Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02437866 2003-08-08
DESCRIPTION
TITLE OF THE INVENTION
NOVEL CLOCK GENE Bma12
Technical Field
The present invention relates to novel proteins BMAL
(Brain -Muscle -Arnt -Like protein) 2 which are involved in
circadian rhythm, their genes, and their use.
Background Art
Life activity is connaturally accompanied with various
cyclic changes ranging from the behavior at the individual level
to the biochemical phenomena at the cellular level. These
rhythmic life activities occurring at certain cycles are called
biorhythm and a periodic length of these phenomena which are
repeated in cycles is often close to a periodic fluctuation of
the environment such as a year, a month or a day. Sleep-wake
rhythm and hormonal-secretion rhythm for such as melatonin and
the adrenal cortex hormone are among those representing
circadian rhythms repeated by an approximately 24-hour cycle,
a daily unit. The circadian rhythms as mentioned have been
observed in almost all the biological species and tissues and
are regulated by the biological clock (Annu. Rev. Physiol. 55,
16-54, 1993). The suprachiasmatic nucleus (SCN) in the
vertebrate central nervous system, pineal gland, specific
neuronal tissues such as retina, etc. are known as tissues
conforming circadian rhythm (Science 203, 1245-1247, 1979,
Science 203, 656-658, 1979, Proc. Natl. Acad. Sci. USA 76,
999-1003, 1979, Brain Res. 245, 198-200, 1982, Neuron 10,
1
CA 02437866 2003-08-08
573-577, 1993, Science 272, 419-421, 1996).
As in the case of the mammalian suprachiasmatic nucleus
(SCN), non-mammalian vertebrate pineal glands produce
melatonin in response to circadian rhythm and light stimuli and
play a central role in the physiological circadian regulation
(Science 203,1245-1247,1979, Science 203, 656-658, 1979, Proc.
Natl. Acad. Sci. USA 76, 999-1003, 1979, Proc. Natl. Acad. Sci.
USA 77, 2319-2322, 1980, Proc. Natl. Acad. Sci. USA 80,
6119-6121, 1983, J. Neurosci. 9, 1943-1950, 1989). The
oscillation mechanism of the above-mentioned circadian rhythm
is said to be characterized by the system wherein oscillation
occurs at the gene level, is then amplified at the cellular level
and finally reaches the individual level (Cell 96, 271-290,
1999). Oscillation at the gene level is brought by a group of
genes called clock genes. Recent studies on the rodent clock
genes have revealed that the circadian oscillator genes in
mammals are positive and negative elements which form the
transcription/translation-based negative feedback loop (Cell
96, 271-290, 1999, Annu. Rev. Neurosci. 23, 713-742, 2000). In
mice, the negative elements include three period gene homologs;
Perl(Cell 90, 1003-1011, 1997, Nature 389, 512-516, 1997), Per2
(Cell 91, 1055-1064, 1997, Neuron 19, 1261-1269, 1997, Genes
Cells 3, 167-176, 1998) and Per3 (EMBO J. 17, 4753-4759, 1998,
Neuron, 20, 1103-1110, 1998) and two cryptochrome homologs;
Cryl and Cry2 (Cell 98, 193-205, 1999, Nature 398, 627-630,
1999).
As for positive elements, BMAL1, CLOCK and the like which
are basic helix-loop-helix (bHLH)-PAS (Per-Arnt-Sim)
transcription elements are known. A CLOCK-BMAL1 complex is
known to activate transcription through an E-box sequence
2
CA 02437866 2003-08-08
(E-box: CACGTG) which is found not only in the negative element
Perl (Science 280, 1564-1569, 1998) but also An clock-
controlled genes such as vasopressin (Cell 96, 57-68, 1999) and
in the albumin D-site binding protein gene (Genes Dev.14,
679-689, 2000). When a protein level of a negative element
mentioned above is increased, its own transactivation for a
promoter induced by a positive element is suppressed, the mRNA
and protein levels of the negative element are down-regulated,
and the molecular cycle is recommenced concomitant with the
transactivation of the negative element gene. Therefore, the
protein and mRNA levels of a negative element display a marked
circadian oscillation. In addition to fluctuations in these
clock genes, Perl and Pert expressions are induced by light
(Cell 91, 1055-1064, 1997, Neuron 19, 1261-1269, 1997, Cell 91,
1043-1053, 1997) and at least photo synchronization of an
oscillator is induced by Perl (J. Neurosci. 19, 1115-1121, 1999).
Further, it has been revealed that mRNA levels of a positive
element Bmall also exhibit circadian oscillation in antiphase
to those of negative elements (Biochem. Biophys. Res. Commun.
250, 83-87, 1998, Biochem. Biophys. Res. Commun. 253, 199-203,
1998). Since its transcriptional rhythm is close to that of
the Drosophila dClock (Science 286, 766-768, 1999), Bmall is
thought to be involved in feedback loop of the negative elements
(Science 286, 2460-2461, 1999, Science 288, 1013-1019, 2000).
On the other hand, the chicken (chick) pineal gland has
been known that it retains the circadian oscillator as well as
photic -input pathway and melatonin-output pathway in the pineal
cell and that these properties can readily be retained under
cultured conditions (Science 203, 1245-1247, 1979, Science 203,
656-658, 1979, Proc. Natl. Acad. Sci. USA 77, 2319-2322, 1980,
3
i d
CA 02437866 2003-08-08
Brain Res.438, 199-215, 1988, Recent Prog. Horm. Res. 45,
279-352, 1989, Nature 372, 94-97, 1994, Proc. Natl. Acad. Sci.
USA 94, 304-309, 1997, Brain Res. 774, 242-245, 1997). On the
basis of these observations, the chick pineal cell is thought
to be a prominent model for the study of the vertebrate circadian
clock systems at the cellular level (Recent Prog. HOrm. Res.
45, 279-352, 1989).
It is known that the biological clock is an auto-
oscillatory system which oscillates autonomically without any
exogenous stimulation and which, at the same time, has a
property of being reset by the exogenous light-stimulation. It
is also known that the vertebrate biological clock (circadian
clock) which autonomically oscillates in a period close to a
day is driven by the auto-feedback-loop consisting of a negative
element and a positive element. Many things, however, still
remain unknown with regard to the molecular clock system and
the like including photic-input and output pathways. The
object of the present invention is to provide novel proteins
BMAL2 (Brain-Muscle-Arnt-Like protein 2) crucial in the clock
oscillation mechanism including photic-input and output
pathways, genes encoding the proteins, a method for screening
a promoter or a suppressor of the promoter transactivation using
the proteins, and the like.
Disclosure of the Invention
The present inventors have made a keen study to solve the
object mentioned above, and isolated cCLOCK, cPER2 and cBMALl
genes from the chicken pineal gland which is a material suitable
for the study of circadian clock, and further isolated cDNA
encoding the novel clock protein cBMAL2 which was homologous
4
CA 02437866 2003-08-08
with cBMALl and sequenced it. The inventors have also isolated
the human, mouse and rat BMAL2 cDNAs respectively from the human
embryonic kidney cell line, the mouse mid brain and the rat early
fibroblast and sequenced them. In the pull-down assay, these
novel clock proteins BMAL2 were found to form heterodimers with
CLOCK, BMAL1 or the like, and to form homodimers among
themselves (BMAL2). Besides, in the luciferase assay, BMAL2
were observed not only to form heterodimers with CLOCK and
activate transcription via E-box but also to form homodimers
and bind to E-box to cooperatively suppress transcription.
Here the present invention is accomplished.
The present invention relates to: DNA encoding a protein
(a) or (b) below,
(a) a protein comprising an amino acid sequence shown by Seq.
ID No. 2, 4, 6 or 8,
(b) a protein which comprises an amino acid sequence wherein
one or a few amino acids are deleted, substituted or added in
the amino acid sequence shown by Seq. ID No. 2, 4, 6 or 8 and
which has the BMAL2 activity (claim 1); DNA containing a base
sequence shown by Seq. ID No. 1, 3, 5 or 7 or its complementary
sequence and part or whole of these sequences (claim 2); DNA
which hybridizes with DNA of claim 2 under a stringent condition
and which encodes a protein having the BMAL2 activity (claim
3); DNA encoding a protein (a) or (b) below,
(a) a protein comprising an amino acid sequence shown by Seq.
ID No. 10,
(b) a protein which comprises an amino acid sequence wherein
one or a few amino acids are deleted, substituted or added in
the amino acid sequence shown by Seq. ID No. 10 and which has
the BMAL2 activity (claim 4); DNA containing a base sequence
CA 02437866 2003-08-08
shown by Seq. ID No. 9 or its complementary sequence and part
or whole of these sequences (claim 5) ; DNA which hybridizes with
DNA of claim 5 under a stringent condition and which encodes
a protein having the BMAL2 activity (claim 6) ; DNA encoding a
protein (a) or (b) below,
(a) a protein comprising an amino acid sequence shown by Seq.
ID No. 12 or 14,
(b) a protein which comprises an amino acid sequence wherein
one or a few amino acids are deleted, substituted or added in
the amino acid sequence shown by Seq. ID No. 12 or 14 and which
has the BMAL2 activity (claim 7) ; DNA containing a base sequence
shown by Seq. ID No. 11 or 13 or its complementary sequence and
part or whole of these sequences (claim 8) ; DNA which hybridizes
with DNA of claim 8 under a stringent condition and which encodes
a protein having the BMAL2 activity (claim 9) ; DNA encoding a
protein (a) or (b) below,
(a) a protein comprising an amino acid sequence shown by Seq.
ID No. 16, 18 or 20,
(b) a protein which comprises an amino acid sequence wherein
one or a few amino acids are deleted, substituted or added in
the amino acid sequence shown by Seq. ID No. 16, 18 or 20 and
which has the BMAL2 activity (claim 10) ; DNA containing a base
sequence shown by Seq. ID No. 15, 17 or 19 or its complementary
sequence and part or whole of these sequences (claim 11) ; and
DNA which hybridizes with DNA'of claim 11 under a stringent
condition and which encodes a protein having the BMAL2 activity
(claim 12).
The present invention further relates to: a protein
comprising an amino acid sequence shown by Seq. ID No. 2, 4,
6 or 8 (claim 13); a protein which comprises an amino acid
6
I I
CA 02437866 2003-08-08
sequence wherein one or a few amino acids are deleted,
substituted or added in the amino acid sequence shown by Seq.
ID No. 2, 4, 6 or 8 and which has the BMAL2 activity (claim 14) ;
a protein comprising an amino acid sequence shown by Seq. ID
No. 10 (claim 15); a protein which comprises an amino acid
sequence wherein one or a few amino acids are deleted,
substituted or added in the amino acid sequence shown by Seq.
ID No. 10 and which has the BMAL2 activity (claim 16) ; a protein
comprising an amino acid sequence shown by Seq. ID No. 12 or
14 (claim 17) ; a protein which comprises an amino acid sequence
wherein one or a few amino acids are deleted, substituted or
added in the amino acid sequence shown by Seq. ID No. 12 or 14
and which has the BMAL2 activity (claim 18); a protein
comprising an amino acid sequence shown by Seq. ID No. 16, 18
or 20 (claim 19); a protein which comprises an amino acid
sequence wherein one or a few amino acids are deleted,
substituted or added in the amino acid sequence shown by Seq.
ID No. 16, 18 or 20 and which has the BMAL2 activity (claim 20) ;
and a peptide which comprises part of the protein of any of claims
13-20 and which has the BMAL2 activity (claim 21).
The present invention still further relates to: a fusion
protein or a fusion peptide wherein the protein of any of claims
13-20 or the peptide of claim 21 is bound with a marker protein
and/or a peptide tag (claim 22) ; an antibody which specifically
binds to the protein of any of claims 13-20 or to the peptide
of claim 21 (claim 23); the antibody according to claim 23,
wherein the antibody is a monoclonal antibody (claim 24); a
recombinant protein or peptide to which the antibody of claim
23 or 24 specifically binds and which has the BMAL2 activity
(claim 25); a host cell comprising an expression system capable
7
i i
CA 02437866 2003-08-08
of expressing the protein of any of claims 13-20 or the peptide
of claim 21 (claim 26); the host cell according to claim 26,
wherein the host cell is further capable of expressing CLOCK
and/or BMAL1 (claim 27); the host cell according to claim 26
or 27, wherein the expression system at least comprises a
promoter having an E-box sequence (CACGTG) (claim 28); the host
cell according to claim 28, wherein the promoter having an E-box
sequence (CACGTG) is a promoter of Per gene, Tim gene, Cry gene,
vasopressin gene or the albumin D-site binding protein gene
(claim 29); a non-human animal which, on its chromosome, is
deficient in the gene function to encode the protein of any of
claims 13-20 or the peptide of claim 21 or which over-expresses
the protein of any of claims 13-20 or the peptide of claim 21
(claim 30); and the non-human animal according to claim 30,
wherein the non-human animal is a mouse or a rat (claim 31).
The present invention also relates to: a method for
screening a promoter or a suppressor for the expression of the
protein of any of claims 13-20/the peptide of claim 21 or a
promoter or a suppressor of the Bma12 activity, wherein a cell
expressing the protein or peptide and a test substance are used
(claim 32) ; the method for screening a promoter or a suppressor
for the expression of the protein/peptide or a promoter or a
suppressor of the Bmal2 activity according to claim 32, wherein
the cell expressing the protein of any of claims 13-20 or the
peptide of claim 21 is the host cell of any of claims 26-29 (claim
33) ; a method for screening a promoter or a suppressor for the
expression of the protein of any of claims 13-20/the peptide
of claim 21 or a promoter or a suppressor of the Bma12 activity,
wherein the non-human animal of claim 30 or 31 and a test
substance are used (claim 34); an expression promoter of the
8
CA 02437866 2003-08-08
protein of any of claims 13-20 or the peptide of claim 21, wherein
the expression promoter is obtained by the screening method
according to any of claims 32-34 (claim 35); an expression
suppressor for the protein of any of claims 13-20 or the peptide
of claim 21, wherein the expression promoter is obtained by the
screening method according to any of claims 32-34 (claim 36);
a promoter of the Bma12 activity obtained by the screening
method according to any of claims 32-34 (claim 37); and a
suppressor for the Bma12 activity obtained by the screening
method according to any of claims 32-34 (claim 38).
The present invention further relates to: a method for
screening a promoter or a suppressor for the promoter
transactivation, wherein a cell which expresses the protein of
any of claims 13-20 or the peptide of claim 21 and which contains
a promoter having an E-box sequence (CACGTG) and a test
substance are used (claim 39); the method for screening a
promoter or a suppressor for the promoter transactivation
according to claim 39, wherein the cell which expresses the
protein of any of claims 13-20 or the peptide of claim 21 and
which contains a promoter having an E-box sequence (CACGTG) is
the host cell of claim 28 or 29 (claim 40) ; a method for screening
a promoter or a suppressor for the transactivation for a
promoter having an E-box sequence (CACGTG) in the non-human
animal of claim 30 or 31, wherein the non-human animal and a
test substance are used (claim 41) ; a promoter of the promoter
transactivation obtained by the screening method according to
any of claims 39-41 (claim 42); a suppressor for the promoter
transactivation obtained by the screening method according to
any of claims 39-41 (claim 43); and a method for diagnosing
diseases associated with the expression or the activity of BMAL2,
9
CA 02437866 2003-08-08
wherein the DNA sequence encoding BMAL2 in a sample is compared
with the DNA sequence encuding the protein of claim 13 or 14
(claim 44).
Brief Description of Drawings
Fig.1 shows the amino acid sequence of cPER2.
Fig. 2 shows the results of the amino acid homologies in
domains among cPER2 and three mouse PER proteins (mPERl-3).
Fig. 3 shows the comparison among the amino acid sequences
of various BMALs.
Fig. 4 shows the results of the amino acid homologies in
domains among various BMAL proteins.
Fig. 5 shows the phylogenetic tree of ARNT-BMAL proteins
and their amino acid homologies with cBMAL2 or hBMAL2.
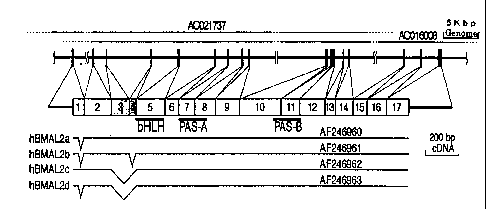
Fig. 6 shows the genomic structure of hBMAL2 gene of the
present invention.
Fig. 7 shows the basic structure of mouse BMAL2 and rat
BMAL2 of the present invention.
Fig. 8 shows the phylogenetic tree of the BMAL-ARNT family
proteins.
Fig. 9 shows the results of the northern blotting for
analyzing the expressions of cBmal2 and cBamll genes of the
present invention.
Fig. 10 shows the results of time-course changes in mRNA
levels of cBmal l , cBmal2, cPer2 and cClock in the chicken pineal
glands of the individuals.
Fig. 11 shows the time-course changes in mRNA levels of
cBmall, cBmal2, cPer2 and cCLOCK in the cultured chicken pineal
cells under LD or DD condition.
Fig. 12 shows the results of the daily fluctuations under
CA 02437866 2003-08-08
LD condition in mRNA expressions of mPer2 ; mClock, mBmall and
mBmal2 in the mouse suprachiasinatic nucleus.
Fig. 13 shows the results of light-dependent changes in
mRNA expressions of cPer2, cBmall and cBmal2 in the chicken
pineal glands.
Fig. 14 shows the results of the in vitro physical
interactions among cBMAL2 of the present invention, cBMALl and
cCLOCK proteins.
Fig. 15 shows the results of the binding between a E-
box sequence and cBMAL1-cCLOCK or cBMAL2-cCLOCK detected by an
electrophoretic mobility shift assay (EMSA).
Fig. 16 shows the results of transcriptional regulation
in the 293EBNA cells induced by cBMAL1, cBMAL2 and cCLOCK.
Fig. 17 shows the cPER2 effect on transactivation
mediated by E-box sequences.
Fig. 18 shows the effect of overexpression of cBMALI or
cBMAL2 on the melatonin-rhythms of the chicken pineal cells.
Best Mode of Carrying Out the Invention
Proteins of the present invention are exemplified by
novel proteins with BMAL2 activity including: human BMAL2 shown
by Seq. ID No. 2, 4, 6 or 8; chicken BMAL2 shown by Seq. ID No.
10; mouse BMAL2 shown by Seq. ID No. 12 or 14; rat BMAL2 shown
by Seq. ID No. 16, 18 or 20; a protein comprising an amino acid
sequence wherein one or a few amino acids are deleted,
substituted or added in the amino acid sequence shown by Seq.
ID No. 2, 4, 6, 8, 10, 12, 14, 16, 18 or 20, and having BMAL2
activity; and the like. Here the BMAL2 activity is taken to
mean an activity to form. a heterodimer with a
transcription-promoting element to promote transcription via
11
I I
CA 02437866 2003-08-08
E-box in the promoter of a clock oscillator gene, and to form
a homodimer to bind to E-box to competitively suppress
transcription. Any peptide comprising part of the above-
mentioned proteins and having BMAL2 activity may serve as a
peptide as an object of. the present invention, however, a
peptide having a basic helix-loop-helix (bHLH) structure or a
PAS (Per-Arnt-Sim) domain is preferable. Proteins and
peptides as objects of the present invention and the recombinant
proteins and peptides to which the antibodies, specifically
binding to these proteins and peptides, bind specifically may
collectively be referred to as the present proteins/peptides
hereinafter. The present proteins/peptides can be prepared in
accordance with known methods base on their DNA sequence
information or the like and there should be no limitation as
to the origin of the proteins/peptides.
Any DNA may be an object of the present invention as long
as the DNA encodes the present proteins/peptides mentioned
above and the specific examples include DNA encoding human BMAL2
shown by Seq. ID No. 2, 4, 6 or 8, DNA encoding chicken BMAL2
shown by Seq. ID No. 10, DNA encoding mouse BMAL2 shown by Seq.
ID No. 12 or 14, DNA encoding rat BMAL2 shown by Seq. ID No.
16, 18 or 20; DNA encoding a protein comprising an amino acid
sequence wherein one or a few amino acids are deleted,
substituted or added in the amino acid sequence shown by Seq.
ID No. 2, 4, 6, 8, 10, 12, 14, 16, 18 or 20 and having BMAL2
activity; and DNA containing the base sequence shown by Seq.
ID No. 1, 3, 5, 7, 9, 11, 13, 15, 17 or 19 or its complementary
sequence and part or whole of these sequences. These can be
prepared by known methods from, for instance, a gene library
or cDNA library and the like of human, chicken, mouse, rat, etc.,
12
CA 02437866 2003-08-08
based on their DNA sequence information or the like.
DNA encoding a protein having BMAL2 activity of the
interest which has the same effect as human BMAL2, chicken BMAL2,
mouse BMAL2, rat BMAL2, etc. can be obtained by hybridization
with various DNA libraries under a stringent condition by using
as a probe the base sequence shown by Seq. ID No. 1, 3, 5, 7,
9, 11, 13, 15, 17 or 19 or its complementary sequence and part
or whole of these sequences, and by subsequent isolation of DNA
which hybridized with the probe. DNAs thus obtained are also
within the scope of the present invention. One example of a
hybridization condition for obtaining DNA of the present
invention is hybridization at 42 C and washing at 42 C in a
buffer solution containing 1 x SSC, 0.1% SDS, and more
preferable example is hybridization at 65 C and washing at 65 C
in a buffer solution containing 0.1 x SSC, 0.1% SDS. There are
number of factors other than the temperature condition
mentioned above that affect the hybridization stringency and
those skilled in the art can actualize the same stringency as
that for the hybridization referred to in the above by
appropriately combining various factors.
Any fusion protein and fusion peptide may be used as a
fusion protein and a fusion peptide for the present invention
as long as the present proteins/peptides are bound with marker
proteins and/or peptide tags. As for a marker protein, there
is no limitation as long as it is a conventionally known marker
protein and the specific examples include alkaline phosphatase,
the Fc region of an antibody, HRP, GFP, etc. Conventionally
known peptide tags including Myc tag, V5 tag, HA tag, His tag,
FLAG tag, S tag, etc. are the specific examples of a peptide
tag for the use in the present invention. Such fusion protein
13
CA 02437866 2003-08-08
can be generated according to ordinary protocols and is useful
for the following: purification of the various BMAL2 or the like
by using affinity of Ni-NTA and His tag; detection of a protein
which interacts with various BMAL2; quantification of an
antibody against various BMAL2 or the like; and use as a
laboratory reagent in this field of art.
Antibodies that. specifically bind to the aforementioned
proteins and peptides of the present invention can be
particularly exemplified by immune-specific antibodies
including monoclonal antibodies, polyclonal antibodies,
chimeric antibodies, single-stranded antibodies, humanized
antibodies, etc. These antibodies can be generated according
to ordinary protocols by using the above-mentioned various
BMAL2 proteins or the like, or part of these proteins as an
antigen. However, monoclonal antibodies are more preferable
than the other sorts of antibodies mentioned because of their
specificity. Antibodies such as the monoclonal antibodies are
useful not only for diagnosis and treatment, such as missile
therapy, for the circadian rhythm sleep disorders or the like
including delayed sleep phase syndrome, non-24-hour sleep-wake
syndrome, advanced sleep phase syndrome, time zone change
syndrome, shift work sleep disorder, etc, but for elucidating
the molecular mechanism of the circadian oscillation system.
Antibodies of the present invention are created by
administering to an animal (preferably non-human) the present
proteins/peptides, their fragments containing epitopes, or the
cells expressing the proteins /peptides on the membrane surf ace,
according to the conventional protocols. The monoclonal
antibodies can be prepared, for instance, by any optional method
that provides antibodies produced by cultured materials of
14
CA 02437866 2003-08-08
continuous cell line such as a hybridoma method (Nature 256,
495-497, 1975), a trioma method, a human B-cell hybridoma method
(Immunology Today 4, 72, 1983), and an EBV-hybridoma method
(MONOCLONAL ANTIBODIES AND CANCER THERAPY, pp. 77-96, Alan R.
Liss, Inc., 1985).
The preparation method for a single chain antibody (US
PAT. No. 4, 946, 778) can be adopted to prepare single-stranded
antibodies to.the present proteins/peptides of the present
invention mentioned above. Transgenic mice, other mammals,
etc. can be used for expressing humanized antibodies. Clones
expressing the present proteins/peptides can be
isolated/ identified using the antibodies mentioned above, and
their polypeptides can be purified by affinity chromatography.
Antibodies to the present proteins/peptides or to peptides
containing their antigenic epitopes can possibly be used for
diagnosis and therapy for circadian rhythm sleep disorders or
the like including delayed sleep phase syndrome, non-24-hour
sleep-wake syndrome, advanced sleep phase syndrome, time zone
change syndrome, shift work sleep disorder, etc, and are useful
for elucidating the molecular mechanism of the circadian
oscillation system. Furthermore, recombinant proteins or
peptides to which these antibodies specifically bind are also
covered by the present proteins/peptides of the present
invention as described earlier.
The functions of the present proteins/peptides can be
analyzed by using, for example, antibodies such as the
aforementioned monoclonal antibodies labeled with fluorescent
materials including FITC (Fluorescein isothiocyanate),
tetramethyirhodamine isothiocyanate, etc., radioisotopes
including 12-1, 32P , 14C , 35S , 3H , etc. , or enzymes including
I I
CA 02437866 2003-08-08
alkaline phosphatase, peroxidase, 0-galactosidase,
phycoerythrin, etc., or fused with fluorescent proteins such
as Green Fluorescent Protein (GFP), BFP, CFP, YFP, RFP, etc.
to serve as fusion proteins. As for immunological detection
methods using the antibodies of the present invention, RIA
method, ELISA method, fluorescent-antibody method, plaque
method, spot method, haemagglutination, Ouchterlony method,
etc. are exemplified.
There is no particular limitation as to a host cell of
the present invention as long as the host cell comprises an
expression system capable of expressing the present
proteins/peptides. However, a preferable host cell is such in
which the genes encoding CLOCK and/or BMALl are incorporated
so that the two proteins can be simultaneously expressed in the
host cell. Even more preferably, the host cell is incorporated
with a DNA fragment which at least contains a promoter having
E-box sequence (CACGTG), e.g. promoters of Per gene, Tim gene,
Cry gene, vasopressin gene, the albumin D-site binding protein
gene, etc., or a promoter introduced with E-box sequence
(CACGTG) or the like. Although there is no particular
limitation as to the above-mentioned DNA fragment so far as the
fragment contains a promoter having E-box sequence (CACGTG),
it is preferable for readily detecting and measuring the
promoter activity that the DNA fragment is linked with a
reporter gene including chloramphenicol acetyltransf erase
(CAT) gene, luciferase gene, etc. , a gene encoding a fluorescent
protein including a short-lived green fluorescent protein
(dlEGFP) , etc. or with a fusion of GFP gene and a clock oscillator
gene, and the like, to the down- stream of the promoter. Further,
as to a promoter introduced with E-box sequence (CACGTG), any
16
I i
CA 02437866 2003-08-08
promoter may be adopted as long as its promoter activity can
be regulated by a promoting element including the present
proteins/peptides, CLOCK, BMAL1, etc. or by a suppressing
element including PER, TIM, CRY, etc. These promoters are
exemplified by RSV promoter, trp promoter, lac promoter, recA
promoter, APL promoter, lpp promoter, SPO1 promoter, SPO2
promoter, penP promoter, PHO5 promoter, PGK promoter, GAP
promoter, ADH promoter, SRQ promoter, SV40 promoter, LTR
promoter, CMV promoter, HSV-TK promoter, etc., but the
promoters will not be limited to these exemplifications alone.
The present proteins /peptides and genes such as CLOCK and
BMAL1 can be introduced into host cells by methods described
in many standard laboratory manuals such as a manual of Davis
et al. (BASIC METHODS IN MOLECULAR BIOLOGY, 1986), of Sambrook
et al. (MOLECULAR CLONING: A LABORATORY MANUAL, 2nd Ed., Cold
Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989)
and the like. The methods include calcium-phosphate
transfection, DEAE-dextran-mediated transfection,
transvection, microinjection, cationic lipid-mediated
transfection, electroporation, transduction, scrape loading,
ballistic introduction, infection, etc. The examples of host
cells include bacterial prokaryotic cells such as E. coli,
Streptomyces, Bacillus subtilis, Streptococcus,
Staphylococcus, etc., eukaryotic cells such as yeast,
aspergillus, etc., insect cells such as Drosophila S2,
Spodoptera Sf9, etc., animal cells such as L cell, CHO cell,
COS cell, HeLa cell, C127 cell, BALB/c3T3 cell (including mutant
strains deficient in dihydrofolate reductase, tymidine kinase,
etc.), BHK21 cell, HEK293 cell, Bowes malignant melanoma cell,
etc. and plant cells or the like.
17
CA 02437866 2003-08-08
There is no limitation to an expression system as long
as the expression system is capable of expressing the present
proteins/peptides described above in a host cell and the
examples include chromosome-, episome- and virus-derived
expression systems, for instance, vectors derived from
bacterial plasmid, yeast plasmid, papovavirus such as SV40,
vaccinia virus, adenovirus, fowlpox virus, pseudorabies virus
and retrovirus, and vectors derived from bacteriophage,
transposon and from the combination of these two, e.g. vectors
derived from genetic factors of plasmid and bacteriophage such
as cosmid and phagemid. Such expression system is not only for
raising the expression and it may contain a regulatory sequence
to regulate the expression.
Host cells comprising the above-mentioned expression
systems and the present proteins /peptides obtained by culturing
the cells can be used in a screening method of the present
invention as described below. Further, the known methods can
be adopted to collect and purify the present proteins/peptides
from the cell culture, where the methods include ammonium
sulfate- or ethanol-precipitation, acid extraction, anion- or
cation-exchange chromatography, phosphocellulose
chromatography, hydrophobic interaction chromatography,
affinity chromatography, hydroxyapatite chromatography and
lectin chromatography, and the high performance liquid
chromatography is preferably used. As a column especially used
for affinity chromatography, for instance, columns to which
antibodies to the present proteins/peptides are bound are used,
and when common peptide tags are added to the present
proteins/peptides mentioned above, columns to which substances
having affinity with the peptide tags are bound are used, in
18
i i
CA 02437866 2003-08-08
order to obtain the present proteins/peptides. The
purification methods for the present proteins/peptides
mentioned above may also be employed for peptide synthesis.
In the present invention, a non-human animal whose gene
function to encode. the present proteins /peptides mentioned
above is deficient on its chromosome means a non-human animal
part or whole of whose gene on its chromosome encoding the
present proteins /peptides is inactivated by gene mutations such
as destruction, deletion, substitutions, etc. and thus whose
function to express the present proteins/peptides is lost.
Further, a non-human animal which over-expresses the present
proteins/peptides is specifically represented by a non-human
animal which produces larger amount of the present
proteins/peptides than a wild-type non-human animal does.
Specific examples of non-human animals in the present invention
include non-human animals such as. rodents including mice, rats,
etc., osteichthyes such as zebra fish, medaka fish, etc.,
arthropods such as Drosophila, silkworm, etc., the non-human
animals should not be limited only to these examples.
Homozygous non-human animals that are born according to
Mendel's Law include the deficient type or the over-expressing
type for the present proteins/peptides as well as their wild
type littermates. By using the deficient type animals or the
over-expressing type animals of these homozygous non-human
animals together with their wild-type littermates at the same
time, accurate comparative experiments can be actualized out
on the individual level. Therefore in performing screening of
the present invention described below, it is, preferable to use
wild type non-human animals, i.e. animals of the same species
as, or even better the littermates of, non-human animals whose
19
CA 02437866 2003-08-08
gene function to encode the present proteins/peptides is
deficient or over-expressed on their chromosomes, in parallel
with the deficient or over-expressed type animals. The method
of generating a non-human animal whose gene function to encode
the present proteins /peptides is deficient or over-expressed
on its chromosome is now explained in the following with
reference to a BMAL2 knockout mouse and a BMAL2 transgenic
mouse.
A mouse, for instance, whose gene function to encode BMAL2
protein is deficient on its chromosome, i.e. a BMAL2 knockout
mouse is generated by the following steps. A gene encoding
mouse BMAL2 is screened by using a gene fragment obtained by
a method such as PCR from a mouse gene library. A gene thus
screened which encodes mouse BMAL2 is subcloned with a viral
vector or the like and is identified by means of DNA sequencing.
Then whole or part of a gene encoding BMAL2 among this clone
is substituted with a pMCl neo gene cassette or the like and
then a gene such as a diphtheria toxin A fragment (DT-A) gene,
a herpes simplex virus tymidine kinase (HSV-tk) gene, etc. is
introduced onto either or both of 5'- or 3'-end, and thus a
targeting vector is constructed.
The targeting vectors thus constructed are linearlized
and introduced into ES cells by electroporation or the like to
cause homologous recombination. Among the homologous
recombinants, ES cells in which homologous recombination have
occurred are selected by the use of antibiotics such as G418,
ganciclovir (GANC) , etc. It is preferable to confirm whether
the ES cells selected are the recombinants of the interest by
Southern blotting or the like. A clone of the ES cells thus
confirmed is microinjected into a mouse blastocyst and which
I I
CA 02437866 2003-08-08
blastocyst is placed back to the recipient mouse to generate
a chimeric mouse. A heterozygous mouse can be obtained by
intercrossing the chimeric mouse and a wild type mouse. By
further intercrossing the heterozygous mice, a BMAL2 knockout
mouse of the present invention can be generated. Whether the
ability of expressing BMAL2 is lost in the BMAL2 knockout mouse
is examined by Northern blotting upon isolating RNA from the
mouse obtained by the above-described method, and by Western
blotting or the like in which the BMAL2 expression in the mouse
can be directly examined.
A BMAL2 transgenic mouse is created by the following steps.
A promoter such as chicken 1i-actin, mouse neurofilament, SV40,
etc. and poly (A) such as rabbit f3-globin, SV40, etc. or introns
are fused with cDNA encoding BMAL2 derived from chicken, mouse,
human, rat, etc., to construct a transgene. This transgene is
microinjected into the pronucleus of a mouse fertilized egg.
After the obtained egg cell is cultured, it is transplanted to
the oviduct of the recipient mouse which was bred thereafter.
Neonatal mice having the aforementioned cDNA were selected from
among all the mice born and thus the transgenic mice are created.
Neonatal mice having the cDNA can be selected by extracting
crude DNA from the mice tails or the like and then by performing
a dot hybridization method using a gene encoding the introduced
BMAL2 as a probe and by PCR method or the like using a specific
primer.
Genes or DNAs encoding the present proteins /peptides, the
present proteins/peptides, fusion proteins in which the present
proteins/peptides and marker proteins and/or peptide tags are
bound, antibodies to the present proteins/peptides, host cells
comprising expression systems capable of expressing the present
21
I I
CA 02437866 2003-08-08
proteins/peptides, CLOCK, BMAL1, etc., non-human animals whose
gene function to encode the present proteins /peptides is
deficient on their chromosome, non-human animals which
over-express the present proteins /peptides and the like make
it possible to elucidate the molecular mechanism of the
circadian oscillation system. In addition to that, these can
be used to screen a promoter or a suppressor for expression of
the present proteins/peptides, a promoter or a suppressor for
the Bmal2 activity, and a promoter or a suppressor for the
promoter transactivation of the clock oscillator genes or the
like. Some among the substances obtained by these screening
methods may possibly be used for therapy of the circadian rhythm
sleep disorders or the like including delayed sleep phase
syndrome, non-24-hour sleep-wake syndrome, advanced sleep
phase syndrome, time zone change syndrome, shift work sleep
disorder, etc.
As for a screening method for a promoter or a suppressor
for expression of the present proteins/peptides, or for a
promoter or a suppressor for the Bmal2 activity of the present
invention, methods are exemplified that use: cells expressing
the present proteins/peptides and a test substance; and a
non-human animal deficient in a gene function to encode the
present proteins /peptides on its chromosome or a non-human
animal overexpressing the present proteins /peptides and a test
substance. A screening method using cells expressing the
present proteins/peptides and a test substance, as mentioned
above, can be exemplified by a method wherein a test substance
is made to contact or introduced into, for instance, the cells
expressing the present proteins/peptides, e.g. cells obtained
from wild-type non-human animals, host cells of the present
22
CA 02437866 2003-08-08
invention, cells obtained from transgenic non-human animals of
the present invention, etc. and wherein the Bmal2 activity and
changes in the expression levels of the present
proteins /peptides are measured and assessed, but the methods
should not be limited to these examples alone.
As for a screening method wherein a non-human animal whose
gene function to encode the aforementioned present
proteins/peptides is deficient on its chromosome or a non-human
animal which over-expresses the present proteins/peptides is
used along with a test substance, the examples specifically
include: a method wherein a non-human animal whose gene function
to encode the aforementioned present proteins /peptides is
deficient on its chromosome or a non-human animal which
over-expresses the present proteins/peptides, as mentioned
above, is administered with a test substance and subsequently
the Bma12 activity and changes in the expression levels of the
present proteins/peptides in the cells obtained from the
non-human animal are measured and assessed; or a method wherein
a non-human animal whose gene function to encode the
aforementioned present proteins/peptides is deficient on its
chromosome or a non-human animal which over-expresses the
present proteins/peptides mentioned above is administered with
a test substance and subsequently the Bmal2activity and changes
in the expression levels of the present proteins /peptides in
the non-human animal are measured and assessed.
An example of a screening method of the present invention
for a promoter or a suppressor of the promoter transactivation
is a method wherein a test substance and a cell expressing either
the present proteins /peptides or the present proteins/peptides
along with CLOCK and/or BMAL1 and containing a promoter which
23
CA 02437866 2003-08-08
has E-box sequence (CACGTG) , more specifically a method in which
a test substance is made to contact or introduced into the
aforementioned cell and the promoter activity mediated by
E-box is then measured and assessed. Another example is a
method wherein a test substance is applied to a non-human animal
whose gene function to encode the present proteins/peptides is
deficient on its chromosome or to a non-human animal which
over-expresses the present proteins/peptides to measure and
assess the change in the promoter activity mediated by E-box.
In addition, it is preferable to have reporter genes or the like,
such as chloramphenicol acetyltransferase (CAT) gene or
luciferase gene, linked to the downstream of a promoter having
E-box sequence (CACGTG), in order to readily analyze the
promoter activity.
The present invention also relates to a diagnostic method
for diseases associated with the activity or expression of BMAL2
protein wherein the method comprises comparing the DNA sequence
encoding BMAL2 protein in a sample with the DNA sequence
encoding BMAL2 protein of the present invention. Mutants of
DNA encoding BMAL2 protein can be detected by finding
individuals with gene mutations at the DNA level, and such
detection is effective for diagnosing with diseases developed
by underexpression, overexpression or mutated expression of
BMAL2 protein. Specific examples of samples used for the
detection include cells of a subject, for example, genomic DNA
obtainable biopsy of blood, urine, saliva, tissue, etc., or RNA
or cDNA. The samples, however, should not be.limited to these
exemplifications and the amplified products of PCR or the like
may also be employed in using the samples. Deletions or
insertion mutations of a base sequence can be detected through
24
CA 02437866 2003-08-08
the changes in size of the amplified products when compared to
that of the normal gene type. Point mutation can be identified
by hybridizing the amplified DNA with a gene encoding a labeled
BMAL2 protein. As described. in the above, the circadian rhythm
sleep disorders or the like including delayed sleep phase
syndrome, non-24-hour sleep-wake syndrome, advanced sleep
phase syndrome, time zone change syndrome, shift work sleep
disorder, etc. can be diagnosed or judged by detecting mutation
in a gene which encodes BMAL2 protein.
The present invention is now further described
specifically with reference to the examples, however, the scope
of the invention should not be limited to these examples alone.
Example 1 (Cloning and sequencing)
1-1 (Cloning and sequencing of cClock cDNA)
cCiock cDNA was amplified with the chicken pineal cDNA
library (AZAPII, 5 x 105 pfu) as a template by PCR using LA-Taq
polymerase (Takara) and a pair of primers [sense primer 1:
5'-ACTAGTCGACTTATGTTTTTTACCATAAGCACC-3' (Seq. ID No. 21),
antisense primer 1: 5'-GTCGACCTGCGCTACTGTGGCTGAGCTTTG- 3' (Seq.
ID No. 22); Each of the primers has a Sall site on its 5'-
terminal] which were designed on the basis of the sequences of
cClock genes deposited in GenBank (GenBank accession nos.
AF132531 and AF144425). The above-mentioned PCR method was
performed four different times and the sequences of the five
clones obtained were determined. One clone with no PCR error
was selected (GenBank accession no. AF246959). The program for
thermal cycles was as follows : degeneration for 1 min at 94 C
only for the first time, followed by 5 repetitive cycles each
consisting of thermal degeneration for 30 sec at 94 C, annealing
for 30 sec at 55 C and extension for 3.5 min at 72 C; followed
CA 02437866 2009-04-29
by 15 repetitive cycles each consisting of thermal degeneration
for 30 sec at 94 C, annealing fur 30 sec at 65 C and extension
for 3. 5 min at 72 C; and finally extension for 6. 5 min at 72 C.
1-2 (Cloning and sequencing of cPer2 cDNA)
A 273 bp fragment of cPer2 cDNA was obtained from a chicken
pineal cDNA library by PCR using Taq-Gold polymerase (PE applied
TM
biosystems) and a pair of degenerate primers [per-F, 5'-
CAGCAGAT(C/G)A(A/G)CTG(C/T)IT(C/G)IGACAG(C/T)(A/G)TC(A/C)TC
AG-3' (Seq. ID No. 23) and per-R, 5'-
GCT(A/G)CACTG(A/G)CTG(A/G)TG(A/C)(C/G)IGAC(A/G)CCAC(A/G)CTC
-3' (Seq. ID No. 24) ] which were designed based on the nucleotide
sequences of dPer and mammalian Per genes. A longer cDNA
fragment (P2-5; 886bp) was amplified from a chicken pineal cDNA
library by the subsequent PCR using cPer2-R1 primer [5'-
TTGCTGTACCAGGCACATTACAAC-3' (Seq. ID No. 25)] synthesized from
the base sequence of the above-obtained fragment, a degenerate
primer [YK-F1; 5'-
(A/G)TICA(C/T)TCIGGITA(C/T)CA(A/G)GCICCI(A/C)GIATICC-3'
(Seq. ID No. 26) ] and LA-Taq polymerase. This fragment was used
as a hybridization probe for the screening of the chicken pineal
cDNA library (AZAPII, 5 x 105 pfu) to isolate a clone Pa (3584
bp) encoding a larger part of cPER2 (Met'-Arg' 14) . This clone
and the cDNA clone obtained by 3'-RACE were ligated together
to generate a full-length clone for cPER2 (Met'-Thr-1344; GenBank
accession no. AF246956). The result is shown in Fig. 1 in which
the DNA sequence and the amino acid sequence are shown as Seq.
ID Nos. 27 and 28 respectively. The bars above the sequence
in Fig. 1 indicate the PAS domains (PAS-A and PAS-B) and the
cytoplasmic localization domain (CLD). Fig. 2 shows the amino
acid homologies in domains between cPER2 obtained as above and
26
CA 02437866 2009-04-29
three mouse PER proteins (mPER1-3). The programming for
thermal cycles of the above was as follows: degeneration for
1 min at 94 C only for the first time; followed by 35 repetitive
cycles each consisting of thermal degeneration for 30 sec at
94 C, annealing for 60 sec at 52 C and extension for 1 min at
72 C; and finally extension for 9 min at 72 C.
1-3 (Cloning and sequencing of cBmal cDNA)
cDNA clones encoding part of cBMAL1 or cBMAL2 were
respectively obtained from the chicken pineal cDNA library by
PCR using LA-Taq polymerase with degenerate primers [BMAL-F,
5'-GTGCT(A/C)(A/C)GGATGGC(A/T)GT(G/T)CAGC-3' (Seq. ID No. 29)
and BMAL-R, 5'-GCG(C/T)CC(A/G)ATTGC(A/C/G)AC(A/G)AGGCAG-3'
(Seq. ID No. 30)] which were designed based on nucleotide
sequences of Bmall of mouse, rat and human and dCycle of
Drosophila. Each amplified fragment and a cDNA clone of the
each amplified fragment obtained by 5' -RACE were used as probes
for screening the chicken pineal cDNA library (AZAPII, 3.5 X
105 pfu) and cDNA clones containing the coding regions for
cBMAL1b' (GenBank accession no. AF246957) and cBMAL2 (GenBank
accession no. AF246958) were respectively isolated and
sequenced (Fig. 3). The bars above the sequences in Fig. 3
indicate the basic helix-loop-helix region (bHLH) and PAS
domains (PAS-A and PAS-B). PCR for the above was performed
TM
using a thermal cycler (Perkin-Elmer) as follows: thermal
degeneration for 1 min at 94 C only for the first time; followed
by 35 repetitive cycles each consisting of thermal degeneration
for 30 sec at 94 C, annealing for 1 min at 50 C and the extension
reaction for 1 min at 72 C; and finally extension for 9 min at
72 C.
The initiation methionine of cBMAL1b' was predicted by
27
CA 02437866 2003-08-08
comparing the cBMAL1b' sequence mentioned above and the BMAL1
sequences of other animal species. The initiation methionine
of the aforementioned cBMAL2 was predicted by the following
three criteria; (i) A nonanucleotide sequence (CCGCCATGG), the
97-105 base sequence of cBmal2 shown as Seq. ID No. 9, fully
matched the Kozak's translation initiation consensus sequence
(Nucleic Acids Res. 12. 857-872, 1984). (ii) The above-
mentioned Bma12 cDNA clone (3.4 kb) and mRNA (3.0, 3.8 kb) were
similar in size to each other. (iii) A promoter region
predicted from its genomic analysis contained the upstream
inframe stop codons.
Next, the amino acid homologies in domains among mBMALlb'
and three novel BMAL proteins (cBMALlb', cBMAL2 and hBMAL2a)
were analyzed and a phylogenetic tree of ARNT and BMAL proteins
was constructed according to Neighbor-joining method using
PHYLIP, v.3.572 as described in the literature (Felsenstein,
J., PHYLIP, Version 3.572, University of Washington, Seattle,
1996). These results are respectively shown in Figs. 4 and 5.
In Fig. 5, since amino acids in cBMAL2 in the amino-terminal
region (Mete-Arg104) and carboxy- terminal region (Gly459-Leu622)
differ in number among animal species, a part corresponding to
this region was omitted from each protein before calculating
the amino acid homologies among the proteins, and then the
phylogenetic tree was constructed. These results demonstrate
that cBMALlb' is 93% homologous to mBMAL1b' to show they are
close to one another (Figs. 3 and 4), while cBMAL2 (ARNT4) is
not particularly close to BMALl (70%; Fig. 5) nor to ARNT1 (41%;
Fig. 5) nor to ARNT2 (40%; Fig. 5) and hence that cBMAL2 is a
novel protein having bHLH-PAS (Fig. 4).
1-4 (Cloning and sequencing of hBmal2 cDNA)
28
CA 02437866 2003-08-08
A partial sequence information of hBmal2 was obtained
from two human EST clones (GenBank accession nos. AA577389 and
A1218390) by in silico screening using cBmal2 as a probe (data
as of October 1999). Several cDNA clones containing the
5'-untranslated region of hBmal2 gene were isolated from cDNA
of 293EBNA cell (a human embryonic kidney cell line) by 5'-
RACE. Then full-length clones were amplified by PCR using hB2F1
and hB2R1 primers [hB2F1, 5'-GACCAAGTGGCTCCTGCGAT-3' (Seq. ID
No. 31) and hB2R1, 5'-GCTAGAGGGTCCACTGGATG (Seq. ID No. 32)].
To eliminate PCR errors, 17 full-length cDNA clones obtained
were sequenced, and all the DNA sequences encoding hBMAL2a-
d (GenBank accession nos. AF246960-AF246963), which were
consistent with the human genomic sequences (GenBank accession
nos. AC021737 and AC016008), were determined. The programming
for the PCR thermal cycles mentioned above was as follows:
degeneration for 1 min at 94 C only for the first time; followed
by 20 repetitive cycles each consisting of thermal degeneration
for 30 sec at 940 C, annealing for 60 sec at 60 C and extension
for 2 min at 72 C; and finally extension for 8 min at 72 C. These
results are shown in Figs. 3, 4 and 5. The arrowheads below
the sequences in Fig. 3 indicate the insertion sites of introns
in hBmal2 gene.
cDNA sequences encoding 4 variants of hBAML2 (hBMAL2a-d)
and obtained from 293EBNA cells as described above, were
compared with the genome sequences registered at GenBank
(accession nos. AC021737 and AC016008). Then the cDNA
sequences were divided into 17 exons as in the case of mBmall
(Biochem. Biophys. Res. Commun. 260, 760-767, 1999) to examine
the genomic organization of hBmal2. The results are shown in
Fig. 6. Bars with GenBank accession numbers in Fig. 6 represent
29
CA 02437866 2003-08-08
genomic and cDNA clone regions and shaded parts are the spliced
regions in the isolated mutants. These results show that the
cDNA clone of hBMAL2b is devoid of Exon 4 (corresponding to
Va196-Arg109 in hBMAL2a) and that of hBMAL2c is devoid of both
Exons 3 and 4 (corresponding to G1n75-Arg109 in hBMAL2a) and
having Exon 1 to which DNA encoding the amino acid sequence of
11 amino acid residues (GEVAGGEATAP) added in-between Gly10 and
Gly" in hBMAL2a is extended. hBMAL2d was revealed to be the
shortest mutant which is devoid of both Exon 1 (as in hBMAL2a/b)
and Exons 3/4 (as in hBMAL2c) in cDNA.
1-5 (Cloning and sequencing of mBmal2 cDNA)
To identify the mouse Bma12 ortholog (mBmal2) expressed
in the suprachiasmatic nuclei (SCN), a 629 bp fragment cDNA was
obtained by RT-PCR for total RNA extracted from the mouse
mid-brain, by using LA-Taq polymerase (Takara) and two primers
synthesized according to the hBmal2 sequence; [hBMAL2-F4: 5'-
GTGCTGGTAGTATTGGAACAGATATTG-3' (Seq. ID No. 33) and hBMAL2-
R1:5'-GCTAGAGGGTCCACTGGATG-3' (Seq. ID No. 34). Subsequently,
several cDNA clones were isolated which contain 5'- or 3'-
untranslated region of mBmal2 cDNA by the method of 5'- and 3'-
rapid amplification of cDNA ends. Based on these sequence
information, two primers [mBMAL2-F1: 5'-
GGTCGACCACCATGGAGTTTTCCAAGGAAACG- 3' (Seq. ID No. 35),
mBMAL2-R1: 5'-GCTAGAGTGCCCACTGGATGTCAC-3' (Seq. ID No. 36)]
were designed that were capable of amplifying full-length
clones covering the total coding sequence of mBMAL2a or mBMAL2b
(Fig. 7; GenBank accession nos. AY005163 and AY014836).
Another RT-PCR was performed using these primers and LA-Taq
polymerase to obtain mBMAL2a comprising 579 amino acid residues.
This amino acid sequence contained bHLH, PAS-A and PAS-B domains
fi
CA 02437866 2003-08-08
and was homologous to hBMAL2 by 74%, cBMAL2 by 63% and zBMAL2
by 48%. On the contrary, mBMAL2b consists of amino acid
residues that are about one third of those of mBMAL2a (199 amino
acid residues) and is devoid of PAS-B domain (Fig. 7). Although
this form of mutation is similar to that previously found in
hBMALlc (a BMAL1 mutant devoid of the C-terminal half in the
BMAL1 comprising a long chain; Biochem. Biophys. Res. Commun.
233, 258-264, 1997), its physiological meaning is yet unknown.
1-6 (Cloning and sequencing of rBmal2 cDNA)
Next, cDNA clone of rat Bmal2 (rBmal2) covering almost
a total coding region was isolated from the rat early fibroblast
rat-1 cells by RT-PCR using two primers [mBMAL2-Fl and
mBMAL2-R1]. Three clones isolated, rBMAL2a-c, were determined
for the amino acid sequences (Fig. 7; respectively registered
to GenBank under GenBank accession nos. AF327071, AY014837,
AY014838). The amino acid sequence at the amino-terminal in
rBmal2 (corresponding to the position of mBMAL2-F1 primer) was
obtained from the in silico screening (GenBank accession no.
AA944306). These results demonstrate that rBMAL2a comprising
the longest sequence among the clones obtained is most similar
to mBMAL2a in its structure. In Fig. 7, dots at the end of the
rBMAL2 sequence indicate the position corresponding to
mBMAL2-R1, a PCR primer. The asterisk indicates the position
of the in-frame stop codon of mBMAL2b and the number at the end
of each line (with "+" on the right shoulder) indicates the
number of amino acid residues for rBMAL2a.
Next, the phylogenetic tree for the BMAL-ARNT family was
constructed according to the amino acid homologies among
various proteins (Fig. 8). Before constructing the
phylogenetic tree, several amino acid sequences of BMAL-ARNT
31
CA 02437866 2003-08-08
proteins that were obtained from GenBank were aligned with Gene
Works (Ver.2.55 , clustal V), then some regions with gaps were
omitted. Since the length of amino acids in amino- and
carboxyl-terminal regions (corresponding to the 1-59 amino acid
sequence and the 413-579 amino acid sequence of mBMAL2a) differ
among mutants, these regions were also omitted. Then the
Neighbor-joining tree was constructed using a PHYLIP 3.572
software package (Felsenstein, J., PHYLIP, Version 3.572,
University of Washington, Seattle, 1996) (Fig. 8), and the
topology of the phylogenetic tree obtained as above was analyzed
by PROTML 2.3 program which adopts a local rearrangement method
for the maximum likelihood analysis and JTT-F model for the
amino acid substitution (Adachi, J. and Hasegawa, M., MOLPHY:
Programs for molecular phylogenetic based on maximum likelihood,
Version 2.3, Institute of Statistical Mathematics, Tokyo, 1996).
Further, in order to assess the reliability of that phylogenetic
tree, a boot strap test was carried out and the boot strap
probabilities of over 70% were respectively shown near the
diversion points in Fig. 8. The diversion points shown by
closed circles indicate the divergence of species and those
shown by open circles indicate gene duplications in Fig. 8.
When the above result is taken into account together with
the fact that there is only a single copy of dCyc gene, a
Bmall/2-like gene, in the Drosophila genome, Bmall and Bma12
genes are likely to be generated from the gene duplication
occurred in their ancestral vertebrates (Diversion point b in
Fig. 8). Besides, branches at the divergence among the members
in the BMAL2 cluster are much longer than those of BMAL1, meaning
that the phylogenetic tree topology in the BMAL2 cluster
reflects the phylogenetic development of vertebrates. It can
32
CA 02437866 2003-08-08
therefore be concluded from these facts that these Bmal2 genes
are in orthologous relationships with each other and have
developed from a highly frequent amino acid substitution. This
conclusion can also be supported by the fact that no m/r/c/z
Bmal2 orthologs other than hBmal2 can be found in the human gene
data base (the htgs database was searched on 9th December, 2000).
Diversion point a in Fig. 8 probably indicates divergence
between ancestors of vertebrates and arthropods and diversion
points c-f indicate divergence among vertebrate species.
Besides, the above-mentioned phylogenetic tree had the same
topology as phylogenetic trees obtained by Parsimony and
Neighbor-joining methods.
Comparison of substitution rates in amino acids among the
members of BMAL1/2 clusters revealed that the amino acid
substation rate of BMAL2 is higher than that of BMAL1 by about
20-fold. This shows that the selective pressure in BMAL2 after
gene duplication is lower than that in BMAL1. What is important
is that there is no any specific region in which the total amino
acid homology among BMAL2 proteins is decreased. Highly
conserved structure of BMAL1 protein which has a higher
selective pressure is thought to include some unrecognized
function which has been lost in BMAL2. BMAL1 is thought to
interact with several essential regulatory factors that have
not yet been characterized, because both BMAL proteins interact
with CLOCK which is a functional heterodimer partner with BMAL
proteins (Science 280, 1564-1569, 1998, Proc. Natl. Acad. Sci.
USA 97, 4339-4344, 2000, J. Neurosci. 20, RC83, 2000, J. Biol.
Chem. 275, 36847-36851, 2000, Proc. Natl. Acad. Sci. USA 95,
5474-5479, 1998, Biochem. Biophys. Res. Commun. 248, 789-794,
1998), with a neuron PAS domain protein 2 (NPAS2 or MOP4) (J.
33
CA 02437866 2003-08-08
Neurosci. 20, RC83, 2000, Proc. Natl. Acad. Sci. USA 95,
5474-5479, 1998), with a hypoxia-inducing factor la (HIFla) (J.
Neurosci. 20, RC83, 2000, Proc. Natl. Acad. Sci. USA 95,
5474-5479, 1998, Biochem. Biophys. Res. Commun. 248, 789-794,
1998), or with HIF2a (HLF or EPAS1) and with the like. Therefore,
analyzing the differences between the functions of BMALl and
BMAL2 is thought to contribute to uncover their unique evolution
processes.
Example 2 (Northern blot analysis)
Total RNA (7.5 Ng) of each tissue from one-week-old chicks
(pineal gland, retina, cerebrum, heart, kidney and skeletal
muscle) was analyzed by Northern blotting in a manner as
described in J. Neurochem. 70, 908-913, 1998. These tissues
were harvested at 0, 6, 12 and 18 hr in Zeitgeber time (ZT),
frozen with liquid nitrogen and mixed before extracting RNA.
Each of total RNA was separated by an agarose gel
electrophoresis and blotted on a nitrocellulose membrane. The
blotting membrane was hybridized with a cBmall probe or a cBmal2
probe and washed (10 min x 3 times) in 0.1 x SSC at 50 C, then
analyzed using a FLA2000 bioimage analyzer (FUJI PHOTO FILM).
The membrane was subsequently hybridized with a chicken histone
H4cDNA probe and analyzed. The chicken histone H4cDNA probe
used was prepared by amplification by PCR with a primer [sense
primer 2; 5'-CATGTCTGGCAGAGGCAAG-3' (Seq. ID No. 37) and
antisense primer2; 5'-TTAGCCGCCGAAGCCGTAG- 3' (Seq. ID No.38)],
which was designed from the chicken pineal cDNA based on the
sequence (accession no. M74533) deposited in GenBank, and by
the subsequent cloning. The results are shown in Fig. 9. These
results demonstrate that two cBmal2 genes (3.8 Kb and 3.0 Kb,
indicated by arrows) and cBmall gene (3.3 Kb) are expressed in
34
CA 02437866 2003-08-08
all the tissues examined at various intensities. It was
confirmed as a result of normalization to histone H4 that heart
and kidney exhibited low transcriptional levels of cBmall and
that no apparent difference was observed in the transcriptional
levels of cBmal2 among the tissues examined.
Example 3 (Expression of chicken Clock genes in the pineal
gland)
One-day-old chicks were entrained to LD cycle (12 hr with
light/ 12 hr in the dark) for 3 weeks, then placed in DD (constant
darkness) condition for 2 days, and the pineal glands were
collected every 4 hr over the last 3 days. Total RNA from each
pineal gland was analyzed by Northern blotting to detect
expression of chicken Clock genes (cBmall, cBmal2, cPer2 and
cClock) in the pineal gland. Total RNA (6 Jg) obtained from
each pineal gland mentioned above was separated by an agarose
gel electrophoresis, blotted on a nitrocellulose membrane.
Two such blotting membranes were prepared. A blot was first
hybridized with a cBmal2 probe or a cPer2 probe and the blotting
membrane was washed in 0.1 x=SSC at 50 C (10 min x 3 times),
which was then analyzed using a FLA2000 bioimage analyzer (FUJI
PHOTO FILM). Next, the blot was hybridized with the histone
H4cDNA and analyzed in the same way. The aforementioned cDNA
fragment P2-5 was used as the cPer2 probe. For another blotting
membrane, the blot was first hybridized with a cBmall probe as
in the above, then with a histone H4cDNA and finally with a cClock
probe, and was analyzed with a FLA2000 bioimage analyzer. These
results are shown in Fig. 10 (bottom lane). Signals for cBmall
(open circles) and cBmal2 (closed circles) were quantified by
MacBAS software (FUJI PHOTO FILM), normalized to those for the
histone H4 cDNA, and the mean value was set as 1 in each case
CA 02437866 2009-04-29
to analyze the time-course changes in transcriptional levels
of the chicken Clock genes. The results are shown in Fig. 10
(top lane). A cross bar above the Northern blotting results
in Fig. 10 indicates light and bright cycles. An open region
indicates a light cycle, closed regions indicate (subjective)
dark cycles and shaded regions indicate subjective light cycles.
Three cPer2 transcripts (9.7 Kb, 7.5 Kb and 4.1 Kb) and a single
cClock transcript (8.5 Kb) were confirmed by these results.
Example 4 (Expression of chicken Clock genes in the pineal cell
culture)
The time course changes in the transcription amounts of
chicken clock genes [cBmall (open circle), cBmal2 (closed
circle), cPer2 (open triangle) and cClock (open square)] in the
pineal cell culture were analyzed by a quantitative RT-PCR
method and the results were compared to those in Example 3 above
(Fig. 11). Pineal cells from one-day-old chicks were plated
on 35 mm dishes (cells from 8 pineal glands per a dish) and
cultured for 5 days under LD cycles in Medium 199 (Life
TM
Technologies) supplemented with 10% fetal bovine serum. On day
6, part of the cultured cells was shifted to culture under
constant darkness (DD, right in Fig. 11). The rest of the
cultured cells remained in the culture under the LD condition
and subjected to a further culture on day 7 under constant
darkness (LD, left in Fig. 11). Then each pineal cell was
harvested every 4 hours. The pineal cells harvested were
suspended in TRIzol reagent (Life Technologies) and stored at
-80 C until total RNA was isolated. 1 pg each of the total RNA
was reverse-transcribed by the SuperScriptll (Life
Technologies) reverse transcriptase and a portion of the
reaction product was used for PCR analysis. First, an optimal
36
CA 02437866 2009-04-29
number of PCR cycle was determined for each primer to give linear
relationships between the amounts of the template cDNA and
amplification products and PCR was carried out under such
condition. The PCR products obtained were separated by a 7.5%
polyacrylamide gel electrophoresis, stained with SYBR Green I
(Molecular Probes), and the transcriptional level of each
chicken clock gene was quantified with a FLA2000 bioimage
TM
analyzer (FUJI PHOTO FILM). Change in the transcriptional
level of GAPDH, as a control, was measured in a similar manner
as the above. Intensity of each signal was normalized to that
of GAPDH, and the mean value for each gene on day 6 was set to
1. Then all the values (mRNA levels) were obtained from three
different culture samples, which were shown by mean SEM.
The primers and number of PCR cycles mentioned above were
set up as follows. For cBmall, cB1F1600 primer; 5'-
TCCAGACATTTCTTCAGCTGG-3' (Seq. ID No. 39) and cBIREND-primer;
5'-GGATGTTGAAGCAAGGTGC-3' (Seq. ID No. 40) were used and 23
cycles were practiced. For cBmal2, cB2F1270-primer; 5'-
ACGAGTACTGCCATCAAGATG-3' (Seq. ID No. 41) and cB2REND-primer;
5'-GAGAGCCCATTGGATGTCAC-3' (Seq. ID No. 42) were used and 23
cycles were practiced. For cClock, cqCF862-primer; 5'-
TTCTTGGATCACAGGGCAC-3' (Seq. ID No. 43) and cgCR1364-primer;
5'-GGAGTGCTAGTGTCCACTGTCA-3' (Seq. ID No. 44) were used and 25
cycles were practiced. For cPer2, cP2RTF primer; 5'-
GGAAGTCCTTGCAGTGCATAC-3' (Seq. ID No. 45) and cP2RTR-primer;
5'-ACAGGAAGCGGATATGCAG-3' (Seq. ID No. 46) were used and 24
cycles were practiced. For GAPDH (GenBank accession no.
K01458), cGAF-primer; 5'-ACCACTGTCCATGCCATCAC-3' (Seq. ID No.
47) and cGAR-primer; 5'-TCCACAACACGGTTGCTGTA-3' (Seq. ID No.
48) were used and 15 cycles were practiced. Taq-Gold was used
37
CA 02437866 2003-08-08
as polymerase. The program of PCR thermal cycler for each clock
gene was as follows : degeneration for 9 min at 95 C only for
the first time; followed by repetitive cycles each consisting
of thermal degeneration for 15 sec at 94 C, annealing for 30
sec at 55 C and extension for 30 sec at 72 C; and finally the
extension reaction for 7 min at 72 C.
Fig. 11 shows the results of the above. It was confirmed
by the result that all four kinds of transcripts that were
expressed in the chick pineal cells displayed daily
fluctuations in abundance with diverged phases and amplitudes
in LD cycles and under DD condition. The fluctuation profiles
in vivo in Example 3 (Fig. 10) are very similar to those in vitro
in Example 4 (Fig. 11), where the cPer2 mRNA levels had a peak
early in the morning and a trough early at night. This result
was similar to the fluctuation profile of mPerl in the mouse
SCN (Cell 90, 1003-1011, 1997, Nature 389, 512-516, 1997). A
high level expression of cPer2 sustained at the early light
phase (Zeitgeber time (ZT) 2-6) under LD condition, as compared
with a rapid decline. in cPer2 expression at circadian time (CT)
2-6 under DD condition, indicated that the pineal
photoreception plays a role in keeping the high level expression
of cPer2 in the morning. The mRNA levels of cBmall and cBmal2
also exhibited clear oscillations and their phases were
opposite to that of cPer2 (Fig. 11). Peak time in the cBmal2
mRNA level was delayed by about 4 hr compared to that in the
in vitro cBmall mRNA level. This tendency was also observed
in the in vivo fluctuation profile. In contrast, the cClock
mRNA level showed a relatively low amplitude with a broad peak
at ZT 10-18 or CT 10-18, and the peak seems to cover the peaks
in expression levels of the two Bmal genes. A similar
38
CA 02437866 2003-08-08
oscillation of cClock mRNA is observed in the chicken retina
(Mol. Brain Res. 70, 253-263, 1999).
Example 5 (Expression of the mouse clock genes in the
suprachiasmatic nuclei)
mRNA levels of mBmal2 and known clock genes (mPer2, mClock
and mBmall) of the mouse suprachiasmatic nuclei under LD cycles
were studied as follows. 5-week-old male C57BL/6 mice were
subjected to LD cycles at 23 C t 1 C (about 200 lux of bright
cycle under a fluorescent lamp) and bred with free access to
feed and water. 3 weeks thereafter, the mice were decapitated
and the brains were rapidly isolated, frozen, and sectioned into
thin strips with 700 pm thickness. Small tissue sections
including SCN on both sides were taken out from the sections
by using a 20-gauge needle, and the mRNA expression levels in
mBmal2, mPer2, mClock, mBmall, etc. in the tissue sections were
quantified by a quantitative RT-PCR. Three independent RNA
samples prepared from six mice (n = 3) were respectively
quantified and each signal intensity thus obtained was
normalized to the signals for mGAPDH and the mean of the three
values (mean t SEM) were calculated. p values in Fig. 12 were
determined by using Student's t test.
The above-mentioned primers and number of PCR cycle were
determined to give linear relationships between the amounts of
the template cDNA and amplification products. For mBmal2,
mBMAL2-F2 primer; 5'-TGGTTGGATGCGAAAGAGG-3' (Seq. ID No. 49)
and mBMAL2-R4 primer; 5'-AGGTTTCTCTCTTGGTGAACC-3' (Seq. ID No.
50) were used and 28 cycles were practiced. For mBmall (GenBank
accession no. AB012600), rmBmall-Fi primer; 5'-
TGGTACCAACATGCAATGC-3' (Seq. ID No. 51) and rmBmall-R1 primer;
5'-AGTGTCCGAGGAAGATAGCTG-3' (Seq. ID No. 52) are used and 28
39
CA 02437866 2003-08-08
cycles were practiced. For mPer2 (GenBank accession no.
AB016532), rmPer2-F1 primer; 5'-GCTCACTGCCAGAACTATCTCC-3'
(Seq. ID No. 53) and rmPer2-R1 primer;. 5'-
CCTCTAGCTGAAGCAGGTTAAG-3' (Seq. ID No. 54) are used and 30
cycles were practiced. For mClock (GenBank accession no.
AB019258), rmClock-F1 primer; 5'-CAAGGTCAGCAACTTGTGACC-3'
(Seq. ID No. 55) and rmClock-R1 primer; 5'-
AGGATGAGCTGTGTCGAAGG-3'(Seq. ID No. 56) were used and 28 cycles
were practiced. For mGAPDH (GenBank accession no. X02231),
rmGAPDH-Fl primer; 5'-CATCACCATCTTCCAGGAGC-3' (Seq. ID No. 57)
and rmGAPDH-R1 primer; 5'-ATTGAGAGCAATGCCAGCC-3' (Seq. ID No.
58) were used and 21 cycles were practiced. Programming for
the PCR thermal cycler for each clock gene was carried out under
the condition described in Example 4.
The results of the above are shown in Fig. 12. In these
results, the mPer2 mRNA level displayed daily fluctuations in
abundance in the SCN region (Fig. 12A) as are reported in the
literatures (Genes Cell 3, 167-176, 1998, Science 288,
1013-1019, 2000). Besides, the mBmall mRNA level showed a faint
oscillation in almost antipahse to mPer2 which is in LD cycles
(Fig. 12C). On the contrary, mRNA level of mBmal2 was almost
constant all day long which was similar to the case of mClock
(Fig. 12B, D), suggesting the difference in transcriptional
regulation between mBmall and mBmal2 genes.
Example 6 (Changes in, the photo-dependency of mRNA levels in
cPer2, cBmall and cBmal2 in the chick pineal glands)
Since the expression level of cBmall/2 in the early
morning was low (Fig. 12), a possible light-dependent down-
regulation of cBmall/2 transcriptions was tested. Chicks were
exposed to light for a time period when both cBmall/2 expression
CA 02437866 2003-08-08
levels were high in the dark (CT14-CT15), as is seen from the
results of Example 4, and changes in mRNA levels were evaluated
at CT15.5 and CT17. One-day-old chicks were entrained to LD
cycle for a week and then placed in DD condition. The chick
pineal glands that were exposed to a 1-hr light-pulse (350 lux)
(CT14-CT15) on the first day of DD condition (Fig. 13A, below)
and the chick pineal glands without exposure to light-pulse (Fig.
13A, top) were respectively isolated at CT15.5 or CT17 and the
total RNA (8 Ng) obtained from each of the pineal glands were
respectively separated by an agarose gel electrophoresis and
blotted onto a nitrocellulose membrane.
Each blotting membrane as aforementioned was cut into two
pieces and one (containing RNA longer than 2.4 Kb) was
hybridized with a cBmall, cBmal2 or cPer2 probe and another with
a histone H4 probe. Then the signals for cBmall (Fig. 13B),
cBmal2 (Fig. 13C), cPer2 (Fig. 13D) and histone H4 were
quantified by MacBAS software (FUJI PHOTO FILM) and the
intensity of all the signals' were normalized to those for
histone H4. The mean value of each gene at CT14 was set to 1
and the mRNA levels were determined. The values were determined
from triplicate experiments practiced in a similar way as in
the above and shown as mean t SEM. Fig. 13 shows the results.
In Fig. 13, "an asterisk" and "double asterisks" mean p < 0.05
and p < 0.02, respectively. p values were determined using
Student's t test. These results demonstrate that mRNA levels
of cBmall and cBmal2 observed in the pineal glands of chicks
exposed to light at CT15.5 were substantially lower than those
of the control animals without exposure to light. On the
contrary, the light-induced cPer2 expression was confirmed at
CT17, two hours after the exposure to light, as was observed
41
CA 02437866 2009-04-29
for mPerl and mPer2 in the SCN of the mice exposed to light (Cell
91, 1055-1064, 1997, Neuron 19, 1261-1269, 1997, Genes Cells
3, 167-176, 1998).
Example 7 (Functional property of cBMAL2; Pull-down assay)
A close kinship between BMAL1 and BMAL2 among ARNT- (aryl
hydrocarbon receptor nuclear translocator) related proteins
(Fig. 5) seems to indicate their functional similarity.
Therefore, relationships among cBMAL1, cBMAL2 and cCLOCK were
tested by a glutathione-S-transferase (GST) pull-down assay
using three kinds of bacterially expressed GST-fusion proteins
[GST-cCLOCKL (a fusion of GST and Met'-Ser466cCLOCK truncated
at the carboxy-terminal region), GST-cBMALl and GST-cBMAL2],
together with [35S] -labeled cBMALli (Met'- Ser449) or [ 35S ] -
labeled cBMAL2L (Met'-Leu458) that were transcribed and
translated in vitro. Because GST-cCLOCK (a fusion protein
composed of GST and the full-length cCLOCK) was not solubilized
TM
by 2% Triton X-100, GST- cCLOCKO mentioned above was used
instead.
A DNA fragment encoding GST-cCLOCKL, GST-BMAL1, GST-
BMAL2 or GST, mentioned above, was introduced into a pGEX5X-1
expression vector and expressed in BL21 E. coli strain. Each
E.coli was subjected to lysis in buffer A [10 mM Na-phosphate
(pH 7.9), 140 mM NaCl, 1 mM MgC121 10 mM EDTA, 5 mM 2-
mercaptoethanol, 2 mM PMSF and one tablet of Complete EDTA-
free protease inhibitor (Roche Diagnostics) per 50 mL], then
each of solubilized fusion proteins or GST was purified by
glutathione-Sepharose column (Amersham Pharmacia Biotech).
On the other hand, [35S]-labeled cBMALlL (Met'-Ser449) and
[ 35S ] -labeled cBMAL2tx (Met'-Leu458) mentioned above were prepared
by the in vitro transcription and translation of an expression
42
CA 02437866 2009-04-29
plasmid containing cDNA fragment of cBMAL1L (Met'-Ser449) or
cB! AL2t (Met'-Leu458) in the presence of [ 35S ] methionine and with
the aid of TNT-T7 Quick Coupled Transcription/Translation
System (Promega). [35S]-labeled luciferase as a control was
similarly transcribed and translated in vitro.
8 (JL each of the [ 35S ] -labeled protein (cBMALli, cBMAL2L
or luciferase protein) solutions was mixed with 40 [!L of
glutathione-sepharose beads, to which GST-cCLOCKLi (0.1 [1g),
GST-cBMALl (1.1 Ng), GST-cBMAL2 (3.3 Ng) or GST (5.6 Ng) had
been bound. Then the mixtures were incubated in 140 pL of buffer
B [20 mM Hepes-NaOH (pH 7.9), 20% (w/v) glycerol, 15 mM KC1,
0.2% Triton X-100, 2.5% skim milk, one tablet of Complete
EDTA-free protease inhibitor per 50 mL] on ice for 1 hr with
gentle rotation. After the incubation the mixtures were washed
four times with buffer C [ 10 mM Tris-HC1 (pH 7. 5) , 0. 2% Triton
X-100, 150 mM NaCl, 2 mM EDTA, 1 mM PMSF, one tablet of Complete
EDTA-free protease inhibitor] and were separated by a SDS-
polyacrylamide (10%) gel electrophoresis, then the gel was
analyzed for autoradiograph by using a FLA2000 bioimage
analyzer (FUJI PHOTO FILM).
The results of the above are shown in Fig. 14. Lanes 16-18
is the results of electrophoresis for [35S] -labeled cBMALlE,
cBMAL2L or luciferase (2. 5% each of the inputs). A faint signal
observed in lane 17 (the upper band) is due to the migration
of luciferase from lane 18. These results revealed that
GST-cCLOCKO specifically bound not only with cBMAL1L but also
with cBMAL21 in vitro (Fig. 14, lanes 1, 2). Interestingly,
GST-cBMAL2 bound with both cBMAL proteins (Fig. 14, lanes 4,
5), and GST-cBMAL1 also showed similar binding profiles (Fig.
14, lanes 7, 8), indicating potential activity of cBMAL proteins
43
CA 02437866 2009-04-29
to forma homodimer as well as a cBMALl-cBMAL2 heterodimer. It
was also demonstrated that a cBMAL protein deficient in the
C-terminal bound more efficiently with a GST-fusion protein
than with a full-length cBMAL protein.
Example 8 (an electrophoretic mobility shift assay using a cPer2
E-box-containing probe)
A binding of cBMAL1-cCLOCK or cBMAL2-cCLOCK to the E-
box sequence was examined by an electrophoretic mobility shift
assay (EMSA) in which an E-box (CACGTG)-containing sequence
present in a promoter region of cPer2 gene was used as a probe.
For preparation of the probe, oligonucleotides [cP2E1-S:
5'-GTGTCACACGTGAGGCTTA-3' (Seq. ID No. 59) and cP2E1-AS:
5'-TAAGCCTCACGTGTGACAC-3' (Seq. ID No. 60)] were synthesized
that correspond to the E-box sequence and its flanking sequences
within a putative promoter/enhancer region of cPer2 gene.
These oligonucleotides synthesized were annealed together and
subcloned into a pCR2.1 vector using TOPO-TA cloning kit
TM
(Invitrogen, CA), from which a 39 bp fragment was excised with
a restriction enzyme EcoRI and used. The above-mentioned
cBMAL1, cBMAL2 and cCLOCK were prepared by being transcribed
and translated in vitro from an expression plasmid containing
the cDNA of cBmall, cBmal2 or cClock with the aid of TNT-T7 Quick
Coupled Transcription/Translation System (Promega). A
pcDNA3.1/V5/His empty vector, an expression vector, alone was
transcribed and translated similarly as in the above and used
as a control.
uL each of the protein mixtures thus obtained (BMAL1
+ BMAL2, BMAL1 + CLOCK, BMAL2 + CLOCK) was added with 32 uL of
buffer [25 mM Hepes-KOH (pH 7 . 6 ) , 100 mM KC1, 0. 1 mM EDTA, 10%
(v/v) glycerol, 7.5 mM MgCl,, 1 mM DTT and lug denatured salmon
44
CA 02437866 2003-08-08
sperm DNA] containing a 32P-labeled probe (33 fmoles, 1.3 x 105
cpm) and was incubated for 20 min at 23 C. After the incubation,
each mixture was separated by a 6% polyacrylamide gel
electrophoresis and analyzed similarly as in Example 7 using
a FLA2000 bioimage analyzer (FUJI PHOTO FILM). Fig. 15 shows
the results. In Fig. 15, lane 1 is the result of the labeled
probe alone, lanes 2-5 are the results of the reactions between
each translation product (control, BMAL1, BMAL2 or CLOCK) and
the labeled probe. In the figure, the asterisk denotes the
position of the free probe, closed arrowheads represent
specific complexes with the bHLH-PAS proteins, and open
arrowheads indicate background. It was confirmed from these
results that in the presence of cCLOCK, cBMAL2 and cBMAL1 had
respectively formed two or three complexes (closed arrowheads
in lanes 7 and 8 in Fig. 15). It is unlikely that these complexes
represent homodimers of any of the PAS proteins examined (cCLOCK,
cBMALl or cBMAL2), because no specific bands were observed when
cCLOCK, cBMALl or cBMAL2 alone was reacted with the probe (lanes
3-6 in Fig. 15). These results suggest that the cPer2 E-box
is one of the in vivo targets of cCLOCK-cBMAL1/2 heteromer.
Example 9 (Transcriptional regulation by cBMAL1, cBMAL2 and
cCLOCK in 293EBNA cells)
Abilities for the transcriptional activation and
suppression of cBMAL1, cBMAL2 and cCLOCK were tested with the
mPer2 E-box or the mPerl promoter as a role model in the
feed-back-loop and the vasopressin gene E-box as a role model
in output pathways. Human embryonic kidney 293EBNA cells
(Invitrogen) were cultured in Dulbecco's modified Eagle's
medium supplemented with 10% fetal bovine serum (Life
Technologies) which cultured cells were then plated at 3 x 105
CA 02437866 2003-08-08
cells per well on six-well plates and transfected by using a
total of 1.0 ug of various expression plasmids (an expression
vector, plasmids containing reporter genes, 0.25 ng of Renilla
luciferase reporter (pRL-CMV; Promega), and plasmids
containing cDNA of each clock gene (cBmall, cBmal2, cClock) with
the amount indicated in Fig. 16] together with Lipofectamine
plus (Life Technologies).
As for the expression vector mentioned above,
pcDNA3. 1 /V5 /His empty vector (Invitrogen) was used. As for the
reporter genes mentioned above, 25 ng of the firefly lucif erase
reporter (cPer2 E-box-luc; a derivative of pGL3-Promoter;
Promega) containing mPer2 E-box, 25 ng of cPer2 mut.E-box-luc,
50 ng of the firefly luciferase reporter containing mPerl
promoter (mPerl-luc; a derivative of pGL3-Basic; Promega) 25
ng of the firefly luciferase reporter containing the mouse
vasopressin E-box (AVP E-box-luc; a derivative of pGL3-
Promoter; Promega), 25 ng of AVP mut. E-box-luc, or 25 ng of
TRE-luc were used. Two days after the transfection, cell
extracts were subjected to dual-luciferase assays by
luminometry (Promega) according to the manufacturer's protocol.
For each extract, the firefly luciferase activity was
normalized by the Renilla lucif erase activity and the mean value
(means SEM) was determined from the values of three independent
culture extracts.
The aforementioned plasmids containing reporter genes
were prepared as follows. The E-box sequence, CACGTG and its
flanking sequences within the promoter/ enhancer region of cPer2
gene were linked in tandem (5'-
GTGTCACACGTGAGGCTTAGTGTCACACGTGAGGCTTAGTGTCACACGTGAGGCTTA-
3'), which was then inserted into a luciferase reporter
46
CA 02437866 2003-08-08
containing SV-40 (pGL3-Promoter, Promega) and thus the cPer2
E-box-luc was constructed. The cPer2 mut.E-box as a reporter
plasmid in the control experiment was constructed by mutating
the E-box sequences into GGACCT in a similar way as previously
reported (Cell 96, 57-68, 1999). mPerl-luc was constructed as
follows; a 2.2 Kb upstream fragment of mPerl was amplified by
PCR using the DNA templates from the mouse genome [sense primer
3; 5'-TCGAGCTCTTTGGTACCTGGCCAGCAACC-3' (Seq. ID No. 61) and
anti- sense primer 3;5'-TCACGACACCTGGCCGTTCGAGG-3' (Seq. ID No.
62) ] and LA-Taq polymerase, base sequences for the six clones
individually obtained by PCR were determined, and then one clone
without PCR error among the six clones was linked to a lucif erase
reporter (pGL3-Basic, Promega). AVP E-box-luc was constructed
by linking E-box sequence (CACGTG) in the promoter/enhancer
region in the mouse vasopressin gene and its flanking sequences ,
and then by inserting the resulting sequence (5'-
TCAGGCCCACGTGTCCCA-3') into the luciferase reporter
containing SV-40 promoter (pGL3-Promoter, Promega). Further,
the AVP mut. E-box-luc (a reporter with a E-box mutation) which
is a reporter plasmid for the control experiment was prepared
in a way previously described (Cell 96, 57-68, 1999). TRE-
luc was prepared as follows; the phorbol ester-responsive
element (TRE) and its flanking sequences within human
collagenase gene were linked in tandem [5'-
CGGCTGACTCATCAAGCTGACTCATCAAGCTGACTCATCAA-3' (Seq. ID No.
63) ] , which was then inserted into the BglII site in a luciferase
reporter in which a BglII-HindIII fragment of pRL-TK vector
(Promega) was ligated to a pGL3-Basic vector (pGL3-TK-promoter
vector).
These results are shown in Fig. 16. The results show that
47
CA 02437866 2003-08-08
cCLOCK binds to not only cBMALl but cBMAL2 and promotes the
transactivation which is mediated by the cPer2E-box (Fig.16A).
Similar results were obtained by using a 2.2-kb mPerl promoter
harboring three E-box sequences (CACGTG) (Fig. 16B).
Interestingly, the transactivation elicited by cBMAL2-cCLOCK
showed a clear peak when a relatively low dose (20 ng) of a cBmal2
expression plasmid and cClock plasmid (250 ng) were coexpressed,
and a higher dose than the above of cBmal2 plasmid suppressed
the transactivation in Fig. 16B (see the left of the figure)
and Fig. 16C (see 10th-16th bars from the left of the figure).
cBmall, however, seems to have no such inhibitory effect.
Endogenous transactivation neither from the TPA-responsive
element (TRE, Fig. 16B) nor from the SV40-promoter was
suppressed by application of a high dose (160 ng) of cBmal2,
which fact suggests that the suppression is due to the specific
effect on E-box or E-box-binding component(s).
Since cBmall and cBmal2 had the slightly shifted
expression profiles as can be seen in Figs. 10 and 11, a
cooperative effect of cBMAL1 and cBMAL2 on the transcriptional
regulation was tested. In the case of a vasopressin gene E-box
as a reporter (Fig. 16C), a low level expression (10 ng) of cBmal2
notably enhanced cBMALl-cCLOCK transactivation (see 17th-23rd
bars from the left in Fig. 16C). A. similar or more pronounced
cooperative effect was observed with a low dose of cBmall
.plasmid (10 ng) for cBMAL2-cCLOCK transactivation (see
24th-30th bars from the left in Fig. 16C). Besides, the
cooperative activation was considerably suppressed by the
application of larger amounts of cBmal2 (80-160 ng) or cBmall
(40-160 ng). Similar results were also observed in the cases
when a cPer2 E-box or a mPerl promoter was used, albeit. with
48
CA 02437866 2003-08-08
less degrees (Fig. 16B).
Example 10 (Effect of cPER2 on transactivation mediated by E-box
sequences)
Next, whether cPER2 negatively acted on the
transactivation elicited by the transactivator cBMAL-cCLOCK
was examined. The experiment described in this Example 10 was
performed in a similar way as in Example 9 except that plasmids
containing a cPer2 cDNA were transfected, with the amounts shown
in Figs. 17A and 17B, to the expression plasmids which were to
be transfected to the human embryonic kidney 293EBNA cells. The
results are shown in Figs. 17A and 17B. The results show that
coexpression of cPer2 plasmid (250 ng) in 293EBNA cells
inhibited the cBMAL2-cCLOCK-dependent transactivation
mediated by cPer2 E-box, and the degree of the inhibitory effect
was stronger than that on cBMALl-cCLOCK-dependent
transactivation under the same conditions (Fig. 17A). Similar
tendency was also observed in the case of cBMAL-cCLOCK-
dependent transactivation mediated by the vasopressin E-box
(Fig. 17B), and the higher degree of inhibitory effect was
observed with the increase in the cPER2 amount.
Then, intrinsic properties of the cPer2 E-box mediated
transactivation were studied in the cultured chick pineal cells.
The pineal cells prepared from one-day-old chicks were plated
at 4 x 10 cells per well on 24-well plates and cultured under
LD cycle. At ZT9 on Day 3 of the culture, the pineal cells were
transfected with 500 ng. of either the aforementioned cPer2
expression plasmid or pcDNA3.1/V5/His (control), 250 ng of
either the cPer2 E-box-luc or the cPer2 mut.E-box-luc, and 5
ng of pRL-CMV (Promega) by using Lipofectamine plus. At ZT6
on the next day of the transfection, the cell extracts were
49
i
CA 02437866 2003-08-08
subjected to a dual-luciferase assay and the results are shown
in Fig. 17C. The results demonstrated that the endogenous
transactivation mediated by cPer2 E-box was markedly decreased
as a result of mutating the E-box sequence and that the
inhibitory effect on transactivation induced by forced
expression of cPER2 was also E-box-dependent. These facts
suggest that the chicken pineal cells express a positive factor
acting on the cPer2 E-box and that this factor exhibits an effect
on the negative regulation by cPER2.
Example 11 (Ablation of melatonin rhythm by the overexpression
of cBMAL1 and cBMAL2)
cBMALl or cBMAL2 was overexpressed in the cultured chick
pineal cells and its effect on the melatonin rhythm was examined
to evaluate the roles of the two PAS proteins in maintenance
of the rhythmicity. The chick pineal cells were cultured in
24-well cloning plates (Greiner Labortechnik, Frickenhausen,
Germany) for 2 days and transfected with 500 ng of either cBMALl
or cBMAL2 expression plasmid mentioned above or pcDNA3. 1/V5/His
(control) by using a combination of Lipofectamine plus (Life
Technologies) and Genefector (VennNova LLc, FL) . 2 days after
the transfection, the cells were subjected to a 4-day culture
in the media containing 200 mg/L G418 (Life Technologies) to
select the transfected cells and the cells selected were further
cultured in the media containing 50 mg/L G418. The culture
media were collected every 4 hours to quantify the released
melatonin by the previously described method (Neurosci 20,
986-991, 2000). Fig. 18 shows the results. Four data in each
panel are the results obtained from the individual cultures
where each value was determined by setting the average of
melatonin production levels during the LD cycles to 1. The bar
CA 02437866 2003-08-08
at the bottom of Fig. 18 represents lighting conditions.
A slight phase-delaying was observed upon studying the
melatonin rhythm in the pineal gland of each cell. This change
was also observed in the untransfected pineal cells, and such
clock oscillation was also observed after culturing control
cells (Fig. 9A) and cells overexpressing proteins unrelated to
clock proteins such as a ml or m2 acetylcholine receptor, under
DD condition for several days. In contrast to these control
cells, cBMAL1- or cBMAL2-overexpressing cells displayed only
a single oscillation in melatonin production under DD condition,
which was thereafter kept at a constant level (Figs. 18B and
18C). Under the LD cycles, daily melatonin fluctuations in
cBMAL1- or cBMAL2-overexpressing cells were quite similar to
those of control cells, indicating that cellular mechanisms for
light-dependent melatonin production were stably maintained by
the overexpressed cBMAL proteins. In spite of this, the
ablation of rhythm under DD condition strongly suggests that
cBMAL1 and cBMAL2 are both indispensable factors for rhythmic
oscillation.
Industrial Applicability
The present invention makes it possible to provide novel
clock proteins having the novel BMAL2 activity crucial for the
clock oscillation mechanism including photic-input pathway and
output pathway, and the gene DNAs encoding the proteins.
Further, with the use of these proteins and the gene DNAs,
substances useful for prevention and therapy of the circadian
rhythm sleep disorders or the like including delayed sleep phase
syndrome, non-24-hour sleep-wake syndrome, advanced sleep
phase syndrome, time zone change syndrome, shift work sleep
51
CA 02437866 2003-08-08
disorder, etc. can be screened, in addition to which a molecular
mechanism of the circadian oscillation system can also be
elucidated. Still further, the proteins of the present
invention having the BMAL2 activity have functions both for
promoting and suppressing transcription and are thought to be
involved in diverse biological functions by binding with
partners other than CLOCK. The proteins are therefore expected
to be applied to specifically inhibit a group of functions in
the transcriptional regulatory regions including that of period
genes by gene-introduction of BMAL2 or the BMAL2-dominant
negative mutants in an excessive amount from the outside.
52
CA 02437866 2003-08-08
SEQUENCE LISTING
<110> JAPAN SCIENCE AND TECHNOLOGY CORPORATION
<120> Novel Clock Gene Bma12
<130> 08-898424CA
<140>
<141> 2001-08-23
<150> JP 2001/35743
<151> 2001-02-13.
<160> 63
<170> Patentln Ver. 2.1
<210> 1
<211> 1930
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (19)..(1926)
<400> 1
gaccaagtgg ctcctgcg atg gcg gcg gaa gag gag get gcg gcg gga ggt 51
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly
1 5 10
aaa gtg ttg aga gag gag aac cag tgc att get cct gtg gtt tcc agc 99
Lys Val Leu Arg Glu Glu Asn Gln Cys Ile Ala Pro Val Val Ser Ser
1/111
i
CA 02437866 2003-08-08
15 20 25
cgc gtg agt cca ggg aca aga cca aca get atg ggg tct ttc agc Ica 147
Arg Val Ser Pro Gly Thr Arg Pro Thr Ala Met Gly Ser Phe Ser Ser
30 35 40
cac atg aca gag ttt cca cga aaa cgc aaa gga agt gat Ica gac cca 195
His Met Thr Glu Phe Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Pro
45 50 55
tee cag Ica gga ate atg aca gaa aaa gtg gtg gaa aag cit tct cag 243
Ser Gin Ser Gly Ile Met Thr Glu Lys Val Val Glu Lys Leu Ser Gln
60 65 70 75
aat ccc ctt ace tat ctt ctt Ica aca agg ata gaa ata Ica gcc tcc 291
Asn Pro Leu Thr Tyr Leu Leu Ser Thr Arg Ile Glu Ile Ser Ala Ser
80 85 90
agt ggc agc aga gtg gaa gat ggt gaa cac caa gtt aaa atg aag gcc 339
Ser Gly Ser Arg Val Glu Asp Gly Glu His Gln Val Lys Met Lys Ala
95 100 105
ttc aga gaa get cat agc caa act gaa aag egg agg aga gat aaa atg 387
Phe Arg Glu Ala His Ser Gln Thr Glu Lys Arg Arg Arg Asp Lys Met
110 115 120
aat aac ctg att gaa gaa ctg tet gca atg ate cct cag tgc aac ccc 435
Asn Asn Leu Ile Glu Glu Leu Ser Ala Met Ile Pro Gln Cys Asn Pro
125 130 135
atg gcg cgt aaa ctg gac aaa ctt aca.gtt tta aga atg get gtt caa 483
Met Ala Arg Lys Leu Asp Lys Leu Thr Val Leu Arg Met Ala Val Gln
~dno -~ - ---_-_ 155
2/111
i
CA 02437866 2003-08-08
cac ttg aga tct tta aaa ggc ttg aca aat tct tat gtg gga agt aat 531
His Leu Arg Ser Leu Lys Gly Leu Thr Asn Ser Tyr Val Gly Ser Asn
160 165 170
tat aga cca tea ttt ctt cag gat aat gag etc aga cat tta ate ctt 579
Tyr Arg Pro Ser Phe Leu Gln Asp Asn Glu Leu Arg His Leu Ile Leu
175 180 185
aag act gca gaa ggc ttc tta ttt gig gtt gga tgt gaa aga gga a 627 Lys Thr Ala
Glu Gly Phe Leu Phe Val Val Gly Cys Glu Arg Gly Lys
190 195 200
att etc ttc gtt tct aag tea gte tce aaa ata ctt aat tat gat cag 675
Ile Leu Phe Val Ser Lys Ser Val Ser Lys Ile Leu Asn Tyr Asp Gin
205 .210 215
get agt ttg act gga caa age tta ttt gac ttc tta cat cca aaa gat 723
Ala Ser Leu Thr Gly Gln Ser Leu Phe Asp Phe Leu His Pro Lys Asp
220 225 230 235
gtt gcc aaa gta aag gaa caa ctt tct tct ttt gat att tea cca aga 771
Val Ala Lys Vat Lys Glu-Gln Leu Ser Ser Phe Asp Ile Ser Pro Arg
240 245 250
gaa aag eta ata gat gcc aaa act ggt ttg caa gtt cac agt aat etc 819
Glu Lys Leu Ile Asp Ala Lys Thr Gly Leu Gin Val His Ser Asn Leu
255 260 265
cac get gga agg aca cgt gtg tat tct ggc tea aga cga tct ttt ttc 867
His Ala Gly Arg Thr Arg Val Tyr Ser Gly Ser Arg Arg Ser Phe Phe
270 275 280
a atcJ-ct_ tr aaa gaa = cat gga tgc 915
Cys Arg Ile Lys Ser Cys Lys Ile Ser Val Lys Glu Glu His Gly Cys
3/111
CA 02437866 2003-08-08
285 290 295
tta ccc aac tca aag aag aaa gag cac aga aaa ttc tat act atc cat 963
Leu Pro Asn Ser Lys Lys Lys Glu His Arg Lys Phe Tyr Thr lie His
300 305 310 315
tgc act ggt tac ttg aga agc tgg cct cca aat att gtt gga atg gaa 1011
Cys Thr Gly Tyr Leu Arg Ser Trp Pro Pro Asn Ile Val Gly Met Glu
320 325 330
gaa gaa agg aac agt aag aaa gac aac agt aat ttt acc tgc ctt.gtg 1059
Glu Glu Arg Asn Ser Lys Lys Asp Asn Ser Asn Phe Thr Cys Leu Val
335 340 345
gcc att gga aga tta cag cca tat att gtt cca cag aac agt gga gag 1107
Ala Ile Gly Arg Leu Gin Pro Tyr Ile Val Pro Gin Asn Ser Gly Glu
350 355 360
att aat gtg aaa cca act gaa ttt ata acc cgg ttt gca gtg aat gga 1155
Ile Asn Vat Lys Pro Thr Glu Phe lie Thr Arg Phe Ala Val Asn Gly
365 370 375
aaa ttt gtc tat gta gat caa agg gca aca gcg att tta gga tat ctg 1203
Lys Phe Val Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr Leu
380 385 390 395
cct cag gaa ctt ttg gga act tct tgt tat gaa tat ttt cat caa gat 1251
Pro Gin Glu Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp
400 405 410
gac cac aat aat ttg act gac aag cac aaa gca gtt cta cag agt aag 1299
Asp His Asn Asn Leu Thr Asp Lys His Lys Ala Val Leu Gin Ser Lys
415 420 425
4/111
CA 02437866 2003-08-08
gag aaa ata ctt aca gat tcc tac aaa tic aga gca aaa gat ggc tct 1347
Glu Lys Ile Leu Thr Asp Ser Tyr Lys Phe Arg Ala Lys Asp Gly Ser
430 435 440
tit gta act tta aaa agc caa tgg tit agt tic aca aat cct tgg aca 1395
Phe Val Thr Leu Lys Ser Gin Trp Phe Ser Phe Thr Asn Pro Trp Thr
445 450 455
aaa gaa ctg gaa tat att gta tct gtc aac act tta gtt ttg gga cat 1443
Lys Glu Leu Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu Gly His
460 465 470 475
agt gag cct gga gaa gca tca tit tta cct tgt agc tct caa tca tca 1491
Ser Glu Pro Gly Glu Ala Ser Phe Leu Pro Cys Ser Ser Gin Ser Ser
480 485 490
gaa gaa tcc tct aga cag tcc tgt atg agt gta cct gga atg tct act 1539
Glu Glu Ser Ser Arg Gin Ser Cys Met Ser Val Pro Gly Met Ser Thr
495 500 505
gga aca gta ctt ggt get ggt agt att gga aca gat att gca aat gaa 1587
Gly Thr Val Leu Gly Ala Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu
510 515 520
att ctg gat tta cag agg tta cag tct tct tca tac ctt gat gat tcg 1635
Ile Leu Asp Leu Gln Arg Leu Gln Ser Ser Ser Tyr Leu Asp Asp Ser
525 530 535
agt cca aca ggt tta atg aaa gat act cat act gta aac.tgc agg agt 1683
Ser Pro Thr Gly Leu Met Lys Asp Thr His Thr Val Asn Cys Arg Ser
540 545 550 555
atg tca apt aag gag ttg tit cca cca agt cct tct gaa atg ggg gag 1731
Met Ser Asn Lys Glu Leu Phe Pro Pro Ser Pro Ser Glu Met Gly Glu
5/111
CA 02437866 2003-08-08
560 565 570
cta gag get ace agg caa aac cag agt act gtt get gtc cac age cat 1779
Lena Glu Ala Thr Arg Gin Asn Gin Ser Thr Val Ala Val His Ser His
575 580 585
gag cca ctc ctc agt gat ggt gca cag ttg gat ttc gat gcc cta tgt 1827
Glu Pro Leu Leu Ser Asp Gly Ala Gin Leu Asp Phe Asp Ala Leu Cys
590 595 600
gac aat gat gac aca gcc atg get gca ttt atg aat tac tta gaa gca 1875
Asp Asn Asp Asp Thr Ala Met Ala Ala Phe Met Asn Tyr Leu Glu Ala
605 610 615
gag ggg ggc ctg gga gac cct ggg gac ttc agt gac ate cag tgg ace 1923
Glu Gly Gly Leu Gly Asp Pro Gly Asp Phe Ser Asp lie Gin Trp Thr
620 625 630 635
ctc tags 1930
Leu
<210> 2
<211> 636
<212> PRT
<213> Homo sapiens
<400> 2
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly Lys Val Leu Arg Glu
1 5 10 15
Glu Asn Gin Cys Ile Ala Pro Val Val Ser Ser Arg Val Ser Pro Gly
20 -25_-------- 30 ---
6/111
1
CA 02437866 2003-08-08
Thr Arg Pro Thr Ala Met Gly Ser Phe Ser Ser His Met Thr Glu Phe
35 40 45
Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Pro Ser Gln Ser Gly Ile
50 55 60
Met Thr Glu Lys Val Val Glu Lys Leu Ser Gln Asn Pro Leu Thr Tyr
65 70 75 80
Leu Leu Ser Thr Arg Ile Glu Ile Ser Ala Ser Ser Gly Ser Arg Val
85 90 95
Glu Asp Gly Glu His Gin Val Lys Met Lys Ala Phe Arg Glu Ala His
100 105 110
Ser Gin Thr Glu Lys Arg Arg Arg Asp Lys Met Asn Asn Leu Ile Glu
115 120 125
Glu Leu Ser Ala Met Ile Pro Gln Cys Asn Pro Met Ala Arg Lys Leu
130 135 140
Asp Lys Leu Thr Val Leu Arg Met. Ala Val Gln His Leu Arg Ser Leu
145 150 155 160
Lys Gly Leu Thr Asn Ser Tyr Val Gly Ser Asn Tyr Arg Pro Ser Phe
165 170 175
Leu Gln Asp Asn Glu Leu Arg His Leu Ile Leu Lys Thr Ala Glu Gly
180 185 190
Phe Leu Phe Val Val Gly Cys Glu Arg Gly Lys Ile Leu Phe Val Ser
195 200 205
Lys Ser Val Ser Lys Ile Leu Asn Tyr Asp Gln Ala Ser Leu Thr Gly
7/111
CA 02437866 2003-08-08
210 215 220
Gin Ser Leu Phe Asp Phe Leu His Pro Lys Asp Val Ala Lys Val Lys
225 230 235 240
Glu Gin Leu Ser Ser Phe Asp Ile Ser Pro Arg Glu Lys Leu Ile Asp
245 250 255
Ala Lys Thr Gly Leu Gin Val His Ser Asn Leu His Ala Gly Arg Thr
260 265 270
Arg Val Tyr Ser Gly Ser Arg Arg Ser Phe Phe Cys Arg Ile Lys Ser
275 280 285
Cys Lys Ile Ser Val Lys Glu Glu His Gly Cys Leu Pro Asn Ser Lys
290 295 300
Lys Lys Glu His Arg Lys Phe Tyr Thr Ile His Cys Thr Gly Tyr Leu
305 310 315 320
Arg Ser Trp Pro Pro Asn Ile Val Gly Met Glu Glu Glu Arg Asn Ser
325 330 335
Lys Lys Asp Asn Ser Asn Phe Thr Cys Leu Val Ala Ile Gly Arg Leu
340 345 350
Gin Pro Tyr Ile Val Pro Gin Asn Ser Gly Glu Ile Asn Val Lys Pro
355 360 365
Thr Glu Phe Ile Thr Arg Phe Ala Val Asn Gly Lys Phe Val Tyr Val
370 375 380
--Asp G1n repo ?1a Tbr Ala I L I eu G1y Tvr Leu Pro Gin Glu Leu Leu
385 390 395 400
8/111
1
CA 02437866 2003-08-08
Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp Asp His Asn Asn Leu
405 410 415
Thr Asp Lys His Lys Ala Val Leu Gin Ser Lys Glu Lys Ile Leu Thr
420 425 430
Asp Ser Tyr Lys Phe Arg Ala Lys Asp Gly Ser Phe Val Thr Leu Lys
435 440 445
Ser Gin Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Glu Leu Glu Tyr
450 455 460
Ile Val Ser Val Asn Thr Leu Val Leu Gly His Ser Glu Pro Gly Glu
465 470 475 480
Ala Ser Phe Leu Pro Cys Ser Ser Gin Ser Ser Glu Glu Ser Ser Arg
485 490 495
Gin Ser Cys Met Ser Val Pro Gly Met Ser Thr Gly Thr Val Leu Gly
500 505 510
Ala Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu Ile Leu Asp Leu Gin
515 520 525
Arg Leu Gin Ser Ser Ser Tyr Leu Asp Asp Ser Ser Pro Thr Gly Leu
530 535 540
Met Lys.Asp Thr His Thr Val Asn Cys Arg Ser Met Ser Asn Lys Glu
545 550 555 560
Leu Phe Pro Pro Ser Pro Ser Glu Met Gly Glu Leu Glu Ala Thr Arg
565 570 575
9111
i i
CA 02437866 2003-08-08
Gln Asn Gin Ser Thr Val Ala Val His Ser His Glu Pro Leu Leu Ser
580 585 590
Asp Gly Ala Gin Leu Asp Phe Asp Ala Leu Cys Asp Asn Asp Asp Thr
595 600 605
Ala Met Ala Ala Phe Met Asn Tyr Leu Glu Ala Glu Gly Gly Leu Gly
610 615 620
Asp Pro Gly Asp Phe Ser Asp Ile Gln Trp Thr Leu
625 630 635
<210> 3
<211> 1888
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (19).. (1884)
<400> 3
gaccaagtgg ctcctgcg atg gcg gcg gaa gag gag get gcg gcg gga ggt 51
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly
1 5 10
aaa gtg ttg aga gag gag aac cag tgc att get cct gtg gtt tcc agc 99
Lys Val Leu Arg Glu Glu Asn Gln Cys Ile Ala Pro Val Val Ser Ser
15 20 25
aca aga cca acagct atg ggg tct ttc agc tca 147
Arg Val Ser Pro Gly Thr Arg Pro Thr Ala Met Gly Ser Phe Ser Ser
10/111
CA 02437866 2003-08-08
30 35 40
cac atg aca gag ttt cca cga aaa cgc aaa gga agt gat tea gac cca 195
His Met Thr Glu Phe Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Pro
45 50 55
tcc cag tea gga atc atg aca gaa aaa gig gtg gaa aag ctt tct cag 243
Ser Gin Ser Gly Ile Met Thr Glu Lys Val Val Glu Lys Leu Ser Gin
60 65 70 75
aat ccc ctt ace tat ctt ctt tea aca agg ata gaa ata tea gcc tcc 291
Asn Pro Leu Thr Tyr Leu Leu Ser Thr Arg Ile Glu Ile Ser Ala Ser
80 85 90
agt ggc agc aga gaa get cat agc caa act gaa aag egg agg aga gat 339
Ser Gly Ser Arg Glu Ala His Ser Gin Thr Glu Lys Arg Arg Arg Asp
95 100 105
aaa atg aat aac ctg att gaa gaa ctg tct gca atg ate cct cag tgc 387
Lys Met Asn Asn Leu Ile Glu Glu Leu Ser Ala Met Ile Pro Gin Cys
110 115 120
aac ccc atg gcg cgt aaa ctg gac aaa ctt aca gtt tta aga atg get 435
Asn Pro Met Ala Arg Lys Leu Asp Lys Leu Thr Val Leu Arg Met Ala
125 130 135
gtt caa cac ttg aga tct tta aaa ggc ttg aca aat tct tat gtg gga 483
Val Gin His Leu Arg Ser Leu Lys Gly Leu Thr Asn Ser Tyr Val Gly
140 145 150 155
agt aat tat aga cca tea ttt ctt cag gat aat gag ctc aga cat tta 531
Ser Asn Tyr Arg Pro Ser Phe Leu Gin Asp Asn Glu Leu Arg His Leu
160 .165 170
11/111
CA 02437866 2003-08-08
ate ctt aag act gca gaa ggc ttc tta ttt gtg gtt gga tgt gaa aga 579
Ile Leu Lys Thr Ala Glu Gly Phe Leu Phe Val Val Gly Cys Glu Arg
175 180 185
gga aaa att ctc ttc gtt tct aag tea gtc tcc aaa ata ctt aat tat 627
Gly Lys Ile Leu Phe Val Ser Lys Ser Val Ser Lys Ile Leu Asn Tyr
190 195 200
gat cag get agt ttg act gga can age tta ttt gac ttc tta cat cca 675
Asp Gin Ala Ser Leu Thr Gly Gin Ser Leu Phe Asp Phe Leu His Pro
205 210 21.5
aaa gat gtt gcc aaa gta aag gaa caa ctt tct tct ttt gat att tea 723
Lys Asp Val Ala Lys Val Lys Glu Gin Leu Ser Ser Phe Asp Ile Ser
220 225 230 235
.cca aga gaa aag cta ata gat gcc aaa act ggt ttg caa gtt cac agt 771
Pro Arg Glu Lys Leu Ile Asp Ala Lys Thr Gly Leu Gin Val His Ser
240 245 250
aat ctc cac get gga agg aca cgt gtg tat tct ggc tea aga cga tct 819
Asn Leu His Ala Gly Arg Thr Arg Val Tyr Ser Gly Ser Arg Arg Ser
255 260 265
ttt ttc tgt egg ata aag agt tgt aaa atc tct gtc aaa.gaa gag cat 867
Phe Phe Cys Arg Ile Lys Ser Cys Lys Ile Ser Val Lys Glu Glu His
270 275 280
gga tgc tta ccc aac tea aag aag aaa gag cac aga aaa ttc tat act 915
Gly Cys Leu Pro Asn Ser Lys Lys Lys Glu His Arg Lys Phe Tyr Thr
285 290 295
rrt rra aat att gtt gga 963
Ile His Cys Thr Gly Tyr Leu Arg Ser Trp Pro Pro Asn Ile Val Gly
12/111
CA 02437866 2003-08-08
300 305 310 315
atg gaa gaa gaa agg aac agt aag aaa gac aac agt aat ttt acc tgc 1011
Met Glu Glu Glu Arg Asn Ser Lys Lys Asp Asn Ser Asn Phe Thr Cys
320 325 330
cit gtg gcc att gga aga tta cag cca tat att gtt cca cag aac agt 1059
Leu Val Ala Ile Gly Arg Leu Gin Pro Tyr Ile Val Pro Gin Asn Ser
335 340 345
gga gag att aat gtg aaa cca act gaa ttt ata acc cgg ttt gca gtg 1107
Gly Glu Ile Asn Val Lys Pro Thr Glu Phe Ile Thr Arg Phe Ala Val
350 355 360
aat gga aaa ttt gtc tat gta gat caa agg gca aca gcg att tta gga 1155
Asn Gly Lys Phe Val Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly
365 370 375
tat ctg cct cag gaa ctt ttg gga act tct tgt tat gaa tat ttt cat 1203
Tyr Leu Pro Gin Glu Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His
380 385 390 395
caa gat gac cac aat aat ttg act gac aag cac aaa gca gtt cta cag 1251
Gin Asp Asp His Asn Asn Leu Thr Asp Lys His Lys Ala Val Leu Gin
400 405 410
agt aag gag aaa ata ctt aca gat tcc tac aaa ttc aga gca aaa gat 1299
Ser Lys Glu Lys Ile Leu Thr Asp Ser Tyr Lys Phe Arg Ala Lys Asp
415 420 425
ggc tct ttt gta act tta aaa agc caa tgg ttt agt ttc aca aat cct 1347
Gly Ser Phe Val Thr Leu Lys Ser Gin Trp Phe Ser Phe Thr Asn Pro
430 435 440
13/111
CA 02437866 2003-08-08
tgg aca aaa gaa ctg gaa tat att gta tct gtc aac act tta gtt ttg 1395
Trp Thr Lys Glu Leu Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu
445 450 455
gga cat agt gag cct gga gaa gca tca ttt tta cct tgt agc tct caa 1443
Gly His Ser Glu Pro Gly Glu Ala Ser Phe Leu Pro Cys Ser Ser Gin
460 465 470 475
tca tca gaa gaa tcc tct aga cag tcc tgt atg agt gta cct gga atg 1491
Ser Ser Glu Glu Ser Ser Arg Gin Ser Cys Met Ser Val Pro Gly Met
480 485 490
tct act gga aca gta ctt ggt get ggt agt att gga aca gat'att gca 1539
Ser Thr Giy Thr Val Leu Gly Ala Gly Ser Ile Gly Thr Asp Ile Ala
495 500 505
aat gaa att ctg gat tta cag agg tta cag tct tct tca tac ctt gat 1587
Asn Glu Ile Leu Asp Leu Gin Arg Leu Gin Ser Ser Ser Tyr Leu Asp
510 515 520
gat tcg agt cca aca ggt tta atg aaa gat act cat act gta aac tgc 1635
Asp Ser Ser Pro Thr Gly Leu Met Lys Asp Thr His Thr Val Asn Cys
525 530 535
agg agt atg tca aat aag gag ttg ttt cca cca agt cct tct gaa atg 1683
Arg Ser Met Ser Asn Lys Gin Leu Phe Pro Pro Ser Pro Ser Glu Met
540 545 550 555
ggg gag eta gag get ace agg caa aac cag agt act gtt get gtc cac 1731
Gly Glu Leu Glu Ala Thr Arg Gin Asn Gin Ser Thr Val Ala Val His
560 565 570
agc cat gag cca ctc ctc agt gat ggt gca cag ttg gat ttc gat gcc 1779
Ser His Glu Pro Leu Leu Ser Asp Gly Ala Gin Leu Asp Phe Asp Ala
14/111
CA 02437866 2003-08-08
575 580 585
eta tgt gac Oat gat gac aca gcc atg get gca ttt atg aat tac tta 1827
Leu Cys Asp Asn Asp Asp Thr Ala Met Ala Ala Phe Met Asn Tyr Leu
590 595 600
gaa gca gag ggg ggc ctg gga gac cct ggg gac ttc agt gac ate cag 1875
Glu Ala Glu Gly Gly Leu Gly Asp Pro Gly Asp Phe Ser Asp Ile Gin
605 610 615
tgg ace ctc tagc 1888
Trp Thr Leu
620
<210> 4
<211> 622
<212> PRT
<213> Homo sapiens
<400> 4
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly Lys Val Leu Arg Glu
1 5 10 15
Glu Asn Gin Cys Ile Ala Pro Val Val Ser Ser Arg Val Ser Pro Gly
20 25 30
Thr Arg Pro Thr Ala Met Gly Ser Phe Ser Ser His Met Thr Glu Phe
35 40 45
Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Pro Ser Gin Ser Gly Ile
50 55 60
Met Thr Glu Lys Val Val Glu Lys Leu Ser Gin Asn Pro Leu Thr Tyr
15/111
CA 02437866 2003-08-08
65 70 75 80
Leu Leu Ser Thr Arg lie Glu lie Ser Ala Ser Ser Gly Ser Arg Glu
85 90 95
Ala His Ser Gin Thr Glu Lys Arg Arg Arg Asp Lys Met Asn Asn Leu
100 105 110
Ile Glu Glu Leu Ser Ala Met Ile Pro Gin Cys Asn Pro Met Ala Arg
115 120 125
Lys Leu Asp Lys Leu Thr Val Leu Arg Met Ala Val Gin His Leu Arg
130 135 140
Ser Leu Lys Gly Leu Thr Asn Ser Tyr Val Gly Ser Asn Tyr Arg Pro
145 150 155 160
Ser Phe Leu Gin Asp Asn Glu Leu Arg His Leu Ile Leu Lys Thr Ala
165 170 175
Glu Gly Phe Leu Phe Val Val Gly Cys Glu Arg Gly Lys Ile Leu Phe
180 185 190
Val Ser Lys Ser Val Ser Lys Ile Leu Asn Tyr Asp Gin Ala Ser Leu
195 200 205
Thr Gly Gin Ser Leu Phe Asp Phe Leu His Pro Lys Asp Val Ala Lys
210 215 220
Val Lys Glu Gin Leu Ser Ser Phe Asp Ile Ser Pro Arg Glu Lys Leu
225 230 235 240
Ile Asp Ala Lys Thr Gly Leu Gin Val His Ser Asn Leu His Ala Gly
245 250 255
16/111
CA 02437866 2003-08-08
Arg Thr Arg Val Tyr Ser Gly Ser Arg Arg Ser Phe Phe Cys Arg lie
260 265 270
Lys Ser Cys Lys Ile Ser Val Lys Glu Glu His Gly Cys Leu Pro Asn
275 280 285
Ser Lys Lys Lys Glu His Arg Lys Phe Tyr Thr Ile His Cys Thr Gly
290 295 300
Tyr Leu Arg Ser Trp Pro Pro Asn Ile Val Gly Met Glu Glu Glu Arg
305 310 315 320
Asn Ser Lys Lys Asp Asn Ser Asn Phe Thr Cys Leu Val Ala Ile Gly
325 330 335
Arg Leu Gin Pro Tyr Ile Val Pro Gin Asn Ser Gly Glu Ile Asn Val
340 345 350
Lys Pro Thr Glu Phe Ile Thr Arg Phe Ala Val Asn Gly Lys Phe Val
355 360 365
Tyr Val Asp Gin Arg Ala Thr Ala lie Leu Gly Tyr Leu Pro Gin Glu
370 375 380
Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp Asp His Asn
385 390 395 400
Asn Leu Thr Asp Lys His Lys Ala Val Leu Gin Ser Lys Gin Lys Ile
405 410 415
Leu Thr Asp Ser Tyr Lys Phe Arg Ala Lys Asp Gly Ser Phe Val Thr
--420 42~-_ 430
17/I11
CA 02437866 2003-08-08
Leu Lys Ser Gin Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Glu Leu
435 440 445
Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu Gly His Ser Glu Pro
450 455 460
Gly Glu Ala Ser Phe Leu Pro Cys Ser Ser Gin Ser Ser Glu Glu Ser
465 470 475 480
Ser Arg Gin Ser Cys Met Ser Val Pro Gly Met Ser Thr Gly Thr Val
485 490 495
Leu Gly Ala Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu Ile Leu Asp
500 505 510
Leu Gin Arg Leu Gin Ser Ser Ser Tyr Leu Asp Asp Ser Ser Pro Thr
515 520 525
Gly Leu Met Lys Asp Thr His Thr Val Asn Cys Arg Ser Met Ser Asn
530 535 540
Lys Glu Leu Phe Pro Pro Ser Pro Ser Glu Met Gly Glu Leu Glu Ala
545 550 555 560
Thr Arg Gin Asn Gin Ser Thr Val Ala Val His Ser His Glu Pro Leu
565 570 575
Leu Ser Asp Gly Ala Gin Leu Asp Phe Asp Ala Leu Cys Asp Asn Asp
580 585 590
Asp Thr Ala Met Ala Ala Phe Met Asn Tyr Leu Glu Ala Glu Gly Gly
595 600 605
Leu Gly Asp Pro Gly Asp Phe Ser Asp Ile Gin Trp Thr Leu
18/111
CA 02437866 2003-08-08
610 615 620
<210> 5
<211> 1819
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (19).. (1815)
<400> 5
gaccaagtgg ctcctgcg atg gcg gcg gaa gag gag get gcg gcg gga ggt 51
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly
1 5 10
gag gtt gcc ggt ggc gag gcg acg gcc cca ggt aaa gtg ttg aga gag 99
Glu Val Ala Gly Gly Glu Ala Thr Ala Pro Gly Lys Val Leu Arg Glu
15 20 25
gag aac cag tgc att get cct gtg gtt tee age cgc gtg agt cca ggg 147
Glu Asn Gln Cys Ile Ala Pro Val Val Set Ser Arg Val Ser Pro Gly
30 35 40
aca aga cca aca get atg ggg tct ttc age tea cac atg aca gag ttt 195
Thr Arg Pro Thr Ala Met Gly Ser Phe Ser Ser His Met Thr Glu Phe
45 50 55
cca cga aaa cgc aaa gga agt gat tea gac cca tcc caa gaa get cat 243
Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Pro Ser Gln Glu Ala His
en
- --5 ------70------ 75
19/111
CA 02437866 2003-08-08
agc caa act gaa aag egg agg aga gat aaa atg aat aac ctg aft gaa 291
Ser Gin Thr Glu Lys Arg Arg Arg Asp Lys Met Asn Asn Leu Ile Glu
80 85 90
gaa ctg tct gca atg ate cct cag tgc aac ccc atg gcg cgt aaa ctg 339
Glu Leu Ser Ala Met Ile Pro Gin Cys Asn Pro Met Ala Arg Lys Leu
95 100 105
gac aaa ctt aca Of tta aga atg get Of caa cac ttg aga tct tta 387
Asp Lys Leu Thr Val Leu Arg Met Ala Val Gin His Leu Arg Ser Leu
110 115 120
aaa ggc ttg aca aat tct tat gtg gga agt aat tat aga cca tca ttt 435
Lys Gly Leu Thr Asn Ser Tyr Val Gly Ser Asn Tyr Arg Pro Ser Phe
125 130 135
ctt cag gat aat gag ctc aga cat tta ate ctt aag act gca gaa ggc 483
Leu Gin Asp Asn Glu Leu Arg His Leu Ile Leu Lys Thr Ala Glu Gly
140 145 150 155
ttc tta ttt gtg gtt gga tgt gaa aga gga aaa aft ctc.ttc gtt tct 531
Phe Leu Phe Val Val Gly Cys Glu Arg Gly Lys Ile Leu Phe Val Ser
160 165 170
aag tca gtc tcc aaa ata ctt aat tat gat cag get agt ttg act gga 579
Lys Ser Val Ser Lys Ile Leu Asn Tyr Asp Gin Ala Ser Leu Thr Gly
175 180 185
caa agc tta ttt gac ttc tta cat cca aaa gat gtt gcc aaa gta aag 627
Gin Ser Leu Phe Asp Phe Leu His Pro Lys Asp Val Ala Lys Val Lys
190 195 200
gaa caa ctt tct tct ttt gat att tca cca aga gaa aag cta ata gat 675
Glu Gin Leu Ser Ser Phe Asp Ile Ser Pro Arg Glu Lys Leu Ile Asp
20/111
I
CA 02437866 2003-08-08
205 210 215
gcc aaa act ggt ttg caa gtt cac agt aat ctc cac get gga agg aca 723
Ala Lys Thr Gly Leu Gin Val His Ser Asn Leu His Ala Gly Arg Thr
220 225 230 235
cgt gtg tat tct ggc tea aga cga tct tit tic tgt cgg ata aag agt 771
Arg Val Tyr Ser Gly Ser Arg Arg Ser Phe Phe Cys Arg Ile Lys Ser
240 245 250
tgt aaa atc tct gtc aaa gaa gag cat gga tgc tta ccc aac tea aag 819
Cys Lys Ile Ser Val Lys Glu Glu His Gly Cys Leu Pro Asn Ser Lys
255 260 265
aag aaa gag cac aga aaa tic tat act ate cat tgc act ggt tac ttg 867
Lys Lys Glu His Arg Lys Phe Tyr Thr Ile His Cys Thr Gly Tyr Leu
270 275 280
aga age tgg cct cca aat att gtt gga atg gaa gaa gaa agg aac agt 915
Arg Ser Trp Pro Pro Asn Ile Val Gly Met Glu Glu Glu Arg Asn Ser
285 290 295
aag aaa gac aac agt aat tit acc tgc ctt gig gcc att gga aga tta 963
Lys Lys Asp Asn Ser Asn Phe Thr Cys Leu Val Ala Ile Gly Arg Leu
300 305 310 315
cag cca tat att gtt cca cag aac agt gga gag att aat gig aaa cca 1011
Gin Pro Tyr Ile Val Pro Gin Asn Ser Gly Glu Ile Asn Val Lys Pro
320 325 330
act gaa tit ata acc cgg tit gca gig aat gga aaa tit gtc tat gta 1059
Thr Glu Phe Ile Thr Arg Phe Ala Val Asn Gly Lys Phe Val Tyr Val
335 340 345
21/111
CA 02437866 2003-08-08
gat caa agg gca aca gcg att tta gga tat ctg cct cag gaa ctt ttg 1107
Asp Gln Arg Ala Thr Ala Ile Leu Gly Tyr Leu Pro Gin Glu Leu Leu
350 355 360
gga act tct tgt tat gaa tat ttt cat caa gat gac.cac aat aat ttg 1155
Gly Thr Ser Cys Tyr Glu Tyr Phe His Gln Asp Asp His Asn An Leu
365 370 375
act gac aag cac aaa gca gtt cta cag agt aag gag aaa ata ctt aca 1203
Thr Asp Lys His Lys Ala Val Leu Gln Ser Lys Glu Lys Ile Leu Thr
380 385 390 395
gat tcc tac aaa ttc aga gca aaa gat ggc tct ttt gta act tta aaa 1251
Asp Ser Tyr Lys Phe Arg Ala Lys Asp Gly Ser Phe Val Thr Leu Lys
400 405 410
agc caa tgg ttt agt ttc aca aat cct tgg aca aaa gaa ctg gaa tat 1299
Ser Gln Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Glu Leu Glu Tyr
415 420 425
att gta tct gtc aac act tta gtt ttg gga cat agt gag cct gga gaa 1347
Ile Val Ser Val Asn Thr Leu Val Leu Gly His Ser Glu Pro Gly Glu
430 435 440
gca tca ttt tta cct tgt agc tct caa.tca tca gaa gaa tcc tct aga 1395
Ala Ser Phe Leu Pro Cys Ser Ser Gln Ser Ser Glu Glu Ser Ser Arg
445 450 455
cag tcc tgt atg agt gta cct gga atg tct act gga aca gta ctt ggt 1443
Gln Ser Cys Met Ser Val Pro Gly Met Ser Thr Gly Thr Val=Leu Gly
460 465 470 475
art ggt agi att aga aca gat afgca aa.t-gaa att ctg gat tta cag 1491 Ala Gly
Ser Ile Gly Thr Asp Ile Ala Asn Glu Ile Leu Asp Leu Gin
22/111
I I
CA 02437866 2003-08-08
480 485 490
agg tta cag tct tct tca tac ctt gat gat tcg agt cca aca ggt tta 1539
Arg Leu Gin Ser Ser Ser Tyr Leu Asp Asp Ser Ser Pro Thr Gly Leu
495 500 505
atg aaa gat act cat act gta aac tgc agg agt atg tca aat aag gag 1587
Met Lys Asp Thr His Thr Val Asn Cys Arg Ser Met Ser Asn Lys Glu
510 515 520
ttg ttt cca cca agt cct tct gaa atg ggg gag cta gag get acc agg 1635
Leu Phe Pro Pro Ser Pro Ser Glu MetGly Glu Leu Glu Ala Thr Arg
525 530 535
caa aac cag agt act gtt get gtc c.ac age cat gag cca ctc ctc agt 1683
Gln Asn Gln Ser Thr Val Ala Val His Ser His Glu Pro Leu Leu Ser
540 545 550 555
gat ggt gca cag ttg gat ttc gat gcc cta tgt gac aat gat gac aca 1731
Asp Gly Ala Gin Leu Asp Phe Asp Ala Leu Cys Asp Asn Asp Asp Thr
560 565 570
gcc atg get gca ttt atg aat tac tta gaa gca gag ggg ggc ctg gga 1779
Ala Met Ala Ala Phe Met Asn Tyr Leu Glu Ala Glu Gly Gly Leu Gly
575 580 585
gac cct ggg gac ttc agt gac ate cag tgg acc ctc tags 1819
Asp Pro Gly Asp Phe Ser Asp Ile Gln Trp Thr Leu
590 595
<210> 6
X211> 599 - - - -
<212> PRT
23/111
CA 02437866 2003-08-08
<213> Homo sapiens
<400> 6
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly Glu Val Ala Gly Gly
1 5 10 15
Glu Ala Thr Ala Pro Gly Lys Val Leu Arg Glu Glu Asn Gin Cys Ile
20 25 30
Ala Pro Val Val Ser Ser Arg Val Ser Pro Gly Thr Arg Pro Thr Ala
35 40 45
Met Gly Ser Phe Ser Ser His Met Thr Glu Phe Pro Arg Lys Arg Lys
50 55 60
Gly Ser Asp Ser Asp Pro Ser Gin Glu Ala His Ser Gin Thr Glu Lys
65 70 75 80
Arg Arg Arg Asp Lys Met Asn Asn Leu Ile Glu Glu Leu Ser Ala Met
85 90 95
Ile Pro Gin Cys Asn Pro Met Ala Arg Lys Leu Asp Lys Leu Thr Val
100 105 110
Leu Arg Met Ala Val Gin His Leu Arg Ser Leu Lys Gly Leu Thr Asn
115 120 125
Ser Tyr Val Gly Ser Asn Tyr Arg Pro Ser Phe Leu Gin Asp Asn Glu
130 135 140
Leu Arg His Leu Ile Leu Lys Thr Ala Glu Gly Phe Leu Phe Val Val
145 150 155 160
Gly Cys Glu Arg Gly Lys Ile Leu Phe Val Ser Lys Ser Val Ser Lys
24/111
CA 02437866 2003-08-08
165 170 175
Ile Leu Asn Tyr Asp Gin Ala Ser Leu Thr Gly Gin Ser Leu Phe Asp
180 185 190
Phe Leu His Pro Lys Asp Val Ala Lys Val Lys Glu Gin Leu Ser Ser
195 200 205
Phe Asp Ile Ser Pro Arg Glu Lys Leu Ile Asp Ala Lys Thr Gly Leu
210 215 220
Gin Val His Ser Asn Leu His Ala Gly Arg Thr Arg Val Tyr Ser Gly
225 230 235 240
Ser Arg Arg Ser Phe Phe Cys Arg Ile Lys Ser Cys Lys Ile Ser Val
245 250 255
Lys Glu Glu His Gly Cys Leu Pro Asn Ser Lys Lys Lys Glu His Arg
260 265 270
Lys Phe Tyr Thr Ile His Cys Thr Gly Tyr Leu Arg Ser Trp Pro Pro
275 280 285
Asn Ile Val Gly Met Glu Glu Glu Arg Asn Ser Lys Lys Asp Asn Ser
290 295 300
Asn Phe Thr Cys Leu Val Ala Ile Gly Arg Leu Gin Pro Tyr Ile Val
305 310 315 320
Pro Gin Asn Ser Gly Glu Ile Asn Val Lys Pro Thr Glu Phe Ile Thr
325 330 335
Arg Phe Ala Val Asn Gly Lys Phe Val Tyr Val Asp Gin Arg Ala Thr
340 345 350
25/111
i I
CA 02437866 2003-08-08
Ala Ile Leu Gly Tyr Leu Pro Gin Glu Leu Leu Gly Thr Ser Cys Tyr
355 360 365
Glu Tyr Phe His Gin Asp Asp His Asn Asn Leu Thr Asp Lys His Lys
370 375 380
Ala Val Leu Gin Ser Lys Glu Lys Ile Leu Thr Asp Ser Tyr Lys Phe
385 390 395 400
Arg Ala Lys Asp Gly Ser Phe Val Thr Leu Lys Ser Gin Trp Phe Ser
405 410 415
Phe Thr Asn Pro Trp Thr Lys Glu Leu Glu Tyr Ile Val Ser Val Asn
420 425 430
Thr Leu Val Leu Gly His Ser Glu Pro Gly Glu Ala Ser Phe Leu Pro
435 440 445
Cys Ser Ser Gin Ser Ser Glu Glu Ser Ser Arg Gin Ser Cys Met Ser
450 455 460
Val Pro Gly Met Ser Thr Gly Thr Val Leu Gly Ala Gly Ser Ile Gly
465 470 475 480
Thr Asp Ile Ala Asn Glu Ile Leu Asp Leu Gin Arg Leu Gin Ser Ser
485 490 495
Ser Tyr Leu Asp Asp Ser Ser Pro Thr Gly Leu Met Lys Asp Thr His
500 505 510
Thr Val Asn Cys Arg Ser Met Ser Asn Lys Glu Leu Phe Pro Pro Ser
515---- 0-- 525
26/111
CA 02437866 2003-08-08
Pro Ser Glu Met Gly Glu Leu Glu Ala Thr Arg Gln Asn Gin Ser Thr
530 535 540
Val Ala Val His Ser His Glu Pro Leu Leu Ser Asp Gly Ala Gln Leu
545 550 555 560
Asp Phe Asp Ala Leu Cys Asp Asn Asp Asp Thr Ala Met Ala Ala Phe
565 570 575
Met Asn Tyr Leu Glu Ala Glu Gly Gly Leu Gly Asp Pro Gly Asp Phe
580 585 590
Ser Asp Ile Gln Trp Thr Leu
595
<210> 7
<211> 1786
<212> DNA
<213> Homo sapiens
<220>
<221> CDS
<222> (19).. (1782)
<400> 7
gaccaagtgg ctcctgcg atg gcg gcg gaa gag gag get gcg gcg gga ggt 51
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly
1 5 10
aaa gtg ttg aga gag gag aac cag tgc att get cct gtg gtt tcc agc 99
Lys Val Leu Arg Glu X11 Asn n_ ys 1le Ala Pro Val Val Ser Ser
15 20 25
2 7/1 11
CA 02437866 2003-08-08
cgc gtg agt cca ggg aca aga cca aca get atg ggg tct tic agc tca 147
Arg Val Ser Pro Gly Thr Arg Pro Thr Ala Met Gly Ser Phe Ser Ser
30 35 40
cac atg aca gag ttt cca cga aaa cgc aaa gga agt gat tca gac cca 195
His Met Thr Glu Phe Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Pro
45 50 55
tcc caa gaa get cat agc caa act gaa aag cgg agg aga gat aaa atg 243
Ser Gin Glu Ala His Ser Gin Thr Glu Lys Arg Arg Arg Asp Lys Met
60 65 70 75
aat aac ctg att gaa gaa ctg tct gca atg atc cct cag tgc aac ccc 291
Asn Asn Leu lie Gin Glu Leu Ser Ala Met Ile Pro Gin Cys Asn Pro
80 85 90
atg gcg cgt aaa ctg gac aaa ctt aca gtt tta aga atg get gtt caa 339
Met Ala Arg Lys Leu Asp Lys Leu Thr Val Leu Arg Met Ala Val Gin
95 100 105
cac ttg aga tct tta aaa ggc ttg aca aat tct tat gtg gga agt aat 387
His Leu Arg Ser Leu Lys Gly Leu Thr Asn Ser Tyr Val Gly Ser Asn
110 115 120
tat aga cca tca ttt ctt cag gat aat gag ctc aga cat tta atc ctt 435
Tyr Arg Pro Ser Phe Leu Gin Asp Asn Glu Leu Arg His Leu Ile Leu
125 130 135
aag act gca gaa ggc ttc tta ttt gtg gtt gga tgt gaa aga gga aaa 483
Lys Thr Ala Glu Gly Phe Leu Phe Val Val Gly Cys Glu Arg Gly Lys-
140 145 150 155
att ctc ttc gtt tct aag tca gtc tcc aaa ata ctt aat tat gat cag 531
28/111
CA 02437866 2003-08-08
Ile Leu Phe Val Ser Lys Ser Val Ser Lys Ile Leu Asn Tyr Asp Gin
160 165 170
get agt ttg act gga caa agc tta ttt gac ttc tta cat cca aaa gat 579
Ala Ser Leu Thr Gly Gin Ser Leu Phe Asp Phe Leu His Pro Lys Asp
175 180 185
gtt gcc aaa gta aag gaa caa ctt tct tct itt gat att tea cca aga 627
Val Ala Lys Val Lys Glu Gin Leu Ser Ser Phe Asp Ile Ser Pro Arg
190 195 200
gaa aag cta ata gat gcc aaa act ggt ttg caa gtt cac agt aat ctc 675
Glu Lys Leu Ile Asp Ala Lys Thr Gly Leu Gin Val His Ser Asn Leu
205 210 215
cac get gga'agg aca cgt gtg tat tct ggc tea aga cga tct ttt ttc 723
His Ala Gly Arg Thr Arg Val Tyr Ser Gly Ser Arg Arg Ser Phe Phe
220 225 230 235
tgt egg ata aag agt tgt aaa ate tct gtc aaa gaa gag cat gga tgc 771
Cys Arg Ile Lys Ser Cys Lys Ile Ser Val Lys Glu Glu His Gly Cys
240 245 250
tta ccc aac tea aag aag aaa gag cac aga aaa He tat act ate cat 819
Leu Pro Asn Ser Lys Lys Lys Glu His Arg Lys Phe Tyr Thr Ile His
255 260 265
tgc act ggt tac ttg aga agc tgg cct cca aat att gtt gga atg gaa 867
Cys Thr Gly Tyr Leu Arg Ser Trp Pro Pro Asn Ile Val Gly Met Glu
270 275 280
gaa gaa agg aac agt aag aaa gac aac agt aat ttt ace tgc ctt gtg 915
- ___ Gln Glu Ara Asn Ser Tys Lys Aap Asn Ser Asn Phe Thr Cys Leu Val
285 290 295
29/111
CA 02437866 2003-08-08
gcc att gga aga tta cag cca tat att gtt cca cag aac agt gga gag 963
Ala Ile Gly Arg Leu Gin Pro Tyr Ile Val Pro Gln Asn Ser Gly Glu
300 305 310 315
att aat gtg aaa cca act gaa ttt ata ace egg ttt gca gtg aat gga 1011
Ile Asn Val Lys Pro Thr Glu Phe Ile Thr Arg Phe Ala Val Asn Gly
320 325 330
aaa ttt gtc tat gta gat caa agg gca aca gcg att tta gga tat ctg 1059
Lys Phe Val Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr Leu
335 340 345
cct cag gaa ctt ttg gga act tct tgt tat gaa tat ttt cat caa gat 1107
Pro Gln Glu Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp
350 355 360
gac cac aat aat ttg act gac aag cac aaa gca gtt cta cag agt aag 1155
Asp His Asn Asn Leu Thr Asp Lys His Lys Ala Val Leu Gln Ser Lys
365 370 375
gag aaa ata ctt aca gat tcc tac aaa ttc aga gca aaa gat ggc tct 1203
Glu Lys Ile Leu Thr Asp Ser Tyr Lys Phe Arg Ala Lys Asp Gly Ser
380 385 390 395
ttt gta act tta aaa agc caa tgg ttt agt ttc aca aat cct tgg aca 1251
Phe Val Thr Leu Lys Ser Gin Trp Phe Ser The Thr Asn Pro Trp Thr
400 405 410
aaa gaa ctg gaa tat att gta tct gtc aac act tta gtt ttg gga cat 1299
Lys Glu Leu Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu Gly His
415 420 425
agt gag cct gga gaa gca tea ttt tta cct tgt agc tct caa tea tea 1347
30/111
CA 02437866 2003-08-08
Ser Glu Pro Gly Glu Ala Ser Phe Leu Pro Cys Ser Ser Gin Ser Ser
430 435 440
gaa gaa tcc tct aga cag tcc tgt atg agt gta cct gga atg tct act 1395
Glu Glu Ser Ser Arg Gin Ser Cys Met Ser Val Pro Gly Met Ser Thr
445 450 455
gga aca gta ctt ggt get ggt agt att gga aca gat att gca aat gaa 1443
Gly Thr Val Leu Gly Ala Gly Ser lie Gly Thr Asp Ile Ala Asn Glu
460 465 470 475
att ctg gat tta cag agg tta cag tct tct tca tac ctt gat gat tcg 1491
lie Leu Asp Leu Gin Arg Leu Gin Ser Ser Ser Tyr Leu Asp Asp Ser
480 485 490
agt cca aca ggt tta atg aaa gat act cat act gta aac tgc agg agt 1539
Ser Pro Thr Gly Leu Met Lys Asp Thr His Thr Val Asn Cys Arg Ser
495 500 505
atg tca aat aag gag ttg ttt cca cca agt cct tct gaa atg ggg gag 1587
Met Ser Asn Lys Glu Leu Phe Pro Pro Ser Pro Ser Glu Met Gly Glu
510 515 520
eta gag get acc agg caa aac cag agt act gtt get gtc cac age cat 1635
Leu Glu Ala Thr Arg Gin Asn Gin Ser Thr Val Ala Val His Ser His
525 530 535
gag cca ctc ctc agt gat ggt gca cag ttg gat ttc gat gcc eta tgt 1683
Glu Pro Leu Leu Ser Asp Gly Ala Gin Leu Asp Phe Asp Ala Leu Cys
540 545 550 555
gac aat gat gac aca gcc atg get gca ttt atg aat tac tta gaa gca 1731
Asp Asn Asp Asp Thr Ala Met Ala Ala Phe Met Asn Tyr Leu Glu Ala
560 565 570
31/111
I I
CA 02437866 2003-08-08
gag ggg ggc ctg gga gac cct ggg gac ttc agt gac ate cag tgg acc 1779
Glu Gly Gly Leu Gly Asp Pro Gly Asp Phe Ser Asp Ile Gin Trp Thr
575 580 585
ctc tags 1786
Leu
<210> 8
<211> 588
<212> PRT
<213> Homo sapiens
<400> 8
Met Ala Ala Glu Glu Glu Ala Ala Ala Gly Gly Lys Val Leu Arg Glu
1 5 10 15
Glu Asn Gin Cys Ile Ala Pro Val Val Ser Ser Arg Val Ser Pro Gly
20 25 30
Thr Arg Pro Thr Ala Met Gly Ser Phe Ser Ser His Met Thr Glu Phe
35 40 45
Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Pro Ser Gin Glu Ala His
50 55 60
Ser Gin Thr Glu Lys Arg Arg Arg Asp Lys Met Asn Asn Leu Ile Glu
65 70 75 80
Glu Leu Ser Ala Met Ile Pro Gin Cys Asn Pro Met Ala Arg Lys Leu
85 90 95
Asp Lys Leu Thr Val Leu Arg Met Ala Val Gin His Leu Arg Ser Leu
32/111
CA 02437866 2003-08-08
100 105 110
Lys Gly Leu Thr Asn Ser Tyr Val Gly Ser Asn Tyr Arg Pro Ser Phe
115 120 125
Leu Gin Asp Asn Glu Leu Arg His Leu Ile Leu Lys Thr Ala Glu Gly
130 135 140
Phe Leu Phe Val Val Gly Cys Glu Arg Gly Lys Ile Leu Phe Val Ser
145 150 155 160
Lys Ser Val Ser Lys Ile Leu Asn Tyr Asp Gin Ala Ser Leu Thr Gly
165 170 175
Gin Ser Leu Phe Asp Phe Leu His Pro Lys Asp Val Ala Lys Val Lys
180 185 190
Glu Gin Leu Ser Ser Phe Asp Ile Ser Pro Arg Glu Lys Leu Ile Asp
195 200 205
Ala Lys Thr Gly Leu Gin Val His Ser Asn Leu His Ala Gly Arg Thr
210 215 220
Arg Val Tyr Ser Gly Ser Arg Arg Ser Phe Phe Cys Arg Ile Lys Ser
225 230 235 240
Cys Lys Ile Ser Val Lys Glu Glu His Gly Cys Leu Pro Asn Ser Lys
245 250 255
Lys Lys Glu His Arg Lys Phe Tyr Thr Ile His Cys Thr Gly Tyr Leu
260 265 270
Arg Ser Trn Pro Pro Asn_LLe_YaL_Gly_ IeLGIu Glu Glu Arg Asn Ser
275 280 285
33/111
CA 02437866 2003-08-08
Lys Lys Asp Asn Ser Asn Phe Thr Cys Leu Val Ala Ile Gly Arg Leu
2~0 295 300
Gln Pro Tyr Ile Val Pro Gln Asn Ser Gly Glu Ile Asn Val Lys Pro
305 310 315 320
Thr Glu Phe Ile Thr Arg Phe Ala Val Asn Gly Lys Phe Val Tyr Val
325 330 335
Asp Gln Arg Ala Thr Ala Ile Leu Gly Tyr Leu Pro Gln Glu Leu Leu
340 345 350
Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp Asp His Asn Asn Leu
355 360 365
Thr Asp Lys His Lys Ala Val Leu Gln Ser Lys Glu Lys Ile Leu Thr
370 375 380
Asp Ser Tyr Lys Phe Arg Ala Lys Asp Gly Ser Phe Val Thr Leu Lys
385 390 395 400
Ser Gln Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Glu Leu Glu Tyr
405 410 415
Ile Val Ser Val Asn Thr Leu Val Leu Gly His Ser Glu Pro Gly Glu
420 425 430
Ala Ser Phe Leu Pro Cys Ser Ser Gln Ser Ser Glu Glu Ser Ser Arg
435 440 445
Gln Ser Cys Met Ser Val Pro Gly Met Ser Thr Gly Thr Val Leu Gly
450 45F
460
34/111
I I'll
CA 02437866 2003-08-08
Ala Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu Ile Leu Asp Leu Gin
465 470 475 480
Arg Leu Gln Ser Ser Ser Tyr Leu Asp Asp Ser Ser Pro Thr Gly Leu
485 490 495
Met Lys Asp Thr His Thr Val Asn Cys Arg Ser Met Ser Asn Lys Glu
500 505 510
Leu Phe Pro Pro Ser Pro Ser Glu Met Gly Glu Leu Glu Ala Thr Arg
515 520 525
Gin Asn Gin Ser Thr Val Ala Val His Ser His Glu Pro Leu Leu Ser
530 535 540
Asp Gly Ala Gin Leu Asp Phe Asp Ala Leu Cys Asp Asn Asp Asp Thr
545 550 555 560
Ala Met Ala Ala Phe Met Asn Tyr Leu Glu Ala Glu Gly Gly Leu Gly
565 570 575
Asp Pro Gly Asp Phe Ser Asp Ile Gin Trp Thr Leu
580 585
<210> 9
<211> 1970
<212> DNA
<213> Gallus gallus
<220>
< 1> CDC - ----
<222> (102).. (1970)
35/111
CA 02437866 2003-08-08
<400> 9
cccccgggcc ggcaggacgg gccgttcctt ctcaccttag ttcacctccc gcatgccgcc 60
ggggccgggg gcgctgtgga gcggggctcg ggccgcccgc c atg gcc gag gca gga 11&
Met Ala Glu Ala Gly
1 5
gtg ggg agc gcc gag ggg gca gag gag gag cgg cgg gcc gtt gaa gag 164
Val Gly Ser Ala Glu Gly Ala Glu Glu Glu Arg Arg Ala Val Glu Glu
15 20
aat ttt cca gta gat gga aac tcg tgc att get tct gga gtc ccc agc 212
Asn Phe Pro Val Asp Gly Asn Ser Cys Ile Ala Ser Gly Val Pro Ser
25 30 35
ctc atg aat cca ata act aag cct get acc act tct tic aac aat tct 260
Leu Met Asn Pro Ile Thr Lys Pro Ala Thr Thr Ser Phe Asn Asn Ser
40 45 50
gtg gtt gag att cca agg aag cgc aaa gga agt gat tct gat aac cag 308
Val Val Glu Ile Pro Arg Lys Arg Lys Gly Ser Asp Ser Asp Asn Gin
55 60 65
gat aca gtt gaa gtt gat.ggg gat cct cag aaa agg aat gaa gat gaa 356
Asp Thr Val Glu Val Asp Gly Asp Pro Gin Lys Arg Asn Glu Asp Glu
70 75 80 85
gaa cat ctt aag ata aaa gat tic aga gag gcc cac agt caa aca gag 404
Glu His Leu Lys Ile Lys Asp Phe Arg Glu Ala His Ser Gln Thr Glu
90 95 100
gaa ttg tct get 452
Lys Arg Arg Arg Asp Lys Met Asn Asn Leu Ile Glu Glu Leu Ser Ala
36/111
j
CA 02437866 2003-08-08
105 110 115
atg ata cct cag tgc aat cc' atg gca cga aag cta gac aag ctt aca 5.00
Met Ile Pro Gin Cys Asn Pro Met Ala Arg Lys Leu Asp Lys Leu Thr
120 125 130
gta tta cgg atg gca gtg caa cac tta aaa tot ttg aaa ggt tcc act 548
Val Leu Arg Met Ala Val Gin His Leu Lys Ser Leu Lys Gly Ser Thr
135 140 145
age tct tac acc gaa gtc cgg tat aaa cct tog ttt tta aag gat gat 596
Ser Ser Tyr Thr Glu Val Arg Tyr Lys Pro Ser Phe Leu Lys Asp Asp
150 155 160 165
gag ctc aga cag tta ate ctt agg get gcg gat gga tic cta ttt gtg 644
Glu Leu Arg Gin Leu Ile Leu Arg Ala Ala Asp Gly Phe Leu Phe Val
170 175 180
gtt gga tgt aac aga gga aaa att ctg ttt gtc tca gaa tca gtt tgc 692
Val Gly Cys Asn Arg Gly Lys Ile Leu Phe Val Ser Glu Ser Val Cys
185 190 195
aaa ata ctt aat tat gat cag acc agt ita att gga caa agt ttg ttt 740
Lys Ile Leu Asn Tyr Asp Gin Thr Ser Leu Ile Gly Gin Ser Leu Phe
200 205 210
gat tac ttg cat cca aaa gat gtt gcc aaa gtt aag gag caa ctt tca 788
Asp Tyr Leu His. Pro Lys Asp Val Ala Lys Val Lys Glu Gin Leu Ser
215 220 225
tct tca gat gtc tct ccc aga gaa aag ctt gta gat ggc aaa act ggc 836
Ser Ser Asp Val Ser Pro Arg Glu Lys Leu Val Asp Gly Lys Thr Gly
230 235 240 245
37/111
CA 02437866 2003-08-08
ttg caa gta cat aca gat ttt caa get gga cca get cga ctg aat tct 884
Leu Gin Val His Thr Asp Phe Gin Ala Gly Pro Ala Arg Leu Asn Ser
250 255 260
ggt get cga cgt tcc ttc ttc tgt cgg ata aaa tgt agt agg acc aca 932
Gly Ala Arg Arg Ser Phe Phe Cys Arg Ile Lys Cys Ser Arg Thr Thr
265 270 275
gtc aaa gaa gag aag gag tgc tta ccc aac cca aag aag aaa gat cac 980
Val Lys Glu Glu Lys Glu Cys Leu Pro Asn Pro Lys Lys Lys Asp His
280 285 290
aga aag tat tgt acc att cac tgt act gga tat atg aag aac tgg cct 1028
Arg Lys Tyr Cys Thr Ile His Cys Thr Gly Tyr Met Lys Asn Trp Pro
295 300 305
cct agc gag gtg gga gtg gaa gag gaa aac gat gta gaa aag aac agt 1076
Pro Ser Glu Val Gly Val Glu Glu Glu Asn Asp Val Glu Lys Asn Ser
310 315 320 325
agt aac ttt aac tgt ctc gtt gca att ggg agg tta cac cct tac att 1124
Ser Asn Phe Asn Cys Leu Val Ala Ile Gly Arg Leu His Pro Tyr Ile
330 335 340
gtt cca caa aag agt gga gag ata aaa gtc aaa gca aca gaa ttt gtt 1172
Val Pro Gin. Lys Ser Gly Glu Ile Lys Val Lys Ala Thr Glu Phe Val
345 350 355
aca cga ttt gcc atg gat gga aaa ttt gtt tat gta gat cag cgt gca 1220
Thr Arg Phe Ala Met Asp Gly Lys Phe Val Tyr Val Asp Gin Arg Ala
360 365 370
ar.a gr.a att tta ggg tat rtg cca caa gag cttsta gga act tct-j-gt 1268
Thr Ala Ile Leu Gly Tyr Lea Pro Gin Glu Leu Leu Gly Thr Ser Cys
38/111
CA 02437866 2003-08-08
375 380 385
tac gag tac tgc cat caa gat gat cac aat cat cta get gaa aaa cat 1316
Tyr Glu Tyr Cys His Gin Asp Asp His Asn His Leu Ala Glu Lys His
390 395 400 405
aaa gaa gig ctg cag aat aaa gaa aaa gta ttt aca aat tcc tac aaa 1364
Lys Glu Val Leu Gin Asn Lys Glu Lys Val Phe Thr Asn Ser Tyr Lys
410 415 420
ttt aga gca aaa gat gga agt ttt att act tta aag agt caa tgg ttt 1412
Phe Arg Ala Lys Asp Gly Ser Phe Ile Thr Leu Lys Ser Gin Trp Phe
425 430 435
agt ttc atg aat ccc tgg ace aag gaa ctg gag tac att gta tca aac 1460
Ser Phe Met Asn Pro Trp Thr Lys Glu Leu Glu Tyr Ile Val Ser Asn
440 445 450
aac act gta gta tta ggt cac aat gag tct get gaa gaa cag gtc tcc 1508
Asn Thr Val Val Leu Gly His Asn Glu Ser Ala Glu Gin Gin Val Ser
455 460 465
tat ggt,tcc cag cct gea gaa ggt get gta aaa cag tct tta gtg agt 1556
Tyr Gly Ser Gin Pro Ala Glu Gly Ala Val Lys Gin Ser Leu Val Ser
470 475 480 485
gta cct gga atg tcc tct gga aca gtt ctt ggt get gga agt ata gga 1604
Val Pro Gly Met Ser Ser Gly Thr Val Leu Gly Ala Gly Ser Ile Gly
490 495 500
act gaa att gca aat gaa ata tta gaa tta caa agg ttg cat tct tca 1652
Thr Glu Ile Ala Asn Glu Ile Leu Glu Leu Gin Arg Leu His Ser Ser
505 510------- 515-------- ----
39/111
CA 02437866 2003-08-08
ccg cct ggg gag tta agt cca tea cat etc ttg aga aag tea cca tct 1700
Pro Pro Gly Glu Leu Ser Pro Ser His Leu Leu Arg Lys Ser Pro Ser
520 525 530
cca get tta act gta aac tgc age aat gtg ccg aat aaa gag ttg att 1748
Pro Ala Leu Thr Val Asn Cys Ser Asn Val Pro Asn Lys Glu Leu lie
535 540 545
cag tta tgt cct tea gaa gca gaa gtt ctg gag act tea gaa caa aac 1796
Gln Leu Cys Pro Ser Glu Ala Glu Val Leu Glu Thr Ser Glu Gln Asn
550 555 560 565
caa ggt get att cca ttc ccc agt aat gag cct etc etc ggt ggt aat 1844
Gln Gly Ala lie Pro Phe Pro Ser Asn Glu Pro Leu Leu Gly Gly Asn
570 575 580
tct cag ctg gac ttt gca ata tgt gaa aat gat gac act gcc atg act 1892
Ser Gln Leu Asp Phe Ala Ile Cys Glu Asn Asp Asp Thr Ala Met Thr
585 590 595
get ctt atg aat tac ttg gag gcc gat gga gga cat ggg gat cca get 1940
Ala Leu Met Asn Tyr Leu Glu Ala Asp Gly Gly Leu Gly Asp Pro Ala
600 605 610
gaa etc agt gac ate caa tgg get etc tag 1970
Glu Leu Ser Asp Ile Gin Trp Ala Leu
615 620
<210> 10
<211> 622
~<2~1g2~> PRT
Callus gal --------------------------------- - -
40/111
CA 02437866 2003-08-08
<400> 10
Met Ala Glu Ala Gly Val Gly Ser Ala Glu Gly Ala Glu Glu Glu Arg
10 15
Arg Ala Val Glu Glu Asn Phe Pro Val Asp Gly Asn Ser Cys Ile Ala
20 25 30
Ser Gly Val Pro Ser Leu Met Asn Pro Ile Thr Lys Pro Ala Thr Thr
35 40 45
Ser Phe Asn Asn Ser Val Val Glu Ile Pro Arg Lys Arg Lys Gly Ser
50 55 60
Asp Ser Asp Asn Gin Asp Thr Val Glu Val Asp Gly Asp Pro Gin Lys
65 70 75 80
Arg Asn Glu Asp Glu Glu His Leu Lys Ile Lys Asp Phe Arg Glu Ala
85 90 95
His Ser Gin Thr Glu Lys Arg Arg Arg Asp Lys Met Asn Asn Leu Ile
100 105 110
Glu Glu Leu Ser Ala Met Ile Pro Gin Cys Asn Pro Met Ala Arg Lys
115 120 125
Leu Asp Lys Leu Thr Val Leu Arg Met Ala Val Gin His Leu Lys Ser
130 135 140
Leu Lys Gly Ser Thr Ser Ser Tyr Thr Glu Val Arg Tyr Lys Pro Ser
145 150 155 160
Phe Leu Lys Asp Asp Glu Leu Arg Gin Leu Ile Leu Arg Ala Ala Asp
165 170 175
Gly Phe Leu Phe Val Val Gly Cys Asn Arg Gly Lys Ile Leu Phe Val
180 185 190
Ser Glu Ser Val Cys Lys Ile Leu Asn Tyr Asp Gin Thr Ser Leu Ile
195 200 205
Gly Gin Ser Leu Phe Asp Tyr Leu His Pro Lys Asp Val Ala Lys Val
210 215 220
Lys Glu Gin Leu Ser Ser Ser Asp Val Ser Pro Arg Glu Lys Leu Val
225 230 235 240
Asp Gly Lys Thr Gly Leu Gin Val His Thr Asp Phe Gin Ala Gly Pro
245 250 255
Ala Arg Leu Asn Ser Gly Ala Arg Arg Ser Phe Phe Cys Arg Ile Lys
41/111
CA 02437866 2003-08-08
260 265 270
Cys Ser Arg Thr Thr Val Lys Glu Glu Lys Glu Cys Leu Pro Asn Pro
275 280 285
Lys Lys Lys Asp His Arg Lys Tyr Cys Thr Ile His Cys Thr Gly Tyr
290 295 300
Met Lys Asn Trp Pro Pro Ser Glu Val Gly Val Glu Glu Glu Asn Asp
305 310 315 320
Val Glu Lys Asn Ser Ser Asn Phe Asn Cys Leu Val Ala Ile Gly Arg
325 330 335
Leu His Pro Tyr Ile Val Pro Gin Lys Ser Gly Glu Ile Lys Val Lys
340 345 350
Ala Thr Glu Phe Val Thr Arg Phe Ala Met Asp Gly Lys Phe Val Tyr
355 360 365
Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr Leu Pro Gin Glu Leu
370 375 380
Leu Gly Thr Ser Cys Tyr Glu Tyr Cys His Gin Asp Asp His Asn His
385 390 395 400
Leu Ala Glu Lys His Lys Glu Val Leu Gin Asn Lys Glu Lys Val Phe
405 410 415
Thr Asn Ser Tyr Lys Phe Arg Ala Lys Asp Gly Ser Phe Ile Thr Leu
420 425 430
Lys Ser Gin Trp Phe Ser Phe Met Asn Pro Trp Thr Lys Glu Leu Glu
435 440 445
Tyr Ile Val Ser Asn Asn Thr Val Val Leu Gly His Asn Glu Ser Ala
450 455 460
Glu Glu Gin Val Ser Tyr Gly Ser Gin Pro Ala Glu Gly Ala Val Lys
465 470 475 480
Gin Ser Leu Val Ser Val Pro Gly Met Ser Ser Gly Thr Val Leu Gly
485 490 495
Ala Gly Ser Ile Gly Thr Glu Ile Ala Asn Glu Ile Leu Glu Leu Gin
500 505 510
Arg Leu His Ser Ser Pro Pro Gly Glu Leu Ser Pro Ser His Leu Leu
515 -520 525
Arg Lys Ser Pro Ser Pro Ala Leu Thr Val Asn Cys Ser Asn Val Pro
42/111
i i
CA 02437866 2003-08-08
530 535 540
Asn Lys Glu Leu lie Gin Leu Cys Pro Ser Glu Ala Glu Val Leu Glu
545 550 555 560
Thr Ser Glu Gin Asn Gin Gly Ala Ile Pro Phe Pro Ser Asn Glu Pro
565 570 575
Leu Leu Gly Gly Asn Ser Gin Leu Asp Phe Ala Ile Cys Glu Asn Asp
580 585 590
Asp Thr Ala Met Thr Ala Leu Met Asn Tyr Leu Glu Ala Asp Gly Gly
595 600 605
Leu Gly.Asp Pro Ala Glu Leu Ser Asp Ile Gin Trp Ala Leu
610 615 620
<210> 11
<211> 1752
<212> DNA
<213> Mus musculus
<220>
<221> CDS
<222> (12). . (1748)
<400> 11
ggtcgaccac c atg gag ttt cca agg aaa cgc aga ggc aga gat tcc cag 50
Met Glu Phe Pro Arg Lys Arg Arg Gly Arg Asp Ser Gin
1 5 10
cca etc cag tca gaa ttc atg aca gac aca aca gtg gaa agt ctt ccc 98
Pro Leu Gin Ser Glu Phe Met Thr Asp Thr Thr Val Glu Ser Leu Pro
15 20 25
cag as-t cc-c t t gc.c- tct -ctt - c - 1 tca aca aga_aia gga gta _tca gcg- _
1.46 - -
Gin Asn Pro Phe Ala Ser Leu Leu Ser Thr Arg Thr Gly Val Ser Ala
43/111
CA 02437866 2003-08-08
30 35 40 45
ccc agt ggc ate agg gaa get cac age cag atg gaa aag cgt egg aga 194
Pro Ser Gly lie Arg Glu Ala His Ser Gin Met Glu Lys Arg Arg Arg
50 55 60
gac aag atg aac cat ctg att cag aaa ctg tca tct atg ate cct cca 242
Asp Lys Met Asn His Leu Ile Gin Lys Leu Ser Ser Met Ile Pro Pro
65 70 75
cac ate ccc acg gcc cac aaa ctg gac aag etc age gtc ttg agg agg 290
His Ile Pro Thr Ala His Lys Leu Asp Lys Leu Ser Val Leu Arg Arg
80 85 90
gcg gtg cag tac ttg agg tct ctg aga ggc atg aca gag ctt tac tta 338
Ala Val Gin Tyr Leu Arg Ser Leu Arg Gly Met Thr Glu Leu Tyr Leu
95 100 105
gga gaa aac tct aaa cct tca ttt att cag gat aag gaa etc agt cac 386
Gly Glu Asn Ser Lys Pro Ser Phe Ile Gin Asp Lys Glu Leu Ser His
110 115 120 125
tta ate etc aag gca gca gaa ggc ttc ctg ttt gtg gtt gga tgc gaa 434
Leu lie Leu Lys Ala Ala Glu Gly Phe Leu Phe Val Val Gly Cys Glu
130 135 140
aga ggg aga att ttt tac gtt tct aag tct gtc tee aaa aca ctg cgt 482
Arg Gly Arg Ile Phe Tyr Val Ser Lys Ser Val Ser Lys Thr Leu Arg
145 150 155
tat gat cag get age ttg ata gga cag aat ttg ttt gac ttc tta cac 530
Tyr Asp Gin Ala Ser Leu Ile Gly Gin Asn Leu Phe Asp Phe Leu His
160 165 170
44/111
CA 02437866 2003-08-08
cca aaa gac gtc gcc aaa gta aag gaa caa ctt tct tgt gat ggt tca 578
Pro Lys Asp Val Ala Lys Val Lys Glu Gin Leu Ser Cys Asp Gly Ser
175 180 185
cca aga gag aaa cct ata gac acc aaa acc tct cag gtt tac agt cac 626
Pro Arg Glu Lys Pro Ile Asp Thr Lys Thr Ser Gin Val Tyr Ser His
190 195 200 205
ccc tac act ggg cga cca cgc atg cat tct ggc tcc aga cga tct ttc 674
Pro Tyr Thr Gly Arg Pro Arg Met His Ser Gly Ser Arg Arg Ser Phe
210 215 220
ttc ttt aga atg aag agc tgt acc gtc cct gtc aaa gaa gag cag cca 722
Phe Phe Arg Met Lys Ser Cys Thr Val Pro Val Lys Glu Glu Gin Pro
225 230 235
tgc tcg tcc tgc tca aag aag aaa gac cat aga aaa ttc cac acc gtc 770
Cys Ser Ser Cys Ser Lys Lys Lys Asp His Arg Lys Phe His Thr Val
240 245 250
cat tgc act gga tac ttg aga agc tgg cct ctg aat gtt gtt ggc atg 818
His Cys Thr Gly Tyr Leu Arg Ser Trp Pro Leu Asn Val Val Gly Met
255 260 265
gag aaa gag tcg ggt ggt ggg aag gac agc ggt cct ctt acc tgc ctt 866
Glu Lys Glu Ser Gly Gly Gly Lys Asp Ser Gly Pro Leu Thr Cys Leu
270 275 280 285
gtg get atg gga cgg ttg cat cca tac att gtc cct caa aag agt ggc 914
Val Ala Met Gly Arg Leu His Pro Tyr Ile Val Pro Gin Lys Ser Gly
290 295 300
aagatc-aacgtg_aga_ccg -&ctgag ttc_ataact_cgc ttc gcaatgaac 962 Lys Ile Asn Val
Arg Pro Ala Glu Phe Ile Thr Arg Phe Ala Met Asn
45/111
I 1 11
CA 02437866 2003-08-08
305 310 315
ggg aaa ttc gtc tat gtt gac caa agg gca acg gca att tta gga tac 1010
Gly Lys Phe Vat Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr
320 325 330
ctg cct cag gaa ctt ttg gga act tca tgt tat gaa tat ttt cat cag 1058
Leu Pro Gin Glu Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin
335 340 345
gat gac cac agt agt ttg act gac aag cac aaa gca gtt ctg cag agt 1106
Asp Asp His Ser Ser Leu Thr Asp Lys His Lys Ala Val Leu Gin Ser
350 355 360 365
aag gag aaa ata ctt aca gac tca tac aaa ttc aga gtg aag gat ggt 1154
Lys Glu Lys Ile Leu Thr Asp Ser Tyr Lys Phe Arg Val Lys Asp Gly
370 375 380
gcc ttc gtg act ctg aag agt gag tgg ttc agc ttc aca aac cct tgg 1202
Ala Phe Val Thr Leu Lys Ser Glu Trp Phe Ser Phe Thr Asn Pro Trp
385 390 395
acc aaa gag ctg gag tac att gtg tct gtc aac aca ttg gtt ttg ggg 1250
Thr Lys Glu Leu Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu Gly
400 405 410
cgc agt gag acc agg ctg tct ttg ctt cat tgc ggc ggc agc agc cag 1298
Arg Ser Glu Thr Arg Leu Ser Leu Leu His Cys Gly Gly Ser Ser Gin
415 420 425
tcc tcc gaa gac tea ttt aga caa tcc tgc ate aat gtg ccc ggt gta 1346
Ser Ser Glu Asp Ser Phe Arg Gin Ser Cys Ile Asn Val Pro Gly Val
- - 43R 435 440 - - 445
46/111
i
CA 02437866 2003-08-08
tcc acg ggg ace gtc ctt ggt get ggg agt att gga aca gat att gca 1394
Ser Thr Gly Thr Val Leu Gly Ala Gly Ser Ile Gly Thr Asp Ile Ala
450 455 460
aat gag gtt ctg agt tta cag aga tta cac tct tca tcc cca gaa gat 1442
Asn Glu Val Leu Ser Leu Gin Arg Leu His Ser Ser Ser Pro Glu Asp
465 470 475
gca agc cct tca gaa gaa gtg aga gat gac tgc agt gta aat ggt ggg 1490
Ala Ser Pro Ser Glu Glu Val Arg Asp Asp Cys Ser Val Asn Gly Gly
480 485 490
aat gcc tat ggg cct gca tcc act agg gag cct ttt gca gtg agc cct 1538
Asn Ala Tyr Gly Pro Ala Ser Thr Arg Glu Pro Phe Ala Val Ser Pro
495 500 505
tct gaa aca gag gtc ctg gag get gcc agg caa cac cag agc act gaa 1586
Ser Glu Thr Glu Val Leu Giu Ala Ala Arg Gin His Gin Ser Thr Glu
510 515 520 525
ccc gcc cac cct cac gga cca ctt ccc ggt gac agt gcc cag ctg ggt 1634
Pro Ala His Pro His Gly Pro Leu Pro Gly Asp Ser Ala Gin Leu Gly
530 535 540
ttt gat gtc ctg tgt gac agt gac agc ata gac atg get gca ttc atg 1682
Phe Asp Val Leu Cys Asp Ser Asp Ser Ile Asp Met Ala Ala Phe Met
545 550 555
aat tac etc gaa gca gag ggg ggc ctg ggt gac cct ggg gac ttc agt 1730
Asn Tyr Leu Glu Ala Glu Gly Giy Leu Gly Asp Pro Gly Asp Phe Ser
560 565 570
gac ate cag .t gg--gca -c t c A age - - - -
- - - - - - - - - - - - - - - -1752_- Asp lie Gin Trp Ala Leu
47/111
CA 02437866 2003-08-08
575
<210> 12
<211> 579
<212> PRT
<213> Mus musculus
<400> 12
Met Glu Phe Pro Arg Lys Arg Arg Gly Arg Asp Ser Gin Pro Leu Gin
1 5 10 15
Ser Glu Phe Met Thr Asp Thr Thr Val Glu Ser Leu Pro Gin Asn Pro
20 25 30
Phe Ala Ser Leu Leu Ser Thr Arg Thr Gly Val Ser Ala Pro Ser Gly
35 40 45
Ile Arg Glu Ala His Ser Gin Met Glu Lys Arg Arg Arg Asp Lys Met
50 55 60
Asn His Leu Ile Gin Lys Leu Ser Ser Met Ile Pro Pro His Ile Pro
65 70 75 80
Thr Ala His Lys Leu Asp Lys Leu Ser Val Leu Arg Arg Ala Val Gin
85 90 95
Tyr Leu Arg Ser Leu Arg Gly Met Thr Glu Leu Tyr Leu Gly Glu Asn
100 105 110
Ser Lys Pro Ser Phe Ile Gin Asp Lys Glu Leu Ser His Leu Ile Leu
115 120 125
Lys Ala Ala Glu Gly Phe Leu Phe Val Val Gly Cys Glu Arg Gly Arg
48/111
CA 02437866 2003-08-08
130 135 140
Ile Phe Tyr Val Ser Lys Ser Val Ser Lys Thr Leu Arg Tyr Asp Gln
145 150 155 160
Ala Ser Leu Ile Gly Gln Asn Leu Phe Asp Phe Leu His Pro Lys Asp
165 170 175
Val Ala Lys Val Lys Glu Gln Leu Ser Cys Asp Gly Ser Pro Arg Glu
180 185 190
Lys Pro Ile Asp Thr Lys Thr Ser Gin Val. Tyr Ser His Pro Tyr Thr
195 200 205
Gly Arg Pro Arg Met His Ser Gly Ser Arg Arg Ser Phe Phe Phe Arg
210 215 220
Met Lys Ser Cys Thr Val Pro Val Lys Glu Glu Gln Pro Cys Ser Ser
225 230 235 240
Cys Ser Lys Lys Lys Asp His Arg Lys Phe His Thr Val His Cys Thr
245 250 255
Gly Tyr Leu Arg Ser Trp Pro Leu Asn Val Val Gly Met Glu Lys Glu
260 265 270
Ser Gly Gly Gly Lys Asp Ser Gly Pro Leu Thr Cys Leu Val Ala Met
275 280 285
Gly Arg Leu His Pro Tyr Ile Val Pro Gin Lys Ser Gly Lys Ile Asn
290 295 300
Val Ar-g -Pro-Al=a-G-lu -Phe-l le Th-r-Arg Phe Ala-Met Asa Gly -Lys -P-he 305
310 315 320
49/111
CA 02437866 2003-08-08
Val Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr Leu Pro Gin
325 330 335
Glu Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp Asp His
340 345 350
Ser Ser Leu Thr Asp Lys His Lys Ala Val Leu Gin Ser Lys Glu Lys
355 360 365
Ile Leu Thr Asp Ser Tyr Lys Phe Arg Val Lys Asp Gly Ala Phe Val
370 375 380
Thr Leu Lys Ser Glu Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Glu
385 390 395 400
Leu Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu Gly Arg Ser Glu
405 410 415
Thr Arg Leu Ser Leu Leu His Cys Giy Gly Ser Ser Gin Ser Ser Glu
420 425 430
Asp Ser Phe Arg Gin Ser Cys Ile Asn Val Pro Gly Val Ser Thr Gly
435 440 445
Thr Val Leu Gly Ala Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu Val
450 455 460
Leu Ser Leu Gin Arg Leu His Ser Ser Ser Pro Glu Asp Ala Ser Pro
465 470 475 480
Ser Glu Glu Val Arg Asp Asp Cys Ser Val Asn Gly Gly Asn Ala Tyr
485 490 495
50/111
i i
CA 02437866 2003-08-08
Gly Pro Ala Ser Thr Arg Glu Pro Phe Ala Val Ser Pro Ser Glu Thr
500 505 510
Glu Val Leu Glu Ala Ala Arg Gin His Gin Ser Thr Glu Pro Ala His
515 520 525
Pro His Gly Pro Leu Pro Gly Asp Ser Ala Gin Leu Gly Phe Asp Val
530 535 540
Leu Cys Asp Ser Asp Ser Ile Asp Met Ala Ala Phe Met Asn Tyr Leu
545 550 555 560
Glu Ala Glu Gly Gly Leu Gly Asp Pro Gly Asp Phe Ser Asp Ile Gin
565 570 575
Trp Ala Leu
<210> 13
<211> 1676
<212> DNA
<213> Mus musculus
<220>
<221> CDS
<222> (12).. (608)
<400> 13
ggtcgaccac c atg gag ttt cca agg aaa cgc aga ggc aga gat tcc cag 50
Met Glu Phe Pro Arg Lys Arg Arg Gly Arg Asp Ser Gin
1- -5-----10
51/111
i
CA 02437866 2003-08-08
cca ctc cag tca gaa ttc atg aca gac aca aca gtg gaa agt ctt ccc 98
Pro Leu Gin Ser Glu Phe Met Thr Asp Thr Thr Val Glu Ser Leu Pro
15 2C 25
cag aat ccc ttt gcc tct ctt ctt tca aca aga aca gga gta tca gcg 146
Gin Asn Pro Phe Ala Ser Leu Leu Ser Thr Arg Thr Gly Val Ser Ala
30 35 40 45
ccc agt ggc ate agg gaa get cac agc cag atg gaa aag cgt egg aga 194
Pro Ser Gly lie Arg Glu Ala His Ser Gin Met Glu Lys Arg Arg Arg
50 55 60
gac aag atg aac cat ctg att cag aaa ctg tca tct atg atc.cct cca 242
Asp Lys Met Asn His Leu Ile Gin Lys Leu Ser Ser Met Ile Pro Pro
65 70 75
cac ate ccc acg gcc cac aaa ctg gac aag ctc agc gtc ttg agg agg 290
His Ile Pro Thr Ala His Lys Leu Asp Lys Leu Ser Val Leu Arg Arg
80 85 90
gcg gtg cag tac ttg agg tct ctg aga ggc atg aca gag ctt tac tta 338
Ala Val Gin Tyr Leu Arg Ser Leu Arg Gly Met Thr Glu Leu Tyr Leu
95 100 105
gga gaa aac tct aaa cct tca ttt att cag gat aag gaa ctc agt cac 386
Gly Glu Asn Ser Lys Pro Ser Phe lie Gin Asp Lys Glu Leu Ser His
110 115 120 125
tta ate ctc aag gca gca gaa ggc ttc ctg ttt gtg gtt gga tgc gaa 434
Leu Ile Leu Lys Ala Ala Glu Gly Phe Leu Phe Val Val Gly Cys Glu
130 135 140
aga ggg_ aga att ttt tac_gtt _tct_aag _tct _gtc_tcc aaa aca ctg cgt 482
Arg Gly Arg Ile Phe Tyr Val Ser Lys Ser Val Ser Lys Thr Leu Arg
52/111
CA 02437866 2003-08-08
145 150 155
tat gat cag get age ttg ata gga cag aat ttg ttt gac ttc tta cac 530
Tyr Asp Gln Ala Ser Leu Ile Gly Gin Asn Leu Phe Asp Phe Leu His
160 165 170
cca aaa gac gtc gcc aaa gta aag gaa caa ctt tct tgt gat ggt tea 578
Pro Lys Asp Val Ala Lys Val Lys Glu Gln Leu Ser Cys Asp Gly Ser
175 180 185
cca aga gag aaa ect ata gac ace aaa aaa tgaagagctg taccgtccct 628
Pro Arg Glu Lys Pro Ile Asp Thr Lys Lys
190 195
gtcaaagaag agcagccatg ctcgtcctgc tcaaagaaga aagaccatag aaaattccac 688
accgtccatt gcactggata cttgagaagc tggcctctga atgttgttgg catggagaaa 748
gagtcgggtg gtgggaagga cagcggtcct cttacctgcc ttgtggctat gggacggttg 808
catccataca ttgtccctca aaagagtggc aagatcaacg tgagaccggc tgagticata 868
actcgcttcg caatgaacgg gaaattcgtc tatgttgacc aaagggcaac ggcaatttta 928
ggatacctgc ctcaggaact tttgggaact tcatgttatg aatattttca tcaggatgac 988
cacagtagtt tgactgacaa gcacaaagca gttctgcaga gtaaggagaa aatacttaca 1048
gactcataca aattcagagt gaaggatggt gccttcgtga ctctgaagag tgagtggttc 1108
agcttcacaa acccttggac caaagagctg gagtacattg tgtctgtcaa cacattggtt 1168
Itggggcgea-gt-gagaccag-getgtct-ttg-cttua-t4gcg-g-cggcagcag ccag-tccicc-12.25
53/111
CA 02437866 2003-08-08
gaagactcat ttagacaatc ctgcatcaat gtgcccggtg tatccacggg gaccgtcctt 1288
ggtgctggga gtattggaac agatattgca aatgagg:tc tgagtttaca gagattacac 1348
tcttcatccc cagaagatgc aagcccttca gaagaagtga gagatgactg cagtgtaaat 1408
ggtgggaatg cctatgggcc tgcatccact agggagcctt ttgcagtgag cccttctgaa 1468
acagaggtcc tggaggctgc caggcaacac cagagcactg aacccgccca ccctcacgga 1528
ccacttcccg gtgacagtgc ccagctgggt tttgatgtcc tgtgtgacag tgacagcata 1588
gacatggctg cattcatgaa ttacctcgaa gcagaggggg gcctgggtga ccctggggac 1648
ttcagtgaca tccagtgggc actctagc 1676
<210> 14
<211> 199
<212> PRT
<213> Mus musculus
<400> 14
Met Glu Phe Pro Arg Lys Arg Arg Gly Arg Asp Ser Gin Pro Leu Gin
1 5 10 15
Ser Glu Phe Met Thr Asp Thr Thr Val Glu Ser Leu Pro Gin Asn Pro
20 25 30
Phe Ala Ser Leu Leu Ser Thr Arg Thr Gly Val Ser Ala Pro Ser Gly
35 40 45
ILe- A-rgGlu -MHis- Ser -Gln- Met Glu Lys -Ar- A-rg Arg Asp tars-Met--
50 55 60
54/111
CA 02437866 2003-08-08
Asn His Leu lie Gin Lys Leu Ser Ser Met Ile Pro Pro His Ile Pro
65 70 75 80
Thr Ala His Lys Leu Asp Lys Leu Ser Val Leu Arg Arg Ala Val Gin
85 90 95
Tyr Leu Arg Ser Leu Arg Gly Met Thr Glu Leu Tyr Leu Gly Glu Asn
100 105 110
Ser Lys Pro Ser Phe Ile Gin Asp Lys Glu Leu Ser His Leu Ile Leu
115 120 125
Lys. Ala Ala Glu Gly Phe Leu Phe Val Val Gly Cys Glu Arg Gly Arg
130 135 140
Ile Phe Tyr Val Ser Lys Ser Val Ser Lys Thr Leu Arg Tyr Asp Gin
145 150 155 160
Ala Ser Leu Ile Gly Gin Asn Leu Phe Asp Phe Leu His Pro Lys Asp
165 170 175
Val Ala Lys Val Lys Glu Gin Leu Ser Cys Asp Gly Ser Pro Arg Glu
180 185 190
Lys Pro Ile Asp Thr Lys Lys
195
<210> 15
<211> 1748
<212> DNA-
<213> Rattus norvegicus
55/111
CA 02437866 2003-08-08
<220>
<221> CDS
<222> (33).. (1745)
<400> 15
tcacagctcc tggacctgtc agctctcttg ca atg gag ttg cca agg aaa cgt 53
Met Glu Leu Pro Arg Lys Arg
1 5
aga aga agt gat tea gag ctg etc cag tea gaa tic agg aca gat gca 101
Arg Arg Ser Asp Ser Glu Leu Leu Gin Ser Glu Phe Arg Thr Asp Ala
15 20
atg gtg gaa aac ctt ccc cgg agt ccc tit ace tct gtt ctt tea aca 149
Met Val Glu Asn Leu Pro Arg Ser Pro Phe Thr Ser Val Leu Ser Thr
25 30 35
aga aca gga gta gca gig ccc aat ggc ate agg gaa get cac age cag 197
Arg Thr Gly Val Ala Val Pro Asn Gly Ile Arg Glu Ala His Ser Gin
40 45 50 55
aca gaa aag cgt egg aga gac aag atg aac cat ctg att tgg aaa ctg 245
Thr Glu Lys Arg Arg Arg Asp Lys Met Asn His Leu Ile Trp Lys Leu
60 65 70
tea tct atg ate cct cca cac ate ccc aca gcc cac aaa ctg gac aaa 293
Ser Ser Met Ile Pro Pro His Ile Pro Thr Ala His Lys Leu Asp Lys
75 80 85
ctg age gtc ctg agg agg gca gig cag tac ttg agg tct cag aga ggc 341
Leu Ser Val Leu Arg Arg Ala Val Gin Tyr Leu Arg Ser Gin Arg Gly
90- - - 95-------------- 100
56/111
I i
CA 02437866 2003-08-08
atg aca gag ttt tat tta gga gaa aat get aaa cct tca ttt att cag 389
Met Thr Glu Phe Tyr Leu Gly Glu Asn Ala Lys Pro Ser Phe Ile Gin
105 110 115
gat aag gaa ctc agc cac tta atc ctc aag gca gca gaa ggc ttc cta 437
Asp Lys Glu Leu Ser His Leu Ile Leu Lys Ala Ala Glu Gly Phe Leu
120 125 130 135
ctt gtg gtt gga tgt gaa gga ggg aga att ctt ttc gtt tct aag tct 485
Leu Val Val Gly Cys Glu Gly Gly Arg Ile Leu Phe Val Ser Lys Ser
140 145 150
gtc tee aaa acg ctg cat tat gat cag get agt ttg atg gga cag aac 533
Val Ser Lys Thr Leu His Tyr Asp Gin Ala Ser Leu Met Gly Gin Asn
155 160 165
ttg ttt gac tic tta cac cca aaa gat gtc gcc aaa gta aag gaa caa 581
Leu Phe Asp Phe Leu His Pro Lys Asp Val Ala Lys Val Lys Glu Gin
170 175 180
ctt tct tgt gat gtt tca ctg aga gag aaa ccc ata ggc acc aaa acc 629
Leu Ser Cys Asp Val Ser Leu Arg Glu Lys Pro Ile Gly Thr Lys Thr
185 190 195
tct cct cag gtt cac agt cac tcc cat att ggg cga tca cgc gtg cat 677
Ser Pro Gin Val His Ser His Ser His Ile Gly Arg Ser Arg Val His
200 205 210 215
tct ggc tcc aga cga tct ttc ttc ttt aga atg aag agc agc tgt aca 725
Ser Gly Ser Arg Arg Ser Phe Phe Phe Arg Met Lys Ser Ser Cys Thr
220 225 230
gtc ccc gtc- aaa gaa gag raa-cga_ 1gc_tcg-tec-tgL Ica -aag_aag_a-aa_ 773 - - -
Val Pro Val Lys Glu Glu Gin Arg Cys Ser Ser Cys Ser Lys Lys Lys
57/111
I
CA 02437866 2003-08-08
235 240 245
gac cag aga aaa ttc cac acc ate cat tgc act gga tac ttg aga age 821
Asp Gin Arg Lys Phe His Thr Ile His Cys Thr Gly Tyr Leu Arg Ser
250 255 260
tgg cca ccg aat gtt gig ggc acg gag aaa gag atg ggc agt ggg aaa 869
Trp Pro Pro Asn Val Val Giy Thr Glu Lys Glu Met Gly Ser Gly Lys
265 270 275
gac agt ggt cct ctt acc tgc ctt gig get atg gga egg tta cag cca 917
Asp Ser Gly Pro Leu Thr Cys Leu Val Ala Met Gly Arg Leu Gin Pro
280 285 290 295
tat act gtc ccc ccg aag aat ggc aag ate aac gig aga ccg get gag 965
Tyr Thr Val Pro Pro Lys Asn Gly Lys Ile Asn Val Arg Pro Ala Glu
300 305 310
tic ata acc cga tic gca atg aac ggg aaa tic gtc tac gtc gac caa 1013
Phe Ile Thr Arg Phe Ala Met Asn Gly Lys Phe Val Tyr Val Asp Gin
315 320 325
agg gca aca gca att tta gga tac ctg cct cag gaa ctt ttg gga act 1061
Arg Ala Thr Ala Ile Leu Gly Tyr Leu Pro Gin Glu Leu Leu Gly Thr
330 335 340
tcg tgt tat gaa tat tit cat cag gat gac cac agt aat ttg agt gac 1109
Ser Cys Tyr Glu Tyr Phe His Gin Asp Asp His Ser Asn Leu Ser Asp
345 350 355
aag cac aaa gca gtt ctg cag agt aag gag aaa ata ctt aca gat tca 1157
Lys His Lys Ala Val Leu Gin Ser Lys Glu Lys Ile Leu Thr Asp Ser
360 365 370 375
.58/111
CA 02437866 2003-08-08
tac aaa tic aga gtg aag gat ggc tcc ttt gtg act ctg aag agc aag 1205
Tyr Lys Phe Arg Val Lys Asp Gly Ser Phe Val Thr Leu Lys Ser Lys
380 385 390
tgg ttc agc ttc act aac cct tgg acc aaa aag ctg gag tac ate gtg 1253
Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Lys Leu Glu Tyr Ile Val
395 400 405
tct gtc aac acg ctg gtt ttg ggg cgc agt gag acc gca gta tee gtg 1301
Ser Val Asn Thr Leu Val Leu Gly Arg Ser Glu Thr Ala Val Ser Val
410 415 420
cct cag tgc cgc agc agc cag tee act gaa gac tea ttt aga caa ccc 1349
Pro Gin Cys Arg Ser Ser Gin Ser Ser Glu Asp Ser Phe Arg Gin Pro
425 430 435
tgc gtc agt gtg ccg ggc ata tcc aca ggg acc tta ctt ggc act ggg 1397
Cys Val Ser Val Pro Gly Ile Ser Thr Gly Thr Leu Leu Gly Thr Gly
440 445 450 455
agt att gga aca gat att gca aat gag gtt ctg agt tta cag agg tea 1445
Ser Ile Gly Thr Asp Ile Ala Asn Glu Val Leu Ser Leu Gin Arg Ser
460 465 470
cac tct tea tee cca gaa gac gca aac cct tea gga gta gtg aga gat 1493
His Ser Ser Ser Pro Glu Asp Ala Asn Pro Ser Gly Val Val Arg Asp
475 480 485
aag cac agt gta aac ttc ggg agc gcc cct gtg ccc gtg tcc act ggg 1541
Lys His Ser Val Asn Phe G1y Ser Ala Pro Val Pro Val Ser Thr Gly
490 495 500
-gag etc tt-t gca-ctg -ag1 cct -gaa- aca gag ggc- ctg gag get -gcc agg 1589
Glu Leu Phe Ala Leu Ser Pro Glu Thr Glu Gly Leu Glu Ala Ala Arg
9/1 11
i I
CA 02437866 2003-08-08
505 510 515
caa cac cag agt tct gag ccc gcc cac tgt cac aaa cca ctc ctc agt 1637
Gin His Gin Ser Ser Glu Pro Ala His Cys His Lys Pro Leu Leu Ser
520 525 530 535
gac agt acc cag ttg ggt ttt gat gcc ctg tgt gac agc gac gac aca 1685
Asp Ser Thr Gin Leu Gly Phe Asp Ala Leu Cys Asp Ser Asp Asp Thr
540 545 550
gcc atg get aca ttc atg aat tac ctc gaa gca gag ggt ggc ctg ggt 1733
Ala Met Ala Thr Phe Met Asn Tyr Leu Glu Ala Glu Gly Gly Leu Gly
555 560 565
gac cct ggg gac ttc 1748
Asp Pro Gly Asp
570
<210> 16
<211> 571
<212> PRT
<213> Rattus norvegicus
<400> 16
Met- Glu Leu Pro Arg Lys Arg Arg Arg Ser Asp Ser Glu Leu Leu Gin
1 5 10 15
Ser Glu Phe Arg Thr Asp Ala Met Val Glu Asn Leu Pro Arg Ser Pro
20 25 30
Phe Thr Ser Val Leu Ser Thr Arg Thr Gly Val Ala Val Pro Asn Gly
35 40 45
60/111
i i
CA 02437866 2003-08-08
Ile Arg Glu Ala His Ser Gln Thr Glu Lys Arg Arg Arg Asp Lys Met
50 55 60
Asn His Leu Ile Trp Lys Leu Ser Ser Met Ile Pro Pro His Ile Pro
65 70 75 80
Thr Ala His Lys Leu Asp Lys Leu Ser Val Leu Arg Arg Ala Val Gin
85 90 95
Tyr Leu Arg Ser Gln Arg Gly Met Thr Glu Phe Tyr Leu Gly Glu Asn
100 105 110
Ala Lys Pro Ser Phe Ile Gln Asp Lys Glu Leu Ser His Leu Ile Leu
115 120 125
Lys Ala Ala Glu Gly Phe Leu Leu Val Val Gly Cys Glu Gly Gly Arg
130 135 140
Ile Leu Phe Val Ser Lys Ser Val Ser Lys Thr Leu His Tyr Asp Gln
145 150 155 160
Ala Ser Leu Met Gly Gln Asn Leu Phe Asp Phe Leu His Pro Lys Asp
165 170 175
Val Ala Lys Val Lys Glu Gln Leu Ser Cys Asp Val Ser Leu Arg Glu
180 185 190
Lys Pro Ile Gly Thr Lys Thr Ser Pro Gln Val His Ser His Ser His
195 200 205
Ile Gly Arg Ser Arg Val His Ser Gly Ser Arg Arg Ser Phe Phe Phe
210 215 220
Arg Met Lys Ser Ser Cys Thr Val Pro Val Lys Glu Glu Gln Arg Cys
61/111
I
CA 02437866 2003-08-08
225 230 235 240
Ser Ser=Cys Ser Lys Lys Lys Asp Gin Arg Lys Phe His Thr Ile His
245 250 255
Cys Thr Gly Tyr Leu Arg Ser Trp Pro Pro Asn Val Val Gly Thr Glu
260 265 270
Lys Glu Met Gly Ser Gly Lys Asp Ser Gly Pro Leu Thr Cys Leu Val
275 280 285
Ala Met Gly Arg Leu Gin Pro Tyr Thr Val Pro Pro Lys Asn Gly Lys
290 295 300
Ile Asn Val Arg Pro Ala Glu Phe Ile Thr Arg Phe Ala Met Asn Gly
305 310 315 320
Lys Phe Val Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr Leu
325 330 335
Pro Gin Gin Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp
340 345 350
Asp His Ser Asn Leu Ser Asp Lys His Lys Ala Val Leu Gin Ser Lys
355 360 365
Glu Lys Ile Leu Thr Asp Ser Tyr Lys Phe Arg Val Lys Asp Gly Ser
370 375 380
Phe Val Thr Leu Lys Ser Lys Trp Phe Ser Phe Thr Asn Pro Trp Thr
385 390 395 400
Lys Lys Leu Glu Tyr. 1le Val Ser Val Asn_Thr Leu Val_Leu Gly Arg
405 410 415
62/111
j j
CA 02437866 2003-08-08
Ser Glu Thr Ala Val Ser Val Pro Gln Cys Arg Ser Ser Gln Ser Ser
420 425 430
Glu Asp Ser Phe Arg Gin Pro Cys Val Ser Val Pro Gly lie Ser Thr
435 440 445
Gly Thr Leu Leu Gly Thr Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu
450 455 460
Val Leu Ser Leu Gln Arg Ser His Ser Ser Ser Pro Glu Asp Ala Asn
465 470 475 480
Pro Ser Gly Val Val Arg Asp Lys His Ser Val Asn Phe Gly Ser Ala
485 490 495
Pro Val Pro Val Ser Thr Gly Glu Leu Phe Ala Leu Ser Pro Glu Thr
500 505 510
Glu Gly Leu Glu Ala Ala Arg Gln His Gln Ser Ser Glu Pro Ala His
515 520 525
Cys His Lys Pro Leu Leu Ser Asp Ser Thr Gln Leu Gly Phe Asp Ala
530 535 540
Leu Cys Asp Ser Asp Asp Thr Ala Met Ala Thr Phe Met Asn Tyr Leu
545 550 555 560
Glu Ala Glu Gly Gly Leu Gly Asp Pro Gly Asp
565 570
<210> 17
63/111
CA 02437866 2003-08-08
<211> 1646
<212> DNA
<213> Rattus norvegicus
<220>
<221> CDS
<222> (33).. (1646)
<400> 17
tcacagctcc tggacctgtc agctctcttg ca atg gag ttg cca agg aaa cgt 53
Met Glu Leu Pro Arg Lys Arg
1 5
aga aga agt gat tea gag ctg ctc cag gaa get cac agc cag aca gaa 101
Arg Arg Ser Asp Ser Glu Leu Leu Gin Glu Ala His Ser Gin Thr Glu
15 20
aag cgt egg aga gac aag atg aac cat ctg att tgg aaa ctg tea tct 149
Lys Arg Arg Arg Asp Lys Met Asn His Leu lie Trp Lys Leu Ser Ser
25 30 35
atg ate cct cca cac ate ccc aca gcc cac aaa ctg gac aaa ctg agc 197
Met Ile Pro Pro His Ile Pro Thr Ala His Lys Leu Asp Lys Leu Ser
40 45 50 55
gtc ctg agg agg gca gtg cag tac ttg agg tct cag aga ggc atg aca 245
Val Leu Arg Arg Ala Val Gin Tyr Leu Arg Ser Gin Arg Gly Met Thr
60 65 70
gag ttt tat tta gga gaa aat get aaa cct tea ttt att cag gat aag 293
Glu Phe Tyr Leu Gly Glu Asn Ala Lys Pro Ser Phe Ile Gin Asp Lys
75 80 85
gaa ctc agc cac tta ate ctc aag gca gca gaa ggc ttc cta ctt gtg 341
64/111
CA 02437866 2003-08-08
Glu Leu Ser His Leu Ile Leu Lys Ala Ala Glu Gly Phe Leu Leu Val
90 95 100
gtt gga tgt gaa gga ggg aga att ctt ttc gtt tct aag tct gtc tcc 389
Val Gly Cys Glu Gly Gly Arg Ile Leu Phe Val Ser Lys Ser Val Ser
105 110 115
aaa acg ctg cat tat gat cag get agt ttg atg gga cag aac ttg ttt 437
Lys Thr Leu His Tyr Asp Gin Ala Ser Leu Met Gly Gin Asn Leu Phe
120 125 130 135
gac ttc tta cac cca aaa gat gtc gcc aaa gta aag gaa caa ctt tct 485
Asp Phe Leu His Pro Lys Asp Val Ala Lys Val Lys Glu Gin Leu Ser
140 145 150
tgt gat gtt tca ctg aga gag aaa ccc ata ggc acc aaa acc tct cct 533
Cys Asp Val Ser Leu Arg Glu Lys Pro Ile Gly Thr Lys Thr Ser Pro
155 160 165
cag gtt cac agt cac tcc cat att ggg cga tca cgc gtg cat tct ggc 581
Gin Val His Ser His Ser His Ile Gly Arg Ser Arg Val His Ser Gly
170 175 180
tcc aga cga tct tic ttc ttt aga atg aag agc agc tgt aca gtc ccc 629
Ser Arg Arg Ser Phe Phe Phe Arg Met Lys Ser Ser Cys Thr Val Pro
185 190 195
gtc aaa gaa gag caa cga tgc tcg tcc tgt tca aag aag aaa gac cag 677
Val Lys Glu Glu Gin Arg Cys Ser Ser Cys Ser Lys Lys Lys Asp Gin
200 205 210 215
aga aaa ttc cac acc atc cat tgc act gga tac ttg aga agc tgg cca 725
Arg Lys Phe His Thr Ile His Cys Thr Gly Tyr Leu_Arg Ser Trp Pro
220 225 230
65/111
I I
CA 02437866 2003-08-08
ccg aat gtt gig ggc acg gag aaa gag atg ggc agt ggg aaa gac agt 773
Pro Asn Val Val Gly Thr Glu Lys Glu Met Gly Ser Gly Lys Asp Ser
235 240 245
ggt cct ctt acc tgc ctt gig get atg gga egg tta cag cca tat act 821
Gly Pro Leu Thr Cys Leu Val Ala Met Gly Arg Leu Gin Pro Tyr Thr
250 255 260
gtc ccc ccg aag aat ggc aag ate aac gig aga ccg get gag tic ata 869
Val Pro Pro Lys Asn Gly Lys Ile Asn Val Arg Pro Ala Glu Phe Ile
265 270 275
acc cga tic gca atg aac ggg aaa tic gtc tac gtc gac caa agg gca 917
Thr Arg Phe Ala Met Asn Gly Lys Phe Val Tyr Val Asp Gin Arg Ala
280 285 290 295
aca gca att tta gga tac ctg cct cag gaa ctt ttg gga act tcg tgt 965
Thr Ala Ile Leu Gly Tyr Leu Pro Gin Glu Leu Leu Gly Thr Ser Cys
300 305 310
tat gaa tat tit cat cag gat gac cac agt aat ttg agt gac aag cac 1013
Tyr Glu Tyr Phe His Gin Asp Asp His Ser Asn Leu Ser Asp Lys His
315 320. 325
aaa gca gtt ctg cag agt aag gag aaa ata ctt aca gat tea tac aaa 1061
Lys Ala Val Leu Gin Ser Lys Glu Lys Ile Leu Thr Asp Ser Tyr Lys
330 335 340
tic aga gig aag gat ggc tcc tit gig act ctg aag agc aag tgg tic 1109
Phe Arg Val Lys Asp Gly Ser Phe Val Thr Leu Lys Ser Lys Trp Phe
345 350 355
agc tic act aac cct tgg acc aaa aag ctg gag tac ate gtg tet gtc 1157
66/111
CA 02437866 2003-08-08
Ser Phe Thr Asn Pro Trp Thr Lys Lys Leu Glu Tyr Ile Val Ser Val
360 365 370 375
aac acg ctg gtt ttg ggg cgc agt gag acc gca gta tcc gtg cct cag 1205
Asn Thr Leu Val Leu Gly Arg Ser Glu Thr Ala Val Ser Val Pro Gin
380 385 390
tgc cgc agc agc cag tcc tct gaa gac tca ttt aga caa ccc tgc gtc 1253
Cys Arg Ser Ser Gin Ser Ser Glu Asp Ser Phe Arg Gin Pro Cys Val
395 400 405
agt gtg ccg ggc ata tcc aca ggg acc tta ctt ggc act ggg agt att 1301
Ser Val Pro Gly Ile Ser Thr Gly Thr Leu Leu Gly Thr Gly Ser Ile
410 415 420
gga aca gat att gca aat gag gtt ctg agt tta cag agg tca cac tct 1349
Gly Thr Asp Ile Ala Asn Glu Val Leu Ser Leu Gin Arg Ser His Ser
425 430 435
tca tcc cca gaa gac gca aac cct tca gga gta gtg aga gat aag cac 1397
Ser Ser Pro Glu Asp Ala Asn Pro Ser Gly Val Val Arg Asp Lys His
440 445 450 455
agt gta aac ttc ggg agc gcc cct gtg ccc gtg tcc act ggg gag etc 1445
Ser Val Asn Phe Gly Ser Ala Pro Val Pro Val Ser Thr Gly Glu Leu
460 465 470
ttt gca ctg agt cct gaa aca gag ggc ctg gag get gcc agg caa cac 1493
Phe Ala Leu Ser Pro Glu Thr Gin Gly Leu Glu Ala Ala Arg Gin His
475 480 485
cag agt tct gag ccc gcc cac tgt cac aaa cca etc etc agt gac agt 1541
Gin Ser Ser Glu_Pro Ala His Cys_His Lys Pro,Leu Leu Ser Asp Ser
490 495 500
67/11.1
CA 02437866 2003-08-08
ace cag ttg ggt ttt gat gcc ctg tgt gac age gac gac aca gcc atg 1589
Thr Gin Leu Gly Phe Asp Ala Leu Cys Asp Ser Asp Asp Thr Ala Met
505 510 515
get aca ttc atg aat tac ctc gaa gca gag ggt ggc ctg ggt gac cct 1637
Ala Thr Phe Met Asn Tyr Leu Glu Ala Glu Gly Gly Leu Gly Asp Pro
520 525 530 535
ggg gac ttc 1646
Gly Asp Phe
<210> 18
<211> 538
<212> PRT
<213> Rattus norvegicus
<400> 18
Met Glu Leu Pro Arg Lys Arg Arg Arg Ser Asp Ser Glu Leu Leu Gin
1 5 10 15
Glu Ala His Ser Gin Thr Glu Lys Arg Arg Arg Asp Lys Met Asn His
20 25 30
Leu Ile Trp Lys Leu Ser Ser Met Ile Pro Pro His Ile Pro Thr Ala
35 40 45
His Lys Leu Asp Lys Leu Ser Val Leu Arg Arg Ala Val Gin Tyr Leu
50 55 60
Arg Ser Gin Arg Gly Met Thr Glu Phe Tyr Leu Gly Glu Asn Ala Lys
65 - - - - -70- - - - ----- -75- - - -- - - - 80
68/111
I
CA 02437866 2003-08-08
Pro Ser Phe Ile Gin Asp Lys Glu Leu Ser His Leu Ile Leu Lys Ala
85 90 95
Ala Glu Gly Phe Leu Leu Val Val Gly Cys Glu Gly Gly Arg Ile Leu
100 105 110
Phe Val Ser Lys Ser Val Ser Lys Thr Leu His Tyr Asp Gin Ala Ser
115 120 125
Leu Met Gly Gin Asn Leu Phe Asp Phe Leu His Pro Lys Asp Val Ala
130 135 140
Lys Val Lys Glu Gin Leu Ser Cys Asp Val Ser Leu Arg Glu Lys Pro
145 150 155 160
Ile Gly Thr Lys Thr Ser Pro Gin Val His Ser His Ser His Ile Gly
165 170 175
Arg Ser Arg Val His Ser Gly Ser Arg Arg Ser Phe Phe Phe Arg Met
180 185 190
Lys Ser Ser Cys Thr Val Pro Val Lys Glu Glu Gin Arg Cys Ser Ser
195 200 205
Cys Ser Lys Lys Lys Asp Gin Arg Lys Phe His Thr Ile His Cys Thr
210 . 215 220
Gly Tyr Leu Arg Ser Trp Pro Pro Asn Val Val Gly Thr Glu Lys Glu
225 230 235 240
Met Gly Ser Gly Lys Asp Ser Gly Pro Leu Thr Cys Leu Val Ala Met
245 250 255
Gly Arg Leu Gin Pro Tyr Thr Val Pro Pro Lys Asn Gly Lys Ile Asn
69/111
CA 02437866 2003-08-08
260 265 270
Val Arg Pro Ala Glu Phe Ile Thr Arg Phe Ala Met Asn Gly Lys Phe
275 280 285
Val Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr Leu Pro Gin
290 295 300
Glu Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp Asp His
305 310 315 320
Ser Asn Leu Ser Asp Lys His Lys Ala Val Leu Gin Ser Lys Glu Lys
325 330 335
Ile Leu Thr Asp Ser Tyr Lys Phe Arg Val Lys Asp Gly Ser Phe Val
340 345 350
Thr Leu Lys Ser Lys Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Lys
355 360 365
Leu Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu Gly Arg Ser Glu
370 375 380
Thr Ala Val Ser Val Pro Gin Cys Arg Ser Ser Gin Ser Ser Glu Asp
385 390 395 400
Ser Phe Arg Gin Pro Cys Val Ser Val Pro Gly Ile Ser Thr Gly Thr
405 410 415
Leu Leu Gly Thr Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu Val Leu
420 425 430
Ser Leu Gln_Arg_ Ser_ His _Ser Ser_ Ser Pro Glu_Asp_Ala As_n Pro_ Ser _
435 440 445
70/111
CA 02437866 2003-08-08
Gly Val Val Arg Asp Lys His Ser Val Asn Phe Gly Ser Ala Pro Val
450 455 460
Pro Val Ser Thr Gly Glu Leu Phe Ala Leu Ser Pro Glu Thr Glu Gly
465 470 475 480
Leu Glu Ala Ala Arg Gin His Gin Ser Ser Glu Pro Ala His Cys His
485 490 495
Lys Pro Leu Leu Ser Asp Ser Thr Gin Leu Gly Phe Asp Ala Leu Cys
500 505 510
Asp Ser Asp Asp Thr Ala Met Ala Thr Phe Met Asn Tyr Leu Glu Ala
515 520 525
Gin Gly Gly Leu Gly Asp Pro Gly Asp Phe
530 535
<210> 19
<211> 1655
<212> DNA
<213> Rattus norvegicus
<220>
<221> CDS
<222> (33).. (1655)
<400> 19
tcacagctcc tggacctgtc agctctcttg ca aig gag ttg cca agg aaa cgt 53
Met Glu Leu Pro Arg Lys Arg
1 5
71/111
I I
CA 02437866 2003-08-08
aga aga agt gat tea gag ctg ctc cag tea gaa ttc agg aca gat gca 101
Arg Arg Ser Asp Ser Glu Leu Leu Gin Ser Glu Phe Arg Thr Asp Ala
15 20
atg gtg gaa aac ctt ccc cgg agt ccc ttt ace tct gtt ctt tea aca 149
Met Val Glu Asn Leu Pro Arg Ser Pro Phe Thr Ser Val Leu Ser Thr
25 30 35
aga aca gga gta gca gtg ccc aat,ggc ate agg gaa get cac agc cag 197
Arg Thr Gly Val Ala Val Pro Asn Gly Ile Arg Glu Ala His Ser Gin
40 45 50 55
aca gaa aag cgt cgg aga gac aag atg aac cat ctg att tgg aaa ctg 245
Thr Glu Lys Arg Arg Arg Asp Lys Met Asn His Leu Ile Trp Lys Leu
60 65 70
tea tct atg ate cct cca cac ate ccc aca gcc cac aaa ctg gac aaa 293
Ser Ser Met Ile Pro Pro His Ile Pro Thr Ala His Lys Leu Asp Lys
75 80 85
ctg agc gtc ctg agg agg gca gtg cag lac ttg agg tct cag aga ggc 341
Leu Ser Val Leu Arg Arg Ala Val Gin Tyr Leu Arg Ser Gin Arg Gly
90 95 100
atg aca gag ttt tat tta gga gaa aat get aaa cct tea ttt att cag 389
Met Thr Glu Phe Tyr Leu Gly Glu Asn Ala Lys Pro Ser Phe Ile Gin
105 110 115
gat aag gaa ctc agc cac tta ate 'ctc aag get agt ttg atg gga cag 437
Asp Lys Glu Leu Ser His Leu Ile Leu Lys Ala Ser Leu Met Gly Gin
120 125 130 135
aac ttg ttt gac ttc tta cac cca aaa gat gtc gcc aaa gta aag gaa 485
72/111
CA 02437866 2003-08-08
Asn Leu Phe Asp Phe Leu His Pro Lys Asp Val Ala Lys Val Lys Glu
140 145 150
caa ctt tct tgt gat gtt tea ctg aga gag aaa ccc ata ggc acc aaa 533
Gin Leu Ser Cys Asp Val Ser Leu Arg Glu Lys Pro Ile Gly Thr Lys
155 160 165
ace tct cct cag gtt cac agt cac tcc cat att ggg cga tea cgc gtg 581
Thr Ser Pro Gin Val His Ser His Ser His Ile Gly Arg Ser Arg Val
170 175 180
cat tct ggc tcc aga cga tct ttc ttc ttt aga atg aag age age tgt 629
His Ser Gly Ser Arg Arg Ser Phe Phe Phe Arg Met Lys Ser Ser Cys
185 190 195
aca gtc ccc gtc aaa gaa gag caa cga tgc tcg tcc tgt tea aag aag 677
Thr Val Pro Val Lys Glu Glu Gin Arg Cys Ser Ser Cys Ser Lys Lys
200 205 210 215
aaa gac cag aga aaa ttc cac ace ate cat tgc act gga tac ttg aga 725
Lys Asp Gin Arg Lys Phe His Thr Ile His Cys Thr Gly Tyr Leu Arg
220 225 230
age tgg cca ccg aat gtt gtg ggc acg gag aaa gag atg ggc agt ggg 773
Ser Trp Pro Pro Asn Val Val Gly Thr Glu Lys Glu Met Gly Ser Gly
235 240 245
aaa gac agt ggt cct ctt ace tgc ctt gtg get atg gga egg tta cag 821
Lys Asp Ser Gly Pro Leu Thr Cys Leu Val Ala Met Gly Arg Leu Gin
250 255 260
cca tat act gtc ccc ccg aag aat ggc aag ate aac gtg aga ccg get 869
Pro Tyr Thr Val Pro Pro_Lys_ Asn Gly Lys _Ile Asn Val Arg Pro Ala
265 270 275
73/111
I I
CA 02437866 2003-08-08
gag ttc ata acc cga ttc gca atg aac ggg aaa ttc gtc tac gtc gac 917
Glu Phe Ile Thr Arg Phe Ala Met Asn Gly Lys Phe Val Tyr Val Asp
280 285 290 295
caa agg gca aca gca att tta gga tac ctg cct cag gaa ctt ttg gga 965
Gin Arg Ala Thr Ala Ile Leu Gly Tyr Leu Pro Gin Glu Leu Leu Gly
300 305 310
act teg tgt tat gaa tat ttt cat cag gat gac cac agt aat ttg agt 1013
Thr Ser Cys Tyr Glu Tyr Phe His Gin Asp Asp His Ser Asn Leu Ser
315 320 325
gac aag cac aaa gca gtt ctg cag agt aag gag aaa ata ctt aca gat 1061
Asp Lys His Lys Ala Val Leu Gin Ser Lys Glu Lys Ile Leu Thr Asp
330 335 340
tca tac aaa ttc aga gtg aag gat ggc tcc tit gtg act ctg aag agc 1109
Ser Tyr Lys Phe Arg Val Lys Asp Gly Ser Phe Val Thr Leu Lys Ser
345 350 355
aag tgg ttc agc ttc act aac cct tgg acc aaa aag ctg gag tac atc 1157
Lys Trp Phe Ser Phe Thr Asn Pro Trp Thr Lys Lys Leu Glu Tyr Ile
360 365 370 375
gtg tct gtc aac acg ctg gtt ttg ggg cgc agt gag acc gca gta tcc 1205
Val Ser Val Asn Thr Leu Val Leu Gly Arg Ser Glu Thr Ala Val Ser
380 385 390
gtg cct cag tgc cgc agc agc cag tcc tct gaa gac tca ttt aga caa 1253
Val Pro Gin Cys Arg Ser Ser Gin Ser Ser Giu Asp Ser Phe Arg Gln
395 400 405
ccc tgc gtc agt gtg ccg ggc ata tcc aca ggg acc tta ctt ggc act 1301
74/111
CA 02437866 2003-08-08
Pro Cys Val Ser Val Pro Gly Ile Ser Thr Gly Thr Leu Leu Gly Thr
410 415 420
ggg agt att gga aca gat att gca aat gag gtt ctg agt tta cag agg 1349
Gly Ser Ile Gly Thr Asp Ile Ala Asn Glu Val Leu Ser Leu Gln Arg
425 430 435
tea cac tct tea tcc cca gaa gac gca aac cct tea gga gta gtg aga 1397
Ser His Ser Ser Ser Pro Glu Asp Ala Asn Pro Ser Gly Val Val Arg
440 445 450 455
gat aag cac agt gta aac ttc ggg agc gcc cct gtg ccc gtg tcc act 1445
Asp Lys His Ser Val Asn Phe Gly Ser Ala Pro Val Pro Val Ser Thr
460 465 470
ggg gag ctc ttt gca ctg agt cct gaa aca gag ggc ctg gag get gcc 1493
Gly Glu Leu Phe Ala Leu Ser Pro Glu Thr Glu Gly Leu Glu Ala Ala
475 480 485
agg caa cac cag agt tct gag ccc gcc cac tgt cac aaa cca ctc ctc 1541
Arg Gin His Gin Ser Ser Glu Pro Ala His Cys His Lys Pro Leu Leu
490 495 500
agt gac agt acc cag ttg ggt ttt gat gcc ctg tgt gac agc gac gac 1589
Ser Asp Ser Thr Gln Leu Gly Phe Asp Ala Leu Cys Asp Ser Asp Asp
505 510 515
aca gcc atg get aca ttc atg aat tac ctc gaa gca gag ggt ggc ctg 1637
Thr Ala Met Ala Thr Phe Met Asn Tyr Leu Glu Ala Glu Gly Gly Leu
520 525 530 535
ggt gac cct ggg gac ttc 1655
Gly Asp Pro Gly Asp Phe
540
75/111
CA 02437866 2003-08-08
<210> 20
<211> 541
<212> PRT
<213> Rattus norvegicus
<400> 20
Met Glu Leu Pro Arg Lys Arg Arg Arg Ser Asp Ser Glu Leu Leu Gin
1 5 10 15
Ser Glu Phe Arg Thr Asp Ala Met Val Glu Asn Leu Pro Arg Ser Pro
20 25 30
Phe Thr Ser Val Leu Ser Thr Arg Thr Gly Val Ala Val Pro Asn Gly
35 40 45
lie Arg Glu Ala His Ser Gin Thr Gin Lys Arg Arg Arg Asp Lys Met
50 55 60
Asn His Leu Ile Trp Lys Leu Ser Ser Met Ile Pro Pro His Ile Pro
65 70 75 80
Thr Ala His Lys Leu Asp Lys Leu Ser Val Leu Arg Arg Ala Val Gin
85 90 95
Tyr Leu Arg Ser Gin Arg Gly Met Thr Glu Phe Tyr Leu Gly Glu Asn
100 105 110
Ala Lys Pro Ser Phe Ile Gin Asp Lys Glu Leu Ser His Leu lie Leu
115 120 125
Lys Ala Ser Leu Met_Gly Gln Asn Leu Phe Asp Phe Leu His Pro Lys
130 135 140
76/111
i i
CA 02437866 2003-08-08
Asp Val Ala Lys Val Lys Glu Gin Leu Ser Cys Asp Val Ser Leu Arg
145 150 155 160
Glu Lys Pro lie Gly Thr Lys Thr Ser Pro Gin Val His Ser. His Ser
165 170 175
His Ile Gly Arg Ser Arg Val His Ser Gly Ser Arg Arg Ser Phe Phe
180 185 190
Phe Arg Met Lys Ser Ser Cys Thr Val Pro Val Lys Glu Glu Gin Arg
195 200 205
Cys Ser Ser Cys Ser Lys Lys Lys Asp Gin Arg Lys Phe His Thr Ile
210 215 220
His Cys Thr Gly Tyr Leu Arg Ser Trp Pro Pro Asn Val Val Gly Thr
225 230 235 240
Glu Lys Glu Met Gly Ser Gly Lys Asp Ser Gly Pro Leu Thr Cys Leu
245 250 255
Val Ala Met Gly Arg Leu Gin Pro Tyr Thr Val Pro Pro Lys Asn Gly
260 265 270
Lys Ile Asn Val Arg Pro Ala Glu Phe Ile Thr Arg Phe Ala Met Asn
275 280 285
Gly Lys Phe Val Tyr Val Asp Gin Arg Ala Thr Ala Ile Leu Gly Tyr
290 295 300
Leu Pro Gin Glu Leu Leu Gly Thr Ser Cys Tyr Glu Tyr Phe His Gin
305 310 315 320
77/111
I I
CA 02437866 2003-08-08
Asp Asp His Ser Asn Leu Ser Asp Lys His Lys Ala Val Leu Gin Ser
325 330 335
Lys Glu Lys Ile Leu Thr Asp Ser Tyr Lys Phe Arg Val Lys Asp Gly
340 345 350
Ser Phe Val Thr Leu Lys Ser Lys Trp Phe Ser Phe Thr Asn Pro Trp
355 360 365
Thr Lys Lys Leu Glu Tyr Ile Val Ser Val Asn Thr Leu Val Leu Gly
370 375 380
Arg Ser Glu Thr Ala Val Ser Val Pro Gin Cys Arg Ser Ser Gin Ser
385 390 395 400
Ser Glu Asp Ser Phe Arg Gin Pro Cys Val Ser Val Pro Gly Ile Ser
405 410 415
Thr Gly Thr Leu Leu Gly Thr Gly Ser Ile Gly Thr Asp Ile Ala Asn
420 425 430
Glu Val Leu Ser Leu Gin Arg Ser His Ser Ser Ser Pro Glu Asp Ala
435 440 445
Asn Pro Ser Gly Val Val Arg Asp Lys His Ser Val Asn Phe Gly Ser
450 455 460
Ala Pro Val Pro Val Ser Thr Gly Glu Leu Phe Ala Leu Ser Pro Glu
465 470 475 480
Thr Glu Gly Leu Glu Ala Ala Arg Gin His Gin Ser Ser Glu Pro Ala
485 490 495
His Cys His Lys Pro Leu Leu Ser Asp Ser Thr Gin Leu Gly Phe Asp
78/111
i I
CA 02437866 2003-08-08
500 505 510
Ala Leu Cys Asp Ser Asp Asp Thr Ala Met Ala Thr Phe Met As,.,. Tyr
515 520 525
Leu Glu Ala Glu Gly Gly Leu Gly Asp Pro Gly Asp Phe
530 535 540
<210> 21
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Sense primer 1
<400> 21
actagtcgac ttatgttttt taccataagc ace 33
<210> 22
<211> 30
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Antisense
primer I
<400> 22
gtcgacctgc gctactgtgg ctgagctttg 30
79/111
CA 02437866 2003-08-08
<210> 23
<211> 33
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:per-F
<220>
<221> modified-base
<222> (16)
<223> i
<220>
<221> modified-base
<222> (19)
<223> i
<400> 23
cagcagatsa rctgyntsng acagyrtcmt cag 33
<210> 24
<211> 31
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:per-R
<220>
<221> modified-base
<222> (19)
80/111
CA 02437866 2003-08-08
<223> i
<400> 24
gctrcactgr ctgrtgmsng acrccacrct c 31
<210> 25
<211> 24
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cPer2-R1 primer
<400> 25
ttgctgtacc aggcacatta caac 24
<210> 26
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:YK-F1
<220>
<221> modified-base
<222> (3)
<223> i
<220>
<221> modified-base
<222> (9)
81/111
i
CA 02437866 2003-08-08
<223> i
<220>
<221> modified-base
<222> (12)
<223> i
<220>
<221> modified-base
<222> (21)
<223> i
<220>
<221> modified-base
<222> (24)
<223> i
<220>
<221> modified-base
<222> (27)
<223> i
<220>
<221> modified-base
<222> (30)
<223> i
<400> 26
rtncaytcng gntaycargc nccnmgnatn cc 32
<210> 27
<211> 4035
<212> DNA
82/111
i I
CA 02437866 2003-08-08
<213> Gallus gallus
<220>
<221> CDS
<222> (1).. (4035)
<400> 27
atg gac tgt atc gag gtc agg ggg ttc tac tct agc act gag gag cag 48
Met Asp Cys Ile Glu Val Arg Gly Phe Tyr Ser Ser Thr Glu Glu Gin
1 5 10 15
aac cct gag cag caa get gat atc agt gaa aac att tct tca ttg ttc 96
Asn Pro Glu Gin Gln Ala Asp Ile Ser Gin Asn Ile Ser Ser Leu Phe
20 25 30
tct tta aaa gag caa cag aaa atg agt gag tat tct gga ctt gca agt 144
Ser Leu Lys Glu Gln Gin Lys Met Ser Glu Tyr Ser Gly Leu Ala Ser
35 40 45
aac cat agc cag atg att get gaa gat tct gaa att cag cca aaa cct 192
Asn His Ser Gln Met Ile Ala Glu Asp Ser Glu Ile Gln Pro Lys Pro
50 55 60
gaa cac tct ccc gaa gtc ctt cag gaa gat att gag atg agc agc gga 240
Glu His Ser Pro Glu Val Leu Gln Glu Asp Ile Glu Met Ser Ser Gly
65 70 75 80
tcc agt gga aat gac ttc agt gga aat gag acg aat gaa aac tac tcc 288
Ser Ser Gly Asn Asp Phe Ser Gly Asn Glu Thr Asn Glu Asn Tyr Ser
85 90 95
agt gga cat gat tct cat ggc cac gaa tct gat gaa aat ggg aaa gat 336
Ser Gly His Asp Ser His Gly His Glu Ser Asp Glu Asn Giy Lys Asp
100 105 110
83/111
i I
CA 02437866 2003-08-08
tea gca atg ctc atg gaa tct tea gac tgt cat aaa agt tea agc tea 384
Ser Ala Met Leu Met Glu Ser Ser Asp Cys His Lys Ser Ser Ser Ser
115 120 125
aat gca ttt agt ctg atg att gcg aac tct gaa cac aat cag tct agc 432
Asn Ala Phe Ser Leu Met Ile Ala Asn Ser Glu His Asn Gin Ser Ser
130 135 140
agt gga tgc age agc gag cag tct act aaa gcc aaa acg caa aag gaa 480
Ser Gly Cys Ser Ser Glu Gin Ser Thr Lys Ala Lys Thr Gin Lys Glu
145 150 155 160
ttg ttg aag aca ttg caa gag ctg aaa get cac ctt cct get gaa aaa 528
Leu Leu Lys Thr Leu Gin Giu Leu Lys Ala His Leu Pro Ala Glu Lys
165 170 175
aga att aaa ggc aaa tcc agt gtc eta aca aca ctg aaa tat gcc ctt 576
Arg Ile Lys Gly Lys Ser Ser Val Leu Thr Thr Leu Lys Tyr Ala Leu
180 185 190
aaa age att aaa caa gtt aaa gcc aat gag gaa tat tac caa ttg ttg 624
Lys Ser Ile Lys Gin Val Lys Ala Asn Glu Glu Tyr Tyr Gin Leu Leu
195 200 205
atg att aat gaa tcc cag cct tct gga ctc aat gtg tea tct tat aca 672
Met Ile Asn Glu Ser Gin Pro Ser Gly Leu Asn Val Ser Ser Tyr Thr
210 215 220
gtg gaa gaa gtt gag act ata acc tea gaa tac ate atg aaa aat gca 720
Val Glu Glu Val Glu Thr Ile Thr Ser Glu Tyr Ile Met Lys Asn Ala
225 230 235 240
gat atg ttt get gta get gtt tct ttg att act ggg aaa att gtg tac 768
84/111
CA 02437866 2003-08-08
Asp Met Phe Ala Val Ala Val Ser Leu Ile Thr Gly Lys Ile Val Tyr
245 250 255
atc tct gat caa get get get att ctg cgc tgt aag agg agt tat tit 816
Ile Ser Asp Gin Ala Ala Ala Ile Leu Arg Cys Lys Arg Ser Tyr Phe
260 265 270
aaa aat gcc aaa tit gig gag tta ttg gca cct caa gat gtc agt gtt 864
Lys Asn Ala Lys Phe Val Glu Leu Leu Ala Pro Gln Asp Val Ser Val
275 280 285
tic tat act tct act acc cca tac aga tta cca tct tgg aat att tgc 912
Phe Tyr Thr Ser Thr Thr Pro Tyr Arg Leu Pro Ser Trp Asn Ile Cys
290 295 300
agc aga get gag tct tcc acc cag gat tgc atg gaa gag aaa tcc tit 960
Ser Arg Ala Glu Ser Ser Thr Gin Asp Cys Met Glu Glu Lys Ser Phe
305 310 315 320
tic tgt cgc atc agt gca gga aag gag cgt gaa aat gag att tgc tat 1008
Phe Cys Arg Ile Ser Ala Gly Lys Glu Arg Glu Asn Glu Ile Cys Tyr
325 330 335
cac cca tit cgg atg act cct tac ctt atc aaa gta caa gat cca gaa 1056
His Pro Phe Arg Met Thr Pro Tyr Leu Ile Lys Val Gin Asp Pro Glu
340 345 350
gta gca gag gac caa ctt tgt tgt gtg ctc ctt gca gaa aaa gtg cac 1104
Val Ala Glu Asp Gln Leu Cys Cys Val Leu Leu Ala Glu Lys Val His
355 360 365
tct ggt tat gaa gca ccc aga aft cct cca gac aaa aga att tit aca 1152
Ser Gly Tyr Glu Ala Pro Arg Ile Pro Pro Asp Lys Arg Ile Phe Thr
370 375 380
85/111
CA 02437866 2003-08-08
aca aca cac aca cca acc tgt ttg tic cag gat gta gat gag aga get 1200
Thr Thr His Thr Pro Thr Cys Leu Phe Gin Asp Val Asp Glu Arg Ala
385 390 395 400
gta cct ctg ttg gga tac cta cct cag gac tta ata gga acg cct gtt 1248
Val Pro Leu Leu Gly Tyr Leu Pro Gin Asp Leu fie Gly Thr Pro Val
405 410 415
ttg gtg cat ctt cac cca aat gac aga ccc tta atg cta gca att cac 1296
Leu Val His Leu His Pro Asn Asp Arg Pro Leu Met Leu Ala lie His
420 425 430
aaa aaa ata ctt caa tat gga gga cag cct ttt gac tat tca cca ate 1344
Lys Lys Ile Leu Gin Tyr Gly Gly Gin Pro Phe Asp Tyr Ser Pro Ile
435 440 445
agg ttt tgc act aga aat gga gat tat ata acc atg gac act age tgg 1392
Arg Phe Cys Thr Arg Asn Gly Asp Tyr Ile Thr Met Asp Thr Ser Trp
450 455 460
tcc agt ttc ate aat cct tgg agt cga aag gtt tca ttt ate att gga 1440
Ser Ser Phe lie Asn Pro Trp Ser Arg Lys Val Ser Phe Ile Ile Gly
465 470 475 480
aga cac aaa gtt agg acg ggt ccc tta aat gaa gat gtt ttt gcc get 1488
Arg His Lys Val Arg Thr Gly Pro Leu Asn Glu Asp Val Phe Ala Ala
485 490 495
ccc aac tat acg gag gac aga ate ctt cac ccc agt gtt cag gag ate 1536
Pro Asn Tyr Thr Glu Asp Arg Ile Leu His Pro Ser Val Gin Glu lie
500 505 510
aca gag caa ata tat egg ctg tta cta cag cct gta cac aac agt gga 1584
86/111
CA 02437866 2003-08-08
Thr Glu Gin lie Tyr Arg Leu Leu Leu Gin Pro Val His Asn Ser Gly
515 520 525
tcc agt ggc tat gga agt cta ggt age aat ggc tea cac gaa cac tta 1632
Ser Ser Gly Tyr Gly Ser Leu Gly Ser Asn Gly Ser His Glu His Leu
530 .535 540
atg agt gtg gca tcc tcc agt gac age aca gga aat aat aat gat gac 1680
Met Ser Val Ala Ser Ser Ser Asp Ser Thr Gly Asn Asn Asn Asp Asp
545 550 555 560
aca caa aag gat aaa aca ata agt caa gat gcc cgt aag gtc aaa act 1728
Thr Gin Lys Asp Lys Thr Ile Ser Gin Asp Ala Arg Lys Val Lys Thr
565 570 575
aaa gga cag cat att ttc act gag aat aaa gga aaa ctg gaa tat aaa 1776
Lys Gly Gin His Ile Phe Thr Glu Asn Lys Gly Lys Leu Glu Tyr Lys
580 585 590
aga gag cct tct gca gaa aaa caa aat ggt cct ggt ggt cag gtg aaa 1824
Arg Glu Pro Ser Ala Glu Lys Gin Asn Gly Pro Gly Gly Gin Val Lys
595 600 605
gat gtg ata gga aag gat acc aca get aca get get cct aaa aat gtg 1872
Asp Val Ile Gly Lys Asp Thr Thr Ala Thr Ala Ala Pro Lys Asn Val
610 615 620
get act gaa gag ttg gca tgg aaa gaa caa cct gta tat tct tat caa 1920
Ala Thr Glu Glu Lev Ala Trp Lys Glu Gin Pro Val Tyr Ser Tyr Gin
625 630 635 640
cag att age tgc ttg gat agt gtc ate agg tat ttg gag agt tgt aat 1968
Gin Ile Ser Cys Leu Asp Ser Val lie Arg Tyr Leu Glu Ser Cys Asn
645 650 655
87/111
i
CA 02437866 2003-08-08
gtg cct ggt aca gca aaa aga aaa tgt gaa cct tea tea agt gtg aat 2016
Val Pro Gly Thr Ala Lys Arg Lys Cys Glu Pro Ser Ser Ser Val Asn
660 665 670
tct agt gtt cac gag caa aaa gca tct gtt aat get ata caa ccc tta 2064:
Ser Ser Val His Glu Gin Lys Ala Ser Val Asn Ala Ile Gin Pro Leu
675 680 685
gga gac tct act gtg ttg aag tea tct ggt aaa tea agt ggt ccc cca 2112
Gly Asp Ser Thr Val Leu Lys Ser Ser Gly Lys Ser Ser Gly Pro Pro
690 695 700
gta gtt ggt get cac tta act tct ttg gcc tta cct ggc aag cct gaa 2160
Val Val Gly Ala His Leu Thr Ser Leu Ala Leu Pro Gly Lys Pro Glu
705 710 715 720
agt gtt gta tcg etc acc agt cag tgc agc tac agt age ace att gtt 2208
Ser Val Val Ser Leu Thr Ser Gin Cys Ser Tyr Ser Ser Thr Ile Val
725 730 735
cat gtt gga gac aaa aaa cca caa cct gaa tta gaa atg ata gaa gat 2256
His Val Gly Asp Lys Lys Pro Gin Pro Glu Leu Giu Met Ile Glu Asp
740 745 750
ggt cca agt gga gca gaa gtc tta gat act caa ctt cct gcc cct cca 2304
Gly Pro Ser Gly Ala Glu Val Leu Asp Thr Gin Leu Pro Ala Pro Pro
755 760 765
ccc age tct acg cat gta aat cag gaa aag gag tea ttt aaa aaa ctg 2352
Pro Ser Ser Thr His Val Asn Gin Glu Lys Glu Ser Phe Lys Lys Leu
770 775 780
gga ctt aca aag gaa gtc ctt gca gtg cat aca caa aaa gaa gag'caa 2400
88/111
CA 02437866 2003-08-08
Gly Leu Thr Lys Glu Val Leu Ala Val His Thr Gin Lys Glu Glu Gin
785 790 795 800
age ttt ttg aat aag ttc aaa gaa ate aag aga ttc aat att ttc cag 2448
Ser Phe Leu Asn Lys Phe Lys Glu Ile Lys Arg Phe Asn Ile Phe Gin
805 810 815
tcc cac tge aat tac tac tta caa gat aaa cct aaa gga agg cct ggt 2496
Ser His Cys Asn Tyr Tyr Leu Gin Asp Lys Pro Lys Gly Arg Pro Gly
820 825 830
gaa cgt ggt ggc cgc gga caa cga aat gga act tctgga atg gat cag 2544
Glu Arg Gly Gly Arg Gly Gin Arg Asn Gly Thr Ser Gly Met Asp Gin
835 840 845
cct tgg aag aaa agt ggg aaa aac agg aaa tea aaa cgc att aaa cca 2592
Pro Trp Lys Lys Ser Gly Lys Asn Arg Lys Ser Lys Arg Ile Lys Pro
850 855 860
cag gag tct tea gac agt aca act tct gga act aaa ttc ccc cat cgc 2640
Gin Glu Ser Ser Asp Ser Thr Thr Ser Gly Thr Lys Phe Pro His Arg
865 870 875 880
ttt cct ctt cag ggt tta aat act acc get tgg tea ccg tea gac act 2688
Phe Pro Leu Gin Gly Leu Asn Thr Thr Ala Trp Ser Pro Ser Asp Thr
885 890 895
tea caa gca age tac tea gcg atg tct ttt cca act gtt atg cct gca 2736
Ser Gin Ala Ser Tyr Ser Ala Met Ser Phe Pro Thr Val Met Pro Ala
900 905 910
tat ccg ctt cct gtt ttt cca gca gca gca gga act gtg cca cca get 2784
Tyr Pro Leu Pro Val Phe Pro Ala Ala Ala.G1y Thr Val Pro Pro Ala
915 920 925
89/111
i i
CA 02437866 2003-08-08
cct gag act tca gtc tct ggt tit aat cag ctg cca gac tcg gga aat 2832
Pro Glu Thr Ser Val Ser Gly PhE Asn Gin Leu Pro Asp Ser Gly Asn
930 935 940
act tgc tct atg caa cca tcc cag tit tct gcc cct ctt atg aca ccc 2880
Thr Cys Ser Met Gin Pro Ser Gin Phe Ser Ala Pro Leu Met Thr Pro
945 950 955 960
gtt gta get ctt gig etc ccc aac tat gtc tac cca gaa atg aac aat 2928
Val Val Ala Leu Val Leu Pro Asn Tyr Val Tyr Pro Glu Met Asn Asn
965 970 975
agc tta cct caa aca ctt tac cac agc caa gcc aat tit ccc ace cat 2976
Ser Leu Pro Gin Thr Leu Tyr His Ser Gin Ala Asn Phe Pro Thr His
980 985 990
cct get tic tct tca cag aca gta ttt cca gcg cag cct cca tic act 3024
Pro Ala Phe Ser Ser Gin Thr Val Phe Pro Ala Gin Pro Pro Phe Thr
995 1000 1005
ace cct age cct tic cca caa cag gcg tit tit cca atg caa cca tic 3072
Thr Pro Ser Pro Phe Pro Gin Gin Ala Phe Phe Pro Met Gin Pro Phe
1010 1015 1020
cat tat aat cca cca gca gaa att gaa aag gtt cct gtc aca gag aca 3120
His Tyr Asn Pro Pro Ala Glu Ile Glu Lys Val Pro Val Thr Glu Thr
1025 1030 1035 1040
cga aac gag cca tcc cgt tcc tgc act cca cag tca gig ggt cct caa 3168
Arg Asn Glu Pro Ser Arg Ser Cys Thr Pro Gin Ser Val Gly Pro Gln
1045 1050 1055
gac cag get tca ccg cct ttg tic caa tca agg tgt agt ict cct ctg 3216
90/111
CA 02437866 2003-08-08
Asp Gin Ala Ser Pro Pro Leu Phe Gin Ser Arg Cys Ser Ser Pro Leu
1060 1065 1070
aat ctt cta cag ttg gaa gaa aac aca aaa act gtg gaa agt gga get 3264
Asn Leu Leu Gin Leu Glu Glu Asn Thr Lys Thr Val Glu Ser Gly Ala
1075 1080 1085
cct gca ggt ttg cat gga get tta aat gag gaa gga acc ata ggc aaa 3312
Pro Ala Gly Leu His Gly Ala Leu Asn Glu Glu Gly Thr lie Gly Lys
1090 1095 1100
atc atg aca act gat get ggt agt gga aag gga tcc cta cca get gag 3360
Ile Met Thr Thr Asp Ala Gly Ser Gly Lys Gly Ser Leu Pro Ala Glu
1105 1110 1115 1120
tct cca atg gat get caa aat agc gat gca ctc tcc atg tcc agt gtc 3408
Ser Pro Met Asp Ala G1nAsn Ser Asp Ala Leu Ser Met Ser Ser Val
1125 1130 1135
ctg ctt gac att tta ctt caa gaa gat gca tgc tca ggc act ggc tca 3456
Leu Leu Asp Ile Leu Leu Gin Glu Asp Ala Cys Ser Gly Thr Gly Ser
1140 1145 1150
get tcc tca ggg agc ggt gta tct gca get get gaa tct ctc ggg tct 3504
Ala Ser Ser Gly Ser Gly Val Ser Ala Ala Ala Glu Ser Leu Gly Ser
1155 1160 1165
gga tct aac ggc tgt gac atg tca ggg agc aga aca ggc agt agt gaa 3552
Gly Ser Asn Gly Cys Asp Met Ser Gly Ser Arg Thr Gly Ser Ser Glu
1170 1175 1180
act agt cat acc agc aaa tac ttt ggg agt atc gat tct tca gaa aac 3600
-Thr Ser His Thr Ser Lys Tyr Phe Giy Ser Ile Asp Ser Ser Glu Asn
1185 1190 1195 1200
91/111
CA 02437866 2003-08-08
cat cat aaa aca aaa atg aag gca gaa ata gaa gaa agt gag cac ttc 3648
His His Lys Thr Lys Met Lys Ala Glu Ile Glu Clu Ser Glu His Phe
1205 1210 1215
att aaa tat gtt ctt cag gat cct ata tgg ctt ttg atg gca aac aca 3696
Ile Lys Tyr Val, Leu Gin Asp Pro Ile Trp Leu Leu Met Ala Asn Thr
1220 1225 1230
gat gac acc gtt atg atg act tac cag tta ccc tct aga gat ttg gaa 3744
Asp Asp Thr Val Met Met Thr Tyr Gin Leu Pro Ser Arg Asp Leu Glu
1235 1240 1245
aca gtt tta aaa gaa gat aag ctg aaa cta aag caa atg cag aaa cta 3792
Thr Val Leu Lys Glu Asp Lys Leu Lys Leu Lys Gin Met Gin Lys Leu
1250 1255 1260
caa cca aaa ttt act gaa gac caa aaa aga gag ctt att gaa gtt cat 3840
Gin Pro Lys Phe Thr Glu Asp Gin Lys Arg Glu Leu Ile Glu Val His
1265 1270 1275 1280
cca tgg atc cag caa ggt gga ctg cca aag act gtt get aac tct gaa 3888
Pro Trp Ile Gin Gin Gly Gly Leu Pro Lys Thr Val Ala Asn Ser Glu
1285 1290 1295
tgt att ttt tgt gag gac aat ata cag agc aat ttt tat aca tcg tac 3936
Cys Ile Phe Cys Glu Asp Asn Ile Gin Ser Asn Phe Tyr Thr Ser Tyr
1300 1305 1310
gat gaa gaa atc cat gaa atg gac ctt aat gaa atg att gaa gac agt 3984
Asp Glu Glu Ile His Glu Met Asp Leu Asn Glu Met Ile Glu Asp Ser
1315 1320 1325
ggg gag aac aat ttg gtt cct ctg agt caa gtc aat gaa gaa caa aca 4032
92/111
I I
CA 02437866 2003-08-08
Gly Glu Asn Asn Leu Val Pro Leu Ser Gin Val Asn Glu Glu Gin Thr
1330 1335 1340
tag 4035
<210> 28
<211> 1344
<212> PRT
<213> Gallus gallus
<400> 28
Met Asp Cys fie Glu Val Arg Gly Phe Tyr Ser Ser Thr Glu Glu Gin
1 5 10 15
Asn Pro Glu Gin Gin Ala Asp Ile Ser Glu Asn Ile Ser Ser Leu Phe
20 25 30
Ser Leu Lys Gin Gin Gin Lys Met Ser Glu Tyr Ser Gly Leu Ala Ser
35 40 45
Asn His Ser Gin Met Ile Ala Glu Asp Ser Glu Ile Gin Pro Lys Pro
50 55 60
Glu His Ser Pro Glu Val Leu Gin Glu Asp Ile Glu Met Ser Ser Gly
65 70 75 80
Ser Ser Gly Asn Asp Phe Ser Gly Asn Glu Thr Asn Glu Asn Tyr Ser
85 90 95
Ser Gly His Asp Ser His Gly His Glu Ser Asp Glu Asn Gly Lys Asp
100 105 110
Ser Ala Met Leu Met Glu Ser Ser Asp Cys His Lys Ser Ser Ser Ser
115 120 125
Asn Ala Phe Ser Leu Met Ile Ala Asn Ser Glu His Asn Gin Ser Ser
130 135 140
Ser Gly Cys Ser Ser Glu Gin Ser Thr Lys Ala Lys Thr Gin Lys Glu
145 150 155 160
Leu Leu Lys Thr Leu Gin Glu Leu Lys Ala His Leu Pro Ala Glu Lys
93/111
CA 02437866 2003-08-08
165 170 175
Arg Ile Lys Gly Lys Ser Ser Val Leu Thr Thr Leu Lys Tyr Ala Leu
180 185 190
Lys Ser Ile Lys Gin Val Lys Ala Asn Glu Glu Tyr Tyr Gin Leu Leu
195 200 205
Met Ile Asn Glu Ser Gin Pro Ser Gly Leu Asn Val Ser Ser Tyr Thr
210 215 220
Val Glu Glu Val Glu Thr Ile Thr Ser Glu Tyr Ile Met Lys Asn Ala
225 230 235 240
Asp Met Phe Ala Val Ala Val Ser Leu Ile Thr Giy Lys Ile Val Tyr
245 250 255
Ile Ser Asp Gin Ala Ala Ala Ile Leu Arg Cys Lys Arg Ser Tyr Phe
260 265 270
Lys Asn Ala Lys Phe Val Glu Leu Leu Ala Pro Gin Asp Val Ser Val
275 280 285
Phe Tyr Thr Ser Thr Thr Pro Tyr Arg Leu Pro Ser Trp Asn Ile Cys
290 295 300
Ser Arg Ala Glu Ser Ser Thr Gin Asp Cys Met Glu Gin Lys Ser Phe
305 310 315 320
Phe Cys Arg Ile Ser Ala Gly Lys Glu Arg Glu Asn Glu Ile Cys Tyr
325 330 335
His Pro Phe Arg Met Thr Pro Tyr Leu Ile Lys Val Gin Asp Pro Glu
340 345 350
Val Ala Glu Asp Gin Leu Cys Cys Val Leu Leu Ala Glu Lys Val His
355 360 365
Ser Gly Tyr Glu Ala Pro Arg Ile Pro Pro Asp Lys Arg Ile Phe Thr
370 375 380
Thr Thr His Thr Pro Thr Cys Leu Phe Gin Asp Val Asp Glu Arg Ala
385 390 395 400
Val Pro Leu Leu Giy Tyr Leu Pro Gin Asp Leu Ile Gly Thr Pro Val
405 410 415
Leu Val His Leu His Pro Asn Asp Arg Pro Leu Met Leu Ala Ile His
420 425 430
Lys Lys Ile Leu Gin Tyr Gly Gly Gin Pro Phe Asp Tyr Ser Pro Ile
94/111
i I
CA 02437866 2003-08-08
435 440 445
Arg Phe Cys Thr Arg Asn Gly Asp Tyr Ile Thr Met Asp Thr Ser Trp
450 455 460
Ser Ser Phe Ile Asn Pro Trp Ser Arg Lys Val Ser Phe Ile Ile Gly
465 470 475 480
Arg His Lys Val Arg Thr Gly Pro Leu Asn Glu Asp Val Phe Ala Ala
485 490 495
Pro Asn Tyr Thr Glu Asp Arg Ile Leu His Pro Ser Val Gin Glu Ile
500 505 510
Thr Glu Gin Ile Tyr Arg Leu Leu Leu Gin Pro Val His Asn Ser Gly
515 520 525
Ser Ser Gly Tyr Gly Ser Leu Gly Ser Asn Gly Ser His Glu His Leu
530 535 540
Met Ser Val Ala Ser Ser Ser Asp Ser Thr Gly Asn Asn Asn Asp Asp
545 550 555 560
Thr Gin Lys Asp Lys Thr Ile Ser Gin Asp Ala Arg Lys Val Lys Thr
565 570 575
Lys Gly Gin His Ile Phe Thr Glu Asn Lys Gly Lys Leu Glu Tyr Lys
580 585 590
Arg Glu Pro Ser Ala Glu Lys Gin Asn Gly Pro Gly Gly Gin Val Lys
595 600 605
Asp Val Ile Gly Lys Asp Thr Thr Ala Thr Ala Ala Pro Lys Asn Val
610 615 620
Ala Thr Glu Glu Leu Ala Trp Lys Glu Gin Pro Val Tyr Ser Tyr Gin
625 630 635 640
Gin Ile Ser Cys Leu Asp Ser Val Ile Arg Tyr Leu Glu Ser Cys Asn
645 650 655
Val Pro Gly. Thr Ala Lys Arg Lys Cys Glu Pro Ser Ser Ser Val Asn
660 665 670
Ser Ser Val His Glu Gin Lys Ala Ser Val Asn Ala Ile Gin Pro Leu
675 680 685
Gly Asp Ser Thr Val Leu Lys Ser Ser Gly Lys Ser Ser Gly Pro Pro
690 695 700
Val Val Gly Ala His Leu Thr Ser Leu Ala Leu Pro Gly Lys Pro Glu
95/111
I I
CA 02437866 2003-08-08
705 710 715 720
Ser Val Val Ser Leu Thr Ser Gin Cys Ser Tyr Ser Ser Thr Ile Val
725 730 735
His Val Gly Asp Lys Lys Pro Gin Pro Glu Leu Glu Met Ile Glu Asp
740 745 750
Gly Pro Ser Gly Ala Glu Val Leu Asp Thr Gin Leu Pro Ala Pro Pro
755 760 765
Pro Ser Ser Thr His Val Asn Gin Glu Lys Glu Ser Phe Lys Lys Leu
770 775 780
Gly Leu Thr Lys Glu Val Leu Ala Val His Thr Gin Lys Giu Glu Gin
785 790 795 800
Ser Phe Leu Asn Lys Phe Lys Glu Ile Lys Arg Phe Asn Ile Phe Gin
805 810 815
Ser His Cys Asn Tyr Tyr Leu Gin Asp Lys Pro Lys Giy Arg Pro Gly
820 825 830
Glu Arg Gly Gly Arg Gly Gin Arg Asn Gly Thr Ser Gly Met Asp Gin
835 840 845
Pro Trp Lys Lys Ser Gly Lys Asn Arg Lys Ser Lys Arg Ile Lys Pro
850 855 860
Gin Glu Ser Ser Asp Ser Thr Thr Ser Gly Thr Lys Phe Pro His Arg
865 870 875 880
Phe Pro Leu Gin Gly Leu Asn Thr Thr Ala Trp Ser Pro Ser Asp Thr
885 890 895
Ser Gin Ala Ser Tyr Ser Ala Met Ser Phe Pro Thr Val Met Pro Ala
900 905 910
Tyr Pro Leu Pro Val Phe Pro Ala Ala Ala Gly Thr Val Pro Pro Ala
915 920 925
Pro Glu Thr Ser Val Ser Gly Phe Asn Gin Leu Pro Asp Ser Gly Asn
930 935 940
Thr Cys Ser Met Gin Pro Ser Gin Phe Ser Ala Pro Leu Met Thr Pro
945 950 955 960
Val Val Ala Leu Val Leu Pro Asn Tyr Val Tyr Pro Glu Met Asn Asn
965 970 975
Ser Leu Pro Gin Thr Leu Tyr His Ser Gin Ala Asn Phe Pro Thr His
96/111
CA 02437866 2003-08-08
980 985 990
Pro Ala Phe Ser Ser Gin Thr Val Phe Pro Ala Gin Pro Pro Phe Thr
995 1000 1005
Thr Pro Ser Pro Phe Pro Gin Gin Ala Phe Phe Pro Met Gin Pro Phe
1010 1015 1020
His Tyr Asn Pro Pro Ala Glu Ile Glu Lys Val Pro Val Thr Glu Thr
1025 1030 1035 1040
Arg Asn Glu Pro Ser Arg Ser Cys Thr Pro Gin Ser Val Gly Pro Gin
1045. 1050 1055
Asp Gin Ala Ser Pro Pro Leu Phe Gin Ser Arg Cys Ser Ser Pro Leu
1060 1065 1070
Asn Leu Leu Gin Leu Glu Glu Asn Thr Lys Thr Val Glu Ser Gly Ala
1075 1080 1085
Pro Ala Gly Leu His Gly Ala Leu Asn Glu Glu Gly Thr Ile Gly Lys
1090 1095 1100
Ile Met Thr Thr Asp Ala Gly Ser Gly Lys Gly Ser Leu Pro Ala Glu
1105 1110 1115 1120
Ser Pro Met Asp Ala Gin Asn Ser Asp Ala Leu Ser Met Ser Ser Val
1125 1130 1135
Leu Leu Asp Ile Leu Leu Gin Glu Asp Ala Cys Ser Gly Thr Gly Ser
1140 1145 1150
Ala Ser Ser Gly Ser Gly Val Ser Ala Ala Ala Glu Ser Leu Gly Ser
1155 1160 1165
Gly Ser Asn Gly Cys Asp Met Ser Gly Ser Arg Thr Gly Ser Ser Glu
1170 1175 1180
Thr Ser His Thr Ser Lys Tyr Phe Gly Ser Ile Asp Ser Ser Glu Asn
1185 1190 1195 1200
His His Lys Thr Lys Met Lys Ala Glu Ile Glu Glu Ser Glu His Phe
1205 1210 1215
Ile Lys Tyr Val Leu Gin Asp Pro Ile Trp Leu Leu Met Ala Asn Thr
1220 1225 1230
Asp Asp Thr Val Met Met Thr Tyr Gin Leu Pro Ser Arg Asp Leu Glu
1235 1240 1245
Thr Val Leu Lys Glu Asp Lys Leu Lys Leu Lys Gin Met Gin Lys Leu
97/111
CA 02437866 2003-08-08
1250 1255 1260
Gin Pro Lys Phe Thr Glu Asp Gin Lys Arg Glu Leu Ile Glu Val His
1265 1270 1275 1280
Pro Trp Ile Gin Gin Gly Gly Leu Pro Lys Thr Val Ala Asn Ser Glu
1285 1290 1295
Cys Ile Phe Cys Glu Asp Asn Ile Gin Ser Asn Phe Tyr Thr Ser Tyr
1300 1305 1310
Asp Glu Glu Ile His Glu Met Asp Leu Asn Glu Met Ile Glu Asp Ser
1315 1320 1325
Gly Glu Asn Asn Leu Val Pro Leu Ser Gin Val Asn Glu Glu Gin Thr
1330 1335 1340
<210> 29
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:BMAL-F
<400> 29
gtgctmmgga tggcwgtkca gc 22
<210> 30
<21,1> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:BMAL-R
98/111
CA 02437866 2003-08-08
<400> 30
gcgyccratt gcvacraggc ag 22
<210> 31
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:hB2F1
<400> 31
gaccaagtgg ctcctgcgat 20
<210> 32
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:hB2R1
<400> 32
gctagagggt ccactggatg 20
<210> 33
<211> 27
<212> DNA
<213> Artificial Sequence
<220>
99/111
CA 02437866 2003-08-08
<223> Description of Artificial Sequence:hBMAL2-F4
<400> 33
gtgctggtag tattggaaca gatattg 27
<210> 34
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:hBMAL2-R1
<400> 34
gctagagggt ccactggatg 20
<210> 35
<211> 32
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:mBMAL2-F1
<400> 35
ggtcgaccac catggagttt tccaaggaaa cg 32
<210> 36
<211> 24
<212> DNA
<213> Artificial Sequence
100/111
i I
CA 02437866 2003-08-08
<220>
<223> Description of Artificial Sequence:mBMAL2-R1
<400> 36
gctagagtgc ccactggatg tcac 24
<210> 37
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Sense primer 2
<400> 37
catgtctggc agaggcaag 19
<210> 38
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Antisense
primer 2
<400> 38
ttagccgccg aagccgtag 19
<210> 39
101/111
CA 02437866 2003-08-08
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cB1F1600-primer
<400> 39
tccagacatt tcttcagctg g 21
<210> 40
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cB1REND-primer
<400> 40
ggatgttgaa gcaaggtgc 19
<210> 41
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cB2F1270-primer
<400> 41
acgagtactg ccatcaagat g 21
102/111
CA 02437866 2003-08-08
<210> 42
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cB2REND-primer
<400> 42
gagagcccat tggatgtcac 20
<210> 43
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cqCF862-primer
<400> 43
ttcttggatc acagggcac 19
<210> 44
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cgCR1364-primer
<400> 44
103/111
I I
CA 02437866 2003-08-08
ggagtgctag tgtccactgt ca 22
<210> 45
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cP2RTF-primer
<400> 45
ggaagtcctt gcagtgcata c 21
<210> 46
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cP2RTR-primer
<400> 46
acaggaagcg gatatgcag 19
<210> 47
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cGAF-primer
104/111
1 I
CA 02437866 2003-08-08
<400> 47
accactgtcc atgccatcac 20
<210> 48
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cGAR-primer
<400> 48
tccacaacac ggttgctgta 20
<210> 49
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:mBMAL2-F2
primer
<400> 49
tggttggatg cgaaagagg 19
<210> 50
<211> 21
<212> DNA
<213> Artificial Sequence
105/111
i
CA 02437866 2003-08-08
<220>
<223> Description of Artificial Sequence:mBMAL2-R4
primer
<400> 50
aggtttctct cttggtgaac c 21
<210> 51
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:rmBmall-F1
primer
<400> 51
tggtaccaac atgcaatgc 19
<210> 52
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:rmBmall-R1
primer
<400> 52
agtgtccgag gaagatagct g 21
106/111
CA 02437866 2003-08-08
<210> 53
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:rmPer2-F1
. primer
<400> 53
gctcactgcc agaactatct cc 22
<210> 54
<211> 22
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:rmPer2-R1
primer
<400> 54
cctctagctg aagcaggtta ag 22
<210> 55
<211> 21
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:rmClock-F1
107/111
CA 02437866 2003-08-08
primer
<400> 55
caaggtcagc aacttgtgac c 21
<210> 56
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:rmClock-R1
primer
<400> 56
aggatgagct gtgtcgaagg 20
<210> 57
<211> 20
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:mGAPDH F1
primer
<400> 57
catcaccatc ttccaggagc 20
<210> 58
<211> 19
108/111
CA 02437866 2003-08-08
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:mGAPDH-R1
primer
<400> 58
attgagagca atgccagcc 19
<210> 59
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cP2E1-S
<400> 59
gtgtcacacg tgaggctta 19
<210> 60
<211> 19
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:cP2El-AS
<400> 60
taagcctcac gtgtgacac 19
109/111
i i
CA 02437866 2003-08-08
<210> 61
<211> 29
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Sense primer 3
<400> 61
tcgagctctt tggtacctgg ccagcaacc 29
<210> 62
<211> 23
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:Antisense
primer 3
<400> 62
tcacgacacc tggccgttcg agg 23
<210> 63
<211> 41
<212> DNA
<213> Artificial Sequence
<220>
<223> Description of Artificial Sequence:TRE and
flanking sequence
110/111
CA 02437866 2003-08-08
<400> 63
cggctgactc atcaagctga ctcatcaagc tgactcatca a 41
111/111