances, to be interpreted or understood to be
encompassed by or
to encompass the phrase "indirectly identifying" or "indirectly identified."
ENDOGENOUS shall mean a material that a mammal naturally produces.
ENDOGENOUS in reference to, for example and not limitation, the term
"receptor," shall
mean that which is naturally produced by a mammal (for example, and not
limitation, a
human) or a virus. By contrast, the term NON-ENDOGENOUS in this context shall
mean
that which is not naturally produced by a mammal (for example, and not
limitation, a
human) or a virus. For example, and not limitation, a receptor which is not
constitutively
active in its endogenous form, but when manipulated becomes constitutively
active, is most
preferably referred to herein as a "non-endogenous, constitutively activated
receptor." Both
terms can be utilized to describe both "in vivo" and "in vitro" systems. For
example, and
not limitation, in a screening approach, the endogenous or non-endogenous
receptor may be
8
CA 02429860 2003-05-26
AREN-0309 PATENT
in reference to an in vitro screening system. As a further example and not
limitation,
where the genome of a mammal has been manipulated to include a non-endogenous
constitutively activated receptor, screening of a candidate compound by means
of an in vivo
system is viable.
G PROTEIN COUPLED RECEPTOR FUSION PROTEIN and GPCR
FUSION PROTEIN, in the context of the invention disclosed herein, each mean a
non-
endogenous protein comprising an endogenous, constitutively activate GPCR or a
non-
endogenous, constitutively activated GPCR fused to at least one G protein,
most preferably
the alpha (oc) subunit ~of such G protein (this being the subunit that binds
GTP), with the G
protein preferably being of the same type as the G protein that naturally
couples with
endogenous orphan GPCR. For example, and not limitation, in an endogenous
state, if the
G protein "Gsoc" is the predominate G protein that couples with the GPCR, a
GPCR Fusion
Protein based upon the specific GPCR would be a non-endogenous protein
comprising the
GPCR fused to Gsa; in some circumstances, as will be set forth below, a non-
predominant
G protein can be fused to the GPCR. The G protein can be fused directly to the
C-terminus
of the constitutively active GPCR or there may be spacers between the two.
HOST CELL shall mean a cell capable of having a Plasmid and/or Vector
incorporated therein. In the case of a prokaryotic Host Cell, a Plasmid is
typically
replicated as a autonomous molecule as the Host Cell replicates (generally,
the Plasmid is
thereafter isolated, for introduction into a eukaryotic Host Cell); in the
case of a eulcaryotic
Host Cell, a Plasmid is integrated into the cellular DNA of the Host Cell such
that when the
eukaryotic Host Cell replicates, the Plasmid replicates. In some embodiments
the Host Cell
is eukaryotic, more preferably, mammalian, and most preferably selected from
the group
consisting of 293, 293T and COS-7 cells.
INDIRECTLY IDENTIFYING or INDIRECTLY IDENTIFIED means the
traditional approach to the drug discovery process involving identification of
an endogenous
ligand specific for an endogenous receptor, screening of candidate compounds
against the
receptor for determination of those which interfere andlor compete with the
ligand-receptor
interaction, and assessing the efficacy of the compound for affecting at least
one second
messenger pathway associated with the activated receptor.
9
CA 02429860 2003-05-26
AREN-0309 PATENT
INHIBIT or INHIBITING, in relationship to the term "response" shall
mean that a response is decreased or prevented in the presence of a compound
as opposed to
in the absence of the compound.
INVERSE AGONISTS shall mean materials (e.g., ligand, candidate compound)
which bind to either the endogenous form of the receptor or to the
constitutively activated
form of the receptor, and which inhibit the baseline intracellular response
initiated by the
active form of the receptor below the normal base level of activity which is
observed in the
absence of agonists, or decrease GTP binding to membranes. Preferably, the
baseline
intracellular response is inhibited in the presence of the inverse agonist by
at least 30%, at
least 50%, at least 60%, at least 70%, at least 75%, at least 80%, at least
85%, at least 90%,
at least 92%, at least 94%, at least 95%, at least 96%, at least 97%, at least
98%, and most
preferably at least 99% as compared with the baseline response in the absence
of the inverse
agonist.
KNOWN RECEPTOR shall' mean an endogenous receptor for which the
endogenous ligand specific for that receptor has been identified.
LIGAND shall~mean a molecule specific for a naturally occurring receptor.
MUTANT or MUTATION in reference to an endogenous receptor's nucleic acid
andlor amino acid sequence shall mean a specified change or changes to such
endogenous
sequences such that a mutated form of an endogenous, non-constitutively
activated receptor
evidences constitutive activation of the receptor. In terms of equivalents to
specific
sequences, a subsequent mutated form of a human receptor is considered to be
equivalent to
a first mutation of the human receptor if (a) the level of constitutive
activation of the
subsequent mutated form of a human receptor is substantially the same as that
evidenced by
the first mutation of the receptor; and (b) the percent sequence (amino acid
and/or nucleic
acid) homology between the subsequent mutated form of the receptor and the
first mutation
of the receptor is at least 80%, at least 85%, at least 90%, at least 92%, at
least 94%, at least
95%, at least 96%, at least 97%, at least 98%, and most preferably at least
99%. In some
embodiments, owing to the fact that some preferred cassettes disclosed herein
for achieving
constitutive activation include a single amino acid and/or codon change
between the
endogenous and the non-endogenous forms of the GPCR, it is preferred that the
percent
sequence homology should be at least 98%.
CA 02429860 2003-05-26
AREN-0309 PATENT
NON-ORPHAN RECEPTOR shall mean an endogenous naturally occurring
molecule specific for an identified ligand wherein the binding of a ligand to
a receptor
activates an intracellular signaling pathway.
ORPHAN RECEPTOR shall mean an endogenous receptor for which the ligand
specific for that receptor has not been identified or is not known.
PHARMACEUTICAL COMPOSITION shall mean a composition comprising at
least one active ingredient, whereby the composition is amenable to
investigation for a
specified, efficacious outcome in a mammal (for example, and not limitation, a
human).
Those of ordinary skill in the art will understand and appreciate the
techniques appropriate
for determining whether an active ingredient has a desired efficacious outcome
based upon
the needs of the artisan.
PLASMID shall mean the combination of a Vector and cDNA. Generally, a
Plasmid is introduced into a Host Cell for the purposes of replication and/or
expression of
the cDNA as a protein.
SECOND MESSENGER shall mean an intracellular response produced as a result
of receptor activation. A second messenger can include, for example, inositol
triphosphate
(1P3), diacycglycerol (DAG), cyclic AMP (cAMP), and cyclic GMP (cGMP). Second
messenger response can be measured for a determination of receptor activation.
1n addition,
second messenger response can be measured for the direct identification of
candidate
compounds, including for example, inverse agonists, agonists, and antagonists.
SIGNAL TO NOISE RATIO shall mean the signal generated in response to
activation, amplification, or stimulation wherein the signal is above the
baclcground noise or
the basal level in response to non-activation, non-amplification, or non-
stimulation.
SPACER shall mean a translated number of amino acids that are located after
the
last codon or last amino acid of a gene, for example a GPCR of interest, but
before the start
codon or beginning regions of the G protein of interest, wherein the
translated number
amino acids are placed in-frame with the beginnings regions of the G protein
of interest.
The number of translated amino acids can be tailored according to the needs of
the skilled
artisan and is generally from about one amino acid, preferably two amino
acids, more
preferably three amino acids, more preferably four amino acids, more
preferably five amino
acids, more preferably six amino acids, more preferably seven amino acids,
more preferably
11
CA 02429860 2003-05-26
AREN-0309 PATENT
eight amino acids, more preferably. nine amino acids, more preferably ten
amino
acids, more preferably eleven amino acids, and even more preferably twelve
amino acids.
STIMULATE or STIMULATING, in relationship to the term "response" shall
mean that a response is increased in the presence of a compound as opposed to
in the
absence of the compound.
SUBSTANTIALLY shall refer to a result which is within 40% of a control result,
preferably within 35%, more preferably within 30%, more preferably within 25%,
more
preferably within 20%, more preferably within 15%, more preferably within 10%,
more
preferably within 5%, more preferably within 2%, and most preferably within 1%
of a
control result. For example, in the context of receptor functionality, a test
receptor may
exhibit substantially similar results to a control receptor if the transduced
signal, measured
using a method taught herein or similar method known to the art-skilled, if
within 40% of
the signal produced by a control signal.
VECTOR in reference to cDNA shall mean a circular DNA capable of
incorporating at least one cDNA and capable of incorporation into a Host Cell.
The order of the following sections is set forth for presentational efficiency
and is
not intended, nor should be construed, as a limitation on the disclosure or
the claims to
follow.
A. Introduction
The traditional study of receptors has typically proceeded from the a priori
assumption (historically based) that the endogenous ligand must first be
identified before
discovery could proceed to find antagonists and other molecules that could
affect the
receptor. Even in cases where an antagonist might have been known first, the
search
immediately extended to looking for the endogenous ligand. This mode of
thinking has
persisted in receptor research even after the discovery of constitutively
activated receptors.
What has not been heretofore recognized is that it is the active state of the
receptor that is
most useful for discovering agonists and inverse agonists of the receptor. For
those diseases
which result from an overly active receptor or an under-active receptor, what
is desired in a
therapeutic drug is a compound which acts to diminish the active state of a
receptor or
enhance the activity of the receptor, respectively, not necessarily a drug
which is an
antagonist to the endogenous ligand. This is because a compound that reduces
or enhances
12
CA 02429860 2003-05-26
AREN-0309 PATENT
the activity of the active receptor state need not bind at the same site as
the endogenous
ligand. Thus, as taught by a method of this invention, any search for
therapeutic
compounds should start by screening compounds against the ligand-independent
active
state.
B. Identification of Human GPCRs
The efforts of the Human Genome project has led to the identification of a
plethora
of information regarding nucleic acid sequences located within the human
genome; it has
been the case in this endeavor that genetic sequence information has been made
available
without an understanding or recognition as to whether or not any particular
genomic
sequence does or may contain open-reading frame information that translate
human
proteins. Several methods of identifying nucleic acid sequences within the
human genome
are within the purview of those having ordinary shill in the art. For example,
and not
limitation, a variety of human GPCRs, disclosed herein, were discovered by
reviewing the
GenBankTM database. Table B, below, lists several endogenous GPCRs that we
have
discovered, along with other GPCRs that are homologous to the disclosed GPCR.
TABLE S
Disclosed Accession Open Reading Reference Per Cent
Human Number Frame To Homology
Orphan GPCRsIdentified (Base Pairs) Homologous To Designated
GPCR GPCR
hRUP28 AC073957 1,002bp hGPR30 34%
liRUP29 AC083865 918bp hGPRl8 27%
hRUP30 AC055863 1,125bp hBlZB1 27%
hRUP31 AL356214 1,086bp hGALR-1 31%
liRUP32 AL513524 1,038bp hPNR 43%
hRUP33 AL513524 1,020bp GPR57 50%
GPR58 51%
hRUP34 AL513524 1,029bp hPNR 45%
hRUP35 AC021089 1,062bp hK-type 27%
3
opioid
hRUP36 AC090099 969bp GPR90 42%
13
CA 02429860 2003-05-26
ARFN-0309 PATENT
hRUP37 ~ AC090099 969bp hMRG 41%
Receptor homology is useful in terms of gaining an appreciation of a role of
the
receptors within the human body. As the patent document progresses, techniques
for
mutating these receptors to establish non-endogenous, constitutively activated
versions of
these receptors will be discussed.
The techniques disclosed herein are also applicable to other human GPCRs known
to the art, as will be apparent to those skilled in the art.
C. Receptor Screening
Screening candidate compounds against a non-endogenous, constitutively
activated
version of the GPCRs disclosed herein allows for the direct identification of
candidate
compounds which act at the cell surface receptor, without requiring use of the
receptor's
endogenous ligand. Using routine, and often commercially available techniques,
one can
determine areas within the body where the endogenous version of human GPCRs
disclosed
herein is expressed and/or over-expressed. The expression location of a
receptor in a
specific tissue provides a scientist with the ability to assign a
physiological functional role
of the receptor. It is also possible using these techniques to determine
related
disease/disorder states which are associated with the expression and/or over-
expression of
the receptor; such an approach is disclosed in this patent document.
Furthermore,
expression of a receptor in diseased organs can assist one in determinilig the
magnitude of
the clinical relevance of the receptor.
Constitutive activation of the GPCRs disclosed herein is based upon the
distance
from the proline residue at which is presumed to be located within TM6 of the
GPCR; this
algorithmic technique is disclosed in co-pending and commonly assigned patent
document
PCT Application Number PCT/US99/2393~, published as WO 00/22129 on April 20,
2000,
wluch, along with the other patent documents listed herein, is incorporated
herein by
reference. The algorithmic technique is not predicated upon traditional
sequence
"alignment" but rather a specified distance from the aforementioned TM6
proline residue
(or, of course, endogenous constitutive substitution for such proline
residue). By mutating
the amino acid residue located 16 amino acid residues from this residue
(presumably
located in the IC3 region of the receptor) to, most preferably, a lysine
residue, constitutive
14
CA 02429860 2003-05-26
AREN-0309 PATENT
activation of the receptor may be obtained. Other amino acid residues may be
useful in
the mutation at this position to achieve this objective and will be discussed
in detail, below.
D. Disease/Disorder Identification and/or Selection
As will be set forth in greater detail below, inverse agonists and agonists to
the non-
endogenous, constitutively activated GPCR can be identified by the
methodologies of this
invention. Such inverse agonists and agonists are ideal candidates as lead
compounds in
drug discovery programs for treating diseases related to this receptor.
Because of the ability
to directly identify inverse agonists to the GPCR, thereby allowing for the
development of
pharmaceutical compositions, a search for diseases and disorders associated
with the GPCR
is relevant. The expression location of a receptor in a specific tissue
provides a scientist
with the ability to assign a physiological function to the receptor. For
example, scanning
both diseased and normal tissue samples for the presence of the GPCR now
becomes more
than an academic exercise or one which might be pursued along the path of
identifying an
endogenous ligand to the specific GPCR. Tissue scans can be conducted across a
broad
range of healthy and diseased tissues. Such tissue scans provide a potential
first step in
associating a specific receptor with a disease and/or disorder. Furthermore,
expression of a
receptor in diseased organs can assist one in determining the magnitude of
clinical
relevance of the receptor.
The DNA sequence of the GPCR can be used to make a probe/primer. In some
preferred embodiments the DNA sequence is used to make a probe for (a) dot-
blot analysis
against tissue-mRNA, and/or (b) RT-PCR identification of the expression of the
receptor in
tissue samples. The presence of a receptor in a tissue source, or a diseased
tissue, or the
presence of the receptor at elevated concentrations in diseased tissue
compared to a normal
tissue, can be used to correlate location to function and indicate the
receptor's physiological
role/function and create a treatment regimen, including but not limited to, a
disease
associated with that function/role. Receptors can also be localized to regions
of organs by
this technique. Based on the known or assumed roles/functions of the specific
tissues to
which the receptor is localized, the putative physiological function of the
receptor can be
deduced. For example and not limitation, proteins located/expressed in areas
of the
thalamus are associated with sensorimotor processing and arousal (see, Goodman
&
Gilman's, The Pharmacological Basis of Therapeutics, 9th Edition, page 465
(1996)).
Proteins expressed in the hippocampus or in Schwann cells are associated with
learning and
CA 02429860 2003-05-26
AREN-0309 PATENT
memory, and myelination of peripheral nerves, respectively (see, Kandel, E. et
al.,
Essentials of Neural Science and Behavior pages 657, 680 and 28, respectively
(1995)).
E. Screening of Candidate Compounds
1. Generic GPCR screening assay techniques
When a G protein receptor becomes constitutively active, it binds to a G
protein
(e.g., Gq, GS, G, GZ, Go) and stimulates the binding of GTP to the G protein.
The G protein
then acts as a GTPase and hydrolyzes the GTP to GDP, whereby the receptor,
under normal
conditions, becomes deactivated. However, constitutively activated receptors
continue to
exchange GDP to GTP. A non-hydrolyzable analog of GTP, [35S]GTPyS, can be.
used to
monitor enhanced binding to membranes which express constitutively activated
receptors.
It is reported that [35S]GTPyS can be used to monitor G protein coupling to
membranes in
the absence and presence of ligand. An example of this monitoring, among other
examples
well-knomn and available to those in the art, was reported by Traynor and
Nahorski in
1995. The use of this assay system is typically for initial screening of
candidate compounds
because the system is generically applicable to all G protein-coupled
receptors regardless of
the particular G protein that interacts with the intracellular domain of the
receptor.
2. Specific GPCR screening assay techniques
Once candidate compounds are identified using the "generic" G protein-coupled
receptor assay (i.e., an assay to select compounds that are agonists or
inverse agonists),
further screening to confirm that the compounds have interacted at the
receptor site is
preferred. For example, a compound identified by the "generic" assay may not
bind to the
receptor, but may instead merely "uncouple" the G protein from the
intracellular domain.
a. GS, GZ atzd G~.
GS stimulates the enzyme adenylyl cyclase. G; (and GZ and Go), on the other
hand,
inhibits adenylyl cyclase. Adenylyl cyclase catalyzes the conversion of ATP to
cAMP;
thus, constitutively activated GPCRs that couple the GS protein are associated
with
increased cellular levels of cAMP. On the other hand, constitutively activated
GPCRs that
couple Gi (or GZ, Go) protein are associated with decreased cellular levels of
cAMP. See,
generally, "Indirect Mechanisms of Synaptic Transmission," Chpt. 8, From
Neuron To
Brain (3rd Ed.) Nichols, J.G. et al eds. Sinauer Associates, Inc. (1992).
Thus, assays that
detect cAMP can be utilized to determine if a candidate compound is, e.g., an
inverse
16
CA 02429860 2003-05-26
AREN-0309 PATENT
agonist to the receptor (i.e., such a compound would decrease the levels of
cAMP). A
variety of approaches known in the art for measuring cAMP can be utilized; a
most
preferred approach relies upon the use of anti-cAMP antibodies in an ELISA-
based format.
Another type of assay that can be utilized is a whole cell second messenger
reporter system
assay. Promoters on genes drive the expression of the proteins that a
particular gene
encodes. Cyclic AMP drives gene expression by promoting the binding of a cAMP-
responsive DNA binding protein or transcription factor (CREB) that then binds
to the
promoter at specific sites (CAMP response elements) and drives the expression
of the gene.
Reporter systems can be constructed which have a promoter containing multiple
cAMP
response elements before the reporter gene, e.g., (3-galactosidase or
luciferase. Thus, a
constitutively activated GS linked receptor causes the accumulation of cAMP
that then
activates the gene and leads to the expression of the reporter protein. The
reporter protein
such as (3-galactosidase or luciferase can then be detected using standard
biochemical
assays (Chen et al. 1995).
b. Go ahd G9.
Gq and Go are associated with activation of the enzyme phospholipase C, which
in
turn hydrolyzes the phospholipid PIPZ, releasing two intracellular messengers:
diacycloglycerol (DAG) and inositol 1,4,5=triphoisphate (IP3). Increased
accumulation of
IP3 is associated with activation of Gq and Go-associated receptors. See,
gehe~ally,
"Indirect Mechanisms of Synaptic Transmission," Chpt. 8, From Neuron To Brain
(3rd Ed.)
Nichols, J.G. et al eds. Siilauer Associates, Inc. (1992). Assays that detect
IP3 accumulation
can be utilized to determine if a candidate compound is, e.g., an inverse
agonist to a Gq or
Go-associated receptor (i. e., such a compound would decrease the levels of
IP3). Gq
associated receptors can also be examined using an AP1 reporter assay wherein
Gq
dependent phospholipase C causes activation of genes containing AP1 elements;
thus,
activated Gg associated receptors will evidence an increase in the expression
of such genes,
whereby inverse agonists thereto will evidence a decrease in such expression,
and agonists
will evidence an increase in such expression. Commercially available assays
for such
detection are available.
3. GPCR Fusion Protein
17
CA 02429860 2003-05-26
AREN-0309 PATENT
The use of an endogenous, constitutively activated GPCR or a
non-endogenous, constitutively activated GPCR, for use in screening of
candidate
compounds for the direct identification of inverse agonists, agoiusts provide
an interesting
screening challenge in that, by definition, the receptor is active even in the
absence of an
endogenous ligand bound thereto. Thus, in order to differentiate between,
e.g., the non-
endogenous receptor in the presence of a candidate compound and the non-
endogenous
receptor in the absence of that compound, twith an aim of such a
differentiation to allow for
an understanding as to whether such compound may be an inverse agonist or
agonist or
have no affect on such a receptor, it is preferred that an approach be
utilized that can
enhance such differentiation. A preferred approach is the use of a GPCR Fusion
Protein.
Generally, once it is determined that a non-endogenous GPCR has been
constitutively activated using the assay techniques set forth above (as well
as others), it is
possible to determine the predominant G protein that couples with the
endogenous GPCR.
Coupling of the G protein to the GPCR provides a signaling pathway that can be
assessed.
In some embodiments it is preferred that screening take place using a
mammalian
expression system, such a system will be expected to have endogenous G protein
therein.
Thus, by definition, in such a system, the non-endogenous, constitutively
activated GPCR
will continuously signal. In some embodiments it is preferred that this signal
be enhanced
such that in the presence of, e.g., an inverse agonist to the receptor, it is
more likely that it
will be able to more readily differentiate, particularly in the context of
screening, between
the receptor when it is contacted with the inverse agonist.
The GPCR Fusion Protein is intended to enhance the efficacy of G protein
coupling
with the non-endogenous GPCR. The GPCR Fusion Protein is preferred for
screening with
either an endogenous, constitutively active GPCR or a non-endogenous,
constitutively
activated GPCR because such an approach increases the signal that is utilized
in such
screening techniques. This is important in facilitating a significant "signal
to noise" ratio;
such a significant ratio is preferred for the screening of candidate compounds
as disclosed
herein.
The construction of a construct useful for expression of a GPCR Fusion Protein
is
within the purview of those having ordinary skill in the art. Commercially
available
expression vectors and systems offer a variety of approaches that can fit the
particular needs
of an investigator. Important criteria on the construction of such a GPCR
Fusion Protein
18
CA 02429860 2003-05-26
A RF N-0309 PATENT
construct include but are not limited to, that the endogenous GPCR sequence
and the G
protein sequence both be in-frame (preferably, the sequence for the endogenous
GPCR is
upstream of the G protein sequence), and that the "stop" codon of the GPCR be
deleted or
replaced such that upon expression of the GPCR, the G protein can also be
expressed.
Other embodiments include constructs wherein the endogenous GPCR sequence and
the G
protein sequence are not in-frame and/or the "stop" codon is not deleted or
replaced. The
GPCR can be linked directly to the G protein, or there can be spacer residues
between the
two (preferably, no more than about 12, although this number can be readily
ascertained by
one of ordinary skill in the art). Based upon convenience it is preferred to
use a spacer.
Preferably, the G protein that couples to the non-endogenous GPCR will have
been
identified prior to the creation of the GPCR Fusion Protein construct. Because
there are
only a few G proteins that have been identified, it is preferred that a
construct comprising
the sequence of the G protein (i.e., a universal G protein construct (see
Examples)) be
available for insertion of an endogenous GPCR sequence therein; this provides
for further
efficiency in the context of large-scale screening of a variety of different
endogenous
GPCRs having different sequences.
As noted above, constitutively activated GPCRs that couple to G, GZ and Go are
expected to inhibit the formation of cAMP making assays based upon these types
of GPCRs
challenging (i.e., the cAMP signal decreases upon activation thus malting the
direct
identification of, e.g., inverse agonists (which would further decrease this
signal),
challenging. As will be disclosed herein, we have ascertained that for these
types of
receptors, it is possible to create a GPCR Fusion Protein that is not based
upon the GPCRs
endogenous G protein, in an effort to establish a viable cyclase-based assay.
Thus, for
example, an endogenous G; coupled receptor can be fused to a GS protein -such
a fusion
construct, upon expression, "drives" or "forces" the endogenous GPCR to couple
with, e.g.,
GS rather than the "natural" G, protein, such that a cyclase-based assay can
be established.
Thus, for G, G~ and Go coupled receptors, in some embodiments it is preferred
that when a
GPCR Fusion Protein is used and the assay is based upon detection of adenylyl
cyclase
activity, that the fusion construct be established with GS (or an equivalent G
protein that
stimulates the formation of the enzyme adenylyl cyclase).
G Effect of CAMP Effect of IP3 Effect of Effect on Il'3
19
CA 02429860 2003-05-26
AREN-0309 PATENT
protein Production Accumulation cAMP Accumulation
upon
Activation upon ActivationProductionupon contact
of
GPCR (i.e., of GPCR (i.e.,upon with an Inverse
constitutive constitutive contact Agonist
activation activation with an
or or
agonist binding)agonist binding)Inverse
Agonist
GS Increase N/A Decrease N/A
G; Decrease N/A Increase N/A
GZ Decrease N/A Increase N/A
Go Decrease Increase Increase Decrease
Gq N/A Increase NIA Decrease
Equally effective is a G Protein Fusion construct that utilizes a Gq Protein
fused
with a GS, G, GZ or Go Protein. In some embodiments a preferred fusion
construct can be
accomplished with a Gg Protein wherein the first six (6) amino acids of the G-
protein a-
subunit ("Gaq") is deleted and the last five (5) amino acids at the C-terminal
end of Gaq is
replaced with the corresponding amino acids of the Ga of the G protein of
interest. For
example, a fusion construct can have a Gq (6 amino acid deletion) fused with a
G; Protein,
resulting in a "Gq/G Fusion Construct". This fusion construct will forces the
endogenous
G; coupled receptor to couple to its non-endogenous G protein, Gg, such that
the second
messenger, for example, inositol triphosphate or diacylgycerol, can be
measured ifZ lieu of
cAMP production.
4. Co-transfection of a Target G; Coupled GPCR with a Signal-
Enhancer GS Coupled GPCR (CAMP Based Assays)
A G; coupled receptor is known to inhibit adenylyl cyclase, and, therefore,
decreases
the level of cAMP production, which can malce assessment of cAMP levels
challenging. An
effective technique in measuring the decrease in production of cAMP as an
indication of
constitutive activation of a receptor that predominantly couples G; upon
activation can be
accomplished by co-transfecting a signal enhancer, e.g., a non-endogenous,
constitutively
activated receptor that predominantly couples with GS upon activation (e.g.,
TSHR-A623I,
disclosed below), with the G; linked GPCR. As is apparent, constitutive
activation of a GS
CA 02429860 2003-05-26
AREN-0309 PATENT
coupled receptor can be determined based upon an increase in production of
cAMP.
Constitutive activation of a G coupled receptor leads to a decrease in
production cAMP.
Thus, the co-transfection approach is intended to advantageously exploit these
"opposite"
affects. For example, co-transfection of a non-endogenous, constitutively
activated GS
coupled receptor (the "signal enhancer") with the endogenous G; coupled
receptor (the
"target receptor") provides a baseline cAMP signal (i. e., although the G;
coupled receptor
will decrease cAMP levels, this "decrease" will be relative to the substantial
increase in
cAMP levels established by constitutively activated GS coupled signal
enhancer). By then
co-transfecting the signal enhancer with a constitutively activated version of
the target
receptor, cAMP would be expected to fiufiher decrease (relative to base line)
due to the
increased functional activity of the G; target (i.e., which decreases cAMP).
Screening of candidate compounds using a cAMP based assay can then be
accomplished, with two 'changes' relative to the use of the endogenous
receptor/G-protein
fusion: first, relative to the G, coupled target receptor, "opposite" effects
will result, i. e., an
inverse agonist of the G, coupled target receptor will increase the measured
cAMP signal,
while an agonist of the G; coupled target receptor will decrease this signal;
second, as would
be apparent, candidate compounds that are directly identified using this
approach should be
assessed iildependently to ensure that these do not target the signal
enhancing receptor (this
can be done prior to or after screening against the co-transfected receptors).
F. Medicinal Chemistry
Generally, but not always, direct identification of candidate compounds is
conducted in conjunction with compounds generated via combinatorial chemistry
techniques, whereby thousands of compounds are randomly prepared for such
analysis.
Generally, the results of such screening will be compounds having unique core
structures; thereafter, these compounds may be subjected to additional
chemical
modification around a preferred core structures) to further enhance the
medicinal
properties thereof. Such techniques are known to those in the art and will not
be
addressed in detail in this patent document.
G. Pharmaceutical compositions
21
CA 02429860 2003-05-26
AREN-0309 PATENT
Candidate compounds selected for further development can be formulated into
pharmaceutical compositions using techniques well known to those in the art.
Suitable
pharmaceutically-acceptable carriers are available to those in the art; for
example, see
Remington's Pharmaceutical Sciences, 16t1' Edition, 1980, Mack Publishing Co.,
(Osol et
al., eds.).
H. Other Utilities
Although a preferred use of the non-endogenous versions of the GPCRs disclosed
herein may be for the direct identification of candidate compounds as inverse
agonists or
agonists (preferably for use as pharmaceutical agents), other uses of these
versions of
GPCRs exist. For example, ih vitro and in vivo systems incorporating GPCRs can
be
utilized to further elucidate and understand the roles these receptors play
i11 the human
condition, both normal and diseased, as well as understanding the role of
constitutive
activation as it applies to understanding the signaling cascade. In some
embodiments it is
preferred that the endogenous receptors be "orphan receptors", i.e., the
endogenous ligand
for the receptor has not been identified. In some embodiments, therefore, the
modified,
non-endogenous GPCRs can be used to understand the role of endogenous
receptors in the
human body before the endogenous ligand therefore is identified. Such
receptors can also
be used to further elucidate known receptors and the pathways through which
they
transduce a signal. Other uses of the disclosed receptors will become apparent
to those in
the art based upon, ifzte~ alia, a review of this patent document.
EXAMPLES
The following examples are presented for purposes of elucidation, and not
limitation, of the present invention. While specific nucleic acid and amino
acid sequences
are disclosed herein, those of ordinary skill in the art are credited with the
ability to make
minor modifications to these sequences while achieving the same or
substantially similar
results reported below. The traditional approach to application or
understanding of
sequence cassettes from one sequence to another (e.g. from rat receptor to
human receptor
or from human receptor A to human receptor B) is generally predicated upon
sequence
alignment techniques whereby the sequences are aligned in an effort to
determine areas of
commonality. The mutational approach disclosed herein does not rely upon this
approach
but is instead based upon an algorithmic approach and a positional distance
from a
conserved proline residue located within the TM6 region of human GPCRs. Once
this
22
CA 02429860 2003-05-26
AREN-0309 PATENT
approach is secured, those in the art are credited with the ability to make
minor
modifications thereto to achieve substantially the same results (i.e.,
constitutive activation)
disclosed herein. Such modified approaches are considered within the purview
of this
disclosure.
Example 1
ENDOGENOUS HUMAN GPCRS
1. Identification of Human GPCRs
The disclosed endogenous human GPCRs were identified based upon a review of
the GenBankTM database information. While searching the database, the
following
cDNA clones were identified as evidenced below (Table C).
TABLE C
DisclosedAccessionOpen Reference Nucleic AcidAmino
Human Number Reading To SEQ.ID. NO. Acid
Orphan IdentifiedFrame Homologous SEQ.H).NO.
GPCRs (Ease GPCR
Pairs)
hRUP28 AC073957 1,002bp hGPR30 1 2
hRUP29 AC083865 918bp hGPRl8 3 4
hRUP30 AC055863 1,125bp hBRBl 5 6
hRUP31 AL356214 1,086bp hGALR-1 7 8
hRUP32 AL513524 1,038bp hPNR 9 l0
hRUP33 AL513524 1,020bp GPR57 1l 12
GPR58
hRUP34 AL513524 1,029bp hPNR 13 14
hRUP35 AC021089 1,062bp hx-type 15 16
3
opioid
hRUP36 AC090099 969bp GPR90 17 18
hRUP37 AC090099 969bp hMRG 19 20
2. Full Length Cloning
a. hRUP28 (Seq. Id. Nos. l & 2)
The disclosed human RUP28 was identified based upon the use of GenBank
database
information. While searching the database, a cDNA clone with Accession Number
AC073957 was identified as a human genomic sequence from chromosome 7.
The full length RUP28 was cloned by PCR using primers:
23
CA 02429860 2003-05-26
AREN-0309 PATENT
5'- CAGAGCTCTGGTGGCCACCTCTGTCC-
3' (SEQ.ID.N0.:21; sense, 5' of initiation codon),
5'-CTGCGTCCACCAGAGTCACGTCTCC-3' (SEQ.ID.N0.:22; antisense, 3' of stop
codon), and human adult liver Marathon-ReadyTM cDNA (Clontech) as template.
AdvantageTM cDNA polymerase (Clontech) was used for the amplification in a
50w1
reaction by the following cycle with step 2 to 4 repeated 35 times:
95°C for 5 min; 94°C
for 30 sec; 58°C for 30 sec; 72°C for 1 min 30 sec; and
72°C for 7 min.
A 1.16kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems). See, SEQ.ID.NO.:l for the nucleic acid sequence and
SEQ.ID.N0.:2
for the putative amino acid sequence.
b. hRUP29 (Seq. Id. Nos. 3 & 4)
The disclosed human RUP29 was identified based upon the use of GenBank
database information. While searching the database, a cDNA clone with
Accession
Number AC0083865 was identified as a human genomic sequence from chromosome 7.
The full length RUP29 was cloned by PCR using primers:
5'-GTATGCCTGGCCACAATACCTCCAGG-3' (SEQ.ID.N0.:23; sense, containing the
initiation codon),
5'-GTTTGTGGCTAACGGCACAAAACACAATTCC-3' (SEQ.117.N0.:24; antisense,
containing the stop codon) and human genomic DNA as template. TaqPlus~
Precision
DNA polymerase (Stratagene) was used for the amplification in a 50,1 reaction
by the
following cycle with step 2 to 4 repeated 35 times: 94°C for 5 min;
94°C for 30 sec; 54°C
for 30 sec; 72°C for 1 min 30 sec; and 72°C for 7 min.
A 930bp PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems).
Rapid amplification of cDNA ends (R.ACE) was performed using human
leukocyte and ovary Marathon-ReadyTM cDNA (Clontech) to determine the precise
5'
end of RUP29 cDNA. RUP29 specific primer (1) having the sequence:
5-GGTACCACA.ATGACAATCACCAGCGTCC-3'(SEQ.ID.N0.:25)
and AP 1 primer (Clontech) were used for the first-round PCR reaction, and
RUP29
specific primer (2) having the following sequence:
24
CA 02429860 2003-05-26
AREN-0309 PATENT
5'-
GGAACGTGAGGTACATGTGGATGTGCAGC-3' (SEQ.ID.N0.:26)
and AP2 primer (Clontech) were used for the second-round PCR reaction. The
products
of the RACE reactions were isolated and cloned into the pCRII-TOPO vector
(Invitrogen) and sequenced. See, SEQ.ID.N0.:3 for the nucleic acid sequence
and
SEQ.ID.N0.:4 for the putative amino acid sequence. '
c. hRUP30 (Seq. Id. Nos. 5 & 6)
The disclosed human RUP30 was identified based upon the use of GenBank
database information. While searching the database, a cDNA clone with
Accession
Number AC055863 was identified as a human genomic sequence from chromosome 17.
The full length RUP30 was cloned by 5'R.ACE -PCR with a human pancreas
Marathon-ReadyTM cDNA (Clontech) as template and the following
oligonucleotide:
5'-GCAGTGTAGCGGTCAACCGTGAGCAGG-3'(SEQ.ID.N0.:27; sense, containing the
initiation codon), and AP1 primer (Clontech) were used for the first round of
RT-PCR and
oligonucleotide:
5'-TGAGCAGGATGGCGATCCAGACTGAGGCGTGG-3'(SEQ.ID.NO.:28; antisense,
containing the stop codon) and AP2 primer (Clontech) were used for the second
round of
PCR. DNA fragments generated by the 5' RACE-PCR were cloned into the pCRII-
TOPO
vector (Invitrogen) and sequenced using the SP6/T7 primers (Stratagene).
Based on the sequence of the 5' RACE products, the full length RUP30 was
cloned
by RT-PCR, using primers:
5'-GAGGTACAGCTGGCGATGCTGACAG-3' (SEQ.ID.N0.:29; sense, ATG as the
initiation codon);
5'-GTGGCCATGAGCCACCCTGAGCTCC-3' (SEQ.D~.N0.:30; antisense, 3' of the stop
codon) and human pancreas Marathon-ReadyTM cDNA (Clontech) as template. Taq
DNA
polymerase (Stratagene) was used for the amplification in 50 ~.l reaction by
the following
cycle with step 2 to step 4 repeated 35 times: 94°C for 40 seconds;
94°C for 20 seconds;
64°C for 20 seconds; 72°C for 2 minutes; and 72°C for 5
minutes.
A 1.2 Kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and several clones were sequenced using the ABI
Big
Dye Terminator kit (P.E. Biosystems). See, SEQ.117.N0.:5 for the nucleic acid
sequence
and SEQ.ID.N0.:6 for the putative amino acid sequence.
CA 02429860 2003-05-26
AREN-0309 PATENT
d. hRUP31 (Seq. Id. Nos. 7 & 8)
The disclosed human RUP31 was identified based upon the use of GenBank
database information. While searclung the database, a cDNA clone with
Accession
Number AL356214 was identified as a human genomic sequence from chromosome 10.
The full length RUP31 was cloned by RT-PCR using primers:
5'-GGAATGTCCACTGAATGCGCGCGG-3' (SEQ.D~.N0.:31; sense, containing the
initiation codon),
5'-AGCTCGCCAGGTGTGAGAAACTCGG-3' (SEQ.ID.NO.:32; antisense, 3' of stop
codon) and human mammary gland Marathon-ReadyTM cDNA (Clontech) as template.
AdvantageTM cDNA polymerase (Clontech) was used for the amplification in 50
~.1 reaction
by the following cycle with step 2 to step 4 repeated 35 times: 94°C
for 40 sec; 94°C for 20
sec; 66°C for 20 sec; 72°C for 1 min 30 sec; and 72°C for
5 min.
A 1.1 kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems). See, SEQ.ID.NO.:7 for the nucleic acid sequence and
SEQ.ID.N0.:8
for the putative amino acid sequence.
e. hRUP32 (Seq. Id. Nos. 9 & 10)
The disclosed human RUP32 was identified based upon the use of GenBank
database information. While searching the database, a cDNA clone with
Accession
Number AL513524 was identified as a human genomic sequence from chromosome 6.
The full length RUP32 was cloned by PCR using primers:
5'-GCGTTATGAGCAGCAATTCATCCCTGCTGG-3' (SEQ.ID.N0.:33; sense,
containing the initiation codon),
5'-GTATCCTGAACTTCGTCTATACAACTGC-3' (SEQ.ID.N0.:34; antisense)
and human genomic DNA (Clontech) as template. TaqPlus~ Precision DNA
polymerase
(Stratagene) was used for the amplification by the following cycle with step 2
to step 4
repeated 35 times: 94°C for 3 min; 94°C for 20 sec; 58°C
for 20 sec; 72°C for 1 min 30 sec;
and 72°C for 7 min.
A 1.06 kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems). See, SEQ.ID.N0.:9 for the nucleic acid sequence and
SEQ.ID.NO.:10 for the putative amino acid sequence.
26
CA 02429860 2003-05-26
AREN-0309 PATENT
f. hRUP33 (Seq. Id. Nos. 1l & 12)
The disclosed human RUP33 was identified based upon the use of GenBank
database information. While searching the database, a cDNA clone with
Accession
Number AL513524 was identified as a human genomic sequence from chromosome 6.
The full length RUP33 was cloned by PCR using primers:
5'-CCCTCAGGAATGATGCCCTTTTGCCACAA-3' (SEQ.ID.N0.:35; sense, containing
the initiation codon),
5'-ATCCATGTGGTTGGTGCATGTGGTTCGT-3' (SEQ.ID.N0.:36; antisense)
and human genomic DNA (Clontech) as template. TaqPlus~ Precision DNA
polymerase
(Stratagene) was used for the amplification by the following cycle with step 2
to step 4
repeated 35 times: 94°C for 3 min; 94°C for 20 sec; 56°C
for 20 sec; 72°C for 1 min 30 sec;
and 72°C for 7 min.
A 1.1 kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems). See, SEQ.ID.NO.:11 for the nucleic acid sequence and
SEQ.ID.NO.:12 for the putative amino acid sequence.
g. hRUP34 (Seq. Id. Nos.13 & 14)
The disclosed human RUP34 was identified based upon the use of GenBank
database information. While searching the database, ' a cDNA clone with
Accession
Number AL513524 was identified as a human genomic sequence from chromosome 6.
The full length RUP34 was cloned by PCR using primers:
5'-AAACAACAAACAGCAGAACCATGACCAGC-3' (SEQ.ID.N0.:37; sense,
containing the initiation codon),
5'-ACATAGAGACAAGTGACATGTGTGAACCAC-3' (SEQ.ID.N0.:38; antisense)
and human genomic DNA (Clontech) as template. TaqPlus~ Precision DNA
polymerase
(Stratagene) was used for the amplification by the following cycle with step 2
to step 4
repeated 35 times: 94°C for 3 min; 94°C for 20 sec; 60°C
for 20 sec; 72°C for 1 min 30 sec;
and 72°C for 7 min.
A 1.27 kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems). See, SEQ.ID.N0.:13 for the nucleic acid sequence and
SEQ.ID.N0.:14 for the putative amino acid sequence.
27
CA 02429860 2003-05-26
AREN-0309 PATENT
h. hRUP35 (Seq. Id. Nos.15 & 16)
The disclosed human RUP35 was identified based upon the use of GenBank
database information. While searching the database, a cDNA clone with
Accession
Number AC021089 was identified as a human genomic sequence from chromosome 16.
The 5' sequence of RUP35 was determined by 5' RACE- PCR with a human fetal
brain Marathon-ReadyTM cDNA (Clontech) as template. Oligonucleotide
5'-GGTATGAGACCGTGTGGTACTTGAGC-3' (SEQ.ID.NO.:39; sense)
and APl primer (Clontech) were used for the first round of RT-PCR and
oligonucleotide
5'-GTGGCAGACAGCGATATACCTGTCAATGG-3' (SEQ.ID N0.:40; antisense)
and AP2 primer (Clontech) were used for the second round of PCR. DNA fragments
generated by the 5' RACE-PCR were cloned into the pCRII-TOPO vector
(Invitrogen) and
sequenced using the SP6/T7 primers (Stratagene).
Based upon the sequence of the 5' RACE products, the full length RUP35 was
cloned by RT-PCR, using primers
5'-GCGCTCATGGAGCACACGCACGCCCAC-3' (SEQ.ID.N0.:41; sense, ATG as the
initiation codon) and
5'-GAGGCAGTAGTTGCCACACCTATGG-3' (SEQ.ID.N0.:42; antisense, 3' of the stop
codon) and human brain Marathon-ReadyTM cDNA (Clontech) as template.
AdvantageTM
cDNA polymerase (Clontech) was used for the amplification in 100 ~,1 reaction
by the
following cycle with step 2 to step 4 repeated 45 times: 95°C for 2
min; 95°C for 20 sec;
60°C for 20 sec; 72°C for 1 min 30 sec; and 72°C for 5
min.
A 1.0 kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems). See, SEQ.ID.NO.:15 for the nucleic acid sequence and
SEQ.ID.N0.:16 for the putative amino acid sequence.
i. hRUP36 (Seq. Id. Nos.17 & 18)
The disclosed human RUP36 was identified based upon the use of GenBank
database information. While searching the database, a cDNA clone with
Accession
Number AC090099 was identified as a human genomic sequence from chromosome 11.
The full length RUP36 was cloned by PCR using primers:
5'- CATCTGGTTTGTGTTCCCAGGGGCACCAG -3' (SEQ.ID.N0.:43; sense, 5' of start
codon),
28
CA 02429860 2003-05-26
AREN-0309 PATENT
5'-
GACAGTGTTGCTCTCAAAGTCCCGTCTGACTG -3' (SEQ.ID.N0.:44; antisense, 3' of
stop codon) and human genomic DNA (Clontech) as template. TaqPlus~ Precision
DNA
polymerase (Stratagene) was used for the amplification in a SOp,I reaction by
the following
cycle with step 2 to step 4 repeated 30 times: 95°C, 5 min; 95°C
for 30 sec; 70°C for 30 sec;
72°C for 1 min 30 sec; and 72°C for 7 min.
A 1.0 kb PCR fragment was isolated from a 1% agarose gel and cloned into the
pCRII-TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator
Kit
(P.E. Biosystems). See, SEQ.ID.N0.:17 for the nucleic acid sequence and
SEQ.ID.NO.:1 ~ for the putative amino acid sequence.
j. hRUP37 (Seq. Id. Nos.19 & 20)
The disclosed human RUP37 was identified based upon the use of GenBank
database information. While searching the database, a cDNA clone with
Accession
Number AC090099 was identified as a human genomic sequence from chromosome 11.
The full length RUP37 was cloned by PCR using primers:
5'-CTGTTTCCAGGGTCATCAGACTGGG-3' (SEQ.ll~.N0.:45; sense);
5'-GCAGCATTGCTCTCAAAGTCCTGTCTG-3' (SEQ.ID.N0.:46; antisense)
and human genomic DNA (Clontech) as template. TaqPlus~ Precision DNA
polymerase
(Stratagene) was used for the amplification by the following cycle with step 2
to step 4
repeated 35 times: 95°C for 5 min; 95°C for 30 sec; 62°C
for 30 sec; 72°C for 1 min 30 sec;
and 72°C for 7 min.
A 969 base pair was isolated from a 1%% agarose gel and cloned into the pCRII-
TOPO vector (Invitrogen) and sequenced using the ABI Big Dye Terminator Kit
(P.E.
Biosystems). See, SEQ.ID.N0.:19 for the nucleic acid sequence and
SEQ.ID.NO.:20 for
the putative amino acid sequence.
Example 2
PREPARATION OF NON-ENDOGENOUS, CONSTITUTIVELY ACTIVATED GPCRS
Those skilled in the art are credited with the ability to select techniques
for
mutation of a nucleic acid sequence. Presented below are approaches utilized
to create
non-endogenous versions of several of the human GPCRs disclosed above. The
mutations disclosed below are based upon an algorithmic approach whereby the
16th
amino acid (located in the IC3 region of the GPCR) from a conserved proline
(or an
29
CA 02429860 2003-05-26
AREN-0309 PATENT
endogenous, conservative substitution therefore) residue (located in the TM6
region of the GPCR, near the TM6/IC3 interface) is mutated, preferably to an
alanine,
histimine, arginine or lysine amino acid residue, most preferably to a lysine
amino acid
residue.
1. Transformer Site-Directed TM Mutagenesis
Preparation of non-endogenous human GPCRs may be accomplished on human
GPCRs using, ihte~ alia, Transformer Site-DirectedTM Mutagenesis Kit
(Clontech)
according to the manufacturer instructions. In some embodiments two
mutagenesis primers
are used, preferably a lysine mutagenesis oligonucleotide that creates the
lysine mutation,
and a selection marker oligonucleotide. For convenience, the codon mutation to
be
incorporated into the human GPCR is also noted, in standard form (Table D):
TABLE D
Receptor Identifier Codon Mutation
hRUP28 V274K
hRUP29 T249K
hRUP30 1Z232K
hRUP31 M294K
hRUP32 F220K
hRUP34 A238K
hRUP35 Y215K
hRUP36 L294K
hRUP37 T219K
Example 3
RECEPTOR EXPRESSION
Although a variety of cells are available to the art-skilled for the
expression of
proteins, it is preferred that mammalian cells be utilized. The primary reason
for this is
predicated upon practicalities, i.e., utilization of, e.g., yeast cells for
the expression of a
GPCR, while possible, introduces into the protocol a non-mammalian cell which
may not
(indeed, in the case of yeast, does not) include the receptor-coupling,
genetic-mechanism
CA 02429860 2003-05-26
AREN-0309 , PATENT
and secretary pathways that have evolved fox mammalian systems - thus, results
obtained in non-mammalian cells, while of potential use, are not as preferred
as those
obtained using mammalian cells. Of the mammalian cells, COS-7, 293 and 293T
cells
are particularly preferred, although the specific mammalian cell utilized can
be
predicated upon the particular needs of the artisan.
a. Transient Transfection
On day one, 6x10 cells/10 cm dish of 293 cells well were plated out. On day
two,
two reaction tubes were prepared (the proportions to follow for each tube are
per plate):
tube A was prepared by mixing 4~.g DNA (e.g., pCMV vector; pCMV vector with
receptor
cDNA, ete.) in 0.5 ml serum free DMEM (Gibco BRL); tube B was prepared by
mixing
24,1 lipofectamine (Gibco BRL) in O.Sml serum free DMEM. Tubes A and B were
admixed by inversion (several times),. followed by incubation at room
temperature for 30-
45min. The admixture is referred to as the "transfection mixture". Plated 293
cells were
washed with 1XPBS, followed by addition of 5 ml serum free DMEM. One ml of the
transfection mixture were added to the cells, followed by incubation for 4hrs
at 37°C/5%
C02. The transfection mixture was removed by aspiration, followed by the
addition of
lOml of DMEM/10% Fetal Bovine Serum. Cells were incubated at 37°C/5%
CO2. After
48hr incubation, cells were harvested and utilized for analysis.
b. Stable Cell Lines
Approximately 12x10 293 cells will be plated on a l5cm tissue culture plate,
and
grown in DME High Glucose Medium containing 10% fetal bovine serum and one
percent
sodium pyruvate, L-glutamine, and antibiotics. Twenty-four hours following
plating of 293
cells (to approximately ~80% confluency), the cells will be transfected using
12~,g of DNA.
The 12~,g of DNA is combined with 60,1 of lipofectamine and 2mL of DME High
Glucose
Medium without serum. The medium will be aspirated from the plates and the
cells washed
once with medium without serum. The DNA, lipofectamine, and medium mixture
will be
added to the plate along with lOmL of medium without serum. Following
incubation at
37°C for four to five hours, the medium will be aspirated and 25m1 of
medium containing
serum will be added. Twenty-four hours following transfection, the medium will
be
aspirated again, and fresh medium with serum will be added. Forty-eight hours
following
transfection, the medium will be aspirated and medium with serum will be added
containing
geneticin (G418 drug) at a final concentration of SOO~,g/mL. The transfected
cells will then
31
CA 02429860 2003-05-26
AREN-0309 PATENT
undergo selection for positively transfected cells containing the G41 ~
resistant gene.
The medium will be replaced every four to five days as selection occurs.
During selection,
cells will be grown to create stable pools, or split for stable clonal
selection.
Example 4
S ASSAYS FOR DETERMINATION OF CONSTITUTIVE ACTIVITY
OF NON-ENDOGENOUS GPCRS
A variety of approaches are available for assessment of constitutive activity
of the
non-endogenous human GPCRs. The following are illustrative; those of ordinary
skill in
the art are credited with the ability to determine those techniques that are
preferentially
beneficial for the needs of the artisan.
1. Membrane Binding Assays: [35S]GTPyS Assay
When a G protein-coupled receptor is in its active state, either as a result
of ligand
binding or constitutive activation, the receptor couples to a G protein and
stimulates the
release of GDP and subsequent binding of GTP to the G protein. The alpha
subunit of the
G protein-receptor complex acts as a GTPase and slowly hydrolyzes the GTP to
GDP, at
which point the receptor normally is deactivated. Constitutively activated
receptors
continue to exchange GDP for GTP. The non-hydrolyzable GTP analog, [35S]GTPyS,
can
be utilized to demonstrate enhanced binding of [35S]GTPyS to membranes
expressing
constitutively activated receptors. Advantages of using [35S]GTPyS binding to
measure
constitutive activation include but are not limited to the following: (a) it
is generically
applicable to all G protein-coupled receptors; (b) it is proximal at the
membrane surface
making it less likely to pick-up molecules which affect the intracellular
cascade.
The assay takes advantage of the ability of G protein coupled receptors to
stimulate
[ssS]GTPyS binding to membranes expressing the relevant receptors. The assay
can,
therefore, be used in the direct identification method to screen candidate
compounds to
constitutively activated G protein-coupled receptors. The assay is generic and
has
application to drug discovery at all G protein-coupled receptors.
The [35S]GTPyS assay is incubated in 20 mM HEPES and between 1 and about
20mM MgCl2 (this amount can be adjusted for optimization of results, although
20mM is
preferred) pH 7.4, binding buffer with between about 0.3 and about 1.2 nM
[35S]GTPyS
32
CA 02429860 2003-05-26
AREN-0309 PATENT
(this amount can be adjusted for optimization of results, although 1.2 is
preferred ) and
12.5 to 75 ~,g membrane protein (e.g., 293 cells expressing the GS Fusion
Protein; this
amount can be adjusted for optimization) and 10 p.M GDP (this amount can be
changed for
optimization) for 1 hour. Wheatgerm agglutinin beads (25 ~.1; Amersham) will
then be
added and the mixture incubated for another 30 minutes at room temperature.
The tubes
will be then centrifuged at 1500 x g for 5 minutes at room temperature and
then counted in
a scintillation counter.
2. Adenylyl Cyclase
A Flash PlateTM Adenylyl Cyclase kit (New England Nuclear; Cat. No. SMP004A)
designed for cell-based assays can be modified for use with crude plasma
membranes. The
Flash Plate wells can contain a scintillant coating which also contains a
specific antibody
recognizing cAMP. The cAMP generated in the wells can be quantitated by a
direct
competition for binding of radioactive cAMP tracer to the cAMP antibody. The
following
serves as a brief protocol for the measurement of changes in CAMP levels in
whole cells
that express the receptors.
Transfected cells will be harvested approximately twenty four hours after
transient
transfection. Media will be carefully aspirated and discarded. Ten ml of PBS
will gently be
added to each dish of cells followed by careful aspiration. One ml of Sigma
cell
dissociation buffer and 3m1 of PBS will be added to each plate. Cells will be
pipetted off
the plate and the cell suspension collected into a SOmI conical centrifuge
tube. Cells will be
centrifuged at room temperature at 1,100 rpm for 5 min. The cell pellet will
be carefully re-
suspended into an appropriate volume of PBS (about 3m1/plate). The cells will
be then
counted using a hemocytometer and additional PBS will be added to give the
appropriate
nmnber of cells (to a final volume of about SOp,I/well).
cAMP standards and Detection Buffer (comprising 1 ~,Ci of tracer [lzsI cAMP
(50
~,1~ to 11 ml Detection Buffer) will be prepared and maintained in accordance
with the
manufacturer's instructions. Assay Buffer will be prepared fresh for screening
and
contained 50,1 of Stimulation Buffer, 3~,1 of test compound (12~,M final assay
concentration) and 50,1 cells, Assay Buffer will be stored on ice until
utilized. The assay
will be initiated by addition of SOwI of cAMP standards to appropriate wells
followed by
addition of 50.1 of PBSA to wells H-11 and H12. Fifty w1 of Stimulation Buffer
will be
added to all wells. DMSO (or selected candidate compounds) will be added to
appropriate
33
CA 02429860 2003-05-26
AREN-0309 PATENT
wells using a pin tool capable of dispensing 3~,1 of compound solution, with a
final assay
concentration of 12~,M test compound and 100.1 total assay volume. The cells
will then be
added to the wells and incubated for 60 min at room temperature. One hundred
~.l of
Detection Mix containing tracer cAMP will then be added to the wells. Plates
will be
incubated for an additional 2 hours followed by counting in a Wallac
MicroBetaTM
scintillation counter. Values of cAMP/well will then be extrapolated from a
standard
cAMP curve which will be contained within each assay plate.
3. Cell-Based CAMP for G; Coupled Target GPCRs
TSHR is a GS coupled GPCR that causes the accumulation of cAMP upon
activation. TSHR will be constitutively activated by mutating amino acid
residue 623 (i.e.,
changing an alanine residue to an isoleucine residue). A G, coupled receptor
is expected to
inhibit adenylyl cyclase, and, therefore, decrease the level of cAMP
production, which can
male assessment of cAMP levels challenging. An effective technique for
measuring the
decrease in production of cAMP as an indication of constitutive activation of
a G, coupled
receptor can be accomplished by co-transfecting, most preferably, non-
endogenous,
constitutively activated TSHR (TSHR-A623I) (or an endogenous, constitutively
active GS
coupled receptor) as a "signal enhancer" with a G, linked target GPCR to
establish a
baseline level of cAMP. TJpon creating a non-endogenous version of the G,
coupled
receptor, this non-endogenous version of the target GPCR is then co-
transfected with the
signal enhancer, and it is this material that can be used for screening. This
approach will be
utilized to effectively generate a signal when a CAMP assay is used; this
approach is
preferably used in the direct identification of candidate compounds against G;
coupled
receptors. It is noted that for a G; coupled GPCR, when this approach is used,
an inverse
agonist of the target GPCR will increase the cAMP signal and an agonist will
decrease the
cAMP signal.
On day one, 2x104 293 cells/well will be plated out. On day two, two reaction
tubes
will be prepared (the proportions to follow for each tube are per plate): tube
A will be
prepared by mixing tug DNA of each receptor transfected into the mammalian
cells, for a
total of 4ug DNA (e.g., pCMV vector; pCMV vector with mutated THSR (TSHR-
A6231);
TSHR-A623I and GPCR, etc.) in 1.2m1 serum free DMEM (Irvine Scientific,
Irvine, CA);
tube B will be prepared by mixing 120.1 lipofectamine (Gibco BRL) in 1.2m1
serum free
34
CA 02429860 2003-05-26
AREN-0309 PATENT
DMEM. Tubes A and B will then be admixed by inversion (several times),
followed by incubation at room temperature for 30-45min. The admixture is
referred to as
the "transfection mixture". Plated 293 cells will be washed with 1XPBS,
followed by
addition of l Oml serum free DMEM. 2.4m1 of the transfection mixture will then
be added
to the cells, followed by incubation for 4hrs at 37°C/5% C02. The
transfection mixture will
then be removed by aspiration, followed by the addition of 25m1 of DMEM/10%
Fetal
Bovine Serum. Cells will then be incubated at 37°C/5% C02. After 24hr
incubation, cells
will be harvested and utilized for analysis.
A Flash PlateTM Adenylyl Cyclase kit (New England Nuclear; Cat. No. SMP004A)
although designed for cell-based assays, can be modified for use with crude
plasma
membranes depending on the need of the skilled artisan. The Flash Plate wells
will contain
a scintillant coating which also contains a specific antibody recognizing
cAMP. The cAMP
generated in the wells can be quantified by a direct competition for binding
of radioactive
cAMP tracer to the cAMP antibody. The following serves as a brief protocol for
the
measurement of changes in cAMP levels in whole cells that express the
receptors.
Transfected cells will be harvested approximately twenty four hours after
transient transfection. Media will be carefully aspirated and discarded. Ten
ml of PBS
will be gently added to each dish of cells followed by careful aspiration. One
ml of
Sigma cell dissociation buffer and 3m1 of PBS will be added to each plate.
Cells will be
pipetted off the plate and the cell suspension will be collected into a SOmI
conical
centrifuge tube. Cells will be centrifuged at room temperature at 1,100 rpm
for 5 min.
The cell pellet will be carefully re-suspended into an appropriate volume of
PBS (about
3ml/plate). The cells will then be counted using a hemocytometer and
additional PBS is
added to give the appropriate number of cells (to a final volume of about
50~,1/well).
cAMP standards and Detection Buffer (comprising 1 ~,Ci of tracer ~l2sI cAMP
(50
~1] to 11 ml Detection Buffer) will be prepared and maintained in accordance
with the
manufacturer's instructions. Assay Buffer should be prepared fresh for
screening and
contained 50,1 of Stimulation Buffer, 3~,1 of test compound (12~,M final assay
concentration) and SOp,I cells, Assay Buffer can be stored on ice until
utilized. The assay
can be initiated by addition of 50,1 of cAMP standards to appropriate wells
followed by
addition of SOpI of PBSA to wells H-11 and H12. Fifty p.1 of Stimulation
Buffer will be
added to all wells'. Selected compounds (e.g., TSH) will be added to
appropriate wells
CA 02429860 2003-05-26
AREN=0309 PATENT
using a pin tool capable of dispensing 3~,1 of compound solution, with a final
assay
concentration of 12~,M test compound and 100,1 total assay volume. The cells
will then be
added to the wells and incubated for 60 min at room temperature. One hundred
p,1 of
Detection Mix containing tracer cAMP will then be added to the wells. Plates
will then be
incubated additional 2 hours followed by counting in a Wallac MicroBeta
scintillation
counter. Values of cAMP/well will then be extrapolated from a standard cAMP
curve
which is contained within each assay plate.
4. Reporter-Based Assays
a. Cup-Luc Reporter Assay (GS -associated receptors)
293 and 293T cells will be plated-out on 96 well plates at a density of 2 x
104
cells per well and will be transfected using Lipofectamine Reagent (BRL) the
following
day according to manufacturer instructions. A DNA/lipid mixture will be
prepared for
each 6-well transfection as follows: 260ng of plasmid DNA in 100,1 of DMEM are
gently mixed with 2~,1 of lipid in 100.1 of DMEM (the 260ng of plasmid DNA
consisted
of 200ng of a 8xCRE-Luc reporter plasmid, SOng of pCMV comprising endogenous
receptor or non-endogenous receptor or pCMV alone, and long of a GPRS
expression
plasmid (GPRS in pcDNA3 (Invitrogen)). The 8XCRE-Luc reporter plasmid is
prepared
as follows: vector SRIF-(3-gal will be obtained by cloning the rat
somatostatin promoter
(-71/+51) at BglV-HindIII site in the p(3gal-Basic Vector (Clontech). Eight
(8) copies of
cAMP response element will be obtained by PCR from an adenovirus template
AdpCF126CCRE8 (see, 7 Human Gene Therapy 1883 (1996)) and cloned into the SRIF-
(3-gal vector at the Kpn-BglV site, resulting in the 8xCRE-(3-gal reporter
vector. The
8xCRE-Luc reporter plasmid will be generated by replacing the beta-
galactosidase gene
in the 8xCRE-(3-gal reporter vector with the luciferase gene obtained from the
pGL3-
basic vector (Promega) at the HindIII-BamHI site. Following 30 min. incubation
at
room temperature, the DNA/lipid mixture will be diluted with 400 ~,1 of DMEM
and
100,1 of the diluted mixture will be added to each well. One hundred p1 of
DMEM with
10% FCS will be added to each well after a 4hr incubation in a cell culture
incubator.
The following day the transfected cells will be changed with 200 ~.1/well of
DMEM with
10% FCS. Eight hours later, the wells will be changed to 100 ~,1 /well of DMEM
without phenol red, after one wash with PBS. Luciferase activity will be
measured the
next day using the LucLiteTM reporter gene assay kit (Packard) following
manufacturer's
36
CA 02429860 2003-05-26
AREN-0309 PATENT
instructions and read on a 1450 MicroBetaTM scintillation and luminescence
counter (Wallac).
b. APl reporter assay (Gq associated receptors)
A method to detect Gq stimulation depends on the known property of Gq
dependent phospholipase C to cause the activation of genes containing AP1
elements in
their promoter. A PathdetectTM AP-1 cis-Reporting System (Stratagene,
Catalogue #
219073) can be utilized following the protocol set forth above with respect to
the CREB
reporter assay, except that the components of the calcium phosphate
precipitate were 410
ng pAPl-Luc, 80 ng pCMV-receptor expression plasmid, and 20 ng CMV-SEAP.
c. S~-Luc Reporter Assay (Gg- associated receptors)
One method to detect Gq stimulation depends on the known property of G9-
dependent phospholipase C to cause the activation of genes containing serum
response
factors in their promoter. A PathdetectTM SRF-Luc-Reporting System
(Stratagene) can be
utilized to assay for Gq coupled activity in, e.g., COS7 cells. Cells are
transfected with
the plasmid components of the system and the indicated expression plasmid
encoding
endogenous or non-endogenous GPCR using a Mammalian TransfectionTM Kit
(Stratagene, Catalogue #200285) according to the manufacturer's instructions.
Briefly,
410 ng SRF-Luc, 80 ng pCMV-receptor expression plasmid and 20 ng CMV-SEAP
(secreted alkaline phosphatase expression plasmid; alkaline phosphatase
activity is
measured in the media of transfected cells to control for variations in
transfection
efficiency between samples) are combined in a calcium phosphate precipitate as
per the
manufacturer's instructions. Half of the precipitate is equally distributed
between 3
wells in a 96-well plate, kept on the cells in a serum free media for 24
hours. The last 5
hours the cells are incubated with lp,M Angiotensin, where indicated. Cells
are then
lysed and assayed for luciferase activity using a LucliteTM Kit (Packard, Cat.
# 6016911)
and "Trilux 1450 Microbeta" liquid scintillation and luminescence counter
(Wallac) as
per the manufacturer's instructions. The data can be analyzed using GraphPad
PrismTM
2.0a (GraphPad Software Inc.).
d. Intracellular IP3 Accumulation Assay (Gq-associated receptors)
37
CA 02429860 2003-05-26
AREN-0309 PATENT
On day 1, cells comprising the receptors (endogenous and/or non-
endogenous) are plated onto 24 well plates, usually 1x105 cells/well (although
his number
can be optimized. On day 2 cells are transfected by firstly mixing 0.25ug DNA
in 50 p,1
serum free DMEM/well and 2 ~.1 lipofectamine in 50 ~,1 serum free DMEM/well.
The
solutions are gently mixed and incubated for 15-30 min at room temperature.
Cells are then
washed with 0.5 ml PBS aizd 400 ~ul of serum free media and then mixed with
the
transfection media and added to the cells. The cells are incubated for 3-4 hrs
at
37°C/5%C02 and then the transfection media is removed and replaced with
lmllwell of
regular growth media. On day 3 the cells are labeled with 3H-myo-inositol.
Briefly, the
media is removed and the cells are washed with 0.5 ml PBS. Then 0.5 ml
inositol-
free/serum free media (GIBCO BRL) are added/well with 0.25 ~,Ci of 3H-myo-
inositol/
well and the cells incubated for 16-18 hrs overnight at 37°C/5%C02. On
Day 4 the cells are
washed with 0.5 ml PBS acid 0.45 ml of assay medium is added containing
inositol-
free/serum free media 10 wM pargyline 10 mM lithium chloride or 0.4 ml of
assay medium
and 50 p.1 of lOx l~etanserin (let) to final concentration of 10~,M. The cells
are then
incubated for 30 min at 37°C. The cells are then washed with 0.5 ml PBS
and 200 p,1 of
fresh/ice cold stop solution (1M I~OH; 18 mM Na-borate; 3.8 mM EDTA) is added
to each
well. The solution is Dept on ice for 5-10 min (or until cells are lysed) and
then neutralized
by 200 ~,1 of fresh/ice cold neutralization solution (7.5 % HCL). The lysate
is then
transferred into 1.5 ml Eppendorf tubes and 1 ml of chloroform/methanol (1:2)
is
added/tube. The solution is vortexed for 15 sec and the upper phase is applied
to a Biorad
AGl-XBTM anion exchange resin (100-200 mesh). First, the resin is washed with
water at
1:1.25 W/V and 0.9 ml of upper phase is loaded onto the column. The column is
then
washed with 10 ml of 5 mM myo-inositol and 10 ml of 5 mM Na-borate/60mM Na-
formate. The inositol tris phosphates are eluted into scintillation vials
containing 10 ml of
scintillation cocktail with 2 ml of 0.1 M formic acid/ 1 M ammonium formate.
The
columns are regenerated by washing with 10 ml of 0.1 M formic acid/3M
axmnonium
formate and rinsed twice with dd HBO and stored at 4°C in water.
Reference is made to Figure 1. In Figure 1, 293 cells were transfected with Gq
protein containing a six amino acid deletion, "Gq(del)"; Gg protein fused to a
G protein,
"Gq(del)/G"; endogenous RUP32; and RUP32 with Gq(del) ("RUP32+ Gq(del)/G").
The
data indicate, based upon measuring IP3 accumulation of RUP32 co-transfection
of
38
CA 02429860 2003-05-26
AREN-0309 PATENT
Gq(del)/G, that RUP32 does not endogenously couple to Gg protein.
However when RUP32 was co-transfected with Gq(del)/G fusion protein, RUP32 was
forced to couple to Gq protein. RUP27+ Gq(del)/G; evidence about a nine (9)
fold increase
in IP3 accumulation when compared to endogenous RUP32. This data demonstrates
that
the Gq(del)/G Fusion Construct can be co-transfected with a GPCR and used to
screen for
agonists or inverse agonists.
Reference is made to Figure 2. In Figure 2, 293 cells were transfected with
RUP35
and RUP36 receptor and compared to the control, pCMV. The data indicate that
both
RUP35 and RUP36 receptor are endogenously, constitutively active. RUP35
evidences
about a six (6) fold increase in intracellular inositol phosphate accumulation
when
compared to pCMV and RUP36 evidences about a four (4) fold increase when
compared to
pCMV.
Example 5
FUSION PROTEIN PREPARATION
a. GPCR: GS Fusion Construct
The design of the constitutively activated GPCR-G protein fusion construct can
be
accomplished as follows: both the 5' and 3' ends of the rat G protein Gsa
(long form; Itoh,
H. et al., 83 PN~1S 3776 (1986)) is engineered to include a HindllI (5'-AAGCTT-
3')
sequence thereon. Following confirmation of the correct sequence (including
the flanking
HindIII sequences), the entire sequence is shuttled into pcDNA3.1(-)
(Invitrogen, cat. no.
V795-20) by subcloning using the HilzdIII restriction site of that vector. The
correct
orientation for the Gsa sequence will be determined after subcloning into
pcDNA3.1(-)
The modified pcDNA3.1 (-) containing the rat Gsa gene at HindIII sequence is
then verified;
this vector will then be available as a "universal" Gsa protein vector. The
pcDNA3.1(-)
vector contains a variety of well-known restriction sites upstream of the
HindIII site, thus
beneficially providing the ability to insert, upstream of the GS protein, the
coding sequence
of an endogenous, constitutively active GPCR. This same approach can be
utilized to
create other "universal" G protein vectors, and, of course, other commercially
available or
proprietary vectors known to the artisan can be utilized. In some embodiments,
the
important criteria is that the sequence for the GPCR be upstream and in-frame
with that of
the G protein.
39
CA 02429860 2003-05-26
AREN-0309 PATENT
Spacers in the restriction sites between the G protein and the GPCR are
optional. The sense and anti-sense primers included the restriction sites for
XbaI and
EcoRV, respectively, such that spacers (attributed to the restriction sites)
exist between the
G protein and the GPCR.
PCR will then be utilized to secure the respective receptor sequences for
fusion
witlvn the Gsa universal vector disclosed above, using the following protocol
for each:
100ng cDNA for GPCR will be added to separate tubes containing 2~,1 of each
primer
(sense and anti-sense), 3~.1 of lOmM dNTPs, 10,1 of lOXTaqPIusTM Precision
buffer, lwl of
TaqPlusTM Precision polymerase (Stratagene: #600211), and 80.1 of water.
Reaction
temperatures and cycle times for the GPCR will be as follows with cycle steps
2 through 4
were repeated 35 times: 94°C for 1 min; 94°C for 30 seconds;
62°C for 20 sec; 72°C 1 min
40sec; and 72°C 5 min. PCR products will be run on a 1% agarose gel and
then purified.
The purified products will be digested with XbaI and EcoRV and the desired
inserts
purified and ligated into the GS unversal vector at the respective restriction
sites. The
positive clones will be isolated following transformation and determined by
restriction
enzyme digestion; expression using 293 cells will be accomplished following
the protocol
set forth infra. Each positive clone for GPCR- GS Fusion Protein will be
sequenced to
verify correctness.
b. Gq(6 amino acid deletion)/G; Fusion Construct
The design of a Gq (del)/G fusion construct was accomplished as follows: the N-
terminal six (6) amino acids (amino acids 2 through 7), having the sequence of
TLESIM
(SEQ.ID.NO.:47) Gaq-subunit was deleted and the C-terminal five (5) amino
acids, having
the sequence EYNLV (SEQ.ID.N0.:48) was replaced with the corresponding amino
acids
of the Gai Protein, having the sequence DCGLF (SEQ.ID.N0.:49). This fusion
construct
was obtained by PCR using the following primers:
5'-gatcAAGCTTCCATGGCGTGCTGCCTGAGCGAGG-3' (SEQ.ID.NO.:50) and
5'-gatcGGATCCTTAGAACAGGCCGCAGTCCTTCAGGTTCAGCTGCAGGATGGTG-3'
(SEQ.ID.N0.:51) and Plasmid 63313 which contains the mouse Gaq-wild type
version with a
hemagglutinin_ tag as template. Nucleotides in lower caps are included as
spacers.
TaqPlus~ Precision DNA polymerase (Stratagene) was utilized for the
amplification by the following cycles, with steps 2 through 4 repeated 35
times: 95°C for
2 min; 95°C for 20 sec; 56°C for 20 sec; 72°C for 2 min;
and 72°C for 7 min. The PCR
CA 02429860 2003-05-26
AREN-0309 PATENT
product will be cloned into a pCRII-TOPO vector (Invitrogen) and sequenced
using the
ABI Big Dye Terminator kit (P.E. Biosystems). Inserts from a TOPO clone
containing
the sequence of the fusion construct will be shuttled into the expression
vector
pcDNA3.1(+) at the HindIIIBamHI site by a 2 step cloning process.
Example 6
TISSUE DISTRIBUTION OF THE DISCLOSED HUMAN GPCRS: RT-PCR
RT-PCR was applied to confine the expression and to detennine the tissue
distribution of several novel human GPCRs. Oligonucleotides utilized were GPCR-
specific and the human multiple tissue cDNA panels (MTC, Clontech) as
templates. Taq
DNA polymerase (Stratagene) were utilized for the amplification in a 40,1
reaction
according to the manufacturer's instructions. Twenty w1 of the reaction will
be loaded on
a 1.5% agarose gel to analyze the RT-PCR products. Table E, below, lists the
receptors,
the cycle conditions and the primers utilized, and also lists exemplary
diseases/disorders
linked to the receptors.
TABLE E
ReceptorCycle 5' Primer 3' Primer DNA Tissue Expression
IdentifierConditions (SEQ.ID.NO.)(SEQ.ID.NO.)Fragment
Min ('),
Sec (")
Cycles 2-4
repeated
35
times
hRUP28 94C for GTCCTCACT CTGCGTCCAC710bp heart; kidney;
5 min; liver; lung
94C for GGTGGCCAT CAGAGTCAC and pancreas
30 sec;
58C for GTACTCC GTCTCC
30 sec, (52) (53)
72C for
1 min,
and 72C
for 7
min
hRUP29 94C for CTTGGATGTT GTTTGTGGCT690bp leukocyte and
5 min; ovary
94C for TGGGCTGCC AACGGCACA
30 sec;
58C for CTTCTGC AAACACAAT
30 sec, (54)
72C for TCC (55)
1 min,
41
CA 02429860 2003-05-26
AREN-0309 PATENT
and 72C for
7
min
hRUP30 94C for 2 CTGCTCACG GTGGCCATG 690bp pancreas
min;
94C for 15 GTTGACCGC AGCCACCCT
sec;
58C for 20 TACACTGC GAGCTCC
sec, (57)
72C for 1 (56)
min,
and 72C for
10
min
hRUP31 95C for 4 CTTCTTCTCCCCAAATCA 516bp colon, lung,
min; pancreas,
95C for 1 GACGTCAAG GTGTGCAA thymus; cerebral
min; cortex,
G (58) hippocampus
52C for 30 ATCG (59) ofbrain,
sec,
and fat cells
72C for 1
min,
and 72C for
7
min
hRUP32 95C for 4 TGAATGGGT CAACGGTCT 527bp thymus
min;
95C for 1 CCTGTGTGA GACAACCTC
min;
52C for 30 ~ (60) CT (61)
sec,
72C for 1
min,
and 72C for
7
min
hRUP34 95C for 4 TTGCTGTGATCAGGAAGCC 534bp peripheral
min; blood
95C for 1 GTGGCATTT"TCATAAAGGC leukocyte
min; ("PBL"),
52C for 30 G (62) ATCAA (63) prostate and
sec, kidney
72C for 1
min,
and 72C for
7
min
hRUP35 95C for 4 ACATCACCT CCAGCATCTT 557bp thalamus
min;
95C for 1 GCTTCCTGA GATGCAGTG
min;
52C for 30 CC (64) T (65)
sec,
72C for 1
min,
and 72C for
7
min
hRUP37 95C for 4 CCATCTCCA GCTGTTAAG 517bp testis, cerebral
min; cortex
95C for 1 AAATCCTCA AGCGGACAG and hippocampus
min;
52C for 30 GTC (66) GAAA (67)
sec,
72C for 1
min,
42
CA 02429860 2003-05-26
AREN-0309 PATENT
and 72°C for 7
min
Diseases and disorders related to receptors located in these tissues or
regions
include, but are not limited to, cardiac disorders and diseases (e.g.
thrombosis, myocardial
infarction; atherosclerosis; cardiomyopathies); kidney disease/disorders
(e.g., renal failure;
renal tubular acidosis; renal glycosuria; nephrogenic diabetes insipidus;
cystinuria;
polycystic kidney disease); eosinophilia; leukocytosis; leukopenia; ovarian
cancer; sexual
dysfunction; polycystic ovarian syndrome; pancreatitis and pancreatic cancer;
irritable
bowel syndrome; colon cancer; Crohn's disease; ulcerative colitis;
diverticulitis; Chronic
Obstructive Pulmonary Disease (COPD); Cystic Fibrosis; pneumonia; pulmonary
hypertension; tuberculosis and lung cancer; Parkinson's disease; movement
disorders and
ataxias; learning and memory disorders; eating disorders (e.g., anorexia;
bulimia, etc.);
obesity; cancers; thymoma; myasthenia gravis; circulatory disorders; prostate
cancer;
prostatitis; kidney diseaseldisorders(e.g., renal failure; renal tubular
acidosis; renal
glycosuria; nephrogenic diabetes insipidus; cystinuria; polycystic l~idney
disease);
sensorimotor processing and arousal disorders; obsessive-compulsive disorders;
testicular
cancer; priapism; prostatitis; hernia; endocrine disorders; sexual
dysfunction; allergies;
depression; psychotic disorders; migraine; reflux; schizophrenia; ulcers;
bronchospasm;
epilepsy; prostatic hypertrophy; anxiety; rhinitis; angina; and glaucoma.
Accordingly, the
methods of the present invention may also be useful in the diagnosis and/or
treatment of
these and other diseases and disorders.
Example 7
Protocol: Direct Identification of Inverse Agonists and Agonists
A. [35S]GTPyS Assay
Although endogenous, constitutively active GPCRs have been used for the direct
identification of candidate compounds as, e.g., inverse agonists, for reasons
that are not
altogether understood, infra-assay variation can become exacerbated. In some
embodiments a GPCR Fusion Protein, as disclosed above, is also utilized with a
non-
endogenous, constitutively activated GPCR. When such a protein is used, infra-
assay
variation appears to be substantially stabilized, whereby an effective signal-
to-noise ratio is
43
CA 02429860 2003-05-26
AREN-0309 PATENT
obtained. This has the beneficial result of allowing for a more robust
identification of
candidate compounds. Thus, in some embodiments it is preferred that for direct
identification, a GPCR Fusion Protein be used and that when utilized, the
following assay
protocols be utilized.
1. Membrane Preparation
Membranes comprising the constitutively active orphan GPCR Fusion Protein of
interest and for use in the direct identification of candidate compounds as
inverse agonists
or agoiusts are preferably prepared as follows:
a. Materials
"Membrane Scrape Buffer" is comprised of 20mM HEPES and lOmM EDTA,
pH 7.4; "Membrane Wash Buffer" is comprised of 20 mM HEPES and 0.1 mM EDTA,
pH 7.4; "Binding Buffer" is comprised of 20mM HEPES, 100 mM NaCl, and 10 mM
MgCl2, pH 7.4
b. Procedure
All materials will be kept on ice throughout the procedure. Firstly, the media
will
be aspirated from a confluent monolayer of cells, followed by rinse with 10m1
cold PBS,
followed by aspiration. Thereafter, Sml of Membrane Scrape Buffer will be
added to scrape
cells; this will be followed by transfer of cellular extract into SOmI
centrifizge tubes
(centrifuged at 20,000 rpm for 17 minutes at 4°C). Thereafter, the
supernatant will be
aspirated and the pellet will be resuspended in 30m1 Membrane Wash Buffer
followed by
centrifugation at 20,000 rpm for 17 minutes at 4°C. The supernatant
will then be aspirated
and the pellet resuspended in Binding Buffer. The resuspended pellet will then
be
homogenized using a Brinkman PolytronTM homogenizes (15-20 second bursts until
the
material is in suspension). This is referred to herein as "Membrane Protein".
2. Bradford Protein Assay
Following the homogenization, protein concentration of the membranes will be
determined, for example, using the Bradford Protein Assay (protein can be
diluted to
about l.Smg/ml, aliquoted and frozen (-80°C) for later use; when
frozen, protocol for use
will be as follows: on the day of the assay, frozen Membrane Protein is thawed
at room
temperature, followed by vortex and then homogenized with a Polytron at about
12 x
1,000 rpm for about 5-10 seconds; it was noted that for multiple preparations,
the
homogenizes is thoroughly cleaned between homogenization of different
preparations).
44
CA 02429860 2003-05-26
AREN-0309 PATENT
a. Materials
Binding Buffer (as discussed above); Bradford Dye Reagent; Bradford Protein
Standard will be utilized, following manufacturer instructions (Biorad, cat.
no. 500-
0006).
~ b. Procedure
Duplicate tubes will be prepared, one including the membrane, and one as a
control "blank". Each contains 800,1 Binding Buffer. Thereafter, lOwl of
Bradford
Protein Standard (lmg/ml) will be added to each tube, and lOwl of membrane
Protein
will then be added to just one tube (not the blank). Thereafter, 200,1 of
Bradford Dye
Reagent will be added to each tube, followed by vortexing. After five minutes,
the tubes
will be re-vortexed and the material therein will be transferred to cuvettes.
The cuvettes
will then be read using a CECIL 3041 spectrophotometer, at wavelength 595.
3. Direct Identification Assay
a. Materials
GDP Buffer consisted of 37.5 ml Binding Buffer and 2mg GDP (Sigma, cat. no. G-
7127), followed by a series of dilutions in Binding Buffer to obtain 0.2 ~,M
GDP (final
concentration of GDP in each well was 0.1 ~,M GDP); each well comprising a
candidate
compound, has a final volume of 200.1 consisting of 100,1 GDP Buffer (final
concentration, 0.1 ~.M GDP), 50,1 Membrane Protein in Binding Buffer, and 50,1
[35S]GTPyS (0.6 nM) in Binding Buffer (2.5 ~,1 [35S]GTPyS per 10m1 Binding
Buffer).
b. Procedure
Candidate compounds will be preferably screened using a 96-well plate format
(these can be frozen at -80°C). Membrane Protein (or membranes with
expression vector
excluding the GPCR Fusion Protein, as control), will be homogenized briefly
until in
suspension. Protein concentration will then be determined using, for example,
the Bradford
Protein Assay set forth above. Membrane Protein (and controls) will then be
diluted to
0.25mg/ml in Binding Buffer (final assay concentration, 12.S~,g/well).
Thereafter, 100 ~,1
GDP Buffer is added to each well of a Wallac ScintistripTM (Wallet). A 5~,1
pin-tool will
then be used to transfer 5 ~,l of a candidate compound into such well (i.e.,
5~,1 in total assay
volume of 200 ~,1 is a 1:40 ratio such that the final screening concentration
of the candidate
compound is 10~,M). Again, to avoid contamination, after each transfer step
the pin tool is
rinsed in three reservoirs comprising water (1X), ethanol (1X) and water (2X) -
excess
CA 02429860 2003-05-26
AREN-0309 PATENT
liquid is shaken from the tool after each rinse and the tool is dried with
paper and Kim__
wipes. Thereafter, 50 p,1 of Membrane Protein will be added to each well (a
control well
comprising membranes without the GPCR Fusion Protein was also utilized), and
pre-
incubated for 5-10 minutes at room temperature. Thereafter, 50 p1 of
[3sS]GTPySo (0.6 nM)
in Binding Buffer will be added to each well, followed by incubation on a
shaker for 60
minutes at room temperature (again, in this example, plates were covered with
foil). The
assay will be stopped by spinnuzg the plates at 4000 RPM for 15 minutes at
22°C. The
plates will then be aspirated with an 8 channel manifold and sealed with plate
covers. The
plates will then be read on a Wallac 1450 using setting "Prot. #37" (as per
manufacturer's
instructions).
B. Cyclic AMP Assay
Another assay approach to directly identify candidate compound will be
accomplished utilizing a cyclase-based assay. In addition to direct
identification, this assay
approach can be utilized as an independent approach to provide confinnation of
the results
from the [3sS]GTPyS approach as set forth above.
A modified Flash PlateTM Adenylyl Cyclase kit (New England Nuclear; Cat. No.
SMP004A) will be preferably utilized for direct identification of candidate
compounds as
inverse agonists and agonists to GPCRs in accordance with the following
protocol.
Transfected cells will be harvested approximately three days after
transfection.
Membranes will be prepared by homogenization of suspended cells in buffer
containing
20mM HEPES, pH 7.4 and lOmM MgCl2. Homogenization will be performed on ice
using
a Brinkman PolytronTM for approximately 10 seconds. The resulting homogenate
will be
centrifuged at 49,000 X g for 15 minutes at 4°C. The resulting pellet
will then be
resuspended in buffer containing 20mM HEPES, pH 7.4 and 0.1 mM EDTA,
homogenized
for 10 seconds, followed by centrifugation at 49,000 X g for 15 minutes at
4°C. The
resulting pellet will then be stored at -80°C until utilized. On the
day of direct identification
screening, the membrane pellet will slowly be thawed at room temperature,
resuspended in
buffer containing 20mM HEPES, pH 7.4 and lOmM MgCl2, to yield a final protein
concentration of 0.60mg/ml (the resuspended membranes will be placed on ice
until use).
cAMP standards and Detection Buffer (comprising 2 ~,Ci of tracer [lzsI cAMP
(100
p.1] to 11 ml Detection Buffer) will be prepared and maintained in accordance
with the
46
CA 02429860 2003-05-26
AREN-0309 PATENT
manufacturer's instructions. Assay Buffer will be prepared fresh for screening
and
contain 20mM HEPES, pH 7.4, lOmM MgCl2, 20mM phosphocreatine (Sigma), 0.1
units/ml creatine phosphokinase (Sigma), 50 p,M GTP (Sigma), and 0.2 mM ATP
(Sigma);
Assay Buffer will be stored on ice until utilized.
Candidate compounds identified as per above (if frozen, thawed .at room
temperature) will be added, preferably, to 96-well plate wells (3~1/well; 12~M
final assay
concentration), together with 40 ~,1 Membrane Protein (30~g/well) and SOp,I of
Assay
Buffer. This admixture will be incubated for 30 minutes at room temperature,
with gentle
shaking.
Following the incubation, 100,1 of Detection Buffer will be added to each
well,
followed by incubation for 2-24 hours. Plates will then be counted in a Wallac
MicroBetaTM plate reader using "Prot. #31" (as per manufacturer instructions).
C. Melanophore Screening Assay
A method for identifying candidate agonists or inverse agonists for a GPCR can
be
preformed by introducing tests cells of a pigment cell line capable of
dispersing or
aggregating their pigment in response to a specific stimulus and expressing an
exogenous
clone coding for the GCPR. A stimulant, e.g., light, sets an initial state of
pigment
disposition wherein the pigment is aggregated within the test cells if
activation of the GPCR
induces pigment dispersion. However, stimulating the cell with a stimulant to
set an initial
state of pigment disposition wherein the pigment is dispersed if activation of
the GPCR
induces pigment aggregation. The tests cells are then contacted with chemical
compounds,
and it is determined whether the pigment disposition in the cells changed from
the initial
state of pigment disposition. Dispersion of pigments cells due to the
candidate compound
coupling to the GPCR will appear dark on a petri dish, while aggregation of
pigments cells
will appear light.
Materials and methods will be followed according to the disclosure of U.S.
Patent
Number 5,462,856 and U.S. Patent Number 6,051,386, each of which are
incorporated by
reference.
Although a variety of expression vectors are available to those in the art,
for
purposes of utilization for both the endogenous and non-endogenous human
GPCRs, in
some embodiments it is preferred that the vector utilized be pCMV. This vector
was
deposited with the American Type Culture Collection (ATCC) on October 13, 1998
(10801
47
CA 02429860 2003-05-26
AREN-0309 PATENT
University Blvd., Manassas, VA 20110-2209 USA) under the provisions of the
Budapest
Treaty for the International Recognition of the Deposit of Microorganisms for
the Purpose
of Patent Procedure. The DNA was tested by the ATCC and determined to be
viable. The
ATCC has assigned the following deposit number to pCMV: ATCC #203351.
References cited throughout this patent document, including co-pending and
related
patent applications, unless otherwise indicated, are fully incorporated herein
by reference.
Modifications and extension of the disclosed inventions that are within the
purview of the
skilled artisan are encompassed within the above disclosure and the claims
that follow.
48
CA 02429860 2003-05-26
SEQUENCE LISTING
<110> Arena Pharmaceuticals, Inc.
<120> Endogenous And Non-Endogenous, Constitutively Activated Human G Protein-
Coupled
Receptors
<130> AREN-0309
<150> 09/170,496
<151> 1998-10-13
<150> PCT/US99/23938
<151> 1998-10-13
<150> 60/253,404
<151> 2000-11-27
<150> 60/255,366
<151> 2000-12-12
<150> 60/270,286
<151> 2001-02-20
<150> 60/282,365
<151> 2001-04-06
<150> 60/270,266
<151> 2001-02-20
<150> 60/282,032
<151> 2001-04-06
<150> 60/282,358
<151> 2001-04-06
<150> 60/282,356
<151> 2001-04-06
<150> 60/290,917
<151> 2001-05-14
<150> 60/309,208
<151> 2001-07-31
<160> 67
<170> Patentln version.3.1
<210> 1
<211> 1002
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 1
atgtggagct gcagctggtt caacggcaca gggctggtgg aggagctgcc tgcctgccag 60
gacctgcagc tggggctgtc actgttgtcg ctgctgggcc tggtggtggg cgtgccagtg 120
ggcctgtgct acaacgccct gctggtgctg gccaacctac acagcaaggc cagcatgacc 180
atgccggacg tgtactttgt caacatggca gtggcaggcc tggtgctcag cgccctggcc 240
Page 1
CA 02429860 2003-05-26
cctgtgcacctgctcggccccccgagctcccggtgggcgctgtggagtgtgggcggcgaa300
gtccacgtggcactgcagatccccttcaatgtgtcctcactggtggccatgtactccacc360
gccctgctgagcctcgaccactacatcgagcgtgcactgccgcggacctacatggccagc420
gtgtacaacacgcggcacgtgtgcggcttcgtgtggggtggcgcgctgctgaccagcttc480
tcctcgctgctcttctacatctgcagccatgtgtccacccgcgcgctagagtgcgccaag540
atgcagaacgcagaagctgccgacgccacgctggtgttcatcggctacgtggtgccagca600
ctggccaccctctacgcgctggtgctactctcccgcgtccgcagggaggacacgcccctg660
gaccgggacacgggccggctggagccctcggcacacaggctgctggtggccaccgtgtgc720
acgcagtttgggctctggacgccacactatctgatcctgctggggcacacggtcatcatc780
tcgcgagggaagcccgtggacgcacactacctggggctactgcactttgtgaaggatttc840
tccaaactcctggccttctccagcagctttgtgacaccacttctctaccgctacatgaac900
cagagcttccccagcaagctccaacggctgatgaaaaagctgccctgcggggaccggcac960
tgctccccggaccacatgggggtgcagcaggtgctggcgtcg 1002
<210> 2
<211> 333
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 2
Met Trp Ser Cys Ser Trp Phe Asn Gly Thr Gly Leu Val Glu Glu Leu
1 5 10 15
Pro Ala Cys Gln Asp Leu Gln Leu Gly Leu Ser Leu Leu Ser Leu Leu
20 25 30
Gly Leu Val Val Gly Val Pro Val Gly Leu Cys Tyr Asn Ala Leu Leu
35 40 45
Val Leu Ala Asn Leu His Ser Lys Ala Ser Met Thr Met Pro Asp Val
50 55 60
Tyr Phe Val Asn Met Ala Val Ala Gly Leu Val Leu Ser Ala Leu Ala
65 70 75 80
Pro Val His Leu Leu Gly Pro Pro Ser Ser Arg Trp Ala Leu Trp Ser
85 90 95
Val Gly Gly Glu Val His Val Ala Leu Gln Ile Pro Phe Asn Val Ser
100 105 110
Ser Leu Val Ala Met Tyr Ser Thr Ala Leu Leu Ser Leu Asp His Tyr
Page 2
CA 02429860 2003-05-26
115 120 125
Ile Glu Arg Ala Leu Pro Arg Thr Tyr Met Ala Ser Val Tyr Asn Thr
l30 135 140
Arg His Val Cys Gly Phe Val Trp Gly Gly A1a Leu Leu Thr Ser Phe
145 150 155 160
Ser Ser Leu Leu Phe Tyr Ile Cys Ser His Val Ser Thr Arg Ala Leu
165 170 175
Glu Cys Ala Lys Met Gln Asn Ala Glu Ala Ala Asp Ala Thr Leu Val
180 185 190
Phe Tle Gly Tyr Val Val Pro Ala Leu Ala Thr Leu Tyr Ala Leu Val
195 200 205
Leu Leu Ser Arg Val Arg Arg Glu Asp Thr Pro Leu Asp Arg Asp Thr
210 215 220
Gly Arg Leu Glu Pro Ser Ala His Arg Leu Leu Val Ala Thr Va1 Cys
225 230 235 240
Thr Gln Phe Gly Leu Trp Thr Pro His Tyr Leu Ile Leu Leu Gly His
245 250 255
Thr Val Ile Ile Ser Arg G1y Lys Pro Val Asp Ala His Tyr Leu Gly
260 265 270
Leu Leu His Phe Val Lys Asp Phe Ser Lys Leu Leu Ala Phe Ser Ser
275 280 285
Ser Phe Val Thr Pro Leu Leu Tyr Arg Tyr Met Asn Gln Ser Phe Pro
290 295 300
Ser Lys Leu Gln Arg Leu Met Lys Lys Leu Pro Cys Gly Asp Arg His
305 310 315 320
Cys Ser Pro Asp His Met Gly Val Gln Gln Val Leu Ala
325 330
<210> 3
<211> 918
<212> DNA
<213> Homo Sapiens
<400> 3
atgcctggcc acaatacctc caggaattcc tcttgcgatc ctatagtgac accccactta 60
atcagcctct acttcatagt gcttattggc gggctggtgg gtgtcatttc cattcttttc 120
ctcctggtga aaatgaacac ccggtcagtg accaccatgg cggtcattaa cttggtggtg 180
Page 3
CA 02429860 2003-05-26
gtccacagcgtttttctgctgacagtgccatttcgcttgacctacctcatcaagaagact240
tggatgtttgggctgcccttctgcaaatttgtgagtgccatgctgcacatccacatgtac300
ctcacgttcctattctatgtggtgatcctggtcaccagatacctcatcttcttcaagtgc360
aaagacaaagtgg attctacagaaaactgcatgctgtggctgccagtgctggcatgtgg420
acgctggtgattgtcattgtggtacccctggttgtctcccggtatggaatccatgaggaa480
tacaatgaggagcactgttttaaatttcacaaagagcttgcttacacatatgtgaaaatc540
atcaactatatgatagtcatttttgtcatagccgttgctgtgattctgttggtcttccag600
gtcttcatcattatgttgatggtgcagaagctacgccactctttactatcccaccaggag660
ttctgggctcagctgaaaaacctattttttataggggtcatccttgtttgtttccttccc720
taccagttctttaggatctattacttgaatgttgtgacgcattccaatgcctgtaacagc780
aaggttgcattttataacgaaatcttcttgagtgtaacagcaattagctgctatgatttg840
cttctctttgtctttgggggaagccattggtttaagcaaaagataattggcttatggaat900
tgtgttttgtgccgttag
918
<210> 4
<211> 305
<2l2> PRT
<213> Homo Sapiens
<400> 4
Met Pro Gly His Asn Thr Ser Arg Asn Ser Ser Cys Asp Pro Ile Val
1 5 10 15
Thr Pro His Leu I1e Ser Leu Tyr Phe Ile Val Leu Ile Gly Gly Leu
20 25 30
Val Gly Val Ile Ser I1e Leu Phe Leu Leu Val Lys Met Asn Thr Arg
35 40 45
Ser Val Thr Thr Met Ala Val Ile Asn Leu Val Val Val His Ser Val
50 55 60
Phe Leu Leu Thr Val Pro Phe Arg Leu Thr Tyr Leu Ile Lys Lys Thr
65 70 75 80
Trp Met Phe Gly Leu Pro Phe Cys Lys Phe Val Ser Ala Met Leu His
85 90 95
Ile His Met Tyr Leu Thr Phe Leu Phe Tyr Val Val Ile Leu Va1 Thr
100 105 110
Arg Tyr Leu Ile Phe Phe Lys Cys Lys Asp Lys Val Glu Phe Tyr Arg
115 120 125
Page 4
CA 02429860 2003-05-26
Lys Leu His Ala Val Ala A1a Ser Ala Gly Met Trp Thr Leu Val Ile
130 135 140
Val Ile Val Val Pro Leu Val Val Ser Arg Tyr Gly Ile His Glu Glu
145 150 155 160
Tyr Asn Glu Glu His Cys Phe Lys Phe His Lys Glu Leu Ala Tyr Thr
165 170 175
Tyr Val Lys Tle Ile Asn Tyr Met Ile Val Ile Phe Val 21e Ala Val
180 185 190
Ala Val Ile Leu Leu Val Phe Gln Val Phe Tle Ile Met Leu Met Val
195 200 205
Gln Lys Leu Arg His Ser Leu Leu Ser His Gln Glu Phe Trp Ala Gln
210 215 220
Leu Lys Asn Leu Phe Phe Ile Gly Val Ile Leu Val Cys Phe Leu Pro
225 230 235 240
Tyr Gln Phe Phe Arg Ile Tyr Tyr Leu Asn Val Val Thr His Ser Asn
245 250 255
Ala Cys Asn Ser Lys Val Ala Phe Tyr Asn Glu Ile Phe Leu Ser Val
260 265 270
Thr Ala Ile Ser Cys Tyr Asp Leu Leu Leu Phe Val Phe Gly Gly Ser
275 280 285
His Trp Phe Lys Gln Lys Ile Ile Gly Leu Trp Asn Cys Val Leu Cys
290 295 300
Arg
305
<210> 5
<211> 1125
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 5
atgctgacag ggagctgcgg ggaccctcag aaaaagccac aggtgaccca ggactcaggg 60
ccccagagca tggggcttga gggacgagag acagctggcc agccacgagt gaccctgctg 120
cccacgccca acgtcagcgg gctgagccag gagtttgaaa gccactggcc agagatcgca 180
gagaggtccc cgtgtgtggc tggcgtcatc cctgtcatct actacagtgt cctgctgggc 240
Page 5
CA 02429860 2003-05-26
ttggggctgcctgtcagcctcctgaccgcagtggccctggcgcgccttgccaccaggacc300
aggaggccctcctactactaccttctggcgctcacagcctcggatatcatcatccaggtg360
gtcatcgtgttcgcgggcttcctcctgcagggagcagtgctggcccgccaggtgccccag420
gctgtggtgcgcacggccaacatcctggagtttgctgccaaccacgcctcagtctggatc480
gccatcctgctcacggttgaccgctacactgccctgtgccaccccctgcaccatcgggcc540
gcctcgtccccaggccggacccgccgggccattgctgctgtcctgagtgctgccctgttg600
accggcatccccttctactggtggctggacatgtggagagacaccgactcacccagaaca660
ctggacgaggtcctcaagtgggctcactgtctcactgtctatttcatcccttgtggcgtg720
ttcctggtcaccaactcggccatcatccaccggctacggaggaggggccggagtgggctg780
cagccccgggtgggcaagagcacagccatcctcctgggcatcaccacactgttcaccctc840
ctgtgggcgccccgggtcttcgtcatgctctaccacatgtacgtggcccctgtccaccgg900
gactggagggtccacctggccttggatgtggccaatatggtggccatgctccacacggca960
gccaacttcggcctctactgctttgtcagcaagactttccgggccactgtccgacaggtc1020
atccacgatgcctacctgccctgcactttggcatcacagccagagggcatggcggcgaag1080
cctgtgatggagcctccgggactccccacaggggcagaagtgtag 1125
<210> 6
<211> 374
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 6
Met Leu Thr Gly Ser Cys Gly Asp Pro Gln Lys Lys Pro Gln Val Thr
1 5 10 15
Gln Asp Ser Gly Pro Gln Ser Met Gly Leu Glu Gly Arg Glu Thr Ala
20 25 30
Gly Gln Pro Arg Val Thr Leu Leu Pro Thr Pro Asn Val Ser G1y Leu
35 40 45
Ser Gln Glu Phe Glu Ser His Trp Pro Glu Ile Ala Glu Arg Ser Pro
50 55 60
Cys Val Ala Gly Val Ile Pro Val Ile Tyr Tyr Ser Val Leu Leu Gly
65 70 75 80
Leu Gly Leu Pro Val Ser Leu Leu Thr Ala Val Ala Leu Ala Arg Leu
85 90 95
Ala Thr Arg Thr Arg Arg Pro Ser Tyr Tyr Tyr Leu Leu Ala Leu Thr
Page 6
CA 02429860 2003-05-26
100 105 110
Ala Ser Asp Ile Ile Ile Gln Val Val Ile Val Phe Ala Gly Phe Leu
115 120 125
Leu Gln Gly Ala Va1 Leu Ala Arg Gln Val Pro Gln Ala Val Val Arg
130 135 140
Thr Ala Asn Ile Leu Glu Phe Ala Ala Asn His Ala Ser Val Trp Ile
145 150 155 160
Ala Ile Leu Leu Thr Val Asp Arg Tyr Thr Ala Leu Cys His Pro Leu
165 170 175
His His Arg Ala Ala Ser Ser Pro Gly Arg Thr Arg Arg Ala Ile Ala
180 185 190
Ala Val Leu Ser Ala Ala Leu Leu Thr Gly Ile Pro Phe Tyr Trp Trp
195 200 205
Leu Asp Met Trp Arg Asp Thr Asp Ser Pro Arg Thr Leu Asp Glu Val
210 215 220
Leu Lys Trp Ala His Cys Leu Thr Val Tyr Phe Ile Pro Cys Gly Val
225 230 235 240
Phe Leu Val Thr Asn Ser Ala Ile Ile His Arg Leu Arg Arg Arg Gly
245 250 255
Arg Ser Gly Leu Gln Pro Arg Val Gly Lys Ser Thr Ala Ile Leu Leu
260 265 270
Gly I12 Thr Thr Leu Phe Thr Leu Leu Trp Ala Pro Arg Val Phe Val
275 280 285
Met Leu Tyr His Met Tyr Val Ala Pro Val His Arg Asp Trp Arg Val
290 295 300
His Leu Ala Leu Asp Va1 Ala Asn Met Val Ala Met Leu His Thr Ala
305 310 315 320
Ala Asn Phe Gly Leu Tyr Cys Phe Val Ser Lys Thr Phe Arg Ala Thr
325 330 335
Val Arg Gln Val Ile His Asp Ala Tyr Leu Pro Cys Thr Leu Ala Ser
340 345 350
Gln Pro Glu Gly Met Ala Ala Lys Pro Val Met Glu Pro Pro Gly Leu
355 360 365
Page 7
CA 02429860 2003-05-26
Pro Thr Gly Ala Glu Val
370
<210> 7
<211> 1086
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400>
7
atgtccactgaatgcgcgcgggcagcgggcgacgcgcccttgcgcagcctggagcaagcc60
aaccgcacccgctttcccttcttctccgacgtcaagggcgaccaccggctggtgctggcc120
gcggtggagacaaccgtgctggtgctcatctttgcagtgtcgctgctgggcaacgtgtgc180
gccctggtgctggtggcgcgccgacgacgccgcggcgcgactgcctgcctggtactcaac240
ctcttctgcgcggacctgctcttcatcagcgctatccctctggtgctggccgtgcgctgg300
actgaggcctggctgctgggccccgttgcctgccacctgctcttctacgtgatgaccctg360
agcggcagcgtcaccatcctcacgctggccgcggtcagcctggagcgcatggtgtgcatc420
gtgcacctgcagcgcggcgtgcggggtcctgggcggcgggcgcgggcagtgctgctggcg480
ctcatatggggctattcggcggtcgccgctctgcctctatgcgtcttcttccgagtcgtc540
ccgcaacggctccccggcgccgaccaggaaatttcgatttgcacactgatttggcccacc600
attcctggagagatctcgtgggatgtctcttttgttactttgaacttcttggtgccagga660
ctggtcattgtgatcagttactccaaaattttacagatcacaaaggcatcaaggaagagg720
ctcacggtaagcctggcctactcggagagccaccagatccgcgtgtcccagcaggacttc780
cggctcttccgcaccctcttcctcctcatggtctccttcttcatcatgtggagccccatc840
atcatcaccatcctcctcatcctgatccagaacttcaagcaagacctggtcatctggccg900
tccctcttcttctgggtggtggccttcacatttgctaattcagccctaaaccccatcctc960
tacaacatgacactgtgcaggaatgagtggaagaaaattttttgctgcttctggttccca1020
gaaaagggagccattttaacagacacatctgtcaaaagaaatgacttgtcgattatttct1080
ggctaa 1086
<210> 8
<211> 361
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 8
Met Ser Thr Glu Cys Ala Arg Ala Ala Gly Asp Ala Pro Leu Arg Ser
1 5 10 15
Page 8
CA 02429860 2003-05-26
Leu Glu Gln Ala Asn Arg Thr Arg Phe Pro Phe Phe Ser Asp Val Lys
20 25 30
Gly Asp His Arg Leu Val Leu Ala Ala Val Glu Thr Thr Val Leu Val
35 40 45
Leu Ile Phe Ala Val Ser Leu Leu Gly Asn Val Cys Ala Leu Val Leu
50 55 60
Val Ala Arg Arg Arg Arg Arg Gly Ala Thr Ala Cys Leu Val Leu Asn
65 70 75 80
Leu Phe Cys Ala Asp Leu Leu Phe Ile Ser A1a Ile Pro Leu Val Leu
85 90 95
Ala Val Arg Trp Thr Glu Ala Trp Leu Leu Gly Pro Val Ala Cys His
100 105 110
Leu Leu Phe Tyr Val Met Thr Leu Ser Gly Ser Val Thr Ile Leu Thr
115 120 125
Leu Ala Ala Val Ser Leu Glu Arg Met Val Cys Ile Val His Leu Gln
130 135 140
Arg Gly Val Arg Gly Pro Gly Arg Arg Ala Arg Ala Val Leu Leu Ala
145 150 155 160
Leu Ile Trp Gly Tyr Ser Ala Val Ala Ala Leu Pro Leu Cys Val Phe
165 170 175
Phe Arg Val Val Pro Gln Arg Leu Pro Gly Ala Asp G1n Glu Ile Ser
180 185 190
Ile Cys Thr Leu Ile Trp Pro Thr Ile Pro Gly Glu Ile Ser Trp Asp
195 200 205
Val Ser Phe Val Thr Leu Asn Phe Leu Val Pro Gly Leu Val Ile Val
210 215 220
Ile Ser Tyr Ser Lys Ile Leu Gln Tle Thr Lys Ala Ser Arg Lys Arg
225 230 235 240
Leu Thr Val Ser Leu Ala Tyr Ser Glu Ser His Gln Ile Arg Val Ser
245 250 255
Gln Gln Asp Phe Arg Leu Phe Arg Thr Leu Phe Leu Leu Met Val Ser
260 265 270
Phe Phe Ile Met Trp Ser Pro Ile Ile Ile Thr Ile Leu Leu Ile Leu
Page 9
CA 02429860 2003-05-26
275 280 285
Ile Gln Asn Phe Lys Gln Asp Leu Val Ile Trp Pro Ser Leu Phe Phe
290 295 300
Trp Val Val Ala Phe Thr Phe Ala Asn Ser Ala Leu Asn Pro Ile Leu
305 310 315 320
Tyr Asn Met Thr Leu Cys Arg Asn Glu Trp Lys Lys Ile Phe Cys Cys
325 330 335
Phe Trp Phe Pro Glu Lys Gly Ala Ile Leu Thr Asp Thr Ser Val Lys
340 345 350
Arg Asn Asp Leu Ser Ile Ile Ser Gly
355 360
<210> 9
<211> 1038
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400>
9
atgagcagcaattcatccctgctggtggctgtgcagctgtgctacgcgaacgtgaatggg60
tcctgtgtgaaaatccccttctcgccgggatcccgggtgattctgtacatagtgtttggc120
tttggggctgtgctggctgtgtttggaaacctcctggtgatgatttcaatcctccatttc180
aagcagctgcactctccgaccaattttctcgttgcctctctggcctgcgctgatttcttg240
gtgggtgtgactgtgatgcccttcagcatggtcaggacggtggagagctgctggtatttt300
gggaggagtttttgtactttccacacctgctgtgatgtggcattttgttactcttctctc360
tttcacttgtgcttcatctccatcgacaggtacattgcggttactgaccccctggtctat420
cctaccaagttcaccgtatctgtgtcaggaatttgcatcagcgtgtcctggatcctgccc480
ctcatgtacagcggtgctgtgttctacacaggtgtctatgacgatgggctggaggaatta540
tctgatgccctaaactgtataggaggttgtcagaccgttgtaaatcaaaactgggtgttg600
acagattttctatccttctttatacctacctttattatgataattctgtatggtaacata660
tttcttgtggctagacgacaggcgaaaaagatagaaaatactggtagcaagacagaatca720
tcctcagagagttacaaagccagagtggccaggagagagagaaaagcagctaaaaccctg780
ggggtcacagtggtagcatttatgatttcatggttaccatatagcattgattcattaatt840
gatgcctttatgggctttataacccctgcctgtatttatgagatttgctgttggtgtgct900
tattataactcagccatgaatcctttgatttatgctttattttacccatggtttaggaaa960
gcaataaaagttattgtaactggtcaggttttaaagaacagttcagcaaccatgaatttg1020
Page 10
CA 02429860 2003-05-26
ttttctgaac atatataa 1038
<210> 10
<211> 345
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 10
Met Ser Ser Asn Ser Ser Leu Leu Val Ala Val Gln Leu Cys Tyr Ala
1 5 10 15
Asn Val Asn Gly Ser Cys Val Lys Ile Pro Phe Ser Pro Gly Ser Arg
20 25 30
Val Ile Leu Tyr Ile Val Phe Gly Phe Gly Ala Val Leu Ala Val Phe
35 40 45
Gly Asn Leu Leu Val Met Ile Ser Ile Leu His Phe Lys Gln Leu His
50 55 60
Ser Pro Thr Asn Phe Leu Val Ala Ser Leu Ala Cys Ala Asp Phe Leu
65 70 75 80
Val Gly Val Thr Val Met Pro Phe Ser Met Val Arg Thr Val Glu Ser
85 90 95
Cys Trp Tyr Phe Gly Arg Ser Phe Cys Thr Phe His Thr Cys Cys Asp
100 105 110
Val Ala Phe Cys Tyr Ser Ser Leu Phe His Leu Cys Phe Ile Ser Ile
115 120 125
Asp Arg Tyr Ile Ala Val Thr Asp Pro Leu Val Tyr Pro Thr Lys Phe
130 135 140
Thr Val Ser Val Ser Gly Ile Cys Ile Ser Val Ser Trp Ile Leu Pro
145 150 155 160
Leu Met Tyr Ser Gly Ala Val Phe Tyr Thr Gly Val Tyr Asp Asp Gly
165 170 175
Leu Glu Glu Leu 5er Asp Ala Leu Asn Cys Ile Gly Gly Cys Gln Thr
180 185 190
Val Val Asn Gln Asn Trp Val Leu Thr Asp Phe Leu Ser Phe Phe Ile
195 200 205
Pro Thr Phe Ile Met Ile Ile Leu Tyr Gly Asn Tle Phe Leu Val Ala
Page 11
CA 02429860 2003-05-26
210 215 220
Arg Arg Gln Ala Lys Lys Ile Glu Asn Thr Gly Ser Lys Thr Glu Ser
225 230 235 240
Ser Ser G1u Ser Tyr Lys Ala Arg Val Ala Arg Arg Glu Arg Lys Ala
245 250 255
Ala Lys Thr Leu Gly Val Thr Val Val Ala Phe Met Ile Ser Trp Leu
260 265 270
Pro Tyr Ser Ile Asp Ser Leu Ile Asp Ala Phe Met Gly Phe Ile Thr
275 280 285
Pro Ala Cys Ile Tyr Glu Ile Cys Cys Trp Cys Ala Tyr Tyr Asn Ser
290 295 300
Ala Met Asn Pro Leu Ile Tyr Ala Leu Phe Tyr Pro Trp Phe Arg Lys
305 310 315 320
Ala Ile Lys Val Ile Val Thr Gly Gln Val Leu Lys Asn Ser Ser Ala
325 330 335
Thr Met Asn Leu Phe Ser Glu His Ile
340 345
<210>
11
<211>
1020
<212>
DNA
<213>
Artificial
Sequence
<220>
<223>
Novel
Sequence
<400>
11
atgatgcccttttgccacaatataattaatatttcctgtgtgaaaaacaactggtcaaat60
gatgtccgtgettccctgtacagtttaatggtgctcataattctgaccacactcgttggc120
aatctgatagttattgtttctatatcacacttcaaacaacttcataccccaacaaattgg180
ctcattcattccatggccactgtggactttcttctggggtgtctggtcatgccttacagt240
atggtgagatctgctgagcactgttggtattttggagaagtcttctgtaaaattcacaca300
agcaccgacattatgctgagctcagcctccattttccatttgtctttcatctccattgac360
cgctactatgctgtgtgtgatccactgagatataaagccaagatgaatatcttggttatt420
tgtgtgatgatcttcattagttggagtgtccctgctgtttttgcatttggaatgatcttt480
ctggagctaaacttcaaaggcgctgaagagatatattacaaacatgttcactgcagagga540
ggttgctctgtcttctttagcaaaatatctggggtactgacctttatgacttctttttat600
atacctggatctattatgttatgtgtctattacagaatatatcttatcgctaaagaacag660
Page 12
CA 02429860 2003-05-26
gcaagattaattagtgatgccaatcagaagctccaaattggattggaaatgaaaaatgga720
atttcacaaagcaaagaaaggaaagctgtgaagacattggggattgtgatgggagttttc780
ctaatatgctggtgccctttctttatctgtacagtcatggacccttttcttcactacatt840
attccacctactttgaatgatgtattgatttggtttggctacttgaactctacatttaat900
ccaatggtttatgcatttttctatccttggtttagaaaagcactgaagatgatgctgttt960
ggtaaaattttccaaaaagattcatccaggtgtaaattatttttggaattgagttcatag1020
<210> 12
<211> 339
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 12
Met Met Pro Phe Cys His Asn Ile Tle Asn Tle Ser Cys Val Lys Asn
1 5 10 15
Asn Trp Ser Asn Asp Val Arg Ala Ser Leu Tyr Ser Leu Met Val Leu
20 25 30
Ile Ile Leu Thr Thr Leu Val Gly Asn Leu Ile Val Ile Val Ser Ile
35 40 45
Ser His Phe Lys Gln Leu His Thr Pro Thr Asn Trp Leu Ile His Ser
50 55 60
Met Ala Thr Val Asp Phe Leu Leu Gly Cys Leu Val Met Pro Tyr Ser
65 70 75 80
Met Val Arg Ser Ala Glu His Cys Trp Tyr Phe Gly Glu Val Phe Cys
85 90 95
Lys Ile His Thr Ser Thr Asp Ile Met Leu Ser Ser Ala Ser Ile Phe
100 105 110
His Leu Ser Phe Ile Ser Ile Asp Arg Tyr Tyr Ala Val Cys Asp Pro
115 120 125
Leu Arg Tyr Lys Ala Lys Met Asn Ile Leu Val Ile Cys Val Met Ile
130 135 140
Phe Ile Ser Trp Ser Val Pro Ala Val Phe Ala Phe Gly Met Ile Phe
145 150 155 160
Leu Glu Leu Asn Phe Lys Gly Ala Glu Glu Ile Tyr Tyr Lys His Val
165 170 175
Page 13
CA 02429860 2003-05-26
His Cys Arg Gly Gly Cys Ser Val Phe Phe Ser Lys Ile Ser Gly Val
180 185 190
Leu Thr Phe Met Thr Ser Phe Tyr Ile Pro Gly Ser I1e Met Leu Cys
195 200 205
Val Tyr Tyr Arg Tle Tyr Leu Ile Ala Lys Glu Gln Ala Arg Leu Ile
210 215 220
Ser Asp Ala Asn Gln Lys Leu Gln Ile Gly Leu Glu Met Lys Asn Gly
225 230 235 240
Ile Ser Gln Ser Lys Glu Arg Lys Ala Val Lys Thr Leu Gly Ile Val
245 250 255
Met Gly Val Phe Leu Ile Cys Trp Cys Pro Phe Phe Ile Cys Thr Val
260 265 270
Met Asp Pro Phe Leu His Tyr Ile Ile Pro Pro Thr Leu Asn Asp Val
275 280 285
Leu Ile Trp Phe Gly Tyr Leu Asn Ser Thr Phe Asn Pro Met Val Tyr
290 295 300
Ala Phe Phe Tyr Pro Trp Phe Arg Lys Ala Leu Lys Met Met Leu Phe
305 310 315 320
Gly Lys Ile Phe Gln Lys Asp Ser Ser Arg Cys Lys Leu Phe Leu Glu
325 330 335
Leu Ser Ser
<210> 13
<211> 1029
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400>
13
atgaccagcaatttttcccaacctgttgtgcagctttgctatgaggatgtgaatggatct60
tgtattgaaactccctattctcctgggtcccgggtaattctgtacacggcgtttagcttt120
gggtctttgctggctgtatttggaaatctcttagtaatgacttctgttcttcattttaag180
cagctgcactctccaaccaattttctcattgcctctctggcctgtgctgacttcttggta240
ggtgtgactgtgatgcttttcagcatggtcaggacggtggagagctgctggtattttgga300
gccaaattttgtactcttcacagttgctgtgatgtggcattttgttactcttctgtcctc360
Page 14
CA 02429860 2003-05-26
cacttgtgcttcatctgcatcgacaggtacattgtggttactgatcccctggtctatgct420
accaagttcaccgtgtctgtgtcgggaatttgcatcagcgtgtcctggattctgcctctc480
acgtacagcggtgctgtgttctacacaggtgtcaatgatgatgggctggaggaattagta540
agtgctctcaactgcgtaggtggctgtcaaattattgtaagtcaaggctgggtgttgata600
gattttctgttattcttcatacctacccttgttatgataattctttacagtaagattttt660
cttatagctaaacaacaagctataaaaattgaaactactagtagcaaagtagaatcatcc720
tcagagagttataaaatcagagtggccaagagagagaggaaagcagctaaaaccctgggg780
gtcacggtactagcatttgttatttcatggttaccgtatacagttgatatattaattgat840
gcctttatgggcttcctgacccctgcctatatctatgaaatttgctgttggagtgcttat900
tataactcagccatgaatcctttgatttatgctctattttatccttggtttaggaaagcc960
ataaaacttattttaagtggagatgttttaaaggctagttcatcaaccattagtttattt1020
ttagaataa 1029
<210> 14
<211> 342
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 14
Met Thr Ser Asn Phe Ser Gln Pro Val Val Gln Leu Cys Tyr Glu Asp
1 5 10 15
Val Asn Gly Ser Cys Ile Glu Thr Pro Tyr Ser Pro Gly Ser Arg Val
20 25 30
Ile Leu Tyr Thr Ala Phe Ser Phe Gly Ser Leu Leu Ala Val Phe Gly
35 40 45
Asn Leu Leu Val Met Thr Ser Val Leu His Phe Lys Gln Leu His Ser
50 55 60
Pro Thr Asn Phe Leu Ile,Ala Ser Leu Ala Cys Ala Asp Phe Leu Val
65 70 75 80
G1y Val Thr Val Met Leu Phe Ser Met Val Arg Thr Val Glu Ser Cys
85 90 95
Trp Tyr Phe Gly Ala Lys Phe Cys Thr Leu His Ser Cys Cys Asp Val
100 105 110
Ala Phe Cys Tyr Ser Ser Va1 Leu His Leu Cys Phe Ile Cys Ile Asp
115 120 125
Page 15
CA 02429860 2003-05-26
Arg Tyr Ile Val Val Thr Asp Pro Leu Val Tyr Ala Thr Lys Phe Thr
130 135 140
Val Ser Val Ser Gly Ile Cys Ile Ser Val Ser Trp Tle Leu Pro Leu
145 150 155 160
Thr Tyr Ser G1y Ala Val Phe Tyr Thr Gly Val Asn Asp Asp Gly Leu
l65 170 175
Glu Glu Leu Val Ser Ala Leu Asn Cys Val Gly Gly Cys Gln Ile Ile
180 185 190
Val Ser Gln Gly Trp Val Leu Ile Asp Phe Leu Leu Phe Phe Ile Pro
195 200 205
Thr Leu Val Met Ile Ile Leu Tyr Ser Lys Ile Phe Leu Ile Ala Lys
210 215 220
Gln Gln Ala Ile Lys Tle Glu Thr Thr Ser Ser Lys Val Glu Ser Ser
225 230 235 240
Ser Glu Ser Tyr Lys Ile Arg Val Ala Lys Arg Glu Arg Lys Ala Ala
245 250 255
Lys Thr Leu Gly Val Thr Val Leu Ala Phe Val Ile Sex Trp Leu Pro
260 265 270
Tyr Thr Val Asp Ile Leu Ile Asp Ala Phe Met G1y Phe Leu Thr Pro
275 280 285
Ala Tyr Ile Tyr Glu Ile Cys Cys Trp Ser Ala Tyr Tyr Asn Ser A1a
290 295 300
Met Asn Pro Leu Ile Tyr Ala Leu Phe Tyr Pro Trp Phe Arg Lys Ala
305 310 315 320
Ile Lys Leu Ile Leu Ser Gly Asp Val Leu Lys Ala Ser Ser Ser Thr
325 330 335
Tle Ser Leu Phe Leu Glu
340
<210> 15
<211> 1062
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 15
Page 26
CA 02429860 2003-05-26
atggagcacacgcacgcccacctcgcagccaacagctcgctgtcttggtggtcccccggc60
tcggcctgcggcttgggtttcgtgcccgtggtctactacagcctcttgctgtgcctcggt120
ttaccagcaaatatcttgacagtgatcatcctctcccagctggtggcaagaagacagaag180
tcctcctacaactatctcttggcactcgctgctgccgacatcttggtcctctttttcata240
gtgtttgtggacttcctgttggaagatttcatcttgaacatgcagatgcctcaggtcccc300
gacaagatcatagaagtgctggaattctcatccatccacacctccatatggattactgta360
ccgttaaccattgacaggtatatcgctgtctgccacccgctcaagtaccacacggtctca420
tacccagcccgcacccggaaagtcattgtaagtgtttacatcacctgcttcctgaccagc480
atcccctattactggtggcccaacatctggactgaagactacatcagcacctctgtgcat540
cacgtcctcatctggatccactgcttcaccgtctacctggtgccctgctccatcttcttc600
atcttgaactcaatcattgtgtacaagctcaggaggaagagcaattttcgtctccgtggc660
tactccacggggaagaccaccgccatcttgttcaccattacctccatctttgccacactt720
tgggccccccgcatcatcatgattctttaccacctctatggggcgcccatccagaaccgc780
tggctggtgcacatcatgtccgacattgccaacatgctagcccttctgaacacagccatc840
aacttcttcctctactgcttcatcagcaagcggttccgcaccatggcagccgccacgctc900
aaggctttcttcaagtgccagaagcaacctgtacagttctacaccaatcataacttttcc960
ataacaagtagcccctggatctcgccggcaaactcacactgcatcaagatgctggtgtac1020
cagtatgacaaaaatggaaaacctataaaagtatccccgtga 1062
<210> 16
<211> 353
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 16
Met Glu His Thr His Ala His Leu Ala Ala Asn Ser Ser Leu Ser Trp
1 5 10 15
Trp Ser Pro Gly Ser Ala Cys Gly Leu Gly Phe Val Pro Val Val Tyr
20 25 30
Tyr,Ser Leu Leu Leu Cys Leu Gly Leu Pro Ala Asn Ile Leu Thr Val
35 40 45
Ile Ile Leu Ser Gln Leu Val Ala Arg Arg Gln Lys Ser Ser Tyr Asn
50 55 60
Tyr Leu Leu Ala Leu Ala Ala Ala Asp Ile Leu Val Leu Phe Phe Ile
65 70 75 _ 80
Page 17
CA 02429860 2003-05-26
Val Phe Val Asp Phe Leu Leu Glu Asp Phe Ile Leu Asn Met Gln Met
85 90 95
Pro Gln Val Pro Asp Lys Ile Ile Glu Val Leu Glu Phe Ser Ser Ile
100 105 110
His Thr Ser Ile Trp Ile Thr Val Pro Leu Thr Ile Asp Arg Tyr Ile
115 120 125
Ala Val Cys His Pro Leu Lys Tyr His Thr Val Ser Tyr Pro Ala Arg
130 235 140
Thr Arg Lys Val Ile Val Ser Val Tyr Ile Thr Cys Phe Leu Thr 5er
145 150 155 160
Ile Pro Tyr Tyr Trp Trp Pro Asn Ile Trp Thr G1u Asp Tyr Ile Ser
165 170 175
Thr Ser Val His His Val Leu Ile Trp Ile His Cys Phe Thr Val Tyr
180 185 190
Leu Val Pro Cys Ser Ile Phe Phe Ile Leu Asn Ser Ile Ile Val Tyr
195 200 205
Lys Leu Arg Arg Lys Ser Asn Phe Arg Leu Arg Gly Tyr Ser Thr Gly
210 215 220
Lys Thr Thr Ala Ile Leu Phe Thr Ile Thr Ser Ile Phe Ala Thr Leu
225 230 235 ' 240
Trp Ala Pro Arg Ile Ile Met Ile Leu Tyr His Leu Tyr Gly A1a Pro
245 250 255
Ile Gln Asn Arg Trp Leu Val His Ile Met Ser Asp Ile Ala Asn Met
260 265 270
Leu Ala Leu Leu Asn Thr Ala Ile Asn Phe Phe Leu Tyr Cys Phe Ile
275 280 285
Ser Lys Arg Phe Arg Thr Met Ala A1a Ala Thr Leu Lys Ala Phe Phe
290 295 300
Lys Cys Gln Lys Gln Pro Val Gln Phe Tyr Thr Asn His Asn Phe Ser
305 310 315 320
Tle Thr 5er Ser Pro Trp Ile Ser Pro Ala Asn Ser His Cys Ile Lys
325 330 335
Met Leu Val Tyr Gln Tyr Asp Lys Asn Gly Lys Pro Ile Lys Val Ser
Page 18
CA 02429860 2003-05-26
340 345 350
Pro
<2l0> 17
<211> 969
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400>
17
atggatccaaccgtcccagtcttcggtacaaaactgacaccaatcaacggacgtgaggag60
actccttgctacaatcagaccctgagcttcacggtgctgacgtgcatcatttcccttgtc120
ggactgacaggaaacgcggtagtgctctggctcctgggctaccgcatgcgcaggaacgct180
gtctccatctacatcctcaacctggccgcagcagacttcctcttcctcagcttccagatt240
atacgttcgccattacgcctcatcaatatcagccatctcatccgcaaaatcctcgtttct300
gtgatgacctttccctactttacaggcctgagtatgctgagcgccatcagcaccgagcgc360
tgcctgtctgttctgtggcccatctggtaccgctgccgccgccccacacacctgtcagcg420
gtcgtgtgtgtcctgctctggggcctgtccctgctgtttagtatgctggagtggaggttc480
tgtgacttcctgtttagtggtgctgattctagttggtgtgaaacgtcagatttcatccca540
gtcgcgtggctgatttttttatgtgtggttctctgtgtttccagcctggtcctgctggtc600
aggatcctctgtggatcccggaagatgccgctgaccaggctgtacgtgaccatcctgctc660
acagtgctggtcttcctcctctgcggcctgcccttcggcattctgggggccctaatttac720
aggatgcacctgaatttggaagtcttatattgtcatgtttatctggtttgcatgtccctg780
tcctctctaaacagtagtgccaaccccatcatttacttcttcgtgggctcctttaggcag840
cgtcaaaataggcagaacctgaagctggttctccagagggctctgcaggacaagcctgag900
gtggataaaggtgaagggcagcttcctgaggaaagcctggagctgtcgggaagcagattg960
gggccatga 969
<210> 18
<211> 322
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 18
Met Asp Pro Thr Val Pro Val Phe Gly Thr Lys Leu Thr Pro Ile Asn
1 5 10 15
Gly Arg Glu Glu Thr Pro Cys Tyr Asn Gln Thr Leu Ser Phe Thr Val
Page 19
CA 02429860 2003-05-26
20 25 30
Leu Thr Cys Ile Ile Ser Leu Val Gly Leu Thr Gly Asn Ala Val Val
35 40 45
Leu Trp Leu~Leu Gly Tyr Arg Met Arg Arg Asn Ala Val Ser Ile Tyr
50 55 60
Ile Leu Asn Leu Ala Ala Ala Asp Phe Leu Phe Leu Ser Phe Gln Ile
65 70 75 80
Ile Arg Ser Pro Leu Arg Leu Ile Asn Ile Ser His Leu Ile Arg Lys
85 90 95
I1e Leu Val Ser Val Met Thr Phe Pro Tyr Phe Thr Gly Leu Ser Met
100 105 110
Leu Ser Ala Ile Ser Thr Glu Arg Cys Leu Ser Val Leu Trp Pro Ile
115 120 125
Trp Tyr Arg Cys Arg Arg Pro Thr His Leu Ser Ala Val Val Cys Val
130 135 140
Leu Leu Trp Gly Leu Ser Leu Leu Phe Ser Met Leu Glu Trp Arg Phe
145 150 155 160
Cys Asp Phe Leu Phe Ser G1y Ala Asp Ser Ser Trp Cys Glu Thr Ser
165 170 175
Asp Phe Ile Pro Val Ala Trp Leu Ile Phe Leu Cys Val Va1 Leu Cys
180 185 190
Val Ser Ser Leu Val Leu Leu Val Arg Ile Leu Cys Gly Ser Arg Lys
195 200 205
Met Pro Leu Thr Arg Leu Tyr Val Thr Ile Leu Leu Thr Val Leu Val
2l0 215 220
Phe Leu Leu Cys Gly Leu Pro Phe Gly Ile Leu Giy Ala Leu Ile Tyr
225 230 235 240
Arg Met His Leu Asn Leu Glu Val Leu Tyr Cys His Val Tyr Leu Val
245 250 255
Cys Met Ser Leu Ser Ser Leu Asn Ser Ser Ala Asn Pro Ile Ile Tyr
260 265 270
Phe Phe Val Gly Ser Phe Arg Gln Arg Gln Asn Arg Gln Asn Leu Lys
275 280 285
Page 20
CA 02429860 2003-05-26
Leu Val Leu Gln Arg Ala Leu Gln Asp Lys Pro Glu Val Asp Lys Gly
290 295 300
Glu Gly Gln Leu Pro Glu Glu Ser Leu Glu Leu Ser Gly Ser Arg Leu
305 310 315 320
Gly Pro
<210> 19
<211> 969
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400>
19
atggattcaaccatcccagtcttgggtacagaactgacaccaatcaacggacgtgaggag60
actccttgctacaagcagaccctgagcttcacggggctgacgtgcatcgtttcccttgtb220
gcgctgacaggaaacgcggttgtgctctggctcctgggctgccgcatgcgcaggaacgct180
gtctccatctacatcctcaacctggtcgcggccgacttcctcttccttagcggccacatt240
atatgttcgccgttacgcctcatcaatatccgccatcccatctccaaaatcctcagtcct300
gtgatgacctttccctactttataggcctaagcatgctgagcgccatcagcaccgagcgc360
tgcctgtccatcctgtggcccatctggtaccactgccgccgccccagatacctgtcatcg420
gtcatgtgtgtcctgctctgggccctgtccctgctgcggagtatcctggagtggatgttc480
tgtgacttcctgtttagtggtgctgattctgtttggtgtgaaacgtcagatttcattaca540
atcgcgtggctggtttttttatgtgtggttctctgtgggtccagcctggtcctgctggtc600
aggattctctgtggatcccggaagatgccgctgaccaggctgtacgtgaccatcctcctc660
acagtgctggtcttcctcctctgtggcctgccctttggcattcagtgggccctgttttcc720
aggatccacctggattggaaagtcttattttgtcatgtgcatctagtttccattttcctg780
tccgctcttaacagcagtgccaaccccatcatttacttcttcgtgggctcctttaggcag840
cgtcaaaataggcagaacctgaagctggttctccagagggctctgcaggacacgcctgag900
gtggatgaaggtggagggtggcttcctcaggaaaccctggagctgtcgggaagcagattg960
gagcagtga 969
<210> 20
<211> 322
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 20
Page 21
CA 02429860 2003-05-26
Met Asp Ser Thr Ile Pro Val Leu Gly Thr Glu Leu Thr Pro Ile Asn
1 5 10 15
Gly Arg Glu Glu Thr Pro Cys Tyr Lys Gln Thr Leu Ser Phe Thr Gly
20 25 30
Leu Thr Cys Ile Val Ser Leu Val Ala Leu Thr Gly Asn Ala Val Val
35 40 45
Leu Trp Leu Leu Gly Cys Arg Met Arg Arg Asn Ala Val Ser Tle Tyr
50 55 60
21e Leu Asn Leu Val Ala Ala Asp Phe Leu Phe Leu Ser Gly His 21e
65 70 75 80
Ile Cys Ser Pro Leu Arg Leu Ile Asn Ile Arg His Pro Ile Ser Lys
85 90 95
Ile Leu Ser Pro Val Met Thr Phe Pro Tyr Phe Ile Gly Leu Ser Met
100 105 110
Leu Ser Ala Ile Ser Thr Glu Arg Cys Leu 5er Ile Leu Trp Pro Tle
115 120 125
Trp Tyr His Cys Arg Arg Pro Arg Tyr Leu Ser Ser Val Met Cys Val
130 135 140
Leu Leu Trp Ala Leu Ser Leu Leu Arg Ser Ile Leu Glu Trp Met Phe
145 150 155 160
Cys Asp Phe Leu Phe Ser Gly Ala Asp Ser Val Trp Cys Glu Thr Ser
165 170 l75
Asp Phe Ile Thr Ile Ala Trp Leu Val Phe Leu Cys Val Val Leu Cys
180 185 l90
Gly Ser Ser Leu Val Leu Leu Val Arg Ile Leu Cys Gly Ser Arg Lys
195 200 205
Met Pro Leu Thr Arg Leu Tyr Val Thr Ile Leu Leu Thr Val Leu Val
210 215 220
Phe Leu Leu Cys Gly Leu Pro Phe Gly Ile Gln Trp Ala Leu Phe Ser
225 230 235 240
Arg Ile His Leu Asp Trp Lys Val Leu Phe Cys His Val His Leu Val
245 250 255
Ser Ile Phe Leu Ser Ala Leu Asn Ser Ser Ala Asn Pro Ile Ile Tyr
Page 22
CA 02429860 2003-05-26
260 265 270
Phe Phe Val Gly Ser Phe Arg Gln Arg Gln Asn Arg Gln Asn Leu Lys
275 280 285
Leu Val Leu Gln Arg Ala Leu Gln Asp Thr Pro Glu Val Asp Glu Gly
290 295 300
Gly Gly Trp Leu Pro Gln Glu Thr Leu Glu Leu Ser Gly Ser Arg Leu
305 310 315 320
Glu Gln
<210> 21
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 21
cagagctctg gtggccacct ctgtcc 26
<210> 22
<21l> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 22
ctgcgtccac cagagtcacg tctcc 25
<210> 23
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 23
gtatgcctgg ccacaatacc tccagg 26
<210> 24
<21l> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence .
<400> 24
gtttgtggct aacggcacaa aacacaattc c 31
Page 23
CA 02429860 2003-05-26
<210> 25
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 25
ggtaccacaa tgacaatcac cagcgtcc 28
<210> 26
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 26
ggaacgtgag gtacatgtgg atgtgcagc 2g
<220> 27
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 27
gcagtgtagc ggtcaaccgt gagcagg 27
<210> 28
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 28
tgagcaggat ggcgatccag actgaggcgt gg 32
<210> 29
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 29
gaggtacagc tggcgatgct gacag 25
<210> 30
<211> 25
<212> DNA
<213> Artificial Sequence
Page 24
CA 02429860 2003-05-26
<220>
<223> Novel Sequence
<400> 30
gtggccatga gccaccctga gctcc 25
<210> 3l
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 31
ggaatgtcca ctgaatgcgc gcgg 24
<210> 32
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 32
agctcgccag gtgtgagaaa ctcgg 25
<210> 33
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 33
gcgttatgag cagcaattca tccctgctgg 30
<210> 34
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 34
gtatcctgaa cttcgtctat acaactgc 28
<210> 35
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 35
ccctcaggaa tgatgccctt ttgccacaa 29
Page 25
CA 02429860 2003-05-26
<210> 36
<211> 28
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 36
atccatgtgg ttggtgcatg tggttcgt 28
<210> 37
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 37
aaacaacaaa cagcagaacc atgaccagc 29
<210> 38
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 38
acatagagac aagtgacatg tgtgaaccac 30
<210> 39
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 39
ggtatgagac cgtgtggtac ttgagc 26
<210> 40
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 40
gtggcagaca gcgatatacc tgtcaatgg 29
<210> 41
<211> 27
<212> DNA
<213> Artificial Sequence
Page 26
CA 02429860 2003-05-26
<220>
<223> Novel Sequence
<400> 41
gcgctcatgg agcacacgca cgcccac 27
<210> 42
<211> 25
<2l2> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 42
gaggcagtag ttgccacacc tatgg 25
<210> 43
<2l1> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 43
catctggttt gtgttcccag gggcaccag 29
<210> 44
<2l1> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 44
gacagtgttg ctctcaaagt cccgtctgac tg 32
<210> 45
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 45
ctgtttccag ggtcatcaga ctggg 25
<210> 46
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 46
gcagcattgc tctcaaagtc ctgtctg 27
Page 27
CA 02429860 2003-05-26
<210> 47
<211> &
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 47
Thr Leu Glu Ser Ile Met
1 5
<210> 48
<211> 5
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 48
Glu Tyr Asn Leu Val
1 5
<210> 49
<211> 5
<212> PRT
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 49
Asp Cys Gly Leu Phe
1 5
<210> 50
<211> 34
<222> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 50
gatcaagctt ccatggcgtg ctgcctgagc gagg 34
<210> 51
<211> 53
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 51
gatcggatcc ttagaacagg ccgcagtcct tcaggttcag ctgcaggatg gtg 53
Page 28
CA 02429860 2003-05-26
<210> 52
<211> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 52
gtcctcactg gtggccatgt actcc 25
<210> 53
<21l> 25
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 53
ctgcgtccac cagagtcacg tctcc 25
<210> 54
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 54
cttggatgtt tgggctgccc ttctgc 26
<210> 55
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 55
gtttgtggct aacggcacaa aacacaattc c 31
<210> 56
<211> 26
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 56
ctgctcacgg ttgaccgcta cactgc 26
<210> 57
<211> 25
<212> DNA
<213> Artificial Sequence
Page 29
CA 02429860 2003-05-26
<220>
<223> Novel Sequence
<400> 57
gtggccatga gccaccctga gctcc 25
<210> 58
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 58 .
cttcttctcc gacgtcaagg 20
<210> 59
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 59
cttcttctcc gacgtcaagg 20
<210> 60
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 60
cttcttctcc gacgtcaagg , 20
<2l0> 61
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 61
caacggtctg acaacctcct 20
<210> 62
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 62
ttgctgtgat gtggcatttt g 21
Page 30
CA 02429860 2003-05-26
<210> 63
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 63
caggaagccc ataaaggcat caa 23
<210> 64
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 64
acatcacctg cttcctgacc 20
<210> 65
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 65
ccagcatctt gatgcagtgt 20
<210> 66
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 66
ccatctccaa aatcctcagt c 21
<210> 67
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Novel Sequence
<400> 67
gctgttaaga gcggacagga as 22
Page 31