Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02636876 2008-08-15
1
TITLE OF THE INVENTION
METHODS AND KITS FOR EXPANDING HEMATOPOIETIC STEM CELLS
CROSS REFERENCE TO RELATED APPLICATIONS
[0001] This application claims priority, under 35 U.S.C. 119(e), of U.S.
provisional
application serial No. 61/031,106 filed on February 25, 2008. All documents
above are
incorporated herein in their entirety by reference.
FIELD OF THE INVENTION
[0002] The present invention relates to hematopoietic stem cells (HSCs). More
specifically,
the present invention is concerned with methods and reagents for expanding
HSCs.
BACKGROUND OF THE INVENTION
[0003] The mature cell contingent of adult hematopoietic tissue is
continuously
replenished in the lifespan of an animal, due to periodical supplies from
hematopoietic
stem cells (HSC) that reside permanently in the niche. To maintain blood
homeostasis,
these primitive cells rely on two critical properties, namely multipotency and
self-renewal
(SR). The former enables differentiation into multiple lineages, the latter
ensures
preservation of fate upon cellular division. By definition, a self-renewal
division implies that
a HSC is permissive to cell cycle entry, while restrained from engaging in
differentiation,
apoptosis or senescence pathways. The transcriptional regulatory network of
HSC self-
renewal still remains largely undefined, an observation that contrasts with
that of
embryonic stem cells (ESC) for which self-renewal and pluripotency are
increasingly
dissected molecularly (1, 2). Only few nuclear factors have been documented as
inducers
of HSC expansion when overexpressed, i.e., Hoxb4 (3) and NF-Ya (4), or
activated, i.e., '&
catenin (5) and STAT5a (6). Of these factors, Hoxb4 and its derivatives
(Hoxa9, NA10HD)
are among the most potent and best documented (7, 8).
[0004] Hematopoietic stem cells (HSCs) are rare cells that have been
identified in fetal
bone marrow, umbilical cord blood, adult bone marrow, and peripheral blood,
which are
capable of differentiating into each of myeloerythroid (red blood cells,
granulocytes,
CA 02636876 2008-08-15
2
monocytes), megakaryocyte (platelets) and lymphoid (T-cells, B-cells, and
natural killer cells
lineages) cells. In addition these celis are long-lived, and are capable of
producing
additional stem cells (self-renewal). Stem cells initially undergo commitment
to lineage
restricted progenitor cells, which can be assayed by their ability to form
colonies in
semisolid media. Progenitor cells are restricted in their ability to undergo
multi-lineage
differentiation and have lost their ability to self-renew. Progenitor cells
eventually
differentiate and mature into each of the functional elements of the blood.
[0005] HSCs are used in clinical transplantation protocols to treat a variety
of diseases
including malignant and non-malignant disorders.
[0006] HSCs obtained directly from the patient (autologous HSCs) are used for
rescuing the
patient from the effects of high doses of chemotherapy or used as a target for
gene-therapy
vectors. HSCs obtained from another person (allogeneic HSCs) are used to treat
haematological malignancies by replacing the malignant haematopoietic system
with normal
cells. Allogeneic HSCs can be obtained from siblings (matched sibling
transplants), parents
or unrelated donors (mismatched unrelated donor transplants). About 45,000
patients each
year are treated by HSC transplantation. Although most of these cases have
involved
patients with haematological malignancies, such as lymphoma, myeloma and
leukemia,
there is growing interest in using HSC transplantation to treat solid tumours
and non-
malignant diseases. For example, erythrocyte disorders such as R-thalassaemia
and sickle-
cell anemia have been successfully treated by transplantation of allogeneic
HSCs.
[0007] Therefore, there is a need for novel methods and reagents for expanding
HSCs.
[0008] The present description refers to a number of documents, the content of
which is
herein incorporated by reference in their entirety.
SUMMARY OF THE INVENTION
[0009] Accordingly, the present invention concerns a novel in vitro--4n vivo
functional
screen which identified a series of HSC regulators (nuclear factors and
asymmetrical cell
division factors) which induce high levels of HSC activity similar to that
previously achieved
with Hoxb4. In total, 22 new determinants have emerged. Eleven of the 18
nuclear factors-
CA 02636876 2008-08-15
3
HSC regulators act in a cell autonomous manner, while the remaining 7 provide
a non-
autonomous influence on HSC activity. Clonal and phenotypic analyses of
hematopoietic
tissues derived from selected recipients confirmed that the majority of the
identified factors
induced HSC expansion in vitro without perturbing their differentiation in
vivo. Epistatic
analyses further revealed that 3 of the most potent candidates, namely Ski,
Prdm16 and
KIf10 may exploit both mechanisms. The present invention thus presents a novel
methodology to screen for determinants of HSC regulatorsas well and methods of
expanding and/or differentiating HSCs.
[0010] More specifically, in accordance with an aspect of the present
invention, there is
provided a method of increasing the expansion and/or differentiation of a
hematopoietic
stem cell (HSC) comprising: (a) increasing the level and/or activity of at
least one HSC
regulator polypeptide encoded by at least one HSC regulator gene selected from
trim27,
xbpl, sox4, smarccl, sfpi1, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10, zfp472,
ap2a2, gpsm2,
gpx3, erdr1, tmod1, pml, cnbp, prdm16, hdacl and ski, or a functional variant
of said
polypeptide, in said cell; (b) increasing the level of a nucleic acid encoding
the HSC
regulator polypeptide or functional variant of (a) in cell; or (c) any
combination of (a) and (b).
[0011] In a specific embodiment of the method, said at least one polypeptide
comprises the
amino acid sequence set forth in Genbank accession Nos: NP_006501 (SEQ ID NO:
2),
NP_005071 (SEQ ID NO: 4), NP_001073007 (SEQ ID NO: 6), NP_003098 (SEQ ID NO:
8),
NP_003065 (SEQ ID NO: 10), NP_001074016 (SEQ ID NO: 12), NP_003111 (SEQ ID NO:
14), NP_005243 (SEQ ID NO: 16), NP_002119 (SEQ ID NO: 18), NP_112740 (SEQ ID
NO:
20), NP_005988 (SEQ ID NO: 22), NP_036384 (SEQ ID NO: 58), NP001018068 (SEQ ID
NO: 60), NP_001027453 (SEQ ID NO: 62), NP_005646 (SEQ ID NO: 64), NP_694703
(SEQ ID NO: 70), NP_036437 (SEQ ID NO: 72), NP_037428 (SEQ ID NO: 74),
NP_002075
(SEQ ID NO: 76), NP_579940 (SEQ ID NO: 78), NP_003266 (SEQ ID NO: 80),
NP_003409
(SEQ ID NO: 82), NP_071397 (SEQ ID NO: 84), NP_955533 (SEQ ID NO: 86),
NP_004955
(SEQ ID NO: 88), NP_003027 (SEQ ID NO: 90), NP_777480 (SEQ ID NO: 24),
NP_775303
(SEQ ID NO: 26), NP_775301 (SEQ ID NO: 28), NP_775300 (SEQ ID NO: 30),
NP_733796
(SEQ ID NO: 32), NP_003235 (SEQ ID NO: 34), NP_775302 (SEQ ID NO: 36),
NP_775299
(SEQ ID NO: 38) NP_150253 (SEQ ID NO: 40), NP_150243 (SEQ ID NO: 42),
NP_150242
(SEQ ID NO: 44), NP_002666 (SEQ ID NO: 46), NP_150252 (SEQ ID NO: 48),
NP_150241
CA 02636876 2008-08-15
4
(SEQ ID NO: 50), NP_150247 (SEQ ID NO: 52), NP_150250 (SEQ ID NO: 54),
NP_150249
(SEQ ID NO: 56),SEQ ID NOs: 93, 94, 95, 96 or 97.
[0012] In a specific embodiment, the method comprises increasing the level of
said nucleic
acid in said cell. In another specific embodiment, said nucleic acid encodes a
HSC regulator
polypeptide comprising the amino acid sequence set forth in Genbank accession
Nos:
NP_006501 (SEQ ID NO: 2), NP_005071 (SEQ ID NO: 4), NP_001073007 (SEQ ID NO:
6),
NP_003098 (SEQ ID NO: 8), NP_003065 (SEQ ID NO: 10), NP_001074016 (SEQ ID NO:
12), NP_003111 (SEQ ID NO: 14), NP_005243 (SEQ ID NO: 16), NP_002119 (SEQ ID
NO:
18), NP_112740 (SEQ ID NO: 20), NP_005988 (SEQ ID NO: 22), NP_036384 (SEQ ID
NO:
58), NP_001018068 (SEQ ID NO: 60), NP_001027453 (SEQ ID NO: 62), NP_005646
(SEQ
ID NO: 64), NP_694703 (SEQ ID NO: 70), NP_036437 (SEQ ID NO: 72), NP_037428
(SEQ
ID NO: 74), NP_002075 (SEQ ID NO: 76), NP_579940 (SEQ ID NO: 78), NP_003266
(SEQ
ID NO: 80), NP_003409 (SEQ ID NO: 82), NP_071397 (SEQ ID NO: 84), NP_955533
(SEQ
ID NO: 86), NP_004955 (SEQ ID NO: 88), NP_003027 (SEQ ID NO: 90), NP_777480
(SEQ
ID NO: 24), NP_775303 (SEQ ID NO: 26), NP_775301 (SEQ ID NO: 28), NP_775300
(SEQ
ID NO: 30), NP_733796 (SEQ ID NO: 32), NP_003235 (SEQ ID NO: 34), NP_775302
(SEQ
ID NO: 36), NP_775299 (SEQ ID NO: 38) NP_150253 (SEQ ID NO: 40), NP_150243
(SEQ
ID NO: 42), NP_150242 (SEQ ID NO: 44), NP_002666 (SEQ ID NO: 46), NP_150252
(SEQ
ID NO: 48), NP_150241 (SEQ ID NO: 50), NP_150247 (SEQ ID NO: 52), NP_150250
(SEQ
ID NO: 54), NP_150249 (SEQ ID NO: 56),SEQ ID NOs: 93, 94, 95, 96 or 97.
[0013] In another specific embodiment, said nucleic acid comprises the coding
region of the
nucleotide sequence set forth in NM_006510 (SEQ ID NOs: 1), NM_005080 (SEQ ID
NOs:
3), NM_001079539 (SEQ ID NOs: 5), NM_003107 (SEQ ID NOs: 7), NM_003074 (SEQ ID
NOs: 9), NM_001080547 (SEQ ID NOs: 11), NM_003120 (SEQ ID NOs: 13), NM_005252
(SEQ ID NOs: 15), NM_002128 (SEQ ID NOs: 17), NM_031372 (SEQ ID NOs: 19),
NM_005997 (SEQ ID NOs: 21), NM_012252 (SEQ ID NOs: 57), NM_001018058 (SEQ ID
NOs: 59), NM_001032282 (SEQ ID NOs: 61), NM_005655 (SEQ ID NOs: 63), NM_153063
(SEQ ID NOs: 69), NM_012305 (SEQ ID NOs: 71), NM_013296 (SEQ ID NOs: 73),
NM_002084 (SEQ ID NOs: 75), NM_133362 (SEQ ID NOs: 77), NM_003275 (SEQ ID NOs:
79), NM_003418 (SEQ ID NOs: 81), NM_022114 (SEQ ID NOs: 83), NM_199454 (SEQ ID
NOs: 85), NM_004964 (SEQ ID NOs: 87), NM_003036 (SEQ ID NOs: 89), NM_174886
CA 02636876 2008-08-15
(SEQ ID NO: 23), NM_173211 (SEQ ID NO: 25), NM_173209 (SEQ ID NO: 27),
NM173208 (SEQ ID NO: 29), NM_170695 (SEQ ID NO: 31), NM_003244 (SEQ ID NO:
33), NM_173210 (SEQ ID NO: 35), NM_173207(SEQ ID NO: 37), NM_033250 (SEQ ID
NO: 39), NM_033240 (SEQ ID NO: 41), NM_033239 (SEQ ID NO: 43), NM_002675 (SEQ
ID NO: 45), NM033249 (SEQ ID NO: 47), NM_033238 (SEQ ID NO: 49), NM_033244
(SEQ ID NO: 51), NM_033247 (SEQ ID NO: 53) or NM_033246 (SEQ ID NO: 55).
[0014] In another specific embodiment, said differentiation is multilineage
differentiation and
said at least one HSC regulator gene is selected from trim27 (SEQ ID NO: 1),
xbpl (SEQ ID
NOs: 3 and 5), sox4 (SEQ ID NO: 7), hnrpdl (SEQ ID NO: 19), vps72 (SEQ ID NO:
21) and
gpx3 (SEQ ID NOs: 75 and 98).
[0015] In another specific embodiment, the method further comprises (a)
increasing the
level and/or activity of at least one further HSC regulator polypeptide; (b)
increasing the
level of a nucleic acid encoding the at least one further HSC regulator
polypeptide or
functional variant of (a) in said cell; or (c) any combination of (a) and (b).
In a specific
embodiment the further HSC regulator polypeptide is selected from Hoxb4,
Hoxa9, Bmil,
NF-YA, R-catenin and STAT5A. In a specific embodiment the HSC regulator
polypeptide
comprises a sequence as set forth in SEQ ID NO: 92 (Hoxb4), SEQ ID NO: 99
(Hoxa9),
SEQ ID NO: 101 (Bmil), SEQ ID NO: 103 (NF-YA), SEQ ID NO: 105 (R-catenin) or
SEQ ID
NO: 107 (STAT5A). In another specific embodiment, said further HSC regulator
polypeptide
is Hoxb4 and comprises the amino acid sequence set forth in Genbank accession
No:
NP_076920 (SEQ ID NO: 92).
[0016] In another specific embodiment, said expansion is multiclonal expansion
and said at
least one HSC regulator gene is selected from trim27, xbpl, sox4, smarccl,
hnrpdl, vps72,
kif10, ap2a2, gpsm2 and gpx3.
[0017] In another specific embodiment, the method comprises transfecting or
transforming
said cell with a vector comprising said nucleic acid. In another specific
embodiment, said
vector is a viral vector. In another specific embodiment, said viral vector is
an adenoviral
vector.
[0018] In accordance with another aspect of the present invention, there is
provided a use
CA 02636876 2008-08-15
6
of an agent capable of: (a) increasing the level and/or activity of at least
one HSC regulator
polypeptide encoded by at least one HSC regulator gene selected from trim27,
xbpl, sox4,
smarccl, sfpil, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10, zfp472, ap2a2, gpsm2,
gpx3, erdrl,
tmodl, cnbpl, prdm16, hdacl, pml and ski, or a functional variant of said
polypeptide; (b)
increasing the level of a nucleic acid encoding the at least one HSC regulator
polypeptide or
functional variant of (a); or (c) any combination of (a) and (b), for
increasing the expansion
and/or differentiation of a hematopoietic stem cell (HSC).
[0019] In accordance with another aspect of the present invention, there is
provided a use
of an agent capable of: increasing the level and/or activity of at least one
HSC regulator
polypeptide encoded by at least one HSC regulator gene selected from trim27,
xbpl, sox4,
smarccl, sfpil, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10, zfp472, ap2a2, gpsm2,
gpx3, erdrl,
tmodl, cnbpl, prdm16, hdacl and ski, or a functional variant of said
polypeptide, in a cell;
increasing the level of a nucleic acid encoding the at least one polypeptide
or functional
variant of (a) in a cell; or any combination of (a) and (b), for the
preparation of a
medicament for increasing the expansion and/or differentiation of a
hematopoietic stem cell
(HSC).
[0020] In a specific embodiment of the use, said polypeptide comprises the
amino acid
sequence set forth in Genbank accession Nos: NP_006501 (SEQ ID NO: 2),
NP_005071
(SEQ ID NO: 4), NP001073007 (SEQ ID NO: 6), NP_003098 (SEQ ID NO: 8),
NP_003065
(SEQ ID NO: 10), NP_001074016 (SEQ ID NO: 12), NP_003111 (SEQ ID NO: 14),
NP_005243 (SEQ ID NO: 16), NP_002119 (SEQ ID NO: 18), NP_112740 (SEQ ID NO:
20),
NP_005988 (SEQ ID NO: 22), NP_036384 (SEQ ID NO: 58), NP_001018068 (SEQ ID NO:
60), NP_001027453 (SEQ ID NO: 62), NP_005646 (SEQ ID NO: 64), NP_694703 (SEQ
ID
NO: 70), NP_036437 (SEQ ID NO: 72), NP_037428 (SEQ ID NO: 74), NP_002075 (SEQ
ID
NO: 76), NP_579940 (SEQ ID NO: 78), NP_003266 (SEQ ID NO: 80), NP_003409 (SEQ
ID
NO: 82), NP_071397 (SEQ ID NO: 84), NP_955533 (SEQ ID NO: 86), NP_004955 (SEQ
ID
NO: 88), NP_003027 (SEQ ID NO: 90), NP_777480 (SEQ ID NO: 24), NP_775303 (SEQ
ID
NO: 26), NP_775301 (SEQ ID NO: 28), NP_775300 (SEQ ID NO: 30), NP_733796 (SEQ
ID
NO: 32), NP_003235 (SEQ ID NO: 34), NP_775302 (SEQ ID NO: 36), NP_775299 (SEQ
ID
NO: 38) NP_150253 (SEQ ID NO: 40), NP_150243 (SEQ ID NO: 42), NP_150242 (SEQ
ID
NO: 44), NP_002666 (SEQ ID NO: 46), NP_150252 (SEQ ID NO: 48), NP_150241 (SEQ
ID
CA 02636876 2008-08-15
7
NO: 50), NP150247 (SEQ ID NO: 52), NP_150250 (SEQ ID NO: 54), NP_150249 (SEQ
ID
NO: 56),SEQ ID NOs: 93, 94, 95, 96, 97 or 98.
[0021] In another specific embodiment, said agent is capable of increasing the
level of said
nucleic acid in said cell. In another specific embodiment, said agent is a
nucleic acid
encoding at least one of trim27, xbpl, sox4, smarccl, sfpil, fos, hmgbl,
hnrpdl, vps72,
tcfec, kIf10, zfp472, ap2a2, gpsm2, gpx3, erdrl, tmodl, cnbpl, prdm16, hdacl,
pml and ski,
or a functional variant thereof. In another specific embodiment, said nucleic
acid encodes a
polypeptide comprising the amino acid sequence set forth in Genbank accession
Nos:
NP_006501 (SEQ ID NO: 2), NP_005071 (SEQ ID NO: 4), NP_001073007 (SEQ ID NO:
6),
NP_003098 (SEQ ID NO: 8), NP_003065 (SEQ ID NO: 10), NP_001074016 (SEQ ID NO:
12), NP003111 (SEQ ID NO: 14), NP_005243 (SEQ ID NO: 16), NP_002119 (SEQ ID
NO:
18), NP_112740 (SEQ ID NO: 20), NP_005988 (SEQ ID NO: 22), NP_036384 (SEQ ID
NO:
58), NP_001018068 (SEQ ID NO: 60), NP_001027453 (SEQ ID NO: 62), NP_005646
(SEQ
ID NO: 64), NP_694703 (SEQ ID NO: 70), NP_036437 (SEQ ID NO: 72), NP_037428
(SEQ
ID NO: 74), NP_002075 (SEQ ID NO: 76), NP_579940 (SEQ ID NO: 78), NP_003266
(SEQ
ID NO: 80), NP_003409 (SEQ ID NO: 82), NP_071397 (SEQ ID NO: 84), NP_955533
(SEQ
ID NO: 86), NP_004955 (SEQ ID NO: 88), NP_003027 (SEQ ID NO: 90), NP_777480
(SEQ
ID NO: 24), NP_775303 (SEQ ID NO: 26), NP_775301 (SEQ ID NO: 28), NP_775300
(SEQ
ID NO: 30), NP_733796 (SEQ ID NO: 32), NP_003235 (SEQ ID NO: 34), NP_775302
(SEQ
ID NO: 36), NP_775299 (SEQ ID NO: 38) NP_150253 (SEQ ID NO: 40), NP_150243
(SEQ
ID NO: 42), NP_150242 (SEQ ID NO: 44), NP_002666 (SEQ ID NO: 46), NP_150252
(SEQ
ID NO: 48), NP_150241 (SEQ ID NO: 50), NP_150247 (SEQ ID NO: 52), NP_150250
(SEQ
ID NO: 54), NP_150249 (SEQ ID NO: 56),SEQ ID NOs: 93, 94, 95, 96, 97 or 98.
[0022] In another specific embodiment, said nucleic acid comprises the coding
region of
nucleotide sequence set forth in Genbank accession Nos: NM_006510 (SEQ ID NOs:
1),
NM_005080 (SEQ ID NOs: 3), NM_001079539 (SEQ ID NOs: 5), NM_003107 (SEQ ID
NOs: 7), NM_003074 (SEQ ID NOs: 9), NM_001080547 (SEQ ID NOs: 11), NM_003120
(SEQ ID NOs: 13), NM_005252 (SEQ ID NOs: 15), NM_002128 (SEQ ID NOs: 17),
NM_031372 (SEQ ID NOs: 19), NM_005997 (SEQ ID NOs: 21), NM_012252 (SEQ ID NOs:
57), NM_001018058 (SEQ ID NOs: 59), NM_001032282 (SEQ ID NOs: 61), NM_005655
(SEQ ID NOs: 63), NM_153063 (SEQ ID NOs: 69), NM_012305 (SEQ ID NOs: 71),
CA 02636876 2008-08-15
8
NM_013296 (SEQ ID NOs: 73), NM_002084 (SEQ ID NOs: 75), NM_133362 (SEQ ID NOs:
77), NM_003275 (SEQ ID NOs: 79), NM_003418 (SEQ ID NOs: 81), NM_022114 (SEQ ID
NOs: 83), NM_199454 (SEQ ID NOs: 85), NM_004964 (SEQ ID NOs: 87), NM_003036
(SEQ ID NOs: 89), NM_174886 (SEQ ID NO: 23), NM_173211 (SEQ ID NO: 25),
NM173209 (SEQ ID NO: 27), NM_173208 (SEQ ID NO: 29), NM_170695 (SEQ ID NO:
31), NM_003244 (SEQ ID NO: 33), NM_173210 (SEQ ID NO: 35), NM_173207(SEQ ID
NO: 37), NM_033250 (SEQ ID NO: 39), NM_033240 (SEQ ID NO: 41), NM_033239 (SEQ
ID NO: 43), NM_002675 (SEQ ID NO: 45), NM_033249 (SEQ ID NO: 47), NM_033238
(SEQ ID NO: 49), NM_033244 (SEQ ID NO: 51), NM_033247 (SEQ ID NO: 53) or
NM_033246 (SEQ ID NO: 55).
[0023] In another specific embodiment, said differentiation is multilineage
differentiation and
said at least one HSC regulator gene is selected from trim27, xbpl, sox4,
hnrpdl, vps72 and
gpx3.
[0024] In another specific embodiment, said expansion is multicional expansion
and said at
least one HSC regulator gene is selected from trim27, xbpl, sox4, smarccl,
hnrpdl, vps72,
kIf10, ap2a2, gpsm2 and gpx3.
[0025] In another specific embodiment, said nucleic acid is comprised within a
vector. In
another specific embodiment, said vector is a viral vector. In another
specific embodiment,
said viral vector is an adenoviral vector.
[0026] In another specific embodiment, the use further comprises (a)
increasing the level
and/or activity of a further HSC regulator polypeptide encoded a further HSC
regulator
gene; (b) increasing the level of a nucleic acid encoding the further HSC
regulator
polypeptide or functional variant of (a) in said cell; or (c) any combination
of (a) and (b). In a
particular embodiment the further HSC regulator is selected from Hoxb4, Hoxa9,
Bmil, NF-
YA, R-catenin and STAT5A. In a specific embodiment, the HSC regulator nucleic
acid
comprises a sequence encoding the sequence as set forth in SEQ ID NO: 92
(Hoxb4), SEQ
ID NO: 100 (Hoxa9), SEQ ID NO: 102 (Bmil), SEQ ID NO: 104 (NF-YA), SEQ ID NO:
106
(0-catenin) or SEQ ID NO: 108 (STAT5A). In another specific embodiment, said
further
HSC regulator polypeptide is Hoxb4 and comprises the amino acid sequence set
forth in
CA 02636876 2008-08-15
9
Genbank accession No: NP_076920 (SEQ ID NO: 92).
[0027] In accordance with another aspect of the present invention, there is
provided a
composition for increasing the expansion and/or differentiation of a
hematopoietic stem cell
(HSC) comprising: (a) an agent capable of: (i) increasing the level and/or
activity of at least
one polypeptide encoded by at least one gene selected from trim27, xbpl, sox4,
smarccl,
sfpil, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10, zfp472, ap2a2, gpsm2, gpx3,
erdrl, tmodl,
cnbpl, prdm16, hdacl, pml and ski, or a functional variant of said
polypeptide, in a cell; (ii)
increasing the level of a nucleic acid encoding the at least one polypeptide
or functional
variant of (a) in a cell; or (iii) any combination of (i) and (ii); and (b) a
pharmaceutically
acceptable carrier or excipient.
[0028] In a specific embodiment, this use comprises (a) an agent capable of
increasing the
level of at least one nucleic acid encoding at least one of trim27, xbpl,
sox4, smarccl,
sfpil, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10, zfp472, ap2a2, gpsm2, gpx3,
erdrl, tmodl,
cnbpl, prdm16, hdacl, pml and ski; and (b) a pharmaceutically acceptable
carrier or
excipient.
[0029] In another specific embodiment, said agent is nucleic acid encoding at
least one of
trim27, xbpl, sox4, smarccl, sfpil, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10,
zfp472, ap2a2,
gpsm2, gpx3, erdrl, tmodl, cnbpl, prdm16, hdacl, pml and ski, or a functional
variant
thereof.
[0030] In another specific embodiment, said nucleic acid encodes a HSC
regulator
polypeptide comprising the amino acid sequence set forth in Genbank accession
Nos:
NP_006501 (SEQ ID NO: 2), NP_005071 (SEQ ID NO: 4), NP_001073007 (SEQ ID NO:
6),
NP003098 (SEQ ID NO: 8), NP_003065 (SEQ ID NO: 10), NP_001074016 (SEQ ID NO:
12), NP003111 (SEQ ID NO: 14), NP_005243 (SEQ ID NO: 16), NP_002119 (SEQ ID
NO:
18), NP_112740 (SEQ ID NO: 20), NP_005988 (SEQ ID NO: 22), NP_036384 (SEQ ID
NO:
58), NP_001018068 (SEQ ID NO: 60), NP_001027453 (SEQ ID NO: 62), NP_005646
(SEQ
ID NO: 64), NP_694703 (SEQ ID NO: 70), NP_036437 (SEQ ID NO: 72), NP_037428
(SEQ
ID NO: 74), NP_002075 (SEQ ID NO: 76), NP_579940 (SEQ ID NO: 78), NP_003266
(SEQ
ID NO: 80), NP_003409 (SEQ ID NO: 82), NP_071397 (SEQ ID NO: 84), NP_955533
(SEQ
ID NO: 86), NP_004955 (SEQ ID NO: 88), NP_003027 (SEQ ID NO: 90), NP_777480
(SEQ
CA 02636876 2008-08-15
ID NO: 24), NP_775303 (SEQ ID NO: 26), NP_775301 (SEQ ID NO: 28), NP_775300
(SEQ
ID NO: 30), NP_733796 (SEQ ID NO: 32), NP_003235 (SEQ ID NO: 34), NP_775302
(SEQ
ID NO: 36), NP_775299 (SEQ ID NO: 38) NP_150253 (SEQ ID NO: 40), NP_150243
(SEQ
ID NO: 42), NP_150242 (SEQ ID NO: 44), NP_002666 (SEQ ID NO: 46), NP_150252
(SEQ
ID NO: 48), NP_150241 (SEQ ID NO: 50), NP_150247 (SEQ ID NO: 52), NP_150250
(SEQ
ID NO: 54), NP_150249 (SEQ ID NO: 56),SEQ ID NOs: 93, 94, 95, 96, 97 or 98.
[0031] In another specific embodiment, said nucleic acid comprises the coding
region of the
nucleotide sequence set forth in Genbank accession Nos: NM_006510 (SEQ ID NOs:
1),
NM_005080 (SEQ ID NOs: 3), NM_001079539 (SEQ ID NOs: 5), NM_003107 (SEQ ID
NOs: 7), NM_003074 (SEQ ID NOs: 9), NM_001080547 (SEQ ID NOs: 11), NM_003120
(SEQ ID NOs: 13), NM_005252 (SEQ ID NOs: 15), NM_002128 (SEQ ID NOs: 17),
NM_031372 (SEQ ID NOs: 19), NM_005997 (SEQ ID NOs: 21), NM_012252 (SEQ ID NOs:
57), NM_001018058 (SEQ ID NOs: 59), NM_001032282 (SEQ ID NOs: 61), NM_005655
(SEQ ID NOs: 63), NM_153063 (SEQ ID NOs: 69), NM_012305 (SEQ ID NOs: 71),
NM_013296 (SEQ ID NOs: 73), NM_002084 (SEQ ID NOs: 75), NM_133362 (SEQ ID NOs:
77), NM_003275 (SEQ ID NOs: 79), NM_003418 (SEQ ID NOs: 81), NM_022114 (SEQ ID
NOs: 83), NM_199454 (SEQ ID NOs: 85), NM_004964 (SEQ ID NOs: 87), NM_003036
(SEQ ID NOs: 89), NM_174886 (SEQ ID NO: 23), NM_173211 (SEQ ID NO: 25),
NM_173209 (SEQ ID NO: 27), NM_173208 (SEQ ID NO: 29), NM_170695 (SEQ ID NO:
31), NM_003244 (SEQ ID NO: 33), NM_173210 (SEQ ID NO: 35), NM_173207(SEQ ID
NO: 37), NM_033250 (SEQ ID NO: 39), NM_033240 (SEQ ID NO: 41), NM_033239 (SEQ
ID NO: 43), NM_002675 (SEQ ID NO: 45), NM_033249 (SEQ ID NO: 47), NM_033238
(SEQ ID NO: 49), NM_033244 (SEQ ID NO: 51), NM_033247 (SEQ ID NO: 53) or
NM_033246 (SEQ ID NO: 55).
[0032] In another specific embodiment, said differentiation is multilineage
differentiation and
said at least one gene is selected from trim27, xbp1, sox4, hnrpdl, vps72 and
gpx3.
[0033] In another specific embodiment, said expansion is multiclonal expansion
and said at
least one gene is selected from trim27, xbp1, sox4, smarccl, hnrpdl, vps72,
kIf10, ap2a2,
gpsm2 and gpx3.
[0034] In another specific embodiment, said agent is a vector comprising said
nucleic acid.
CA 02636876 2008-08-15
11
In another specific embodiment, said vector is a viral vector. In another
specific
embodiment, said viral vector is an adenoviral vector.
[0035] In another specific embodiment, the composition comprises a further
agent capable
of:(a) increasing the level and/or activity of at least one further HSC
regulator polypeptide ;
(b) increasing the level of a nucleic acid encoding the HSC regulator
polypeptide or
functional variant of (a) in a cell; or (c) any combination of (a) and (b). In
another specific
embodiment said at least one further HSC regulator polypeptide is selected
from Hoxb4,
Hoxa9, Bmil, NF-YA, R-catenin and STAT5A. In another specific embodiment, said
further
agent is a Hoxb4 nucleic acid encoding the amino acid sequence set forth in
Genbank
accession No: NP_076920 (SEQ ID NO: 92).
[0036] In accordance with another aspect of the present invention, there is
provided an
hematopoietic stem cell transformed or transduced with a vector comprising a
nucleic acid
encoding at least one HSC regulator selected from trim27, xbpl, sox4, smarccl,
sfpil, fos,
hmgb1, hnrpdl, vps72, tcfec, kIf10, zfp472, ap2a2, gpsm2, gpx3, erdrl, tmod1,
cnbpl,
prdm16, hdac1, pml and ski, and a functional variant thereof.
[0037] In a specific embodiment of the cell, said nucleic acid encodes a HSC
regulator
polypeptide comprising the amino acid sequence set forth in Genbank accession
Nos:
NP_006501 (SEQ ID NO: 2), NP_005071 (SEQ ID NO: 4), NP_001073007 (SEQ ID NO:
6),
NP_003098 (SEQ ID NO: 8), NP_003065 (SEQ ID NO: 10), NP_001074016 (SEQ ID NO:
12), NP_003111 (SEQ ID NO: 14), NP_005243 (SEQ ID NO: 16), NP_002119 (SEQ ID
NO:
18), NP_112740 (SEQ ID NO: 20), NP_005988 (SEQ ID NO: 22), NP_036384 (SEQ ID
NO:
58), NP_001018068 (SEQ ID NO: 60), NP_001027453 (SEQ ID NO: 62), NP_005646
(SEQ
ID NO: 64), NP_694703 (SEQ ID NO: 70), NP_036437 (SEQ ID NO: 72), NP_037428
(SEQ
ID NO: 74), NP_002075 (SEQ ID NO: 76), NP_579940 (SEQ ID NO: 78), NP_003266
(SEQ
ID NO: 80), NP_003409 (SEQ ID NO: 82), NP_071397 (SEQ ID NO: 84), NP_955533
(SEQ
ID NO: 86), NP_004955 (SEQ ID NO: 88), NP_003027 (SEQ ID NO: 90), NP_777480
(SEQ
ID NO: 24), NP_775303 (SEQ ID NO: 26), NP_775301 (SEQ ID NO: 28), NP_775300
(SEQ
ID NO: 30), NP_733796 (SEQ ID NO: 32), NP_003235 (SEQ ID NO: 34), NP_775302
(SEQ
ID NO: 36), NP_775299 (SEQ ID NO: 38) NP_150253 (SEQ ID NO: 40), NP_150243
(SEQ
ID NO: 42), NP_150242 (SEQ ID NO: 44), NP_002666 (SEQ ID NO: 46), NP_150252
(SEQ
CA 02636876 2008-08-15
12
ID NO: 48), NP_150241 (SEQ ID NO: 50), NP_150247 (SEQ ID NO: 52), NP_150250
(SEQ
ID NO: 54), NP_150249 (SEQ ID NO: 56),SEQ ID NOs: 93, 94, 95, 96 97 or 98.
[0038] In another specific embodiment, said nucleic acid comprises the coding
region of the
nucleotide sequence set forth in Genbank accession Nos: NM_006510 (SEQ ID NOs:
1),
NM_005080 (SEQ ID NOs: 3), NM_001079539 (SEQ ID NOs: 5), NM_003107 (SEQ ID
NOs: 7), NM_003074 (SEQ ID NOs: 9), NM_001080547 (SEQ ID NOs: 11), NM_003120
(SEQ ID NOs: 13), NM_005252 (SEQ ID NOs: 15), NM_002128 (SEQ ID NOs: 17),
NM_031372 (SEQ ID NOs: 19), NM_005997 (SEQ ID NOs: 21), NM_012252 (SEQ ID NOs:
57), NM_001018058 (SEQ ID NOs: 59), NM_001032282 (SEQ ID NOs: 61), NM_005655
(SEQ ID NOs: 63), NM_153063 (SEQ ID NOs: 69), NM_012305 (SEQ ID NOs: 71),
NM_013296 (SEQ ID NOs: 73), NM_002084 (SEQ ID NOs: 75), NM_133362 (SEQ ID NOs:
77), NM_003275 (SEQ ID NOs: 79), NM_003418 (SEQ ID NOs: 81), NM_022114 (SEQ ID
NOs: 83), NM_199454 (SEQ ID NOs: 85), NM_004964 (SEQ ID NOs: 87), NM_003036
(SEQ ID NOs: 89), NM_174886 (SEQ ID NO: 23), NM_173211 (SEQ ID NO: 25),
NM_173209 (SEQ ID NO: 27), NM_173208 (SEQ ID NO: 29), NM_170695 (SEQ ID NO:
31), NM_003244 (SEQ ID NO: 33), NM_173210 (SEQ ID NO: 35), NM_173207(SEQ ID
NO: 37), NM_033250 (SEQ ID NO: 39), NM_033240 (SEQ ID NO: 41), NM_033239 (SEQ
ID NO: 43), NM_002675 (SEQ ID NO: 45), NM_033249 (SEQ ID NO: 47), NM_033238
(SEQ ID NO: 49), NM_033244 (SEQ ID NO: 51), NM_033247 (SEQ ID NO: 53) or
NM_033246 (SEQ ID NO: 55).
[0039] In another specific embodiment, said vector is a viral vector. In
another specific
embodiment, said viral vector is an adenoviral vector. In another specific
embodiment, the
vector further comprises a nucleic acid encoding a further HSC regulator
selected from
Hoxb4, Hoxa9, Bmi1, NF-YA, R-catenin and STAT5A. In another specific
embodiment, the
further HSC regulator is Hoxb4. In another specific embodiment, said nucleic
acid encodes
a Hoxb4 polypeptide comprising the amino acid sequence set forth in Genbank
accession
No: NP_076920 (SEQ ID NO: 92).
[0040] In accordance with another aspect of the present invention, there is
provided a
method for increasing the number of blood cells in a subject comprising
administering to
said subject the hematopoietic stem cell of the present invention.
CA 02636876 2008-08-15
13
[0041] In accordance with another aspect of the present invention, there is
provided a
method for reconstituting the hematopoietic system or tissue of a subject
comprising
administering to said subject the hematopoietic stem cell of the present
invention.
[0042] In accordance with another aspect of the present invention, there is
provided a use
of the hematopoietic stem cell of the present invention for hematopoietic stem
cell
transplantation.
[0043] In accordance with another aspect of the present invention, there is
provided a use
of the hematopoietic stem cell of the present invention for reconstituting the
hematopoietic
system or tissue of a subject.
[0044] In accordance with another aspect of the present invention, there is
provided a use
of the hematopoietic stem cell of the present invention for the preparation of
a medicament
for reconstituting the hematopoietic system or tissue of a subject.
[0045] In accordance with another aspect of the present invention, there is
provided a use
of the hematopoietic stem cell of the present invention for increasing the
number of blood
cells in a subject.
[0046] In accordance with another aspect of the present invention, there is
provided a use
of the hematopoietic stem cell of the present invention for the preparation of
a medicament
for increasing the number of blood cells in a subject.
[0047] In accordance with another aspect of the present invention, there is
provided a
method for increasing the number of blood cells in a subject comprising
administering to
said subject the composition of the present invention.
[0048] In accordance with another aspect of the present invention, there is
provided a
method for reconstituting the hematopoietic system or tissue of a subject
comprising
administering to said subject the composition of the present invention.
[0049] In accordance with another aspect of the present invention, there is
provided a use
of the composition of the present invention for hematopoietic stem cell
transplantation.
[0050] In accordance with another aspect of the present invention, there is
provided a use
CA 02636876 2008-08-15
14
of the composition of the present invention for reconstituting the
hematopoietic system or
tissue of a subject.
[0051] In accordance with another aspect of the present invention, there is
provided a use
of the composition of the present invention for the preparation of a
medicament for
reconstituting the hematopoietic system or tissue of a subject.
[0052] In accordance with another aspect of the present invention, there is
provided a use
of the composition of the present invention for increasing the number of blood
cells in a
subject.
[0053] In accordance with another aspect of the present invention, there is
provided a use
of the composition of the present invention for the preparation of a
medicament for
increasing the number of blood cells in a subject.
[0054] In accordance with another aspect of the present invention, there is
provided a
method of increasing the expansion and/or differentiation of a hematopoietic
stem cell
(HSC) comprising: (a) increasing the level and/or activity of at least one HSC
regulator
polypeptide encoded by at least one HSC regulator gene selected from erdrl,
tmodl,
cnbpl, prdm16, hdacl and ski, or a functional variant of said polypeptide, in
said cell; (b)
increasing the level of at least one nucleic acid encoding the at least one
polypeptide or
functional variant of (a) in said cell; or (c) any combination of (a) and (b).
[0055] In accordance with another aspect of the present invention, there is
provided an
hematopoietic stem cell transformed or transduced with a vector comprising a
nucleic acid
encoding at least one of erdrl, tmodl, cnbpl, prdm16, hdacl and ski, or a
functional variant
thereof.
[0056] As used herein, "expansion" and "self-renewal" are used interchangeably
and refer
to the propagation of a cell or cells without terminal differentiation and
"differentiation" refers
to the developmental process of lineage commitment. A "lineage" refers to a
pathway of
cellular development, in which precursor or "progenitor" cells undergo
progressive
physiological changes to become a specified cell type having a characteristic
function (e.g.,
a T cell, a macrophage). Differentiation occurs in stages, whereby cells
gradually become
CA 02636876 2008-08-15
more specified until they reach full maturity.
[0057] Accordingly, the methods of the invention can be used to treat a
disease or disorder
in which it is desirable to increase the number of HSCs or their progenitors.
Frequently,
subjects in need of the inventive treatment methods will be those undergoing
or expecting
to undergo a blood cell (e.g., an immune cell) depleting treatment, such as
chemotherapy.
[0058] Thus, methods of the invention can be used, for example, to treat
patients requiring
a bone marrow transplant or a hematopoietic stem cell transplant (e.g., to
reconstitute the
hematopoietic system/tissue), such as cancer patients undergoing chemo and/or
radiation
therapy. Disorders treated by methods of the invention can be the result of an
undesired
side effect or complication of another primary treatment, such as radiation
therapy,
chemotherapy, or treatment with a bone marrow suppressive drug. Methods of the
invention
can further be used as a means to increase the number of mature cells derived
from HSCs
(e.g., erythrocytes, lymphocytes). For example, disorders or diseases
characterized by a
lack of, or low levels of, blood cells, or a defect in blood cells, can be
treated by increasing
the pool of HSCs. Such conditions include, for example, thrombocytopenia,
anemias and
lymphopenia. The disorder to be treated may also be the result of an infection
causing
damage to blood/lymphoid cells and/or stem cells.
[0059] Hematopoietic stem cell progenitors include virtually any cell capable
of giving rise to
a hematopoietic stem cell (e.g., mesenchymal stem cells, embryonic stem
cells). The
hematopoietic stem cell, which may be isolated from bone marrow, blood,
umbilical cord
blood, peripheral blood, fetal liver and yolk sac for example, is the
progenitor cell that
generates blood cells or following transplantation reinitiates multiple
hematopoietic lineages
and can reinitiate hematopoiesis for the life of a recipient. When
transplanted into lethally
irradiated subjects (e.g., animals, humans), hematopoietic stem cells can
repopulate the
erythroid, neutrophil-macrophage, megakaryocyte and/or lymphoid hematopoietic
cell pool.
[0060] It is well known in the art that hematopoietic cells include
pluripotent stem cells,
multipotent progenitor cells (e.g., a Iymphoid stem cell), and/or progenitor
cells committed to
specific hematopoietic lineages. The progenitor cells committed to specific
hematopoietic
lineages maybe of T cell lineage, B cell lineage, dendritic cell lineage,
Langerhans cell
lineage and/or lymphoid tissue-specific macrophage cell lineage. Where the
stem cells to be
CA 02636876 2008-08-15
16
provided to a subject in need of such treatment are hematopoietic stem cells,
they are most
commonly obtained from the bone marrow of the subject (autologous) or a
compatible
donor (heterologous). Bone marrow cells can be easily isolated using methods
known in the
art.
[0061] Hematopoietic stem cells can also be obtained from biological samples
(e.g., blood
products). A "blood product" as used in the present invention defines a
product obtained
from the body or an organ of the body containing cells of hematopoietic
origin. Such
sources include unfractionated bone marrow, umbilical cord, peripheral blood,
liver such as
fetal liver, thymus, lymph, spleen and yolk sac. It will be apparent to those
of ordinary skill in
the art that all of the aforementioned crude or unfractionated blood products
can be
enriched for cells having "hematopoietic stem cell" characteristics in a
number of ways. For
example, the blood product can be depleted from the more differentiated
progeny. The
more mature, differentiated cells can be selected against, via cell surface
molecules they
express (e.g., by FACS). Unfractionated blood products can be obtained
directly from a
donor or retrieved from cryopreservative storage.
[0062] Once obtained from a desired source, contacting of HSCs with a
polypeptide and/or
nucleic acid molecule and/or agent may, if desired, occur in culture (e.g., ex
vivo or in vitro).
Employing the polypeptides or nucleic acid molecules of the present invention,
it is possible
to stimulate the expansion and/or differentiation of hematopoietic stem cells.
The media
used is that which is conventional for culturing cells. Appropriate culture
media can be a
chemically defined serum-free media, such as the chemically defined media
RPMI, DMEM,
Iscove's, etc or so-called "complete media". Typically, serum-free media are
supplemented
with human or animal plasma or serum. Such plasma or serum can contain small
amounts
of hematopoietic growth factors. If desired, a hematopoietic or other stem
cell may be
treated with additional agents that promote stem cell maintenance and
expansion. It is well
within the level of ordinary skill in the art for practitioners to vary the
parameters
accordingly. The growth agents of particular interest in connection with the
present
invention are hematopoietic growth factors. By hematopoietic growth factors,
it is meant
factors that influence the survival or proliferation of hematopoietic stem
cells. Growth agents
that affect only survival and proliferation, but are not believed to promote
differentiation,
include the interleukins 3, 6 and 11, stem cell factor and FLT- 3 ligand. The
foregoing
CA 02636876 2008-08-15
17
factors are well known to those of ordinary skill in the art and most are
commercially
available. They can be obtained by purification, by recombinant methodologies
or can be
derived or synthesized synthetically.
[0063] By the term "HSC regulator polypeptide" is meant to include any
polypeptide of the
present invention which increases directly or indirectly (e.g., cell-
autonomous vs non-cell
autonomous) HSC expansion and/or differentiation. These include trim27, xbpl,
sox4,
smarccl, sfpil, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10, zfp472, ap2a2, gpsm2,
gpx3, erdrl,
tmodl, pml, cnbp, prdm16, hdacl and ski, or a functional variant of thereof.
In a specific
embodiment, the HSC regulator polypeptide of the present invention comprise a
sequence
comprise a sequence as set forth in SEQ ID NOs: 2, 4, 6, 8, 10, 12, 14, 16,
18, 20, 22, 24,
26, 28, 30, 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, 62,
64, 66, 68, 70, 72,
74, 76, 78, 80, 84, 86, 88, 90, and 98) Similarly, the term "HSC regulator
gene" or "HSC
regulator nucleic acid" includes any gene or nucleic acid which when expressed
in cells
increases directly or indirectly (e.g., cell-autonomous vs non-cell
autonomous) HSC
expansion and/or differentiation. These include nucleic acids encoding trim27,
xbpl, sox4,
smarccl, sfpil, fos, hmgbl, hnrpdl, vps72, tcfec, kIf10, zfp472, ap2a2, gpsm2,
gpx3, erdrl,
tmodl, pml, cnbp, prdm16, hdacl and ski, or a functional variant of thereof.
In a specific
embodiment, HSC regulator nucleic acids of the present invention comprise a
sequence as
se forth in SEQ ID NOs: 1, 3, 5, 7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29,
31, 33, 35, 57,
39, 41, 43, 45, 47, 49, 51, 53, 55, 57, 59, 61, 63, 65, 67, 69, 71, 73, 75,
77, 79, 81, 83, 85,
87 or 89).
[0064] Thus, the present invention includes HSC regulator polypeptides having
altered
amino acid sequences (e.g., functional variants) as compared to those of the
"natural" or
"wild-type" polypeptides due to the artificial or natural substitution,
deletion, addition, and/or
insertion of amino acids as long as they have the activity of the natural
polypeptides (i.e.,
can promote the expansion and/or differentiation of a HSC). Preferably, an
amino acid can
be substituted with the one having similar property to that of the amino acid
to be
substituted. It has been shown that recombinant TAT-HOXB4 protein, when added
to the
HSC culture, could penetrate the cell membrane and provides significant HSC
expansion
stimuli ((24); US 2004/0082003) and similar effect of stroma cell derived
HOXB4 on human
HSC has also been reported (10). Human HSCs, assessed with NOD/SCID SRC assay,
CA 02636876 2008-08-15
18
can be efficiently and significantly expanded ex vivo using TAT-HOXB4 protein
(11). The
present invention thus encompasses recombinant polypeptides comprising a
protein
encoded by the genes of Table II below or functional variants thereof and a
motif enhancing
penetration of the protein into the HSC cell membranes, and their use for
administration to
HSC culture.
[0065] The present invention also includes polypeptides variants comprising an
amino acid
sequence having at least 50% identity, preferably at least 60%, preferably at
least 75%
identity, more preferably at least 90%; at least 95% and at least 98% identity
to the
polypeptides of the present invention (e.g., polypeptides comprising the
sequence set forth
in NP_006501 (SEQ ID NO: 2), NP_005071 (SEQ ID NO: 4), NP_001073007 (SEQ ID
NO:
6), NP_003098 (SEQ ID NO: 8), NP_003065 (SEQ ID NO: 10), NP_001074016 (SEQ ID
NO: 12), NP_003111 (SEQ ID NO: 14), NP_005243 (SEQ ID NO: 16), NP_002119 (SEQ
ID
NO: 18), NP_112740 (SEQ ID NO: 20), NP_005988 (SEQ ID NO: 22), NP_036384 (SEQ
ID
NO: 58), NP_001018068 (SEQ ID NO: 60), NP_001027453 (SEQ ID NO: 62), NP_005646
(SEQ ID NO: 64), NP_694703 (SEQ ID NO: 70), NP_036437 (SEQ ID NO: 72),
NP_037428
(SEQ ID NO: 74), NP_002075 (SEQ ID NO: 76), NP_579940 (SEQ ID NO: 78),
NP003266
(SEQ ID NO: 80), NP_003409 (SEQ ID NO: 82), NP_071397 (SEQ ID NO: 84),
NP_955533
(SEQ ID NO: 86), NP_004955 (SEQ ID NO: 88), NP_003027 (SEQ ID NO: 90),
NP_777480
(SEQ ID NO: 24), NP_775303 (SEQ ID NO: 26), NP_775301 (SEQ ID NO: 28),
NP_775300
(SEQ ID NO: 30), NP_733796 (SEQ ID NO: 32), NP_003235 (SEQ ID NO: 34),
NP_775302
(SEQ ID NO: 36), NP_775299 (SEQ ID NO: 38) NP_150253 (SEQ ID NO: 40),
NP_150243
(SEQ ID NO: 42), NP_150242 (SEQ ID NO: 44), NP_002666 (SEQ ID NO: 46),
NP_150252
(SEQ ID NO: 48), NP_150241 (SEQ ID NO: 50), NP_150247 (SEQ ID NO: 52),
NP_150250
(SEQ ID NO: 54), NP_150249 (SEQ ID NO: 56),SEQ ID NOs: 93, 94, 95, 96 97 or
98.
[0066] The term functional variants also includes fragment of the polypeptides
of the
invention. Such fragments may be truncated at the N-terminus or C-terminus, or
may lack
internal residues, for example, when compared with a full length native
polypeptide. Certain
fragments lack amino acid residues that are not essential for a desired
biological activity of
the polypeptides. For examples, when several functional variants of a
polypeptide exists,
one skilled in the art can readily identify residues which are not essential
for a given
biological activity by aligning the variants and identifying the residues
which are different
CA 02636876 2008-08-15
19
(see for example Figure 28). Alternatively, residues that can be modified
without affecting
the biological activity of a gene can be identified by comparing the
polypeptide sequences
of several species (e.g., mouse, rats, human, pigs, primates, cats dogs, cows
etc) and
determining the residues which are different. Residues which are not conserved
between
the species are those that are likely not to affect the biological activity of
the gene if
modified. When relating to a protein sequence, the substituting amino acid
generally has
chemico-physical properties which are similar to that of the substituted amino
acid. The
similar chemico-physical properties include, similarities in charge,
bulkiness, hydrophobicity,
hydrophylicity and the like as well known by the skilled artisan.
[0067] Preferred variants of the present invention are those which retain
their biological
activity (e.g., promoting expansion/self-renewal and/or differentiation into
blood cells) and
whose nucleic acid sequence can specifically hybridize under high stringency
conditions to
HSC regulator nucleic acid sequences of the present invention (e.g., SEQ ID
NOs: 1, 3, 5,
7, 9, 11, 13, 15, 17, 19, 21, 23, 25, 27, 29, 31, 33, 35, 57, 39, 41, 43, 45,
47, 49, 51, 53, 55,
57, 59, 61, 63, 65, 67, 69, 71, 73, 75, 77, 79, 81, 83, 85, 87 and ,89).
Hybridization to filter-
bound sequences under stringent conditions may, for example, be performed in
0.5 M
NaHPO4, 7% SDS, 1 mM EDTA at 65 C, and washing in 0.1 x SSC/0.1% SDS at 68 C
(see Ausubel, et al. (eds), 1989). Hybridization conditions may be modified in
accordance
with known methods depending on the sequence of interest (28). Generally,
stringent
conditions are selected to be about 5 C lower than the thermal melting point
for the specific
sequence at a defined ionic strength and pH.
[0068] The present invention also relates to a nucieic acid molecule encoding
the above-
mentioned polypeptides or functional variants thereof. The type of the nucleic
acid molecule
encoding the polypeptides of this invention is not limited as long as they are
capable of
encoding the polypeptides, and includes cDNA, genomic DNA, RNA (e.g., mRNA),
synthetic
or recombinantly produced nucleic acid, and nucleic acids comprising
nucleotide sequences
resulted from the degeneracy of genetic codes, all of which can be prepared by
methods
that are well-known in the art. The nucleic acid molecules of the present
invention also
encompass those having nucleotide sequences altered from those of the natural
nucleic
acids due to the insertions, deletions, or substitutions of nucleotide, as
long as the
polypeptides encoded by these altered nucleic acids encode polypeptides having
the
CA 02636876 2008-08-15
activity of the natural polypeptides (e.g., promoting expansion or
differentiation of HSCs).
[0069] In an embodiment, the above-mentioned nucleic acid encodes a
polypeptide
comprising the sequence set forth in NP_006501 (SEQ ID NO: 2), NP_005071 (SEQ
ID
NO: 4), NP_001073007 (SEQ ID NO: 6), NP_003098 (SEQ ID NO: 8), NP_003065 (SEQ
ID
NO: 10), NP_001074016 (SEQ ID NO: 12), NP_003111 (SEQ ID NO: 14), NP_005243
(SEQ ID NO: 16), NP_002119 (SEQ ID NO: 18), NP_112740 (SEQ ID NO: 20),
NP_005988
(SEQ ID NO: 22), NP_036384 (SEQ ID NO: 58), NP_001018068 (SEQ ID NO: 60),
NP001027453 (SEQ ID NO: 62), NP005646 (SEQ ID NO: 64), NP_694703 (SEQ ID NO:
70), NP_036437 (SEQ ID NO: 72), NP_037428 (SEQ ID NO: 74), NP_002075 (SEQ ID
NO:
76), NP_579940 (SEQ ID NO: 78), NP_003266 (SEQ ID NO: 80), NP_003409 (SEQ ID
NO:
82), NP_071397 (SEQ ID NO: 84), NP_955533 (SEQ ID NO: 86), NP_004955 (SEQ ID
NO:
88), NP_003027 (SEQ ID NO: 90), NP_777480 (SEQ ID NO: 24), NP_775303 (SEQ ID
NO:
26), NP_775301 (SEQ ID NO: 28), NP_775300 (SEQ ID NO: 30), NP_733796 (SEQ ID
NO:
32), NP_003235 (SEQ ID NO: 34), NP_775302 (SEQ ID NO: 36), NP_775299 (SEQ ID
NO:
38) NP_150253 (SEQ ID NO: 40), NP_150243 (SEQ ID NO: 42), NP_150242 (SEQ ID
NO:
44), NP_002666 (SEQ ID NO: 46), NP_150252 (SEQ ID NO: 48), NP_150241 (SEQ ID
NO:
50), NP_150247 (SEQ ID NO: 52), NP_150250 (SEQ ID NO: 54), NP_150249 (SEQ ID
NO:
56),SEQ ID NOs: 93, 94, 95, 96 97 or 98.
[0070] In a further embodiment, the above-mentioned nucleic acid comprises the
coding
region of nucleotide sequence set forth in Genbank accession Nos: NM_006510
(SEQ ID
NOs: 1), NM_005080 (SEQ ID NOs: 3), NM_001079539 (SEQ ID NOs: 5), NM_003107
(SEQ ID NOs: 7), NM_003074 (SEQ ID NOs: 9), NM_001080547 (SEQ ID NOs: 11),
NM_003120 (SEQ ID NOs: 13), NM_005252 (SEQ ID NOs: 15), NM_002128 (SEQ ID NOs:
17), NM_031372 (SEQ ID NOs: 19), NM_005997 (SEQ ID NOs: 21), NM_012252 (SEQ ID
NOs: 57), NM_001018058 (SEQ ID NOs: 59), NM_001032282 (SEQ ID NOs: 61),
NM_005655 (SEQ ID NOs: 63), NM_153063 (SEQ ID NOs: 69), NM_012305 (SEQ ID NOs:
71), NM_013296 (SEQ ID NOs: 73), NM_002084 (SEQ ID NOs: 75), NM_133362 (SEQ ID
NOs: 77), NM_003275 (SEQ ID NOs: 79), NM_003418 (SEQ ID NOs: 81), NM_022114
(SEQ ID NOs: 83), NM_199454 (SEQ ID NOs: 85), NM_004964 (SEQ ID NOs: 87),
NM_003036 (SEQ ID NOs: 89), NM_174886 (SEQ ID NO: 23), NM_173211 (SEQ ID NO:
25), NM_173209 (SEQ ID NO: 27), NM_173208 (SEQ ID NO: 29), NM_170695 (SEQ ID
CA 02636876 2008-08-15
21
NO: 31), NM_003244 (SEQ ID NO: 33), NM_173210 (SEQ ID NO: 35), NM_173207(SEQ
ID NO: 37), NM_033250 (SEQ ID NO: 39), NM_033240 (SEQ ID NO: 41), NM033239
(SEQ ID NO: 43), NM_002675 (SEQ ID NO: 45), NM_033249 (SEQ ID NO: 47),
NM_033238 (SEQ ID NO: 49), NM_033244 (SEQ ID NO: 51), NM_033247 (SEQ ID NO:
53) or NM_033246 (SEQ ID NO: 55).
[0071] The nucleic acid molecules encoding the above-mentioned polypeptides
may also
be applied to the gene therapy of disorders caused by lack of expression of
the
polypeptides (e.g., a disease or condition associated with altered expansion
and/or
differentiation of HSCs), or in gene therapy applications where expansion
and/or
differentiation of HSCs is desirable (e.g., bone marrow/stem cell
transplantion). Examples of
vectors used for the gene therapy are viral vectors such as retroviral vector,
adenoviral
vector, adeno-associated viral vector, vaccinia viral vector, lentiviral
vector, herpes viral
vector, alphaviral vector, EB viral vector, papillomaviral vector, and
foamyviral vector, and
non-viral vector such as cationic liposome, ligand DNA complex, and gene gun.
Gene
transduction may be carried out in vivo and ex vivo, and also co-transduction
with one or
more gene of interest may be carried out. In an embodiment, the above-
mentioned gene
transduction is performed ex vivo and the transduced cells (i.e., expressing
one or more of
the polypeptide(s)) are administered to a subject.
[0072] Hematopoietic stem cells, progenitor cells, or a mixture comprising
such cell types
may be administered to a subject according to methods known in the art. Such
compositions may be administered by any conventional route, including
injection or by
gradual infusion over time. The administration may, depending on the
composition being
administered, for example, be, pulmonary, intravenous, intraperitoneal,
intramuscular,
intracavity, subcutaneous, or transdermal. The stem cells are administered in
"effective
amounts", or the amounts that either alone or together with further doses
produce the
desired therapeutic response. Administered cells of the invention can be
autologous ("self')
or heterologous/non-autologous ("non-self," e.g., allogeneic, syngeneic or
xenogeneic).
Generally, administration of the cells can occur within a short period of time
following the
induction of an increase in polypeptide activity/expression (or of increase in
expression of a
nucleic acid encoding the polypeptide (e.g., 1, 2, 5, 10, 24, 48 hours, 1 week
or 2 weeks
after the induction/increase)) and according to the requirements of each
desired treatment
CA 02636876 2008-08-15
22
regimen. For example, where radiation or chemotherapy is conducted prior to
administration, treatment, and transplantation of stem cells of the invention
should optimally
be provided within about one month of the cessation of therapy. However,
transplantation at
later points after treatment has ceased can be done with derivable clinical
outcomes.
[0073] Following harvest and treatment with a suitable agent, polypeptide or
nucleic acid,
hematopoietic stem cells or their progenitors, or a mixture of cells that
include these cells
may be combined with pharmaceutical carriers/excipients known in the art to
enhance
preservation and maintenance of the cells prior to administration. In some
embodiments,
cell compositions of the invention can be conveniently provided as sterile
liquid
preparations, e.g., isotonic aqueous solutions, suspensions, emulsions,
dispersions, or
viscous compositions, which may be buffered to a selected pH. Liquid
preparations are
normally easier to prepare than gels, other viscous compositions, and solid
compositions.
Additionally, liquid compositions are somewhat more convenient to administer,
especially by
injection. Viscous compositions, on the other hand, can be formulated within
the appropriate
viscosity range to provide longer contact periods with specific tissues.
Liquid or viscous
compositions can comprise carriers, which can be a solvent or dispersing
medium
containing, for example, water, saline, phosphate buffered saline, polyol (for
example,
glycerol, propylene, glycol, liquid polyethylene glycol, and the like) and
suitable mixtures
thereof.
[0074] Sterile injectable solutions can be prepared by incorporating the cells
utilized in
practicing the present invention in the required amount of the appropriate
solvent with
various amounts of the other ingredients, as desired. Such compositions may be
in
admixture with a suitable carrier, diluent, or excipient such as sterile
water, physiological
saline, glucose, dextrose, or the like. The compositions can also be
lyophilized. The
compositions can contain auxiliary substances such as wetting, dispersing, or
emulsifying
agents (e.g., methylcellulose), pH buffering agents, gelling or viscosity
enhancing additives,
preservatives, flavoring agents, colors, and the like, depending upon the
route of
administration and the preparation desired. Standard texts, such as
"Remington's
Pharmaceutical Science", 17th edition, 1985, incorporated herein by reference,
may be
consulted to prepare suitable preparations, without undue experimentation.
CA 02636876 2008-08-15
23
[0075] Various additives which enhance the stability and sterility of the
compositions,
including antimicrobial preservatives, antioxidants, chelating agents, and
buffers, can be
added. Prevention of the action of microorganisms can be ensured by various
antibacterial
and antifungal agents, for example, parabens, chlorobutanol, phenol, sorbic
acid, and the
like.
[0076] The compositions can be isotonic, i.e., they can have the same osmotic
pressure as
blood and lacrimal fluid. The desired isotonicity of the compositions of this
invention may be
accomplished using sodium chloride, or other pharmaceutically acceptable
agents such as
dextrose, boric acid, sodium tartrate, propylene glycol or other inorganic or
organic solutes.
Sodium chloride is preferred particularly for buffers containing sodium ions.
[0077] A method to potentially increase cell survival when introducing the
cells (e.g., the
HSCs) into a subject in need thereof is to incorporate the cells of interest
into a biopolymer
or synthetic polymer. Depending on the subject's condition, the site of
injection might prove
inhospitable for cell seeding and growth because of scarring or other
impediments.
Examples of biopolymer include, but are not limited to, cells mixed with
fibronectin, fibrin,
fibrinogen, thrombin, collagen, and proteoglycans. This could be constructed
with or without
included expansion or differentiation factors. Additionally, these could be in
suspension, but
residence time at sites subjected to flow would be nominal. Another
alternative is a three-
dimensional gel with cells entrapped within the interstices of the cell
biopolymer admixture.
Again, expansion or differentiation factors could be included with the cells.
These could be
deployed by injection via various routes described herein. Those skilled in
the art will
recognize that the components of the compositions should be selected to be
chemically
inert and will not affect the viability or efficacy of the stem cells or their
progenitors as
described in the present invention.
[0078] The quantity of cells to be administered will vary for the subject
being treated. The
precise determination of what would be considered an effective dose may be
based on
factors individual to each patient, including their size, age, sex, weight,
and condition of the
particular patient. As few as 100-1000 cells can be administered for certain
desired
applications among selected patients. Therefore, dosages can be readily
ascertained by
those skilled in the art from this disclosure and the knowledge in the art.
The skilled artisan
CA 02636876 2008-08-15
24
can readily determine the amount of cells and optional additives, vehicles,
and/or carrier in
compositions and to be administered in methods of the invention.
[0079] The pharmaceutical composition of the present invention (e.g.,
comprising an agent
capable of increasing the expression and/or activity of at least one
polypeptide encoded by
at least one gene selected from trim27, xbpl, pml, sox4, smarccl, sfpil, fos,
hmgbl, hnrpdl,
vps72, tcfec, kIf10, zfp472, ap2a2, pml, gpsm2, gpx3, erdrl, tmodl, cnbpl,
prdm16, hdacl
and ski) is administered in a manner compatible with the dosage formulation,
and in a
therapeutically effective amount, for example intravenously,
intraperitoneally,
intramuscularly, subcutaneously, and intradermally. It may also be
administered by any of
the other numerous techniques known to those of skill in the art, see for
example the latest
edition of Remington's Pharmaceutical Science, the entire teachings of which
are
incorporated herein by reference. For example, for injections, the
pharmaceutical
composition of the present invention may be formulated in adequate solutions
including but
not limited to physiologically compatible buffers such as Hank's solution,
Ringer's solution,
or a physiological saline buffer. The solutions may contain formulatory agents
such as
suspending, stabilizing, and/or dispersing agents. Alternatively, the
pharmaceutical
composition of the present invention may be in powder form for combination
with a suitable
vehicle, e.g., sterile pyrogen-free water, before use. Further, the
composition of the present
invention may be administered per se or may be applied as an appropriate
formulation
together with pharmaceutically acceptable carriers, diluents, or excipients
that are well-
known in the art. In addition, other pharmaceutical delivery systems such as
liposomes and
emulsions that are well-known in the art, and a sustained-release system, such
as semi-
permeable matrices of solid polymers containing the therapeutic agent, may be
employed.
Various sustained-release materials have been established and are well-known
to one
skilled in the art. Further, the composition of the present invention can be
administered
alone or together with another therapy conventionally used for the treatment
of a
disease/condition associated with poor expansion and/or differentiation of
HSCs, or in which
expansion and/or differentiation of HSCs is desirable.
[0080] The quantity to be administered and timing may vary within a range
depending on
the formulation, the route of administration, and the tissue or subject to be
treated, e.g., the
patient's age, body weight, overall health, and other factors. The dosage of
protein or
CA 02636876 2008-08-15
nucleic acid of the present invention preferably will be in the range of about
0.01 ug/kg to
about 10 g/kg of patient weight, preferably 0.01 mg/kg to 100 mg/kg. When
using the
pharmaceutical composition of the invention as a gene therapeutic agent, the
pharmaceutical composition may be administered directly by injection or by
administering a
vector integrated with the nucleic acid. For the nucleic acid molecule, the
amount
administered depends on the properties of the expression vector, the tissue to
be treated,
and the like. For viral vectors, the dose of the recombinant virus containing
such viral
vectors will typically be in the range of between about 0.1 to about 100
pfu/kg per kg of
body weight, in an embodiment between about 1 to about 50 pfu/kg per kg of
body weight
(e.g., about 10 pfu/kg per kg of body weight).
[0081] The agent useful for the method of the present invention includes, but
is not limited
to, that which directly or indirectly modifies the activity of the protein and
that which
modulates the production (i.e., expression) and/or stability of the protein
(e.g., at the level of
transcription, translation, maturation, post-translational modification,
phosphorylation and
degradation). In general, compounds/agents capable of modulating (e.g.,
increasing) the
expression or activity of one or more polypeptide and/or nucleic acid of the
present
invention may be identified from large libraries of both natural product or
synthetic (or semi-
synthetic) extracts or chemical libraries or from polypeptide or nucleic acid
libraries,
according to methods known in the art. Those skilled in the field of drug
discovery and
development will understand that the precise source of test extracts or
compounds is not
critical to the screening procedure(s) of the invention. Compounds used in
screens may
include known compounds (for example, known therapeutics used for other
diseases or
disorders).
[0082] Other objects, advantages and features of the present invention will
become more
apparent upon reading of the following non-restrictive description of specific
embodiments
thereof, given by way of example only with reference to the accompanying
drawings.
BRIEF DESCRIPTION OF THE DRAWINGS
[0083] In the appended drawings:
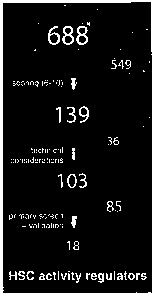
[0084] Figure 1 shows the experimental design of the nuclear factors screening
strategy of
CA 02636876 2008-08-15
26
the present invention. (A) A list of candidate genes was generated combining
data from
available stem cell databases, literature searches, and expression profiling
results of a
HoxA9-Meisl induced fetal liver leukemia (FLA2) highly enriched in Leukemia
Repopulating
Cells (LRC, frequency of -1:1.5) available from the inventors laboratory. The
putative
nuclear factors were subsequently ranked based on an algorithm that would
stratify them
according to self-renewal properties. Highest scoring candidates (n=139) were
further
selected for functional assessment using a retroviral over-expression
approach. 103 of the
139 genes selected were tested. (B) The coding sequence of each candidate was
PCR-
amplified, FLAG-tagged and subcloned into 1 out of 3 modified MSCV vectors
containing a
different reading frame (pKOF-1, -2 and -3). Respective retroviral producers
were plated in
a single well of a 96-well dish and co-cultured for 5 days with freshly sorted
(CD150+CD48-
Lin) bone marrow cells. Immediately upon infection (Day 0), a fraction of each
well was
transplanted into sublethally irradiated congenic recipient mice along with
competitor cells
(Ly5.2+ helpers). A similar assay was performed following an additional week
of ex vivo
culture (Day 7). (C) Expression of candidate proteins in retroviral producing
cells (GP+E86)
was confirmed by western immunobloting and revealed using an anti-FLAG
antibody.
Corresponding molecular weights are shown on Figure 4. (D) Range of retroviral
gene
transfer efficiencies of sampled gene candidates based on GFP epifluorescence
assessment
of (Day 0) cultured BM cells;
[0085] Figure 2 shows the scoring system used in candidate selection.
Candidates were
selected using microarray gene expression profiling from a pure stem cell
leukemia (FLA2,
submitted), the Stem Cell Database (SCDb) of Princeton University available
online
(http://stemcell.princeton.edul) and expression profiles performed on enriched
stem cells
populations (9, 13-23). The 103 genes in grey have been tested in the screen;
[0086] Figure 3 shows the subcloning strategy and protein expression of
candidates. The
accession number corresponding to each cDNA used as a template for PCR
amplification are
shown in addition to the sequences of forward and reverse primers used, with
restriction sites
used for subcloning underlined;
[0087] Figure 4 shows white blood cell chimerism data obtained from
competitive repopulation
assays with 103 genes. The second column shows gene transfer efficiencies for
each nuclear
gene candidate during the primary screen based on the level of GFP+ HSC
derivatives at day 4.
p.8
CA 02636876 2008-08-15
27
The other values represent the reconstitution levels of Ly5.1+ cells in each
independent
experiment presented as the mean of 2 (day 0) or 3 (day 7) mice per
experiment. Some mice
were already eliminated from the screen at 8 or 12 weeks because they did not
meet our
selection criteria for positive outcome, mainly based on peripheral blood
reconstitution by Ly5.1+
cells above 10% at 8 weeks and 30% at Z 12 weeks (see Figure 3A for
competitive repopulation
assays). Exp= experiment; w=weeks; inf. lev.=infection level; blank in
exp1=mice died or were
sacrificed (low level of Ly5.1+ cells); blank in exp2-5=genes eliminated after
the primary screen;
not all data are shown for vector (which has n=8);
[0088] Figure 5 shows competitive repopulation assays as a measurement of HSC
activity. (A)
Graft-derived hematopoiesis was evaluated at 4-week intervals in primary
recipient mice of
cultured BM cells by determining the percentage of Ly5.1 positive cells (donor-
derived) in
peripheral blood (PB) using FACS analysis. As a set of reference values, left
panel indicates PB
reconstitution levels from cultures initiated with a positive regulator of
self-renewal (Hoxb4) after
a 7-day expansion, in relation to values observed with an empty vector (mean
of pKOF-1, -2
and -3) at initiation (day 0) and termination of cultures (day 7). Day 7
values for the screened
103 candidates are compiled and presented in the middle panel, with the
established cut-off
level for a gain-of-function readout. Values from one experiment, presented as
mean SD for left
panel: n=2 mice for Hoxb4, n=6 mice for day 0 empty vector, n=9 mice for day 7
empty vector;
and as mean for middle panel: n=3 mice for each candidate cDNA. (B) Values are
reported as
peripheral blood reconstitution of Ly5.1+ cells following a 7 day of ex vivo
culture (solid line)
compared to empty vector (dashed line=day 7). Number of independent experiment
per
candidate gene equals 4 except for pKOF (vector control) n=8; Hoxb4, Cnbp and
Prdm16: n=3,
Ski: n=1. Each experiments: mean 3 mice per gene. Values=meantSEM except for
Ski:
mean SD. WBC=white blood cells;
[0089] Figure 6 shows nuclear candidates providing net increase in HSC
activity in vitro. (A)
When using vector control at day 0, a peripheral blood reconstitution at 14.4
2.2% in recipients
transplanted 16 weeks earlier was observed, which provides a reliable
estimation of the level of
HSC activity present at the initiation of the 7-day culture. Based on this,
genes that provide a
significantly net increase in HSC activity (black solid lines) above that
measured at the initiation
of the culture (black dotted line) from those which do not (grey solid lines)
were identified. (B)
Table with p values for day 0 and day 7 data from vector in comparison with
day 7 data from
each validated hit. Framed values correspond to genes that provide a
significantly net increase
CA 02636876 2008-08-15
28
in HSC activity (black solid lines in (A)). WBC=white blood cells;
[0090] Figure 7 shows that enhanced HSC activity is supported by intrinsic and
extrinsic groups
of effectors. (A) Southern blot analysis showing the presence of the expected
proviral DNA in
the BM of selected recipients that were highly reconstituted (between 10 to
85% of Ly5.1+ cells)
at 20 weeks post-transplantation. For 11 of the 18 genes identified in the
screen proviral DNA
was observed in 58 of the 65 recipients that were analyzed at this late time
point (46 are shown
in the 2 upper panels, presented as the cell autonomous group). The analysis
of proviral DNA
integration patterns in selected hematopoietic tissues from these mice
revealed that several
different clones with long-term reconstitution ability contributed to
hematopoiesis for each of
these 11 genes. This was true for different recipients within the same
experiments and,
obviously from different experiments, thus supporting that insertional
mutagenesis is not
responsible for these results. In several instance, the same proviral
integrations in the DNA from
2 different mice reconstituted by cells derived from the same culture could be
identified,
demonstrating that LT-HSC self-renewal has occurred in the cultures (a-i).
Bottom panel shows
the other 7 of the 18 validated genes, namely Fos, Hmgbl, Tcfec, Sfpil,
Zfp472, Hdacl and
Pml, and that only a minority of the highly reconstituted recipients (between
10 to 85% of Ly5.1+
cells) at 20 weeks post-transplantation contained integrated proviral DNA in
their BM raising the
possibility of non-cell autonomous activity in the cultures in which these
HSCs were kept prior to
transplantation (non-cell autonomous group). Each blot was systematically
exposed for the
same period of time (3 days). To ensure the absence of bands in bottom panel,
the brightness
and contrast of the images was enhanced. Below each blot is presented the
level of peripheral
blood Ly5.1+ or GFP+ cell reconstitution of recipient mice 20 weeks post-
transplantation. (B)
Compiled features of newly identified HSC self-renewal determinants. From left
to right:
individual gene candidates were evaluated for gene transfer efficiencies (mean
SD of %GFP+
HSC in culture at day 4) in experiments containing selected mice mentioned in
A (3rd column),
followed by peripheral blood cell reconstitution of the same mice (mean SD of
%Ly5.1+ cells,
4th column). Proportion of mice containing proviral DNA in their BM on the
total of selected mice
analysed is indicated in the 5th column, and the number of independent clones
identified per
gene is shown in the 6th column. In the 7th and 8th column, the peripheral
blood cell
reconstitution of every mice transplanted for each gene at day 0 and day
7(meantSEM of
%Ly5.1+ cells) is shown. Finally, the last column indicates the conclusion
about the cell
autonomous or non-autonomous effect of each gene on enhanced HSC
activity.X=GFP
expression not reliable for these constructs/clones; PBR=peripheral blood
reconstitution;
CA 02636876 2008-08-15
29
n/a=not applicable; CA=cell autonomous; NCA=non-cell autonomous;
[0091] Figure 8 shows the morphological analysis by Wright staining of
derivatives of HSC
populations overexpressing self-renewal candidates (upper-left inserts) at day
7 of ex vivo
culture. Proportions of immature blasts vs terminally differentiated cells
(neutrophils, monocytes
and masts cells: black arrows in upper-left inserts) for respective cultures
are depicted in right
panel. Values are presented as mean SD and a field comprising 100 cells were
examined per
independent experiment (n); n=3, except for vector: n=6; Ski, Hoxb4, Tcfec,
Sfpil and Hmgbl:
n=1; *p<_0.05 in right panel (relative to vector). (B) In vivo differentiation
potential along the
lympho-myeloid lineages was assessed in long-term recipients (20 weeks post-
transplantation)
of HSC transduced with Trim27 used as an example: immnophenotypic analysis by
flow
cytometry was performed using specific antibodies against B, T and myeloid
cell surface
markers (B220, CD3 and CD11b, respectively) on Ly5.1+ cells derived from the
peripheral
blood, bone marrow and thymus of these mice (and on Ly5.1+/GFP+ cells in
Figure 28A). (C)
Summary of results obtained in B for most of the validated candidates. Values
are presented as
mean SD of different selected mice (n) for each gene; n=2, except for vector:
n=6; NA10HD,
Trim27, Prdm16, Erdrl, Zfp472, Cnbp, Xbpl and Hdacl: n=3. Only Pml is absent.
Dashed lines
are presented to compare values of each gene with those of vector in different
hematopoietic
tissues. (D) Southern blots showing the proviral DNA integrations in the BM
(left panel) and in
the thymus (right panel) of mice transplanted with Trim27-overexpressing HSCs
indicating that
the same clones have contributed to repopulation of these two different
hematopoietic tissues.
Note that these mice are the same mice presented in Figure 3A. The same
analysis for other
validated hits is available in Figure 9B;
[0092] Figure 9 shows the in vivo differentiation of HSCs transduced with
newly identified
cell autonomous genes. (A) Differentiation potential along the lympho-myeloid
lineages in
long-term recipients (20 weeks post-transplantation) of HSC transduced with
cell
autonomous hits. Immnophenotypic analysis by flow cytometry was performed
using
specific antibodies against B, T and myeloid cell surface markers (B220, CD3
and CD1 1 b,
respectively) and gated on Ly5.1+/GFP+ populations derived from the peripheral
blood, bone
marrow and thymus of these mice. These data are not available for few genes
(Smarccl
and Prdm16 in all tissues analysed; Ski, KIf10, and Erdrl in the thymus) due
to absence of
EGFP expression in the transduced cells. Values represent mean SD and the
number of
mice analyzed (n) per candidate gene was n=2 except for vector: n=5; NA10HD
and Cnbp:
CA 02636876 2008-08-15
n=3; Trim27 and KIf10: n=1. (B) Southern blot analysis showing the proviral
DNA in the BM
(upper panel) and in the thymus (bottom panel) of selected recipients that
were highly
reconstituted at 20 weeks post-transplantation, corresponding to the cell
autonomous
group. Transduced HSCs remain competent in T cell differentiation although
they displayed
enhanced reconstitution activity for each gene except for Ski, Prdm16 and
Erdrl. n/a=not
available;
[0093] Figure 10 shows a schematic representation of the network of HSC
activity (A)
Quantitative analysis of gene-expression levels in HSC enriched population
singly
overexpressing all 18 newly identified nuclear HSC activity regulators
determined by Q-RT-
PCR. RNA was extracted from CD150+CD48'Lin"Kit+Sca+ bone marrow cells co-
cultured with
retroviral producers for 5 days, and sorted for the GFP positive fraction.
Average ACt values
were determined with R-actin serving as endogenous control to normalize levels
of target gene
expression. Relative fold differences (RQ) were determined and corresponding
empty vector
(mean of pKOF-1, -2, and -3) was used as reference calibrator to assess
relative fold
differences in expression levels of each candidate in HSC. Reactions were done
in triplicate,
and average values were calculated for each independent experiment (n); n=3,
except for Ski
and Sfpil: n=1. Relative fold differences were determined using the oOCt
method. ND (not
determined) values are shown in white. The legend colouring is based on the
scaled values of
each row for ACt heatmap, and on the log2 of all values in the plot with a
maximum value of 13.5
for RQ heatmap. (B) An integrative diagram is presented, correlating mRNA
transcript
upregulation by overexpression of a hit (black solid arrows) and cell fate
determination (grey
dotted arrows). Numbers indicate relative fold differences (z3-fold) observed
in (A);
[0094] Figure 11 shows two different forms of Trim27 with different potential.
(A) Two
different forms of Trim27 have been tested, i.e., one containing a frame-shift
error
(truncated; accession number BC085503; upper panel) preserving intact only the
RING, B-
box and first Coiled-coil domains, and another full-length form (accession
number
BC003219; bottom panel) containing moreover the second Coiled-coil and the
SPRY
domains. (B) Competitive repopulation assays reporting the differential
reconstitution level
of recipient mice by HSCs transduced with the different forms of Trim27. Note
that the
SPRY domain within the full-length form of Trim27 seems to limit the potential
of this gene
in HSC expansion. WBC=white blood cells;
CA 02636876 2008-08-15
31
[0095] Figure 12 shows HSCs depletion following transformation with empty
vectors (A) and
vectors expressing control genes (B);
[0096] Figure 13 shows HSC expansion following transformation with an empty
vector and
a vector expressing different genes (A) sfbl, xbp, fos, trim27, ap2a2, sox4;
and B) kif1;
[0097] Figure 14 shows the differentiation of HSCs transformed with a vector
expressing
different genes (xbp, trim27, sox4 and hnrpdl) into various cell
types/lineages in different
tissues (blood, bone marrow and thymus). B220 is a B-cell lineage marker, CD11
b is a
myeloid lineage marker and CD4/CD8 are T-cell lineage markers;
[0098] Figure 15 shows the differentiation of HSCs transformed with a vector
expressing
different genes (xbpl, trim27, sox4, pbx2, meis, klf10, spnsl and cbfb) into
various cell
types/lineages in the blood. PKOF = empty vector;
[0099] Figure 16 shows the differentiation of HSCs transformed with a vector
expressing
different genes (xbpl, trim27, sox4, pbx2, meis, kIf10, spnsl and cbfb) into
various cell
types/lineages in the bone marrow. PKOF = empty vector;
[00100] Figure 17 shows the differentiation of HSCs transformed with a vector
expressing different genes (xbpl, trim27, sox4, pbx2, meis, kIf10, spnsl and
cbfb) into
various cell types/lineages in the thymus. PKOF = empty vector;
[00101] Figure 18 shows the clonality of the differentiated HSCs transformed
with a
vector expressing different genes (xbpl, sox4, hnrpdl, gpsm2 and ap2a2) in the
bone
marrow (BM) and the thymus;
[00102] Figure 19 shows the differentiation (20 weeks post-transplantation) of
HSCs
transformed with a vector expressing xbpl into various cell types/lineages;
[00103] Figure 20 shows the differentiation (20 weeks post-transplantation) of
HSCs
transformed with a vector expressing trim27 into various cell types/lineages;
[00104] Figure 21 shows the differentiation (20 weeks post-transplantation) of
HSCs
transformed with a vector expressing sox4 into various cell types/lineages;
CA 02636876 2008-08-15
32
[00105] Figure 22 shows the differentiation (20 weeks post-transplantation) of
HSCs
transformed with a vector expressing cbfb into various cell types/lineages;
[00106] Figure 23 shows the differentiation (20 weeks post-transplantation) of
HSCs
transformed with a vector expressing pbx2 into various cell types/lineages;
[00107] Figure 24 shows the differentiation (20 weeks post-transplantation) of
HSCs
transformed with a vector expressing kIf10 into various cell types/lineages;
[00108] Figure 25 shows the expansion of HSCs transduced with a vector
expressing
ap2a2 as compared to HSCs transduced with an empty vector;
[00109] Figure 26 shows that screening strategy improves the signal to noise
ratio of the
results after ex vivo culture (A) WBC chimerism showing that HSCs
overexpressing Hoxb4
transplanted without ex vivo culture gives reconstitution levels similar to
that observed with
vector alone, making signal and noise difficult to separate. (B) WBC chimerism
showing that a
7-day ex vivo culture prior to transplantation enhances considerably the
signal to noise ratio due
to a better reconstitution ability of HSCs overexpressing Hoxb4 coupled with a
depletion of HSC
activity with control vector. These results, from which the screen has been
planned, have been
previously obtained using whole (not sorted) BM cells from 5-fluorouracil-pre-
treated donor
mice, which differ from Figure 5A; WBC=white blood cells;
[00110] Figure 27 shows the expression of ap2a2 protein in the virus
producers. (A)
HSCs transformed with a vector expressing ap2a2; (B) the clonality; and (C)
the
differentiation into various cell types/lineages at twenty weeks are shown;
and
[00111] Figure 28 shows nucleic acids and proteins sequence alignments
performed
using Clustal WT"" between variants of HSC regulators of the present
invention. (A) Xbpl;
(B) tgif; (C) Pml; (D) tcfec; (E) KIf10 (F)cbfb; and (G) Prdm16.
DESCRIPTION OF ILLUSTRATIVE EMBODIMENTS
[00112] The present invention is illustrated in further details by the
following non-
limiting examples.
EXAMPLE 1
CA 02636876 2008-08-15
33
Experimental model
Selection and ranking of candidates
[00113] As a corollary to ESC studies, it can be stipulated that HSC fate is
controlled
by a series of master regulators, analogous to Oct4, and several subordinate
effectors,
providing sound basis to the generation of a stem cell nuclear factors
database. Towards
this end, we created a database consisting of 688 nuclear factors (see
www.132.204.81.89:8088; Figure 1A), considered candidate regulators of HSC
activity.
This list was mostly derived from microarray gene expression profiling of
normal and
leukemia stem cells including our recently generated FLA2 leukemia (1 in 1.5
cells are
leukemia stem cells). This database was also enriched by genes obtained
following a
review of the literature on HSC self-renewal (15-21). A similar approach was
used to
identify candidate genes which are asymmetrical cell division regulators.
[00114] Candidate genes were next ranked from 1(lowest priority) to 10
(highest
priority) based on 3 factors: 1) differential expression between primitive and
more mature
cellular fractions (e.g., LT-HSC-enriched); 2) expression levels (high,
highest priority); and
3) consistency of findings between datasets.
[00115] Rank 1=Factors expressed only in one database/report and at relatively
low
level; Rank 2=Factors expressed in two different contexts (e.g., 2 probesets
or 2 libraries);
Rank 3=Factors expressed in three different contexts; Rank 4=Factors selected
for their
function (e.g., stem cell regulator); Rank 5=Factors highly expressed in a
given
database/report (i.e., top 10%); Rank 6=(Rank 4 or Rank 5) + (Rank 2 or Rank
3); Rank
7=Factors expressed in 2 independent databases/reports or [Rank 3 + (2 x (Rank
4 or Rank
5))]; Rank 8=[Factors expressed in 4 different contexts + (3 x (Rank 4 or Rank
5))] or (Rank 7 +
Rank 2); Rank 9=Rank 7 + [(Rank 4 or Rank 5) or (Rank 2 + (Rank 4 or Rank 5))
or Rank 3];
Rank 10=Factors expressed in 3 independent databases/reports or [Rank 7 +
((Rank 2 + (2 x
(Rank 4 or Rank 5))) or (Rank 3 + (Rank 4 or Rank 5)))]. Genes with a score of
6 and above
(n=139) were selected for functional studies, of which 103 were tested. See
Figure 2.
EXAMPLE 2
Primary screen
CA 02636876 2008-08-15
34
[00116] As a primary screen, a competitive repopulation assay was used for
measurement of HSC activity to validate candidates previously identified.
[00117] The ability of the 139 highest scored candidates to affect
hematopoietic stem
cell (HSC) self-renewal and/or proliferation in vitro and in vivo was
evaluated.
[00118] The screening protocol is outlined in Figure 1 B. In brief, the cDNA
corresponding to the open reading frames for each of these genes was amplified
by PCR,
FLAG-tagged and subcloned into 1 out of 3 modified MSCV vectors containing a
different
reading frame (pKOF-1, pKOF-2 and pKOF-3) that includes a GFP reporter
cassette (Figure
1 B). High-titer retroviruses were produced in 96 well plates seeded with
viral producer cells
using a procedure optimized locally. Protein extracts derived from producer
cells in each of
the 103 wells were analyzed by western blotting which confirmed the presence
of a FLAG-
protein in 88% of the cases (Figures 1 C and 3), with 92% of these proteins
showing the
expected molecular size (Figure 3). Respective retroviral producers were
plated in a single
well of a 96-well dish and co-cultured for 5 days with freshly sorted
(CD150+CD48'Lin") bone
marrow cells. Immediately upon infection (Day 0), half of each well was
transplanted into
sublethally irradiated congenic recipient mice along with competitor cells
(Ly5.2+ helpers). A
similar assay was performed following an additional week of ex vivo culture
(Day 7).
Figures 1 D and 4 show the retroviral gene transfer efficiencies of sampled
gene candidates
based on GFP epifluorescence assessment of (Day 0) cultured BM cells. A List
of predicted
and observed molecular weights for most proteins tested in the present
invention is shown
in Figure 3. Retroviral gene transfer to freshly isolated mouse bone marrow
cells enriched
for HSC activity (Lin"CD150+CD48-) varied significantly with an average of
50.0% 31%
(Figures 1D and 4). For each gene analyzed, a proportion of the transduced
cells was
transplanted into lethally irradiated recipients along with competitor cells
immediately at the
end of retroviral gene transfer (day 0) and after an additional 7 days of ex
vivo culture (day
7) (Figure 1 B). Peripheral blood cell reconstitution was then assessed after
short (4 and 8
weeks) and long periods of time (12 and 16 weeks) post-transplantation to
evaluate the
impact of each candidate to affect in vivo (day 0) and ex vivo (day 7)
expansion of short and
long-term repopulating cells. MSCV-Hoxb4-GFP was used as a positive control in
these
experiments and 3 different MSCV-GFP viruses were used as negative controls.
CA 02636876 2008-08-15
[00119] As indicated above, graft-derived hematopoiesis was evaluated at 4-
week
intervals in primary recipient mice of cultured BM cells by determining the
percentage of
Ly5.1 positive cells (donor-derived) in peripheral blood (PB) using FACS
analysis (Figure 5).
Day 7 values for the screened 103 candidates are compiled and presented in
Figure 5A,
with the established cut-off level for a gain-of-function readout. Criteria
used for hit selection
were: peripheral blood reconstitution by Ly5.1+ cells above 10% at 8 weeks and
30% at 12
weeks. Candidates clustering above this level were selected for confirmatory
experiments,
while those below were disregarded (see right panels). One hit (Hesl) was
eliminated based
on the marked reduction in repopulation noted between early and late time
points (upper
line in Figure 5A, right lower panel).
[00120] Recipients of HSCs transduced with Hoxb4 (positive control) or with
the
backbone vectors in all 3 frames (pKOF1, 2, 3: negative controls) were thus
used to set the
cut off for selecting the candidates needing further validation. As expected
from previous
results (13), depletion of HSC activity was verified during 7 day cultures
since peripheral
blood reconstitution of recipients transplanted 16 weeks earlier with pKOF-
transduced cells
decreased from 13.3 6.2% (day 0 ce115, dotted line in Figure 5A) to 4.9 4.8%
(day 7 cells,
dashed line in Figure 5A). This led to estimate that approximately 3 Ly5.11'
HSCs were
competing with 20 Ly5.2+ HSCs to repopulate each mouse transplanted with day
0 cells
and approximately 1 HSC after 7 days of ex vivo culture. These data also
suggests that
reconstitution from non-infected Ly5.1+ cells, even at 0% infection rate,
would be
consistently below 10% (see dashed line in Figure 5A) indicating that the 30%
cut off used
in the primary screen (Figure 5A, middle panel) was stringent enough to
identify genes that
confer enhanced in vitro/in vivo activity to transduced HSCs. Notably, the 30%
cut off value
represents the average reconstitution observed in recipients of Hoxb4-
transduced cells in
these conditions (see Figure 5A, left panel). Based on these criteria, we
expect that the
newly identified candidates should be equivalent to -or more potent than-
Hoxb4 in inducing
enhanced HSC activity.
[00121] In total, 18 nuclear factor genes hits were identified in this primary
screen for a
frequency of 17% (18/103) (Figure 5A, upper right panel; and Figure 4, Figures
12-26 as
well as Tables 1 and 2). These included Cnbp, Erdrl, Fos, Hdacl, Hmgbl,
Hnrpdl, K1fIO,
Pml, Prdm16, Sfpil (PU.1), Ski, Smarcci (Baf155), Sox4, Tcfec, Trim27, Vps72,
Xpbl, and
CA 02636876 2008-08-15
36
Zfp472.
[00122] Using the same approach as described above, 4 additional genes
encoding
factors controlling assymetrical cell division (ap2a2, tmdol, gpsm2 and gpx3)
were also
identified. Together, these 22 selected candidate genes provided competitive
advantage
(i.e., promoting expansion) to transduced HSC to levels similar to those
observed with
Hoxb4-transduced HSCs.
[00123] Table I presents expansion results for genes providing competitive
advantage
to transduced HSCs.
inf. rate Da 0 Da 0 Da 0 Day 0 Da 7 Da 7 Da 7 Da 7
4w 8w 12w 16w 4w 8w 12w 16w
vector
ex 4 KOF1 73 23.8 17.5 14.7 14.9 7.7 5.5 2.7 2.7
ex 4 KOF2 85 14.8 14.4 11.6 8.1 8.4 6.6 4.2 4.4
ex 4 KOF3 99 24.2 19.3 16.4 16.8 15.7 13.8 9.0 7.5
ex 8 KOF1 48.2 3.9 9.7 21.9 15.5 4.1 2.1 3.7 2.0
ex 10 KOF1 30.9 12.6 16.6 20.5 22.6 6.4 5.5 3.4 4.0
exp11 apKOF
1 30.1 13.3 13.2 9.1 9.5
exp11 bpKOF
1 30.1 2.7 4.0 2.4 2.0
mean 56.6 15.9 15.5 17.0 15.6 8.3 7.2 4.9 4.6
SEM 3.79 1.65 1.89 2.31 1.77 1.71 1.08 1.09
hoxb4
exp6 15 38.4 41.2 43.8 42.4 37.7 39.7 34.2 29.6
exp7 18.6 20.0 30.3 34.9 28.5 40.0 41.3 43.3 42.6
exp8 5.3 17.5 21.3 24.5 24.0 20.6 19.9 17.8 21.3
mean 13.0 25.3 30.9 34.4 31.6 32.8 33.6 31.8 31.2
SEM 6.60 5.75 5.58 5.55 6.10 6.87 7.48 6.18
NA10hd
explO 37.6 64.6 74.7 84.8 84.8 75.1 79.0 91.0 90.4
ex 11 a 82.2 69.9 89.1 90.7 89.1
ex 11 b 82.2 75.1 88.5 90.6 89.0
mean 67.3 64.6 74.7 84.8 84.8 73.4 85.5 90.7 89.5
SEM 0.00 0.00 0.00 0.00 1.73 3.28 0.12 0.46
smarccl
exp5 13.2 46.2 61.4 56.8 49.7 26.8 37.2 40.7 39.9
explO 6.2 13.8 13.9 14.1 14.1 17.3 33.7 35.6 34.3
ex 11 a 3.3 11.1 8.9 7.5 7.0
ex 11 b 3.3 29.0 37.6 38.2 41.1
mean 6.5 30.0 37.7 35.5 31.9 21.0 29.3 30.5 30.6
CA 02636876 2008-08-15
37
SEM 16.20 23.74 21.36 17.80 4.17 6.86 7.74 8.00
xbpl
exp4 80 13.1 9.3 7.4 7.2 40.7 45.4 50.0 39.8
explO 75.7 18.3 12.7 7.2 7.2 16.7 11.8 6.8 6.8
ex 11 a 80.4 8.1 8.3 5.5 6.0
ex 11 b 80.4 25.8 26.3 23.7 22.9
mean 79.1 15.7 11.0 7.3 7.2 22.8 23.0 21.5 18.9
SEM 2.57 1.73 0.11 0.00 6.98 8.44 10.36 7.99
fos
exp4 80 11.4 6.0 4.7 4.6 18.3 30.6 35.1 33.9
explO 63.2 7.5 6.7 5.9 5.9 8.1 13.0 10.4 10.6
ex 11 a 63.3 24.2 32.2 28.2 29.5
ex 11 b 63.3 48.4 50.8 47.5 48.4
mean 67.5 9.5 6.4 5.3 5.2 24.8 31.7 30.3 30.6
SEM 1.93 0.38 0.63 0.67 8.54 7.73 7.75 7.79
hmgbl
exp4 40 19.7 18.4 12.0 9.4 31.7 32.1 38.7 42.2
explO 7 12.0 10.5 9.1 9.1 5.4 4.2 2.6 2.6
explla 17.3 8.0 8.4 6.0 6.0
ex 11 b 17.3 36.5 33.5 32.9 30.9
mean 20.4 15.9 14.5 10.5 9.2 20.4 19.6 20.0 20.4
SEM 3.85 3.91 1.47 0.19 7.98 7.72 9.20 9.63
tcfec
exp4 76 13.9 17.3 17.0 17.0 29.2 30.4 36.4 41.5
explO 53.4 1.1 3.8 6.5 6.5 7.6 13.4 7.2 7.6
ex 11 a 27.7 26.6 26.5 26.1 25.2
ex 11 b 27.7 9.2 13.0 12.5 12.5
mean 46.2 7.5 10.6 11.7 11.7 18.2 20.8 20.6 21.7
SEM 6.38 6.78 5.26 5.26 5.67 4.49 6.60 7.57
kIf10
exp4 68 19.0 23.7 24.5 23.8 35.9 35.6 32.8 31.2
explO 47.4 7.7 7.5 7.4 7.4 25.1 43.4 56.4 55.3
ex 11 a 47 20.0 29.4 32.0 33.4
ex 11 b 47 17.1 23.0 23.9 21.6
mean 52.4 13.4 15.6 16.0 15.6 24.5 32.8 36.3 35.4
SEM 5.69 8.09 8.54 8.16 4.15 4.35 7.01 7.10
trim27
exp4 97 12.3 5.6 3.5 4.3 18.5 48.4 59.1 61.3
explO 73 14.1 13.4 12.7 13.4 22.2 34.7 35.5 32.4
ex 11 a 44.1 31.5 38.5 37.3 39.0
ex 11 b 44.1 14.1 11.8 9.2 9.2
mean 64.6 13.2 9.5 8.1 8.9 21.6 33.4 35.3 35.5
SEM 0.87 3.88 4.59 4.55 3.70 7.74 10.20 10.72
ap2a2
exp4 62 20.6 25.4 27.1 27.9 64.1 69.4 75.0 73.6
CA 02636876 2008-08-15
38
explO 48.1 20.0 18.8 17.6 17.6 27.3 33.8 37.4 40.8
ex 11 a 52.1 23.8 41.9 44.7 46.8
ex 11 b 52.1 56.9 70.3 71.8 75.4
mean 53.6 20.3 22.1 22.4 22.8 43.0 53.8 57.2 59.2
SEM 0.31 3.30 4.76 5.14 10.22 9.40 9.48 8.97
gpsm2
exp5 33.6 32.9 35.5 23.6 19.3 25.2 35.1 45.8 43.6
explO 18.6 10.2 9.6 8.9 8.9 5.7 4.2 2.4 2.4
ex 11 a 4.9 42.8 56.5 59.1 60.9
ex 11 b 4.9 37.3 42.8 39.3 40.0
mean 15.5 21.6 22.5 16.3 14.1 27.8 34.7 36.7 36.7
SEM 11.36 12.96 7.35 5.17 8.22 11.07 12.14 12.32
sox4
exp4 97 12.2 13.1 11.6 10.4 18.0 23.4 28.5 30.7
explO 50.6 20.9 25.3 29.7 31.4 38.7 36.7 40.8 42.3
ex 11 a 72.2 52.6 63.8 60.7 57.0
ex 11 b 72.2 36.3 35.9 32.4 29.2
mean 73.0 16.5 19.2 20.6 20.9 36.4 39.9 40.6 39.8
SEM 4.33 6.09 9.03 10.49 7.11 8.52 7.17 6.44
hnrpdl
exp6 83 18.3 6.0 4.2 4.2 12.0 18.6 29.2 35.4
explO 94.4 5.5 4.3 3.2 3.2 6.5 4.8 2.5 2.5
ex 11 a 74.5 13.3 17.5 14.5 14.4
ex 11 b 74.5 36.3 36.0 31.0 28.9
mean 81.6 11.9 5.2 3.7 3.7 17.0 19.2 19.3 20.3
SEM 6.37 0.82 0.50 0.53 6.60 6.40 6.71 7.38
vps72
exp6 53 24.7 15.4 12.1 8.8 20.0 30.4 40.3 41.4
explO 64.8 27.2 21.0 14.9 14.9 15.1 17.5 17.7 20.2
ex 11 a 49.6 34.5 35.8 34.9 34.9
ex 11 b 49.6 13.5 12.2 8.5 8.5
mean 54.3 25.9 18.2 13.5 11.8 20.8 24.0 25.3 26.2
SEM 1.23 2.78 1.39 3.05 4.79 5.49 7.39 7.38
gpx3
exp6 96 21.3 28.5 36.6 41.0 17.3 27.9 33.3 30.1
explO 76.8 15.4 10.5 5.6 5.5 12.6 7.4 5.5 5.5
ex 11 a 71.8 57.7 72.4 76.3 77.3
ex 11b 71.8 17.6 13.8 10.1 10.1
mean 79.1 18.3 19.5 21.1 23.2 26.3 30.4 31.3 30.8
SEM 2.96 9.00 15.52 17.79 10.54 14.64 16.18 16.40
sfpil
exp4 57 16.2 9.4 7.9 8.3 44.9 48.8 56.0 57.3
ex 10 48.9 10.1 7.1 4.2 4.2 16.8 15.4 9.6 9.6
ex 11 a 17.8 12.7 11.4 8.3 8.3
ex 11 b 17.8 34.2 30.0 28.6 29.8
CA 02636876 2008-08-15
39
mean 35.4 13.1 8.3 6.1 6.2 27.1 26.4 25.6 26.2
SEM 3.09 1.15 1.88 2.06 7.52 8.46 11.15 11.47
erdrl
explO 31.8 14.3 13.1 11.9 11.9 11.1 7.0 3.6 3.6
ex 11 a 31.6 44.6 44.8 45.1 45.1
ex 11 b 31.6 9.5 7.6 5.4 5.4
mean 31.7 14.3 13.1 11.9 11.9 21.8 19.8 18.0 18.0
SEM 0 0 0 0 11.45 12.50 13.52 13.54
zfp472
exp8 2.8 7.3 5.5 11.4 11.4 17.0 26.0 27.4 37.9
explO 3.1 23.7 23.7 23.7 23.2 3.2 2.7 1.5 1.5
ex 11 a 2.7 30.4 24.8 20.5 19.2
ex 11 b 2.7 23.4 19.0 15.9 15.9
mean 2.8 15.5 14.6 17.6 17.3 18.5 18.1 16.3 18.6
SEM 8.23 9.10 6.17 5.91 5.78 5.36 5.47 7.48
tmodl
explO 50.1 5.2 5.5 5.9 5.9 31.1 39.7 43.5 43.2
cnbpl
explO 75.9 20.1 27.3 34.5 31.6 40.1 34.3 37.3 37.4
rdm16
explO 3.2 33.5 34.2 34.9 36.1 52.2 45.2 40.5 43.5
hdacl
explO 38.1 12.8 9.1 5.5 5.5 39.7 36.7 31.1 30.8
ski
exp6 5 36.0 47.1 52.0 39.4 23.2 21.5 29.9 36.6
ctrl neg (rela) 20.96 10.625 6.54 5 9.1 5.1 2.9 2.4
[00124] Table II present the Genbank accession numbers for the genes providing
competitive advantage to transduced HSC.
Genbank accession Genbank accession SEQ ID NO:
Gene name number (nucleic number Nucleic
acid) (polypeptide) acid/polypeptide
trim27 NM 006510 NP 006501 1/2
N M_005080 N P_005071 3/4
Xbpl N M 001079539 N P 001073007 5/6
Sox4 NM_003107 NP_003098 7/8
Smarccl NM 003074 NP 003065 9/10
CA 02636876 2008-08-15
sfpi 1 NM_001080547 NP 001074016 11/12
NM_003120 NP003111 13/14
fos NM_005252 NP_005243 15/16
hmgbl NM_002128 NP_002119 17/18
hnrpdl NM_031372 NP_112740 19/20
vps72 N M_005997 N P_005988 21/22
N M_174886 N P_777480 23/24
N M173211 N P_775303 25/26
N M_173209 N P_775301 27/28
tgif N M 173208 N P775300 29/30
N M_170695 N P_733796 31/32
N M_003244 N P_003235 33/34
NM_173210 NP_775302 35/36
NM_173207 NP_775299 37/38
Consensus 93
NM_033250 NP_150253 39/40
N M033240 N P_150243 41/42
NM_033239 NP_150242 43/44
NM_002675 NP_002666 45/46
pml NM_033249 NP_150252 47/48
N M_033238 N P_150241 49/50
N M_033244 N P_150247 51/52
N M_033247 N P_150250 53/54
N M_033246 N P_150249 55/56
57/58
tcfec NM_012252 NP_036384 59/60
NM 001018058 NP 001018068
Consensus: 94
61/62
kif 10 N M001032282 N P001027453 63/64
N M 005655 N P 005646
Consensus 95
CA 02636876 2008-08-15
41
65/66
cbfb N M022845 N P074036 67/68
NM_001755 NP 001746
- Consensus: 96
zfp472 N M_153063 N P_694703 69/70
ap2a2 NM_012305 NP_036437 71/72
gpsm2 NM_013296 NP_037428 73/74
Gpx3 NM_002084 NP_002075 75/76, 98
erdrl NM 133362 NP 579940 77/78
tmodl N M_003275 N P_003266 79/80
cn bp 1 N M_003418 N P_003409 81/82
83/84
N M 022114 N P071397
Prdm 16 85/86
NM 199454 NP 955533
Consensus: 97
hdacl NM 004964 NP 004955 87/88
ski NM 003036 NP 003027 89/90
Hoxb4 NM 024015 NP 076920 91/92#
EXAMPLE 3
Validation
[00125] To validate the candidate genes identified in the above primary
screen, additional
independent experiments (n=4, unless indicated) were performed using the same
96 well plate
protocol described in Figure 1 B. A summary of these results is provided in
Figure 5B. From left
to right and top to bottom, genes are presented based on the level of
statistical significance at
16 weeks (from highest to lowest) reached in these experiments: Hoxb4
(p=9.5x10-9) (control);
Ski (p=1.6x10-10); Hoxb4 (p=9.5x10-9); Smarccl (p=8.5x10$); Vps72
(p=2.4x10"7); Fos
(p=3.2x10-'); Trim27 (p=5.1x10-7); Sox4 (p=1.0x10~); KIflO (p=1.8x10-6);
Prdm16 (p=4.0x'0-6);
Erdrl, Tcfec, Sfpil, Zfp472 and Hmgbl (all between p=1.1 to 8.8x10-4 ); Cnbp,
Pml and Xbpl
(p=0.001); Hnrpdl (p=0.002) and Hdacl (p=0.015). Thus, all of the 18
candidates were
confirmed (p:50.05), for a positive predictive value (PPV) of 100%.
[00126] The design of the screen and validation protocol included an
assessment of the
CA 02636876 2008-08-15
42
reconstitution activity of HSCs isolated at the end of the infection -prior to
the initiation of the 7
day culture-the so-called "day 0" time point (Figure 1 B). In the case of the
negative control
experiments, performed with the pKOF vectors alone, peripheral blood
reconstitution was
observed at 14.4 2.2% in recipients transplanted 16 weeks earlier. This value
provides a
reliable estimation of the level of HSC activity present at the initiation of
the 7 day culture. Based
on this, it is possible to identify genes that provide a net increase in HSC
activity above that
measured at the initiation of the culture from those which do not. In that
respect, Hoxb4 is a
prototype since transduced HSCs show a net expansion of 1 to 2 logs in short
term cultures.
The following genes were significantly higher than vector at day 0: Ski, Sox4,
Smarccl, Vps72,
Fos, Trim27, KIflO and Prdm16 (Figure 6, dotted lines on panel A and boxed
values in panel B),
indicating a possible ex vivo expansion of HSCs to levels above input numbers,
as does Hoxb4.
EXAMPLE 4
Evidence that some candidates operates in a non-cell autonomous manner
[00127] The 7-day ex vivo culture inherent to the screening strategy (Figure 1
B) should,
provide sufficient time for extrinsic factors to impact on HSC expansion (13).
Based on this, it is
possible that non-transduced HSCs would respond favorably to a series of
factors secreted by -
or present on- adjacent cells (e.g., viral producers or other progenitors)
thereby conferring a
competitive advantage to all (transduced and untransduced) HSCs in these
cultures (Ly5.1+). To
address this possibility, we analyzed the hematopoietic system of selected
recipients that were
highly reconstituted (between 10 to 85% of Ly5.1+ cells) at 20 weeks post-
transplantation, a
time point deemed sufficient such that reconstitution is strictly derived from
so-called long-term
HSCs (LT-HSCs) (27). The presence of the expected proviral DNA in the
appropriate
reconstituted tissues was first verified. This constitutes a necessary
attribute for cell
autonomous effects. For 11 of the 18 nuclear genes identified in the present
screen, namely Ski,
Smarccl, Vps72, Trim27, Sox4, KIf10, Prdm16, Erdrl, Cnbp, Xbpl and Hnrpdl,
proviral DNA
was observed in 58 of the 65 recipients (89%) that were analyzed at this late
timepoint (Figure
7A, 2 upper panels; Figure 7B, 5th column). Considering that gene transfer
efficiency was on
average at 50% for the entire gene set and 55% for these 11 genes (Figure 4,
2nd column) and
that a limiting number of transduced HSCs were transferred to each recipient,
this observation
on its own is compatible with these genes intrinsically enhancing HSC
activity. Furthermore, the
analysis of proviral DNA integration patterns in selected hematopoietic
tissues from these mice
revealed that several different clones with long-term reconstitution ability
contributed to
CA 02636876 2008-08-15
43
hematopoiesis for each of these 11 nuclear factor genes (Figure 7A). This was
true for different
recipients within the same experiments and, obviously from different
experiments, thus
supporting that insertional mutagenesis is not responsible for these results.
In several instances,
the same proviral integrations in the DNA from 2 different mice reconstituted
by cells derived
from the same culture could be identified, demonstrating that LT-HSC self-
renewal has occurred
in these cultures (see a-i in Figure 7A).
[00128] Interestingly for 7 of the 18 validated nuclear genes, namely Fos,
Hmgbl, Tcfec,
Sfpil, Zfp472, Hdacl and Pml, it was found that only a minority of the highly
reconstituted
recipients (between 10 to 85% of Ly5.1+ cells at 20 weeks post
transplantation; Figure 7A third
panel) contained integrated proviral DNA in their hematopoietic tissues. This
observation raises
the possibility of a non-cell autonomous activity in cultures in which these
HSCs were kept prior
to transplantation. A detailed evaluation of these recipients is provided in
Figure 7B to stand
comparison with mice that were reconstituted with cells transduced with each
of the 11 genes
described in the previous paragraph (also presented in this Figure as the
"cell autonomous"
group). First and foremost, gene transfer efficiencies were similar between
both groups or
around 40-50% (mean values). Second, the repopulation activity for 4 of the 7
genes with
presumed non-cell autonomous activity was enhanced by the 7-day culture prior
to
transplantation described in Figure 1 B [Figure 7B, compare %Ly5.1 day 0 (7th
column) vs day 7
(8th column) for Fos, Tcfec, Sfpil and Hmgbl]. Fos represents a notable
example for this: with
an initial gene transfer above 70%, it was found that recipients reconstituted
with HSCs prior to
the 7-day culture were repopulated by Ly5.1+ cells at 5 1% whereas, following
the 7-day
culture, this number increased to 31 8% in 4 independent experiments with 2
mice per
experiment at day 0, and 3 at day 7 (Figures 4 and 7B). As presented in Figure
5B, Tcfec,
Sfpi1and Hmgb1 show a similar trend.
[00129] Thus, the combination of results from proviral integrations and
hematopoietic
reconstitution analyses support the existence of 2 broad groups of effectors
for the nuclear gene
candidates, one which includes 7 genes that appear to extrinsically support
enhanced HSC
activity and another of 11 genes which seem to provide intrinsic contribution.
EXAMPLE 5
Impact of validated candidates on HSC differentiation
CA 02636876 2008-08-15
44
[00130] There is growing evidence to suggest that HSC self-renewal involves
the active
repression of a differentiation program that is coupled to cell division (14).
In support of this, the
present inventors recently found that Hoxb4 or NA10HD-transduced HSCs, which
actively
undergo in vitro self-renewal divisions, show evidence of differentiation
arrest [Figure 8A; (14)].
The newly validated candidates were investigated to determine if they behaved
similarly. To
achieve this, the cytological characteristics of transduced and sorted HSCs
was analyzed after a
7-day in vitro culture period (prior to their transplantation). In this
context, cultures initiated with
control vector-infected HSCs contained differentiated cells in a proportion of
70 8%. These
included neutrophils, monocytes and mast cells (Figure 8A, arrows in upper
left panel with
summary of results in histogram: grey bars=undifferentiated cells or blasts,
and dark grey
bars=differentiated cells). Conversely, cellular differentiation was reduced
in cultures initiated
with HSCs transduced with most of the newly validated candidates (Figure 8A).
The increase in
the proportion of undiffentiated to differentiated cells was most important
for Ski, Vps72, Fos,
Sox4, KIf10, Prdm16, Erdrl, Hnrpdl and Hdacl when compared to cultures
initiated with HSCs
infected with the control virus.
[00131] The in vitro differentiation arrest displayed by Hoxb4 or NA10HD-
transduced
HSCs is eventually reverted following their transplantation in vivo. Thus,
depending on the
environment, these 2 genes can either interfere (e.g., in vitro in the
presence of growth factors)
or not (e.g., in vivo under steady state conditions) with HSC differentiation.
To determine if the
newly identified regulators of HSC activity are similarly permissive to HSC
differentiation in vivo,
4 different approaches were used. First, the general health, spleen size and
bone phenotype
(white vs red) of each recipient was evaluated. Except for recipient of Prdm16-
transduced cells,
which eventually developed splenomegaly, white femurs and myeloproliferation
at 20 weeks
(data not shown), none of the mice transplanted with cells expressing the 17
other nuclear
genes ever presented this, or any other, hematological phenotype. Second,
microscopic
evaluation of bone marrow and spleen cytological preparations derived from
representative
mice for each gene was performed. Results from these analyses were normal for
all groups,
except for the Prdm16 cohort, which showed an excess of poorly differentiated
myeloid cells in
their bone marrow and for the Ski cohort in which the number of lymphocytes in
the bone
marrow was reduced. Besides recipients of Prdm16-transduced cells, spleens
were never
infiltrated with myeloid cells nor did they include enhanced numbers of
erythroblasts. To confirm
this, a third approach consisting in performing FACS analysis on donor-derived
(Ly5.1+) cells
from selected recipients in which reconstitution was well above background
values (see Figure
CA 02636876 2008-08-15
7A for values) was devised. The results, presented in Figure 8B for the
peripheral blood, bone
marrow and thymus of a representative mouse (Trim27) and summarized in Figure
8C for all
groups, largely confirmed microscopic evaluation. Indeed, except for
recipients of Ski
transduced cells which showed a marked reduction in B lymphocytes in their
peripheral blood
and marrow, with a compensatory increase in other cell types, most groups of
mice showed
either normal FACS profiles or presented some minor variations (detailed in
Figure 8C). This
analysis was further extended by gating only on Ly5.1+/GFP+ cells with genes
for which this was
possible and ended with the same conclusion, except that KiflO tended to act
like Ski (Figure
9). Finally, clonal analyses of recipients that were reconstituted with
retrovirally marked cells
(mostly from the 11 "cell autonomous geryes") were performed on bone marrow
(mostly myeloid,
erythoid and B cells) and thymus (less than 5% non-T cells). A representative
result is
presented in Figure 8D for Trim27 which shows that identical clones
contributed to the
reconstitution of these 2 tissues, thus reinforcing the finding that these
transduced HSCs remain
competent in T cell differentiation although they displayed enhanced
reconstitution activity. This
finding with Trim27 can be extended to all other genes but Ski, Prdm16 and
Erdrl where it
cannot be certain that the same clone contributed to thymic and bone marrow
reconstitution
(see Figure 9B).
[00132] Together, these results confirm that the majority of the genes
identified in the
screen conferred enhanced HSC activity without causing hematological disease
nor profoundly
altering cell differentiation at least until 20 weeks post-transplantation.
Prdm16 was a notable
exception.
EXAMPLE 6
Building a network of HSC activity
[00133] Epistatic studies were performed by analyzing transcription levels of
all 18 nuclear
genes identified in addition to known regulators of HSC SR, i.e., Hoxb4,
Hoxa9, Bmil while
overexpressing each of them individually, in a matrix-like manner to find any
cross-regulation
between these genes. Surprisingly, few genes significantly affected transcript
levels of tested
genes (_3-fold; black solid arrows in Figure 10B). Among them, Prdm16 was the
most influent
as it upregulated the expression of Hoxb4, known SR inducer, and Vps72, a
newly identified
HSC activity regulator with cell autonomous effect.
CA 02636876 2008-08-15
46
[00134] Moreover, some of these interactions occurred in the 2 groups of
autonomy
effectors mentioned above, e.g., Ski, Prdm16 and KIf10 have cell autonomous
effect on HSC
activity but also regulate factors that have a non-cell autonomous effect,
i.e., Fos and Sfpil.
EXAMPLE 7
Two forms of trim27
[00135] Two different forms of Trim27 have been tested in the competitive
repopulation
assay of this study. The first one, used in the primary screen, contains a
frame-shift error
(truncated form; accession number BC085503; Figure 11A, upper panel)
preserving intact only
the RING, B-box and first Coiled-coil domains of the entire protein. The other
form, latter
recognized as the full-length form (accession number BC003219; Figure 11A
bottom panel) also
contains the second Coiled-coil and the SPRY domains. The 2 FLAG-Trim27
polypeptides were
detected at the expected size and the competitive repopulation assays revealed
a different
reconstitution potential by the different forms, the highest potential being
held by the truncated
form (Figure 11 B). Based on this, the second part of the second Coiled-coil
domain in
combination with the SPRY domain, seem to limit the potential of this gene in
HSC expansion.
EXAMPLE 8
Clonal analysis
[00136] Additional clonal analyses of hematopoietic tissues (bone marrow,
blood and
thymus) derived from selected recipients sacrificed at 20 weeks post-
transplantation
confirmed the multi-potentiality and clonality of repopulation, thus
indicating that the newly
identified genes (nuclear or asymmetrical cell division factors) affect HSC
self-renewal or
proliferation. Data showing the expansion and/or differentiation of cells
transduced with
nuclear factors as well as asymmetrical cell division regulators (xbpl,
trim27, sox4, fos,
pbx2, kIf10, hes1, hnrpdl, gpsm2, ap2a2 and cbfb) are presented in Figures 13
to 26.
Similar experiments were performed using HSCs transformed with smarccl, sfpil,
hmgbl,
vps72, tcfec, zfp472, gpx3, erdrl, tmodl, cnbpl, prdm16, hdac1, erdrl, tmodl,
cnbpl,
prdm16, hdacl and ski (Figure 26).
CA 02636876 2008-08-15
47
[00137] Thus, the following genes for instance were shown to provide
competitive
advantage to transduced HSC (e.g., increasing their expansion and/or
differentiation) (Table
II): trim27, xbpl, sox4, smarccl, sfpil, fos, hmgbl, hnrpdl, vps72, tcfec,
kIf10, zfp472,
ap2a2, gpsm2, gpx3, erdrl, tmodl, cnbpl, prdm16, pml and hdacl and ski. Among
these
genes, trim27, xbpl, sox4, hnrpdl, vps72, gpx3, tmodl, cnbpl and hdacl
promoted
multilineage differentiation. Among these genes, trim27, xbpl, sox4, smarccl,
hnrpdl,
vps72, kIf10, ap2a2, gpsm2 and gpx3 promoted multiclonal expansion.
EXAMPLE 9
Materials and Methods
Retroviral vectors
[00138] Generation of MSCV-Hoxb4-IRES-GFP was described before (25) and MSCV-
NUP98-HOXA10HD-IRES-GFP (NA10HD) was a gift from Dr. Keith Humphries (26) ORF
from each candidate gene was amplified by PCR using primers containing
restriction sites
(underlined in Figure 3) and template cDNA (Figure 3; BC accession numbers
come from
ATCC, Manassas, VA, USA and AK accession numbers come from Riken DNABook,
Japan). Digested amplicons were then subcloned into I of 3 modified MSCV-PGK-
GFP
(pKOF-1, -2 or -3, containing different reading frames) according to the
reading frame
needed for each candidate and sequenced for correct integrity and reading
frame.
Animals
[00139] Recipients were C57BU6 J(B6) mice that express Ly5.2 and transplant
donors were C57B1/6Ly-Pep3b (Pep3b) congenic mice that express Ly5.1. All
animals
were housed in ventilated cages and provided with sterilized food and
acidified water at a
specific pathogen-free (SPF) animal facility at the Institute for Research in
Immunology and
Cancer in Montreal.
Purification of CD150+CD48-Lin" and CD150+CD48'Lin'Kit+Sca+ cells
[00140] Bone marrow cells were stained with allophycocyanin (APC)-labeled anti-
Gr-1,
-B220, - Ter119, and depleted using anti-APC magnetic beads and AUTO-MACS
system
CA 02636876 2008-08-15
48
(Becton-Dickinson, San Jose, CA, USA). Depleted cells were then stained with
fluorescein
isothiocyanate (FITC)-labeled anti-CD48, and phycoerythrin (PE)-labeled anti-
CD150 for
CRAs, or in addition to PE-Cy7-labeled c-Kit and PE-Cy5-labeled Sca for q-RT-
PCR
(BioLegend, San Diego, CA). Sorting was performed on a FACSAria system
(Becton-
Dickinson, San Jose, CA, USA).
Retroviral infection, cell culture and transplantation
[00141] Generation of retrovirus-producing GP+E-86 cells were performed as
previously described (9), in 96-well plate, producing one different candidate
gene/well. 1500
CD150+CD48-Lin" sorted Ly5.1+ cells/well were cocultured with irradiated (1500
cGy of137Cs
gamma radiation) GP+E-86 virus producer cells during 5 days in Dulbecco's
modified
Eagle's medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 10 ng/mL
human interleukin-6 (IL-6), 6 ng/mL murine interleukin-3 (IL-3), 100 ng/mL
murine stem cell
factor (SF), and 6 g/ml polybrene, 10 g/ml ciprofloxacin and 10"4M R-
mercaptoethanol.
After trypsinization, 3/8 of each well was prepared for transplantation of 2
sublethally
irradiated (800 cGy of137Cs gamma radiation) B6 mice (1/8 per mouse) along
with 2 x 105
whole bone marrow Ly5.2+ competitor/helper cells per mouse (Day 0). Also, 1/2
of each well
was kept in culture for an additional 7 days before being prepared for
transplantation of 3
sublethally irradiated (800 cGy of 137Cs gamma radiation) B6 mice (1/4 per
mouse) along
with 2 x 105 whole bone marrow Ly5.2+ competitor/helper cells per mouse (Day
7). The
remaining 1/8 of each well at Day 0 was kept in culture for an additional 4
days before being
analyzed by FACS to assess the infection efficiency based on the proportion of
GFP+ bone
marrow cells.
Competitive repopulation assay and flow cytometry
[00142] To determine the contributions of the transplanted donor-derived HSCs
to
hematopoietic reconstitution at various intervals posttransplantation, 50 pL
of blood
obtained from the tail vein were incubated with excess ammonium chloride
(StemCell
Technologies, Vancouver, BC, Canada) to lyse erythrocytes, and the proportions
of Ly5.1+
white blood cells were determined by flow cytometry using a PE-labeled anti-
Ly5.1
antibody, and differentiation analysis were determined on whole bone marrow
cells 20
weeks post-transplantation using APC-Cy7-labeled anti-B220, PE-Cy5-labeled
anti-CD11 b
CA 02636876 2008-08-15
49
and PE-Cy5.5-labeled anti-CD38 antibodies. Data were acquired using BD LSR II
flow
cytometer (BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo
software
(Tree Star Inc., Ashland, OR, USA).
Southern blot analysis of genomic DNA
[00143] Genomic DNA from 20 week old mice was isolated with DNAzoI reagent
(Invitrogen, Carlsbad, CA, USA), as recommended by the manufacturer. Southern
blot
analysis was performed as previously described (9). Unique proviral
integrations were
identified by digestion of DNA with EcoRl, which cleaves once within the
provirus and at
various distances within the genome. 15 g of digested whole genomic DNA was
then
separated in 1% agarose gel by electrophoresis and transferred to zeta-probe
membranes
(Bio-Rad, Mississauga, ON, Canada) and a and a 710 bp [32P]dCTP EGFP probe,
digested
from pEYFP-N1 (Clontech Laboratories Inc., Palo Alto, CA, USA) with
EcoRl/Hindlll (Invitrogen,
Burlington, ON, Canada), was used to reveal the integration pattern.
Western blot analysis
[00144] Protein expression of cloned cDNAs was assessed in retroviral
producing cell
lines. Protein extracts were obtained from transfected GP+E-86 or BOSC cells
grown in 96-
well plates by incubation with a 30 uL volume of lx Laemli (1/60 [3-
mercaptoethanol)
solution per well, followed by a 10 min boiling step. Western blots analyses
were performed
as described (9). A mouse anti-FLAG primary antibody used to reveal the
presence of the
candidate protein, followed by a goat horseradish peroxidase-conjugated anti-
mouse
secondary antibody (Biolegend San Diego, CA).
Q-RT-PCR expression studies
[00145] For gene expression profiles analyses of retrovirally transduced BM
cells, co-
cultures were initiated as described above, but the number of sorted CD150+
Sca1 +cKit+CD48-Lin" cells plated per well increased to 5000. After 5 days of
infection, cells
were again harvested using trypsinization and individual well contents
resubmitted to cell
sorting (FACSAria cell sorter, Becton-Dickinson, San Jose, CA, USA). Gates
were set to
positively seiect for GFP+ cells, excluding GP+E-86 retroviral producers by
forward- and
CA 02636876 2008-08-15
side-scatter criteria. Cells were directly collected in TrizolT"" solution to
isolate total RNA,
according to the manufacturer's protocol (Invitrogen). Reverse transcription
of total RNA
was performed using the MMLV-reverse transcriptase (RT) and random hexamers
according to manufacturer's guidelines (Invitrogen). Resulting cDNA was pre-
amplified
using a TaqMan PreAmp (Applied Biosystems, Foster City, CA) algorithm in
which
candidate genes specific oligos were added to the PreAmp Master mix (final
concentration
of 50nM). PCR conditions for the pre-amplification reactions were as follows:
95 C for 10
minutes, followed by 12 cycles of 95 C / 15 sec and 60 C / 4 min. The ABI Gene
Expression Assay was performed to measure gene expression levels using primer
and
probe sets from Applied Biosystems (primer and probe sequences are available
on
request). Q-RT-PCR reactions were done on a high-throughput ABI 7900HTTM.
Fast Real-Time PCR System (Applied Biosystems)
[00146] Briefly, the Ct (threshold cycle) values of each gene were normalized
to the
endogenous control gene R-actin (Applied Biosystems; pCt = Cttarger-
Ctendogenous) and
compared with the mean of our 3 corresponding empty vectors transduced tissue
(calibrator
sample) using the AOCt method (AACt = ACtsamp,e - OCtCalibra,or). Relative
fold difference
(RQ) and ACt values are provided in Figure 10. Q-RT-PCR cycling conditions
were 2
minutes at 50 C and 10 minutes at 95 C, followed by 40 cycles of 15 seconds at
95 C and
1 minute at 59 C. All reactions were done in triplicate. Average values were
used for
quantification.
[00147] Although the present invention has been described hereinabove by way
of
specific embodiments thereof, it can be modified, without departing from the
spirit and
nature of the subject invention as defined in the appended claims.
CA 02636876 2008-08-15
51
REFERENCES
1. L. A. Boyer et al., Ce11122, 947 (Sep 23, 2005).
2. J. Kim, J. Chu, X. Shen, J. Wang, S. H. Orkin, Ce11132, 1049 (Mar 21,
2008).
3. J. Antonchuk, G. Sauvageau, R. K. Humphries, Cel1109, 39 (Apr 5, 2002).
4. J. Zhu, Y. Zhang, G. J. Joe, R. Pompetti, S. G. Emerson, Proc Natl Acad Sci
U S A
102, 11728 (Aug 16, 2005).
5. T. Reya et al., Nature 423, 409 (May 22, 2003).
6. J. J. Schuringa, K. Y. Chung, G. Morrone, M. A. Moore, J Exp Med 200, 623
(Sep 6,
2004).
7. U. Thorsteinsdottir et al., Blood 99, 121 (Jan 1, 2002).
8. H. Ohta et al., Exp Hemato135, 817 (May, 2007).
9. J. Krosl, N. Beslu, N. Mayotte, R. K. Humphries, G. Sauvageau, Immunity 18,
561
(Apr, 2003).
10. S. Amsellem, F. Pflumio, D. Bardinet, B. Izac, P. Charneau, PH. Romeo, A.
Dubart-
Kupperschmitt and S. Fichelson Nat Med 9(11):1423-7 (Nov. 2003).
11. J. Krosl Hematol J.;5 Suppl 3:S118-21 (2004).
12. Kros12005b.
13. Sauvageau, G., Iscove, N.N., and Humphries, R.K. (2004). In vitro and in
vivo
expansion of hematopoietic stem cells. Oncogene 23, 7223-7232.
14. S. Cellot et al., Exp Hemato135, 802 (May, 2007).
15. B. Bhattacharya et al., Blood 103, 2956 (Apr 15, 2004).
16. R. W. Georgantas, 3rd et al., Cancer Res 64, 4434 (Jul 1, 2004).
17. N. B. Ivanova et al., Science 298, 601 (Oct 18, 2002).
18. R. L. Phillips et al., Science 288, 1635 (Jun 2, 2000).
19. M. Ramalho-Santos, S. Yoon, Y. Matsuzaki, R. C. Mulligan, D. A. Melton,
Science
298, 597 (Oct 18, 2002).
20. M. H. Shim, A. Hoover, N. Blake, J. G. Drachman, J. A. Reems, Exp Hematol
32, 638
(Jul, 2004).
21. F. Shojaei, L. Gallacher, M. Bhatia, Blood 103, 2530 (Apr 1, 2004).
22. A. V. Terskikh et al., Proc Natl Acad Sci USA 98, 7934 (Jul 3, 2001).
23. O. P. Pinto do, E. Wandzioch, A. Kolterud, L. Carlsson, Exp Hemato129,
1019 (Aug,
2001).
24. J. Krosl, P. Austin, N. Beslu, E. Kroon, RK. Humphries and G. Sauvageau,
Nat
Med 9 (11), 1428-32 (Nov, 2003).
25. Cell. 2002 Apr 5;109(1):39-45.
26. Exp Hematol. 2007 May;35(5):817-30.
27. Cheshier, S.H., et al. (1999). Proc Natl Acad Sci U S A 96, 3120-3125.
28. Tijssen, 1993, Laboratory Techniques in Biochemistry and Molecular Biology
--
Hybridization with Nucleic Acid Probes, Part I, Chapter 2 "Overview of
principles of
hybridization and the strategy of nucleic acid probe assays", Elsevier, New
York.
29. Ausubel, et al. (eds), 1989, Current Protocols in Molecular Biology, Vol.
1, Green
Publishing Associates, Inc., and John Wiley & Sons, Inc., New York, at p.
2.10.3