Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
BIOSYNTHETIC PATHWAY AND GENES REQUIRED FOR TROPODITHIETIC
ACID BIOSYNTHESIS IN SILICIBACTER TM1040
GOVERNMENT RIGHTS IN INVENTION
[0001] Work related to the invention was conducted in the performance of
National Science
Foundation Grant MCB0446001. The United States Government has certain rights
in the
invention.
CROSS-REFERENCE TO RELATED APPLICATION
[0002] This application claims the benefit of priority of U.S. Provisional
Patent Application
60/861,117 filed November 27, 2006 in the names of Robert Belas, et al. for
"BIOSYNTHETIC
PATHWAY AND GENES REQUIRED FOR TROPODITHIETIC ACID BIOSYNTHESIS IN
SILICIBACTER TM1040." The disclosure of U.S. Provisional Patent Application
60/861,117 is
hereby incorporated herein by reference in its entirety, for all purposes.
BACKGROUND OF THE INVENTION
Field of the Invention
[0003] This invention relates to Roseobacter bacteria and to the production of
antibiotics by use
of such microbial species.
Description of the Related Art
[0004] Bacteria of the Roseobacter clade of marine alpha-Proteobacteria stand
out as some of
the most critical players in the oceanic sulfur cycle due to the ability of
several genera to degrade
dimethylsulfoniopropionate (DMSP). While roseobacters are wide-spread
throughout the marine
ecosystem, their abundance is significantly correlated with DMSP-producing
algae, especially
prymnesiophytes and dinoflagellates, such as Prorocentrum, Alexandrium and
Pfiesteria species.
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0005] Roseobacters have abundant and diverse transporters, complex regulatory
systems,
multiple pathways for acquiring carbon and energy in seawater, with the
potential to produce
secondary, biologically active metabolites.
SUMMARY OF INVENTION
[0006] The present invention relates to Roseobacter bacteria and to the
production of antibiotic
tropodithietic acid (TDA) by use of such microbial species.
[0007] In one aspect, the invention relates to an isolated nucleic acid
encoding a megaplasmid
(pSTM3) of Silicibacter sp. TM1040, wherein the nucleic acid comprises genes
involved in
tropodithietic acid biosynthesis of Roseobacter bacteria.
[0008] Another aspect of the invention relates to a protein encoded by a
nucleic acid sequence
represented by SEQ. ID. 1; wherein the protein is involved in the biosynthesis
of tropodithietic
acid by Roseobacter bacteria.
[0009] Yet another aspect of the invention relates to a protein encoded by a
nucleic acid
sequence represented by SEQ. ID. 2; wherein the protein is involved in the
biosynthesis of
tropodithietic acid by Roseobacter bacteria.
[0010] In another aspect, the invention relates to a protein encoded by a
nucleic acid sequence
represented by SEQ. ID. 3; wherein the protein is involved in the biosynthesis
of tropodithietic
acid by Roseobacter bacteria.
[0011] An additional aspect of the invention relates to a protein encoded by a
nucleic acid
sequence represented by SEQ. ID. 4; wherein the protein is involved in the
biosynthesis of
tropodithietic acid by Roseobacter bacteria.
[0012] A further aspect of the invention relates to an antibacterial
composition comprising
tropodithietic acid isolated from bacteria of the Roseobacter clade
[0013] Another aspect of the invention relates to a method for producing an
antibacterial
composition comprising tropodithietic acid, the method comprising:
a) culturing Silicibacter sp. TM1040 in a culture medium supporting growth of
the
bacterium and production of tropodithietic acid; and
2
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
b) separating the tropodithietic acid from the culture medium; and
c) purifying the tropodithietic acid by high performance liquid
chromatography.
[0014] Still another aspect of the invention relates to a method for producing
an antibacterial
composition comprising tropodithietic acid, the method comprising:
a) culturing Roseobacter sp.27-4 in a culture medium supporting growth of the
bacterium
and production of tropodithietic acid; and
b) separating the tropodithietic acid from the culture medium; and
c) purifying the tropodithietic acid by high performance liquid
chromatography.
[0015] A further aspect of the invention relates to a method of treating or
preventing bacterial
disease in a subject in need of such treatment or prevention, comprising
administering to said
subject an antibacterial composition comprising tropodithietic acid isolated
from bacteria of the
Roseobacter clade.
[0016] Yet another aspect of the invention relates to a plasmid pSTM3.
[0017] Another aspect of the invention relates to a compound selected from the
group consisting
of:
1, 2-dihydro-phenylacetyl-CoA;
2-hydroxy-7-oxo-cyclohepta-3,5-dienecarboxylic acid;
2,7-; dihydroxy-cyclohepta-1,3,5-trienecarboxylic acid;
2,7-dihydroxy-3-oxo-cyclohepta-1,4,6-trienecarboxylic acid;
2,7-dihydroxy-3-thioxo-cyclohepta-1,4,6-trienecarboxylic acid; and
7-hydroxy-2-mercapto-3-thioxo-cyclohepta-1,4,6-trienecarboxylic acid.
[0018] Other aspects, features and advantages of the invention will be more
fully apparent from
the ensuing disclosure and appended claims.
BRIEF DESCRIPTION OF THE DRAWINGS
[0019] FIG. 1. When grown in static liquid media, Silicibacter sp. strain
TM1040 produces a
yellow-brown pigment and has a large amount of antibacterial activity, which
was measured by a
3
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
well diffusion assay using V. anguillarum as the target organism (Materials
and Methods). In
contrast, pigment and antibacterial compound is very low under 30 C shaking
conditions.
[0020] FIG. 2. Tropodithietic acid. C18 reverse phase HPLC chromatograms of
ethyl acetate
extracts from TM1040 and Phaeobacter 27-4. Insets show the UV spectra of the
HPLC peak
corresponding to the antibiotic activity. For 27-4, the peak is TDA.
[0021] FIG. 3. Genes required for synthesis of TDA in TM1040.The black boxes
indicate the
ORF interrupted by the transposon. Arrows indicate ORFs transcriptional
orientations and hatch
marks indicate a break in the region. (A) Sulfur assimilation genes, tdaH,
malY, cysl, are located
in the TM1040 chromosome. Phenylacetate catabolism genes are in the
megaplasmid pSTM1.
(B) tdaA-tdaF genes reside on a cryptic plasmid, with their closest homologues
found on the
chromosome of P. denitrificans PD1222. tdaH encodes sulfite oxidase domain
protein; hik2
encodes two-component hybrid sensor and regulator; malY encodes (3-C-S lyase
(cystathionase);
asnC encodes transcriptional regulator AsnC family; cysG encodes siroheme
synthesis; hypo
encodes hypothetical protein; cysl encodes sulfite reductase beta (siroheme-
dependent); cysH
encodes adenylylsulfate reductase; gntR encodes GntR family transcriptional
regulator; paaG,
paaH, paa7, paaJ, paaK encode respectively phenylacetic acid degradation
protein complex
protein 1,2,3,4,5;tdaA encodes LysR substrate binding domain protein; tdaB
encodes (3-etherase;
glutathione-S-transferase; tdaC encodes prephenate dehydratase; tdaD, 4-
hydroxybenzoyl-CoA
thioesterase; tdaE encodes Acyl-CoA dehydrogenase; tdaF encodes
phosphopantothenoylcysteine decarboxylase. P. denitrificans PD1222 genome
contains two
chromosomes and one plasmid, whereas tdaAB, tdaCDE and tdaF homologue genes
located
discretely in a 19 kb region of chromosome 1.
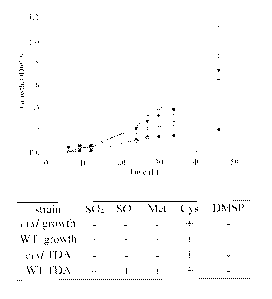
[0022] FIG. 4. Growth and TDA synthesis is affected by mutations in cysl.
TM1040 (inverted
triangles) and the cysl mutant (HG1220; circles) were grown in minimal medium
containing
either methionine (closed symbols) or methionine (open symbols), and growth
was measured
optically at 600 nm. Unlike the wild-type, the Cyst mutant cannot grow
methionine, but does
utilize cysteine. Measurement of antibiotic activity indicates that the cysl
defect also affects
TDA synthesis, which is corrected by the addition of cysteine to the medium,
but not
methionine, DMSP, sulfite, or sulfate addition (table).
4
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0023] FIG. 5. TM1040 tda genes reside on a cryptic plasmid that undergoes a
low frequency
spontaneous loss. (A). Pigment synthesis. TM1040 (wt) produces a yellow-brown
extracellular
pigment that is correlated with TDA synthesis. In contrast, a tdaE:Tn mutant
(strain HG1265)
and a spontaneous mutant (sm; TM1040SM) are nonpigmented and have lost the
ability to
produce both TDA and pigment. (B). Spontaneous loss of pigment and antibiotic
activity results
from a loss of tda genes. PCR amplification of tdaE results in a band from wt
and tdaE:Tn
DNA, respectively, with the additional 2 kb in size of the tdaE:Tn product
resulting from
insertion of the transposon. No product was amplified from the spontaneous
nonpigment mutant
(sm). (C). PFGE separation of total DNA obtained from TM1040 (wt), the
spontaneous
nonpigmented mutant (sm), and the tdaE:Tn mutant. All three strains show a
fuzzy band at ca.
130 kb, with a slight increases in the width of the wt and tdaE bands. (D).
Southern blot
hybridization of the PFGE gel to labeled tdaE DNA. The tdaE probe hybridized
to the band
migrating at ca. 130 kb in both wt and tdaE:Tn (first and third lanes), but
failed to hybrid to the
DNA obtained from the spontaneous nonpigmented mutant. (E). Ncol digestion of
plasmid DNA
isolated from TM1040 (wt), the spontaneous nonpigmented mutant (sm), and
HG1265
(tdaE:Tn), respectively. The digested DNAs were separated by electrophoresis
and the band
patterns compared to each other and to an in silico Ncol digestion of pSTM2
(supplemental
data). The pattern of fragments from sm DNA matched the predicted pSTM2 Ncol
digestion,
while both wt and tdaE DNA patterns showed evidence of additional restriction
fragments. (F).
Southern blot hybridization of Ncol-digested plasmid DNA to tdaE. A tdaE probe
hybridizes to
one fragment in wt and tdaE:Tn DNA cut with Ncol, but to any fragments
produced from Ncol
digestion of plasmid. The increase in the size of the fragment in tdaE:Tn
results from the
insertion of the 2 kb transposon.
[0024] FIG. 6. Ncol digestion patterns of pSTM3 transformed into E. coli. A
mixture of plasmid
pSTM3-1265 (pSTM3 harboring a transposon in tdaE) and pSTM2 was isolated from
HG1265
and the DNAs used to transform E. coli (Materials and Methods). Each of the
plasmids harbored
in the resulting Kanr transformants was purified, digested with Ncol, and the
resulting DNA
fragments separated by agarose gel electrophoresis. Of the total DNAs
examined, four types of
band patterns emerged and are shown in lanes 1-4, respectively.
5
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0025] FIG. 7. DNA from other roseobacter species hybridizes to tda DNA. Total
DNA was
extracted from 13 roseobacters, TM1040, and a non-roseobacter control species
(V.
anguillarum), and used in a slot blot hybridization with labeled tda DNA.
Positive hybridization
was strongly correlated with measurable antibiotic activity (indicated by an
asterick). The strains
used were: ISM: Roseovarius strain ISM; TM1038: Roseobacter sp. strain TM1038;
TM1039,
Roseobacter sp. strain TM1039; 33942, Roseobacter denitrificans ATCC 33942;
SE62,
Sulfitobacter strain SE62; 49566, Roseobacter litoralis ATCC 49566; DSS-3,
Silicibacter
pomeroyi DSS-3; EE36, Sulfitobacter strain EE36; 1921, Sulfitobacter strain
1921; TM1040,
Silicibacter sp. TM1040; V. a, Vibrio anguillarum; 51442, Roseobacter algicola
ATCC 51442;
27-4, Phaeobacter 27-4; TM1035, Roseovarius sp. strain TM1035; and, TM1042,
Roseovarius
sp. strain TM1042.
[0026] FIG. 8. The presence and relative abundance of each of the Tda proteins
identified in
TM1040 (rows) in the GOS metagenomic database (via the internet website at
hypertext transfer
protocol address, camera.calit2.net/). The relative abundance is based on the
total BLASTP
matching sequences in the individual filters using a cutoff E value of 1E-20
(48). The
distribution of Tda proteins harbored on pSTM3 (TdaA-F) in the sample is
remarkably different
from the distribution of Paa and sulfur metabolism proteins (Cysl, Ma1Y, and
TdaH), which have
a more even distribution throughout the series of samples. Relative abundance
is indicated by the
size of the circle. GOS sample numbers are indicated on the horizontal axis.
[0027] FIG. 9. A putative model of the TDA biosynthetic pathway based on the
present genetic
analysis. The suggested pathway involved phenylacetate derivation from
shikimate-chorismate
and degradation pathway providing precursors (step 1-6) and an core oxidative
ring-expansion
pathway forming the seven carbon tropolone skeleton (step7-10) followed by
sulfur-oxygen
exchange (step11-15), consistent with the proposed TDA synthesis based on
chemical labeling
studies in Pseudomonas CB-104 (14). The protein assignment was based on
predicted functions.
DETAILED DESCRIPTION OF THE INVENTION
[0028] The present invention relates to Roseobacter bacteria and to the
production of
tropodithietic acid (TDA) by use of such microbial species.
6
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0029] The symbiotic association between the roseobacter Silicibacter sp.
TM1040 and the
dinoflagellate Pfiesteria piscicida involves bacterial chemotaxis to
dinoflagellate-produced
dimethylsulfoniopropionate (DMSP), DMSP demethylation, and ultimately a
biofilm on the
surface of the host. Biofilm formation is coincident with the production of an
antibiotic and a
yellow-brown pigment. The antibiotic is a sulfur-containing compound,
tropodithietic acid
(TDA). Using random transposon insertion mutagenesis, 12 genes were identified
as critical for
TDA biosynthesis by the bacteria, and mutation in any one of these results in
loss of antibiotic
activity (Tda ) and pigment production. Unexpectedly, six of the genes,
referred to as tdaA-F,
could not be found on the annotated TM1040 genome and were instead located on
a previously
unidentified cryptic megaplasmid (ca. 130 kB; pSTM3) that exhibited a low
frequency of
spontaneous loss. Homologs of tdaA and tdaB from Silicibacter sp. TM1040 were
identified by
mutagenesis in another TDA-producing roseobacter, Phaeobacter 27-4, which also
possesses
two large plasmids (ca. 60 and ca. 70 kb, respectively), and tda genes were
found by DNA:DNA
hybridization in 88 % of a diverse collection of 9 roseobacters with known
antibiotic activity.
These data suggest that roseobacters employ a common pathway for TDA
biosynthesis that
involves plasmid-encoded proteins. Using metagenomic library databases and a
bioinformatics
approach, a pronounced difference in the biogeographical distribution between
the critical TDA
synthesis genes was observed, implying substantial environmental preference
differences among
these genes.
[0030] The present invention in another specific aspect relates to the
interaction of a roseobacter,
Silicibacter sp. TM1040, and Pfiesteria piscicida. Silicibacter sp. TM1040
(hereafter referred to
as TM1040) is isolatable from laboratory microcosm culture of heterotrophic
DMSP-producing
dinoflagellate P. piscicida. Marine algae are major producers of DMSP in the
marine
environment while members of the Roseobacter clade are capable of DMSP
catabolism.
TM1040 degrades DMSP via a demethylation pathway producing 3-
methylmercaptopropionate
(MMPA) as a major breakdown product. The bacteria respond via chemotaxis to
dinoflagellate
homogenates, and are specifically attracted to DMSP, methionine, and valine.
TM1040 motility
is important in the initial phases of the symbiosis. Once the bacteria are in
close proximity to
their host, TM1040 forms a biofilm on the surface of the dinoflagellate. The
symbiosis includes
7
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
two parts: one that involves chemotaxis and motility, and a second step in
which a biofilm
predominates.
[0031] Specific phenotypes, e.g., the ability to produce antibacterial
compounds and biofilm
formation, may give members of the Roseobacter clade a selective advantage,
and help to
explain the dominance of members of this clade in association with marine
algae. The
production of an antibiotic activity is observed in roseobacters and is
hypothesized to provide an
advantage when colonizing phytoplanktonic hosts, such as dinoflagellates. The
genome of
TM1040 consists of a 3.2 Mb chromosome and two plasmids, pSTM1 (823 Kb) and
pSTM2
(131 Kb) (41). A comparison between TM1040 and two other roseobacters
(Silicibacter
pomeroyi DSS-3 and Jannaschia sp. CSS-1) suggests that roseobacters have
abundant and
diverse transporters, complex regulatory systems, multiple pathways for
acquiring carbon and
energy in seawater, with the potential to produce secondary, biologically
active metabolites.
[0032] Biologically active metabolites, including antibacterial compounds, are
obtainable from
roseobacters. A sulfur-containing antibiotic compound, tropodithietic acid
(TDA), has been
isolated and chemically characterized from Phaeobacter 27-4, hereafter called
27-4, and
Roseobacter T5. The chemical backbone of TDA (shown in Fig. 2) is a seven
member aromatic
tropolone ring, which is highly significant as tropolone derivatives, notably
hydroxylated forms,
are widely seen as medically important sources of antibacterial, antifungal,
antiviral, and
antiparasitic agents. Thiotropocin, another tropothione derivative closely
related to TDA, can be
synthesized from shikimate by an oxidative ring expansion of phenylacetic
acid.
[0033] We have used both genomic and genetic techniques to identify the genes
and proteins
required for TDA synthesis in TM1040 and 27-4 as models for the Roseobacter
clade. In the
process of locating these genes, we discovered a megaplasmid critical for TDA
biosynthesis that
is part of the TM1040 genome, as hereinafter more fully described.
[0034] Materials and Methods
[0035] Bacteria and media
[0036] The strains used in our study are listed in (Table 1). Silicibacter sp.
TM1040,
Phaeobacter 27-4 and Vibrio anguillarum 90-11-287 were grown and maintained in
2216 marine
broth or 2216 agar as recommended by the manufacturer (BD Biosciences,
Franklin Lakes, NJ).
A marine basal minimal medium (MBM; per liter: 8.47g Tris HC1, 0.37 g of
NH4C1, 0.0022 g of
8
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
K2HPO4, 11.6 g NaC1, 6 g MgSO4, 0.75 g KC1, 1.47 g CaC12=2H20, 2.5 mg FeEDTA;
pH 7.6, 1
ml of RPMI-1640 vitamins [Sigma R7256]) was used for determining carbon and
sulfur
requirements. Sole carbon sources were added at a final concentration of 1
g/1. Escherichia coli
strains were grown in Luria-Bertani (LB) broth or on LB agar containing 1.5%
Bacto Agar
(Becton Dickinson, Franklin Lakes, N.J.). As appropriate, kanamycin was used
at 120 g per ml
for Roseobacter strains and 50 g per ml for E. coli.
[0037] Characterization of antibiotic
[0038] Bacterial spent medium was either injected directly (up to 10 L) or
purified by mixed
phase anion-exchange reversed phase mini column chromatography on Oasis MAX
columns as
previously described. Tropodithietic acid was analyzed by reverse phase liquid
chromatography
(LC) on an Agilent 1100 HPLC system equipped with a diode array detector
(DAD). Separation
was conducted using a Phenomenex (Torrance, CA) Curosil PFP 15 cm, 2 mm, 3 m
column
using a water-acetonitrile (ACN) gradient system. Both solvents contained 200
L/L
trifluoroacetic acid, and started 35% ACN increasing this linear to 60% in 6
min. The
wavelength 304 4 nm was used for detection. LC-DAD with online high
resolution mass
spectrometry (HR-MS) using positive and negative electrospray was used for
validation of
tropodithietic acid detection as previous described.
[0039] Transposon mutagenesis and Tda screening
[0040] Electrocompetent roseobacter strains were prepared following the method
described by
Garg et al. as modified by Miller and Belas. Random transposon insertion
libraries were
constructed in TM1040 and 27-4 using the EZ-Tn5<R6Kyori/KAN-2>Tnp
TransposomeTM Kit
(Epicentre, Madison, WI). Strains were spread onto 2216 plates containing
kanamycin and
incubated for 1 day at 30 C. Individual Kanr transposon insertion strains
were transferred to
7x7- arrays on 2216 marine agar plus kanamycin to facilitate further
screening. To screen for
loss-of-function, antibiotic-negative (Tda ) mutants, a modification of the
method described by
Bruhn et al. was used. Bacteria were replicated, as a 7x7 array, to a lawn of
Vibrio anguillarum
strain 90-11-287, and incubated at 20 C for 24 h, after which a zone of
clearing indicative of
antibiotic production was measured and compared to the parental strain (TM1040
or 27-4). For
purposes of this study, Tda is defined as a strain lacking a detectable zone
of clearing on V.
anguillarum. Strains determined to be Tda- by the modified well-diffusion
assay were further
9
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
tested by incubation at 30 C for 48 h in 2216 marine broth without shaking.
Bacteria were
removed by filtering through a 0.22 m MCE membrane (Millex Millipore,
Bedford, MA) and
the antibacterial activity of the supernatant measured using the V.
anguillarum well diffusion
assay, as described by Bruhn et al.
[0041] Sole carbon and sulfur source growth
[0042] Bacterial utilization of sole carbon sources was determined by
measuring growth in
MBM broth that was modified by replacing glycerol with the carbon source to be
tested. Carbon
compounds tested included amino acids (alanine, arginine, aspartic acid,
cysteine, glutamic acid,
glycine, histidine, isoleucine, leucine, methionine, phenylalanine, proline,
serine, threonine,
tryptophan, tyrosine, valine); sugars (arabinose, fructose, galactose glucose,
lactose, maltose,
mannose, N-acetylglucosamine, ribose, sucrose, xylose); tricarboxylic acid
cycle (TCA)
intermediates (citrate, fumurate, succinate); as well as phenylacetic acid and
sodium
phenylpyruvate.
[0043] Sulfur utilization was tested by growth in MBM containing different
sulfur sources:
DMSP, cysteine, methionine, sodium sulfate, and sodium sulfite.
[0044] Bioinformatics analysis
[0045] Approximately 1 g of genomic DNA isolated from the candidate mutant
was digested
with Nco I, self-religated with T4 DNA ligase, and electroporated into DH5a
Qpir). Following
selection for kanamycin resistance, Kanr colonies were picked and the plasmid
isolated for
bidirectional sequencing with transposon-specific primers as recommended by
the supplier
(Epicentre, Madison, WI). Nucleotide sequence thus obtained was analyzed by
BLAST analyses
using DNA-DNA homology searches again the Silicibacter sp. TM1040 genome
(Accession
numbers: NC_008044 , NC_008043 , NC_008042). The genes identified are listed
in Table 2
for TM1040 and Table 3 for 27-4.
Table 2. Silicibacter sp. TM1040 genes and encoded proteins required for the
regulation
and synthesis of tropodithietic acid.
GenBank
Gene Number Assession Gene
Number Designation Function Best Hit
Ortholog / E
score
...Y.......... ..;,.... ...Y.... ..............
...,...........................................................................
:...............................................:~
e~~rs~..~ .~.....~...~.... ..a...~......... .~~.. o~
..............::::::::.......:::::::::.......:::::::::.......:::::::::.......::
:::::::
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
----------------------------------------------------------------------- -------
--------------------------- ---------------------------------------------------
------------------------------------------------------------------------
TM1040_3728 CP000376 paaK Phenylacetate oxidoreductase Roseobacter sp.
MED 193
phenylacetic acid
degradation
oxidoreductase
PaaK / 8e-161
---------------------------------------- ----------------------------- --------
-- ------------------------------------------------------------------- --------
--------------------------------------
TM1040_3726 CP000376 paal Phenylacetate oxygenase Roseobacter sp.
MED 193
phenylacetic acid
degradation
protein Paal / 4e-
110
---------------------------------------- -----------------------------;--------
--------------------------_----------------------------------------------------
---------------------- ----------------------------------------------
TM1040_3727 CP000376 paaJ Phenylacetate oxygenase Roseobacter sp.
MED193
phenylacetic acid
degradation
protein PaaJ / 2e-
69
- ----:---------- - - - - - - - - - -------
---------------------------------- --------------------------------------------
--------------------------
EF139203 EF139203 tdaD 4-hydroxybenzoyl-CoA Paracoccus.
thioesterase denitrificans
PD1222
conserved
hypothetical
protein / 2e-45
---------------------------------------- -----------------------------
............ ------------------------------------------------------------------
-------------------------------- ---------
EF139204 EF139204 tdaE Acyl-CoA dehydrogenase Paracoccus
denitrificans
PD 1222 acyl-
CoA
dehydrogenase /
9e-120
_____
---------------------------------- ------- - - - - ---
_____________________________
EF139201 EF139201 tdaB (3 etherase, glutathione S Paracoccus
transferase denitrificans
PD 1222 putative
(3-etherase ((3-aryl
ether cleaving
enzyme) protein /
6e-56
-----------------------------------------
---------
=
EF130202 EF130202 tdaC Prephenate dehydratase Paracoccus
denitrificans
PD1222
hypothetical
.::::~ protein: ~-.2e.: 45-.::::::
11~~~1:1':::~~~~~
~::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>:::
:>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::
::>::::>::::>::::>::::>::::>::::>::::>::::>::::>
:::::::::::::::::::::::::::::::.:.:?
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . ..
11
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
----------------------------------------------------------------------- -------
--------------------------- ---------------------------------------------------
------------------------------------------------------------------------
TM1040_2581 CP000377 malY (3-C-S lyase (cystathionase); Roseobacter sp.
amino transferase MED193
aminotransferase,
classes I and II /
0.0
---------------------------------------- ----------
--------------------------------------------------------------
TM10400961 CP000377 tdaH Sulfite oxidase domain Sulfitobacter sp.
protein NAS-14.1
hypothetical
protein / 7e 34
---------------------------------------- ----------------------------- --------
--- ---------------------------------------------------------------
-------------------------------- ----------
TM1040_1758 CP000377 cysl Sulfite reductase Roseobacter sp.
MED 193 sulfite
reductase / 0.0
~::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>
::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::
>::::> ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: :::::
::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::::: ::>:
:~~::::::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:
::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>:::>::::::>::::::>::::::>::::::>
::::::>::::::>::::::>::::
~ :::::::::
;::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
:::::::::::::::::::::::::::::::
~~ EF139205 EF139205 tdaF Phosphopantothenoylcysteine Paracoccus
decarboxylase denitrificans
PD1222
flavoprotein / 2e-
:
>......... ......... ;
>;
:
.........; .........
........ ....... ........ ......... ........ ........ ........ ......... .
.:.,
EF139200 EF139200 tdaA LysR substrate binding Paracoccus
domain protein denitrificans
PD1222
regulatory
protein,
LysR:LysR,
substrate-binding
/ le-29
.................................................. ___________________
________
_______________________________________________________________________________
____________________________________________
Table 3. Sole carbon source tested for TM1040 and mutants.
Gene Cy Trp Phe Phenylaceti Sodium Sodium 2216 Other
s c acid phenylpyruvat phenylbutyra Amino
e te acid
WT + + + + + + + +
paai + - - - - - + +
paaJ + - - - - - + +
paaK + - - - - - + +
tdaA + + + + + + + +
tdaB + + + + + + + +
tdaC + + + + + + + +
tdaD + + + + + + + +
tdaE + + + + + + + +
tdaF + + + + + + + +
cysl + - - - - - + -
12
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
malY + + + + + + + +
tdaH + + + + + + + +
[0046] Signature amino acid domains in the deduced amino acid sequence of the
respective
ORFs were identified using BLASTP, Pfam, SMART, and the Conserved Domains
Database
(CDD; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Homologs in
roseobacters were
identified using BLASTP analysis of Roseobase (http://www.roseobase.org/) and
Gordon and
Betty Moore Foundation Marine Microbial Genome databases
(https://research.venterinstitute.org/moore/) with respective predicted
protein sequence as the
query sequence and a maximum E value of 1E-30. Homologs in the Global Ocean
Sampling
Expedition metagenomic libraries (http://camera.calit2.net/index) were
identified by BLASTP
analysis using a cutoff E value 1E-20.
[0047] DNA extraction and separation
[0048] Chromosomal DNA was extracted from bacterial cells by routine methods
or by the
DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA). Plasmid DNA was prepared by
the
alkaline lysis method, digested with Ncol (New England Biolabs, Beverly, MA),
and the
resulting restriction fragments were separated by agarose gel electrophoresis
in Tris-acetate-
EDTA (TAE) buffer.
[0049] Pulsed Field Gel Electrophoresis (PFGE) was performed using a CHEF DR-
III clamped
homogeneous electric field system (Bio-Rad, Richmond, Calif.) with a 1%
agarose gel, a 3- to
15-s pulse ramp, an electrophoresis rate of 6.0 V/cm with an included angle of
120 at a constant
temperature of 14 C, and a run time of 26 h. Gels were stained with ethidium
bromide (EB) and
visualized with a Typhoon 9410 (Amersham Biosciences, Piscataway, N.J.).
[0050] PCR amplification
[0051] Multiplex PCR amplification was used to screen for the presence of tda
genes in Tda
mutants. A 716-bp sequence internal to tdaE was amplified using primers 5'-
CAGATGATGGTGCCAAAGGACTAT-3'and 5' -GGTCAGTTTCTTCTGCACATACTGG-3' ,
while (in the same reaction), an interna1401-bp fragment of flaA (accession
number: CP000377,
locus tag: TM1040_2952) was also amplified using primers 5'-
TTGCAGTATCCAATGGTCGTG-3' and 5'-TGAATTGCGTCAGAGTTTGCC-3' as a control.
Standard PCR amplification conditions were 100 M dNTP each, 0.2 M of each
primer, 1 U
13
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
Taq DNA polymerase (New England Biolabs, Beverly, MA) in lx reaction buffer
(New England
BioLabs) with an initial denaturing step at 94 C for 3 min, followed by 30
cycles of 94 C for 1
min each, annealing at 55 C for 30 s, and an elongation at 72 C for 1 min.
[0052] To detect the tdaA-E locus, PCR amplification was conducted with a
forward primer
complementary to tdaA (5'-CGCTTTCCGGAACTGGAGAT-3') and a reverse primer
complementary to tdaE (5' -GGCTGCCGTATAGTTTCAGCA-3' ) using the. Expand Long
Template PCR System (Roche Applied Science, Indianapolis, IN) and the PCR
program
conditions and cycle parameters as described by the supplier.
[0053] DNA hybridization
[0054] DNA:DNA hybridization by Southern `slot' blot (3) was used to detect
the presence of
tda genes in other roseobacters. The roseobacter strains used were:
Phaeobacter strain 27-4,
Roseobacter algicola ATCC 51442, Roseobacter denitrificans ATCC 33942,
Roseobacter
litoralis ATCC 49566, Roseobacter sp. strain TM1038, Roseobacter sp. strain
TM1039,
Roseovarius sp. strain TM1035, Roseovarius sp. strain TM1042, Roseovarius
strain ISM,
Silicibacter pomeroyi DSS-3, Silicibacter sp. strain TM1040, Sulfitobacter
strain EE36,
Sulfitobacter strain 1921, Sulfitobacter strain SE62, and Vibrio anguillarum
90-11-287.
[0055] Following extraction, 100 ng of total genomic DNA purified from each
strain was
spotted onto a positively charged nylon membrane (Roche). The DNA was cross-
linked to the
membrane with ultraviolet light using a Stratalinker UV Crosslinker
(Stratagene, La Jolla, CA),
followed by prehybridization of the membrane at 25 C for 30 min, using the DIG
High Prime
DNA Labeling and Detection Starter Kit II (Roche) as described by the
manufacturer. The
membrane was incubated at 25 C overnight with a double-stranded DNA probe
prepared by
Hind III digestion of a plasmid bearing tdaA cloned from strain HG1310 that
was labeled with
digoxigenin-dUTP using random priming as recommended by the manufactures
(Roche).
Unbound labeled DNA was removed from the membrane by 2 x5 min in 2xSSC, 0.1%
SDS
followed by 2 x15 min in 0.2xSSC, 0.1% SDS (3). In the southern blot, the
membrane was
prehybridized for 30 min in the same buffer to which was added a tdaD gene
probe, and the
probe allowed to hybridize overnight at 42 C. The blots were washed under high
stringency
conditions following the manufacturer's protocol (Roche applied science) and
exposed to Lumi-
14
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
film chemiluminescent detection film (Roche) for subsequent detection of the
hybridization
signal.
[0056] RESULTS
[0057] TM1040 produces the sulfur-containing antibiotic tropodithietic acid
[0058] TM1040 produces an extracellular broad spectrum antibacterial compound
capable of
inhibiting or killing many bacteria. We found that greater antibacterial
activity occurred when
the bacteria were grown in a nutrient broth culture under static conditions,
i.e., no shaking,
compared to shaking conditions (11 mm; Fig. 1). Under static conditions,
TM1040 cells attached
to one another forming rosettes and produced a very distinct yellow-brown
pigment (Fig. 1).
These phenotypes are consistent with Phaeobacter 27-4 and other roseobacters.
During the
course of this investigation, non-pigmented colonies were sometimes seen after
TM1040 was
incubated on nutrient agar, and subsequent analysis revealed that these `white
spontaneous
mutants' also had lost antibacterial activity as well.
[0059] TM1040 produces an antibiotic and shares common phenotypic traits with
other
roseobacters, notably Phaeobacter 27-4 whose antibiotic is tropodithietic acid
(TDA). We
therefore hypothesized that the antibacterial compound produced by TM1040 may
also be
tropodithietic acid. Cell-free supernatants were collected independently from
both TM1040 and
27-4, ethyl acetate extraction of the supernatants was used to separate TDA
from other
compounds, and the concentrated extract was analyzed by HPLC. The resulting
elution
chromatograms and subsequent UV spectra of the putative peak of TDA from
TM1040 and 27-4
are shown in Fig. 2. Both chromatograms and UV spectra are nearly identical,
indicating
chemically similar metabolites are produced by both strains. A compound with a
retention time
of 4.2 min (indicated in Fig. 2) is observed in both chromatograms and has
been positively
identified as TDA in 27-4. The equivalent `TDA peak' from TM1040 has a UV
spectrum that
overlaps with that of published spectrum of TDA obtained from 27-4, with four
major
absorptions at 210 nm, 304 nm, 355 nm and 452 nm. Mass spectroscopy of the
TM1040 `TDA
peak' was used to confirm the efficacy of the compound as TDA. Taken together,
the data
corroborate TDA as the antibacterial metabolite produced by TM1040.
[0060] Identification of genes involved in the synthesis of TDA
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0061] With the exception of genes involved in shikimate and phenylacetate
metabolism, an
analysis of the genome of TM1040 does not provide much insight into genes
likely to participate
in the biosynthesis and regulation of TDA. To determine the genes required for
TDA synthesis, a
genome-wide random-insertion transposon bank of 11,284 Kanr colonies was
generated in
TM1040 and screened for antibiotic loss-of-function mutants (Tda phenotype).
Approximately
0.7% of the transposon insertions (81 out of 11,284) were Tda mutants, all of
which were
defective in TDA synthesis as well as in pigment formation.
[0062] The location of the transposon insertion site in each of the 81 Tda
mutants was
determined by sequencing TM1040 DNA adjacent to the transposon. The pair of
sequences
(both sides of the transposon insertion point) obtained from each mutant was
used to search the
annotated TM1040 genome to identify the mutated gene. Surprisingly, we were
unable to find
homologs in the genome for 32 or nearly 40% of the Tda mutants, yet these DNAs
overlapped
permitting assembly into one large contiguous DNA fragment of 4.5 kb harboring
at least 6
ORFs that we have called tdaA-F (Table 2 and Fig. 3A). It is clear that tdaA-F
represent DNA
that is not part of the original annotation of the genome, suggesting that
this DNA may have
been lost from the sequenced variant of TM1040. A thorough analysis of these
`orphan' genes is
presented below (TDA biosynthesis genes resided on a 130 kb cryptic plasmid).
[0063] Forty nine Tda mutants had transposon insertions in genes found in one
of the three
DNAs that make up the genome. Due to the observation of a low frequency
spontaneous loss of
TDA synthesis and knowledge of the existence of tdaA-F, we analyzed each of
the 49 genomic
Tda strains for the presence of tdaA-F. Nearly 90% (43 out of 49) did not
harbor tdaA-F, as
determined by PCR amplification with primers to tdaE, and had lost this DNA
presumably
resulting in their Tda phenotype. The transposon insertion in these strains
may contribute to the
Tda phenotype.
[0064] The sequences obtained from the remaining 6 Tda mutants were highly
informative
(Table 2).
[0065] An analysis of the genes identified from the 6`genomic' TDA- mutants
revealed that
the phenylacetate catabolism (paa) pathway is required for TDA synthesis (Fig.
3A). Transposon
insertions were identified in homologs of paa7, paaJ, and paaK. The deduced
amino acid
sequence from each of these ORFs had strong homology to similar proteins
encoded by other
16
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
roseobacters. For example, TM1040 paal is 79% similar to paal of Silicibacter
pomeroyi DSS-
3, TM1040 paaT is 74% similar to an ORF of Roseobacter sp. MED193, and paaK is
77% to
Roseobacter sp. MED193 paaK (Table 2). In other bacteria, paaGHIJK encodes a
ring-
hydroxylating complex of proteins that is responsible for the first step in
the aerobic catabolism
of phenylacetate involving Coenzyme A(CoA) activation, producing 1, 2-dihydro-
phenylacetate-CoA. The finding that mutations in paa genes affects TDA
synthesis is consistent
with the biochemical evidence of phenylacetate metabolism in thiotropocin
synthesis.
[0066] Mutants with defects in phenylacetate metabolism were also unable to
grow on
phenylalanine, phenylacetic acid, tryptophan, sodium phenylpyruvate or
phenylbutyrate as a sole
carbon source (Table 3).
[0067] TDA is a disulfide-modified tropolone compound, indicating that sulfur
metabolism
must be involved in TDA synthesis. This hypothesis is supported by the
identification of 3 Tda
mutants (Table 2) each with a transposon inserted in a gene whose product is
involved in sulfur
metabolism: cysl, malY, and an ORF (tdaH) with homology to sulfite oxidase
(Table 2). The
identification of these genes suggests that sulfur from reductive sulfur
pathways is used and
incorporated into TDA, which was tested by observing growth of the sulfur-
metabolism mutants
on a minimal medium containing a sole sulfur source (Materials and Methods).
The results are
shown in Fig. 4. The cysl mutant grew when provided complex sulfur sources or
cysteine and
was unable to utilize DMSP, S032 , S042 , or methionine. The addition of
cysteine to the
medium resulted in enhanced growth of the cysl mutant as well as increased
synthesis of TDA
(Fig. 4).
[0068] TDA biosynthesis genes resided on a 130 kb crXptic plasmid
[0069] As previously described, tdaA-F genes were not part of the annotated
TM1040 genome
and were absent in spontaneous Tda mutants. We conducted a series of
bioinformatic analyses
to elucidate the potential function of these genes (Table 2) and their
proteins. Interestingly, these
genes share their strongest homology with a similar set of genes in Paracoccus
denitrificans
PD1222 chromosome 1 (Accession number: NC_008686), a non-motile
alphaproteobacterium
first isolated from soil by Beijerinck. As shown in Fig. 3B, the orientation
and spacing between
tdaA and tdaB suggests that these genes form a bicistronic message while tdaC-
E are likely to
compose an operon separate from tdaAB. tdaF is in a different operon (Fig. 3).
17
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0070] Amino acid domain identification was useful in assigning potential
functions to the
encoded proteins. For example, TdaA (Table 2) has homology with LysR
regulatory proteins,
possessing a helix-turn-helix and a LysR substrate-binding domain (57). TdaA
is the only
regulatory protein uncovered in this study, perhaps indicating that it is the
sole regulator of TDA
synthesis. The remaining ORFs encode putative enzymes. TdaB contains a
glutathione S-
transferase (GST) domain and belongs to the bacterial GST protein family
(Table 1). TdaC has
an amino acid domain with homology to prephenate dehydratase (PheA), an enzyme
involved in
the conversion of chorismate to prephenate, a step in the pathway leading to
phenylacetate
synthesis.
Table 1. Bacterial strains and plasmids used.
Strain/plasmid Genotype/phenotype Source of reference
Escherichia coli
F- endAl hsdR17 (rK mK ) supE44 thi-1 (1)
DH5a
recAl gyrA96 relAl 080dlacZ4M15
DH5a(a,pir) DH5cc transduced with Apir (2, 3)
EC100D pir+ F- mcrA D(mrr-hsdRMS-mcrBC) EpicentreTm
f80dlacZDM15 DlacX74 recAl endAl
araD139 D(ara, leu)7697 galU galK 1- rpsL
nupG pir+(DHFR).
Roseobacters
Silicibacter sp. Wild type, antibacterial activity (4)
TM 1040
Mutants derived from TM1040
Silicibacter sp. None pigment and tda spontaneous strain current study
1 Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a
laboratory manual., 2nd ed. Cold
Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
2 Hanahan, D. 1983. Studies on transformation of Escherichia coli with
plasmids. J. Mol. Biol. 166:557-580.
3 Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Transcomplementation-
dependent replication of a low
molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208.
' Miller, T. R., and R. Belas. 2004. Dimethylsulfoniopropionate metabolism by
Pfiesteria-associated Roseobacter
spp. Appl. Environ. Microbiol. 70:3383-3391
18
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
TM1040 SM
HG1005 paaK::EZ-Tn5,Kan "
HG1015 tdaB::EZ-Tn5,Kan "
HG1050 tdaF::EZ-Tn5,Kan "
HG1056 paaJ::EZ-Tn5,Kan "
HG1080 tdaC::EZ-Tn5,Kan "
HG1110 tdaD::EZ-Tn5,Kan "
HG1213 malY::EZ-Tn5,Kan "
HG1220 cysl::EZ-Tn5,Kan "
HG1244 tdaH::EZ-Tn5,Kan "
HG1265 tdaE::EZ-Tn5,Kan "
HG1299 paal::EZ-Tn5,Kan "
HG1310 tdaA::EZ-Tn5,Kan "
Phaeobacter sp. Wild type, antibacterial activity (5, 6)
27-4
Mutants derived form 27-4
JBB1001 tdaB::EZ-Tn5,Kan current study
JBB1003 tdbC::EZ-Tn5,Kan "
JBB1005 tral::EZ-Tn5,Kan "
JBB1006 clpX::EZ-Tn5,Kan "
JBB1007 tdbF::EZ-Tn5,Kan "
JBB1009 tdbA::EZ-Tn5,Kan "
JBB1011 tdbD::EZ-Tn5,Kan "
JBB1029 tdbE::EZ-Tn5,Kan "
JBB1030 tdaA::EZ-Tn5,Kan "
JBB 1044 metF::EZ-Tn5,Kan "
s Bruhn, J. B., L. Gram, and R. Belas. 2007. Production of antibacterial
compounds and biofilm formation by
Roseobacter species are influenced by culture conditions. Appl. Environ.
Microbiol. 73:442-450.
6 Hjelm, M., O. Bergh, A. Riaza, J. Nielsen, J. Melchiorsen, S. Jensen, H.
Duncan, P. Ahrens, H. Birkbech,
and L. Gram. 2004. Selection and identification of autochthonous potential
probiotic bacteria from turbot larvae
(Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27:360-371.
19
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
JBB1045 tdbB::EZ-Tn5,Kan
Other
Roseobacters
Roseobacter Wild type, none antibacterial activity (5, 7)
algicola 51442
Roseobacter Wild type, none antibacterial activity (5, 8)
denitrificans
33942
Roseobacter Wild type, none antibacterial activity (5, 8)
litoralis 49566
Roseobacter sp. Wild type, antibacterial activity (5, 4)
TM1038
Roseobacter sp. Wild type, antibacterial activity (5, 4)
TM1039
Roseovarius sp. Wild type, antibacterial activity
ISM
Roseovarius sp. Wild type, antibacterial activity (5, 4)
TM1035
Roseovarius sp. Wild type, antibacterial activity (5, 4)
TM1042
Silicibacter Wild type, antibacterial activity (5, 9)
pomeroyi DSS-
3
Sulfitobacter Wild type, none antibacterial activity (5)
7 Lafay, B., R. Ruimy, C. Rausch de Traubenberg, V. Breittmayer, M. J.
Gauthier, and R. Christen. 1995.
Roseobacter algicola sp. nov., a new marine bacterium isolated from the
phycosphere of the toxin-producing
dinoflagellate Prorocentrum lima. Int. J. Syst. Bacteriol. 45:290-296.
8 Shiba, T. 1991. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter
denitrificans sp. nov., aerobic pink-
pigmented bacteria which contain bacteriochlorophyll a. Syst. Appl. Microbiol.
14:140-145.
9 Gonzalez, J. M., J. S. Covert, W. B. Whitman, J. R. Henriksen, F. Mayer, B.
Scharf, R. Schmitt, A. Buchan,
J. A. Fuhrman, R. P. Kiene, and M. A. Moran. 2003. Silicibacterpomeroyi sp.
nov. and Roseovarius
nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from
marine environments. Int. J. Syst.
Evol. Microbiol. 53:1261-1269.
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
sp.1921
Sulfitobacter sp. Wild type, antibacterial activity (5, 10)
EE36
Sulfitobacter sp. Wild type, none antibacterial activity (5, 11)
SE62
Vibrio Wild type, serotype 01, susceptible to (5, 12)
anguillarum 90- tropodithietic acid
11-287
Plasmid
pSTM3 Harboring tda genes current study
pSTM3-1265 pSTM3 carrying a Tn5 insertion in tdaE, current study
derived from HG1265
[0071] The involvement of CoA metabolism, addition, or modification is evident
from the
functional domains on TdaD and TdaE. TdaD is anticipated to be a member of the
thioesterase
superfamily of acyl-CoA thioesterases (Table 2), TdaE encodes a putative acyl-
CoA
dehydrogenase (ACAD), and TdaF has homology to aldehyde dehydrogenase.
[0072] The secondary evidence suggests that tdaA-F resides on a cryptic
plasmid that may be
spontaneously lost. To develop a means to test the hypothesis, we used three
strains, TM1040, a
spontaneous Tda nonpigmented strain of TM1040 (TM1040SM), and HG1265
(tdaE::Tn) (Fig.
5A and Table 1), along with a PCR amplification using primers for tdaA-E,
predicted to generate
a 3.8 kb product from wild-type DNA. As shown in Fig. 5B, PCR amplification of
wild-type
10 Buchan, A., L. S. Collier, E. L. Neidle, and M. A. Moran. 2000. Key
aromatic-ring-cleaving enzyme,
protocatechuate 3,4-dioxygenase, in the ecologically important marine
roseobacter lineage. Appl. Environ.
Microbiol. 66:4662-4672.
11 Buchan, A., E. L. Neidle, and M. A. Moran. 2001. Diversity of the ring-
cleaving dioxygenase gene pcaH in a
salt marsh bacterial community. Appl. Environ. Microbiol. 67:5801-5809.
12 Skov, M. N., K. Pedersen, and J. L. Larsen. 1995. Comparison of pulsed-
field gel electrophoresis, ribotyping,
and plasmid profiling for typing of Vibrio anguillarum serovar 01. Appl.
Environ. Microbiol. 61:1540-1545.
21
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
DNA gave the predicted 3.8 kb band, a 5.7 kb product when tdaE:Tn DNA was used
as a
template, and no product when the DNA from the SM strain was amplified
indicating that the
SM strain had lost the tdaA-E locus.
[0073] Total DNA from TM1040, TM1040SM, and HG1265 (tdaE:Tn) was separated by
PFGE.
As observed in Fig. 5C, all three strains had high molecular weight DNA,
presumably a mixture
of chromosomal and pSTM1 and a band or bands at ca. 130 kb, corresponding to
the size of
pSTM2 (132 kb) (41). Close inspection of this region and comparison between
the SM DNA
lane (middle, Fig. 5C) and either the TM1040 or tdaE:Tn DNA (left and right
lanes,
respectively) shows that the SM band is thinner than either TM1040 or tdaE:Tn
hinting that SM
DNA is missing a DNA species in this size range that overlaps with pSTM2.
Repeated attempts
to change PFGE conditions did not resolve this region. To overcome this
limitation, a Southern
blot (Fig. 5D) using a tdaD DNA probe was performed on the gel shown in Fig.
5C, and the
results confirmed that the SM DNA, while possessing a 130 kb band, fails to
hybridize to tdaD.
In contrast, both wild-type DNA and tdaE:Tn DNA hybridize to the expected band
(ca. 130 kb).
This confirms the loss of tda DNA in SM and adds evidence supporting the
hypothesis that the
missing tda DNA is on a plasmid. It does not rule out the unlikely possibility
that tda genes
reside on pSTM2 and are somehow deleted from that known molecule.
[0074] To resolve the issue, we isolated plasmids from each of the three
strains (TM1040,
TM1040SM, and HG1265) and subjected each mixture to Ncol digestion (Fig. 5E),
chosen
because an in silico Ncol digestion of pSTM2 provided a recognizable pattern
of DNA
fragments. As shown in Fig. 5E, the TM1040SM DNA digest had much fewer bands
than wild-
type DNA or DNA from tdaE:Tn. This would be expected if the TM1040SM strain
lost a large
plasmid. Consistent with this hypothesis, Southern blotting showed that a tdaD
probe hybridized
to a 4.5 kb fragment in wild-type plasmid DNA and to a 6.4 kb fragment from
plasmids isolated
from the tdaE:Tn strain (Fig. 5E).
[0075] We reasoned that it is possible to transform a cryptic tda plasmid
bearing a selectable
marker into a suitable host and thereby provide proof of the existence of this
plasmid. The
transposon that we used, EZ:Tn, contains a kanamycin-resistance gene as well
as the oriR6K
origin of replication permitting replication in permissive hosts carrying the
pir gene. Thus, the
plasmid from tdaE:Tn was used to transform E. coli EC100D (Table 1) with a
subsequent
22
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
selection for kanamycin resistance (Materials and Methods). This
transformation was successful
despite a very low transformation efficiency resulting in 7 Colony Forming
Units (CFUs) per g
of mixed plasmid DNA, and provides strong evidence for the existence of a
cryptic ca. 130 kb
plasmid harboring tda genes. This new plasmid was called pSTM3.
[0076] Twelve random colonies were chosen from the transformation with pSTM3
and the
Ncol-digestion pattern of each compared. Fig. 6 shows the four common patterns
resulting from
this analysis. Although each plasmid was PCR positive for the tda genes (data
not shown) and
the set of four shared many common bands, they had remarkably different
patterns indicating
deletion and/or rearrangements had occurred during or after transfer of pSTM3
to E. coli. The
reason and molecular mechanism underlying these band pattern differences is
not known;
however, the sum of the results indicates that TM1040 harbors a ca. 130 kb
plasmid, pSTM3,
which is essential for TDA and pigment biosynthesis and which may be
spontaneously lost in
laboratory culture.
[0077] Distribution of tda genes in other Roseobacters
[0078] The Roseobacter clade produce an antibacterial activity. In light of
the current findings,
we sought confirmation that other roseobacters had tda genes as well, and
chose Phaeobacter
27-4 as a suitable candidate.
[0079] We used the same transposon to construct a 6,321-member library and
screened these
mutants for the Tda phenotype. 37 Tda mutants were found of which 12 were
analyzed further.
Two of the 12 ORFs mutated were similar to TdaA (identity 38%) and TdaB
(identity 55%) from
TM1040 (Table 4), suggesting that these two roseobacters share a common TDA
biosynthesis
and regulation scheme. The remaining 9 genes were not identified as important
to TDA synthesis
in TM1040 and had varying degrees of homology to genes in the annotated TM1040
genome,
but, unlike TM1040, were not part of the phenylacetate or reductive sulfur
pathways. The one
exception is 27-4 metF (Table 4), which may possibly be involved in sulfur
metabolism.
[0080] We also used DNA:DNA hybridization to measure hybridization of a tdaA-F
gene
probe to DNA from 14 Roseobacter clade species (Fig. 7). The tda probe
hybridized to 8 of the
9 roseobacters that have been established as producing antibacterial activity
(Fig. 7), with the
ninth, Silicibacter pomeroyi DSS-3, showing a low amount of hybridization.
Three of 6 non-
antibiotic-producing roseobacters also positively hybridized to the tda DNA.
This false positive
23
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
may have resulted from a strain that has very low tda expression and
antibiotic activity below the
detection limits of the well diffusion assay, or from spurious hybridization
to non-tda DNA. The
tda probe did not hybridize with DNA from V. anguillarum, implying that the
second possibility
is the more likely scenario.
Table 4. Phaeobacter 27-4 genes and encoded proteins required for the
regulation and
synthesis of tropodithietic acid.
---------------------------- --------------------------------------------------
--------------- ------------------------------------------------------------ --
-------------------------------------------------------------------------
GenBank
Mutant Assession Gene
Number Number Designation Function Best Hit Ortholog / E score
~:~:<i~i.i..i..i..i..i..i..i..i..i..i..i..i..i`..i..i~~i~~i~~i~~i~~i~~i..i..i..
i..i..i..i..i..i..i..i...i..i..i..i..
:~ ~re.e~irs~.rs'':c~a~ d~~~~ :a~d:~
::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::
::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::::>::::
::>::::::>:::::>
~ ::::::::::::::::::::::::::: N....
JBB1001 EF139212 (3 etherase, glutathione Sinorhizobium meliloti
/ tdaB S transferase putative (3-etherase ((3-aryl
JBB 1030 ether cleaving enzyme / 4e-52
. ~ 0' r 1~~~~a.1~~~~[;~ :Ai~di~~ o~[:
;
... ;...
JBB1044 EF139218 metF 5- Silicibacter sp. TM1040
methyltetrahydrofolate- MetF protein / 2e-77
-homocysteine S-
methltransferase
-------------------------- ----------------------------------------------------
------------- _________________
*
~.:.e~a.~
:::>::::::::::>::::::::::>::::::::::>:::::<:::::::::::>::::::::::>::::::::::>::
::::::::>::::::::::>::::::::::>::::::::::>::::::::::>::::::::::>::::::::::>`:::
::::::::>::::::::::>::::::::::>::::::::::>::::::::::>::::::::::>::::::::::>::::
::::::>:::::::>::::::::::>::::::::::> :::::;
:::: ...............
~ ::::::;::::::::::: <;: ::::::: ,
JBB1009 EF139215 tdbA D-(3-hydroxybutyrate Roseovarius sp. 217 D-(3-
dehydrogenase hydroxybutyrate
dehydrogenase / 2e-32
-------------------------------------------------------- ----------------------
------------ ------------------------------------------------------------- ----
------------------------------------------------------------------------
JBB1045 EF139216 tdbB Phosphate Roseobacter sp. MED193
acetyltransferase phosphate acetyltransferase
8e-81
--------------------------------------------------------- ---------------------
--------------------------------------------------------------------------- ---
--------------------------------------------------------------------------
-------------------------------------------------------------------------------
---------- ------------------------------------------------------------ -------
---------------------------------------------------------------------
>
;::;:::,r:::~ :::::.:;
~~a~~ ~rt::>~~ ....r~':a~~i~:::~
.o~:::>:::::>:::::>:::::>:::::>:::::>:::::>:::::>:::::>:::::>:::::>:::::>:::::>
:::::>:::::>:::::>:::::>:::::>:::::>:::::>::::::::::>::::::::::>::::::::::>::::
::::::>::::::::::>::
::::::<::::::? :
JBB1003 EF139213 tdbC Lytic transglycosylase, Roseobacter sp. MED193
__________________hypothetical protein / 6e__ 85_____
-------------------------- ----------------------------------------------------
------------- ____________ peptidase C14
JBB1005 EF139221 tral Tral, Type IV (Vir- Rhodobacter sphaeroides
like) secretion 2.4.1 Tral / 5e-58
------------- ------------- ------------------------------
JBB1011 EF139222 tdbD Type I secretion target Roseobacter sp. MED193
repeat protein type I secretion target repeat
protein / 8e-54
--------------------------- ------------------------------ --------------------
--------------- ---------------------------------- ------ ---------------------
----------------------------------------------------
-------------------
JBB1029 EF139216 tdbE Oligopeptide/dipeptide Silicibacter sp. TM1040
ABC transporter binding-protein-dependent
transport systems inner
membrane component / 6e-
124
------------------------- ----------------------------- -----------------------
----------- ----------------------------------------------------------- -------
-----------------------------------------------------------------------=
24
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
. . . . .
::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: :::
::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: ::: :::
::: ::>
: .............. ::I::>: >:~ ~.>::>: ~ . ~r.
~::>::>::>::>::>:::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::
>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>:::
:>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::::>::
::>::>::>::>::>::>::::>
~:::a::::
:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
:::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
~ .............. .Y...........................
..................................
............................................................
............................................................................,
JBB1006 EF139220 clpX ATP-dependent Clp Silicibacter sp. TM1040
protease ATP-binding subunit C1pX
le-47
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
- - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - - -
- - - - - - - - - - - - - - - - - - - - - - - - - - - - -
=
JBB1007 EF139214 tdbF Ribonuclease D Roseobacter sp. MED193
ribonuclease D / 6e-49
-------------------------- -----------------------------= ---------------------
------------- ------------------------------------------------------------ ----
-----------------------------------------------------------------------
JBB 1030 EF139217 tdaA LysR substrate binding Paracoccus denitrificans
domain protein PD 1222 regulatory protein,
LysR:LysR, substrate-binding
/ 3e-51
-------------------------- ~---------------------------- ~---------------------
------------ ~------------------------------------------------------------~----
-----------------------------------------------------------------------
[0081] Distribution of tda genes in the environment
[0082] Marine genome and metagenomic databases were searched for sequences
with
homology to one of the 12 genes (Table 2) required for TDA synthesis by
TM1040. While
homologs to the proteins involved in phenylacetate and reductive sulfur
metabolism were found
within the 14 selected roseobacter genomes in Roseobase
(http://www.roseobase.org/) and the
Gordon and Betty Moore Foundation Marine Microbial Genome databases
(https://research.venterinstitute.org/moore/), close homologs of TdaA-F were
absent (at a
BLASTP E value cutoff of 1E-30). While the reason for the absence of homologs
is not known,
it is possible, although unlikely, that all 14 roseobacters do not produce
TDA, produce an
antibacterial activity that involves another compound, or lost their tda
plasmid. The last
possibility is most likely to have resulted from laboratory culturing,
therefore we searched for
Tda homologs in environmental metagenomic libraries
(http://camera.calit2.net/) that should
contain abundant uncultivated roseobacter DNA.
[0083] The data gathered from searching the CAMERA marine metagenomic GOS
dataset
database are shown graphically in Fig. 8, where a circle and its relative size
indicates the
presence and abundance (respectively) of a given protein. As was observed with
the roseobacter
genomes, phenylacetate and reductive sulfur metabolism proteins were found at
numerous sites,
with the greatest abundance of PaaIJK and Cysl at site GSOOa, a Sargasso Sea
sample (3132'6"
N, 63 35' 42" W). Positive Tda protein `hits' were also recorded in a
hypersaline pond sample
(GS033) and a sample obtained from Lake Gatun, Panama Canal (Fig. 8). In no
sample did we
find hits to all 12 proteins involved in TDA biosynthesis.
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0084] Various members of the Roseobacter clade, whose genomes reveal a great
potential for
the synthesis of bioactive molecules, produce TDA. Many marine bacteria
produce an antibiotic
activity, including antibacterial activity from roseobacters, e.g., a compound
that produces a
probiotic effect on scallop larvae and is antagonistic to y-Proteobacteria
strains, as well as a
compound that is antagonistic against fish larval bacterial pathogens. From
our data, it is likely
that much of the antibiotic activity seen in roseobacters is due to plasmid-
borne tda genes that
can be difficult to maintain in laboratory conditions.
[0085] There is a direct link between the spontaneous appearance of non-
pigmented Tda
colonies and the loss of pSTM3 of TM1040. Over 40 of the mutants initially
screened as Tda
were ultimately found to have lost pSTM3. This suggests that loss of pSTM3 is
a relatively
frequent event during laboratory cultivation of TM1040. Instability of the
Tda+ phenotype is not
unique to TM1040. The appearance of spontaneous nonpigmented Tda mutants or
variants is
characteristic of other roseobacters, including Phaeobacter 27-4 and
Roseobacter gallaeciensis
sp. T5. One possible explanation for the cause of these spontaneous mutants is
a loss of a
plasmid carrying one or more critical genes required for TDA synthesis.
Indeed, 27-4 possesses
at least two plasmids of ca. 60 kb and 70 kb respectively. One or both of
these plasmids may be
involved in TDA biosynthesis of 27-4 and tdaA and tdaB, identified by
transposon insertion
mutagenesis in 27-4 Tda mutants, reside on one of these plasmids.
[0086] Instability of pSTM3 is also apparent when the plasmid is transformed
into a
nonroseobacter host, e.g., E. coli. As shown in Fig. 6, at least four unique
Ncol-restriction
fragment patterns were observed from pSTM3 that had been successfully
transformed into a new
host. As a cause of this instability, it seems improbable that TDA
biosynthesis is to blame,
because the pSTM3 used to transform E. coli does not confer a TDA+ phenotype
due to the
presence of a transposon in tdaE. It is possible that, despite absence of
TdaE, some other
protein(s) encoded by other tda genes (tdaABCD or -F) may be detrimental when
expressed in
E. coli. While there is no evidence to directly link instability of pSTM3 in
E. coli with
spontaneous loss of the plasmid in TM1040, these phenomena may share a common
cause. We
have initiated efforts to sequence and annotate pSTM3 and compare it to the
pSTM3 species
obtained from E. coli. Preliminary evidence indicates that pSTM3 harbors a
repC homolog
upstream of tdaA. RepC forms a complex along with RepAB and is required for
plasmid
26
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
replication and maintenance. It will be determined if the pSTM3 plasmid
species obtained from
E. coli transformation have defects in repABC.
[0087] The ability of pSTM3 to replicate in E. coli, albeit with significant
alteration in the
plasmid, suggests that pSTM3 also may transferred to other marine bacteria,
perhaps other
roseobacters, or even to higher organisms, e.g., dinoflagellates. TM1040
possesses varied
capabilities to achieve horizontal gene transfer, including the presence of
several prophage
genomes in the bacterium's genome, one of which is homologous to the gene
transfer agent of
other alphaproteobacteria, and many of genes on pSTM2 are homologs of the vir
system of
Agrobacterium tumefaciens. The A. tumefaciens Ti plasmid, transferred by Vir
Type IV
secretion, requires RepABC, suggesting that a similar mechanism may allow
pSTM3 transfer to
other organisms. Plasmids similar to pSTM3, such as pSymA of Sinorhizobium
meliloti and the
Ti plasmid, are important for the proper interaction of those bacteria and
their respective hosts,
and TM1040 pSTM3 and pSTM2 may correspondingly serve to enhance the TM1040-
dinoflagellate symbiosis.
[0088] It is important to note that TDA activity and biosynthesis depend on
culture conditions
and the physiology of TM1040. TDA activity is significantly enhanced when
TM1040 is
cultured in a static nutrient broth, a condition that accentuates biofilm
formation. The symbiosis
includes two phases: the motile phase in which TM1040 cells actively respond
to dinoflagellate-
derived molecules by swimming towards the host, and sessile phase, whereupon
having located
the zoospore, the bacteria cease to be motile and form a biofilm on the
surface of the
dinoflagellate. Thus, there is a direct correlation between biofilm formation
and TDA
biosynthesis.
[0089] Biosynthesis of TDA has several potentially beneficial effects on the
TM1040-
dinoflagellate symbiosis. TDA is likely to benefit the dinoflagellate by
acting as a probiotic with
antibacterial activity whose action prevents the growth and colonization of
bacteria on the
surface of the dinoflagellate that could potentially harm the zoospore. In
turn, the antibacterial
activity of TDA may enhance the growth of TM1040 cells attached to the
zoospore by warding
off other biofilm-forming bacteria that compete with TM1040 for space on the
surface of and
nutrients from P. piscicida. Interestingly, DMSP appears not to be a primary
source of the sulfur
27
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
atoms of TDA. One or more non-DMSP sulfur-containing metabolites produced by
the
dinoflagellate may be used by TM1040 in the biosynthesis of TDA.
[0090] One of the unexpected results from our study is the paucity of
homologous Tda proteins
in either the genomes of other sequenced roseobacters or in the CAMERA
metagenomic library
(Fig. 8). There are several reasons why Tda proteins were not found. For
example, amino acid
sequence divergence between Tda proteins of TM1040 and other roseobacters
could result in
BLASTP E values greater than our chosen cutoff (1E-20). This argument may also
be applied to
the metagenomics search. In focusing on just the search for Tda homologs in
roseobacter
genomes, it is possible that, in culturing these roseobacter species in
preparation for isolation and
purification of their genomic DNAs, the bacteria lost a pSTM3-like plasmid
harboring tda genes.
Equally feasible is the possibility that TDA is but one of many antibiotic
compounds produced
by roseobacters or that more than one biochemical pathway exists to produce
TDA. Both
arguments may help explain the lack of Tda protein homologs in roseobacter
genomes.
[0091] The lack of Tda protein homologs in the marine metagenomics database
presents a much
more difficult problem to interpret, especially in the context of PaaIJK and
Cysl searches that
frequently identified their respective homologs in numerous samples within the
database (Fig.
8). While the data do not provide definitive answers to this question, our
data show that stability
and retention of pSTM3 by TM1040 is greatest when the bacteria are directly
associated with the
dinoflagellate, i.e., the plasmid may be lost when TM1040 is grown in
laboratory culture, yet
retained when cultivated as part of the Pfiesteria piscicida mesocosm from
which the bacteria
were isolated. Close association of TM1040 with P. piscicida provides a
selection to maintain
the pSTM3; that selective pressure is lost when the bacteria are taken away
from their host (as
happens under laboratory culture). The CAMERA metagenomic samples analyzed
were prepared
after filtration to remove 0.8 m particles, which may have removed the
portion of the
roseobacter population harboring a tda plasmid like pSTM3.
[0092] The two metagenomic samples that showed relatively good Tda homolog
hits were from
a site in the Sargasso Sea and a hypersaline pond, respectively. DMSP is
potentially useful by
algae as an osmolyte that protects the cells against changes in salinity. Our
results suggest that
DMSP is not used as a sole sulfur source in the biosynthesis of TDA, and show
that there is a
correlation between salinity, DMSP, and the presence of Tda homologs.
28
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
[0093] The genetic data from the current study, specifically the
identification of paaIJK and
tdaC (prephenate dehydratase), indicate that TDA biosynthesis originates from
the shikimate
pathway and proceeds through phenylacetate (Fig. 9). The results also show
that phenylacetate-
CoA and CoA metabolism is vital to TDA production and are consistent with TdaD-
F
involvement in a ring expansion reaction that converts PAA-CoA to a seven-
member tropolone
ring (step 8 in Fig. 9). TdaB, a homolog of glutathione S-transferase, is a
potential agent in the
addition of sulfur to the nascent TDA molecule.
[0094] The compounds shown in Fig. 9 include the following:
Table 5
Compound
Produced from IUPAC13 name
Reaction:
6 1, 2-dihydro-phenylacetyl-CoA
8 2-hydroxy-7-oxo-cyclohepta-3,5-dienecarboxylic acid
9 2,7-dihydroxy-cyclohepta-1,3,5-trienecarboxylic acid
2,7-dihydroxy-3-oxo-cyclohepta-1,4,6-trienecarboxylic acid
11 2,7-dihydroxy-3-thioxo-cyclohepta-1,4,6-trienecarboxylic acid
13 7-hydroxy-2-mercapto-3-thioxo-cyclohepta-1,4,6-trienecarboxylic acid
[0095] Identification of a LysR homolog in TdaA is consistent with the
regulation of TDA
biosynthesis involving a cofactor. In other bacteria, LysR cofactors can
function as precursor
molecules required to synthesize the final product, implicating molecules in
the shikimate
pathway, phenylacetate, or other TDA precursors as being required for maximal
expression of
the tda genes. Consistent therewith, modifications of the broth by addition of
phenylalanine and
histidine significantly increase production of TDA from Phaeobacter T5.
[0096] We therefore disclose the genes and proteins required for TDA synthesis
by roseobacters,
and the occurrence of tda genes on a previously unknown megaplasmid (pSTM3) of
TM1040, as
aspects of the present invention. The backbone of TDA is a seven member
aromatic tropolone
ring, which is highly significant as tropolone derivatives, notably
hydroxylated forms, are
medically important sources of antibacterial, antifungal, antiviral, and
antiparasitic agents.
13 International Union of Pure and Applied Chemistry
29
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
Chemical synthesis of tropolone and derivatives can be difficult, making
natural sources of
tropolone precursors often the preferred choice as starting material for the
synthesis of new
tropolone antibiotics. The mutants obtained in this study may lead to the
development of
bacterial sources of medically important tropolone compounds and a suite of
new antimicrobial
agents based on TDA.
[0097] Sequencing of the 130 kb pSTM3 plasmid bearing genes required for
tropodithietic acid
biosynthesis has been carried out with determination of the following
sequences.
pSTM3 partial sequence:
contig tdaA-tdaE
GCCCCCCGGGGGGGGGCCCGGGCCAGGTAAATTCGCCCGGGGTTTTACGGGGGGGT
TTTTTTTCCCGAAAGGATGACGCAAAATTCCACCCAGTTTCCTGGCCCCGGAAATAG
AAGCCCCCCGGTTCGGGGGGTGAACTCGGGGGGAGGGGGCCTTTGCCCATCCCAGA
TGCAGCTTGCGCAGATAGGCCGTCGGTTGACCCCCCAAGAGCCAAGCCGCCTCGCCG
GGAGGTGAACTTGCGCTCCCCTTGGCGCTCGGGGGGAAAGGAGGCTTTCGCGTTGAT
TGTGCAATGTGCGCCCAGCCATTCGAAATGCTCCCGAATAAGCTGGTTGAGATCCTC
ATGCAGCGCTTCTGCTGCTGCCGGAGCCTTGGTCGTTGCATGCCGTCCTGCCCTTCGT
ATCCTCTGTGACGGTTCCACTGTGACGGTGGCGATGGCGCAAGGAGCCGCCTCAGAT
CGGGCGTTTTCTCTTCAGCCTGCCCGCCGTGTTCACGGAAATCGACGTTTTTGTATCT
TTCCGTGACTATTTACCGCCGAGCGGGATTCGTGCAAGGGTTTTCTGCCCAAGTTAT
CCACAGGATGCGCAATTTTTGAGCCCCGCAGACGCGGTGATGGCCTCTGGGGGCGG
AGAAGTTGCCTGTCATACCCGTGACACGAGACTAAAGGCATTCTGCAATAGCCAGC
CGCCCAGTCCGGTCTCTCTGTGACCTTTGGCATCCGGGACGGCGCCGCCAAACCGGC
CCCATGTCAGCGCCGCATTGCGGGAAACGCCAGGGCGCAGAAGACCCGAAGACGGC
CCGCAAACCGCCGGATGCCGCGTGGGACAGGGGCGGGACAGGATGGAGAACCGTG
GTGGCCTCGCCCTTTCTGGCGAGGGATTTTCGCGCGTAACCCGTGTGGCGAAACCCC
GCCGAAGCGCGTAAGTCTCAGAAAAAATGACTAAATTATCGGCTTGATAAAATCTG
TAGACGACATAACCTATAGGAGATTCGTTTGCCAGTGTTTTACCCTTGGTTGTTGAG
GGCACATTAATAAGACCGCGGTTCCGGCCAGCTATTGACCGCCGCCGTTGCAGACCC
CCTGCAATGCGCGCCGTCCAGCGAGAGAGACCGACTTTCCCAAAACCCAACCCAAG
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
ACCAGATATGGTGCACTGTGCGTCATCAGTGAGTGGGAGCGAGATTAGATTTGGAC
ATTCAACAGCTAAGAGTCTTTGTCACCGTTGCAAAACATGGCAGCATCACCCGTGCT
TCTGACATTCTGTGGCAGCCAGCCCTCGGTGAGCGCGCAGATCAAGAGCCTGGAGA
CGACACTCGGGATCACGCTGTTTGAGCGCACCTCGCGGGGCATGGTGGTCACGCAG
GGGGGCGAGCGCCTTCTGGATGAGGCGACCGCTTGTGGATCGGCACAAACAGTTCA
TGCAGGAGGCCTCGCGACTGAAGGGCAGTGTCTCGGGGCTGTTTGCCATGGGCGCA
GGGCGGCATTCGGGCAACGGCTTTGTCAGCTCTTTCCTGCATTGTCTCGGAACGCTT
TCCGGAACTGGAGATCGAGCTCAAACACCTGGCCTCGGCGCAGGTGATCGAGGGGC
TGCGCGATCAGTCGCTGGACATGGGATTTTTCACCGAAACCGAAAGCGACACCTCG
ATTGACGCTGGTGGAGGTGGCCAGTTTCGGCATCTACCTTGCGGCGCCGCGCGGGAT
GATCCGTTGTTCAGAGACCCCTGACTGGGCGCGTCTTCAGGATCAGATCTGGATTGT
CTCGTCTCATGTGGCGCTGCGGTCGCTGGGCCAATGCCCTCATGGAGCAGCATGACA
TTCGCCCAAGGCGGGTGATCAAGGTTGATGACGAGGCGGTGACGCGGACGCTGGTG
GCAAGCGGAGCCGGGGTCGGGCTGTGCATTCTCGGGTGATGAAGCGCTGACGCCGC
CCGATGACATCGACCTGTTGCACCGGGTGCGCAAGACCGCGCGGATCATGTGCGGCT
ATCTCGAAGCGCGCAGCGATGATCCCTCTATTCGCGCGTAGATAATTGGTTCTGGAT
TTGCTGAAATCTCAACAAAAAGGCGAAACACCCTCCTTGTTGCAATTGGCGTAATCA
CAATTTCATTTGAGAATCCCCAAATAAGGGAATACGTCATTCGAGAGTGTTATTTTG
GAGTTGTCATGATTACGATTTATAGCCTCTGTGGCAAAGACGATATTCATTATTCCC
CGCATGTTTGGAAAGTCATTATGGCCCTGCATCACAAAGGGCTTTCATTTGACGTGG
TGCCGTGGATTTTCGACGATCCGCGACATCGAGGGCGGGGCGTTCAACAGCGTGCC
GGTGCTGCGCGATGGCGACCGGGTGATCGGGGACAGCTTCGAGATCTGCACCTATCT
GGATGCCGCCTACCCGGTGCCCCGGCCTGTTTGCCGGTGCGGGCAGTGAGGCGCAG
GTGCGGTTCCTGGAAAGCTATTGCCTGACGGCGCTGCACCCACCGCTCGCGGTGATC
GCGGTGATGGCGATGCATGACATCATGCATCGGGCGATCAGCCTATTTCCGGGCCAA
ACGCGAAGAGCGTTTTGGCGTGTCCATCGAGGCGCTGGCGGAAACCGCGCCCGCCG
AGCGCGCGCGATTGCAGGAGCGGCTGGCGCCGGTGCGCGCCGTCTTACGCATCACA
CTGGCTTGCGGGCGATGCCCCGGCGATGGCCGATTACGTGGTGTTCAGCGCCTTGCA
GTGGTGCTGGGTCGTGGGGCTGCGCGATCTTCTGTCCCCCGACGATTCGGTGGCGCG
TGGTTCAGCCGTGTCAGGCCCTGTTTGGAGGGGCGGCGCAAAAGCCTGCTGGAGCC
CCGCGCTAAGCCTGAGCTGAATCTGCGCGAACAAACCGGCAAAACCCGGCCCAAAT
TCATCTGATGCGCCCCCGATCGGGGCCGCTTTTTGTTGGTTTTGGGGCATTTACGGCT
GTGTCACCAAAGCCGATAGCTGACCTCAGTTTTTCCGAATTGCGACAAAGCGCGTCA
TTGGATCATATGAGTCCCAAGGTTCGATACGTCCTGAGCGAATTGATTTTTGAAACG
GTTGGAAATGAACAAGTAAATGGTTGCGTATCCGAAATTGAATTTCAGTCAATTGAT
GATGCCATTGGAGGACTCTTGAATGGACGTCGCGCTATGGACGGTCCCAGAACCAA
CGCAGTGAAGACATATCCAAAACCTATGACTGGGGTGCGCCATGTTCATACCCTGGG
ACCGGCTGGCACCAACTGTGAAAAGGCGGCGCTGAAATGGGCGGCGCTCAGTGCCG
CAATGCTGCCTGGTCCTGCATGACTCGATGGAGGAGGCCGCAGAGCAGGTCGCGGC
CTGCGGCTGTTCGGTGCTTCTGAGCGTGGTGGCCTACCCGCAGCTGCATTCGATCAT
CTACGACATATCGCGCATCTGGGCTTCTGGATGTGTTCATCATGAAGACCGACGACA
TGGTGCTGGCCTCGGTGAGCGGCGCCATGCCGACGCTGTGCCAGACCC
ACCCGGCGCCGGAAAAGCTGCTGCCGCCCGAGATGCAGCGGATCTATGCGACGAGC
AATTCCCACGCGGCCTCTGAGGTGGCGGCAGGGCGGGGCGATGGCTGCATCACCAC
GCGTGCCGCCGCCGAAGCACGGGCTTTTGGTGGTGCAGACCTTTGGCCAGGTGCCGA
TGGGGTTCACCATTCACGGCCCGCTCAAGCATGCGGGCTGCGCGGACACCGCCTTTG
ACGTTTCAGCACCAGATCACAACAGGATTTTCCCAATGACCCAACGCGCATTTGAGA
CCCGGATCGAAGTCCGCTACCGCGACACCGACTCGATGGGCCATATCAGCAGCCCG
31
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
ATCTACTACGACTACATGCAGTCGGCCTATCTGGAATACAGCCGCGCTGCTGGAGCT
GCCGAAGTCCGAAAAGCTGCCGCATATCATGGTGAAAACCGCCTGCGAGTACATCA
GCCAGGCCTATTACGGCGATACCGTGGTGGTGCTGAGCAAAGTGTCGAAATTCGCG
CAAGAGTTTCGAGATCGACCATGAGATCCGCCTTGGCAGCGCGGACGGCCGGGTGG
TGGCAAAGCTACAGTCGGTGCATGTGATGTTCGATTACGAAAAGCAGAGCACCTAC
CCGGTTCCGGAGATTTCGCAGCCGCGTCGCCGATTTTCAGGACGCCGCCTGAGCGCG
CGCCACGGTCCAGAGAGGGAGAATGCAATGGATTTGAGTTGGAGCACGCAGCAGCA
GTCGATCCGGGCGGAGTTTGCCTCCTCGGAGCCGCACAGACCGCGATGAGCTGCGTC
TTGGACGGCGCGCCTTTGACCAGCAGACCTGGGATCAGCTGGGAGAGGCGGGCCTG
TGGCAGATGATGGTGCCAAAGGACTATGGTGGCACCGGGGCGGACCGGGGCTTGCT
GGTGGGATGTCACCGCCGCCCTTGAGGGGCTGGCCTCGACCATCCGCGCGCCGGGG
CTGTTGCTGTCGGTGATCGCCCAAGCGGGTATGGCCTACGCGCTGGAGCTCTTGGCA
CCCGGCGCAGAAATCCGACTATTTCCGCCGCATCCTGCGCGGCGCGCTGAGCGCCAC
GGCCATCGCGGACCCCGACACCGGCACCGATGTCCGCGCCAGCTCCACTTACCTCAG
CCCGCGCCGAACGAACCTTTGTGCTCAACGGGAAGAAATACAACATCGCCCATGCG
CCGGTGGCGAATTTCACTCTGGTGGTCTGCAAGCTCGAAGGCCATGCCCGCGACGGC
ATCTCCCTGGTTCTGGTGGTCAGGACAGAAGGGCGTCACCATCGGTGCCAAGGATCG
CAAGCTTGGAAACCTAGATTTGCCGACGGGGGCGCTCTCGTTTGAGAATGTGCCGCT
GCACTATGGGCATATTCTGGGGGTGCCGGGCAAGGGCTGCGAACCTTGTGCGGTTTG
TCTCGCTGGGGCGGATCTATTACGGGCTGGTGGCGGCGACCCTGTGCGGCCCGATGC
TTGCGGAGGCGCTGTCTTATGCCAAGGCCCGGCAGACCTTTGGCAGCCCATCGTGAT
CACCAGTATGTGCAGAAGAAACTGACCGATATGCGCATCGCGGCAGAGACCGCCAA
ATGGACCTCTTATGGGGCGTTGCACCAGTTGCTGAGCGGCGCGCCCGAGGCGGTGA
GAGCTGTTCGATGCCAAGCTGGCCGGAGCCAGCGCGATCACCGATGGGGCCGTGGA
CCTGCTGAAACTATACGGCAGCCGGGGCTATCACGTAGGGCGAGGTGTCCACGTTCC
TGCGCGATGCGCTGCCTTTTGCAGCGTGGCGGCACCGAGGAAATGCATCGGCGCAA
CATCATGAACCAGATGATGCGAGAGGCCCGCCCGGCCAAGTCCAAGCCCGCCGCCC
CGGCGCGGGATCTGGAAACCGTCTGAGGCCGCCTTTATTGATTGGAGACAATCATGT
TAAAAGATTTCAACAGCTTGCGCTGTCTCGCGCATGGTGCTGCACTAGGCGCTGTAC
TGGGCGCGATGCCGCTTGCGGCGGGTGCCGCAGAAGAGGGATCCTGAGCGAGGCCA
GATCGACTGGGCCGCCGCAGAGGACTCGGCGGTGGCAACCGCTACAGCAGAAGCCG
CACTCACGGAGGCGTTTCTGGCCCTGCCCGCGAGCGCCGAGCCCACCGGTTTGCGGT
GATGCTCTTTGGGCGGCGGCGGATCTGCCCGAGCCGGGCTTTGTCAGTCAGGGCAGC
GCCTATGTGGCCTATTACGCGCAGGACGATATTCAGCTTTCGATCTCGGGGTCAAAG
GCGGTGGTCAGGCGGGGGATGCGCTGTTCTGCACCATGCCCCAAGCGCGTGGGAAA
GCATCGGAACGGGCGCGGATTACAAT
32
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
#is# tda A 4d:"i B tda C t~~ D tdaE
.~
.......... .......... .......... .................
:iiiii< ;
tdaF and membrane protein gene:
GCGCCGGTGGAGAGAACAAACATCCTCGCGGCTTGCTGGAGTTCTACCTCGAGGAA
GCGAGATCACGGCCCGCCACCTGCTGACACAAGTCTCGGTATCTGAGTCGAAGGTGT
GCCGGTGCGCGAGGGAACACCCGTTTCGCAAACCGCTCTCCCGGCTCAGGGTGGGG
GATCACCTGCGCAGCGCCGACCGCATCGTGCGGCCGAGGACGTCGAAGCCTTTGGC
CACCTCACGGGCGATCTGTTTTATGCCCTTGGACGAGGCCGCCGCGCGCAATCATCC
GTTCTTTGACGGGCGCGTCGCGCATGGGCAATACATCATGGCGCTGGCCAACGGCCT
CTTTGTGGACCCCGAGCCCGGCCCCGTGCTGGCCAATCTCGCCCCGCGATCTGCGTT
TTTTTGCGCCGGTCTATTTCGACACCGCGCTCTATGTGACGCTCACCTGCTGCTGCAT
CGGCCCCCTCAACAGGTCGGGCGCGGCCGAAGTGCAATGGAGCTGCCAGGTCGGGC
AGCGATGACGACACCCGCGTGGCCCAGTTCGACCTGCTGACCCTTGTCGCCGCCCAA
TGGCCGCCCCAGCCCGCCCCCCGCGCCTGAGAGGCCCGAGAGGCCTGAGACGCCGG
GTACGCCAATGCCCCTTCCCCTGCTATTGAAACAAAAGGATTTCCAATGACCTCTGC
TCCAAAGCCCCGCATCCTGATCGGTGCCTGCGGCTCGCTCGACCTGCTGATGCTGCC
GCAGCACCTGCGCGCCATCAGGACACATCGACTGCACGCTGAGCCTGATGCTCACGC
CGACGGCGGTGAAATTTGTCAACACGGATGCGCTGGCCCTGCTGGTGGACCGGCTG
ATCCACGGCGACCGCCCCGACGACTGGCCACGCACAAGCCGGACGCCTTGCCGCCG
ATCACGATCTTCTTGCGGTGCTGCCGACAACCGCAAACACCCTCAGTGCCGTGGCCA
ACGGCAGCTCGCAGAACCGCCTCACCACGGTGATCCTCGCAGCGGATTTCCGGACTG
TTCTTTCCCGTGATGGGCGGGCCGATGTGGGACAAGGCCTCGGTGCAGCGCAATGTG
AGCCAGATCCGCGCCGACGGGTATGAGGTGTTTCAGCCGGTCTGGCGCGAACACAG
CGCCCGCATCGCAAAAGGTCCACGGCCATCATTCGCTGCCGGACCCCGCGGATGTGG
TCGACATCCTCCAAAGCCGCCTGCCCGCGCAGCGCTGACCCGGCCTACCCGCCTGCC
CCCCCTGCCCCACAGATCTGTCCAAACAGGAAACGCCGCCGGATCTCCTCCGGCGGC
GTTTCTCGTGGTCTCTTTGCCTTTGGCCCTAGCCGGTCACATCACGCAGGCCGGGGC
GCAGCATGGGCCACAGCCGCGCCTGCCCAGCGCCGCAGATAGAGCCCCACCCCAAA
GGAGAGATGCGTCATCGCGCTCTTCAGCTGCGCAAAGGTCGGATCGGGCTTGTTGCT
GGCCATGATGCCCGCACCCATCGCCGGCTGCATCACAAAAAGGGAAAGACATGGTG
CCGAGCCCCACCACAAGCGCCAGCAACACCTGCGGCCGTTGCAGCTGCCCGACGCC
CCCGATCGCCACAAAGAGCGCCGCAAAGACCACGCCCACCGCATAATGTACCGCCA
GCCAAGCGCGCCTCACCCGCCACCGGCGCCGCCGCGCGAATGCTCTCATGGGCGAA
CACGCCCTCGGGCATATGGCCGACCCAACGCCCCACCAGGGCAAAGTTGCTCTGCG
GAATCGCAAACAGCGCCTCCGCCAGCACGGCCCAGAGATCCATCACCACCGTGGCA
CCCACGCCCAAGCAGTACGCGAAAAACAATCTGTGCAGGAGACATCCTGAGAACCC
TCTCTCTCGCTCACAGGCGCCAGCCGCCTGGCCAGCCCCTTTTGAAGATGCGCCCGA
CCCATCAAAATGCGGGCCGGGCGGTACGCGTTACGCCCTTTACTGGGACACCATCGT
33
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
GCTATCGAACCCGGCACAGGTCTGGGGCGCGGCGGCCTCGGCGCTGAGGGGTTGTA
GCGCGGTTGCAGCGACGTGATGCCCGATCTGGTAGCCACATATCGCCCACCGCGTTC
GAGCGCCGGTAATGCACGCCGCCAATCAGGCGCGATTCCGATTCCTCTTCGGCGAGG
TGCTGCAGACTGTCGTAAGACTTGCTGATGTTGGCGGCCCCCTCAAAGGGACGGGA
AAGGCGCGCTGCCAAAGACCGAGGTCATCACCTCAAGCGCTGCGCTGCCGCTGGTG
CAATGCTGACAGGGGTATTCGGGATGCATCGGCGCAGGAATGCGCGATTCCCAGAC
CAGACCGGCTCAGGTCGGGGTGGTTGTAGGTTTTGCCCAACGCCTCCACGGCGGTCT
GTGGCCGCCAGAGCGCGTAGCTGTACTTGGCGTTGAACCCGGCCACCAACGCATCGC
TCAGGGCGACATTCAACACGCCACATGCGCGTTTCCTCGAGAAGAGGCAGCGCGCC
AGGCCGCGCAGAG
F uta3iwe memÃarance Proteita
'{Kb trfaF Puiative PhosphOgsterase
~ \ : ._
<<' ~;;:?;??>::?>::?>::?>:::>??>:<<
s~ f i 3:i 1030+cQ ltEg 1377 071119,
lipoprotein:
GAATTCGCCCGCCTTGATCGCGGCGCGCACCTCTTCCATCGGGGCGATGGCCTCTTC
GGGCACCATCGACGCGGTAAAGGAGATGTCGTTGCCGCCCTCTTTCATGAGGCCAA
AGCGGGTGTAGTCCCCCGCTGGAATTGCCCGCCGCGCGATCCGCCAGCGCCGCCCCA
AGGATGGGGCCAAAGTTCCAGATCGCATTCGAGAACACGGTCTCGGGGTAGCGCTC
GGTATAATCGGTGAGCGAGCCCACCGCTGATGCCGCGTTCCTTGGCGGCGTCGGCGG
TGCCGATCCGCTCGCCAAAGAGGATGTCGGCGCCGGAATCGATCTGCGCCAGACCG
GCCTCGCGGGCCTTGGGCGGATCAAAGAAGGTGCCGATAAGTGATGAGATGGGTCG
CATCGGGACGCACCGCGTCGACACCCTGACGGAACCCGTTGATGAGCATGTTGACCT
CGGGGATCGGGATCGCCCCCACCGCGCCAAAGACGCCGGACTGGCTCATCTTGCCG
CAGCATGCCGCACAGATAGGCCGCCTCGTGGTTCCAGGTGCCAAAGGTGCCAAAGT
TGTCGCCCGCGGGCTTGCCGCTGGAGCCCATCACGAAGCGCGTGTCGGGATAGTCGC
CCGCCACCTGCGCGCTCGCGCTCCACCGCATAGGCTTCGCCCACGATCACATCCGCG
CCCTGCTCGGCATATTCACGCATCGCGCGCGCATAGTCGGTGCCCGCGATCCCCTCG
GAAAAGACATATTCGATCTCGCGCGCTTGCCGCTTCGAGCATCGCCACATGCAGACG
CGAGTTCCACGCGTTCTCCACCGGCGAGGCATGCACCCCTGCCACCTTGATCGGCGC
TTGCGCCAGCACCATCGGCGCCGGCAGCAACGAGGCCGCCCCAAGTGCTGCGCCGC
TCTTCAGGATCGATCTTCGGGTTCATACC
repC:
GAATTCCTCGGCGCGGACATCCTCTGCCAGCATTTCGATCTCTTGCGCACGCAGCCG
CAGGGGAGAGAGATCAAAGCCATAGGCGACCTTCTCGGTGCCATAGCGCCGCGCAT
AGCGTTTGCCATTGGCTGTCGCGTCTGAGCAGCAAACCGGCCTCGACCAGGCGGGCG
AGATAGCGGCGCATGGTGGAATTGGCCATGCCGTTGAGCCGCTCGCAGATGCTCTG
GTTCGAGGGGTGAATGACAAGGTCGCGTCACGGGCAGCTCGGCCCCGGACCAGAAG
CTCAAGAGCGCTTGCAGCACTGAGAGATCCCGGTCGCTGAGGCCAAAATGATGCCG
34
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
GGCGGTCGCGAGATCGCGCAGCACATTCCACTTGCTGACAGTGAAGTGCGGTCGTG
GGCTGAGACCTCGGGGCCTCGGGATCGGTGGATGCGGCAGGCATACGGGAGCTGGC
CGCCTGAAGCTGCGTCTGACGTTTGATCAGAACAGCATCAACGGTGCGCCCAAAAG
CGGACAGGATGATACCCCATGTTTCATTCACGAAGACAAAGAAATCCCGTTCTGCGA
ATCACATTTGACTTGCAGTTTCAGGCTCCTGACACTAGCTTGATGGTGCTAAACACA
AGTCAGGGTCTGTGGGCGATGTCTTTGCGGGACCTTTTCTTTTGTCTGCTCGTGCCTC
CTTTCTTAG
[0098] Another aspect of the invention relates to a methodology for
purification of TDA and
intermediate compounds, including the use of solid phase extraction techniques
to obtain
tropodithietic acid from Silicibacter sp. TM1040.
[0099] A still further aspect of the invention relates to a method of
purification of TDA by
HPLC techniques.
[00100] An illustrative purification technique is set out below.
Purification of Compound.
1. Roseobacter 27-4 was grown in 500 ml MB in a 5 liter volumetric flask at 25
C for 4
days.
2. The cells were removed by centrifugation (10,000 x g for 10 min).
3. The pH of the supernatant was adjusted to 3.5
4. Extraction was carried out with 3 times 500 ml ethyl acetate acidified with
0.1 Io formic
acid (FA)
5. The organic phase was transferred to a vessel and evaporated to dryness
under nitrogen
flow.
6. The dry ethyl acetate extract was redissolved in 3 x 3 ml acetonitrile
(CAN)-water (1:19)
containing 1% FA
7. The redissolved extract was sequentially applied to two 60 mg Oasis MAX
columns
(Waters, Milford, MA) which had previously been sequentially conditioned with
4 ml
methanol (HPLC grade) and 3 ml CAN-water (1:19) containing 1 Io FA.
CA 02670626 2009-05-26
WO 2008/067338 PCT/US2007/085681
8. After loading the samples by gravity the columns were washed with 4 ml PBS
buffer (pH
7).
9. 3.5 ml CAN-water (1:1) was passed through the column and collected
(fraction 1)
10. 3.5 ml CAN-water (9:1) (fraction 2)
11. 3.5 ml CAN-water (1:1) with 2% FA (fraction 3)
12. 3.5 ml CAN-water (9:1) with 2% FA (fraction 4)
13. The solvents were then removed in vacuo on a SpeedVac (ThemoSavant,
Holbrook,
NY).
INDUSTRIAL APPLICABILITY
[00101] The invention provides an effective and useful biosynthetic capability
for the
production of tropodithietic acid (TDA) by use of Roseobacter bacteria. TDA is
a useful sulfur-
containing antibiotic compound. The biosynthetic route of the present
invention enables
scalable production of TDA and TDA derivatives.
36