Note : Les descriptions sont présentées dans la langue officielle dans laquelle elles ont été soumises.
WO 2022/081702
PCT/US2021/054775
TITLE OF THE INVENTION
In vivo targeting of CD4+-T cells for mRNA therapeutics
CROSS REFERENCE TO RELATED APPLICATIONS
This application claims priority to U.S. Provisional Application No.
63/091,010, filed
October 13, 2020, which is incorporated by reference herein in its entirety.
STATEMENT REGARDING FEDERALLY SPONSORED RESEARCH OR
DEVELOPMENT
This invention was made with government support under A1045 008 awarded by
National
Institutes of Health. The government has certain rights in the invention.
BACKGROUND OF THE INVENTION
Modulation of immune cells through activation, inhibition, or modification to
alter their
properties has become a popular and high-demand class of therapy, called
immunotherapeutics.
Today's immunotherapeutics largely rely on biological protein-based agents,
which are expensive
and challenging to manufacture (Pranchevicius et al., 2013, Bioengineered,
4:305-312; Liu et al.,
2018, Precision Clinical Medicine, 1:65-74), or require ex vivo modification
of immune cells
(Schultz et al., 2018, Immunotherapy in Translational Cancer Research; Maugeri
et al., 2019,
Nature Communications, 10:4333). Some examples include antibodies or cytokines
for
modulating immune cell function, monoclonal antibodies for redirecting immune
function,
genetic editing of T cells for preventing viral infections, and chimeric
antigen receptor (CAR) T
cell therapy (Schlake et al., 2019, Cellular and Molecular Life Sciences,
76:301-328; Wraith,
2017, Frontiers in immunology 8, 1668-1668; Whilding et al., 2015, Mol Oncol,
9:1994-2018).
One of the most relevant applications of cancer immunotherapeutics are CAR T
cell
therapies Currently, CART cells are generated ex vivo, which is costly as it
requires extended
cell culture in GMP cell processing facilities. Additionally, it is not a
treatment option for patients
with highly malignant cancers, very low T cell counts, or settings requiring
large scale use
(Schmidts et al., 2018, Frontiers in immunology, 9:2593-2593; Zhao et al.,
2019, Front. Immunol,
10:2250; Junghans, 2017, Cancer Gene Therapy, 24:89-99). There is a vital need
for development
of in vivo T cell-targeted mRNA delivery systems for robust and rapid
generation of CAR T cells.
mRNA-based CAR T cell therapeutics could also provide a safer platform by
reducing the risk of
CART cell-induced toxicities, because of their transient nature, as well as
avoiding the risk of
genomic integration, when so desired (Foster et al., 2019, Molecular Therapy,
27:747-756; Foster
1
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
et al., 2019, Hum Gene Ther, 30:168-178; Kowalski et al., 2019, Molecular
Therapy, 27:710-728;
Pardi etal., 2018, Nat Rev Drug Discov, 17:261-279). Moreover, mRNA-based
therapeutics
could offer gene editing tools for treating viral infections and cancer or
correcting genetic defects,
such as knocking out the C-C chemokine receptor 5 (CCR5) gene for preventing
HIV infection of
T cells (Didigu et al., 2014, Blood, 123:61-69; Liu et al., 2017, Cell Biosci,
7:47), or knocking
out the programmed cell death-1 (PD-1) gene for engineering superior tumor
infiltrating
lymphocytes (TILs) (Bailet etal., 2019, Nature Biotechnology, 37:1425-1434).
One of the key
obstacles in development of mRNA-based immunotherapeutics is efficient in vivo
delivery.
Thus there is a need in the art for an efficient, safe, and immune cell-
specific mRNA-
delivery system for the introduction and wide scale use of current and the
generation of a new
class of robust mRNA-based immunotherapeutics. The present invention satisfies
this unmet
need.
BRIEF DESCRIPTION OF THE DRAWINGS
The following detailed description of embodiments of the invention will be
better
understood when read in conjunction with the appended drawings. It should be
understood that
the invention is not limited to the precise arrangements and instrumentalities
of the embodiments
shown in the drawings.
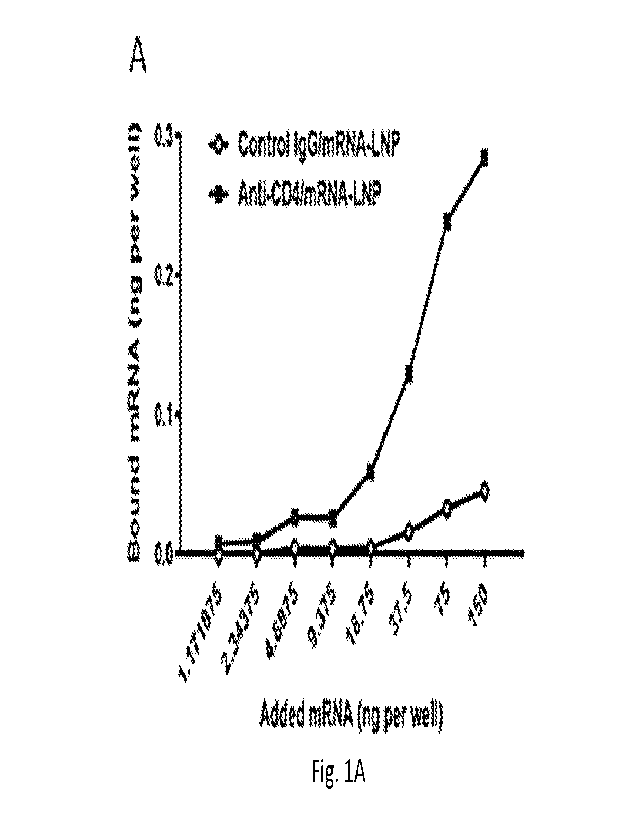
Figure lA through Figure 1C depicts the binding and functional activity of CD4-
targeted
particles in vitro. Figure lA depicts specific in vitro binding of anti-human
CD4/125I-labeled
mRNA-LNP to human CD4+ T cells after a 1 hour incubation at room temperature
(RT). Figure
1B depicts the binding of anti-CD4/mRNA-LNP and control IgG/mRNA-LNP to human
CD4+ T
cells, with increasing mRNA-LNP doses, and their corresponding mean
fluorescence intensity
(MFI). Figure 1C depicts the Luc activity measured in human CD4+ T cells
treated with anti-
human CD4/mRNA-LNP or control IgG/mRNA-LNP.
Figure 2A and Figure 2B depict the results of example experiments
demonstrating Cre
mRNA-mediated genetic recombination in vitro. Figure 2A depicts Cre mRNA-
induced genetic
recombination and consequent reporter gene expression presented as % of
ZsGreen1+ cells among
CD3+CD8- cells. Splenocytes were harvested from Ai6 mice and incubated with
Cre mRNA-LNP
at doses of 1, 3, 6 or 9 [is per 2 million cells. %ZsGreen1+ cells upon anti-
CD4/mRNA-LNP
administration was compared to control IgG/mRNALNP and unconjugated mRNA-LNP
administration (****P < 0.0001, two-way ANOVA with Bonferroni correction).
Figure 2B
depicts the gating strategy to identify ZsGreen1 positive cells among CD3+CD8-
cells.
2
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Figure 3 depicts the results of example experiments demonstrating the flow
cytometric
analysis of mRNA-LNP-treated CD3/CD4 cell populations. Splenocytes were
harvested from Ai6
mice and incubated with Cre mRNA-LNP at a dose of 3 lag per 2 million cells
per well in 6-well
plates overnight. Transient disappearance of CD4 staining is observed upon
administration of
anti-CD4/mRNA-LNP, while non-targeted LNP treated cells are unaffected.
Figure 4A through Figure 4D depicts the results of example experiments
demonstrating
targeting of mRNA-LNP to CD4 + T cells in vivo. Figure 4A depicts the
biodistribution of125I-
labeled anti-CD4/ and control IgG/poly(C) mRNA-LNP in mice at 0.5 hours.
Tissue uptake is
indicated as mean SEM (****P < 0.0001). Figure 4B depicts the localization
ratio, calculated as
the ratio of %ID/g of a given organ to that in the blood of mice treated with
either 125I-labeled anti
CD4/ or control IgG/mRNA-LNP at 30 minutes post-injection. Mean SEM is
shown. In vivo
mRNA-LNP-binding as quantitative measurement of the percentage of radiolabeled
anti-
CD4/mRNA-LNP in selected organs (Figure 4C) and localization ratios in spleens
(Figure 4D),
after intravenous injection of mRNA-LNP. Group size is 3 animals. Statistical
analysis was
performed by two-way ANOVA with Bonferroni correction (****P <0.0001).
Figure 5A and Figure 5B depict the results of example experiments
demonstrating
targeting of Poly(C) RNA-LNP to CD4 in vivo. Figure 5A depicts the in vivo
kinetics of anti-
CD4/mRNA-LNP-binding in spleen and liver as immuno-specificity index.
Immunospecificity
index was calculated as the ratio of %ID/g of selected organs in mice treated
with anti- CD4/ vs.
control IgG/mRNA-LNP, normalized to blood levels. Figure 5B depicts the in
vivo kinetics of
control IgG/mRNA-LNP-binding as quantitative measurement of the percentage of
radiolabeled
mRNALNP in selected organs after intravenous injection of mRNA-LNP. Group size
is 3
animals.
Figure 6A through Figure 6D depict the results of example experiments
demonstrating
biodistribution of targeted mRNA-LNP expression in vivo. Mice were IV injected
with 8 lag of
mRNA-LNP. Organ distribution of Luc mRNA expression 5 hours after
administration of anti-
CD4/ and control IgG/Luc mRNA-LNP was evaluated by measuring Luc activity in
lysed tissues
(Figure 6A) and by luminescence imaging (Figure 6B and Figure 6C). Figure 6A
depicts a
quantitative expression of Luc as light unit (LU)/mg protein. A representative
sample set of
dissected mouse organs (Figure 6B) and whole carcasses after organ removal
(showing
luminescing lymph nodes) (Figure 6C) were analyzed 5 minutes after the
administration of D-
luciferin. Figure 6D depicts a quantitative expression of Luc as LU/mg protein
values in CD3+
cell preparation obtained from the spleens of mice injected with the mRNA-LNP.
For Figure 6A
and Figure 6D, the error bars indicate SEM. Group size is 3 animals.
Statistical analysis was
3
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
performed by two-way ANOVA with Bonferroni correction, (*P < 0.05, **P< 0.01,
and ***P<
0.001).
Figure 7A through Figure 7E depicts the results of example experiments
demonstrating
Cre-mediated genetic recombination upon in vivo administration of CD4-
targeted Cre mRNA-
LNP. Figure 7A depicts a schematic diagram depicting targeted delivery of anti-
CD4/mRNA-
LNP for selective genetic recombination in CD4 T cells, and the principle of
the Ai6 reporter
allele: Cre-mediated excision of a loxP-flanked STOP cassette allows robust
expression of
ZsGreenl, a fluorescent protein. Ai6 mice received Cre mRNA-LNP at doses of 3,
10, and 30 ng
via IV administration. Spleens and lymph nodes were harvested at 24 hours post
treatment and %
of ZsGreen1+ cells in the CD3+CD8- cell population were determined in splenic
(Figure 7B) and
lymph node (Figure 7C) single cell suspensions using flow cytometry. Changes
in the number of
ZsGreenl-expressing CD4+ T cells in spleens (Figure 7D) and lymph nodes
(Figure 7E) over time
were monitored after IV injection of 10 ttg of mRNA-LNP. Group size is 8 or 9
(Figure 7B and
Figure 7C) or 6 (Figure 7D and Figure 7E) animals in a total of three
independent experiments.
Each symbol represents one animal and horizontal lines show the mean with SEM.
Statistical
analysis was performed by two-way ANOVA with Bonferroni correction. %ZsGreenr
cells after
injection of different doses of anti-CD4/mRNA-LNP [*P < 0.05, ""P< 0.0001] and
unconjugated mRNA-LNP [<figref></figref>P< 0.0001] were compared.
Figure 8A and Figure 8B depict the results of example experiments
demonstrating Cre-
mediated genetic recombination in non-T cells upon in vivo administration of
Cre mRNA-LNP.
Ai6 mice received Cre mRNA-LNP at doses of 3, 10, and 30 jig via IV
administration. At 24
hours post treatment, %ZsGreen1+ cells in the dendritic cells (MTICIrCD11c+)
(Figure 8A) and
macrophages (MTICIFF4/80 ) (Figure 8B) of spleen were determined in splenic
single cell
suspensions using flow cytometry. Group size is 8-9 animals in a total of
three independent
experiments. Each symbol represents one animal and horizontal lines show the
mean with SEM.
Figure 9A through Figure 9C depicts the results of example experiments
demonstrating in
vivo uptake of Cre mRNA-LNP by different T cell subtypes. Spleens were
harvested at 24 hours
post-treatment with 10 tug of Cre mRNA-LNP, and % of ZsGreen1+ cells in CD4+ T
cell
subpopulations (Figure 9A) and vs. CD25 marker (Figure 9B) were determined
using flow
cytometry. Naive CD4+ T cells are considered as CD44-CD62L-, central memory T
cells as
CD44 CD62L+, and effector memory T cells as CD44+CD62L-. Group size is 3-11
animals. Each
symbol represents one animal and horizontal lines show the mean with SEM.
Statistical analysis
was performed by two-way ANOVA with Bonferroni correction comparing T cell
subtypes.
4
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Figure 9C depicts the gating strategy to identify ZsGreen1 positive cells
among different CD4+ T
cell subtypes.
Figure 10A and Figure 10B depict the results of example experiments
demonstrating
mRNA-LNP targeting efficiency using multiple administrations. Ai6 mice
received 10 jig (0.4
mg/kg) of anti-CD4/, control IgG/ or =conjugated Cre mRNA-LNP via IV
administration as
daily injections for 3 or 5 days. Spleens and lymph nodes were harvested after
three or five
sequential injections, and the % of ZsGreen1+ cells in the CD3 CD8- cell
population was
determined in splenic (Figure 10A) and lymph node (Figure 10B) single cell
suspensions using
flow cytomety. Group size is 9 animals. Each symbol represents one animal and
horizontal lines
show the mean. Error bars indicate SEM. Statistical analysis was performed by
two-way ANOVA
with Bonferroni correction. %ZsGreen1+ cells after different number of
injections of anti-
CD4/mRNA-LNP [**P <0.01, ****P< 0.00011 were compared.
DETAILED DESCRIPTION
The present invention relates to compositions comprising a delivery vehicle
conjugated to
a CD4+ T cell targeting domain, wherein the delivery vehicle comprises at
least one agent. In one
embodiment, the targeting domain specifically binds to CD4.
In certain embodiments, the delivery vehicle is a lipid nanoparticle
comprising a PEG-
lipid conjugated to the targeting domain. In some embodiments, the at least
one agent is a nucleic
acid. In some embodiments, the at least one agent is an mRNA molecule. In one
embodiment, the
mRNA molecule is a nucleoside-modified mRNA. The present invention also
relates to methods
of treating a disease or disorder using the compositions described herein.
Definitions
Unless defined otherwise, all technical and scientific terms used herein have
the same
meaning as commonly understood by one of ordinary skill in the art to which
this invention
belongs.
As used herein, each of the following terms has the meaning associated with it
in this
section.
The articles "a" and "an" are used herein to refer to one or to more than one
(i. e. , to at
least one) of the grammatical object of the article. By way of example, -an
element- means one
element or more than one element.
-About" as used herein when referring to a measurable value such as an amount,
a
temporal duration, and the like, is meant to encompass variations of 20%,
10%, 5%, 1%, or
5
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0.1% from the specified value, as such variations are appropriate to perform
the disclosed
methods.
The term -antibody,- as used herein, refers to an immunoglobulin molecule,
which
specifically binds with an antigen or epitope. Antibodies can be intact
immunoglobulins derived
from natural sources or from recombinant sources and can be immunoreactive
portions of intact
immunoglobulins. Antibodies are typically tetramers of immunoglobulin
molecules. The
antibodies in the present invention may exist in a variety of forms including,
for example,
polyclonal antibodies, monoclonal antibodies, Fv, Fab and F(ab)2, as well as
single chain
antibodies and humanized antibodies (Harlow et al., 1999, In: Using
Antibodies: A Laboratory
Manual, Cold Spring Harbor Laboratory Press, NY; Harlow et al., 1989, In:
Antibodies: A
Laboratory Manual, Cold Spring Harbor, New York; Houston et al., 1988, Proc.
Natl. Acad. Sci.
USA 85:5879-5883; Bird et al., 1988, Science 242:423-426).
The term antibody fragment" refers to a portion of an intact antibody and
refers to the
antigenic determining variable regions of an intact antibody. Examples of
antibody fragments
include, but are not limited to, Fab, Fab', F(ab')2, and Fv fragments, linear
antibodies, scFy
antibodies, and multispecific antibodies formed from antibody fragments.
An "antibody heavy chain," as used herein, refers to the larger of the two
types of
polypeptide chains present in all antibody molecules in their naturally
occurring conformations.
An "antibody light chain," as used herein, refers to the smaller of the two
types of
polypeptide chains present in all antibody molecules in their naturally
occurring conformations. k
and 1 light chains refer to the two major antibody light chain isotypes.
By the term "synthetic antibody" as used herein, is meant an antibody, which
is generated
using recombinant DNA technology, such as, for example, an antibody expressed
by a
bacteriophage. The term should also be construed to mean an antibody which has
been generated
by the synthesis of a DNA molecule encoding the antibody and which DNA
molecule expresses
an antibody protein, or an amino acid sequence specifying the antibody,
wherein the DNA or
amino acid sequence has been obtained using synthetic DNA or amino acid
sequence technology
which is available and well known in the art. The term should also be
construed to mean an
antibody, which has been generated by the synthesis of an RNA molecule
encoding the antibody.
The RNA molecule expresses an antibody protein, or an amino acid sequence
specifying the
antibody, wherein the RNA has been obtained by transcribing DNA (synthetic or
cloned) or other
technology, which is available and well known in the art.
A -disease" is a state of health of an animal wherein the animal cannot
maintain
homeostasis, and wherein if the disease is not ameliorated then the animal's
health continues to
6
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
deteriorate. In contrast, a "disorder" in an animal is a state of health in
which the animal is able to
maintain homeostasis, but in which the animal's state of health is less
favorable than it would be
in the absence of the disorder. Left untreated, a disorder does not
necessarily cause a further
decrease in the animal's state of health.
An "effective amount" as used herein, means an amount which provides a
therapeutic or
prophylactic benefit.
"Encoding" refers to the inherent property of specific sequences of
nucleotides in a
polynucleotide, such as a gene, a cDNA, or an mRNA, to serve as templates for
synthesis of other
polymers and macromolecules in biological processes having either a defined
sequence of
nucleotides (i.e., rRNA, tRNA and mRNA) or a defined sequence of amino acids
and the
biological properties resulting therefrom. Thus, a gene encodes a protein if
transcription and
translation of mRNA corresponding to that gene produces the protein in a cell
or other biological
system. Both the coding strand, the nucleotide sequence of which is identical
to the mRNA
sequence and is usually provided in sequence listings, and the non-coding
strand, used as the
template for transcription of a gene or cDNA, can be referred to as encoding
the protein or other
product of that gene or cDNA.
"Expression vector" refers to a vector comprising a recombinant polynucleotide
comprising expression control sequences operatively linked to a nucleotide
sequence to be
expressed. An expression vector comprises sufficient cis-acting elements for
expression; other
elements for expression can be supplied by the host cell or in an in vitro
expression system.
Expression vectors include all those known in the art, such as cosmids,
plasmids (e.g., naked or
contained in liposomes) RNA, and viruses (e.g., lentiviruses, retrovinises,
adenoviruses, and
adeno-associated viruses) that incorporate the recombinant polynucleotide.
"Homologous" refers to the sequence similarity or sequence identity between
two
polypeptides or between two nucleic acid molecules. When a position in both of
the two
compared sequences is occupied by the same base or amino acid monomer subunit,
e.g., if a
position in each of two DNA molecules is occupied by adenine, then the
molecules are
homologous at that position. The percent of homology between two sequences is
a function of the
number of matching or homologous positions shared by the two sequences divided
by the number
of positions compared X 100. For example, if 6 of 10 of the positions in two
sequences are
matched or homologous then the two sequences are 60% homologous. By way of
example, the
DNA sequences ATTGCC and TATGGC share 50% homology. Generally, a comparison is
made
when two sequences are aligned to give maximum homology.
7
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
"Isolated" means altered or removed from the natural state. For example, a
nucleic acid
or a peptide naturally present in a living animal is not "isolated," but the
same nucleic acid or
peptide partially or completely separated from the coexisting materials of its
natural state is
"isolated." An isolated nucleic acid or protein can exist in substantially
purified form, or can
exist in a non-native environment such as, for example, a host cell.
In the context of the present invention, the following abbreviations for the
commonly
occurring nucleosides (nucleobase bound to ribose or deoxyribose sugar via N-
glycosidic
linkage) are used. -A" refers to adenosine, -C" refers to cytidine, -G" refers
to guanosine, "T"
refers to thymidine, and "U" refers to uridine.
Unless otherwise specified, a -nucleotide sequence encoding an amino acid
sequence"
includes all nucleotide sequences that are degenerate versions of each other
and that encode the
same amino acid sequence. The phrase nucleotide sequence that encodes a
protein or an RNA
may also include introns to the extent that the nucleotide sequence encoding
the protein may in
some version contain an intron(s).
By the term "modulating," as used herein, is meant mediating a detectable
increase or
decrease in the level of a response in a subject compared with the level of a
response in the
subject in the absence of a treatment or compound, and/or compared with the
level of a response
in an otherwise identical but untreated subject. The term encompasses
perturbing and/or affecting
a native signal or response thereby mediating a beneficial therapeutic
response in a subject,
preferably, a human.
Unless otherwise specified, a "nucleotide sequence encoding an amino acid
sequence"
includes all nucleotide sequences that are degenerate versions of each other
and that encode the
same amino acid sequence. Nucleotide sequences that encode proteins and RNA
may include
introns. In addition, the nucleotide sequence may contain modified nucleosides
that are capable of
being translation by translational machinery in a cell. For example, in some
aspects the nucleotide
sequence comprises an mRNA where some or all of the uridines have been
replaced with
pseudouridine, 1-methyl psuedouridine, or another modified nucleoside.
The term "operably linker refers to functional linkage between a regulatory
sequence
and a heterologous nucleic acid sequence resulting in expression of the
latter. For example, a first
nucleic acid sequence is operably linked with a second nucleic acid sequence
when the first
nucleic acid sequence is placed in a functional relationship with the second
nucleic acid sequence.
For instance, a promoter is operably linked to a coding sequence if the
promoter affects the
transcription or expression of the coding sequence. Generally, operably linked
DNA or RNA
8
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
sequences are contiguous and, where necessary to join two protein coding
regions, in the same
reading frame.
The terms "patient," "subject," "individual," and the like are used
interchangeably herein,
and refer to any animal, or cells thereof whether in vitro or in situ,
amenable to the methods
described herein. In certain non-limiting embodiments, the patient, subject or
individual is a
human.
The term -polynucleotide" as used herein is defined as a chain of nucleotides.
Furthermore, nucleic acids are polymers of nucleotides. Thus, nucleic acids
and polynucleotides
as used herein are interchangeable. One skilled in the art has the general
knowledge that nucleic
acids are polynucleotides, which can be hydrolyzed into the monomeric
"nucleotides." The
monomeric nucleotides can be hydrolyzed into nucleosides. As used 'herein
polynucleotides
include, but are not limited to, all nucleic acid sequences which are obtained
by any means
available in the art, including, without limitation, recombinant means, i.e.,
the cloning of nucleic
acid sequences from a recombinant library or a cell genome, using ordinary
cloning technology
and PCRTM, and the like, and by synthetic means.
In certain instances, the polynucleotide or nucleic acid of the invention is a
-nucleoside-
modified nucleic acid," which refers to a nucleic acid comprising at least one
modified
nucleoside. A "modified nucleoside" refers to a nucleoside with a
modification. For example,
over one hundred different nucleoside modifications have been identified in
RNA (Rozenski, et
al., 1999, The RNA Modification Database: 1999 update. Nucl Acids Res 27: 196-
197).
In certain embodiments, "pseudouridine" refers, in another embodiment, to
mlacp3Y (1-
methy1-3-(3-amino-3-carboxypropyl) pseudouridine. In another embodiment, the
term refers to
(1-methylpseudouridine). In another embodiment, the term refers to Ym (2'-0-
methvlpseudouridine. In another embodiment, the term refers to m5D (5-
methyldihydrouridine).
In another embodiment, the term refers to m3Y (3-methylpseudouridine). In
another embodiment,
the term refers to a pseudouridine moiety that is not further modified. In
another embodiment, the
term refers to a monophosphate, diphosphate, or triphosphate of any of the
above pseudouridines.
In another embodiment, the term refers to any other pseudouridine known in the
art. Each
possibility represents a separate embodiment of the present invention.
As used herein, the terms "peptide," "polypeptide," and "protein" are used
interchangeably, and refer to a compound comprised of amino acid residues
covaIently linked by
peptide bonds. A protein or peptide must contain at least two amino acids, and
no limitation is
placed on the maximum number of amino acids that can comprise a protein's or
peptide's
sequence. Polypeptides include any peptide or protein comprising two or more
amino acids joined
9
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
to each other by peptide bonds. As used herein, the term refers to both short
chains, which also
commonly arc referred to in the art as peptides, oligopeptidcs and oligomcrs,
for example, and to
longer chains, which generally are referred to in the art as proteins, of
which there are many
types. "Polypeptides" include, for example, biologically active fragments,
substantially
homologous polypeptides, oligopeptides, homodimers, heterodimers, variants of
polypeptides,
modified polypeptides, derivatives, analogs, fusion proteins, among others.
The polypeptides
include natural peptides, recombinant peptides, synthetic peptides, or a
combination thereof
The term -promoter" as used herein is defined as a DNA sequence recognized by
the
synthetic machinery of the cell, or introduced synthetic machinery, required
to initiate the specific
transcription of a polynucleotide sequence. For example, the promoter that is
recognized by
bacteriophage RNA polymerase and is used to generate the mRNA by in vitro
transcription.
By the term "specifically binds," as used herein with respect to an affinity
ligand, in
particular, an antibody, is meant an antibody which recognizes a specific
antigen, but does not
substantially recognize or bind other molecules in a sample. For example, an
antibody that
specifically binds to an antigen from one species may also bind to that
antigen from one or more
other species. But, such cross-species reactivity does not itself alter the
classification of an
antibody as specific. In another example, an antibody that specifically binds
to an antigen may
also bind to different allelic forms of the antigen. However, such cross
reactivity does not itself
alter the classification of an antibody as specific. In some instances, the
terms "specific binding"
or "specifically binding," can be used in reference to the interaction of an
antibody, a protein, or a
peptide with a second chemical species, to mean that the interaction is
dependent upon the
presence of a particular stmcture (e.g., an antigenic determinant or epitope)
on the chemical
species; for example, an antibody recognizes and binds to a specific protein
structure rather than
to proteins generally. If an antibody is specific for epitope "A-, the
presence of a molecule
containing epitope A (or free, unlabeled A), in a reaction containing labeled
"A" and the
antibody, will reduce the amount of labeled A bound to the antibody.
The term -therapeutic- as used herein means a treatment and/or prophylaxis. A
therapeutic effect is obtained by suppression, diminution, remission, or
eradication of at least one
sign or symptom of a disease or disorder.
The term "therapeutically effective amount" refers to the amount of the
subject
compound that will elicit the biological or medical response of a tissue,
system, or subject that is
being sought by the researcher, veterinarian, medical doctor or other
clinician. The term
"therapeutically effective amount" includes that amount of a compound that,
when administered,
is sufficient to prevent development of, or alleviate to some extent, one or
more of the signs or
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
symptoms of the disorder or disease being treated. The therapeutically
effective amount will vary
depending on the compound, the disease and its severity and the age, weight,
etc., of the subject
to be treated.
To "treat" a disease as the term is used herein, means to reduce the frequency
or severity
of at least one sign or symptom of a disease or disorder experienced by a
subject.
The term -transfected" or "transformed" or "transduced" as used herein refers
to a
process by which exogenous nucleic acid is transferred or introduced into the
host cell. A
-transfected" or -transformed" or -transduced" cell is one which has been
transfected,
transformed or transduced with exogenous nucleic acid. The cell includes the
primary subject cell
and its progeny.
The phrase "under transcriptional control- or "operatively linked" as used
herein means
that the promoter is in the correct location and orientation in relation to a
polynucleotide to
control the initiation of transcription by RNA polymerase and expression of
the polynucleotide.
A "vector' is a composition of matter which comprises an isolated nucleic acid
and
which can be used to deliver the isolated nucleic acid to the interior of a
cell. Numerous vectors
are known in the art including, but not limited to, linear polynucleotides,
polynucleotides
associated with ionic or amphiphilic compounds, plasmids, and viruses. Thus,
the term "vector"
includes an autonomously replicating plasmid or a virus. The term should also
be construed to
include non-plasmid and non-viral compounds which facilitate transfer of
nucleic acid into cells,
such as, for example, polylysine compounds, liposomes, and the like. Examples
of viral vectors
include, but are not limited to, adenoviral vectors, adeno-associated virus
vectors, retroviral
vectors, and the like.
"Alkyl- refers to a straight or branched hydrocarbon chain radical consisting
solely of
carbon and hydrogen atoms, which is saturated or unsaturated (i.e., contains
one or more double
and/or triple bonds), having from one to twenty-four carbon atoms (Ci-C,4
alkyl), one to twelve
carbon atoms (CI-C12 alkyl), one to eight carbon atoms (C1-Cs alkyl) or one to
six carbon atoms
(CI-C6 alkyl) and which is attached to the rest of the molecule by a single
bond, e.g., methyl,
ethyl, n propyl, 1-methylethyl (iso propyl), n butyl, n pentyl, 1,1
dimethylethyl (t butyl), 3
methylhexyl, 2 methylhexyl, ethenyl, prop 1 enyl, but-l-enyl, pent-l-enyl,
penta-1,4-dienyl,
ethynyl, propynyl, butynyl, pentynyl, hexynyl, and the like. Unless
specifically stated otherwise,
an alkyl group is optionally substituted.
"Alkylene" or "alkylene chain" refers to a straight or branched divalent
hydrocarbon
chain linking the rest of the molecule to a radical group, consisting solely
of carbon and
hydrogen, which is saturated or unsaturated (i.e., contains one or more double
(alkenylene) and/or
11
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
triple bonds (alkynylene)), and having, for example, from one to twenty-four
carbon atoms (CI-
C24 alkylcne), one to fifteen carbon atoms (C1-C15 alkylcnc),onc to twelve
carbon atoms (C1-C12
alkylene), one to eight carbon atoms (CI-Cs alkylene), one to six carbon atoms
(CI-C6alkylene),
two to four carbon atoms (C2-C4 alkylene), one to two carbon atoms (C1-C2
alkylene), e.g.,
methylene, ethylene. propylene, n-butylene, ethenylene, propenylene, n-
butenylene, propynylene,
n-butynylene, and the like. The alkylene chain is attached to the rest of the
molecule through a
single or double bond and to the radical group through a single or double
bond. The points of
attachment of the alkylene chain to the rest of the molecule and to the
radical group can be
through one carbon or any two carbons within the chain. Unless stated
otherwise specifically in
the specification, an alkylene chain may be optionally substituted.
"Cycloalkyl" or "carbocyclic ring- refers to a stable non aromatic monocyclic
or
polycyclic hydrocarbon radical consisting solely of carbon and hydrogen atoms,
which may
include fused or bridged ring systems, having from three to fifteen carbon
atoms, preferably
having from three to ten carbon atoms, and which is saturated or unsaturated
and attached to the
rest of the molecule by a single bond. Monocyclic radicals include, for
example, cyclopropyl,
cyclobutyl, cyclopentyl, cyclohexyl, cycloheptyl, and cyclooctyl. Polycyclic
radicals include, for
example, adamantyl, norbomyl, decalinyl, 7,7 dimethyl bicyclo[2.2.1Theptanyl,
and the like.
Unless specifically stated otherwise, a cycloalkyl group is optionally
substituted.
"Cycloalkylene" is a divalent cycloalkyl group. Unless otherwise stated
specifically in
the specification, a cycloalkylene group may be optionally substituted.
"Heterocyclyr or "heterocyclic ring" refers to a stable 3-to 18-membered non-
aromatic
ring radical which consists of two to twelve carbon atoms and from one to six
heteroatoms
selected from the group consisting of nitrogen, oxygen and sulfur. Unless
stated otherwise
specifically in the specification, the heterocyclyl radical may be a
monocyclic, bicyclic, tricyclic
or tetracyclic ring system, which may include fused or bridged ring systems;
and the nitrogen,
carbon or sulfur atoms in the heterocyclyl radical may be optionally oxidized;
the nitrogen atom
may be optionally quatemized; and the heterocyclyl radical may be partially or
fully saturated.
Examples of such heterocyclyl radicals include, but are not limited to,
dioxolanyl,
thienyl[1,3]dithianyl, decahydroisoquinolyl, imidazolinyl, imidazolidinyl,
isothiazolidinyl,
isoxazolidinyl, morpholinyl, octahydroindolyl, octahydroisoindolyl, 2-
oxopiperazinyl,
2-oxopiperidinyl, 2-oxopyrrolidinyl, oxazolidinyl, piperidinyl, piperazinyl, 4-
piperidonyl,
pyrrolidinyl, pyrazolidinyl, quinuclidinyl, thiazolidinyl, tetrahydrofuryl,
trithianyl,
tetrahydropyranyl, thiomorpholinyl, thiamorpholinyl, 1-oxo-thiomorpholinyl,
and
12
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
1,1-dioxo-thiomorpholinyl. Unless specifically stated otherwise, a
heterocyclyl group may be
optionally substituted.
The term -substituted" used herein means any of the above groups (e.g., alkyl,
cycloalkyl
or heterocycly1) wherein at least one hydrogen atom is replaced by a bond to a
non-hydrogen
atoms such as, but not limited to: a halogen atom such as F, Cl, Br, and I;
oxo groups (=0);
hydroxyl groups (-OH); alkoxy groups (-01ta, where IV is Ct-C12 alkyl or
cycloalkyl); carboxyl
groups (-0C(=0)Ra or -C(=0)012a, where Ra is H, CI-Ct2 alkyl or cycloalkyl);
amine groups
(-NRaRb, where Ra and Rb are each independently H. Ci-C 12 alkyl or
cycloalkyl); CI-Ct2alkyl
groups; and cycloalkyl groups. In some embodiments the substituent is a C1-C12
alkyl group. In
other embodiments, the substituent is a cycloalkyl group. In other
embodiments, the substituent is
a halo group, such as fluoro. In other embodiments, the substituent is a oxo
group. In other
embodiments, the substituent is a hydroxyl group. In other embodiments, the
substituent is an
alkoxy group. In other embodiments, the substituent is a carboxyl group. In
other embodiments,
the substituent is an amine group.
"Optional" or "optionally" (e.g., optionally substituted) means that the
subsequently
described event of circumstances may or may not occur, and that the
description includes
instances where said event or circumstance occurs and instances in which it
does not. For
example, -optionally substituted alkyl" means that the alkyl radical may or
may not be substituted
and that the description includes both substituted alkyl radicals and alkyl
radicals having no
substitution.
Ranges: throughout this disclosure, various aspects of the invention can be
presented in a
range format. It should be understood that the description in range format is
merely for
convenience and brevity and should not be construed as an inflexible
limitation on the scope of
the invention. Accordingly, the description of a range should be considered to
have specifically
disclosed all the possible subranges as well as individual numerical values
within that range. For
example, description of a range such as from 1 to 6 should be considered to
have specifically
disclosed subranges such as from 1 to 3, from 1 to 4, from 1 to 5, from 2 to
4, from 2 to 6, from 3
to 6 etc., as well as individual numbers within that range, for example, 1, 2,
2.7, 3, 4, 5, 5.3, and
6. This applies regardless of the breadth of the range.
Descrintion
The present invention relates in part to compositions and methods for targeted
delivery of
a delivery vehicle. In one aspect, the present invention relates to
composition comprising a
delivery vehicle conjugated to a targeting domain. In one embodiment, the
delivery vehicle
13
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
comprises at least one agent. In one embodiment, the delivery vehicle
comprises an RNA
molecule, including but not limited to mRNA, nucleoside-modified RNA, siRNA,
miRNA,
shRNA, or antisense RNA. In one embodiment, the delivery vehicle comprises a
therapeutic
agent. In one embodiment, the therapeutic agent is a nucleoside-modified RNA.
In various embodiments, the targeting domain binds to a cell surface molecule
of a T cell
antigen. In one embodiment, the T cell antigen is a surface antigen of a CD4+
T cell. In one
embodiment, the T cell surface antigen is CD4.
In one embodiment, the composition comprises a delivery vehicle conjugated to
a
targeting domain that binds CD4 or a surface antigen of a CD4+ T cell, thereby
directing the
composition to CD4+ T cells.
In one embodiment, the delivery vehicle comprises or encapsulates an agent for
modulation of CD4+ T cells. In some embodiments the agent is nucleic acid
molecule. In some
embodiments the nucleic acid molecule is a nucleoside modified mRNA.
In one embodiment, the delivery vehicle comprises or encapsulates a
therapeutic agent
for modulation of CD4+ T cells. In some embodiments the therapeutic agent is a
nucleoside-
modified mRNA. In some embodiments the therapeutic agent is an mRNA-based
immunotherapeutic.
The present invention also relates in part to methods of treating diseases or
disorders in
subjects in need thereof, the method comprising the administration of a
composition including a
delivery vehicle comprising an agent conjugated to a targeting domain that
binds CD4 or a
surface antigen of a CD4+ T cell.
Exemplary diseases and disorders that can be treated using the CD4 T cell
targeted
therapeutic compositions of the invention include, but are not limited to,
cancers, infectious
diseases, and immunological disorders.
As a non-limiting example, in one embodiment, the CD4+ T cell-targeted
delivery
vehicle of the invention comprises or encapsulates a nucleoside-modified 1086C
Env mRNA,
encoding the clade C transmitted/founder human immunodeficiency virus (HIV)-1
envelope
(Env) 1086C, for the treatment or prevention of HIV infection or a disease or
disorder associated
therewith.
Delivery vehicle cargo
In various embodiments, the delivery vehicle comprises a cargo of one or more
nucleic
acid molecules (e.g., mRNA, expression vector, or genome editing vector) which
genetically
modify the immune cell. After cellular uptake of the delivery vehicle by the
target immune cell
14
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
(e.g., by endocytosis), the cargo nucleic acid becomes released from the
endosome. Once
released, the cargo nucleic acid modifies the target immune cell of the
subject to express one or
more surface moieties (e.g., a cell receptor).
In one embodiment, the delivery vehicle comprises at least one agent. In some
embodiments, the agent is a therapeutic agent, an imaging agent, diagnostic
agent, a contrast
agent, a labeling agent, a detection agent, or a disinfectant. The agent may
also include substances
with biological activities which are not typically considered to be active
ingredients, such as
fragrances, sweeteners, flavorings and flavor enhancer agents, pH adjusting
agents, effervescent
agents, emollients, bulking agents, soluble organic salts, permeabilizing
agents, anti-oxidants,
colorants or coloring agents, and the like.
In one embodiment, the delivery vehicle comprises at least one therapeutic
agent. The
present invention is not limited to any particular therapeutic agent, but
rather encompasses any
suitable therapeutic agent that can be included within the delivery vehicle.
Exemplary therapeutic
agents include, but are not limited to, anti-viral agents, anti-bacterial
agents, anti-oxidant agents,
thrombolytic agents, chemotherapeutic agents, anti-inflammatory agents,
immunogenic agents,
antiseptics, anesthetics, analgesics, pharmaceutical agents, small molecules,
peptides, nucleic
acids, and the like. In one embodiment, the agent is an mRNA molecule (e.g., a
nucleoside
modified mRNA molecule) as described elsewhere herein.
Nucleic acid agents
In one aspect, the present disclosure provides delivery vehicles comprising a
nucleic acid
cargo (e.g., DNA or RNA), including, but not limited to, an mRNA, expression
vector, genome
editing vector, siRNA, shRNA, an miRNA for use in inhibiting, inactivating,
and/or destroying
activated fibroblasts. In various embodiments, the nucleic acid cargo may be a
nucleoside
modified nucleic acid molecule (e.g., a nucleoside modified mRNA molecule). In
various
embodiments, the agent is an isolated nucleic acid. In some embodiments, the
isolated nucleic
acid molecule is a cDNA, mRNA, siRNA, shRNA or miRNA molecule. In some
embodiments,
the isolated nucleic acid molecule is a nucleoside modified RNA molecule. In
some
embodiments, the nucleoside modified RNA molecule is an siRNA, miRNA, shRNA,
or an
antisense molecule.
In various embodiments, that delivery vehicles comprise a cargo of one or more
nucleic
acid molecules (e.g., mRNA, expression vector, or genome editing vector, DNA,
or RNA) which
genetically modify the immune cell. After cellular uptake of the delivery
vehicle by the target
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
immune cell (e.g., by endocytosis), the cargo nucleic acid becomes released
from the endosome.
Once released, the cargo nucleic acid modifies the target immune cell of the
subject.
In one embodiment, the nucleic acid comprises a promoter/regulatory sequence
such that
the nucleic acid is capable of directing expression of the nucleic acid. Thus,
the invention
encompasses expression vectors and methods for the introduction of exogenous
nucleic acid into
cells with concomitant expression of the exogenous nucleic acid in the cells
such as those
described, for example, in Sambrook et al. (2012, Molecular Cloning: A
Laboratory Manual,
Cold Spring Harbor Laboratory, New York), and in Ausubel et al. (1997, Current
Protocols in
Molecular Biology, John Wiley & Sons, New York) and as described elsewhere
herein.
Nucleoside-modified RNA affenis
In one embodiment, the composition of the present invention comprises a
nucleoside-
modified nucleic acid (e.g., a nucleoside-modified mRNA molecule). In one
embodiment, the
composition of the invention comprises a nucleoside-modified RNA encoding a
protein, such as a
therapeutic protein.
For example, in one embodiment, the composition comprises a nucleoside-
modified
RNA. In one embodiment, the composition comprises a nucleoside-modified mRNA.
Nucleoside-
modified mRNA have particular advantages over non-modified mRNA, including for
example,
increased stability, low or absent innate immunogenicity, and enhanced
translation. Nucleoside-
modified mRNA useful in the present invention is further described in U.S.
Patent Nos.
8,278,036,8,691,966, and 8,835,108, each of which is incorporated by reference
herein in its
entirety.
In certain embodiments, nucleoside-modified mRNA does not activate any
pathophysiologic pathways, translates very efficiently and almost immediately
following
delivery, and serve as templates for continuous protein production in vivo
lasting for several days
(Kariko et al., 2008, Mol Ther 16:1833-1840; Kariko et al., 2012, Mol Ther
20:948-953). The
amount of mRNA required to exert a physiological effect is small and that
makes it applicable for
human therapy.
In certain instances, expressing a protein by delivering the encoding mRNA has
many
benefits over methods that use protein, plasmid DNA or viral vectors. During
mRNA
transfection, the coding sequence of the desired protein is the only substance
delivered to cells,
thus avoiding all the side effects associated with plasmid backbones, viral
genes, and viral
proteins. More importantly, unlike DNA- and viral-based vectors, the mRNA does
not carry the
risk of being incorporated into the genome and protein production starts
immediately after mRNA
16
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
delivery. For example, high levels of circulating proteins have been measured
within 15 to 30
minutes of in vivo injection of the encoding mRNA. In certain embodiments,
using mRNA rather
than the protein also has many advantages. Half-lives of proteins in the
circulation are often short,
thus protein treatment would need frequent dosing, while mRNA provides a
template for
continuous protein production for several days. Purification of proteins is
problematic and they
can contain aggregates and other impurities that cause adverse effects
(Kromminga and
Schellekens, 2005, Ann NY Acad Sci 1050:257-265).
In certain embodiments, the nucleoside-modified RNA comprises the naturally
occurring
modified-nucleoside pseudouridine. In certain embodiments, inclusion of
pseudouridine makes
the mRNA more stable, non-immunogenic, and highly translatable (Kariko et al.,
2008, Mol Ther
16:1833-1840; Anderson et al., 2010, Nucleic Acids Res 38:5884-5892; Anderson
et al., 2011,
Nucleic Acids Research 39:9329-9338; Kariko et al., 2011, Nucleic Acids
Research 39:e142;
Kariko et al., 2012, Mol Ther 20:948-953; Kariko et al., 2005, Immunity 23:165-
175).
It has been demonstrated that the presence of modified nucleosides, including
pseudouridines in RNA suppress their innate immunogenicity (Kariko et al.,
2005, Immunity
23:165-175). Further, protein-encoding, in vitro-transcribed RNA containing
pseudouridine can
be translated more efficiently than RNA containing no or other modified
nucleosides (Kariko et
al., 2008, Mol Ther 16:1833-1840). Subsequently, it is shown that the presence
of pseudouridine
improves the stability of RNA (Anderson et al., 2011, Nucleic Acids Research
39:9329-9338) and
abates both activation of PKR and inhibition of translation (Anderson et al.,
2010, Nucleic Acids
Res 38:5884-5892).
Similar effects as described for pseudouridine have also been observed for RNA
containing 1-methyl-pseudouridine.
In some embodiments, the nucleoside-modified nucleic acid molecule is a
purified
nucleoside-modified nucleic acid molecule. For example, in some embodiments,
the composition
is purified to remove double-stranded contaminants. In some instances, a
preparative HPLC
purification procedure is used to obtain pseudouridine-containing RNA that has
superior
translational potential and no innate immunogenicity (Kariko et al., 2011,
Nucleic Acids
Research 39:e142). Administering HPLC-purified, pseudourine-containing RNA
coding for
erythropoietin into mice and macaques resulted in a significant increase of
serum EPO levels
(Kariko et al., 2012, Mol Ther 20:948-953), thus confirming that pseudouridine-
containing
mRNA is suitable for in vivo protein therapy. In some embodiments, the
nucleoside-modified
nucleic acid molecule is purified using non-HPLC methods. In some instances,
the nucleoside-
modified nucleic acid molecule is purified using chromatography methods,
including but not
17
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
limited to HPLC and fast protein liquid chromatography (FPLC). An exemplary
FPLC-based
purification procedure is described in Wcissman et al., 2013, Methods Mol
Biol, 969: 43-54.
Exemplary purification procedures are also described in U.S. Patent
Application Publication No.
US2016/0032316, which is hereby incorporated by reference in its entirety.
The present invention encompasses RNA, oligoribonucleotide, and
polyribonucleotide
molecules comprising pseudouridine or a modified nucleoside. In certain
embodiments, the
composition comprises an isolated nucleic acid, wherein the nucleic acid
comprises a
pseudouridine or a modified nucleoside. In certain embodiments, the
composition comprises a
vector, comprising an isolated nucleic acid, wherein the nucleic acid
comprises a pseudouridine
or a modified nucleoside.
In one embodiment, the nucleoside-modified RNA of the invention is IVT RNA, as
described elsewhere herein. For example, in certain embodiments, the
nucleoside-modified RNA
is synthesized by T7 phage RNA polymerase. In another embodiment, the
nucleoside-modified
mRNA is synthesized by SP6 phage RNA polymerase. In another embodiment, the
nucleoside-
modified RNA is synthesized by T3 phage RNA polymerase.
In one embodiment, the modified nucleoside is mlacp3IP (1-methy1-3-(3-amino-3-
carboxypropyl) pseudouridine. In another embodiment, the modified nucleoside
is in' tlf (1-
methylpseudouridine). In another embodiment, the modified nucleoside is Win
(2'-0-
methylpseudouridine. In another embodiment, the modified nucleoside is m5D (5-
methyldihydrouridine). In another embodiment, the modified nucleoside is m3tP
(3-
methylpseudouridine). In another embodiment, the modified nucleoside is a
pseudouridine moiety
that is not further modified. In another embodiment, the modified nucleoside
is a monophosphate,
diphosphate, or triphosphate of any of the above pseudouridines. In another
embodiment, the
modified nucleoside is any other pseudouridine-like nucleoside known in the
art.
In another embodiment, the nucleoside that is modified in the nucleoside-
modified RNA
the present invention is uridine (U). In another embodiment, the modified
nucleoside is cytidine
(C). In another embodiment, the modified nucleoside is adenosine (A). In
another embodiment,
the modified nucleoside is guanosine (G).
In another embodiment, the modified nucleoside of the present invention is m5C
(5-
methylcytidine). In another embodiment, the modified nucleoside is m5U (5-me
thyluridine). In
another embodiment, the modified nucleoside is m6A (N6-methyladenosine). In
another
embodiment, the modified nucleoside is s2U (2-thiouridine). In another
embodiment, the modified
nucleoside is LI' (pseudouridine). In another embodiment, the modified
nucleoside is Um (2'-0-
methyluridine).
18
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In other embodiments, the modified nucleoside is m'A (1-methyladenosine); in2A
(2-
methyladenosine); Am (2'-0-methyladenosinc); ms2m6A (2-methylthio-N6-
methyladenosine); i6A
(N6-isopentenyladenosine); ms2i6A (2-methylthio-N6isopentenyladenosine); io6A
(N6-(cis-
hydroxyisopentenyl)adenosine); ms2io6A (2-methylthio-N6-(cis-
hydroxyisopentenyl) adenosine);
g6A (N6-glycinylcarbamoyladenosine); t6A (N6-threonylearbamoyladenosine);
ms2t6A (2-
methylthio-N6-threonyl carbamoyladenosine); m6t6A (N6-methyl-N6-
threonylcarbamoyladenosine); hn6A(N6-hydroxynorvalylcarbamoyladenosine);
ms2hn6A (2-
methylthio-N6-hydroxynorvaly1 carbamoyladenosine); Ar(p) (2'-0-
ribosyladenosine (phosphate));
I (inosine); mII (1-methylinosine); m'Im (1,2'-0-dimethylinosine); m3C (3-
methylcytidine); Cm
(2'-0-methylcytidine); s2C (2-thiocytidine); ac4C (N4-acetylcytidine); f5C (5-
formylcytidine);
m5Cm (5,2'-0-dimethylcytidine); ac4Cm (N4-acetyl-2'-0-methylcytidine); k2C
(lysidine); miG (1-
methylguanosine); m2G (N2-methylguanosine); m7G (7-methylguanosine); Gin (2'-0-
methylguanosine); m22G (N2,N2-dimethylguanosine); m2Gm (N2,2'-0-
dimethylguanosine);
m22Gm (N2,N2,2'-0-trimethylguanosine); Gr(p) (2'-0-ribosylguanosine
(phosphate)); yW
(wybutosine); o2yW (peroxywybutosine); OHyW (hydroxywybutosine); OHyW*
(undermodified
hydroxywybutosine); imG (wyosine); mimG (methylwyosine); Q (queuosine); oQ
(epoxyquettosine); galQ (galactosyl-queuosine); manQ (mannosyl-queuosine);
preQ0 (7-cyano-7-
deazaguanosine); preQi (7-aminomethy1-7-deazaguanosine); Cr+ (archaeosine); D
(dihydrouridine); m5Um (5,2'-0-dimethyluridine); s4U (4-thiouridine); m5s2U (5-
methyl-2-
thiouridine); s2Um (2-thio-2'-0-methyluridine); acp3U (3-(3-amino-3-
carboxypropyl)uridine);
ho5U (5-hydroxyuridine); mo5U (5-methoxyuridine); cmo51J (uridine 5-oxyacetic
acid); mcmo5U
(uridine 5-oxyacetic acid methyl ester); chm5U (5-
(carboxyhydroxymethyl)uridine)); mchm5U (5-
(carboxyhydroxymethyl)uridine methyl ester); mcm5U (5-
methoxycarbonylmethyluridine);
mcm5Um (5-methoxycarbonylmethy1-2'-0-methyluridine); mcm5s2U (5-
methoxycarbonylmethyl-
2-thiouridine); nm5s2U (5-aminomethy1-2-thiouridine); mnm5U (5-
methylaminomethyluridine);
mnm5S2U (5-methylaminomethy1-2-thiouridine); mnm5se2U (5-methylaminomethy1-2-
selenouridine); ncm5U (5-carbamoylmethyluridine); ncm5Um (5-carbamoylmethy1-2'-
0-
methyluridine); cmnm5U (5-carboxymethylaminomethyluridine); cmnm5Um (5-
carboxymethylaminomethy1-2'-0-methyluridine); emnm5s2U (5-
carboxymethylaminomethyl-2-
m62A (N6,N6-dimethyladenosine); Im (2'-0-methylinosine); m4C (N4-
methylcytidine); m4Cm (N4,2'-0-dimethylcytidine); hm5C (5-
hydroxymethylcytidine); m5-15 (3-
methyluridine); cm5U (5-carboxymethyluridine); m6Am (N6,2'-0-
dimethyladenosine); m62Am
(N6,N6,0-2'-trimethyladenosine); m2'7G (N2,7-dimethylguanosine); m2,2,7G
(N2,N2,7_
trimethylguanosine); m3Um (3,2'-0-dimethyluridine); m5D (5-
methyldihydrouridine); f5Cm
19
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
(5-formy1-2'-0-methylcytidine); m'Gm (1,21-0-dimethylguanosine); m 'Am (1,2'4)-
dimethyladenosinc); fin5U (5-taurinomethyluridinc), rm5s2U (5-taurinomethy1-2-
thiouridinc));
imG-14 (4-demethylwyosine); imG2 (isowyosine); or ac6A (N6-acetyladenosine).
In another embodiment, a nucleoside-modified RNA of the present invention
comprises a
combination of 2 or more of the above modifications. In another embodiment,
the nucleoside-
modified RNA comprises a combination of 3 or more of the above modifications.
In another
embodiment, the nucleoside-modified RNA comprises a combination of more than 3
of the above
modifications.
In another embodiment, between 0.1% and 100% of the residues in the nucleoside-
modified of the present invention are modified (e.g. either by the presence of
pseudouridine or a
modified nucleoside base). In another embodiment, 0.1% of the residues are
modified. In another
embodiment, the fraction of modified residues is 0.2%. In another embodiment,
the fraction is
0.3%. In another embodiment, the fraction is 0.4%. In another embodiment, the
fraction is 0.5%.
In another embodiment, the fraction is 0.6%. In another embodiment, the
fraction is 0.8%. In
another embodiment, the fraction is 1%. In another embodiment, the fraction is
1.5%. In another
embodiment, the fraction is 2%. In another embodiment, the fraction is 2.5%.
In another
embodiment, the fraction is 3%. In another embodiment, the fraction is 4%. In
another
embodiment, the fraction is 5%. In another embodiment, the fraction is 6%. In
another
embodiment, the fraction is 8%. In another embodiment, the fraction is 10%. In
another
embodiment, the fraction is 12%. In another embodiment, the fraction is 14%.
In another
embodiment, the fraction is 16%. In another embodiment, the fraction is 18%.
In another
embodiment, the fraction is 20%. In another embodiment, the fraction is 25%.
In another
embodiment, the fraction is 30%. In another embodiment, the fraction is 35%.
In another
embodiment, the fraction is 40%. In another embodiment, the fraction is 45%.
In another
embodiment, the fraction is 50%. In another embodiment, the fraction is 60%.
In another
embodiment, the fraction is 70%. In another embodiment, the fraction is 80%.
In another
embodiment, the fraction is 90%. In another embodiment, the fraction is 100%.
In another embodiment, the fraction is less than 5%. In another embodiment,
the fraction
is less than 3%. In another embodiment, the fraction is less than 1%. In
another embodiment, the
fraction is less than 2%. In another embodiment, the fraction is less than 4%.
In another
embodiment, the fraction is less than 6%. In another embodiment, the fraction
is less than 8%. In
another embodiment, the fraction is less than 10%. In another embodiment, the
fraction is less
than 12%. In another embodiment, the fraction is less than 15%. In another
embodiment, the
fraction is less than 20%. In another embodiment, the fraction is less than
30%. In another
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
embodiment, the fraction is less than 40%. In another embodiment, the fraction
is less than 50%.
In another embodiment, the fraction is less than 60%. In another embodiment,
the fraction is less
than 70%.
In another embodiment, 0.1% of the residues of a given nucleoside (i.e.,
uridine, cytidine,
guanosine, or adenosine) are modified. In another embodiment, the fraction of
the given
nucleotide that is modified is 0.2%. In another embodiment, the fraction is
0.3%. In another
embodiment, the fraction is 0.4%. In another embodiment, the fraction is 0.5%.
In another
embodiment, the fraction is 0.6%. In another embodiment, the fraction is 0.8%.
In another
embodiment, the fraction is 1%. In another embodiment, the fraction is 1.5%.
In another
embodiment, the fraction is 2%. In another embodiment, the fraction is 2.5%.
In another
embodiment, the fraction is 3%. In another embodiment, the fraction is 4%. In
another
embodiment, the fraction is 5%. In another embodiment, the fraction is 6%. In
another
embodiment, the fraction is 8%. In another embodiment, the fraction is 10%. In
another
embodiment, the fraction is 12%. In another embodiment, the fraction is 14%.
In another
embodiment, the fraction is 16%. In another embodiment, the fraction is 18%.
In another
embodiment, the fraction is 20%. In another embodiment, the fraction is 25%.
In another
embodiment, the fraction is 30%. In another embodiment, the fraction is 35%.
In another
embodiment, the fraction is 40%. In another embodiment, the fraction is 45%.
In another
embodiment, the fraction is 50%. In another embodiment, the fraction is 60%.
In another
embodiment, the fraction is 70%. In another embodiment, the fraction is 80%.
In another
embodiment, the fraction is 90%. In another embodiment, the fraction is 100%.
In another embodiment, the fraction of the given nucleotide that is modified
is less than
8%. In another embodiment, the fraction is less than 10%. In another
embodiment, the fraction is
less than 5%. In another embodiment, the fraction is less than 3%. In another
embodiment, the
fraction is less than 1%. In another embodiment, the fraction is less than 2%.
In another
embodiment, the fraction is less than 4%. In another embodiment, the fraction
is less than 6%. In
another embodiment, the fraction is less than 12%. In another embodiment, the
fraction is less
than 15%. In another embodiment, the fraction is less than 20%. In another
embodiment, the
fraction is less than 30%. In another embodiment, the fraction is less than
40%. In another
embodiment, the fraction is less than 50%. In another embodiment, the fraction
is less than 60%.
In another embodiment, the fraction is less than 70%.
In some embodiments, the composition comprises a purified preparation of
single-
stranded nucleoside modified RNA. For example, in some embodiments, the
purified preparation
of single-stranded nucleoside modified RNA is substantially free of double
stranded RNA
21
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
(dsRNA). In some embodiments, the purified preparation is at least 90%, or at
least 91%, or at
least 92%, or at least 93 % or at least 94%, or at least 95%, or at least 96%,
or at least 97%, or at
least 98%, or at least 99%, or at least 99.5%, or at least 99.9% single
stranded nucleoside
modified RNA, relative to all other nucleic acid molecules (DNA, dsRNA, etc.).
In another embodiment, a nucleoside-modified RNA of the present invention is
translated
in the cell more efficiently than an unmodified RNA molecule with the same
sequence. In another
embodiment, the nucleoside-modified RNA exhibits enhanced ability to be
translated by a target
cell. In another embodiment, translation is enhanced by a factor of 2-fold
relative to its
unmodified counterpart. In another embodiment, translation is enhanced by a 3-
fold factor. In
another embodiment, translation is enhanced by a 5-fold factor. In another
embodiment,
translation is enhanced by a 7-fold factor. In another embodiment, translation
is enhanced by a
10-fold factor. In another embodiment, translation is enhanced by a 15-fold
factor. In another
embodiment, translation is enhanced by a 20-fold factor. In another
embodiment, translation is
enhanced by a 50-fold factor. In another embodiment, translation is enhanced
by a 100-fold
factor. In another embodiment, translation is enhanced by a 200-fold factor.
In another
embodiment, translation is enhanced by a 500-fold factor. In another
embodiment, translation is
enhanced by a 1000-fold factor. In another embodiment, translation is enhanced
by a 2000-fold
factor. In another embodiment, the factor is 10-1000-fold. In another
embodiment, the factor is
10-100-fold. In another embodiment, the factor is 10-200-fold. In another
embodiment, the factor
is 10-300-fold. In another embodiment, the factor is 10-500-fold. In another
embodiment, the
factor is 20-1000-fold. In another embodiment, the factor is 30-1000-fold. In
another
embodiment, the factor is 50-1000-fold. In another embodiment, the factor is
100-1000-fold. In
another embodiment, the factor is 200-1000-fold. In another embodiment,
translation is enhanced
by any other significant amount or range of amounts.
In another embodiment, the nucleoside-modified RNA of the present invention
exhibits
significantly less innate immunogenicity than an unmodified in vitro-
synthesized RNA molecule
of the same sequence. In another embodiment, the modified RNA molecule
exhibits an innate
immune response that is 2-fold less than its unmodified counterpart. In
another embodiment,
innate immunogenicity is reduced by a 3-fold factor. In another embodiment,
innate
immunogenicity is reduced by a 4-fold factor. In another embodiment, innate
immunogenicity is
reduced by a 5-fold factor. In another embodiment, innate immunogenicity is
reduced by a 6-fold
factor. In another embodiment, innate immunogenicity is reduced by a 7-fold
factor. In another
embodiment, innate immunogenicity is reduced by a 8-fold factor. In another
embodiment, innate
immunogenicity is reduced by a 9-fold factor. In another embodiment, innate
immunogenicity is
22
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
reduced by a 10-fold factor. In another embodiment, innate immunogenicity is
reduced by a 15-
fold factor. In another embodiment, innate immunogenicity is reduced by a 20-
fold factor. In
another embodiment, innate immunogenicity is reduced by a 50-fold factor. In
another
embodiment, innate immunogeni city is reduced by a 100-fold factor. In another
embodiment,
innate immunogenicity is reduced by a 200-fold factor. In another embodiment,
innate
immunogenicity is reduced by a 500-fold factor. In another embodiment, innate
immunogenicity
is reduced by a 1000-fold factor. In another embodiment, innate immunogenicity
is reduced by a
2000-fold factor. In another embodiment, innate immunogenicity is reduced by
another fold
difference.
In another embodiment, -exhibits significantly less innate immunogenicity"
refers to a
detectable decrease in innate immunogenicity. In another embodiment, the term
refers to a fold
decrease in innate immunogenicity (e.g., 1 of the fold decreases enumerated
above). In another
embodiment, the term refers to a decrease such that an effective amount of the
nucleoside-
modified RNA can be administered without triggering a detectable innate immune
response. In
another embodiment, the term refers to a decrease such that the nucleoside-
modified RNA can be
repeatedly administered without eliciting an innate immune response sufficient
to detectably
reduce production of the protein encoded by the modified RNA. In another
embodiment, the
decrease is such that the nucleoside-modified RNA can be repeatedly
administered without
eliciting an innate immune response sufficient to eliminate detectable
production of the protein
encoded by the modified RNA.
RNA interference agents
In one embodiment, siRNA is used to decrease the level of a targeted protein.
RNA
interference (RNAi) is a phenomenon in which the introduction of double-
stranded RNA
(dsRNA) into a diverse range of organisms and cell types causes degradation of
the
complementary mRNA. In the cell, long dsRNAs are cleaved into short 21-25
nucleotide small
interfering RNAs, or siRNAs, by a ribonuclease known as Dicer. The siRNAs
subsequently
assemble with protein components into an RNA-induced silencing complex (RISC),
unwinding in
the process. Activated RISC then binds to complementary transcript by base
pairing interactions
between the siRNA antisense strand and the mRNA. The bound mRNA is cleaved and
sequence
specific degradation of mRNA results in gene silencing. See, for example, U.S.
Patent No.
6,506,559; Fire et al., 1998, Nature 391(19):306-311; Timmons et al., 1998,
Nature 395:854;
Montgomery et al., 1998, T1G 14 (7):255-258; David R. Engelke, Ed., RNA
Interference (RNAi)
Nuts & Bolts of RNAi Technology, DNA Press, Eagleville, PA (2003); and Gregory
J. Hannon,
23
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Ed., RNAi A Guide to Gene Silencing, Cold Spring Harbor Laboratory Press, Cold
Spring
Harbor, NY (2003). Soutschck et al. (2004. Naturc 432:173-178) describe a
chemical
modification to siRNAs that aids in intravenous systemic delivery. Optimizing
siRNAs involves
consideration of overall G/C content, C/T content at the termini, Tin and the
nucleotide content of
the 3' overhang. See, for instance, Schwartz et al., 2003, Cell, 115:199-208
and Khvorova et al.,
2003, Cell 115:209-216. Therefore, the present invention also includes methods
of decreasing
levels of PTPN22 using RNAi technology.
In one aspect, the invention includes a vector comprising an siRNA or an
antisense
polynucleotide. Preferably, the siRNA or antisense polynucleotide is capable
of inhibiting the
expression of a target polypeptide. The incorporation of a desired
polynucleotide into a vector
and the choice of vectors are well-known in the art as described in, for
example, Sambrook et al.
(2012), and in Ausubel et al. (1997), and elsewhere herein.
In certain embodiments, the expression vectors described herein encode a short
hairpin
RNA (shRNA) agents. shRNA molecules are well known in the art and are directed
against the
mRNA of a target, thereby decreasing the expression of the target. In certain
embodiments, the
encoded shRNA is expressed by a cell, and is then processed into siRNA. For
example, in certain
instances, the cell possesses native enzymes (e.g., dicer) that cleave the
shRNA to form siRNA.
In order to assess the expression of the siRNA, shRNA, or antisense
polynucleotide, the
expression vector to be introduced into a cell can also contain either a
selectable marker gene or a
reporter gene or both to facilitate identification of expressing cells from
the population of cells
sought to be transfected or infected using the delivery vehicle of the
invention. In other
embodiments, the selectable marker may be carried on a separate piece of DNA
and also be
contained within the delivery vehicle. Both selectable markers and reporter
genes may be flanked
with appropriate regulatory sequences to enable expression in the host cells.
Useful selectable
markers are known in the art and include, for example, antibiotic-resistance
genes, such as
neomycin resistance and the like.
Therefore, in one aspect, the delivery vehicle may contain a vector,
comprising the
nucleotide sequence or the construct to be delivered. The choice of the vector
will depend on the
host cell in which it is to be subsequently introduced. In a particular
embodiment, the vector of
the invention is an expression vector. Suitable host cells include a wide
variety of prokaryotic and
eukaryotic host cells. In specific embodiments, the expression vector is
selected from the group
consisting of a viral vector, a bacterial vector and a mammalian cell vector.
Prokaryote- and/or
eukaryote-vector based systems can be employed for use with the present
invention to produce
24
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
polynucleotides, or their cognate polypeptides. Many such systems are
commercially and widely
available.
By way of illustration, the vector in which the nucleic acid sequence is
introduced can be
a plasrnid, which is or is not integrated in the genome of a host cell when it
is introduced in the
cell. Illustrative, non-limiting examples of vectors in which the nucleotide
sequence of the
invention or the gene construct of the invention can be inserted include a tet-
on inducible vector
for expression in eukaryote cells.
The vector may be obtained by conventional methods known by persons skilled in
the art
(Sambrook et al., 2012). In a particular embodiment, the vector is a vector
useful for transforming
animal cells.
In one embodiment, the recombinant expression vectors may also contain nucleic
acid
molecules, which encode a peptide or peptidomimetic.
A promoter may be one naturally associated with a gene or polynucleotide
sequence, as
may be obtained by isolating the 5' non-coding sequences located upstream of
the coding
segment and/or exon. Such a promoter can be referred to as "endogenous."
Similarly, an
enhancer may be one naturally associated with a polynucleotide sequence,
located either
downstream or upstream of that sequence. Alternatively, certain advantages
will be gained by
positioning the coding polynucleotide segment under the control of a
recombinant or
heterologous promoter, which refers to a promoter that is not normally
associated with a
polynucleotide sequence in its natural environment. A recombinant or
heterologous enhancer
refers also to an enhancer not normally associated with a polynucleotide
sequence in its natural
environment. Such promoters or enhancers may include promoters or enhancers of
other genes,
and promoters or enhancers isolated from any other prokaryotic, viral, or
eukaryotic cell, and
promoters or enhancers not "naturally occurring," i.e., containing different
elements of different
transcriptional regulatory regions, and/or mutations that alter expression. In
addition to producing
nucleic acid sequences of promoters and enhancers synthetically, sequences may
be produced
using recombinant cloning and/or nucleic acid amplification technology,
including PCRTM, in
connection with the compositions disclosed herein (U.S. Patent 4,683,202, U.S.
Patent
5,928,906). Furthermore, it is contemplated the control sequences that direct
transcription and/or
expression of sequences within non-nuclear organelles such as mitochondria,
chloroplasts, and
the like, can be employed as well.
Naturally, it will be important to employ a promoter and/or enhancer that
effectively
directs the expression of the DNA segment in the cell type, organelle, and
organism chosen for
expression. Those of skill in the art of molecular biology generally know how
to use promoters,
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
enhancers, and cell type combinations for protein expression, for example, see
Sambrook et al.
(2012). The promoters employed may be constitutive, tissue-specific,
inducible, and/or useful
under the appropriate conditions to direct high level expression of the
introduced DNA segment,
such as is advantageous in the large-scale production of recombinant proteins
and/or peptides.
The promoter may be heterologous or endogenous.
The recombinant expression vectors may also contain a selectable marker gene,
which
facilitates the selection of host cells. Suitable selectable marker genes are
genes encoding proteins
such as G418 and hygromycin, which confer resistance to certain drugs, fl-
galactosidase,
chloramphenicol acetyltransferase, firefly luciferase, or an immunoglobulin or
portion thereof
such as the Fe portion of an immunoglobulin preferably IgG. The selectable
markers may be
introduced on a separate vector from the nucleic acid of interest.
Following the generation of the siRNA polynucleotide, a skilled artisan will
understand
that the siRNA polynucleotide will have certain characteristics that can be
modified to improve
the siRNA as a therapeutic compound. Therefore, the siRNA polynucleotide may
be further
designed to resist degradation by modifying it to include phosphorothioate, or
other linkages,
methylphosphonate, sulfone, sulfate, ketyl, phosphorodithioate,
phosphoramidate, phosphate
esters, and the like (see, e.g., Agrawal et al., 1987, Tetrahedron Lett.
28:3539-3542; Stec et al.,
1985 Tetrahedron Lett. 26:2191-2194; Moody et al., 1989 Nucleic Acids Res.
12:4769-4782;
Eckstein, 1989 Trends Biol. Sci. 14:97-100; Stein, In: Oligodeoxynucleotides.
Antisense
Inhibitors of Gene Expression, Cohen, ed., Macmillan Press, London, pp. 97-117
(1989)).
Any polynucleotide may be further modified to increase its stability in vivo.
Possible
modifications include, but are not limited to, the addition of flanking
sequences at the 5 and/or 3'
ends; the use of phosphorothioate or 2' 0-methyl rather than phosphodiester
linkages in the
backbone; and/or the inclusion of nontraditional bases such as inosine,
queuosine, and
wybutosine and the like, as well as acetyl- methyl-, thio- and other modified
forms of adenine,
cytidine, guanine, thymine, and uridine.
In one embodiment of the invention, an antisense nucleic acid sequence, which
is
expressed by a plasmid vector is used as an agent to inhibit the expression of
a target protein. The
antisense expressing vector is used to transfect a mammalian cell or the
mammal itself, thereby
causing reduced endogenous expression of the target protein.
Antisense molecules and their use for inhibiting gene expression are well
known in the
art (see, e.g., Cohen, 1989, In: Oligodeoxyribonucleotides, Antisense
Inhibitors of Gene
Expression, CRC Press). Antisense nucleic acids are DNA or RNA molecules that
are
complementary, as that term is defined elsewhere herein, to at least a portion
of a specific mRNA
26
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
molecule (Weintraub, 1990, Scientific American 262:40). In the cell, antisense
nucleic acids
hybridize to the corresponding mRNA, forming a double-stranded molecule
thereby inhibiting the
translation of genes.
The use of antisense methods to inhibit the translation of genes is known in
the art, and is
described, for example, in Marcus-Sakura (1988, Anal. Biochem. 172:289). Such
antisense
molecules may be provided to the cell via genetic expression using DNA
encoding the antisense
molecule as taught by Inoue, 1993, U.S. Patent No. 5,190,931.
Alternatively, antisense molecules of the invention may be made synthetically
and then
provided to the cell. Antisense oligomers of between about 10 to about 30, and
more preferably
about 15 nucleotides, are preferred, since they are easily synthesized and
introduced into a target
cell. Synthetic antisense molecules contemplated by the invention include
oligonucleotide
derivatives known in the art which have improved biological activity compared
to unmodified
oligonucleotides (see U.S. Patent No. 5,023,243).
In one embodiment of the invention, a ribozyme is used as an agent to inhibit
expression
of a target protein. Ribozymes useful for inhibiting the expression of a
target molecule may be
designed by incorporating target sequences into the basic ribozyme structure,
which are
complementary, for example, to the mRNA sequence encoding the target molecule.
Ribozymes
targeting the target molecule, may be synthesized using commercially available
reagents (Applied
Biosystems, Inc., Foster City, CA) or they may be genetically expressed from
DNA encoding
them.
In one embodiment, the agent may comprise one or more components of a CRISPR-
Cas
system, where a guide RNA (gRNA) targeted to a gene encoding a target
molecule, and a
CR1SPR-associated (Cas) peptide form a complex to induce mutations within the
targeted gene.
In one embodiment, the agent comprises a gRNA or a nucleic acid molecule
encoding a gRNA.
In one embodiment, the agent comprises a Cas peptide or a nucleic acid
molecule encoding a Cas
peptide.
microRNA cwents
In one embodiment, the agent comprises a miRNA or a mimic of a miRNA. In one
embodiment, the agent comprises a nucleic acid molecule that encodes a miRNA
or mimic of a
miRNA.
MiRNAs are small non-coding RNA molecules that are capable of causing post-
transcriptional silencing of specific genes in cells by the inhibition of
translation or through
degradation of the targeted mRNA. A miRNA can be completely complementary or
can have a
27
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
region of noncomplementarity with a target nucleic acid, consequently
resulting in a "bulge" at
the region of non-complcmcntarity. A miRNA can inhibit gene expression by
repressing
translation, such as when the miRNA is not completely complementary to the
target nucleic acid,
or by causing target RNA degradation, which is believed to occur only when the
miRNA binds its
target with perfect complementarity. The disclosure also can include double-
stranded precursors
of miRNA. A miRNA or pri-miRNA can be 18- 100 nucleotides in length, or from
18-80
nucleotides in length. Mature miRNAs can have a length of 19-30 nucleotides,
or 21-25
nucleotides, particularly 21, 22, 23, 24, or 25 nucleotides. MiRNA precursors
typically have a
length of about 70-100 nucleotides and have a hairpin conformation. miRNAs are
generated in
vivo from pre- miRNAs by the enzymes Dicer and Drosha, which specifically
process long pre-
miRNA into functional miRNA. The hairpin or mature microRNAs, or pri-microRNA
agents
featured in the disclosure can be synthesized in vivo by a cell-based system
or in vitro by
chemical synthesis.
In various embodiments, the agent comprises an oligonucleotide that comprises
the
nucleotide sequence of a disease-associated miRNA. In certain embodiments, the
oligonucleotide
comprises the nucleotide sequence of a disease-associated miRNA in a pre -
microRNA, mature or
hairpin form. In other embodiments, a combination of oligonucleotides
comprising a sequence of
one or more disease-associated miRNAs, any pre -miRNA, any fragment, or any
combination
thereof is envisioned.
MiRNAs can be synthesized to include a modification that imparts a desired
characteristic. For example, the modification can improve stability,
hybridization
thermodynamics with a target nucleic acid, targeting to a particular tissue or
cell -type, or cell
permeability, e.g., by an endocytosis-dependent or -independent mechanism.
Modifications can also increase sequence specificity, and consequently
decrease off-site
targeting. Methods of synthesis and chemical modifications are described in
greater detail below.
If desired, miRNA molecules may be modified to stabilize the miRNAs against
degradation, to
enhance half-life, or to otherwise improve efficacy. Desirable modifications
are described, for
example, in U.S. Patent Publication Nos. 20070213292, 20060287260,
20060035254.
20060008822. and 2005028824, each of which is hereby incorporated by reference
in its entirety.
For increased nuclease resistance and/or binding affinity to the target, the
single- stranded
oligonucleotide agents featured in the disclosure can include 2'-0-methyl, 2'-
fluorine, 2'-0-
methoxyethyl, 2'-0-aminopropyl, 2'-amino, and/or phosphorothioate linkages.
Inclusion of locked
nucleic acids (LNA), ethylene nucleic acids (ENA), e.g., 2'-4'-ethylene-
bridged nucleic acids,
and certain nucleotide modifications can also increase binding affinity to the
target. The inclusion
28
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
of pyranose sugars in the oligonucleotide backbone can also decrease
endonucleolytic cleavage.
An oligonucicotidc can be further modified by including a 3' cationic group,
or by inverting the
nucleoside at the 3'-terminus with a 3 -3' linkage. In another alternative,
the 3 '-terminus can be
blocked with an aminoalkyl group. Other 3' conjugates can inhibit 3'-5'
exonucleolytic cleavage.
While not being bound by theory, a 3 may inhibit exonucleolytic cleavage by
sterically blocking
the exonuclease from binding to the 3' end of the oligonucleotide. Even small
alkyl chains, aryl
groups, or heterocyclic conjugates or modified sugars (D-ribose, deoxyribose,
glucose etc.) can
block 3'-5'-exonucleases.
In one embodiment, the miRNA includes a 2'-modified oligonucleotide containing
oligodeoxynucleotide gaps with some or all internucleotide linkages modified
to
phosphorothioates for nuclease resistance. The presence of methylphosphonate
modifications
increases the affinity of the oligonucleotide for its target RNA and thus
reduces the IC5Q. This
modification also increases the nuclease resistance of the modified
oligonucleotide. It is
understood that the methods and reagents of the present disclosure may be used
in conjunction
with any technologies that may be developed to enhance the stability or
efficacy of an inhibitory
nucleic acid molecule.
miRNA molecules include nucleotide oligomers containing modified backbones or
non-
natural internucleoside linkages. Oligomers having modified backbones include
those that retain a
phosphorus atom in the backbone and those that do not have a phosphorus atom
in the backbone.
For the purposes of this disclosure, modified oligonucleotides that do not
have a phosphorus atom
in their internucleoside backbone are also considered to be nucleotide
oligomers. Nucleotide
oligomers that have modified oligonucleotide backbones include, for example,
phosphorothioates,
chiral phosphorothioates, phosphorodithioates, phosphotriesters, aminoalkyl-
phosphotriesters,
methyl and other alkyl phosphonates including 3'-alkylene phosphonates and
chiral phosphonates,
phosphinates, phosphoramidates, thionophosphoramidates,
thionoalkylphosphonates,
thionoalkylphosphotriest- ers, and boranophosphates. Various salts, mixed
salts and free acid
forms are also included.
A miRNA described herein, which may be in the mature or hairpin form, may be
provided as a naked oligonucleotide. In some cases, it may be desirable to
utilize a formulation
that aids in the delivery of a miRNA or other nucleotide oligomer to cells
(see, e.g., U.S. Pat.
Nos. 5,656,61 1, 5,753,613, 5,785,992, 6,120,798, 6,221,959, 6,346,613, and
6,353,055, each of
which is hereby incorporated by reference).
In some examples, the miRNA composition is at least partially crystalline,
uniformly
crystalline, and/or anhydrous (e.g., less than 80, 50, 30, 20, or 10% water).
In another example,
29
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
the miRNA composition is in an aqueous phase, e.g., in a solution that
includes water. The
aqueous phase or the crystalline compositions can be incorporated into a
delivery vehicle, e.g., a
liposome (particularly for the aqueous phase), or a particle (e.g., a
microparticle as can be
appropriate for a crystalline composition). Generally, the miRNA composition
is formulated in a
manner that is compatible with the intended method of administration. A miRNA
composition
can be formulated in combination with another agent, e.g., another therapeutic
agent or an agent
that stabilizes an oligonucleotide agent, e.g., a protein that complexes with
the oligonucleotide
agent. Still other agents include chelators, e.g., EDTA (e.g., to remove
divalent cations such as
Mg), salts, and RNAse inhibitors (e.g., a broad specificity RNAse inhibitor).
In one embodiment,
the miRNA composition includes another miRNA, e.g., a second miRNA composition
(e.g., a
microRNA that is distinct from the first). Still other preparations can
include at least three, five,
ten, twenty, fifty, or a hundred or more different oligonucleotide species.
In certain embodiments, the composition comprises an oligonucleotide
composition that
mimics the activity of a miRNA. In certain embodiments, the composition
comprises
oligonucleotides having nucleobase identity to the nucleobase sequence of a
miRNA, and are thus
designed to mimic the activity of the miRNA. In certain embodiments, the
oligonucleotide
composition that mimics miRNA activity comprises a double-stranded RNA
molecule which
mimics the mature miRNA hairpins or processed miRNA duplexes.
In one embodiment, the oligonucleotide shares identity with endogenous miRNA
or
miRNA precursor nucleobase sequences. An oligonucleotide selected for
inclusion in a
composition of the present invention may be one of a number of lengths. Such
an oligonucleotide
can be from 7 to 100 linked nucleosides in length. For example, an
oligonucleotide sharing
nucleobase identity with a miRNA may be from 7 to 30 linked nucleosides in
length. An
oligonucleotide sharing identity with a miRNA precursor may be up to 100
linked nucleosides in
length. In certain embodiments, an oligonucleotide comprises 7 to 30 linked
nucleosides. In
certain embodiments, an oligonucleotide comprises 7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19,
20, 21, 22, 23, 24, 25, 26, 28, 29, or 30 linked nucleotides. In certain
embodiments, an
oligonucleotide comprises 19 to 23 linked nucleosides. In certain embodiments,
an
oligonucleotide is from 40 up to 50, 60, 70, 80, 90, or 100 linked nucleosides
in length.
In certain embodiments, an oligonucleotide has a sequence that has a certain
identity to a
miRNA or a precursor thereof Nucleobase sequences of mature miRNAs and their
corresponding
stem-loop sequences described herein are the sequences found in miRBase, an
online searchable
database of miRNA sequences and annotation. Entries in the miRBase Sequence
database
represent a predicted hairpin portion of a miRNA transcript (the stem-loop),
with information on
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
the location and sequence of the mature miRNA sequence. The miRNA stem-loop
sequences in
the database are not strictly precursor miRNAs (pre-miRNAs), and may in some
instances include
the pre-miRNA and some flanking sequence from the presumed primary transcript.
The miRNA
nucleobase sequences described herein encompass any version of the miRNA,
including the
sequences described in Release 10.0 of the miRBase sequence database and
sequences described
in any earlier Release of the miRBase sequence database. A sequence database
release may result
in the re-naming of certain miRNAs. A sequence database release may result in
a variation of a
mature miRNA sequence. The compositions of the present invention encompass
oligomeric
compound comprising oligonucleotides having a certain identity to any
nucleobase sequence
version of a miRNAs described herein.
In certain embodiments, an oligonucleotide has a nucleobase sequence at least
60%, 65%,
70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical to the miRNA over a
region of 7,
8,9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27,
28, 29, or 30
nucleobases. Accordingly, in certain embodiments the nucleobase sequence of an
oligonucleotide
may have one or more non-identical nucleobases with respect to the miRNA.
In certain embodiments, the composition comprises a nucleic acid molecule
encoding a
miRNA, precursor, mimic, or fragment thereof. For example, the composition may
comprise a
viral vector, plasmid, cosmid, or other expression vector suitable for
expressing the miRNA,
precursor, mimic, or fragment thereof in a desired mammalian cell or tissue.
In vitro transcribed RNA agents
In one embodiment, the agent of the invention comprises in vitro transcribed
(IVT) RNA.
In one embodiment, the agent of the invention comprises in vitro transcribed
(IVT) RNA
encoding a therapeutic protein. In one embodiment, the agent of the invention
comprises IVT
RNA encoding a plurality of therapeutic proteins.
In one embodiment, an IVT RNA can be introduced to a cell as a form of
transient
transfection. The RNA is produced by in vitro transcription using a plasmid
DNA template
generated synthetically. DNA of interest from any source can be directly
converted by PCR into a
template for in vitro mRNA synthesis using appropriate primers and RNA
polymerase. The
source of the DNA can be, for example, genomic DNA, plasmid DNA, phage DNA,
cDNA,
synthetic DNA sequence or any other appropriate source of DNA. In one
embodiment, the
desired template for in vitro transcription is a therapeutic protein, as
described elsewhere herein.
In one embodiment, the DNA to be used for PCR contains an open reading frame.
The
DNA can be from a naturally occurring DNA sequence from the genome of an
organism. In one
31
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
embodiment, the DNA is a full-length gene of interest of a portion of a gene.
The gene can
include some or all of the 5' and/or 3' untranslatcd regions (UTRs). The gene
can include exons
and introns. In one embodiment, the DNA to be used for PCR is a human gene. In
another
embodiment, the DNA to be used for PCR is a human gene including the 5 and 3'
UTRs. In
another embodiment, the DNA to be used for PCR is a gene from a pathogenic or
commensal
organism, including bacteria, viruses, parasites, and fungi. In another
embodiment, the DNA to be
used for PCR is from a pathogenic or commensal organism, including bacteria,
viruses, parasites,
and fungi, including the 5' and 3' UTRs. The DNA can alternatively be an
artificial DNA
sequence that is not normally expressed in a naturally occurring organism. An
exemplary
artificial DNA sequence is one that contains portions of genes that are
ligated together to form an
open reading frame that encodes a fusion protein. The portions of DNA that are
ligated together
can be from a single organism or from more than one organism.
Genes that can be used as sources of DNA for PCR include genes that encode
polypeptides that induce or enhance an adaptive immune response in an
organism. Preferred
genes are genes which are useful for a short-term treatment, or where there
are safety concerns
regarding dosage or the expressed gene.
In various embodiments, a plasmid is used to generate a template for in vitro
transcription
of RNA which is used for transfection.
Chemical stmctures with the ability to promote stability and/or translation
efficiency may
also be used. The RNA preferably has 5' and 3' UTRs. In one embodiment, the 5'
UTR is between
zero and 3000 nucleotides in length. The length of 5' and 3' UTR sequences to
be added to the
coding region can be altered by different methods, including, but not limited
to, designing
primers for PCR that anneal to different regions of the UTRs. Using this
approach, one of
ordinary skill in the art can modify the 5' and 3' UTR lengths required to
achieve optimal
translation efficiency following transfection of the transcribed RNA.
The 5' and 3' UTRs can be the naturally occurring, endogenous 5' and 3' UTRs
for the
gene of interest. Alternatively, UTR sequences that are not endogenous to the
gene of interest can
be added by incorporating the UTR sequences into the forward and reverse
primers or by any
other modifications of the template. The use of UTR sequences that are not
endogenous to the
gene of interest can be useful for modifying the stability and/or translation
efficiency of the RNA.
For example, it is known that AU-rich elements in 3' UTR sequences can
decrease the stability of
RNA. Therefore, 3' UTRs can be selected or designed to increase the stability
of the transcribed
RNA based on properties of UTRs that are well known in the art.
32
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In one embodiment, the 5' UTR can contain the Kozak sequence of the endogenous
gene.
Alternatively, when a 5' UTR that is not endogenous to the gene of interest is
being added by
PCR as described above, a consensus Kozak sequence can be redesigned by adding
the 5' UTR
sequence. Kozak sequences can increase the efficiency of translation of some
RNA transcripts,
but does not appear to be required for all RNAs to enable efficient
translation. The requirement
for Kozak sequences for many RNAs is known in the art. In other embodiments
the 5' UTR can
be derived from an RNA virus whose RNA genome is stable in cells. In other
embodiments
various nucleotide analogues can be used in the 3' or 5' UTR to impede
exonuclease degradation
of the RNA.
To enable synthesis of RNA from a DNA template without the need for gene
cloning, a
promoter of transcription should be attached to the DNA template upstream of
the sequence to be
transcribed. When a sequence that functions as a promoter for an RNA
polymerase is added to the
5' end of the forward primer, the RNA polymerase promoter becomes incorporated
into the PCR
product upstream of the open reading frame that is to be transcribed. In one
preferred
embodiment, the promoter is a T7 RNA polymerase promoter, as described
elsewhere herein.
Other useful promoters include, but are not limited to, T3 and SP6 RNA
polymerase promoters.
Consensus nucleotide sequences for T7, T3 and SP6 promoters are known in the
art.
In a preferred embodiment, the RNA has both a cap on the 5' end and a 3'
poly(A) tail
which determine ribosome binding, initiation of translation and stability mRNA
in the cell. On a
circular DNA template, for instance, plasmid DNA, RNA polymerase produces a
long
concatameric product which is not suitable for expression in eukaryotic cells.
The transcription of
plasmid DNA linearized at the end of the 3' UTR results in normal sized RNA
which is effective
in eukaryotic transfection when it is polyadenylated after transcription.
On a linear DNA template, phage T7 RNA polymerase can extend the 3' end of the
transcript beyond the last base of the template (Schenbom and Mierendorf, Nue
Acids Res.,
13:6223-36 (1985); Nacheva and Berzal-Herranz, Eur. J. Biochem., 270:1485-65
(2003).
The conventional method of integration of polyA/T stretches into a DNA
template is
molecular cloning. However polyA/T sequence integrated into plasmid DNA can
cause plasmid
instability, which can be ameliorated through the use of recombination
incompetent bacterial cells
for plasmid propagation.
Poly(A) tails of RNAs can be further extended following in vitro transcription
with the
use of a poly(A) polymerase, such as E. coli polyA polymerase (E-PAP) or yeast
polyA
polymerase. In one embodiment, increasing the length of a poly(A) tail from
100 nucleotides to
between 300 and 400 nucleotides results in about a two-fold increase in the
translation efficiency
33
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
of the RNA. Additionally, the attachment of different chemical groups to the
3' end can increase
RNA stability. Such attachment can contain modified/artificial nucleotides,
aptamers and other
compounds. For example, ATP analogs can be incorporated into the poly(A) tail
using poly(A)
polyrnerase. ATP analogs can further increase the stability of the RNA.
5' caps on also provide stability to RNA molecules. In a preferred embodiment,
RNAs
produced by the methods to include a 5' capl structure. Such capl structure
can be generated
using Vaccinia capping enzyme and 2'-0-methyltransferase enzymes (CellScript,
Madison, WI).
Alternatively, 5' cap is provided using techniques known in the art and
described herein (Cougot,
et al., Trends in Biochem. Sci., 29:436-444 (2001); Stepinski, et al.. RNA,
7:1468-95 (2001);
Elango, et al., Biochim. Biophys. Res. Commun., 330:958-966 (2005)).
Polvpeptide agents
In other related aspects, the agent includes an isolated peptide that
modulates a target. For
example, in one embodiment, the peptide of the invention inhibits or activates
a target directly by
binding to the target thereby modulating the normal functional activity of the
target. In one
embodiment, the peptide of the invention modulates the target by competing
with endogenous
proteins. In one embodiment, the peptide of the invention modulates the
activity of the target by
acting as a transdominant negative mutant.
The variants of the polypeptide agents may be (i) one in which one or more of
the amino
acid residues are substituted with a conserved or non-conserved amino acid
residue (preferably a
conserved amino acid residue) and such substituted amino acid residue may or
may not be one
encoded by the genetic code, (ii) one in which there are one or more modified
amino acid
residues, e.g., residues that are modified by the attachment of substituent
groups, (iii) one in
which the polypeptide is an alternative splice variant of the polypeptide of
the present invention,
(iv) fragments of the polypeptides and/or (v) one in which the polypeptide is
fused with another
polypeptide, such as a leader or secretory sequence or a sequence which is
employed for
purification (for example, His-tag) or for detection (for example, Sy5 epitope
tag). The fragments
include polypeptides generated via proteolytic cleavage (including multi-site
proteolysis) of an
original sequence. Variants may be post-translationally, or chemically
modified. Such variants are
deemed to be within the scope of those skilled in the art from the teaching
herein.
Antibody agents
34
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
The invention also contemplates a delivery vehicle comprising an antibody, or
antibody
fragment, specific for a target. That is, the antibody can inhibit a target to
provide a beneficial
effect.
The antibodies may be intact monoclonal or polyclonal antibodies, and
immunologically
active fragments (e.g., a Fab or (Fab)2 fragment), an antibody heavy chain, an
antibody light
chain, humanized antibodies, a genetically engineered single chain FV molecule
(Ladner et al,
U.S. Pat. No. 4,946,778), or a chimeric antibody, for example, an antibody
which contains the
binding specificity of a murine antibody, but in which the remaining portions
are of human
origin. Antibodies including monoclonal and polyclonal antibodies, fragments
and chimeras, may
be prepared using methods known to those skilled in the art.
Antibodies can be prepared using intact polypeptides or fragments containing
an
immunizing antigen of interest. The polypeptide or oligopeptide used to
immunize an animal may
be obtained from the translation of RNA or synthesized chemically and can be
conjugated to a
carrier protein, if desired. Suitable carriers that may be chemically coupled
to peptides include
bovine serum albumin and thyroglobulin, keyhole limpet hemocyanin. The coupled
polypeptide
may then be used to immunize the animal (e.g., a mouse, a rat, or a rabbit).
CAR agents
In one embodiment, the agent comprises a recombinant nucleic acid sequence
encoding a
chimeric antigen receptor (CAR). In one embodiment, the agent comprises a mRNA
molecule
encoding a CAR. In one embodiment, the agent comprises a nucleoside modified
mRNA
molecule encoding a CAR.
The term -chimeric antigen receptor" or "CAR," as used herein, refers to an
artificial T
cell receptor that is engineered to be expressed on an immune effector cell
and specifically bind
an antigen. CARS may be used as a therapy with adoptive cell transfer. T cells
are removed from
a patient and modified so that they express the receptors specific to a
particular form of antigen.
In some embodiments, the CARS have specificity to a selected target. CARS may
also comprise
an intracellular activation domain, a transmembrane domain and an
extracellular domain
comprising an antigen binding region that specifically binds to a selected
target. In some aspects,
CARS comprise an extracellular domain comprising an anti-B cell binding domain
fused to
CD3-zeta transmembrane and intracellular domain.
In one embodiment, the invention relates to a delivery vehicle comprising an
agent,
wherein the agent comprises a recombinant nucleic acid sequence (e.g., an
mRNA) encoding a
chimeric antigen receptor (CAR). In one embodiment, the agent comprises an
mRNA molecule
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
(e.g., a modified nucleoside mRNA molecule) encoding a chimeric antigen
receptor (CAR). In
one embodiment, agent comprises an mRNA molecule encoding a CAR. In one
embodiment,
agent comprises a nucleoside modified mRNA molecule encoding a CAR.
In various embodiments, the CARs contemplated herein comprise an extracellular
domain, a transmembrane domain, and an intracellular domain. The extracellular
domain
comprises a target-specific binding element otherwise referred to as an
antigen binding domain.
In some embodiments, the extracellular domain also comprises a hinge domain.
In certain
embodiments, the intracellular domain or otherwise the cytoplasmic domain
comprises, a
costimulatory signaling region and a zeta chain portion. The costimulatory
signaling region refers
to a portion of the CAR comprising the intracellular domain of a costimulatory
molecule.
Costimulatory molecules are cell surface molecules other than antigens
receptors or their ligands
that are required for an efficient response of lymphocytes to antigen.
Between the extracellular domain and the transmembrane domain of the CAR, or
between the cytoplasmic domain and the transmembrane domain of the CAR, there
may be
incorporated a spacer domain. As used herein, the term "spacer domain"
generally means any
oligo- or polvpeptide that functions to link the transmembrane domain to,
either the extracellular
domain or, the cytoplasmic domain in the polypeptide chain. A spacer domain
may comprise up
to 5 amino acids, or 10 amino acids, or 20 amino acids, or 30 amino acids, or
40 amino acids, or
50 amino acids, or 60 amino acids, or 70 amino acids, or 80 amino acids, or 90
amino acids, or
100 amino acids, or 110 amino acids, or 120 amino acids, or 130 amino acids,
or 140 amino
acids, or 150 amino acids, or 160 amino acids, or 170 amino acids, or 180
amino acids, or 190
amino acids, or 200 amino acids, or 210 amino acids, or 220 amino acids, or
230 amino acids, or
240 amino acids, or 250 amino acids, or 260 amino acids, or 270 amino acids,
or 280 amino
acids, or 290 amino acids, or 300 amino acids.
The extracellular domain. transmembrane domain, and intracellular domain can
be
derived from any desired source of such domains.
CAR antigen binding domain
The antigen binding domain may be obtained from any of the wide variety of
extracellular domains or secreted proteins associated with ligand binding
and/or signal
transduction. In one embodiment, the antigen binding domain may consist of an
Ig heavy chain
which may in turn be covalently associated with Ig light chain by virtue of
the presence of CHI
and hinge regions, or may become covalently associated with other 1g
heavy/light chain
complexes by virtue of the presence of hinge, CH2 and CH3 domains. In the
latter case, the
36
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
heavy/light chain complex that becomes joined to the chimeric construct may
constitute an
antibody with a specificity distinct from the antibody specificity of the
chimeric construct.
Depending on the function of the antibody, the desired structure and the
signal transduction, the
entire chain may be used or a truncated chain may be used, where all or a part
of the CHI, CH2,
or CH3 domains may be removed or all or part of the hinge region may be
removed.
In various embodiments, the CAR antigen binding domain may be humanized or
comprise a fully human sequence.
CAR transmemhrane domain
With respect to the transmembrane domain, a CAR of the disclosure can be
designed to
comprise a transmembrane domain that is fused to the extracellular domain of
the CAR. In one
embodiment, the transmembrane domain that naturally is associated with one of
the domains in
the CAR is used. In some instances, the transmembrane domain can be selected
or modified by
amino acid substitution to avoid binding of such domains to the transmembrane
domains of the
same or different surface membrane proteins to minimize interactions with
other members of the
receptor complex.
The transmembrane domain may be derived either from a natural or from a
synthetic
source. Where the source is natural, the domain may be derived from any
membrane-bound or
transmembrane protein. Transmembrane regions of particular use in this
invention may be
derived from (i.e., comprise at least the transmembrane region(s) of) the
alpha, beta or zeta chain
of the T-cell receptor, CD28, CD3 epsilon, CD45, CD4, CD5, CD8, CD9, CD16,
CD22, CD33,
CD37, CD64, CD80, CD86, CD134, CD137, or CD 154. Alternatively, the
transmembrane
domain may be synthetic, in which case it will comprise predominantly
hydrophobic residues
such as leucine and valine. In one embodiment, a triplet of phenylalanine,
tryptophan and valine
can be found at each end of a synthetic transmembrane domain. Optionally, a
short oligo- or
polypeptide linker, for example, but not limited to between 2 and 10 amino
acids in length, may
form the linkage between the transmembrane domain and the cytoplasmic
signaling domain of
the CAR. In another embodiment, the linker comprises a glycine-serine doublet.
CAR intracellular domain
In various embodiments, the cytoplasmic domain or otherwise the intracellular
domain of
a CAR may be responsible for activation of at least one of the normal effector
functions of the
immune cell in which the CAR is expressed. The term "effector function" refers
to a specialized
function of a cell. Effector function of a T cell, for example, may be
cytolytic activity or helper
37
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
activity, including the secretion of cytokines. The term "intracellular
signaling domain" refers to
the portion of a protein which transduces the effector function signal and
directs the cell to
perform a specialized function. While usually the entire intracellular domain
can be employed, in
many cases it is not necessary to use the entire chain. To the extent that a
truncated portion of the
intracellular domain is used, such truncated portion may be used in place of
the intact chain as
long as it transduces the effector function signal. The term intracellular
domain is thus meant to
include any truncated portion of the intracellular domain sufficient to
transduce the effector
function signal.
Preferred examples of intracellular domains for use in the CARs of the
disclosure include
the cytoplasmic sequences of the T cell receptor (TCR) and co-receptors that
act in concert to
initiate signal transduction following antigen receptor engagement, as well as
any derivative or
variant of these sequences and any synthetic sequence that has the same
functional capability.
It is known that signals generated through the TCR alone are insufficient for
full
activation of the T cell and that a secondary or co-stimulatory signal is also
required. Thus, T cell
activation can be said to be mediated by two classes of intracellular
signaling sequences: those
that initiate antigen-dependent primary activation through the TCR (primary
cytoplasmic
signaling sequences) and those that act in an antigen-independent manner to
provide a secondary
or co-stimulatory signal (secondary cytoplasmic signaling sequences).
Primary intracellular signaling sequences regulate primary activation of the
TCR
complex either in a stimulatory way, or in an inhibitory way. Primary
intracellular signaling
sequences that act in a stimulatory manner may contain signaling motifs which
are known as
immunoreceptor tyrosine-based activation motifs or ITAMs.
Examples of ITAMs containing primary intracellular signaling sequences that
are of
particular use in the invention include those derived from TCR zeta, FcR
gamma, FcR beta, CD3
gamma, CD3 delta, CD3 epsilon, CD5, CD22, CD79a, CD79b, and CD66d. In one
embodiment,
the intracellular signaling molecule in the CAR of the invention comprises an
intracellular
signaling sequence derived from CD3 zeta.
In another embodiment, the intracellular domain of the CAR can be designed to
comprise
the CD3-zeta signaling domain by itself or combined with any other desired
cytoplasmic
domain(s) useful in the context of the CAR of the invention. For example, the
intracellular
domain of the CAR can comprise a CD3 zeta chain portion and a costimulatory
signaling region.
The costimulatory signaling region refers to a portion of the CAR comprising
the intracellular
domain of a costimulatory molecule. A costimulatory molecule is a cell surface
molecule other
than an antigen receptor or their ligands that is required for an efficient
response of lymphocytes
38
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
to an antigen. Examples of such molecules include CD2, CD27, CD28, 4-1BB
(CD137), 0x40,
CD30, CD40, PD-1, ICOS, lymphocyte function-associated antigen-1 (LFA-1), CD2,
CD7,
LIGHT, NKG2C, B7-H3, and a ligand that specifically binds with CD83, and the
like.
The intracellular signaling sequences within the intracellular domain of the
CAR of the
invention may be linked to each other in a random or specified order.
Optionally, a short oligo- or
polypeptide linker, for example, between 2 and 10 amino acids in length may
form the linkage. A
glycine-serine doublet provides a suitable linker in some embodiments.
In one embodiment, the intracellular domain is designed to comprise the
signaling
domain of CD3-zeta and the signaling domain of CD28. In yet another
embodiment, the
intracellular domain is designed to comprise the signaling domain of CD3-zeta
and the signaling
domain of 4- IBB.
The antigen binding domain can be any domain that binds to the antigen
including but
not limited to monoclonal antibodies, polyclonal antibodies, synthetic
antibodies, scFvs, human
antibodies, humanized antibodies, and fragments thereof. In one non-limiting
embodiment, the
antigen binding region specifically binds to a selected target, e.g., an
activated fibroblast cell
surface receptor (such as CD90, FAP, FSP-1, CD140a, CD140b, CD49b, CD87, CD95,
a smooth
muscle actin (aSMA), or platelet derived growth factor 13 (PDGFR13.
In various embodiments, the CAR can be a "first generation," "second
generation," "third
generation," "fourth generation" or "fifth generation" CAR (see, for example,
Sadelain et al.,
Cancer Discov. 3(4):388-398 (2013); Jensen et al., Immunol. Rev. 257:127-133
(2014); Sharpe et
al., Dis. Model Mech. 8(4):337-350 (2015); Brentjens et al., Clin. Cancer Res.
13:5426-5435
(2007); Gade et al., Cancer Res. 65:9080-9088 (2005); Maher et al., Nat.
Biotechnol. 20:70-75
(2002); Kershaw et al., J. Immunol. 173:2143-2150 (2004); Sadelain et al.,
Curr. Opin. Immunol.
(2009); Hollyman et al., J. Immunother. 32:169-180 (2009), each of which are
incorporated by
reference in its entirety).
"First generation" CARS for use in the invention comprise an antigen binding
domain, for
example, a single-chain variable fragment (scFv), fused to a transmembrane
domain, which is
fused to a cytoplasmic/intracellular domain of the T cell receptor chain.
"First generation" CARS
typically have the intracellular domain from the CD3C-chain, which is the
primary transmitter of
signals from endogenous T cell receptors (TCRs). "First generation" CARs can
provide de novo
antigen recognition and cause activation of both CD4+ and CD8+ T cells through
their CD3C
chain signaling domain in a single fusion molecule, independent of HLA-
mediated antigen
presentation.
39
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
"Second-generation" CARs for use in the invention comprise an antigen binding
domain,
for example, a single-chain variable fragment (scFv), fused to an
intracellular signaling domain
capable of activating T cells and a co-stimulatory domain designed to augment
T cell potency and
persistence (Sadelain et al., Cancer Discov. 3:388-398 (2013)). CAR design can
therefore
combine antigen recognition with signal transduction, two functions that are
physiologically
borne by two separate complexes, the TCR heterodimer and the CD3 complex.
"Second
generation" CARs include an intracellular domain from various co-stimulatory
molecules, for
example, CD28, 4-1BB, ICOS, 0X40, and the like, in the cytoplasmic tail of the
CAR to provide
additional signals to the cell.
-Second generation" CARs provide both co-stimulation, for example, by CD28 or
4-1BB
domains, and activation, for example, by a CD3 signaling domain. Preclinical
studies have
indicated that "Second Generation" CARs can improve the anti-tumor activity of
T cells. For
example, robust efficacy of "Second Generation" CAR modified T cells was
demonstrated in
clinical trials targeting the CD19 molecule in patients with chronic
lymphoblastic leukemia
(CLL) and acute lymphoblastic leukemia (ALL) (Davila et al., Oncoimmunol.
1(9):1577-1583
(2012)).
"Third generation" CARs provide multiple co-stimulation, for example, by
comprising
both CD28 and 4- IBB domains, and activation, for example, by comprising a CD3
activation
domain.
"Fourth generation' CARs provide co-stimulation, for example, by CD28 or 4-1BB
domains, and activation, for example, by a CD3 signaling domain in addition to
a constitutive or
inducible chemokine component.
"Fifth generation" CARs provide co-stimulation, for example, by CD28 or 4-1BB
domains, and activation, for example, by a CD3 signaling domain, a
constitutive or inducible
chemokine component, and an intracellular domain of a cytokine receptor, for
example, IL-2R43.
In various embodiments, the CAR can be included in a multivalent CAR system,
for
example, a DualCAR or "TandemCAR" system. Multivalent CAR systems include
systems or
cells comprising multiple CARs and systems or cells comprising
bivalent/bispecific CARs
targeting more than one antigen.
In the embodiments disclosed herein, the CARs generally comprise an antigen
binding
domain, a transmembrane domain and an intracellular domain, as described
above. In a particular
non-limiting embodiment, the antigen-binding domain is an scFv.
In one embodiment, the antigen binding domain is a targeting domain, wherein
the
targeting domain directs the T cell expressing the CAR to a specific cell or
tissue of interest. For
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
example, in one embodiment, the targeting domain comprises an antibody,
antibody fragment, or
peptide that specifically binds to an antigen (e.g., a salef-antigen or a
foreign antigen) thereby
directing the T cell expressing the CAR to a cell or tissue expressing the
antigen.
The antigen binding domain of-the CAR Ell 01 ccule of the invention can be
generated to be
reactive to any desirable antigen of interest, or fragment thereof, including,
but not limited to a
tumor antigen, a foreign antigen (e.g, a bacterial antigen, or a viral
antigen) or a self-antigen.
Tumor antigens are proteins that are produced by tumor cells that elicit an
immune
response. The selection of the antigen binding domain of the V M-domain
containing fusion
molecule of the invention will depend on the particular type of cancer to be
treated. Tumor
antigens are well known in the art and include, for example, a glioma-
associated antigen,
carcinoembryonic antigen (CEA), I3-human chorionic gonadotropin,
alphafetoprotein (AFP),
lectin-reactive AFP, thyroglobulin, RAGE-1, MN-CA IX, human telomerase reverse
transcriptase, RUI, RU2 (AS), intestinal carboxyl esterase, mut hsp70-2, M-
CSF, prostase,
prostate-specific antigen (PSA), PAP, NY-ESO-1, LAGE- la, p53, prostein, PSMA,
Her2/neu,
survivin and telomerase, prostate-carcinoma tumor antigen-1 (PCTA-1), MAGE,
ELF2M,
neutrophil elastase, ephrinB2, CD22, insulin growth factor (IGF)-I, IGF-II,
IGF-I receptor and
mesothelin. Another exemplary tumor antigen is chondroitin sulfate
proteoglycan 4 (CSPG4)
(also referred to as melanoma-associated chondroitin sulfate proteoglycan
(MCSP), high-
molecular-weight melanoma-associated antigen (HMVV-MAA), or neuron-glial
antigen 2 (NG2)).
In one embodiment, the tumor antigen comprises one or more antigenic cancer
epitopes
associated with a malignant tumor. Malignant tumors express a number of
proteins that can serve
as target antigens for an immune attack. These molecules include but are not
limited to tissue-
specific antigens such as MART-1, tyrosinase and GP 100 in melanoma and
prostatic acid
phosphatase (PAP) and prostate-specific antigen (PSA) in prostate cancer.
Other target molecules
belong to the group of transformation-related molecules such as the oncogene
HER-2/Neu/ErbB-
2. Yet another group of target antigens are onco-fetal antigens such as
carcinoembryonic antigen
(CEA). In B-cell lymphoma the tumor-specific idiotype immunoglobulin
constitutes a truly
tumor-specific immunoglobulin antigen that is unique to the individual tumor.
B-cell
differentiation antigens such as CD19, CD20 and CD37 are other candidates for
target antigens in
B-cell lymphoma. Some of these antigens (CEA, HER-2, CD19, CD20, idiotype)
have been used
as targets for passive immunotherapy with monoclonal antibodies with limited
success.
The type of tumor antigen referred to in the invention may also be a tumor-
specific
antigen (TSA) or a tumor-associated antigen (TAA). A TSA is unique to tumor
cells and does not
occur on other cells in the body. A TAA associated antigen is not unique to a
tumor cell and
41
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
instead is also expressed on a normal cell under conditions that fail to
induce a state of
immunologic tolerance to the antigen. The expression of the antigen on the
tumor may occur
under conditions that enable the immune system to respond to the antigen. TAAs
may be antigens
that are expressed on normal cells during fetal development when the immune
system is
immature and unable to respond or they may be antigens that are normally
present at extremely
low levels on normal cells but which are expressed at much higher levels on
tumor cells.
Non-limiting examples of TSA or TAA antigens include the following:
differentiation
antigens such as MART-1/MelanA (MART-1), gp100 (Pmel 17), tyrosinase, TRP-1,
TRP-2 and
tumor-specific multilineage antigens such as MAGE-1, MAGE-3, BAGE, GAGE-1,
GAGE-2,
p15; overexpressed embryonic antigens such as CEA; overexpressed oncogenes and
mutated
tumor-suppressor genes such as p53, Ras, HER-2/neu; unique tumor antigens
resulting from
chromosomal translocations; such as BCR-ABL, E2A-PRL, H4-RET, IGH-IGK, MYL-
RAR; and
viral antigens, such as the Epstein Barr virus antigens EBVA and the human
papillomavirus
(HPV) antigens E6 and E7. Other large, protein-based antigens include TSP-180,
MAGE-4,
MAGE-5, MAGE-6, RAGE, NY-ESO, p185erbB2, p180erbB-3, c-met, nm-23H1, PSA, TAG-
72,
CA 19-9, CA 72-4, CAM 17.1, NuMa, K-ras, beta-Catenin, CDK4, Mum-1, p 15, p
16, 43-9F,
5T4, 791Tgp72, alpha-fetoprotein, beta-HCG, BCA225, BTAA, CA 125, CA 15-3\CA
27.29\BCAA, CA 195, CA 242, CA-50, CAM43, CD68\PI, CO-029, FGF-5, G250,
Ga733\EpCAM, HTgp-175, M344, MA-50, MG7-Ag, M0V18, NB/70K, NY-CO-1, RCAS1,
SDCCAG16, TA-90\Mac-2 binding protein\cyclophilin C-associated protein, TAAL6,
TAG72,
TLP, and TPS.
A foreign antigen can be a viral antigen, a bacterial antigen, a fungal
antigen, a parasitic
antigen or fragment thereof, or variant thereof. Exemplary viruses, bacterium,
fungi and parasites
that can be targeted using the compositions and methods of the invention are
discussed elsewhere
herein.
Bispeeifie T-cell engager (e.g., BiTE) agents
In still another embodiment, the nucleic acid cargo molecule (e.g., mRNA,
expression
vector, CR1SPR genome editing system, or nucleoside modified mRNA molecule) of
the
disclosure may encode a bispecific T-cell engager that specifically binds to
both an antigen on an
immune cell (e.g., a CD4+ T cell) and an antigen on a cell of interest, e.g.,
an pathogen.
Bispecific T-cell engagers are bispecific molecules that are created by
linking the
targeting regions (i.e., antigen binding domains) of two antibodies as a
single molecule. One arm
of the molecule is engineered to bind with a protein found on the surface of
CD4+ T cells, and the
42
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
other arm is designed to bind to a specific protein found primarily on a
target cell. When both
targets are engaged, the bispecific T-cell engager (i.e., a BiTE molecule)
forms a bridge between
the CD4+ T cell and the target cell. For example, in one embodiment, a target
cell is an activated
fibroblast and the BiTE molecule comprises a binding arm specific for binding
to a fibroblast-
specific marker. Fibroblast-specific markers, include, but are not limited to,
CD9O, FAP, FSP-1,
CD140a, CD140b, CD49b, CD87, CD95, a smooth muscle actin (aSMA), and platelet
derived
growth factor P. (PDGFR13). Further reference may be made to Diego Ellerman, -
Bispecific T-
cell engagers: Towards understanding variable influencing the in vitro potency
and tumor
selectivity and their modulation to enhance their efficacy and safety,"
Methods, Vol.154, Feb.
2019, pp.102-117, which is incorporated herein by reference.
The term -bispecific- means that the bispecific molecule (e.g., a bispecific T-
cell
engager) is able to specifically bind to at least two distinct antigenic
determinants (e.g., one from
a CD4+ T cell and another from a target cell, such as a pathogen). Typically,
a bispecific antigen
binding molecule comprises two antigen binding sites, each of which is
specific for a different
antigenic determinant. In certain embodiments the bispecific antigen binding
molecule is capable
of simultaneously binding two antigenic determinants, particularly two
antigenic determinants
expressed on two distinct cells.
The present disclosure is not limited to the BiTE format but contemplates the
use of any
suitable bispecific format suitable for T cell redirection, including
diabodies (Holliger et al, Prot
Eng 9, 299-305 (1996)) and derivatives thereof, such as tandem diabodies
(Kipriyanov et al, J
Mol Biol 293, 41-66 (1999)), DART (dual affinity retargeting) molecules, which
are based on the
diabody format but feature a C-terminal disulfide bridge for additional
stabilization (Moore et al,
Blood 117, 4542-51(2011)), and triomabs, which are whole hybrid mouse/rat IgG
molecules and
also currently being evaluated in clinical trials, represent a larger sized
format (reviewed in
Seimetz et al, Cancer Treat Rev 36, 458-467 (2010)). Each of the
aforementioned references are
incorporated herein by reference.
Methods for making bispecific antibodies are known in the art. (See, e.g.,
Millstein et al.,
Nature, 305:537-539 (1983); Traunecker et al., EMBOJ., 10:3655-3659 (1991);
Suresh et al.,
Methods in Enzymology, 121:210 (1986); Kostelny et al., J. Immunol.
148(5):1547-1553 (1992);
Hollinger et al., PNAS USA, 90:6444-6448 (1993); Gruber et al., J. Immunol.
152:5368 (1994);
U.S. Pat. Nos. 4,474,893; 4,714,681; 4,925,648; 5,573,920; 5,601,81;
95,731,168; 4,676,980; and
4,676,980, WO 94/04690; WO 91/00360; WO 92/200373; WO 93/17715; WO 92/08802;
and EP
03089.) Each of these aforementioned references relating to making bispecific
antibodies,
including BiTEs, are incorporated herein by reference.
43
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Exemplary bispecific antibody molecules useful in practicing the methods
described
herein contain (i) two antibodies, a first antibody with a binding specificity
to an antigen
expressed on the surface of a target cell and a second antibody with a binding
specificity for an
antigen expressed on the surface of an immune cell (e.g., a CD4+ T cell), (ii)
a single antibody
that has one chain or arm with a binding specificity to an antigen expressed
on the surface of a
target cell and a second chain or arm with a binding specificity to an immune
cell (e.g., a CD4+ T
cell), (iii) a single chain antibody that has binding specificity to an
antigen expressed on the
surface of a target cell and also binding specificity to an immune cell (e.g.,
a CD4+ T cell), e.g.,
via two scFvs linked in tandem by an extra peptide linker; (iv) a dual-
variable-domain antibody
(DVD-Ig), where each light chain and heavy chain contains two variable domains
in tandem
through a short peptide linkage; (v) a chemically-linked bispecific (Fab')2
fragment; (vi) a
Tandab, which is a fusion of two single chain diabodies resulting in a
tetravalent bispecific
antibody that has two binding sites for each of the target antigens: (vii) a
flexibody (a
combination of scFvs with a diabody resulting in a multivalent molecule);
(viii) a so called "dock
and lock" molecule (an adaptation of the "dimerization and docking domain" in
Protein Kinase
A, that can be applied to Fabs to generate a trivalent bispecific binding
protein containing two
identical Fab fragments linked to a different Fab fragment; (ix) a so-called
"Scorpion" molecule,
containing for example, two scFvs fused to both termini of a human Fe-region;
(x) a diabody; and
(xi) a so-called "ImmTAU molecule (Immune mobilising mTCR Against Cancer; see
e.g., Liddy
et al., Nat. Med. 18:980-987 (2012)).
Imcmin,g Agents
In one embodiment, the delivery vehicle comprises an imaging agent. Imaging
agents are
materials that allow the delivery vehicle to be visualized after exposure to a
cell or tissue.
Visualization includes imaging for the naked eye, as well as imaging that
requires detecting with
instruments or detecting information not normally visible to the eye, and
includes imaging that
requires detecting of photons, sound or other energy quanta. Examples include
stains, vital dyes,
fluorescent markers, radioactive markers, enzymes or plasmid constructs
encoding markers or
enzymes. Many materials and methods for imaging and targeting that may be used
in the delivery
vehicle are provided in the Handbook of Targeted delivery of Imaging Agents,
Torchilin, ed.
(1995) CRC Press, Boca Raton, Fla.
Visualization based on molecular imaging typically involves detecting
biological
processes or biological molecules at a tissue, cell, or molecular level.
Molecular imaging can be
used to assess specific targets for gene therapies, cell-based therapies, and
to visualize
44
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
pathological conditions as a diagnostic or research tool. Imaging agents that
are able to be
delivered intracellularly are particularly useful because such agents can be
used to assess
intracellular activities or conditions. Imaging agents must reach their
targets to be effective; thus,
in some embodiments, an efficient uptake by cells is desirable. A rapid uptake
may also be
desirable to avoid the RES, see review in Allport and Weissleder, Experimental
Hematology
1237-1246 (2001).
Further, imaging agents preferably should provide high signal to noise ratios
so that they
may be detected in small quantities. whether directly, or by effective
amplification techniques
that increase the signal associated with a particular target. Amplification
strategies are reviewed
in Allport and Weissleder, Experimental Hematology 1237-1246 (2001), and
include, for
example, avidin-biotin binding systems, trapping of converted ligands, probes
that change
physical behavior after being bound by a target, and taking advantage of
relaxation rates.
Examples of imaging technologies include magnetic resonance imaging,
radionuclide imaging,
computed tomography, ultrasound, and optical imaging.
Delivery vehicles as set forth herein may advantageously be used in various
imaging
technologies or strategies, for example by incorporating imaging agents into
delivery vehicles.
Many imaging techniques and strategies are known, e.g., see review in Allport
and Weissleder,
Experimental Hematology 1237-1246 (2001); such strategies may be adapted to
use with delivery
vehicles. Suitable imaging agents include, for example, fluorescent molecules,
labeled antibodies,
labeled avidin:biotin binding agents, colloidal metals (e.g., gold, silver),
reporter enzymes (e.g.,
horseradish peroxidase), superparamagnetic transferrin, second reporter
systems (e.g., tyrosinase),
and paramagnetic chelates.
In some embodiments, the imaging agent is a magnetic resonance imaging
contrast agent.
Examples of magnetic resonance imaging contrast agents include, but are not
limited to, 1,4,7,10-
tetraazacyclododecane-N,N1,N"N'"-tetracetic acid (DOTA),
diethylenetriaminepentaacetic
(DTPA), 1,4,7,10-tetraazacyclododecane-N,N', N",N'"-tetraethylphosphorus
(DOTEP), 1,4,7,10-
tetraazacyclododecane-N,N',N"-triacetic acid (DOTA) and derivatives thereof
(see U.S. Pat. Nos.
5,188,816, 5,219,553, and 5,358,704). In some embodiments, the imaging agent
is an X-Ray
contrast agent. X-ray contrast agents already known in the art include a
number of halogenated
derivatives, especially iodinated derivatives, of 5-amino-isophthalic acid.
Small molecule agents
In various embodiments, the agent is a small molecule. When the agent is a
small
molecule, a small molecule may be obtained using standard methods known to the
skilled artisan.
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Such methods include chemical organic synthesis or biological means.
Biological means include
purification from a biological source, recombinant synthesis and in vitro
translation systems,
using methods well known in the art. In one embodiment, a small molecule
agents comprises an
organic molecule, inorganic molecule, biomolecule, synthetic molecule, and the
like.
Combinatorial libraries of molecularly diverse chemical compounds potentially
useful in
treating a variety of diseases and conditions are well known in the art, as
are method of making
the libraries. The method may use a variety of techniques well-known to the
skilled artisan
including solid phase synthesis, solution methods, parallel synthesis of
single compounds,
synthesis of chemical mixtures, rigid core structures, flexible linear
sequences, deconv-olution
strategies, tagging techniques, and generating unbiased molecular landscapes
for lead discovery
vs. biased structures for lead development. In some embodiments of the
invention, the agent is
synthesized and/or identified using combinatorial techniques.
In a general method for small library synthesis, an activated core molecule is
condensed
with a number of building blocks, resulting in a combinatorial library of
covalently linked, core-
building block ensembles. The shape and rigidity of the core determines the
orientation of the
building blocks in shape space. The libraries can be biased by changing the
core, linkage, or
building blocks to target a characterized biological structure ("focused
libraries") or synthesized
with less structural bias using flexible cores. In some embodiments of the
invention, the agent is
synthesized via small library synthesis.
The small molecule and small molecule compounds described herein may be
present as
salts even if salts are not depicted, and it is understood that the invention
embraces all salts and
solvates of the agents depicted here, as well as the non-salt and non-solvate
form of the agents, as
is well understood by the skilled artisan. In some embodiments, the salts of
the agents of the
invention are pharmaceutically acceptable salts.
Where tautomeric forms may be present for any of the agents described herein,
each and
every tautomeric form is intended to be included in the present invention,
even though only one
or some of the tautomeric forms may be explicitly depicted. For example, when
a 2-
hydroxypyridyl moiety is depicted, the corresponding 2-pyridone tautomer is
also intended.
The invention also includes any or all of the stereochemical forms, including
any
enantiomeric or diastereomeric forms of the agents described. The recitation
of the structure or
name herein is intended to embrace all possible stereoisomers of agents
depicted. All forms of the
agents are also embraced by the invention, such as crystalline or non-
crystalline forms of the
agent. Compositions comprising an agent of the invention are also intended,
such as a
composition of substantially pure agent, including a specific stereochemical
form thereof, or a
46
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
composition comprising mixtures of agents of the invention in any ratio,
including two or more
stereochcmical forms, such as in a raccmic or non-raccmic mixture.
The invention also includes any or all active analog or derivative, such as a
prodrug, of
any agent described herein. In one embodiment, the agent is a prodrug. In one
embodiment, the
small molecules described herein are candidates for derivatization. As such,
in certain instances,
the analogs of the small molecules described herein that have modulated
potency, selectivity, and
solubility are included herein and provide useful leads for drug discovery and
drug development.
Thus, in certain instances, during optimization new analogs are designed
considering issues of
drug delivery, metabolism, novelty, and safety.
In some instances, small molecule agents described herein are derivatives or
analogs of
known agents, as is well known in the art of combinatorial and medicinal
chemistry. The analogs
or derivatives can be prepared by adding and/or substituting functional groups
at various
locations. As such, the small molecules described herein can be converted into
derivatives/analogs using well known chemical synthesis procedures. For
example, all of the
hydrogen atoms or substituents can be selectively modified to generate new
analogs. Also, the
linking atoms or groups can be modified into longer or shorter linkers with
carbon backbones or
hetero atoms. Also, the ring groups can be changed so as to have a different
number of atoms in
the ring and/or to include hetero atoms. Moreover, aromatics can be converted
to cyclic rings, and
vice versa. For example, the rings may be from 5-7 atoms, and may be
carbocyclic or
heterocyclic.
As used herein, the term "analog," "analogue," or "derivative" is meant to
refer to a
chemical compound or molecule made from a parent compound or molecule by one
or more
chemical reactions. As such, an analog can be a structure having a structure
similar to that of the
small molecule agents described herein or can be based on a scaffold of a
small molecule agents
described herein, but differing from it in respect to certain components or
structural makeup,
which may have a similar or opposite action metabolically. An analog or
derivative of any of a
small molecule inhibitor in accordance with the present invention can be used
to treat a disease or
disorder.
In one embodiment, the small molecule agents described herein can
independently be
derivatized, or analogs prepared therefrom, by modifying hydrogen groups
independently from
each other into other substituents. That is, each atom on each molecule can be
independently
modified with respect to the other atoms on the same molecule. Any traditional
modification for
producing a derivative/analog can be used. For example. the atoms and
substituents can be
independently comprised of hydrogen, an alkyl, aliphatic, straight chain
aliphatic, aliphatic
47
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
having a chain hetero atom, branched aliphatic, substituted aliphatic, cyclic
aliphatic, heterocyclic
aliphatic having one or more hetero atoms, aromatic, heteroaromatic,
polyaromatic, polyamino
acids, peptides, polypeptides, combinations thereof, halogens, halo-
substituted aliphatics, and the
like. Additionally, any ring group on a compound can be derivatized to
increase and/or decrease
ring size as well as change the backbone atoms to carbon atoms or hetero
atoms.
Delivery Vehicle
In some embodiments, the invention relates to composition comprising delivery
vehicles
for delivery of one or more agent. In some embodiments, the agent comprises an
mRNA
molecule (e.g., a nucleoside modified mRNA molecule) of the invention.
In some embodiments, the delivery vehicle is a colloidal dispersion system,
such as
macromolecule complexes, nanocapsules, microspheres, beads, and lipid-based
systems including
oil-in-water emulsions, micelles, mixed micelles, and liposomes. An exemplary
colloidal system
for use as a delivery vehicle in vitro and in vivo is a liposome (e.g., an
artificial membrane
vesicle).
The use of lipid formulations is contemplated for the introduction of the at
least one agent
into a host cell (in vitro, ex vivo or in vivo). In another aspect, the at
least one agent may be
associated with a lipid. The at least one agent associated with a lipid may be
encapsulated in the
aqueous interior of a liposome, interspersed within the lipid bilayer of a
liposome, attached to a
liposome via a linking molecule that is associated with both the liposome and
the oligonucleotide,
entrapped in a liposome, complexed with a liposome, dispersed in a solution
containing a lipid,
mixed with a lipid, combined with a lipid, contained as a suspension in a
lipid, contained or
complexed with a micelle, or otherwise associated with a lipid. Lipid,
lipid/nucleic acid or
lipid/expression vector associated compositions are not limited to any
particular structure in
solution. For example, they may be present in a bilayer structure, as
micelles, or with a
"collapsed" structure. They may also simply be interspersed in a solution,
possibly forming
aggregates that are not uniform in size or shape. Lipids are fatty substances
which may be
naturally occurring or synthetic lipids. For example, lipids include the fatty
droplets that naturally
occur in the cytoplasm as well as the class of compounds which contain long-
chain aliphatic
hydrocarbons and their derivatives, such as fatty acids, alcohols, amines,
amino alcohols, and
aldehydes.
Lipids and their derivatives
In various embodiments, the delivery vehicle may comprise lipids or a
derivative thereof.
48
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Lipids are fatty substances which may be naturally occurring or synthetic
lipids. For
example, lipids include the fatty droplets that naturally occur in the
cytoplasm as well as the class
of compounds which contain long-chain aliphatic hydrocarbons and their
derivatives, such as
fatty acids, alcohols, amines, amino alcohols, aldehydes, and polymers (e.g.
PEGylated lipids).
Lipids suitable for use can be obtained from commercial sources. For example,
dimyristyl
phosphatidylcholine ("DMPC") can be obtained from Sigma, St. Louis, MO;
dicetyl phosphate
("DCP") can be obtained from K & K Laboratories (Plainview, NY); cholesterol
("Chol") can be
obtained from Calbiochem-Behring; dimyristyl phosphatidylglycerol (-DMPG") and
other lipids
may be obtained from Avanti Polar Lipids, Inc. (Birmingham, AL). Stock
solutions of lipids in
chloroform or chloroform/methanol can be stored at about -20 C. Chloroform is
used as the only
solvent since it is more readily evaporated than methanol.
In some embodiments, cationic lipids are preferred. In certain embodiments,
the cationic
lipid comprises any of a number of lipid species which carry a net positive
charge at a selective
pH, such as physiological pH. Such lipids include, but are not limited to, N,N-
dioleyl-N,N-
dimethylammonium chloride (DODAC); N-(2,3-dioleyloxy)propy1)-N,N,N-
trimethylammonium
chloride (DOTMA); N,N-distearyl-N,N-dimethylammonium bromide (DDAB); N-(2,3-
dioleoyloxy)propy1)-N,N,N-trimethylammonium chloride (DOTAP); 3-(N¨(N',N1-
dimethylaminoethane)-carbamoyl)cholesterol (DC-Chol), N-(1-(2,3-
dioleoyloxy)propy1)-N-2-
(sperminecarboxamido)ethyl)-N,N-dimethylammonium trifluoracetate (DOSPA),
dioctadecylamidoglycyl carboxyspermine (DOGS), 1,2-dioleoy1-3-dimethylammonium
propane
(DODAP), N,N-dimethy1-2,3-dioleoyloxy)propylamine (DODMA), and N-(1,2-
dimyristyloxyprop-3-y1)-N,N-dimethyl-N-hydroxy-ethyl ammonium bromide (DMR1E).
Additionally, a number of commercial preparations of cationic lipids are
available which can be
used in the present invention. These include, for example, LIPOFECTIN
(commercially
available cationic liposomes comprising DOTMA and 1,2-dioleoyl-sn-3-
phosphoethanolamine
(DOPE), from GIBCO/BRL, Grand Island, N.Y.); LIPOFECTAMINER (commercially
available
cationic liposomes comprising N-(1-(2,3-dioleyloxy)propy1)-N-(2-
(sperminecarboxamido)ethyl)-
N,N-dimethylammonium trifluoroacetate (DOSPA) and (DOPE), from GIBCO/BRL); and
TRANSFECTAM (commercially available cationic lipids comprising
dioctadecylamidoglycyl
carboxyspennine (DOGS) in ethanol from Promega Corp., Madison, Wis.). The
following lipids
are cationic and have a positive charge at below physiological pH: DODAP,
DODMA, DMDMA,
1,2-dilinoleyloxy-N,N-dimethylaminopropane (DLinDMA), 1,2-dilinolenyloxy-N,N-
dimethylaminopropane (DLenDMA).
49
CA 03195281 2023- 4- 11
WO 2022/081702
PCT/US2021/054775
In one embodiment, the cationic lipid is an amino lipid. Suitable amino lipids
useful in
the invention include those described in WO 2012/016184, incorporated herein
by reference in its
entirety. Representative amino lipids include, but are not limited to, 1,2-
dilinoleyoxy-3-
(dimethylamino)acetoxypropane (DLin-DAC), 1,2-dilinoleyoxy-3-morpholinopropane
(DLin-
MA), 1,2-dilinoleoy1-3-dimethylaminopropane (DLinDAP), 1,2-dilinoleylthio-3-
dimethylaminopropane (DLin-S-DMA), 1-linoleoy1-2-linoleyloxy-3-
dimethylaminopropane
(DLin-2-DMAP), 1,2-dilinoleyloxy-3-trimethylaminopropane chloride salt (DLin-
TMA.C1), 1,2-
dilinoleoy1-3-trimethylaminopropane chloride salt (DLin-TAP.C1), 1,2-
dilinoleyloxy-3-(N-
methylpiperazino)propane (DLin-MPZ), 3-(N,N-dilinoleylamino)-1,2-propanediol
(DLinAP), 3-
(N,N-dioleylamino)-1,2-propanediol (DOAP), 1,2-dilinoleyloxo-3-(2-N,N-
dimethylamino)ethoxypropane (DLin-EG-DMA), and 2,2-dilinoley1-4-
dimethylaminomethyl-
[1,31-dioxolane (DLin-K-DMA).
Suitable amino lipids include those having the formula:
re.1:7 1:1
R4 ¨N CH217 -i-
t
R3 'N*7 -6
wherein Ri and R2 are either the same or different and independently
optionally
substituted Cio-C24alkyl, optionally substituted Cio-C24alkenyl, optionally
substituted Cui-C24
alkynyl, or optionally substituted C10-C24acyl;
R3 and R4 are either the same or different and independently optionally
substituted C1-C6
alkyl, optionally substituted C2-C6alkenyl, or optionally substituted C2-
C6alkynyl or R3 and R4
may join to form an optionally substituted heterocyclic ring of 4 to 6 carbon
atoms and 1 or 2
heteroatoms chosen from nitrogen and oxygen;
R5 is either absent or present and when present is hydrogen or C1-C6 alkyl;
m, n, and p are either the same or different and independently either 0 or 1
with the
proviso that m, n, and p are not simultaneously 0;
q is 0, 1,2, 3, or 4; and
Y and Z are either the same or different and independently 0, S. or NH.
In one embodiment, Ri and R2 are each linoleyl, and the amino lipid is a
dilinoleyl amino
lipid. In one embodiment, the amino lipid is a dilinoleyl amino lipid.
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
A representative useful dilinoley1 amino lipid has the formula:
(C112-1:r--C) (CII-2)5
Min-IC-DMA
wherein n is 0, 1, 2, 3, or 4.
In one embodiment, the cationic lipid is a DLin-K-DMA. In one embodiment, the
cationic lipid is DLin-KC2-DMA (DLin-K-DMA above, wherein n is 2).
In one embodiment, the cationic lipid component of the LNPs has the structure
of
Formula (I):
R1 a R2a R3a R4a
J-k /H\ J-.)\
R5 'a Li b N C L2 d
Rib R2b R3b R4b
R7 eN R5
R9
(1)
or a pharmaceutically acceptable salt, tautomer, prodrug or stereoisomer
thereof,
wherein:
Li and L2 are each independently -0(C=0)-, -(C=0)0- or a carbon-carbon double
bond;
Ria and Rib are, at each occurrence, independently either (a) H or Ci-C12
alkyl, or (b) Rh
is H or Ci-C12 alkyl, and Rib together with the carbon atom to which it is
bound is taken together
with an adjacent Rib and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
R2a and R2b are, at each occurrence, independently either (a) H or C i-C12
alkyl, or (b) R2a
is H or CI-Cu alkyl, and R2b together with the carbon atom to which it is
bound is taken together
with an adjacent R2b and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
51
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
R38 and R3b are, at each occurrence, independently either (a) H or C1-C12
alkyl, or (b) R38
is H or Ci-C12 alkyl, and R3b together with the carbon atom to which it is
bound is taken together
with an adjacent R3b and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
R4a and R4b are, at each occurrence, independently either (a) H or CI-Cu
alkyl, or (b) R4a
is H or C1-C12 alkyl, and R4b together with the carbon atom to which it is
bound is taken together
with an adjacent R4b and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
R8 and le are each independently methyl or cycloalkyl;
R7 is, at each occurrence, independently H or Ci-C12 alkyl;
R8 and R9 are each independently C1-C12 alkyl; or R8 and R9, together with the
nitrogen
atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring
comprising one
nitrogen atom;
a and d are each independently an integer from 0 to 24;
b and c are each independently an integer from 1 to 24; and
e is 1 or 2.
a
In certain embodiments of Formula (I), at least one of R", ¨2, R8a or R4a is
C1-C12 alkyl,
or at least one of Li or L2 is ¨0(C=0)- or ¨(C=0)0-. In other embodiments, Ria
and Rib are not
isopropyl when a is 6 or n-butyl when a is 8.
¨a
In still further embodiments of Formula (I), at least one of Ria, K2, R3a or
R48 is Ci-C12
alkyl, or at least one of L' or L2 is -0(C=0)- or -(C=0)0-; and
Ria and Rib are not isopropyl when a is 6 or n-butyl when a is 8.
In other embodiments of Formula (1), R8 and R9 are each independently
unsubstituted C1-
C12 alkyl; or R8 and R9, together with the nitrogen atom to which they arc
attached, form a 5, 6 or
7-membered heterocyclic ring comprising one nitrogen atom;
In certain embodiments of Formula (1), any one of Li or L2 may be -0(C=0)- or
a
carbon-carbon double bond. Li and L2 may each be -0(C=0)- or may each be a
carbon-carbon
double bond.
In some embodiments of Formula (I), one of Li or L2 is -0(C=0)-. In other
embodiments,
both Li and L2 are -0(C=0)-.
52
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In some embodiments of Formula (I), one of L' or L2 is -(C=0)0-. In other
embodiments,
both Li and L2 are -(C=0)0-.
In some other embodiments of Formula (1), one of L' or L2 is a carbon-carbon
double
bond. In other embodiments, both L' and L2 are a carbon-carbon double bond.
In still other embodiments of Formula (I), one of L' or L2 is -0(C=0)- and the
other of L'
or L2 is -(C=0)0-. In more embodiments, one of L' or L2 is -0(C=0)- and the
other of L' or L2 is
a carbon-carbon double bond. In yet more embodiments, one of Li or L2 is -
(C=0)0- and the
other of L' or L2 is a carbon-carbon double bond.
It is understood that "carbon-carbon" double bond, as used throughout the
specification,
refers to one of the following structures:
Rb
Ra Rb \
"71q" jsrsjj' \ or Ra
wherein Ra and Rb are, at each occurrence, independently H or a substituent.
For
example, in some embodiments Ra and Rb are, at each occurrence, independently
H, C1-C12 alkyl
or cycloalkyl, for example H or Ci-C12 alkyl.
In other embodiments, the lipid compounds of Formula (I) have the following
structure
(Ia):
R1 a R2a R3a R4a
R6a
Rib R2b R3b R4b
R7 e
R9
(Ia)
In other embodiments, the lipid compounds of Formula (I) have the following
structure
(Ib):
53
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0 R2a R3a
R1a R4a
IR5a N 0E.yR6a
a R2b R3b
Rib jckR R4b
R7 e
R9
(Ib)
In yet other embodiments, the lipid compounds of Formula (I) have the
following
structure (lc):
R2a R3a
RI a R4a
5a
R
a R2b R3b
Rib 0 0 R4ID
R7 e 8
R
R9
(Ic)
In certain embodiments of the lipid compound of Formula (I), a, b, c and d are
each
independently an integer from 2 to 12 or an integer from 4 to 12. In other
embodiments, a, b, c
and d are each independently an integer from 8 to 12 or 5 to 9. In some
certain embodiments, a is
0. In some embodiments, a is 1. In other embodiments, a is 2. In more
embodiments, a is 3. In yet
other embodiments, a is 4. In some embodiments, a is 5. In other embodiments,
a is 6. In more
embodiments, a is 7. In yet other embodiments, a is 8. In some embodiments, a
is 9. In other
embodiments, a is 10. In more embodiments, a is 11. In yet other embodiments,
a is 12. In some
embodiments, a is 13. In other embodiments, a is 14. In more embodiments, a is
15. In yet other
embodiments, a is 16.
In some other embodiments of Formula (I), b is 1. In other embodiments, b is
2. In more
embodiments, b is 3. In yet other embodiments, b is 4. In some embodiments, b
is 5. In other
embodiments, b is 6. In more embodiments, b is 7. In yet other embodiments, b
is 8. In some
embodiments, b is 9. In other embodiments, b is 10. In more embodiments, b is
11. In yet other
embodiments, b is 12. In some embodiments, b is 13. In other embodiments, b is
14. In more
embodiments, b is 15. In yet other embodiments, b is 16.
54
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In some more embodiments of Formula (I), c is 1. In other embodiments, c is 2.
In more
embodiments, c is 3. In yet other embodiments, c is 4. In some embodiments, c
is 5. In other
embodiments, c is 6. In more embodiments, c is 7. In yet other embodiments, c
is 8. In some
embodiments, c is 9. In other embodiments, c is 10. In more embodiments, c is
11. In yet other
embodiments, c is 12. In some embodiments, c is 13. In other embodiments, c is
14. In more
embodiments, c is 15. In yet other embodiments, c is 16.
In some certain other embodiments of Formula (I), d is 0. In some embodiments,
d is 1.
In other embodiments, d is 2. In more embodiments, d is 3. In yet other
embodiments, d is 4. In
some embodiments, d is 5. In other embodiments, d is 6. In more embodiments, d
is 7. In yet
other embodiments, d is 8. In some embodiments, d is 9. In other embodiments,
d is 10. In more
embodiments, d is 11. In yet other embodiments, d is 12. In some embodiments,
d is 13. In other
embodiments, d is 14. In more embodiments, d is 15. In yet other embodiments,
d is 16.
In some other various embodiments of Formula (1), a and dare the same. In some
other
embodiments, b and c are the same. In some other specific embodiments, a and d
are the same
and b and care the same.
The sum of a and b and the sum of c and d in Formula (I) are factors which may
be varied
to obtain a lipid of Formula (I) having the desired properties. In one
embodiment, a and b are
chosen such that their sum is an integer ranging from 14 to 24. In other
embodiments, c and d are
chosen such that their sum is an integer ranging from 14 to 24. In further
embodiment, the sum of
a and b and the sum of c and d are the same. For example, in some embodiments
the sum of a and
b and the sum of c and d are both the same integer which may range from 14 to
24. In still more
embodiments, a. b, c and d are selected such the sum of a and band the sum of
c and d is 12 or
greater.
In some embodiments of Formula (I), c is 1. In other embodiments, c is 2.
The substituents at R la, R2a, R3a and R4a of Fomiula (1) are not particularly
limited. In
certain embodiments RIG, R2a, R3a and R4a are H at each occurrence. In certain
other embodiments
at least one of Rla, R2a, R3a and Ria is L -I-
C12 alkyl. In certain other embodiments at least one of
R2a, R3a and R4a is C1-05
alkyl. In certain other embodiments at least one of Rla, R2a, 3a and
R4a is C1-C6
alkyl. In some of the foregoing embodiments, the CI-Cs alkyl is methyl, ethyl,
n-
propyl, iso-propyl, n-butyl, iso-butyl, tert-butyl, n-hexyl or n-octyl.
CA 03195281 2023-4-11
WO 2022/081702
PCT/US2021/054775
In certain embodiments of Formula (I), Ria, Rib, Raa and Rab are u ¨1-
Co alkyl at each
occurrence.
In further embodiments of Formula (1), at least one of Rib, R2b, R3b and K.-
.41)
is H or Rib,
R2b, R3b and R4b are H at each occurrence.
In certain embodiments of Formula (I), Rib together with the carbon atom to
which it is
bound is taken together with an adjacent Rib and the carbon atom to which it
is bound to form a
carbon-carbon double bond. In other embodiments of the foregoing R4b together
with the carbon
atom to which it is bound is taken together with an adjacent R41 and the
carbon atom to which it is
bound to form a carbon-carbon double bond.
The substituents at R8 and R6 of Formula (I) are not particularly limited in
the foregoing
embodiments. In certain embodiments one or both of R8 or R6 is methyl. In
certain other
embodiments one or both of R5 or R6 is cycloalkyl for example cyclohexyl. In
these embodiments
the cycloalkyl may be substituted or not substituted. In certain other
embodiments the cycloalkyl
is substituted with CI-C12 alkyl, for example tert-butyl.
The sub stituents at R7 are not particularly limited in the foregoing
embodiments of
Formula (1). In certain embodiments at least one R7 is H. In some other
embodiments, R7 is H at
each occurrence. In certain other embodiments R7 is CI-C12 alkyl.
In certain other of the foregoing embodiments of Formula (I), one of R8 or R9
is methyl.
In other embodiments, both R8 and R9 are methyl.
In some different embodiments of Formula (I), R8 and R9, together with the
nitrogen
atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring.
In some
embodiments of the foregoing, R8 and R9, together with the nitrogen atom to
which they are
attached, form a 5-membered heterocyclic ring, for example a pyrrolidinyl
ring.
In various different embodiments, exemplary lipid of Formula (1) can include
56
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0
-y0
0
0 0
N
HT,0
0
0
0
N
0
o
0
0
0
N
0
57
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0 0
0...T.,
0
0
0
0
0 0
0
0
0 0
0
0
0 O--N.,/\./
0
0r_0_________
0
0
0
58
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
N N
0
0
0
N N
0
0
0 0 n
N N
0 213(C.
0
0
N N
0
0
0
N N
0
0
0 0
N N
0
0
N N
0
59
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0
0
I o0
0
0
0 0
0
0
0
0
0
0
0
I o0
NWy
0
0
0
0
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0
0
0 0
0
0
0
0
NrC)
H,Ir0
0
0
./W
0 N N
0
0
0
N N 0
0
0
61
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
o
0
0
0
0 0
N
o
0
N N
j(D
0
0
0
0
0
N N 0
0
0
62
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0
0
w 0
In some embodiments, the LNPs comprise a lipid of Formula (I), at least one
agent, and
one or more excipients selected from neutral lipids, steroids and pegylated
lipids. In some
embodiments the lipid of Formula (I) is compound 1-5. In some embodiments the
lipid of
Formula (I) is compound 1-6.
In some other embodiments, the cationic lipid component of the LNPs has the
structure
of Formula (II):
Ri a R2a R3a R4a
R5 a Ll b ft'L2 R6
Rib R2b R3b R4b
G1
-R7
R5
R9
(11)
or a pharmaceutically acceptable salt, tautomer, prodrug or stereoisomer
thereof,
wherein:
Li and L2 are each independently -0(C=0)-, -(C=0)0-, -C(=0)-, -0-,
- , S S C(=0)S-, -SC(=0)-, -NRaC(=0)-, -C(=0)NRa-, -NRaC(=0)NRa,
-0C(=0)NR3-, -NR3C(=0)0-, or a direct bond;
GI is Ci-C2 alkylene, -(C=0)-, -0(C=0)-, -SC(=0)-, -NRaC(=0)- or a direct
bond;
G2 is -C(=0)-, -(C=0)0-, -C(=0)S-, -C(=0)NRa or a direct bond;
G3 is CI-C6 alkylene;
Ra is H or C1-C12 alkyl;
RI' and Rib are, at each occurrence, independently either: (a) H or C1-C12
alkyl; or (b)
is H or Ci-C12 alkyl, and Rib together with the carbon atom to which it is
bound is taken together
63
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
with an adjacent Rib and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
R2 and R2b arc, at each occurrence, independently- either: (a) H or Ci-C12
alkyl; or (b) R2'
is H or CI-Cu alkyl, and R2b together with the carbon atom to which it is
bound is taken together
with an adjacent R2b and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
R3 a and 113b are, at each occurrence, independently either: (a) H or CI-C12
alkyl; or (b)
is H or CI-Cu alkyl, and R3b together with the carbon atom to which it is
bound is taken together
with an adjacent Feb and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
Wia and R46 are, at each occurrence, independently either: (a) H or C1-C12
alkyl; or (b)
is H or CI-Cu alkyl, and R4b together with the carbon atom to which it is
bound is taken together
with an adjacent R4b and the carbon atom to which it is bound to form a carbon-
carbon double
bond;
R5 and R6 are each independently H or methyl;
R7 is C4-C20 alkyl;
R5 and R9 are each independently CI-Cu alkyl; or R5 and R9, together with the
nitrogen
atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring;
a, b, c and d are each independently an integer from 1 to 24; and
x is 0, 1 or 2.
In some embodiments of Formula (II), L' and L2 are each independently
¨0(C=0)-, -(C=0)0- or a direct bond. In other embodiments, Gi and G2 are each
independently -(C=0)- or a direct bond. In some different embodiments, Li and
L2 are each
independently ¨0(C=0)-, -(C=0)0- or a direct bond; and Gi and G2 are each
independently ¨
(C=0)- or a direct bond.
In some different embodiments of Formula (II), Li and L2 are each
independently -C(=0)-, -0-, -S(0)2,-, -S-S-, -C(=0)S-, -SC(=0)-, -NRa-, -
NRaC(=0)-,
-C(=0)NRa-, -NWC(=0)NRa, 40C(=0)NRa-, -NRaC(=0)0-, -NWS(0),NRa-,
-NR3S(0)õ- or -S(0)õNR3-.
64
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In other of the foregoing embodiments of Formula (II), the lipid compound has
one of the
following structures (IA) or (JIB):
R1 a R2a R3a R4a
R1 a R2a R3a R4a
R5 Li L24 R6
444
Rib R2b R3b R4b
R5 Li L2 R6 R7
Rib R2b R3b R4b ON
,N
G3 R7
0
R9 R8 or R8
(IA) (IIB)
In some embodiments of Formula (II), the lipid compound has structure (IA). In
other
embodiments, the lipid compound has structure (IIB).
In any of the foregoing embodiments of Formula (II), one of Li or L2 is -
0(C=0)-. For
example, in some embodiments each of Li and L2 are -0(C=0)-.
In some different embodiments of Formula (II), one of Li or L2 is -(C=0)0-.
For
example, in some embodiments each of Li and L2 is -(C=0)0-.
In different embodiments of Formula (II), one of Li or L2 is a direct bond. As
used
herein, a "direct bond" means the group (e.g., Li or L2) is absent. For
example, in some
embodiments each of Li and L2 is a direct bond.
In other different embodiments of Formula (II), for at least one occurrence of
R' and Rib,
Ria is H or C1-C12 alkyl, and Rib together with the carbon atom to which it is
bound is taken
together with an adjacent Rib and the carbon atom to which it is bound to form
a carbon-carbon
double bond.
In still other different embodiments of Formula (II), for at least one
occurrence of R' and
4b
K Wia is H or C1-C12 alkyl, and R4b together with the carbon
atom to which it is bound is taken
together with an adjacent R4b and the carbon atom to which it is bound to form
a carbon-carbon
double bond.
In more embodiments of Formula (II), for at least one occurrence of R23 and
R2b, R2a is H
or Ci-C12 alkyl, and R2b together with the carbon atom to which it is bound is
taken together with
an adjacent R2b and the carbon atom to which it is bound to form a carbon-
carbon double bond.
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In other different embodiments of Formula (II), for at least one occurrence of
R3a and R3b,
R3 is H or C1-C12 alkyl, and R3b together with the carbon atom to which it is
bound is taken
together with an adjacent R3b and the carbon atom to which it is bound to form
a carbon-carbon
double bond.
In various other embodiments of Formula (II), the lipid compound has one of
the
following structures (IIC) or (IID):
Rla R2a R3a R4a
R5 e g
R1 b R2b R3b R4b
,N
G3- R7
0
R9 R8 or
(TIC)
R 1 a R2a R3a R4a
R5 e
g
h R6
Rib R2b R3b R4b
ON
R9 /G3
R6
(IID)
wherein e, f, g and h are each independently an integer from 1 to 12.
In some embodiments of Formula (11), the lipid compound has structure (TIC).
In other
embodiments, the lipid compound has structure (IID).
In various embodiments of structures (TIC) or (IID), e, f, g and h are each
independently
an integer from 4 to 10.
In certain embodiments of Formula (II), a, b, c and d are each independently
an integer
from 2 to 12 or an integer from 4 to 12. In other embodiments, a, b, c and d
are each
independently an integer from 8 to 12 or 5 to 9. In some certain embodiments,
a is 0. In some
embodiments, a is 1. In other embodiments, a is 2. In more embodiments, a is
3. In yet other
embodiments, a is 4. In some embodiments, a is 5. In other embodiments, a is
6. In more
66
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
embodiments, a is 7. In yet other embodiments, a is 8. In some embodiments, a
is 9. In other
embodiments, a is 10. In more embodiments, a is 11. In yet other embodiments,
a is 12. In some
embodiments, a is 13. In other embodiments, a is 14. In more embodiments, a is
15. In yet other
embodiments, a is 16.
In some embodiments of Formula (II), b is 1. In other embodiments, b is 2. In
more
embodiments, b is 3. In yet other embodiments, b is 4. In some embodiments, b
is 5. In other
embodiments, b is 6. In more embodiments, b is 7. In yet other embodiments, b
is 8. In some
embodiments, b is 9. In other embodiments, b is 10. In more embodiments, b is
11. In yet other
embodiments, b is 12. In some embodiments, b is 13. In other embodiments, b is
14. In more
embodiments, b is 15. In yet other embodiments, b is 16.
In some embodiments of Formula (II), c is 1. In other embodiments, c is 2. In
more
embodiments, c is 3. In yet other embodiments, c is 4. In some embodiments, c
is 5. In other
embodiments, c is 6. In more embodiments, c is 7. In yet other embodiments, c
is 8. In some
embodiments, c is 9. In other embodiments, c is 10. In more embodiments, c is
11. In yet other
embodiments, c is 12. In some embodiments, c is 13. In other embodiments, c is
14. In more
embodiments, c is 15. In yet other embodiments, c is 16.
In some certain embodiments of Formula (II), d is 0. In some embodiments, d is
1. In
other embodiments, d is 2. In more embodiments, d is 3. In yet other
embodiments, d is 4. In
some embodiments, d is 5. In other embodiments, d is 6. In more embodiments, d
is 7. In yet
other embodiments, d is 8. In some embodiments, d is 9. In other embodiments,
d is 10. In more
embodiments, d is 11. In yet other embodiments, d is 12. In some embodiments,
d is 13. In other
embodiments, d is 14. In more embodiments, d is 15. In yet other embodiments,
d is 16.
In some embodiments of Formula (II), e is 1. In other embodiments, e is 2. In
more
embodiments, c is 3. In yet other embodiments, c is 4. In some embodiments, c
is 5. In other
embodiments, e is 6. In more embodiments, e is 7. In yet other embodiments, e
is 8. In some
embodiments, e is 9. In other embodiments, e is 10. in more embodiments, e is
11. In vet other
embodiments, e is 12.
In some embodiments of Formula (II), f is 1. In other embodiments, f is 2. In
more
embodiments, f is 3. In yet other embodiments, f is 4. In some embodiments, f
is 5. In other
embodiments, f is 6. In more embodiments, f is 7. In yet other embodiments, f
is 8. In some
67
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
embodiments, f is 9. In other embodiments, f is 10. In more embodiments, f is
11. In yet other
embodiments, f is 12.
In some embodiments of Formula (11), g is 1. In other embodiments, g is 2. In
more
embodiments, g is 3. In yet other embodiments, g is 4. In some embodiments, g
is 5. In other
embodiments, g is 6. In more embodiments, g is 7. In yet other embodiments, g
is 8. In some
embodiments, g is 9. In other embodiments, g is 10. In more embodiments, g is
11. In yet other
embodiments, g is 12.
In some embodiments of Formula (II), h is I. In other embodiments, e is 2. In
more
embodiments, h is 3. In yet other embodiments, h is 4. In some embodiments, e
is 5. In other
embodiments, h is 6. In more embodiments, h is 7. In yet other embodiments, h
is 8. In some
embodiments, h is 9. In other embodiments, h is 10. In more embodiments, h is
11. In yet other
embodiments, h is 12.
In some other various embodiments of Formula (11), a and d are the same. In
some other
embodiments, b and c are the same. In some other specific embodiments and a
and d are the same
and band care the same.
The sum of a and b and the sum of c and d of Formula (II) are factors which
may be
varied to obtain a lipid having the desired properties. In one embodiment, a
and b are chosen such
that their sum is an integer ranging from 14 to 24. In other embodiments, c
and d are chosen such
that their sum is an integer ranging from 14 to 24. In further embodiment, the
sum of a and b and
the sum of c and d are the same. For example, in some embodiments the sum of a
and b and the
sum of c and dare both the same integer which may range from 14 to 24. In
still more
embodiments, a. b, c and d are selected such that the sum of a and b and the
sum of c and d is 12
or greater.
The substituents at R1a, R2a, R3a and ¨4a
of Formula (11) are not particularly limited. In
sonic embodiments, at least one of R R2a, R3a and R4a
is H. In certain embodiments Ria, 2R a, R3a
and R4a are H at each occurrence. In certain other embodiments at least one of
R R2a, R3a and
R4a is Cl-C12 alkyl. In certain other embodiments at least one of lea, R2a,
R3a and R4a is ci-C8
alkyl. In certain other embodiments at least one of Rla, R2a, R3a and R4a is C
-Cfi alkyl. In some of
the foregoing embodiments, the CI-Cs alkyl is methyl, ethyl, n-propyl, iso-
propyl, n-butyl, iso-
butyl, tert-butyl, n-hexyl or n-octyl.
68
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In certain embodiments of Formula (II), Rh, R, R4a and Rib are t ¨i-
Cp alkyl at each
occurrence.
In further embodiments of Formula (11), at least one of Rib, R21, R3b and R41
is H or Rib,
R2b, R3b and R4b are H at each occurrence.
In certain embodiments of Formula (II), Rib together with the carbon atom to
which it is
bound is taken together with an adjacent Rib and the carbon atom to which it
is bound to form a
carbon-carbon double bond. In other embodiments of the foregoing R4b together
with the carbon
atom to which it is bound is taken together with an adjacent R41 and the
carbon atom to which it is
bound to form a carbon-carbon double bond.
The sub stituents at R5 and R6 of Formula (II) are not particularly limited in
the foregoing
embodiments. In certain embodiments one of R5 or R6 is methyl. In other
embodiments each of
R5 or R6 is methyl.
The sub stituents at R7 of Formula (11) are not particularly limited in the
foregoing
embodiments. In certain embodiments leis C6-C16 alkyl. In some other
embodiments, R7 is C6-C9
alkyl. In some of these embodiments, R7 is substituted with -(C=0)0Rb, ¨
0(C=0)Rb, -C(=0)Rb, -ORb, -S(0)xRb, -S-SRb, -C(=0)SRb,
-SC(=0)Rb, -NRaRb, -NRaC(=0)Rb, -C(=0)NRaRb, -NRaC(=0)NRaRb,
-0C(=0)NRaRb, -NR1C(=0)0Rb, -NWS(0)xNR1Rb, -N WS(0)R" or -S(0)õ1\11taRb,
wherein: Ra is H or CI-Cu alkyl; Rb is Ci-C15 alkyl; and xis 0, 1 or 2. For
example, in some
embodiments R7 is substituted with -(C=0)0Rb or ¨O(CO)R'.
In various of the foregoing embodiments of Formula (II), Rh is branched C-Cs
alkyl. For
example, in some embodiments Rb has one of the following structures:
or
)z,W
=
69
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In certain other of the foregoing embodiments of Formula (II), one of R8 or R9
is methyl.
In other embodiments, both R8 and R9 are methyl.
In some different embodiments of Formula (11), R8 and R9, together with the
nitrogen
atom to which they are attached, form a 5, 6 or 7-membered heterocyclic ring.
In some
embodiments of the foregoing, R8 and R9, together with the nitrogen atom to
which they are
attached, form a 5-membered heterocyclic ring, for example a pyrrolidinyl
ring. In some different
embodiments of the foregoing, R8 and R9, together with the nitrogen atom to
which they are
attached, form a 6-membered heterocyclic ring, for example a piperazinyl ring.
In still other embodiments of the foregoing lipids of Formula (II), G-3 is C2-
C4 alkylene,
for example C3 alkylene.
In various different embodiments, the lipid compound has one of the following
structures:
N N
I
N N
0
N N
0
0
CA 03195281 2023-4- 11
WO 2022/081702
PC T/US2021/054775
0
0
N N
0
N N -
0
0
N
0
0
0
0
N N
0 0
0
0
0
0 0
N N 0
W.,
0 0
0
0
N N
0
0-NO
71
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0
-y0
0
0 /\./"\-/
0
0
-y0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
0
72
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0 0
0
0
0
0
0
0
0
0
0
0
0-- -0
0
0 0
0 0
0
0
0 0
73
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0
0
0 0
0
0
0
0
0
0 0
0
0 0
0 0
0 0
ON N
0
74
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0 0
0
0
0
In some embodiments, the LNPs comprise a lipid of Formula (II), at least one
agent, and
one or more excipient selected from neutral lipids, steroids and pegylated
lipids, in some
embodiments, the lipid of Formula (II) is compound 11-9. In some embodiments,
the lipid of
Formula (II) is compound II-10. In some embodiments, the lipid of Formula (II)
is compound II-
11. In some embodiments, the lipid of Formula (II) is compound 11-12. In some
embodiments, the
lipid of Formula (II) is compound 11-32.
In some other embodiments, the cationic lipid component of the LNPs has the
structure
of Formula (III):
R3,
G3
ii
1_2õ,
1- -le- -R2
R G
(III)
or a pharmaceutically acceptable salt, tautomer, prodrug or stereoisomer
thereof,
wherein:
one of L' or L2 is ¨0(C=0)-, -(C=0)0-, -C(=0)-, -0-, -S(0)õ-, -S-S-,
-C(=0)S-, SC(=0)-, -NRaC(=0)-, -C(=0)NRa-, NRaC(=0)NW-, -0C(=0)NRa- or
-NWC(-0)0-, and the other of L' or L2 is ¨0(C=0)-, -(C=0)0-, -C(=0)-, -0-, -
S(0)-,
-S-S-, -C(=0)S-, SC(=0)-, -NWC(=0)-, -C(=0)NW-õNWC(=0)NW-, -0C(=0)NW- or
-NRaC(=0)0- or a direct bond;
GI and G2 are each independently unsubstituted Ci-C12 alkylene or Ci-C12
alkenylene,
G3 is Ci-C24 alkylene, Ci-C24 alkenylene, C3-C8 cycloalkylene, C3-C8
cycloalkenylene;
W is H or Ci-C12 alkyl;
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
RI and R2 are each independently C6-C24 alkyl or C6-C24 alkenyl;
R3 is H, OR5, CN, -C(=0)0R4, -0C(=0)R4 or ¨NR5C(=0)R4;
R4 is C1-C12 alkyl;
R5 is H or C1-C6 alkyl; and
x is 0, 1 or 2.
In some of the foregoing embodiments of Formula (III), the lipid has one of
the following
structures (IIIA) or (IIIB):
R3 R6
R3 R6 A
,L2
R1G1G2R2 or R" -G1-- -G2 -R2
(IIIA) (IIIB)
wherein:
A is a 3 to 8-membered cycloalkyl or cycloalkylene ring;
R6 is, at each occurrence, independently H, OH or CI-C24 alkyl;
n is an integer ranging from I to 15.
In some of the foregoing embodiments of Formula (III), the lipid has structure
(IIIA), and
in other embodiments, the lipid has structure (MB).
In other embodiments of Formula (III), the lipid has one of the following
structures (IIIC)
or (IIID):
R3 R6
R3.y R6 A
R1 N Li L 2 L2
NõY'
ly iz = ,y = /z
or N R2
(IIIC) (IIID)
wherein y and z are each independently integers ranging from 1 to 12.
In any of the foregoing embodiments of Formula (III), one of 1_,1 or L2 is -
0(C=0)-. For
example, in some embodiments each of L' and L2 are -0(C=0)-. In some different
embodiments
of any of the foregoing, L1 and L2 are each independently -(C=0)0- or -0(C=0)-
. For example,
in some embodiments each of LI and L2 is -(C=0)0-.
76
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In some different embodiments of Formula (III), the lipid has one of the
following
structures (IIIE) or (IIIF):
R3
I R3
-... 3
R1 0 ,N 0 0 G
0
G1 G2R2
I
0 0
0 G1 G2 0
or
.
(IIIE) (IIIF)
In some of the foregoing embodiments of Formula (III), the lipid has one of
the following
structures (IIIG), (11TH), (IIII), or (IIIJ):
R3 R6
n R3 R6
1 0 N 0 0 -
ii:-; 0
R R2
\/-
y R1
....,...---,....4,..N..i.....y.:=-= /R2
0
0
0 0 = Y
-
'
n
(JIG) (11TH)
R3 R6
A R3 R6
A
0 0
R1 0 N 0
\/ R2
y or N R1
'0-'("r -.. R2
0 0 Y
.
(IIII) (IIIJ)
In some of the foregoing embodiments of Formula (111), n is an integer ranging
from 2 to
12, for example from 2 to 8 or from 2 to 4. For example, in some embodiments,
n is 3, 4, 5 or 6.
In some embodiments, n is 3. In some embodiments, n is 4. In some embodiments,
n is 5. In some
embodiments, n is 6.
In some other of the foregoing embodiments of Formula (III), y and z are each
independently an integer ranging from 2 to 10. For example, in some
embodiments, y and z are
each independently an integer ranging from 4 to 9 or from 4 to 6.
In some of the foregoing embodiments of Formula (III), R6 is H. In other of
the foregoing
embodiments, R6 is C1-C24 alkyl. In other embodiments, R6 is OH.
77
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In some embodiments of Formula (III), G3 is unsubstituted. In other
embodiments, G3 is
substituted. In various different embodiments, G3 is linear C1-C24 alkylene or
linear C1-C24
alkenylcne.
In some other foregoing embodiments of Formula (III), RI or R2, or both, is C6-
C24
alkenyl. For example, in some embodiments, R' and R2 each, independently have
the following
structure:
R7a
H ____________________________________________________
a
R7b
wherein:
R7a and RTh are, at each occurrence, independently H or (211212 alkyl; and
a is an integer from 2 to 12,
wherein R7a, RTh and a arc each selected such that RI and R2 each
independently comprisc
from 6 to 20 carbon atoms. For example, in some embodiments a is an integer
ranging from 5 to 9
or from 8 to 12.
In some of the foregoing embodiments of Formula (III), at least one occurrence
of R7a is
H. For example, in some embodiments, R7a is H at each occurrence. In other
different
embodiments of the foregoing, at least one occurrence offeb is Ci-C8 alkyl.
For example, in some
embodiments, C1-C8 alkyl is methyl, ethyl, n-propyl, iso-propyl, n-butyl, iso-
butyl, tert-butyl, n-
hexyl or n-octyl.
In different embodiments of Formula (III). RI or R2, or both, has one of the
following
structures:
.siss
= ')22.
78
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In some of the foregoing embodiments of Formula (III), 12_3 is OH,
CN, -C(=0)0R4, -0C(=0)R4 or ¨NHC(=0)R4. In some embodiments, R4 is methyl or
ethyl.
In various different embodiments, the cationic lipid of Formula (Ill) has one
of the
following structures:
HLcc
0o
0
0
0
H 0
0
HO
0
0
0
HO
0
CO
0
H 0
0
0
0
HO NJ
0
79
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
HO N
0
0
HOOO
0
HON
HO
0 H 0
0
0
0
0
N 0
0
HO N
0
0
0 0
Lo
0
H _
0
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
HO
0
HO
,y0
0
0
0
HO N
oOOC
0
H 0
0
0
H NO
0
0
0 H
0
0
0
0
H 0
LI,Two 0
81
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
0
0
H 0N
0
0
0
0
H 0 N
0
0
H NO
0
0
HO N0
OO
0
0
0
0
HO
0
0
0 H 0
0
cO
82
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
H O No
fl
0
cc
H
H 0
JiiIiiiXiO
0
OO
0 0
0
0
N o
0 0
0
OO
NN
0
0
0 0
0
In some embodiments, the LNPs comprise a lipid of Formula (III), at least one
agent, and
one or more excipient selected from neutral lipids, steroids and pegylated
lipids. In some
embodiments, the lipid of Formula (III) is compound 111-3. In some
embodiments, the lipid of
Formula (111) is compound 111-7.
In certain embodiments, the cationic lipid is present in the LNP in an amount
from about
30 to about 95 mole percent. In one embodiment, the cationic lipid is present
in the LNP in an
83
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
amount from about 30 to about 70 mole percent. In one embodiment, the cationic
lipid is present
in the LNP in an amount from about 40 to about 60 mole percent. In one
embodiment, the
cationic lipid is present in the LNP in an amount of about 50 mole percent. In
one embodiment,
the LNP comprises only cationic lipids.
In certain embodiments, the LNP comprises one or more additional lipids which
stabilize
the formation of particles during their formation.
Suitable stabilizing lipids include neutral lipids and anionic lipids.
The term -neutral lipid" refers to any one of a number of lipid species that
exist in either
an uncharged or neutral zwitterionic form at physiological pH. Representative
neutral lipids
include diacylphosphatidylcholines, diacylphosphatidylethanolamines,
ceramides,
sphingomyelins, dihydro sphingomyelins, cephalins, and cerebrosides.
Exemplary neutral lipids include, for example, distearoylphosphatidylcholine
(DSPC),
dioleoylphosphatidylcholine (DOPC), dipalmitoylphosphatidylcholine (DPPC),
dioleoylphosphatidylglycerol (DOPG), dipalmitoylphosphatidylglycerol (DPPG),
dioleoyl-
phosphatidylethanolamine (DOPE), palmitoyloleoylphosphatidylcholine (POPC),
palmitoyloleoyl-phosphatidylethanolamine (POPE) and dioleoyl-
phosphatidylethanolamine 4-(N-
maleimidomethyl)-cyclohexane-1-carboxylate (DOPE-mal), dipalmitoylphosphatidyl
ethanolamine (DPPE), dimyristoylphosphoethanolamine (DMPE), distearoyl-
phosphatidylethanolamine (DSPE), 16-0-monomethyl PE, 16-0-dimethyl PE, 18-1-
trans PE, 1-
stearioy1-2-oleoyl-phosphatidyethanol amine (SOPE), and 1,2-dielaidoyl-sn-
glycero-3-
phophoethanolamine (transDOPE). In one embodiment, the neutral lipid is 1,2-
distearoyl-sn-
glycero-3-phosphocholine (DSPC).
In some embodiments, the LNPs comprise a neutral lipid selected from DSPC,
DPPC,
DMPC, DOPC, POPC, DOPE and SM. In various embodiments, the molar ratio of the
cationic
lipid (e.g., lipid of Formula (I)) to the neutral lipid ranges from about 2:1
to about 8:1.
In various embodiments, the LNPs further comprise a steroid or steroid
analogue. A
"steroid" is a compound comprising the following carbon skeleton:
84
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In certain embodiments, the steroid or steroid analogue is cholesterol. In
some of these
embodiments, the molar ratio of thc cationic lipid (e.g., lipid of Formula
(1)) to cholesterol ranges
from about 2:1 to 1:1.
The term "anionic lipid" refers to ally lipid that is negatively charged at
physiological pH.
These lipids include phosphatidylglycerol, cardiolipin,
diacylphosphatidylserine,
diacylphosphatidic acid, N-dodecanoylphosphatidylethanolamines, N-
succinylphosphatidylethanolamines, N-glutarylphosphatidylethanolamines,
lysylphosphatidylglycerols, palmitoyloleyolphosphatidylglycerol (POPG), and
other anionic
modifying groups joined to neutral lipids.
In certain embodiments, the LNP comprises glycolipids (e.g.,
monosialoganglioside
GM1). In certain embodiments, the LNP comprises a sterol, such as cholesterol.
In some embodiments, the LNPs comprise a polymer conjugated lipid. The term
-polymer conjugated lipid" refers to a molecule comprising both a lipid
portion and a polymer
portion. An example of a polymer conjugated lipid is a pegylated lipid. The
term "pegylated
lipid" refers to a molecule comprising both a lipid portion and a polyethylene
glycol portion.
Pegylated lipids are known in the art and include
1-(monomethoxy-polyethyleneglycol)-2,3-dimyristoylglycerol (PEG-s- DMG) and
the like.
In certain embodiments, the LNP comprises an additional, stabilizing -lipid
which is a
polyethylene glycol-lipid (pegylated lipid). Suitable polyethylene glycol-
lipids include PEG-
modified phosphatidylethanolamine, PEG-modified phosphatidic acid, PEG-
modified ceramides
(e.g., PEG-CerC14 or PEG-CerC20), PEG-modified dialkylamines, PEG-modified
diacylglycerols, PEG-modified dialkylglycerols. Representative polyethylene
glycol-lipids
include PEG-c-DOMG, PEG-c-DMA, and PEG-s-DMG. In one embodiment, the
polyethylene
glycol-lipid is N-[(methoxy poly(ethylene glycol)2000)carbamy11-1,2-
dimyristyloxlpropyl-3-amine
(PEG-c-DMA). In one embodiment, the polyethylene glycol-lipid is PEG-c-DOMG).
In other
embodiments, the LNPs comprise a pegylated diacylglyccrol (PEG-DAG) such as
1-(monomethoxy-polyethyleneglycol)-2,3-dimyristoylglycerol (PEG-DMG), a
pegylated
phosphatidylethanoloamine (PEG-PE), a PEG succinate diacylglycerol (PEG-S-DAG)
such as 4-
0-(2',3' -di(tetradecanoyloxy)propy1-1-0-(co-
methoxy(polyethoxy)ethypbutanedioate (PEG-S-
DMG), a pegylated ceramide (PEG-cer), or a PEG dialkoxypropylcarbamate such as
(0-
methoxy(polyethoxy)ethyl-N-(2,3-di(tetradecanoxy)propyl)carbamate or 2,3-
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
di(tetradecanoxy)propyl-N-(co-methoxy(polyethoxy)ethyecarbamate. In various
embodiments,
the molar ratio of the cationic lipid to the pegylated lipid ranges from about
100:1 to about 25:1.
In some embodiments, the LNPs comprise a pegylated lipid having thc following
structure (IV):
0
R1
R
0 \z
(IV)
or a pharmaceutically acceptable salt, tautomer or stereoisomer thereof,
wherein:
R1 and R11 are each independently a straight or branched, saturated or
unsaturated alkyl
chain containing from 10 to 30 carbon atoms, wherein the alkyl chain is
optionally interrupted by
one or more ester bonds; and
z has mean value ranging from 30 to 60.
In some of the foregoing embodiments of the pegylated lipid (IV), R" and R"
are not
both n-octadecyl when z is 42. In some other embodiments, R1 and R11 are each
independently a
straight or branched, saturated or unsaturated alkyl chain containing from 10
to 18 carbon atoms.
In some embodiments, R" and R" are each independently a straight or branched,
saturated or
unsaturated alkyl chain containing from 12 to 16 carbon atoms. In some
embodiments, R" and
R" are each independently a straight or branched, saturated or unsaturated
alkyl chain containing
12 carbon atoms. In some embodiments, Rio and R" are each independently a
straight or
branched, saturated or unsaturated alkyl chain containing 14 carbon atoms. In
other embodiments,
R" and R" are each independently a straight or branched, saturated or
unsaturated alkyl chain
containing 16 carbon atoms. In still more embodiments, R1 and R11 are each
independently a
straight or branched, saturated or unsaturated alkyl chain containing 18
carbon atoms. In still
other embodiments, R" is a straight or branched, saturated or unsaturated
alkyl chain containing
12 carbon atoms and R" is a straight or branched, saturated or unsaturated
alkyl chain containing
14 carbon atoms.
86
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In various embodiments, z spans a range that is selected such that the PEG
portion of (II)
has an average molecular weight of about 400 to about 6000 g/mol. In some
embodiments, the
average z is about 45.
In other embodiments, the pegylated lipid has one of the following structures:
0 0
N
N
13 0 \ in
15
(IVa)
(IVb)
13
15
0 0
\
.'*\/ N
kNk,
0 \ 11 0 \ /n
13
(IVc) (IVd)
11 11
wherein n is an integer selected such that the average molecular weight of the
pegylated
lipid is about 2500 g/mol.
In certain embodiments, the additional lipid is present in the LNP in an
amount from
about 1 to about 10 mole percent. In one embodiment, the additional lipid is
present in the LNP in
an amount from about 1 to about 5 mole percent. In one embodiment, the
additional lipid is
present in the LNP in about 1 mole percent or about 1.5 mole percent.
In some embodiments, the LNPs comprise a lipid of Formula (I), a nucleoside-
modified
RNA, a neutral lipid, a steroid and a pegylated lipid. In some embodiments the
lipid of Formula
(I) is compound 1-6. In different embodiments, the neutral lipid is DSPC. In
other embodiments,
the steroid is cholesterol. In still different embodiments, the pegylated
lipid is compound IVa.
In certain embodiments, the LNP comprises one or more targeting moieties that
targets
the LNP to a cell or cell population. For example, in one embodiment, the
targeting domain is a
ligand which directs the LNP to a receptor found on a cell surface.
In certain embodiments, the LNP comprises one or more internalization domains.
For
example, in one embodiment, the LNP comprises one or more domains which bind
to a cell to
induce the internalization of the LNP. For example, in one embodiment, the one
or more
internalization domains bind to a receptor found on a cell surface to induce
receptor-mediated
uptake of the LNP. In certain embodiments, the LNP is capable of binding a
biomolecule in vivo,
87
CA 03195281 2023-4- 11
WO 2022/081702 PCT/US2021/054775
where the LNP-bound biomolecule can then be recognized by a cell-surface
receptor to induce
internalization. For example, in one embodiment, the LNP binds systemic ApoE,
which leads to
the uptake of the LNP and associated cargo.
Other exemplary LNPs and their manufacture are described in the art, for
example in
U.S. Patent Application Publication No. US20120276209, Semple et al., 2010,
Nat Biotechnol.,
28(2):172-176; Akinc et al., 2010, Mol Ther., 18(7): 1357-1364; Basha et al.,
2011, Mol Ther,
19(12): 2186-2200; Leung et al., 2012, J Phys Chem C Nanomater Interfaces,
116(34): 18440-
18450; Lee et al., 2012, Int J Cancer., 131(5): E781-90; Belliveau et al.,
2012, Mol Ther nucleic
Acids, 1: e37; Jayaraman et al., 2012, Angew Chem Int Ed Engl., 51(34): 8529-
8533; Mui et al.,
2013, Mol Ther Nucleic Acids. 2, 0139; Maier et al., 2013, Mol Ther., 21(8):
1570-1578; and
Tam et al., 2013, Nanomedieine, 9(5): 665-74, each of which are incorporated
by reference in
their entirety.
The following Reaction Schemes illustrate methods to make lipids of Formula
(I), (II) or
(III).
GENERAL REACTION SCHEME 1
0 OR
H2
0 ROH
0n
"m
Br<õ).A...,OH A-2 Br
A-4
m NrOR
0
A-1 A-3
A-5
Embodiments of the lipid of Formula (1) (e.g., compound A-5) can be prepared
according
to General Reaction Scheme 1 ("Method A-), wherein R is a saturated or
unsaturated Ci-C24 alkyl
or saturated or unsaturated cycloalkyl, in is 0 or 1 and n is an integer from
1 to 24. Referring to
General Reaction Scheme 1, compounds of structure A-1 can be purchased from
commercial
sources or prepared according to methods familiar to one of ordinary skill in
the art. A mixture of
A-1, A-2 and DMAP is treated with DCC to give the bromide A-3. A mixture of
the bromide A-
3, a base (e.g., N,N-diisopropylethylamine) and the N,N-dimethyldiamine A-4 is
heated at a
temperature and time sufficient to produce A-5 after any necessarily workup
and or purification
step.
88
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
GENERAL REACTION SCHEME 2
0 0
RACI 01R
HO-q0H B-2
in-1
B-1
B-3
0
OAR
"m
B-4''in (L)_ n
11\
B-5
Other embodiments of the compound of Fommla (1) (e.g., compound B-5) can be
prepared according to General Reaction Scheme 2 ("Method B"), wherein R is a
saturated or
unsaturated C1-C24 alkyl or saturated or unsaturated cycloalkyl, m is 0 or 1
and n is an integer
from 1 to 24. As shown in General Reaction Scheme 2, compounds of structure B-
1 can be
purchased from commercial sources or prepared according to methods familiar to
one of ordinary
skill in the art. A solution of B-1 (1 equivalent) is treated with acid
chloride B-2 (1 equivalent)
and a base (e.g., triethylamine). "[he crude product is treated with an
oxidizing agent (e.g.,
pyridinum chlorochromate) and intermediate product B-3 is recovered. A
solution of crude B-3,
an acid (e.g., acetic acid), and N,N-dimethylaminoa.mine B-4 is then treated
with a reducing agent
(e.g., sodium triacetoxyborohydride) to obtain B-5 after any necessary work up
and/or
purification.
It should be noted that although starting materials A-1 and B-1 arc depicted
above as
including only saturated methylene carbons, starting materials which include
carbon-carbon
double bonds may also be employed for preparation of compounds which include
carbon-carbon
double bonds.
89
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
GENERAL REACTION SCHEME 3
0...,,OR
0 HO
'('-----''-\ /HI NI-12 HO+-...(-,N,0 SOCl2
13NqLOR
C-2 m (10R _____ ..-
n
m (yoR
n ' n
n
0
01 0
C-3 C-5
----7
R I C-8
HNR'
NH
-----./ I
C-6
0 OR 0.,,OR
===:,-' '
i
N) N
n m (I m ),TrOR (I)rOR
n n
0
0
C-7 C-9
Different embodiments of the lipid of Formula (I) (e g , compound C-7 or C9)
can be
prepared according to General Reaction Scheme 3 ("Method C"), wherein R is a
saturated or
unsaturated CI-C24 alkyl or saturated or unsaturated cycloalkyl, m is 0 or 1
and n is an integer
from 1 to 24. Referring to General Reaction Scheme 3, compounds of structure C-
1 can be
purchased from commercial sources or prepared according to methods familiar to
one of ordinary
skill in the art.
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
GENERAL REACTION SCHEME 4
R1 a R2a R3a R4a
R1 a R2a
R3a R4a
IM-4.) L2 ('-')(- R6
Rib R2b R3b R4b
R5 4-)1_14.)(4R6
G30
D-2 Rib R2b
R3b Rab
R8., ,..- ....
N NH2 Po-
HN,..... 3
I G
R9 D-3
I
D-1 R8.--
N .... ,
R'
Ri a R2a R3a R4a
0
Li 1.-8561_24R6
..õ........õ Y R7, Rib R2b R3b R4b
LiAl F14
D-4 0
___________________________ ).-- N G3 0-6 ,...-
Y=C1 or OH I
R7' _N,,..
R8 R9
D-5
Ri a R2a R3a R4a
R5 (''') Li 1E33'.(/'') L2 41 R6
Rib R2b R3b R41
I
RT N
R8- -R9
D-7
Embodiments of the compound of Formula (11) (e.g., compounds D-5 and D-7) can
be
prepared according to General Reaction Scheme 4 ("Method D"), wherein Ria,
R113, R2a, R2b, R3a,
R31', R4a, R46, R5, R6, R8, R9, Li, L2, GI, G2, G3, a, b,
c and d are as defined herein, and R7
represents R7 or a C3-C19 alkyl. Referring to General Reaction Scheme 1,
compounds of structure
D-1 and D-2 can be purchased from commercial sources or prepared according to
methods
familiar to one of ordinary skill in the art. A solution of D-1 and D-2 is
treated with a reducing
agent (e.g., sodium triacetoxyborohydride) to obtain D-3 after any necessary
work up. A solution
of D-3 and a base (e.g. trimethylamine, DMAP) is treated with acyl chloride D-
4 (or carboxylic
acid and DCC) to obtain D-5 after any necessary work up and/or purification. D-
5 can be reduced
with LiA1H4 D-6 to give D-7 after any necessary work up and/or purification.
91
CA 03195281 2023-4-11
WO 2022/081702
PCT/US2021/054775
GENERAL REACTION SCHEME 5
R1 a R2a R3a
R4a
R5 Li 1
L24 R6
Rib R2 b
R3b R4b
XR7
0
R8 G3 E-2 R8 G3
N H2 ___________________________________________ NHR7 E-4
R9 X=CI, Br or I R9 Y= CI or OH
E-1 E-3
R1 a R2a R3a R4a
R5 Li 4R6
Rib R2b R3b L2 R4b
0 N R7
E-5 G3
R8
Embodiments of the lipid of Formula (II) (e.g., compound E-5) can be prepared
according to General Reaction Scheme 5 (-Method E-), wherein Ria,
R2a, R2b, R3a, R3b, R4a,
R.
R. R6, R7, R, R9, 1,1, L2, G', a, b, c and d are as defined herein. Referring
to General
Reaction Scheme 2, compounds of structure E-1 and E-2 can be purchased from
commercial
sources or prepared according to methods familiar to one of ordinary skill in
the art. A mixture of
E-1 (in excess), E-2 and a base (e.g., potassium carbonate) is heated to
obtain E-3 after any
necessary work up. A solution of E-3 and a base (e.g. trimethylamine, DMAP) is
treated with acyl
chloride E-4 (or carboxylic acid and DCC) to obtain E-5 after any necessary
work up and/or
purification.
92
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
GENERAL REACTION SCHEME 6
0 0
HO¨G1-0H
F-2 [0]
R1.../\, OH R1 0 OH
F-1 F-3
0
G3
H H2N F-5 R'
R1 0 (III)
F-4 0
General Reaction Scheme 6 provides an exemplary method (Method F) for
preparation of
Lipids of Formula (III). GI, G3, RI and R3 in General Reaction Scheme 6 are as
defined herein for
Formula (III), and G1' refers to a one-carbon shorter homologue of Gl.
Compounds of structure
F-1 are purchased or prepared according to methods known in the art. Reaction
of F-1 with diol
F-2 under appropriate condensation conditions (e.g., DCC) yields ester/alcohol
F-3, which can
then be oxidized (e.g., PCC) to aldehyde F-4. Reaction of F-4 with amine F-5
under reductive
amination conditions yields a lipid of Formula (III).
It should be noted that various alternative strategies for preparation of
lipids of Formula
(III) are available to those of ordinary skill in the art. For example, other
lipids of Formula (III)
wherein LI and L2 arc other than ester can be prepared according to analogous
methods using the
appropriate starting material. Further, General Reaction Scheme 6 depicts
preparation of a lipids
of Formula (III), wherein GI and G2 arc the same; however, this is not a
required aspect of the
invention and modifications to the above reaction scheme are possible to yield
compounds
wherein GI and G2 are different.
It will be appreciated by those skilled in the art that in the process
described herein the
functional groups of intermediate compounds may need to be protected by
suitable protecting
groups. Such functional groups include hydroxy, amino, mercapto and carboxylic
acid. Suitable
protecting groups for hydroxy include trialkylsilyl or diarylalkylsilyl (for
example, t-
butyldimethylsilyl, t-butyldiphenylsilyl or trimethylsilyl),
tetrahydropyranyl, benzyl, and the like.
Suitable protecting groups for amino, amidino and guanidino include t-
butoxycarbonyl,
benzyloxycarbonyl, and the like. Suitable protecting groups for mercapto
include -C(0)-R"
(where R" is alkyl, aryl or arylalkyl), p-methoxybenzyl, trityl and the like.
Suitable protecting
93
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
groups for carboxylic acid include alkyl, aryl or arylalkyl esters. Protecting
groups may be added
or removed in accordance with standard techniques, which are known to one
skilled in the art and
as described herein. The usc of protecting groups is described in detail in
Green, T.W. and
P.G.M. Wutz, Protective Groups in Organic Synthesis (1999), 3rd Ed., Wiley. As
one of skill in
the art would appreciate, the protecting group may also be a polymer resin
such as a Wang resin,
Rink resin or a 2-chlorotrityl-chloride resin.
Delivery Vehicle Embodiments
Any suitable delivery vehicle format is contemplated.
In some embodiments, the delivery vehicle is a colloidal dispersion system,
such as
macromolecule complexes, nanocapsules, microspheres, beads, and lipid-based
systems including
oil-in-water emulsions, micelles, mixed micelles, liposomes, and lipid
nanoparticles. Exemplary
colloidal systems for use as delivery vehicles in vitro and in vivo include
liposomes (e.g., an
artificial membrane vesicle) and lipid nanoparticles.
The use of lipid formulations, as described above, is contemplated for the
introduction of
the at least one agent into the host cell (in vitro, ex vivo, or in vivo). In
another aspect, the at
least one agent may bc associated with a lipid. The at least one agent
associatcd with a lipid may
be encapsulated in the aqueous interior of a liposome, interspersed within the
lipid bilayer of a
liposome, attached to a liposome via a linking molecule that is associated
with both the liposome
and the oligonucleotide, entrapped in a liposome, complexed with a liposome,
dispersed in a
solution containing a lipid, mixed with a lipid, combined with a lipid,
contained as a suspension
in a lipid, complexed with a lipid, contained or complexed with a micelle, or
otherwise associated
with a lipid. Lipid, lipid/nucleic acid or lipid/expression vector associated
compositions are not
limited to any particular structure in solution. For example, they may be
present in a bilayer
structure, as micelles, or with a "collapsed" structure. They may also simply
be interspersed in a
solution, possibly forming aggregates that are not uniform in size or shape.
In one embodiment, delivery of the at least one agent comprises any suitable
delivery
method, including exemplary delivery methods described elsewhere herein. In
certain
embodiments, delivery of the at least one agent to a subject comprises mixing
the at least one
agent with a transfection reagent prior to the step of contacting. In another
embodiment, a method
of the present invention further comprises administering the at least one
agent together with the
transfection reagent. In another embodiment, the transfection reagent is a
cationic lipid reagent.
94
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In another embodiment, the transfection reagent is a lipid-based transfection
reagent. In
another embodiment, the transfection reagent is a protein-based transfection
reagent. In another
embodiment, the transfection reagent is a polyethyleneimine based transfection
reagent. In
another embodiment, the transfection reagent is calcium phosphate. In another
embodiment, the
transfection reagent is Lipofectin , Lipofectamineg, or TransITIT. In another
embodiment, the
transfection reagent is any other transfection reagent known in the art.
In some embodiments, delivery of the at least one agent comprises liposomes.
"Liposome" is a generic term encompassing a variety of single and
multilamellar lipid vehicles
formed by the generation of enclosed lipid bilayers or aggregates. Liposomes
can be
characterized as having vesicular structures with a phospholipid bilayer
membrane and an inner
aqueous medium. Multilamellar liposomes have multiple lipid layers separated
by aqueous
medium. They form spontaneously when phospholipids are suspended in an excess
of aqueous
solution. The lipid components undergo self-rearrangement before the formation
of closed
structures and entrap water and dissolved solutes between the lipid bilayers
(Ghosh et al., 1991
Glycobiology 5: 505-10). However, compositions that have different structures
in solution than
the normal vesicular structure are also encompassed. For example, the lipids
may assume a
micellar structure or merely exist as nonuniform aggregates of lipid
molecules.
The at least one agent associated with a lipid may be encapsulated in the
aqueous interior
of a liposome, interspersed within the lipid bilayer of a liposome, attached
to a liposome via a
linking molecule that is associated with both the liposome and the
oligonucleotide, entrapped in a
liposome, complexed with a liposome, dispersed in a solution containing a
lipid, mixed with a
lipid, combined with a lipid, contained as a suspension in a lipid, contained
or complexed with a
micelle, or otherwise associated with a lipid. Lipid, lipid/nucleic acid or
lipid/expression vector
associated compositions are not limited to any particular structure in
solution. For example, they
may be present in a bilayer structure, as micelles, or with a "collapsed"
structure. They may also
simply be interspersed in a solution, possibly forming aggregates that are not
uniform in size or
shape.
In another embodiment, the transfection reagent forms a liposome. Liposomes,
in another
embodiment, increase intracellular stability, increase uptake efficiency and
improve biological
activity. In another embodiment, liposomes are hollow spherical vesicles
composed of lipids
arranged in a similar fashion as those lipids which make up the cell membrane.
In some
embodiments, the liposomes comprise an internal aqueous space for entrapping
water-soluble
compounds. In another embodiment, liposomes can deliver the at least one agent
to cells in an
active form.
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In one embodiment, the composition comprises a lipid nanoparticle (LNP) and at
least
one agent.
The term Lipid nanoparticle" refers to a particle having at least one
dimension on the
order of nanometers (e.g., 1-1000nm) which includes one or more lipids. In
some embodiments,
LNPs comprise at least one agent that is either organized within inverse lipid
micelles and
encased within a lipid monolayer envelope or intercalated between adjacent
lipid bilayers (e.g.
lipid bilayer-agent-lipid bilayer). In some embodiments, the morphology of the
LNPs are not like
a traditional liposome, which are characterized by a lipid bilayer surrounding
an aqueous core, as
they possess an electron-dense core, where the cationic/ionizable lipids are
organized into
inverted micelles around the encapsulated agent (e.g. mRNA molecules)(Cullis
and Hope, 2017;
Guevara et al., 2019b). In various embodiments, the particle includes a lipid
of Formula (I), (II)
or (III). In some embodiments, lipid nanoparticles are included in a
formulation comprising at
least one agent as described herein. In some embodiments, such lipid
nanoparticles comprise a
cationic lipid (e.g., a lipid of Formula (I), (II) or (III)) and one or more
excipients selected from
neutral lipids, charged lipids, steroids and lipid-anchored polyethylene
glycol (e.g., a pegylated
lipid such as a pegylated lipid of structure (IV), such as compound IVa). In
some embodiments,
the at least one agent is encapsulated in the lipid portion of the lipid
nanoparticle or an aqueous
space enveloped by some or all of the lipid portion of the lipid nanoparticle,
thereby protecting it
from enzymatic degradation or other undesirable effects induced by the
mechanisms of the host
organism or cells e.g. an adverse immune response.
In various embodiments, the lipid nanoparticles have a mean diameter of from
about 30
nm to about 150 nm, from about 40 nm to about 150 nm, from about 50 nm to
about 150 nm,
from about 60 nm to about 130 nm, from about 70 nm to about 110 nm, from about
70 nm to
about 100 nm, from about 80 nm to about 100 nm, from about 90 nm to about 100
nm, from
about 70 to about 90 nm, from about 80 nm to about 90 nm, from about 70 nm to
about 80 nm, or
about 30 nm, 35 nm, 40 nm, 45 nm, 50 mu, 55 nm, 60 nm, 65 nm, 70 nm, 75 nm, 80
nm, 85 nm,
90 nm, 95 nm, 100 nm, 105 nm, 110 nm, 115 nm, 120 nm, 125 nm, 130 nm, 135 nm,
140 nm,
145 nm, or 150 nm. In one embodiment, the lipid nanoparticles have a mean
diameter of about 83
nm. In one embodiment, the lipid nanoparticles have a mean diameter of about
102 mu. In one
embodiment, the lipid nanoparticles have a mean diameter of about 103 nm. In
some
embodiments, the lipid nanoparticles are substantially non-toxic. In certain
embodiments, the at
least one agent, when present in the lipid nanoparticles, is resistant in
aqueous solution to
degradation by intra- or intercellular enzymes
96
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
The LNP may comprise any lipid capable of forming a particle to which the at
least one
agent is attached, or in which the at least one agent is encapsulated or
complexcd. The term
"lipid" refers to a group of organic compounds that are derivatives of fatty
acids (e.g., esters) and
are generally characterized by being insoluble in water but soluble in many
organic solvents.
Exemplary lipids are shown elsewhere herein.
In one embodiment, the LNP comprises one or more cationic lipids, and one or
more
stabilizing lipids. Stabilizing lipids include neutral lipids, anionic lipids
and pegylated lipids.
In one embodiment, the LNP comprises a cationic lipid. As used herein, the
term
"cationic or ionizabe lipid" refers to a lipid that is cationic or becomes
cationic (protonated) as the
pH is lowered below the pKa of the ionizable group of the lipid, but is
progressively more neutral
at higher pH values. At pH values below the pKa, the lipid is then able to
associate with
negatively charged nucleic acids. In certain embodiments, the cationic lipid
comprises a
zwitterionic lipid that assumes a positive charge on pH decrease.
In various embodiments, the LNP comprises a cationic or ionizable lipids,
stabilizing
lipids, sterol, and a lipid-anchored polyethylene glycol (i.e PEGylated
lipids).
In some embodiments, the LNPs comprise an ionic lipid of Formula (I), at least
one
agent, and one or more excipients selected from neutral lipids, steroids and
pegylated lipids. In
some embodiments the lipid of Formula (I) is compound I-5. In some embodiments
the lipid of
Formula (I) is compound 1-6.
In some embodiments, the LNPs comprise an ionic lipid of Fonnula (II), at
least one
agent, and one or more excipient selected from neutral lipids, steroids and
pegylated lipids. In
some embodiments, the lipid of Formula (II) is compound 11-9. In some
embodiments, the lipid of
Formula (II) is compound II-10. In some embodiments, the lipid of Formula (II)
is compound II-
11. In some embodiments, the lipid of Formula (II) is compound 11-12. In some
embodiments, the
lipid of Formula (II) is compound 11-32.
In some embodiments, the LNPs comprise an ionic lipid of Formula (III), at
least one
agent, and one or more excipient selected from neutral lipids, steroids and
pegylated lipids. In
some embodiments, the lipid of Formula (III) is compound 111-3. In some
embodiments, the lipid
of Formula (III) is compound III-7.
In certain embodiments, the cationic lipid is present in the LNP in an amount
from about
30 to about 95 mole percent. In one embodiment, the cationic lipid is present
in the LNP in an
amount from about 30 to about 70 mole percent. In one embodiment, the cationic
lipid is present
in the LNP in an amount from about 40 to about 60 mole percent. In one
embodiment, the
97
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
cationic lipid is present in the LNP in an amount of about 50 mole percent. In
one embodiment,
the LNP comprises only cationic lipids.
In certain embodiments, the LNP comprises one or more stabilizing lipids (e.g.
neutral or
anionic lipids) which help to encapsulate the cargo and stabilize the
formation of particles during
their formation. Exemplary neutral lipids include, for example,
distearoylphosphatidylcholine
(DSPC), dioleoylphosphatidyleholine (DOPC), dipalmitoylphosphatidylcholine
(DPPC),
dioleoylphosphatidylglycerol (DOPG), dipalmitoylphosphatidylglycerol (DPPG),
dioleoyl-
phosphatidylethanolamine (DOPE). palmitoyloleoylphosphatidylcholine (POPC),
palmitoyloleoyl-phosphatidylethanolamine (POPE) and dioleoyl-
phosphatidylethanolamine 4-(N-
maleimidomethyl)-cyclohexane-l-carboxylate (DOPE-mal), dipalmitoyl
phosphatidyl
ethanolamine (DPPE), dimyristoylphosphoethanolamine (DMPE), distearoyl-
phosphatidylethanolamine (DSPE), 16-0-monomethyl PE, 16-0-dimethyl PE, 18-1-
trans PE, 1-
stearioy1-2-oleoyl-phosphatidyethanol amine (SOPE), and 1,2-dielaidoyl-sn-
glycero-3-
phophoethanolamine (transDOPE). In one embodiment, the neutral lipid is 1,2-
distearoyl-sn-
glycero-3-phosphocholine (DSPC). In various embodiments, the molar ratio of
the cationic lipid
(e.g., lipid of Formula (I)) to the neutral lipid ranges from about 2:1 to
about 8:1.
In various embodiments, the LNPs further comprise a steroid or a steroid
analogue. In
certain embodiments, the steroid or steroid analogue is cholesterol. In some
of these
embodiments, the molar ratio of the cationic lipid (e.g., lipid of Formula
(I)) to cholesterol ranges
from about 2:1 to 1:1.
In certain embodiments, the LNP comprises glycolipids (e.g..
monosialoganglioside
GM1).
In certain embodiments, the LNP comprises an additional lipid which is a
polyethylene
glycol-lipid (pegylated lipid) to reduce immune system recognition and improve
biodistribution.
In one embodiment, the polyethylene glycol-lipid is PEG-c-DOMG. In other
embodiments, the
LNPs comprise a pegylated diacylglycerol (PEG-DAG) such as 1 (monomethoxy
polyethyleneglycol) 2,3 dimyristoylglyeerol (PEG-DMG), a pegylated
phosphatidylethanoloamine (PEG-PE), a PEG succinate diacylglycerol (PEG-S-DAG)
such as 4-
0-(2',3'-di(tctradecanoyloxy)propy1-1-0-(co-
methoxy(polycthoxy)cthyl)butancdioatc (PEG-S-
DMG), a pegylated ceramide (PEG-cer), or a PEG dialkoxypropylcarbamate such as
(0-
methoxy(polyethoxy)ethyl-N-(2,3-di(tetradecanoxy)propyl)carbamate or 2,3-
di(tetradecanoxy)propyl-N-(0)¨methoxy(polyethoxy)ethyl)carbamate. In various
embodiments,
the molar ratio of the cationic lipid to the pegylated lipid ranges from about
100:1 to about 25:1.
98
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In certain embodiments, the PEGylated lipid is present in the LNP in an amount
from
about 1 to about 10 mole percent. In one embodiment, the PEGylated lipid is
present in the LNP
in an amount from about 1 to about 5 mole percent. In one embodiment, the
PEGylated lipid is
present in the LNP in about 1 mole percent or about 1.5 mole percent.
In some embodiments, the LNPs comprise a lipid of Formula (I), a nucleoside-
modified
RNA, a neutral lipid, a steroid and a pegylated lipid. In some embodiments the
lipid of Formula
(I) is compound 1-6. In different embodiments, the neutral lipid is DSPC. In
other embodiments,
the steroid is cholesterol. In still different embodiments, the pegylated
lipid is compound 1Va.
In certain embodiments, the LNP comprises one or more targeting moieties that
targets
the LNP to a cell or cell population. For example, in one embodiment, the
targeting domain is a
ligand which directs the LNP to a receptor found on a cell surface. Exemplary
targeting domains
include CD4.
In certain embodiments, the LNP comprises one or more internalization domains.
For
example, in one embodiment, the LNP comprises one or more domains which bind
to a cell to
induce the internalization of the LNP. For example, in one embodiment, the one
or more
internalization domains bind to a receptor found on a cell surface to induce
receptor-mediated
uptake of the LNP. In certain embodiments, the LNP is capable of binding a
biomolecule in vivo,
where the LNP-bound biomolecule can then be recognized by a cell-surface
receptor to induce
internalization. For example, in one embodiment, the LNP binds systemic ApoE,
which leads to
the uptake of the LNP and associated cargo.
Other exemplary LNPs and their manufacture are described in the art, for
example in
U.S. Patent Application Publication No. US20120276209, Semple et al., 2010,
Nat Biotechnol.,
28(2):172-176; Akinc et al., 2010, Mol Ther., 18(7): 1357-1364; Basha et al.,
2011, Mol Ther,
19(12): 2186-2200; Leung et al., 2012, J Phys Chem C Nanomater Interfaces,
116(34): 18440-
18450; Lee et al., 2012, Int J Cancer., 131(5): E781-90; Belliveau et al.,
2012, Mol Ther nucleic
Acids, 1: e37; Jayaraman et al., 2012, Angew Chem Int Ed Engl., 51(34): 8529-
8533; Mui et al.,
2013, Mol Ther Nucleic Acids. 2, e139; Maier et al., 2013, Mol Ther., 21(8):
1570-1578; and
Tarn et al., 2013, Nanomedicine, 9(5): 665-74, each of which are incorporated
by reference in
their entirety.
Targeting moieties
As taught above, the delivery vehicles contemplated herein¨which may include
various
formats, such as, but not limited to, macromolecule complexes, nanocapsules,
microspheres,
beads, and lipid-based systems including oil-in-water emulsions, micelles,
mixed micelles,
99
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
liposomes, and lipid nanoparticles (LNPs)¨may comprise one or more targeting
moieties (or
equivalently "targeting domains" or "targeting ligands") which function to
target the delivery
vehicle (e.g., LNP) to a cell or cell population. Targeting moieties may
include any suitable
binding agent which is capable of specifically interacting with and bind to a
target cell ligand on
the surface of a target cell or tissue. The targeting moieties may be
naturally occurring or
engineered. Targeting moieties may include, but are not limited to, proteins,
peptides, antibodies
or antibody fragments, immunoglobulins or immunoglobulin fragments, small
molecules,
aptamers, vitamins, nucleic acid molecules, and the like. No limit is meant to
be placed on the
targeting moieties contemplated herein so long as any particular targeting
moiety may be (a)
coupled to a delivery vehicle (either covalently or non-covalently) and (b) is
capable of causing
or facilitating the localization or targeting of the delivery vehicle to a
target cell or tissue by the
binding or otherwise interaction between the targeting moiety on the delivery
vehicle and a target
cell ligand on a target cell or tissue.
The target cell ligand may include endogenous ligands occurring on the surface
of a cell
or in the extracellular space outside of a cell, such as carbohydrates,
lipids, polysaccharides,
proteins, glycoproteins, glycolipids, peptides, cell membrane components
(e.g., cholesterol) or the
like. In certain embodiments, the endogenous ligands on the target cell are
specific for the target
cell, i.e., are expressed and/or are contained only on the target cell, or at
least, are minimally
present in cells that are not the target cells. For example, the endogenous
ligand on the target cell
could be a disease-associated protein, e.g., a cancer cell protein cell
surface protein that are not
typically expressed in healthy cells. In other embodiments, the target ligand
on the target cells
can be an engineered or otherwise non-naturally occurring ligand, e.g., a
genetically modified
target cell that expresses a non-naturally occurring surface cell protein.
Suitable targeting ligands
can be selected so that the unique properties of the target cell are utilized,
thus allowing the
composition to differentiate between target and non-target cells.
This aspect may be referred to as -selective delivery" of a delivery vehicle
to a target cell
of interest (e.g., a lymphocyte, such as a T-cell). The term "selective
delivery" means that
delivery vehicles are localized by binding covalently or non-covalently to a
target cell (e.g., a
particular T-cell subpopulation) through the binding interaction between the
targeting moiety of
the delivery vehicle and the target cell ligand on the target cell of interest
(e.g., a particular T-cell
subpopulation), but wherein the delivery vehicles do not bind, or bind
minimally, to cells that do
not express the target cell ligand (i.e., such cells may be referred to as
"non-target cells"). By
"bind minimally," it is meant that binding of the delivery vehicle to non-
target cells ranges
between undetected to less than 1%, or less than 2%, or less than 3%, or less
than 4%, or less than
100
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
5%, or less than 6%, or less than 7%, or less than 8%, or less than 9%, or
less than 10% increased
binding relative to a negative control (which can be a cell type known not to
bind to the delivery
vehicle).
Thus, the delivery vehicles of the present disclosure may be localized or
targeted to a
particular type of cell (e.g., a particular type of T cell) by utilizing a
targeting moiety which is
attached (either covalently or non-covalently) to a delivery vehicle.
Preferably, the targeting
moiety is attached such that the targeting moiety is presented or otherwise
exposed on the outer
surface of the delivery vehicle such that the moiety may interact with a
cognate binding domain
or ligand on the surface of a target cell or tissue (e.g., a particular CD4
antigen), thereby
promoting or facilitating the binding of the delivery vehicle to the target
cell or tissue (such as,
CD4+ T cells, where it would then become internalized (e.g., through active
internalization, such
as endocytosis) with the concomitant release of the agent (e.g., mRNA) carried
by the delivery
vehicle once inside the cell.
It will be appreciated that a targeting moiety can be linked to the surface of
a delivery
vehicle during or after preparation. In some embodiments, the targeting moiety
is attached to the
surface of a delivery vehicle after the vehicles has been prepared. In other
embodiments, the
targeting moiety is attached to a component (e.g., a lipid) of an unassembled
delivery vehicle
before the vehicles has been prepared. Such attachment means may be carried
out by any known
means in the art, including any suitable conjugation chemistry already well
known in the art and
discussed herein.
In some other embodiments, the delivery vehicles or compositions comprising
the
delivery vehicles may further include one or more additional agents that
enhance the localization
of the delivery vehicles to a target cell. Such additional agents may include
other peptides,
aptamers, oligonucleotides, vitamins or other molecules that facilitate the
localization of a
delivery vehicle to a target cell, but which are not necessarily directly
coupled to the delivery
vehicle.
In one embodiment, the delivery vehicles of the present disclosure comprise
one or more
targeting moieties that are capable of targeting the delivery vehicle to a
leukocyte, which
generally include myeloid and lymphoid classes of immune system cells. Myeloid
cells can
include, for example, neutrophils, eosinophils, mast cells, basophils, and
monocytes. Monocytes
are further classified into dendritic cells and macrophages. Lymphoid cells
(or lymphocytes)
include T cells (subdivided into helper T cells, memory T cells, and cytotoxic
T cells), B cells
(subdivided into plasma cells and memory B cells), and natural killer cells.
101
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
One or ordinary skill in the art will be able to identify appropriate cell
target ligands on
each of these types of leukocytes that may be utilized as a means to localize
the delivery vehicles
described herein by installing an appropriately matching targeting moiety on
the delivery vehicle,
e.g., an antibody, peptide, protein, oligonucleotide, small molecule, vitamin,
or aptamer which is
coupled (covalently or non-covalently to the delivery vehicle) such that the
delivery vehicle
becomes localized to the target cell due to specific, and preferably,
selective interaction between
the targeting moiety and the cell target ligand.
In various embodiments, the disclosure contemplates delivery vehicles that
target and/or
localize to leukocytes, and in particular, to a particular lymphocyte, such as
a CD4+ T cell. In
particular embodiments, the delivery vehicles comprise one or more targeting
moieties that are
capable of targeting the delivery vehicles to T cells, including helper T
cells.
One of ordinary skill in the art will appreciate that leukocytes comprise cell
surface
antigens known as CD antigens which are characteristic of different types of
leukocytes and help
define various subpopulations of leukocytes.
The cluster of differentiation (CD) is a nomenclature system conceived to
identify and
classify antigens found on the cell surface of leukocytes. Initially, surface
antigens were named
after the monoclonal antibodies that bound to them. As there were often
multiple monoclonal
antibodies raised against each antigen in different labs, the need arose to
adopt a consistent
nomenclature. The current system was adopted in 1982 through the 1st
International Workshop
and Conference on Human Leukocyte Differentiation Antigens (HLDA). The Human
Cell
Differentiation Molecules organization continues to hold HLDA conferences to
maintain and
develop the list of known CD markers.
Under this naming system, antigens that are well characterized are assigned an
arbitrary
number (e.g., CD1, CD2, CD3, CD4, CD5, CD8 etc.) whereas molecules that are
recognized by
just one monoclonal antibody are given the provisional designation "CDw" e.g.,
CDw50. Lower
class letters are also added after the assigned number to indicate larger
molecules that share a
common chain, for example CD la or CD 1d. Physiologically, CD molecules do not
belong in any
particular class, with their functions ranging widely from cell surface
receptors to adhesion
molecules. Although initially used just for human leukocytes, the CD molecule
naming
convention has now been expanded to cover different species (e.g., mouse) as
well as other cell
types. As of April 2016, human CD antigens are numbered up to CD371.
The presence or absence of a specific antigen from the surface of a particular
cell
population is denoted with -+" or
respectively. Varying cellular expression levels are also
marked as hi or low, for example central memory T-cells are CD62Lhi whereas
effector memory
102
CA 03195281 2023- 4- 11
WO 2022/081702
PCT/US2021/054775
T-cells are CD62Llow. Monitoring the expression profiles of different CD
antigens has permitted
the identification, isolation and phcnotyping of cell types according to their
function in various
immune processes.
The delivery vehicles of the present disclosure may include one or more
targeting
moieties that bind to or otherwise associate with a CD4 antigen. In various
embodiments, the
targeting moieties for targeting the delivery vehicles to a target cell (e.g.,
leukocytes) are
antibodies or an antibody binding fragments. Such antibodies or antibody
binding fragments may
include, but are not limited to, anti-CD4 antibodies or antigen binding
fragments.
As used herein, "antibody" refers to a polypeptide of the immunoglobulin
family that is
capable of binding a corresponding antigen non-covalently, reversibly, and in
a specific manner
(e.g., a CD4 antigen). For example, a naturally occurring IgG antibody is a
tetramer comprising at
least two heavy (H) chains and two light (L) chains inter-connected by
disulfide bonds. Each
heavy chain is comprised of a heavy chain variable region (abbreviated herein
as VH) and a
heavy chain constant region. The heavy chain constant region is comprised of
three domains,
CH1, CH2 and CH3. Each light chain is comprised of a light chain variable
region (abbreviated
herein as VL) and a light chain constant region. The light chain constant
region is comprised of
one domain, CL. The VH and VL regions can be further subdivided into regions
of
hypervariability, termed complementarity determining regions (CDR),
interspersed with regions
that are more conserved, termed framework regions (FR). Each VH and VL is
composed of three
CDRs and four FRs arranged from amino-terminus to carboxy-tenninus in the
following order:
FR1, CDR1, FR2, CDR2, FR3, CDR3, and FR4. The variable regions of the heavy
and light
chains contain a binding domain that interacts with an antigen. The constant
regions of the
antibodies may mediate the binding of the immunoglobulin to host tissues or
factors, including
various cells of the immune system (e.g., effector cells) and the first
component (C lq) of the
classical complement system.
Antibodies of the present disclosure include, but are not limited to,
monoclonal
antibodies, human antibodies, humanized antibodies, camelid antibodies,
chimeric antibodies, and
anti-idiotypic (anti-Id) antibodies (including, e.g., anti-1d antibodies to
antibodies of the present
disclosure). The antibodies can be of any isotype/class (e.g., IgG, IgE, IgM,
IgD, IgA and IgY),
or subclass (e.g., IgGl, IgG2, IgG3, IgG4, IgAl and IgA2).
As used herein, "complementarity-determining domains- or "complementary-
determining regions" ("CDRs") interchangeably refer to the hypervariable
regions of VL and VH.
The CDRs are the target protein-binding site of the antibody chains that
harbors specificity for
such target protein. There are three CDRs (CDR1-3, numbered sequentially from
the N-terminus)
103
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
in each human VL or VH, constituting about 15-20% of the variable domains.
CDRs can be
referred to by their region and order. For example, "VHCDR1" or "HCDR1" both
refer to the
first CDR of the heavy chain variable region. The CDRs are structurally
complementary to the
epitope of the target protein and are thus directly responsible for the
binding specificity. The
remaining stretches of the VL or VH, the so-called framework regions, exhibit
less variation in
amino acid sequence (Kuby, Immunology, 4th ed., Chapter 4. W.H. Freeman & Co.,
New York,
2000).
The positions of the CDRs and framework regions can be determined using
various well
known definitions in the art, e.g., Kabat, Chothia, and AbM (see, e.g.,
Johnson et al., Nucleic
Acids Res., 29:205-206 (2001); Chothia and Lesk, J. Mol. Biol., 196:901-917
(1987); Chothia et
al., Nature, 342:877-883 (1989); Chothia et al., J. Mol. Biol., 227:799-817
(1992); Al-Lazikani et
al., J. Mol. Biol., 273:927-748 (1997)). Definitions of antigen combining
sites are also described
in the following: Ruiz et al., Nucleic Acids Res., 28:219-221 (2000); and
Lefranc, M. P., Nucleic
Acids Res., 29:207-209 (2001); MacCallum et al., J. Mol. Biol., 262:732-745
(1996); and Martin
et al., Proc. Natl. Acad. Sci. USA, 86:9268-9272 (1989); Martin et al.,
Methods Enzymol.,
203:121-153 (1991); and Rees et al., In Sternberg M. J. E. (ed.), Protein
Structure Prediction,
Oxford University Press, Oxford, 141-172 (1996).). In a combined Kabat and
Chothia numbering
scheme, in some embodiments, the CDRs correspond to the amino acid residues
that are part of a
Kabat CDR, a Chothia CDR, or both. For instance, in some embodiments, the CDRs
correspond
to amino acid residues 26-35 (HC CDR1), 50-65 (HC CDR2), and 95-102 (HC CDR3)
in a VH,
e.g., a mammalian VH, e.g., a human VH; and amino acid residues 24-34 (LC
CDR1). 50-56 (LC
CDR2), and 89-97 (LC CDR3) in a VL, e.g., a mammalian VL, e.g., a human VL.
Both the light and heavy chains are divided into regions of structural and
functional
homology. The terms "constant" and "variable" are used functionally. In this
regard, it will be
appreciated that the variable domains of both the light (VL) and heavy (VH)
chain portions
determine antigen recognition and specificity. Conversely, the constant
domains of the light chain
(CL) and the heavy chain (CH1, CH2 or CH3) confer important biological
properties such as
secretion, transplacental mobility, Fc receptor binding, complement binding,
and the like. By
convention, the numbering of the constant region domains increases as they
become more distal
from the antigen binding site or amino-terminus of the antibody. The N-
tenninus is a variable
region and at the C-terminus is a constant region; the CH3 and CL domains
actually comprise the
carboxy-terminal domains of the heavy and light chain, respectively.
As used herein, -antigen binding fragment" refers to one or more portions of
an antibody
that retain the ability to specifically interact with (e.g., by binding,
steric hindrance,
104
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
stabilizing/destabilizing, spatial distribution) an epitope of a CD4 antigen
of a leukocyte_
Examples of binding fragments include, but are not limited to, single-chain
Fvs (scFv), disulfide-
linked Fvs (sdFv), Fab fragments; F(ab') fragments, a monovalent fragment
consisting of the VL,
VH, CL and CHI domains; a F(ab)2 fragment, a bivalent fragment comprising two
Fab fragments
linked by a disulfide bridge at the hinge region; a Fd fragment consisting of
the VH and CH1
domains; a Fv fragment consisting of the VL and VH domains of a single arm of
an antibody; a
dAb fragment (Ward et al., Nature 341:544-546, 1989), which consists of a VH
domain; and an
isolated complementarity determining region (CDR), or other epitope-binding
fragments of an
antibody.
Furthermore, although the two domains of the Fv fragment, VL and VH, are coded
for by
separate genes; they can be joined, using recombinant methods, by a synthetic
linker that enables
them to be made as a single protein chain in which the VL and VH regions pair
to form
monovalent molecules (known as single chain FAT ("scFv"); see, e.g., Bird et
al., Science 242:423-
426, 1988; and Huston et al., Proc. Natl. Acad. Sci. 85:5879-5883, 1988). Such
single chain
antibodies are also intended to be encompassed within the term "antigen-
binding fragment."
These antigen binding fragments are obtained using conventional techniques
known to those of
skill in the art, and the fragments are screened for utility in the same
manner as are intact
antibodies.
Antigen binding fragments can also be incorporated into single domain
antibodies,
maxibodies, minibodies, nanobodies, intrabodies, diabodies, triabodies,
tetrabodies, v-NAR and
bis-scFv (see, e.g., Hollinger and Hudson, Nature Biotechnology 23:1126-
1136,2005). Antigen
binding fragments can be grafted into scaffolds based on polypeptides such as
fibronectin type III
(Fn3) (see U.S. Pat. No. 6,703,199, which describes fibronectin polypeptide
monobodies).
Accordingly, the antibodies and antigen binding fragments herein (e.g., anti-
CD4 antigen binding
fragments) can be a variety of structures, including, but not limited to
bispecific antibodies,
minibodies, domain antibodies, synthetic antibodies, antibody mimetics,
chimeric antibodies,
antibody fusions (sometimes referred to as "antibody conjugates-), and
fragments of each;
respectively. Specific antibody fragments (or antigen binding fragments)
include, but are not
limited to, (i) the Fab fragment consisting of VL, VH, CL and CH1 domains,
(ii) the Fd fragment
consisting of the VH and CH1 domains, (iii) the Fv fragment consisting of the
VL and VH
domains of a single antibody; (iv) the dAb fragment, which consists of a
single variable region,
(v) isolated CDR regions, (vi) F(ab')2 fragments, a bivalent fragment
comprising two linked Fab
fragments (vii) single chain Fv molecules (scFv), wherein a VH domain and a VL
domain are
linked by a peptide linker which allows the two domains to associate to form
an antigen binding
105
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
site (viii) bispecific single chain Fv dimers and (ix) "diabodies" or
"triabodies", multivalent or
multispccific fragments constructed by gene fusion. The antibody fragments may
be modified.
For example, the molecules may be stabilized by the incorporation of disulfide
bridges linking the
VH and VL domains. Examples of antibody formats and architectures are
described in Carter,
2006, Nature Reviews Immunology 6:343-357 and references cited therein, all
expressly
incorporated by reference.
Antigen binding fragments can be incorporated into single chain molecules
comprising a
pair of tandem FA/ segments (VH-CH1-VH-CH1) which, together with complementary
light chain
polypeptides, form a pair of antigen binding regions (Zapata et al., Protein
Eng. 8:1057-1062,
1995; and U.S. Pat. No. 5,641,870).
As used herein, "monoclonal antibody" refers to polypeptides, including
antibodies and
antigen binding fragments that have substantially identical amino acid
sequence or are derived
from the same genetic source. This term also includes preparations of antibody
molecules of
single molecular composition. A monoclonal antibody composition displays a
single binding
specificity and affinity for a particular epitope.
As used herein, a "human antibody" includes antibodies having variable regions
in which
both the framework and CDR regions are derived from sequences of human origin.
Furthermore,
if the antibody contains a constant region, the constant region also is
derived from such human
sequences, e.g., human germline sequences, or mutated versions of human
germline sequences or
antibody containing consensus framework sequences derived from human framework
sequences
analysis, for example, as described in Knappik et al.. J. Mol. Biol. 296:57-
86, 2000).
In some embodiments, the antibody is a chimeric antibody or antigen-binding
fragment
thereof. A chimeric antibody is an antibody comprising amino acid sequences
from different
genetic sources. In some embodiments, the chimeric antibody comprises amino
acid sequences
from a mouse and amino acid sequences from a human. In some embodiments a
chimeric
antibody comprises a variable domain derived from a mouse and constant domains
derived from a
human.
In some embodiments, the antibody is a humanized antibody or antigen-binding
fragment
thereof. By "humanized" antibody as used herein is meant an antibody
comprising a human
framework region (FR) and one or more complementarily determining regions
(CDRs) from a
non-human (usually mouse or rat) antibody. The non-human antibody providing
the CDRs is
called the -donor" and the human immunoglobulin providing the framework is
called the
acceptor". Humanization relies principally on the grafting of donor CDRs onto
acceptor (human)
VL and VH frameworks (Winter U.S. Pat. No. 5,225,539, incorporated entirely by
reference).
106
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
This strategy is referred to as "CDR grafting." "Backmutation" of selected
acceptor framework
residues to the corresponding donor residues is often required to regain
affinity that is lost in the
initial grafted construct (U.S. Pat. No. 5,693,762, incorporated entirely by
reference). The
humanized antibody optimally also will comprise at least a portion of an
immunoglobulin
constant region, typically that of a human immunoglobulin, and thus will
typically comprise a
human Fc region. A variety of techniques and methods for humanizing and
reshaping non-human
antibodies are well known in the art (See Tsurushita cYz Vasquez, 2004,
Humanization of
Monoclonal Antibodies, Molecular Biology of B Cells, 533-545, Elsevier Science
(USA), and
references cited therein, all incorporated entirely by reference).
Humanization or other methods of
reducing the immunogenicity of nonhuman antibody variable regions may include
resurfacing
methods, as described for example in Roguska et al., 1994, Proc. Natl. Acad.
Sci. USA 91:969-
973, incorporated entirely by reference. In one embodiment, selection-based
methods may be
employed to humanize and/or affinity mature antibody variable regions, that
is, to increase the
affinity of the variable region for its target antigen. Other humanization
methods may involve the
grafting of only parts of the CDRs, including but not limited to methods
described in U.S. Ser.
No. 09/810,502; Tan et al., 2002, J. Immunol. 169:1119-1125; De Pascalis et
al., 2002, J.
Immunol. 169:3076-3084, incorporated entirely by reference. Structure-based
methods may be
employed for humanization and affinity maturation, for example as described in
U.S. Ser. No.
10/153,159 and related applications, all incorporated entirely by reference.
In some embodiments, the antibody is a human engineered antibody. A human
engineered antibody refers to an antibody derived from a non-human source,
such as mouse, in
which one or more substitutions have been made to improve a desired
characteristic of the
antibody, such as to increase stability or reduce immunogenicity when the
antibody is
administered to a subject. In some embodiments, the substitutions are made at
low-risk positions
(e.g. exposed to solvent but not contributing to antigen binding or antibody
structure). Such
substitutions mitigate the risk that a subject will generate an immune
response against the
antibody following its administration, without affecting the ability of the
antibody to bind to a
desired epitope or antigen (see, e.g,. Studnicka et al. Protein Eng. 1994.
7(6):805-814).
In some embodiments, the antibody is a single chain antibody or antigen-
binding
fragment. A single chain antibody, or single chain variable fragment (scFV) is
a protein or
polypeptide comprising a VH domain and a VL domain joined together, such as by
a synthetic
linker, to form a single protein or polypeptide (see, e.g., Bird et al.,
Science. 242:423-426, 1988;
and Huston et al., Proc. Natl. Acad. Sci. 85:5879-5883, 1988).
107
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In some embodiments, the antibody is an antibody fragment or antigen-binding
fragment.
An antibody fragment is protein or polypcptidc derived from an antibody. An
antigen-binding
fragment is a protein or polypeptide derived from an antibody that is capable
of binding to the
same epitope or antigen as the antibody from which it was derived.
In some embodiments, the antibody has reduced glycosylation, no glycosylation,
or is
hypofucosylated. Glycosylation refers to the covalent attachment of sugar,
monosaccharide,
disaccharide, oligosaccharide, polysaccharide, or glycan moieties to a
molecule, such as a
polypeptide or protein. These sugar or glycan moieties are generally attached
to an antibody in a
post-translational matter, prior to secretion by a B cell. An antibody with
reduced glycosylation
has fewer of these attached sugar or glycan moieties than the number that are
typically attached to
an antibody with a substantially identical amino acid sequence, such as when
the antibody is
produced by a B cell in vitro or in vivo in a mouse or human. An antibody with
no glycosylation
has no attached sugar or glycan moieties. An antibody that is hypofucosylated
has fewer fucosyl
residues than the number that are typically attached to an antibody with a
substantially identical
amino acid sequence, such as when the antibody is produced by a B cell in
vitro or in vivo in a
mouse or human.
In still other embodiments, the antibodies and antigen binding fragments
discussed herein
may be modified in a manner that reduces immunogenicity. Modifications to
reduce
immunogenicity may include modifications that reduce binding of processed
peptides derived
from the parent sequence to MHC proteins. For example, amino acid
modifications would be
engineered such that there are no or a minimal number of immune epitopes that
are predicted to
bind, with high affinity, to any prevalent MHC alleles. Several methods of
identifying MHC-
binding epitopes in protein sequences are known in the art and may be used to
score epitopes in
an antibody of the present invention. See, for example, U.S. Ser. No.
09/903,378, U.S. Ser. No.
10/754,296, U.S. Ser. No. 11/249,692, and references cited therein, all
expressly incorporated by
reference.
CD4 is a type I transmembrane protein in which four immunoglobulin superfamily
domains (designated in order as D lto D4 from the N terminal to the cell
membrane side) are
present on the outside of the cells, and two N-linked sugar chains in total
are bound to the
domains D3 to D4. CD4 binds to a major histocompatibility complex (MHC) class
II molecule
through D1 and D2 domains, and then activates the T cells. Further, it is also
known that CD4
polymerizes through D3 and D4 domains. The D1 domain of CD4 is known to serve
as a receptor
for a human immunodeficiency virus (HIV) (Anderson et al, Clinical Immunology
and
Immunopathology, 84(1):73-84), 1997).
108
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
CD4 comprises the following amino acid sequence as per entry No P01730-1
(UniParc):
MNRGVPFRHLLLVLQLALLPAATQGKKVVLGKKGDTVELTCTASQKKSIQFHWKNSNQ
IKILGNQGSFLTKGPSKLNDRADSRRSLWDQGNFPLIIKNLKIEDSDTYICEVEDQKEEVQ
LLVFGLTANSDTHLLQGQSLTLTLESPPGS SPSVQCRSPRGKNIQGGKTLSVSQLELQDSG
TWTCTVLQNQKKVEFKIDIVVLAFQKASSIVYKKEGEQVEFSFPLAFTVEKLTGSGELW
WQAERASSSKSWITFDLKNKEVSVKRVTQDPKLQMGKKLPLHLTLPQALPQYAGSGNL
TLALEAKTGKLHQEVNLVVMRATQLQKNLTCEVWGPTSPKLMLSLKLENKEAKVSKRE
KAVWVLNPEAGMWQCLLSDSGQVLLESN1KVLPTW STPVQPMALIVLGGVAGLLLFIGL
GIFFCVRCRHRRRQAER_MSQIKRLLSEKKTCQCPHRFQKTCSPI (SEQ ID NO:1).
The anti-CD4 antibodies of the present invention may be any antibody that
binds to CD4,
e.g., may comprise the variable regions (e.g., the CDRs) of any known or
undiscovered anti-CD4
antibody. Antibodies of the invention may display selectivity for CD4.
Examples include full-
length versus splice variants, cell-surface vs. soluble forms, selectivity for
various polymorphic
variants, or selectivity for specific conformational forms of a target. An
antibody of the present
invention may bind any epitope or region on CD4 and may be specific for
fragments, mutant
forms, splice forms, or aberrant forms of said antigens. Examples of CD4-
positive cells include
CD4-positive T cells such as a Thl cell, a Th2 cell, a Th17 cell, a regulatory
T cell (Treg), and a
y6T cell. Further, CD4-positive cells are associated with diseases including
cancer and
inflammatory diseases (e.g., autoimmune disease or an allergic disease).
Numerous anti-CD4 antibodies and antigen binding fragments are known in the
art and/or
are available commercially, all of which may find use in the present
invention.
Table 1 provides a list of various commercially-sourced anti-CD4 antibodies
that may be
used in the present disclosure.
Table 1: Exemplary anti-CD4 antibodies available commercially
Antibody Name/Description Source
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat
#12-0049-42
PE, eBiosciencelm
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat
#11-0049-80
FITC, eBioscience TM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat
#12-0049-80
PE, eBiosciencelm
CD4 Monoclonal Antibody (OKT4 ThermoFisher Scientific Cat it
11-0048-42
(OKT-4)), FITC, eBioscienceTM
CD4 Monoclonal Antibody (OKT4 ThermoFisher Scientific Cat ft
11-0048-80
(OKT-4)), FITC, eBioscienceTM
109
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
CD4 Monoclonal Antibody (OKT4 ThermoFisher Scientific Cat #
56-0048-82
(OKT-4)), Alexa Fluor 700,
eBioscience TM
CD4 Monoclonal Antibody (OKT4 ThermoFisher Scientific Cat #
53-0048-42
(OKT-4)), Alexa Fluor 488,
eBioscience TM
CD4 Monoclonal Antibody (OKT4 ThennoFisher Scientific Cat #
56-0048-41
(OKT-4)), Alexa Fluor 700,
eBioscience TM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat #
14-0049-82
eBioscience TM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat 14
17-0049-42
APC, eBioscience TM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat #
45-0049-42
PerCP-Cyanine5 .5, eBiosci en ce TM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat /4
25-0049-42
PE-Cyanine7, eBioscience TM
CD4 Monoclonal Antibody (RPA-T4), ThennoFisher Scientific Cat #
15-0049-42
PE-Cyanine5, eBioscience TM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat #
48-0049-42
eFluor 450, eBioscienceTM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat It
56-0049-42
Alexa Fluor 700, eBioscienceTM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat #
13-0049-82
Biotin, eBioscience TM
CD4 Monoclonal Antibody (RPA-T4), ThermoFisher Scientific Cat 14
16-0049-85
Functional Grade, eBioscienceTM
CD4 Monoclonal Antibody (RPA-T4), ThennoFisher Scientific Cat #
61-0049-42
PE-eFluor 610, eBioscienceTM
In addition to commercial source, a large number of monoclonal antibodies
against CD4
have been reported in the literature. Various anti-CD4 mAbs are under clinical
development for
the purpose of treating cancers, immune diseases, and infections. For example,
based on the fact
that the binding between CD4 and HIV is essential for the infection of HIV, an
antibody which
recognizes D1 domain of CD4 can inhibit the infection of HIV, under the
development as an HIV
therapeutic agent. Examples of anti-CD4 mAbs developed as a therapeutic agent
for cancers or
immune diseases include zanolimumab (6G5), ibalizumab, tregalizumab, and
keliximab (CE9.1).
These antibodies are antibodies which exert their medicinal efficacy by
specifically attacking
CD4-expressing cells which are target cells, and it is considered that the
mechanism of medicinal
efficacy is mainly due to an ADCC activity (Kim eral., Blood, 109(11):4655-
4662, 2007).
In addition, the present disclosure contemplates the use of any of the anti-
CD4 antibodies
or antibody fragments thereof disclosed in the following references:
U57338658B2;
U55741488A; US5871732A; U59758581B2; Delmonico et al., "Anti-CD4 monoclonal
antibody
110
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
therapy," Clin Transplant, 1996, Oct 10(5): pp. 397-403; Konig et al.,
"Tregalizumab ¨ A
Monoclonal Antibody to Target Regulatory T Cells," Front lmmunol. 2016,
Vol.7:11; and JF
Bach, "Therapeutic monoclonal antibodies, Ann Pharm Fr., 2006, 64(5): 308-11,
each of which
are incorporated herein by reference in their entireties.
All of the above-noted commercially-available and anti-CD4 antibodies known in
the
literature can be used in the instant disclosure.
In certain other embodiments, the targeting moieties for targeting the
delivery vehicles to
a target cell (e.g., leukocytes) are anti-CD8 antibodies or antigen binding
fragments.
CD8 is a surface glycoprotein that functions as a co-receptor for TCR
recognition of
peptide antigen complexed with MHC Class I molecule (pMHCI). It is expressed
either as an aa
homodimer or as an a43 heterodimer (Zamoyska, Immunity, 1 :243-6, 1994), both
chains
expressing a single extracellular Ig superfamily (IgSF) V domain, a membrane
proximal hinge
region, a transmembrane domain, and a cytoplasmic tail. CD8 interacts with im
and the a2 and a3
domains of MHC Class I molecules using its 13 strands and the complementary
determining
regions (CDRs) within the extracellular IgSF V domain. This association
increases the
adhesion/avidity of the T cell receptor with its Class I target.
In addition, an internal signaling cascade mediated by the CD8a chain
associated tyrosine
protein kinase p561ck4'5 leads to T cell activation. Lck is required for
activation and expansion
of naive CD8+ T cells; however its expression is not essential for responses
of memory CD8+ T
cells to secondary antigenic stimulation in vivo or in vitro (Bachman et al, J
Exp Med, 189: 1521-
30, 1999). As shown by either CD8a or CD8B gene targeted mice, CD8 plays an
important role in
the maturation and function of MHC Class I-restricted T lymphocytes (Nakayama
et al, Science,
263: 1131-3, 1984). One patient suffering from repeated bacterial infections
was found to display
a CD8 deficiency due to a single mutation in the CD8a gene. The lack of CD8
did not appear to
be essential for either CD8+ T cell lineage commitment or peripheral cytolytic
function (de la
Calle-Martin et al, J Clin Invest, 108: 117-23, 2001).
The human CD8 molecule is a glycoprotein and cell surface marker expressed on
cytotoxic T-cells (CTLs). These are a subset of T-lymphocytes and play an
important role in the
adaptive immune system of vertebrates. They are responsible for the
elimination of virus-
infected cells or other abnormal cells such as some tumor cells. These cells
are specifically
recognized via the T-cell receptor (TCR), which interacts with the certain
antigen presented via
MHC (major histocompatibility complex) class I on target cells.
An exemplary CD8 amino acid sequence is represented by P01732-1 (UniParc),
which is
known as the canonical sequence:
111
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
MALPVTALLLP LALLLHAARP S QFRV SPLDRTWNLGETVELKC QVLL SNPTS GC SWLFQ
PRGAAA S PTFLLY LS Q N KPKAAEGLD TQRF SGKRLGDTFVLTL S DFRREN EGY YFC SAL S
NSIMYFSHFVPVFLPAKPTTTPAPRPPTPAPTIASQPLSLRPEACRPAAGGAVHTRGLDFA
CDIYIWAPLAGTCGVLLLSLVITLYCNHRNRRRVCKCPRPVVK SGDKPSLSARYV (SEQ
ID NO:2).
Numerous anti-CD8 antibodies and antigen binding fragments are known in the
art and/or
are available commercially, all of which may find use in the present
invention.
Table 2 provides a list of various commercially-sourced anti-CD8 antibodies
that may be
used in the present disclosure.
Table 2: Exemplary anti-CD8 antibodies available commercially
Antibody Name/Description Source
CD8a Monoclonal Antibody (RPA-T8), PE, ThermoFisher Scientific Cat
# 12-0088-42
eBioscience TM
CD8a Monoclonal Antibody (RPA-T8), PE, ThermoFisher Scientific Cat
14 12-0088-80
eBioscience TM
CD8a Monoclonal Antibody (OKT8 (OKT-8)), ThermoFisher Scientific Cat # 53-0086-
42
Alexa Fluor 488, eBioscience TM
CD8a Monoclonal Antibody (53-6.7), ThenuoFisher Scientific Cat
# 14-0081-82
eBioscience TM
CD8a Monoclonal Antibody (53-6.7), FITC, ThermoFisher Scientific Cat
# 11-0081-82
eBioscience TM
CD8a Monoclonal Antibody (53-6.7), PE, ThermoFisher Scientific Cat
# 12-0081-82
cBioscienceTm
CD8a Monoclonal Antibody (53-6.7), eFluor ThermoFisher Scientific Cat
# 48-0081-82
450, eBioscience TM
CD8a Monoclonal Antibody (53-6.7), Biotin, ThermoFisher Scientific Cat
# 13-0081-82
eBioscience TM
CD8a Monoclonal Antibody (53-6.7), Alexa ThermoFisher Scientific Cat
14 56-0081-82
Fluor 700, eBioscienceTM
CD8a Monoclonal Antibody (53-6.7), PE- ThermoFisher Scientific Cat
# 15-0081-82
Cyanine5, eBioscienceTM
CD8a Monoclonal Antibody (53-6.7), Alexa ThermoFisher Scientific Cat
/4 53-0081-82
Fluor 488, eBioscienceTM
CD8a Monoclonal Antibody (53-6.7), ThermoFisher Scientific Cat
/4 16-0081-82
Functional Grade, eBioscienceTM
CD8a Monoclonal Antibody (53-6.7), eFluor ThermoFisher Scientific Cat
# 50-0081-82
660, eBiosciencem
CD8a Monoclonal Antibody (53-6.7), Alexa ThermoFisher Scientific Cat
# 58-0081-80
Fluor 532, eBioscienceTM
CD8a Monoclonal Antibody (RPA-T8), PE, ThermoFisher Scientific Cat
# 12-0088-42
eBioscience TM
CD8a Monoclonal Antibody (OKT8 (OKT-8)), ThermoFisher Scientific Cat ft 12-
0088-80
Alexa Fluor 488, eBioscience TM
112
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
CD8a Monoclonal Antibody (RPA-T8), APC, ThermoFisher Scientific Cat
# 53-0086-42
eBioscience TM
CD8a Monoclonal Antibody (RPA-T8), ThermoFisher Scientific Cat
# 17-0088-42
PerCP-Cyanine5 .5, eBiosciencc TM
CD8a Monoclonal Antibody (RPA-T8), Alexa ThermoFisher Scientific Cat # 45-0088-
42
Fluor 532, eBioscienceTM
CD8a Monoclonal Antibody (RPA-T8), APC- ThermoFisher Scientific Cat # 58-0088-
42
eFluor 780, eBioscienceTM
There are numerous anti-CD8 antibodies, including monoclonal antibodies, known
in the
art, including: 2D2; 4D12.1; 7B12 1G1 1; 8E-1.7; 8G5; 14; 21Thy; 51.1 ; 66.2;
109-2D4; 138-
17; 143-44; 278F24; 302F27; AICD8.1 ; anti-T8; B9.1.1; B9.2.4; B9.3.1 ; B9.4.1
; B9.7.6;
B9.8.6; B9.1 1; B9.1 1.10; BE48; BL15; BL-TS8; BMAC8; BU88; BW135/80; C1-11G3;
CIO;
C12/D3; CD8-4C9; CLB-T8/1 CTAG-CD8, 3B5; F80-1D4D11 ; F101-87 (S-T8a); GIO-I;
G10-
1.1; HI208; HI209; HI212; HIT8a; HIT8b; HIT8d; IC0-31 ; IC0-122; IP48; ITI-
5C2; ITM8-1;
JML-H7; JML-H8; L2; L533; Leu-2a; LT8; LY17.2E7; LY19.3B2; M236; M-T122; M-
T415; M-
T805; M-T806; M- T807; M-T808; M-T809; M-T1014; MCD8; MEM-31; MEM- 146; NU-
Ts/c;
OKT8; OKT8f; P218; RPA-T8; SM4; T8; T8 /2T8-19; T8 /2T8-2A1 ; T8 /2T8- 1B5; T8
/2T8-
ICI ; T8 /7Pt3F9; T8 /21thy2D3; T8 /21 thy; T8 /TPE3FP; T8b; T41D8; T811 ;
TU68; TU102;
UCHT4; VIT8; VIT8b; WuT8-1; X107; YTC141.1; and/or YTC 182.20. In addition to
commercial sources, a large number of monoclonal antibodies against CD8 have
been reported in
the literature. For example, the anti-CD8 antibodies or fragments thereof
described in the
following publications are contemplated herein: AU2014249243B2; 10,746,726;
9,790,279;
9,758,581; 9,587,022; 8,877,913; 8,685,651; 8,673,304; 8,586,715; 8,440,806;
8,399,621;
7,541,443; 7,482,000; 7,452,981; 7,452,534; 7,338,658; 6,136,310; 6,056,956;
5,871,732; and
5,741,488, each of which are incorporated herein by reference in their
entireties.
All of the above-noted commercially-available and anti-CD8 antibodies known in
the
literature can be used in the instant disclosure.
In certain other embodiments, the targeting moieties for targeting the
delivery vehicles to
a target cell (e.g., leukocytes) are anti-CD3 antibodies or antigen binding
fragments.
CD3 antigen is associated with the T-cell receptor complex on T-cells.
Multispecific
antigen binding proteins having specificity to CD3 and an antigen of a target
cell can trigger the
cytotoxic activity of T-cells on target cells. Namely, by multispecific
binding of the antigen
binding protein to CD3 and to a target cell, e.g. a tumor cell, cell lysis of
the target cell may be
induced. Antigen binding proteins with a CD3 binding site and their production
are known in the
art (and described for example in Kipriyanov et al., 1999, Journal of
Molecular Biology 293:41-
56, Le Gall et al., 2004, Protein Engineering, Design & Selection, 17/4:357-
366).
113
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
The CD3 antigen is a complex of 5 invariable polypeptide chains: y, 6, E, and
r, whose
molecular weights arc respectively 25-28, 21, 20, 16 and 22 kDa. The CD3
chains arc clustered in
a group of two invariant dimers, y/s and 6/s associated with a variable dimer
which consists of
homodimers, or t; /it or Cy FcR heterodimers (y FcR being the y chain of the
Fe receptors), or
yFcR homodimers. The CD3 is part of a bigger complex which includes the T cell
receptor
(TCR). CD3 complex associated with the TCR is involved in the recognition of
peptides bound to
the major histocompatibility complex class I and II during the immune
response. T cell activation
may be induced when a foreign antigen is presented to the TCR through MI-IC
complex. The CD3
antigen is expressed by mature T lymphocytes and by a subset of thymocytes.
Numerous anti-CD3 antibodies and antigen binding fragments are known in the
art and/or
are available commercially, all of which may find use in the present
invention.
Table 3 provides a list of various commercially-sourced anti-CD8 antibodies
that may be
used in the present disclosure.
Table 3: Exemplary anti-CD3 antibodies available commercially
Antibody Name/Description Source
CD3 Monoclonal Antibody (17A2), 'ThermoFisher Scientific
Cat # 14-0032-82
eBioscience TM
CD3 Monoclonal Antibody (17A2), ThermoFisher Scientific
Cat # 48-0037-42
eBioscience TM
CD3 Monoclonal Antibody (OKT3), Biotin, ThermoFisher Scientific
Cat # 13-0037-82
eBioscience TM
CD3 Monoclonal Antibody (OKT3), Alexa ThermoFisher Scientific
Cat # 53-0037-42
Fluor 48%, eRioscienceTM
CD3 Monoclonal Antibody (OKT3), Alexa ThermoFisher Scientific
Cat # 56-0037-42
Fluor 700, eBioscienceTM
CD3 Monoclonal Antibody (OKT3), eFluor ThermoFisher Scientific
Cat # 50-0037-42
660, eBioscience TM
CD3 Monoclonal Antibody (OKT3), Super ThermoFisher Scientific
Cat # 63-0037-42
Bright 600, eBioscienceTM
CD3 Monoclonal Antibody (UCHT1), Alexa ThermoFisher Scientific
Cat # 58-0038-42
Fluor 532, eBioscienceTM
114
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
CD3 Monoclonal Antibody (UCHT1), FITC, ThermoFisher Scientific
Cat # 11-0038-42
eBioscience TM
CD3 Monoclonal Antibody (UCHT1), FITC, ThermoFisher Scientific
Cat # 11-0038-80
eBioscience TM
CD3 Monoclonal Antibody (UCHT1), ThermoFisher Scientific
Cat # 14-0038-82
eBioscience TM
Anti-CD3 antibodies, including monoclonal antibodies, are known in the art,
including:
those disclosed in U52018/005 7593, US11007267B2, US10865251B2, US10759858B2,
U510906978B2, U520210253701A1, US20200123255A1, U520210095027A1,
US20210147561A1, US10544220B2, US20190284278A1, US20190263904A1, and
US20200048348A1, each of which are incorporated herein by reference in their
entireties.
All of the above-noted commercially-available and anti-CD3 antibodies known in
the
literature can be used in the instant disclosure.
In addition, the antibodies of the present disclosure which may be used as
targeting
moieties on the delivery vehicle used herein may also be made by any suitable
conventional
means. In one approach, following immunization with an antigen of interest
(e.g., CD4), somatic
cells with the potential for producing antibodies, specifically B lymphocytes
(B cells), are
selected for use in the MAb generating protocol. These cells may be obtained
from biopsied
spleens or lymph nodes, or from circulating blood. The antibody-producing B
lymphocytes from
the immunized animal are then fused with cells of an immortal myeloma cell,
generally one of the
same species as the animal that was immunized or human or human/mouse chimeric
cells.
Myeloma cell lines suited for use in hybridoma-producing fusion procedures
preferably are non-
antibody-producing, have high fusion efficiency, and enzyme deficiencies that
render then
incapable of growing in certain selective media which support the growth of
only the desired
fused cells (hybridomas). Any one of a number of myeloma cells may be used, as
are known to
those of skill in the art (Goding, pp. 65-66, 1986; Campbell, pp. 75-83,
1984).
Methods for generating antibodies
Methods for generating hybrids of antibody-producing spleen or lymph node
cells and
myeloma cells usually comprise mixing somatic cells with myeloma cells in the
presence of an
agent or agents (chemical or electrical) that promote the fusion of cell
membranes. Fusion
methods using Sendai virus have been described by Kohler and Milstein (1975;
1976), and those
115
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
using polyethylene glycol (PEG), such as 37% (v/v) PEG, by Gefter et al.
(1977). The use of
electrically induced fusion mcthods also is appropriate (Goding, pp. 71-74,
1986).
Culturing provides a population of hybridomas from which specific hybridomas
are
selected. Typically, selection of hybridomas is performed by culturing the
cells by single-clone
dilution in microtiter plates, followed by testing the individual clonal
supernatants (after about
two to three weeks) for the desired reactivity. The assay should be sensitive,
simple and rapid,
such as radioimmunoassays, enzyme immunoassays, cytotoxicity assays, plaque
assays dot
immunobinding assays, and the like. The selected hybridomas are then serially
diluted or single-
cell sorted by flow cytometric sorting and cloned into individual antibody-
producing cell lines,
which clones can then be propagated indefinitely to provide mAbs.
The cell lines may be exploited for MAb production in two basic ways. A sample
of the
hybridoma can be injected (often into the peritoneal cavity) into an animal
(e.g., a mouse).
Optionally, the animals are primed with a hydrocarbon, especially oils such as
pristane
(tetramethylpentadecane) prior to injection. When human hybridomas are used in
this way, it is
optimal to inject immunocompromised mice, such as SCID mice, to prevent tumor
rejection. The
injected animal develops tumors secreting the specific monoclonal antibody
produced by the
fused cell hybrid. The body fluids of the animal, such as serum or ascites
fluid, can then be
tapped to provide MAbs in high concentration. The individual cell lines could
also be cultured in
vitro, where the MAbs are naturally secreted into the culture medium from
which they can be
readily obtained in high concentrations. Alternatively, human hybridoma cells
lines can be used
in vitro to produce immunoglobulins in cell supernatant. The cell lines can be
adapted for growth
in serum-free medium to optimize the ability to recover human monoclonal
immunoglobulins of
high purity.
MAbs produced by either means may be further purified, if desired, using
filtration,
centrifugation and various chromatographic methods such as FPLC or affinity
chromatography.
Fragments of the monoclonal antibodies of the disclosure can be obtained from
the purified
monoclonal antibodies by methods which include digestion with enzymes, such as
pepsin or
papain, and/or by cleavage of disulfide bonds by chemical reduction.
Alternatively, monoclonal
antibody fragments encompassed by the present disclosure can be synthesized
using an automated
peptide synthesizer.
It also is contemplated that a molecular cloning approach may be used to
generate
monoclonals. For this, RNA can be isolated from the hybridoma line and the
antibody genes
obtained by RT-PCR and cloned into an immunoglobulin expression vector.
Alternatively,
combinatorial immunoglobulin phagemid libraries are prepared from RNA isolated
from the cell
116
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
lines and phagemids expressing appropriate antibodies are selected by panning
using viral
antigens. The advantages of this approach over conventional hybridoma
techniques are that many
more antibodies can be produced and screened in a single round, and that new
specificities are
generated by H and L chain combination which further increases the chance of
finding
appropriate antibodies.
U.S. patents, each incorporated herein by reference, that teach the production
of
antibodies useful in the present disclosure include U.S. Patent 5,565,332,
which describes the
production of chimeric antibodies using a combinatorial approach; U.S. Patent
4,816,567 which
describes recombinant immunoglobulin preparations; and U.S. Patent 4,867,973
which describes
antibody-therapeutic agent conjugates.
Combinations
In one embodiment, the invention provides a combination of T cell targeted
delivery
vehicles, targeting two or more T cell antigens. In one embodiment, the two or
more T cell
antigens are selected from CD1, CD2, CD3, CD4, CD5, CD7, CD8, CD16, CD25,
CD26, CD27,
CD28, CD30, CD38, CD39, CD4OL, CD44, CD45, CD62L, CD69, CD73, CD80, CD83,
CD86,
CD95, CD103, CD119, CD126, CD150, CD153, CD154, CD161, CD183, CD223, CD254,
CD275, CD45RA, CXCR3, CXCR5, FasL, IL18R1, CTLA-4, 0X40, GITR, LAG3, ICOS, PD-
1,
leu-12, TCR, TLR1, TLR2, TLR3, TLR4, TLR6, NKG2D, CCR, CCR1, CCR2, CCR4, CCR6,
or
CCR7. In one embodiment, the combination comprises one or more T cell targeted
delivery
vehicles, targeting a surface antigen of a CD4-I- T cell and a surface antigen
of a CD8+ T cell. In
one embodiment, the combination comprises two or more T cell targeted delivery
vehicles,
targeting CD4 and CD8.
In one embodiment, the combination of T cell targeted delivery vehicles
delivers the
same agent to T cells expressing different surface antigens. In one
embodiment, the combination
of T cell targeted delivery vehicles delivers a first agent to one subset of T
cells expressing a first
surface antigen and a second, different agent to a second subset of T cells
expressing a second
surface antigen. Therefore, in various embodiments, the combinations of the
invention can be
used deliver 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more than 10 different agents to
T cells.
In one embodiment, the composition of the present invention comprises a
combination of
agents described herein. In certain embodiments; a composition comprising a
combination of
agents described herein has an additive effect, wherein the overall effect of
the combination is
approximately equal to the sum of the effects of each individual agent. In
other embodiments, a
composition comprising a combination of agents described herein has a
synergistic effect,
117
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
wherein the overall effect of the combination is greater than the sum of the
effects of each
individual agent.
A composition comprising a combination of agents comprises individual agents
in any
suitable ratio. For example, in one embodiment, the composition comprises a
1:1 ratio of two
individual agents. However, the combination is not limited to any particular
ratio. Rather any
ratio that is shown to be effective is encompassed.
Conjugation
In various embodiments of the invention, the delivery vehicle is conjugated to
a targeting
domain. Exemplary methods of conjugation can include, but are not limited to,
covalent bonds,
electrostatic interactions, and hydrophobic ("van der Waals") interactions. In
one embodiment,
the conjugation is a reversible conjugation, such that the delivery vehicle
can be disassociated
from the targeting domain upon exposure to certain conditions or chemical
agents. In another
embodiment, the conjugation is an irreversible conjugation, such that under
normal conditions the
delivery vehicle does not dissociate from the targeting domain.
In some embodiments, the conjugation comprises a covalent bond between an
activated
polymer conjugated lipid and the targeting domain. The term "activated polymer
conjugated
lipid" refers to a molecule comprising a lipid portion and a polymer portion
that has been
activated via functionalization of a polymer conjugated lipid with a first
coupling group. In one
embodiment, the activated polymer conjugated lipid comprises a first coupling
group capable of
reacting with a second coupling group. In one embodiment, the activated
polymer conjugated
lipid is an activated pegylated lipid. In one embodiment, the first coupling
group is bound to the
lipid portion of the pegylated lipid. In another embodiment, the first
coupling group is bound to
the polyethylene glycol portion of the pegylated lipid. In one embodiment, the
second functional
group is covalently attached to the targeting domain.
The first coupling group and second coupling group can be any functional
groups known
to those of skill in the art to together form a covalent bond, for example
under mild reaction
conditions or physiological conditions. In some embodiments, the first
coupling group or second
coupling group are selected from the group consisting of maleimides, N-
hydroxysuccinimide
(NHS) esters, carbodiimides, hydrazide, pentafluorophenyl (PFP) esters,
phosphines,
hydroxymethyl phosphines, psoralen, imidoesters, pyridyl disulfide,
isocyanates, vinyl sulfones,
alpha-haloacetyls, aryl azides, acyl azides, alkyl azides, diazirines,
benzophenone, epoxides,
carbonates, anhydrides, sulfonyl chlorides, cyclooctyne, aldehydes, and
sulfhydryl groups. In
some embodiments, the first coupling group or second coupling group is
selected from the group
118
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
consisiting of free amines (-NH2), free sulfhydryl groups (-SH), free
hydroxide groups (-OH),
carboxylates, hydrazides, and alkoxyamines. In some embodiments, the first
coupling group is a
functional group that is reactive toward sulfhydryl groups, such as maleimide,
pyridyl disulfide,
or a haloacetyl. In one embodiment, the first coupling group is a 'mien-nide.
In one embodiment, the second coupling group is a sulfhydryl group. The
sulfhydryl
group can be installed on the targeting domain using any method known to those
of skill in the
art. In one embodiment, the sulfhydryl group is present on a free cysteine
residue. In one
embodiment, the sulfhydryl group is revealed via reduction of a disulfide on
the targeting domain,
such as through reaction with 2-mercaptoethylamine. In one embodiment, the
sulfhydryl group is
installed via a chemical reaction, such as the reaction between a free amine
and 2-iminothilane or
N-succinimidyl S-acetylthioacetate (SATA).
In some embodiments, the polymer conjugated lipid and targeting domain are
functionalized with groups used in "click" chemistry. Bioorthogonal -click"
chemistry comprises
the reaction between a functional group with a 1,3-dipole, such as an azide, a
nitrile oxide, a
nitrone, an isocyanide, and the link, with an alkene or an alkyne
dipolarophiles. Exemplary
dipolarophiles include any strained cycloalkenes and cycloalkynes known to
those of skill in the
art, including, but not limited to, cyclooctynes, dibenzocyclooctynes,
monofluorinated
cycicooctynes, difluorinated cyclooctynes, and biarylazacyclooctynone.
Peptide targeting moiety
In one embodiment, the targeting domain of the invention comprises a peptide.
In certain
embodiments, the peptide targeting domain specifically binds to a target of
interest.
The peptide of the present invention may be made using chemical methods. For
example,
peptides can be synthesized by solid phase techniques (Roberge J Y et al
(1995) Science 269:
202-204), cleaved from the resin, and purified by preparative high performance
liquid
chromatography. Automated synthesis may be achieved, for example, using the
ABI 431 A
Peptide Synthesizer (Perkin Elmer) in accordance with the instructions
provided by the
manufacturer.
The peptide may alternatively be made by recombinant means or by cleavage from
a
longer polypeptide. The composition of a peptide may be confirmed by amino
acid analysis or
sequencing.
The variants of the peptides according to the present invention may be (i) one
in which
one or more of the amino acid residues are substituted with a conserved or non-
conserved amino
acid residue (preferably a conserved amino acid residue) and such substituted
amino acid residue
119
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
may or may not be one encoded by the genetic code, (ii) one in which there are
one or more
modified amino acid residues, e.g., residues that are modified by the
attachment of substitucnt
groups, (iii) one in which the peptide is an alternative splice variant of the
peptide of the present
invention, (iv) fragments of the peptides and/or (v) one in which the peptide
is fused with another
peptide, such as a leader or secretory sequence or a sequence which is
employed for purification
(for example, His-tag) or for detection (for example, Sy5 epitope tag). The
fragments include
peptides generated via proteolytic cleavage (including multi-site proteolysis)
of an original
sequence. Variants may be post-translationally, or chemically modified. Such
variants are deemed
to be within the scope of those skilled in the art from the teaching herein.
As known in the art the -similarity" between two peptides is determined by
comparing
the amino acid sequence and its conserved amino acid substitutes of one
peptide to a sequence of
a second peptide. Variants are defined to include peptide sequences different
from the original
sequence, preferably different from the original sequence in less than 40% of
residues per
segment of interest, more preferably different from the original sequence in
less than 25% of
residues per segment of interest, more preferably different by less than 10%
of residues per
segment of interest, most preferably different from the original protein
sequence in just a few
residues per segment of interest and at the same time sufficiently homologous
to the original
sequence to preserve the functionality of the original sequence. The present
invention includes
amino acid sequences that are at least 60%, 65%, 70%, 72%, 74%, 76%, 78%, 80%,
90%, or 95%
similar or identical to the original amino acid sequence. The degree of
identity between two
peptides is determined using computer algorithms and methods that are widely
known for the
persons skilled in the art. The identity between two amino acid sequences is
preferably
determined by using the BLASTP algorithm [BLAST Manual, Altschul, S., et al.,
NCBI NLM
NIH Bethesda, Md. 20894, Altschul, S., et al., J. Mol. Biol. 215: 403-410
(1990)1.
The peptides of the invention can be post-translationally modified. For
example, post-
translational modifications that fall within the scope of the present
invention include signal
peptide cleavage, glycosylation, acetylation, isoprenylation, proteolysis,
myristoylation, protein
folding and proteolytic processing, etc. Some modifications or processing
events require
introduction of additional biological machinery. For example, processing
events, such as signal
peptide cleavage and core glycosylation, are examined by adding canine
microsomal membranes
or Xenopus egg extracts (U.S. Pat. No. 6,103,489) to a standard translation
reaction.
The peptides of the invention may include unnatural amino acids formed by post-
translational modification or by introducing unnatural amino acids during
translation.
120
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Nucleic acid targeting moiety
In one embodiment, the targeting domain of the invention comprises an isolated
nucleic
acid, including for example a DNA oligonucleotide and a RNA oligonucleotide.
In certain
embodiments, the nucleic acid targeting domain specifically binds to a target
of interest. For
example, in one embodiment, the nucleic acid comprises a nucleotide sequence
that specifically
binds to a target of interest.
The nucleotide sequences of a nucleic acid targeting domain can alternatively
comprise
sequence variations with respect to the original nucleotide sequences, for
example, substitutions,
insertions and/or deletions of one or more nucleotides, with the condition
that the resulting
nucleic acid functions as the original and specifically binds to the target of
interest.
In the sense used in this description, a nucleotide sequence is "substantially
homologous"
to any of the nucleotide sequences describe herein when its nucleotide
sequence has a degree of
identity with respect to the nucleotide sequence of at least 60%,
advantageously of at least 70%,
preferably of at least 85%, and more preferably of at least 95%. Other
examples of possible
modifications include the insertion of one or more nucleotides in the
sequence, the addition of
one or more nucleotides in any of the ends of the sequence, or the deletion of
one or more
nucleotides in any end or inside the sequence. The degree of identity between
two
polynucleotides is determined using computer algorithms and methods that are
widely known for
the persons skilled in the art. The identity between two amino acid sequences
is preferably
determined by using the BLASTN algorithm [BLAST Manual, Altschul, S., et al.,
NCBI NLM
NIH Bethesda, Md. 20894, Altschul, S., et al., J. Mol. Biol. 215: 403-410
(1990)1.
Antibody ta1i2etin2 moiety
In one embodiment, the targeting domain of the invention comprises an
antibody, or
antibody fragment. In certain embodiments, the antibody targeting domain
specifically binds to a
target of interest. Such antibodies include polyclonal antibodies, monoclonal
antibodies, Fab and
single chain FA,' (scFv) fragments thereof, bispecific antibodies,
heteroconjugates, human and
humanized antibodies.
The antibodies may be intact monoclonal or polyclonal antibodies, and
immunologically
active fragments (e.g., a Fab or (Fab)2 fragment), an antibody heavy chain, an
antibody light
chain, humanized antibodies, a genetically engineered single chain Fv molecule
(Ladner et al,
U.S. Pat. No. 4,946,778), or a chimeric antibody, for example, an antibody
which contains the
binding specificity of a murine antibody, but in which the remaining portions
are of human
121
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
origin. Antibodies including monoclonal and polyclonal antibodies, fragments
and chimeras, may
be prepared using methods known to those skilled in the art.
Such antibodies may be produced in a variety of ways, including hybridoma
cultures,
recombinant expression in bacteria or mammalian cell cultures, and recombinant
expression in
transgenic animals. The choice of manufacturing methodology depends on several
factors
including the antibody structure desired, the importance of carbohydrate
moieties on the
antibodies, ease of culturing and purification, and cost. Many different
antibody structures may be
generated using standard expression technology, including full-length
antibodies, antibody
fragments, such as Fab and Fv fragments, as well as chimeric antibodies
comprising components
from different species. Antibody fragments of small size, such as Fab and FAT
fragments, having
no effector functions and limited pharmokinetic activity may be generated in a
bacterial
expression system. Single chain Fv fragments show low immunogenicity.
In one embodiment, the targeting domain of the instant invention is an
antibody that
specifically binds to a surface antigen of a CD4+ T cell. In one embodiment,
the targeting domain
of the instant invention is an antibody that specifically binds to CD4.
T cells
In various embodiments, the target of the delivery vehicles can be any type of
cell in the
body (i.e., "target cells-). In preferred embodiments, target cells are immune
cells, such as, but
not limited to, any class of myeloid cell (e.g., neutrophils, eosinophils,
mast cells, basophils, and
monoc_vtes) or any class of lymphocyte (e.g., T cells (e.g., cytotoxic T
cells, helper T cells, or
memory T cells), B cells (e.g., plasma cells and memory B cells), and natural
killer cells).
In some embodiments, T cells that can be targeted using the compositions of
the
invention immunostimulatory cells, i.e., cells that mediate an immune
response. In some
embodiments, T cells that can be targeted using the compositions of the
invention can include,
but are not limited to, T helper cells (CD4+) and CD4+ cytotoxic T cells (also
referred to as
cytotoxic T lymphocytes, CD4+ CTL). In certain embodiments, the target cells
are T cells. In
some embodiments, T cells that can be targeted using the compositions of the
invention can be
CD4+ or CD8+ and can include, but are not limited to, T helper cells (CD4+),
cytotoxic T cells
(also referred to as cytotoxic T lymphocytes, CTL; CD8¨ T cells), and memory T
cells, including
central memory T cells (TCM), stem memory T cells (TSCM), stem-cell-like
memory T cells (or
stem-like memory T cells), and effector memory T cells, for example, TEM cells
and TEMRA
(CD45RA+) cells, effector T cells, Thl cells. Th2 cells, Th9 cells, Th17
cells, Th22 cells, Tfh
(follicular helper) cells, T regulatory cells, natural killer T cells, mucosal
associated invariant T
122
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
cells (MAIT), and 7.5 T cells. Major T cell subtypes include TN (naive), TSCM
(stem cell
memory), TCM (central memory), TTM (Transitional Memory), TEM (Effector
memory), and
TTE (Terminal Effector), TCR-transgenic T cells, T-cells redirected for
universal cytokine-
mediated killing (TRUCK), Tumor infiltrating T cells (TIL), CAR-T cells or any
T cell that can
be used for treating a disease or disorder. In some embodiments, the T cells
are CD4+ T cells.
Therapeutic Methods
The present invention provides methods of delivering at least one agent for
diagnosis,
treatment or prevention of a disease, or a disease or disorder to a CD4+ T
cell. In one
embodiment, the CD4+ T cell targeted delivery vehicle comprises or
encapsulates an agent to be
administered to a subject. In some embodiments, the agent is a nucleoside
modified mRNA. The
present invention therefore provides methods of delivering at least one agent
to a CD4+ T cell.
In one embodiment, the CD4+ T cell targeted delivery vehicle comprises or
encapsulates
a therapeutic agent for the treatment of a disease or disorder. In some
embodiments, the
therapeutic agent is a nucleoside modified mRNA. The present invention
therefore provides
methods of delivering at least one therapeutic agent to a CD4 T cell. In
certain embodiments, the
method is used to treat or prevent a disease or disorder in a subject.
Exemplary diseases or
disorders include, but are not limited to, cancers, infectious diseases, and
immunological
disorders.
The following are non-limiting examples of cancers that can be treated or
prevented by
the disclosed methods: acute lymphoblastic leukemia, acute myeloid leukemia,
adrenocortical
carcinoma, appendix cancer, basal cell carcinoma, bile duct cancer, bladder
cancer, bone cancer,
brain and spinal cord tumors, brain stem glioma, brain tumor, breast cancer,
bronchial tumors,
burkitt lymphoma, carcinoid tumor, central nervous system atypical
teratoid/rhabdoid tumor,
central nervous system embryonal tumors, central nervous system lymphoma,
cerebellar
astrocytoma, cerebral astrocytoma/malignant glioma, cerebral
astrocytotna/malignant glioma,
cervical cancer, childhood visual pathway tumor, chordoma, chronic lymphoeytic
leukemia,
chronic myelogenous leukemia, chronic myeloproliferative disorders, colon
cancer, colorectal
cancer, craniopharyngioma, cutaneous cancer, cutaneous t-cell lymphoma,
endometrial cancer,
ependymoblastoma, ependymoma, esophageal cancer, ewing family of tumors,
extracranial
cancer, extragonadal germ cell tumor, extrahepatic bile duct cancer,
extrahepatic cancer, eye
cancer, fungoides, gallbladder cancer, gastric (stomach) cancer,
gastrointestinal cancer,
gastrointestinal carcinoid tumor, gastrointestinal stromal tumor (gist), germ
cell tumor,
gestational cancer, gestational trophoblastic tumor, glioblastoma, glioma,
hairy cell leukemia,
123
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
head and neck cancer, hepatocellular (liver) cancer, histiocytosis, hodgkin
lymphoma,
hypopharyngcal cancer, hypothalamic and visual pathway glioma, hypothalamic
tumor,
intraocular (eye) cancer, intraocular melanoma, islet cell tumors, kaposi
sarcoma, kidney (renal
cell) cancer, langerhans cell cancer, langerhans cell histiocytosis, laryngeal
cancer, leukemia, lip
and oral cavity cancer, liver cancer, lung cancer, lymphoma,
macroglobulinemia, malignant
fibrous histiocvtoma of bone and osteosarcoma, medulloblastoma,
medulloepithelioma,
melanoma, merkel cell carcinoma, mesothelioma, metastatic squamous neck cancer
with occult
primary, mouth cancer, multiple endocrine neoplasia syndrome, multiple
myeloma, mycosis,
myelodysplastic syndromes, myelodysplastic/myeloproliferative diseases,
myelogenous
leukemia, myeloid leukemia, myeloma, myeloproliferative disorders, nasal
cavity and paranasal
sinus cancer, nasopharyngeal cancer, neuroblastoma, non-hodgkin lymphoma, non-
small cell
lung cancer, oral cancer, oral cavity cancer, oropharyngeal cancer,
osteosarcoma and malignant
fibrous histiocytoma, osteosarcoma and malignant fibrous histiocytoma of bone,
ovarian, ovarian
cancer, ovarian epithelial cancer, ovarian germ cell tumor, ovarian low
malignant potential tumor,
pancreatic cancer, papillomatosis, paraganglioma, parathyroid cancer, penile
cancer, pharyngeal
cancer, pheochromocytoma, pineal parenchymal tumors of intermediate
differentiation,
pineoblastoma and supratentorial primitive neuroectodermal tumors, pituitary
tumor, plasma cell
neoplasm, plasma cell neoplasm/multiple myeloma, pleuropulmonary blastoma,
primary central
nervous system cancer, primary central nervous system lymphoma, prostate
cancer, rectal cancer,
renal cell (kidney) cancer, renal pelvis and ureter cancer, respiratory tract
carcinoma involving
the nut gene on chromosome 15, retinoblastoma, rhabdomyo sarcoma, salivary
gland cancer,
sarcoma, sezary syndrome, skin cancer (melanoma), skin cancer (nonmelanoma),
skin carcinoma,
small cell lung cancer, small intestine cancer, soft tissue cancer, soft
tissue sarcoma, squamous
cell carcinoma, squamous neck cancer, stomach (gastric) cancer, supratentorial
primitive
neuroectodermal tumors, supratentorial primitive neuroectodermal tumors and
pineoblastoma, T-
cell lymphoma, testicular cancer, throat cancer, thymoma and thymic carcinoma,
thyroid cancer,
transitional cell cancer, transitional cell cancer of the renal pelvis and
ureter, trophoblastic tumor,
urethral cancer, uterine cancer, uterine sarcoma, vaginal cancer, visual
pathway and hypothalamic
glioma, vulvar cancer, waldenstrom macroglobulinemia, and wilms tumor.
In some embodiments, the present invention features methods for treating or
preventing
autoimmune diseases, including, but not limited to, rheumatoid
arthritis/seronegative
arthropathies, osteoarthritis, inflammatory bowel disease, systemic lupus
erythematosis,
iridoeyelitis/uveitistoptic neuritis, idiopathic pulmonary fibrosis, systemic
vasculitis/Wegener's
gramilomatosis, sarcoidosis, including, but not limited to, rheumatoid
arthritis/seronegative
124
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
arthropathies, osteoarthritis, inflammatory bowel disease, systemic lupus
erythematosis,
iridocyclitis/uvcitistoptic neuritis, idiopathic pulmonary fibrosis, systemic
vasculitis/Wcgcncr's
gramilornatosis, sarcoidosis, myocarditis, postmyocardial infarction syndrome,
postpericardiotomy syndrome, subacute bacterial endocarditis (SBE), anti-
glomerular basement
membrane nephritis, interstitial cystitis, lupus nephritis, autoimmune
hepatitis, primary biliary
cholangitis(PBC), primary sclerosing cholangitis, antisynthetase syndrome,
alopecia areata,
autoimmune angioedema, autoimmune progesterone dermatitis, autoimmune
urticaria, bullous
pemphigoid, cicatricial pemphigoid, dermatitis herpetiformis, discoid lupus
erythematosus,
epidermolysis bullosa acquisita, erythema nodosum, gestational pemphigoid,
hidradenitis
suppurativa, lichen planus, lichen sclerosus, linear IgA disease (LAD),
morphea, pemphigus
vulgaris, pityriasis lichenoides et varioliformis acuta, Mucha-Habermann
disease, psoriasis,
systemic scleroderma, vitiligo, Addison's disease, autoimmune polyendocrine
syndrome (APS)
type 1, autoimmune polyendocrine syndrome (APS) type 2, autoimmune
polyendocrine syndrome
(APS) type 3, autoimmune pancreatitis (AIP), diabetes mellitus type 1,
autoimmune thyroiditis,
Ord's thyroiditis, Graves' disease, autoimmune oophoritis, endometriosis,
autoimmune orchitis,
Sjogren's syndrome, autoimmune enteropathy, Coeliac disease, Crohn's disease,
microscopic
colitis, ulcerative colitis, antiphospholipid syndrome(APS, APLS), aplastic
anemia, autoimmune
hemolytic anemia, autoimmune lymphoproliferative syndrome, autoimmune
neutropenia,
autoimmune thrombocytopenic purpura, cold agglutinin disease, essential mixed
cryoglobulinemia, Evans syndrome, pernicious anemia, pure red cell aplasia,
thrombocytopenia,
adiposis dolorosa, adult-onset Still's disease, ankylosing spondylitis, CREST
syndrome, drug-
induced lupus, enthesitis-related arthritis, eosinophilic fasciitis Felty
syndrome, IgG4-related
disease, juvenile arthritis, Lyme disease (chronic), mixed connective tissue
disease (MCTD),
palindromic rheumatism, Parry Romberg syndrome, Parsonage-Turner syndrome,
psoriatic
arthritis, reactive arthritis, relapsing polychondritis, retroperitoneal
fibrosis, rheumatic fever,
Schnitzler syndrome, undifferentiated connective tissue disease (UCTD),
dermatomyositis,
fibromyalgia, inclusion body myositis, myositis, myasthenia gravis,
neuromyotonia,
paraneoplastic cerebellar degeneration, polymyositis, acute disseminated
encephalomyelitis
(ADEM), acute motor axonal neuropathy, anti-N-methyl-D-aspartate (Anti-NMDA)
receptor
encephalitis, balo concentric sclerosis, Bickerstaffs encephalitis, chronic
inflammatory
demyelinating polyneuropathy, Guillain¨Barre syndrome, Hashimoto's
encephalopathy,
idiopathic inflammatory demyelinating diseases, Lambert-Eaton myasthenic
syndrome, multiple
sclerosis, pattern 11, Oshtoran Syndrome, pediatric autoimmune
neuropsychiatric disorder
associated with streptococcus (PANDAS), progressive inflammatory neuropathy,
restless leg
125
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
syndrome, stiff person syndrome, sydenham chorea, transverse myelitis,
autoimmune retinopathy,
autoimmune uvcitis, Cogan syndrome, Graves ophthalmopathy, intermediate
uveitis, ligneous
conjunctivitis, Mooren's ulcer, neuromyelitis optica, opsoclonus myoclonus
syndrome, optic
neuritis, scleritis, Susac's syndrome, sympathetic ophthalmia, Tolosa-Hunt
syndrome,
autoimmune inner ear disease(AIED), Meniere's disease, Behcet's disease,
eosinophilic
granulomatosis with polyangiitis (EGPA), giant cell arteritis, granulomatosis
with polyangiitis
(GPA), IgA vasculitis (IgAV), Kawasaki's disease, leukocytoclastic vasculitis,
lupus vasculitis,
rheumatoid vasculitis, microscopic polyangiitis (MPA), polyarteritis nodosa
(PAN), polymyalgia
rheumatic, urticarial vasculitis, vasculitis, and primary immune deficiency.
In some embodiments, the therapeutic agent is an agent for the treatment or
prevention of
an infection or an infectious disease. In one embodiment, the therapeutic
agent is an agent for the
treatment or prevention of a bacterial infection or a disease or disorder
associated therewith. The
bacterium can be from any one of the following phyla: Acidobacteria,
Actinobacteria, Aquificae,
Bacteroidetes, Caldiserica, Chlamydiae, Chlorobi, Chloroflexi, Chrysiogenetes,
Cyanobacteria,
Deferribacteres, Deinococcus-Thermus, Dictyoglomi, Elusimicrobia,
Fibrobacteres, Firmicutes,
Fusobacteria, Gemmatimonadetes, Lentisphaerae, Nitrospira, Planctomycetes,
Proteobacteria,
Spirochaetes, Synergistetes, Tenericutes, Thermodesulfobacteria, Thermotogae,
and
Verrucomicrobia.
The bacterium can be a gram-positive bacterium or a gram-negative bacterium.
The
bacterium can be an aerobic bacterium or an anerobic bacterium. The bacterium
can be an
autotrophic bacterium or a heterotrophic bacterium. The bacterium can be a
mesophile, a
neutrophile, an extremophile, an acidophile, an alkaliphile, a thermophile, a
psychrophile, a
halophile, or an osmophile.
The bacterium can be an anthrax bacterium, an antibiotic resistant bacterium,
a disease-
causing bacterium, a food poisoning bacterium, an infectious bacterium,
Salmonella bacterium,
Staphylococcus bacterium, Streptococcus bacterium, or tetanus bacterium. The
bacterium can be a
mycobacteria, Clostridium tetani, Yersinia pestis, Bacillus anthracis,
methicillin-resistant
Staphylococcus aureus (MRSA), or Clostridium difficile.
In one embodiment, the therapeutic agent is an agent for the treatment or
prevention of a
viral infection, or a disease or disorder associated therewith. In some
embodiments, the virus is
from one of the following families: Adenoviridae, Arenaviridae, Bunyaviridae,
Caliciviridae,
Coronaviridae (including SARS and SARS-CoV-2), Filoviridae, Hepadnaviridae,
Herpesviridae,
Orthomyxoviridae, Papovaviridae, Paramyxoviridae, Parvoviridae,
Picornaviridae, Poxviridae,
Reoviridae, Retroviridae, Rhabdoviridae, or Togaviridae. The viral antigen can
be from human
126
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
immunodeficiency virus (HIV), Chikungunya virus (CHIKV), dengue fever virus,
papilloma
viruses, for example, human papillomoa virus (HPV), polio virus, hcpatitis
viruses, for example,
hepatitis A virus (HAV), hepatitis B virus (HBV), hepatitis C virus (HCV),
hepatitis D virus
(HDV), and hepatitis E virus (HEV), smallpox virus (Van ola major and minor),
vaccinia virus,
influenza virus, rhinoviruses, equine encephalitis viruses, rubella virus,
yellow fever virus,
Norwalk virus, hepatitis A virus, human T-cell leukemia virus (HTLV-I), hairy
cell leukemia virus
(HTLV-II), California encephalitis virus, Hanta virus (hemorrhagic fever),
rabies virus, Ebola
fever virus, Marburg virus, measles virus, mumps virus, respiratory syncytial
virus (RSV), herpes
simplex 1 (oral herpes), herpes simplex 2 (genital herpes), herpes zoster
(varicella-zoster, a.k.a.,
chickenpox), cytomegalovirus (CMV), for example human CMV, Epstein-Barr virus
(EBV),
flavivirus, foot and mouth disease virus, lassa virus, arenavirus, or a cancer
causing virus.
In one embodiment, the therapeutic agent is an agent for the treatment or
prevention of a
parasitic infection, or a disease or disorder associated therewith. In some
embodiments, the
parasite is a protozoa, helminth, or ectoparasite. The helminth (i.e., worm)
can be a flatworm (e.g.,
flukes and tapeworms), a thorny-headed worm, or a round worm (e.g., pinworms).
The
ectoparasite can be lice, fleas, ticks, and mites.
The parasite can be any parasite causing any one of the following diseases:
Acanthamoeba
keratitis, Amoebiasis, Ascariasis, Babesiosis, Balantidiasis,
Baylisascariasis, Chagas disease,
Clonorchiasis, Cochliomyia, Cryptosporidiosis, Diphyllobothriasis,
Dracunculiasis,
Echinococcosis, Elephantiasis, Enterobiasis, Fascioliasis, Fasciolopsiasis,
Filariasis, Giardiasis,
Gnathostomiasis, Hymenolepiasis, Isosporiasis, Katayama fever, Leishmaniasis,
Lyme disease,
Malaria, Metagonimiasis, Myiasis, Onchocerciasis, Pediculosis, Scabies,
Schistosomiasis,
Sleeping sickness, Strongyloidiasis, Taeniasis, Toxocariasis, Toxoplasmosis,
Trichinosis, and
Trichuriasis.
The parasite can be Acanthamoeba, Anisakis, Ascaris lumbricoides, Botfly,
Balantidium
coli, Bedbug, Cestoda (tapeworm), Chiggers, Cochliomyia hominivorax, Entamoeba
histolytica,
Fasciola hepatica, Giardia lamblia, Hookworm, Leishmania, Linguatula serrata,
Liver fluke, Loa
loa, Paragonimus - lung fluke, Pinworm, Plasmodium falciparum, Schistosoma,
Strongyloides
stercoralis, Mite, Tapeworm, Toxoplasma gondii, Trypanosoma, Whipworm, or
Wuchereria
bancrofti.
In one embodiment, the therapeutic agent is an agent for the treatment or
prevention of a
fungal infection, or a disease or disorder associated therewith. In some
embodiments, the fungus is
Aspergillus species, Blastomyces dermatitidis, Candida yeasts (e.g., Candida
albicans),
Coccidioides, Cryptococcus neoformans, Cryptococcus gattii, dermatophyte,
Fusarium species,
127
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Histoplasma capsulatum, Mucoromycotina, Pneumocystis jirovecii, Sporothrix
schenckii,
Exserohilum, or Cladosporium.
By way of a non-limiting example, in one embodiment, the invention provides a
CD4+ T
cell-targeted delivery vehicle comprising or encapsulating a nucleoside-
modified 1086C Env
mRNA, encoding the clade C transmitted/founder human immunodeficiency virus
(HIV)-1
envelope (Env) 1086C, for the treatment or prevention of HIV infection or a
disease or disorder
associated therewith.
It will be appreciated by one of skill in the art, when armed with the present
disclosure
including the methods detailed herein, that the invention is not limited to
treatment of diseases or
disorders that are already established. Particularly, the disease or disorder
need not have
manifested to the point of detriment to the subject; indeed, the disease or
disorder need not be
detected in a subject before treatment is administered. That is, significant
signs or symptoms of
diseases or disorders do not have to occur before the present invention may
provide benefit.
Therefore, the present invention includes a method for preventing diseases or
disorders, in that a
composition, as discussed previously elsewhere herein, can be administered to
a subject prior to
the onset of diseases or disorders, thereby preventing diseases or disorders.
One of skill in the art, when armed with the disclosure herein, would
appreciate that the
prevention of a disease or disorder, encompasses administering to a subject a
composition as a
preventative measure against the development of, or progression of, a disease
or disorder. As
more fully discussed elsewhere herein, methods of modulating the level or
activity of a gene, or
gene product, encompass a wide plethora of techniques for modulating not only
the level and
activity of polypeptide gene products, but also for modulating expression of a
nucleic acid,
including either transcription, translation, or both.
The invention encompasses delivery of a delivery vehicle, comprising at least
one agent,
conjugated to a targeting domain. To practice the methods of the invention;
the skilled artisan
would understand, based on the disclosure provided herein, how to formulate
and administer the
appropriate composition to a subject. The present invention is not limited to
any particular
method of administration or treatment regimen.
One of skill in the art will appreciate that the compositions of the invention
can be
administered singly or in any combination. Further, the compositions of the
invention can be
administered singly or in any combination in a temporal sense, in that they
may be administered
concurrently, or before, and/or after each other. One of ordinary skill in the
art will appreciate,
based on the disclosure provided herein, that the compositions of the
invention can be used to
prevent or to treat a disease or disorder, and that a composition can be used
alone or in any
128
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
combination with another composition to affect a therapeutic result. In
various embodiments, any
of the compositions of the invention described herein can be administered
alone or in
combination with other modulators of other molecules associated with diseases
or disorders.
Administration of the compositions of the invention (e.g., the delivery
vehicles) to a
human patient can be by any route, including but not limited to intravenous,
intranodal,
intradermal, transdermal, subcutaneous, intramuscular, inhalation (e.g., via
an aerosol), buccal
(e.g., sub-lingual), topical (i.e., both skin and mucosal surfaces, including
airway surfaces),
intrathecal, intraarticular, intraplural, intracerebral, intra-arterial,
intraperitoneal, oral,
intralymphatic, intranasal, rectal or vaginal administration, by perfusion
through a regional
catheter, or by direct intralesional injection. In one embodiment, the
compositions of the
invention (e.g. the delivery vehicles) are administered by intravenous push or
intravenous
infusion given over defined period (e.g., 0.5 to 2 hours). The compositions of
the invention can be
delivered by peristaltic means or in the form of a depot, although the most
suitable route in any
given case will depend, as is well known in the art, on such factors as the
species, age, gender and
overall condition of the subject, the nature and severity of the condition
being treated and/or on
the nature of the particular composition (i.e., dosage, formulation) that is
being administered. In
particular embodiments, the route of administration is via bolus or continuous
infusion over a
period of time, once or twice a week. In other particular embodiments, the
route of administration
is by subcutaneous injection given in one or more sites (e.g. thigh, waist,
buttocks, arm),
optionally once or twice weekly. In one embodiment, the compositions, and/or
methods of the
invention are administered on an outpatient basis.
In one embodiment, the invention includes a method comprising administering a
combination of compositions described herein. In certain embodiments, the
method has an
additive effect, wherein the overall effect of the administering a combination
of compositions is
approximately equal to the sum of the effects of administering each individual
inhibitor. In other
embodiments, the method has a synergistic effect, wherein the overall effect
of administering a
combination of compositions is greater than the sum of the effects of
administering each
individual composition.
The method comprises administering a combination of composition in any
suitable ratio.
For example, in one embodiment, the method comprises administering two
individual
compositions at a 1:1 ratio. However, the method is not limited to any
particular ratio. Rather any
ratio that is shown to be effective is encompassed.
129
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Pharmaceutical Compositions
The formulations of the pharmaceutical compositions described herein may be
prepared
by any method known or hereafter developed in the art of pharmacology. In
general, such
preparatory methods include the step of bringing the active ingredient into
association with a
carrier or one or more other accessory ingredients, and then, if necessary or
desirable, shaping or
packaging the product into a desired single- or multi-dose unit.
Although the description of pharmaceutical compositions provided herein are
principally
directed to pharmaceutical compositions which are suitable for ethical
administration to humans,
it will be understood by the skilled artisan that such compositions are
generally suitable for
administration to animals of all sorts. Modification of pharmaceutical
compositions suitable for
administration to humans in order to render the compositions suitable for
administration to
various animals is well understood, and the ordinarily skilled veterinary
pharmacologist can
design and perform such modification with merely ordinary, if any,
experimentation. Subjects to
which administration of the pharmaceutical compositions of the invention is
contemplated
include, but are not limited to, humans and other primates, mammals including
commercially
relevant mammals such as non-human primates, cattle, pigs, horses, sheep,
cats, and dogs.
Pharmaceutical compositions that are useful in the methods of the invention
may be
prepared, packaged, or sold in formulations suitable for ophthalmic, oral,
rectal, vaginal,
parenteral, topical, pulmonary, intranasal, buccal, intravenous,
intracerebroventricular,
intradennal, intramuscular, or another route of administration. Other
contemplated formulations
include projected nanoparticles, liposomal preparations, resealed erythrocytes
containing the
active ingredient, and immunogenic-based formulations.
A pharmaceutical composition of the invention may be prepared, packaged, or
sold in
bulk, as a single unit dose, or as a plurality of single unit doses. As used
herein, a -unit dose" is
discrete amount of the pharmaceutical composition comprising a predetermined
amount of the
active ingredient. The amount of the active ingredient is generally equal to
the dosage of the
active ingredient which would be administered to a subject or a convenient
fraction of such a
dosage such as, for example, one-half or one-third of such a dosage.
The relative amounts of the active ingredient, the pharmaceutically acceptable
carrier.
and any additional ingredients in a pharmaceutical composition of the
invention will vary,
depending upon the identity, size, and condition of the subject treated and
further depending upon
the route by which the composition is to be administered. By way of example,
the composition
may comprise between 0.1% and 100% (w/vv) active ingredient.
130
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
In addition to the active ingredient, a pharmaceutical composition of the
invention may
further comprise one or more additional pharmaceutically active agents.
Controlled- or sustained-release formulations of a pharmaceutical composition
of the
invention may be made using conventional technology.
As used herein, "parenteral administration" of a pharmaceutical composition
includes any
route of administration characterized by physical breaching of a tissue of a
subject and
administration of the pharmaceutical composition through the breach in the
tissue. Parenteral
administration thus includes, but is not limited to, administration of a
pharmaceutical composition
by injection of the composition, by application of the composition through a
surgical incision, by
application of the composition through a tissue-penetrating non-surgical
wound, and the like. In
particular, parenteral administration is contemplated to include, but is not
limited to, intraocular,
intravitreal, subcutaneous, intraperitoneal, intramuscular, intradennal,
intrastemal injection,
intratumoral, intravenous, intracerebroventricular and kidney dialytic
infusion techniques.
Formulations of a pharmaceutical composition suitable for parenteral
administration
comprise the active ingredient combined with a pharmaceutically acceptable
carrier, such as
sterile water or sterile isotonic saline. Such formulations may be prepared,
packaged, or sold in a
form suitable for bolus administration or for continuous administration.
Injectable formulations
may be prepared, packaged, or sold in unit dosage form, such as in ampules or
in multi-dose
containers containing a preservative. Formulations for parenteral
administration include, but are
not limited to, suspensions, solutions, emulsions in oily or aqueous vehicles,
pastes, and
implantable sustained-release or biodegradable formulations. Such formulations
may further
comprise one or more additional ingredients including, but not limited to,
suspending, stabilizing,
or dispersing agents. In one embodiment of a formulation for parenteral
administration, the active
ingredient is provided in dry (i.e., powder or granular) form for
reconstitution with a suitable
vehicle (e.g., sterile pyrogen-free water) prior to parenteral administration
of the reconstituted
composition.
The pharmaceutical compositions may be prepared, packaged, or sold in the form
of a
sterile injectable aqueous or oily suspension or solution. This suspension or
solution may be
formulated according to the known art, and may comprise, in addition to the
active ingredient,
additional ingredients such as the dispersing agents, wetting agents, or
suspending agents
described herein. Such sterile injectable formulations may be prepared using a
non-toxic
parenterallv-acceptable diluent or solvent, such as water or 1,3-butane diol,
for example. Other
acceptable diluents and solvents include, but are not limited to, Ringer's
solution, isotonic sodium
chloride solution, and fixed oils such as synthetic mono- or di-glycerides.
Other parentally-
131
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
administrable formulations which are useful include those which comprise the
active ingredient in
microcrystalline form, in a liposomal preparation, or as a component of a
biodegradable polymer
systems. Compositions for sustained release or implantation may comprise
pharmaceutically
acceptable polymeric or hydrophobic materials such as an emulsion, an ion
exchange resin, a
sparingly soluble polymer, or a sparingly soluble salt.
A pharmaceutical composition of the invention may be prepared, packaged, or
sold in a
formulation suitable for pulmonary administration via the buccal cavity. Such
a formulation may
comprise dry particles which comprise the active ingredient and which have a
diameter in the
range from about 0.5 to about 7 nanometers, and preferably from about 1 to
about 6 nanometers.
Such compositions are conveniently in the form of dry powders for
administration using a device
comprising a dry powder reservoir to which a stream of propellant may be
directed to disperse the
powder or using a self-propelling solvent/powder-dispensing container such as
a device
comprising the active ingredient dissolved or suspended in a low-boiling
propellant in a sealed
container. Preferably, such powders comprise particles wherein at least 98% of
the particles by
weight have a diameter greater than 0.5 nanometers and at least 95% of the
particles by number
have a diameter less than 7 nanometers. More preferably, at least 95% of the
particles by weight
have a diameter greater than 1 nanometer and at least 90% of the particles by
number have a
diameter less than 6 nanometers. Dry powder compositions preferably include a
solid fine powder
diluent such as sugar and are conveniently provided in a unit dose form.
Low boiling propellants generally include liquid propellants having a boiling
point of
below 65 F at atmospheric pressure. Generally the propellant may constitute 50
to 99.9% (w/w)
of the composition, and the active ingredient may constitute 0.1 to 20% (w/w)
of the composition.
The propellant may further comprise additional ingredients such as a liquid
non-ionic or solid
anionic surfactant or a solid diluent (preferably having a particle size of
the same order as
particles comprising the active ingredient).
Formulations of a pharmaceutical composition suitable for parenteral
administration
comprise the active ingredient combined with a pharmaceutically acceptable
carrier, such as
sterile water or sterile isotonic saline. Such formulations may be prepared,
packaged, or sold in a
form suitable for bolus administration or for continuous administration.
Injectable formulations
may be prepared, packaged, or sold in unit dosage form, such as in ampules or
in multi-dose
containers containing a preservative. Formulations for parenteral
administration include, but are
not limited to, suspensions, solutions, emulsions in oily or aqueous vehicles,
pastes, and
implantable sustained-release or biodegradable formulations. Such formulations
may further
comprise one or more additional ingredients including, but not limited to,
suspending, stabilizing,
132
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
or dispersing agents. In one embodiment of a formulation for parenteral
administration, the active
ingredient is provided in dry (i.e., powder or granular) form for
reconstitution with a suitable
vehicle (e.g., sterile pyrogen-free water) prior to parenteral administration
of the reconstituted
composition.
The pharmaceutical compositions may be prepared, packaged, or sold in the form
of a
sterile injectable aqueous or oily suspension or solution. This suspension or
solution may be
formulated according to the known art, and may comprise, in addition to the
active ingredient,
additional ingredients such as the dispersing agents, wetting agents, or
suspending agents
described herein. Such sterile injectable formulations may be prepared using a
non-toxic
parenterally-acceptable diluent or solvent, such as water or 1,3-butane diol,
for example. Other
acceptable diluents and solvents include, but are not limited to, Ringer's
solution, isotonic sodium
chloride solution, and fixed oils such as synthetic mono- or di-glycerides.
Other parentally-
administrable formulations that are useful include those that comprise the
active ingredient in
microcrystalline form, in a liposomal preparation, or as a component of a
biodegradable polymer
system. Compositions for sustained release or implantation may comprise
pharmaceutically
acceptable polymeric or hydrophobic materials such as an emulsion, an ion
exchange resin, a
sparingly soluble polymer, or a sparingly soluble salt.
As used herein, -additional ingredients" include, but are not limited to, one
or more of the
following: excipients; surface active agents; dispersing agents; inert
diluents; granulating and
disintegrating agents; binding agents; lubricating agents; sweetening agents;
flavoring agents;
coloring agents; preservatives; physiologically degradable compositions such
as gelatin; aqueous
vehicles and solvents; oily vehicles and solvents; suspending agents;
dispersing or wetting agents;
emulsifying agents, demulcents; buffers; salts; thickening agents; fillers;
emulsifying agents;
antioxidants; antibiotics; antifungal agents; stabilizing agents; and
pharmaceutically acceptable
polymeric or hydrophobic materials. Other "additional ingredients" which may
be included in the
pharmaceutical compositions of the invention are known in the art and
described, for example in
Remington's Pharmaceutical Sciences (1985, Genaro, ed., Mack Publishing Co.,
Easton, PA),
which is incorporated herein by reference.
EXPERIMENTAL EXAMPLES
The invention is further described in detail by reference to the following
experimental
examples. These examples are provided for purposes of illustration only, and
are not intended to
be limiting unless otherwise specified. Thus, the invention should in no way
be construed as
133
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
being limited to the following examples, but rather, should be construed to
encompass any and all
variations which become evident as a result of the teaching provided herein.
Without further description, it is believed that one of ordinary skill in the
art can, using
the preceding description and the following illustrative examples, make and
utilize the present
invention and practice the claimed methods. The following working examples
therefore are not to
be construed as limiting in any way the remainder of the disclosure.
Example 1: Highly efficient CD4+ T cell targeting and genetic recombination
using
engineered CD4+ T cell-homing mRNA-LNPs
mRNA-based therapeutics offer numerous advantages that address challenges with
the
current protein- or viral-based immunotherapy approaches, such as difficult
manufacturing,
instability, lack of control over amount and duration of expression, high
toxicity, and in some
cases, genomic integration or off-site effects (Goswami et al., 2019, Front.
Oncol, 9:297; Das et
al., 2015, J. Cell. Physiol, 230:259-271; Chames et al., 2009, Br. J.
Pharmacol, 157:220-233).
Efficient in vivo delivery has been the key obstacle in development of mRNA-
based
immunotherapeutics. To date, T cell modification for clinical application has
required extraction
of autologous T cells, expansion, and genomic editing ex vivo, which is
expensive and time-
consuming, and precludes widespread use for more common diseases, such as HIV
or sickle cell
anemia. T cells are known as hard-to-transfeet cells (Peer et al., 2010,
Journal of Controlled
Release, 148:63-68; Gust et al., 2008, Cell Commun Signal, 6:3; Goffinet et
al., 2006, The
FASEB Journal, 20:500-502). Here, it is demonstrated that targeting human T
cells with an anti-
CD4 antibody-conjugated Luc mRNA containing LNP, but not with control IgG-
conjugated LNP,
resulted in strong binding and luciferase expression in human CD4+ T cells in
a dose-dependent
manner (Figure lA through Figure 1C). Similarly, when injected systemically
into C57BL/6
mice, anti-mouse CD4/mRNA-LNP specifically accumulated and the mRNA was
translated in T
cell-enriched tissues, such as spleen and lymph nodes (Figure 3 and Figure 4A
through Figure
4C). Furthermore, the potential of the T cell-targeted mRNA-LNP system to
mediate genome
editing using a Cre/loxP reporter system was functionally evaluated. Cre-
mediated genetic
recombination was induced in CD4+ T cells in vivo. Interestingly, the signal
from nontargeted
mRNA-LNP in both splenic and lymphatic tissues at high dose (30 jag per mouse)
was not zero,
as observed in untreated mice (28% ZsGreen1+ cells among CD3+CD8- cells). This
observation
is likely due to expression of an ApoE receptor by some T cells (Sundqvist,
2018, Front.
lmmunol, 9:974; Panezai et at, 2017, Immunology 152:308-327), as LNP bind ApoE
and
typically target the liver 31. The percentage of gene edited cells was further
increased by multiple
134
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
injections of anti-CD4/mRNA-LNP. This high level of in vivo T cell targeted
genetic
rccombination has not been reported elsewhere. An important finding for
potential gene editing
therapies was that similar levels of gene recombination were observed in
resting and activated
CD4+ T cells. Evaluation of the presence of successfully targeted T cells over
time showed a
gradual decrease of ZsGreen1 signal in spleen during one-week post-treatment
with anti-
CD4/mRNA-LNP. This trend was expected considering the lifespan of circulatory
T cells, which
are a predominant population of T cells in spleen (Langeveld et al., 2006,
Journal of Clinical
Investigation, 36:250-256; Bronte et al., 2013, Immunity, 39:806-818).
However, ZsGreen1
expression in lymph nodes remained minimally changed over the 7-day experiment
time after
treatment with anti- CD4/mRNA-LNP. In a report analyzing the migration of Si
Cr-labeled
thoracic duct lymphocytes (TDLs) in major lymphoid and non-lymphoid tissues of
rats revealed
that lymphocytes have longer residence time in the lymph nodes than the spleen
24. Comparable
residence times were reported for mice as well (Mandl et al., 2012, Proc Natl
Acad Sci U S A,
109:18036-18041). Finally, whether various T cell subtypes differed in being
targeted was
evaluated. There was no significant difference in uptake and recombination
among naive,
memory, and effector memory subtypes when treated with anti-CD4/Cre mRNA-LNP.
The anti-
CD4 mRNA-LNP system allows for CD4+ T cell targeting in the tissues, such as
spleen and
lymph nodes, which is critical for T cell therapies. The current T cell
targeting platform has great
potential for many in vivo T cell manipulation-based applications by making T
cell targeted
therapeutic mRNA delivery possible. In vivo delivery to specific cell types
(e.g. T lymphocytes,
among others) is an intensely developing field, evidenced by many recent
studies (Veiga et al.,
2020, Adv Drug Deliv Rev, 159:364-376; Mizrahv et al., 2017, Mol Ther, 25:1491-
1500; Fenton
et al., 2017, Adv Mater, 29; Ramishetti et al., 2016, J Drug Target, 24:780-
786; Veiga et al.,
2018, Nat Commun, 9:4493). LNP modified with antibodies have been used for
delivery of
siRNA to lymphocytes for gene silencing purposes (Ramishetti et al., 2015, ACS
Nano, 9:6706-
6716). Ramishetti et al. surface modified siRNA-loaded LNP with anti- CD4
monoclonal
antibodies for targeting CD4+ T lymphocytes. They observed gene silencing in
approximately
30% of CD4+ T cells isolated from spleen, which is only half of the targeted
functional activity
that was observed with the CD4-targeted Cre mRNA-LNP (-60% of CD4+ T cells in
spleen). It
is of note that because of their use of siRNA-LNP and the non-binary readout
of their
experiments, direct comparison of targeting efficiencies of the two platforms
is not
straightforward. Other attempts have been made for lymphocyte targeting with
other lipid- and
polymer-based carriers. McKinlay et al. (McKinlay et al., 2018, Proceedings of
the National
Academy of Sciences 115, E5859-E5866) reported on a combinatorial chemical
approach of
135
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
mRNA delivery using hybrid lipid-based amphiphilic charge-altering releasable
transporters
(CARTs), achieving approximately 1.5% T lymphocyte transfection efficiency in
mice. Similarly,
Fenton et al. (Fenton et al., 2017, Advanced Materials, 29:1606944) described
specific LNP
design for delivering mRNA to B lymphocytes without using active targeting
ligand. They
showed an enhanced luminescence signal from the B cell targeted-Luc mRNA-LNP
formulation
in spleen compared to other non-selective formulations of their LNP
formulation library. Veiga et
al. (Veiga et al., 2018, Nature communications, 9:4493-4493) delivered mRNA in
surface
modified LNP to inflammatory Ly6C-h leukocytes using their ASSET platform,
which also
employs monoclonal antibody targeting. Delivery and expression of mRNA
encoding IL-10
showed significant therapeutic effect in a colitis model. While some T cells
also express Ly6c, as
do monocytes, macrophages and neutrophils, it was not determined which
populations of
leukocytes take up and express Ly6c-targeted LNP, and to what extent. The
targeted mRNA-LNP
platform is the first report of an LNP-based mRNA delivery system for
selective and functional
CD4+ targeting. Overall, the T cell-targeted mRNA-LNP platform presented here
offers
tremendous opportunity for a wide range of in vivo T cell manipulations. The
great potential of
this system to reach all T cell subtypes in difficult-to-access-tissues such
as lymph nodes, will
make the targeting platform available for many types of T cell manipulation in
vivo. The
application potentials include delivering mRNA therapeutics to T cells for
potential HIV cure. In
particular, targeted delivery of engineered genomic editing enzymes have the
potential to cure
HIV, by excising the HIV integrated provirus from the genome of infected cells
(Karpinski et al.,
2016, Nat Biotechnol, 34:401-409). Additionally, targeted modification of
lymphocytes have
numerous applications for development of fast-acting and cost-effective
immunotherapeutics for
a range of cancers, infectious diseases, and immunological disorders.
The materials and methods used for the experiments are now described.
Mice
C57BL/6J mice. Equal numbers of male and female C57BL/61 mice were purchased
from Jackson laboratories. Ai6(RCL-ZsGreen) mice. Ai6 (RCL-ZsGreen) mice on
C57BL/61
background were purchased from Jackson Laboratory (stock no: 007906) and bred
homozygous
in-house. Ai6 is a Cre reporter allele with a loxP-flanked STOP cassette
preventing transcription
of a CAG promoterdriven enhanced green fluorescent protein variant (ZsGreen1) -
all inserted
into the Gt(ROSA)26Sor locus. Upon Cre-mediated recombination, Ai6 mice
express robust
ZsGreen1 fluorescence.
136
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
mRNA production and LNP preparation
Coding sequences of Cre recombinase or firefly luciferase were codon-
optimized,
synthesized and cloned into the mRNA production plasmid (pUC-ccTEV-Cre-A101
and pUC-
ccTEV-Luc2-A101, respectively). mRNAs were produced using T7 RNA polymerase
(Megascript, Ambion) on linearized plasmids. mRNAs were transcribed to contain
101
nucleotide-long poly(A) tails. m PP-5'-triphosphate (TriLink) instead of UTP
was used to
generate modified nucleoside-containing mRNA. Capping of the in vitro
transcribed mRNAs was
performed co-transcriptionally using the trinucleotide capl analog, CleanCap
(TriLink). mRNA
was purified by cellulose purification, as described (Baiersdorfer et al.,
2019, Mol Ther Nucleic
Acids, 15:26-35). All mRNAs were analyzed by native agarose gel
electrophoresis and were
stored frozen at -20 C.
m1y-containing mRNAs were encapsulated in LNP using a self-assembly process in
which an aqueous solution of mRNA at pH = 4.0 is rapidly mixed with a solution
of lipids
dissolved in ethanol (Maier et al., 2013, Mol Ther, 21:1570-1578). LNP used in
this study were
similar in composition to those described previously (Maier et al., 2013, Mol
Ther, 21:1570-
1578; Jayaraman et al., 2012, Angew Chem Int Ed Engl, 51:8529-8533), which
contain an
ionizable cationic lipid (Acuitas)/ phosphatidylcholine/cholesterol/PEG-lipid
(50:10:38.5:1.5
mol/mol) and were encapsulated at an RNA to total lipid ratio of ¨0.04
(wt/wt). The diameter of
the nanoparticles was ¨80 nm as measured by dynamic light scattering using a
Zetasizer Nano ZS
(Malvern Instruments Ltd., Malvern, UK) instrument. mRNA-LNP formulations were
stored at
¨80 'V at a concentration of mRNA of ¨1 Rg/RL.
Monoclonal antibody-conjugated lipid nanoparticles
LNP were conjugated with mAbs specific for CD4. Purified NA/LE Rat anti-mouse
CD4
(BD PharmingenTM), purified rat anti-human CD4 antibody, clone A161A1
(BioLegend), and
control isotype-matched IgG were coupled to LNP via SATA¨maleimide conjugation
chemistry,
as described earlier 18. Briefly, LNP were modified with DSPE-PEG-maleimide by
a post-
insertion technique. The antibody was modified with SATA (N-succinimidyl S-
acetylthioacetate)
(Sigma-Aldrich) to introduce sulfhydryl groups allowing conjugation to
maleimide. SATA was
deprotected using 0.5 M hydroxylamine followed by removal of the unreacted
components by G-
25 Sephadex Quick Spin Protein columns (Roche Applied Science, Indianapolis,
IN). The
reactive sulfhydryl group on the antibody was then conjugated to maleimide
moieties using
thioether conjugation chemistry. Purification was performed using Sepharose CL-
4B gel filtration
137
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
columns (Sigma-Aldrich). mRNA content was calculated by performing a modified
Quant-iT
RiboGreen RNA assay (Invitrogen). Size and surface charge of the targeted
lipid nanoparticles
were determined using dynamic light scattering (DLS) and laser doppler
velocimetry (LDV),
respectively on a Malvern Zetasizer Nano ZS (Malvern instruments,
Worcestershire, UK), Both
size and zeta potential measurements were carried out in PBS pH 7.4 at 25 C in
relevant
disposable capillary cells. A non-invasive back scatter system (NIBS) with a
scattering angle of
173' was used for size measurements. Diameters of unconjugated and antibody-
modified mRNA-
LNP were interpreted as normalized intensity size distribution as well as z-
average values for
particle preparations.
In vitro cell binding studies
For cell binding studies using radioactivity measurements, LNP were first
radiolabeled
with Na1251 using Iodination Beads (Pierce) as described earlier (Khoshnejad
et al., 2016,
Bioconjug Chem, 27:628-637). Human CD4+ T cells were then incubated with
increasing
quantities of either Anti-CD4/ or Control IgG/mRNA-LNP for one hour at room
temperature.
Incubation medium was then removed and cells were washed with PBS buffer three
times to
remove the unbound nanoparticles from the cell surface. The cells were lysed
with 1% Triton
X100 in 1 N NaOH and the cell-associated radioactivity was measured by a
Wallac 1470 Wizard
gamma counter (Gaithersburg, MD) and compared to total added activity.
For cell binding studies using flow cytometry, human CD4+ T cells were seeded
at
150,000 cells per well in 24-well plates. LNP carrying Poly(C) RNA were added
to the media at
increasing quantities of mRNA per well, and cells were incubated for one hour
at room
temperature. Incubation medium was then removed and cells were washed with PBS
buffer three
times to remove the unbound nanoparticles from the cell surface. FITC-tagged
anti-rat IgG
(Abeam, Cambridge, UK) was used to monitor binding of antibody-conjugated LNP
on a BD
LSR II flow cytometer.
In vitro cell transfection studies
For cell transfection studies using firefly luciferase mRNA, human CD4+ T
cells were
plated in 48-well plates. After 18 hours, LNP carrying reporter luciferase
mRNA were added at
increasing concentrations to the cells, and incubated for 1.5 hours. Plates
were then washed three
times with PBS and complete medium was added to the cells. After culturing for
24 hours in
complete media, cells were washed with PBS, lysed in luciferase cell culture
lysis reagent
(Promega, Madison, WI) and the luciferase enzymatic activity as luminescence
(Luciferase assay
138
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
system, Promega) was measured 18. Transfections were performed in triplicate.
For cell
transfection studies using Cre rccombinase mRNA, spleens from two Ai6 mice
were harvested,
and a pooled single cell suspension was produced. 2 million splenocytes were
then plated in each
well of 6-well plates. Cells were incubated with 1,3, 6 or 9 itg of CD4-
targeted or non-targeted
(unconjugated or control IgG-conjugated) Cre mRNA-LNP overnight. Cells were
then collected
and stained with Live/Dead Aqua (Thermo Fisher Sci, L34966) and antibodies
against CD3 and
CD8 (and CD4, which was omitted from later experiments), and the percentage of
ZsGreenl-
expressing CD3+CD8- cells was determined using flow cytometry.
Biodistribution of anti-CD4/mRNA-LNP in C57BL/6J mice; tissue uptake
T251-radiolabeled mRNA-LNP were administered by IV (retro-orbital) injection
into
C57BL/6 mice (The Jackson Laboratory, Bar Harbor, ME). Blood was collected at
0.5, 1, and 24
hours post-injection from the inferior vena cava. Specific organs (liver,
spleen, lung, kidney and
heart) were also harvested at the same time-points, rinsed with saline,
blotted dry, and weighed.
The amount of radioactivity in each organ as well as in 100-1,11_, samples of
blood was measured in
a gamma counter (Wallac 1470 Wizard gamma counter, Gaithersburg, MD). Tissue
uptake as
percent of injected dose per gram tissue (%ID/g), and localization ratio (LR)
as organ-to-blood
ratio were calculated using radioactivity values and weight of the samples.
Immunospecificity
index (ISI) was also calculated as the ratio of the LR of CD4-targeted mRNA-
LNP to that of
Control IgG-modified ones.
Biodistribution of anti-CD4/mR1NA-LNP in C57BL/6J mice; luciferase mRNA
translation at tissue and cellular level
C57BL/6J mice were IV (retro-orbital) injected with anti-CD4/mRNA-LNP or
control
IgG/mRNA-LNP formulations. At 5 hours after injection, animals were euthanized
and selected
organs (liver, spleen, lung, kidney and heart) were harvested, rinsed with
PBS, and stored at ¨80
C until analysis. When thawed, tissue samples were homogenized in appropriate
volumes of cell
lysis buffer (1X) (Promega Corp, Madison, WI) containing protease inhibitor
cocktail (1X) and
mixed gently at 4 C for one hour. The homogenates were then subjected to
cycles of freeze/thaw
in dry ice/37 C and centrifuged for 10 minutes at 16,000 g at 4 C.
Luciferase activity was then
measured in the supernatant using a Victor 3 1420 Multilabel Plate Counter
(Perkin Elmer,
Wellesley, MA). Further, the mRNA expression in the CD3+ cell population was
evaluated.
CD3+ cells were isolated from the spleens or lymph nodes of injected mice
using the
MagniSortTM Mouse CD3+ Selection Kit (ThermoFisher Scientific, Waltham, MA)
based on
139
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
manufacturer's instructions. Briefly, a biotinylated Anti-Mouse CD3 antibody
and
streptavidincoated magnetic beads were utilized for CD3+ cell isolation. CD3+
cells were bound
to the antibody and then magnetic beads. When placed in a magnetic field, the
undesired cells
were separated from CD3+ cells by decanting. Luciferase activity measurements
were performed
on the cell lysate of the CD3+-enriched cell population.
Bioluminescence imaging
C57BL/6J mice were IV (retro-orbital) injected with anti- CD4/mRNA-LNP or
control
IgG/mRNA-LNP formulations. At 5 hours after injection, bioluminescence imaging
was carried
out as described previously 18 using an IVIS Spectrum imaging system (Caliper
Life Sciences,
Waltham, MA). D-luciferin was administered to mice intraperitoneally at a dose
of 150 mg/kg.
After 5 minutes, the mice were euthanized; desired tissues were harvested, and
immediately
placed on the imaging platform. Tissue luminescence was measured on the IVIS
imaging system
using an exposure time of 5 seconds or longer to ensure that the signal
obtained was within
operative detection range. Bioluminescence values were also quantified by
measuring photon flux
(photons/second) in the region of interest using LivingImage software provided
by Caliper.
Determination of targeting efficiency of anti-CD4/mRNA-LNP using a Cre/loxP
reporter
system
To analyze delivery efficiency to targeted cell populations within the spleen
and lymph
nodes, mRNA translation was tracked with single-cell resolution. The targeted
and nontargeted
LNP containing Cre recombinase mRNA were IV (retro-orbital) injected into Ai6
mice carrying a
Cre reporter allele with a loxP-flanked STOP cassette preventing transcription
of a green
fluorescent protein variant (ZsGreen1). Cre recombinase excises the loxP-
flanked STOP cassette,
therefore allowing the transcription of ZsGreenl. At desired time points after
injection, animals
were euthanized and spleens and lymph nodes were harvested. The number of
CD3+CD8- cells
emitting green fluorescent signal in organ single cell suspensions was
evaluated using flow
cytometry.
Single cell suspension preparation and flow cytometry
Single cell suspensions were prepared from spleens and lymph nodes. Briefly,
the tissues
were crushed using the frosted end of glass microscope slides and then passed
through a 70-1.tm
filter. Following centrifugation and removal of supernatant, cells were
resuspended in RPM1 +
10% FBS medium, and were first stained with Live/Dead Aqua cell stain (Thermo
Fisher Sci.,
140
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
Cat# L34957), then a mixture of anti-mouse antibodies (Figure 11). The stained
single cell
populations were characterized on a BD LSR 11 flow cytometer (BD Biosciences).
500,000 events
were collected per sample. Compensation of multicolor flow was carried out
using ArCTM Amine
Reactive beads (Thermo Fisher Sci.) for Live/Dead Aqua, Compbead anti Rat and
anti-Hamster
Ig x/Negative Control Compensation Particles set (BD Biosciences) for all
antibodies, and GFP
BrightComp eBeads (Thermo Fisher Sci., Cat# A10514) for ZsGreenl. Data were
analyzed with
FlowJo software (Ashland, OR). The materials and methods used in these
experiments are now
described.
The results of the experiments are now described.
mRNA-LNP targeting CD4+ T cells in vitro
Considering that T cells do not naturally endocytose nanoparticles, initially
CD4+ T cell
surface antigens were sought that endocytose after mAb binding. CD4-targeted
receptor mediated
endocytosis was selected to achieve CD4+ T cell targeted delivery19. CD4
receptor targeting has
also been shown to be capable of uptake and internalization upon nanoparticle
binding 20, 21.
The binding capacity of the targeted mRNA-LNP was first evaluated on human
CD4+ T cells
obtained from healthy donors. Anti-CD4 IgG antibody (anti-CD4/mRNA-LNP) or non-
specific
isotype control IgG (control IgG/mRNA-LNP) was conjugated to mRNA-LNP. As
shown in
Figure 1A, radiolabeled anti-CD4/mRNA-LNP selectively bound to human CD4+ T
cells, while
control IgG counterparts did not. Selective targeting to CD4+ T cells was also
confirmed using
flow cytomeny (Figure 1B ¨ Figure 1C). Human CD4+ T cells were incubated with
either anti-
CD4/Poly(C) RNA-LNP or control IgG/Poly(C) RNA-LNP, and FITC-tagged anti-rat
IgG was
used to monitor binding of antibody-conjugated LNP to cells. Dose-responsive
binding of anti-
CD4/Poly(C) RNA-LNP was observed. In order to determine internalization and
functional
activity (mRNA translation) of the targeted mRNA-LNP, anti-CD4 antibody- or
control IgG-
conjugated LNP carrying firefly luciferase (Luc)-encoding mRNA were incubated
with human
CD4+ T cells. Efficient translation of the mRNA in anti-CD4/mRNA-LNP was
demonstrated
compared to control IgG/mRNA-LNP. Incubation of CD4+ T cells with higher doses
of Luc
mRNA-LNP yielded higher Luc activity, demonstrating a dose-response
correlation (Figure 1C).
In order to directly assess targeting efficiency on the single cell level, and
to test the
targeting platform for gene editing purposes, splenocytes were harvested from
mice harboring
Ai6 (a Cre reporter allele with a loxP-flanked STOP cassette, which upon Cre-
mediated
recombination, expresses robust ZsGreen1 fluorescence), and treated with
different amounts of
141
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
targeted or non-targeted (unconjugated or control IgG-conjugated) Cre mRNA-
LNP. The cells
were then collected and stained with antibodies against CD3 and CD8
(antibodies are listed in
Figure 11) and analyzed by flow cytometry. Gating strategy to identify
ZsGreenl positive cells
among CD3+CD8- population is presented on Figure 2B., with the corresponding
ZsGreenl
positive cells shown on Figure 2A. CD3+CD8- staining was used instead of
direct CD4 staining
to identify CD4+ T cells because of the transient disappearance of CD4 upon
administration of
the anti-CD4 antibody-conjugated nanoparticles (Figure 3). A very low
percentage of CD3+CD8-
cells exhibited positive ZsGreen1 signal when non-targeted LNP were used,
while approximately
80% of this cell population took up and translated Cre mRNA delivered in anti-
CD4 antibody-
conjugated LNP, even at the lowest amount of mRNA-LNP administered. The same
experiment
was performed in splenocytes harvested from Ai9 mice, which express robust
tdTomato
fluorescence following Cre-mediated recombination, and similar results were
obtained. This
shows the potential of the targeted anti-CD4/mRNA-LNP to efficiently transfect
CD4+ T cells in
vitro.
Anti-CD4/mRNA-LNP target CD4+ T cells in vivo
Next, the biodistribution of anti-CD4/mRNA-LNP in mice was analyzed after
retro-
orbital intravenous (IV) administration. LNP were directly labeled with 1251
prior to conjugation
with anti-mouse CD4 or control IgG, therefore, measured radioactivity only
showed distribution
of particles without any detached targeting antibodies affecting the outcome.
To measure tissue
uptake, the amount of radioactivity in various tissues (percent of injected
dose per gram of tissue-
%ID/g) was calculated. As expected, a substantial amount of control IgG/mRNA-
LNP particles
were still circulating in the blood (19.35+2.2 %ID/g) 0.5 hour (h) post
injection, representing a
significant change in the biodistribution with a reduction in liver targeting
of control IgG LNP
(Figure 4A). For anti- CD4/mRNA-LNP, lower amounts of particles were
circulating (10.84+0.42
%ID/g). The majority of the anti-CD4/mRNA-LNP uptake occurred in the spleen
(131.59+9.71
% ID/g), representing a 3.5-fold increase in splenic uptake compared to the
control IgG/mRNA-
LNP (37.6+8.67 %ID/g). The localization ratio (LR), defined as the ratio of
%ID/g of a given
organ to that in the blood, was also calculated for both CD4-targeted and
control IgG/mRNA-
LNP. The spleen being part of the reticuloendothelial system contributes to
non-specific splenic
uptake that is observed with the untargeted mRNA-LNP. Anti-CD4/mRNA-LNP were
localized
in spleen at 6-fold higher level than their control IgG counterparts (Figure
4B, Figure 5A). To
further explore and quantitate the kinetics of in vivo tissue uptake of anti-
CD4/mRNALNP,
targeted and non-targeted 125I-labeled poly(C) RNA-LNP were injected IV into
mice. Groups of
142
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
animals were sacrificed at 0.5, 1, and 24 hours after injection, and selected
tissues (blood, spleen,
liver, lung, and kidneys) were harvested. The highest circulating amount of
targeted mRNA-LNP
was 10.840.42 %ID/g in blood at the earliest time point tested (Figure 4C). At
later time points,
the concentration of targeted particles in blood quickly dropped to a %ID/g of
6.86+1.34 and
0.51+0.07 at 1 and 24 hours, respectively. Specific splenic uptake of targeted
particles peaked at
0.5 h post injection (131.59+9.71 % ID/g) (Figure 4C) and the localization
ratio increased over
time reaching to 61.15 at the last time point tested, 24 hours (Figure 4D).
mRNA in targeted LNP is efficiently delivered to CD4+ T cells in vivo
mRNA translation after IV administration of Luc mRNA-LNP was then analyzed.
Control IgG/Luc mRNA-LNP and anti-CD4/Luc mRNA-LNP were first administered at
a dose of
8 jig (0.32 mg/kg) mRNA. Five hours after injection, various organs were
harvested, and
luciferase activity was either measured from tissue lysates, or was detected
by direct luminescent
imaging of whole organs (Figure 6A-Figure 6C). The Luc expression pattern
showed a marked
difference between anti-CD4 and control IgG/Luc mRNA-LNP-treated mice, as the
luminescence
signal decreased significantly in liver with CD4- targeting. Most importantly,
Luc activity for
anti-CD4/Luc-mRNA-LNP was ¨7 fold higher compared to the control IgG-modified
mRNA-
LNP in the spleen (Figure 6A and Figure 6B). After removal of the spleen,
kidneys, lungs, heart,
and liver - which exhibits high uptake of both unconjugated and antibody-
conjugated LNP -
lymph node luciferase expression was observed in the anti- CD4/Luc mRNA-LNP-
treated mice
(Figure 6C). This shows the capacity of targeted LNP to traverse endothelial
membranes and
functionally access cells in tissues, such as lymph nodes. To demonstrate that
mRNA was
delivered specifically to the T cell population, CD3+ T cells were isolated
(as CD4 selection
could not be performed) from the spleen of mice treated as above. Luc activity
of the CD3+
population after anti-CD4/Luc mRNA-LNP administration was 33-fold higher than
in control
IgG/Luc mRNA-LNP-treated samples. It was concluded that with Luc activity
concentrated in T
cells (Figure 6D), CD4+ T cells are being specifically and efficiently
targeted after IV delivery of
targeted nanoparticles.
To confirm the targeting efficiency of the CD4-targeted mRNALNP platform with
LNP
formulations other than ALC-0307 LNPs, the same antibody conjugation strategy
was applied on
the Acuitas LNP formulation containing the ionizable lipid 0315 (ALC-0315
LNP), which is the
LNP formulation in the recently US Food and Drug Administration (FDA)-approved
Pfizer/BioNTech COVID vaccine (Thran et al., 2017, EMBO Mol. Med. 9, 1434-
1447;
W02017075531A1). The list of ingredients in this LNP formulation includes (4-
143
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
hydroxybutyl)azanediy1)bis(hexane-6,1-diy1)bis(2-hexyldecanoate) or ALC-0315
ionizable lipid,
2[(polyethylcne glycol-2000J-N,N-ditctradecylacctamide, 1,2-distcaroyl-sn-
glyccro-3-
phosphocholine and cholesterol. Five hours after injection of anti-CD4-
targeted Luc mRNA-
ALC-0315 LNPs, a very similar Luc expression pattern to CD4-targeted ALC-0307
LNPs (i.e., a
higher luminescence signal in the spleen and a lower signal in liver) was
observed when
compared to control IgG counterparts. These data prove that similar targeting
efficiency is
achieved with CD4 targeting of other LNP formulations, such as ALC-0315 LNP.
CD4-targeted Cre-mRNA-LNP effect genetic recombination in CD4+ T cells in vivo
One use for targeted mRNA therapy is gene editing and insertion. To evaluate
the
efficiency of delivery of mRNA using CD4-targeted LNP at the cellular level
for in vivo genetic
modification, Cre mRNA-LNP was administered to Ai6 mice (Figure 7A). In these
mice, the
Cre/loxPmediated expression of a reporter gene encoding the fluorescent
protein ZsGreen1
allows for easy readout of successfully transfected and LoxP recombined target
cells using flow
cytometry (Figure 7B and Figure 7C). A wide range of doses (3, 10, 30, and 90
jag) were tested.
Mice were injected IV, then spleens and lymph nodes were harvested the next
day, and single cell
suspensions were prepared from each tissue. Cells were stained for flow
cytometry using
antibodies against CD3 and CD8 to identify CD4+ T cells (Figure 11). No signal
was observed in
non-treated animals, indicating no leakage of the reporter construct.
Administration of control
IgG/Cre mRNA-LNP led to low efficiency of transfection, similar in level over
the range of
mRNA doses used, in both tissues tested, i.e. spleens (Figure 7B) and lymph
nodes (Figure 7C).
As expected, a significant increase in the number of ZsGreenl-expressing cells
was observed
with anti-CD4/Cre mRNALNP treatment at all tested mRNA doses when compared to
control
IgG- and unconjugated mRNA-LNP counterparts (Figure 7B and Figure 7C). In mice
treated with
unconjugated mRNA-LNP, a substantial increase in mRNA delivery and subsequent
Cre/loxP
recombination was observed (up to approximately 20% of ZsGreen1+ cells in the
CD3+CD8- cell
population) when the dose was increased to 30 jag. This is still well below
the strong response
observed with targeted mRNA-LNP at all tested doses, and is likely due to
expression of an ApoE
receptor by some T cells (Sundqvist, 2018, Front. Immunol, 9:974; Panezai et
al., 2017,
Immunology 152, 308-327). While administration of 90 lag of anti-CD4/Cre mRNA-
LNP resulted
in an even higher percentage of ZsGreen1+ CD4+ T cells, this amount of LNP
proved to be toxic
in all groups (both unconjugated and control IgG-conjugated, and CD4-targeted
LNP treatments),
thus that dose was eliminated from further experiments. Selective CD4
targeting versus control of
untargeted LNP did not increase the uptake of nanoparticles in macrophages and
dendritic cells
144
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
(Figure 8), likely due to their extensive natural phagocytic uptake of
nanoparticles, whereas with
CD4+ T cells, there is significant increase in targeted mRNA-LNP uptake
compared to
untargeted control mRNA-LNP. The number of ZsGreenl-expressing cells in non-T
cell
splenocytes, such as dendritic cells and macrophages, did not differ among the
range of doses in
this study (Figure 8).
A similar experiment was performed using CD4-targeted ALC-0315 LNPs. When Ai6
mice were iv. injected with these targeted LNPs carrying Cre mRNA, targeting
efficiency
comparable to CD4-targeted ALC-0307 LNP-Cre mRNA was observed (increase in the
number
of ZsGreenl-expressing cells in mice treated with anti-CD4/ALC-0315 LNP-Cre
mRNA
treatment compared to control IgG counterparts).
CD4+ T cell-targeting with anti-CD4/mRNA-LNP is not T cell subtype specific
Next, whether the uptake of the targeted LNP were favored by certain T cell
subtypes
was investigated. One day after the administration of a dose 10 jig of Cre
mRNA-LNP, spleens
were harvested, and single cell suspensions were stained with antibodies
against CD3, CD8,
CD44 and CD62L to identify naive, memory, and effector memory T cell
subpopulations. No
significant preference was found for the CD4-targeted mRNA-LNP to be taken up
and expressed
by any specific CD4+ T cell subpopulation examined: CD4+ naive T cells (C1144-
CD62L-),
central memory T cells (CD44+CD62L-P), and effector memory T cells (CD44+CD62L-
) (Figure
9A). The majority of T cells in vivo are not activated. The expression of the
T cell activation
marker (CD25) was analyzed on the CD4+ T cells receiving Cre mRNA-LNP.
Notably, CD4-
targeted mRNA-LNP induced Cre recombination in ¨57% of resting (CD25-),
compared to ¨40%
of activated (CD25 ) CD4+ T cells (Figure 9B), thus demonstrating efficient
targeting,
transfection, and gene recombination in resting CD4+ T cells.
Recombined cells decrease over time after CD4-targeted delivery in the spleen
ZsGreen1 expression was tracked for seven days after a single IV
administration of 10 jig
of Cre mRNALNP. Spleens and lymph nodes were harvested one, four or seven days
post-
injection, and single cell preparations were stained for flow cytometric
analysis as above. Four
days after administration of 10 jig of anti-CD4/Cre mRNA-LNP, the number of
ZsGreenl-
expressing splenic CD4+ T cells dropped significantly (from ¨ 50% at day 1 to
¨ 32% at day 4).
However, it held at a similar level of around 26% at the last time point
tested (day 7), still
significantly above the values observed with IgG and unconjugated counterparts
(Figure 711).
Blood and spleen are sites of transient-recirculating T cells, which this data
reflects (Ganusov et
145
CA 03195281 2023-4- 11
WO 2022/081702
PCT/US2021/054775
al., 2014, PLoS Comput Biol, 10:e1003586; Mandl et al. 2012, Proc Nat! Acad
Sci U S A,
109:18036-18041). The ZsGreen1 expression for all treatments did not
significantly change over
7 days in T cells extracted from lymph nodes (Figure 7E). This reflects the
longer residence time
of T cells in lymph nodes (Ganusov et al., 2014, PLoS Comput Biol,
10:e1003586; Mandl et al.
2012, Proc Natl Acad Sci USA, 109:18036-18041).
In vivo targeted mRNA-LNP-induced specific genetic recombination shows an
additive
effect
The potential additive effect of targeted mRNA delivery was tested by serial
administrations of mRNA-LNP (Figure 10A and Figure 10B). Mice received three
or five IV
injections of 10 jig doses of Cre mRNA-LNP, one injection every 24 hours.
Spleens and lymph
nodes were harvested the day after the last injection. Five injections
resulted in a significantly
higher number of ZsGreen1+ cells when compared to three injections.
Interestingly, a steady
increase in ZsGreenl- expressing CD4+ T cell numbers was observed for both
control IgG/ and
unconjugated mRNALNP. However, the expression increased to 28% in the
unconjugated group,
still relatively lower than the anti-CD4/mRNA-LNP at any number of injections
tested. Overall,
the sequential administrations of the targeted mRNA-LNP resulted in increasing
Cre-induced
genetic recombination with increased number of injections in both the spleen
and lymph nodes.
The disclosures of each and every patent, patent application, and publication
cited herein
are hereby incorporated herein by reference in their entirety. While this
invention has been
disclosed with reference to specific embodiments, it is apparent that other
embodiments and
variations of this invention may be devised by others skilled in the art
without departing from the
true spirit and scope of the invention. The appended claims are intended to be
construed to
include all such embodiments and equivalent variations.
146
CA 03195281 2023-4- 11