Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
CA 03032506 2019-01-30
Anti-PCSK9 Monoclonal Antibody
Technical Field
The disclosure relates to the technical field of antibody engineering, and in
particular to a fully human anti-Proprotein Convertase Subtilisin/Kexin Type 9
(PCSK9)
monoclonal antibody, obtaining method and application thereof.
Background
Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9) belongs to a proteinase
K
subfamily of proprotein convertase. The human PCSK9 gene is located at
chromosome 1p32.3, has a length about 22kb, has 12 exons and is capable of
encoding a protein having 692 amino acid residues. The PCSK9 protein is
composed
of a signal peptide, a front structural domain, a catalytic domain and a
carboxyl
terminal structural domain (V structural domain), is synthesized as a soluble
precursor
of 74kDa, and is capable of generating propeptide of 14kDa and mature protease
of
60kDa by means of catalytic cracking of self in an endoplasmic reticulum. The
PCSK9
is mainly expressed in livers, intestinal tracts and kidneys, and is also
slightly
expressed in skin and nerve systems, but only the PCSK9 in the livers can be
secreted into blood circulation systems.
Research shows that the PCSK9 is capable of mediating degradation of a Low
Density Lipoprotein Receptor (LDLR) to regulate the level of Low-Density
Lipoprotein-Cholesterol (LDL-C) in plasma, and the LDL endocytosis process
mediated with LDLR in liver is a main way for eliminating LDL from circulatory
system.
The LDLR is a protein having multiple structural domains, and its
extracellular domain
is tightly connected with epidermal growth factor precursor homologous
structural
domains EGF-A, EGF-B and EGF-C. When degradation of the LDLR is mediated with
the PCSK9, the PCSK9 firstly needs to be bound with the LDLR, the LDLR mainly
has
a binding site which is mainly EGF-A, and a composition of the PCSK9 and the
EGF-A
is formed. Research shows that the PCSK9 is also capable of regulating
cholesterol
metabolism by means of a very low density lipoprotein receptor, an
apolipoprotein B
receptor and an apolipoprotein E receptor, but molecular mechanisms therein
are not
1
CA 03032506 2019-01-30
clear.
Basic study and clinical test show that inhibiting activity of the PCSK9 by
means
of ectogenic interference measures, elimination of Low Density Lipoprotein
(LDL) in
the plasma can be accelerated, and thus a blood fat reduction function can be
achieved. At present, PCSK9 inhibitors mainly include monoclonal antibodies,
antisense nucleotides, small interfering Ribonucleic Acid (RNA), mimic
peptides,
small-molecule inhibitors, and the like.
The monoclonal antibody medicine is a research and development hotspot of a
biomedicine field in the year, which has characteristics of being good in
targeting
property, high in specificity, low in toxic or side effect, and the like, This
represents a
latest development direction of a medicine treatment field. A monoclonal
antibody
having the PCSK9 as a target can be specifically combined with the PCSK9 and
is
capable of interdicting interactions of the PCSK9 and the LDLR and retarding a
degradation process of the LDLR so as to take an effect of reducing the level
of LDL-C.
The clinical experimental data showed the safety, effectiveness and unique
clinical
disclosure values of anti-PCSK9 monoclonal antibody medicine.
A fully human antibody is a main direction of the development of therapeutic
antibodies. Due to an antibody library technique, a good technical platform is
provided
for preparation and screening of human antibodies. Due to the antibody library
technique, an essential hybridoma process in a conventional monoclonal
antibody
research process is avoided, and even various antibody genes and antibody
molecular
fragments can be obtained without an immunologic process. The phage antibody
library is the earliest and most widely used antibody library at present.
According to the
source of antibody genes, the phage antibody library is divided into an immune
library
and a nonimmune library, and the nonimmune library also includes a natural
library, a
semisynthesis library and a complete synthesis library. An antibody affinity
maturation
process is simulated in screening of the phage antibody library, generally an
antigen is
coated by a solid phase medium, a phage antibody library to be screened is
added,
and multiple rounds of processes "adsorption, washing, elution and
amplification" (that
is, elutriation) are carried out till an antibody having high affinity
specificity is screened.
At present, multiple pharmaceutical companies are actively developing
2
CA 03032506 2019-01-30
monoclonal antibody medicine targeting at PCSK9. Repatha (evolocumab) of Amgen
and Praluent (alirocumab) of Sanofi/ Regeneron are both fully human
antibodies, are
approved to sell in 2015 successively and are applied to treat on primary
hypercholesterolemia and familial hypercholesterolemia (heterozygote and
homozygote). On the basis of statin, the LDL-C of a patient suffering from
primary
hypercholesterolemia can be reduced by 77% together with Evolocumab, the LDL-C
of
a patient suffering from heterozygote familial hypercholesterolemia can be
reduced by
68%, and the LDL-C of a patient suffering from homozygote familial
hypercholesterolemia can be reduced by 31%. The evolocunnab has good tolerance
and has no conspicuous security problem at present. A human monoclonal
antibody
bococizumab of Pfizer is at phase-III clinical test, and a human monoclonal
antibody
lodelcizumab of Novartis is at phase-II clinical test. Roche and Merck are
also having
clinical test.
At present, China is still in lack of self-developed anti-PCSK9 fully human
antibodies having high affinity in the field.
Summary
The disclosure provides an anti-Proprotein Convertase Subtilisin/Kexin Type 9
(PCSK9) monoclonal antibody. Anti-PCSK9 monoclonal antibodies are screened
from
a complete synthesis antibody library; a small-capacity synthetic phage
antibody light
chain library is established by means of computer aided design and analysis; a
library
of mutations of light chain Complementarity-Determining Regions CDR1, 2, 3 of
the
anti-PCSK9 monoclonal antibodies is obtained by means of screening; after
screening,
monoclonal antibodies having high affinity are selected; a library is
established to
screen mutations at heavy chain regions CDR1, 2, 3 of the monoclonal
antibodies; and
finally an anti-PCSK9 monoclonal antibody having high affinity is obtained by
means of
screening. The anti-PCSK9 monoclonal antibody has completely new sequences,
has
good functions in vitro, particularly at a cellular level, and has very good
medicinal
disclosure prospects.
In order to achieve above purposes, the disclosure provides the anti-PCSK9
monoclonal antibody, including:
3
CA 03032506 2019-01-30
light chains and heavy chains; Light Complementarity-Determining Regions CDR1,
CDR2 and CDR3 of the light chains are represented by LCDR1, LCDR2 and LCDR3
respectively; in addition, Heavy Complementarity-Determining Regions CDR1,
CDR2
and CDR3 of the heavy chains are represented by HCDR1, HCDR2 and HCDR3
respectively; LCDR1 includes any one of RASQSIDNRLT(SEQ ID NO.22),
RASQSVRNWLD(SEQ ID NO.23), RASQGINSWLN(SEQ ID NO.24),
RASQNVNNWLN(SEQ ID NO.25), RASQNINSWLN(SEQ ID NO.26),
RASQN I N NWLN(SEQ ID NO.27), RASQG I H NWLN (SEQ ID
NO.28),
RASQDVDSWLT(SEQ ID NO.29), RASQSVRNWLN(SEQ ID NO.30),
RASQDVRNWLT(SEQ ID NO.31) or RASQSIRSYLN(SEQ ID NO.32); LCDR2
includes any one of DASSRQS(SEQ ID NO.33), GASTLES(SEQ ID NO.34),
AASTRET(SEQ ID NO.35), GASSRQS(SEQ ID NO.36), GASTRPT(SEQ ID NO.37),
DASNRQS(SEQ ID NO.38), GASNLAS(SEQ ID NO.39), DASNLQS(SEQ ID NO.40) or
DASSRPT(SEQ ID NO.41); LCDR3 includes any one of QQPENDPTT(SEQ ID NO.42),
QQDNDIPLT(SEQ ID NO.43), QQWNNTPNT(SEQ ID NO.44), QQDNDMPLT(SEQ ID
NO.45), QQWFDVPTT(SEQ ID NO.46), QQWDDTPNT(SEQ ID NO.47),
QQNSNIPLT(SEQ ID NO.48), QQDSKIPLT(SEQ ID NO.49), QQWTDTPLT(SEQ ID
NO.50), QQDDSTPPT(SEQ ID NO.51) or QQGDSMPMT(SEQ ID NO.52); HCDR1
includes any one of GGTFTNNA(SEQ ID NO.53), GYTVTSYG(SEQ ID NO.54) or
GYSLTSYG(SEQ ID NO.55); HCDR2 includes any one of RIIPMFGMA(SEQ ID
NO.56), WLSFYNGNT(SEQ ID NO.57), WVTFYNGNT(SEQ ID NO.58),
VVVSFYQGNT(SEQ ID NO.59), VVVSFYNGQT(SEQ ID NO.60) or VVVSFYNGNS(SEQ
ID NO.61); HCDR3 includes AREGIPMI(SEQ ID NO.62), ARGYSLDV(SEQ ID NO.63),
ARGYGMSI(SEQ ID NO.64), ARGFGMDR(SEQ ID NO.65), ARGYGMTV(SEQ ID
NO.66) or ARGFGLSV(SEQ ID NO.67).
Herein, a light chain variable region amino acid sequence is preferably
selected
from any one SEQ ID NO.11, SEQ ID NO.12, SEQ ID NO.13, SEQ ID NO.14, SEQ ID
NO.15, SEQ ID NO.16, SEQ ID NO.17, SEQ ID NO.18, SEQ ID NO.19, SEQ ID
NO.20 or SEQ ID NO.21.
Herein, a heavy chain variable region amino acid sequence is preferably
selected
from any one SEQ ID NO.1, SEQ ID NO.2, SEQ ID NO.3, SEQ ID NO.4, SEQ ID NO.5,
4
CA 03032506 2019-01-30
SEQ ID NO.6, SEQ ID NO.7, SEQ ID NO.8, SEQ ID NO.9 or SEQ ID NO.10.
Herein, a heavy chain variable region HCDR1 sequence is selected from any one
of GYTVTSYG(SEQ ID NO.54) or GYSLTSYG(SEQ ID NO.55); a light chain variable
region LCDR1 sequence is selected from any one amino acid sequence of
RASQSVRNWLD(SEQ ID NO.23), RASQNVNNWLN(SEQ ID NO.25),
RASQNINSWLN(SEQ ID NO.26), RASQNINNWLN(SEQ ID NO.27) or
RASQDVDSWLT(SEQ ID NO.29); a heavy chain variable region HCDR2 sequence is
selected from any one amino acid sequence of WVSFYQGNT(SEQ ID NO.59),
\M/SFYNGQT(SEQ ID NO.60) or VVVSFYNGNS(SEQ ID NO.61); a light chain
variable region LCDR2 sequence is selected from any one amino acid sequence of
GASTLES(SEQ ID NO.34), AASTRET(SEQ ID NO.35), GASSRQS(SEQ ID NO.36),
GASTRPT(SEQ ID NO.37) or GASNLAS(SEQ ID NO.39); a heavy chain variable
region HCDR3 sequence is selected from any one amino acid sequence of
ARGYSLDV(SEQ ID NO.63), ARGYGMSI(SEQ ID NO.64), ARGFGMDR(SEQ ID
NO.65) or ARGYGMTV(SEQ ID NO.66); a light chain variable region LCDR3
sequence is selected from any one amino acid sequence of QQDNDIPLT(SEQ ID
NO.43), QQDNDMPLT(SEQ ID NO.45), QQWFDVPTT(SEQ ID NO.46),
QQWDDTPNT(SEQ ID NO.47) or QQDSKIPLT(SEQ ID NO.49).
Herein, the disclosure further provides multiple antibodies, polypeptides or
proteins having the above light chain or the heavy chain.
Herein, the disclosure further provides multiple antibodies having the above
light
chain or the heavy chain, the antibodies are capable of specifically binding
with the
PCSK9, interdicting binding of the PCSK9 with the LDLR, increasing the number
of the
LDLR on a cell surface or a level of the LDLR in a blood circulation system,
and
reducing the level of the LDL or the LDL-C in the blood circulation system.
Herein, the disclosure further provides a polynucleotide sequence or a
combination having the above light chain or the heavy chain.
Herein, a heavy chain constant region of the anti-PCSK9 monoclonal antibody
includes IgG1, IgG2, IgG3 and IgG4; and the light chain constant region
includes Ck or
.
CA 03032506 2019-01-30
Herein, the heavy chain constant region preferably includes IgG4 or IgG2, and
preferably an eukaryotic expression vector or a procaryotic organism
expression
vector of the heavy chain.
Herein, the light chain constant region preferably includes Ck, and preferably
an
eukaryotic expression vector or a procaryotic organism expression vector of
the light
chain.
Herein, the disclosure further provides a recombinant DNA expression vector
having the polynucleotide sequence or the combination; a DNA sequence of the
recombinant DNA expression vector includes a DNA sequence for encoding the
anti-PCSK9 antibody in the above heavy chain variable region, the heavy chain
constant region, the light chain variable region and the light chain constant
region.
Herein, the disclosure further provides host cells transfected with the
recombinant
DNA expression vector, and the host cells include prokaryotic cell, yeast and
insect
cell or mammalian cell.
Herein, the prokaryotic cell is preferably escherichia coli.
Herein, the mammalian cell is preferably Human Embryonic Kidney 293 (HEK293)
cell, Chinese Hamster Ovary (CHO) cell or Myeloma (NSO)
cell.
Herein, the disclosure further provides multiple whole-length antibodies,
single-chain antibodies, single domain antibodies, bispecific antibodies and
antibody-drug conjugates.
Herein, the disclosure further provides multiple monoclonal antibodies,
artificial
vectors, drugs or drug compositions having the above light chain or the heavy
chain.
Herein, the monoclonal antibodies include whole-length antibodies and
fragments
of the anti-PCSK9 monoclonal antibody, and the fragments include, but not
limited,
Fab, Fab', F(ab1)2, Fv or ScFv.
Herein, the disclosure further provides a detection reagent or a kit having
the
above light chain or the heavy chain.
The antibody of the disclosure may be applied to diseases which are
alleviated,
relieved, inhibited or prevented by eliminating, inhibiting or reducing
activity of the
PCSK9, and the diseases include dyslipidemia, cardiovascular and
cerebrovascular
6
CA 03032506 2019-01-30
diseases and thrombotic occlusive diseases.
Herein, the dyslipidemia includes cholesterol increase, triglyceride increase,
low-density lipoprotein increase and high-density lipoprotein reduction. The
cardiovascular and cerebrovascular diseases include coronary arteriosclerotic
heart
diseases, acute myocardial infarction, atherosclerosis, stroke and peripheral
artery
occlusive diseases.
A method for obtaining the anti-PCSK9 monoclonal antibody, including:
(1) carrying out biopanning for an anti-PCSK9 single-chain antibody, and
carrying
out three rounds of enrichment screening of an antibody library to obtain an
antibody
sequence DFSK9-1 having high affinity from a completely synthetic ScFv phage
library,
a heavy chain DFSK9-H1 includes an amino acid sequence of SEQ ID NO.1, and a
light chain DFSK9-L1 includes an amino acid sequence of SEQ ID NO.11;
(2) based on DFSK9-1, by means of Tertiary structure simulation of a computer,
designing and establishing a antibody library of mutations light chain
Complementarity-Determining Regions CDR1, CDR2 and CDR3, carrying out
biopanning and positive cloning screening and identification on the antibody
library of
the mutations to obtain 10 single-chain antibody sequences having different
light
chains which are respectively named as DFSK9-2, DFSK9-3, DFSK9-4, DFSK9-5,
DFSK9-6, DFSK9-7, DFSK9-8, DFSK9-9, DFSK9-10 and DFSK9-11, corresponding
light chain variable regions are respectively named as DFSK9-L2, DFSK9-L3,
DFSK9-L4, DFSK9-L5, DFSK9-L6, DFSK9-L7, DFSK9-L8, DFSK9-L9, DFSK9-L10
and DFSK9-L11, and corresponding amino acid sequences of the regions are
respectively shown in SEQ ID NO.12, SEQ ID NO.13, SEQ ID NO.14, SEQ ID NO.15,
SEQ ID NO.16, SEQ ID NO.17, SEQ ID NO.18, SEQ ID NO.19, SEQ ID NO.20 and
SEQ ID NO.21; comparing affinity of the single-chain antibodies at a phage
level;
(3) selecting five cloning DFSK9-2, DFSK9-4, DFSK9-5, DFSK9-6 and DFSK9-8,
designing and establishing a mutation antibody library of heavy chain
Complementarity-Determining Regions CDR1, CDR2 and CDR3, carrying out
biopanning and positive cloning screening and identification on the heavy
chain of the
mutations of the heavy chain to obtain 10 different single-chain antibody
sequences
which are respectively named as DFSK9-12, DFSK9-13, DFSK9-14, DFSK9-15,
7
CA 03032506 2019-01-30
DFSK9-16, DFSK9-17, DFSK9-18, DFSK9-19, DFSK9-20 and DFSK9-21; herein,
DFSK9-12, DFSK9-13, DFSK9-14, DFSK9-15, DFSK9-16, DFSK9-17, DFSK9-18,
DFSK9-19, DFSK9-20 and DFSK9-21 respectively have light chain variable region
sequences of DFSK9-L2, DFSK9-L8, DFSK9-L5, DFSK9-L6, DFSK9-L5, DFSK9-L6,
DFSK9-L5, DFSK9-L4, DFSK9-L6 and DFSK9-L6; corresponding amino acid
sequences of the sequences are respectively shown in SEQ ID NO.12, SEQ ID
NO.18,
SEQ ID NO.15, SEQ ID NO.16, SEQ ID NO.15, SEQ ID NO.16, SEQ ID NO.15, SEQ
ID NO.14, SEQ ID NO.16 and SEQ ID NO.16; DFSK9-12, DFSK9-13, DFSK9-14,
DFSK9-15, DFSK9-16, DFSK9-17, DFSK9-18, DFSK9-19, DFSK9-20 and DFSK9-21
respectively have heavy chain variable region sequences of DFSK9-H2, DFSK9-H8,
DFSK9-H7, DFSK9-H3, DFSK9-H6, DFSK9-H4, DFSK9-H4, DFSK9-H9, DFSK9-H5
and DFSK9-H10; corresponding amino acid sequences of these sequences are
respectively shown in SEQ ID NO.2, SEQ ID NO.8, SEQ ID NO.7, SEQ ID NO.3, SEQ
ID NO.6, SEQ ID NO.4, SEQ ID NO.4, SEQ ID NO.9, SEQ ID NO.5 and SEQ ID
NO.10; comparing affinity of the single-chain antibodies at a phage level;
(4) cloning a heavy chain variable region gene and a light chain variable gene
of
the clones in (3) into an eukaryotic expression vector, transfecting a host
cell, and
obtaining a complete antibody of an anti-PCSK9 monoclonal antibody.
Herein, the CDR is a complementarity-determining region; the ScFv is
single-chain fragment variable; the ADCs are antibody-drug conjugates; the
LDLR is a
Low Density Lipoprotein Receptor; the LDL-C is low Density Lipoprotein-
Cholesterol;
the HEK293E cell is a human embryonic kidney 293E cell; the CHO cell is a
Chinese
hamster ovary cell; the NSO cell is a mouse NSO thymoma cell.
Compared with conventional art, the disclosure has the beneficial effects
that:
The disclosure provides multiple completely novel anti-PCSK9 antibodies which
have high binding affinity with substrates, are capable of well interdicting
binding of the
PCSK9 with the LDLR, and in addition influences distribution and expression of
the
LDLR on the cell surface. Therefore, binding performance of the LDL-R with the
LDL
on the cell surface is improved, intake and degradation of the LDL by a cell
are
improved, and the purpose of reducing extracellular contents of the LDL and
the
LDL-C and reducing total cholesterol of the LDL and the LDL-C is achieved.
CA 03032506 2019-01-30
The monoclonal antibody provided by the disclosure may be used for
eliminating,
inhibiting or reducing activity of the PCSK9 to alleviate, relieve, inhibit or
prevent
diseases; the diseases include dyslipidemia, cardiovascular and
cerebrovascular
diseases and thrombotic occlusive diseases; the dyslipidemia includes
cholesterol,
triglyceride increase, low-density lipoprotein increase, high-density
lipoprotein
reduction, and the like; the cardiovascular and cerebrovascular diseases
include
coronary arteriosclerotic heart diseases, acute myocardial infarction,
atherosclerosis,
stroke, peripheral artery occlusive diseases, and the like.
Brief Description of the Drawings
FIG. 1 shows a pScFvDisb-S1 plasmid profile;
FIG. 2 shows relative affinity of the single-chain antibodies from positive
cloning
phage of a mutant light chain antibody library by Enzyme Linked Immunosorbent
Assay (ELISA);
FIG. 3 shows relative affinity comparison of the single-chain antibody of the
antibody library of the mutant light chain by means of positive cloning phage
monoclonal gradient diluted ELISA;
FIG. 4 shows identification of relative affinity of the single-chain antibody
of an
antibody library of the mutant heavy chain by means of positive cloning phage
monoclonal (ELISA);
FIG. 5 shows relative affinity comparison of the single-chain antibody of the
antibody library of the mutant heavy chain by means of positive cloning phage
monoclonal gradient diluted ELISA;
FIG. 6 shows a pTSEG4 plasmid profile;
FIG. 7 shows a pTSEK plasmid profile;
FIG. 8 shows binding test of the complete anti-Proprotein Convertase
Subtilisin/Kexin Type 9 (PCSK9) antibody with PCSK9 at the molecular level;
FIG. 9 shows the anti-PCSK9 antibody competitively inhibiting the binding
between Low Density Lipoprotein Receptor (LDLR) and the PCSK9;
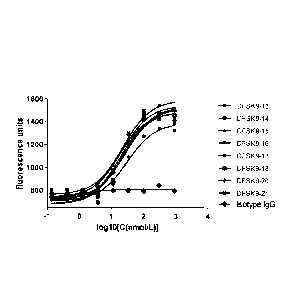
FIG. 10 shows biological activity test of the complete anti-PCSK9 antibody.
9
CA 03032506 2019-01-30
Detailed Description of the Embodiments
Detailed implementation methods of the disclosure are shown in embodiments.
The experimental methods and reagents described in the embodiments are
conventional experimental methods and reagents without special description.
The
following are used only to illustrate and interpret the present disclosure,
rather than
limiting it in any way.
The disclosure provides one type of monoclonal antibody which is specifically
binding with Proprotein Convertase Subtilisin/Kexin Type 9 (PCSK9), the heavy
chain
variable region sequence includes any one of SEQ ID NO.1, 2, 3, 4, 5, 6, 7, 8,
9 and
10, and the light chain variable region sequence includes any one of SEQ ID
NO.11,
12, 13, 14, 15, 16, 17, 18, 19,20 and 21.
Preferably, the heavy chain variable region sequence of the monoclonal
antibody
which is specifically binding with the PCSK9 is selected from any one of SEQ
ID NO.2,
3, 4, 5, 6, 7, 8, 9 and 10, and the light chain variable region sequence is
selected from
any one of SEQ ID NO.12, 14, 15, 16 and 18.
By means of screening of a light chain phage library , the amino acid sequence
of
light chain Complementarity-Determining Regions LCDR1, LCDR2 and LCDR3 froman
antibody light chain or functional fragments of the antibody light chain is
selected from
any one combination as follows (as shown in table 1):
Table 1 Amino acid sequences of different CDRs of the light chain
No. LCDR1 LCDR2 LCDR3
A RASQSIDNRLT(SEQ ID DASSRQS(SEQ ID QQPENDPTT(SEQ ID
NO.22) NO.33) NO.42)
RASQSVRNWLD(SEQ ID GASTLES(SEQ ID QQDNDIPLT(SEQ ID
NO.23) NO.34) NO.43)
RASQGINSWLN(SEQ ID AASTRET(SEQ ID QQWNNTPNT(SEQ ID
NO.24) NO.35) NO.44)
RASQNVNNWLN(SEQ ID AASTRET(SEQ ID QQDNDMPLT(SEQ ID
NO.25) NO.35) NO.45)
RASQNINSWLN(SEQ ID GASSRQS(SEQ ID QQWFDVPTT(SEQ ID
NO.26) NO.36) NO.46)
CA 03032506 2019-01-30
RASQNINNWLN(SEQ ID GASTRPT(SEQ ID QQWDDTPNT(SEQ ID
NO.27) NO.37) NO.47)
RASQGIHNWLN(SEQ ID DASNRQS(SEQ ID QQNSNIPLT(SEQ ID
NO.28) NO.38) NO.48)
RASQDVDSWLT(SEQ ID GASNLAS(SEQ ID QQDSKIPLT(SEQ ID
NO.29) NO.39) NO.49)
RASQSVRNWLN(SEQ ID DASNLQS(SEQ ID QQVVTDTPLT(SEQ ID
NO.30) NO.40) NO.50)
RASQDVRNVVLT(SEQ ID GASNLAS(SEQ ID QQDDSTPPT(SEQ ID
NO.31) NO.39) NO.51)
RASOSIRSYLN(SEQ ID DASSRPT(SEQ ID QQGDSMPMT(SEQ ID
NO.32) NO.41) NO.52)
By means of screening of a heavy chain phage library, theCDR1, CDR2 and
CDR3 of the heavy chain from an antibody or a functional fragment are
respectively
represented by HCDR1, HCDR2 and HCDR3: HCDR1 is any one of GGTFTNNA(SEQ
ID NO.53), GYTVTSYG(SEQ ID NO.54) or GYSLTSYG(SEQ ID NO.55); HCDR2 is
any one of RIIPMFGMA(SEQ ID NO.56), WLSFYNGNT(SEQ ID NO.57),
VVVIFYNGNT(SEQ ID NO.58), VVVSFYQGNT(SEQ ID NO.59), VVVSFYNGQT(SEQ
ID NO.60) or VVVSFYNGNS(SEQ ID NO.61); HCDR3 is any one of AREGIPMI(SEQ
ID NO.62), ARGYSLDV(SEQ ID NO.63), ARGYGMSI(SEQ ID NO.64),
ARGFGMDR(SEQ ID NO.65), ARGYGMTV(SEQ ID NO.66) or ARGFGLSV(SEQ ID
NO.67).
Preferably, by means of screening of the heavy chain phage library, the
monoclonal antibody which is specifically binding with the PCSK9 includes
heavy
chain variable regions of HCDR1, HCDR2 and HCDR3 and light chain variable
regions
of LCDR1, LCDR2 and LCDR3; herein, a sequence of the heavy chain variable
region
HCDR1 is an amino acid sequence selected from GYTVTSYG(SEQ ID NO.54) or
GYSLTSYG(SEQ ID NO.55); a sequence of the light chain variable region LCDR1 is
any one amino acid sequence selected from RASQSVRNWLD(SEQ ID NO.23),
RASQNVNNWLN(SEQ ID NO.25), RASQNINSWLN(SEQ ID NO.26),
RASQNINNWLN(SEQ ID NO.27) or RASQDVDSWLT(SEQ ID NO.29); a sequence of
the heavy chain variable region HCDR2 is any one amino acid sequence selected
CA 03032506 2019-01-30
from VVVSFYQGNT(SEQ ID NO.59), VVVSFYNGQT(SEQ ID NO.60) or
WVSFYNGNS(SEQ ID NO.61); a sequence of the light chain variable region LCDR2
is
any one amino acid sequence selected from GASTLES(SEQ ID NO.34),
AASTRET(SEQ ID NO.35), GASSRQS(SEQ ID NO.36), GASTRPT(SEQ ID NO.37) or
GASNLAS(SEQ ID NO.39); a sequence of the heavy chain variable region HCDR3 is
any one amino acid sequence selected from ARGYSLDV(SEQ ID NO.63),
ARGYGMSI(SEQ ID NO.64), ARGFGMDR(SEQ ID NO.65) or ARGYGMTV(SEQ ID
NO.66); a sequence of the light chain variable region LCDR3 is any one amino
acid
sequence selected from QQDNDIPLT(SEQ ID NO.43), QQDNDMPLT(SEQ ID NO.45),
QQWFDVPTT(SEQ ID NO.46), QQWDDTPNT(SEQ ID NO.47) or QQDSKIPLT(SEQ
ID NO.49).
A method for obtaining a specific antibody by means of completely synthesizing
a
ScFv single-chain phage antibody library, the fully human monoclonal antibody
which
is specifically binding with the PCSK9 is obtained by means of screening by
using a
phage antibody library technique, including:
(1) carrying out biopanning for an anti-PCSK9 single-chain antibody, and
carrying
out three rounds of enrichment screening of an antibody library to obtain an
antibody
sequence DFSK9-1 having high affinity;
(2) based on DFSK9-1, by means of computer aided design, establishing a light
chain CDR123 mutation library, and carrying out biopanning and positive clone
screening and identification on the antibody library to obtain 10 different
light chain
antibody sequences including clones DFSK9-2, DFSK9-3, DFSK9-4, DFSK9-5,
DFSK9-6, DFSK9-7, DFSK9-8, DFSK9-9, DFSK9-10 and DFSK9-11 ; Affinity
comparison of the above 10 single chain antibodies at phage level;
(3) selecting five clones having high affinity, and establishing a heavy chain
CDR123 library, carrying out biopanning and positive clone screening and
identification on the antibody library to obtain single-chain antibodies with
different
sequences including DFSK9-12, DFSK9-13, DFSK9-14, DFSK9-15, DFSK9-16,
DFSK9-17, DFSK9-18, DFSK9-19, DFSK9-20 and DFSK9-21 ; Affinity comparison of
the single-chain antibodies at phage level;
(4) cloning a heavy chain variable region gene and a light chain variable gene
of
12
CA 03032506 2019-01-30
the clones in step (3) into an eukaryotic expression vector, transfected into
a host cell
to obtain a complete anti-PCSK9 monoclonal antibody.
Preferably, affinity and bioactivity test on the complete anti-PCSK9
monoclonal
antibody of step (4) is further carried out.
Specific Embodiments
The present disclosure is described in detail below in connection with the
drawings and embodiments.
Embodiment 1, biopanning of an anti-Proprotein Convertase Subtilisin/Kexin
Type 9 (PCSK9) monoclonal antibody
Modifying a p0om3 vector by means of gene cloning, naming the modified vector
as pScFvDisb-S1 (as shown Fig. 1), and establishing a complete synthetic phage
antibody library based on the vector.
Coating an antigen PCSK9-His 10pg/1m1/ tube by an immune tube and overnight
at 4 C. Blocking the immune tube and the phage antibody library (the amount
of
phage is about 109-1012) respectively by using PBST-4% milk for one hour at 37
C,
putting the blocked phage antibody library into the immune tube to carry out
antigen-antibody binding, and reacting for 1 hour at 3700; washing off unbound
phage
by using a Phosphate Buffered Solution-Phosphate Buffer Saline (PBST-PBS),
eluting
by using 0.1M of Glycine-HCl of pH2.2, and neutralizing by using 1.5M of a
Tris-HCI of
pH8.8 neutral eluant till about pH7.0; infecting 10m1 of the eluant to grow
into an
XL1-Blue bacterial liquid with an OD value is about 0.5-0.8, firstly leaving
to stand for
30 minutes at 37 C, and carrying out shaking table oscillation culture for 1
hour at
150rpm; carrying out gradient dilution on 1% of the bacterial liquid, coating
a 2YTATG
small plate, and calculating a yield of the phage; centrifuging the rest
bacterial liquid,
coating a 2YTATG large plate, and culturing overnight at 37 C; transferring
the
bacterium cultured overnight into 2YTATG liquid culture medium, shaking till a
logarithmic phase, adding M13K07 to assist phage infection, culturing at 28 C
overnight to amplify the phage, carrying out sedimentation purification on the
phage
with PEG6000-NaCl for a next round of screening, and carrying out three rounds
of
13
CA 03032506 2019-01-30
phage library enrichment and screening in all.
Embodiment 2, screening of positive clones of anti-Proprotein Convertase
Subtilisin/Kexin Type 9 (PCSK9) single chain antibody
After three rounds of screening, selecting well partitioned monoclonal
bacterial
colonies, inoculating in a deep 96-well plate with 2YTATG liquid culture
medium,
culturing for 5 hours at 220rpm and 37 C till a logarithmic phase, putting
about 1010 of
helper phage M13K07 into each well, leaving to stand for 30 minutes at 37 C,
and
carrying out oscillation culture for 1 hour at 150rpm; centrifuging for 15
minutes at
4000rpm, resuspending precipitate in the 2YTATKA liquid culture medium, and
culturing overnight at 220rpm and 28 C; centrifuging for 15 minutes at
4000rpm at 4
C, and carrying out monoclonal Enzyme Linked Immunosorbent Assay (ELISA)
identification on phage-containing supernate; screening an single-chain
antibody
DFSK9-1 having high affinity, the heavy chain variable region of the antibody
is named
as DFSK9-H1, and the amino acid sequence of the antibody is shown in SEQ ID
NO.1;
the light chain variable region of the antibody is named as DFSK9-L1, and the
amino
acid sequence of the antibody is shown in SEQ ID NO.11;
SEQ ID NO.1 (DFSK9-H1 heavy chain variable region sequence):
QVQLVQSGAEVKRPGASVKVSCKASGGTFTN NAI SVVVRQAPGQGLEWMG RI I P
MFGMANYAQKLQGRGTMTTDPSTSTAYMELRSLRSDDTAVYYCAREGI PM IWGQGT
TVTVSS
SEQ ID NO.11 (DFSK9-Lllight chain variable region sequence):
DI QMTQS PSS LSASVG DRVTITCRASQSI DN RLTVVYQQ KPG KAPKLLIYDASSRQ
SGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQPEN DPTTFGQGTKVEIK
Embodiment 3, in-vitro affinity maturation of anti-Proprotein Convertase
Subtilisin/Kexin Type 9 (PCSK9) single chain antibody DFSK9-1
3.1 Establishment of a DFSK9-1 light chain CDR123 mutation library
Carrying out double-digestion on pScFvDisb-DFSK9-1 plasma by using Nhel and
Notl, carrying out agarose gel electrophoresis on a digestion product, cutting
gel, and
recycling strips of 5.5kb; carrying out double-digestion on a synthetic light
chain
mutation library gene VLCDR123M by using Nhel and Notl, and recycling a
product by
using a universal product recycling kit; connecting the mutation library gene
with a
14
CA 03032506 2019-01-30
vector for 4 hours by using T4 Deoxyribonucleic Acid (DNA) ligase at 16 C
according
to a mole ratio of 3:1; transforming a connection product into XL1-Blue
electrocompetent cells by using an electroporation method; at 37 C, carrying
out
vibration culture for one hour at 150rpm to achieve anabiosis; diluting 1%
bacterial
liquid, coating a small plate, and calculating a library capacity;
centrifuging other
bacterial liquid for 15 minutes at 4000rpnn, coating precipitate to a 2YTATG
large plate,
inverting and culturing at 37 C overnight; the library capacity of
established antibody
library is about 108, randomly selecting 20 clones to carry out sequence
analysis, and
both the sequence accuracy rate and diversity are greater than 90%.
3.2 Biopanning of phage antibody libraries and screening of positive clones
Carrying out biopanning and positive clone screening by using methods of the
embodiment 1 and the embodiment 2, sequencing clones having high affinity to
obtain
different single chain antibody sequences which are respectively named as
DFSK9-2, DFSK9-3, DFSK9-4, DFSK9-5, DFSK9-6, DFSK9-7, DFSK9-8, DFSK9-9,
DFSK9-10 and DFSK9-11, corresponding light chain variable region are named as
DFSK9-L2, DFSK9-L3, DFSK9-L4, DFSK9-L5, DFSK9-L6, DFSK9-L7, DFSK9-L8,
DFSK9-L9, DFSK9-L10 and DFSK9-L11, and corresponding amino acid sequences
are respectively shown in SEQ ID NO.12, SEQ ID NO.13, SEQ ID NO.14, SEQ ID
NO.15, SEQ ID NO.16, SEQ ID NO.17, SEQ ID NO.18, SEQ ID NO.19, SEQ ID
NO.20 and SEQ ID NO.21; light chain variable region sequences SEQ ID NO.12-SEQ
ID NO.21 as shown as follows:
SEQ ID NO.12 ( DFSK9-L2 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQSVRNWLDWYQQKPGKAPKWYGASTL
ESGVPSR FSGSGSGTDFTLTISSLQ PE DFATYYCQQ DN DI PLIFGQGTKVEI K
SEQ ID NO.13 ( DFSK9-L3light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQGINSWLNVVYQQKPGKAPKLLIYAASTR
ETGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQWNNTPNTFGQGTKVEIK
SEQ ID NO.14 ( DFSK9-L4 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQNVNNWLNVVYQQKPGKAPKLLIYAASTR
CA 03032506 2019-01-30
ETGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQDNDMPLTFGQGTKVEIK
SEQ ID NO.15 ( DFSK9-L5 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQNINSWLNVVYQQKPGKAPKWYGASSR
QSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQWFDVPTTFGOGTKVEIK
SEQ ID NO.16 ( DFSK9-L6 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASONINNWLNVVYQQKPGKAPKWYGASTR
PTGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQWDDTPNTFGQGTKVEIK
SEQ ID NO.17 ( DFSK9-L7 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQGIHNWLNVVYQQKPGKAPKWYDASNR
QSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQNSNIPLTFGQGTKVEIK
SEQ ID NO.18 ( DFSK9-L8 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQDVDSWLTVVYQQKPGKAPKWYGASNL
ASGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQDSKIPLTFGQGTKVEIK
SEQ ID NO.19 ( DFSK9-L9 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQSVRNWLNINYQQKPGKAPKWYDASNL
QSGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQVVTDTPLTFGQGTKVEIK
SEQ ID NO.20 ( DFSK9-L10 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQDVRNWLTWYQQKPGKAPKWYGASNL
ASGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQDDSTPPTFGQGTKVEIK
SEQ ID NO.21 ( DFSK9-L11 light chain variable region sequence ) :
DIQMTQSPSSLSASVGDRVTITCRASQSIRSYLNWYQQKPGKAPKWYDASSRP
TGVPSRFSGSGSGTDFTLTISSLQPEDFATYYCQQGDSMPMTFGQGTKVEIK
Monoclonal phage ELISA identification results are shown in Fig. 2.
3.3 Identification of relative affinity of the anti-PCSK9 single-chain
antibody by
means of phage horizontal gradient diluted Enzyme Linked Immunosorbent Assay
(ELISA)
Carrying out monoclonal phage display and purification on the clones obtained
in
16
CA 03032506 2019-01-30
the embodiment 3.2, and carrying out phage horizontal gradient diluted ELISA
identification on relative affinity of the monoclonal antibody.
Coating PCSK9-His (300ng/wel1/100p1) by 0.01M of Phosphate Buffer Saline
(PBS) of pH7.2, and coating overnight at 4 C; washing with Polybutylene
Terephthalate (PBST) for three times, and sealing with PBST-4% milk at 37 C
for one
hour; adding a purified phage sample (100pl/well) which is diluted with PBST-
4%milk
for a 5-time gradient, and leaving to stand for one hour at 37 C; washing
with PBST
for five times, adding an anti-M13-HRP monoclonal antibody (100pl/well) which
is
diluted with PBST-4%milk in a ratio of 1:5000, and leaving to stand for one
hour at 37
C; developing by using a Tetramethylbenzidine (TMB) developing kit
(100p1/well) for
minutes at a room temperature, and terminating developing with 2M H2SO4
(50p1/well); reading numbers by using a microplate reader at wavelengths of
450nm
and 630nm; analyzing data and drawing pictures by using software GraphPad
Prism
5Demo, and results are shown in Fig. 3. Results show that the screened phage
single-chain antibodies all are capable of binding with PCSK9. The affinities
of
DFSK9-2, DFSK9-4, DFSK9-5, DFSK9-6 and DFSK9-8 with PCSK9 are remarkably
higher than that of other clones, and five of the single-chain antibodies are
selected for
next test.
Embodiment 4, in-vitro affinity maturation of screened anti-Proprotein
Convertase Subtilisin/Kexin Type 9 (PCSK9) single chain antibodies for another
time
4.1 Establishment of heavy chain CDR123 mutation libraries by using a chain
displacement method
Carrying out double-digestion on mixed plasmid of five single-chain antibodies
including DFSK9-2, DFSK9-4, DFSK9-5, DFSK9-6 and DFSK9-8 in the embodiment
3.3 by using Ncol-HF and Kpnl, cutting glue, and recycling a strip of 5,5kb;
carrying
out double-digestion on a synthetic heavy chain mutation library gene
VHCDR123M by
using Ncol-HF and Kpnl, and recycling a digestion product by using a universal
recycling agent kit; connecting the mutation library gene with a vector for 4
hours by
using T4 Deoxyribonucleic Acid (DNA) ligase at 16 C according to a mole ratio
of 3:1;
transforming connection product into XL1-Blue electroconnpetent cells by using
an
17
CA 03032506 2019-01-30
electroporation method; at 37 C, carrying out vibration culture for one hour
at 150rpm
to achieve anabiosis; diluting a 1% bacterial liquid, coating a small plate,
and
calculating a library capacity; centrifuging other bacterial liquid for 15
minutes at
4000rpm, coating precipitate to a 2YTATG large plate, inverting and culturing
at 37 C
overnight; the established antibody library has a library capacity about
5*108, randomly
selecting 20 clones to carry out sequence analysis, and both a sequence
accuracy
rate and diversity are greater than 90%.
4.2 Biopanning of phage antibody libraries and screening of positive clones
Carrying out phage display and purification on the antibody library in the
embodiment 4.1; panning anti-PCSK9 single-chain antibodies; a biopanning
method of
the phage antibody library and screening of the positive clones are identical
to those of
the embodiment 1 and the embodiment 2; sequencing to screen 10 different
anti-PCSK9 antibody sequences which are respectively named as DFSK9-12,
DFSK9-13, DFSK9-14, DFSK9-15, DFSK9-16, DFSK9-17, DFSK9-18, DFSK9-19,
DFSK9-20 and DFSK9-21; herein, DFSK9-12 has a light chain variable region
sequence of DFSK9-L2, and the corresponding amino acid sequence is shown in
SEQ
ID NO.12; DFSK9-13 has a light chain variable region sequence of DFSK9-L8, and
the
corresponding amino acid sequence is shown in SEQ ID NO.18; DFSK9-14,
DFSK9-16 and DFSK9-18 have a light chain variable region sequence of DFSK9-L5,
and the corresponding amino acid sequence is shown in SEQ ID NO.15; DFSK9-15,
DFSK9-17, DFSK9-20 and DFSK9-21 have a light chain variable region sequence of
DFSK9-L6, and the corresponding amino acid sequence is shown in SEQ ID NO.16;
DFSK9-19 has a light chain variable region sequence of DFSK9-L4, and the
corresponding amino acid sequence is shown in SEQ ID NO.14. DFSK9-12 has a
heavy chain variable region sequence of DFSK9-H2, and corresponding amino acid
sequence is shown in SEQ ID NO.2; DFSK9-13 has a heavy chain variable region
sequence of DFSK9-H8, and the corresponding amino acid sequence is shown in
SEQ ID NO.8; DFSK9-14 has a heavy chain variable region sequence of DFSK9-H7,
and the corresponding amino acid sequence is shown in SEQ ID NO.7; DFSK9-15
has
a heavy chain variable region sequence of DFSK9-H3, and the corresponding
amino
acid sequence is shown in SEQ ID NO.3; DFSK9-16 has a heavy chain variable
region
18
CA 03032506 2019-01-30
sequence of DFSK9-H6, and the corresponding amino acid sequence is shown in
SEQ ID NO.6; DFSK9-17 and DFSK9-18 have a heavy chain variable region
sequence of DFSK9-H4, and the corresponding amino acid sequence is shown in
SEQ ID NO.4; DFSK9-19 has a heavy chain variable region of DFSK9-H9, and the
corresponding amino acid sequence is shown in SEQ ID NO.9; DFSK9-20 has a
heavy chain variable region of DFSK9-H5, and the corresponding amino acid
sequence is shown in SEQ ID NO.5; DFSK9-21 has a heavy chain variable region
sequence of DFSK9-H10, and the corresponding amino acid sequence is shown in
SEQ ID NO.10. Light chain variable region sequences are shown in amino acid
sequence SEQ ID NO.12, SEQ ID NO.14, SEQ ID NO.15, SEQ ID NO.16 and SEQ ID
NO.18 in the embodiment 3.2, and heavy chain variable region amino acid
sequences
SEQ ID NO.2-SEQ ID NO.10 are as follows:
SEQ ID NO. 2 ( DFSK9-H2 heavy chain variable region sequence ) :
QVQ LVQSGAEVKKPGASVKVSC KASGYTVTSYG I SVVVRQAPGQG LEWM GWLS
FYNGNTNYAQKLQGRGTMTTDPSTSTAYMELRSLRSDDTAVYYCARGYSLDVWG
QGTTVTVSS
SEQ ID NO.3 ( DFSK9-H3 heavy chain variable region sequence ) :
QVQ LVQSGAEVKKPGASVKVSC KASGYTVTSYG I SVVVRQAPGQ
GLEWMGVVVSFYNGQTNYAQKLQGRGTMTTDPSTSTAYMELRSLRSDDTAVYYCAR
GYSLDVWGQGTTVTVSS
SEQ ID NO.4 ( DFSK9-H4 heavy chain variable region sequence ) :
QVQLVQSGAEVKKPGASVKVSCKASGYTVTSYGISVVVRQAPGQ
GL EWM GWVSFYNG NSNYAQ KLQGRGTMTTDPSTSTAYM EL RSLRSDDTAVYYCAR
GYSLDVWGQGTTVTVSS
SEQ ID NO.5 ( DFSK9-H5 heavy chain variable region sequence ) :
QVQ LVQSGAEVKKPGASVKVSC KASGYS LTSYG I SVVVRQAPGQ
GLEWMG \ANSFYNG NS NYAQKLQGRGTMTTDPSTSTAYM ELRS LRSDDTAVYYCAR
GFGMDRWGQGTIVTVSS
19
CA 03032506 2019-01-30
SEQ ID NO.6 ( DFSK9-H6 heavy chain variable region sequence ) :
QVQLVQSGAEVKKPGASVKVSCKASGYTVTSYGISWVRQAPGQGLEWMGVVVS
FYQGNTNYAQKLQGRGTMTTDPSTSTAYMELRSLRSDDTAVYYCARGFGMDRWGQ
GTTVTVSS
SEQ ID NO.7 ( DFSK9-H7 heavy chain variable region sequence ) :
QVQLVQSGAEVKKPGASVKVSCKASGYSLTSYGISVVVRQAPGQGLEWMGVVVS
FYNGQTNYAQKLQGRGTMTTDPSTSTAYM ELRSLRSDDTAVYYCARGYGMS IWGQ
GTTVTVSS
SEQ ID NO.8 ( DFSK9-H8 heavy chain variable region sequence ) :
QVQLVQSGAEVKKPGASVKVSCKASGYSLTSYGISWVRQAPGQGLEWMGVVVS
FYQGNTNYAQKLQGRGTMTTDPSTSTAYMELRSLRSDDTAVYYCARGYGMTVVVGQ
GTTVTVSS
SEQ ID NO.9 ( DFSK9-H9 heavy chain variable region sequence ) :
QVQLVQSGAEVKKPGASVKVSCKASGYSLTSYGISVVVRQAPGQGLEWMGVVVT
FYNGNTNYAQKLQGRGTMTTDPSTSTAYMELRSLRSDDTAVYYCARGFGLSVWGQ
GTTVTVSS
SEQ ID NO.10 ( DFSK9-H10 heavy chain variable region sequence ) :
QVQLVQSGAEVKKPGASVKVSCKASGYSLTSYGISVVVRQAPGQGLEWMGVVVS
FYNGQTNYAQKLQGRGTMTTDPSTSTAYMELRSLRSDDTAVYYCARGFGMDRWGQ
GTTVTVSS
Monoclonal phage ELISA identification on relative affinity of phage-Abs is
shown
in Fig. 4.
4.3 Gradient diluted phage ELISA identification on affinity of the PCSK9
single
chains antibodies
Carrying out monoclonal phage display and purification on the clones obtained
in
the embodiment 4.2, and carrying out phage horizontal gradient diluted ELISA
identification on affinity of the single chain antibody, by using the same
methods as
those in the embodiment 3.3 of the embodiment 3. Results are shown in Fig. 5.
The
CA 03032506 2019-01-30
ten different screened single-chain antibodies all are capable of well binding
with the
PCSK9 and have higher affinity than that of primary single-chain DFSK9-1.
Embodiment 5, affinity identification of the completeanti-Proprotein
Convertase Subtilisin/Kexin Type 9 (PCSK9) antibodies
5.1 Preparation of the complete antibodies of the anti-PCSK9
Cloning the heavy chain VH gene of the antibody screened in the embodiment 4
into the vector pTSEG4 (Fig. 6) which having a heavy chain constant region
gene (y4),
cloning the light chain VK gene into the vector pTSEK (Fig. 7) which having
a light chain constant region gene (k chain), and both the vectors pTSEG4 and
pTSEK are obtained by means of transformation on the basis of the PTT vector.
The
preparation process of the PTT vector is specifically described in a reference
(Yves.Durocher,Sylvie.Perret and Amine. Kamen Nucleic Acids Research,
2002Vol.30,
No.2e9). Carrying out transient transfection on the HEK293E cell, carrying out
complete antibody expression, and purifying by using an AKTA protein a
affinity
column to obtain complete antibody proteins.
5.2 Binding test of complete antibodies with PCSK9
Coating PCSK9-His(300ng/well/100p1) by 0.01M PBS buffer at pH 7.2 overnight at
4 C, washing three times with PBST (1%0Tween 20) of 300p1/well, further
adding
PBST-4%milk, and blocking for one hour at 37 C; adding complete antibodies
DFSK9-1, DFSK9-12, DFSK9-13, DFSK9-14, DFSK9-15, DFSK9-16, DFSK9-17,
DFSK9-18, DFSK9-19, DFSK9-20 and DFSK9-21 of different dilution degrees; the
highest concentration of the 11 complete antibodies is 50pg/ml, diluting at a
five-time
gradient, diluting for eight gradients for each complete antibody, and
incubating for one
hour at 37 C; washing 5 times with PBST of 300p1/well, further adding a
goat-anti-human IgG-HRP secondary antibody diluted by PBST-4%milk at a ratio
of
1:5000, and incubating for one hour at 37 C; washing 5 times with PBST of
300p1/well,
developing by using a Tetramethylbenzidine (TMB) developing kit (100pl/well),
developing for 10 minutes at a room temperature, and terminating developing
with 2M
H2S0.4 (50pl/well); reading numbers by using a microplate reader at
wavelengths of
450nm and 630nm; analyzing data and drawing pictures by using software
GraphPad
21
CA 03032506 2019-01-30
Prism 5 Demo, and results are shown in Fig. 8 and table 2. Results show that
all
antibodies are capable of well binding with PCSK9, and DFSK9-14, DFSK9-15,
DFSK9-16, DFSK9-17, DFSK9-18, DFSK9-20 and DFSK9-21 have high affinity.
Table 2 Affinity EC50 value of complete antibodies
No. Sample EC50(ng/m1) No. Sample E050(ng/m1)
1 DFSK9-1 170.8 7 DFSK9-17 31.41
2 DFSK9-12 64.65 8 DFSK9-18 33.9
3 DFSK9-13 83.51 9 DFSK9-19 83.16
4 DFSK9-14 42.22 10 DFSK9-20 33.73
DFSK9-15 39.74
11 DFSK9-21 28.2
6 DFSK9-16 56.21
5.3 The test of complete antibodies inhibiting the binding between Low Density
Lipoprotein Receptor (LDLR) and PCSK9
Coating LDLR-Fc(10Ong/well/100p1) by 0.01M PBS buffer at pH 7.2 overnight at 4
C, washing three times with PBST, further adding PBST-4%milk, and blocking for
one
hour at 37 C; further adding 2pg/m1 PCSK9-His(100p1/well) diluted by PBST-
4%milk,
and incubating to one hour at 37 C; adding complete antibodies DFSK9-1, DFSK9-
12,
DFSK9-13, DFSK9-14, DFSK9-15, DFSK9-16, DFSK9-17, DFSK9-18, DFSK9-19,
DFSK9-20 and DFSK9-21 of different dilution degrees; the highest concentration
of
the 11 complete antibodies is 100pg/ml, diluting at a five-time gradient,
diluting for
eight gradients for each complete antibody, and incubating for 2 hours at 37
C;
washing 5 times with PBST, further adding a mouse anti-His IgG-HRP secondary
antibody diluted by PBST-4%milk, and incubating for 1 hour at 37 C;
developing by
using the TMB developing kit (100pl/well), developing for 10 minutes at the
room
temperature, and terminating developing with 2M H2504 (50pl/well); reading
numbers
by using the microplate reader at 450nm and 630nm; analyzing data and drawing
pictures by using software GraphPad Prism 5 Demo, and results are shown in
Fig. 9
and table 3. Results show that all antibodies can effectively inhibit binding
of the
PCSK9 with the LDLR, and DFSK9-14, DFSK9-15, DFSK9-16, DFSK9-17, DFSK9-18,
DFSK9-20 and DFSK9-21 have higher inhibition capabilities.
22
CA 03032506 2019-01-30
Table 3 IC50 value of competitive testing of complete antibodies
No. Sample IC50(ng/m I) No. Sample IC50(ng/m1)
1 DFSK9-1 847.4 7 DFSK9-17 88.98
2 DFSK9-12 266.2 8 DFSK9-18 107.5
3 DFSK9-13 382.8 9 DFSK9-19 533.5
4 DFSK9-14 145.9 10 DFSK9-20 216.3
DFSK9-15 113.9
11 DFSK9-21 161.3
6 DFSK9-16 281.7
5.4 Affinity test of complete antibodies by means of BlAcore X100
Affinity test of complete antibodies by using a capturing method; coupling
goat-anti-human IgG to a surface of a CM5 chip, respectively diluting DFSK9-1,
DFSK9-12, DFSK9-13, DFSK9-14, DFSK9-15, DFSK9-16, DFSK9-17, DFSK9-18,
DFSK9-19, DFSK9-20 and DFSK9-21, and ensuring that about 200RU of the
antibodies are captured by the goat-anti-human IgG; setting a series of
concentration
gradients (200nM, 100nM, 50nM, 25nM, 12.5nM, 6.25nM, 3.125nM, 1.5625nM,
0.78125nM) for PCSK9, flowing through a surface of a stationary phase, and
testing
the affinity of the antibodies. Results show that the screened antibodies all
have high
affinity (see table 4), and eight complete antibodies having the highest
affinity are
selected for bioactivity test.
Table 4 Constant test values of affinity of anti-PCSK9 complete antibodies
No. Sample ka(1/Ms) kd ( 1/s ) KD (M)
1 DFSK9-1 5.178E+5 9.191E-5 1.755E-10
2 DFSK9-12 4.180E+5 4.975E-5 1.190E-10
3 DFSK9-13 5.747E+5 2.990E-5 5.202E-11
4 DFSK9-14 2.700E+6 7.901E-5 2.926E-11
5 DFSK9-15 5.425E+5 4.746E-6 8.749E-12
6 DFSK9-16 1.217E+6 5.613E-5 4.613E-11
7 DFSK9-17 1.184E+6 1.844E-6 1.557E-12
23
CA 03032506 2019-01-30
8 DFSK9-18 5.117E+5 1.992E-6 3.893E-12
9 DFSK9-19 3.952E+5 7.339E-5 1.857E-10
DFSK9-20 9.252E+5 1.451E-5 1.568E-11
11 DFSK9-21 5.918E+5 4.239E-6 7.162E-12
Embodiment 6, bioactivity test of complete anti-Proprotein Convertase
Subtilisin/Kexin Type 9 (PCSK9) antibodies
Inoculating HepG2 cell into 96 wells at a ratio of 2.5X105 cells/ml; on a next
day,
replacing a 10% FBS Mem Ebss Minimum Essential Medium (EMEM) growth culture
medium by a 80p1 analysis culture medium of 5% Fetal Bovine Serum (FBS), and
culturing for 24 hours at 37 C; on a next day, putting complete antibodies
(10pl/well)
diluted by the analysis culture medium with different dilution degrees into an
inoculated
HepG2 cell culture plate. Eight complete antibody samples have an initial
concentration of 900nmo1/L, diluting at a three-time gradient, and diluting
each
complete antibody for eight gradients; further adding a 30nmo1/L PCSK9
solution
(10pl/well), uniformly mixing, and culturing for 4 hours at 37 C in the
presence of 5%
CO2; adding 10u1 of an Low Density Lipoprotein (LDL) solution marked by
0.1mg/m L
Boron Dipyrromethene (BODIPY), uniformly mixing, and continuously culturing
for
15-20 hours in the presence of 5% CO2 at 37 C; adding 200p1 of the PBS into
each
well, washing twice, adding 100p1 of the PBS into each well, reading Relative
Fluorescent Unit (RFU) values by using a microplate reader at wavelengths of
490nm
and 520nm; analyzing data and drawing diagrams by using software GraphPad
Prism
5 Demo, and demonstrating results in Fig.10. As negative control with
homologous
IgG , the diagrams show that the eight antibodies all can block binding of the
PCSK9
with the LDLR in a dose-dependent manner and increase an intake rate of the
LDL in
the HepG2 cell, and the antibodies have approximate bioactivity.
Table 5 E050 value of bioactivity testing
No. Sample EC50(nmol/L) No. Sample EC50(n
mol/L)
1 DFSK9-13 29.61 7 DFSK9-17 18.51
2 DFSK9-14 16.59 8 DFSK9-18 23.91
24
CA 03032506 2019-01-30
3 DFSK9-15 15.31 9 DFSK9-20 17.95
4 DFSK9-16 23.80 10 DFSK9-21 20.33
For ordinary persons skilled in the art, the embodiments only exemplarily
describe
the disclosure, and obviously the specific implementation of the present
disclosure is
not limited by the above-mentioned methods. Any non-substantive improvement on
the basis of method conception and technical schemes of the disclosure, or
disclosures of the method conception and the technical schemes of the
disclosure
without improvement to other situations shall fall within the scope of
protection of the
disclosure.